- 1Guangdong Provincial Key Laboratory of Animal Molecular Design and Precise Breeding, College of Life Science and Engineering, Foshan University, Foshan, China
- 2School of Life Science and Engineering, Foshan University, Foshan, China
- 3Animal Husbandry and Fisheries Technology Extension Station, Chongqing, China
- 4Farm Animal Genetic Resources Exploration and Innovation Key Laboratory of Sichuan Province, Sichuan Agricultural University, Chengdu, China
Human skin microbes play critical roles in skin health and diseases. Microbes colonizing on the skin of Tibetans living in the high-altitude area for generations may have a stronger ability to resist the harsh environment, such as high ultraviolet radiation (UV). Isolation of a potential probiotic from Tibetans skin is beneficial for resistance of skin disease for humans in the world. In this study, the signature microbiota for Tibetan skin were characterized compared to low-altitude humans. Next, using culture-omics, 118 species were isolated. The culturability of high-altitude of Tibetan skin microbiome reached approximate 66.8%. Next, we found that one strain, Pantoea eucrina, had the greatest ability to repair UV damage to the skin as the lowest pathological score was observed in this group. Interestingly, another animal trial found this bacterium resisted UV rather than its metabolites. Using whole genome sequencing, this strain P. eucrina KBFS172 was confirmed, and its functions were annotated. It might involve in the metabolic pathway of carotenoid biosynthesis with anti-oxidative stress properties, which plays critical roles in UV-damage repair. In conclusion, we characterized the signature microbes of skin in high-altitude Tibetans, isolated a skin bacterium of Pantoea eucrina KBFS172 which could repair UV damage via involving the metabolic pathway of carotenoid biosynthesis. Our results provide a new potential skin probiotic for skin disease prevention or sunburn.
Introduction
Skin, the largest organ of the human body, has over 30 m2 surface area, and serves as the first line to defense against the harsh external environment (Yoshio et al., 2004; Ross et al., 2017). Variable microbes colonize on the surface of skin (Gallo, 2017). Advances in sequencing and bioinformatics have unraveled the mysteries of the skin microbiome (Gao et al., 2008; Grice et al., 2008), including the characterization of commensal and pathogenic microorganisms on human skin (Roszkowski et al., 1990; Tang and Stratton, 2010). Numerous studies have demonstrated the crucial roles of skin commensal bacteria in maintaining the overall health of the skin and the host (Shu et al., 2013; Wang et al., 2014; Knackstedt et al., 2020). However, the specific roles of skin microbiota associated with common diseases (e.g., skin cancer) are still unclear.
Qinghai-Tibet Plateau (QTP), known as “the roof of the world,” is the highest plateau on earth, with a resident population exceeding 12 million as early as 2006 (Li et al., 2019). The living environment in QTP is extreme, with high ultraviolet radiation (UV), hypobaric pressure, and hypoxia (Jablonski and Chaplin, 2010). In spite prolonged exposure to high-intensity UV may cause skin cancer (Armstrong et al., 2001), the adaptation of Tibetans may have ability to resist extreme environment (e.g., UV) or even skin cancer. In term of this, some studies started to investigate the skin microbiota in subjects living in high and low altitudes. Previous studies have shown that high altitudes impacted the composition and structure skin microbes (Muletz Wolz et al., 2018; Li et al., 2019). Our previous study suggested that altitudes have a significant effect on the skin microbiome of Tibetans (Zeng et al., 2017). However, whether commensal bacteria on the skin of Tibetans could resist UV and what is the mechanism are still unknown.
Thus, we hypothesized that skin microbiota isolated from humans living in QTP may have the ability to resist and repair the UV damage. In this study, we first compared the human skin microbiomes form high and low altitude by re-analyzing our previous published data. Then we isolated and purified high-altitude Tibetan skin microbes using the culturomics method, and validated the potential functions of bacterial isolations on the repair of the UV damage by a mice model. Furthermore, we utilized whole-genome sequencing to unveil the mechanism of this isolate to resist UV. This current isolated a potential skin probiotic successfully, which benefits the protection of disease, such as skin cancer and sunburn.
Materials and methods
The experiment protocol was approved by the Animal Ethics and Humane Animal Care of the Foshan University (protocol#: FOSU2021010).
Sample collection
Ten healthy Tibetans (Supplementary Table S1), living in the Daocheng area (high altitude, 3,750 masl, latitude 27°58’ N, longitude 99°56′ E) in China and having long-term outdoor activities (e.g., agricultural production), were recruited for this study. All human subjects had no skin disease and free access of antibiotics within 3 months when sampled. All the participants were given the written informed consent. Relevant characteristics, including gender, age, and sampling site, were recorded to exclude individuals with interfering factors in this experiment. Skin samples were collected by swabbing the forehead of the Tibetans. Regarding sampling, all sterile swabs were pre-wet with SCF-1 buffer [50 mM Tris buffer [pH 7.6], 1 mM EDTA [pH 8.0], 0.5% Tween-20], and were rubbed vigorously on the forehead over 30 times to collect skin microbes. Then, the forehead swabs were cut off and placed into 2 mL centrifuge tubes, snap-frozen in liquid nitrogen, and stored at −80°C until further analysis.
Pure culture
The samples were thawed from −80°C and transferred to 4°C for 30 min. After the microorganisms were revived, the swab heads were immersed in sterile PBS buffer in the EP tube to shake out the bacteria. The resulting bacterial suspension was used for further pure culture experiments.
Five culture media, namely Nutrient Agar (Van der Weele et al., 2000), R-2A Agar (Reasoner et al., 1985; Massa et al., 1998), TSBYS (Peeters et al., 2011; Nicholson et al., 2013), BHIA (Osawa, 1990) and Agar medium J (Pagnanelli et al., 2000), were selected for pure culture. Each medium had six replicates. The bacterial suspension was mixed well with sterile PBS buffer (pH = 7.0) at a ratio of 0.1:9.9. Then, 200 μL of the bacterial suspension was spread evenly onto each prepared culture medium plate using a coating rod. The plates were then incubated in an inverted position at 28°C for microbial growth. The plates were observed every 12 h until no new colonies appeared. Three plates of each culture medium for each sample were randomly selected for subculture, and the plates were eluted with sterile PBS buffer. The eluted liquid was then sequenced directly to obtain the V3-V4 region of the 16S rRNA gene.
DNA extraction, sequencing, and bioinformatics for pure culture sample
For pure culture samples, the DNA of all the single strain were extracted using the boiling method (Dashti et al., 2009). The full-length sequence of 16S rRNA gene was amplified using the 27f/1492r primers (27f: 5’-AGAGTTTGATCCTGGCTCAG-3′, 1492r: 5’-TACGGYACCTTGTTACGACTT-3′). The quality of the PCR products was assessed by 1% gel electrophoresis, and qualified samples were sequenced on an Illumina MiSeq platform (Sangon Biotech Co., Ltd., Shanghai, China). Raw sequencing reads were quality controlled using Seqman software (Swindell and Plasterer, 1997). The high-quality sequences were aligned using Blastn in the NCBI database, and similar sequences of genes of single bacteria and their similarities were obtained. The isolated strains were then preliminarily divided into groups.
For the non-culture group that was sequenced directly from the swab heads, the total bacterial DNA was extracted using a Mo Bio PowerFecal DNA isolation kit according to the manufacturer’s recommendations. DNA concentration (μg/μl) was quantified using a NanoDrop 2000C ultra-micro spectrophotometer. The general primers 341F (5’-CCTAYGGGRBGCASCAG-3′) and 806R (5’-GGACTACNNGGGTATCTAAT-3′) were used to amplify the V3-V4 region of the 16S RNA gene. The quality of the PCR products was checked by 1% gel electrophoresis, and qualified samples were sequenced on an Illumina MiSeq platform (NovogeneCo, Ltd., Beijing, China). The raw sequences were denoised, dereplicated, and filtered for chimeras using QIIME2 (Bolyen et al., 2019) (dada2 Package). The previous data (Zeng et al., 2017) (16S rRNA, v4 region, 128 samples) were also quality controlled using QIIME2 (deblur Package) (Bolyen et al., 2019). The taxonomy was annotated using the Greengenes database (gg_13_8) (Cannone et al., 2002).
UV irradiation trial #1
Five-week-old SPF-class female ICR mice (n = 38, body weight = 23 g) were used for this trial. The mice were divided into eight groups: a blank control group (n = 3) that received no treatment, negative control group (n = 5), and experimental groups A–E (n = 5 for each group) that received bacterial therapy with different candidates (A: Arthrobacter gandavensis; B: Bacillus psychrosaccharolyticus; C: Pantoea eucrina; D: Paenibacillus amylolyticus; E: Paenibacillus terrae) (Supplementary Table S3). After a preparation period, the area on the back of each mouse, measuring 2 cm × 3 cm, was depilated and smeared with the corresponding bacterial suspension for 3 days.
Before the trial, the mice were pre-feed with SPF pellet feed for one week. All feeding utensils were sterilized twice a week, and the ambient temperature was controlled at 20–25°C. Each mouse was housed in an individual cage with ad libitum access to food and water. After the mice were adaptively fed for a week, and bacterial colonization of each mouse was checked by bacterial culture of the back. Further details regarding the grouping and treatment conditions can be found in Supplementary Table S3. All mice were exposed to the same combination-rays light, and the changes in the skin morphology of each group were observed and recorded daily throughout the trial. Two types of UV light (UVA, UVB) boxes were used to irradiate the depilated mice.
