
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 November 2023
Sec. Microbe and Virus Interactions with Plants
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1302167
This article is part of the Research Topic Microbiome Assembly and Functions in Non-Soil-Borne Plant Diseases View all 8 articles
Introduction: Ecological underpinnings of the invasion success of exotic plants may be found in their interactions with microbes, either through the enemy release hypothesis and the enhanced mutualism hypothesis. Whereas recent high-throughput sequencing techniques have significantly expanded our understanding of plant-associated microbiomes and their functional guilds, few studies to date have used these techniques to compare the microbiome associated with invasive plants between their native and exotic ranges.
Methods: We extracted fungal and bacterial DNA within leaf endosphere, root endosphere and soil of an invasive plant, Ardisia crenata, sampled from their native range Japan and exotic range Florida, USA. Using Illumina sequencing data, we compared microbial community compositions and diversity between the native and exotic ranges, and tested whether abundance of pathogenic or mutualistic microbes differ between the native or exotic ranges in accordance to the enemy release hypothesis or the enhanced mutualism hypothesis.
Results: Fungal and bacterial community compositions differed among leaves, roots and soil, and between the native and exotic ranges. Despite a higher microbial diversity in the soil in the exotic range than in the native range, the microbial diversity within leaf and root was lower in the exotic range compared to the native range. In addition, leaves in the native range harbored a greater number of plant pathogenic fungi compared to those in the exotic range.
Discussion: These patterns suggest plant controls over what microbes become associated with leaves and roots. The higher abundance of leaf pathogenic fungi, including the pathogen which is known to cause specific disease in A. crenata in the exotic range than in the native range, support the enemy release hypothesis and highlighted potential importance of examining microbial communities both above- and below-ground.
Plant-microbe interactions are increasingly recognized to play a role for the reason why some exotic plants become invasive (Dawson and Schrama, 2016; Dickie et al., 2017; Egidi and Franks, 2018; Pearson et al., 2018). Since Elton (1958) published the concept of the “enemy release hypothesis,” many consider that the presence/absence of host-specific natural enemies is key to explain the increased invasiveness of organisms (Keane and Crawley, 2002; Catford et al., 2009; Jeschke et al., 2012; Heger and Jeschke, 2014). According to the enemy release hypothesis applied to plant-microbe interactions, a plant population may be kept at check by host-specific natural enemies in its original range, but the lack of these microbes in the exotic range endows a competitive advantage to non-native plant species (Adams et al., 2009; Chiuffo et al., 2015; Aldorfová et al., 2020; Huang et al., 2020; Kuźniar et al., 2020). Not only pathogenic microbes, but also differences in the geographical distribution of mutualistic microbes may explain invasion success by plants, for example, via novel associations with mutualistic microbes that enhance plant defense, growth and stress tolerance (“enhanced mutualism hypothesis”; Callaway and Ridenour, 2004; Traveset and Richardson, 2014; Aslani et al., 2019).
Despite the relevance of the enemy release and enhanced mutualism hypotheses in plant-microbe interactions between native and exotic ranges, only a few studies have compared the biogeography of microbial communities associated with invasive species (Harrison and Griffin, 2020). In such comparative studies, two aspects need to be considered. Firstly, in a given region, a plant species interact with many different microbes that interact with each other. Secondly, what microbes become associated with a given plant species in a given place depends not only on the microbial community composition in the environment, but also on specific parts where microbes may harbor (e.g., leaves, roots, and soil). If done properly, a comparison of how a given invasive plant species interacts with microbes between its native and exotic ranges in leaves, roots, and soil should shed light on the basic ecology of plant-microbial interactions, beyond the search for the key ecological process that explains its invasiveness.
To date, many studies on interactions between invasive plants and microbes have focused on soil microbiomes (Chiuffo et al., 2015; Aldorfová et al., 2020). Certain mutualistic microbes in the roots and rhizosphere are known to enhance stress tolerance to salt, heat, or resistance to plant pathogens (Rodriguez et al., 2009; Jogaiah et al., 2013), subsequently leading to successful plants invasion (Kamutando et al., 2017). Soil microbial communities vary geographically (Castellanos et al., 2009), and so do root endophytes (Brigham et al., 2023). Yet, few have investigated geographical differences in root-associated microbial communities of invasive plant species between its native vs. exotic ranges. Plants can recruit beneficial microbes to the rhizosphere with root exudates (Upadhyay et al., 2022), and control which microbes in the rhizosphere can penetrate the roots through immune responses and/or biofilm formation (Vieira et al., 2020). It is postulated that such selectiveness exerted by plants involves species-specific genetic factors (Bulgarelli et al., 2013; Edwards et al., 2015; Yu and Hochholdinger, 2018). Hence, if an exotic plant species has a strong coevolved relationship with certain soil microbes in its native range, a reduction in root-associated microbial diversity might be observed in the exotic range due to a scarcity of microbes that can enter roots. Additionally, a local monodominance of an invasive plant could lead to a decrease of local plant diversity, resulting in a reduction of microbial species richness in the rhizosphere (Lu-Irving et al., 2019).
Similarly to root-associated microbes, certain leaf endophytes are shown to benefit the host plant performance by protecting against pathogens and increasing resistance to insect herbivory (Arnold et al., 2003; Tanaka et al., 2005). Yet, leaf endophytes receive much less attention than soil microbes do in invasive plant research. Hence, understanding how differences in microbial composition and diversity between native and exotic ranges influence plant invasiveness remains largely unexplored (but see Lu-Irving et al., 2019; Pan et al., 2023). Experimental evidence from field and manipulative studies suggest that the leaf microbial composition is influenced more by the surrounding environmental microbial pool than by plant genetic factors (Whitaker et al., 2018; Pan et al., 2023). Therefore, deciphering how leaf microbial communities associated with leaves and roots of an invasive plant species differ between its native and exotic ranges will help fill a research gap relevant for both the enemy release hypothesis and the enhanced mutualism hypothesis.
In recent years, amplicon sequencing technologies allow high-throughput analyses of microbial taxa/species belonging to diverse functional guilds such as pathogens and mutualists (Nguyen et al., 2016). This approach has certain advantages over the more traditional approach of comparing differences in plant growth between sterilized and non-sterilized soils (Dawson and Schrama, 2016). Whereas sterilization experiments can provide useful hints as to the importance of microbial communities in the soil and leaf litter (which is the major spore source of leaf endophytic fungi), the effect detected is a net effect of negative and positive effects from pathogens and mutualists (e.g., Pizano et al., 2019). Comparisons of microbial functional guild compositions between their native and exotic ranges can be informative as to how the abundance of pathogens and mutualists differ between the geographic ranges.
In this study, we chose Ardisia crenata, a shade tolerant shrub native to East Asia that acts as an aggressive invader in North America (Dozier, 2000; Kitajima et al., 2006). It can form a mutualistic association with arbuscular mycorrhizal (AM) fungi found in the mesic forests that it invades in North Central Florida, USA (Bray et al., 2003). Although A. crenata occurs at low densities in its native range in Japan (no more than a few adults within 5 m of each other), it forms a dense monodominant understory in Florida (Supplementary Table S1), reducing the diversity of native plant species. The lack of noticeable herbivores or seed predators in both ranges (Kitajima et al., 2006) suggests that differences in plant-microbe interactions between native and exotic populations may be an important factor underpinning its invasion success. A. crenata is widely cultivated in Japan, but cultivation at high density often results in an onset of heavy mortality within Japan. These pieces of background information make A. crenata to be an ideal candidate for studying microbiome differences between the native range (Japan) and exotic range (Florida). We described and compared the diversity and structure of fungal and bacterial communities in leaves, roots, and soil with high-throughput Illumina sequencing. Furthermore, we assigned microbes to functional guilds (pathogens, mutualists) and compared taxonomic composition within each microbial functional guild. We hypothesized that (1) microbial community structure would differ by plant parts and geographical ranges, (2) microbial α diversity would be lower in the exotic range than in the native range, because of the monodominance of A. crenata in the former, (3) the observed differences in functional taxa between native and exotic ranges would align with either the enemy release hypothesis or the enhanced mutualism hypothesis. The results would be corroborative of the enemy release hypothesis if putative pathogens are more prominent in the native range than in the exotic range. Conversely, the enhanced mutualism hypothesis is supported if we detect positive patterns supportive of the possibility that A. crenata in the exotic range has formed novel associations with beneficial microbes.
