- 1Department of Microbiology and Molecular Genetics, Oklahoma State University, Stillwater, OK, United States
- 2Department of Chemistry, Oklahoma State University, Stillwater, OK, United States
Cryptococcus neoformans is an opportunistic fungal pathogen that causes meningitis in >152,000 immunocompromised individuals annually, leading to 112,000 yearly deaths. The four classes of existing antifungal agents target plasma membrane sterols (ergosterol), nucleic acid synthesis, and cell wall synthesis. Existing drugs are not highly effective against Cryptococcus, and antifungal drug resistance is an increasing problem. A novel antimicrobial compound, a eumelanin-inspired indoylenepheyleneethynylene, EIPE-1, was synthesized and has antimicrobial activity against Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus (MSRA), but not towards Gram-negative organisms. Based on EIPE-1’s antibacterial activity, we hypothesized that EIPE-1 could have antifungal activity. For these studies, we tested EIPE-1 against C. neoformans strain H99 and 6 additional cryptococcal clinical isolates. We examined antifungal activity, cytotoxicity, effects on fungal gene expression, and mechanism of action of EIPE-1. Results showed that EIPE-1 has fungicidal effects on seven cryptococcal strains with MICs ranging from 1.56 to 3.125 μg/mL depending on the strain, and it is non-toxic to mammalian cells. We conducted scanning and transmission electron microscopy on the exposed cells to examine structural changes to the organism following EIPE-1 treatment. Cells exposed displayed structural changes to their cell wall and membranes, with internal contents leaking out of the cells. To understand the effect of EIPE-1 on fungal gene expression, RNA sequencing was conducted. Results showed that EIPE-1 affects several processes involved stress response, ergosterol biosynthesis, capsule biosynthesis, and cell wall attachment and remodeling. Therefore, our studies demonstrate that EIPE-1 has antifungal activity against C. neoformans, which affects both cellular structure and gene expression of multiple fungal pathways involved in cell membrane stability and viability.
Introduction
Cryptococcus neoformans is an encapsulated fungal pathogen, transmitted frequently via inhalation of environmental spores found in soil, pigeon excrement, and decomposing wood (Levitz, 1991; Kwon-Chung et al., 2014). Infection by the pathogen can result in cryptococcosis, which manifests initially as a pulmonary disease but can also disseminate to the central nervous system (CNS) leading to cryptococcal meningitis (Chang et al., 2004; Shi et al., 2010; Kwon-Chung et al., 2014). HIV patients are primarily at risk of the development of cryptococcosis. They make up 95% of all cases reported in low-to-middle income countries, such as those in Sub-Saharan Africa, and 80% of all cases in high-income countries (Dhana, 2013; Sloan and Parris, 2014). This is a result of a decrease in their immune system’s ability to clear pathogens, due to the HIV suppression of their CD4+ T-cell count (Pappas et al., 2010; Kwon-Chung et al., 2014; Gibson and Johnston, 2015). In an immunocompetent host, a Th-1 type CD4+ T-cell response would typically clear the infection relatively quickly, with little to no symptoms (Pappas et al., 2010; Gibson and Johnston, 2015). However, in an HIV or immunocompromised host, their immune systems are unable to remove the fungal pathogens from their lungs, leading to the development of a cryptococcal infection (Pappas et al., 2010; Gibson and Johnston, 2015; Garelnabi and May, 2018). After initial infection, C. neoformans can traffic from the lungs to the host’s central nervous system (Garelnabi and May, 2018). This leads to the development of cryptococcal meningitis, which has a 40% fatality rate, even with the use of antifungal treatments with ideal conditions (Rajasingham et al., 2017; Patel et al., 2018). For many individuals with severely compromised immune systems, complete removal of the cryptococcal infection is impossible, resulting in a lifelong commitment to the use of antifungal therapies to keep the infection at bay (Coelho and Casadevall, 2016). Current estimates show in AIDS patients, roughly 152,000 cases of cryptococcal meningitis occur each year, with an average of 112,000 yearly deaths (Rajasingham et al., 2022). To help prevent fatalities from cryptococcal infections, early diagnosis is crucial. However, the use of potent fungicidal drugs in combination with fungistatic drugs are still important for the treatment of the disease. Without them, cryptococcal meningitis is fatal (Chen et al., 2010; Dhana, 2013; Sloan and Parris, 2014; Guo et al., 2016). The World Health Organization (WHO) recommended treatment regimen for cryptococcal meningitis in AIDS patients involves three phases: (a) Induction (2 weeks of treatment), (b) Consolidation (8 weeks of treatment), and (c) Maintenance (6–12 weeks or until HIV is controlled by Highly Active Antiretroviral Therapy (HAART)) (Sloan and Parris, 2014). This treatment requires a combination of antifungal therapies over a course of 6–12 months depending on the drug availability. Unfortunately, reduced availability of the drugs for treatment of cryptococcal meningitis continues to be an issue in most Asian and African countries where the disease is most prevalent (Sloan and Parris, 2014).
As the prevalence of dangerous fungal infections continues to rise, the importance of antifungal drugs has risen significantly (Perlin et al., 2017; Fuentefria et al., 2018). Despite many advances in antifungal therapies over the past few decades, the antifungal treatment options are currently limited to only four structural classes of drugs – polyenes, azoles, 5-fluorocytosine, and echinocandins (Perlin et al., 2017; Van Daele et al., 2019). These classes are divided into their respective group based on their mechanism of action (Perea and Patterson, 2002; Pappas et al., 2010). Each of the above-mentioned classes of antifungal therapies have limitations in relation to effectiveness, toxicity and/or the development of drug resistance (Fuentefria et al., 2018). Moreover, the emergence of intrinsic resistance and the ongoing evolution of drug resistant strains has put weight on the limited selection of antifungals available and contributes to the challenge of treating these infections (Perfect, 2017; Wiederhold, 2017; Geddes-McAlister and Shapiro, 2019; Bermas and Geddes-McAlister, 2020). Therefore, the discovery of novel antifungal therapies is critical for fighting these deadly infections (Perlin et al., 2017).
Melanins are dark, negatively charged, hydrophobic pigments that are naturally produced by a multitude of microbes, including bacteria and fungi (Casadevall et al., 2000; Nosanchuk and Casadevall, 2006; Eisenman and Casadevall, 2012; Garcia-Rubio et al., 2020). Eumelanin is a black-brown variety of melanin synthesized by phenoloxidases within a select number of microbes from 3,4-dihydroxyphenyalanine (DOPA) (Hogan et al., 1996; Nosanchuk and Casadevall, 2006; Eisenman and Casadevall, 2012). This variety of melanin is comprised of two monomers, 5,6-dihydroxyindole (DHI) and 5,6-dihydroxyindole-2-carboxylic acid (DHICA) (Selvaraju et al., 2015). The unique properties of the pigments are suggested to have a potential application in the field of medicine (Eisenman and Casadevall, 2012). Melanin production has been associated with increased virulence for various pathogenic microorganisms due to its ability to reduce host defenses by means of antimicrobial mechanisms, including protection from oxidative stress (Nosanchuk and Casadevall, 2006). The ability to protect microbes from the defenses of the host could be relevant to the development of antimicrobial therapies since the use of antimicrobials in tandem with the host immune defenses can increase the effectiveness of some antimicrobial therapies (Nosanchuk and Casadevall, 2006; Adhikari et al., 2022). As such, melanin could be a potential target for the discovery of future antimicrobial therapies. A recent study by Adhikari et al. utilized vanillin for the synthesis of a eumelanin-inspired indolyenepheneethylene synthetic compound, EIPE-1 (Adhikari et al., 2022). Through the application of synthetic approaches for derivatization of the methyl-4,7-dibromo-5,6-dimethoxy-1-methyl-1H-indole-2-carboxylate (DBI), a eumelanin-inspired indole core decorated at the 4- and 7- positions, EIPE-1 was prepared as a potential new antimicrobial agent (Selvaraju et al., 2015; Adhikari et al., 2022; Reed et al., 2023). EIPE-1 has two bactericidal moieties ligated to the DBI core that were intended to exhibit similar antibiotic mechanisms to cationic cell-wall disrupting compounds (Baker et al., 1941; Velkov et al., 2013; Adhikari et al., 2022). This compound demonstrated antimicrobial effects against 13 strains of gram-positive bacteria, including two methicillin resistant strains (Adhikari et al., 2022; Reed et al., 2023). Thus, we hypothesized that EIPE-1 may be effective as an antifungal treatment. In this article, we report the antifungal activity of EIPE-1 and its effects on the medically relevant fungal pathogen, Cryptococcus neoformans.
Materials and methods
Reagents
Unless otherwise stated, chemical reagents and plasticware were obtained from Fisher Scientific (St. Louis, MO). PBS used in washing of cryptococcal cells was obtained at a 10X concentration and diluted 1:10 with deionized water, then sterilized before use. The medium used in Minimum Inhibitory Concentration (MIC) and Minimum Fungicidal Concentration (MFC) Assays was RPMI 1640 supplemented with 0.165 M morpholinepropanesulfonic acid (MOPS), pH 6.9–7, filter-sterilized using a 0.22 μm filter. The cell culture medium used in cytotoxicity experiments was DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS), 10% NCTC-109, 1% non-essential amino acids, 100 U penicillin/ml, and 100 μg streptomycin/ml, filter-sterilized using a 0.22 μm filter. All mammalian cell cultures were incubated at 37° C, 5% CO2 in humidified environments.
Cryptococcus cultures
Cryptococcus neoformans strains H99 (serotype A, mating type α) (gift of John Perfect, Duke University Medical Center, Durham, NC), Cn145a (serotype A), and Cryptococcus gattii strains R265 (serotype B), R272 (serotype B), R4247 (serotype C), and WSA87 (serotype C) (gift of Brian Wickes, University of Texas Health Science Center, San Antonio, TX), and Cryptococcus deneoformans strain 52D (serotype D) were stored at −80°C in 15% glycerol stocks and were plated on yeast extract peptone-dextrose (YPD) (BD Difco; Franklin Lakes, NJ) agar plates. Prior to experiments, individual cryptococcal strains were incubated with shaking in YPD broth for 18 h at 30°C. Cells were collected through centrifugation and washed three times in sterile phosphate-buffered saline (PBS). The cells were quantified using Trypan blue exclusion in a hemacytometer and were resuspended in required medium at the concentration needed for each experiment.
