
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 November 2023
Sec. Microbe and Virus Interactions with Plants
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1305899
This article is part of the Research Topic Women in Microbe and Virus Interactions with Plants: 2022/2023 View all 6 articles
The plant pathogenic bacterium Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) causes disease in tomato, in the model plant Arabidopsis thaliana, and conditionally in Nicotiana benthamiana. The pathogenicity of Pst DC3000 is mostly due to bacterial virulence proteins, known as effectors, that are translocated into the plant cytoplasm through the type III secretion system (T3SS). Bacterial type III secreted effectors (T3SEs) target plants physiological processes and suppress defense responses to enable and support bacterial proliferation. The Pst DC3000 T3SE HopD1 interferes with plant defense responses by targeting the transcription factor NTL9. This work shows that HopD1 also targets the immune protein AtNHR2B (Arabidopsis thaliana nonhost resistance 2B), a protein that localizes to dynamic vesicles of the plant endomembrane system. Live-cell imaging of Nicotiana benthamiana plants transiently co-expressing HopD1 fused to the epitope haemagglutinin (HopD1-HA) with AtNHR2B fused to the red fluorescent protein (AtNHR2B-RFP), revealed that HopD1-HA interferes with the abundance and cellular dynamics of AtNHR2B-RFP-containing vesicles. The results from this study shed light into an additional function of HopD1 while contributing to understanding how T3SEs also target vesicle trafficking-mediated processes in plants.
The bacterium Pseudomonas syringae is a ubiquitous and diverse bacterium species complex that can be isolated from a wide range of habitats, including agricultural environments (Morris et al., 2013). P. syringae strains isolated from agricultural habitats have been intensely investigated as they cause diseases in a wide range of plant species (Mansfield et al., 2012). Among the pathogenic strains of P. syringae, Pseudomonas syringae pv. tomato DC3000 has acquired particular importance for its ability to cause disease in the model plant Arabidopsis thaliana and conditionally on Nicotiana benthamiana (Wei et al., 2007; Xin and He, 2013).
All the plant pathogenic strains of P. syringae possess a hrp/hrc gene cluster that encodes a type III secretion system (T3SS), a complex of proteins assembled in the inner and outer bacterial membranes; the T3SS extends outwards with an extracellular filament termed the Hrp pilus, through which bacterial proteins, known as type III secreted effectors (T3SEs), are translocated directly into the plant cytoplasm (Alfano and Collmer, 2004). Translocation of T3SEs results in two opposite outcomes depending on the specific P. syringae strain and plant combination. In nonhost plants, T3SE are recognized directly or indirectly by plants nucleotide-binding leucine-rich repeat receptors (NLRs), triggering Effector-Trigger Immunity (ETI) that culminates in a process of programmed cell death called the hypersensitive response (HR). In host plants, T3SEs promote bacterial parasitism by interfering with plant defense responses, altering essential biological functions and promoting pathogen proliferation, ultimately resulting in disease development (Wei et al., 2018).
The complexity, diversity and abundance of T3SEs has been uncovered by analyzing the genomic sequence of 494 P. syringae strains (Dillon et al., 2019). Interestingly, while each P. syringae strain has a unique number and repertoire of effectors, four core effectors: AvrE, HopB1, HopM1 and HopAA1, were identified in more than 95% of the strains, highlighting their importance in basic interactions with plants, or in pathogenesis (Bundalovic-Torma et al., 2022). The function of the remaining effectors in each strain is difficult to assess for the following reasons: (i) a given effector can have several targets, (ii) multiple effectors are redundant, having the same function, or targeting the same plant component, and (iii) effectors can interact with other effectors in multiple combinations. The generation of Pst DC3000 polymutants with sequential deletion of its well-expressed 28 effectors and further combinatorial reassembly of effectors has significantly contributed to untangle the complexity of Pst DC3000 by evaluating their growth in N. benthamiana (Kvitko et al., 2009; Cunnac et al., 2011). With that approach, eight effectors were identified as the minimal repertoire required for pathogenesis of Pst DC3000 in N. benthamiana: AvrPtoB, HopM1, HopE1, HopG1, HopAM1-1, AvrE1, HopAA1-1 and HopN1 (Kvitko et al., 2009; Cunnac et al., 2011). In addition to those effectors, HopD1 was also identified as an important effector to enable Pst DC3000 growth in N. benthamiana and Arabidopsis (Kvitko et al., 2009; Block et al., 2014), likely due to its ability to suppress ETI (Block et al., 2014). Moreover, transgenic plants expressing HopD1 enhanced the growth of the avirulent Pst DC3000 strains: Pst DC3000 (avrRpm1) and Pst DC3000 (avrRpt2), which in non-transgenic plants are detected by the plant immune system preventing their proliferation (Block et al., 2014).
Previously, AtNHR2B was identified as a protein participating in plant immunity, as the Atnhr2b mutant was susceptible to the non-adapted pathogen P. syringae pv. tabaci, which is unable to cause disease and proliferate in wild-type Arabidopsis plants (Singh et al., 2018). Moreover, AtNHR2B fused to the green fluorescent protein (AtNHR2B-GFP) localizes to cytoplasm, chloroplasts and small dynamic vesicles of unknown identity, which represent compartments of the endomembrane system that participate in vesicle trafficking-mediated events such as endocytosis and secretion (Singh et al., 2018). Interestingly, inoculation of Arabidopsis AtNHR2B-GFP-transgenic lines with the adapted pathogen Pseudomonas syringae pv. tomato DC3000 (Pst DC3000), which is capable of causing disease in Arabidopsis, alters the localization and abundance of AtNHR2B-GFP (Rodriguez-Puerto et al., 2022). This finding led us to hypothesize that Pst DC3000 type III secreted effectors (T3SE) acting as virulence proteins target AtNHR2B. Given the importance of HopD1 in Pst DC3000 virulence (Kvitko et al., 2009; Block et al., 2014), we decided to investigate whether AtNHR2B could be an additional target for HopD1. The results showed that HopD1 physically interacts and partially co-localizes with AtNHR2B in N. benthamiana, and that HopD1 reduces the abundance and cellular dynamics of AtNHR2B-containing vesicles. These results provide direct evidence that Pst DC3000 hijacks the plant immune system by interfering with vesicle trafficking events.
N. benthamiana seeds were sown on potting soil (PRO mix LP15), kept at 4°C for 2 days in the dark to break dormancy and transferred to a growth chamber set up at 21°C with a photoperiod of 8 h of light and 16 h of darkness. Seeds were grown for 2 weeks, and single seedlings were transplanted to individual pots and grown for two more weeks.
Full length AtNHR2B in the entry vector pDONR201 was recombined into vector pK7RWG2 to generate AtNHR2B-RFP, pK7FWG2::HopD1-GFP and pPZ212::HopD1-HA (Block et al., 2014), were obtained from Dr. Jim Alfano, University of Nebraska-Lincoln.
Plasmid pK7RWG2::AtNHR2B, pK7FWG2::HopD1-GFP and pPZ212::HopD1-HA were transformed into Agrobacterium tumefaciens GV2260 and plated on Luria Bertani (LB) supplemented with rifampicin (50 μg/mL) and streptomycin (50 μg/mL). Glycerol stocks were generated from individual transformants obtained.
AtNHR2B and HopD1 were cloned into bait and prey vectors of the ProQuest™ Two-Hybrid System (Invitrogen, Carlsbad, CA). Full length AtNHR2B in the entry vector pDONR201 was recombined into the bait vector pDEST22 to generate a fusion to the GAL4 transcriptional activation domain. HopD1 fused to the GAL4 DNA binding domain in pDEST32 vector was synthesized by Gene Universal (Newark, DE).