We set three and four sampling sites each in the native range (Honshu, Japan) and exotic range (Florida, United States), respectively (Figure 1). Sampling sites in the native range were Kamigamo Experimental Station (KA) of Kyoto University, Tokuyama Experimental Station (TO) of Kyoto University, and Yanagido Experimental Station (YA) of Gifu University, all of which have a warm temperate climate (mean annual temperature of 15.7–17.7°C), mean annual precipitation of 1522.9–2625.5 mm (from observation from1991 to 2020, Japanese meteorological agency). All four sites in the exotic range were in Alachua County, Florida (mean annual temperature of 20.7°C, mean annual precipitation of 1227.1 mm) (1991–2020, Florida Climate Center). They were Bivens Arm Nature Park (BA), Evergreen Cemetery (EC), Hawthorne Trail (HT), and Newnan’s Lake (NL). See Supplementary Table S1 for geographical coordinates and vegetation characteristics of these sites. Kitajima et al. (2006) has reported genetic differences between the invasive population in Florida and wild populations of A. crenata in Kyushu and Okinawa. In contrast, the wild populations of A. crenata in Honshu sampled in the current study are genetically close to the invading populations in Florida both in terms of morphological traits and DNA sequences analyzed with ddRAD-seq (Wataru Noyori, unpublished data). Hence, differences in microbial communities associated with A. crenata individuals in the current study are unlikely to reflect differences in plant genotype, but likely due to geographical differences in the background microbial communities and/or differences in local density of A. crenata individuals (low vs. high density in Japan vs. Florida).
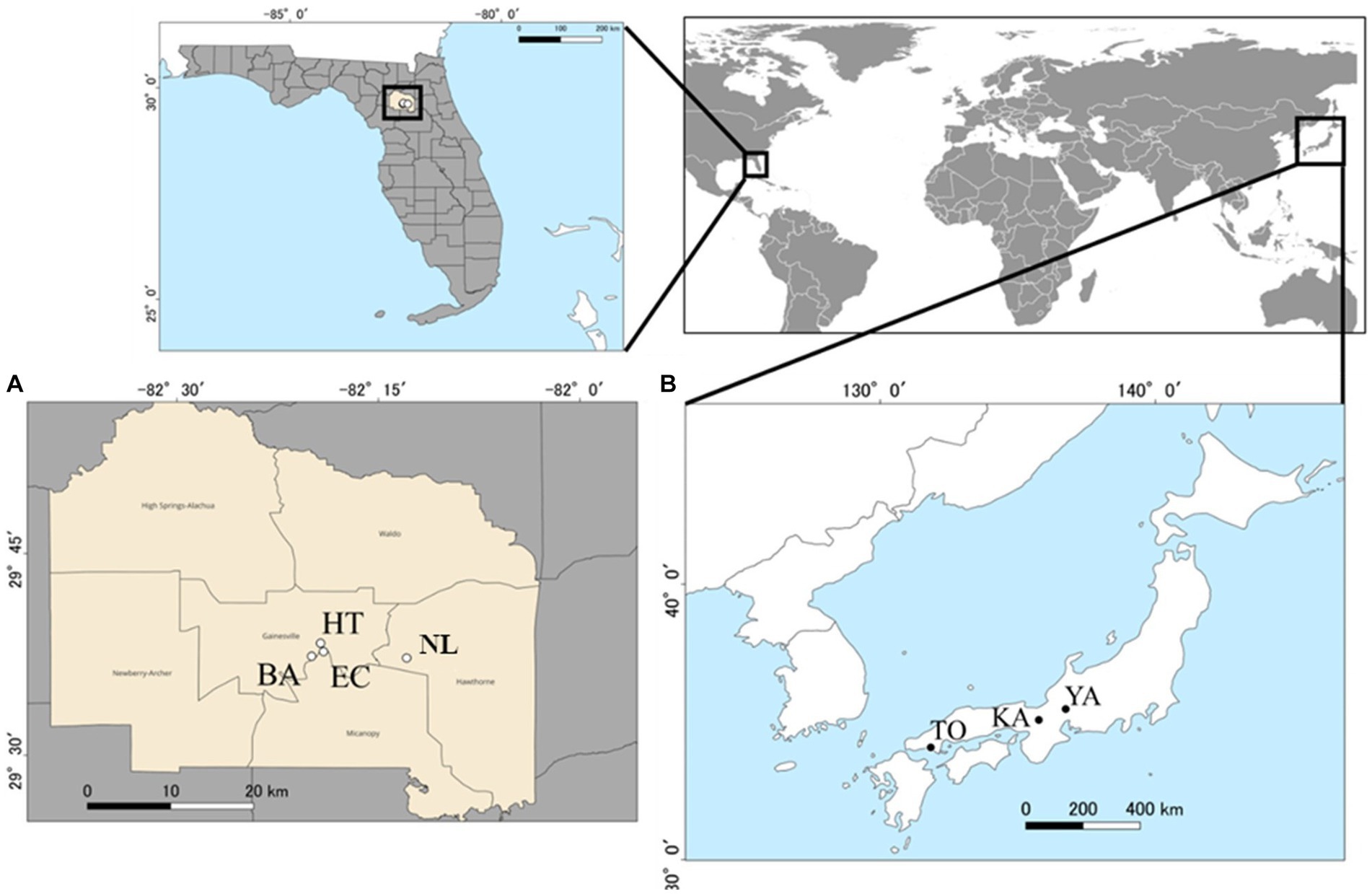
Figure 1. Sampling sites (circles) for this study within the exotic range (4 locations in Alachua County, Florida) [(A) open circles], and the native range (three locations in Japan) [(B) closed circles]. See Supplementary Table S1 for the full names of locations abbreviated in two letter codes. County layers map (TIGER/Line Shapefile, 2016, state, Florida, Current County Subdivision State-based) and city locations (TIGER/Line Shapefile, Current, State, Florida, Places) were accessed from the United States Census Bureau, https://catalog.data.gov/dataset.
At each sampling site, two transects, 1 m wide and 15 m long each, were set to encompass the highest local density of A. crenata. The two transects were separated by a minimum of 10 m from each other. Each transect was divided into 1 m × 1 m quadrats, from which we collected a healthy individual of A. crenata with height less than 15 cm (a total of 30 per site). From each plant sampled, we collected three leaves without disease, five fine roots as well as ca.10 g soil from its vicinity (within 5 cm). The total number of plants sampled was 210 (30 individuals × 7 sites). In the field, samples were individually sealed in plastic bags, and kept immediately in a cooler box with ice packs until further processing in the laboratory. In the lab, leaf and root samples were surface sterilized for 1 min in water with an ultrasonic cleaner, followed by sequentially soaking in 70% ethanol for 1 min, 0.5% NaClO for 1 min, and sterile water for 1 min. Afterward, all samples were sealed in plastic bags with silica gel to dry and store at the −20°C freezer until DNA extraction.
From each leaf sample, we cut a 1-cm2 piece to include leaf edge with a pair of sterile scissors. For root samples, three 2 cm long pieces of fine root were cut with a pair of sterile scissors. These samples were pulverized with Qiagen Tissue Lyser II (at 25/s, for 2 min, Qiagen) with two 4 mm zirconium beads inside 1 mL lysis buffer (20 mmol/L Tris, pH 8.0, 2.5 mmol/L EDTA, 0.4 mol/L NaCl, 0.05% SDS). For extraction of DNA from soil, 0.25 mL of each sample was placed in a 2 mL microcentrifuge tube containing 900 μL lysis buffer and 100 μL skim milk, and 0.25 mL of 0.5 mm zirconium beads, then pulverized with Qiagen Tissue Lyser II (at 25/s, for 2 min). After centrifugation at 4,400 rpm for 5 min, we collected the supernatant containing extracted DNA for PCR intended for amplifying the 16S region for procaryotes. Because this supernatant could not yield successful PCR for fungal ITS, we also used the phenol-chloroform extraction method (Wilson, 2001) for PCR intended for the ITS region of fungi.
The prokaryotic 16S rRNA and fungal internal transcribed space 1 (ITS1) regions were PCR-amplified following the protocol detailed elsewhere (Toju et al., 2019) with some modifications. Briefly, for the prokaryotic 16S rRNA region, the primer set 515f/806rB (515f, Caporaso et al., 2011; 806rB, Apprill et al., 2015) was fused with the Illumina sequencing primer region and 3-6-mer Ns for improving sequencing quality (Lundberg et al., 2013). Likewise, the fungal ITS1 region was amplified using the primer set ITS1-F_KYO1/ITS2_KYO2 (Toju et al., 2012) fused with the Illumina sequencing primer region and 3-6-mer Ns. We conducted PCR with the DNA polymerase system of Ampdirect Plus (Shimadzu, Kyoto, Japan) with a temperature profile of 35 cycles consisting of denaturation at 98°C for 10 s, annealing at 55°C for 60 s, extension at 72°C for 60 s, and a final extension at 72°C for 7 min, for both primers. Fusion primers with P5/P7 Illumina adapters and 8-mer index sequences for sample identification were added to the PCR products. In the reaction, the DNA polymerase system of KOD One (Toyobo) was used with a temperature profile of 8 cycles at 98°C for 10 s, 55°C for 5 s, 68°C for 30 s, and a final extension at 68°C for 2 min. The amplified PCR fragments were purified and equalized using AMpure XP Kit (Beckman Coulter), and equal volumes of all specimens were pooled together. The pooled library was sequenced with the Illumina MiSeq sequencer with 10% PhiX spike-in (Center for Ecological Research, Kyoto, Japan, 2 × 300 cycles).
