- 1Institute for Poultry Science and Health, Guangxi University, Nanning, China
- 2Guangxi Key Laboratory for Polysaccharide Materials and Modifications, School of Marine Sciences and Biotechnology, Guangxi Minzu University, Nanning, China
Infectious bursal disease (IBD) classical virus strain (cIBDV) can cause morbidity and mortality in young chickens with severe long-term immunosuppression. However, since the emergence and widespread prevalence of very virulent strain (vvIBDV) in China from 1991, reports of cIBDV have become rare. A novel reassortant and recombinant strain GXYL211225 (genotype A1aB1a) with segment A originating from the classical strain (A1a) and segment B from the attenuated vaccine strain (B1a) was characterized in the study. Notably, segment A resulted from recombination between the cIBDV strains 150127-0.2 and Faragher52-70, expressing as a backbone from 150127-0.2, where a fragment located at the position of nucleotide (nt) 519-1 410 was replaced by the corresponding region of Faragher52-70. The infection of GXYL211225 caused mortality in SPF chicken embryos, despite lacking the critical amino acid (aa) residues 253H, 279 N and 284A associated with the cellular tropism, and induced significant cytopathic effect (CPE) on a wide range of cells, confirming its natural cell-adapted character. Furthermore, the challenge experiment of GXYL211225 was performed on the commercial Three-yellow chickens of 4-week-old, and with the vvIBDV HLJ-0504-like strain NN1172 and the novel variant (nv) IBDV strain QZ191002 as the comparison. All the challenged birds experienced reduced body-weight gain. QZ191002 infected birds showed no obvious clinical symptoms or mortality, while those of NN1172 and GXYL211225 showed typical IBD symptoms and resulted in 20% (2/10) and 10% (1/10) of mortality rates, respectively. At 7 days post-challenge (dpc), the damages of bursal of Fabricius (BF) varied among groups, with NN1172 causing the most severe lesions, followed by GXYL211225, and then QZ191002. It was also found that the pathogenicity was correlated positively with the viral load, aligning with the histopathological severity in BF. The study confirms the rapid and diverse evolution of the re-emerged classical strains in the field and emphasizes the need to monitor the changes of IBDV on both the genetic and pathogenic aspects for the effective control of the disease.
Highlights
- A naturally occurring reassortant and recombinant classical strain GXYL211225 (genotype A1aB1a) was isolated and characterized for the first time.
- Segment A of GXYL211225 is a result of recombination between the classical strains 150127-0.2 (major parent) and Faragher52-70 (minor parent), and with segment B reasserting with the attenuated vaccine strain.
- GXYL211225 exhibits an extensive cell-tropism and has been confirmed to be a naturally occurring cell-adapted strain.
- The virulence of GXYL211225 was intermediate between that of the vvIBDV NN1172 and nvIBDV QZ191002, causing a 10% mortality rate and severe immune organ damages in chickens.
1 Introduction
Infectious bursal disease (IBD) is an acute, highly contagious, immunosuppressive poultry disease caused by infectious bursal disease virus (IBDV) (Muller et al., 2003; Alkie and Rautenschlein, 2016). IBDV mainly damages the avian central immune organ, the bursa of Fabricius (BF), and causes varying degrees of lesions in other immune organs (Sharma et al., 2000; Dulwich et al., 2018). IBDV belongs to the Birnaviridae family of Avibirnavirus and is a non-enveloped icosahedral virus composed of two segments (segment A and segment B) of double-stranded RNA (Dobos et al., 1979; Muller et al., 1979). Segment A contains two partially overlapping open reading frames (ORFs), where the small ORF encodes the VP5 protein, and the large ORF encodes the polyproteins VP2-VP4-VP3, which is processed into VP2, VP4, and VP3 through autolytic cleavage (Birghan et al., 2000; Coulibaly et al., 2005). VP2 serves as a major structural protein and a host-protective immunogen that is closely related to IBDV cell tropism, antigenic variation and virulence (Brandt et al., 2001; Letzel et al., 2007; Jackwood et al., 2008; Alfonso-Morales et al., 2013). Segment B encodes only the VP1 protein possessing RNA-dependent RNA polymerase activity, which plays an important role in viral genome replication, translation and virulence (Boot et al., 2005; Wang W. et al., 2021).
Currently, IBDV is classified into two serotypes (I and II), but only serotype I has been confirmed to be pathogenic in chickens (Jackwood et al., 1982; Ismail et al., 1988). Traditionally, serotype I IBDV can be further divided into classical strain (cIBDV), very virulent strain (vvIBDV), antigenic variant IBDV (avIBDV), and attenuated vaccine strain (attIBDV) (Rautenschlein et al., 2003; Jackwood et al., 2018; Islam et al., 2021; Wang et al., 2022b). The cIBDV in China was first reported in 1979, causing huge economic losses in the poultry industry at that time (Zhou et al., 1982). Since the emergence and widespread prevalence of vvIBDV in 1991, reports of classical strains in China have become rare (Zhang et al., 2022). In 2016, novel variant strain (nvIBDV) emerged in multiple regions of China, causing subclinical symptoms in chicken flocks without apparent morbidity and mortality (Fan et al., 2019; Xu et al., 2020; Hou et al., 2022; Huang et al., 2022; Wang et al., 2022a,b). Notably, since 2020, cIBDV infections have been re-emerged in several poultry-producing provinces in southern China, with their prevalence increasing year by year (Author unpublished data). Up to now, China’s field situations demonstrate the co-circulation of various strains including vvIBDV, nvIBDV and cIBDV, posing new challenges to the diagnosis, prevention and control of IBD in clinical practice.
The evolution of IBDV (segmented dsRNA viruses) primarily involves gene mutations, segment reassortment and intra-segment homologous recombination, which play a crucial role in the emergence and dissemination of the novel variant/reassortant/recombinant strains (He et al., 2009, 2014, 2016). The co-circulation of different types of strains in the field and the widespread use of live attenuated vaccines have increased the potential for reassortment /homologous recombination among different IBDV strains (He et al., 2009; Wang et al., 2022b). Studies suggest that when two different types of viruses infect the same cell, their genome segments can mispair and exchange, resulting in the generation of new types viruses (McDonald et al., 2016). This process allows viruses to acquire new pathogenic characteristics and antigenic combinations, facilitating their immune escape and cross-species transmission (Pérez-Losada et al., 2015). In this study, we comprehensively characterized, for the first time, the molecular characters of a naturally occurring reassortant and recombinant cIBDV strain GXYL211225, and evaluated its pathogenicity in Three-yellow chickens, number one breed of Yellow-chicken in southern China.
2 Materials and methods
2.1 Clinical case, virus isolation, and identification
In December 2021, a poultry farm in Guangxi province experienced suspected IBDV infection in a vaccinated flock of 35-day-old Yellow-chicken. Clinical signs in the affected birds included severe depression, ruffled feathers, and watery or white diarrhea. Necropsy revealed severe hemorrhaging of BF and displayed typical IBD pathological features like muscular hemorrhaging, purple grape shaped BF. Laboratory examinations were conducted on the BF tissue samples collected from the diseased birds. Out of 12 samples detected, a total of 5 samples were found positive (5/12) of IBDV by the RT-PCR detection (Wei et al., 2004). The tissue suspensions of the positive sample were sterile-filtered through a 0.22 μm filter and then used to inoculate SPF chicken embryos for virus isolation as the previous description (He et al., 2012). The isolates, using the tissue suspensions of the infected embryos were identified through the amplification and sequencing analysis of the VP1 and VP2 genes of IBDV, following the method descripted previously (He et al., 2014).
2.2 Cell culture, purification, propagation, growth characteristics, and virus titer determination
The isolate was inoculated onto the monolayers of chicken embryo fibroblast (CEF), chicken fibroblast cell line (DF1) and chicken macrophages cell line (HD11) cultured cells, and continually passaged for 3 passages to assess the virus’s growth characteristics, such as cytopathic effects (CPE) and plaque formation (He et al., 2019). Following the method outlined by Wang et al. (2022b), the virus was purified by the plaque technique and then propagated in CEF. Briefly, CEF cells were seeded onto a six-well plate at a density of 5 × 105 cells per well. After 80–85% cells confluent, the medium was removed and the monolayer of cells were infected with 500 μL of each of the 10-fold series virus diluted in DMEM without fetal bovine serum (FBS). After 1 h of virus adsorption at 37°C, the cells were washed twice and then add 2 mL of overlay medium (1 volume of DMEM with 10% FBS and 1 volume of 2% agar) to each well. The overlay medium in the plates were allowed to set for 30 min to solidify, and then inverted the plates and continue incubating at 37°C. The second overlay (containing 0.03% neutral red) was added at 1–2 days post-infection (dpi) and further incubated at 37°C for 24 h to observe the formation of plaques. The titer of the harvested viruses of the purified clone was determined and expressed as 50% tissue culture infectious dose (TCID50) (He et al., 2016).
