
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 10 January 2024
Sec. Virology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1290302
This article is part of the Research TopicAdvances and Insights in the Diagnosis of Viral Infections and Vaccines Development in AnimalsView all 24 articles
Introduction: The detection of African swine fever virus (ASFV) is commonly performed using quantitative real-time PCR (qPCR), a widely used virological method known for its high sensitivity and specificity. However, qPCR has a limitation in distinguishing between infectious and inactivated virus, which can lead to an overestimation of viral targets.
Methods: To provide insights into ASFV infectivity, we evaluated the suitability of PMAxx, an improved version of propidium monoazide (PMA), as a means to differentiate between infectious and non-infectious ASFV. Pre-treatment with 50 μM PMAxx for 15 min significantly reduced the qPCR signal of ASFV in the live vaccine. Additionally, thermal treatment at 85°C for 5 min effectively inactivated the live ASFV in the vaccine. Based on a standard curve, the sensitivity of the PMAxx-qPCR assay was estimated to be approximately 10 copies/μL. Furthermore, we observed a strong agreement between the results obtained from PMAxx-qPCR and pig challenge experiments. Moreover, we utilized the PMAxx-qPCR assay to investigate the persistence of ASFV, revealing a close relationship between viral persistence and factors such as temperature and type of piggery materials.
Conclusion: The findings of this study suggest that pre-treating viruses with PMAxx prior to qPCR is a reliable method for distinguishing between infectious and non-infectious ASFV. Thus, integrating of PMAxx-qPCR into routine diagnostic protocols holds potential for improving the interpretation of positive ASFV results obtained through qPCR.
African swine fever (ASF) is a devastating disease that affects domestic pigs and wild boars. ASF outbreaks are currently occurring in Africa, Eastern Europe and Asia, causing significant economic losses globally (Costard et al., 2009; Norbert Mwiine et al., 2019). The African swine fever virus (ASFV), the pathogen responsible for ASF, is the sole member of the Asfarviridae family. It has a double-stranded DNA genome of approximately 180–190 kb and encodes over 150 open reading frames (ORFs) (Dixon et al., 2013). ASFV can survive in the environment for extended periods and can be transmitted through infected tick bites, direct contact with infected pigs, and contaminated materials (Gaudreault et al., 2020; Pereira De Oliveira et al., 2020). Due to the lack of commercial vaccines in the past few decades, preventing severe ASF outbreaks heavily relies on restricting animal movements and culling infected herds (Zhang et al., 2020; Borca et al., 2021). Recently, a promising recombinant vaccine candidate, ASFV-G-1I177L, has been developed by deleting the I177L gene from the genome of the highly virulent ASFV Georgia strain (Borca et al., 2020, 2021; Tran et al., 2021, 2022). This attenuated vaccine has been authorized as the first commercial gene-modified live vaccine in Vietnam and has shown no residual toxicity in long-term clinical studies (Borca et al., 2023). However, the vaccine can be only given to pigs aged between 8 and 10 weeks according to the directions. The residual infectious particles in the environment poses significant challenges to ASF risk management in the clearance of ASFV.
Quantitative real-time polymerase chain reaction (qPCR) is highly sensitive and specific for detecting the presence of viral genomes (Choi and Jiang, 2005; Hamza et al., 2011). However, qPCR cannot differentiate between infectious and inactivated viruses to directly indicate infectivity (Fittipaldi et al., 2010). Methods that can rapidly provide information about viral infectivity are of interest, given that only active viruses pose a public health threat (Knight et al., 2013). Various methods have been employed to detect infectious viruses, including cytopathic effect, fluorescent microscopy, flow cytometry, and detection of genome or envelope integrity (Zeng et al., 2022). Detection methods involving cell culture are considered the gold standard for quantifying certain viral infectivity. However, ASFV cultivation requires costly primary porcine alveolar macrophages and is constrained to biosafety level 3 laboratories (Blackmer et al., 2000; Rodríguez et al., 2009; Hamza et al., 2011). Additionally, the proposal to analyze the integrity of viral genomes through PCR amplification of long target regions may be related to viral infectivity (Li et al., 2002; Simonet and Gantzer, 2006). Nevertheless, viral inactivation can occur without damaging the viral genome, limiting the general applicability of long target region PCR as a surrogate marker for viral infectivity (Hamza et al., 2011).
Cellular or envelope integrity is one of the characteristics used to distinguish between live and inactivated cells or enveloped viruses. One promising strategy to overcome the limitations of qPCR is pre-treating samples with photosensitizing dyes such as ethidium monoazide (EMA) and propidium monoazide (PMA) before qPCR (Parshionikar et al., 2010). This approach has been successfully used to differentiate infectious and non-infectious bacteria, protozoa, nematode eggs, fungi and viruses (Brescia et al., 2009; Fittipaldi et al., 2010; Graiver et al., 2010; Kim et al., 2011; Dreo et al., 2014). Theoretically, the dyes are membrane-impermeant and bind irreversibly to nucleic acids by photoactivation, leaving the DNA in viable cells intact (Nocker et al., 2006). Modified nucleic acid structures interfere with PCR amplification, resulting in reduced signal intensity in subsequent qPCR (Rudi et al., 2005; Nocker and Camper, 2009). Moreover, light exposure leads to the reaction of unbound excess dye with water molecules, preventing the purified DNA from being further modified in cells with intact cell membranes (Nocker and Camper, 2009). Some studies have shown significant DNA loss in the genomic DNA of live bacteria induced by EMA (Flekna et al., 2007), while PMA has been demonstrated to be more selective, only penetrating dead bacterial cells and not cells with intact membranes (Nocker et al., 2006). PMAxx, an improved version of PMA with a higher molecular charge, inhibits PCR amplification of modified DNA templates through a combination of removal of modified DNA during purification and inhibition of template amplification by DNA polymerases (Figure 1). In experimental bacterial strains, PMAxx increased the difference between live and dead bacteria by an additional 3 to 7 CT values compared to PMA (Nocker et al., 2006). Recently, Liu et al. conducted a study to investigate the addition of Triton X-100 for enhancing the penetration of PMAxx into inactivated ASFV virions, which may be helpful to interpret the results, though the infectivity of samples was still distinguishable by PMAxx-qPCR without the assistance of Triton X-100 (Liu et al., 2022). Moreover, these experiments were carried out solely under laboratory conditions and the validity of the results was not confirmed through animal inoculation.
