- 1Department of Life Science and Biochemical Engineering, Graduate School, SunMoon University, Asan, Republic of Korea
- 2Genome-Based BioIT Convergence Institute, Asan, Republic of Korea
- 3Division of Computer Science and Engineering, SunMoon University, Asan, Republic of Korea
- 4Department of Pharmaceutical Engineering and Biotechnology, SunMoon University, Asan, Republic of Korea
Burkholderia is a versatile strain that has expanded into several genera. It has been steadily reported that the genome features of Burkholderia exhibit activities ranging from plant growth promotion to pathogenicity across various isolation areas. The objective of this study was to investigate the secondary metabolite patterns of 366 Burkholderia species through comparative genomics. Samples were selected based on assembly quality assessment and similarity below 80% in average nucleotide identity. Duplicate samples were excluded. Samples were divided into two groups using FastANI analysis. Group A included B. pseudomallei complex. Group B included B. cepacia complex. The limitations of MLST were proposed. The detection of genes was performed, including environmental and virulence-related genes. In the pan-genome analysis, each complex possessed a similar pattern of cluster for orthologous groups. Group A (n = 185) had 14,066 cloud genes, 2,465 shell genes, 682 soft-core genes, and 2,553 strict-core genes. Group B (n = 181) had 39,867 cloud genes, 4,986 shell genes, 324 soft-core genes, 222 core genes, and 2,949 strict-core genes. AntiSMASH was employed to analyze the biosynthetic gene cluster (BGC). The results were then utilized for network analysis using BiG-SCAPE and CORASON. Principal component analysis was conducted and a table was constructed using the results obtained from antiSMASH. The results were divided into Group A and Group B. We expected the various species to show similar patterns of secondary metabolite gene clusters. For in-depth analysis, a network analysis of secondary metabolite gene clusters was conducted, exemplified by BiG-SCAPE analysis. Depending on the species and complex, Burkholderia possessed several kinds of siderophore. Among them, ornibactin was possessed in most Burkholderia and was clustered into 4,062 clans. There was a similar pattern of gene clusters depending on the species. NRPS_04014 belonged to siderophore BGCs including ornibactin and indigoidine. However, it was observed that each family included a similar species. This suggests that, besides siderophores being species-specific, the ornibactin gene cluster itself might also be species-specific. The results suggest that siderophores are associated with environmental adaptation, possessing a similar pattern of siderophore gene clusters among species, which could provide another perspective on species-specific environmental adaptation mechanisms.
1 Introduction
Burkholderia classified in the phylum Proteobacteria have been isolated from various sources worldwide (Radua et al., 2000; Levy et al., 2008; Hall et al., 2015; Peddayelachagiri et al., 2016), including cystic fibrosis patients (Medina-Pascual et al., 2015) and environmental sources such as soil (Hall et al., 2015), rice (Luo et al., 2007), and lichen (Han et al., 2016). The genus Burkholderia was named in 1992. It initially consisted of seven pathogenic strains affecting humans, animals, and plants (Yabuuchi et al., 1992). It has been categorized into two major groups: Burkholderia cepacia complex (BCC) and environmental Burkholderia (Compant et al., 2008; Sousa et al., 2011). However, some studies have focused on the beneficial effects of Burkholderia in plants, leading to its reclassification into new genera such as Paraburkholderia or Caballeronia (Kaur et al., 2017). Expansion of the Burkholderia genus is ongoing (Euzéby, 1997; Estrada-de Los Santos et al., 2018). The multi-genus complex that includes Paraburkholderia and Caballeronia is referred to as B. sensu lato which encompasses Robbsia, Pararobbsia, Burkholderia, Trinickia, and Mycetohabitans. Species reclassified within Burkholderia now fall under B. sensu lato, which also includes Burkholderia. B. sensu stricto encompasses BCC, Burkholderia pseudomallei complex (BPC), and rice pathogenic Burkholderia.
Recent reports of Burkholderia have highlighted the use of genome-based classification (Jin et al., 2020), genomic diversity analysis (Gee et al., 2021), and pan-genome analysis (Lee et al., 2021). After the report on the genome-based classification of BCC, the taxonomic position of the inherent Taxon K has been reevaluated (Vanlaere et al., 2009). Recent reports have focused on the reclassification and comparison analysis of genomics due to the registration of various genomes in databases. It is possible to study genome-based classification as well as secondary metabolites (Bach et al., 2022a) and evolutionary comparisons (Yu et al., 2006).
In the multi-genus complex known as Burkholderia sensu lato, antimicrobial compounds have been reported (Petrova and Mahenthiralingam, 2022; Rodríguez-Cisneros et al., 2023). This study primarily compiles the antimicrobial compounds identified in Burkholderia. Research in the field of discovery has progressed, including studies of new antimicrobial secondary metabolites associated with Burkholderia, and antimicrobial secondary metabolites are related to taxonomy (Depoorter et al., 2016; Deng et al., 2023). In Burkholderia, a wide range of natural products have been reported, including antimicrobial compounds and metabolites related to biocontrol and agriculture (Sulochana et al., 2014; Esmaeel et al., 2016, 2018; Kunakom and Eustáquio, 2019). For example, the antimicrobial compound bactobolin has been identified in Burkholderia thailandensis E264 (Seyedsayamdost et al., 2010). In addition, siderophores known for their roles in plant growth promotion have been detected in pathogenic Burkholderia.
Despite ongoing genomic and metabolite studies on Burkholderia, studies on B. sensu stricto have been limited and treated as a secondary aspect within the broader context of B. sensu lato study. In this study, we aimed to understand the characteristics of BCC and BPC complexes within B. sensu stricto. Different from previous studies focusing on the prevalence of BGCs according to individual genera within B. sensu lato (Mullins and Mahenthiralingam, 2021), we conducted a more detailed analysis of BGCs within Burkholderia, considering species variations. Furthermore, we aimed to uncover the interrelationships among different species and metabolites through network analysis.
2 Materials and methods
2.1 Preparation of Burkholderia genome
Complete genome sequences of 458 Burkholderia available in the NCBI genome database were downloaded on 17 February 2023 (Benson et al., 2017). Duplicated assemblies were removed prior to analysis. Information about strain isolation was searched in the database at NCBI (Barrett et al., 2012) and ENA (Cummins et al., 2022). Samples were categorized into three groups: none (unknown sample), PATH (pathological sample), and ENV (environmental sample) based on isolation information. We confirmed validated Burkholderia species in the LPSN database (Parte et al., 2020). The assembly quality of these genomes was determined using CheckM (Parks et al., 2015) and QUAST v5.2.0 (Gurevich et al., 2013) with default parameters. Assemblies were selected based on criteria of at least 90% completeness, less than 10% contamination, and a calculated value of completeness minus 5 times contamination greater than 50%. For further analysis, all genomes were annotated with prokka using default parameters (Seemann, 2014).
2.2 Phylogenomic analysis
Average Nucleotide Identity (ANI) was analyzed using FastANI (Jain et al., 2018) with default settings (K-mer size = 16, threads count for parallel execution = 1, fragment length default = 3,000). We analyzed multiple genomes using a genome list with command options of “—ql” and “--al.” For visualization, we used “-matrix” command. For subsequent analysis, genomes with over 80% similarity were selected using FastANI. We visualized ANI results as a heatmap using ggplot2 package in R (Wickham, 2016). Referring to the study of Wallner et al. (2019), we analyzed genes listed as environmental-related genes (nthAB, oxd, lipA, faeB, prnA-D, and uxaAB) and human virulence genes (clab, adhA, esnR, amil, ccil, cciR, opcl, kdgR, baiE, taruX, xsc, telA, terCEF, narG-J, narLM, narX, and lxa) using blastp (Camacho et al., 2009).
2.3 Pan-genome analysis
Pan-genome and core-genome were analyzed using PEPPAN (Zhou et al., 2020). Prokka results in “.gff” file format were compiled for pan-genome analysis. PEPPAN uses a reference-based approach to generate an alignment for each gene group, which is then used to reconstruct a neighbor-joining gene tree using RapidNJ. In the case of PEPPAN results, “.gff” files were created as results. We generated them using “PEPPAN_parser.” Results of the “PEPPAN_parser” displayed summaries, curve information, and gene presence-absence. We used curve information for visualization with a law model (Tettelin et al., 2008). All these processes were performed for group A and group B separately. The pan-genome value, referred to as gamma, was calculated using the Heaps’ law model (Lü et al., 2010). Values of pan-genome and core-genome were calculated as alpha using the Power law model (Tettelin et al., 2008). We analyzed orthologues of core-genes and pan-genes using the web version of EGGNOG-mapper v2.1.9 (Cantalapiedra et al., 2021). Clusters of Orthologous Groups (COGs) results were visualized and preprocessed using R. We excluded no-detected COGs and duplicated COGs.
2.4 Secondary metabolite gene cluster analysis
Gene cluster analysis was conducted using the local version of antiSMASH (Blin et al., 2019) with command options of “--mibig.” To visualize the results of antiSMASH, we conducted PCA based on BGC composition. We measured covariance and used scaled data for PCA analysis. These analyses obtained eigenvalues and eigenvectors using scikit-learn in Python. We visualized PCA data using Matplotlib (Hunter, 2007).
2.5 Network analysis in BGCs
Advanced analysis was performed using antiSMASH with the “--cc-mibig” command for comparison with the MIBiG database (Terlouw et al., 2023). For a more detailed analysis of BGCs, output results from antiSMASH were subjected to network and phylogeny analyses using BiG-SCAPE and CORASON (Navarro-Muñoz et al., 2020). We collected genes involved in BGCs already known and present in antiSMASH results. These genes were confirmed using blastp. We extracted the results of antiSMASH and information about similar gene clusters in MIBiG using Python. Gene clusters found to be similar to reference gene clusters were labeled in the network analysis. BiG-SCAPE offers advantages for conducting large-scale analyses and predicting domains using the pfam database (Mistry et al., 2021) and hmmscan from HMMER (Mistry et al., 2013). The process starts with calculating sequence similarity, followed by measuring pairwise distances between BGCs using sequence similarity. Network analysis was visualized using Cytoscape v3.10.0 (Shannon et al., 2003). Gene cluster visualization and comparison were conducted using Easyfig v2.2.5 (Sullivan et al., 2011).
2.6 Statistical analysis
Statistical analysis for the basic genome was performed using QUAST and tabulated. We conducted a Kruskal-Wallis test and post hoc Dunn test for multiple comparisons between species. Results were visualized using ggplot2. Pan-genome analysis was conducted using Heaps’ law and Power law model for each Gamma value and Kappa value to check significant differences using PEPPAN. To compare genome features, we performed statistical analysis using Mann–Whitney in R. For group comparisons, we used the Mann–Whitney test. Data were visualized using the ggbetweenstats package in R.
3 Results and discussion
3.1 Curation and reclassification of genomic dataset
To ensure data integrity, we implemented two exclusion criteria. Firstly, we removed duplicated samples sourced from NCBI. Additionally, we excluded samples that did not meet the criteria after assessing their completeness and contamination using CheckM. To accurately analyze species of Burkholderia, the frequent occurrence of genus expansion, as reported in Burkholderia, was considered (Lin et al., 2020). We excluded samples with an ANI value below 83%, following the recommendation provided in the reference of fastANI. We excluded samples based on fastANI analysis, where species with ANI values below 83% were considered inter-species. This distinction was explained by Konstantinidis and Tiedje (2005), who confirmed that an ANI value above 94% corresponded to a DNA–DNA hybridization of around 70%. Excluded samples based on these results were documented in Supplementary Table 1. Species displaying ANI values lower than 83% included B. glumae, B. gladioli, and B. plantarii known to be plant pathogens belonging to B. sensu stricto (Kim et al., 2021; Bach et al., 2022b). B. glumae, B. gladioli, and B. plantarii are known as rice pathogenic bacteria belonging to B. sensu stricto. According to Bach et al. (2022b), B. sensu stricto shared a low number of conserved genes with Burkholderia spp. and BCC.
