- 1Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China
- 2College of Horticulture, Gansu Agricultural University, Lanzhou, China
- 3Huasheng Seed Group Co., Ltd, Qingzhou, China
Clubroot disease, caused by Plasmodiophora brassicae, is a serious soil-borne disease in Brassica crops worldwide. It seriously occurs in conducive soils of southern China, while never happens in some areas of northern China with suppressive soils. To understanding the differences, we measured the soil suppressiveness, chemical properties, and microbial communities in suppressive and conducive soils by bioassay and sequencing of 16S and 18S rRNA amplicons. The biological basis of clubroot suppressiveness was supported by the ability to remove it by pasteurization. The pH value and calcium content in the suppressive soils were higher than those in the conducive soils. Suppressive soils were associated with higher fungal diversity and bacterial abundance. The fungal phyla Chytridiomycota, Olpidiomycota, and Mucoromycota and the bacterial phyla Acidobacteriota and Gemmatimonadota were enriched in suppressive soils. More abundant beneficial microbes, including Chaetomium and Lysobacter, were found in the suppressive soils than in the conducive soils. Molecular ecological network analysis revealed that the fungal network of suppressive soils was more complex than that of conducive soils. Our results indicate that plant health is closely related to soil physicochemical and biological properties. This study is of great significance for developing strategies for clubtroot disease prevention and control.
1 Introduction
Many crops are attacked by soil-borne pathogens worldwide, and the resulting diseases are difficult to control due to the complexity of the soil. Clubroot, which is caused by Plasmodiophora brassicae, is an important disease of plants of the family Brassicaceae (Dixon, 2009a). The disease is found throughout the world wherever crucifers (e.g., Brassica napus) are grown (Wallenhammar, 1996; Donald and Porter, 2009; Feng et al., 2013) and causes 10−15% of yield losses worldwide (Dixon, 2009b; Faggian and Strelkov, 2009). In China, clubroot disease has occurred in many provinces except Shānxi, Qinghai Province, Ningxia Hui Autonomous Region, and Inner Mongolia Autonomous Region. Clubroot infection is much more severe in Shandong, Liaoning, Jilin, Shānxi, Jiangxi, Yunnan, Sichuan, Guizhou, Chongqing, Hunan, Hubei Provinces, and the Tibet Autonomous Region of China (Supplementary Figure 1). Field studies have indicated that the pathogen has a half-life of at least 3.6 years, and some spores may exist for at least 17.3 years in soil in the absence of suitable hosts before spore populations are eroded to undetectable levels (Wallenhamar, 2010). Although many methods have been proposed, it is still difficult to control the disease (Niwa et al., 2008).
Plant diseases are determined by the interactions among host, pathogen, and the environment. It is the balance of these interactions among three elements that determines whether or not disease develops to destructive levels in a particular situation (Keane and Kerr, 1997). Among them, environmental factors, including biotic and abiotic factors have been considered to play an important role in the development of plant diseases, especially for soil-borne diseases (Keane and Kerr, 1997; Husson et al., 2021). Several studies have confirmed that soil abiotic factors can influence these interactions and thereby disease incidence or severity (Nerey et al., 2010; Tamm et al., 2010; Mehmood et al., 2013; You et al., 2017). For instance, soil pH and calcium showed a significantly negative correlation with the wilt infection rate (Yang et al., 2017). For root rot, Calcisol is always the most disease-conducive soil, and Vertisol is the most disease-repressive soil (Nerey et al., 2010). For clubroot, soil type, pH, and calcium, boron and nitrogen concentrations are known to have substantial effects on the infection and development of P. brassicae, but specific information on these factors is often surprisingly weak and incomplete (Gossen et al., 2014). Root hair infection and subsequent symptom development were limited when the soil pH was higher than 7.2, but infection of root hairs was not affected by high pH in the absence of calcium (Myers and Campbell, 1985; Rashid et al., 2013). Previous studies have shown that boron slows the development of P. brassicae during infection of root hairs and the root cortex, and the effect of boron on clubroot was related to soil type (Deora et al., 2011, 2014).
Moreover, soil and plant health also depend largely on the composition and diversity of the soil microbial community (van Elsas et al., 2012). Different microbial species in soil microbial communities have complex interactions (such as symbioses, parasitism, competition, or predation), forming a complex interrelated ecological network (Poudel et al., 2016). For example, healthy soils contained higher microbial diversity and more abundant beneficial microbes than bacterial wilt-infected soils (Wang et al., 2017). A higher abundance of Fusarium was observed in Fusarium wilt-diseased soils, whereas the phylum Firmicutes and the genera Bacillus, Lactococcus, and Pseudomonas were enriched in disease-free soils (Zhou et al., 2019). The healthy soil network contained more interacting species, more key microorganisms, and better high-order organization than bacterial wilt-susceptible soil (Qi et al., 2019). A recent study found that initial soil microbiome composition and functioning predetermine future plant health (Wei et al., 2019).
Disease suppressive soils have been defined as those in which disease development is minimal even in the presence of a virulent pathogen and a susceptible host (Mazzola, 2010). Classically, soil suppressiveness has been considered either general or specific (Janvier et al., 2007). General suppression is linked with the total microbial biomass in soil or related to physical or chemical attributes of the soil (Weller et al., 2002; Mazzola, 2010). Specific suppression is superimposed over the background of general suppression and is due, at least in part, to the effects of individual or select groups of microorganisms during some stage in the life cycle of a pathogen (Weller et al., 2002). The biotic factors that contribute to specific soil suppressiveness have been elucidated for a number of plant-pathogen systems (Rosenzweig et al., 2012; Penton et al., 2014; Giné et al., 2016; Lee et al., 2021). The soil suppressiveness of clubroot disease has been studied, and it has been suggested that both abiotic and biotic factors play an important role in disease suppression (Murakami et al., 2000). However, the differences in soil properties, microbial community composition and structure, and microbial networks between clubroot-suppressive and clubroot-conducive soils are still unclear.
In this study, we hypothesized that the chemical properties, microbial community and structure, and microbial network of suppressive soils are different from those of conducive soils. Therefore, the objectives of this study were to (1) compare the chemical properties between suppressive soils and conducive soils, (2) determine the relationships between soil microbial properties and plant health, and (3) investigate key microorganisms associated with plant health in networks. This study will present novel insight into understanding the differences of soil characteristics and microbiota between clubroot-suppressive and conducive soils, which will contribute to the development of new strategies for the control of clubroot disease.
2 Materials and methods
2.1 Site description and sample collection
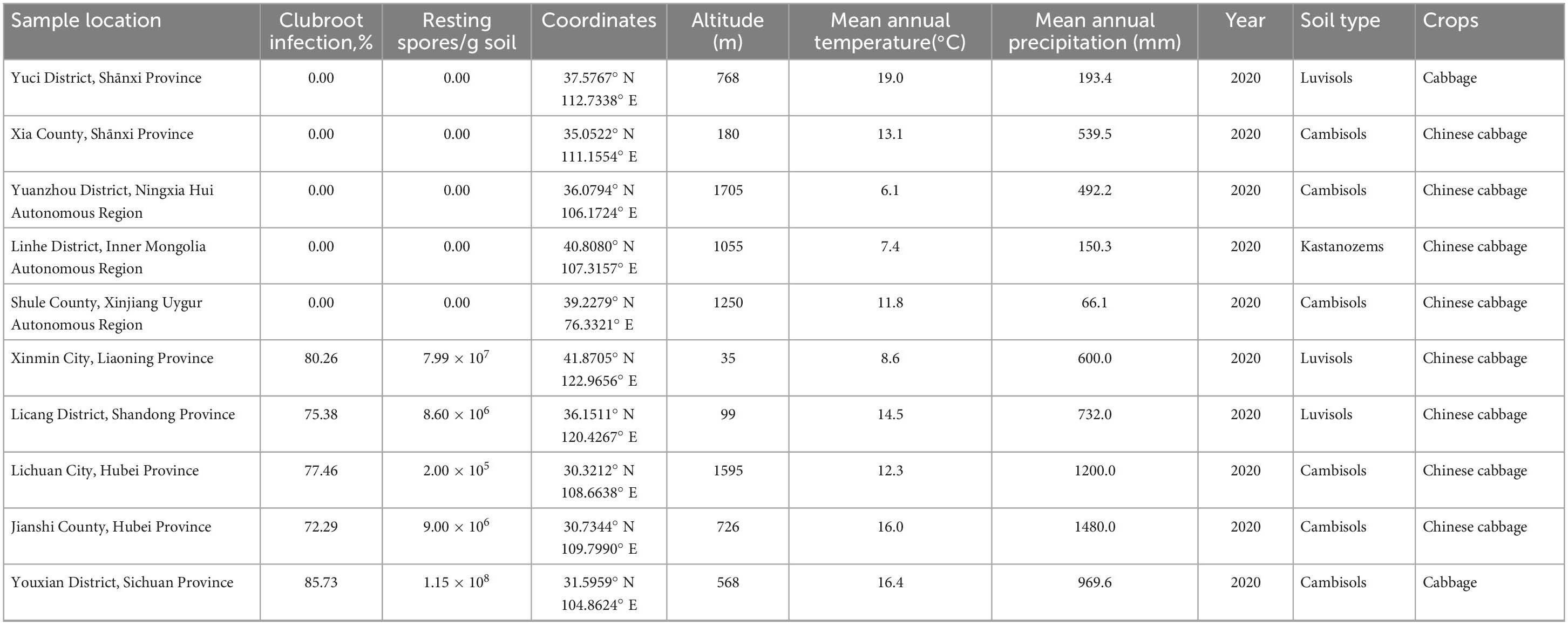
Soil samples were collected from ten different sites (Supplementary Figure 1) with cultivated cruciferous vegetables, and detailed information is listed in Table 1. Among the 10 sites, five sites (Xinmin City, Liaoning Province; Licang District, Shandong Province; Jianshi County, Hubei Province; Lichuan City, Hubei Province; Youxian District, Sichuan Province) had a clubroot infection rate of more than 70%. The soil samples, with infestation levels of approximately 105−108 spores⋅g–1 were determined to be conducive soils, and classified into two soil types Luvisols and Cambisols. Clubroot disease was not observed in the other five sites (Yuci District, Shānxi Province; Xia County, Shānxi Province; Yuanzhou District, Ningxia Hui Autonomous Region; Shule County, Xinjiang Uygur Autonomous Region; Linhe District, Inner Mongolia Autonomous Region), and the soils were confirmed to be free of P. brassicae by qPCR and tentatively classified as suppressive soils, which were classified into Cambisols, Luvisols, and Kastanozems.
For each site, 3 random subplots (approximately 60 m2) were chosen, and soil samples from approximately 10 healthy (suppressive soil) or 10 clubroot-diseased (conducive soil) plants from each subplot were collected using the checkerboard sampling method during August 2020. Briefly, each subplot was divided into 10 areas and rhizosphere soil of plant in the central point of each area was collected (Niu et al., 2017). The 10 soil samples from each subplot were mixed to form one composite sample. The composite samples were placed into sterile bags and transported to the laboratory at 0°C. Samples were then divided into two subsamples: one portion (50 g) was stored at −80°C for DNA extraction, and another (300 g) was used for chemical property analysis after thorough homogenization through a 2-mm sieve and air-drying. The soil (200 kg) used for the pot experiments was collected with a shovel from each subplot and thoroughly mixed for each site.
2.2 Quantification of P. brassicae in soils
For each composite soil, total DNA was extracted from 0.5 g of soil using the FastDNA® Spin kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA) according to the manufacturer’s instructions. The abundance of P. brassicae in the soil was quantified using quantitative polymerase chain reaction (qPCR) according to established methods (Chai et al., 2016). Fluorescence was detected after each cycle, and the concentration of pathogen was determined via qPCR according to the protocol described in our previous work (Chai et al., 2016).
2.3 Bioassay to assess the disease suppressiveness of the soils
To determine the suppressiveness of the soils, pot experiments were performed based on Mendes et al. (2011) with some modifications in a greenhouse in Qingzhou City, Weifang City, Shandong Province, in September 2020. In brief, three treatments were compared: (i) natural soil without the addition of P. brassicae, (ii) natural soil with the addition of P. brassicae, and (iii) pasteurized soil (80°C for 1 h) with the addition of P. brassicae. All 10 soil samples were used for pot experiments.