Yeast strain MaV203 competent cells (Thermo Fisher Scientific, Waltham, MA) were transformed with yeast two-hybrid constructs using the Frozen-EZ Yeast Transformation II Kit (Zymo Research, Irvine, CA). The following combinations of constructs were co-transformed into yeast: pDEST22 + pDEST32::HopD1 and pDEST22::AtNHR2B + pDEST32::HopD1. The combination of constructs (pEXP22™ /RalGDS-wt + pEXP32™/Krev1) and (pEXP22™ /RalGDS-m1 + pEXP32™/Krev1), provided by the manufacturer (Thermo Fisher Scientific, Waltham, MA), were used as positive and negative controls, respectively. Transformed yeast cells were plated on Double Drop Out (DDO) selection plates lacking amino acids leucine and tryptophan and grown at 30°C for 4 days. Single colonies were picked from the plates and cultured in 15 mL DDO broth at 30°C overnight. The overnight culture was diluted to an OD600nm of 0.2 and plated on Triple Drop Out (TDO) selection plates lacking amino acids leucine, tryptophan and histidine and supplemented with 15 mM 3-Amino-1,2,4-Triazole (3-AT) and grown at 30°C for 4 days.
Agrobacterium tumefaciens strains harboring constructs of interest, depending on the experiment, were streaked out on LB plates containing rifampicin (50 μg/mL) and streptomycin (50 μg/mL); single colonies were transferred to LB broth with respective antibiotics, and grown with constant aeration for 18 h. Cultures were harvested by centrifugation at 6,000 rpm for 10 min and supernatants were discarded. Bacterial pellets were resuspended in 5 mL induction buffer (20 mM MES pH 5.5; Mannitol; 200 mM acetosyringone) and incubated for 4 h at room temperature with slow agitation. After induction, bacteria were harvested by centrifugation at 6,000 rpm for 10 min, and pellets resuspended in 5 mL resuspension buffer (10 mM MES pH 5.5). Bacterial concentration was adjusted to a final concentration of OD600 of 0.3 and infiltrated into fully expanded leaves of four-week-old N. benthamiana plants.
A. tumefaciens harboring HopD1-HA was co-infiltrated either with A. tumefaciens strains harboring AtNHR2B-GFP or A. tumefaciens harboring free GFP into fully expanded leaves of four-week-old N. benthamiana plants. Infiltrated leaves were collected at 3 days post-infiltration and tissue was ground in liquid nitrogen for protein extraction. Tissue powder was homogenized in 5 mL of co-immunoprecipitation extraction buffer (100 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10 mM MgCl2, 10% Glycerol, 0.2% Nonidet P-40, 1 mM PMSF, 5 mM DTT, 1X Proteinase inhibitor cocktail) (Sigma Aldrich, St. Louis, MO) and incubated on ice for 30 min. Samples were centrifuged at 4°C for 30 min at 13,000 rpm and supernatants containing extracted proteins were collected in a pre-chilled 50 mL falcon tube.
Before co-immunoprecipitation, protein concentration was measured by Bradford Assay (BioRad, Hercules, CA) and proteins of interest were confirmed by running protein samples into a SDS-PAGE gel followed by Western blot with Anti-GFP-HRP (1:1000 dilution; Miltenyi Biotec, Auburn, CA) and Anti-HA-HRP (1:1000 dilution; Thermo Fisher Scientific Inc., Carlsbad, CA) and detected by luminol solution (ImmunoCruz, SantaCruz Biotechnology Inc., Dallas, TX).
For co-immunoprecipitation, 1 mg of total protein extract was mixed with 20 μL of PierceTM HA Epitope Tag Antibody conjugated to agarose beads (Thermo Fisher Scientific, Waltham, MA) and incubated overnight at 4°C with end-to-end rocking. After incubation, protein complexes bound to beads were washed three times with 1X TBS buffer (50 mM Tris–HCl, 150 mM NaCl, pH 7.5). Protein complexes bound to the beads were eluted in 2× SDS protein loading buffer, loaded and ran into an SDS-PAGE gel and transferred to nitrocellulose membranes. Proteins were detected by Western Blot with anti-GFP-HRP or anti-HA-HRP antibodies.
N. benthamiana plants transiently co-expressing AtNHR2B-RFP and HopD1-GFP were imaged at 2 days post-inoculation (dpi) using the Leica Stellaris 8 laser scanning confocal microscope at 20× magnification. Images were acquired in sequential scanning mode in the GFP (excitation wavelength: 496 nm, emission wavelength: 549 nm) and RFP (excitation wavelength: 570 nm, emission wavelength: 657 nm) channels.
Five independent images were chosen for colocalization analysis. Images in microscope-specific format (.lif) files were imported into FIJI, converted into TIFF files and pre-processed to remove background noise and to adjust brightness and contrast using automatic function. Green and read channels were split into separate images, and the overlapped image was selected for analysis. BIOP’s (Bioimaging and optics Platform) JACoP (Just Another Colocalization Plugin) plugin was used to evaluate co-localization of AtNHR2B-RFP and HopD1-GFP and to calculate Pearson’s Correlation coefficient.
N. benthamiana plants transiently expressing AtNHR2B-RFP alone or AtNHR2B-RFP in combination with HopD1-HA were imaged at 2 dpi using the Leica Stellaris 8 laser scanning confocal microscope. Images were acquired using the XYT image acquisition mode, to generate 4 videos of 120 frames for each treatment. Each video was taken using identical parameters: 1024 × 512, 400 bps, with line averages of 1, line accuracies of 1, frame averages of 1, and frame accuracies of 1.
Tracking of AtNHR2B-RFP-containing vesicles in each treatment was done with the software IMARIS. Video files in microscope-specific format (.lif) were converted into IMARIS-specific format (.ims) files which were used for subsequent data processing and analysis. Using the Spot Objects option and the circle selection mode, vesicles of 0.75 μm in diameter were chosen for analysis. A total of 400 vesicles for each treatment (AtNHR2B-RFP or AtNHR2B-RFP/HopD1-HA) were tracked in automated mode for speed and displacement using software’s default parameters.
Data obtained from speed and displacement was imported into R version 4.2.1. and sorted into 30 bins of equal range. Speed was sorted into bins ranging from 0.00738 μm/s to 2.56 μm/s and displacement was sorted into bins ranging from 0.00518 to 13.93 μm.
The previous findings that HopD1 in is important in Pst DC3000 virulence (Kvitko et al., 2009; Block et al., 2014), led us to hypothesize that HopD1 targets AtNHR2B. To start testing this hypothesis, we initially evaluated the physical interaction between both proteins in yeast given its simplicity. The interaction in yeast was evaluated by co-transforming AtNHR2B and HopD1 into yeast. The results show that all the co-transformations grew in DDO, lacking leucine and tryptophan, demonstrating the presence of both bait and prey vectors. Plating on the highly selective media TDO lacking leucine, tryptophan and histidine and supplemented with 15 mM 3 AT, showed growth of yeast strains co-expressing AtNHR2B and HopD1, demonstrating that HopD1 interacts with AtNHR2B in yeast. Yeast growth on TDO + 3AT was also observed with the positive controls, as expected. The observed growth when yeast was co-transformed with AtNHR2B and HopD1 represents a true interaction, and not the result of auto-activation as yeast strains co-transformed with HopD1 and the empty vector pDEST22 were not able to grow in the highly selective media (Figure 1A).