The bcl2fastq 1.8.4 program distributed by Illumina was used to convert the raw sequence data into FASTAQ files. The FASTAQ files were demultiplexed with the program Claident v0.2.2018.05.29 (Tanabe and Toju, 2013). Chimeric and low quality-score reads (< 20) were subsequently discarded and the reads that passed the filtering process were clustered using VSEARCH (Rognes et al., 2016) with 97% clustering threshold as implemented in Claident. Then, operational taxonomic units (OTUs) were obtained, resulting in a total read number of 4,040,099 and 3,556,515 from the 16S and ITS1 primers, respectively. Taxonomic assignment of OTUs was performed with a combination of the query-centric auto-k-nearest neighbor (QCauto) method (Tanabe and Toju, 2013) and the lowest common ancestor (LCA) algorithm (Huson et al., 2007) as implemented in Claident. Afterward, unclassified bacterial and fungal OTUs at the kingdom level were subjected to a blastn search, and OTUs matching of plant-derived sequences (chloroplast-derived 16S) were removed. Note that the 16S 515f/806rB primers used in this study also amplified host-derived sequences from leaf and root samples (around 60% of all 16S reads). For 16S reads of leaf samples, about 90% of the post-filter reads clustered into closely related OTUs affiliated with Burkhorderia crenata, a known symbiont in the leaf-edge nodules of A. crenata (Carlier et al., 2016). Many of these OTUs could be identified with nonuniform descriptions like “symbiont bacteria” in the public database. Thus, we attempted a phylogenetic approach to conclude that these OTUs as “Burkholderia sp.” for use in subsequent analysis after confirming the blastn results of those OTUs were within monophyletic group of B. crenata (Supplementary Figure S1). Finally, we excluded all OTUs that could not be assigned to either bacteria or fungi at the kingdom level (i.e., archaea or unidentified taxa), and then proceeded with the following statistical analyses.
To minimize the effect of PCR/sequencing errors, we removed the OTUs that represented less than 0.1% of the total reads in each sample (Peay et al., 2015). This resulted in high quality reads of 982,788 and 3,264,380 for 16S and ITS primers, respectively. The dataset was rarefied at 200 reads and 1,000 reads per sample for bacteria and fungi, respectively, with the “rrarefy” function of the “vegan” package (Oksanen et al., 2022) of R version 4.2.1 (R Core Team, 2022). Samples that yielded less than these read numbers were discarded, leaving bacterial 499 and fungal 469 samples. See Supplementary Table S2 for the sample size for each site. The rarefaction curves for each group reached to an asymptote (except for 16S in the soil), indicating that we had adequate levels of sampling (Supplementary Figure S2). After rarefaction, 4,819 and 3,998 unique OTUs of bacteria and fungi were included in the final analysis. Of these, 4,021 (83.4%) and 3,933 (98.4%) could be classified at the phylum level, and 1,603 (33.3%) and 1,653 (41.3%) were classified at the genus level for 16S and ITS reads, respectively. Further blastn analysis based on the NCBI database was performed on several OTUs to identify them at the species level. Fungal OTUs were assigned their ecological functional guilds including pathogens and mutualists (i.e., plant pathogen, AM fungi) based on their taxonomic assignment, with the FUNGuild (Nguyen et al., 2016). Only OTUs with a confidence rank of “High Probability” or “Probability” were retained in the analysis, but those with “Possible” and those assigned to more than one guild were treated as “Unidentified.” This resulted in 1,213 of 2,785 OTUs assigned guilds (30.5%), with remaining 69.5% being unidentified. Finally, for fungal and bacterial OTUs that account for more than 2% of the total relative abundance at the genus level, a literature survey based on the published literature was conducted on functional taxonomic guilds.
We calculated Shannon diversity of fungal and bacterial communities was calculated as the exponent of Shannon entropy (i.e., ; where Pi is the proportional abundance of species i, Shannon, 1948) calculated from the R package “vegan.” To test differences among plant parts and between the native and exotic ranges, ANOVA was conducted and a post hoc comparison with a nonparametric Kruskal-Wallis rank sum test. To examine how microbial compositions differed by plant parts and geographical ranges, permutational multivariate analysis of variance (PerMANOVAs, 9,999 permutations; Anderson, 2006) was conducted based on Bray-Curtis distance values with the “adonis2” function in “vegan.” We used ‘strata’ in adonis2 to control for site-to-site variation, as a random effect included in the PERMANOVA to restrict permutations solely within each country for range comparison. With Non-metric multidimensional scaling (NMDS) based on Bray–Curtis dissimilarity was also conducted within “vegan” and “ggplot2” (Wickham, 2016). In addition, Permutational analysis for multivariate homogeneity of dispersions was conducted for bacterial and fungal communities (PERMDISP, 9,999 permutations; Anderson, 2006). We also used sites as strata when as mentioned above. To compare the abundance of putative pathogens and mutualists between the two ranges, Welch’s t-test was conducted for the relative abundance of OTUs belonging to each FUNGuild. To find microbial genera biased to either the native or exotic range, we performed paired comparisons of genera that constituted more than 2% of the total relative abundance with Mann–Whitney U tests using the R package “exactRankTests” (Hothorn and Hornik, 2022).
The total numbers of fungal OTUs derived from leaves, roots and soil were 992, 1,244, and 2,415, respectively. The number of fungal OTUs detected only in the native range was 1,459 (36.5%), while 1,684 (42.1%) OTUs were specific to the exotic range, with 885 (21.4%) OTUs shared between the two regions. The leaf endophytic fungi were dominated by Ascomycota (90%; an average of the two ranges combined; Supplementary Figure S3A), whereas Basidiomycota accounted for a much smaller proportion (10%). The abundance of Ascomycota in the leaf was higher in the exotic range than in the native range (95 and 75%, respectively). Ascomycota was also dominant in the root and soil samples (57.4 and 38.3%, respectively), followed by Basidiomycetes (24 and 35%, respectively), and Mucoromycota (18 and 22%, respectively).
Shannon diversity of fungal OTUs showed a significant interaction effect between 2 factors: plant parts and geographical range (ANOVA, p < 0.001, Supplementary Table S3). Additionally, there were significant differences among plant parts (ANOVA, p < 0.001), and small overall differences between the two geographical ranges (ANOVA, p = 0.685). Shannon diversity of fungi within leaves and roots were higher in the native range than in the exotic range, (the top two panels of Figure 2A), but it was higher in the exotic range than in the native range for soil (the bottom panel of Figure 2A).
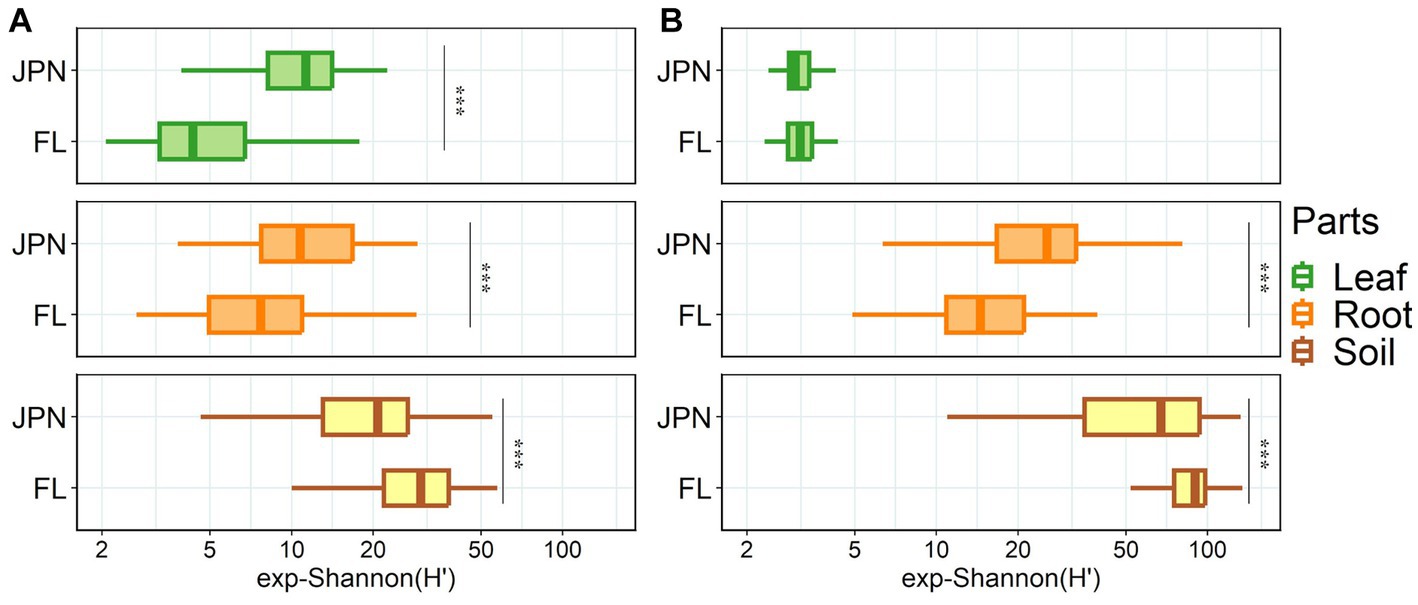
Figure 2. Shannon diversity of fungal (A) and bacterial (B) communities in Japan (JPN) and Florida (FL) sampled from leaves (top), roots (middle), and soil (bottom). Significant difference between locations as indicated by asterisks, determined by the non-parametric Kruskal-Wallis rank sum test (*p < 0.05, **p < 0.01, ***p < 0.001). See Supplementary Table S2 for each sample size.
