
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 06 December 2023
Sec. Microbiotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1275553
This article is part of the Research TopicPharmaceutically Active Micropollutants – How Serious is the Problem and is there a Microbial Way Out?View all 6 articles
 Elena Tyumina1,2
Elena Tyumina1,2 Grigory Bazhutin1,2
Grigory Bazhutin1,2 Nadezhda Kostrikina3
Nadezhda Kostrikina3 Vladimir Sorokin3
Vladimir Sorokin3 Andrey Mulyukin3
Andrey Mulyukin3 Irina Ivshina1,2*
Irina Ivshina1,2*Introduction: The increasing use of non-steroidal anti-inflammatory drugs (NSAIDs) has raised concerns regarding their environmental impact. To address this, understanding the effects of NSAIDs on bacteria is crucial for bioremediation efforts in pharmaceutical-contaminated environments. The primary challenge in breaking down persistent compounds lies not in the biochemical pathways but in capacity of bacteria to surmount stressors.
Methods: In this study, we examined the biodegradative activity, morphological and physiological changes, and ultrastructural adaptations of Rhodococcus cerastii strain IEGM 1243 when exposed to ibuprofen, diclofenac, and their mixture.
Results and Discussion: Our findings revealed that R. cerastii IEGM 1243 exhibited moderate biodegradative activity towards the tested NSAIDs. Cellular respiration assay showed higher metabolic activity in the presence of NSAIDs, indicating their influence on bacterial metabolism. Furthermore, catalase activity in R. cerastii IEGM 1243 exposed to NSAIDs showed an initial decrease followed by fluctuations, with the most significant changes observed in the presence of DCF and the NSAID mixture, likely influenced by bacterial growth phases, active NSAID degradation, and the formation of multicellular aggregates, suggesting potential intercellular synergy and task distribution within the bacterial community. Morphometric analysis demonstrated alterations in size, shape, and surface roughness of cells exposed to NSAIDs, with a decrease in surface area and volume, and an increase in surface area-to-volume ratio (SA/V). Moreover, for the first time, transmission electron microscopy confirmed the presence of lipid inclusions, polyphosphates, and intracellular membrane-like structures in the ibuprofen-treated cells.
Conclusion: These results provide valuable insights into the adaptive responses of R. cerastii IEGM 1243 to NSAIDs, shedding light on the possible interaction between bacteria and pharmaceutical compounds in the environment.
Chemical pollution of the environment, alongside climate change and catastrophic biodiversity loss, is recognized as a major global environmental problem demanding urgent attention and resolution (Wang et al., 2021; Brack et al., 2022). Of particular concern is the continuous introduction of active pharmaceutical ingredients (APIs) into ecosystems. Due to their biological activity and stable chemical structure, APIs have adverse effects on non-target organisms and pose a significant threat to the delicate balance of aquatic and terrestrial environments (Mulkiewicz et al., 2021; Wilkinson et al., 2022).
Among the various APIs detected in environmental samples, nonsteroidal anti-inflammatory drugs (NSAIDs) have garnered considerable attention owing to their widespread use and persistence (Richards et al., 2011; Domaradzka et al., 2015; Izadi et al., 2020; Mejía-García et al., 2020; Parolini, 2020; Herzig et al., 2021; Mulkiewicz et al., 2021; Petrie and Camacho-Muñoz, 2021; Rastogi et al., 2021; Wojcieszyńska et al., 2022). Specifically, diclofenac (DCF) and ibuprofen (IBP), which consistently rank among the most frequently detected pharmaceuticals in environmental matrices, have drawn considerable interest. The incomplete decomposition of DCF and IBP in the human body leads to their migration through sewage and wastewater treatment plants to surface water (Lonappan et al., 2016; Chopra and Kumar, 2020). The improper disposal of unused or expired DCF and IBP significantly contributes to their environmental contamination, as these medications are often flushed down toilets or disposed of in landfills, ultimately entering wastewater treatment plants or leaching into groundwater (Sadutto et al., 2021). Additionally, the manufacturing process of DCF and IBP can release these compounds into the environment, further contributing to their presence in ecosystems (Bibi et al., 2023).
The main environmental hazard of these NSAIDs lies in their biological activity, even at low (μg/L) concentrations. The toxic effects of DCF and IBP on birds, aquatic vertebrates and invertebrates, microalgae, and bacterial consortia have been well-documented (Cleuvers, 2004; Swan et al., 2006; Ericson et al., 2010; Parolini et al., 2011; De Felice et al., 2012; Memmert et al., 2013; González-González et al., 2014; Gamarra et al., 2015; Davids et al., 2017; Mezzelani et al., 2018; Aguilar-Romero et al., 2020; Chopra and Kumar, 2020; Parolini, 2020; Jan-Roblero and Cruz-Maya, 2023). Exposure to these NSAIDs can disrupt reproductive processes in living organisms, impair growth, and even cause mortality (Ortiz de García et al., 2014; Lonappan et al., 2016). DCF and IBP have the potential to bioaccumulate in the food chain, meaning they can concentrate in the tissues of organisms at higher levels than in the surrounding environment (Xie et al., 2015; Mezzelani et al., 2018). This accumulation can lead to adverse effects in higher trophic levels, including fish-eating birds and mammals. Moreover, the toxicity of DCF and IBP can have cascading effects on aquatic ecosystems, altering the balance of species and disrupting crucial ecosystem functions (Świacka et al., 2021).
In addition to addressing the environmental impact of these NSAIDs, extensive research has elucidated the microbial biodegradation pathways of NSAIDs, as comprehensively reviewed by Guzik and Wojcieszyńska (2019). However, despite significant efforts to examine the effects of NSAIDs on various organisms, our understanding of their specific impacts on microorganisms, particularly with respect to response mechanisms and adaptation patterns, remains limited (Jiang et al., 2017; Ivshina et al., 2019; Tyumina et al., 2019, 2020; Żur et al., 2020; Ivshina et al., 2021b; Żur et al., 2021).
Rhodococci are intriguing members of microbial communities, renowned for their remarkable survival strategies and exceptional ability to metabolize xenobiotic compounds, including pharmaceuticals (Yam et al., 2010; de Carvalho, 2012; Cheremnykh et al., 2018; Kuyukina and Ivshina, 2019; Rodrigues and de Carvalho, 2019; Pátek et al., 2021; Ivshina et al., 2021a, 2022c). These survival mechanisms encompass cell aggregation, production of extracellular polymeric substances, changes in lipid content, reduction of reactive oxygen species, and accumulation of intracellular storage compounds (Pátek et al., 2021). Despite their potential as valuable bioremediation agents, our understanding of how rhodococci respond to pharmaceuticals, particularly NSAIDs, remains scarce.
In our previous studies, we provided evidence of the biodegradation potential of rhodococci, demonstrating their ability to degrade NSAIDs such as DCF, paracetamol, IBP, and ketoprofen (Ivshina et al., 2006, 2019, 2021b; Bazhutin et al., 2022). Building on these foundational investigations, our current research aims to explore the individual and combined effects of IBP and DCF on Rhodococcus cerastii strain IEGM 1243 (GenBank # JAJNDD010000001-JAJNDD010000295; http://www.iegmcol.ru/strains/rhodoc/cerastii/r_cerastii1243.html accessed on 31 May 2023). Understanding the response mechanisms and adaptive behavior of rhodococci to pharmaceutical exposure is vital for assessing the potential risks posed by the widespread occurrence of NSAIDs and for developing effective strategies to mitigate their environmental impact.
In light of the aforementioned gaps in knowledge, this investigation aims to address the following research questions: (1) How do NSAIDs, specifically IBP and DCF, affect the biodegradative activity of R. cerastii strain IEGM 1243? (2) What are the morphological and physiological changes in R. cerastii IEGM 1243 cells when exposed to IBP, DCF, and their mixture? (3) Are there ultrastructural adaptations in R. cerastii IEGM 1243 cells in response to NSAID exposure?
This investigation contributes to the expanding body of knowledge concerning the ecotoxicity of pharmaceuticals and the intricate interactions between microorganisms and environmental contaminants. The findings of this study will provide important information on the effect of NSAIDs on rhodococci. Understanding how biodegraders adapt can help to build more efficient biocatalysts, allowing for more informed decision-making and encouraging sustainable management of pharmaceutical pollution in ecosystems.
R. cerastii strain IEGM 1243 from the Regional Specialized Collection of Alkanotrophic Microorganisms (IEGM; WFCC-WDCM 768; UNU/CKP 73559/480868; www.iegmcol.ru accessed on 31 May 2023) was used in this study. The strain IEGM 1243 was isolated from soil, lake shore Kumnylor, Tyumen region, Russia. In preliminary studies it showed IBP-degrading (100 mg/L) activity in the presence of 0.1% n-hexadecane (data not shown). Moreover, IEGM 1243 uses n-hexadecane as a sole carbon source and is resistant to Mo6+ (5.0 mM). The experiments were conducted in 250 mL Erlenmeyer flasks containing 100 mL of “RS” (Rhodococcus Surfactant) mineral salt medium. The composition of the medium per liter included: KNO3 1.0 g, K2HPO4 1.0 g, KH2PO4 1.0 g, NaCl 1.0 g, MgSO4 × 7H2O 0.2 g, CaCl2 × 2H2O 0.02 g, FeCl3 0.001 g, and 0.1% (v/v) trace element solution (1.5 g/L FeCl3 × 7H2O, 0.1 g/L H3BO3, 0.01 g/L ZnSO4 × 7H2O, 0.05 g/L Co (NO3)2 × 6H2O, 0.005 g/L CuSO4 × 5H2O, and 0.005 g/L MnCl2 × 4H2O). IBP (C13H17O2Na; CAS: 31121-93-4; (RS)-2-(4-(2-methylpropyl)phenyl)propanoic acid) and DCF (C14H10Cl2NNaO2, CAS: 15307–79-6, 2-(2-(2,6-dichlorophenylamino)phenyl)acetic acid) were used as sodium salts (Sigma-Aldrich, St. Louis, MO, United States; Glentham Life Sciences, Corsham, United Kingdom) and added to the mineral salt medium, each at a final concentration of 50 mg/L. Stock solutions of IBP and DCF (50 mg/mL) were prepared individually using water and stored at 4°C before use. We opted for this high concentration of NSAIDs due to the potential application of Rhodococcus-based biocatalysts in local post-treatment technologies during drug manufacturing, where influents contain elevated pharmaceutical levels. Moreover, previous research on biodegradation and the diverse effects of pharmaceuticals on bacteria supports the use of higher dosages (Alobaidi et al., 2021; Żur et al., 2021; Mohamed et al., 2023; Suleiman et al., 2023). D-glucose was employed as an additional carbon and energy source at a concentration of 0.5 g/L. Cells of R. cerastii IEGM 1243 grown for 3 days in LB broth (Himedia Laboratories Pvt. Limited, Maharashtra, India) and washed twice with phosphate buffer (pH 7.0) were added to the medium to a concentration of 0.96 × 107 cells/mL. The experiments were conducted at 160 rpm and 28°C.
The following treatments were used in the experiments: (1) mineral salt medium + R. cerastii IEGM 1243 + 0.5 g/L glucose (biotic control), (2) mineral salt medium + R. cerastii IEGM 1243 + 0.5 g/L glucose + 50 mg/L IBP (positive control 1), (3) mineral salt medium + R. cerastii IEGM 1243 + 0.5 g/L glucose +50 mg/L DCF (positive control 2), (4) mineral salt medium + R. cerastii IEGM 1243 + 0.5 g/L glucose + NSAID mixture (50 mg/L IBP + 50 mg/L DCF) (positive control 3), (5) mineral salt medium +0.5 g/L glucose + 50 mg/L NSAID (abiotic controls), (6) mineral salt medium + autoclaved R. cerastii IEGM 1243 + 0.5 g/L glucose + 50 mg/L NSAID (sorption controls). The abiotic controls were necessary to elucidate the abiotic degradation of the pharmaceuticals, while the sorption controls were essential for assessing the removal of the pharmaceuticals by sorbing them onto bacterial cells. The biotic (negative) control was essential to differentiate the effects of NSAIDs on bacterial cells from the natural variations in their life cycle.
To determine the viability of cells, the bacterial suspension was stained using a LIVE/DEAD® BacLight™ Bacterial Viability Kit following the manufacturer’s protocol (Invitrogen, Waltham, MA, United States). The staining process allowed for differentiation between live and dead cells. Visualization of the stained cells was carried out using an Axio Imager M2 microscope (Carl Zeiss Microscopy GmbH, Jena, Germany) in fluorescence mode (Luchnikova et al., 2022). Images were captured by an Axoicam 506 Color camera (Carl Zeiss Microscopy GmbH, Jena, Germany) and Zen Blue 3.1 software (Carl Zeiss Microscopy GmbH, Jena, Germany).
