
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 04 January 2024
Sec. Infectious Agents and Disease
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1333500
This article is part of the Research Topic Salmonella spp.- Transmission, Pathogenesis, Host-pathogen interaction, Prevention and Treatment View all 20 articles
Programmed cell death (PCD) is the collective term for the intrinsically regulated death of cells. Various types of cell death are triggered by their own programmed regulation during the growth and development of organisms, as well as in response to environmental and disease stresses. PCD encompasses apoptosis, pyroptosis, necroptosis, autophagy, and other forms. PCD plays a crucial role not only in the growth and development of organisms but also in serving as a component of the host innate immune defense and as a bacterial virulence strategy employed by pathogens during invasion. The zoonotic pathogen Salmonella has the ability to modulate multiple forms of PCD, including apoptosis, pyroptosis, necroptosis, and autophagy, within the host organism. This modulation subsequently impacts the bacterial infection process. This review aims to consolidate recent findings regarding the mechanisms by which Salmonella initiates and controls cell death signaling, the ways in which various forms of cell death can impede or restrict bacterial proliferation, and the interplay between cell death and innate immune pathways that can counteract Salmonella-induced suppression of host cell death. Ultimately, these insights may contribute novel perspectives for the diagnosis and treatment of clinical Salmonella-related diseases.
Programmed cell death (PCD) is a pervasive phenomenon in the growth and development of organisms, characterized by a genetically regulated process of active and organized cell death. Specifically, PCD involves the initiation of a protective mechanism in response to cytokine stimulation from either the internal or external environment. This mechanism entails the activation of gene coding and protein metabolism. Initially, apoptosis was acknowledged as the prevailing form of cell death in the early stages of PCD research, while necrosis was predominantly viewed as an incidental consequence of external stimuli leading to cell death. Necrosis is commonly defined as the uncontrolled death of a cell, typically occurring after a significant injury, leading to the release of cellular contents into neighboring tissues and subsequent harm (D'Arcy, 2019). As investigations into this genetically regulated, self-contained, and organized form of cell death progress, an increasing number of cell death modalities have been unveiled. In the context of pathogenic bacterial infections, these cell deaths are commonly classified into various prevalent forms of PCD, such as apoptosis, pyroptosis, necroptosis, autophagy, and ferroptosis, based on discernible morphological alterations, essential biochemical factors, immune pathway components, and effector proteins (Table 1).
Host cell death is a prevalent characteristic observed in numerous bacterial infections, serving as a means to restrict bacterial replication. Conversely, bacteria employ a diverse range of virulence factors or effector proteins to counteract or manipulate the signaling pathways associated with cell death, thereby evading or impeding cell death induction. For instance, the T3SS effectors of Salmonella Typhimurium have been identified to interact with apoptotic, necroptotic, and pyroptotic cell death cascades, thereby impeding the efficient elimination of the bacteria and the recruitment of neutrophils or dendritic cells to the site of infection (LaRock et al., 2015). PCD is widely recognized as an integral component of the innate immune defense in host organisms. Salmonella, a zoonotic pathogen, is a facultative intracellular pathogen that poses a significant threat to both global human and animal health, resulting in substantial economic burdens. This pathogen consistently influences the progression and resolution of infections by stimulating the production of cytokines with potent immunomodulatory properties and the secretion of various virulence effector proteins via the bacterial type III secretion system (T3SS), which governs the regulation of PCD (Wemyss and Pearson, 2019). However, the type of cell death induced during a bacterial infection is contingent upon various factors, including the infecting pathogen, its virulence factors, the specific tissues and cell types affected, and the host's inflammatory response. This review aims to provide a comprehensive overview of the molecular mechanisms governing PCD in Salmonella infections, with a particular emphasis on apoptosis, pyroptosis, necroptosis, and autophagy, as well as their interplay in the containment or eradication of Salmonella during infections. The insights presented herein offer novel targets and ideas for the treatment of diseases associated with Salmonella infection.
Apoptosis is a regulated process of cellular death, wherein the activation of Caspases serves as the principal mechanism (Stringer et al., 2023). It is triggered in cells that have been subjected to either internal or external signaling cascades due to damage or stress. The initiation of apoptosis is contingent upon the activation of apoptotic Caspases and is marked by various cellular changes, including the formation of apoptotic vesicles, cell membrane blebbing, cell shrinkage, DNA fragmentation, and nucleoplasmic condensation (Guiney, 2005). The pathways of Caspase dependency encompass both the extrinsic death pathway mediated by death receptors and the intrinsic death pathway mediated by mitochondria. The extrinsic death pathway is triggered when ligands or cytokines bind to transmembrane death receptors located on the cell surface, thereby regulating apoptosis through the activation of the death-inducing signaling complex (DISC) and Caspases-8. Specifically, the death receptor Fas binds to the ligand FasL and interacts with the Death domain (DD) on the protein, leading to the recruitment of the Fas-associated death domain protein (FADD). FADD possesses a Death effector domain (DED) that facilitates the interaction between FADD and other DED-containing proteins, such as Caspase-8. FADD undergoes aggregation with Caspase-8 to form the DISC. Upon activation, Caspase-8 undergoes a conformational change from a single-stranded zymogen to a biologically active double-stranded protein. This activated Caspase-8 then further activates Caspase-3, initiating the extrinsic death pathway (Grassme et al., 2001). On the other hand, the intrinsic death pathway is triggered by the release of apoptotic proteins due to internal stimulation. Various factors such as microbial infection, DNA damage, cytotoxic stimulation, and other influences lead to the oligomerization of B cell lymphoma 2 (Bcl-2) family proteins Bax and Bak (Stringer et al., 2023). This oligomerization increases the permeability of the mitochondrial membrane, resulting in the formation of annular lipid pores in the intermembrane space of the mitochondria. Cytochrome C (CytC) and other pro-apoptotic factors are released into the cytoplasm via perforated mitochondria. In the presence of dATP, CytC and apoptotic protease-activating factor 1 (Apaf-1) with CARD domain bind to Caspase-9, forming an apoptotic complex. This complex can activate Caspase-9, which in turn triggers the hydrolysis and processing activation of apoptosis execution proteins Caspase-3 and Caspase-7 into their mature forms. Ultimately, this cascade leads to intrinsic cell death (Ketelut-Carneiro and Fitzgerald, 2022) (Figure 1).