A UV facility comprises a UVA lamp (40 W, with a wavelength range of 320 to 400 nm and a peak wavelength of 365 nm, from Philips, Germany) and a UVB lamp (20 W, with a wavelength range of 290 to 320 nm and a peak wavelength of 297 nm, also from Philips, Germany) positioned side by side. The distance between the lamps and the backs of the mice was set at 30 to 40 cm, and the irradiation intensity was measured using a UV-radiometer (UVAB/ST-513, SENTRY, Taiwan, China). We followed the subacute light injury model used in animal experiments (Wei et al., 2002) and optimized it based on our specific experimental conditions. After preheating the tubes for 20 min, the mouse cages were placed under the lamp box. During the trial, the position of the mouse cages was rotated to ensure equal irradiation dose for all mice. In the first week of irradiation, we performed daily UV irradiation, with a daily UVA dose of 3.96 J/cm2 and a UVB dose of 252 mJ/cm2. During the second and third weeks of irradiation, we performed UV irradiation every other day for a duration of 6 h, with UVA and UVB doses of 10.8 J/cm2 and 1.08 J/cm2, respectively. After each irradiation, we applied the corresponding bacterial suspension on the depilated area of the mice’s backs. Overall, the total amount of UVA irradiation in this trial was 114.12 J/cm2, and for UVB, it was 8.964 J/cm2.
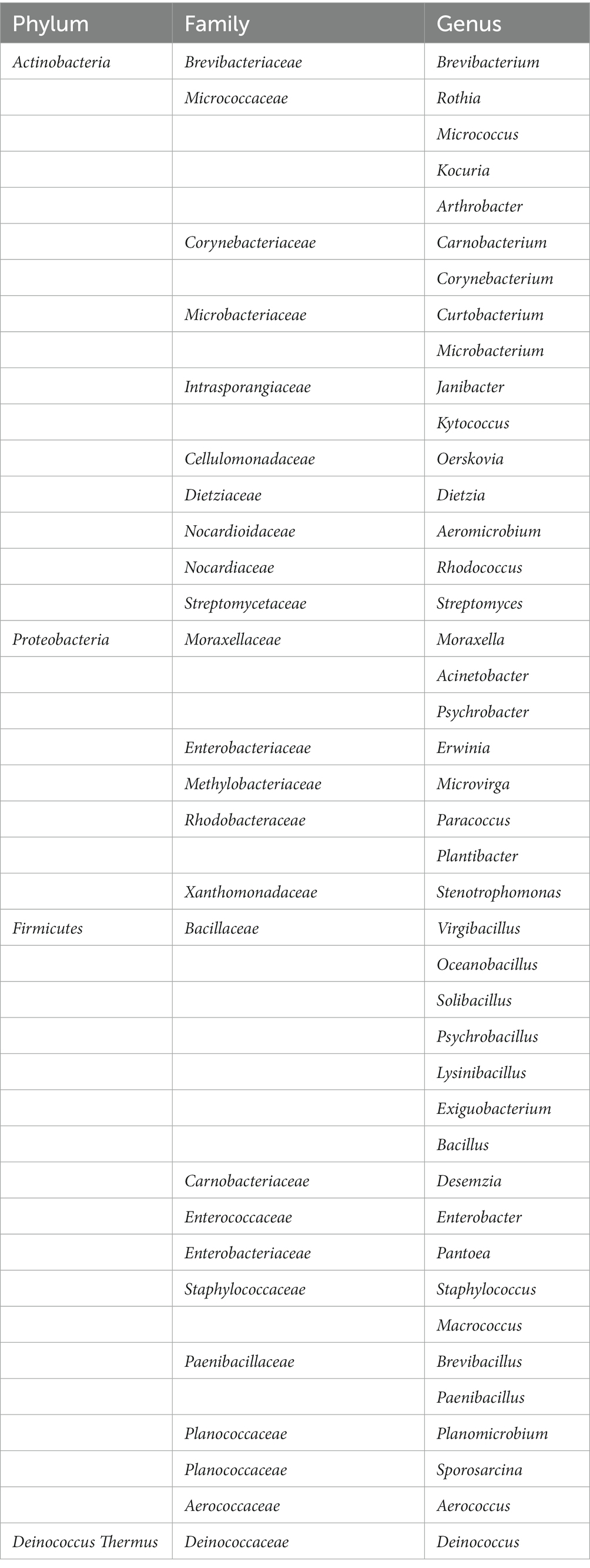
At the end of the trial, the mice were euthanized by cervical dislocation. The skin on their backs was immediately peeled off and immersed in a 4% formaldehyde solution, and stored at 4°C. The skin was then cut into 5 μm thickness sections, stained with H&E (hematoxylin–eosin staining), and observed under a microscope to record the histopathological changes (Figures 1A–D).
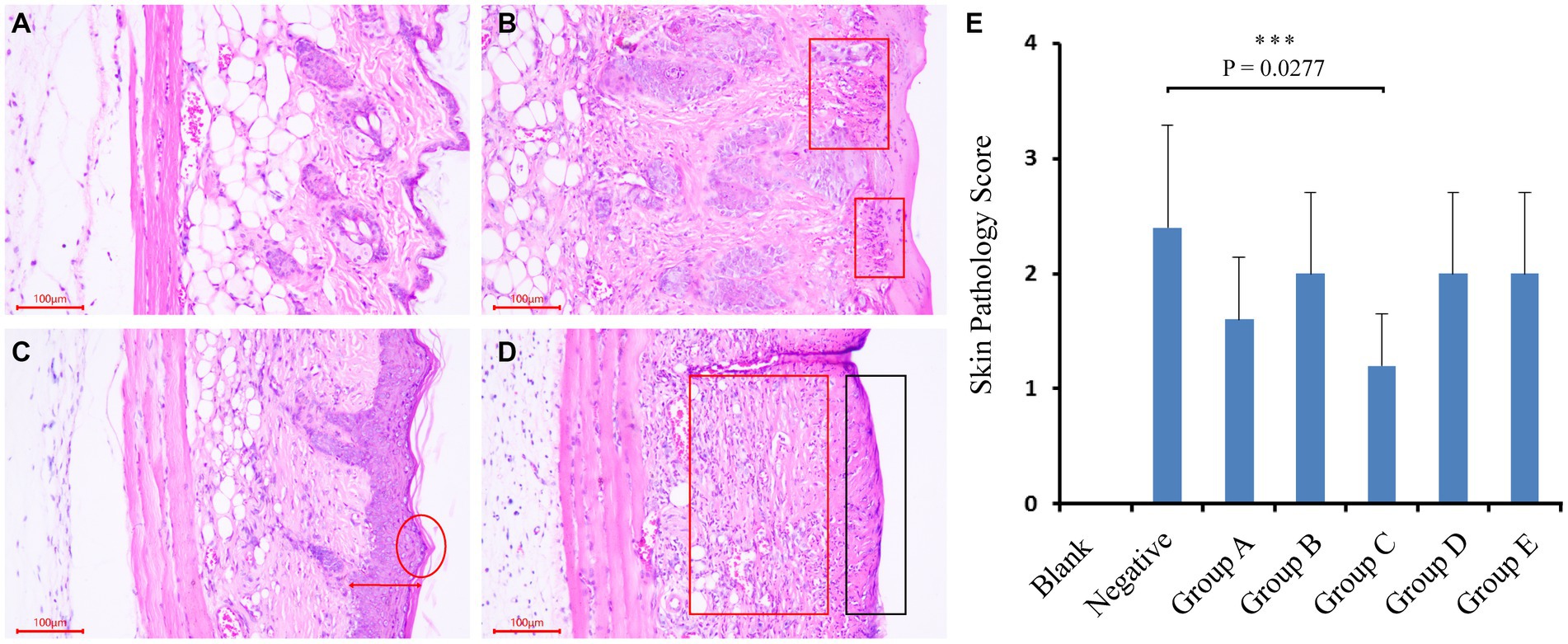
Figure 1. Mice trial #1: pathological changes of skin and sections, and scoring of skin pathological changes. (A–D) As followed, no obvious pathological changes in mouse skin; mild lesions, a small amount of inflammatory cell infiltration; moderate lesions, epidermal thickening, inflammatory cell infiltration and severe lesions, exposed subcutaneous tissue, and a large number of inflammatory cell infiltration. (E) Scores of pathological changes in skin sections of mice in each group. Blank (blank control group): without UV and smear bacteria treatment; Negative (negative control group): only UV treatment without smear bacteria suspension treatment; Group A–E: separately smear the suspension of Arthrobacter gandavensis, Bacillus psychrosaccharolyticus, Pantoea eucrina, Paenibacillus amylolyticus, and Paenibacillus terrae.
Mice UV irradiation trial #2
Pantoea eucrina has been shown to significantly repaired the UV damage by the first trial, we thus designed the second validation trial. Five-week-old SPF-class female ICR mice (n = 40) were used for this trial. The mice were divided into five groups, with eight mice per group: BS group (bacterial suspension: Suspension of Pantoea eucrina), BL group (bacterial lysate: lysate of Pantoea eucrina), NC group (negative control), and VE group (vitamin E). To prepare the bacterial suspension, the bacteria were activated and then incubated overnight in nutrient broth with shaking at 28°C. The concentration of the suspension was adjusted to 1 × 108 CFU/mL by counting on the plate. To prepare the bacterial lysate, the bacterial suspension was taken into a centrifuge tube and subjected to three cycles of freeze-thawing (−20°C, 30 min; 20°C, 20 min). The bacteria were then lysed using an ultrasonic cell disruptor (JY96-IIN, ShangHai JinXin). The lysate was then cooled in an ice-water bath for 30 min, filtered through a 0.22 μm filter, and stored at 4°C. For further details on the experimental grouping and treatment conditions, please refer to Supplementary Table S4. The trial procedures were the same as those of the previous trial (Mice UV Irradiation Trial #1).
After the trial, the skin on the back of the mice was stained using the same method as in the previous trial. The observations and recordings were conducted in a similar manner.
The whole genome sequencing
The bacterial specie (Pantoea eucrina) in group C were isolated to perform whole genome sequencing in order to identify the bacterial strain and its potential functions. The sequencing was performed on a single strain using the Nanopore sequencing technology platform. The raw data was subjected to format conversion and filtering. Canu v1.5 (Koren et al., 2017) was utilized for assembling the filtered subreads and Pilon (Walker et al., 2014) was used to correct any errors in the assembled genomes. Prodigal (Hyatt et al., 2010) was employed to predict the encoded genes in the assembled genome. The default parameters for these three softwares were chosen. Finally, the assembled contig sequences were aligned with the NT database to determine the chromosome type.
