
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 20 December 2023
Sec. Food Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1301325
This article is part of the Research TopicMicrobial Communities in Fermented Products: Current Knowledge and Future ProspectsView all 10 articles
The selection of native yeast for alcoholic fermentation in wine focuses on ensuring the success of the process and promoting the quality of the final product. The purpose of this study was firstly to create a large collection of new yeast isolates and categorize them based on their oenological potential. Additionally, the geographical distribution of the most dominant species, Saccharomyces cerevisiae, was further explored. Towards this direction, fourteen spontaneously fermented wines from different regions of Greece were collected for yeast typing. The yeast isolates were subjected in molecular analyses and identification at species level. RAPD (Random Amplified Polymorphic DNA) genomic fingerprinting with the oligo-nucleotide primer M13 was used, combined with Matrix Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry (MALDI-TOF MS) technique. All yeast isolates were scrutinized for their sensitivity to killer toxin, production of non-desirable metabolites such as acetic acid and H2S, β-glucosidase production and resistance to the antimicrobial agent; SO2. In parallel, S. cerevisiae isolates were typed at strain level by interdelta – PCR genomic fingerprinting. S. cerevisiae strains were examined for their fermentative capacity in laboratory scale fermentation on pasteurized grape must. Glucose and fructose consumption was monitored daily and at the final point a free sorting task was conducted to categorize the samples according to their organoleptic profile. According to our results, among the 190 isolates, S. cerevisiae was the most dominant species while some less common non-Saccharomyces species such as Trigonopsis californica, Priceomyces carsonii, Zygosaccharomyces bailii, Brettanomyces bruxellensis and Pichia manshurica were identified in minor abundancies. According to phenotypic typing, most isolates were neutral to killer toxin test and exhibited low acetic acid production. Hierarchical Cluster Analysis revealed the presence of four yeast groups based on phenotypic fingerprinting. Strain level typing reported 20 different S. cerevisiae strains from which 65% indicated fermentative capacity and led to dry wines. Sensory evaluation results clearly discriminated the produced wines and consequently, the proposed yeast categorization was confirmed. A novel approach that employs biostatistical tools for a rapid screening and classification of indigenous wine yeasts with oenological potential, allowing a more efficient preliminary selection or rejection of isolates is proposed.
The principal metabolic process in wine production is alcoholic fermentation (AF), wherein grape sugars are transformed into ethanol, carbon dioxide through the action of yeast and in parallel a plethora of secondary metabolites are produced (Ribéreau-Gayon et al., 2006; Fleet, 2008; Querol et al., 2018). Although the traditional function of wine yeasts is carrying out alcoholic fermentation, the advent of modern wine microbiology targets to unravel the yeasts properties, in order not only to improve fermentation performance but also wine quality (Barata et al., 2012; Suárez-Lepe and Morata, 2012; Cordero-Bueso et al., 2013; Maicas, 2020).
During fermentation process, the consortium of yeasts is rapidly evolving and shaped by biotic and abiotic factors (Ciani et al., 2004; Jolly et al., 2014; Belda et al., 2016; Sha et al., 2018; Comitini et al., 2021; Dimopoulou et al., 2022). Non-Saccharomyces (NS) yeasts dominate the early stages of fermentation, but the gradual production of ethanol allows the prevalence of the more adaptable species with the most dominant; Saccharomyces cerevisae (Lambrechts and Pretorius, 2000; Soden et al., 2000; Clemente-Jimenez et al., 2005; Sadoudi et al., 2012; Liu et al., 2016; Gobert et al., 2017). Other factors besides ethanol, which create a stressful environment, are high sugar concentration (osmotic stress), low pH (acid stress), decreasing oxygen (hypoxia), presence of numerous microorganisms that compete for nutrients or produce inhibitory compounds and also presence of sulfur dioxide (Mateo et al., 2001; Benito et al., 2015; Roudil et al., 2020; Reiter et al., 2021). Nowadays grapes during harvest contain even higher concentrations of sugars due to climate change, rendering the role of yeast even harder and increasing the possibility of stuck or delayed fermentation. Yeast inoculation in wine industry is the key to ensure fermentation flow and sugar depletion (Bely et al., 2008; Benito et al., 2015; Ciani and Comitini, 2015; Dimopoulou et al., 2020). The commercialization of selected autochthonous strains of S. cerevisiae to drive alcoholic fermentation is justified by their remarkable adaptability to wine stressors (Fleet, 2008; Rossouw et al., 2012; Reiter et al., 2021). Commercial S. cerevisiae strains assure fermentation completion and enhance the standardization and reproducibility of the final product. However, they often lack some unique characteristics linked to biodiversity parameters, and therefore the final wines may lack complexity and typicity (Comitini et al., 2017; Parapouli et al., 2020; Sidari et al., 2021; Christofi et al., 2022).
Targeting the success of alcoholic fermentation and the production of high value wines, producers have focused on the selection of indigenous S. cerevisiae strains which have been previously evaluated for their oenological properties to drive AF (Caridi et al., 2002; Le Jeune et al., 2006; Pulcini et al., 2022). Numerous studies focus on the selection of “novel” S. cerevisiae with main concern, their improved technological and organoleptic properties; such as high yields of productivity, stress tolerance, unique aromatic characteristics and positive sensory attributes (Capece et al., 2010, 2019; Suárez-Lepe and Morata, 2012; Basa et al., 2022; Tronchoni et al., 2022). Some beneficial oenological traits are alcohol tolerance, lower production of acetic acid and H2S, SO2 tolerance, neutral killer character and resistance to high concentrations of sugars (de Ullivarri et al., 2011; Comitini et al., 2017; Pulcini et al., 2022). Furthermore, some yeasts possess the enzymes of β-glucosidases, whose activity results in releasing aglycones and this procedure directly affects beneficially the produced aroma (Mansfield et al., 2002). All the abovementioned characteristics are criteria for the selection of starter cultures, resulting in wines with controlled quality and attractive organoleptic profile (Christofi et al., 2022; Pulcini et al., 2022).
The autochthonous yeast strains, which drive and survive until the end of alcoholic fermentation, are usually characterized by high fermentation rate and alcohol tolerance (Suárez-Lepe and Morata, 2012; Gutiérrez et al., 2013; Garofalo et al., 2018). Numerous researchers have previously isolated a large collection of native yeasts and by means of molecular biology, culture-based methods and mini-vinifications have concluded in some strains with oenological perspective (Caridi et al., 2002; Mestre Furlani et al., 2017; Garofalo et al., 2018; Binati et al., 2019). However, this procedure demands time, advanced analysis and special equipment. The rapid elimination of some isolates from a big yeast collection can result in a more practical and cost-efficient way to select new autochthonous strains with oenological interest. The main objective of this research was to classify a large collection of yeast isolates from spontaneously fermented wines produced from various cultivars and regions in Greece, based on their technological properties with oenological interest. A simple and applicable phenotypic-based methodology for rapid preselection of wine autochthonous yeast with oenological potential is proposed. The qualitative data were transformed accordingly and treated by various biostatistical tools in order to achieve a classification method. The proposed HCA on selected phenotypic tests was validated by wine micro-fermentation trials of the 20 isolated S. cerevisiae strains and their corresponding sensory attributes.
Fourteen samples of spontaneously fermented wines were obtained, from four geographical areas in Greece, namely Goumenissa in northern Greece, Pelion in central Greece, Nemea in southern Peloponnese and the island of Santorini (Table 1). The varieties and the vintage of the wines are noted in Table 1. All samples were collected from dry wines (before SO2 addition), with an alcohol level from 12.5% vol to 14% vol. The majority of the wineries have never used commercial S. cerevisiae strains to drive alcoholic fermentation, whatsoever for the wineries that do use, the profile of the commercial strains (Supplementary Table S1) has been compared with the isolated strains of the present study.
All wines were collected from the wineries at the end of the fermentation process.
For yeast isolation, 100 μL of wine was directly and aseptically spreaded on WL agar plates (Condalab, Madrid Spain). Plates were incubated at 28°C for 48 h. Each sample was analyzed in duplicate. When there were noted more than 20 colonies by plate, a representative selection of yeast colonies was made from WL plates in accordance with the method described by Harrigan and McCance (1976). Colonies were purified by streaking on YPD agar plates [(g/L): Yeast extract 10, Bacteriological peptone 20, Dextrose (D-Glucose) 20, Agar 20]. Plates were incubated at 28°C for 48 h. Each sample was analyzed in duplicate. Additionally, the cultures were maintained at −20°C in YPD broth supplemented with 30% (v/v) glycerol (Serva, Heidelberg, Germany). Before experimental use each isolate was subcultured twice in YPD broth (at 28°C) for 48 h.
Total genomic DNA from the yeast isolates was extracted according to the protocol described by Ercolini et al. (2001) modified by adding lyticase at 2.5 U/mL (Lyticase from Arthrobacter luteus, Sigma–Aldrich, Germany) for yeast cell lysis (Bonatsou et al., 2018). Moreover, quantification and quality control of DNA extract was performed by spectrophotometer (Epoch, Biotek, USA) at wavelengths of 260, 280, and 230 nm.
RAPD-PCR analysis was initially used for clustering the isolates, employing the primer M13 (5′-GAGGGTGGCGGTTCT-3′), according to the protocol of Lieckfeldt et al. (1993). PCR amplification was conducted in 20 μL final reaction volumes, containing 5 μL of One Taq Quick-Load Reaction Buffer (New England Biolabs, USA), 1 U of One Taq Quick-Load DNA Polymerase (New England Biolabs, USA), 100 μM of dNTP’s (10 mM), 10 μM of M13 oligonucleotide primer and 20 ng of template DNA. The amplification program consisted of: 30 s of initial denaturation at 94°C, 3 cycles of 30 s at 94°C, 5 min at 35°C, 5 min at 68°C and then 32 cycles of 30 s at 95°C, 2 min at 53°C, 3 min at 68°C, concluding with 3 min at 68°C.
Genetic diversity within S. cerevisiae isolates was assessed by interdelta analysis proposed by Legras and Karst (2003) with minor modifications. Briefly, PCR amplifications were carried out in 20 μL reaction containing 2.5 μL of Buffer A 10 X, 0.25 μL of Taq DNA Polymerase (5 U/μL, Kapa Biosystems, USA), 100 μM of each dNTP, 10 μM of each oligonucleotide primer [delta 12 (5′- TCAACAATGGAATCCCAAC-3′) and delta 21 (5′-CATCTTAACACCGTATATGA-3′)]. Amplification reactions were performed with the following conditions: 4 min at 95°C followed by 40 cycles of 30s at 95°C, 30 s at 46°C and 90s at 72°C and a finishing step of 10 min at 72°C. In addition to the indigenous S. cerevisiae strains, the commercial S. cerevisiae strains (Supplementary Table S1) were also examined. The commercial strains were treated as all the other isolates subjected in the fingerprint analysis. All amplifications were carried out in a thermocycler (T100, Biorad, United States).
