
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 23 November 2023
Sec. Virology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1269869
This article is part of the Research TopicRecent highlights in the development of therapeutic antiviral strategiesView all 12 articles
 Joshua Wiggins1,2†
Joshua Wiggins1,2† Ngan Nguyen1,3†
Ngan Nguyen1,3† Wenzhong Wei1,3
Wenzhong Wei1,3 Leah Liu Wang1,3
Leah Liu Wang1,3 Haley Hollingsead Olson1,3
Haley Hollingsead Olson1,3 Shi-Hua Xiang1,3*
Shi-Hua Xiang1,3*Scytovirin (SVN) is a lectin from cyanobacteria which has a strong inhibitory activity against Ebola virus infection. We engineered scytovirin as the inhibitor for surface display of lactic acid bacteria to block Ebola virus infection. Two different bacterial strains (Lactobacillus casei and Lactococcus lactis) were successfully engineered for scytovirin expression on the bacterial surface. These bacteria were found to be effective at neutralizing pseudotyped Ebolavirus in a cell-based assay. This approach can be utilized for prophylactic prevention, as well as for treatment. Since lactic acid bacteria can colonize the human body, a long-term efficacy could be achieved. Furthermore, this approach is also simple and cost-effective and can be easily applied in the regions of Ebola outbreaks in the developing countries.
Ebola virus (EBOV) is an enveloped negative-stranded RNA virus which can cause severe Ebola viral disease (EVD) in humans and nonhuman primates (Jacob et al., 2020). EBOV belongs to the family Filoviridae along with Marburg virus that also causes a similar disease. Ebolavirus in Zaire was first identified in 1976 in Africa (Bowen et al., 1980), now five Ebola species have been recognized (Kuhn et al., 2019): Zaire ebolavirus (ZEBOV or EBOV), Sudan ebolavirus (SUDV), Tai Forest ebolavirus (TAFV), Reston ebolavirus (RESTV) and Bundibugyo ebolavirus (BDBV). Ebolavirus has produced more than 20 outbreaks in humans with high mortality rates from 25 to 90% (Feldmann and Geisbert, 2011;WHO, n.d.). The recent large Ebola outbreak in 2014 in West Africa infected more than 28,000 people and more than 11,000 died (Cenciarelli et al., 2015). It is evident that this emerging and reemerging viral pathogen represents a great threat to human health. However, we do not have any medicines to treat this lethal viral disease until December 2019, when a vaccine (rVSVΔG-ZEBOV-GP) was approved by FDA for limited use against Zaire Ebolavirus, and in 2020, two antibody drugs (mAb114 and REGN-EB3) were approved for treating this viral infection. It is apparent that more medicines are required for fighting against this deadly, infectious viral disease.
Scytovirin (SVN) is a small protein of 95 amino acids which was first identified from cyanobacteria of Scytonema varium (Bokesch et al., 2003). Scytovirin is a type of lectin which are known carbohydrate-binding proteins with non-immunologic nature (Fernandez Romero et al., 2021). Because lectins recognize the carbohydrates on the glycoproteins of viral particles, they usually exhibit antiviral activities (Akkouh et al., 2015; Naik and Kumar, 2022). Lectins from algae, plants and cyanobacteria show strong inhibitory activity against HIV infection (Li et al., 2008; Akkouh et al., 2015). Due to their inhibitory effect and immunosuppressive properties, lectins such as cyanovirin-N (CV-N) (Colleluori et al., 2005; Li et al., 2011; O'Keefe et al., 2015; Vamvaka et al., 2016) and griffithsin (GRFT) (Emau et al., 2007; Girard et al., 2018; Alexandre et al., 2020), have been used as microbicides for treating viral diseases (Abdool Karim and Baxter, 2012; Huskens and Schols, 2012; Koharudin and Gronenborn, 2014) and have been investigated for pre-exposure prophylaxis (PrEP) against HIV-1 infection. Like the well-studied lectins CV-N and GRFT in HIV research, SVN has also shown highly specific activity to the high mannose moieties that exist on the surface of glycoproteins of HIV, Ebola, and Marburg viruses (Barrientos and Gronenborn, 2005, Mori et al., 2005, Alexandre et al., 2010). More studies have demonstrated that scytovirin has much stronger activity against Ebola than HIV in vitro and in vivo (Garrison et al., 2014).