The predicted gene sequences were aligned using the BLAST algorithm (Altschul et al., 1997) to the Non-Redundant Protein Database (Nr database). Subsequently, the functional annotation of the Gene Ontology (GO) database (Ashburner et al., 2000) was performed using the Blast2GO software (Conesa et al., 2005) based on the alignment results with the Nr database. In addition, KEGG (Kanehisa et al., 2004) metabolic pathway enrichment analysis and GO functional enrichment analysis were carried.
Macroscopic and microcosmic evaluation
The changes in the mice’s skin were observed and recorded daily, and the degree of skin damage on their backs was evaluated based on Supplementary Table S5. After the UV Irradiation Trial, the mice were sacrificed, and pathological sections of their backs were made. The degree of pathological changes was evaluated using Supplementary Table S6 as a reference.
Statistical analyses
The algorithm of Linear Discriminant Analysis (LDA) coupled with effect size (LEfSe) (Paulson et al., 2013) was employed to determine features with significantly different abundances between high and low altitude human groups by re-analyzing our previous data (SRP065099, Sequence Read Archive (SRA) in NCBI).
The alpha and beta diversity of the high-altitude Tibetan skin microbiome of the 16S sequencing data were measured using the QIIME2 platform. The differences in alpha and beta diversity between groups were calculated using Student’s t-test. Statistical significance was determined at p < 0.05 for all analyses. All figures were generated using the R (Wickham, 2011).
Results
Skin microbial differences in high- and low-altitude humans
Our previously published 16S rRNA data (Zeng et al., 2017) (128 samples) of skin samples from high and low- altitude humans were re-analyzed at the ASV level to identify signature bacteria between groups using linear discriminant analysis (LDA) effect size (LEfSe) (Figure 2). The main skin-featured bacteria at high altitude were Aerococcus (ASV6), Staphylococcus (ASV2, ASV10, ASV35), Stenotrophomonas (ASV14), Acinetobacter (ASV4), Arthrobacter (ASV22, ASV46), Sanguibacter (ASV20), Pantoea (ASV31), Bacillus (ASV25), Paenibacillus (ASV18, ASV29), Pseudomonas (ASV47) and Enterobacteriaceae unclassified (ASV1). In the low altitude group, the main signature included Burkholderia (ASV9), Staphylococcaceae (ASV16), Enhydrobacter (ASV11, ASV13), Sphingomonas (ASV23), Acinetobacter (ASV28), Lactococcus (ASV26, ASV30), Brevundimonas (ASV37, ASV48), Corynebacterium (ASV40, ASV51, ASV53), Micrococcus (ASV56) and Neisseriaceae unclassified (ASV69).
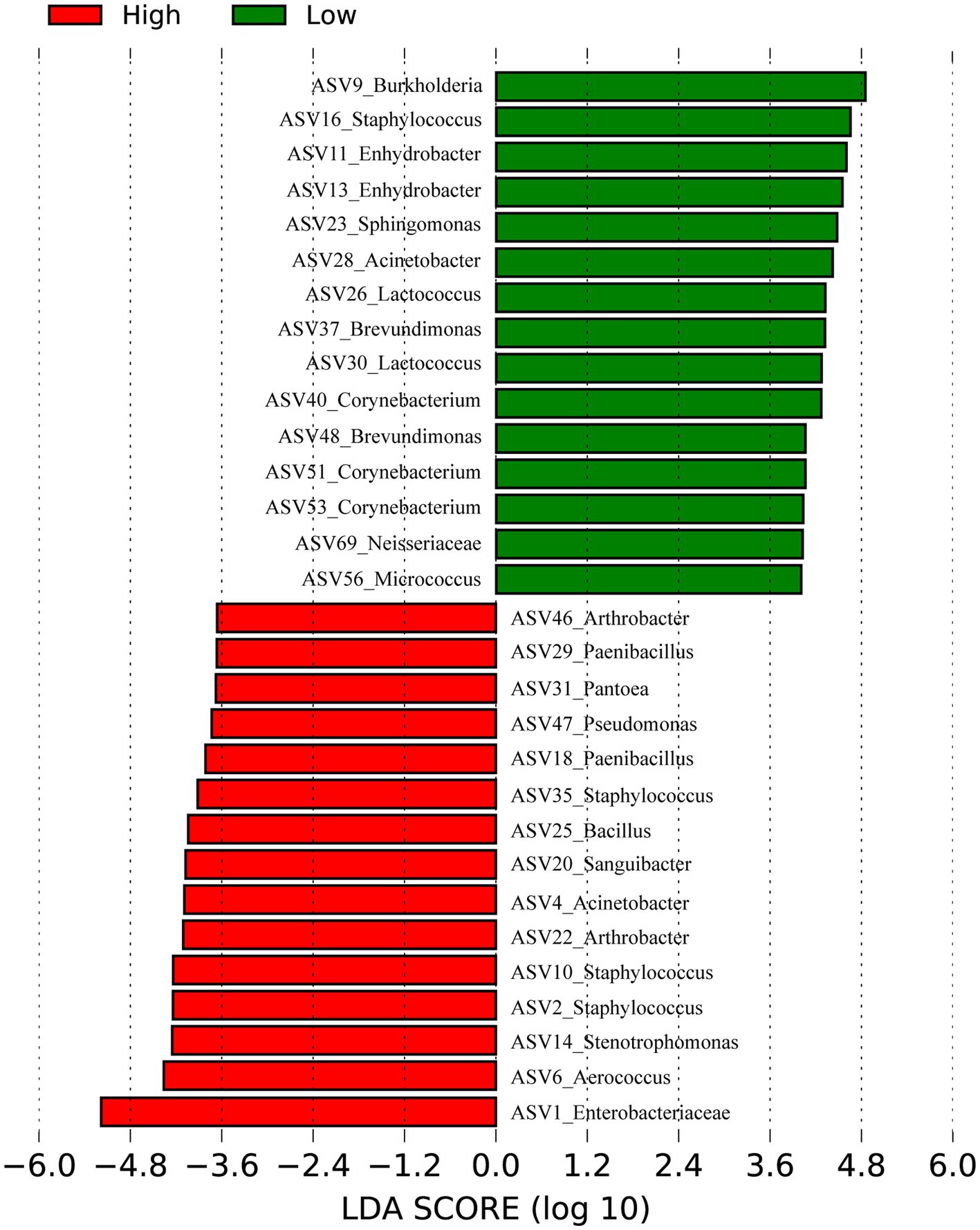
Figure 2. Bacteria differentiating high and low altitudes Humans by Linear discriminant analysis (LDA) coupled with effect size (LEfSe). Red represents the high-altitude group and green represents the low-altitude group.
Culturability of high-altitude human skin microbes
Next, to achieve the goal of bacterial isolation from high-altitude human skin that may resist and repair UV damage, the cultivation method using five different medias were performed. Compared to 1,438 ASVs from next-generation sequencing method (non-culture group), 1,054 ASVs were observed in the cultivation methods (pure culture group). In the meantime, 961 shared ASVs between the cultivation and next-generation sequencing methods (Supplementary Figure S1A), and then the culturability of high-altitude human skin microbes is 66.83% (961/1,438) at the ASV level. After annotation, we obtained 628 and 502 genera in the non-culture and pure culture groups, respectively, with 475 genera co-existing in both groups (Supplementary Figure S1B). Furthermore, non-culture group had higher alpha diversity than pure culture group (Figure 3). Correspondingly, beta diversity based on Bray-Curtis distance showed significant differences between pure culture (cult) and non-culture (skin).
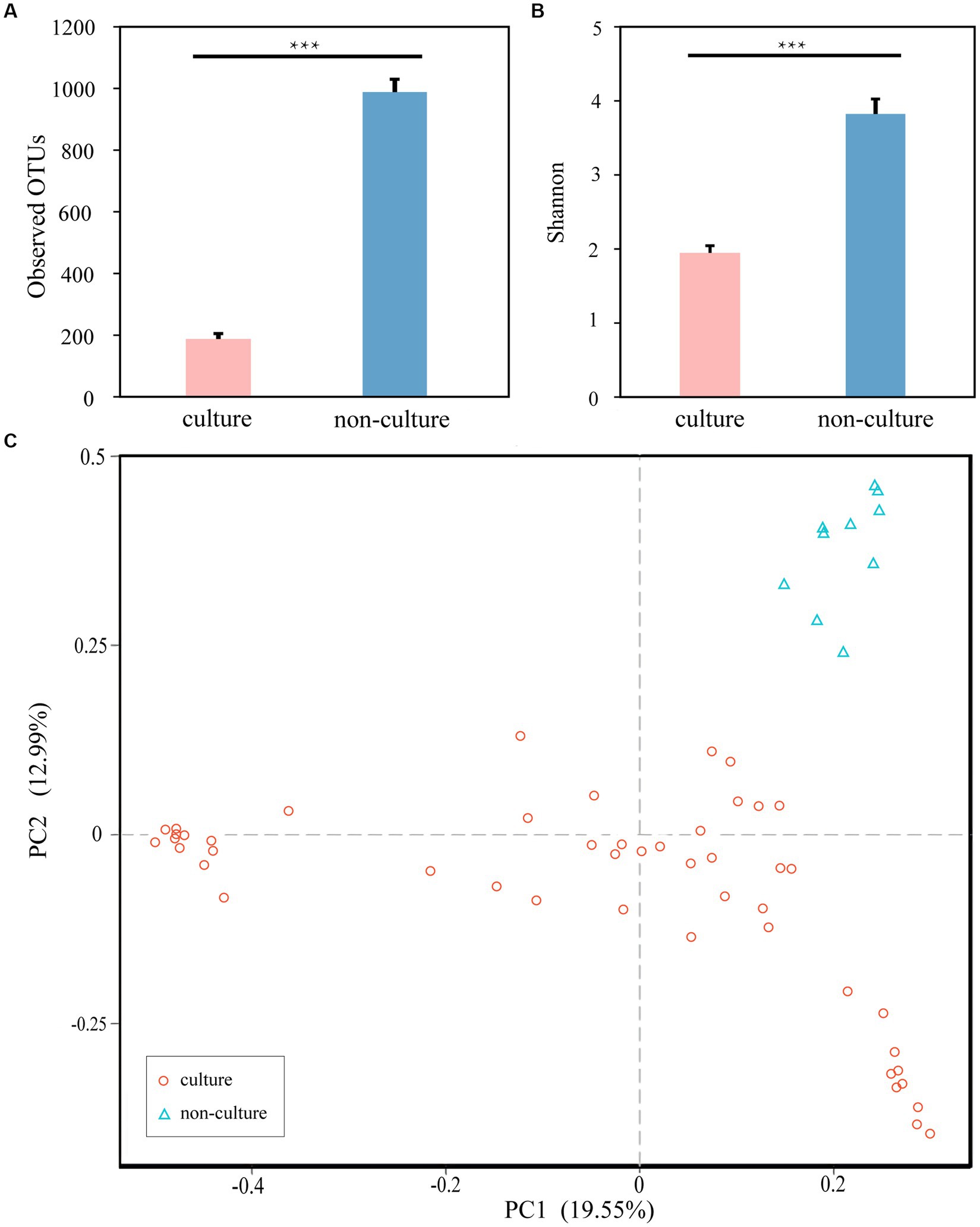
Figure 3. Microbial differences between non-culture and pure culture groups. (A) Observed OTUs. (B) Shannon Index. (C) PCoA plot based Bray-Curtis distance. Colors of red and blue represent samples on pure culture group and non-culture group. Pure culture group: using cultivation methods; non-culture group: using next-generation sequencing methods (***p < 0.05).
