- 1Department of Gastroenterology and Hepatology, University of Groningen, University Medical Center Groningen, Groningen, Netherlands
- 2Department of Medical Microbiology and Infection Prevention, University of Groningen, University Medical Center Groningen, Groningen, Netherlands
- 3School of Medicine and Medical Science and the Conway Institute, University College Dublin, Dublin, Ireland
Introduction: Intestinal epithelial cells produce interleukin-18 (IL-18), a key factor in promoting epithelial barrier integrity. Here, we analyzed the potential role of gut bacteria and the hypoxia-inducible factor 1α (HIF1α) pathway in regulating mucosal IL18 expression in inflammatory bowel disease (IBD).
Methods: Mucosal samples from patients with IBD (n = 760) were analyzed for bacterial composition, IL18 levels and HIF1α pathway activation. Wild-type Caco-2 and CRISPR/Cas9-engineered Caco-2-HIF1A-null cells were cocultured with Faecalibacterium prausnitzii in a “Human oxygen-Bacteria anaerobic” in vitro system and analyzed by RNA sequencing.
Results: Mucosal IL18 mRNA levels correlated positively with the abundance of mucosal-associated butyrate-producing bacteria, in particular F. prausnitzii, and with HIF1α pathway activation in patients with IBD. HIF1α-mediated expression of IL18, either by a pharmacological agonist (dimethyloxallyl glycine) or F. prausnitzii, was abrogated in Caco-2-HIF1A-null cells.
Conclusion: Butyrate-producing gut bacteria like F. prausnitzii regulate mucosal IL18 expression in a HIF1α-dependent manner that may aid in mucosal healing in IBD.
1 Introduction
The human intestinal lumen harbors a complex ecosystem of microbial species, collectively named microbiota, that play a crucial role in gut homeostasis and are increasingly recognized as controllers of human health and disease (Manor et al., 2020). The intestinal epithelium is the barrier between the gut microbiota and the surrounding tissue. Interactions between the luminal bacteria and the adjacent epithelium control the homeostatic balance between the host and intestinal microbes. Disturbances in the composition of the microbiota are collectively named dysbiosis (Schippa and Conte, 2014; Ananthakrishnan, 2015) and have been associated with a variety of gastrointestinal and metabolic diseases, including type-2 diabetes, colon carcinoma and inflammatory bowel diseases (IBD) (Kostic et al., 2014; Schippa and Conte, 2014; Vich Vila et al., 2018). Such diseases are typically associated with a reduction in the dominant commensal Faecalibacterium prausnitzii, which is a strict anaerobic bacterium and a prominent producer of the short-chain fatty acid butyrate (Sokol et al., 2008; Cao et al., 2014).
The intestinal epithelium is in a constant state of physiological hypoxia due to its proximity to the anaerobic gut lumen that, in turn, drives the activation of the hypoxia-inducible factors 1α and 2α (HIF1α/HIF2α) (Fagundes and Taylor, 2017). HIF1α activation in the intestine transcriptionally promotes expression of genes encoding proteins that are involved in metabolic processes, barrier function and immune cell regulation, playing a key role in controlling intestinal homeostasis (Colgan et al., 2020). Under normoxic conditions, HIF1α protein is repressed and rapidly degraded through the action of asparaginyl- and prolyl-hydroxylases (FIH and PHD1-3, respectively) that utilizes oxygen as a cofactor for the hydroxylation of HIF1α. The activity of these hydroxylases is impaired during hypoxia, thereby preventing HIF1α repression and degradation that then translocate to the nucleus to activate gene transcription. Intestinal inflammation, characteristic in IBD, additionally creates a pathophysiological state of hypoxia that further activates HIF1α signaling to restore intestinal homeostasis (Colgan and Taylor, 2010; Cummins et al., 2016).
A variety of pro- and anti-inflammatory cytokines are produced to fight inflammation and promote tissue regeneration. Several cytokines are produced and secreted by the intestinal epithelium itself (Mahapatro et al., 2021). Interleukin-18 (IL-18; previously known as interferon-γ activating factor) is such a cytokine expressed by intestinal epithelial cells that stimulates epithelial barrier integrity, repair and tolerance of neighboring immune cells in the intestinal mucosa, thus acting as an anti-inflammatory cytokine (Zaki et al., 2010; Elinav et al., 2011; Normand et al., 2011; Mantovani et al., 2019; Yasuda et al., 2019). Balancing mucosal IL-18 levels is, however, of crucial importance, as it is a pro-inflammatory factor at sites of inflammation (Nakamura et al., 1989; Okamura et al., 1995; Tsutsumi et al., 2019). IL-18 is an IL-1-type cytokine produced as pro-IL-18 and processed intracellularly by the nucleotide-binding oligomerization domain (NLRP) inflammasome prior to secretion (Sellin et al., 2015). The NLRP inflammasome is composed of three subunits: the NLRP3 (or 6) protein as a platform, the adaptor protein ASC (apoptosis-associated speck-like protein containing a CARD) and CASP1 (Caspase-1) as the effector/proteolytic domain. Mice deficient in either NLRP3/6, CASP1 or IL-18 show exacerbated response to DSS-induced colitis (Zaki et al., 2010). Moreover, single nucleotide polymorphisms (SNPs) in the IL-18 receptor (IL18R1) and accessory protein (IL18RAP) loci are associated with both adult and severe early-onset IBD (Kim et al., 2014; Andreoletti et al., 2015; Soderquest et al., 2017). On the other hand, plasma levels IL-18 are elevated in ulcerative colitis patients (Wiercinska-Drapalo et al., 2005); and murine models revealed a paradoxical role for IL-18 in colitis, depending on disease stage (Nowarski et al., 2015; Pu et al., 2019). Interestingly, IL-18 was also shown to affect the gut microbiome composition directly (Levy et al., 2015; Liu et al., 2018).
We hypothesize that HIF1α transcriptional activation regulates the intestinal epithelial IL-18 production, stimulated by butyrate-producing bacteria and a potential target to promote mucosal healing in IBD. Here, we investigated the causal and functional connection between epithelial IL18 expression, gut bacteria and HIF1α activation.
2 Materials and methods
2.1 Study population and ethical considerations
All patients included in this study were recruited by the IBD center of the of the Department of Gastroenterology and Hepatology of the University Medical Center Groningen (UMCG). Patients consented to participate in the 1000IBD project and Dutch parelsnoer IBD Biobank (Spekhorst et al., 2017; Imhann et al., 2019), and were at least 18-years old and had an established pre-existing diagnosis of IBD for at least 1 year. The study was approved by the Institutional Review Board (IRB) of the University Medical Center Groningen (UMC Groningen) (in Dutch: “Medisch Ethische Toetsingscommissie,” METc; IRB no. 2008/338 and 2016/424). The study was performed in accordance with the principles of the Declaration of Helsinki (2013). Diagnosis was based upon clinical, laboratory, endoscopic and histopathological criteria, the latter criteria also used for determining the inflammatory status of collected tissues (Lennard-Jones, 1989).
2.2 Intestinal biopsies collection
Intestinal biopsies were collected within the context of the 1000IBD project and Dutch parelsnoer IBD Biobank at time of colonoscopy procedures, which were part of standard clinical care. Intestinal biopsies were snap-frozen in liquid nitrogen shortly after the colonoscopy procedure. Biopsies were stored at −80°C until further processing. In total, 760 intestinal biopsies from 371 patients with IBD were analyzed in this study. During the endoscopic procedure, the inflammatory status of the biopsy was macroscopically categorized by assessing the mucosal morphology. Macroscopic inflammation was characterized by redness and edema, with or without ulceration of the intestinal mucosa. Additionally, histopathological evaluation later confirmed the macroscopic inflammation status.
2.3 DNA/RNA isolation and RNA sequencing
Isolation of DNA and RNA was performed using the AllPrep DNA/RNA mini kit (Qiagen; reference number: 80204) according to the manufacturer’s protocol. Homogenization of intestinal biopsies was performed in RLT lysis buffer including β-mercaptoethanol using the Qiagen Tissue Lyser with stainless steel beads (diameter of 5 mm; reference number: 69989). Sample preparation was performed using the BioScientific NextFlex mRNA sample preparation kit. RNA sequencing and data processing were performed as described earlier (Hu et al., 2021). Briefly, sequencing was executed with the Illumina NextSeq500 sequencer. Sampling and sequencing were performed in two different batches. RNA samples were pseudo-randomized on plates to mitigate potential batch effects. On average, 25 million reads were generated per sample. The quality of the raw reads was checked using FastQC at default parameters (ref v0.11.7). The adaptors identified by FastQC were clipped using Cutadapt (ref v1.1) with default settings. Sickle (ref v1.200) was used to trim low-quality ends from the reads (length < 25 nucleotides, quality < 20). Reads were aligned to the human genome (human_g1k_v37) using HISAT (ref v0.1.6) (2 mismatches allowed) and read sorting was done by SAMtools (ref v0.1.19). SAMtools flagstat and Picard tools (ref v2.9.0) were used to obtain mapping statistics. Seven samples with low percentage read alignment (<90%) were removed. Finally, gene expression was estimated through HTSeq (ref 0.9.1) based on Ensemble version 75 annotation. Gene-level expression data were normalized using a trimmed mean of M values, and log2 normalization was applied. From the RNA sequencing data, expression levels of IL18, HIF1A, HIF2A, HIF-hydroxylases (EGLN1, EGLN2 and EGLN3), VHL and HIF1AN were selected for analysis. In addition, HIF1α and HIF2α scores (quotients calculated using the mRNA levels of either HIF-1α or HIF-2α divided by the sum total of the negative regulators of these factors), were calculating as described before (Brown et al., 2020; Fagundes et al., 2022).
2.4 16S rRNA gene sequencing
Biopsy-adherent bacteria abundances was assessed by 16S rRNA sequencing of human intestinal biopsies. Amplification of DNA extracted from biopsies was performed by PCR using modified 341F and 806R primers with a six-nucleotide barcode on the 806R primer on V3-V4 hypervariable region of the 16S rRNA gene. Then it was subjected to Illumina MiSeq paired-end sequencing as described previously (Swarte et al., 2020). The raw reads were trimmed and filtered using Trimmomatic v0.33 to obtain an average quality of 25 and a minimum length of 50 bases. Taxonomic assignment was following the pipeline https://benjjneb.github.io/dada2/. Dada2 R package (v1.03) was used to get the amplicon sequence variants (ASVs). ASVs that were not present in 10% of all samples were filtered out. The rest were classified against SILVA database (release 132). The taxa with present rate > 10% was kept and centered log-ratio (clr) transformed for further analysis.
2.5 Human cell culture and CRISPR/Cas9 editing
Human epithelial colon adenocarcinoma cells (Caco-2, male, ATCC®, HTB-37TM, Manassas, United States) tested negative for mycoplasma infection and were used as in vitro representatives of intestinal epithelial cells. Single guide RNAs for HIF1A (sequence 5′-GATGGTAAGCCTCATCACAG-3′) were designed via Benchling© online platform (https://www.benchling.com/) and cloned in pLenti-sgRNA backbone (Addgene, 71409). For lentivirus production, HEK293T cells in T25 were transfected using 15 μL Fugene HD (Promega, E2311), 250 μL Opti-MEM, 1.5 μg Lenti-iCas9-neo (Addgene, 85400) or pLenti-sgRNA (see above), 1 μg pMD2.G (Addgene, 12259) and 2.5 μg psPAX2 (Addgene 12260). Medium was refreshed 24 h after transfection and changed for Glutamax™ Dulbecco’s Modified Eagle Medium (DMEM, ThermoFisher Scientific Inc), supplemented with 10% fetal calf serum (FCS, Invitrogen), 1% non-essential amino acids (NEAA, Gibco) and 1% PSF antibiotic cocktail [penicillin (10 U/mL), streptomycin sulfate (100 μg/mL) and fungi zone; Lonza, Bazel, Switzerland]. Lentivirus suspension was collected 48 h after transfection, filtered through a 0.45 μm filter, aliquoted and stored at −80°C.
