
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 23 January 2024
Sec. Food Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1333696
This article is part of the Research Topic How the Application of Antimicrobial Hurdles in Meat Processing Facilities Shapes Microbial Ecology View all 6 articles
Meat production is a complex system, continually receiving animals, water, air, and workers, all of which serve as carriers of bacteria. Selective pressures involved in different meat processing stages such as antimicrobial interventions and low temperatures, may promote the accumulation of certain residential microbiota in meat cutting facilities. Bacteria including human pathogens from all these sources can contaminate meat surfaces. While significant advancements have been made in enhancing hygienic standards and pathogen control measures in meat plants, resulting in a notable reduction in STEC recalls and clinical cases, STEC still stands as a predominant contributor to foodborne illnesses associated with beef and occasionally with pork. The second-and third-generation sequencing technology has become popular in microbiota related studies and provided a better image of the microbial community in the meat processing environments. In this article, we reviewed the potential factors influencing the microbial ecology in commercial meat processing facilities and conducted a meta-analysis on the microbiota data published in the last 10 years. In addition, the mechanisms by which bacteria persist in meat production environments have been discussed with a focus on the significant human pathogen E. coli O157:H7 and generic E. coli, an indicator often used for the hygienic condition in food production.
The process of converting livestock to wholesale/retail-ready packages of meat products in commercial practice is complex and systematic. Meat processing facilities are open systems in that there is a constant intake of animals, water, air, and workers, all of which serve as carriers of bacteria (Cobo-Díaz et al., 2021). On the other hand, certain locations in the facilities may be conducive to the accumulation of residential microbiota, due to the selective pressures associated with the microenvironment that favors subgroups of the incoming microbiota. During the meat production process, bacteria carried by animals at slaughter and from the processing facility environment can be deposited on meat surfaces, a fraction of which may be pathogenic. For example, STEC does not cause overt disease in cattle but is a significant human pathogen, with some strains having an infectious dose as low as a few cells (Thorpe, 2004). STEC infections can lead to symptoms ranging from mild gastrointestinal discomfort to more serious conditions such as bloody diarrhea and, in some cases, life-threatening complications like hemolytic uremic syndrome (HUS), particularly for vulnerable populations such as young children, the elderly, and individuals with compromised immune systems (Tarr et al., 2005; CDC, 2014). In North America, 7 serogroups of E. coli, namely O157, O103, O26, O111, O121, O45, and O145, are more frequently associated with severe clinical outcomes than other serogroups and are thus referred to as Top 7 E. coli (Heiman et al., 2015). Beef is the most common vehicle in STEC transmission in North America (WHO, 2018). In recent years, there have also been reports on STEC in pork processing plants in North America (Essendoubi et al., 2020; Haque et al., 2022). To reduce microbial contamination and ensure the safety of meat, antimicrobial interventions are routinely implemented in meat processing facilities. Even though the measures are intended to target pathogenic bacteria, they inevitably have consequences on commensal microorganisms, due to the broad spectrum of their activities (Youssef et al., 2014). In addition to biocides, common operational conditions such as low temperatures and desiccation from drying equipment or carcass surfaces in meat processing facilities also play a role in shaping the microbial ecology in such environments. The wide adoption of various sequencing tools has resulted in a much higher resolution of the microbial population structure. The present work aimed to review factors shaping the microbial ecology in commercial meat processing facilities, the bacteria persisting in meat production environment with a focus on microbiome data published in the last 10 years (2013–2023), and the mechanisms applied by bacteria to persist.
The gastrointestinal (GI) tracts of mammalians harbor trillions of microorganisms. It is estimated that the bacterial populations of cattle and pigs’ intestines often exceed 1011 CFU/gram feces (Gaskins et al., 2002; Dowd et al., 2008). Not only are they high in number, but also these bacterial communities are very diverse. In a study of Mao et al. (2015) more than 542 genera belonging to 23 phyla were found in the GI tracts of cattle, with Prevotella, Treponema, Succiniclasticum, Ruminococcus, Acetitomaculum, Mogibacterium, Butyrivibrio and Acinetobacter, as well as those unclassified on genus level but derived from higher taxonomic ranks Peptostreptococcaceae, Ruminococcaceae, Enterobacteriaceae, Prevotellaceae, Clostridiales, Rikenellaceae and Bacteroidales being most abundant. Similarly, a shotgun metagenomic analysis of pig feces revealed 2,797 bacterial genera, with the most abundant ones being Clostridium, Clostridioides, Escherichia, Prevotella, Bacteroides, and Treponema (Quan et al., 2020).
Both husbandry practices and the inherent host factors of animals affect the gut microbiome (Bessegatto et al., 2017; Zhang et al., 2021; Lin et al., 2022). A change in environment especially those eliciting stressors also affects the gut microbiome. Fecal shedding of E. coli O157 often increases in summer seasons as reviewed by Kempf et al. (2022). Placing animals in a feedlot decreased the diversity in individual animals and the similarity between animals after 2 days, but these changes diminished at 7 and 14 days after the placement, suggesting the native microbiome in a cattle tends to be robust (Maslen et al., 2022). However, changes in minor or rare taxa may not always be sufficiently captured by metagenomic based studies. For example, when considering cattle exhibiting a fecal carriage of E. coli O157 at a concentration of 104 CFU/g, they are classified as “super shedders” (Stephens et al., 2009). However, even at this high shedding level, E. coli O157 remains a minute fraction within the vast landscape of the total fecal bacterial load, which typically reaches 1011 CFU/g. In addition, it is also challenging to assemble metagenomes of different strains of the same species in a diverse microbial population (Meziti et al., 2021). Khaitsa and co-workers examined the fecal shedding of naturally occurring E. coli O157:H7 in steers from the same calf cohort and fed the same diet, by culture-dependent methods (Khaitsa et al., 2003). They found the prevalence of E. coli O157:H7 varied with sampling time for animals in the same pen and varied among pens at the same sampling time, both from 0% to 100%. To capture these changes and have a holistic picture of the microbiome, some form of enrichment would be necessary.
The bacteria on a particular animal hide could originate from feces of animals sharing the same space, through cross-contamination from fecal origin but are better equipped to survive in secondary environments other than the host, and from the rearing environments such as soil, bedding, water etc (Narvaez-Bravo et al., 2013). The fecal microbiota are primarily mesophilic microorganisms, and can be very different from those that inhabits the animal rearing environments (Zaheer et al., 2019). In contrast with the commonly found 1011 CFU/gram feces bacterial load in cattle intestines, the number of bacteria on hides varies largely, from 104 to 1010 CFU/cm2 in various studies from several countries and with different ratios of Enterobacteriaceae and coliforms to the total bacterial population, likely reflecting the differences in climate and animal husbandry practices (Yang, 2017). A systematic review and meta-analysis of the prevalence and concentration of E. coli O157 in cattle in North America showed an overall fecal prevalence of 10.7% and hide prevalence at processing plants of 56.4% (Ekong et al., 2015). The percentage of cattle carrying high concentration of E. coli O157:H7 on hides was much higher at processing plants (23.81%) than at feedlots (1.74%) for fed beef, likely resulting from much closer contact between animals and stress during transportation. Even so, different and diverse genotypes as determined by pulse field gel electrophoresis were observed for E. coli O157 on hides of cattle processed on the same day and on different days at the same processing facility (Arthur et al., 2014), suggesting the microbial diversity on hide of incoming animals is also reflected by within species diversity.
The process of converting cattle to meat products in commercial slaughter plants can involve more than 50 different operations (Supplementary Table S1), which can be divided into four major stages: slaughtering of animals, carcass dressing, carcass chilling and carcass breaking (Yang, 2017). Most packing plants perform the operations in that order, however, carcass breaking can also happen before carcass is completely chilled, through a hot boning process (Keenan et al., 2016). Food safety programs such as standard sanitation procedures, good manufacturing practices and hazard analysis critical control point (HACCP) systems are implemented especially in state-inspected processing facilities by which microbiological effects of operations are analyzed and control measures implemented to minimize such effects (Gill, 2005). The microbiological impact of slaughtering (stunning and bleeding) on meat can be negligible as contamination is localized around the wound. Muscle tissues of healthy animals are literally free of bacteria; however, microorganisms will be introduced to the previously sterile meat surface upon exposure. For instance, the initial cuts of the skinning process where knives work from outside to inside of the skin and bioaerosols generated by flapping of the hide during the dehiding operations inevitably deposit bacteria on carcass surfaces. Leakage of gut contents could also serve as sources of contamination. However, with improvement in dressing operations in modern meat processing facilities, gut leakage has become a rare event during the removal of viscera (Blagojevic et al., 2012). Most bacteria contaminating skinned carcass surfaces came from the hide of animals (Arthur et al., 2004). Pork dressing can differ from that for cattle in that it includes scalding, dehairing, singeing and polishing (black scraping) of carcasses before evisceration and the dressing process does not always involve hide/skin removal (Nastasijevic et al., 2020).
Temperature plays a crucial role in microbial growth and can significantly impact the rate at which microorganisms proliferate. Temperature can affect enzymatic activity and membrane fluidity leading to reduced cellular activity. The minimum growth temperature for E. coli and associated mesophilic organisms is around 8°C (Shaw et al., 1971; Smith, 1985). Regulatory agencies in some countries require the surface temperature of carcasses (beef and pork) be reduced to 7°C or below within 24 h of carcass dressing, and an internal temperature (the warmest part) of 7°C be reached before further processing (CFIA, 2010). In commercial practice, carcasses are chilled either by spray chilling where carcasses are intermittently sprayed with water, or by dry chilling where they are exposed to a flow of refrigerated air (Savell et al., 2005). Air chilling with appropriate parameters can be an effective antimicrobial intervention step, resulting in >2 log reductions of the carcass bacterial population, likely by the desiccating effect from the rapid evaporation of water from warm carcass surfaces in a refrigerated environment (Greig et al., 2012; Liu et al., 2016; Zdolec et al., 2022). In contrast, no consistent antimicrobial effects have been reported for spray chilling, with increase, decrease and no change in bacterial numbers on carcass surfaces have been reported (Yang, 2017). Psychrobacter and Pseudomonas, respectively, were the dominant taxa among the bacterial population on spray chilled and dry chilled carcasses, likely resulting from differences in carcass surface water activity by the two chilling methods (Yang et al., 2017b).
Carcass breaking is the process by which carcass sides are made into cuts with predetermined specifications, and trimmed off pieces (trimmings) are collected into large containers. At large facilities, the process can be very complex, involving the progressive removal of portions from the hanging side and each portion being passed to a separate collection or a processing line. In this process, multiple tools such as saws, knives, cutting boards and conveyor belts are used and workers often wear gloves (cotton, mesh and or plastic), aprons and other protective gears. The air temperature at fabrication facility is maintained at below 10°C to minimize growth of mesophilic bacteria (CFIA, 2010).
In spite of the potential sources of bacterial contamination described above, antimicrobial interventions implemented in commercial plants can result in significant reductions of the bacterial populations and may also affect the composition of the microbial populations. The commonly used antimicrobial interventions in beef processing facilities during dressing process include hide-on carcass wash with NaOH (Yang et al., 2015b), spraying carcasses (pre-evisceration) and carcass sides (post-evisceration) with solutions of lactic acid, acetic acid, peroxyacetic acid or organic acid mixtures and pasteurization of carcass sides with hot water or steam (Algino et al., 2007; Gill, 2009; Yang et al., 2012; Scott et al., 2015). Spraying carcasses with ozone has also been reported (Casas et al., 2021). The application of spray/wash is carried out either in large spray cabinets through which carcass sides sequentially pass or using small handheld sprayers, depending on the line speed, space availability and cost. Even for the same antimicrobial intervention type, different application parameters such as concentrations of chemicals, the way the spray is applied, water temperature as well the topographical/physiochemical properties of the treatment surface may vary from study to study and consequently their antimicrobial efficacies. A meta-analysis of popular interventions used in cattle processing plants to reduce E. coli contamination revealed that the initial microbial concentrations and timing of extra water washes were the most important predictors of intervention effectiveness (Zhilyaev et al., 2017). The mean reduction of E. coli on hides by water, acetic acid, lactic acid, and sodium hydroxide wash were 0.08, 2.21, 3.02, 3.66 log units, while for carcasses, the mean reductions by water, acetic acid and lactic acid were 1.90, 1.44, and 2.07 log units. In addition, applications toward targeted areas such as trimming, vacuum cleaning with or without spraying with hot water, spraying the initial opening cut lines with lactic acid are also in use. Their effects on the microbiological conditions of carcasses would be localized (Gill, 2009). Intermittent application of low concentrations of oxidizing agents such as chlorine and peroxyacetic acid during spray chilling showed up to 4-log reductions of both Salmonella and E. coli in laboratory settings (Kocharunchitt et al., 2020), likely resulting from the synergistic effect of low temperature and oxidative stress on these enteric pathogens (King et al., 2016). In some facilities, cuts and trimmings are misted with 200 ppm peroxyacetic acid over the conveyor belt through the spray cabinet (Yang et al., 2012, 2021). This treatment is ineffective for reducing the number of bacteria on meat surface but may affect the survival and growth of subpopulations during extended storage under chilled and vacuum packaged conditions. In addition to antimicrobial interventions, bacteria on pork carcasses during dressing can also be significantly reduced by scalding and singeing but not by polishing or dehairing, all of which are unique to pork carcass dressing (Zdolec et al., 2022).