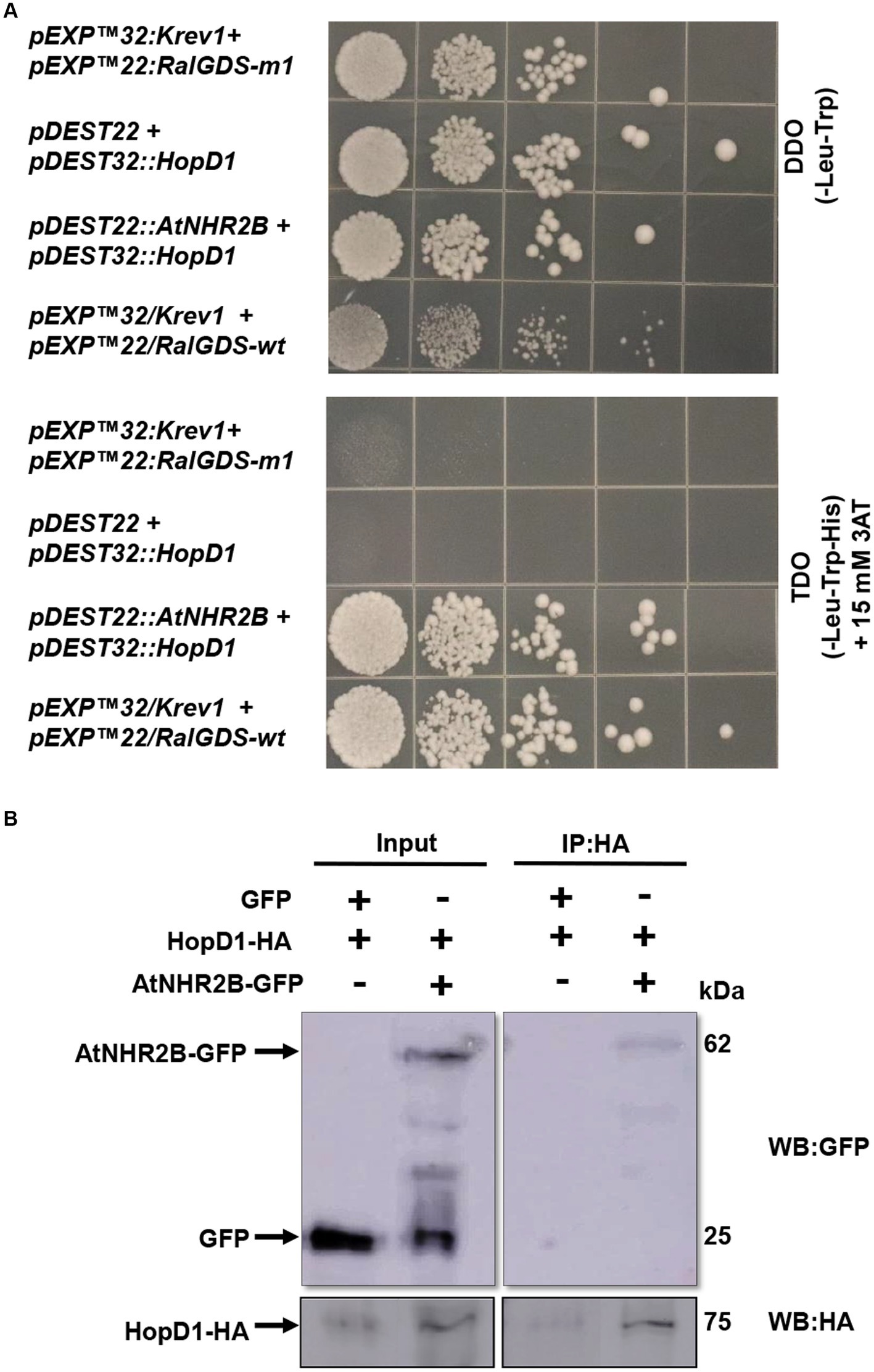
Figure 1. HopD1 interacts AtNHR2B in yeast and in planta. Yeast two-hybrid system was used to evaluate the interaction between HopD1 and AtNHR2B. Yeast strain MaV203 was co-transformed with yeast two-hybrid constructs and plated on double drop out (DDO – Leucine-Tryptophan) (top) or triple drop out (TDO – Leucine-Tryptophan-Histidine) and supplemented with 3-Amino-1,2,4-Triazole (3-AT) (bottom) (A). Agrobacterium tumefaciens strains harboring HopD1-HA and AtNHR2B-GFP were co-infiltrated into four-weeks N. benthamiana plants for transient expression. As control Agrobacterium tumefaciens harboring HopD1-HA was co-infiltrated with Agrobacterium tumefaciens harboring a construct expressing 35S-GFP. Infiltrated leaves were harvested for protein extraction followed by immunoprecipitation using anti-HA antibodies. Immunoprecipitated samples were separated by SDS-PAGE electrophoresis and transferred to a nitrocellulose membrane for Western Blot analysis using anti-HA and anti-GFP antibodies (B).
To validate the yeast interaction in planta, we transiently co-expressed HopD1-HA with either AtNHR2B-GFP or GFP in N. benthamiana. Infiltrated leaves were collected for protein extraction followed by immunoprecipitation with anti-HA antibodies. The results show that HA antibodies immunoprecipitated HopD1-HA as expected, and co-immunoprecipitated AtNHR2B-GFP. HA antibodies did not co-immunoprecipitate free GFP, demonstrating that HopD1 physically interacts with AtNHR2B and not with the GFP tag (Figure 1B).
The physical interaction between HopD1 and AtNHR2B in yeast and in planta, prompted our interest to start dissecting how HopD1 targets AtNHR2B. For that purpose, we evaluated the co-localization in situ between HopD1 and AtNHR2B by transiently co-expressing HopD1-GFP with AtNHR2B-RFP in N. benthamiana and monitoring the co-localization by live-cell imaging. The results showed that AtNHR2B-RFP localizes to cytoplasm and small vesicles as previously described (Singh et al., 2018), whereas HopD1-GFP predominantly localizes to cytoplasm, and occasionally to small vesicles (Figure 2). Interestingly, overlaying the images obtained from the green and red channels revealed co-localization areas (yellow signal) mostly in small vesicles (Figure 2, Supplementary Figure S1). The co-localization of HopD1 and AtNHR2B is not complete and Pearson’s correlation coefficient analysis calculated for 5 images ranged between 0.361 and 0.633 implying a positive but partial co-localization (Figure 2, Supplementary Figure S1). The differences in the Pearson’s correlation coefficients might be related to the dynamic nature of AtNHR2B-RFP transitioning through subcellular compartments.
