- 1Donghai Laboratory, Zhoushan, Zhejiang, China
- 2Key Laboratory of Marine Ecosystem Dynamics, Second Institute of Oceanography, Ministry of Natural Resources, Hangzhou, Zhejiang, China
- 3Ocean College, Zhejiang University, Hangzhou, Zhejiang, China
- 4School of Oceanography, Shanghai Jiao Tong University, Shanghai, China
Ammonia-oxidizing archaea (AOA) and bacteria (AOB), nitrite-oxidizing bacteria (NOB), and complete ammonia oxidizers (comammox) are responsible for nitrification in nature; however, some groups have been reported to utilize labile-dissolved organic nitrogen (LDON) for satisfying nitrogen demands. To understand the universality of their capacity of LDON metabolism, we collected 70 complete genomes of AOA, AOB, NOB, and comammox from typical environments for exploring their potentials in the metabolism of representative LDON (urea, polyamines, cyanate, taurine, glycine betaine, and methylamine). Genomic analyses showed that urea was the most popular LDON used by nitrifiers. Each group harbored unique urea transporter genes (AOA: dur3 and utp, AOB: utp, and NOB and comammox: urtABCDE and utp) accompanied by urease genes ureABC. The differentiation in the substrate affinity of these transporters implied the divergence of urea utilization efficiency in nitrifiers, potentially driving them into different niches. The cyanate transporter (cynABD and focA/nirC) and degradation (cynS) genes were detected mostly in NOB, indicating their preference for a wide range of nitrogen substrates to satisfy high nitrogen demands. The lack of genes involved in the metabolism of polyamines, taurine, glycine betaine, and methylamines in most of nitrifiers suggested that they were not able to serve as a source of ammonium, only if they were degraded or oxidized extracellularly as previously reported. The phylogenetic analyses assisted with comparisons of GC% and the Codon Adaptation Index between target genes and whole genomes of nitrifiers implied that urea metabolic genes dur3 and ureC in AOA evolved independently from bacteria during the transition from Thaumarchaeota to AOA, while utp in terrestrial AOA was acquired from bacteria via lateral gene transfer (LGT). Cyanate transporter genes cynS and focA/nirC detected only in a terrestrial AOA Candidadus Nitrsosphaera gargensis Ga9.2 could be gained synchronously with Nitrospira of NOB by an ancient LGT. Our results indicated that LDON utilization was a common feature in nitrifiers, but metabolic potentials were different among nitrifiers, possibly being intensely interacted with their niches, survival strategies, and evolutions.
1 Introduction
Labile-dissolved organic nitrogen (LDON) compounds are the groups with low-molecule weights and rapid turnover rates in the environments (Sipler et al., 2013). They are generally produced from the degradation of proteins or released from primary producers (e.g., phytoplankton in the ocean), and preferentially taken up by heterotrophic bacteria as nitrogen sources (e.g., Liu et al., 2016, 2022b; Damashek et al., 2019). LDON compounds generally include dissolved free amino acids, urea, polyamines, methylamines, taurine, cyanate, and glycine betaine. Their uptake contributes significantly to bacterial nitrogen demands and enhances nitrogen cycling (e.g., Jørgensen, 2006; Liu et al., 2015, 2022a; Clifford et al., 2019). In addition, photoautotrophic phytoplankton in the ocean can also assimilate or oxidize LDON for acquiring nitrogen and energy, especially in N-limiting environments (Glibert et al., 2016). In recent years, more studies reveal that the chemoautotrophic prokaryotes involved in nitrification (e.g., Nitrospinae and Thaumarchaeota) may be capable of utilizing LDON for enhancing or sustaining the growth as well (Koch et al., 2015; Qin et al., 2017; Kitzinger et al., 2019, 2020).
The nitrification process is one of the most important steps in the nitrogen cycle driven by a complex microbial consortium (Voss et al., 2013), including ammonia-oxidizing archaea (AOA) and bacteria (AOB), nitrite-oxidizing bacteria (NOB), and complete ammonia oxidizer (comammox; He et al., 2018). They are key players in global nitrogen and carbon cycles (Bayer et al., 2019). AOA and AOB perform ammonia oxidation, the first and rate-limiting step of nitrification (Könneke et al., 2005), and NOB catalyze the second step of nitrification by oxidizing nitrite to nitrate (Daims et al., 2016). Comammox are capable of converting ammonia to nitrate in one step (Hu and He, 2017). AOA usually outcompete AOB for ammonia and play a major role in controlling ammonia oxidation in most environments due to their relatively higher affinity for ammonia (Stahl and de la Torre, 2012). They are mainly categorized into four phylogenetic lineages, namely, Nitrosopumilales (Group I.1a), “Ca. Nitrosotaleales” (Group I.1a-associated), Nitrososphaerales (Group I.1b), and “Ca. Nitrosocaldales” (Jung et al., 2022). AOB are commonly detected in ammonia-rich environments, such as sewage treatment plants, eutrophic freshwater, coastal waters, and soil (Soliman and Eldyasti, 2018). A total of five genera have been identified as AOB, in which Nitrosomonas, Nitrosospira, Nitrosovibrio, and Nitrosolobus belong to the subclass β-Proteobacteria and Nitrosococcus to the subclass γ-Proteobacteria. Among all known NOB, the genus Nitrospira appears to be most widespread and phylogenetically diverse in different habitats (Koch et al., 2015). Nitrospira strains are well adapted to low nitrite concentrations and form at least six phylogenetic lineages that are globally distributed in soils, oceans, freshwater, hot springs, etc. (Koch et al., 2015). Nitrospinae are the dominant marine NOB and can reach high abundances (up to ∼10% of the microbial community) in mesopelagic zones, oxygen minimum zones (OMZs), deep-sea waters, and sediments (Daims et al., 2016). The comammox Nitrospira are abundant in natural and engineered habitats. It is reported that comammox may functionally outcompete other canonical nitrifiers under highly oligotrophic conditions (Hu and He, 2017).
The capability of utilizing extracellular LDON may increase nitrogen assimilation and be beneficial for the production of energy and biomass of nitrifiers (Kitzinger et al., 2019). Ammonia oxidizers (AOM) have been suggested to utilize extracellular LDON as an alternative source of ammonia under the situation of ammonia limitation (Sliekers et al., 2004; Qin et al., 2017), while NOB use them for reciprocal feeding with AOM (Palatinszky et al., 2015). The urea utilization has been detected in verified experiments for AOA strains Ca. Nitrososphaera gargensis Ga9.2, N. viennensis EN76, and Nitrosopumilus ureiphilus PS0 (Wetzel et al., 2011; Qin et al., 2014; Damashek et al., 2019), AOB strains Nitrosomonas oligotropha and N. ureae (Tourna et al., 2011; Spang et al., 2012; Qin et al., 2014), and NOB Nitrospira moscoviensis (Sliekers et al., 2004). The field samples collected from marine environments also reveal that AOA and Nitrospinae of NOB can incorporate urea-and cyanate-derived nitrogen at significantly higher rates than other microorganisms (Kitzinger et al., 2018, 2020). A recent study in the Gulf of Mexico found that AOA mainly used ammonium, while most of the cellular nitrogen-demand of Nitrospinae was met by the assimilation of urea and cyanate (Kitzinger et al., 2020). The alternative utilization of LDON for avoiding the competition with ammonia-oxidizing microbes may be a key factor for ecological success of NOB (Kitzinger et al., 2020). Moreover, the metagenome-assembled genomes (MAGs) of Nitrospinae encode ABC-type transporter of spermidine, amino acids, and peptides, an indication for their additional nitrogen sources for growth (Kitzinger et al., 2020). Therefore, the potential of nitrifiers in utilizing LDON could be related to their survival strategies.
Although a few studies have showed that nitrifying microbes are capable of utilizing LDON based on both laboratory experiments or genomic analysis (Tourna et al., 2011; Palatinszky et al., 2015; Qin et al., 2017), as more strains are identified from different habitats, little has been done to systematically catalog the metabolic potential of LDON of these nitrifiers from different environments for understanding their utilization mechanisms and strategies. Whether it is a common metabolic process or only occurs in certain environments needs more investigation. Moreover, since the availability of LDON increases rapidly as a consequence of anthropogenic impact, especially in estuary and coastal waters (Seitzinger et al., 2002), assessing the potentials of nitrifiers in the utilization of LDON can further explore the ecological role of LDON in the ecosystem. To fill this gap, we compared metabolic potentials of LDON among AOA, AOB, NOB, and comammox, and between marine and terrestrial taxa based on genomic analyses, to discuss mechanisms and strategies of LDON utilization by nitrifiers in different environments.
2 Materials and methods
2.1 Genomic information collection
The complete genome sequences of representative AOA (n = 46), AOB (n = 10), NOB (n = 12), and comammox (n = 2) strains were collected from National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/), Joint Genome Institute (JGI; https://jgi.doe.gov/), or Beijing Institute of Genomics Data Center (BIGD; https://ngdc.cncb.ac.cn/) according to the accession number (Supplementary Table S1). The dataset included all available genomes from isolated and enriched AOA strains, representatives of available genomes of AOB and NOB, and selected metagenomic-assembled genomes (MAGs, completeness >70%, low completeness could result in non-detection of target genes) of nitrifiers from extreme marine environments (Supplementary Table S1). Diverse habitats from both marine (e.g., sediment, estuary, coastal seawater, and deep-sea) and terrestrial (e.g., soil, freshwater, and wastewater treatment plant) environments were covered for the subsequent analysis and comparison (Supplementary Table S1).
2.2 Cell volume estimation
We recorded the shape, width, and length of cells for collected AOA, AOB, NOB, and comammox according to the description in the literature (Supplementary Table S2). The ratio of surface area and cell volume (SA/V) was estimated assuming a spherical cell based on equation (1) (Pachiadaki et al., 2017). The SA/V ratio of a rod-shaped cell was calculated with equation (2) (Boyde and Williams, 1971). Cell widths and lengths used in the formula were means of values in corresponding references (Supplementary Table S2).
where V is the cell volume, d is the cell diameter, h is the cell length, and SA is the surface area.
2.3 Gene collection and genome annotation
Amino acid sequences of key functional genes related to ammonia oxidation, and transport, biosynthesis, and degradation of selected LDON compounds (urea, polyamines, cyanate, taurine, glycine betaine, and methylamines) were collected from the NCBI (Figure 1; Supplementary Table S3) for the subsequent alignment with genomes of AOA, AOB, NOB, and comammox strains. They included genes encoding ammonia monooxygenase (amoABC, K10944-K10946; Zhang et al., 2023), bacteria-type urea ABC transporter (urtABCDE, K11959-K11963; Veaudor et al., 2019), a prokaryote-origin mammalian urea transporter (utp, K08717; Minocha et al., 2003; Levin et al., 2009; Spang et al., 2012), urea active transporter (dur3, K20989) generally detected in marine unicellular photosynthetic eukaryotes (Solomon et al., 2010), urease (ureABC, K01428-K01430; ureDFG, K03188-K03190; ureE, K03187; Veaudor et al., 2019), polyamine ABC transporter (spermidine-preferential ABC transporter, potABCD, K11069-K11072; putrescine ABC transporter, potFGHI, K11073-K11076; Mou et al., 2010), cyanate ABC transporter (cynABD, K15576, K15577, and K15579; Maeda and Omata, 2009), a formate–nitrite transporter (FNT, focA/nirC, K21990) functioning in cyanate assimilation in cyanobacteria (Maeda and Omata, 2009), cyanase (cynS, K01725; Taubert et al., 2017), taurine ABC transporter (tauACB, K15551, K15552, and K10831; Cook and Denger, 2006; Rohwerder, 2020), glycine betaine transporter (opuD, K05020; Wetzel et al., 2011; Boysen et al., 2022), trimethylamine (TMA) monooxygenase (tmm, K18277), glutamate-methylamine (GMA) synthetase (gmaS, K01949), N-Methyl-L-glutamate (NMG) synthase (mgsABC, K22081-K22083), NMG dehydrogenase (mgdABCD, K22084-K22087), methylamine dehydrogenase (mauAB, K15228, K15229) in methylamines metabolism (Chen, 2012; Taubert et al., 2017), and enzymes involved in polyamine biosynthesis (speA, K01585; speB, K01480; speC, K01581; speE, K00797; aguA, K10536; aguB, K12251) and catabolism (puuA, K09470; spuC, K12256; kauB, K12254; gabT, K07250; spdH, K00316; Mou et al., 2010, 2011), taurine degradation (tauD, K03119; tpa, K03851; xsc, K03852; pta, K13788; tauXY, K07255 and K07256; Cook and Denger, 2006; Rohwerder, 2020), glycine betaine synthesis (betAB, K00108, and K00130), and catabolism (gbcAB, K00479, and K21832; bhmt, K00544; grdHI, K21579, and K21578; cdh, K17735; Wetzel et al., 2011; Boysen et al., 2022). The genome-wide gene annotation was carried out through the website of Rapid Annotation using Subsystem Technology (RAST, https://rast.nmpdr.org/) with all collected genomes of AOA, AOB, NOB, and comammox. RAST annotation results and the NCBI database were used to identify homologs of targeted genes (Supplementary Table S3). All functional gene sequences were subjected to BLASTp. The amino acid sequence with a percent identity to the reference gene (Supplementary Table S3) greater than 40% was considered to be the homolog of the target gene (Pearson, 2013).
