- 1Key Laboratory of Systems Bioengineering (Ministry of Education), Tianjin University, Tianjin, China
- 2SynBio Research Platform, Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), School of Chemical Engineering and Technology, Tianjin University, Tianjin, China
- 3Frontiers Science Center for Synthetic Biology (Ministry of Education), Tianjin University, Tianjin, China
Fengycin is a multifunctional peptide antibiotic produced mainly by Bacillus species and the purpose of this research was to construct a Bacillus subtilis strain that can produce fengycin with the xylose as the substrate with CRSIPR-Cas9. Hence, at the beginning of this study, functional sfp and degQ were expressed in B. subtilis 168 strain to give the strain the ability to produce the fengycin with the titer of 71.21 mg/L. Subsequently, the native promoter PppsA of the cluster responsible for the fengycin synthesis was replaced by the Pveg promoter, resulting in a further 5.22-fold increase in fengycin titer. To confer xylose utilization capacity to B. subtilis, deletion of araR and constitutive overexpression of araE were performed, and the xylose consumption rate of the engineered strain BSUY06 reached 0.29 g/L/h, which is about 6.25-fold higher than that of the parent strain BSUY04-1. In the final phase of this study, the fermentation characteristics were observed and the initial xylose concentration was optimized. In this study, 40 g/L xylose was proved to be the most suitable initial concentration for growth and fengycin fermentation, which leading to a fengycin titer of 430.86 mg/L. This study demonstrated that lignocellulose, the clean and sustainable substrate with xylose as the second largest sugar, is a potential substrate for the production of fengycin.
1 Introduction
Bacillus subtilis has received considerable attention as one of the safest industrial microorganisms and has been widely used in the production of high-value chemicals. With the intensive study of genomic information, the potential of B. subtilis for the production of antibiotics is gradually being discovered, with peptide antibiotics being the predominant category and surfactin, iturin and fengycin are the main members of the cycle lipopeptides produced by B. subtilis (Hu et al., 2019). Fengycin, a cycle antimicrobial decapeptide consisting of a β-hydroxyl fatty acid containing a carbon number in the range of 14–18 (Villegas-Escobar et al., 2013), has aroused considerable interest in recent years owing to its lower toxicity, higher biodegradability and better environmental compatibility (Zhang et al., 2019b). Fengycin shows more pronounced antagonism against filamentous fungi than other lipopeptides, so it can be used to treat various diseases in plants without the adverse effects of chemical pesticides (Sur et al., 2018; Ding et al., 2019; Zhang et al., 2022). Fengycin also has potential applications in the food (Liu et al., 2012; Tang et al., 2014; Ahmad et al., 2023) and cosmetics industries (Lu et al., 2016; Zhao et al., 2016). It’s worth noting that its anti-cancer activity in inhibiting the growth of human lung cancer cells (Yin et al., 2013), human colon cancer cells (Cheng et al., 2016) and human leukemia cell (Ma et al., 2012) has also been unveiled.
However, one of the major obstacles for the large-scale application of fengycin is its low productivity, and many studies have attempted to increase the productivity of engineered strains to meet the growing market demand for fengycin, including optimization the fermentation conditions and genetic engineering. Among these, promoter engineering seems to be one of the most effective methods to increase fengycin production by directly enhancing the transcription level of the fen (pps) operon responsible for fengycin production. B. subtilis Bs2508, a derivative strain in which the weak fengycin promoter was replaced by the promoter of amyQ and fengycin production was increased from undetectable to 434 mg/L (Ongena et al., 2007), while replacing the Ppps promoter of B. subtilis BBG111 with the Pfen promoter from BBG21 increased fengycin production by approximately 10-fold (Yaseen et al., 2016). Although promoter replacement has been proved to be effective in increasing fengycin production, few promoters have been studied. Thus, it is essential to investigate the effect of more promoters on transcription level of the fen operon as well as fengycin titer.
In addition to low productivity, another limitation to the large-scale application of fengycin is the relatively high production cost of feedstock. Nowadays, glucose is still the main carbon source for fengycin production, which is not only uneconomical but also faces the competition issues between global food supply and industrial biotechnology (Zhang et al., 2019a). To solve this issue as well as to develop clean and long-term renewable sources for the production of more high-value compounds, lignocellulosic, the most abundant bioresource on the earth, is attracting increasing attention as a more promising feedstock (Wei et al., 2015; Zhang M. et al., 2019). Lignocellulose is basically composed of cellulose (30%–50%), hemicellulose (25%–30%) and lignin (15%–20%) (Zhao et al., 2020), and the lignocellulose hydrolysate contains abundant hexoses and pentoses. In practice applications, however, the majority of fermentation microorganisms are unable to efficiently convert the hydrolysate of lignocellulosic biomass, especially xylose, which is a major limiting factor for the large-scale utilization of lignocellulose. Since xylose is the main part of hemicellulose and the second most abundant sugar in lignocellulose (Wei et al., 2015), the effective utilization of xylose is a prerequisite for the efficient conversion of lignocellulose into valuable chemicals, including fengycin.
To this end, in this study, a B. subtilis strain for fengycin production from xylose was obtained in three steps (Figure 1). First, we expressed functional sfp and degQ in B. subtilis 168 strain to obtain a fengycin-producing B. subtilis strain. Then, we increased fengycin production by replacement of the weak promoter of pps gene cluster. In addition, we enhanced the expression of AraE to endow B. subtilis with the ability to utilize xylose. At the end of this study, we optimized the initial xylose concentration in the fermentation medium to ensure an adequate carbon source while avoiding cell growth inhibition and waste of xylose caused by high substrate concentrations. This study confirms the feasibility of producing fengycin from lignocellulose.
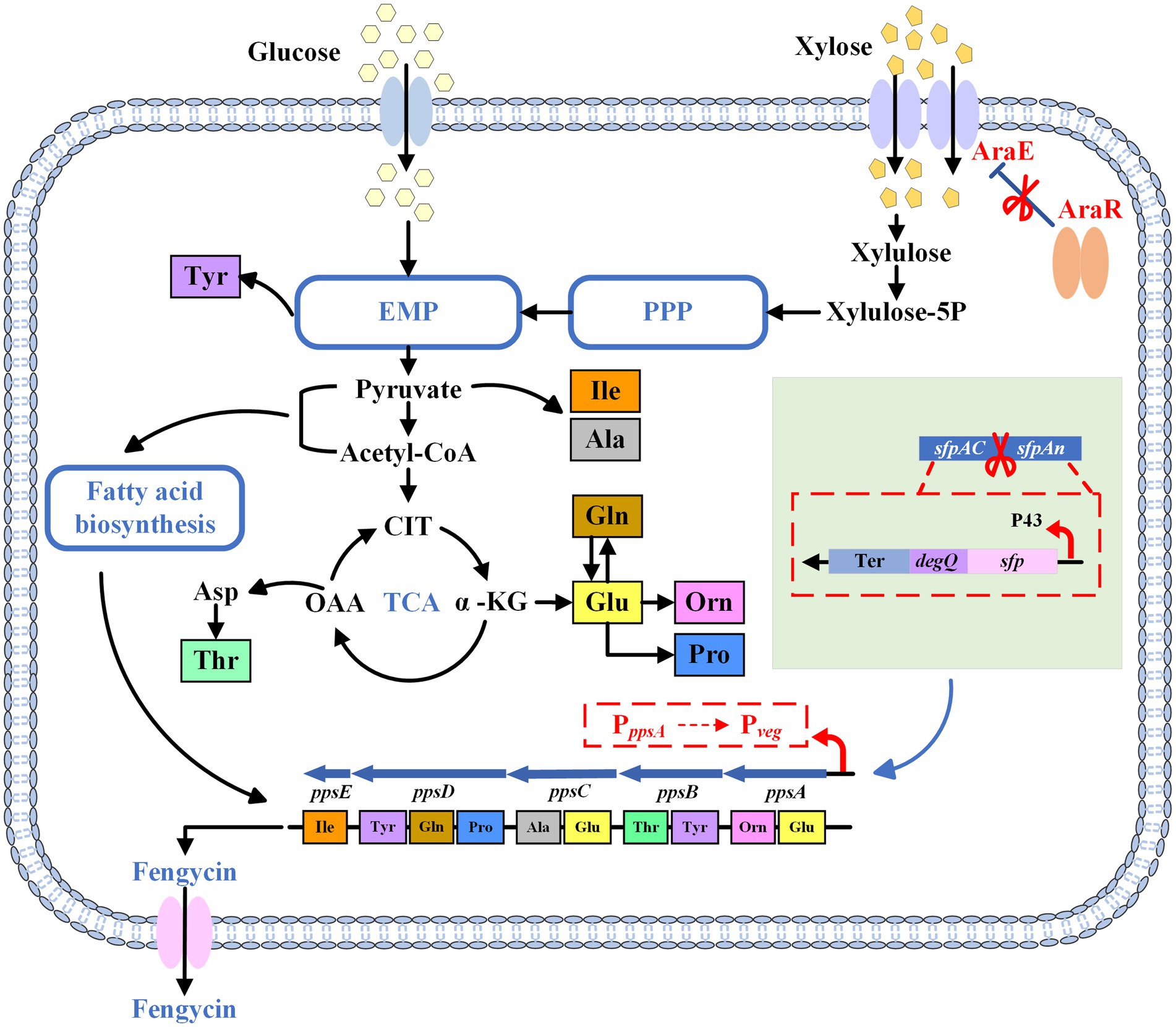
Figure 1. The schematic diagram of the metabolic pathway of fengycin. PPP, pentose phosphate pathway; EMP, Embden-Meyerhof pathway; TCA, tricarboxylic acid cycle; Ter, terminator; CIT, citrate; OAA, oxaloacetate; α-KG, α-ketoglutarate; Xylulose-5P, xylulose-5-phosphate.
