
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 January 2024
Sec. Microbial Symbioses
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1336345
This article is part of the Research Topic The Regulation of Environmental Factors on the Microbial Cooperation View all 13 articles
Introduction: Bacterial symbionts are prevalent in arthropods globally and play a vital role in the fitness and resistance of hosts. While several symbiont infections have been identified in the white-backed planthopper Sogatella furcifera, the impact of environmental factors on the microbiota within S. furcifera remains elusive.
Methods: In this study, a total of 142 S. furcifera individuals from 18 populations were collected from 14 locations across six countries (China, Thailand, Myanmar, Cambodia, Vietnam, and Laos) analyzed with 2bRAD-M sequencing, to examine the effects of symbionts on the microbiota in the S. furcifera population, as well as the vital effects of environmental factors on the bacterial communities.
Results and discussion: Based on the results, in S. furcifera, the presence of symbionts Wolbachia and Cardinium negatively influenced the abundance of other bacteria, including Enterobacter, Acinetobacter, and Lysinibacillus, while Wolbachia infection significantly decreased the diversity of the microbial community. Moreover, several environmental factors, including longitude, latitude, temperature, and precipitation, affected the abundance of symbionts and microbiota diversity in S. furcifera. These results collectively highlight the vital role of Wolbachia in S. furcifera microbiota, as well as the intricate effects of environmental factors on the bacterial communities of S. furcifera.
As a significant pest in China and South Asia, the white-backed planthopper (WBPH), scientifically referred to as Sogatella furcifera (Hemiptera: Delphacidae), causes substantial losses on agricultural fields, particularly rice paddies (Savary et al., 2012; Huang and Qin, 2018). S. furcifera causes damage by sucking phloem sap from rice plants and transmitting viruses, including the southern rice black-streaked dwarf virus, to rice hosts (Zhou et al., 2008, 2018). Over the past decade, the frequency of S. furcifera outbreaks has dramatically increased, making it a destructive pest in rice production (Hu et al., 2015; Wang et al., 2018). With its high adaptability, long-distance migratory abilities, and the development of resistance to existing pesticides, managing S. furcifera outbreaks has become increasingly challenging (Wang et al., 2019). Coupled with the growing problem of pesticide residue, there is an urgent need to develop novel and efficient methods to mitigate the damage elicited by pest insects, including S. furcifera.
Microbial communities, especially bacteria, including endosymbionts, play a vital role in the development, fitness, fecundity, and resistance of host arthropods (Douglas, 2009; McFall-Ngai et al., 2013). These symbiotic bacteria fall into two categories: primary symbionts (obligate symbionts) and secondary symbionts (facultative symbionts), residing predominantly in bacteriocytes within hosts and exerting complex influences on host arthropods (Werren et al., 2008; Zeng et al., 2018; Yang et al., 2022). The former is essential for the development and survival of host invertebrates. For instance, the symbiont Portiera provides essential amino acids (EAA) and B vitamins to whitefly hosts (Wang et al., 2022), whilst the symbiont Buchnera supplies aphid hosts with various nutritional elements and protects them from heat stress (Dunbar et al., 2007; Douglas, 2009).
Wolbachia, the most renowned symbiont in invertebrates, was first discovered in mosquitoes in 1924 (Hertig and Wolbach, 1924). Interestingly, it can infect at least 66% of all arthropod species (Werren, 1997; Miao et al., 2020). In filarial nematodes, Wolbachia is regarded as an obligate mutualist, essential for the development and fertility of host nematodes (Taylor et al., 2005; Wasala et al., 2019). Previously considered reproductive parasitism in invertebrates, capable of manipulating host reproduction through phenomena such as cytoplasmic incompatibility, feminization, parthenogenesis, and male-killing (Werren et al., 2008), recent studies have established the benefits conferred by Wolbachia (Zug and Hammerstein, 2015). Another study corroborated the influence of Wolbachia on microbial communities in small planthoppers (Duan et al., 2020). However, the intricate interaction between Wolbachia and the bacterial community in arthropods warrants further investigations.
Microbial communities, including symbionts in arthropods, are influenced by various biotic and abiotic factors. For example, high temperatures significantly influence symbiont titer and the diversity of the bacterial community in whiteflies (Yang et al., 2022). Similarly, the diet of invertebrate hosts significantly impacts the structure of microbial communities (Santo Domingo et al., 1998; Colman et al., 2012), and both temperature and humidity affect bacterial communities (Behar et al., 2008). Furthermore, earlier research has documented the crucial impact of host genetic backgrounds on microbiota (Suzuki et al., 2019; Gupta and Nair, 2020). Interestingly, the presence of symbionts exerts a significant influence on microbial communities. Symbionts occupy a relatively large space and engage in competition with other bacteria for nutrients. According to a prior study, Spiroplasma infection significantly decreases the titer of Wolbachia in the same host (Goto et al., 2006). Additionally, endosymbionts such as Wolbachia and Cardinium can lower the microbiome diversity of the host Sogatella furcifera (Li et al., 2022). The complex effects of symbionts on the microbiota of S. furcifera require further investigation.
2bRAD-M sequencing is a novel method that is efficient in detecting low-biomass microbiomes at the species level with high fidelity (Sun et al., 2022). In recent years, the damage caused by S. furcifera has drammatically rapidly, particularly in Asia and China. However, the complex interactions between the bacterial community of S. furcifera, including bacterial symbionts and environmental factors, remain poorly understood. Moreover, the potential application of symbionts in the biocontrol of S. furcifera is an area that warrants exploration. In this study, 18 S. furcifera populations sourced from six Asian countries were subjected to 2bRAD-M sequencing in order to unravel the vital factors contributing to shaping microbial communities in S. furcifera. Moreover, this comprehensive study explored the effects of symbionts and environmental factors on bacterial communities.
To determine the relationships among symbionts, bacterial communities in S. furcifera, and environmental factors, a total of 142 S. furcifera individuals from 18 populations were collected from 14 locations across six countries (China, Thailand, Myanmar, Cambodia, Vietnam, and Laos) analyzed in this study. Each S. furcifera population comprised at least six independent samples. All S. furcifera samples were preserved in 100% alcohol and subsequently dispatched for 2bRAD-M sequencing. Information about the samples is detailed in Table 1 and Supplementary Table S1, wherein each replicate represents an independent S. furcifera adult.
Bacterial communities in S. furcifera were analyzed using the 2bRAD-M sequencing methods. 2bRAD-M sequencing and library construction were performed by Qingdao OE Biotech Co., Ltd. (Qingdao, China), following previously established protocols (Wang et al., 2012) with marginal modifications. Briefly, the DNA (100 ng) from each sample was digested using 4 U of the BcgI enzyme (NEB) at 37°C for 3 h. Next, the resulting DNA fragments were ligated with specific adaptors. A mix of 5 μL of digested DNA and 10 μL of ligation master mix, containing 0.2 μM of each adaptor and 800 U of T4 DNA ligase (NEB), underwent a ligation reaction at 4°C for 12 h.
Following ligation, the products were PCR amplified, and the resulting DNA was subjected to electrophoresis on an 8% polyacrylamide gel. An approximately 100-bp band was excised, and the DNA fragments were then diffused in DEPC water at 4°C for 12 h. A PCR test was conducted using primers bearing platform-specific barcodes to introduce sample-specific barcodes. Each PCR sample, totaling 20 μL, contained 25-ng of gel-extracted PCR product, 0.2 μM each of forward and reverse primers, 0.3 mM dNTP mix, 0.4 U Phusion high-fidelity DNA polymerase (NEB), and 1 × Phusion HF buffer. After PCR amplification and electrophoresis, the PCR products were purified using the QIAquick PCR purification kit (50) (Qiagen) and sequenced on the Illumina Nova PE150 platform.
To conduct the 2bRAD-M analysis, microbial genome data from the NCBI database, consisting of 173,165 species of fungi, bacteria, and archaea, were employed. Restriction enzymes of 16 type 2B were utilized to fragment the samples, and Perl scripts were used in this process. Thereafter, the RefSeq (GCF) number was used to assign 2bRAD-M tags with microbial genome information, including taxonomic data. Unique 2bRAD tags occurring only once in every GCF were selected as species-specific 2bRAD-M markers, forming a reference database. A detection threshold of 0.0001 (0.01%) relative abundance was set as the default (Franzosa et al., 2018).
To calculate the relative abundance of each bacterium, 2bRAD tags from all samples, post-quality control, were mapped against the 2bRAD marker database containing tags unique to the 26,163 microbial species using a built-in Perl script. To mitigate false positives in species identification, a G score was calculated for each identified species within a given sample. This score, derived from a harmonic means of read coverage of 2bRAD markers belonging to a species and the total number of possible 2bRAD markers for this species, was employed identify species while minimizing errors. A threshold G score of 5 was set to prevent false-positive microbial species discovery (Sun et al., 2022).
S: the number of reads assigned to all 2bRAD markers belonging to species i within a sample.
T: number of all 2bRAD markers of species i that have been sequenced within a sample.
The average read coverage of all 2bRAD markers for each species was computed to represent the number of individuals of a species present within a sample at a given sequencing depth. The relative abundance of a species was then calculated as the ratio of the number of microbial individuals belonging to that species against the total number of individuals from known species detected within a sample.
S: the number of reads assigned to all 2bRAD markers of species i within a sample.
T: the number of all theoretical 2bRAD markers of species i.