At the phylum level, the dominant bacteria in the non-culture group were Proteobacteria (44.48%), Firmicutes (29.19%), Actinobacteria (13.04%), Bacteroidetes (9.57%) and Acidobacteria (0.92%) (Figure 4). The dominant genera were Pseudomonas (10.20%), Enhydrobacter (8.76%), Chryseobacterium (6.90%), Staphylococcus (5.85%), Acinetobacter (3.68%), Clostridium sensu stricto 1 (3.32%), Bacillus (2.89%), Sphingomonas (2.37%), Peptoclostridium (2.35%), Psychrobacter (2.26%), Glutamicibacter (1.42%), Kocuria (1.31%) and Arthrobacter (1.12%) from the non-culture group (Figure 5). In the pure culture group, the dominant phyla were Firmicutes (55.82%), Proteobacteria (22.14%) and Actinobacteria (21.02%). Then, we identified 10 genera with relative abundance greater than 1%, which were considered to be the main genera that could be cultivated from all culture samples. These genera were Staphylococcus (30.81%), Bacillus (15.19%), Arthrobacter (5.65%), Psychrobacter (5.51%), Glutamicibacter (5.27%), Kocuria (4.09%), Acinetobacter (3.88%), Psychrobacillus (3.19%), Enhydrobacter (2.08%) and Micrococcus (1.27%).
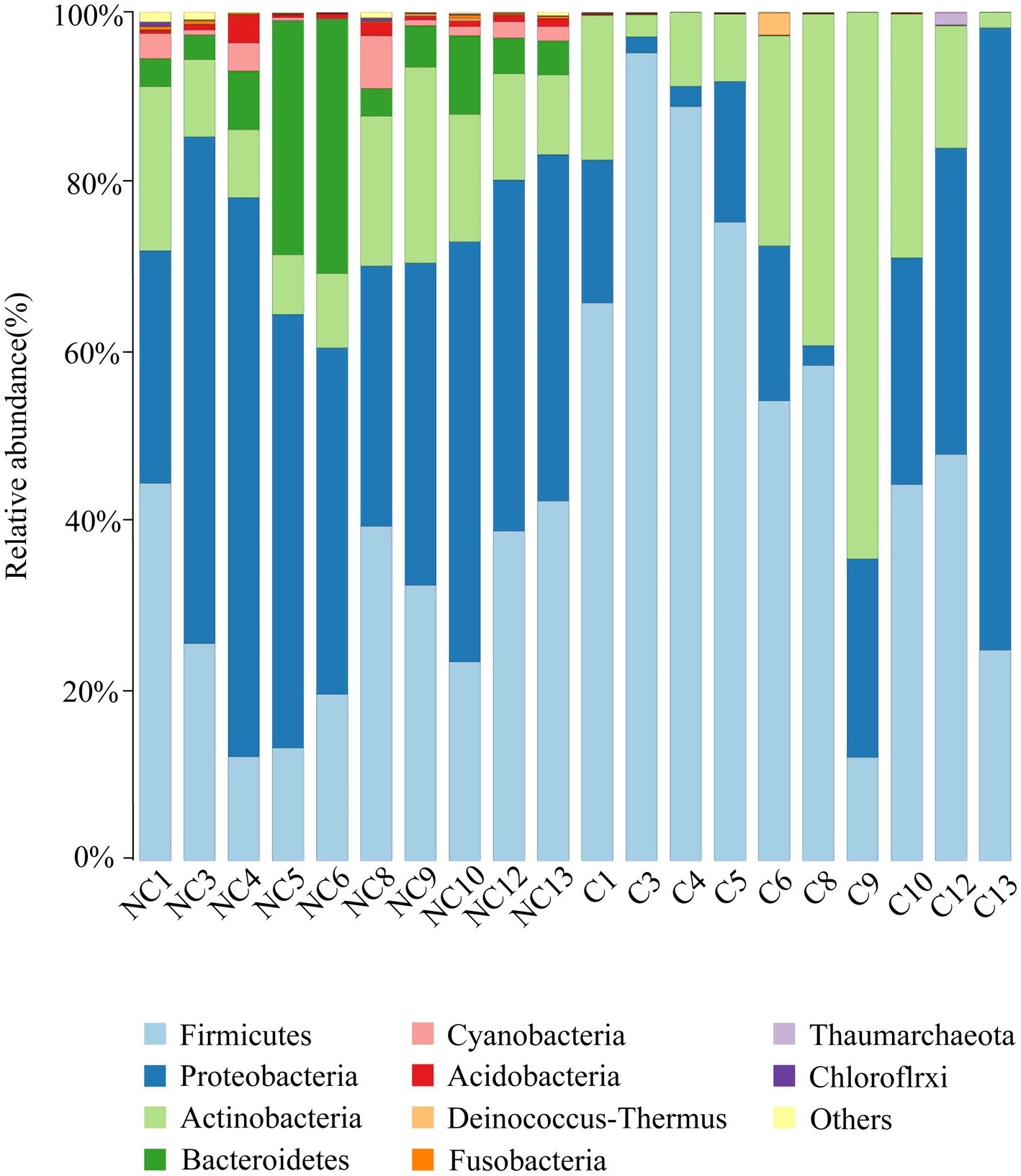
Figure 4. Microbial composition at the phylum level of each sample (top 10 taxon). NC (non-culture group): using next-generation sequencing methods; C (pure culture group): using cultivation methods. The same number represents from the same sample.
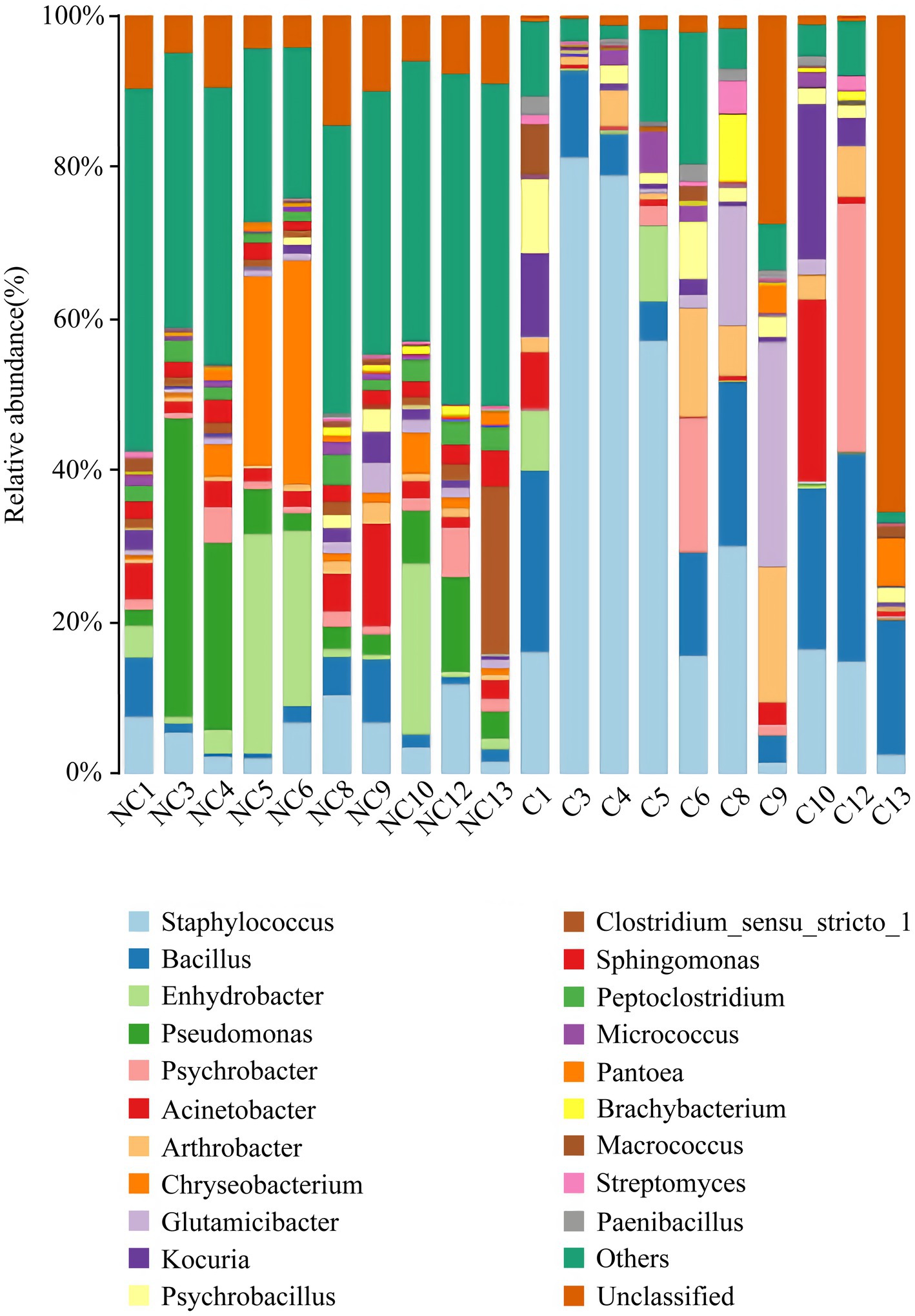
Figure 5. Microbial composition at the genus level of each sample (top 20 taxon). NC (non-culture group): using next-generation sequencing methods; C (pure culture group): using cultivation methods. The same number represents from the same sample.