2 Martial and methods
2.1 Strains, plasmids and culture conditions
All the strains and plasmids used in this work are listed in Table 1. Luria-Bertani (LB) medium (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl, pH = 7.0) was used for the growth of Escherichia coli and the construction of B. subtilis strains under a shaking condition at 200 rpm and 37°C, during which appropriate antibiotics (ampicillin 100 μg/mL, kanamycin 50 μg/mL for E. coli and 25 μg/mL for B. subtilis) was added into the medium for strains with antibiotic resistance.
To prepare seed cultures, B. subtilis 168 and its derivative strains were cultivated in LB medium at 200 rpm and 37°C for about 9 h, after which OD600 reached 5.5–6.0. To produce fengycin and detect various indexes during the fermentation, the seed cultures were inoculated into the fermentation medium (glucose/xylose 20 g/L, peptone 20 g/L, KH2PO4 2 g/L, K2HPO4 4 g/L, KCl 0.5 g/L, MgSO4·7H2O 0.5 g/L, MnSO4 5.0 mg/L, CuSO4 0.16 mg/L, FeSO4 0.15 mg/L, pH = 7.0–7.2) with the initial OD600 of 0.4 and the fermentation broth was shaken at 180 rpm and 30°C.
To measure the efficiency of xylose utilization, cultivation was performed in M9 medium with xylose as the sole carbon source (xylose 20 g/L, Na2HPO4 6.8 g/L, KH2PO4 3 g/L, NH4Cl 1 g/L, NaCl 0.5 g/L, tryptophan 5 g/L, MgSO4·7H2O 1 mol/L, CaCl2 0.1 mol/L, MnCl4·4H2O 1 mg/L, CuCl2·2H2O 0.43 mg/L, ZnCl2 1.7 mg/L, CoCl2·2H2O 0.6 mg/L, NaMoO4 0.6 mg/L, FeCl3·6H2O 13.5 mg/L) at 180 rpm and 30°C with the initial OD600 of 0.4.
2.2 The construction of plasmids and strains through CRISPR-Cas9
Oligonucleotide primers mentioned in this study are listed in Table 2. The general isolation and manipulation of recombinant DNA followed standard techniques (Sambrook, 2001). The plasmids used for gene deletion and integrated overexpression in this work were all derived from the CRISPR-Cas9 plasmid of B. subtilis named PJOE8999 (Altenbuchner and Müller, 2016). The gRNAs were designed in the website1 and inserted into PJOE8999 to give a series of plasmids named PJOE-sgRNA as described previously (Tan et al., 2019). To obtain the plasmids for knockout, the upstream and downstream fragments were connected to PJOE-sgRNA plasmids by seamless cloning kit. While to construct the plasmids used for integrated overexpression, upstream regions, promoter regions, target genes as well as downstream regions were amplified from the corresponding templates, and then fragments were inserted into corresponding PJOE-sgRNA plasmids by seamless cloning kit or ligase as required. Unless otherwise specified, the genes were amplified from B. subtilis 168. After introducing the correctly constructed plasmids into B. subtilis strains, the selection of engineering strains was carried out as previous description (Altenbuchner and Müller, 2016; Tan et al., 2019). The correct mutants were confirmed by PCR and DNA sequencing.
2.3 Determination of fengycin
To determine the concentration of fengycin produced by the B. subtilis 168 and its derivative strains constructed in this study, the cultures were centrifuged at 12,000 rpm and 4°C for 30 min, and the cell-free supernatant was collected and adjusted to pH 2.0 with hydrochloric acid and kept it at 4°C for 6–8 h. Then the precipitate was collected and dissolved with methanol. After adjusting the mixture to a final pH of 7.0, the samples were kept at 4°C for 12–24 h for fengycin extraction. Finally, the supernatant was collected and filtered through a 0.22 μm filter, which can be quantified by high performance liquid chromatography (HPLC) 1200 series equipped with an Agilent Eclipse XDB-C18 column (5 μm, 4.6 × 150 mm, United States) at an operating temperature of 30°C (Tan et al., 2021; Jin et al., 2022). The mobile phase consisting of acetonitrile, water and trifluoroacetate (50:50:0.1, v/v/v) was used at a flow rate of 0.8 mL/min, and the detection wavelength was 210 nm (Li and Wen, 2023).
2.4 Determination of the concentration of glucose, xylose, lactate and acetate
In this study, glucose concentrations in culture broth were determined enzymatically by a bioanalyzer (SBA-40C, Shandong, China). To measure the concentration of xylose, lactate as well as acetate of the fermentation broth, HPLC (Agilent, USA) equipped with the HPX-87H column (300 mm × 7.8 mm, Cosmosil, United States) and the differential refractive index detector (SFD GmbH, Schambeck, Germany) was used with a mobile phase of 5 mM H2SO4 at a flow rate of 0.6 mL/min. The column temperature was 30°C.
2.5 Determination of fluorescence intensity of the green fluorescent protein
In order to compare the fluorescence intensity of green fluorescent protein (GFP) under different promoter expression, samples were collected from the fermentation broth at different times and the precipitate was collected and resuspended in 0.9% (w/v) NaCl solution after washing three times. The fluorescence intensity of GFP was measured at an excitation wavelength of 480 nm and an emission wavelength of 527 nm. Samples were assayed in triplicate.
3 Results
3.1 Construction of fengycin-produced Bacillus subtilis
Due to the frameshift mutation in sfp and the single base substitutional mutations at the −10 position of degQ promoter, B. subtilis 168 lost its ability for fengycin production (Tsuge et al., 2007), which is a major problem that must be solved first. Therefore, the strain BSUY00 was constructed by the mean of replacing mutated sfp gene of B. subtilis 168 with normally-worked sfp from Bacillus amyloliquefaciens FZB42 and degQ from B. subtilis 168 under the control of P43 promoter. As shown in Figure 2A, both B. subtilis 168 and BSUY00 grew well in the fermentation medium, indicating that the expression of sfp and degQ, as well as the synthesis of fengycin, did not have a negative effect on the growth of B. subtilis. Surprisingly, the biomass of BSUY00 was significantly higher than that of B. subtilis 168 during the whole fermentation process. In addition, the strain BSUY00 could consume approximately 20.33 g/L glucose after 24 h of fermentation while the strain B. subtilis 168 required 32 h to consume all of the glucose. And we speculated that the earlier glucose consumption of strain BSUY00 might be due to the significant increase in biomass.
