- Department of Plant Sciences, University of Hyderabad, Hyderabad, India
Papaya ringspot virus (PRSV) is a devastating Potyvirus that causes papaya ringspot disease in Carica papaya plantations globally. In this study, the complete genome sequence of a PRSV isolate from Shankarpalli, Telangana, India, was reported and designated as PRSV-HYD (KP743981.1). The genome is a single-stranded positive-sense RNA comprising 10,341 nucleotides. Phylogenetic analysis revealed that PRSV-HYD is closely related to PRSV Pune (Aundh) isolate with 92 and 95% nucleotide and amino acid sequence identity, respectively. To develop infectious cDNA (icDNA), the complete nucleotide sequence of PRSV-HYD was cloned between the right and left borders in the binary vector pCB301 using BglII and XmaI restriction sites. Cauliflower mosaic virus (CaMV) double promoter (35S) was fused at the 5′-end and Avocado sunblotch viroid (ASBVd) ribozyme (RZ) sequence was fused to the 3′ end to generate an authentic 3′ viral end in the transcribed mRNAs. The icDNA generated was mobilized into the Agrobacterium tumefaciens EHA 105, and the agrobacterial cultures were infiltrated into the natural host C. papaya and a non-host Nicotiana benthamiana plants; both did not show any symptoms. In RT-PCR analysis of RNAs isolated from N. benthamiana, we could detect viral genes as early as 3 days and continued up to 28 days post infiltration. Alternatively, virion particles were purified from agroinfiltrated N. benthamiana plants and introduced into C. papaya by mechanical inoculation as well as by pinprick method. In both cases, we could see visible systemic symptoms similar to that of wild type by 40 days. Additionally, we studied the expression patterns of the genes related to plant defense, transcription factors (TFs), and developmental aspects from both C. papaya and N. benthamiana.
1 Introduction
Carica papaya is an extensively cultivated, highly valued fruit crop due to its manifold nutritional and medicinal advantages (de Oliveira and Vitória, 2011). According to the FAO statistical database, 2020, India is the highest papaya-producing country in the world, with an annual yield of 5.7 million metric tonnes. However, like most crops, C. papaya farming is vulnerable to several pathogens, including bacteria, fungi, and viruses. Various viral pathogens from diverse families, such as Potyviridae, Alphaflexiviridae, Geminiviridae, Tospoviridae, and Solemoviridae, are commonly known to infect C. papaya plants. The Potyviridae family alone has 244 plant viruses classified into 12 genera, with most species belonging to the Potyvirus genus (Inoue-Nagata et al., 2022) infecting several economically significant crops, such as banana, beans, peanuts, chili, maize, watermelon, papaya, potato, and tobacco.
Papaya ringspot disease (PRSD) is the most destructive viral disease affecting C. papaya plants worldwide, as it poses a significant threat to the economies of the countries that produce papaya (Tripathi et al., 2008). The causative agent, Papaya ringspot virus (PRSV), is a Potyvirus, that can infect its host at any stage of growth in a systematic manner, resulting in a severe chlorotic and mosaic pattern on leaves, water-soaked streaks on leaf petioles and trunks, deformed fruits with ring-like spots, distorted and shoestring-like appearance of leaves along with reduced photosynthetic efficiency (Gonsalves et al., 2010). PRSV can infect Caricaceae, Chenopodiaceae, and Cucurbitaceae family members experimentally. Based on the host range, it has two subtypes: PRSV-P and PRSV-W (Yeh, 1984). More than 24 species of aphids are involved in the transmission of PRSV in a non-persistent manner, with Myzus persicae, Aphis gossypii, and A. craccivora being the most efficient ones (Kalleshwaraswamy and Kumar, 2008). The PRSV genome is comprised of a 10.3 kb positive-sense single-stranded RNA containing a single open reading frame (ORF), which encodes a single large polyprotein and a ribosomal frameshifting product PIPO (Chung et al., 2008; Gonsalves et al., 2010). The polyprotein gets cleaved into 10 different mature proteins by three of its own proteases: P1 pro, HC-Pro, and NIa-Pro (Yeh et al., 1992; Tripathi et al., 2008).
Infectious cDNA clones of plant viruses have been proven to be highly effective tools for the confirmation of Koch’s postulates, reverse genetic approaches, selecting plant in-breeding programs, and investigating the intricate interaction between viruses and their hosts (Navas-Hermosilla et al., 2021). This approach offers invaluable insights into the viral life cycle and pathogenesis, ultimately facilitating a better understanding of such processes (Boyer and Haenni, 1994). Since the inception of the first infectious clone of the Brome mosaic virus (BMV) (Ahlquist et al., 1984), agrobacterium-mediated icDNAs have been engineered for several plant viruses, paved the way for molecular manipulations and functional characterization studies in planta and greatly expanded the scope of research in this field (Díaz-Cruz et al., 2018).
The study of plant viruses has been primarily directed toward their detrimental impact on crop plants (Anderson et al., 2004; Nelson and Citovsky, 2005). To safeguard themselves from these threats, plants employ pre-existing defense mechanisms to recognize elicitor(s) linked to the attacker and initiate the appropriate defense responses (Zhou and Zhang, 2020). However, many viruses encode RNA-silencing suppressor proteins to prevent the recognition of small interfering RNA (siRNA), inhibiting the RNA-silencing pathway (Zhao et al., 2016). Plant hypersensitive response against abiotic stresses includes the formation of reactive oxygen species (ROS), induction of salicylic acid (SA) and jasmonic acid (JA) signaling, and PR gene response (Akbudak et al., 2020). Transcription factors (TFs) are highly sought after for genetic engineering because they regulate stress-related genes (Baillo et al., 2019). Manipulating TFs has become a popular research topic, as many respond to stress and control several downstream genes, making them promising candidates for improving plant stress tolerance (Hoang et al., 2017). Many TF families, such as MYB, bZIP, NAC, ERF, and many more, have been identified to be involved in biotic stress response in plants (Ng et al., 2018).
Viral infections significantly impact plant growth, regenerative ability, and physiological metabolism (Jiang and Zhou, 2023). A recent study highlighted the crucial role of cytokinin(s) in regulating the transcriptional expression of downstream genes during various stages of leaf development (Wu et al., 2021). Jasmonates (JA) is another type of phytohormone that regulates defenses against herbivores and pathogens and plays a crucial role in plant development (Goossens et al., 2016). When viruses invade plants, they undergo various physiological and molecular changes, including stunted growth, poorly developed leaves, chlorophyll degradation and consequently leaf senescence, programmed cell death (PCD), and autophagy (Espinoza et al., 2007; Park et al., 2007; Tang and Bassham, 2018; Wu et al., 2021).
The unavailability of a significant number of complete PRSV genome sequences from different geographical regions of India has been a substantial limitation in understanding the molecular basis of papaya ringspot disease severity at the national and global levels. It is essential to surveil the circulating PRSV strains to keep track of the genotypic and phenotypic traits of the host plant. A comparative study regarding the agroinfectivity of the infectious cDNA in the natural host and non-host plants was conducted to monitor plant response(s) based on the expression change of essential plant genes and TFs.
2 Materials and methods
2.1 Field survey and virus isolate
During field surveys, a papaya orchard was identified at the Shankarpalli mandal (17° 28′ 7.04″ N, 78° 7′ 54.16″ E) of Rangareddy district, Telangana, India, where the leaves of several plants appeared yellowish, distorted and shoestring-like. The leaf samples from different plants of that orchard were collected and brought to the laboratory for further analysis. The presence of PRSV was confirmed by performing DAC-ELISA and RT-PCR analysis (Supplementary Figure S1A). Based on these analyses, one sample (Shankarpalli 4) was chosen for further analysis.
2.2 Total RNA extraction, cDNA synthesis, and cloning of cDNA fragments
Total RNA was isolated from the infected leaf samples using TRIzol® reagent (Invitrogen) following the manufacturer’s instructions. First-strand cDNA synthesis was carried out using primers designed (Supplementary Table S1) based on the complete genome sequence of the PRSV-Del isolate (Accession no. EF017707.1) with the help of SuperScript™ III Reverse Transcriptase (Invitrogen). This cDNA mixture was diluted up to 50 μL to amplify the full-length viral genome in overlapping fragments using several combinations of sense and antisense primers and Taq DNA polymerase (NEB). The PCR products were purified using a QIAquick gel extraction kit (QIAGEN) and ligated in pGEM-T Easy (Promega) TA cloning vector following the manufacturer’s instructions. Nine cDNA clones spanning the entire genome length were designated PK1.6- PK9.2 and selected for sequencing analysis (Figure 1B).
