- 1The School of Basic Medical Science and Public Center of Experimental Technology, Southwest Medical University, Luzhou, Sichuan, China
- 2Department of Clinical Laboratory, Hospital of Chengdu Office of People’s Government of Tibetan Autonomous Region, Chengdu, Sichuan, China
- 3Department of Clinical Laboratory, The Affiliated Traditional Chinese Medicine Hospital of Southwest Medical University, Luzhou, Sichuan, China
Introduction: The tigecycline-resistant Enterobacterales have emerged as a great public concern, and the mobile tet(X) variants and tmexCD-toprJ efflux pump are mainly responsible for the spread of tigecycline resistance. Hospital sewage is considered as an important reservoir of antimicrobial resistance, while tigecycline resistance in this niche is under-researched.
Methods: In this study, five Escherichia coli and six Klebsiella pneumoniae strains were selected from a collection of tigecycline-resistant Enterobacterales for further investigation by antimicrobial susceptibility testing, conjugation, whole-genome sequencing, and bioinformatics analysis.
Results: All five E. coli strains harbored tet(X4), which was located on different plasmids, including a novel IncC/IncFIA(HI1)/IncHI1A/IncHI1B(R27) hybrid structure. In addition, tet(X4)-bearing plasmids were able to transfer by conjugation and be stabilized in the recipient in the absence of antibiotics. tmexCD1-toprJ1 was identified in two K. pneumoniae (LZSFT39 and LZSRT3) and it was carried by a novel multidrug-resistance transposon, designated Tn7368, on a novel IncR/IncU hybrid plasmid. In addition, we found that two K. pneumoniae (LZSFZT3 and LZSRT3) showed overexpression of efflux genes acrB and oqxB, respectively, which was most likely to be caused by mutations in ramR and oqxR.
Discussion: In conclusion, the findings in this study expand our knowledge of the genetic elements that carry tigecycline resistance genes, which establishes a baseline for investigating the structure diversity and evolutionary trajectories of human, animal, and environmental tigecycline resistomes.
1. Introduction
Antibiotic resistance poses a serious threat to global public health. Tigecycline, the third-generation tetracycline antibiotic, is a last-resort drug to treat serious infections, especially those caused by carbapenem-resistant Enterobacterales and Acinetobacter spp., which have been classified as critical-priority bacteria by the World Health Organization (Tacconelli et al., 2018). Tigecycline resistance has emerged over the years, while the mechanism is complex and has not yet been fully elucidated. Previous studies have reported that overexpression of efflux pumps (such as AcrAB and OqxAB; Sheng et al., 2014; He et al., 2015), or mutation in the tet(A) gene (coding for Tet(A) efflux protein) (Xu et al., 2021) often underlies the resistance mechanisms of tigecycline. High-level expression of acrAB can result from the up-regulation of ramA, which can be caused by a mutation in ramR, a local transcriptional repressor of ramA (Wang et al., 2015). rarA overexpression upregulates the oqxAB efflux pump while oqxR, a transcriptional repressor, can downregulate the oqxAB (Veleba et al., 2012). In addition, the mutation in the rpsJ (encoding the S10 ribosomal subunit; Beabout et al., 2015), might be also involved in tigecycline resistance among Klebsiella pneumoniae isolates.
tet(X) has been shown to encode a flavin-dependent monooxygenase that degrades tigecycline (Forsberg et al., 2015). The recent emergence and dissemination of plasmid-mediated tet(X) variants [tet(X3)-tet(X6)] that confers high-level tigecycline resistance raise the concern that the efficacy of this last-resort antibiotic may be compromised, further limiting clinical treatment choices. Among them, tet(X3) and tet(X4) are frequently detected in Acinetobacter and Enterobacterales isolates from different origins, especially animals, and their meat products in China (Sun et al., 2019, 2020; Chen et al., 2020). In addition to tet(X) genes, the emergence of plasmid-borne RND-type efflux pump tmexCD-toprJ gene clusters that confers resistance to multiple drugs, including tigecycline, also poses a huge risk to public health. Six tmexCD-toprJ variants have been identified in different bacterial species, and they are frequently detected in food-producing animals (Wang C. Z. et al., 2021; Wang Q. et al., 2021; Gao et al., 2022; Liu et al., 2022; Wang C. et al., 2023; Wang J. et al., 2023). Data from nationwide surveillance in China showed that tmexCD-toprJ-positive bacteria are rare (0.64%, 48/7517) in clinical settings, with Pseudomonas and Klebsiella serving as the main reservoir of tmexCD-toprJ variants (Dong et al., 2022).
Plasmid-mediated horizontal gene transfer plays a vital role in the dissemination of tigecycline resistance. Tet(X4) has been reported to be carried on plasmids with a variety of replicon types, including IncQ1, IncX1, IncHI1, IncFIB, and untypeable plasmids (Fang et al., 2019; Sun et al., 2020; Yu et al., 2021), and the mobilization of tet(X4) seems to be closely related to ISCR2 (He et al., 2019). IncR, IncFIA, IncFIB, and IncFIB/IncHI1B hybrid plasmids are mainly vectors for efflux pump gene cluster tmexCD-toprJ, and different mobile elements, including site-specific integrases and IS elements, such as IS26 and IS6100, might facilitate its transmission (Peng et al., 2021; Dong et al., 2022). Although food animals are the principal sources of mobile tigecycline resistance determinates, it is also crucial to monitor the expanding antibiotic resistance in the environment under the ‘One Health’ framework, particularly in hospital sewage, as it is a hotspot for horizontal gene transfer for antimicrobial resistance genes (ARGs) between bacterial communities, and also a reflection of the ARGs and pathogens that are prevalent in the hospital (Cahill et al., 2019). A recent investigation demonstrated that tmexCD-toprJ coexists with mcr-3 and carbapenemase genes in bacteria in hospital sewage in Zhejiang province in China, which highlighted the potential risks of antimicrobial resistance development and spread in water systems (Wu et al., 2023). Yet reports are currently lacking on the prevalence and genetic characteristics of tigecycline resistance in hospital sewage.
In the present study, we aimed to determine the genetic contexts of tigecycline resistance genes and diversity of resistance plasmids in six tigecycline-resistant Escherichia coli and eight tigecycline-resistant K. pneumoniae isolates from hospital sewage in Southwest China, where the prevalence of tigecycline resistance has rarely been reported (Wu et al., 2023).
2. Materials and methods
2.1. Bacterial isolates
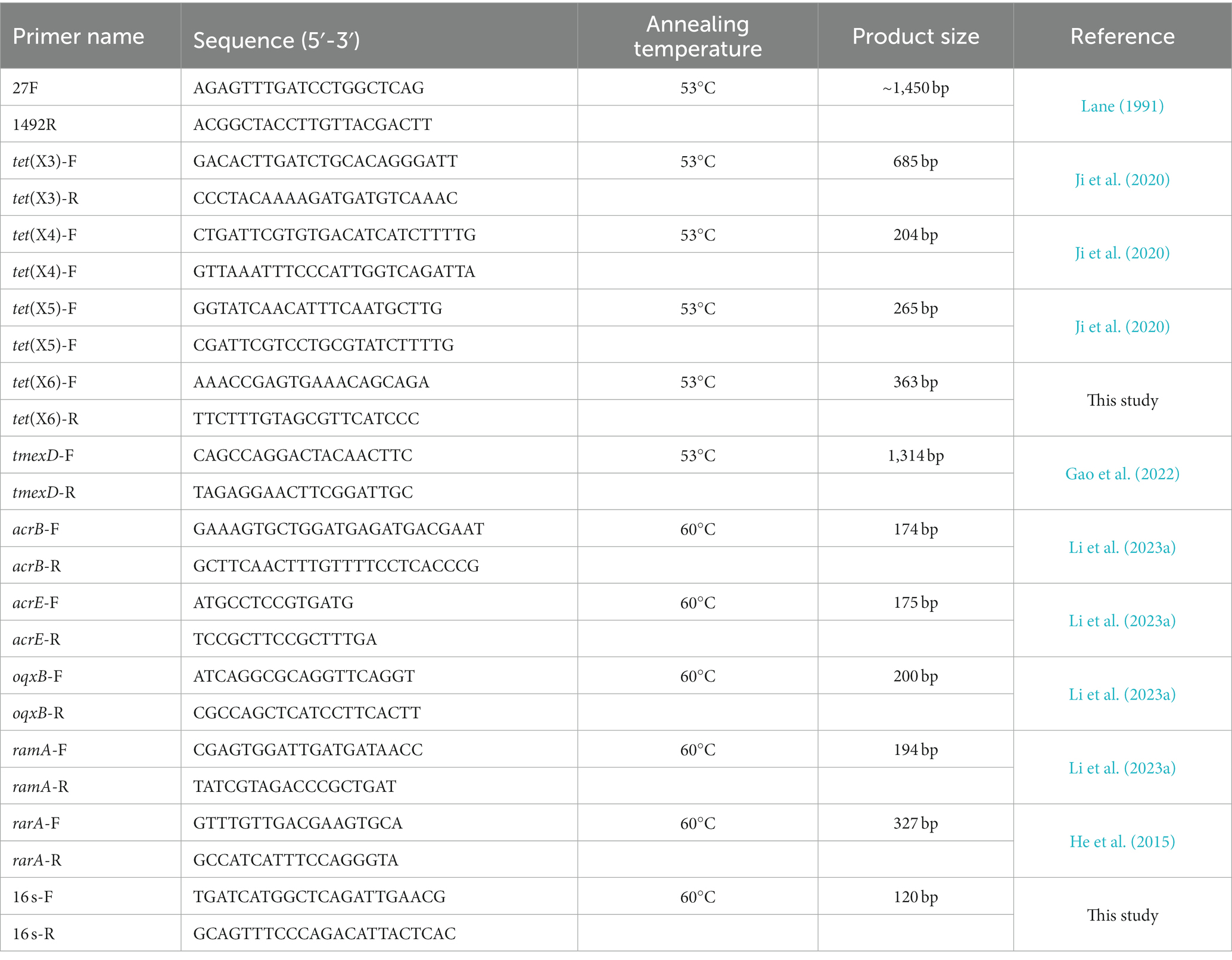