There was a total of 72 genera with a relative abundance greater than 0.1% in the non-cultivation group, and except for Endobacter, we were able to acquire 71 genera through cultivation. Among these, 29 genera (Supplementary Table S3) of strains were obtained through isolated and purified culture. Additionally, we also obtained strains of 9 genera through isolated and purified culture, whose relative abundance was less than 0.1% in the non-culture group.
Isolation of skin bacterial candidates to repair UV damage
We subsequently cultivated and isolated 340 single strains from these medias. Through sequencing the full-length of the 16S genes, a taxonomic map was generated. All isolations of strains were preliminarily divided into 4 phyla, 25 families, 42 genera, and 118 species (Table 1). We were able to cultivate 3 phyla (Firmicutes, Proteobacteria, Actinobacteria; n = 339), with Firmicutes (n = 175) and Actinobacteria (n = 148) being the most dominant (Figure 5). At the genus level, the majority of the isolated strains were Kocuria (n = 64), Staphylococcus (n = 54), Bacillus (n = 39), Micrococcus (n = 28), Macrococcus (n = 19), Pantoea (n = 16), Rhodococcus (n = 12), and Paenibacillus (n = 12). Furthermore, combining with results of Figure 2 and references, several bacterial candidates that may repair UV damage were selected for the below animal trial (Supplementary Table S3).
Animal trial to test UV-damage repair isolations
To test UV damage repair functions of isolations, bacterial suspension of Arthrobacter gandavensis, Bacillus psychrosaccharolyticus, Pantoea eucrina, Paenibacillus amylolyticus and Paenibacillus terrae were applied to the mice skin hurt by UV lamp. The skin of all the negative control mice visually thickened, and numerous inflammatory cells infiltrated in the dermis and subcutaneous tissue layers. The skin pathological changes in experimental groups (A, B, D, E) were similar to those in the negative control group (Supplementary Figure S2). We scored the skin pathological changes of each mouse in the groups (Figures 1A–D) and found that experimental group C (Pantoea eucrina) had the lowest pathological score except blank control group, indicating the lowest degree of skin damage (for detailed score statistics, Supplementary Table S7) (Figure 1E). Notably, there was a significant difference (p = 0.027) in the skin pathological section scores between the experimental group C and the negative control group, with only a small amount of inflammatory cell infiltration in the dermis and subcutaneous tissue layers (Figure 1B). Therefore, the bacteria in group C (Pantoea eucrina) have the potential to repair skin UV damage.
Validation of UV-resistant functions of Pantoea eucrina KBFS172
To further investigate whether the Pantoea eucrina strain or its metabolites could repair skin UV damage, a mice trial #2 was conducted. Firstly, in visual observation, the skin of the mice in the BS group (bacterial suspension: Suspension of Pantoea eucrina) appeared chapped erythema, while other control groups (NC group: negative control, BL group: bacterial lysate: lysate of Pantoea eucrina, and VE group: vitamin E, Supplementary Table S4) showed varying degrees of ulceration, dandruff production, and scarring. From the appearance of the mice skin, it appeared that the skin damage situation of the mice in the BC group was indeed better than that in the NC group (Figures 6B,C). The appearance and tissue pathological damage scores in BS groups were the best compared to NC Group, BL Group, and VE group (Figure 6), and the trial details were documented in Supplementary Tables S4–S6, S8, S9, and Supplementary Figure S2. In addition, we observed that BS groups had a better UV-resistant effect than NC and BL groups, indicating that Pantoea eucrina strain plays critical roles in UV-resistant rather than its metabolites.
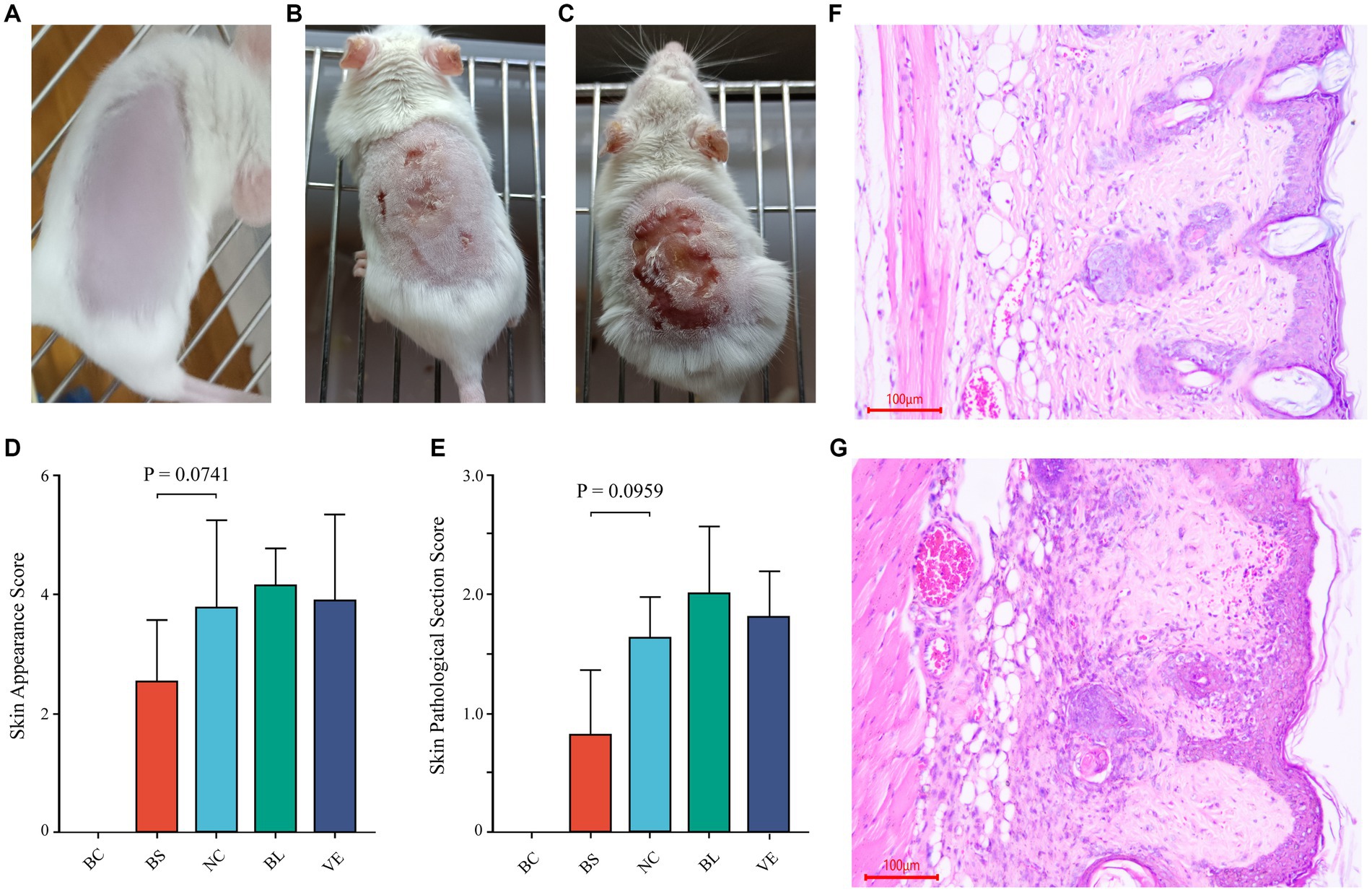
Figure 6. Mice trial #2: pathological changes of skin and sections, and scoring of skin pathological changes. (A) The healthy mouse skin appearance (BC). (B) Appearance of the skin of the mice smeared with BS. (C) Appearance of the skin of the mice in NC. (D, E) Statistical bar graph of skin appearance and skin pathological damage score in each group. (F) Representative diagram of skin section in the BS. (G) Representative diagram of skin section in the NC. BC, blank control; BS, bacterial suspension: Suspension of Pantoea eucrina; NC, negative control; BL, bacterial lysate: lysate of Pantoea eucrina; VE, vitamin E.