For generating stable Caco-2-iCas9 cell line, Caco-2 cells were seeded in a 6-well plate and incubated with 1 mL lentivirus suspension, supplemented with 8 μg/mL polybrene (Sigma-Aldrich, TR-1003). Cells were washed 24 h after infection with PBS and selection was started with Neomycin 2 mg/mL for 7 days with one cell passage in between. For introducing sgRNA, Caco-2-iCas9 cells were seeded in a 6-well plate and incubated with 1 mL lentivirus containing sgRNA (empty vector or against HIF1A – described above), supplemented with 8 μg/mL polybrene. Cells were washed 24 h after infection with PBS and selection was started 20 μg/mL puromycin for 7 days. Once puromycin treatment was finished, the resulting mutant Caco-2 cell line was cultured at low density on Glutamax™ DMEM, as described above. Twenty one clones were picked from the culture dish and cultured individually. Clones were expanded and DNA, RNA and protein collected for sequencing and functional validation of stable transfection. Treatments with control or Dimethyloxalylglycine (DMOG; D3695, Sigma Aldrich), at the concentrations of 1 or 5 mM, were carried out on selected clones for 24 h, followed by RNA isolation using TRIzol method, cDNA synthesis and gene expression analysis by RT-PCR, as described by Sadaghian Sadabad et al. (2015). Sequences for primers and probes are shown on Supplementary Table S1.
2.6 Gaussia luciferase assay
A HIF1α-responsive element (HRE)-Gaussia Luciferase reporter system (Cavadas et al., 2018) was used to assess HIF1α functional activity in Caco-2-HIF1A-null cells and empty vector controls. Caco-2 cells were transfected with 1 mg of the HRE-Gaussia Luciferase reporter construct in antibiotic-free media using Lipofectamine 2000 for 24 h, after which cells were treated with 1 mM DMOG for up to 48 h. Secreted luciferase was indicative of HRE-activity. Bioluminescence was quantified in media samples using the Pierce™ Gaussia Luciferase Glow Assay Kit (Thermo Scientific, 16161), carried out in a 96-well plate as per manufacturer’s instructions. Luminescence was determined using the CLARIOstar microplate reader (BMG Labtech, Ortenberg, Germany). Relative Luminescence Units (RLU) are presented normalized to total protein content (μg) for comparison of HIF1α functional activity levels.
2.7 Bacterial culture and HoxBan coculture system
HoxBan coculturing was performed essentially as described before (Sadaghian Sadabad et al., 2015): anaerobically-grown F. prausnitzii strain A2-165 (provided by S. Duncan, Aberdeen, UK) was inoculated in liquid broth containing yeast extract, casitone, and fatty acids and supplemented with 25 mM glucose as carbon and energy source (YCFAG). One (1) ml of an overnight F. prausnitzii culture was used to inoculate 1 liter of autoclaved agar (1.0%)-based-YCFAG (pH = 6.5) and cooled-down to approximately 40°C. In an anaerobic cabinet, 40 mL of this mixture was added to a 50 mL Falcon test tube and the bacteria-containing YCFAG-agar was allowed to solidify. After transfer to a laminar flow cabinet, 10 mL of 37°C -pre-warmed PSF-free DMEM medium was added to each tube. Next, the Caco-2 cells (Caco-2-HIF1A-null or Caco-2-empty vector) pre-grown on coverslips were laid upside-down on the top of the bacteria-containing YCFAG agar medium. The screw caps of the Falcon tubes were kept loosely tightened, to allow oxygen entry into the system for the Caco-2 cells. Caco-2/F. prausnitzii coculturing took place for 18 h at 37°C and 5% CO2, after which coverslips were collected and processed for downstream analyses.
2.8 RNA sequencing of cultured cell lines
Isolation of RNA from Caco-2 cells after HoxBan (co)culturing was performed using the RNeasy Plus Mini Kit (Qiagen; reference number: 74134) according to the manufacturer’s protocol. Homogenization of cells was performed in RLT lysis buffer containing β-mercaptoethanol. Sample preparation was performed using the QuantSeq 3’mRNA-Seq Library Prep Kit sample preparation kit (Lexogen; reference number 015.95). RNA sequencing was executed with the Illumina NextSeq500 sequencer. RNA samples were pseudo-randomized on plates.
2.9 Statistics
The associations between gene expressions and microbial taxa in 760 intestinal biopsies from 371 IBD patients were assessed using mixed linear models adjusting for batch, age, sex, BMI, smoking, tissue location and inflammation status. Patient IDs were included as random effect to account for repeat measurements. Data were presented as mean ± standard deviation (SD). Assessment of normality was performed using histograms and normal probability plots (Q-Q plots). Differential gene expression analysis between groups was analyzed using independent sample t-tests with the EdgeR package on RStudio (version 1.4.1717; Rstudio, Boston, MA, United States) or ordinary One-way ANOVA using GraphPad Prism 9.0 (GraphPad software, San Diego, CA, United States). Gene expression was normalized using the trimmed mean of M-values normalization method (TMM) and then 2log-transformation was applied. Finally, gene expression means were centered to zero and standard deviations scaled to one. Data visualization was performed using GraphPad Prism 9.0 and Rstudio. Two-tailed p-values ≤ 0.05 were considered statistically significant.
3 Results
3.1 Gene expression levels of IL18 correlate with bacterial abundance in the intestinal mucosa of IBD patients
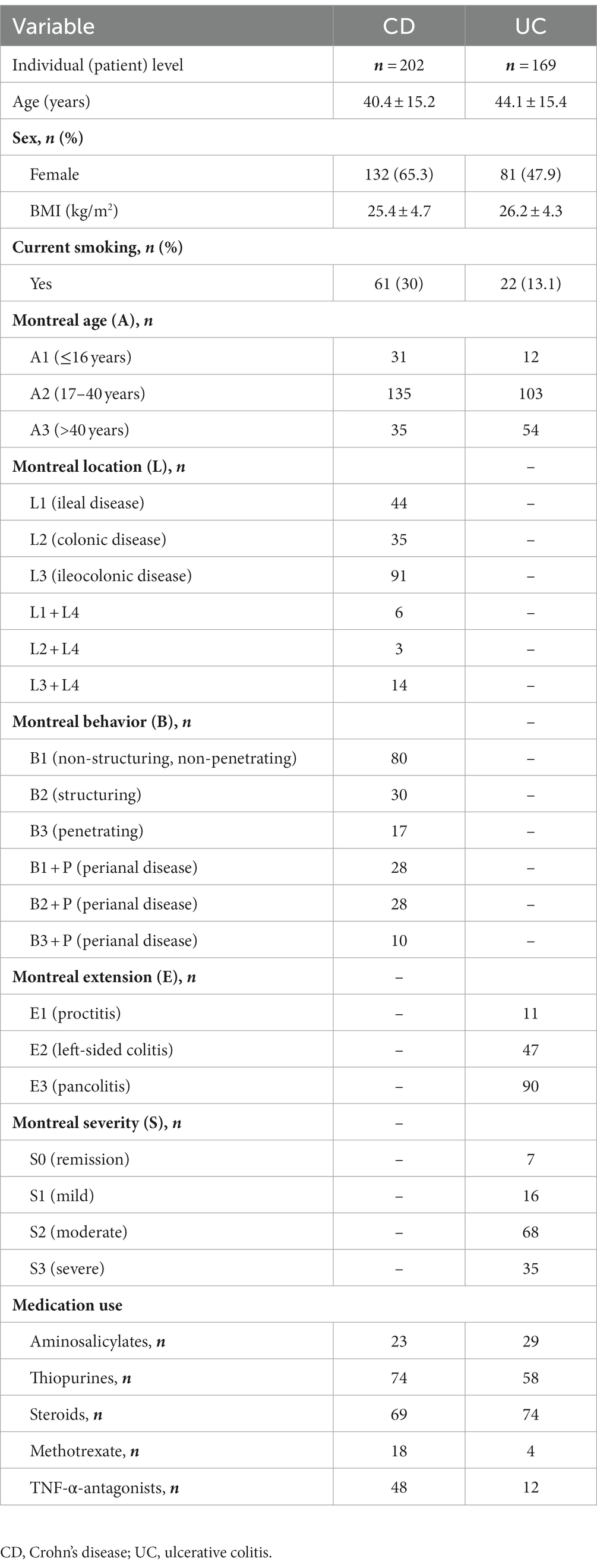
Descriptive statistics of the patient cohort is described in Table 1. Mucosal IL18 expression in 760 intestinal mucosa biopsies from 371 IBD patients was obtained from an in-house dataset of RNA sequencing dataset (Hu et al., 2022) and was correlated to the abundance of bacterial genera, as established by 16S rRNA gene sequencing of the same biopsies. A significant positive correlation between IL18 expression and mucosal levels of prominent butyrate-producing bacteria was observed, including Ruminococcaceae, Ruminococcus species, Agathobacter, and Faecalibacterium (Table 2). In contrast, IL18 expression correlated negatively with GCA-900066575, Sutterella and Bacteroides abundances (Table 2). Species from the Sutterella genus have been described as potential pathogens associated with inflammatory diseases (Gryaznova et al., 2021). At the species level, a significant positive correlation was observed between IL18 expression and F. prausnitzii (Figure 1, effect size = 0.227, p = 7.18 × 10−4). While no significant differences were observed in mucosal-associated F. prausnitzii abundances between inflamed and non-inflamed tissue (Table 3), we identified correlations between the abundance of F. prausnitzii and certain bacterial genera outlined in Table 2 (Supplementary Figure S2). Given the positive association of IL18 expression with butyrate-producing bacteria, and since butyrate activates the epithelial HIF1α pathway (Kelly et al., 2015), we next investigated whether mucosal IL18 expression correlates with HIF1α pathway activation.
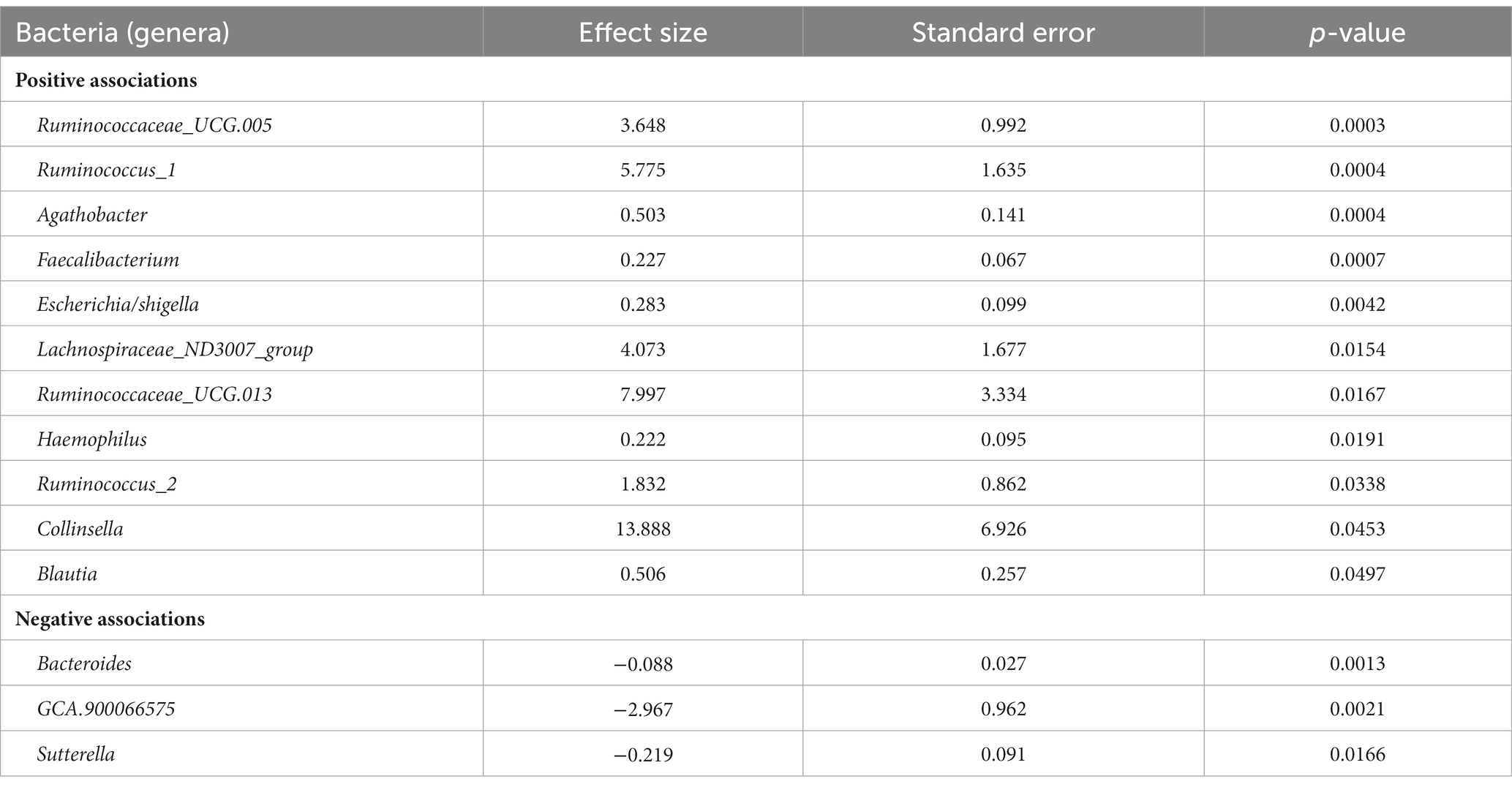
Table 2. Bacteria significantly correlated with IL18 expression in intestinal mucosa of IBD patients ordered by significance.
