
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 06 November 2023
Sec. Biology of Archaea
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1296008
This article is part of the Research TopicThe Methane Moment – Cross-Boundary Significance of MethanogensView all 20 articles
 Bela Haifa Khairunisa1†
Bela Haifa Khairunisa1† Christian Heryakusuma2,3†
Christian Heryakusuma2,3† Kelechi Ike4
Kelechi Ike4 Biswarup Mukhopadhyay2,3,5*
Biswarup Mukhopadhyay2,3,5* Dwi Susanti1*
Dwi Susanti1*Production of methane by methanogenic archaea, or methanogens, in the rumen of ruminants is a thermodynamic necessity for microbial conversion of feed to volatile fatty acids, which are essential nutrients for the animals. On the other hand, methane is a greenhouse gas and its production causes energy loss for the animal. Accordingly, there are ongoing efforts toward developing effective strategies for mitigating methane emissions from ruminant livestock that require a detailed understanding of the diversity and ecophysiology of rumen methanogens. Rumen methanogens evolved from free-living autotrophic ancestors through genome streamlining involving gene loss and acquisition. The process yielded an oligotrophic lifestyle, and metabolically efficient and ecologically adapted descendants. This specialization poses serious challenges to the efforts of obtaining axenic cultures of rumen methanogens, and consequently, the information on their physiological properties remains in most part inferred from those of their non-rumen representatives. This review presents the current knowledge of rumen methanogens and their metabolic contributions to enteric methane production. It also identifies the respective critical gaps that need to be filled for aiding the efforts to mitigate methane emission from livestock operations and at the same time increasing the productivity in this critical agriculture sector.
Livestock production in the US emitted close to 200 million metric tons of CO2-equivalent (MMT CO2–e) of methane, mainly originating from enteric fermentation in beef and dairy cattle representing 72 and 25% of emissions from livestock, respectively (EPA, 2022). The corresponding value at the global scale is approximately 2,500 MMT CO2-e (EPA, 2023a), and it is estimated to rise substantially due to an increase in demand for milk and meat to feed the 9.8 billion global population by 2050 (FAO, 2018; Henchion et al., 2021).
Methane is 28 times more potent greenhouse gas (GHG) with a much shorter shelf-life than CO2 (EPA, 2023b). In the rumen, it is produced as a by-product of microbial fermentation, and methanogenic archaea or methanogens are the only microorganisms that are known to produce methane anaerobically (Smith and Hungate, 1958; Ramanathan et al., 1985; Wolfe, 1992). In addition to contributing to global warming, methane emission from the rumen causes a loss of 2–12% of the energy provided by the feed (Johnson and Johnson, 1995; Janssen, 2010). Hence, a reduction of methane emission from cattle would have a greater near-term contribution to the effort toward mitigating global climate change and improving animal productivity (Janssen, 2010; Beauchemin et al., 2020).
For the above-mentioned importance, the metabolism of rumen microbes including methanogens has been investigated for almost eight decades (Barker, 1936; Elsden, 1945; Hungate, 1950; Beijer, 1952; Hungate, 1966; Henderson et al., 2015; Seshadri et al., 2018). These studies yielded a plethora of basic and applied science information about rumen methanogens including their role in facilitating microbial fermentation in the rumen (Hungate, 1966; Beauchemin et al., 2020). These details have been leveraged for developing tools for mitigating methane emission in the livestock industry and some of these can provide an average of 30% reduction in methane production with acceptable safety in both beef and dairy cattle (Yu et al., 2021). However, the outcome varies greatly (Patra et al., 2017; Arndt et al., 2022). What causes such variabilities? Which methanogens escape such intervention and how could one target them effectively? What factors drive the composition of a rumen methanogen community over another, spatially and temporally? Answering these questions requires a deeper understanding of the metabolic diversity and in situ physiology of rumen methanogens, which sorely remains incomplete even after close to eight decades of interrogation. It is because the current knowledge base for this field has mostly been built on studies with pure culture isolates from the rumen, which are a few, and inferences from the properties of non-rumen methanogen isolates (Jeyanathan, 2010; Seshadri et al., 2018). The technical hurdles of working with strict anaerobes and the absence of clues to specific auxotrophies have limited the isolation efforts, which could have allowed useful in vitro studies.
The culture-independent approaches leveraging high throughput omics are beginning to fill the above-mentioned gap in terms of phylogenetic diversity and metabolic potentials. The discovery of species from the Methanomassiliicoccales order that provide an additional route for removing the hydrogen-based thermodynamic block on ruminal fermentation (Borrel et al., 2013) and key genomic features that allow rumen methanogens to associate with other organisms (Leahy et al., 2010; Ng et al., 2016) and battle the toxicity of plant product (i.e., tannin) are examples of such advances (Kelly et al., 2016c; Loh et al., 2020). However, the absence of information on the metabolic and physiological properties of individual rumen methanogens that are generally obtained from studies on pure culture isolates or even low complexity enrichments has prevented making a clear sense of physiological data originating from in vivo or whole animal-based measurements.
This review presents a summary and analysis of the past and evolving knowledge of rumen methanogens (Figure 1) including the ongoing and upcoming research that would fill the above-mentioned gaps and help the efforts to mitigate enteric methane emissions while bringing sustainability to the livestock industry.
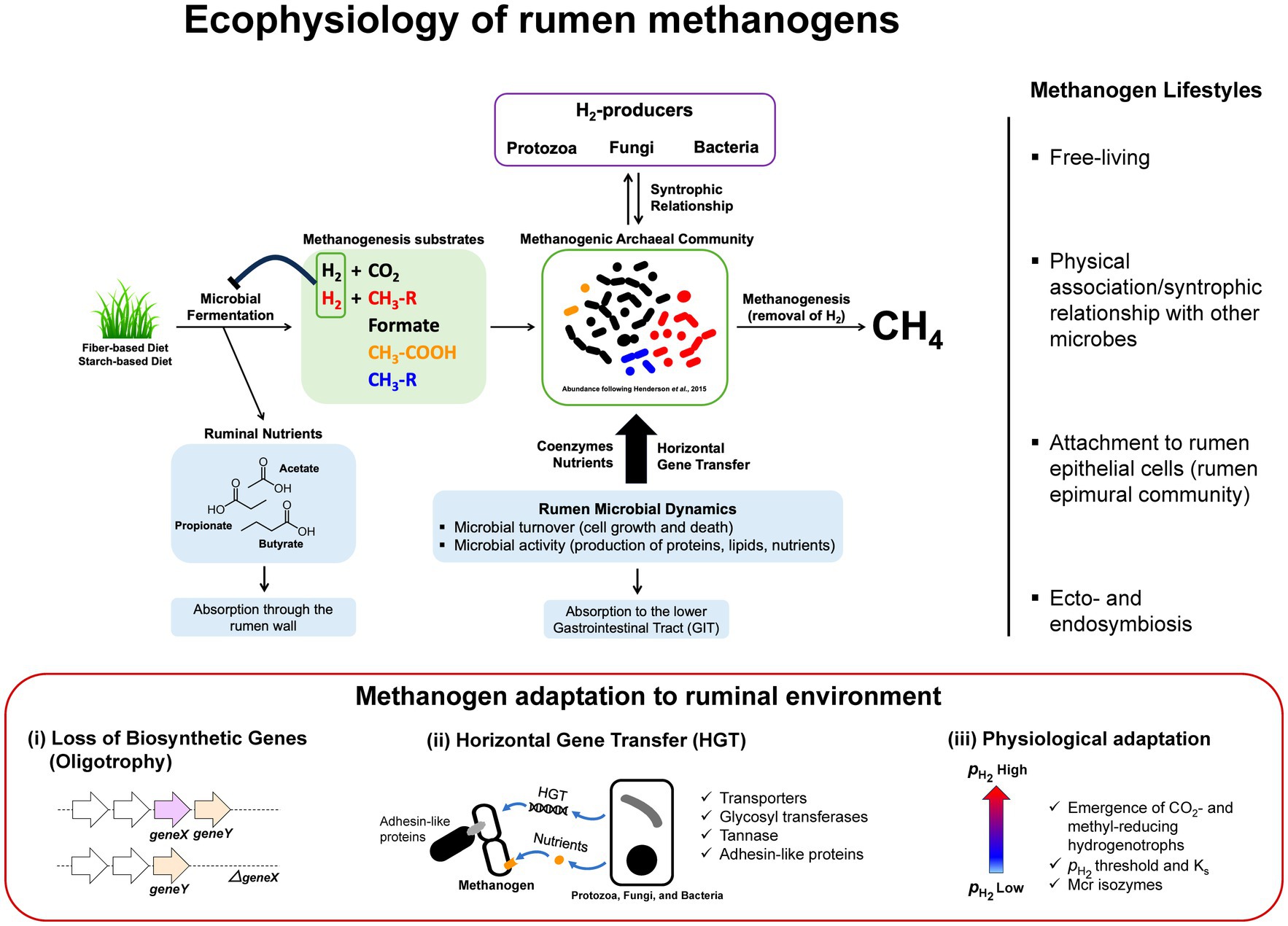
Figure 1. Ecophysiology and metabolic adaptation of rumen methanogens. A schematic diagram illustrating functional roles of methanogens that facilitate the continuation of rumen microbial fermentation by removal of H2 from microbial fermentation to generate methane. In the process, methanogens interact with different functional guilds via syntrophic associations and cross-feedings. Uptakes of nutrients and genetic materials via horizontal gene transfer (HGT) are shaping rumen methanogen metabolism, physiology, and lifestyle resulting in better adaptations and competitiveness in the rumen environment. Interactions between methanogens and other rumen microbiota are diverse and complex where methanogens are found as free-living, in a physical association or syntrophic relationship with other microbes, attach to the rumen epithelial cells as part of rumen epimural community, or ecto−/endosymbiosis with protozoa (right panel). Metabolic adaptation of methanogens in rumen environment (lower panel) results in loss of biosynthetic genes generating oligotrophy, acquisition of new functions through HGT, and physiological adaptation to methanogenic substrate fluctuations in the rumen (i.e., high and low conditions following feeding) that have significant impacts on the emergence of CO2- and methyl-reducing hydrogenotrophs (i.e., Ks and the deployment of different Mcr isozymes).
Ruminants gain 70% of their energy from microbial activities that degrade feed materials in the first two compartments of the digestive tract, the rumen and the reticulum, which collectively called reticulorumen and hereafter is referred to as rumen (Flint and Bayer, 2008; Yeoman and White, 2014). The rumen microbial community is composed of bacteria, protozoa, archaea, and fungi in the order of the most abundant to the least (Hungate, 1966; Henderson et al., 2015; Seshadri et al., 2018); highly abundant and diverse virus populations are also important components in the rumen even though it has not been studied significantly (Gilbert et al., 2020). These individual members of rumen microbial community have been co-evolving with the ruminants for about 50 million years (Webb and Taylor, 1980; Hackmann and Spain, 2010; Jiang et al., 2014), making them resilient to environmental perturbation through their overlapping metabolic functionality (Weimer, 2015). Their concerted actions convert fermentable carbohydrates and amino acids anaerobically via fermentation into volatile fatty acids that provide energy to the animals and surplus reducing equivalents in the forms of hydrogen and formate, with most products coming from carbohydrates. If unutilized, excess H2 blocks the progress of fermentation thermodynamically, and in the rumen and many other anaerobic biodegradation systems, this block is removed by methanogens that utilize the excess H2 and generate methane (Figures 1, 2; Zinder, 1993).
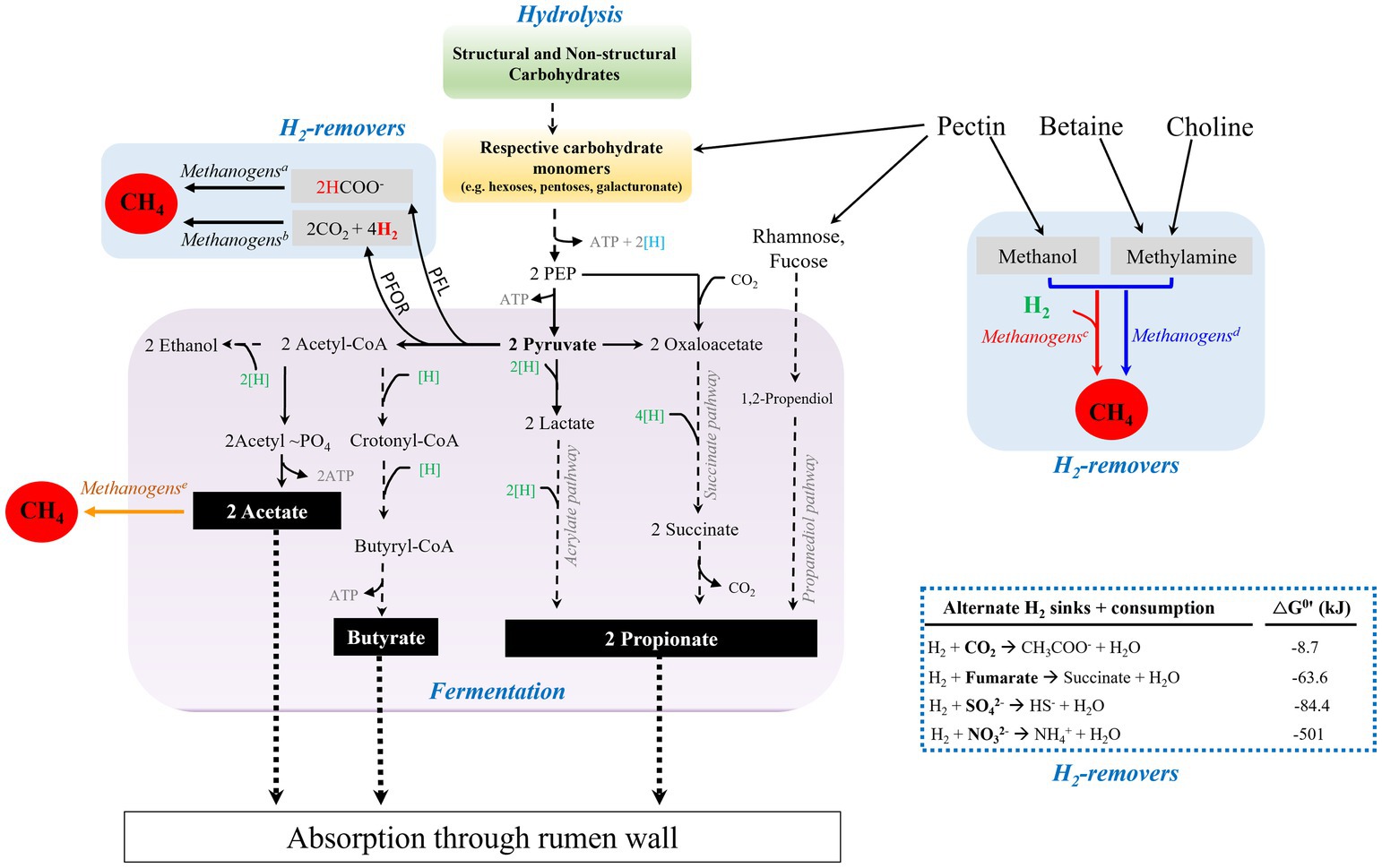
Figure 2. Carbohydrate degradation in the reticulorumen (rumen) of ruminants. Microbial degradation of structural carbohydrates and fermentation of resulting sugar monomers generate fatty acids, acetate, propionate, and butyrate. H2 is generated from the electron confurcation of NADH and Fdxred during decarboxylation of pyruvate to generate acetyl-CoA. Generation of acetyl CoA from pyruvate can be performed by the actions of Pyruvate:Ferredoxin OxidoReductase (PFOR) or Pyruvate Formate Lyase (PFL). The H2 level is kept low by formate-dependent methanogenesis, CO2- and methyl-reducing hydrogenotrophs (superscripts a, b, and c, respectively), thus relieving the thermodynamic block on reoxidation of NADH and fermentation. Methyl-dismutating and acetoclastic methanogenesis are not commonly found in the rumen (superscripts d and e, respectively). When available, sulfate, nitrate, and fumarate can be used as alternate hydrogen sinks, blue dashed-lined box. Blue and green [H], production and consumption of reducing equivalent or (NAD(P)H), respectively; black dashed-lines, multi-step pathway; black solid lines, process in the rumen; black dotted-lines, absorption of volatile fatty acids by rumen wall.
Working synergistically, a group of bacteria, fungi, and protozoa hydrolyze cellulose and hemicellulose fibers into respective sugar monomers, and ferment these products into primarily three major volatile fatty acids, namely acetate, propionate, and butyrate that are absorbed by rumen epithelial walls (Hungate, 1966; Czerkawski and Breckenridge, 1973; Prins and van der Meer, 1976; Wolin, 1979; Williams and Coleman, 1997; Ragsdale, 2003; Sawers and Clark, 2004; Reichardt et al., 2014; Henderson et al., 2015; Hackmann et al., 2017; Gruninger et al., 2019; Ungerfeld, 2020; Williams et al., 2020; Pereira et al., 2022). In addition, lactate, ethanol, and succinate are produced as reduced intermediates (Gottschalk, 1986; Hackmann et al., 2017), where lactate and succinate are further converted to propionate (Gottschalk, 1986; Weimer, 1998; Reichardt et al., 2014; Hackmann et al., 2017; Moraïs and Mizrahi, 2019; Ungerfeld, 2020). Figure 2 summarizes this overall process. Acetate, propionate, and butyrate account for 40–75%, 15–40%, and 10–20% of the total rumen VFAs, respectively (Wolin, 1960; Bergman, 1990; DeFrain et al., 2004). Propionate serves as a major precursor for the biosynthesis of glucose through gluconeogenesis in the liver, which in turn is used as an energy source for the animal (Young, 1977). Acetate and butyrate can be used as precursors in lipid biogenesis by the host (Black et al., 1961; Hanson and Ballard, 1967; Moran, 2005).
Production of acetate and butyrate from glucose is associated with more negative ΔGo’ values than is propionate production (van Lingen et al., 2016). In addition, the production of propionate is associated with a net consumption of two moles of H2 per mole of glucose utilized, whereas that of acetate and butyrate lead to net productions of four and two moles of H2, respectively (van Lingen et al., 2016; Leahy et al., 2022). Accordingly, despite a higher thermodynamic feasibility of acetate and butyrate production from glucose under standard conditions, the generation of these VFAs is less favored as it leads to H2 accumulation and consequent thermodynamic inhibition of microbial fermentation.
The above-mentioned fermentation process generates pyruvate, ATP, and NADH (Figure 2). To allow unimpeded continuation of the fermentation process, NAD+ must be regenerated (Baldwin and Allison, 1983; Stams and Plugge, 2009). Depending on the prevailing cellular redox status (i.e., NAD+/NADH ratio) of the cells, it can be done through the production of reduced fermentation products such as ethanol, lactate, and propionate, and/or hydrogen generation via NADH:Ferredoxin oxidoreductase coupled with a hydrogenase or via electron confurcation reaction involving NADH and reduced ferredoxin (Fdxred) (Schut and Adams, 2009; Stams and Plugge, 2009). Processing of pyruvate via Pyruvate Formate Lyase (PFL) provides acetyl-CoA and formate, and the latter can be excreted or oxidized to H2 by formate hydrogen lyase (Baldwin and Allison, 1983, Stams and Plugge, 2009). As H2 accumulates, elevating its partial pressure or , it blocks NADH oxidation thermodynamically (Baldwin and Allison, 1983; Gottschalk, 1986; Stams and Plugge, 2009); thus, H2 is a central regulator and called the ‘currency’ of rumen fermentation (Czerkawski, 1986). Hydrogenotrophic methanogens remove this block on fermentation by consuming H2 via CO2 and methyl group reduction to methane (4H2 + CO2 → CH4 + 2H2O; H2 + CH3-X → CH4 + HX) and allowing NAD+ regeneration (Baldwin and Allison, 1983; Stams and Plugge, 2009). Excretion of formate, as mentioned above, lowers pH and its sequential oxidation to H2 imposes a thermodynamic block and methanogens alleviate these problems via formate methanogenesis (4HCOO− + 4H+ → CH4 + 3CO2 + 2H2O) (Figure 2).
The process of electron transfer from a hydrogen producer to a methanogen via hydrogen as a vehicle was the first recognized case of interspecies electron transfer (IET) (Bryant et al., 1967). Direct IET (DIET) occurring via conducting pili or nanowires, or IET employing extracellular cytochromes that occur in other ecological systems (Lovley and Holmes, 2022) remains to be investigated for rumen microbiome (Kelly et al., 2022). With fiber digestion by protozoa, a unique reductant transfer process is seen. Here, protozoa release excess reductant as H2 through hydrogenosome, a mitochondria-type organelle representing an ancient bacterial endosymbiont (Lewis et al., 2020), which is captured directly by methanogens living syntrophically as protozoal endo- and ecto-symbiont (Vogels et al., 1980; Stumm and Zwart, 1986; Belanche et al., 2014). These symbiotic methanogens representing 10–20% of rumen methanogens contribute to 15–35% of ruminal methane production (Hegarty, 1999; Morgavi et al., 2008, 2012). This association is non-specific in terms of a methanogen’s selectivity for protozoa type (Henderson et al., 2015).
Figure 2 shows alternate routes for hydrogen removal in the rumen with the respective thermodynamic potentials. Except for acetogenesis (4H2 + 2CO2 → CH3COO− + 2H2O + H+), which utilizes readily available CO2, these alternate avenues are used only if the respective electron acceptors are available in the rumen. For example, the sulfate reduction pathway occurs only when the sulfate concentration in the rumen is sufficient (Huisingh et al., 1974).
Methanogens account for less than 3.3% of the total rRNA gene sequences in bovine rumen (Patra et al., 2017) and the dominant rumen methanogens are rather conserved across geographical regions (Henderson et al., 2015). Despite this relatively low abundance, methanogens have a major impact on microbial metabolism in this ecosystem for the reasons mentioned above. In this section, the diversity and methanogenesis or energy conservation processes of rumen methanogens are summarized and discussed.
According to the taxonomic classification of the Genome Taxonomy Database (GTDB; Parks et al., 2022), the methanogen phyla represented in the rumen microbiome are Halobacteriota (H), Methanobacteriota (M), and Thermoplasmatota (T) (Janssen and Kirs, 2008; Henderson et al., 2015; Parks et al., 2022). These methanogens belong to four orders (phyla): Methanobacteriales (M), Methanomicrobiales (H), Methanosarcinales (H), Methanomassiliicoccales (T). The reports of Methanococcales (M) especially from Methanocaldococcaceae family and Methanopyrales (M) phyla, representing hyperthermophiles, in rumen samples (Janssen and Kirs, 2008; Henderson et al., 2015; Tan et al., 2021) are likely artifactual, and Methanocellales (H) have never been found in rumen. The identification of Methanomassiliicoccales in the rumen as major utilizers of hydrogen via a non-CO2 reduction route reshaped the concept of hydrogenotrophy (Borrel et al., 2013, 2014; Li et al., 2016; Kelly et al., 2016a,b; Garcia et al., 2022).
The rumen methanogen community is dominated by members of Methanobacteriales, especially from Methanobrevibacter and Methanosphaera genera, and those of Methanomassiliicoccales, with small contributions from Methanomicrobium and Methanosarcina genera (Henderson et al., 2015). Methanobrevibacter gottschalkii, Methanobrevibacter ruminantium, Methanosphaera sp., and two Methanomassiliicoccaceae (formerly grouped as the rumen cluster C or RCC) comprise close to 90% of the total rumen methanogen rRNA gene sequences with Methanobrevibacter covering 74% of the total sequences and the rest 16% belonging to Methanosphaera sp. and Methanomassiliicoccaceae (Janssen and Kirs, 2008; Henderson et al., 2015). These abundance values, however, are dynamic and vary across hosts and diets, even though the core methanogen players are rather conserved (Henderson et al., 2015). Table 1 describes all known pure culture isolates of rumen methanogens and their key cellular characteristics. Some of these features are discussed below; energy metabolism is covered in Section 3.2.
Members of this order reduce CO2 with H2 and some use formate, CO, and secondary alcohols as reductants (Liu, 2010a); Methanosphaera, an exception, reduce methanol with H2 (Miller and Wolin, 1985). Their cell walls contain an archaeal-type peptidoglycan composed of N-acetyltalosaminuronic acid with β-1,3 glycosidic bonds and L-amino acid peptide crosslinks (König and Kandler, 1979; Sprott and Beveridge, 1993). Most members are mesophiles, and the respective genera occur in the ruminant digestive tract (Liu and Whitman, 2008; Liu, 2010a; Lyu and Liu, 2019).
These methanogens are major contributors in rumen methane production (Janssen and Kirs, 2008, Henderson et al., 2015). Approximately 74% of the 16S rRNA amplicons of rumen methanogens from rumen samples are affiliated with Mbb. gottschalkii and Mbb. ruminantium (Janssen and Kirs, 2008; Henderson et al., 2015). Thus far, only a few rumen Methanobrevibacter species have been isolated from the rumen (Table 1) and they form two phylogenetic clades, smithii-gottschalkii-millerae-thaurei (SGMT) and ruminantium-olleyae (RO) (Table 1 and Figure 3; King et al., 2011). These clades’ abundance and distribution vary over hosts and diets (St-Pierre et al., 2015), with generally one clade dominating over the other (Wright et al., 2007; Yeoman and White, 2014; Seedorf et al., 2015), and in only a few instances these exhibiting balanced abundances (Wright et al., 2007; St-Pierre et al., 2015). From a phylogenetic analysis that included Mbb. woesei, Mbb. wolinii, and Mbb. boviskoreani (Figure 3), we propose to expand the SGMT into the woesei-smithii-gottschalkii-millerae-thaurei (WSGMT) and form a new clade of boviskoreani-wolinii (BW), while retaining the RO clade (Figure 3).