Functional annotation of Pantoea eucrina KBFS172 using whole genome sequencing
Through whole genome sequencing technology, a total of 4,053,112 bases of Pantoea eucrina KBFS172 was generated, which included 1 chromosome sequence and 4 plasmid sequences, and predicted 3,871 genes (Supplementary Table S10). A total of 107 potential functional pathways were recognized and quantified, which included the metabolic pathways of carotenoid synthesis, ascorbate metabolism, geraniol degradation and streptomycin biosynthesis, etc. Carotenoids have been shown to act as an antioxidant against oxidative stress and increase bacterial tolerance to UV radiation (Ordenes-Aenishanslins et al., 2016; Portero et al., 2019; Reis-Mansur et al., 2019; Sedlacek et al., 2019). In this study, Pantoea eucrina KBFS172 had a large number of genes enriched in the metabolic pathway of carotenoid biosynthesis (Supplementary Figure S4, S5), among which beta-Carotene biosynthesis module was particularly prominent (Figure 7A). In addition, 6 enzymes were annotated to be closely related to carotenoid biosynthesis, including geranylgeranyl pyrophosphate synthase (crt_E; GE03214), zeaxanthin glucosyltransferase (crt_X; GE03215), lycopene cyclase (crt_Y; GE03216), phytoene dehydrogenase (crt_I; GE03217), phytoene synthase (crt_B; GE03218), and beta-carotene hydroxylase (crt_Z; GE03219). Through consultation of the key gene set for carotenoid synthesis in Pantoea (Wang et al., 2007; Mohammadi et al., 2012; Choi et al., 2021), we speculated that these 6 enzymes were involved in the pathway of carotenoid biosynthesis in the order shown in Figure 7B.
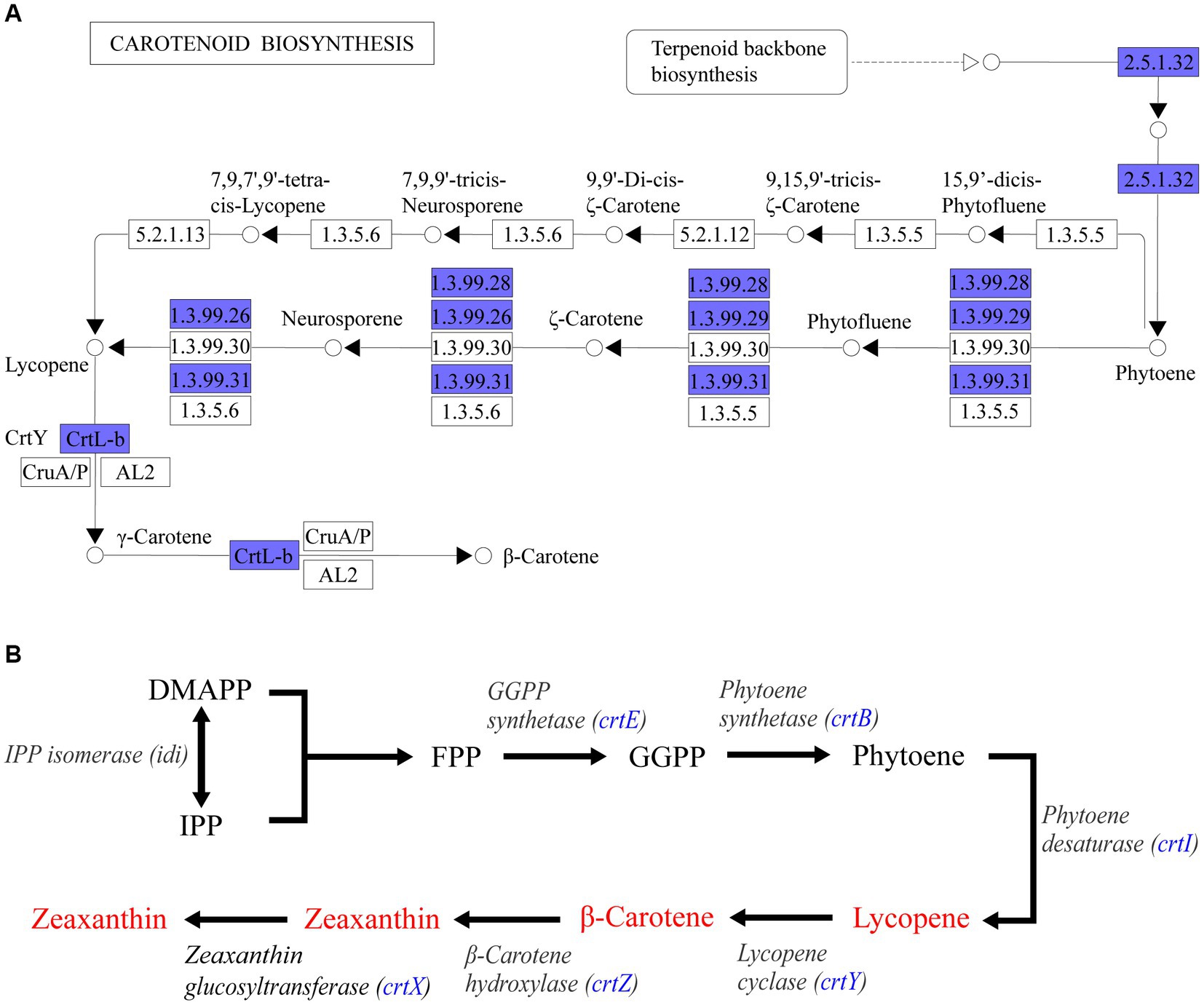
Figure 7. KEGG pathway enrichment analysis annotation results. (A) Beta-Carotene biosynthesis module (M00097) in carotenoid biosynthesis metabolic pathway. Blue filled were the annotated genes in Pantoea eucrina KBFS172. (B) Sketch of carotenoid synthesis pathway inferred from P. agglomerans KFS-9 and Pantoea ananatis. The blue filled was the enzyme annotated by Pantoea eucrina KBFS172.
Discussion
Although many studies focus on isolations of beneficial bacteria of gut microbiota (Deng et al., 2023; Xu et al., 2023; Zhao et al., 2023), the crucial role of skin microbes in humans have been highlighted. Interactions between certain skin microbiome and host cells can result in functional changes in the latter. Past research has shown that skin microbes are involved in anti-inflammatory processes, maintenance of skin homeostasis, and regulation of the immune system in both humans and animals (Ridaura et al., 2018).
Four dominant bacterial phyla in the human skin microbiota had been identified, including Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes (Grice et al., 2008; Li et al., 2019). Changes in the living environment could affect the skin microbial composition. A previous study found that the relative abundance of Bacteroidetes and Firmicutes was higher at high altitudes (Li et al., 2019) than at low altitudes (Grice et al., 2008). This difference suggests that altitude may be an important driving force for skin microbiota composition (Gao et al., 2007; Fierer et al., 2008; Grice et al., 2009). Our own results showed that the relative abundance of continued to increase compared to the previous study. Therefore, Firmicutes might be beneficial for humans living at high altitudes. At the genus level, previous studies have found the resident microbiota in human skin, such as Propionibacterium, Corynebacterium, Staphylococcus, Streptococcus, Acinetobacter, Colwiebacteria, and Enhydrobacter (Gao et al., 2007; Fierer et al., 2008; Grice et al., 2009; Leung et al., 2015). Propionibacterium and Staphylococcus have been found to be the predominant species in sebaceous sites (Grice et al., 2008). However, in our study, Propionibacterium was not found to be predominant and its relative abundance was less than 1%, suggesting that it may not be able to tolerate the high-altitude environment. Interestingly, we found two genera, Pseudomonas (10.20%, Proteobacteria) and Chryseobacterium (6.90%, Bacteroidetes), with high relative abundance that had rarely been found at such levels in previous studies on human skin. This indicates that these two genera may be tolerant to high-altitude environments. Previous studies (Seelam et al., 2021) have demonstrated that Pseudomonas has the potential ability to provide protective effects mediated by melanin as a sunscreen agent against UV-B radiation. However, Pseudomonas has also been reported as an opportunistic pathogen of human skin (Spernovasilis et al., 2021), Chryseobacterium has been less studied on human skin, but a few studies (Gurav and Jadhav, 2013; Venil et al., 2015; Hui et al., 2019) have reported that its metabolite flexirubins can treat chronic skin diseases, while other studies (Nulens et al., 2001; Cascio et al., 2005) have shown that it is closely associated with some human diseases. Therefore, further research is needed to elucidate the specific functions of these two genera at the species level.
In our study, we employed the cultivation method using five different media and were able to obtain approximately 66.83% (961/1,438) of operational taxonomic units (OTUs). However, in the non-cultured group, we observed that five genera, including Pseudomonas, Chryseobacterium, Clostridium sensu stricto 1, Sphingomonas, and Peptoclostridium, had a relative abundance greater than 1% but were uncultured. These genera need to be further cultivated using other media, particularly Pseudomonas (isolated referencing (Ravi et al., 2018)) and Chryseobacterium (isolated referencing (Nishioka et al., 2016)), which are speculated from our results to be closely associated with high altitude. The skin bacteria that we were able to cultivate were dominated by Firmicutes, with most isolates belonging to Staphylococcus (30.81%) followed by Bacillus (15.19%). These bacteria were relatively common (Timm et al., 2020; Fleming et al., 2021). Certainly, culturability is closely related to the culture conditions. Thus, more culture media and different culture conditions can be employed to further cultivate uncultivated bacteria.
Until now, there have been relatively few studies conducted on the skin microbiota of Tibetans living on the plateau. Our study has provided insight into the structure of the skin microbiota of Tibetans living on the plateau, with a focus on the culturomics of extremely rare high-altitude human skin microbes. This study serves as a reference for future investigations into human skin microbes. Additionally, we observed that approximately 66% of plateau Tibetan skin microbes could be cultivated using five different media (Supplementary Table S2). Although some of the isolated strains may be more commonly associated with soil or environmental sources, they were nonetheless found on the skin of healthy individuals. The contribution of these strains to the function of the skin microbiome is unknown, and further research in this area is warranted.