A total of 366 assembly samples were processed for a subsequent study, and samples were well divided by BCC and BPC. A total of 366 strains were validated by Burkholderia in the LPSN database. These samples were confirmed as contigs, total length, GC contents, and N50 using QUAST. We computed statistics for each species. The results are summarized in Table 1. The information for all samples is also included in Supplementary Table 2. The average values were 2.7 contigs, with a 7.18 Gb total length, and 67.43% GC content with a mean and average N50 length of 3.62 Gb. We confirmed 30 types of species, which contained BCC and BPC (Vanlaere et al., 2008; Price et al., 2013; Beukes et al., 2017; Tuanyok et al., 2017; Depoorter et al., 2020; Hall et al., 2022; Morales-Ruíz et al., 2022). The total length and GC contents were compared for species using the Kruskal-Wallis test in R (value of p < 2.2e − 16; Figure 1). Genome sizes of Burkholderia were observed to range from a minimum of 5.2 Gb to a maximum of 9.4 Gb. GC contents ranged from 66% to 69%. Statistically significant variations of genome features were also detected among the species. In the post hoc Dunn test, if the value of p was less than or equal to alpha/2 (where alpha = 0.05), the null hypothesis was rejected. We visualized the length of the genome and GC content of species showing significant differences, along with their corresponding p-values as shown in Figure 1.
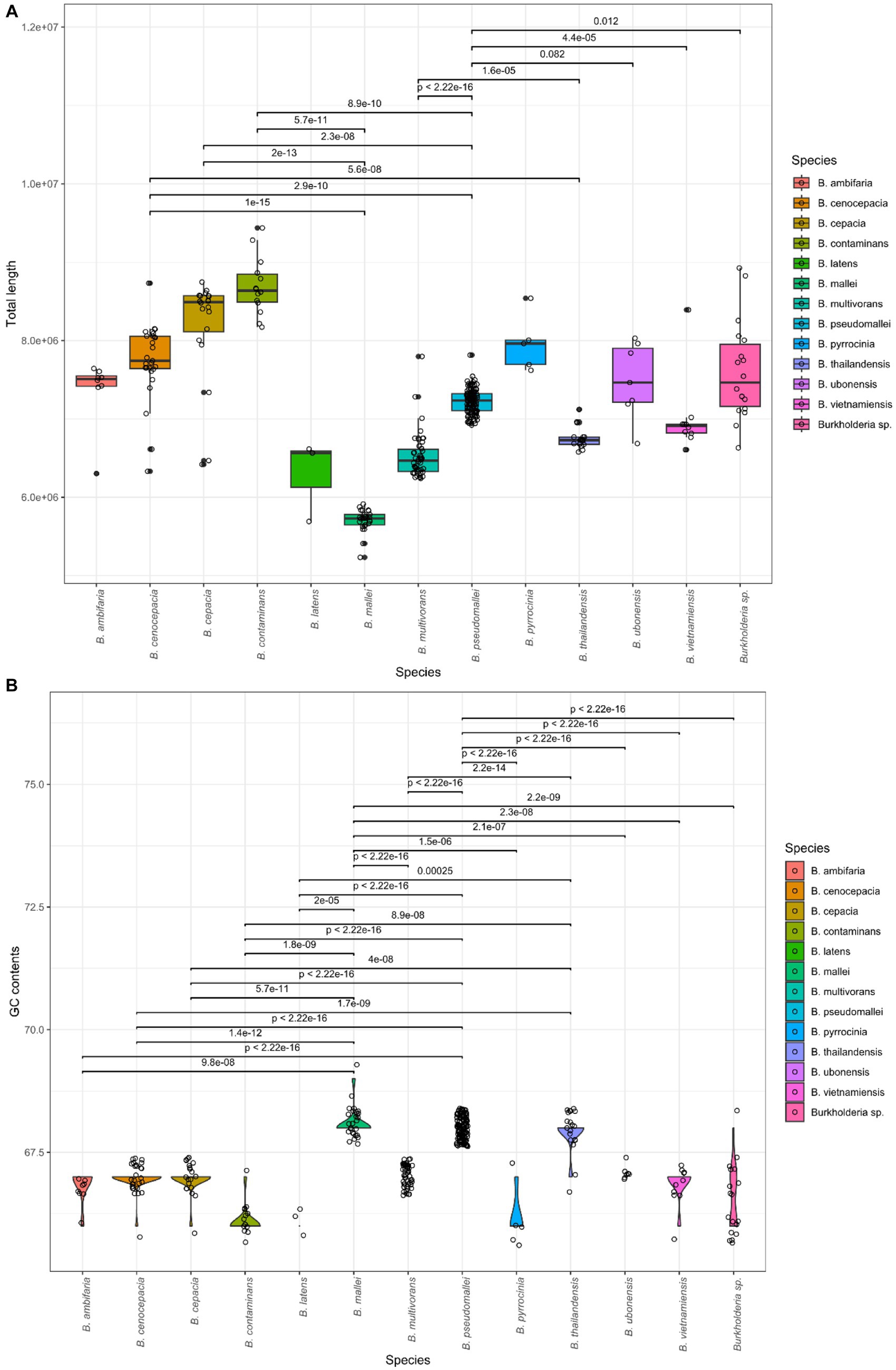
Figure 1. Comparison of genomic features in Burkholderia spp. (A) Average total length per species. (B) Average GC content per species.
3.2 Phylogenomic analysis for BCC and BPC
We calculated pairwise ANI for 366 Burkholderia species (Figure 2A). Based on ANI analysis results, the 366 Burkholderia species were divided into two groups denoted as group A and group B. Group A included B. humptydooensis, B. mallei, B. mayonis, B. oklahomensis, B. pseudomallei, B. savannae, B. sp., and B. thailandensis. The group B were contained B. aenigmatica, B. ambifaria, B. anthina, B. arboris, B. cenocepacia, B. cepacia, B. contaminans, B. diffusa, B. dolosa, B. lata, B. latens, B. metallica, B. multivorans, B. orbicola, B. pseudomultivorans, B. pyrrocinia, B. seminalis, B. sp. B. stabilis, B. stagnalis, B. territorii, B. ubonensis, and B. vietnamiensis. The high similarity observed between B. pseudomallei and B. mallei in ANI results was consistent with merging results reported by Wallner et al. (2019). To determine differences between groups, the Mann–Whitney test was used for comparisons of contigs, length, GC contents, and N50 (Figure 2). Group B contained BCC such as B. cepacia, B. multivorans, B. cenocepacia, and B. stabilis (Lipuma, 2005). Burkholderia species with similarities over 99% were reclassified as similar species (Supplementary Table 3).
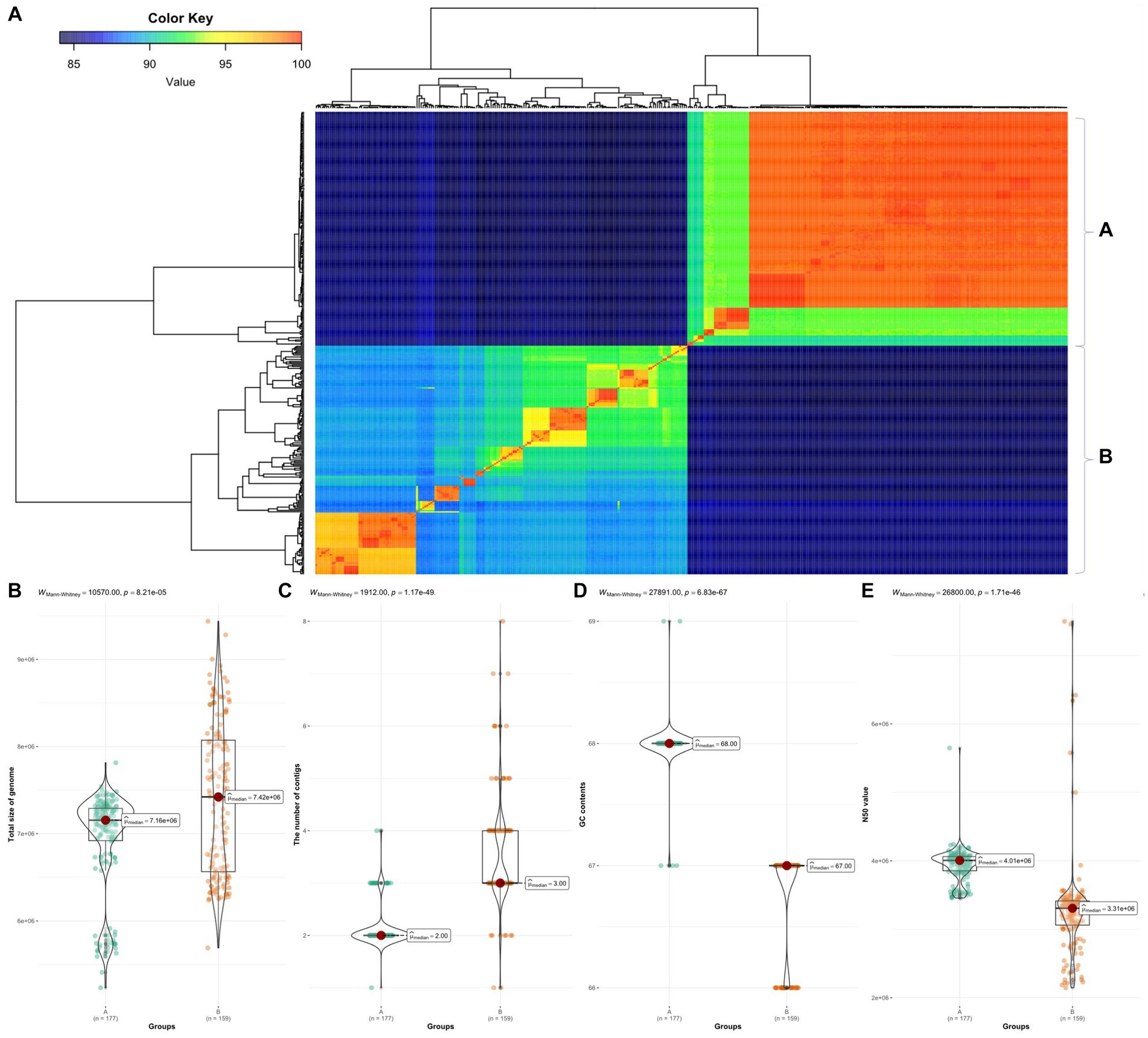
Figure 2. ANI in 366 Burkholderia spp. (A) ANI results using fastANI visualized using ggplot2. We divided samples into groups A and B. (B) Comparison of total length between group A and group B. (C) Comparison of contigs between group A and group B. (D) Comparison of GC contents between group A and group B. (E) Comparison of N50 between group A and group B. All comparisons were visualized using ggbetweenstats package in R.
We visualized the data using Minimum Spanning Tree analysis based on curated MLST genes for Burkholderia, which included aptD, gltB, gyrB, recA, lepA, phaC, and trpB known in BCC, as well as ace, gltB, gmhD, lepA, lipA, narK, and ndh known in B. mallei and B. pseudomallei. However, no notable peculiarities or distinctive patterns were identified (Supplementary Figure 1). Following the findings of Mullins et al. (2019), who concluded the possibility of expandability due to the limitations of curated genes, we collected genes and visualized their presence using BLAST analysis (Supplementary Table 4). However, selected genes did not separate each isolation, species, or group. Genes obtained from the Uniprot database included virulence-related genes (such as adhA, kdgR, narG, narH, narX, telA, terC, xsc, and amiI) and environmental-related genes including oxd, PrnA, prnC, PrnD, and uxaA. We constructed a database with these acquired genes and confirmed them using blastp. Results did not match the separation information recorded in NCBI or ENA. However, it was important to note that confirmed genes varied by species, indicating that these genes could serve as additional factors for confirming species, addressing limitations of MLST with only BCC, B. mallei, and B. pseudomallei available in pubMLST. The distinction between pathogenic and non-pathogenic strains can be confirmed by the presence of virulence factors. Taxonomic analysis may not serve as a clear indicator of their pathogenic potential. In comparative genomics studies, it remains a challenge to determine a suitable indicator for phenotypic characteristic analysis (Eberl and Vandamme, 2016). Investigating pathogenicity and non-pathogenicity is not straightforward.