It is commonly believed that bacteria on meat products are primarily from chilled carcasses and contribution from other sources, if at all, would be negligible. If so, the number of bacteria on meat products would be lower than that on chilled carcass surface, with the area extension of newly cut surfaces during the fabrication process. A study of Youssef et al. shows that the numbers of aerobes and E. coli on cuts and trimmings are >10 times the number for chilled carcasses and the number of coliforms on products are >100 times the number for chilled carcasses (Youssef et al., 2013). Genotyping of E. coli on beef products, incoming chilled carcasses and surfaces of various equipment involved with fabrication has found the majority of E. coli on products share the same genotype with E. coli on equipment surfaces, rather than chilled carcasses (Yang et al., 2015a, 2017a). The same genotypes of STEC O157:H7 across “high-event periods” over time and of generic E. coli on beef trimmings produced on different days at the same processing facility have been reported (Arthur et al., 2014; Visvalingam et al., 2016). In addition, some E. coli strains persisted, evidenced by repeated recovery of the same genotype on multiple sampling times from equipment at the same facility (Yang et al., 2017c). Even though processing facilities and equipment involved with meat fabrication are cleaned and sanitized daily, the goal of sanitation is to achieve visibly clean equipment rather than sterility of equipment, and standards on maximum number of bacteria on post sanitization are often lacking.
As we have discussed above, the meat processing environment, e.g., fabrication equipment surfaces, is an important source of bacteria contaminating meat. We performed a meta-analysis on the potential persistent microbiota in meat production environment. Literature search was conducted in Google Scholar and Scopus with “microbiota”/“microbiome”/“metagenomics”/“sequencing” and “meat processing/cutting plant” as keywords. The literature which was published between 2013 and 2023 and investigated the microbiota in meat production environment using high-throughput sequencing technology was used in the analysis in the present study.
A total of 22 relevant references were found (Supplementary Table S2). In these studies, metagenome DNA, 16S rRNA gene DNA or RNA using long- or short-read sequencing technology were sequenced, mainly identifying bacteria at the genus level. We were able to retrieve sequencing data from 10 of these studies. The raw sequencing reads for relevant samples were downloaded from NCBI1 using Faster-dump of SRA toolkit v3.0.3.2
For the 16S rRNA gene amplicon of a hypervariable region sequenced under Illumina platform, the primers in sequencing reads were removed using Cutadapt v4.4 (Martin, 2011). For the sequencing of full-length 16S rRNA gene amplicon using PacBio platform, the primers were removed using removePrimers function of Dada2 v1.26.0 in R v4.2.3 (Callahan et al., 2016). Dada2 workflow was used to process trimmed reads. Both forward and reverse reads were, respectively, truncated to 220 and 220 bases for Illumina sequencing. The reads < 1,000 bp for PacBio sequencing were removed. Taxa of sequencing reads were assigned using SILVA 16S rRNA gene database (v138.1; Quast et al., 2013).
For metagenomics sequencing data, Fastp v0.23.2 was used to remove adapters and bases with mean quality score < 15 at the beginning and end of the reads (Chen et al., 2018). Bowtie2 v2.5.1 was used to remove Phix contamination and animal DNA contamination with Phix (accession in NCBI, NC_001422.1) and cattle (GCA_002263795.2) or pig (GCA_000003025.6) genomes as references (Langmead and Salzberg, 2012). Taxonomy assignment for sequencing reads was performed using Kraken2 v1.1.1 and improved using Bracken v2.8 (Lu et al., 2017; Wood et al., 2019). The output was converted to Biom format using Kraken-biom v1.2.03 and imported into R using package Phyloseq v1.30.0 (McMurdie and Holmes, 2013).
The relative abundance of taxa in each sample was summarized and plotted using Phyloseq and ggplot2 v3.4.2 (Wickham, 2016). The alpha and beta diversity were analyzed using both Phyloseq and Vegan v2.6.4.4
We used the sequencing data for samples collected from various meat plant environmental surfaces after cleaning and/or sanitation to identify persistent bacteria. A total of 447 samples collected from environmental surfaces such as conveyor belt, chopping board, equipment surfaces and other surfaces at eight meat processing plants were included (Kang et al., 2019, 2020; Botta et al., 2020; Cobo-Díaz et al., 2021; Cherifi et al., 2022; Yang et al., 2023b; Table 1; Supplementary Table S3). These plants were, respectively, located in geographical regions including Canada (Cherifi et al., 2022; Yang et al., 2023b), Italy (Botta et al., 2020), Spain (Cobo-Díaz et al., 2021), and Australia (Kang et al., 2019, 2020). One functional area/room was investigated in most of these plants except for the plant in Spain (Cobo-Díaz et al., 2021), in which four rooms including pork carcass chilling room, pork cutting room, pork packing or storage room, and Trotter’s washing room were studied. For the convenience of data presentation, we assigned labels for the rooms based on their function and origin. There were 9 rooms in total (RA01-RA09; Table 1).
A total of 17 genera each accounted for >5% of the microbiota in at least one of nine functional rooms (Figure 1A). On average, Pseudomonas (19.5%), Acinetobacter (10%), Psychrobacter (9.3%), Sphingomonas (3.3%), Enterococcus (2.8%), Proteus (2.8%), Staphylococcus (1.9%), Burkholderia-Caballeronia-Paraburkholderia (1.8%), Acidovorax (1.6%), and Brevundimonas (1.5%) were the top 10 most predominant genera in all functional rooms included (Supplementary Table S4). Rooms RA01-04 were at a pork cutting plant in Spain (Cobo-Díaz et al., 2021). No significant (p > 0.05) difference in either alpha (genus richness) or beta diversity (genus composition) was observed for these rooms (Figures 1B,D). However, variations in both alpha diversity and beta diversity were observed among the rooms from different plants (Figures 1B,D). The beef slaughter room (RA08) in an Australian plant and the pork cutting room (RA09) in a Canadian plant showed relative larger alpha diversity compared to other rooms (Figure 1B). In terms of beta diversity, all the rooms from different plants showed significant difference. Sequencing methods seem to affect the beta diversity, which may have contributed to the further distance between rooms RA01-04/RA05 and others (Figures 1D,E). RA01-04, RA05, and RA06-09 were investigated by sequencing metagenome DNA, amplicon of V3 region of 16S rRNA and amplicon of V4 region of 16S rRNA gene DNA in collected samples, respectively. Sequencing of cDNA of 16S rRNA focuses on the live bacteria in a sample, while direct DNA sequencing was not able to distinguish between live and dead bacterial cells. This also supports the observation in this review that RNA based method showed lower alpha diversity compared to DNA based methods (Figure 1C).
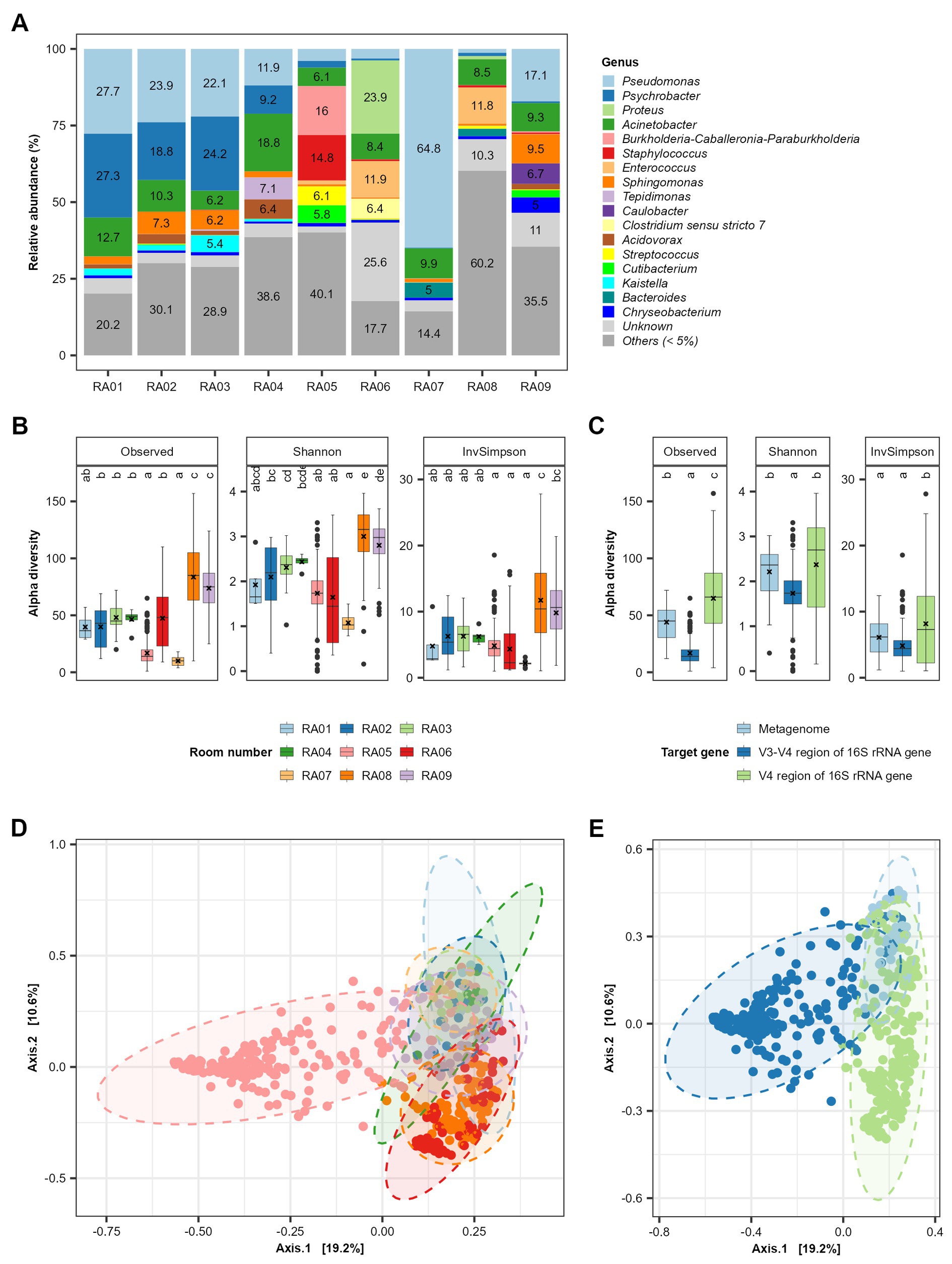
Figure 1. Alpha and beta diversity of persistent bacterial genera in meat processing plants. (A) The relative abundance of bacterial genera in the combined samples in each functional room. The room function is shown in Table 1. Panels (B,C) show the comparison of alpha diversity among different functional rooms and sequencing methods, respectively. The sequencing methods includes sequencing of metagenome DNA, V4 region of 16S rRNA gene DNA and V3-V4 region of 16S rRNA. Panels (D,E) show the PCoA plot distinguished with room numbers and sequencing methods, respectively.