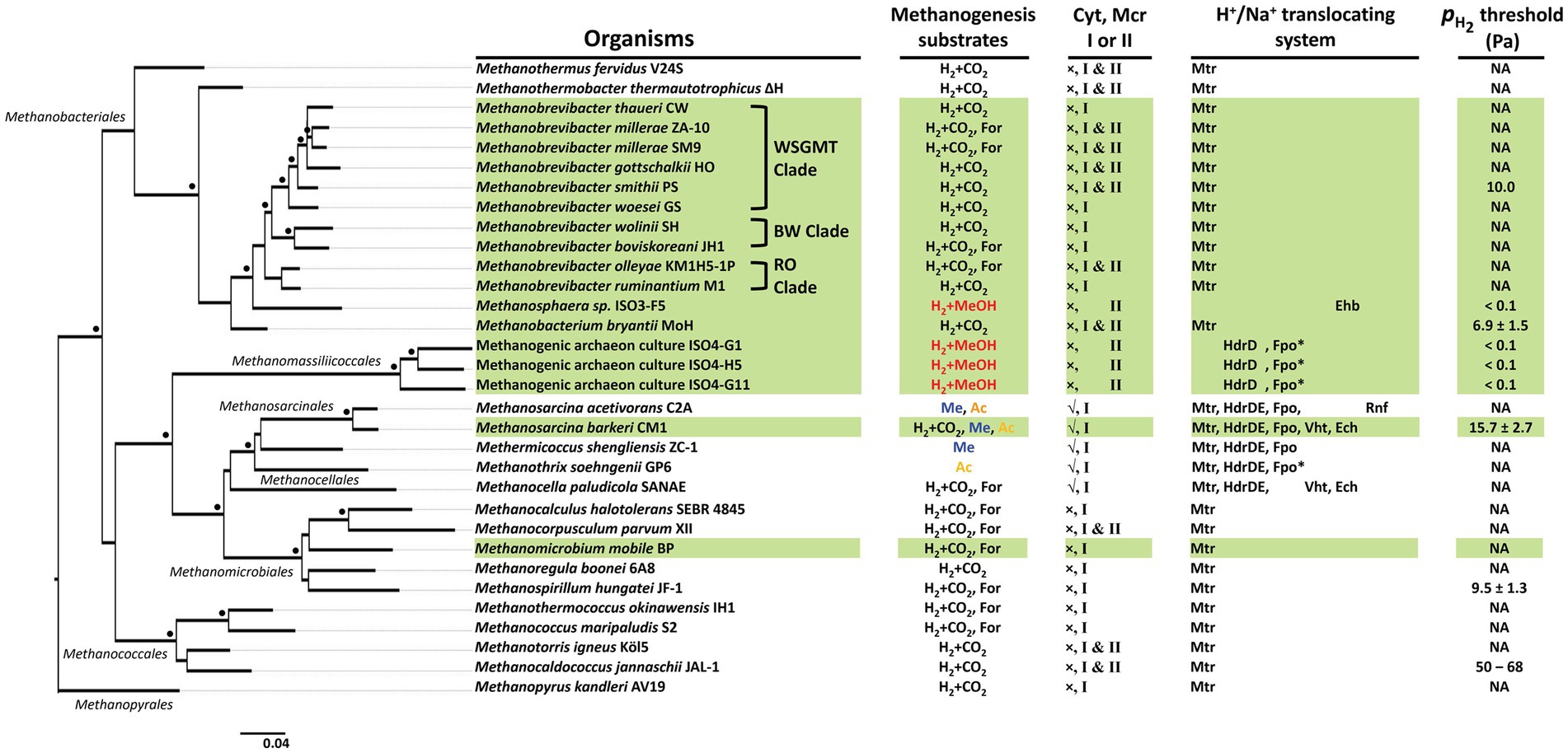
Figure 3. Phylogeny of rumen methanogenic archaea. A 16S ribosomal RNA (rRNA) gene sequence-based phylogenetic tree was constructed via a distance-based phylogeny inference algorithm at NGPhylogeny webserver (https://ngphylogeny.fr/) (Desper and Gascuel, 2002; Criscuolo and Gribaldo, 2010; Junier and Zdobnov, 2010; Katoh and Standley, 2013; Lefort et al., 2015; Lemoine et al., 2018) with Desulfurococcus amylolyticus Z-1312 16S rRNA gene sequence as an outgroup (not shown). Black dots at the branches, confidence values of ≥700 (out of 1,000 replicates). Scale bar, number of base substitutions per site. Mode of methanogenesis substrate use (as shown in Figure 5): black, CO2-reduction or formate-dependent; red, methyl-reduction with H2; blue, Methyl-dismutating; yellow, acetoclastic. Highlighted in green, rumen-associated methanogens. Sources of information in the H+/Na+ translocating system and threshold: (Deppenmeier, 2004; Thauer et al., 2008; Sakai et al., 2011; Ver Eecke et al., 2012; Welte and Deppenmeier, 2014; Kulkarni et al., 2018; Kröninger et al., 2019; Mand and Metcalf, 2019; Feldewert et al., 2020; Kurth et al., 2020, 2021; Downing et al., 2023). For, formate; MeOH, methanol; Me, methanol and mono-, di-, and trimethylamines; Ac, acetate; Mtr, methyl-tetrahydromethanopterin:coenzyme M methyltransferase; HdrDE, membrane-bound heterodisulfide reductase; Ech, energy-conserving hydrogenase; Ehb, a homolog of energy-conserving hydrogenase; Fpo, F420H2:phenazine oxidoreductase; Fpo*, Fpo in the absence of the F and O subunits; Vht, [NiFe] hydrogenase; Rnf, an equivalent of Rhodobacter nitrogen fixation complex; Mcr, methyl-coenzyme M reductase; cyt, cytochrome; √, cytochrome-containing; ×, cytochrome-non-containing.
Within the WSGMT clade, the presence of Mbb. smithii in the rumen system is questionable (Janssen and Kirs, 2008; Table 1 and Supplementary Table S1), as it was originally isolated from a sewage digester (Balch et al., 1979) and others were isolated from human feces and large intestine (Miller and Wolin, 1981; Miller et al., 1982). Rare detection of Mbb. smithii-like organisms in rumen have been based on the 16S rRNA sequence analysis (Supplementary Table S1). Methanobrevibacter species can produce methane from CO2-reduction with H2 and formate. The genomes of rumen methanogens often lack essential biosynthetic genes, such as those for coenzyme M, perhaps due to gene loss from prototrophic ancestors (Figure 4, Section 3.3), and in the rumen, resulting auxotrophies are supported with supplements from other organisms, including other methanogens (Hazlewood and Dawson, 1977). These auxotrophies often make the laboratory cultivation of rumen Methanobrevibacter species quite tedious, since growth factor(s), such as coenzyme M, short-chain fatty acids, amino acids, acetate, and vitamins need to be provided (Bryant et al., 1971; Balch and Wolfe, 1976; Balch et al., 1979; Miller and Lin, 2002; Rea et al., 2007; Lee et al., 2013; Table 1). Branched-chain volatile fatty acids, especially 2-methylbutyrate and isovalerate, are used for amino acid synthesis of isoleucine and leucine, respectively (Whitman et al., 1982; Shieh et al., 1988). In some cases, because of the unknown type auxotrophies, supplementation with rumen fluid is necessary (Bryant et al., 1971; Balch and Wolfe, 1976; Balch et al., 1979; Miller and Lin, 2002; Rea et al., 2007; Lee et al., 2013). Certain Methanobrevibacter species express adhesin-like proteins that likely allow symbiosis with ciliates and other hydrogen producers (Figure 4; Ng et al., 2016; Patra et al., 2017).
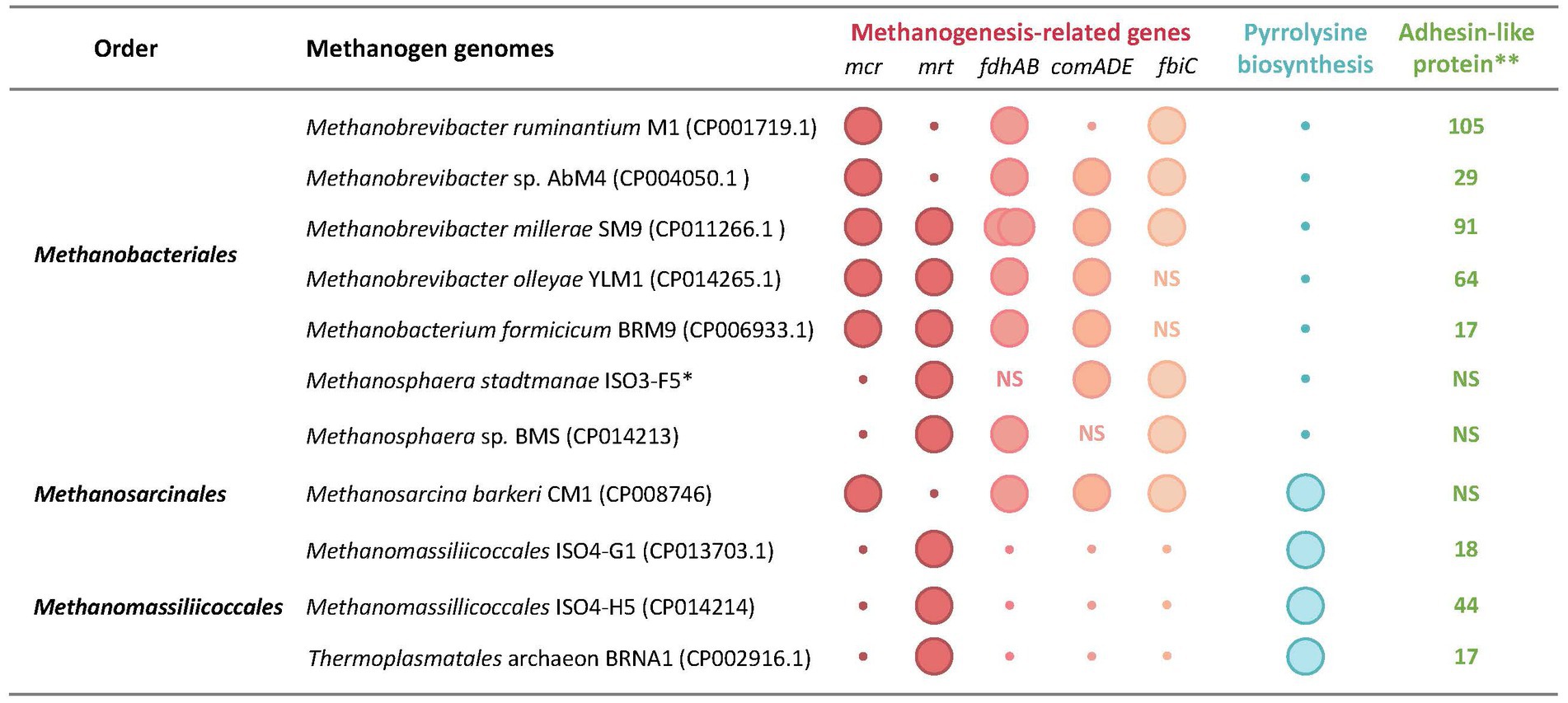
Figure 4. Unique genomic features of rumen methanogens. Genomic features: mcr and mrt, McrI and McrII isozymes of methyl-CoM reductase, respectively; fdhAB, two subunits of formate dehydrogenase as utilized for formate utilization; comADE and fbiC, CoM and F420 biosynthetic genes; pylBCD, genes encoding pyrrolysine biosynthesis enzymes; pylT, transfer RNA for pyrrolysine (tRNAPyl); pylS, pyrrolysyl-tRNA synthetase (PylRS); *, total number of adhesin-like protein; NS, not specified. The data are from Jeyanathan (2010), Leahy et al. (2010, 2013), Kelly et al. (2014, 2016a,b,c), Lambie et al. (2015), Li (2016), and Li et al. (2016).
Msp. stadtmanae ISO3-F5 and Msp. BMS are the sole rumen isolates of the Methanosphaera genus (Jeyanathan, 2010; Hoedt et al., 2018) and the former is closely related to the human fecal isolate Msp. stadtmanae MCB-3 (Miller and Wolin, 1985) with a 16S rRNA sequence similarity of 96% (Jeyanathan, 2010). Methanosphaera species are obligate H2-dependent methylotrophs (Miller and Wolin, 1985; Jeyanathan, 2010). The genome sequence of Msp. BMS but not Msp. stadtmanae ISO3-F5 is available (Jeyanathan, 2010; Hoedt et al., 2018). Rumen isolates require several growth factors, such as yeast extract, acetate, and fatty acids (Table 1). Msp. stadtmanae occurs in a free state as well as a symbiont of the rumen protozoa, Eudiplodinium and Entodinium (Tymensen et al., 2012; Xia et al., 2014).
Mb. formicicum BRM9 is the rumen representative of this genus, and this cow rumen isolate uses H2 + CO2 and formate for methanogenesis (Jarvis et al., 2000). It requires yeast extract and fatty acids for growth (Table 1).
Methanomassiliicoccales order of the more recently recognized phyla of Candidatus Thermoplasmatota represent the second most abundant methanogen group after Methanobrevibacter in the rumen (Henderson et al., 2015). It is currently represented by five families, four genera, and one pure culture isolate, Methanomassiliicoccus luminyensis B10 of Methanomassiliicoccaceae family obtained from human feces (Dridi et al., 2012). Methanomassiliicoccales are mesophiles and mostly associated with animal gastrointestinal tracts (Dridi et al., 2012; Li et al., 2016; Söllinger et al., 2016; Kelly et al., 2016a,b; Cozannet et al., 2020). The strain B10 derives energy from H2-dependent methanogenesis from methylated compounds, such as methanol, methyl-, dimethyl-, and trimethylamine (Dridi et al., 2012; Li et al., 2016; Kelly et al., 2016a,b). Similar to Mycoplasma, which are cell wall deficient bacteria (Brown et al., 2018), Methanomassiliicoccales lack the archaeal S-layer cell wall and possess a bi-layer cell membrane (Dridi et al., 2012; Li et al., 2016), which in strain B10 contains unusual butane- and pentanetriol-based tetraether lipids (Becker et al., 2016).
There are reports on the enrichment of rumen Methanomassiliicoccales, and ISO4-H5, ISO4-G1, ISO4-G11, RumEn M1, and RumEn M2 are such examples (Kelly et al., 2016a,b; Li et al., 2016; Söllinger et al., 2016). These isolates rely exclusively on H2-dependent methyl-reducing methanogenesis for energy production (Kelly et al., 2016a,b; Li et al., 2016; Söllinger et al., 2016), and genome analysis suggests that ISO4-H5 and ISO4-G1 are coenzyme M auxotrophs (Li et al., 2016; Kelly et al., 2016a,b); the genome sequence of ISO4-G11 is not available (Jeyanathan, 2010) and that of RumEn M1 and RumEn M2 are incomplete (Söllinger et al., 2016). Further investigations on the physiology of these methanogens will require isolation in pure cultures.
The species of this order representing eight families perform methanogenesis from CO2 with H2, formate, and secondary alcohol as electron sources (Zellner and Winter, 1987; Liu, 2010b). Methanomicrobium mobile BP, a bovine isolate that uses H2 + CO2 and formate (Paynter and Hungate, 1968) and belongs to the Methanomicrobiaceae family, is the only rumen representative of this order (Table 1). It constitutes only a small fraction of rumen methanogen population (Henderson et al., 2015) and forms symbioses with ciliates via an unknown mechanism (Regensbogenova et al., 2004). Mm. mobile has the most complex growth factor requirements among methanogens (Table 1); the nature of a factor that is called mobile element and could be provided from boiled cell extract of Methanothermobacter thermautotrophicus remains unknown (Tanner and Wolfe, 1988; Kuhner et al., 1991; Table 1).
The rumen representatives of this order are Methanosarcina barkeri CM1 and Methanosarcina thermophila Ms97 that belong to the Methanosarcinaceae family (Table 1), and like other Methanosarcina, they are metabolically versatile and can utilize H2 and CO2, methylated compounds, such as methanol, methylamines, and methanethiol, and acetate, for methane production (Table 1; Rospert et al., 1990; Reeve, 1992; Morgan et al., 1997; Reeve et al., 1997; Deppenmeier et al., 2002; Galagan et al., 2002; Maeder et al., 2006; Lambie et al., 2015). The rumen isolates require yeast extract and rumen fluids for growth (Table 1). While co-culture experiments show symbiotic interactions of a non-rumen isolate of Ms. barkeri with ruminal fungi and ciliates (Mountfort et al., 1982; Hillman et al., 1988; Ushida et al., 1997), no such information is available for a rumen Methanosarcina (Lambie et al., 2015).
Methanothrix species are the sole members of this order (Garrity et al., 2011) and are known to obtain energy solely from acetoclastic methanogenesis (Lyu and Liu, 2019; Akinyemi et al., 2021), although their genomes suggest a capability of CO2-reduction with H2 and CO as electron sources (Smith and Ingram-Smith, 2007). A low abundance of 16S rRNA gene sequences representing Methanothrix concilii have been detected in rumen samples (Henderson et al., 2015).
For energy production, methanogens rely on methanogenesis, and based on the methanogenic substrates utilized, these archaea are divided into three groups (substrate, group name): hydrogen and formate as electron donor for CO2 reduction (hydrogenotrophic and formate-dependent, respectively); methyl-containing compounds and acetate as sources of both methyl group and electron source (methylotrophic and acetoclastic, respectively) (Wolfe, 1992). However, for the recently emphasized role of methanogens that remove H2 via methyl group reduction in the rumen, human gut, and many other ecological niches, the definition of hydrogenotrophic methanogenesis has been expanded to the following (hydrogenotrophic pathway, associated methanogens): CO2-reducing hydrogenotrophy (CO2-reducing hydrogenotrophs) and methyl-reducing hydrogenotrophy (methyl-reducing hydrogenotrophs) (Garcia et al., 2022; Bueno de Mesquita et al., 2023). Similarly, methanogenesis from CO2 with formate and secondary alcohols as reductants, where the electrons are recovered from the primary donor as F420H2 (Thauer et al., 2008; Yan and Ferry, 2018) could be called formate-dependent and secondary alcohol-dependent methanogenesis, respectively; for the former, CO2-reducing formatotrophic name has also been proposed (Garcia et al., 2022). In the following subsections, each of the methanogenesis pathways and the corresponding energy conservation strategies are described and linked to the rumen methanogens that employ them.
CO2-reducing hydrogenotrophy (Figure 5) is one of the most ancient respiratory metabolisms on Earth (Leigh, 2002; Teske et al., 2003). Here, CO2 is first reduced to a formyl group, which is dehydrated to methenyl and then sequentially reduced to methylene and methyl groups and finally, to methane (Figure 5); three coenzymes, methanofuran (MFR), tetrahydromethanopterin (H4MPT), and coenzyme M (CoM-SH or CoM) act as carriers for the carbon units at four oxidation states (+4, +2, 0, and − 2) (Wolfe, 1992). Reduced coenzyme F420 (F420H2), generated by an F420-reducing [NiFe]-hydrogenase (Frh) with H2, serves as a direct electron donor for the reduction of methenyl and methylene forms. Coenzyme B (CoB-SH or CoB) helps to reduce the methyl group of CH3-S-CoM to CH4, and this process generates heterodisulfide of CoM and CoB (CoM-S-S-CoB) (Wolfe, 1992; Thauer et al., 2010; Thauer, 2012).
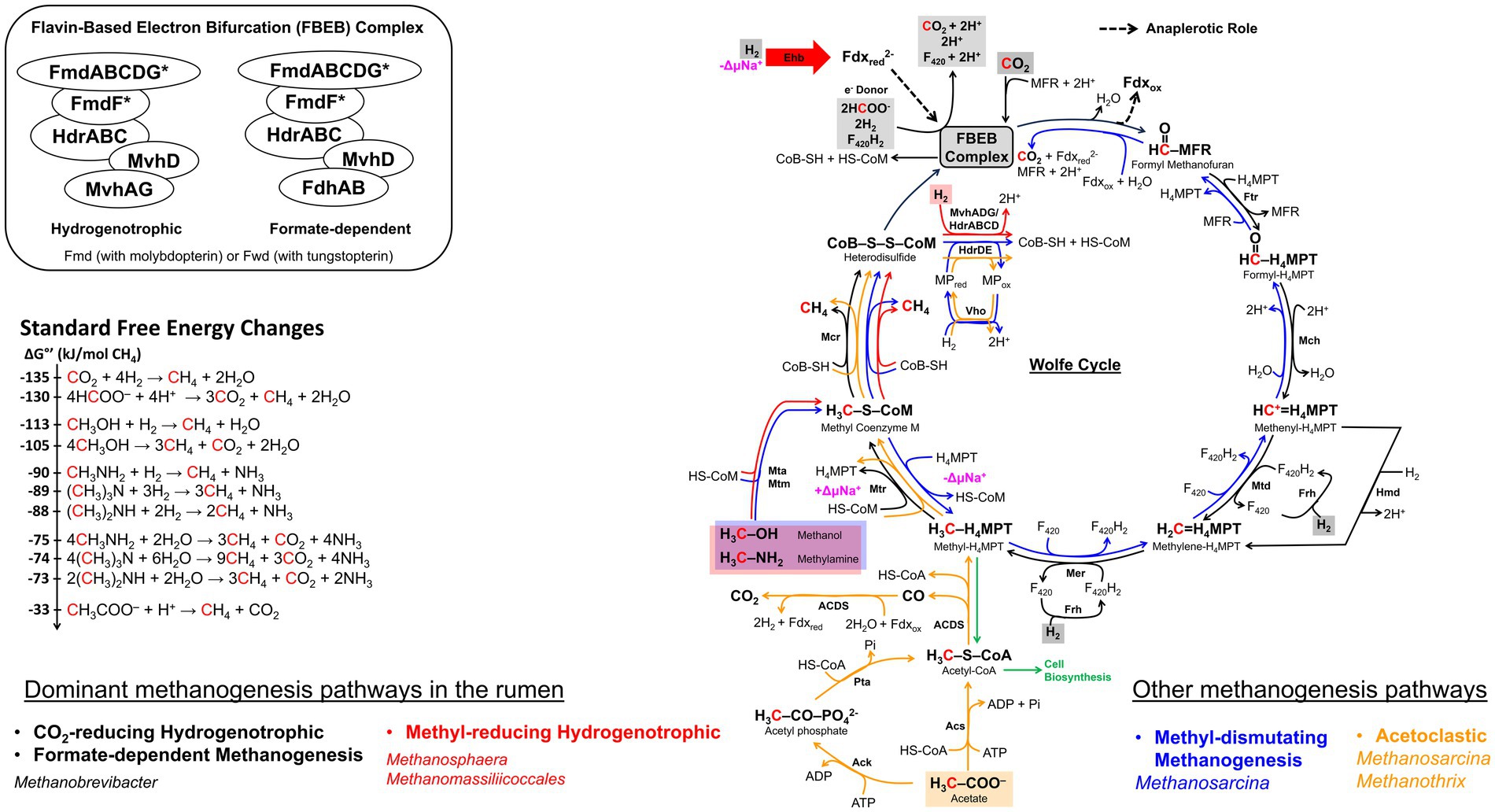
Figure 5. The methanogenesis cycle, energy conservation, and associated standard free energy changes. The cycle is first proposed by Rouvière and Wolfe (1988) and subsequent discovery in flavin-based electron bifurcation (FBEB) makes the cycle full circle (Thauer, 2012; Watanabe et al., 2021). Pathway arrow colors: black, CO2-reducing hydrogenotrophic or formate-dependent methanogenesis (Wood et al., 2003; Costa et al., 2010; Thauer, 2012); red, blue, and orange, methyl-reducing hydrogenotrophic, methyl-dismutating, and acetoclastic, respectively (Welander and Metcalf, 2005; Kurth et al., 2020). The FBEB complex which consists of Mvh-Hdr-Fmd/Fwd or Fdh-Hdr-Fmd/Fwd is proposed to be a common structure in CO2-reducing or formate-dependent methanogens without cytochromes (Watanabe et al., 2021). Values for standard free energy changes are taken from Liu and Whitman (2008) or calculated from Thauer et al. (1977). Fdxred, reduced ferredoxin; Fdxox, oxidized ferredoxin; MFR, methanofuran; H4MPT, tetrahydromethanopterin; MP, methanophenazine; CoB-SH or CoB, coenzyme B; CoM-SH or CoM, coenzyme M; Fmd, molydopterin containing formyl-MFR dehydrogenase; Ftr, formyl-MFR:H4MPT formyltransferase; Mch, methenyl-H4MPT cyclohydrolase; Mtd, methylene-H4MPT dehydrogenase; Hmd, H2-dependent methylene-H4MPT dehydrogenase; Mer, methylene-H4MPT reductase; Mtr, methyl-H4MPT:CoM methyltransferase; Mcr, methyl-CoM reductase; Hdr, electron bifurcating hydrogenase-heterodisulfide reductase complex; HdrABC, soluble heterodisulfide reductase; HdrDE, membrane-bound heterodisulfide reductase; Mvh, non-F420-reducing hydrogenase; Frh, F420-reducing hydrogenase; Mta, methylcobamide:CoM methyltransferase; Mtm, monomethylamine methyltransferase; Acs, acetyl-CoA synthase; Ack, acetate kinase; Pta, phosphotransacetylase; ACDS, acetyl-CoA decarbonylase/synthase; Fdh, formate dehydrogenase; Eha, a homolog of energy-conserving hydrogenase; Vho, methanophenazine-dependent hydrogenase; ΔμNa+, electrochemical sodium ion potential.
In CO2-hydrogenotrophic methanogens without cytochromes such as Methanobrevibacter, the only site for energy conservation is the sodium translocating membrane-associated methyl-H4MPT:CoM methyltransferase complex composed of MtrA–H subunits (Figure 5; Thauer et al., 2008) and heterodisulfide reduction occurs as follows. A cytoplasmic complex composed of heterodisulfide reductase (HdrABC), non-F420-reducing hydrogenase (MvhADG), and formyl-methanofuran dehydrogenase (FmdABCDFG or FwdABCDFG) retrieves electrons from H2 (Eo’, −420 mV), and bifurcates these using the FAD unit of HdrA to provide high potential electrons for the reduction of CoM-S-S-CoB (Eo’, −140 mV) at HdrB and low potential electrons for formyl-MFR synthesis from CO2 (Eo’, −500 mV) (Costa et al., 2010; Yan and Ferry, 2018; Watanabe et al., 2021; Figure 5); Fmd and Fwd are molybdo- and tungsto-pterin carrying isoenzymes of formyl-methanofuran dehydrogenase, respectively (Schmitz et al., 1992). For methanogenesis with formate, as discussed below, the MvhADG unit is replaced with F420-reducing formate dehydrogenase (FdhAB) that can obtain electrons from either formate using FdhA or F420H2 via FdhB (Costa et al., 2010; Watanabe et al., 2021). The direct electronic coupling of the first (formyl-methanofuran synthesis) and last (CoM-S-S-CoB reduction) steps of methanogenesis generates a cyclic system that has been called the Wolfe Cycle, named after Ralph Wolfe (Rouvière and Wolfe, 1988; Thauer, 2012; Figure 5). When the electron bifurcation falls short, an energy-converting hydrogenase (Eha) provides low potential electrons for formyl-MFR synthesis via a ferredoxin, serving an anaplerotic function (Figure 5; Lie et al., 2012).