Synthesis of EIPE-1
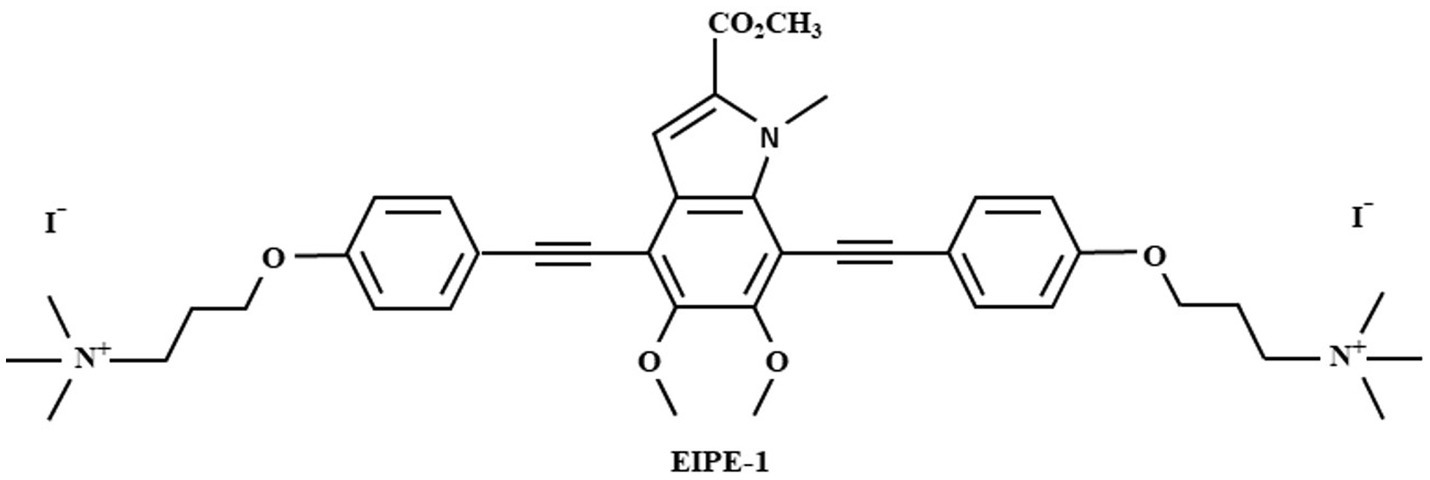
3,3′-(((5,6-dimethoxy-2-(methoxyxarbonyl)-1-methyl-1H-indole-4,7-diyl)bis(ethyne-2,1-diyl))bis(4,1-phenylene)bis(oxy))bis(N,N,N-trimethylpropan-1-aminium) iodide (EIPE-1) (Figure 1) was synthesized and provided by Dr. Nelson’s laboratory (Adhikari et al., 2022; Reed et al., 2023). EIPE-1 powder was then reconstituted with dimethyl sulfoxide (DMSO) to a stock concentration of 5 mg/mL. Dilutions to working concentrations for experiments were made into the media used for each experiment.
Minimum inhibitory concentration and minimum fungicidal concentration assays of Cryptococcus
MIC assays were conducted according to CLSI guidelines (CLSI, 2017 #3098). Briefly, either EIPE-1 or Amphotericin B (AmB) was diluted in RPMI-MOPS, pH 7.0 and evaluated in a two-fold dilution in a concentration range of 100 μg/mL to 0.0488 μg/mL. Dilutions were made in RPMI-MOPS, in a 96-well microtiter plate. A single cryptococcal strain was added to all wells at 0.5×103/ml. Growth controls included the cryptococcal strain grown in media alone. Plates were incubated at 35°C in a humidified incubator for 48 h. The optical densities at 490 nm were measured with a Synergy HTX multi-mode plate reader (BioTek, Winooski, VT), and plates were also visually inspected for turbidity (indicating growth). For MFC assays, dilutions including and above the determined MIC concentration were plated on YPD plates and incubated at 30°C in for 48 h. MFC was defined as the concentration that permitted less than three colony forming units (CFUs), or no growth on the plates, indicating a reduction in 99.9% of the initial inoculum (fungicidal). In other words, the compound has a 99.9% fungicidal activity against the yeast cells (Ernst et al., 1996; Graybill et al., 1997; Espinel-Ingroff et al., 2002; Leite et al., 2014).
Checkerboard assay using EIPE-1 in combination with AmB against Cryptococcus neoformans
Checkerboard assays were conducted using a method previously described, to determine the antifungal activity of EIPE-1 in combination with AmB (Bonifácio et al., 2019; Nelson et al., 2021). EIPE-1 or AmB were evaluated in a two-fold dilution as described in the MIC protocol above. Dilutions were made in RPMI-MOPS, in a 96-well microtiter plate. A single cryptococcal strain was added to all wells at 0.5×103/ml for evaluation of efficacy of combinations. Controls used were EIPE-1 only (row H), AmB only (Column 10), growth control (column 11), and media control (column 12). Plates were incubated at 35°C in a humidified incubator for 48 h. The optical densities at 490 nm were measured with a Synergy HTX multi-mode plate reader (BioTek, Winooski, VT). Results were analyzed using the Fractional Inhibitory Concentration Index (FICI), a non-parametric model based on the Loewe additivity theory to determine the interaction of the combination of EIPE-1 and AmB, where FICI ≤ 0.5 is synergistic, FICI 0.5–4 is indifferent, and FICI ≥ 4 is antagonistic. FICIs were defined as the sum of individual FICs (FICI = FICAm B + FICEIPE-1), with FICs being defined as the MIC derived from the combination therapy divided by their MIC alone (FIC = MICCombination/MICAlone). Off-scale MICs were considered to be the highest or lowest concentration tested in the microdilution assay (Bonifácio et al., 2019; Nelson et al., 2021).
Cytotoxicity assay with EIPE-1
To test the cytotoxicity of EIPE-1 on mammalian cells, individual cell lines, including the human cervical epithelial cell line HeLa, murine fibroblast cell line McCoy, and human lung epithelial cell line A549 (all acquired from ATCC), were tested using the CyQUANT™ LDH Cytotoxicity Assay, fluorescence (Invitrogen). For this, cells were grown in cell culture medium according to ATTC guidelines at 37° C with 5% CO2. The Cytotoxicity Kit was used according to the manufacturer’s instructions. Briefly, mammalian cells were added in triplicate to wells of a 96-well plate (1 × 106 cells/ml in 100 μL). EIPE-1 was prepared similarly to the MIC assay (1X MIC, 2X MIC, and 10X MIC) except cell culture media was used for dilutions and was added at 10 μL per well. Negative controls included media alone and untreated cells, and the positive control included fully lysed cells. Plates were incubated for 24 h at 37°C, 5% CO2. After incubation, 50 μL of reaction mixture was added and incubated for 10 min at room temperature. Following incubation, 50 μL of stop solution was added to each sample. Fluorescence was measured on a Synergy HTX multi-mode plate reader (BioTek) with filters for 560/25 (excitation) and 590/20 (emission). Cytotoxicity of EIPE-1 was conducted in two independent experiments (n = 2) with each cell line, with each condition performed in triplicate. Percent cytotoxicity was defined as the fluorescence of experimental wells (cell line and EIPE-1) divided by negative control untreated cells. Greater than 30% cytotoxicity is considered cytotoxic, whereas lower percentages (<30%) were considered non-toxic (ISO10993-5, 2009).
Electron microscopy
In order to visualize fungal cells using electron microscopy, a higher quantity of fungal cells (10×106 cells) was used. Prior to conducting electron microscopy experiments, we determined the MIC of EIPE-1 using a higher number of cryptococcal cells (strain H99). We followed the same MIC protocol above and determined the MIC was 6.25 μg/mL for this number of cells. Cryptococcus neoformans cells were resuspended in RPMI-MOPS, pH 7.0, at a concentration of 10 × 106 cells/ml. For compound treated samples, EIPE-1 was added at 6.25 μg/mL. Negative controls included untreated C. neoformans strain H99 cells incubated under the same conditions for each time point. The fungal cells were incubated at 35°C in humidity for 4 h, 6 h, 8 h, or 12 h to detect changes in cell morphology over time. The cells were collected by centrifugation. The pellet was resuspended in 2.0% glutaraldehyde in 0.1 M cacodylate buffer at a volume of 1 mL for a minimum of 2 h and processed for scanning electron microscopy (SEM) or transmission electron microscopy (TEM) at the Oklahoma State University (OSU) Microscopy Laboratory (Stillwater, OK) using their provided protocols.
Examination of Cryptococcus via scanning electron microscopy
Fixed C. neoformans cells were collected by centrifugation and transferred to a 12-well plate. Cells were rinsed three times in a buffered wash (30% cacodylate buffer, and 6.15% sucrose) at fixed intervals of 15 min. C. neoformans were incubated in osmium tetroxide (1% OsO4) for 1 h in a 36-well plate with a clear coverslip. Following incubation, C. neoformans was rinsed three times in a buffered wash at fixed intervals of 15 min. Cryptococcus neoformans were dehydrated in ethanol (50, 70, 90, 95, and 100%) in increasing percentages three times at fixed intervals of 15 min. Cryptococcus neoformans were washed two times with hexamethyldisilane at a time interval of 5 min. Coverslips were removed and placed on a clear sheet for 12 h until dried. Coverslips were mounted on stubs using silver paint. Sample mounts were covered in an Au-Pd coat by the OSU Microscopy Laboratory. Images were examined with a FEI Quanta 600 field-emission gun Environmental Scanning Electron Microscope with a Bruker EDS X-ray microanalysis system and HKL EBSD system. Images were examined at 10000X and 20,000x for SEM. At least 8 fields per condition were examined.
Examination of Cryptococcus via transmission electron microscopy
Fixed C. neoformans cells were collected by centrifugation. Media was removed and C. neoformans was rinsed 3 times in a buffered wash at fixed intervals of 15 min. Rinsed cells were resuspended in 1% OsO4 at room temperature for 1 h. 1% OsO4 was removed. Cryptococcus neoformans was rinsed 3 times in a buffered wash at fixed intervals of 15 min. Cryptococcus neoformans were dehydrated in ethanol (50, 70, 90, 95, and 100%) in increasing concentrations three times at fixed intervals of 15 min. Cryptococcus neoformans were washed three times in propylene oxide for fixed intervals of 15 min. Cells were placed in 1:1 propylene oxide and Poly/Bed for 12 h. Cryptococcus neoformans cells were embedded (100% embedding medium) and sliced to 80 nm in thickness by the staff of the OSU Microscopy Laboratory. Images were examined with a JEOL JEM-2100 with Bruker EDS at 8000X for TEM. At least 8 fields per condition per time point were examined.
RNA purification of Cryptococcus neoformans
Cryptococcus neoformans at a concentration of 10 × 106 cells/ml was incubated with EIPE-1 and RPMI MOPS using the minimum inhibitory concentration from the previously described MIC assays for this number of cells. Cells were incubated at 35°C in a humid incubator for 6 h, which correlated to the time point we observed morphological changes in the fungal cells by SEM and TEM. Untreated cryptococcal cells incubated under the same conditions were used as controls. RNA was purified using AllPrep© Fungal DNA/RNA/Protein kit (Qiagen) and quantified using the Take3 plate on a Synergy HTX multi-mode plate reader (BioTek). RNA was determined pure at a 260/280 ratio of 2.0 (ISO20395, 2019). RNA experiments were conducted in triplicate.
RNA analysis
RNA was sent for sequencing to Novogene Corp (Sacramento, CA). Fungal RNA-sequencing was conducted using SMARTer Stranded V2 library prep and samples were sequenced on the Illumina Platform (PE150 Q30 ≥ 80%) (Novogene Corp). Gene expression was compared between each untreated C. neoformans strain H99 incubated for 6 h compared to H99 treated with EIPE-1 for 6 h. This time point was chosen because the fungal cells were still alive, but initial microsocopy studies showed changes in cell wall/membrane were observed starting at 4 h incubation. Statistics were performed by Novogene, and statistically significant differentially-expressed genes (DEG) in the treated vs. untreated cells were reported. Differentially-regulated genes and their reported functions were examined using FungiDB – Fungal & Oomycete Informatics Resources (Amos et al., 2022).
Galleria mellonella infection
Galleria mellonella larvae (Carolina Biological Supply, Burlington, NC) were briefly examined for melanization before storage in groups of ten. Prior to experiments, G. mellonella were removed from food for 24 h. Larvae were washed in 70% ethanol and ampicillin (1 mg/mL) or Rifampicin (1 mg/mL). Galleria mellonella larvae were given an injection into the last proleg with 10 μL of C. neoformans H99 (1×104 cells/ml), heat-killed C. neoformans H99 (1×104 cells/ml), or PBS (Mylonakis et al., 2005; Fuchs et al., 2010; Kay et al., 2019). Following a 2 h incubation period at room temperature, the larvae were injected with 10 μL of EIPE-1 at 15 μg/mL, 20 μg/mL, or 25 μg/mLdiluted in PBS (treatment) or with 10 μL PBS (control) in the second to last proleg (Mylonakis et al., 2005; Tsai et al., 2016; Kay et al., 2019). Galleria mellonella were incubated at 37°C and were examined every 12 h for 10 days. Every 12 h, survival was checked and cocoons were removed to arrest the G. mellonella in their larval stage (Sprynski et al., 2014). Galleria mellonella larvae were considered dead following full-body melanism (turning brown/black) and immobility (Kay et al., 2019).