Additionally, five environmental factors, namely annual mean temperature, annual precipitation, longitude, latitude, and altitude, were analyzed in the study. Data for these factors were downloaded from the WorldClim website.1 The impacts of climate and geographical factors on diversity and abundance indexes and symbiont infections were described using two structural equation models (SEM) with Satorra-Bentler correction. To limit heteroscedasticity, the log value of the “precipitation” values was used in the SEMs. SEM models were deemed acceptable when p value >0.05 and CFI > 0.95, after systematically excluding redundant pathways based on a lower AIC value. To standardize each parameter and eliminate variance, SEM coefficients were estimated through standardized transformation. Prior to analysis, data on Cardinium abundance (shown in Figure 1 left panel), Wolbachia abundance (shown in Figure 1 right panel), and microbial diversities of samples (shown in Figure 2) underwent normality testing using the Kolmogorov–Smirnov test and Levene’s test to assess the homogeneity of group variances. Data exhibiting a normal distribution were analyzed using one-way ANOVA with post-hoc Tukey HSD. In cases where the Cardinium abundance, Wolbachia abundance, and diversity data did not conform to normal distribution, they were analyzed using the Kruskal-Wallis test and Dunn’s test with Bonferroni correction for multiple comparisons. All statistical analyzes were conducted using SPSS 21.0. PCoA plots illustrating Bray-Curtis intersample distances and classification probabilities were generated using QIIME software (Caporaso et al., 2010). Pearson correlation analysis, performed using SPSS 21.0, was used to explore the relationships between different symbionts and diversity indexes. Furthermore, all graphical representations were generated using GraphPad Prism 9.0.0.
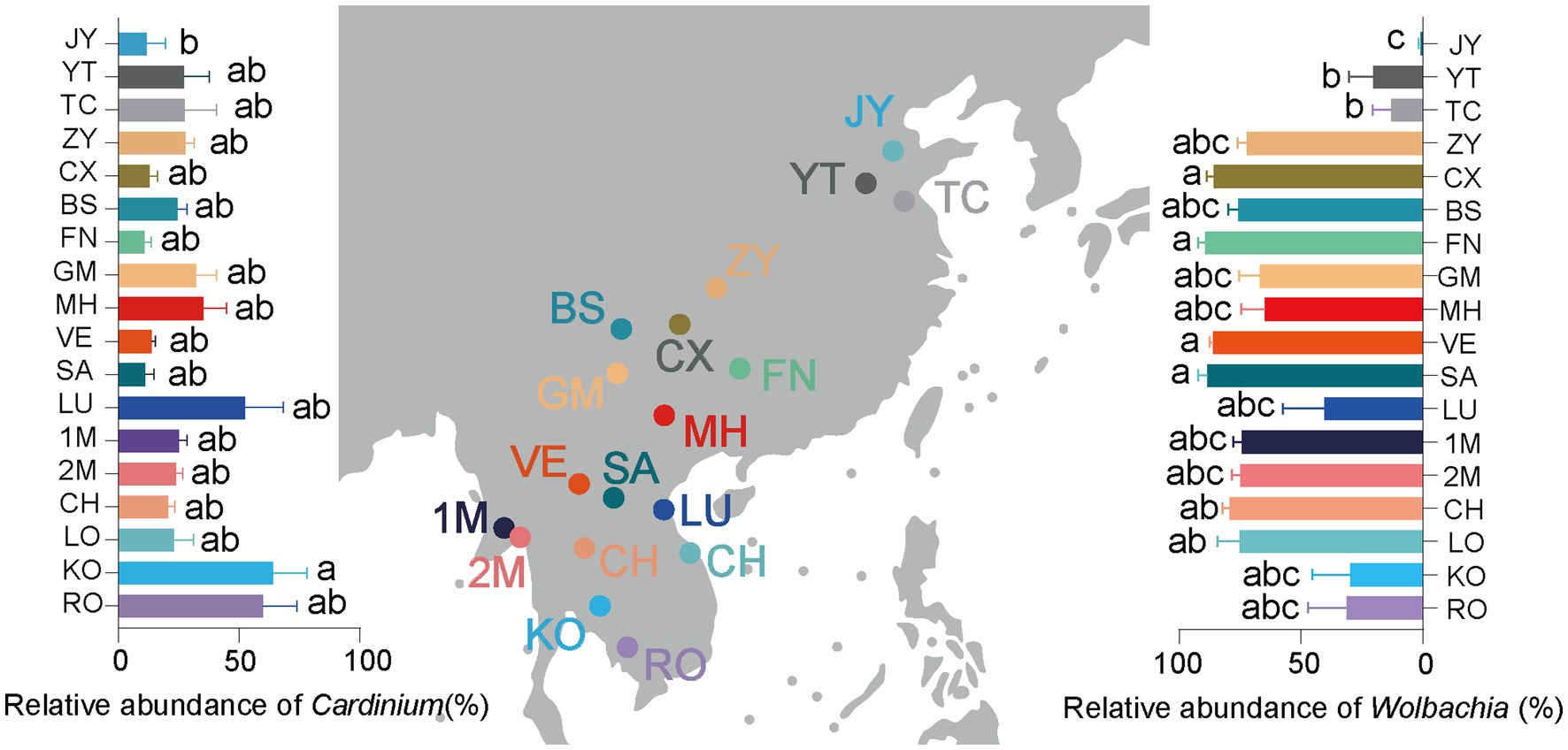
Figure 1. Relative infection abundance of Cardinium (left panel), Wolbachia (right panel) and collection sites (middle panel) of Sogatella furcifera across 18 geographical locations by 2bRAD-M sequencing. As the symbiont infection data do not follow a normal distribution, the difference of Wolbachia abundance in various S. furcifera populations was analyzed by Kruskal–Wallis test and Dunn’s test with Bonferroni correction for multiple comparisons by SPSS 21.0, different letters represent significant difference.
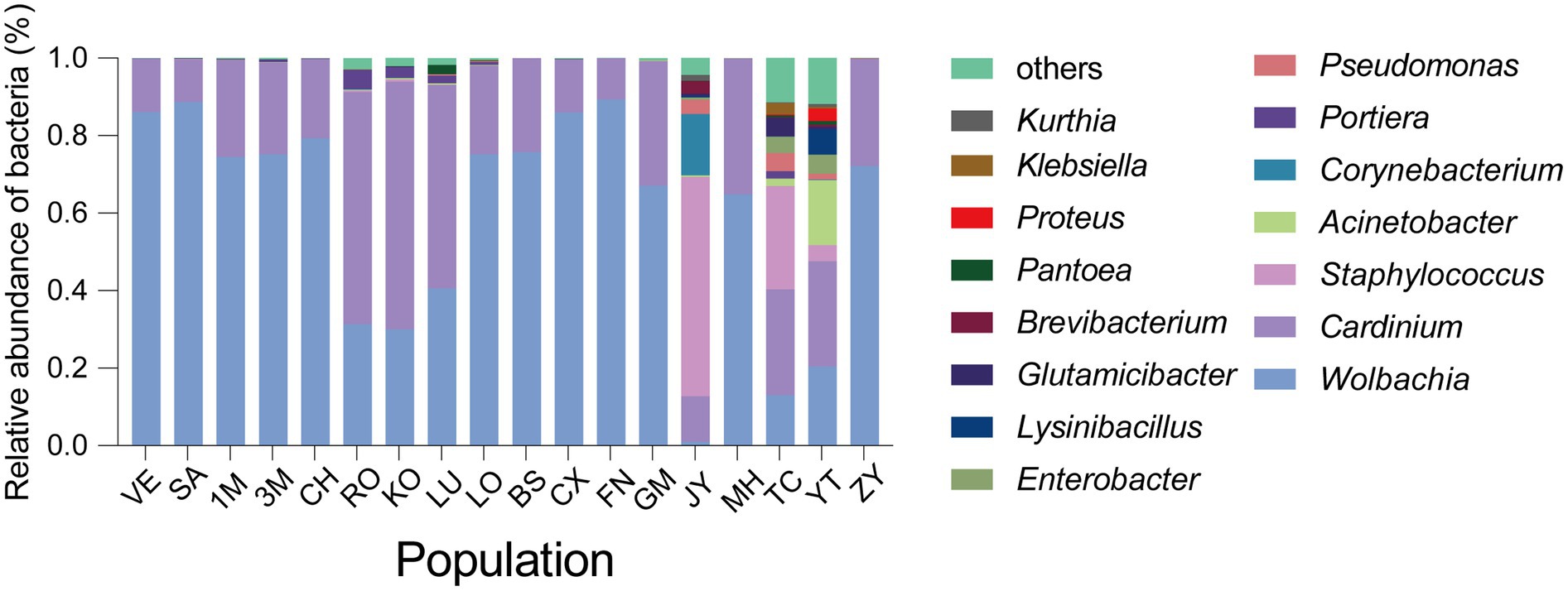
Figure 2. The abundance of microbial communities of Sogatella furcifera across 18 populations collected in Asia based on 2bRAD-M sequencing results. The relative abundance of top 15 abundant bacteria at the genus level was shown with different populations.