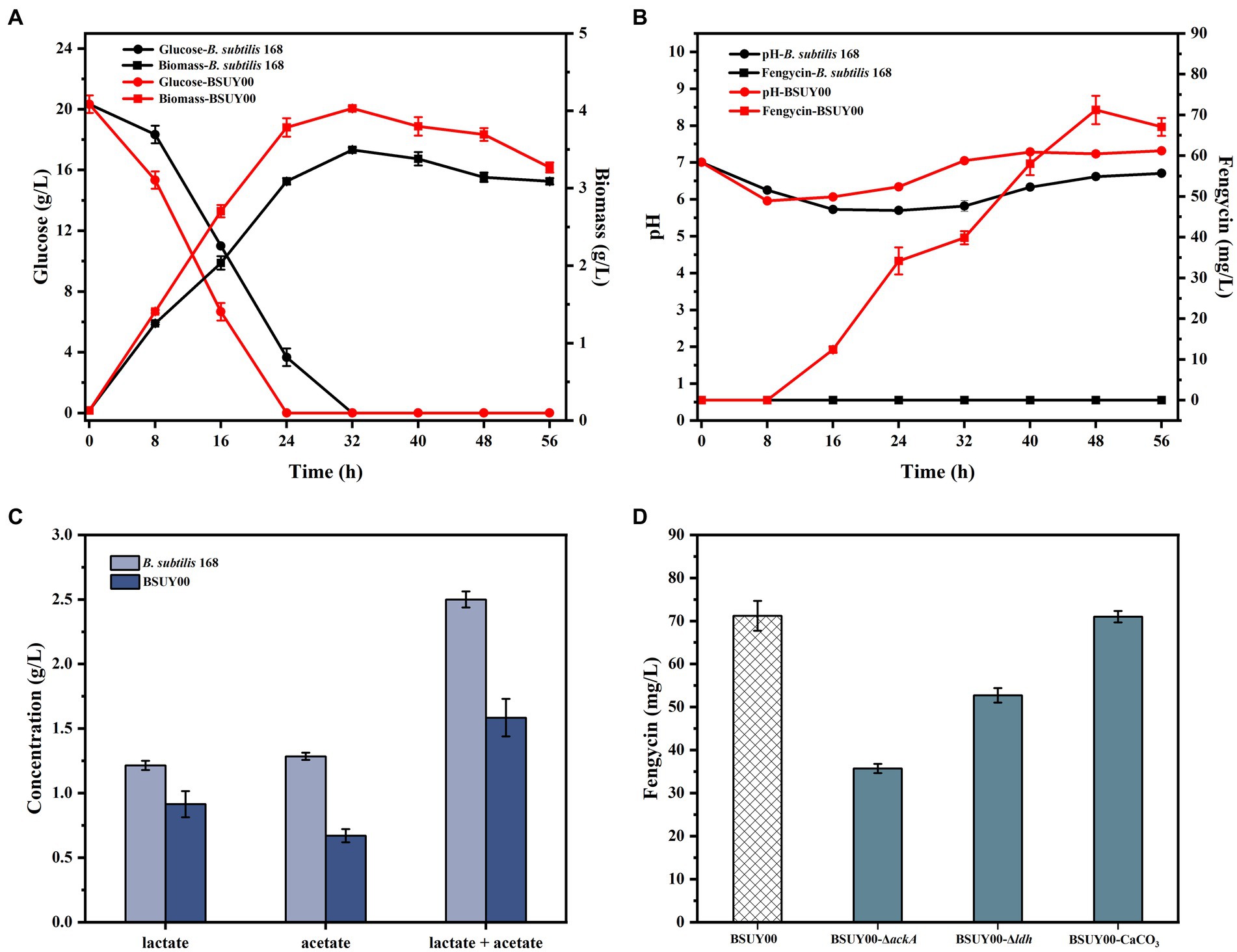
Figure 2. Comparison of fermentation characteristics between the parent strain B. subtilis 168 and constructed fengycin-producing strain BSUY00. (A) The comparison of residual glucose concentration and biomass between B. subtilis 168 and BSUY00. (B) The comparison of fengycin titer and pH of fermentation broth between B. subtilis 168 and BSUY00. (C) The comparison of the concentration of lactate and acetate between B. subtilis 168 and BSUY00. (D) The fengycin titer of BSUY00, BSUY00-ΔackA, BSUY00-Δldh and BSUY00 with addition of 10 g/L CaCO3. The values shown represent the means of three independent experiments and the error bars represent standard deviations of three values.
The results of the fengycin fermentation are shown in Figure 2B. As we suspected, the strain B. subtilis 168 was unable to accumulate fengycin throughout the fermentation process. In the strain BSUY00, fengycin accumulation was began at the mid-exponential phase (about 16 h) and the titer of fengycin was increased rapidly in the mid-exponential and stationary phases. After 48 h of fermentation, the titer of fengycin in BSUY00 reached 71.21 mg/L. The fermentation results mean that the expression of sfp and degQ enabled the BSUY00 strain to successfully gain the ability to synthesize fengycin using glucose as a substrate. However, the titer of fengycin of BSUY00 strain was extremely low and it is necessary to carry out further research based on this strain.
By examining the pH of the fermentation broth of the two strains at different times (Figure 2B), it was found that the pH of the fermentation broth of the two strains decreased significantly during both the delayed and pre-exponential phases (0–8 h), which indicated that the strains started to synthesize organic acids with the glucose uptake, and the phosphate buffer system in the fermentation medium was not sufficient for rapid neutralization of the produced organic acids. While during the phase when fengycin began to accumulate rapidly, the pH of the fermentation broth of the strain BSUY00 was higher than that of the fermentation broth of the strain B. subtilis 168. By determining the acetate and lactate contents of fermentation broth of B. subtilis 168 and BSUY00, it was found that the strain BSUY00 accumulated 0.91 g/L lactate and 0.67 g/L acetate after 16 h of fermentation, which were 24.79% and 47.66% lower than those of B. subtilis 168 (1.21 g/L lactate and 1.28 g/L acetate), respectively (Figure 2C). Based on this result, it was speculated that the synthesis of fengycin led to a reduction in the synthesis of organic acids and a redirection of intracellular metabolic fluxes in the BSUY00 strain, which increased the metabolic flux of intracellular precursors for fengycin synthesis as well as made the pH environment in the culture medium more suitable for the growth of B. subtilis.
It has been proved that pH of fermentation medium plays an important role in cycle lipopeptides production (Dang et al., 2019; Gao et al., 2022). Thus, combined with aforementioned findings, ackA, which encodes acetate kinase, and ldh, which encodes lactate dehydrogenase, were subsequently knocked out in the BSUY00 strain, respectively. Contrary to our expectations, the knockout of ackA and ldh resulted in a 49.84% and 25.95% decrease in fengycin production, respectively (Figure 2D). It was speculated that although the production of acetate and lactate may compete with fengycin production for precursors and energy, while affect the pH of the fermentation system, the knockout of ackA and ldh has a negative effect on fengycin production, possibly because it dramatically disrupts the metabolic flux balance in the cell. In particular, it has previously been shown that the lactate dehydrogenase encoded by the ldh gene oxidizes NADPH to NADP+, and the knockout of ldh resulted in the overaccumulation of NADPH in the cell, which led to a severe inhibition on the growth of B. subtilis (Romero et al., 2007). Subsequently, the addition of 10 g/L CaCO3 was attempted in this study to adjust the pH of the fermentation system, however, no significant changes in fengycin titer were detected, which could be attributed to the regulation of three two-component signal-transduction systems ResD/ResE, PhoP/PhoR and DegU/DegS caused by released Ca2+ (Chen et al., 2022).