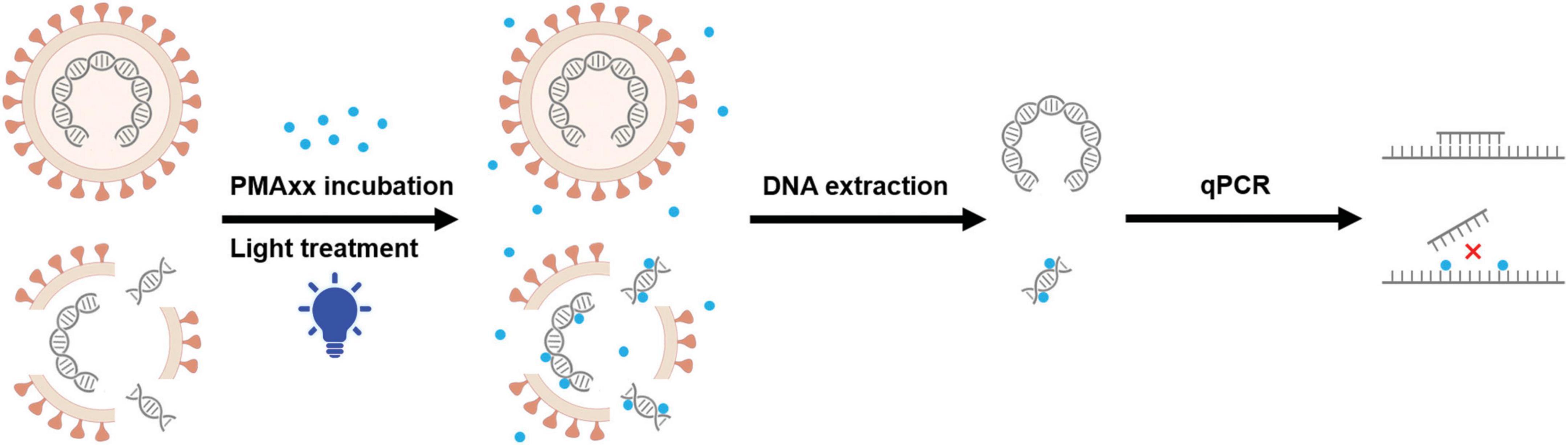
Figure 1. Proposed mechanism of PMAxx modification of dead viruses. The envelope impermeant PMAxx dye (purple dot) selectively penetrates dead viruses with compromised membranes. Upon exposure to light, PMAxx covalently modifies dissociated DNA. Genomic DNA was extracted from these samples, while some of the PMAxx-modified DNA became insoluble and was lost during DNA extraction. Subsequent qPCR amplification of modified DNA templates is inhibited, allowing selective quantification of DNA from viable viruses.
The objective of this study was to evaluate the applicability of pure PMAxx pre-treatment for differentiating infectious and non-infectious ASFV and the correlation between PMAxx-qPCR detection results and animal challenges. We also used PMAxx-qPCR to assess the persistence of ASFV on commonly encountered materials in pig farms. Our data demonstrate that PMAxx-qPCR detection can improve the interpretation of ASFV qPCR-positive results and provide better risk management strategies.
NAVET-ASFVAC vaccine containing live-attenuated ASFV-G-ΔI177L strain was purchased from Navetco National Veterinary Joint Stock Company (NAVETCO). The vaccine was diluted with saline at a twofold dilution ratio (ranging from 1:10 to 1:2560), aliquoted and stored at −80°C until use. The experiments of animal inoculation and viral persistence using NAVET-ASFVAC vaccine were performed in a fattening pig farm in Binh Phuoc province, Vietnam.
Each sample was divided into three portions: one portion was heat-inactivated at 85°C for 10 min, while the other two portions were kept at room temperature. PMAxx (Biotium, Inc., Hayward, CA, USA) was dissolved in deionized water to obtain a stock solution of 1 mM. Then, 10 μL of the PMAxx solution was added to 190 μL aliquots of both the non-heated and heat-treated samples, resulting in a final concentration of 50 μM. Additionally, 10 μL of deionized water was added to an untreated aliquot as a control template for standard qPCR. These three aliquots were incubated in the dark at 37°C for 15 min with occasional mixing to allow reagent penetration. Subsequently, the samples were irradiated with a PMA-Lite™ LED photoactivator (E90002, Biotium) at room temperature for 15 min. The experiment was repeated in triplicate, and DNA extraction of the samples was performed. Only when the CT values of the heat-treated-PMAxx treated aliquots were significantly higher than those of the PMAxx treated aliquots, the presence of ASFV viral particles was considered to be existed.
Samples were vortexed and centrifuged at 8,000 × g for 2 min. Genomic DNA from the samples (200 μL) was extracted using the Virus DNA Extraction Kit II (Geneaid, Taiwan) according to the instructions provided. The extracted nucleic acid was eluted in 50 μL elution buffer and stored at 4°C for subsequent qPCR analysis.
African swine fever virus-specific primers (forward: 5′-AAAATGATACGCAGCGAAC-3′, reverse: 5′-TTGTTTACCT GCTGTTTGGAT-3′) and a probe (5′-FAM-TTCACAGCATT TTCCCGAGAACT-BHQ1-3′) targeting the B646L gene were used for qPCR detection. A qPCR reaction mixture of 20 μL was prepared, containing 10 μL PerfectStart R II Probe qPCR SuperMix (TransGen Biotech, China), 0.5 μM primers and probe, 5 μL DNA template, and PCR-grade water. The reaction consisted of an initial denaturation step at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 15 s, and extension at 72°C for 30 s. qPCR results were recorded using the Step One Plus™ Real-Time PCR System (ABI, 4376600).
A standard plasmid containing the ASFV B646L gene was constructed as described previously (Li et al., 2022). Briefly, partial sequences of the B646L gene were amplified by PCR. The products and pMD18-T plasmids (D101A, Takara, Japan) were digested with the same restriction enzymes. The fragment was gel-purified and ligated to the vector using DNA ligase (C301-01, Vazyme) following standard procedures. Positive clones were screened and identified by sequencing. The standard curve was constructed using logarithmic 10-fold dilutions ranging from 2.5 × 107 to 2.5 genome copies. The ASFV genome copy numbers corresponding to the copy numbers of the standard plasmid were calculated using the following formula:
where m (/g) is the amount of plasmids measured using a BioSpec-nano Micro-volume UV-Vis Spectrophotometer (Shimadzu, Kyoto, Japan), 6.022 × 1023 is the Avogadro number, 1,840,105.22 (Da) is the molecular weight of standard plasmids calculated using the Sequence Manipulation Suite (Stothard, 2000), and 1 × 109 is used to convert the molecular weight of the plasmids to nanograms. A fresh dilution set was prepared to construct the standard curve for each qPCR run, which could convert CT values obtained from qPCR analysis to ASFV genome equivalents.
