
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 16 November 2023
Sec. Evolutionary and Genomic Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1287491
This article is part of the Research Topic Unlocking the Value of Wastewater: Innovative Biotechnologies and Bioprocesses for Resource Recovery in a Circular Economy Approach View all 8 articles
Recovering nitrogen (N) from municipal wastewater is a promising approach to prevent nutrient pollution, reduce energy use, and transition toward a circular N bioeconomy, but remains a technologically challenging endeavor. Existing N recovery techniques are optimized for high-strength, low-volume wastewater. Therefore, developing methods to concentrate dilute N from mainstream wastewater will bridge the gap between existing technologies and practical implementation. The N-rich biopolymer cyanophycin is a promising candidate for N bioconcentration due to its pH-tunable solubility characteristics and potential for high levels of accumulation. However, the cyanophycin synthesis pathway is poorly explored in engineered microbiomes. In this study, we analyzed over 3,700 publicly available metagenome assembled genomes (MAGs) and found that the cyanophycin synthesis gene cphA was ubiquitous across common activated sludge bacteria. We found that cphA was present in common phosphorus accumulating organisms (PAO) Ca. ‘Accumulibacter’ and Tetrasphaera, suggesting potential for simultaneous N and P bioconcentration in the same organisms. Using metatranscriptomic data, we confirmed the expression of cphA in lab-scale bioreactors enriched with PAO. Our findings suggest that cyanophycin synthesis is a ubiquitous metabolic activity in activated sludge microbiomes. The possibility of combined N and P bioconcentration could lower barriers to entry for N recovery, since P concentration by PAO is already a widespread biotechnology in municipal wastewater treatment. We anticipate this work to be a starting point for future evaluations of combined N and P bioaccumulation, with the ultimate goal of advancing widespread adoption of N recovery from municipal wastewater.
Recovering nitrogen (N) from municipal wastewater is a promising method to circularize anthropogenic N use. Traditionally, fertilizer manufacturing and other industries synthetically fix N from the atmosphere through the energy intensive Haber Bosch process, which accounts for 1–2% of global energy use (Batstone et al., 2015). A significant portion of this reactive N is ultimately lost to municipal and industrial wastewater or agricultural drainage water. This reactive N harms aquatic environments and impacts public health (Galloway et al., 2008). Therefore, reducing N emissions to the environment is a key goal of wastewater treatment plants. Reactive N is typically removed from municipal wastewater through microbially driven redox reactions back to inert N2, which is dissipated back to the atmosphere. N recovery from wastewater is appealing because it can reduce N release to the environment while rerouting reactive N back to food or chemical production, promoting a transition from a linear to a circular anthropogenic N cycle and reducing reliance on Haber Bosch. However, existing N recovery techniques are optimized for high-strength, low-volume wastewater and are not feasible to apply to low-strength, high-volume mainstream municipal wastewater (Beckinghausen et al., 2020). Therefore, a partition-release-recovery (PRR) approach has been proposed to sequester nutrients from mainstream wastewater to a highly concentrated sidestream (Batstone et al., 2015). An efficient partition step to concentrate dilute N is an essential yet poorly explored element of this approach for N recovery.
A promising lead for N bioconcentration is cyanophycin, an intracellular biopolymer composed of amino acids aspartate and arginine. Cyanophycin is polymerized via cyanophycin synthetase, cphA. Cyanophycin can be synthesized de novo by cyanophycin synthetase, or existing cyanophycin can be used as a primer compound (Ziegler et al., 1998). Cyanophycin synthesis results in ATP hydrolysis to ADP (Berg et al., 2000). Cyanophycin is broken down into dipeptides by cyanophycinase, cphB (Richter et al., 1999; Supplementary Figure S1). Cyanophycin dipeptides may be hydrolyzed by isoaspartyl peptidase iaaA, though other enzymes can perform this function (Sharon et al., 2023). Cyanophycin was originally characterized in cyanobacteria and has also been studied in a limited number of non-phototrophic bacteria (Füser and Steinbüchel, 2007). In phototrophic and diazotrophic cyanobacteria, cyanophycin is most likely used as a nitrogen storage compound under alternating light conditions, during periods where nitrogen is in excess, and during periods where sulfate and phosphate are limited (Flores et al., 2019). In non-phototrophic bacteria, cyanophycin synthesis, prevalence, and function is poorly understood. It is not known if these selective pressures for cyanophycin synthesis are generalizable to other taxa, particularly to as-yet-uncultivated heterotrophs that are prevalent in wastewater treatment bioreactors. Studies on axenic Acinetobacter cultures have found that cyanophycin accumulation occurs under phosphate or sulfate limitation, as well as in the presence of ammonium or arginine in excess of what is needed for growth (Elbahloul et al., 2005).
In isolate cultures, cyanophycin can reach up to 40% of cell dry weight (Elbahloul et al., 2005). Cyanophycin granules can be selectively by manipulating pH, enabling straightforward purification (Füser and Steinbüchel, 2005). Given the attractive solubility properties, industrial biotechnology research has focused on maximizing cyanophycin production in recombinant bacteria, yeasts, and transgenic plants (Nausch et al., 2016; Du et al., 2019). Cyanophycin has been explored as a starter compound for a variety of downstream applications, including animal feedstock supplementation (Nausch et al., 2020), polyelectrolyte multilayer production for biomedical applications (Uddin et al., 2020), and feedstock for biodegradable plastic production (Neumann et al., 2005).
Cyanophycin production in mixed microbial communities, particularly in wastewater bioprocesses, has not been well-documented to date. Recent work has shown that cyanophycin can be produced unintentionally in activated sludge (Zou et al., 2022). Other meta-omic studies have incidentally identified cyanophycin synthetase genes in microbes commonly found in activated sludge (Füser and Steinbüchel, 2007; Singleton et al., 2022) but have not systematically searched for the cyanophycin pathway in wastewater bioprocesses. Cyanophycin accumulation could add immense value to existing biological nutrient removal practices, namely the enhanced biological phosphorus removal (EBPR) process. EBPR processes enrich phosphorus accumulating organisms (PAO), heterotrophs that release and uptake P under alternating redox conditions and substrate availability. Existing EBPR processes already use the PRR approach for P recovery, where P-rich biomass is bioconcentrated and physically separated from the dilute liquid stream. Therefore, integrating cyanophycin accumulation with existing P removal practices offers a lower barrier to entry for N recovery.
To better understand the potential role of cyanophycin as an N-rich biopolymer for the PRR approach, we assessed over 3,700 publicly available metagenome assembled genomes (MAGs) to understand the prevalence of the cyanophycin biosynthetic pathway in activated sludge microbiomes. We also curated MAGs and isolate genomes of key functional groups known to contribute to N cycling and P accumulation to determine their capability for cyanophycin accumulation. Finally, we analyzed gene expression data of known PAO to understand whether PAO could utilize cyanophycin synthesis genes. We found that genes enabling cyanophycin accumulation were ubiquitous amongst activated sludge communities, which may be leveraged in the future to partition and recover N as cyanophycin.