For methanogens with cytochromes, electrons derived from H2 by the action of a membrane-bound and proton pumping VhoAGC hydrogenase complex are channeled to HdrDE for CoM-S-S-CoB reduction, and the low-potential Fdxred that are needed for formyl-MFR synthesis are generated via another membrane-bound hydrogenase complex (EchA-F) that is aided by a proton-motive force (Thauer et al., 2008). Thus, a cytochrome carrying CO2-hydrogenotroph such as Methanosarcina has two sites of energy conservation, Mtr and VhoAGC (Thauer et al., 2008).
Under standard conditions, the hydrogenotrophic mode is the most exergonic of all methanogenesis systems (ΔG°′ = −135 kJ/mol CH4) (Figure 5). However, under the rumen conditions (hydrogen partial pressure or ), 162 Pa (Barry et al., 1977; Ungerfeld and Kohn, 2006), the prevailing ΔG’ value of the hydrogenotrophic methane formation reaction is only −67.4 kJ/mol CH4 (Ungerfeld and Kohn, 2006) and yet, Mbb. ruminantium and Mbb. gottschalkii together represent as high as 74% of the total archaeal community (Henderson et al., 2015). It has been suggested that the flavin-dependent bifurcation system producing a low potential reduced Fdx pool is a key tool for a methanogen living under low (Yan and Ferry, 2018).
The genome of Mbb. ruminantium strain M1 carries all the genes necessary for methane production from H2 and CO2 (Leahy et al., 2010). It also carries a locus with genes for a formate transporter (fdhC, mru_0332), a formate dehydrogenase (fdhAB, mru_0333 and mru_0334), and genes encoding molybdopterin-guanine dinucleotide biosynthesis (moa, mru_0335 and mru_0336), enabling the organism to transport and oxidize formate; formate dehydrogenase contains a molybdopterin cofactor (May et al., 1986; Reeve, 1993).
There are indications that various groups of Methanobrevibacter use two different isoenzymes of methyl-coenzyme M reductase (Mcr) which catalyzes methane production from CH3-S-CoM (Figure 5). This difference has a major implication for their hydrogen metabolism (Reeve, 1992; Bonacker et al., 1993). The Mcr isoenzymes, Mcr I and McrII, encoded by the mcr and mrt genes, respectively, are considered physiologically adapted to function at low and high values (Rospert et al., 1990; Reeve et al., 1997). Mbb. ruminantium M1, a RO group organism, with mcrBCDGA genes encodes only McrI (Leahy et al., 2010). Of the rumen isolates from the WSGMT group, only for Mbb. millerae SM9’s complete genome sequence is available, and it carries both mcr and mrt genes (Kelly et al., 2016c).
In this process, the methyl group from methylated compounds, such as methanol, methylamine, and methanethiol are transferred to CoM to form methyl-CoM which is then reduced to methane by Mcr (Figure 5), and H2 serves as the primary electron source for CoM-S-S-CoB reduction (Figure 5; Keltjens and Vogels, 1993). In the rumen, only methanol and methylamines (either mono-, di-, or trimethylamines), but not methanethiol, are available for this metabolism (Miller and Wolin, 1985; Fricke et al., 2006; Jeyanathan, 2010; Henderson et al., 2015; Kelly et al., 2016a,b; Li et al., 2016).
The ΔGo’ value for this process under standard conditions is −113 kJ/mol CH4, making it the second most exergonic methanogenesis process (Figure 5). The ΔG’ values under rumen conditions, however, have not been reported and would be variable as methanol and methylamine concentrations are dependent on the host diet, and also varies (Henderson et al., 2015). The methyl-hydrogenotrophs constitute about 16% of the rumen archaeal community and along with CO2-hydrogenotrophs and formate-utilizers, these organisms cover close to 90% of the methanogens in this habitat (Henderson et al., 2015). These methyl-hydrogenotrophic rumen methanogens belong to Methanosphaera sp. and two Methanomassiliicoccales-affiliated groups (Miller and Wolin, 1985; Fricke et al., 2006; Jeyanathan, 2010; Henderson et al., 2015; Li et al., 2016; Kelly et al., 2016a,b); Methanosarcina, which also can perform methyl-hydrogenotrophy (Mukhopadhyay et al., 1993) are rarely encountered in the rumen (Henderson et al., 2015).
The metabolic potential of Msp genus was inferred from the genome sequence analyses of a human fecal isolate, Msp. stadtmanae MCB-3 (Fricke et al., 2006), and that of rumen strain BMS (Hoedt et al., 2018). Genome analysis of MCB-3 showed that it lacks the genes for the biosynthesis of molybdopterin, an essential prosthetic group of formylmethanofuran dehydrogenase (Fmd), making the organism incapable of activating CO2 to the formyl stage and performing CO2-hydrogenotrophic methanogenesis (Fricke et al., 2006; Figure 5). The organism also lacks the genes for the synthesis of acetyl-CoA decarbonylase/synthase complex, which explains the requirement of acetate for its growth and its inability to utilize acetate for methanogenesis (Miller and Wolin, 1985). All these phenotypes have been observed in the rumen strains ISO3-F5 and BMS (Jeyanathan, 2010; Hoedt et al., 2018) which relies solely on H2 and methanol for methane production (Jeyanathan, 2010; Hoedt et al., 2018; Figure 5). Based on the presence of mrt and absence of mcr in the genome of MCB-3, it is inferred that the ISO3-F5 strain uses McrII (Fricke et al., 2006; Jeyanathan, 2010), which likely operates at high values (Rospert et al., 1990). The growth of BMS strain was also greatly enhanced at high , suggesting a dependence on McrII as well (Figure 5; Hoedt et al., 2018). The energy conservation system in Methanosphaera relies on the generation of reduced Fdx by the electron bifurcating HdrABC/MvhADG complex, and the free energy of the reduced Fdx is used for sodium ion translocation via membrane-bound energy-conserving hydrogenase (Ehb) complex (Fricke et al., 2006; Thauer et al., 2008; Yan and Ferry, 2018).
The genomes of Methanomassiliicoccales strains ISO4-H5, ISO4-G1, RumEn M1, and RumEn M2 (Jeyanathan, 2010; Li et al., 2016; Söllinger et al., 2016; Kelly et al., 2016a,b) lack the genes for many of the enzymes that are required to reduce CO2 to the methyl stage or to oxidize the methyl group of methyl-CoM to CO2 that could provide reductants for methyl-coenzyme M reduction (Lang et al., 2015; Li et al., 2016; Söllinger et al., 2016; Kelly et al., 2016a,b; Figure 5). Consequently, members of the Methanomassiliicoccales order are restricted to methyl-hydrogenotrophy; as mentioned above, for Methanosphaera species, such a restriction is due to a narrower reason, an inability to biosynthesize the molybdopterin cofactor for Fmd.
Above-mentioned rumen methanogens of the Methanomassiliicoccales order contain a F420H2:MP oxidoreductase-like (Fpo-like) complex and this could translocate protons for energy conservation (Lang et al., 2015; Li et al., 2016; Söllinger et al., 2016; Kelly et al., 2016a,b). However, they lack the genes for coenzyme F420, cytochrome, MP biosynthesis, and FpoF and FpoO subunits (Li et al., 2016; Söllinger et al., 2016; Kelly et al., 2016a,b) which in a Fpo complex of Methanosarcina species interact with F420H2 and MP, respectively (Welte and Deppenmeier, 2011); RumEn M2 strain also lacks the FpoA subunit (Söllinger et al., 2016). These genomes encode MvhADG, HdrABC, and HdrD but not HdrE (Lang et al., 2015; Li et al., 2016; Kelly et al., 2016a,b). Thus, it is possible that in rumen representatives of the Methanomassiliicoccales order, the Fpo-like complex couples the oxidation of bifurcation-derived reduced Fdx to the formation of a proton gradient (Kröninger et al., 2019). Methanomassiliicoccales carry mrt genes and lack the mcr system (Li et al., 2016; Kelly et al., 2016a,b), and therefore, utilize McrII which is known to operate at higher values (Rospert et al., 1990; Reeve, 1992; Reeve et al., 1997); as mentioned above, a similar situation exists with the Methanosphaera spp.
Recently the term methyl-dismutating methanogenesis has been proposed as an alternate for methylotrophic methanogenesis (Garcia, Gribaldo et al., 2022). As the former embodies the mechanism of the process (Wolfe, 1992), we use this term for the rest of the narrative. Methyl-dismutating and acetoclastic methanogenesis are not significant processes in the rumen and the associated methanogens, Methanosarcina and Methanothrix species, are rarely encountered in this system (Hungate et al., 1970; Janssen and Kirs, 2008; Henderson et al., 2015; Seshadri et al., 2018); Ms. barkeri CM1 and Ms. thermophila Ms97 are the two rumen isolates (Lambie et al., 2015; Zhou et al., 2021). Methanosarcina species carry the mcr system and lack mrt genes (Deppenmeier et al., 2002; Galagan et al., 2002; Maeder et al., 2006; Lambie et al., 2015), hence these methanogens employ McrI that has been postulated to operate under low conditions (Rospert et al., 1990; Reeve, 1992; Morgan et al., 1997; Reeve et al., 1997). Methanothrix spp. carry the mrt system that generates Mcr II (Barber et al., 2011; Zhu et al., 2012).
In methyl-dismutating methanogenesis, one-fourth of the available methyl groups are oxidized, generating F420H2 and reduced Fdx which in turn allows the reduction of the rest of the methyl groups to methane, (4CH3X + H2O → 3CH4 + CO2; X = −OH, −NH3, and −SH) (Figure 5; Deppenmeier et al., 1996; Deppenmeier, 2004; Buan and Metcalf, 2010; Yan and Ferry, 2018). Following are the ΔGo’ values (kJ/mol CH4) for this process with the indicated substrates: −105 (methanol), −74 (trimethylamine), and − 49 (dimethylsulfide) [Figure 5; see reference (Liu and Whitman, 2008) for a comprehensive list]. The electrons from reduced Fdx, originating from the oxidation of formyl-MFR, are bifurcated to reduce CoM-S-S-CoB and to generate F420H2 (Deppenmeier et al., 1996; Deppenmeier, 2004; Buan and Metcalf, 2010; Yan and Ferry, 2018); F420H2 is also generated from the oxidation of methyl- and methylene-H4SPT (H4SPT, tetrahydrosarcinapterin, a variation of H4MPT). Then a FpoA-O complex couples the oxidation of F420H2 to proton translocation and also provides additional reductants for CoM-S-S-CoB reduction via methanophenazine (MP) and HdrDE (Deppenmeier et al., 1996; Deppenmeier, 2004; Buan and Metcalf, 2010; Yan and Ferry, 2018). Additional energy is generated by a Frh-based H2 cycling system that retrieves electrons from F420H2 via Frh and produces H2 and H+ gradient; the internally produced H2 diffuses out and is oxidized at the extra cytoplasmic location via VhtAGC to generate electrons that are transported to HdrDE via MP for heterodisulfide reduction (Kulkarni et al., 2018; Mand and Metcalf, 2019).
Of all types of methanogenesis, the acetoclastic mode has the least negative ΔGo’ value (−33 kJ/mol CH4) (Figure 5). Here, the methyl group of acetate is transferred to H4SPT for further processing, generating methane, CoM-S-S-CoB, and a Na+-motive force, and the oxidation of the carboxyl group provides reduced Fdx (Yan and Ferry, 2018). The reduced Fdx is utilized by the Rnf complex (equivalent of Rhodobacter nitrogen fixation complex) in two ways: first, Fdx2− is oxidized employing cytochrome and the above-described MP- and HdrDE-mediated steps causing proton translocation and CoM-S-S-CoB reduction (Yan and Ferry, 2018; Mand and Metcalf, 2019); second, two Fdx2− are processed with two oxidized F420, promoting sodium ion translocation and the resulting F420H2 are used by HdrA2B2C2 for the reduction of CoM-S-S-CoB and the production of Fdx2− (Yan et al., 2017; Buckel and Thauer, 2018; Yan and Ferry, 2018). In both cases, a Na+/H+ antiporter adjusts the respective gradients for optimal ATP synthesis (Deppenmeier et al., 2002; Galagan et al., 2002; Maeder et al., 2006; Lambie et al., 2015; Yan and Ferry, 2018). A H2 cycling system similar to that described above for methyl-dismutating methanogenesis but with electrons derived from Fdx2− via Ech, provides an additional avenue for energy production (Barber et al., 2011; Zhu et al., 2012; Kulkarni et al., 2018; Mand and Metcalf, 2019).
The source of H2, formate, and acetate is predominantly carbohydrate fermentation as detailed above (Figure 2). Methanol is generated from de-esterification of methoxylated form of pectin, which is a polysaccharide component of the plant cell wall composed of alpha-1,4-galacturonic acid (Mitchell et al., 1979; Patterson and Hespell, 1979; Pol and Demeyer, 1988). This reaction is catalyzed by pectinase produced by Butyrivibrio, Prevotella, Bacteroides, Ruminococcus, and Fibrobacter species (Comtet-Marre et al., 2017; Sollinger et al., 2018; Kelly et al., 2019); fungi, protozoa or associated bacteria also hydrolyze pectin (Wright, 1960). Degradation of choline and betaine, that are present in the feed (Mitchell et al., 1979; Patterson and Hespell, 1979; Pol and Demeyer, 1988) by choline-TMA lyase and betaine reductase, respectively, provides trimethylamine (TMA) (Craciun and Balskus, 2012; Rath et al., 2019). In the rumen, the choline-TMA lyase gene occurs in Desulfovibrio, Clostridia, Streptococcus, Klebsiella, and Proteus species (Craciun and Balskus, 2012) and betaine reductase is likely provided by Eubacterium, Clostridium, and various members of the Firmicutes (Naumann et al., 1983; Hormann and Andreesen, 1989; Rath et al., 2019). A recent study has provided the following values for the methyl-group containing substrate concentrations (μM) in bovine rumen fluid (Bica et al., 2022): methanol, 23–26; methylamine, 12–16; dimethylamine, 1.8–2.1; and trimethylamine, 1.6–2; the values were not significantly different across different diets. An earlier study in cattle and sheep rumens reported that the concentration of methylamine increases steadily during the 6–8 h period post-feeding and then decreases rapidly (Hill and Mangan, 1964). After an additional 5 h, methylamine was absent from the rumen and this status remained for a 24 h period that followed (Hill and Mangan, 1964). These data are consistent with a rapid utilization of methyl-group containing substrates by the methyl-hydrogenotrophs under the high condition following feeding (Sollinger et al., 2018).
Identification of several gastrointestinal tract (GIT)- and rumen-associated microbes with reduced genome sizes that are smaller than that of the same species from non-host-associated niches suggest that nutrient-abundant nature of animal digestive tracts have facilitated genome streamlining events in these organisms (Walter and Ley, 2011; Söllinger et al., 2016). In some cases, GIT and rumen microorganisms gained additional genes (Leahy et al., 2013; Kelly et al., 2016c; Söllinger et al., 2016). For example, Mbb. smithii PS, a human gut-associated Methanobrevibacter species, can be distinguished from the rumen-associated Methanobrevibacter sp. Abm4 based on the presence of the mtaABC operon encoding methanol:cobalamin methyltransferase genes in the latter (Leahy et al., 2013). This is a surprise as Methanobrevibacter species are not known to utilize methanol (Boone et al., 1993) and the roles of mtaABC in strain Abm4 are unclear (Leahy et al., 2010, 2013). If these genes indeed allow H2-dependent methanogenesis from methanol in Abm4 similar to Methanosphaera and Methanomassiliicoccales or only on methanol as seen in Methanosarcina, these capabilities will introduce a major change in the concept of rumen methanogenesis. Remarkably, a comparison of genomes of rumen methanogens with those of closely related species originating from amoeba-associated and freshwater isolates has revealed higher metabolic versatility in the rumen methanogens (Kelly et al., 2014; Lambie et al., 2015).
Currently, for rumen methanogens at least 15 complete assembled genome sequences are available in public repositories (Supplementary Table S2), and these include that of Mbb. boviskoreani JH1 (Lee et al., 2013), Methanoculleus bourgensis KOR-2 (Battumur et al., 2019), and Methanomassiliicoccales RuMen M1 and M2 (Söllinger et al., 2016). The number increases further if those submitted as drafts or scaffolds are considered (Chen et al., 2023). Some of the genomes have been reported with corresponding publications (Jeyanathan, 2010; Leahy et al., 2010, 2013; Lee et al., 2013; Kelly et al., 2014, 2016a,b,c; Lambie et al., 2015; Li, 2016; Li et al., 2016; Söllinger et al., 2016; Battumur et al., 2019) and several, such as that for the Thermoplasmatales BRNA1 genome, have been deposited to the GenBank (accession number, CP002916) and not yet been reported in a publication.
Analyses of the methanogen genomes pinpoint specific gene markers that can be used to infer their metabolic capabilities. These markers include methanogenesis-related and cofactor biosynthesis genes (Leahy et al., 2010; Roehe et al., 2016; Sollinger et al., 2018; López-García et al., 2022; Figures 4, 5). Genes fmdB and mtrA that encode formylmethanofuran dehydrogenase subunit B and methyl-H4MPT:HS-CoM methyltransferase subunit A, respectively, for example, are effective markers for CO2-reducing hydrogenotrophs, whereas for methyl-reducing hydrogenotrophs, such as Methanosphaera and Methanomassiliicoccales, the markers are methanol- and methylamine-specific methyltransferase genes, mtaB and mtMA, respectively (Sollinger et al., 2018); mtMA represents a combination of mono-, di- and trimethylamine methyltransferase genes. An alignment of the sequences of the following seven core methanogenesis proteins extracted from whole genome sequences has been used in a taxonomic characterization of various methanogens from diverse ecological niches: four subunits of methyl-H4MPT:HS-CoM methyltransferase (MtrB, -C, -D, and -E); F420-dependent methylene tetrahydromethanopterin dehydrogenase, Mtd (Mukhopadhyay et al., 1995); coenzyme M biosynthesis enzyme, ComD (Graupner et al., 2000); and FO synthase subunit 1, CofG (Choi et al., 2002; Graham et al., 2003; Anderson et al., 2009) where FO is a core unit of coenzyme F420 (Eirich et al., 1979).
Even the limited amount of data that are available for the relevant metabolic and genome characteristics clearly show evidence for the evolutionary developments that are specific to rumen methanogens as a member of a rumen microbial consortium. In the following sections, methanogen colonization and adaptation processes in the rumen are summarized.
Calves are born with undeveloped rumens and function as monogastric animals. This development stage is also called the pre-ruminant phase (Church, 1988; Davis and Drackley, 1998). The reflective closure of the reticular groove bypasses the rumen and directs the feed, mostly milk or milk replacer, directly to the abomasum and then to small and large intestines (Van Soest, 1994). The rumen is established through three sequential steps, namely the development of rumen anatomy, fermentation capacity and function, and microbial colonization (Yáñez-Ruiz et al., 2015). This development occurs within the first several weeks or months of a calf’s life with a fully mature rumen forming following a major diet transition from colostrum in neonatal, and milk and a concentrate/grain-based feed for pre-weaned calves to solid feed in post-weaned calves.
Consumption of solid feed such as roughage or grains stimulates the development of rumen papillae for nutrient absorption, muscular structure for rumination, expansion of rumen capacity, and production of saliva (Tamate et al., 1962, Stobo et al., 1966; Lane and Jesse, 1997; Baldwin et al., 2004). In concert with these anatomical and feed changes, the rumen microbial community develops. Initial microbial colonization in the rumen occurs immediately after birth by diverse aerobes and facultative anaerobes (Fonty et al., 1987; Li et al., 2012; Jami et al., 2013). Several studies suggested that microbial colonization in the rumen may occur in utero between 5 and 7 months gestation or even much earlier such as at the end of the first trimester, although the mechanism of this transfer from mother to fetus is unclear (Guzman et al., 2020; Husso et al., 2021; Zhu et al., 2021; Amat et al., 2022).
These early occupants consume O2, and thus, provide an anoxic environment for obligate anaerobes that colonize by the second day of life (Fonty et al., 1987). Intriguingly, a study with euthanized Holstein bull calves detected a typical rumen microbial community comprised of methanogens, fibrolytic bacteria, and Geobacter spp. belonging to Proteobacteria phylum in the rumen fluid of dairy calves 20 min after their birth, suggesting that these microbes present in the GIT right after birth and long before the introduction of solid feed (Guzman et al., 2015). This finding is somewhat surprising given that these neonatal calves solely depend on colostrum and suckle milk for energy, and here, the rumen is bypassed. Thus, these observations are raising the question about the roles of these early microbial communities in the under-developed rumen.
Most studies of methanogen community in fully developed rumens point to the major abundance of CO2-reducing hydrogenotrophic methanogens (Janssen and Kirs, 2008). The information on methanogen community composition in pre-ruminants is scarce. Methanomicrobium mobile, Methanococcus voltae, and Methanobrevibacter sp., which are capable of utilizing H2 and formate, have been found in neonatal calves (Guzman et al., 2015). However, hydrogen is not considered to be the most prevalent electron source for methanogenesis at this stage. Instead, methanol and methylamine are used for methanogenesis in young animals, and species from Methanosarcinales order have been found to occur primarily in young and developing calves (Friedman et al., 2017).
This selection could be due to the presence of other hydrogen utilizers such as acetogens and sulfate reducers, which outcompete methanogens (Fonty et al., 1987, Morvan et al., 1994; Fonty et al., 2007). A study with gnotobiotically-reared lambs that were inoculated with functional methanogen-free rumen microbiota and then placed on solid feed has demonstrated that it is possible to establish a rumen system with hydrogenotrophic acetogens and sulfate-reducing bacteria as the main hydrogen sink (Fonty et al., 2007); this system persisted for 12 months after the initiation. It is noteworthy that the composition of the rumen methanogen community early in a calf’s life is also determined by the route of delivery and a lower abundance of methanogens is seen in vaginally delivered animals (Furman et al., 2020).
In addition to animal development stage, rumen microbial composition is influenced by factors such as host genetics and diets. Host genetics play roles in shaping the rumen microbiome and determining the efficiency of energy harvest from feed and extent of methane emission (Carberry et al., 2012; Jami et al., 2014; Kittelmann et al., 2014; McCann et al., 2014; Wallace et al., 2015; Roehe et al., 2016; Sasson et al., 2017; Difford et al., 2018; Zhang et al., 2020; Martínez-Álvaro et al., 2022). A link of the host genetics to the selection of twenty heritable microbes belonging to exclusively Bacteroidetes and Firmicutes phyla has been established (Sasson et al., 2017). However, the mechanisms underlying this observation remain to be clearly defined.
Of all factors influencing microbial community, diet composition and its physical characteristics such as particle size are considered as main drivers (Li et al., 2009; Henderson et al., 2015). A fiber-rich diet containing structural carbohydrates and large particles enriches fiber-degraders such as Fibrobacter succinogenes, Ruminococcus flavifaciens, and Ruminococcus albus (Johnson and Johnson, 1995). This type of diet also decreases feed digestion rate due to the presence of cell wall components that are less rapidly degraded than a starch-based diet, hence reducing feed passage rate and resulting in relatively higher methane emission (Janssen, 2010). Non-structural carbohydrate-rich diets, such as grains, concentrates, and readily fermented and small particle feed, shift the microbial community to one with Butyrivibrio spp. and Succinivibrionaceae as predominant members, increasing feed digestion and passage rate and resulting in lower methane emission (Tajima et al., 2001a; Luton et al., 2002; Tatsuoka et al., 2004; Friedrich, 2005; Janssen and Kirs, 2008; King et al., 2011; Henderson et al., 2015). Supplementary Table S3 summarized data on the methanogen communities in cattle fed various diets.
The rumen is rich in nutrients and metabolites that are generated from the degradation of plant materials and microbial activities. Additionally, internal rumen environment is dynamic, due to the constant efflux of feed, ruminal passage rate, and nutrients absorption by the animals (Saleem et al., 2012; Ungerfeld, 2020; Malheiros et al., 2021; Bica et al., 2022). Such features encourage members of an ecosystem to interact and provide a fertile ground for horizontal gene transfer or native gene modification-driven development of capabilities to transport externally available metabolites into the cells and utilize these (Cui et al., 2023). It could also allow the loss of certain de novo biosynthesis capabilities through genomic mutations and deletions, as the resultant strain would be supported with supplements from the community (Li et al., 2016; Kelly et al., 2016a,b,c). The need to protect the cells from toxic products released from plant material biodegradation and to leverage physical association with a donor for better efficiency of nutrient acquisition is also likely a promoter of genomic changes. Genome evolution in the face of temporal changes in nutrient availability could make an organism either a specialist, thriving at a specific time or under specific physiochemical conditions, or a generalist.
The most striking case is the loss of components of the methanogenesis system, causing both simple and complex impacts on the energy metabolism of the organisms. The genomes of Mbb. ruminantium, Methanomassiliicoccales isolates ISO4-G1 and ISO4-H5, and Thermoplasmatales archaeon BRNA1 lack coenzyme M biosynthetic genes (comADE) (Figure 4), causing a need for exogenous supply of CoM for the growth of these organisms (Li, 2016); no such information is available for the Methanomassiliicoccales isolates ISO4-G11, RumEn M1, and RumEn M2 (Jeyanathan, 2010; Söllinger et al., 2016). Almost all methanogens carry CoM transporter genes, ssuABC, as reflected in their sensitivities to bromoethane sulfonate (BES), an analog of CoM (Santoro and Konisky, 1987; Zhang et al., 2000). The requirement for CoM for rumen methanogens has been known for a long time (Balch et al., 1979; Balch and Wolfe, 1979a,b; Lovley et al., 1984), and a CoM auxotroph has been used in a bioassay for this coenzyme (Balch et al., 1979; Balch and Wolfe, 1979a,b).