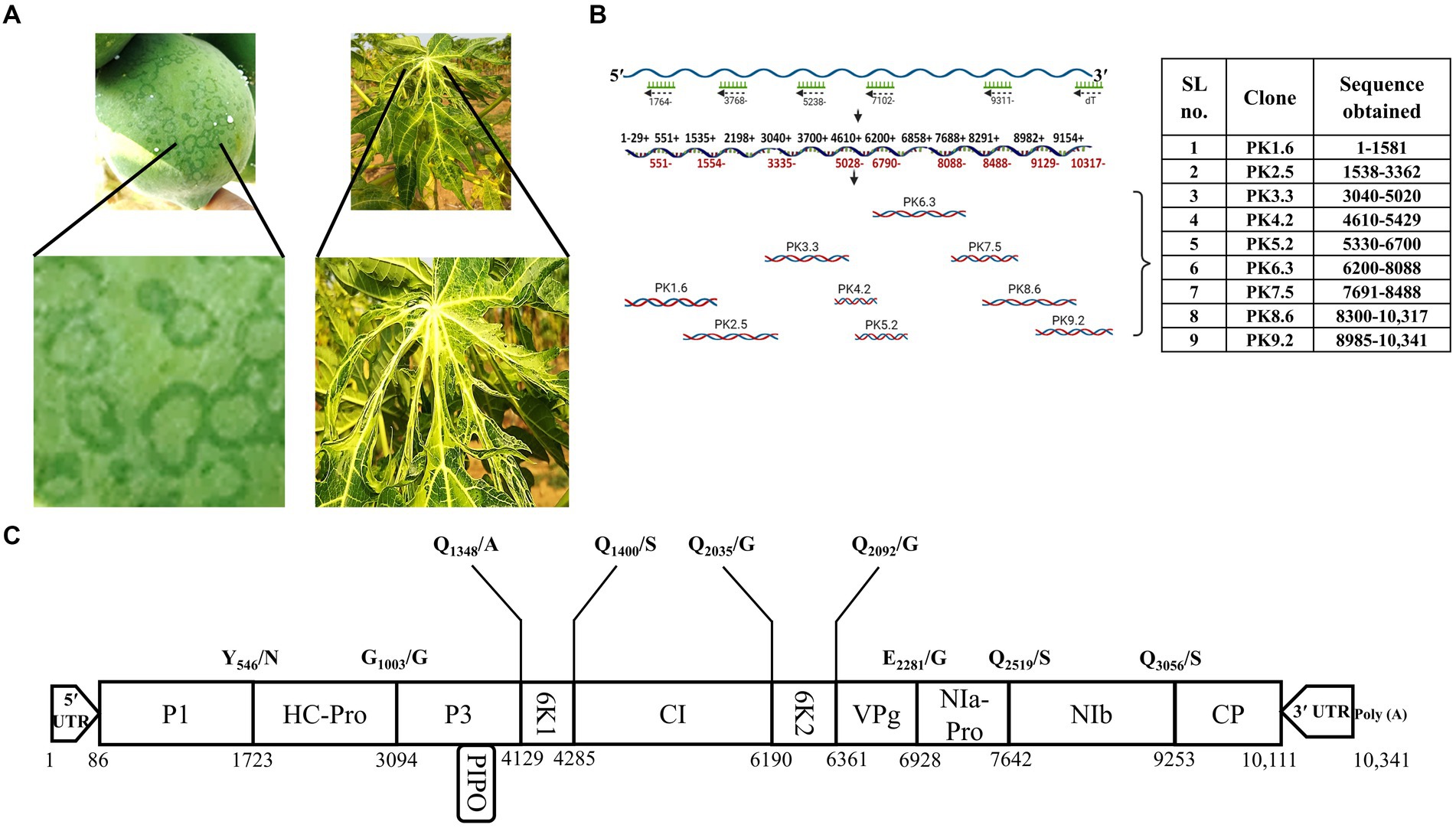
Figure 1. Symptomatology, complete genome sequencing, and the putative genome organization of PRSV-HYD isolate. (A) The infected leaf and fruit samples were collected based on the visible symptoms- ringspots on fruits and distorted leaves. (B) The full-length PRSV-HYD genome was converted into nine overlapping cDNA fragments, amplified, and cloned into the pGEM-T Easy vector (Promega). These nine TA clones were sequenced on both strands, with three biological replicates of each. Sequences obtained from each clone are as follows: PK1.6- nucleotide 1–1,581, PK2.5–1,538-3362, PK3.3–3,040-5020, PK4.2–4,610-5429, PK5.2–5,330-6700, PK6.3–6,200-8088, PK7.5–7,691-8488, PK8.6–8,300-10,317, and PK9.2–8,985-10,341. (C) The deduced genomic map of PRSV-HYD with the exact position of individual genes and putative cleavage sites of the proteins.
2.3 Complete genome sequencing and in silico analysis of PRSV-HYD
To obtain the complete genome sequence of PRSV-HYD, we sequenced a minimum of 3 positive clones for each of the nine cDNA clones on both strands with the help of sequencing primers (Supplementary Table S1) using the Sanger’s dideoxy method by 3,500 XL Genetic analyzers (ThermoFisher Scientific). Authentic 5′- and 3′- terminal sequences of PRSV cDNA were confirmed by using SMARTer® 5′ RACE and 3′ RACE kits (TaKaRa Bio) by following the manufacturer’s instructions. A minimum of 5 independent clones were sequenced to confirm the termini. The obtained raw sequences from 9 different pGEM-T easy clones were trimmed and assembled using PRSV-Del (Accession no. EF017707.1) genome sequence as a template. To identify ORFs, the ORF finder tool1 was used. In silico translation was performed using the translate tool by ExPASy (Gasteiger et al., 2003). The full-length nucleotide sequence and the in silico translated polyprotein sequences were used for blastn and blastp analysis, respectively. This helped us obtain closely related PRSV sequences from the NCBI database based on sequence homology. Phylogenetic analysis was carried out using MEGA-X version 10.1.8 with the Maximum-likelihood method with the Jones-Taylor-Thornton (JTT) model (for polyprotein sequences) with 1,000 bootstrap replicates (Kumar et al., 2018).
2.4 Infectious cDNA (icDNA) construction for PRSV-HYD
To construct PRSV-HYD icDNA, TRIzol (Invitrogen)-extracted total RNA was sequentially reverse transcribed using SuperScript™ III Reverse Transcriptase (Invitrogen) and three sequence-specific primers (Supplementary Table S1) as shown in Figure 2A. Further, using Phusion™ High-Fidelity DNA Polymerase (NEB), the cDNAs were converted into three overlapping PCR amplified fragments designated as PG1, PG2, and PG3 (Figure 2A), encompassing the full-length PRSV-HYD genome with a minimum of 50 nucleotide overlap. By performing primer overlap extension PCR, Cauliflower mosaic virus (CaMV) 35S double promoter sequence containing BglII restriction site was fused to the 5′ end of PG1 fragment. Similarly, a ribozyme (RZ) sequence from Avocado sunblotch viroid (ASBVd) followed by XmaI restriction site was added at the 3′ end of PG3 fragment (Gopinath et al., 2005). PG1, PG2, and PG3 fragments were double digested with BglII-MluI, MluI-StuI, and StuI-XmaI, respectively. The fragments were gel-purified using a QIAquick gel extraction kit (QIAGEN) and ligated into the vector backbone (binary vector pCB301) digested with BglII/XmaI (Figure 2A). The ligation mix was transformed into competent ElectroMAX™ Stbl4™ cells (Invitrogen) by electroporation (25 μF, 200 Ω, 1200 V) in a Gene pulser XCell (Bio-Rad). A total of six clones were confirmed by colony PCR and restriction digestion. All six positive clones were sequenced on both strands using sequence-specific primers and matched entirely with the full-length PRSV-HYD sequence. A minimum of three positive clones (pCB301-icPRSV-HYD) were mobilized into the Agrobacterium tumefaciens EHA105 strain by electroporation using the parameters mentioned above.
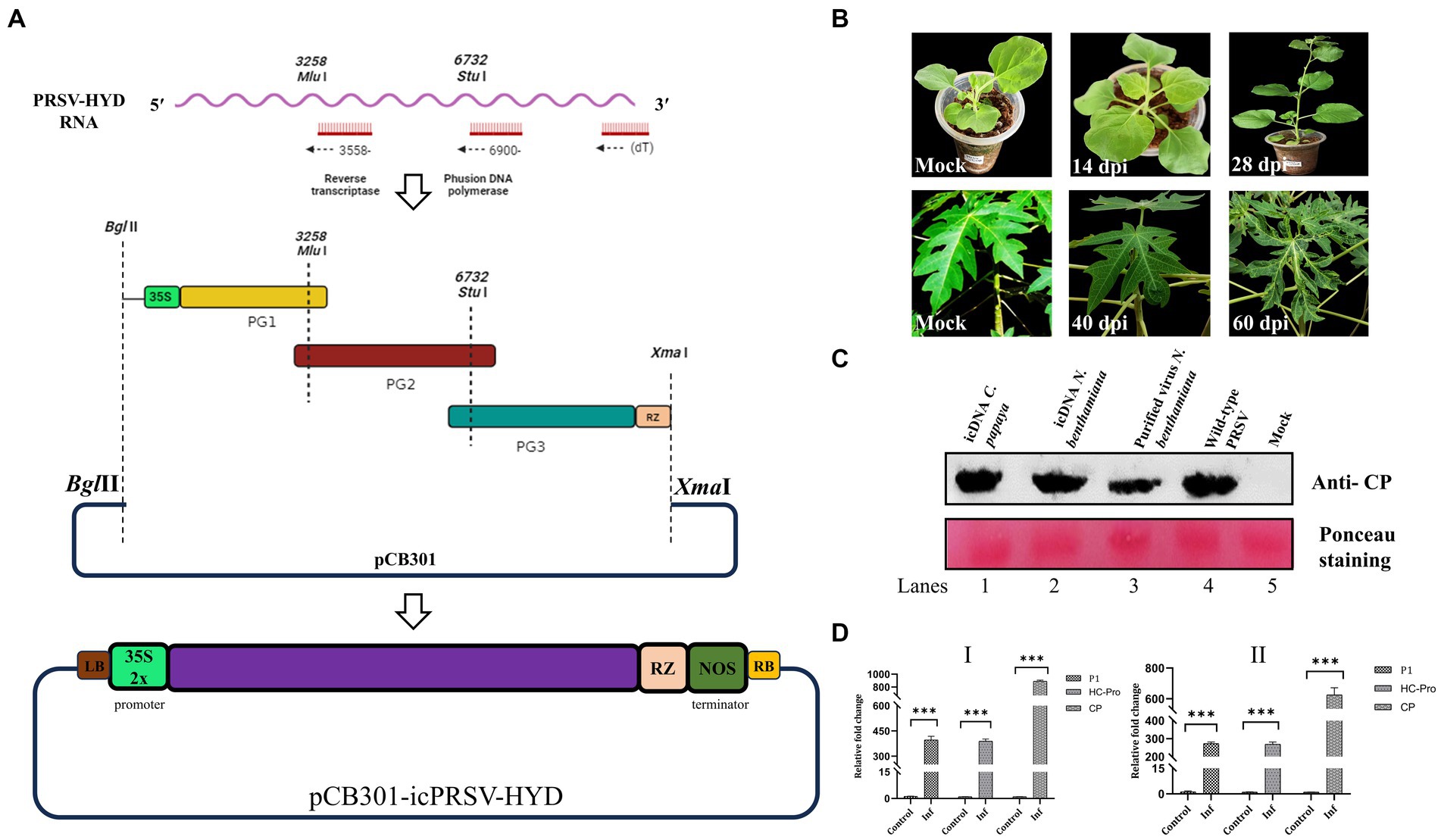
Figure 2. PRSV-HYD infectious clone preparation and its infectivity in different hosts. (A) The schematic representation of the PRSV-HYD infectious clone preparation; three sequence-specific primers were used for reverse transcription, followed by amplification using Phusion DNA polymerase (NEB). BglII restriction site followed by CaMV 35S double promoter was fused to the 5′ terminus by primer overlap extension PCR. Similarly, the ASBVd RZ sequence followed by the XmaI restriction site was added to the 3′ terminus. Then, restriction digestion was performed using BglII-MluI (PG1), MluI-StuI (PG2), and StuI-XmaI (PG3). These three double-digested fragments were ligated into the pCB301 vector backbone. (B) Carica papaya leaves showed mild leaf distortion symptoms at 40 dpi, and the symptom severity was observed at 60 dpi. In the case of N. benthamiana, no symptoms were observed on the inoculated leaves (14 dpi) or systemic leaves (28 dpi). Empty vector pCB301 in A. tumefaciens served as control. (C) Immunoblot analysis using PRSV coat protein (CP) specific antibody showed expression of CP in C. papaya leaves (60 dpi) inoculated with the purified virus particles from agroinfiltrated N. benthamiana plants (Lane-1), agroinfiltrated N. benthamiana leaves (28 dpi) (Lane-2), and purified virus particles from the agroinfiltrated N. benthamiana plants (28 dpi) (Lane-3). Wild-type PRSV-infected C. papaya leaves served as a positive control (Lane-4), and empty vector pCB301 in A. tumefaciens served as mock (Lane-5). (D) Real-time PCR analysis showed significant upregulation of P1, HC-Pro, and CP genes in both C. papaya (I) and N. benthamiana (II) plants.