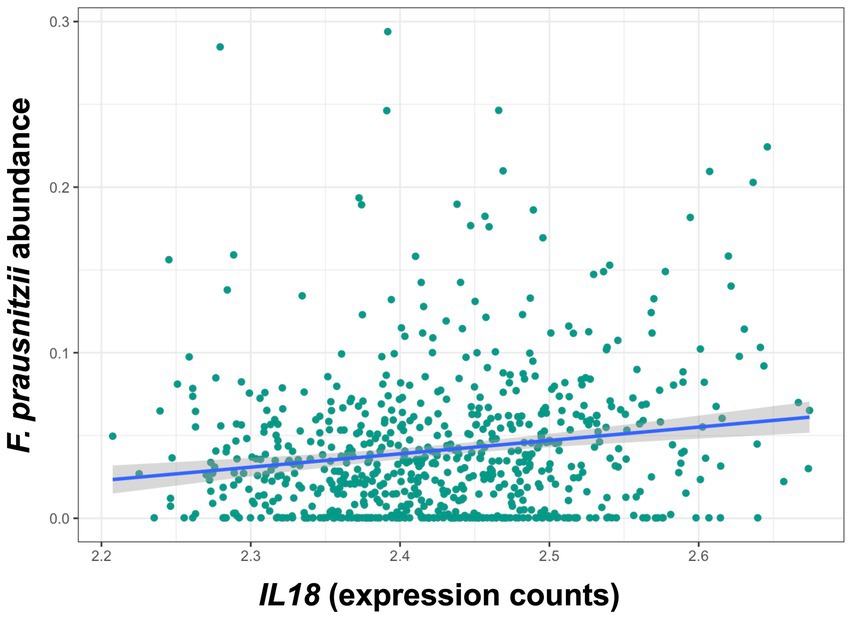
Figure 1. IL18 expression levels are significantly associated with abundances of F. prausnitzii in human intestinal mucosa. F. prausnitzii abundance is positively correlated to IL18 expression in intestinal mucosa. Patient cohort study performed in N = 760 biopsies.
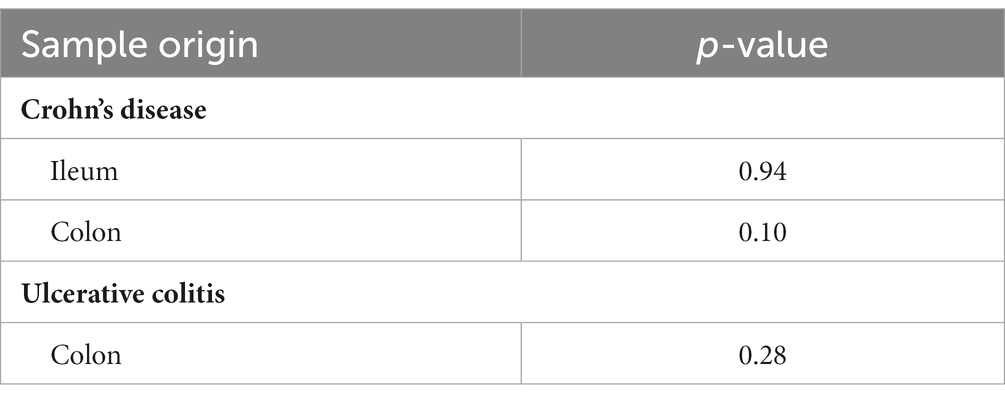
Table 3. Comparison between F. prausnitzii abundances between inflamed and non-inflamed mucosa tissue of IBD patients.
3.2 IL18 expression correlates with HIF1α activity in intestinal mucosa of IBD patients
IL18 mRNA levels were significantly enhanced in inflamed intestinal mucosa when compared to non-inflamed tissue, both in colon and in ileum (Figure 2A). Similarly, HIF1α scores [representing HIF1α activation capacity (Brown et al., 2020)] were increased in inflamed mucosa in both locations, when compared to non-inflamed mucosa (Figure 2B). In line, Spearman correlation analysis revealed a positive association between IL18 and HIF1α scores in the intestinal mucosa of IBD patients (Figure 2C; r = 0.2052 and p < 0.0001).
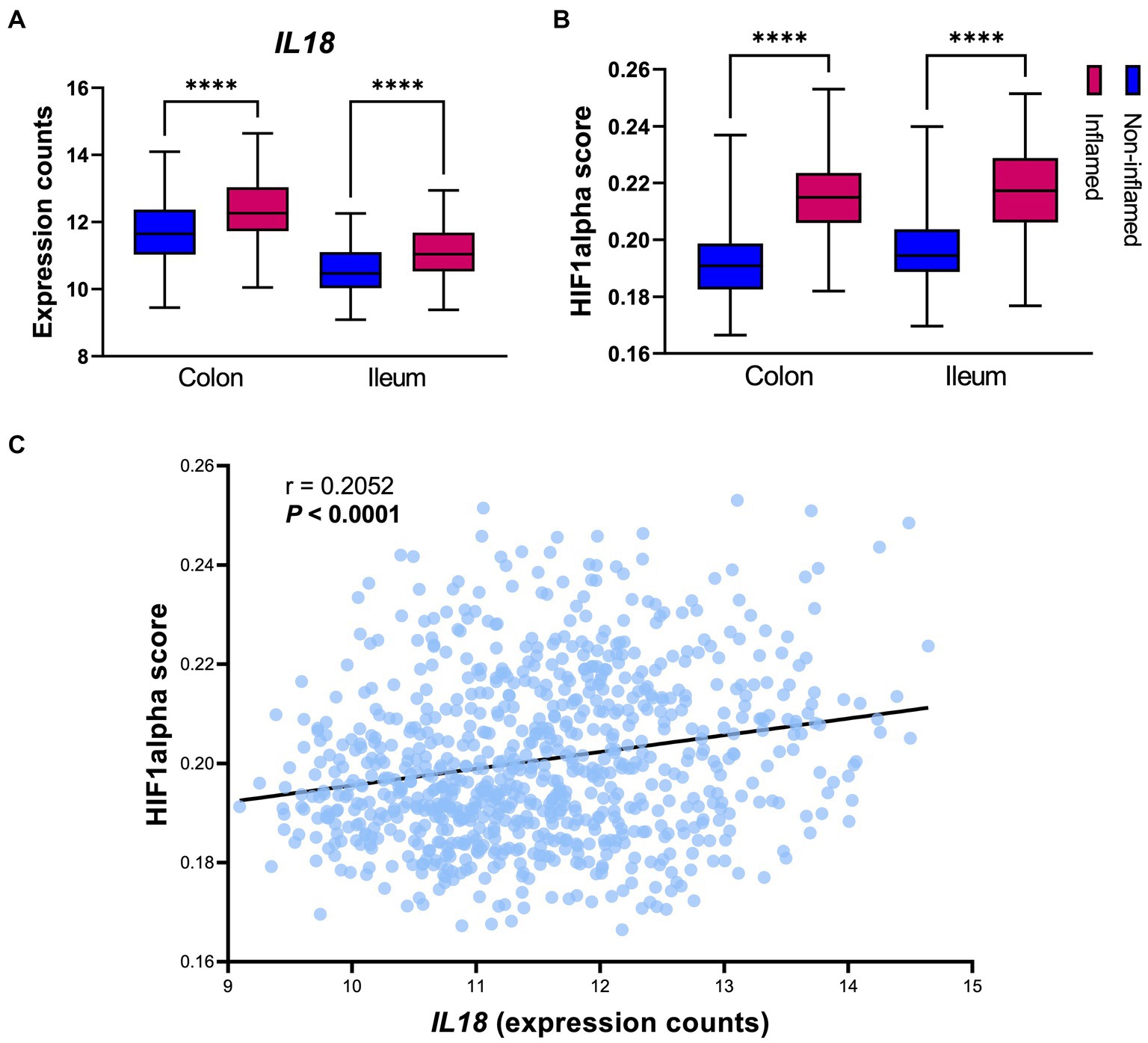
Figure 2. IL18 expression strongly associates to HIF1α activation capacity in the human intestinal mucosa. (A) Gene expression levels of IL18 in intestinal mucosa of IBD patients, divided by location (colon and ileum). (B) HIF1α activation capacity (calculated by HIF1α scores) in intestinal mucosa of IBD patients. (C) Spearman correlation analysis shows significant positive association between IL18 and HIF1α scores in intestinal mucosa of IBD patients. Patient cohort study performed in n = 760 samples (biopsies), segregated by their inflammatory statues and biopsy location as colonic inflamed (n = 195), colonic non-inflamed (n = 307), ileal inflamed (n = 78) and ileal non-inflamed (n = 180) biopsies. Data presented as box and whiskers (min to max); ****p < 0.0001.
3.3 HIF1A knock out in intestinal epithelial cells
In order to establish whether IL18 expression is under direct control of HIF1α, we inactivated the HIF1A gene in Caco-2 cells using CRISPR/Cas9 technology (Figure 3). We functionally tested the loss of HIF1α activation in the Caco-2-HIF1A-null cells using a secreted Gaussia luciferase driven by HIF1α responsive elements (HRE) over 48 h exposition to 1 mM of the prolyl-hydroxylase inhibitor DMOG (Figure 3A). We observed an almost complete loss of HIF1α response in Caco-2-HIF1A-null cells, compared to Caco-2(-iCas9-empty vector) control cells, which had HRE-luciferase signal up to 10-fold upregulated of over 48 h exposition to DMOG. Because of the mutation introduced by the guide RNA, HIF1A mRNA remained detectable in Caco-2-HIF1A-null cells, with no significant difference compared to Caco-2-empty vector control cells (Figure 3B). Pharmacological activation of HIF1α (using DMOG) dose-dependently enhanced mRNA levels of typical HIF1α target genes EGLN3 and PGK1 in Caco-2 control cells, which was blunted in the Caco-2-HIF1A-null cells (Figures 3C,D). Similarly, IL18 expression was slightly, but significantly induced by DMOG in Caco-2 control cells, which was blocked in the absence of HIF1A (Figure 3E). Interestingly, basal IL18 levels were already significantly lower in Caco-2-HIF1A-null cells compared to Caco-2 control cells, which was also observed for PGK1 (Figures 3D,E, respectively).