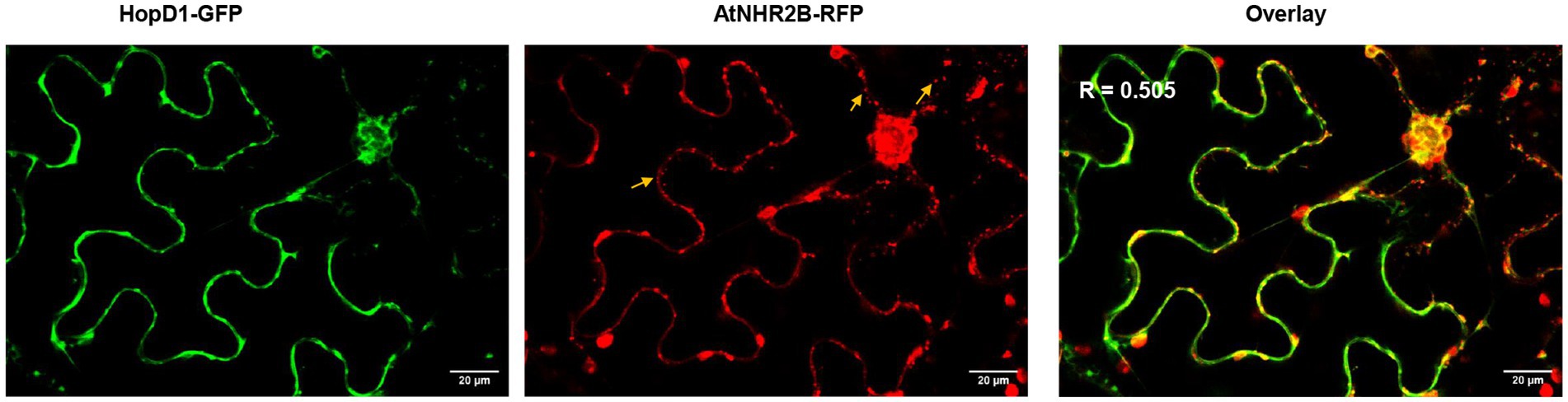
Figure 2. HopD1 partially colocalizes with AtNHR2B. Four-week-old Nicotiana benthamiana plants were co-infiltrated with two A. tumefaciens strains harboring AtNHR2B-RFP and HopD1-GFP. At 2dpi, plants were imaged on a Leica Stellaris 8 laser scanning confocal microscope at 20 X magnification by sequential scanning using GFP and RFP channels. Yellow areas in the overlay reveal areas of co-localization. Yellow arrows show AtNHR2B-RFP-containing vesicles. Pearson’s coefficient analysis (R) was calculated by Image J.
The finding that HopD1 co-localizes with AtNHR2B, together with our previous findings that AtNHR2B localizes to highly dynamic vesicles, led us to hypothesize that HopD1 interferes with cellular processes mediated by AtNHR2B. To test this hypothesis, we evaluated the effect of HopD1 on the dynamics of AtNHR2B-RFP-containing vesicles by live-cell imaging in N. benthamiana. For that purpose, AtNHR2B-RFP alone or in combination with HopD1-HA were transiently expressed in N. benthamiana. Live-cell imaging of N. benthamiana plants transiently expressing AtNHR2B-RFP alone confirmed its localization to cytoplasm and to small, highly dynamic vesicles, as reported previously (Singh et al., 2018). In contrast, N. benthamiana plants transiently co-expressing AtNHR2B-RFP with HopD1-HA, showed a decrease in the fluorescent signal and a dramatic reduction in the overall number of AtNHR2B-RFP-containing vesicles (Figure 3A). To confirm these results, time-lapse images of N. benthamiana plants expressing AtNHR2B-RFP alone, and plants co-expressing AtNHR2B-RFP and HopD1-HA were taken to quantify the number of AtNHR2B-RFP-containing vesicles in plants expressing AtNHR2B-RFP alone in comparison with plants co-expressing AtNHR2B-RFP with HopD1-HA. The results from 120 frames for 4 independent videos for each of the treatments, showed that plants expressing AtNHR2-RFP alone had on average number of 415 AtNHR2B-RFP-containing vesicles, whereas plants co-expressing AtNHR2B-RFP with HopD1-HA had on average of 277 (Figure 3B). Unfortunately, although the videos revealed differences between both experimental conditions, the difference in quantification was not statistically significant, likely due to the large margin of error and the relatively small sample size.
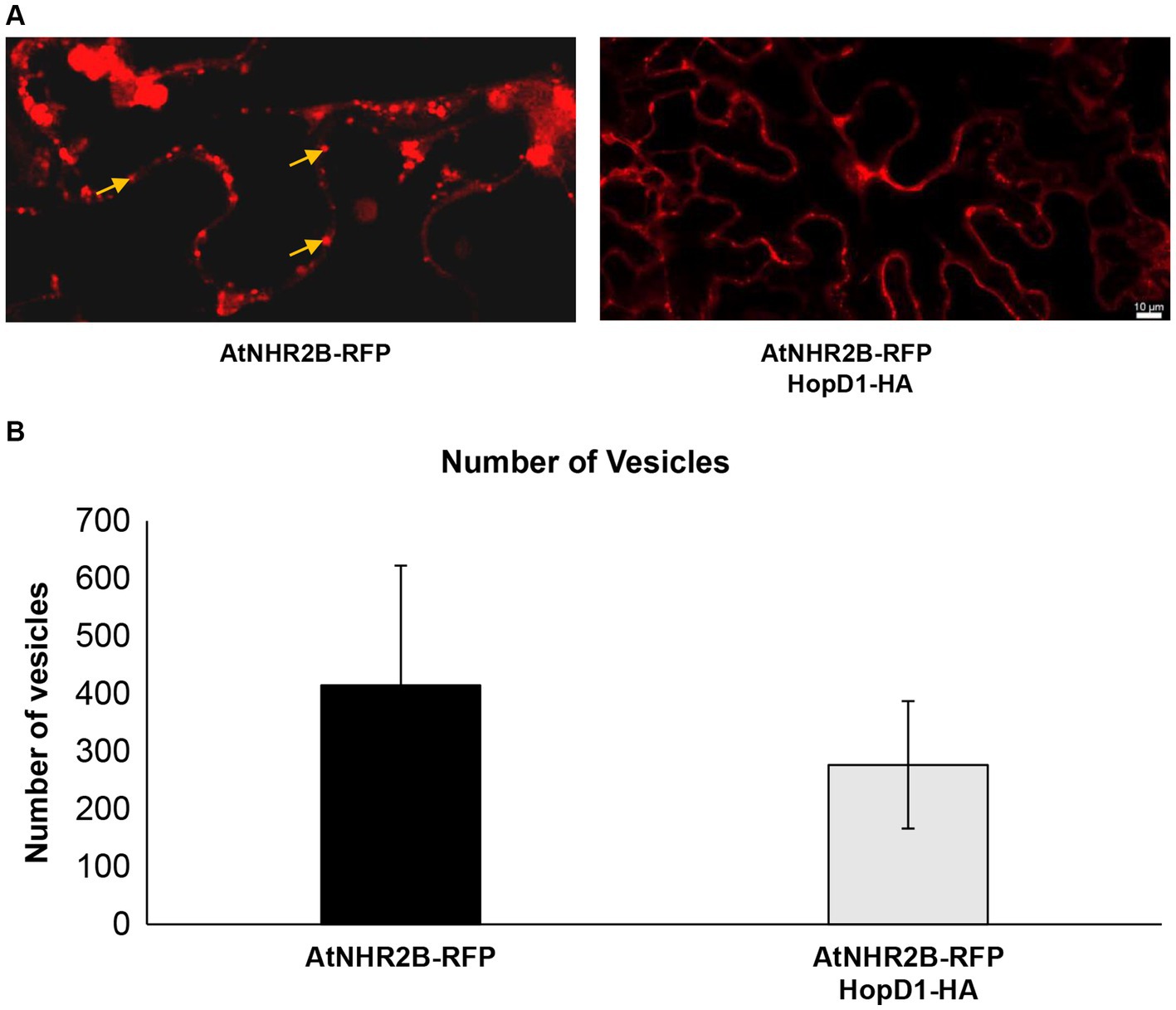
Figure 3. HopD1 interferes with the abundance of AtNHR2B-containing vesicles. Four-week-old Nicothiana benthamiana plants were infiltrated with A. tumefaciens harboring AtNHR2B-RFP or co-infiltrated with two A. tumefaciens strains harboring AtNHR2B-RFP and HopD1-HA. Plants were imaged at 2 days post-infiltration using a Leica Stellaris 8 laser scanning confocal microscope at 20× magnification (A). Images collected from 120 independent frames from three experiments were used to quantify the number of vesicles using software Imaris. Bars indicate average number of vesicles per treatment. (B). Arrows in (A) show AtNHR2B-RFP-containing vesicles. No statistically significant difference according to Mann–Whitney test were observed in the number of vesicles in (B).