In the current study, a total of 18 S. furcifera populations were analyzed, comprising 9 populations from China and 9 populations from the South Asian region. The predominant bacterial symbionts identified in all S. furcifera populations were Wolbachia and Cardinium. Interestingly, the former was the most abundant bacteria in nearly all S. furcifera populations, being present in every S. furcifera sample. Notably, the relative abundance of Wolbachia exceeded 80% in two Chinese populations, specifically CS (85.95 ± 2.92%, Mean ± SEM) and FN (89.26 ± 3.01%, Mean ± SEM), as well as in all Laos populations, including VE (86.02 ± 1.57%, Mean ± SEM) and SA (86.02 ± 3.58%, Mean ± SEM). Nevertheless, significant variations in Wolbachia abundance were observed among different populations. Meanwhile, 3 S. furcifera populations from China (JY, YT, and TC) exhibited the lowest Wolbachia abundance, significantly lower than the Wolbachia-abundant populations such as FN, VE, and SA (Figure 1 right panel). Cardinium emerged as the second most abundant bacterium in S. furcifera (Figure 1 left panel). In several South Asia populations, including KO (63.88 ± 14.38%, Mean ± SEM), RO (59.96 ± 14.04%, Mean ± SEM), and LU (52.50 ± 15.74%, Mean ± SEM) as well as one Chinese population, YT (27.18 ± 10.38%, Mean ± SEM) Cardinium was the dominant symbiont, with its abundance in the KO population being significantly higher than that in the JY population (11.72 ± 7.78%, Mean ± SEM) (p < 0.05, Kruskal-Wallis test). Besides, Wolbachia abundance was less than 1% in the JY population, while Cardinium abundance was the highest (56.59 ± 11.90%, Mean ± SEM). Additionally, the primary symbiont Portiera, typically associated with whiteflies, was also detected in several S. furcifera populations, including TC, KO, VE, etc. (Figure 3). Principal Component Analysis was conducted to explore differences among bacterial communities of all S. furcifera populations, revealing distinct microbial communities in S. furcifera individuals from FN and JY populations compared to other samples (Supplementary Figure S1).
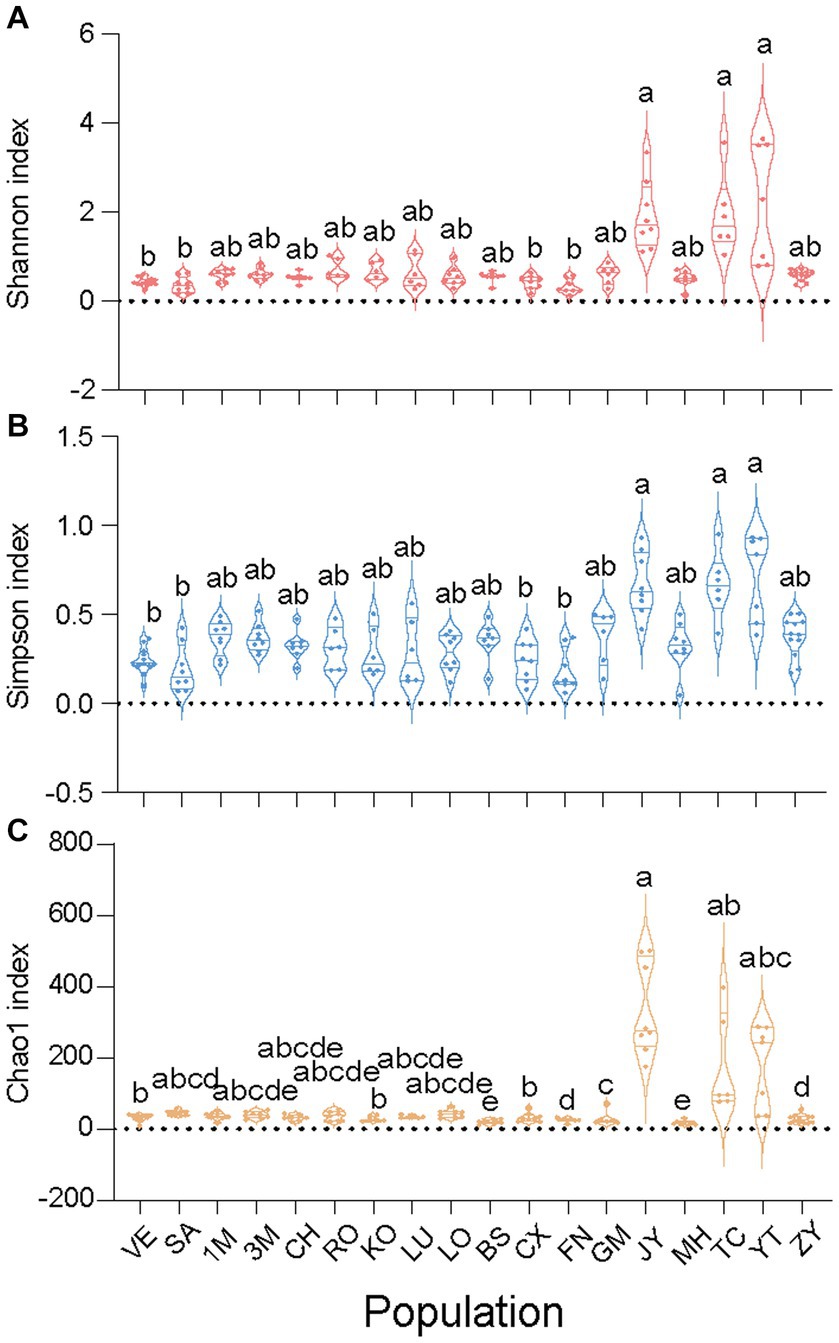
Figure 3. Alpha diversities of all 18 Sogatella furcifera MED populations collected in Asia. The Shannon index (A), Simpson index (B) and Chao1 index (C) were present based on 2bRAD-M sequencing results, respectively. As the diversity data do not follow a normal distribution, they were analyzed by Kruskal–Wallis test and Dunn’s test with Bonferroni correction for multiple comparisons by SPSS 21.0, different letters represent significant difference.
To elucidate variations in microbial communities among different S. furcifera populations, alpha diversity indices, including Shannon, Simpson, and Chao1, were calculated for each S. furcifera population. Considering that the three alpha indices of all populations did not follow a normal distribution, the Kruskal–Wallis test, and Dunn’s test with Bonferroni correction for multiple comparisons were adopted (Figure 2). Intriguingly, populations with low Wolbachia abundance exhibited higher alpha diversities in bacterial communities compared to those with high Wolbachia abundance. Specifically, populations with low Wolbachia abundance, such as JY, TC, and YT, displayed significantly higher Shannon and Simpson indices compared to populations with high Wolbachia abundance, including VE, SA, CX, and FN (Figure 2). The UPGMA hierarchical cluster diagram of different Sogatella furcifera populations with 2bRAD-M sequencing data was shown (Supplementary Figure S2), and the phylogenetic tree of different Sogatella furcifera populations with 2bRAD sequencing results was constructed (Supplementary Figure S3).
Pearson analysis was used to assess the correlations between symbionts and other bacteria in S. furcifera. As illustrated in Figure 4A, the presence of Wolbachia negatively impacted the abundance of various bacteria, encompassing Enterobacter, Acinetobacter, and Lysinibacillus. Notably, Wolbachia significantly and negatively influenced the abundance of Portiera in S. furcifera (r = −0.518, p < 0.05, Pearson analysis). While the abundance of Wolbachia was negatively correlated with that of Cardinium, the correlation was not statistically significant (r = −0.451, p = 0.06, Pearson analysis). Consistently, Cardinium significantly positively influenced the abundance of Pseudomonas (r = −0.804, p < 0.001, Pearson analysis) and that of various other bacteria, albeit the correlation was not statistically significant (Figure 4B).
