- Laboratory of Natural Drug Discovery and Research, College of Pharmacy, Binzhou Medical University, Yantai, China
Introduction: Alkaloidal natural products are attractive for their broad spectrum of pharmaceutical bioactivities. In the present work, the highly productive saline soil derived fungus, Penicillium raistrichii, was subjected to the strategy of OSMAC (one strain many compounds) with changes of cultivation status. Then, the work-flow led to the expansion of the alkaloid chemical diversity and subsequently induced the accumulation of four undescribed alkaloids, named raistrimides A–D (1–4), including three β-carbolines (1–3), one 2-quinolinone (4), and one new natural product, 2-quinolinone (5), along with five known alkaloid chemicals (6–10).
Methods: A set of NMR techniques including 1H, 13C, HSQC and HMBC, along with other spectroscopic data of UV-Vis, IR and HRESIMS, were introduced to assign the plain structures of compounds 1–10. The absolute configuration of 1–3 were elucidated by means of X-ray crystallography or spectroscopic analyses on optical rotation values and experimental electronic circular dichroism (ECD) data. In addition, it was the first report on the confirmation of structures of 6, 7 and 9 by X-ray crystallography data. The micro-broth dilution method was applied to evaluate antimicrobial effect of all compounds towards Staphylococcus aureus, Escherichia coli, and Candida albicans.
Results and discussion: The results indicated compounds 1, 3 and 4 to be bioactive, which may be potential for further development of anti-antimicrobial agents. The finding in this work implied that OSMAC strategy was a powerful and effective tool for promotion of new chemical entities from P. raistrichii.
Introduction
It is clearly understood that the secondary metabolite (SM) production capacity of microbes lies on biosynthetic gene clusters (BGCs) (Bills and Gloer, 2016; Genilloud, 2019; Zhang J. J. et al., 2019; Ke and Yoshikuni, 2020; Alexander, 2023). Yet, the traditional workflow involving laboratory cultivation of microbes, followed by extraction of fermentation, SMs purification, and structure elucidation, often results in the silent or weak expression of BGCs and thus confines the occurrences and number of the new chemical entities, due to failure of resembling the nature environment with intense microbial competition. There is a growing consensus that unlocking the silent BGCs is a key channel to counteract the dramatic decline in the quantity of novel naturally-produced chemical structures (Reen et al., 2015; Zhang X. et al., 2019).
During the recent several decades, in order to tap the biosynthetic potential and strengthen silent gene expression, molecular methodologies (Reen et al., 2015; Palys et al., 2020; Wang et al., 2020; Han et al., 2020a,b) like gene manipulation, including knocking down, introduction or heterologous expression, regulation of promoters, and induction of mutations, as well as approaches based on cultivation conditions variation, have been employed and have achieved great successes in mining cryptic SMs. Of these, the one strain many compounds (OSMAC) strategy, covering medium variation, cultivation condition change, co-cultivation, or addition of epigenetic modifiers, has been efficiently applied to promote new therapeutic agents discovery, highlighting it as an easy and productive approach to trigger the reaction of the crypt BGCs and enhance the natural SMs production according to the extensive literature reports (Bode et al., 2002; Romano et al., 2018; Pan et al., 2019).
During our ongoing efforts in pursuit of therapeutic pharmaceuticals from saline soil-derived fungi, a talent genus P. raistrichii isolated from saline sediment in Bohai Bay, entered into our vision. Chemical investigations on the metabolic profile resulted in the discovery of several different novel scaffolds including polyketides and alkaloids with biological behaviors of antiinsectan, antimicrobial, radical scavenging, cytotoxic, or anti-HCV activities (Belofsky et al., 1998; Ma et al., 2016; Jia et al., 2017; Liu et al., 2018; Li et al., 2019). In recognition of the superior ability for the production of intriguing molecules from the talent strain, and also enlightened by OSMAC strategy of expansion on the fungal metabolome, the strain was subjected to culture under static conditions in a liquid medium. Excitingly, the change of culture conditions, which may have exerted influences on the biochemical reactions, caused the strain to produce four new alkaloids, named raistrimides A-D (1–4), one new natural product 2-quinolinone, 3-hydroxy-4-(4′-methoxyphenyl)-2(1H)-quinolinone (5) (He et al., 2015), and five known alkaloids, pesimquinolone R (6) (Dai et al., 2021), peniprequinolone (7) (Hayashi et al., 1997; Uchida et al., 2006), 4,5-dihydroxy-3,4-dihydro-3-methoxy-4-(4′-methoxyphenyl)-2(1H)-quinolinone (8) (Framm et al., 1973), dehydrocyclopeptin (9) (Ishikawa et al., 2014), and tunicoidine F (10) (Wen et al., 2014; Zhang et al., 2015). Detailed information about the isolation, purification, and structure elucidation, as well as the antimicrobial effect assessment of these compounds (Figure 1) is presented in this work.
Materials and methods
General experimental procedures
An uncorrected XRC-1 micro melting point apparatus was used to measure the melting points. Uv–Vis and IR data were performed on a TU-1091 spectrophotometer and a Thermo Nicolet 6,700 infrared spectrophotometer, respectively. A Rudolph Autopol V Plus digital polarimeter, a Bruker Smart 1,000 CCD X-ray diffractometer, and an Applied Photophysics Chirascan spectropolarimeter were used for the determination of optical rotations, X-ray crystal data, and electron circular dichroism (ECD) spectra, respectively. Varian Inova 600 and Bruker AV-400/AVIII 500 spectrometers were used to measure 1D and 2D NMR data in different deuterium solvents using tetramethylsilane as an internal standard (IS). HRESIMS spectra were recorded using an Agilent 1200RRLC-6520 Accurate-Mass Q-TOF LC/MS mass spectrometer or a Waters Q-TOF Ultima GLOBAL GAA076 LC. A Shimadzu LC-6 AD and LC-20A Liquid Chromatography were applied for the preparative isolation and analysis on HyperClone C18 columns (5 μm, 10 × 250 mm, 5 μm, 4.6 × 250 mm, respectively). Open column chromatography (CC) isolations were performed on silica gel (200–300 mesh, Qingdao Marine Chemical, China), reverse phase (RP) C18 material (Pharmacia Fine Chemical Co., Sweden), and Sephadex LH-20 (Ge Healthcare Bio-Sciences AB, Sweden).
Fungal material
Saline soil samples were collected from the coast in the Circum-Bohai-Sea region in Zhanhua county, from which a talent strain of Penicillium raistrickii with GenBank Accession NO. HQ717799 was isolated in August 2008. The voucher specimen was preserved at the laboratory of Natural Drug Discovery and Research, Binzhou Medical University.
Cultivation and extraction
The spores of P. raistrickii growing well on potato dextrose agar (PDA) were incubated in 500-mL Erlenmeyer flasks which contained 180 mL of culture medium comprising 2% glucose, 1% maltose, 0.03% yeast extract, 0.003% MgSO4·7H2O, and 0.005% KH2PO4 with the natural pH value in seawater gathered from offshore areas near Yantai, and fermented under static condition at about 28°C for 60 days. A whole volume of 40 L culture broth was isolated into supernatant and mycelia through cheesecloth. The supernatant was extracted with ethyl acetate (3 × 30 L), and the mycelia was extracted with methanol (3 × 10 L) to afford crude aqueous extract followed by partitioning with ethyl acetate, which yielded an extract of 45 g.