To investigate whether a bacterial genus has significant correlation with a specific plant/functional room, we performed correlation analysis using a R package MaAsLin2 (Mallick et al., 2021). The rooms (RA06-RA09) for which the same sequencing method was used, i.e., sequencing of V4 region of 16S rRNA gene, were included for comparison. RA06 (beef cutting room in an Australian meat plant) was used as a reference. A total of 127 bacterial genera each showed a significant (Benjamini-Hochberg corrected p-value < 0.001) positive or negative correlation with at least one functional room/plant (Figure 2). Clostridium sensu stricto 7, Klebsiella, Proteus were more abundant in RA06, or negatively associated with RA07-09 with correlation coefficient (effect estimate by MaAsLin2) ≤ −3.3, ≤ −2.3, and ≤−4.8, respectively (Figure 2). RA07 (beef cutting room) and RA09 (pork cutting room) were both located in Canada, the microbiota of which were more predominated with Pseudomonas, Janthinobacterium, and Flavobacterium but less predominated with Enterococcus, compared to other functional rooms. Bacteroides and UCG-005 were only positively associated with the beef slaughter room (RA08) in the Australia plant with coefficients >3. In addition, Caulobacter, Sphingomonas, Cutibacerium, Mycobacterium, Methylobacterium-Methylorubrum, Acidovorax, and Afipia were only positively correlated with RA09 with coefficients >3, the pork cutting room in a Canadian plant. The correlation between a specific genus with a meat plant found in this review may be attributable to several factors including different production practices, geographical regions and meat processing stages/functions of/in the included meat plants. Our previous study has shown meat products produced at different meat plants are predominated with different strains of Carnobacterium, a bacterial genus associated with the storage life of vacuum-packaged meat (Zhang et al., 2018). Whole genome sequencing data showed the meat production environment was the likely source of the contaminating Carnobacterium strains (Zhang et al., 2018). A better understanding of the persistent bacterial genera/species/strain in a meat plant will likely help to better control/predict the shelf life of produced meat products.
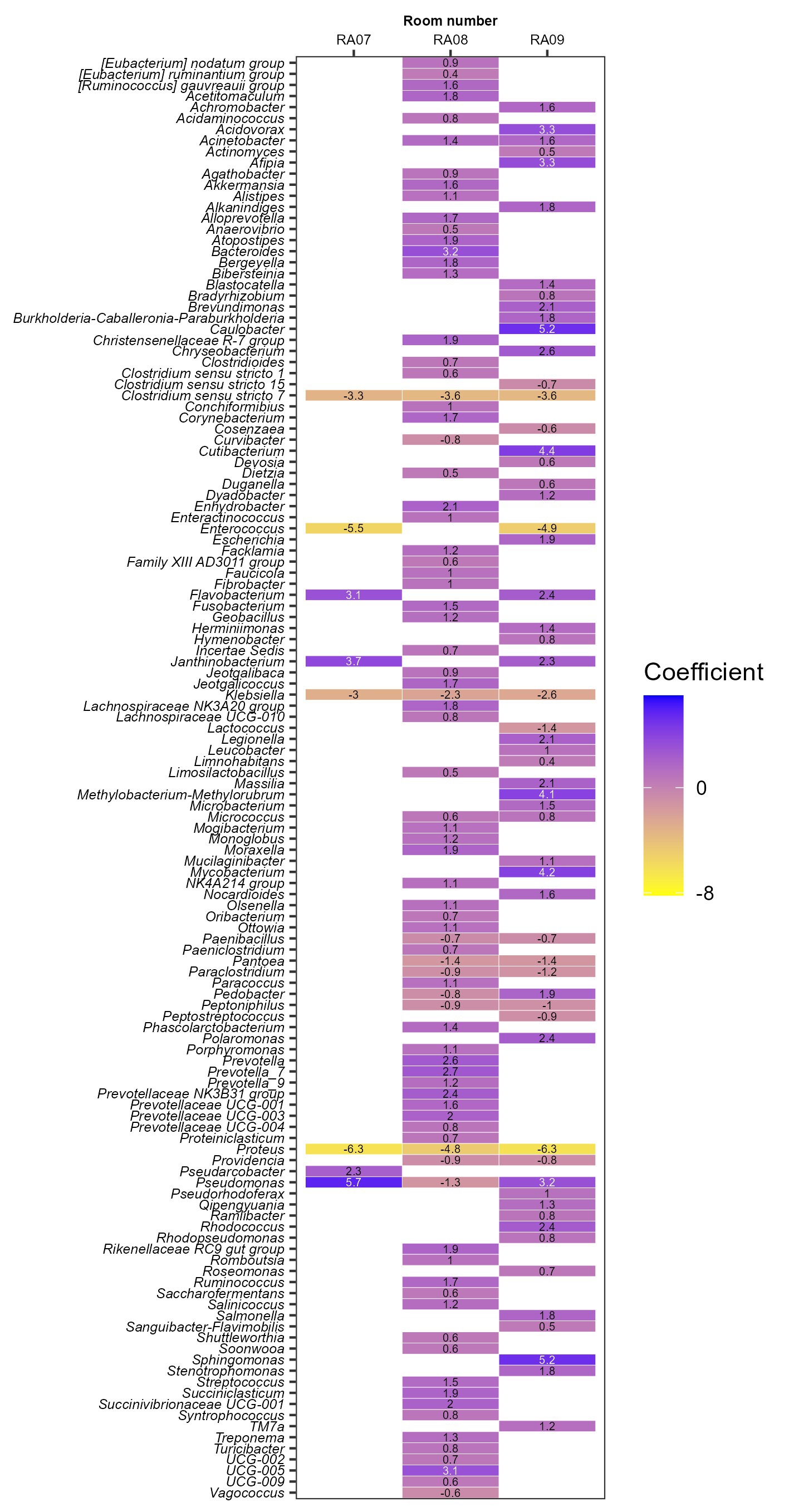
Figure 2. The association of persistent bacterial genera with each functional room investigated by sequencing of V4 region of 16S rRNA gene DNA. Room RA06 was used as a reference.
We investigated the correlation between the relative abundance of bacterial genera, to explore the potential bacterial co-existence network in meat plant environments. A positive correlation indicates the possible co-existence between two bacterial genera, which may be attributed to their interaction in a micro-environment/biofilm, the preference for the same growth condition, or the same contamination pattern. For this purpose, we only included the functional rooms with ≥10 samples, i.e., RA02-03, RA05-06, and RA08-09. We first rarefied the sequencing data to 1,000 reads for each sample and then calculated the relative abundance of bacterial genera. Therefore, the co-existence network analysis in this review only focused on the bacterial genera with a relative abundance ≥0.1% in each functional room. Theoretically, the bacterial genus which accounts for <0.1% of the microbiota will get a zero as its relative abundance after data rarefaction. For statistical analysis, 0.01% was arbitrarily assigned to those zeros. A log transformation with 10 as the base was then performed for the percentage values. Therefore, the log values for relative abundance of <0.1, 0.1, 1, 10, and 100% were − 2, −1, 0, 1, and 2, respectively. Spearman’s rank correlation was performed for each combination of two genera for each functional room. The correlation with a p value <0.001 and coefficient > 0.5 or <−0.5 was regarded as significant. We looked at the combinations with significant correlation found in more than one functional room.
Three and 42 combinations of bacterial genera were found to have significant correlation in three and two functional rooms, respectively (Figure 3A; Supplementary Figure S1). No significant correlations were found in >3 functional rooms. All the correlations were found positive except for Escherichia and Psychrobacter which were negatively correlated with a coefficient of −0.58 in RA09 (the pork cutting room in a Canadian plant; Supplementary Figure S1). However, the two genera had positive correlation in RA06 (a beef cutting room in an Australian plant). The significant correlations found in three functional rooms were between Acinetobacter and Psychrobacter, Chryseobacterium and Flavobacterium, and Sphingomonas and Methylobacterium-Methylorubrum (Supplementary Figure S1). A network based on the correlations was constructed (Figure 3A), which showed that Acinetobacter correlated with the greatest number of genera (n = 5) followed by Psychrobacter (n = 4), Chryseobacterium (n = 4), Pedobacter (n = 3), and Prevotella (n = 3).
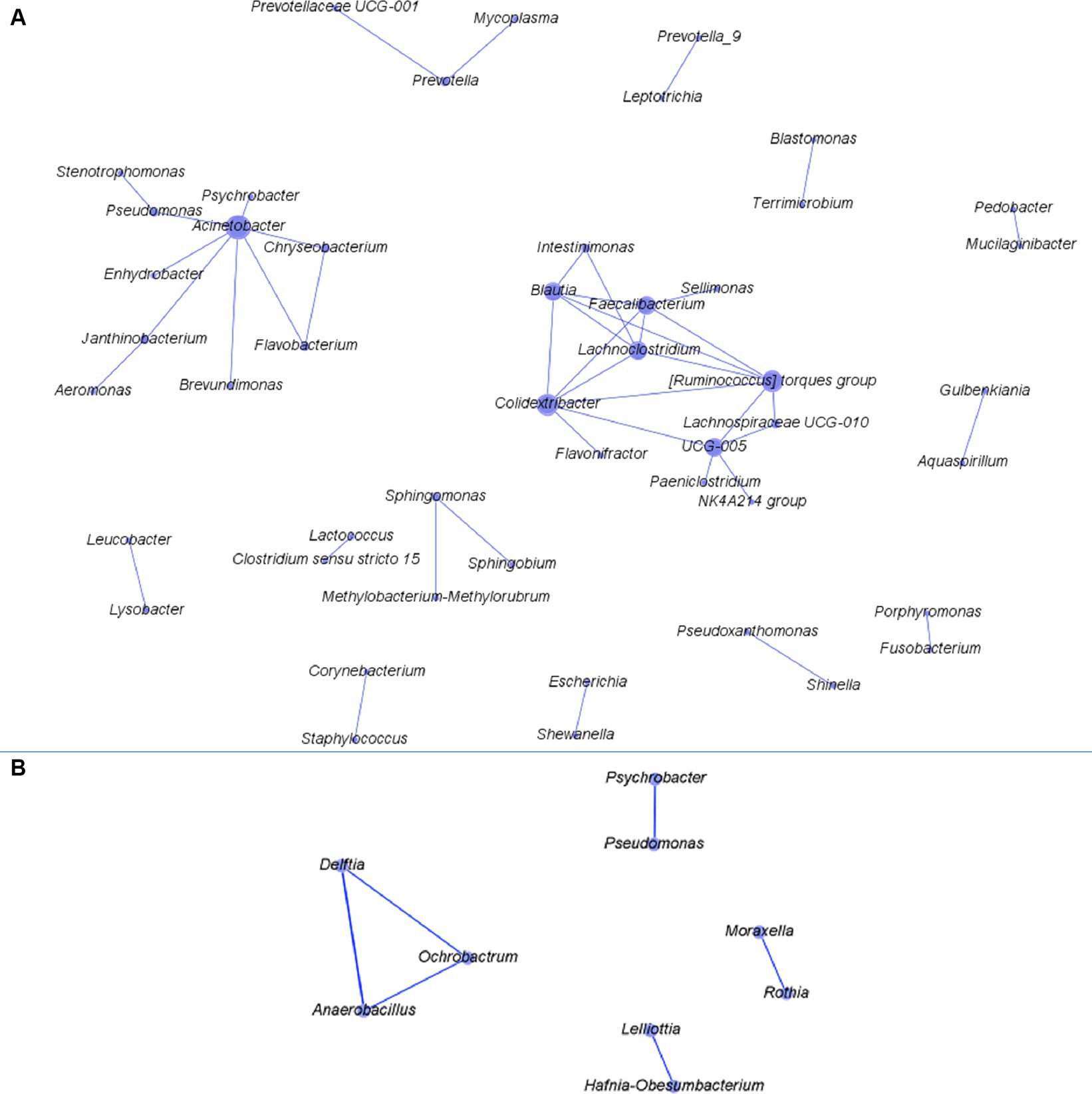
Figure 3. The network constructed based on the correlations between two bacterial genera found in more than two of the functional rooms with >10 samples. Panels (A,B) are networks constructed for bacteria from after-cleaning and before-cleaning samples.
The meat plant samples in some studies included those collected before the routine cleaning or during meat production. We also analyzed the bacteria in these samples (n = 439), which were from conveyor belts, equipment surfaces, knives, aprons, gloves, etc. (Supplementary Table S3) in plants in Austria, USA, Italy, and Canada (Yang et al., 2016; Botta et al., 2020; Zwirzitz et al., 2020, 2021; Shedleur-Bourguignon et al., 2023; Supplementary Table S2). Like after-cleaning samples, three bacterial genera including Pseudomonas (9.7%), Psychrobacter (9.6%), and Acinetobacter (6.2%) also had average relative abundance of >5% in before-cleaning samples (Supplementary Table S4). Delftia was not predominant (≤0.6%) in any of functional rooms after cleaning but had relative abundance > 10% in two functional rooms before cleaning, including the pig slaughter room (RB01, 16.7%) and pork cutting room (RB02, 15.7%) in a meat plant in Austria (Supplementary Table S4). As the before- and after-cleaning samples were sequenced using different methods and were from different functional rooms in different geographical regions, we did not perform statistical comparison.