The NAVET-ASFVAC vaccine was diluted with saline (1:2560) and divided into four portions, with three portions (Groups B, C, and D) subjected to water bath heating at 85°C for 0.5, 1, and 10 min, respectively. The infectiousness of ASFV particles in the samples was quantified using PMA-qPCR. Then, each sample was injected into the muscles of three ASFV-negative fattening pigs. Throat swab samples were collected from each fattening pig 7 days later as previously described (Li et al., 2022) and ASFV DNA was measured using qPCR.
The vaccine was diluted with saline to a CT value of 25. Latex gloves and packaging bags from the pig farm were cut into squares with sides measuring 2 cm and soaked in the vaccine. Dry feed particles of approximately 1 cm in length were selected, and 50 μL of vaccine was added to each particle. The foam plastic heads of fertilization tubes were dipped into the vaccine. These materials were then thoroughly air-dried and placed in an incubator at temperatures of 4, 15, or 25°C for several days. The samples’ ASFV infectivity was measured by PMAxx-qPCR in three separate experiments after dissolution in 2 mL of saline. The experiments were performed in a fattening pig farm in Binh Phuoc province, Vietnam.
The significance of differences between CT values was evaluated by unpaired Student’s t-test using GraphPad Prism v8.3.0. In all cases, a value of P < 0.05 was considered significant.
To determine the appropriate working concentration, various doses of PMAxx were added to ASFV-positive vaccine samples and exposed to light for 15 min. Subsequently, genomic DNA was extracted and subjected to qPCR using ASFV-specific primers and probes. The results displayed in Figure 2A demonstrated that increasing concentrations of PMAxx led to higher CT values, indicating greater inhibition of qPCR amplification. Concentrations ranging from 25 to 50 μM led to substantial inhibition, whereas higher concentrations did not entirely eliminate qPCR signals in samples with high ASFV content.
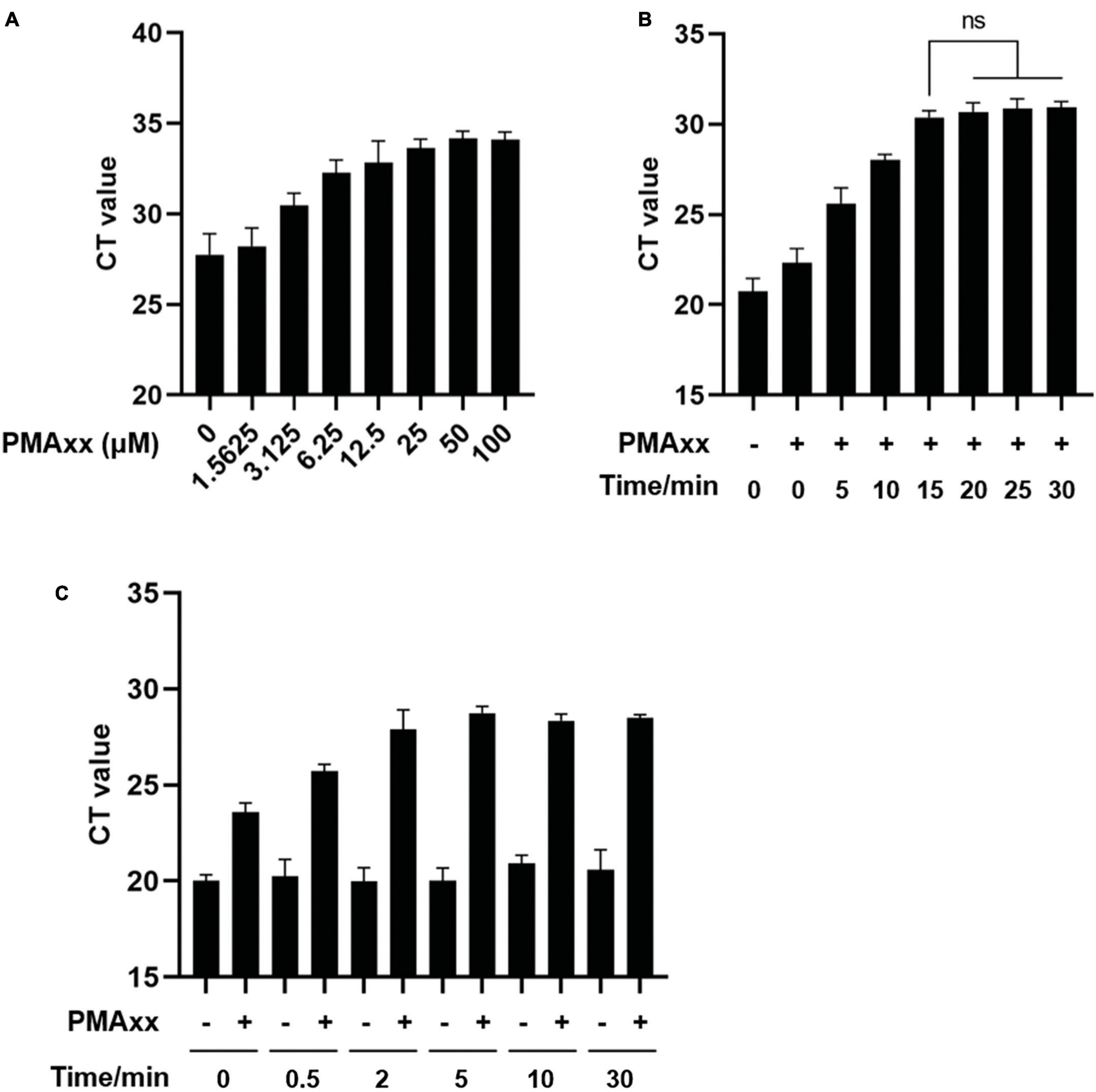
Figure 2. Optimization of PMAxx-qPCR assay conditions. (A) Vaccine samples containing ASFV-G-ΔI177L were treated with different concentrations of PMAxx-qPCR at 37°C for 15 min. Genomic DNA content was detected by qPCR using ASFV B646L gene-specific primers and probe. (B) Vaccine samples containing ASFV-G-ΔI177L were incubated with 50 μM PMAxx at 37°C for different times. ASFV DNA was detected by qPCR. (C) Vaccine samples containing ASFV-G-ΔI177L were heat-killed at 85°C for various times and then incubated with 50 μM PMAxx at 37°C for 15 min. ASFV DNA was detected by qPCR. All assays were performed in triplicate.