To identify intracellular lipid inclusions, the rhodococci were stained with Nile Red (Nanjing Dulai Biotechnology Co., Nanjing, China) (Spiekermann et al., 1999) with slight modifications (Luchnikova et al., 2022). Briefly, the cell suspension was centrifuged at 12,000 rpm for 5 min to obtain a cell pellet. The cell pellet was then resuspended in 1 mL of distilled water and mixed with 40 μL of a 0.08% solution of Nile Red in dimethyl sulfoxide. This suspension was incubated at 28°C with shaking at 160 rpm for 40 min. After incubation, cells were harvested by centrifugation (12,000 rpm for 5 min), resuspended in 1 mL of distilled water and visualized using the Axio Imager M2 microscope (Carl Zeiss Microscopy GmbH, Jena, Germany). Two different spectral settings were utilized: yellow-gold fluorescence with a 450–500 nm band pass exciter filter and red fluorescence with a 515–560 nm band pass exciter filter.
The influence of NSAIDs on the morphology and surface topography of cells was investigated using a combined scanning system, comprising an atomic force microscope (AFM) MFP-3D-BIO™ (Asylum Research Inc., Santa Barbara, CA, United States) and a confocal laser scanning microscope (CLSM) Olympus Fluo View 1000 (Olympus Corporation, Tokyo, Japan). Sample preparation and scanning procedures followed the methods outlined in previous studies (Kuyukina et al., 2014; Ivshina et al., 2019). Image processing and analysis, including the determination of root mean square roughness of the cell surface, cell length, and cell width, were performed using Igor Pro 6.22A software (WaveMetrics, Portland, OR, United States). Furthermore, cell volume and surface area were calculated using the formulas provided in a study (Neumann et al., 2005).
The IEGM 1243 cells grown for 72 h on solid media, including nutrient agar (NA), NA + 50 mg/L IBP, and mineral salt agar (MSA) + 50 mg/L IBP, were harvested, fixed in a solution containing 2.5% glutaraldehyde (w/v) in 0.1 M sodium cacodylate buffer (pH 7.2) for 2.5 h. Subsequently, the fixed cells underwent post-fixation using a 1% (w/v) osmium tetroxide solution in the same buffer. To prepare the samples for microscopy, the fixed cells were dehydrated by a series of ethanol solutions, including absolute ethanol saturated with uranyl acetate, followed by embedding in araldite (Ivshina et al., 2022a). Thin sections were generated using an 8800 Ultrotome III (LKB-Produkter, Stockholm, Sweden), and these sections were stained with lead citrate. The resulting ultrathin sections were examined using a JEM-1400 electron microscope (Japanese Electron Optics Laboratory, Tokyo, Japan).
The harvested IEGM 1243 cell suspensions (as described in section 2.2.4) were directly placed onto copper grids coated with formvar and carbon reinforcement, without the addition of any fixatives. These grids were then allowed to air-dry. For the analysis, transmission electron microscopy combined with energy-dispersive X-ray analysis (TEM-EDX) was performed using the JEM-1400 microscope (Japanese Electron Optics Laboratory, Tokyo, Japan) equipped with an EDXA system (Inca Energy-350, Oxford Instruments, Abingdon, United Kingdom). The microscope operated at an accelerating voltage of 80 keV with a tilt angle of 15° (Ivshina et al., 2022a). The acquisition of EDX spectra and elemental maps was carried out using AZtec software (Oxford Instruments, Abingdon, United Kingdom). In this analysis, only the main Kα peaks of carbon (C), oxygen (O), nitrogen (N), phosphorous (P), potassium (K), sodium (Na), copper (Cu), calcium (Ca), magnesium (Mg), sulfur (S), chlorine (Cl), silicon (Si), and iron (Fe) were taken into consideration.
The catalase activity of R. cerastii IEGM 1243 was determined spectrophotometrically following the protocol described by Gogoleva et al. (2012). Bacterial cells cultured with glucose with and without NSAIDs were centrifuged at 3,000 rpm for 5 min, washed with phosphate buffer (pH 7.0), and resuspended in the same buffer to obtain an optical density (OD492) of 0.2. Next, a 0.00125 M hydrogen peroxide (H2O2) solution (1 mL) was added to 200 μL of the cell suspension, and the mixture was incubated for 10 min at room temperature. To stop the decomposition of H2O2 by catalase, 100 μL of 2 M HCl solution was added. Subsequently, a 0.025 M potassium iodide solution (1 mL) was added to the resulting mixture, gently mixed, and centrifuged at 3,000 rpm for 15 min. Distilled water was used in control samples instead of cell suspensions. Finally, the absorbance of the supernatant was measured at 492 nm using a Lambda EZ201 spectrophotometer (Perkin-Elmer, Waltham, MA, United States).
The catalase activity was calculated using the equation:
where Acat represents the catalase activity in μM/min × OD; ODv is the optical density of the supernatant in the experimental sample (in arbitrary units of optical density); ODc is the optical density of the supernatant in the control sample (in arbitrary units of optical density), T is the incubation time of bacteria in the presence of H2O2 (10 min); and ODb is the optical density of the bacterial suspension (OD492 0.2). The catalase activity was expressed as a percentage of the initial level.
Cell respiration activity was evaluated using a 10-channel Micro-Oxymax® respirometer (Columbus Instruments, Columbus, OH, United States) (Ivshina et al., 2019). The experiments were conducted in 300 mL Micro-Oxymax glass flasks containing 100 mL of mineral salt medium with constant agitation (160 rpm, 28 ± 2°C). The rate (μL/h) and accumulation (μL) of oxygen consumption were measured. Automated recording of respiratory activity was performed every 30 min over a period of 7 days.
The removal rates of IBP and DCF were determined using high-performance liquid chromatography (HPLC) on an LC Prominence chromatograph (Shimadzu Corporation, Kyoto, Japan) equipped with a reverse-phase C18 column, 25 cm × 4.6 mm, 5 μm (Supelco Inc., Bellefonte, PA, United States) and a diode array detector. Mobile phase consisted of a phosphate buffer solution (pH 5.0) and acetonitrile, mixed in a 60:40 ratio. Elution was performed isocratically at a flow rate of 0.5 mL/min, and detection was carried out at a wavelength of 254 nm. The injection volume was 20 μL, and the column thermostat temperature was set at 40°C. Under these conditions, the retention times for IBP and DCF were determined to be 9.28 ± 0.03 and 18.70 ± 0.02 min, respectively. Prior to analysis, samples were prepared by centrifuging for 5 min at 10,000 rpm. The resulting supernatants were then filtered using a 0.20 μm nylon membrane filter (Nantong FilterBio Membrane Co., Ltd., Nantong, China). The concentrations of DCF and IBP were calculated by comparing the areas under the peaks with those of the standard solutions.
For cell analysis and quantification, ImageJ software (US National Institutes of Health, Bethesda, Maryland, United States) was employed. A detailed protocol for image processing can be found here (https://www.allevi3d.com/livedead-assay-quantification-fiji/ accessed on 31 May 2023).
Statistical analyses were performed to determine the differences in the experimental data using one-way ANOVA followed by two-sided Dunnett’s t-test. The software used for conducting the statistical tests was SPSS 23.0 (IBM, Armonk, New York, United States). The significance level for the results was set at p < 0.05. Each experiment was performed in triplicate to ensure accuracy and reproducibility of the results. For presenting the data from repeated experiments, the values were expressed as mean ± standard deviation.
According to our data, R. cerastii IEGM 1243 exhibited moderate biodegradative activity towards the tested pharmaceutical compounds (Figure 1). When considering individual NSAIDs, the residual content of IBP after a 7 days period was 68.4%, while that of DCF was 75.9%. Interestingly, when using the combination of NSAIDs, the degradation of IBP was higher, with a residual content of 40.4% after 7 days. However, the degradation of DCF in this case was negligible, with a residual content of 89.7%. No losses of substances were observed in abiotic and sorption controls, indicating the biocatalytic nature of the degradation process.
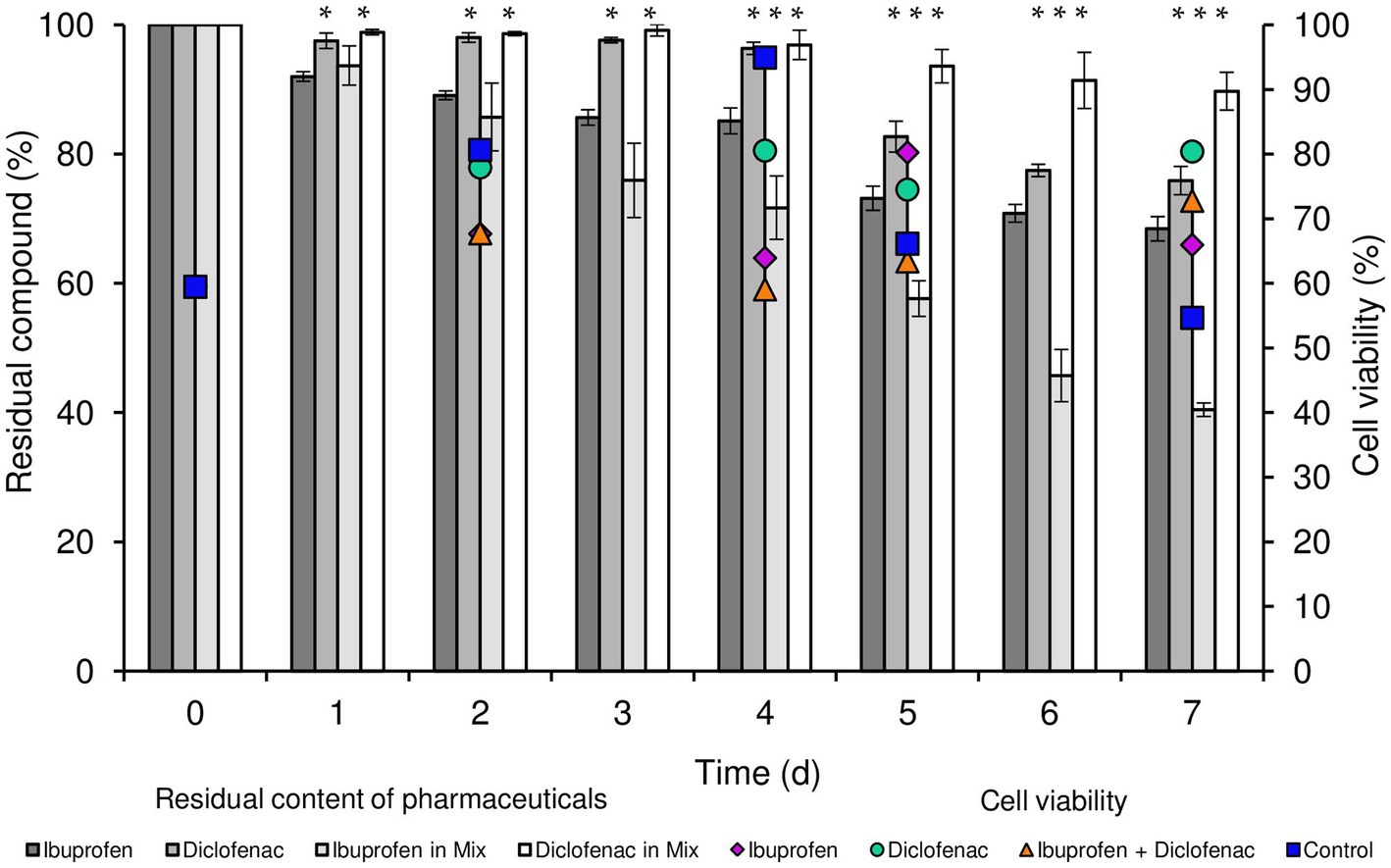
Figure 1. Residual content of NSAIDs and cell viability of R. cerastii IEGM 1243 during the biodegradation process. Cells were cultivated in mineral salt medium supplemented with 0.5 g/L glucose (control) and 50 mg/L IBP, 50 mg/L DCF or their mixture (50 mg/L IBP + 50 mg/L DCF). Asterisks indicate significant differences between control and treatments (p < 0.05).
In our previous studies, we confirmed that R. cerastii strain IEGM 1278 was capable of completely cometabolizing 100 mg/L IBP within 6 days in the presence of n-hexadecane (Ivshina et al., 2021b). Other research showed that IBP could be efficiently degraded at high concentrations (200–1,000 mg/L) by both Gram-negative strains (Sphingomonas sp. Ibu-2, Variovorax sp. Ibu-1) and Gram-positive strains [Bacillus thuringiensis B1(2015b), Citrobacter freundii PYI-2, C. portucalensis YPI-2, Nocardia sp. NRRL 5646, Patulibacter sp. I11] (Jan-Roblero and Cruz-Maya, 2023). On the other hand, DCF is more recalcitrant to biodegradation. Previously, we isolated R. ruber strain IEGM 346, which was able to degrade only 50% of 50 mg/L DCF within 60 days (Ivshina et al., 2019). In other reports, only Gram-negative Klebsiella sp. KSC exhibited the capability to degrade high concentrations of DCF (70 mg/L) within three days, while other strains such as Pseudomonas moorei KB4, Labrys portucalensis F11, Raoultella sp. DD4 degraded much lower concentrations (ranging from 0.5 to 7 mg/L) over up to 28 days (Wojcieszyńska et al., 2023). The results obtained in our study indicate that further optimization of biodegradation conditions for IEGM 1243 is necessary, as our findings do not stand out in this regard.