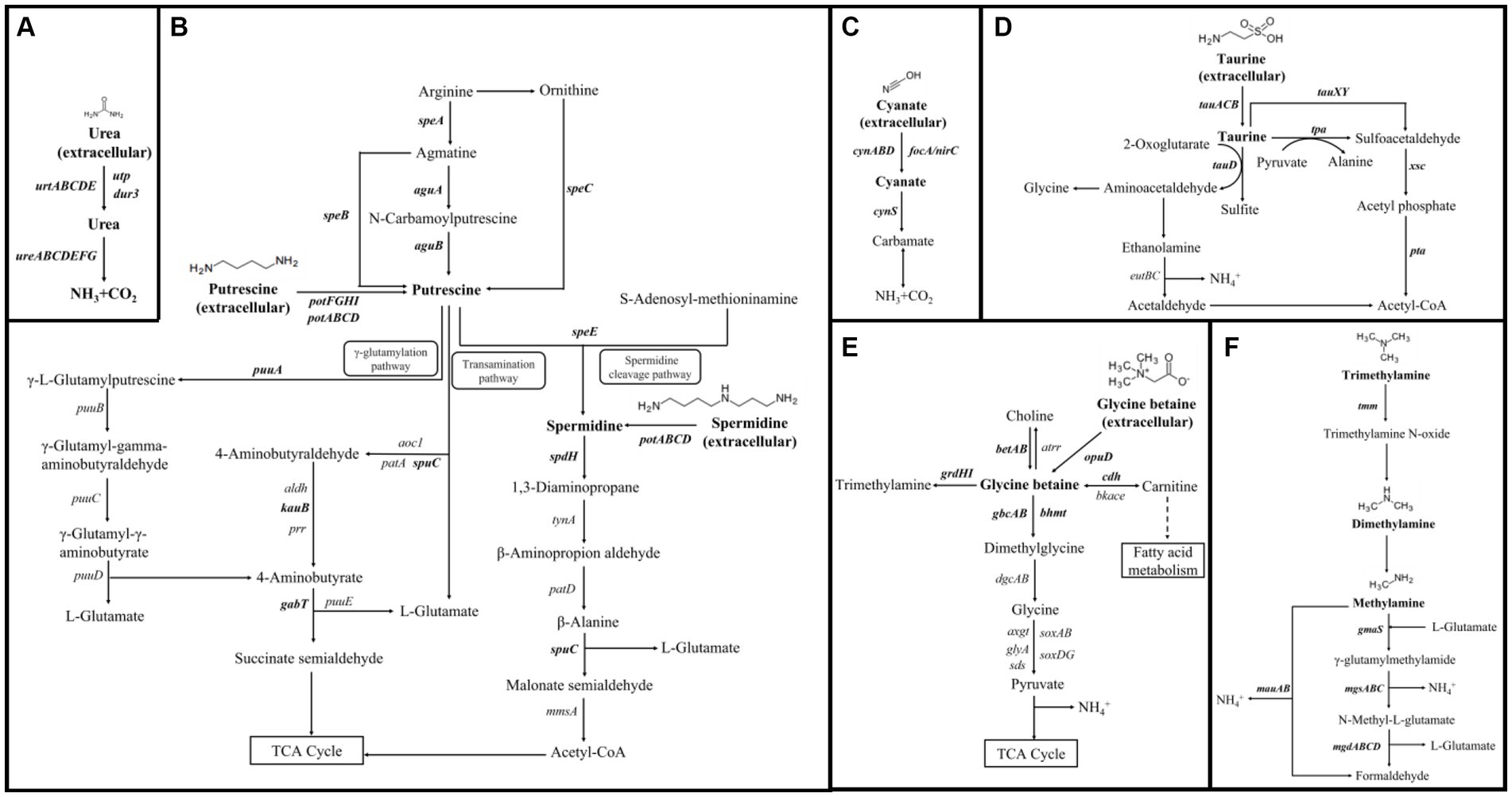
Figure 1. Metabolic pathways and products of representative labile-dissolved organic nitrogen (LDON), including (A) urea (Spang et al., 2012; Veaudor et al., 2019), (B) polyamines (Igarashi and Kashiwagi, 2010; Mou et al., 2011), (C) cyanate (Maeda and Omata, 2009; Palatinszky et al., 2015), (D) taurine (Cook and Denger, 2006; Engelberts et al., 2020; Rohwerder, 2020), (E) glycine betaine (Li et al., 2021; Boysen et al., 2022), and (F) methylamine (Chen, 2012; Taubert et al., 2017). Gene involved in metabolic processes are listed in italics. The LDON compounds and genes investigated in this study are highlighted in bold. Black dotted arrows represent the potential fates of molecules.
2.4 Phylogenetic analysis of key functional genes
To demonstrate the evolution of functional genes involved in LDON metabolism, phylogenetic trees were constructed with amino acid sequences for key genes by maximum likelihood based on the model of Le and Gascuel (2008) with 1,000 bootstrap replications using the software MEGA 7.0 (Kumar et al., 2016). The amino acid sequences were aligned using Clustal W (Thompson et al., 1994). The best model was used after the alignment (Hall, 2013). Models with the lowest Bayesian information criterion (BIC) score were considered to best describe the substitution pattern (Bast, 2013). A discrete gamma distribution was used to model differences of evolutionary rates among sites [five categories (+G)]. For utp and focA/nirC, the rate variation model allowed for some sites to be evolutionarily invariable ([+I]). All phylogenetic trees were drawn to scale with branch lengths measured in the number of substitutions per site.
The GC contents of key genes of selected AOA, AOB, NOB, and comammox, and outgroup species in phylogenetic analyses were calculated using GC Content Calculator,1 while those of whole genomes were obtained from the NCBI database.2 The values of the Codon Adaptation Index (CAI) of the same group of genes and whole genomes of selected nitrifiers and outgroup species were calculated using an online CAI calculator.3
2.5 Data and material availability
Genome sequence data are available in NCBI, JGI, or BIGD databases, and their accession numbers are listed in Supplementary Table S1. All other data products associated with this study are available from the corresponding authors upon request.
3 Results
3.1 Genomic and phenotypic characteristics of collected nitrifiers
Collected strains of AOA belonging to the genera Nitrosopumilus, Nitrosopelagicus, Nitrosomarinus, and Cenarchaeum were all of marine origin (n = 26). The genera Nitrosarchaeum, Nitrosotenuis, Nitrosotalea, Nitrososphaera, Nitrosocosmicus, and Nitrosocaldus were mostly from terrestrial environments, including hot springs, lakes, and soil (n = 20; Supplementary Table S1). Only strains Ca. Nitrosarchaeum limnium SFB1 and BG20 were enriched from marine environments (Supplementary Table S1). In AOB, all three Nitrosococcus strains were enriched from marine environments, and Nitrosomonas strains and Ca. Nitrosacidococcus tergens sp. RJ19 were terrestrial (Supplementary Table S1). Nitrospina gracilis, Nitrospira marina Nb-295, Ca. Nitrohelix vancouverensis, and Ca. Nitronauta litoralis of NOB were inhabited in marine environments, and two Nitrobacter strains, Ca. Nitrotoga arctica, and the rest of Nitrospira species including two comammox Ca. N. inopinata and Ca. N. kreftii were terrestrial origin (Supplementary Table S1).
Since functional and genomic characteristics of the comammox were similar to NOB, we grouped them into NOB for the subsequent analysis. The total genome length of AOB (1.81–4.08 Mb, median: 3.16 Mb, n = 10) was smaller than NOB (3.08–4.69 Mb, median: 3.91 Mb, n = 14; one-way ANOVA and Dunn’s method, p < 0.01) but larger than AOA (1.05–3.43 Mb, median: 1.85 Mb, n = 46; p < 0.01; Figure 2A; Supplementary Table S1). The GC contents of NOB ranging from 47.2 to 62.0% (median: 56.1%, n = 14) were greater than AOB ranging from 37.0 to 51.6% (47.1%, n = 10; one-way ANOVA and Dunn’s method, p < 0.01), and they were both greater than those of AOA (31.4–57.4%, 36.7%, n = 46; p < 0.01). Only the GC contents of marine AOA Cenarchaeum symbiosum A and two terrestrial species Nitrososphaera viennensis EN76 and Ca. N. evergladensis SR1 were over 50%. Marine AOA exhibited significantly smaller total genome lengths and GC contents than terrestrial ones (p < 0.05, Student’s t-test), but there was no difference observed between marine and terrestrial AOB or NOB (p > 0.05). In addition, it showed that bacterial or archaeal strains of the same genus had similar GC contents and total genome lengths (Figure 2A; Supplementary Table S1). The SA/V ratios of AOA (4.62–23.4 μm−1, 13.8 μm−1, n = 26) were larger than those of AOB (2.79–24.0 μm−1, 8.30 μm−1, n = 8; one-way ANOVA and Dunn’s method, p < 0.05), but the ratios of NOB (0.93–24.2 μm−1, 12.9 μm−1, n = 10) were not significantly different from those of AOA and AOB (p > 0.05; Figure 2B). The SA/V ratios of collected marine AOA (15.4–23.4 μm−1, 19.6 μm−1, n = 10) were larger than those of terrestrial ones (4.62–22.9 μm−1, 10.13 μm−1, n = 16; one-way ANOVA and Dunn’s method, p < 0.01), but the ratios of AOB and NOB did not show the same trend (p > 0.05; Figure 2B). The GC content and the SA/V ratio were positively (R = 0.783) and negatively (R = 0.326) correlated with the genome length, respectively (Pearson’s correlation, p < 0.05; Figure 2).
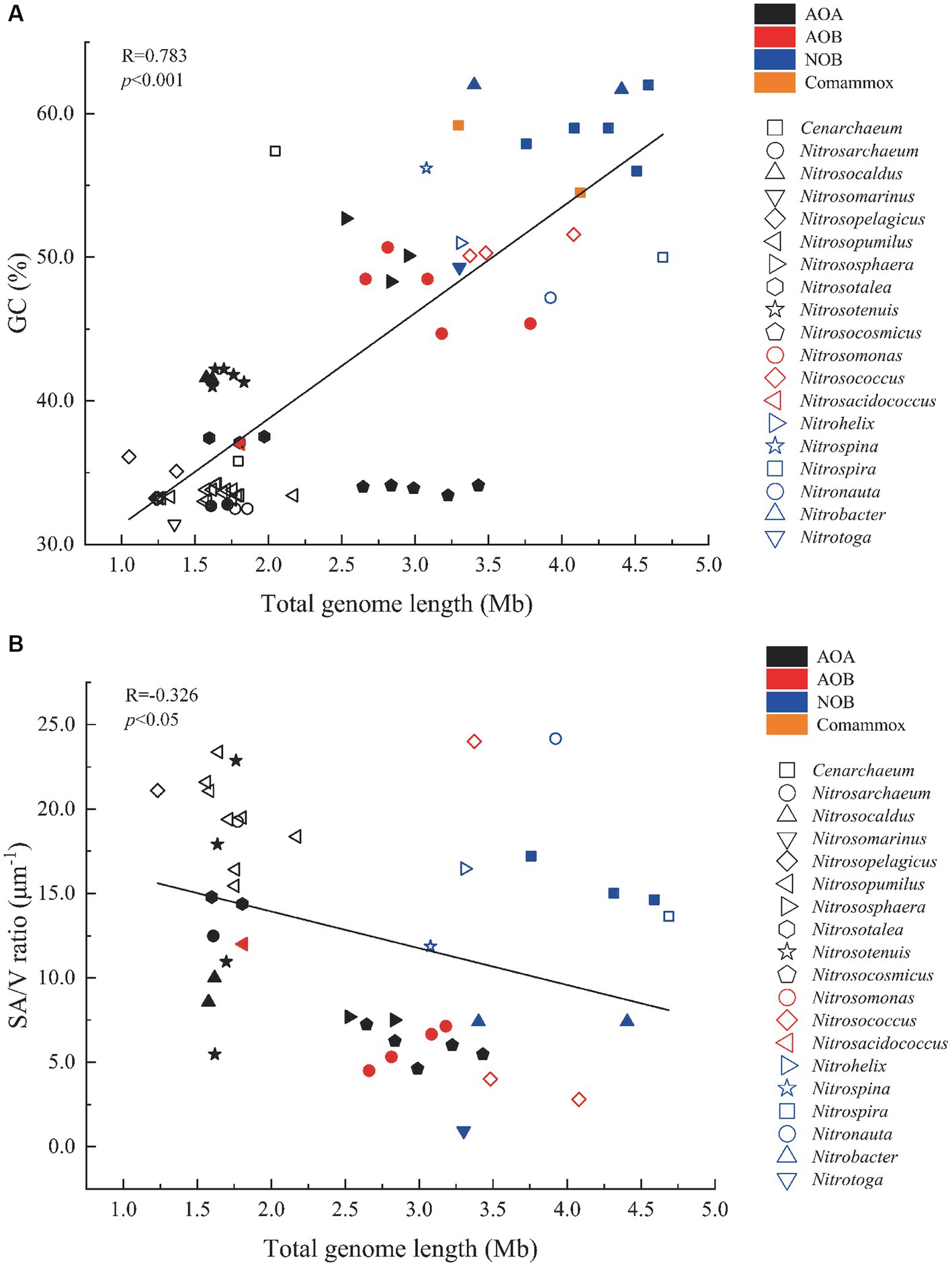
Figure 2. Scatter plots of correlations between (A) GC content (%) and total genome length (Mb) and between (B) SA/V ratio (μm−1) and total genome length (Mb) of selected nitrifiers. The Pearson correlation coefficient (R) of each plot is listed. Filled and open symbols represent strains from terrestrial and marine environments, representatively. AOA, Ammonia-oxidizing archaea; AOB, Ammonia-oxidizing bacteria; NOB, Nitrite-oxidizing bacteria; and Comammox, Complete ammonia oxidizers.