Isolation and purification
The whole extract was subjected to a VLC column with silica gel eluting by a step gradient of petroleum ether/CHCl3 (from 1:2 to 0:1, v/v) to give five fractions (Fr.s 1–5), and then followed by a step gradient of CHCl3/MeOH (from 1:2 to 0:1, v/v) to afford another five fractions (Fr.s 6–10). Among these, Fr. 3 (4.1 g) and Fr. 4 (3.3 g) were separated with MeOH/H2O (20:80 to 100:0, v/v) on ODS to afford nine (Fr.s 3.1–3.9) and six (Fr.s 4.1–4.6) subfractions, respectively. Then, compound 1 (9.2 mg) was purified by a Sephadex LH-20 CC from Fr. 4.3 (0.2 g) with MeOH as the eluent. Compounds 2 (3.6 mg) and 10 (16.0 mg) were purified by semipreparative HPLC (MeOH:H2O, 70:30, v:v) with a retention time (t R ) of 20.8 min and 17.3 min, respectively. Compound 3 (22.0 mg) was also obtained from semipreparative HPLC (MeOH:H2O, 65:35, v:v) with a t R value of 17.1 min. Compounds 4 (12.6 mg) and 5 (9.3 mg) were given from Fr. 3.8 (0.5 g) which was chromatographed on a silica gel column, eluting with CHCl3/MeOH (30:1, v/v). Compounds 6 (14.6 mg) and 8 (32.6 mg) were obtained from Fr. 4.2 (0.7 g) and Fr. 5 (1.7 g) by crystalization in MeOH. Compounds 7 (15.8 mg) and 9 (10.8 mg) were purified by semipreparative HPLC (MeOH:H2O, 60:70, v:v) from Fr. 3.3 (0.9 g) and Fr. 4.6 (0.7 g) with values of 26.2 min and 13.8 min, respectively.
Raistrimide A (1)
Yellowish needle (MeOH); mp 278°C (dec.); UV (MeOH) λmax (log ε): 377 (3.79), 286 (4.61), 220 (4.56) nm; IR (ATR) νmax 3,352, 2,869, 1,728, 1,633, 1,537, 1,493, 1,444, 1,336, 1,195, 1,115, 963, 887, 750, 682 cm−1; 1H and 13C NMR data, see Table 1; HRESIMS (m/z): 324.0993 [M – H]− (calcd for C17H14N3O4, 324.0990). + 10.8 (c 0.062, MeOH); ECD (MeOH) λmax (Δε) 229 (− 45.21), 204 (+ 120.47) nm.
Raistrimide B (2)
yellowish needle (MeOH); mp 196–199°C; UV (MeOH) λmax (log ε): 377 (3.67), 287 (4.53), 220 (4.44) nm; IR (ATR) νmax 3,329, 2,964, 1,740, 1,663, 1,626, 1,515, 1,491, 1,459, 1,204, 1,179, 963, 847, 738, 683 cm−1; 1H and 13C NMR data, see Table 1; HRESIMS (m/z): 382.1762 [M + H]+ (calcd for C21H24N3O4, 382.1761). + 4.4 (c 0.069, MeOH); ECD (MeOH) λmax (Δε) 280 (+ 0.88), 268 (− 0.02), 240 (+ 0.84), 229 (− 0.50), 204 (+ 3.99) nm.
Raistrimide C (3)
yellowish needle (MeOH); mp 280°C (dec.); UV (MeOH) λmax (log ε): 377 (3.73), 286 (4.53), 220 (4.46) nm; IR (ATR) νmax 3,394, 2,969, 2,542, 1,745, 1,680, 1,622, 1,542, 1,494, 1,361, 1,302, 1,202, 1,147, 965, 899, 788, 742, 681 cm−1; 1H and 13C NMR data, see Table 1; HRESIMS (m/z): 352.1302 [M – H]− (calcd for C19H18N3O4, 352.1303). + 12.0 (c 0.055, MeOH); ECD (MeOH) λmax (Δε) 280 (+ 0.73), 264 (− 0.24), 232 (+ 0.30), 224 (− 0.21), 204 (+ 3.68) nm.
Raistrimide D (4)
colorless needle (MeOH); mp 230–232°C; UV (MeOH) λmax (log ε): 337 (3.18), 324 (3.36), 311 (3.35), 298 (3.33), 278 (3.37), 218 (4.03) nm; IR (ATR) νmax 1,647, 1,609, 1,553, 1,515, 1,500, 1,434, 1,282, 1,219, 1,175, 1,146, 1,017, 899, 836, 747, 699 cm−1; 1H and 13C NMR data, see Table 2; HRESIMS (m/z): 280.0973 [M – H]− (calcd for C17H14NO3, 280.0968).
3-hydroxy-4-(4′-methoxyphenyl)-2(1H)-quinolinone (5)
Colorless blocks (MeOH); UV (MeOH) λmax (log ε): 331 (3.51), 318 (3.65), 306 (3.60), 286 (3.58), 243 (3.86), 222 (4.16) nm; IR (ATR) νmax 2,918, 1,652, 1,608, 1,574, 1,515, 1,501, 1,455, 1,403, 1,291, 1,245, 1,031, 950, 881, 753, 713 cm−1; 1H and 13C NMR data, see Table 2; HRESIMS (m/z): 266.0815 [M – H]− (calcd for C16H12NO3, 266.0812).
Dehydrocyclopeptin (9)
colorless blocks (MeOH); 97–99°C; UV (MeOH) λmax (log ε): 288 (4.28), 214 (4.59), 220 (4.46), 205 (4.43) nm; IR (ATR) νmax 3,064, 1,670, 1,622, 1,602, 1,512, 1,478, 1,385, 1,252, 1,175, 1,028, 825, 754, 699 cm−1; 1H and 13C NMR data, see Table 3.