In four functional rooms with ≥10 samples, we found one (Anaerobacillus vs. Delftia) and five (Rothia vs. Moraxella; Lelliottia vs. Hafnia-Obesumbacterium; Anaerobacillus vs. Ochrobactrum; Delftia vs. Ochrobactrum; Pseudomonas vs. Psychrobacter) combinations of bacterial genera that were positively correlated in three and two functional rooms, respectively (Supplementary Figure S2). Among the six combinations, Delftia, Anaerobacillus, and Ochrobactrum all had significant correlation with two genera and others with one genus (Figure 3B). Studies on the co-existence network among meat plant microbiota are very scarce. Botta et al. recently explored the co-occurrence network for bacteria both on beef carcass and in slaughterhouse environments (Botta et al., 2022, 2023). The mechanisms mediating the pairwise co-existence/occurrence in both studies and those in our analysis are not clear. However, further relevant studies may help us better understand the micro-ecology of bacteria in meat production chain. Considering the co-existence network constructed for both before- and after-cleaning samples together, the bacterial genera with more connections in the network tended to have larger relative abundance than others. This finding suggests the co-existence relationship between predominant bacteria is easier to capture than their scarce counterparts. The contamination level of pathogens in meat processing plants is normally very low. Nevertheless, the information on its synergistic or antagonistic interaction with commensal bacteria in meat plant would be meaningful to food safety. For this purpose, a culture-dependent method is necessary.
In meat processing environments, the survival of bacteria could be mediated by one or more of the following ways: inherent/increased resistance to biocides, shielded by surfaces (e.g., hard to reach spots) or meat debris, and protected by biofilms. Generic E. coli has long been used as an indicator to evaluate the hygienic condition in meat processing plant, the sensitivity of which to biocides have been reported by various researchers. Aarestrup and Hasman (2004) compared the susceptibility of 202 E. coli isolates from Danish cattle, broilers, and pigs to a number of biocides including quaternary compounds (QAC), the most commonly used sanitizer in food settings. Most E. coli had minimum inhibitory concentration (MIC) values of 64 ppm and the highest MIC observed for E. coli was 128 ppm. The mean MIC values for QAC and sodium hypochlorite of both persisting (n = 50, genotype recovered more than twice) and transient (n = 50, genotype recovered once) E. coli strains collected from fabrication equipment at a beef packing plant did not differ significantly and were well below the in-use concentration for both sanitizers (QAC, 200 ppm; sodium hypochlorite, 200 ppm active chlorine; Yang et al., 2018). Lavilla Lerma et al. (2015) tested the sensitivity of pseudomonads isolated from a goat and lamb slaughterhouse to biocides including triclosan, cetrimide, benzalkonium chloride. They found the included Pseudomonas isolates were all highly susceptible to industry formulations of these biocides. There is very limited information on the resistance of other bacterial species to the sanitizers often used in meat plants. However, the available studies suggest the likelihood of biocide resistance of planktonic cells of meat plant bacteria being the main mechanism of persistence in meat processing environment would be low.
The inability to remove biofilms poses a risk for ongoing microbial contamination in meat processing facilities. Biofilms are complex structures with bacterial cells embedded in extracellular polymeric substances (EPS) also known as the glycocalyx, and are widely acknowledged as the dominant mode of microorganism existence (Watnick and Kolter, 2000; White et al., 2011). The EPS is a mixture of polysaccharides, proteins, lipids, and highly hydrated nucleic acids that acts as a shield and source of water, protecting the microorganisms within it against desiccation, biocides, and other environmental stressors (Coughlan et al., 2016; Yin et al., 2019). EPS also aids in the retention and concentration of essential nutrients (White et al., 2011). Sessile cells exhibit altered physiological characteristics compared to their planktonic counterparts, including changes in gene expression, metabolism, and resistance to antimicrobial agents (Costerton et al., 1999). Thus, the biofilm mode of living provides a better fitness to bacterial survival through the physical barrier provided by EPS, efflux systems, differentiation of bacterial cells into a dormant state, and the modification of the micro-environment. This can render a particular sanitizer less effective (Giaouris et al., 2014), which is certainly relevant in meat processing environments. The persistent E. coli population in the study by Yang et al. (2018) had a large fraction of biofilm formers when tested at 15°C for 6 days. Wang et al. (2016) also reported strong biofilm forming ability and higher tolerance to sanitizers in biofilms of “high-event period” E. coli O157:H7 strains.
The attachment and subsequent biofilm formation of bacteria depends on interactions between bacteria and the environment (Garrett et al., 2008). During meat processing operations, the wetting of processing surfaces such as conveyor belts by meat juice and the adsorption of food residues to surfaces provide a conditioning layer which modifies surface properties favorably for bacterial attachment and subsequent growth (Yin et al., 2019; Carrascosa et al., 2021). In addition, biofilms formed initially under high humidity conditions can dehydrate, resulting in prolonged bacterial survival (Adator et al., 2018; Nan et al., 2022).
Numerous studies have investigated the biofilm forming ability of E. coli. A larger proportion of generic E. coli tend to be biofilm formers compared to pathogenic E. coli. Uhlich et al. (2014) examined the biofilm formation of clinical E. coli O157 strains on polystyrene surface under various conditions and found that most (49/54) strains did not form any measurable biofilms. Of the five biofilm formers, only one was considered to be strong. Similarly, Wang et al. (2012) reported that two out of 30 STEC strains from various sources were biofilm formers when cultured on polystyrene surface for 24 h. Generic E. coli strains (n = 700) collected from carcasses along the dressing process and during chilling, from meat products and fabrication equipment surfaces were compared with Top 7 E. coli strains (n = 745) recovered from cattle in their biofilm formation under equivalent conditions (Stanford et al., 2021). Biofilm formers accounted for 7.1% of the total Top 7 strains and 42.9% of the total generic E. coli population. Comparative genomic analysis of Top 7 E. coli strains revealed that more virulence factors were associated with the non-biofilm forming population and acid resistant population, while they were least present in populations where heat resistance genes and metal resistance genes were enriched (Fang et al., 2023). These findings suggest that there is a divergence between environmental fitness and virulence of E. coli. A number of factors could have driven this divergence: 1. The much lower temperature in the meat fabrication environment (mostly ≤ 10°C during operation and ≤ 15°C during downtime) compared to the host’s intestines (35°C); 2. The potential encounter of physical (desiccation from equipment drying) and chemical (sanitizers/cleaners) bactericides.
Much less attention has been drawn to spoilage or resident microbiota in meat processing plants. Among the most predominant persistent bacterial genera revealed by our meta-analysis on meat plant microbiota, Pseudomonas strains have been proven to be biofilm-formers under chilled conditions simulating meat production environment (Morimatsu et al., 2012; Liu et al., 2013; Visvalingam et al., 2019; Wickramasinghe et al., 2020; Wagner et al., 2021). Acinetobacter can be a strong or weak biofilm-former depending on the species/strain identity of the isolates (Liu et al., 2013; Visvalingam et al., 2019). Yang et al. (2023b) reported the predominance of Pseudomonas and Acinetobacter isolates in biofilms formed by meat plant microbiota on stainless steel coupons at 15°C. Both species also predominated in the multi-species biofilms which formed on conveyor belts under conditions simulating meat processing environments and retained its stability even when conveyor belts were rinsed by QAC, peracetic acid or H2O (Fagerlund et al., 2017). Psychrobacter does not seem to be a strong biofilm-former although it often predominates in meat packing plants. A Psychrobacter strain did not form measurable biofilms quantified using crystal violet staining (Visvalingam et al., 2019). Wagner et al. (2021) reported the lowest bacterial load of the tested Psychrobacter strains on stainless steel slides compared to other meat plant microbiota (e.g., Pseudomonas fragi, Acinetobacter harbinensis, Microbacterium sp., Carnobacterium maltaromaticum, etc.). Factors such as the initial load from animal intestines and their ability to grow at low temperatures, rather than their ability to form biofilms, may have contributed to the predominance and persistence of Psychrobacter in meat processing environments. Sphingomonas is a group of strictly aerobic Gram-negative bacteria and ranked among the top four most predominant genus in our meta-analysis. It was a moderate biofilm former in the study by Liu et al. (2013) and did not form measurable biofilms in the study by Visvalingam et al. (2019), respectively. Castaño-Arriba et al. (2020) tested 200 Enterococcus spp. recovered from red meat and poultry products, and found they all produced weak, moderate or strong biofilms on polystyrene microwell plates depending on the isolates. Very little information has been found for other bacterial species reported in meat plant microbiota.
The diverse species of bacteria found on/in processing equipment/environments may affect biofilm formation by E. coli (Møretrø et al., 2013; Wang et al., 2018; Fagerlund et al., 2021). When co-cultured with bacteria recovered from processing environments in dual-species cultures, STEC strains showed interactions in a STEC-strain and companion-strain dependent manner, with both synergistic and antagonistic effects being observed (Fang et al., 2022; Nan et al., 2022). For example, biofilms formed by Pseudomonas aeruginosa were found to be antagonistic against STEC O103 strains (Nan et al., 2022). The Gram-positive bacterium Microbacterium phyllosphaerae was synergistic for biofilm formation with various STEC strains in dual-species cultures (Fang et al., 2022). As such, the presence of background bacteria must be considered when evaluating the biofilm formation of E. coli. A recent study reported that a STEC O157:H7 strain was able to co-develop biofilms with post-sanitation process equipment surface microbiota and insert into biofilms developed by such microbial communities, but could not form biofilms on its own when evaluated under the same incubation conditions (Yang et al., 2023b). This particular O157:H7 strain also lacks the ability to produce curli or cellulose at the temperature (15°C) used for biofilm formation. A similar phenomenon was observed for generic E. coli strain, PHL565, which was not able to adhere to a glass surface on its own, but it could do so when co-cultured with P. putida MT2 (Castonguay et al., 2006). The survival of STEC strains could be enhanced by background microbiota through multispecies biofilms in meat plants. The importance of effective cleaning and sanitation of fabrication environments cannot be overstated. Targeting the commensal background microbiota in meat processing environment rather than individual pathogen strains based on their biofilm forming ability may be a more rewarding approach, from a pathogen control standpoint. In addition, studies on commensal bacteria in meat plants have been mainly on genus level and the species identity of recovered isolates is largely unknown let alone the strain level identification. Phenotypic characterization of these background bacteria combining genomic analysis will help with the development of effective biofilm control/removal measures.
The development of biofilms can be divided into five discrete steps: initial reversible attachment of planktonic cells to a surface; irreversible attachment; microcolony growth; maturation (macrocolony); and dissolution (dispersal) which releases bacterial cells back into planktonic state and a new cycle may start (Van Houdt and Michiels, 2005). Adhesins and fimbriae are involved in the attachment of bacteria to the surfaces, allowing bacterial cells to form microcolonies (White et al., 2011). For E. coli, the fimbrial adhesin curli has been reported to be essential for biofilm formation (Uhlich et al., 2013). In addition to adhesins, cellulose, flagella, poly-β-1,6-N-acetyl-D-glucosamine (PGA), and colanic acid could also be involved in E. coli biofilm formation at different stages (Beloin et al., 2008). Consequently, biofilm formation is an orchestrated process of the work of many genes including those encoding for attachment apparatus, EPS, and their respective regulatory genes, as well as global regulatory genes for quorum sensing and the stationary phase sigma factor. Secretion systems have also been reported to play a crucial role in biofilm formation by facilitating the transport of various molecules, including proteins and polysaccharides, across the bacterial cell envelope and into the extracellular matrix (O’Toole and Kolter, 1998; Costa et al., 2015; Römling and Galperin, 2015).
Escherichia coli O157:H7 is the top ranked pathogenic STEC serotype associated with human outbreaks. To date, there has been no published literature inquiring into genomic features related to biofilm formation of E. coli O157:H7 on a population perspective. We hence scrutinized the presence of biofilm formation related genes in a number of O157:H7 strains for which both the genomes and origin information were available. A total of 98 genomes of O157:H7 strains were included, which were derived from cattle (n = 25), clinical samples/humans (n = 43), package lettuce (n = 1) beef products (n = 11). Eighteen strains of unknown origin were also included. Those strains were provided by the Canadian Food Inspection Agency (CFIA), the Food and Drug Administration (FDA) or the United State Department of Agriculture (USDA), and thus the likely origin would be food or animals. These strains originated from USA, UK, Japan, China, Mexico, Argentina, Canada, Germany, Denmark, and Brazil. We also included two non-STEC E. coli strains as references, E. coli K12 substr. MG1655 and ATCC 11775 (O1:K1:H7). Strain MG1655 is non-pathogenic and closely resembles wild-type E. coli, while ATCC 11775 was isolated from a patient with urinary infection and positive for cellulose and fimbria production. We utilized a comparative systems approach to analyze protein families across selected genomes. The genome annotation was carried out using RASTtk4, focusing on intra-genus comparisons (PLfams; Overbeek et al., 2005, 2014).