To optimize the efficacy of PMAxx-DNA cross-linking, ASFV nucleic acids were incubated with 50 μM PMAxx at 37°C for varying durations. As depicted in Figure 2B, inhibition increased with longer incubation times. Pre-treatment with PMAxx for 15 min yielded similar results as a 30-min pre-treatment, suggesting that a 15-min period achieved complete cross-linking of PMAxx with ASFV DNA.
For the preparation of an inactive ASFV control, genomic DNA solutions were subjected to thermal inactivation using water baths set at 85°C for different durations. Subsequently, PMAxx was added at a final concentration of 50 μM and incubated at 37°C for 15 min with periodic mixing. Figure 2C indicated that heat treatment of infectious ASFV for up to 30 min did not affect the qPCR CT values. However, the signal from PMAxx-treated DNA was reduced compared to untreated DNA at all-time points. Based on the degree of inhibition, a thermal inactivation period of 5–30 min efficiently inactivated ASFV and released DNA for subsequent reaction with PMAxx at 85°C.
To guarantee efficient cross-linking, a PMAxx concentration of 50 μM, an incubation time of 15 min, and the thermal inactivation at 85°C for 10 min were selected for subsequent experiments.
To assess the sensitivity of the PMAxx-qPCR assay, a standard curve was generated using a 10-fold serial dilution of standard plasmids containing ASFV B646L partial sequences. The CT values were plotted against the logarithm of the standard plasmid copies to perform linear regression analysis (Figure 3A). The experimental points aligned in a straight line with a high correlation coefficient (R2 = 0.9983), indicating accurate prediction. The limit of quantification for the PMAxx-qPCR assay was determined by testing serially diluted vaccine samples containing ASFV-G-ΔI177L. As shown in Figure 3B, samples with a mean CT value of 34.38 were identified as having infectious ASFV by PMAxx-qPCR, corresponding to approximately 10.57 copies of total ASFV according to the linear relationship in Figure 3A. Therefore, the detection limit of this assay was approximately 10 copies/μL when testing for infectious ASFV in this vaccine.
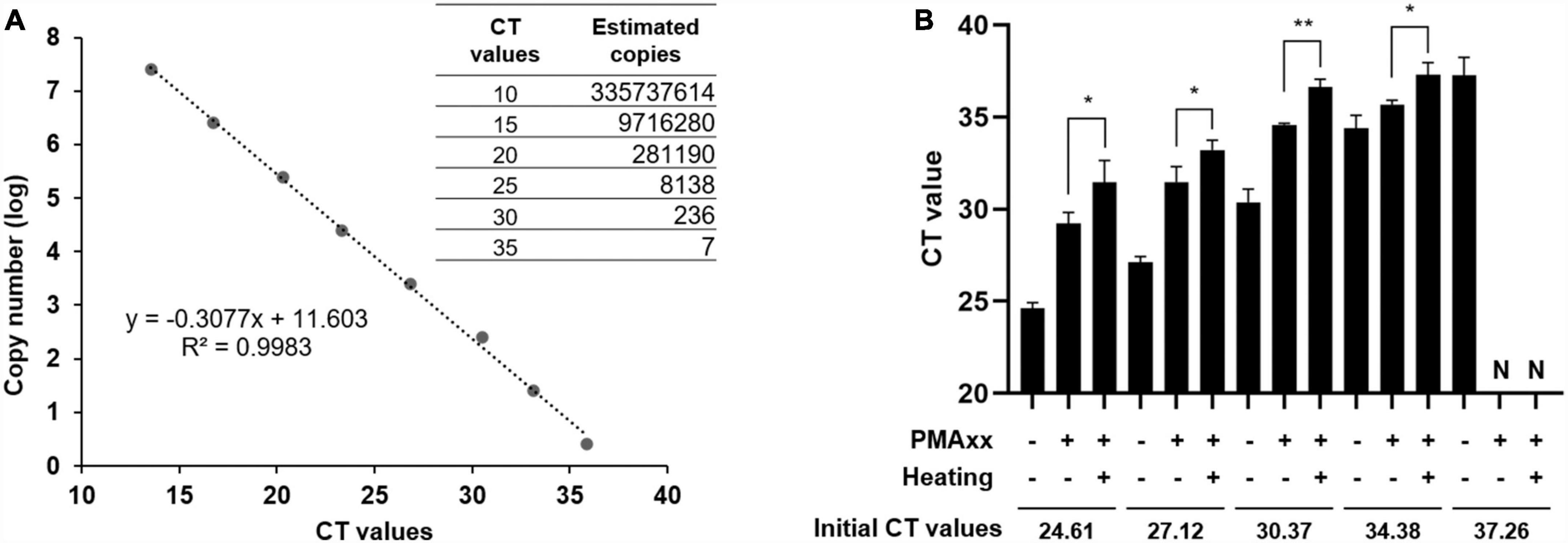
Figure 3. PMAxx-qPCR sensitivity estimated from the standard curve. (A) Correlation between ASFV copy numbers and CT values was established using ASFV B646L gene standard plasmids. (B) Vaccine samples containing ASFV-G-ΔI177L were subjected to PMAxx-qPCR assay. All assays were performed in triplicate. N, negative. *P < 0.05, **P < 0.01.
To validate the reliability of the PMAxx-qPCR assay, the diluted vaccine containing viable ASFV was subjected to different durations of heat treatment at 85°C. The PMAxx-qPCR assay was then performed to detect the ASFV activity in all samples. Figure 4A demonstrated that heat treatment for 0.5 min (B) and 1 min (C) did not completely inactivate ASFV, while samples heated for 10 min (D) showed no infectious ASFV. Each sample was intramuscularly injected into three ASFV-negative fattening pigs, and their throat swab samples was collected after 7 days. ASFV genomic DNA was quantified using specific primers and probes targeting the B646L gene. Figure 4B revealed that pigs challenged with sample A (positive control), B, or C were successfully infected with ASFV, while pigs injected with sample D remained ASFV-negative throughout the experiment. These results demonstrated a strong agreement between the PMAxx-qPCR assay and pig challenge results.
