
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 23 January 2024
Sec. Virology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1296179
 Ilana S. Fratty1,2
Ilana S. Fratty1,2 Menucha Jurkowicz1,3
Menucha Jurkowicz1,3 Neta Zuckerman1,3
Neta Zuckerman1,3 Ital Nemet1
Ital Nemet1 Nofar Atari1
Nofar Atari1 Limor Kliker1
Limor Kliker1 Lea Gur-Arie2
Lea Gur-Arie2 Alina Rosenberg2
Alina Rosenberg2 Aharona Glatman-Freedman2,3
Aharona Glatman-Freedman2,3 Yaniv Lustig1,3
Yaniv Lustig1,3 Michal Mandelboim1,3*
Michal Mandelboim1,3*Introduction: Following the significant decrease in SARS-CoV-2 cases worldwide, Israel, as well as other countries, have again been faced with a rise in seasonal influenza. This study compared circulating influenza A and B in hospitalized patients in Israel with the influenza strains in the vaccine following the 2021–2022 winter season which was dominated by the omicron variant.
Methods: Nasopharyngeal samples of 16,325 patients were examined for the detection of influenza A(H1N1)pdm09, influenza A(H1N1)pdm09 and influenza B. Phylogenetic trees of hemagglutinin were then prepared using sanger sequencing. Vaccine immunogenicity was also performed using the hemagglutination inhibition test.
Results: Of the 16,325 nasopharyngeal samples collected from hospitalized patients between September 2021 (Week 40) and April 2023 (Week 15), 7.5% were found to be positive for influenza. Phylogenetic analyses show that in the 2021–2022 winter season, the leading virus subtype was influenza A(H3N2), belonging to clade 3C.2a1b.2a.2. However, the following winter season was dominated by influenza A(H1N1)pdm09, which belongs to clade 6B.aA.5a.2. The circulating influenza A(H1N1)pdm09 strain showed a shift from the vaccine strain, while the co-circulating influenza A(H3N2) and influenza B strains were similar to those of the vaccine. Antigenic analysis coincided with the sequence analysis.
Discussion: Influenza prevalence during 2022–2023 returned to typical levels as seen prior to the emergence of SARS-CoV-2, which may suggest a gradual viral adaptation to SARS-CoV-2 variants. Domination of influenza A(H1N1)pdm09 was observed uniquely in Israel compared to Europe and USA and phylogenetic and antigenic analysis showed lower recognition of the vaccine with the circulating influenza A(H1N1)pdm09 in Israel compared to the vaccine.
In temperate and continental climates, influenza outbreaks occur every year, most commonly in the winter when humidity and temperature levels are low. In tropical and sub-tropical climates, it can occur year-round (Uyeki et al., 2022). Influenza infection is associated with respiratory and non-respiratory complications such as pneumonia, bacterial co-infection, and cardiac complications that can lead to hospitalization and morbidity. An estimated 300,000 influenza-related deaths occur annually worldwide (Petrova and Russell, 2018).
Annual vaccination is considered the most effective approach to preventing influenza (Uyeki et al., 2022). The composition of the annual vaccine is determined by the World Health Organization (WHO) based on worldwide surveillance data (WHO, 2023a). However, studies show low vaccine effectiveness (VE), especially in vaccines containing the influenza A(H3N2) strain, which drifts and adapts new mutations (Huddleston et al., 2020). Vaccine preparation also poses significant challenges. Generally, influenza vaccines are produced using an egg-based manufacturing process. However, the virus can acquire mutations and drift during egg adaptation, which further contributes to low VE. Hence, the vaccine composition is evaluated every year and updated according to the current circulating strains (WHO, 2023a,b).
Prior to the COVID-19 pandemic, influenza seasons in temperate areas were generally dominated by a single strain, with other influenza strains circulating at lower levels. As SARS-CoV-2 cases decreased in Israel in September 2021, influenza cases increased and were dominated by influenza A(H3N2), with no other strains circulating, while in the United States and Europe, other influenza strains were circulating [Grohskopf et al., 2016; Centers for Disease Control and Prevention (CDC), 2023; Europe Centers for Disease Control and Prevention (ECDC), 2023; Price et al., 2023]. In this study, we characterized the influenza seasons 2021–2022 and 2022–2023, during which SARS-CoV-2 circulation transitioned from the pandemic to the post-pandemic phase, and examined the compatibility of the influenza vaccine with circulating strains in Israel as compared to the United States and Europe.
Nasopharyngeal samples of patients hospitalized at Sheba Medical Center (SMC), the largest tertiary medical center in Israel (1,619 hospital beds), were received at the Central Virology Laboratory of the Ministry of Health located on the premises of SMC. In total, 16,325 samples were tested for the influenza virus between week 40 of 2021 (which started October 2, 2021) and week 15 of 2023 (which started April 9, 2023). The distribution of samples by patients’ age, sex, and critical hospitalization is presented in the Supplementary Tables S1, S2.