3.2 Distributions of amo and metabolic genes of representative LDON in genomes of nitrifiers
In AOA, Ca. Nitrosotalea okcheonensis CS and Nitrosopumilus piranensis D3C had two copies of amoA and amoB, respectively, and Nitrosopumilus ureiphilus PS0, Ca. Nitrosotenuis uzonensis N4, Ca. Nitrososphaera gargensis Ga9.2, and Ca. Nitrosocosmicus agrestis SS contained two copies of amoC. Two terrestrial AOA, namely, Nitrososphaera viennensis EN76 and Ca. N. evergladensis SR1, had six and seven copies of amoC gene, respectively (Figure 3). In AOB, all strains in genus Nitrosococcus (n = 3) and Ca. Nitrosacidococcus tergens sp. RJ19 only contained one copy of amoABC, in contrary to multiple ones in the genus Nitrosomonas (Figure 3). Two Nitrospira strains of the comammox contained one copy of amoABC as well.
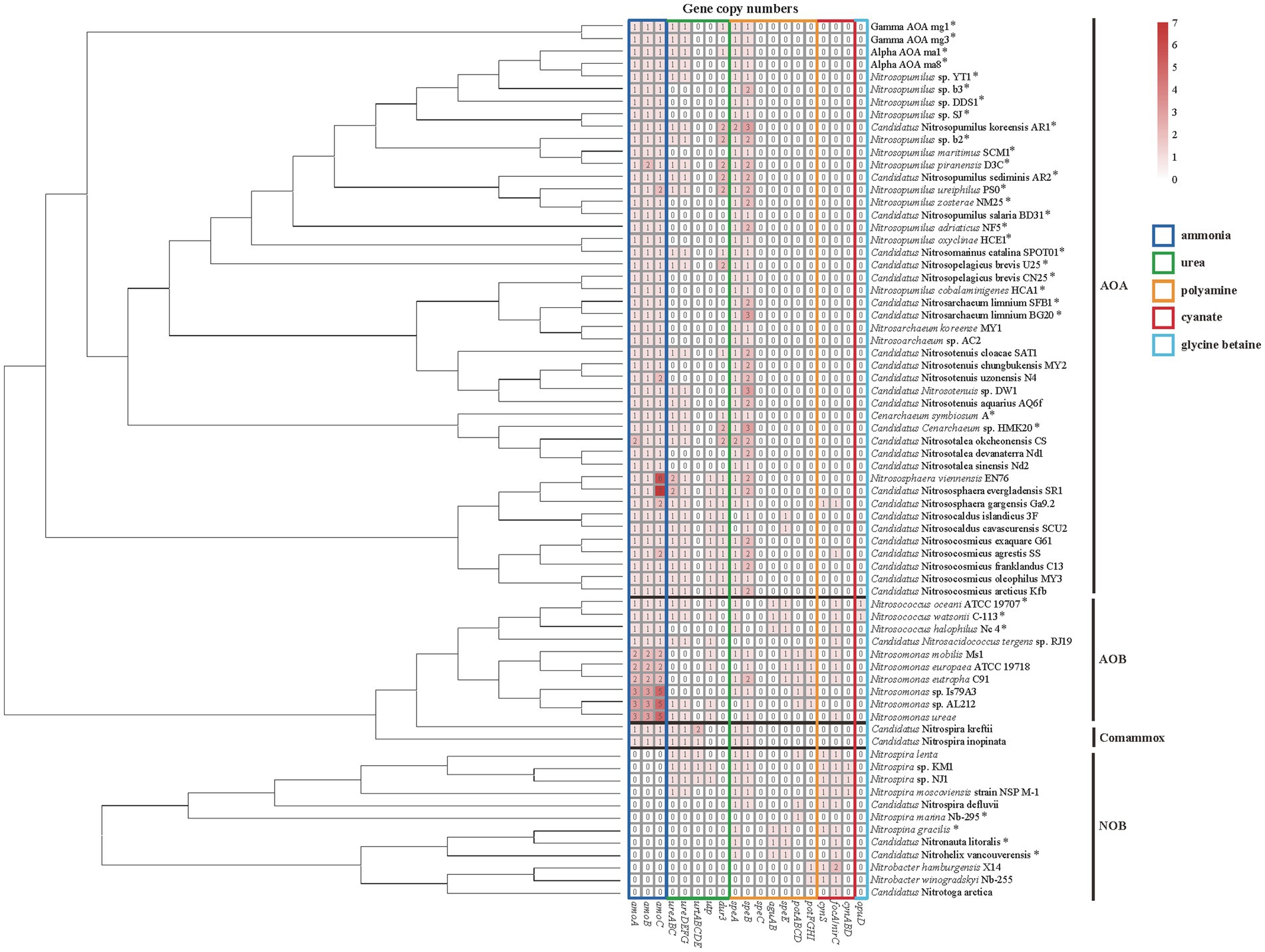
Figure 3. Heatmap of copy numbers of metabolic genes of ammonia (dark blue box), urea (green box), polyamine (orange box), cyanate (red box), and glycine betaine (GBT; light blue box). The strain with an asterisk represents the marine origin. The phylogenetic tree of amino acid sequences of amoA is used for the classification of ammonia-oxidizing archaea (AOA), ammonia-oxidizing bacteria (AOB), and complete ammonia oxidizers (Comammox) and that of 16S rRNA gene is used for the classification of nitrite-oxidizing bacteria (NOB).
In AOA, none of the collected genomes contains bacteria-type urea ABC transporter genes urtABCDE. Instead, most harbored the gene dur3 accompanied by urease-encoding genes ureABC and ureDEFG (Figure 3). In addition, the genomes of terrestrial genera Nitrososphaera, Nitrosocosmicus, and Nitrosocaldus also contained the gene utp (Figure 3). The marine AOA including Nitrosopumilus strains which contained the gene dur3, Ca. Nitrosopelagicus brevis U25 and Ca. Cenarchaeum sp. HMK 20, and the terrestrial one Ca. Nitrosotalea okcheonensis CS had two copies of dur3. Two Nitrososphaera strains, namely, Ca. N. evergladensis SR1 and N. viennensis EN76, harbored two copies of ureABC (Figure 3). The genomes of Nitrosopumilus sp. YT1, Alpha AOA ma8, Gamma AOA mg3, Ca. Nitrosotenuis sp. DW1, and Ca. N. aquarius AQ6f harbored urease genes but were not detected with any type of urea transporter genes (Figure 3). All collected strains in the genus Nitrosarchaeum did not contain genes related to urea utilization (Figure 3). The ordering of ureABC and ureEFGD in AOA was contiguous or spaced by a small number of genes (Figure 4A). Except Alpha AOA ma1, Gamma AOA mg1, and Ca. Nitrosotenuis cloacae SAT1, the genes utp and dur3 were in close proximity to ure in AOA, and utp was closer to ure than dur3 when both genes were present (Figure 4A). In AOB, genomes of two Nitrosococcus strains, N. oceani ATCC 19707 and N. watsonii C-113, four Nitrosomonas strains, N. mobilis Ms1, N. europaea ATCC 19718, Nitrosomonas sp. AL212 and N. ureae, and Ca. Nitrosacidococcus tergens sp. RJ19 harbored the gene utp, but N. mobilis Ms1 and N. europaea ATCC 19718 did not carry ure genes (Figure 3). The gene ureD was divided from ureEFG by ureABC, and utp was adjacent to ureD or ureG (Figure 4A). The complete set of urea transporter genes urtABCDE was only found in genomes of two comammox, and Nitrospira sp. KM1, Nitrospira sp. NJ1, and N. lenta of NOB in the neighbor of ureABC and ureDFG. The comammox Ca. Nitrospira kreftii contained a second copy of urtABCDE that was distantly away from ure (2236 interval open reading frames; Figures 3, 4A). The gene utp was present close to ureA in genomes of Nitrospira sp. KM1 and Nitrospira sp. NJ1 (Figures 3, 4A). The gene ureE was absent in genomes of NOB and comammox except in that of Nitrospira sp. NJ1 (Figure 4A).
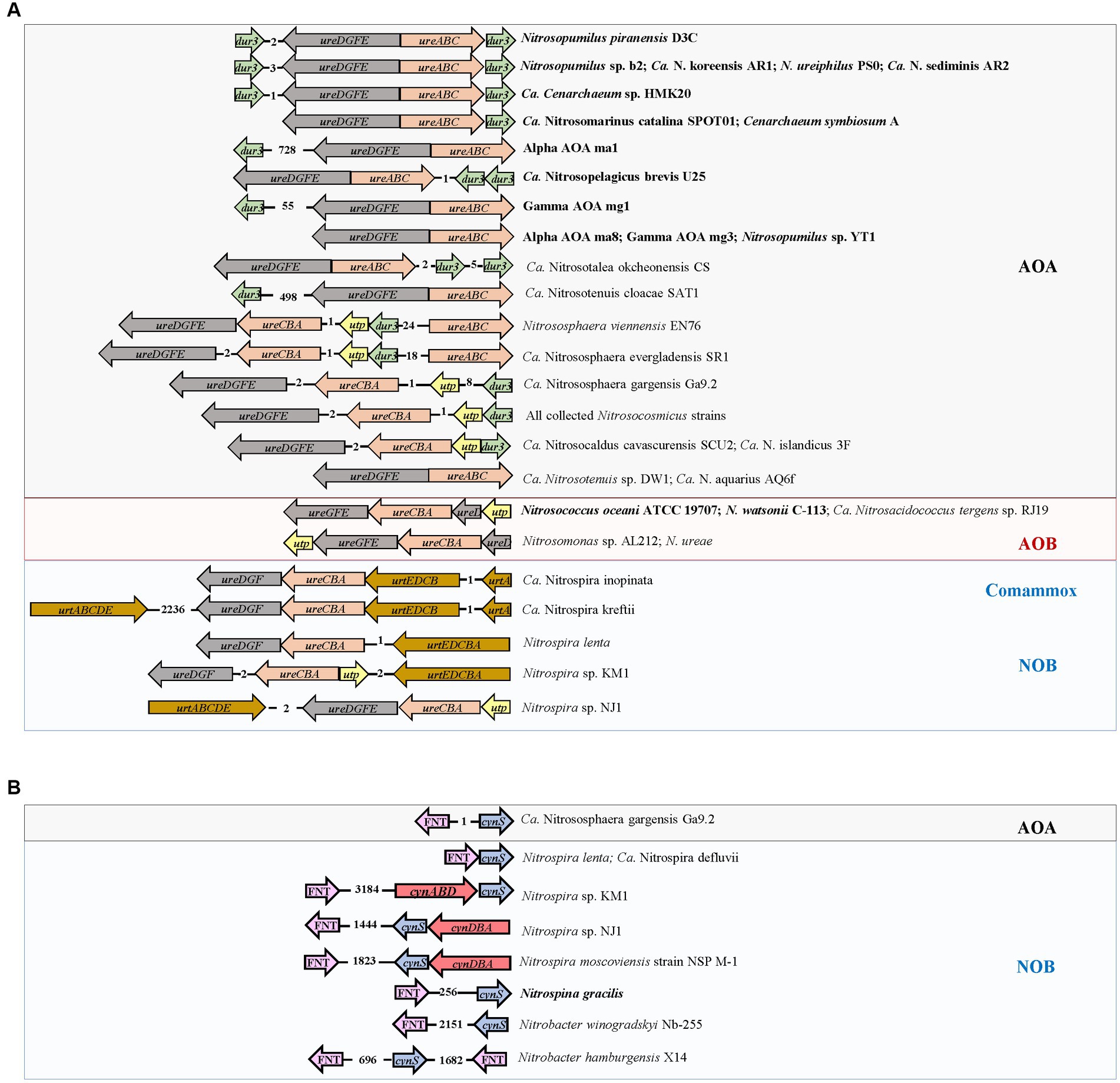
Figure 4. Organization of (A) genes encoding urease (ure) and urea transporter (utp, dur3, and urt) in genomes of ammonia-oxidizing archaea (AOA) and bacteria (AOB), nitrite-oxidizing bacteria (NOB), and complete ammonia oxidizers (Comammox), and (B) genes encoding cyanase (cynS), the FNT family (focA/nirC), and cyanate ABC transporter (cynABD) in genomes of AOA and NOB. The numbers between two genes indicate the number of interval open reading frames (ORFs). The strains highlighted in bold represent marine origin.