Time-lapse images of AtNHR2B-RFP-containing vesicles showed the dynamic movement patterns of AtNHR2B-RFP-containing vesicles (Supplementary Video S1). Those dynamic patterns of movement of AtNHR2B-RFP-containing vesicles are significantly altered and reduced in the presence of HopD1 (Supplementary Video S2). To evaluate this phenomenon in more detail, AtNHR2B-RFP-containing vesicles were tracked to quantify their speed under two experimental conditions: in N. benthamiana plants expressing AtNHR2B-RFP alone, and in N. benthamiana co-expressing AtNHR2B-RFP and HopD1-HA. To make meaningful comparisons and to prevent biased results arising from the differences in the number of AtNHR2B-RFP-containing vesicles between experimental conditions, the same number of vesicles were evaluated for each of the 4 videos per experimental condition, tracking a total of 400 vesicles for each experimental condition.
Sorting the AtNHR2B-RFP-containing vesicles by speed into 30 bins ranging from 0.00738 to 2.56 μm/s revealed that AtNHR2B-RFP-containing vesicles from N. benthamiana plants expressing AtNHR2B-RFP alone span 22 of the 30 bins, and their abundance in each of those bins ranged from 1 to 85. More than 10 vesicles per bin were observed in bins 1–10, that cumulatively cover speeds from 0.00738 to 0.859 μm/s (Table 1). The largest number of AtNHR2B-RFP-containing vesicles (85) underwent selective sorting into the bin corresponding to speeds between 0.18 μm/s and 0.265 μm/s (Table 1). In contrast to AtNHR2B-RFP-containing vesicles from N. benthamiana plants expressing AtNHR2B-RFP alone, N. benthamiana plants co-expressing AtNHR2B-RFP and HopD1-HA were sorted into 7 of the 30 different bins spanning a range of 0.00738 to 0.604 μm/s. In this experimental condition, only bins 1–4 have more than 10 vesicles, and the bin containing the largest number of AtNHR2B-RFP-containing vesicles was observed in the bin corresponding to speeds between 0.0949 μm/s and 0.18 μm/s (Table 1). Altogether the data shows that expression of AtNHR2B-RFP alone generate a diverse population of AtNHR2B-RFP-containing vesicles with a wider range of speeds, include fast moving vesicles (Figure 4A), whereas the co-expression of AtNHR2B-RFP with HopD1-HA generates fewer types of AtNHR2B-RFP-containing vesicles sorted into slow speeds (Figure 4B) and slowest total mean speed. In addition, the mean speed of AtNHR2B-GFP-containing vesicles in N. benthamiana plants expressing AtNHR2B-RFP alone was 0.425 μm/s, whereas the mean speed for AtNHR2B-RFP-containing vesicles in N. benthamiana plants co-expressing AtNHR2B-RFP and HopD1-HA was 0.171 μm/s (Figure 4C). These findings demonstrate that HopD1 significantly interferes with the speed of AtNHR2B-RFP-containing vesicles.
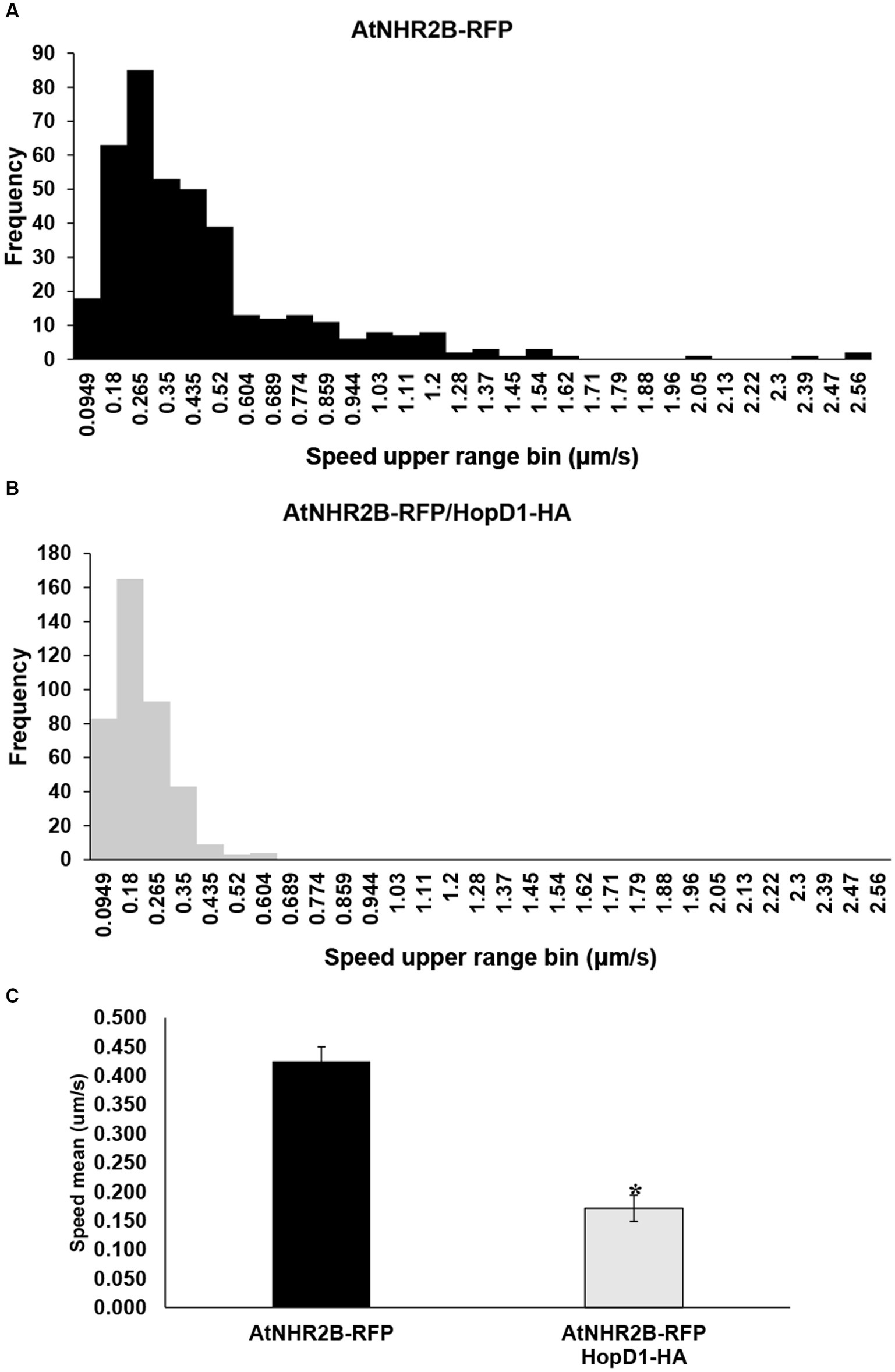
Figure 4. The type III effector HopD1 interferes with the speed of AtNHR2B-containing vesicles. Four-week-old N. benthamiana plants were infiltrated with with A. tumefaciens harboring AtNHR2B-RFP or co-infiltrated with two A. tumefaciens strains harboring AtNHR2B-RFP and HopD1-HA. Infiltrated leaves were imaged at 2 dpi, and four videos of 120 frames were captured for each treatment using a Leica Stellaris 8 laser scanning confocal microscope. Videos were analyzed with Imaris Microscopy Image Analysis software to sort out the AtNHR2B-RFP-containing vesicles into different populations (bins) based on speed and with or without HopD1-HA (A,B), and to calculate the average speed with and without HopD1-HA (C). Bars in (C) represent average and standard deviation for each of the measurements for each treatment. Asterisk in (C) indicates statistically significant difference by Student’s t-test.