However, it is important to note that the biodegradation pattern was not the primary focus of our research. We previously outlined the putative biodegradation pathways of DCF and IBP in Ivshina et al. (2019, 2021b). Additionally, Guzik and Wojcieszyńska’s (2019) thorough review covered the microbiological degradation pathways of NSAIDs. Instead, our study primarily concentrated on the physiological and morphological responses of rhodococci when exposed to NSAIDs, which are presented below.
Concurrently with the biodegradation experiments, we conducted quantitative assessments of total cell count and cell viability percentage (Figure 1; Supplementary Figures S1, S2). The initial cell count was determined to be 0.96 cells/mL × 107. Throughout the duration of the experiment, an increase in biomass was consistently observed across all experimental conditions, with the most significant growth recorded in both the control and in the presence of IBP. The highest cell count was attained when IBP was present, peaking at 3.32 cells/mL × 107 after 4 days of the experiment. In parallel, the viability of rhodococci in the control sample exhibited an upward trend until day 4, with viability levels rising from 59.5 to 94.9%, followed by a decline to 54.7% on day 7. In the presence of IBP, the maximum cell viability was reached on day 5 (80.2%), gradually decreasing to 65.9% on day 7. Notably, the overall cell count in the presence of DCF and the NSAID mixture on day 7 of the experiment amounted to 2.1 and 1.59 cells/mL × 107, respectively. Interestingly, these conditions exhibited the highest proportion of viable cells at the end of the experiment.
Analysis of cellular respiration in the presence of NSAIDs revealed the following patterns (Figure 2). Both in the biotic control and in the presence of IBP, the lag phase of IEGM 1243 cells was less than 24 h (Figure 2A). The maximum rate of oxygen consumption in the control was reached after 1.5 days of the experiment, amounting to 4753.7 μL/h. In contrast, in the presence of IBP, this parameter was recorded after 2.5 days of the experiment, with a significantly higher value of 13125.4 μL/h.
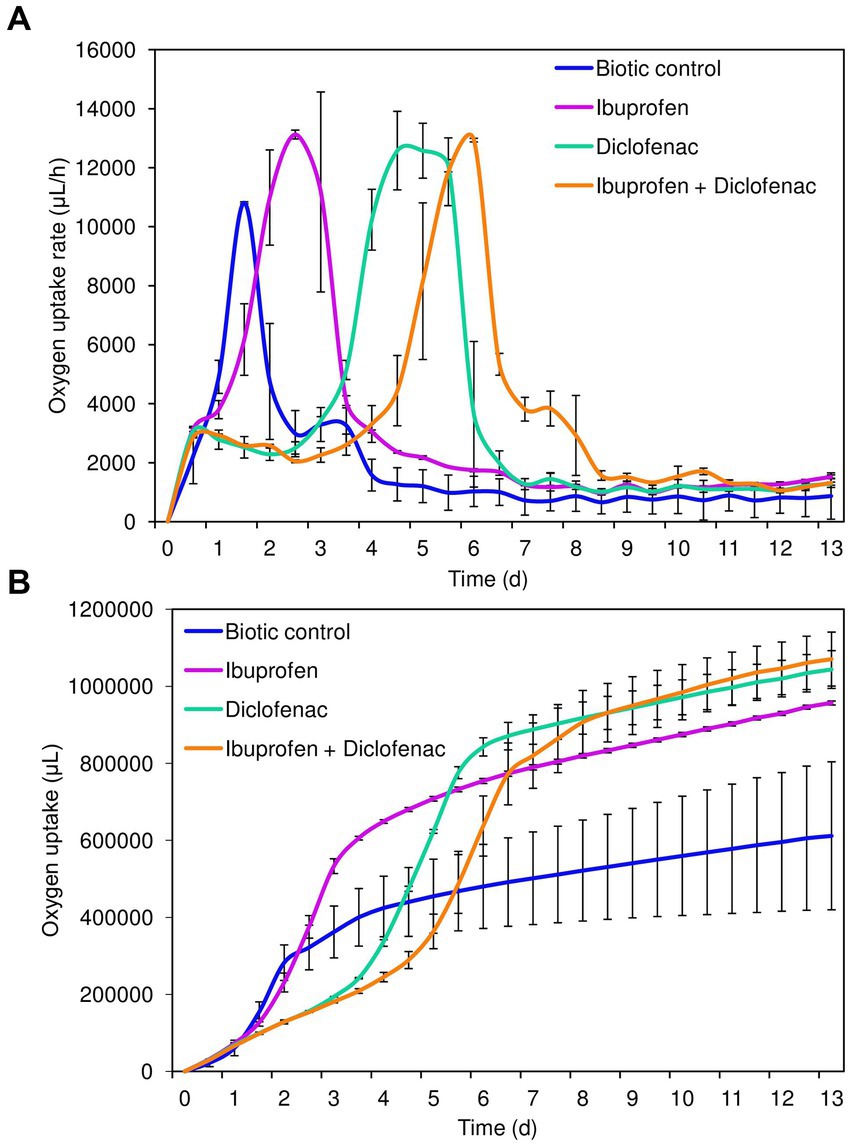
Figure 2. Oxygen uptake rate (A) and cumulative oxygen consumption (B) by R. cerastii IEGM 1243 cells exposed to NSAIDs. Cells were cultivated in mineral salt medium supplemented with 0.5 g/L glucose (control) and 50 mg/L IBP, 50 mg/L DCF or their mixture (50 mg/L IBP + 50 mg/L DCF).
The transition of rhodococci to the stationary growth phase (Figure 2B) correlated with a slowdown in the biodegradation of NSAIDs (Figure 1). DCF and the mixture exerted a suppressive effect on the metabolic activity of rhodococci for a period of 3–4 days. The maximum rate of oxygen consumption in the presence of DCF was 12580.9 μL/h after 4.5 days of the experiment. Under the combination of IBP and DCF, the peak oxygen consumption was observed on day 6, measuring 12936.4 μL/h.
Cumulative oxygen uptake analysis demonstrated that cells exhibited higher metabolic activity in the presence of NSAIDs (Figure 2B). For instance, on day 9, the total amount of oxygen consumed in the control was 611911.4 μL, whereas in the presence of IBP, DCF, and their mixture, these values were 956805.2 μL, 1,043,623 μL, and 1,070,803 μL, respectively. The increased amount of oxygen consumed in the presence of DCF and the NSAID mixture after 7 days of the experiment correlates with data on higher cell viability under these conditions (Figure 1; Supplementary Figure S2).
Catalase, as a vital antioxidant enzyme, plays a crucial role in the reduction of hydrogen peroxide (H2O2) to oxygen (O2) and water (H2O). This enzyme holds significant importance in bacterial stress response and the degradation of xenobiotics (Wang et al., 2000, 2020; Kaushal et al., 2018). Rhodococcus spp. are well-known for their high abundance of catalases (Yuan et al., 2021), which makes them robust and adaptable microorganisms. In the context of xenobiotic degradation, including the biodegradation of NSAIDs, the high levels of catalases in rhodococci provide significant benefits. The presence of NSAIDs can lead to the production of harmful reactive oxygen species in the bacteria. However, thanks to their abundant catalases, Rhodococcus spp. may efficiently neutralize these ROS, ensuring their metabolic activity and successful degradation of the xenobiotics.
The initial catalase activity of R. cerastii IEGM 1243 was 0.66 relative units corresponding to 100%. Within the first day, a decrease in catalase activity was observed in all tested variants. However, the most significant (p < 0.05) changes in catalase activity of rhodococci were observed in the presence of DCF and the NSAID mixture (Figure 3). The decrease in catalase activity under these conditions during the first 3 days can be attributed to the cells being in the lag phase of growth (refer to Figure 2A). Interestingly, on the second day, there was an increase in catalase activity in the presence of NSAIDs, while in the control group, the activity continued to decrease. The opposite pattern was observed on the third day. The increase in catalase activity in the samples with NSAIDs on day 4 can be explained by the bacteria entering the exponential growth phase, active degradation of NSAIDs (as seen in Figure 1), and possible accumulation of metabolites. The decrease in catalase activity in all samples on day 7 could be associated with the transition to the stationary growth phase, slowing down of cellular metabolism, and the formation of multicellular aggregates (Supplementary Figure S2) (Wood and Sørensen, 2001). Moreover, it is hypothesized that cell aggregation enhances intercellular synergy, leading to a reduced need for cells to secrete the necessary enzymes (D’Souza et al., 2023). By forming multicellular aggregates, bacterial cells can establish communication networks that facilitate the exchange of genetic material and metabolic by-products. This can lead to a more efficient distribution of tasks among the cells, allowing some individuals to focus on enzyme production while others specialize in different functions. Consequently, this division of labor within the aggregated community can reduce the overall demand for enzyme secretion by individual cells.
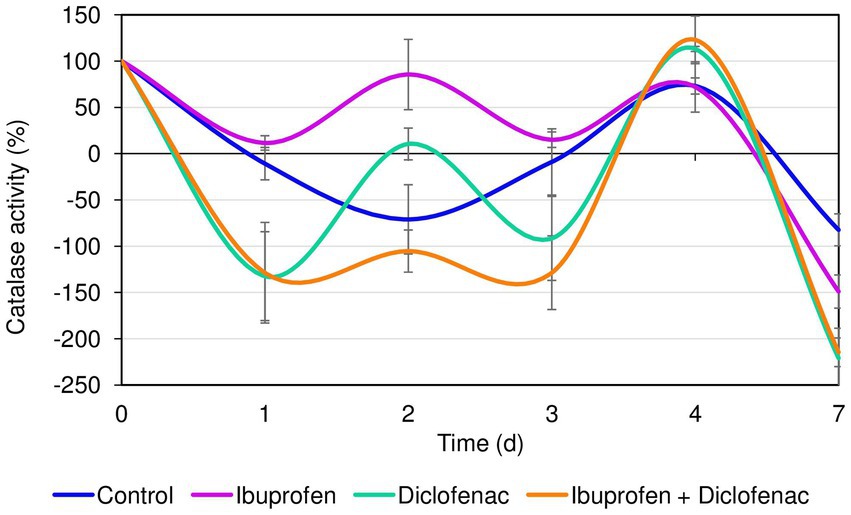
Figure 3. Catalase activity of R. cerastii IEGM 1243 in the presence of NSAIDs. Cells were cultivated in mineral salt medium supplemented with 0.5 g/L glucose (control) and 50 mg/L IBP, 50 mg/L DCF or their mixture (50 mg/L IBP + 50 mg/L DCF).
Studies on cell morphology and morphogenesis are of great importance for understanding cell growth, reproduction, and adaptation to the environment (Young, 2006; Levin and Angert, 2015; Ojkic et al., 2019; Shi et al., 2021; Govers et al., 2023). In this study, we analyzed the morphometric parameters of live cells of R. cerastii IEGM 1243 exposed to NSAIDs, and the results are presented in Figures 4–7, Table 1, and Supplementary Figures S3–S5. Initially, in their native state, the R. cerastii IEGM 1243 cells exhibited an elongated rod-shaped morphology (Figures 4A,B) with distinct peaks and valleys (Figure 4D), as well as a rough cell surface.