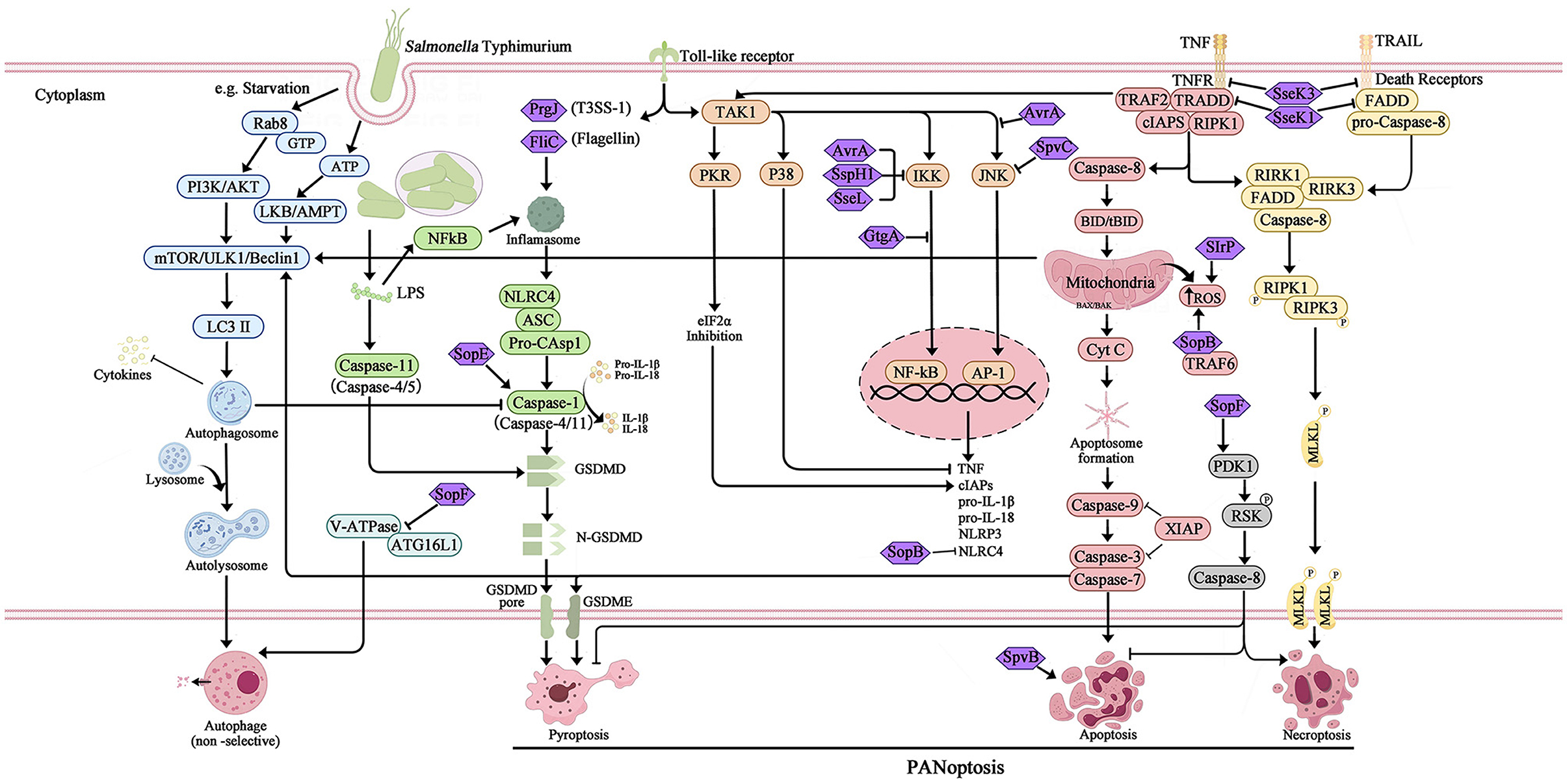
Figure 1. Overview of the different type of programmed cell death and the interference of T3SS effector proteins during Salmonella Typhimurium infection. The activation and inhibition of apoptosis, pyroptosis, necroptosis, and canonical autophagy are mediated by the SPI-1 and SPI-2 T3SS effector proteins, which interact with host innate immune pathways during Salmonella Typhimurium infection. The extensive overlap between apoptosis, pyroptosis, and necroptosis results in a phenomenon known as PANoptosis was also induced during Salmonella Typhimurium infection.
The foodborne pathogen Salmonella is acquired through the ingestion of contaminated food and subsequently enters the digestive tract. During the initial phase of Salmonella infection, the pathogen infiltrates intestinal epithelial cells by employing SPI-1 effectors, which induce alterations in intestinal folds and actin structures. This process facilitates the uptake of Salmonella by non-phagocytic cells. Alternatively, Salmonella may traverse M cells that cover the Peyer's patches, allowing entry into the lymphoid follicles and lamina propria, where they are captured by phagocytes such as dendritic cells and macrophages. Upon internalization by host cells and residing within specialized membrane-bound blebs known as Salmonella-containing vacuoles (SCVs), the expression of SPI-1 T3SS and its associated effectors is down-regulated, while SPI-2 is up-regulated (LaRock et al., 2015; Wemyss and Pearson, 2019). This regulatory shift promotes the formation of SCVs and facilitates the replication of Salmonella (Figure 2). Within these vacuoles, the bacteria employ the T3SS to deliver various effector proteins into the cytoplasm of phagocytes, thereby triggering Caspase-1 activation and subsequent cell death. The release of SipB leads to rapid cell death, exhibiting characteristics of both necrosis and apoptosis, along with the release of the proinflammatory cytokines IL-1β and IL-18 (Boise and Collins, 2001). The second mechanism primarily involves the activation of Toll-like receptor 4 (TLR4) by Salmonella through various components such as lipopolysaccharide (LPS) in the cell wall, intracellular TLR4 adapter proteins, and kinase pathways. This activation initiates a complex cascade of signal transduction, leading to the induction of both pro- and anti-apoptotic factors. Specifically, IKK and JNK activate the transcription factors NF-κB and AP-1, which are pivotal in the stimulation of pro-inflammatory cytokine genes, including TNF-α. Additionally, p38-MAPK and AKT play crucial roles in the induction of anti-apoptotic proteins (Guiney, 2005). On the other hand, the kinase PKR plays a role in facilitating Salmonella-induced apoptosis through its ability to impede protein synthesis via eIF2a phosphorylation. Additionally, PKR and type I interferons stimulate the activation of interferon response factor 3 (IRF3), which further triggers the production of pro-apoptotic factors within phagocytes (Hsu et al., 2004). The equilibrium between these apoptotic regulators ultimately dictates the fate of a cell, determining whether it will endure or undergo PCD. Notably, Caspase-3 assumes a pivotal role as the principal executor of these distinctive apoptotic alterations. However, Salmonella is known to produce several effector proteins that have the potential to influence the apoptotic pathway. Specifically, the SPI-1 T3SS facilitates the secretion of SptP, AvrA, and SspH1 (also secreted by SPI-2), which in turn promote apoptosis by inhibiting NF-κB, down-regulating MAPK signaling, or facilitating the secretion of SigD (SopB) to activate the AKT effect and delay apoptosis. Additionally, the SPI-2 T3SS mediates the secretion of SpvB, which leads to the depolymerization of the actin cytoskeleton in macrophages, thereby contributing to SPI-2-dependent apoptosis (Schleker et al., 2012; Wemyss and Pearson, 2019) (Figure 1).