2.3 RNA isolation, full-genome amplification, and sequence analysis
Total RNA was isolated from purified viruses using Spin Column Animal Total RNA Purification Kit (Sangon Biotech, China) according to the manufacturer’s instructions and the RNA concentration was determined using a BioDrop ultramicro spectrophotometer (BioDrop, England). Approximately 2 μg RNA was reverse-transcribed into cDNA for each sample using the All-In-One 5X RT MasterMix Kit (abm, Canada) according to the manufacturer’s protocol. Using the obtained cDNA as a template, PCR amplification of the full-genome was carried out with primers as described in previously study (Wang et al., 2022b). The PCR products were purified by gel recovery and ligated into the pMD18-T vector, and then transformed into the E. coli DH5α competent cells. At least three individual positive clones of each fragment were selected for sequencing (BGI, Guangzhou, China). Analysis of the nt homology and amino acid (aa) alignment for both segment A and B of the isolate were conducted using MEGA 6.0 software. Furthermore, maximum likelihood (ML) phylogenetic trees were constructed using the MEGA 6.0 program with the best-fit model (GTR + I + G). The full-genome of the segments A and B of the isolate (named GXYL211225) were submitted to GenBank (accession No. OR523680 and OR523681).
2.4 Genome recombination analysis
Two different recombination detection methods were used to identify the potential recombination events in the full-genome of GXYL211225 as the previous report (Wang et al., 2020a, 2022a). Initially, both the two datasets, A and B, were tested by RDP software, which is regarded as reliable when at least four out of seven distinct algorithms (RDP, GENECONV, Bootscan, MaxChi, Chimera, SiScan, and 3Seq) detect significant recombination signals (p < 10E-6). Subsequently, potential recombination events were further validated using similarity plots in Simplot.
2.5 Pathogenesis experiments
A total of 40 1-day-old non-vaccinated Three-yellow chickens were purchased from a local commercial poultry farm. The birds were randomly divided into 4 groups (Groups 1, 2, 3, 4; n = 10) and housed in separate isolators, where they had unrestricted access to water and feed. At 28 days, serum samples were collected from all birds for ELISA to determine the antibody levels and ensure the exclusion of the maternal antibody interference during the subsequent infection (Wang et al., 2020b). The birds of Group 1, 2, and 3 were orally administered with 105 TCID50/0.5 mL viruses of vvIBDV strain NN1172 (genotype A2B3) (He et al., 2014), nvIBDV strain QZ191102 (genotype A2dB1b) (Wang et al., 2022b) and the isolate GXYL211225 (genotype A1aB1a) (cIBDV strain), respectively, while Group 4 received 0.5 mL of PBS buffer. Daily clinical symptom observations were recorded, and clinical symptom were quantified by the mean symptom index (MSI) score (Nouen et al., 2012), which is based on four degrees: 0 = “lack of signs,” 1 = “typical IBD signs (ruffled feathers) conspicuous in quiet bird only, the bird stimulated by a sudden change in environment (light, noise, or vicinity of experiment observer) appears normal, motility is not reduced”; 2 = “typical IBD signs conspicuous even when bird is stimulated, dehydration is apparent, motility may be slightly reduced” and 3 = “typical severe IBD signs with prostration or death.” At 7 days post-challenge (dpc), all birds were euthanized and necropsied. The BF was taken and weighed, and the bursa/body weight ratio index (BBIX) was calculated, a BBIX value of less than 0.7 was considered as atrophy. Half of the bursa tissue from the birds was fixed in 4% neutral buffered formaldehyde for histopathological examination and quantified according to Skeeles’ scale for histopathological bursal lesion scores (Skeeles et al., 1979), which is based on five degrees: 0 = no lesions; 1 = mild scattered cell depletion in a few follicles; 2 = moderate, one-third to one half of the follicles have atrophy or depletion of cells; 3 = diffuse, atrophy of all follicles; and 4 = acute inflammation and acute necrosis typical of IBD. While the other half of the bursa tissue was used for RNA extraction.
2.6 Quantitative RT-PCR assay targeting the IBDV
qRT-PCR assay targeting the IBDV was conducted on the extracted RNA samples using the previously descripted method (He et al., 2016). Briefly, the qRT-PCR reaction was conduct with the primer IBDV-F 5′-GGAGCCTTCTGATGCCACA-3′ and IBDV-R 5′-ATTGTAGGTCGAGGTCTCTG-3′. Firstly, total RNA was isolated from each sample using Spin Column Animal Total RNA Purification Kit (Sangon Biotech, China) according to the manufacturer’s instructions and the concentration was determined using a BioDrop ultramicro spectrophotometer (BioDrop, England). Secondly, approximately 2 μg RNA was reverse-transcribed into cDNA for each sample using the All-In-One 5X RT MasterMix Kit (abm, Canada) according to the manufacturer’s protocol. Finally, the qRT-PCR analysis was performed in a reaction volume of 20 μL containing 10 μL of 2 × ChamQ Universal SYBR qPCR Master Mix, 0.4 μL of each primer, 7.2 μL of ddH2O and 2 μL of cDNA. Thermal cycling was performed by using two-step real-time PCR at 95°C for 30 s, followed by 40 cycles of 95°C for 10 s and 55°C for 30 s. Fluorescence values were read and recorded for RT-PCR analysis.
2.7 Statistical analysis
All data in this study were processed using GraphPad Prism 8.0 software and presented as mean ± standard deviation (SD). Statistical differences among the groups were evaluated using one-way analysis of variance (ANOVA). To compare the differences among treatments, Tukey’s multiple comparisons test was used. Results with p values of < 0.05 *, p < 0.01 **, p < 0.001 ***, p < 0.0001 **** were considered statistically significant.
3 Results
3.1 Isolation and identification of a cIBDV strain from clinical case
The isolate demonstrated the capability to induce mortality in SPF chicken embryos as early as the first passage, showing severe subcutaneous hemorrhaging within 36 h post-inoculation. Amplification and sequencing analysis of the VP1 and VP2 genes of the isolate confirmed that the isolated strain in the clinical samples belonged to cIBDV (Figure 1) and was named GXYL211225.
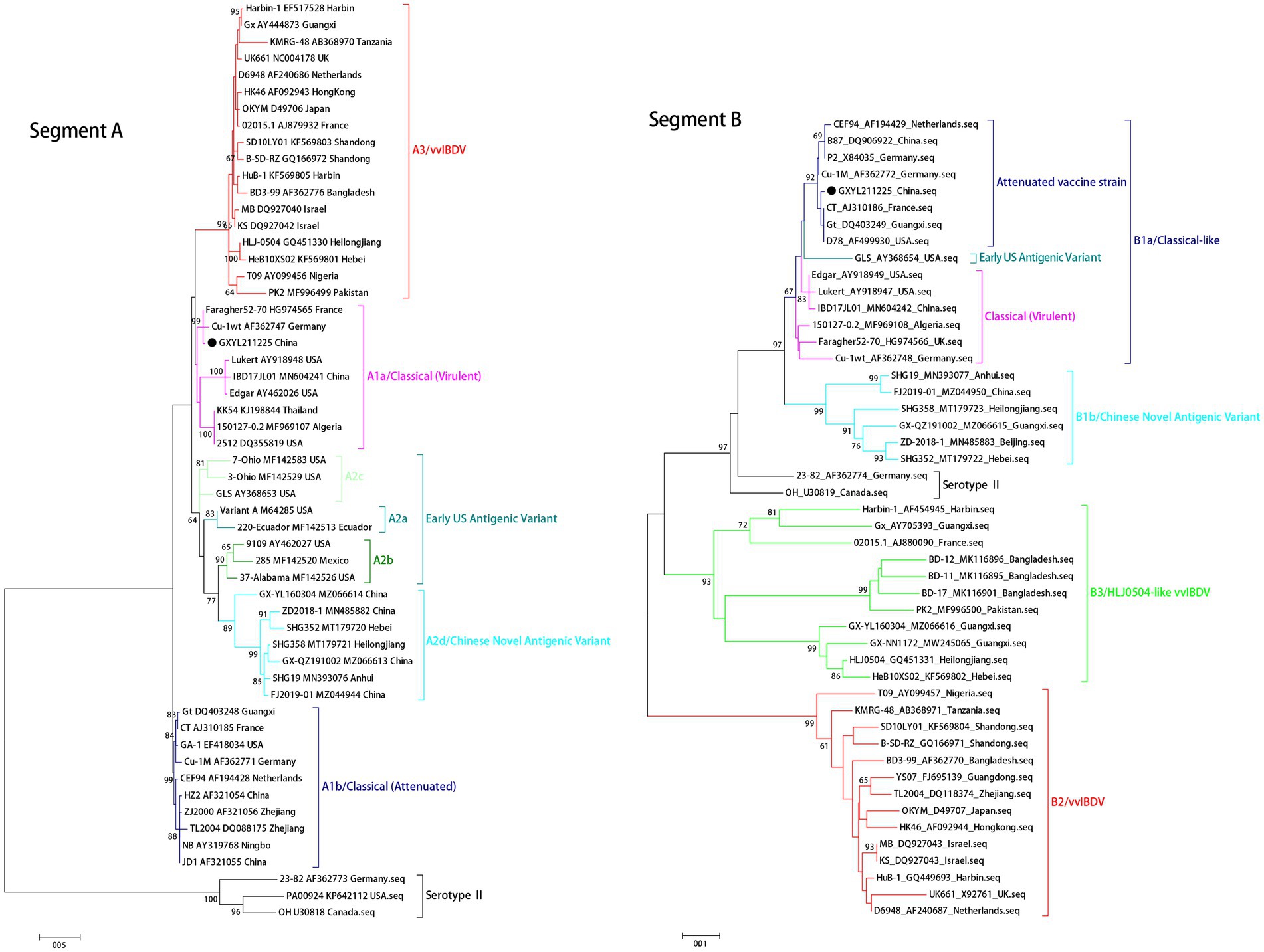
Figure 1. Maximum likelihood tree (ML tree) was constructed using the datasets of 55 vVP2 sequence (474 bp, nt 737–1,210) and 48 VP1 sequence (767 bp, nt 756–1,522) by the MEGA 6.0 software with the best-fit model (GTR + I + G) with 1,000 ultrafast bootstrap replicates. The strain GXYL211225 was labeled with ● mark in the figure.