The products were run on a 1.5% (w/v) agarose gel in 1 × TAE buffer, stained with ethidium bromide (20 min) at 110 V for 140 min and scanned under ultraviolet light (MiniBIS, DNr, Israel). A 100 bp and 1Kb DNA ladder (Nippon Genetics, Germany) served as size standard in RAPD-M13 and interdelta PCR products, respectively. The resulting fingerprints were digitally captured, converted, normalized and analyzed using the Dice coefficient with Bionumerics software version 6.1 (Applied Maths, Sint-Martens-Latem, Belgium). Means of the Unweighted Pair Group Method using the Arithmetic Average (UPGMA) clustering algorithm led to the formation of the species- and strain specific dendrogram. Furthermore, species identification was achieved since two to five representative strains from each different cluster (distance >90%) were selected to species identification by MALDI-TOF MS method as described by Windholtz et al. (2022).
Important technological characteristics such as production and sensitivity to killer toxin, acetic acid production, β-glucosidase production, resistance to SO2 and H2S production were tested for the yeast screening (Rodríguez et al., 2004; Comitini et al., 2011; Domizio et al., 2011; Konate et al., 2014). All assays were replicated twice. Precultures were grown in YPD broth at 28°C for 48 h.
The killer character determination was performed using the plate assay described by Domizio et al. (2011), with positive activity (K+) recognized by inhibition of growth of the sensitive strain (S. cerevisiae SO classic, Martin Vialatte, France), seen as a clear zone surrounding the seeded strain. The S. cerevisiae killer strain VIN13, (Anchor, France), showing killer activity, was used as positive control. Sensitive character was observed when colonies could not grow onto agar substrate which was poured with a killer yeast strain (S. cerevisiae VIN13, Anchor, France); the isolate was designated as sensitive (K-, R-). The S. cerevisiae sensitive strain (SO classic, Martin Vialatte, France), was used as positive control. Yeasts with negative reaction to the killer character (K-) and negative sensitivity (R+) were characterized as neutral (de Ullivarri et al., 2011). The S. cerevisiae killer strain VIN13, (Anchor, France), showing killer activity, was used as positive control.
Acetic acid production was noticed by formation of clear zones around colonies of the strains which were implemented and spotted on Hestrin-Schramm CaCO3 agar (Aydin, 2009). This medium was composed of [g/L: CaCO3 5.0, Yeast extract 3.0, Agar 15.0 and Dextrose 15.0] (Konate et al. 2014). Cultures were incubated at 28°C for 5 days. The ability of the colonies to form clear zones through the hydrolysis of the white salt was considered as positive reaction to this test.
The β-glucosidase activity was evaluated as described by Rodríguez et al. (2004) on agar plates containing arbutin as substrate. Screening was carried out on agar plates with arbutin as substrate [g/L: Yeast Nitrogen Base/YNB (Condalab, Madrid Spain) 6.7, arbutin (Sigma Aldrich, USA), 5, agar, 20]. The pH was adjusted to 5.0 and after sterilization 2 mL of a sterile 1% (w/v) ferric ammonium citrate solution was added to 100 mL of melted medium. Each plate was inoculated by spot assay, incubated at 28°C and examined after 8 and 15 days. Enzymatic activity was noticed visually when brown color develops in the agar.
The SO2 resistance was determined by screening on plates with synthetic substrate. Based on the protocol described in detail by Comitini et al. (2011), the isolates were inoculated onto YPD agar plates at pH 3 (with citrate–phosphate buffer), added with increasing doses of K2S2O5 in different concentration corresponding to 0, 100, 200, 300, 400, 500 mg/L of free SO2 and incubated at 28°C. The yeast growth was observed after 2, 5 and 8 days after the inoculation.
H2S potential production was estimated by spreading the yeasts onto Biggy Agar (Condalab, Madrid, Spain) (Domizio et al., 2011). On this medium, H2S-positive isolates create brown colonies, while H2S negative isolates create white colonies. The H2S effect was noticed after 2, 5 and 8 days of incubation at 28°C when the color of the colonies was recorded. The following arbitrary scale was used: 0 = white (no production); 1 = light brown; 2 = brown; 3 = dark brown.
The fermentation potential of the yeast strains was evaluated in micro-fermentation trials. Fermentations were carried out at 18°C in 50 mL of pasteurized (72°C, 10 min) Assyrtiko must which was provided by Gaia (Sanotrini, Greece) winery (vintage 2021) under static conditions. The initial pasteurized grape must (pH = 3.2, total acidity = 5.77 g tartaric acid/L, YAN = 609 mgN2/L, 16.6 mg SO2, 5.1 free SO2) contained 119.5 g/L glucose and 120.1 g/L fructose. Precultures were grown in YPD broth at 28°C for 48 h, and then used to inoculate each fermentation (106 cell/mL). Residual sugar (glucose and fructose) determination was performed on the wines in daily basis using Enzytec kit-liquid Glucose-Fructose (r-biopharm, Germany). All fermentations were assessed duplicated and analysis was conducted twice.
In the end of the fermentation all produced wines were categorized based on their aromatic profile (odor) by means of a free sorting task test (Sáenz-Navajas et al., 2012). A total of 10 experienced panelists participated in the pilot study. Participants were provided with the produced dry wine samples (15 mL) in ISO approved wine glasses coded with different three-digit numbers and arranged in random order. Participants were asked to sort the 13 wines on the basis of similarity attending to the global sensations perceived in nose (p.e. intensity, floral, fruity, off odor characteristics). Panelists could make as many groups as they wished. Upon completion, they recorded the three-digit codes of the samples of each group on a paper sheet. All wines were served at room temperature. The sessions took place in a ventilated and air-conditioned tasting room (at around 20°C). Panelists were not informed about the nature of the samples.
Data obtained from the phenotypic tests, were converted into numerical from character data (+, −) as it is illustrated in Table 2. Consequently, data are further investigated by means of statistical analysis. Firstly, MANOVA was attempted but the scaled data of H2S experiment during the MANOVA application process could not satisfy its application assumptions. MANOVA assumes multivariate normality and homogeneity of variance–covariance tables between groups. These assumptions were not met, and we considered a univariate analysis using ANOVA more appropriate for each variable. Univariate method may offer simpler and more straightforward interpretations of the results. Using ANOVA for each dependent variable separately allows focusing on each variable’s unique response to the independent variables. More specifically, the possibility of having statistically important differences was examined at origin and species level. Therefore we apply the model Yij = μi + εij, i = 1, 2, 3, …where with i we denote the levels of the H2S factor and j = 1, 2, … the observations we have for each level (Koutras and Evagelaras, 2010). In the current analysis the average level of H2S production per origin or species was considered as a dependent variable Y and time of incubation as variable X. To obtain safe statistical conclusions, (a) the assumption of equality of dispersions at the levels of factor at the level of significance of 5% and (b) the test of the normality and independence of errors at the level of significance of 5% were carried out. To check the equality of variations, Levene’s test was used, where we do not reject the zero hypothesis to be checked and therefore ensure homoscedasedality. Then, to check the normality and independence of the errors, Studentized residuals were used. Utilizing the non-parametric test of Kolmogorov–Smirnov does not reject the null hypothesis that errors follow Normal Distribution. Regarding the test of independence of errors, the non-parametric, Run’s test was used and the null hypothesis that errors are independent cannot be rejected. Having ensured the above conditions, the test for whether there are significant differences between the levels of the factor is of the form:
The value of p of the test is less than 0.05 and therefore at a significance level of 5%, zero assumption that there are no significant differences in the levels of the factor is rejected.
Hiercharchical Cluster Analysis (HCA) of the different phenotypes based on the results of the five phenotypic tests was performed under R (3.6.2) software using Euclidean distance and Ward method. Among various hierarchical clustering methods such as Single Linkage Method, Weighted Average Linkage Method, Centroid Method, Flexible Strategy Method etc., the Ward’s Method was chosen as it is the most effective. This method differs from others and is designed to minimize variance within groups. In particular, the method has some very good properties and usually creates groups with a similar number of observations. The development of logical rules that lead to finding the optimal number of groups of a dataset has occupied several researchers active in the field of cluster analysis, since it is obvious that this problem is of great practical interest. Thorndike (1953) proposed a graphical approach to the problem whereby an axis is first depicted on one axis of the average within-cluster distance and on a second axis of the number of groups. With each increase in the number k of groups there is a corresponding decrease in the average distances within the groups. In most cases a position appears where we have a sharp decrease in the average distances within the groups and then “leveling” the graph. In order to find the number of groups the datagram resulting from a hierarchical cumulative method was examined and from it determine the optimal number. More specifically, at that point of the dendrogram where the greatest change in the quantity recorded on the horizontal axis (distance) is observed, we can bring a parallel line to the vertical axis and see at how many points the datagram intersects. The number k for which we observe large concatenation distances relative to the previous one (k-1 groups) is a reasonable value for the optimal number of groups. For this reason, 4 groups were selected.
Based on the phenotypic test results, we encoded the positive and negative responses to the microbiological phenotypic assays. The data were organized according to oenological significance to establish an overall phenotype. Priority was given to the production of killer toxin, followed by H2S production, acetic acid production, SO2 resistance, and, finally, β-glycosidase activity. All parameters were considered, with particular attention to the sequence of data. This arrangement was determined with oenological requirements in mind, aiming for yeast strains that are insensitive to killer toxin, are low producers of H2S or acetic acid, exhibit resistance to SO2, and possess desirable β-glucosidase activity. Α detailed description is provided of the development of the proposed selection method (Figure 1).