Viral RNA was extracted using the STARMag Viral DNA/RNA 200C universal kit (Seegene Inc., Korea). Reverse transcription and RT-PCR assays were performed using the AllplexTM RV essential assay (Seegene Inc., Korea) in a CFX real-time PCR system (BIO-RAD, United States; Folgueira et al., 2019). Influenza typing for A(H1N1) and A(H3N2) was performed as previously described (Meningher et al., 2014; Pando et al., 2021). Briefly, the master mix was prepared with the SensiFAST kit (Meridian Bioscience, United Kingdom) using specific primers for influenza A(H3N2) and influenza A (H1N1pdm) that are listed in Supplementary Table S3 (Pando et al., 2021). The lineage of influenza B was determined by RT-PCR The qRT-PCR assay was performed on a CFX real-time PCR system (BIO-RAD, United States). The influenza B lineage was determined by Sanger sequencing of the hemagglutinin, as described below.
Hemagglutinins (HA) from 77 laboratory-confirmed influenza viruses were randomly selected for sequencing. Sequencing was performed using the Sanger method and analyzed using the Sequencher software (Gene Codes Corporation, United States). In brief, qRT-PCR was performed on positive influenza samples using the One-Step RT-PCR kit (Qiagen, Germany), separated on a 2% agarose gel, and visualized by agarose gel electrophoresis. The primers that were used for the sequencing are listed in Supplementary Table S4. PCR products were purified with the EPPiC Fast enzyme (A&A Biotechnology, Poland). The DNA templates were sequenced using the BigDye Terminator v1.1 kit on an ABI Prism 3100 automated sequencer (Applied Biosystems, United States). Phylogenetic analysis of each influenza strain was performed by Geneious software (Dotmatics) using the neighbor-joining method. All ID numbers of sequences that were retrieved for phylogenetic analysis are listed in Supplementary Index 2.
To analyze how well the vaccine recognized currently circulating viruses, a hemagglutination inhibition (HI) test was performed. The HI test was performed by the WHO Collaborating Center, London, according to the standard WHO procedure using ferret post-infection antisera raised against the vaccine strains (WHO, 1997).
The HA protein structure was visualized using PyMOL software (Schrodinger, United States). Influenza sequences were retrieved from the EpiFluTM database of the Global Initiative on Sharing All Influenza Data (GISAID) (GISAID, 2023).
The predominant influenza virus types and subtypes detected among SMC hospitalized patients were compared to those of other regions in the Northern Hemisphere (WHO, 2023c).
The data on new SARS-CoV-2 cases in Israel are published daily by the Israel Ministry of Health (IMoH) and follow the rate of COVID-19 since February 2020 (WHO, 2023d). The data on SARS-CoV-2 variants are attributed to the World Consortium of SARS-CoV-2 sequences, which updates the variants of concern daily (Nextstrain, 2023).
During the 2021–2022 winter season, an increased percentage of influenza viruses was detected in November 2021 among samples from hospitalized patients that were abruptly interrupted by the arrival of the omicron variants. As shown in Figure 1A, only the influenza A(H3N2) subtype was detected in the winter season of 2021–2022. However, in the 2022–2023 winter season, influenza A(H1N1)pdm09 cases emerged with a lower percentage of influenza A(H3N2) and influenza B/Victoria lineages. The predominant strain in the 2022–2023 winter season was influenza A(H1N1)pdm09. At the end of the influenza season (March–April 2023), the percentage of influenza A cases decreased, while the percentage of influenza B-positives remained stable. Figure 1B represents the percentage of influenza rate alongside SARS-CoV-2 variants from September 2021 to March–April 2023. With the introduction of the omicron BA.1 variant, the influenza rate decreased immediately in January 2022. However, BA.2, BA.4, BA.5, BQ, and XBB variants that were present in Israel during 2022 and 2023 were not accompanied by a decrease in the influenza rate. Additionally, a cross-checking analysis of double infection by SARS-CoV-2 and influenza was also performed and showed only five cases of double infection (0.08%). The comparison of critical cases between patients with positive influenza A(H1N1)pdm09 vs. influenza A(H3N2) showed no significant difference (Supplementary Table S2).