3.2 GXYL211225 was further confirmed to be a natural reassortant strain by the full-length genome sequencing and analysis
The ML phylogenetic tree of the full-length segments A and B of GXYL211225 were shown in Figure 2, providing further evidence that this isolated is a natural reassortant strain (A1aB1a) according to the newly proposed unified classification and nomenclature system (Jackwood et al., 2018; Islam et al., 2021; Wang Y. L. et al., 2021; Wang et al., 2022b). The phylogenetic tree of the segment A revealed that GXYL211225 clustered with the classical virulent strains (A1a) like Faragher52-70, 150127–0.2 and STC, demonstrating the typical characteristics of the A1a classical strains. Notably, key aa residues associated with cIBDV were observed in the VP5 protein, where critical sites 3G, 45G, 74I, 87G, 125S matched those of the classical strain 150127–0.2 originating from Algeria (Table 1). Similar cases can be observed in the polyproteins VP2-VP4-VP3, with the exception of seven unique aa residues mutations 10R, 76G, 142S, 242I, 294 L, 802 V, and 942 T, all the characteristic aa residues in VP2-VP4-VP3 were also completely consistent with the classical strain 150127–0.2 (Table 1).
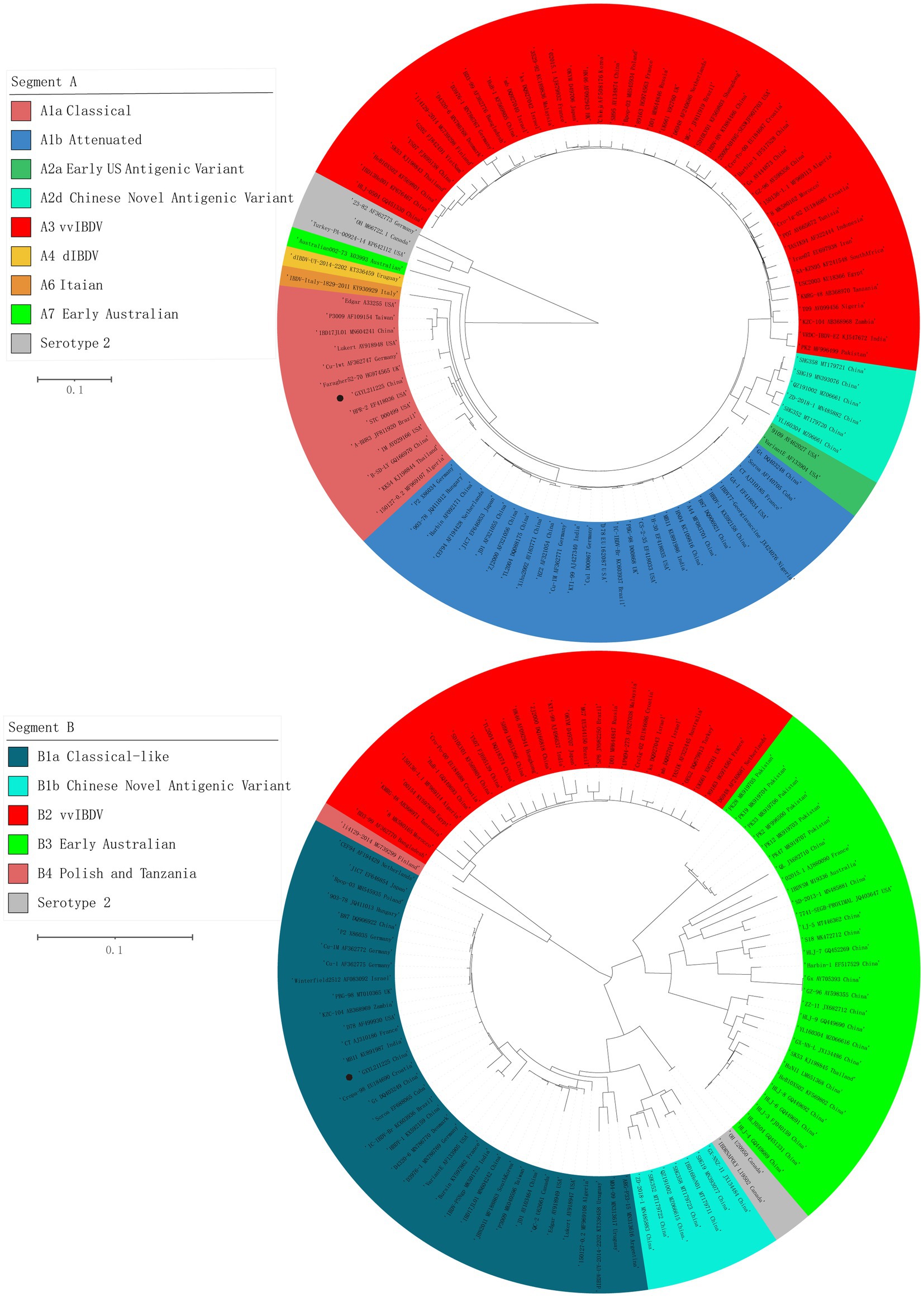
Figure 2. Maximum likelihood tree (ML tree) was constructed using the datasets of 102 strains of IBDV with both the full-length segments A (3,260 bp) and B (2,827 bp) by the MEGA 6.0 software with the best-fit model (GTR + I + G) with 1,000 ultrafast bootstrap replicates. The strain GXYL211225 was classified as A1aB1a and was labeled with ● mark in the figure.

Table 1. The differences of the aa residues among the cIBDV, attIBDV, vvIBDV, early US avIBDV and nvIBDV isolates in VP5, polyprotein and VP1.
The phylogenetic tree of segment B sequence showed that GXYL211225 clustered with the classical-like (B1a) attenuated vaccine strain (Figure 2). The segment B exhibited close similarity to the commonly used attenuated vaccine strains in China, such as B87 and CT, sharing all characteristic aa residues 4I, 13 T, 61 V, 145 N, 146E, 147G, 287 T, 508R, 511R, 546 L, 646G, and 687S, except for two unique aa residues 251R and 492D (Table 1). Comprehensive analysis of the full-genome segments A and B suggested that GXYL211225 is a naturally occurring reassortant strain, which segment A originates from the classical strains (A1a) and segment B originates from the attenuated vaccine strains (B1a).
3.3 Wide cell tropism and prominent cytopathic effects in different cells
The CPEs induced by the GXYL211225 strain on the commonly used cell lines CEF, DF1, and HD11 were depicted in Figure 3. GXYL211225 inoculated on the CEF, DF1, and HD11 monolayer cells all grown well after three blind passages, and prominent CPEs were evident as early as 36 h post-inoculation. The hallmark characteristics of CPEs included rounding, enhanced refractive index, cell rupture, and the detaching of the cells from the monolayer. As expected, the cells in the control group growth normally and no CPE was observed. It is noteworthy that significant CPEs were observed in the first passage of CEF monolayer infected with the isolate, and this pronounced effect persisted through the subsequent passages.