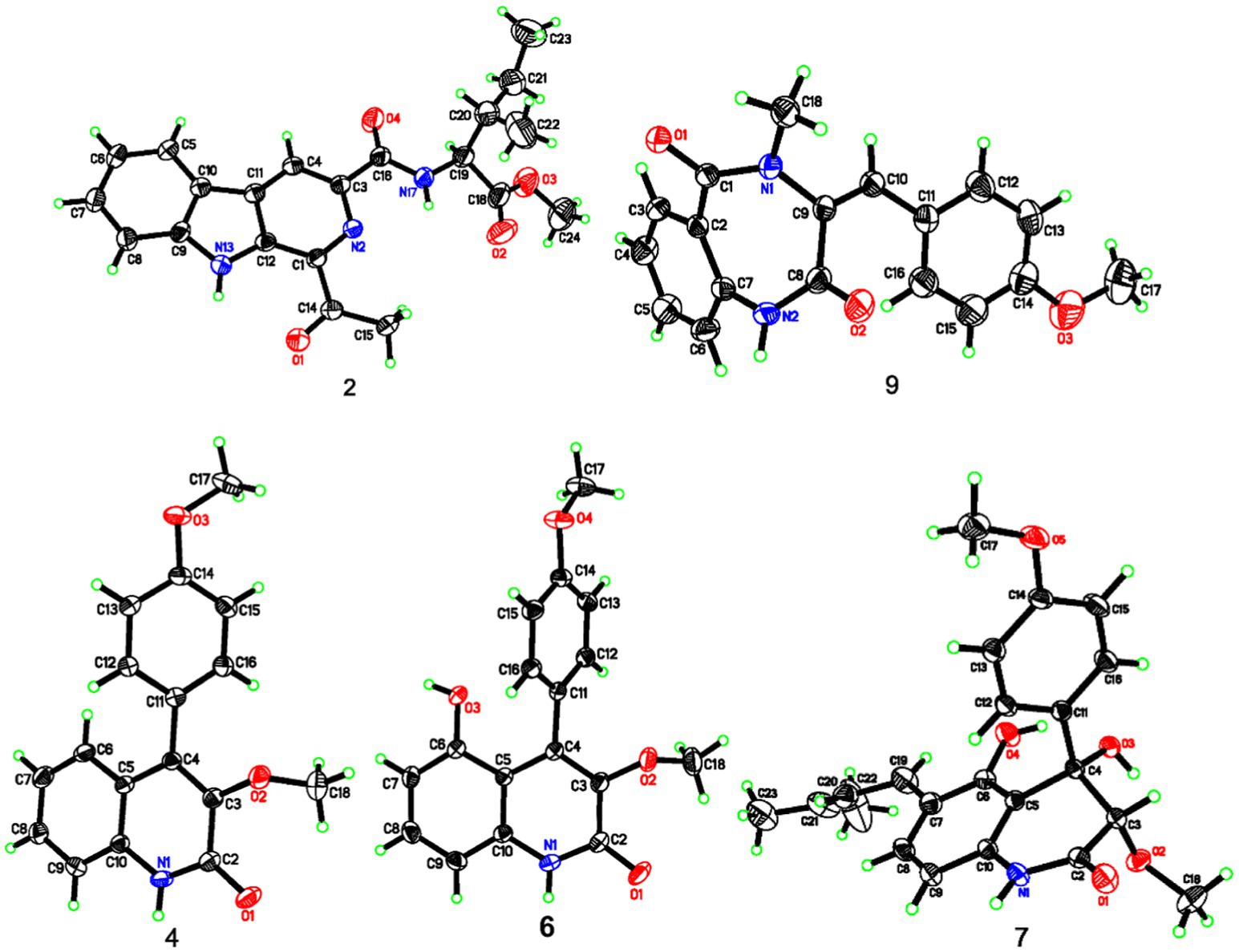
X-ray single-crystal structure determinations of 2, 4, 6, 7, and 9
All of the determined compounds were acquired from the solvent of MeOH by vapor diffusion. The structure elucidation of these compounds was settled by the Sheldrick (2014) software package through direct methods and were refined using least squares minimization. The crystallographic data of compounds 2 (CCDC 2002826), 4 (CCDC 2002827), 6 (CCDC 2002870), 7 (CCDC 2002825), and 9 (CCDC 2002821) have been already deposited in the Cambridge Crystallographic Data Centre.1
Crystal data for 2
C21H23N3O4, Mr = 381.42, monoclinic, space group C2, a = 15.8834 (11) Å, b = 6.8528 (7) Å, c = 18.4959 (17) Å, V = 2013.0 (3) Å3, T = 293 (2) K, Z = 4, μ(Cu Kα) = 0.723 mm−1, Dcalcd = 1.259 g/cm3. α = γ = 90°, β = 90.565 (8)°. In total, 3,289 reflections were measured (9.56 ≤ 2θ ≤ 132.08), with 2,354 independent unique reflections (Rint = 0.0466). The final refinement [I ≥ 2σ (I)] gave R1 = 0.0633 and wR2 = 0.1351. The final refinement presented R1 = 0.1103 and wR2 = 0.1650 (all data). Flack parameter = 0.0 (9).
Crystal data for 4
C17H15NO3, Mr = 281.30, monoclinic, space group C2/c, a = 26.002 (2) Å, b = 7.4221 (8) Å, c = 14.8450 (13) Å, V = 2787.6 (4) Å3, T = 293 (2) K, Z = 8, μ(Cu Kα) = 0.092 mm−1, Dcalcd = 1.341 g/cm3. α = γ = 90°, β = 103.343 (2)°. In total, 5,145 reflections were measured (5.64 ≤ 2θ ≤50.02), with 2,453 independent unique reflections (Rint = 0.1063). The final refinement [I ≥ 2σ (I)] gave R1 = 0.1063, and wR2 = 0.1351. The final refinement presented R1 = 0.0951 and wR2 = 0.1400 (all data).
Crystal data for 6
C17H15NO4, Mr = 297.30, monoclinic crystal, space group P21/n, a = 8.7113 (5) Ǻ, b = 9.5303 (7) Ǻ, c = 17.3892 (12) Ǻ, α = γ = 90°, β = 103.465 (2)°, V = 1403.99 (16) Ǻ3, T = 293 (2) K, Z = 4, μ(Cu Kα) = 0.834 mm−1, Dcalc = 1.407 g/cm3. In total, 4,326 reflections were measured (10.46 ≤ 2θ ≤ 132.18), with 2,454 independent unique reflections (Rint = 0.0221). The final refinement [I ≥ 2σ (I)] gave R1 = 0.0483, and wR2 = 0.1334. The final refinement presented R1 = 0.0608 and wR2 = 0.1451 (all data).
Crystal data for 7
C22H24NO5, Mr == 382.42, orthorhombic crystal, space group P212121, a = 9.6170 (4) Ǻ, b = 13.3217 (5) Ǻ, c = 15.7153 (6) Ǻ, α = β = γ = 90°, V = 2013.36 (14) Ǻ3, T = 293 (2) K, Z = 4, μ(Cu Kα) = 0.732 mm−1, Dcalc = 1.262 g/cm3. In total, 4,449 reflections were measured (8.70 ≤ 2θ ≤ 132.78), with 2,991 independent unique reflections (Rint = 0.0261). The final refinement [I ≥ 2σ (I)] gave R1 = 0.0498, and wR2 = 0.1377. The final refinement gave R1 = 0.0538, and wR2 = 0.1422 (all data). Flack parameter = −0.27 (18).
Crystal data for 9
C18H16N2O3, Mr = 308.33, orthorhombic crystal, space group Pbca, a = 15.8992 (14) Ǻ, b = 8.5824 (8) Ǻ, c = 23.758 (2) Ǻ, α = β = γ = 90°, V = 3241.8 (5) Ǻ3, T = 298 (2) K, Z = 8, μ(Mo Kα) = 0.087 mm−1, Dcalc = 1.263 g/cm3. In total, 14,810 reflections were measured (5.12 ≤ 2θ ≤ 50.04), with 2,861 independent unique reflections (Rint = 0.0842). The final refinement [I ≥ 2σ (I)] gave R1 = 0.0516, and wR2 = 0.1024. The final refinement presented R1 = 0.1094 and wR2 = 0.1146 (all data).
Biological assay
The antimicrobial activity of the isolated compounds was examined by the broth microdilution method according to the method as previously described (Ma et al., 2019). Microorganisms evaluated in this work included Staphylococcus aureus, Escherichia coli, and Candida albicans, and chloramphenicol or ketoconazole was applied as positive control.
