- 1Bioproduction Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), Tsukuba, Japan
- 2Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Tokyo, Japan
- 3Graduate School of Life and Environmental Sciences, University of Tsukuba, Tsukuba, Japan
Insect–microbe endosymbiotic associations are omnipresent in nature, wherein the symbiotic microbes often play pivotal biological roles for their host insects. In particular, insects utilizing nutritionally imbalanced food sources are dependent on specific microbial symbionts to compensate for the nutritional deficiency via provisioning of B vitamins in blood-feeding insects, such as tsetse flies, lice, and bedbugs. Bat flies of the family Nycteribiidae (Diptera) are blood-sucking ectoparasites of bats and shown to be associated with co-speciating bacterial endosymbiont “Candidatus Aschnera chinzeii,” although functional aspects of the microbial symbiosis have been totally unknown. In this study, we report the first complete genome sequence of Aschnera from the bristled bat fly Penicillidia jenynsii. The Aschnera genome consisted of a 748,020 bp circular chromosome and a 18,747 bp circular plasmid. The chromosome encoded 603 protein coding genes (including 3 pseudogenes), 33 transfer RNAs, and 1 copy of 16S/23S/5S ribosomal RNA operon. The plasmid contained 10 protein coding genes, whose biological function was elusive. The genome size, 0.77 Mbp, was drastically reduced in comparison with 4–6 Mbp genomes of free-living γ-proteobacteria. Accordingly, the Aschnera genome was devoid of many important functional genes, such as synthetic pathway genes for purines, pyrimidines, and essential amino acids. On the other hand, the Aschnera genome retained complete or near-complete synthetic pathway genes for biotin (vitamin B7), tetrahydrofolate (vitamin B9), riboflavin (vitamin B2), and pyridoxal 5'-phosphate (vitamin B6), suggesting that Aschnera provides these vitamins and cofactors that are deficient in the blood meal of the host bat fly. Similar retention patterns of the synthetic pathway genes for vitamins and cofactors were also observed in the endosymbiont genomes of other blood-sucking insects, such as Riesia of human lice, Arsenophonus of louse flies, and Wigglesworthia of tsetse flies, which may be either due to convergent evolution in the blood-sucking host insects or reflecting the genomic architecture of Arsenophonus-allied bacteria.
Introduction
Insect–microbe endosymbiotic associations are omnipresent in nature, wherein the symbiotic microbes often play pivotal biological roles for their host insects (Buchner, 1965; Bourtzis and Miller, 2003). In particular, insects utilizing nutritionally imbalanced food sources are dependent on specific microbial symbionts to compensate for the nutritional deficiency via provisioning of essential amino acids in plant sap-feeding insects, such as aphids, leafhoppers, and cicadas (Moran et al., 2008; Douglas, 2009), or supply of B vitamins in blood-feeding insects, such as tsetse flies, lice, and bedbugs (Rio et al., 2016; Husnik, 2018).
Vertebrate blood is certainly nutritious, which comprises a rich protein source mainly in the form of hemoglobin, but it entails disproportionally low levels of lipids and carbohydrates, high levels of iron that is potentially toxic, and deficiency in some important nutritional elements such as B vitamins (Sterkel et al., 2017). Therefore, many insects and other invertebrates nutritionally dependent solely on vertebrate blood all through their life stages, including tsetse flies, louse flies, bat flies, lice, bedbugs, kissing bugs, ticks, and leeches, are obligatorily associated with specific symbiotic bacteria whose main biological roles are suggested as provisioning of B vitamins (Rio et al., 2016; Husnik, 2018; Duron and Gottlieb, 2020; Fukatsu et al., 2023).
Bat flies of the family Nycteribiidae (Diptera) are blood-sucking ectoparasites of bats of peculiar spider-like appearance with highly specialized morphological traits, represented by lack of wings, reduced head and eyes, long and powerful legs with grasping claws, and flexible and tough exoskeleton (Figures 1A, B). The nycteribiid bat flies (Nycteribiidae) constitute the superfamily Hippoboscoidea together with tsetse flies (Glossinidae), louse flies (Hippoboscidae), and streblid bat flies (Streblidae) (Petersen et al., 2007), which share such remarkable traits as adult feeding solely on vertebrate blood, adenotrophic viviparity with highly developed uterus and milk glands, and larval uterine growth with no free life until pupation outside the maternal body (Lehane, 2005; Dick and Patterson, 2006; Nikoh et al., 2011). Reflecting their obligatory blood-feeding lifestyle, these insect groups rely on specific symbiotic bacteria and develop specialized cells and organs, called bacteriocytes and bacteriomes, for retaining the microbial partners (Roubaud, 1919; Zacharias, 1928; Aschner, 1931; Buchner, 1965). All tsetse flies are associated with the monophyletic endosymbiotic bacteria Wigglesworthia glossinidea (Aksoy, 2000; Rio et al., 2012; Wang et al., 2013). All nycteribiid bat flies are associated with and co-speciating with the monophyletic endosymbiotic bacteria “Candidatus Aschnera chinzeii” (hereafter called Aschnera) (Hosokawa et al., 2012; Duron et al., 2014). Among louse flies, their Arsenophonus endosymbionts do not necessarily exhibit host-symbiont co-speciation and are thought to be of multiple evolutionary origins (Dale et al., 2006; Trowbridge et al., 2006; Duron et al., 2014; Nováková et al., 2015; Ríhová et al., 2023). As for streblid bat flies, while an array of bacterial associates has been identified, the main symbiotic bacteria are elusive (Trowbridge et al., 2006; Morse et al., 2012, 2013).