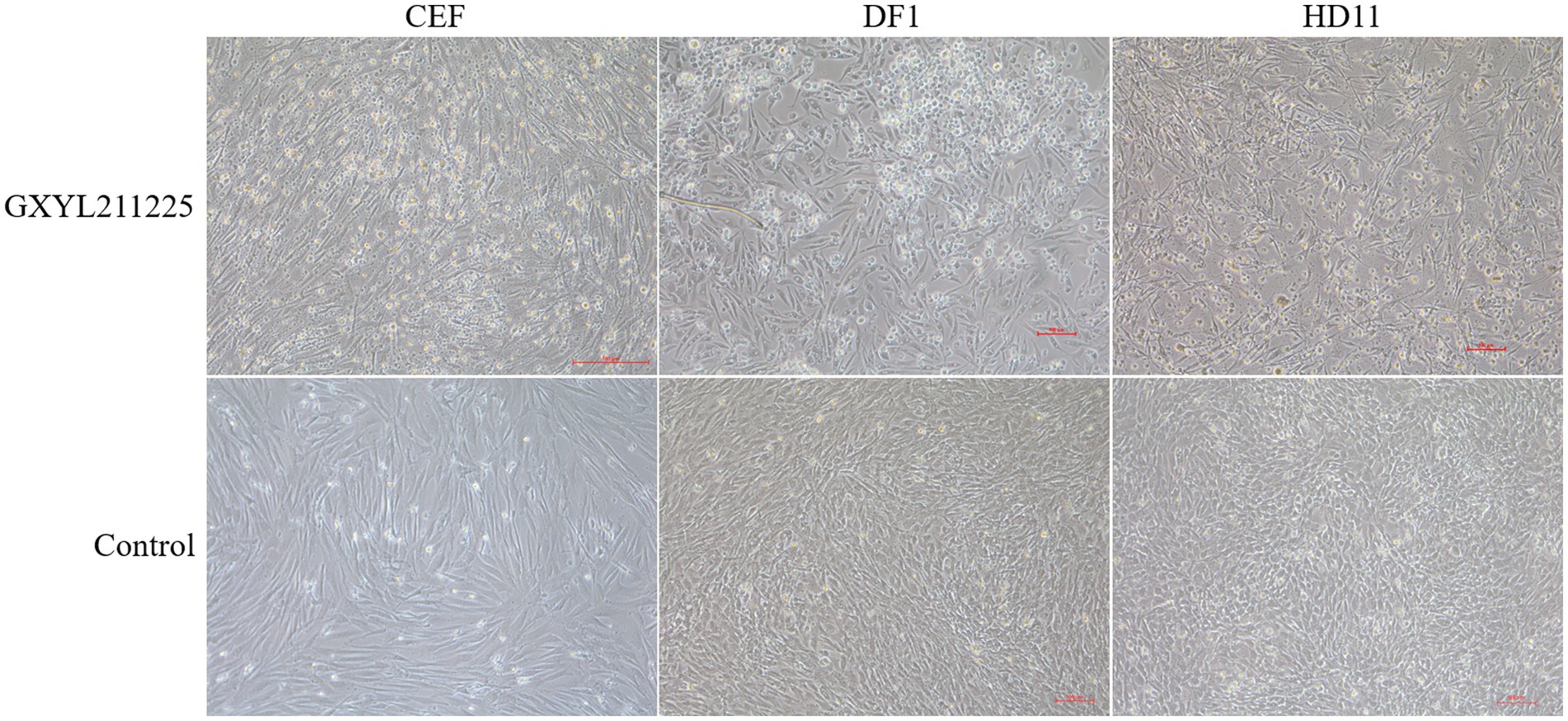
Figure 3. The CPEs of CEF, DF1, and HD-11 monolayer cells inoculated with the isolate GXYL211225 at 36 h post-infection.
3.4 Detection of significant recombination events in segment A of GXYL211225
Five out of the seven different algorithms (Figure 4D) detected a significant recombination event (p < 10E-6) within the segment A of GXYL211225. Segment A is a result of recombination between the classical strain 150127–0.2 (major parent) and Faragher52-70 (minor parent) (Figures 4A,B). The recombination breakpoints were identified at the position of nt 519–1,410, and manifested as a backbone originating from the classical strain 150127–0.2, wherein a fragment located at the position of nt 519–1410 is replaced by the corresponding part from Faragher52-70 strain (Figure 4C).
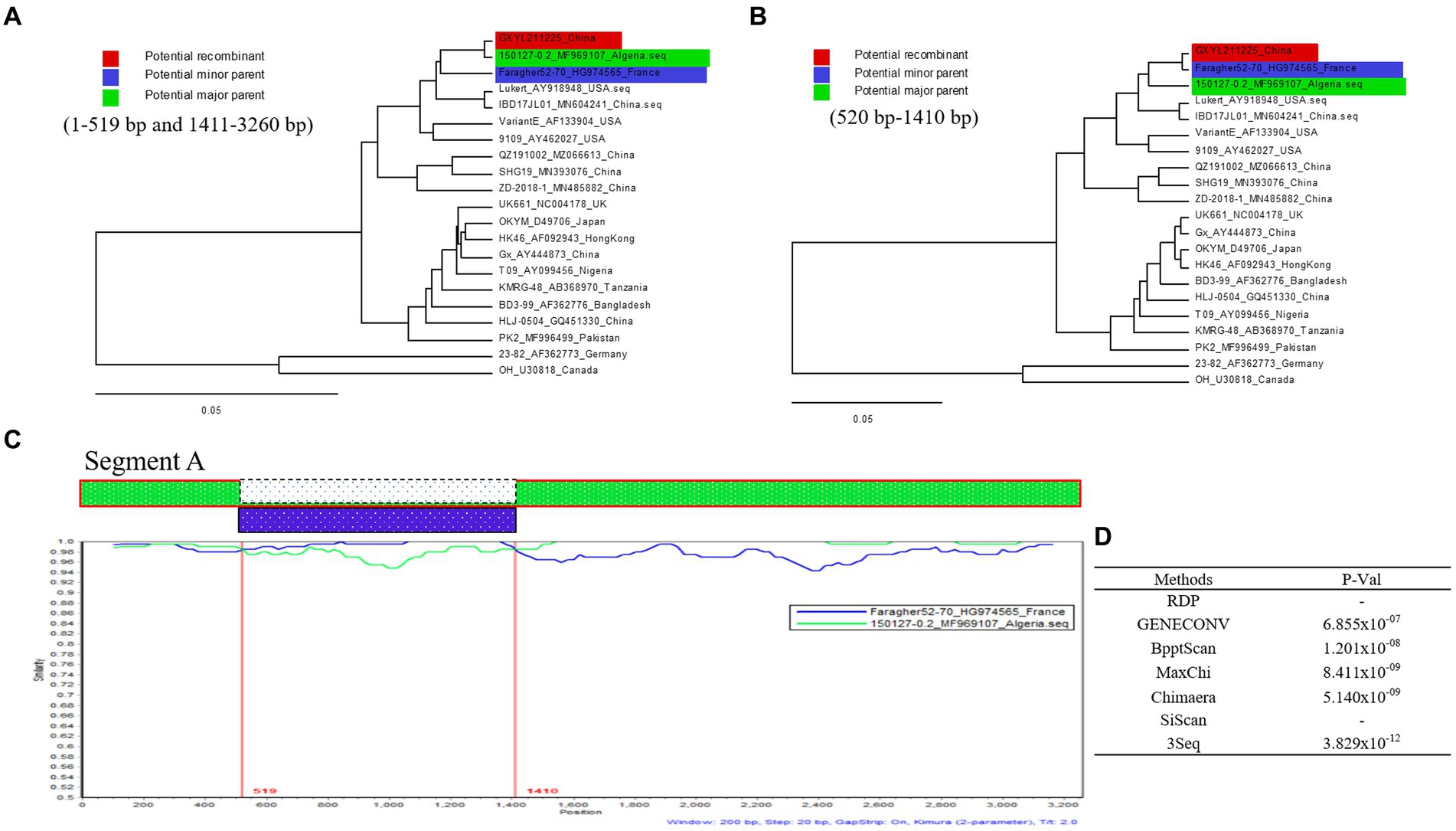
Figure 4. Recombination analysis of the GXYL211225 strain. (A,B) Maximum-likelihood phylogenetic profiles of separate regions in the segment A of reference strain (of 21 strains, 3,260 bp). Panel (A) represents the 5′ region (nt 1–519) and 3′ region (nt 1,411–3,260) sections of segment A. Panel (B) represents the region (nt 520–1,410) section of segment A; (C) Comparison of the similarity plots of segment A scales (query) with 150127-0.2 (green) and Faragher52-70 (blue); (D) The P-Val of seven different algorithms.