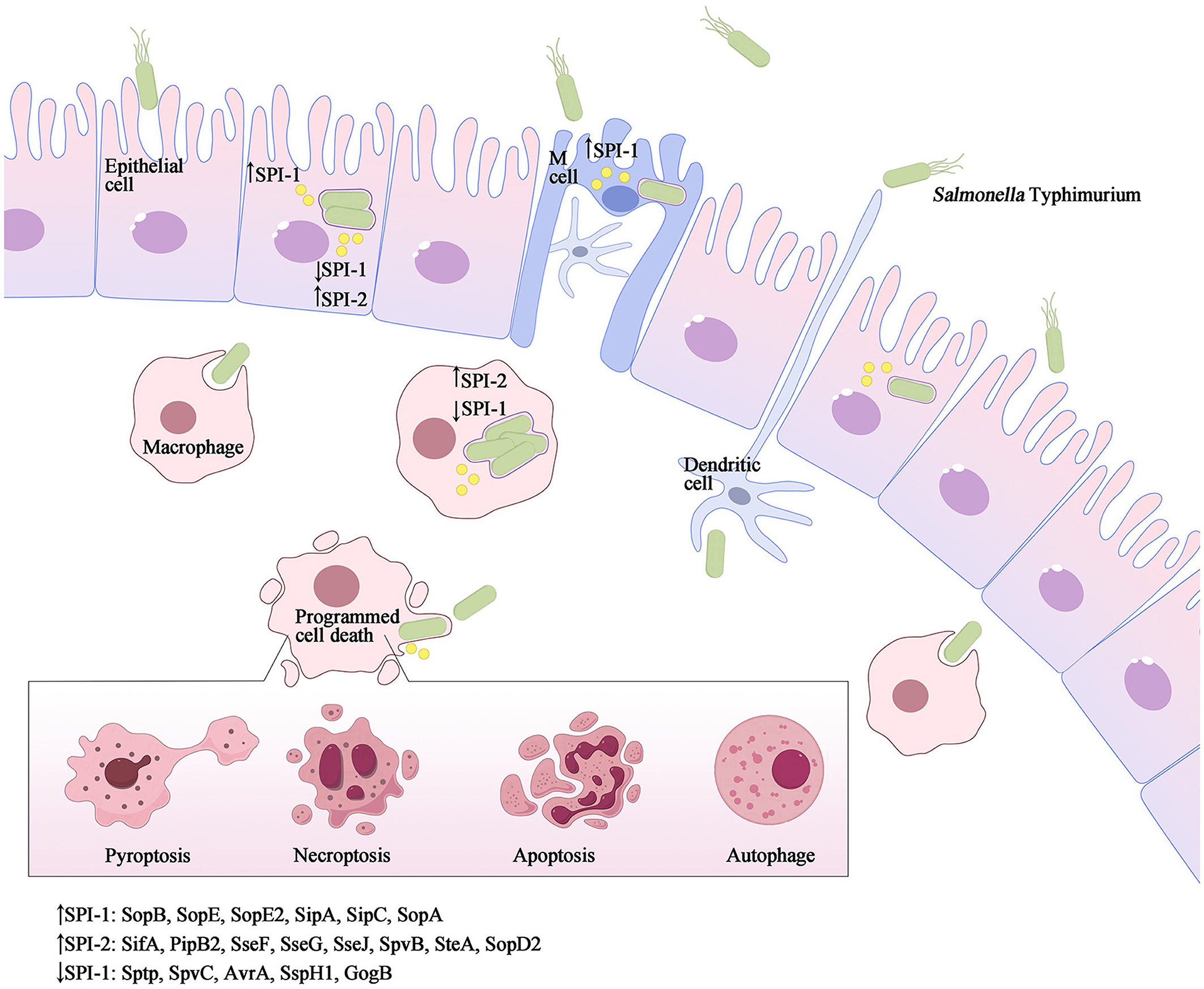
Figure 2. Salmonella Typhimurium invasion elicits various forms of programmed cell death. As a facultative intracellular pathogen, Salmonella Typhimurium can infect a range of cell types such as macrophages, dendritic cells, and epithelial cells. Salmonella Typhimurium accomplishes intestinal epithelium traversal by invading M-cells located above Peyer's patches, and it can also be captured by phagocytic cells such as dendritic cells or macrophages from the intestinal lumen. Once internalized, the SPI-1 T3SS and its effectors are downregulated, while SPI-2 is upregulated to promote the formation of Salmonella-containing vacuoles (SCVs) and facilitate Salmonella replication. Throughout the infection process, both SPI-1 and SPI-2 effector proteins interact with host innate immune pathways, either activating or inhibiting inflammatory responses and inducing different types of programmed cell death, such as apoptosis, pyroptosis, necroptosis, autophagy and others.
Pyroptotic cell death is characterized by the progressive enlargement of cells until the rupture of the plasma membrane, leading to the formation of a pore. This pore allows for the release of intracellular contents, including the cytokines IL-1β and IL-18, resulting in a robust inflammatory response (Ketelut-Carneiro and Fitzgerald, 2022). The classical pathway of pyroptosis is regulated by Caspase-1, which undergoes processing and activation within the inflammasomes. These inflammasomes are supramolecular complexes that consist of NLRs, typically featuring C-terminal leucine-rich repeat sequences (LRRs), and are responsible for sensing signals from bacteria (Frank and Vince, 2019). Furthermore, nucleotide-binding domain and leucine-rich repeat containing receptors (NLRs) possess a variable N-terminal region housing the pyrin structural domain (PYD). Upon stimulation of the host by signals emanating from bacterial infection, inducible pattern recognition receptors (PRRs) engage with pathogen-associated molecular patterns (PAMPs) and assemble an inflammasome alongside the pyroptosis adapter protein ASC. This assembly catalyzes the activation of Caspase-1, which subsequently cleaves the precursors of IL-1β and IL-18, resulting in the formation of biologically active inflammatory cytokines (Fink and Cookson, 2007). IL-1β is an endogenous pyrogen that elicits fever, stimulates the expression of cytokines and chemokines, and facilitates the migration of leukocytes. Additionally, IL-18 triggers the production of IFN-γ, which plays a crucial role in the activation of T cells and macrophages (Guiney, 2005). Furthermore, the activation of the inflammasome-associated Caspase-1 can cleave Gasdermin D (GSDMD), leading to the formation of structural domain proteins containing the active fragment of GSDMD-N (Fink and Cookson, 2007). This active fragment induces the perforation and rupture of the cell membrane, resulting in the rapid release of IL-1β and IL-18 into the extracellular environment, thereby instigating inflammatory responses and pyroptosis (Brokatzky and Mostowy, 2022) (Figure 3).
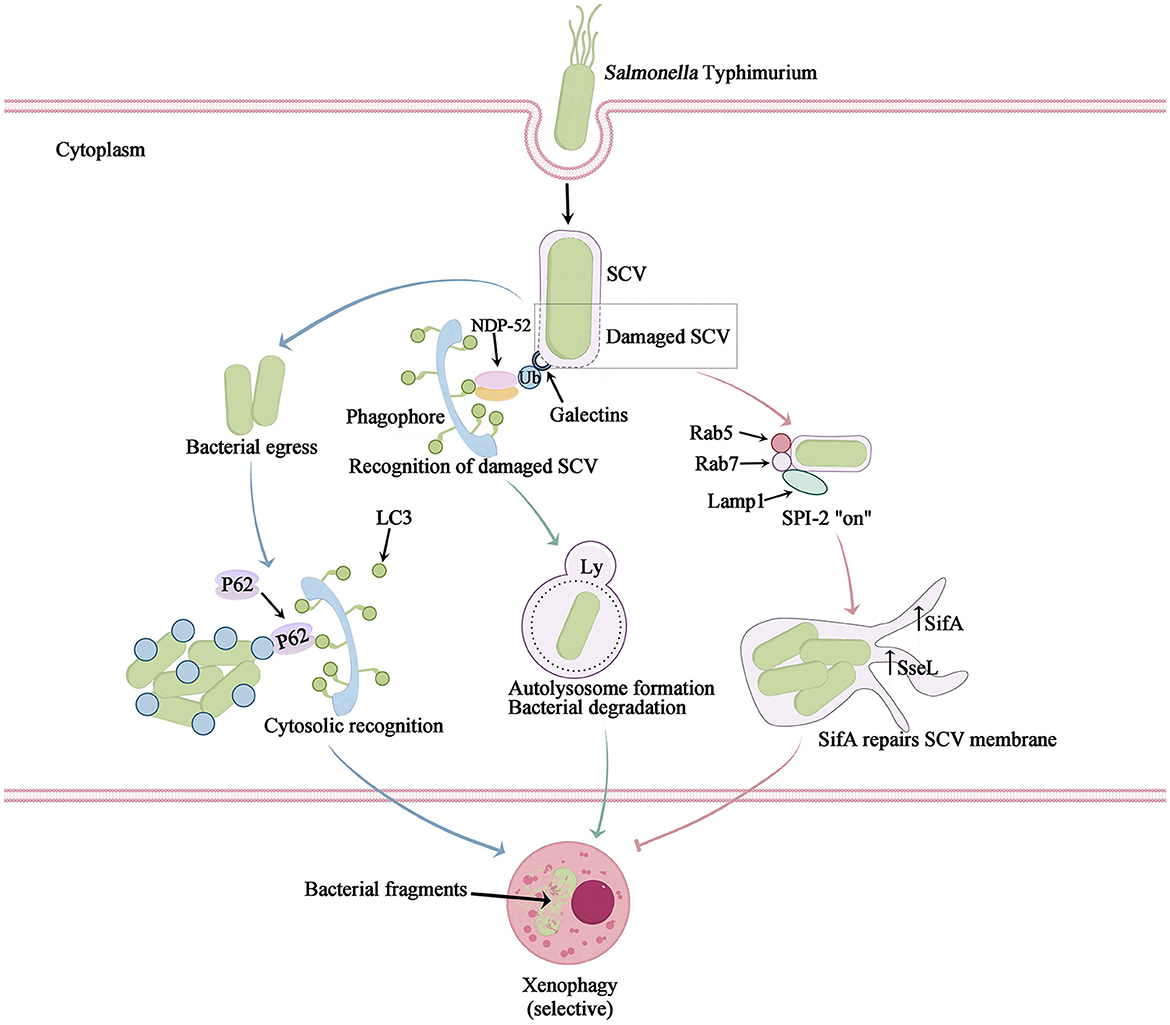
Figure 3. The mechanism of intracellular Salmonella Typhimurium-induced xenophagy. Salmonella Typhimurium infection not only causes cell nutrient deprivation or stimulation leading to non-selective autophagy, but also triggers selective autophagy to clear bacteria, also known as xenophagy. After their escape from a Salmonella-containing vacuole (SCV), Salmonella bacteria are marked with ubiquitin. Autophagy adaptors then bind to the ubiquitylated bacteria and link them to LC3-II on the initial autophagosome membrane (blue arrow). The SCV is damaged by T3SS-1, leading to the recruitment of galectin-8 to bind ubiquitin and the recruitment of NDP52, which connects LC3 and promotes autophagosome maturation. This process exposes internal proteins of the SCV membrane to the cytosol and causes sharp changes in ion concentration, potentially resulting in the formation of an ubiquitinated SCV membrane. The autophagic machinery recognizes this ubiquitinated membrane (green arrow). Autophagy facilitates the restoration of the SCV membrane damaged by T3SS-1 and subsequently induces acidification to generate SIFs, thereby enabling bacterial survival and replication within (pink arrow).