To further evaluate the effect of HopD1 on the dynamics of AtNHR2B-RFP-containing vesicles, the same time-lapse images were used to evaluate displacement of AtNHR2B-RFP-containing vesicles. AtNHR2B-RFP-containing vesicles from plants expressing AtNHR2B-RFP alone and from plants co-expressing AtNHR2B-RFP and HopD1-HA were sorted into 30 bins ranging from 0.00518 μm to 13.93 μm. The results revealed that plants expressing AtNHR2B-RFP alone were sorted into 22 of the 30 bins and their abundance in each bin ranges from 1 to 82. Bins 1–9 contain more than 10 vesicles; the largest number of AtNHR2B-RFP-containing vesicles were observed in bin 1 corresponding to displacement ranging from 0.00518 μm to 0.473 μm (Table 2). The displacement of AtNHR2B-RFP-containing vesicles in N. benthamiana plants co-expressing AtNHR2B-RFP and HopD1-HA were sorted into only 13 of the 30 available and their abundance in each bin ranged from 1 to 178. More than 10 vesicles/bin were observed in bins 1–6 and the largest number of vesicles (178) were found bin 1 corresponding to displacements between 0.00518 μm and 0.473 μm (Table 2). When comparing the bins in common for both experimental conditions (AtNHR2B-RFP alone versus AtNHR2B-RFP/HopD1-HA), the results show a similar pattern of distribution to the pattern observed in speed, wherein AtNHR2B-RFP-containing vesicles obtained from plants expressing AtNHR2B-RFP alone showed a wider range of displacement but less abundance (Figure 5A), whereas AtNHR2B-RFP-containing vesicles obtained from plants co-expressing AtNHR2B-RFP with HopD1-HA, were aggregated into a narrower range of bins, corresponding to lower displacement, and with larger numbers of vesicles per bin (Figure 5B). Moreover, the mean displacement of AtNHR2B-RFP-containing vesicles was 2.072 μm in N. benthamiana plants expressing AtNHR2B-RFP alone, whereas the mean displacement for AtNHR2B-RFP-containing vesicles in N. benthamiana plants co-expressing AtNHR2B-RFP and HopD1-HA was 0.876 μm. Altogether, these findings demonstrate that HopD1 actively reduces the displacement of AtNHR2B-RFP-containing vesicles (Figure 5C).
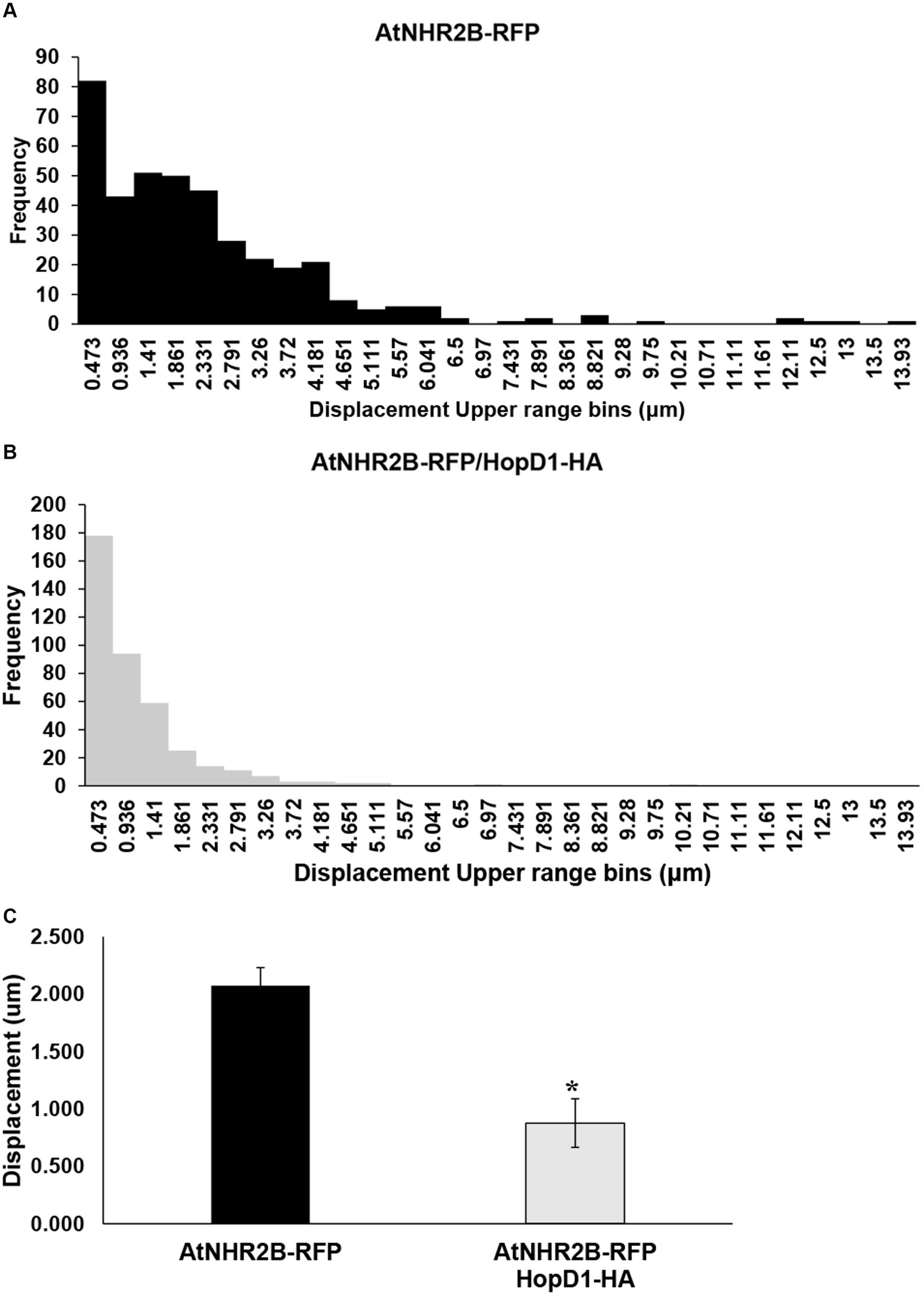
Figure 5. The type III effector HopD1 interferes with the displacement of AtNHR2B-containing vesicles. Four-week-old N. benthamiana plants were infiltrated with A. tumefaciens harboring AtNHR2B-RFP or co-infiltrated with two A. tumefaciens strains harboring AtNHR2B-RFP and HopD1-HA. Infiltrated leaves were imaged at 2 dpi, and four videos of 120 frames were captured for each treatment using a Leica Stellaris 8 laser scanning confocal microscope. Videos were analyzed with Imaris Microscopy Image Analysis software to sort out the AtNHR2B-RFP-containing vesicles into different populations (bins) based on displacement and with or without HopD1-HA (A,B), and to calculate the average speed with and without HopD1-HA (C). Bars in (C). represent average and standard deviation for each of the measurements for each treatment. Asterisk in (B) indicates statistically significant difference by Student’s t-test.