Because scytovirin demonstrated potent antiviral activity against Ebola infection, we used it to develop a novel live microbicide for Ebola infection control. This approach displayed scytovirin on the surface of Lactic acid bacteria (LAB) for delivery against Ebola virus infection. These bacteria can be delivered into mucosal surfaces of the body (e.g., mouth, nose, and GI tract) that are the ports of viral entry where they can colonize and replicate. Previous studies using LAB expressing the antiviral lectin CV-N demonstrated that the bacteria can pass through the GI tract unharmed and stably produce lectins to inhibit HIV (Lagenaur et al., 2011, Li et al., 2011, Brichacek et al., 2013). Therefore, this approach for application of viral inhibition should be safe, long-lasting and effective. Furthermore, this approach is cost-effective and easy-to-use, so it is especially valuable for use in those outbreak regions in Africa. Here, we reported the in vitro data we have achieved successfully in this research direction.
Escherichia coli DH5α and E. coli BL21 DE3 were used for cloning and initial protein verification, respectively. Both were cultured in LB media (FisherSci, BP1427-500) overnight in a shaker at 250 rpm and 37°C. The antibiotics used for plasmid selection were 50 μg/mL kanamycin for pET28a and 250 μg/mL erythromycin for pLSVN3 and pLSVN7 (Table 1). L. casei and L. lactis were cultured statically in MRS media (FisherSci, CM0359) at 37°C and 5% CO2 for 2–3 days until reaching log phase. For engineered bacterial culturing, 5 μg/mL erythromycin was added to the MRS media for applying selection pressure. HEK 293 T and TZM-bl mammalian cell lines were grown in Dulbecco Minimum Essential Media (DMEM) (Gibco, 11,965,092) supplemented with 10% FBS (Gibco, 10,082), 1 mM L-glutamine (Gibco, 25,030), and 100 μg/mL penicillin/streptomycin (Gibco, 15,140). Cells were cultured in a humidified cell culture incubator at 37°C with 5% CO2 using T-75 flasks.
The scytovirin (SVN) gene sequence (GenBank: P86041.1) (Bokesch et al., 2003) with an added E-tag sequence (total 348 bp) was synthesized with SacI/XhoI restriction sites by GenScript and inserted into the vector pET28a (Novagen, Inc.) to test protein expression in E. coli BL21 (DE3) bacterial cells. SVN was overexpressed in E. coli previously by Xiong et al. (2006). SVN expression was induced by IPTG (1 mM) to increase protein production which was verified by Western blotting using an anti-E-tag (ab3397, Abcam), anti-His-tag (HRP-66005, Proteintech) or anti-SVN (A64238-050, Epigentek) polyclonal antibodies.
To express the SVN protein on the surface of LAB, constructs were created which included the Lactate dehydrogenase promoter (Pldh), signal peptide (SP), cell membrane anchor protein (ANC), E-tag marker (E), and the protein marker (GFP). The constructs were built up based on our previous plasmid pWZ486 (Wei et al., 2019) derived from pTRKH3-ldhGFP (Addgene). The SVN-E-tag-GFP fusion sequence replaced the CD4 gene sequence in pWZ486 using the SacI/XhoI restriction sites. The constructs pLSVN3 and pLSVN7 were verified by DNA sequencing and PCR using the specific primers (Table 1) marked in the construct maps.
Plasmids were transformed into the Lactic acid bacteria (LAB) by electroporation as previously described (Wei et al., 2019; Welker et al., 2019). Briefly, overnight cultures of LAB cells were diluted (1:50) into fresh MRS media with 1% glycine and incubated at 37°C without shaking for 2 h. Cells were harvested and treated with 50 mM EDTA (pH 8.0) for 15 min, followed by two washes with ice-cold electroporation buffer (0.5 M sucrose) and resuspended in electroporation buffer (1/100 volume of the initial culture). 50 μL of cells were mixed with plasmid DNA and incubated on ice for 15 min. The mixture was added to an ice-cold 0.2 cm GenePulser (Biorad) cuvette and pulse was immediately applied at the conditions of 10 KV/cm, 200 Ω, and 25 μF. Cells were suspended in 1 mL MRS broth with 2 mM CaCl2 and 20 mM MgCl2 and then incubated at 37°C for 4 h. Cells were pooled on MRS plates with 5 μg/mL erythromycin and cultured as described above. To verify transformation, single colonies were picked and added to 25 μL PCR master mix (Promega, M791B) containing 1 μM forward and reverse primers (Table 1). PCR amplification occurred in a SimpliAmp thermocycler (Applied BioSystems, A24811) under the following conditions: Stage 1; 95°C for 5 min, Stage 2 (35 cycles); 95°C for 30 s, 55°C for 30 s, 72°C for 1 min, Stage 3; 72°C for 7 min. PCR products were visualized on 1% agarose gel electrophoresis.