2.5 Agroinoculation of plants
Agroinoculation of plants with pCB301-icPRSV-HYD in A. tumefaciens EHA105 cells was carried out following the method described by Gopinath et al. (2005). Briefly, A. tumefaciens EHA105 cells containing pCB301-icPRSV-HYD were allowed to grow in Luria broth (LB) supplemented with Rifampicin (50 μg/mL) and Kanamycin (50 μg/mL) at 28°C with 180 rpm of continuous shaking. 1 mL of the pre-culture was used to inoculate 100 mL of LB-Rifampicin-Kanamycin supplemented freshly with 10 mM sterile 2-(N-Morpholino) ethanesulphonic acid (MES) pH 5.85 and 20 mM of acetosyringone, and the culture was grown for 36 h at 28°C incubator with 180 rpm shaking. Cells were harvested by centrifugation at 6000 rpm for 10 min and then resuspended in 10 mL of infiltration buffer (10 mM MgCl2 and 10 mM MES pH 5.85) with 100 μM acetosyringone. This suspension was incubated at room temperature for 3 h. A. tumefaciens carrying empty vector pCB301 served as a negative control. The suspension was diluted to OD595 0.1 with infiltration buffer and infiltrated at the abaxial surface of 25 Nicotiana benthamiana plants using a 2 mL syringe without a needle. Infiltrated leaves were harvested from all these plants at a regular interval of 3 days. Then, virion particles were purified from this batch of N. benthamiana plants at 28 dpi according to the method explained by Moghal and Francki (1976). The purified virus particles were mechanically inoculated onto the C. papaya leaves (3-leaf stage) and pinpricked the young stems by sterile needles (Zhang et al., 2021). All the agro-inoculated plants were maintained inside an insect-free climate chamber (25°C/21°C Day/night temperature, 70% relative humidity, 16 h photoperiod at 150 μmol s−1 m−2 light intensity).
2.6 Analysis of agro-inoculated icDNAs in Carica papaya and Nicotiana benthamiana
To check the functionality of the icDNA, two approaches were used. Initially, total RNAs were isolated from N. benthamiana leaves infiltrated with PRSV icDNA from 3 to 28 days at regular intervals. RT-PCRs were performed for P1, HC-Pro, and CP genes individually. However, we have relied on the development of symptoms in the case of C. papaya. Secondly, total soluble proteins were isolated from the N. benthamiana and C. papaya following the protocol described by Gopinath et al. (2000). Immunoblot analysis was performed using PRSV CP-specific antiserum using chemiluminescent detection system (Thermo Fisher Scientific) according to the manufacturer’s instructions.
2.7 Quantitative analysis of virus and host genes from Nicotiana benthamiana and Carica papaya
Total RNA isolated from the agroinfiltrated plants were quantified using NanoDrop 2000 UV–Vis Spectrophotometer (ThermoFisher Scientific). Primers targeted for the genes encoding three viral proteins (P1, HC-Pro, and CP) and host genes encoding pathogenesis-related gene-1 and 10 (PR1a and PR10), defensin-like protein 1.2 (PDF1.2), Ran-binding protein 1a (RanBP1); host transcription factors basic leucine zipper 60 (bZIP60), NbNAC042, Ethylene responsive transcription factor-5 (ERF5), MYB44, Isopentenyl transferase-1 (IPT1), Lonely Guy-1 (LOG1), Allene oxide cyclase-1 (AOC1), 12- oxo-phytodienoic acid reductase-2 (OPR2), Bax inhibitor-1 (BI-1) Staygreen1 (SGR1), senescence-associated gene-12 (SAG12), autophagy 8f (ATG8f) were synthesized using GenScript (Supplementary Table S2). cDNA was synthesized from the total RNA, as described earlier. We performed Real-time PCR on Mastercycler Realplex (Eppendorf, Germany), as reported previously (Supriya et al., 2022). The actin genes of C. papaya and N. benthamiana (AY179605.1) were used as an internal controls for respective plants. The relative expression levels of the undertaken genes were estimated using the ΔΔCT method (Livak and Schmittgen, 2001). Three biological replicates were used for every individual gene.
2.8 Statistical analysis
The data presented represents the mean value of three treatments, each containing three replicates. The data was analyzed using a two-way ANOVA. The error bars on the graph represent the standard deviation (± SD) of the mean values. The Duncan multiple range test (p ≤ 0.05) was used to determine significant treatment differences.
3 Results
3.1 Complete genome sequencing of PRSV-HYD
In this study, we collected C. papaya leaves and fruits suspected to be infected with PRSV (Figure 1A). The presence of PRSV was confirmed by DAC-ELISA and RT-PCR analysis (Supplementary Figure S1). We generated nine overlapping PCR amplified fragments by performing RT-PCR using different combinations of primers, followed by successful TA cloning of these fragments. Positive clones were confirmed through colony PCR (data not shown). The positive clones were designated as PK1.6, PK2.5, PK3.3, PK4.2, PK5.2, PK6.3, PK7.5, PK8.6, and PK9.2 (Figure 1B). From the sequencing data of these nine overlapping TA clones (Figure 1B), complete genome sequence was deduced (Figure 1C). We determined the terminal nucleotide sequences using 5′- and 3′- RACE techniques (Supplementary Figure S2). The complete genome is 10,341 nucleotides in length. A single large putative ORF was identified, starting from the 86th nucleotide and ending at the 10,111th nucleotide of the genome. Putative sizes of the coding and non-coding regions were identified (Table 1). 3,342 amino acids long polyprotein sequence was identified by in silico analysis. We identified the putative cleavage sites of the functional proteins (Figure 1C) by aligning them with other PRSV sequences from the NCBI GenBank database. We deposited the deduced full-length genome sequence in the NCBI GenBank database with accession number KP743981.1. We have also identified conserved regions based on previous reports of the known potyviruses available in the NCBI databases (Supplementary Figure S3). Some of those notable conserved motifs are G496SSG in P1, F726RNK in HC-Pro, N2868GDDL in NIb, D3063AG in CP, and a stretch of KE amino acids at the N-terminus of the CP.
Table 1. Putative sizes of individual genes and mature proteins of PRSV-HYD were identified and documented.
3.2 Phylogenetic analysis of PRSV-HYD
A maximum likelihood tree was created with 1,000 bootstrap replicates using the in silico-translated PRSV-HYD polyprotein sequence (Accession no. AKQ98195.1) and other available PRSV polyprotein sequences from different regions worldwide. PRSV Pune (Aundh) and PRSV-Del isolates (Accession no. ASV48700.1 and ABJ74175.1, respectively) were present within the same clade as PRSV-HYD, with PRSV Pune (Aundh) being the closest neighbor. The isolates have been grouped into distinct clusters based on their geographical locations, and individual clusters have been denoted with distinct color codes (Figure 3).