Six samples were collected from the influx of the wastewater treatment plant of three tertiary care hospitals, the Affiliated Hospital of Southwest Medical University (2,200 beds), the Affiliated Traditional Chinese Medicine Hospital of Southwest Medical University (3,000 beds), Luzhou People’s Hospital (1,000 beds)in Luzhou City, Sichuan province, Southwest China in 2021. Each hospital was sampled twice at 3-month intervals. These three hospitals are located in the north, east and middle of the city, respectively, and account for most of the city’s medical care. Sewage samples (5 mL) were collected at a depth of ~10 cm below the water surface using a sterile centrifuge tube during weekdays in the morning (9 am to 11 am). The samples were stored on ice before being taken to the laboratory for subsequent testing within the following 1 h. After sufficient mixing, 100 μL of water sample were spread on MacConkey agar supplemented with 2 μg/mL tigecycline before overnight incubation at 37°C. One to three colonies of each type with different colors and morphology from each sample were picked. Species identification of the colonies was performed by partial amplification and sequencing of the 16 s rRNA gene as described previously with the primes 27F/1492R (Table 1; Lane, 1991). All the isolates were examined for the presence of tet(X) variants, and tmexCD-toprJ variants by PCR using specific primers (Table 1). All the PCR products were analyzed on 1.5% agarose gel by electrophoresis. The positive PCR products were determined using Sanger sequencing by Tsingke Bioinformatics Technology Co. Ltd. (Beijing, China) and were compared with reported sequences by BLASTn.1
2.2. Susceptibility to antibiotics
Antimicrobial susceptibility testing was performed using the Kirby Bauer disk diffusion method on Mueller-Hinton agar as recommended by Clinical and Laboratory Standards Institute (CLSI) and interpreted according to CLSI guidelines (CLSI, 2023). The following antibiotics were tested: amikacin (AMK, 30 μg), gentamicin (GEN, 10 μg), tetracycline (TET, 30 μg), ciprofloxacin (CIP, 5 μg), tigecycline (TIG, 15 μg), cefoxitin (FOX, 30 μg), chloramphenicol (CHL, 30 μg), cefotaxime (CTX, 30 μg), meropenem (MEM, 10 μg), sulfamethoxazole-trimethoprim (SXT, 25 μg). The diameter of inhibition zones was interpreted following the CLSI recommendations for Enterobacteriaceae. E. coli ATCC 25922 served as the control strain. MICs of tigecycline was determined using the microdilution broth method and interpreted according to the FDA criteria (susceptible, ≤2 μg/mL; intermediate, 4 μg/mL; resistant, ≥8 μg/mL).2
2.3. Genomic DNA sequencing and data analysis
Genomic DNA of selected strains was extracted using Rapid Bacterial Genomic DNA Isolation Kit (Sangon Biotech, Shanghai, China) according to the manufacturer’s protocol. Purified DNA was subjected to short-read sequencing on an Illumina HiSeq 2000 platform (Illumina, San Diego, CA, United States) with the 150-bp paired-end approach by the Tsingke Biotech (Beijing, China). Clean reads were de novo assembled into contigs using SPAdes with the careful mode (Bankevich et al., 2012). Four isolates (LZSFT34, LZSFT39, LZSFZT33, and LZSRT11) were additionally sequenced on the long-read MinION sequencer (Nanopore, Oxford, United Kingdom). Both the long MinION reads and short Illumina reads were de novo assembled by using Unicycler v0.4.3 (Wick et al., 2017). Pilon was used to correct the assembled contigs with Illumina reads (Walker et al., 2014). Annotation was carried out using RAST v2.0(Aziz et al., 2008) and BLASTp/BLASTn searches against the UniProtKB/SwissProt database (Boutet et al., 2016). The core genome alignment was performed using Roary and single nucleotide polymorphisms (SNPs) were extracted using snp-sites v2.3.2 (Page et al., 2015). Multilocus sequence typing (MLST) of strains, plasmid replicons, and ARGs were determined using the Center for Genomic Epidemiology3 web tools MLST v2.0, PlasmidFinder v2.1, and ResFinder v4.1, respectively. Insertion elements (IS) and integrons were annotated using online databases IS Finder (Siguier et al., 2006) and INTEGRALL (Moura et al., 2009). BRIG and Easyfig were employed to generate the genetic comparison figures (Alikhan et al., 2011; Sullivan et al., 2011).
2.4. Plasmid transferability and stability
Conjugation experiments were performed using broth-based method with the E. coli EC600 (rifampin-resistance) as the recipient, as described previously with minor modification (Li et al., 2022a). After the donors and recipients were grown to the exponential stage when the optical density at 600 nm (OD600) reaches ~0.5, mix them at a donor/recipient ratio of 1:1 before incubation at 37°C for 24 h. Transconjugants were selected on Luria-Bertani (LB) agar plates containing tigecycline (2 μg/mL) plus rifampin (400 μg/mL). The presence of tet(X4) in transconjugants was confirmed by PCR using the primers tet(X4)-F/R in Table 1.
The plasmid stability was studied by serial passage in antibiotic-free LB broth as previously described (Lv et al., 2020; Li et al., 2023b). Briefly, three separate cultures of E. coli transconjugants carrying the target plasmid were grown overnight in antibiotic-free LB broth, followed by dilution in fresh LB medium at a ratio of 1:102. Serial passaging of the overnight culture to new LB broth was performed daily (approximately 10 generations of growth per passage), lasting for 14 days. 48 single clones of 14th passages were randomly selected from each culture, and the presence of tet(X4) was confirmed by PCR using primers tet(X4)-F/R.
2.5. Mutation analysis
The sequences of ramR, oqxR, and rpsJ in the tested strains were aligned with the reference sequence of tigecycline-susceptible isolate K. pneumoniae MGH 78578 (GenBank accession no. CP000647). The tet(A) variant was identified by comparing to the original tet(A) in plasmid pUUH239.2 (Accession no. NC_016966).
2.6. Real-time relative quantitative PCR assays
The RT-qPCR experiments were performed as previously described (He et al., 2015). Overnight bacterial cultures were diluted 1/100 into fresh LB broth and grown to the mid-exponential stage (OD600 ~ 0.5) at 37°C. The total bacterial RNA was harvested using a TaKaRa MiniBEST universal RNA extraction kit (TaKaRa, Dalian, Japan). The quantity and purity were evaluated using a NanoDrop 1,000 spectrophotometer (Thermo Scientific, Hvidovre, Denmark). RNA was reverse transcribed into cDNA using the PrimeScript RT reagent Kit with gDNA Eraser RR047A (TaKaRa, Dalian, China) according to the manufacturer’s instructions. RT-qPCR was performed using Taq Pro Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) on an FGD-96A real-time system (BIOER, Hangzhou, China) with 40 cycles of 30s at 95°C, 10s at 95°C, 30s at 60°C, 15 s at 95°C, 60s at 65°C, and 15 s at 97°C. Primers for the efflux pump genes (acrB, acrE, and oqxB) and the regulator genes (ramA, and rarA) were presented in Table 1. The relative expression levels were normalized against the 16 s rRNA gene, and the fold changes were calculated using the 2-ΔΔCT method. A tigecycline-susceptible K. pneumoniae clinical isolate SCNJ10 (MIC ≤0.5 μg/mL) was used as a reference strain for the gene expression analysis. This experiment was repeated three times independently with triplicate samples. Data were analyzed with GraphPad Prism version 6.0 (GraphPad Software, San Diego, CA). Values returning a p value of <0.05 from a Student t test were taken as significant and indicated by an asterisk.