A total of 150 biofilm formation related genes were examined in these genomes, as listed in Figure 4. Associated proteins were grouped together in the figure, such as FlhB, FlhC, FlhD, and FlhD. Interestingly, the majority of those genes were found in all the genomes regardless of their isolation source. These included genes encoding proteins associated with quorum sensing (TqsA, LuxS, and TnaA), dedicated to biofilm regulation (BssS, BssR, and TabA), and related to biosynthesis and regulation of EPS matrix (PgaABCD, BcsEFGQ), curli (CsgABCD), and colanic acid (WcaABCDFIKLM, RcsA; Barnhart and Chapman, 2006; Domka et al., 2006; Itoh et al., 2008; Kim et al., 2009; Zhang and Poh, 2018). The genes encoding bacterial appendages, such as type-1 fimbriae (FimBEFGH) and type-4 fimbriae (pili, PilABCMNOPQT), and flagella (FlgA-N, FlgJ, Flk, FlhA) were found in all the genomes as well (O’Toole et al., 2000; Paranjpye and Strom, 2005; Charbonneau and Mourez, 2007; Kim et al., 2009; Belas, 2014). It has been hypothesized that 95% of O157:H7 isolates are non-biofilm-formers due to an insertion in the gene encoding a transcription factor (MlrA) and variation in a gene encoding a sigma regulator (RpoS), which limit curli expression and biofilm formation in O157:H7 (Uhlich et al., 2013). However, other research has shown that STEC that does not produce curli or cellulose phenotypically are able to form biofilms (Adator et al., 2018; Nan et al., 2022). The biofilm forming ability of the O157:H7 strains in this review needs further study. Nevertheless, the presence of these biofilm related genes in all O157:H7 strains underscores their fundamental importance in the ecological success of a diverse group of O157:H7 strains.
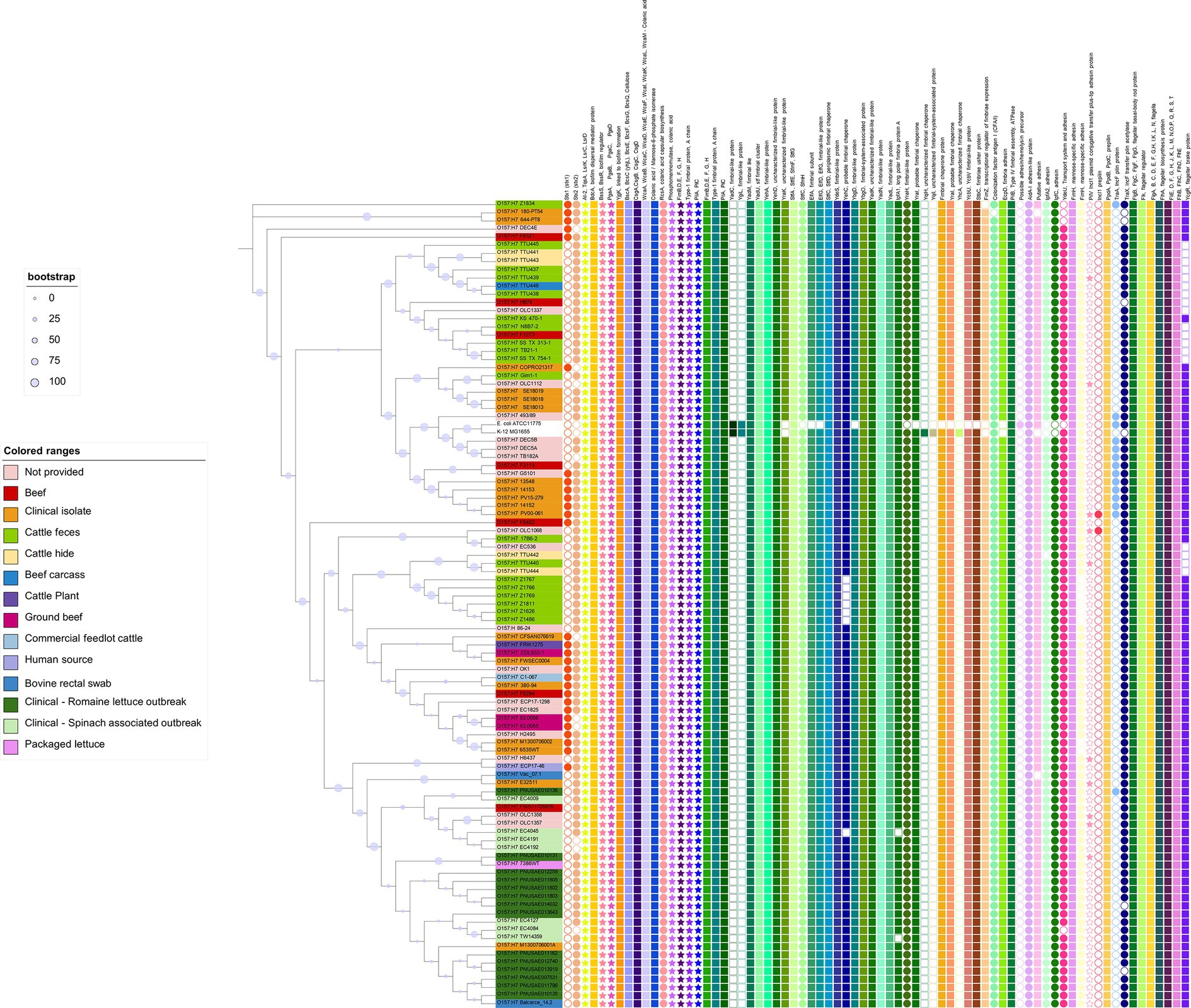
Figure 4. Biofilm related genes in Escherichia coli O157:H7 strains. The phylogenetic tree was constructed using the core genomes of the included genomes. Escherichia coli K12 and ATCC 11775 were included as references. The encoded gene products of relevant genes are annotated on the top of the figure. Solid-colored shapes represent the presence of relevant genes, while non-colored filled shapes indicate absence.
Plasmids have been reported to carry accessory genes influencing biofilm formation, particularly those related to surface attachment (Gama et al., 2020). Our analysis identified gene encoding proteins associated with conjugative plasmids in some selected genomes. Specifically, pilV (encoding IncI1 plasmid conjugative transfer pilus-tip adhesin protein), pilS (IncI1 plasmid conjugative transfer prepilin), traA (IncF plasmid conjugative transfer pilin protein), and traX (IncF plasmid conjugative transfer pilin acetylase) were more prevalent in O157:H7 and ATCC11775 strains. Notably, no plasmid-related proteins were found in E. coli K12.
Simply comparing the O157:H7 strains to two generic E. coli strains may not provide a comprehensive understanding of the underlying patterns. Consequently, we conducted a second comparative analysis that involved a broader spectrum of generic E. coli strains. This expanded analysis encompassed 32 isolates of generic E. coli obtained from meat processing equipment (Yang et al., 2023a). Our analysis revealed the consistent presence of genes associated with biofilm formation across all the generic E. coli genomes examined (Figure 5). Furthermore, it is noteworthy that generic E. coli strains originating from beef processing facilities exhibited a shared genetic repertoire related to biofilm formation, mirroring similar patterns observed in the O157:H7 strains. However, differences emerged in genes associated with adhesins, fimbria-like proteins, as well as the presence of adhesins or pili carried by plasmids.
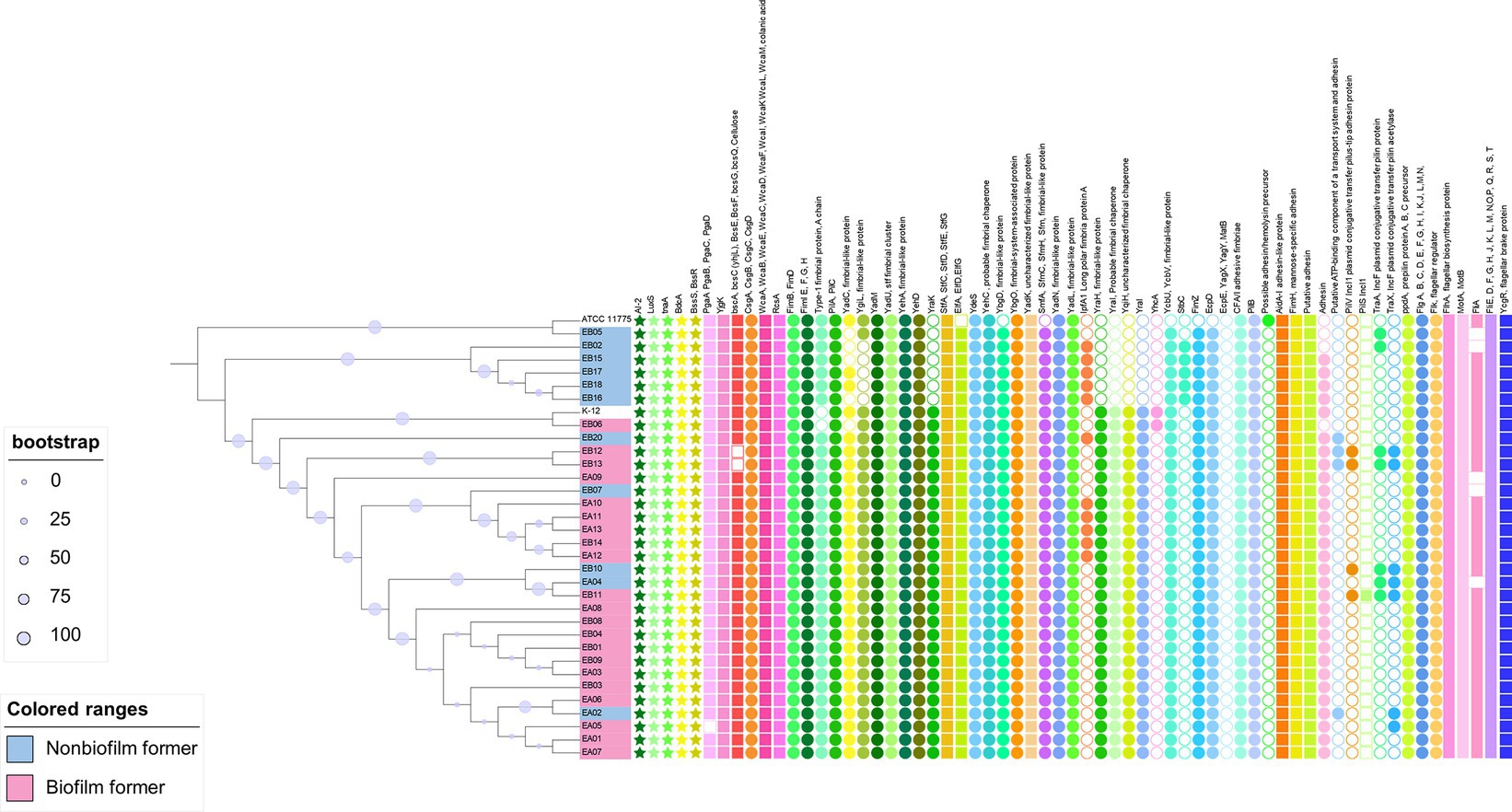
Figure 5. Biofilm related genes in genetic Escherichia coli strains recovered from meat cutting plants. The phylogenetic tree was constructed using the core genomes of the included genomes. Escherichia coli K12 and ATCC 11775 were included as references. The encoded gene products of relevant genes are annotated on the top of the figure. Solid-colored shapes represent the presence of relevant genes, while non-colored filled shapes indicate absence.
Information regarding biofilm formation phenotype of meat plant E. coli at 15°C was available and highlighted in Figure 5. However, no clear patterns were observed regarding the association between the absence of presence of relevant genes and their ability to form biofilms at 15°C. Neither group showed overrepresentation of genes according to the original genomic analysis performed by Yang et al. (2023a). It is important to acknowledge that the ability to form biofilms at 15°C, as depicted in our data, may not be indicative of the strains’ complete phenotypic biofilm-forming capabilities, given the variability in environmental conditions within meat processing facilities. Further research is needed to explore the full spectrum of biofilm formation in these strains under different conditions.