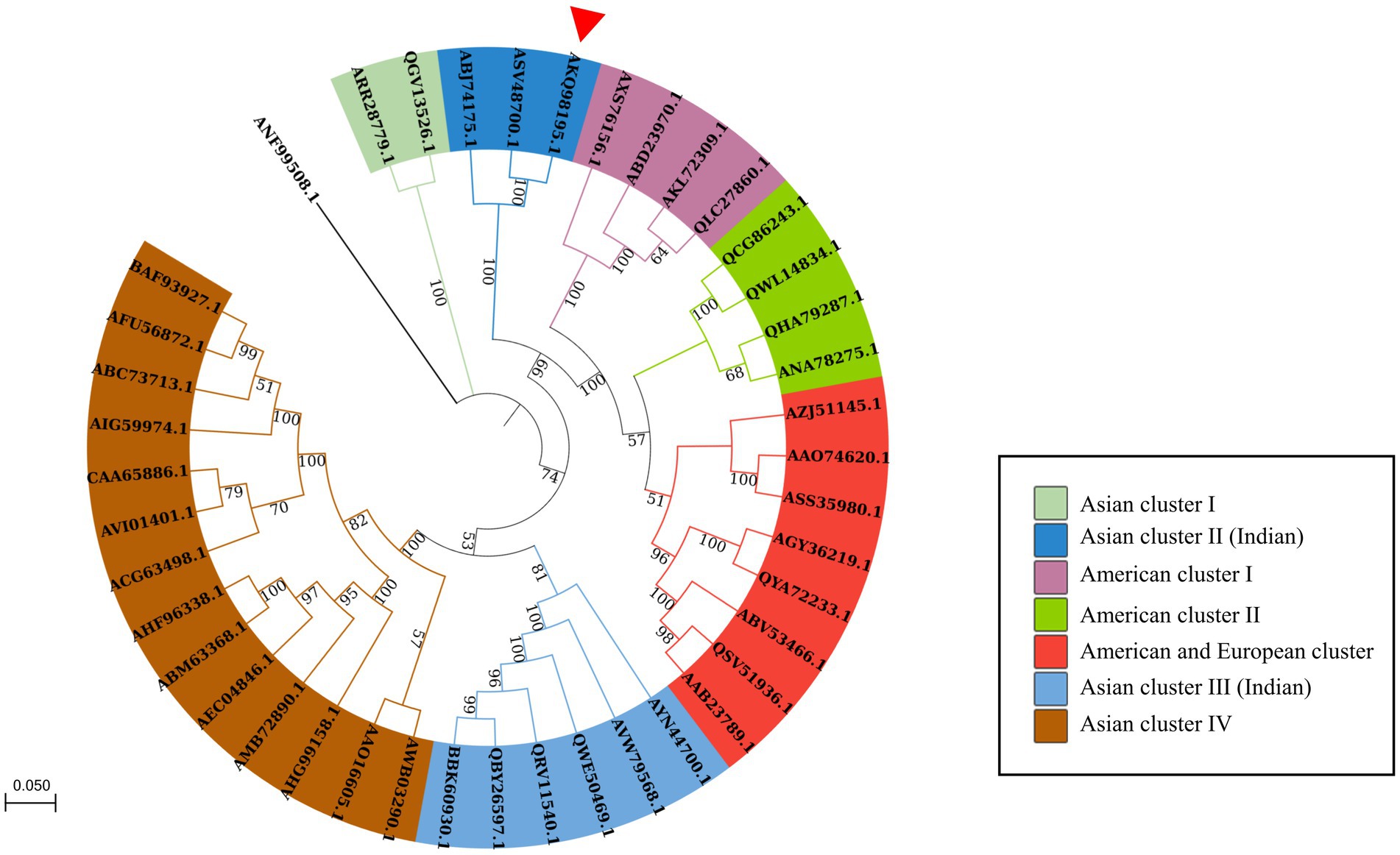
Figure 3. Phylogenetic analysis of the polyprotein sequence of PRSV-HYD isolate (accession no. AKQ98195.1) along with 40 closely related PRSV polyprotein sequences retrieved from the NCBI database and a Moroccan watermelon mosaic virus (accession no. ANF99508.1) isolate as an outgroup. The sequences were aligned using ClustalW, and a maximum-likelihood phylogenetic tree was constructed with MEGA-X [14]. The numbers at the nodes represent the percentage of 1,000 bootstraps. Bootstrap values less than 50% are not shown. PRSV-Pune (Aundh) and PRSV-Del isolate (accession no. ASV48700.1 and ABJ74175.1) are present within the same clade as PRSV-HYD, with PRSV Pune isolate being the closest neighbor to PRSV-HYD, has been shown with a red triangle.
Another maximum likelihood tree was constructed with 1,000 bootstrap replicates involving PRSV-HYD and 25 other Potyviral polyprotein sequences. Zucchini tigre mosaic virus (ZTMV) USA isolate (ATY37425.1), Papaya leaf distortion mosaic virus (PLDMV) Hainan isolate (AGC54443.1), and Cucurbit vein banding virus (CVBV) Argentina isolate (ASB15795.1) and PRSV-HYD (AKQ98195.1) are present within the same clade. ZTMV is the closest neighbor to PRSV-HYD (Supplementary Figure S4).
Furthermore, PRSV-HYD complete nucleotide and amino sequence shows up to 92 and 95% of sequence homology, respectively, with other Asian isolates. The 5′- non-coding region and P1 coding region showed the highest sequence diversity of up to 21 and 33%, respectively, which is the maximum compared to other genomic regions. P1 amino acid sequence showed up to 41% of sequence diversity compared to the Asian isolates (Table 1).
3.3 Construction of PRSV-HYD icDNA and its expression patterns in planta
To Construct PRSV-HYD icDNA, the three amplified cDNA fragments PG1, PG2, and PG3 were (3.7 kb, 3.2 kb, and 3.4 kb in size, respectively) used. By employing diverse molecular manipulations, PRSV-HYD infectious cDNA was constructed in the binary vector pCB301 under the right and left borders of the T-DNA region. Cauliflower mosaic virus 35S double promoter was fused at the 5′ end of the genome for efficient transcription. Care was taken that no additional sequences are introduced in the transcribed RNAs. Similarly, at the 3′ end, Avocado sunblotch viroid RZ sequence was fused for cis preferential cleavage and generation of authentic viral ends. The resultant clones were sequence-confirmed and designated as pCB301-icPRSV-HYD (Figure 2A). Upon agroinfiltration, no visible symptoms were observed up to 28 dpi in N. benthamiana plants, indicating this could be an asymptomatic host. However, RT-PCR analysis demonstrated the presence of P1, HC-Pro, and CP gene products in all the N. benthamiana plants infiltrated from 3 dpi up to 28 dpi (Supplementary Figure S5). This is clear evidence of the efficient transcription of the icDNAs introduced under the CaMV 35S double promoter.
We have not observed any visible symptoms up to 120 dpi in C. papaya plants infiltrated with icDNAs. The plants are not as conducive to infiltrations as N. benthamiana. Alternatively, we have purified virus particles from N. benthamiana plants (28 dpi) infiltrated with the icDNAs using the protocol of Moghal and Francki (1976). This purified virus was introduced into the C. papaya plants by mechanical inoculation, and the virus particles were introduced into the young stems by the pinprick method.14 out of 25 mechanically inoculated plants and 20 out of 25 plants inoculated by the pinprick method, started showing symptoms 40 days post-inoculation, indicating the infectivity of infiltration-generated virus particles and the symptom severity increased by 60 dpi (Figure 2B). These results were confirmed in immunoblot analysis using PRSV-Del CP polyclonal antibody (Figure 2C). The results were further substantiated in Real-time PCR, where we observed the upregulation of the three viral gene products CP, P1, and HC-Pro in both N. benthamiana and C. papaya (Figure 2D). The newly emerging leaves of both C. papaya and N. benthamiana were analyzed for the presence of viral CP gene by RT-PCR analysis to confirm the systemic movement of the virus particles. We could detect the PRSV CP gene in the systemic leaves of C. papaya indicating its systemic spread beyond the infiltrated leaves (Supplementary Figure S6A). However, we failed to detect PRSV CP gene from the systemic leaves of N. benthamiana indicating its inability to go systemic (Supplementary Figure S6B).
3.4 Comparison of host-specific gene expression patterns between natural host and non-host of PRSV-HYD
All the real-time PCR analyses were performed using total RNA extracted from 28 dpi N. benthamiana and 60 dpi C. papaya plants. In C. papaya, PR1a showed no change, PR10 and RanBP1 were upregulated by 1.4-fold and 2.4-fold, respectively, whereas PDF1.2 was significantly downregulated (Figure 4A). PR1a, PR10, and RanBP1 genes were significantly upregulated in N. benthamiana, but PDF1.2 showcased unchanged expression compared to control plants (Figure 4B). The expression levels of some transcription factors, such as MYB44, basic Leucine zipper-60 (bZIP60), NAC042, and Ethylene response factor-5 (ERF5), were checked. In the case of C. papaya, all four transcription factors showed significant upregulation, with bZIP60 and EFR5 showing the highest level of upregulation by 42- and 13-fold, respectively (Figure 4C). N. benthamiana showed 1.26-, 1.25- and 1.20-fold upregulation of MYB44, NAC042, and ERF5, respectively, but there was no change in bZIP60 expression (Figure 4D). Some genes that are directly related to the physiology of plants, such as Isopentenyl transferase-1 (IPT1), Lonely Guy-1 (LOG1), Allene oxide cyclase-1 (AOC1), 12- oxo-phytodienoic acid reductase (OPR2), and Autophagy-related protein 8f (ATG8f) were also checked. The host plants showed significant downregulation of all these five genes, whereas the non-host N. benthamiana exhibited no change in expression levels. When we checked the expression levels of Bax inhibitor-1 (BI-1), Staygreen-1 (SGR1), and senescence-associated gene-12 (SAG12), these genes were found to be upregulated by 2.6, 7.3, and 3.4-fold, respectively in C. papaya (Figure 4E). On the other hand, BI-1 and SGR1 showed a 1.17- and 1.13-fold change in N. benthamiana, whereas SAG12 showed 1.1-fold downregulation (Figure 4F).