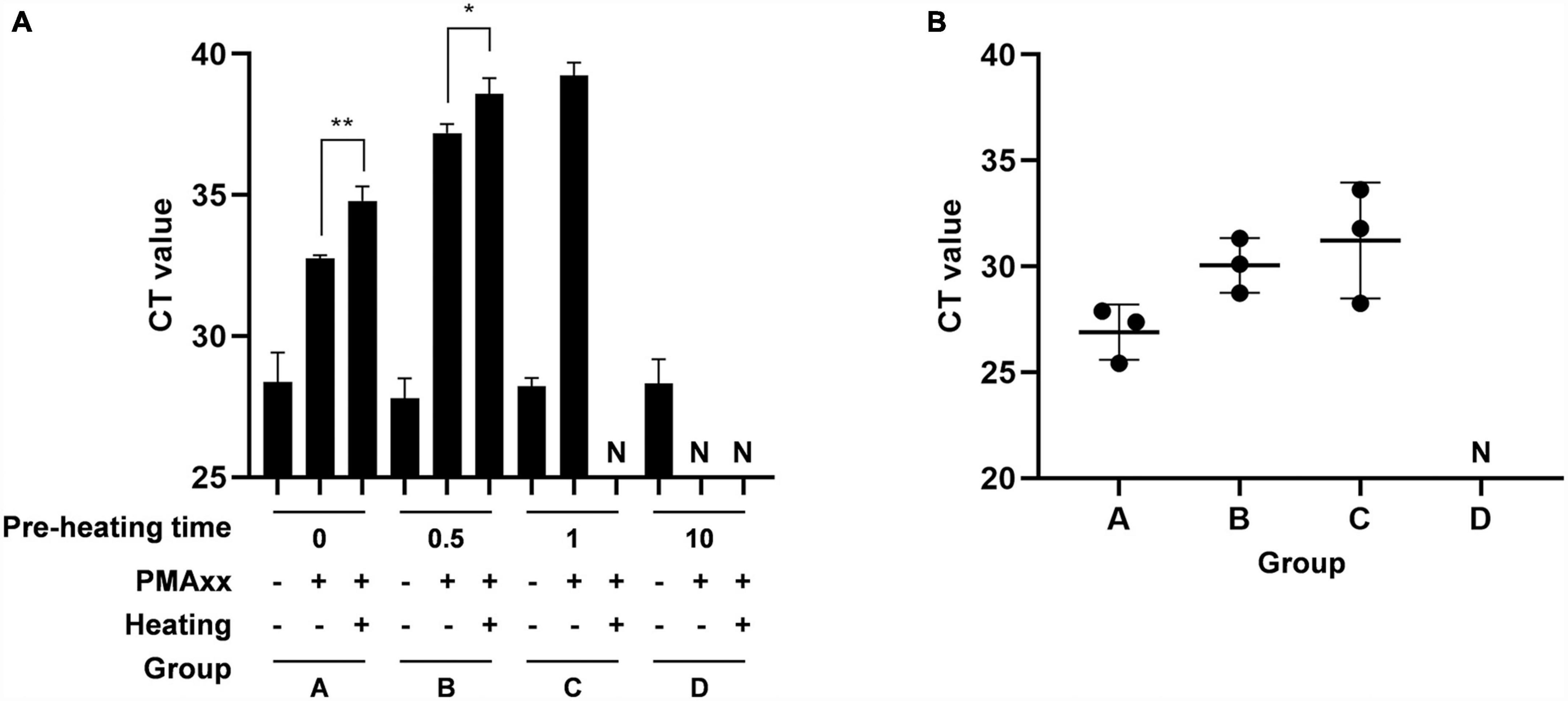
Figure 4. Comparison of agreement between PMAxx-qPCR and inoculation of animals. The vaccine containing ASFV-G-ΔI177L was heated at 85°C for different times. (A) The infectivity of samples was quantified by PMAxx-qPCR. (B) Each sample was intramuscularly injected into the neck of three ASFV-negative fatting pigs. Throat swab samples were collected from all pigs after 7 days and ASFV DNA levels were determined by qPCR. N, negative. *P < 0.05, **P < 0.01.
African swine fever virus can be transmitted to pigs through close contact with contaminated piggery supplies (Gaudreault et al., 2020). In this study, the persistence of ASFV particles on the surface of piggery materials at different temperatures was estimated.
Different materials, including dry feed, the head of fertilization tubes, latex gloves, and packaging bags were soaked in ASFV vaccine, air-dried, and stored at different temperatures for several days. ASFV on the materials was eluted with saline, and viral infectivity was measured using PMAxx-qPCR (Table 1). Overall, the survival time of infectious ASFV on the four different matrices decreased with increasing temperature (from 20–45 days at 4°C to 7–15 days at 25°C). The fertilization tubes showed the greatest delay in ASFV survival time from 25 to 4°C (30 days), while latex gloves exhibited the shortest change of survival time (13 days) when the temperature decreased. Infectious ASFV persisted for 20 days on latex gloves at 4°C, whereas the survival time on fertilization tubes was 45 days at the same temperature. Significant differences in ASFV survival time were observed between latex gloves and fertilization tubes at 25°C and 15°C, but no significant difference was found between dry feeds and packaging bags at all temperatures. These findings indicate that the persistence of ASFV infectivity varied depending on the type of piggery material and temperature.
Quantitative PCR-based methods are commonly used for risk assessment in pig production. However, samples from farms often contain a mixture of infectious and non-infectious causative agents, limiting the significance of qPCR without information on residual infectivity. Furthermore, incomplete or excessive disinfection is a prevalent issue due to the lack of information on disinfection efficiency, resulting in residual live virus and failed reproduction, or wasted resources and environmental pollution. A technically easy, highly sensitive, widely applicable, and cost-effective method is required to determine virus infectivity. To address this, alternative methods have been proposed, such as using photoactivatable dyes to eliminate signal interference from inactivated viruses during PCR amplification. This technique has demonstrated success in detecting infectivity in many viruses (Parshionikar et al., 2010; Hamza et al., 2011; Sanchez et al., 2012; Leifels et al., 2015; Prevost et al., 2016; Randazzo et al., 2016).
One photoactivatable dye that has garnered significant attention, particularly for its potential use in assessing ASFV infectivity, is PMAxx. A recent review highlighted the promising application prospects of a rapid infectious ASFV detection technology based on PMA pre-treatment, which could greatly enhance various aspects of ASF prevention and control, including epidemic surveillance, disinfection treatment, and drug development (Zeng et al., 2022). In our study, we aimed to demonstrate the potential of PMAxx pre-treatment in distinguishing between infectious and non-infectious ASFV strains in vaccine. We successfully detected as few as 10 copies/μL of infectious ASFV using PMAxx-qPCR. Furthermore, there was a strong correlation between the results of PMAxx-qPCR and animal inoculation (Figure 4). Hence, the PMAxx-qPCR assay may serve as a rapid and cost-effective analytical tool for assessing the efficacy of virus-inactivating disinfectants by monitoring capsid damage. By employing PMAxx-qPCR, the appropriate working concentration of disinfectants could be determined, ensuring effective and economical usage. It should be noted we utilized a vaccine containing ASFV-G-ΔI177L as the sole material to develop the method. This choice was made in accordance with biosafety management requirements. However, for potential clinical application, it is crucial to assess the applicability of the developed method to clinical samples such as blood, saliva, and environmental samples in future studies. What’s more, PMA is not suitable for monitoring UV irradiation of bacteria and viruses. This is because viability dyes rely on membrane integrity as a viability criterion, whereas UV light primarily damages viral nucleic acids (Nocker et al., 2006; Leifels et al., 2015). Similarly, PMAxx-qPCR is not applicable to non-enveloped viruses.