Pyroptosis serves as a vital innate immune response against Salmonella, influencing the progression of bacterial infection by inducing pyroptosis in both immune and non-immune cells. The activation of distinct inflammasomes in Salmonella-induced pyroptosis is contingent upon the virulence of the Salmonella strain and the response of the infected cell. During the Salmonella Typhimurium infection, the activation of host cell NLR family apoptosis inhibitory proteins (NAIP) occurs upon sensing of flagellin (FliC and FljB) or PrgJ (an SPI-1 secreted protein). These proteins subsequently interact with protein 4 (NLRC4), which possesses the NLR family Caspase recruitment domain (CARD), resulting in the formation of a complex with ASC. This complex then catalyzes the hydrolytic activation of Caspase-1, thereby mediating cellular pyroptosis (Fink and Cookson, 2007; Wemyss and Pearson, 2019). Additionally, NLRP3 (which detects increased ROS or K+ efflux) and pyrin (which senses inhibition of RhoA GTPase activity) can also induce pyroptosis through ASC Caspase-1 inflammasomes (Wemyss and Pearson, 2019). Both NLRC4 and NLRP3 are involved in the maturation of IL-1β and IL-18, as well as the induction of pyroptosis, in macrophages infected with Salmonella Typhimurium (Vladimer et al., 2013). In the non-classical pathway of pyroptosis, the LPS from Salmonella directly interacts with Caspase-4/5/11 in macrophages, leading to the activation of Caspase-4/5/11 (Knodler et al., 2014). This activated Caspase then cleaves GSDMD protein, triggering the activation of NLRP3 inflammasomes and subsequent activation of Caspase-1. This cascade ultimately results in the release of IL-1β, IL-18, and the occurrence of pyroptosis (Ma et al., 2021). Additionally, NLRC4 and the activation of non-classical inflammasomes are implicated in the response of epithelial cells to Salmonella infection, potentially contributing to the containment of bacterial dissemination within the intestinal mucosa.
Salmonella is known to generate various effector proteins that have the potential to impact the pyroptosis pathway. In the case of Salmonella Typhimurium-infected HeLa and RAW264.7 cells, as well as in vivo infection of mouse enterocytes, the Salmonella Typhimurium type III effector protein (SopE) was observed to stimulate the activation of host cell Rac1, which is a subunit of the Rho GTPase. This activation subsequently induces Caspase-1 activation and the secretion of IL-1β (Müller et al., 2009; Hoffmann et al., 2010). SopB has been found to be linked with the downregulation of NLRC4 in Salmonella Typhimurium-infected macrophages and B cells (Hu et al., 2017). The absence of NLRC4, a crucial inflammasome, hinders the pyroptosis response to Salmonella Typhimurium infection, thereby providing bacteria with an enhanced opportunity for replication prior to evading the host cell (García-Gil et al., 2018). SipB which interacts with SipC to form a translocon pore, facilitating SPI-1 effector translocation into the host cell is reportedly sufficient to induce Caspase-1-mediated “apoptosis” and IL-18 maturation in SipB transfected or Salmonella Typhimurium-infected (Hersh et al., 1999; van der Velden et al., 2003). Overall, Salmonella infection process is capable of activating different inflammasomes, causing cellular focalization and enhancing bacterial invasion and immune escape from the host cell.
Necrosis is characterized as a passive and unregulated form of cell death, whereas necroptosis adheres to intracellular signaling regulation, exhibiting Caspase activity-independent characteristics. Necroptosis is visually manifested through cell swelling, mitochondrial dysfunction, plasma rupture, and subsequent release of intracellular contents, which incites an inflammatory response in the surrounding tissue (Blériot and Lecuit, 2016). The canonical pathway of necroptosis, known as the tumor necrosis factor (TNF) signaling pathway, necessitates the participation of various kinases, including receptor-interacting serine/threonine protein kinase 1/3 (RIPK1/RIPK3)and mixed-lineage kinase domain-like protein (MLKL). Furthermore, the process of necroptosis necessitates the involvement of specialized receptors and ligands located on the plasma membrane, primarily mediated by tumor necrosis factor-α (TNF-α) and tumor necrosis factor receptor 1 (TNFR1). Upon ligation of TNF-α and TNFR1, RIPK1 and TNFR1-associated death domain protein (TRADD) are recruited, leading to the formation of a multimeric complex 1. This multimeric complex I comprises not only RIPK1 and TRADD, but also cellular inhibitors of apoptosis 1/2 (cIAP1/2), tumor necrosis factor receptor-associated factor 2/5 (TRAF2/5). Additionally, cIAP1/2 is capable of inducing the ubiquitination of RIPK1 and facilitating the up-regulation of anti-apoptotic genes. When the deubiquitinase CYLD deubiquitinates RIPK1, it results in a reduction in the stability of multimeric complex I. This reduction leads to the formation of a multimeric complex consisting of RIPK1, FADD, TRADD, RIPK3, and Caspase-8. The activation of multimeric complex II triggers the downstream signaling pathway of apoptosis. Caspase-8 then cleaves and inactivates RIPK1 and RIPK3, initiating the Caspase cascade reaction, ultimately resulting in cell apoptosis (Hu and Zhao, 2013; Wemyss and Pearson, 2019). However, bacterial pathogens hinder the activity of Caspase-8, preventing the cleavage and inactivation of RIPK1 and RIPK3 by Caspase-8. RIPK1 and RIPK3 form a complex and initiate phosphorylation, leading to the recruitment of the downstream necrosis execution protein MLKL and the subsequent formation of necrosomes (Hu and Zhao, 2013). Within these bodies, RIPK3 phosphorylates MLKL, resulting in the oligomerization and translocation of phosphorylated MLKL to the plasma membrane. At the plasma membrane, phosphorylated MLKL interacts with phosphatidylinositol phosphate (PIP), thereby increasing the permeability of the cytoplasmic membrane and facilitating the release of highly inflammatory damage-associated molecular patterns (DAMPs) into the extracellular space (Frank and Vince, 2019). This process ultimately leads to cell rupture and the induction of cell necroptosis (Figure 1).
During Salmonella Typhimurium infection, virulence effector proteins are involved in facilitating the host necroptotic response. Specifically, Salmonella secretes effector proteins SseK1 and SseK3, which glycosylate TNFR superfamily members and TRADD or FADD. This glycosylation process inhibits TNF-mediated NF-κB signaling and prevents cell death through apoptosis or necroptosis. Mass spectrometry analysis has identified TNFR1 and TRAIL-R as the glycosylation targets of SseK3, while TRADD is the preferred binding target of SseK1 (Günster et al., 2017; Xue et al., 2020). In addition, SopF was found could activate phosphoinositide-dependent protein kinase-1 (PDK1) to phosphorylate p90 ribosomal S6 kinase (RSK) which down-regulated Caspase-8 activation, resulting in inhibition of pyroptosis and apoptosis, but promotion of necroptosis (Yuan et al., 2023).