3.5 Naturally occurring reassortant and homologous recombinant strain GXYL211225 cause mortality and immune organ damages in three-yellow chickens
The pathogenicity of GXYL211225 in Three-yellow chickens was assessed and compared to the HLJ-0504-like vvIBDV strain NN1172 and the nvIBDV strain QZ191002 (Figure 5). All serum samples tested negative by ELISA, indicating that the maternal antibody has been decreased to a very low level at 28d. During the whole experiment, no obvious clinical symptoms or mortality was observed in the control group and the QZ191002 group (Figure 5A). In the NN1172 and GXYL211225 groups, typical IBD symptoms such as disheveled feathers, depression, diarrhea were observed, with mortality rates of 20% (2/10) and 10% (1/10), respectively (Figures 5B,D). Compared to the control group, birds in all the challenged groups showed significant body-weight loss (Figure 5C). Upon necropsy, bursal atrophy in all the challenged groups (Figures 5E,H), and typical hemorrhages in pectoral and thigh muscle were observed (Figure 5D). Histopathological examination of bursa sections revealed the most severe lesions happened in the NN1172 group, characterized by loss of follicular structural contour and replacement by the proliferated connective tissue and reticular cells; followed by the GXYL211225 group, showing moderate to severe lymphocyte necrosis and loss of follicular structural contours; and then the QZ191002 group with moderate lesions (Figures 5F,H,I). Viral loads (virus copy numbers) detected in the BF tissue were consistent with the histopathological changes (Figures 5F,G). These data taken together confirm that the naturally occurring reassortant and recombinant strain GXYL211225 exhibited lower pathogenicity in chickens when compared to vvIBDV, and higher pathogenicity when compared to nvIBDV.
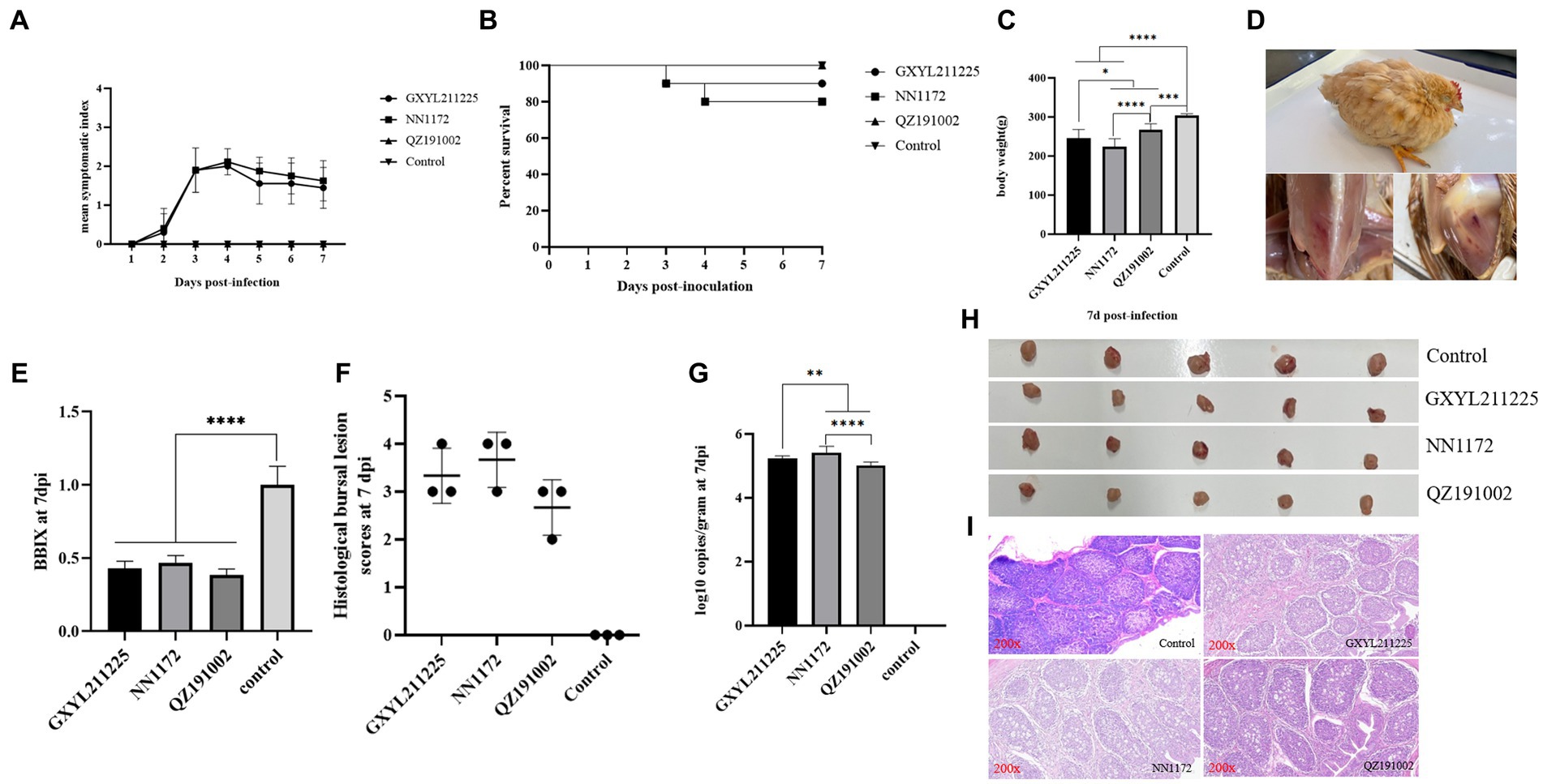
Figure 5. Pathogenic analysis of GXYL211225, QZ191002, and NN1172 strains in Three-yellow chickens. (A) Mean symptom index (MSI) scores; (B) Survival rates; (C) Body weights; (D) Typical clinical symptoms and pathological changes challenged with GXYL211225; (E) The bursa/body weight index (BBIX) ratio; (F) Histopathological bursal lesion scores (HBLS); (G) Viral loads of IBDV in the BF tissues; (H) The BF images at 7 dpc; (I) Histopathological changes of BF at 7 dpc (200×). Results with p values of < 0.05 *, p < 0.01 **, p < 0.001 ***, p < 0.0001 **** were considered significant different.
4 Discussion
In China, the infection caused by cIBDV strain was first reported in 1979, resulting in significant economic losses to the poultry industry at that time. However, with the emergence of vvIBDV in 1991 (Zhang et al., 2022), there has been an increasing focus on these strains in China, while reports related to cIBDV have been scarce. The research conducted by our group revealed that since 2020, classical strains have gradually re-emerged in southern China and have undergone different types of reassortment events with locally prevalent strains (unpublished data). Subsequently, classical strains were also reported in Hubei province of central China (Lian et al., 2022). These classical strains have caused significant economic losses due to their widespread circulation in the field. Furthermore, the use of attenuated live vaccines has substantially increased the likelihood of reassortment and recombination events involving the classical strains. Therefore, considering the re-emergence of classical strains and their potential to undergo reassortment and recombination with locally circulating strains, a continuous and in-depth understanding of the genetic variation and pathogenic characteristics of this type of strains is of paramount importance for better control of the disease in the poultry industry.
Reassortment is a crucial mechanism in the evolution of viruses, contributing to the generation of new strains in viruses with segmented genomes (Hon et al., 2008; He et al., 2009; Pérez-Losada et al., 2015; McDonald et al., 2016; Feng et al., 2022; Wang et al., 2022a,b). Studies confirmed that isolate GXYL211225 was a naturally occurring reassortant and recombinant strain of cIBDV. Its segment A belongs to the classical strain (A1a), while segment B derives from the classical-like (B1a) attenuated vaccine strain (Figure 2). Further analysis of the recombination in its segment A reveals a significant recombination event was found. Specifically, segment A is a result of recombination between the classical strain 150127–0.2 (major parent) and Faragher52-70 (minor parent), characterized by a backbone originating from the classical strain 150127–0.2 (Abed et al., 2018), where a fragment located at nt 519–1410 is replaced by the corresponding part of the Faragher52-70 strain (Figure 4). Two factors can explain this case: firstly, the gradual prevalence of strains like Faragher52-70 in southern China since 2020, including isolates such as NN200430, NN200519, and YL200617 etc., have provided opportunities for the recombination; Secondly, it’s possible that the 150127–0.2 strain, belonging to cIBDV, may have altered its antigenicity after recombination with Faragher52-70 strain at nt 519–1410, resulting from the mutations of the critical antigenic activity residues V242I and I294L, thereby escaped the immune protections from the cIBDV vaccines, and also the widely used vvIBDV vaccines (resulting from the mutations of the critical antigenic activity residues A222P, I256V, I294L, and S299N) over the past two decades (Table 1). Additionally, the segment B of the GXYL211225 isolate originated from the attenuated vaccine strain, exhibiting a high homology with the commonly used vaccine strains in China (Figure 1). Considering that the isolate GXYL211225 was obtained from a vaccinated flock, it is speculated that this strain may have resulted from the reassortment between the re-emerging classical strains, observed in recent years, with the widely used attenuated vaccine strains in clinical practice. The identification of this strain indicated that classical strains are undergoing rapid evolution through multiple mechanisms, including not only the common site mutations but also the simultaneous processes of genetic reassortment and homologous recombination. The emergence of these novel-featured strains may contribute to their ability to escape the protection offered by current vaccines, and further immunization-challenge experiments will be necessary to validate this in the future.