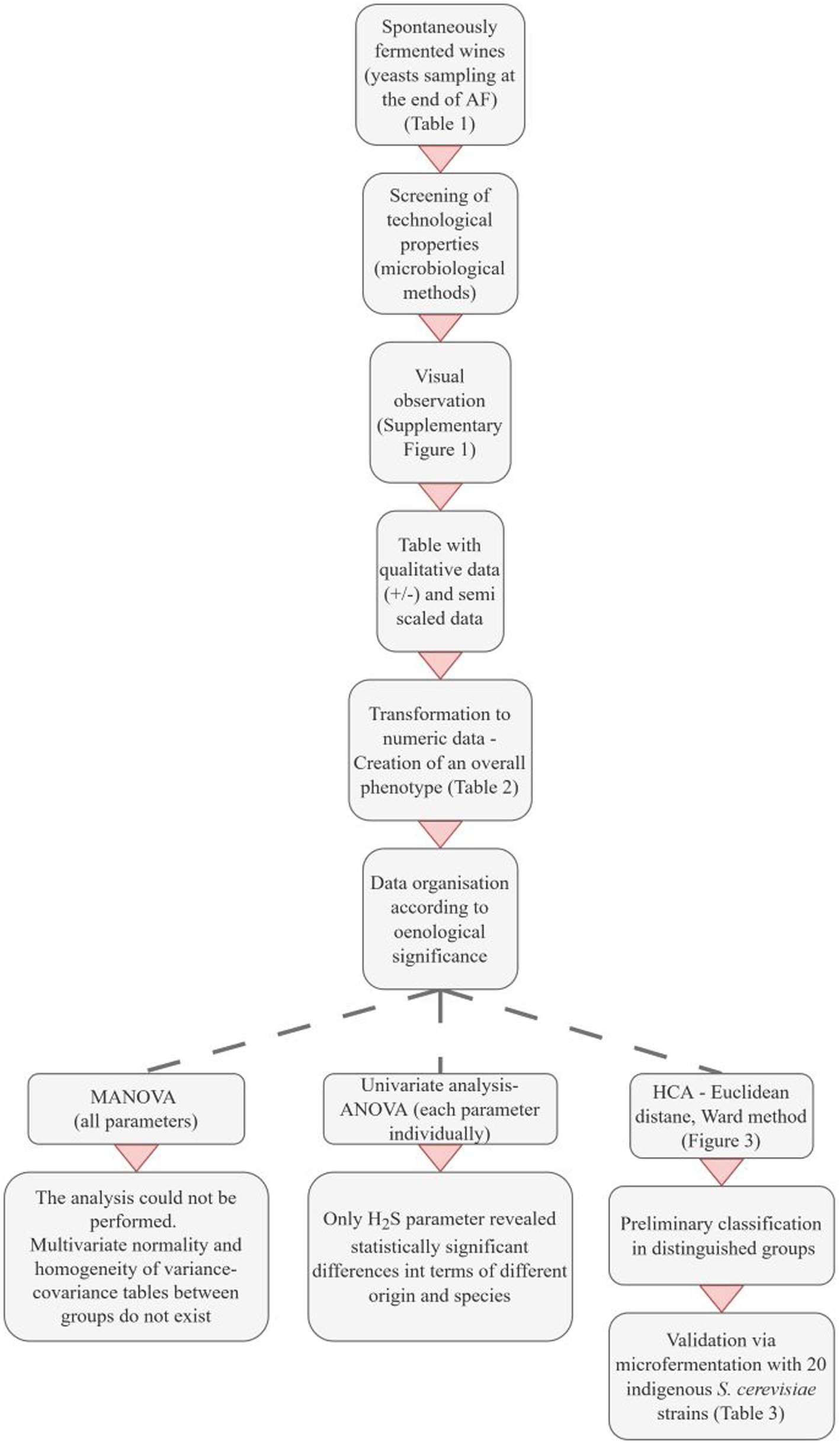
Figure 1. Overview of the development of the proposed phenotypic classification method. AF, Alcoholic fermentation; HCA, Hierarchical cluster analysis.
Encoding free sorting data was the key to categorize wine samples based on the results of the sensory assessment. For each group, results are encoded in an individual similarity matrix (wines × wines), in which 1 stand for two wines set in the same group and 0 for two wines put in different groups. These individual matrices are summed across subjects; the resulting co-occurrence matrix represents the global similarity matrix where larger numbers indicate higher similarity between samples. The assumption underlying this method is that samples grouped together are more similar than samples sorted into different groups. The resulting cooccurrence matrix was submitted to HCA (Ward method) in order to derive a spatial arrangement of wines with R (3.6.2) software analysis.
The community structure of yeast in wine samples collected directly from wineries was determined at the end of alcoholic fermentation (AF). A total of 14 wine samples were collected, including two from Santorini, five from Pelion in central Greece, five from Nemea in southern Peloponnese, and two from Goumenissa in northern Greece (Table 1). A total of 190 yeast isolates were obtained, and their geographic origins are shown in Figure 2. The obtained RAPD-M13 PCR fingerprints were clustered using UPGMA analysis with Dice as a coefficient, and a representative number of isolates from each cluster were identified using MALDI-TOF MS. Six different species were identified, namely Saccharomyces cerevisiae (168 isolates), Trigonopsis californica (1 isolate), Brettanomyces bruxellensis (5 isolates), Zygosaccharomyces bailii (8 isolates), Priceomyces carsonii (1 isolate), and Pichia manshurica (7 isolates). Specifically, S. cerevisiae was the most dominant species with the isolation frequency exceeding 88.4% (data not shown). Although 7 isolates of P. manshurica were found, all of them were obtained from a single sample and there was no repetition across samples. Thus, it is not possible to make assumptions based on the presence of a random spoilage yeast/fungus species in just one sample. Conversely, Z. bailii was detected in minor amounts in 5 different samples from 3 different regions (Nemea, Goumenissa, and Pelion).
All 190 isolates were subjected to some phenotypic tests to monitor their technological properties. These yeasts were analyzed for different characteristics, such as killer activities, acetic acid production, β-glucosidase activity, hydrogen sulphide production and sulphur dioxide resistance, revealing both inter-generic and intra-generic biodiversity.
Among our observations, only 12% of the yeasts (23 isolates) were characterized as sensitive to killer toxin and the rest were noted as neutral. No killer yeast was reported. Among these 23 yeasts the only isolate of T. californica is sensitive to killer toxin, 20 isolates belong to S. cerevisiae and 2 isolates of Z. bailii. 137 yeasts were observed to produce clearly acetic acid and thus forming clear zones around the colony in CaCO3 agar; whereas 53 did not produce acetic acid. From these isolates, T. californica made the only exception since the rest 52 isolates belonged to S. cerevisiae group. All yeasts showed low or absence of β-glucosidase activity because no color change from white to brown/dark brown was noticed. However, 12 colonies (8 S. cerevisiae, 1 T. californica and 3 Z. bailii) were slightly darker, compared to the others, revealing low enzymatic activity (6,3% of the total isolates). Moreover, regarding the potential H2S production at species level B. bruxellensis followed by T. californica and P. carsonii perceived significantly the highest levels of H2S. All isolates of Z. bailii proved to be low H2S producers while S. cerevisiae isolates expressed great variability. Sulphur dioxide resistance, a very desirable oenological characteristic, was determined to be a common trait to almost all tested isolates. Only 6, 12 and 11 isolates were sensitive at the concentrations of 100 mg/L, 200 and 300 mg/L, respectively. It is noteworthy that SO2 inhibited the growth of most Z. bailii isolates and 82% of the isolates were resistant to the extreme concentrations of 400 and 500 mg/L.
Hydrogen sulphide production was the only phenotypic characteristic which exhibited statistically significant differences in terms of origin and species characterization. The production of H2S was measured at 2, 5 and 8 days and color grading indicated its quantity [0 = white (no production); 1 = light brown; 2 = brown; 3 = dark brown]. In the current analysis the average level of H2S production (0,1,2,3) was considered as a dependent variable Y per origin and species (data not shown), and time of incubation as variable X that takes the values 1: for H2S that was produced after 2 days of incubation, 2: for H2S that was produced after 5 days of incubation, 3: for H2S that was produced after 8 days of incubation. Test of homogenicity of variances, one sample Kolmogorov–Smirnov test and test off between-subjects effects proved that significant differences between the levels of factor H2S-production exist in terms of origin and species. Isolates from the regions of Nemea and Pelion revealed lower levels of H2S production, regardless of the species parameter. Moreover, B. bruxellensis followed by T. californica and P. carsonii perceived significantly the highest levels of H2S. More specifically, B. bruxellensis released more H2S at 5th and 8th day, whereas T. californica and P. carsonii emissions did not change after the 2nd day. Z. bailii proved to be low H2S producers, P. manshurica fair producer and S. cerevisiae strain-dependent.
A novel approach that employs biostatistical tools for rapid screening and classification of large collection of indigenous wine yeasts, allowing for efficient isolate selection is introduced. Following the results of the screening tests, the positive or negative responses to the phenotypic assays were coded (Table 2) and an overall phenotype has been created. In total, 29 different phenotypes were observed. Hierarchical cluster analysis was performed and four main clusters/groups were finally obtained (Figure 3). The algorithm could discriminate the phenotypes based on the 5 tested different parameters. The first group (yellow) consists of isolates which were characterized as sensitive to killer toxin and consequently the yeasts of this group cannot be proposed as starter cultures. Additionally, the second group (grey) is characterized by neutral, no acetic acid production, low resistance to SO2, no β-glucosidase activity and varies regarding H2S production. The third group (green) includes neutral yeasts, with low H2S production, positive acetic acid productivity and absence of β-glucosidase. Finally, in the last group (pink) belong neutral to Killer yeasts (with one exception), high production of H2S and acetic acid but resistant to SO2 and possible β-glucosidase activity. Therefore, the most preferred groups are the 2nd – grey and the 3rd – green. Additionally, species allocation among phenotypes was also examined (Figure 4). The isolates of S. cerevisiae and Z. bailli were distributed in the four created phenotypic groups. On the contrary P. manshurica and B. bruxellensis only in the second and fourth group, respectively.