While we cannot specifically discriminate individual species, our results suggest that for group B, which comprises species belonging to BCC, it would be possible to determine whether the target species is part of the BCC or not. Genomes of Burkholderia used in this study displayed significant similarity in cases where samples were obtained from the same hospital, which led to time-consuming analyses. Therefore, we propose that Burkholderia is the most valuable sample for making a pangenome graph (Hickey et al., 2023). By clustering samples and excluding similar regions, it would be possible to create a pangenome graph for comparison not only within Burkholderia, but also for a broader range of B. sensu lato and other related microorganisms.
3.3 Pan-genome analysis for each group of Burkholderia
We compared genomic patterns among 366 species of Burkholderia which were collected from NCBI. Prokka analysis was performed using species of Burkholderia, while pan-genome and core-genome analyses were performed using PEPPAN with the Prokka output (Figure 3). In group A (n = 185) we observed, 14,066 cloud genes, 2,465 shell genes, 682 soft-core genes, and 2,553 strict-core genes (Figure 3A). The pan-genome results of group A of Burkholderia showed 18.53% of total core genes. Group A was confirmed to have 5,770 genes per genome with 20,292 pan-genes and 2,553 core-genes. In group A, Heaps’ law showed a Gamma value of 0.289 ± 0.002 and a Kappa value of 4493.245 ± 46.171. In the Power law model, an Alpha value of 0.638 ± 0.013 and a Kappa value of 964.425 ± 32.586 were observed. The power law model for core genomes showed an Alpha value of 0.149 ± 0.001 and a Kappa value of 5591.983 ± 20.623. In the results for group B (n = 181), 39,867 cloud genes, 4,986 shell genes, 324 soft-core genes, 222 core genes, and 2,949 strict-core genes were observed (Figure 3B). Ran-genome results for group B of Burkholderia showed 7.22% of total core genes. In group B, Heaps’ law showed a Gamma value of 0.389 ± 0.001 and a Kappa value of 6444.550 ± 30.636. In the Power law model, an Alpha value of 0.629 ± 0.003 and a Kappa value of 2719.814 ± 21.208 were observed. The power law model for core genomes showed an Alpha value of 0.099 ± 0.007 and a Kappa value of 4795.235 ± 131.235. Strict core genes showed 100% similarity, while core genes showed similarities ranging from over 99% to less than 100%. Soft-core genes showed similarities ranging from over 95% to less than 99%. Genes with similarities ranging from 0% to 15% were classified as cloud genes. Those with similarities ranging from 15% to 95% were categorized as shell genes. Pangenomes in both groups showed Alpha values under one. The pan-genome analysis of Burkholderia began in 2009, where 56 species of Burkholderia were found to possess 4,000 genes, with a core-genome consisting of 1,000 genes (Ussery et al., 2009). Subsequently, Bochkareva et al. (2018) analyzed 127 Burkholderia species using the micropan tool in the R environment. They conducted an analysis employing the binomial mixture model and Chao’s lower bound. In this study, we attempted to conduct pan-genome analysis using the micropan tool, similar to previous research methods. However, we were unable to complete the analysis due to insufficient computer performance. Because of the large number of samples, we divided our samples into group A and group B based on ANI results. For pan-genome analysis, we utilized PEPPAN, which employs representative genes obtained through clustering to expedite the analysis process. When comparing our research findings to those of Bach et al. (2022a), we observed that group B, which included BCC, showed a slightly higher number of 2,949 strict core genes compared to ROARY results.
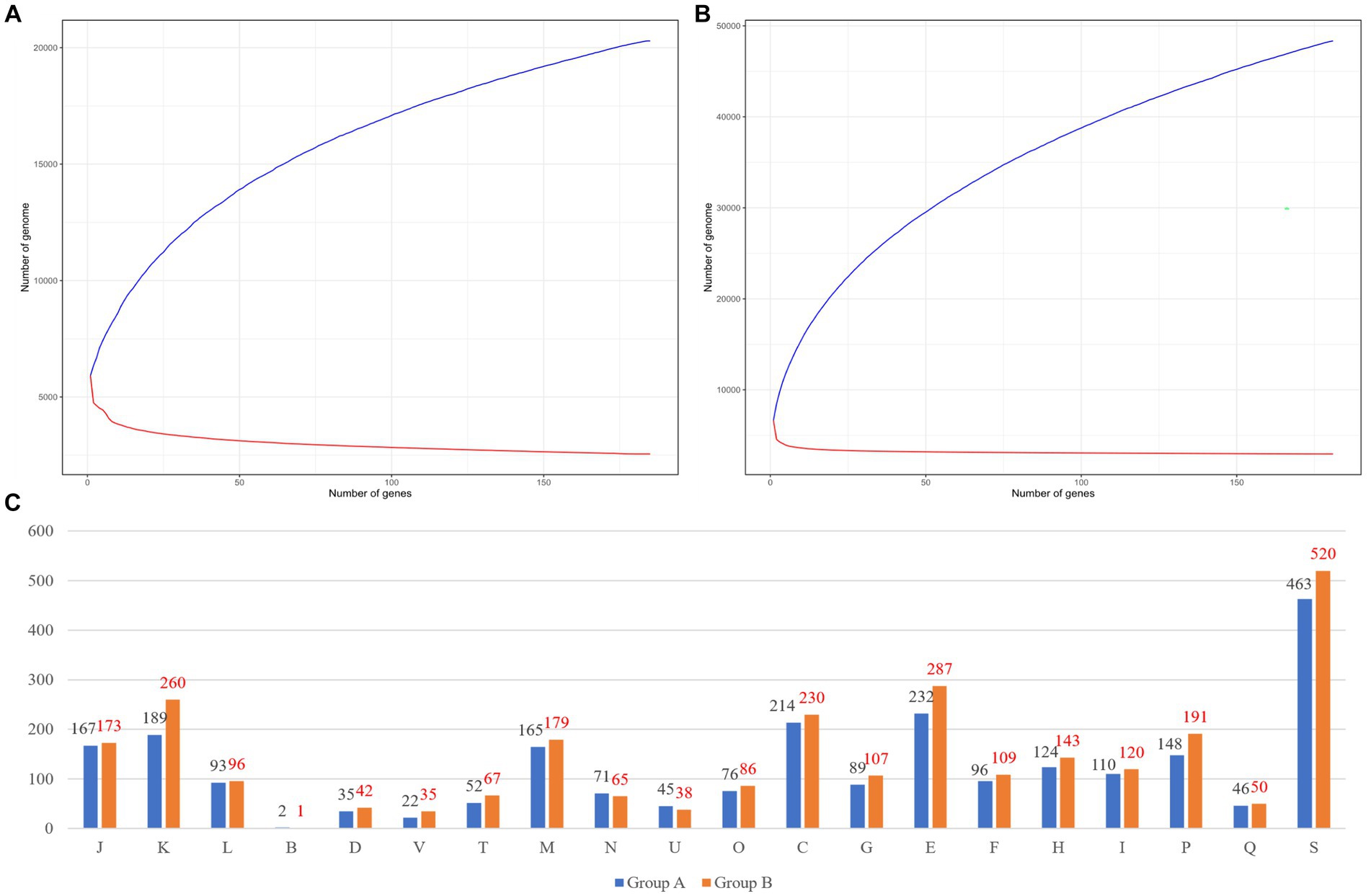
Figure 3. Pan-genome results of each group. (A) Group A’s pan-genome and core-genome results. (B) Group B’s pan-genome and core-genome results. (C) Differences between group A and group B. The number of COGs in group A is indicated by a black color and the number of COGs in group B is indicated by a red color. COGs categories are as follows: J, translation, ribosomal structure and biogenesis; (A), RNA processing and modification; (K), transcription; (L), replication, recombination, and repair; (B), chromatin structure and dynamics; D, cell cycle control, cell division, and chromosome partitioning; Y, nuclear structure; V, defense mechanisms; T, signal transduction mechanisms; (M), cell wall/membrane/envelope biogenesis; (N), cell motility; (Z), cytoskeleton; (W), extracellular structures; (U), intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, and chaperones; (X), mobilome, prophages, and transposons; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport, and catabolism; R, general function prediction only; and S, function unknown.
Ortholog analyses of core genes in group A and group B were performed using eggNOG. We visualized COGs. We analyzed functional annotations for 2,553 core-genes from group A. We visualized the results of COGs with both group A and group B (Figure 3C). Most differences were shown in V (37%) and K (27%), respectively. Pan-genome analyses have been steadily conducted for diverse Burkholderia species based on genomics of B. contaminans (Kim et al., 2023) or B. cepacia (Ahmad and Azam, 2020; which showed similar results with ours) and BCC classification (Jin et al., 2020). Recently, Bach et al. (2022b) reported a pan-genome focused study that analyzed not only BCC but also B. sensu lato and B. sensu stricto. Through pan-genome analysis, Lood et al. (2021) confirmed patient diversity using genomics. The present study has identified unique attributes exclusive to Burkholderia using a pattern analysis of BGCs based on a comparison between BCC and BPC of Burkholderia. Our results provide unique insights and can be used to perform further genome analysis for pathogenic Burkholderia.
3.4 Analysis of BGC
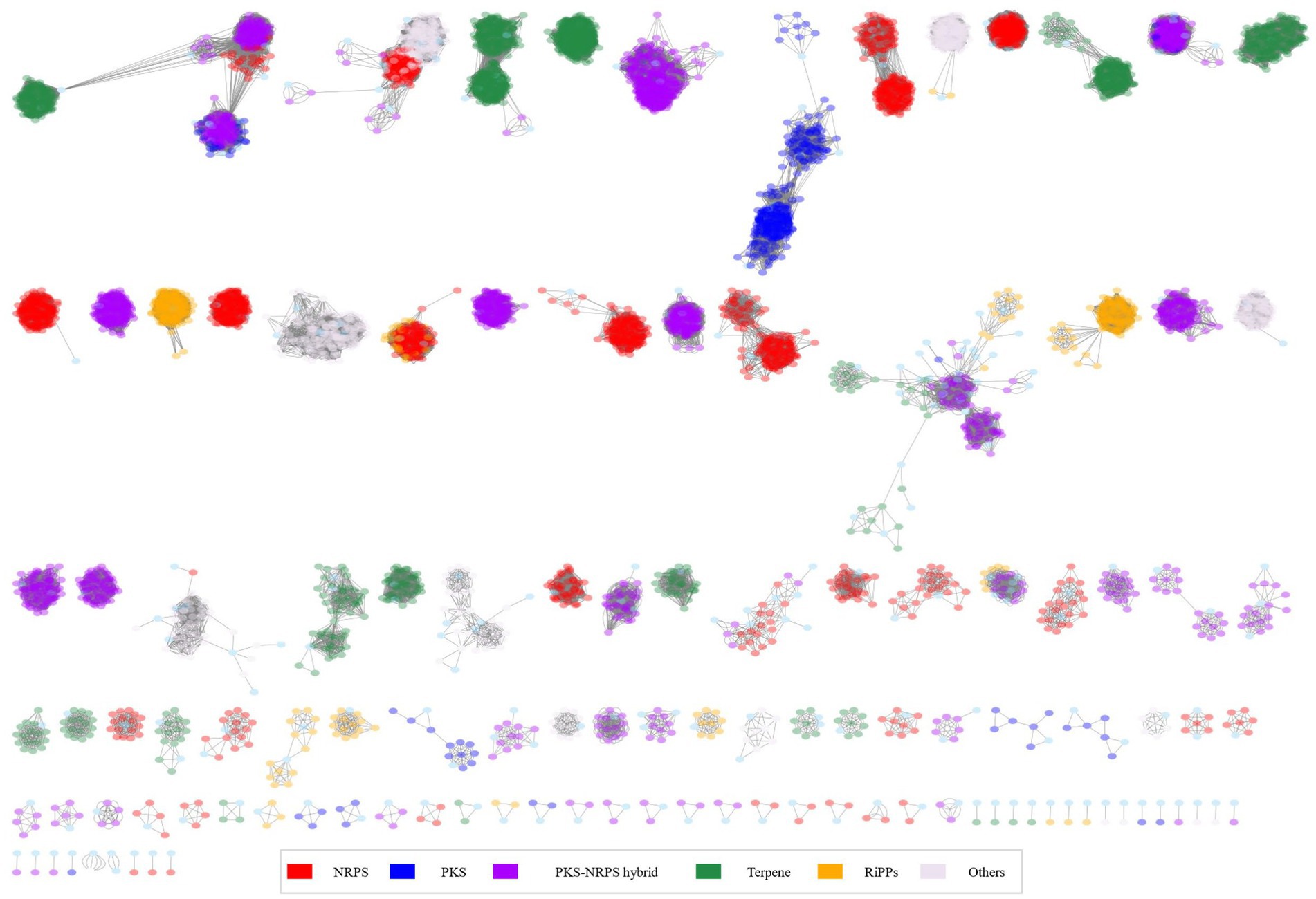
Using antiSMASH, this study detected and categorized 6,666 BGCs belonging to 113 kinds of BGCs and 30 classes including polyketide synthase (PKS), terpene, and siderophore. We analyzed BGCs using PCA (Figure 4A). During the process of visualizing PCA results, we examined both environmental and pathological samples. However, no significant differences were observed. Instead, PCA results revealed a more distinct separation based on species and groups (data not shown).