3. Results and discussion
3.1. Bacterial isolates and their phenotypic and genotypic resistance
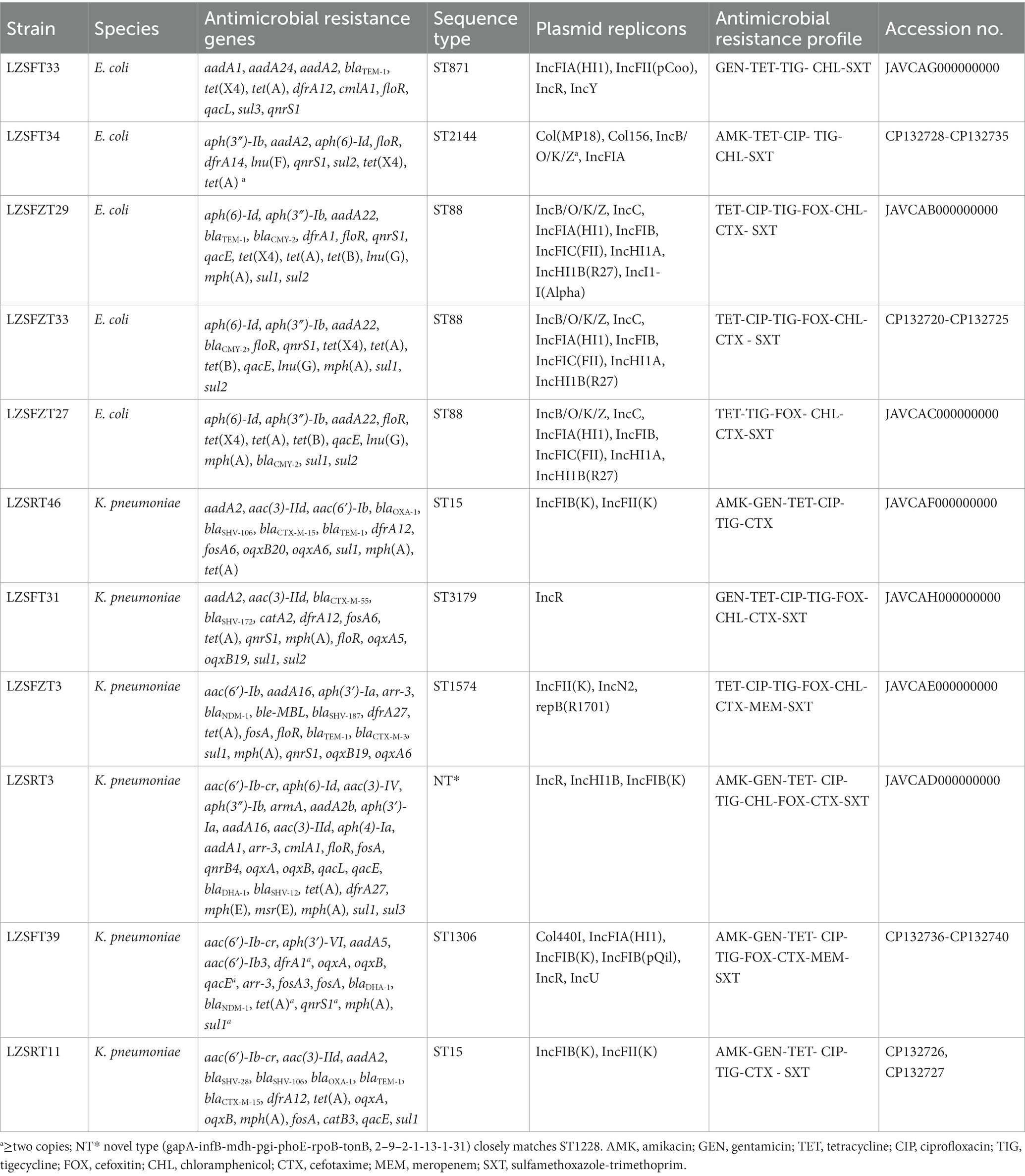
During a survey evaluating the prevalence of tigecycline-resistant strains from the hospital sewage, a total of 113 tigecycline-resistant Enterobacterales isolates were collected, including 16 E. coli, 65 Klebsiella spp., 26 Enterobacter spp., and 6 Citrobacter spp., as revealed by 16 s rRNA gene analysis. Among these, six E. coli were positive for tet(X4), and three K. pneumoniae carried tmexD (Table 2). 14 strains were selected for further analysis in detail by whole genome sequencing (WGS) using the Illumina HiSeq platform, including six tet(X4)-bearing E. coli, three tmexD-bearing K. pneumoniae, and five additional tigecycline-resistant K. pneumoniae from different samples (Table 2), which all exhibited resistance to tigecycline with MICs of 8 to16 μg/mL. Of them, LZSFT34 (E. coli, carrying tet(X4)), LZSFT39 (K. pneumoniae, carrying tmexCD-toprJ1), LZSFZT33 (E. coli, carrying tet(X4)), and LZSRT11 (K. pneumoniae) were further sequenced using the Nanopore technology.
According to the WGS data, three strains were identified as redundant (i.e., isolated from the same location, belonging to the same species and the same sequence type [ST], and carrying the same ARGs and plasmid replicons). We excluded these redundant isolates, leaving 11 isolates, including five E. coli strains (LZSFT34, LZSFZT27, LZSFZT29, LZSFZT33, and LZSFT33), and six K. pneumoniae (LZSFT39, LZSRT11, LZSRT46, LZSFT31, LZSFZT3, and LZSRT3) (Table 2). Antimicrobial susceptibility testing showed that most of these tigecycline-resistant strains exhibited co-resistance to tetracycline (n = 11), cefotaxime (n = 9), ciprofloxacin (n = 9), and sulfamethoxazole-trimethoprim (n = 10). Some of them were also resistant to chloramphenicol (n = 8), cefoxitin (n = 7), or gentamicin (n = 6). Only three strains (LZSRT11, LZSRT46, and LZSRT3) showed resistance to amikacin. Alarmingly, two isolates (LZSFT39, and LZSFZT3) were also resistant to meropenem. Genome analysis showed that 63 different ARGs were detected in the 11 tigecycline-resistant strains, and each of them carried 10 to 29 ARGs (Table 2). Thirteen types of β-lactam resistance genes were detected, including blaCTX-M-15, blaCTX-M-55, blaCMY-2, and blaSHV-12 that were reported to be common ESBL genes in clinical isolates (Zhang et al., 2013, 2014; Feng et al., 2015). Of note, the carbapenem resistance gene blaNDM-1 was found in two strains (LZSFZT3 and LZSFT39). The coexistence of blaNDM-1 and tigecyclineresistance determinant tmexCD-toprJ would pose a serious threat to the treatment of complicated infections by multidrug-resistant (MDR) Gram-negative bacterial infections.
3.2. Genomic characteristics of tet(X4)-bearing Escherichia coli isolates
Strain LZSFT34 consists of a 4,945,961-bp chromosome with a GC content of 50.93%, and seven plasmids ranging in size from 1,166 to 114,852 bp. It has 10 ARGs mediating resistance to aminoglycosides (aph(3″)-Ib, aadA2, and aph(6)-Id), trimethoprim (dfrA1), quinolones (qnrS1), phenicol (floR), sulfonamides (sul2), tetracyclines (tet(A), two copies), lincosamide (lnu(F)), and tigecycline (tet(X4)). All these ARGs are located on the tet(X4)-bearing plasmid pTetX4_FT34, suggesting that tigecycline resistance could be co-selected by other antimicrobial resistance determinants. pTetX4_FT34 is 112,510 bp in size with an average GC content of 51.75% and belongs to the IncFIA group. The plasmid backbone was composed of regions for replication (repA), maintenance (parAB), and conjugal transfer (tra, trb genes) (Figure 1). It also carried a ~ 31 kb accessory resistance region, which contained all the ARGs and abundant mobile genetic elements that are responsible for the formation of this mosaic MDR region. BLASTn comparison of pTetX4_FT34 with plasmids in the NCBI database showed that it is most similar (91% coverage, 99.96% nucleotide identity) to pSX5G-122 k (MZ367885, E. coli, pork, China), and partial similar (>60% coverage, >99.9% identity) to pRHB17-C03_2 (CP057706, E. coli, pig, United Kingdom) and pN16EC0140–1 (CP043748, E. coli, pork, United States). Of note, tet(X4) is absent from these similar plasmids, indicating a stepwise integration of horizontally acquired tet(X4) in pTetX4_FT34.
