
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 November 2023
Sec. Microbial Physiology and Metabolism
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1285796
This article is part of the Research Topic Advances in Molecular Plant Pathology, Plant Abiotic and Biotic Stress View all 19 articles
 Jiale Qiu
Jiale Qiu Jie Zhang
Jie Zhang Huihui Zhao
Huihui Zhao Cuiping Wu
Cuiping Wu Caoliang Jin
Caoliang Jin Xiangdong Hu
Xiangdong Hu Jian Li
Jian Li Xiuling Cao
Xiuling Cao Shenkui Liu
*
Shenkui Liu
* Xuejiao Jin
*
Xuejiao Jin
*Carbonate stress has profound impacts on both agricultural and industrial production. Although a number of salinity-tolerant genes have been reported and applied in plants, there is a lack of research on the role of cell wall-related genes in resistance to carbonate. Likewise, in industry, current strategies have not been able to more effectively address the conflict between stress-induced microalgal biofuel accumulation and microalgal growth inhibition. It is of great significance to study the adaptation mechanism of carbonate-tolerant organisms and to explore related genes for future genetic modification. In this study, the role of the cell wall in the NaHCO3-tolerant chlorella JB17 was investigated. We found that JB17 possesses a relatively thick cell wall with a thickness of 300–600 nm, which is much higher than that of the control chlorella with a thickness of about 100 nm. Determination of the cell wall polysaccharide fractions showed that the cellulose content in the JB17 cell wall increased by 10.48% after NaHCO3 treatment, and the decrease in cellulose levels by cellulase digestion inhibited its resistance to NaHCO3. Moreover, the saccharide metabolome revealed that glucose, rhamnose, and trehalose levels were higher in JB17, especially rhamnose and trehalose, which were almost 40 times higher than in control chlorella. Gene expression detection identified an up-regulated expressed gene after NaHCO3 treatment, JbKOBITO1, overexpression of which could improve the NaHCO3 tolerance of Chlamydomonas reinhardtii. As it encodes a glycosyltransferase-like protein that is involved in cellulose synthesis, the strong tolerance of JB17 to NaHCO3 may be partly due to the up-regulated expression of JbKOBITO 1 and JbKOBITO 1-mediated cellulose accumulation. The above results revealed a critical role of cellulose in the NaHCO3 resistance of JB17, and the identified NaHCO3-tolerance gene will provide genetic resources for crop breeding in saline-alkali soils and for genetic modification of microalgae for biofuel production.
Carbonate is an important limiting factor for biological growth and for the development of industry and agriculture. On the one hand, carbonate stress not only causes ionic toxicity and osmotic stress, but also further increase the pH of saline-alkaline soils, which seriously affects crop growth and yield (Wang Y. et al., 2020). On the other hand, microalgae, as a potential feedstock for biofuel production (Khoo et al., 2020), has been limited in the commercialisation of microalgae biofuels due to the contradiction between microalgal growth under environmental stress (including carbonate stress) and biofuel accumulation (Ji et al., 2018). Therefore, it is of great significance to study the tolerance mechanism of organisms to carbonate stress and excavate carbonate-tolerant genes to improve the tolerance in plants and industrial microbial strains.
Current research on saline–alkali soils has screened a number of carbonate-tolerant plant resources, with the more familiar ones including Suaeda corniculata, Puccinellia tenuiflora, oats, etc. (Pang et al., 2016; Bai et al., 2018; Qin et al., 2018; Ye et al., 2019). A range of genes for carbonate tolerance have also been reported. For instance, SlWRKY28, a gene cloned from the WRKY family of Salix linearistipular, improves the saline-alkali tolerance of transgenic lines by inducing regulatory responses of enzyme genes in the reactive oxygen scavenging pathway (Wang X. et al., 2020). Zheng et al. (2021) reported that CrPIP23, a membrane Aquaporins (AQPs) protein gene from Canavalia rosea, enhanced alkaline tolerance of transgenic plants by promoting water transport. Liu et al. (2020) reported that a chitinase gene LcCHI2 from Leymus chinensis helped reduce osmotic stress other than Na+ ion toxicity or improved water-use-efficiency, improves salt-alkali tolerance in transgenic maize. However, the cell wall, as the first barrier against stress, and its regulatory gene, which has been extensively studied in plant resistance to salt and cadmium stress in recent years (Jia et al., 2021; Dabravolski and Isayenkov, 2023), has not been strongly implicated in resistance to carbonate stress.
Carbonate stress also limits the growth of certain engineered strains for biofuel production. In the industrial field, microalgae are regarded as an important resource for biofuel production. Previous study reported that many environmental stress conditions can promoted cell wall thinning and significantly increased lipid production in Chlorella vulgaris (Nadzir et al., 2023). For example, the study by Li et al. (2018) reported that treatment with high concentrations of NaHCO3 (160 mM) stimulated the accumulation of Chlorella vulgaris lipids, but inhibited cell growth. Therefore, the conflict, between the use of stress to promote biofuel accumulation and stress-induced microbial growth inhibition, limits the industrial production of biofuels in large quantities. Although certain study has reported that product content can be effectively increased by two-stage cultivation of algae, it is simpler and more effective to search for carbonate-tolerant genes and apply them to improve stress resistance (Yun et al., 2019). A recent review summarised the use of genetic modification strategies to improve lipid production in microalgae but these studies have focused on genes that promote lipid synthesis in microalgae (Khoo et al., 2023). Since many genes in algae can be involved in stress response (Hong et al., 2023), the mining of carbonate-tolerant genes is important to optimise the balance between microalgal growth and lipid accumulation.
JB17, is a member of the Nannochloris species that we previously isolated from extremely saline-alkali soil in northeastern China. It showed higher tolerance to NaCl and NaHCO3, surviving in NaHCO3 medium at concentrations up to 1 M (Qiao et al., 2015). JB17, as a eukaryotic microalga, is a unicellular microorganism with both plant and microbial properties. Specifically, it has a structure similar to plant cells, such as a nucleus, chloroplasts and mitochondria, as well as other organelles surrounded by cell membranes, which enable it to photosynthesise to produce high-value bioactive substances (Khoo et al., 2023). In addition, its characteristic of rapid microbial growth enables the response to carbonate stress to be observed in a short period of time, and the unicellular structure makes it easier to study its physiological and molecular responses at the cellular level. Therefore, studying the mechanism of JB17 NaHCO3 tolerance and identifying carbonate-tolerant genes will not only benefit agricultural development but also the commercialisation of microalgal biofuels.
In this study, we analyzed the changes in the composition of the cell wall of the saline-alkali tolerant JB17 before and after NaHCO3 exposure, and identified a key cell wall gene involved in resistance to NaHCO3 through transcriptome analysis and transgenic experiments. Our results not only revealed the mechanism of NaHCO3 tolerance in JB17, but also the identified cell wall-related gene would provide valuable genetic resources for agricultural and industrial production.
The control chlorella (Chlorella variabilis) and Chlamydomonas reinhardtii were obtained from the Freshwater Algae Culture Collection (Institute of Hydrobiology, Chinese Academy of Science, Wuhan, China). The saline-alkali-tolerant Chlorella JB17 was previously isolated from extremely alkaline-saline soil (pH > 10) from the Songnen Plain (46°27′N, 125°22′E, Heilongjiang Province, China) (Qiao et al., 2015). For algal growth, JB17 and C. variabilis were cultured using Blue-Green Medium (BG11) medium, while C. reinhardtii was cultured using Tris-Acetate-Phosphate (TAP) medium. The medium was prepared according to the information provided by the Freshwater Algae Culture Collection at the Institute of Hydrobiology (http://algae.ihb.ac.cn/, accessed on 5 March 2023). All algae were cultured at 23 ± 1°C, with illumination at 40 μM photons m−1 s −1 under 16 h light/8 h dark photoperiod (Wang M. et al., 2020).
Transmission electron microscopy (TEM) was performed according to previously described methods (Jin et al., 2018). 15 mL algae culture in logarithmic growth phase (at approximately 0.5 × 108 cells/mL) were collected and centrifugated at 800 g for 10 min, and fixed overnight at 4°C in 1 mL fixation buffer (2.5% glutaraldehyde, 0.05 M phosphate, pH 7.2). The glutaraldehyde-fixed algae were washed three times with fixation buffer and then fixed in 2% osmium tetroxide (OsO4) at 4°C for 2 h in the dark. Then the algae were dehydrated through a series of graded ethanol (70, 80, 90, 95, and 100%) for 10 min each time, and then embedded in Spurr’s resin. Ultrathin sections (70 nm) were cut with an ultramicrotome (EM UC7; Leica, Germany) and sequentially stained with uranyl acetate for 20 min and Reynolds’ lead citrate for 5 min. The specimens were observed using a Hitachi H-7650 (Hitachi, Japan) or a JEM-1230 (JEOL Co. Ltd., Japan) transmission electron microscope operated at 80 kV.
