- 1School of Life Sciences and Engineering, Foshan University, Foshan, China
- 2Honghe Animal Disease Prevention and Control Center, Mengzi, China
- 3Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, China
- 4Institute of Special Economic Animal and Plant Sciences, Chinese Academy of Agricultural Sciences, Changchun, China
- 5Eco-Engineering Department, Guangdong Eco-Engineering Polytechnic, Guangzhou, China
Getah Virus (GETV) is an RNA virus that is transmitted by mosquitoes and can cause disease or death in a variety of vertebrates. Its prevalence is increasingly severe in Asia. This study conducted a GETV epidemiological investigation on 1,300 bovine sera collected in the Honghe Prefecture of Yunnan Province on the China-Myanmar border from 2022 to 2023. The positive rate of GETV antibodies in bovine serum in Honghe Prefecture was determined to be 20.25% through indirect Enzyme-linked immunosorbent test (ELISA) methods. Using Real-time PCR methods to detect GETV RNA in bovine serum, the positive rate was 0.23% (3/1300), and viral nucleic acids were only detected in three bovine sera in Jianshui area in 2022. The YN2305 strain was successfully isolated from mouse neuroblastoma (N2a) cells and the complete gene sequence was obtained. All the above results indicate the existence of GETV infection in cattle in Honghe Prefecture, Yunnan Province. Homology and genetic evolution analysis found that the isolated strain has a high homology with the JL1808 strain isolated from cattle in 2018, with a nucleotide identity of 100%, and a nucleotide identity of 99.8% with the SD17-09 strain isolated from foxes in 2017. Compared with the nucleotides of 44 virus strains published in Genbank, YN2305 has multiple nucleotide site mutations in the structural gene E2 and non-structural gene NS. The nucleotide and amino acid identity of the E2 gene are 94.2−100% and 96.4−100%, respectively. Genetic evolution analysis found that this virus strain is most closely related to the bovine origin JL1808, and it is in gene group III with HuN1, Kochi-01, SD17-09, MI-110-C1, and MI-110-C2 strains that causes significant clinical symptoms in Chinese pig, fox and horse populations, belonging to the same evolutionary branch. This study determined the infection rate, genotype, and main prevalence areas of GETV in bovine sera in the China-Myanmar border area. Therefore, the epidemiological investigation of GETV infection in multiple animal hosts should be further expanded, and research on its pathogenicity and vectors should be carried out.
1 Introduction
Getah virus (GETV) is an arbovirus, belonging to the Alphavirus genus of the Togaviridae family, along with Chikungunya virus (CHIKV) and Venezuelan equine encephalitis virus (VEEV) (Zhai et al., 2008; Shi et al., 2022a). GETV was first isolated from Culex gelidus mosquitoes in Malaysia in 1955 (Zhai et al., 2008; Ren et al., 2020; Xing et al., 2020; Shi et al., 2022a; Sun et al., 2022). Since then, GETV has rapidly expanded its host range and geographical distribution (Zhai et al., 2008). Currently, it has appeared and become prevalent in 12 countries including China, Russia, South Korea, Japan, Thailand, and Australia (Zhai et al., 2008; Yang et al., 2018; Rattanatumhi et al., 2022). Since its first isolation from mosquitoes in Hainan Province, China in 1964, it has broken out and become prevalent in 17 provinces and cities (Zhai et al., 2008). So far, the virus RNA or antibodies have been detected in horses, pigs, cattle, sheep, blue foxes, kangaroos, monkeys, red pandas, and some poultry (Nemoto et al., 2015; Li et al., 2017, 2019; Yang et al., 2018; Ren et al., 2020; Xing et al., 2020; Rattanatumhi et al., 2022; Shi et al., 2022a; Zhao et al., 2022). Of particular note is that neutralizing antibodies against GETV have been found in human serum in Malaysia, Australia, and China (Zhai et al., 2008; Shi et al., 2022b), and the specific antibody titers in febrile populations are significantly higher than in healthy populations, indicating that GETV infection is related to human diseases. Yunnan Province, located in southwestern China, borders Myanmar, Laos, and Vietnam. Frequent tourism and animal trade have accelerated the spread of arboviruses locally, which may pose a threat to livestock and human health. In recent years, most GETV strains in China have come from four genera and seven species of mosquito specimens in Yunnan Province (Li et al., 2019). Cattle are the primary reservoir hosts of arboviruses, however, there have been no reports on cattle infection with GETV in the border areas of Yunnan Province. Therefore, this study conducted a GETV epidemiological investigation on bovine serum samples in the China-Myanmar border area from 2022-2023 using indirect ELISA and real-time PCR methods, to provide data support for disease prevention and control.