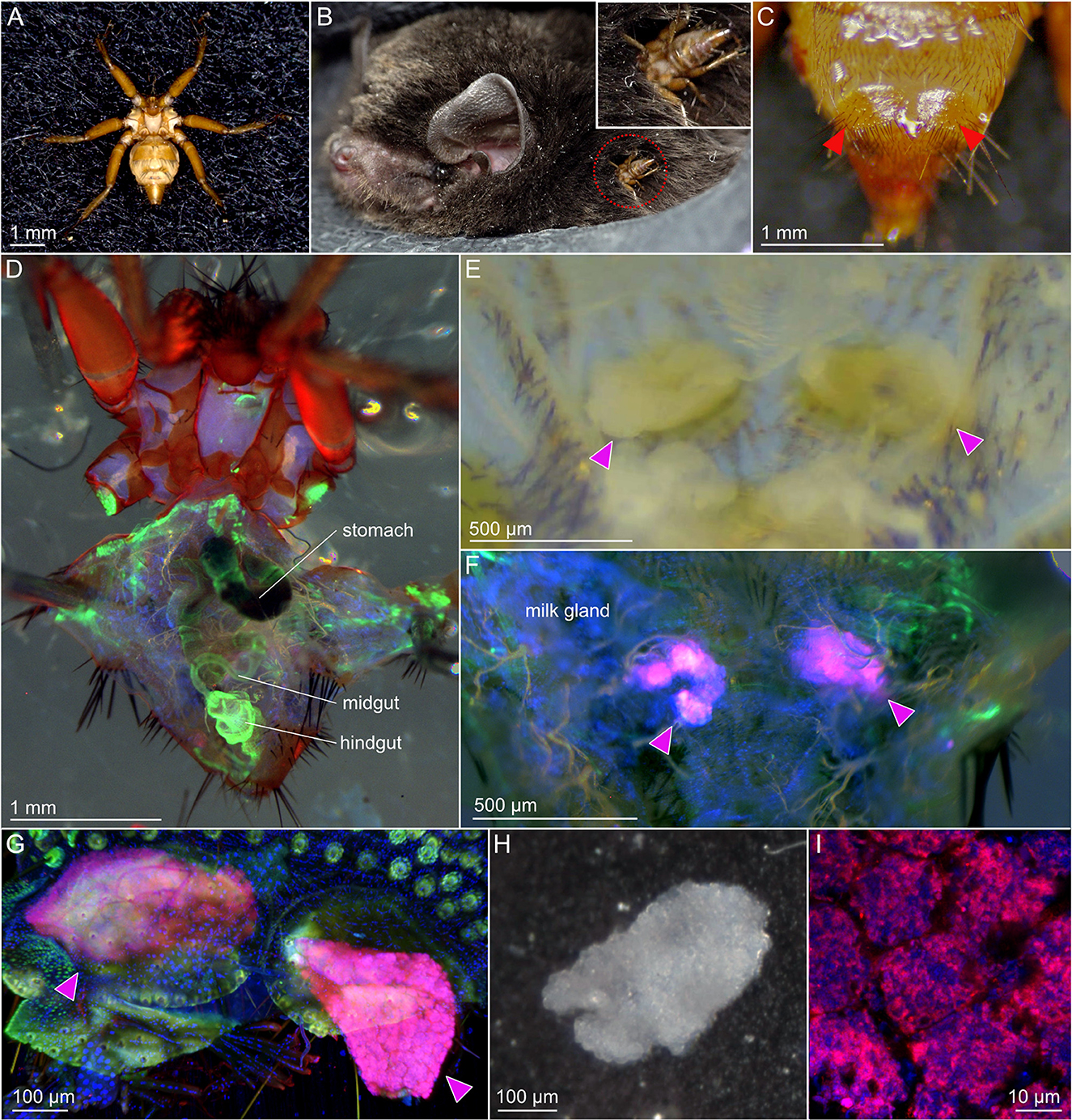
Figure 1. The bat fly Penicillidia jenynsii and its Aschnera-harboring bacteriomes. (A) Dorsal view of a female insect. (B) An insect parasitizing the host bat Miniopterus fuliginosus. (C) Ventral view of a female insect's abdomen, on which the projections housing the bacteriomes inside are seen (arrowheads). (D) Dorsal view of a female insect whose abdomen is cut open at the dorsal midline. Internal organs, such as stomach, midgut and hindgut are seen. (E) Enlarged bright-field image of the abdomen from which the internal organs are removed. Whitish bacteriomes packed inside the projections on the ventral plate are exposed (arrowheads). (F) Fluorescent image of the abdomen from which the internal organs are removed. Strong bacterial signals due to Aschnera localization are detected in the bacteriomes (arrowheads). (G) Enlarged fluorescent image of the bacteriomes to which Aschnera is localized (arrowheads). (H) Bright-field image of an isolated bacteriome. (I) Confocal fluorescent microscopic image of an isolated bacteriome in which Aschnera cells are visualized. In (D, F, G, I), magenta, blue, and green indicate Alexa Fluor 555-labeled FISH signals of bacterial 16S rRNA, DAPI signals of DNA, and Alexa Fluor 488 phalloidin signals of filamentous actin, respectively.
Except for several tsetse species (Mews et al., 1977), laboratory rearing and experiments are very challenging for the blood-sucking ectoparasitic flies. Hence, to gain insight into functional aspects of the symbiotic bacteria, their genomic information is very important. As for the Glossinidae, the Wigglesworthia genomes have been determined for the tsetse flies Glossinidia brevipalpis (Akman et al., 2002) and G. morsitans (Rio et al., 2012). As for the Hippoboscidae, the genomes of Arsenophonus melophagi from the sheep ked Melophagus ovinus (Nováková et al., 2015) and Arsenophonus lipoptenae from the deer louse fly Lipoptena cervi (Nováková et al., 2016) have been reported. As for the Nycteribiidae, no symbiont genome has been reported thus far.
In this study, we report the first genome sequencing and analysis of Aschnera endosymbiont of the bat fly Penicillidia jenynsii parasitizing the bat Miniopterus fuliginosus.
Materials and methods
Insect materials
The bat flies P. jenynsii were collected from the host eastern bent-winged bats M. fuliginosus caught at Kawabe County, Hyogo Prefecture, Japan, on 14 May and 16 September 2022 in field bat monitoring conducted by Nobutaka Urano and Shunji Fujita (permission number 環近地野許第 2107301 号, the Ministry of the Environment, Japan). The collected bat flies were placed in plastic tubes with wet tissue paper, kept in a cooling box, brought to laboratory, and used for experiments.