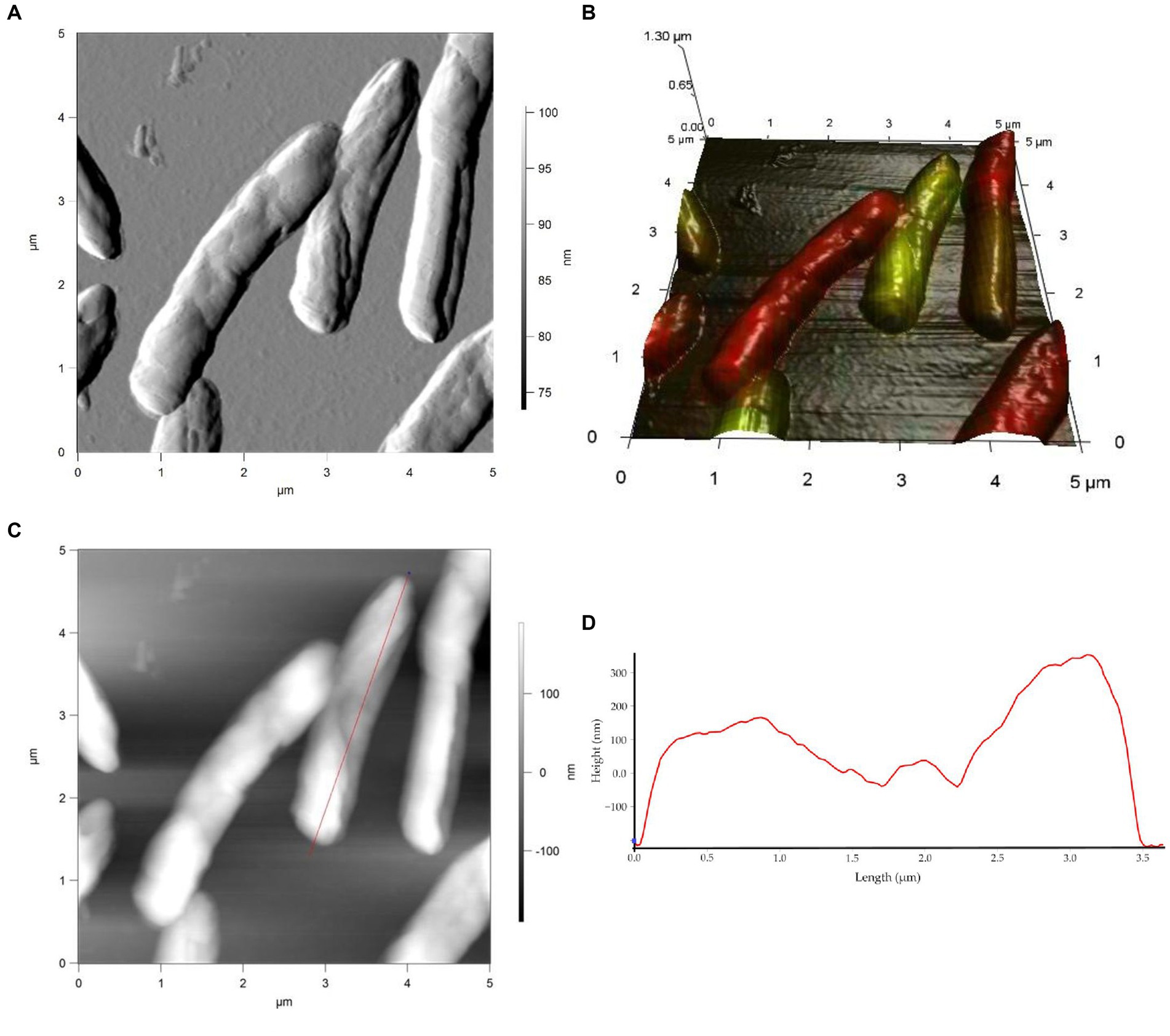
Figure 4. AFM (A), AFM-CLSM (B) images, height (C) and profile (D) of R. cerastii IEGM 1243 cells on day 0.
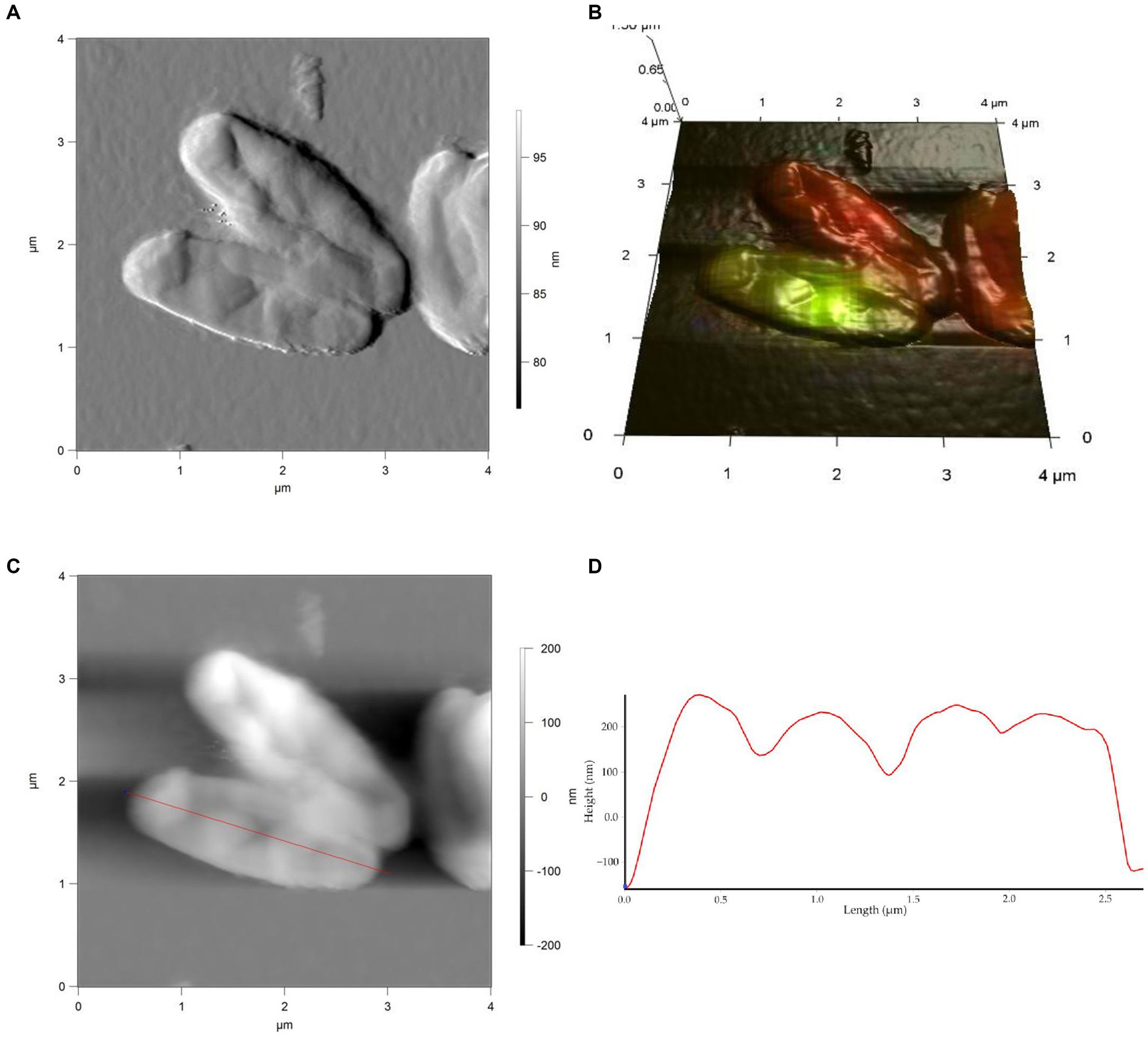
Figure 5. AFM (A), AFM-CLSM (B) images, height (C) and profile (D) of R. cerastii IEGM 1243 cells grown in mineral salt medium supplemented with 0.5 g/L glucose for 4 days.
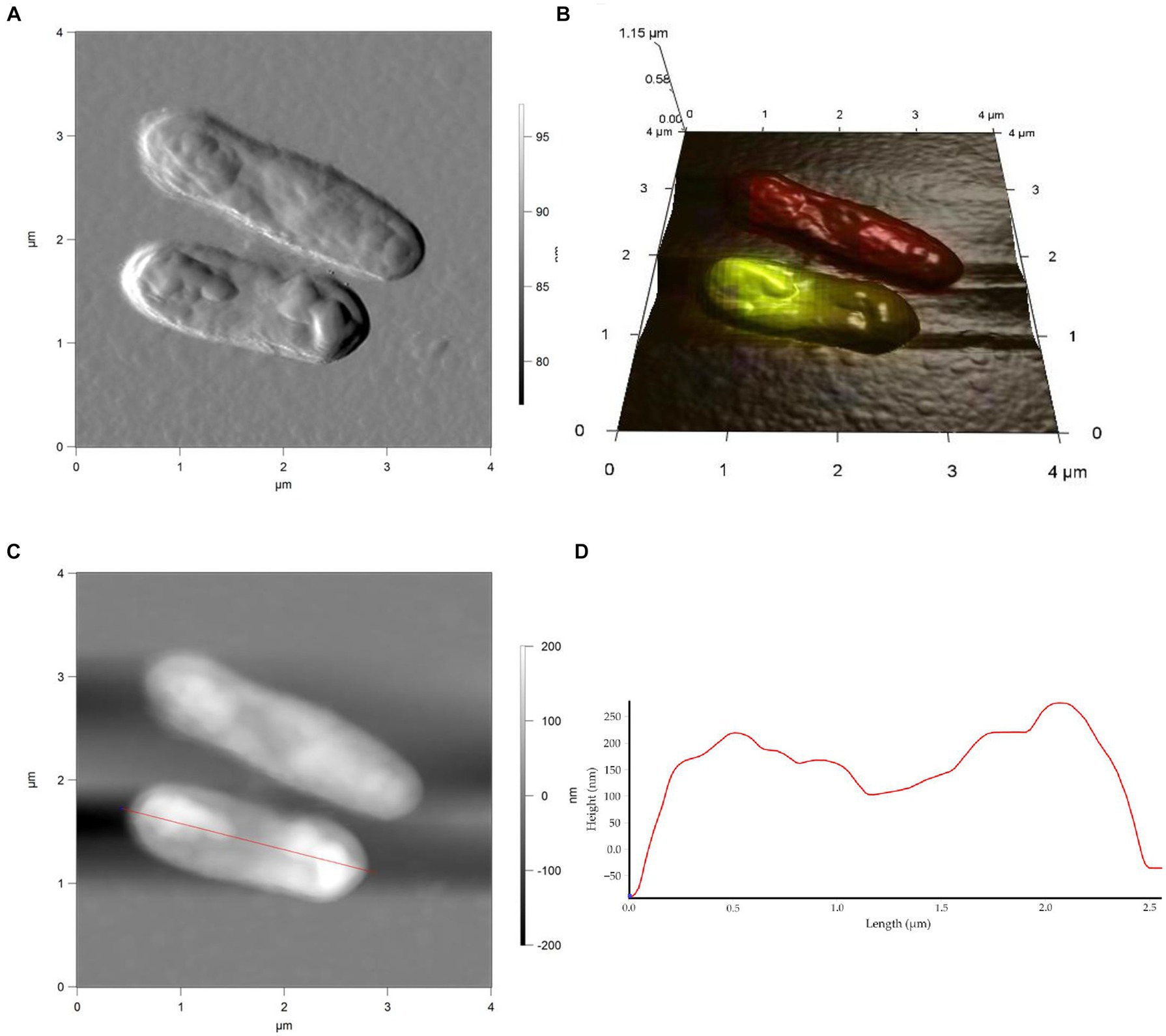
Figure 6. AFM (A), AFM-CLSM (B) images, height (C) and profile (D) of R. cerastii IEGM 1243 cells grown in mineral salt medium supplemented with 0.5 g/L glucose and 50 mg/L IBP for 4 days.

Figure 7. AFM of R. cerastii IEGM 1243 cells grown in mineral salt medium supplemented with 0.5 g/L glucose and 50 mg/L DCF (A) or 50 mg/L IBP + 50 mg/L DCF (B) for 4 days.
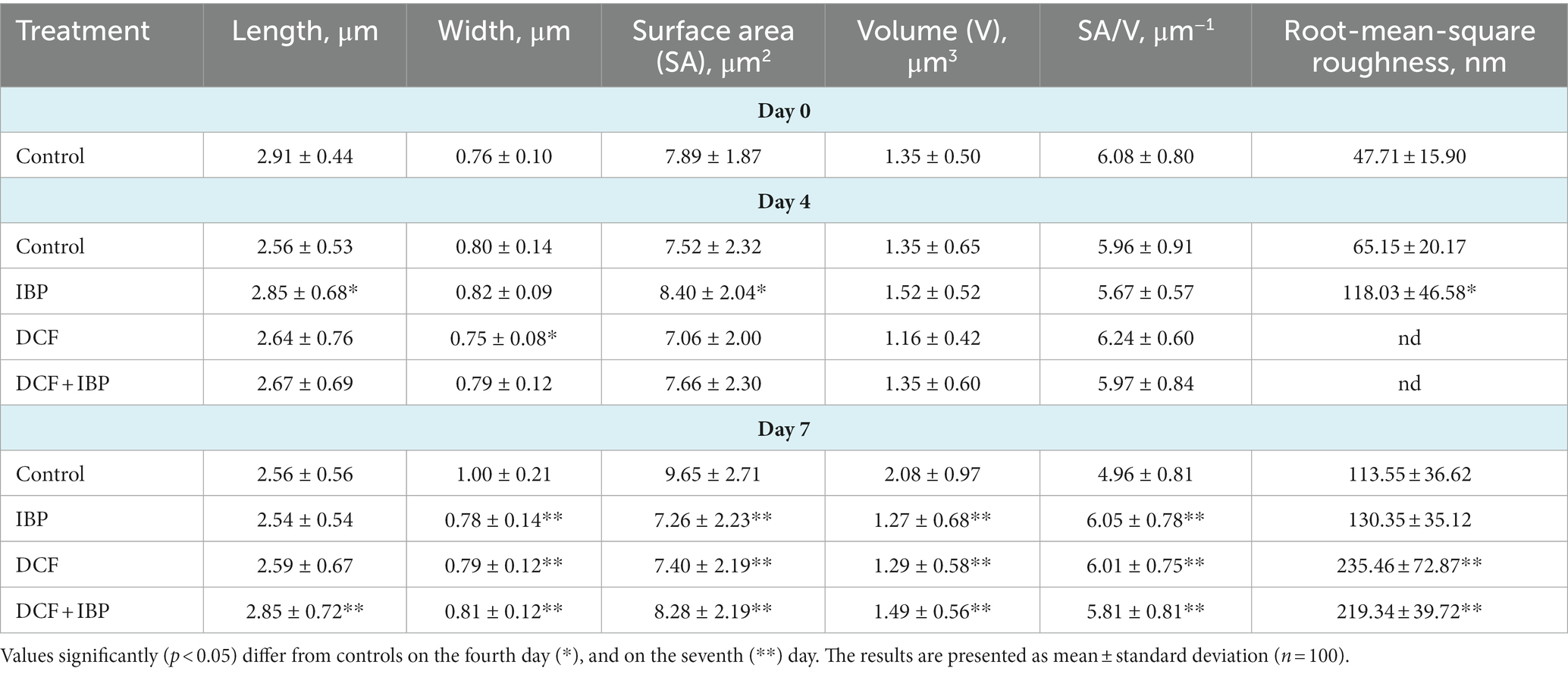
Table 1. Morphometric features of R. cerastii IEGM 1243 cells grown in mineral salt medium supplemented 0.5 g/L with glucose (control) and 50 mg/L NSAIDs.