Statistical analysis
Data analyses were conducted using GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA). Depending on the data collected and interaction observed between cryptococcal cells and the compound, the one-way ANOVA with the Tukey’s multiple comparison test was used to compare the data. For G. mellonella studies, the log-rank test was used to compare survival rates.
Results
EIPE-1 inhibits cryptococcal growth
To determine the antifungal activity of the compound EIPE-1 against cryptococcal strains (H99, Cn145a, R272, R2625, R4247, 52D, and WSA87), we conducted minimum inhibitory concentration (MIC) assays. AmB is an established antifungal drug used against C. neoformans in immunocompromised patients, therefore it was used as a control compound for MIC value comparison against EIPE-1 (Perfect et al., 2010; Sloan and Parris, 2014). Statistical analysis showed a significant difference (p < 0.05) in antifungal activity following incubation with EIPE-1 compared to C. neoformans alone or AmB in RMPI-MOPS (Figure 2). The AmB MIC value had high variation between cryptococcal strains ranging from 0.39 to 6.25 μg/ml. The MIC of EIPE-1 in our assay was 3.125 μg/mL against C. neoformans strain H99, C. gattii strains R272, R625, and R4247. The MIC value of EIPE-1 was 1.56 μg/mL against C. neoformans strain Cn145a, the C. deneoformans strain 52D, and the C. gattii strainWSA87 (Figure 2). However, despite the variation in the EIPE-1 MIC data, it demonstrates that EIPE-1 can inhibit cryptococcal growth of multiple cryptococcal strains at low concentrations.
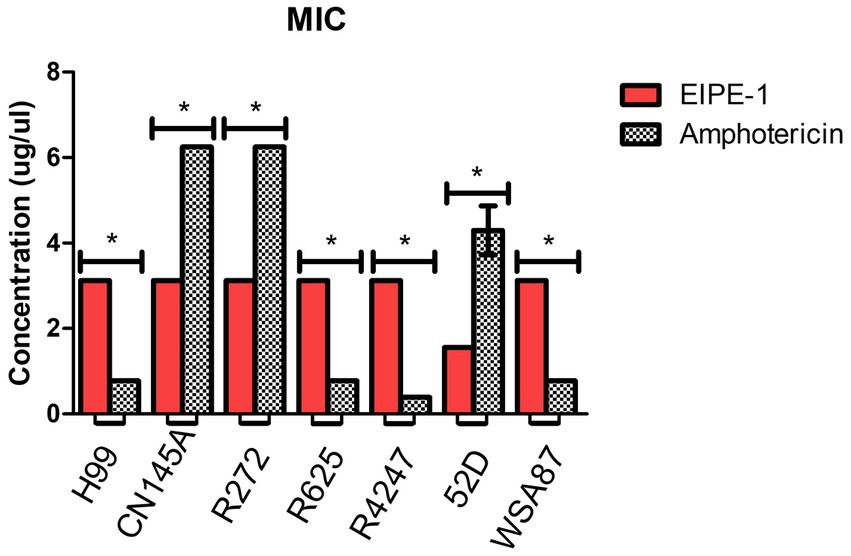
Figure 2. Minimum Inhibition Concentration of EIPE-1 and Amphotericin B against Cryptococcal Strains. Cryptococcal yeast cells (strains H99, Cn145a, R265, R272, R4247, WSA87, or 52D) were incubated in RMPI-MOPs alone, in RPMI-MOPS with either EIPE-1, or Amphotericin B in a two-fold dilution for 48 h at 35°C, with humidity. Optical Densities were determined using a multi-mode plate reader. Data shown are from two independent experiments with each cryptococcal strain and means ± SEM are shown. Statistical significance (p < 0.05) is shown with an asterisk *. Some strains had no variation between experiments and do not have an error bar.
To determine whether the antifungal activity demonstrated by EIPE-1 is fungistatic or fungicidal, we conducted minimum fungicidal concentration (MFC) assays. The results of our YPD plates displayed no visible CFUs present after 48 h incubation. This indicates that EIPE-1 is fungicidal at the MIC concentration for each cryptococcal strain.
The in vitro interaction of antifungal therapy combinations can have a greater efficacy than the sum of their individual actions, such as seen in the current cryptococcal meningitis treatment guidelines that advises treatment via combination drug therapy (Perfect et al., 2010; Sloan and Parris, 2014; Nelson et al., 2021). Therefore, we tested the synthetic compound EIPE-1 in combination with AmB against C. neoformans H99 using a checkerboard assay and categorized the results by the FICI. Both drugs maintained their individual MICs as determined above. Each EIPE-1/AmB combination had an average FICI between 1.17–1.19, placing them in the indifferent category (0.5–4.0). This result is not dependent on the concentration of EIPE-1 used.
EIPE-1 is non-cytotoxic to mammalian cells
In order to determine the relative cytotoxicity of EIPE-1 to mammalian cells, the CyQUANT™ Cytotoxicity Assay Kit was used with three different mammalian cell lines, including the murine fibroblast cell line McCoy, human lung epithelial cell line A549, and the human cervical epithelial cell line HeLa. EIPE-1 was shown to have a cytotoxicity of <30% (non-toxic) at the MIC concentration tested in all cell lines (Figures 3A–C). At 2X concentration, EIPE-1 was non-toxic for two of the three cell lines, but at 10X concentration, EIPE-1 was toxic (>30%) for all three cell lines. However, since the compound cytotoxicity was less than 30% at the 1X and 2X MIC concentration in most cell lines, the compound was determined to be non-toxic to mammalian cells (ISO10993-5, 2009).
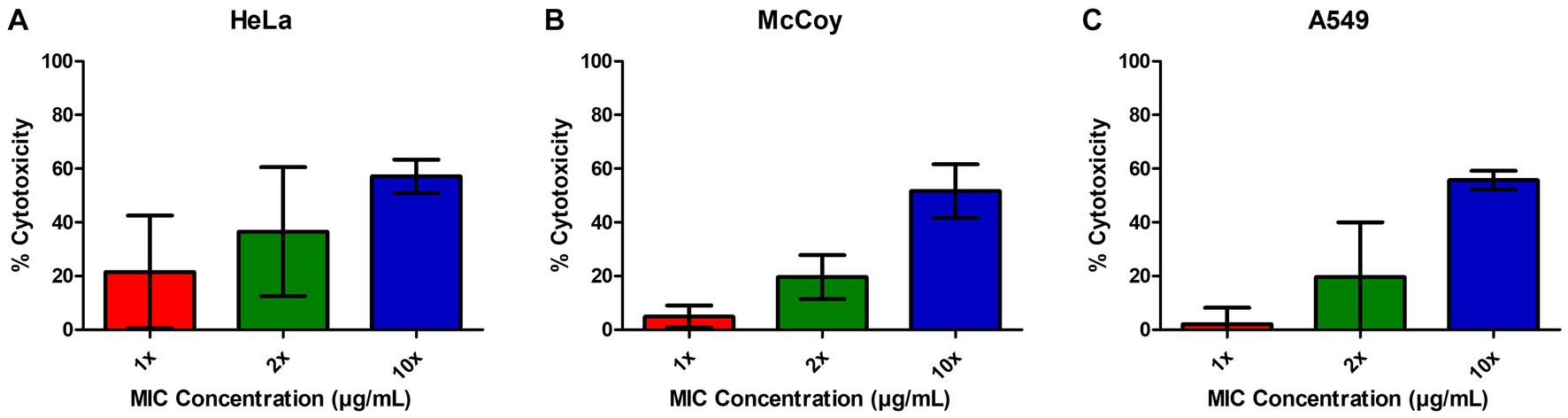
Figure 3. EIPE-1 is not cytotoxic to mammalian cells. (A) Human cervical epithelial cell line HeLa, (B) Murine fibroblast cell line McCoy, and (C) human lung epithelial cell line A549 were incubated in RPMI-MOPS, in RPMI-MOPS with either 6.25 µg/ml EIPE-1, 12.5 µg/ml EIPE-1, or 62.5 µg/ml EIPE-1 for 24 h at 37°C, 5% CO2. Cytotoxicity was defined as greater than 30%. Fluorescence was measured on a Synergy HTX multi-mode plate reader. Data displayed are the mean ± SEM of results of 2 independent experiments.
Electron microscopy reveals structural changes to cryptococcal cells
To understand the mechanism of action of EIPE-1, SEM and TEM analyses were conducted. SEM and TEM can provide vital information about the surface and internal structures of cells (Nixon, 1964; Koga et al., 2021). Cryptococcus neoformans strain H99 cells were incubated with EIPE-1 at 4 h, 8 h, or 12 h, following which cells were prepared for electron microscopy. SEM images displayed cell wall/membrane damage as early as 4 h post-incubation (Figure 4A). Damage was indicated by c-shaped cells, mis-shaped cells, etc. (see arrows Figure 4A), indicative of dead/dying cryptococcal cells (Hole et al., 2012). To determine further the effects of EIPE-1 on the cell wall and membrane of the cryptococcal cells, TEM was conducted. TEM allows the internal structures of the cell to be imaged by sectioning of the sample (Winey et al., 2014). Cryptococcus neoformans cells were incubated with EIPE-1 at 4 h, 8 h, or 12 h. Time points remained the same as with the SEM to provide a comparison between the SEM and TEM images. The TEM images confirmed that the compound is affecting the cell wall and cell membrane of the fungal cells. Four morphologies were identified within the images (Figure 4C). We observed damage to the cell wall and membrane at 8 h and 12 h (Figure 4B). In addition, it appears that the membrane damage results in a leakage of internal contents into the surrounding media (Figures 4B,C). The TEM results confirmed that the compound is affecting these specific cellular structures on the fungal cells. Additionally, TEM showed two other cell morphologies that represented dying or dead cells, including c-shaped cells. The images also include cells with degraded membranes and a black smudge. TEM incorporates the use of heavy metals to prevent electrons from passing through the prepared sample. These metals bind to regions concentrated with DNA and proteins, or components of the cell that are rich in lipids. In a bright-field TEM, these regions, and regions high in mass density tend to appear dark in the imaging to allow contrast. Thus, the black region (smudge) observed in the TEM images could be a representation of regions rich in DNA, protein or lipids (Dempster, 1960; Belazi et al., 2009; Klein et al., 2015; Lange et al., 2021). Nucleic acid or DNA, protein and lipids make up a majority of the internal macromolecules of a living eukaryotic cell. Due to the composition of the internal cellular components of C. neoformans, it is likely the black region observed in the TEM images was comprised of internal structures leaking into the media from a pore present in the cellular wall or membrane of the fungal cell (Schie et al., 2016). As assessed by both the SEM and TEM images, the compound appears to affect the cell wall and membrane of the C. neoformans cells, leading to cell lysis.