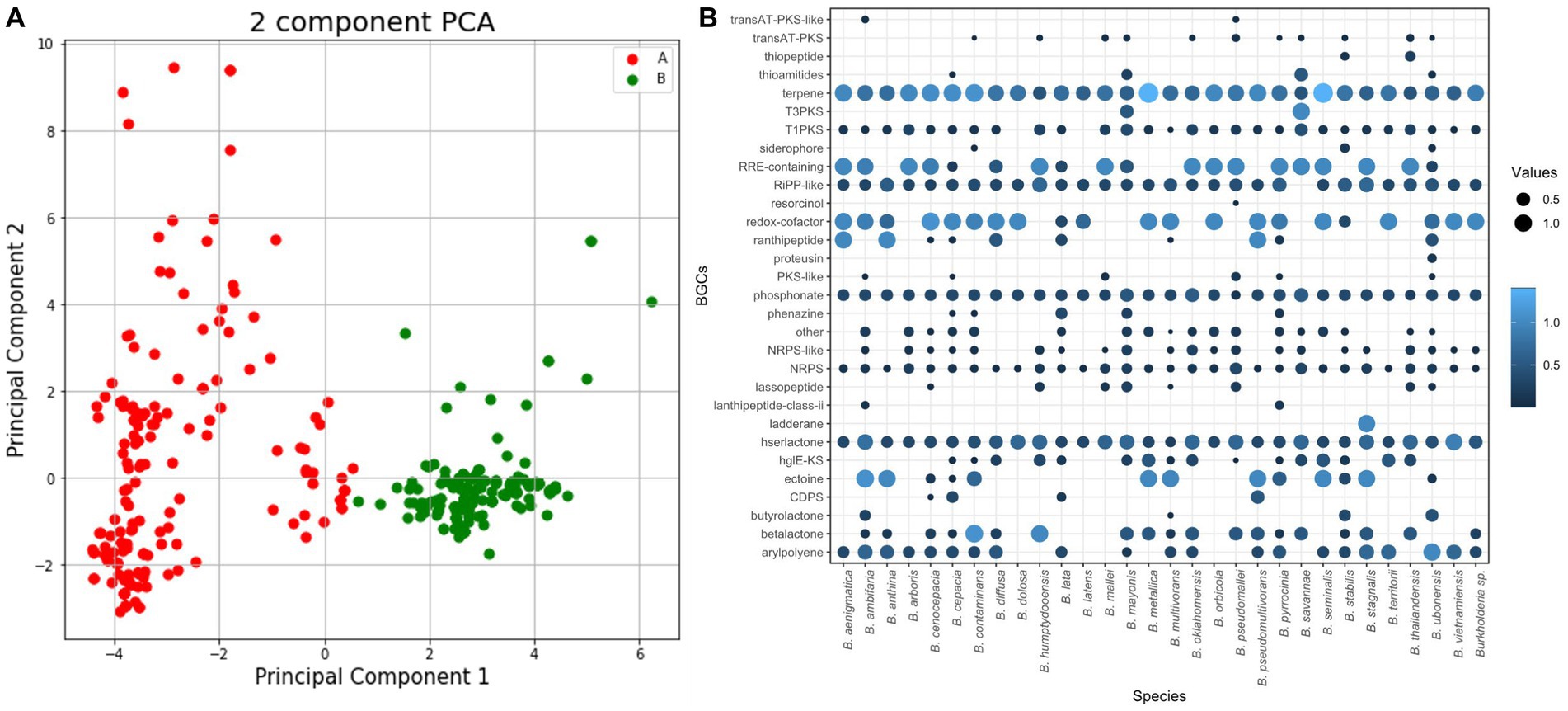
Figure 4. BGC analysis for 366 Burkholderia spp. (A) AntiSMASH results of Burkholderia spp. We visualized BGCs in each species and combined similar BGCs in one group. (B) PCA analysis using antiSMASH results matrix and color by groups.
When visualizing the results of PCA for each species, we observed distinct clusters corresponding to B. pseudomallei and B. mallei. B. pseudomallei was the most frequently analyzed species with 128 samples, which might have influenced PCA results, showing a distinct clustering. Despite the high similarity observed between B. pseudomallei and B. mallei in ANI results, they exhibited clear separation in the PCA analysis (Supplementary Figure 2). Investigation of 366 Burkholderia strains revealed an average of 17 BGCs per strain. B. latens AU0505 was found to possess 7 BGCs, while B. pseudomallei 1710b, B. pseudomallei BSR, B. pseudomallei 406e, and B. mayonis BDU8 were found to harbor 26 BGCs each. In this study, we calculated the number of BGCs in each species. BCC and BPC showed higher numbers of candidate BGCs than previously known BGCs (Mullins and Mahenthiralingam, 2021; Figure 4B). We found three novel BGCs from B. cenocepacia J2315, B. pseudomallei 3,000,047,530, and B. pyrrocinia MS455. A study on genomic diversity and metabolic capabilities of B. sensu lato, has found that Burkholderia possesses a more diverse set of BGCs than other genera (Mullins and Mahenthiralingam, 2021; Petrova and Mahenthiralingam, 2022). Alam et al. (2021) have also confirmed genome mining of BGCs in Burkholderia including B. latens, B. cenocepacia, B. cepacia, B. ambifaria, and B. lata. Our study showed similar patterns of BGC possession and confirmed novel species recently assigned to BCC and BPC.
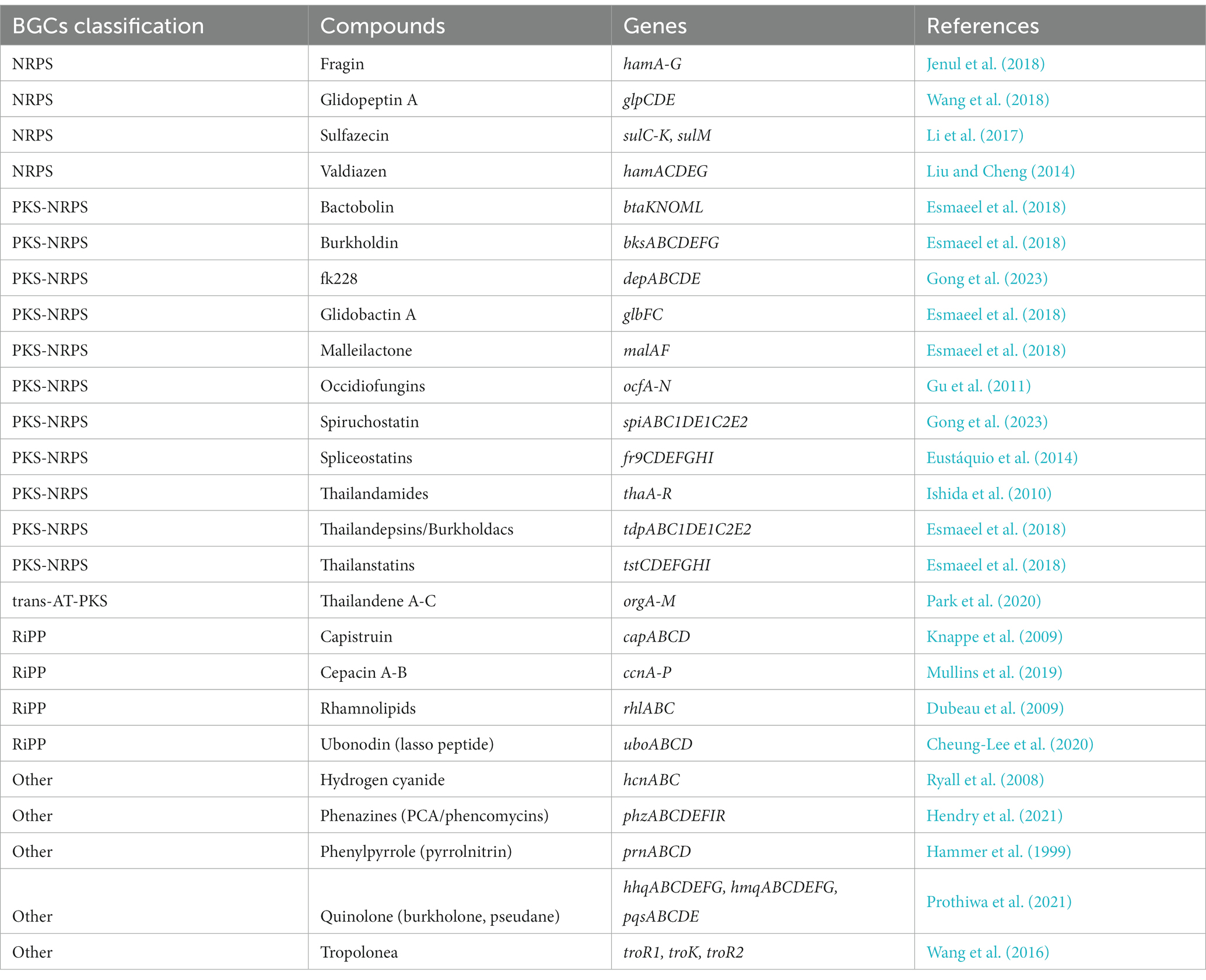
Based on these results, we hypothesized that there would be distinct metabolite patterns depending on the species of Burkholderia. BiG-SCAPE results were then subjected to network analysis, categorizing antiSMASH outcomes into NRPS, terpene, PKSI, RiPPs, PKSother, PKS-NRPS hybrids, and others (Figure 5; Supplementary Table 5). A total of 192 types of BGCs were found and compared with the MiBIG database (Supplementary Tables 6, 7). Two types of alkaloids, 48 types of NRPS (including one kind of NRPS-alkaloid and 13 kinds of NRPS-polyketide BGCs), 30 kinds of polyketide, 28 kinds of RiPP, 12 kinds of saccharide, 17 kinds of terpene, and 55 BGCs of others were found. We confirmed the most popular 667 terpene-related BGCs known as carotenoids-related BGCs from Myxococcus xanthus. We found 44 novel BGCs in distinct samples. These novel BGCs contained 13 other BGCs classes and followed polyketide. For each BGC, known compounds were compiled and core gene information was organized to detect gene clusters (Table 2; Liu and Cheng, 2014; Kunakom and Eustáquio, 2019) and undertake a more detailed analysis, described below.