P. brassicae-infected Chinese cabbage samples were collected from a natural field in Beijing and stored at −20°C. The resting spores were prepared by the method of Castlebury et al. (1994). The soils were sprayed with suspensions of resting spores (25 mL kg–1 soil) and mixed thoroughly to obtain 108 resting spores g–1 soil.
Chinese cabbage (cv. Sushenglvxiu) seeds were sown in sterilized plastic pots (19 cm × 14 cm × 11.5 cm). Four seedlings were planted in each pot. Treatments were arranged in a randomized complete block design and replicated three times with five pots serving as a replicate. Plants were incubated in a greenhouse (25°C; 80% relative humidity and 16 h light and 8 h dark). Roots of each Chinese cabbage plant were removed 35−40 days after sowing and examined for gall formation. Disease severity was scored according to the following classes (Wallenhammar et al., 2012): 0, no galls; 1, enlarged lateral roots; 2, enlarged taproot; 3, enlarged napiform taproot, lateral roots healthy; and 4, enlarged napiform taproot, lateral roots infected. The disease severity index (DSI) was calculated according to the method of Chai et al. (2016). The experiments were independently repeated three times.
2.4 Soil chemical analysis
Electrical conductivity (EC) was measured using a conductivity meter in a soil water suspension (1:5 w/v) after shaking for 30 min. The soil pH was determined with a pH meter (Shanghai Sanxin Instrumentation, Inc., China) in a 1:5 (m/m) soil to water ratio, and the suspension was shaken at 200 r min–1 for 15 min at 25°C. The cation exchange capacity (CEC) was measured by the method of Mulvaney et al. (2004). Available K (AK) was extracted with ammonium acetate and determined by flame photometry using an FP640 Flame Photometer (Shanghai Instruments Group Co., Ltd., China). Available phosphorus (AP) was determined using the sodium hydrogen carbonate solution-Mo-Sb anti-spectrophotometric method (Shen et al., 2011). Soil organic matter (SOM) and available N (AN) were determined using the potassium dichromate external heating method (Ciavatta et al., 1991) and the alkaline-hydrolyzable diffusion method (Li et al., 2014), respectively. The total nitrogen (TN) level was determined using the semimicro Kjeldahl method. The exchangeable calcium (Ca) was measured by the method of Bao (2000). The available boron (B) level was determined as previously described (Bhering et al., 2017).
2.5 Amplicon sequencing and data processing
Total DNA of soil was extracted using a PowerSoil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA) according to the manufacturer’s protocols. Each composite soil sample was extracted in triplicate, and the extracted DNA solutions were pooled. The primer set ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) (Gardes and Bruns, 1993) and ITS2 (5′-GCTGCGTTCT TCATCGATGC-3′) was selected to target the fungal ITS1 region. 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′) were used to amplify the V4 hypervariable regions of the bacterial 16S rRNA gene. Amplicons were sequenced on the MiSeq platform at Allwegene CO., LIMITED (Beijing, China).
To obtain high-quality sequences, quality filtering of the raw sequences was performed using the QIIME package (Quantitative Insights Into Microbial Ecology) (v1.2.1). After detecting and removing the chimeric sequences using UCHIME V4.2, the effective sequences were obtained and used to perform operational taxonomic unit (OTU) and species annotation. Qualified reads were clustered into operational taxonomic units (OTUs) at a similarity level of 97% using the Uparse algorithm of Vsearch (v2.7.1) software (Edgar, 2013). QIIME (v1.8.0) was used to calculate the richness and diversity indices based on the OTU information. Sequences were compared to the entries in the SILVA database (Release 128/132/138,1 bacteria) and Unite database (Release 8.2,2 fungi) with a confidence threshold of 80% and phylogenetically assigned to taxonomic classifications using the Ribosomal Database Project naïve Bayesian classifier3 at a distance of 0.03.
2.6 Construction and analysis of the microbial network
We used the phylogenetic molecular ecological network (pMEN) method to construct interaction networks for disease-suppressive and disease-conducive soils (Deng et al., 2012; Qi et al., 2019). Random matrix theory (RMT) was used to automatically identify the appropriate similarity threshold prior to network construction. All analyses were performed using the Molecular Ecological Network Analyses Pipeline,4 and network graphs were visualized using Cytoscape 2.8.2 software (Su et al., 2014).
2.7 Statistical analysis
Soil physicochemical characteristics, Chinese cabbage clubroot DSI, and alpha diversity indices between suppressive and conducive soils were compared using Tukey’s HSD multiple range test. A Tukey HSD test was performed using SPSS Statistics 20.0 software (IBM, New York, USA). To examine the similarity between different samples, partial least squares discrimination analysis was performed by R (v3.6.0) (R Core Team, 2018) based on the OTU information. Redundancy analysis (RDA) using the vegan package in R version 3.6.0 was performed to analyze the relationships between microbial community structure and environmental variables. The significant differences in bacterial and fungal species between suppressive and conducive soils were compared using the Wilcoxon rank-sum test (P < 0.05) in R version 3.6.0. The linear discriminant analysis (LDA) effect size (LEfSe) method (LDA score > 3.0, P < 0.05) was used to analyse the association of the top 50 abundant fungal and bacterial genera with the disease-suppressive and -conducive soils.
3 Results
3.1 P. brassicae abundance at different sites
The abundance of P. brassicae was analyzed in each soil sample, and the results are presented in Table 1. P. brassicae was not detected in any suppressive soils. Quantifiable levels of P. brassicae were detected in all five conducive soil samples, ranging from 2.00 × 105−1.15 × 108 resting spores per g of soil. The highest level of resting spore infestation, 1.15 × 108 resting spores per g of soil, was found in conducive soil collected from Youxian District.
3.2 Identification of soils suppressive to P. brassicae
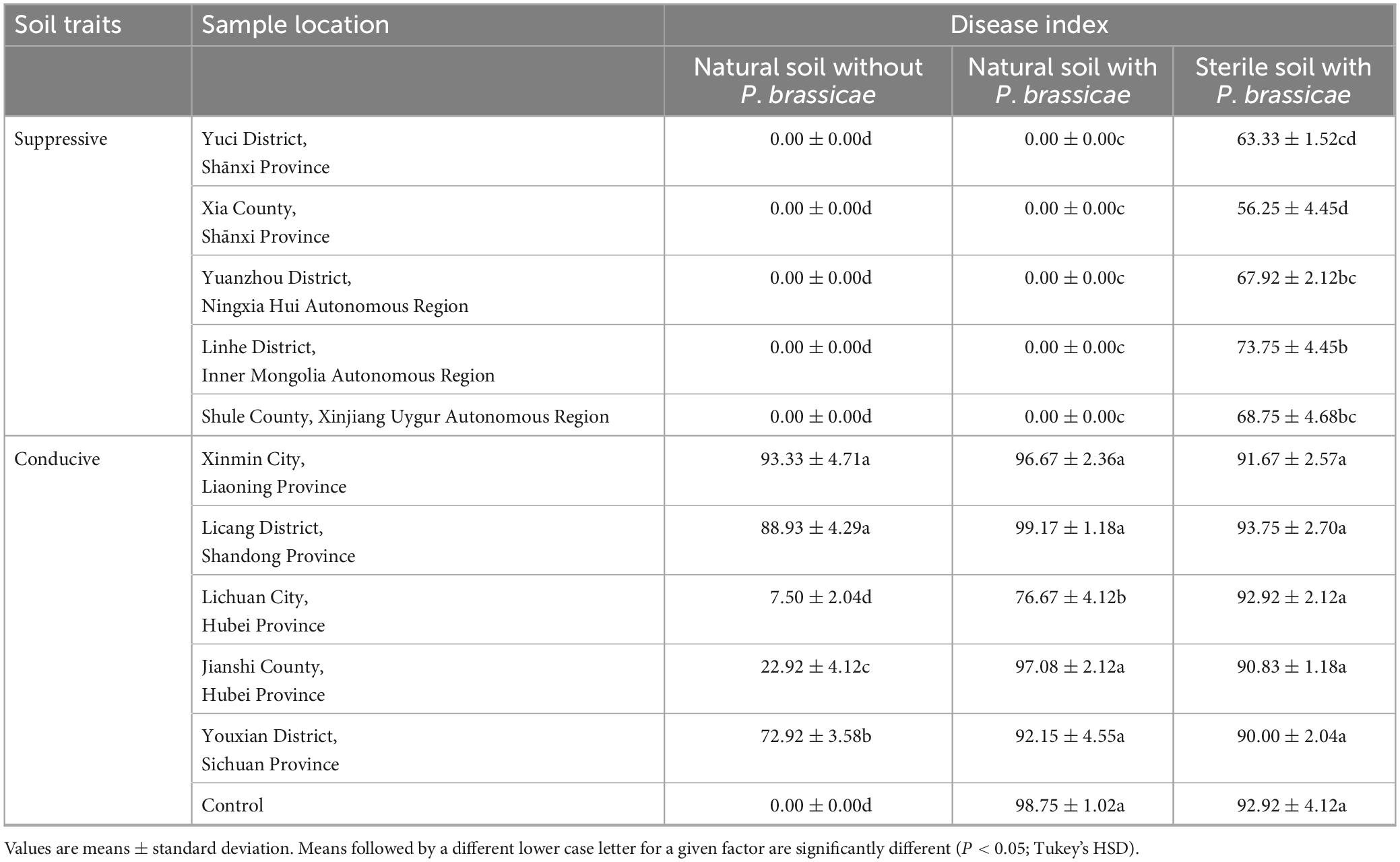
A collection of 10 soils of diverse geographical origins was used for disease suppressiveness against P. brassicae in greenhouse pot experiments. No symptoms of clubroot disease were observed in plants planted to suppressive soils in treatment with or without the addition of P. brassicae (Table 2). The suppressive soils lost clubroot suppressiveness after sterilization (80°C 1 h), and the DSIs of plants grown in sterilized suppressive soil ranged from 56.25 ± 4.45−73.75 ± 4.45, which were lower than those of the conducive soils (P < 0.05; Tukey’s HSD). The plants exposed to each conducive soil always developed clubroot symptoms in all three treatments. Altogether, these results indicate that soil suppressiveness was correlated with soil microbial communities.
3.3 Soil chemical properties
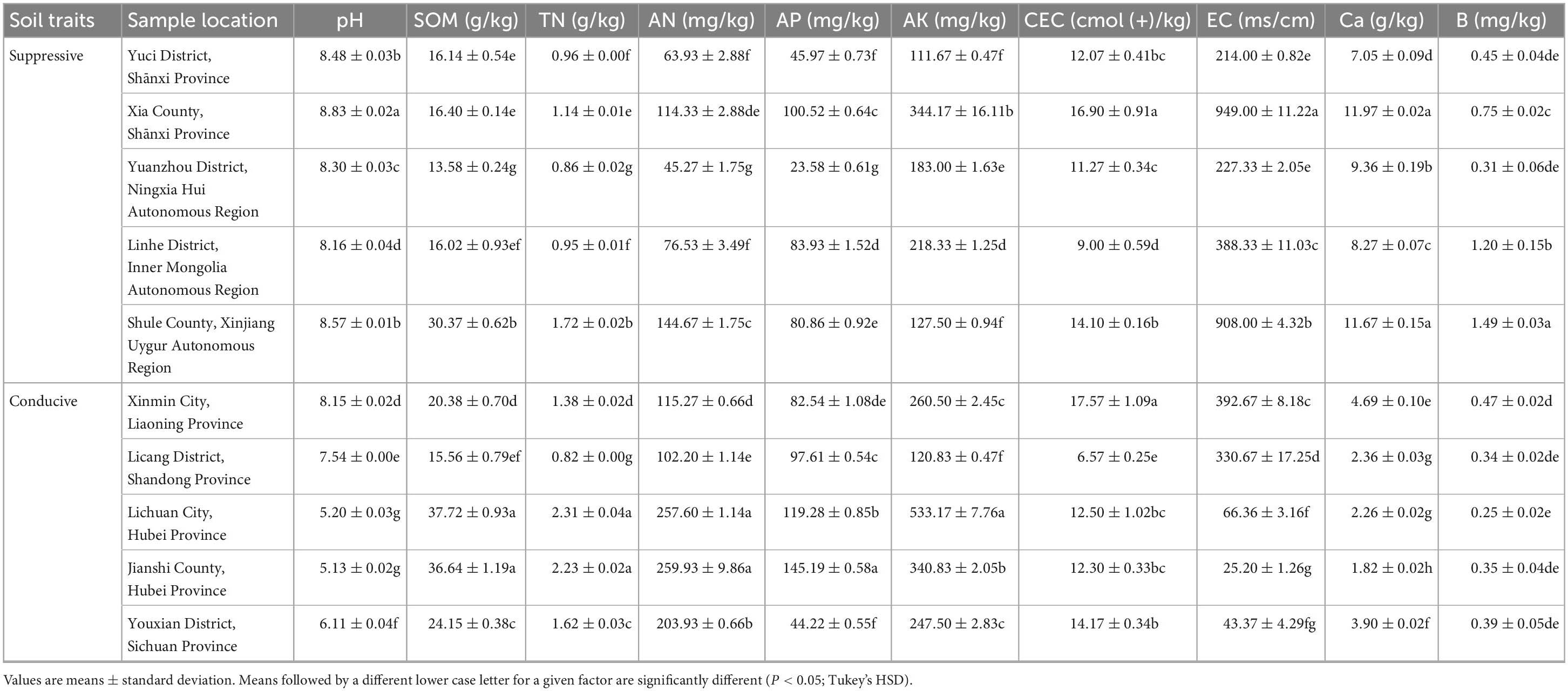
The soil pH values of all the suppressive soils were greater than ≥8.16 and higher than those of the conducive soils (P < 0.05; Tukey’s HSD). Among the conducive soils, the pH of soil collected from Xinmin City and Licang District was significantly higher than that of soil collected from Lichuan City, Jianshi County and Youxian District (P < 0.05; Tukey’s HSD). Similar to pH, the contents of exchangeable calcium in the suppressive soils were significantly higher than those in the conducive soils (P < 0.05; Tukey’s HSD). No significant association was observed for SOM, TN, AN, AP, AK, CEC, EC, and B with the presence of the pathogen or with the suppressive phenotype (Table 3). The above results indicated that the occurrence of clubroot may be related to soil pH and available calcium content.