2 Materials and methods
2.1 Animal serum collection
From 2022 to 2023, a total of 1,300 bovine sera were collected in Honghe Prefecture, Yunnan Province, including Luxi (400), Jianshui (420), and Mile (480). All samples were stored in a −80°C freezer.
2.2 RNA detection
RNA was extracted from the bovine serum samples using the QIAamp Viral RNA Mini Kit (Qiagen). The RNA was then converted to cDNA using the Vazyme HiScript II First Strand cDNA Synthesis Kit (Vazyme Biotech Co., Ltd, China). The detection was carried out using the previously established quantitative reverse transcription PCR (RT-qPCR) detection method for GETV non-structural protein 1 (nsP1) (Shi et al., 2018).
2.3 Enzyme-linked immunosorbent test (ELISA)
The amino acid sequence of the E2 protein of GETV virus strain in Genbank was used, and after codon optimization, the gene was artificially synthesized and directionally cloned into the pGEX-6P-1 vector. After correct identification by enzyme cleavage and sequencing, it was transformed into the host bacterium BL-21 for induced expression. The identification of the recombinant E2 (rE2) protein was achieved by employing Western blot and SDS-PAG techniques subsequent to the completion of GST tag purification. Subsequently, the rE2 protein was employed as a diagnostic antigen in order to establish an indirect ELISA method for the detection of GETV antibodies, in accordance with prior literature (Sun et al., 2022). The cross-reactivity of the rE2-based indirect ELISA was assessed by examining the anti-sera from various common bovine and porcine viruses, such as BVDV, FMDV, PRRSV, BATV, JEV, and PEDV. Additionally, the sensitivity of the Indirect ELISA was analyzed by testing different dilutions of bovine GETV positive serum.
2.4 Virus isolation and amplification of whole gene sequence
The serum with GETV nucleic acid positive was filtered through a 0.22°μm filter (Millipore, Billerica, MA, USA) and inoculated into N2a monolayer cells in a six-well plate. After incubating at 37°C for 1°h, the cells were washed twice with PBS, and preserved in 2% FBS (Gibco) MEM in a 5% CO2 incubator. The cytopathic effect (CPE) of the cells was observed daily. After five continuous cell passages, the cell suspension was collected and the isolated GETV strain was sequenced for the whole genome (Ren et al., 2020).
2.5 Alignment and phylogenetic analysis of GETV genome sequences
To analyze the homology relationship between GETV YN2305 isolate and other various GETV strains, the MegAlign software was used to perform homology analysis on the full gene and E2 gene sequences of the YN2305 isolate. Other GETV strains are from those registered in Genbank. MEGA 7 software was used to perform phylogenetic analysis on 44 full genes and 58 E2 gene groups of GETV that have been published in Genbank and YN2305. The Tamura-Nei model and the maximum likelihood method of gamma distribution rate heterogeneity were used in MEGA 7.
2.6 Statistical analysis
The binom.confint function in R version 3.5.0 was used to calculate the seroprevalence and 95% confidence interval (95% CI).