Wastewater bioprocess MAGs were obtained from two primary sources that used the minimum information about a metagenome-assembled genome (MIMAG) standard, where high-quality MAGs met ≥90% completeness and ≤ 5% contamination and medium-quality MAGs met ≥50% completion and ≤ 10% contamination (Bowers et al., 2017). The first dataset is Genomes from Earth’s Microbiomes, a dataset assembled by the IGM/M Data Consortium from a variety of natural and engineered systems (Nayfach et al., 2021). Out of the entire collection of 52,515 medium and high-quality MAGs, MAGs with the metadata field “ecosystem_category” matching the query “wastewater” were selected for this analysis, resulting in a subset of 2,627 MAGs. We annotated the MAGs for coding regions and function using prokka v1.14.6 with default e-value threshold of 1e-06 (Seemann, 2014). The wastewater MAGs from this dataset were primarily represented by anaerobic digester samples, so a second set of MAGs representing activated sludge was also used and accessed through NCBI BioProject PRJNA629478. In this study, 1,083 high-quality MAGs were recovered through a combination of short-read and long-read sequencing (Singleton et al., 2021). In total, we analyzed 3,710 MAGs from activated sludge and wastewater bioreactors.
Isolate genomes and high-quality MAGs from activated sludge functional groups were curated from NCBI. As a point of comparison, genomes of cyanobacteria and Acinetobacter with known cyanophycin metabolic pathways were also included. A complete list of genomes used is available in Supplementary Table S1. To understand patterns among genes clustered near cphA, conserved gene clusters with gene synteny 6,000 bp upstream and downstream of cphA were identified and binned using GeneGrouper, with search settings of > = 20% identity and > =80% coverage of the seed gene (McFarland et al., 2022).
High-quality Candidatus ‘Accumulibacter’ (referred to herein as Accumulibacter) MAGs and associated metatranscriptomic data were used to examine cphA expression in Accumulibacter PAO. Our group previously operated a lab-scale denitrifying PAO reactor enriched in Accumulibacter (Gao et al., 2017; Wang et al., 2021). Three high quality Ca. Accumulibacter MAGs were assembled from this work belonging to clades IA, IC, and IF based on polyphosphate kinase (ppk1) gene phylogeny. MAGs and time-series metatranscriptomic data from this study were accessed through NCBI BioProject PRJNA576469. Raw RNA reads were filtered for quality with fastp (Chen et al., 2018) and reads mapping to rRNA were removed using BBMap against the SILVA database (Quast et al., 2012).1 Cleaned reads were aligned against Accumulibacter MAGs using kallisto (Bray et al., 2016). Expression levels of each mapped gene were normalized to Transcript per Million Reads (TPM). We also examined MAGs and expression levels in TPM from a lab-scale EBPR reactor where Tetrasphaera MAGs were recovered (McDaniel et al., 2022). Further operational details of the EBPR reactors can be found in their respective publications.
We searched for cphA in broad collections of wastewater-associated MAGs to understand the prevalence of the cyanophycin synthesis function. The first dataset we examined was primarily represented by MAGs recovered from anaerobic digester sludge (Figure 1). Around 10% of the MAGs, 271 out of 2,627, possessed the cphA gene. While nearly three quarters of the MAGs were derived from anaerobic digesters, only one third of these MAGs possessed a cphA gene (Figure 1A). On the other hand, the nutrient removal and activated sludge categories had greater proportions of MAGs with cphA compared to the original dataset. Examining the MAGs with cphA more closely, we found that key nutrient cycling organisms possessed a cphA gene, including known PAO Accumulibacter and Tetrasphaera, as well as ammonia oxidizing bacteria affiliated with the genus Nitrosomonas (Figure 1B). This result was surprising, because cphA had not been documented in Ca. Accumulibacter genomes to our knowledge.
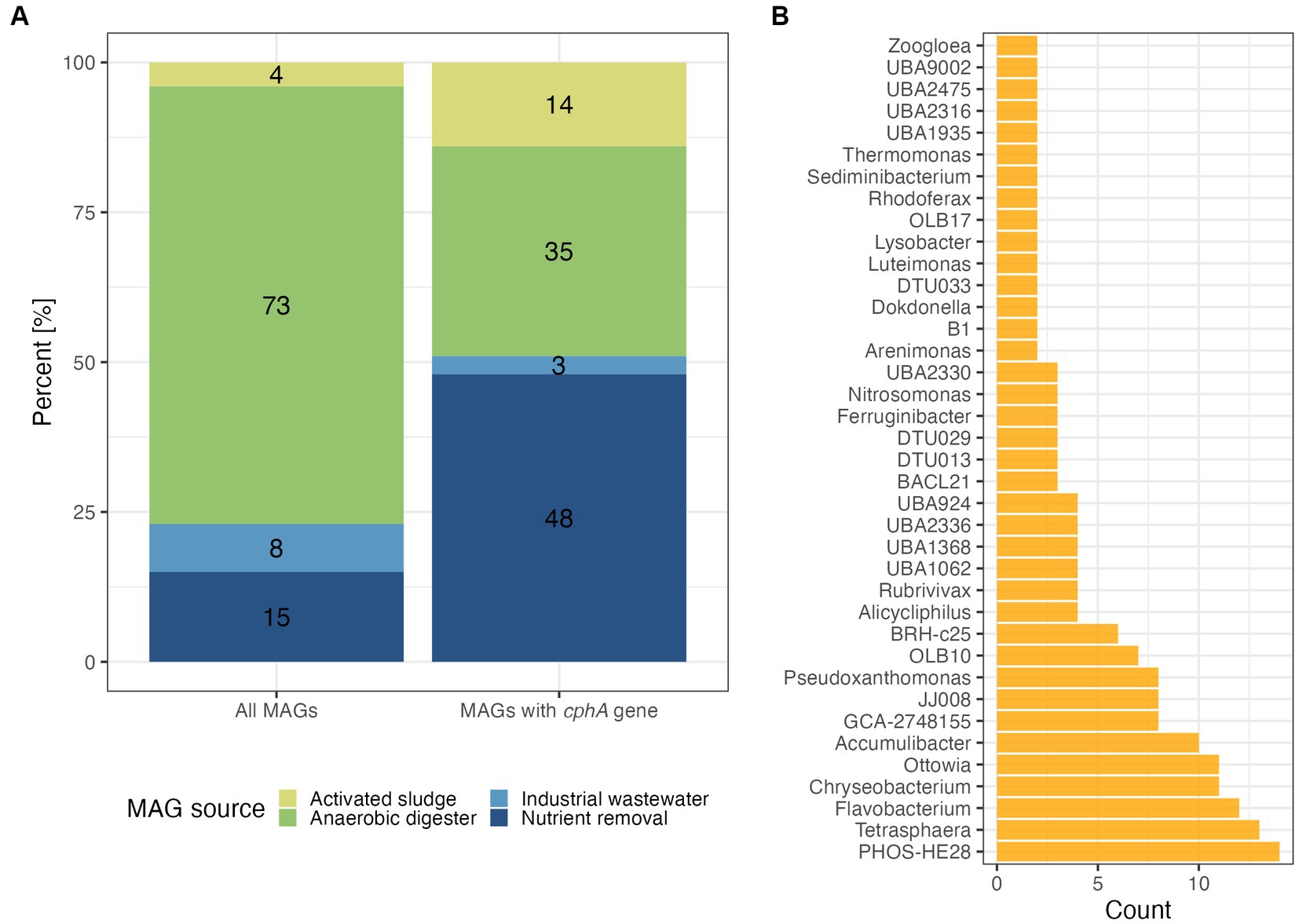
Figure 1. Distribution of MAGs from the Genomes from Earth’s Microbiomes collection (Nayfach et al., 2021) based on their identified ecosystem type with percent of total count labeled (A) and genus classifications of MAGs that had a cphA gene (B). Data in panel (B) are shown for genus-level classifications of two or more MAGs based on GTDB taxonomy.