3.4 Microbial diversity composition and structure
3.4.1 Fungal composition and structure
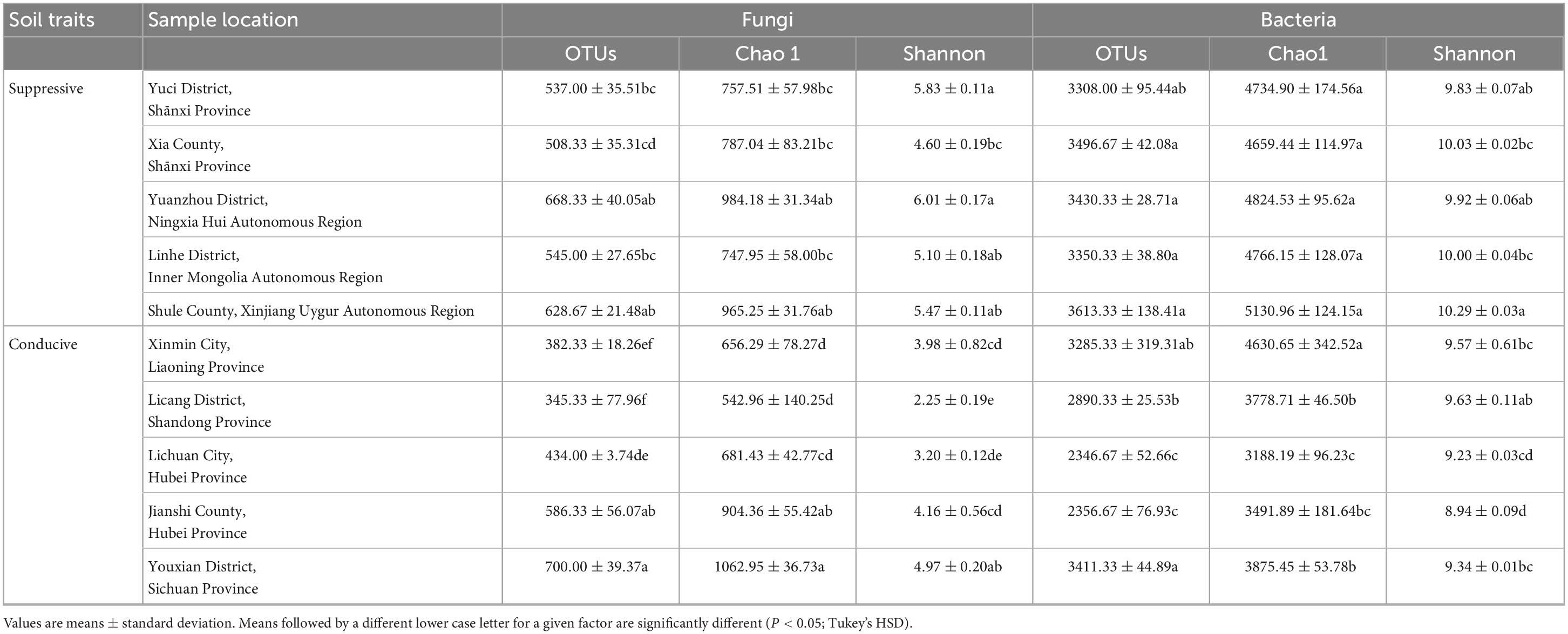
A total of 4, 160, 237 reads were obtained from 10 soil samples, and 4, 962 OTUs were identified from the reads. The analysis of the estimated Shannon indices revealed that the suppressive soils harbored higher fungal diversity (P < 0.05; Tukey’s HSD), whereas the richness indices (Chao 1) and OTU numbers showed no significant difference between suppressive soils and conducive soils (Table 4). Fourteen fungal phyla were identified in all soil samples. Ascomycota was the most abundant phylum (67.45%), followed by Mortierellomycota (8.89%), Basidiomycota (7.80%), and Chytridiomycota (2.30%). The phylum Ascomycota was more abundant in conducive soils than in suppressive soils (P < 0.05; Tukey’s HSD), whereas the phyla Olpidiomycota, Chytridiomycota, and Mucoromycota showed the opposite trend (Figure 1A). The difference in fungal abundance between suppressive and conducive soils was analyzed by linear discriminant analysis. The fungal genera Botryotrichum, Chaetomium, Cystofilobasidium, Pichia, Lophotrichus, Acremonium, Gamsia, Conocybe, Stachybotrys, Metarhizium, Podospora, Pseudeurotium, Coprinopsis, Remersonia, Corynascella, Schizothecium, Clonostachys, Bipolaris, Lecythophora, and Preussia were more abundant in the suppressive soils than in the conducive soils. Botryotrichum was the most dominant genus, comprising 12.01% of the total fungal genera in suppressive soils, whereas Verticillium was the most dominant genus in conducive soils, accounting for 18.39% of the total fungal genera (only 0.06% in suppressive soils) (Figure 2A).
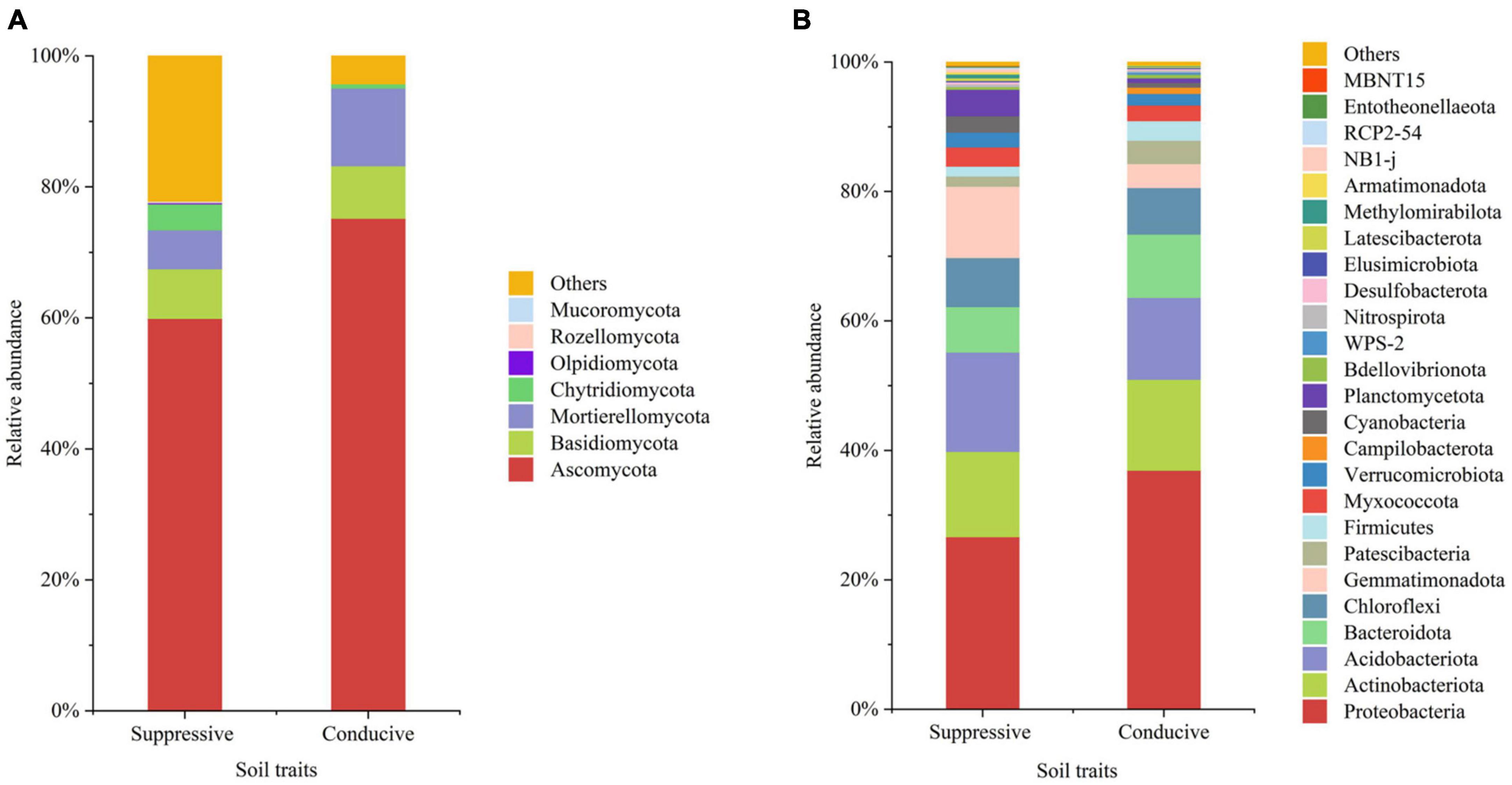
Figure 1. The average relative abundances in the top 20 fungal (A) and bacterial (B) phyla from disease-suppressive and disease-conducive soils. Others represent phyla of low relative abundance that rank lower than 20.