3.2 Enhancement of fengycin titer through promoter optimization
As the most direct and effective way to regulate gene expression levels, promoter optimization has been widely used in the regulation of carbon flux distribution and the expression of exogenous proteins. Hence, in order to improve the transcription level of pps cluster and to achieve the purpose of increasing the production of fengycin, four promoters (including P43 promoter, promoter of veg, promoter of spoVG and promoter of lytR) were selected and their expression strength was analyzed through fluorescence intensity of GFP reporter system, which gave the strains named BSUY00-P43-gfp, BSUY00-Pveg-gfp, BSUY00-PspoVG-gfp and BSUY00-PlytR-gfp, respectively. As shown in Figures 3A,B, although the growth of BSUY00-Pveg-gfp was slightly lower than that of the other strains, which probably due to a metabolic stress on the strain caused by the high expression of the GFP fluorescent protein, it showed the highest fluorescence intensity throughout the fermentation process. And the maximum relative fluorescence intensity of BSUY00-Pveg-gfp was about 3.08 times, 8.19 times and 13.25 times that of BSUY00-P43-gfp, BSUY00-PspoVG-gfp and BSUY00-PlytR-gfp, respectively (Figure 3C). The results demonstrate that the four promoters selected in this study exhibit different strength of gene expression, which can be applied for the subsequent replacement of the original promoters in the BSUY00 strain to regulate the expression level of the target genes and modify its metabolic pathway. Subsequently, fengycin production of different strains constructed by replacement of native PppsA promoter with different promoters, which were named BSUY04, BSUY04-1, BSUY04-2 and BSUY04-3, respectively, were compared with the fengycin titer of BSUY00 strain as a control (Figure 3D). The replacement of native weak PppsA promoter with all four promoters selected in this study improved fengycin titer, in which the strain BSUY04-1 constructed with Pveg showed the highest fengycin production, reaching 371.63 mg/L, which was approximately 5.2 times that of BSUY00.
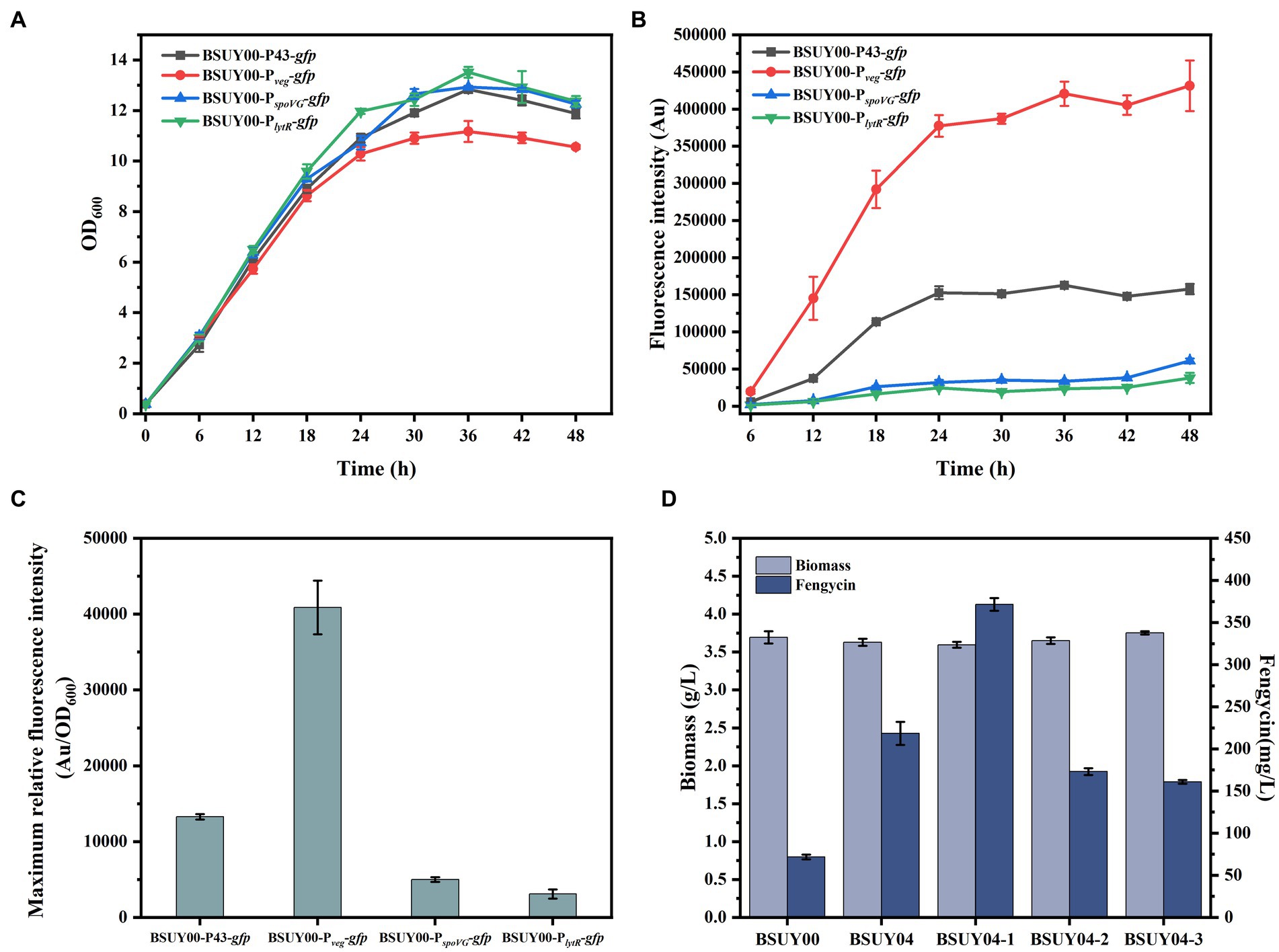
Figure 3. Characterization of GFP expression systems and fermentation characteristics of BSUY00 and its derivative strains constructed by replacement of PppsA with different promoters. (A) Cell growth curves of the strains BSUY00-P43-gfp, BSUY00-Pveg-gfp, BSUY00-PspoVG-gfp and BSUY00-PlytR-gfp. (B) Fluorescence intensity of the strains BSUY00-P43-gfp, BSUY00-Pveg-gfp, BSUY00-PspoVG-gfp and BSUY00-PlytR-gfp. (C) Maximum relative fluorescence intensity of the strains BSUY00-P43-gfp, BSUY00-Pveg-gfp, BSUY00-PspoVG-gfp and BSUY00-PlytR-gfp. (D) Biomass and fengycin titer of the strains BSUY00, BSUY04, BSUY04-1, BSUY04-2 and BSUY04-3. The values shown represent the means of three independent experiments and the error bars represent standard deviations of three values.
3.3 Construction of Bacillus subtilis strains for xylose utilization
In B. subtilis, xylose transport is in the charge of the AraE protein (Krispin and Allmansberger, 1998). Once xylose is successfully taken up into cells, xylose is first isomerised to xylulose by xylulose kinase encoded by xylA, and then xylulose is subsequently phosphorylated to form xylulose-5-phosphate by xylulokinase encoded by xylB, after which the xylose metabolic pathway is channeled to glycolysis via the pentose phosphate pathway (PPP). However, the expression of AraE was negative regulated by the protein encoded by araR (Park et al., 2012). And the low efficiency of xylose transport in B. subtilis is the problem needed to be addressed first.
To visualize the effect of the knockout of the araR, the BSUY04-1 and BSUY05 strains were cultured in M9 medium with xylose as the sole carbon source and the results were shown in Figure 4A. As we suspected, the strain BSUY04-1 grew very slowly, with growth rate of only 0.04 h−1, while the BSUY05 strain can grow rapidly at a growth rate of 0.14 h−1. After 48 h of incubation, the BSUY04-1 strain consumed only 2.11 g/L xylose with the consumption rate of 0.04 g/L/h and the BSUY05 strain could consume about 12.71 g/L xylose with xylose consumption rate of 0.26 g/L/h, which is 5.5-fold higher than that of BSUY04-1. These results show that knockout of the araR gene can significantly improve the xylose uptake capacity.