Firstly, total RNA was extracted from JB17, and mRNA was reverse transcribed into cDNA using M-MLV reverse transcription kit (Thermo Fisher, Shanghai, China). The full-length coding sequence (CDS) of JbKOBITO 1 was amplified from cDNA and inserted into the Hpa I-linearized plasmid pLM006 by homologous recombination methods to generate a vector for electro-transformation of C. reinhardtii. The pLM006 plasmid was expressed under the control of the PASD promoter, and mCherry protein was expressed at the C-terminus of the target gene. The primers used for plasmid construction are listed in Supplementary Table S1. The correctness of the plasmid was verified by DNA sequencing.
The expression changes of JbKOBITO 1 before and after NaHCO3 treatment were detected by RT–qPCR as described previously (Li et al., 2023). The primers used are listed in Supplementary Table S1.
The generation of transgenic algae lines overexpressing JbKOBITO 1 was performed as described previously (Li et al., 2023). After the colonies had grown, different primers were designed to identify the mcherry fusion fluorescent protein gene, the PASD promoter and the target gene by PCR, to obtain the positive strains into which the plasmid had been successfully transferred. Three overexpression lines were analyzed for their NaHCO3 tolerance, and the results showed the same trends.
Log-phase cells were aspirated and added to a modified TAP liquid medium containing 0 mM and 300 mM NaHCO3 for 3 days. Then, the cells were cultured in the dark for 12 h to consume starch before being rapidly frozen in liquid nitrogen, after which the cells were disrupted to obtain the cell wall by milling using a steel bead and FastPrep-24™ 5G bead-beating grinder (MP Biomedicals, United States). All the cells were examined under a microscope for disruption to ensure that the cell wall preparation was as clean as possible. The cell samples were added to 80% (vol/vol) ethanol, extracted on ice and the supernatant was removed by centrifugation. This was repeated until a clean sample was obtained. Samples were then extracted with acetone for 10 min at room temperature, and the supernatant was removed by centrifugation. Methanol was extracted for 10 min, and the supernatant was removed by centrifugation to obtain the final white substance as a cell wall. The samples were dried in an oven, the weights were recorded, and the dried cell walls were sent to Wuhan ProNets Biotechnology Co., Ltd. (Wuhan, China, http://www.pronetsbio.com, accessed on 20 March 2023) for detection. The experiments were repeated three times.
Chlorella variabilis and JB17 were cultivated in BG11 for 7 days, and then diluted to the fresh BG11 medium with 0 mM or 300 mM NaHCO3, respectively. After 3 days of treatment, the cells were treated in the dark for 12 h to consume starch before collection, and then sent to Wuhan Metwell Biotechnology Co., Ltd. (Wuhan, China, http://www.metware.cn, accessed on 25 March 2023) for monosaccharide or disaccharide detection.
JB17 was cultured in BG11 liquid medium for 7 days and then were collected by centrifugation. Each gram of algae was treated with 0, 50 and 100 U cellulase (Solarbio, Beijing, China) for 3 days. The cultures with a cell density of approximately 1 × 108 cells/mL, were then gradient diluted in 96-well plates with sterile water and 4 μL of the dilutions were spotted in modified BG11 solid medium supplemented with 0, 50 and 100 mM NaHCO3, respectively. After 16 days of culture, algal growth was observed.
Log-phase C. reinhardtii cells (at approximately 0.5 × 108 cells/mL) were collected, serially diluted and spotted onto the modified TAP solid medium containing 0 or 40 mM NaHCO3, respectively. After growth for about 10 days, algal growth was observed. For the growth curve determination, the log-phase C. reinhardtii cells were diluted to an OD680 of 0.15 in TAP medium containing 0 mM, 35 mM, and 40 mM NaHCO3, respectively, and the cell density of the cultures was determined using a spectrophotometer (Ultrospec 2,100 Pro, Biochrom, St. Albans, UK), at different time points from 0 to 120 h.
Sequence data of glycosyltransferase-like KOBITO 1 from different organisms in this work can be found in NCBI under the following accession number: Nannochloris sp. JB17 (OR486971), Arabidopsis thaliana (NP187467.1), Chlorella vulgaris (KAI3434462.1), Micractinium conductrix (PSC67501.1), Chlorella variabilis (XP005843966.1), Chlorella sorokiniana (PRW20664.1), Coccomyxa sp. Obi (BDA46538.1), Oryza sativa Japonica Group (NP001393036), and Capsicum annuum (PHT71472.1).
In order to study the ultrastructural changes of JB17 before and after treatment with high concentrations of NaHCO3, the morphology was observed using TEM. It showed that under normal conditions, the control chlorella and JB17 had intact cellular structures (Figures 1A,B). Interestingly, the cell wall of JB17 was thicker, with a thickness of about 300–600 nm, while that of the control chlorella was only about 100 nm thick (Figures 1A,B). After 300 mM NaHCO3 treatment, the cell wall morphology of JB17 was similar to that before stress, whereas the cell wall of the control chlorella was deformed and damaged, and the internal structure of the cell was affected to some extent (Figures 1C,D). These results suggest that the stable and thick cell walls of JB17 may be involved in its tolerance to high concentrations of NaHCO3.
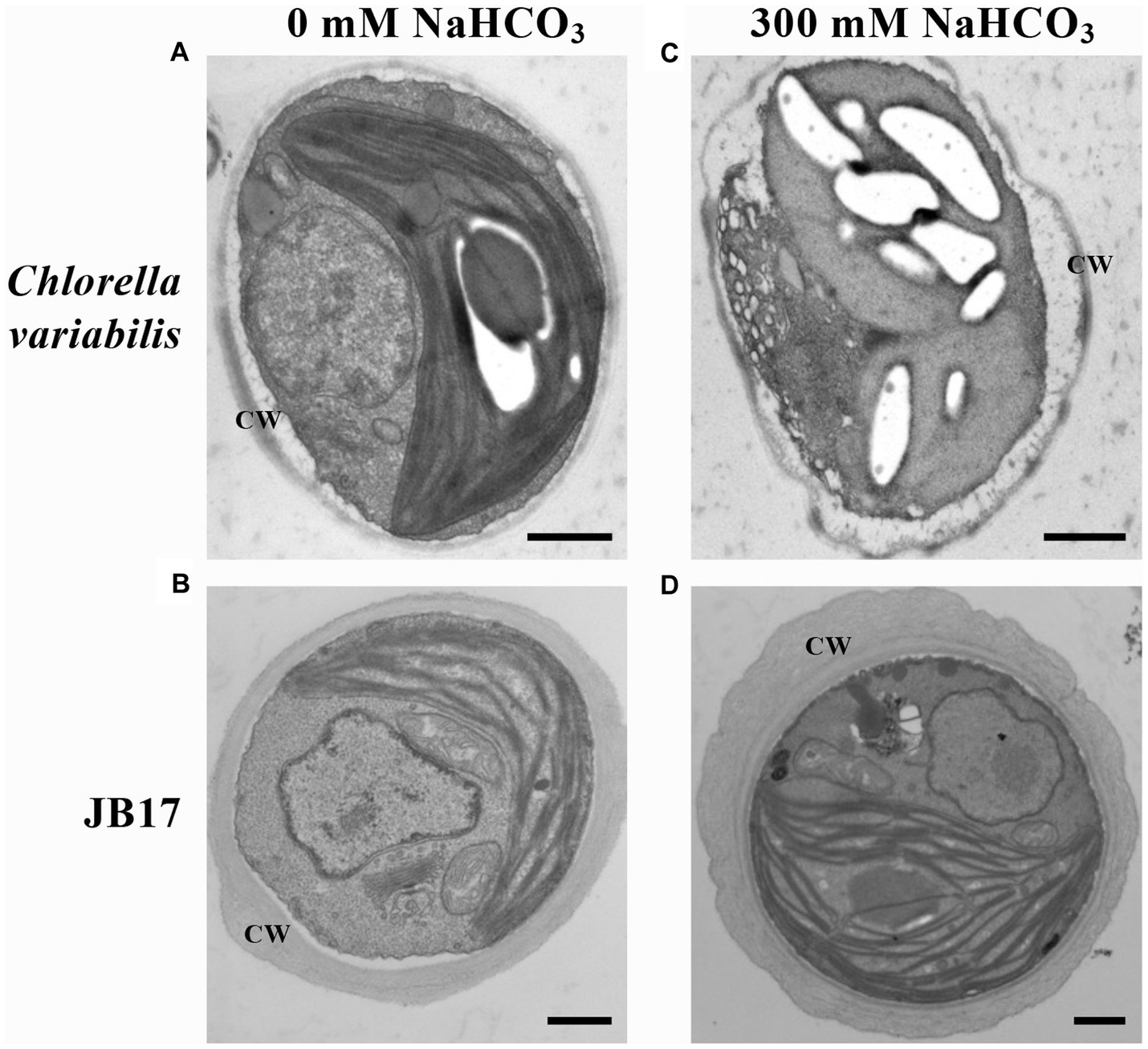
Figure 1. Electron microscopic visualization of C. variabilis and JB17 exposed to NaHCO3 stress. (A,B) Untreated C. variabilis and JB17 were used as controls. (C,D) Representative images of the ultrastructure of C. variabilis and JB17 treated with NaHCO3. CW, cell wall; scale bars, 500 nm.