Methanomassiliicoccales ISO4-G1 genome lacks the uroporphyrinogen-III C-methyltransferase (corA) gene that is involved in F430 biosynthesis and the organism likely requires F430 for growth (Li, 2016; Figure 4). Mbb. millerae SM9 and Mbb. olleyae YLM1 genomes do not carry any of the biotin biosynthesis genes (Figure 4). However, both genomes encode a biotin transporter, BioY (Kelly et al., 2016a,b,c), suggesting an ability of biotin uptake from the environment; rumen fluid contains biotin (Midla et al., 1998; Fitzgerald et al., 2000; Zimmerly and Weiss, 2001; Bergsten et al., 2003). In pure cultures, methanogens harboring CoM biosynthetic genes grow faster than the respective CoM auxotrophic strains (Lovley et al., 1984). On the other hand, auxotrophy could give a competitive advantage to methanogen in the rumen, as it would not have to invest energy for biosynthesis activities.
The instances of horizontal gene transfer (HGT) from bacteria to methanogens have been reported in numerous studies (Deppenmeier et al., 2002; Fournier and Gogarten, 2008; Lurie-Weinberger et al., 2012; Garushyants et al., 2015) though the transfer of methanogenesis genes to non-methanogenic species has not yet been reported (Gribaldo and Brochier-Armanet, 2006). A highly visible case of the former is the transfer of acetate kinase (ackA) and phosphotransacetylase (pta) genes from clostridia that provided acetoclastic methanogenesis capability in Methanosarcina (Fournier and Gogarten, 2008). In Methanobrevibacter smithii, a human gut-abundant methanogen species, over 15% of the genomic coding regions have bacterial characteristics (Lurie-Weinberger et al., 2012). For rumen methanogens, most of the transferred genes likely originated from organisms belonging to the Firmicutes phylum (Leahy et al., 2010; Kelly et al., 2016c). We describe below two examples of HGT events that likely helped methanogens to adapt to the rumen ecosystem.
As many as 294 genes of Mbb. ruminantium M1 have been postulated to be HGT-derived (Leahy et al., 2010), and most of these are for glycosyl transferases and adhesin-like proteins, which likely support Mbb. ruminantium to adapt in this environment (Samuel et al., 2007; Lurie-Weinberger et al., 2012; Shterzer and Mizrahi, 2015). In terms of the number of adhesin-like proteins encoded by the genome, this organism ranks first among the rumen methanogens, followed by Mbb. millerae SM9 (Figure 4). These values are consistent with the observed overall fitness in the rumen environment and the roles of adhesins in facilitating interaction with other ruminal guilds (Leahy et al., 2010; Ng et al., 2016; Wei et al., 2017).
In a co-culture experiment where Mbb. ruminantium was found to form aggregates with Butyrivibrio proteoclasticus, a Gram-positive rumen bacterium that degrades plant polysaccharides and forms butyrate, acetate, and hydrogen (Kelly et al., 2010), the levels of six adhesin-like proteins were enhanced in the methanogen (Leahy et al., 2010). A similar interaction of Mbb. ruminantium with rumen protozoa Epidinium and Entodinium (Ng et al., 2016) and rumen anaerobic fungi of the Piromyces genus has been documented, and in both cases, cell-to-cell attachments were clearly visualized (Wei et al., 2017). For the interaction with the protozoa, Mbb. ruminantium employs Mru_1499, an adhesin (Ng et al., 2016), and its association with Piromyces facilitates a high degree of biomass degradation, and methane and acetate formation (Wei et al., 2017). These findings call for further studies on the functional roles as well as the bacterial or protozoan targets for a large number of genes for adhesin-like proteins that have been bioinformatically identified in rumen methanogen genomes (Figure 4).
Tannins, which are water-soluble polyphenols and originate from plants, denature and precipitate proteins, thereby preventing their degradation by microbes in the rumen (Westendarp, 2006). This action facilitates the passage of proteins to the small intestine, wherein the free proteins, detached from the tannin, are hydrolyzed to generate amino acids for use by the host animal. Tannins are not significantly toxic to ruminants but possess antimicrobial properties, and accordingly, have been used to treat diarrhea and control parasite infection (Westendarp, 2006; Cardoso-Gutierrez et al., 2021).
An observed post-feeding decrease in the methanogen population in the rumen has been thought to be due to the tannins (Fagundes et al., 2020), and direct inhibition of methanogens by these compounds have also been reported (Tavendale et al., 2005; Goel and Makkar, 2011). Yet, some of the rumen methanogens tolerate tannins, and this is likely due to HGT-derived genes for tannin-modifying enzymes (Kelly et al., 2016c). An example of such an enzyme is the tannin acyl hydrolase of Mbb. millerae SM9 which hydrolyzes the galloyl ester bond in tannins releasing gallic acid and glucose (Banerjee et al., 2012). This hydrolase occurs mostly in bacteria and fungi (Banerjee et al., 2012) and represents the first known tannase in a methanogen (Kelly et al., 2016c). It is highly homologous to the Lactobacillus plantarum enzyme (Kelly et al., 2016c).
The early studies on the methanogens’ contributions to the conversion of feed into nutrients in ruminants were based on isolation, cultivation, and functional characterizations of rumen isolates. These efforts revolutionized the field of anaerobic microbiology and provided a first look into the rumen microbiome metabolism and respective roles in host physiology (Bryant and Burkey, 1953; Hungate, 1969; Henderson et al., 2015; Seshadri et al., 2018; Zehavi et al., 2018). However, the challenges of culturing strict anaerobes and the multiple auxotrophies of many of the rumen microbes and their metabolic dependence on community members hindered progress in the culture-dependent approach (Bryant and Burkey, 1953; Hungate, 1969; Henderson et al., 2015; Seshadri et al., 2018; Zehavi et al., 2018). Then, omics technologies brought a culture-independent approach toward an advanced assessment of the composition, metabolic potentials, and more importantly, in situ contributions of rumen methanogens (Tajima et al., 2001a,b; Luton et al., 2002; Tatsuoka et al., 2004; Friedrich, 2005; Janssen and Kirs, 2008; King et al., 2011; Henderson et al., 2015). We summarize below the progress and the gaps in these efforts.
The development of small subunit rRNAs, 16S and 18S, as universal genomic markers for taxonomic identification of prokaryotes and eukaryotes, respectively, has revolutionized the field of microbial ecology (Pace, 1997). The community structure and relative abundance of each taxon in a rumen sample could be analyzed by amplifying and sequencing the hypervariable regions of 16S or 18S rRNA genes and comparing the sequence information with a reference database (Janssen and Kirs, 2008; Henderson et al., 2015). Then, the resultant community structure information could be associated with the observed events and phenotypes such as methane emission, VFA profile, and high- versus low-efficiency animals (Danielsson et al., 2017). Such analyses could help to identify and target the methanogens that contribute to high methane emissions for developing highly specific anti-methanogen interventions while limiting the effects on ruminant’s feed utilization efficiency and health.
Cattle with higher feed efficiencies, as measured in terms of the amount of milk produced or weight gain per kilogram of dry matter intake (DMI), emit about 30% less methane than others (Hernandez-Sanabria et al., 2012). A strong relationship also exists between methane production and residual feed index (RFI) (Herd and Arthur, 2009; Muro-Reyes et al., 2011). An RFI value, which is independent of animal production parameters, is calculated from the difference between an animal’s actual and predicted feed intake values where the prediction is based on the animal’s body weight and growth rate over a specified period (Nkrumah et al., 2006). Cattle with low and high RFI values are categorized as “efficient” and “inefficient,” respectively. The efficient animals eat less than the predicted average and produce less methane (Hegarty et al., 2007; Waghorn and Hegarty, 2011). Since methane emissions cause energy loss from the feed, high and low-methane-emitting animals are also classified as inefficient and efficient, respectively.
Supplementary Table S1 presents the observed relationships between methanogen abundance and methane emission phenotypes. In general, Methanobrevibacter spp. and Methanosphaera spp. were detected in higher abundance, numerically, in the rumen of high and low methane-emitting cattle, respectively (Kittelmann et al., 2014; Shi et al., 2014; Stepanchenko et al., 2023). High abundances of Mbb. ruminantium and unclassified Methanomassiliicoccales have been correlated to low emitting phenotype while that of Methanobrevibacter gottschalkii was associated with high methane phenotype (Danielsson et al., 2017). In contrast to the above findings, Wallace et al. (2015) reported that both Methanobrevibacter spp. and Methanosphaera spp. were enriched in the high methane emitter.
While CO2-hydrogenotrophs were found in both high and low methane emitters, total methanogen abundance was double in high methane emitters than in the low methane emitters (Auffret et al., 2018). An instance with a 7 times higher abundance of Candidatus Methanomethylophilus, a methyl-dismutating methanogen, in low-emitting animals than in high-emitting animals, has been reported (Auffret et al., 2018; Supplementary Table S1). In another case, the rumen microbiomes of both high and low methane emitters were found to exhibit similar abundances of methanogens, with Mbb. gottschalkii and Mbb. ruminantium as dominant members (Kittelmann et al., 2014); Methanosphaera spp. and members of Methanomassiliicoccales order were present at lower abundances. A higher value for the abundance of Methanomassiliicoccaceae has been recorded for the rumen of barley-fed beef steers with low RFI than with high RFI (Li and Guan, 2017). This mixed picture originates from the complexity of the rumen microbiome, variable feed composition, animal production systems, and sampling times, as well as the uncertainties in the 16S rRNA-based genotype assessments as detailed below. Of the available ruminant datasets, those pertaining to agriculturally important ruminants other than cattle (e.g., buffalo, yak, goat) and ruminants from the low- and middle-income countries are still limited, and this area needs more attention for further studies (Xie et al., 2021; Arndt et al., 2022).
Although the 16S rRNA amplicon sequence-based method is widely used in microbial community analysis and offers several advantages, it is important to consider the following limitations. First, the choice of a particular hypervariable region of 16S rRNA as the target of amplification influences the results’ accuracy. The often-used hypervariable region 4 (16S rRNA-V4) underrepresents methanogen species in the amplicons due to poor sequence homology (Supplementary Figure S1; Gilbert et al., 2014), and the V6-V8 regions, as well as archaeal-specific or degenerate primers (A109F/958R or 1Af/1100Ar), have been suggested as more effective tools for capturing rumen archaeome diversity (Janssen and Kirs, 2008; Tymensen and McAllister, 2012; Snelling et al., 2014; Li et al., 2016; Bahram et al., 2018). Accordingly, to analyze both ruminal bacterial and archaeal communities, the 16S rRNA primer set combinations targeting bacterial V1-V3 or V4 and archaeal V6-V8 regions have been used (De Mulder et al., 2016; Lopes et al., 2021; Tan et al., 2021).
Second is the accuracy of the reference taxonomy that determines the quality of classification (Schloss and Westcott, 2011), as a noticeable fraction of the sequences in the commonly used databases, RDP (Cole et al., 2014), SILVA (Quast et al., 2013) and Greengenes (DeSantis et al., 2006), lack informative annotation beyond the genus level. Consequently, the highest taxonomic confidence for the amplicon-based approach reaches only the genus level (Schloss and Westcott, 2011; Johnson et al., 2019). The outcomes can be improved by using curated niche-specific databases (Henderson et al., 2019). Such databases are available for rumen and bovine GIT (Kittelmann et al., 2014; Seedorf et al., 2014; Shi et al., 2014; Ritari et al., 2015), insect gut (Newton and Roeselers, 2012; Mikaelyan et al., 2015), freshwater (Rohwer et al., 2018) and marine ecosystems (Tangherlini et al., 2018), and wastewater treatment units (McIlroy et al., 2017). Third, DNA-based analyses cannot distinguish between active community members and non-active or even non-viable members. Lastly, a marker gene-based analysis does not provide information on the full genomes, and consequently, fails to reveal information on the metabolic capabilities of individual organisms, especially those lost through mutations or gained horizontally.
Even then, the 16S rRNA-based approach serves as an affordable and powerful tool for the initial analysis, providing encouragement for higher-resolution omics analyses toward a holistic picture of rumen microbiome processes that contribute to methane emissions from ruminants. A hopeful development is that the full-length rRNA gene sequences recovered from ecological samples are increasing the resolution for phylogenetic profiling (Matsuo et al., 2021). With latest advancements in the next-generation DNA sequencing technology, which substantially lowers the sequencing costs, shallow shotgun metagenomic sequencing could provide an alternative and effective method for characterizing microbiome samples. It offers both taxonomic and functional information at a cost comparable to amplicon-based 16S rRNA analysis (Hillmann et al., 2018; Xu et al., 2021; Stothart et al., 2022; La Reau et al., 2023).
Shotgun metagenome and metatranscriptome sequencing, and metaproteomic and metabolomic analyses, stable-isotope probing, as well as full genomes of the isolates, have made it possible to perform thorough and precise in situ assessments of the structures and metabolic functions of the rumen microbiome (Shi et al., 2014; Estes et al., 2018; Stewart et al., 2018; Shakya et al., 2019; Wilkinson et al., 2020; van Cleef et al., 2021, 2022). The recently developed technology to rapidly generate full genome sequences from metagenomic DNA samples, namely metagenome-assembled genomes or MAGs (Tyson et al., 2004; Almeida et al., 2019; Nayfach et al., 2019; Youngblut et al., 2020; Haryono et al., 2022) has been extended to studies on rumen microbiome (Solden et al., 2018; Stewart et al., 2018, 2019; Wilkinson et al., 2020; Xie et al., 2021) and it allows the assignment of potential metabolic capabilities and in situ roles to microbes that have not even been obtained in pure or enrichment cultures.
Thousands of microbial MAGs have been recovered from the rumen samples (Solden et al., 2018; Stewart et al., 2018, 2019; Wilkinson et al., 2020; Xie et al., 2021). Two studies delivered >10,000 MAGs even from short-read sequences (Wilkinson et al., 2020; Xie et al., 2021). The genome sequences are facilitating not only the predictions of systems’ metabolic capabilities but also the strategy for genetic manipulations in situ (Roehe et al., 2016). Additionally, pangenome analysis from the MAG datasets is helping to identify environment-signature genes that could shed more insight into specific organism’s lifestyles and roles in an ecosystem, such as the rumen (Hansen et al., 2011; de la Cuesta-Zuluaga et al., 2021).
A major caveat of metagenomic analysis is its inability to distinguish between dead, dormant, and living cells (Shakya et al., 2019; Weinroth et al., 2022). It also fails to offer a complete assessment of the true in situ metabolic activities of the consortia (Shakya et al., 2019). It is only a combination of the genome and MAG sequences and metatranscriptomic, metaproteomic, and metabolomic data helps to assign comprehensive potential metabolic capabilities and capture real-time community metabolic activities and responses toward environmental changes such as feeding for the animals, and following are some of the examples of such studies (Shi et al., 2014; Li and Guan, 2017; Ma et al., 2018; Sollinger et al., 2018; Stewart et al., 2019; Wilkinson et al., 2020; Pitta et al., 2021; Xie et al., 2021; Pitta et al., 2022).
There are reports showing disagreements between the findings about rumen methanogens’ metabolic activities from DNA- and RNA-based characterizations (Shi et al., 2014; Li et al., 2016; Pitta et al., 2022) and the ratio of the number of transcripts and copies of the corresponding DNA (mRNA:DNA) has been proposed as an indicator of in situ metabolic activity (Pitta et al., 2022). Methanogens of the Methanobacteriales order account for more than 61% of methanogen DNA sequences followed by Methanomassiliicoccales which contributes 15.8% of the sequences (Janssen and Kirs, 2008). In a study where the CO2-hydrogenotrophic Methanobacteriales were highly represented in both the metagenomic and metatranscriptomic datasets, the respective mRNA:DNA value for formyl-MFR dehydrogenase, a CO2-reduction methanogenesis gene, was 1.5:1 (Pitta et al., 2022). In contrast, the mRNA:DNA value for mtaB of Methanosphaera species, which are methyl-hydrogenotrophs, was 6:1. A positive correlation between Methanomassiliicoccales rRNA and mtMA transcripts with the CH4 emission rate over time following feeding has also been recorded (Sollinger et al., 2018). These observations suggest that Methanomassiliicoccales and Methanosphaera species are more active than previously thought (Shi et al., 2014; Li et al., 2016; Sollinger et al., 2018; Söllinger and Urich, 2019; Pitta et al., 2021, 2022).
There have been efforts to determine the metabolic responses of specific methanogens to temporal changes in the rumen and link these to the organisms’ thresholds or hydrogen affinities and deployment of specific enzymes (Sollinger et al., 2018; Feldewert et al., 2020; Pitta et al., 2022). Such details are needed for judicial targeting of methanogens for mitigating enteric methane emission. In this effort, the methyl-CoM reductase isozymes (McrI and McrII), and methanol and methylamine-specific methyl transferases have particularly been in focus.
In Methanothermobacter species, McrI and McrII are encoded by the mcrBDCGA and mrtBDGA genes and expressed under low and high hydrogen availabilities, respectively (Rospert et al., 1990; Reeve, 1992; Morgan et al., 1997; Reeve et al., 1997), and these two systems are readily identified in methanogen genomes via protein primary sequence-based homology searches (Deppenmeier et al., 2002; Galagan et al., 2002; Fricke et al., 2006; Maeder et al., 2006; Jeyanathan, 2010; Leahy et al., 2010; Lambie et al., 2015; Li et al., 2016; Söllinger et al., 2016; Kelly et al., 2016a,b,c). As presented below and summarized in Figure 6, the reported data that links these factors together presents a complex and at times apparently contradicting picture.
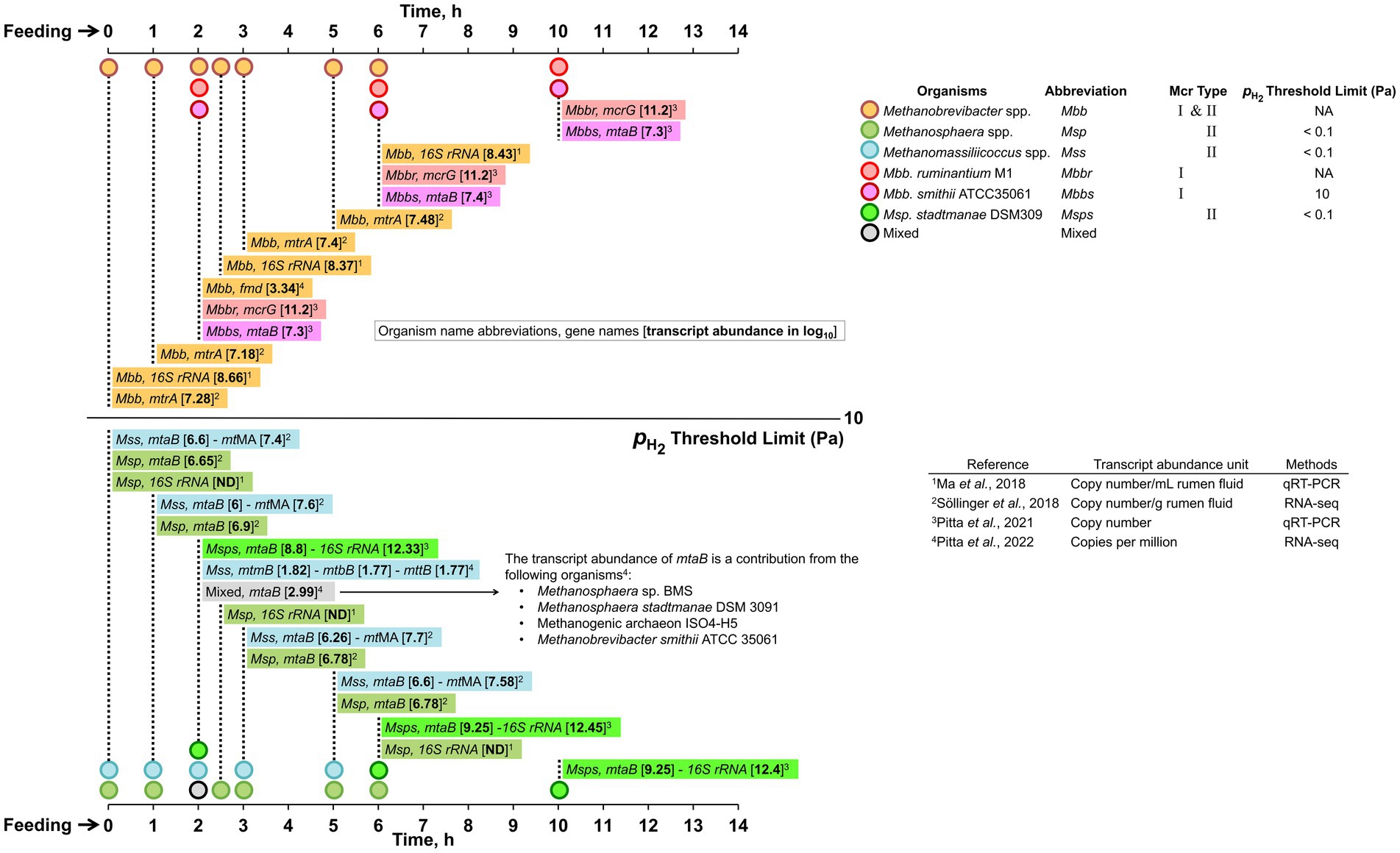
Figure 6. Temporal dynamics of rumen methanogen following feeding, a takeaway from transcriptomic studies. Transcript abundance of hydrogenotrophic and methyl-dismutating methanogenesis-related genes of rumen methanogens post-feeding. The abundance is shown in log10 values based on (Ma et al., 2018) or estimated from Sollinger et al. (2018) and Pitta et al. (2021, 2022), respectively. The threshold limit for Mbb. smithii (Feldewert et al., 2020). fmd, formylmethanofuran dehydrogenase; mcrG, methyl-CoM reductase subunit G; mtaB, methylcobamide:CoM methyltransferase; mtmB, monomethylamine methyltransferase; mtbB, dimethylamine methyltransferase; mttB, trimethylamine methyltransferase; mtMA, summarizes mono-, di-, and trimethylamine-specific methyltransferase (mtmB, mtbB, and mttB) transcripts whereas mttB transcripts constitute >70% of the mtMA transcripts; ND, not determined.
Following feeding, the concentrations of fermentation products such as CO2, H2, and VFAs increase, and methyl-containing compounds (i.e., methanol, mono-, di-, and trimethylamines) become available in the rumen due to the resident microbiome’s metabolic activities (Sollinger et al., 2018; Kelly et al., 2019). This situation sets up a competition among various functional guilds of ruminal methanogens (Rooke et al., 2014; Sollinger et al., 2018; Martínez-Álvaro et al., 2020; Ungerfeld, 2020). For example, in one case, it was found that the abundances of Methanosphaera and Methanomassiliicoccales transcripts increased immediately after feeding (1–3 h), suggesting that the methanogenesis activity of methyl-hydrogenotrophs spikes following increased availability of hydrogen, methanol, and methylamines (Sollinger et al., 2018; Pitta et al., 2021). Methanogenesis activity of CO2-hydrogenotrophs such as Methanobrevibacter spp., however, remained constant (Sollinger et al., 2018; Pitta et al., 2021). In another case, significant activity of CO2-hydrogenotrophs has been observed at 6–10 h post-feeding (Pitta et al., 2021).
A theoretical analysis assuming all reactants, except H2, being under standard conditions, suggested that the thermodynamic equilibrium (Figure 5) of the CO2-hydrogenotrophic methanogenesis will be reached at a of 0.18 Pa, and for the methyl-hydrogenotrophic system, it would occur at a much lower of 8 × 10−15 Pa, suggesting competitive advantages of methyl-reducing members over CO2-reducing hydrogenotrophs at low . However, since in a typical rumen, is relatively high (162 Pa), the ecological success of the methyl-reducing hydrogenotrophs is more likely determined by their ability to utilize methyl-group containing substrates and the availability of these substrates in the rumen (Thauer et al., 2008; Feldewert et al., 2020).
Further adaptation has been observed following this common response toward , where methyl-hydrogenotrophs developed intra-guild competition and substrate preference. For example, a study documented that immediately after feeding, when rumen is high, the abundance of Methanosphaera mtaB mRNA abundance soared, while a similar response was seen for the Methanomassiliicoccales mtMA and not mtaB (Sollinger et al., 2018). This finding indicates that Methanosphaera and Methanomassiliicoccales were positioned to utilize two different methyl-group containing substrates, methanol, and methylamines, respectively, although both groups can utilize all these compounds.
A detailed understanding of the diversity, lifestyle, and metabolism of rumen methanogens is key to developing strategies for achieving a substantial reduction of methane emissions from ruminants. The following section lays out key findings as well as challenges, research questions, and outlooks to guide future research toward the stated goal.
Like other host-associated relatives, rumen methanogens have evolved from free-living ancestors through genome-size reduction, mutations, and gene acquisitions through horizontal gene transfer (HGT) (Shterzer and Mizrahi, 2015; Söllinger et al., 2016; Thomas et al., 2021). This genome streamlining process has provided competitive advantages to the rumen methanogens, allowing them to: (1) conserve energy through auxotrophies and transform into oligotrophic metabolic lifestyles and become metabolically efficient; (2) increase fitness through acquisitions of new metabolic capabilities; and (3) develop syntrophic interactions with hydrogen-producing bacteria and protozoa for effective transfer of H2. These very factors pose serious challenges to the isolation of methanogens for use in in vitro physiological studies (Seshadri et al., 2018; Zehavi et al., 2018). A report on Methanomassiliicoccales showcases potential of bias when employing artificial laboratory culturing conditions as these tend to enrich the metabolically versatile, free-living environmental members over the auxotrophic gut-associated species (Söllinger et al., 2016). For instance, the Methanomassiliicoccus luminyensis and Methanomassiliicoccus intestinalis, which were isolated from human feces, belong to an environmental clade and non-gut-associated cluster (Dridi et al., 2012; Borrel et al., 2013; Söllinger et al., 2016). Thus, future isolation efforts for rumen methanogens must leverage information on their metabolic dependencies and syntrophic lifestyles gathered from microbial community analyses (Seshadri et al., 2018; Zehavi et al., 2018).