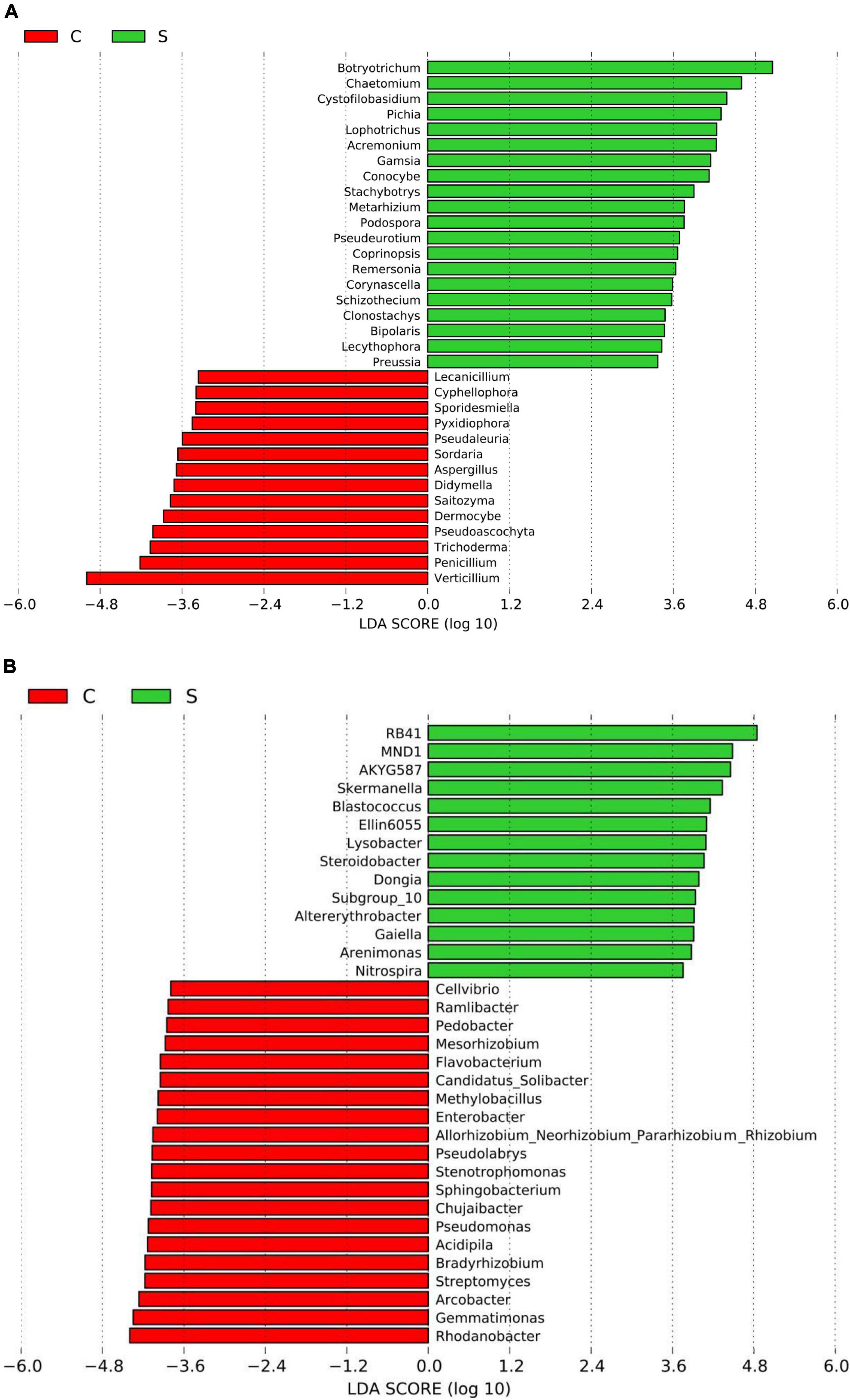
Figure 2. Histogram of the LDA scores computed for differentially abundant fungal (A) and bacterial (B) genera between the disease-suppressive and disease-conducive soils. C, conducive soil; S, suppressive soil.
Partial least squares discrimination analysis (PLS-DA) was used to analyze the β-diversity of fungal communities. PC1 and PC2 represented 4.80 and 8.37%, respectively, of the variance, and the contribution of the cumulative variance of the two principal coordinates (PC1 and PC2) accounted for 13.17% (Figure 3A). The results showed that bacterial communities from suppressive soils were clearly separated from those from conducive soils.
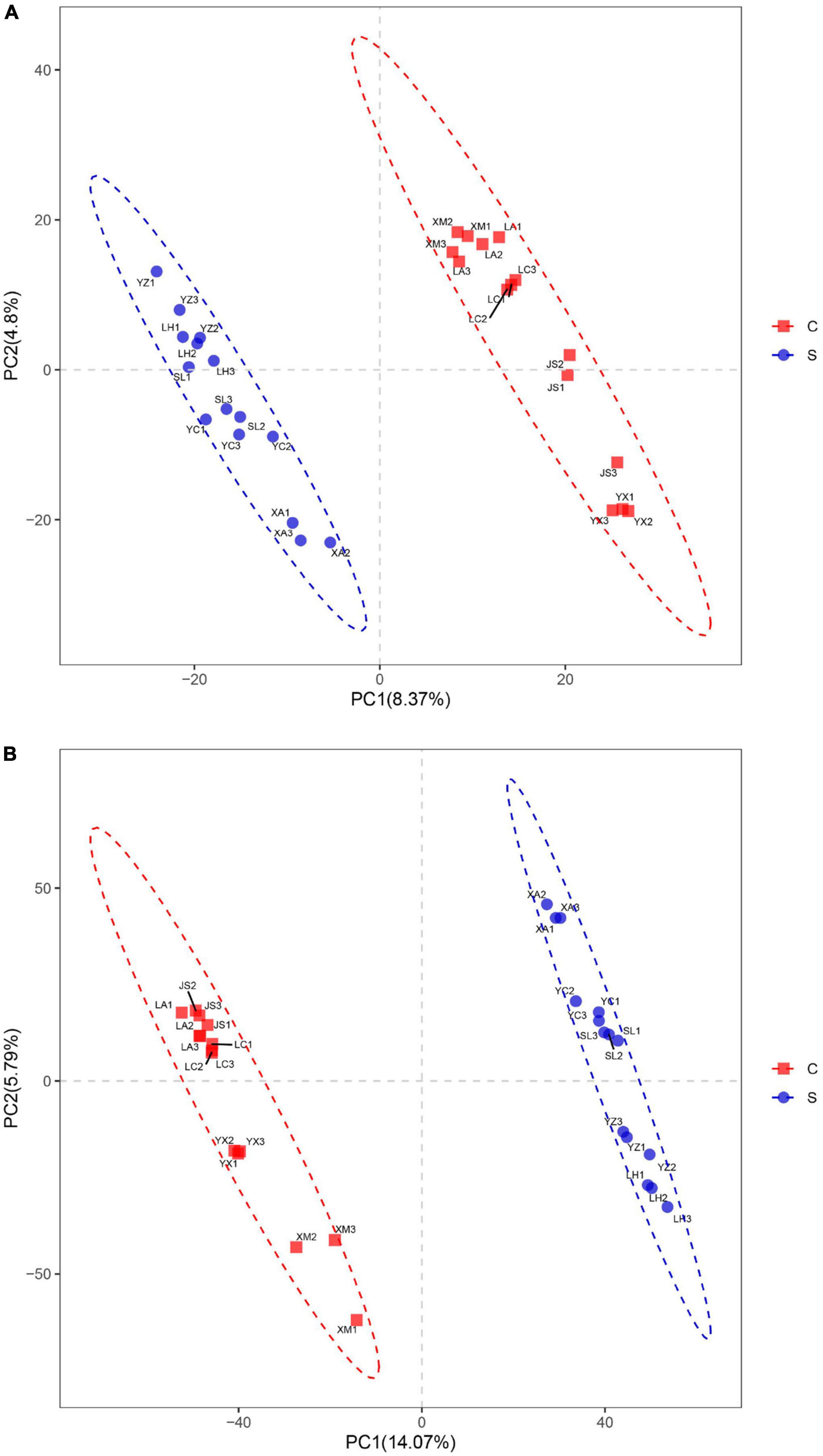
Figure 3. Partial least squares discrimination analysis of fungal (A) and bacterial (B) community structures in disease-suppressive and disease-conducive soils. C, conducive soil; S, suppressive soil. YC, Yuci District, Shānxi Province; XA, Xia County, Shānxi Province; YZ, Yuanzhou District, Ningxia Hui Autonomous Region; SL, Shule County, Xinjiang Uygur Autonomous Region; LH, Linhe District, Inner Mongolia Autonomous Region; XM, Xinmin City, Liaoning Province; LA, Licang District, Shandong Province; LC, Lichuan City, Hubei Province; JS, Jianshi County, Hubei Province; YX, Youxian District, Sichuan Province.
3.4.2 Bacterial composition and structure
A total of 2, 398, 352 reads were obtained from 10 soil samples, and 13, 650 OTUs were identified from the reads. The bacterial OTU number in the suppressive soils was higher than that in the conducive soils (P < 0.05; Tukey’s HSD), except for soil from Youxian District (Table 4). Analysis by Chao1 revealed a higher richness of bacteria in the suppressive soils. The Shannon index indicated that no significant difference was observed in the bacterial diversity of suppressive and conducive soils. In all soil samples, Proteobacteria, Acidobacteriota, Actinobacteriota, Bacteroidota, Chloroflexi, Gemmatimonadota, Myxococcota, Patescibacteria, Planctomycetota, Firmicutes, Verrucomicrobiota, and Cyanobacteria were the dominant phyla, accounting for 96.19% of the total bacterial sequences (Figure 1B). Acidobacteriota and Gemmatimonadota were more abundant in suppressive soils than in conducive soils (P < 0.05; Tukey’s HSD). The abundances of Proteobacteria, Patescibacteria, and Firmicutes were higher in conducive soils than in suppressive soils (P < 0.05; Tukey’s HSD). The LEfSe analytical method was used to visualize the representative taxa between suppressive and conducive soils. In the suppressive soils, the relative abundances of RB41, MND1, AKYG587, Skermanella, Blastococcus, Ellin6055, Dongia, Lysobacter, Subgroup_10, Altererythrobacter, Gaiella, Arenimonas, Nitrospira, and Steroidobacter were higher in suppressive soils than in conducive soils, whereas 19 genera were more abundant in conducive soils, such as Rhodanobacter, Gemmatimonas, and Arcobacter (Figure 2B).
The bacterial communities within the suppressive and conducive soil were separately clustered at the PC1 axis according to partial least squares discrimination analysis (PLS-DA). PC1 and PC2 explained 19.86% of the total bacterial community (Figure 3B).
3.5 Network analysis
In the fungal phylogenetic molecular ecological networks (pMENs), 120 and 76 nodes were constructed from suppressive (Figure 4A) and conducive (Figure 4B) soil samples, respectively. For suppressive and conducive networks, the average connectivity [Connectivity is the number of links (edges) of a node with other nodes] was 4.50 and 4.39, respectively. Suppressive and conducive soils harbored 8 and 6 modules with modularity values (Modularity measures the degree to which a network is organized into clearly delimited modules) of 0.44 and 0.41, respectively, (Table 5). Suppressive soils (270 links) had higher link numbers than conducive soils (167 links). With respect to bacteria, networks with 189 and 161 nodes were constructed from suppressive (Supplementary Figure 2A) and conducive (Supplementary Figure 2B) soil samples, respectively. The average connectivity was 2.77 and 9.44, and the average clustering coefficient value was 0.20 and 0.19 for suppressive and conducive networks, respectively, (Table 5). The suppressive and conducive soil networks had different modularity values (0.79 for suppressive and 0.25 for conducive soil). The total link numbers in suppressive and conducive soils were 262 and 760, respectively.