A key limitation of the GEM dataset is the metadata categories. For example, separate nutrient removal and activated sludge categories are not representative of real wastewater treatment systems. Nutrient removal is often performed in activated sludge systems, where redox conditions are controlled to achieve N and P removal, such as the anaerobic-anoxic-oxic (A2O) process. One specific example from this dataset of potential category overlap is from Taxon Object ID 3300009540. This study was marked as activated sludge in the metadata, but further examination of the study (GOLD ID Gs0103597) shows that the samples were collected from an activated sludge system performing nitrification.
Given the metadata limitations of the GEM dataset, as well as the presence of cphA in nutrient cycling microbes, we next searched for cphA in a set of MAGs from activated sludge systems performing EBPR and N removal (Singleton et al., 2021). Out of 1,083 MAGs from this study, 552 possessed a cphA gene copy. Similar to the GEM dataset, we found that N and P cycling microbes harbored the cphA genes, including Ca. Accumulibacter, Dechloromonas, Nitrosomonas, and Propionivibrio (Supplementary Figure S2).
We also found that common filamentous bacteria possessed a cphA gene copy, including Zoogloea and Kouleothrix. Although filamentous bacteria are undesirable in large quantities in activated sludge systems due to their contribution to sludge bulking and poor settling, they are ubiquitous throughout activated sludge systems and can improve floc strength in balance with other microbes (Burger et al., 2017). The presence of cphA in filamentous bacteria may improve the viability of future cyanophycin applications, as filamentous bacteria can represent over 25% of sludge biomass in well-functioning systems (Mielczarek et al., 2012; Araújo Dos Santos et al., 2015).
A prominent cphA harboring genus in both large-scale datasets was PHOS-HE28. These organisms are poorly characterized members of the Flavobacteriales order. PHOS-HE28 have been identified in activated sludge and are related to bacteria isolated from saline environments, primarily seawater (Bowman, 2020). PHOS-HE28 and other related bacteria possess a ppk1 gene, so it is possible that these bacteria can store phosphate like PAO (Lucena et al., 2022), though more examination of ppk1 phylogeny and other phosphate transport genes is necessary to infer this function. Further investigation of PHOS-HE28 may be a promising avenue for integrating cyanophycin accumulation into existing treatment facilities given its ubiquity in activated sludge.
Overall, the unexpectedly high prevalence cphA genes in activated sludge MAGs is a positive sign that cyanophycin accumulation could integrate with existing wastewater treatment practices. This finding agrees with recent work that studied cyanophycin gene abundance and production in two full-scale wastewater treatment facilities using biofilm reactors for N and P removal (Zou et al., 2022). This work successfully identified cphA genes in biomass samples and found an association between cphA abundance and Accumulibacter marker gene abundance. However, they used read-based analysis for their metagenomic work rather than assembly-based analysis, while we analyzed MAGs to directly associate cphA genes with particular taxa. We next focused our analysis on specific N and P cycling organisms to better understand their potential for cyanophycin accumulation.
Since cphA genes were widespread among wastewater treatment microbiomes, we next examined a wider suite of complete genomes and near-complete MAGs obtained from NCBI of N and P cycling microbes to determine their potential for cyanophycin accumulation. We selected genomes of ammonia oxidizing bacteria (AOB), nitrite oxidizing bacteria (NOB), denitrifiers, PAO, and GAO to search for the cphA gene. A complete list of genomes examined is available in Supplementary Table S1.
Out of 68 genomes searched, 34 possessed at least one copy of the cphA gene. Notably, nearly all PAO genomes possessed a copy of the cphA gene. All Accumulibacter and Tetrasphaera genomes had a copy, as well as Ca. ‘Dechloromonas phosphorivorans’. This finding is consistent with previous research; cphA has been identified in Tetrasphaera (Singleton et al., 2022), and correlations were found between Accumulibacter phylogenetic markers and cphA gene abundance (Zou et al., 2022). The only PAO genome without a cphA gene copy was one Ca. ‘Dechloromonas phosphorivorans’ genome. This result was surprising since the six other Dechloromonas species analyzed possessed a cphA gene. It is unclear whether this discrepancy is due to a true lack of cphA in this particular species or limitations in sequencing and metagenome assembly. Regardless, given that PAO are already harnessed for their affinity for P bioconcentration, the potential for simultaneous N recovery via synthesis of cyanophycin in the same organism may be a promising avenue for combined P and N recovery.
Another notable finding was that no NOB genome possessed a cphA gene, while multiple AOB genomes possessed a cphA gene copy. This finding was surprising since AOB and NOB are similar metabolically as chemolithoautotrophs. Furthermore, some Nitrospira-affiliated taxa previously thought to be NOB are capable of complete ammonia oxidation (comammox), resulting in even more metabolic similarities to AOB with the ability to oxidize ammonia (Daims et al., 2016). We analyzed a known comammox genome of Ca. Nitrospira nitrosa (van Kessel et al., 2015) and did not find a cphA gene. Among the AOB genera, we found cphA in all Nitrosospira genomes and seven out of 12 Nitrosomonas genomes. A previous study also identified cphA in a Nitrosospira originally isolated from soil (Norton et al., 2008). Of the five Nitrosomonas genomes that did not possess cphA, four were originally isolated from marine or brackish environments, not activated sludge, and required a salt-enriched medium for growth (Koops et al., 1991). The other Nitrosomonas genome that did not possess cphA was originally isolated from cattle manure (Nakagawa and Takahashi, 2015). The presence of cphA in common activated sludge AOB, such as N. europaea and N. nitrosa, is promising for future study and integration of cyanophycin accumulation with existing nitrification bioprocesses.
We also assessed the presence of cphB in all functional group genomes. Interestingly, out of all analyzed genomes, only Tetrasphaera species possessed a cphB gene. This was unlike the four cyanobacterial genomes, which all possessed a cphB gene copy. The lack of a cphB gene does not guarantee that an organism is incapable of depolymerizing cyanophycin, which has been confirmed in isolate cultures of Pseudomonas aeruginosa (Sharon et al., 2023). To better understand how cyanophycin could fit into a broader biosynthetic pathway, particularly in genomes lacking a cphB gene, we next examined the genes upstream and downstream of cphA in each genome.