Bacterial pathogens such as Pst DC3000 employ T3SE to interfere with two main branches of plant immunity: Pathogen-Associated Molecular Patterns (PAMP)-triggered immunity (PTI) and Effector-Triggered Immunity (ETI) (Jones and Dangl, 2006). PTI is initiated when plants recognize common features present in the surface of microbes/pathogens and known as Microbe-Associated Molecular Patterns (MAMPs)/ Pathogen-Associated Molecular Patterns (PAMPs) (Zhang and Zhou, 2010). PAMP recognition is achieved by Pathogen Recognition Receptors (PRRs) located on the plant surface (Saijo et al., 2018), and subsequent signal transduction requires the phosphorylation of PRRs and interaction with co-receptors such as BAK1 (Couto and Zipfel, 2016; Yu et al., 2017; Saijo et al., 2018). To counteract PTI, Pst DC3000 T3SE redundantly target PRRs as well as proteins that participate in PTI signaling. For example, T3SE AvrPto and AvrPtoB target PRRs such as FLS2, EFR and CERK1 (Xiang et al., 2008; Gimenez-Ibanez et al., 2009), and prevent the binding of flg22 to FLS2 (Shan et al., 2008), HopAO1 with tyrosine phosphatase activity prevents phosphorylation of EFR (Underwood et al., 2007; Macho et al., 2014), and HopB1 targets BAK1 (Li et al., 2016). T3SE also redundantly suppress ETI (Jamir et al., 2004; Guo et al., 2009), and one of those effectors is HopD1 (Block et al., 2014).
HopD1 localizes to the endoplasmic reticulum (ER) where it interacts with the ER-localized transcription factor NTL9, which regulates the expression of ETI-related genes (Block et al., 2014). This work showed that HopD1 also interacts with the immune-related protein AtNHR2B. The co-localization of HopD1-GFP with AtNHR2B-RFP was found to be in cytoplasm and in small subcellular compartments (vesicles) of the endomembrane system where AtNHR2B was previously found to be localized (Singh et al., 2018). The localization of AtNHR2B to compartments of the endomembrane systems suggests a function in vesicle trafficking which mediates cellular processes such as endocytosis and secretion (Jeon and Segonzac, 2023). During plant immunity, endocytosis mediates the internalization of PAMPs-activated PRR which in turn induces downstream signaling pathways involved in plant defense (Robatzek et al., 2006; Chinchilla et al., 2007; Ben Khaled et al., 2015; Mbengue et al., 2016; Gu et al., 2017).Vesicle trafficking events also mediate secretory processes, which in the context of plant immunity are responsible for the remodeling of the cell wall, the release of antimicrobials to the apoplast and the replenishing of PRR to the plasma membrane (Kwon et al., 2008; Kim and Brandizzi, 2014).
While it is still not known if AtNHR2B participates in endocytosis or secretion, the results from this work clearly demonstrate that HopD1 alters the speed and displacement of AtNHR2B-RFP carrying vesicles. Targeting endocytosis and secretion by T3SE is not unexpected given their importance in plant immunity (Gu et al., 2017), and examples of effectors targeting those processes are starting to emerge. For example, HopM1 localizes to endomembrane compartments, specifically to trans-Golgi network/early endosomes (Nomura et al., 2011), and targets AtMIN7 (Arabidopsis thaliana HopM1 interactor 7), a vesicle trafficking component required for callose deposition (Nomura et al., 2006). AvrPtoB targets EXO70B1 a subunit of the exocyst complex which plays a vital role in the tethering of secretory vesicles to the plasma membrane for subsequent exocytosis (Wang et al., 2019). HopG1 and HopW1 bind to actin and disrupt actin cytoskeleton and endocytosis interfering with vesicle trafficking (Jelenska et al., 2014; Kang et al., 2014; Shimono et al., 2016), whereas HopZ1a, an acetyltransferase destroys microtubule networks interfering with secretion (Lee et al., 2012). HopE1 also interferes with the microtubule network by interacting with MAP65 (microtubule-associated protein 65) causing its dissociation from the microtubule network and affecting secretion as well (Guo et al., 2016). The results from this work provide compelling evidence that HopD1 interferes with vesicle trafficking processes mediated by AtNHR2B but the specific processes affected still remain to be characterized.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
LM-P: Formal analysis, Methodology, Writing – original draft, Writing – review & editing. CR-P: Investigation, Methodology, Writing – review & editing. PR-L: Investigation, Methodology, Writing – review & editing. CR: Conceptualization, Funding acquisition, Investigation, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Science Foundation CAREER award Grant (IOS # 1842970) and by Equipment Grants Program, award #2021-70410-35293, from the US Department of Agriculture.
The authors would like to thank Jim Alfano (University of Nebraska-Lincoln), for providing the HopD1-HA and HopD1-GFP constructs, Martin Egan (University of Arkansas, Fayetteville) for assistance with quantification of vesicle movement and Alejandro Rojas (University of Arkansas, Fayetteville), for assistance with vesicle movement data analysis. The authors also thank Thiago Maia and Conner Chu (University of Nebraska-Lincoln) for valuable comments on the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1305899/full#supplementary-material
Alfano, J. R., and Collmer, A. (2004). Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414. doi: 10.1146/annurev.phyto.42.040103.110731
Ben Khaled, S., Postma, J., and Robatzek, S. (2015). A moving view: subcellular trafficking processes in pattern recognition receptor-triggered plant immunity. Annu. Rev. Phytopathol. 53, 379–402. doi: 10.1146/annurev-phyto-080614-120347
Block, A., Toruno, T. Y., Elowsky, C. G., Zhang, C., Steinbrenner, J., Beynon, J., et al. (2014). The Pseudomonas syringae type III effector HopD1 suppresses effector-triggered immunity, localizes to the endoplasmic reticulum, and targets the Arabidopsis transcription factor NTL9. New Phytol. 201, 1358–1370. doi: 10.1111/nph.12626
Bundalovic-Torma, C., Lonjon, F., Desveaux, D., and Guttman, D. S. (2022). Diversity, evolution, and function of Pseudomonas syringae effectoromes. Annu. Rev. Phytopathol. 60, 211–236. doi: 10.1146/annurev-phyto-021621-121935
Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nurnberger, T., Jones, J. D., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500. doi: 10.1038/nature05999
Couto, D., and Zipfel, C. (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552. doi: 10.1038/nri.2016.77
Cunnac, S., Chakravarthy, S., Kvitko, B. H., Russell, A. B., Martin, G. B., and Collmer, A. (2011). Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc. Natl. Acad. Sci. U. S. A. 108, 2975–2980. doi: 10.1073/pnas.1013031108
Dillon, M. M., Almeida, R. N. D., Laflamme, B., Martel, A., Weir, B. S., Desveaux, D., et al. (2019). Molecular evolution of Pseudomonas syringae type III secreted effector proteins. Front. Plant Sci. 10:418. doi: 10.3389/fpls.2019.00418
Gimenez-Ibanez, S., Hann, D. R., Ntoukakis, V., Petutschnig, E., Lipka, V., and Rathjen, J. P. (2009). AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19, 423–429. doi: 10.1016/j.cub.2009.01.054
Gu, Y., Zavaliev, R., and Dong, X. (2017). Membrane trafficking in plant immunity. Mol. Plant 10, 1026–1034. doi: 10.1016/j.molp.2017.07.001