Relevant studies have shown the presence of more functional genes related to fimbria, flagella, curli, cellulose production, adhesins in E. coli could improve biofilm formation (Niba et al., 2007; Wang, 2019). On the other hand, an engineered E. coli strain with 17.6% of the parental genome removed including the genes involved in the synthesis of various cell structures such as type I fimbriae, curli, EPS, and the quorum sensing molecule autoinducer-2 (AI-2) is able to develop mature biofilms (May and Okabe, 2011). A study of pellicle (air-liquid biofilms) and non-pellicle forming E. coli found differences in the regulatory region of curli biosynthesis, but not in the absence or presence of genes in the two groups (Xu et al., 2023). Collectively, these findings suggest that three aspects could all be relevant for biofilm formation in E. coli: genetic determinants of cellular apparatus involved in biofilm formation, overall genetic background of individual strains, and variations in regulatory genes.
The process of converting livestock to meat products is very complex. Throughout the whole process, many factors can affect the microbial ecology in meat facilities and the final meat products. The antimicrobial intervention type, chemical concentration, spraying method, and temperature of the antimicrobial strategies applied in meat plants may have different decontamination effects and lead to different compositions of bacteria on the applied surfaces. For example, Psychrobacter and Pseudomonas predominated among bacterial population on spray chilled and dry chilled carcasses, respectively (Yang et al., 2017b). In fact, the collective incoming microbiota, and any condition that differentially impacts members of the microbial communities along the entire process may have consequences on transient and residential microbiota.
The bacteria residing on meat fabrication equipment and other environmental surfaces have been regarded as an important source contaminating meat products. Diverse bacteria can persist in meat plants, among which Pseudomonas, Acinetobacter, Psychrobacter, Sphingomonas, Enterococcus, Proteus, Staphylococcus, Burkholderia-Caballeronia-Paraburkholderia, Acidovorax, and Brevundimonas are the top 10 most predominant genera. However, information on the predominant bacteria identified at species/strains level is largely lacking. Biofilm is believed to be an important mechanism through which bacteria persist in meat plants. The commensal/residential bacteria may have synergistic or antagonistic effects for pathogenic bacteria such as STEC to form biofilm. They may also enhance the biofilm formation of pathogens who otherwise do not form biofilms on their own. Targeted cleaning and sanitizing efforts against residential microbiota may be rewarding in both safety and storage stability of meat products.
XY: Conceptualization, Funding acquisition, Resources, Supervision, Writing – original draft, Writing – review & editing. CN -B: Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Resources, Writing – original draft, Writing – review & editing. PZ: Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Resources, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. PZ acknowledges the library funding for publication provided by Colorado State University. XY acknowledges funding support from Beef Cattle Research Council and Results Driven Agricultural Research (FOS.01.21 and 2022F027R). CN-B acknowledges funding support from Beef Cattle Research Council (FOS.04.18).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1333696/full#supplementary-material
1. ^https://www.ncbi.nlm.nih.gov/
2. ^https://github.com/ncbi/sra-tools
Aarestrup, F. M., and Hasman, H. (2004). Susceptibility of different bacterial species isolated from food animals to copper sulphate, zinc chloride and antimicrobial substances used for disinfection. Vet. Microbiol. 100, 83–89. doi: 10.1016/j.vetmic.2004.01.013
Adator, E. H., Cheng, M., Holley, R., Mcallister, T., and Narvaez-Bravo, C. (2018). Ability of Shiga toxigenic Escherichia coli to survive within dry-surface biofilms and transfer to fresh lettuce. Int. J. Food Microbiol. 269, 52–59. doi: 10.1016/j.ijfoodmicro.2018.01.014
Algino, R. J., Ingham, S. C., and Zhu, J. (2007). Survey of antimicrobial effects of beef carcass intervention treatments in very small state-inspected slaughter plants. J. Food Sci. 72, M173–M179. doi: 10.1111/j.1750-3841.2007.00386.x
Arthur, T. M., Bono, J. L., and Kalchayanand, N. (2014). Characterization of Escherichia coli O157:H7 strains from contaminated raw beef trim during “high event periods”. Appl. Environ. Microbiol. 80, 506–514. doi: 10.1128/AEM.03192-13
Arthur, T. M., Bosilevac, J. M., Nou, X., Shackelford, S. D., Wheeler, T. L., Kent, M. P., et al. (2004). Escherichia coli O157 prevalence and enumeration of aerobic bacteria, Enterobacteriaceae, and Escherichia coli O157 at various steps in commercial beef processing plants. J. Food Prot. 67, 658–665. doi: 10.4315/0362-028X-67.4.658
Barnhart, M. M., and Chapman, M. R. (2006). Curli biogenesis and function. Annu. Rev. Microbiol. 60, 131–147. doi: 10.1146/annurev.micro.60.080805.142106
Belas, R. (2014). Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol. 22, 517–527. doi: 10.1016/j.tim.2014.05.002
Beloin, C., Roux, A., and Ghigo, J. M. (2008). Escherichia coli biofilms. Curr. Top. Microbiol. Immunol. 322, 249–289. doi: 10.1007/978-3-540-75418-3_12
Bessegatto, J. A., Paulino, L. R., Lisboa, J. A. N., Alfieri, A. A., Montemor, C. H., Medeiros, L. P., et al. (2017). Changes in the fecal microbiota of beef cattle caused by change in management and the use of virginiamycin as a growth promoter. Res. Vet. Sci. 114, 355–362. doi: 10.1016/j.rvsc.2017.06.011
Blagojevic, B., Antic, D., Ducic, M., and Buncic, S. (2012). Visual cleanliness scores of cattle at slaughter and microbial loads on the hides and the carcases. Vet. Rec. 170:563. doi: 10.1136/vr.100477
Botta, C., Ferrocino, I., Pessione, A., Cocolin, L., and Rantsiou, K. (2020). Spatiotemporal distribution of the environmental microbiota in food processing plants as impacted by cleaning and sanitizing procedures: the case of slaughterhouses and gaseous ozone. Appl. Environ. Microbiol. 86, e01861–e01820. doi: 10.1128/AEM.01861-20
Botta, C., Franciosa, I., Alessandria, V., Cardenia, V., Cocolin, L., and Ferrocino, I. (2022). Metataxonomic signature of beef burger perishability depends on the meat origin prior grinding. Food Res. Int. 156:111103. doi: 10.1016/j.foodres.2022.111103
Botta, C., Franciosa, I., Coisson, J. D., Ferrocino, I., Colasanto, A., Arlorio, M., et al. (2023). Beef carcass microbiota after slaughtering and primary cooling: a metataxonomic assessment to infer contamination drivers. Food Res. Int. 174:113466. doi: 10.1016/j.foodres.2023.113466
Callahan, B. J., Mcmurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., and Holmes, S. P. (2016). Dada2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Carrascosa, C., Raheem, D., Ramos, F., Saraiva, A., and Raposo, A. (2021). Microbial biofilms in the food industry-a comprehensive review. Int. J. Environ. Res. Public Health 18:14. doi: 10.3390/ijerph18042014
Casas, D. E., Vargas, D. A., Randazzo, E., Lynn, D., Echeverry, A., Brashears, M. M., et al. (2021). In-plant validation of novel on-site ozone generation technology (bio-safe) compared to lactic acid beef carcasses and trim using natural microbiota and Salmonella and E. coli O157:H7 surrogate enumeration. Foods 10:1002. doi: 10.3390/foods10051002
Castaño-Arriba, A., González-Machado, C., Igrejas, G., Poeta, P., Alonso-Calleja, C., and Capita, R. (2020). Antibiotic resistance and biofilm-forming ability in Enterococcal isolates from red meat and poultry preparations. Pathogens 9:1021. doi: 10.3390/pathogens9121021
Castonguay, M. H., Van Der Schaaf, S., Koester, W., Krooneman, J., Van Der Meer, W., Harmsen, H., et al. (2006). Biofilm formation by Escherichia coli is stimulated by synergistic interactions and co-adhesion mechanisms with adherence-proficient bacteria. Res. Microbiol. 157, 471–478. doi: 10.1016/j.resmic.2005.10.003
CDC. (2014). E. coli (Escherichia coli). Available at: https://www.cdc.gov/ecoli/general/index.html (Accessed 25 October 2023).
CFIA (2010). “Ante and post-mortem procedures, dispositions, monitoring and controls—meat species, ostriches, rheas and emus” in Meat Hygiene Manual of Procedures, https://epe.lac-bac.gc.ca/100/206/301/cfia-acia/2011-09-21/inspection.gc.ca/english/fssa/meavia/man/mane.shtml
Charbonneau, M. E., and Mourez, M. (2007). Functional organization of the autotransporter adhesin involved in diffuse adherence. J. Bacteriol. 189, 9020–9029. doi: 10.1128/JB.01238-07
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one Fastq preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Cherifi, T., Arsenault, J., Quessy, S., and Fravalo, P. (2022). Co-occurrence of L. monocytogenes with other bacterial genera and bacterial diversity on cleaned conveyor surfaces in a swine slaughterhouse. Microorganisms 10:613. doi: 10.3390/microorganisms10030613
Cobo-Díaz, J. F., Alvarez-Molina, A., Alexa, E. A., Walsh, C. J., Mencía-Ares, O., Puente-Gómez, P., et al. (2021). Microbial colonization and resistome dynamics in food processing environments of a newly opened pork cutting industry during 1.5 years of activity. Microbiome 9, 1–19. doi: 10.1186/s40168-021-01131-9
Costa, T. R. D., Felisberto-Rodrigues, C., Meir, A., Prevost, M. S., Redzej, A., Trokter, M., et al. (2015). Secretion systems in gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 13, 343–359. doi: 10.1038/nrmicro3456
Costerton, J. W., Stewart, P. S., and Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Coughlan, L. M., Cotter, P. D., Hill, C., and Alvarez-Ordóñez, A. (2016). New weapons to fight old enemies: novel strategies for the (bio)control of bacterial niofilms in the food industry. Front. Microbiol. 7:1641. doi: 10.3389/fmicb.2016.01641
Domka, J., Lee, J., and Wood, T. K. (2006). YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl. Environ. Microbiol. 72, 2449–2459. doi: 10.1128/AEM.72.4.2449-2459.2006
Dowd, S. E., Callaway, T. R., Wolcott, R. D., Sun, Y., Mckeehan, T., Hagevoort, R. G., et al. (2008). Evaluation of the bacterial diversity in the feces of cattle using 16S rdna bacterial tag-encoded Flx amplicon pyrosequencing (btefap). BMC Microbiol. 8:125. doi: 10.1186/1471-2180-8-125
Ekong, P. S., Sanderson, M. W., and Cernicchiaro, N. (2015). Prevalence and concentration of Escherichia coli O157 in different seasons and cattle types processed in North America: a systematic review and meta-analysis of published research. Prev. Vet. Med. 121, 74–85. doi: 10.1016/j.prevetmed.2015.06.019
Essendoubi, S., Yang, X., King, R., Keenliside, J., Bahamon, J., Diegel, J., et al. (2020). Prevalence and characterization of Escherichia coli O157:H7 on pork carcasses and in swine colon contents from provincially licensed abattoirs in Alberta, Canada. J. Food Prot. 83, 1909–1917. doi: 10.4315/JFP-20-146
Fagerlund, A., Langsrud, S., and Møretrø, T. (2021). Microbial diversity and ecology of biofilms in food industry environments associated with Listeria monocytogenes persistence. Curr. Opin. Food Sci. 37, 171–178. doi: 10.1016/j.cofs.2020.10.015