Results and discussion
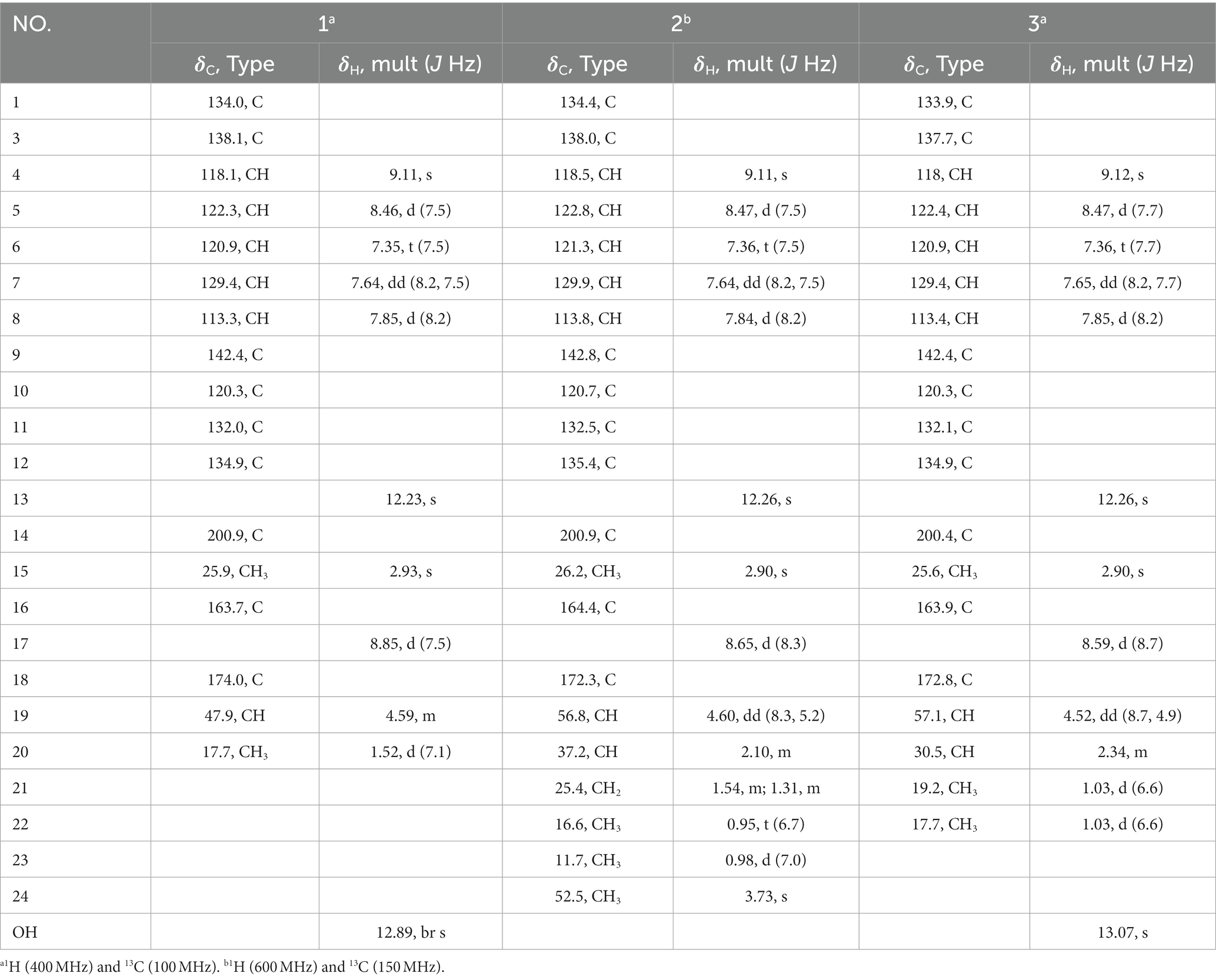
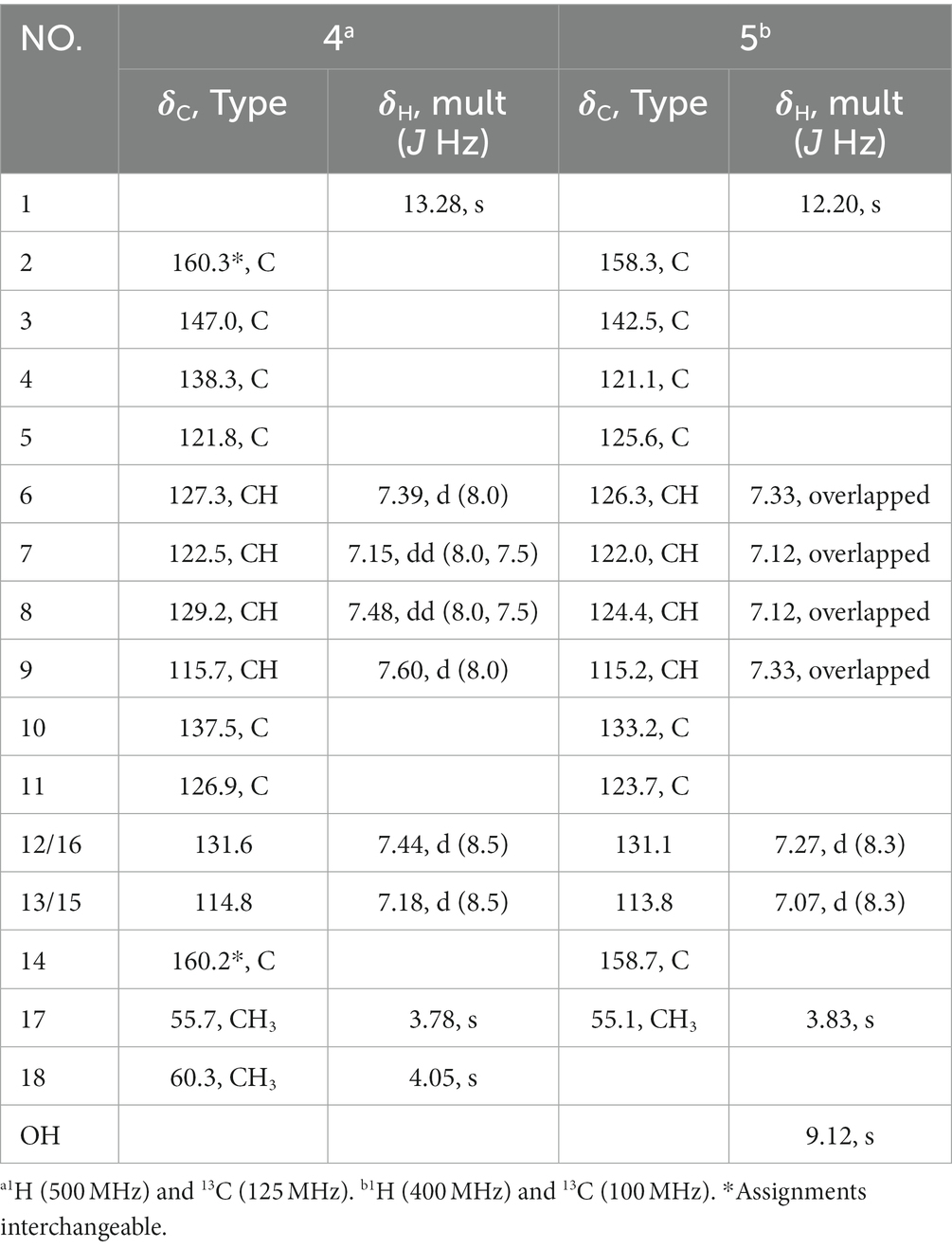
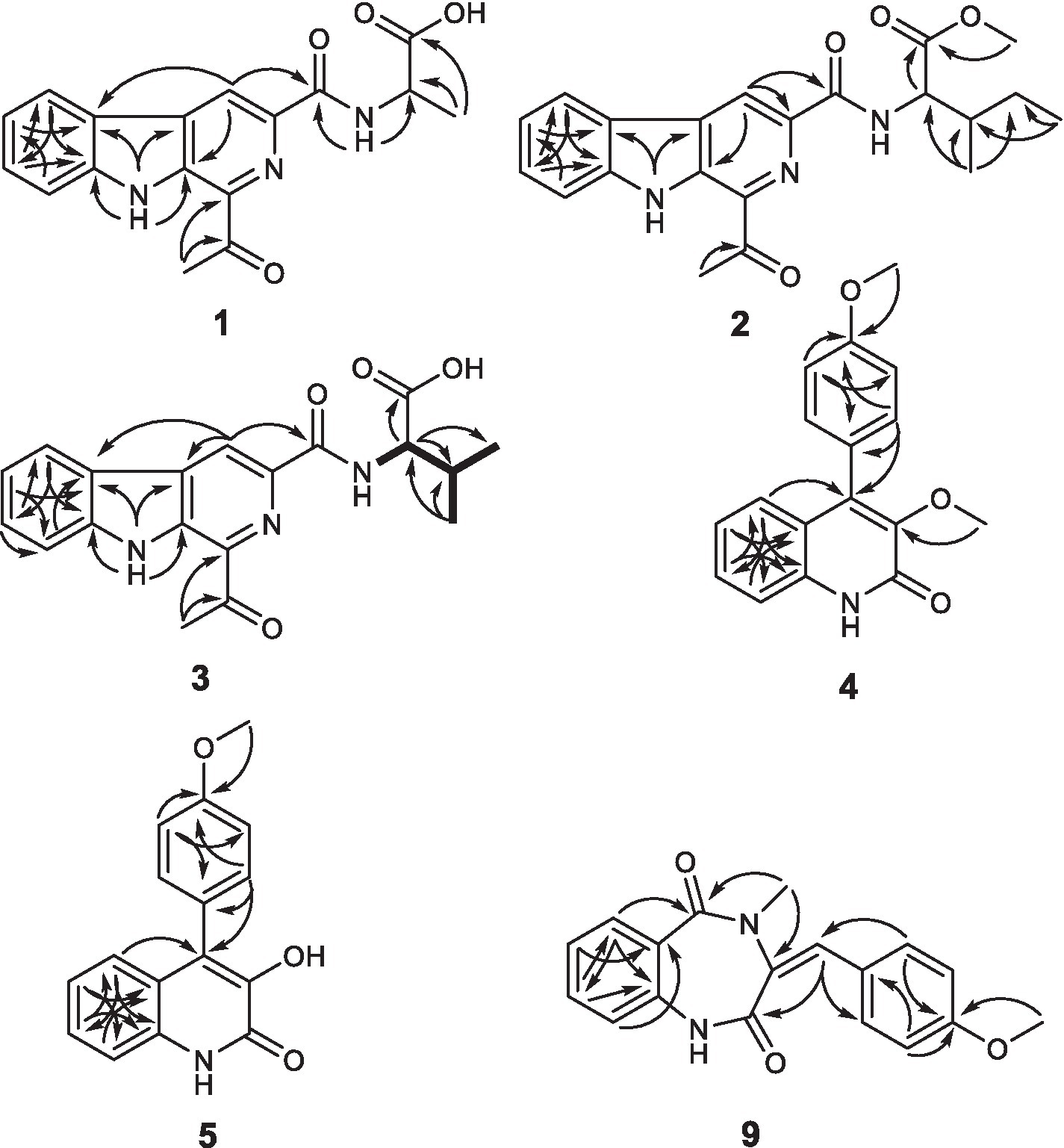
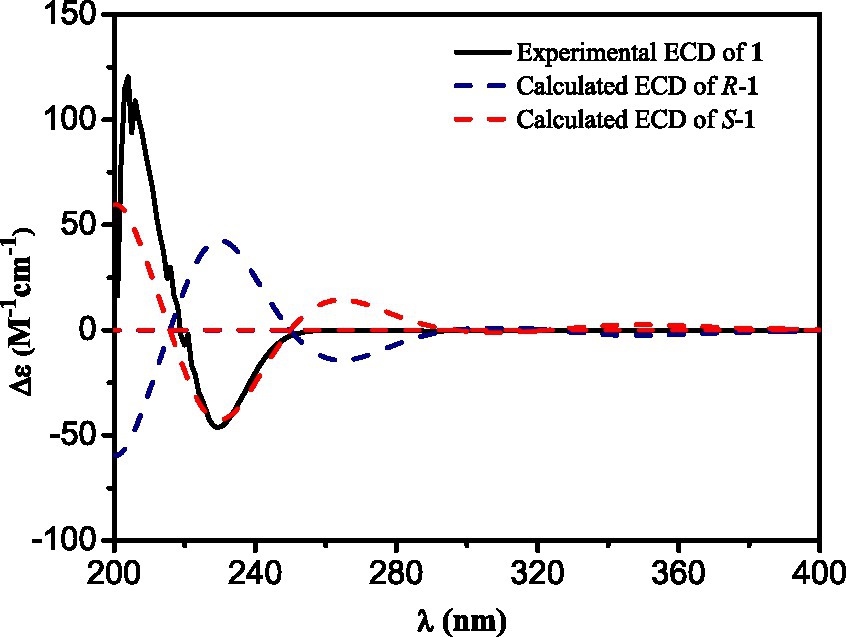
Compound 1 was obtained as a yellowish needle. The molecular formula of C17H15N3O4 was established from the deprotonated-ion HRESIMS at m/z 324.0993 (calcd for C17H14N3O4, 324.0990). It indicated a β-carboline chromophore by the feature of the UV absorptions at 377, 286 and 220 nm (Chen et al., 2010). The 1H NMR spectrum in Table 1 showed four aromatic hydrogen signals at δH 8.46 (d, J = 7.5 Hz), 7.85 (d, J = 8.2 Hz), 7.64 (t, J = 7.5 Hz), and 7.35 (t, J = 7.5 Hz). The coupling patterns of these four aromatic hydrogens indicated the characteristics of an ο-disubstituted benzene ring in the indole moiety (Tangella et al., 2018). The doublet methyl at δH 1.52, multiplet methine at δH 4.59, and the doublet imino proton at δH 8.85 were suggestive of an alanine residue functionality. The 13C NMR data showed eleven aromatic carbon signals from δC 142.4 to 113.3 (Table 1), which featured the β-carboline skeleton. The signals of keto carbonyl at δC 200.9 and the methyl at δC/H 25.9/2.93, along with the HMBC crosspeaks from the methyl to the carbonyl, implied the existence of an acetyl group (Figure 2). It was located at C-1 on the basis of correlation from H-15 to C-1. Further interactions from H-17 to C-16 and C-19, and from H-4 to C-16 suggested the linkage of C-3 and the alanine residue through C-16. Thus, the plain structure of 1 was accomplished. Considering that alanine moiety plays a crucial role in the specific optical rotation in compound 1 (the same condition also applied to compounds 2, 3, and 10), the consistent specific rotation behaviors of 1 {[ + 10.8 (c 0.062, MeOH)}and oldhamiaine A {[ + 14.0 (c 0.062, MeOH)}suggested the absolute configuration of C-19 to be S (Zhang et al., 2015). Additionally, the experimental and calculated ECD curves (Figure 3) further supported the absolute configuration assignment. The absolute stereochemistry of 1 was established to be 19S.