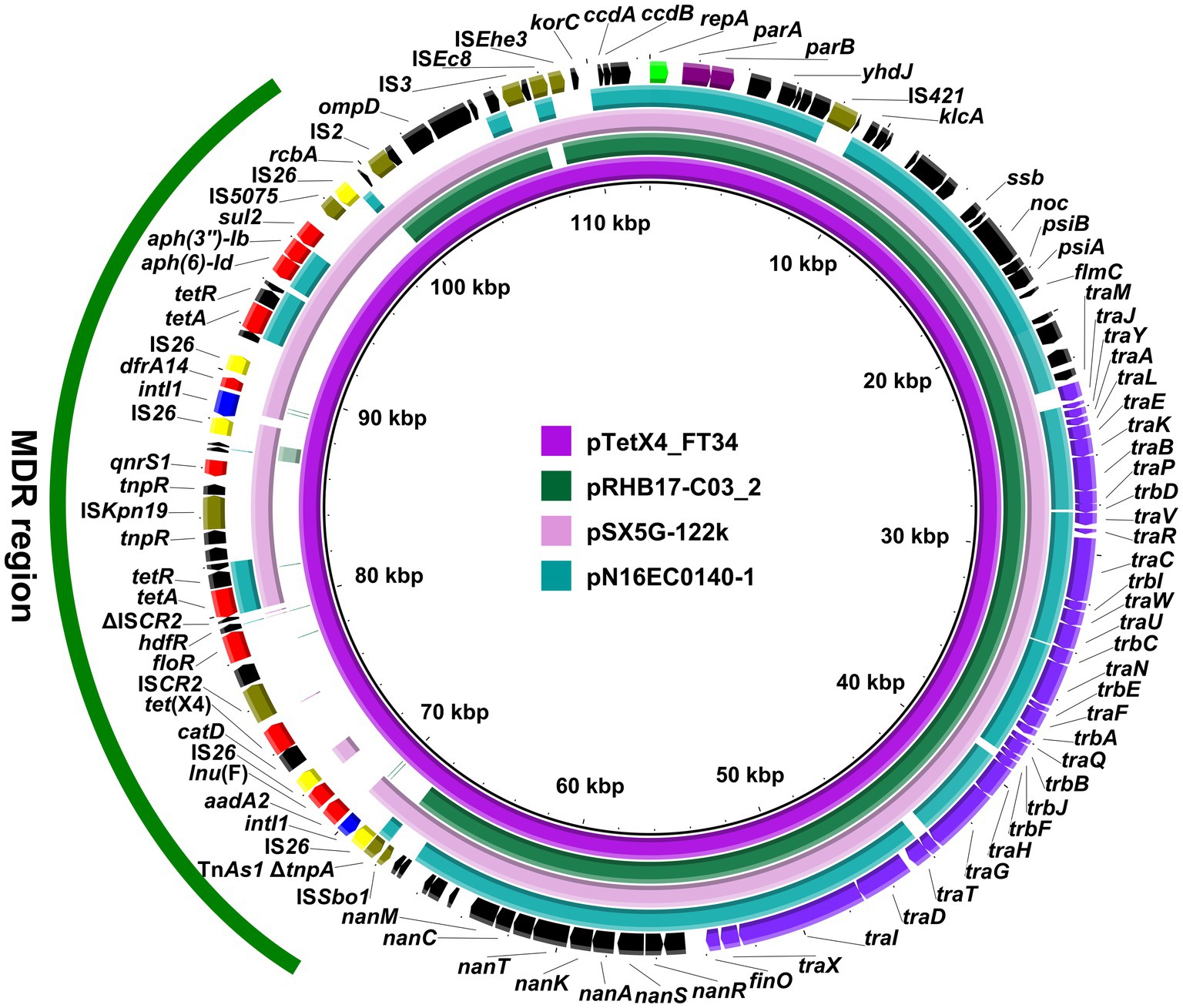
Figure 1. Circular comparison of pTetX4_FT34 with related plasmids. The complete sequence of pTetX4_FT34 was used as the reference. Arrows on the outer ring indicate deduced ORFs and their orientations. Genes for replication (repA), maintenance (parAB), and conjugal transfer (tra, trb genes) are indicated in green, purple, and violet. The accessory resistance region is indicated by green curve. Genes for resistance, integrase, IS26, and other transposase genes are highlighted in red, blue, yellow, and olive, respectively.
According to the draft genome sequences, E. coli LZSFZT27, LZSFZT29, and LZSFZT33 are all assigned to ST88 by MLST analysis. While they are different strains, with at least 258 SNPs between each other in their core genomes. LZSFZT33 was selected as a representative strain and was further sequenced using Nanopore to obtain the whole-genome sequences. According to the WGS data, LZSFZT33 has a 5,056,393-bp chromosome with an average GC content of 50.69%, four plasmids (5,058 to 127,405 bp), and one unclosed contig. Fourteen ARGs were identified in LZSFZT33, including aph(6)-Id, aph(3″)-Ib, aadA22, blaCMY-2, floR, qnrS1, tet(X4), tet(A), tet(B), qacE, lnu(G), mph(A), sul1, and sul2. All of them are distributed on the unclosed contig (designated pTetX4_FZT33), except for the chromosomally located tet(B). The pTetX4_FZT33 is 309,872 bp in length with an average GC content of 48.71%. It is a novel hybrid structure that contains four different replicons, IncC, IncFIA(HI1), IncHI1A, and IncHI1B(R27). Sequence analysis showed that pTetX4_FZT33 consists of a 175-kb region (2,871 to 177,389 bp) which is almost identical (99.95% identity) to pRW7-1_235k (MT219825, E. coli, wastewater, China), and a 30-kb region (216,124 to 246,496 bp) that is highly similar (99.99% identity) to IncC type plasmid pVA833-165 (CP093454, K. pneumoniae, patient, Chile) (Figure 2). According to the genetic organization, ISCR2 downstream of tet(X4), IS26 upstream of mphA, and IS26 downstream of qnrS1 are most likely to be the recombination junctions of the hybrid plasmid (Figure 2).
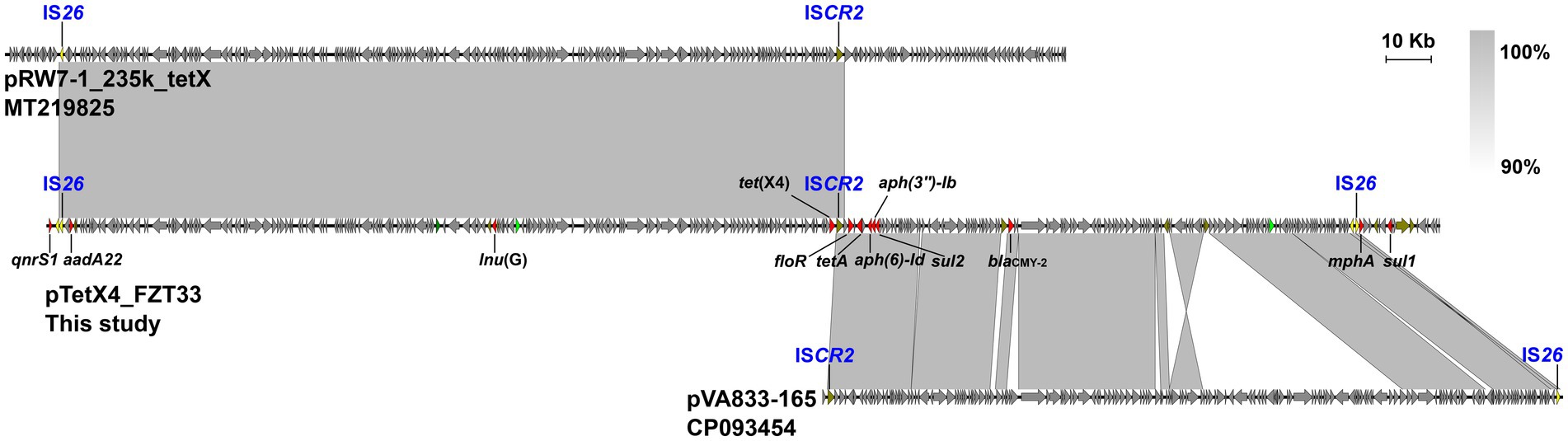
Figure 2. Organization of pTetX4_FZT33, and comparisons to related plasmids. Genes are denoted by arrows. ARGs, replication genes, IS26, and other transposase genes are indicated in red, green, yellow, and olive, respectively. The presumed recombination junctions (ISCR2 and IS26) are highlighted. Regions of >90% homology are indicated by grey shading.
It has been known that ISCR2 plays a vital role in tet(X4) transmission by rolling-circle transposition (Fang et al., 2020). In this study, an intact ISCR2 was found downstream of the catD-tet(X4) cassette, leaving the structure catD-tet(X4)-terlS-ISCR2-orilS (Figure 3), which was the reported tet(X4)-bearing circular intermediate (He et al., 2019). Instead of another ISCR2 located upstream of the catD-tet(X4)-ISCR2 cassette, as had been reported previously in other plasmids (He et al., 2019), an IS1R or IS26 was identified in our cases. A similar structure was also identified in IncHI1 and IncX1 type plasmids from animal-derived E. coli strains (Yu et al., 2021). The findings highlight the diversity of tet(X4)-positive plasmids and tet(X4)-bearing genetic contexts in E. coli clones from different ecological niches.
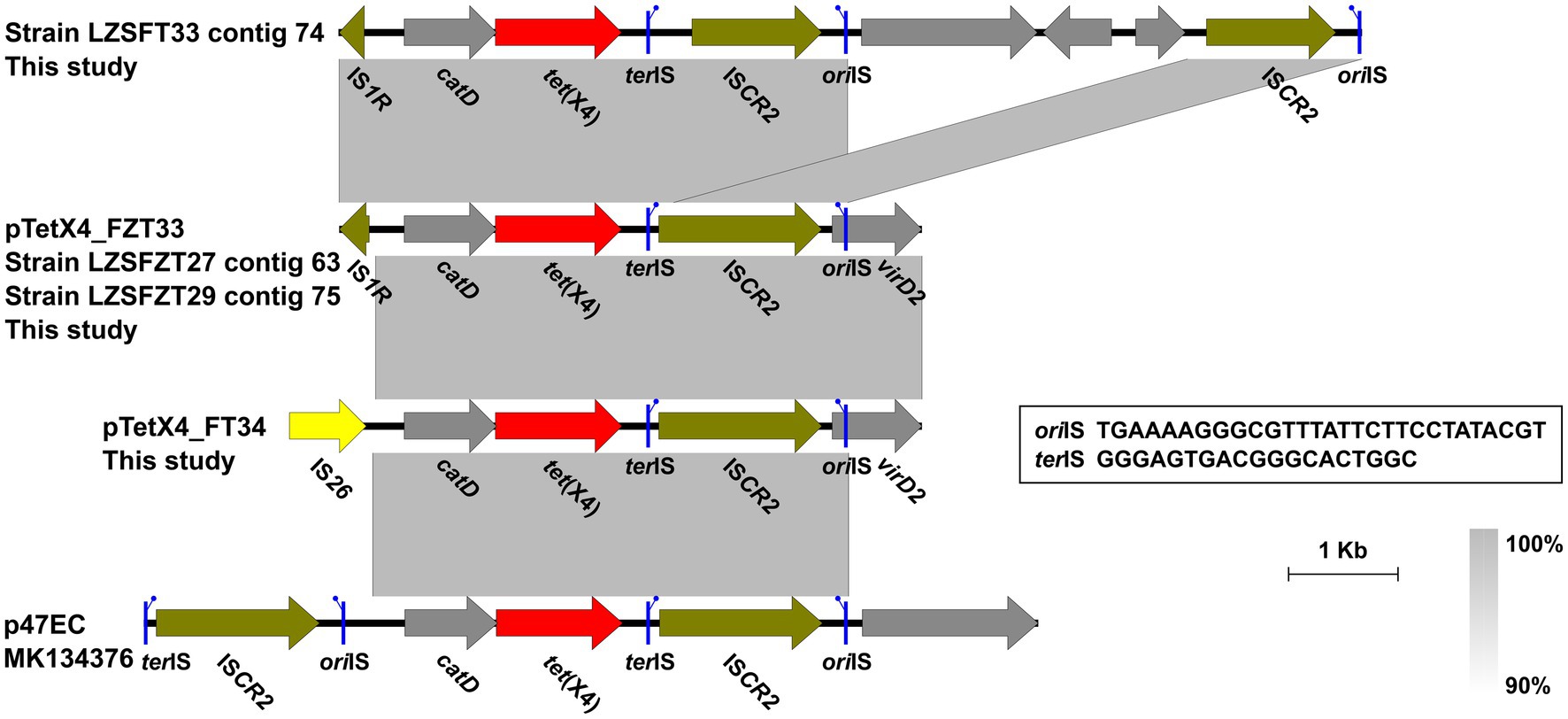
Figure 3. Genetic contexts of the tet(X4) in isolates from this study and the reported plasmid p47EC. Genes are denoted by arrows. ARGs, IS26, and other transposase genes are indicated in red, yellow, and olive, respectively. The putative oriIS (left-facing sticks in blue) and terIS (right-facing sticks in blue) are marked. Regions of >90% homology are indicated by grey shading. Δ represents truncated genes.