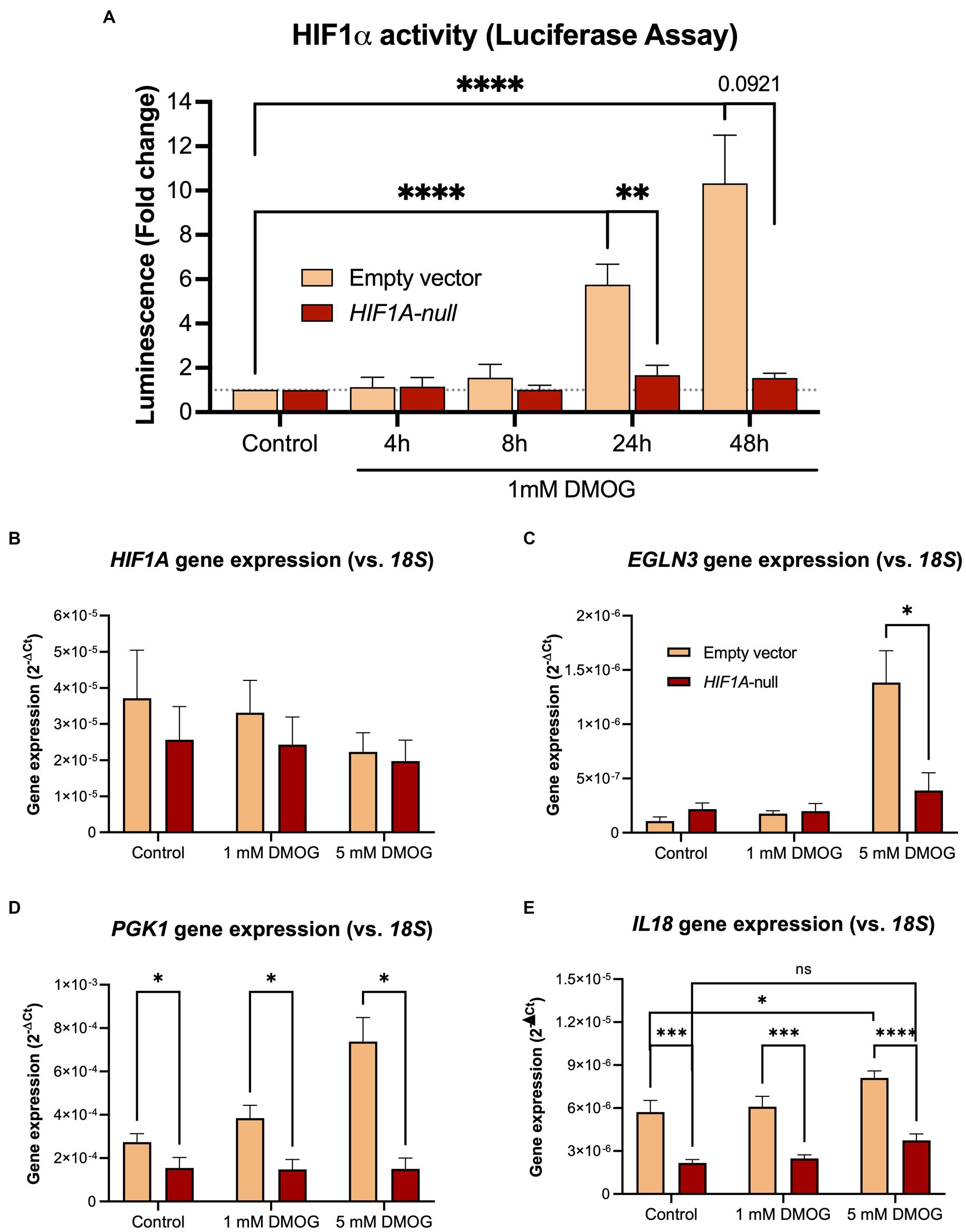
Figure 3. Stable knockout of HIF1A in Caco-2 cells leads to decreased HIF1α signaling upon pathway activation. (A) Time-dependent HIF1α responsive element (HRE)-luciferase assay over 48 h treatment with 1 mM DMOG. Gene expression levels of (B) HIF1A and the HIF1α pathway targets (C) PGK1, (D) EGLN3, and (E) IL18 in Caco-2-HIF1A-null and control cells treated with 1 or 5 mM DMOG for 24 h. Data presented as mean ± SD. All experiments were performed with n = 3; *p < 0.05, **p < 0.01, ****p < 0.0001.
3.4 Faecalibacterium prausnitzii regulates gene transcription in a HIF1α-dependent manner in intestinal epithelial cells
To analyze the putative role of HIF1α in regulating F. prausnitzii-mediated transcriptional responses, Caco-2 control (empty vector) and Caco-2-HIF1A-null cells were cultured for 18 h in the absence and presence of the strict anaerobic bacterium F. prausnitzii using the “Human-oxygen Bacteria-anaerobic” (HoxBan) coculture system (schematically drawn in Figure 4A), followed by RNA sequencing of the Caco-2 cells. In the absence of F. prausnitzii, a total of 149 genes were significantly differentially expressed between Caco-2 control and Caco-2-HIF1A-null cells (Figure 4B), while 187 genes were significantly differentially expressed between those 2 cell lines in the presence of F. prausnitzii (Figure 4C). The absence of HIF1α significantly enhanced expression of 64 genes, and reduced levels of 85 genes (Figures 4D,E), when grown in the absence F. prausnitzii in the HoxBan setup. In the presence of F. prausnitzii, expression levels of 67 genes were enhanced, and of 120 genes reduced (Figures 4D,E) in the absence of HIF1α. Overlaying these effects, we observed enhanced expression of 32 genes (Table 4), and reduced expression of 51 genes (Table 5), specifically in F. prausnitzii-cocultured Caco-2-HIF1A-null cells, when compared to F. prausnitzii-cocultured Caco-2 control cells (Figures 4D,E). Among the 32 upregulated genes, two (NRP2 and CPT1A) were previously reported to be suppressed by HIF1α signaling (Coma et al., 2011; Du et al., 2017; Tan and Welford, 2020; Ezzeddini et al., 2021), whereas no established HIF1α-upregulated targets were detected among these genes that were enhanced by F. prausnitzii in Caco-2-HIF1A-null cells. On the other hand, the 51 downregulated genes were enriched for known HIF1α targets, including PFKB3 (Minchenko et al., 2002), FOS (Ebersole et al., 2018), IER3 (Ravaud et al., 2015), ANXA1 (Bizzarro et al., 2017), HK2 (Mathupala et al., 2001), KDM3A (Mimura et al., 2012), MT2A (Mo et al., 2020), LBH (Jiang et al., 2019), DDIT4 (Gharibi et al., 2016), and TGM2 (Jang et al., 2010). Interestingly, IL18 was among these 51 genes (Table 5), of which the expression appears to be controlled through F. prausnitzii-mediated activation of HIF1α.
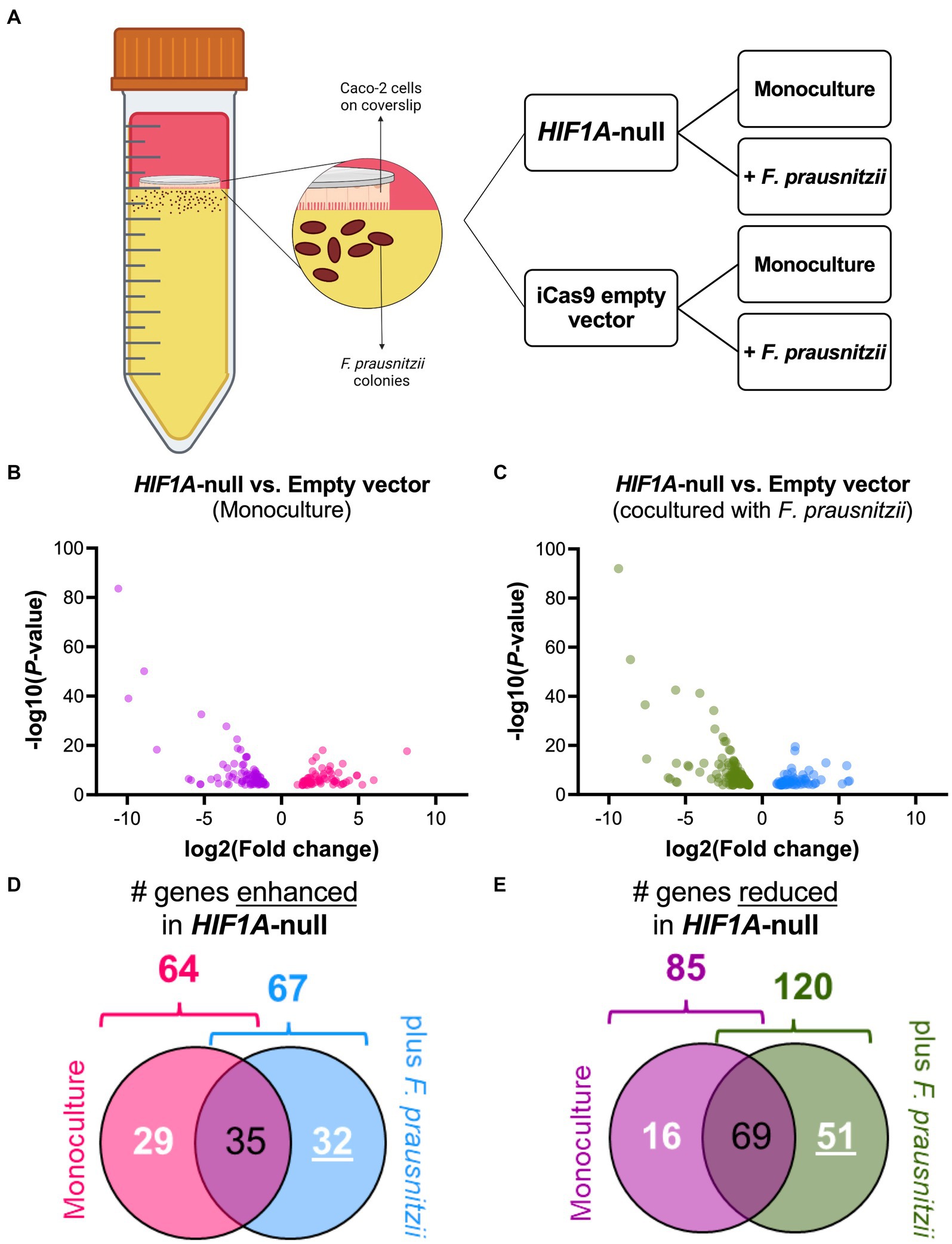
Figure 4. Differential gene expression (DGE) analysis of Caco-2-HIF1A-null upon coculture with F. prausnitzii, compared to Caco-2 empty vector controls. (A) Schematic representation of the HoxBan coculture system, including experiment layout. Caco-2-HIF1A-null cells, or appropriate control (iCas9 empty vector), were cultured in the HoxBan system for 18 h in the absence or presence of F. prausnitzii inoculum (in figure, mono and co, respectively). Volcano plots showing differential gene expression analysis comparing Caco-2-HIF1A-null cells and Caco-2-empty vector in monoculture (B; purple and pink dots represent downregulated and upregulated genes, respectively) and coculture with F. prausnitzii (C; green and blue dots represent downregulated and upregulated genes, respectively). All dots display significant observations with p < 0.05. Veen diagrams were used to discover uniquely (D) upregulated and (E) downregulated DGE (numbers underlined inside blue and green diagrams, respectively) in Caco-2-HIF1A-null cultured with F. prausnitzii, compared to Caco-2-empty vector in same condition. All experiments performed in n = 3.