The diversity-driven evolution of viruses contributes to changes in aspects such as pathogenicity and cell tropism, allowing newly emerging viruses to adapt to the new hosts and environments for better survival (Pérez-Losada et al., 2015; McDonald et al., 2016). The study revealed that GXYL211225 was able to cause death in all SPF chicken embryos within 36 h post-inoculation even just in the first passage, with severe subcutaneous hemorrhage lesion. Compared to the previous reported strains that required 2–3 continuous passages on chicken embryos to induce significant lesions and death (He et al., 2016; Chen et al., 2018), the ability of GXYL211225 to cause embryos pathogenic lesions and death without the need of adaptation suggests the presence of new and uncharacterized characters. Previous studies have confirmed that the crucial aa residues 253H, 279 N and 284A in VP2 are associated with IBDV’s cell tropism (Brandt et al., 2001; Qi et al., 2009; Ben Abdeljelil et al., 2014; Noor et al., 2014), considered as determining factors for the cell tropism. Notably, GXYL211225 exhibited mutations H253Q, N279D, and A284T at these crucial cell tropism-related aa residues in VP2, yet still displayed wide cell tropism (Figure 3). This suggests the presence of other crucial aa residues impacting IBDV’s cell tropism that have yet not to be full characterized. Increasing evidence indicates that IBDV’s cell tropism is a result of interactions among multiple genes or crucial aa residues within its genome. Besides 253H, 284A, and 279 N in VP2, aa residues like 222P and 256 V have also been implicated in IBDV’s cell tropism (Qi et al., 2013, 2016), which is consistent with the characteristics of GXYL211225. Additionally, mutations at critical aa residues Y680C, N685K, S715P, D751H, V990A, and A1005T in VP4 and VP3 proteins of GXYL211225 also contribute to the virus adaptation to cells. This is consistent with the previous reports of aa mutations associated with cell adaptation to the attenuation Gt strain obtained by continuous passage in the embryos and cells from vvIBDV GX strain (Wang et al., 2007, 2010). Furthermore, VP5 protein has also been found to impact the replication efficiency and pathogenicity of IBDV in vitro (Qin et al., 2009, 2010). Recent studies suggest that VP1 protein contributes to IBDV’s replication and virulence (Boot et al., 2005; Escaffre et al., 2013; Gao et al., 2018), with mutations like V4I and P687S enhancing the replication capacity in vitro (Yu, 2011; Yu et al., 2013; Wang W. et al., 2021). Factors associated with cell tropism remain intricate, and the extensive cell tropism exhibited by this newly emerged recombinant strain requires further investigation in the future.
Pathogenicity experiments confirmed that GXYL211225 exhibited stronger pathogenicity in the SPF chicken embryos and the commercial Three-yellow chickens compared with the nvIBDV strain QZ191002. Throughout the experiment period, the birds in QZ191002 challenged group and the control group showed no obvious clinical symptoms, as the previous reports (Fan et al., 2019; Chen et al., 2022; Wang et al., 2022b). In contrast, in the vvIBDV strain NN1172 and GXYL211225 challenged groups, birds exhibited mortality rates of 20% (2/10) and 10% (1/10), respectively (Figure 5). Infection of GXYL211225 resulted in similar symptoms as those of NN1172, characterized by ruffled feathers, watery white feces, reduced weight, hemorrhage in pectoral and thigh muscle, and BF atrophy (BBIX<0.7). Previous studies have shown that the virulence can also change when IBDV undergoes reassortant and/or recombination occurs in the field (Hon et al., 2008; He et al., 2009; Jackwood et al., 2011; Lu et al., 2015; Wu et al., 2020; Feng et al., 2022; Nooruzzaman et al., 2022; Wang et al., 2022b). Compared to the earlier cIBDV strain Faragher52-70 as previously reported (Bulow et al., 1985), the isolated GXYL211225 exhibited a lower mortality rate and significantly less BF damage. This may be attributed to the fact that the B-segment of this isolated strain is derived from an attenuated vaccine strain (He et al., 2016; Wang W. et al., 2021). With the emergence of GXYL211225 and the strains of similar recombination and reassortment, this type of strains may have the potential to escape the protections of the currently used vaccines (vvIBDV or/and attIBDV), posing a significant threat to the industry in the future.
In conclusion, this study isolated and characterized a naturally occurring classical IBDV strain GXYL2111225 (genotype A1aB1a) with unique recombination and reassortment events for the first time. This classical IBDV strain with distinct molecular characteristics exhibits extensive cell-tropism. Its virulence was intermediate between those of the vvIBDV strain NN1172 and the nvIBDV strain QZ191002, causing a 10% mortality rate and severe immune organ damages in the commercial chickens. Furthermore, our research findings confirmed the rapid and diverse evolution of the re-emerged classical strains in China and emphasizes the need to monitor and characterize the evolution of IBDV in the field, including in the aspects of both genetics and pathogenicity, for the effective control of the disease.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov/; OR523680-OR523681.
Ethics statement
Animal experiments in this study were approved by the Animal Experimental Ethical Committee of Guangxi University (GXU-2022-314). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
WW: Data curation, Writing – original draft. JS: Data curation, Writing – original draft. YZ: Investigation, Writing – original draft. YQ: Investigation, Writing – original draft. WZ: Formal analysis, Writing – original draft. ZW: Formal analysis, Writing – original draft. TN: Formal analysis, Writing – original draft. SH: Formal analysis, Writing – original draft. YC: Formal analysis, Writing – original draft. RK: Formal analysis, Writing – original draft. XH: Writing – review & editing. PW: Conceptualization, Funding acquisition, Methodology, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (31660717, 32160824, 31560706), the Guangxi Program for Modern Agricultural Industry Technical System Construction-Chicken Industry (nycytxgxcxtd-19-03), the Innovation Project of Guangxi Graduate Education (YCBZ2022034), and the Guangxi Special Funding on Science and Technology Research (AA17204057).
Acknowledgments
The manuscript was kindly reviewed by Richard Roberts, Aurora, CO 80014, USA.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abed, M., Soubies, S., Courtillon, C., Briand, F. X., Allée, C., Amelot, M., et al. (2018). Infectious bursal disease virus in Algeria: detection of highly pathogenic reassortant viruses. Infect. Genet. Evol. 60, 48–57. doi: 10.1016/j.meegid.2018.01.029
Alfonso-Morales, A., Martínez-Pérez, O., Dolz, R., Valle, R., Perera, C. L., Bertran, K., et al. (2013). Spatiotemporal phylogenetic analysis and molecular characterisation of infectious bursal disease viruses based on the VP2 hyper-variable region. PLoS One 8:e65999. doi: 10.1371/journal.pone.0065999
Alkie, T. N., and Rautenschlein, S. (2016). Infectious bursal disease virus in poultry: current status and future prospects. Vet. Med. (Auckl) 7, 9–18. doi: 10.2147/VMRR.S68905
Ben Abdeljelil, N., Khabouchi, N., Kassar, S., Miled, K., Boubaker, S., Ghram, A., et al. (2014). Simultaneous alteration of residues 279 and 284 of the VP2 major capsid protein of a very virulent infectious bursal disease virus (vvIBDV) strain did not lead to attenuation in chickens. Virol. J. 11:199. doi: 10.1186/s12985-014-0199-7
Birghan, C., Mundt, E., and Gorbalenya, A. E. (2000). A non-canonical lon proteinase lacking the ATPase domain employs the ser-Lys catalytic dyad to exercise broad control over the life cycle of a double-stranded RNA virus. EMBO J. 19, 114–123. doi: 10.1093/emboj/19.1.114
Boot, H. J., Hoekman, A. J., and Gielkens, A. L. (2005). The enhanced virulence of very virulent infectious bursal disease virus is partly determined by its B-segment. Arch. Virol. 150, 137–144. doi: 10.1007/s00705-004-0405-9
Brandt, M., Yao, K., Liu, M., Heckert, R. A., and Vakharia, V. N. (2001). Molecular determinants of virulence, cell tropism, and pathogenic phenotype of infectious bursal disease virus. J. Virol. 75, 11974–11982. doi: 10.1128/JVI.75.24.11974-11982.2001
Bulow, V. V., Fuchs, B., and Rudolph, R. (1985). Avian infectious anaemia caused by chicken anaemia agent (CAA). Martinus Nijhoff Publishers, Dordrecht.