The total numbers of bacterial OTUs derived from leaf, root and soil samples were 490, 1,575 and 3,687, respectively. Among bacterial OTUs, 1,571 OTUs (32.6%) were detected only from the native range, whereas 1,300 OTUs (27.0%) were detected only from the exotic range, with 1,948 OTUs (40.4%) shared between the two regions. As to bacterial taxonomic composition, Proteobacteria was dominant in the leaf (Supplementary Figure S3B). Burkholderia crenata, an obligate symbiont taxon detected from Ardisia crenata previously (Carlier et al., 2016), was the most abundant Proteobacteria. In the root and soil, Proteobacteria and Actinobacteria dominated and accounted for more than half of the total OTUs.
Shannon diversity of bacterial OTUs had significant interaction effect between 2 factors: plant parts and geographical range (ANOVA, p < 0.001, Supplementary Table S3). Shannon diversity varied between the two geographical ranges (ANOVA, p = 0.002), and varied among the three plant parts (ANOVA, p < 0.001). Shannon diversity of the root-associated bacteria was higher in the native range than in the exotic range (middle panel, Figure 2B). In contrast, Shannon diversity in the -soil in the exotic range was higher than in the native range (bottom panel, Figure 2B).
Non-metric multidimensional scaling (NMDS) plots showed strong differentiation of fungal and bacterial community structures among leaves, roots and soil, as well as between native and exotic regions (Figures 3A,B). The results of the PerMANOVA showed that plant parts and ranges had significant effects with strong interactions (Supplementary Table S4). Leaf endophytic fungi showed stronger differences between the native and exotic ranges than fungal communities in roots and soil (Figure 3A; Supplementary Table S5). Interactive effects between plant parts and geographical regions were also significant for bacterial communities (PerMANOVA, Supplementary Table S4). For bacteria, however, the regional difference was less pronounced in leaves compared to roots and soil (Supplementary Table S5), as visualized by the NMDS plot (Figure 3B). PERMADISP, which test whether the heterogeneity of OUT compositions differed between regions, showed significant differences (Supplementary Table S5). Greater community heterogeneity in the native range than in the exotic range was significant for fungal communities associated with leaves and roots (but not with soil), and for bacterial communities associated with rotos and soils (but not with leaves).
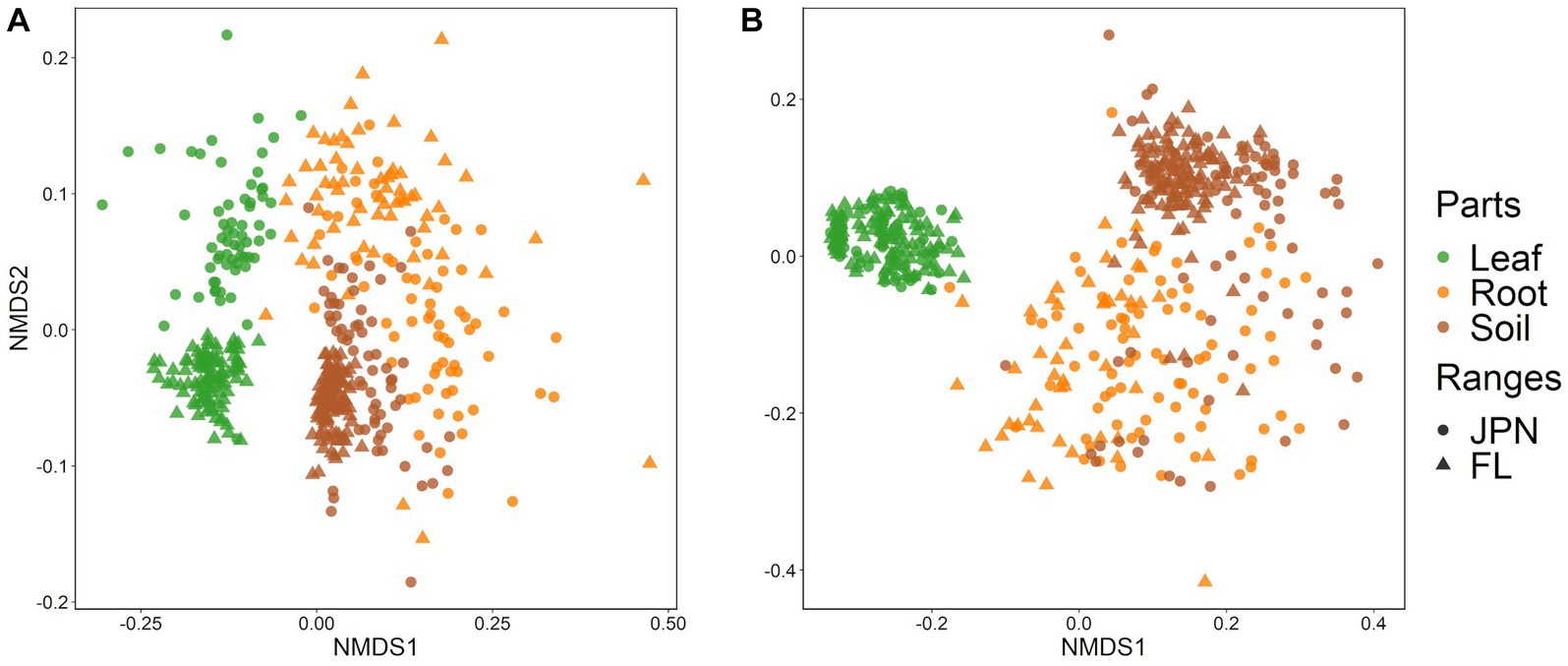
Figure 3. Nonmetric multidimensional scaling (NMDS) plots based on Bray–Curtis distances of (A) fungal and (B) bacterial communities sampled from leaves (green), roots (orange), and soil (brown). Each point represents individual of A. crenata sampled from Japan (〇) and Florida (△). Small numbers of obvious outliers were excluded from the figure (4 root samples of the fungal community, and 1 leaf sample and 4 root samples of the bacterial community). See Supplementary Table S4 for the results of PerMANOVA test on difference among parts and geographical ranges.
Leaf endophytic fungi showed similar order-level diversity between the native and exotic ranges, with noticeable abundance of Capnodiales in the exotic range (Supplementary Figure S5A). At the genus level, Pallidocercospora dominated in the exotic range (62.5%, Figure 4C; Table 1), but Pallidocercospora was also common in the native range along with Pestalotiopsis, Phyllosticta, and Carlosrosaea. Order-level diversity of root-associated fungi was similar between the native and exotic ranges, with the same five orders dominant (Figure 4A). At the genus level, Melanconiella and Russula, and Glomus were the most common in the native range, whereas Russula, Glomus, and Mortierella were the most common in the exotic range (Figure 4C). Within the soil, Mortierellales was the most common in the native range, whereas its abundance was similar to those of Hypocreales (Figure 4A). In the exotic range, Russulales was also common. This reflects the abundance of Russula, Saitozyma, and Metarhizium in the exotic range, which was more pronounced than their abundance in the native range (Figure 4C). Order-level and genus-level fungal community compositions for each site were shown in Supplementary Figures S4A,C.

Figure 4. Taxonomic compositions of fungal OTUs (relative abundance) at the order (A) and genus (C) levels (left), and bacterial OTUs at the order (B) and genus (D) levels (right). The top 20 taxa are indicated, and all remaining taxa are consolidated into the ‘Others’ category.

Table 1. List of fungal and bacterial genera exhibiting a prevalence of 2% or more in either native (Japan) or exotic range (Florida).