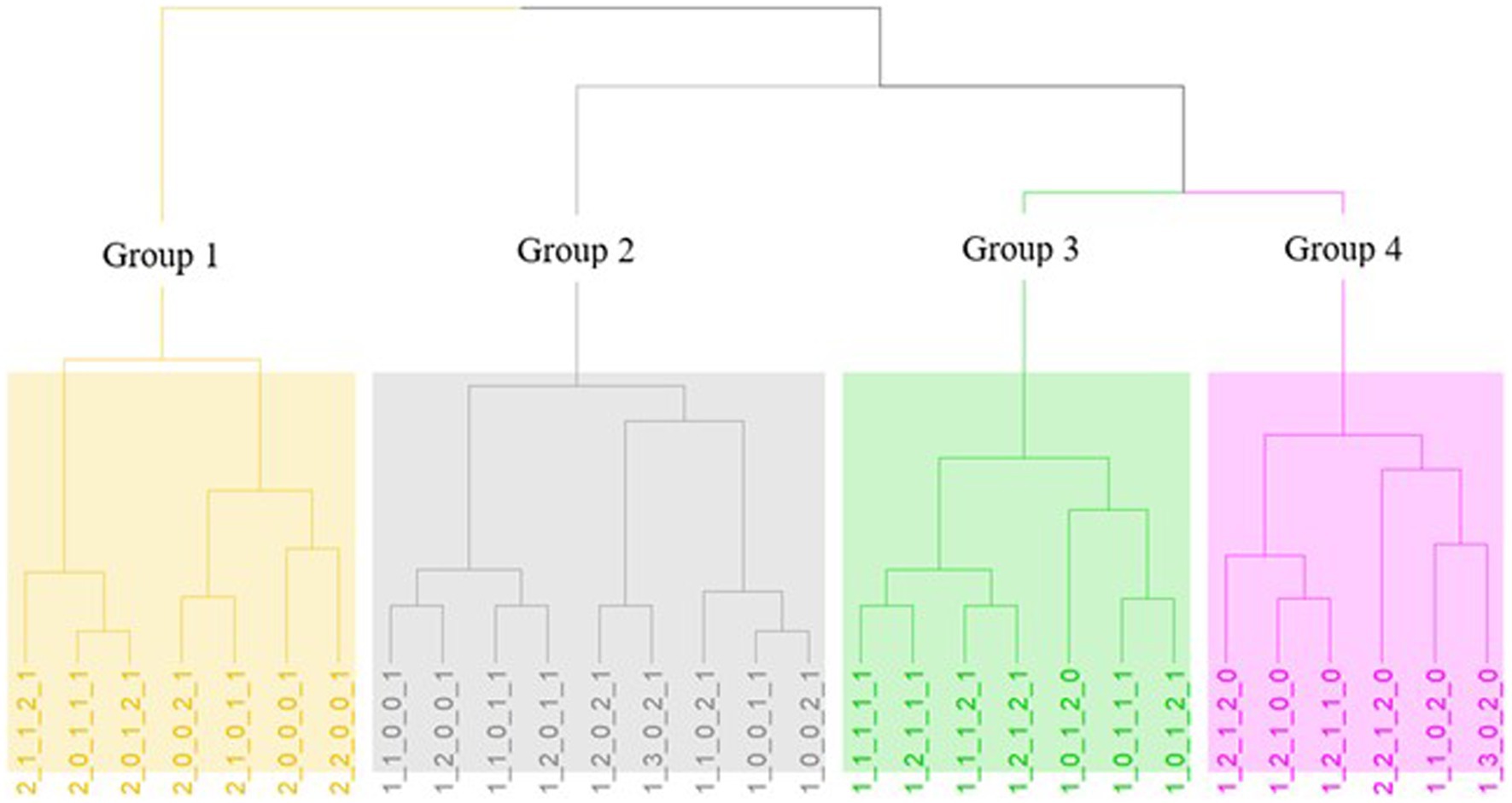
Figure 3. Hierarchical cluster analysis (HCA) of the different phenotypes with Eudlidean distance and Ward method. The four groups are distinct by colors: Group 1 = yellow, Group 2 = grey, Group 3 = green and Group 4 = pink.
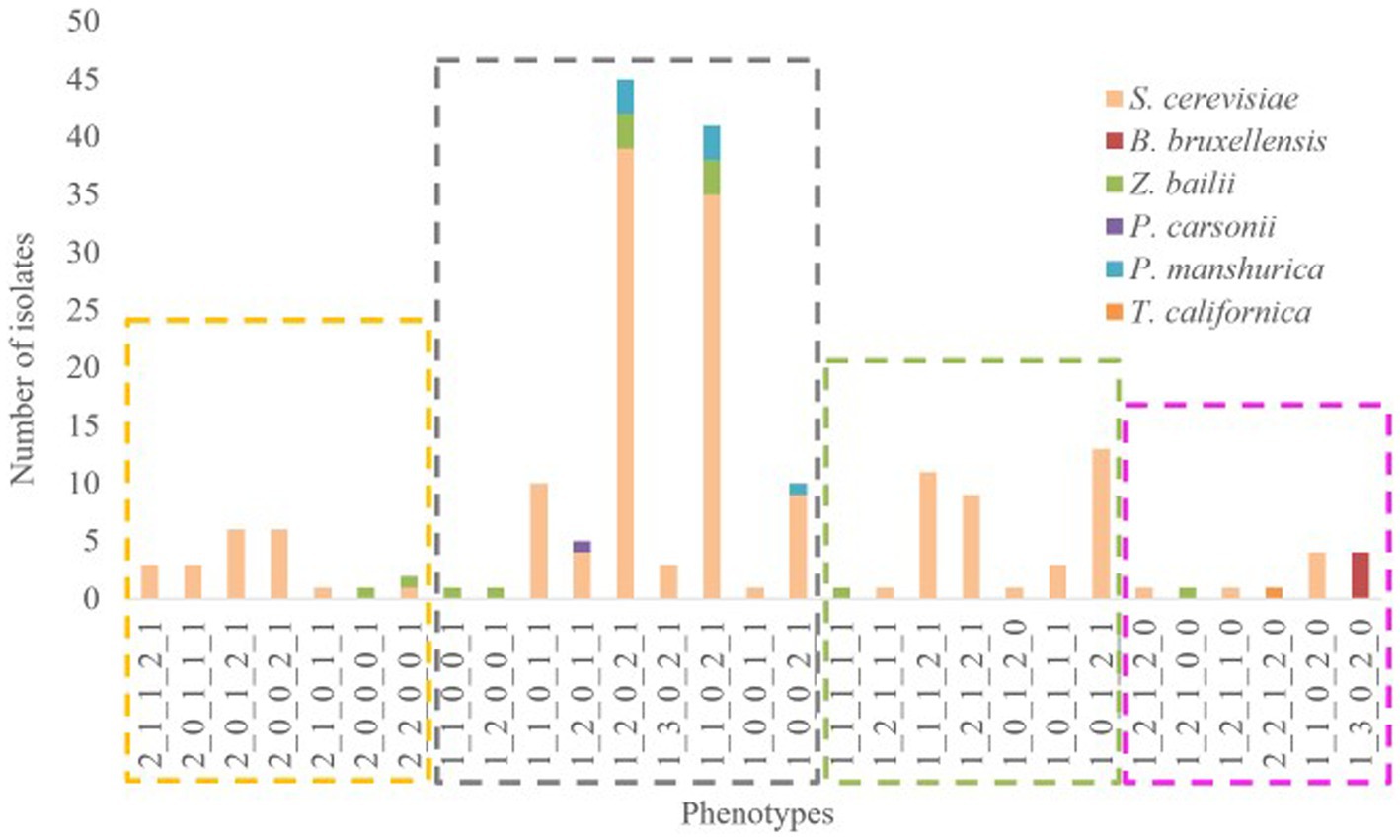
Figure 4. Composition plot of species among the different phenotypes. The captions distinguish the groups which were obtained from HCA (Group 1 = yellow, Group 2 = grey, Group 3 = green and Group 4 = pink).
During spontaneous fermentation process, non-Saccharomyces yeasts dominate at the beginning of AF and the conversion of sugars into ethanol is completed by S. cerevisiae yeasts (Di Maro et al., 2007; Garofalo et al., 2016). Consequently, validation of the proposed categorization and also the interest for additional insights into the geographical distribution was focused on the isolates which were identified as S. cerevisiae. Strain typing of S. cerevisiae revealed the existence of 20 distinct strains performing interdelta PCR, namely S1 to S20. To assess the performance of each S. cerevisiae strain under fermentative conditions, laboratory-scale fermentations were conducted, and sugar consumption was measured on a daily basis. It was observed that only 35% of the inoculated S. cerevisiae strains (S2, S3, S4, S7, S8, S13, S19) had a lower ability to catabolize sugars, resulting in wines with residual sugar levels of greater than 10 g/L. Therefore, those strains are not suggested to be used as starter cultures. Based on the sensory results of free sorting task, four main clusters were identified (Figure 5). The wine samples that were clustered in groups C and D based on Hierarchical Cluster Analysis (HCA) were deemed as undesirable wines (S). On the other hand, Group B exhibited floral and fruity characteristics, while Group A presented a more complex aromatic profile, which was highly preferable (Figure 6). Hence, the proposed strains are those belonging to groups A and B and it is noteworthy that S1, S10, S14, and S20 produced the most desirable wines with no problematic catabolism of sugars and different organoleptic perception.
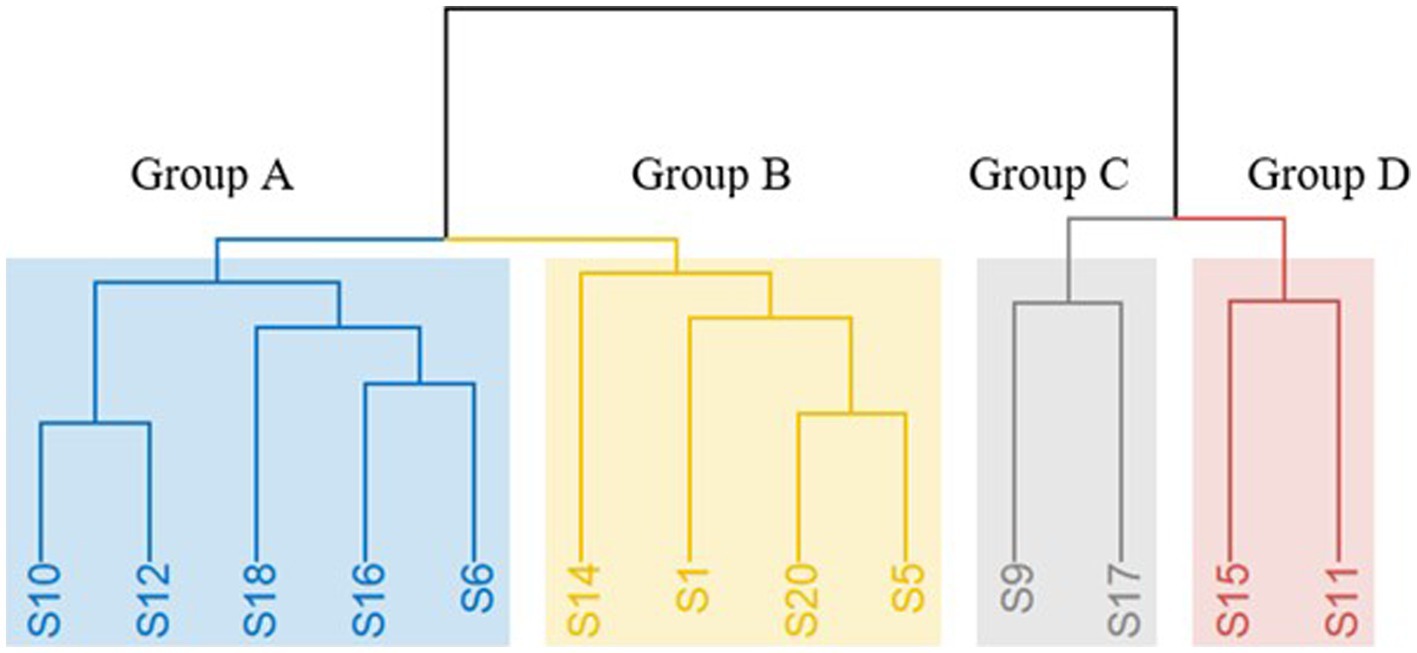
Figure 5. Hierarchical cluster analysis (HCA) of the different aromatic profiles of the 13 produced dry wines from different S. cerevisiae strains, based on the results of the free sorting task with Ward method. The four groups are distinct by colors: Group A = yellow, Group B = grey, Group C = green and Group D = pink.