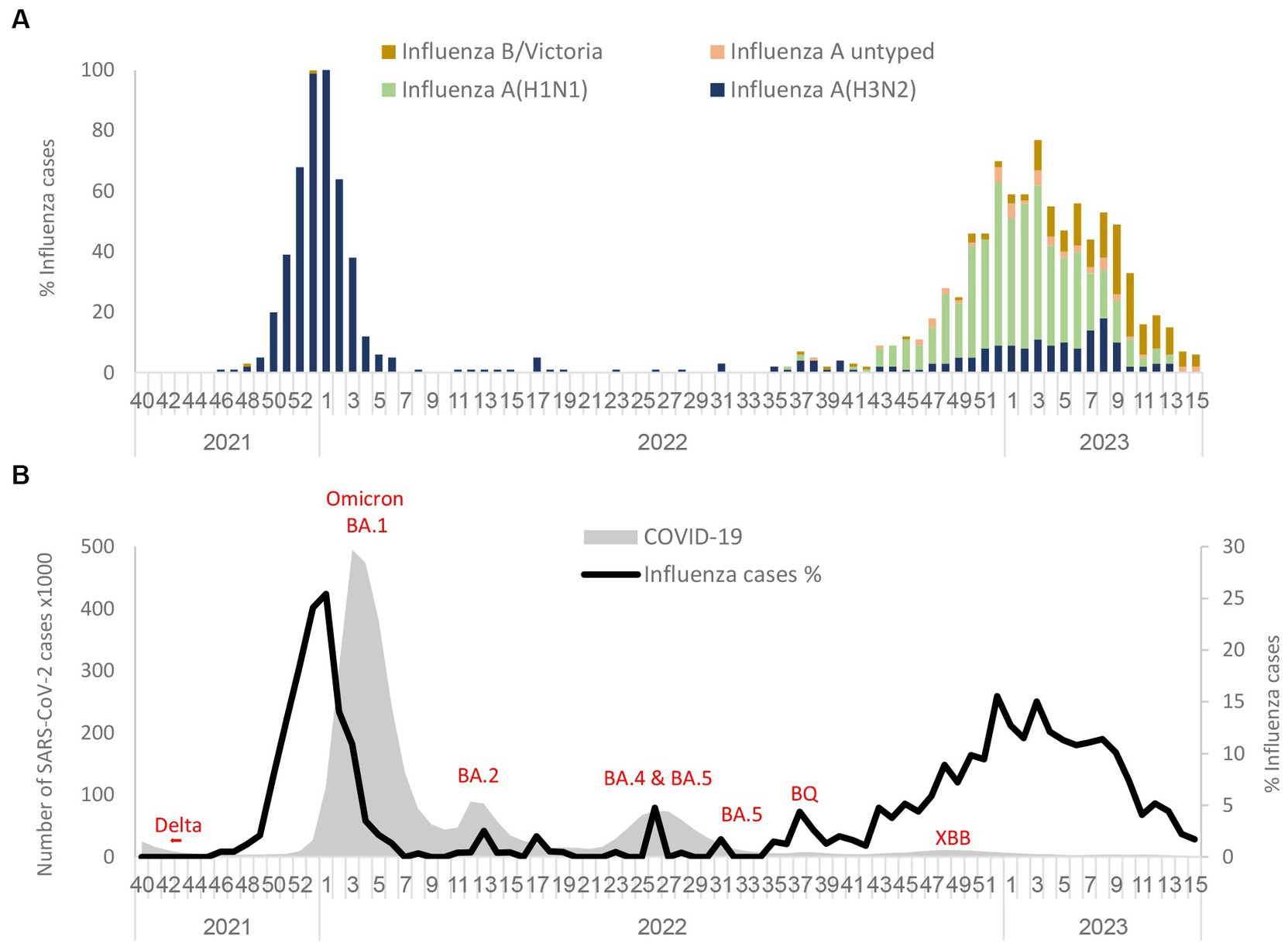
Figure 1. Circulation of influenza subtypes (A) and SARS-CoV2 (B) from 2021 to April 2023 in Israel. Influenza A(H1N1)pdm09, influenza A(H3N2), influenza B (Victoria), and influenza A untyped were collected and analyzed in the period of 2021 to April 2023 in SMC hospitalized patients (A). Number of SARS-CoV-2 detected in Israel from 2021 to 2023 along with the percentage of influenza rate in SMC hospitalized patients (B).
During the 2021–2022 season, influenza A(H3N2) was the predominant influenza virus in Central Asia, Europe, and North America in addition to Israel (Table 1). In the 2022–2023 season, influenza A(H1N1)pdm09 was predominant in Central Asia and Eastern Europe in addition to Israel. Influenza A(H3N2) was predominant in Southern Asia, Southwest Europe, and North America. Influenza B(Victoria) was predominant in Eastern Asia during both seasons.
Influenza A(H1N1)pdm09 detected during the 2022–2023 season were of the 6B.1A.5a.2a and 6B.1A.5a.2a.1 clades (Figure 2A). The vaccine strains (A/Wisconsin/588/2019 and A/Victoria/2570/2019) belong to 6B.1A.5a.2, which are located in a different branch from the samples detected in Israel, as shown in Figure 2A. Antigenic analysis is also shown in Figure 2A, in the right corner. The samples that were analyzed are indicated in the phylogenetic tree with an asterisk. Of the six samples, three (A/Israel/R12781/2022, A/Israel/R663/2023, and A/Israel/R11323/2022) were located in the upper branches and showed high hemagglutination inhibition (HI) titer (>1,280), while the samples A/Israel/R13228/2022, A/Israel/R463/2023, and A/Israel/R10810/2022 show low HI titer (<640) and were located in a different branch.