None of the collected AOA and comammox genomes harbored polyamine transporter genes (potABCD and potFGHI; Figure 3). Nitrosomonas strains of AOB contained potABCD and potFGHI except N. ureae. Nitrospira lenta, Ca. N. defluvii, and N. marina Nb-295 of NOB harbored potABCD, while two Nitrobacter strains owned potFGHI (Figure 3). Genes encoding enzymes for complete pathways of putrescine catabolism were absent in all genomes of collected nitrifiers (Figures 1B, 3). The genomic evidence showed that all AOA, Nitrosomonas of AOB, and most Nitrospira of NOB and comammox might be capable of synthesizing polyamines intracellularly by arginine decarboxylase [EC: 4.1.1.19] and agmatinase [EC: 3.5.5.11] encoded by speA and speB, respectively (Figures 1B, 3). Most of AOA contained 2–3 copies of speB (Figures 1B, 3). Nitrosococcus of AOB, and NOB strains Nitrospina gracilis, Ca. Nitrohelix vancouverensis, and Ca. Nitronauta litoralis were lack of speB but owned aguAB encoding agmatine deiminase [EC: 3.5.3.12] and N-carbamoylputrescine amidase [EC: 3.5.1.53] for an alternative pathway to synthesize putrescine from agmatine (Figures 1B, 3). The gene speC encoding ornithine decarboxylase which could convert ornithine to putrescine was not found in these bacteria and archaea (Figures 1B, 3).
The cyanate ABC-type transporter encoding genes cynABD were only detected in Nitrospira moscoviensis strain NSP M-1, Nitrospira sp. KM1, and Nitrospira sp. NJ1 of NOB, accompanied by the gene cynS encoding cyanase (Figure 3). The genes cynABD were in the neighbor of cynS (Figure 4B). In addition, the gene cynS was also present in Ca. Nitrososphaera gargensis Ga9.2 of AOA, and Nitrospira lenta, Ca. N. defluvii, Nitrobacter hamburgensis X14, and N. winogradskyi Nb-255 of NOB (Figure 3), but they only contained the FNT family gene focA/nirC, which were more common in AOB and NOB (Figure 3). Among the nine strains possessing cynS, the gene focA/nirC was adjacent to cynS in Ca. N. gargensis Ga9.2, N. lenta, and Ca. N. defluvii (Figure 4B) but was distantly away from cynS (over 256 interval open reading frames) in remaining genomes of NOB (Figure 4B).
The GBT transporter gene opuD was only detected in genomes of two AOB strains Nitrosococcus oceani ATCC 19707 and N. watsonii C-113 (Figure 3), but genes functioning in GBT synthesis and degradation, such as betAB, gbcAB, bhmt, grdHI, and cdh (Figure 1E), were absent in all genomes of collected nitrifiers. In additions, genes encoding enzymes involved in the utilization of taurine (tauACB, tauD, tpa, tauXY, xsc, and pta) and methylamine (tmm, mauAB, gmaS, mgsABC, and mgdABCD; Figures 1D,F) were not found in any genome of collected nitrifiers.
3.3 Phylogenetic relationships of key genes involved in urea and cyanate utilization
Thirty-one amino acid sequences of dur3 from AOA strains were analyzed for phylogenetic relationship (Figure 5A). The sequence of dur3 in the genome of Micromonas commoda was selected as the out-group because dur3 was originally detected in eukaryotic organisms (Coimbra, 2022). In AOA with two copies of dur3, we considered the copy closer to ure genes as Copy 1 and the other as Copy 2 for subsequential phylogenetic analysis (Figures 4A, 5A). Basically, the sequences from terrestrial and marine AOA were well divided into two clusters except that Copy 2 of dur3 from marine AOA strains formed a close relationship with those from terrestrial genera Nitrososphaera and Nitrosocaldus (Figure 5A; Supplementary Table S4). The Copy 1 sequences of dur3 from marine AOA genus were homogeneous to that of M. commoda, which also included Copy 2 of dur3 from a terrestrial strain Ca. Nitrosotalea okcheonensis CS (Figure 5A). The phylogenetic analysis grouped 10 amino acid sequences of the gene utp from AOAs, 7 from AOBs, and 2 from NOBs. The sequence of utp of Deltaproteobacteria Desulfovibrio vulgaris DP4 was used as an out-group because it possessed a homologous urea transporter gene (utp) found in mammals (Levin et al., 2009; Figure 5B). The sequences of utp from NOB and AOB were homologous to that of D. vulgaris DP4 and distinguished from the cluster of AOA (Figure 5B). The amino acid sequence of ureC from a marine ɑ-proteobacterium Ruegeria pomeroyi DSS-3 was used as an out-group for constructing a phylogenetic tree of ureC (Figure 5C). Ruegeria pomeroyi DSS-3 is a heterotrophic bacterium ubiquitous in marine environments and is capable of degrading urea with urease (Ferrer-González et al., 2023). Similarly, the sequences of ureC from archaea and bacteria were distinctively divided into two groups (Figure 5C). In AOA, the sequences from genera Nitrosocosmicus and Nitrososphaera belonging to the order of Nitrososphaerales (Group I.1b) were clustered with Nitrosocaldus of the order Ca. Nitrosocaldales except for the second copies of ureC in genomes of Nitrososphaera viennensis EN76 and Ca. N. evergladensis SR1. They were more phylogenetically close to sequences from the genus Nitrosotalea belonging to the order Ca. Nitrosotaleales (Group I.1a-associated) and the order Nitrosopumilales mostly comprised of marine AOA (Figure 5C). The amino acid sequences of ureC from AOB, NOB, and comammox were tightly clustered and homologous to that of R. pomeroyi DSS-3 (Figure 5C).

Figure 5. Phylogenetic trees of amino acid fragments of (A) dur3, (B) utp, and (C) ureC identified from ammonia-oxidizing archaea (AOA) and bacteria (AOB), nitrite-oxidizing bacteria (NOB) and complete ammonia oxidizers (Comammox). The trees are constructed with the maximum likelihood method. Bootstrap values based on 1,000 replicates are indicated for the major branches, and the values >50 are shown as black dots. The numbers in brackets indicate the second copy of dur3 or ureC from the same strain. The strains highlighted in bold represent marine origin.
Amino acid sequences of cynS and the FNT family gene focA/nirC from Ca. Nitrososphaera gargensis Ga9.2 and NOB strains were individually analyzed for the phylogenetic relationship with corresponding sequences from the out-group Prochlorococcus marinus since the genes were mostly found in marine cyanobacterial strains and their functions had been verified (Maeda and Omata, 2009; Maeda et al., 2015; Figure 6). The cynS sequence of Ca. N. gargensis Ga9.2 was clustered with those of Nitrospira and distinguished from two Nitrobacter strains and the marine AOB Nitrospina gracilis, which were phylogenetically close to P. marinus (Figure 6A). Differently, in the phylogenetic tree of the gene focA/nirC, in addition to three Nitrospira strains, Nitrospira sp. NJ1, Nitrospira sp. KM1, and N. Moscoviensis strain NSP M-1, the sequence of Ca. N. gargensis Ga9.2 was also clustered with those of two Nitrobacter strains, which were closely related to that of P. marinus (Figure 6B). The gene focA/nirC of the marine AOB N. gracilis grouped with those from Nitrospira lenta and Ca. N. defluvii and the second copy of focA/nirC in Nitrobacter hamburgensis X14 (Figure 6B).
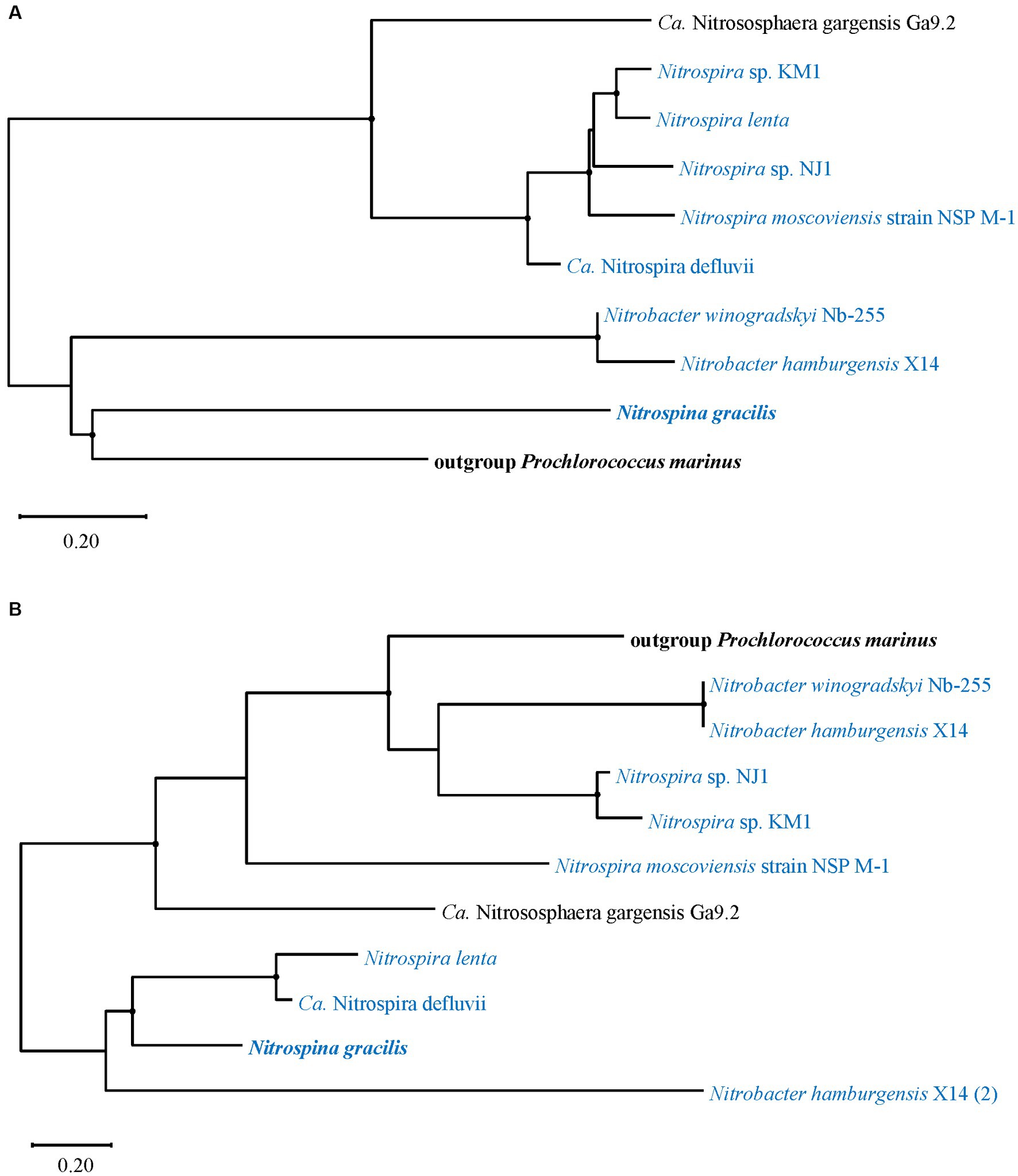
Figure 6. Phylogenetic trees of amino acid fragments of (A) cynS and (B) the FNT family gene focA/nirC identified from ammonia-oxidizing archaea (AOA) and nitrite-oxidizing bacteria (NOB). The strains shown in panel (B) are selected as ones which also contain cynS. The trees are constructed with the maximum likelihood method. Bootstrap values based on 1,000 replicates are indicated for the major branches, and the values >50 are shown as black dots. The numbers in brackets indicate the second copy of focA/nirC from the same strain. The strains highlighted in bold represent marine origin.
4 Discussion
4.1 Genomic and phenotypic characteristics of nitrifiers reflecting nitrogen availability and affinity of nitrifiers
Although we did not collect genomes of all nitrifiers, such as those identified by MAG and single-cell sequencing, the genomes of isolated and enriched nitrifier strains as well as several MAGs from extreme environments could act as representatives of typical habitats (Supplementary Table S1). The genomic analysis and comparison were believed to be vigorous to gain insights into differences in physiological and metabolic characteristics among AOA, AOB, NOB, and comammox.
Generally, AOA have smaller genome lengths and lower GC contents than AOB and NOB (Figure 2A; Supplementary Table S1). The environmental differences in ecological niches may lead to this differentiation, in line with survival strategy and nitrogen metabolic capacity (Kitzinger et al., 2020). Previous studies on bacteria showed that those with large chromosomes usually had higher GC contents (Guo et al., 2009). Genomes lacking GC may be beneficial to the survival in nitrogen-limiting environments because AT pairs use one less nitrogen than GC pairs (Luo et al., 2015). Thus, low GC contents in AOA indicated nitrogen limitation in their niches, especially those in marine environments. The limited capacity of using LDON may also prohibit the access of nitrogen to AOA (Figure 3). However, great SA/V ratios of marine AOA suggested a high affinity of ammonia for compensating low concentrations in marine environments. More copies of urea transporter gene dur3 corresponding to higher SA/V ratios might be a strategy for conquering nitrogen limitation as well (R = 0.754, p = 0.003, Spearman rank correlation; Supplementary Figure S1). The group Nitrosocosmicus had a large genome size (2.64–3.63 Mb) but a low GC content (33.4–34.1%; Figure 2A; Supplementary Table S1) probably due to its extreme low affinity of NH3 plus NH4+ (Jung et al., 2022), although the surrounding environment might not be limited by nitrogen (e.g., waste water plant treatment; Sauder et al., 2017). NOB generally had larger GC contents in response to greater genome lengths (Figure 2A), suggesting either nitrogen sufficiency in their habitats or their great efficiency to absorb substrates from the environment. The larger SA/V ratio of most collected NOB (e.g., Nitronauta, Nitrohelix, and Nitrospira) implied the latter circumstance, but the potential of using diverse nitrogen sources (e.g., urea and cyanate; Figure 3) might enhance their versatility of acquiring nitrogen from the environment, resulting in increased GC content and genome length. The relatively greater GC contents but lower SA/V ratios of AOB compared to AOA could be the consequence of AOB survival in substrate-rich environments with increased nitrogen availability (Soliman and Eldyasti, 2018). Thus, the genomic (e.g., GC%) and phenotypic characteristics (e.g., SA/V) of nitrifiers might reveal their niche partitioning as the basis of their capability and efficiency of nitrogen utilization for environmental adaptation.