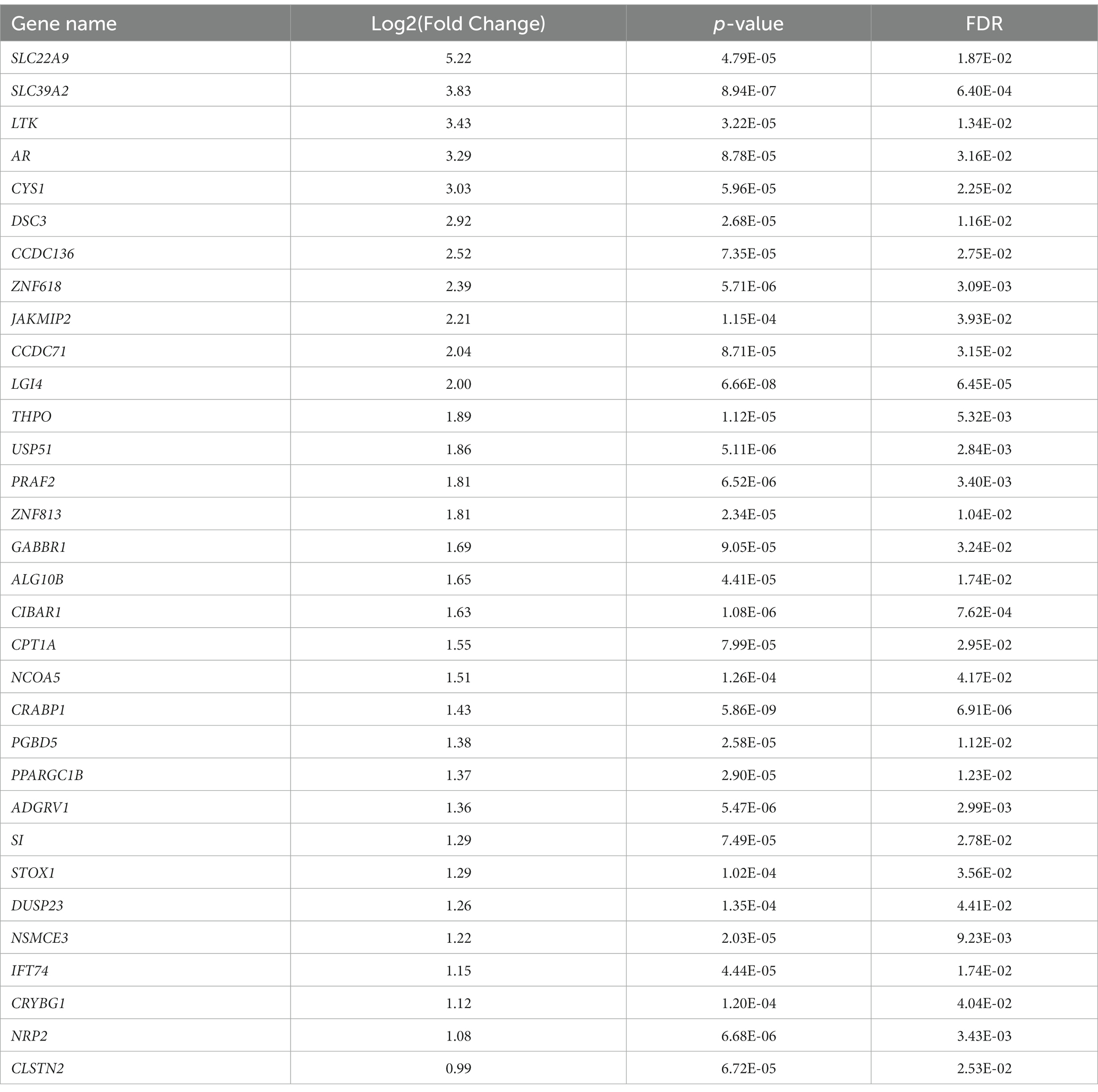
Table 4. Uniquely upregulated genes in F. prausnitzii-cocultured Caco-2-HIF1A-null cells, compared to F. prausnitzii-cocultured Caco-2 control cells.
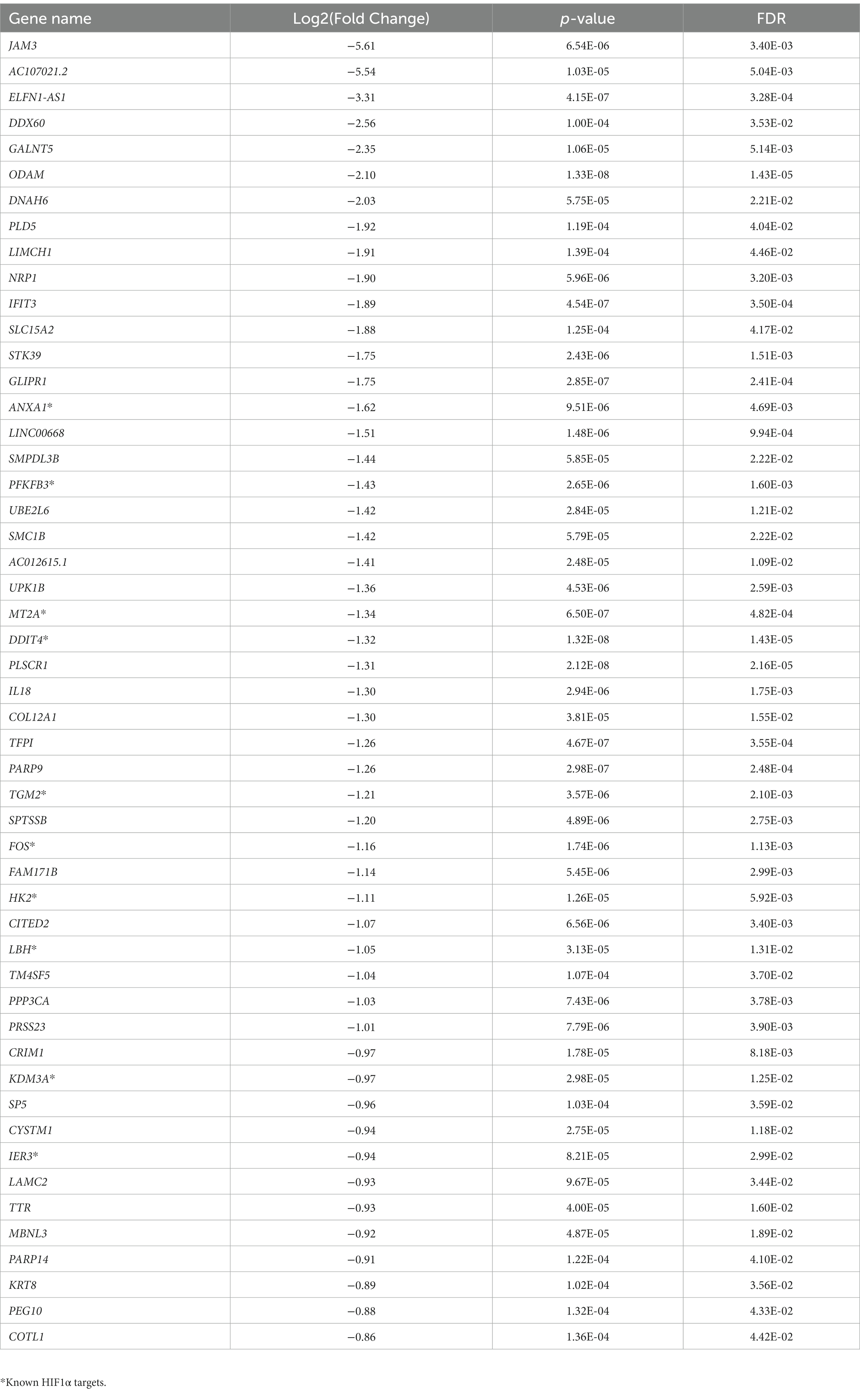
Table 5. Uniquely downregulated genes in F. prausnitzii-cocultured Caco-2-HIF1A-null cells, compared to F. prausnitzii-cocultured Caco-2 control cells.
3.5 Faecalibacterium prausnitzii enhances IL18 expression in Caco-2 cells in a HIF1α-dependent manner
Specifically analyzing the HIF1α and -2α scores from the RNA sequencing results, we observed a (non-significant) enhancement of HIF1α scores in Caco-2 control cells when comparing monocultures (Caco-2 cells without F. prausnitzii) to Caco-2-F. prausnitzii cocultures (Figure 5A). F. prausnitzii did not enhance the HIF1α score in Caco-2-HIF1A-null cells (Figure 5A, red bars). Notably, the HIF1α score was significantly lower in Caco-2-HIF1A-null cells cocultured with F. prausnitzii when compared to similarly-cultured Caco-2 control cells (Figure 5A; p < 0.0001). HIF2α scores were lower than HIF1α scores in all conditions, with no significant changes between different conditions (Figure 5B). IL18 mRNA levels exactly followed the HIF1α score: F. prausnitzii enhanced IL18 levels non-significantly in Caco-2 control cells and were significantly lower in F. prausnitzii-cocultured Caco-2-HIF1A-null cells (Figure 5C, p < 0.001). In line, a highly significant positive correlation between IL18 gene expression and HIF1α scores was observed (r = 0.6516 and p = 0.0002; Figure 5D). Taken together, these data point to a F. prausnitzii-mediated regulation of IL18 in the intestinal epithelium that is controlled by HIF1α.
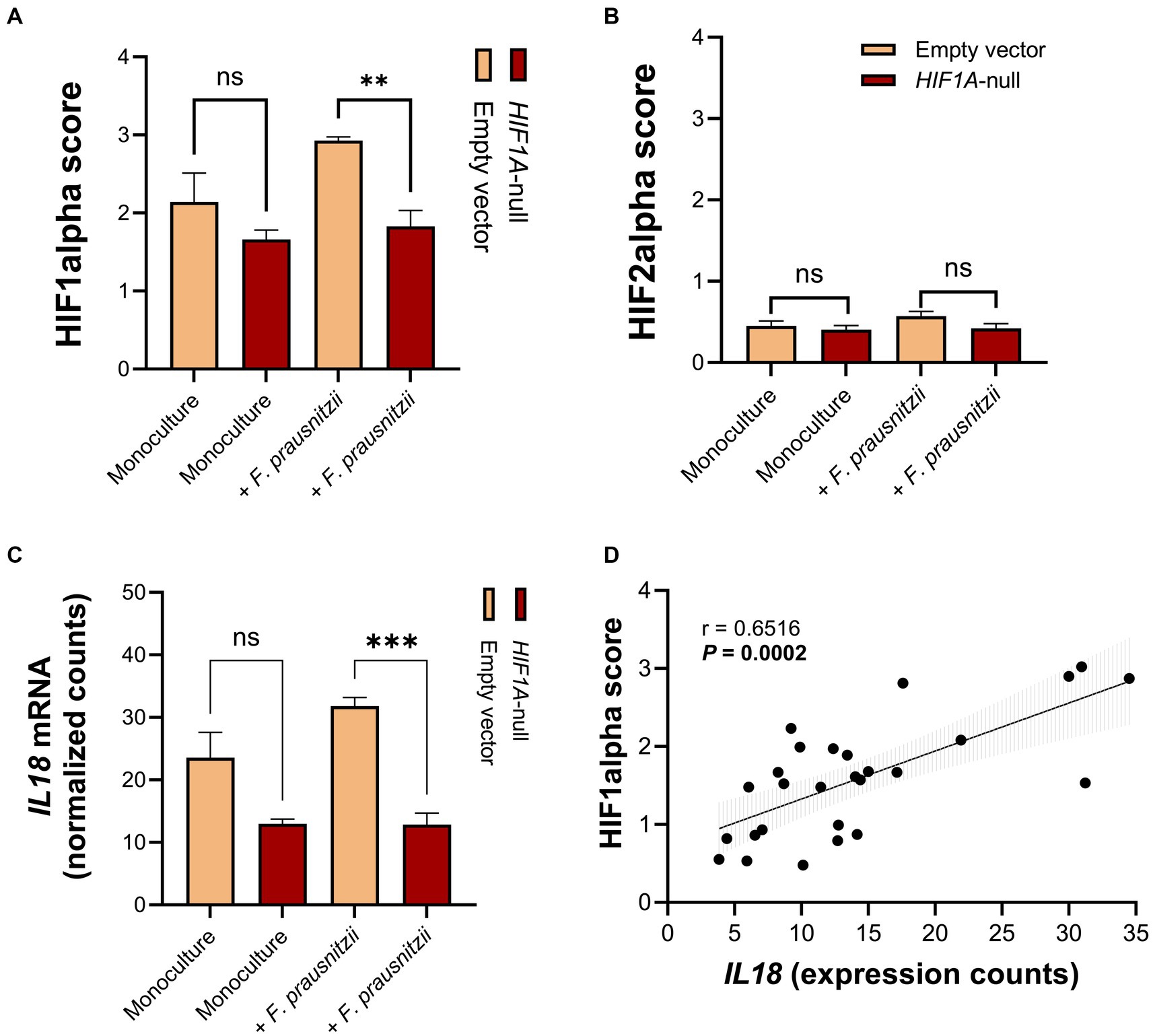
Figure 5. RNA sequencing analysis reveals that HIF1A ablation prevents upregulation of IL18 by F. prausnitzii in intestinal epithelial cells. (A,B) HIF1α and HIF2α scores shows a decrease in HIF1α, but not HIF2α, activation capacity in Caco-2-HIF1A-null, compared to Caco-2-empty vector control in coculture with F. prausnitzii. (C) Normalized gene expression counts of IL18 in Caco-2-HIF1A-null cells, compared to Caco-2-empty vector in monoculture and coculture with F. prausnitzii. (D) Spearman correlation analysis between IL18 gene expression and HIF1α scores on Caco-2 cells in the HoxBan system. All experiments performed in n = 3 (ns = not significant, **p < 0.001 and ***p < 0.0001).