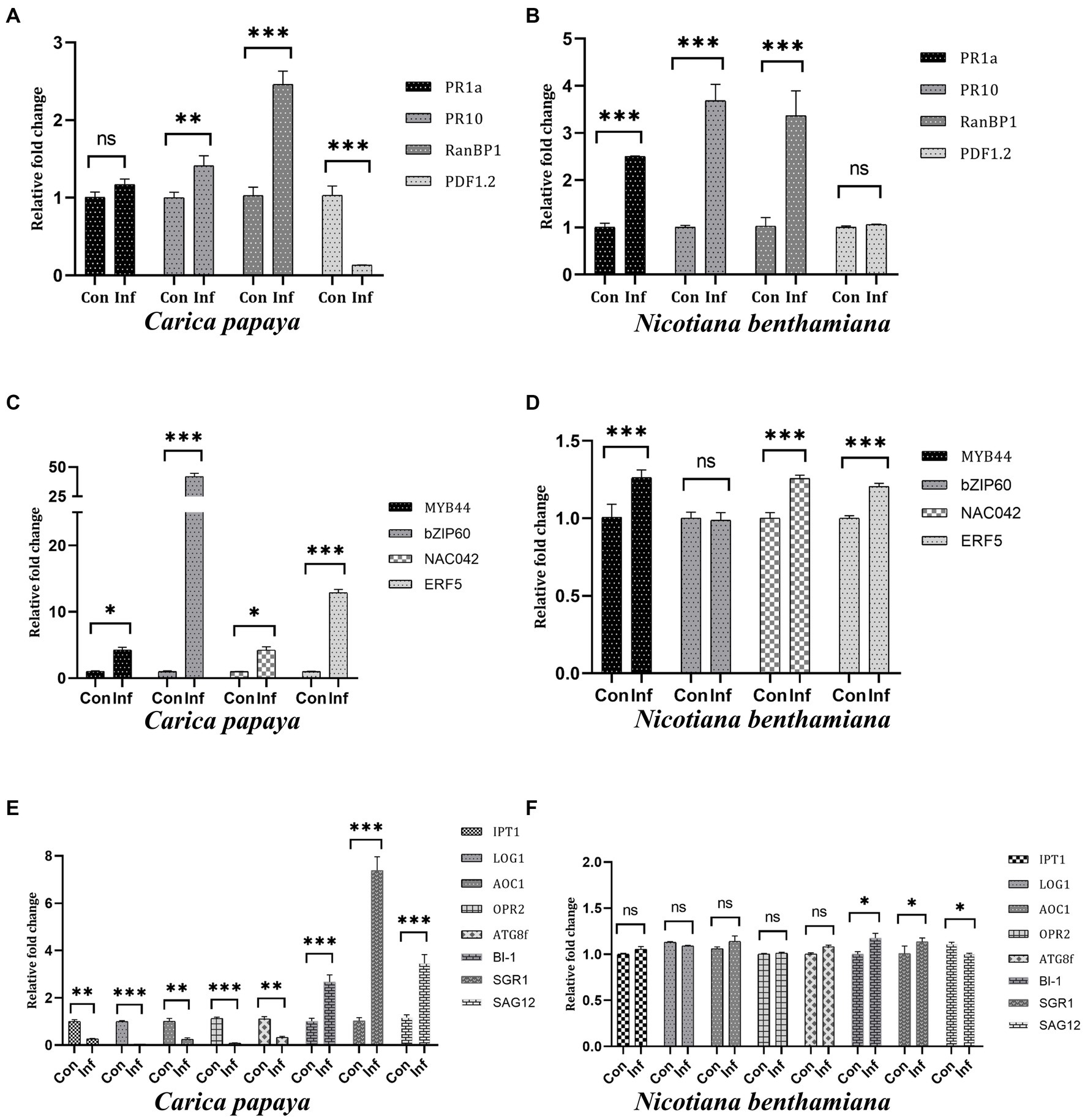
Figure 4. Relative expression levels of different plant-specific genes from host (C. papaya) (A,C,E) and non-host (N. benthamiana) (B,D,F) plants. (A) It was observed that the genes PR1a showed no change, whereas PR10 and RanBP1 showed 1.4-fold and 2.4-fold upregulation. Meanwhile, PDF1.2 was significantly downregulated. (B) Significant upregulation of PR1a, PR10, and RanBP1 was observed in non-host plants, while PDF1.2 expression remained at a regular level. (C) All four TFs were found to be upregulated in host plants, with bZIP60 showing the highest upregulation of 42-fold. (D) MYB44, NAC042, and ERF5 showed 1.26-, 1.25- and 1.20-fold upregulation in expression levels in non-host plants, while bZIP60 was expressed at a regular level. (E) Cytokinin biosynthesis-related genes IPT1 and LOG1, Jasmonate biosynthesis genes AOC1 and OPR2, and autophagy-related gene ATG8f were significantly downregulated in host plants, while BI-1, SGR1, and SAG12 genes were upregulated significantly. (F) There were no significant changes in the expression levels of IPT1, LOG1, AOC1, OPR2, and ATG8f genes in non-host plants. BI-1 and SGR1 showed a 1.17- and 1.13-fold change, while SAG12 showed no change in expression.
4 Discussion
The severity of PRSV infection in papaya orchards is rising alarmingly with time (Premchand et al., 2023). The lack of availability of a significant number of full-length genomic sequences is one of the major drawbacks in viral disease management. We sequenced and confirmed PRSV infection in the samples from an orchard mentioned earlier (Figure 1). This full-length genome sequence provides us with information about the diverse nature of this virus, conserved regions or motifs, mutation-prone genomic regions, and many more. The presence of some of the conserved motifs is essential for successful PRSV infection, as different motifs have different functions such as GDSG motif is required for the protease activity of P1 (Adams et al., 2005), FRNK within HC-Pro is involved in symptom development (Gal-On, 2000), GDD motif of NIb is important for RNA-dependent RNA polymerase activity (Liu et al., 2021), coat protein DAG motif is required for aphid transmission (Bravo et al., 2008), a region of KE repeats at the N-terminal region of coat protein with unknown function (Shukla et al., 1994), and many others. All these conserved motifs with minor modifications have been identified in PRSV-HYD, and the modifications could be due to the natural diversities that did not hinder the functionality. P1 is the most diverse functional protein among potyviruses (Tordo et al., 1995; Mishra et al., 2019). Our study showed the same outcome, with P1 showing maximum nucleotide and amino acid sequence diversities compared to other PRSV isolates. The 5′ UTR was also more diverse than the 3′ UTR (Table 1). We can presume that the 5′ terminal of the PRSV genome might be more prone to random mutations than the 3′ terminal. This complete genome sequence data will contribute to the further understanding of PRSV epidemiology and sequence diversity studies.
ZTMV, PLDMV, CVBV, and PRSV-HYD sharing the same clade signifies their similar evolutionary origin. ZTMV-USA isolate (Accession no. ATY37425.1) is the closest to PRSV-HYD, and these two isolates share 67.2% of amino acid sequence homology, suggesting they might have a common or closely related ancestor. PRSV Pune (Aundh) (Accession no. ASV48700.1) is the closest relative to PRSV-HYD, with a 95% amino acid sequence similarity, suggesting they might have a similar evolutionary origin (Figure 3).
The importance of having stable infectious cDNA clones for any RNA virus is well-known in the field of plant virology. Several infectious clones available for many potyviruses, such as Watermelon mosaic virus (WMV) (Desbiez et al., 2012), Potato virus Y (PVY) (Chikh Ali et al., 2011), Tobacco vein mottling virus (TVMV) (Domier et al., 1989), Zucchini yellow mosaic virus (ZYMV) (Gal-On et al., 1991), Soybean mosaic virus (SMV) (Bao et al., 2020), Plum pox virus (PPV) (Riechmann et al., 1990), and many more. We have successfully developed icDNA for PRSV-HYD, which was agroinfectious in N. benthamiana; however, no visible symptoms appeared in the infiltrated plants, indicating this as an asymptomatic host (Figure 2). We have some limitations in the infiltration protocol of C. papaya. However, the infiltration-generated virus particles from N. benthamiana were infecting the C. papaya plants efficiently, with systemic movement of the virus particles. As C. papaya is the natural host of PRSV-P, there might be certain essential factors that are present only in C. papaya plants that aid in the establishment and multiplication of PRSV. These factors may play a crucial role in promoting PRSV systemic infection. So, the absence of such factors can be a reason for non-systematic infection in N. benthamiana.
The importance of PR genes is invaluable regarding the host response during any biotic or abiotic stress condition. PR1a gene is an activator of the Salicylate (SA) pathway and, thereby, plays an essential role in systemic acquired resistance (SAR) (Ali et al., 2018). PR10 genes have been shown to possess anti-microbial activities during biotic stress (Chen et al., 2006). RanBP1 is another crucial gene of plants as it accumulates terpenoids that show resistance against several pathogens (Mizuno et al., 2019). The expression levels of the PR genes and RanBP1 were relatively lesser in the case of C. papaya plants compared to that in N. benthamiana, indicating that the natural host resistance is not strong enough against PRSV-HYD to overcome its symptoms. However, real-time PCR and immunoblot analysis proved that the virus replicates inside the non-host. However, the non-host plant has exerted resistance against the invasion of the virus, but the virus can still replicate without any symptoms. PDF1.2 is a gene of the plant defensin family. It is known to be a part of the innate immune system and is involved in jasmonate (JA)- dependent defense response (Manners et al., 1998; Brown et al., 2003). Downregulation of PDF1.2 in the host but unchanged in the non-host can be described as the ability of the virus to overcome the innate immune barrier of the natural host but not the non-host (Figures 4A,B).
Transcription factors (TFs) are essential as they regulate several genes involved in different plant growth and development stages, metabolic pathways, and many other molecular events. MYB44 TF gets activated by ethylene, which has multiple roles in growth, development, defense responses, etc. (Adie et al., 2007; Iqbal et al., 2017). Higher levels of MYB44 expression in C. papaya compared to N. benthamiana indicate that the latter has managed to minimize the effect of the virion particles on the developmental stages of the plant. Another important TF is NAC042, which was shown to be a modulator of defense response in N. benthamiana during viral infections (Ke et al., 2022). It was upregulated in both host and non-host, but like MYB44, the expression was higher in the host plant, signifying infection severity. Another important TF is bZIP60, which is involved in ER stress response as it regulates the genes involved in unfolded protein response (Xu et al., 2019). Its significant upregulation in host plants designates that these plants are under severe stress due to the presence of virion particles. However, no significant change of bZIP60 expression in the non-host plant may be pointing toward a better resistance mechanism against the virus. ERF5 is necessary for plant innate immunity (Son et al., 2012). Elevated viral load has resulted in the expression of ERF5 being upregulated by 12-fold in the natural host, whereas just 1.2-fold increased expression in the non-host. This signifies that the non-host system has dealt with the adversity of the infection (Figures 4C,D).