4.2 Strategy of urea and cyanate utilization being intensely to characteristics of environmental niches and nitrogen demand of nitrifiers
Most of the collected AOA harbor urease genes (ureABC and ureDEFG; Figure 3); however, urea ABC transporter genes urtABCDE commonly detected in bacterial genomes (Veaudor et al., 2019) were not found in any collected AOA. Instead, two types of urea transporter genes dur3 (Solomon et al., 2010) and utp (Bossé et al., 2001) were present in AOA harboring ure genes except Ca. Nitrosotenuis sp. DW1, Ca. N. aquarius AQ6f, Nitrosopumilus sp. YT1, Alpha AOA ma8, and Gamma AOA mg3 (Figure 3). Since nitrite production has been detected in the enrichment culture of Ca. N. aquarius AQ6f with urea (Sauder et al., 2018), it suggests that these five strains may contain unknown urea transporter proteins, or urea could diffuse across the cell membrane without active transports (Sauder et al., 2018). The genes utp and dur3 are both present in genera belonging to Group I.1b (Nitrososphaera and Nitrosocosmicus) and Nitrosocaldus, while dur3 is the only urea-transporter gene in genomes of Group I.1a (Nitrosopumilus, Nitrosopelagicus, and Nitrosotenuis) and Group I.1a-associated (Nitrosotalea) strains (Figures 3, 4A). The verified growth of two Nitrososphaera strains and Nitrosopumilus ureiphilus PS0 in media added with urea suggests that the urea transporter encoded by dur3 is functional; however, whether dur3 is functional requires further experimental validation because some AOA lacking dur3 or utp can also hydrolyze urea without the known transporters (Sauder et al., 2018). In addition, the presence of two copies of dur3 in the genus Nitrosopumilus except Nitrosopumilus sp. YT1, and Ca. Nitrosopelagicus brevis U25 from marine environments (Figure 3) could be the result of the low availability of urea, which triggers marine AOA to produce more transporter proteins for efficiently utilizing urea (Offre et al., 2014). The protein Dur3 has been demonstrated to encode a high-affinity urea active transporter in marine unicellular photosynthetic eukaryotes (Solomon et al., 2010). Thus, AOA may be advantageous in urea uptake, corresponding to their higher urea uptake rates in several marine ecosystem (e.g., polar waters, the Gulf of Mexico, and coastal Georgia; Alonso-Sáez et al., 2012; Tolar et al., 2016; Kitzinger et al., 2019). The homogeneity of Copy 2 of dur3 in marine AOA to those in genomes of terrestrial ones indicates lateral gene transfer (LGT) of genes from terrestrial AOA (Figure 5A). The gene utp identified in genomes of some terrestrial AOA appears simultaneously with dur3 (Figures 3, 4A). Since the protein Utp has been considered as a low-affinity urea transporter (Bossé et al., 2001; Raunser et al., 2009), the presence of utp in terrestrial AOA could be due to the complex terrestrial environments (wastewater treatment plant, agricultural soils, mud, etc.) with the detection of high urea concentrations (Rittstieg et al., 2001; Wang et al., 2012); however, whether the urea transporter protein encoded by utp functions in AOA as an alternative way of urea transport by Dur3 still needs further experimental verification.
The urea transporter encoding gene dur3 was not detected in genomes of selected AOB and NOB, but instead it was replaced by utp and urt, respectively (Figures 3, 4A). Genomes of Nitrosococcus oceani ATCC 19707, N. watsonii C-113, Nitrosomonas sp. AL212, N. ureae and Ca. Nitrosacidococcus tergens sp. RJ19 of AOB processed both utp and ure, while terrestrial AOB Nitrosomonas mobilis Ms1 and N. europaea ATCC 19718 only possessed a copy of utp (Figure 3). Nitrite has been found rapidly produced from N. oceani ATCC19707 cultured in the medium with urea replacing ammonia (Koper et al., 2004), suggesting that utp functions as a urea transporter. Assimilated urea could be efficiently degraded and used as a source of NH4+ by AOB. Nitrosococcus oceani is an AOB species distributed ubiquitously in the oceanic environment and is an important nitrifier in the OMZ (Lam et al., 2009). It incorporates with anammox bacteria and is responsible for nitrogen loss (Woebken et al., 2008). Thus, N. oceani may be responsible for urea hydrolyzation and subsequential oxidization of ammonia in the OMZ. Urease activity in N. europaea ATCC 19718 has been experimentally proven absent (de Boer and Laanbroek, 1989; Sliekers et al., 2004), consistent with the genomic evidence (Figure 3). The presence of utp without the co-occurrence of urease genes is probably due to its non-specificity for urea. It may also facilitate the diffusion of urea analogs along their concentration gradients (Levin et al., 2009). The detection of utp only in AOB implies the advantage of AOA with both utp and dur3 in utilizing urea in different environments because they may alternate the urea transporter depending on urea concentrations.
Genomes of collected NOB and comammox have a full set of urt and ure genes (Figures 3, 4A), indicating that they are capable of urea utilization. The ureolytic activity has been observed in the culture of Nitrospira moscoviensis, Ca. N. nitrosa, and Ca. N. nitrificans with urea-containing media (Koch et al., 2015; Van Kessel et al., 2015; Vijayan et al., 2021). The gene cluster urt is a high-affinity urea transporter (Valladares et al., 2002), suggesting that Nitrospira has a competitive advantage in urea uptake in environments with low urea concentrations. Since Nitrospira occur ubiquitously in different terrestrial and aquatic habitats (Latocheski et al., 2022), it is tempting to speculate that reciprocal feeding between Nitrospira and AOM could be a common phenomenon in nature, but their contribution to total nitrification in different ecosystems remains to be determined. The marine NOB (Nitrospira marina Nb-295, Ca. Nitronauta litoralis, Ca. Nitrohelix vancouverensis, and Nitrospina gracilis) do not contain any urea-related gene (Figure 3); however, although the genome of the type strain N. gracilis of the genus Nitrospinae does not contain ureases genes, other clades of Nitrospinae (e.g., Nitrospinae Clade 2 in the Gulf of Mexico, Kitzinger et al., 2020) representing the major groups of NOB in marine environments commonly contain complete sets of urea transporter and urease genes, and form an intense relationship of reciprocal feeding with AOM (Koch et al., 2015; Pachiadaki et al., 2017). In addition, in NOB, only Nitrospira sp. KM1 and Nitrospira sp. NJ1 own utp (Figures 3, 4A). The reason that both AOB and NOB lack the gene dur3 could be due to the fact that dur3 of AOA was derived from the same ancestor as that of eukaryotes (Figure 5A; Levin et al., 2009; Spang et al., 2012). The gene utp of terrestrial AOA may evolve from bacteria through LGT (Figure 5B). Overall, the genes dur3 and utp may be functional and encode proteins for urea transport from extracellular environments; however, as evidence shows that AOM harboring these two genes have a lower urea uptake rate than those with urt, such as Nitrospira or other heterotrophic bacteria (Vijayan et al., 2021), it suggests that urea may be the alternative energy and nitrogen source of AOM, which still use ammonia or ammonium as their major substrate (Kitzinger et al., 2020). It is noted that most of Nitrospira genomes lack ureE that serves as a bridge to acquire nickel from hydrogenase maturation factor HypA, which is subsequently donated to UreG. HypA is a metallochaperone and selectively delivers the nickel to the active site (Xiong et al., 2023). The absence of ureE may result in low urease activity in NOB (Carter et al., 2009; Fujitani et al., 2020).
The cyanate degradation gene cynS was only detected in NOB and one AOA strain Ca. Nitrososphaera gargensis Ga9.2 (Figure 3). Although both comammox strains do not contain cynS in this study, it has been found that a comammox MAG LK70 owns cynS (Yang et al., 2020). The genes cynABD encoding a high-affinity cyanate ABC transporter (Maeda and Omata, 2009) were only detected in Nitrospira sp. KM1, Nitrospira sp. NJ1, and N. moscoviensis strain NSP M-1 of NOB (Figures 3, 4B), and N. moscoviensis was proven to use cyanate (Palatinszky et al., 2015). The remaining strains with cynS without cynABD contained the FNT family encoded by focA/nirC (Figures 3, 4B), which was hypothesized as the low-affinity cyanate transporter (Rycovska et al., 2012; Spang et al., 2012; Palatinszky et al., 2015). FNT proteins are found in most phyla of bacteria, archaea, and lower eukaryotes (Falke et al., 2010) and are key regulators of the metabolic flow in microorganisms (Wiechert and Beitz, 2017). The formate transporter (FocA) fuels the energy-generating formate hydrogen lyase reaction. Nitrite derived from chemical reduction of nitrate or oxidation of nitrogen monoxide is transported via NirC (Wiechert and Beitz, 2017). Formate/nitrite transporter is also presumed to be permeable for cyanate due to the proximity of genes for transporter and enzymatic degradation (Spang et al., 2012). Thus, the adjacent relationship between cynS and focA/nirC in genomes of Ca. Nitrososphaera gargensis Ga9.2, Nitrospira lenta, Ca. N. defluvii, Nitrobacter hamburgensis X14, and N. winogradskyi Nb-255 (Figure 4B) may suggest the involvement of FNT family gene encoding protein in cyanate transport. However, there is no experimental evidence that those five nitrifiers can use cyanate. The role of FNT family gene as the cyanate transporter is still uncertain with only genomic evidence (Spang et al., 2012). Although it is lack of evidence that the AOM can assimilate and break down cyanate from the environment, a previous study has shown that the pure culture of Nitrosopumilus maritimus SCM1 added with 15N-cyanate produces 15N-ammonium and 15N-nitrite, which suggests a process of extracellular breakdown of cyanate by AOM (Kitzinger et al., 2019). It is also reported that NOB supply cyanase-lacking AOM with ammonium from cyanate. The ammonium can be fully nitrified by this microbial consortium through reciprocal feeding in co-culture experiments (Palatinszky et al., 2015). If the FNT family was a cyanate transporter, NOB could be the dominant nitrifier in cyanate utilization and play a key role in reciprocal feeding in nature. Thus, NOB that have high GC contents and SA/V ratios may have more versatility to utilize different extracellular nitrogen to satisfy their nitrogen demands.
4.3 Metabolic potentials of other LDON compounds
Polyamines are the primary amines consisting of two or more amine substitutions (Liu et al., 2015; Damashek et al., 2019). They are ubiquitous in cells of all lives and are essential for integral cellular processes, such as nucleic acid synthesis and stabilization, cellular growth, protein synthesis, biofilm formation, and siderophore production (Michael, 2018). They are de novo synthesized intracellularly (Figure 1B) and can be directly released from living and dead cells or from protein degradation into environments (Liu et al., 2016; Michael, 2016). Eukaryotic phytoplankton and heterotrophic bacteria (e.g., Roseobacter and SAR11) have been detected to utilize extracellular polyamines (Mou et al., 2010; Liu et al., 2016; Noell et al., 2021). Polyamines can be used by bacterioplankton as a nitrogen source (Figure 1B) and contribute to over 4% of bacterial nitrogen demand in aquatic environments (Liu et al., 2015; Krempaska et al., 2018; Madhuri et al., 2019; Liu et al., 2022a). Previous studies observed that putrescine-N could be oxidized and contribute to a significant fraction of total nitrification in coastal waters, and putrescine-N oxidation rate even exceeded that of urea (Damashek et al., 2019). Moreover, the faster polyamine-N oxidation rate than its uptake rate in the same water region as well as the production of 15N-NO2− in pure cultures of some AOA with 15N-putrescine suggest that AOM directly oxidize amine groups of polyamines resembling the pathway of ammonia oxidation (Liu et al., 2015; Damashek et al., 2021) since ammonia monooxygenase can co-metabolize a variety of organic compounds (Rasche et al., 1991; Wright et al., 2020). Thus, it is reasonable that genes homologous to pot-encoding polyamine transporters are not detected in selected AOA strains. The protein Dur3 has been found to be capable of transporting polyamines along with urea (Uemura et al., 2007), raising the potential that AOA may also assimilate polyamines; however, since all AOA, Nitrosomonas of AOB, and Nitrospira lenta, Ca. N. defluvii, N. marina Nb-295, Nitrobacter hamburgensis X14, and N. winogradskyi Nb-255 of NOB, which harbor pot genes, lack genes involved in polyamine catabolism (Figures 1B, 3), polyamines may not be degraded into ammonium intracellularly for the following oxidation processes. Instead, assimilated polyamines may serve for other physiological purposes as mentioned above (Kim et al., 2016).