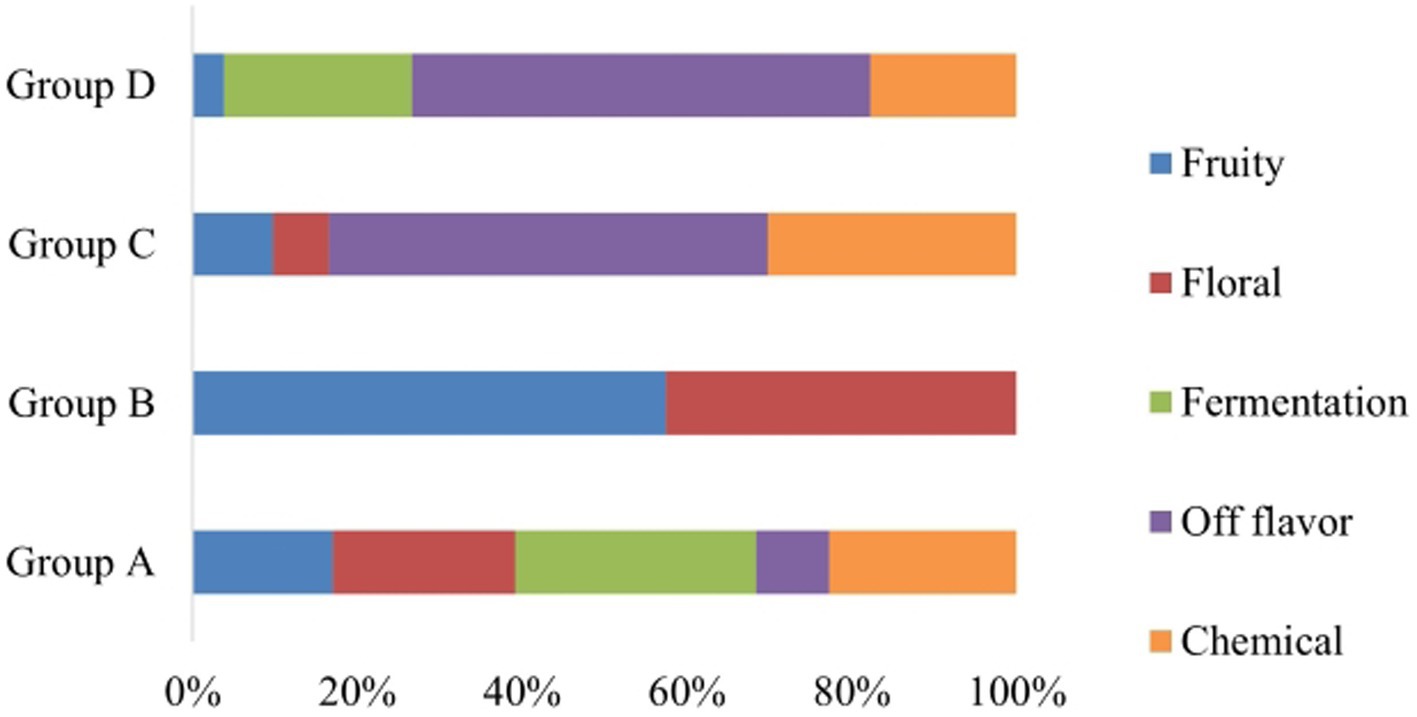
Figure 6. % Composition plot of the descriptors defining each cluster (Group A, Group B, Group C, and Group D) according to the panelists of the sensory assessment.
Table 3 clarifies that the current findings are in line with the preliminary selection of yeasts, as all isolates from the same strain were also grouped in the same cluster based on their phenotypic characteristics. More specifically, from the preliminary selection, the green cluster (Group 3) and the grey cluster (Group 2) consisted mostly of strains from Group A and B, along with some strains that could not catabolize all sugars, namely S1, S7, S10, S13, S18, and S20 for the green cluster, and S5, S6, S8, S12, S14, S16, and S19 for the grey cluster. Additionally, strains that resulted in abnormal fermentations with off-odor characteristics were clustered in the less beneficial groups: 5 strains in the yellow/Group 1 (S2, S3, S4, S9, and S17) and two in the pink/Group 4 (S11 and S15).

Table 3. Fermentation capacity, sensory profile, preliminary group classification, and validation of the classification of the 20 different S. cerevisiae strains.
Finally, the geographical distribution of the isolated S. cerevisiae strains was examined. Three distinct types of S. cerevisiae communities were identified among the investigated samples. The first type consisted of wine samples (GB, A9, K16, A19) with up to two different strains, indicating the prevalence of only one or two strains. The second type consisted of samples that were observed with three to four different strains, while the third type included samples A6, K29, K32, and K33, which exhibited the most complex S. cerevisiae community structure. In these samples, seven to eight different strains interacted and survived until the end of the alcoholic fermentation process. Additionally, how the contribution of geographical origin, type of wine, and vintage effects to the dispersion of S. cerevisiae strains was investigated. Venn diagrams are illustrated in Figure 7, revealing that one unique strain (S10) was isolated from all tested regions, while three strains were found only in Santorini (S15, S17, S18), three only in Nemea (S11, S12, S13) and two Pelion region (S1, S2). Based on the current results, 10 strains (S2, S3, S4, S5, S6, S7, S8, S9, S10, S20) were found in both red and white wines, while 6 strains (S1, S11, S12, S13, S14 and S19) and 4 strains (S15, S16, S17 and S18) were isolated only from red and white wines, respectively.
Nowadays climate change leads to even more stressful conditions for wine yeast, due to the higher concentrations of sugars on grapes during harvest. Consequently, commercial yeasts, which have been isolated in the past, may not be able to adapt to this new challenging environment (Fleet, 2008; Rossouw et al., 2012; Reiter et al., 2021). The commercialization of new selected indigenous strains to drive alcoholic fermentation is a necessity for the wine industry. Therefore, time efficient methods are crucial when isolating new strains. The process of screening and selecting wine yeasts typically involves several sequential steps (Sidari et al., 2021; Pulcini et al., 2022). However, it is necessary to initially exclude certain isolates when dealing with a vast collection of yeast isolates during the selection process. Assessing technological properties through phenotypic plating methods is a well-established approach. However, manual selection and rejection may not always be feasible especially when no ideal combination of technological properties is discernible, as suggested in previous studies (Mestre Furlani et al., 2017; Sidari et al., 2021).
Yeast selection is a very interesting field not only in wine microbiology, but also in food microbiology. Previous research on indigenous strain selection focused on examining similar characteristics (Caridi et al., 2002; Settanni et al., 2012; Aponte and Blaiotta, 2016). Numerous researchers promote the isolation and selection of indigenous microbiota from various fermented products such as table olives and cheese in order to promote the quality of the final product (del Castillo et al., 2007; Bleve et al., 2015; Bonatsou et al., 2015). In the present study, a similar selection process for indigenous strains was followed, but additionally all the phenotypic results were coded analyzed and further categorized. To the best of our knowledge, no other relevant work managed to transform the qualitative data into numeric in order to perform HCA. Up to now phenotypic results are mostly depicted by tables with negative and positive response and the selection was resulting manually. The proposed idea is to find a way to allow the preliminary rejection of some isolates among a plethora of isolates where no perfect or worst combination is noticed. Qualitative data were transformed into numerical values, and the phenotypic characteristics were arranged based on their impact on the predominance of the selected strain and on the quality of the final product. All isolates were obtained from wines that reflect tolerance to high alcohol and sugar concentrations. Even though no perfect phenotypic combination was noticed, this classification allows the categorization of a large collection of isolates under more than one parameter in a more efficient, low-cost and rapid way.