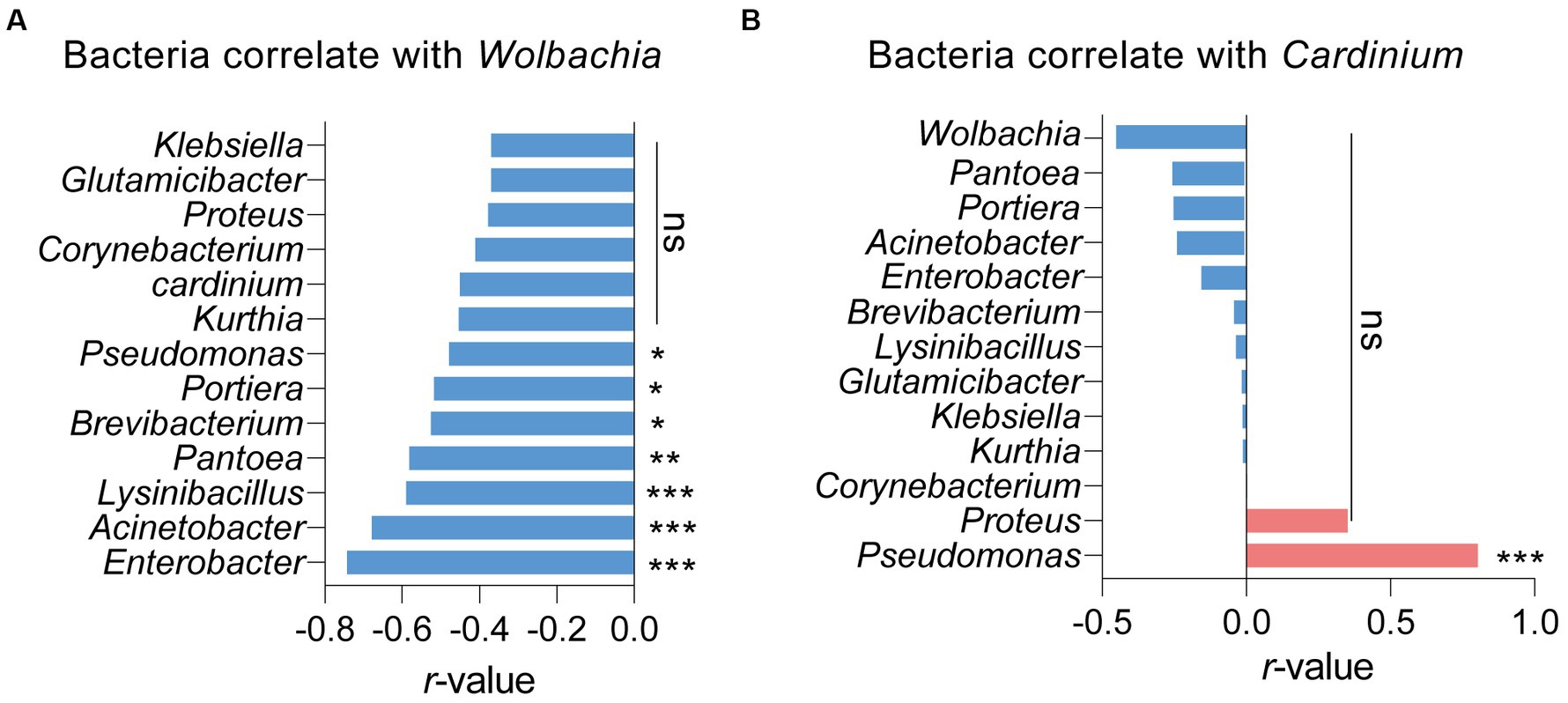
Figure 4. Relationships between the proportions of other main 13 bacteria and the proportions of Wolbachia (A) or the proportions of Cardinium (B) among all 18 Sogatella furcifera populations by Pearson correlation analysis (SPSS 21.0) based on 2bRAD-M sequencing results. r-values and p values of each linear regression plots are provided. “ns” means no significant; asterisks indicate significant difference the two compared group, *p < 0.05; **p < 0.01; ***p < 0.001.
Noteworthily, Pearson analysis exposed that the presence of Wolbachia significantly and negatively affected the three diversity indexes of S. furcifera, namely the Shannon index (r = −0.834, p < 0.001, Pearson analysis), Simpson index (r = −775, p < 0.001, Pearson analysis), and Chao1 index (r = −0.750, p < 0.001, Pearson analysis) (Figure 5). Cardinium infection also had a negative impact on the diversity indexes, although the correlations were not significant (Supplementary Figure S4).
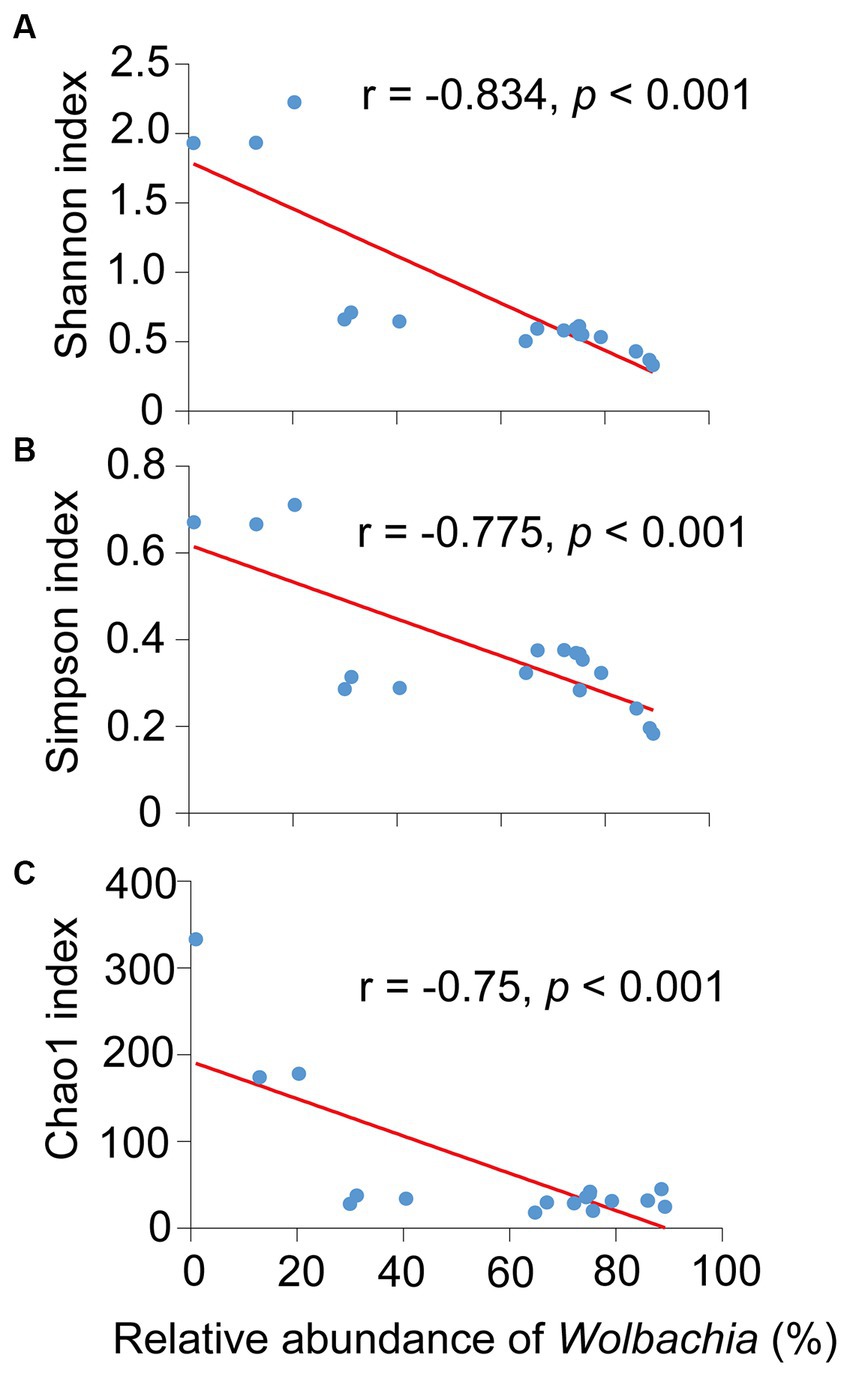
Figure 5. Relationships between the abundance of Wolbachia and the Shannon diversity index (A), Simpson index (B) and Chao1 index (C) among all 18 Sogatella furcifera populations by Pearson correlation analysis (SPSS 21.0) based on 2bRAD-M sequencing results. r-values and p values of each linear regression plots are provided.
The effects of five environmental factors (annual mean temperature, annual precipitation, latitude, longitude, and altitude) on S. furcifera microbiota were determined through Pearson analysis. While both Wolbachia and Cardinium abundance in S. furcifera increased with annual mean temperature, there was no significant correlation between temperature and the abundance of these two symbionts. Notwithstanding, temperature had a significantly negative impact on the abundance of numerous bacteria, such as Enterobacter, Lysinibacillus, and Acinetobacter (Figure 6A). Precipitation had a significant positive influence on Wolbachia abundance (r = 0.489, p < 0.05, Pearson analysis) (Figure 6B). Latitude was significantly and positively correlated with the abundance of many bacteria and was negatively correlated with that of Cardinium and Wolbachia, although the difference was not significant for the latter (Figure 6C). On the other hand, longitude was significantly and negatively correlated with the abundance of Wolbachia (r = 0.750, p < 0.001, Pearson analysis) and positively correlated with the abundance of other bacteria in S. furcifera (Figure 6D). Finally, altitude did not significantly affect bacterial abundance in S. furcifera (Supplementary Figure S5).