Dissection and DNA extraction
Each adult female P. jenynsii was fixed to a dissection stage assembled with a silicon rubber disk and a plastic Petri dish by pinning at the center of its flat thorax. Legs of the insect were removed using ophthalmological microscissors in a phosphate buffered saline (PBS: 0.8% NaCl, 0.02% KCl, 0.115% Na2HPO4, 0.02% KH2PO4, pH 7.4), and their dorsal abdomen was cut open along the midline to expose the internal organs of the insect. Fat bodies, milk glands, and digestive tract were all carefully removed not to break the stomach full of blood meal, by which whitish bacteriomes packed in the paired projections on the fifth ventral plate were exposed and carefully collected using fine forceps. In total, 60 bacteriomes from 30 insects were collected and subjected to DNA extraction using a DNA tissue mini kit (Qiagen).
Genome sequencing and assembly
The DNA sample extracted from the bacteriomes was subjected to the library preparation for long-read sequencing by using Ligation sequencing kit SQK-LSK109 (Oxford Nanopore Technologies). The library prepared was analyzed on a MinION sequencer implemented with the flow cell R9.4.1 (Oxford Nanopore Technologies). The DNA sample was also subjected to short-read sequencing. The short-read library was prepared using NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolab), and 150 bp paired-end reads were obtained by NovaSeq 6000 (Illumina). Because excessive short-read data were obtained, a 10% subset was extracted from the original read library. Furthermore, the subset was quality-trimmed on CLC genomics workbench version 23 (Qiagen). A short- and long-read hybrid assembling was conducted by using Unicycler version 0.5.0 (Wick et al., 2017). Assemble errors in the resultant draft genome sequence were searched and manually corrected with the assistance of CLC genomics workbench. The circularity of the assembled genome was confirmed by mapping the long reads to the rotated contigs and checking the resultant read maps. Annotation of the corrected genome was performed using DFAST version 1.2.18 (Tanizawa et al., 2018). The BUSCO scores were calculated with the enterobacterales_odb10 database using BUSCO version 5.5.0 (Manni et al., 2021). The genome map was drawn using DNAplotter in Artemis version 18.0.0 (Carver et al., 2009).
Phylogenetic analysis
Deduced protein sequences inferred from the Aschnera genome and 31 bacterial genomes retrieved from DNA databases (GenBank, accessed in July, 2023) were prepared from the annotated genome sequences using CLC genomic workbench. BCGtree pipeline (Ankenbrand and Keller, 2016) was used to assemble and concatenate the multiple alignments of 107 bacterial core protein genes from 32 bacterial genomes with the following parameter: –min-proteomes = 2. The resultant concatenated alignment and corresponding partition information were used to infer the maximum likelihood tree with 1,000 bootstrap replicates using IQ-TREE version 2.2.2.7 (Minh et al., 2020) with the following parameter: -m MFP+MERGE. The phylogenetic tree was visualized using TreeViewer version 2.1.0 (Bianchini and Sánchez-Baracaldo, 2023).
Metabolic pathway analysis
The metabolic capability of Aschnera was predicted based on the genome sequence using GhostKOALA web service (https://www.kegg.jp/ghostkoala/) (Kanehisa et al., 2016). The metabolic models of the endosymbiont of human louse, Riesia pediculicola strain USDA (T number of the KEGG database: T01218), the endosymbiont of louse fly Arsenophonus lipoptenae strain CB (T04248), the endosymbiont of tsetse fly Wigglesworthia glossinidia (T00101), the endosymbiont of bedbug Wolbachia sp. strain wCle (T03628), the endosymbiont of aphid Buchnera aphidicola strain APS (T00036), the gut symbiont of plataspid stinkbug Ishikawaella capsulata (T04149), and Escherichia coli strain K-12 MG1655 (T00007) were acquired from the KEGG database (https://www.kegg.jp/) (Kanehisa et al., 2022). The metabolic pathways were compared essentially based on KEGG modules with references to the EcoCyc database (Keseler et al., 2021).
Whole-mount fluorescence in situ hybridization (wFISH)
The abdomen of each adult female P. jenynsii was cut to make a slit at the midline using ophthalmological microscissors in PBS. Then, the insects were fixed in PBS containing 4% paraformaldehyde at 4°C overnight. After several washes with PBS, the specimens were subjected to wFISH as previously described (Koga et al., 2009). The specimens were hybridized with Al555-EUB338 (5'-Alexa Fluor 555- GCT GCC TCC CGT AGG AGT-3') targeting bacterial 16S ribosomal RNA. After overnight hybridization at room temperature, the specimens were washed with PBT (PBS containing 0.1% Tween-20) containing 165 nM Alexa Fluor 488 phalloidin and 2 μg/mL 4',6-diamidino-2-phenylindole (DAPI) for 30 min at room temperature and subsequently washed with PBT three times. The hybridized specimens were dissected and pictured in PBS under a fluorescence dissection microscope M165 controlled by LAS version 4.13.0 (Leica). The bacteriomes on the abdominal cuticle were mounted in PBS-50% glycerol and observed with a laser scanning microscope LSM700 controlled with Zen 2011 version 7.0 (Zeiss). Digital images were manually manipulated using Affinity Photo version 2.1.1 (Affinity).
Results and discussion
Endosymbiotic system of P. jenynsii
The bat fly P. jenynsii (Figure 1A) is a blood-sucking ectoparasite of the eastern bent-winged bat M. fuliginosus (Figure 1B). On the ventral side of the abdomen of a female insect, the fifth ventral plate bears paired projections on both sides (Figure 1C), each of which contains the bacteriome consisting of the bacteriocytes harboring the symbiotic bacteria (Hosokawa et al., 2012). When the abdomen of a female insect was dissected from the dorsal side (Figure 1D) and deprived of the internal organs (alimentary tract, gonad, fat body, etc.), the bacteriome was seen as a whitish cell mass inside each of the projections (Figure 1E). FISH targeting bacterial 16S rRNA detected the symbiont signals within the bacteriomes in the abdominal projections (Figures 1F, G). The bacteriomes were carefully isolated (Figure 1H), wherein the symbiotic bacteria within numerous bacteriocytes were visualized using FISH (Figure 1I).