One necessary trait for a strain, in order to be used as a starter culture, is tolerance to killer toxin (Liu et al., 2015). In the current study, the first digit reveals the killer character and the majority of the isolates were classified as neutral. Previous research has also indicated that autochthonous yeasts are predominantly sensitive or neutral (Comitini et al., 2011; Domizio et al., 2011; de Ullivarri et al., 2014; Velázquez et al., 2016). The lack of killer properties in the isolated yeasts of the current survey, justifies the strain biodiversity among wine samples at the end of AF (Puyo et al., 2023). Furthermore, a brief overview of the literature over the past few decades supports that the most preferable strains should be characterized by non or low production of H2S in order to be used as starter cultures (Caridi et al., 2002; Settanni et al., 2012; Aponte and Blaiotta, 2016). Thus, H2S production is evaluated as the second most important oenological characteristic of those examined. From another point of view, organic winemaking process emphasizes on the higher risks of oxidation, microbial contamination and H2S production. Hence, new starters ‘low H2S – SO2 – acetaldehyde producers’ are desired (Comitini et al., 2017). Additionally, acetic acid screening was performed because some non-Saccharomyces yeasts produce undesirable concentrations of acetic acid and ethyl acetate from sugars, regarded as unsuitable for winemaking (Caridi et al., 2002; Rodríguez et al., 2004). Moreover, recently S. cerevisiae and Z. bailii adaptive response and tolerance to acetic acid have been investigated based on functional and comparative genomics strategies (Palma et al., 2018; Capece et al., 2022). Sulphur dioxide resistance is a very desirable oenological characteristic and most studies have been conducted in autochthonous S. cerevisiae strains, showing high resistance of this species to SO2 and differentiation at strain level (Divol et al., 2012; Settanni et al., 2012). Although legacy allows up to 150–200 mg/L SO2 addition for dry wines, while in exceptional cases it can reach up to 400 mg/L for some sweet wines, in the present study the isolates were tested in more extreme concentrations such as 500 mg/L of free SO2 (OIV, 2023). Consequently, higher concentrations of SO2 can delay the growth of these isolates, but we have to take into account that the experiment is designed in vitro and intermediate concentrations were not examined. Yeasts are the main producers of β-glucosidase which is an important enzyme for the hydrolysis of grape glycosides during winemaking. The importance of glycoside hydrolysis in aroma, flavor, color, and color stability was underlined previously (Mansfield et al., 2002; Settanni et al., 2012; Zhang et al., 2021). Modern winemaking techniques often use specific strains of Saccharomyces or non-Saccharomyces yeasts with known β-glucosidase activity to compensate the insufficient enzyme activity in grapes (Aponte and Blaiotta, 2016). Almost all of the abovementioned results are in line with previous studies, in which it is stated that β-glucosidase activity of the species S. cerevisiae, P. carsonii and Z. bailii is mostly low or even absent (Rodríguez et al., 2004; Cordero-Bueso et al., 2013; Aponte and Blaiotta, 2016; Vilela, 2020; Zhang et al., 2021). On the contrary, our results do not align with those of previous investigations regarding P. manshurica, where many species in the Pichia genus have been characterized for their moderate to high β-glucosidase producing ability and the enhance of beneficial volatile compounds in the final product (Zhang et al., 2021; Perpetuini et al., 2020).
The isolates’ classification was validated by the species and strain dispersion. After pilot fermentations assay was conducted, the majority of S. cerevisiae strains completed successfully AF and led to wines with exceptional sensory characteristics. Based on recent literature, it is also mentioned that B. bruxellensis, in general, intensifies the off-flavor characteristic by producing high concentrations of H2S (Avramova et al., 2018). According to the results presented in this study, B. bruxellensis isolates were all clustered in the group with the highest H2S production. Notably, there is significant intra-species variability, particularly between S. cerevisiae and Z. bailii species. The validation of the proposed categorization is further validated by the fact that all isolates belonging to the same strain of S. cerevisiae exhibited the same phenotypes without exceptions. It is important to highlight that the strains with the most preferable sensorial characteristics were clustered in the 2nd and 3rd group and the opposite. The evaluation of the alcoholic fermentation and produced wines was achieved by monitoring the sugar consumption and the basic sensory evaluation of the final product. The aim was to be able to discriminate the wines based on their basic organoleptic characteristics, that are also examined by the clustering method [pe off odor aromas (acetic acid, H2S), fruity/floral aromas (β- glucosidase)], in order to examine the correctness of the proposed classification. The kinetics of alcoholic fermentation reveals the possibility of having a stacked or delayed fermentation. A comparative sensory analysis was chosen in order to evaluate the final product in a more global point of view. According to previous research the free sorting task is an efficient technique for assessing the perception of a set of products by a panel of subjects (Courcoux et al., 2015). This holistic and non-verbal task is an effective tool to be used in product development. Moreover, is a technique that is widespread also in the wine science (Rossouw and Bauer, 2016; Binati et al., 2020).
Hence, the proposed coding and classification method offers the advantage of not requiring expensive molecular techniques and provides valid results within a short timeframe of only two days. Biostatistical tools enhance the categorization of a large collection of yeasts based on their phenotype and allow a preliminary selection of the isolates. This preliminary rejection is time and cost efficient and therefore a very useful tool not only for wineries but also for yeast suppling companies. In the current study the selection some yeast strains and their direct application in the wine industry is not possible since more analysis of oenological, biochemical and aromatic point of view should be implied.
Wine samples collected at the end of AF revealed the great predominance of S. cerevisiae, with a high intraspecific biodiversity (Garofalo et al., 2016). Furthermore, S. cerevisiae strains are mostly selected as starter cultures due to their unique biotechnological characteristics, such as fermentation capacity, the production of alcohol and CO2 and its resilience to adverse conditions of low pH and osmolality (Dimopoulou et al., 2020). The strain collection created in this research verifies the abovementioned observations. The identified different strains varied in richness and evenness among the wine samples, indicating the complex microbial interactions that occur during spontaneous fermentation. Microbial interactions play a dominant and complex role during AF. Despite the presence of multiple S. cerevisiae strains, some of which exhibited off-flavor characteristics during the final fermentation stage, none of the wine samples exhibited an off-flavor odors. It is well known that some indigenous strains persist in wineries for multiple years and are referred to as resident strains (Le Jeune et al., 2006). Particularly intriguing is the isolation of a single strain from samples from all regions, vintages and different varieties. This strain could be considered as a universal S. cerevisiae strain in Greek terroir. The genetic evaluation of this strain and its expansion all over Greece would be rather intriguing and further research is proposed. Martínez et al. (2004) propose a procedure that could be used as a tool for evaluating if a native isolate derives from the region where it was collected or if it is a strain derived from a commercial strain by microevolution. It is important to highlight that this strain showed different genomic fingerprint based on interdelta analysis among 5 different commercial strains and also it was isolated from a winery which has never used commercial strains. According to a previous study, two S. cerevisiae strains were isolated from the final stage of fermentation from different wineries of Beijing and possess important region-specific oenological characteristics (Sun et al., 2009). Finally, it is worth noting that 50% of the isolated strains were found in both white and red wines, suggesting that the assertion of certain strains being exclusively suitable for white or red vinification is questionable.
The aim of this study was to develop a novel, rapid and applicable method for preliminary yeast preliminary selection of alcoholic fermentation starters in wine. The proposed phenotypic classification method was validated by the results of fermentation kinetics and sensory evaluation of the tested S. cerevisiae strains. A future perspective is to focus on some of the strains that are presented in the current survey, perform bigger volume laboratory fermentations, large scale pilot fermentations and afterwards to propose new functional indigenous yeast to the wine industry. Additionally, there is no doubt that it would be quite interesting in the future to test the proposed classification test in a larger collection. Interestingly, the geographical distribution of the species revealed the presence of one ubiquitous strain with great oenological potential. Further research work could be done for the evaluation of this unique strain under large scale fermentation in order to examine the commercialization potential by the wine industry.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
AT: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. VT: Data curation, Formal analysis, Methodology, Software, Writing – original draft. IP: Funding acquisition, Project administration, Resources, Supervision, Visualization, Writing – review & editing. MD: Conceptualization, Investigation, Methodology, Project administration, Supervision, Visualization, Writing – review & editing, Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research has been co-financed by the European Union and Greek national funds through the Operational Program Competitiveness, Entrepreneurship and Innovation, under the call “Greece – Israel Call for Proposals for Joint R&D Projects 2019” (project code: T10ΔΙΣ-00060).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1301325/full#supplementary-material
Aponte, M., and Blaiotta, G. (2016). Potential role of yeast strains isolated from grapes in the production of Taurasi DOCG. Front. Microbiol. 7:809. doi: 10.3389/fmicb.2016.00809