The composition of cell wall polymers usually determines the cell wall structure. Plant cell walls are mainly composed of cellulose, hemicellulose, pectin, and lignin (Lampugnani et al., 2018). Many studies have reported that changes in these components are involved in plant resistance to salt stress (Dabravolski and Isayenkov, 2023), but the role of these components in resistance to carbonate stress is unclear. Therefore, in order to elucidate the changes in JB17 cell wall components in response to NaHCO3 stress, the contents of major polysaccharide components including cellulose, hemicellulose, lignin, and pectin of JB17 cell walls before and after NaHCO3 treatment were measured. When exposed to 300 mM NaHCO3, the levels of all four components changed, with the exception of lignin, which did not change significantly (Table 1). Of these, cellulose content was the highest as a percentage of dry weight and showed the greatest percentage change in response to NaHCO3 treatment. The cellulose content was significantly increased by 10.48% compared to that of 0 mM NaHCO3 (Table 1). These results suggest that JB17 may adapt to NaHCO3 stress by altering the polysaccharide composition of the cell wall.
Previous study has found that the cell wall cellulose content of rice increased by 10.64% under drought stress, thereby enhancing the drought tolerance of rice (Sun et al., 2022). Therefore, we followed up with a cellulase digestion assay to test whether the accumulated cellulose in JB17 cell wall is critical for NaHCO3 tolerance. Under the treatment of different concentrations of cellulase, there were no significant differences in the growth of JB17 on 0 and 50 mM NaHCO3 plates, indicating that cellulase digestion did not affect the normal growth of algae (Figure 2). When JB17 was grown on 100 mM NaHCO3 plates, the growth of 50 U/g cellulase-treated algae showed a slight degree of inhibition, and the growth defects were more pronounced when the concentration of cellulase reached 100 U/g (Figure 2). In addition, the results of cellulose content determination showed that the cellulose content of JB17 cell wall decreased significantly by 18.472% after 100 U/g cellulase treatment (Supplementary Figure S1). These results indicate that the decrease in cellulose content of JB17 affects its tolerance to high concentrations of NaHCO3 stress, which confirms our hypothesis that NaHCO3-induced cellulose accumulation in the cell wall contributes to the tolerance of JB17 to NaHCO3.
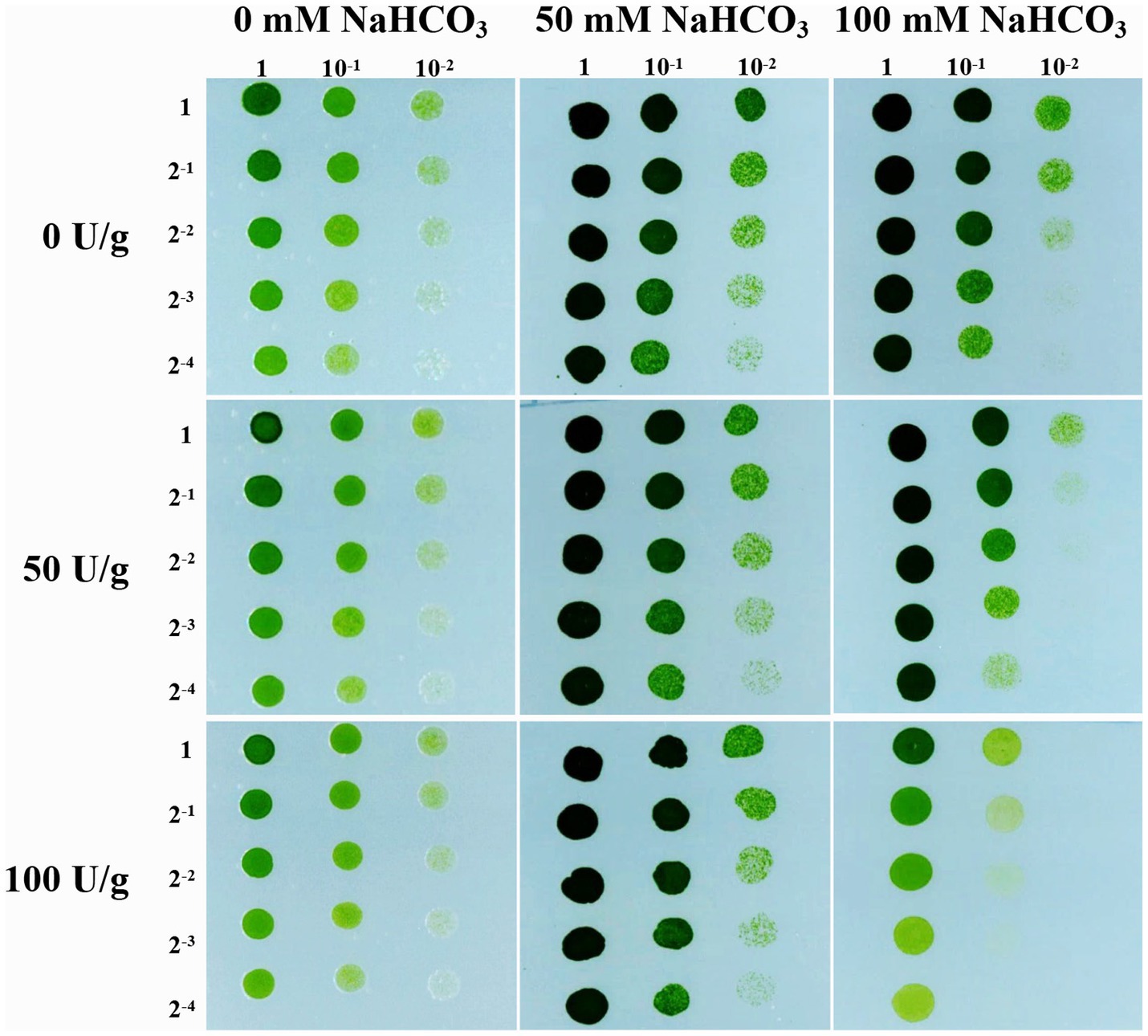
Figure 2. Cellulose content affects JB17 tolerance to NaHCO3. The treatment without cellulase addition was used as control. Each treatment was carried out in three biological replicates. The numbers labelled in the figure are the dilution multiples for the gradient dilution assays.