Genome sequence of Aschnera
The isolated bacteriomes collected from 30 adult female P. jenynsii were subjected to DNA extraction, library construction, long- and short-read DNA sequencing, and genome assembly, whereby the complete Aschnera genome was determined as a 748,020 bp circular chromosome and a 18,747 bp circular plasmid. The latter was identified as the plasmid because of the presence of the replication initiation protein A (repA) gene in this contig. The genome size approximately agreed with a previous estimation of the genome size of Aschnera as 0.76 Mbp by pulsed field gel electrophoresis (Hosokawa et al., 2012). The 0.75 Mbp chromosome, consisting of 23.1% G + C, encoded 603 putative protein-coding sequences (CDSs), of which 3 were predicted as pseudogenes, 33 transfer RNAs, and 1 copy of 16S/23S/5S ribosomal RNA operon (Figure 2; Table 1; Supplementary Table S1). The 18 kbp plasmid, consisting of 22.3% G + C, encoded only 10 genes, whose biological relevance was elusive (Figure 2; Table 1; Supplementary Table S2). The Aschnera genome, 0.77 Mbp in size, is around 1/5–1/7 of the E. coli genomes of 4–6 Mbp (Lukjancenko et al., 2010). The BUSCO score of A. chinzeii was 81.1%. This score was higher than those of R. pediculicola USDA (66.8%) and R. sp. GBBU (61.8%). These results confirmed that Aschnera has experienced reductive genome evolution in the process of intimate co-evolution with nycteribiid bat flies (Hosokawa et al., 2012).
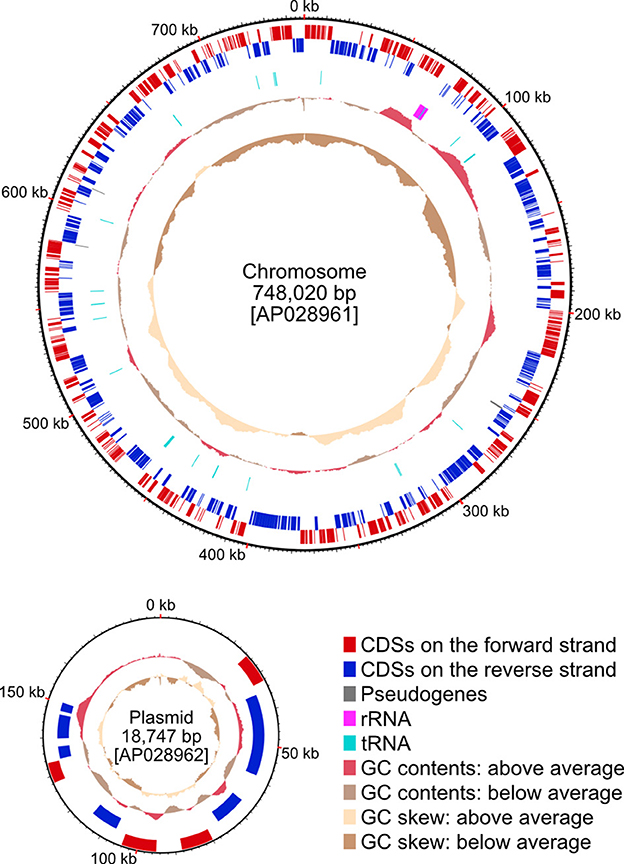
Figure 2. The genome map of Aschnera. The chromosome is oriented to make the reading frame of dnaA gene the first CDS on the forward strand. The plasmid is oriented arbitrarily. The outer-most circle represents the positions of CDSs on the forward strand while the second circle represents those on the reverse strand. The third circle represents the position of three pseudogenes. The fourth circle represents the positions of ribosomal and transfer RNA genes. Red, blue, gray, magenta, and cyan boxes represent CDSs on the forward strand, CDSs on the reverse strand, pseudogenes, and RNA-coding genes, respectively. The fifth and sixth circles represent GC contents and GC skews, respectively.
Phylogenomic placement of Aschnera
Molecular phylogenetic analysis based on 109 bacterial core protein sequences revealed that Aschnera is placed in the γ-proteobacterial clade consisting of Arsenophonus and allied bacterial lineages that embraces many insect-associated endosymbiotic bacteria (Nováková et al., 2009) (Figure 3). Among them, Aschnera was closely related to the endosymbionts of primate lice Riesia spp. (Kirkness et al., 2010; Boyd et al., 2014) and other endosymbionts of blood-sucking insects, such as Lightella of rodent lice (Ríhová et al., 2022), Puchtella of monkey lice (Boyd et al., 2017), and Wigglesworthia of tsetse flies (Akman et al., 2002; Rio et al., 2012). Arsenophonus endosymbionts of louse flies (Nováková et al., 2016) were placed outside of them in the phylogenetic tree. These phylogenetic patterns strongly suggested that although tsetse flies (Glossinidae), louse flies (Hippoboscidae), and bat flies (Nycteribiidae) belong to the same well-defined superfamily Hippoboscoidea (Petersen et al., 2007), their current endosymbiotic bacterial associates are likely of independent evolutionary origins presumably via symbiont acquisitions and replacements, as previously suggested (Nováková et al., 2009; Hosokawa et al., 2012; Duron et al., 2014; Ríhová et al., 2023).