Rod-shaped bacteria have the ability to readily adapt their shape in response to changes in the surrounding environment (Chang and Huang, 2014; Ojkic et al., 2019). In our study, we observed that the control cells underwent shape alterations on day 4, with a reduction in length and an increase in width (Figure 5 and Table 1). However, according to height measurements, no significant differences in cell sizes were observed between the NSAID-treated cells and the control (Figures 6, 7 and Table 1). Notably, when exposed to DCF and the NSAID mixture, cells produced a significant amount of extracellular polymeric substances, which hindered the assessment of cell profile and height (Figure 7).
On the seventh day, significant differences in morphometric parameters of cells in the presence of NSAIDs were observed, particularly a decrease in surface area, volume, and an increase in the surface area-to-volume ratio (SA/V) (Table 1). SA/V is an important physical parameter that regulates the influx and efflux of substances and metabolites (Ojkic et al., 2022). The cell size, shape, and SA/V are interconnected, and individual cells can change their size and shape to achieve an optimal SA/V (Harris and Theriot, 2018; Shi et al., 2021). Previously, we observed alterations in the SA/V ratio in rhodococci exposed to pharmaceuticals (Ivshina et al., 2019; Tyumina et al., 2019; Ivshina et al., 2021b, 2022b). For example, when exposed to toxic substrates such as DCF or naproxen, cells tend to reduce their SA/V to minimize the exposed cell surface for contact with the stressor (Ivshina et al., 2019, 2022b). Studies by other authors have shown that in the presence of ampicillin, Escherichia coli cells maintain an ellipsoidal shape and a low SA/V to reduce their metabolic activity, conserve energy, and prevent division (Uzoechi and Abu-Lail, 2019). The reduction in metabolic activity, in turn, leads to cell persistence and resistance (Ojkic et al., 2022). On the other hand, in the presence of less toxic compounds or under carbon source limitation, SA/V ratio may increase for more efficient contact between cells and a substrate (Ivshina et al., 2019, 2021b). In the short term, a cell is capable of regulating its shape by adjusting turgor pressure. However, over longer time scales, changes in cell size are predominantly determined by alterations in cell width (Oldewurtel et al., 2021). In our case, on day 7, the NSAID-treated cells reduced their width (p < 0.05), which ultimately resulted in smaller cell volume and larger SA/V. Possible reasons for the change in SA/V ratio include the availability of precursors for peptidoglycan biosynthesis and the presence of inhibitors of fatty acid biosynthesis (Harris and Theriot, 2018). Additionally, cell size is influenced by the levels of transport proteins, metabolic activity, and the accumulation of cell division protein (FtsZ) (Ojkic et al., 2019; Bertaux et al., 2020).
The effect of IBP on the ultrastructure of R. cerastii IEGM 1243 cells was investigated using transmission electron microscopy (TEM) analysis. TEM provides high-resolution imaging of cellular structures, allowing for detailed observations of cellular ultrastructure. Comparative analysis of ultrathin sections revealed that the morphological traits of rhodococci were similar in cultures developed under both control and IBP-containing conditions (Figure 8; Supplementary Figures S6–S8). As evident from the Figure 8E, strain IEGM 1243 exhibits an apical mode of cell elongation, resulting in the increased accumulation of new cell wall material at the cell poles (Donovan and Bramkamp, 2014). The cells exhibited characteristic features, including the presence of an outer capsular layer, which was visually discernible as protrusions. Furthermore, they displayed a stratified cell wall with an outer layer that appeared electron-dense, indicating the existence of a well-defined cell envelope. Additionally, the cytoplasmic membrane remained intact, indicating the overall structural integrity of the cells.
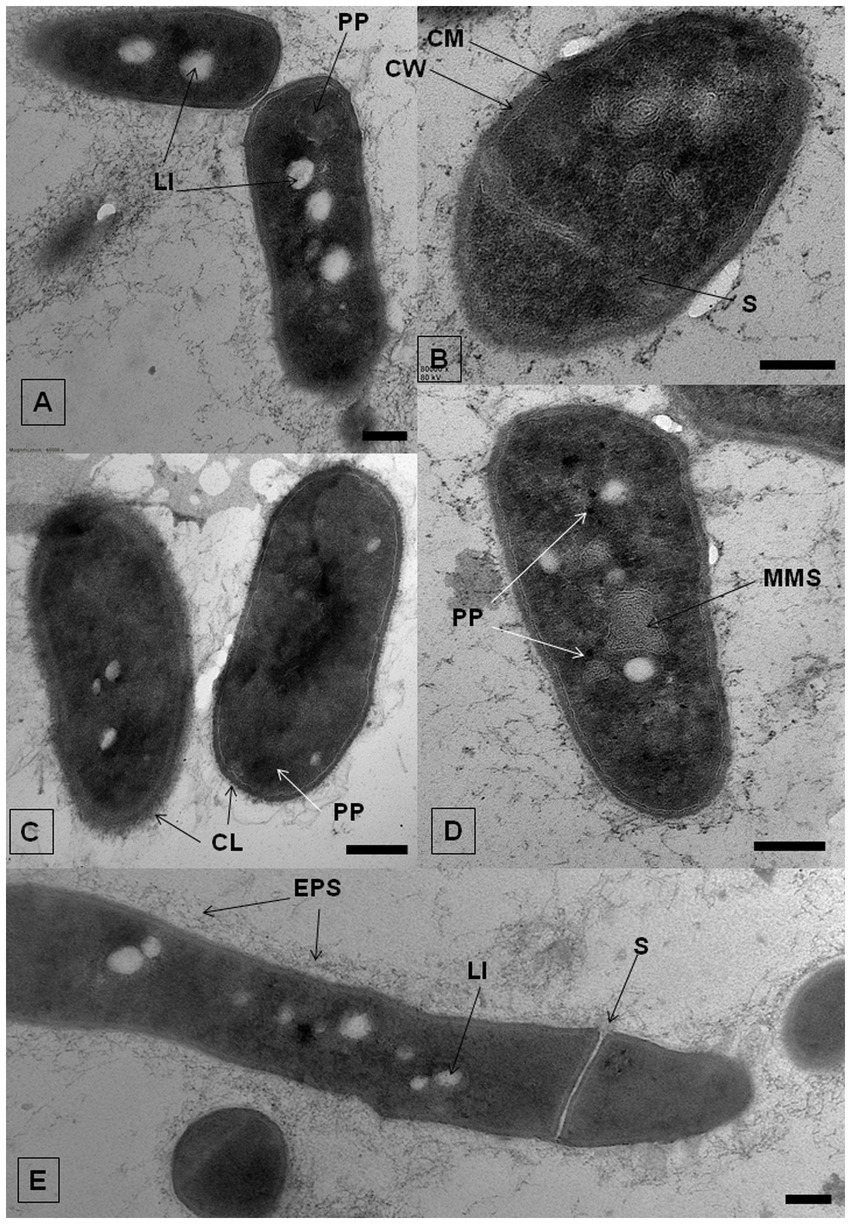
Figure 8. TEM-images of R. cerastii IEGM 1243 grown on nutrient agar (control, A,E) supplemented with 50 mg/L IBP (B,D) or on minimal salt agar supplemented with 50 mg/L IBP (C). Cells were cultivated for 3 days. CL, capsular layer; CM, cytoplasmatic membrane; CW, cell wall; EPS, extracellular polymeric substances; LI, lipid inclusions; MLS, membrane-like structures; PP, polyphosphate granules; S, septum. Scale bars correspond to 200 nm.
In both the control and IBP-treated cells, TEM revealed the presence of two distinct types of structures: electron-transparent bodies, likely lipid inclusions, and electron-dense bodies, believed to be polyphosphates. Intracellular lipid inclusions important energy storage compounds that fulfill carbon requirements and maintain redox homeostasis, enabling bacteria to survive for extended periods (Alvarez et al., 1996, 2000; Mallick et al., 2021). Furthermore, they play a crucial role in the pathogenesis of mycobacteria (Mallick et al., 2021). Rhodococcus spp. commonly utilize polyhydroxyalkanoates, triacylglycerols, and glycogen as intracellular carbon reserves (Hernández et al., 2008; Cappelletti et al., 2020).
Nile Red staining of the bacteria revealed the presence of a small amount of lipid inclusions, mainly located at the periphery of the cells, at the initial time point (Figure 9). By day 4 of the experiment, there was a tendency for the accumulation of lipid inclusions in all treatments, particularly evident in the control and in the presence of IBP. The number of inclusions reached up to 10 per cell, predominantly localized in the central part of the cell. In the case of DCF and the NSAID mixture, the quantity and size of lipids were smaller, and the inclusions were often located at the periphery of the cells. After 7 days of observation, the control cells maintained a significant amount of lipid inclusions; however, individual cells without intracellular lipids were also observed. When exposed to NSAIDs, cells primarily depleted their lipid reserves, with individual lipid granules located at the periphery of the cells (IBP and NSAID-mixture) or at the center of the cells (DCF). The red fluorescence around the cell perimeter could be associated with the lipids of the cell membranes (Presentato et al., 2018).
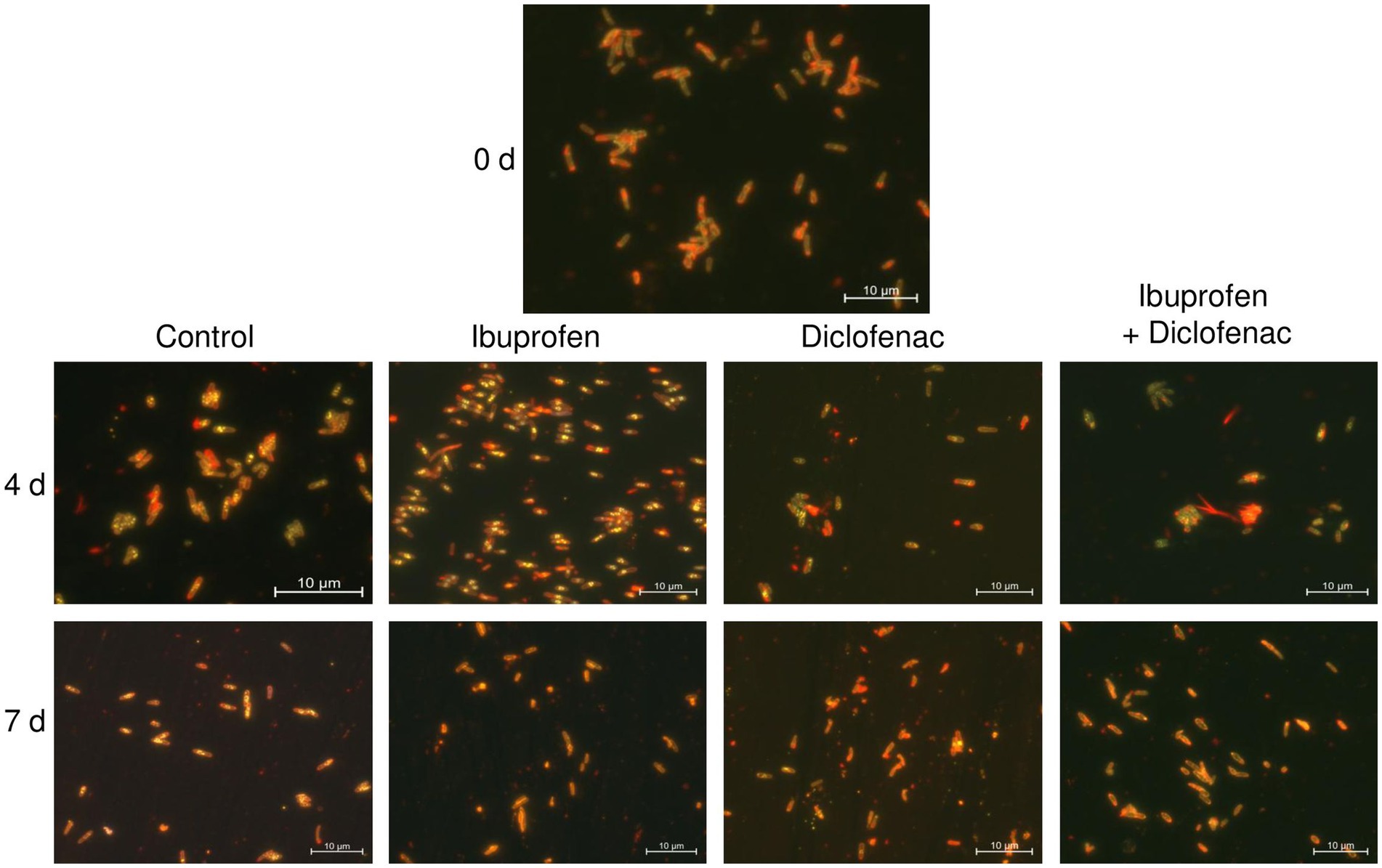
Figure 9. Fluorescent microscopy images of R. cerastii IEGM 1243. Cells were cultivated in mineral salt medium supplemented with 0.5 g/L glucose (control) and 50 mg/L IBP, 50 mg/L DCF or their mixture (50 mg/L IBP + 50 mg/L DCF). Yellow dots are lipid inclusions.