In the case of thermally inactivated ASFV, theoretically, the addition of PMAxx should completely eliminate the qPCR signal. However, even with relatively high levels of PMAxx, CT values could still be detected in the presence of high viral genome concentrations (Figure 2). These findings align with observations in viability qPCR, where complete prevention of PCR amplification of thermally inactivated viruses is challenging (Leifels et al., 2015; Moreno et al., 2015; Randazzo et al., 2016). The incomplete inhibition could be attributed to several factors. Firstly, it may stem from the limited concentration of PMAxx, which aims to effectively eliminate non-infectious ASFV genomic DNA while avoiding potential DNA loss due to overconcentration. Moreover, the background levels could be influenced by the target sequence, amplicon size, incubation temperature, and secondary structure of the viral genome (Contreras et al., 2011; Soejima et al., 2011; Schnetzinger et al., 2013; Leifels et al., 2015; Prevost et al., 2016). Therefore, further studies should focus on optimizing the working conditions of PMAxx to maximize the differentiation between signals from infectious and inactivated viruses. This includes determining the optimal concentration, incubation time, and considering the addition of Triton, PMA enhancer, and protease K (Randazzo et al., 2016).
The estimated survival time of infectious ASFV is affected by several environmental factors, including pH, temperature, type of fomite, light exposure, and the presence of viral aggregates (Arzumanyan et al., 2021; Nuanualsuwan et al., 2022). Previous studies have evaluated the persistence of infectious ASFV on different materials under ambient temperatures. At 20°C, infectious ASFV was detected in complete feed until 1 dpi, in soybean meal until at least 21 dpi and in corncob particles until 1 dpi (Niederwerder et al., 2022). ASFV survived longer in pork (18–83 days) than in tissues (9–17 days) and plasma (14 days) at room temperature (Petrini et al., 2019; Mazur-Panasiuk and Wozniakowski, 2020; Fischer et al., 2021). Additionally, porous materials, such as rubber and cellulose paper, supported ASFV viability for longer periods (14–22 days) than non-porous materials, like glass and metal (11–17 days) at 25°C (Nuanualsuwan et al., 2022). Infectious ASFV has also been detected in sterile sand for at least 3 weeks, beach sand for up to 2 weeks, yard soil for 1 week, and swamp soil for 3 days (Carlson et al., 2020). However, farmers are particularly concerned about the survival time of live ASFV on daily input materials accessible to pigs. In our study, we estimated that ASFV remained infectious on the surface of piggery materials for 7–15 days at 25°C (Table 1), which aligns with previous research on other materials. Furthermore, we observed variations in the persistence of ASFV on four matrices. The causes of these differences remain unclear, but it is likely linked to differences in their physicochemical properties, such as the micropore size in the head of fertilization tubes and the watertightness of latex gloves. Nonetheless, it is essential to disinfect piggery materials thoroughly as they are significant carriers of ASFV between domestic pigs. For those materials that cannot be conventionally disinfected, sufficient static storage before transport to pig farms is a viable option. The duration of static storage should increase as the ambient temperature decreases.
This study aimed to develop and optimize the PMAxx-qPCR assay for effective discrimination of infectious ASFV. The authenticity of the PMAxx-qPCR assay was verified by animal inoculation experiments. Furthermore, the persistence of ASFV was assessed on various surfaces, including latex gloves, dry feed, packaging bags, and fertilization tubes. The results revealed that the longevity of infectious ASFV decreased with rising temperatures on the four different matrices tested. Notably, the porous head of the fertilization tube exhibited higher stability than the waterproof latex gloves. Based on these findings, it can be concluded that the PMAxx-qPCR assay holds promise as an alternative method for assessing ASFV infectivity. Its integration into routine diagnostics can significantly enhance the interpretation of positive ASFV results, leading to improved accuracy in identifying infected cases.
The original contributions presented in this study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
The animal study was approved by the Ethics Committee at Northwest A&F University. The study was conducted in accordance with the local legislation and institutional requirements.
YL: Writing – original draft, Project administration, Methodology, Visualization. ZW: Methodology, Visualization, Writing – review and editing. JQ: Methodology, Visualization, Writing – review and editing. DH: Resources, Supervision, Validation, Writing – review and editing. HV: Data curation, Writing – review and editing. KT: Data curation, Writing – review and editing. XW: Supervision, Writing – review and editing. XL: Conceptualization, Funding acquisition, Writing – review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Taishan Industry Leadership Talent Project of Shandong Province in China.