Avramova, M., Cibrario, A., Peltier, E., Coton, M., Coton, E., Schacherer, J., et al. (2018). Brettanomyces bruxellensis population survey reveals a diploid-triploid complex structured according to substrate of isolation and geographical distribution. Sci. Rep. 8, 4136–4113. doi: 10.1038/s41598-018-22580-7
Aydin, Y. (2009). Isolation of cellulose producing Bacteria from wastes of vinegar fermentation. Proceedings of the world congress on, I, 20–23. Available at: http://www.iaeng.org/publication/WCECS2009/WCECS2009_pp99-102.pdf
Barata, A., Malfeito-Ferreira, M., and Loureiro, V. (2012). The microbial ecology of wine grape berries. Int. J. Food Microbiol. 153, 243–259. doi: 10.1016/j.ijfoodmicro.2011.11.025
Basa, K., Papanikolaou, S., Dimopoulou, M., Terpou, A., Kallithraka, S., and Nychas, G. J. E. (2022). Trials of commercial-and wild-type Saccharomyces cerevisiae strains under aerobic and microaerophilic/anaerobic conditions: ethanol production and must fermentation from grapes of Santorini (Greece) native varieties. Fermentation 8:249. doi: 10.3390/fermentation8060249
Belda, I., Ruiz, J., Alastruey-Izquierdo, A., Navascués, E., Marquina, D., and Santos, A. (2016). Unraveling the enzymatic basis of wine “Flavorome”: a phylo-functional study of wine related yeast species. Front. Microbiol. 7, 1–13. doi: 10.3389/fmicb.2016.00012
Bely, M., Stoeckle, P., Masneuf-Pomarède, I., and Dubourdieu, D. (2008). Impact of mixed Torulaspora delbrueckii – Saccharomyces cerevisiae culture on high-sugar fermentation. Int. J. Food Microbiol. 122, 312–320. doi: 10.1016/j.ijfoodmicro.2007.12.023
Benito, S., Hofmann, T., Laier, M., Lochbühler, B., Schüttler, A., Ebert, K., et al. (2015). Effect on quality and composition of Riesling wines fermented by sequential inoculation with non-Saccharomyces and Saccharomyces cerevisiae. Eur. Food Res. Technol. 241, 707–717. doi: 10.1007/s00217-015-2497-8
Binati, R. L., Innocente, G., Gatto, V., Celebrin, A., Polo, M., Felis, G. E., et al. (2019). Exploring the diversity of a collection of native non-saccharomyces yeasts to develop co-starter cultures for winemaking. Food Res. Int. 122, 432–442. doi: 10.1016/j.foodres.2019.04.043
Binati, R. L., Lemos Junior, W. J. F., Luzzini, G., Slaghenaufi, D., Ugliano, M., and Torriani, S. (2020). Contribution of non-Saccharomyces yeasts to wine volatile and sensory diversity: a study on Lachancea thermotolerans, Metschnikowia spp. and Starmerella bacillaris strains isolated in Italy. Int. J. Food Microbiol 318. doi: 10.1016/j.ijfoodmicro.2019.108470
Bleve, G., Tufariello, M., Durante, M., Grieco, F., Ramires, F. A., Mita, G., et al. (2015). Physico-chemical characterization of natural fermentation process of Conservolea and Kalamàta table olives and developement of a protocol for the pre-selection of fermentation starters. Food Microbiol. 46, 368–382. doi: 10.1016/j.fm.2014.08.021
Bonatsou, S., Benítez, A., Rodríguez-Gómez, F., Panagou, E. Z., and Arroyo-López, F. N. (2015). Selection of yeasts with multifunctional features for application as starters in natural black table olive processing. Food Microbiol. 46, 66–73. doi: 10.1016/j.fm.2014.07.011
Bonatsou, S., Paramithiotis, S., and Panagou, E. Z. (2018). Evolution of yeast consortia during the fermentation of Kalamata natural black olives upon two initial acidification treatments. Front. Microbiol. 8, 1–13. doi: 10.3389/fmicb.2017.02673
Capece, A., Pietrafesa, A., Pietrafesa, R., Garrigós, V., Tedesco, F., Romano, P., et al. (2022). Impact of Starmerella bacillaris and Zygosaccharomyces bailii on ethanol reduction and Saccharomyces cerevisiae metabolism during mixed wine fermentations. Food Res. Int. 159:111649. doi: 10.1016/j.foodres.2022.111649
Capece, A., Pietrafesa, R., Siesto, G., Romaniello, R., Condelli, N., and Romano, P. (2019). Selected indigenous saccharomyces cerevisiae strains as profitable strategy to preserve typical traits of primitivo wine. Fermentation 5:87. doi: 10.3390/fermentation5040087
Capece, A., Romaniello, R., Siesto, G., Pietrafesa, R., Massari, C., Poeta, C., et al. (2010). Selection of indigenous Saccharomyces cerevisiae strains for Nero d’Avola wine and evaluation of selected starter implantation in pilot fermentation. Int. J. Food Microbiol. 144, 187–192. doi: 10.1016/j.ijfoodmicro.2010.09.009
Caridi, A., Cufari, A., and Ramondino, D. (2002). Isolation and clonal pre-selection of enological Saccharomyces. J. Gen. Appl. Microbiol. 48, 261–267. doi: 10.2323/jgam.48.261
Christofi, S., Papanikolaou, S., Dimopoulou, M., Terpou, A., Cioroiu, I. B., Cotea, V., et al. (2022). Effect of yeast Assimilable nitrogen content on fermentation kinetics, wine chemical composition and sensory character in the production of Assyrtiko wines. Appl. Sci. 12:1405. doi: 10.3390/app12031405
Ciani, M., and Comitini, F. (2015). Yeast interactions in multi-starter wine fermentation. Curr. Opin. Food Sci. 1, 1–6. doi: 10.1016/j.cofs.2014.07.001
Ciani, M., Mannazzu, I., Marinangeli, P., Clementi, F., and Martini, A. (2004). Contribution of winery-resident Saccharomyces cerevisiae strains to spontaneous grape must fermentation. Anton. Leeuw. Int. J. Gen. Mol. Microbiol. 85, 159–164. doi: 10.1023/B:ANTO.0000020284.05802.d7
Clemente-Jimenez, J. M., Mingorance-Cazorla, L., Martínez-Rodríguez, S., Las Heras-Vázquez, F. J., and Rodríguez-Vico, F. (2005). Influence of sequential yeast mixtures on wine fermentation. Int. J. Food Microbiol. 98, 301–308. doi: 10.1016/j.ijfoodmicro.2004.06.007
Comitini, F., Agarbati, A., Canonico, L., and Ciani, M. (2021). Yeast interactions and molecular mechanisms in wine fermentation: a comprehensive review. Int. J. Mol. Sci. 22:7754. doi: 10.3390/ijms22147754
Comitini, F., Capece, A., Ciani, M., and Romano, P. (2017). New insights on the use of wine yeasts. Curr. Opin. Food Sci. 13, 44–49. doi: 10.1016/j.cofs.2017.02.005
Comitini, F., Gobbi, M., Domizio, P., Romani, C., Lencioni, L., Mannazzu, I., et al. (2011). Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 28, 873–882. doi: 10.1016/j.fm.2010.12.001
Cordero-Bueso, G., Esteve-Zarzoso, B., Cabellos, J. M., Gil-Díaz, M., and Arroyo, T. (2013). Biotechnological potential of non-Saccharomyces yeasts isolated during spontaneous fermentations of Malvar (Vitis vinifera cv. L.). Eur. Food Res. Technol. 236, 193–207. doi: 10.1007/s00217-012-1874-9
Courcoux, P., Qannari, E. M., and Faye, P. (2015). “Free sorting as a sensory profiling technique for product development” in Rapid sensory profiling techniques and related methods: Applications in new product development and consumer research (Sawston: Woodhead Publishing Limited)
de Ullivarri, M. F., Mendoza, L. M., and Raya, R. R. (2014). Killer activity of Saccharomyces cerevisiae strains: partial characterization and strategies to improve the biocontrol efficacy in winemaking. Anton. Leeuw. Int. J. Gen. Mol. Microbiol. 106, 865–878. doi: 10.1007/s10482-014-0256-7
de Ullivarri, M. F., Mendoza, L. M., Raya, R. R., and Farías, M. E. (2011). Killer phenotype of indigenous yeasts isolated from Argentinian wine cellars and their potential starter cultures for winemaking. Biotechnol. Lett. 33, 2177–2183. doi: 10.1007/s10529-011-0674-9
del Castillo, L., Lozano, M., Tâche, R., Bonnarme, P., and Landaud, S. (2007). Evaluation of a quantitative screening method for hydrogen sulfide production by cheese-ripening microorganisms: the first step towards l-cysteine catabolism. J. Microbiol. Methods 69, 70–77. doi: 10.1016/j.mimet.2006.12.001
Di Maro, E., Ercolini, D., and Coppola, S. (2007). Yeast dynamics during spontaneous wine fermentation of the Catalanesca grape. Int. J. Food Microbiol. 117, 201–210. doi: 10.1016/j.ijfoodmicro.2007.04.007
Dimopoulou, M., Goulioti, E., Troianou, V., Toumpeki, C., Paramithiotis, S., Gosselin, Y., et al. (2022). Effect of Saccharomyces cerevisiae and Saccharomyces pastorianus co-inoculation on alcoholic fermentation behavior and aromatic profile of sauvignon blanc wine. Fermentation 8:539. doi: 10.3390/fermentation8100539
Dimopoulou, M., Troianou, V., Toumpeki, C., Gosselin, Y., Dorignac, É., and Kotseridis, Y. (2020). Effect of strains from different Saccharomyces species used in different inoculation schemes on chemical composition and sensory characteristics of sauvignon blanc wine. Oeno One 54, 745–759. doi: 10.20870/OENO-ONE.2020.54.4.3240
Divol, B., Du Toit, M., and Duckitt, E. (2012). Surviving in the presence of Sulphur dioxide: strategies developed by wine yeasts. Appl. Microbiol. Biotechnol. 95, 601–613. doi: 10.1007/s00253-012-4186-x
Domizio, P., Romani, C., Lencioni, L., Comitini, F., Gobbi, M., Mannazzu, I., et al. (2011). Outlining a future for non-Saccharomyces yeasts: selection of putative spoilage wine strains to be used in association with Saccharomyces cerevisiae for grape juice fermentation. Int. J. Food Microbiol. 147, 170–180. doi: 10.1016/j.ijfoodmicro.2011.03.020
Ercolini, D., Moschetti, G., Blaiotta, G., and Coppola, S. (2001). The potential of a polyphasic PCR-DGGE approach in evaluating microbial diversity of natural whey cultures for water-buffalo mozzarella cheese production: Bias of culture-dependent and culture-independent analyses. Syst. Appl. Microbiol. 24, 610–617. doi: 10.1078/0723-2020-00076
Fleet, G. H. (2008). Wine yeasts for the future. FEMS Yeast Res. 8, 979–995. doi: 10.1111/j.1567-1364.2008.00427.x
Garofalo, C., Berbegal, C., Grieco, F., Tufariello, M., Spano, G., and Capozzi, V. (2018). Selection of indigenous yeast strains for the production of sparkling wines from native Apulian grape varieties. Int. J. Food Microbiol. 285, 7–17. doi: 10.1016/j.ijfoodmicro.2018.07.004
Garofalo, C., Tristezza, M., Grieco, F., Spano, G., and Capozzi, V. (2016). From grape berries to wine: population dynamics of cultivable yeasts associated to “Nero di Troia” autochthonous grape cultivar. World J. Microbiol. Biotechnol. 32, 59–10. doi: 10.1007/s11274-016-2017-4
Gobert, A., Tourdot-Maréchal, R., Morge, C., Sparrow, C., Liu, Y., Quintanilla-Casas, B., et al. (2017). Non-Saccharomyces yeasts nitrogen source preferences: impact on sequential fermentation and wine volatile compounds profile. Front. Microbiol. 8:2175. doi: 10.3389/fmicb.2017.02175
Gutiérrez, A., Beltran, G., Warringer, J., and Guillamón, J. M. (2013). Genetic basis of variations in nitrogen source utilization in four wine commercial yeast strains. PLoS One 8, e67166–e67113. doi: 10.1371/journal.pone.0067166
Harrigan, W. F., and McCance, M. E. (1976). Laboratory methods in food and dairy microbiology. Cambridge, MA: Academic Press Inc.
Jolly, N. P., Varela, C., and Pretorius, I. S. (2014). Not your ordinary yeast: non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 14, 215–237. doi: 10.1111/1567-1364.12111
Konate, M., Akpa, E. E., Koffi, L. B., Kra, K. A. S., Megnanou, R. M., and Niamke, S. (2014). Isolation of thermotolerant and high acetic acid-producing Acetobacter pasteurianus from Ivorian palm wine. Emir J Food Agric. 26, 773–785. doi: 10.9755/ejfa.v26i9.18122
Koutras, M., and Evagelaras, C. (2010). Regression analysis (theory and applications), Stamoulis edition, 656–686.
Lambrechts, M. G., and Pretorius, I. S. (2000). Yeast and its importance to wine aroma. Understanding Wine Chem. 21, 97–129.