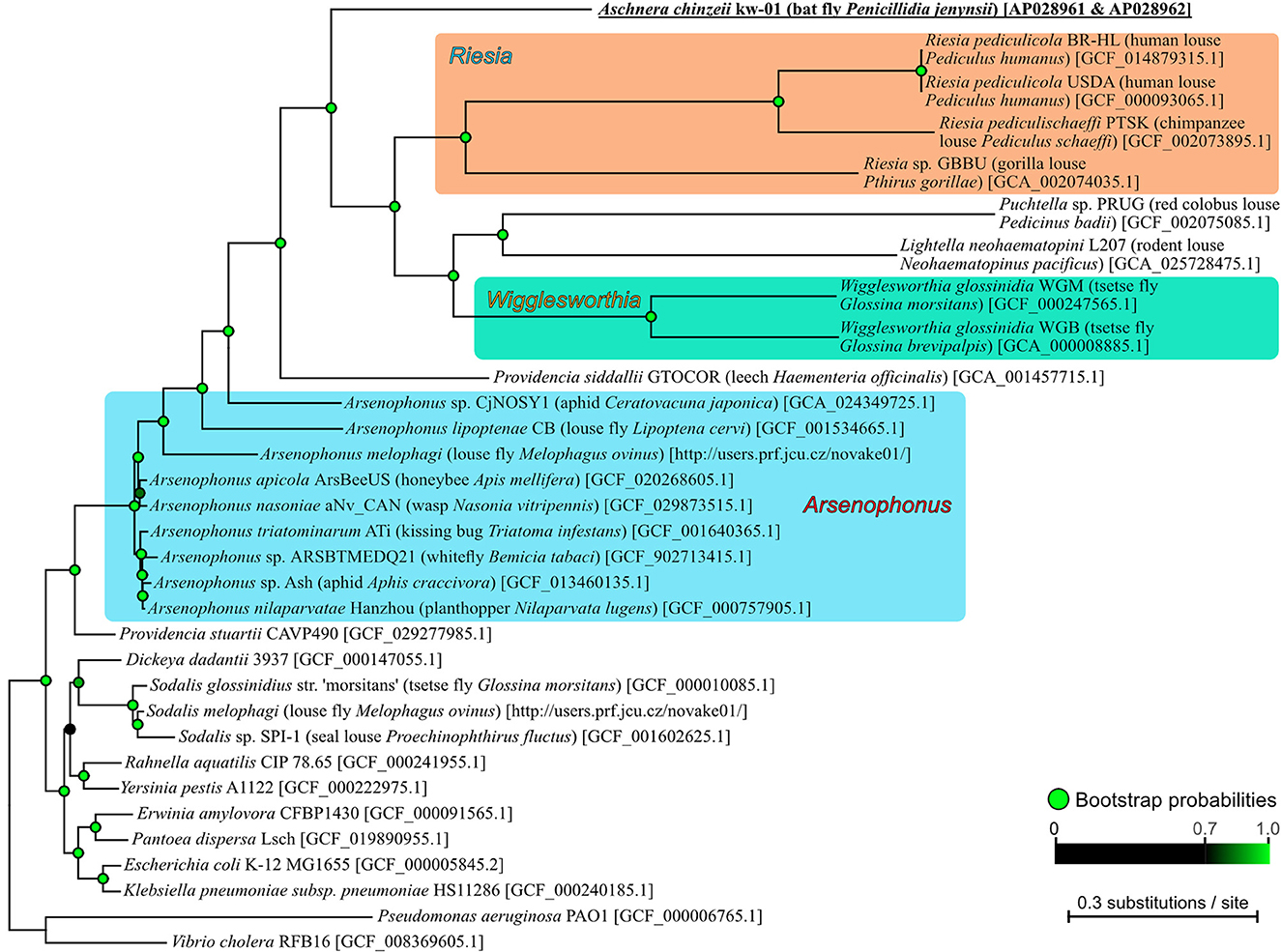
Figure 3. Phylogenetic placement of Aschnera in the γ-Proteobacteria. The maximum likelihood phylogeny was inferred from 33,136 amino acid sites inferred from concatenated 107 bacterial core protein genes. Generic, specific, and strain names of the bacteria are indicated on the right side of the phylogeny with host names in parentheses and sequence accession numbers in brackets. The bootstrap probabilities of the nodes were obtained from 1,000 resamplings, which were shown by color scale in the figure.
Gene repertoire of Aschnera
In accordance with the drastic genome reduction, the Aschnera genome has lost many important metabolic pathways (Figure 4). For example, synthetic pathway genes for purines and pyrimidines are mostly missing. Strikingly, synthetic pathway genes for essential amino acids are completely lost from the Aschnera genome, which was also observed in the primary endosymbionts of other blood-sucking insects, including Riesia of human louse, Arsenophonus of louse fly, Wigglesworthia of tsetse fly, and Wolbachia of bedbug (Figure 4). These patterns may be relevant to the feeding habit of the host bat fly P. jenynsii as an obligatory blood-sucking ectoparasite of bats (Funakoshi, 1977; Lehane, 2005), considering that the blood meal is very rich in proteins and nucleic acids.

Figure 4. Comparison of metabolic gene repertoire between the Aschnera genome and other reduced endosymbiont genomes associated with blood-sucking and plant-sucking insects. The typical number of genes for each metabolic pathway is shown on the left side, whereas number of genes for each metabolic pathway encoded in the E. coli genome is shown on the right side. Colors indicate the completeness of the pathway in the genomes: green, complete; orange, 1 gene missing; gray, 2 or more genes missing.
Synthetic pathways for B vitamins in Aschnera genome
On the other hand, the blood meal is deficient in some nutrients, such as B vitamins. Therefore, obligatory blood-feeding insects, such as tsetse flies, lice, and bedbugs, rely on their specific bacterial symbiont for supply of B vitamins (Akman et al., 2002; Kirkness et al., 2010; Nikoh et al., 2014). Consequently, such symbiont genomes retain the synthetic pathway genes for some, if not all, vitamins and cofactors (Rio et al., 2016; Husnik, 2018; Duron and Gottlieb, 2020; Fukatsu et al., 2023). The Aschnera genome retained complete or near-complete synthetic pathway genes for biotin (vitamin B7), tetrahydrofolate (vitamin B9), riboflavin (vitamin B2), and pyridoxal 5'-phosphate (vitamin B6) (Figures 5A–E). The Aschnera genome also encoded synthetic pathway genes for such cofactors as coenzyme A (from pantothenate) and nicotinamide adenine dinucleotide (NAD) (from nicotinate) (Figures 5A, F, G). On the other hand, the Aschnera genome was incapable of synthesizing thiamine (vitamin B1) and pantothenate (vitamin B5) (Figures 5A, F, H). Based on these results, it seems plausible that Aschnera provides the host insect with biotin, pyridoxal, riboflavin, and tetrahydrofolate to compensate for nutritional deficiency of the blood meal. It is notable that not only Aschnera of the bat fly but also Riesia of human lice, Arsenophonus of louse flies, and Wigglesworthia of tsetse flies exhibit similar retention patterns of the synthesis pathway genes for B vitamins (Figure 5A), which may be either due to convergent evolution in the blood-sucking host insects or reflecting the genomic architecture of γ -proteobacterial Arsenophonus-allied bacteria. In this context, it seems meaningful that the α-proteobacterial mutualistic Wolbachia endosymbiont of bedbug retains synthesis pathway genes for biotin and riboflavin only (Figure 5A).