Compound 2 was purified as light yellow needles with a molecular formula of C21H23N3O4 given by the positive HRESIMS at m/z 382.1762 [M + H]+ (calcd for C21H24N3O4, 382.1761). The UV and IR spectra of 2 also showed the absorption features of a β-carboline skeleton. The 1H and 13C NMR spectroscopic data of 2 (Table 1) in the downfield regions were almost identical with those of 1, indicative of the same β-carboline moiety. Furthermore, the signals in the upfield of two methines at δH 4.60 and 2.10, one diastereotopic methylene at δH 1.54 and 1.31, two methyls at δH 0.98 and 0.95, and the carbon resonances at δC 56.8, 37.2, 25.4, 16.6, 11.7 (Table 1), suggested the fragment of an isoleucine residue, which was supported by the HMBC correlations (Figure 2). Finally, the X-ray analyses unambiguously determined the absolute configuration of 2 (Figure 4).
Compound 3, light yellow needles, was determined to have a molecular formula of C19H19N3O4 by the negative HRESIMS data (m/z 352.1302, calcd for C19H18N3O4, 352.1303). Its UV and IR absorption bands were almost the same as those of compounds 1 and 2. The spectroscopic data covering 1H and 13C NMR (Table 1) closely resembled those of 1, except for additional appearances of one methine and one methyl signals. Careful analyses of the chemical shifts and the splitting behaviors of a double doublets methine at δH 4.52, a multiplet methine at δH 2.34, two doublet methyl groups at δH 1.03, and a doublet NH at δH 8.59 were suggestive of a valine motif, which was supported by the corresponding 1H-1H COSY data (Figure 2). The position of the valine motif was accomplished according to the HMBC interactions from H-4 and H-17 to C-16 (Figure 2). The acetyl group linked at C-1 was elucidated by the correlations from H-15 to C-1 and C-14. The similar ECD absorptions (ESI Supplementary Figure S24) together with evidence of the same optical rotation direction with that of 10 confirmed the absolute configuration of 3.
Compound 4, colorless needles, exhibited a deprotonated ion signal at m/z 280.0973 (calcd for C17H14NO3, 280.0968) in negative HRESIMS, which corresponded with its molecular formula of C17H15NO3. In 1H NMR spectroscopic data, two aromatic protons at δH 7.44 and 7.18 (Table 2) which were presented in an ortho-coupled doublet fashion, indicated the presence of a p-disubstituted ring fragment. Four aromatic protons at δH 7.60, 7.48, 7.39, and 7.15 (Table 2) represented the existence of an o-disubstituted phenyl ring moiety. Evidence of the HMBC data from H-17 to C-14 and from H-12 to C-4 and C-14 revealed the methoxyl and C-4 anchored at the p-disubstituted phenyl ring (Figure 2). The NH group and C-4 were located at the o-disubstituted phenyl ring, which was supported by HMBC cross peaks from H-6 to C-4 and C-10, along with the chemical shift of C-10. The remaining three carbon signals, including a carbonyl at δC 160.3, a quaternary carbon at δC 147.0, and a methoxyl at δC 60.3 (Table 2) were assembled to complete the whole structure. Finally, the proposed structure of 3 was further undoubtedly defined by the X-ray diffraction analysis (Figure 4).
The structure of 5 was finally determined by 1D and 2D NMR data (Figure 2) along with the data from a previously reported synthetic intermediate (He et al., 2015; Zhang et al., 2015), and it was purified from natural for the first time.
Compound 9 was named dehydrocyclopeptin (Hayashi et al., 1997; Ma et al., 2012). It was first isolated from Penicillium cyclopium (Framm et al., 1973), but the chemical shift assignments of some signals and the configuration of its double band needed to be discussed. In the current report, HSQC and HMBC spectra (Figure 2) helped to provide the 1H and 13C NMR data (Table 3), and X-ray diffraction analysis unambiguously built up an E configuration of the double band (Figure 4).
The rest of the compounds, namely, 6, 7, 8, and 10, were determined to be pesimquinolone R; peniprequinolone; 4,5-dihydroxy-3,4-dihydro-3-methoxy-4-(4′-methoxyphenyl)-2(1H)-quinolinone; and tunicoidine F, respectively, through comparison of the corresponding spectroscopic data with the literature. Among them, we first reported the X-ray data for compounds 6, 7, and 9.
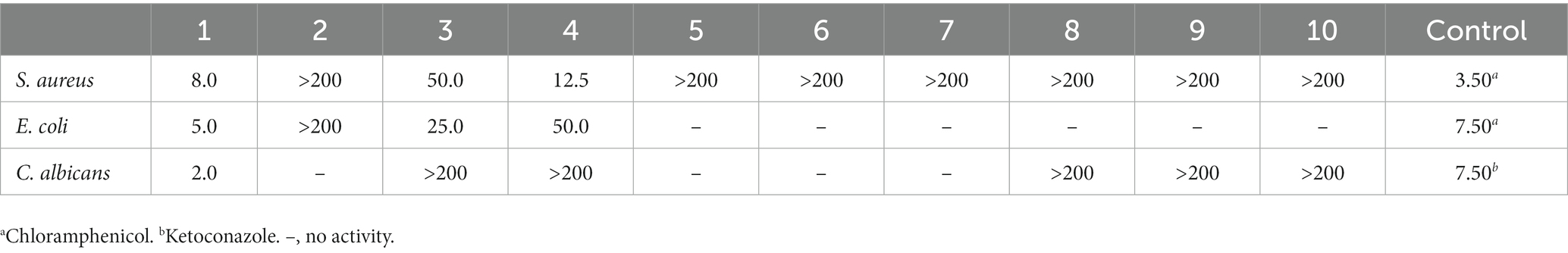
The antimicrobial activity of compounds 1–10 was examined against S. aureus, E. coli, and C. albicans with chloramphenicol or ketoconazole as the positive control. As the results show in Table 4, compound 1 demonstrated an obvious antimicrobial effect, while 3 and 4 exhibited activity toward S. aureus and E. coli.
Conclusion
In summary, an OSMAC-based strategy applied to P. raistrickii of changing cultivation status from shaking fermentation to a static condition in a liquid medium expanded the alkaloidal SM diversity and thus induced the accumulation of four unreported alkaloids, including three β-carbolines (1–3) and one 2-quinolinone (4), along with one new natural compound 2-quinolinone (5) and five known alkaloids (6–10). The outcome indicated a big effect from a small change in the OSMAC strategy application, showing it to be a powerful tool in unlocking the fungal cryptic BGCs to obtain novel natural products.