In this study, all selected nitrifiers have the potential to synthesize putrescine either from arginine or agmatine except Ca. Nitrosacidococcus tergens sp. RJ19 of AOB and Nitrospira marina Nb-295, Nitrobacter hamburgensis X14, N. winogradskyi Nb-255, and Ca. Nitrotoga arctica of NOB (Figures 1B, 3). Archaea have been mentioned to form branched or long-chain polyamines and induce structural changes to DNA that can facilitate growth in extreme environments (Michael, 2018), or polyamines could be used as a donation of aminobutyl group for the growth of some archaeal halophiles and some methanogens (Michael, 2018). Polyamine synthesis and excretion are significantly up-regulated in AOA grown in environments with high levels of ammonia, which is thought to be one of the reasons for the ammonia tolerance of AOA in terrestrial environments (Liu et al., 2021). AOA may also use polyamines for detoxification (e.g., H2O2) or form biofilm for substrate uptake (Michael, 2018). More copies of speB detected in AOA may be the evolutionary consequence for polyamine synthesis (Magadum et al., 2013). Thus, instead of utilizing polyamine-N as an energy source, AOA might be a significant source of polyamines and contribute to polyamine cycling in different environments.
Taurine dissimilation could be an important source of nitrogen (Denger et al., 2004). Thaumarchaeota and Euryarchaeota have been reported to assimilate taurine in the upper water column of the northern Adriatic Sea identified by MICRO-CARD-FISH, and the uptake by Thaumarchaeota is even beyond that of SAR11 and Roseobacter clade in fall when the release of taurine is enhanced by zooplankton (Clifford et al., 2019); however, in this study, none of collected nitrifiers possesses taurine transporter genes tauACB and catabolic genes (Figure 1D), suggesting that nitrifiers either assimilate taurine using other transporters (e.g., those for amino acids) or directly break down taurine extracellularly (Damashek et al., 2019, 2021). In this study, only two marine AOB strains Nitrosococcus oceani ATCC 19707 and N. watsonii C-113 own glycine betaine (GBT) transport gene opuD. GBT has been found common in bacteria as osmotic molecules (Shakhman and Harries, 2021). A variety of soil and aquatic bacteria have catabolic pathways that convert choline to glycine in multiple steps via GBT (Figure 1E), using both choline and GBT as the sole carbon and nitrogen sources (Wargo, 2013). In addition, bacteria and methanogenic archaea in the cold spring are able to synergistically convert GBT to methane (Li et al., 2021). The lack of GBT transport, synthesis, and degradation genes in most of selected nitrifiers suggests that GBT is not a nitrogen source to nitrifiers but may function in regulating osmotic pressure (Csonka, 1989). Similarly, methylamines which have been considered as important nitrogen and energy sources for heterotrophic bacteria in natural environments (Chen, 2012) may not be utilized by nitrifiers due to the lack of functional genes.
4.4 Evolution of LDON metabolism in nitrifiers
It is reported that archaeal ammonia monooxygenases share a more recent evolutionary history with actinobacterial monooxygenases than with those of AOB or comammox (Alves et al., 2018). The amoA of the comammox Nitrospira was transferred to AOB or that of both bacteria was derived from an unknown third donor (Palomo et al., 2018). The ureC gene of these bacteria and archaea probably evolved independently after an early gene duplication event, as did cyanobacteria and eukaryotes (Glass et al., 2009). The acquisition of urease genes may coincide with the gain of ammonia monooxygenase genes during the transition from Thaumarchaeota to AOA (Sheridan et al., 2020). Therefore, the potential LGT between bacteria and archaea may not exist in the evolutions of amoA and ureC (Figures 3, 5C). However, a previous study suggested that a certain amount of thaumarchaeotal gene clusters were recruited from bacteria for overcoming stresses and facilitating the environmental adaptation of Thaumarchaeota (Ren et al., 2019). According to the phylogenetic relationships of utp (Figure 5B), and two cyanate metabolic genes cynS and focA/nirC (Figure 6) between nitrifiers, it seems that LGT affects these genes and mostly happens between terrestrial AOA and bacteria (Figures 5B, 6). It has been proposed that the UT family is prokaryotic origin, and the encoding gene utp in groups of terrestrial AOA (Nitrososphaerales and Ca. Nitrosocaldales) is probably either transmitted vertically or horizontally acquired from a bacterium (Figure 5B; Minocha et al., 2003). Since the GC content of a newly acquired gene differs from that of the whole genome, a significant difference in the GC content between the gene utp (41.6–57.0%, median: 43.5%) and whole genomes of terrestrial AOA (33.4–52.7, 37.8%; p = 0.04, Mann–Whitney rank sum test; Supplementary Table S5) indicates a LGT event of utp (Lal and Lal, 2010). The adaptation of bacterial utp to their respective genomes as depicted by GC contents (utp: 35.0–59.0, 48.1%, genome: 37.0–59.0, 50.1%; p > 0.05; Supplementary Table S5) suggests that utp is bacterial origin and acquired by terrestrial AOA via LGT. GC contents of cynS (53.5–59.9, 56.3%) and focA/nirC (59.4–66.6, 60.3%) in bacteria which contain both genes did not show significant differences from those of whole genomes (56.0–62.0, 59.0%; p < 0.05). Similarly, GC contents of the two genes of Ca. Nitrososphaera gargensis Ga9.2 follow the same pattern as Nitrospira with a relatively lower and higher GC content of cynS and focA/nirC than that of the whole genome, respectively (Supplementary Table S5). It implies that Nitrososphaera and Nitrospira may acquire them via an ancient LGT event, which was also supported by good bootstrap values (Figure 6). The significantly greater CAI values of cynS (0.65–0.75, 0.66) and focA/nirC (0.65–0.76, 0.70) than those of bacterial whole genomes (0.51–0.62, 0.56; p < 0.05) further support the idea of LGT and indicate high gene expression in bacteria (Supplementary Table S6). A synchronous acquisition of genes via LGT by Nitrospira and terrestrial AOA genera Nitrsosphaera could also be verified by speA (Supplementary Figure S2) that is involved in the key process of putrescine synthesis (Figure 1B).
It is reported that the emerged AOA progress through an adaptive pathway from terrestrial hot springs to mesophilic soil (∼652 Ma) and then to shallow and deep oceans (∼509 Ma; Yang et al., 2021). The glaciation triggers the evolution of AOA diverging into two groups, one having the mesophilic terrestrial AOA group (genera Nitrosocosmicus and Nitrososphaera) and the other including marine AOA and acidic soil AOA group (genus Nitrosotalea; Yang et al., 2021). The driver for the evolutionary divergence of marine AOA from acidic soil AOA is oxygenation (Yang et al., 2021). The cluster of amino acid sequences of amoA, dur3, or ureC of marine AOA (Figures 3, 5B,C) confirms that marine AOA evolve intimately. However, dur3 in genomes of AOA, on the other hand, differs from other genes in that the two copies of marine AOA are of different origins according to both phylogenetic relationships and GC contents (Figures 4A, 5A; Supplementary Table S5). It is observed that both copies of dur3 of marine AOA exhibit higher GC contents (Copy 1: 40.1–43.9, 40.6%; Copy 2: 36.3–40.6, 37.5%) than those of whole genomes (33.2–35.8, 33.6%; calculations only count marine AOA with both copies of dur3; p < 0.05; Supplementary Table S5), while GC contents of dur3 of terrestrial AOA (36.3–56.4, 38.9%) are in the same range to those of respective genomes (33.4–52.7, 39.3%; p > 0.05). It suggests that dur3 may originate around the same period of time of ure acquisition during the transition from Thaumarchaeota to AOA and then diverge into two groups during glaciation events (Yang et al., 2021). The close clustering of Copy 1 of dur3 of marine AOA and Copy 2 of Nitrosotalea verifies this evolutionary process (Figure 5A). Copy 1 of dur3 of Nitrosotalea and Copy 2 of marine AOA are apparently not gained from gene duplication; instead, it could be transferred from terrestrial AOA through LGT. Dur3 orthologues have been detected in higher plants, algae, and fungi (Solomon et al., 2010). The lack of dur3 in genomes of bacterial nitrifiers, and the clustering of dur3 sequences of marine AOA and a marine alga M. commoda (Figure 5A) imply that dur3 in eukaryotic may evolve from AOA.
5 Conclusion
The analysis of diversities and phylogenetic relationships of genes involved in LDON metabolisms in genomes of representative AOA, AOB, NOB, and comammox develops a more holistic understanding of the potentials of LDON metabolism by nitrifiers and sheds light on evolutionary relationships of functional genes involved in these processes. Our data suggest that GC contents, genome sizes, and SA/V ratios of nitrifiers may reflect the availability of nitrogen in their environmental niches and their capability of nitrogen assimilation for environmental adaptation. Our finding reinforces that nitrifiers tend to assimilate and degrade LDON for acquiring nitrogen or reciprocal feeding (e.g., urea and cyanate). They may also directly oxidize amine groups in LDON (e.g., polyamines and taurine) extracellularly to increase their competitive advantage when facing the substrate limitation. They could acquire this capability from early genetic evolution or LGT. Within different groups of nitrifiers, NOB are more advantageous and versatile in nitrogen assimilation than AOM due to their high affinity of ammonia and urea, and potentials in cyanate utilization. They may share similar environmental niches with AOA and form intense reciprocal feeding relationships. In marine environments, AOA could be more efficient in using urea than AOB, which only dominate in environments with high urea concentration. In terrestrial environments, AOA may adjust the protein expression of the urea transporter (Utp or Dur3) to adapt to different urea concentrations, but again AOB only use urea at high concentrations. Our comparative analysis of LDON metabolic genes in different nitrifiers will guide future studies on the isolation and culture of new strains, providing a theoretical basis for their survival strategies in diverse environments. Moreover, it will contribute to model systems to study reciprocal or competitive interactions, which can severely affect matter and energy flows of ecosystems.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
QL: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. YC: Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. X-WX: Project administration, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (no. 42176038), Scientific Research Fund of the Second Institute of Oceanography, MNR (no. HYGG1901), the Project of State Key Laboratory of Satellite Ocean Environment Dynamics, Second Institute of Oceanography (no. SOEDZZ2204), and Science Foundation of Donghai Laboratory (no. DH-2022KF0211).
Acknowledgments
We thank Dr. Hong Chen and Dr. Yue-Hong Wu from the Second Institute of Oceanography, MNR China for providing hardware facilities.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1273211/full#supplementary-material
Footnotes
References
Alonso-Sáez, L., Waller, A. S., Mende, D. R., Bakker, K., Farnelid, H., Yager, P. L., et al. (2012). Role for urea in nitrification by polar marine Archaea. Proc. Natl. Acad. Sci. 109, 17989–17994. doi: 10.1073/pnas.1201914109
Alves, R. J. E., Minh, B. Q., Urich, T., von Haeseler, A., and Schleper, C. (2018). Unifying the global phylogeny and environmental distribution of ammonia-oxidizing archaea based on amoA genes. Nat. Commun. 9, 1–17. doi: 10.1038/s41467-018-03861-1
Bast, F. (2013). Sequence similarity search, multiple sequence alignment, model selection, distance matrix and phylogeny reconstruction. Nat Protoc Exchang. doi: 10.1038/protex.2013.065
Bayer, B., Vojvoda, J., Reinthaler, T., Reyes, C., Pinto, M., and Herndl, G. J. (2019). Nitrosopumilus adriaticus sp. nov. and Nitrosopumilus piranensis sp. nov., two ammonia-oxidizing archaea from the Adriatic Sea and members of the class Nitrososphaeria. Int. J. Syst. Evol. Microbiol. 69, 1892–1902. doi: 10.1099/ijsem.0.003360
Bossé, J. T., Gilmour, H. D., and MacInnes, J. I. (2001). Novel genes affecting urease activity in Actinobacillus pleuropneumoniae. J. Bacteriol. 183, 1242–1247. doi: 10.1128/JB.183.4.1242-1247.2001
Boyde, A., and Williams, R. (1971). Estimation of the volumes of bacterial cells by scanning electron microscopy. Arch. Oral Biol. 16, 259–267. doi: 10.1016/0003-9969(71)90019-7
Boysen, A. K., Durham, B. P., Kumler, W., Key, R. S., Heal, K. R., Carlson, L., et al. (2022). Glycine betaine uptake and metabolism in marine microbial communities. Environ. Microbiol. 24, 2380–2403. doi: 10.1111/1462-2920.16020
Carter, E. L., Flugga, N., Boer, J. L., Mulrooney, S. B., and Hausinger, R. P. (2009). Interplay of metal ions and urease. Metallomics 1, 207–221. doi: 10.1039/b903311d
Chen, Y. (2012). Comparative genomics of methylated amine utilization by marine Roseobacter clade bacteria and development of functional gene markers (tmm, gmaS). Environ. Microbiol. 14, 2308–2322. doi: 10.1111/j.1462-2920.2012.02765.x
Clifford, E. L., Varela, M. M., Corte, D. D., Bode, A., Ortiz, V., Herndl, G. J., et al. (2019). Taurine is a major carbon and energy source for marine prokaryotes in the North Atlantic Ocean off the Iberian Peninsula. Microb. Ecol. 78, 299–312. doi: 10.1007/s00248-019-01320-y
Coimbra, T.A.V.P. (2022). Bioinformatic strategies to explore iodine transport in plants and its potential application in biofortification. Master's thesis. University of Minho, Braga (MP).