There is a great need for basic information for deciphering the mechanisms driving hydrogen removal and methane formation in the rumen in the face of temporal fluctuations in and availability of methanol and methylamines and time of deployment of two types of hydrogenotrophs. The values for individual methanogen’s threshold of and affinity (Ks values) for CO2 and methyl-group containing substrates, respectively, and the efficiency of harvesting electrons from bacterial and eukaryotic syntrophic partners are in this list (Feldewert et al., 2020).
Methyl-reducing hydrogenotrophs belonging to the poorly characterized Methanomassiliicoccales order are of particular interest (Dridi et al., 2012; Gorlas et al., 2012; Borrel et al., 2013). These organisms have lost the genes for all enzymes catalyzing the first six steps of the CO2-reducing methanogenesis pathway, an unprecedented phenomenon that has not been encountered in any other methanogen order (Dridi et al., 2012; Li et al., 2016; Kelly et al., 2016a,b; Lyu and Liu, 2019). However, they carry genes for the utilization of a diversity of methylated compounds, suggesting their metabolic limitation on one side and versatility on the other side as a way of adaptation to a nutrient-rich environment (Söllinger et al., 2016; Thomas et al., 2021).
An anticipated greater contribution of the previously underestimated Methanomassiliicoccales to methane production in the rumen (Pitta et al., 2022) is potentially driven by two factors. First, their lower threshold for H2 as mentioned above allows them to function at lower than that of the CO2-reducing hydrogenotrophs. Second, by utilizing a diversity of methyl groups containing methanogenesis substrates effectively, they prevail over other methyl-reducing hydrogenotrophs and methyl-dismutating methanogens. Nevertheless, a more definitive assessment of such relative capabilities requires information on the affinities (Ks) for methyl-group containing substrates of methanogens that utilize methyl groups for methanogenesis.
There is a lack of sufficient data for assigning the conditions under which an organism will deploy a particular Mcr isoenzyme. As a result, the reported assignments do not always match with an observed physiological response of the respective organisms toward hydrogen availability. For example, as mentioned above, Methanomassiliicoccales and Methanosphaera spp. employ Mcr II that is thought to be expressed under high conditions (Rospert et al., 1990; Reeve, 1992; Morgan et al., 1997; Reeve et al., 1997), and this functional association contradicts the observed lower threshold of these organisms (Feldewert et al., 2020). This discrepancy illuminates a major gap in studies on an enzyme that is the ultimate biological producer of methane (Wolfe, 1992).
The information on Mcr isoenzymes originated from investigations with two Methanothermobacter species, which are thermophiles (Rospert et al., 1990; Reeve, 1992; Morgan et al., 1997; Reeve et al., 1997), and these may not apply to other methanogens. The suggestion that even the activity of a Mcr could be under or redox-based regulation (Susanti et al., 2014) has also not been tested. As a result, a primary sequence homology-based identification of mcrA and mrt, which is the norm in ecological work, cannot indicate with certainty if the enzymes encoded by these genes are expressed or active under a particular condition.
The possibility that certain Methanobrevibacter species (i.e., Mbb. ruminantium M1) may perform methyl-hydrogenotrophy employing an HGT-derived methyl transferase (Leahy et al., 2013) brings a new dimension to the roles of these organisms in the rumen. Also, right after feeding, the H2 production rate far exceeds rumen methanogens’ available capacities to utilize this energy source (Rooke et al., 2014; Ungerfeld, 2020). This situation sets a lag between H2 production and CH4 emission (Rooke et al., 2014; van Lingen et al., 2017), and the suggestion that this effect is mainly due to a delayed expression of methanogenesis genes needs a detailed interrogation (Sollinger et al., 2018; Ungerfeld, 2020).
Early studies showed that formate is not a major precursor of methane in the rumen (Carroll and Hungate, 1955; Hungate et al., 1970) and this conclusion has recently been supported by the observation that the rumen samples lack transcripts for formate dehydrogenase; Fdh (Pitta et al., 2022) and formate were not detected at most timepoints following feeding (Sollinger et al., 2018). These findings contrast the observation that Methanobrevibacter species represent 60–80% of the rumen methanogen community (Henderson et al., 2015), and as mentioned above, these organisms carry the fdhABC genes (Schauer and Ferry, 1982; Nölling and Reeve, 1997). Fdh is encoded by an fdhABC operon that provides FdhAB and FdhC as a catalytic unit and formate transporter, respectively (Baron and Ferry, 1989). FdhAB oxidizes formate to CO2 and utilizes the electrons so generated for the reduction of F420 to F420H2.
Indeed, in Mbb. ruminantium M1, the abundance of fdhAB rRNA is enhanced when this methanogen is grown in a co-culture with Butyrivibrio proteoclasticus B316, an H2 and formate producer, indicating formate utilization by this methanogen during this syntrophic growth (Leahy et al., 2010). Also, formate as a methanogenesis substrate supports the growth of Mbb. ruminantium (Smith and Hungate, 1958). The sheep rumen microbiome has been found to exhibit poor expression of bacterial formate hydrogen lyases and other formate dehydrogenases (Greening et al., 2019). This suggests that formate produced in the rumen would be available for formate utilizers like Methanobrevibacter. The formate metabolism could also bring ecological fitness to the Methanobrevibacter spp. and Mb. formicicum.
The absence of formate dehydrogenase in the methyl-hydrogenotrophs gives free rein to formate utilizing methanogens for this substrate. In addition, being soluble, formate is an excellent vehicle for interspecies electron transfer and planktonic metabolism (Thiele and Zeikus, 1988; Leng, 2014), and removal of formate would prevent the acidification of the system as the pKa of the formic acid/formate pair is 3.75. The reported low levels of formate and fdh transcripts in the rumen (Sollinger et al., 2018; Pitta et al., 2022) could be rationalized by the high abundance of the Methanobrevibacter population. Also, the reported data were collected 2 h after feeding (Pitta et al., 2022), where the formate level would have dropped substantially, obviating the need for high-level fdh transcripts. The identification of formate utilizing methanogens in early colonizers in calves (Guzman et al., 2015) is intriguing, raising a question of whether formate is the substrate for methanogenesis at this stage. Further study on the formate and dissolved H2 levels in the undeveloped foregut of pre-ruminants could give insights into the role of methanogenesis from formate at this stage of the animals.
Thus, formate methanogenesis is an important yet less appreciated area of rumen microbial metabolism research. It needs to be studied with consideration that acetogenic bacteria with their ability to perform acetogenesis with formate would compete for this substrate (Greening and Leedle, 1989; Doré and Bryant, 1990; Schink et al., 2017; Greening et al., 2019; Moon et al., 2021).
As mentioned above, there is a need to strengthen the 16S rRNA sequence database as it would allow effective use of the most affordable route to community composition analysis that employs sequencing and analysis of short (~100–200 bp) amplicons of 16S rRNA gene (Johnson et al., 2019; Weinroth et al., 2022). Under the current situation, the results of such analyses need to be considered with caution as it often provides only low-resolution identities, an over-simplification of the diversity and incomplete metabolic inferences for methanogens in the rumen (Pitta et al., 2022). There are instances where metatranscriptomic and 16S rRNA amplicon sequences from rumen samples detected the presence of Methanocaldococcus spp. and Methanopyrus spp., which are obligate hyperthermophiles (Lyu and Liu, 2019), and Mbb. smithii, a human-associated organism (Zhou et al., 2009; Kong et al., 2013; Auffret et al., 2018; Mann et al., 2018; Tan et al., 2021).
Comprehensive and effective comparative genomic studies and analysis of metatranscriptomic and metaproteomic data with rumen methanogens are limited by the inadequate number of well-annotated reference genomes of pure culture isolates and MAGs. Even the Hungate 1,000 Project which generated sequences of 501 genomes, covering 480 ruminal bacteria and 21 archaea species, represents only 15 rumen methanogens (Seshadri et al., 2018). The number of isolate genome and MAG sequences for rumen methanogens that are publicly available are only 14 (Supplementary Table S2) and 206, respectively (Söllinger et al., 2016; Stewart et al., 2018, 2019; Wilkinson et al., 2020; Glendinning et al., 2021; Xie et al., 2021).
The gap in reference data extends beyond the molecular data. In most cases there is little information on the association of isolate genomes, MAGs, and even sometimes the metatranscriptomic and metaproteomic data for the methanogens with the following key parameters: (i) details of the feed; (ii) spatial location within the rumen, namely, fiber-associated, planktonic, and epimural microbiome; (iii) co-occurrence, such as association with the syntrophic partners, protozoa, and bacteria; (iv) timing of sampling with respect to feeding; and (v) comparison with free-living counterparts. In a recent study with cattle grazing tall fescue, a major perturbation of the microbiome by a toxic version of the grass was detected only when the sessile and planktonic fractions were analyzed separately (Khairunisa et al., 2022) and similar observations have been reported by others (Pitta et al., 2014).
The developmental stage of the animal host is a key factor (Fonty et al., 1987; Morvan et al., 1994; Fonty et al., 2007; Guzman et al., 2015; Friedman et al., 2017; Furman et al., 2020), as rumen microbiome modulation at this early stage of the animals is being considered as a potential methane mitigation strategy (Meale et al., 2021). The metabolism of methanogens that colonize the gut of the pre-ruminant phase and its influence on the development of the rumen remains to be investigated critically.
This review shows that the current knowledge of rumen methanogens cannot adequately support the efforts for designing measures that will mitigate methane emissions from ruminants and preserve rumen function in the absence or in reduced methanogenic activity. Even after ~80 years of research, it is not known why Methanobrevibacter spp. dominate the rumen microbiome and what their specific contributions are. Filling these gaps requires significant isolation efforts, especially for those members with very few or no pure culture representatives (e.g., Methanomassiliicoccales) and the generation of more well-annotated genomes and MAG sequences.
A culturomic approach leveraging both undefined media containing rumen fluid and defined media showed that 23% of the rumen microbiota is cultivable with these technologies (Zehavi et al., 2018). However, it provided a relatively low coverage for the rumen methanogens. For example, of the prokaryotes in the Hungate 1,000 culture collection, methanogens represent only 4.1% of the total (Seshadri et al., 2018). Thus, for an isolation effort to be productive will require innovative approaches. If the unknown growth requirements make it difficult to generate axenic cultures, attempts could be made to obtain low-complexity mixed cultures. Since 16S rRNA provides an affordable and amenable route for rapid assessment of microbiome diversity, the respective database needs to be strengthened.
With more reference isolates, comprehensive physiological studies could occur with a focus on newly recognized genomic features that promote colonization of the rumen and high-level methane production. One high-value area is the cellular interactions of rumen methanogens with their syntrophic partners such as protozoa, fungi, and bacteria where the following questions are key. What governs such interactions? What defines the specificity and recognition by interacting partners? What are the mechanisms of interspecies electron transport? Co-occurrence analysis that could reveal metabolic differences between host-associated and free-living methanogens would also be valuable. Detailed information on methanol and methylamine concentrations in the rumen of animals fed various diets, thresholds for these substrates and of various rumen methanogens, and catalytic properties and expression conditions of the Mcr isoenzymes are needed to make the analysis and interpretation of in situ observations more reliable. The information on the Mcr isoenzymes is also needed for correct functional annotations of mcr and mrt homologs. The diversity and metabolic activities of methanogens residing in various locations of the rumen as mentioned above could give insights into true activities driving in situ methane production.
BK: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. CH: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. KI: Formal analysis, Writing – original draft. BM: Formal analysis, Funding acquisition, Supervision, Writing – review & editing. DS: Conceptualization, Formal analysis, Writing – original draft, Funding acquisition, Supervision, Writing – review & editing, Data curation.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The relevant research on methanogens in BM’s laboratory has been supported in part by the National Aeronautics and Space Administration Astrobiology: Exobiology and Evolutionary Biology grant NNX13AI05G, Virginia Tech Agricultural Experiment Station Hatch Program (CRIS project VA-160021), and a Pratt Endowment grant of the Virginia Tech College of Agriculture and Life Sciences (CALS). CH received a Graduate Fellowship from the GBCB Ph.D. Program of Virginia Tech. KI received a Summer Internship at Elanco Animal Health.
We thank Drs. German Plata, Luke Bown, Dharanesh Gangaiah, and Arvind Kumar for providing thorough reviews and invaluable inputs.
DS and BK were employed by BiomEdit, an animal health biotechnology company that leverages a unique platform combining the leading science of the microbiome with synthetic biology to innovate novel animal health products to address challenges in livestock production and pet health.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1296008/full#supplementary-material
Akinyemi, T. S., Shao, N., Whitman, W. B., and Oren, A. (2021). “Methanotrichales ord. nov.” in Bergey’s manual of systematics of archaea and bacteria. Eds. W. B. Whitman, P. DeVos, S. Dedysh, B. Hedlund, P. Kämpfer, and F. Rainey, et al. (New York: John Wiley & Sons), 1–2.
Almeida, A., Mitchell, A. L., Boland, M., Forster, S. C., Gloor, G. B., Tarkowska, A., et al. (2019). A new genomic blueprint of the human gut microbiota. Nature 568, 499–504. doi: 10.1038/s41586-019-0965-1
Amat, S., Holman, D. B., Schmidt, K., McCarthy, K. L., Dorsam, S. T., Ward, A. K., et al. (2022). Characterization of the microbiota associated with 12-week-old bovine fetuses exposed to divergent in utero nutrition. Front. Microbiol. 12:771832. doi: 10.3389/fmicb.2021.771832
Anderson, I., Ulrich, L. E., Lupa, B., Susanti, D., Porat, I., Hooper, S. D., et al. (2009). Genomic characterization of Methanomicrobiales reveals three classes of methanogens. PLoS One 4:e5797. doi: 10.1371/journal.pone.0005797
Arndt, C., Hristov, A. N., Price, W. J., McClelland, S. C., Pelaez, A. M., Cueva, S. F., et al. (2022). Full adoption of the most effective strategies to mitigate methane emissions by ruminants can help meet the 1.5 °C target by 2030 but not 2050. Proc. Natl. Acad. Sci. 119:e2111294119. doi: 10.1073/pnas.2111294119
Auffret, M. D., Stewart, R., Dewhurst, R. J., Duthie, C.-A., Rooke, J. A., Wallace, R. J., et al. (2018). Identification, comparison, and validation of robust rumen microbial biomarkers for methane emissions using diverse Bos taurus breeds and basal diets. Front. Microbiol. 8:2642. doi: 10.3389/fmicb.2017.02642
Bahram, M., Anslan, S., Hildebrand, F., Bork, P., and Tedersoo, L. (2018). Newly designed 16S rRNA metabarcoding primers amplify diverse and novel archaeal taxa from the environment. Environ. Microbiol. Rep. 11, 487–494. doi: 10.1111/1758-2229.12684
Balch, W. E., Fox, G. E., Magrum, L. J., Woese, C. R., and Wolfe, R. S. (1979). Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43, 260–296. doi: 10.1128/mr.43.2.260-296.1979
Balch, W. E., and Wolfe, R. S. (1976). New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl. Environ. Microbiol. 32, 781–791. doi: 10.1128/aem.32.6.781-791.1976
Balch, W. E., and Wolfe, R. S. (1979a). Specificity and biological distribution of coenzyme M (2-mercaptoethanesulfonic acid). J. Bacteriol. 137, 256–263. doi: 10.1128/jb.137.1.256-263.1979
Balch, W. E., and Wolfe, R. S. (1979b). Transport of coenzyme M (2-mercaptoethanesulfonic acid) in Methanobacterium ruminantium. J. Bacteriol. 137, 264–273. doi: 10.1128/jb.137.1.264-273.1979
Baldwin, R. L., McLeod, K. R., Klotz, J. L., and Heitmann, R. N. (2004). Rumen development, intestinal growth and hepatic metabolism in the pre- and postweaning ruminant. J. Dairy Sci. 87, E55–E65. doi: 10.3168/jds.S0022-0302(04)70061-2
Banerjee, A., Jana, A., Pati, B. R., Mondal, K. C., and Das Mohapatra, P. K. (2012). Characterization of tannase protein sequences of bacteria and fungi: an in silico study. Protein J. 31, 306–327. doi: 10.1007/s10930-012-9405-x
Barber, R. D., Zhang, L., Harnack, M., Olson, M. V., Kaul, R., Ingram-Smith, C., et al. (2011). Complete genome sequence of Methanosaeta concilii, a specialist in aceticlastic methanogenesis. J. Bacteriol. 193, 3668–3669. doi: 10.1128/JB.05031-11
Barker, H. A. (1936). On the biochemistry of the methane fermentation. Arch. Mikrobiol. 7, 404–419. doi: 10.1007/BF00407413
Baron, S. F., and Ferry, J. G. (1989). Reconstitution and properties of a coenzyme F420-mediated formate hydrogenlyase system in Methanobacterium formicicum. J. Bacteriol. 171, 3854–3859. doi: 10.1128/jb.171.7.3854-3859.1989
Barry, T. N., Thompson, A., and Armstrong, D. G. (1977). Rumen fermentation studies on two contrasting diets. 1. Some characteristics of the in vivo fermentation, with special reference to the composition of the gas phase, oxidation/reduction state and volatile fatty acid proportions. J. Agric. Sci. 89, 183–195. doi: 10.1017/S0021859600027362
Battumur, U., Lee, M., Bae, G. S., and Kim, C.-H. (2019). Isolation and characterization of a new Methanoculleus bourgensis strain KOR-2 from the rumen of Holstein steers. Asian Australas. J. Anim. Sci. 32, 241–248. doi: 10.5713/ajas.18.0409
Beauchemin, K. A., Ungerfeld, E. M., Eckard, R. J., and Wang, M. (2020). Review: fifty years of research on rumen methanogenesis: lessons learned and future challenges for mitigation. Animal 14, s2–s16. doi: 10.1017/S1751731119003100
Becker, K. W., Elling, F. J., Yoshinaga, M. Y., Söllinger, A., Urich, T., and Hinrichs, K.-U. (2016). Unusual butane- and pentanetriol-based tetraether lipids in Methanomassiliicoccus luminyensis, a representative of the seventh order of methanogens. Appl. Environ. Microbiol. 82, 4505–4516. doi: 10.1128/AEM.00772-16
Beijer, W. H. (1952). Methane fermentation in the rumen of cattle. Nature 170, 576–577. doi: 10.1038/170576a0
Belanche, A., de la Fuente, G., and Newbold, C. J. (2014). Study of methanogen communities associated with different rumen protozoal populations. FEMS Microbiol. Ecol. 90, 663–677. doi: 10.1111/1574-6941.12423
Bergman, E. N. (1990). Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70, 567–590. doi: 10.1152/physrev.1990.70.2.567
Bergsten, C., Greenough, P. R., Gay, J. M., Seymour, W. M., and Gay, C. C. (2003). Effects of biotin supplementation on performance and claw lesions on a commercial dairy farm. J. Dairy Sci. 86, 3953–3962. doi: 10.3168/jds.S0022-0302(03)74005-3
Bica, R., Palarea-Albaladejo, J., Lima, J., Uhrin, D., Miller, G. A., Bowen, J. M., et al. (2022). Methane emissions and rumen metabolite concentrations in cattle fed two different silages. Sci. Rep. 12:5441. doi: 10.1038/s41598-022-09108-w
Black, A. L., Kleiber, M., and Brown, A. M. (1961). Butyrate metabolism in the lactating cow. J. Biol. Chem. 236, 2399–2403. doi: 10.1016/S0021-9258(18)64010-4
Bonacker, L. G., Baudner, S., Morschel, E., Bocher, R., and Thauer, R. K. (1993). Properties of the two isoenzymes of methyl-coenzyme M reductase in Methanobacterium thermoautotrophicum. Eur. J. Biochem. 217, 587–595. doi: 10.1111/j.1432-1033.1993.tb18281.x
Boone, D. R., Whitman, W. B., and Rouviere, P. (1993). “Diversity and taxonomy of methanogens” in Methanogenesis: ecology, physiology, biochemistry and genetics. ed. J. G. Ferry (New York, NY: Chapman and Hall), 35–80.
Borrel, G., Harris, H. M., Parisot, N., Gaci, N., Tottey, W., Mihajlovski, A., et al. (2013). Genome sequence of "Candidatus Methanomassiliicoccus intestinalis" issoire-Mx1, a third thermoplasmatales-related methanogenic archaeon from human feces. Genome Announc. 1:e00453-13. doi: 10.1128/genomeA.00453-13
Borrel, G., O’Toole, P. W., Harris, H. M. B., Peyret, P., Brugère, J.-F., and Gribaldo, S. (2013). Phylogenomic data support a seventh order of methylotrophic methanogens and provide insights into the evolution of methanogenesis. Genome Biol. Evol. 5, 1769–1780. doi: 10.1093/gbe/evt128
Borrel, G., Parisot, N., Harris, H. M. B., Peyretaillade, E., Gaci, N., Tottey, W., et al. (2014). Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics 15:679. doi: 10.1186/1471-2164-15-679
Brown, D. R., May, M., Bradbury, J. M., Balish, M. F., Calcutt, M. J., Glass, J. I., et al. (2018). “Mycoplasma” in Bergey’s manual of systematics of archaea and bacteria. Eds. W. B. Whitman, P. DeVos, S. Dedysh, B. Hedlund, P. Kämpfer, and F. Rainey, et al. 1–78.
Bryant, M. P., and Burkey, L. A. (1953). Cultural methods and some characteristics of some of the more numerous groups of bacteria in the bovine rumen. J. Dairy Sci. 36, 205–217. doi: 10.3168/jds.S0022-0302(53)91482-9
Bryant, M. P., Tzeng, S. F., Robinson, I. M., and Joyner, A. E. Jr. (1971). Nutrient requirements of methanogenic bacteria. Anaerobic Biol. Treat. Process. 105, 23–40. doi: 10.1021/ba-1971-0105.ch003
Bryant, M. P., Wolin, E. A., Wolin, M. J., and Wolfe, R. S. (1967). Methanobacillus omelianskii, a symbiotic association of two species of bacteria. Arch. Mikrobiol. 59, 20–31. doi: 10.1007/BF00406313
Buan, N. R., and Metcalf, W. W. (2010). Methanogenesis by Methanosarcina acetivorans involves two structurally and functionally distinct classes of heterodisulfide reductase. Mol. Microbiol. 75, 843–853. doi: 10.1111/j.1365-2958.2009.06990.x
Buckel, W., and Thauer, R. K. (2018). Flavin-based electron bifurcation, a new mechanism of biological energy coupling. Chem. Rev. 118, 3862–3886. doi: 10.1021/acs.chemrev.7b00707
Bueno de Mesquita, C. P., Wu, D., and Tringe, S. G. (2023). Methyl-based methanogenesis: an ecological and genomic review. Microbiol. Mol. Biol. Rev. 87:e0002422. doi: 10.1128/mmbr.00024-22
Carberry, C. A., Kenny, D. A., Han, S., McCabe, M. S., and Waters, S. M. (2012). Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle. Appl. Environ. Microbiol. 78, 4949–4958. doi: 10.1128/AEM.07759-11
Cardoso-Gutierrez, E., Aranda-Aguirre, E., Robles-Jimenez, L. E., Castelán-Ortega, O. A., Chay-Canul, A. J., Foggi, G., et al. (2021). Effect of tannins from tropical plants on methane production from ruminants: a systematic review. Vet. Anim. Sci. 14:100214. doi: 10.1016/j.vas.2021.100214
Carroll, E. J., and Hungate, R. E. (1955). Formate dissimilation and methane production in bovine rumen contents. Arch. Biochem. Biophys. 56, 525–536. doi: 10.1016/0003-9861(55)90272-1
Chen, I. M. A., Chu, K., Palaniappan, K., Ratner, A., Huang, J., Huntemann, M., et al. (2023). The IMG/M data management and analysis system v.7: content updates and new features. Nucleic Acids Res. 51, D723–D732. doi: 10.1093/nar/gkac976
Choi, K.-P., Kendrick, N., and Daniels, L. (2002). Demonstration that fbiC is required by Mycobacterium bovis BCG for coenzyme F420 and FO biosynthesis. J. Bacteriol. 184, 2420–2428. doi: 10.1128/JB.184.9.2420-2428.2002
Church, D. C. (1988). The ruminant animal: digestive physiology and nutrition. Englewood Cliffs, NJ: Prentice-Hall, Inc.