For the pLSVN3 construct transformed into L. casei, the fusion protein was detected based on the GFP fluorescence. Bacteria were washed three times with PBS and analyzed on a BD FACSAria using a 488 nm laser. For the pLSVN7 construct transformed into L. lactis, the bacteria were first stained for 1 h with two primary antibodies: mouse monoclonal anti E-tag (Novus NBP2-67081) and rabbit polyclonal anti SVN (A64238-050, Epigentek). Goat anti mouse conjugated with AlexaFluor488 (A-11011, ThermoFisher) and goat anti rabbit conjugated with AlexaFluor594 (A-11012, ThermoFisher) were used as secondary antibodies, respectively. Bacteria were analyzed on a Beckman Coulter CytoFLEX FX at 488 nm and 561 nm. Unstained bacteria and stained wild-type bacteria were used in all experiments for an appropriate gating strategy.
Confocal microscopy was performed using the same fluorescent fusion protein or antibody combinations as described for flow cytometry (above). Following the antibody staining, bacteria were pelleted, resuspended in 10 μL PBS, and transferred to a microscope slide with cover slip. Images were captured using a Nikon A1R-Ti2 (Nikon Instruments, NY, USA) inverted confocal system.
The Ebola pseudotyped viruses were made from a HIV-1 backbone plasmid, pSG3ΔEnv (NIH HIV Reagent Program). The Ebola Envelope gene (GP, Zaire ebolavirus, GenBank: AIO11753.1), was synthesized by GenScript and cloned into pcDNA3.1(+). Both plasmids were co-transfected into 293 T cells in a 10 cm plate using transfection reagent polyethyleneimine (PEI). Three days post transfection, the medium was harvested and centrifuged at 500 g to remove cell debris, and then the supernatants were stored at −80°C (Platt et al., 2009; Wang et al., 2022). The viral titers were determined by reverse transcriptase assay.
The titers of pseudotyped viruses were determined by Reverse transcriptase assay (RTA) (Wei et al., 2019). 500 μL of pseudotyped virus stock was spun at 14,000 g for 2 h at 4°C to precipitate the virus. The viral pellet was resuspended in a Triton X-100-based suspension buffer and vortexed, followed by three rapid freeze–thaw cycles to lyse the viruses. 50 μL of reaction mix [Oligo-dT Poly-A and 3H-dTTP (PerkinElmer)] was added, and the samples were incubated at 37°C for 1 h in a heating block. Then, the samples were pipetted onto DEAE Filter mat circle papers (PerkinElmer), followed by three 10 min washes in 2X SSC buffer, and one 10 s wash in 100% ethanol. The filters were dried at room temperature and analyzed using a scintillation counter to measure the incorporation of 3H-dTTP into cDNA. The average CPM values from duplicates were determined.
Pseudotyped Ebola virus stocks were mixed with wild-type bacteria or engineered bacteria (~5×107/mL) in 1.5 mL micro-centrifuge tubes. The mixtures of bacteria and viruses were incubated for 1 h at room temperature. Then the tubes were spun for 1 min at a 13,000 g to remove the bacteria and bound pseudotyped virus. The supernatants were collected, and the viral titers determined by RTA.
For the neutralization assay, pseudotyped Ebola virus was mixed with wild-type or engineered bacteria in the same manner as the adsorption assay described above. After centrifugation to remove the bacteria and bound pseudovirus, the remaining supernatants were applied to TZM-bl cells which were used as the target cells due to their ability to express luciferase when infected (Platt et al., 2009). The TZM-bl cells were set at a density of 6.0 × 103 per well in a 96-well plate. Each neutralization assay was performed in triplicate with 5,000 RT units of pseudovirus per well used as the starting titer. Two days post-infection, the supernatants were removed, the cells were washed once with PBS, lysed in 1x Passive Lysis Buffer, and frozen at −80°C. The plates were then thawed, and luciferase activity was measured using beetle luciferin substrate (Promega) in a Veritas Luminometer.