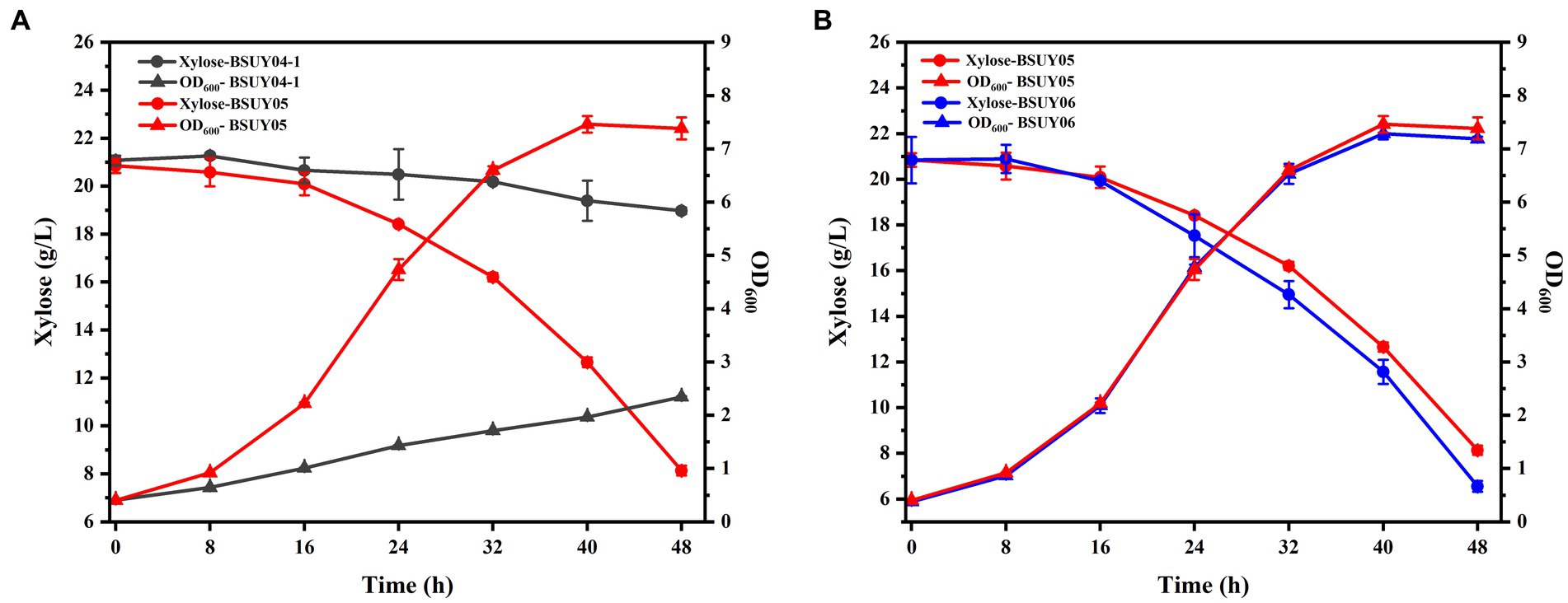
Figure 4. Comparison of fermentation characteristic between the parent strain BSUY04-1 and its derivative strains. (A) The comparison of residual xylose concentration and OD600 between BSUY04-1 and BSUY05. (B) The comparison of residual xylose concentration and OD600 between BSUY05 and BSUY06. The values shown represent the means of three independent experiments and the error bars represent standard deviations of three values.
To enhance the expression of the xylose transport protein AraE, and thus further improve the efficiency of xylose transportation into the cell to participate in metabolic process, the araE gene was intensively constitutive expressed in subsequent studies, and the engineered strain BSUY06 was cultured in M9 medium to visualize the effect with the BSUY05 strain as a control. As shown in Figure 4B, the growth rate of BSUY06 was not significantly different from that of BSUY05 during 48 h of incubation period. However, the xylose consumption rate of BSUY06 reached 0.29 g/L/h, which was 11.54% higher than that of BSUY05. These results suggested that the enhancement of araE gene expression on the basis of the knockdown of araR gene still promotes an increase in xylose uptake rate.
3.4 The fermentation characteristics of the strain BSUY06 with xylose as the substrate
Through the previous studies, we had successfully obtained a B. subtilis strains BSUY06, which can efficiently utilize xylose. To give the fermentation characteristics for fengycin production, the strain BSUY06 was cultured in fermentation medium for 64 h and the results were shown in Figure 5A. Under the fermentation conditions, strain BSUY06 was able to consume approximately 20.41 g/L xylose within 40 h, with a xylose consumption rate of 0.51 g/L/h. No fengycin accumulation was observed during both the delayed and pre-exponential phases (0–8 h), and fengycin production in BSUY06 strain was began after about 16 h of incubation, with a final titer of 376.58 mg/L. The aforementioned results suggested that the strain BSUY06 displays a formidable capability to product fengycin with xylose as the substrate.
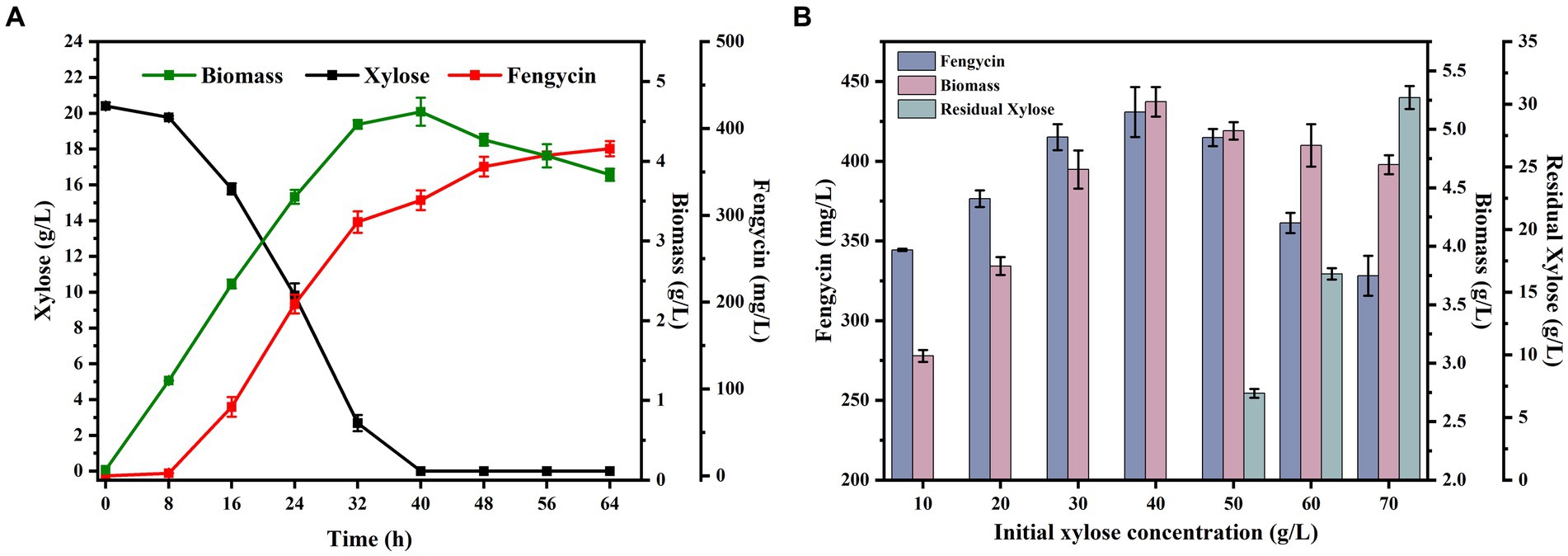
Figure 5. Fermentation characteristics of strain BSUY06. (A) Fermentation characteristics of the strain BSUY06 with 20 g/L xylose as the carbon source during 64 h of fermentation. (B) Fermentation characteristics of strain BSUY06 with different initial xylose concentrations (10–70 g/L) after 64 h of fermentation. The values shown represent the means of three independent experiments and the error bars represent standard deviations of three values.