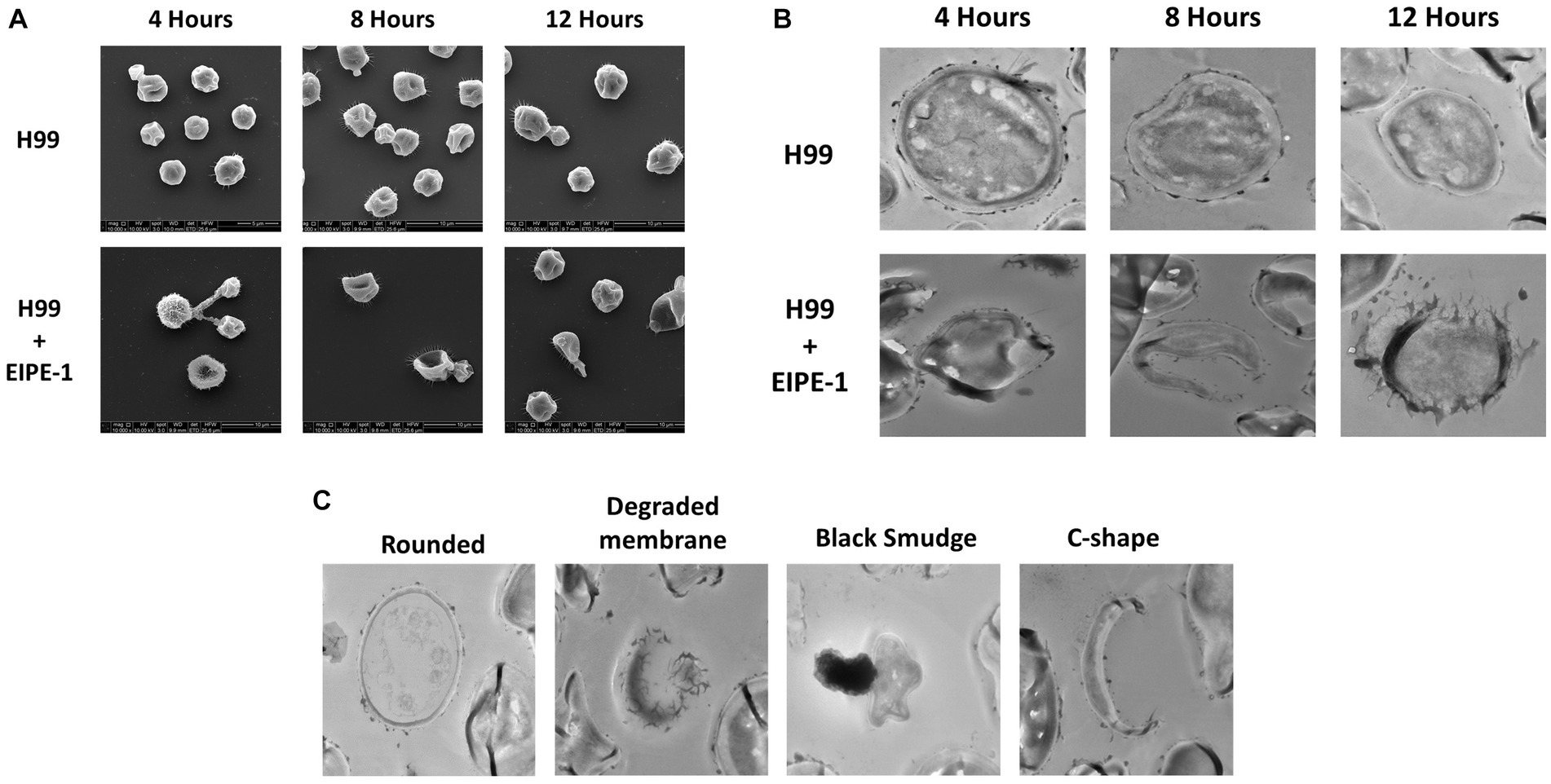
Figure 4. Electron Microscopy of C. neoformans with EIPE-1 shows structural changes. C. neoformans H99 were grown in the presence of EIPE-1 at the calculated MIC of 1.749 μg/mL, for 4, 6, 8, and 12 h, fixed with 2.0% glutaraldehyde in 0.1 M cacodylate buffer, prepared for electron microscopy, and examined for TEM or SEM. (A) SEM of C. neoformans displayed structural changes (as indicated by the arrows) to the cells incubated with EIPE-1 for 4, 8, and 12 h, but not in the untreated cells. (B) TEM of C. neoformans displayed structural changes (arrows) to cells treated with EIPE-1 for 4, 8 and 12 h, but not in the untreated cells. (C) TEM of C. neoformans cells treated with EIPE-1 for 12 h cells displayed four variations in structure – rounded cell (undamaged), degraded membrane, black smudge present over the cell, and c-shape. Magnification is 10,000X for SEM and 8,000X for TEM. Images are representative of at least 8 fields per condition per time point examined.
EIPE-1 treatment affects cryptococcal gene expression
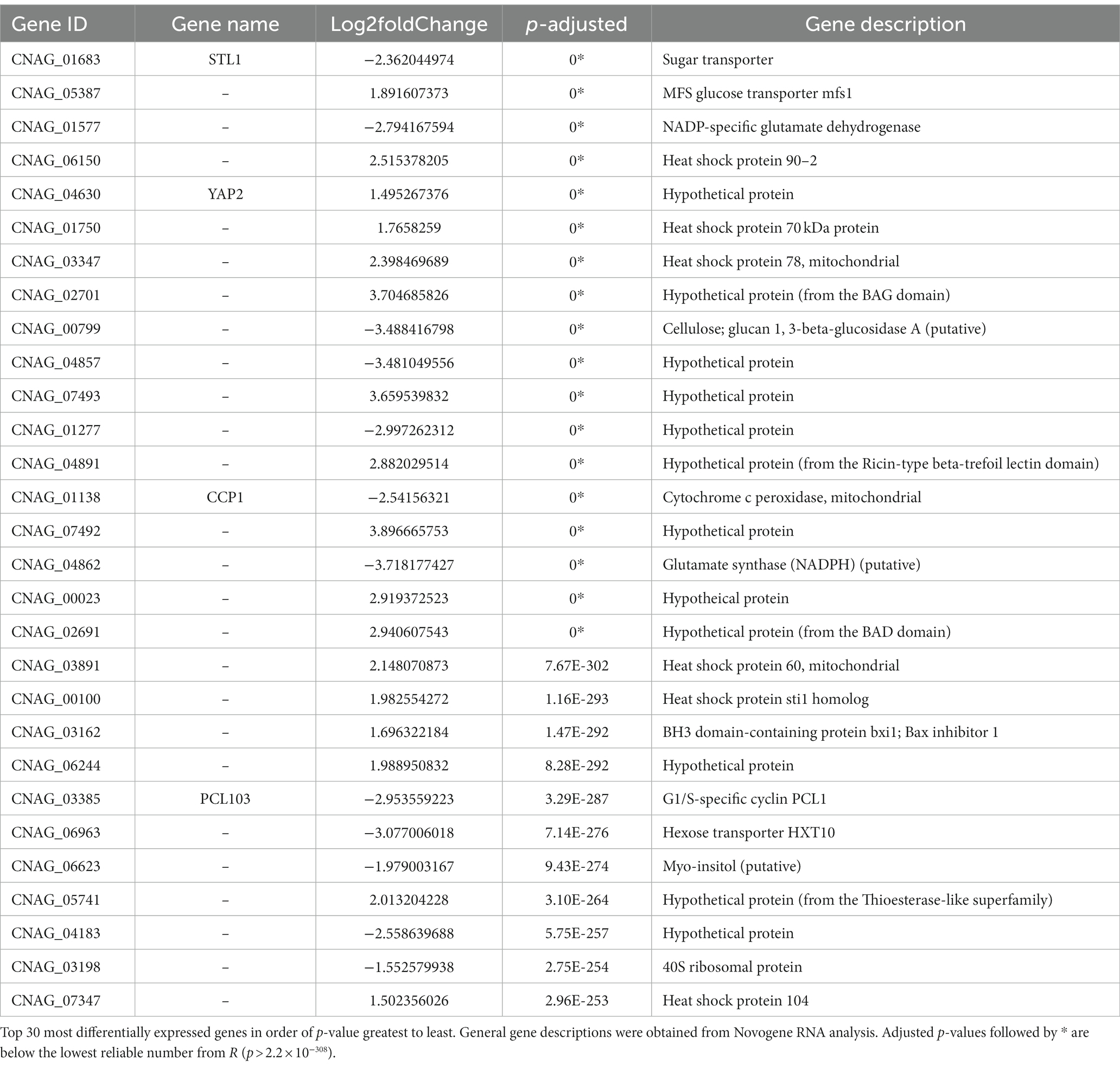
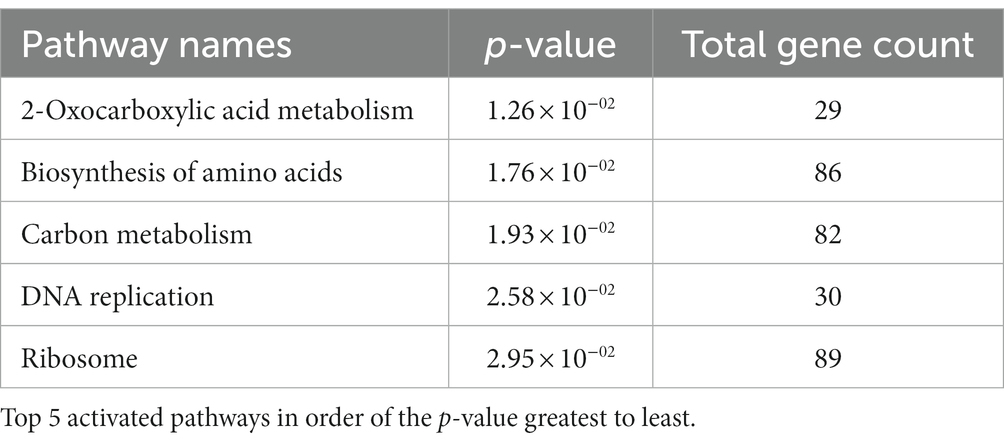
To understand the effect of EIPE-1 on fungal gene expression, we were interested in identifying differential gene expression between C. neoformans incubated with EIPE-1 compared to control. Purified RNA was sent for sequencing at Novogene (Novogene Corp, Sacramento, CA). The analyses identified 4,936 statistically significantly differentially expressed genes (DEG) between untreated and EIPE-1 treated cryptococcal cells. Of these genes, 2,486 were significantly upregulated and 2,450 genes were significantly down-regulated. Due to a limitation on the information available for C. neoformans strain H99, one third of the greatest DEGs listed (Table 1) have unknown functions. However, our analyses of FungiDB showed that the genes with available information have roles in metabolic processes, stress response, and virulence of the cells (Amos et al., 2022). Descriptions of the top thirty differentially-regulated genes and their putative functions are shown (Table 2). Furthermore, analyses identified 91 enriched pathways through the KEGG online database. Of these 91 pathways, none were statistically significant. The top five active pathways and their p-values are shown (Table 2). These pathways are involved in amino acid biosynthesis, carbon metabolism, ribosome formation, and the replication of DNA (Amos et al., 2022). We further filtered through the list of genes and identified several DEGs involved in C. neoformans viability, capsule biosynthesis, capsule attachment and remodeling and ergosterol biosynthesis (Table 3).