3 Results
3.1 Results of the serological tests
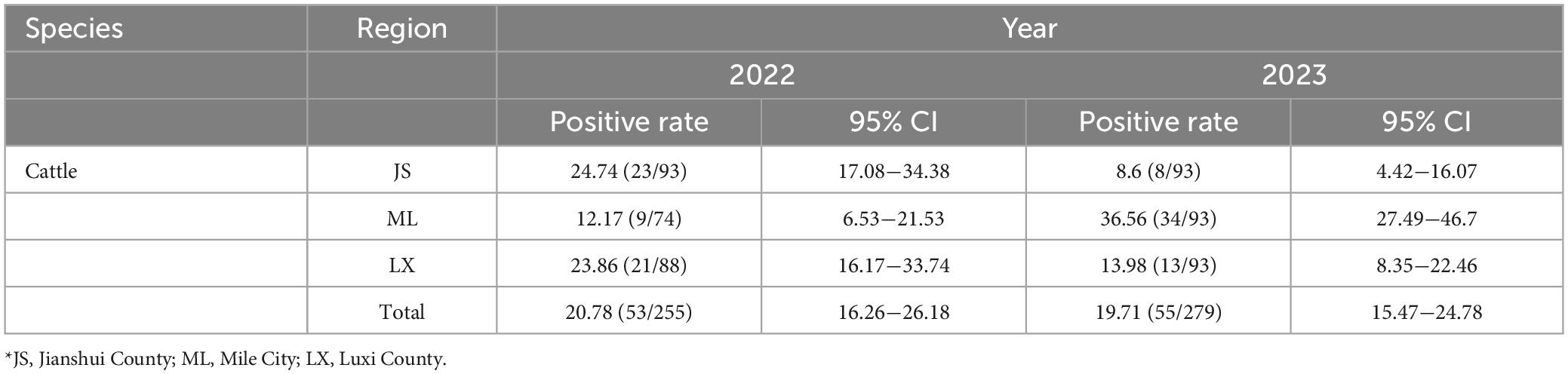
From 2022 to 2023, a total of 1,300 bovine sera were collected in Honghe Prefecture, Yunnan Province, with a total positive rate of 20.25% (108/534, 95% CI: 17.04−27.84). Among them, 1,000 bovine serum samples collected in Jianshui, Mile, and Luxi regions of Honghe Prefecture in Yunnan Province in 2022 were tested, with a positive rate of 20.78% (53/255, 95% CI: 16.26−26.18). The positive rates in each region were 24.74% (23/93, 95% CI: 17.08−34.38), 12.17% (9/74, 95% CI: 6.53−21.53), and 23.86% (21/88, 95% CI: 16.17−33.74), respectively. In 2023, the positive rate of GETV antibodies in 300 bovine serum samples collected in Honghe Prefecture was 19.71% (55/279, 95% CI: 15.47−24.78), with Jianshui, Mile, and Luxi counties at 8.6% (8/93, 95% CI: 4.42−16.07), 36.56% (34/93, 95% CI:27.49−46.7), and 13.98% (13/93, 95% CI: 8.35−22.46), respectively, (Table 1).
3.2 GETV RNA test results
All bovine serum samples were tested for GETV RNA using RT-qPCR method, with a positive rate of 0.23% (3/1300).
3.3 Enzyme-linked immunosorbent test (ELISA)
Following the process of GST label purification, the E2 protein underwent identification through SDS-PAGE and Western blot techniques, which revealed a protein size of 74.6 kDa, aligning with the anticipated value (Technical Appendix Supplementary Figure 1). The average outcomes of the antisera for BVDV, FMDV, PRRSV, BATV, JEV, and PEDV indicated negativity, thereby confirming the absence of cross-reactivity with the established rE2 indirect ELISA. Consequently, the indirect ELISA exhibited a significant of specificity. In addition, the serum from bovines infected with GETV were subjected to testing at various dilutions, ranging from 1:100 to 1:51200. The results indicated that the lowest detectable dilution using this methodology was 1:6400.