Both apoptosis and autophagy are activated in response to metabolic stress. Growth factor deprivation, limitation of nutrients and energy metabolism, activate the mTOR/ULK1/Beclin1 pathway, the multiple effects of some key proteins such as Caspase-3, Beclin1, and p53 lead to the complex interplay between these two pathways (Nikoletopoulou et al., 2013). However, Salmonella infection not only causes cell nutrient deprivation or stimulation leading to non-selective autophagy, but also triggers selective autophagy to clear bacteria, also known as xenophagy. Salmonella infection elicits non-selective autophagy, primarily triggered by amino acid deficiency, resulting in the suppression of the cell growth regulator mTOR. The PI3K-Akt-mTOR signaling pathway plays a pivotal role in this process (Miller et al., 2020) (Figure 1). Furthermore, xenophagy which is the activation of selective autophagy, is widely recognized as a promising strategy to combat bacterial infections. Xenophagy exerts its influence on Salmonella infection through three distinct pathways, two of which function as antibacterial defense mechanisms, effectively targeting invading Salmonella, eradicating pathogens, and subsequently safeguarding host cells. Additionally, Salmonella-infected cells employ an alternative pathway that not only facilitates the restoration of the damaged SCV, but also fosters an environment conducive to the replication and survival of the pathogen itself (Owen and Casanova, 2015; Zheng et al., 2022) (Figure 3). In the initial route, a portion of Salmonella bacteria possess the ability to evade this vacuole. Once these bacteria infiltrate the cytoplasm, they become tagged with unbound ubiquitin and undergo selective autophagy. Adapter proteins, namely SLRs (such as SQSTM1/p62, NDP52, and OPTN proteins), are recognized for their capacity to bind to ubiquitin-labeled bacteria. Additionally, these adapter proteins function as autophagy substrates, binding to LC3 in order to facilitate the mediation of autophagosomes. Specifically, it accomplishes this by means of the ubiquitin-binding domain and ubiquitin-labeled bacteria. It establishes a bond with characterized bacteria and engages with the autophagosome molecular marker Atg8/LC3 as well as the autophagy substrate SQSTM1/p62, thereby facilitating its association with the developing autophagosome. Consequently, it encapsulates Salmonella within the autophagosome and subsequently breaks down the bacteria. Another mechanism by which cells defend against Salmonella autophagy involves the recognition of damage to the SCV membrane caused by the pinhole apparatus of SPI-1 T3SS (Mostowy, 2013). In this scenario, Salmonella is found within the damaged SCV. The perforation of the SCV membrane, induced by SPI-1 T3SS, attracts various cytosolic components, including galectin-8. Galectin-8 serves as a marker protein for intact membrane structure and also acts as an inducer of autophagy. The galectin-8-labeled SCV then recruits LC3 and NDP52 to the cell membrane, thereby promoting the generation of autophagy signals. Consequently, this process leads to the autophagic capture and degradation of Salmonella (Wu et al., 2020).
Salmonella, being a parthenogenetic intracellular bacterium, has the ability to enhance its own survival through the induction of cellular autophagy. Unlike autophagy, which serves as a natural cellular immune defense mechanism, Salmonella can secrete certain effector proteins to evade fusion with lysosomes during autophagy, allowing for persistent proliferation within the SCV (Wang et al., 2018). For instance, Salmonella Typhimurium possesses a virulence plasmid that encompasses a conserved region of approximately 8 kb, which plays a crucial role in bacterial serum resistance, adhesion, and colonization. The spv region of the Salmonella plasmid virulence gene encompasses three genes associated with virulence, namely the transcriptional regulator spvR, as well as the structural genes spvB and spvC. Among these, spvB plays a crucial role in determining the extent of Salmonella proliferation within macrophages, while also inducing late apoptosis in host cells during Salmonella infection. The SPV proteins are essential for ADP-ribosyltransferase activity, which serves as an endotoxin and covalently modifies monomeric actin, thereby creating a favorable growth environment for Salmonella infection (Shintani and Klionsky, 2004; Xie et al., 2020). The upregulation of the autophagosome molecular marker Atg8/LC3 facilitates the repair and maturation of the SCV membrane. This process is accompanied by the recruitment of essential membrane-loading molecules Rab5 and Rab7, which ensure cellular maturation and enable the expression of the SPI-2 T3SS. Consequently, the effector protein filamentous body SifA is produced to maintain the integrity of the SCV membrane, further promoting SCV maturation and facilitating the replication of Salmonella within the host cell, thereby enhancing its survival (Birmingham and Brumell, 2006). During this temporal phase, the SPI-1 T3SS-mediated damage to the SCV leads to the liberation of ubiquitin and galectin-8 signaling molecules, thereby facilitating the internalization of crucial antimicrobial autophagic constituents (Owen and Casanova, 2015; Xie et al., 2020). Concurrently, the SPI-2 T3SS effector protein, SseL, is activated, culminating in the fulfillment of intracellular Salmonella replication through the reduction of autophagic fluxes. Consequently, this reduction instigates apoptosis in macrophages (Mesquita et al., 2012) (Figure 3).
In the initial stages of autophagic activation, the bacteria secrete effector proteins to impede the recognition of PAMPs, thereby evading cellular autophagy (Wang et al., 2018). During the process of autophagy, Salmonella fluid releases multiple effector proteins through either simulation or direct covalent modification of host proteins, thus facilitating their escape mechanisms (Table 2). Furthermore, Salmonella Typhimurium counteracts host SLRs and releases the effector protein SopA, which promotes the ubiquitination and subsequent degradation of TRIM56 and TRIM65, thereby suppressing IFN-γ-driven autophagy (Zhang et al., 2006). Salmonella Typhimurium has developed the membrane proteinase IcsP as a means of evading recognition by the C3 complement system and escaping cell-autonomous autophagy (Sorbara et al., 2018). The effector protein SrrB plays a role in disrupting the activation of the AMPK pathway, leading to the degradation of the AMPK/Sirt1/LBK1 complex. This disruption by SrrB results in an increase in mTOR levels and the inhibition of ULK complex formation (Ganesan et al., 2017). Recent studies have revealed that Salmonella effector proteins SseF and SseG disrupt the interaction between the guanine nucleotide exchange factor of GTPase Rab1A and transporter protein particle III, impede the assembly of the ULK1 complex, and reduce the synthesis of PI3P. These actions ultimately inhibit the nucleation of autophagosomes and membrane elongation. Additionally, it has been discovered that the Salmonella effector protein SopF can specifically modify V-ATPase, a crucial protein involved in sensing bacterial infection and recruiting autophagy protein ATG16L1 to initiate xenophagy. This alteration inhibits the V-ATPase-ATG16L1 pathway, thereby promoting the proliferation and spread of Salmonella within the host (Xu et al., 2019, 2022). In a typical physiological state, mannose-6-phosphate receptors (M6PR) facilitate the transportation of synthetic hydrolases from the trans-Golgi network (TGN) to endocytic lysosomes. However, the Salmonella-secreted effector SifA, in conjunction with the small guanosine triphosphatase Rab9, obstructs M6PR transport and reduces hydrolase activity (McGourty et al., 2012). Consequently, this enhances bacterial survival within the host cell.