We used GeneGrouper to identify genes surrounding cphA in each genome analyzed in section 3.2 and bin the gene clusters into homologous groups. We also included four cyanobacterial genomes and an Acinetobacter species with a well-characterized cphA gene to understand whether cphA gene clusters from the activated sludge functional groups were similar to known cyanophycin producers. This gene cluster grouping approach is useful for determining the potential role of cyanophycin in a broader bacterial metabolism; genes that cluster together in a genome often constitute a specific biosynthetic pathway or operon (Fischbach and Voigt, 2010), and operons have been shown to be conserved across bacterial classes (Brandis et al., 2019).
We found two distinct gene clusters with gene synteny 6,000 bp upstream and downstream of cphA, shown in Figure 2. The cphA gene clusters from Tetrasphaera formed their own distinct group (Group 2), while the remainder of the genomes formed another group (Group 1). The reference cyanobacteria and Acinetobacter genomes did not form their own cluster nor cluster with the activated sludge taxa. The complexity of the Group 2 gene cluster was notable. The cluster consisted of a copy of cphB directly upstream of cphA, similar to cphA gene clusters of cyanobacteria (Krehenbrink et al., 2002; Füser and Steinbüchel, 2007). Furthermore, the Group 2 gene cluster harbored genes for glycogen synthesis and storage including glycogen synthetase glgA and glucose-1-adenylyltransferase glgC, as well as the amino acid utilization gene phosphoserine phosphatase serB. Glycogen is an important carbon reserve for Tetrasphaera, as they utilize glycogen under anaerobic conditions and replenish stores during aerobic conditions (Close et al., 2021; Yu et al., 2023).
The clustering of cphA and cphB with glycogen synthesis genes suggests that cyanophycin could be an active storage compound for Tetrasphaera. Genes in biosynthetic pathways with increasingly complex metabolites or related functions often cluster together, such as complex electron transport chains (Simon et al., 2004), protection against bacterial host immune response (Fischbach et al., 2006), and intracellular carbon storage (Kutralam-Muniasamy et al., 2017). Further analysis of carbon storage and amino acid utilization in Tetrasphaera can illuminate the role of cyanophycin as a storage compound.
The other cphA gene cluster group, Group 1, included a variety of activated sludge taxa. This gene cluster consisted of two copies of cphA, an unclassified transmembrane transport gene (ATP-binding ABC transporter), and two insertion sequences (IS). Notably, the gene cluster did not include a cphB gene copy. The presence of flanking IS indicates that this cluster could be a composite transposon, a type of mobile genetic element that facilitates movement of genetic material within a genome and between bacteria. Flanking IS around functional genes are a hallmark of composite transposons (Siguier et al., 2009). Composite transposons have been studied extensively for facilitating the spread of antibiotic resistance and xenobiotic resistance genes via horizontal gene transfer between taxa in diverse microbiomes (Top and Springael, 2003; Bennett, 2008). Mobile genetic elements have previously been identified as important vectors for horizontal gene transfer of antibiotic resistance genes in activated sludge microbiomes (Petrovich et al., 2018; Razavi et al., 2020), and recent work has also highlighted the role of composite transposons in transferring micropollutant degradation genes between bacteria (Bonatelli et al., 2023).
Further analysis of the cphA gene cluster in the Group 1 organisms would greatly improve our understanding of whether the gene cluster is a composite transposon or another type of mobile genetic element, which may have important implications for gene mobilization and transfer in complex microbial communities that are typical in wastewater bioprocesses. Future work could validate our findings through targeted PCR and long read sequencing to compare cphA gene clusters within activated sludge samples collected over time. Furthermore, the role of cyanophycin in the absence of cphB can be interrogated in future work that focuses on potential cphB analogs and cyanophycin transport genes.
Since PAO would be an excellent candidate for combined N and P bioconcentration, we wanted to determine whether cphA could be expressed in-situ by these bacteria. We first examined gene expression by mapping metatranscriptomic reads against three high quality Ca. Accumulibacter MAGs assembled from a lab-scale denitrifying P removal bioreactor (Gao et al., 2019; Wang et al., 2021). The samples were obtained over complete reactor cycles consisting of three redox phases: anaerobic, anoxic (N supplied as nitrite), and aerobic. The reactor was fed with either acetate or propionate as a carbon source in equivalent concentrations on a COD basis. The three Accumulibacter MAGs affiliated with different clades (IA, IC, and IF) based on ppk1 phylogeny and will be referred to hereafter by their clades. All three MAGs had two neighboring copies of cphA present in the genome. Neighboring copies of cphA have also been observed in other non-cyanobacterial genomes (Füser and Steinbüchel, 2007).
Each of the Accumulibacter MAGs exhibited different expression patterns across redox conditions; IA and IF had the greatest cphA expression during the aerobic phase, while IC had the greatest cphA expression during the anoxic phase (Supplementary Figure S3). Overall, IF had the greatest cphA expression, which agrees with previous findings that IF was the most transcriptionally active of all three MAGs (Wang et al., 2021). There were no apparent differences in cphA expression as a result of different carbon sources in the feed (Supplementary Figure S3).
In addition to analyzing the cphA expression of each MAG, we compared cphA expression to other key functional genes shown in Table 1. We used ppk1, phaC, and glgC as points of comparison against the expression of cphA, These genes are part of important phosphate transport and carbon storage functions and have been identified in a majority of Ca. Accumulibacter genomes (Petriglieri et al., 2022). The ppk1 gene is an essential biomarker for PAO phosphate cycling activity and is expressed actively in EBPR processes (He et al., 2007; He and McMahon, 2011). Both phaC and glgC are used in carbon storage pathways. Accumulibacter PAO assimilate biodegradable substrate under anaerobic conditions and store the carbon intracellularly using polyhydroxyalkanoates (PHA), while simultaneously depleting glycogen reserves. Under anoxic or aerobic conditions, Accumulibacter replenish glycogen reserves while depleting PHA reserves (Lanham et al., 2014). As shown in Figure 3, phaC had the highest level of expression across all MAGs. Surprisingly, cphA expression was significantly higher than ppk1 and glgC expression in IF. While gene expression does not directly indicate microbial activity or kinetics, the level of cphA expression relative to ppk1 and glgC points to the possibility of a highly active cyanophycin synthesis pathway.