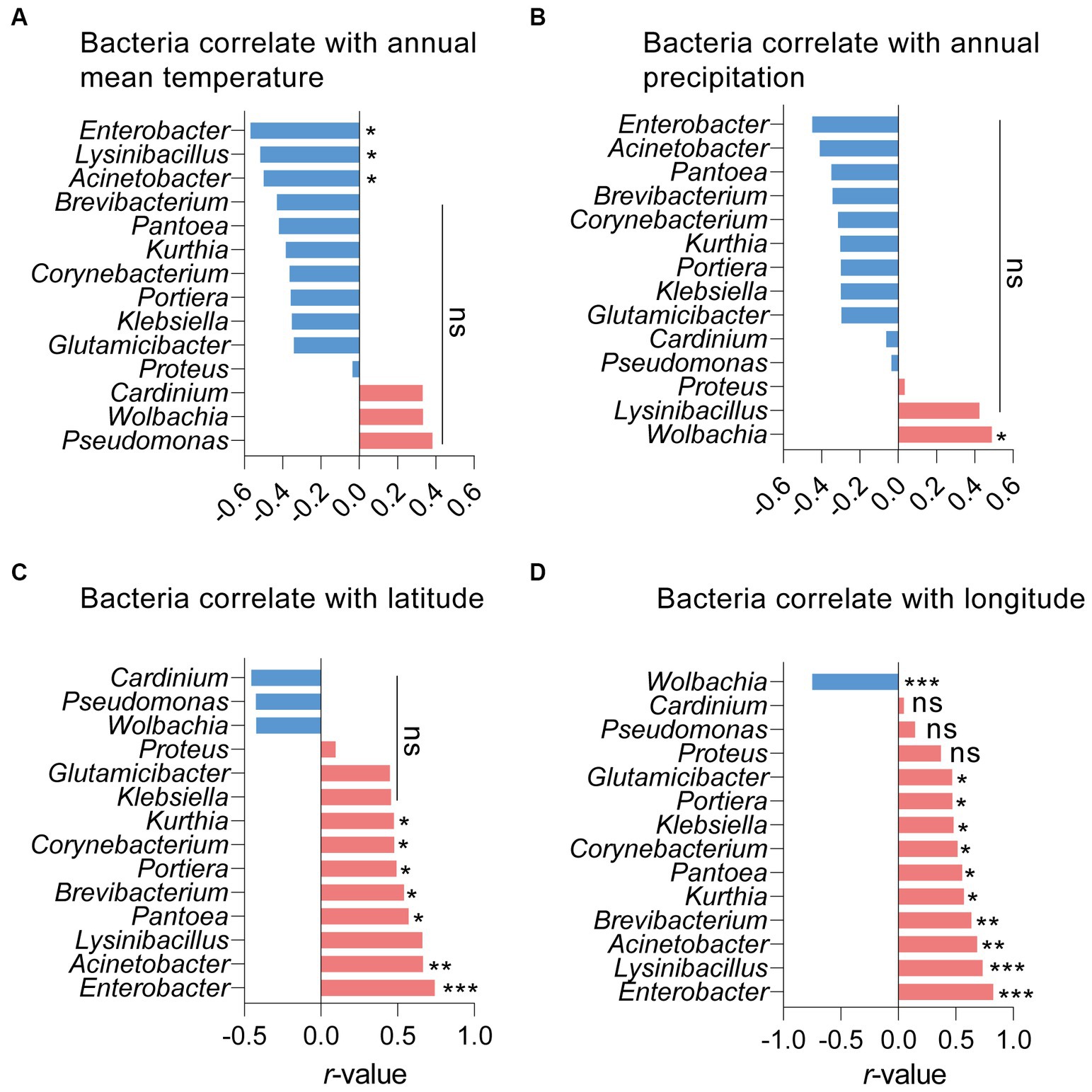
Figure 6. Relationships between the proportions of main 14 bacteria and the annual mean temperature (A), annual precipitation (B), latitude (C) and longitude (D) among all 18 Sogatella furcifera populations by Pearson correlation analysis (SPSS 21.0) based on 2bRAD-M sequencing results. r-values and p values of each linear regression plots are provided. “ns” means no significant; asterisks indicate significant difference the two compared group, *p < 0.05; **p < 0.01; ***p < 0.001.
SEM analysis validated the significant influence of precipitation on Wolbachia abundance (r = 0.400, z = 1.967, p < 0.05, SEM) and the negative effect of latitude on Cardinium abundance (r = −3.021, z = −2.498, p < 0.05, SEM). In contrast to Pearson analysis, SEM analysis revealed a significant negative correlation between Wolbachia and Cardinium (r = −0.87, z = −2.785, p < 0.01, SEM). Lastly, temperature had a numerically positive effect on Wolbachia abundance and a negative effect on Cardinium abundance (Figure 7).
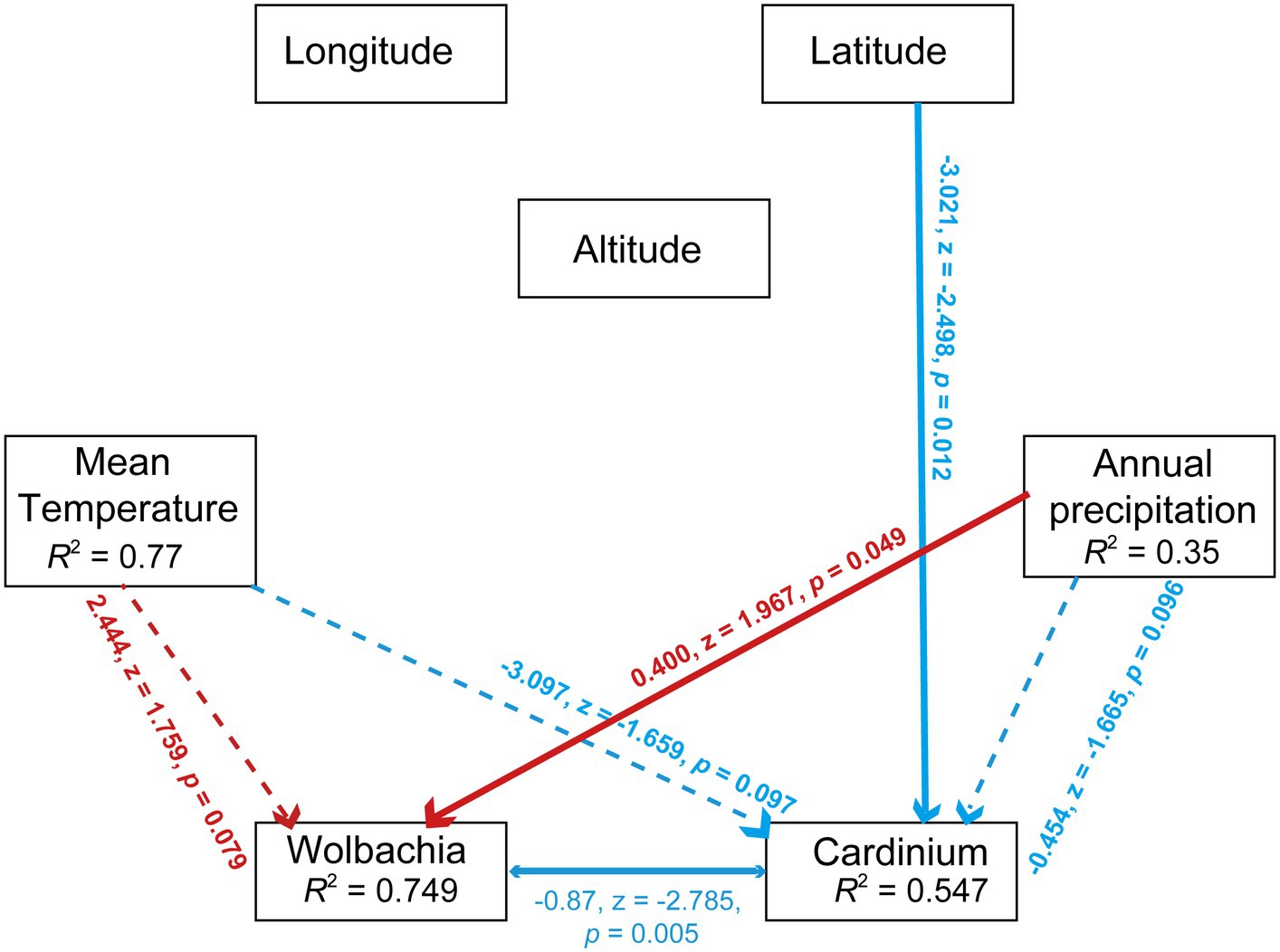
Figure 7. Path diagram for the structural equation model (SEM) for environmental factors (including annual mean temperature, annual precipitation, longitude, altitued and latitude) and symbionts’ abundance (including Wolbachia and Cardinium). Statistically significant negative paths are indicated by blue arrows, while positive paths are indicated by red arrows. The r values in each box indicate the amount of variation in that variable explained by the input arrows. Numbers next to arrows are unstandardized slopes.
Furthermore, the effects of environmental factors on microbiota diversity in S. furcifera were explored. According to Pearson analysis, both latitude and longitude exerted a positive influence on all three diversity indexes (Shannon, Simpson, and Chao1 indexes) of bacterial communities in S. furcifera, whereas altitude had a marginal negative effect on microbiota diversity (Figure 8). In contrast, SEM analysis identified that altitude negatively impacted the Shannon index (r = −0.806, z = −2.096, p < 0.05, SEM), which differed from the results of Pearson analysis (Figure 8G). SEM analysis uncovered that temperature had a negative influence on both the Shannon index (r = −1.879, z = −2.527, p < 0.05, SEM) and Simpson index (r = −3.189, z = −3.493, p < 0.001, SEM) (Figure 9).