Guo, M., Kim, P., Li, G., Elowsky, C. G., and Alfano, J. R. (2016). A bacterial effector co-opts calmodulin to target the plant microtubule network. Cell Host Microbe 19, 67–78. doi: 10.1016/j.chom.2015.12.007
Guo, M., Tian, F., Wamboldt, Y., and Alfano, J. R. (2009). Majority of the type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol. Plant-Microbe Interact. 22, 1069–1080. doi: 10.1094/MPMI-22-9-1069
Jamir, Y., Guo, M., Oh, H. S., Petnicki-Ocwieja, T., Chen, S. R., Tang, X. Y., et al. (2004). Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J. 37, 554–565. doi: 10.1046/j.1365-313X.2003.01982.x
Jelenska, J., Kang, Y., and Greenberg, J. T. (2014). Plant pathogenic bacteria target the actin microfilament network involved in the trafficking of disease defense components. BioArchitecture 4, 149–153. doi: 10.4161/19490992.2014.980662
Jeon, H., and Segonzac, C. (2023). Manipulation of the host endomembrane system by bacterial effectors. Mol. Plant-Microbe Interact. 36, 208–217. doi: 10.1094/MPMI-09-22-0190-FI
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kang, Y., Jelenska, J., Cecchini, N. M., Li, Y., Lee, M. W., Kovar, D. R., et al. (2014). HopW1 from Pseudomonas syringae disrupts the actin cytoskeleton to promote virulence in Arabidopsis. PLoS Pathog. 10:e1004232. doi: 10.1371/journal.ppat.1004232
Kim, S. J., and Brandizzi, F. (2014). The plant secretory pathway: an essential factory for building the plant cell wall. Plant Cell Physiol. 55, 687–693. doi: 10.1093/pcp/pct197
Kvitko, B. H., Park, D. H., Velasquez, A. C., Wei, C. F., Russell, A. B., Martin, G. B., et al. (2009). Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5:e1000388. doi: 10.1371/journal.ppat.1000388
Kwon, C., Neu, C., Pajonk, S., Yun, H. S., Lipka, U., Humphry, M., et al. (2008). Co-option of a default secretory pathway for plant immune responses. Nature 451, 835–840. doi: 10.1038/nature06545
Lee, A. H., Hurley, B., Felsensteiner, C., Yea, C., Ckurshumova, W., Bartetzko, V., et al. (2012). A bacterial acetyltransferase destroys plant microtubule networks and blocks secretion. PLoS Pathog. 8:e1002523. doi: 10.1371/journal.ppat.1002523
Li, L., Kim, P., Yu, L., Cai, G., Chen, S., Alfano, J. R., et al. (2016). Activation-dependent destruction of a co-receptor by a Pseudomonas syringae effector dampens plant immunity. Cell Host Microbe 20, 504–514. doi: 10.1016/j.chom.2016.09.007
Macho, A. P., Schwessinger, B., Ntoukakis, V., Brutus, A., Segonzac, C., Roy, S., et al. (2014). A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science 343, 1509–1512. doi: 10.1126/science.1248849
Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
Mbengue, M., Bourdais, G., Gervasi, F., Beck, M., Zhou, J., Spallek, T., et al. (2016). Clathrin-dependent endocytosis is required for immunity mediated by pattern recognition receptor kinases. Proc. Natl. Acad. Sci. U. S. A. 113, 11034–11039. doi: 10.1073/pnas.1606004113
Morris, C. E., Monteil, C. L., and Berge, O. (2013). The life history of Pseudomonas syringae: linking agriculture to earth system processes. Annu. Rev. Phytopathol. 51, 85–104. doi: 10.1146/annurev-phyto-082712-102402
Nomura, K., Debroy, S., Lee, Y. H., Pumplin, N., Jones, J., and He, S. Y. (2006). A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313, 220–223. doi: 10.1126/science.1129523
Nomura, K., Mecey, C., Lee, Y. N., Imboden, L. A., Chang, J. H., and He, S. Y. (2011). Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 108, 10774–10779. doi: 10.1073/pnas.1103338108
Robatzek, S., Chinchilla, D., and Boller, T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20, 537–542. doi: 10.1101/gad.366506
Rodriguez-Puerto, C., Chakraborty, R., Singh, R., Rocha-Loyola, P., and Rojas, C. M. (2022). The Pseudomonas syringae type III effector HopG1 triggers necrotic cell death that is attenuated by AtNHR2B. Sci. Rep. 12:5388. doi: 10.1038/s41598-022-09335-1
Saijo, Y., Loo, E. P., and Yasuda, S. (2018). Pattern recognition receptors and signaling in plant-microbe interactions. Plant J. 93, 592–613. doi: 10.1111/tpj.13808
Shan, L., He, P., Li, J., Heese, A., Peck, S. C., Nürnberger, T., et al. (2008). Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4, 17–27. doi: 10.1016/j.chom.2008.05.017
Shimono, M., Lu, Y. J., Porter, K., Kvitko, B. H., Henty-Ridilla, J., Creason, A., et al. (2016). The Pseudomonas syringae type III effector HopG1 induces actin remodeling to promote symptom development and susceptibility during infection. Plant Physiol. 171, 2239–2255. doi: 10.1104/pp.16.01593
Singh, R., Lee, S., Ortega, L., Ramu, V. S., Senthil-Kumar, M., Blancaflor, E. B., et al. (2018). Two chloroplast-localized proteins: AtNHR2A and AtNHR2B, contribute to Callose deposition during nonhost disease resistance in Arabidopsis. Mol. Plant-Microbe Interact. 31, 1280–1290. doi: 10.1094/MPMI-04-18-0094-R
Underwood, W., Zhang, S., and He, S. Y. (2007). Pseudomonas syringae type III effector tyrosine phosphatase HopAO1 suppresses innate immunity in Arabidopsis thaliana. Plant J. 52, 658–672. doi: 10.1111/j.1365-313X.2007.03262.x
Wang, W., Liu, N., Gao, C., Rui, L., and Tang, D. (2019). The Pseudomonas syringae effector AvrPtoB associates with and ubiquitinates Arabidopsis exocyst subunit EXO70B1. Front. Plant Sci. 10:1027. doi: 10.3389/fpls.2019.01027
Wei, C. F., Kvitko, B. H., Shimizu, R., Crabill, E., Alfano, J. R., Lin, N. C., et al. (2007). A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J. 51, 32–46. doi: 10.1111/j.1365-313X.2007.03126.x
Wei, H. L., Zhang, W., and Collmer, A. (2018). Modular study of the type III effector repertoire in Pseudomonas syringae pv. tomato DC3000 reveals a matrix of effector interplay in pathogenesis. Cell Rep. 23, 1630–1638. doi: 10.1016/j.celrep.2018.04.037
Xiang, T., Zong, N., Zou, Y., Wu, Y., Zhang, J., Xing, W., et al. (2008). Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol 18, 74–80. doi: 10.1016/j.cub.2007.12.020
Xin, X. F., and He, S. Y. (2013). Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51, 473–498. doi: 10.1146/annurev-phyto-082712-102321
Yu, X., Feng, B., He, P., and Shan, L. (2017). From Chaos to harmony: responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 55, 109–137. doi: 10.1146/annurev-phyto-080516-035649
Keywords: Pseudomonas syringae pv. tomato DC3000, type III secreted effectors, HopD1, AtNHR2B, cellular dynamics, vesicle trafficking
Citation: Marín-Ponce LF, Rodríguez-Puerto C, Rocha-Loyola P and Rojas CM (2023) The Pseudomonas syringae pv. tomato DC3000 effector HopD1 interferes with cellular dynamics associated with the function of the plant immune protein AtNHR2B. Front. Microbiol. 14:1305899. doi: 10.3389/fmicb.2023.1305899
Received: 02 October 2023; Accepted: 10 November 2023;
Published: 23 November 2023.
Edited by:
Nuria Montes, Hospital Universitario de La Princesa, SpainReviewed by:
Jose Antonio Lucas Lucas Garcia, CEU San Pablo University, SpainCopyright © 2023 Marín-Ponce, Rodríguez-Puerto, Rocha-Loyola and Rojas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Clemencia M. Rojas, Y3JvamFzMkB1bmwuZWR1
†Present addresses: Clemencia M. Rojas, Department of Plant Pathology, University of Nebraska-Lincoln, Lincoln, NE, United States; Center for Plant Science Innovation, University of Nebraska-Lincoln, Lincoln, NE, United States Catalina Rodríguez-Puerto, Cell and Molecular Biology Program, University of Arkansas, Fayetteville, AR, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.