Metabolites are downstream products of the genome and final products (Qiu et al., 2023). They reflect the final changes in biological systems when they are subjected to genetic and environmental alterations (Alawiye and Babalola, 2021). Changes in cell wall polysaccharides are closely related to saccharide metabolism. Therefore, we further studied the changes in saccharide metabolism in JB17 before and after NaHCO3 treatment. We determined nine monosaccharide and disaccharide components in C. variabilis without NaHCO3 treatment, and JB17 treated with 0 mM, 100 mM, and 300 mM NaHCO3, including galactose, fucose, sucrose, glucose, inositol, xylitol, maltose, rhamnose, and trehalose. Under untreated NaHCO3 conditions, sucrose was the major component of both JB17 and the control chlorella, and galactose, fucose, xylitol, and sucrose were lower in JB17 cell walls than the control chlorella. In contrast, glucose, rhamnose, and trehalose in JB17 were higher, especially rhamnose and trehalose, which were nearly 40-fold higher than the control chlorella (Figure 3). After NaHCO3 treatment, the sucrose in JB17 did not change significantly, while other eight saccharide components, including galactose, fucose, glucose, inositol, xylitol, maltose, rhamnose, and trehalose, showed significant decreases, among which rhamnose and trehalose showed the most significant changes with a decrease of about 100-fold (Figure 3). These saccharide fractions were able to undergo significant changes in response to NaHCO3, and their metabolic processes may be involved in the cell wall polysaccharide changes and NaHCO3 adaptation in JB17.
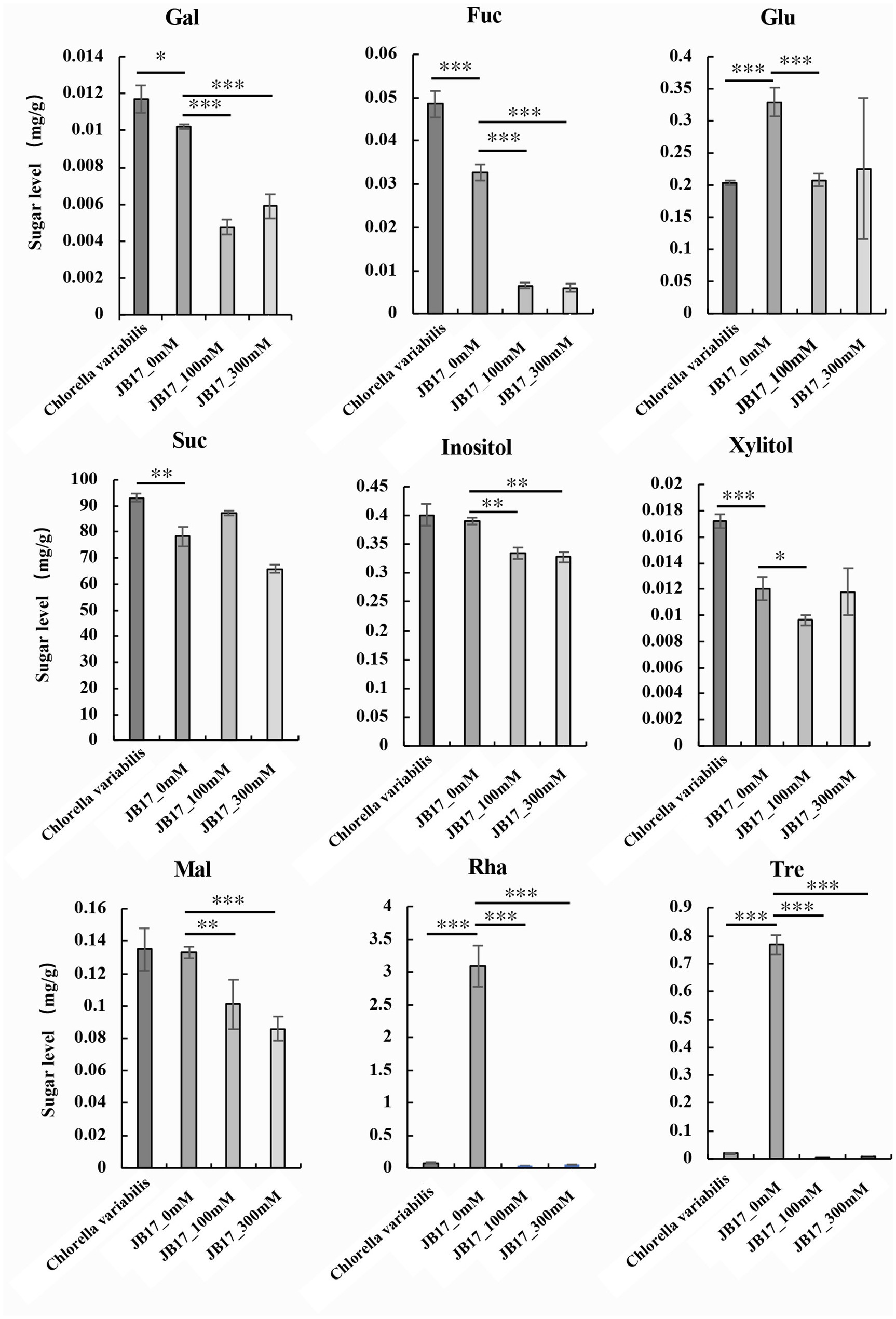
Figure 3. Changes in saccharide metabolism in JB17 before and after NaHCO3 treatment. Gal, galactose; Fuc, fucose; Glu, glucose; Suc, sucrose; Mal, maltose; Rha, rhamnose; Tre, trehalose. Error indicates standard deviation (SD) from three independent experiments. ***p < 0.001, **p < 0.01, *0.01 < p < 0.05, Student’s t-test.
To search for key genes that respond to NaHCO3 stress and improve NaHCO3 tolerance, we analyzed the transcriptome sequencing data of JB17 cells after 0 mM and 300 mM NaHCO3 treatment (data not shown). Among the genes whose expressions were up-regulated by 300 mM NaHCO3, a gene involved in saccharide metabolism and related to cell wall synthesis was identified. And a search for its homologous proteins with known functions in the Uniprot protein database (https://www.uniprot.org/, accessed on 15 July 2023) revealed that it shares 55% amino acid sequence identity with the A. thaliana glycosyltransferase-like protein KOBITO 1 (Gen Bank NP 187467.1). Therefore, it was tentatively named JbKOBITO 1.
The amino acid sequence of the glycosyltransferase-like KOBITO 1 from Nannochloris sp. JB17, C. vulgaris, C. variabilis, C. sorokiniana, and A. thaliana were analyzed using NCBI Batch CD-search (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, accessed on 15 July 2023). MEME software (https://meme-suite.org/meme/tools/, accessed on 15 July 2023) was used to identify conserved motifs of glycosyltransferase-like KOBITO 1 from different organisms. All these proteins shared a conserved Glyco_tranf_GTA_type superfamily domain (Figure 4A), which is involved in synthesizing and modifying of saccharide molecules. Comparing with JbKOBITO 1 in JB17, the sequences of the homologous proteins in C. variabilis and C. sorokiniana were much shorter in size, and the homologous protein sequences in A. thaliana and C. vulgaris were similar in size, but lacked the motif 7, 8, 9, 10, 12 in A. thaliana (Figure 4B).