3.5 Pattern analysis for NRPS and PKS
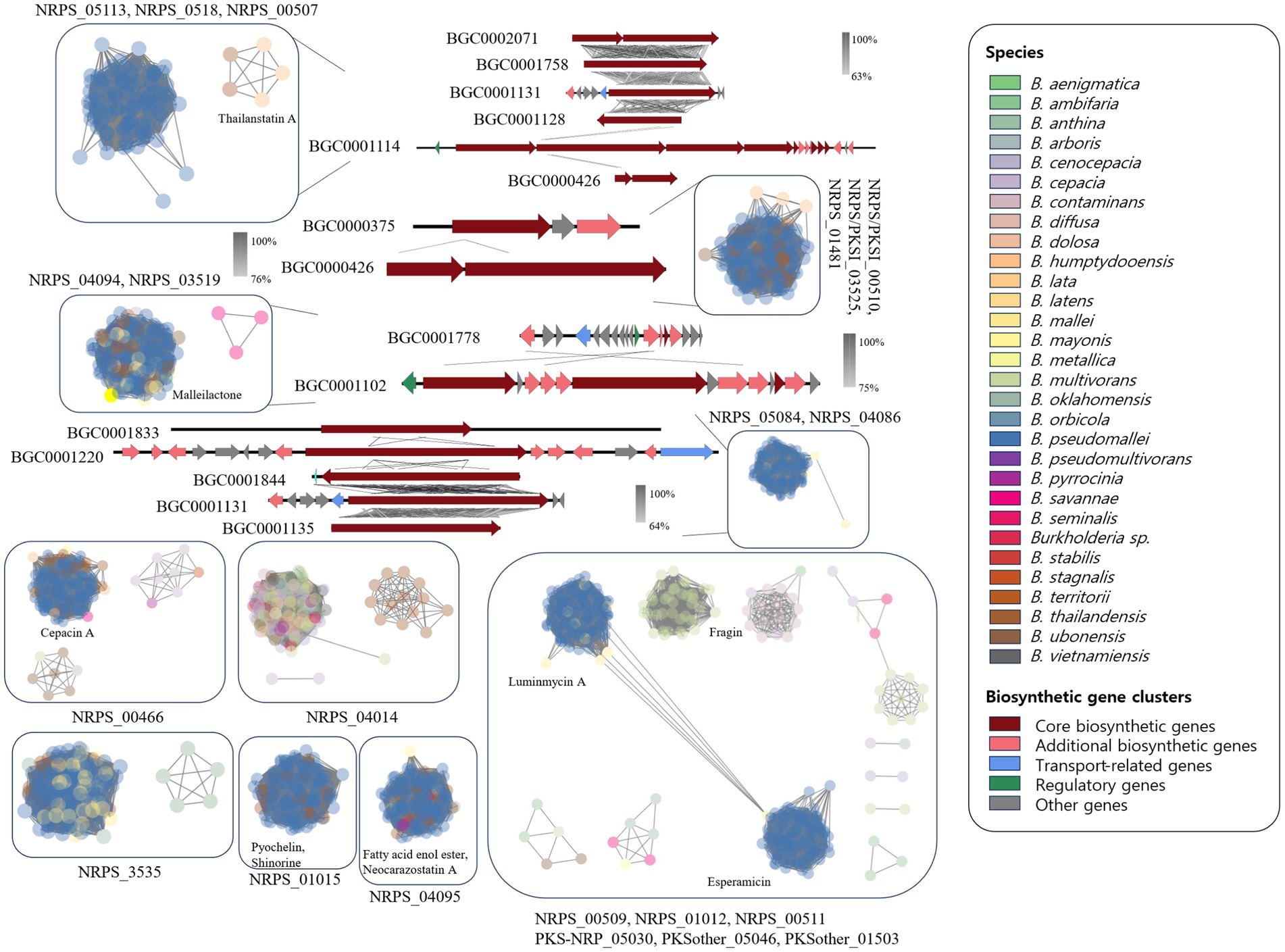
The NRPS network yielded 2,345 BGCs with 138,894 links and 118 families, including 28 singletons. Many compounds have been reported in Burkholderia. The following compounds were identified in NRPS gene clusters: valdiazen, glidopeptin A, rhizomide A, occidiofungins, fragin, sulfazecin, and icosalide A/B. PKSI network yielded 459 BGCs with 22,730 links, and 30 families, including 7 singletons. PKSother network showed 642 BGCs, 33,515 links, 48 families, and 15 singletons. The network of PKS-NRPS hybrids yielded 524 BGCs with 29,440 links and 31 families, including 6 singletons. PKS-NRPS hybrid BGCs could be categorized as follows based on their gene cluster types: NRPS-T1PKS, PKS-NRPS, and transAT-PKS. Network analysis was visualized with reference gene clusters (Figure 6). We found 44 types of BGCs, and BGCs were known from Burkholderia, Paraburkholderia, and Pseudomonas. In the network analysis, we visualized the node as the sample species and the edge as the link between samples. Each node color indicates a species. We visualized the reference for BGC from the MiBIG database in each dependent network. We grouped each clan based on the results of BiG-SCAPE. In the results of network analysis, some BGCs were calculated in the part of PKS-NRPS hybrid, NRPS, and PKSother. We visualized the main clans in NRPS, PKS-NRPS hybrid, PKSI, and PKSother. We also visualized BGCs known as Burkholderia-related species such as Paraburkholderia and Pseudomonas. Although network analyses of BGCs through genome mining and broader global analyses have been reported in previous studies, only B. ambifaria has been found to focus on the secondary metabolite (Mullins et al., 2019).
Clan members within NRPS_05113 were confirmed to include BGC0002071, BGC0001758, BGC0001131, and BGC0001128, which are known to generate virginiafactin, rhizomide A, ambactin, and luminmide, respectively. The similarity between each BGC was visualized using Easyfig. Reference BGCs identified in each network were compared and visualized as shown in Figure 6. The representative compounds identified in each clan are also indicated. Although the comparison of BGCs between BCC and the BPC was based on gene cluster similarity analysis and network analysis, a greater diversity of BGCs was observed than anticipated. Similar BGCs were also identified in other plant species, fungi, and beyond Burkholderia. These unique BGC-specific features across different species have expanded the scope of Burkholderia research, enabling more extensive investigations. Most clans revealed that B. pseudomallei was the main species excluding NRPS_04014. The NRPS_04014 clan contained ornibactin and indigoidine-related BGCs. Ornibactin was known as a siderophore from B. cenocepacia. In 44 types of BGCs, we confirmed several types of siderophore. Siderophore-related BGCs were confirmed. Ornibactin are known to possess Pseudomonas and siderophore-like compounds (Anthoni et al., 1995).
3.6 Identification of siderophore and pattern analysis for BCC and BPC
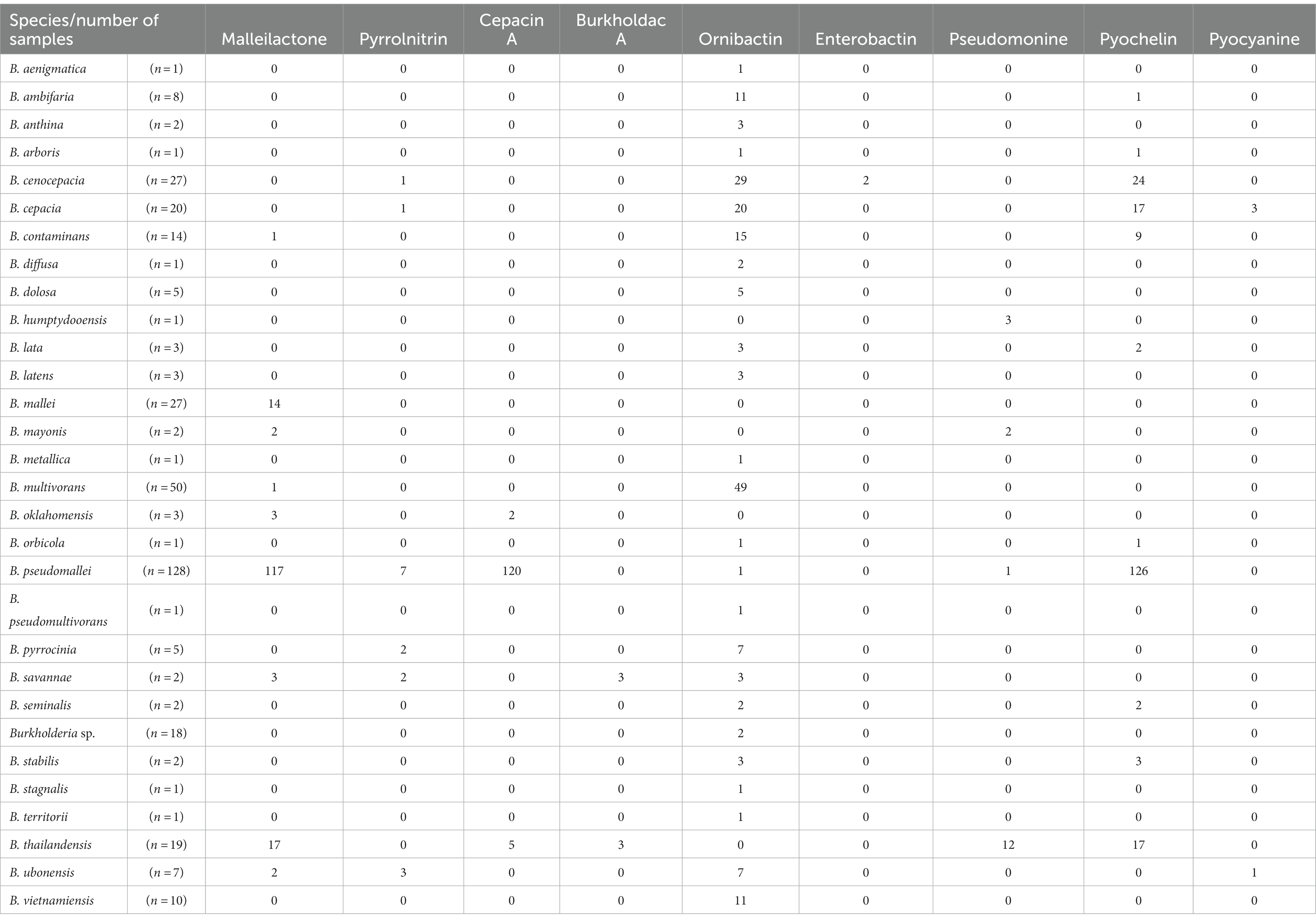
We predicted that complex and/or species would have different types of siderophore-related BGCs. To test our hypothesis, we performed additional analysis. Siderophores have been reported to exist in BCC. A comparative transcriptome study of B. pseudomallei reported on the induction of siderophores that were able to adapt and survive in the host (Ghazali et al., 2023). However, in antiSMASH results, species belonging to group B did not show the presence of siderophore-related clans. Since Burkholderia are known to produce various siderophores that might not be detected by antiSMASH, we conducted further analysis using the results of MiBIG similarity to explore siderophore gene clusters (Table 3). In Burkholderia, the following types of siderophores have been reported: ornibactin, malleobactin, cepaciachelin, pyochelin, and cepabactin (Butt and Thomas, 2017). For ornibactin biosynthesis, NRPS genes such as orbI and orbJ are known, and we used them to search for the corresponding BGCs (Agnoli et al., 2006). Pyochelin biosynthesis involves pchE and pchF known as NRPS (Quadri et al., 1999). We used these genes to search for the corresponding BGCs with malleobactin related genes as well (Alice et al., 2006). To find cepaciachelin, we collected genes such as cphA, cphB, and cphC (Esmaeel et al., 2016). Cepabactin-related genes have not been described. Malleonitrone is a compound formed by the combination of malleobactins and pyochelin (Trottmann et al., 2019). Other siderophores, burkholdac A and pseudomonine, were identified in group A, which belonged to the BPC, but not in group B. Burkholdac A was exclusively found in B. savannae and B. thailandensis, while pseudomonine was detected in B. humptydooensis, B. mayonis, B. pseudomallei, and B. thailandensis only. Siderophores are known to be species-specific. Our investigation revealed that species within the same complex in Burkholderia commonly possessed siderophore-related BGCs (Sandy and Butler, 2009). Alongside siderophores, compounds related to terpenes and RiPPs have been reported. Additionally, certain strains known as plant pathogens, such as B. glumae, have been reported to contain a variety of secondary metabolites. For instance, toxoflavin (Kim et al., 2004), gladiofungin A (gladiostatin; Niehs et al., 2020), and gladiolin from B. gladioli (Song et al., 2017) have been identified. Furthermore, compounds such as enacyloxin, which has not been confirmed due to limited known genes, have been reported in B. ambifaria. They are believed to contain PKS-related modules (Mahenthiralingam et al., 2011).