Furthermore, the interplay between Salmonella effector proteins and the phenomenon of cross-regulation among different cell death pathways gives rise to the manifestation of hybrid forms of PCD. The interconnectedness of apoptosis and necroptosis pathways is primarily facilitated by the involvement of Caspase-8, whereby the activation of extrinsic death receptors mediated by the RIPK1/FADD/Caspase-8 complex can yield diverse signaling outcomes. In the absence of inhibition of RIPK or Caspase, the dominant mechanism of cell survival was through the activation of anti-apoptotic programs mediated by NF-κB. Suppression of RIPK resulted in the initiation of extrinsic apoptosis through Caspase-8 homodimerization. Conversely, inhibition of Caspase-8 facilitated necroptosis by promoting the oligomerization of RIPK1 and RIPK3 (Demarco et al., 2020). Necroptosis interacts with focal death signaling pathways via the effector protein MLKL. Additionally, the activation of cytoplasmic ZBP1 leads to the phosphorylation of MLKL, which in turn forms a pore in cellular membranes, including the plasma membrane. The activation of the NLRP3 inflammasome, triggered by subsequent K+ efflux mediated by MLKL, leads to the assembly of inflammasomes and cleavage of Caspase-1, resulting in the formation of GSDM-D pores and the release of IL-1β and IL-18 (Frank and Vince, 2019). Similarly, both apoptosis and pyroptosis involve the activation of Caspase proteases, which may have common evolutionary origins. As a result, these processes can interact at various levels. For example, the pyroptotic Caspase-1 protease can cleave the Bcl-2 family member Bid, leading to mitochondrial outer membrane permeabilization (MOMP) and subsequent activation of apoptotic signaling (Bock and Tait, 2020). Apoptosis-induced Caspase-8 has the capability to interact with ASC, resulting in Caspase-8-dependent pyroptotic activation (Fink and Cookson, 2007). Additionally, the transcription of NLRC4 and NLRP3 inflammasomes driven by apoptosis can facilitate Caspase-1 activation, leading to the hydrolysis of the pyroptotic execution protein GSDMD and subsequent formation of gasdermin pores, thereby inducing morphological characteristics associated with pyroptosis (Wemyss and Pearson, 2019). Emerging evidence indicates that autophagy and apoptosis can also engage in interactions, exhibiting antagonistic or cooperative effects, thereby exerting distinct influences on cellular destiny. Regulators involved in the interplay between autophagy and apoptosis have been identified, including the Bcl-2 protein family, Caspases, Beclin1, NF-κB, and certain microRNAs. These regulators exhibit dual coordinated roles at the transcriptional level. Additionally, kinase signaling pathways such as JNK and PI3K/Akt/mTOR have been demonstrated to play a significant role in mediating the interaction between autophagy and apoptosis (Wemyss and Pearson, 2019; Snyder and Oberst, 2021). Notably, recent research has revealed a significant finding that Salmonella infection has the capability to induce PANoptosis, a complex interplay involving apoptosis, pyroptosis, and necroptosis. Moreover, it has been discovered that the Salmonella effector SopF plays a crucial role in regulating the PANoptosis of intestinal epithelial cells, thereby exacerbating systemic infection (Christgen et al., 2020; Yuan et al., 2023). The intricate molecular network and the pleiotropic nature displayed by these regulators are expected to be influenced by the cellular milieu and diverse downstream targets. Alternatively, inhibition of specific proteins in the pathway may activate multiple PCD pathways, thereby enabling the organism to accomplish its defense objectives through alternative mechanisms.
During Salmonella infection, bacteria manipulate PCD in immune and non-immune cells through multiple mechanisms, including interference with cell signaling pathways and regulation of cellular metabolism. The interaction between these pathways of cell death implies that, similar to numerous immune pathways, cell death signals have undergone evolutionary adaptation and diversification in response to the selective pressure imposed by Salmonella infection. The flexible utilization and interconnectedness of various cell death pathways serve as a defense mechanism against intracellular infection. Through extensive research on the mechanism of action of Salmonella and PCD, our understanding of the intricate complexity and diverse nature of Salmonella and cell programmed has significantly advanced. However, numerous unanswered questions persist. For instance, which PCD pathway is predominantly activated during Salmonella infection? How do the multiple virulence factors produced by Salmonella synchronize and regulate these PCD mechanisms? What are the principal regulatory effector proteins implicated in these processes? Do effector proteins possess comparable sites for bulk activation or inhibition, facilitating precise regulation of PCD processes through ubiquitination modification? Furthermore, what is the safety profile of clinical drugs targeting PCD in the treatment of Salmonella infections, considering the escalating issue of antibiotic resistance? In an era of increasing antibiotic resistance, it is imperative to comprehend the significance of PCD within the innate immune system, as well as to unravel the intricate molecular mechanisms employed by bacteria to evade and exploit PCD. This comprehension is of utmost importance in the regulation of bacterial infections through PCD, the identification of novel drug targets for intracellular bacterial infections, and the formulation of innovative strategies for the prevention and control of bacterial proliferation.
YZ: Supervision, Writing—original draft. MX: Investigation, Writing—original draft. YG: Visualization, Writing—review & editing. LC: Writing—review & editing. WV: Supervision, Writing—review & editing. QX: Conceptualization, Writing—review & editing. LL: Formal analysis, Investigation, Writing—review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Funds of China (32372863), Jiangsu Province Agricultural Science and Technology Independent Innovation Fund Project (CX (22) 3033), and State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products (2021DG700024-KF202KF202319).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Bernal-Bayard, J., Cardenal-Munoz, E., and Ramos-Morales, F. (2010). The Salmonella type III secretion effector, Salmonella leucine-rich repeat protein (SlrP), targets the human chaperone ERdj3. J. Bio. Chem. 285, 16360–16368. doi: 10.1074/jbc.M110.100669
Birmingham, C. L., and Brumell, J. H. (2006). Autophagy recognizes intracellular Salmonella enterica serovar Typhimurium in damaged vacuoles. Autophagy 2, 156–158. doi: 10.4161/auto.2825
Blériot, C., and Lecuit, M. (2016). The interplay between regulated necrosis and bacterial infection. Cell. Mol. Life Sci. 73, 2369–2378. doi: 10.1007/s00018-016-2206-1
Bock, F. J., and Tait, S. W. G. (2020). Mitochondria as multifaceted regulators of cell death. Nat. Rev. Mol. Cell Biol. 21, 85–100. doi: 10.1038/s41580-019-0173-8