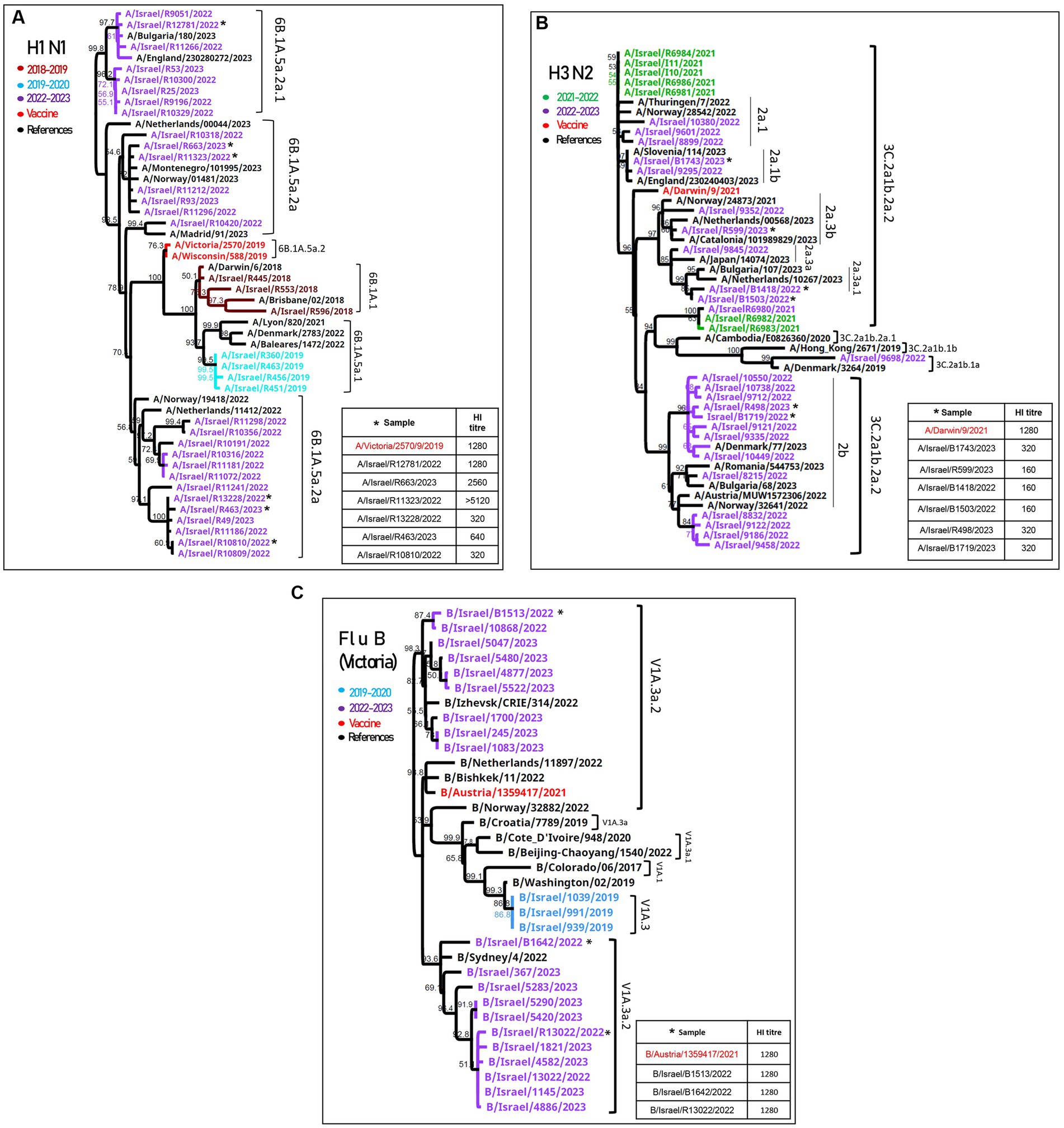
Figure 2. Phylogenetic and antigenic analyses of influenza A(H1N1)pdm09, influenza A(H3N2), and influenza B. Phylogenetic trees of patients’ samples positive for influenza in SMC hospitalized patients in the 2022–2023 season of (H1N1)pdm09 (A), influenza (H3N2) (B), and influenza B/Victoria (C). Antigenic analysis of each strain is presented in the right corner of each figure. The samples that were tested for hemagglutination inhibition (HI) are indicated with an asterisk.
The HA of influenza A(H3N2) samples from both seasons (2021–2022 and 2022–2023) were found to belong to the 3C.2a1b.2a.2 clade, adjacent to the vaccine clade (A/Drawin/9/2021) (Figure 2B). The antigenic analysis of six more samples shown in the right corner of Figure 2B had a low HI titer compared to the vaccine.