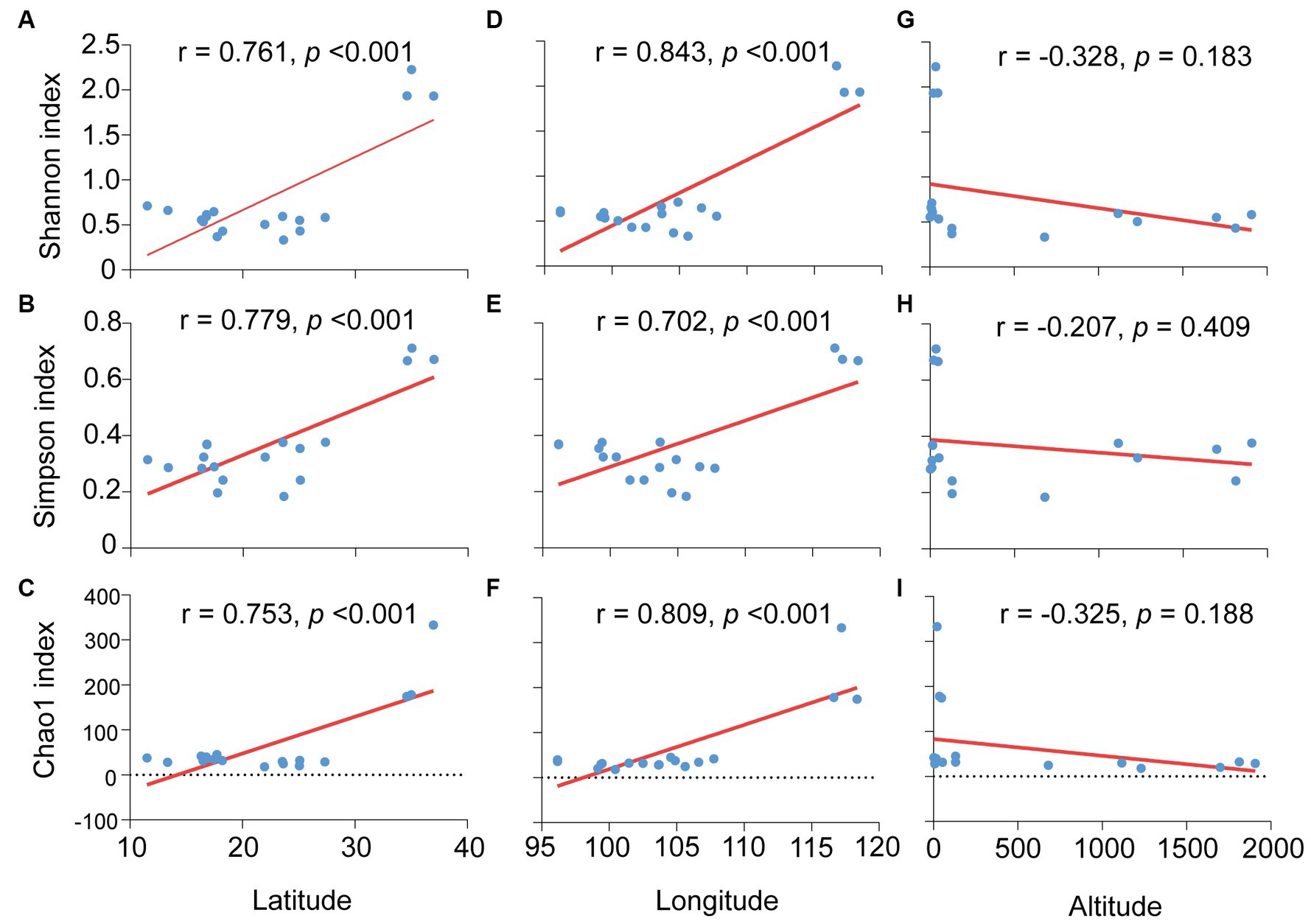
Figure 8. Relationships between the microbiota diverisity indexes and environmental factors in Sogatella furcifera. The relationships between latitude and Shannon diversity index (A), Simpson index (B) and Chao1 index (C), between longitude and Shannon diversity index (D), Simpson index (E) and Chao1 index (F), between altitude and Shannon diversity index (G), Simpson index (H) and Chao1 index (I) were shown, respectively, all analysis were used by Pearson correlation analysis (SPSS 21.0) based on 2bRAD-M sequencing results. r-values and p values of each linear regression plots are provided.
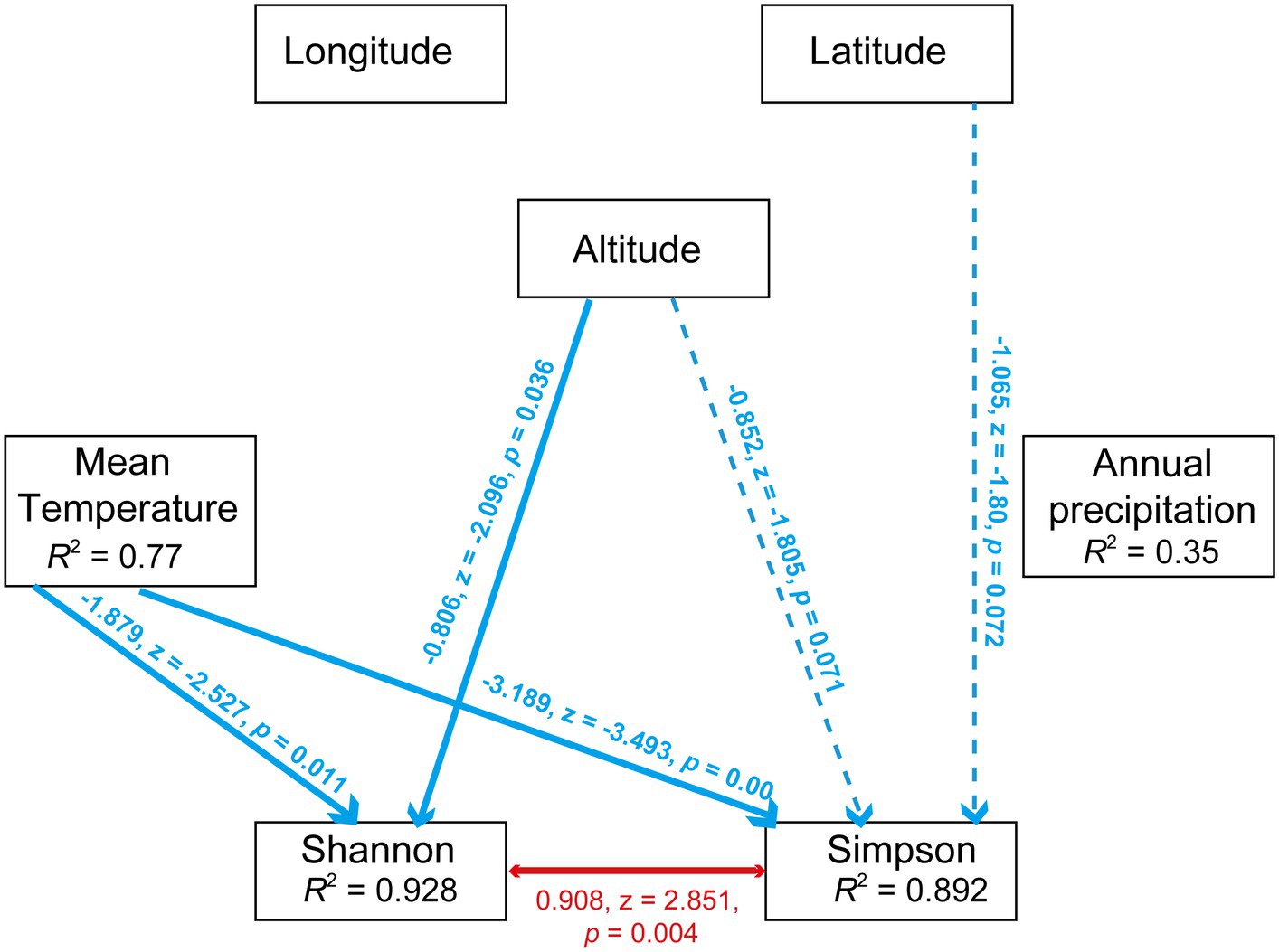
Figure 9. Path diagram for the structural equation model (SEM) for environmental factors (including annual mean temperature, annual precipitation, longitude, altitude and latitude) and diversity indexes (including Shannon index and Simpson index). Statistically significant negative paths are indicated by blue arrows, while positive paths are indicated by red arrows. The r values in each box indicate the amount of variation in that variable explained by the input arrows. Numbers next to arrows are unstandardized slopes.
Symbiotic bacteria play a critical role in the biology, ecology, and evolution of hosts, and various environmental factors significantly impact bacterial communities in invertebrate hosts. Herein, the effects of symbionts on the microbiota in S. furcifera populations and the significant effects of environmental factors on bacterial communities in S. furcifera were analyzed. Our findings revealed that the presence of symbionts Wolbachia and Cardinium in S. furcifera negatively influenced the abundance of numerous other bacteria. Additionally, Wolbachia infection significantly reduced the diversity of microbial communities in S. furcifera. Several environmental factors, including longitude, latitude, temperature, and precipitation, were found to impact the abundance of symbionts and the microbial diversity in S. furcifera.
In the current study, the bacterial community composition in S. furcifera was abundant, primarily consisting of two dominant symbionts, namely Wolbachia and Cardinium (Figure 3). These results are consistent with the findings of previous studies on S. furcifera microbiota (Li et al., 2020, 2021, 2022). Notably, the presence of a primary symbiont, Portiera, in the S. furcifera microbiota, with significant infection rates was noted in various populations such as RO (5.03 ± 10.38%, Mean ± SEM) and KO (2.73 ± 1.40%, Mean ± SEM) (Figure 3). The horizontal transmission of symbionts between different arthropod species has been well-documented (Werren et al., 2008; Zhang et al., 2016). Research has established that horizontal transmission can occur between different phloem sap-feeding insect species, accounting for the presence of Portiera within S. furcifera (Gonella et al., 2015). Previous studies confirmed a vital influence of genetic backgroud on the microbiome in invertebrates (Suzuki et al., 2019; Gupta and Nair, 2020), however, no direct relationship between S. furcifera genetic phylogenetic tree and UPGMA hierarchical cluster diagram of microbiome in S. furcifera discovered (Supplementary Figures S2, S3).