According to a previous study (Wältermann et al., 2005), the process of lipid-body synthesis involves the formation of small lipid droplets that remain attached to membrane-associated enzymes. Over time, these droplets aggregate and merge, leading to the formation of larger structures known as membrane-bound lipid prebodies. Eventually, these lipid prebodies are released into the cytoplasm, becoming mature entities within the cell. Our current study corroborates these findings (Supplementary Figure S9). For instance, at the beginning of the experiment, IEGM 1243 cells grown in the presence of glucose (control) exhibited small inclusions at the periphery, which likely represent small lipid droplets. As time progressed, these droplets transitioned into the cytoplasm, forming larger lipid prebodies in the early stages and eventually maturing into large lipid inclusions on days 2 and 3. The formation of lipid inclusions in the presence of IBP followed a similar scenario, while under the influence of DCF and the NSAID mixture, the maturation of lipid inclusions occurred on day 4.
When IBP was present, cells exhibited the formation of intracellular membrane-like structures (Figure 8D; Supplementary Figure S7). These structures appeared as loop-like formations and are believed to be primarily involved in the transportation of complex compounds and their subsequent degradation through the action of membrane-bound enzymes (Royes et al., 2020; Thi Mo et al., 2022). Similar structures have been observed in previous studies involving rhodococci grown in the presence of benzoate (Solyanikova et al., 2017), oleanolic acid (Luchnikova et al., 2022), as well as liquid and solid n-hexadecane (Thi Mo et al., 2022). This suggests a common mechanism or adaptive response in the formation of these membrane-like structures across different environmental conditions and compound exposures.
Polyphosphates were represented as compartments mainly located at the cell poles (Figures 8A,C). Similar observations of polyphosphates were made in cells of R. rhodochrous IEGM 1362 grown in a nutrient-rich medium and in the presence of (−)-isopulegol (Ivshina et al., 2022a), R. rhodochrous IEGM 757 in the presence of oleanolic acid (Luchnikova et al., 2022), and R. erythropolis N9T-4 in a basal medium without any additional carbon, nitrogen, sulfur, and energy sources (Yoshida et al., 2017). In the latter case, the authors referred to these compartments as oligobodies and confirmed their high phosphorus and potassium content using X-ray spectroscopy. Phosphorus-rich granules are composed of linear chains of polyphosphate and cations such as magnesium, potassium, and calcium (Keim et al., 2005). Polyphosphate inclusions serve as an internal phosphate reserve in cells and play an important role in the adaptive mechanisms of bacteria under suboptimal environmental conditions. In our case, particularly significant accumulations of phosphorus were observed in cells grown on mineral salt agar supplemented with IBP. TEM-EDX analysis showed distinct zones of increased phosphorus accumulation at the cell poles, along with significant amounts of potassium and magnesium (Figure 10; Supplementary Figures S10–S13). Furthermore, the elevated potassium content in the external environment in the presence of IBP may indicate its efflux from the cells due to the disruption of cell membrane permeability (Luchnikova et al., 2022). However, it is essential to note that further investigations are needed to fully understand the underlying mechanisms behind these observations.
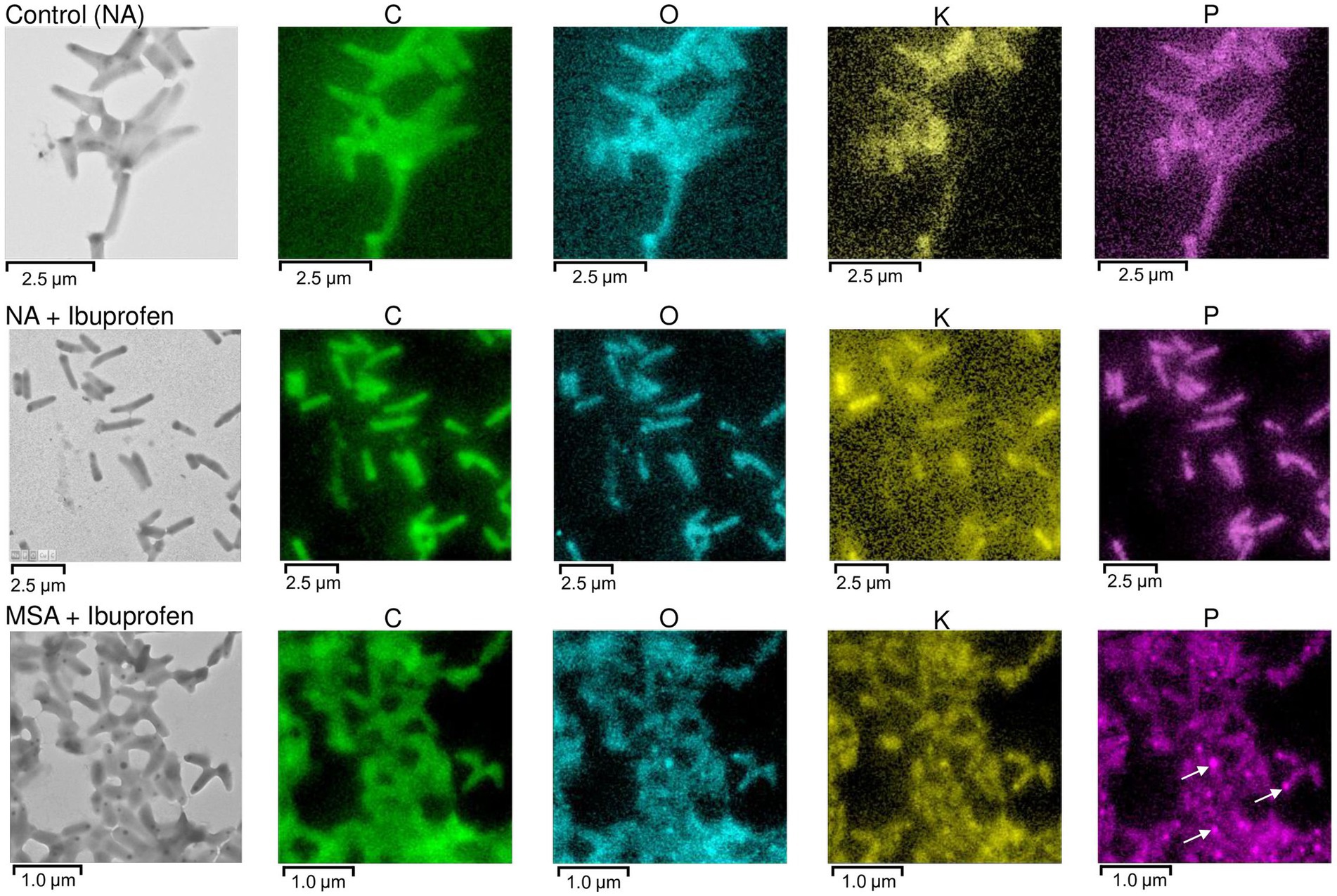
Figure 10. TEM-EDX analysis: Images and elemental mapping (C, O, K, P) of R. cerastii IEGM 1243 cells. Cells were grown on nutrient agar (Сontrol, NA) supplemented with IBP (NA + Ibuprofen) or on minimal salt agar supplemented with IBP (MSA + Ibuprofen) for 3 days. Other elements are presented in Supplementary Figures S11–S13. The white arrows indicate polyphosphate inclusions.
In this work, we investigated how individual and combined NSAIDs affect Rhodococcus cerastii strain IEGM 1243. Among the key findings, we observed significant alterations in catalase activity and a noticeable depletion of lipid inclusions in bacterial cells exposed to IBP, DCF, and their combination. These changes in catalase activity are indicative of adjustments in the stress response mechanisms employed by R. cerastii strain IEGM 1243 when confronted with NSAID-induced stressors. Furthermore, the observed depletion of lipid inclusions suggests potential modifications in the lipid metabolism pathways, which may play a pivotal role in adapting to NSAID exposure.
Morphometric analysis has revealed significant differences in cell size and surface area-to-volume ratio in the presence of NSAIDs. These alterations in cellular morphology might be influenced by factors related to peptidoglycan biosynthesis and fatty acid biosynthesis inhibitors.
For the first time, we employed high-precision electron microscopy to uncover ultrastructural changes and element mapping of R. cerastii IEGM 1243 cells treated with IBP. This advanced technique revealed the structural integrity of the cells, even in the presence of the pharmaceutical. The identification of lipid inclusions, polyphosphates, and intracellular membrane-like structures in IBP-treated cells provides a deeper understanding of the cellular adaptations that occur in response to NSAIDs. These ultrastructural insights open new avenues for research into the mechanisms of bacterial resistance and adaptation at the nanoscale level.
Looking ahead, future prospects in this field should incorporate advanced omics techniques to unravel the underlying genetic, transcriptomic, proteomic, and metabolomic changes in R. cerastii IEGM 1243 under NSAID exposure. Such sophisticated analyses can provide a holistic understanding of the bacterial response, shedding light on the intricate molecular mechanisms driving adaptation to pharmaceutical pollution. Additionally, the knowledge gained from understanding the adaptive reactions of bacteria towards pharmaceuticals can be harnessed in the development of biocatalysts for pharmaceutical waste disposal, aligning with the growing need for sustainable and environmentally friendly approaches to address pharmaceutical contamination.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
ET: Investigation, Visualization, Writing – original draft. GB: Investigation, Writing – original draft. NK: Investigation, Visualization, Writing – review & editing. VS: Investigation, Writing – review & editing. AM: Methodology, Visualization, Writing – review & editing. II: Conceptualization, Data curation, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Russian Science Foundation, Grant number 21-14-00132.
The work was carried out using the equipment of The Core Facilities Centers “Regional Specialised Collection of Alkanotrophic Microorganisms” and “Research of materials and matter” at PFRC UB RAS. TEM microscopy and EDX analysis were performed in the Core Facility Center “UNIQEM Collection” at Research Center of Biotechnology, Russian Academy of Sciences.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1275553/full#supplementary-material
Aguilar-Romero, I., Romero, E., Wittich, R. M., and van Dillewijn, P. (2020). Bacterial ecotoxicity and shifts in bacterial communities associated with the removal of ibuprofen, diclofenac and triclosan in biopurification systems. Sci. Total Environ. 741:140461. doi: 10.1016/J.SCITOTENV.2020.140461
Alobaidi, R. A. K., Ulucan-Altuntas, K., Mhemid, R. K. S., Manav-Demir, N., and Cinar, O. (2021). Biodegradation of emerging pharmaceuticals from domestic wastewater by membrane bioreactor: the effect of solid retention time. Int. J. Environ. Res. Public Health 18:3395. doi: 10.3390/ijerph18073395
Alvarez, H. M., Kalscheuer, R., and Steinbüchel, A. (2000). Accumulation and mobilization of storage lipids by Rhodococcus opacus PD630 and Rhodococcus ruber NCIMB 40126. Appl. Microbiol. Biotechnol. 54, 218–223. doi: 10.1007/s002530000395
Alvarez, H. M., Mayer, F., Fabritius, D., and Steinbüchel, A. (1996). Formation of intracytoplasmic lipid inclusions by Rhodococcus opacus strain PD630. Arch. Microbiol. 165, 377–386. doi: 10.1007/s002030050341
Bazhutin, G. A., Polygalov, M. A., Tyumina, E. A., Tyan, S. M., and Ivshina, I. B. (2022). “Cometabolic bioconversion of ketoprofen by Rhodococcus erythropolis IEGM 746” in Science and global challenges of the 21st-century—science and technology. eds. A. Rocha and E. Isaeva (Cham: Springer), 404–410.