3.4 GETV homology and genetic evolution analysis
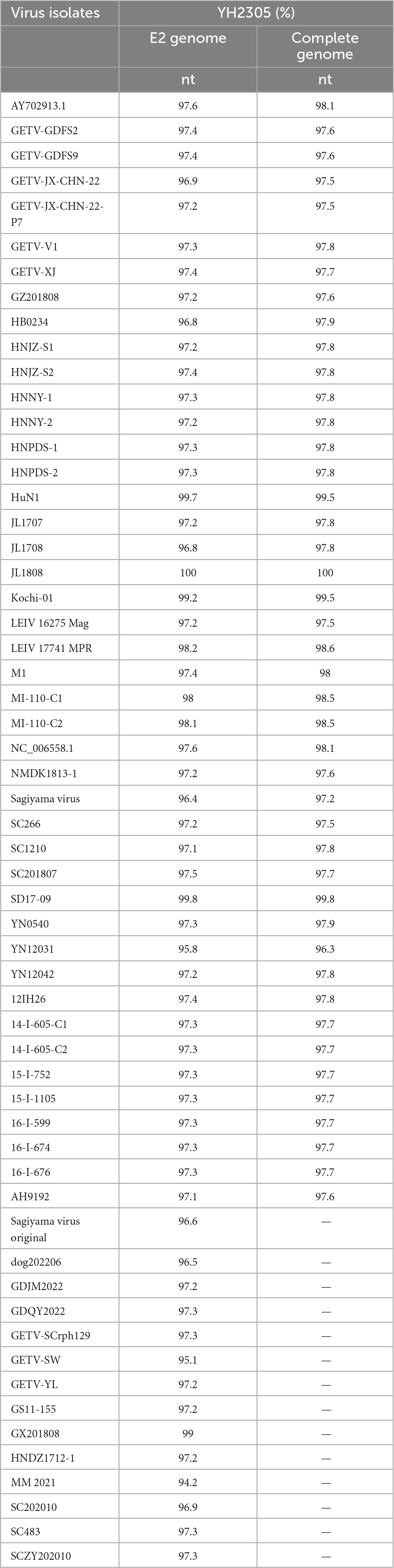
The full gene sequence of GETV YN2305 strain was obtained by PCR method with a length of 11,689 nt and submitted to Genbank (Genbank number: OR371719). Homology analysis was carried out with 44 GETV strains registered in Genbank, revealing that the YN2305 strain has the highest nucleotide homology with the bovine-derived JL1808 strain, with an identity of 100%. The nucleotide identity with the fox-derived SD17-09 strain and pig-derived Kochi-01 strain were 99.8 and 99.5%, respectively. Similarly, comparison with 58 E2 gene nucleotide sequences revealed that the identity of YN2305 strain with JL1808 was 100%, while the identity with E2 genes of other strains was 94.2−99.8% (Table 2). Through the phylogenetic analysis of the full gene and E2, YN2305 belongs to the GETV gene group III, and is most closely related to the bovine-derived JL1808 in genetic evolution, belonging to the same branch (Li et al., 2017; Figures 1A, B).