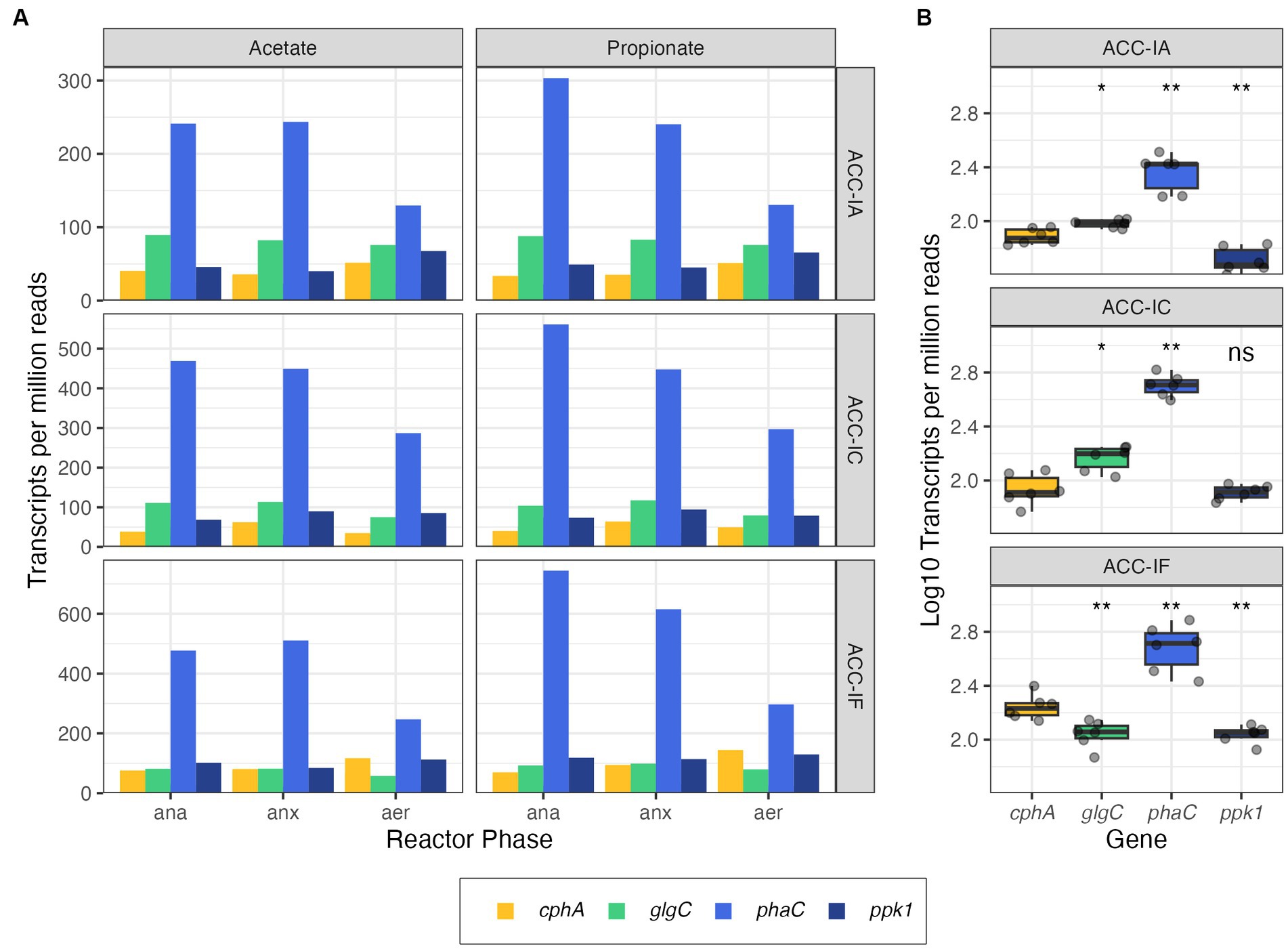
Figure 3. Gene expression profiles from metatranscriptomic reads of cphA (expression levels of both copies summed), glgC, phaC, and ppk1 in Accumulibacter MAGs plotted by the reactor phase (ana = anaerobic, anx = anoxic, aer = aerobic) and carbon source (A) and summarized per MAG (B). Significance levels in panel (B) are based on the Wilcox rank sum test, where glgC, phaC, and ppk are compared against cphA in each subpanel (ns, not significant, *p < =0.05, **p < = 0.01).
We also examined gene expression data of two Tetrasphaera MAGs, TET1 and TET2, recovered from a time-series study of an EBPR bioreactor (McDaniel et al., 2022). These MAGs contained two copies of the cphA gene in series, similar to Accumulibacter and the Group 1 cluster from section 3.3. TET1 and TET2 had similar genes surrounding cphA as the other Tetrasphaera genomes examined in section 3.3, with a neighboring cphB as well as other carbon storage and cycling genes (Supplementary Figure S4). Furthermore, the cphA copies in TET1 and TET2 were not surrounded by flanking IS.
Unlike the cphA expression in Accumulibacter, where both copies were expressed evenly, there was a notable difference in expression levels between the two cphA copies in the Tetrasphaera MAGs (Supplementary Figure S5). Notably, in both MAGs, the longer copy of cphA exhibited higher expression levels than the shorter copy; since the gene length is included in the normalization technique of calculating TPM, the gene length should not impact the reported expression levels. The combined gene expression of both cphA copies was relatively close to that of glgC and ppk1 in the Tetrasphaera bins across the sampling period (Figure 4). Again, gene expression does not directly indicate degree of function or activity, but the comparable level of expression of cphA compared to well-understood genes of the PAO phenotype is a positive indication that cyanophycin is an active storage polymer for Tetrasphaera. We also observed gene expression of cphB in the Tetrasphaera MAGs (Figure 4), further increasing the likelihood that cyanophycin is actively produced and utilized in these bacteria.
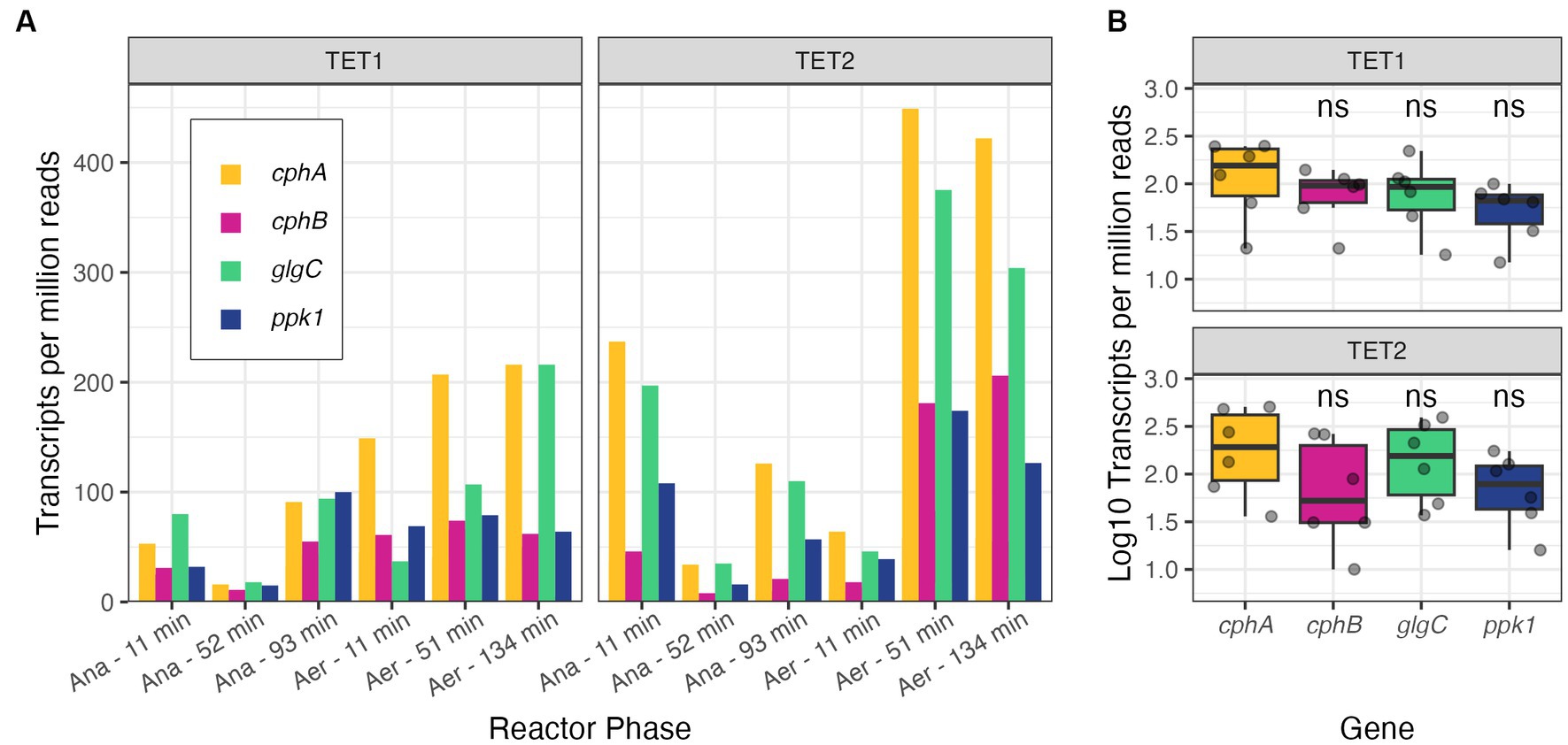
Figure 4. Gene expression profiles of cphA (expression levels of both copies summed), cphB, glgC, and ppk1 in Tetrasphaera MAGs plotted by the reactor phase (Ana = anaerobic, Aer = aerobic) (A) and summarized per MAG (B) of TET1 and TET2 from McDaniel et al. (2022). Significance levels in panel (B) are based on the Wilcox rank sum test, where cphB, glgC, and ppk1 are compared against cphA in each subpanel (ns, not significant).