Statistical analyses were conducted for virus adsorption and virus neutralization data using GraphPad Prism software (version 9.0). The significances were determined by using unpaired two-tailed Student’s t-test at p-value ≤0.05.
The scytovirin gene was initially synthesized by adding an E-tag at the N-terminus and cloned into pET28a with SacI/XhoI sites. The SVN plasmid was transformed to E. coli BL21 (DE3) cells for protein expression. The SVN fusion protein was observed under induction of 1 mM IPTG by Coomassie blue staining. The induced band of ~16kD fusion protein was noticed clearly (Figure 1A). To further confirm this protein, Western blotting was carried out by using E-tag, His-tag, and SVN specific antibodies. Positive bands were observed from all three different specific antibodies (Figure 1B), suggesting that the ~16kD SVN-E-tag fusion protein is expressed correctly. Figure 1C shows the construct design including the two His-tags encoded by the pET28a vector, as well the SVN ribbon structure with two highly similar domains (D1 and D2) for carbohydrate binding (Moulaei et al., 2007). Thus, the SVN-E-tag expressing construct can be further utilized for following bacterial engineering.
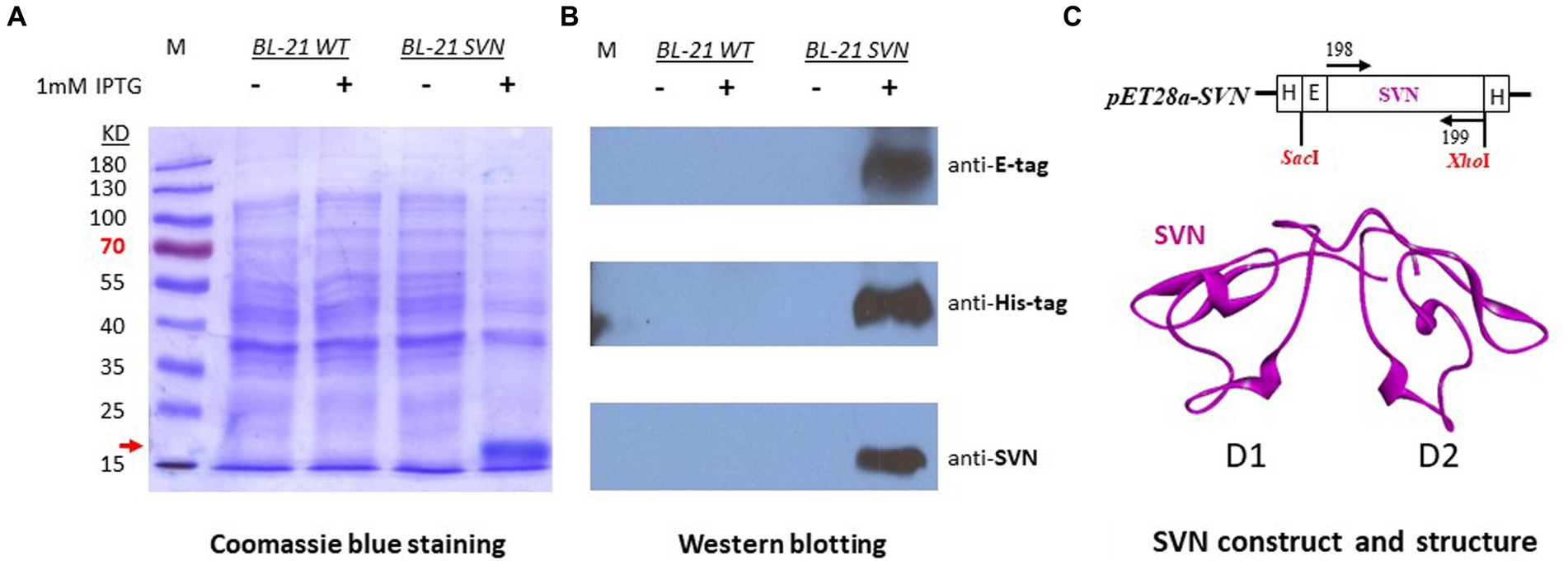
Figure 1. Expression of SVN-E-tag-His-tag in bacterial Escherichia coli K12 BL21 cells. (A) Coomassie blue staining, the induced fusion protein band was marked with the red arrow. (B) Western blots showing the specific positive bands. The specific antibody for anti-SVN is a scytovirin polyclonal antibody. (C) The SVN construct in pET28a and the SVN 3D structure, showing the two similar domains (D1 and D2) in magenta. H, His-tag, E, E-tag.