Interactions among different bacteria, especially symbionts, are complex in arthropods. Specifically, the presence of one symbiont generally influences the infection patterns of other symbionts (Evans and Armstrong, 2006; Zhao et al., 2018; Fromont et al., 2019; Duan et al., 2020). This phenomenon is not surprising, given that symbionts are primarily restricted to bacteriocytes and reproductive tissues within invertebrate hosts, leading to competition for limited nutrition and space (Engel and Moran, 2013; Douglas, 2015; Yang et al., 2019). For example, in Laodelphax striatellus, Wolbachia infection negatively affects the abundance of 154 other bacterial genera within hosts (Duan et al., 2020). Conflicting interactions between symbionts, such as Cardinium and Hamiltonella in whiteflies, have been observed (Zhao et al., 2018), while other bacteria in honeybees decrease the development rate of the primary symbiont Paenibacillus larvae (Evans and Armstrong, 2006). Comparable results were obtained in our study; Wolbachia negatively influenced the abundance of 13 primary genera of bacteria, while Cardinium negatively influenced the abundance of 11 other genera (Figure 4). This observation suggests a competitive relationship among bacteria in S. furcifera. However, Cardinium infection was positively correlated with the abundance of Pseudomonas and Proteus, indicating a cooperative interaction between Cardinium and these two bacteria (Figure 4B). Similar interactions have been described in other studies. For example, the presence of Wolbachia in the fruit fly Drosophila neotestacea promotes the infection of Spiroplasma (Fromont et al., 2019). In Laodelphax striatellus, Wolbachia infection increases the abundance of other bacteria, such as Spiroplasma and Ralstonia (Duan et al., 2020). These complex interactions among bacteria, especially symbionts, necessitate further exploration.
Symbiont plays a pivotal role in shaping the microbial community of invertebrates and invariably influences the host’s microbiota structure. Symbiont infections frequently lead to a reduction in bacterial diversities in arthropod hosts (Duan et al., 2020; Li et al., 2020, 2022). In the present study, Wolbachia infection significantly decreased the diversity of the bacterial community in S. furcifera (Figure 5). This finding is in line with the result of previous studies in Aedes aegypti, wherein the presence of Wolbachia reduced the diversity of resident bacteria in mosquitoes (Audsley et al., 2018). Similar reductions in microbial diversities have been observed in the small brown planthopper Laodelphax striatellus (Zhang et al., 2020) and Drosophila melanogaster (Ye et al., 2017). These studies collectively underscore the negative influence of symbionts on the bacterial communities of arthropods.
The influence of environmental factors on arthropod microbial communities has been explored in previous research. Studies on global invertebrates indicated that temperature significantly affects the occurrence of Wolbachia and Cardinium infections in host arthropods (Charlesworth et al., 2019). Similar temperature effects on symbionts were corroborated in other studies (Corbin et al., 2016). However, in this study, the impact of temperature on symbionts (Cardinium and Wolbachia) was not significant in S. furcifera (Figures 6A, 7), suggesting weak temperature effects on symbiont infections in S. furcifera. Other environmental factors also influence symbiont infections in hosts. For example, in the spider mite Tetranychus truncatus, Wolbachia infection was significantly influenced by annual mean temperatures, whereas the rates of Cardinium and Spiroplasma infections were correlated with altitude (Zhu et al., 2018). Additionally, bacterial communities were significantly affected by geographical distances in stoneflies (Zhu et al., 2022). Herein, the abundance of numerous bacteria in S. furcifera were affected by environmental factors, with Wolbachia abundance significantly influenced by precipitation and longitude and Cardinium abundance not significantly affected by the environment (Figure 6 and Supplementary Figure S3). The differential impact of the environment on Wolbachia and Cardinium requires further exploration.
In summary, our 2bRAD-M sequencing analysis of 18 S. furcifera populations offers a novel and comprehensive perspective of the relationship between bacterial community structure, environmental factors, and symbionts in Asian S. furcifera populations. Our observations provide credible evidence that the Wolbachia and Cardinium symbionts within host whiteflies influence each other and negatively impact the abundance of other bacteria. Importantly, Wolbachia’s presence negatively affected microbial diversity, with both symbiont and microbiota diversities being significantly influenced by various environmental factors. However, further studies are necessary to elucidate the mechanisms by which environmental factors influence the diversity of bacterial communities in different S. furcifera populations across global ecological timescales.
The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJNA1055281.
The manuscript presents research on animals that do not require ethical approval for their study.
KY: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Writing – original draft, Writing – review & editing. H-YZ: Investigation, Methodology, Validation, Writing – review & editing. PW: Supervision, Writing – original draft. G-XJ: Formal analysis, Validation, Conceptualization, Writing – original draft, Writing – review & editing. DC: Conceptualization, Formal analysis, Funding acquisition, Supervision, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Taishan Scholar Foundation of Shandong Province of China (tstp20221135), and Shandong Modern Agricultural Technology & Industry System (SDAIT-17-07), and the Qingdao Agricultural University High-level Talent Fund (663–1121025).
We are grateful to Aidong Chen (Agriculture Environment and Resources Institute, Yunnan Academy of Agricultural Sciences, Kunming 650205, China) for providing the insect samples.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1336345/full#supplementary-material
Audsley, M. D., Seleznev, A., Joubert, D. A., Woolfit, M., O'Neill, S. L., and McGraw, E. A. (2018). Wolb-achia infection alters the relative abundance of resident bacteria in adult Aedes aegypti m-osquitoes, but not larvae. Mol. Ecol. 27, 297–309. doi: 10.1111/mec.14436
Behar, A., Yuval, B., and Jurkevitch, E. (2008). Community structure of the Mediterranean fruit fly microbiota: seasonal and spatial sources of variation. Isr. J. Ecol. Evol. 54, 181–191. doi: 10.1080/15659801.2008.10639612
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Charlesworth, J., Weinert, L. A., Araujo, J. E. V., and Welch, J. J. (2019). Wolbachia, Cardinium and climate: an analysis of global data. Biol. Lett. 15:20190273. doi: 10.1098/rsbl.2019.0273
Colman, D. R., Toolson, E. C., and Takacs-Vesbach, C. D. (2012). Do diet and taxonomy influence insect gut bacterial communities? Mol. Ecol. 21, 5124–5137. doi: 10.1111/j.1365-294X.2012.05752.x
Corbin, C., Heyworth, E. R., Ferrari, J., and Hurst, G. D. D. (2016). Heritable symbionts in a world of varying temperature. Heredity 118, 10–20. doi: 10.1038/hdy.2016.71
Douglas, A. E. (2009). The microbial dimension in insect nutritional ecology. Funct. Ecol. 23, 38–47. doi: 10.1111/j.1365-2435.2008.01442.x
Douglas, A. E. (2015). Multiorganismal insects: diversity and function of resident microorganims. Annu. Rev. Entomol. 60, 17–34. doi: 10.1146/annurev-ento-010814-020822
Duan, X. Z., Sun, J. T., Wang, L. T., Shu, X. H., Guo, Y., Keiichiro, M., et al. (2020). Recent infection by Wolbachia alters microbial communities in wild Laodelphax striatellus populations. Microbiome 8:104. doi: 10.1186/s40168-020-00878-x
Dunbar, H. E., Wilson, A. C., Ferguson, N. R., and Moran, N. A. (2007). Aphid thermal tolerance is gov-erned by a point mutation in bacterial symbionts. PLoS Biol. 5:e96. doi: 10.1371/journal.pbio.0050096
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects-diversity in structure and functi-on. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Evans, J. D., and Armstrong, T. N. (2006). Antagonistic interactions between honey bee bacterial sy-mbionts and implications for disease. BMC Ecol. 6:4. doi: 10.1186/1472-6785-6-4
Franzosa, E. A., McIver, L. J., Rahnavard, G., Thompson, L. R., Schirmer, M., Weingart, G., et al. (2018). Species-level function-al profiling of metagenomes and metatranscriptomes. Nat. Methods 15, 962–968. doi: 10.1038/s41592-018-0176-y
Fromont, C., Adair, K. L., and Douglas, A. E. (2019). Correlation and causation between the microbio-me, Wolbachia and host functional traits in natural populations of drosophilid flies. Mol. Ecol. 28, 1826–1841. doi: 10.1111/mec.15041
Gonella, E., Pajoro, M., Marzorati, M., Crotti, E., Mandrioli, M., Pontini, M., et al. (2015). Plant-mediated interspecific horizontal transmission of an intracellular symbiont in insects. Sci. Rep. 5:15811. doi: 10.1038/srep15811
Goto, S., Anbutsu, H., and Fukatsu, T. (2006). Asymmetrical interactions between Wolbachia and Spiroplasma endosymbionts coexisting in the same insect host. Appl. Environ. Microbiol. 72, 4805–4810. doi: 10.1128/AEM.00416-06
Gupta, A., and Nair, S. (2020). Dynamics of insect-microbiome interaction influence host and microbial symbiont. Front. Microbiol. 11:1357. doi: 10.3389/fmicb.2020.01357
Hertig, M., and Wolbach, S. B. (1924). Studies on rickettsia-like microorganisms in insects. J. Med. Res. 44, 329–374.7.