The influenza B/Victoria sequences were of the V1a.3a.2 lineage, similar to the vaccine clade (B/Austria/1359417/2021), and the antigenic analysis showed a similar HI titer compared to the vaccine (Figure 2C).
A comparison of the structure models of the vaccine homo-trimer (A/Wisconsin/588/2019) HA of influenza can be seen in Figure 2C. The main mutations that were detected in Israel are highlighted by different colors (Figure 3A). The mutations found for influenza A(H1N1)pdm09 were K54Q, S88P, D94N, V132I, A186T, Q189E, T216A, E224A, R259N H273Q, K308R, and F330X. A list of all the amino acid substitutions is presented in Supplementary Index 3. The HA modeling of influenza A(H3N2) in Figure 3B emphasizes the mutations detected in the positive samples of influenza A(H3N2) in the 2022–2023 season. The mutations found in Israel were E50K, F79V, I140K, S156H, N186D, and G225D. The influenza B mutations identified in Israel were E183K, A209E, D249E, and S260P (Figure 3C).
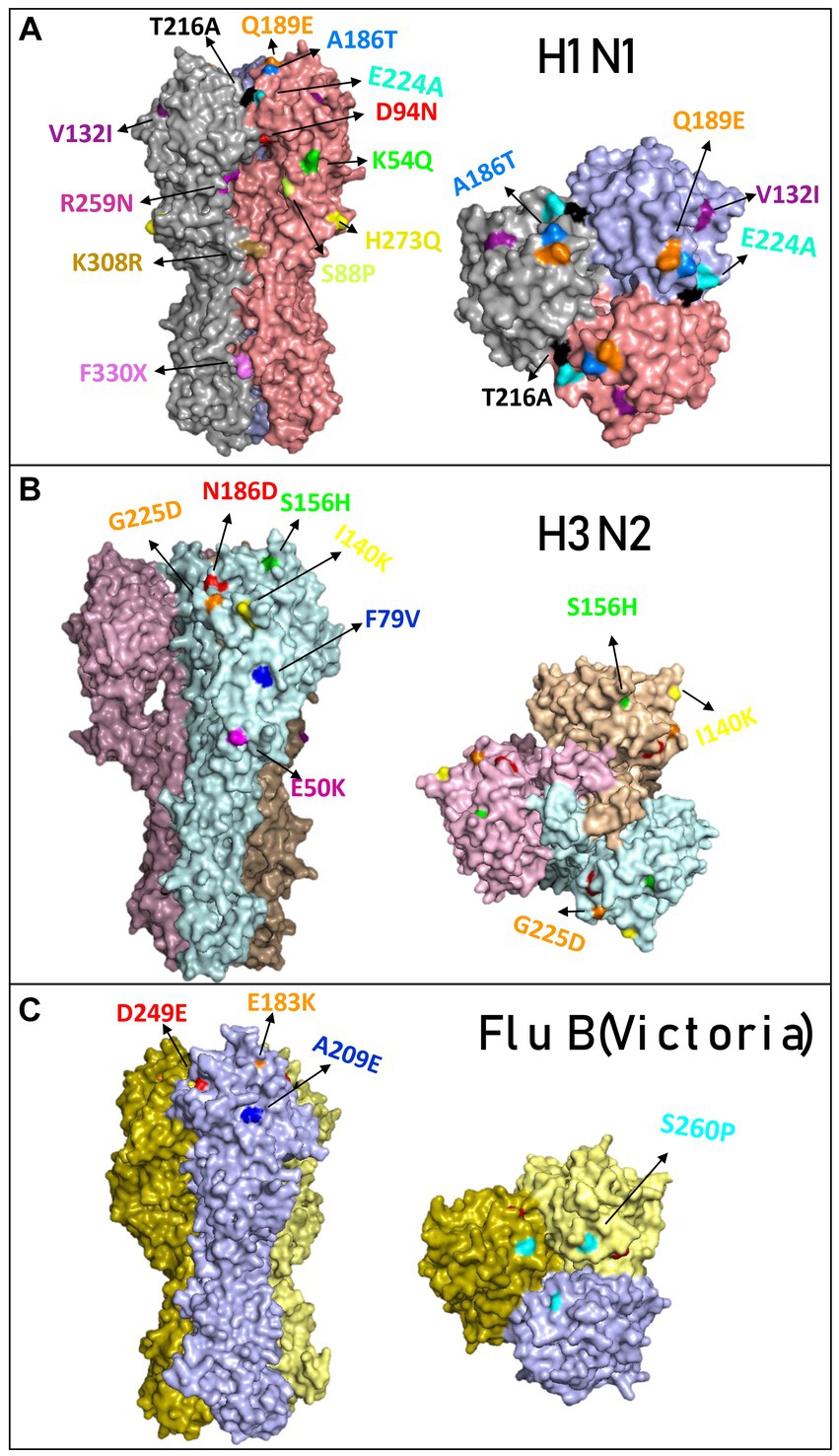
Figure 3. Overview of the hemagglutinin amino acid substitutions of the influenza isolated from hospitalized patients compared to the vaccine. Modeling of hemagglutinin (HA) and its amino acid substitutions compared to the vaccine. The samples were compared to the vaccines for influenza A(H1N1)pdm09 (compared with sample A/Israel/R13228/2022) (A), influenza (H3N2) (compared with sample A/Israel/R8215/2022) (B), and influenza B/Victoria (compared with sample B/Israel/R13022/2022) (C). Mutations found in the 2022–2023 season are colored differently.