Le Jeune, C., Erny, C., Demuyter, C., and Lollier, M. (2006). Evolution of the population of Saccharomyces cerevisiae from grape to wine in a spontaneous fermentation. Food Microbiol. 23, 709–716. doi: 10.1016/j.fm.2006.02.007
Legras, J. L., and Karst, F. (2003). Optimisation of interdelta analysis for Saccharomyces cerevisiae strain characterisation. FEMS Microbiol. Lett. 221, 249–255. doi: 10.1016/S0378-1097(03)00205-2
Lieckfeldt, E., Meyer, W., and Börner, T. (1993). Rapid identification and differentiation of yeasts by DNA and PCR fingerprinting. J. Basic Microbiol. 33, 413–425. doi: 10.1002/jobm.3620330609
Liu, G. L., Chi, Z., Wang, G. Y., Wang, Z. P., Li, Y., and Chi, Z. M. (2015). Yeast killer toxins, molecular mechanisms of their action and their applications. Crit. Rev. Biotechnol. 35, 222–234. doi: 10.3109/07388551.2013.833582
Liu, P. T., Lu, L., Duan, C. Q., and Yan, G. L. (2016). The contribution of indigenous non-Saccharomyces wine yeast to improved aromatic quality of cabernet sauvignon wines by spontaneous fermentation. Lwt 71, 356–363. doi: 10.1016/j.lwt.2016.04.031
Maicas, S. (2020). The role of yeasts in fermentation processes. Microorganisms 8, 1–8. doi: 10.3390/microorganisms8081142
Mansfield, A. K., Zoecklein, B. W., and Whiton, R. S. (2002). Quantification of glycosidase activity in selected strains of Brettanomyces bruxellensis and Oenococcus oeni. Am. J. Enol. Vitic. 53, 303–307. doi: 10.5344/ajev.2002.53.4.303
Martínez, C., Gac, S., Lavín, A., and Ganga, M. (2004). Genomic characterization of Saccharomyces Cerevisiae strains isolated from wine-producing areas in South America. J. Appl. Microbiol. 96, 1161–1168. doi: 10.1111/j.1365-2672.2004.02255.x
Mateo, J. J., Jiménez, M., Pastor, A., and Huerta, T. (2001). Yeast starter cultures affecting wine fermentation and volatiles. Food Res. Int. 34, 307–314. doi: 10.1016/S0963-9969(00)00168-X
Mestre Furlani, M. V., Maturano, Y. P., Combina, M., Mercado, L. A., Toro, M. E., and Vazquez, F. (2017). Selection of non-Saccharomyces yeasts to be used in grape musts with high alcoholic potential: a strategy to obtain wines with reduced ethanol content. FEMS Yeast Res. 17, 1–10. doi: 10.1093/femsyr/fox010
OIV . (2023). International organization of vine and wine. Available at: https://www.oiv.int/ (Assessed July 20, 2023)
Palma, M., Guerreiro, J. F., and Sá-Correia, I. (2018). Adaptive response and tolerance to acetic acid in Saccharomyces cerevisiae and Zygosaccharomyces bailii: a physiological genomics perspective. Front. Microbiol. 9, 1–16. doi: 10.3389/fmicb.2018.00274
Parapouli, M., Vasileiadis, A., Afendra, A. S., and Hatziloukas, E. (2020). Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 6, 1–31. doi: 10.3934/microbiol.2020001
Perpetuini, G., Tittarelli, F., Battistelli, N., Suzzi, G., and Tofalo, R. (2020). Contribution of Pichia manshurica strains to aroma profile of organic wines. Eur. Food Res. Technol. 246, 1405–1417. doi: 10.1007/s00217-020-03499-8
Pulcini, L., Gamalero, E., Costantini, A., Tommaso Vaudano, E., Tsolakis, C., and Garcia-Moruno, E. (2022). An overview on Saccharomyces cerevisiae indigenous strains selection methods. Grapes Wine. doi: 10.5772/intechopen.99095
Puyo, M., Simonin, S., Bach, B., Klein, G., Alexandre, H., and Tourdot-Maréchal, R. (2023). Bio-protection in oenology by Metschnikowia pulcherrima: from field results to scientific inquiry. Front. Microbiol. 14, 1–11. doi: 10.3389/fmicb.2023.1252973
Querol, A., Pérez-Torrado, R., Alonso-del-Real, J., Minebois, R., Stribny, J., Oliveira, B. M., et al. (2018). New trends in the uses of yeasts in oenology. Adv. Food Nutr. Res. 85:2. doi: 10.1016/bs.afnr.2018.03.002
Reiter, T., Montpetit, R., Byer, S., Frias, I., Leon, E., Viano, R., et al. (2021). Saccharomyces Cerevisiae gene expression during fermentation of pinot noir wines at an industrially relevant scale. Appl. Environ. Microbiol. 87, 1–21. doi: 10.1128/AEM.00036-21
Ribéreau-Gayon, Pascal, Denis Dubourdieu, B., and Donèche, A. L. (2006). Handbook of enology, volume 1: The microbiology of wine and Vinifications.
Rodríguez, M. E., Lopes, C. A., Van Broock, M., Valles, S., Ramón, D., and Caballero, A. C. (2004). Screening and typing of Patagonian wine yeasts for glycosidase activities. J. Appl. Microbiol. 96, 84–95. doi: 10.1046/j.1365-2672.2003.02032.x
Rossouw, D., Jolly, N., Jacobson, D., and Bauer, F. F. (2012). The effect of scale on gene expression: commercial versus laboratory wine fermentations. Appl. Microbiol. Biotechnol. 93, 1207–1219. doi: 10.1007/s00253-011-3564-0
Rossouw, D., and Bauer, E. F. (2016). Exploring the phenotypic space of non-Saccharomyces wine yeast biodiversity. Food Microbiol. 55, 32–46. doi: 10.1016/j.fm.2015.11.017
Roudil, L., Russo, P., Berbegal, C., Albertin, W., Spano, G., and Capozzi, V. (2020). Non-Saccharomyces commercial starter cultures: scientific trends, recent patents and innovation in the wine sector. Recent Patents Food Nutr. Agric. 11, 27–39. doi: 10.2174/2212798410666190131103713
Sadoudi, M., Tourdot-Maréchal, R., Rousseaux, S., Steyer, D., Gallardo-Chacón, J. J., Ballester, J., et al. (2012). Yeast-yeast interactions revealed by aromatic profile analysis of sauvignon blanc wine fermented by single or co-culture of non-Saccharomyces and Saccharomyces yeasts. Food Microbiol. 32, 243–253. doi: 10.1016/j.fm.2012.06.006
Sáenz-Navajas, M. P., Campo, E., Avizcuri, J. M., Valentin, D., Fernández-Zurbano, P., and Ferreira, V. (2012). Contribution of non-volatile and aroma fractions to in-mouth sensory properties of red wines: wine reconstitution strategies and sensory sorting task. Anal. Chim. Acta 732, 64–72. doi: 10.1016/j.aca.2011.12.042
Settanni, L., Sannino, C., Francesca, N., Guarcello, R., and Moschetti, G. (2012). Yeast ecology of vineyards within Marsala wine area (western Sicily) in two consecutive vintages and selection of autochthonous Saccharomyces cerevisiae strains. J. Biosci. Bioeng. 114, 606–614. doi: 10.1016/j.jbiosc.2012.07.010
Sha, S. P., Suryavanshi, M. V., Jani, K., Sharma, A., Shouche, Y., and Tamang, J. P. (2018). Diversity of yeasts and molds by culture-dependent and culture-independent methods for Mycobiome surveillance of traditionally prepared dried starters for the production of Indian alcoholic beverages. Front. Microbiol. 9:2237. doi: 10.3389/fmicb.2018.02237
Sidari, R., Ženišová, K., Tobolková, B., Belajová, E., Cabicarová, T., Bučková, M., et al. (2021). Wine yeasts selection: laboratory characterization and protocol review. Microorganisms 9:2223. doi: 10.3390/microorganisms9112223
Soden, A., Francis, I. L., Oakey, H., and Henschke, P. A. (2000). Effects of co-fermentation with Candida stellata and Saccharomyces cerevisiae on the aroma and composition of chardonnay wine. Aust. J. Grape Wine Res. 6, 21–30. doi: 10.1111/j.1755-0238.2000.tb00158.x
Suárez-Lepe, J. A., and Morata, A. (2012). New trends in yeast selection for winemaking. Trends Food Sci. Technol. 23, 39–50. doi: 10.1016/j.tifs.2011.08.005
Sun, H., Ma, H., Hao, M., Pretorius, I. S., and Chen, S. (2009). Identification of yeast population dynamics of spontaneous fermentation in Beijing wine region, China. Ann. Microbiol. 59, 69–76. doi: 10.1007/BF03175601
Thorndike, R. L. (1953). Who belongs in the family? Psychometrika 18, 267–276. doi: 10.1007/BF02289263
Tronchoni, J., Gonzalez, R., Guindal, A. M., Calleja, E., and Morales, P. (2022). Exploring the suitability of Saccharomyces cerevisiae strains for winemaking under aerobic conditions. Food Microbiol. 101:103893. doi: 10.1016/j.fm.2021.103893
Velázquez, R., Zamora, E., Álvarez, M., Álvarez, M. L., and Ramírez, M. (2016). Using mixed inocula of Saccharomyces cerevisiae killer strains to improve the quality of traditional sparkling-wine. Food Microbiol. 59, 150–160. doi: 10.1016/j.fm.2016.06.006
Vilela, A. (2020). Modulating wine pleasantness throughout wine-yeast co-inoculation or sequential inoculation. Fermentation 6:22. doi: 10.3390/fermentation6010022
Windholtz, S., Dutilh, L., Lucas, M., Maupeu, J., Vallet-Courbin, S., Farris, L. M., et al. (2022). Population dynamics and yeast diversity in early winemaking stages without sulfites revealed by three complementary approaches applied sciences population dynamics and yeast diversity in early winemaking stages. Appl. Sci. 11:2494. doi: 10.20870/IVES-TR.2022.5534
Keywords: Greek terroir, phenotypic diversity, indigenous yeast, wine yeast selection, biostatistics tools
Citation: Tzamourani AP, Taliadouros V, Paraskevopoulos I and Dimopoulou M (2023) Developing a novel selection method for alcoholic fermentation starters by exploring wine yeast microbiota from Greece. Front. Microbiol. 14:1301325. doi: 10.3389/fmicb.2023.1301325
Received: 24 September 2023; Accepted: 05 December 2023;
Published: 20 December 2023.
Edited by:
Maria Aspri, Cyprus University of Technology, CyprusReviewed by:
Shankar Prasad Sha, Kurseong College, IndiaCopyright © 2023 Tzamourani, Taliadouros, Paraskevopoulos and Dimopoulou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Dimopoulou, bWRpbW9wb3Vsb3VAdW5pd2EuZ3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.