Further conjugation assays showed that tet(X4) could be successfully transferred from E. coli LZSFZT27, LZSFZT29, and LZSFZT33 to laboratory strain E. coli EC600, leading to an increased MIC of tigecycline in EC600 by 16-fold (from 0.5 to 8 μg/mL). This result indicates that tet(X4)-bearing plasmids in these three strains were self-transmissible. However, the acquisition of tet(X4) from LZSFT33 and LZSFT34 failed to confer tigecycline resistance in EC600, despite that a 4-fold (0.5 versus 2 μg/mL) increase in MIC of tigecycline was detected. After 14d (approximately 140 generations) of serial passage without antibiotic treatment, tet(X4)-bearing plasmids from LZSFZT27, LZSFZT29, LZSFZT33, LZSFT33 and LZSFT34 were all stably maintained in the transconjugants host, with 93.8–100% retention. The transferability and stability of plasmids containing tet(X4) have serious public health implications.
3.3. Characterization of tigecycline-resistant Klebsiella pneumoniae strains
Two non-redundant tigecycline-resistant K. pneumoniae LZSFT39 and LZSRT3 were identified to carry the tmexCD1-toprJ1. LZSFT39 contains a chromosome of 5,138,005 bp (GC content of ~57.57%), and four plasmids ranging in size from 13,871 to 355,922 bp. LZSFT39 carries 17 different ARGs, including aac(6′)-Ib-cr, aph(3′)-VI, aadA5, aac(6′)-Ib3, dfrA1 (two copies), oqxA, oqxB, qacE (five copies), arr-3, fosA3, fosA, blaDHA-1, blaNDM-1, tet(A) (two copies), qnrS1 (two copies), mph(A), sul1 (five copies). Of them, fosA, oqxA, and oqxB were located on the chromosome, while the remaining ARGs were all distributed on the tmexCD1-toprJ1-bearing plasmid pTmexCD-FT39. It is an IncR/IncU hybrid plasmid, with 355,922-bp in length and an average GC content of 49.38%. BLASTn analysis showed that pTmexCD-FT39 is most similar (77% coverage, 99.87% identity) to p7_SCLZS62, a tmexCD1-toprJ1-bearing plasmid from Raoultella planticola isolated from the same sample collection site in November 2019 (Li et al., 2022b). This finding highlights the dissemination of tigecycline resistance mediated by plasmids between bacterial communities in hospital sewage. Sequence analysis showed that the backbone of pTmexCD-FT39 (nt 1 to 213,922 bp and 353,771 to 355,922 bp) is highly similar (86% coverage, 99.7% identity) to pKOX26_3 (CP089402, Klebsiella oxytoca, patient, Australia), and its accessory region that contained all the ARGs and tmexCD1-toprJ1 gene cluster was inserted into a site between relB and orf113 of pKOX26_3, revealing that pTmexCD-FT39 is likely derived from a pKOX26_3-like plasmid (Figure 4A).
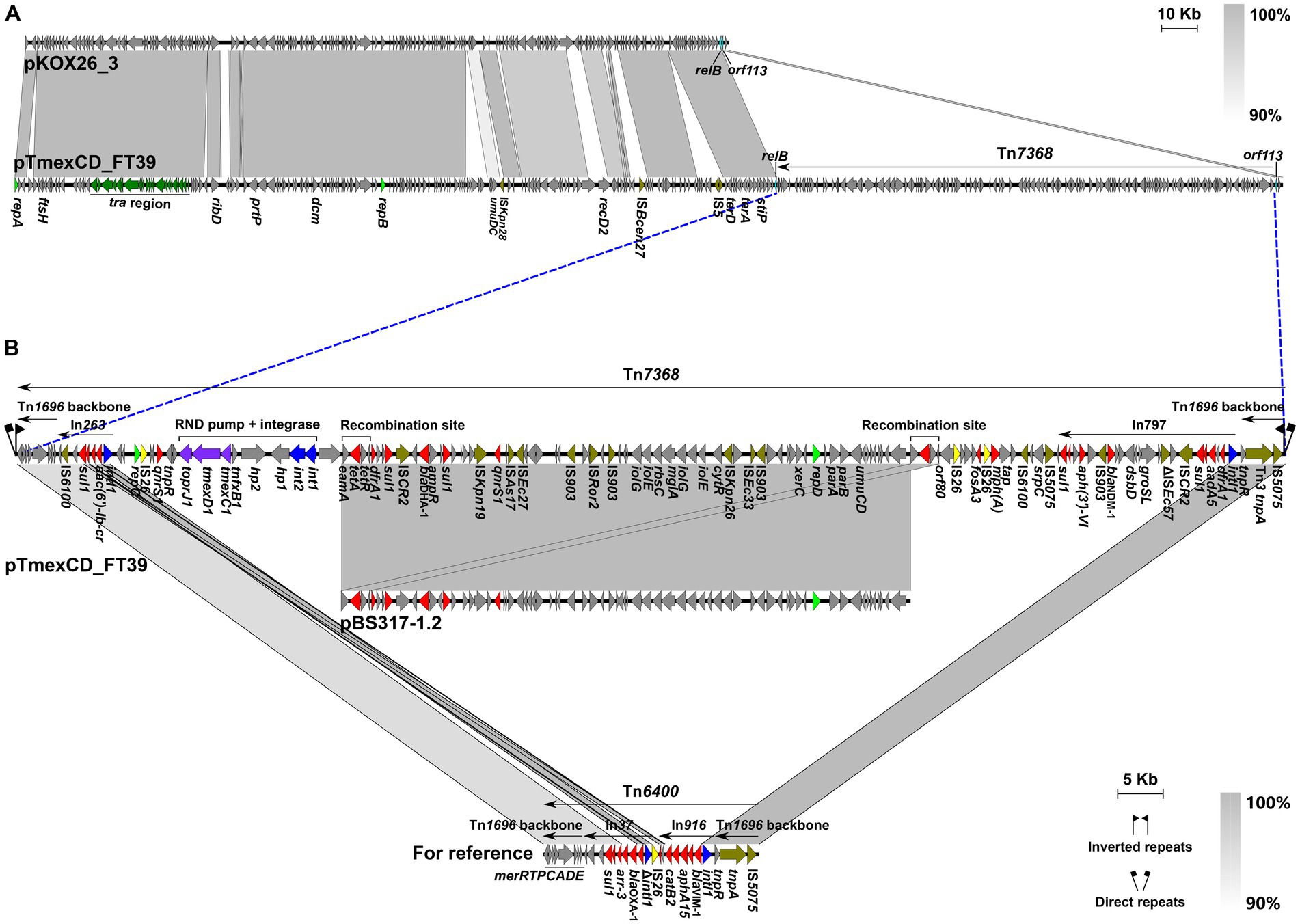
Figure 4. Genetic features of pTmexCD_FT39. (A) Comparison of pTmexCD_FT39 with pKOX26_3. The novel transposon Tn7368 was inserted into a site between relB and orf113. (B) Organization of Tn7368, and comparisons to related regions. Genes are denoted by arrows. ARGs, integrase genes, replication genes, IS26, and other transposase genes are indicated in red, green, yellow, and olive, respectively. The tra region is highlighted in dark green, and the tmexCD1-toprJ1 gene cluster is in purple. Regions of >90% homology are indicated by grey shading. Δ represents truncated genes.