The expression of cphA in PAO Ca. Accumulibacter and Tetrasphaera is promising for future applications of combined P and N accumulation. In particular, the location of the cphA gene in Tetrasphaera near other key carbon cycling genes, such glgA and glgC for glycogen synthesis, increases the likelihood that cyanophycin is an actively used biopolymer. Further analyses of cyanophycin pathway activity in response to operational variables and measurements of cyanophycin in real biomass would increase our confidence in successful simultaneous N and P bioconcentration in PAO.
In this study, we examined the prevalence of cyanophycin synthesis genes in wastewater bioprocess microbiomes. We observed a high prevalence of the cphA gene across a broad phylogenetic spectrum of common bacterial taxa in wastewater bioprocesses. The capacity for cyanophycin accumulation seems widespread given the presence of cphA in common PAO Accumulibacter, Tetrasphaera, and Dechloromonas and nitrifiers Nitrosomonas and Nitrosospira. We also used metatranscriptomic profiling to determine whether cphA genes were expressed by PAO under typical operating conditions, and found expression levels of cphA similar to other important P and carbon cycling genes. We also observed expression of cphB in Tetrasphaera, indicating that Tetrasphaera can actively produce and utilize cyanophycin. Overall, the presence of cyanophycin synthetase in nutrient cycling taxa suggests that cyanophycin cycling may already be occurring in existing biological nutrient removal processes.
Further research will expand on the findings of this work to advance N bioconcentration and fundamental microbial ecology questions. First, the feasibility of integrating cyanophycin accumulation into existing nutrient removal processes will largely depend on the ability to modulate cyanophycin production in concert with other desired functions, particularly P accumulation. Although we found that PAO harbor and express cphA, it is not clear whether cyanophycin accumulation occurs simultaneously with P accumulation. Second, fundamental understanding of cyanophycin accumulation by wastewater bioprocess taxa will improve with further examination of the cphA gene cluster as a possible mobile genetic element. While mobile genetic elements are intensely studied as a means of transferring antibiotic resistance genes in wastewater-associated microbiomes, their role in transferring nutrient cycling genes is less clear. Overall, our findings provide evidence that cyanophycin accumulation is a widespread function in nutrient removal bioprocesses and opens possibilities for accelerating nutrient recovery from wastewater through N bioconcentration.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
MF: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. RR: Conceptualization, Data curation, Writing – review & editing. WT: Conceptualization, Funding acquisition, Writing – review & editing. KT: Conceptualization, Funding acquisition, Investigation, Writing – review & editing. GW: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This material is based upon work supported by the National Science Foundation under Award No. 2033793.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1287491/full#supplementary-material.
Araújo Dos Santos, L., Ferreira, V., Neto, M. M., Pereira, M. A., Mota, M., and Nicolau, A. (2015). Study of 16 Portuguese activated sludge systems based on filamentous bacteria populations and their relationships with environmental parameters. Appl. Microbiol. Biotechnol. 99, 5307–5316. doi: 10.1007/s00253-015-6393-8
Batstone, D. J., Hülsen, T., Mehta, C. M., and Keller, J. (2015). Platforms for energy and nutrient recovery from domestic wastewater: a review. Chemosphere 140, 2–11. doi: 10.1016/j.chemosphere.2014.10.021
Beckinghausen, A., Odlare, M., Thorin, E., and Schwede, S. (2020). From removal to recovery: an evaluation of nitrogen recovery techniques from wastewater. Appl. Energy 263:114616. doi: 10.1016/j.apenergy.2020.114616
Bennett, P. M. (2008). Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria: plasmid-encoded antibiotic resistance. Br. J. Pharmacol. 153, S347–S357. doi: 10.1038/sj.bjp.0707607
Berg, H., Ziegler, K., Piotukh, K., Baier, K., Lockau, W., and Volkmer-Engert, R. (2000). Biosynthesis of the cyanobacterial reserve polymer multi-L-arginyl-poly-L-aspartic acid (cyanophycin): mechanism of the cyanophycin synthetase reaction studied with synthetic primers. Eur. J. Biochem. 267, 5561–5570. doi: 10.1046/j.1432-1327.2000.01622.x
Bonatelli, M. L., Rohwerder, T., Popp, D., Liu, Y., Akay, C., Schultz, C., et al. (2023). Recently evolved combination of unique sulfatase and amidase genes enables bacterial degradation of the wastewater micropollutant acesulfame worldwide. Front. Microbiol. 14:1223838. doi: 10.3389/fmicb.2023.1223838
Bowers, R. M., Kyrpides, N. C., Stepanauskas, R., Harmon-Smith, M., Doud, D., Reddy, T. B. K., et al. (2017). Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731. doi: 10.1038/nbt.3893
Bowman, J. P. (2020). Out from the shadows – resolution of the taxonomy of the family cryomorphaceae. Front. Microbiol. 11:795. doi: 10.3389/fmicb.2020.00795
Brandis, G., Cao, S., and Hughes, D. (2019). Operon concatenation is an ancient feature that restricts the potential to rearrange bacterial chromosomes. Mol. Biol. Evol. 36, 1990–2000. doi: 10.1093/molbev/msz129
Bray, N. L., Pimentel, H., Melsted, P., and Pachter, L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. doi: 10.1038/nbt.3519
Burger, W., Krysiak-Baltyn, K., Scales, P. J., Martin, G. J. O., Stickland, A. D., and Gras, S. L. (2017). The influence of protruding filamentous bacteria on floc stability and solid-liquid separation in the activated sludge process. Water Res. 123, 578–585. doi: 10.1016/j.watres.2017.06.063