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., McGarrell, D. M., Sun, Y., et al. (2014). Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642. doi: 10.1093/nar/gkt1244
Comtet-Marre, S., Parisot, N., Lepercq, P., Chaucheyras-Durand, F., Mosoni, P., Peyretaillade, E., et al. (2017). Metatranscriptomics reveals the active bacterial and eukaryotic fibrolytic communities in the rumen of dairy cow fed a mixed diet. Front. Microbiol. 8:67. doi: 10.3389/fmicb.2017.00067
Costa, K. C., Wong, P. M., Wang, T., Lie, T. J., Dodsworth, J. A., Swanson, I., et al. (2010). Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc. Natl. Acad. Sci. U. S. A. 107, 11050–11055. doi: 10.1073/pnas.1003653107
Cozannet, M., Borrel, G., Roussel, E., Moalic, Y., Allioux, M., Sanvoisin, A., et al. (2020). New insights into the ecology and physiology of Methanomassiliicoccales from terrestrial and aquatic environments. Microorganisms 9:30. doi: 10.3390/microorganisms9010030
Craciun, S., and Balskus, E. P. (2012). Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl. Acad. Sci. 109, 21307–21312. doi: 10.1073/pnas.1215689109
Criscuolo, A., and Gribaldo, S. (2010). BMGE (block mapping and gathering with entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 10:210. doi: 10.1186/1471-2148-10-210
Cui, X., Wang, Z., Guo, P., Li, F., Chang, S., Yan, T., et al. (2023). Shift of feeding strategies from grazing to different forage feeds reshapes the rumen microbiota to improve the ability of Tibetan sheep (Ovis aries) to adapt to the cold season. Microbiol. Spectr. 11:e0281622. doi: 10.1128/spectrum.02816-22
Czerkawski, J. W., and Breckenridge, G. (1973). Dissimilation of 1,2-propanediol by rumen micro-organisms. Br. J. Nutr. 29, 317–330. doi: 10.1079/BJN19730106
Danielsson, R., Dicksved, J., Sun, L., Gonda, H., Müller, B., Schnürer, A., et al. (2017). Methane production in dairy cows correlates with rumen methanogenic and bacterial community structure. Front. Microbiol. 8:226. doi: 10.3389/fmicb.2017.00226
Davis, C. L., and Drackley, J. K. (1998). The development, nutrition, and management of the Young Calf. Ames, IA: Iowa State Univeristy Press.
de la Cuesta-Zuluaga, J., Spector, T. D., Youngblut, N. D., and Ley, R. E. (2021). Genomic insights into adaptations of trimethylamine-utilizing methanogens to diverse habitats, including the human gut. mSystems 6:e00939-20. doi: 10.1128/mSystems.00939-20
De Mulder, T., Goossens, K., Peiren, N., Vandaele, L., Haegeman, A., De Tender, C., et al. (2016). Exploring the methanogen and bacterial communities of rumen environments: solid adherent, fluid and epimural. FEMS Microbiol. Ecol. 93:fiw251. doi: 10.1093/femsec/fiw251
DeFrain, J. M., Hippen, A. R., Kalscheur, K. F., and Schingoethe, D. J. (2004). Feeding lactose increases ruminal butyrate and plasma beta-hydroxybutyrate in lactating dairy cows. J. Dairy Sci. 87, 2486–2494. doi: 10.3168/jds.S0022-0302(04)73373-1
Deppenmeier, U. (2004). The membrane-bound electron transport system of Methanosarcina species. J. Bioenerg. Biomembr. 36, 55–64. doi: 10.1023/B:JOBB.0000019598.64642.97
Deppenmeier, U., Johann, A., Hartsch, T., Merkl, R., Schmitz, R. A., Martinez-Arias, R., et al. (2002). The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4, 453–461.
Deppenmeier, U., Müller, V., and Gottschalk, G. (1996). Pathways of energy conservation in methanogenic archaea. Arch. Microbiol. 165, 149–163. doi: 10.1007/BF01692856
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Desper, R., and Gascuel, O. (2002). Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. J. Comput. Biol. 9, 687–705. doi: 10.1089/106652702761034136
Difford, G. F., Plichta, D. R., Løvendahl, P., Lassen, J., Noel, S. J., Højberg, O., et al. (2018). Host genetics and the rumen microbiome jointly associate with methane emissions in dairy cows. PLoS Genet. 14:e1007580. doi: 10.1371/journal.pgen.1007580
Doré, J., and Bryant, M. P. (1990). Metabolism of one-carbon compounds by the ruminal acetogen Syntrophococcus sucromutans. Appl. Environ. Microbiol. 56, 984–989. doi: 10.1128/aem.56.4.984-989.1990
Downing, B. E., Gupta, D., and Nayak, D. D. (2023). The dual role of a multi-heme cytochrome in methanogenesis: MmcA is important for energy conservation and carbon metabolism in Methanosarcina acetivorans. Mol. Microbiol. 119, 350–363. doi: 10.1111/mmi.15029
Dridi, B., Fardeau, M.-L., Ollivier, B., Raoult, D., and Drancourt, M. (2012). Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int. J. Syst. Evol. Microbiol. 62, 1902–1907. doi: 10.1099/ijs.0.033712-0
Eirich, L. D., Vogels, G. D., and Wolfe, R. S. (1979). Distribution of coenzyme F420 and properties of its hydrolytic fragments. J. Bacteriol. 140, 20–27. doi: 10.1128/jb.140.1.20-27.1979
Elsden, S. R. (1945). The fermentation of carbohydrates in the rumen of the sheep. J. Exp. Biol. 22, 51–62. doi: 10.1242/jeb.22.1-2.51
EPA (2023b). Understanding global warming potentials. Available at: https://www.epa.gov/ghgemissions/understanding-global-warming-potentials.
Estes, K. A., White, R. R., Yoder, P. S., Pilonero, T., Schramm, H., Lapierre, H., et al. (2018). An in vivo stable isotope-based approach for assessment of absorbed amino acids from individual feed ingredients within complete diets. J. Dairy Sci. 101, 7040–7060. doi: 10.3168/jds.2017-13447
Fagundes, G. M., Benetel, G., Welter, K. C., Melo, F. A., Muir, J. P., Carriero, M. M., et al. (2020). Tannin as a natural rumen modifier to control methanogenesis in beef cattle in tropical systems: friend or foe to biogas energy production? Res. Vet. Sci. 132, 88–96. doi: 10.1016/j.rvsc.2020.05.010
FAO (2018). The future of food and agriculture—alternative pathways to 2050. Food and Agriculture Organization of the United Nations. Rome, Italy.
Feldewert, C., Lang, K., and Brune, A. (2020). The hydrogen threshold of obligately methyl-reducing methanogens. FEMS Microbiol. Lett. 367:fnaa137. doi: 10.1093/femsle/fnaa137
Fitzgerald, T., Norton, B. W., Elliott, R., Podlich, H., and Svendsen, O. L. (2000). The influence of long-term supplementation with biotin on the prevention of lameness in pasture fed dairy cows. J. Dairy Sci. 83, 338–344. doi: 10.3168/jds.S0022-0302(00)74884-3
Flint, H. J., and Bayer, E. A. (2008). Plant cell wall breakdown by anaerobic microorganisms from the mammalian digestive tract. Ann. N. Y. Acad. Sci. 1125, 280–288. doi: 10.1196/annals.1419.022
Fonty, G., Gouet, P., Jouany, J.-P., and Senaud, J. (1987). Establishment of the microflora and anaerobic fungi in the rumen of lambs. Microbiology 133, 1835–1843. doi: 10.1099/00221287-133-7-1835
Fonty, G., Joblin, K., Chavarot, M., Roux, R., Naylor, G., and Michallon, F. (2007). Establishment and development of ruminal hydrogenotrophs in methanogen-free lambs. Appl. Environ. Microbiol. 73, 6391–6403. doi: 10.1128/AEM.00181-07
Fournier, G. P., and Gogarten, J. P. (2008). Evolution of acetoclastic methanogenesis in Methanosarcina via horizontal gene transfer from cellulolytic Clostridia. J. Bacteriol. 190, 1124–1127. doi: 10.1128/JB.01382-07
Fricke, W. F., Seedorf, H., Henne, A., Krüer, M., Liesegang, H., Hedderich, R., et al. (2006). The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J. Bacteriol. 188, 642–658. doi: 10.1128/JB.188.2.642-658.2006
Friedman, N., Jami, E., and Mizrahi, I. (2017). Compositional and functional dynamics of the bovine rumen methanogenic community across different developmental stages. Environ. Microbiol. 19, 3365–3373. doi: 10.1111/1462-2920.13846
Friedrich, M. W. (2005). Methyl-coenzyme M reductase genes: unique functional markers for methanogenic and anaerobic methane-oxidizing archaea. Environ. Microbiol. 397, 428–442. doi: 10.1016/S0076-6879(05)97026-2
Furman, O., Shenhav, L., Sasson, G., Kokou, F., Honig, H., Jacoby, S., et al. (2020). Stochasticity constrained by deterministic effects of diet and age drive rumen microbiome assembly dynamics. Nat. Commun. 11:1904. doi: 10.1038/s41467-020-15652-8
Galagan, J. E., Nusbaum, C., Roy, A., Endrizzi, M. G., Macdonald, P., FitzHugh, W., et al. (2002). The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12, 532–542. doi: 10.1101/gr.223902
Garcia, P. S., Gribaldo, S., and Borrel, G. (2022). Diversity and evolution of methane-related pathways in Archaea. Annu. Rev. Microbiol. 76, 727–755. doi: 10.1146/annurev-micro-041020-024935
Garrity, G. M., Labeda, D. P., and Oren, A. (2011). Judicial Commission of the International Committee on systematics of prokaryotes XIIth international (IUMS) congress of bacteriology and applied microbiology. Int. J. Syst. Evol. Microbiol. 61, 2775–2780. doi: 10.1099/ijs.0.037366-0
Garushyants, S. K., Kazanov, M. D., and Gelfand, M. S. (2015). Horizontal gene transfer and genome evolution in Methanosarcina. BMC Evol. Biol. 15:102. doi: 10.1186/s12862-015-0393-2
Gilbert, J. A., Jansson, J. K., and Knight, R. (2014). The earth microbiome project: successes and aspirations. BMC Biol. 12:69. doi: 10.1186/s12915-014-0069-1
Gilbert, R. A., Townsend, E. M., Crew, K. S., Hitch, T. C. A., Friedersdorff, J. C. A., Creevey, C. J., et al. (2020). Rumen virus populations: technological advances enhancing current understanding. Front. Microbiol. 11:450. doi: 10.3389/fmicb.2020.00450
Glendinning, L., Genç, B., Wallace, R. J., and Watson, M. (2021). Metagenomic analysis of the cow, sheep, reindeer and red deer rumen. Sci. Rep. 11:1990. doi: 10.1038/s41598-021-81668-9
Goel, G., and Makkar, H. P. S. (2011). Methane mitigation from ruminants using tannins and saponins. Trop. Anim. Health Prod. 44, 729–739. doi: 10.1007/s11250-011-9966-2
Gorlas, A., Robert, C., Gimenez, G., Drancourt, M., and Raoult, D. (2012). Complete genome sequence of Methanomassiliicoccus luminyensis, the largest genome of a human-associated Archaea species. J. Bacteriol. 194:4745. doi: 10.1128/JB.00956-12
Graham, D. E., Xu, H., and White, R. H. (2003). Identification of the 7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase required for coenzyme F 420 biosynthesis. Arch. Microbiol. 180, 455–464. doi: 10.1007/s00203-003-0614-8
Graupner, M., Xu, H., and White, R. H. (2000). Identification of the gene encoding sulfopyruvate decarboxylase, an enzyme involved in biosynthesis of coenzyme M. J. Bacteriol. 182, 4862–4867. doi: 10.1128/JB.182.17.4862-4867.2000
Greening, C., Geier, R., Wang, C., Woods, L. C., Morales, S. E., McDonald, M. J., et al. (2019). Diverse hydrogen production and consumption pathways influence methane production in ruminants. ISME J. 13, 2617–2632. doi: 10.1038/s41396-019-0464-2
Greening, R. C., and Leedle, J. A. Z. (1989). Enrichment and isolation of Acetitomaculum ruminis, gen. nov., sp. nov.: acetogenic bacteria from the bovine rumen. Arch. Microbiol. 151, 399–406. doi: 10.1007/BF00416597
Gribaldo, S., and Brochier-Armanet, C. (2006). The origin and evolution of Archaea: a state of the art. Philos. Trans. R. Soc. B Biol. Sci. 361, 1007–1022. doi: 10.1098/rstb.2006.1841
Gruninger, R. J., Ribeiro, G. O., Cameron, A., and McAllister, T. A. (2019). Invited review: application of meta-omics to understand the dynamic nature of the rumen microbiome and how it responds to diet in ruminants. Animal 13, 1843–1854. doi: 10.1017/S1751731119000752
Guzman, C. E., Bereza-Malcolm, L. T., De Groef, B., and Franks, A. E. (2015). Presence of selected methanogens, fibrolytic bacteria, and proteobacteria in the gastrointestinal tract of neonatal dairy calves from birth to 72 hours. PLoS One 10:e0133048. doi: 10.1371/journal.pone.0133048
Guzman, C. E., Wood, J. L., Egidi, E., White-Monsant, A. C., Semenec, L., Grommen, S. V. H., et al. (2020). A pioneer calf foetus microbiome. Sci. Rep. 10:17712. doi: 10.1038/s41598-020-74677-7
Hackmann, T. J., Ngugi, D. K., Firkins, J. L., and Tao, J. (2017). Genomes of rumen bacteria encode atypical pathways for fermenting hexoses to short-chain fatty acids. Environ. Microbiol. 19, 4670–4683. doi: 10.1111/1462-2920.13929
Hackmann, T. J., and Spain, J. N. (2010). Invited review: ruminant ecology and evolution: perspectives useful to ruminant livestock research and production. J. Dairy Sci. 93, 1320–1334. doi: 10.3168/jds.2009-2071
Hansen, E. E., Lozupone, C. A., Rey, F. E., Wu, M., Guruge, J. L., Narra, A., et al. (2011). Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc. Natl. Acad. Sci. 108, 4599–4606. doi: 10.1073/pnas.1000071108
Hanson, R. W., and Ballard, F. J. (1967). The relative significance of acetate and glucose as precursors for lipid synthesis in liver and adipose tissue from ruminants. Biochem. J. 105, 529–536. doi: 10.1042/bj1050529
Haryono, M. A. S., Law, Y. Y., Arumugam, K., Liew, L. C. W., Nguyen, T. Q. N., Drautz-Moses, D. I., et al. (2022). Recovery of high quality metagenome-assembled genomes from full-scale activated sludge microbial communities in a tropical climate using longitudinal metagenome sampling. Front. Microbiol. 13:869135. doi: 10.3389/fmicb.2022.869135
Hazlewood, G., and Dawson, R. M. C. (1977). Acylgalactosylglycerols as a source of Long-Chain fatty acids for a naturally occurring rumen auxotroph. Biochem. Soc. Trans. 5, 1721–1723. doi: 10.1042/bst0051721
Hegarty, R. S. (1999). Reducing rumen methane emissions through elimination of rumen protozoa. Crop Pasture Sci. 50, 1321–1327. doi: 10.1071/AR99008
Hegarty, R. S., Goopy, J. P., Herd, R. M., and McCorkell, B. (2007). Cattle selected for lower residual feed intake have reduced daily methane production 1, 2. J. Anim. Sci. 85, 1479–1486. doi: 10.2527/jas.2006-236
Henchion, M., Moloney, A. P., Hyland, J., Zimmermann, J., and McCarthy, S. (2021). Review: trends for meat, milk and egg consumption for the next decades and the role played by livestock systems in the global production of proteins. Animal 15:100287. doi: 10.1016/j.animal.2021.100287
Henderson, G., Cox, F., Ganesh, S., Jonker, A., Young, W., Global Rumen Census Collaborators, et al. (2015). Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 5:14567. doi: 10.1038/srep14567
Henderson, G., Yilmaz, P., Kumar, S., Forster, R. J., Kelly, W. J., Leahy, S. C., et al. (2019). Improved taxonomic assignment of rumen bacterial 16S rRNA sequences using a revised SILVA taxonomic framework. PeerJ 7:e6496. doi: 10.7717/peerj.6496
Herd, R. M., and Arthur, P. F. (2009). Physiological basis for residual feed intake1. J. Anim. Sci. 87, E64–E71. doi: 10.2527/jas.2008-1345
Hernandez-Sanabria, E., Goonewardene, L. A., Wang, Z., Durunna, O. N., Moore, S. S., and Guan, L. L. (2012). Impact of feed efficiency and diet on adaptive variations in the bacterial community in the rumen fluid of cattle. Appl. Environ. Microbiol. 78, 1203–1214. doi: 10.1128/AEM.05114-11
Hill, K. J., and Mangan, J. L. (1964). The formation and distribution of methylamine in the ruminant digestive tract. Biochem. J. 93, 39–45. doi: 10.1042/bj0930039
Hillman, K., Lloyd, D., and Williams, A. G. (1988). Interactions between the methanogen Methanosarcina barkeri and rumen holotrich ciliate protozoa. Lett. Appl. Microbiol. 7, 49–53. doi: 10.1111/j.1472-765X.1988.tb01250.x
Hillmann, B., Al-Ghalith, G. A., Shields-Cutler, R. R., Zhu, Q., Gohl, D. M., Beckman, K. B., et al. (2018). Evaluating the information content of shallow shotgun metagenomics. mSystems 3:e00069-18. doi: 10.1128/mSystems.00069-18
Hoedt, E. C., Parks, D. H., Volmer, J. G., Rosewarne, C. P., Denman, S. E., McSweeney, C. S., et al. (2018). Culture- and metagenomics-enabled analyses of the Methanosphaera genus reveals their monophyletic origin and differentiation according to genome size. ISME J. 12, 2942–2953. doi: 10.1038/s41396-018-0225-7
Hormann, K., and Andreesen, J. R. (1989). Reductive cleavage of sarcosine and betaine by Eubacterium acidaminophilum via enzyme systems different from glycine reductase. Arch. Microbiol. 153, 50–59. doi: 10.1007/BF00277541
Huisingh, J., McNeill, J. J., and Matrone, G. (1974). Sulfate reduction by a desulfovibrio species isolated from sheep rumen. Appl. Microbiol. 28, 489–497. doi: 10.1128/am.28.3.489-497.1974
Hungate, R. E. (1950). The anaerobic mesophilic cellulolytic bacteria. Bacteriol. Rev. 14, 1–49. doi: 10.1128/br.14.1.1-49.1950
Hungate, R. E. (1969). “Chapter IV a roll tube method for cultivation of strict anaerobes” in Methods in microbiology. eds. J. R. Norris and D. W. Ribbons, vol. 3 (Cambridge, MA, USA: Academic Press), 117–132.
Hungate, R. E., Smith, W., Bauchop, T., Yu, I., and Rabinowitz, J. C. (1970). Formate as an intermediate in the bovine rumen fermentation. J. Bacteriol. 102, 389–397. doi: 10.1128/jb.102.2.389-397.1970
Husso, A., Lietaer, L., Pessa-Morikawa, T., Grönthal, T., Govaere, J., Van Soom, A., et al. (2021). The composition of the microbiota in the full-term fetal gut and amniotic fluid: a bovine cesarean section study. Front. Microbiol. 12:626421. doi: 10.3389/fmicb.2021.626421
Jami, E., Israel, A., Kotser, A., and Mizrahi, I. (2013). Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 7, 1069–1079. doi: 10.1038/ismej.2013.2
Jami, E., White, B. A., and Mizrahi, I. (2014). Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS One 9:e85423. doi: 10.1371/journal.pone.0085423
Janssen, P. H. (2010). Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim. Feed Sci. Technol. 160, 1–22. doi: 10.1016/j.anifeedsci.2010.07.002
Janssen, P. H., and Kirs, M. (2008). Structure of the archaeal community of the rumen. Appl. Environ. Microbiol. 74, 3619–3625. doi: 10.1128/AEM.02812-07
Jarvis, G. N., Strömpl, C., Burgess, D. M., Skillman, L. C., Moore, E. R. B., and Joblin, K. N. (2000). Isolation and identification of ruminal methanogens from grazing cattle. Curr. Microbiol. 40, 327–332. doi: 10.1007/s002849910065
Jeyanathan, J. (2010). Investigation of rumen methanogens in New Zealand livestock: a thesis presented in partial fulfillment of the requirements for the degree of doctor of philosophy in animal science. Massey University, Palmerston North, New Zealand. Doctor of Philosophy (Ph.D.) Doctoral, Massey University.
Jiang, Y., Xie, M., Chen, W., Talbot, R., Maddox, J. F., Faraut, T., et al. (2014). The sheep genome illuminates biology of the rumen and lipid metabolism. Science 344, 1168–1173. doi: 10.1126/science.1252806
Johnson, K. A., and Johnson, D. E. (1995). Methane emissions from cattle. J. Anim. Sci. 73, 2483–2492. doi: 10.2527/1995.7382483x
Johnson, J. S., Spakowicz, D. J., Hong, B.-Y., Petersen, L. M., Demkowicz, P., Chen, L., et al. (2019). Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 10:5029. doi: 10.1038/s41467-019-13036-1
Junier, T., and Zdobnov, E. M. (2010). The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 26, 1669–1670. doi: 10.1093/bioinformatics/btq243
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kelly, W. J., Leahy, S. C., Altermann, E., Yeoman, C. J., Dunne, J. C., Kong, Z., et al. (2010). The glycobiome of the rumen bacterium Butyrivibrio proteoclasticus B316T highlights adaptation to a polysaccharide-rich environment. PLoS One 5:e11942. doi: 10.1371/journal.pone.0011942
Kelly, W. J., Leahy, S. C., Kamke, J., Soni, P., Koike, S., Mackie, R., et al. (2019). Occurrence and expression of genes encoding methyl-compound production in rumen bacteria. Anim. Microbiome 1:15. doi: 10.1186/s42523-019-0016-0
Kelly, W. J., Leahy, S. C., Li, D., Perry, R., Lambie, S. C., Attwood, G. T., et al. (2014). The complete genome sequence of the rumen methanogen Methanobacterium formicicum BRM9. Stand. Genomic Sci. 9:15. doi: 10.1186/1944-3277-9-15
Kelly, W. J., Li, D., Lambie, S. C., Cox, F., Attwood, G. T., Altermann, E., et al. (2016a). Draft genome sequence of the rumen methanogen Methanobrevibacter olleyae YLM1. Genome Announc. 4:e00232-16. doi: 10.1128/genomeA.00232-16
Kelly, W. J., Li, D., Lambie, S. C., Jeyanathan, J., Cox, F., Li, Y., et al. (2016b). Complete genome sequence of methanogenic archaeon ISO4-G1, a member of the Methanomassiliicoccales, isolated from a sheep rumen. Genome Announc. 4:e00221-16. doi: 10.1128/genomeA.00221-16
Kelly, W. J., Mackie, R. I., Attwood, G. T., Janssen, P. H., McAllister, T. A., and Leahy, S. C. (2022). Hydrogen and formate production and utilisation in the rumen and the human colon. Anim. Microbiome 4:22. doi: 10.1186/s42523-022-00174-z
Kelly, W. J., Pacheco, D. M., Li, D., Attwood, G. T., Altermann, E., and Leahy, S. C. (2016c). The complete genome sequence of the rumen methanogen Methanobrevibacter millerae SM9. Stand. Genomic Sci. 11:49. doi: 10.1186/s40793-016-0171-9
Keltjens, J. T., and Vogels, G. D. (1993). “Conversion of methanol and methylamines to methane and carbon dioxide” in Methanogenesis. ed. J. G. Ferry (New York: Springer Science & Business Media), 253–303.
Khairunisa, B. H., Susanti, D., Loganathan, U., Teutsch, C. D., Campbell, B. T., Fiske, D., et al. (2022). Dominant remodelling of cattle rumen microbiome by Schedonorus arundinaceus (tall fescue) KY-31 carrying a fungal endophyte. Access Microbiol. 4:000322. doi: 10.1099/acmi.0.000322
King, E. E., Smith, R. P., St-Pierre, B., and Wright, A.-D. G. (2011). Differences in the rumen methanogen populations of lactating Jersey and Holstein dairy cows under the same diet regimen. Appl. Environ. Microbiol. 77, 5682–5687. doi: 10.1128/AEM.05130-11
Kittelmann, S., Pinares-Patiño, C. S., Seedorf, H., Kirk, M. R., Ganesh, S., McEwan, J. C., et al. (2014). Two different bacterial community types are linked with the low-methane emission trait in sheep. PLoS One 9:e103171. doi: 10.1371/journal.pone.0103171
Kong, Y., Xia, Y., Seviour, R., Forster, R., and McAllister, T. A. (2013). Biodiversity and composition of methanogenic populations in the rumen of cows fed alfalfa hay or triticale straw. FEMS Microbiol. Ecol. 84, 302–315. doi: 10.1111/1574-6941.12062
König, H., and Kandler, O. (1979). The amino acid sequence of the peptide moiety of the pseudomurein from Methanobacterium thermoautotrophicum. Arch. Microbiol. 121, 271–275. doi: 10.1007/BF00425067
Kröninger, L., Steiniger, F., Berger, S., Kraus, S., Welte, C. U., and Deppenmeier, U. (2019). Energy conservation in the gut microbe Methanomassiliicoccus luminyensisis based on membrane-bound ferredoxin oxidation coupled to heterodisulfide reduction. FEBS J. 286, 3831–3843. doi: 10.1111/febs.14948
Kuhner, C. H., Smith, S. S., Noll, K. M., Tanner, R. S., and Wolfe, R. S. (1991). 7-Mercaptoheptanoylthreonine phosphate substitutes for heat-stable factor (mobile factor) for growth of Methanomicrobium mobile. Appl. Environ. Microbiol. 57, 2891–2895. doi: 10.1128/aem.57.10.2891-2895.1991
Kulkarni, G., Mand, T. D., and Metcalf, W. W. (2018). Energy conservation via hydrogen cycling in the methanogenic archaeon Methanosarcina barkeri. MBio 9:e01256-18. doi: 10.1128/mBio.01256-18
Kurth, J. M., Nobu, M. K., Tamaki, H., de Jonge, N., Berger, S., Jetten, M. S. M., et al. (2021). Methanogenic archaea use a bacteria-like methyltransferase system to demethoxylate aromatic compounds. ISME J. 15, 3549–3565. doi: 10.1038/s41396-021-01025-6
Kurth, J. M., Op den Camp, H. J. M., and Welte, C. U. (2020). Several ways one goal—methanogenesis from unconventional substrates. Appl. Microbiol. Biotechnol. 104, 6839–6854. doi: 10.1007/s00253-020-10724-7
La Reau, A. J., Strom, N. B., Filvaroff, E., Mavrommatis, K., Ward, T. L., and Knights, D. (2023). Shallow shotgun sequencing reduces technical variation in microbiome analysis. Sci. Rep. 13:7668. doi: 10.1038/s41598-023-33489-1
Lambie, S. C., Kelly, W. J., Leahy, S. C., Li, D., Reilly, K., McAllister, T. A., et al. (2015). The complete genome sequence of the rumen methanogen Methanosarcina barkeri CM1. Stand. Genomic Sci. 10:57. doi: 10.1186/s40793-015-0038-5
Lane, M. A., and Jesse, B. W. (1997). Effect of volatile fatty acid infusion on development of the rumen epithelium in neonatal sheep. J. Dairy Sci. 80, 740–746. doi: 10.3168/jds.S0022-0302(97)75993-9
Lang, K., Schuldes, J., Klingl, A., Poehlein, A., Daniel, R., and Brune, A. (2015). New mode of energy metabolism in the seventh order of methanogens as revealed by comparative genome analysis of “Candidatus Methanoplasma termitum”. Appl. Environ. Microbiol. 81, 1338–1352. doi: 10.1128/AEM.03389-14
Leahy, S. C., Janssen, P. H., Attwood, G. T., Mackie, R. I., McAllister, T. A., and Kelly, W. J. (2022). Electron flow: key to mitigating ruminant methanogenesis. Trends Microbiol. 30, 209–212. doi: 10.1016/j.tim.2021.12.005
Leahy, S. C., Kelly, W. J., Altermann, E., Ronimus, R. S., Yeoman, C. J., Pacheco, D. M., et al. (2010). The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS One 5:e8926. doi: 10.1371/journal.pone.0008926
Leahy, S. C., Kelly, W. J., Li, D., Li, Y., Altermann, E., Lambie, S. C., et al. (2013). The complete genome sequence of Methanobrevibacter sp. AbM4. Stand. Genomic Sci. 8, 215–227. doi: 10.4056/sigs.3977691
Lee, J.-H., Kumar, S., Lee, G.-H., Chang, D.-H., Rhee, M.-S., Yoon, M.-H., et al. (2013). Methanobrevibacter boviskoreani sp. nov., isolated from the rumen of Korean native cattle. Int. J. Syst. Evol. Microbiol. 63, 4196–4201. doi: 10.1099/ijs.0.054056-0
Lefort, V., Desper, R., and Gascuel, O. (2015). FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program: Table 1. Mol. Biol. Evol. 32, 2798–2800. doi: 10.1093/molbev/msv150
Lemoine, F., Domelevo Entfellner, J. B., Wilkinson, E., Correia, D., Dávila Felipe, M., De Oliveira, T., et al. (2018). Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature 556, 452–456. doi: 10.1038/s41586-018-0043-0
Leng, R. A. (2014). Interactions between microbial consortia in biofilms: a paradigm shift in rumen microbial ecology and enteric methane mitigation. Anim. Prod. Sci. 54:519. doi: 10.1071/AN13381
Lewis, W. H., Lind, A. E., Sendra, K. M., Onsbring, H., Williams, T. A., Esteban, G. F., et al. (2020). Convergent evolution of Hydrogenosomes from mitochondria by gene transfer and loss. Mol. Biol. Evol. 37, 524–539. doi: 10.1093/molbev/msz239
Li, Y. (2016). Comparative genomics of rumen methanogens: a thesis presented in partial fulfillment of the requirements for the degree of doctor of philosophy in biochemistry at Massey University, Palmerston North, New Zealand. Doctor of Philosophy (PhD) Doctoral, Massey University.