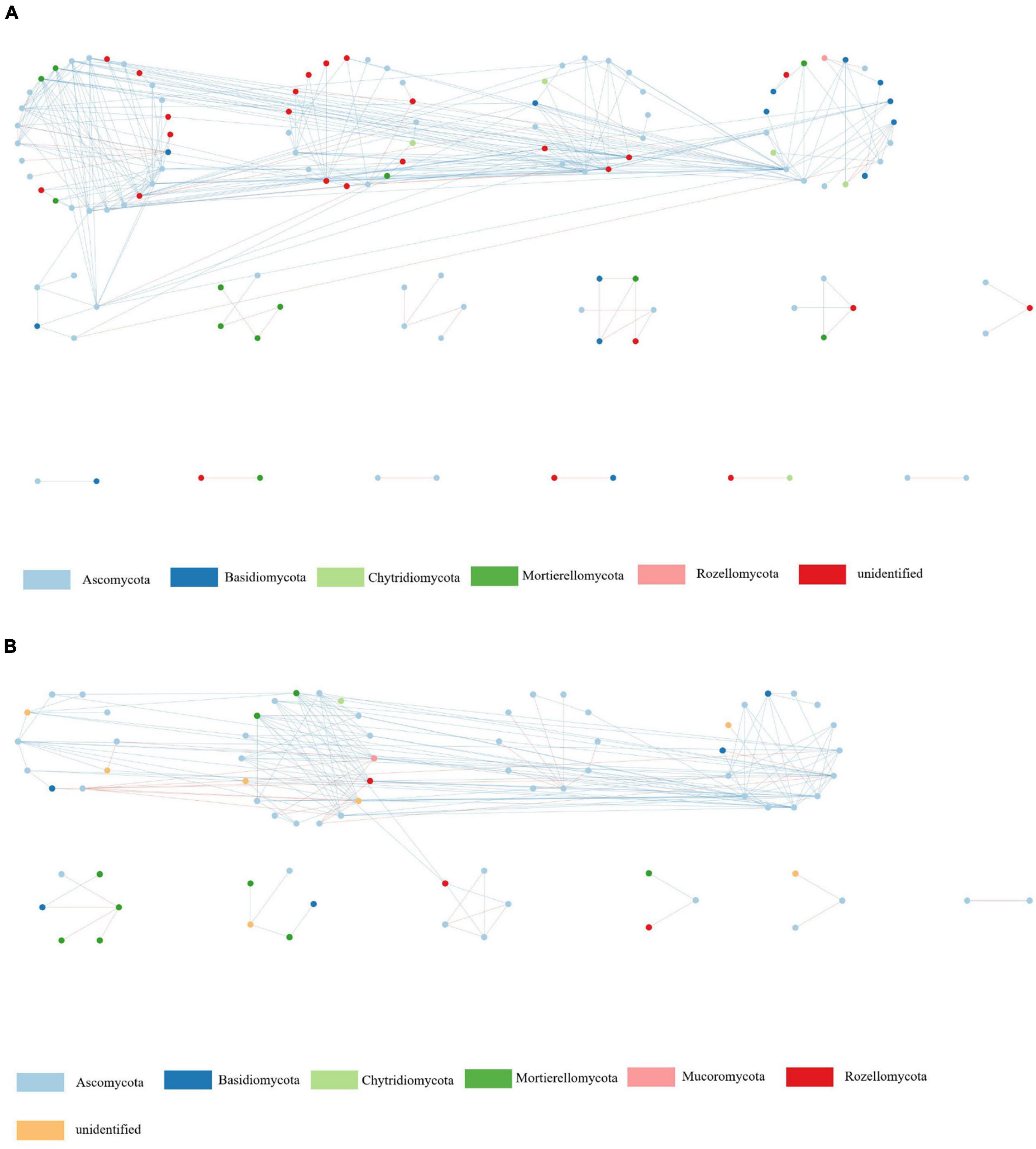
Figure 4. Network analyses of soil fungal communities in suppressive (A) and conducive (B) soil. Nodes of different colors belong to different bacterial phyla. Blue edges represent negative interactions between nodes. Red edges represent positive interactions.
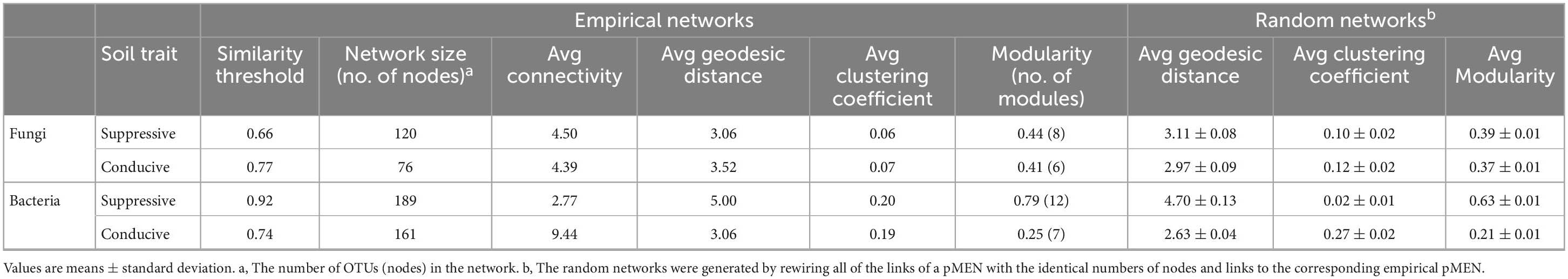
Table 5. Major topological properties of the empirical phylogenetic Molecular Ecological Networks (pMENs) of fungal and bacterial communities in disease-suppressive and disease-conducive soils and their associated random pMENs.
The topological role of each node (OTU) was defined by the within-module connectivity (Zi) and among-module connectivity (Pi). In fungal networks, the majority of nodes (88.5%) were peripherals, while 11.5% were connectors (Figure 5 and Supplementary Table 1). No network hub or module hub was found in the fungal networks. More connectors (21) were found in the suppressive network than in the conducive network (19). Four connectors (OTU_2101, OTU_3841, OTU_403, and OTU_471) in the suppressive network are closely related to Ascomycota, Ascomycota, Olpidiomycota, and Chytridiomycota, respectively. The two connectors (OTU_4194 and OTU_4714) in the conducive network belonged to Mortierellomycota and Basidiomycota, respectively, (Supplementary Table 1). With respect to bacterial networks, the conducive network had more connectors (twenty) than the suppressive network (thirteen). In the conducive networks, OTU_10229, OTU_1140, OTU_1313, OTU_1477, and OTU_765 belonging to Proteobacteria and the other two connectors (OTU_7005 and OTU_9010) were closely related to Gemmatimonadota and Chloroflexi, respectively (Supplementary Figure 3 and Table 1).
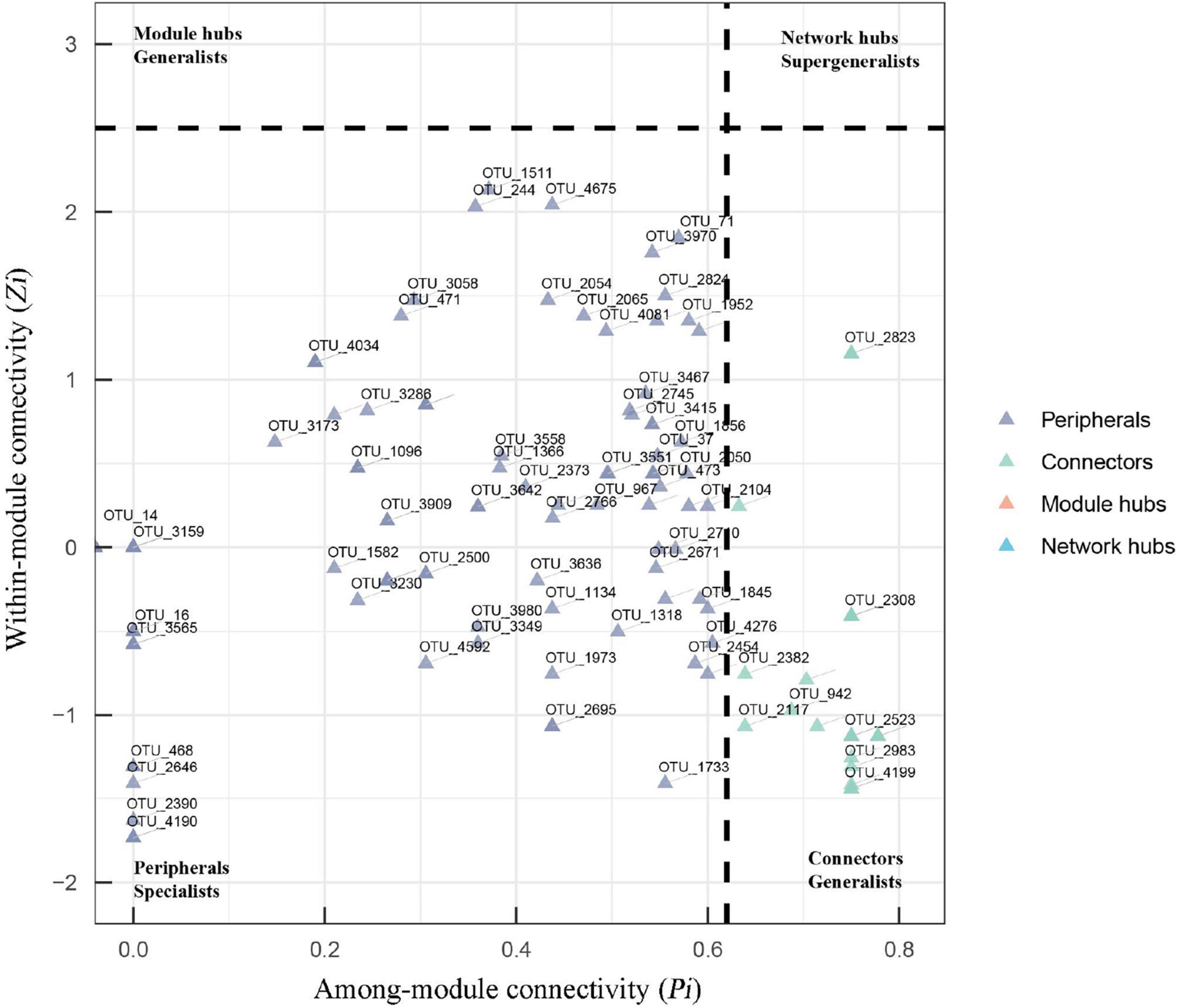
Figure 5. Zi-Pi plots showing the distribution of OTUs based on their topological roles in fungal networks. The threshold values of Zi and Pi for categorizing OTUs were 2.5 and 0.62, respectively. Nodes are defined as peripherals (Pi ≤ 0.62, Zi ≤ 2.5), module hubs (Pi ≤ 0.62, Zi > 2.5), connectors (Pi > 0.62, Zi ≤ 2.5) and network hubs (Pi > 0.62, Zi > 2.5).
3.6 Correlations between microbial communities and environmental factors
The relationships between microbial community structure and soil chemical properties were analyzed with redundancy analysis (RDA) (Figure 6). The first and second RDA components explained 56.18 and 72.83% of the total fungal and bacterial variation, respectively. Redundancy analysis indicated that high abundances of Chaetomium, Botryotrichum, Gamsia, Lophotrichus, and Acremonium in the suppressive soils were positively correlated with pH, EC, Ca, and B but negatively correlated with AN, AP, AK, TN, and SOM. The relative abundances of Fusarium, Penicillium, Pseudoascochyta, Trichoderma, Didymella, Lectera, and Mortierella were positively correlated with AN, AP, AK, TN, and SOM but negatively correlated with pH, EC, Ca, and B (Figure 6A).
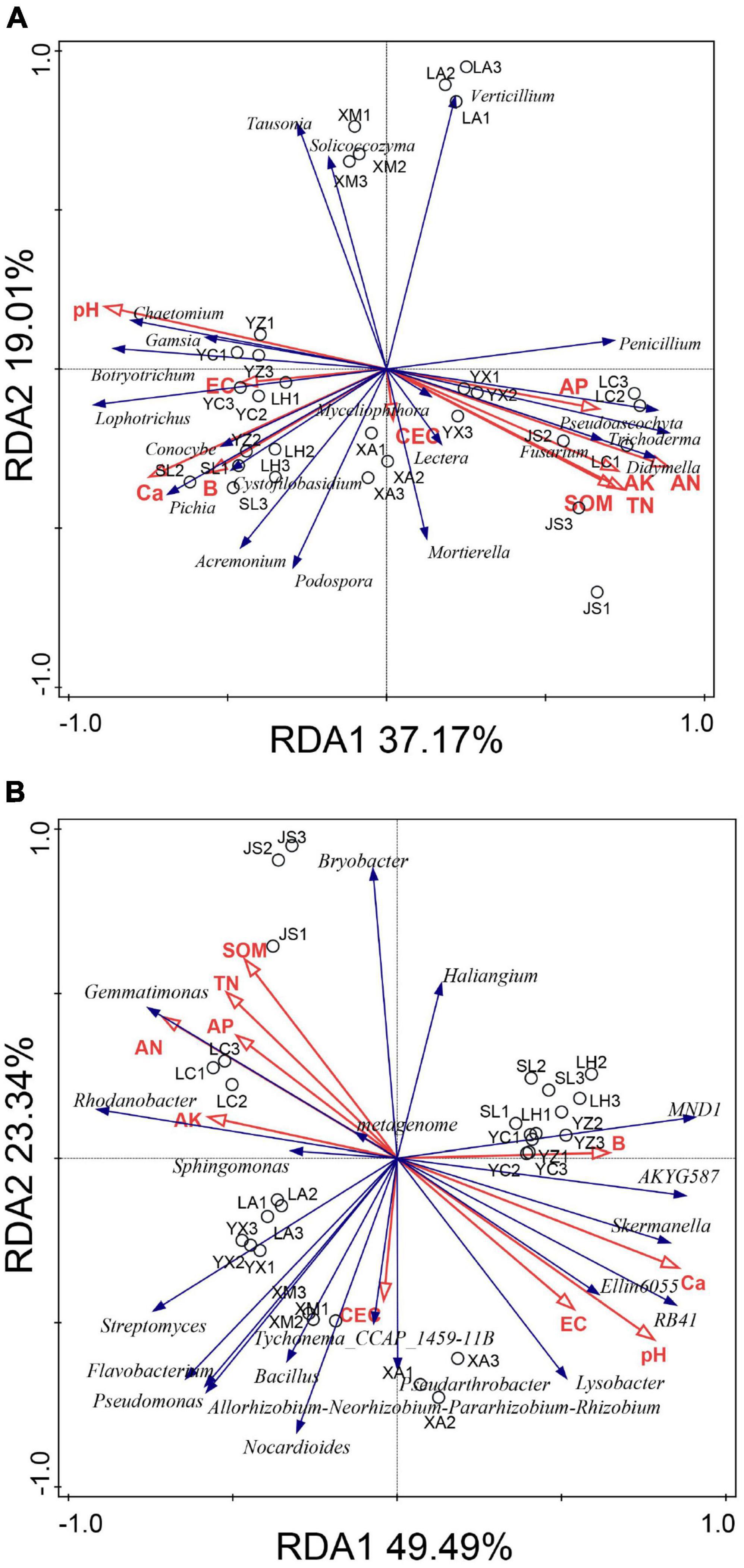
Figure 6. Redundancy analysis (RDA) of the fungal (A) and bacterial (B) communities based on genera distribution. YC, Yuci District, Shānxi Province; XA, Xia County, Shānxi Province; YZ, Yuanzhou District, Ningxia Hui Autonomous Region; SL, Shule County, Xinjiang Uygur Autonomous Region; LH, Linhe District, Inner Mongolia Autonomous Region; XM, Xinmin City, Liaoning Province; LA, Licang District, Shandong Province; LC, Lichuan City, Hubei Province; JS, Jianshi County, Hubei Province; YX, Youxian District, Sichuan Province.