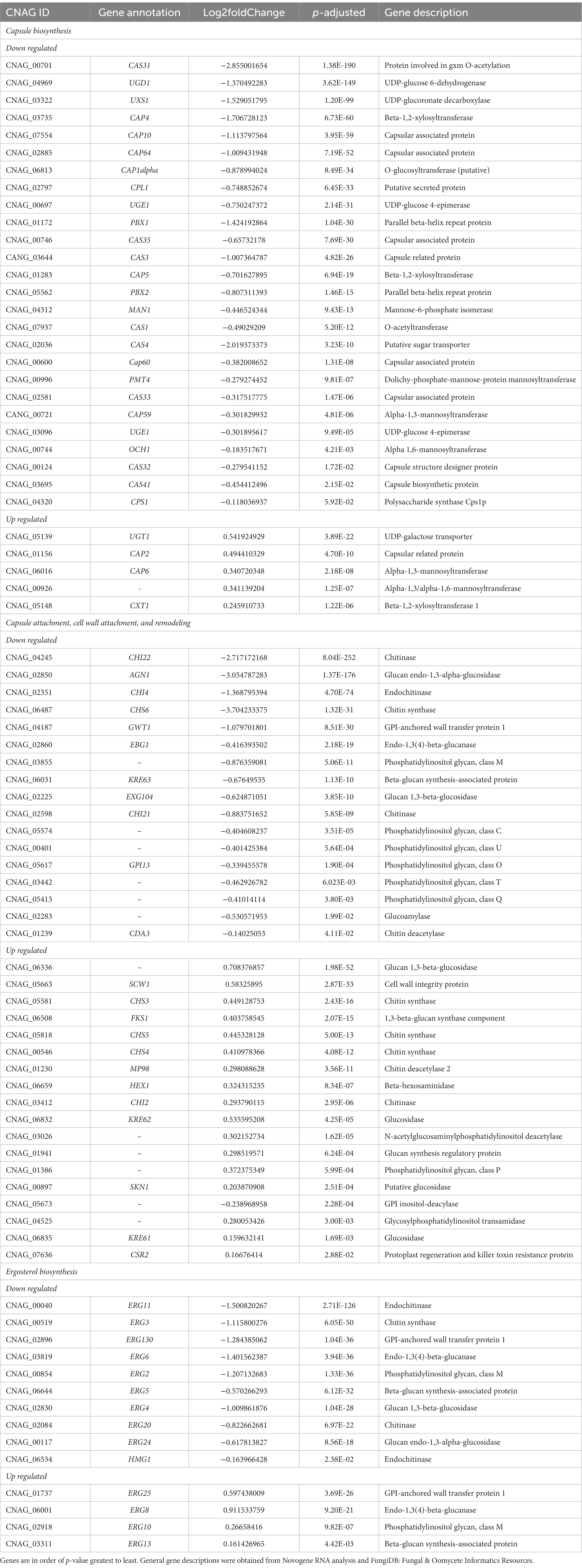
Table 3. Differently regulated genes associated with C. neoformans capsule and cellular wall biosynthesis.
EIPE-1 does not clear cryptococcal infection in Galleria mellonella
To determine the efficacy of EIPE-1 in a living infection model, G. mellonella larvae were infected with C. neoformans H99 and treated with various concentrations of EIPE-1 as mentioned in the methods. As shown in Figure 5, larvae of the G. mellonella inoculated with C. neoformans H99 experienced rapid death by day five of infection. Additionally, larvae inoculated with C. neoformans and treated with EIPE-1 experienced death at similar time points to H99 alone, regardless of the concentration of EIPE-1 (Figure 5).
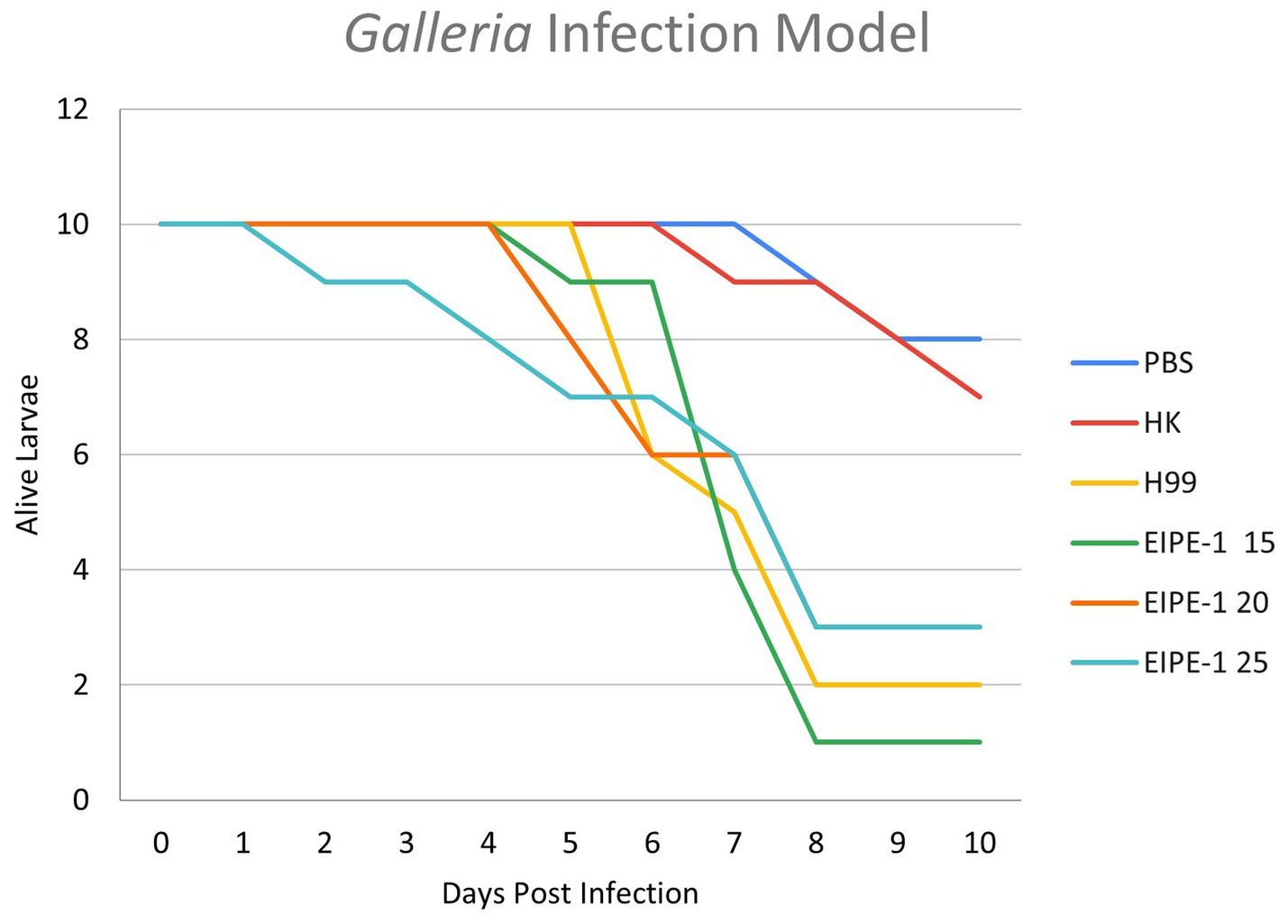
Figure 5. EIPE-1 does not provide antifungal protection during in-vivo infection of Galleria mellonella. G. mellonella larvae were inoculated with either PBS alone or EIPE-1 at the concentration of 15 μg/mL, 20 μg/mL, 25 μg/mL, 50 μg/mL, 100 μg/mL, 150 μg/mL, or 200 μg/mL diluted in PBS. Infected G. mellonella were incubated at 37°C and were examined every 12 h for mortality for 10 days. Larvae were considered dead following full-body melanism and immobility. Data are representative of three independent experiments.
Discussion
Despite advances in antifungal therapies over the decades, antifungals are limited to only 14 individual agents (in 4 classes) that have been approved by the U.S. Federal Drug and Food Administration (FDA) for use in the treatment of fungal infections (Dismukes, 2000; Nett and Andes, 2016). The goal of this study is to explore the potential of EIPE-1 as an effective and non-toxic antifungal for the purpose of increasing the current therapies on the market for the treatment of fungal infections, in particular the infection caused by C. neoformans (Perlin et al., 2017; Van Daele et al., 2019). While its original purpose was as a building block for organic semiconductors, it was later discovered that this core has the interesting capability of serving as a foundation for the integration of antibactericidal moieties and possesses intrinsic antimicrobial activity that causes cytoplasmic membrane disruption in gram positive bacteria. Additionally, it was revealed that thirteen different strains from eight gram-positive bacteria, including two methicillin resistant strains were found to be susceptible to EIPE-1 (Adhikari et al., 2022; Reed et al., 2023). While not previously studied in fungal organisms, we found enough evidence to support studying this compound as an antifungal against Cryptococcus isolates. Therefore, we decided to study this compound to determine its use as a novel antifungal drug against Cryptococcus.
Cells come into contact with environmental stressors (Cowen and Steinbach, 2008). A microbe’s ability to adapt to these stresses present in its surrounding environment is crucial for survival in their biological niches. One key attribute of Cryptococcus is its ability to survive in harsh environments via sensing, responding, and adapting to changes for its survival and proliferation. During antimicrobial treatments, the fungal organism senses and initiates stress signal pathways which allows them to adapt (Dismukes, 2000; Cowen and Steinbach, 2008; Fuchs and Mylonakis, 2009; Nett and Andes, 2016). A stress response can be seen during treatment with EIPE-1, as several stress-associated genes recognized in previous literature have been identified in the RNAseq analysis. These genes include but are not limited to the Ricin Beta Lectin superfamily, ATPases associated with diverse cellular activities (AAA+) superfamily, Bcl-2-associated athanogen (BAG)-family proteins and SLC2A (GLUT) family (Doong et al., 2002; Ishikawa et al., 2011; Mueckler and Thorens, 2013; Gallegos et al., 2014; Khan et al., 2022). In addition, we saw a reduction in ribosomal protein translation in our treated populations. Regulation of translation is crucial for C. neoformans to adapt to the environmental stressors (Knowles et al., 2021).
This capability of C. neoformans to adapt to stressors demonstrates not only its cellular mechanisms, but also its plasticity of its cell wall, which plays a key role in the defense of the cell from environmental stress and maintains integrity of the cell (Rodrigues et al., 2008; Garcia-Rubio et al., 2020; Upadhya et al., 2023). Disruption of the fungal cellular wall by interfering with glucosidases and chitinase may be an important mechanism by which EIPE-1 exerts its antifungal effects. Specifically, during fungal growth, chitinase is involved in the breakdown of chitin and chitosan by hydrolyzing polymers of chitin at the beta-(1-4) linkages. Chitin and chitosan are vital components of the fungal cell wall and have been shown to contribute to the general stability of the cellular wall (Banks et al., 2005; Baker et al., 2009).
All this is an expected response of the cell when exposed to stress and/or apoptotic stimuli of an agent with antifungal capabilities. However, while we believe these genes and transcriptional pathways are the most important for the morphological changes and cellular death observed in the EIPE-1 treated population, we must note that there may be important genes involved that were not identified during initial analyses. Additionally, whereas the C. neoformans genome for strain H99 has been previously sequenced, not all the genes have been annotated to determine the function. Moreover, many of the annotated genes of fungal species are generated by comparison of genomes and by automatic sequence analysis pipelines. Therefore, it is possible that important genes were excluded from our analyses, due to these limitations (Janbon et al., 2014). In the future, follow-up studies need to be done to validate the gene expression data.