We further found that the accessory region was a novel MDR transposon that was designated Tn7368, according to the nomenclature of transposons.4 Tn7368 is 139,848 bp (corresponding to bases 213,923 to 353,770 in GenBank accession number CP132737) with an average GC content of 55.56%, which differs from that of the rest of the plasmid (GC content, ~45.37%). It was identified as Tn6400-derivative with similar tnpAR (99.97% identity) and mer (90.70% identity) modules, and was bracketed by 5-bp direct repeats (DRs, TTTCA) (Figure 4B). Tn7368 is a mosaic structure composed of multiple class I transposons (such as In797 and In263) and insertion sequences (such as IS26 and IS6100), and it carries the int1-int2-hp1-hp2-tnfxB1-tmexCD1-toprJ1 core genetic structure as described in reference plasmid pHNAH8I-1 (MK347425) (Lv et al., 2020). Of note, a 62.6-kb region (nt 249,836 to 312,442 bp) inside Tn7368 is almost identical (>99.9% identity) to the IncR-type plasmid pBS317-1.2 (CP063938) that was found in a K. pneumoniae isolate from the human fecal sample in China. Sequence analysis reveals that homologous recombination mediated by the 2,983-bp eamA-tet(A)-tetR-orf80 module is most likely to contribute to the formation of such a structure (Figure 4B).
In LZSRT3, the tmexCD1-toprJ1 genes coexist with 29 ARGs, including aac(6′)-Ib-cr, aph(6)-Id, aac(3)-IV, aph(3″)-Ib, armA, aadA2b, aph(3′)-Ia, aadA16, aac(3)-IId, aph(4)-Ia, aadA1, arr-3, cmlA1, floR, fosA, qnrB4, oqxA, oqxB, qacL, qacE, blaDHA-1, blaSHV-12, tet(A), dfrA27, mph(E), msr(E), mph(A), sul1, and sul3. It has been suggested that site-specific integrases and Tn5393-like transposon are responsible for the capture and transmission of tmexCD1-toprJ1 in Klebsiella (Dong et al., 2022). In LZSRT3, the tnfxB1-tmexCD1-toprJ1 gene cluster was identical to that in the pHNAH8I-1, while the int1-int2-hp1 and the adjacent ΔTn5393-5′ was lost, which was most likely to be caused by the recombination event of IS26 that was inserted into hp2 (Figure 5). Of note, compared to the Tn5393 residue downstream of the tmexCD1-toprJ1 gene cluster in pHNAH8I-1, LZSRT3 had intact Tn5393-3′. Unlike the scenario in LZSRT3, the int1-int2-hp1-hp2-tnfxB1-tmexCD1-toprJ1 segment was intact in strain LZSFT39, while the ΔTn5393-3′ was truncated and replaced by the ΔtnpA-tnpR-qnrS1-IS26 module (Figure 5). The findings suggest a parallel diversification and evolution of tmexCD1-toprJ1-bearing genetic contexts in K. pneumoniae. The transfer ability of tmexCD1-toprJ1-bearing plasmids was not determined because their host strains LZSFT39 and LZSRT3 were highly resistant to sodium azide (MIC ≥300 μg/mL, used for selection of E.coli J53) and rifampicin (MIC ≥1,000 μg/mL, used for selection of E.coli EC600). Given that the backbone of pTmexCD-FT39 is almost identical (99.87% identity) to p7_SCLZS62, which was previously identified to be self-transmissible (Li et al., 2022b), the transfer of pTmexCD-FT39 seems likely.
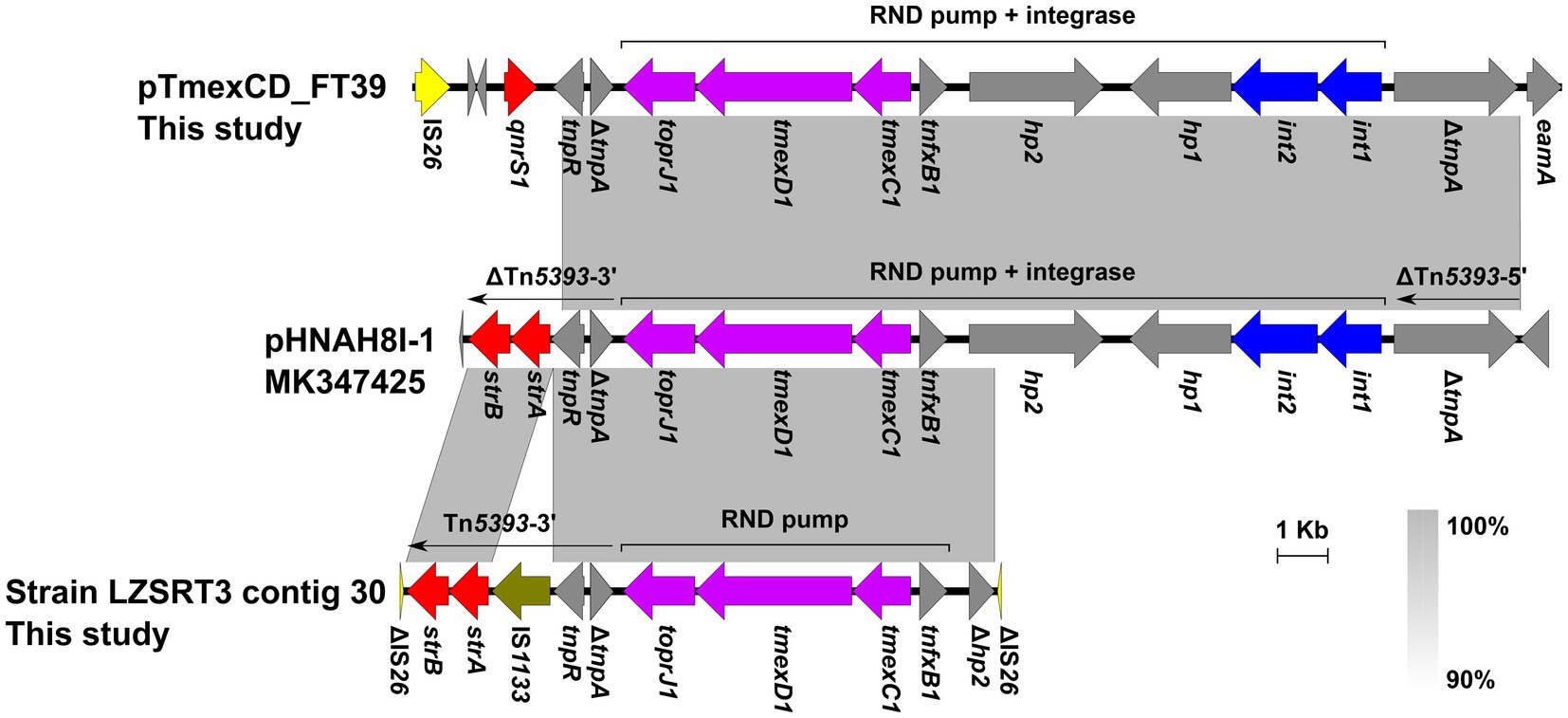
Figure 5. Genetic contexts of the tmexCD1-toprJ1 in isolates from this study and the reported plasmid pHNAH8I-1. Genes are denoted by arrows. ARGs, integrase genes, IS26, and IS1133 are indicated in red, blue, yellow, and olive, respectively. The tmexCD1-toprJ1 gene cluster is highlighted in purple. Regions of >90% homology are indicated by grey shading. Δ represents truncated genes.
LZSRT11 comprises a 5,266,515-bp chromosome (GC content, ~57.47%), and one 221,089-bp plasmid p1_RT11. In LZSRT11, ARGs including oqxA, oqxB, fosA, blaSHV-28, blaSHV-106, and aac(3)-IId are located on the chromosome, while aac(6′)-Ib-cr, aadA2, blaOXA-1, blaTEM-1, blaCTX-M-15, dfrA12, tet(A), mph(A), catB3, qacE, and sul1 are distributed on the p1_RT11. This plasmid has two replicons, IncFIB(K) and IncFII(K). BLASTn analysis showed that it is highly similar (>90% coverage, >99% identity) to several plasmids from clinical K. pneumoniae isolates in China (Figure S1), such as pXHKP75-1 (CP066896, Shanghai), p1_CRKP_11(CP107469, Wuhan), and pC2660-2 (CP039809, Beijing), revealing the circulation and transmission of this MDR plasmid across China. Of note, we found that LZSRT11 as well as LZSRT46 belongs to ST15, which is a high-risk clone with frequent hospital outbreaks in China and has emerged carrying virulence-resistant heterozygous plasmids associated with carbapenemases and ESBL genes (Huang et al., 2022; Zhao et al., 2022). The identification of ST15 tigecycline-resistant K. pneumoniae highlights that close surveillance is urgently needed to monitor the prevalence of ST15 K. pneumoniae in the local clinical settings.