With respect to bacteria, SOM, TN, AN, AP, and AK were positively correlated with Rhodanobacter, Gemmatimonas, and Sphingomonas but negatively correlated with Skermanella, Ellin6055, RB41, and Lysobacter (Figure 6B).
4 Discussion
4.1 Soil suppressiveness is related to microorganisms in soil
Disease suppressive soils have been identified for many plant pathogens, and there is plenty of evidence for the role of both biotic and abiotic factors of the soil in disease suppression (Mazzola, 2002). Physical and chemical attributes of soil, such as texture and micronutrients, can operate in the suppression of plant diseases directly or indirectly through their impact on the activity of soil microorganisms (Mazzola, 2002). Although these abiotic characteristics of soil are beneficial to disease suppression, suppressiveness is fundamentally a function of microbiological activity of soil microorganisms or microbial metabolites. Higher abundances and diversity of microbes that harbor the non-ribosomal peptide (NRPS) gene were observed in Fusarium wilt disease-suppressive soil samples than in Fusarium wilt disease-conducive soil samples (Zhao et al., 2020). The study of Fusarium wilt-suppressive soils from Korea indicated that the suppressiveness has been attributed mainly to natural antibiotics secreted by Streptomyces sp. S4-7. In the present study, severe symptoms of clubroot were observed in plants exposed to sterilized suppressive soils, which indicated that the suppression of soil was related to soil microorganisms. This result follows the results of previous reports (Mendes et al., 2011; Cha et al., 2016; Xiong et al., 2017). However, the DSIs of plants grown in pasteurized suppressive soils were lower than those grown in control soil, which inferred that abiotic factors may also be critical in the suppressiveness of soils. Severe symptoms of clubroot were consistently observed on plants exposed to conducive soils, suggesting that soil biotic and abiotic factors were conducive to the occurrence of the disease, which is supported by previous studies (Murakami et al., 2000). Given that 80°C cannot completely eliminate all microorganisms, the soil microbiome left after heat treatment may still have pathogen inhibition.
4.2 Influence of soil chemical properties on plant disease
Various soil chemical properties not only affect pathogen growth and survival in soils but also influence the growth and health of plants. It has been reported that labile carbon is important for the maintenance of an abundant soil microbial community and the expression of soil suppressiveness (Bongiorno et al., 2019). Previous studies have shown that higher pH and AP may enhance the inhibition of banana Fusarium wilt disease. Reduction in root hair infection and subsequent reduction in clubroot development in brassicas at alkaline pH has been reported in some crops (Webster and Dixon, 1991a; Donald and Porter, 2004). Application of calcium, magnesium or boron to raise soil pH above 7.2 under controlled conditions reduced root hair infection and subsequent symptom development (Webster and Dixon, 1991a,b; Rashid et al., 2013). However, the pattern of low clubroot at neutral or slightly alkaline pH is not consistently observed in field situations (Karling, 1968; McDonald et al., 2004). In this study, all five suppressive soils were alkaline and had a higher pH than conducive soils. Consequently, higher pH values in soils probably indicate a healthier plant status. Notably, the pH values of the conducive soils collected from the Xinming and Licang sites were greater than 7.2, which may indicate that slightly alkaline soil pH alone did not eliminate the risk of clubroot when other conditions were conducive to pathogen infection and development. Boron application in brassica vegetables generally reduced clubroot incidence by up to 50% and increased yield by 30% (Dixon, 1996; Nott et al., 1999; Ruaro et al., 2009). However, there was no significant correlation between boron content and clubroot severity in this study. High concentrations of calcium in suppressive soils may suppress the occurrence of clubroot, which was in line with previous reports (Webster and Dixon, 1991b; Donald and Porter, 2004). Many studies have indicated that plant disease incidence is negatively correlated with the soil TON, AP, and AK contents (Oyarzun et al., 1998; Shen et al., 2014), but there were no significant correlations between these chemical properties and plant health in this study.
Based on the results of RDA, the beneficial microorganisms (e.g., Chaetomium, Lysobacter, and RB41) were positively correlated with the soil pH and Ca. These findings indicated that soil pH or calcium may enhance potential functional taxa to suppress clubroot.
4.3 Correlation between plant disease and soil microbial diversity or community structure
Soil microorganisms have profound effects on the growth, nutrition and health of plants in natural and agricultural ecosystems (Garbeva et al., 2004; Almario et al., 2013). The diversity and composition of the rhizosphere microbial community is a function of both plant species and soil properties (Ling et al., 2022). Previous investigations have shown that disruption of protective rhizobacteria abundance in the tomato rhizosphere led to the occurrence of bacterial wilt disease (Lee et al., 2021). The suppressive soils harbored a higher fungal abundance than the conducive soils in this study, which is consistent with the study of Wang et al. (2019). The bacterial diversity was higher in suppressive soils than in conducive soils, which is in line with the findings of Wang et al. (2017) and Rosenzweig et al. (2012) but in contrast with those of Xiong et al. (2017). Many studies have demonstrated that a diverse microbial community is often less prone to pathogen invasion than a simpler microbial community (Fargione and Tilman, 2005; van Elsas et al., 2012; Bonanomi et al., 2014; Chen et al., 2019). However, Meng et al. (2019) proposed that microbial diversity and abundance might not be consistent indicators for soil-supporting plant health. Consequently, the importance of soil microbial diversity varies in different systems.
Soil community structure is as important as community diversity in affecting plant health and the invasion of pathogens (Garbeva et al., 2004; Janvier et al., 2007). Liu et al. (2021) found that the fungal community outperformed the bacterial community in predicting plant health status. The occurrence of potato common scab has been reported to be related to community composition (Shi et al., 2019). The microbial community composition and structure were distinct between the suppressive soil and conducive soil in the present study. Ascomycota was more abundant in conducive soils than in suppressive soils, which was in agreement with a previous study (Xiong et al., 2017). The phyla Acidobacteriota and Gemmatimonadota were more abundant in our suppressive soils and are known to produce high levels of secondary metabolites (Crits-Christoph et al., 2018). This result was in agreement with the results of a previous study in which Acidobacteriota and Gemmatimonadota were more abundant in all Fusarium wilt-suppressive soils (Cha et al., 2016). Previous studies have shown that the relative abundance of several bacterial taxa was a more important indicator of disease suppression than the exclusive presence of specific bacterial taxa (Mendes et al., 2011; Cha et al., 2016). Our results suggested that members of these phyla may collectively play an important role in clubroot disease suppression.
The linear discriminant analysis (LDA) effect size (LEfSe) method revealed some of the specific microbial groups associated with clubroot disease suppression. At the genus level, Botryotrichum, Chaetomium, and Acremonium were more abundant in suppressive soils than in conducive soils. Acremonium. sp. was reported to exert biocontrol activity against plant pathogens such as Aspergillus flavus, Fusarium verticillioides, and Meloidogyne incognita (Donald et al., 2005; Yao et al., 2015). Indeed, A. alternatum has been developed for the control of clubroot (Jäschke et al., 2010). Chaetomium spp. belong to Ascomycota and have been reported as antagonists against several plant pathogens (Dhingra et al., 2003; Aggarwal et al., 2004; Park et al., 2005). Many species of Chaetomium with the potential to be biological control agents suppress the growth of bacteria and fungi through competition (for substrate and nutrients), mycoparasitism, antibiosis, or various combinations of these (Zhang and Yang, 2007). Interestingly, Verticillium was the most dominant genus in conducive soil. Verticillium is the pathogenic genus in the soil of Verticillium wilt-infected plants and can also infect cabbage (Dumin et al., 2021). Higher abundances of RB41 and Lysobacter were found in suppressive soils. A recent report found that RB41 plays an important role in controlling the carbon cycle (Stone et al., 2021). Lysobacter is a gram-negative bacterium widely distributed in diverse ecosystems, including soil and rhizosphere, and can promote plant growth and control soil-borne diseases (Expósito et al., 2015). Therefore, we infer that these beneficial bacteria are positively correlated with soil quality and plant health.
4.4 The fungal network was more complex in suppressive soil than in conducive soil
Microbial interactions are a vital part of the soil microbiome (Zhang et al., 2014), and the interactions among different microbial groups are important for determining ecosystem functioning/stability (Zhou et al., 2011); thus, perhaps a more integrated, complex network is more difficult for a pathogen or other intruder to enter. Many studies have demonstrated that microbial networks in healthy soils are more complex and stable than those in diseased soils (Yang et al., 2017). In this study, the suppressive soil showed a higher number of links than the conducive soil for fungal networks, whereas the conducive soil had higher link numbers than the suppressive soil for bacterial networks. This result is in agreement with a previous study (Xiong et al., 2017) and indicated that the fungal network of suppressive soils was more complex and stable than that of conducive soils. We also observed that the nodes of Ascomycota were the most numerous in the fungal network, which may be related to its high relative abundance in soils.
Furthermore, network analysis provides characterization not only of taxa with direct associations with important outcomes such as disease suppression but also of taxa with indirect associations via their association with other key taxa (Poudel et al., 2016). Connectors are essential microorganisms in the microbial network (Qi et al., 2019). More connectors possibly made more frequent exchanges of materials and information among the bacterial species in the conducive network than in the suppressive network. This result was also in agreement with a previous study in which the diseased soil had more connectors than the healthy soil (Jia et al., 2022). Indeed, we found that several potential beneficial fungi, such as Botryotrichum, Chaetomium, and Acremonium, were enriched in the suppressive soil, which may be the core taxa of the fungal network. Acidibacter, Mitsuaria, and Phycicoccus were enriched in the conducive soil, and they are often rare in agricultural soils. Therefore, we speculate that the host plant might selectively regulate the community abundance of some species under pathogen stress.
5 Conclusion
We found that the soil abiotic and biotic factors were distinct between clubroot-suppressive and clubroot-conducive soils. Specifically, the pH value and calcium content in the suppressive soils were higher than those in the conducive soils. The suppressive soils harbored higher fungal diversity and bacterial abundance than the conducive soils. Microorganisms beneficial to plants such as Botryotrichum and Chaetomium were more abundant in suppressive soils. The suppressive soil fungal network was more complex and stable and contained more interacting microbial species than the conducive soil fungal network. To demonstrate the inhibitory effect of microorganisms on clubroot, further studies identifying hyperdominant taxon isolates with effective clubroot disease suppression ability and revealing their interactions may open new avenues for the development of informed biocontrol strategies.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA982587.