Li, R. W., Connor, E. E., Li, C., Baldwin Vi, R. L., and Sparks, M. E. (2012). Characterization of the rumen microbiota of pre-ruminant calves using metagenomic tools. Environ. Microbiol. 14, 129–139. doi: 10.1111/j.1462-2920.2011.02543.x
Li, F., and Guan, L. L. (2017). Metatranscriptomic profiling reveals linkages between the active rumen microbiome and feed efficiency in beef cattle. Appl. Environ. Microbiol. 83:e00061-17. doi: 10.1128/AEM.00061-17
Li, F., Henderson, G., Sun, X., Cox, F., Janssen, P. H., and Guan, L. L. (2016). Taxonomic assessment of rumen microbiota using Total RNA and targeted amplicon sequencing approaches. Front. Microbiol. 7:2097. doi: 10.3389/fmicb.2016.02097
Li, Y., Leahy, S. C., Jeyanathan, J., Henderson, G., Cox, F., Altermann, E., et al. (2016). The complete genome sequence of the methanogenic archaeon ISO4-H5 provides insights into the methylotrophic lifestyle of a ruminal representative of the Methanomassiliicoccales. Stand. Genomic Sci. 11:59. doi: 10.1186/s40793-016-0183-5
Li, M., Penner, G. B., Hernandez-Sanabria, E., Oba, M., and Guan, L. L. (2009). Effects of sampling location and time, and host animal on assessment of bacterial diversity and fermentation parameters in the bovine rumen. J. Appl. Microbiol. 107, 1924–1934. doi: 10.1111/j.1365-2672.2009.04376.x
Lie, T. J., Costa, K. C., Lupa, B., Korpole, S., Whitman, W. B., and Leigh, J. A. (2012). Essential anaplerotic role for the energy-converting hydrogenase Eha in hydrogenotrophic methanogenesis. Proc. Natl. Acad. Sci. U. S. A. 109, 15473–15478. doi: 10.1073/pnas.1208779109
Lin, C., and Miller, T. L. (1998). Phylogenetic analysis of Methanobrevibacter isolated from feces of humans and other animals. Arch. Microbiol. 169, 397–403. doi: 10.1007/s002030050589
Liu, Y. (2010a). “Methanobacteriales” in Handbook of hydrocarbon and lipid microbiology. ed. K. N. Timmis (Springer, Berlin Heidelberg: Berlin/Heidelberg), 559–571.
Liu, Y. (2010b). “Methanomicrobiales” in Handbook of hydrocarbon and lipid microbiology. ed. K. N. Timmis (Springer, Berlin Heidelberg: Berlin/Heidelberg), 583–593.
Liu, Y., and Whitman, W. B. (2008). Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N. Y. Acad. Sci. 1125, 171–189. doi: 10.1196/annals.1419.019
Loh, Z. H., Ouwerkerk, D., Klieve, A. V., Hungerford, N. L., and Fletcher, M. T. (2020). Toxin degradation by rumen microorganisms: a review. Toxins 12:664. doi: 10.3390/toxins12100664
Lopes, D. R. G., de Souza Duarte, M., La Reau, A. J., Chaves, I. Z., de Oliveira Mendes, T. A., Detmann, E., et al. (2021). Assessing the relationship between the rumen microbiota and feed efficiency in Nellore steers. J. Anim. Sci. Biotechnol. 12:79. doi: 10.1186/s40104-021-00599-7
López-García, A., Saborío-Montero, A., Gutiérrez-Rivas, M., Atxaerandio, R., Goiri, I., García-Rodríguez, A., et al. (2022). Fungal and ciliate protozoa are the main rumen microbes associated with methane emissions in dairy cattle. GigaScience 11:giab088. doi: 10.1093/gigascience/giab088
Lovley, D. R., Greening, R. C., and Ferry, J. G. (1984). Rapidly growing rumen methanogenic organism that synthesizes coenzyme M and has a high affinity for formate. Appl. Environ. Microbiol. 48, 81–87. doi: 10.1128/aem.48.1.81-87.1984
Lovley, D. R., and Holmes, D. E. (2022). Electromicrobiology: the ecophysiology of phylogenetically diverse electroactive microorganisms. Nat. Rev. Microbiol. 20, 5–19. doi: 10.1038/s41579-021-00597-6
Lurie-Weinberger, M. N., Peeri, M., and Gophna, U. (2012). Contribution of lateral gene transfer to the gene repertoire of a gut-adapted methanogen. Genomics 99, 52–58. doi: 10.1016/j.ygeno.2011.10.005
Luton, P. E., Wayne, J. M., Sharp, R. J., and Riley, P. W. (2002). The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148, 3521–3530. doi: 10.1099/00221287-148-11-3521
Lyu, Z., and Liu, Y. (2019). “Diversity and taxonomy of methanogens” in Biogenesis of hydrocarbons. eds. A. Stams and D. Sousa (Cham: Springer), 19–77.
Ma, Z., Wang, R., Wang, M., Zhang, X., Mao, H., and Tan, Z. (2018). Short communication: variability in fermentation end-products and methanogen communities in different rumen sites of dairy cows. J. Dairy Sci. 101, 5153–5158. doi: 10.3168/jds.2017-14096
Maeder, D. L., Anderson, I., Brettin, T. S., Bruce, D. C., Gilna, P., Han, C. S., et al. (2006). The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within methanosarcinal genomes. J. Bacteriol. 188, 7922–7931. doi: 10.1128/JB.00810-06
Malheiros, J. M., Correia, B. S. B., Ceribeli, C., Cardoso, D. R., Colnago, L. A., Junior, S. B., et al. (2021). Comparative untargeted metabolome analysis of ruminal fluid and feces of Nelore steers (Bos indicus). Sci. Rep. 11:12752. doi: 10.1038/s41598-021-92179-y
Mand, T. D., and Metcalf, W. W. (2019). Energy conservation and hydrogenase function in methanogenic archaea, in particular the genus Methanosarcina. Microbiol. Mol. Biol. Rev. 83:e00020-19. doi: 10.1128/MMBR.00020-19
Mann, E., Wetzels, S. U., Wagner, M., Zebeli, Q., and Schmitz-Esser, S. (2018). Metatranscriptome sequencing reveals insights into the gene expression and functional potential of Rumen Wall Bacteria. Front. Microbiol. 9:43. doi: 10.3389/fmicb.2018.00043
Martínez-Álvaro, M., Auffret, M. D., Duthie, C.-A., Dewhurst, R. J., Cleveland, M. A., Watson, M., et al. (2022). Bovine host genome acts on rumen microbiome function linked to methane emissions. Commun. Biol. 5:350. doi: 10.1038/s42003-022-03293-0
Martínez-Álvaro, M., Auffret, M. D., Stewart, R. D., Dewhurst, R. J., Duthie, C.-A., Rooke, J. A., et al. (2020). Identification of complex rumen microbiome interaction within diverse functional niches as mechanisms affecting the variation of methane emissions in bovine. Front. Microbiol. 11:659. doi: 10.3389/fmicb.2020.00659
Matsuo, Y., Komiya, S., Yasumizu, Y., Yasuoka, Y., Mizushima, K., Takagi, T., et al. (2021). Full-length 16S rRNA gene amplicon analysis of human gut microbiota using MinION™ nanopore sequencing confers species-level resolution. BMC Microbiol. 21:35. doi: 10.1186/s12866-021-02094-5
May, H. D., Schauer, N. L., and Ferry, J. G. (1986). Molybdopterin cofactor from Methanobacterium formicicum formate dehydrogenase. J. Bacteriol. 166, 500–504. doi: 10.1128/jb.166.2.500-504.1986
McCann, J. C., Wiley, L. M., Forbes, T. D., Rouquette, F. M. Jr., and Tedeschi, L. O. (2014). Relationship between the rumen microbiome and residual feed intake-efficiency of Brahman bulls stocked on bermudagrass pastures. PLoS One 9:e91864. doi: 10.1371/journal.pone.0091864
McIlroy, S. J., Kirkegaard, R. H., McIlroy, B., Nierychlo, M., Kristensen, J. M., Karst, S. M., et al. (2017). MiDAS 2.0: an ecosystem-specific taxonomy and online database for the organisms of wastewater treatment systems expanded for anaerobic digester groups. Database 2017:bax016. doi: 10.1093/database/bax016
Meale, S. J., Popova, M., Saro, C., Martin, C., Bernard, A., Lagree, M., et al. (2021). Early life dietary intervention in dairy calves results in a long-term reduction in methane emissions. Sci. Rep. 11:3003. doi: 10.1038/s41598-021-82084-9
Midla, L. T., Hoblet, K. H., Weiss, W. P., and Moeschberger, M. L. (1998). Supplemental dietary biotin for prevention of lesions associated with aseptic subclinical laminitis (pododermatitis aseptica diffusa) in primiparous cows. Am. J. Vet. Res. 59, 733–738.
Mikaelyan, A., Köhler, T., Lampert, N., Rohland, J., Boga, H., Meuser, K., et al. (2015). Classifying the bacterial gut microbiota of termites and cockroaches: a curated phylogenetic reference database (DictDb). Syst. Appl. Microbiol. 38, 472–482. doi: 10.1016/j.syapm.2015.07.004
Miller, T. L., and Lin, C. (2002). Description of Methanobrevibacter gottschalkii sp. nov., Methanobrevibacter thaueri sp. nov., Methanobrevibacter woesei sp. nov. and Methanobrevibacter wolinii sp. nov. Int. J. Syst. Evol. Microbiol. 52, 819–822. doi: 10.1099/00207713-52-3-819
Miller, T. L., and Wolin, M. J. (1981). Fermentation by the human large intestine microbial community in an in vitro semicontinuous culture system. Appl. Environ. Microbiol. 42, 400–407. doi: 10.1128/aem.42.3.400-407.1981
Miller, T. L., and Wolin, M. J. (1985). Methanosphaera stadtmaniae gen. nov., sp. nov.: a species that forms methane by reducing methanol with hydrogen. Arch. Microbiol. 141, 116–122. doi: 10.1007/BF00423270
Miller, T. L., Wolin, M. J., de Macario, E. C., and Macario, A. J. (1982). Isolation of Methanobrevibacter smithii from human feces. Appl. Environ. Microbiol. 43, 227–232. doi: 10.1128/aem.43.1.227-232.1982
Mitchell, A. D., Chappell, A., and Knox, K. L. (1979). Metabolism of betaine in the ruminant. J. Anim. Sci. 49, 764–774. doi: 10.2527/jas1979.493764x
Moon, J., Dönig, J., Kramer, S., Poehlein, A., Daniel, R., and Müller, V. (2021). Formate metabolism in the acetogenic bacterium Acetobacterium woodii. Environ. Microbiol. 23, 4214–4227. doi: 10.1111/1462-2920.15598
Moraïs, S., and Mizrahi, I. (2019). The road not taken: the rumen microbiome, functional groups, and community states. Trends Microbiol. 27, 538–549. doi: 10.1016/j.tim.2018.12.011
Moran, J. (2005). Tropical dairy farming: feeding management for small holder dairy farmers in the humid tropics. Collingwood: Landlinks Press.
Morgan, R. M., Pihl, T. D., Nölling, J., and Reeve, J. N. (1997). Hydrogen regulation of growth, growth yields, and methane gene transcription in Methanobacterium thermoautotrophicum ΔH. J. Bacteriol. 179, 889–898. doi: 10.1128/jb.179.3.889-898.1997
Morgavi, D. P., Jouany, J.-P., and Martin, C. (2008). Changes in methane emission and rumen fermentation parameters induced by refaunation in sheep. Aust. J. Exp. Agric. 48, 69–72. doi: 10.1071/EA07236
Morgavi, D. P., Martin, C., Jouany, J. P., and Ranilla, M. J. (2012). Rumen protozoa and methanogenesis: not a simple cause-effect relationship. Br. J. Nutr. 107, 388–397. doi: 10.1017/S0007114511002935
Morvan, B., Dore, J., Rieu-Lesme, F., Foucat, L., Fonty, G., and Gouet, P. (1994). Establishment of hydrogen-utilizing bacteria in the rumen of the newborn lamb. FEMS Microbiol. Lett. 117, 249–256. doi: 10.1111/j.1574-6968.1994.tb06775.x
Mountfort, D. O., Asher, R. A., and Bauchop, T. (1982). Fermentation of cellulose to methane and carbon dioxide by a rumen anaerobic fungus in a Triculture with Methanobrevibacter sp. strain RA1 and Methanosarcina barkeri. Appl. Environ. Microbiol. 44, 128–134. doi: 10.1128/aem.44.1.128-134.1982
Mukhopadhyay, B., Purwantini, E., and Daniels, L. (1993). Effect of methanogenic substrates on coenzyme F420-dependent N5,N10-methylene-H4MPT dehydrogenase, N5,N10-methenyl-H4MPT cyclohydrolase and F420-reducing hydrogenase activities in Methanosarcina barkeri. Arch. Microbiol. 159, 141–146. doi: 10.1007/BF00250274
Mukhopadhyay, B., Purwantini, E., Pihl, T. D., Reeve, J. N., and Daniels, L. (1995). Cloning, sequencing, and transcriptional analysis of the coenzyme F420-dependent methylene-5,6,7,8-tetrahydromethanopterin dehydrogenase gene from Methanobacterium thermoautotrophicum strain marburg and functional expression in Escherichia coli. J. Biol. Chem. 270, 2827–2832. doi: 10.1074/jbc.270.6.2827
Muro-Reyes, A., Gutierrez, H., Diaz-Garci, L. H., Gutierrez, F. J., Escareno-S, L. M., Banuelos-V, R., et al. (2011). Potential environmental benefits of residual feed intake as strategy to mitigate methane emissions in sheep. J. Anim. Vet. Adv. 10, 1551–1556. doi: 10.3923/javaa.2011.1551.1556
Naumann, E., Hippe, H., and Gottschalk, G. (1983). Betaine: new oxidant in the stickland reaction and methanogenesis from betaine and l-alanine by a Clostridium sporogenes-Methanosarcina barkeri coculture. Appl. Environ. Microbiol. 45, 474–483. doi: 10.1128/aem.45.2.474-483.1983
Nayfach, S., Shi, Z. J., Seshadri, R., Pollard, K. S., and Kyrpides, N. C. (2019). New insights from uncultivated genomes of the global human gut microbiome. Nature 568, 505–510. doi: 10.1038/s41586-019-1058-x
Newton, I. L. G., and Roeselers, G. (2012). The effect of training set on the classification of honey bee gut microbiota using the Naïve Bayesian classifier. BMC Microbiol. 12:221. doi: 10.1186/1471-2180-12-221
Ng, F., Kittelmann, S., Patchett, M. L., Attwood, G. T., Janssen, P. H., Rakonjac, J., et al. (2016). An adhesin from hydrogen-utilizing rumen methanogen Methanobrevibacter ruminantium M1 binds a broad range of hydrogen-producing microorganisms. Environ. Microbiol. 18, 3010–3021. doi: 10.1111/1462-2920.13155
Nkrumah, J. D., Okine, E. K., Mathison, G. W., Schmid, K., Li, C., Basarab, J. A., et al. (2006). Relationships of feedlot feed efficiency, performance, and feeding behavior with metabolic rate, methane production, and energy partitioning in beef cattle1. J. Anim. Sci. 84, 145–153. doi: 10.2527/2006.841145x
Nölling, J., and Reeve, J. N. (1997). Growth- and substrate-dependent transcription of the formate dehydrogenase (fdhCAB) operon in Methanobacterium thermoformicicum Z-245. J. Bacteriol. 179, 899–908. doi: 10.1128/jb.179.3.899-908.1997
Pace, N. R. (1997). A molecular view of microbial diversity and the biosphere. Science 276, 734–740. doi: 10.1126/science.276.5313.734
Parks, D. H., Chuvochina, M., Rinke, C., Mussig, A. J., Chaumeil, P.-A., and Hugenholtz, P. (2022). GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 50, D785–D794. doi: 10.1093/nar/gkab776
Patra, A., Park, T., Kim, M., and Yu, Z. (2017). Rumen methanogens and mitigation of methane emission by anti-methanogenic compounds and substances. J. Anim. Sci. Biotechnol. 8:13. doi: 10.1186/s40104-017-0145-9
Patterson, J. A., and Hespell, R. B. (1979). Trimethylamine and methylamine as growth substrates for rumen bacteria and Methanosarcina barkeri. Curr. Microbiol. 3, 79–83. doi: 10.1007/BF02602436
Paynter, M. J. B., and Hungate, R. E. (1968). Characterization of Methanobacterium mobilis, sp. n., isolated from the bovine rumen. J. Bacteriol. 95, 1943–1951. doi: 10.1128/jb.95.5.1943-1951.1968
Pereira, A. M., de Lurdes Nunes, M., Dapkevicius, E., and Borba, A. E. S. (2022). Alternative pathways for hydrogen sink originated from the ruminal fermentation of carbohydrates: which microorganisms are involved in lowering methane emission? Anim. Microbiome 4:5. doi: 10.1186/s42523-021-00153-w
Pitta, D. W., Indugu, N., Melgar, A., Hristov, A., Challa, K., Vecchiarelli, B., et al. (2022). The effect of 3-nitrooxypropanol, a potent methane inhibitor, on ruminal microbial gene expression profiles in dairy cows. Microbiome 10:146. doi: 10.1186/s40168-022-01341-9
Pitta, D. W., Melgar, A., Hristov, A. N., Indugu, N., Narayan, K. S., Pappalardo, C., et al. (2021). Temporal changes in total and metabolically active ruminal methanogens in dairy cows supplemented with 3-nitrooxypropanol. J. Dairy Sci. 104, 8721–8735. doi: 10.3168/jds.2020-19862
Pitta, D. W., Pinchak, W. E., Dowd, S., Dorton, K., Yoon, I., Min, B. R., et al. (2014). Longitudinal shifts in bacterial diversity and fermentation pattern in the rumen of steers grazing wheat pasture. Anaerobe 30, 11–17. doi: 10.1016/j.anaerobe.2014.07.008
Pol, A., and Demeyer, D. I. (1988). Fermentation of methanol in the sheep rumen. Appl. Environ. Microbiol. 54, 832–834. doi: 10.1128/aem.54.3.832-834.1988
Prins, R. A., and van der Meer, P. (1976). On the contribution of the acrylate pathway to the formation of propionate from lactate in the rumen of cattle. Antonie Van Leeuwenhoek 42, 25–31. doi: 10.1007/BF00399446
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ragsdale, S. W. (2003). Pyruvate ferredoxin oxidoreductase and its radical intermediate. Chem. Rev. 103, 2333–2346. doi: 10.1021/cr020423e
Ramanathan, V., Cicerone, R. J., Singh, H. B., and Kiehl, J. T. (1985). Trace gas trends and their potential role in climate change. J. Geophys. Res. Atmos. 90, 5547–5566. doi: 10.1029/JD090iD03p05547
Rath, S., Rud, T., Pieper, D. H., and Vital, M. (2019). Potential TMA-producing bacteria are ubiquitously found in mammalia. Front. Microbiol. 10:2966. doi: 10.3389/fmicb.2019.02966
Rea, S., Bowman, J. P., Popovski, S., Pimm, C., and Wright, A.-D. G. (2007). Methanobrevibacter millerae sp. nov. and Methanobrevibacter olleyae sp. nov., methanogens from the ovine and bovine rumen that can utilize formate for growth. Int. J. Syst. Evol. Microbiol. 57, 450–456. doi: 10.1099/ijs.0.63984-0
Reeve, J. N. (1992). Molecular biology of methanogens. Annu. Rev. Microbiol. 46, 165–191. doi: 10.1146/annurev.mi.46.100192.001121
Reeve, J. N. (1993). “Structure and organization of genes” in Methanogenesis: ecology, physiology, biochemistry, and genetics. ed. J. G. Ferry (New York: Chapman and Hall), 493–527.