Combining all the data from the RNA sequencing and the electron microscopy, we composed a putative model based on the effects of EIPE-1 against C. neoformans as displayed in Figure 6. As found in all living eukaryotic cells, beneath the fungal cell wall, C. neoformans possesses a plasma membrane that consists of a phospholipid bilayer (Rodrigues et al., 2008; Agustinho et al., 2018; Upadhya et al., 2023; Zhukov and Popov, 2023). This membrane maintains the viability of a cell and prevents the free exchange of molecules from the cytoplasm to the cell’s environment and vice versa (Upadhya et al., 2023). Previously, microorganism membrane permeability to hydrophobic molecules was identified as being pertinent for susceptibility to the molecule (Reed et al., 2023). Since C. neoformans has a high cellular surface hydrophobicity due to the presence of mannoproteins, lipids, glucan, and chitin molecules, it could allow the hydrophobic EIPE-1 molecule to passively diffuse across the membrane into the cell (van der Rest et al., 1995; Danchik and Casadevall, 2020; Vij et al., 2020). Once within the cell, it interacts with the cell’s ability to synthesize the cell wall and the membrane. During this process, the cell responds to the presence of EIPE-1 by the up-regulation of genes involved in stress response, including efflux pumps, heat shock proteins, etc. (Doong et al., 2002; Cowen and Steinbach, 2008; Ishikawa et al., 2011; Mueckler and Thorens, 2013; Gallegos et al., 2014; Holmes et al., 2016; Kurop et al., 2021; Khan et al., 2022). Specifically, efflux pumps allow the organism to regulate its internal environment by the removal of antimicrobial substances (Holmes et al., 2016). While the overall function of EIPE-1 is still relatively unknown, it has been shown to interfere with several pathways involved in the biosynthesis of ergosterol, GPI-anchored proteins, GXM/GalXM, chitin, and chitosan. The result of this interference, the cell’s membrane and wall are damaged leading to the formation of a pore and/or breakdown of the cellular membrane. When this occurs, the cell is no longer able to maintain the internal environment and may allow leakage of cellular organelles, as seen in Figure 4C, leading to cellular death.
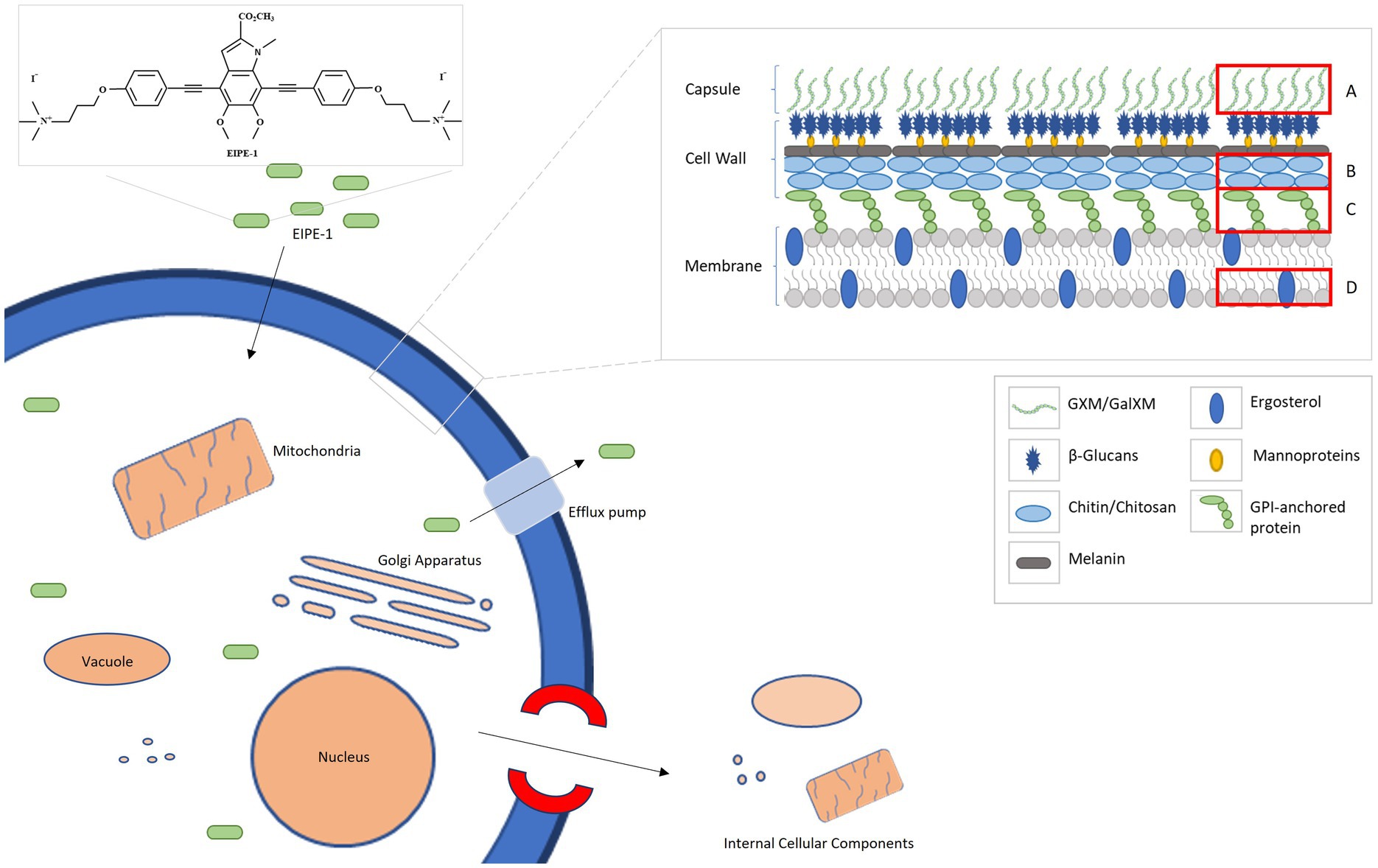
Figure 6. A putative model for the mechanism of antifungal activity of EIPE-1. C. neoformans H99 have a complex cell wall comprised of chitin, chitosan, a-1,3 glucan, B-1,3 glucan, B-1,6 glucan, mannoproteins and GPI-anchored proteins. Additionally, they possess a capsule that is consistently maintained on the outer cell wall and composed of GXM and GalXM. EIPE-1, a hydrophobic eumelanin inspired molecule, may enter the cell through a passive method and alters components of the cell by a downregulation in genes involved with capsule biosynthesis, cell wall attachment and remodeling, and ergosterol biosynthesis. (A) Glucuronoxylomannan (GXM) and Galactoxylomannan (GalXM). (B) Chitin and Chitosan. (C) GPI-anchored proteins. (D) Ergosterol.
This model provides perhaps the clearest illustration of the dynamics of EIPE-1 against C. neoformans. However, we must recognize that the true mechanism of the synthetic molecule’s antifungal activity against Cryptococcus is still largely unknown. It is possible that the damage we are observing in the treated populations could be a downstream effect of the true target of EIPE-1. Future investigations involving the use of C. neoformans mutant libraries to pinpoint the molecular target of EIPE-1 are currently underway in our laboratory. While these data suggest that EIPE-1 may have potential as a novel antifungal against C. neoformans, we are aware that we do not know the true efficiency of the synthetic compound within an in vivo model. There are various factors that can impact the efficiency of a therapeutic treatment in a living model over time from host-pathogen interactions to the distribution, metabolism, and elimination of EIPE-1 from the host’s body. All these can impact the bioavailability and efficiency of the administered drug since less of our drug may remain active or as potent at the target sites of infection (Gillette, 1971; Ekins et al., 2000; Adepu and Ramakrishna, 2021). Additionally, during pathogenesis, C. neoformans cells typically interfere with immune cell recognition and phagocytosis with its virulence factors, including melanin production and a capsule composed of galactoxylomannan (GalXM) and glucuronoxylomannan (GXM). These factors add protective features to the fungal cells. For example, the components of the capsule have an anti-phagocytic influence on immune phagocytes, allowing the pathogen to evade phagocytosis (Kozel and Gotschlich, 1982; Kozel et al., 1988; Yauch et al., 2006; Zaragoza et al., 2009; Vecchiarelli et al., 2013; Conn and Wozniak, 2023). Interestingly, our EIPE-1 treated C. neoformans cells have a reduction in genes that regulate capsule biosynthesis. While this pathway is not typically required for viability of the yeast since acapsular mutants can survive and replicate in vitro (Grijpstra et al., 2009; Tefsen et al., 2014), it is required for virulence in vivo (Chang and Kwon-Chung, 1994; Chang et al., 1996), and the absence of capsule results in a reduction in virulence. This indicates that during treatment our C. neoformans may remain in a less virulent state, which may also aid our immune cells during pathogen clearance when the correct bioavailability and potency is maintained (Gillette, 1971; Ekins et al., 2000; Adepu and Ramakrishna, 2021). It is important to learn the role our innate immune cells will play during pathogen clearance of C. neoformans during EIPE-1 treatment. Studies are ongoing in our lab to understand how treatment with EIPE-1 may affect immune-mediated clearance of C. neoformans.
Finally, C. neoformans is capable of producing its own melanin in the host in the presence of L-DOPA. This plays an important role in protecting C. neoformans from host induced damage due to reactive oxygen species. Melanin is also capable of binding and impacting the effect of antifungal treatments on the fungal cells (Wang and Casadevall, 1994; Zaragoza et al., 2009; Lee et al., 2019). This leads to the question of whether EIPE-1 will be effective against cryptococcal cells when in a melanized form. As previously mentioned, the structure of the EIPE-1 indole core is inspired by eumelanin molecular structure (Adhikari et al., 2022). We do not know if structural similarities will contribute to stronger binding or decrease the susceptibility of C. neoformans to the novel compound. Future studies are being conducted on the potential of EIPE-1 on melanized C. neoformans.
Data availability statement
The transcriptome datasets that support the findings of this article are available to the public in the NCBI BioSample database (ncbi.nlm.nih.gov/biosample/) under the accession number PRJNA1052015, samples SAMN38810541, SAMN38810542, SAMN38810543, SAMN38810544, SAMN38810545, and SAMN38810546. Further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
BC: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. JL: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. PC: Data curation, Formal analysis, Investigation, Writing – review & editing. KC: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. MaE: Writing – review & editing, Investigation, Methodology. MoE: Investigation, Methodology, Writing – review & editing. TN: Conceptualization, Formal analysis, Funding acquisition, Methodology, Writing – review & editing. KW: Conceptualization, Data curation, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported in part by Oklahoma State University start-up funds (KW) and funds from Cowboy Technologies (TN and KW). In addition, BC was supported by 5T32GM140953-03 from the National Institutes of Health (NIH) institute of General Medical Sciences (NIGMS) and KW was supported by 5P20GM134973 from NIH NIGMS. The funders had no involvement in the study design, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Acknowledgments
We would like to acknowledge the assistance of members of the OSU Microscopy Facility for their work with the electron microscopy studies, especially Lisa Whitworth and Brent Johnson.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adepu, S., and Ramakrishna, S. (2021). Controlled drug delivery systems: current status and future directions. Molecules 26:5905. doi: 10.3390/molecules26195905
Adhikari, S., Essandoh, M. A., Starr, W. C., Sah, P., la Force, C. N., Eleshy, R. G., et al. (2022). Eumelanin-inspired antimicrobial with biocidal activity against methicillin-resistant Staphylococcus aureus. ACS Appl. Bio Mater. 5, 545–551. doi: 10.1021/acsabm.1c01036
Agustinho, D. P., Miller, L. C., Li, L. X., and Doering, T. L. (2018). Peeling the onion: the outer layers of Cryptococcus neoformans. Mem. Inst. Oswaldo Cruz 113:e180040. doi: 10.1590/0074-02760180040
Amos, B., Aurrecoechea, C., Barba, M., Barreto, A., Basenko, E. Y., Bażant, W., et al. (2022). VEuPathDB: the eukaryotic pathogen, vector and host bioinformatics resource center. Nucleic Acids Res. 50, D898–D911. doi: 10.1093/nar/gkab929
Baker, Z., Harrison, R. W., and Miller, B. F. (1941). ACTION OF SYNTHETIC DETERGENTS ON THE METABOLISM OF BACTERIA. J. Exp. Med. 73, 249–271. doi: 10.1084/jem.73.2.249
Baker, L. G., Specht, C. A., and Lodge, J. K. (2009). Chitinases are essential for sexual development but not vegetative growth in Cryptococcus neoformans. Eukaryot. Cell 8, 1692–1705. doi: 10.1128/EC.00227-09
Banks, I. R., Specht, C. A., Donlin, M. J., Gerik, K. J., Levitz, S. M., and Lodge, J. K. (2005). A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell 4, 1902–1912. doi: 10.1128/EC.4.11.1902-1912.2005
Belazi, D., Solé-Domènech, S., Johansson, B., Schalling, M., and Sjövall, P. (2009). Chemical analysis of osmium tetroxide staining in adipose tissue using imaging ToF-SIMS. Histochem. Cell Biol. 132, 105–115. doi: 10.1007/s00418-009-0587-z
Bermas, A., and Geddes-McAlister, J. (2020). Combatting the evolution of antifungal resistance in Cryptococcus neoformans. Mol. Microbiol. 114, 721–734. doi: 10.1111/mmi.14565
Bonifácio, B. V., Vila, T. V. M., Masiero, I. F., da Silva, P. B., da Silva, I. C., de Oliveira Lopes, É., et al. (2019). Antifungal activity of a Hydroethanolic extract from Astronium urundeuva leaves against Candida albicans and Candida glabrata. Front. Microbiol. 10:2642. doi: 10.3389/fmicb.2019.02642
Casadevall, A., Rosas, A. L., and Nosanchuk, J. D. (2000). Melanin and virulence in Cryptococcus neoformans. Curr. Opin. Microbiol. 3, 354–358. doi: 10.1016/S1369-5274(00)00103-X
Chang, Y. C., and Kwon-Chung, K. J. (1994). Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 14, 4912–4919.