Plant phenotypes can be significantly affected by viral infections, which can cause various symptoms that hinder growth, development, and productivity (Roossinck, 2010). The symptoms occur due to intricate molecular interactions between the virus and the host plant (Jiang and Zhou, 2023). Cytokinins (CKs) are plant hormones that stimulate cell growth and elongation and are instrumental in several developmental processes, such as transmitting nutritional signals and postponing senescence (Sakakibara, 2006). Additionally, CKs have been found to contribute to plant resistance against viral infections (Choi et al., 2011). Two essential genes involved in the cytokinin biosynthesis pathway are IPT1 and LOG1 (Takei et al., 2001; Kurakawa et al., 2007). The significantly decreased transcript expressions of these two genes in the host plant might suggest the hindrance in CK production due to virus assembly, which ultimately contributes to the distorted phenotype and wilted leaves in plants. Contrarily, unchanged expressions of these two genes in non-host plants point toward its unaffected CK biosynthesis, correlated with unaltered phenotype and non-chlorotic leaves. During pathogen invasion, Jasmonic acid (JA) acts as a vital phytohormone in defense mechanisms (Sun et al., 2011). In the absence of JA, symptoms of PVY-PVX co-infection develop at an accelerated rate, whereas treatment with JA enhances resistance against double infection (García-Marcos et al., 2013). The biosynthesis pathway of JA involves the AOC1 and OPR2 genes (Eng et al., 2021). Significant downregulation of these genes indicates the impeded JA biosynthesis and, consequently, compromised virus resistance. Nevertheless, despite the occurrence of viral replication, maintained JA biosynthesis as proved by unaltered expressions of said genes, justifying the futile impact of the virus on the physiology of non-host plants.
Autophagy delivers non-functional intracellular components to vacuoles to regulate the host’s response to pathogens like viruses (Shoji-Kawata and Levine, 2009). ATG8 proteins are crucial in forming autophagosomes and selecting cargo for degradation. (Nakatogawa et al., 2007; Johansen and Lamark, 2011). The expression of ATG8f is significantly suppressed during viral infection, indicating that the host is unable to undergo autophagy. However, maintained ATG8f expression in the non-host suggesting the occurrence of autophagy. Plants use PCD to defend themselves against different pathogens (Jorgensen et al., 2017). BI-1 is a well-known suppressor of PCD (Watanabe and Lam, 2009). The virus has likely overcome the host’s resistance through PCD, as shown by the significant increase in BI-1 expression in the host. Conversely, while there is a change in BI-1 expression in the non-host, it is relatively less pronounced as compared to the host plant, suggesting effective usage of PCD.
The accumulation of viral CP inside chloroplasts was related to the induction of chlorotic symptoms during Tobacco mosaic virus (TMV) infection (Lehto et al., 2003). It has been reported that Sugarcane mosaic virus (SCMV) HC-Pro has a specific interaction with the maize chloroplast precursor protein of Fd V, ultimately resulting in disturbances to the structure and function of chloroplasts (Cheng et al., 2008). Staygreen-1 (Sgr-1) regulates chlorophyll degradation during leaf senescence (Park et al., 2007). Based on current research, SAG12 is considered the most effective molecular marker for identifying senescence (Weaver et al., 1998). The chlorotic spots on the symptomatic leaves of the host plants are justified by the upregulated SGR1 and SAG12 gene expressions. On the other hand, the absence of any chlorotic symptom or leaf senescence in the non-host could be due to the considerably lower expression levels of SGR1 and SAG12 compared to the host plant (Figures 4E,F). This entire study has been represented as a schematic diagram (Figure 5).
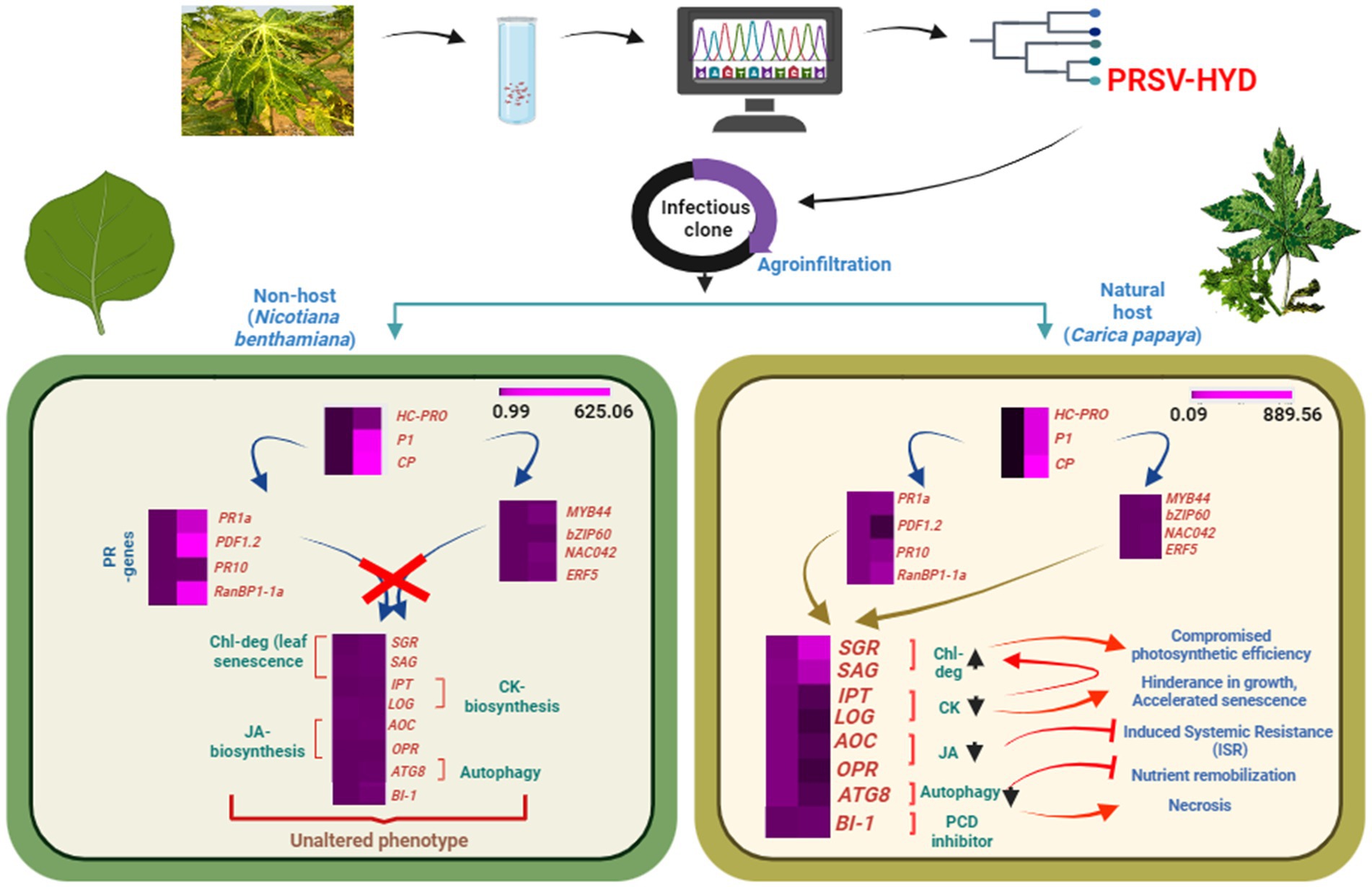
Figure 5. Schematic representation of the study. Depiction of complete genome sequencing and icDNA clone preparation of PRSV-HYD isolate, followed by agroinoculation in host (Carica papaya) and non-host (Nicotiana benthamiana) plants. Putative expression analysis of essential genes of both host and non-host plants showcasing the indispensability of host factors for effective systemic infection in host plant.
5 Conclusion
This study was conducted to characterize the PRSV-HYD isolate through complete genome sequencing. To our knowledge, it is the first report of PRSV isolate from South India to be fully sequenced. The PRSV-HYD genome differs significantly from other PRSV isolates in certain genomic regions, suggesting its evolving nature. It is essential to monitor the different circulating strains of PRSV to understand the phenotypic and genotypic characteristics of the infected host plants. Our comprehensive genome sequence data will aid future PRSV epidemiology and sequence diversity research. Moreover, this study is the first of its kind to report a complete infectious cDNA clone of PRSV from the Indian subcontinent. Despite the agroinfectivity of pCB301-icPRSV-HYD in both the host (C. papaya) and non-host (N. benthamiana) plants, only the host exhibited systemic infection with compromised expressions of genes related to cytokinin, jasmonic acid, and autophagy. Additionally, the virulence also affected the photosynthetic ability of the host by elevating chlorophyll degradation and consequently increasing chlorosis. Although the ability of this virus to be replicated, due to some unknown mechanism, the infectivity was subdued in non-host, proving the essential requirement of host factors for a successful systemic PRSV infection. This icDNA clone can be aided in the further detailed study of this virus at the molecular level, investigate virus-vector interactions, or use it as a viral vector to express heterologous proteins.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
PG: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Validation, Writing – original draft. PP: Conceptualization, Investigation, Methodology, Writing – original draft. LS: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft, Software, Validation. HS: Investigation, Methodology, Writing – original draft. GP: Investigation, Validation, Writing – review & editing. KG: Conceptualization, Formal analysis, Funding acquisition, Investigation, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. PG acknowledges the Department of Science and Technology (DST) for providing the Junior Research Fellowship [IF160758, August 2017 to February 2021] and Senior Research Fellowship [IF160758, March 2021 to August 2022]. KG gratefully acknowledges the financial support from the University of Hyderabad Institute of Eminence, UGC-SAP (DRS-I), and DBT-Builder project.