Boise, L. H., and Collins, C. M. (2001). Salmonella-induced cell death: apoptosis, necrosis or programmed cell death? Trends Microbiol. 9, 64–67. doi: 10.1016/S0966-842X(00)01937-5
Brokatzky, D., and Mostowy, S. (2022). Pyroptosis in host defence against bacterial infection. Dis. Model. Mech. 15, 414. doi: 10.1242/dmm.049414
Browne, S. H., Lesnick, M. L., and Guiney, D. G. (2002). Genetic requirements for Salmonella-induced cytopathology in human monocyte-derived macrophages. Infect. Immun. 70, 7126–7135. doi: 10.1128/IAI.70.12.7126-7135.2002
Christgen, S., Zheng, M., Kesavardhana, S., Karki, R., Malireddi, R. K. S., Banoth, B., et al. (2020). Identification of the PANoptosome: a molecular platform triggering pyroptosis, apoptosis, and necroptosis (PANoptosis). Front. Cell. Infect. Microbiol. 10, 237. doi: 10.3389/fcimb.2020.00237
Collier-Hyams, L. S., Zeng, H., Sun, J., Tomlinson, A. D., Bao, Z. Q., Chen, H., et al. (2002). Cutting edge: Salmonella AvrA effector inhibits the key proinflammatory, anti-apoptotic NF-κB pathway. J. Immunol. 169, 2846–2850. doi: 10.4049/jimmunol.169.6.2846
D'Arcy, M. S. (2019). Cell death: a review of the major forms of apoptosis, necrosis and autophagy. Cell Biol. Int. 43, 582–592. doi: 10.1002/cbin.11137
Demarco, B., Chen, K. W., and Broz, P. (2020). Cross talk between intracellular pathogens and cell death. Immunol. Rev. 297, 174–193. doi: 10.1111/imr.12892
Feng, Z. Z., Jiang, A. J., Mao, A. W., Feng, Y. H., Wang, W. N., Li, J. J., et al. (2018). The Salmonella effectors SseF and SseG inhibit Rab1A-mediated autophagy to facilitate intracellular bacterial survival and replication. J. Biol. Chem. 293, 9662–9673. doi: 10.1074/jbc.M117.811737
Fink, S. L., and Cookson, B. T. (2007). Pyroptosis and host cell death responses during Salmonella infection. Cell Microbiol. 9, 2562–2570. doi: 10.1111/j.1462-5822.2007.01036.x
Frank, D., and Vince, J. E. (2019). Pyroptosis versus necroptosis: similarities, differences, and crosstalk. Cell Death Differ. 26, 99–114. doi: 10.1038/s41418-018-0212-6
Ganesan, R., Hos, N. J., Gutierrez, S., Fischer, J., Stepek, J. M., Daglidu, E., et al. (2017). Salmonella typhimurium disrupts Sirt1/AMPK checkpoint control of mTOR to impair autophagy. PLoS Pathog. 13, e1006227. doi: 10.1371/journal.ppat.1006227
García-Gil, A., Galán-Enríquez, C. S., Pérez-López, A., Nava, P., Alpuche-Aranda, C., Ortiz-Navarrete, V., et al. (2018). SopB activates the Akt-YAP pathway to promote Salmonella survival within B cells. Virulence 9, 1390–1402. doi: 10.1080/21505594.2018.1509664
Grassme, H., Jendrossek, V., and Gulbins, E. (2001). Molecular mechanisms of bacteria induced apoptosis. Apoptosis 6, 441–445. doi: 10.1023/A:1012485506972
Guiney, D. G. (2005). The role of host cell death in Salmonella infections. Curr. Top Microbiol. Immunol. 289, 131–150. doi: 10.1007/3-540-27320-4_6
Günster, R. A., Matthews, S. A., Holden, D. W., and Thurston, T. L. M. (2017). SseK1 and SseK3 type III secretion system effectors inhibit NF-κB signaling and necroptotic cell death in Salmonella-infected macrophages. Infect. Immun. 85, e00010–17. doi: 10.1128/IAI.00010-17
Hersh, D., Monack, D. M., Smith, M. R., Ghori, N., Falkow, S., Zychlinsky, A., et al. (1999). The Salmonella invasin SipB induces macrophage apoptosis by binding to Caspase-1. Proc. Natl. Acad. Sci. U.S.A. 96, 2396–2401. doi: 10.1073/pnas.96.5.2396
Hoffmann, C., Galle, M., Dilling, S., Käppeli, R., Müller, A. J., Songhet, P., et al. (2010). In macrophages, Caspase-1 activation by SopE and the type III secretion system-1 of S. Typhimurium can proceed in the absence of flagellin. PLoS ONE 5, e12477. doi: 10.1371/journal.pone.0012477
Hsu, L. C., Park, J. M., Zhang, K. Z., Luo, J. L., Maeda, S., Kaufman, R. J., et al. (2004). The protein kinase PKR is required for macrophage apoptosis after activation of Toll-like receptor 4. Nature 428, 341–345. doi: 10.1038/nature02405
Hu, G. Q., Song, P. X., Chen, W., Qi, S., Yu, S. X., Du, C. T., et al. (2017). Cirtical role for Salmonella effector SopB in regulating inflammasome activation. Mol. Immunol. 90, 280–286. doi: 10.1016/j.molimm.2017.07.011
Hu, Z. Q., and Zhao, W. H. (2013). Type 1 interferon-associated necroptosis: a novel mechanism for Salmonella enterica Typhimurium to induce macrophage death. Cell. Mol. Immunol. 10, 10–12. doi: 10.1038/cmi.2012.54
Jiao, Y., Zhang, Y. G., Lin, Z. J., Lu, R., Xia, Y. L., Meng, C., et al. (2020). Salmonella Enteritidis effector AvrA suppresses autophagy by reducing Beclin-1 protein. Front. Immunol. 11, 686. doi: 10.3389/fimmu.2020.00686
Jones, R. M., Wu, H. X., Wentworth, C., Luo, L. P., Collier-Hyams, L., Neish, A. S., et al. (2008). Salmonella AvrA coordinates suppression of host immune and apoptotic defenses via JNK pathway blockade. Cell Host Microbe. 3, 233–244. doi: 10.1016/j.chom.2008.02.016
Kamanova, J., Sun, H., Lara-Tejero, M., and Galán, J. E. (2016). The Salmonella effector protein SopA modulates innate immune responses by targeting TRIM E3 ligase family members. PLoS Pathog. 12, e1005552. doi: 10.1371/journal.ppat.1005552
Ketelut-Carneiro, N., and Fitzgerald, K. A. (2022). Apoptosis, pyroptosis, and necroptosis-oh my! The many ways a cell can die. J Mol Biol. 434, 167378. doi: 10.1016/j.jmb.2021.167378
Knodler, L.eigh A., Crowley, S.hauna M., Sham, H.o P., Yang, H., Wrande, M., Ma, C., et al. (2014). Noncanonical inflammasome activation of caspase-4/caspase-11 mediates epithelial defenses against enteric bacterial pathogens. Cell Host Microbe. 16, 249–256. doi: 10.1016/j.chom.2014.07.002
LaRock, D. L., Chaudhary, A., and Miller, S. I. (2015). Salmonellae interactions with host processes. Nat. Rev. Microbiol. 13, 191–205. doi: 10.1038/nrmicro3420
Lesnick, M. L., Reiner, N. E., Fierer, J., and Guiney, D. G. (2001). The Salmonella spvB virulence gene encodes an enzyme that ADP-ribosylates actin and destabilizes the cytoskeleton of eukaryotic cells. Mol. Microbiol. 39, 1464–1470. doi: 10.1046/j.1365-2958.2001.02360.x
Luis, L. B., Ana, G. T., Carlos, G. E., Abraham, G. G., Iris, E. G., Martha, M. L., et al. (2022). Salmonella promotes its own survival in B cells by inhibiting autophagy. Cells 11, 2061. doi: 10.3390/cells11132061
Ma, X. Y., Li, Y. J., Shen, W. X., Oladejo, A. O., Yang, J., Jiang, W., et al. (2021). LPS mediates bovine endometrial epithelial cell pyroptosis directly through both NLRP3 classical and non-classical inflammasome pathways. Front. Immunol. 12, 676088. doi: 10.3389/fimmu.2021.676088
McGourty, K., Thurston, T. L., Matthews, S. A., Pinaud, L., Mota, L. J., Holden, D. W., et al. (2012). Salmonella inhibits retrograde trafficking of mannose-6-phosphate receptors and lysosome function. Science 338, 963–967. doi: 10.1126/science.1227037
Mesquita, F. S., Thomas, M., Sachse, M., Santos, A. J. M., Figueira, R., Holden, D. W., et al. (2012). The Salmonella deubiquitinase SseL inhibits selective autophagy of cytosolic aggregates. PLoS Pathog. 8, e1002743. doi: 10.1371/journal.ppat.1002743
Miller, D. R., Cramer, S. D., and Thorburn, A. (2020). The interplay of autophagy and non-apoptotic cell death pathways. Cell Death Dis. 352, 159–187. doi: 10.1016/bs.ircmb.2019.12.004