Order-level and genus-level composition of leaf endophytic bacteria were similar between native and exotic ranges, with a dominance of Burkholderia crenata, a symbiont in the leaf-edge nodules of A. crenata (Figures 4B,D; Table 1). At the order level, the composition of root-associated bacteria was dominated by Burkholderiales and Rhizobiales in both the native and exotic ranges. However, Mycoplasmatales were more common in the native range, while Streptosporangiales were more common in the exotic range (Figure 4B). At the genus level, Candidatus.Moeniiplasma, Burkholderia, and Bradyrhizoblium were most commonly found in the native range, while Burkholderia, Mycobacterium, and Halomonas dominated in the exotic range (Figure 4D). Within the soil, the order-level composition was similar between the native and exotic ranges, with Rhizobiales dominant in both. At the genus level, Rhodoplanes was the most common in both ranges, although its abundance was more pronounced in the exotic range. Order-level and genus-level fungal community compositions for each site were shown in Supplementary Figures S4B,D.
The relative abundance of plant pathogens differed significantly in leaf samples, with much higher relative abundance in the native range than in the exotic range (Figure 5, Welch’s t-test, p < 0.0001). However, there was no significant difference in the relative abundance of plant pathogens in roots (p = 0.349; Figure 5), and the relative abundance of pathogens in soil was similar but significantly higher in the exotic range than in the native range (p < 0.001; Figure 5). Among taxa that constituted more than 2% of the total relative abundance, Pallidocercospora, Pestalotiopsis, and Phyllosticta were assigned to plant pathogens (Table 1). Pallidocercospora was more common in the exotic range than in the native range, while Pestalotiopsis and Phyllosticta were unique in the native range (Figure 4). For Phyllosticta, 80% of the reads were confirmed to be Phyllosticta ardisiicola by the blastn top hit sequence in the NCBI database (Percent identity = 100%; accession number; NR136952.1). P. ardisiicola is reported to be a pathogen discovered from A. crenata, causing leaf spots in A. crenata (Motohashi et al., 2008). A literature review was conducted on microbial taxa with high abundance (> 2% of the total), but no candidate pathogenic microbes were found.
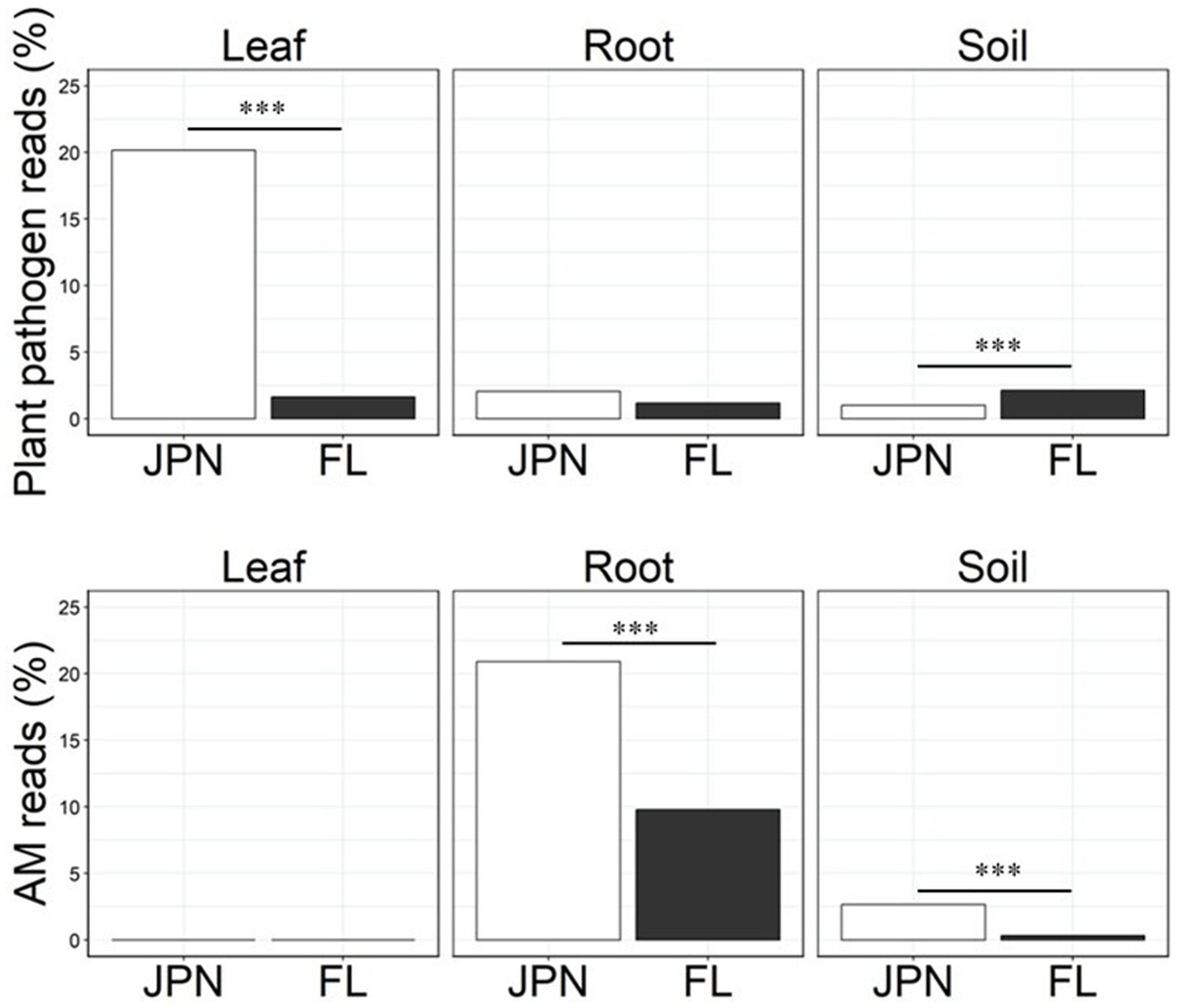
Figure 5. Relative abundance of fungal guilds, i.e., % of sequence reads that could be classified to plant pathogens (top) and arbuscular mycorrhizal (AM) fungi (bottom) within Japan (JPN) and Florida (FL) populations of A. crenata detected from leaf, root and soil samples. The guild classification was based on the FUNGuild at genus or family level. Asterisks indicates the results of Welch’s t-test (*p < 0.05, **p < 0.01, ***p < 0.001).
The relative abundance of AM fungi in roots and soil was significantly higher in the native range than in the exotic range by 114 and 706%, respectively (Welch’s t-test, p < 0.0001 each, Figure 5, bottom). Glomus and Russula were identified as mutualistic microbes among taxa that constituted more than 2% of the total relative abundance (Table 1). Glomus is a major group of arbuscular mycorrhizal fungi (Morton and Benny, 1990), and Russula is considered to be ectomycorrhizal fungi (Smith and Read, 2010). Glomus was more common in the native range than in the exotic range, while the genus Russula was less common in the native range than in the exotic range. Our literature review of microbial taxa with high abundance (> 2% of the total) found no clear candidates that may act as mutualists in leaves, except for Carlosrosaea. This genus was more common in the native range than in the exotic range, and it has been reported as a potentially mutualistic endophyte that improves seedling growth of Bromeliaceae (Marques et al., 2021).
To explore potential differences in microbial communities associated with the invasion success of A. crenata, we compared fungal and bacterial community structures within the leaf, root, and soil between native and exotic ranges. Two trends stood out. Firstly, the microbial diversity inside leaves and roots was lower in the exotic range than that in the native range, despite higher microbial diversity in the soil in the former. Secondly, leaves harbored a higher number of plant pathogenic fungal genera in the native range than in the exotic range, which is corroborative of the enemy release hypothesis. These findings underscore the importance of microbial diversity including potential pathogens in keeping check on the host plant population in the native range.
Previous studies that examined either fungi or bacteria associated with invasive plants reported differences in microbial community between plant parts or population ranges (Gundale et al., 2016; Pickett et al., 2022). Our study that examined both fungi and bacteria is more comprehensive in revealing differences among leaves, roots and soils, as well as between geographical ranges (Figures 3A,B). Lu-Irving et al. (2019) compared bacterial communities of invasive grass between its native and exotic ranges, and reported lower bacterial diversity in the leaf endosphere, root endosphere, and root surface in the exotic range than in the native range, while some research indicates the opposite, or shows no clear trend (Pickett et al., 2022; Pan et al., 2023). These inconsistent results might be dependent on the invasive plant species under consideration.
The composition and diversity of leaf endophytic bacteria were similar between the native and exotic ranges, with strong dominance of Burkholderia crenata, which is known to be an obligate symbiont that is vertically transmitted from mother plants (Ku and Hu, 2014). Perhaps, this bacterium is maintained throughout the process of cultivation and subsequent naturalization of A. crenata in the US. In contrast, the composition of leaf endophytic fungi showed wide differences by geographical ranges compared to leaf endophytic bacteria and fungi in roots and soil (Figures 3A,B; Supplementary Table S5), suggesting that A. crenata interacts with distinct fungal communities in its exotic range as opposed to its native range. We found a lower diversity of leaf endophytic fungi in the exotic range than in the native range, possibly tied to the dominance of a latent plant pathogen, Pallidocercospolla, in the exotic range (Figure 4C). Previous studies have reported cases of accumulation of specific plant pathogens in invasive plants in their exotic ranges (Stricker et al., 2016; Anthony et al., 2017). Pallidocercospolla, which is obviously non-lethal, may have accumulated in the leaf of A. crenata as its population density increased within the exotic range.