Fagerlund, A., Møretrø, T., Heir, E., Briandet, R., and Langsrud, S. (2017). Cleaning and disinfection of biofilms composed of Listeria monocytogenes and background microbiota from meat processing surfaces. Appl. Environ. Microbiol. 83:17. doi: 10.1128/AEM.01046-17
Fang, Y., Tran, F., Stanford, K., and Yang, X. (2023). Stress resistance and virulence gene profiles associated with phylogeny and phenotypes of Escherichia coli from cattle. J. Food Prot. 86:100122. doi: 10.1016/j.jfp.2023.100122
Fang, Y., Visvalingam, J., Zhang, P., and Yang, X. (2022). Biofilm formation by non-O157 Shiga toxin-producing Escherichia coli in monocultures and co-cultures with meat processing surface bacteria. Food Microbiol. 102:103902. doi: 10.1016/j.fm.2021.103902
Gama, J. A., Fredheim, E. G. A., Cléon, F., Reis, A. M., Zilhão, R., and Dionisio, F. (2020). Dominance between plasmids determines the extent of biofilm formation. Front. Microbiol. 11:2070. doi: 10.3389/fmicb.2020.02070
Garrett, T. R., Bhakoo, M., and Zhang, Z. (2008). Bacterial adhesion and biofilms on surfaces. Prog. Nat. Sci. 18, 1049–1056. doi: 10.1016/j.pnsc.2008.04.001
Gaskins, H. R., Collier, C. T., and Anderson, D. B. (2002). Antibiotics as growth promotants: mode of action. Anim. Biotechnol. 13, 29–42. doi: 10.1081/ABIO-120005768
Giaouris, E., Heir, E., Hébraud, M., Chorianopoulos, N., Langsrud, S., Møretrø, T., et al. (2014). Attachment and biofilm formation by foodborne bacteria in meat processing environments: causes, implications, role of bacterial interactions and control by alternative novel methods. Meat Sci. 97, 298–309. doi: 10.1016/j.meatsci.2013.05.023
Gill, C. O. (2005). “Haccp in the processing of fresh meat” in Improving the safety of fresh meat. ed. J. N. Sofos (Cambridge, U.K.: Crc/Woodhead Publishing Limited)
Gill, C. O. (2009). Effects on the microbiological condition of product of decontaminating treatments routinely applied to carcasses at beef packing plants. J. Food Prot. 72, 1790–1801. doi: 10.4315/0362-028X-72.8.1790
Greig, J. D., Waddell, L., Wilhelm, B., Wilkins, W., Bucher, O., Parker, S., et al. (2012). The efficacy of interventions applied during primary processing on contamination of beef carcasses with Escherichia coli: a systematic review-meta-analysis of the published research. Food Control 27, 385–397. doi: 10.1016/j.foodcont.2012.03.019
Haque, M., Bosilevac, J. M., and Chaves, B. D. (2022). A review of Shiga-toxin producing Escherichia coli (Stec) contamination in the raw pork production chain. Int. J. Food Microbiol. 377:109832. doi: 10.1016/j.ijfoodmicro.2022.109832
Heiman, K. E., Mody, R. K., Johnson, S. D., Griffin, P. M., and Gould, L. H. (2015). Escherichia coli O157 outbreaks in the United States, 2003-2012. Emerg. Infect. Dis. 21, 1293–1301. doi: 10.3201/eid2108.141364
Itoh, Y., Rice, J. D., Goller, C., Pannuri, A., Taylor, J., Meisner, J., et al. (2008). Roles of pgaabcd genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J. Bacteriol. 190, 3670–3680. doi: 10.1128/JB.01920-07
Kang, S., Ravensdale, J., Coorey, R., Dykes, G. A., and Barlow, R. (2019). A comparison of 16S rrna profiles through slaughter in Australian export beef abattoirs. Front. Microbiol. 10:2747. doi: 10.3389/fmicb.2019.02747
Kang, S., Ravensdale, J. T., Coorey, R., Dykes, G. A., and Barlow, R. S. (2020). Bacterial community analysis using 16S rrna amplicon sequencing in the boning room of Australian beef export abattoirs. Int. J. Food Microbiol. 332:108779. doi: 10.1016/j.ijfoodmicro.2020.108779
Keenan, D. F., Hayes, J. E., Kenny, T. A., and Kerry, J. P. (2016). Effect of hot boning and elevated brine temperature on the processing, storage and eating quality of cured beef hindquarter (M. Biceps femoris) and forequarter (M. pectoralis profundus) muscles. J. Food Qual. 39, 126–139. doi: 10.1111/jfq.12179
Kempf, F., La Ragione, R., Chirullo, B., Schouler, C., and Velge, P. (2022). Super shedding in enteric pathogens: a review. Microorganisms 10:2101. doi: 10.3390/microorganisms10112101
Khaitsa, M. L., Smith, D. R., Stoner, J. A., Parkhurst, A. M., Hinkley, S., Klopfenstein, T. J., et al. (2003). Incidence, duration, and prevalence of Escherichia coli O157:H7 fecal shedding by feedlot cattle during the finishing period. J. Food Prot. 66, 1972–1977. doi: 10.4315/0362-028X-66.11.1972
Kim, Y., Wang, X., Ma, Q., Zhang, X. S., and Wood, T. K. (2009). Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J. Bacteriol. 191, 1258–1267. doi: 10.1128/JB.01465-08
King, T., Kocharunchitt, C., Gobius, K., Bowman, J. P., and Ross, T. (2016). Physiological response of Escherichia coli O157:H7 Sakai to dynamic changes in temperature and water activity as experienced during carcass chilling. Mol. Cell. Proteomics 15, 3331–3347. doi: 10.1074/mcp.M116.063065
Kocharunchitt, C., Mellefont, L., Bowman, J. P., and Ross, T. (2020). Application of chlorine dioxide and peroxyacetic acid during spray chilling as a potential antimicrobial intervention for beef carcasses. Food Microbiol. 87:103355. doi: 10.1016/j.fm.2019.103355
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lavilla Lerma, L., Benomar, N., Casado Muñoz, M. d. C., Gálvez, A., and Abriouel, H. (2015). Correlation between antibiotic and biocide resistance in mesophilic and psychrotrophic Pseudomonas spp. isolated from slaughterhouse surfaces throughout meat chain production. Food Microbiol. 51, 33–44. doi: 10.1016/j.fm.2015.04.010
Lin, Y., Yu, C., Ma, Z., Che, L., Feng, B., Fang, Z., et al. (2022). Effects of yeast culture supplementation in wheat-rice-based diet on growth performance, meat quality, and gut microbiota of growing-finishing pigs. Animals 12:2177. doi: 10.3390/ani12172177
Liu, N. T., Lefcourt, A. M., Nou, X., Shelton, D. R., Zhang, G., and Lo, Y. M. (2013). Native microflora in fresh-cut produce processing plants and their potentials for biofilm formation. J. Food Prot. 76, 827–832. doi: 10.4315/0362-028X.JFP-12-433
Liu, Y., Youssef, M. K., and Yang, X. (2016). Effects of dry chilling on the microflora on beef carcasses at a Canadian beef packing plant. J. Food Prot. 79, 538–543. doi: 10.4315/0362-028X.JFP-15-476
Lu, J., Breitwieser, F. P., Thielen, P., and Salzberg, S. L. (2017). Bracken: estimating species abundance in metagenomics data. PeerJ Computer Science 3:e104. doi: 10.7717/peerj-cs.104
Mallick, H., Rahnavard, A., Mciver, L. J., Ma, S., Zhang, Y., Nguyen, L. H., et al. (2021). Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 17:e1009442. doi: 10.1371/journal.pcbi.1009442
Mao, S., Zhang, M., Liu, J., and Zhu, W. (2015). Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: membership and potential function. Sci. Rep. 5:16116. doi: 10.1038/srep16116
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 3:10. doi: 10.14806/ej.17.1.200
Maslen, B. N., Gray, L. A., Ghorashi, S. A., White, J. D., Campbell, M. A., and Pant, S. D. (2022). Temporal changes in the faecal microbiota of beef cattle on feedlot placement. Animals 12:2500. doi: 10.3390/ani12192500
May, T., and Okabe, S. (2011). Enterobactin is required for biofilm development in reduced-genome Escherichia coli. Environ. Microbiol. 13, 3149–3162. doi: 10.1111/j.1462-2920.2011.02607.x
Mcmurdie, P. J., and Holmes, S. (2013). Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8:e61217. doi: 10.1371/journal.pone.0061217
Meziti, A., Rodriguez, R. L., Hatt, J. K., Pena-Gonzalez, A., Levy, K., and Konstantinidis, K. T. (2021). The reliability of metagenome-assembled genomes (mags) in representing natural populations: insights from comparing mags against isolate genomes derived from the same fecal sample. Appl. Environ. Microbiol. 87:20. doi: 10.1128/AEM.02593-20
Møretrø, T., Langsrud, S., and Heir, E. (2013). Bacteria on meat abattoir process surfaces after sanitation: characterisation of survival properties of Listeria monocytogenes and the commensal bacterial flora. Adv. Microbiol. 3, 255–264. doi: 10.4236/aim.2013.33037
Morimatsu, K., Eguchi, K., Hamanaka, D., Tanaka, F., and Uchino, T. (2012). Effects of temperature and nutrient conditions on biofilm formation of Pseudomonas putida. Food Sci. Technol. Res. 18, 879–883. doi: 10.3136/fstr.18.879
Nan, Y., Rodas-Gonzalez, A., Stanford, K., Nadon, C., Yang, X., Mcallister, T., et al. (2022). Formation and transfer of multi-species biofilms Containing E. coli O103:H2 on food contact surfaces to beef. Front. Microbiol. 13:863778. doi: 10.3389/fmicb.2022.863778
Narvaez-Bravo, C., Miller, M. F., Jackson, T., Jackson, S., Rodas-Gonzalez, A., Pond, K., et al. (2013). Salmonella and Escherichia coli O157:H7 prevalence in cattle and on carcasses in a vertically integrated feedlot and harvest plant in Mexico. J. Food Prot. 76, 786–795. doi: 10.4315/0362-028X.JFP-12-079
Nastasijevic, I., Schmidt, J. W., Boskovic, M., Glisic, M., Kalchayanand, N., Shackelford, S. D., et al. (2020). Seasonal prevalence of Shiga toxin-producing Escherichia coli on pork carcasses for three steps of the harvest process at two commercial processing plants in the United States. Appl. Environ. Microbiol. 87:20. doi: 10.1128/AEM.01711-20
Niba, E. T., Naka, Y., Nagase, M., Mori, H., and Kitakawa, M. (2007). A genome-wide approach to identify the genes involved in biofilm formation in E. coli. DNA Res. 14, 237–246. doi: 10.1093/dnares/dsm024
O’Toole, G., Kaplan, H. B., and Kolter, R. (2000). Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79. doi: 10.1146/annurev.micro.54.1.49
O’Toole, G. A., and Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304. doi: 10.1046/j.1365-2958.1998.01062.x
Overbeek, R., Begley, T., Butler, R. M., Choudhuri, J. V., Chuang, H. Y., Cohoon, M., et al. (2005). The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33, 5691–5702. doi: 10.1093/nar/gki866
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The seed and the rapid annotation of microbial genomes using subsystems technology (Rast). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Paranjpye, R. N., and Strom, M. S. (2005). A Vibrio vulnificus type iv pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect. Immun. 73, 1411–1422. doi: 10.1128/IAI.73.3.1411-1422.2005
Quan, J., Wu, Z., Ye, Y., Peng, L., Wu, J., Ruan, D., et al. (2020). Metagenomic sharacterization of tntestinal regions in pigs with contrasting feed efficiency. Front. Microbiol. 11:32. doi: 10.3389/fmicb.2020.00032
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The Silva ribosomal Rna gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Römling, U., and Galperin, M. Y. (2015). Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol. 23, 545–557. doi: 10.1016/j.tim.2015.05.005
Savell, J. W., Mueller, S. L., and Baird, B. E. (2005). The chilling of carcasses. Meat Sci. 70, 449–459. doi: 10.1016/j.meatsci.2004.06.027
Scott, B. R., Yang, X., Geornaras, I., Delmore, R. J., Woerner, D. R., Adler, J. M., et al. (2015). Antimicrobial efficacy of a lactic acid and citric acid blend against Shiga toxin-producing Escherichia coli, Salmonella, and nonpathogenic Escherichia coli biotype I on inoculated prerigor beef carcass surface tissue. J. Food Prot. 78, 2136–2142. doi: 10.4315/0362-028X.JFP-15-194
Shaw, M. K., Marr, A. G., and Ingraham, J. L. (1971). Determination of the minimal temperature for growth of Escherichia coli. J. Bacteriol. 105, 683–684. doi: 10.1128/jb.105.2.683-684.1971