Acknowledgments
We thank Dr. R. K. Jain (Former Dean of the Indian Agricultural Research Institute, New Delhi) for providing us with the PRSV CP antisera. We also thank Dr. Valerian V. Dolja, Professor of Botany and Plant Pathology, Oregon State University, USA for providing us with the pCB301 vector.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1310236/full#supplementary-material
Footnotes
References
Adams, M. J., Antoniw, J. F., and Beaudoin, F. (2005). Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol. Plant Pathol. 6, 471–487. doi: 10.1111/j.1364-3703.2005.00296.x
Adie, B., Chico, J. M., Rubio-Somoza, I., and Solano, R. (2007). Modulation of plant defenses by ethylene. J. Plant Growth Regul. 26, 160–177. doi: 10.1007/s00344-007-0012-6
Ahlquist, P., French, R., Janda, M., and Loesch-Fries, L. S. (1984). Multicomponent RNA plant virus infection derived from cloned viral cDNA. Proc. Natl. Acad. Sci. U. S. A. 81, 7066–7070. doi: 10.1073/pnas.81.22.7066
Akbudak, M. A., Yildiz, S., and Filiz, E. (2020). Pathogenesis related protein-1 (PR-1) genes in tomato (Solanum lycopersicum L.): bioinformatics analyses and expression profiles in response to drought stress. Genomics 112, 4089–4099. doi: 10.1016/j.ygeno.2020.07.004
Ali, A., Shah, L., Rahman, S., Riaz, M. W., Yahya, M., Xu, Y. J., et al. (2018). Plant defense mechanism and current understanding of salicylic acid and NPRs in activating SAR. Physiol. Mol. Plant Pathol. 104, 15–22. doi: 10.1016/j.pmpp.2018.08.001
Anderson, P. K., Cunningham, A. A., Patel, N. G., Morales, F. J., Epstein, P. R., and Daszak, P. (2004). Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 19, 535–544. doi: 10.1016/j.tree.2004.07.021
Baillo, E. H., Kimotho, R. N., Zhang, Z., and Xu, P. (2019). Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 10:771. doi: 10.3390/genes10100771
Bao, W., Yan, T., Deng, X., and Wuriyanghan, H. (2020). Synthesis of full-length cDNA infectious clones of soybean mosaic virus and functional identification of a key amino acid in the silencing suppressor Hc-pro. Viruses 12:886. doi: 10.3390/v12080886
Boyer, J. C., and Haenni, A. L. (1994). Infectious transcripts and cDNA clones of RNA viruses. Virology 198, 415–426. doi: 10.1006/viro.1994.1053
Bravo, E., Calvert, L. A., and Morales, F. J. (2008). The complete nucleotide sequence of the genomic RNA of bean common mosaic virus strain NL4. Rev. Acad. Colomb. Cienc. 32, 37–46.
Brown, R. L., Kazan, K., McGrath, K. C., Maclean, D. J., and Manners, J. M. (2003). A role for the GCC-box in Jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 132, 1020–1032. doi: 10.1104/pp.102.017814
Chen, Z.-Y., Brown, R. L., Rajasekaran, K., Damann, K. E., and Cleveland, T. E. (2006). Identification of a maize kernel pathogenesis-related protein and evidence for its involvement in resistance to aspergillus flavus infection and aflatoxin production. Phytopathology 96, 87–95. doi: 10.1094/PHYTO-96-0087
Cheng, Y.-Q., Liu, Z.-M., Xu, J., Zhou, T., Wang, M., Chen, Y.-T., et al. (2008). HC-pro protein of sugar cane mosaic virus interacts specifically with maize ferredoxin-5 in vitro and in planta. J. Gen. Virol. 89, 2046–2054. doi: 10.1099/vir.0.2008/001271-0
Chikh Ali, M., Said Omar, A., and Natsuaki, T. (2011). An infectious full-length cDNA clone of potato virus Y(NTN-NW), a recently reported strain of PVY that causes potato tuber necrotic ringspot disease. Arch. Virol. 156, 2039–2043. doi: 10.1007/s00705-011-1062-4
Choi, J., Choi, D., Lee, S., Ryu, C.-M., and Hwang, I. (2011). Cytokinins and plant immunity: old foes or new friends? Trends Plant Sci. 16, 388–394. doi: 10.1016/j.tplants.2011.03.003
Chung, B. Y.-W., Miller, W. A., Atkins, J. F., and Firth, A. E. (2008). An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. 105, 5897–5902. doi: 10.1073/pnas.0800468105
de Oliveira, J. G., and Vitória, A. P. (2011). Papaya: nutritional and pharmacological characterization, and quality loss due to physiological disorders. An overview. Food Res. Int. 44, 1306–1313. doi: 10.1016/j.foodres.2010.12.035
Desbiez, C., Chandeysson, C., Lecoq, H., and Moury, B. (2012). A simple, rapid and efficient way to obtain infectious clones of potyviruses. J. Virol. Methods 183, 94–97. doi: 10.1016/j.jviromet.2012.03.035
Díaz-Cruz, G. A., Smith, C. M., Wiebe, K. F., Charette, J. M., and Cassone, B. J. (2018). First report of brome mosaic virus infecting soybean, isolated in Manitoba, Canada. Plant Dis. 102:460. doi: 10.1094/PDIS-07-17-1012-PDN
Domier, L. L., Franklin, K. M., Hunt, A. G., Rhoads, R. E., and Shaw, J. G. (1989). Infectious in vitro transcripts from cloned cDNA of a potyvirus, tobacco vein mottling virus. Proc. Natl. Acad. Sci. U. S. A. 86, 3509–3513. doi: 10.1073/pnas.86.10.3509
Eng, F., Marin, J. E., Zienkiewicz, K., Gutiérrez-Rojas, M., Favela-Torres, E., and Feussner, I. (2021). Jasmonic acid biosynthesis by fungi: derivatives, first evidence on biochemical pathways and culture conditions for production. PeerJ 9:e10873. doi: 10.7717/peerj.10873
Espinoza, C., Medina, C., Somerville, S., and Arce-Johnson, P. (2007). Senescence-associated genes induced during compatible viral interactions with grapevine and Arabidopsis. J. Exp. Bot. 58, 3197–3212. doi: 10.1093/jxb/erm165
Gal-On, A. (2000). A point mutation in the FRNK motif of the Potyvirus helper component-protease gene alters symptom expression in cucurbits and elicits protection against the severe homologous virus. Phytopathology 90, 467–473. doi: 10.1094/PHYTO.2000.90.5.467
Gal-On, A., Antignus, Y., Rosner, A., and Raccah, B. (1991). Infectious in vitro RNA transcripts derived from cloned cDNA of the cucurbit potyvirus, zucchini yellow mosaic virus. J. Gen. Virol. 72, 2639–2643. doi: 10.1099/0022-1317-72-11-2639
García-Marcos, A., Pacheco, R., Manzano, A., Aguilar, E., and Tenllado, F. (2013). Oxylipin biosynthesis genes positively regulate programmed cell death during compatible infections with the synergistic pair potato virus X-potato virus Y and tomato spotted wilt virus. J. Virol. 87, 5769–5783. doi: 10.1128/JVI.03573-12
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Gonsalves, D., Tripathi, S., Carr, J. B., and Suzuki, J. Y. (2010). Papaya Ringspot virus. Plant Health Instr. 10:1094. doi: 10.1094/PHI-I-2010-1004-01
Goossens, J., Fernández-Calvo, P., Schweizer, F., and Goossens, A. (2016). Jasmonates: signal transduction components and their roles in environmental stress responses. Plant Mol. Biol. 91, 673–689. doi: 10.1007/s11103-016-0480-9
Gopinath, K., Dragnea, B., and Kao, C. (2005). Interaction between brome mosaic virus proteins and RNAs: effects on RNA replication, protein expression, and RNA stability. J. Virol. 79, 14222–14234. doi: 10.1128/JVI.79.22.14222-14234.2005
Gopinath, K., Wellink, J., Porta, C., Taylor, K. M., Lomonossoff, G. P., and van Kammen, A. (2000). Engineering cowpea mosaic virus RNA-2 into a vector to express heterologous proteins in plants. Virology 267, 159–173. doi: 10.1006/viro.1999.0126
Hoang, X. L. T., Nhi, D. N. H., Thu, N. B. A., Thao, N. P., and Tran, L.-S. P. (2017). Transcription factors and their roles in signal transduction in plants under abiotic stresses. Curr. Genomics 18, 483–497. doi: 10.2174/1389202918666170227150057
Inoue-Nagata, A. K., Jordan, R., Kreuze, J., Li, F., López-Moya, J. J., Mäkinen, K., et al. (2022). ICTV Virus Taxonomy Profile: Potyviridae 2022. J. Gen. Virol. 103:001738. doi: 10.1099/jgv.0.001738
Iqbal, N., Khan, N. A., Ferrante, A., Trivellini, A., Francini, A., and Khan, M. I. R. (2017). Ethylene role in plant growth, development and senescence: interaction with other Phytohormones. Front. Plant Sci. 8:475. doi: 10.3389/fpls.2017.00475
Jiang, T., and Zhou, T. (2023). Unraveling the mechanisms of virus-induced symptom development in plants. Plan. Theory 12:2830. doi: 10.3390/plants12152830
Johansen, T., and Lamark, T. (2011). Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296. doi: 10.4161/auto.7.3.14487
Jorgensen, I., Rayamajhi, M., and Miao, E. A. (2017). Programmed cell death as a defence against infection. Nat. Rev. Immunol. 17, 151–164. doi: 10.1038/nri.2016.147
Kalleshwaraswamy, C. M., and Kumar, N. K. K. (2008). Transmission efficiency of papaya ringspot virus by three aphid species. Phytopathology 98, 541–546. doi: 10.1094/PHYTO-98-5-0541