β-Carboline alkaloids possess the fundamental framework of tricyclic pyrido [3,4-b] indole and are mainly obtained from plants and marine organisms, with only a few from microbes to our knowledge. 2-quinolinones, or named 2-quinolones, are quinoline derivatives with a carbonyl group inserted in the 2 position, most of which are produced by fungal genera (Neff et al., 2012). Benzodiazepine alkaloids are seven-membered cyclodipeptides composed of an anthranilic acid and another α-amino acid or its deoxygenated analogs. They are metabolites of filamentous fungi or actinomycetes, with one exception from sea hare up to now, and have not been found in higher plants (Roos, 1990; Ojika et al., 1993). These three kinds of alkaloids have a strong attraction to scientists due to their structural diversities and extensive scope of pharmacological effects, such as antitumor, antidepressant, antimicrobial, antimalarial, anti-inflammatory, antioxidant, insecticidal and analgesic effects (Roos, 1990; Cao et al., 2007; Cui et al., 2009; Wang et al., 2014; Simonetti et al., 2016; Reddy et al., 2018).
The structures of 1–10 were constructed by comprehensive analyses of the spectroscopic data. Measures including X-ray diffraction data, ECD, and optical rotation values helped to solve the absolute configurations of the new compounds. In addition, the configuration of the double band in compound 9 was unambiguously revised by X-ray diffraction data, and its 1H and 13C NMR data were reassigned in this work. Furthermore, it was the first report of the X-ray diffraction analyses on the absolute stereochemistry of compounds 7.
All of the compounds (1–10) in the work were screened for their antimicrobial activity against microorganisms like S. aureus, E. coli, and C. albicans. Compounds 1, 3, and 4 showed antimicrobial effects. As for the β-carbolines obtained in the work, compound 1 exhibited an obvious pharmaceutical effect against tested microorganisms with MIC values of 8.0, 5.0, and 2.0 μg/mL, and compound 3 also showed some activity. The results implied that the amino acid residues and a free carboxyl group in this type of β-carboline alkaloids might play a pivotal part in the antimicrobial effect.
Data availability statement
The datasets generated for this study can be found in the online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
MZ: Data curation, Investigation, Writing – original draft. HK: Investigation, Writing – original draft, Formal analysis. WL: Writing – original draft, Resources, Supervision, Validation, Visualization, Writing – review & editing. LM: Investigation, Writing – original draft, Formal analysis, Methodology. DL: Funding acquisition, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The National Natural Science Foundation of China (No. 31270082), Science and Technology Development Plan of Yantai (No. 2020XDRH111) gave financial support to this work.
Acknowledgments
We are thankful to Tianjiao Zhu who helped to identify strain of P. raistrichii (School of Medicine and Pharmacy, Ocean University of China).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1279140/full#supplementary-material
Footnotes
References
Alexander, A. Z. (2023). Fungal BGCs for production of secondary metabolites: main types, central roles in strain improvement, and regulation according to the piano principle. Int. J. Mol. Sci. 24:11184. doi: 10.3390/ijms241311184
Belofsky, G. N., Gloer, K. B., Gloer, J. B., Wicklow, D. T., and Dowd, P. F. (1998). New p-terphenyl and polyketide metabolites from the sclerotia of Penicillium raistrickii. J. Nat. Prod. 61, 1115–1119. doi: 10.1021/np980188o
Bills, G. F., and Gloer, J. B. (2016). Biologically active secondary metabolites from the fungi. Microbiol. Spectr. 4, 1089–1119. doi: 10.1128/microbiolspec.FUNK-0009-2016
Bode, H. B., Bethe, B., Höfs, R., and Zeeck, A. (2002). Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem 3, 619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9
Cao, R. H., Peng, W. L., Wang, Z. H., and Xu, A. L. (2007). beta-Carboline alkaloids: biochemical and pharmacological functions. Curr. Med. Chem. 14, 479–500. doi: 10.2174/092986707779940998
Chen, Y. F., Kuo, P. C., Chan, H. H., Kuo, I. J., Lin, F. W., Su, C. R., et al. (2010). β-Carboline alkaloids from Stellaria dichotoma var. lanceolata and their anti-inflammatory activity. J. Nat. Prod. 73, 1993–1998. doi: 10.1021/np1003627
Cui, C. G., Li, X. M., Li, C. S., Sun, H. F., Gao, S. S., and Wang, B. G. (2009). Benzodiazepine alkaloids from marine-derived endophytic fungus aspergillus ochraceus. Helv. Chim. Acta 92, 1366–1370. doi: 10.1002/hlca.v92:7
Dai, C., Li, X., Zhang, K., Li, X. N., Wang, W., Zang, Y., et al. (2021). Pesimquinolones I-S, eleven new quinolone alkaloids produced by Penicillium simplicissimum and their inhibitory activity on NO production. Bioorg. Chem. 108:104635. doi: 10.1016/j.bioorg.2021.104635
Framm, J., Nover, L., El-Azzouny, A., Richter, H., Winter, K., Werner, S., et al. (1973). Cyclopeptin and dehydrocyclopeptin. Intermediates in the biosynthesis of cyclopeptin-viridicatin alkaloids in Penicillium cyclopium Westling. Eur. J. Biochem. 37, 78–85. doi: 10.1111/j.1432-1033.1973.tb02959.x
Genilloud, O. (2019). Natural products discovery and potential for new antibiotics. Curr. Opin. Microbiol. 51, 81–87. doi: 10.1016/j.mib.2019.10.012
Han, J. Y., Liu, M. M., Jenkins, I. D., Liu, X. T., Zhang, L. X., Quinn, R. J., et al. (2020a). Genome-inspired chemical exploration of marine fungus Aspergillus fumigatus MF071. Mar. Drugs 18:352. doi: 10.3390/md18070352
Han, J. Y., Zhang, J. Y., Song, Z. J., Zhu, G. L., Liu, M. M., Dai, H. Q., et al. (2020b). Genome-based mining of new antimicrobial meroterpenoids from the phytopathogenic fungus Bipolaris sorokiniana strain 11134. Appl. Microbiol. Biotechnol. 104, 3835–3846. doi: 10.1007/s00253-020-10522-1
Hayashi, H., Nakatani, T., Inoue, Y., Nakayama, M., and Nozaki, H. (1997). New dihydroquinolinone toxic to artemia salina produced by Penicillium sp. NTC-47. Biosci. Biotechnol. Biochem. 61, 914–916. doi: 10.1271/bbb.61.914
He, J., Lion, U., Sattler, I., Gollmick, F. A., Grabley, S., Cai, J. M., et al. (2015). Diastereomeric quinolinone alkaloids from the marine-derived fungus Penicillium janczewskii. J. Nat. Prod. 68, 1397–1399. doi: 10.1021/np058018g