This study utilized ClusterBlast to extract similar gene clusters. We structured them into a table using Python. Among confirmed siderophore-related compounds, we identified nine, namely malleilactone, pyrrolnitrin, cepacin A, malleobactin, burkholdac A, ornibactin, enterobactin, pseudomonine, pyochelin, quinolobactin, and pyocyanine. Among these, ornibactin was the most commonly found across various species. The NRPS_4602 clan, which harbored the ornibactin BGC, encompassed gene clusters for ornibactin, pyocyanine, indigoidine, burkholderic acid, anabaenopeptin, burkholdac A, depudecin, and fellutamide B. We visualized ornibactin BGCs within this clan using CORASON (Figure 7). Within the NRPS_4602 clan, ornibactin BGCs were distributed across families FAM_1520, FAM_2544, FAM_2559, FAM_4014, FAM_4564, FAM_4602, and FAM_5592. Each family’s gene clusters exhibited similarity, further confirming their species-specific nature.
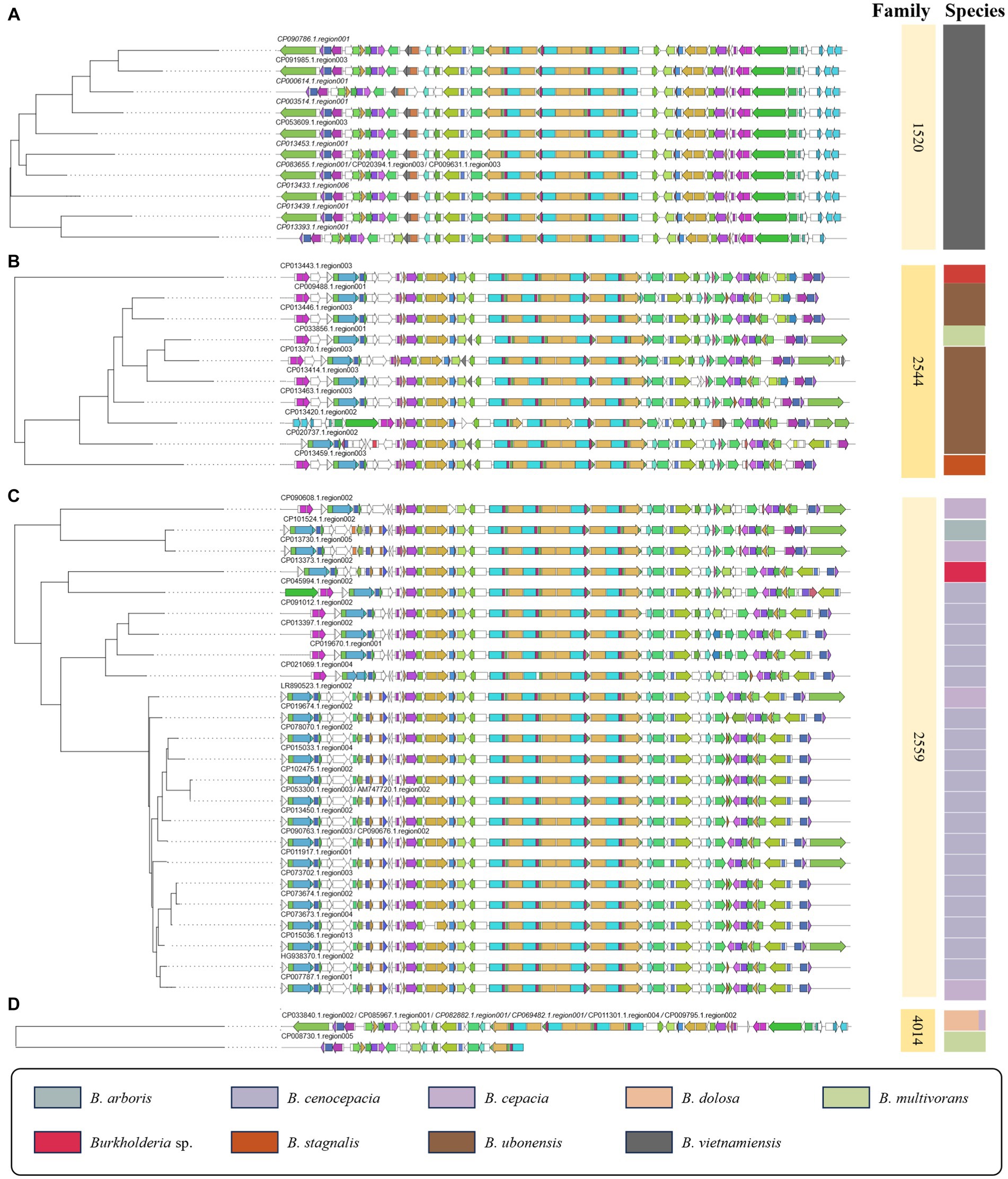
Figure 7. Visualization of ornibactin-related biosynthetic genes in NRPS_4602 clan. (A) The phylogeny of 1520 family. (B) The phylogeny of 2544 family. (C) The phylogeny of 2559 family. (D) The phylogeny of 4014 family.
In the case of FAM_1520, it mainly corresponded to ornibactin BGCs identified in B. vietnamiensis, while FAM_2544 was primarily associated with B. ubonensis. For all other families, we visualized gene clusters as shown in Figure 7 and Supplementary Figures 2, 3. The observed similarity in gene clusters within the same species suggested their potential utility in distinguishing between BCC and BPC. Previous studies on siderophores have been limited to a small number of Burkholderia strains (Esmaeel et al., 2016). Our research presents a new direction for the genome mining of the secondary metabolism, with an expanded sample size and an analytical approach.
4 Conclusion
To date, Burkholderia has been known for its diversity in species, but more recent reclassification efforts have associated all complexes within the Burkholderia genus as having pathogenic characteristics. In this study, we aimed to analyze features of Burkholderia through ANI analysis and reference genes, categorizing them into BCC and BPC for analysis. Through PCA analysis based on antiSMASH results, BCC and BPC were revealed to be distinct. This indicates that each complex is likely to possess a different pattern of BGCs. The network analysis, BiG-SCAPE analysis, and comparative analysis using the MiBIG database revealed that BGCs differed by complex. Through network analysis and visualization of siderophores specific to each species, we also demonstrated unique siderophore patterns for each species and/or complex. Furthermore, this study explored known BGCs reported not only in Burkholderia, but also in Pseudomonas and Paraburkholderia. By visualizing the gene cluster of ornibactin, the siderophore found in the highest number of species, we anticipate that pattern analysis could be further advanced from a broader perspective. This research became possible due to the increasing number of Burkholderia genomes and the identification of various BGCs in Burkholderia. However, identifying novel BGCs remains challenging. We could only confirm that results for just three BGCs were not detected from the known MiBIG database. This study serves as a comprehensive investigation into NRPS and PKS. It contributes to future research on secondary metabolites in Burkholderia.
Author contributions
BK: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Validation, Writing – original draft, Writing – review & editing. S-RH: Data curation, Methodology, Software, Writing – original draft, Writing – review & editing. HL: Formal analysis, Methodology, Software, Writing – original draft, Writing – review & editing. T-JO: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Technology Innovation Program (20018705, entitled “Development of masking and commercialization of biodegradable technology in an urban residential environment using rancid odor-reducing microorganisms and its fragrances”) funded by the Ministry of Trade, Industry and Energy (MOTIE, Republic of Korea). This research was also supported by a project entitled “Development of potential antibiotic compounds using polar organism resources (20200610)” funded by the Ministry of Oceans and Fisheries, Republic of Korea.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1302236/full#supplementary-material
References
Agnoli, K., Lowe, C. A., Farmer, K. L., Husnain, S. I., and Thomas, M. S. (2006). The ornibactin biosynthesis and transport genes of Burkholderia cenocepacia are regulated by an extracytoplasmic function sigma factor which is a part of the Fur regulon. J. Bacteriol. 188, 3631–3644. doi: 10.1128/JB.188.10.3631-3644.2006
Ahmad, F., and Azam, S. S. (2020). From pan-genome to protein dynamics: a computational hierarchical quest to identify drug target in multi-drug resistant Burkholderia cepacia. J. Mol. Liq. 317:113904. doi: 10.1016/j.molliq.2020.113904
Alam, K., Islam, M. M., Gong, K., Abbasi, M. N., Li, R., Zhang, Y., et al. (2021). In silico genome mining of potential novel biosynthetic gene clusters for drug discovery from Burkholderia bacteria. Comput. Biol. Med. 140:105046. doi: 10.1016/j.compbiomed.2021.105046
Alice, A. F., López, C. S., Lowe, C. A., Ledesma, M. A., and Crosa, J. H. (2006). Genetic and transcriptional analysis of the siderophore malleobactin biosynthesis and transport genes in the human pathogen Burkholderia pseudomallei K96243. J. Bacteriol. 188, 1551–1566. doi: 10.1128/JB.188.4.1551-1566.2006
Anthoni, U., Christophersen, C., Nielsen, P. H., Gram, L., and Petersen, B. O. (1995). Pseudomonine, an isoxazolidone with siderophoric activity from Pseudomonas fluorescens AH2 isolated from Lake Victorian Nile perch. J. Nat. Prod. 58, 1786–1789. doi: 10.1021/np50125a026
Bach, E., Passaglia, L. M. P., Jiao, J., and Gross, H. (2022a). Burkholderia in the genomic era: from taxonomy to the discovery of new antimicrobial secondary metabolites. Crit. Rev. Microbiol. 48, 121–160. doi: 10.1080/1040841X.2021.1946009
Bach, E., Sant'Anna, F. H., Seger, G. D. D. S., and Passaglia, L. M. P. (2022b). Pangenome inventory of Burkholderia sensu lato, Burkholderia sensu stricto, and the Burkholderia cepacia complex reveals the uniqueness of Burkholderia catarinensis. Genomics 114, 398–408. doi: 10.1016/j.ygeno.2021.11.011
Barrett, T., Clark, K., Gevorgyan, R., Gorelenkov, V., Gribov, E., Karsch-Mizrachi, I., et al. (2012). BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 40, D57–D63. doi: 10.1093/nar/gkr1163
Benson, D. A., Cavanaugh, M., Clark, K., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., et al. (2017). GenBank. Nucleic Acids Res. 45, D37–D42. doi: 10.1093/nar/gkw1070
Beukes, C. W., Palmer, M., Manyaka, P., Chan, W. Y., Avontuur, J. R., van Zyl, E., et al. (2017). Genome data provides high support for generic boundaries in Burkholderia sensu lato. Front. Microbiol. 8:1154. doi: 10.3389/fmicb.2017.01154
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87. doi: 10.1093/nar/gkz310
Bochkareva, O. O., Moroz, E. V., Davydov, I. I., and Gelfand, M. S. (2018). Genome rearrangements and selection in multi-chromosome bacteria Burkholderia spp. BMC Genomics 19:965. doi: 10.1186/s12864-018-5245-1