Mostowy, S. (2013). Autophagy and bacterial clearance: a not so clear picture. Cell Microbiol. 15, 395–402. doi: 10.1111/cmi.12063
Müller, A. J., Hoffmann, C., Galle, M., Van Den Broeke, A., Heikenwalder, M., Falter, L., et al. (2009). The S. typhimurium effector SopE induces Caspase-1 activation in stromal cells to initiate gut inflammation. Cell Host Microbe 6, 125–136. doi: 10.1016/j.chom.2009.07.007
Nikoletopoulou, V., Markaki, M., Palikaras, K., and Tavernarakis, N. (2013). Crosstalk between apoptosis, necrosis and autophagy. Biochim. Biophys. Acta. 1833, 3448–3459. doi: 10.1016/j.bbamcr.2013.06.001
Owen, K. A., and Casanova, J. E. (2015). Salmonella manipulates autophagy to “serve and protect”. Cell Host Microbe. 18, 517–519. doi: 10.1016/j.chom.2015.10.020
Paesold, G., Guiney, D. G., Eckmann, L., and Kagnoff, M. F. (2002). Genes in the Salmonella pathogenicity island 2 and the Salmonella virulence plasmid are essential for Salmonella-induced apoptosis in intestinal epithelial cells. Cell. Microbiol. 4, 771–781. doi: 10.1046/j.1462-5822.2002.00233.x
Rao, S., Schieber, A. M. P., O'Connor, C. P., Leblanc, M., Michel, D., Ayres, J. S., et al. (2017). Pathogen-mediated inhibition of anorexia promotes host survival and transmission. Cell 168, 503–516. doi: 10.1016/j.cell.2017.01.006
Rayamajhi, M., Zak, D. E., Chavarria-Smith, J., Vance, R. E., and Miao, E. A. (2013). Cutting edge: mouse NAIP1 detects the type III secretion system needle protein. J. Immunol. 191, 3986–3989. doi: 10.4049/jimmunol.1301549
Ruan, H. H., Li, Y., Zhang, X. X., Liu, Q., Ken, H., Zhang, K. S., et al. (2014). Identification of TRAF6 as a ubiquitin ligase engaged in the ubiquitination of SopB, a virulence effector protein secreted by Salmonella typhimurium. Biochem. Biophys. Res. Commun. 447, 172–177. doi: 10.1016/j.bbrc.2014.03.126
Ruan, H. H., Zhang, Z., Tian, L., Wang, S. Y., Hu, S. Y., Qiao, J. J., et al. (2016). The Salmonella effector SopB prevents ROS-induced apoptosis of epithelial cells by retarding TRAF6 recruitment to mitochondria. Biochem. Biophys. Res. Commun. 478, 618–623. doi: 10.1016/j.bbrc.2016.07.116
Schleker, S., Sun, J., Raghavan, B., Srnec, M., Muller, N., Koepfinger, M., et al. (2012). The current Salmonella-host interactome. Proteomics Clin Appl 6, 117–133. doi: 10.1002/prca.201100083
Shintani, T., and Klionsky, D. J. (2004). Autophagy in health and disease: a double-edged sword. Science 306, 990–995. doi: 10.1126/science.1099993
Snyder, A. G., and Oberst, A. (2021). The antisocial network: cross talk between cell death programs in host defense. Annu. Rev. Immunol. 39, 77–101. doi: 10.1146/annurev-immunol-112019-072301
Sorbara, M. T., Foerster, E. G., Tsalikis, J., Abdel-Nour, M., Mangiapane, J., Sirluck-Schroeder, I., et al. (2018). Complement C3 drives autophagy-dependent restriction of cyto-invasive bacteria. Cell Host Microbe 23, 644–652. doi: 10.1016/j.chom.2018.04.008
Stringer, J. M., Alesi, L. R., Winship, A. L., and Hutt, K. J. (2023). Beyond apoptosis: evidence of other regulated cell death pathways in the ovary throughout development and life. Hum Reprod. Update 29, 434–456. doi: 10.1093/humupd/dmad005
van der Velden, A. W. M., Velasquez, M., and Starnbach, M. N. (2003). Salmonella rapidly kill dendritic cells via a Caspase-1-dependent mechanism. J. Immunol. 171, 6742–6749. doi: 10.4049/jimmunol.171.12.6742
Vladimer, G. I., Marty-Roix, R., Ghosh, S., Weng, D., and Lien, E. (2013). Inflammasomes and host defenses against bacterial infections. Curr. Opin. Microbiol. 16, 23–31. doi: 10.1016/j.mib.2012.11.008
Wang, L. D., Yan, J., Niu, H., Huang, R., and Wu, S. Y. (2018). Autophagy and ubiquitination in Salmonella infection and the related inflammatory responses. Front. Cell. Infect. Microbiol. 8, 78. doi: 10.3389/fcimb.2018.00078
Wemyss, M. A., and Pearson, J. S. (2019). Host cell death responses to non-typhoidal Salmonella infection. Front. Immunol. 10, 1758. doi: 10.3389/fimmu.2019.01758
Wu, S., Shen, Y. R., Zhang, S., Xiao, Y. Q., and Shi, S. R. (2020). Salmonella interacts with autophagy to offense or defense. Front. Microbiol. 11, 721. doi: 10.3389/fmicb.2020.00721
Xie, Z., Zhang, Y., and Huang, X. (2020). Evidence and speculation: the response of Salmonella confronted by autophagy in macrophages. Future Microbiol. 15, 1277–1286. doi: 10.2217/fmb-2020-0125
Xu, Y., Cheng, S., Zeng, H., Zhou, P., Ma, Y., Li, L., et al. (2022). ARF GTPases activate Salmonella effector SopF to ADP-ribosylate host V-ATPase and inhibit endomembrane damage-induced autophagy. Nat. Struct. Mol. Biol. 29, 67–77. doi: 10.1038/s41594-021-00710-6
Xu, Y., Zhou, P., Cheng, S., Lu, Q., Nowak, K., Hopp, A. K., et al. (2019). A bacterial effector reveals the V-ATPase-ATG16L1 axis that initiates xenophagy. Cell 178, 552–566.e520. doi: 10.1016/j.cell.2019.06.007
Xue, J., Hu, S., Huang, Y., Zhang, Q., Yi, X., Pan, X., et al. (2020). Arg-GlcNAcylation on TRADD by NleB and SseK1 is crucial for bacterial pathogenesis. Front. Cell Dev. Biol. 8, 641. doi: 10.3389/fcell.2020.00641
Yang, Z., Soderholm, A., Lung, T. W. F., Giogha, C., Hill, M. M., Brown, N. F., et al. (2015). SseK3 is a Salmonella effector that binds TRIM32 and modulates the host's NF-κB signalling activity. PLoS ONE 10, e0138529. doi: 10.1371/journal.pone.0138529
Ye, Z. D., Petrof, E. O., Boone, D., Claud, E. C., and Sun, J. (2007). Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. Am. J. Pathol. 171, 882–892. doi: 10.2353/ajpath.2007.070220
Yuan, H. B., Zhou, L. T., Chen, Y. L., You, J. Y., Hu, H. Y., Li, Y. Y., et al. (2023). Salmonella effector SopF regulates PANoptosis of intestinal epithelial cells to aggravate systemic infection. Gut Microbes 15, 2180315. doi: 10.1080/19490976.2023.2180315
Zhang, Y., Higashide, W. M., McCormick, B. A., Chen, J., and Zhou, D. G. (2006). The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol. Microbiol. 62, 786–793. doi: 10.1111/j.1365-2958.2006.05407.x
Zheng, L., Wei, F., and Li, G. L. (2022). The crosstalk between bacteria and host autophagy: host defense or bacteria offense. J. Microbiol. 60, 451–460. doi: 10.1007/s12275-022-2009-z
Zouhir, S., Bernal-Bayard, J., Cordero-Alba, M., Cardenal-Muñoz, E., Guimaraes, B., Lazar, N., et al. (2014). The structure of the Slrp-Trx1 complex sheds light on the autoinhibition mechanism of the type III secretion system effectors of the NEL family. Biochem. J. 464, 135–144. doi: 10.1042/BJ20140587
Keywords: programmed cell death, salmonella, apoptosis, pyroptosis, necroptosis, autophagy
Citation: Zhang Y, Xu M, Guo Y, Chen L, Vongsangnak W, Xu Q and Lu L (2024) Programmed cell death and Salmonella pathogenesis: an interactive overview. Front. Microbiol. 14:1333500. doi: 10.3389/fmicb.2023.1333500
Received: 06 November 2023; Accepted: 12 December 2023;
Published: 04 January 2024.
Edited by:
Sébastien Holbert, INRA Centre Val de Loire, FranceReviewed by:
Qiuhe Lu, Cleveland Clinic, United StatesCopyright © 2024 Zhang, Xu, Guo, Chen, Vongsangnak, Xu and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qi Xu, eHVxaUB5enUuZWR1LmNu; Lizhi Lu, bHVsaXpoaWJveEAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.