Chen, G., He, X., Yang, L., and Wei, P. (2018). Antigenicity characterization of four representative natural reassortment IBDVs isolated from commercial three-yellow chickens from southern China reveals different subtypes co-prevalent in the field. Vet. Microbiol. 219, 183–189. doi: 10.1016/j.vetmic.2018.04.024
Chen, Z., Lian, J., Liang, Z., Leng, M., Lin, W., and Chen, F. (2022). Characterization and pathogenicity of infectious bursal disease virus in southern China. Poult. Sci. 101:102018. doi: 10.1016/j.psj.2022.102018
Coulibaly, F., Chevalier, C., Gutsche, I., Pous, J., Navaza, J., Bressanelli, S., et al. (2005). The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cells 120, 761–772. doi: 10.1016/j.cell.2005.01.009
Dobos, P., Hill, B. J., Hallett, R., Kells, D. T., Becht, H., and Teninges, D. (1979). Biophysical and biochemical characterization of five animal viruses with bisegmented double-stranded RNA genomes. J. Virol. 32, 593–605. doi: 10.1128/jvi.32.2.593-605.1979
Dulwich, K. L., Asfor, A. S., Gray, A. G., Nair, V., and Broadbent, A. J. (2018). An ex vivo chicken primary bursal-cell culture model to study infectious bursal disease virus pathogenesis. J. Vis. Exp. 4:58489. doi: 10.3791/58489
Escaffre, O., Le Nouen, C., Amelot, M., Ambroggio, X., Ogden, K. M., Guionie, O., et al. (2013). Both genome segments contribute to the pathogenicity of very virulent infectious bursal disease virus. J. Virol. 87, 2767–2780. doi: 10.1128/JVI.02360-12
Fan, L., Wu, T., Hussain, A., Gao, Y., Zeng, X., Wang, Y., et al. (2019). Novel variant strains of infectious bursal disease virus isolated in China. Vet. Microbiol. 230, 212–220. doi: 10.1016/j.vetmic.2019.01.023
Feng, X., Zhu, N., Cui, Y., Hou, L., Zhou, J., Qiu, Y., et al. (2022). Characterization and pathogenicity of a naturally reassortant and recombinant infectious bursal disease virus in China. Transbound. Emerg. Dis. 69, e746–e758. doi: 10.1111/tbed.14347
Gao, L., Li, K., Qi, X., Gao, Y., Wang, Y., Gao, H., et al. (2018). N-terminal domain of the RNA polymerase of very virulent infectious bursal disease virus contributes to viral replication and virulence. Sci. China Life Sci. 61, 1127–1129. doi: 10.1007/s11427-017-9297-8
He, X., Chen, G., Yang, L., Xuan, J., Long, H., and Wei, P. (2016). Role of naturally occurring genome segment reassortment in the pathogenicity of IBDV field isolates in three-yellow chickens. Avian Pathol. 45, 178–186. doi: 10.1080/03079457.2016.1139687
He, C. Q., Ma, L. Y., Wang, D., Li, G. R., and Ding, N. Z. (2009). Homologous recombination is apparent in infectious bursal disease virus. Virology 384, 51–58. doi: 10.1016/j.virol.2008.11.009
He, X., Wang, W., Chen, G., Jiao, P., Ji, Z., Yang, L., et al. (2019). Serological study reveal different antigenic IBDV strains prevalent in southern China during the years 2000-2017 and also the antigenic differences between the field strains and the commonly used vaccine strains. Vet. Microbiol. 239:108458. doi: 10.1016/j.vetmic.2019.108458
He, X., Wei, P., Yang, X., Guan, D., Wang, G., and Qin, A. (2012). Molecular epidemiology of infectious bursal disease viruses isolated from southern China during the years 2000-2010. Virus Genes 45, 246–255. doi: 10.1007/s11262-012-0764-3
He, X., Xiong, Z., Yang, L., Guan, D., Yang, X., and Wei, P. (2014). Molecular epidemiology studies on partial sequences of both genome segments reveal that reassortant infectious bursal disease viruses were dominantly prevalent in southern China during 2000-2012. Arch. Virol. 159, 3279–3292. doi: 10.1007/s00705-014-2195-z
Hon, C. C., Lam, T. T., Yip, C. W., Wong, R. T., Shi, M., Jiang, J., et al. (2008). Phylogenetic evidence for homologous recombination within the family Birnaviridae. J. Gen. Virol. 89, 3156–3164. doi: 10.1099/vir.0.2008/004101-0
Hou, B., Wang, C. Y., Luo, Z. B., and Shao, G. Q. (2022). Commercial vaccines used in China do not protect against a novel infectious bursal disease virus variant isolated in Fujian. Vet. Rec. 191:e1840. doi: 10.1002/vetr.1840
Huang, Y., Shu, G., Huang, C., Han, J., Li, J., Chen, H., et al. (2022). Characterization and pathogenicity of a novel variant infectious bursal disease virus in China. Front. Microbiol. 13:1039259. doi: 10.3389/fmicb.2022.1039259
Islam, M. R., Nooruzzaman, M., Rahman, T., Mumu, T. T., Rahman, M. M., Chowdhury, E. H., et al. (2021). A unified genotypic classification of infectious bursal disease virus based on both genome segments. Avian Pathol. 50, 190–206. doi: 10.1080/03079457.2021.1873245
Ismail, N. M., Saif, Y. M., and Moorhead, P. D. (1988). Lack of pathogenicity of five serotype 2 infectious bursal disease viruses in chickens. Avian Dis. 32, 757–759. doi: 10.2307/1590995
Jackwood, D. J., Saif, Y. M., and Hughes, J. H. (1982). Characteristics and serologic studies of two serotypes of infectious bursal disease virus in turkeys. Avian Dis. 26, 871–882. doi: 10.2307/1589875
Jackwood, D. J., Schat, K. A., Michel, L. O., and de Wit, S. (2018). A proposed nomenclature for infectious bursal disease virus isolates. Avian Pathol. 47, 576–584. doi: 10.1080/03079457.2018.1506092
Jackwood, D. J., Sommer-Wagner, S. E., Crossley, B. M., Stoute, S. T., Woolcock, P. R., and Charlton, B. R. (2011). Identification and pathogenicity of a natural reassortant between a very virulent serotype 1 infectious bursal disease virus (IBDV) and a serotype 2 IBDV. Virology 420, 98–105. doi: 10.1016/j.virol.2011.08.023
Jackwood, D. J., Sreedevi, B., LeFever, L. J., and Sommer-Wagner, S. E. (2008). Studies on naturally occurring infectious bursal disease viruses suggest that a single amino acid substitution at position 253 in VP2 increases pathogenicity. Virology 377, 110–116. doi: 10.1016/j.virol.2008.04.018
Letzel, T., Coulibaly, F., Rey, F. A., Delmas, B., Jagt, E., van Loon, A. A., et al. (2007). Molecular and structural bases for the antigenicity of VP2 of infectious bursal disease virus. J. Virol. 81, 12827–12835. doi: 10.1128/JVI.01501-07
Lian, J., Wang, Z., Xu, Z., Pang, Y., Leng, M., Tang, S., et al. (2022). Pathogenicity and molecular characterization of infectious bursal disease virus in China. Poult. Sci. 101:101502. doi: 10.1016/j.psj.2021.101502
Lu, Z., Zhang, L., Wang, N., Chen, Y., Gao, L., Wang, Y., et al. (2015). Naturally occurring reassortant infectious bursal disease virus in northern China. Virus Res. 203, 92–95. doi: 10.1016/j.virusres.2015.04.003
McDonald, S. M., Nelson, M. I., Turner, P. E., and Patton, J. T. (2016). Reassortment in segmented RNA viruses: mechanisms and outcomes. Nat. Rev. Microbiol. 14, 448–460. doi: 10.1038/nrmicro.2016.46
Muller, H., Islam, M. R., and Raue, R. (2003). Research on infectious bursal disease--the past, the present and the future. Vet. Microbiol. 97, 153–165. doi: 10.1016/j.vetmic.2003.08.005
Muller, H., Scholtissek, C., and Becht, H. (1979). The genome of infectious bursal disease virus consists of two segments of double-stranded RNA. J. Virol. 31, 584–589. doi: 10.1128/jvi.31.3.584-589.1979
Noor, M., Mahmud, M. S., Ghose, P. R., Roy, U., Nooruzzaman, M., Chowdhury, E. H., et al. (2014). Further evidence for the association of distinct amino acid residues with in vitro and in vivo growth of infectious bursal disease virus. Arch. Virol. 159, 701–709. doi: 10.1007/s00705-013-1885-2
Nooruzzaman, M., Hossain, I., Rahman, M. M., Uddin, A. J., Mustari, A., Parvin, R., et al. (2022). Comparative pathogenicity of infectious bursal disease viruses of three different genotypes. Microb. Pathog. 169:105641. doi: 10.1016/j.micpath.2022.105641