Shedleur-Bourguignon, F., Duchemin, T., Thériault, W., Longpré, J., Thibodeau, A., Hocine, M. N., et al. (2023). Distinct microbiotas are associated with different production lines in the cutting room of a swine slaughterhouse. Microorganisms 11:133. doi: 10.3390/microorganisms11010133
Smith, M. G. (1985). The generation time, lag time, and minimum temperature of growth of coliform organisms on meat, and the implications for codes of practice in abattoirs. J. Hyg. 94, 289–300. doi: 10.1017/S0022172400061519
Stanford, K., Tran, F., Zhang, P., and Yang, X. (2021). Biofilm-forming capacity of Escherichia coli isolated from cattle and beef packing plants: relation to virulence attributes, stage of processing, antimicrobial interventions, and heat tolerance. Appl. Environ. Microbiol. 87:e0112621. doi: 10.1128/AEM.01126-21
Stephens, T. P., Mcallister, T. A., and Stanford, K. (2009). Perineal swabs reveal effect of super shedders on the transmission of Escherichia coli O157:H7 in commercial feedlots. J. Anim. Sci. 87, 4151–4160. doi: 10.2527/jas.2009-1967
Tarr, P. I., Gordon, C. A., and Chandler, W. L. (2005). Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365, 1073–1086. doi: 10.1016/S0140-6736(05)71144-2
Thorpe, C. M. (2004). Shiga toxin-producing Escherichia coli infection. Clin. Infect. Dis. 38, 1298–1303. doi: 10.1086/383473
Uhlich, G. A., Chen, C. Y., Cottrell, B. J., Hofmann, C. S., Dudley, E. G., Strobaugh, T. P., et al. (2013). Phage insertion in mlrA and variations in rpoS limit curli expression and biofilm formation in Escherichia coli serotype O157: H7. Microbiology 159, 1586–1596. doi: 10.1099/mic.0.066118-0
Uhlich, G. A., Chen, C.-Y., Cottrell, B. J., and Nguyen, L.-H. (2014). Growth media and temperature effects on biofilm formation by serotype O157:H7 and non-O157 Shiga toxin-producing Escherichia coli. FEMS Microbiol. Lett. 354, 133–141. doi: 10.1111/1574-6968.12439
Van Houdt, R., and Michiels, C. W. (2005). Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res. Microbiol. 156, 626–633. doi: 10.1016/j.resmic.2005.02.005
Visvalingam, J., Wang, H., Youssef, M. K., Devos, J., Gill, C. O., and Yang, X. (2016). Spatial and temporal distribution of Escherichia coli on beef trimmings obtained from a beef packing plant. J. Food Prot. 79, 1325–1331. doi: 10.4315/0362-028X.JFP-15-598
Visvalingam, J., Zhang, P., Ells, T. C., and Yang, X. (2019). Dynamics of biofilm formation by Salmonella typhimurium and beef processing plant bacteria in mono- and dual-species cultures. Microb. Ecol. 78, 375–387. doi: 10.1007/s00248-018-1304-z
Wagner, E. M., Fischel, K., Rammer, N., Beer, C., Palmetzhofer, A. L., Conrady, B., et al. (2021). Bacteria of eleven different species isolated from biofilms in a meat processing environment have diverse biofilm forming abilities. Int. J. Food Microbiol. 349:109232. doi: 10.1016/j.ijfoodmicro.2021.109232
Wang, R. (2019). Biofilms and meat safety: a mini-review. J. Food Prot. 82, 120–127. doi: 10.4315/0362-028X.JFP-18-311
Wang, R., Bono, J. L., Kalchayanand, N., Shackelford, S., and Harhay, D. M. (2012). Biofilm formation by Shiga toxin-producing Escherichia coli O157:H7 and non-O157 strains and their tolerance to sanitizers commonly used in the food processing environment. J. Food Prot. 75, 1418–1428. doi: 10.4315/0362-028X.JFP-11-427
Wang, H., He, A., and Yang, X. (2018). Dynamics of microflora on conveyor belts in a beef fabrication facility during sanitation. Food Control 85, 42–47. doi: 10.1016/j.foodcont.2017.09.017
Wang, R., Luedtke, B. E., Bosilevac, J. M., Schmidt, J. W., Kalchayanand, N., and Arthur, T. M. (2016). Escherichia coli O157:H7 strains isolated from high-event period beef contamination have strong biofilm-forming ability and low sanitizer susceptibility, which are associated with high pO157 plasmid copy number. J. Food Prot. 79, 1875–1883. doi: 10.4315/0362-028X.JFP-16-113
Watnick, P., and Kolter, R. (2000). Biofilm, City of microbes. J. Bacteriol. 182, 2675–2679. doi: 10.1128/JB.182.10.2675-2679.2000
White, D., Drummond, J., and Fuqua, C. (2011). The physiology and biochemistry of prokaryotes. Oxford, United Kingdom: Oxford University Press.
WHO (2018). Shiga toxin-producing Escherichia coli (Stec) and food. Microbiological risk assessment series ; 31. Rome: World Health Organization.
Wickramasinghe, N. N., Hlaing, M. M., Ravensdale, J. T., Coorey, R., Chandry, P. S., and Dykes, G. A. (2020). Characterization of the biofilm matrix composition of psychrotrophic, meat spoilage pseudomonads. Sci. Rep. 10:16457. doi: 10.1038/s41598-020-73612-0
Wood, D. E., Lu, J., and Langmead, B. (2019). Improved metagenomic analysis with kraken 2. Genome Biol. 20:257. doi: 10.1186/s13059-019-1891-0
Xu, Z. S., Zhu, T., Wang, Z., Yang, X., and Gänzle, M. G. (2023). Socializing at the air-liquid tnterface: a functional genomic analysis on biofilm-related genes during pellicle formation by Escherichia coli and its interaction with Aeromonas australiensis. Appl. Environ. Microbiol. 89:e0045623. doi: 10.1128/aem.00456-23
Yang, X. (2017). “Microbial ecology of beef carcasses and beef products” in Quantitative Microbiology Food Processing. John Wiley & Sons, Ltd, the Atrium, Southern Gate, Chichester, West Sussex, PO19 8SQ, UK.
Yang, X., Badoni, M., Tran, F., and Gill, C. O. (2015b). Microbiological effects of a routine treatment for decontaminating hide-on carcasses at a large beef packing plant. J. Food Prot. 78, 256–263. doi: 10.4315/0362-028X.JFP-14-226
Yang, X., Badoni, M., Youssef, M. K., and Gill, C. O. (2012). Enhanced control of microbiological contamination of product at a large beef packing plant. J. Food Prot. 75, 144–149. doi: 10.4315/0362-028X.JFP-11-291
Yang, X., He, A., Badoni, M., Tran, F., and Wang, H. (2017a). Mapping sources of contamination of Escherichia coli on beef in the fabrication facility of a commercial beef packing plant. Food Control 75, 153–159. doi: 10.1016/j.foodcont.2016.12.004
Yang, X., Noyes, N. R., Doster, E., Martin, J. N., Linke, L. M., Magnuson, R. J., et al. (2016). Use of metagenomic shotgun sequencing technology to detect foodborne pathogens within the microbiome of the beef production chain. Appl. Environ. Microbiol. 82, 2433–2443. doi: 10.1128/AEM.00078-16
Yang, X., Tran, F., and Wolters, T. (2017b). Microbial ecology of decontaminated and not decontaminated beef carcasses. J. Food Res. 6, 85–91. doi: 10.5539/jfr.v6n5p85
Yang, X., Tran, F., Youssef, M. K., and Gill, C. O. (2015a). Determination of sources of Escherichia coli on beef by multiple-locus variable-number tandem repeat analysis. J. Food Prot. 78, 1296–1302. doi: 10.4315/0362-028X.JFP-15-014
Yang, X., Tran, F., and Zhang, P. (2023a). Comparative genomic analyses of Escherichia coli from a meat processing environment in relation to their biofilm formation and persistence. Microbiol Spectr 11:e0018323. doi: 10.1128/spectrum.00183-23
Yang, X., Wang, H., He, A., and Tran, F. (2017c). Microbial efficacy and impact on the population of Escherichia coli of a routine sanitation process for the fabrication facility of a beef packing plant. Food Control 71, 353–357. doi: 10.1016/j.foodcont.2016.07.016
Yang, X., Wang, H., He, A., and Tran, F. (2018). Biofilm formation and susceptibility to biocides of recurring and transient Escherichia coli isolated from meat fabrication equipment. Food Control 90, 205–211. doi: 10.1016/j.foodcont.2018.02.050
Yang, X., Wang, H., Hrycauk, S., Holman, D. B., and Ells, T. C. (2023b). Microbial dynamics in mixed-culture biofilms of Salmonella typhimurium and Escherichia coli O157: H7 and bacteria surviving sanitation of conveyor belts of meat processing plants. Microorganisms 11:421. doi: 10.3390/microorganisms11020421
Yang, X., Wang, H., Hrycauk, S., and Klassen, M. D. (2021). Effects of peroxyacetic acid spray and storage temperature on the microbiota and sensory properties of vacuum-packed subprimal cuts of meat. Appl. Environ. Microbiol. 87:20. doi: 10.1128/AEM.03143-20
Yin, W., Wang, Y., Liu, L., and He, J. (2019). Biofilms: the microbial “protective clothing” in extreme environments. Int. J. Mol. Sci. 20:423. doi: 10.3390/ijms20143423
Youssef, M. K., Badoni, M., Yang, X., and Gill, C. O. (2013). Sources of Escherichia coli deposited on beef during breaking of carcasses carrying few E. coli at two packing plants. Food Control 31, 166–171. doi: 10.1016/j.foodcont.2012.09.045
Youssef, M. K., Gill, C. O., and Yang, X. (2014). Storage life at 2oC or −1.5oC of vacuum-packaged boneless and bone-in cuts from decontaminated beef carcasses. J. Sci. Food Agric. 94, 3118–3124. doi: 10.1002/jsfa.6659
Zaheer, R., Lakin, S. M., Polo, R. O., Cook, S. R., Larney, F. J., Morley, P. S., et al. (2019). Comparative diversity of microbiomes and Resistomes in beef feedlots, downstream environments and urban sewage influent. BMC Microbiol. 19:197. doi: 10.1186/s12866-019-1548-x
Zdolec, N., Kotsiri, A., Houf, K., Alvarez-Ordonez, A., Blagojevic, B., Karabasil, N., et al. (2022). Systematic review and meta-analysis of the efficacy of interventions applied during primary processing to reduce microbial contamination on pig carcasses. Foods 11:110. doi: 10.3390/foods11142110
Zhang, P., Badoni, M., Gänzle, M., and Yang, X. (2018). Growth of Carnobacterium spp. isolated from chilled vacuum-packaged meat under relevant acidic conditions. Int. J. Food Microbiol. 286, 120–127. doi: 10.1016/j.ijfoodmicro.2018.07.032
Zhang, J., and Poh, C. L. (2018). Regulating exopolysaccharide gene wcaF allows control of Escherichia coli biofilm formation. Sci. Rep. 8:13127. doi: 10.1038/s41598-018-31161-7
Zhang, Z., Yang, L., He, Y., Luo, X., Zhao, S., and Jia, X. (2021). Composition of fecal microbiota in grazing and feedlot angus beef cattle. Animals 11:3167. doi: 10.3390/ani11113167
Zhilyaev, S., Cadavez, V., Gonzales-Barron, U., Phetxumphou, K., and Gallagher, D. (2017). Meta-analysis on the effect of interventions used in cattle processing plants to reduce Escherichia coli contamination. Food Res. Int. 93, 16–25. doi: 10.1016/j.foodres.2017.01.005
Zwirzitz, B., Wetzels, S. U., Dixon, E. D., Fleischmann, S., Selberherr, E., Thalguter, S., et al. (2021). Co-occurrence of Listeria spp. and spoilage associated microbiota during meat processing due to cross-contamination events. Front. Microbiol. 12:632935. doi: 10.3389/fmicb.2021.632935
Keywords: microbiota, meat processing facilities, beef, pork, persistent bacteria, sequencing, Shiga toxin-producing Escherichia coli, biofilm
Citation: Yang X, Narvaez-Bravo C and Zhang P (2024) Driving forces shaping the microbial ecology in meat packing plants. Front. Microbiol. 14:1333696. doi: 10.3389/fmicb.2023.1333696
Received: 05 November 2023; Accepted: 22 December 2023;
Published: 23 January 2024.
Edited by:
Mehran Moradi, Urmia University, IranReviewed by:
Eleftherios H. Drosinos, Agricultural University of Athens, GreeceCopyright © 2024 Claudia Narvaez, and His Majesty the King in Right of Canada, as represented by the Minister of Agriculture and Agri-Food Canada for the contribution of Xianqin Yang and Peipei Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianqin Yang, eGlhbnFpbi55YW5nQGFnci5nYy5jYQ==; Peipei Zhang, cGVpcGVpLnpoYW5nQGNvbG9zdGF0ZS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.