To ensure a sufficient carbon source and avoid the inhibition of cell growth and waste of residual xylose caused by high concentration substrate, a series fermentation of the BSUY06 strain was performed under different initial xylose concentrations (10, 20, 30, 40, 50, 60 and 70 g/L xylose) in this study. As shown in Figure 5B, at the end of the fermentation, the biomass of BSUY06 strain was increased from 3.06 g/L to 5.23 g/L when the initial xylose concentration was increased from 10 g/L to 40 g/L. In contrast, the decrease in biomass could be observed when the initial xylose concentration continued to increase to 70 g/L. This result indicated that the BSUY06 strain demonstrated favorable growth under low concentration of xylose, while high initial xylose concentration (50–70 g/L) can considerably impede cell growth due to substrate inhibition and increased osmotic pressure. Notably, the fengycin titer increased from 344.35 mg/L to 430.86 mg/L when the initial xylose concentration was increased from 10 g/L to 40 g/L while the opposite trend could be observed when the initial xylose concentration continued to increase to 70 g/L, which showed a similar trend with the biomass. In addition, when the initial concentration of xylose reached 50 g/L, a residual xylose concentration of 6.93 g/L was detected in the broth at the end of fermentation, and as the initial xylose concentration continued to increase to 70 g/L, the level of residual xylose rose gradually from 6.93 g/L to 30.53 g/L, leading to significant wastage of xylose. Based on the aforementioned results, it can be concluded that the most optimal initial concentration of xylose for fengycin fermentation is 40 g/L, as this concentration resulted in the highest biomass and fengycin titer, with no residual waste of xylose at the end of the fermentation.
4 Discussion
Bacillus subtilis, a safety model strain approved by the U.S. Food and Drug Administration (FDA), own a clear genetic background and it has been elucidated to contain three gene clusters devoted to non-ribosomal synthesis of fengycin (synonymous to plipastatin), surfactin and siderophore bacillibactin, making it a potential productivity for peptide antibiotics (Tosato et al., 1997; Chen et al., 2009; Medeot et al., 2017; Yang et al., 2017). However, no fengycin or surfactin synthesis was observed. Previous studies have shown that gene clusters are not the only elements required for fengycin synthesis, but that the sfp and degQ genes also play an irreplaceable role (Tsuge et al., 1999; Wang et al., 2015; Yaseen et al., 2016). The sfp gene encodes 4′-phosphopantetheinyl transferase that converts inactive apo-enzyme peptide synthetases to their active holo-enzyme forms by posttranslational transfer of the 4′-phosphopantetheinyl moiety of coenzyme A to the synthetases (Tsuge et al., 1999, 2007). The DegQ may function as a regulator for the expression of pps operon (Tsuge et al., 1999, 2007). In B. subtilis, DegU/DegS is an important two-component system and previous studies had proved that DegU is a positive regulator of fengycin synthesis (Wang et al., 2015; Lilge et al., 2021). Since DegQ plays a direct role in activating of the DegU response regulator, it may serve as a regulatory key point for the DegU-mediated production of either surfactin or plipastatin, and an unaffected degQ expression resulted in a sixfold increase in plipastatin, whereas degQ deletion in DSM10T reduced plipastatin production by fivefold (Lilge et al., 2021). However, due to the frameshift mutation in sfp and the single-base substitutional mutations at the −10 position of the degQ promoter, B. subtilis 168 lost its ability (Tsuge et al., 2007), which is a major problem need to be solved first. Therefore, in this study, functional sfp and degQ were expressed in B. subtilis 168, resulting in a fengycin production of 71.21 mg/L. However, in the previous study, a functional sfp from B. subtilis ATCC21332 was also expressed in B. subtilis 168 and led to a fengycin titer of merely 15 mg/L after 48 h of fermentation in the derivative strain BBG111 (Coutte et al., 2010), which is much lower than that of the strain BSUY00 in our study. We hypothesized that the different fengycin titers may due to the fact that sfp from different sources may exhibit different protein activities in B. subtilis 168. On the other hand, the lack of functional degQ gene expression and the difference in fermentation medium also contributed to the differences in fengycin titers.
Promoter engineering is considered to be a powerful strategy to regulate gene expression levels and increasing the titer of high-value chemicals. And till now on, many promoters have been identified through transcriptome data as well as related databases and applied for the optimization of biological system and metabolic engineering. According to the previous study, Pveg showed the higher mRNA expression level as well as the protein production than P43 promoter, the most widely used strong promoter for the cytidine deaminase gene (cdd) in B. subtilis (Jin et al., 2019). Besides, the replacement of native PsrfA with Pveg led to an increase of surfactin titer from 0.07 g/L to 0.26 g/L (Willenbacher et al., 2016), which makes Pveg promoter one of the preferred promoters for increasing the expression level of pps cluster. As the most of the endogenous promoters of B. subtilis were not well explored and comprehensively analyzed, 114 promoters were selected and characterized in the published study, in which PspoVG (class II) and PlytR (class III) showed the highest relative fluorescent intensity, which were higher than those of the P43 promoter and the Pveg promoter, respectively, and resulted the highest activities of keratinase (Yang et al., 2017). However, different trends were observed in this study, which indicated that the expression levels of promoters might be influenced by culture conditions. And the similar results can also be obtained in previous study that different culture temperature and pH could lead to a different magnitudes and trends in relative fluorescent intensity (Yang et al., 2017). Therefore, to better achieve our objective, it is necessary to employ reporter genes and systematically analyze promoter functions on the basis of analyzing transcriptomic data. In the present study, it was demonstrated that Pveg was proved to be the most effective promoter, leading to the highest fengycin titer.
Over the last few decades, more and more researches are devoted to the development of alternative raw materials in response to economic and environmental challenges. Lignocellulosic biomass, derived from agricultural and forestry residues, exceeds 200 billion tons and has been widely recognized as a highly promising feedstock due to its renewability, abundant availability and great contribution to the development of rural economies. The efficient utilization of xylose, the second most abundant sugar in lignocellulosic biomass after glucose, is crucial for its application (Jojima et al., 2010; Wei et al., 2015; Zhang M. et al., 2019). However, the lack of a highly efficient xylose transport system has limited the xylose utilization of many microorganisms, including B. subtilis, which remains a significant bottleneck to economical utilization of the hydrolysis product of lignocellulosic hydrolysates (Jojima et al., 2010; Xiao et al., 2012). To address this issue, corresponding studies have been carried out in microorganisms such as yeast and Corynebacterium glutamicum, and the results have further demonstrated the importance of xylose transporters for xylose utilization (Sasaki et al., 2009; Jojima et al., 2010; Li et al., 2019). In B. subtilis, xylose is taken up through arabinose transporter AraE. Nevertheless, the expression of araE was regulated by the negative regulator AraR, leading to a low growth rate when the xylose was been used as the sole carbon source (Park et al., 2012). Therefore, in this study, both the deletion of araR and constitutive overexpression of araE were performed to improve the efficiency of xylose transportation and leading to a 6.25-fold increase in xylose consumption, which allows the efficient production of fengycin from xylose with higher efficiency and titers. However, contrary to our expectations, expression of xylAB, the genes in the xylose metabolism pathway, resulted in a significant decrease in the OD600 at the later stages of growth while the overexpression of tal and tktA, the key genes in pentose phosphate pathway, did not contribute to alleviation of growth inhibition (data not shown), which may account for the blockage of downstream metabolic flux. Therefore, more exploration is needed for further increase the xylose consumption rate of xylose.