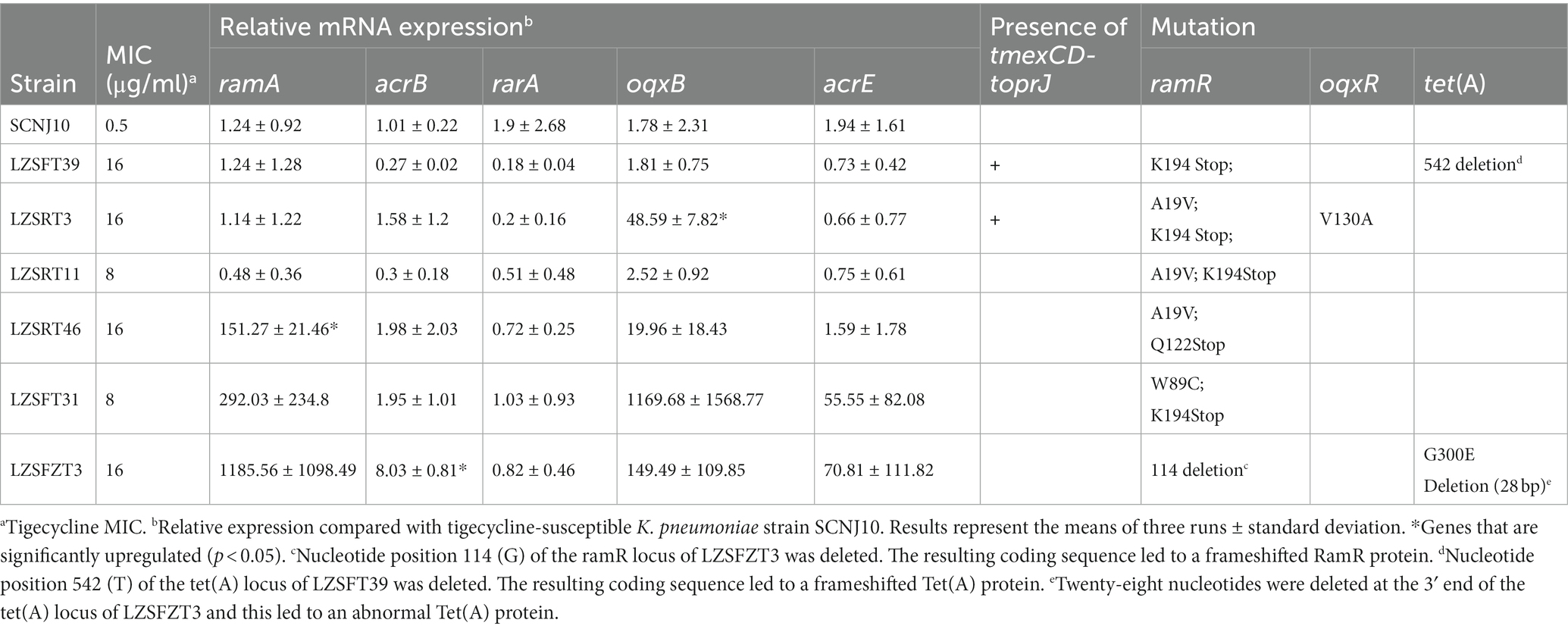
We did not identify tmexCD-toprJ or any genes belonging to the tet(X) family in LZSRT11 and the remaining three tigecycline-resistant K. pneumoniae strains (LZSRT46, LZSFT31, and LZSFZT3). It has been suggested that tigecycline resistance was mainly caused by mutations in ramR or oxqR and the associated overexpression of efflux pumps in K. pneumoniae (He et al., 2015). To confirm this, mRNA expression and sequences of several related genes were analyzed. All the strains contained mutations in ramR, among which LZSFZT3 had a frameshift mutation and the remaining five strains had point substitutions (Table 3). Consistent with the finding by a previous study that not all of the changes within ramR resulted in ramA overexpression (Rosenblum et al., 2011), upregulation of ramA was only identified in strain LZSRT46 in this study (Table 3). Besides, the upregulated ramA did not lead to a higher expression level of acrB in LZSRT46. And, strain LZSFZT3 had an increased expression level of acrB but with baseline expression of ramA. These results suggest that the expression level of the ramA was not always correlated with that of the acrB gene. Only one isolate, LZSRT3, harbored a point mutation (V130A) in the oqxR gene (Table 3). The V130A mutation had been identified to be associated with increased transcript level of rarA and accounted for oqxAB overexpression (Cheng et al., 2020). In LZSRT3, the overexpression of oqxAB was also observed but the transcription of rarA was not significantly changed. The findings highlight that further research is needed to clarify the regulatory networks involved in tigecycline resistance in K. pneumoniae (He et al., 2015).
A previous study suggested that the increased expression of acrEF plays a role in tigecycline resistance in K. pneumoniae (Li et al., 2023a). However, no strains displayed overexpression of acrE in this study. Also, the rpsJ mutation was not detected in all strains. In addition, LZSFT39 and LZSFZT3 had mutations of tet(A) that have not been reported before. Whether these variants tet(A) contribute to tigecycline resistance warrants further study. In all, tmexCD1-toprJ1 and overexpression of efflux pump oqxAB or acrAB caused by mutations in oxqR or ramR may explain the tigecycline resistance in LZSFT39, LZSRT3, and LZSFZT3. While, for strains LZSRT11, LZSRT46, and LZSFT31, novel alternative mechanisms are presumed to exist.
4. Conclusion
In summary, to our knowledge, this is the first detailed report to describe the genome characteristics of tigecycline-resistant E. coli and K. pneumoniae from hospital sewage. Our study revealed novel hybrid plasmids and transposon in the dissemination of tet(X4) or tmexCD1-toprJ1 and also provided insight into the oqxAB/acrAB-encoding tigecycline resistance. Considering the potential threats of the tigecycline resistance genes to public health, continuous monitoring is needed to understand their evolution and transmissible pathways of these high-risk genes. Further researches are also required to investigate the epidemiological links between resistant isolates from the natural environment and humans.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA1004410 https://www.ncbi.nlm.nih.gov/genbank/, CP132725 https://www.ncbi.nlm.nih.gov/genbank/, CP132730 https://www.ncbi.nlm.nih.gov/genbank/, CP132737 https://www.ncbi.nlm.nih.gov/genbank/, CP132727.
Author contributions
YL: Conceptualization, Formal analysis, Writing – original draft. YF: Formal analysis, Methodology, Resources, Writing – review & editing. YQ: Formal analysis, Methodology, Resources, Writing – review & editing. QL: Resources, Software, Writing – review & editing. MY: Formal analysis, Methodology, Resources, Writing – review & editing. LZ: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Scientific and technological project in Sichuan Province (2022JDRC0144). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1282988/full#supplementary-material
Footnotes
1. ^https://blast.ncbi.nlm.nih.gov/Blast.cgi
2. ^http://www.eucast.org/clinical_breakpoints/
References
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Beabout, K., Hammerstrom, T. G., Perez, A. M., Magalhães, B. F., Prater, A. G., Clements, T. P., et al. (2015). The ribosomal S10 protein is a general target for decreased Tigecycline susceptibility. Antimicrob. Agents Chemother. 59, 5561–5566. doi: 10.1128/aac.00547-15
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Methods Mol. Biol. 1374, 23–54. doi: 10.1007/978-1-4939-3167-5_2
Cahill, N., O'Connor, L., Mahon, B., Varley, A., McGrath, E., Ryan, P., et al. (2019). Hospital effluent: a reservoir for carbapenemase-producing Enterobacterales? Sci. Total Environ. 672, 618–624. doi: 10.1016/j.scitotenv.2019.03.428
Chen, C., Cui, C. Y., Yu, J. J., He, Q., Wu, X. T., He, Y. Z., et al. (2020). Genetic diversity and characteristics of high-level tigecycline resistance Tet(X) in Acinetobacter species. Genome Med. 12:111. doi: 10.1186/s13073-020-00807-5
Cheng, Y. H., Huang, T. W., Juan, C. H., Chou, S. H., Tseng, Y. Y., Chen, T. W., et al. (2020). Tigecycline-non-susceptible hypervirulent Klebsiella pneumoniae strains in Taiwan. J. Antimicrob. Chemother. 75, 309–317. doi: 10.1093/jac/dkz450
CLSI. (2023). Performance standards for antimicrobial susceptibility testing 33th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Dong, N., Zeng, Y., Wang, Y., Liu, C., Lu, J., Cai, C., et al. (2022). Distribution and spread of the mobilised RND efflux pump gene cluster tmexCD-toprJ in clinical gram-negative bacteria: a molecular epidemiological study. Lancet Microbe 3, e846–e856. doi: 10.1016/S2666-5247(22)00221-X
Fang, L. X., Chen, C., Cui, C. Y., Li, X. P., Zhang, Y., Liao, X. P., et al. (2020). Emerging high-level Tigecycline resistance: novel tetracycline Destructases spread via the Mobile Tet(X). BioEssays 42:e2000014. doi: 10.1002/bies.202000014
Fang, L. X., Chen, C., Yu, D. L., Sun, R. Y., Cui, C. Y., Chen, L., et al. (2019). Complete nucleotide sequence of a novel plasmid bearing the high-level Tigecycline resistance gene tet(X4). Antimicrob. Agents Chemother. 63:e01373. doi: 10.1128/AAC.01373-19
Feng, Y., Yang, P., Xie, Y., Wang, X., McNally, A., and Zong, Z. (2015). Escherichia coli of sequence type 3835 carrying blaNDM-1, blaCTX-M-15, blaCMY-42 and blaSHV-12. Sci. Rep. 5:12275. doi: 10.1038/srep12275
Forsberg, K. J., Patel, S., Wencewicz, T. A., and Dantas, G. (2015). The tetracycline Destructases: a novel family of tetracycline-inactivating enzymes. Chem. Biol. 22, 888–897. doi: 10.1016/j.chembiol.2015.05.017
Gao, X., Wang, C., Lv, L., He, X., Cai, Z., He, W., et al. (2022). Emergence of a novel plasmid-mediated Tigecycline resistance gene cluster, tmexCD4-toprJ4, in Klebsiella quasipneumoniae and Enterobacter roggenkampii. Microbiol Spectr 10:e0109422. doi: 10.1128/spectrum.01094-22
He, F., Fu, Y., Chen, Q., Ruan, Z., Hua, X., Zhou, H., et al. (2015). Tigecycline susceptibility and the role of efflux pumps in tigecycline resistance in KPC-producing Klebsiella pneumoniae. PLoS One 10:e0119064. doi: 10.1371/journal.pone.0119064
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564-019-0445-2
Huang, J., Chen, X., Yang, J., Zhao, Y., Shi, Y., Ding, H., et al. (2022). Outbreak of KPC-producing Klebsiella pneumoniae ST15 strains in a Chinese tertiary hospital: resistance and virulence analyses. J. Med. Microbiol. 71:1494. doi: 10.1099/jmm.0.001494
Ji, K., Xu, Y., Sun, J., Huang, M., Jia, X., Jiang, C., et al. (2020). Harnessing efficient multiplex PCR methods to detect the expanding Tet(X) family of tigecycline resistance genes. Virulence 11, 49–56. doi: 10.1080/21505594.2019.1706913
Lane, D. J. (1991). “16S/23S rRNA sequencing” in Nucleic acid techniques in bacterial systematics. eds. E. Stackebrandt and M. Goodfellow (New York: Wiley), 115–175.