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Close, K., Marques, R., Carvalho, V. C. F., Freitas, E. B., Reis, M. A. M., Carvalho, G., et al. (2021). The storage compounds associated with Tetrasphaera PAO metabolism and the relationship between diversity and P removal. Water Res. 204:117621. doi: 10.1016/j.watres.2021.117621
Daims, H., Lücker, S., and Wagner, M. (2016). A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 24, 699–712. doi: 10.1016/j.tim.2016.05.004
Du, J., Li, L., and Zhou, S. (2019). Microbial production of cyanophycin: from enzymes to biopolymers. Biotechnol. Adv. 37:107400. doi: 10.1016/j.biotechadv.2019.05.006
Elbahloul, Y., Krehenbrink, M., Reichelt, R., and Steinbüchel, A. (2005). Physiological conditions conducive to high cyanophycin content in biomass of Acinetobacter calcoaceticus strain ADP1. Appl. Environ. Microbiol. 71, 858–866. doi: 10.1128/AEM.71.2.858-866.2005
Fischbach, M. A., Lin, H., Zhou, L., Yu, Y., Abergel, R. J., Liu, D. R., et al. (2006). The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc. Natl. Acad. Sci. 103, 16502–16507. doi: 10.1073/pnas.0604636103
Fischbach, M., and Voigt, C. A. (2010). Prokaryotic gene clusters: a rich toolbox for synthetic biology. Biotechnol. J. 5, 1277–1296. doi: 10.1002/biot.201000181
Flores, E., Arévalo, S., and Burnat, M. (2019). Cyanophycin and arginine metabolism in cyanobacteria. Algal Res. 42:101577. doi: 10.1016/j.algal.2019.101577
Füser, G., and Steinbüchel, A. (2005). Investigations on the solubility behavior of cyanophycin. Solubility of cyanophycin in solutions of simple inorganic salts. Biomacromolecules 6, 1367–1374. doi: 10.1021/bm049371o
Füser, G., and Steinbüchel, A. (2007). Analysis of genome sequences for genes of cyanophycin metabolism: identifying putative cyanophycin metabolizing prokaryotes. Macromol. Biosci. 7, 278–296. doi: 10.1002/mabi.200600207
Galloway, J. N., Townsend, A. R., Erisman, J. W., Bekunda, M., Cai, Z., Freney, J. R., et al. (2008). Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320, 889–892. doi: 10.1126/science.1136674
Gao, H., Liu, M., Griffin, J. S., Xu, L., Xiang, D., Scherson, Y. D., et al. (2017). Complete nutrient removal coupled to nitrous oxide production as a bioenergy source by denitrifying polyphosphate-accumulating organisms. Environ. Sci. Technol. 51, 4531–4540. doi: 10.1021/acs.est.6b04896
Gao, H., Mao, Y., Zhao, X., Liu, W.-T., Zhang, T., and Wells, G. (2019). Genome-centric metagenomics resolves microbial diversity and prevalent truncated denitrification pathways in a denitrifying PAO-enriched bioprocess. Water Res. 155, 275–287. doi: 10.1016/j.watres.2019.02.020
He, S., Gall, D. L., and McMahon, K. D. (2007). “Candidatus Accumulibacter” population structure in enhanced biological phosphorus removal sludges as revealed by polyphosphate kinase genes. Appl. Environ. Microbiol. 73, 5865–5874. doi: 10.1128/AEM.01207-07
He, S., and McMahon, K. D. (2011). ‘Candidatus Accumulibacter’ gene expression in response to dynamic EBPR conditions. ISME J. 5, 329–340. doi: 10.1038/ismej.2010.127
Koops, H. P., Böher, B., Mör, U. C., Pommerening-Röser, A., and Stehr, G. (1991). Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov. and Nitrosomonas halophila sp. nov. Microbiology 137, 1689–1699. doi: 10.1099/00221287-137-7-1689
Krehenbrink, M., Oppermann-Sanio, F.-B., and Steinbüchel, A. (2002). Evaluation of non-cyanobacterial genome sequences for occurrence of genes encoding proteins homologous to cyanophycin synthetase and cloning of an active cyanophycin synthetase from Acinetobacter sp. strain DSM 587. Arch. Microbiol. 177, 371–380. doi: 10.1007/s00203-001-0396-9
Kutralam-Muniasamy, G., Corona-Hernandez, J., Narayanasamy, R.-K., Marsch, R., and Pérez-Guevara, F. (2017). Phylogenetic diversification and developmental implications of poly-(R)-3-hydroxyalkanoate gene cluster assembly in prokaryotes. FEMS Microbiol. Lett. 364:135. doi: 10.1093/femsle/fnx135
Lanham, A. B., Oehmen, A., Saunders, A. M., Carvalho, G., Nielsen, P. H., and Reis, M. A. M. (2014). Metabolic modelling of full-scale enhanced biological phosphorus removal sludge. Water Res. 66, 283–295. doi: 10.1016/j.watres.2014.08.036
Lucena, T., Sánchez, O., Sanz-Saez, I., Acinas, S. G., Garrido, L., Mas, J., et al. (2022). Parvicella tangerina gen. nov., sp. nov. (Parvicellaceae fam. nov., Flavobacteriales), first cultured representative of the marine clade UBA10066, and Lysobacter luteus sp. nov., from activated sludge of a seawater-processing wastewater treatment plant. Int. J. Syst. Evol. Microbiol. 72:5498. doi: 10.1099/ijsem.0.005498
McDaniel, E. A., Van Steenbrugge, J. J. M., Noguera, D. R., McMahon, K. D., Raaijmakers, J. M., Medema, M. H., et al. (2022). TbasCO: trait-based comparative ‘omics identifies ecosystem-level and niche-differentiating adaptations of an engineered microbiome. ISME Commun. 2:111. doi: 10.1038/s43705-022-00189-2
McFarland, A. G., Kennedy, N. W., Mills, C. E., Tullman-Ercek, D., Huttenhower, C., and Hartmann, E. M. (2022). Density-based binning of gene clusters to infer function or evolutionary history using GeneGrouper. Bioinformatics 38, 612–620. doi: 10.1093/bioinformatics/btab752
Mielczarek, A. T., Kragelund, C., Eriksen, P. S., and Nielsen, P. H. (2012). Population dynamics of filamentous bacteria in Danish wastewater treatment plants with nutrient removal. Water Res. 46, 3781–3795. doi: 10.1016/j.watres.2012.04.009
Nakagawa, T., and Takahashi, R. (2015). Nitrosomonas stercoris sp. nov., a chemoautotrophic ammonia-oxidizing bacterium tolerant of high ammonium isolated from composted cattle manure. Microbes Environ. 30, 221–227. doi: 10.1264/jsme2.ME15072
Nausch, H., Dorn, M., Frolov, A., Hoedtke, S., Wolf, P., and Broer, I. (2020). Direct delivery of health promoting β-asp-arg dipeptides via stable co-expression of cyanophycin and the cyanophycinase CphE241 in tobacco plants. Front. Plant Sci. 11:842. doi: 10.3389/fpls.2020.00842