4 Discussion
In this study, we found that mucosal IL18 expression associates positively with gut commensal and butyrate-producing bacteria in a cohort of 371 IBD patients, while it negatively associates with potential pathogens. At the bacterial strain level, mucosal IL18 gene expression positively associated with the mucosal abundance of the gut commensal F. prausnitzii. Using the HoxBan in vitro coculture model, we showed that this mechanistic relationship is directly regulated by activation of the transcription factor HIF1α in intestinal epithelial cells. Moreover, we described transcriptional modulations that are triggered in a HIF1α-dependent manner by F. prausnitzii on intestinal epithelial cells, also involving the regulation of IL18 expression.
In our study, we provided evidence that epithelial expression levels of IL18 are regulated in a HIF1α-dependent manner and associates positively to bacterial species directly linked to epithelial health. However, the absence of protein quantification in our study represents a limitation, and further investigations may provide a more comprehensive understanding of the molecular mechanisms involved. Nonetheless, in line with our results, previous studies using germ-free mouse models showed a decrease in IL18 gene expression and secretion, when compared to conventionally raised mice (Singh et al., 2014; Levy et al., 2015). Additionally, other murine models have pointed to a tissue-protective role of IL-18, following injury to the intestinal epithelium. IL-18 may act via its cognate membrane receptor, IL18R1, that prevents the differentiation of CD4+ T helper 17 (Th17) and promoting the expression of key Foxp3+ regulatory T cell (Treg) effectors (Harrison et al., 2015; Mahapatro et al., 2021). Furthermore, studies have shown that inflammasome (NLRP6)-deficient mice, part of the inflammasome complex essential for processing of pro-IL-18 to its active form, showed reduced levels of IL-18 and alterations in the gut microbiota composition, including increased abundance of potential pathogens such as Prevotella and bacteria from the phylum Saccharibacteria (previously known as TM7) (Elinav et al., 2011; Levy et al., 2015; Chen, 2017). On the other hand, there is also data that support that IL-18 contributes to intestinal inflammation (Nowarski et al., 2015; Pu et al., 2019). Indeed, we observed increased levels of IL18 in the inflamed intestinal mucosa of patients with IBD. Therefore, there appears to be a fine balance between the anti-inflammatory and pro-inflammatory actions of IL-18 in the intestinal mucosa. This equilibrium may be maintained through the direct influence of the soluble decoy receptor IL-18 binding protein (IL-18BP), which effectively neutralizes IL-18 activity in vivo, preventing excessive NF-κB activation and inflammation (Fantuzzi et al., 2003; Nowarski et al., 2015). Our findings indicate that mucosa-adherent bacteria, particularly F. prausnitzii, play a role in regulating IL18 gene expression, primarily within the intestinal epithelium. This regulation may enhance barrier function and act as an anti-inflammatory factor, suggesting potential benefits for IBD patients in preventing disease flares.
F. prausnitzii plays an important role in maintaining healthy gut homeostasis, which is often linked to its capacity to produce short-chain fatty acids (SCFA), in particularly butyrate (Sokol et al., 2008; Cao et al., 2014; Laval et al., 2015; Moens and de Vuyst, 2017). Indeed, prophylactic F. prausnitzii administration reduced colonic paracellular permeability and neutrophil infiltration in a rat model of pelvic radiation disease, which was attributed to F. prausnitzii-mediated induction of intestinal epithelial IL-18 expression and secretion (Lapiere et al., 2020). In line, IL18 gene and protein expression in vitro and in vivo have been shown to be increased by treatment of mice with SCFA or niacin (Kalina et al., 2002; Singh et al., 2014). Also, a positive significant correlation between F. prausnitzii abundance and mRNA and protein expression of NLRP6 was found in rats in an induced model for irritable bowel syndrome (IBS) (Bao et al., 2019). Similarly, activity of the NLRP3-containing inflammasome is induced in mice fed a fiber-rich diet and induced the production of IL-18 through the activation of intestinal epithelial SCFA receptors (GPR43 and GPR109A) (Macia et al., 2015). All these studies in animal models support our findings in IBD patients that F. prausnitzii abundance positively correlates to mucosal IL18 gene expression, which is very likely maneuvered via activation of the HIF1α pathway. We thus provided first evidence of the interrelationship between butyrate-producing bacteria and HIF1α-dependent intestinal IL18 expression in humans. We have, however, not yet identified butyrate per se as the F. prausnitzii-derived factor that induces IL18 in the intestinal epithelium. However, we have established earlier that F. prausnitzii effectively produces butyrate when co-cultured with Caco-2 cells in the HoxBan system (Sadaghian Sadabad et al., 2015) and others have shown that butyrate activates HIF1α pathway in Caco-2 cells (Kelly et al., 2015; Zheng et al., 2017). Still, further studies are needed to fully understand this interrelationship.
We propose that the increase in gene expression of both HIF1A and IL18 in inflamed biopsies is primarily driven by pathophysiological hypoxia, which is installed during inflammation in the intestinal mucosa (Fagundes and Taylor, 2017; Brown et al., 2020; Fagundes et al., 2022). Nonetheless, activation of the HIF1α pathway has been mechanistically linked to gut mucosal health, both in the context of prevention and recovery from intestinal inflammation. Blocking HIF1α in mouse models, especially on intestinal epithelial cells, impairs epithelial barrier integrity, because of dysregulation of HIF1α targets, such as tight junction proteins, intestinal trefoil factors and beta-defensins (Furuta et al., 2001; Synnestvedt et al., 2002; Kelly et al., 2013; Saeedi et al., 2015). Moreover, conditional knockout of HIF1α exacerbated inflammation in TNBS-induced colitis in mice, while increased HIF1α activation via knock-out of the von Hippel–Lindau (VHL) gene was protective (Karhausen et al., 2004). In our study, we show that stable knockout of HIF1A in Caco-2 cells decreased the expression of several well-known HIF1α target genes upon coculture with F. prausnitzii. Additionally, this led to suppression of 6 Gene Ontology biological processes, all involving the HIF1α-dependent regulation of IL18. In line, we detected 2 hypoxia-responsive elements (HRE) in the IL-18 promoter region, located 548 and 1,218 base pairs upstream of the first exon (Supplementary Figure S1). To the best of our knowledge, this is the first study to show HIF1α-mediated regulation of IL18, which may contribute to the protective role of HIF1α in the intestine.
Pharmacological activation of the HIF1α pathway promotes mucosal regeneration and intestinal barrier function in several mouse models of colitis (Cummins et al., 2008; Robinson et al., 2008; Gupta et al., 2014), as recently reviewed by Colgan & Taylor (Colgan et al., 2020). Prolyl-hydroxylase inhibitors (PHI) are a class of HIF-agonistic drugs that promote HIF1α activation by blocking the PHD-mediated degradation of HIF1α protein (Joharapurkar et al., 2018). Some of these drugs have already entered phase 2 and 3 clinical trials for the treatments of anemia of chronical renal disease, being well-tolerated by patients and healthy individuals (Haase, 2017). Thus, we postulate that PHIs may also be beneficial for IBD patients suffering from disease flares. Moreover, a beneficial effect may be expected from strategies to enhance the abundance of F. prausnitzii in the gut, thereby strengthening the fine epithelial HIF1α-IL18 crosstalk. However, the applications of live F. prausnitzii as a probiotic therapeutic option remain a challenge, mostly because of its strict anaerobic nature. Alternatively, dietary fibers (prebiotics, such as inulin) could be used to increase F. prausnitzii abundance in the human gut (Verhoog et al., 2019). Although the anti-inflammatory effects of prebiotics in IBD patients remains controversial, co-treatment with PHI may be an interesting strategy, which would combine the regenerative potential of HIF1α activation with stimulation of growth of commensal bacteria.
In conclusion, our study reveals the interrelationship between F. prausnitzii, HIF1α activation and IL18 expression in the intestinal mucosa. These factors are all relevant therapeutics to restore intestinal homeostasis in patients with IBD.
Data availability statement
The patient-derived data from the Groningen 1000IBD cohort presented in this study are deposited in the European Genome-Phenome Archive (EGA) repository, with the accession number EGAS00001002702 (https://ega-archive.org/studies/EGAS00001002702; IDs: EGAD00001003991, EGAD00001008214, and EGAD00001008215). The transcriptomics data from HoxBan experiments presented in this study are deposited in National Library of Medicine (NHI) repository, with the accession number PRJNA954196 (https://www.ncbi.nlm.nih.gov/bioproject/954196; BioSample: SAMN34136424).
Ethics statement
The studies involving humans were approved by the Institutional Review Board (IRB) of the University Medical Center Groningen (UMC Groningen) (in Dutch: “Medisch Ethische Toetsingscommissie,” METc; IRB no. 2008/338 and 2016/424). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
RF: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. GB-R: Formal analysis, Methodology, Writing – original draft, Writing – review & editing. SH: Data curation, Formal analysis, Software, Visualization, Writing – review & editing. SK: Methodology, Validation, Writing – review & editing. RW: Conceptualization, Funding acquisition, Project administration, Resources, Software, Supervision, Writing – review & editing. CT: Conceptualization, Investigation, Supervision, Writing – original draft, Writing – review & editing. GD: Conceptualization, Funding acquisition, Investigation, Resources, Supervision, Writing – original draft, Writing – review & editing. HH: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing. KF: Conceptualization, Formal analysis, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. RW is supported by a research grant from the Seerave Foundation and by the collaborative TIMID project (LHSM18057-SGF) financed by the PPP allowance made available by Top Sector Life Sciences & Health Samenwerkende Gezondheidsfondsen (SGF). The research position of RF was supported by the Graduate School of Medical Sciences of the University of Groningen. The funders had no role in the design of the study, collection, analysis, interpretation of data or writing the manuscript.