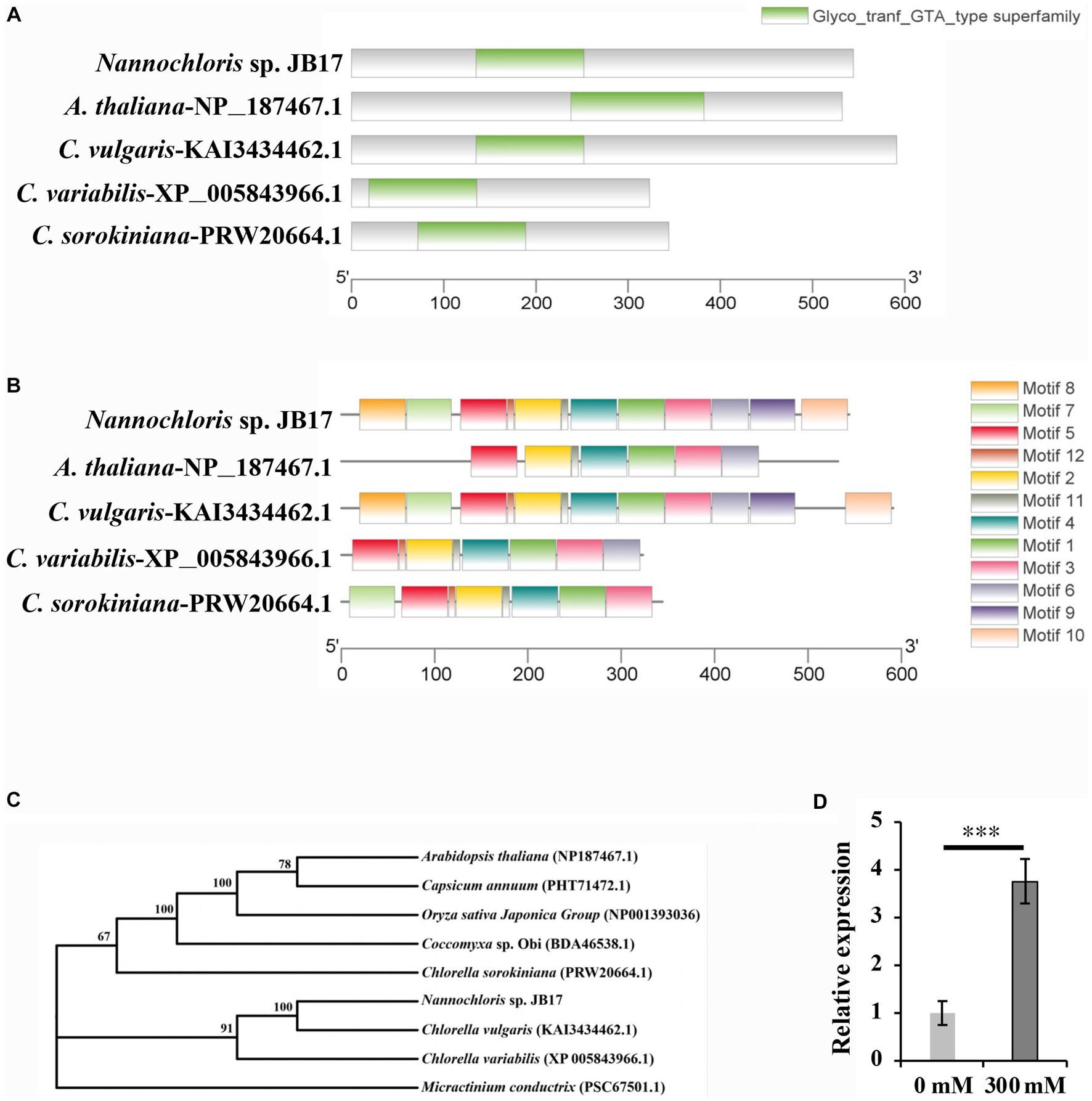
Figure 4. Sequence analysis and expression detection of JbKOBITO 1. (A) JbKOBITO 1 possesses a conserved Glyco_tranf_GTA_type superfamily domain. (B) Conserved motif analysis of glycosyltransferase-like protein KOBITO 1 from algae and A. thaliana. The different colored boxes represent different motifs and their position in each protein sequence. All motifs were identified by MEME. (C) Neighbor-joining (NJ) phylogenetic relationships of the KOBITO 1 protein from JB17 and predicted homologs from other species or organisms. Bootstrap values were calculated 1,000 times; values <50% are not shown. (D) The expression of JbKOBITO 1 was induced by NaHCO3 treatment. Error bars indicate standard deviation (SD). ***, p < 0.001.
Evolutionary tree was constructed using MEGA 11 software. It was clear that the evolutionary relationship between JbKOBITO 1 and C. vulgaris hypothetical protein (D9Q98_002539) was close (Figure 4C). Sequence alignment was performed using the software Jalview (http://www.jalview.org/, accessed on 16 July 2023), and Muscle with Defaults was selected to align. Some highly conserved sites were found (Supplementary Figure S2). Therefore, JbKOBIOT 1 may have a similar function to the Arabidopsis glycosyltransferase-like protein KOBITO 1, which is involved in the coordination between cell elongation and cellulose synthesis by promoting the expression of related genes (Pagant et al., 2002). Moreover, to elucidate whether NaHCO3 stress up-regulated the expression of JbKOBITO 1, the transcript levels of JbKOBITO 1 were tested by RT-qPCR. And the results showed that JbKOBITO 1 was significantly up-regulated in the context of 300 mM NaHCO3 treatment, which was consistent with the results of the transcriptome (Figure 4D). It prompted us to speculate that the NaHCO3-induced up-regulation of JbKOBITO 1 may be involved in the synthesis of accumulated cellulose in the JB17 cell wall.
In order to verify whether JbKOBITO 1 is involved in NaHCO3 tolerance, the recombinant plasmid expressing JbKOBITO 1 was electroporated into C. reinhardtii. Then, the NaHCO3 tolerance of the positively transformed strains was monitored. Gradient dilution assays showed no significant difference in the growth of C. reinhardtii overexpressing the empty vector and overexpressing the target gene on 0 mM NaHCO3 plate, indicating that JbKOBITO 1 overexpression did not affect the growth of algae (Figure 5A). With the increase of NaHCO3 concentration, the growth of the control was significantly inhibited, whereas the growth of C. reinhardtii overexpressing JbKOBITO 1 was significantly better than that of the control (Figure 5A). Similarly, the sensitivity of transgenic C. reinhardtii to different concentrations of NaHCO3 in liquid medium was consistent with the results from gradient dilution assays. The cell density of C. reinhardtii expressing the empty vector was not significantly different from that of C. reinhardtii expressing JbKOBITO 1 in the absence of NaHCO3 (Figure 5B). However, at NaHCO3 concentrations of 35 mM, the cell density of C. reinhardtii expressing JbKOBITO 1 was significantly higher than that of the control group from 60 h, with final cell densities (120 h) of 0.903 and 0.740, respectively (Figure 5B). When the concentration of NaHCO3 was 40 mM, the difference was more obvious, with final cell densities (120 h) of 0.802 and 0.594, respectively (Figure 5B). These results demonstrated that the introduction of JbKOBITO 1 into algae could improve the ability of C. reinhardtii to resist NaHCO3.
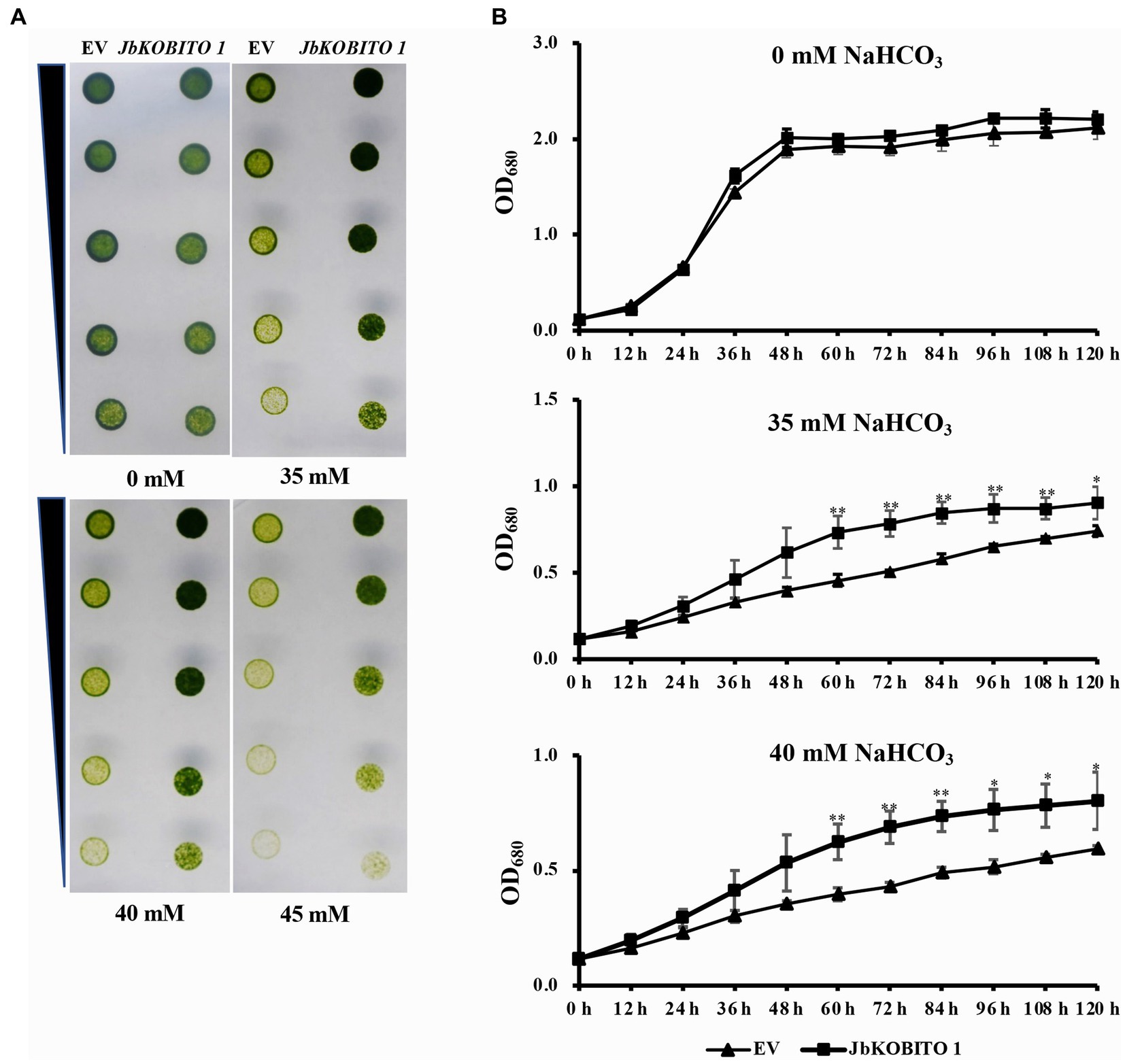
Figure 5. Overexpression of JbKOBITO 1 enhanced C. reinhardtii NaHCO3 tolerance. (A) Gradient dilution assays of transgenic C. reinhardtii on solid medium with different concentrations of NaHCO3. (B) Growth curve of transgenic C. reinhardtii in liquid medium with different concentrations of NaHCO3. **p < 0.01, *0.01 < p < 0.05, Student’s t-test.