Bertaux, F., von Kügelgen, J., Marguerat, S., and Shahrezaei, V. (2020). A bacterial size law revealed by a coarse-grained model of cell physiology. PLoS Comput. Biol. 16:e1008245. doi: 10.1371/JOURNAL.PCBI.1008245
Bibi, M., Rashid, J., Malik, M., Iqbal, A., and Xu, M. (2023). Physicochemical analysis and detection of exceptionally high diclofenac concentration in the pharmaceutical wastewaters collected from the production units of national industrial zone, Rawat, Pakistan. Appl. Water Sci. 13:157. doi: 10.1007/S13201-023-01954-X
Brack, W., Barcelo Culleres, D., Boxall, A. B. A., Budzinski, H., Castiglioni, S., Covaci, A., et al. (2022). One planet: one health. A call to support the initiative on a global science-policy body on chemicals and waste. Environ. Sci. Eur. 34:21. doi: 10.1186/S12302-022-00602-6
Cappelletti, M., Presentato, A., Piacenza, E., Firrincieli, A., Turner, R. J., and Zannoni, D. (2020). Biotechnology of Rhodococcus for the production of valuable compounds. Appl. Microbiol. Biotechnol. 104, 8567–8594. doi: 10.1007/s00253-020-10861-z
Chang, F., and Huang, K. C. (2014). How and why cells grow as rods. BMC Biol. 12:54. doi: 10.1186/S12915-014-0054-8
Cheremnykh, K. M., Luchnikova, N. A., Grishko, V. V., and Ivshina, I. B. (2018). Bioconversion of ecotoxic dehydroabietic acid using Rhodococcus actinobacteria. J. Hazard. Mater. 346, 103–112. doi: 10.1016/j.jhazmat.2017.12.025
Chopra, S., and Kumar, D. (2020). Ibuprofen as an emerging organic contaminant in environment, distribution and remediation. Heliyon 6:e04087. doi: 10.1016/j.heliyon.2020.e04087
Cleuvers, M. (2004). Mixture toxicity of the anti-inflammatory drugs diclofenac, ibuprofen, naproxen, and acetylsalicylic acid. Ecotoxicol. Environ. Saf. 59, 309–315. doi: 10.1016/S0147-6513(03)00141-6
D’Souza, G., Ebrahimi, A., Stubbusch, A., Daniels, M., Keegstra, J., Stocker, R., et al. (2023). Cell aggregation is associated with enzyme secretion strategies in marine polysaccharide-degrading bacteria. ISME J. 17, 703–711. doi: 10.1038/s41396-023-01385-1
Davids, M., Gudra, D., Radovica-Spalvina, I., Fridmanis, D., Bartkevics, V., and Muter, O. (2017). The effects of ibuprofen on activated sludge: shift in bacterial community structure and resistance to ciprofloxacin. J. Hazard. Mater. 340, 291–299. doi: 10.1016/j.jhazmat.2017.06.065
de Carvalho, C. C. C. R. (2012). Adaptation of Rhodococcus erythropolis cells for growth and bioremediation under extreme conditions. Res. Microbiol. 163, 125–136. doi: 10.1016/j.resmic.2011.11.003
De Felice, B., Copia, L., and Guida, M. (2012). Gene expression profiling in zebrafish embryos exposed to diclofenac, an environmental toxicant. Mol. Biol. Rep. 39, 2119–2128. doi: 10.1007/s11033-011-0959-z
Domaradzka, D., Guzik, U., and Wojcieszyńska, D. (2015). Biodegradation and biotransformation of polycyclic non-steroidal anti-inflammatory drugs. Rev. Environ. Sci. Biotechnol. 14, 229–239. doi: 10.1007/s11157-015-9364-8
Donovan, C., and Bramkamp, M. (2014). Cell division in Corynebacterineae. Front. Microbiol. 5:132. doi: 10.3389/FMICB.2014.00132
Ericson, H., Thorsén, G., and Kumblad, L. (2010). Physiological effects of diclofenac, ibuprofen and propranolol on Baltic Sea blue mussels. Aquat. Toxicol. 99, 223–231. doi: 10.1016/j.aquatox.2010.04.017
Gamarra, J. S., Godoi, A. F. L., de Vasconcelos, E. C., de Souza, K. M. T., and Ribas de Oliveira, C. M. (2015). Environmental risk assessment (ERA) of diclofenac and ibuprofen: a public health perspective. Chemosphere 120, 462–469. doi: 10.1016/j.chemosphere.2014.08.020
Gogoleva, O. A., Nemtseva, N. V., and Bukharin, O. V. (2012). Catalase activity of hydrocarbon-oxidizing bacteria. Appl. Biochem. Microbiol. 48, 552–556. doi: 10.1134/s0003683812060051
González-González, E. D., Gómez-Oliván, L. M., Galar-Martínez, M., Vieyra-Reyes, P., Islas-Flores, H., García-Medina, S., et al. (2014). Metals and nonsteroidal anti-inflammatory pharmaceuticals drugs present in water from Madín Reservoir (Mexico) induce oxidative stress in gill, blood, and muscle of common carp (Cyprinus carpio). Arch. Environ. Contam. Toxicol. 67, 281–295. doi: 10.1007/s00244-014-0048-0
Govers, S. K., Campos, M., Tyagi, B., Laloux, G., and Jacobs-Wagner, C. (2023). Apparent simplicity and emergent robustness in bacterial cell cycle control. bioRxiv. Available at: https://doi.org/10.1101/2023.01.16.524295 [Epub ahead of preprint]
Guzik, U., and Wojcieszyńska, D. (2019). “Biodegradation of non-steroidal anti-inflammatory drugs and their influence on soil microorganisms” in Microbes and enzymes in soil health and bioremediation. eds. A. Kumar and S. Sharma (Springer: Singapore), 379–401.
Harris, L. K., and Theriot, J. A. (2018). Surface area to volume ratio: a natural variable for bacterial morphogenesis. Trends Microbiol. 26, 815–832. doi: 10.1016/J.TIM.2018.04.008
Hernández, M. A., Mohn, W. W., Martínez, E., Rost, E., Alvarez, A. F., and Alvarez, H. M. (2008). Biosynthesis of storage compounds by Rhodococcus jostii RHA1 and global identification of genes involved in their metabolism. BMC Genomics 9:600. doi: 10.1186/1471-2164-9-600
Herzig, S. J., Anderson, T. S., Jung, Y., Ngo, L., Kim, D. H., and McCarthy, E. P. (2021). Relative risks of adverse events among older adults receiving opioids versus NSAIDs after hospital discharge: a nationwide cohort study. PLoS Med. 18:e1003804. doi: 10.1371/JOURNAL.PMED.1003804
Ivshina, I. B., Bazhutin, G., Tyan, S., Polygalov, M., Subbotina, M., and Tyumina, E. (2022b). Cellular modifications of rhodococci exposed to separate and combined effects of pharmaceutical pollutants. Microorganisms 10:1101. doi: 10.3390/microorganisms10061101
Ivshina, I. B., Bazhutin, G., and Tyumina, E. (2022c). Rhodococcus strains as a good biotool for neutralizing pharmaceutical pollutants and obtaining therapeutically valuable products: through the past into the future. Front. Microbiol. 13:3861. doi: 10.3389/fmicb.2022.967127
Ivshina, I. B., Kuyukina, M. S., Krivoruchko, A. V., and Tyumina, E. A. (2021b). Responses to ecopollutants and pathogenization risks of saprotrophic Rhodococcus species. Pathogens 10:974. doi: 10.3390/PATHOGENS10080974
Ivshina, I. B., Luchnikova, N. A., Maltseva, P. Y., Ilyina, I. V., Volcho, K. P., Gatilov, Y. V., et al. (2022a). Biotransformation of (−)-isopulegol by Rhodococcus rhodochrous. Pharmaceuticals 15:964. doi: 10.3390/PH15080964
Ivshina, I. B., Rychkova, M. I., Vikhareva, E. V., Chekryshkina, L. A., and Mishenina, I. I. (2006). Catalysis of the biodegradation of unusable medicines by alkanotrophic rhodococci. Appl. Biochem. Microbiol. 42, 392–395. doi: 10.1134/S0003683806040090
Ivshina, I. B., Tyumina, E. A., Bazhutin, G. A., and Vikhareva, E. V. (2021b). Response of Rhodococcus cerastii IEGM 1278 to toxic effects of ibuprofen. PLoS One 16:e0260032. doi: 10.1371/JOURNAL.PONE.0260032
Ivshina, I. B., Tyumina, E. A., Kuzmina, M. V., and Vikhareva, E. V. (2019). Features of diclofenac biodegradation by Rhodococcus ruber IEGM 346. Sci. Rep. 9:9159. doi: 10.1038/s41598-019-45732-9
Izadi, P., Izadi, P., Salem, R., Papry, S. A., Magdouli, S., Pulicharla, R., et al. (2020). Non-steroidal anti-inflammatory drugs in the environment: where were we and how far we have come? Environ. Pollut. 267:115370. doi: 10.1016/j.envpol.2020.115370
Jan-Roblero, J., and Cruz-Maya, J. A. (2023). Ibuprofen: toxicology and biodegradation of an emerging contaminant. Molecules 28:2097. doi: 10.3390/MOLECULES28052097
Jiang, C., Geng, J., Hu, H., Ma, H., Gao, X., and Ren, H. (2017). Impact of selected non-steroidal anti-inflammatory pharmaceuticals on microbial community assembly and activity in sequencing batch reactors. PLoS One 12:e0179236. doi: 10.1371/journal.pone.0179236
Kaushal, J., Mehandia, S., Singh, G., Raina, A., and Arya, S. K. (2018). Catalase enzyme: application in bioremediation and food industry. Biocatal. Agric. Biotechnol. 16, 192–199. doi: 10.1016/j.bcab.2018.07.035
Keim, C. N., Solórzano, G., Farina, M., and Lins, U. (2005). Intracellular inclusions of uncultured magnetotactic bacteria. Int. Microbiol. 8, 111–117. doi: 10.1111/1462-2920.13388
Kuyukina, M. S., and Ivshina, I. B. (2019). “Bioremediation of contaminated environments using Rhodococcus” in Biology of Rhodococcus. ed. H. M. Alvarez (Cham: Springer), 231–270.
Kuyukina, M. S., Ivshina, I. B., Korshunova, I. O., and Rubtsova, E. V. (2014). Assessment of bacterial resistance to organic solvents using a combined confocal laser scanning and atomic force microscopy (CLSM/AFM). J. Microbiol. Methods 107, 23–29. doi: 10.1016/j.mimet.2014.08.020
Levin, P. A., and Angert, E. R. (2015). Small but mighty: cell size and bacteria. Cold Spring Harb. Perspect. Biol. 7:a019216. doi: 10.1101/cshperspect.a019216
Lonappan, L., Brar, S. K., Das, R. K., Verma, M., and Surampalli, R. Y. (2016). Diclofenac and its transformation products: environmental occurrence and toxicity—a review. Environ. Int. 96, 127–138. doi: 10.1016/j.envint.2016.09.014
Luchnikova, N. A., Grishko, V. V., Kostrikina, N. A., Sorokin, V. V., Mulyukin, A. L., and Ivshina, I. B. (2022). Biotransformation of oleanolic acid using Rhodococcus rhodochrous IEGM 757. Catalysts 12:1352. doi: 10.3390/CATAL12111352
Mallick, I., Santucci, P., Poncin, I., Point, V., Kremer, L., Cavalier, J. F., et al. (2021). Intrabacterial lipid inclusions in mycobacteria: unexpected key players in survival and pathogenesis? FEMS Microbiol. Rev. 45:fuab029. doi: 10.1093/FEMSRE/FUAB029
Mejía-García, A., Islas-Flores, H., Gómez-Oliván, L. M., SanJuan-Reyes, N., Ortega-Olvera, J. M., and Hernández-Navarro, M. D. (2020). “Overview of non-steroidal anti-inflammatory drugs as emerging contaminants” in Non-steroidal anti-inflammatory drugs in water. ed. L. M. Gómez-Oliván (Cham: Springer), 41–53.