Hu, S. J., Liu, X. F., Fu, D. Y., Huang, W., Wang, X. Y., Liu, X. J., et al. (2015). Projecting distribution of the overwintering population of Sogatella furcifera (Hemiptera: Delphacidae), in Yunn-an, China with analysis on key influencing climatic factors. J. Insect Sci. 15:148. doi: 10.1093/jisesa/iev131
Huang, Y. X., and Qin, D. Z. (2018). First mitogenome for the tribe Saccharosydnini (Hemiptera: D-elphacidae: Delphacinae) and the phylogeny of three predominant rice planthoppers. Eur. J. Entomol. 115:242. doi: 10.14411/eje.2018.023
Li, F., Li, P., Hua, H., Hou, M., and Wang, F. (2020). Diversity, tissue localization, and infection pa-ttern of bacterial symbionts of the white-backed blanthopper, Sogatella furcifera (Hemiptera: Delphacidae). Microb. Ecol. 79, 720–730. doi: 10.1007/s00248-019-01433-4
Li, T. P., Zha, S. S., Zhou, C. Y., Xia, X., Hoffmann, A. A., and Hong, X. Y. (2021). Two newly introduced Wolbachia endosymbionts induce cell host differences in competitiveness and metabolic re-sponses. Appl. Environ. Microbiol. 87:e0147921. doi: 10.1128/AEM.01479-21
Li, T. P., Zhou, C. Y., Wang, M. K., Zha, S. S., Chen, J., Bing, X. L., et al. (2022). Endosymbionts reduce microbiome diversity and modify host metabolism and fecundity in the planthopper Sogatella furcifera. Msystems 7:e0151621. doi: 10.1128/msystems.01516-21
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C. G., Carey, H. V., Domazet-Lošo, T., Douglas, A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U. S. A. 110, 3229–3236. doi: 10.1073/pnas.1218525110
Miao, Y.-H., Xiao, J.-H., and Huang, D.-W. (2020). Distribution and evolution of the bacteriophage WO and its antagonism with Wolbachia. Front. Microbiol. 11:595629. doi: 10.3389/fmicb.2020.595629
Santo Domingo, J. W., Kaufman, M. G., Klug, M. J., Holben, W. E., Harris, D., and Tiedje, J. M. (1998). In-fluence of diet on the structure and function of the bacterial hindgut community of cricke-ts. Mol. Ecol. 7, 761–767. doi: 10.1046/j.1365-294x.1998.00390.x
Savary, S., Horgan, F., Willocquet, L., and Heong, K. L. (2012). A review of principles for sustainab-le pest management in rice. Crop Prot. 32, 54–63. doi: 10.1016/j.cropro.2011.10.012
Sun, Z., Huang, S., Zhu, P., Tzehau, L., Zhao, H., Lv, J., et al. (2022). Species-resolved sequencing of low-biomass or degraded microbiomes using 2bRAD-M. Genome Biol. 23:36. doi: 10.1186/s13059-021-02576-9
Suzuki, T. A., Phifer-Rixey, M., Mack, K. L., Sheehan, M. J., Lin, D., Bi, K., et al. (2019). Host genetic determinants of the gut microbiota of wild mice. Mol. Ecol. 28, 3197–3207. doi: 10.1111/mec.15139
Taylor, M. J., Bandi, C., and Hoerauf, A. (2005). Wolbachia bacterial endosymbionts of filarial nem-atodes. Adv. Parasitol. 60, 245–284. doi: 10.1016/S0065-308X(05)60004-8
Wang, Y. B., Li, C., Yan, J. Y., Wang, T. Y., Yao, Y. L., Ren, F. R., et al. (2022). Autophagy regulates whitefly-symbiont metabolic interactions. Appl. Environ. Microbiol. 88:e0208921. doi: 10.1128/AEM.02089-21
Wang, S., Meyer, E., McKay, J., and Matz, M. V. (2012). 2b-RAD: a simple and flexible method for genome-wide genotyping. Nat. Methods 9, 808–810. doi: 10.1038/nmeth.2023
Wang, Z., Yang, H., Zhou, C., Yang, W. G., Jin, D.-C., and Long, G.-Y. (2019). Molecular cloning, expr-ession, and functional analysis of the chitin synthase 1 gene and its two alternative splici-ng variants in the white-backed planthopper, Sogatella furcifera (Hemiptera: Delphacidae). Sci. Rep. 9:1087. doi: 10.1038/s41598-018-37488-5
Wang, Z., Zhou, C., Long, G. Y., Yang, H., and Jin, D. C. (2018). Sublethal effects of buprofezin on d-evelopment, reproduction, and chitin synthase 1 gene (SfCHS1) expression in the white-bac-ked planthopper, Sogatella furcifera (Hemiptera: Delphacidae). J. Asia Pac. Entomol. 21, 585–591. doi: 10.1016/j.aspen.2018.03.009
Wasala, S. K., Brown, A. M. V., Kang, J., Howe, D. N. K., Peetz, A. B., Zasada, I. A., et al. (2019). Variable abundance and distribution of Wolbachia and Cardinium endosymbionts in plantp-arasitic nematode field populations. Front. Microbiol. 10:964. doi: 10.3389/fmicb.2019.00964
Werren, J. H. (1997). Biology of wolbachia. Annu. Rev. Entomol. 42, 587–609. doi: 10.1146/annurev.ento.42.1.587
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate bio-logy. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
Yang, K., Qin, P.-H., Yuan, M.-Y., Chen, L., Zhang, Y.-J., and Chu, D. (2022). Infection density pattern of Cardinium affects the responses of bacterial communities in an invasive whitefly under heat conditions. Insect Sci. 30, 1149–1164. doi: 10.1111/1744-7917.13141
Yang, K., Xie, K., Zhu, Y. X., Huo, S. M., Hoffmann, A. A., and Hong, X. Y. (2019). Wolbachia dominate Spiroplasma in the co–infected spider mite Tetranychus truncatus. Insect Mol. Biol. 29, 19–37. doi: 10.1111/imb.12607
Ye, Y. H., Seleznev, A., Flores, H. A., Woolfit, M., and McGraw, E. A. (2017). Gut microbiota in Droso-phila melanogaster interacts with Wolbachia but does not contribute to Wolbachia-mediated antiviral protection. J. Invertebr. Pathol. 143, 18–25. doi: 10.1016/j.jip.2016.11.011
Zeng, Z., Fu, Y., Guo, D. Y., Wu, Y. X., Ajayi, O. E., and Wu, Q. F. (2018). Bacterial endosymbiont Cardi-nium cSfur genome sequence provides insights for understanding the symbiotic relationshi-p in Sogatella furcifera host. BMC Genomics 19:688. doi: 10.1186/s12864-018-5078-y
Zhang, Y. K., Chen, Y. T., Yang, K., and Hong, X. Y. (2016). A review of prevalence and phylogeny of the bacterial symbiont Cardinium in mites (subclass: Acari). Syst. Appl. Acarol. 21, 978–990. doi: 10.11158/saa.21.7.11
Zhang, X., Li, T. P., Zhou, C. Y., Zhao, D. S., Zhu, Y. X., Bing, X. L., et al. (2020). A-ntibiotic exposure perturbs the bacterial community in the small brown planthopper Laodelphax striatellus. Insect Sci. 27, 895–907. doi: 10.1111/1744-7917.12675
Zhao, D. X., Hoffmann, A. A., Zhang, Z. C., Niu, H. T., and Guo, H. F. (2018). Interactions between facult-ative symbionts Hamiltonella and Cardinium in Bemisia tabaci (Hemiptera: Aleyrodoidea): cooperation or conflict? J. Econ. Entomol. 111, 2660–2666. doi: 10.1093/jee/toy261
Zhou, G. H., Wen, J. J., Cai, D. J., Xu, D. L., and Zhang, S. G. (2008). Southern rice black-streaked dwarf virus: a new proposed Fijivirus species in the family Reoviridae. Chin. Sci. Bull. 53, 3677–3685. doi: 10.1007/s11434-008-0467-2
Zhou, C., Yang, H., Wang, Z., Long, G.-Y., and Jin, D.-C. (2018). Comparative transcriptome analysis of Sogatella furcifera (Horváth) exposed to different insecticides. Sci. Rep. 8:8773. doi: 10.1038/s41598-018-27062-4
Zhu, Y. X., Huo, Q. B., Wen, T., Wang, X. Y., Zhao, M. Y., and Du, Y. Z. (2022). Mechanisms of fungal c-ommunity assembly in wild stoneflies moderated by host characteristics and local environ-ment. NPJ Biofilms Microb. 8:31. doi: 10.1038/s41522-022-00298-9
Zhu, Y. X., Song, Y. L., Zhang, Y. K., Hoffmann, A. A., Zhou, J. C., Sun, J. T., et al. (2018). Incidence of facultative bacterial endosymbionts in spider mites associated with local environments and host plants. Appl. Environ. Microbiol. 84, e02546–e02517. doi: 10.1128/AEM.02546-17
Keywords: Sogatella furcifera, Wolbachia, Cardinium, 2bRAD-M sequencing, environmental factors
Citation: Yang K, Zhang H-Y, Wang P, Jin G-X and Chu D (2024) Both symbionts and environmental factors contribute to shape the microbiota in a pest insect, Sogatella furcifera. Front. Microbiol. 14:1336345. doi: 10.3389/fmicb.2023.1336345
Received: 10 November 2023; Accepted: 26 December 2023;
Published: 24 January 2024.
Edited by:
Zhenlin Han, University of Hawaii at Manoa, United StatesReviewed by:
Vipin Rana, University of Maryland, College Park, United StatesCopyright © 2024 Yang, Zhang, Wang, Jin and Chu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong Chu, Y2hpbmFjaHVkb25nQHFhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.