Reeve, J. N., Nölling, J., Morgan, R. M., and Smith, D. R. (1997). Methanogenesis: genes, genomes, and who's on first? J. Bacteriol. 179, 5975–5986. doi: 10.1128/jb.179.19.5975-5986.1997
Regensbogenova, M., McEwan, N. R., Javorsky, P., Kisidayova, S., Michalowski, T., Newbold, C. J., et al. (2004). A re-appraisal of the diversity of the methanogens associated with the rumen ciliates. FEMS Microbiol. Lett. 238, 307–313. doi: 10.1111/j.1574-6968.2004.tb09771.x
Reichardt, N., Duncan, S. H., Young, P., Belenguer, A., McWilliam Leitch, C., Scott, K. P., et al. (2014). Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 8, 1323–1335. doi: 10.1038/ismej.2014.14
Ritari, J., Salojärvi, J., Lahti, L., and de Vos, W. M. (2015). Improved taxonomic assignment of human intestinal 16S rRNA sequences by a dedicated reference database. BMC Genomics 16:1056. doi: 10.1186/s12864-015-2265-y
Roehe, R., Dewhurst, R. J., Duthie, C.-A., Rooke, J. A., McKain, N., Ross, D. W., et al. (2016). Bovine host genetic variation influences rumen microbial methane production with best selection criterion for low methane emitting and efficiently feed converting hosts based on metagenomic gene abundance. PLoS Genet. 12:e1005846. doi: 10.1371/journal.pgen.1005846
Rohwer, R. R., Hamilton, J. J., Newton, R. J., and McMahon, K. D. (2018). TaxAss: leveraging a custom freshwater database achieves fine-scale taxonomic resolution. mSphere 3:e00327-18. doi: 10.1128/mSphere.00327-18
Rooke, J. A., Wallace, R. J., Duthie, C.-A., McKain, N., de Souza, S. M., Hyslop, J. J., et al. (2014). Hydrogen and methane emissions from beef cattle and their rumen microbial community vary with diet, time after feeding and genotype. Br. J. Nutr. 112, 398–407. doi: 10.1017/S0007114514000932
Rospert, S., Linder, D., Ellermann, J., and Thauer, R. K. (1990). Two genetically distinct methyl-coenzyme M reductases in Methanobacterium thermoautotrophicum strain Marburg and ΔH. Eur. J. Biochem. 194, 871–877. doi: 10.1111/j.1432-1033.1990.tb19481.x
Rouvière, P. E., and Wolfe, R. S. (1988). Novel biochemistry of methanogenesis. J. Biol. Chem. 263, 7913–7916. doi: 10.1016/S0021-9258(18)68417-0
Sakai, S., Takaki, Y., Shimamura, S., Sekine, M., Tajima, T., Kosugi, H., et al. (2011). Genome sequence of a mesophilic hydrogenotrophic methanogen Methanocella paludicola, the first cultivated representative of the order Methanocellales. PLoS One 6:e22898. doi: 10.1371/journal.pone.0022898
Saleem, F., Bouatra, S., Guo, A. C., Psychogios, N., Mandal, R., Dunn, S. M., et al. (2012). The bovine ruminal fluid metabolome. Metabolomics 9, 360–378. doi: 10.1007/s11306-012-0458-9
Samuel, B. S., Hansen, E. E., Manchester, J. K., Coutinho, P. M., Henrissat, B., Fulton, R., et al. (2007). Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc. Natl. Acad. Sci. 104, 10643–10648. doi: 10.1073/pnas.0704189104
Santoro, N., and Konisky, J. (1987). Characterization of bromoethanesulfonate-resistant mutants of Methanococcus voltae: evidence of a coenzyme M transport system. J. Bacteriol. 169, 660–665. doi: 10.1128/jb.169.2.660-665.1987
Sasson, G., Ben-Shabat, S. K., Seroussi, E., Doron-Faigenboim, A., Shterzer, N., Yaacoby, S., et al. (2017). Heritable bovine rumen bacteria are phylogenetically related and correlated with the cow’s capacity to harvest energy from its feed. mBio 8:e00703-17. doi: 10.1128/mBio.00703-17
Sawers, R. G., and Clark, D. P. (2004). Fermentative pyruvate and acetyl-coenzyme a metabolism. EcoSal Plus 1. doi: 10.1128/ecosalplus.3.5.3
Schauer, N. L., and Ferry, J. G. (1982). Properties of formate dehydrogenase in Methanobacterium formicicum. J. Bacteriol. 150, 1–7. doi: 10.1128/jb.150.1.1-7.1982
Schink, B., Montag, D., Keller, A., and Muller, N. (2017). Hydrogen or formate: alternative key players in methanogenic degradation. Environ. Microbiol. Rep. 9, 189–202. doi: 10.1111/1758-2229.12524
Schloss, P. D., and Westcott, S. L. (2011). Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 77, 3219–3226. doi: 10.1128/AEM.02810-10
Schmitz, R. A., Albracht, S. P., and Thauer, R. K. (1992). A molybdenum and a tungsten isoenzyme of formylmethanofuran dehydrogenase in the thermophilic archaeon Methanobacterium wolfei. Eur. J. Biochem. 209, 1013–1018. doi: 10.1111/j.1432-1033.1992.tb17376.x
Schut, G. J., and Adams, M. W. W. (2009). The Iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J. Bacteriol. 191, 4451–4457. doi: 10.1128/JB.01582-08
Seedorf, H., Kittelmann, S., Henderson, G., and Janssen, P. H. (2014). RIM-DB: a taxonomic framework for community structure analysis of methanogenic archaea from the rumen and other intestinal environments. PeerJ 2:e494. doi: 10.7717/peerj.494
Seedorf, H., Kittelmann, S., and Janssen, P. H. (2015). Few highly abundant operational taxonomic units dominate within rumen methanogenic archaeal species in New Zealand sheep and cattle. Appl. Environ. Microbiol. 81, 986–995. doi: 10.1128/AEM.03018-14
Seshadri, R., Leahy, S. C., Attwood, G. T., Teh, K. H., Lambie, S. C., Cookson, A. L., et al. (2018). Cultivation and sequencing of rumen microbiome members from the Hungate1000 collection. Nat. Biotechnol. 36, 359–367. doi: 10.1038/nbt.4110
Shakya, M., Lo, C.-C., and Chain, P. S. G. (2019). Advances and challenges in metatranscriptomic analysis. Front. Genet. 10:904. doi: 10.3389/fgene.2019.00904
Shi, W., Moon, C. D., Leahy, S. C., Kang, D., Froula, J., Kittelmann, S., et al. (2014). Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res. 24, 1517–1525. doi: 10.1101/gr.168245.113
Shieh, J., Mesbah, M., and Whitman, W. B. (1988). Pseudoauxotrophy of Methanococcus voltae for acetate, leucine, and isoleucine. J. Bacteriol. 170, 4091–4096. doi: 10.1128/jb.170.9.4091-4096.1988
Shterzer, N., and Mizrahi, I. (2015). The animal gut as a melting pot for horizontal gene transfer. Can. J. Microbiol. 61, 603–605. doi: 10.1139/cjm-2015-0049
Smith, P. H., and Hungate, R. E. (1958). Isolation and characterization of Methanobacterium ruminantium n. sp. J. Bacteriol. 75, 713–718. doi: 10.1128/jb.75.6.713-718.1958
Smith, K. S., and Ingram-Smith, C. (2007). Methanosaeta, the forgotten methanogen? Trends Microbiol. 15, 150–155. doi: 10.1016/j.tim.2007.02.002
Snelling, T. J., Genç, B., McKain, N., Watson, M., Waters, S. M., Creevey, C. J., et al. (2014). Diversity and community composition of methanogenic archaea in the rumen of Scottish upland sheep assessed by different methods. PLoS One 9:e106491. doi: 10.1371/journal.pone.0106491
Solden, L. M., Naas, A. E., Roux, S., Daly, R. A., Collins, W. B., Nicora, C. D., et al. (2018). Interspecies cross-feeding orchestrates carbon degradation in the rumen ecosystem. Nat. Microbiol. 3, 1274–1284. doi: 10.1038/s41564-018-0225-4
Söllinger, A., Schwab, C., Weinmaier, T., Loy, A., Tveit, A. T., Schleper, C., et al. (2016). Phylogenetic and genomic analysis of Methanomassiliicoccales in wetlands and animal intestinal tracts reveals clade-specific habitat preferences. FEMS Microbiol. Ecol. 92:fiv149. doi: 10.1093/femsec/fiv149
Sollinger, A., Tveit, A. T., Poulsen, M., Noel, S. J., Bengtsson, M., Bernhardt, J., et al. (2018). Holistic assessment of rumen microbiome dynamics through quantitative metatranscriptomics reveals multifunctional redundancy during key steps of anaerobic feed degradation. mSystems 3:e00038-18. doi: 10.1128/mSystems.00038-18
Söllinger, A., and Urich, T. (2019). Methylotrophic methanogens everywhere — physiology and ecology of novel players in global methane cycling. Biochem. Soc. Trans. 47, 1895–1907. doi: 10.1042/BST20180565
Sprott, G. D., and Beveridge, T. J. (1993). “Microscopy” in Methanogenesis. ed. J. G. Ferry, Chapman & Hall Microbiology Series (Boston, MA: Springer), 81–127.
Stams, A. J., and Plugge, C. M. (2009). Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat. Rev. Microbiol. 7, 568–577. doi: 10.1038/nrmicro2166
Stepanchenko, N., Stefenoni, H., Hennessy, M., Nagaraju, I., Wasson, D. E., Cueva, S. F., et al. (2023). Microbial composition, rumen fermentation parameters, enteric methane emissions, and lactational performance of phenotypically high and low methane-emitting dairy cows. J. Dairy Sci. 106, 6146–6170. doi: 10.3168/jds.2022-23190
Stewart, R. D., Auffret, M. D., Warr, A., Walker, A. W., Roehe, R., and Watson, M. (2019). Compendium of 4,941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery. Nat. Biotechnol. 37, 953–961. doi: 10.1038/s41587-019-0202-3
Stewart, R. D., Auffret, M. D., Warr, A., Wiser, A. H., Press, M. O., Langford, K. W., et al. (2018). Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen. Nat. Commun. 9:870. doi: 10.1038/s41467-018-03317-6
Stobo, I. J., Roy, J. H., and Gaston, H. J. (1966). Rumen development in the calf. 1. The effect of diets containing different proportions of concentrates to hay on rumen development. Br. J. Nutr. 20, 171–188. doi: 10.1079/BJN19660021
Stothart, M. R., McLoughlin, P. D., and Poissant, J. (2022). Shallow shotgun sequencing of the microbiome recapitulates 16S amplicon results and provides functional insights. Mol. Ecol. Resour. 23, 549–564. doi: 10.1111/1755-0998.13713
St-Pierre, B., Cersosimo, L. M., Ishaq, S. L., and Wright, A.-D. G. (2015). Toward the identification of methanogenic archaeal groups as targets of methane mitigation in livestock animals. Front. Microbiol. 6:776. doi: 10.3389/fmicb.2015.00776
Stumm, C. K., and Zwart, K. B. (1986). Symbiosis of protozoa with hydrogen-utilizing methanogens. Microbiol. Sci. 3, 100–105.
Susanti, D., Wong, J. H., Vensel, W. H., Loganathan, U., DeSantis, R., Schmitz, R. A., et al. (2014). Thioredoxin targets fundamental processes in a methane-producing archaeon, Methanocaldococcus jannaschii. Proc. Natl. Acad. Sci. 111, 2608–2613. doi: 10.1073/pnas.1324240111
Tajima, K., Aminov, R. I., Nagamine, T., Matsui, H., Nakamura, M., and Benno, Y. (2001a). Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl. Environ. Microbiol. 67, 2766–2774. doi: 10.1128/AEM.67.6.2766-2774.2001
Tajima, K., Nagamine, T., Matsui, H., Nakamura, M., and Aminov, R. I. (2001b). Phylogenetic analysis of archaeal 16S rRNA libraries from the rumen suggests the existence of a novel group of archaea not associated with known methanogens. FEMS Microbiol. Lett. 200, 67–72. doi: 10.1111/j.1574-6968.2001.tb10694.x
Tamate, H., McGilliard, A. D., Jacobson, N. L., and Getty, R. (1962). Effect of various dietaries on the anatomical development of the stomach in the Calf1. J. Dairy Sci. 45, 408–420. doi: 10.3168/jds.S0022-0302(62)89406-5
Tan, R. S. G., Zhou, M., Li, F., and Guan, L. L. (2021). Identifying active rumen epithelial associated bacteria and archaea in beef cattle divergent in feed efficiency using total RNA-seq. Curr. Res. Microb. Sci. 2:100064. doi: 10.1016/j.crmicr.2021.100064
Tangherlini, M., Miralto, M., Colantuono, C., Sangiovanni, M., Anno, A. D.’., Corinaldesi, C., et al. (2018). GLOSSary: the GLobal Ocean 16S subunit web accessible resource. BMC Bioinformatics 19:443. doi: 10.1186/s12859-018-2423-8
Tanner, R. S., and Wolfe, R. S. (1988). Nutritional requirements of Methanomicrobium mobile. Appl. Environ. Microbiol. 54, 625–628. doi: 10.1128/aem.54.3.625-628.1988
Tatsuoka, N., Mohammed, N., Mitsumori, M., Hara, K., Kurihara, M., and Itabashi, H. (2004). Phylogenetic analysis of methyl coenzyme-M reductase detected from the bovine rumen. Lett. Appl. Microbiol. 39, 257–260. doi: 10.1111/j.1472-765X.2004.01566.x
Tavendale, M. H., Meagher, L. P., Pacheco, D., Walker, N., Attwood, G. T., and Sivakumaran, S. (2005). Methane production from in vitro rumen incubations with Lotus pedunculatus and Medicago sativa, and effects of extractable condensed tannin fractions on methanogenesis. Anim. Feed Sci. Technol. 123-124, 403–419. doi: 10.1016/j.anifeedsci.2005.04.037
Teske, A., Dhillon, A., and Sogin, M. L. (2003). Genomic markers of ancient anaerobic microbial pathways: sulfate reduction, methanogenesis, and methane oxidation. Biol. Bull. 204, 186–191. doi: 10.2307/1543556
Thauer, R. K. (2012). The Wolfe cycle comes full circle. Proc. Natl. Acad. Sci. 109, 15084–15085. doi: 10.1073/pnas.1213193109
Thauer, R. K., Jungermann, K., and Decker, K. (1977). Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41, 100–180. doi: 10.1128/br.41.1.100-180.1977
Thauer, R. K., Kaster, A.-K., Goenrich, M., Schick, M., Hiromoto, T., and Shima, S. (2010). Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu. Rev. Biochem. 79, 507–536. doi: 10.1146/annurev.biochem.030508.152103
Thauer, R. K., Kaster, A.-K., Seedorf, H., Buckel, W., and Hedderich, R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591. doi: 10.1038/nrmicro1931
Thiele, J. H., and Zeikus, J. G. (1988). Control of interspecies electron flow during anaerobic digestion: significance of formate transfer versus hydrogen transfer during syntrophic methanogenesis in flocs. Appl. Environ. Microbiol. 54, 20–29. doi: 10.1128/aem.54.1.20-29.1988
Thomas, C. M., Taib, N., Gribaldo, S., and Borrel, G. (2021). Comparative genomic analysis of Methanimicrococcus blatticola provides insights into host adaptation in archaea and the evolution of methanogenesis. ISME Commun. 1:47. doi: 10.1038/s43705-021-00050-y
Tymensen, L. D., Beauchemin, K. A., and McAllister, T. A. (2012). Structures of free-living and protozoa-associated methanogen communities in the bovine rumen differ according to comparative analysis of 16S rRNA and mcrA genes. Microbiology 158, 1808–1817. doi: 10.1099/mic.0.057984-0
Tymensen, L. D., and McAllister, T. A. (2012). Community structure analysis of methanogens associated with rumen protozoa reveals bias in universal archaeal primers. Appl. Environ. Microbiol. 78, 4051–4056. doi: 10.1128/AEM.07994-11
Tyson, G. W., Chapman, J., Hugenholtz, P., Allen, E. E., Ram, R. J., Richardson, P. M., et al. (2004). Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428, 37–43. doi: 10.1038/nature02340
Ungerfeld, E. M. (2020). Metabolic hydrogen flows in rumen fermentation: principles and possibilities of interventions. Front. Microbiol. 11:589. doi: 10.3389/fmicb.2020.00589
Ungerfeld, E. M., and Kohn, R. A. (2006). “The role of thermodynamics in the control of ruminal fermentation” in Ruminant physiology: digestion, metabolism and impact of nutrition on gene expression, immunology and stress. eds. K. Sejrsen, T. Hvelplund, and M. O. Nielsen (Netherlands: Wageningen Academic Publishers)
Ushida, K., Newbold, C. J., and Jouany, J.-P. (1997). Interspecies hydrogen transfer between the rumen ciliate Polyplastron multivesiculatum and Methanosarcina barkeri. J. Gen. Appl. Microbiol. 43, 129–131. doi: 10.2323/jgam.43.129
van Cleef, F. O. S., Dubeux, J. C. B., Naumann, H. D., Santos, E. R. S., Sollenberger, L. E., Vendramini, J. M. B., et al. (2021). Methane emissions and delta13C composition from beef steers consuming increasing proportions of sericea lespedeza hay on bermudagrass hay diets. J. Anim. Sci. 99:skab224. doi: 10.1093/jas/skab224
van Cleef, F. O. S., Dubeux, J. C. B., Wheeler, C. S., V García, C. C., Ruiz-Moreno, M., and Sollenberger, L. E. (2022). Stable isotopes provide evidence that condensed tannins from Sericea lespedeza are degraded by ruminal microbes. Sci. Rep. 12:14318. doi: 10.1038/s41598-022-18566-1
van Lingen, H. J., Edwards, J. E., Vaidya, J. D., van Gastelen, S., Saccenti, E., van den Bogert, B., et al. (2017). Diurnal dynamics of gaseous and dissolved metabolites and microbiota composition in the bovine rumen. Front. Microbiol. 8:425. doi: 10.3389/fmicb.2017.00425
van Lingen, H. J., Plugge, C. M., Fadel, J. G., Kebreab, E., Bannink, A., and Dijkstra, J. (2016). Thermodynamic driving force of hydrogen on rumen microbial metabolism: a theoretical investigation. PLoS One 11:e0161362. doi: 10.1371/journal.pone.0161362
Ver Eecke, H. C., Butterfield, D. A., Huber, J. A., Lilley, M. D., Olson, E. J., Roe, K. K., et al. (2012). Hydrogen-limited growth of hyperthermophilic methanogens at deep-sea hydrothermal vents. Proc. Natl. Acad. Sci. 109, 13674–13679. doi: 10.1073/pnas.1206632109
Vogels, G. D., Hoppe, W. F., and Stumm, C. K. (1980). Association of methanogenic bacteria with rumen ciliates. Appl. Environ. Microbiol. 40, 608–612. doi: 10.1128/aem.40.3.608-612.1980
Waghorn, G. C., and Hegarty, R. S. (2011). Lowering ruminant methane emissions through improved feed conversion efficiency. Anim. Feed Sci. Technol. 166-167, 291–301. doi: 10.1016/j.anifeedsci.2011.04.019
Wallace, R. J., Rooke, J. A., McKain, N., Duthie, C. A., Hyslop, J. J., Ross, D. W., et al. (2015). The rumen microbial metagenome associated with high methane production in cattle. BMC Genomics 16:839. doi: 10.1186/s12864-015-2032-0
Walter, J., and Ley, R. (2011). The human gut microbiome: ecology and recent evolutionary changes. Annu. Rev. Microbiol. 65, 411–429. doi: 10.1146/annurev-micro-090110-102830
Watanabe, T., Pfeil-Gardiner, O., Kahnt, J., Koch, J., Shima, S., and Murphy, B. J. (2021). Three-megadalton complex of methanogenic electron-bifurcating and CO(2)-fixing enzymes. Science 373, 1151–1156. doi: 10.1126/science.abg5550
Webb, S. D., and Taylor, B. E. (1980). The phylogeny of hornless ruminants and a description of the cranium of Archaeomeryx. Bull. Am. Mus. Nat. Hist. 167, 3–57.
Wei, Y.-Q., Yang, H.-J., Long, R.-J., Wang, Z.-Y., Cao, B.-B., Ren, Q.-C., et al. (2017). Characterization of natural co-cultures of Piromyces with Methanobrevibacter ruminantium from yaks grazing on the Qinghai-Tibetan plateau: a microbial consortium with high potential in plant biomass degradation. AMB Express 7:160. doi: 10.1186/s13568-017-0459-1
Weimer, P. J. (1998). Manipulating ruminal fermentation: a microbial ecological perspective. J. Anim. Sci. 76, 3114–3122. doi: 10.2527/1998.76123114x
Weimer, P. J. (2015). Redundancy, resilience, and host specificity of the ruminal microbiota: implications for engineering improved ruminal fermentations. Front. Microbiol. 6:296. doi: 10.3389/fmicb.2015.00296
Weinroth, M. D., Belk, A. D., Dean, C., Noyes, N., Dittoe, D. K., Rothrock, M. J., et al. (2022). Considerations and best practices in animal science 16S ribosomal RNA gene sequencing microbiome studies. J. Anim. Sci. 100:skab346. doi: 10.1093/jas/skab346
Welander, P. V., and Metcalf, W. W. (2005). Loss of the mtr operon in Methanosarcina blocks growth on methanol, but not methanogenesis, and reveals an unknown methanogenic pathway. Proc. Natl. Acad. Sci. 102, 10664–10669. doi: 10.1073/pnas.0502623102
Welte, C., and Deppenmeier, U. (2011). Re-evaluation of the function of the F420 dehydrogenase in electron transport of Methanosarcina mazei. FEBS J. 278, 1277–1287. doi: 10.1111/j.1742-4658.2011.08048.x
Welte, C., and Deppenmeier, U. (2014). Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim. Biophys. Acta 1837, 1130–1147. doi: 10.1016/j.bbabio.2013.12.002
Westendarp, H. (2006). Effects of tannins in animal nutrition. Dtsch. Tierarztl. Wochenschr. 113, 264–268.
Whitman, W. B., Ankwanda, E., and Wolfe, R. S. (1982). Nutrition and carbon metabolism of Methanococcus voltae. J. Bacteriol. 149, 852–863. doi: 10.1128/jb.149.3.852-863.1982
Wilkinson, T., Korir, D., Ogugo, M., Stewart, R. D., Watson, M., Paxton, E., et al. (2020). 1200 high-quality metagenome-assembled genomes from the rumen of African cattle and their relevance in the context of sub-optimal feeding. Genome Biol. 21:229. doi: 10.1186/s13059-020-02144-7
Williams, A. G., and Coleman, G. S. (1997). The rumen protozoa. London: Blackie Academic & Professional.
Williams, C. L., Thomas, B. J., McEwan, N. R., Rees Stevens, P., Creevey, C. J., and Huws, S. A. (2020). Rumen protozoa play a significant role in fungal predation and plant carbohydrate breakdown. Front. Microbiol. 11:720. doi: 10.3389/fmicb.2020.00720
Wolin, M. J. (1960). A theoretical rumen fermentation balance. J. Dairy Sci. 43, 1452–1459. doi: 10.3168/jds.S0022-0302(60)90348-9
Wolin, M. J. (1979). “The rumen fermentation: a model for microbial interactions in anaerobic ecosystems” in Advances in microbial ecology. ed. M. Alexander, vol. 3 (Boston, MA: Springer), 49–77.
Wood, G. E., Haydock, A. K., and Leigh, J. A. (2003). Function and regulation of the formate dehydrogenase genes of the methanogenic archaeon Methanococcus maripaludis. J. Bacteriol. 185, 2548–2554. doi: 10.1128/JB.185.8.2548-2554.2003
Wright, D. E. (1960). Pectic enzymes in rumen protozoa. Arch. Biochem. Biophys. 86, 251–254. doi: 10.1016/0003-9861(60)90413-6
Wright, A.-D. G., Auckland, C. H., and Lynn, D. H. (2007). Molecular diversity of methanogens in feedlot cattle from Ontario and Prince Edward Island, Canada. Appl. Environ. Microbiol. 73, 4206–4210. doi: 10.1128/AEM.00103-07
Xia, Y., Kong, Y. H., Seviour, R., Forster, R. J., Kisidayova, S., and McAllister, T. A. (2014). Fluorescence in situ hybridization probing of protozoal Entodinium spp. and their methanogenic colonizers in the rumen of cattle fed alfalfa hay or triticale straw. J. Appl. Microbiol. 116, 14–22. doi: 10.1111/jam.12356
Xie, F., Jin, W., Si, H., Yuan, Y., Tao, Y., Liu, J., et al. (2021). An integrated gene catalog and over 10,000 metagenome-assembled genomes from the gastrointestinal microbiome of ruminants. Microbiome 9:137. doi: 10.1186/s40168-021-01078-x
Xu, W., Chen, T., Pei, Y., Guo, H., Li, Z., Yang, Y., et al. (2021). Characterization of shallow whole-metagenome shotgun sequencing as a high-accuracy and low-cost method by complicated mock microbiomes. Front. Microbiol. 12:820380. doi: 10.3389/fmicb.2021.820380
Yan, Z., and Ferry, J. G. (2018). Electron bifurcation and confurcation in methanogenesis and reverse methanogenesis. Front. Microbiol. 9:1322. doi: 10.3389/fmicb.2018.01322
Yan, Z., Wang, M., and Ferry, J. G. (2017). A ferredoxin- and F420H2-dependent, electron-bifurcating, heterodisulfide reductase with homologs in the domains bacteria and archaea. mBio 8:e02285-16. doi: 10.1128/mBio.02285-16
Yáñez-Ruiz, D. R., Abecia, L., and Newbold, C. J. (2015). Manipulating rumen microbiome and fermentation through interventions during early life: a review. Front. Microbiol. 6:1133. doi: 10.3389/fmicb.2015.01133
Yeoman, C. J., and White, B. A. (2014). Gastrointestinal tract microbiota and probiotics in production animals. Annu. Rev. Anim. Biosci. 2, 469–486. doi: 10.1146/annurev-animal-022513-114149
Young, J. W. (1977). Gluconeogenesis in cattle: significance and methodology. J. Dairy Sci. 60, 1–15. doi: 10.3168/jds.S0022-0302(77)83821-6
Youngblut, N. D., de la Cuesta-Zuluaga, J., Reischer, G. H., Dauser, S., Schuster, N., Walzer, C., et al. (2020). Large-scale metagenome assembly reveals novel animal-associated microbial genomes, biosynthetic gene clusters, and other genetic diversity. mSystems 5:e01045-20. doi: 10.1128/mSystems.01045-20
Yu, G., Beauchemin, K. A., and Dong, R. (2021). A review of 3-Nitrooxypropanol for enteric methane mitigation from ruminant livestock. Animals 11:3576. doi: 10.3390/ani11123576
Zehavi, T., Probst, M., and Mizrahi, I. (2018). Insights into culturomics of the rumen microbiome. Front. Microbiol. 9:1999. doi: 10.3389/fmicb.2018.01999
Zellner, G., and Winter, J. (1987). Secondary alcohols as hydrogen donors for CO2-reduction by methanogens. FEMS Microbiol. Lett. 44, 323–328. doi: 10.1111/j.1574-6968.1987.tb02309.x
Zhang, Q., Difford, G., Sahana, G., Løvendahl, P., Lassen, J., Lund, M. S., et al. (2020). Bayesian modeling reveals host genetics associated with rumen microbiota jointly influence methane emission in dairy cows. ISME J. 14, 2019–2033. doi: 10.1038/s41396-020-0663-x
Zhang, J. K., Pritchett, M. A., Lampe, D. J., Robertson, H. M., and Metcalf, W. W. (2000). In vivo transposon mutagenesis of the methanogenic archaeon Methanosarcina acetivorans C2A using a modified version of the insect mariner-family transposable element Himar1. Proc. Natl. Acad. Sci. U. S. A. 97, 9665–9670. doi: 10.1073/pnas.160272597
Zhou, M., Hernandez-Sanabria, E., and Guan, L. L. (2009). Assessment of the microbial ecology of ruminal methanogens in cattle with different feed efficiencies. Appl. Environ. Microbiol. 75, 6524–6533. doi: 10.1128/AEM.02815-08
Zhou, J., Holmes, D. E., Tang, H.-Y., and Lovley, D. R. (2021). Correlation of key physiological properties of Methanosarcina isolates with environment of origin. Appl. Environ. Microbiol. 87:e0073121. doi: 10.1128/AEM.00731-21
Zhu, H., Yang, M., Loor, J. J., Elolimy, A., Li, L., Xu, C., et al. (2021). Analysis of cow-calf microbiome transfer routes and microbiome diversity in the newborn Holstein dairy calf hindgut. Front. Nutr. 8:736270. doi: 10.3389/fnut.2021.736270
Zhu, J., Zheng, H., Ai, G., Zhang, G., Liu, D., Liu, X., et al. (2012). The genome characteristics and predicted function of methyl-group oxidation pathway in the obligate aceticlastic methanogens, Methanosaeta spp. PLoS One 7:e36756. doi: 10.1371/journal.pone.0036756
Zimmerly, C. A., and Weiss, W. P. (2001). Effects of supplemental dietary biotin on performance of Holstein cows during early lactation. J. Dairy Sci. 84, 498–506. doi: 10.3168/jds.S0022-0302(01)74500-6
Keywords: rumen, methanogen, methane, greenhouse gas, archaea, ruminants, microbiome
Citation: Khairunisa BH, Heryakusuma C, Ike K, Mukhopadhyay B and Susanti D (2023) Evolving understanding of rumen methanogen ecophysiology. Front. Microbiol. 14:1296008. doi: 10.3389/fmicb.2023.1296008
Received: 17 September 2023; Accepted: 12 October 2023;
Published: 06 November 2023.
Edited by:
Zhe Lyu, North Carolina State University, United StatesReviewed by:
Vivek Fellner, North Carolina State University, United StatesCopyright © 2023 Khairunisa, Heryakusuma, Ike, Mukhopadhyay and Susanti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dwi Susanti, ZHN1c2FudGlAYmlvbWVkaXQuY29t; Biswarup Mukhopadhyay, Ymlzd2FydXBAdnQuZWR1
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.