Chang, Y. C., Penoyer, L. A., and Kwon-Chung, K. J. (1996). The second capsule gene of cryptococcus neoformans, CAP64, is essential for virulence. Infect. Immun. 64, 1977–1983. doi: 10.1128/iai.64.6.1977-1983.1996
Chang, Y. C., Stins, M. F., McCaffery, M. J., Miller, G. F., Pare, D. R., Dam, T., et al. (2004). Cryptococcal yeast cells invade the central nervous system via transcellular penetration of the blood-brain barrier. Infect. Immun. 72, 4985–4995. doi: 10.1128/IAI.72.9.4985-4995.2004
Chen, S. C., Playford, E. G., and Sorrell, T. C. (2010). Antifungal therapy in invasive fungal infections. Curr. Opin. Pharmacol. 10, 522–530. doi: 10.1016/j.coph.2010.06.002
Coelho, C., and Casadevall, A. (2016). Cryptococcal therapies and drug targets: the old, the new and the promising. Cell. Microbiol. 18, 792–799. doi: 10.1111/cmi.12590
Conn, B. N., and Wozniak, K. L. (2023). Innate pulmonary phagocytes and their interactions with pathogenic Cryptococcus species. J. Fungi (Basel) 9:617. doi: 10.3390/jof9060617
Cowen, L. E., and Steinbach, W. J. (2008). Stress, drugs, and evolution: the role of cellular signaling in fungal drug resistance. Eukaryot. Cell 7, 747–764. doi: 10.1128/EC.00041-08
Danchik, C., and Casadevall, A. (2020). Role of cell surface hydrophobicity in the pathogenesis of medically-significant Fungi. Front. Cell. Infect. Microbiol. 10:594973. doi: 10.3389/fcimb.2020.594973
Dempster, W. T. (1960). Rates of penetration of fixing fluids. Am. J. Anat. 107, 59–72. doi: 10.1002/aja.1001070105
Dhana, A. (2013). Diagnosis of Cryptococcosis and prevention of Cryptococcal meningitis using a novel point-of-care lateral flow assay. Case Rep. Med. 2013:640216. doi: 10.1155/2013/640216
Dismukes, W. E. (2000). Introduction to antifungal drugs. Clin. Infect. Dis. 30, 653–657. doi: 10.1086/313748
Doong, H., Vrailas, A., and Kohn, E. C. (2002). What’s in the “BAG”?–a functional domain analysis of the BAG-family proteins. Cancer Lett. 188, 25–32. doi: 10.1016/S0304-3835(02)00456-1
Eisenman, H. C., and Casadevall, A. (2012). Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 93, 931–940. doi: 10.1007/s00253-011-3777-2
Ekins, S., Ring, B. J., Grace, J., McRobie-Belle, D. J., and Wrighton, S. A. (2000). Present and future in vitro approaches for drug metabolism. J. Pharmacol. Toxicol. Methods 44, 313–324. doi: 10.1016/S1056-8719(00)00110-6
Ernst, M. E., Klepser, M. E., Wolfe, E. J., and Pfaller, M. A. (1996). Antifungal dynamics of LY 303366, an investigational echinocandin B analog, against Candida ssp. Diagn. Microbiol. Infect. Dis. 26, 125–131. doi: 10.1016/S0732-8893(96)00202-7
Espinel-Ingroff, A., Chaturvedi, V., Fothergill, A., and Rinaldi, M. G. (2002). Optimal testing conditions for determining MICs and minimum fungicidal concentrations of new and established antifungal agents for uncommon molds: NCCLS collaborative study. J. Clin. Microbiol. 40, 3776–3781. doi: 10.1128/JCM.40.10.3776-3781.2002
Fuchs, B. B., and Mylonakis, E. (2009). Our paths might cross: the role of the fungal cell wall integrity pathway in stress response and cross talk with other stress response pathways. Eukaryot. Cell 8, 1616–1625. doi: 10.1128/EC.00193-09
Fuchs, B. B., O’Brien, E., Khoury, J. B. E., and Mylonakis, E. (2010). Methods for using galleria mellonella as a model host to study fungal pathogenesis. Virulence 1, 475–482. doi: 10.4161/viru.1.6.12985
Fuentefria, A. M., Pippi, B., Dalla Lana, D. F., Donato, K. K., and de Andrade, S. F. (2018). Antifungals discovery: an insight into new strategies to combat antifungal resistance. Lett. Appl. Microbiol. 66, 2–13. doi: 10.1111/lam.12820
Gallegos, B., Martínez, R., Pérez, L., del Socorro Pina, M., Perez, E., and Hernández, P. (2014). Lectins in human pathogenic fungi. Rev. Iberoam. Micol. 31, 72–75. doi: 10.1016/j.riam.2013.09.010
Garcia-Rubio, R., de Oliveira, H. C., Rivera, J., and Trevijano-Contador, N. (2020). The fungal Cell Wall: Candida, Cryptococcus, and aspergillus species. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02993
Garelnabi, M., and May, R. C. (2018). Variability in innate host immune responses to cryptococcosis. Mem. Inst. Oswaldo Cruz 113:e180060. doi: 10.1590/0074-02760180060
Geddes-McAlister, J., and Shapiro, R. S. (2019). New pathogens, new tricks: emerging, drug-resistant fungal pathogens and future prospects for antifungal therapeutics. Ann. N. Y. Acad. Sci. 1435, 57–78. doi: 10.1111/nyas.13739
Gibson, J. F., and Johnston, S. A. (2015). Immunity to Cryptococcus neoformans and C. Gattii during cryptococcosis. Fungal Genet. Biol. 78, 76–86. doi: 10.1016/j.fgb.2014.11.006
Gillette, J. R. (1971). Factors affecting drug metabolism. Ann. N. Y. Acad. Sci. 179, 43–66. doi: 10.1111/j.1749-6632.1971.tb46890.x
Graybill, J. R., Burgess, D. S., and Hardin, T. C. (1997). Key issues concerning fungistatic versus fungicidal drugs. Eur. J. Clin. Microbiol. Infect. Dis. 16, 42–50. doi: 10.1007/BF01575120
Grijpstra, J., Tefsen, B., van Die, I., and de Cock, H. (2009). The Cryptococcus neoformans cap10 and cap59 mutant strains, affected in glucuronoxylomannan synthesis, differentially activate human dendritic cells. FEMS Immunol. Med. Microbiol. 57, 142–150. doi: 10.1111/j.1574-695X.2009.00587.x
Guo, X. S., Bu, H., He, J. Y., Zou, Y. L., Zhao, Y., Li, Y. Y., et al. (2016). Current diagnosis and treatment of cryptococcal meningitis without acquired immunodeficiency syndrome. Neuroimmunol. Nueroinflamm. 3, 249–256. doi: 10.20517/2347-8659.2016.10
Hogan, L. H., Klein, B. S., and Levitz, S. M. (1996). Virulence factors of medically important fungi. Clin. Microbiol. Rev. 9, 469–488. doi: 10.1128/CMR.9.4.469
Hole, C. R., Bui, H., Wormley, F. L. Jr., and Wozniak, K. L. (2012). Mechanisms of dendritic cell lysosomal killing of Cryptococcus. Sci. Rep. 2:739. doi: 10.1038/srep00739
Holmes, A. R., Cardno, T. S., Strouse, J. J., Ivnitski-Steele, I., Keniya, M. V., Lackovic, K., et al. (2016). Targeting efflux pumps to overcome antifungal drug resistance. Future Med. Chem. 8, 1485–1501. doi: 10.4155/fmc-2016-0050
Ishikawa, T., Watanabe, N., Nagano, M., Kawai-Yamada, M., and Lam, E. (2011). Bax inhibitor-1: a highly conserved endoplasmic reticulum-resident cell death suppressor. Cell Death Differ. 18, 1271–1278. doi: 10.1038/cdd.2011.59
ISO10993-5. Part 5: tests for in vitro cytoxicity. Vol. biological evalulation of medical devices (2009).
ISO20395. Biotechnology – requirements for evaluating the performance of quantification methods for nucleic acid target sequences – qPCR and dPCR. (2019).
Janbon, G., Ormerod, K. L., Paulet, D., Byrnes, E. J. 3rd, Yadav, V., Chatterjee, G., et al. (2014). Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 10. doi: 10.1371/journal.pgen.1004261
Kay, S., Edwards, J., Brown, J., and Dixon, R. (2019). Galleria mellonella infection model identifies both high and low lethality of Clostridium perfringens toxigenic strains and their response to antimicrobials. Front. Microbiol. 10:1281. doi: 10.3389/fmicb.2019.01281
Khan, Y. A., White, K. I., and Brunger, A. T. (2022). The AAA+ superfamily: a review of the structural and mechanistic principles of these molecular machines. Crit. Rev. Biochem. Mol. Biol. 57, 156–187. doi: 10.1080/10409238.2021.1979460
Klein, N. D., Hurley, K. R., Feng, Z. V., and Haynes, C. L. (2015). Dark field transmission electron microscopy as a tool for identifying inorganic nanoparticles in biological matrices. Anal. Chem. 87, 4356–4362. doi: 10.1021/acs.analchem.5b00124
Knowles, C. M., McIntyre, K. M., and Panepinto, J. C. (2021). Tools for assessing translation in Cryptococcus neoformans. J. Fungi (Basel) 7:159. doi: 10.3390/jof7030159
Koga, D., Kusumi, S., Shibata, M., and Watanabe, T. (2021). Applications of scanning Electron microscopy using secondary and backscattered Electron signals in neural structure. Front. Neuroanat. 15:759804. doi: 10.3389/fnana.2021.759804
Kozel, T. R., and Gotschlich, E. C. (1982). The capsule of cryptococcus neoformans passively inhibits phagocytosis of the yeast by macrophages. J. Immunol. 1950, 1675–1680.