5 Conclusion
In summary, the expression of functional sfp and degQ in B. subtilis 168 enabled the realization of fengycin production from unproductive level to 71.21 mg/L, while concurrently increasing both the biomass and glucose consumption rates. To increase the titer of fengycin through promoter engineering, the Pveg was proved to be the most powerful promoter among the four selected promoters through fluorescence intensity and fengycin production. To address the low transport efficiency in B. subtilis, it was demonstrated that knockdown of araR and overexpression of araE had a synergistic effect on the increasing of xylose uptake rate, resulting in a remarkable increase in cell growth and xylose consumption. At the end of this study, it was found that a high initial concentration of xylose could decrease fengycin production and result in a significant concentration of xylose remaining in the fermentation broth. The initial concentration of 40 g/L xylose was proved to be optimal for fengycin production, with a final fengycin titer of 430.86 mg/L. This study demonstrated that lignocellulose, the clean and sustainable substrate with xylose as the second largest sugar, is a potential substrate for the production of fengycin.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
YY: Formal Analysis, Methodology, Data curation, Writing – original draft, Writing – review & editing. PW: Methodology, Writing – review & editing. XW: Formal Analysis, Writing – review & editing. JW: Conceptualization, Funding acquisition, Project administration.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China (No. 2018YFA0902200) and the Frontier Science Center of the Ministry of Education and Tianjin University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Ahmad, T., Xing, F., Nie, C., Cao, C., Xiao, Y., Yu, X., et al. (2023). Biocontrol potential of lipopeptides produced by the novel Bacillus subtilis strain Y17B against postharvest Alternaria fruit rot of cherry. Front. Microbiol. 14:1150217. doi: 10.3389/fmicb.2023.1150217
Altenbuchner, J., and Müller, V. (2016). Editing of the Bacillus subtilis genome by the CRISPR-Cas 9 system. Appl. Environ. Microbiol. 82, 5421–5427. doi: 10.1128/AEM.01453-16
Chen, X. H., Koumoutsi, A., Scholz, R., Schneider, K., Vater, J., Süssmuth, R., et al. (2009). Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J. Biotechnol. 140, 27–37. doi: 10.1016/j.jbiotec.2008.10.011
Chen, M., Zheng, M., Chen, Y., Xiao, R., Zheng, X., Liu, B., et al. (2022). Effect of metal ions on lipopeptide secretion from Bacillus subtilis strain FJAT-4: negative regulation by Ca2+. J. Appl. Microbiol. 132, 2167–2176. doi: 10.1111/jam.15347
Cheng, W., Feng, Y., Ren, J., Jing, D., and Wang, C. (2016). Anti-tumor role of Bacillus subtilis fmbJ-derived fengycin on human colon cancer HT29 cell line. Neoplasma 63, 215–222. doi: 10.4149/206_150518N270
Coutte, F., Leclère, V., Béchet, M., Guez, J. S., Lecouturier, D., Chollet-Imbert, M., et al. (2010). Effect of pps disruption and constitutive expression of srfA on surfactin productivity, spreading and antagonistic properties of Bacillus subtilis 168 derivatives. J. Appl. Microbiol. 109, 480–491. doi: 10.1111/j.1365-2672.2010.04683.x
Dang, Y., Zhao, F., Liu, X., Fan, X., Huang, R., Gao, W., et al. (2019). Enhanced production of antifungal lipopeptide iturin A by Bacillus amyloliquefaciens LL3 through metabolic engineering and culture conditions optimization. Microb. Cell Factories 18:68. doi: 10.1186/s12934-019-1121-1
Ding, L., Guo, W., and Chen, X. (2019). Exogenous addition of alkanoic acids enhanced production of antifungal lipopeptides in Bacillus amyloliquefaciens Pc3. Appl. Microbiol. Biotechnol. 103, 5367–5377. doi: 10.1007/s00253-019-09792-1
Gao, W., Yin, Y., Wang, P., Tan, W., He, M., and Wen, J. (2022). Production of fengycin from D-xylose through the expression and metabolic regulation of the Dahms pathway. Appl. Microbiol. Biotechnol. 106, 2557–2567. doi: 10.1007/s00253-022-11871-9
Hu, F., Liu, Y., and Li, S. (2019). Rational strain improvement for surfactin production: enhancing the yield and generating novel structures. Microb. Cell Factories 18:42. doi: 10.1186/s12934-019-1089-x
Jin, L., Li, L., Zhou, L., Zhang, R., Xu, Y., and Li, J. (2019). Improving expression of bovine lactoferrin N-lobe by promoter optimization and codon engineering in Bacillus subtilis and its antibacterial activity. J. Agric. Food Chem. 67, 9749–9756. doi: 10.1021/acs.jafc.9b02350
Jin, J., Yin, Y., Wang, X., and Wen, J. (2022). Metabolic engineering of Bacillus subtilis 168 for the utilization of arabinose to synthesize the antifungal lipopeptide fengycin. Biochem. Eng. J. 185:108528. doi: 10.1016/j.bej.2022.108528
Jojima, T., Omumasaba, C. A., Inui, M., and Yukawa, H. (2010). Sugar transporters in efficient utilization of mixed sugar substrates: current knowledge and outlook. Appl. Microbiol. Biotechnol. 85, 471–480. doi: 10.1007/s00253-009-2292-1
Krispin, O., and Allmansberger, R. (1998). The Bacillus subtilis AraE protein displays a broad substrate specificity for several different sugars. J. Bacteriol. 180, 3250–3252. doi: 10.1128/JB.180.12.3250-3252.1998
Li, X., Chen, Y., and Nielsen, J. (2019). Harnessing xylose pathways for biofuels production. Curr. Opin. Biotechnol. 57, 56–65. doi: 10.1016/j.copbio.2019.01.006
Li, Y., and Wen, J. (2023). Metabolomic analysis of the effect glutamate on fengycin-overproducing Bacillus subtilis ATCC 21332 with an enhanced fatty acid synthesis pathway. Biochem. Eng. J. 196:108957. doi: 10.1016/j.bej.2023.108957
Lilge, L., Vahidinasab, M., Adiek, I., Becker, P., Kuppusamy Nesamani, C., Treinen, C., et al. (2021). Expression of degQ gene and its effect on lipopeptide production as well as formation of secretory proteases in Bacillus subtilis strains. Microbiologyopen 10:e1241. doi: 10.1002/mbo3.1241
Liu, H., Sun, L., Wang, Y., Lei, X., and Xu, D. (2012). Modeling antimicrobial activity of lipopeptides from Bacillus amyloliquefaciens ES-2 against Shewanella putrefaciens in shrimp meat using a response surface method. J. Food Prot. 75, 1855–1858. doi: 10.4315/0362-028X.JFP-12-073
Lu, H., Qian, S., Muhammad, U., Jiang, X., Han, J., and Lu, Z. (2016). Effect of fructose on promoting fengycin biosynthesis in Bacillus amyloliquefaciens fmb-60. J. Appl. Microbiol. 121, 1653–1664. doi: 10.1111/jam.13291
Ma, Z., Wang, N., Hu, J., and Wang, S. (2012). Isolation and characterization of a new iturinic lipopeptide, mojavensin A produced by a marine-derived bacterium Bacillus mojavensis B0621A. J. Antibiot. 65, 317–322. doi: 10.1038/ja.2012.19
Medeot, D. B., Bertorello-Cuenca, M., Liaudat, J. P., Alvarez, F., Flores-Cáceres, M. L., and Jofré, E. (2017). Improvement of biomass and cyclic lipopeptides production in Bacillus amyloliquefaciens MEP218 by modifying carbon and nitrogen sources and ratios of the culture media. Biol. Control 115, 119–128. doi: 10.1016/j.biocontrol.2017.10.002
Ongena, M., Jourdan, E., Adam, A., Paquot, M., Brans, A., Joris, B., et al. (2007). Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 9, 1084–1090. doi: 10.1111/j.1462-2920.2006.01202.x
Park, Y.-C., Jun, S. Y., and Seo, J.-H. (2012). Construction and characterization of recombinant Bacillus subtilis JY123 able to transport xylose efficiently. J. Biotechnol. 161, 402–406. doi: 10.1016/j.jbiotec.2012.07.192
Romero, S., Merino, E., Bolívar, F., Gosset, G., and Martinez, A. (2007). Metabolic engineering of Bacillus subtilis for ethanol production: lactate dehydrogenase plays a key role in fermentative metabolism. Appl. Environ. Microbiol. 73, 5190–5198. doi: 10.1128/AEM.00625-07
Sambrook, D. R. (2001). Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press: New York.