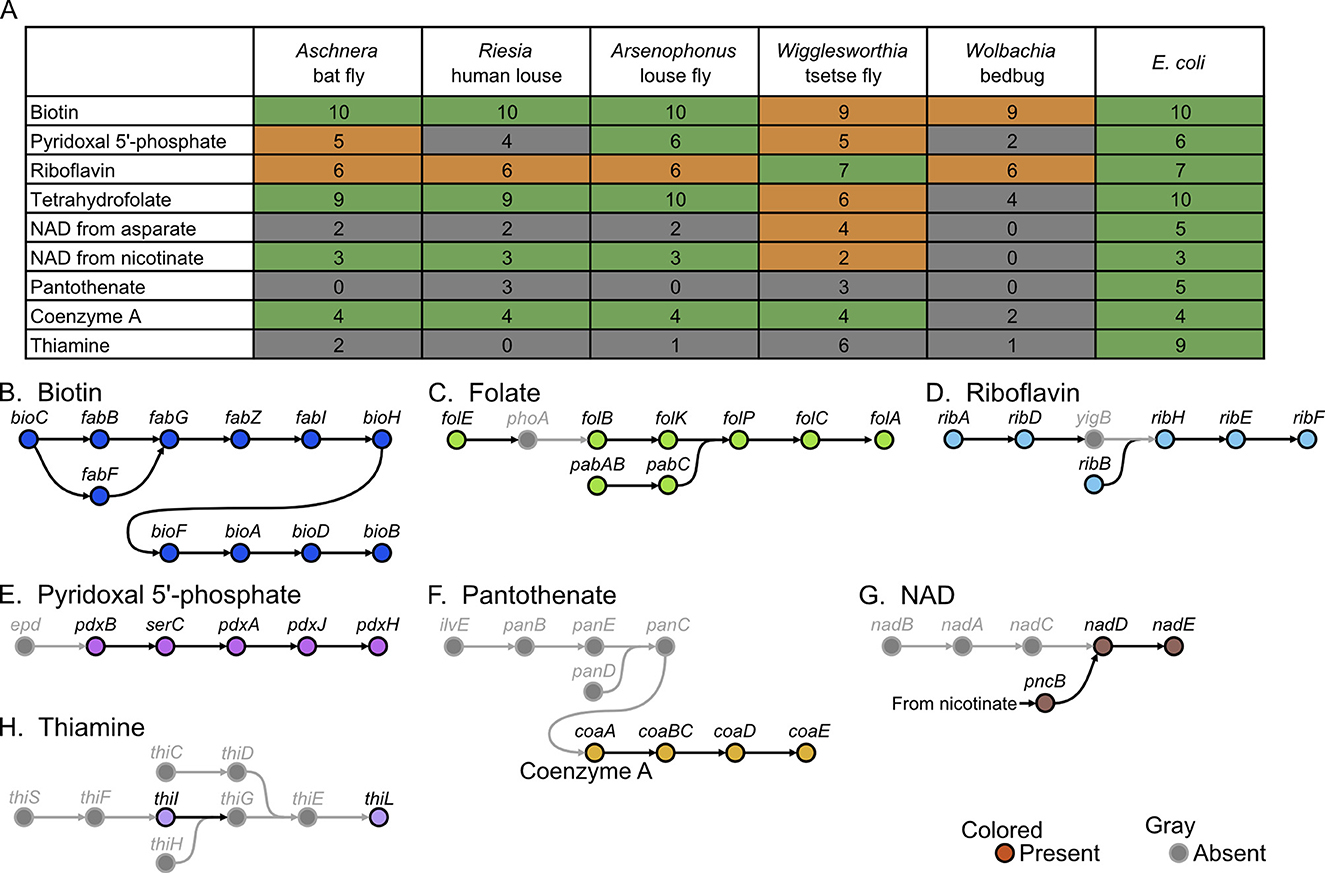
Figure 5. Synthesis pathway genes for B vitamins and cofactors encoded in the Aschnera genome. (A) Comparison of synthetic capabilities of B vitamins and other cofactors among Aschnera and other endosymbionts of blood-feeding insects. The typical number of genes for each metabolic pathway is shown on the right side represented by E. coli. Colors indicate the completeness of the pathway in the genomes: green, complete; orange, 1 gene missing; gray, 2 or more genes missing. (B–H) Presence and absence of genes in the synthesis pathways for B vitamins and cofactors. (B) Biotin (vitamin B7). (C) Tetrahydrofolate (vitamin B9). (D) Riboflavin (vitamin B2). (E) Pyridoxal 5'-phosphate (vitamin B6). (F) Coenzyme A (from pantothenate). (G) Nicotinamide adenine dinucleotide (NAD) (from nicotinate). (H) Thiamine (vitamin B1).
Conclusion
In this study, we reported the complete genome sequence of Aschnera, the obligatory bacterial endosymbiont of the bat fly, and demonstrated that the symbiont genome has been streamlined for provisioning of B vitamins and cofactors that are deficient in the blood meal of the host bat fly. The bat fly is a very peculiar and experimentally intractable insect, being capable of surviving only on the body surface of specific bat species. Both the bat fly and the host bat are very difficult, or practically almost impossible, to maintain in the laboratory for experimental purposes. Therefore, the approach from the symbiont genomics provides invaluable information on the nature of the intimate microbial endosymbiosis in the non-model blood-sucking insect group Nycteribiidae. Transcriptomic, proteomic, and metabolomic approaches would further contribute to understanding the physiological, metabolic, and molecular interactions ongoing in the bat fly endosymbiosis.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://ddbj.nig.ac.jp/search/en, DRA017199, DRA017200, AP028961, AP028962, SAMD00647857, SAMD00647858, PRJDB16712.
Author contributions
RK: Conceptualization, Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. MM: Investigation, Writing – review & editing. TN: Investigation, Writing – review & editing. TF: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Japan Science and Technology Agency (JST) ERATO Grant Number JPMJER1902 to RK and TF and by the Japan Society for the Promotion of Science (JSPS) KAKENHI grant number JP17H06388 to TF.