YL, ZW, JQ, and XL are employed by Xiajin New Hope Liuhe Agriculture and Animal Husbandry Co., Ltd. DH, HV, KT, and XL are employed by New Hope Binh Phuoc livestock Co., Ltd.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Arzumanyan, H., Hakobyan, S., Avagyan, H., Izmailyan, R., Nersisyan, N., and Karalyan, Z. (2021). Possibility of long-term survival of African swine fever virus in natural conditions. Vet. World 14, 854–859. doi: 10.14202/vetworld.2021.854-859
Blackmer, F., Reynolds, K. A., Gerba, C. P., and Pepper, I. L. (2000). Use of integrated cell culture-PCR to evaluate the effectiveness of poliovirus inactivation by chlorine. Appl. Environ. Microbiol. 66, 2267–2268. doi: 10.1128/aem.66.5.2267-2268.2000
Borca, M. V., Ramirez-Medina, E., Silva, E., Rai, A., Espinoza, N., Velazquez-Salinas, L., et al. (2023). ASF vaccine candidate ASFV-G-ΔI177L does not exhibit residual virulence in long-term clinical studies. Pathogens 12:805. doi: 10.3390/pathogens12060805
Borca, M. V., Ramirez-Medina, E., Silva, E., Vuono, E., Rai, A., Pruitt, S., et al. (2021). ASFV-G-ΔI177L as an effective oral nasal vaccine against the Eurasia strain of Africa swine fever. Viruses 13:765. doi: 10.3390/v13050765
Borca, M. V., Ramirez-Medina, E., Silva, E., Vuono, E., Rai, A., Pruitt, S., et al. (2020). Development of a highly effective African swine fever virus vaccine by deletion of the I177L gene results in sterile immunity against the current epidemic Eurasia strain. J. Virol. 94:e02017-19. doi: 10.1128/jvi.02017-19
Brescia, C. C., Griffin, S. M., Ware, M. W., Varughese, E. A., Egorov, A. I., and Villegas, E. N. (2009). Cryptosporidium propidium monoazide-PCR, a molecular biology-based technique for genotyping of viable Cryptosporidium oocysts. Appl. Environ. Microbiol. 75, 6856–6863. doi: 10.1128/aem.00540-09
Carlson, J., Fischer, M., Zani, L., Eschbaumer, M., Fuchs, W., Mettenleiter, T., et al. (2020). Stability of African swine fever virus in soil and options to mitigate the potential transmission risk. Pathogens 9:977. doi: 10.3390/pathogens9110977
Choi, S., and Jiang, S. C. (2005). Real-time PCR quantification of human adenoviruses in urban rivers indicates genome prevalence but low infectivity. Appl. Environ. Microbiol. 71, 7426–7433. doi: 10.1128/aem.71.11.7426-7433.2005
Contreras, P. J., Urrutia, H., Sossa, K., and Nocker, A. (2011). Effect of PCR amplicon length on suppressing signals from membrane-compromised cells by propidium monoazide treatment. J. Microbiol. Methods 87, 89–95. doi: 10.1016/j.mimet.2011.07.016
Costard, S., Wieland, B., de Glanville, W., Jori, F., Rowlands, R., Vosloo, W., et al. (2009). African swine fever: how can global spread be prevented? Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 2683–2696. doi: 10.1098/rstb.2009.0098
Dixon, L. K., Chapman, D. A., Netherton, C. L., and Upton, C. (2013). African swine fever virus replication and genomics. Virus Res. 173, 3–14. doi: 10.1016/j.virusres.2012.10.020
Dreo, T., Pirc, M., Ramšak, Ž, Pavšič, J., Milavec, M., Zel, J., et al. (2014). Optimising droplet digital PCR analysis approaches for detection and quantification of bacteria: a case study of fire blight and potato brown rot. Anal. Bioanal. Chem. 406, 6513–6528. doi: 10.1007/s00216-014-8084-1
Fischer, M., Pikalo, J., Beer, M., and Blome, S. (2021). Stability of African swine fever virus on spiked spray-dried porcine plasma. Transbound. Emerg. Dis. 68, 2806–2811. doi: 10.1111/tbed.14192
Fittipaldi, M., Rodriguez, N. J., Codony, F., Adrados, B., Peñuela, G. A., and Morató, J. (2010). Discrimination of infectious bacteriophage T4 virus by propidium monoazide real-time PCR. J. Virol. Methods 168, 228–232. doi: 10.1016/j.jviromet.2010.06.011
Flekna, G., Stefanic, P., Wagner, M., Smulders, F. J., Mozina, S. S., and Hein, I. (2007). Insufficient differentiation of live and dead Campylobacter jejuni and Listeria monocytogenes cells by ethidium monoazide (EMA) compromises EMA/real-time PCR. Res. Microbiol. 158, 405–412. doi: 10.1016/j.resmic.2007.02.008
Gaudreault, N. N., Madden, D. W., Wilson, W. C., Trujillo, J. D., and Richt, J. A. (2020). African swine fever virus: an emerging DNA arbovirus. Front. Vet. Sci. 7:215. doi: 10.3389/fvets.2020.00215
Graiver, D. A., Saunders, S. E., Topliff, C. L., Kelling, C. L., and Bartelt-Hunt, S. L. (2010). Ethidium monoazide does not inhibit RT-PCR amplification of nonviable avian influenza RNA. J. Virol. Methods 164, 51–54. doi: 10.1016/j.jviromet.2009.11.024
Hamza, I. A., Jurzik, L., Überla, K., and Wilhelm, M. (2011). Methods to detect infectious human enteric viruses in environmental water samples. Int. J. Hyg. Environ. Health 214, 424–436. doi: 10.1016/j.ijheh.2011.07.014
Kim, K., Katayama, H., Kitajima, M., Tohya, Y., and Ohgaki, S. (2011). Development of a real-time RT-PCR assay combined with ethidium monoazide treatment for RNA viruses and its application to detect viral RNA after heat exposure. Water Sci. Technol. 63, 502–507. doi: 10.2166/wst.2011.249
Knight, A., Li, D., Uyttendaele, M., and Jaykus, L. A. (2013). A critical review of methods for detecting human noroviruses and predicting their infectivity. Crit. Rev. Microbiol. 39, 295–309. doi: 10.3109/1040841x.2012.709820
Leifels, M., Jurzik, L., Wilhelm, M., and Hamza, I. A. (2015). Use of ethidium monoazide and propidium monoazide to determine viral infectivity upon inactivation by heat, UV- exposure and chlorine. Int. J. Hyg. Environ. Health 218, 686–693. doi: 10.1016/j.ijheh.2015.02.003
Li, J. W., Xin, Z. T., Wang, X. W., Zheng, J. L., and Chao, F. H. (2002). Mechanisms of inactivation of hepatitis a virus by chlorine. Appl. Environ. Microbiol. 68, 4951–4955. doi: 10.1128/aem.68.10.4951-4955.2002
Li, X., Li, Y., Fan, M., Fan, S., Gao, W., Ren, J., et al. (2022). Inguinal lymph node sample collected by minimally invasive sampler helps to accurately diagnose ASF in dead pigs without necropsy. Front. Vet. Sci. 9:1000969. doi: 10.3389/fvets.2022.1000969
Liu, H., Meng, F., Nyaruaba, R., He, P., Hong, W., Jiang, M., et al. (2022). A triton X-100 assisted PMAxx-qPCR assay for rapid assessment of infectious African swine fever virus. Front. Microbiol. 13:1062544. doi: 10.3389/fmicb.2022.1062544
Mazur-Panasiuk, N., and Wozniakowski, G. (2020). Natural inactivation of African swine fever virus in tissues: influence of temperature and environmental conditions on virus survival. Vet. Microbiol. 242:108609. doi: 10.1016/j.vetmic.2020.108609
Moreno, L., Aznar, R., and Sánchez, G. (2015). Application of viability PCR to discriminate the infectivity of hepatitis A virus in food samples. Int. J. Food Microbiol. 201, 1–6. doi: 10.1016/j.ijfoodmicro.2015.02.012
Niederwerder, M. C., Khanal, P., Foland, T., Constance, L. A., Stoian, A. M. M., Deavours, A., et al. (2022). Stability of African swine fever virus in feed during environmental storage. Transbound. Emerg. Dis. 69, 3216–3224. doi: 10.1111/tbed.14666
Nocker, A., and Camper, A. K. (2009). Novel approaches toward preferential detection of viable cells using nucleic acid amplification techniques. FEMS Microbiol. Lett. 291, 137–142. doi: 10.1111/j.1574-6968.2008.01429.x
Nocker, A., Cheung, C. Y., and Camper, A. K. (2006). Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 67, 310–320. doi: 10.1016/j.mimet.2006.04.015
Norbert Mwiine, F., Nkamwesiga, J., Ndekezi, C., and Ochwo, S. (2019). Molecular characterization of African swine fever viruses from outbreaks in Peri-Urban Kampala, Uganda. Adv. Virol. 2019:1463245. doi: 10.1155/2019/1463245
Nuanualsuwan, S., Songkasupa, T., Boonpornprasert, P., Suwankitwat, N., Lohlamoh, W., and Nuengjamnong, C. (2022). Persistence of African swine fever virus on porous and non-porous fomites at environmental temperatures. Porcine Health Manag. 8:34. doi: 10.1186/s40813-022-00277-8
Parshionikar, S., Laseke, I., and Fout, G. S. (2010). Use of propidium monoazide in reverse transcriptase PCR to distinguish between infectious and noninfectious enteric viruses in water samples. Appl. Environ. Microbiol. 76, 4318–4326.