Sasaki, M., Jojima, T., Kawaguchi, H., Inui, M., and Yukawa, H. (2009). Engineering of pentose transport in Corynebacterium glutamicum to improve simultaneous utilization of mixed sugars. Appl. Microbiol. Biotechnol. 85, 105–115. doi: 10.1007/s00253-009-2065-x
Sur, S., Romo, T. D., and Grossfield, A. (2018). Selectivity and mechanism of fengycin, an antimicrobial lipopeptide, from molecular dynamics. J. Phys. Chem. B 122, 2219–2226. doi: 10.1021/acs.jpcb.7b11889
Tan, W., Yin, Y., and Wen, J. (2021). Increasing fengycin production by strengthening the fatty acid synthesis pathway and optimizing fermentation conditions. Biochem. Eng. J. 177:108235. doi: 10.1016/j.bej.2021.108235
Tan, T.-T., Zhang, X.-D., Miao, Z., Yu, Y., Du, S.-L., Hou, X.-Y., et al. (2019). A single point mutation in hmgA leads to melanin accumulation in Bacillus thuringiensis BMB181. Enzym. Microb. Technol. 120, 91–97. doi: 10.1016/j.enzmictec.2018.10.007
Tang, Q., Bie, X., Lu, Z., Lv, F., Tao, Y., and Qu, X. (2014). Effects of fengycin from Bacillus subtilis fmbJ on apoptosis and necrosis in Rhizopus stolonifer. J. Microbiol. 52, 675–680. doi: 10.1007/s12275-014-3605-3
Tosato, V., Albertini, A. M., Zotti, M., Sonda, S., and Bruschi, C. V. (1997). Sequence completion, identification and definition of the fengycin operon in Bacillus subtilis 168. Microbiology 143, 3443–3450. doi: 10.1099/00221287-143-11-3443
Tsuge, K., Ano, T., Hirai, M., Nakamura, Y., and Shoda, M. (1999). The genes degQ, pps, and lpa-8 (sfp) are responsible for conversion of Bacillus subtilis 168 to plipastatin production. Antimicrob. Agents Chemother. 43, 2183–2192. doi: 10.1128/AAC.43.9.2183
Tsuge, K., Matsui, K., and Itaya, M. (2007). Production of the non-ribosomal peptide plipastatin in Bacillus subtilis regulated by three relevant gene blocks assembled in a single movable DNA segment. J. Biotechnol. 129, 592–603. doi: 10.1016/j.jbiotec.2007.01.033
Villegas-Escobar, V., Ceballos, I., Mira, J. J., Argel, L. E., Orduz Peralta, S., and Romero-Tabarez, M. (2013). Fengycin C produced by Bacillus subtilis EA-CB 0015. J. Nat. Prod. 76, 503–509. doi: 10.1021/np300574v
Wang, P., Guo, Q., Ma, Y., Li, S., Lu, X., Zhang, X., et al. (2015). DegQ regulates the production of fengycins and biofilm formation of the biocontrol agent Bacillus subtilis NCD-2. Microbiol. Res. 178, 42–50. doi: 10.1016/j.micres.2015.06.006
Wei, L., Liu, J., Qi, H., and Wen, J. (2015). Engineering Scheffersomyces stipitis for fumaric acid production from xylose. Bioresour. Technol. 187, 246–254. doi: 10.1016/j.biortech.2015.03.122
Willenbacher, J., Mohr, T., Henkel, M., Gebhard, S., Mascher, T., Syldatk, C., et al. (2016). Substitution of the native srfA promoter by constitutive Pveg in two B. subtilis strains and evaluation of the effect on surfactin production. J. Biotechnol. 224, 14–17. doi: 10.1016/j.jbiotec.2016.03.002
Xiao, H., Li, Z., Jiang, Y., Yang, Y., Jiang, W., Gu, Y., et al. (2012). Metabolic engineering of D-xylose pathway in Clostridium beijerinckii to optimize solvent production from xylose mother liquid. Metab. Eng. 14, 569–578. doi: 10.1016/j.ymben.2012.05.003
Yang, S., Du, G., Chen, J., and Kang, Z. (2017). Characterization and application of endogenous phase-dependent promoters in Bacillus subtilis. Appl. Microbiol. Biotechnol. 101, 4151–4161. doi: 10.1007/s00253-017-8142-7
Yaseen, Y., Gancel, F., Drider, D., Béchet, M., and Jacques, P. (2016). Influence of promoters on the production of fengycin in Bacillus spp. Res. Microbiol. 167, 272–281. doi: 10.1016/j.resmic.2016.01.008
Yin, H., Guo, C., Wang, Y., Liu, D., Lv, Y., Lv, F., et al. (2013). Fengycin inhibits the growth of the human lung cancer cell line 95D through reactive oxygen species production and mitochondria-dependent apoptosis. Anti-Cancer Drugs 24, 587–598. doi: 10.1097/cad.0b013e3283611395
Zhang, B., Gao, G., Chu, X.-H., and Ye, B.-C. (2019a). Metabolic engineering of Corynebacterium glutamicum S9114 to enhance the production of L-ornithine driven by glucose and xylose. Bioresour. Technol. 284, 204–213. doi: 10.1016/j.biortech.2019.03.122
Zhang, B., Li, Y., Zhang, Y., Qiao, H., He, J., Yuan, Q., et al. (2019b). High-cell-density culture enhances the antimicrobial and freshness effects of Bacillus subtilis S1702 on table grapes (Vitis vinifera cv. Kyoho). Food Chem. 286, 541–549. doi: 10.1016/j.foodchem.2019.02.050
Zhang, M., Puri, A. K., Wang, Z., Singh, S., and Permaul, K. (2019). A unique xylose reductase from Thermomyces lanuginosus: effect of lignocellulosic substrates and inhibitors and applicability in lignocellulosic bioconversion. Bioresour. Technol. 281, 374–381. doi: 10.1016/j.biortech.2019.02.102
Zhang, D., Qiang, R., Zhou, Z., Pan, Y., Yu, S., Yuan, W., et al. (2022). Biocontrol and action mechanism of Bacillus subtilis lipopeptides’ fengycins against Alternaria solani in potato as assessed by a transcriptome analysis. Front. Microbiol. 13:861113. doi: 10.3389/fmicb.2022.861113
Zhao, Z., Xian, M., Liu, M., and Zhao, G. (2020). Biochemical routes for uptake and conversion of xylose by microorganisms. Biotechnol. Biofuels 13:21. doi: 10.1186/s13068-020-1662-x
Keywords: Bacillus subtilis , fengycin, promoter replacement, xylose transport, initial concentration optimization
Citation: Yin Y, Wang P, Wang X and Wen J (2024) Construction of Bacillus subtilis for efficient production of fengycin from xylose through CRISPR-Cas9. Front. Microbiol. 14:1342199. doi: 10.3389/fmicb.2023.1342199
Edited by:
Eduardo J. Gudiña, University of Minho, PortugalReviewed by:
Surekha K. Satpute, Savitribai Phule Pune University, IndiaVivek Rangarajan, Birla Institute of Technology and Science, India
Murat Ozdal, Atatürk University, Türkiye
Copyright © 2024 Yin, Wang, Wang and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianping Wen, anB3ZW5AdGp1LmVkdS5jbg==
 Ying Yin1,2,3
Ying Yin1,2,3 Jianping Wen
Jianping Wen