Kozel, T. R., Pfrommer, G. S., Guerlain, A. S., Highison, B. A., and Highison, G. J. (1988). Role of the capsule in phagocytosis of Cryptococcus neoformans. Rev. Infect. Dis. 10, S436–S439. doi: 10.1093/cid/10.Supplement_2.S436
Kurop, M. K., Huyen, C. M., Kelly, J. H., and Blagg, B. S. J. (2021). The heat shock response and small molecule regulators. Eur. J. Med. Chem. 226:113846. doi: 10.1016/j.ejmech.2021.113846
Kwon-Chung, K. J., Fraser, J. A., Doering, T. L., Wang, Z. A., Janbon, G., Idnurm, A., et al. (2014). Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb. Perspect. Med. 4:a019760. doi: 10.1101/cshperspect.a019760
Lange, F., Agüi-Gonzalez, P., Riedel, D., Phan, N. T. N., Jakobs, S., and Rizzoli, S. O. (2021). Correlative fluorescence microscopy, transmission electron microscopy and secondary ion mass spectrometry (CLEM-SIMS) for cellular imaging. PLoS One 16:e0240768. doi: 10.1371/journal.pone.0240768
Lee, D., Jang, E. H., Lee, M., Kim, S. W., Lee, Y., Lee, K. T., et al. (2019). Unraveling melanin biosynthesis and signaling networks in Cryptococcus neoformans. MBio 10:e02267-19. doi: 10.1128/mbio.02267-19
Leite, M. C. A., Bezerra, A. P. B., Sousa, J. P., Guerra, F. Q. S., and Lima, E. O. (2014). Evaluation of antifungal activity and mechanism of action of Citral against Candida albicans. Evid. Based Complement. Alternat. Med. 2014:378280, 1–9. doi: 10.1155/2014/378280
Levitz, S. M. (1991). The ecology of Cryptococcus neoformans and the epidemiology of cryptococcosis. Rev. Infect. Dis. 13, 1163–1169. doi: 10.1093/clinids/13.6.1163
Mueckler, M., and Thorens, B. (2013). The SLC2 (GLUT) family of membrane transporters. Mol. Asp. Med. 34, 121–138. doi: 10.1016/j.mam.2012.07.001
Mylonakis, E., Moreno, R., El Khoury, J. B., Idnurm, A., Heitman, J., Calderwood, S. B., et al. (2005). Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect. Immun. 73, 3842–3850. doi: 10.1128/IAI.73.7.3842-3850.2005
Nelson, B. N., Beakley, S. G., Posey, S., Conn, B., Maritz, E., Seshu, J., et al. (2021). Antifungal activity of dendritic cell lysosomal proteins against Cryptococcus neoformans. Sci. Rep. 11:13619. doi: 10.1038/s41598-021-92991-6
Nett, J. E., and Andes, D. R. (2016). Antifungal agents: Spectrum of activity, pharmacology, and clinical indications. Infect. Dis. Clin. N. Am. 30, 51–83. doi: 10.1016/j.idc.2015.10.012
Nixon, W. C. (1964). Scanning Electron microscopy. J. R. Microsc. Soc. 83, 213–216. doi: 10.1111/j.1365-2818.1964.tb00531.x
Nosanchuk, J. D., and Casadevall, A. (2006). Impact of melanin on microbial virulence and clinical resistance to antimicrobial compounds. Antimicrob. Agents Chemother. 50, 3519–3528. doi: 10.1128/AAC.00545-06
Pappas, P. G., Alexander, B. D., Andes, D. R., Hadley, S., Kauffman, C. A., Freifeld, A., et al. (2010). Invasive fungal infections among organ transplant recipients: results of the transplant-associated infection surveillance network (TRANSNET). Clin. Infect. Dis. 50, 1101–1111. doi: 10.1086/651262
Patel, R. K. K., Leeme, T., Azzo, C., Tlhako, N., Tsholo, K., Tawanana, E. O., et al. (2018). High mortality in HIV-associated Cryptococcal meningitis patients treated with amphotericin B-based therapy under routine care conditions in Africa. Open forum. Infect. Dis. 5:ofy267. doi: 10.1093/ofid/ofy267
Perea, S., and Patterson, T. F. (2002). Antifungal resistance in pathogenic fungi. Clin. Infect. Dis. 35, 1073–1080. doi: 10.1086/344058
Perfect, J. R. (2017). The antifungal pipeline: a reality check. Nat. Rev. Drug Discov. 16, 603–616. doi: 10.1038/nrd.2017.46
Perfect, J. R., Dismukes, W. E., Dromer, F., Goldman, D. L., Graybill, J. R., Hamill, R. J., et al. (2010). Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the infectious diseases society of america. Clin. Infect. Dis. 50, 291–322. doi: 10.1086/649858
Perlin, D. S., Rautemaa-Richardson, R., and Alastruey-Izquierdo, A. (2017). The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect. Dis. 17, e383–e392. doi: 10.1016/S1473-3099(17)30316-X
Rajasingham, R., Govender, N. P., Jordan, A., Loyse, A., Shroufi, A., Denning, D. W., et al. (2022). The global burden of HIV-associated cryptococcal infection in adults in 2020: a modelling analysis. Lancet Infect. Dis. 22, 1748–1755. doi: 10.1016/S1473-3099(22)00499-6
Rajasingham, R., Smith, R. M., Park, B. J., Jarvis, J. N., Govender, N. P., Chiller, T. M., et al. (2017). Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect. Dis. 17, 873–881. doi: 10.1016/S1473-3099(17)30243-8
Reed, D. R., Nehmzow, K., Essandoh, M. A., Ebqa'ai, M. A., Nelson, T. L., Lutter, E. I., et al. (2023). Relationship between cell envelope ultrastructure and the antibacterial properties of a novel hydrophobic eumelanin-inspired derivative. Front. Bacteriol. 2. doi: 10.3389/fbrio.2023.1253097
Rodrigues, M. L., Nakayasu, E. S., Oliveira, D. L., Nimrichter, L., Nosanchuk, J. D., Almeida, I. C., et al. (2008). Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot. Cell 7, 58–67. doi: 10.1128/EC.00370-07
Schie, I. W., Kiselev, R., Krafft, C., and Popp, J. (2016). Rapid acquisition of mean Raman spectra of eukaryotic cells for a robust single cell classification. Analyst 141, 6387–6395. doi: 10.1039/C6AN01018K
Selvaraju, S., Niradha Sachinthani, K. A., Hopson, R. A. A., McFarland, F. M., Guo, S., Rheingold, A. L., et al. (2015). Eumelanin-inspired core derived from vanillin: a new building block for organic semiconductors. Chem. Commun. (Camb.) 51, 2957–2959. doi: 10.1039/C4CC09011J
Shi, M., Li, S. S., Zheng, C., Jones, G. J., Kim, K. S., Zhou, H., et al. (2010). Real-time imaging of trapping and urease-dependent transmigration of Cryptococcus neoformans in mouse brain. J. Clin. Invest. 120, 1683–1693. doi: 10.1172/JCI41963
Sloan, D. J., and Parris, V. (2014). Cryptococcal meningitis: epidemiology and therapeutic options. Clin. Epidemiol. 6, 169–182. doi: 10.2147/CLEP.S38850
Sprynski, N., Valade, E., and Neulat-Ripoll, F. (2014). Galleria mellonella as an infection model for select agents. Methods Mol. Biol. 1197, 3–9. doi: 10.1007/978-1-4939-1261-2_1
Tefsen, B., Grijpstra, J., Ordonez, S., Lammers, M., van Die, I., and de Cock, H. (2014). Deletion of the CAP10 gene of Cryptococcus neoformans results in a pleiotropic phenotype with changes in expression of virulence factors. Res. Microbiol. 165, 399–410. doi: 10.1016/j.resmic.2014.04.001
Tsai, C. J.-Y., Loh, J. M. S., and Proft, T. (2016). Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7, 214–229. doi: 10.1080/21505594.2015.1135289
Upadhya, R., Lam, W. C., Hole, C. R., Vasselli, J. G., and Lodge, J. K. (2023). Cell wall composition in Cryptococcus neoformans is media dependent and alters host response, inducing protective immunity. Front. Fungal Biol. 4:1183291. doi: 10.3389/ffunb.2023.1183291
Van Daele, R., Spriet, I., Wauters, J., Maertens, J., Mercier, T., Van Hecke, S., et al. (2019). Antifungal drugs: what brings the future? Med. Mycol. 57, S328–S343. doi: 10.1093/mmy/myz012
van der Rest, M. E., Kamminga, A. H., Nakano, A., Anraku, Y., Poolman, B., and Konings, W. N. (1995). The plasma membrane of Saccharomyces cerevisiae: structure, function, and biogenesis. Microbiol. Rev. 59, 304–322.
Vecchiarelli, A., Pericolini, E., Gabrielli, E., Kenno, S., Perito, S., Cenci, E., et al. (2013). Elucidating the immunological function of the Cryptococcus neoformans capsule. Future Microbiol. 8, 1107–1116. doi: 10.2217/fmb.13.84
Velkov, T., Roberts, K. D., Nation, R. L., Thompson, P. E., and Li, J. (2013). Pharmacology of polymyxins: new insights into an “old” class of antibiotics. Future Microbiol. 8, 711–724. doi: 10.2217/fmb.13.39
Vij, R., Danchik, C., Crawford, C., Dragotakes, Q., and Casadevall, A. (2020). Variation in cell surface hydrophobicity among Cryptococcus neoformans strains influences interactions with amoebas. mSphere 5:e00310-20. doi: 10.1128/mSphere.00310-20
Wang, Y., and Casadevall, A. (1994). Growth of Cryptococcus neoformans in presence of L-dopa decreases its susceptibility to amphotericin B. Antimicrob. Agents Chemother. 38, 2648–2650. doi: 10.1128/AAC.38.11.2648
Wiederhold, N. P. (2017). Antifungal resistance: current trends and future strategies to combat. Infect. Drug Resist. 10, 249–259. doi: 10.2147/IDR.S124918
Winey, M., Meehl, J. B., O’Toole, E. T., and Giddings, T. H. Jr. (2014). Conventional transmission electron microscopy. Mol. Biol. Cell 25, 319–323. doi: 10.1091/mbc.e12-12-0863
Yauch, L. E., Lam, J. S., and Levitz, S. M. (2006). Direct inhibition of T-cell responses by the Cryptococcus capsular polysaccharide glucuronoxylomannan. PLoS Pathog. 2:e120. doi: 10.1371/journal.ppat.0020120
Zaragoza, O., Rodrigues, M. L., de Jesus, M., Frases, S., Dadachova, E., and Casadevall, A. (2009). The capsule of the fungal pathogen Cryptococcus neoformans. Adv. Appl. Microbiol. 68, 133–216. doi: 10.1016/S0065-2164(09)01204-0
Keywords: Cryptococcus neoformans , antifungal, novel antifungal compound, EIPE-1, eumelanin
Citation: Conn BN, Lieberman JA, Chatman P, Cotton K, Essandoh MA, Ebqa’ai M, Nelson TL and Wozniak KL (2024) Antifungal activity of eumelanin-inspired indoylenepheyleneethynylene against Cryptococcus neoformans. Front. Microbiol. 14:1339303. doi: 10.3389/fmicb.2023.1339303
Edited by:
Santi M. Mandal, Indian Institute of Technology Kharagpur, IndiaReviewed by:
Zhangyong Song, Southwest Medical University, ChinaFausto Almeida, University of São Paulo, Brazil
Copyright © 2024 Conn, Lieberman, Chatman, Cotton, Essandoh, Ebqa’ai, Nelson and Wozniak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karen L. Wozniak, S2FyZW4uV296bmlha0Bva3N0YXRlLmVkdQ==
 Brittney N. Conn
Brittney N. Conn Jacob A. Lieberman
Jacob A. Lieberman Priscilla Chatman
Priscilla Chatman Kaitlyn Cotton1
Kaitlyn Cotton1 Mohammad Ebqa’ai
Mohammad Ebqa’ai Toby L. Nelson
Toby L. Nelson Karen L. Wozniak
Karen L. Wozniak