Li, Y., Li, D., Liang, Y., Cui, J., He, K., He, D., et al. (2023a). Characterization of a Tigecycline-resistant and Bla(CTX-M)-bearing Klebsiella pneumoniae strain from a peacock in a Chinese zoo. Appl. Environ. Microbiol. 89:e0176422. doi: 10.1128/aem.01764-22
Li, Y., Liu, Q., Qiu, Y., Fang, C., Zhou, Y., She, J., et al. (2022a). Genomic characteristics of clinical multidrug-resistant Proteus isolates from a tertiary care hospital in Southwest China. Front. Microbiol. 13:977356. doi: 10.3389/fmicb.2022.977356
Li, Y., Qiu, Y., Gao, Y., Chen, W., Li, C., Dai, X., et al. (2022b). Genetic and virulence characteristics of a Raoultella planticola isolate resistant to carbapenem and tigecycline. Sci. Rep. 12:3858. doi: 10.1038/s41598-022-07778-0
Li, Y., Xu, L., Li, Y., Wang, M., He, T., Bai, L., et al. (2023b). Genomic and functional analysis of high-level tigecycline resistant Klebsiella michiganensis co-carrying tet(X4) and tmexCD2-toprJ2 from pork. Int. J. Food Microbiol. 391-393:110138. doi: 10.1016/j.ijfoodmicro.2023.110138
Liu, Y. Y., Gao, X., He, X., Lv, L., Jiao, Y., Yu, R., et al. (2022). Emergence of the tigecycline resistance gene cluster tmexCD1-toprJ1 in an IncC plasmid and Citrobacter portucalensis. J. Antimicrob. Chemother. 77, 2030–2033. doi: 10.1093/jac/dkac136
Lv, L., Wan, M., Wang, C., Gao, X., Yang, Q., Partridge, S. R., et al. (2020). Emergence of a plasmid-encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including Tigecycline, in Klebsiella pneumoniae. MBio 11:19. doi: 10.1128/mBio.02930-19
Moura, A., Soares, M., Pereira, C., Leitao, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Peng, K., Wang, Q., Yin, Y., Li, Y., Liu, Y., Wang, M., et al. (2021). Plasmids shape the current prevalence of tmexCD1-toprJ1 among Klebsiella pneumoniae in food production chains. mSystems 6:21. doi: 10.1128/mSystems.00702-21
Rosenblum, R., Khan, E., Gonzalez, G., Hasan, R., and Schneiders, T. (2011). Genetic regulation of the ramA locus and its expression in clinical isolates of Klebsiella pneumoniae. Int. J. Antimicrob. Agents 38, 39–45. doi: 10.1016/j.ijantimicag.2011.02.012
Sheng, Z. K., Hu, F., Wang, W., Guo, Q., Chen, Z., Xu, X., et al. (2014). Mechanisms of tigecycline resistance among Klebsiella pneumoniae clinical isolates. Antimicrob. Agents Chemother. 58, 6982–6985. doi: 10.1128/AAC.03808-14
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference Centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Sun, J., Chen, C., Cui, C. Y., Zhang, Y., Liu, X., Cui, Z. H., et al. (2019). Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 4, 1457–1464. doi: 10.1038/s41564-019-0496-4
Sun, C., Cui, M., Zhang, S., Liu, D., Fu, B., Li, Z., et al. (2020). Genomic epidemiology of animal-derived tigecycline-resistant Escherichia coli across China reveals recent endemic plasmid-encoded tet(X4) gene. Comms Biol. 3:1148. doi: 10.1038/s42003-020-01148-0
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Veleba, M., Higgins, P. G., Gonzalez, G., Seifert, H., and Schneiders, T. (2012). Characterization of RarA, a novel AraC family multidrug resistance regulator in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 56, 4450–4458. doi: 10.1128/AAC.00456-12
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Wang, X., Chen, H., Zhang, Y., Wang, Q., Zhao, C., Li, H., et al. (2015). Genetic characterisation of clinical Klebsiella pneumoniae isolates with reduced susceptibility to tigecycline: role of the global regulator RamA and its local repressor RamR. Int. J. Antimicrob. Agents 45, 635–640. doi: 10.1016/j.ijantimicag.2014.12.022
Wang, C., Gao, X., Liang, X., Lv, L., Lu, L., Yue, C., et al. (2023). Pseudomonas acts as a reservoir of novel Tigecycline resistance efflux pump tmexC6D6-toprJ1b and tmexCD-toprJ variants. Microbiol Spectr 11:e0076723. doi: 10.1128/spectrum.00767-23
Wang, C. Z., Gao, X., Yang, Q. W., Lv, L. C., Wan, M., Yang, J., et al. (2021). A novel transferable resistance-nodulation-division pump gene cluster, tmexCD2-toprJ2, confers Tigecycline resistance in Raoultella ornithinolytica. Antimicrob. Agents Chemother. 65:2229. doi: 10.1128/AAC.02229-20
Wang, J., Jiang, Y., Lu, M. J., Wang, Z. Y., and Jiao, X. (2023). Emergence of a novel tigecycline resistance gene cluster tmexC3-tmexD5-toprJ2b in Oceanimonas sp. from chicken, China. J. Antimicrob. Chemother. 78, 1311–1313. doi: 10.1093/jac/dkad080
Wang, Q., Peng, K., Liu, Y., Xiao, X., Wang, Z., and Li, R. (2021). Characterization of TMexCD3-TOprJ3, an RND-type efflux system conferring resistance to tigecycline in Proteus mirabilis, and its associated integrative conjugative element. Antimicrob. Agents Chemother. 65:e0271220. doi: 10.1128/AAC.02712-20
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wu, Y., Dong, N., Cai, C., Zhang, R., and Chen, S. (2023). Hospital wastewater as a reservoir for the tigecycline resistance gene cluster tmexCD-toprJ. Lancet Microbe 4:e134. doi: 10.1016/S2666-5247(22)00326-3
Xu, J., Zhu, Z., Chen, Y., Wang, W., and He, F. (2021). The plasmid-borne tet(a) gene is an important factor causing Tigecycline resistance in ST11 Carbapenem-resistant Klebsiella pneumoniae under selective pressure. Front. Microbiol. 12:644949. doi: 10.3389/fmicb.2021.644949
Yu, Y., Cui, C. Y., Kuang, X., Chen, C., Wang, M. G., Liao, X. P., et al. (2021). Prevalence of tet(X4) in Escherichia coli from duck farms in Southeast China. Front. Microbiol. 12:716393. doi: 10.3389/fmicb.2021.716393
Zhang, L., Lu, X., and Zong, Z. (2013). The emergence of blaCTX-M-15-carrying Escherichia coli of ST131 and new sequence types in Western China. Ann. Clin. Microbiol. Antimicrob. 12:35. doi: 10.1186/1476-0711-12-35
Zhang, J., Zheng, B., Zhao, L., Wei, Z., Ji, J., Li, L., et al. (2014). Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect. Dis. 14:659. doi: 10.1186/s12879-014-0659-0
Keywords: tet(X4), tmexCD-toprJ, tigecycline resistance, plasmid, efflux pumps
Citation: Li Y, Fu Y, Qiu Y, Liu Q, Yin M and Zhang L (2023) Genomic characterization of tigecycline-resistant Escherichia coli and Klebsiella pneumoniae isolates from hospital sewage. Front. Microbiol. 14:1282988. doi: 10.3389/fmicb.2023.1282988
Edited by:
Sulagna Basu, National Institute of Cholera and Enteric Diseases (ICMR), IndiaReviewed by:
Subhasree Roy, National Institute of Cholera and Enteric Diseases (ICMR), IndiaTuhina Banerjee, Banaras Hindu University, India
Daichi Morita, Hiroshima University, Japan
Copyright © 2023 Li, Fu, Qiu, Liu, Yin and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luhua Zhang, emhsdWh1YUBzd211LmVkdS5jbg==
 Ying Li
Ying Li Yu Fu1
Yu Fu1 Luhua Zhang
Luhua Zhang