Differences in the root fungal and bacterial taxonomic composition were observed between the native and exotic ranges, although these variations were less pronounced than those in the leaves or soil (Supplementary Table S5). This could be attributed to a higher microbial diversity in roots (i.e., wider scatter Figures 3A,B), compared to that in leaf and soil samples. There are two potential factors for explaining such pattern: (1) colonization of microbes into roots could vary among individual plants; (2) microbial colonization of roots varies greatly among fine roots within individuals (Rüger et al., 2021). We cannot distinguish these two possibilities as we sampled only a few fine root samples per plant. Overall, the diversity of root-associated microbes in the exotic range was lower than in the native range, despite more diverse soil microbial pool than in the native range (Figures 2A,B). Considering this, one possibility is that root-associated microbes are selectively accumulated by A. crenata rather than passively recruited from the locally available species pool in the soil. This could be due to the host plants imposing selection or because fewer microbes in exotic ranges can overcome host resistance.
Soil microbial communities differed significantly between native and exotic ranges for both bacteria and fungi, similar to the findings of a previous study comparing geographical ranges of invasive grasses (Lu-Irving et al., 2019). These differences could merely reflect biogeographical differences between Japan and Florida, and/or the possible influence of invasive plants on soil microbial communities (Trognitz et al., 2016; Rodríguez-Caballero et al., 2020). The local monodominance of invasive exotic plants may influence microbial communities along with decreasing species richness of aboveground plant communities (Anthony et al., 2017; Zhang et al., 2020). Contrary to our hypotheses, both fungal and bacterial diversity of soil were higher in the exotic range than in the native range. According to Ramirez et al. (2019), invasive species can produce more root exudates when they are in environments without their natural predators. This increased production of root exudates might contribute to a rise in the diversity of microbes in the soil. These possibilities are worth testing to improve mechanistic understanding of the geographical variances of microbial composition and diversity.
Many studies examining the enemy release hypothesis have focused on belowground pathogens or aboveground herbivores (Adams et al., 2009; Chiuffo et al., 2015; Aldorfová et al., 2020; Huang et al., 2020). Whereas less attention has been given by the research community to leaf endophytic microbes in the context of the enemy release hypothesis, the results of our study suggest the potential importance of leaf endophytic fungi. Indeed, we found a sign of enemy release only in leaf endophytic fungi, but not in bacteria or root-associated fungi (Figure 5; Table 1). Notably, the pathogenic fungi observed in the leaf endosphere of the native population were diverse, including Pestalotiopsis with a wide host range (Maharachchikumbura et al., 2014), and Phyllosticta ardisiicola known to cause specific diseases in A. crenata (Motohashi et al., 2008). These deleterious fungi were found only in the native range (Japan) but absent in the exotic range (Florida). Hence, they deserve further study as potential candidates that may limit the local density of A. crenata in Japan. On the other hand, Pallidocercospora, a known pathogenic genus, was more abundant in leaves sampled in the exotic range (Crous et al., 2013). However, Pallidocercospora is often detected in healthy leaves and hence it has been suggested to be a latent pathogen (Napitupulu et al., 2021). We suspect that it is only weakly deleterious and the high local density of A. crenata in the exotic range may promote its abundance. To explore this possibility, we are currently conducting another study to examine the effect of local population density of A. crenata within the exotic range of Florida. Overall, we detected patterns in support of the enemy release hypothesis with leaf endophytic fungi than with leaf endophytic bacteria, or fungi and bacteria associated found in roots or soils.
We did not observe microbial patterns that are supportive of the enhanced mutualism hypothesis either for fungi, as the relative abundance of arbuscular mycorrhizal (AM) fungi within roots was higher in the native range than in the exotic range (Figure 5). Several other studies on the acquisition of new AM fungi report higher AM fungal colonization rates and greater AM fungi diversity in the exotic range compared to the native range (Yang et al., 2013; Soti et al., 2014; Sheng et al., 2022). A. crenata is reported to be capable of acquiring genotypes of AM fungi that enhance growth in Florida (Bray et al., 2003). Even though many AM fungi are considered generalist, their effectiveness depends on the combination of host and fungal species and genotypes. Our study found greater abundance of AM fungi within the roots of plants from the native range than those from the exotic range (Figure 5; Table 1), but it is not clear whether they are necessarily effective mutualists. Although ectomycorrhizal fungi (EcM) were more common in the exotic range soils (largely attributable to the genus Russula), the relative abundance of OTUs assigned to EcM in the roots was similar between native and exotic ranges (Table 1). A. crenata is not known to form symbiotic relationships with EcM fungi: there are no reports suggesting an interaction between A. crenata and EcM, and we do not detect EcM hyphae inside the roots under microscopes (data not shown). The high relative abundance of EcM in exotic range soils is likely to reflect differences in the overstory vegetation. Whereas ectomycorrhizal pines and oaks were dominant in Florida sites, AM fungi dependent conifers (cedars and cypress) were mixed with oaks in Japan (Supplementary Table S1). For bacterial communities, we did not find any notable differences in abundance of potentially beneficial bacteria between native and exotic ranges, with ubiquitously high abundance of Burkholderia in leaves.
Whereas many recent studies addressed positive and negative feedbacks between plants and soil microbial communities, our results suggest that it is essential to simultaneously examine leaf-associated microbial communities. A vast diversity of microbes were found to interact with A. crenata in both native and exotic ranges, including mutualistic, commensalistic, to pathogenic fungi and bacteria. While functional guilds were estimated from the database, a given microbe may act differently depending on environmental and host conditions. Furthermore, these microbes interact with each other in addition to their direct interaction with their host plant. We did not evaluate the interactions between hosts and microbes, but narrowed down candidates that may cause ecologically significant interactions with A. crenata in its native and exotic ranges. Specifically, the results suggest a potential importance of leaf pathogenic fungi in explaining the local density of A. crenata in Japan vs. Florida. Manipulative experimental study that employs density manipulation and inoculation tests with these putative pathogens within the native range of A. crenata will prove whether these are the key density-dependent agents, the lack of which explains the invasive population growth in the exotic range.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://ddbj.nig.ac.jp/public/ddbj_database/dra/fastq/, DRA017027.
NN: Investigation, Writing - original draft, Conceptualization, Validation. HT: Writing - review & editing, Conceptualization, Validation. KK: Writing - review & editing, Conceptualization, Validation.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Research and publication of this article was supported by the Management Expense Grant of Kyoto University.
We would like to extend our gratitude to F. E. (Jack) Putz for allowing us to use his private property as a sampling site. We also thank Kamigamo Experimental Station (Kyoto University), Tokuyama Experimental Station (Kyoto University) and Yanagido Experimental Station (Gifu University) for allowing us to sample in their fields. Gerardo Celis gave critical assistance in selecting sites in Florida, and Jeremy Lichstein, Luke Flory, Seth Robinson, and Keiko Ohira provided logistic support to the fieldwork. Shogo Kato and Yi-Shen Chen provided logistic support to the fieldwork in Gifu, Japan. Hiroaki Fujita, Sayaka Suzuki, and Yoshie Hori gave technical supports for a bunch of bioinformatics. Kohmei Kadowaki gave statistical advice and helped improve the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1302167/full#supplementary-material
Adams, J. M., Fang, Æ. W., Callaway, Æ. R. M., Cipollini, Æ. D., Newell, E., Acer, Æ. T., et al. (2009). A cross-continental test of the enemy release hypothesis: leaf herbivory on Acer platanoides (L.) is three times lower in North America than in its native Europe. Biol. Invasions 11, 1005–1016. doi: 10.1007/s10530-008-9312-4
Aldorfová, A., Knobová, P., and Münzbergová, Z. (2020). Plant–soil feedback contributes to predicting plant invasiveness of 68 alien plant species differing in invasive status. Oikos 129, 1257–1270. doi: 10.1111/oik.07186
Anderson, M. J. (2006). Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62, 245–253. doi: 10.1111/j.1541-0420.2005.00440.x
Anthony, M. A., Frey, S. D., and Stinson, K. A. (2017). Fungal community homogenization, shift in dominant trophic guild, and appearance of novel taxa with biotic invasion. Ecosphere 8:1951. doi: 10.1002/ecs2.1951
Apprill, A., McNally, S., Parsons, R., and Weber, L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753
Arnold, A. E., Mejia, L. C., Kyllo, D., Rojas, E. I., Maynard, Z., Robbins, N., et al. (2003). Fungal endophytes limit pathogen damage in a tropical tree. Proc. Natl. Acad. Sci. 100, 15649–15654. doi: 10.1073/pnas.2533483100
Aslani, F., Juraimi, A. S., Ahmad-hamdani, M. S., and Alam, M. A. (2019). The role of arbuscular mycorrhizal fungi in plant invasion trajectory. 1–14. Springer: Berlin.