Plant cell walls play an important role in sensing and responding to stress. Recent studies have suggested that the plant cell wall is highly dynamic, with changes in structure and composition occurring during plant growth and development or in response to abiotic stresses to maintain normal life activities (Le Gall et al., 2015; Jia et al., 2021). In order to investigate the role of the JB17 cell wall in carbonate tolerance, we first observed its ultrastructure by TEM, and found that it has a thicker cell wall than the control chlorella, and the morphology and structure of the cell are still stable under the high concentration of carbonate stress. Previous studies have found that salt-adapted cells of A. thaliana acquire a thicker cell wall structure than control cells, which physically strengthens the cells, provides better mechanical strength, reduces salt ion invasion and enhances tolerance to salt stress (Chun et al., 2019). Nadzir et al. (2023) also found that the cell wall thickness of Chlorella vulgaris increased from 175 nm to 321 nm at higher salt salinity. Therefore, the ability of JB17 to tolerate high concentrations of NaHCO3 may be closely related to its cell wall.
Subsequently, we examined the changes in the main components of the cell wall before and after treatment with NaHCO3, which showed a significant increase in cellulose content, and a significant decrease in hemicellulose, soluble pectin and protopectin content. But of these, the change in cellulose content is the most pronounced, as cellulose content was the highest as a percentage of dry weight and showed the greatest percentage change in response to NaHCO3 treatment. This suggests that carbonate stress induces complex cell wall polysaccharide modifications and that the ability of JB17 to tolerate carbonate stress may be closely related to changes in cellulose. It is consistent with recent research advances which have demonstrated the impact of cellulose synthesis on plant tolerance to abiotic stresses. For example, DROUGHT1 (DROT1) encodes a COBRA-like protein that confers drought resistance in rice. It has been shown that under drought stress, DROT1 enhances drought tolerance in rice by regulating cell structure through increasing cellulose content by 10.64% and maintaining cellulose crystallinity (Sun et al., 2022). Moreover, Liu et al. reported that the cellulose content in the cell wall of the halophyte Suaeda salsa was significantly higher than that of the glycophyte Spinacia oleracea in a high-salinity environment, giving the cell wall a relatively high hardness, which resisted the deformation of the cell wall under this pressure, and protected the stability of the cell wall structure under salinity stress (Liu et al., 2022). Our results of a cellulase digestion assay also indicated that the decrease in cellulose content of JB17 affects its tolerance to high concentrations of NaHCO3.
Previous studies have also reported on the relationship between the content of pectin and the resistance to stress. An et al. (2014) observed that salt-tolerant soybean varieties contained higher pectin content than salt-sensitive varieties. Jin et al. (2020) reported that OsEIL2 (ethylene insensitive 3-like2) regulates the β-subunit of the polygalacturonase (PG1β-like) subfamily of genes in rice OsBURP16. Overexpression of OsEIL2 in rice resulted in lower pectin content and reduced salt stress tolerance. In these studies, higher pectin levels appeared to favor plant salt tolerance. In addition, pectin was also found to undergo structural changes under the action of modifying enzymes, affecting cell wall characteristics and further affecting its ability to cope with salt stress (Yan et al., 2018). However, in our study, protopectin and soluble pectin content decreased under NaHCO3 stress. This may be due to the different stress response mechanisms of pectin in higher plants and microorganisms. Alternatively, changes in pectin content in JB17 did not play an important role in stress tolerance, as evidenced by the fact that the pectin content in higher plant cell walls is up to 40% (Dabravolski and Isayenkov, 2023), whereas the JB17 cell wall contained very little pectin.
In order to identify the key genes involved in NaHCO3 tolerance, transcriptome data of JB17 before and after NaHCO3 treatment were analyzed. JbKOBITO 1, whose expression was up-regulated in response to NaHCO3, caught our attention. It is homologous to the A. thaliana glycosyltransferase-like protein KOBITO 1 and has a conserved Glyco_tranf_GTA_type superfamily domain. Previous studies have reported that the functions of this protein include coordinating cell elongation and cellulose synthesis by promoting the expression of genes involved in cell elongation and cellulose synthesis, acting as a regulator of intercellular junction filament permeability, and mediating the saccharide response essential for ABA and growth (Pagant et al., 2002; Dinneny et al., 2008; Kong et al., 2012; Wang et al., 2015). We found that both the expression of JbKOBITO 1 and the cellulose content in JB17 cell wall were significantly up-regulated after NaHCO3 treatment, and the overexpression of JbKOBITO 1 in C. reinhardtii can enhance the tolerance of C. reinhardtii to NaHCO3. Therefore, we speculated that the strong tolerance of JB17 to NaHCO3 may be partially attributed to the JbKOBITO 1-mediated cellulose accumulation under NaHCO3 stress. Of course, it cannot be excluded that other genes are also involved in this process. For example, Tang et al. (2022) reported a gene OsUGE3 in rice, which exhibits UDP-galactose/glucose epimerase activity and provides substrate for cellulose synthesis, thereby promoting cellulose accumulation. OsUGE3-mediated increase in cellulose accumulation enhances rice tolerance to salt stress by means of cellular bulking that maintains the integrity of the cell wall. Therefore, there may be other genes involved in JB17 cellulose synthesis, although no other cellulose synthesis-related genes with significant up-regulation in expression were found in our transcriptome data, they may function through post-translational modification or changes in enzyme activity.
Changes in polysaccharides are closely related to saccharide metabolism, which prompted us to investigate JB17 saccharide metabolism. Our results showed higher levels of JB17 glucose, rhamnose and trehalose compared to the NaHCO3 low-tolerance control chlorella, especially rhamnose and trehalose, which are nearly 40 times higher than the control chlorella. Since the discovery of the gene family encoding active trehalose phosphate synthase (TPS; EC 2.4.1.15) and trehalose phosphatases (TPP; EC 3.1.3.12) in A. thaliana, the metabolism of trehalose has been recognized as playing an essential and pervasive role in the life of plants (Lunn et al., 2014). Trehalose (Tre), a non-reducing disaccharide, is an excellent osmolyte for inducing salt tolerance (Ye et al., 2023). Joshi et al. (2020) reported that transgenic lines overexpressing trehalose biosynthetic fusion gene (TPSP) in rice had higher trehalose accumulation and enhanced tolerance to salt and alkali stress. Exogenously applied Tre increases endogenous Tre, another strategy to reduce the adverse effects of salt stress (Kosar et al., 2019). Several studies have reported that exogenous application of trehalose improves salt tolerance in plants (Yuan et al., 2022; Yang et al., 2022a,b). Siddiqui et al. (2020) reported that endogenous glucose content increases when plants are subjected to various abiotic stresses as well as exogenous supply of glucose. Ghorbani Javid et al. (2011) suggested that glucose has a direct role in osmoregulation and radical scavenging in rice grains under salt stress. All these studies suggest that the accumulation of trehalose and glucose may play an important role in plant salt tolerance. This is consistent with our study in which JB17 had a higher trehalose and glucose content. Although there is a lack of research on the role of rhamnose in plant resistance to saline and alkaline stress, it has been found in some studies that rhamnose is beneficial in maintaining osmoregulation and enhancing tolerance to osmotic stress in plants (Ghouili et al., 2021).