Acknowledgments
All authors declare their gratitude toward all participants of the 1000IBD cohort. We thank Mathilde J. C. Broekhuis and Floris Foijer from the iPSC and CRISPR/Cas9 Facilities at the Laboratory of Ageing Biology and Stem Cells, European Research Institute for Biology of Ageing (ERIBA), University Medical Center Groningen, University of Groningen (Groningen, The Netherlands) for help with generating Caco-2-HIF1A-null cell line.
Conflict of interest
GD received research grant from Royal DSM and speaker’s fees from Janssen Pharmaceuticals, Pfizer and Abbvie. RW acted as consultant for Takeda and received unrestricted grants from Takeda, Johnson & Johnson, Tramedico and Ferring, and received speaker’s fee from MSD, Abbvie and Janssen Pharmaceuticals.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1298304/full#supplementary-material
References
Ananthakrishnan, A. N. (2015). Epidemiology and risk factors for IBD. Nat. Rev. Gastroenterol. Hepatol. 12, 205–217. doi: 10.1038/nrgastro.2015.34
Andreoletti, G., Ashton, J. J., Coelho, T., Willis, C., Haggarty, R., Gibson, J., et al. (2015). Exome analysis of patients with concurrent pediatric inflammatory bowel disease and autoimmune disease. Inflamm. Bowel Dis. 21, 1–1236. doi: 10.1097/MIB.0000000000000381
Bao, C. H., Wang, C. Y., Li, G. N., Yan, Y. L., Wang, D., Jin, X. M., et al. (2019). Effect of mild moxibustion on intestinal microbiota and NLRP6 inflammasome signaling in rats with post-inflammatory irritable bowel syndrome. World J. Gastroenterol. 25, 4696–4714. doi: 10.3748/wjg.v25.i32.4696
Bizzarro, V., Belvedere, R., Migliaro, V., Romano, E., Parente, L., and Petrella, A. (2017). Hypoxia regulates ANXA1 expression to support prostate cancer cell invasion and aggressiveness. Cell Adhes. Migr. 11, 247–260. doi: 10.1080/19336918.2016.1259056
Brown, E., Rowan, C., Strowitzki, M. J., Fagundes, R. R., Faber, K. N., Güntsch, A., et al. (2020). Mucosal inflammation downregulates PHD1 expression promoting a barrier-protective HIF-1α response in ulcerative colitis patients. FASEB J. 34, 3732–3742. doi: 10.1096/fj.201902103R
Cao, Y., Shen, J., and Ran, Z. H. (2014). Association between Faecalibacterium prausnitzii reduction and inflammatory bowel disease: A Meta-analysis and systematic review of the literature. Gastroenterol. Res. Pract. 2014:872725, 1–7. doi: 10.1155/2014/872725
Cavadas, M. A. S., Taylor, C. T., and Cheong, A. (2018). Acquisition of temporal HIF transcriptional activity using a secreted luciferase assay. Methods Mol. Biol. 1742, 37–44. doi: 10.1007/978-1-4939-7665-2_4/COVER
Chen, G. Y. (2017). Regulation of the gut microbiome by inflammasomes. Free Radic. Biol. Med. 105, 35–40. doi: 10.1016/J.FREERADBIOMED.2016.11.011
Colgan, S. P., Furuta, G. T., and Taylor, C. T. (2020). Hypoxia and innate immunity: keeping up with the HIFsters. Annu. Rev. Immunol. 38, 341–363. doi: 10.1146/annurev-immunol-100819-121537
Colgan, S. P., and Taylor, C. T. (2010). Hypoxia: an alarm signal during intestinal inflammation. Nat. Rev. Gastroenterol. Hepatol. 7, 281–287. doi: 10.1038/nrgastro.2010.39
Coma, S., Shimizu, A., and Klagsbrun, M. (2011). Hypoxia induces tumor and endothelial cell migration in a semaphorin 3F- and VEGF-dependent manner via transcriptional repression of their common receptor neuropilin 2. Cell Adhes. Migr. 5, 266–275. doi: 10.4161/CAM.5.3.16294
Cummins, E. P., Keogh, C. E., Crean, D., and Taylor, C. T. (2016). The role of HIF in immunity and inflammation. Mol. Asp. Med. 47-48, 24–34. doi: 10.1016/j.mam.2015.12.004
Cummins, E. P., Seeballuck, F., Keely, S. J., Mangan, N. E., Callanan, J. J., Fallon, P. G., et al. (2008). The hydroxylase inhibitor dimethyloxalylglycine is protective in a murine model of colitis. Gastroenterology 134, 156–165.e1. doi: 10.1053/j.gastro.2007.10.012
Du, W., Zhang, L., Brett-Morris, A., Aguila, B., Kerner, J., Hoppel, C. L., et al. (2017). HIF drives lipid deposition and cancer in ccRCC via repression of fatty acid metabolism. Nat. Commun. 8:1769. doi: 10.1038/S41467-017-01965-8
Ebersole, J. L., Novak, M. J., Orraca, L., Martinez-Gonzalez, J., Kirakodu, S., Chen, K. C., et al. (2018). Hypoxia-inducible transcription factors, HIF1A and HIF2A, increase in aging mucosal tissues. Immunology 154, 452–464. doi: 10.1111/IMM.12894
Elinav, E., Strowig, T., Kau, A. L., Henao-Mejia, J., Thaiss, C. A., Booth, C. J., et al. (2011). NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cells 145, 745–757. doi: 10.1016/J.CELL.2011.04.022
Ezzeddini, R., Taghikhani, M., Farrokhi, A. S., Somi, M. H., Samadi, N., Esfahani, A., et al. (2021). Downregulation of fatty acid oxidation by involvement of HIF-1α and PPARγ in human gastric adenocarcinoma and related clinical significance. J. Physiol. Biochem. 77, 249–260. doi: 10.1007/S13105-021-00791-3
Fagundes, R. R., Bourgonje, A. R., Hu, S., Barbieri, R., Jansen, B. H., Sinnema, N., et al. (2022). HIF1α-dependent induction of TFRC by a combination of intestinal inflammation and systemic Iron deficiency in inflammatory bowel disease. Front. Physiol. 13:889091. doi: 10.3389/FPHYS.2022.889091/BIBTEX
Fagundes, R. R., and Taylor, C. T. (2017). Determinants of hypoxia-inducible factor activity in the intestinal mucosa. J. Appl. Physiol. 123, 1328–1334. doi: 10.1152/japplphysiol.00203.2017
Fantuzzi, G., Banda, N. K., Guthridge, C., Vondracek, A., Kim, S.-H., Siegmund, B., et al. (2003). Generation and characterization of mice transgenic for human IL-18-binding protein isoform a. J. Leukoc. Biol. 74, 889–896. doi: 10.1189/JLB.0503230
Furuta, G. T., Turner, J. R., Taylor, C. T., Hershberg, R. M., Comerford, K., Narravula, S., et al. (2001). Hypoxia-inducible factor 1-dependent induction of intestinal trefoil factor protects barrier function during hypoxia. J. Exp. Med. 193, 1027–1034. doi: 10.1084/jem.193.9.1027
Gharibi, B., Ghuman, M., and Hughes, F. J. (2016). DDIT4 regulates mesenchymal stem cell fate by mediating between HIF1α and mTOR signalling. Sci. Rep. 6, 1–9. doi: 10.1038/srep36889
Gryaznova, M. V., Solodskikh, S. A., Panevina, A. V., Syromyatnikov, M. Y., Dvoretskaya, Y. D., Sviridova, T. N., et al. (2021). Study of microbiome changes in patients with ulcerative colitis in the central European part of Russia. Heliyon 7:e06432. doi: 10.1016/J.HELIYON.2021.E06432
Gupta, R., Jadhav, A., Zambad, S., Gupta, R., Deshpande, S., Chauthaiwale, V., et al. (2014). Therapeutic treatment with a novel hypoxia-inducible factor hydroxylase inhibitor (TRC160334) ameliorates murine colitis. Clin. Exp. Gastroenterol. 7:13. doi: 10.2147/CEG.S51923
Haase, V. H. (2017). HIF-prolyl hydroxylases as therapeutic targets in erythropoiesis and iron metabolism. Hemodial. Int. 21, S110–S124. doi: 10.1111/hdi.12567
Harrison, O. J., Srinivasan, N., Pott, J., Schiering, C., Krausgruber, T., Ilott, N. E., et al. (2015). Epithelial-derived IL-18 regulates Th17 cell differentiation and Foxp3+ Treg cell function in the intestine. Mucosal Immunol. 8, 1226–1236. doi: 10.1038/mi.2015.13
Hu, S., Bourgonje, A. R., Gacesa, R., Jansen, B. H., Björk, J. R., Bangma, A., et al. (2022). Mucosal host–microbe interactions associate with clinical phenotypes in inflammatory bowel disease. bioRxiv 2022.06.04.494807. doi: 10.1101/2022.06.04.494807
Hu, S., Venema, W. T. U., Westra, H.-J., Vila, A. V., Barbieri, R., Voskuil, M. D., et al. (2021). Inflammation status modulates the effect of host genetic variation on intestinal gene expression in inflammatory bowel disease. Nat. Commun. 12, 1–10. doi: 10.1038/s41467-021-21458-z
Imhann, F., Van der Velde, K. J., Barbieri, R., Alberts, R., Voskuil, M. D., Vich Vila, A., et al. (2019). The 1000IBD project: multi-omics data of 1000 inflammatory bowel disease patients; data release 1. BMC Gastroenterol. 19:5. doi: 10.1186/s12876-018-0917-5
Jang, G. Y., Jeon, J. H., Cho, S. Y., Shin, D. M., Kim, C. W., Jeong, E. M., et al. (2010). Transglutaminase 2 suppresses apoptosis by modulating caspase 3 and NF-kappaB activity in hypoxic tumor cells. Oncogene 29, 356–367. doi: 10.1038/ONC.2009.342
Jiang, Y., Zhou, J., Zou, D., Hou, D., Zhang, H., Zhao, J., et al. (2019). Overexpression of limb-bud and Heart (LBH) promotes angiogenesis in human glioma via VEGFA-mediated ERK signalling under hypoxia. EBioMedicine 48, 36–48. doi: 10.1016/J.EBIOM.2019.09.037
Joharapurkar, A. A., Pandya, V. B., Patel, V. J., Desai, R. C., and Jain, M. R. (2018). Prolyl hydroxylase inhibitors: A breakthrough in the therapy of Anemia associated with chronic diseases. J. Med. Chem. 61, 6964–6982. doi: 10.1021/acs.jmedchem.7b01686
Kalina, U., Koyama, N., Hosoda, T., Nuernberger, H., Sato, K., Hoelzer, D., et al. (2002). Enhanced production of IL-18 in butyrate-treated intestinal epithelium by stimulation of the proximal promoter region. Eur. J. Immunol 32, 2635–2643. doi: 10.1002/1521-4141
Karhausen, J., Furuta, G. T., Tomaszewski, J. E., Johnson, R. S., Colgan, S. P., and Haase, V. H. (2004). Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. J. Clin. Investig. 114, 1098–1106. doi: 10.1172/jci21086
Kelly, C. J., Glover, L. E., Campbell, E. L., Kominsky, D. J., Ehrentraut, S. F., Bowers, B. E., et al. (2013). Fundamental role for HIF-1α in constitutive expression of human β defensin-1. Mucosal Immunol. 6, 1110–1118. doi: 10.1038/mi.2013.6
Kelly, C. J., Zheng, L., Campbell, E. L., Saeedi, B., Scholz, C. C., Bayless, A. J., et al. (2015). Crosstalk between microbiota-derived short-Chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 17, 662–671. doi: 10.1016/j.chom.2015.03.005
Kim, S., Becker, J., Bechheim, M., Kaiser, V., Noursadeghi, M., Fricker, N., et al. (2014). Characterizing the genetic basis of innate immune response in TLR4-activated human monocytes. Nat. Commun. 5:5236. doi: 10.1038/ncomms6236