Bray, S. R., Kitajima, K., and Sylvia, D. M. (2003). Mycorrhizae differentially alter growth, physiology, and competitive ability of an invasive shrub. Ecol. Appl. 13, 565–574. doi: 10.1890/1051-0761(2003)013[0565:MDAGPA]2.0.CO;2
Brigham, L. M., Bueno De Mesquita, C. P., Spasojevic, M. J., Farrer, E. C., Porazinska, D. L., Smith, J. G., et al. (2023). Drivers of bacterial and fungal root endophyte communities: understanding the relative influence of host plant, environment, and space. FEMS Microbiol. Ecol. 99, 1–12. doi: 10.1093/femsec/fiad034
Bulgarelli, D., Schlaeppi, K., Spaepen, S., Van Themaat, E. V. L., and Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Callaway, R. M., and Ridenour, W. M. (2004). Novel weapons: invasive success and the evolution of increased competitive ability. Front. Ecol. Environ. 2, 436–443. doi: 10.1890/1540-9295(2004)002[0436:NWISAT]2.0.CO;2
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Carlier, A., Fehr, L., Pinto-Carbó, M., Schäberle, T., Reher, R., Dessein, S., et al. (2016). The genome analysis of Candidatus Burkholderia crenata reveals that secondary metabolism may be a key function of the Ardisia crenata leaf nodule symbiosis. Environ. Microbiol. 18, 2507–2522. doi: 10.1111/1462-2920.13184
Castellanos, T., Dohrmann, A. B., Imfeld, G., Baumgarte, S., and Tebbe, C. C. (2009). Search of environmental descriptors to explain the variability of the bacterial diversity from maize rhizospheres across a regional scale. Eur. J. Soil Biol. 45, 383–393. doi: 10.1016/j.ejsobi.2009.07.006
Catford, J. A., Jansson, R., and Nilsson, C. (2009). Reducing redundancy in invasion ecology by integrating hypotheses into a single theoretical framework. Divers. Distrib. 15, 22–40. doi: 10.1111/j.1472-4642.2008.00521.x
Chiuffo, M. C., MacDougall, A. S., and Hierro, J. L. (2015). Native and non-native ruderals experience similar plant–soil feedbacks and neighbor effects in a system where they coexist. Oecologia 179, 843–852. doi: 10.1007/s00442-015-3399-y
Crous, P. W., Braun, U., Hunter, G. C., Wingfield, M. J., Verkley, G. J. M., Shin, H. D., et al. (2013). Phylogenetic lineages in Pseudocercospora. Stud. Mycol. 75, 37–114. doi: 10.3114/sim0005
Dawson, W., and Schrama, M. (2016). Identifying the role of soil microbes in plant invasions. J. Ecol. 104, 1211–1218. doi: 10.1111/1365-2745.12619
Dickie, I. A., Cooper, J. A., Bufford, J. L., Hulme, P. E., and Bates, S. T. (2017). Loss of functional diversity and network modularity in introduced plant-fungal symbioses. AoB Plants 9:84. doi: 10.1093/aobpla/plw084
Dozier, H. (2000). Plant introductions to invasion: History, public awareness, and the case of Ardisia crenata. University of Florida: Gainesville, FL.
Edwards, J., Johnson, C., Santos-Medellín, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. 112, E911–E920. doi: 10.1073/pnas.1414592112
Egidi, E., and Franks, A. E. (2018). Incorporating fungal community ecology into invasion biology: Challenges and opportunities. Microbiol. Australia 39, 56–60. doi: 10.1071/ma18015
Gundale, M. J., Almeida, J. P., Wallander, H., Wardle, D. A., Kardol, P., Nilsson, M. C., et al. (2016). Differences in endophyte communities of introduced trees depend on the phylogenetic relatedness of the receiving forest. J. Ecol. 104, 1219–1232. doi: 10.1111/1365-2745.12595
Harrison, J. G., and Griffin, E. A. (2020). The diversity and distribution of endophytes across biomes, plant phylogeny and host tissues: how far have we come and where do we go from here? Environ. Microbiol. 22, 2107–2123. doi: 10.1111/1462-2920.14968
Heger, T., and Jeschke, J. M. (2014). The enemy release hypothesis as a hierarchy of hypotheses. Oikos 123, 741–750. doi: 10.1111/j.1600-0706.2013.01263.x
Hothorn, T., and Hornik, K. (2022). exactRankTests: exact distributions for rank and permutation tests. Available at: https://cran.r-project.org/package=exactRankTests (Accessed February 17, 2022).
Huang, K., Kong, D. L., Lu, X. R., Feng, W. W., Liu, M. C., and Feng, Y. L. (2020). Lesser leaf herbivore damage and structural defense and greater nutrient concentrations for invasive alien plants: evidence from 47 pairs of invasive and non-invasive plants. Sci. Total Environ. 723:137829. doi: 10.1016/j.scitotenv.2020.137829
Huson, D. H., Auch, A. F., Qi, J., and Schuster, S. C. (2007). MEGAN analysis of metagenomic data. Genome Res. 17, 377–386. doi: 10.1101/gr.5969107
Jeschke, J., Gómez Aparicio, L., Haider, S., Heger, T., Lortie, C., Pyšek, P., et al. (2012). Support for major hypotheses in invasion biology is uneven and declining. NeoBiota 14, 1–20. doi: 10.3897/neobiota.14.3435
Jogaiah, S., Abdelrahman, M., Tran, L. S. P., and Shin-Ichi, I. (2013). Characterization of rhizosphere fungi that mediate resistance in tomato against bacterial wilt disease. J. Exp. Bot. 64, 3829–3842. doi: 10.1093/jxb/ert212
Kamutando, C. N., Vikram, S., Kamgan-Nkuekam, G., Makhalanyane, T. P., Greve, M., Roux, J. J. L., et al. (2017). Soil nutritional status and biogeography influence rhizosphere microbial communities associated with the invasive tree Acacia dealbata. Sci. Rep. 7, 6472–6479. doi: 10.1038/s41598-017-07018-w
Keane, R. M., and Crawley, M. J. (2002). Exotic plant invasions and the enemy release hypothesis. Trends Ecol. Evol. 17, 164–170. doi: 10.1016/S0169-5347(02)02499-0
Kitajima, K., Fox, A. M., Sato, T., and Nagamatsu, D. (2006). Cultivar selection prior to introduction may increase invasiveness: evidence from Ardisia crenata. Biol. Invasions 8, 1471–1482. doi: 10.1007/s10530-005-5839-9
Ku, C., and Hu, J. M. (2014). Phylogenetic and cophylogenetic analyses of the leaf-nodule symbiosis in Ardisia subgenus Crispardisia (Myrsinaceae): evidence from nuclear and chloroplast markers and bacterial RRN operons. Int. J. Plant Sci. 175, 92–109. doi: 10.1086/673306
Kuźniar, A., Włodarczyk, K., Grządziel, J., Woźniak, M., Furtak, K., Gałązka, A., et al. (2020). New insight into the composition of wheat seed microbiota. Int. J. Mol. Sci. 21, 1–18. doi: 10.3390/ijms21134634
Lu-Irving, P., Swope, S. M., and Baltrus, D. A. (2019). Native and Invading Yellow Starthistle (Centaurea solstitialis) Microbiomes Differ in Composition and Diversity of Bacteria. ASM Journals 4:19. doi: 10.1128/msphere.00088-19
Lundberg, D. S., Yourstone, S., Mieczkowski, P., Jones, C. D., and Dangl, J. L. (2013). Practical innovations for high-throughput amplicon sequencing. Nat. Methods 10, 999–1002. doi: 10.1038/nmeth.2634
Maharachchikumbura, S. S. N., Hyde, K. D., Groenewald, J. Z., Xu, J., and Crous, P. W. (2014). Pestalotiopsis revisited. Stud. Mycol. 79, 121–186. doi: 10.1016/j.simyco.2014.09.005
Marques, A. R., Resende, A. A., Gomes, F. C. O., Santos, A. R. O., Rosa, C. A., Duarte, A. A., et al. (2021). Plant growth–promoting traits of yeasts isolated from the tank bromeliad Vriesea minarum L.B. Smith and the effectiveness of Carlosrosaea vrieseae for promoting bromeliad growth. Brazilian J. Microbiol. 52, 1417–1429. doi: 10.1007/s42770-021-00496-1
Morton, J. B., and Benny, G. L., and others (1990). Revised classification of arbuscular mycorrhizal fungi (Zygomycetes): a new order, Glomales, two new suborders, Glomineae and Gigasporineae, and two new families, Acaulosporaceae and Gigasporaceae, with an emendation of Glomaceae. Mycotaxon 37, 471–491.