To further explore the function of skin microbes in plateau Tibetans and identify strains that can repair UV damage, we conducted mice trials using five selected strains from pure culture based on previous studies. Ultimately, we identified a strain (Pantoea eucrina KBFS172) that showed ability in repairing photodamage. Previous studies have demonstrated the photodamage repair function of skin bacteria, and they have multiple repair mechanisms, which have different repair mechanisms. Some studies have found that the metabolites of certain microorganisms can achieve the function of photodamage repair or alleviation, such as bioactive peptides (Wang et al., 2022), Vitamin C, ferulic acid, and phloretin, etc. (Oresajo et al., 2008). In our study, Pantoea eucrina was identified to associate with the metabolism of carotenoids. Carotenoids have been shown to have anti-oxidative stress properties, increase bacterial UV radiation tolerance (Ordenes-Aenishanslins et al., 2016; Reis-Mansur et al., 2019), and found that co-cultivation of carotenoids with bacteria can enhance the UV radiation tolerance of sensitive bacteria (Portero et al., 2019; Sedlacek et al., 2019). Additionally, we also found other species of the genus Pantoea, which could also produce carotenoids, such as Pantoea ananatis, Pantoea stewartii subsp. Stewartii, and P. agglomerans Eho10 (Mohammadi et al., 2012; Sedlacek et al., 2019; Choi et al., 2021). This indicated that the Pantoea genus may have excellent potential for repairing photodamage. Moreover, Micrococcus (Hofer et al., 2011), cyanobacteria (Stege, 2001), and Lactobacillus acidophilus (Im et al., 2018) can also repair skin UV damage by synthesizing DNA damage repair enzymes or photolytic enzymes. Therefore, skin microbes can reduce the damage of ultraviolet rays to human skin in a variety of ways.
Conclusion
In conclusion, we characterized the skin microbiota of humans living in high altitude regions of China using 16S rRNA gene sequencing and culturomics methods. We identified and isolated strains that were effective in repairing UV damage through mouse trials, and obtained a potentially functional strain. The function of the strain was then verified by analyzing the whole-genome sequencing results. These findings gave the insights for further studies to explore the potential of skin-associated microbial communities in high altitude adaptation of humans, and offer new insights into the development of human skin probiotic products to resist skin diseases, such as skin cancer or sunburn.
Data availability statement
The rumen metagenome sequences were deposited into NCBI Sequence Read Archive (SRA) under the accession number PRJNA1002829. The other sequences data in the study are included in the article/Supplementary material, and further inquiries can be directed to the corresponding authors.
Ethics statement
The experiment protocol was approved by the Animal Ethics and Humane Animal Care of the Foshan University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
ZZ: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. HR: Investigation, Writing – original draft. YH: Investigation, Writing – review & editing. FD: Resources, Visualization, Writing – review & editing. BZ: Investigation, Resources, Validation, Visualization, Writing – original draft. JC: Supervision, Validation, Visualization, Writing – review & editing. YL: Conceptualization, Funding acquisition, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project was supported by National Natural Science Foundation of China (32170430), Guangdong Provincial Key Laboratory of Animal Molecular Design and Precise Breeding (2019B030301010), and Key Laboratory of Animal Molecular Design and Precise Breeding of Guangdong Higher Education Institutes (2019KSYS011).
Acknowledgments
We greatly appreciate Tao Zhang, Xingyin Zhang, Donghao Liu, and Peini Wang for participating in animal trials and animal sample collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1273902/full#supplementary-material
References
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Armstrong, B. K., Kricker, A., and Biology, P. B. (2001). The epidemiology of UV induced skin cancer. J. Photochem. Photobiol. B Biol. 63, 8–18. doi: 10.1016/S1011-1344(01)00198-1
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Cannone, J. J., Subramanian, S., Schnare, M. N., Collett, J. R., D'souza, L. M., Du, Y., et al. (2002). The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformat. 3, 1–31. doi: 10.1186/1471-2105-3-2
Cascio, A., Stassi, G., Costa, G. B., Crisafulli, G., Rulli, I., Ruggeri, C., et al. (2005). Chryseobacterium indologenes bacteraemia in a diabetic child. J. Med. Microbiol. 54, 677–680. doi: 10.1099/jmm.0.46036-0
Choi, O., Kang, B., Lee, Y., Lee, Y., and Kim, J. (2021). Pantoea ananatis carotenoid production confers toxoflavin tolerance and is regulated by Hfq-controlled quorum sensing. Microbiology 10:e1143. doi: 10.1002/mbo3.1143
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. J. B. (2005). Blast2go: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Dashti, A. A., Jadaon, M. M., Abdulsamad, A. M., and Dashti, H. (2009). Heat treatment of bacteria: a simple method of DNA extraction for molecular techniques. Kuwait Med. J. 41, 117–122.
Deng, F., Wang, C., Li, D., Peng, Y., Deng, L., Zhao, Y., et al. (2023). The unique gut microbiome of giant pandas involved in protein metabolism contributes to the host’s dietary adaption to bamboo. Microbiome 11:180. doi: 10.1186/s40168-023-01603-0
Fierer, N., Hamady, M., Lauber, C. L., and Knight, R. (2008). The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. U. S. A. 105, 17994–17999. doi: 10.1073/pnas.0807920105
Fleming, E., Pabst, V., Scholar, Z., Xiong, R., Voigt, A. Y., Zhou, W., et al. (2021). Cultivation of common bacterial species and strains from human skin, oral, and gut microbiota. BMC Microbiol. 21, 278–216. doi: 10.1186/s12866-021-02314-y
Gallo, R. L. (2017). Human skin is the largest epithelial surface for interaction with microbes. J. Invest. Dermatol. 137, 1213–1214. doi: 10.1016/j.jid.2016.11.045
Gao, Z., Tseng, C. H., Pei, Z., and Blaser, M. J. (2007). Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. U. S. A. 104, 2927–2932. doi: 10.1073/pnas.0607077104
Gao, Z., Tseng, C. H., Strober, B. E., Pei, Z., and Blaser, M. J. (2008). Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS One 3:e2719. doi: 10.1371/journal.pone.0002719
Grice, E. A., Kong, H. H., Conlan, S., Deming, C. B., Davis, J., Young, A. C., et al. (2009). Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192. doi: 10.1126/science.1171700
Grice, E. A., Kong, H. H., Renaud, G., Young, A. C., Program, N. C. S., Bouffard, G. G., et al. (2008). A diversity profile of the human skin microbiota. Genome Res. 18, 1043–1050. doi: 10.1101/gr.075549.107
Gurav, R. G., and Jadhav, J. P. (2013). Biodegradation of keratinous waste by Chryseobacterium sp. RBT isolated from soil contaminated with poultry waste. J. Basic Microbiol. 53, 128–135. doi: 10.1002/jobm.201100371
Hofer, A., Legat, F. J., Gruber-Wackernagel, A., Quehenberger, F., and Wolf, P. (2011). Topical liposomal Dna-repair enzymes in polymorphic light eruption. Photochem. Photobiol. Sci. 10, 1118–1128. doi: 10.1039/c1pp05009e
Hui, N., Gronroos, M., Roslund, M. I., Parajuli, A., Vari, H. K., Soininen, L., et al. (2019). Diverse environmental microbiota as a tool to augment biodiversity in urban landscaping materials. Front. Microbiol. 10:536. doi: 10.3389/fmicb.2019.00536
Hyatt, D., Chen, G.-L., Locascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformat. 11, 1–11. doi: 10.1186/1471-2105-11-119
Im, A. R., Lee, B., Kang, D. J., and Chae, S. (2018). Skin moisturizing and Antiphotodamage effects of Tyndallized Lactobacillus acidophilus IDCC 3302. J. Med. Food 21, 1016–1023. doi: 10.1089/jmf.2017.4100
Jablonski, N. G., and Chaplin, G. (2010). Colloquium paper: human skin pigmentation as an adaptation to UV radiation. Proc. Natl. Acad. Sci. U. S. A. 107, 8962–8968. doi: 10.1073/pnas.0914628107
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., and Hattori, M. (2004). The Kegg resource for deciphering the genome. Nucleic Acids Res. 32, 277D–2280D. doi: 10.1093/nar/gkh063
Knackstedt, R., Knackstedt, T., and Gatherwright, J. (2020). The role of topical probiotics in skin conditions: a systematic review of animal and human studies and implications for future therapies. Exp. Dermatol. 29, 15–21. doi: 10.1111/exd.14032
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Leung, M. H., Wilkins, D., and Lee, P. K. (2015). Insights into the pan-microbiome: skin microbial communities of Chinese individuals differ from other racial groups. Sci. Rep. 5:11845. doi: 10.1038/srep11845
Li, H., Wang, Y., Yu, Q., Feng, T., Zhou, R., Shao, L., et al. (2019). Elevation is associated with human skin microbiomes. Microorganisms 7:611. doi: 10.3390/microorganisms7120611
Massa, S., Caruso, M., Trovatelli, F., and Tosques, M. (1998). Comparison of plate count agar and R2A medium for enumeration of heterotrophic bacteria in natural mineral water. World J. Microbiol. Biotechnol. 14, 727–730. doi: 10.1023/A:1008893627877
Mohammadi, M., Burbank, L., and Roper, M. C. (2012). Biological role of pigment production for the bacterial phytopathogen Pantoea stewartii subsp. stewartii. Appl. Environ. Microbiol. 78, 6859–6865. doi: 10.1128/Aem.01574-12
Muletz Wolz, C. R., Yarwood, S. A., Campbell Grant, E. H., Fleischer, R. C., and Lips, K. R. (2018). Effects of host species and environment on the skin microbiome of plethodontid salamanders. J. Anim. Ecol. 87, 341–353. doi: 10.1111/1365-2656.12726
Nicholson, W. L., Krivushin, K., Gilichinsky, D., and Schuerger, A. C. (2013). Growth of Carnobacterium spp. from permafrost under low pressure, temperature, and anoxic atmosphere has implications for earth microbes on Mars. Proc. Natl. Acad. Sci. 110, 666–671. doi: 10.1073/pnas.1209793110
Nishioka, T., Elsharkawy, M. M., Suga, H., Kageyama, K., Hyakumachi, M., and Shimizu, M. (2016). Development of culture medium for the isolation of Flavobacterium and Chryseobacterium from rhizosphere soil. Microbes Environ. 31, 104–110. doi: 10.1264/jsme2.Me15144
Nulens, E., Bussels, B., Bols, A., Gordts, B., and Van Landuyt, H. W. (2001). Recurrent bacteremia by Chryseobacterium indologenes in an oncology patient with a totally implanted intravascular device. Clin. Microbiol. Infect. 7, 391–393. doi: 10.1046/j.1198-743x.2001.00273.x
Ordenes-Aenishanslins, N., Anziani-Ostuni, G., Vargas-Reyes, M., Alarcon, J., Tello, A., and Perez-Donoso, J. M. (2016). Pigments from UV-resistant Antarctic bacteria as photosensitizers in dye sensitized solar cells. J. Photochem. Photobiol. B 162, 707–714. doi: 10.1016/j.jphotobiol.2016.08.004
Oresajo, C., Stephens, T., Hino, P. D., Law, R. M., Yatskayer, M., Foltis, P., et al. (2008). Protective effects of a topical antioxidant mixture containing vitamin C, ferulic acid, and phloretin against ultraviolet-induced photodamage in human skin. J. Cosmet. Dermatol. 7, 290–297. doi: 10.1111/j.1473-2165.2008.00408.x
Osawa, R. (1990). Formation of a clear zone on tannin-treated brain heart infusion agar by a Streptococcus sp. isolated from feces of koalas. Appl. Environ. Microbiol. 56, 829–831.