Analysis of influenza incidence prior to and during the COVID-19 pandemic identified a significantly lower number of cases alongside the emergence of the SARS-CoV-2 alpha and delta variants, as seen in other reports (Olsen et al., 2021; Lauring et al., 2022). As with other SARS-CoV-2 variants, the emergence of the omicron variant in December 2021 was followed by a dramatic reduction in influenza cases among hospitalized patients in Israel (Figure 1). The results shown in this study (Figure 1) demonstrate that during the 2022–2023 season, influenza activity among hospitalized patients and the epidemic curve were similar to those of the pre-pandemic seasons. This “typical” influenza pattern among hospitalized patients occurred, while the circulation of the SARS-CoV-2 variants was low throughout the season (WHO, 2023c,d). The possibility of dual infection of SARS-CoV-2 and influenza was also examined in this study, as dual infection may lead to severe cases and increased mortality (Bai et al., 2021; Tang et al., 2022). However, a very low percentage of such cases were found (0.08%), presumably as a result of SARS-CoV-2 circulation at that time.
In the 2021–2022 season, the influenza virus detected among hospitalized patients belonged to influenza A(H3N2), clade 3C.2a1b.2a.2. This clade was the predominant influenza virus throughout the world in the 2021–2022 season (WHO, 2023a). Subsequently, in 2022–2023, clade 3C.2a1b.2a.2 was still circulating in the world; however, influenza A(H3N2) was less frequent in hospitalized patients in Israel. The predominant virus in the 2022–2023 season was influenza (H1N1)pdm09, which was not detected in 2021–2022, while in Southern Asia, most of Europe, and North America, influenza A(H3N2) predominated [Centers for Disease Control and Prevention (CDC), 2023; Europe Centers for Disease Control and Prevention (ECDC), 2023]. Influenza B/Victoria was also detected but less frequently in hospitalized patients, and its positive rate remained stable at the end of the 2022–2023 season, while influenza A decreased, corresponding with its typical end-of-the-season upticks (Uyeki et al., 2022). Influenza B/Yamagata was not detected among hospitalized patients in Israel, similar to global findings during the COVID-19 pandemic (Koel et al., 2013).
The phylogenetic analysis of the 2022–2023 influenza A(H1N1)pdm09 strain found a lack of compatibility between the vaccine and the circulating influenza A(H1N1)pdm09 viruses among hospitalized patients in Israel. While the vaccine virus belongs to 6B.1A.5a.1, the influenza A(H1N1)pdm09 virus detected in patients evolved into two clades, 6B.1A.5a.2a and 6B.1A.5a.2a.1, similar to influenza A(H1N1)pdm09 viruses detected in Europe. This result coincides with the immunogenicity experiments, which showed a low HI titer that indicates a lack of compatibility between the circulating strain and the vaccine. Reduced compatibility between the vaccine and the circulating influenza viruses is attributed to the amino acid substitutions in hemagglutinin positions K54Q, A186T, Q189E, E224A, R259K, and K308R detected in hospitalized patients in Israel (Supplementary Index 3). Mutations in positions 186 and 189 have been found to belong to the patches responsible for major antigenic change (Koel et al., 2013; Kratsch et al., 2016). Influenza A(H3N2) also evolved into smaller clades from the 3C.2a1b.2a.2 clade, but reports from the WHO showed that compatibility with the influenza A(H3N2) vaccine remained (WHO, 2023a).
Despite the match between the vaccine and the circulating influenza A(H3N2), the United States was still burdened by more influenza A(H3N2) infections, possibly due to the low vaccine coverage in the population (Uyeki et al., 2022). In Israel in 2022–2023, vaccination rates were relatively low (17.9%). Nevertheless, the influenza A(H3N2) infection rate was lower in Israel compared with the United States and Europe, which may have been due to immunity gained in the 2021–2022 winter season when influenza A(H3N2) circulated. In contrast, influenza A(H1N1)pdm09 last circulated in Israel in the winter season of 2019–2020, which may explain its dominance in Israel in 2022–2023 (Fratty et al., 2022; Omer et al., 2022; Malosh et al., 2023). Overall, the prevalence of influenza A strains tends to shift each year between influenza A(H1N1)pdm09 and influenza A(H3N2), as seen prior to the emergence of SARS-CoV-2 in Israel [Europe Centers for Disease Control and Prevention (ECDC), 2023].