Author contributions
HK: Conceptualization, Writing – original draft. ZL: Conceptualization, Data curation, Writing – review and editing, Supervision. YS: Writing – review and editing. XX: Writing – review and editing. XY: Writing – review and editing. LL: Writing – review and editing. TF: Writing – review and editing. BL: Conceptualization, Writing – review and editing. AC: Conceptualization, Writing – review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key R&D Program of China (2022YFD1401200), Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, P. R. China, China Agriculture Research System of MOF and MARA (CARS-23).
Acknowledgments
We thank SPRINGER NATURE (https://www.aje.cn/) for the expert linguistic services provided.
Conflict of interest
XY was employed by the Huasheng Seed Group Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1293360/full#supplementary-material
Footnotes
- ^ http://www.arb-silva.de
- ^ http://unite.ut.ee/index.php
- ^ http://www.mothur.org/wiki/Classify.seqs
- ^ http://ieg2.ou.edu/MENA/main.cgi
References
Aggarwal, R., Tewari, A. K., Srivastava, K. D., and Singh, D. V. (2004). Role of antibiosis in the biological control of spot blotch (Cochliobolus sativus) of wheat by Chaetomium globosum. Mycopathologia 157, 369–377. doi: 10.1023/B:MYCO.0000030446.86370.14
Almario, J., Moenne-Loccoz, Y., and Muller, D. (2013). Monitoring of the relation between 2,4-diacetylphloroglucinol-producing Pseudomonas and Thielaviopsis basicola populations by real-time PCR in tobacco black root-rot suppressive and conducive soils. Soil Biol. Biochem. 57, 144–155. doi: 10.1016/j.soilbio.2012.09.003
Bhering, A. D. S., do Carmo, M. G. F., Matos, T. D. S., Lima, E. S., and do Amaral Sobrinho, N. M. (2017). Soil factors related to the severity of clubroot in Rio de Janeiro. Brazil. Plant Dis. 101, 1345–1353. doi: 10.1094/PDIS-07-16-1024-SR
Bonanomi, G., Capodilupo, M., Incerti, G., Gaglione, S. A., and Scala, F. (2014). Fungal diversity increases soil fungistasis and resistance to microbial invasion by a nonresident species. Biol. Control 72, 38–45. doi: 10.1016/j.biocontrol.2014.02.005
Bongiorno, G., Postma, J., Bünemann, E. K., Brussaard, L., de Goede, R. G. M., Mäder, P., et al. (2019). Soil suppressiveness to Pythium ultimum in ten European long-term field experiments and its relation with soil parameters. Soil Biol. Biochem. 133, 174–187. doi: 10.1016/j.soilbio.2019.03.012
Castlebury, L. A., Maddox, J. V., and Glawe, D. A. (1994). A technique for the extraction and purification of viable Plasmodiophora brassicae resting spores from host root tissue. Mycologia 86, 458–460. doi: 10.1080/00275514.1994.12026435
Cha, J. Y., Han, S., Hong, H. J., Cho, H., Kim, D., Kwon, Y., et al. (2016). Microbial and biochemical basis of a Fusarium wilt-conducive soils. ISME J. 10, 119–129. 2015.95 doi: 10.1038/ismej
Chai, A. L., Li, J. P., Xie, X. W., Shi, Y. X., and Li, B. J. (2016). Dissemination of Plasmodiophora brassicae in livestock manure detected by qPCR. Plant Pathol. 65, 137–144. doi: 10.1111/ppa.12391
Chen, Q. L., An, X. L., Zheng, B. X., Gillings, M., Peñuelas, J., Cui, L., et al. (2019). Loss of soil microbial diversity exacerbates spread of antibiotic resistance. Soil Ecol. Lett. 1, 3–13. doi: 10.1007/s42832-019-0011-0
Ciavatta, C., Govi, M., Antisari, L. V., and Sequi, P. (1991). Determination of organic carbon in aqueous extracts of soils and fertilizers. Commun. Soil Sci. Plant. Anal. 22, 795–807. doi: 10.1080/00103629109368455
Crits-Christoph, A., Diamond, S., Butterfield, C. N., Thomas, B. C., and Banfield, J. F. (2018). Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature 558, 440–444. doi: 10.1038/s41586-018-0207-y
Deng, Y., Jiang, Y. H., Yang, Y., He, Z., Luo, F., and Zhou, J. (2012). Molecular ecological network analyses. BMC Bioinformatics 13:113. doi: 10.1186/1471-2105-13-113
Deora, A., Gossen, B. D., Hwang, S. F., Pageau, D., Howard, R. J., Walley, F., et al. (2014). Effect of boron on clubroot of canola in organic and mineral soils and on residual toxicity to rotational crops. Can. J. Plant. Sci. 94, 109–118. doi: 10.4141/cjps2013-00
Deora, A., Gossen, B. D., Walley, F., and McDonald, M. R. (2011). Boron reduces development of clubroot in canola. Can. J. Plant Sci. 33, 475–484. doi: 10.1080/07060661.2011.620630
Dhingra, O. D., Mizubuti, E. S. G., and Santana, F. M. (2003). Chaetomium globosum for reducing primary inoculum of Diaporthe phaseolorum f.sp. meridionalis in soil surface soybean stubble in field conditions. Biol. Control 26, 302–310. doi: 10.1016/S1049-9644(02)00167-6
Dixon, G. R. (1996). Repression of the morphogenesis of Plasmodiophora brassicae Wor. by boron-a review. Acta Hort. 407, 393–401. doi: 10.17660/ActaHortic.1996.407.50
Dixon, G. R. (2009a). Plasmodiophora brassicae in its environment. J. Plant Growth Regul. 28, 212–228. doi: 10.1007/s00344-009-9098-3
Dixon, G. R. (2009b). The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J. Plant Growth Regul. 28, 194–202. doi: 10.1007/s00344-009-9090-y
Donald, E. C., and Porter, I. J. (2004). A sand-solution culture technique used to observe the effect of calcium and pH on root hair and cortical stages of infection by Plasmodiophora brassicae. Australas. Plant Path. 33, 585–589. doi: 10.1071/ap04068
Donald, E. C., and Porter, I. J. (2009). Integrated control of clubroot. J. Plant Growth Regul. 28, 289–303. doi: 10.1007/s00344-009-9094-7
Donald, T., Shoshannah, R., Deyrup, S. T., and Deyrup, S. T. (2005). A protective endophyte of maize: Acremonium zeae antibiotics inhibitory to Aspergillus flavus and Fusarium verticillioides. Mycol. Res. 109, 610–618. doi: 10.1017/S0953756205002820
Dumin, W., Park, M. J., Park, J. H., Han, K. S., Choi, H. W., and Back, C. G. (2021). First report of Verticillium wilt caused by Verticillium dahliae infection on Chinese cabbage in Korea. Plant Dis. 105, 489–489. doi: 10.1094/PDIS-05-20-1132-PDN
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/NMETH.2604
Expósito, R. G., Postma, J., Raaijmakers, J. M., and De Bruijn, I. (2015). Diversity and activity of Lysobacter species from disease conducive soils. Front. Microbiol. 6:1243. doi: 10.3389/fmicb.2015.01243
Faggian, R., and Strelkov, S. E. (2009). Detection and measurement of Plasmodiophora brassicae. J. Plant Growth Regul. 28, 282–288. doi: 10.1007/s00344-009-9092-9
Fargione, J. E., and Tilman, D. (2005). Diversity decreases invasion via both sampling and complementarity effects. Ecol. Lett. 8, 604–611. doi: 10.1111/j.1461-0248.2005.00753.x
Feng, J., Hwang, S. F., and Strelkov, S. E. (2013). Studies into primary and secondary infection processes by Plasmodiophora brassicae on canola. Plant Pathol. 62, 177–183. doi: 10.1111/j.1365-3059.2012.02612.x
Garbeva, P., van Veen, J. A., and van Elsas, J. D. (2004). Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu. Rev. Phytopathol. 42, 243–270. doi: 10.1146/annurev.phyto.42.012604.135455
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Giné, A., Carrasquilla, M., Martínez-Alonso, M., Gaju, N., and Sorribas, F. J. (2016). Characterization of soil suppressiveness to root-knot nematodes in organic horticulture in plastic greenhouse. Front. Plant Sci. 7:164. doi: 10.3389/fpls.2016.00164
Gossen, B. D., Deora, A., Peng, G., Hwang, S. F., and McDonald, M. R. (2014). Effect of environmental parameters on clubroot development and the risk of pathogen spread. Can. J. Plant Pathol. 36, 37–48. doi: 10.1080/07060661.2013.859635
Husson, O., Sarthou, J. P., Bousset, L., Ratnadass, A., Schmidt, H. P., Kempf, J., et al. (2021). Soil and plant health in relation to dynamic sustainment of Eh and pH homeostasis: A review. Plant Soil 466, 391–447. doi: 10.1007/s11104-021-05047-Z
Janvier, C., Villeneuve, F., Alabouvette, C., Edel-Hermann, V., Mateille, T., and Steinberg, C. (2007). Soil health through soil disease suppression: which strategy from descriptors to indicators? Soil Biol. Biochem. 39, 1–23. doi: 10.1016/j.soilbio.2006.07.001
Jäschke, D., Dugass-Gobena, D., Karlovsky, P., Vidal, S., and Ludwig-Müller, J. (2010). Suppression of clubroot (Plasmodiophora brassicae) development in Arabidopsis thaliana by the endophytic fungus Acremonium alternatum. Plant pathol. 59, 100–111. doi: 10.1111/j.1365-3059.2009.02199.x
Jia, M., Sun, X., Chen, M., Liu, S., Zhou, J., and Peng, X. (2022). Deciphering the microbial diversity associated with healthy and wilted Paeonia suffruticosa rhizosphere soil. Front. Microbiol. 13:967601. doi: 10.3389/fmicb.2022.967601
Keane, P., and Kerr, A. (1997). “Factors affecting disease development,” in Plant Pathogens and Plant Diseases, eds J. F. Brown and H. J. Ogle (Armidale: Rockvale Publication), 287–298.
Lee, S. M., Kong, H. G., Song, G. C., and Ryu, C. M. (2021). Disruption of Firmicutes and Actinobacteria abundance in tomato rhizosphere causes the incidence of bacterial wilt disease. The ISME J. 15, 330–347. doi: 10.1038/s41396-020-00785-x
Li, J. G., Ren, G. D., Jia, Z. J., and Dong, Y. H. (2014). Composition and activity of rhizosphere microbial communities associated with healthy and diseased greenhouse tomatoes. Plant Soil 380, 337–347. doi: 10.1007/s11104-014-2097-6
Ling, N., Wang, T., and Kuzyakov, Y. (2022). Rhizosphere bacteriome structure and functions. Nat. Commun. 13:836. doi: 10.1038/s41467-022-28448-9
Liu, L., Yan, Y., Ding, H., Zhao, J., Cai, Z., and Dai, C. (2021). The fungal community outperforms the bacterial community in predicting plant health status. Appl. Microbiol. Biot. 105, 6499–6513. doi: 10.1007/s00253-021-11486-6
Mazzola, M. (2002). Mechanisms of natural soil suppressiveness to soilborne diseases. Anton. Leeuwenh. 81, 557–564. doi: 10.1023/a:1020557523557
Mazzola, M. (2010). Management of resident soil microbial community structure and function to suppress soilborne disease development. Clim. Change Crop Produc. 1, 200–218. doi: 10.1079/9781845936334.0200
McDonald, M. R., Kornatowska, B., and McKeown, A. W. (2004). Management of clubroot of Asian brassica crops grown in organic soils. Acta Hortic. 635, 25–30. 635.3 doi: 10.17660/ActaHortic.2004
Mehmood, Y., Khan, M. A., Javed, N., and Arif, M. J. (2013). Effect of soil and environmental factors on chickpea wilts disease caused by Fusarium oxysporum f.sp. ciceris. Pak. J. Phytopathol. 25, 52–58.