Cook, A. M., and Denger, K. (2006). Metabolism of taurine in microorganisms. Taurine 583, 3–13. doi: 10.1007/978-0-387-33504-9_1
Csonka, L. N. (1989). Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 53, 121–147. doi: 10.1128/mr.53.1.121-147.1989
Daims, H., Lücker, S., and Wagner, M. (2016). A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 24, 699–712. doi: 10.1016/j.tim.2016.05.004
Damashek, J., Bayer, B., Herndl, G.J., Wallsgrove, N.J., Allen, T., Popp, B.N., et al. (2021). Limited accessibility of nitrogen supplied as amino acids, amides, and amines as energy sources for marine Thaumarchaeota. bioRxiv. doi: 10.1101/2021.07.22.453390
Damashek, J., Tolar, B. B., Liu, Q., Okotie-Oyekan, A. O., Wallsgrove, N. J., Popp, B. N., et al. (2019). Microbial oxidation of nitrogen supplied as selected organic nitrogen compounds in the South Atlantic bight. Limnol. Oceanogr. 64, 982–995. doi: 10.1002/lno.11089
de Boer, W., and Laanbroek, H. J. (1989). Ureolytic nitrification at low pH by Nitrosospira spec. Arch. Microbiol. 152, 178–181. doi: 10.1007/BF00456098
Denger, K., Weinitschke, S., Hollemeyer, K., and Cook, A. M. (2004). Sulfoacetate generated by Rhodopseudomonas palustris from taurine. Arch. Microbiol. 182, 254–258. doi: 10.1007/s00203-004-0678-0
Engelberts, J. P., Robbins, S. J., de Goeij, J. M., Aranda, M., Bell, S. C., and Webster, N. S. (2020). Characterization of a sponge microbiome using an integrative genome-centric approach. ISME J. 14, 1100–1110. doi: 10.1038/s41396-020-0591-9
Falke, D., Schulz, K., Doberenz, C., Beyer, L., Lilie, H., Thiemer, B., et al. (2010). Unexpected oligomeric structure of the FocA formate channel of Escherichia coli: a paradigm for the formate–nitrite transporter family of integral membrane proteins. FEMS Microbiol. Lett. 303, 69–75. doi: 10.1111/j.1574-6968.2009.01862.x
Ferrer-González, F. X., Hamilton, M., Smith, C. B., Schreier, J. E., Olofsson, M., and Moran, M. A. (2023). Bacterial transcriptional response to labile exometabolites from photosynthetic picoeukaryote Micromonas commoda. ISME Commun. 3:5. doi: 10.1038/s43705-023-00212-0
Fujitani, H., Momiuchi, K., Ishii, K., Nomachi, M., Kikuchi, S., Ushiki, N., et al. (2020). Genomic and physiological characteristics of a novel nitrite-oxidizing Nitrospira strain isolated from a drinking water treatment plant. Front. Microbiol. 11:545190. doi: 10.3389/fmicb.2020.545190
Glass, J. B., Wolfe-Simon, F., and Anbar, A. (2009). Coevolution of metal availability and nitrogen assimilation in cyanobacteria and algae. Geobiology 7, 100–123. doi: 10.1111/j.1472-4669.2009.00190.x
Glibert, P. M., Wilkerson, F. P., Dugdale, R. C., Raven, J. A., Dupont, C. L., Leavitt, P. R., et al. (2016). Pluses and minuses of ammonium and nitrate uptake and assimilation by phytoplankton and implications for productivity and community composition, with emphasis on nitrogen-enriched conditions. Limnol. Oceanogr. 61, 165–197. doi: 10.1002/lno.10203
Guo, F.-B., Lin, H., and Huang, J. (2009). A plot of G+ C content against sequence length of 640 bacterial chromosomes shows the points are widely scattered in the upper triangular area. Chromosom. Res. 17, 359–364. doi: 10.1007/s10577-009-9024-3
Hall, B. G. (2013). Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 30, 1229–1235. doi: 10.1093/molbev/mst012
He, H., Zhen, Y., Mi, T., Fu, L., and Yu, Z. (2018). Ammonia-oxidizing Archaea and Bacteria differentially contribute to ammonia oxidation in sediments from adjacent waters of Rushan Bay, China. Front. Microbiol. 9:116. doi: 10.3389/fmicb.2018.00116
Hu, H.-W., and He, J.-Z. (2017). Comammox—a newly discovered nitrification process in the terrestrial nitrogen cycle. J. Soils Sediments 17, 2709–2717. doi: 10.1007/s11368-017-1851-9
Igarashi, K., and Kashiwagi, K. (2010). Characteristics of cellular polyamine transport in prokaryotes and eukaryotes. Plant Physiol. Biochem. 48, 506–512. doi: 10.1016/j.plaphy.2010.01.017
Jørgensen, N. O. G. (2006). Uptake of urea by estuarine bacteria. Aquat. Microb. Ecol. 42, 227–242. doi: 10.3354/ame042227
Jung, M.-Y., Sedlacek, C. J., Kits, K. D., Mueller, A. J., Rhee, S.-K., Hink, L., et al. (2022). Ammonia-oxidizing archaea possess a wide range of cellular ammonia affinities. ISME J. 16, 272–283. doi: 10.1038/s41396-021-01064-z
Kim, J.-G., Park, S.-J., Sinninghe Damsté, J. S., Schouten, S., Rijpstra, W. I. C., Jung, M.-Y., et al. (2016). Hydrogen peroxide detoxification is a key mechanism for growth of ammonia-oxidizing archaea. Proc. Natl. Acad. Sci. 113, 7888–7893. doi: 10.1073/pnas.1605501113
Kitzinger, K., Koch, H., Lücker, S., Sedlacek, C. J., Herbold, C., Schwarz, J., et al. (2018). Characterization of the first “Candidatus Nitrotoga” isolate reveals metabolic versatility and separate evolution of widespread nitrite-oxidizing bacteria. MBio 9:e01186. doi: 10.1128/mBio.01186-18
Kitzinger, K., Marchant, H. K., Bristow, L. A., Herbold, C. W., Padilla, C. C., Kidane, A. T., et al. (2020). Single cell analyses reveal contrasting life strategies of the two main nitrifiers in the ocean. Nat. Commun. 11, 1–12. doi: 10.1038/s41467-020-14542-3
Kitzinger, K., Padilla, C. C., Marchant, H. K., Hach, P. F., Herbold, C. W., Kidane, A. T., et al. (2019). Cyanate and urea are substrates for nitrification by Thaumarchaeota in the marine environment. Nat. Microbiol. 4, 234–243. doi: 10.1038/s41564-018-0316-2
Koch, H., Lücker, S., Albertsen, M., Kitzinger, K., Herbold, C., Spieck, E., et al. (2015). Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc. Natl. Acad. Sci. 112, 11371–11376. doi: 10.1073/pnas.1506533112
Könneke, M., Bernhard, A. E., de La Torre, J. R., Walker, C. B., Waterbury, J. B., and Stahl, D. A. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546. doi: 10.1038/nature03911
Koper, T. E., El-Sheikh, A. F., Norton, J. M., and Klotz, M. G. (2004). Urease-encoding genes in ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 70, 2342–2348. doi: 10.1128/AEM.70.4.2342-2348.2004
Krempaska, N., Horňák, K., and Pernthaler, J. (2018). Spatiotemporal distribution and microbial assimilation of polyamines in a mesotrophic lake. Limnol. Oceanogr. 63, 816–832. doi: 10.1002/lno.10672
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lal, D., and Lal, R. (2010). Evolution of mercuric reductase (merA) gene: a case of horizontal gene transfer. Microbiology 79, 500–508. doi: 10.1134/S0026261710040120
Lam, P., Lavik, G., Jensen, M. M., van de Vossenberg, J., Schmid, M., Woebken, D., et al. (2009). Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc. Natl. Acad. Sci. 106, 4752–4757. doi: 10.1073/pnas.0812444106
Latocheski, E. C., da Rocha, M. C. V., and Braga, M. C. B. (2022). Nitrospira in wastewater treatment: applications, opportunities and research gaps. Rev. Environ. Sci. Biotechnol. 21, 905–930. doi: 10.1007/s11157-022-09634-z
Le, S. Q., and Gascuel, O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. doi: 10.1093/molbev/msn067
Levin, E. J., Quick, M., and Zhou, M. (2009). Crystal structure of a bacterial homologue of the kidney urea transporter. Nature 462, 757–761. doi: 10.1038/nature08558
Li, L., Zhang, W., Zhang, S., Song, L., Sun, Q., Zhang, H., et al. (2021). Bacteria and archaea synergistically convert glycine betaine to biogenic methane in the Formosa cold seep of the South China Sea. Msystems 6, e00703–e00721. doi: 10.1128/mSystems.00703-21
Liu, L., Liu, M., Jiang, Y., Lin, W., and Luo, J. (2021). Production and excretion of polyamines to tolerate high ammonia, a case study on soil ammonia-oxidizing archaeon “Candidatus Nitrosocosmicus agrestis”. Msystems 6, e01003–e01020. doi: 10.1128/mSystems.01003-20
Liu, Q., Lu, X., Tolar, B. B., Mou, X., and Hollibaugh, J. T. (2015). Concentrations, turnover rates and fluxes of polyamines in coastal waters of the South Atlantic bight. Biogeochemistry 123, 117–133. doi: 10.1007/s10533-014-0056-1
Liu, Q., Lu, Y., Xu, J., Zhu, Z.-Y., Yuan, Y., Ma, W.-C., et al. (2022a). Dissolved free amino acids and polyamines are two major dissolved organic nitrogen sources for marine bacterioplankton in the northern slope of the South China Sea. Biogeochemistry 157, 109–126. doi: 10.1007/s10533-021-00860-1
Liu, Q., Nishibori, N., and Hollibaugh, J. T. (2022b). Sources of polyamines in coastal waters and their links to phytoplankton. Mar. Chem. 242:104121. doi: 10.1016/j.marchem.2022.104121
Liu, Q., Nishibori, N., Imai, I., and Hollibaugh, J. T. (2016). Response of polyamine pools in marine phytoplankton to nutrient limitation and variation in temperature and salinity. Mar. Ecol. Prog. Ser. 544, 93–105. doi: 10.3354/meps11583
Luo, H., Thompson, L. R., Stingl, U., and Hughes, A. L. (2015). Selection maintains low genomic GC content in marine SAR11 lineages. Mol. Biol. Evol. 32, 2738–2748. doi: 10.1093/molbev/msv149
Madhuri, S., Wang, K., Bade, D., and Mou, X. (2019). Concentration and turnover of dissolved free polyamines on the south coast of Lake Erie. Limnol. Oceanogr. 64, 1641–1650. doi: 10.1002/lno.11141
Maeda, S. I., Murakami, A., Ito, H., Tanaka, A., and Omata, T. (2015). Functional characterization of the FNT family nitrite transporter of marine picocyanobacteria. Lifestyles 5, 432–446. doi: 10.3390/life5010432
Maeda, S.-I., and Omata, T. (2009). Nitrite transport activity of the ABC-type cyanate transporter of the cyanobacterium Synechococcus elongatus. J. Bacteriol. 191, 3265–3272. doi: 10.1128/JB.00013-09
Magadum, S., Banerjee, U., Murugan, P., Gangapur, D., and Ravikesavan, R. (2013). Gene duplication as a major force in evolution. J. Genet. 92, 155–161. doi: 10.1007/s12041-013-0212-8
Michael, A. J. (2016). Biosynthesis of polyamines and polyamine-containing molecules. Biochem. J. 473, 2315–2329. doi: 10.1042/BCJ20160185
Michael, A. J. (2018). Polyamine function in archaea and bacteria. J. Biol. Chem. 293, 18693–18701. doi: 10.1074/jbc.TM118.005670
Minocha, R., Studley, K., Saier, J., and Milton, H. (2003). The urea transporter (UT) family: bioinformatic analyses leading to structural, functional, and evolutionary predictions. Recept. Channels 9, 345–352. doi: 10.3109/714041015
Mou, X., Sun, S., Rayapati, P., and Moran, M. A. (2010). Genes for transport and metabolism of spermidine in Ruegeria pomeroyi DSS-3 and other marine bacteria. Aquat. Microb. Ecol. 58, 311–321. doi: 10.3354/ame01367
Mou, X., Vila-Costa, M., Sun, S., Zhao, W., Sharma, S., and Moran, M. A. (2011). Metatranscriptomic signature of exogenous polyamine utilization by coastal bacterioplankton. Environ. Microbiol. Rep. 3, 798–806. doi: 10.1111/j.1758-2229.2011.00289.x
Noell, S. E., Barrell, G. E., Suffridge, C., Morré, J., Gable, K. P., Graff, J. R., et al. (2021). SAR11 cells rely on enzyme multifunctionality to metabolize a range of polyamine compounds. MBio 12:e0109121. doi: 10.1128/mBio.01091-21