For each coming winter season, a composition for the influenza vaccine is recommended by the WHO after examining the recent circulating subtypes (WHO, 2023a). Since the influenza A(H1N1)pdm09 included in the vaccine was incompatible in 2022–2023, the WHO recommended the inclusion of influenza with stronger clade-specific recognition (WHO, 2023a,b). Hence, we suggest removing the B/Yamagata component from the vaccine, using a higher antigen content in targeted populations (such as older adults), and using cell culture for propagation, a procedure that introduces fewer molecular changes (Uyeki et al., 2022). Alternatively, other technologies, such as mRNA technology, which was proven to be a game-changer during the COVID-19 pandemic, can be used.
In conclusion, higher rates of influenza activity were measured in the 2022–2023 season as compared with the 2021–2022 season. The influenza strain that dominated the 2022–2023 season was unique in Israel compared to Europe and the United States. The majority of influenza samples in hospitalized patients in Israel in 2022–2023 belonged to influenza A(H1N1)pdm09, but influenza A(H3N2) and influenza B/Victoria were also detected. Phylogenetic and antigenic analyses of two out of the three influenza types detected in Israel showed a resemblance to the vaccine recommended by the WHO, with the exception of influenza A(H1N1)pdm09.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The studies involving humans were approved by the institutional review board (IRB) of the Sheba Medical Center (Helsinki Number 7688-20-SMC). The studies were conducted in accordance with the local legislation and institutional requirements. This study was a retrospective using anonymized data, so did not require written informed consent.
IF: Investigation, Writing – original draft, Formal analysis. MJ: Writing – review & editing. NZ: Writing – review & editing. IN: Investigation, Writing – review & editing. NA: Investigation, Writing – review & editing. LK: Investigation, Writing – review & editing. LG-A: Writing – review & editing, Formal analysis. AR: Writing – review & editing, Formal analysis. AG-F: Supervision, Writing – review & editing. YL: Supervision, Writing – review & editing. MM: Supervision, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors gratefully acknowledge Nicola S. Lewis and Ruth Harvey from the WHO Collaborating Center for Reference and Research on Influenza at the Francis Crick Institute in the United Kingdom for the HI data. Additionally, the authors are thankful for the originating and submitting laboratories of the sequences from GISAID’s Influenza Database used in the phylogenetic analysis and also grateful for the professional assistance of David Fratty in this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1296179/full#supplementary-material
Bai, L., Zhao, Y., Dong, J., Liang, S., Guo, M., Liu, X., et al. (2021). Coinfection with influenza a virus enhances SARS-CoV-2 infectivity. Cell Res. 31, 395–403. doi: 10.1038/s41422-021-00473-1
Centers for Disease Control and Prevention (CDC) (2023). Weekly U.S. influenza surveillance report. Available at: https://www.cdc.gov/flu/weekly/index.htm
Europe Centers for Disease Control and Prevention (ECDC) (2023). Season overview. Available at: https://flunewseurope.org/SeasonOverview
Folgueira, L., Moral, N., Pascual, C., and Delgado, R. (2019). Comparison of the panther fusion and Allplex assays for the detection of respiratory viruses in clinical samples. PLoS One 14:e0226403. doi: 10.1371/journal.pone.0226403
Fratty, I. S., Reznik-Balter, S., Nemet, I., Atari, N., Kliker, L., Sherbany, H., et al. (2022). Outbreak of influenza and other respiratory viruses in hospitalized patients alongside the SARS-CoV-2 pandemic. Front. Microbiol. 13:902476. doi: 10.3389/fmicb.2022.902476
GISAID (2023). GISAID. Available at: https://platform.epicov.org/epi3/frontend#355366
Grohskopf, L. A., Sokolow, L. Z., Broder, K. R., Olsen, S. J., Karron, R. A., Jernigan, D. B., et al. (2016). Prevention and control of seasonal influenza with vaccines. MMWR Recomm. Rep. 65, 1–54. doi: 10.15585/mmwr.rr6505a1
Huddleston, J., Barnes, J. R., Rowe, T., Xu, X., Kondor, R., Wentworth, D. E., et al. (2020). Integrating genotypes and phenotypes improves long-term forecasts of seasonal influenza A/H3N2 evolution. elife 9:60067. doi: 10.7554/eLife.60067
Koel, B. F., Burke, D. F., Bestebroer, T. M., van der Vliet, S., Zondag, G. C., Vervaet, G., et al. (2013). Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 342, 976–979. doi: 10.1126/science.1244730