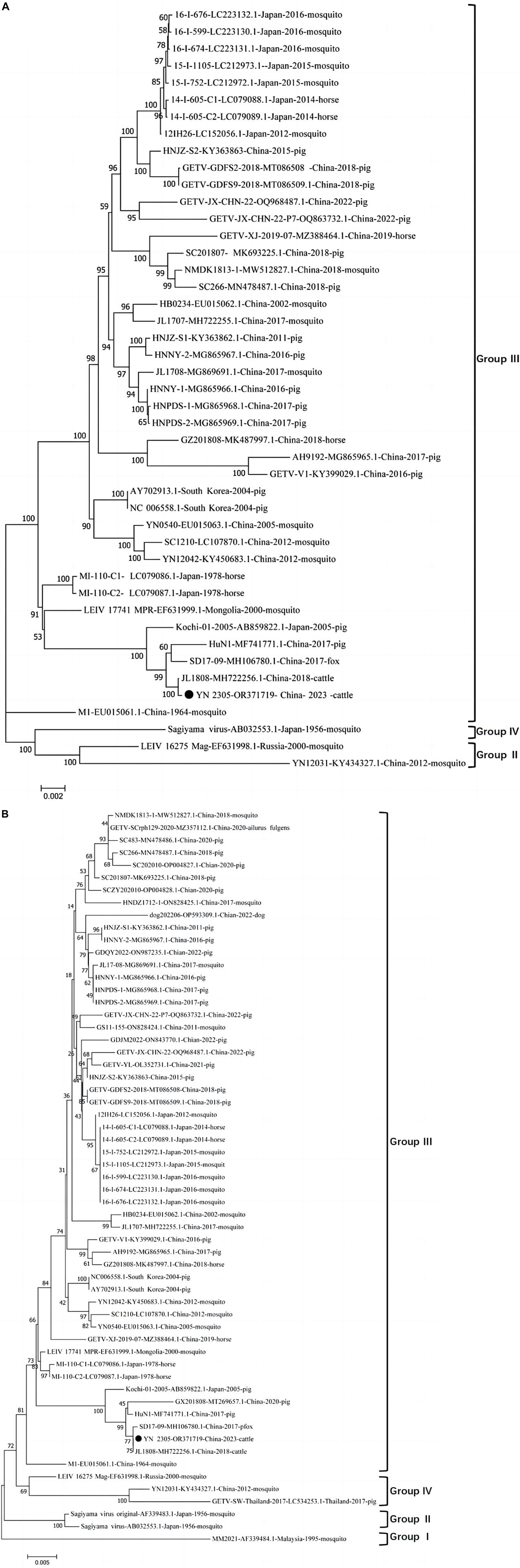
Figure 1. (A) Diagrammatic representation of a tree based on the complete genome sequence of GETV YN2305 and the available complete GETV genomes from the GenBank. (B) Diagrammatic representation of a tree based on the complete E2 gene nucleotide sequences of GETVs.
4 Discussion
Getah virus (GETV) was first discovered in Hainan Province, China in 1964 (Li et al., 2019; Lu et al., 2019). In recent years, the widespread distribution of GETV has been found in mosquitoes and animals in 21 provinces and cities including Yunnan, Sichuan, Guizhou, Gansu, Jilin, Hebei, and Shanxi. Prior to 2006, only six provinces in China were known to be affected by GETV (Zhai et al., 2008), but by 2018, the number of provinces affected had dramatically increased to 15 (Lu et al., 2019; Xia et al., 2021). Yunnan Province, bordering Laos, Myanmar, and Vietnam, has frequent trade and a climate conducive to mosquito breeding, facilitating the transmission of GETV. Reports indicate that there are high titers of GETV neutralizing antibodies in many livestock in Yunnan Province, and the positive antibody rate in pigs is high (Li et al., 2019), indicating that cattle, as livestock, face the risk of infection from pig-derived GETV. This study detected the highest positive rate of GETV antibodies in bovine serum in Jianshui County, Honghe Prefecture, Yunnan Province in 2022 at 24.74% using indirect ELISA methods. In 2023, the positive rate in Jianshui decreased by 16.14%, which related to the implementation of local pest and mosquito control measures and the seasonal prevalence of vector-borne viruses. While the positive rate in Mile County increased by 24.39% compared to the previous year. Based on these serological survey results, we speculate that there is a trend for GETV virus to spread from border areas to inland areas. The YN2305 strain was successfully isolated from cattle in Yunnan Province (Genbank Number: OR371719), and this isolate has the highest nucleotide identity and the closest genetic evolutionary relationship with the previously isolated bovine-derived JL1808 strain in Jilin Province. It also has high identity and close phylogenetic relationships with the fox-derived SD17-09 strain and the pig-derived HuN1 strain, which causes fever, anorexia, and neurological symptoms in host animals, but there are multiple mutations in the E2 gene and NS gene of GETV, which may be related to host differences (Shi et al., 2022b). In addition, phylogenetic analysis shows that YN2305, along with Kochi-01, MI-110-C1, and MI-110-C2 strains that causes significant clinical symptoms in Chinese pig and horse populations, belongs to gene group III and the same evolutionary branch (Nemoto et al., 2015). These results indicate that GETV is widely present in various animal hosts with differences in nucleotide sequences. As cattle are in close contact with humans and other animals, the bovine-derived isolate YN2305 may pose a threat to other livestock, wildlife, and even humans. To reduce the impact of GETV virus on humans and animals, it is necessary to further expand the scope of monitoring and carry out long-term serological surveys.