Mendes, R., Kruijt, M., De Bruijn, I., Dekkers, E., van der Voort, M., Schneider, J. H. M., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
Meng, T., Wang, Q., Abbasi, P., and Ma, Y. (2019). Deciphering differences in the chemical and microbial characteristics of healthy and Fusarium wilt-infected watermelon rhizosphere soils. Appl. Microbiol. Biot. 103, 1497–1509. doi: 10.1007/s00253-018-9564-6
Mulvaney, R. L., Yaremych, S. A., Khan, S. A., Swiader, J. M., and Horgan, B. P. (2004). Use of diffusion to determine soil cation-exchange capacity by ammonium saturation. Commun. Soil Sci. Plant Anal. 35, 51–67. doi: 10.1081/CSS-120027634
Murakami, H., Tsushima, S., and Shishido, Y. (2000). Soil suppressiveness to clubroot disease of Chinese cabbage caused by Plasmodiophora brassicae. Soil Biol. Biochem. 32, 1637–1642. doi: 10.1016/S0038-0717(00)00079-1
Myers, D. F., and Campbell, R. N. (1985). Lime and the control of clubroot of crucifers: effects of pH, calcium, magnesium, and their interactions. Phytopathology 75, 670–673.
Nerey, Y., Beneden, S. V., Fran, S. C., Jimenez, A., Cupull, R., Herrera, L., et al. (2010). Influence of soil type and indigenous pathogenic fungi on bean hypocotyl rot caused by Rhizoctonia solani AG4 HGI in Cuba. Soil Biol. Biochem. 42, 797–803. doi: 10.1016/j.soilbio.2010.01.015
Niu, J., Chao, J., Xiao, Y., Chen, W., Zhang, C., Liu, X., et al. (2017). Insight into the effects of different cropping systems on soil bacterial community and tobacco bacterial wilt rate. J. Basic Microbiol. 57, 3–11. doi: 10.1002/jobm.201600222
Niwa, R., Nomura, Y., Osaki, M., and Ezawa, T. (2008). Suppression of clubroot disease under neutral pH caused by inhibition of spore germination of Plasmodiophora brassicae in the rhizosphere. Plant Pathol. 57, 445–452. doi: 10.1111/j.1365-3059.2007.01817.x
Nott, H., Falloon, R., and Cheah, L. H. (1999). Clubroot control from safe chemicals. N.Z. Commer. Grower 54, 24–26.
Oyarzun, P. J., Gerlagh, M., and Zadoks, J. C. (1998). Factors associated with soil receptivity to some fungal root rot pathogens of peas. Appl. Soil Ecol. 10, 151–169. doi: 10.1016/S0929-1393(98)00042-0
Park, J. H., Choi, G. J., Jang, K. S., Lim, H. K., Kim, H. T., Cho, K. Y., et al. (2005). Antifungal activity against plant pathogenic fungi of chaetoviridins isolated from Chaetomium globosum. FEMS Microbiol. Lett. 252, 309–313. doi: 10.1016/j.femsle.2005.09.013
Penton, C. R., Gupta, V., Tiedje, J. M., Neate, S. M., Ophel-Keller, K., Gillings, M., et al. (2014). Fungal community structure in disease conducive soils assessed by 28S LSU gene sequencing. PLoS One 9:e93893. doi: 10.1371/journal.pone.0093893
Poudel, R., Jumpponen, A., Schlatter, D. C., Paulitz, T. C., Gardener, B. B. M., Kinkel, L. L., et al. (2016). Microbiome networks: a systems framework for identifying candidate microbial assemblages for disease management. Phytopathology 106, 1083–1096. doi: 10.1094/PHYTO-02-16-0058-FI
Qi, G., Ma, G., Chen, S., Lin, C., and Zhao, X. (2019). Microbial network and soil properties are changed in bacterial wilt-susceptible soil. Appl. Environ. Microb. 85, e162–e119. doi: 10.1128/AEM.00162-19
R Core Team (2018). A language and environment for statistical computing, 3.6.0 edn. Geneva: R Foundation for Statistical Computing.
Rashid, A., Ahmed, H. U., Xiao, Q., Hwang, S. F., and Strelkov, S. E. (2013). Effects of root exudates and pH on Plasmodiophora brassicae resting spore germination and infection of canola (Brassica napus L.) root hairs. Crop Prot. 48, 16–23. doi: 10.1016/j.cropro.2012.11.025
Rosenzweig, N., Tiedje, J. M., Quensen, J. F., Meng, Q., and Hao, J. J. (2012). Microbial communities associated with potato common scab-conducive soils determined by pyrosequencing analyses. Plant Dis. 96, 718–725. doi: 10.1094/PDIS-07-11-0571
Ruaro, L., Lima Neto, V. D. C., and Ribeiro Júnior, P. J. (2009). Influence of boron, nitrogen sources and soil pH on the control of club root of crucifers caused by Plasmodiophora brassicae. Trop. Plant Pathol. 34, 231–238. doi: 10.1590/S1982-56762009000400005
Shen, Z., Wang, D., Ruan, Y., Xue, C., Zhang, J., Li, R., et al. (2014). Deep 16SrRNA pyrosequencing reveals a bacterial community associated with Banana Fusarium wilt disease suppression induced by bioorganic fertilizer application. PLoS One 9:e98420. doi: 10.1371/journal.pone.0098420
Shen, Z. Q., Zhang, Q., Liu, L. J., and Qiu, Y. (2011). Determination of available phosphorus in soil by sodium bicarbonate extraction Mo-Sb anti-spectrophotometry method. Environ. Monit. Forewarn. 3, 12–15. doi: 10.3969/j.issn.1674-6732.2011.05.004
Shi, W., Li, M., Wei, G., Tian, R., Li, C., Wang, B., et al. (2019). The occurrence of potato common scab correlates with the community composition and function of the geocaulosphere soil microbiome. Microbiome 7, 1–18. doi: 10.1186/s40168-019-0629-2
Stone, B. W., Li, J., Koch, B. J., Blazewicz, S. J., Dijkstra, P., Hayer, M., et al. (2021). Nutrients cause consolidation of soil carbon flux to small proportion of bacterial community. Nat. Commun. 12:3381. doi: 10.1038/s41467-021-23676-x
Su, G., Morris, J. H., Demchak, B., and Bader, G. D. (2014). Biological network exploration with Cytoscape 3. Curr. protoc. in bioinformatics 47, 8.13.1–8.13.24.
Tamm, L., Thürig, B., Bruns, C., Fuchs, J. G., Köpke, U., Laustela, M., et al. (2010). Soil type, management history, and soil amendments influence the development of soil-borne (Rhizoctonia solani, Pythium ultimum) and air-borne (Phytophthora infestans, Hyaloperonospora parasitica) diseases. Eur. J. Plant Pathol. 127, 465–481. doi: 10.1007/s10658-010-9612-2
van Elsas, J. D., Chiurazzi, M., Mallon, C. A., and Salles, J. F. (2012). Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. U.S.A. 109, 1159–1164. doi: 10.1073/pnas.11093261
Wallenhamar, A. C. (2010). Monitoring and control of Plasmodiophora brassicae in spring oilseed brassica crops. Acta Hort. 867, 181–190. doi: 10.17660/ActaHortic.2010.867.23
Wallenhammar, A. C. (1996). Prevalence of Plasmodiophora brassicae in a spring oilseed rape growing area in central Sweden and factors influencing soil infestation levels. Plant Pathol. 45, 710–719. doi: 10.1046/j.1365-3059.1996.d01-173.x
Wallenhammar, A. C., Almquist, C., Soderstroma, M., and Jonsson, A. (2012). Infield distribution of Plasmodiophora brassicae measured using quantitative real-time PCR. Plant Pathol. 61, 16–28. doi: 10.1111/j.1365-3059.2011.02477.x
Wang, R., Zhang, H., Sun, L., Qi, G., Chen, S., and Zhao, X. (2017). Microbial community composition is related to soil biological and chemical properties and bacterial wilt outbreak. Sci. Rep. 7:343. doi: 10.1038/s41598-017-00472-6
Wang, T., Hao, Y., Zhu, M., Yu, S., Ran, W., Xue, C., et al. (2019). Characterizing differences in microbial community composition and function between Fusarium wilt diseased and healthy soils under watermelon cultivation. Plant Soil 438, 421–433. doi: 10.1007/s11104-019-04037-6
Webster, M. A., and Dixon, G. R. (1991a). Boron, pH and inoculum concentration influencing colonization by Plasmodiophora brassicae. Mycol. Res. 95, 74–79. doi: 10.1016/S0953-7562(09)81363-4
Webster, M. A., and Dixon, G. R. (1991b). Calcium, pH and inoculum concentration influencing colonization by Plasmodiophora brassicae. Mycol. Res. 195, 64–73. doi: 10.1016/S0953-7562(09)81362-2
Wei, Z., Gu, Y., Friman, V. P., Kowalchuk, G. A., Xu, Y., Shen, Q., et al. (2019). Initial soil microbiome composition and functioning predetermine future plant health. Sci. Adv. 5:eaaw0759. doi: 10.1126/sciadv.aaw07
Weller, D. M., Raaijmakers, J. M., Gardener, B. B. M., and Thomashow, L. S. (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. phytopathol. 40, 309–348. doi: 10.1146/annurev.phyto.40.030402.110010
Xiong, W., Li, R., Ren, Y., Liu, C., Zhao, Q., Wu, H., et al. (2017). Distinct roles for soil fungal and bacterial communities associated with the suppression of vanilla Fusarium wilt disease. Soil Biol. Biochem. 107, 198–207. doi: 10.1016/j.soilbio.2017.01.010
Yang, H., Li, J., Xiao, Y., Gu, Y., Liu, H., Liu, Y., et al. (2017). An integrated insight into the relationship between soil microbial community and tobacco bacterial wilt disease. Front. Microbiol. 8:2179. doi: 10.3389/fmicb.2017.02179
Yao, Y. R., Tian, X. L., Shen, B. M., Mao, Z. C., Chen, G. H., and Xie, B. Y. (2015). Transformation of the endophytic fungus Acremonium implicatum with GFP and evaluation of its biocontrol effect against Meloidogyne incognita. World J. Microb. Biot. 31, 549–556. doi: 10.1007/s11274-014-1781-2
You, M. P., Rensing, K., Renton, M., and Barbetti, M. (2017). Modeling effects of temperature, soil, moisture, nutrition and variety as determinants of severity of Pythium damping-off and root disease in subterranean clover. Front. Microbiol. 8:2223. doi: 10.3389/fmicb.2017.02223
Zhang, H. Y., and Yang, Q. (2007). Expressed sequence tags-based identification of genes in the biocontrol agent Chaetomium cupreum. Appl. Microbiol. Biot. 74, 650–658. doi: 10.1007/s00253-006-0701-2
Zhang, Y., Cong, J., Lu, H., Yang, C., Yang, Y., Zhou, J., et al. (2014). An integrated study to analyze soil microbial community structure and metabolic potential in two forest types. PLoS ONE 9:e93773. doi: 10.1371/journal.pone.0093773
Zhao, M., Yuan, J., Zhang, R., Dong, M., Deng, X., Zhu, C., et al. (2020). Microflora that harbor the NRPS gene are responsible for Fusarium wilt disease-suppressive soil. Appl. Soil Ecol. 132, 83–90. doi: 10.1016/j.apsoil.2018.08.022
Zhou, D., Jing, T., Chen, Y., Wang, F., Qi, D., Feng, R., et al. (2019). Deciphering microbial diversity associated with Fusarium wilt-diseased and disease-free banana rhizosphere soil. BMC microbiol. 19:6. doi: 10.1186/s12866-019-1531-6
Keywords: clubroot, suppressive soil, soil chemical properties, microbial community composition, microbial network
Citation: Kang H, Lin Z, Yuan X, Shi Y, Xie X, Li L, Fan T, Li B and Chai A (2024) The occurrence of clubroot in cruciferous crops correlates with the chemical and microbial characteristics of soils. Front. Microbiol. 14:1293360. doi: 10.3389/fmicb.2023.1293360
Received: 13 September 2023; Accepted: 11 December 2023;
Published: 08 January 2024.
Edited by:
Md. Motaher Hossain, Bangabandhu Sheikh Mujibur Rahman Agricultural University, BangladeshReviewed by:
Andreas Westphal, University of California, Riverside, United StatesQiuxia Wang, Institute of Special Animal and Plant Sciences, Chinese Academy of Agricultural Sciences, China
Copyright © 2024 Kang, Lin, Yuan, Shi, Xie, Li, Fan, Li and Chai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ali Chai, Y2hhaWFsaUBjYWFzLmNu; Baoju Li, bGliYW9qdWl2ZkAxNjMuY29t
 Huajun Kang
Huajun Kang Zihan Lin1
Zihan Lin1 Xiaowei Yuan
Xiaowei Yuan Yanxia Shi
Yanxia Shi Lei Li
Lei Li Baoju Li
Baoju Li