Kratsch, C., Klingen, T. R., Mumken, L., Steinbruck, L., and McHardy, A. C. (2016). Determination of antigenicity-altering patches on the major surface protein of human influenza a/H3N2 viruses. Virus Evol. 2:25. doi: 10.1093/ve/vev025
Lauring, A.S., Tenforde, M.W., Chappell, J.D., Gaglani, M., Ginde, A.A., McNeal, T., et al. (2022). Clinical severity and mRNA vaccine effectiveness for omicron, delta, and alpha SARS-CoV-2 variants in the United States: a prospective observational study. medRxiv [Preprint]. doi: 10.1101/2022.02.06.22270558
Malosh, R. E., McGovern, I., and Monto, A. S. (2023). Influenza during the 2010-2020 decade in the United States: seasonal outbreaks and vaccine interventions. Clin. Infect. Dis. 76, 540–549. doi: 10.1093/cid/ciac653
Meningher, T., Hindiyeh, M., Regev, L., Sherbany, H., Mendelson, E., and Mandelboim, M. (2014). Relationships between a(H1N1)pdm09 influenza infection and infections with other respiratory viruses. Influenza Other Respir. Viruses 8, 422–430. doi: 10.1111/irv.12249
Nextstrain (2023). Genomic epidemiology of SARS-CoV-2 with subsampling focused globally over the past 6 months. Available at: https://nextstrain.org/ncov/gisaid/global/6m
Olsen, S. J., Winn, A. K., Budd, A. P., Prill, M. M., Steel, J., Midgley, C. M., et al. (2021). Changes in influenza and other respiratory virus activity during the COVID-19 pandemic-United States, 2020-2021. Am. J. Transplant. 21, 3481–3486. doi: 10.1111/ajt.16049
Omer, I., Rosenberg, A., Sefty, H., Pando, R., Mandelboim, M., Mendelson, E., et al. (2022). Lineage-matched versus mismatched influenza B vaccine effectiveness following seasons of marginal influenza B circulation. Vaccine 40, 880–885. doi: 10.1016/j.vaccine.2021.12.056
Pando, R., Stern, S., Nemet, I., Glatman-Freedman, A., Sefty, H., Zuckerman, N. S., et al. (2021). Diversity in the circulation of influenza a(H3N2) viruses in the northern hemisphere in the 2018-19 season. Vaccine 9:375. doi: 10.3390/vaccines9040375
Petrova, V. N., and Russell, C. A. (2018). The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 16:60. doi: 10.1038/nrmicro.2017.146
Price, A. M., Flannery, B., Talbot, H. K., Grijalva, C. G., Wernli, K. J., Phillips, C. H., et al. (2023). Influenza vaccine effectiveness against influenza a(H3N2)-related illness in the United States during the 2021-2022 influenza season. Clin. Infect. Dis. 76, 1358–1363. doi: 10.1093/cid/ciac941
Tang, C. Y., Boftsi, M., Staudt, L., McElroy, J. A., Li, T., Duong, S., et al. (2022). SARS-CoV-2 and influenza co-infection: a cross-sectional study in Central Missouri during the 2021-2022 influenza season. Virology 576, 105–110. doi: 10.1016/j.virol.2022.09.009
Uyeki, T. M., Hui, D. S., Zambon, M., Wentworth, D. E., and Monto, A. S. (2022). Influenza. Lancet 400, 693–706. doi: 10.1016/S0140-6736(22)00982-5
WHO (1997). Committee for Proprietary Medicinal Products (CPMP). Note for guidance on harmonization of requirements for influenza vaccines. Available at: http://www.ema.europa.eu/docs/en_GB/docu
WHO (2023a). Influenza virus characterization: Summary report, Europe. Available at: https://www.who.int/europe/publications/i/item/WHO-EURO-2023-6189-45954-68789
WHO (2023b). Recommended composition of influenza virus vaccines for use in the 2023–2024 northern hemisphere influenza season.
WHO (2023c). Comparison of number of influenza detection by subtype. World Health Organization. Available at: https://app.powerbi.com/
WHO (2023d). World health organization. WHO coronavirus (COVID-19) dashboard. Available at: https://covid19.who.int/
Keywords: influenza A, influenza B, vaccine, hemagglutinin, SARS-CoV-2
Citation: Fratty IS, Jurkowicz M, Zuckerman N, Nemet I, Atari N, Kliker L, Gur-Arie L, Rosenberg A, Glatman-Freedman A, Lustig Y and Mandelboim M (2024) Influenza vaccine compatibility among hospitalized patients during and after the COVID-19 pandemic. Front. Microbiol. 14:1296179. doi: 10.3389/fmicb.2023.1296179
Received: 18 September 2023; Accepted: 29 December 2023;
Published: 23 January 2024.
Edited by:
Wenjun Song, Guangzhou National Laboratory, ChinaReviewed by:
Lalit Batra, University of Louisville, United StatesCopyright © 2024 Fratty, Jurkowicz, Zuckerman, Nemet, Atari, Kliker, Gur-Arie, Rosenberg, Glatman-Freedman, Lustig and Mandelboim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michal Mandelboim, TWljaGFsLk1hbmRlbGJvaW1Ac2hlYmEuaGVhbHRoLmdvdi5pbA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.