Taken together, our study provides an experimental basis for understanding the role of the cell wall polysaccharide, especially the role of cellulose in resistance to carbonate stress. The obtained NaHCO3 tolerance gene JbKOBITO 1 will provide genetic resources for crop breeding in saline-alkali soils and for genetic modification of strains for biofuel production. In the future, genetic modification strategies can be used to increase the cellulose content in the cell walls of crops to increase tolerance to saline-alkali stress for agricultural development, as well as to solve the problem of microalgae that accumulate more lipids under NaHCO3 stress but whose growth is inhibited.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
JQ: Formal analysis, Investigation, Methodology, Writing – original draft. JZ: Formal analysis, Investigation, Methodology, Writing – review & editing. HZ: Formal analysis, Investigation, Methodology, Writing – review & editing. CW: Formal analysis, Investigation, Methodology, Writing – review & editing. CJ: Formal analysis, Investigation, Methodology, Writing – review & editing. XH: Formal analysis, Investigation, Methodology, Writing – review & editing. JL: Writing – review & editing, Data curation, Investigation. XC: Writing – review & editing, Funding acquisition. SL: Conceptualization, Writing – review & editing, Resources. XJ: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Zhejiang Provincial Natural Science Foundation of China (LY23C060001): experimental reagent purchase the Scientific Research Foundation of Zhejiang A&F University (2021LFR053): labour costs of researchers the National Natural Science Foundation of China (32000387): measurement of cell wall composition.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1285796/full#supplementary-material
Alawiye, T. T., and Babalola, O. O. (2021). Metabolomics: current application and prospects in crop production. Biologia 76, 227–239. doi: 10.2478/s11756-020-00574-z
An, P., Li, X., Zheng, Y., Matsuura, A., Abe, J., Eneji, A. E., et al. (2014). Effects of NaCl on root growth and cell wall composition of two soya bean cultivars with contrasting salt tolerance. J. Agron. Crop Sci. 200, 212–218. doi: 10.1111/jac.12060
Bai, J. H., Yan, W. K., Wang, Y. Q., Yin, Q., Liu, J. H., Wight, C., et al. (2018). Screening oat genotypes for tolerance to salinity and alkalinity. Front. Plant Sci. 9:1302. doi: 10.3389/fpls.2018.01302
Chun, H. J., Baek, D., Cho, H. M., Jung, H. S., Jeong, M. S., Jung, W. H., et al. (2019). Metabolic adjustment of Arabidopsis root suspension cells during adaptation to salt stress and mitotic stress memory. Plant Cell Physiol. 60, 612–625. doi: 10.1093/pcp/pcy231
Dabravolski, S. A., and Isayenkov, S. V. (2023). The regulation of plant cell wall organisation under salt stress. Front. Plant Sci. 14:1118313. doi: 10.3389/fpls.2023.1118313
Dinneny, J. R., Long, T. A., Wang, J. Y., Jung, J. W., Mace, D., Pointer, S., et al. (2008). Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320, 942–945. doi: 10.1126/science.1153795
Ghorbani Javid, M., Sorooshzadeh, A., Modarres Sanavy, S. A. M., Allahdadi, I., and Moradi, F. (2011). Effects of the exogenous application of auxin and cytokinin on carbohydrate accumulation in grains of rice under salt stress. Plant Growth Regul. 65, 305–313. doi: 10.1007/s10725-011-9602-1
Ghouili, E., Sassi, K., Jebara, M., Hidri, Y., Ouertani, R. N., Muhovski, Y., et al. (2021). Physiological responses and expression of sugar associated genes in faba bean (Vicia faba L.) exposed to osmotic stress. Physiol. Mol. Biol. Plants 27, 135–150. doi: 10.1007/s12298-021-00935-1
Hong, D. D., Hien, H. T. M., Thom, L. T., Ha, N. C., Huy, L. A., Thu, N. T. H., et al. (2023). Transcriptome analysis of Spirulina platensis sp. at different salinity and nutrient compositions for sustainable cultivation in Vietnam. Sustainability 15:11906. doi: 10.3390/su151511906
Ji, C. L., Mao, X., Hao, J. Y., Wang, X. D., Xue, J. N., Cui, H. L., et al. (2018). Analysis of bZIP transcription factor family and their expressions under salt stress in. Int. J. Mol. Sci. 19:2800. doi: 10.3390/ijms19092800
Jia, H. L., Wang, X. H., Wei, T., Wang, M., Liu, X., Hua, L., et al. (2021). Exogenous salicylic acid regulates cell wall polysaccharides synthesis and pectin methylation to reduce cd accumulation of tomato. Ecotoxicol. Environ. Saf. 207:111550. doi: 10.1016/j.ecoenv.2020.111550
Jin, J., Duan, J., Shan, C., Mei, Z., Chen, H., Feng, H., et al. (2020). Ethylene insensitive3-like2 (OsEIL2) confers stress sensitivity by regulating OsBURP16, the β subunit of polygalacturonase (PG1β-like) subfamily gene in rice. Plant Sci. 292:110353. doi: 10.1016/j.plantsci.2019.110353
Jin, X., Jiang, Z., Zhang, K., Wang, P., Cao, X., Yue, N., et al. (2018). Three-dimensional analysis of chloroplast structures associated with virus infection. Plant Physiol. 176, 282–294. doi: 10.1104/pp.17.00871
Joshi, R., Sahoo, K. K., Singh, A. K., Anwar, K., Pundir, P., Gautam, R. K., et al. (2020). Enhancing trehalose biosynthesis improves yield potential in marker-free transgenic rice under drought, saline, and sodic conditions. J. Exp. Bot. 71, 653–668. doi: 10.1093/jxb/erz462
Khoo, K. S., Ahmad, I., Chew, K. W., Iwamoto, K., Bhatnagar, A., and Show, P. L. (2023). Enhanced microalgal lipid production for biofuel using different strategies including genetic modification of microalgae: a review. Prog. Energy Combust. Sci. 96:101071. doi: 10.1016/j.pecs.2023.101071
Khoo, K. S., Chew, K. W., Yew, G. Y., Leong, W. H., Chai, Y. H., Show, P. L., et al. (2020). Recent advances in downstream processing of microalgae lipid recovery for biofuel production. Bioresour. Technol. 304:122996. doi: 10.1016/j.biortech.2020.122996
Kong, D., Karve, R., Willet, A., Chen, M. K., Oden, J., and Shpak, E. D. (2012). Regulation of plasmodesmatal permeability and stomatal patterning by the glycosyltransferase-like protein KOBITO1. Plant Physiol. 159, 156–168. doi: 10.1104/pp.112.194563
Kosar, F., Akram, N. A., Sadiq, M., Al-Qurainy, F., and Ashraf, M. (2019). Trehalose: a key organic osmolyte effectively involved in plant abiotic stress tolerance. J. Plant Growth Regul. 38, 606–618. doi: 10.1007/s00344-018-9876-x
Lampugnani, E. R., Khan, G. A., Somssich, M., and Persson, S. (2018). Building a plant cell wall at a glance. J. Cell Sci. 131:jcs207373. doi: 10.1242/jcs.207373
Le Gall, H., Philippe, F., Domon, J. M., Gillet, F., Pelloux, J., and Rayon, C. (2015). Cell wall metabolism in response to abiotic stress. Plan. Theory 4, 112–166. doi: 10.3390/plants4010112
Li, J. Y., Li, C. H., Lan, C. Q., and Liao, D. K. (2018). Effects of sodium bicarbonate on cell growth, lipid accumulation, and morphology of Chlorella vulgaris. Microb. Cell Factories 17:111. doi: 10.1186/s12934-018-0953-4
Li, W. J., Zhang, Y. T., Ren, H. M., Wang, Z., OuYang, Y. H., Wang, S., et al. (2023). Identification of potassium transport proteins in algae and determination of their role under salt and saline-alkaline stress. Algal Res. 69:102923. doi: 10.1016/j.algal.2022.102923