Butt, A. T., and Thomas, M. S. (2017). Iron acquisition mechanisms and their role in the virulence of Burkholderia species. Front. Cell. Infect. Microbiol. 7:460. doi: 10.3389/fcimb.2017.00460
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics. 10:421. doi: 10.1186/1471-2105-10-421
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P., and Huerta-Cepas, J. (2021). eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829. doi: 10.1093/molbev/msab293
Cheung-Lee, W. L., Parry, M. E., Zong, C., Cartagena, A. J., Darst, S. A., Connell, N. D., et al. (2020). Discovery of ubonodin, an antimicrobial lasso peptide active against members of the Burkholderia cepacia complex. ChemBioChem 21, 1335–1340. doi: 10.1002/cbic.201900707
Compant, S., Nowak, J., Coenye, T., Clément, C., and Ait Barka, E. (2008). Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 32, 607–626. doi: 10.1111/j.1574-6976.2008.00113.x
Cummins, C., Ahamed, A., Aslam, R., Burgin, J., Devraj, R., Edbali, O., et al. (2022). The European nucleotide archive in 2021. Nucleic Acids Res. 50, D106–D110. doi: 10.1093/nar/gkab1051
Deng, P., Jia, J., Foxfire, A., Baird, S. M., Smith, L. J., and Lu, S. E. (2023). A polyketide synthetase gene cluster is responsible for antibacterial activity of Burkholderia contaminans MS14. Phytopathology 113, 11–20. doi: 10.1094/PHYTO-03-22-0106-R
Depoorter, E., Bull, M. J., Peeters, C., Coenye, T., Vandamme, P., and Mahenthiralingam, E. (2016). Burkholderia: an update on taxonomy and biotechnological potential as antibiotic producers. Appl. Microbiol. Biotechnol. 100, 5215–5229. doi: 10.1007/s00253-016-7520-x
Depoorter, E., De Canck, E., Peeters, C., Wieme, A. D., Cnockaert, M., Zlosnik, J. E. A., et al. (2020). Burkholderia cepacia complex taxon K: where to split? Front. Microbiol. 11:1594. doi: 10.3389/fmicb.2020.01594
Dubeau, D., Déziel, E., Woods, D. E., and Lépine, F. (2009). Burkholderia thailandensis harbors two identical rhl gene clusters responsible for the biosynthesis of rhamnolipids. BMC Microbiol. 9:263. doi: 10.1186/1471-2180-9-263
Eberl, L., and Vandamme, P. (2016). Members of the genus Burkholderia: good and bad guys. F1000Res 5:F1000. doi: 10.12688/f1000research.8221.1
Esmaeel, Q., Pupin, M., Jacques, P., and Leclère, V. (2018). Nonribosomal peptides and polyketides of Burkholderia: new compounds potentially implicated in biocontrol and pharmaceuticals. Environ. Sci. Pollut. Res. Int. 25, 29794–29807. doi: 10.1007/s11356-017-9166-3
Esmaeel, Q., Pupin, M., Kieu, N. P., Chataigné, G., Béchet, M., Deravel, J., et al. (2016). Burkholderia genome mining for nonribosomal peptide synthetases reveals a great potential for novel siderophores and lipopeptides synthesis. Microbiology 5, 512–526. doi: 10.1002/mbo3.347
Estrada-de Los Santos, P., Palmer, M., Chávez-Ramírez, B., Beukes, C., Steenkamp, E. T., Briscoe, L., et al. (2018). Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. Nov., and Trinickia gen. Nov.): implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes 9:389. doi: 10.3390/genes9080389
Eustáquio, A. S., Janso, J. E., Ratnayake, A. S., O'Donnell, C. J., and Koehn, F. E. (2014). Spliceostatin hemiketal biosynthesis in Burkholderia spp. is catalyzed by an iron/α-ketoglutarate-dependent dioxygenase. Proc. Natl. Acad. Sci. U. S. A. 111, E3376–E3385. doi: 10.1073/pnas.1408300111
Euzéby, J. P. (1997). List of bacterial names with standing in nomenclature: a folder available on the internet. Int. J. Syst. Evol. Microbiol. 47, 590–592. doi: 10.1099/00207713-47-2-590
Gee, J. E., Gulvik, C. A., Castelo-Branco, D. S. C. M., Sidrim, J. J. C., Rocha, M. F. G., Cordeiro, R. A., et al. (2021). Genomic diversity of Burkholderia pseudomallei in Ceara, Brazil. mSphere. 6, e01259–e01220. doi: 10.1128/mSphere.01259-20
Ghazali, A. K., Firdaus-Raih, M., Uthaya Kumar, A., Lee, W. K., Hoh, C. C., and Nathan, S. (2023). Transitioning from soil to host: comparative transcriptome analysis reveals the Burkholderia pseudomallei response to different niches. Microbiol Spectr. 11:e0383522. doi: 10.1128/spectrum.03835-22
Gong, K., Wang, M., Duan, Q., Li, G., Yong, D., Ren, C., et al. (2023). High-yield production of FK228 and new derivatives in a Burkholderia chassis. Metab. Eng. 75, 131–142. doi: 10.1016/j.ymben.2022.12.002
Gu, G., Smith, L., Liu, A., and Lu, S. E. (2011). Genetic and biochemical map for the biosynthesis of occidiofungin, an antifungal produced by Burkholderia contaminans strain MS14. Appl. Environ. Microbiol. 77, 6189–6198. doi: 10.1128/AEM.00377-11
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hall, C. M., Baker, A. L., Sahl, J. W., Mayo, M., Scholz, H. C., Kaestli, M., et al. (2022). Expanding the Burkholderia pseudomallei complex with the addition of two novel species: Burkholderia mayonis sp. nov. and Burkholderia savannae sp. nov. Appl. Environ. Microbiol. 88:e0158321. doi: 10.1128/AEM.01583-21
Hall, C. M., Busch, J. D., Shippy, K., Allender, C. J., Kaestli, M., Mayo, M., et al. (2015). Diverse Burkholderia species isolated from soils in the southern United States with no evidence of B. pseudomallei. PloS One 10:e0143254. doi: 10.1371/journal.pone.0143254
Hammer, P. E., Burd, W., Hill, D. S., Ligon, J. M., and van Pée, K. (1999). Conservation of the pyrrolnitrin biosynthetic gene cluster among six pyrrolnitrin-producing strains. FEMS Microbiol. Lett. 180, 39–44. doi: 10.1111/j.1574-6968.1999.tb08775.x
Han, S. R., Yu, S. C., Ahn, D. H., Park, H., and Oh, T. J. (2016). Complete genome sequence of Burkholderia sp. strain PAMC28687, a potential octopine-utilizing bacterium isolated from Antarctica lichen. J. Biotechnol. 226, 16–17. doi: 10.1016/j.jbiotec.2016.03.043
Hendry, S., Steinke, S., Wittstein, K., Stadler, M., Harmrolfs, K., Adewunmi, Y., et al. (2021). Functional analysis of phenazine biosynthesis genes in Burkholderia spp. Appl. Environ. Microbiol. 87, e02348–e02320. doi: 10.1128/AEM.02348-20
Hickey, G., Monlong, J., Ebler, J., Novak, A. M., Eizenga, J. M., and Gao, Y. (2023). Pangenome graph construction from genome alignments with Minigraph-Cactus. Nat. Biotechnol. doi: 10.1038/s41587-023-01793-w
Hunter, J. D. (2007). Matplotlib: a 2D graphics environment. Comput Sci Eng. 9, 90–95. doi: 10.1109/MCSE.2007.55
Ishida, K., Lincke, T., Behnken, S., and Hertweck, C. (2010). Induced biosynthesis of cryptic polyketide metabolites in a Burkholderia thailandensis quorum sensing mutant. J. Am. Chem. Soc. 132, 13966–13968. doi: 10.1021/ja105003g
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Jenul, C., Sieber, S., Daeppen, C., Mathew, A., Lardi, M., Pessi, G., et al. (2018). Biosynthesis of fragin is controlled by a novel quorum sensing signal. Nat. Commun. 9:1297. doi: 10.1038/s41467-018-03690-2
Jin, Y., Zhou, J., Zhou, J., Hu, M., Zhang, Q., Kong, N., et al. (2020). Genome-based classification of Burkholderia cepacia complex provides new insight into its taxonomic status. Biol. Direct 15:6. doi: 10.1186/s13062-020-0258-5
Kaur, C., Selvakumar, G., and Ganeshamurthy, A. N. (2017). “Burkholderia to Paraburkholderia: the journey of a plant-beneficial-environmental bacterium” in Recent Advances in Applied Microbiology. ed. P. Shukla (Springer Nature Singapore).