Ishikawa, N., Tanaka, H., Koyama, F., Noguchi, H., Wang, C. C., Hotta, K., et al. (2014). Non-heme dioxygenase catalyzes atypical oxidations of 6, 7-bicyclic systems to form the 6, 6-quinolone core of viridicatin-type fungal alkaloids. Angew. Chem. Int. Ed. Engl. 53, 12880–12884. doi: 10.1002/anie.201407920
Jia, L. G., Dong, J. Z., Wang, R. J., Mao, S. H., Lu, F., Singh, S., et al. (2017). Identification and characterization of the steroid 15α-hydroxylase gene from Penicillium raistrickii. Appl. Microbiol. Biotechnol. 101, 6409–6418. doi: 10.1007/s00253-017-8377-3
Ke, J., and Yoshikuni, Y. (2020). Multi-chassis engineering for heterologous production of microbial natural products. Curr. Opin. Biotechnol. 62, 88–97. doi: 10.1016/j.copbio.2019.09.005
Li, J. L., Hu, Y. Y., Hao, X. M., Tan, J. L., Li, F., Qiao, X. R., et al. (2019). Raistrickindole a, an anti-hcv oxazinoindole alkaloid from Penicillium raistrickii IMB17-034. J. Nat. Prod. 82, 1391–1395. doi: 10.1021/acs.jnatprod.9b00259
Liu, D. S., Rong, X. G., Kang, H. H., Ma, L. Y., Hamann, M. T., and Liu, W. Z. (2018). Raistrickiones A-E from a highly productive strain of Penicillium raistrickii generated through thermo change. Mar. Drugs 16:213. doi: 10.3390/md16060213
Ma, L. Y., Liu, D. S., Li, D. G., Huang, Y. L., Kang, H. H., Wang, C. H., et al. (2016). Pyran rings containing polyketides from Penicillium raistrickii. Mar. Drugs 15:2. doi: 10.3390/md15010002
Ma, L. Y., Liu, W. Z., Shen, L., Huang, Y. L., Rong, X. G., Xu, Y. Y., et al. (2012). Spiroketals, isocoumarin, and indoleformic acid derivatives from saline soil derived fungus Penicillium raistrickii. Tetrahedron 68, 2276–2282. doi: 10.1016/j.tet.2012.01.054
Ma, L. Y., Zhang, H. B., Kang, H. H., Zhong, M. J., Liu, D. S., Ren, H., et al. (2019). New butenolides and cyclopentenones from saline soil-derived fungus aspergillus Sclerotiorum. Molecules 24:2642. doi: 10.3390/molecules24142642
Neff, S. A., Lee, S. U., Asami, Y., Ahn, J. S., Oh, H., Baltrusaitis, J., et al. (2012). Aflaquinolones A-G: secondary metabolites from marine and fungicolous isolates of aspergillus spp. J. Nat. Prod. 75, 464–472. doi: 10.1021/np200958r
Ojika, M., Yoshida, T., and Yamada, K. (1993). Aplysepine, a novel 1, 4-benzodiazepine alkaloid from the sea hare Aplysia kurodai. Tetrahed. Lett. 34, 5307–5308. doi: 10.1016/S0040-4039(00)73981-5
Palys, S., Pham, T. T. M., and Tsang, A. (2020). Biosynthesis of alkylcitric acids in aspergillus Niger involves both co-localized and unlinked genes. Front. Microbiol. 11:1378. doi: 10.3389/fmicb.2020.01378
Pan, R., Bai, X. L., Chen, J. W., Zhang, H. W., and Wang, H. (2019). Exploring structural diversity of microbe secondary metabolites using OSMAC strategy: a literature review. Front. Microbiol. 10:294. doi: 10.3389/fmicb.2019.00294
Reddy, T. R., Reddy, D. N., Reddy, B. K., Kasturaiah, C., and Yadagiri, K. (2018). An innovative approach for the synthesis of 7-hydroxyquinolin-2(1H)-one: a key intermediate of brexpiprazole. Asian J. Chem. 30, 834–836. doi: 10.14233/ajchem.2018.21092
Reen, F. J., Romano, S., Dobson, A. D., and O'Gara, F. (2015). The sound of silence: activating silent biosynthetic gene clusters in marine microorganisms. Mar. Drugs 13, 4754–4783. doi: 10.3390/md13084754
Romano, S., Jackson, S. A., Patry, S., and Dobson, A. D. W. (2018). Extending the "one strain many compounds" (OSMAC) principle to marine microorganisms. Mar. Drugs 16:244. doi: 10.3390/md16070244
Simonetti, S. O., Larghi, E. L., and Kaufman, T. S. (2016). Correction: a convenient approach to an advanced intermediate toward the naturally occurring, bioactive 6-substituted 5-hydroxy-4-aryl-1H-quinolin-2-ones. Org. Biomol. Chem. 14:3333. doi: 10.1039/c6ob90030e
Tangella, Y., Manasa, K. L., Krishna, N. H., Sridhar, B., Kamal, A., and Nagendra, B. (2018). Regioselective ring expansion of isatins with in situ generated α-aryldiazomethanes: direct access to viridicatin alkaloids. Org. Lett. 20, 3639–3642. doi: 10.1021/acs.orglett.8b01417
Uchida, R., Imasato, R., Yamaguchi, Y., Masuma, R., Shiomi, K., Tomoda, H., et al. (2006). Yaequinolones, new insecticidal antibiotics produced by Penicillium sp. FKI-2140. I. Taxonomy, fermentation, isolation and biological activity. J. Antibiot. (Tokyo) 59, 646–651. doi: 10.1038/ja.2006.86
Wang, K. B., Di, Y. T., Bao, Y., Yuan, C. M., Chen, G., Li, D. H., et al. (2014). Peganumine a, a β-carboline dimer with a new octacyclic scaffold from Peganum harmala. Org. Lett. 16, 4028–4031. doi: 10.1021/ol501856v
Wang, L., Zhang, X. Y., Zhang, K. J., Zhang, X. M., Zhu, T. J., Che, Q., et al. (2020). Overexpression of Global regulator Pbrlae a leads to the discovery of new polyketide in fungus Penicillium Brocae HDN-12-143. Front. Chem. 8:270. doi: 10.3389/fchem.2020.00270
Wen, B., Yuan, X., Zhang, W. D., Chen, B. Y., Fu, K. L., Li, B., et al. (2014). Chemical constituents from the aerial parts of Psammosilene tunicoides. Phytochem. Lett. 9, 59–66. doi: 10.1016/j.phytol.2014.04.006
Zhang, X., Hindra,, and Elliot, M. A. (2019). Unlocking the trove of metabolic treasures: activating silent biosynthetic gene clusters in bacteria and fungi. Curr. Opin. Microbiol. 51, 9–15. doi: 10.1016/j.mib.2019.03.003
Zhang, J. J., Tang, X. Y., and Moore, B. S. (2019). Genetic platforms for heterologous expression of microbial natural products. Nat. Prod. Rep. 36, 1313–1332. doi: 10.1039/c9np00025a
Keywords: alkaloidal secondary metabolites, antimicrobial activity, β-carboline, OSMAC strategy, Penicillium raistrichii , 2-quinolinone
Citation: Zhong M, Kang H, Liu W, Ma L and Liu D (2023) Alkaloid diversity expansion of a talent fungus Penicillium raistrichii through OSMAC-based cultivation. Front. Microbiol. 14:1279140. doi: 10.3389/fmicb.2023.1279140
Edited by:
Yongbo Xue, Sun Yat-sen University, ChinaReviewed by:
Gui-Hua Tang, Sun Yat-sen University, ChinaXi-Tao Yan, Northwest A&F University, China
Said Hassan, Bacha Khan University, Pakistan
Copyright © 2023 Zhong, Kang, Liu, Ma and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liying Ma, NjAxNzYzMDkwQHFxLmNvbQ==; Desheng Liu, ZGVzaGVuZ19saXVAc2luYS5jb20=
 Meijia Zhong
Meijia Zhong Desheng Liu
Desheng Liu