Liu, J., Shao, Y., Feng, X., Otie, V., Matsuura, A., Irshad, M., et al. (2022). Cell wall components and extensibility regulate root growth in Suaeda salsa and Spinacia oleracea under salinity. Plan. Theory 11:900. doi: 10.3390/plants11070900
Liu, X. G., Yu, Y., Liu, Q., Deng, S. R., Jin, X. B., Yin, Y. J., et al. (2020). A Na2CO3-responsive chitinase gene from leymus chinensis improve pathogen resistance and saline-alkali stress tolerance in transgenic tobacco and maize. Front. Plant Sci. 11:504. doi: 10.3389/fpls.2020.00504
Lunn, J. E., Delorge, I., Figueroa, C. M., Van Dijck, P., and Stitt, M. (2014). Trehalose metabolism in plants. Plant J. 79, 544–567. doi: 10.1111/tpj.12509
Nadzir, S. M., Yusof, N., Kamari, A., and Nordin, N. (2023). “Effect of environmental stress on biomolecules production and cell wall degradation in Chlorella vulgaris” in Proceedings of the 8th international conference on the applications of science and mathematics. EduTA 2022, springer proceedings in physics. eds. A. Mustapha, N. Ibrahim, H. Basri, M. S. Rusiman, and S. Zuhaib Haider Rizvi, vol. 294 (Singapore: Springer)
Pagant, S., Bichet, A., Sugimoto, K., Lerouxel, O., Desprez, T., McCann, M., et al. (2002). KOBITO1 encodes a novel plasma membrane protein necessary for normal synthesis of cellulose during cell expansion in Arabidopsis. Plant Cell 14, 2001–2013. doi: 10.1105/tpc.002873
Pang, Q. Y., Zhang, A. Q., Zang, W., Wei, L., and Yan, X. F. (2016). Integrated proteomics and metabolomics for dissecting the mechanism of global responses to salt and alkali stress in Suaeda corniculata. Plant Soil 402, 379–394. doi: 10.1007/s11104-015-2774-0
Qiao, K., Takano, T., and Liu, S. (2015). Discovery of two novel highly tolerant NaHCO3 Trebouxiophytes: identification and characterization of microalgae from extreme saline–alkali soil. Algal Res. 9, 245–253. doi: 10.1016/j.algal.2015.03.023
Qin, Y., Bai, J. H., Wang, Y. Q., Liu, J. H., Hu, Y., Dong, Z., et al. (2018). Comparative effects of salt and alkali stress on photosynthesis and root physiology of oat at anthesis. Arch. Biol. Sci. 70, 329–338. doi: 10.2298/Abs171124050q
Qiu, S., Guo, S. F., Yang, Q., Xie, Y. Q., Tang, S. Q., and Zhang, A. H. (2023). Innovation in identifying metabolites from complex metabolome-highlights of recent analytical platforms and protocols. Front. Chem. 11:1129717. doi: 10.3389/fchem.2023.1129717
Siddiqui, H., Sami, F., and Hayat, S. (2020). Glucose: sweet or bitter effects in plants-a review on current and future perspective. Carbohydr. Res. 487:107884. doi: 10.1016/j.carres.2019.107884
Sun, X., Xiong, H., Jiang, C., Zhang, D., Yang, Z., Huang, Y., et al. (2022). Natural variation of DROT1 confers drought adaptation in upland rice. Nat. Commun. 13:4265. doi: 10.1038/s41467-022-31844-w
Tang, Y., Wang, M., Cao, L., Dang, Z., Ruan, N., Wang, Y., et al. (2022). OsUGE3-mediated cell wall polysaccharides accumulation improves biomass production, mechanical strength, and salt tolerance. Plant Cell Environ. 45, 2492–2507. doi: 10.1111/pce.14359
Wang, X., Ajab, Z., Liu, C., Hu, S., Liu, J., and Guan, Q. (2020). Overexpression of transcription factor SlWRKY28 improved the tolerance of Populus davidiana × P. Bolleana to alkaline salt stress. BMC Genet. 21:103. doi: 10.1186/s12863-020-00904-9
Wang, X., Jing, Y. J., Zhang, B. C., Zhou, Y. H., and Lin, R. C. (2015). Glycosyltransferase-like protein ABI8/ELD1/KOB1 promotes Arabidopsis hypocotyl elongation through regulating cellulose biosynthesis. Plant Cell Environ. 38, 411–422. doi: 10.1111/pce.12395
Wang, M., Liu, H., Qiao, K., Ye, X., Takano, T., Liu, S., et al. (2020). Exogenous NaHCO3 enhances growth and lipid accumulation of the highly NaHCO3-tolerant Nannochloris sp. JB17. J. Appl. Phycol. 33, 241–253. doi: 10.1007/s10811-020-02293-z
Wang, Y., Wang, M., Ye, X. X., Liu, H., Takano, T., Tsugama, D., et al. (2020). Biotin plays an important role in Arabidopsis thaliana seedlings under carbonate stress. Plant Sci. 300:110639. doi: 10.1016/j.plantsci.2020.110639
Yan, J. W., He, H., Fang, L., and Zhang, A. (2018). Pectin methylesterase31 positively regulates salt stress tolerance in Arabidopsis. Biochem. Biophys. Res. Commun. 496, 497–501. doi: 10.1016/j.bbrc.2018.01.025
Yang, Y., Xie, J. M., Li, J., Zhang, J., Zhang, X. D., Yao, Y. D., et al. (2022a). Trehalose alleviates salt tolerance by improving photosynthetic performance and maintaining mineral ion homeostasis in tomato plants. Front. Plant Sci. 13:974507. doi: 10.3389/fpls.2022.974507
Yang, Y., Yao, Y. D., Li, J., Zhang, J., Zhang, X. D., Hu, L. X., et al. (2022b). Trehalose alleviated salt stress in tomato by regulating ROS metabolism, photosynthesis, osmolyte synthesis, and trehalose metabolic pathways. Front. Plant Sci. 13:772948. doi: 10.3389/fpls.2022.772948
Ye, X., Wang, H., Cao, X., Jin, X., Cui, F., Bu, Y., et al. (2019). Transcriptome profiling of Puccinellia tenuiflora during seed germination under a long-term saline-alkali stress. BMC Genomics 20:589. doi: 10.1186/s12864-019-5860-5
Ye, N. H., Wang, Y. X., Yu, H. H., Qin, Z. E., Zhang, J. H., Duan, M. J., et al. (2023). Abscisic acid enhances trehalose content via OsTPP3 to improve salt tolerance in rice seedlings. Plan. Theory 12:2665. doi: 10.3390/plants12142665
Yuan, G. P., Sun, D. X., An, G. L., Li, W. H., Si, W. J., Liu, J. P., et al. (2022). Transcriptomic and metabolomic analysis of the effects of exogenous trehalose on salt tolerance in watermelon (Citrullus lanatus). Cells 11:2338. doi: 10.3390/cells11152338
Yun, C. J., Hwang, K. O., Han, S. S., and Ri, H. G. (2019). The effect of salinity stress on the biofuel production potential of freshwater microalgae Chlorella vulgaris YH703. Biomass Bioenergy 127:105277. doi: 10.1016/j.biombioe.2019.105277
Zheng, J. X., Lin, R. Y., Pu, L., Wang, Z. F., Mei, Q. M., Zhang, M., et al. (2021). Ectopic expression of CrPIP2;3, a plasma membrane intrinsic protein gene from the halophyte Canavalia rosea, enhances dought and salt-alkali stress tolerance in Arabidopsis. Int. J. Mol. Sci. 22:565. doi: 10.3390/ijms22020565
Keywords: algae, cell wall, Chlamydomonas reinhardtii , JB17, saccharide metabolism
Citation: Qiu J, Zhang J, Zhao H, Wu C, Jin C, Hu X, Li J, Cao X, Liu S and Jin X (2023) Cellulose and JbKOBITO 1 mediate the resistance of NaHCO3-tolerant chlorella to saline-alkali stress. Front. Microbiol. 14:1285796. doi: 10.3389/fmicb.2023.1285796
Received: 04 September 2023; Accepted: 31 October 2023;
Published: 15 November 2023.
Edited by:
Takashi Osanai, Meiji University, JapanReviewed by:
Pau Loke Show, University of Nottingham Malaysia Campus, MalaysiaCopyright © 2023 Qiu, Zhang, Zhao, Wu, Jin, Hu, Li, Cao, Liu and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shenkui Liu, c2hlbmt1aWxpdUBuZWZ1LmVkdS5jbg==; Xuejiao Jin, amlueHVlamlhbzE5OTFAY2F1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.