Memmert, U., Peither, A., Burri, R., Weber, K., Schmidt, T., Sumpter, J. P., et al. (2013). Diclofenac: new data on chronic toxicity and bioconcentration in fish. Environ. Toxicol. Chem. 32, 442–452. doi: 10.1002/etc.2085
Mezzelani, M., Gorbi, S., Fattorini, D., D’Errico, G., Consolandi, G., Milan, M., et al. (2018). Long-term exposure of Mytilus galloprovincialis to diclofenac, ibuprofen and ketoprofen: insights into bioavailability, biomarkers and transcriptomic changes. Chemosphere 198, 238–248. doi: 10.1016/j.chemosphere.2018.01.148
Mohamed, M. S. M., Asair, A. A., Fetyan, N. A. H., and Elnagdy, S. M. (2023). Complete biodegradation of diclofenac by new bacterial strains: postulated pathways and degrading enzymes. Microorganisms 11:1445. doi: 10.3390/microorganisms11061445
Mulkiewicz, E., Wolecki, D., Świacka, K., Kumirska, J., Stepnowski, P., and Caban, M. (2021). Metabolism of non-steroidal anti-inflammatory drugs by non-target wild-living organisms. Sci. Total Environ. 791:148251. doi: 10.1016/J.SCITOTENV.2021.148251
Neumann, G., Veeranagouda, Y., Karegoudar, T. B., Sahin, Ö., Mäusezahl, I., Kabelitz, N., et al. (2005). Cells of Pseudomonas putida and Enterobacter sp. adapt to toxic organic compounds by increasing their size. Extremophiles 9, 163–168. doi: 10.1007/s00792-005-0431-x
Ojkic, N., Serbanescu, D., and Banerjee, S. (2019). Surface-to-volume scaling and aspect ratio preservation in rod-shaped bacteria. eLife 8:e47033. doi: 10.7554/ELIFE.47033
Ojkic, N., Serbanescu, D., and Banerjee, S. (2022). Antibiotic resistance via bacterial cell shape-shifting. mBio 13, 00659–00622. doi: 10.1128/MBIO.00659-22
Oldewurtel, E. R., Kitahara, Y., and van Teeffelen, S. (2021). Robust surface-to-mass coupling and turgor-dependent cell width determine bacterial dry-mass density. Proc. Natl. Acad. Sci. U. S. A. 118:e2021416118. doi: 10.1073/PNAS.2021416118
Ortiz de García, S. A., Pinto Pinto, G., García-Encina, P. A., and Irusta-Mata, R. (2014). Ecotoxicity and environmental risk assessment of pharmaceuticals and personal care products in aquatic environments and wastewater treatment plants. Ecotoxicology 23, 1517–1533. doi: 10.1007/s10646-014-1293-8
Parolini, M. (2020). Toxicity of the non-steroidal anti-inflammatory drugs (NSAIDs) acetylsalicylic acid, paracetamol, diclofenac, ibuprofen and naproxen towards freshwater invertebrates: a review. Sci. Total Environ. 740:140043. doi: 10.1016/j.scitotenv.2020.140043
Parolini, M., Binelli, A., and Provini, A. (2011). Chronic effects induced by ibuprofen on the freshwater bivalve Dreissena polymorpha. Ecotoxicol. Environ. Saf. 74, 1586–1594. doi: 10.1016/j.ecoenv.2011.04.025
Pátek, M., Grulich, M., and Nešvera, J. (2021). Stress response in Rhodococcus strains. Biotechnol. Adv. 53:107698. doi: 10.1016/j.biotechadv.2021.107698
Petrie, B., and Camacho-Muñoz, D. (2021). Analysis, fate and toxicity of chiral non-steroidal anti-inflammatory drugs in wastewaters and the environment: a review. Environ. Chem. Lett. 19, 43–75. doi: 10.1007/S10311-020-01065-Y
Presentato, A., Cappelletti, M., Sansone, A., Ferreri, C., Piacenza, E., Demeter, M. A., et al. (2018). Aerobic growth of Rhodococcus aetherivorans BCP1 using selected naphthenic acids as the sole carbon and energy sources. Front. Microbiol. 9:672. doi: 10.3389/FMICB.2018.00672
Rastogi, A., Tiwari, M. K., and Ghangrekar, M. M. (2021). A review on environmental occurrence, toxicity and microbial degradation of non-steroidal anti-inflammatory drugs (NSAIDs). J. Environ. Manag. 300:113694. doi: 10.1016/J.JENVMAN.2021.113694
Richards, N. L., Cook, G., Simpson, V., Hall, S., Harrison, N., and Scott, K. S. (2011). Qualitative detection of the NSAIDs diclofenac and ibuprofen in the hair of Eurasian otters (Lutra lutra) occupying UK waterways with GC-MS. Eur. J. Wildl. Res. 57, 1107–1114. doi: 10.1007/s10344-011-0513-2
Rodrigues, C. J. C., and de Carvalho, C. C. C. R. (2019). Phenotypic adaptations help Rhodococcus erythropolis cells during the degradation of paraffin wax. Biotechnol. J. 14:e1800598. doi: 10.1002/biot.201800598
Royes, J., Biou, V., Dautin, N., Tribet, C., and Miroux, B. (2020). Inducible intracellular membranes: molecular aspects and emerging applications. Microb. Cell Factories 19:176. doi: 10.1186/S12934-020-01433-X
Sadutto, D., Andreu, V., Ilo, T., Akkanen, J., and Picó, Y. (2021). Pharmaceuticals and personal care products in a Mediterranean coastal wetland: impact of anthropogenic and spatial factors and environmental risk assessment. Environ. Pollut. 271:116353. doi: 10.1016/j.envpol.2020.116353
Shi, H., Hu, Y., Odermatt, P. D., Gonzalez, C. G., Zhang, L., Elias, J. E., et al. (2021). Precise regulation of the relative rates of surface area and volume synthesis in bacterial cells growing in dynamic environments. Nat. Commun. 12:1975. doi: 10.1038/s41467-021-22092-5
Solyanikova, I. P., Suzina, N. E., Emelyanova, E. V., Polivtseva, V. N., Pshenichnikova, A. B., Lobanok, A. G., et al. (2017). Morphological, physiological, and biochemical characteristics of a benzoate-degrading strain Rhodococcus opacus 1CP under stress conditions. Microbiology 86, 202–212. doi: 10.1134/S0026261717020199
Spiekermann, P., Rehm, B. H. A., Kalscheuer, R., Baumeister, D., and Steinbüchel, A. (1999). A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch. Microbiol. 171, 73–80. doi: 10.1007/S002030050681
Suleiman, M., Demaria, F., Zimmardi, C., Kolvenbach, B. A., and Corvini, P. F. X. (2023). Analyzing microbial communities and their biodegradation of multiple pharmaceuticals in membrane bioreactors. Appl. Microbiol. Biotechnol. 107, 5545–5554. doi: 10.1007/s00253-023-12677-z
Swan, G. E., Cuthbert, R., Quevedo, M., Green, R. E., Pain, D. J., Bartels, P., et al. (2006). Toxicity of diclofenac to Gyps vultures. Biol. Lett. 2, 279–282. doi: 10.1098/rsbl.2005.0425
Świacka, K., Michnowska, A., Maculewicz, J., Caban, M., and Smolarz, K. (2021). Toxic effects of NSAIDs in non-target species: a review from the perspective of the aquatic environment. Environ. Pollut. 273:115891. doi: 10.1016/j.envpol.2020.115891
Thi Mo, L., Irina, P., Natalia, S., Irina, N., Lenar, A., Andrey, F., et al. (2022). Hydrocarbons biodegradation by Rhodococcus: assimilation of hexadecane in different aggregate states. Microorganisms 10:1594. doi: 10.3390/microorganisms10081594
Tyumina, E. A., Bazhutin, G. A., Cartagena Gómez, A., Cartagena Gómez, A. d. P., and Ivshina, I. B. (2020). Nonsteroidal anti-inflammatory drugs as emerging contaminants. Microbiology 89, 148–163. doi: 10.1134/S0026261720020125
Tyumina, E. A., Bazhutin, G. A., Vikhareva, E. V., Selyaninov, A. A., and Ivshina, I. B. (2019). Diclofenac as a factor in the change of Rhodococcus metabolism. IOP Conf. Ser. Mater. Sci. Eng. 487:012027. doi: 10.1088/1757-899X/487/1/012027
Uzoechi, S. C., and Abu-Lail, N. I. (2019). The effects of β-lactam antibiotics on surface modifications of multidrug-resistant Escherichia coli: a multiscale approach. Microsc. Microanal. 25, 135–150. doi: 10.1017/S1431927618015696
Wältermann, M., Hinz, A., Robenek, H., Troyer, D., Reichelt, R., Malkus, U., et al. (2005). Mechanism of lipid-body formation in prokaryotes: how bacteria fatten up. Mol. Microbiol. 55, 750–763. doi: 10.1111/J.1365-2958.2004.04441.X
Wang, Z., Altenburger, R., Backhaus, T., Covaci, A., Diamond, M. L., Grimalt, J. O., et al. (2021). We need a global science-policy body on chemicals and waste. Science 371, 774–776. doi: 10.1126/science.abe9090
Wang, C., Chen, Y., Zhou, H., Li, X., and Tan, Z. (2020). Adaptation mechanisms of Rhodococcus sp. CNS16 under different temperature gradients: physiological and transcriptome. Chemosphere 238:124571. doi: 10.1016/J.CHEMOSPHERE.2019.124571
Wang, R.-F., Wennerstrom, D., Cao, W.-W., Khan, A. A., and Cerniglia, C. E. (2000). Cloning, expression, and characterization of the katG gene, encoding catalase-peroxidase, from the polycyclic aromatic hydrocarbon-degrading bacterium Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 66, 4300–4304. doi: 10.1128/AEM.66.10.4300-4304.2000
Wilkinson, J. L., Boxall, A. B. A., Kolpin, D. W., Leung, K. M. Y., Lai, R. W. S., Galban-Malag, C., et al. (2022). Pharmaceutical pollution of the world’s rivers. Proc. Natl. Acad. Sci. U. S. A. 119:e2113947119. doi: 10.1073/pnas.2113947119
Wojcieszyńska, D., Guzik, H., and Guzik, U. (2022). Non-steroidal anti-inflammatory drugs in the era of the COVID-19 pandemic in the context of the human and the environment. Sci. Total Environ. 834:155317. doi: 10.1016/j.scitotenv.2022.155317
Wojcieszyńska, D., Łagoda, K., and Guzik, U. (2023). Diclofenac biodegradation by microorganisms and with immobilised systems—a review. Catalysts 13:412. doi: 10.3390/catal13020412
Wood, N. J., and Sørensen, J. (2001). Catalase and superoxide dismutase activity in ammonia-oxidising bacteria. FEMS Microbiol. Ecol. 38, 53–58. doi: 10.1111/J.1574-6941.2001.TB00881.X
Xie, Z., Lu, G., Liu, J., Yan, Z., Ma, B., Zhang, Z., et al. (2015). Occurrence, bioaccumulation, and trophic magnification of pharmaceutically active compounds in Taihu Lake, China. Chemosphere 138, 140–147. doi: 10.1016/j.chemosphere.2015.05.086
Yam, K. C., van der Geize, R., and Eltis, L. D. (2010). “Catabolism of aromatic compounds and steroids by Rhodococcus” in Biology of Rhodococcus. ed. H. Alvarez (Berlin: Springer)
Yoshida, N., Yano, T., Kedo, K., Fujiyoshi, T., Nagai, R., Iwano, M., et al. (2017). A unique intracellular compartment formed during the oligotrophic growth of Rhodococcus erythropolis N9T-4. Appl. Microbiol. Biotechnol. 101, 331–340. doi: 10.1007/S00253-016-7883-Z
Young, K. D. (2006). The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 70, 660–703. doi: 10.1128/MMBR.00001-06
Yuan, F., Yin, S., Xu, Y., Xiang, L., Wang, H., Li, Z., et al. (2021). The richness and diversity of catalases in bacteria. Front. Microbiol. 12:645477. doi: 10.3389/FMICB.2021.645477
Żur, J., Marchlewicz, A., Piński, A., Guzik, U., and Wojcieszyńska, D. (2021). Degradation of diclofenac by new bacterial strains and its influence on the physiological status of cells. J. Hazard. Mater. 403:124000. doi: 10.1016/J.JHAZMAT.2020.124000
Keywords: rhodococci, NSAIDs, ibuprofen, diclofenac, stress responses, AFM/CLSM, TEM, TEM-EDX
Citation: Tyumina E, Bazhutin G, Kostrikina N, Sorokin V, Mulyukin A and Ivshina I (2023) Phenotypic and metabolic adaptations of Rhodococcus cerastii strain IEGM 1243 to separate and combined effects of diclofenac and ibuprofen. Front. Microbiol. 14:1275553. doi: 10.3389/fmicb.2023.1275553
Received: 10 August 2023; Accepted: 21 November 2023;
Published: 06 December 2023.
Edited by:
Srinivasan Ramanathan, Prince of Songkla University, ThailandReviewed by:
Huaming Qin, Jinan University, ChinaCopyright © 2023 Tyumina, Bazhutin, Kostrikina, Sorokin, Mulyukin and Ivshina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Irina Ivshina, aXZzaGluYUBpZWdtLnJ1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.