Nausch, H., Hausmann, T., Ponndorf, D., Hühns, M., Hoedtke, S., Wolf, P., et al. (2016). Tobacco as platform for a commercial production of cyanophycin. New Biotechnol. 33, 842–851. doi: 10.1016/j.nbt.2016.08.001
Nayfach, S., Roux, S., Seshadri, R., Udwary, D., Varghese, N., Schulz, F., et al. (2021). A genomic catalog of earth’s microbiomes. Nat. Biotechnol. 39, 499–509. doi: 10.1038/s41587-020-0718-6
Neumann, K., Stephan, D. P., Ziegler, K., Hühns, M., Broer, I., Lockau, W., et al. (2005). Production of cyanophycin, a suitable source for the biodegradable polymer polyaspartate, in transgenic plants: production of cyanophycin in transgenic plants. Plant Biotechnol. J. 3, 249–258. doi: 10.1111/j.1467-7652.2005.00122.x
Norton, J. M., Klotz, M. G., Stein, L. Y., Arp, D. J., Bottomley, P. J., Chain, P. S. G., et al. (2008). Complete genome sequence of Nitrosospira multiformis, an ammonia-oxidizing bacterium from the soil environment. Appl. Environ. Microbiol. 74, 3559–3572. doi: 10.1128/AEM.02722-07
Petriglieri, F., Singleton, C. M., Kondrotaite, Z., Dueholm, M. K. D., McDaniel, E. A., McMahon, K. D., et al. (2022). Reevaluation of the phylogenetic diversity and global distribution of the genus “Candidatus Accumulibacter.”. mSystems 7:e0001622. doi: 10.1128/msystems.00016-22
Petrovich, M., Chu, B., Wright, D., Griffin, J., Elfeki, M., Murphy, B. T., et al. (2018). Antibiotic resistance genes show enhanced mobilization through suspended growth and biofilm-based wastewater treatment processes. FEMS Microbiol. Ecol. 94:fiy041. doi: 10.1093/femsec/fiy041
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Razavi, M., Kristiansson, E., Flach, C.-F., and Larsson, D. G. J. (2020). The association between insertion sequences and antibiotic resistance genes. mSphere 5:e00418-20. doi: 10.1128/mSphere.00418-20
Richter, R., Hejazi, M., Kraft, R., Ziegler, K., and Lockau, W. (1999). Cyanophycinase, a peptidase degrading the cyanobacterial reserve material multi-L-arginyl-poly-L-aspartic acid (cyanophycin). Molecular cloning of the gene of Synechocystis sp. PCC 6803, expression in Escherichia coli, and biochemical characterization of the purified enzyme. Eur. J. Biochem. 263, 163–169. doi: 10.1046/j.1432-1327.1999.00479.x
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sharon, I., McKay, G. A., Nguyen, D., and Schmeing, T. M. (2023). Discovery of cyanophycin dipeptide hydrolase enzymes suggests widespread utility of the natural biopolymer cyanophycin. Proc. Natl. Acad. Sci. 120:e2216547120. doi: 10.1073/pnas.2216547120
Siguier, P., Gagnevin, L., and Chandler, M. (2009). The new IS1595 family, its relation to IS1 and the frontier between insertion sequences and transposons. Res. Microbiol. 160, 232–241. doi: 10.1016/j.resmic.2009.02.003
Simon, J., Einsle, O., Kroneck, P. M. H., and Zumft, W. G. (2004). The unprecedented nos gene cluster of Wolinella succinogenes encodes a novel respiratory electron transfer pathway to cytochrome c nitrous oxide reductase. FEBS Lett. 569, 7–12. doi: 10.1016/j.febslet.2004.05.060
Singleton, C. M., Petriglieri, F., Kristensen, J. M., Kirkegaard, R. H., Michaelsen, T. Y., Andersen, M. H., et al. (2021). Connecting structure to function with the recovery of over 1000 high-quality metagenome-assembled genomes from activated sludge using long-read sequencing. Nat. Commun. 12:2009. doi: 10.1038/s41467-021-22203-2
Singleton, C. M., Petriglieri, F., Wasmund, K., Nierychlo, M., Kondrotaite, Z., Petersen, J. F., et al. (2022). The novel genus, ‘Candidatus Phosphoribacter’, previously identified as Tetrasphaera, is the dominant polyphosphate accumulating lineage in EBPR wastewater treatment plants worldwide. ISME J. 16, 1605–1616. doi: 10.1038/s41396-022-01212-z
Top, E. M., and Springael, D. (2003). The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr. Opin. Biotechnol. 14, 262–269. doi: 10.1016/S0958-1669(03)00066-1
Uddin, Z., Fang, T., Siao, J., and Tseng, W. (2020). Wound healing attributes of polyelectrolyte multilayers prepared with multi- L -arginyl-poly- L -aspartate pairing with hyaluronic acid and γ-polyglutamic acid. Macromol. Biosci. 20:2000132. doi: 10.1002/mabi.202000132
van Kessel, M. A. H., Speth, D. R., Albertsen, M., Nielsen, P. H., Camp, H. J. M. O., Kartal, B., et al. (2015). Complete nitrification by a single microorganism. Nature 528:555+. doi: 10.1038/nature16459
Wang, Y., Gao, H., and Wells, G. (2021). Integrated omics analyses reveal differential gene expression and potential for cooperation between denitrifying polyphosphate and glycogen accumulating organisms. Environ. Microbiol. 23, 3274–3293. doi: 10.1111/1462-2920.15486
Yu, Z., Xu, S., Wang, P., Liu, D., and Lu, H. (2023). Phosphorus removal and storage polymer synthesis by Tetrasphaera -related bacteria with different carbon sources. ACS EST Water 3, 1243–1421. doi: 10.1021/acsestwater.3c00046
Ziegler, K., Diener, A., Herpin, C., Richter, R., Deutzmann, R., and Lockau, W. (1998). Molecular characterization of cyanophycin synthetase, the enzyme catalyzing the biosynthesis of the cyanobacterial reserve material multi- L-arginyl-poly- L-aspartate (cyanophycin). Eur. J. Biochem. 254, 154–159. doi: 10.1046/j.1432-1327.1998.2540154.x
Keywords: nitrogen recovery, microbial ecology, cyanophycin, activated sludge, phosphorus accumulating organisms
Citation: Farmer M, Rajasabhai R, Tarpeh W, Tyo K and Wells G (2023) Meta-omic profiling reveals ubiquity of genes encoding for the nitrogen-rich biopolymer cyanophycin in activated sludge microbiomes. Front. Microbiol. 14:1287491. doi: 10.3389/fmicb.2023.1287491
Received: 01 September 2023; Accepted: 01 November 2023;
Published: 16 November 2023.
Edited by:
Nídia Dana Lourenço, New University of Lisbon, PortugalReviewed by:
Paula Maza Marquez, University of Granada, SpainCopyright © 2023 Farmer, Rajasabhai, Tarpeh, Tyo and Wells. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: George Wells, Z2VvcmdlLndlbGxzQG5vcnRod2VzdGVybi5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.