Offre, P., Kerou, M., Spang, A., and Schleper, C. (2014). Variability of the transporter gene complement in ammonia-oxidizing archaea. Trends Microbiol. 22, 665–675. doi: 10.1016/j.tim.2014.07.007
Pachiadaki, M. G., Sintes, E., Bergauer, K., Brown, J. M., Record, N. R., Swan, B. K., et al. (2017). Major role of nitrite-oxidizing bacteria in dark ocean carbon fixation. Science 358, 1046–1051. doi: 10.1126/science.aan8260
Palatinszky, M., Herbold, C., Jehmlich, N., Pogoda, M., Han, P., von Bergen, M., et al. (2015). Cyanate as an energy source for nitrifiers. Nature 524, 105–108. doi: 10.1038/nature14856
Palomo, A., Pedersen, A. G., Fowler, S. J., Dechesne, A., Sicheritz-Pontén, T., and Smets, B. F. (2018). Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira. ISME J. 12, 1779–1793. doi: 10.1038/s41396-018-0083-3
Pearson, W. R. (2013). An introduction to sequence similarity (“homology”) searching. Curr Protoc Bioinform 42, 3.1.1–3.1.8. doi: 10.1002/0471250953.bi0301s42
Qin, W., Amin, S. A., Martens-Habbena, W., Walker, C. B., Urakawa, H., Devol, A. H., et al. (2014). Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc. Natl. Acad. Sci. 111, 12504–12509. doi: 10.1073/pnas.1324115111
Qin, W., Heal, K. R., Ramdasi, R., Kobelt, J. N., Martens-Habbena, W., Bertagnolli, A. D., et al. (2017). Nitrosopumilus maritimus gen. Nov., sp. nov., Nitrosopumilus cobalaminigenes sp. nov., Nitrosopumilus oxyclinae sp. nov., and Nitrosopumilus ureiphilus sp. nov., four marine ammonia-oxidizing archaea of the phylum Thaumarchaeota. Int. J. Syst. Evol. Microbiol. 67, 5067–5079. doi: 10.1099/ijsem.0.002416
Rasche, M. E., Hyman, M. R., and Arp, D. J. (1991). Factors limiting aliphatic chlorocarbon degradation by Nitrosomonas europaea: cometabolic inactivation of ammonia monooxygenase and substrate specificity. Appl. Environ. Microbiol. 57, 2986–2994. doi: 10.1128/aem.57.10.2986-2994.1991
Raunser, S., Mathai, J. C., Abeyrathne, P. D., Rice, A. J., Zeidel, M. L., and Walz, T. (2009). Oligomeric structure and functional characterization of the urea transporter from Actinobacillus pleuropneumoniae. J. Mol. Biol. 387, 619–627. doi: 10.1016/j.jmb.2009.02.005
Ren, M., Feng, X., Huang, Y., Wang, H., Hu, Z., Clingenpeel, S., et al. (2019). Phylogenomics suggests oxygen availability as a driving force in Thaumarchaeota evolution. ISME J. 13, 2150–2161. doi: 10.1038/s41396-019-0418-8
Rittstieg, K., Robra, K.-H., and Somitsch, W. (2001). Aerobic treatment of a concentrated urea wastewater with simultaneous stripping of ammonia. Appl. Microbiol. Biotechnol. 56, 820–825. doi: 10.1007/s002530100696
Rohwerder, T. (2020). New structural insights into bacterial sulfoacetaldehyde and taurine metabolism. Biochem. J. 477, 1367–1371. doi: 10.1042/BCJ20200079
Rycovska, A., Hatahet, L., Fendler, K., and Michel, H. (2012). The nitrite transport protein NirC from Salmonella typhimurium is a nitrite/proton antiporter. Biochim Biophys Acta Biomemb 1818, 1342–1350. doi: 10.1016/j.bbamem.2012.02.004
Sauder, L. A., Albertsen, M., Engel, K., Schwarz, J., Nielsen, P. H., Wagner, M., et al. (2017). Cultivation and characterization of Candidatus Nitrosocosmicus exaquare, an ammonia-oxidizing archaeon from a municipal wastewater treatment system. ISME J. 11, 1142–1157. doi: 10.1038/ismej.2016.192
Sauder, L. A., Engel, K., Lo, C.-C., Chain, P., and Neufeld, J. D. (2018). “Candidatus Nitrosotenuis aquarius,” an Ammonia-oxidizing archaeon from a freshwater aquarium biofilter. Appl. Environ. Microbiol. 84, e01430–e01418. doi: 10.1128/AEM.01430-18
Seitzinger, S. P., Sanders, R., and Styles, R. (2002). Bioavailability of DON from natural and anthropogenic sources to estuarine plankton. Limnol. Oceanogr. 47, 353–366. doi: 10.4319/lo.2002.47.2.0353
Shakhman, Y., and Harries, D. (2021). How glycine betaine modifies lipid membrane interactions. Chem Syst Chem 3:e2100010. doi: 10.1002/syst.202100010
Sheridan, P. O., Raguideau, S., Quince, C., Holden, J., Zhang, L., Consortium, T., et al. (2020). Gene duplication drives genome expansion in a major lineage of Thaumarchaeota. Nat. Commun. 11:5494. doi: 10.1038/s41467-020-19132-x
Sipler, R. E., Bronk, D. A., Seitzinger, S. P., Lauck, R. J., McGuinness, L., Kirkpatrick, G. J., et al. (2013). Trichodesmium-derived dissolved organic matter is a source of nitrogen capable of supporting the growth of toxic red tide Karenia brevis. Mar. Ecol. Prog. Ser. 483, 31–45. doi: 10.3354/meps10258
Sliekers, A. O., Haaijer, S., Schmid, M., Harhangi, H., Verwegen, K., Kuenen, J. G., et al. (2004). Nitrification and anammox with urea as the energy source. Syst. Appl. Microbiol. 27, 271–278. doi: 10.1078/0723-2020-00259
Soliman, M., and Eldyasti, A. (2018). Ammonia-oxidizing Bacteria (AOB): opportunities and applications—a review. Rev. Environ. Sci. Biotechnol. 17, 285–321. doi: 10.1007/s11157-018-9463-4
Solomon, C. M., Collier, J. L., Berg, G. M., and Glibert, P. M. (2010). Role of urea in microbial metabolism in aquatic systems: a biochemical and molecular review. Aquat. Microb. Ecol. 59, 67–88. doi: 10.3354/ame01390
Spang, A., Poehlein, A., Offre, P., Zumbrägel, S., Haider, S., Rychlik, N., et al. (2012). The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ. Microbiol. 14, 3122–3145. doi: 10.1111/j.1462-2920.2012.02893.x
Stahl, D. A., and de la Torre, J. R. (2012). Physiology and diversity of ammonia-oxidizing archaea. Annu. Rev. Microbiol. 66, 83–101. doi: 10.1146/annurev-micro-092611-150128
Taubert, M., Grob, C., Howat, A. M., Burns, O. J., Pratscher, J., Jehmlich, N., et al. (2017). Methylamine as a nitrogen source for microorganisms from a coastal marine environment. Environ. Microbiol. 19, 2246–2257. doi: 10.1111/1462-2920.13709
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tolar, B. B., Ross, M. J., Wallsgrove, N. J., Liu, Q., Aluwihare, L. I., Popp, B. N., et al. (2016). Contribution of ammonia oxidation to chemoautotrophy in Antarctic coastal waters. ISME J. 10, 2605–2619. doi: 10.1038/ismej.2016.61
Tourna, M., Stieglmeier, M., Spang, A., Könneke, M., Schintlmeister, A., Urich, T., et al. (2011). Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. 108, 8420–8425. doi: 10.1073/pnas.1013488108
Uemura, T., Kashiwagi, K., and Igarashi, K. (2007). Polyamine uptake by DUR3 and SAM3 in Saccharomyces cerevisiae. J. Biol. Chem. 282, 7733–7741. doi: 10.1074/jbc.M611105200
Valladares, A., Montesinos, M. L., Herrero, A., and Flores, E. (2002). An ABC-type, high-affinity urea permease identified in cyanobacteria. Mol. Microbiol. 43, 703–715. doi: 10.1046/j.1365-2958.2002.02778.x
Van Kessel, M. A., Speth, D. R., Albertsen, M., Nielsen, P. H., Op den Camp, H. J., Kartal, B., et al. (2015). Complete nitrification by a single microorganism. Nature 528, 555–559. doi: 10.1038/nature16459
Veaudor, T., Cassier-Chauvat, C., and Chauvat, F. (2019). Genomics of urea transport and catabolism in cyanobacteria: biotechnological implications. Front. Microbiol. 10:2052. doi: 10.3389/fmicb.2019.02052
Vijayan, A., Vattiringal Jayadradhan, R. K., Pillai, D., Prasannan Geetha, P., Joseph, V., and Isaac Sarojini, B. S. (2021). Nitrospira as versatile nitrifiers: taxonomy, ecophysiology, genome characteristics, growth, and metabolic diversity. J. Basic Microbiol. 61, 88–109. doi: 10.1002/jobm.202000485
Voss, M., Bange, H. W., Dippner, J. W., Middelburg, J. J., Montoya, J. P., and Ward, B. (2013). The marine nitrogen cycle: recent discoveries, uncertainties and the potential relevance of climate change. Philos Transac Roy Soc B Biol Sci 368:20130121. doi: 10.1098/rstb.2013.0121
Wang, W. H., Köhler, B., Cao, F. Q., Liu, G. W., Gong, Y. Y., Sheng, S., et al. (2012). Rice DUR3 mediates high-affinity urea transport and plays an effective role in improvement of urea acquisition and utilization when expressed in Arabidopsis. New Phytol. 193, 432–444. doi: 10.1111/j.1469-8137.2011.03929.x
Wargo, M. J. (2013). Homeostasis and catabolism of choline and glycine betaine: lessons from Pseudomonas aeruginosa. Appl. Environ. Microbiol. 79, 2112–2120. doi: 10.1128/AEM.03565-12
Wetzel, K. J., Bjorge, D., and Schwan, W. R. (2011). Mutational and transcriptional analyses of the Staphylococcus aureus low-affinity proline transporter OpuD during in vitro growth and infection of murine tissues. FEMS Immunol Med Microbiol 61, 346–355. doi: 10.1111/j.1574-695X.2011.00781.x
Wiechert, M., and Beitz, E. (2017). Mechanism of formate–nitrite transporters by dielectric shift of substrate acidity. EMBO J. 36, 949–958. doi: 10.15252/embj.201695776
Woebken, D., Lam, P., Kuypers, M. M., Naqvi, S. W. A., Kartal, B., Strous, M., et al. (2008). A microdiversity study of anammox bacteria reveals a novel Candidatus Scalindua phylotype in marine oxygen minimum zones. Environ. Microbiol. 10, 3106–3119. doi: 10.1111/j.1462-2920.2008.01640.x
Wright, C. L., Schatteman, A., Crombie, A. T., Murrell, J. C., and Lehtovirta-Morley, L. E. (2020). Inhibition of ammonia monooxygenase from ammonia-oxidizing archaea by linear and aromatic alkynes. Appl. Environ. Microbiol. 86, e02388–e02419. doi: 10.1128/AEM.02388-19
Xiong, Z., Zhang, N., Xu, L., Deng, Z., Limwachiranon, J., Guo, Y., et al. (2023). Urease of aspergillus fumigatus is required for survival in macrophages and virulence. Microbiol Spectr 11, e03508–e03522. doi: 10.1128/spectrum.03508-22
Yang, Y., Daims, H., Liu, Y., Herbold, C. W., Pjevac, P., Lin, J.-G., et al. (2020). Activity and metabolic versatility of complete ammonia oxidizers in full-scale wastewater treatment systems. MBio 11, e03175–e03119. doi: 10.1128/mBio.03175-19
Yang, Y., Zhang, C., Lenton, T. M., Yan, X., Zhu, M., Zhou, M., et al. (2021). The evolution pathway of ammonia-oxidizing archaea shaped by major geological events. Mol. Biol. Evol. 38, 3637–3648. doi: 10.1093/molbev/msab129
Keywords: ammonia-oxidizing archaea, ammonia-oxidizing bacteria, nitrite-oxidizing bacteria, comammox, dissolved organic nitrogen, urea, polyamine, cyanate
Citation: Liu Q, Chen Y and Xu X-W (2023) Genomic insight into strategy, interaction and evolution of nitrifiers in metabolizing key labile-dissolved organic nitrogen in different environmental niches. Front. Microbiol. 14:1273211. doi: 10.3389/fmicb.2023.1273211
Edited by:
Long Jin, Nanjing Forestry University, ChinaReviewed by:
Xuesong Luo, Huazhong Agricultural University, ChinaYantao Liang, Ocean University of China, China
Copyright © 2023 Liu, Chen and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Liu, bGl1cWlhbkBzaW8ub3JnLmNu
†These authors have contributed equally to this work and share first authorship
 Qian Liu
Qian Liu Yuhao Chen2,4†
Yuhao Chen2,4† Xue-Wei Xu
Xue-Wei Xu