Acknowledgments
We thank Nobutaka Urano and Shunji Fujita for helping collection of bat fly samples, and Song Kiyomiya for technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1336919/full#supplementary-material
References
Akman, L., Yamashita, A., Watanabe, H., Oshima, K., Shiba, T., Hattori, M., et al. (2002). Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32, 402–407. doi: 10.1038/ng986
Aksoy, S. (2000). Tsetse – a haven for microorganisms. Parasitol. Today 16, 114–118. doi: 10.1016/S0169-4758(99)01606-3
Ankenbrand, M. J., and Keller, A. (2016). bcgTree: automatized phylogenetic tree building from bacterial core genomes. Genome 59, 783–791. doi: 10.1139/gen-2015-0175
Aschner, M. (1931). Die Bakterienflora der Pupiparen (Diptera). Eine Symbiosestudie an blutsaugenden Insekten. Z. Morphol. Ökol. Tiere 20, 368–442. doi: 10.1007/BF00444102
Bianchini, G., and Sánchez-Baracaldo, P. (2023). TreeViewer-Cross-Platform Software to Draw Phylogenetic Trees (Version 2.1. 0). Geneva: Zenodo.
Boyd, B. M., Allen, J. M., de Crécy-Lagard, V., and Reed, D. L. (2014). Genome sequence of Candidatus Riesia pediculischaeffi, endosymbiont of chimpanzee lice, and genomic comparison of recently acquired endosymbionts from human and chimpanzee lice. G3: GenesGenomesGenet. 4, 2189–2195. doi: 10.1534/g3.114.012567
Boyd, B. M., Allen, J. M., Nguyen, N. P., Vachaspati, P., Quicksall, Z. S., Warnow, T., et al. (2017). Primates, lice and bacteria: speciation and genome evolution in the symbionts of hominid lice. Mol. Biol. Evol. 34, 1743–1757. doi: 10.1093/molbev/msx117
Carver, T., Thomson, N., Bleasby, A., Berriman, M., and Parkhill, J. (2009). DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25, 119–120. doi: 10.1093/bioinformatics/btn578
Dale, C., Beeton, M., Harbison, C., Jones, T., and Pontes, M. (2006). Isolation, pure culture, and characterization of “Candidatus Arsenophonus arthropodicus,” an intracellular secondary endosymbiont from the hippoboscid louse fly Pseudolynchia canariensis. Appl. Environ. Microbiol. 72, 2997–3004. doi: 10.1128/AEM.72.4.2997-3004.2006
Dick, C. W., and Patterson, B. D. (2006). “Bat flies: obligate ectoparasites of bats.,” in Micromammals and Macroparasites: From Evolutionary Ecology to Management, eds. S. Morand, B. R. Krasnov, and R. Poulin (Cham : Springer), 179–194.
Douglas, A. E. (2009). The microbial dimension in insect nutritional ecology. Funct. Ecol. 23, 38–47. doi: 10.1111/j.1365-2435.2008.01442.x
Duron, O., and Gottlieb, Y. (2020). Convergence of nutritional symbioses in obligate blood feeders. Trends Parasitol. 36, 816–825. doi: 10.1016/j.pt.2020.07.007
Duron, O., Schneppat, U. E., Berthomieu, A., Goodman, S. M., Droz, B., Paupy, C., et al. (2014). Origin, acquisition and diversification of heritable bacterial endosymbionts in louse flies and bat flies. Mol. Ecol. 23, 2105–2117. doi: 10.1111/mec.12704
Fukatsu, T., Gottlieb, Y., Duron, O., and Graf, J. (2023). Editorial: Microbial associates of blood-sucking arthropods and other animals: relevance to their physiology, ecology and evolution. Front. Microbiol. 14, 1256275. doi: 10.3389/fmicb.2023.1256275
Funakoshi, K. (1977). Ecological studies on the bat fly, Penicillidia jenynsii (Diptera : Nycteribiidae), infested on the Japanese long-fingered bat : with special reference to the adaptability to their hosts from the viewpoint of life history. Jpn J. Ecol. 27, 125–140.
Hosokawa, T., Nikoh, N., Koga, R., Satô, M., Tanahashi, M., Meng, X. Y., et al. (2012). Reductive genome evolution, host-symbiont co-speciation and uterine transmission of endosymbiotic bacteria in bat flies. ISME J. 6, 577–587. doi: 10.1038/ismej.2011.125
Husnik, F. (2018). Host–symbiont–pathogen interactions in blood-feeding parasites: nutrition, immune cross-talk and gene exchange. Parasitology 145, 1294–1303. doi: 10.1017/S0031182018000574
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M., and Ishiguro-Watanabe, M. (2022). KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592. doi: 10.1093/nar/gkac963
Kanehisa, M., Sato, Y., and Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Keseler, I. M., Gama-Castro, S., Mackie, A., Billington, R., Bonavides-Martínez, C., Caspi, R., et al. (2021). The EcoCyc database in 2021. Front. Microbiol. 12, 711077. doi: 10.3389/fmicb.2021.711077
Kirkness, E. F., Haas, B. J., Sun, W., Braig, H. R., Perotti, M. A., Clark, J. M., et al. (2010). Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc. Natl. Acad. Sci. U. S. A. 107, 12168–12173. doi: 10.1073/pnas.1003379107
Koga, R., Tsuchida, T., and Fukatsu, T. (2009). Quenching autofluorescence of insect tissues for in situ detection of endosymbionts. Appl. Entomol. Zool. 44, 281–291. doi: 10.1303/aez.2009.281
Lehane, M. J. (2005). The Biology of Blood-Sucking in Insects. Cambridge: Cambridge University Press.