Kim, E., Jung, H. I., Park, S. H., Kim, H. Y., and Kim, S. K. (2023). Comprehensive genome analysis of Burkholderia contaminans SK875, a quorum-sensing strain isolated from the swine. AMB Exp 13:30. doi: 10.1186/s13568-023-01537-8
Kim, J., Kim, J. G., Kang, Y., Jang, J. Y., Jog, G. J., Lim, J. Y., et al. (2004). Quorum sensing and the LysR-type transcriptional activator ToxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae. Mol. Microbiol. 54, 921–934. doi: 10.1111/j.1365-2958.2004.04338.x
Kim, N., Mannaa, M., Kim, J., Ra, J. E., Kim, S. M., Lee, C., et al. (2021). The in vitro and in planta interspecies interactions among rice-pathogenic Burkholderia species. Plant Dis. 105, 134–143. doi: 10.1094/PDIS-06-20-1252-RE
Knappe, T. A., Linne, U., Robbel, L., and Marahiel, M. A. (2009). Insights into the biosynthesis and stability of the lasso peptide capistruin. Chem. Biol. 16, 1290–1298. doi: 10.1016/j.chembiol.2009.11.009
Konstantinidis, K. T., and Tiedje, J. M. (2005). Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 102, 2567–2572. doi: 10.1073/pnas.0409727102
Kunakom, S., and Eustáquio, A. S. (2019). Burkholderia as a source of natural products. J. Nat. Prod. 82, 2018–2037. doi: 10.1021/acs.jnatprod.8b01068
Lee, H. H., Park, J., Jung, H., and Seo, Y. S. (2021). Pan-genome analysis reveals host-specific functional divergences in Burkholderia gladioli. Microorganisms. 9:1123. doi: 10.3390/microorganisms9061123
Levy, A., Merritt, A. J., Aravena-Roman, M., Hodge, M. M., and Inglis, T. J. (2008). Expanded range of Burkholderia species in Australia. Am. J. Trop. Med. Hyg. 78, 599–604. doi: 10.4269/ajtmh.2008.78.599
Li, R., Oliver, R. A., and Townsend, C. A. (2017). Identification and characterization of the sulfazecin monobactam biosynthetic gene cluster. Cell Chem Biol. 24, 24–34. doi: 10.1016/j.chembiol.2016.11.010
Lin, Q. H., Lv, Y. Y., Gao, Z. H., and Qiu, L. H. (2020). Pararobbsia silviterrae gen. Nov., sp. nov., isolated from forest soil and reclassification of Burkholderia alpina as Pararobbsia alpina comb. nov. Int. J. Syst. Evol. Microbiol. 70, 1412–1420. doi: 10.1099/ijsem.0.003932
Lipuma, J. J. (2005). Update on the Burkholderia cepacia complex. Curr. Opin. Pulm. Med. 11, 528–533. doi: 10.1097/01.mcp.0000181475.85187.ed
Liu, X., and Cheng, Y. Q. (2014). Genome-guided discovery of diverse natural products from Burkholderia sp. J. Ind. Microbiol. Biotechnol. 41, 275–284. doi: 10.1007/s10295-013-1376-1
Lood, C., Peeters, C., Lamy-Besnier, Q., Wagemans, J., De Vos, D., Proesmans, M., et al. (2021). Genomics of an endemic cystic fibrosis Burkholderia multivorans strain reveals low within-patient evolution but high between-patient diversity. PLoS Pathog. 17:e1009418. doi: 10.1371/journal.ppat.1009418
Lü, L., Zhang, Z. K., and Zhou, T. (2010). Zipf's law leads to Heaps' law: analyzing their relation in finite-size systems. PloS One 5:e14139. doi: 10.1371/journal.pone.0014139
Luo, J., Xie, G., Li, B., and Lihui, X. (2007). First report of Burkholderia glumae isolated from symptomless rice seeds in China. Plant Dis. 91:1363. doi: 10.1094/PDIS-91-10-1363B
Mahenthiralingam, E., Song, L., Sass, A., White, J., Wilmot, C., Marchbank, A., et al. (2011). Enacyloxins are products of an unusual hybrid modular polyketide synthase encoded by a cryptic Burkholderia ambifaria genomic island. Chem. Biol. 18, 665–677. doi: 10.1016/j.chembiol.2011.01.020
Medina-Pascual, M. J., Valdezate, S., Carrasco, G., Villalón, P., Garrido, N., and Saéz-Nieto, J. A. (2015). Increase in isolation of Burkholderia contaminans from Spanish patients with cystic fibrosis. Clin. Microbiol. Infect. 21, 150–156. doi: 10.1016/j.cmi.2014.07.014
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Mistry, J., Finn, R. D., Eddy, S. R., Bateman, A., and Punta, M. (2013). Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41:e121. doi: 10.1093/nar/gkt263
Morales-Ruíz, L. M., Rodríguez-Cisneros, M., Kerber-Díaz, J. C., Rojas-Rojas, F. U., Ibarra, J. A., Santos, E.-d. L., et al. (2022). Burkholderia orbicola sp. nov., a novel species within the Burkholderia cepacia complex. Arch. Microbiol. 204:178. doi: 10.1007/s00203-022-02778-0
Mullins, A. J., and Mahenthiralingam, E. (2021). The hidden genomic diversity, specialized metabolite capacity, and revised taxonomy of Burkholderia sensu lato. Front. Microbiol. 12:726847. doi: 10.3389/fmicb.2021.726847
Mullins, A. J., Murray, J. A. H., Bull, M. J., Jenner, M., Jones, C., Webster, G., et al. (2019). Genome mining identifies cepacin as a plant-protective metabolite of the biopesticidal bacterium Burkholderia ambifaria. Nat. Microbiol. 4, 996–1005. doi: 10.1038/s41564-019-0383-z
Navarro-Muñoz, J. C., Selem-Mojica, N., Mullowney, M. W., Kautsar, S. A., Tryon, J. H., Parkinson, E. I., et al. (2020). A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 16, 60–68. doi: 10.1038/s41589-019-0400-9
Niehs, S. P., Kumpfmüller, J., Dose, B., Little, R. F., Ishida, K., Flórez, L. V., et al. (2020). Insect-associated bacteria assemble the antifungal butenolide gladiofungin by non-canonical polyketide chain termination. Angew. Chem. Int. Ed. Engl. 59, 23122–23126. doi: 10.1002/anie.202005711
Park, J. D., Moon, K., Miller, C., Rose, J., Xu, F., Ebmeier, C. C., et al. (2020). Thailandenes, cryptic polyene natural products isolated from Burkholderia thailandensis using phenotype-guided transposon mutagenesis. ACS Chem. Biol. 15, 1195–1203. doi: 10.1021/acschembio.9b00883
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Parte, A. C., Sardà Carbasse, J., Meier-Kolthoff, J. P., Reimer, L. C., and Göker, M. (2020). List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612. doi: 10.1099/ijsem.0.004332
Peddayelachagiri, B. V., Paul, S., Nagaraj, S., Gogoi, M., Sripathy, M. H., and Batra, H. V. (2016). Prevalence and identification of Burkholderia pseudomallei and near-neighbor species in the Malabar coastal region of India. PLoS Negl. Trop. Dis. 10:e0004956. doi: 10.1371/journal.pntd.0004956
Petrova, Y. D., and Mahenthiralingam, E. (2022). Discovery, mode of action and secretion of Burkholderia sensu lato key antimicrobial specialised metabolites. Cell Surf. 8:100081. doi: 10.1016/j.tcsw.2022.100081
Price, E. P., Sarovich, D. S., Webb, J. R., Ginther, J. L., Mayo, M., Cook, J. M., et al. (2013). Accurate and rapid identification of the Burkholderia pseudomallei near-neighbour, Burkholderia ubonensis, using real-time PCR. PloS One 8:e71647. doi: 10.1371/journal.pone.0071647
Prothiwa, M., Filz, V., Oehler, S., and Böttcher, T. (2021). Inhibiting quinolone biosynthesis of Burkholderia. Chem. Sci. 12, 6908–6912. doi: 10.1039/d0sc06167k
Quadri, L. E., Keating, T. A., Patel, H. M., and Walsh, C. T. (1999). Assembly of the Pseudomonas aeruginosa nonribosomal peptide siderophore pyochelin: in vitro reconstitution of aryl-4,2-bisthiazoline synthetase activity from PchD, PchE, and PchF. Biochemistry 38, 14941–14954. doi: 10.1021/bi991787c
Radua, S., Ling, O. W., Srimontree, S., Lulitanond, A., Hin, W. F., Yuherman, L., et al. (2000). Characterization of Burkholderia pseudomallei isolated in Thailand and Malaysia. Diagn. Microbiol. Infect. Dis. 38, 141–145. doi: 10.1016/s0732-8893(00)00189-9
Rodríguez-Cisneros, M., Morales-Ruíz, L. M., Salazar-Gómez, A., Rojas-Rojas, F. U., and Santos, E.-d. L. (2023). Compilation of the antimicrobial compounds produced by Burkholderia sensu stricto. Molecules 28:1646. doi: 10.3390/molecules28041646
Ryall, B., Lee, X., Zlosnik, J. E., Hoshino, S., and Williams, H. D. (2008). Bacteria of the Burkholderia cepacia complex are cyanogenic under biofilm and colonial growth conditions. BMC Microbiol. 8:108. doi: 10.1186/1471-2180-8-108
Sandy, M., and Butler, A. (2009). Microbial iron acquisition: marine and terrestrial siderophores. Chem. Rev. 109, 4580–4595. doi: 10.1021/cr9002787
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Seyedsayamdost, M. R., Chandler, J. R., Blodgett, J. A., Lima, P. S., Duerkop, B. A., Oinuma, K., et al. (2010). Quorum-sensing-regulated bactobolin production by Burkholderia thailandensis E264. Org. Lett. 12, 716–719. doi: 10.1021/ol902751x
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Song, L., Jenner, M., Masschelein, J., Jones, C., Bull, M. J., Harris, S. R., et al. (2017). Discovery and biosynthesis of gladiolin: a Burkholderia gladioli antibiotic with promising activity against Mycobacterium tuberculosis. J. Am. Chem. Soc. 139, 7974–7981. doi: 10.1021/jacs.7b03382
Sousa, S. A., Ramos, C. G., and Leitão, J. H. (2011). Burkholderia cepacia complex: emerging multihost pathogens equipped with a wide range of virulence factors and determinants. Int J Microbiol. 2011:607575. doi: 10.1155/2011/607575
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Sulochana, M. B., Jayachandra, S. Y., Kumar, S. A., Parameshwar, A. B., Reddy, K. M., and Dayanand, A. (2014). Siderophore as a potential plant growth-promoting agent produced by Pseudomonas aeruginosa JAS-25. Appl. Biochem. Biotechnol. 174, 297–308. doi: 10.1007/s12010-014-1039-3
Terlouw, B. R., Blin, K., Navarro-Muñoz, J. C., Avalon, N. E., Chevrette, M. G., Egbert, S., et al. (2023). MIBiG 3.0: a community-driven effort to annotate experimentally validated biosynthetic gene clusters. Nucleic Acids Res. 51, D603–D610. doi: 10.1093/nar/gkac1049
Tettelin, H., Riley, D., Cattuto, C., and Medini, D. (2008). Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 11, 472–477. doi: 10.1016/j.mib.2008.09.006
Trottmann, F., Franke, J., Ishida, K., García-Altares, M., and Hertweck, C. (2019). A pair of bacterial siderophores releases and traps an intercellular signal molecule: an unusual case of natural nitrone bioconjugation. Angew. Chem. Int. Ed. Engl. 58, 200–204. doi: 10.1002/anie.201811131
Tuanyok, A., Mayo, M., Scholz, H., Hall, C. M., Allender, C. J., Kaestli, M., et al. (2017). Burkholderia humptydooensis sp. nov., a new species related to Burkholderia thailandensis and the fifth member of the Burkholderia pseudomallei complex. Appl. Environ. Microbiol. 83, e02802–e02816. doi: 10.1128/AEM.02802-16
Ussery, D. W., Kiil, K., Lagesen, K., Sicheritz-Pontén, T., Bohlin, J., and Wassenaar, T. M. (2009). The genus Burkholderia: analysis of 56 genomic sequences. Genome Dyn. 6, 140–157. doi: 10.1159/000235768
Vanlaere, E., Baldwin, A., Gevers, D., Henry, D., De Brandt, E., LiPuma, J. J., et al. (2009). Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int. J. Syst. Evol. Microbiol. 59, 102–111. doi: 10.1099/ijs.0.001123-0
Vanlaere, E., Lipuma, J. J., Baldwin, A., Henry, D., De Brandt, E., Mahenthiralingam, E., et al. (2008). Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int. J. Syst. Evol. Microbiol. 58, 1580–1590. doi: 10.1099/ijs.0.65634-0
Wallner, A., King, E., Ngonkeu, E. L. M., Moulin, L., and Béna, G. (2019). Genomic analyses of Burkholderia cenocepacia reveal multiple species with differential host-adaptation to plants and humans. BMC Genomics 20:803. doi: 10.1186/s12864-019-6186-z
Wang, M., Tachibana, S., Murai, Y., Li, L., Lau, S. Y., Cao, M., et al. (2016). Indole-3-acetic acid produced by Burkholderia heleia acts as a phenylacetic acid antagonist to disrupt tropolone biosynthesis in Burkholderia plantarii. Sci. Rep. 6:22596. doi: 10.1038/srep22596
Wang, X., Zhou, H., Chen, H., Jing, X., Zheng, W., Li, R., et al. (2018). Discovery of recombinases enables genome mining of cryptic biosynthetic gene clusters in Burkholderiales species. Proc. Natl. Acad. Sci. U. S. A. 115, E4255–E4263. doi: 10.1073/pnas.1720941115
Yabuuchi, E., Kosako, Y., Oyaizu, H., Yano, I., Hotta, H., Hashimoto, Y., et al. (1992). Proposal of Burkholderia gen. Nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 36, 1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x
Yu, Y., Kim, H. S., Chua, H. H., Lin, C. H., Sim, S. H., Lin, D., et al. (2006). Genomic patterns of pathogen evolution revealed by comparison of Burkholderia pseudomallei, the causative agent of melioidosis, to avirulent Burkholderia thailandensis. BMC Microbiol. 6:46. doi: 10.1186/1471-2180-6-46
Keywords: Burkholderia genus, comparative genomics, biosynthetic gene cluster, pattern analysis, network analysis
Citation: Kim B, Han S-R, Lee H and Oh T-J (2024) Insights into group-specific pattern of secondary metabolite gene cluster in Burkholderia genus. Front. Microbiol. 14:1302236. doi: 10.3389/fmicb.2023.1302236
Edited by:
Ragini Bodade, Savitribai Phule Pune University, IndiaReviewed by:
Julian Ferreras, CONICET Institute of Subtropical Biology (IBS), ArgentinaRohit Sharma, IES University, India
Copyright © 2024 Kim, Han, Lee and Oh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tae-Jin Oh, dGpvaDM3ODJAc3VubW9vbi5hYy5rcg==
 Byeollee Kim
Byeollee Kim So-Ra Han
So-Ra Han Hyun Lee2,3
Hyun Lee2,3 Tae-Jin Oh
Tae-Jin Oh