Nouen, C. L., Toquin, D., Muller, H., Raue, R., Kean, K. M., Langlois, P., et al. (2012). Different domains of the RNA polymerase of infectious bursal disease virus contribute to virulence. PLoS One 7:e28064. doi: 10.1371/journal.pone.0028064
Pérez-Losada, M., Arenas, M., Galán, J. C., Palero, F., and González-Candelas, F. (2015). Recombination in viruses: mechanisms, methods of study, and evolutionary consequences. Infect. Genet. Evol. 30, 296–307. doi: 10.1016/j.meegid.2014.12.022
Qi, X., Gao, H., Gao, Y., Qin, L., Wang, Y., Gao, L., et al. (2009). Naturally occurring mutations at residues 253 and 284 in VP2 contribute to the cell tropism and virulence of very virulent infectious bursal disease virus. Antivir. Res. 84, 225–233. doi: 10.1016/j.antiviral.2009.09.006
Qi, X., Gao, X., Lu, Z., Zhang, L., Wang, Y., Gao, L., et al. (2016). A single mutation in the PBC loop of VP2 is involved in the in vitro replication of infectious bursal disease virus. Sci. China Life Sci. 59, 717–723. doi: 10.1007/s11427-016-5054-1
Qi, X., Zhang, L., Chen, Y., Gao, L., Wu, G., Qin, L., et al. (2013). Mutations of residues 249 and 256 in VP2 are involved in the replication and virulence of infectious bursal disease virus. PLoS One 8:e70982. doi: 10.1371/journal.pone.0070982
Qin, L., Qi, X., Gao, H., Gao, Y., Bu, Z., and Wang, X. (2009). Exchange of the VP5 of infectious bursal disease virus in a serotype I strain with that of a serotype II strain reduced the viral replication and cytotoxicity. J. Microbiol. 47, 344–350. doi: 10.1007/s12275-009-0028-7
Qin, L., Qi, X., Gao, Y., Gao, H., Lu, X., Wang, Y., et al. (2010). VP5-deficient mutant virus induced protection against challenge with very virulent infectious bursal disease virus of chickens. Vaccine 28, 3735–3740. doi: 10.1016/j.vaccine.2010.02.102
Rautenschlein, S., Yeh, H.-Y., and Sharma, J. M. (2003). Comparative Immunopathogenesis of mild, intermediate, and virulent strains of classic infectious bursal disease virus. Avian Dis. 47, 66–78. doi: 10.1637/0005-2086(2003)047[0066:CIOMIA]2.0.CO;2
Sharma, J. M., Kim, I. J., Rautenschlein, S., and Yeh, H. Y. (2000). Infectious bursal disease virus of chickens: pathogenesis and immunosuppression. Devel. Comp. Immunol. 24, 223–235. doi: 10.1016/S0145-305X(99)00074-9
Skeeles, J. K., Lukert, P. D., Fletcher, O. J., and Diseases, J. D. L. J. A. (1979). Immunization studies with a cell-culture-adapted infectious bursal disease virus. Avian Dis. 23, 456–465. doi: 10.2307/1589576
Wang, Y.-L., Fan, L.-J., Jiang, N., Gao, L., Li, K., Gao, Y.-L., et al. (2021). An improved scheme for infectious bursal disease virus genotype classification based on both genome-segments a and B. J. Integr. Agr. 20, 1372–1381. doi: 10.1016/S2095-3119(20)63424-4
Wang, W., He, X., Zhang, Y., Qiao, Y., Shi, J., Chen, R., et al. (2022a). Analysis of the global origin, evolution and transmission dynamics of the emerging novel variant IBDV (A2dB1b): the accumulation of critical aa-residue mutations and commercial trade contributes to the emergence and transmission of novel variants. Transbound. Emerg. Dis. 69, e2832–e2851. doi: 10.1111/tbed.14634
Wang, W., Huang, Y., Ji, Z., Chen, G., Zhang, Y., Qiao, Y., et al. (2021). The full region of N-terminal in polymerase of IBDV plays an important role in viral replication and pathogenicity: either partial region or single amino acid V4I substitution does not completely Lead to the virus attenuation to three-yellow chickens. Viruses 13:107. doi: 10.3390/v13010107
Wang, W., Huang, Y., Zhang, Y., Qiao, Y., Deng, Q., Chen, R., et al. (2022b). The emerging naturally reassortant strain of IBDV (genotype A2dB3) having segment a from Chinese novel variant strain and segment B from HLJ 0504-like very virulent strain showed enhanced pathogenicity to three-yellow chickens. Transbound. Emerg. Dis. 69, e566–e579. doi: 10.1111/tbed.14336
Wang, W., Liang, J., Shi, M., Chen, G., Huang, Y., Zhang, Y., et al. (2020a). The diagnosis and successful replication of a clinical case of duck spleen necrosis disease: an experimental co-infection of an emerging unique reovirus and Salmonella Indiana reveals the roles of each of the pathogens. Vet. Microbiol. 246:108723. doi: 10.1016/j.vetmic.2020.108723
Wang, W., Liu, T., Chen, G., Ji, Z., Zhao, Z., Huang, Y., et al. (2020b). Prokaryotic expression of VP2, VP1 and VP2-VP1 multimer genes of infectious bursal disease virus and its preliminary clinical application. Anim. Husband. Vet. Med. 52, 73–80. (In Chinese).
Wang, Y., Qi, X., Kang, Z., Yu, F., Qin, L., Gao, H., et al. (2010). A single amino acid in the C-terminus of VP3 protein influences the replication of attenuated infectious bursal disease virus in vitro and in vivo. Antivir. Res. 87, 223–229. doi: 10.1016/j.antiviral.2010.05.004
Wang, X., Zhang, H., Gao, H., Fu, C., Gao, Y., and Ju, Y. (2007). Changes in VP3 and VP5 genes during the attenuation of the very virulent infectious bursal disease virus strain Gx isolated in China. Virus Genes 34, 67–73. doi: 10.1007/s11262-006-0002-y
Wei, P., Long, J., Yang, X., and Li, K., (2004). The development of rapid diagnosis and Pathotyping techniques for infectious bursal disease viruses. Chin. J. Vet. Med. 4, 313–316. (In Chinese).
Wu, T., Wang, Y., Li, H., Fan, L., Jiang, N., Gao, L., et al. (2020). Naturally occurring homologous recombination between novel variant infectious bursal disease virus and intermediate vaccine strain. Vet. Microbiol. 245:108700. doi: 10.1016/j.vetmic.2020.108700
Xu, A., Pei, Y., Zhang, K., Xue, J., Ruan, S., and Zhang, G. (2020). Phylogenetic analyses and pathogenicity of a variant infectious bursal disease virus strain isolated in China. Virus Res. 276:197833. doi: 10.1016/j.virusres.2019.197833
Yu, F., (2011). The effect of VP1of infectious bursal disease virus on replication and pathogenicity. Beijing: Chinese Academy of Agricultural Sciences (CASA). (In Chinese).
Yu, F., Ren, X., Wang, Y., Qi, X., Song, J., Gao, Y., et al. (2013). A single amino acid V4I substitution in VP1 attenuates virulence of very virulent infectious bursal disease virus (vvIBDV) in SPF chickens and increases replication in CEF cells. Virology 440, 204–209. doi: 10.1016/j.virol.2013.02.026
Zhang, W., Wang, X., Gao, Y., and Qi, X. (2022). The Over-40-years-epidemic of infectious bursal disease virus in China. Viruses 14. doi: 10.3390/v14102253
Keywords: infectious bursal disease virus, classical IBDV, full-length genome, reassortment, recombination, cell tropism, pathogenicity
Citation: Wang W, Shi J, Zhang Y, Qiao Y, Zuo W, Wang Z, Nong T, Hu S, Chen Y, Kong R, He X and Wei P (2023) Genetic and pathogenic characterizations of a naturally occurring reassortant and homologous recombinant strain of the classical infectious bursal disease virus re-emerging in chickens in southern China. Front. Microbiol. 14:1293072. doi: 10.3389/fmicb.2023.1293072
Edited by:
Mohammed Rohaim, Lancaster University, United KingdomReviewed by:
Zaib Ur Rehman, Pir Mehr Ali Shah Arid Agriculture University, PakistanMohammed Nooruzzaman, Bangladesh Agricultural University, Bangladesh
Copyright © 2023 Wang, Shi, Zhang, Qiao, Zuo, Wang, Nong, Hu, Chen, Kong, He and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiumiao He, eGl1bWlhb2hlMDgzOUBzaW5hLmNvbQ==; Ping Wei, cGluZ3dlaThAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Weiwei Wang
Weiwei Wang Jun Shi1†
Jun Shi1† Ping Wei
Ping Wei