5 Conclusion
In conclusion, this study indicates that GETV is present and prevalent in cattle in the China-Vietnam border region, with the highest positive rate of bovine serum antibodies in Jianshui and Mile counties, and a trend to spread inland. In addition, GETV YN2305 strain has the highest homology with bovine isolate JL1808, but there are genetic differences in the E2 and NS genes with closely related fox and pig derived isolates. The isolate may pose a threat to other livestock and humans. This study fills the gap in information on the infection rate and prevalence of GETV in livestock in the Yunnan border region, providing data support for the prevention and control of the epidemic.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/; OR371719.
Ethics statement
The animal studies were approved by the Institutional Animal Care and Use Ethics Committee (IACUC) of the Chinese Academy of Military Medical Science. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
HL: Funding acquisition, Investigation, Writing – original draft. JH: Investigation, Writing – original draft, Data curation, Formal Analysis. L-XL: Software, Validation, Writing – review & editing. Z-SL: Software, Validation, Writing – review & editing. X-TS: Resources, Writing – review & editing. H-JL: Resources, Writing – review & editing. N-YJ: Resources, Supervision, Writing – review & editing. LZ: Supervision, Writing – review & editing. L-NZ: Supervision, Writing – review & editing, Funding acquisition.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Sciences Foundation of China (32273097), Key Laboratory for prevention and control of Avian Influenza and Other Major Poultry Diseases, Ministry of Agriculture and Rural Affairs, P.R. China; Key Laboratory of Livestock Disease Prevention of Guangdong Province (YDWS202205).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1309650/full#supplementary-material
References
Li, Y., Fu, S., Guo, X., Li, X., Li, M., Wang, L., et al. (2019). Serological survey of getah virus in domestic animals in Yunnan Province, China. Vector Borne Zoonot. Dis. 19, 59–61. doi: 10.1089/vbz.2018.2273
Li, Y.-Y., Liu, H., Fu, S.-H., Li, X.-L., Guo, X.-F., Li, M.-H., et al. (2017). From discovery to spread: The evolution and phylogeny of Getah virus. Infect. Genet. Evol. 55, 48–55. doi: 10.1016/j.meegid.2017.08.016
Lu, G., Ou, J., Ji, J., Ren, Z., Hu, X., Wang, C., et al. (2019). Emergence of Getah virus infection in horse with fever in China, 2018. Front. Microbiol. 10:1416.