Lukjancenko, O., Wassenaar, T. M., and Ussery, D. W. (2010). Comparison of 61 sequenced Escherichia coli genomes. Microb. Ecol. 60, 708–720. doi: 10.1007/s00248-010-9717-3
Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A., and Zdobnov, E. M. (2021). BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 38, 4647–4654. doi: 10.1093/molbev/msab199
Mews, A. R., Langley, P. A., Pimley, R. W., and Flood, M. E. T. (1977). Large-scale rearing of tsetse flies (Glossina spp.) in the absence of a living host. Bull. Entomol. Res. 67, 119–128. doi: 10.1017/S0007485300010944
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Moran, N. A., McCutcheon, J. P., and Nakabachi, A. (2008). Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42, 165–190. doi: 10.1146/annurev.genet.41.110306.130119
Morse, S. F., Bush, S. E., Patterson, B. D., Dick, C. W., Gruwell, M. E., Dittmar, K., et al. (2013). Evolution, multiple acquisition, and localization of endosymbionts in bat flies (Diptera: Hippoboscoidea: Streblidae and Nycteribiidae). Appl. Environ. Microbiol. 79, 2952–2961. doi: 10.1128/AEM.03814-12
Morse, S. F., Dick, C. W., Patterson, B. D., and Dittmar, K. (2012). Some like it hot: evolution and ecology of novel endosymbionts in bat flies of cave-roosting bats (Hippoboscoidea, Nycterophiliinae). Appl. Environ. Microbiol. 78, 8639–8649. doi: 10.1128/AEM.02455-12
Nikoh, N., Hosokawa, T., Moriyama, M., Oshima, K., Hattori, M., Fukatsu, T., et al. (2014). Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc. Natl. Acad. Sci. U. S. A. 111, 10257–10262. doi: 10.1073/pnas.1409284111
Nikoh, N., Sato, M., Kondo, N., and Fukatsu, T. (2011). Phylogenetic comparison between nycteribiid bat flies and their host bats. Med. Entomol. Zool. 62, 185–194. doi: 10.7601/mez.62.185
Nováková, E., Husník, F., Šochová, E., and Hypša, V. (2015). Arsenophonus and Sodalis symbionts in louse flies: an analogy to the Wigglesworthia and Sodalis system in tsetse flies. Appl. Environ. Microbiol. 81, 6189–6199. doi: 10.1128/AEM.01487-15
Nováková, E., Hypša, V., and Moran, N. A. (2009). Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution. BMC Microbiol. 9, 143. doi: 10.1186/1471-2180-9-143
Nováková, E., Hypša, V., Nguyen, P., Husník, F., and Darby, A. C. (2016). Genome sequence of Candidatus Arsenophonus lipopteni, the exclusive symbiont of a blood sucking fly Lipoptena cervi (Diptera: Hippoboscidae). Stand. Genom. Sci. 11, 72. doi: 10.1186/s40793-016-0195-1
Petersen, F. T., Meier, R., Kutty, S. N., and Wiegmann, B. M. (2007). The phylogeny and evolution of host choice in the Hippoboscoidea (Diptera) as reconstructed using four molecular markers. Mol. Phylogenet. Evol. 45, 111–122. doi: 10.1016/j.ympev.2007.04.023
Ríhová, J., Bell, K. C., Nováková, E., and Hypša, V. (2022). Lightella neohaematopini: A new lineage of highly reduced endosymbionts coevolving with chipmunk lice of the genus Neohaematopinus. Front. Microbiol. 13, 900312. doi: 10.3389/fmicb.2022.900312
Ríhová, J. M., Gupta, S., Darby, A. C., Nováková, E., and Hypša, V. (2023). Arsenophonus symbiosis with louse flies: multiple origins, coevolutionary dynamics, and metabolic significance. mSystems 8, e00706–e00723. doi: 10.1101/2023.07.13.548870
Rio, R. V. M., Attardo, G. M., and Weiss, B. L. (2016). Grandeur alliances: symbiont metabolic integration and obligate arthropod hematophagy. Trends Parasitol. 32, 739–749. doi: 10.1016/j.pt.2016.05.002
Rio, R. V. M., Symula, R. E., Wang, J., Lohs, C., Wu, Y., Snyder, A. K., et al. (2012). Insight into the transmission biology and species-specific functional capabilities of tsetse (Diptera: Glossinidae) obligate symbiont Wigglesworthia. mBio 3, e00240–e00211. doi: 10.1128/mBio.00240-11
Roubaud, B. (1919). Les particularites de la nutrition et de la vie symbiotique chez les mouches tsetse. Ann. Inst. Pasteur. 33, 489–537.
Sterkel, M., Oliveira, J. H. M., Bottino-Rojas, V., Paiva-Silva, G. O., and Oliveira, P. L. (2017). The dose makes the poison: nutritional overload determines the life traits of blood-feeding arthropods. Trends Parasitol. 33, 633–644. doi: 10.1016/j.pt.2017.04.008
Tanizawa, Y., Fujisawa, T., and Nakamura, Y. (2018). DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 34, 1037–1039. doi: 10.1093/bioinformatics/btx713
Trowbridge, R. E., Dittmar, K., and Whiting, M. F. (2006). Identification and phylogenetic analysis of Arsenophonus- and Photorhabdus-type bacteria from adult Hippoboscidae and Streblidae (Hippoboscoidea). J. Invertebr. Pathol. 91, 64–68. doi: 10.1016/j.jip.2005.08.009
Wang, J., Weiss, B. L., and Aksoy, S. (2013). Tsetse fly microbiota: form and function. Front. Cell. Infect. Microbiol. 3, 69. doi: 10.3389/fcimb.2013.00069
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Keywords: Aschnera chinzeii, nycteribiid bat fly, Penicillidia jenynsii, symbiotic bacteria, genome reduction, B vitamin provisioning, blood-sucking insect
Citation: Koga R, Moriyama M, Nozaki T and Fukatsu T (2024) Genome analysis of “Candidatus Aschnera chinzeii,” the bacterial endosymbiont of the blood-sucking bat fly Penicillidia jenynsii (Insecta: Diptera: Nycteribiidae). Front. Microbiol. 14:1336919. doi: 10.3389/fmicb.2023.1336919
Received: 12 November 2023; Accepted: 26 December 2023;
Published: 22 January 2024.
Edited by:
Amparo Latorre, University of Valencia, SpainReviewed by:
Xiao-Li Bing, Nanjing Agricultural University, ChinaArturo Vera-Ponce de León, Norwegian University of Life Sciences, Norway
Copyright © 2024 Koga, Moriyama, Nozaki and Fukatsu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ryuichi Koga, ci1rb2dhQGFpc3QuZ28uanA=; Takema Fukatsu, dC1mdWthdHN1QGFpc3QuZ28uanA=
 Ryuichi Koga
Ryuichi Koga Minoru Moriyama
Minoru Moriyama Tomonari Nozaki
Tomonari Nozaki Takema Fukatsu
Takema Fukatsu