Pereira De Oliveira, R., Hutet, E., Lancelot, R., Paboeuf, F., Duhayon, M., Boinas, F., et al. (2020). Differential vector competence of Ornithodoros soft ticks for African swine fever virus: what if it involves more than just crossing organic barriers in ticks? Parasit. Vectors 13:618. doi: 10.1186/s13071-020-04497-1
Petrini, S., Feliziani, F., Casciari, C., Giammarioli, M., Torresi, C., and De Mia, G. M. (2019). Survival of African swine fever virus (ASFV) in various traditional Italian dry-cured meat products. Prev. Vet. Med. 162, 126–130.
Prevost, B., Goulet, M., Lucas, F. S., Joyeux, M., Moulin, L., and Wurtzer, S. (2016). Viral persistence in surface and drinking water: suitability of PCR pre-treatment with intercalating dyes. Water Res. 91, 68–76. doi: 10.1016/j.watres.2015.12.049
Randazzo, W., López-Gálvez, F., Allende, A., Aznar, R., and Sánchez, G. (2016). Evaluation of viability PCR performance for assessing norovirus infectivity in fresh-cut vegetables and irrigation water. Int. J. Food Microbiol. 229, 1–6. doi: 10.1016/j.ijfoodmicro.2016.04.010
Rodríguez, R. A., Pepper, I. L., and Gerba, C. P. (2009). Application of PCR-based methods to assess the infectivity of enteric viruses in environmental samples. Appl. Environ. Microbiol. 75, 297–307. doi: 10.1128/aem.01150-08
Rudi, K., Moen, B., Drømtorp, S. M., and Holck, A. L. (2005). Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl. Environ. Microbiol. 71, 1018–1024. doi: 10.1128/aem.71.2.1018-1024.2005
Sanchez, G., Elizaquivel, P., and Aznar, R. (2012). Discrimination of infectious hepatitis A viruses by propidium monoazide real-time RT-PCR. Food Environ. Virol. 4, 21–25. doi: 10.1007/s12560-011-9074-5
Schnetzinger, F., Pan, Y., and Nocker, A. (2013). Use of propidium monoazide and increased amplicon length reduce false-positive signals in quantitative PCR for bioburden analysis. Appl. Microbiol. Biotechnol. 97, 2153–2162. doi: 10.1007/s00253-013-4711-6
Simonet, J., and Gantzer, C. (2006). Degradation of the Poliovirus 1 genome by chlorine dioxide. J. Appl. Microbiol. 100, 862–870. doi: 10.1111/j.1365-2672.2005.02850.x
Soejima, T., Schlitt-Dittrich, F., and Yoshida, S. (2011). Polymerase chain reaction amplification length-dependent ethidium monoazide suppression power for heat-killed cells of Enterobacteriaceae. Anal. Biochem. 418, 37–43. doi: 10.1016/j.ab.2011.06.027
Stothard, P. (2000). The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28, 1102, 1104. doi: 10.2144/00286ir01
Tran, X. H., Le, T. T. P., Nguyen, Q. H., Do, T. T., Nguyen, V. D., Gay, C. G., et al. (2021). African swine fever virus vaccine candidate ASFV-G-ΔI177L efficiently protects European and native pig breeds against circulating Vietnamese field strain. Transbound. Emerg. Dis. 69, e497–e504. doi: 10.1111/tbed.14329
Tran, X. H., Phuong, L. T. T., Huy, N. Q., Thuy, D. T., Nguyen, V. D., Quang, P. H., et al. (2022). Evaluation of the safety profile of the ASFV vaccine candidate ASFV-G-ΔI177L. Viruses 14:896. doi: 10.3390/v14050896
Zeng, D., Qian, B., Li, Y., Zong, K., Peng, W., Liao, K., et al. (2022). Prospects for the application of infectious virus detection technology based on propidium monoazide in African swine fever management. Front. Microbiol. 13:1025758. doi: 10.3389/fmicb.2022.1025758
Keywords: African swine fever virus, propidium monoazide, quantitative PCR, viability, persistence
Citation: Li Y, Wang Z, Qing J, Hu D, Vo HT, Thi KT, Wang X and Li X (2024) Application of propidium monoazide quantitative PCR to discriminate of infectious African swine fever viruses. Front. Microbiol. 14:1290302. doi: 10.3389/fmicb.2023.1290302
Received: 07 September 2023; Accepted: 18 December 2023;
Published: 10 January 2024.
Edited by:
Jingqiang Ren, Wenzhou University, ChinaReviewed by:
Lauro Velazquez-Salinas, Agricultural Research Service (USDA), United StatesCopyright © 2024 Li, Wang, Qing, Hu, Vo, Thi, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinglong Wang, d3hsb25nQG53c3VhZi5lZHUuY24=; Xiaowen Li, bHh3ODI3MkAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.