Ke, Y.-D., Huang, Y.-W., Viswanath, K. K., Hu, C.-C., Yeh, C.-M., Mitsuda, N., et al. (2022). NbNAC42 and NbZFP3 transcription factors regulate the virus inducible NbAGO5 promoter in Nicotiana benthamiana. Front. Plant Sci. 13:924482. doi: 10.3389/fpls.2022.924482
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kurakawa, T., Ueda, N., Maekawa, M., Kobayashi, K., Kojima, M., Nagato, Y., et al. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655. doi: 10.1038/nature05504
Lehto, K., Tikkanen, M., Hiriart, J.-B., Paakkarinen, V., and Aro, E.-M. (2003). Depletion of the photosystem II core complex in mature tobacco leaves infected by the flavum strain of tobacco mosaic virus. Mol. Plant Microbe Interact. 16, 1135–1144. doi: 10.1094/MPMI.2003.16.12.1135
Liu, X., Chen, X., Liu, S., Du, K., Wang, P., Jiang, T., et al. (2021). A new potyvirus isolated from Pennisetum alopecuroides with the potential to infect cereal crops. Phytopathol. Res. 3:6. doi: 10.1186/s42483-021-00082-1
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Manners, J. M., Penninckx, I. A. M. A., Vermaere, K., Kazan, K., Brown, R. L., Morgan, A., et al. (1998). The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol. Biol. 38, 1071–1080. doi: 10.1023/A:1006070413843
Mishra, R., Patil, S., Patil, A., and Patil, B. L. (2019). Sequence diversity studies of papaya ringspot virus isolates in South India reveal higher variability and recombination in the 5′-terminal gene sequences. Virusdisease 30, 261–268. doi: 10.1007/s13337-019-00512-x
Mizuno, Y., Ohtsu, M., Shibata, Y., Tanaka, A., Camagna, M., Ojika, M., et al. (2019). Nicotiana benthamiana RanBP1-1 is involved in the induction of disease resistance via regulation of nuclear-cytoplasmic transport of small GTPase ran. Front. Plant Sci. 10:222. doi: 10.3389/fpls.2019.00222
Moghal, S. M., and Francki, R. I. B. (1976). Towards a system for the identification and classification of potyviruses: I. Serology and amino acid composition of six distinct viruses. Virology 73, 350–362. doi: 10.1016/0042-6822(76)90396-2
Nakatogawa, H., Ichimura, Y., and Ohsumi, Y. (2007). Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cells 130, 165–178. doi: 10.1016/j.cell.2007.05.021
Navas-Hermosilla, E., Fiallo-Olivé, E., and Navas-Castillo, J. (2021). Infectious clones of tomato chlorosis virus: toward increasing efficiency by introducing the Hepatitis Delta virus ribozyme. Front. Microbiol. 12:693457. doi: 10.3389/fmicb.2021.693457
Nelson, R. S., and Citovsky, V. (2005). Plant viruses. Invaders of cells and pirates of cellular pathways. Plant Physiol. 138, 1809–1814. doi: 10.1104/pp.104.900167
Ng, D. W.-K., Abeysinghe, J. K., and Kamali, M. (2018). Regulating the regulators: the control of transcription factors in plant defense signaling. Int. J. Mol. Sci. 19:3737. doi: 10.3390/ijms19123737
Park, S.-Y., Yu, J.-W., Park, J.-S., Li, J., Yoo, S.-C., Lee, N.-Y., et al. (2007). The senescence-induced Staygreen protein regulates chlorophyll degradation. Plant Cell 19, 1649–1664. doi: 10.1105/tpc.106.044891
Premchand, U., Mesta, R. K., Devappa, V., Basavarajappa, M. P., Venkataravanappa, V., Narasimha Reddy, L. R. C., et al. (2023). Survey, detection, characterization of papaya ringspot virus from southern India and management of papaya ringspot disease. Pathogens 12:824. doi: 10.3390/pathogens12060824
Riechmann, J., Laín, S., and Garcia, J. A. (1990). Infectious in vitro transcripts from a plum pox potyvirus cDNA clone. Virology 177, 710–716. doi: 10.1016/0042-6822(90)90537-2
Roossinck, M. J. (2010). Lifestyles of plant viruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 1899–1905. doi: 10.1098/rstb.2010.0057
Sakakibara, H. (2006). CYTOKININS: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57, 431–449. doi: 10.1146/annurev.arplant.57.032905.105231
Shoji-Kawata, S., and Levine, B. (2009). Autophagy, antiviral immunity, and viral countermeasures. Biochim. Biophys. Acta. 1793, 1478–1484. doi: 10.1016/j.bbamcr.2009.02.008
Son, G. H., Wan, J., Kim, H. J., Nguyen, X. C., Chung, W. S., Hong, J. C., et al. (2012). Ethylene-responsive element-binding factor 5, ERF5, is involved in chitin-induced innate immunity response. Mol. Plant Microbe Interact. 25, 48–60. doi: 10.1094/MPMI-06-11-0165
Sun, J.-Q., Jiang, H.-L., and Li, C.-Y. (2011). Systemin/Jasmonate-mediated systemic defense signaling in tomato. Mol. Plant 4, 607–615. doi: 10.1093/mp/ssr008
Supriya, L., Durgeshwar, P., Muthamilarasan, M., and Padmaja, G. (2022). Melatonin mediated differential regulation of drought tolerance in sensitive and tolerant varieties of upland cotton (Gossypium hirsutum L.). Front. Plant Sci. 13:821353. doi: 10.3389/fpls.2022.821353
Takei, K., Sakakibara, H., and Sugiyama, T. (2001). Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 276, 26405–26410. doi: 10.1074/jbc.M102130200
Tang, J., and Bassham, D. C. (2018). Autophagy in crop plants: what’s new beyond Arabidopsis? Open Biol. 8:180162. doi: 10.1098/rsob.180162
Tordo, V. M., Chachulska, A. M., Fakhfakh, H., Le Romancer, M., Robaglia, C., and Astier-Manifacier, S. (1995). Sequence polymorphism in the 5’NTR and in the P1 coding region of potato virus Y genomic RNA. J. Gen. Virol. 76, 939–949. doi: 10.1099/0022-1317-76-4-939
Tripathi, S., Suzuki, J. Y., Ferreira, S. A., and Gonsalves, D. (2008). Papaya ringspot virus-P: characteristics, pathogenicity, sequence variability and control. Mol. Plant Pathol. 9, 269–280. doi: 10.1111/j.1364-3703.2008.00467.x
Watanabe, N., and Lam, E. (2009). Bax Inhibitor-1, a conserved cell death suppressor, is a key molecular switch downstream from a variety of biotic and abiotic stress signals in plants. Int. J. Mol. Sci. 10, 3149–3167. doi: 10.3390/ijms10073149
Weaver, L. M., Gan, S., Quirino, B., and Amasino, R. M. (1998). A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol. Biol. 37, 455–469. doi: 10.1023/a:1005934428906
Wu, W., Du, K., Kang, X., and Wei, H. (2021). The diverse roles of cytokinins in regulating leaf development. Hortic. Res. 8, 118–113. doi: 10.1038/s41438-021-00558-3
Xu, Z., Song, N., Ma, L., and Wu, J. (2019). IRE1-bZIP60 pathway is required for Nicotiana attenuata resistance to fungal pathogen Alternaria alternata. Front. Plant Sci. 10:263. doi: 10.3389/fpls.2019.00263
Yeh, S.-D. (1984). Evaluation of induced mutants of papaya ringspot virus for control by cross protection. Phytopathology 74:1086. doi: 10.1094/Phyto-74-1086
Yeh, S.-D., Jan, F.-J., Chiang, C.-H., Doong, T.-J., Chen, M.-C., Chung, P.-H., et al. (1992). Complete nucleotide sequence and genetic organization of papaya ringspot virus RNA. J. Gen. Virol. 73, 2531–2541. doi: 10.1099/0022-1317-73-10-2531
Zhang, F., Liu, S., Zhang, T., Ye, Z., Han, X., Zhong, K., et al. (2021). Construction and biological characterization of an infectious full-length cDNA clone of a Chinese isolate of wheat yellow mosaic virus. Virology 556, 101–109. doi: 10.1016/j.virol.2021.01.018
Zhao, J.-H., Hua, C.-L., Fang, Y.-Y., and Guo, H.-S. (2016). The dual edge of RNA silencing suppressors in the virus-host interactions. Curr. Opin. Virol. 17, 39–44. doi: 10.1016/j.coviro.2015.12.002
Keywords: papaya ringspot virus, infectious cDNA, double promoter, ribozyme, PR genes, transcription factors
Citation: Gupta P, Parupudi PLC, Supriya L, Srivastava H, Padmaja G and Gopinath K (2023) Complete genome sequencing and construction of full-length infectious cDNA clone of papaya ringspot virus-HYD isolate and its efficient in planta expression. Front. Microbiol. 14:1310236. doi: 10.3389/fmicb.2023.1310236
Edited by:
Prabu Gnanasekaran, Washington State University, United StatesReviewed by:
Deepjyoti Singh, North Carolina State University, United StatesJose Trinidad Ascencio-Ibáñez, North Carolina State University, United States
Ved Prakash, Kansas State University, United States
Copyright © 2023 Gupta, Parupudi, Supriya, Srivastava, Padmaja and Gopinath. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kodetham Gopinath, a2duc2xAdW9oeWQuYWMuaW4=
 Prodosh Gupta
Prodosh Gupta Pavani L. C. Parupudi
Pavani L. C. Parupudi Laha Supriya
Laha Supriya Harshal Srivastava
Harshal Srivastava Gudipalli Padmaja
Gudipalli Padmaja