Pagnanelli, F., Petrangeli Papini, M., Toro, L., Trifoni, M., and Veglio, F. (2000). Biosorption of metal ions on Arthrobacter sp.: biomass characterization and biosorption modeling. Environ. Sci. Technol. 34, 2773–2778. doi: 10.1021/es991271g
Paulson, J. N., Stine, O. C., Bravo, H. C., and Pop, M. (2013). Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–1202. doi: 10.1038/nmeth.2658
Peeters, K., Hodgson, D. A., Convey, P., and Willems, A. (2011). Culturable diversity of heterotrophic bacteria in Forlidas pond (Pensacola Mountains) and Lundström Lake (Shackleton range), Antarctica. Microb. Ecol. 62, 399–413. doi: 10.1007/s00248-011-9842-7
Portero, L. R., Alonso-Reyes, D. G., Zannier, F., Vazquez, M. P., Farias, M. E., Gartner, W., et al. (2019). Photolyases and Cryptochromes in UV-resistant Bacteria from high-altitude Andean lakes. Photochem. Photobiol. 95, 315–330. doi: 10.1111/php.13061
Ravi, K., García-Hidalgo, J., Nöbel, M., Gorwa-Grauslund, M. F., and Lidén, G. (2018). Biological conversion of aromatic monolignol compounds by a Pseudomonas isolate from sediments of the Baltic Sea. AMB Express 8, 32–14. doi: 10.1186/s13568-018-0563-x
Reasoner, D. J., Geldreich, E. J. A., and Microbiology, E. (1985). A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49, 1–7. doi: 10.1128/aem.49.1.1-7.1985
Reis-Mansur, M., Cardoso-Rurr, J. S., Silva, J., De Souza, G. R., Cardoso, V. D. S., Mansoldo, F. R. P., et al. (2019). Carotenoids from UV-resistant Antarctic Microbacterium sp. Lemmj01. Sci. Rep. 9:9554. doi: 10.1038/s41598-019-45840-6
Ridaura, V. K., Bouladoux, N., Claesen, J., Chen, Y. E., Byrd, A. L., Constantinides, M. G., et al. (2018). Contextual control of skin immunity and inflammation by Corynebacterium. J. Exp. Med. 215, 785–799. doi: 10.1084/jem.20171079
Ross, A. A., Doxey, A. C., and Neufeld, J. D. (2017). The skin microbiome of cohabiting couples. mSystems 2:43. doi: 10.1128/mSystems.00043-17
Roszkowski, W., Roszkowski, K., Ko, H., Beuth, J., and Jeljaszewicz, J. (1990). Immunomodulation by propionibacteria. Zentralblatt für Bakteriologie 274, 289–298. doi: 10.1016/S0934-8840(11)80686-9
Sedlacek, I., Pantucek, R., Kralova, S., Maslanova, I., Holochova, P., Stankova, E., et al. (2019). Hymenobacter amundsenii sp. nov. resistant to ultraviolet radiation, isolated from regoliths in Antarctica. Syst. Appl. Microbiol. 42, 284–290. doi: 10.1016/j.syapm.2018.12.004
Seelam, S. D., Agsar, D., Halmuthur, M. S., Reddy Shetty, P., Vemireddy, S., Reddy, K. M., et al. (2021). Characterization and photoprotective potentiality of lime dwelling Pseudomonas mediated melanin as sunscreen agent against UV-B radiations. J. Photochem. Photobiol. B 216:112126. doi: 10.1016/j.jphotobiol.2021.112126
Shu, M., Wang, Y., Yu, J., Kuo, S., Coda, A., Jiang, Y., et al. (2013). Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PLoS One 8:e55380. doi: 10.1371/journal.pone.0055380
Spernovasilis, N., Psichogiou, M., and Poulakou, G. (2021). Skin manifestations of Pseudomonas aeruginosa infections. Curr. Opin. Infect. Dis. 34, 72–79. doi: 10.1097/qco.0000000000000717
Stege, H. (2001). Effect of xenogenic repair enzymes on photoimmunology and photocarcinogenesis. J. Photochem. Photobiol. B Biol. 65, 105–108. doi: 10.1016/s1011-1344(01)00246-9
Swindell, S. R., and Plasterer, T. N. (1997). Seqman. Sequence data analysis guidebook. Berlin: Springer.
Tang, Y. W., and Stratton, C. W. (2010). Staphylococcus aureus: an old pathogen with new weapons. Clin. Lab. Med. 30, 179–208. doi: 10.1016/j.cll.2010.01.005
Timm, C. M., Loomis, K., Stone, W., Mehoke, T., Brensinger, B., Pellicore, M., et al. (2020). Isolation and characterization of diverse microbial representatives from the human skin microbiome. Microbiome 8, 58–12. doi: 10.1186/s40168-020-00831-y
Van Der Weele, C. M., Spollen, W. G., Sharp, R. E., and Baskin, T. I. (2000). Growth of Arabidopsis thaliana seedlings under water deficit studied by control of water potential in nutrient-agar media. J. Exp. Bot. 51, 1555–1562. doi: 10.1093/jexbot/51.350.1555
Venil, C. K., Zakaria, Z. A., and Ahmad, W. A. (2015). Optimization of culture conditions for flexirubin production by Chryseobacterium artocarpi Cect 8497 using response surface methodology. Acta Biochim. Pol. 62, 185–190. doi: 10.18388/abp.2014_870
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Wang, H., Jiang, X., Mu, H., Liang, X., and Guan, H. (2007). Structure and protective effect of exopolysaccharide from P. agglomerans strain KFS-9 against UV radiation. Microbiol. Res. 162, 124–129. doi: 10.1016/j.micres.2006.01.011
Wang, Y., Kuo, S., Shu, M., Yu, J., Huang, S., Dai, A., et al. (2014). Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: implications of probiotics in acne vulgaris. Appl. Microbiol. Biotechnol. 98, 411–424. doi: 10.1007/s00253-013-5394-8
Wang, S., Yang, M., Yin, S., Zhang, Y., Zhang, Y., Sun, H., et al. (2022). A new peptide originated from amphibian skin alleviates the ultraviolet B-induced skin photodamage. Biomed. Pharmacother. 150:112987. doi: 10.1016/j.biopha.2022.112987
Wei, H., Zhang, X., Wang, Y., and Lebwohl, M. (2002). Inhibition of ultraviolet light-induced oxidative events in the skin and internal organs of hairless mice by isoflavone genistein. Cancer Lett. 185, 21–29. doi: 10.1016/S0304-3835(02)00240-9
Wickham, H. J. W. I. R. C. S. (2011). ggplot2. Computational Statistics 3, 180–185. doi: 10.1002/wics.147
Xu, W., Yu, J., Yang, Y., Li, Z., Zhang, Y., Zhang, F., et al. (2023). Strain-level screening of human gut microbes identifies Blautia producta as a new anti-hyperlipidemic probiotic. Gut Microbes 15:2228045. doi: 10.1080/19490976.2023.2228045
Yoshio, H., Lagercrantz, H., Gudmundsson, G. H., and Agerberth, B. (2004). First line of defense in early human life. Semin. Perinatol. 28, 304–311. doi: 10.1053/j.semperi.2004.08.008
Zeng, B., Zhao, J., Guo, W., Zhang, S., Hua, Y., Tang, J., et al. (2017). High-altitude living shapes the skin microbiome in humans and pigs. Front. Microbiol. 8:1929. doi: 10.3389/fmicb.2017.01929
Keywords: skin microbiota, skin disease, UV damage, high altitude, culturomics, probiotic
Citation: Zhang Z, Ran H, Hua Y, Deng F, Zeng B, Chai J and Li Y (2023) Screening and evaluation of skin potential probiotic from high-altitude Tibetans to repair ultraviolet radiation damage. Front. Microbiol. 14:1273902. doi: 10.3389/fmicb.2023.1273902
Edited by:
Yijuan Xu, South China Agricultural University, ChinaReviewed by:
Yongyi Shen, South China Agricultural University, ChinaDaniela Pinto, Giuliani S.p.A., Italy
Copyright © 2023 Zhang, Ran, Hua, Deng, Zeng, Chai and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Li, eWluZ2xpQGZvc3UuZWR1LmNu; Jianmin Chai, amNoYWlAdWFyay5lZHU=; Bo Zeng, YXBvbGxvYm92ZXlAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Zhihao Zhang
Zhihao Zhang Haixia Ran3†
Haixia Ran3† Feilong Deng
Feilong Deng Bo Zeng
Bo Zeng Jianmin Chai
Jianmin Chai Ying Li
Ying Li