Nemoto, M., Bannai, H., Tsujimura, K., Kobayashi, M., Kikuchi, T., Yamanaka, T., et al. (2015). Getah virus infection among racehorses, Japan, 2014. Emerg. Infect. Dis. 21, 883–885. doi: 10.3201/eid2105.141975
Rattanatumhi, K., Prasertsincharoen, N., Naimon, N., Kuwata, R., Shimoda, H., Ishijima, K., et al. (2022). A serological survey and characterization of Getah virus in domestic pigs in Thailand, 2017–2018. Transbound. Emerg. Dis. 69, 913–918. doi: 10.1111/tbed.14042
Ren, T., Mo, Q., Wang, Y., Wang, H., Nong, Z., Wang, J., et al. (2020). Emergence and phylogenetic analysis of a Getah virus isolated in Southern China. Front. Vet. Sci. 7:552517. doi: 10.3389/fvets.2020.552517
Shi, N., Liu, H., Li, L., Hu, B., Zhang, L., Zhao, C., et al. (2018). Development of a TaqMan probe-based quantitative reverse transcription PCR assay for detection of Getah virus RNA. Arch. Virol. 163, 2877–2881. doi: 10.1007/s00705-018-3927-2
Shi, N., Qiu, X., Cao, X., Mai, Z., Zhu, X., Li, N., et al. (2022a). Molecular and serological surveillance of Getah virus in the Xinjiang Uygur Autonomous Region, China, 2017–2020. Virol. Sin. 37, 229–237. doi: 10.1016/j.virs.2022.02.004
Shi, N., Zhu, X., Qiu, X., Cao, X., Jiang, Z., Lu, H., et al. (2022b). Origin, genetic diversity, adaptive evolution and transmission dynamics of Getah virus. Transbound. Emerg. Dis. 69, e1037–e1050. doi: 10.1111/tbed.14395
Sun, Q., Xie, Y., Guan, Z., Zhang, Y., Li, Y., Yang, Y., et al. (2022). Seroprevalence of Getah virus in pigs in Eastern China determined with a recombinant E2 protein-based indirect ELISA. Viruses 14:2173. doi: 10.3390/v14102173
Xia, Y., Shi, Z., Wang, X., Li, Y., Wang, Z., Chang, H., et al. (2021). Development and application of SYBR Green I real-time quantitative reverse transcription PCR assay for detection of swine Getah virus. Mol. Cell. Probes 57:101730. doi: 10.1016/j.mcp.2021.101730
Xing, C., Jiang, J., Lu, Z., Mi, S., He, B., Tu, C., et al. (2020). Isolation and characterization of Getah virus from pigs in Guangdong province of China. Transbound. Emerg. Dis. doi: 10.1111/tbed.13567 [Epub ahead of print].
Yang, T., Li, R., Hu, Y., Yang, L., Zhao, D., Du, L., et al. (2018). An outbreak of Getah virus infection among pigs in China, 2017. Transbound. Emerg. Dis. 65, 632–637. doi: 10.1111/tbed.12867
Zhai, Y., Wang, H.-Y., Sun, X., Fu, S., Wang, H., Attoui, H., et al. (2008). Complete sequence characterization of isolates of Getah virus (genus Alphavirus, family Togaviridae) from China. J. Gen. Virol. 89, 1446–1456. doi: 10.1099/vir.0.83607-0
Keywords: Getah virus, virus Isolation, sequence determination and analysis, phylogenetic analysis, seroepidemiological investigation, cattle
Citation: Liu H, Hu J, Li L-X, Lu Z-S, Sun X-T, Lu H-J, Jin N-Y, Zhang L and Zhang L-N (2023) Seroepidemiological investigation of Getah virus in the China-Myanmar border area from 2022-2023. Front. Microbiol. 14:1309650. doi: 10.3389/fmicb.2023.1309650
Received: 08 October 2023; Accepted: 20 November 2023;
Published: 14 December 2023.
Edited by:
James Weger-Lucarelli, Virginia Tech, United StatesReviewed by:
Shandian Gao, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, ChinaHongzhuan Wu, Alabama State University, United States
Copyright © 2023 Liu, Hu, Li, Lu, Sun, Lu, Jin, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Na Zhang, emxuX3RpYW50YW5nQDE2My5jb20=; Lei Zhang, emhhbmdsZWlfdGNzQDEyNi5jb20=
†These authors have contributed equally to this work and share first authorship
 Hao Liu
Hao Liu Jin Hu
Jin Hu Li-Xia Li
Li-Xia Li Zi-Shuo Lu
Zi-Shuo Lu Xiu-Tao Sun
Xiu-Tao Sun Hui-Jun Lu
Hui-Jun Lu Ning-Yi Jin
Ning-Yi Jin Lei Zhang
Lei Zhang Li-Na Zhang
Li-Na Zhang