Kostic, A. D., Xavier, R. J., and Gevers, D. (2014). The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146, 1489–1499. doi: 10.1053/j.gastro.2014.02.009
Lapiere, A., Geiger, M., Robert, V., Demarquay, C., Auger, S., Chadi, S., et al. (2020). Prophylactic Faecalibacterium prausnitzii treatment prevents the acute breakdown of colonic epithelial barrier in a preclinical model of pelvic radiation disease. Gut Microbes 12, 1–15. doi: 10.1080/19490976.2020.1812867
Laval, L., Martin, R., Natividad, J. N., Chain, F., Miquel, S., de Maredsous, C. D., et al. (2015). Lactobacillus rhamnosus CNCM I-3690 and the commensal bacterium faecalibacterium prausnitzii A2-165 exhibit similar protective effects to induced barrier hyper-permeability in mice. Gut Microbes 6, 1–9. doi: 10.4161/19490976.2014.990784
Lennard-Jones, J. E. (1989). Classification of inflammatory bowel disease. Scand. J. Gastroenterol. 24, 2–6. doi: 10.3109/00365528909091339
Levy, M., Thaiss, C. A., Zeevi, D., Dohnalová, L., Zilberman-Schapira, G., Mahdi, J. A., et al. (2015). Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 Inflammasome signaling. Cells 163, 1428–1443. doi: 10.1016/J.CELL.2015.10.048
Liu, X., Lu, J., Liu, Z., Zhao, J., Sun, H., Wu, N., et al. (2018). Intestinal epithelial cell-derived LKB1 suppresses Colitogenic microbiota. J. Immunol. 200, 1889–1900. doi: 10.4049/JIMMUNOL.1700547
Macia, L., Tan, J., Vieira, A. T., Leach, K., Stanley, D., Luong, S., et al. (2015). Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 6, 1–15. doi: 10.1038/ncomms7734
Mahapatro, M., Erkert, L., and Becker, C. (2021). Cytokine-mediated crosstalk between immune cells and epithelial cells in the gut. Cells 10:111. doi: 10.3390/cells10010111
Manor, O., Dai, C. L., Kornilov, S. A., Smith, B., Price, N. D., Lovejoy, J. C., et al. (2020). Health and disease markers correlate with gut microbiome composition across thousands of people. Nat. Commun. 11, 1–12. doi: 10.1038/s41467-020-18871-1
Mantovani, A., Dinarello, C. A., Molgora, M., and Garlanda, C. (2019). Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity 50, 778–795. doi: 10.1016/j.immuni.2019.03.012
Mathupala, S. P., Rempel, A., and Pedersen, P. L. (2001). Glucose catabolism in cancer cells: identification and characterization of a marked activation response of the type II hexokinase gene to hypoxic conditions. J. Biol. Chem. 276, 43407–43412. doi: 10.1074/JBC.M108181200
Mimura, I., Nangaku, M., Kanki, Y., Tsutsumi, S., Inoue, T., Kohro, T., et al. (2012). Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible factor 1 and KDM3A. Mol. Cell. Biol. 32, 3018–3032. doi: 10.1128/MCB.06643-11
Minchenko, A., Leshchinsky, I., Opentanova, I., Sang, N., Srinivas, V., Armstead, V., et al. (2002). Hypoxia-inducible factor-1-mediated expression of the 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3) gene. Its possible role in the Warburg effect. J. Biol. Chem. 277, 6183–6187. doi: 10.1074/JBC.M110978200
Mo, Z., Yu, L., Cao, Z., Hu, H., Luo, S., and Zhang, S. (2020). Identification of a hypoxia-associated signature for lung adenocarcinoma. Front. Genet. 11:647. doi: 10.3389/FGENE.2020.00647
Moens, F., and de Vuyst, L. (2017). Inulin-type fructan degradation capacity of clostridium cluster IV and XlVa butyrate- producing colon bacteria and their associated metabolic outcomes. Benef Microbes 8, 473–490. doi: 10.3920/BM2016.0142
Nakamura, K., Okamura, H., Wada, M., Nagata, K., and Tamura, T. (1989). Endotoxin-induced serum factor that stimulates gamma interferon production. Infect. Immun. 57, 590–595. doi: 10.1128/IAI.57.2.590-595.1989
Normand, S., Delanoye-Crespin, A., Bressenot, A., Huot, L., Grandjean, T., Peyrin-Biroulet, L., et al. (2011). Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc. Natl. Acad. Sci. U. S. A. 108, 9601–9606. doi: 10.1073/pnas.1100981108
Nowarski, R., Jackson, R., Gagliani, N., De Zoete, M. R., Palm, N. W., Bailis, W., et al. (2015). Epithelial IL-18 equilibrium controls barrier function in colitis. Cells 163, 1444–1456. doi: 10.1016/j.cell.2015.10.072
Okamura, H., Tsutsui, H., Komatsu, T., Yutsudo, M., Hakura, A., Tanimoto, T., et al. (1995). Cloning of a new cytokine that induces IFN-γ production by T cells. Nature 378, 88–91. doi: 10.1038/378088a0
Pu, Z., Che, Y., Zhang, W., Sun, H., Meng, T., Xie, H., et al. (2019). Dual roles of IL-18 in colitis through regulation of the function and quantity of goblet cells. Int. J. Mol. Med. 43, 2291–2302. doi: 10.3892/ijmm.2019.4156
Ravaud, C., Esteve, D., Villageois, P., Bouloumie, A., Dani, C., and Ladoux, A. (2015). IER3 promotes expansion of adipose progenitor cells in response to changes in distinct microenvironmental effectors. Stem Cells 33, 2564–2573. doi: 10.1002/STEM.2016
Robinson, A., Keely, S., Karhausen, J., Gerich, M. E., Furuta, G. T., and Colgan, S. P. (2008). Mucosal protection by hypoxia-inducible factor prolyl hydroxylase inhibition. Gastroenterology 134, 145–155. doi: 10.1053/j.gastro.2007.09.033
Sadaghian Sadabad, M., von Martels, J. Z. H., Khan, M. T., Blokzijl, T., Paglia, G., Dijkstra, G., et al. (2015). A simple coculture system shows mutualism between anaerobic faecalibacteria and epithelial Caco-2 cells. Sci. Rep. 5:17906. doi: 10.1038/srep17906
Saeedi, B. J., Kao, D. J., Kitzenberg, D. A., Dobrinskikh, E., Schwisow, K. D., Masterson, J. C., et al. (2015). HIF-dependent regulation of claudin-1 is central to intestinal epithelial tight junction integrity. Mol. Biol. Cell 26, 2252–2262. doi: 10.1091/mbc.E14-07-1194
Schippa, S., and Conte, M. (2014). Dysbiotic events in gut microbiota: impact on human health. Nutrients 6, 5786–5805. doi: 10.3390/nu6125786
Sellin, M. E., Maslowski, K. M., Maloy, K. J., and Hardt, W. D. (2015). Inflammasomes of the intestinal epithelium. Trends Immunol. 36, 442–450. doi: 10.1016/j.it.2015.06.002
Singh, N., Gurav, A., Sivaprakasam, S., Brady, E., Padia, R., Shi, H., et al. (2014). Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139. doi: 10.1016/j.immuni.2013.12.007
Soderquest, K., Hertweck, A., Giambartolomei, C., Henderson, S., Mohamed, R., Goldberg, R., et al. (2017). Genetic variants alter T-bet binding and gene expression in mucosal inflammatory disease. PLoS Genet. 13:e1006587. doi: 10.1371/journal.pgen.1006587
Sokol, H., Pigneur, B., Watterlot, L., Lakhdari, O., Bermúdez-Humarán, L. G., Gratadoux, J. J., et al. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U. S. A. 105, 16731–16736. doi: 10.1073/pnas.0804812105
Spekhorst, L. M., Imhann, F., Festen, E. A. M., De Bodegraven, A. D. V., De Boer, N. K. H., Bouma, G., et al. (2017). Cohort profile: design and first results of the Dutch IBD biobank: A prospective, nationwide biobank of patients with inflammatory bowel disease. BMJ Open 7:e016695. doi: 10.1136/bmjopen-2017-016695
Swarte, J. C., Douwes, R. M., Hu, S., Vila, A. V., Eisenga, M. F., van Londen, M., et al. (2020). Characteristics and Dysbiosis of the gut microbiome in renal transplant recipients. J. Clin. Med. 9:386. doi: 10.3390/JCM9020386
Synnestvedt, K., Furuta, G. T., Comerford, K. M., Louis, N., Karhausen, J., Eltzschig, H. K., et al. (2002). Ecto-5′-nucleotidase (CD73) regulation by hypoxia-inducible factor-1 mediates permeability changes in intestinal epithelia. J. Clin. Investig. 110, 993–1002. doi: 10.1172/jci15337
Tan, S. K., and Welford, S. M. (2020). Lipid in renal carcinoma: queen bee to target? Trends Cancer 6, 448–450. doi: 10.1016/J.TRECAN.2020.02.017
Tsutsumi, N., Yokota, A., Kimura, T., Kato, Z., Fukao, T., Shirakawa, M., et al. (2019). An innate interaction between IL-18 and the propeptide that inactivates its precursor form. Sci. Rep. 9, 1–9. doi: 10.1038/s41598-019-42661-5
Verhoog, S., Taneri, P. E., Díaz, Z. M. R., Marques-Vidal, P., Troup, J. P., Bally, L., et al. (2019). Dietary factors and modulation of bacteria strains of akkermansia muciniphila and faecalibacterium prausnitzii: A systematic review. Nutrients 11:1565. doi: 10.3390/nu11071565
Vich Vila, A., Imhann, F., Collij, V., Jankipersadsing, S. A., Gurry, T., Mujagic, Z., et al. (2018). Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci. Transl. Med. 10:eaap8914. doi: 10.1126/scitranslmed.aap8914
Wiercinska-Drapalo, A., Flisiak, R., Jaroszewicz, J., and Prokopowicz, D. (2005). Plasma interleukin-18 reflects severity of ulcerative colitis. World J. Gastroenterol. 11, 605–608. doi: 10.3748/wjg.v11.i4.605
Yasuda, K., Nakanishi, K., and Tsutsui, H. (2019). Interleukin-18 in health and disease. Int. J. Mol. Sci. 20:649. doi: 10.3390/ijms20030649
Zaki, M. H., Boyd, K. L., Vogel, P., Kastan, M. B., Lamkanfi, M., and Kanneganti, T. D. (2010). The NLRP3 Inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 32, 379–391. doi: 10.1016/j.immuni.2010.03.003
Keywords: HIF1α, IL-18, intestine, epithelial-bacteria interaction, HoxBan in vitro coculture system, Faecalibacterium , IBD
Citation: Fagundes RR, Bravo-Ruiseco G, Hu S, Kierans SJ, Weersma RK, Taylor CT, Dijkstra G, Harmsen HJM and Faber KN (2023) Faecalibacterium prausnitzii promotes intestinal epithelial IL-18 production through activation of the HIF1α pathway. Front. Microbiol. 14:1298304. doi: 10.3389/fmicb.2023.1298304
Edited by:
Thi Thu Hao Van, RMIT University, AustraliaReviewed by:
Andrey Kruglov, German Rheumatism Research Center (DRFZ), GermanyFederica Rubbino, Humanitas Research Hospital, Italy
Copyright © 2023 Fagundes, Bravo-Ruiseco, Hu, Kierans, Weersma, Taylor, Dijkstra, Harmsen and Faber. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Klaas Nico Faber, ay5uLmZhYmVyQHVtY2cubmw=
 Raphael R. Fagundes
Raphael R. Fagundes Gabriela Bravo-Ruiseco
Gabriela Bravo-Ruiseco Shixian Hu
Shixian Hu Sarah J. Kierans3
Sarah J. Kierans3 Cormac T. Taylor
Cormac T. Taylor Gerard Dijkstra
Gerard Dijkstra Hermie J. M. Harmsen
Hermie J. M. Harmsen Klaas Nico Faber
Klaas Nico Faber