Motohashi, K., Araki, I., and Nakashima, C. (2008). Four new species of Phyllosticta, one new species of Pseudocercospora, and one new combination in Passalora from Japan. Mycoscience 49, 138–146. doi: 10.1007/S10267-007-0395-Z
Napitupulu, T. P., Ramadhani, I., Kanti, A., and Sudiana, I. M. (2021). Diversity, phosphate solubilizing, and IAA production of culturable fungi associated with healthy and wilt banana. Arch. Phytopathol. Plant Protect. 54, 2306–2332. doi: 10.1080/03235408.2021.1983362
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Oksanen, J., Simpson, G. L., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., et al. (2022). Vegan: community ecology package. Available at: https://cran.r-project.org/package=vegan (Accessed August 23, 2023).
Pan, Y., Liu, M., Sosa, A., Li, B., Shi, M., and Pan, X. (2023). Hierarchical metacommunity structure of fungal endophytes. New Phytol. 239, 1464–1474. doi: 10.1111/nph.19065
Pearson, D. E., Ortega, Y. K., Villarreal, D., Lekberg, Y., Cock, M. C., Eren, Ö., et al. (2018). The fluctuating resource hypothesis explains invasibility, but not exotic advantage following disturbance. Ecology 99, 1296–1305. doi: 10.1002/ecy.2235
Peay, K. G., Russo, S. E., Mcguire, K. L., Lim, Z., Chan, J. P., Tan, S., et al. (2015). Lack of host specificity leads to independent assortment of dipterocarps and ectomycorrhizal fungi across a soil fertility gradient. Ecol. Lett. 18, 807–816. doi: 10.1111/ele.12459
Pickett, B., Carey, C. J., Arogyaswamy, K., Botthoff, J., Maltz, M., Catalán, P., et al. (2022). Enriched root bacterial microbiome in invaded vs native ranges of the model grass allotetraploid Brachypodium hybridum. Biol. Invasions 24, 1097–1116. doi: 10.1007/s10530-021-02692-4
Pizano, C., Kitajima, K., Graham, J. H., and Mangan, S. A. (2019). Negative plant–soil feedbacks are stronger in agricultural habitats than in forest fragments in the tropical Andes. Ecology 100, e02850–e02812. doi: 10.1002/ecy.2850
R Core Team. (2022). R: A Language and Environment for Statistical Computing. Available at: https://www.r-project.org/ (Accessed August 23, 2023).
Ramirez, K. S., Snoek, L. B., Koorem, K., Geisen, S., Bloem, L. J., ten Hooven, F., et al. (2019). Range-expansion effects on the belowground plant microbiome. Nat. Ecol. Evol. 3, 604–611. doi: 10.1038/s41559-019-0828-z
Rodriguez, R. J., White, J. F., Arnold, A. E., and Redman, R. S. (2009). Fungal endophytes: diversity and functional roles: Tansley review. New Phytol. 182, 314–330. doi: 10.1111/j.1469-8137.2009.02773.x
Rodríguez-Caballero, G., Roldán, A., and Caravaca, F. (2020). Invasive Nicotiana glauca shifts the soil microbial community composition and functioning of harsh and disturbed semiarid Mediterranean environments. Biol. Invasions 22, 2923–2940. doi: 10.1007/s10530-020-02299-1
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Rüger, L., Feng, K., Dumack, K., Freudenthal, J., Chen, Y., Sun, R., et al. (2021). Assembly patterns of the rhizosphere microbiome along the longitudinal root Axis of maize (Zea mays L.). Front. Microbiol. 12, 1–14. doi: 10.3389/fmicb.2021.614501
Shannon, C. E. (1948). A mathematical theory of communication. Bell Syst. Tech. J. 27, 623–656. doi: 10.1002/j.1538-7305.1948.tb00917.x
Sheng, M., Rosche, C., Al-Gharaibeh, M., Bullington, L. S., Callaway, R. M., Clark, T., et al. (2022). Acquisition and evolution of enhanced mutualism—an underappreciated mechanism for invasive success? ISME J. 16, 2467–2478. doi: 10.1038/s41396-022-01293-w
Soti, P. G., Jayachandran, K., and Purcell, M. (2014). Mycorrhizal symbiosis and Lygodium microphyllum invasion in South Florida — a biogeographic comparison. Symbiosis 62, 81–90. doi: 10.1007/s13199-014-0272-4
Stricker, K. B., Harmon, P. F., Goss, E. M., Clay, K., and Luke Flory, S. (2016). Emergence and accumulation of novel pathogens suppress an invasive species. Ecol. Lett. 19, 469–477. doi: 10.1111/ele.12583
Tanabe, A. S., and Toju, H. (2013). Two new computational methods for universal DNA barcoding: a benchmark using barcode sequences of Bacteria, Archaea, animals, Fungi, and land plants. PLoS One 8:e76910. doi: 10.1371/journal.pone.0076910
Tanaka, A., Tapper, B. A., Popay, A., Parker, E. J., and Scott, B. (2005). A symbiosis expressed non-ribosomal peptide synthetase from a mutualistic fungal endophyte of perennial ryegrass confers protection to the symbiotum from insect herbivory. Mol. Microbiol. 57, 1036–1050. doi: 10.1111/j.1365-2958.2005.04747.x
Toju, H., Kurokawa, H., and Kenta, T. (2019). Factors influencing leaf- and root-associated communities of bacteria and fungi across 33 plant orders in a grassland. Front. Microbiol. 10, 1–14. doi: 10.3389/fmicb.2019.00241
Toju, H., Tanabe, A. S., Yamamoto, S., and Sato, H. (2012). High-coverage ITS primers for the DNA-based identification of Ascomycetes and Basidiomycetes in environmental samples. PLoS One 7:e40863. doi: 10.1371/journal.pone.0040863
Traveset, A., and Richardson, D. M. (2014). Mutualistic interactions and biological invasions. Annu. Rev. Ecol. Evol. Syst. 45, 89–113. doi: 10.1146/annurev-ecolsys-120213-091857
Trognitz, F., Hackl, E., Widhalm, S., and Sessitsch, A. (2016). The role of plant – microbiome interactions in weed establishment and control. FEMS Microbiol. Ecol. 92:fiw138. doi: 10.1093/femsec/fiw138
Upadhyay, S. K., Srivastava, A. K., Rajput, V. D., Chauhan, P. K., Bhojiya, A. A., Jain, D., et al. (2022). Root exudates: mechanistic insight of plant growth promoting Rhizobacteria for sustainable crop production. Front. Microbiol. 13:6488. doi: 10.3389/fmicb.2022.916488
Vieira, S., Sikorski, J., Dietz, S., Herz, K., Schrumpf, M., Bruelheide, H., et al. (2020). Drivers of the composition of active rhizosphere bacterial communities in temperate grasslands. ISME J. 14, 463–475. doi: 10.1038/s41396-019-0543-4
Whitaker, B. K., Reynolds, H. L., and Clay, K. (2018). Foliar fungal endophyte communities are structured by environment but not host ecotype in Panicum virgatum (switchgrass). Ecology 99, 2703–2711. doi: 10.1002/ecy.2543
Wilson, K. (2001). Preparation of genomic DNA from Bacteria. Curr. Protoc. Mol. Biol. 56:2.4.1-2.4.5. doi: 10.1002/0471142727.mb0204s56
Yang, Q., Carrillo, J., Jin, H., Shang, L., Hovick, S. M., Nijjer, S., et al. (2013). Plant-soil biota interactions of an invasive species in its native and introduced ranges: implications for invasion success. Soil Biol. Biochem. 65, 78–85. doi: 10.1016/j.soilbio.2013.05.004
Yu, P., and Hochholdinger, F. (2018). The role of host genetic signatures on root–microbe interactions in the rhizosphere and endosphere. Front. Plant Sci. 9, 1–5. doi: 10.3389/fpls.2018.01896
Keywords: bacteria, fungi, exotic invasive species, leaf endophytes, functional guilds, next generation sequencing, plant-microbe interactions, soil microbial community
Citation: Nakamura N, Toju H and Kitajima K (2023) Leaf, root, and soil microbiomes of an invasive plant, Ardisia crenata, differ between its native and exotic ranges. Front. Microbiol. 14:1302167. doi: 10.3389/fmicb.2023.1302167
Received: 26 September 2023; Accepted: 08 November 2023;
Published: 22 November 2023.
Edited by:
Hancheng Wang, Independent Researcher, Guiyang, ChinaReviewed by:
Anna Liisa Ruotsalainen, University of Oulu, FinlandCopyright © 2023 Nakamura, Toju and Kitajima. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Naoto Nakamura, bmFrYW11cmEubmFvdG8uMzVjQHN0Lmt5b3RvLXUuYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.