The SVN-E-tag construct was cloned into our previous plasmid (pWZ486) through SacI/XhoI sites to produce a new construct designated as pLSVN3 (Figure 2A). This GFP-E-tag-SVN fusion protein with the anchor has been modeled and shown in Figure 2B, which should be flexible for capturing viral particles. The construct was created first in E. coli DH5α cells and demonstrated the PCR fragment of ~1800 bp of GFP-E-tag-SVN sequence was correctly amplified using primers 154 and 037 (Figure 2C). To engineer the L. casei strain, the SVN construct pLSVN3 was transformed by electroporation into the L. casei for protein expression. The transformants were verified by PCR. A ~ 1800 bp PCR band from the amplification with primers 154 and 037 revealed that the pLSVN3 plasmid was transformed into the L. casei cells (Figure 2D). A ~ 75kD band corresponding to the fusion protein size (GFP-E-SVN-ANC) was detected by Western blotting using specific GFP and SVN antibodies (Figure 2E), suggesting that the SVN-GFP based fusion protein was produced from the L. casei cells. The smaller size band (~43kD) that appeared is the partial fragment of the fusion protein complex without the anchor domain (ANC). Flow cytometry analysis revealed the rate of SVN-GFP positive cells was 33.5% (Figure 2F a and b). To determine whether these protein inhibitors are displayed on the bacterial surface, confocal microscopy was used to visualize these SVN-GFP fused proteins. The pictures from confocal microscopy exhibited the fusion protein in green (GFP) displayed on the surface of bacteria (Figure 2F b). The biological functional studies were conducted for virus binding and neutralization. The engineered bacteria showed a moderate binding activity to the pseudotyped Ebola particles, reduced 37.2% of virus load, but the wild-typed (WT) bacteria also showed weak binding to the pseudotyped viruses because of the unspecific binding, reduced 21.8% (Figure 3A). The virus inhibition assay indicated that SVN-expressing bacteria had a moderate inhibition activity against pseudotyped Ebola virus infection which was 39.1%, but the WT-bacteria also had 23% inhibition (Figure 3B). These moderate functions may be due to the lower positive rate of SVN-expressing cells and the GFP interference of SVN binding in the SVN-GFP-ANC fusion protein complex.

Figure 2. Plasmid constructs and characterizations of SVN-fusion protein expression in Lactobacillus casei. (A) Maps of scytovirin (SVN) plasmid constructs. Promoter, Pldh; SP, signal peptide; GFP, green fluorescent protein; E, E-tag; SVN, scytovirin; ANC, anchor domain. (B) Three-dimensional (3D) model of fusion protein of ANC-SVN-E-tag-GFP. (C) DNA gels showing the PCR bands with the pair of primers 154/037, indicating the total 1797 bp band from the positive colonies of E. coli: 1. pLSVN3 plasmid, 2. pLSVN3 C1 (colony 1), 3. pLSVN3 C2 (colony 2). 4. E. coli WT. (D) Verification of pLSVN3 plasmid in L. casei after electroporation using PCR primers 154/037. wt, wild-type L. casei; 1. pLSVN3 C1 (colony 1); 2. pLSVN3 C2 (colony 2). (E) Western blot for detecting the SVN-GFP fusion protein. 1. L. casei wild-type bacteria only. 2. Engineered L. casei (using anti-GFP antibody). 3. Engineered L. casei (using anti-SVN HRP conjugated polyclonal antibody, MBS7005164). (F) Flow cytometry analysis: (a), Wild-type L. casei (negative control), (b). Engineered L. casei and the images of SVN-GFP surface displayed bacteria.
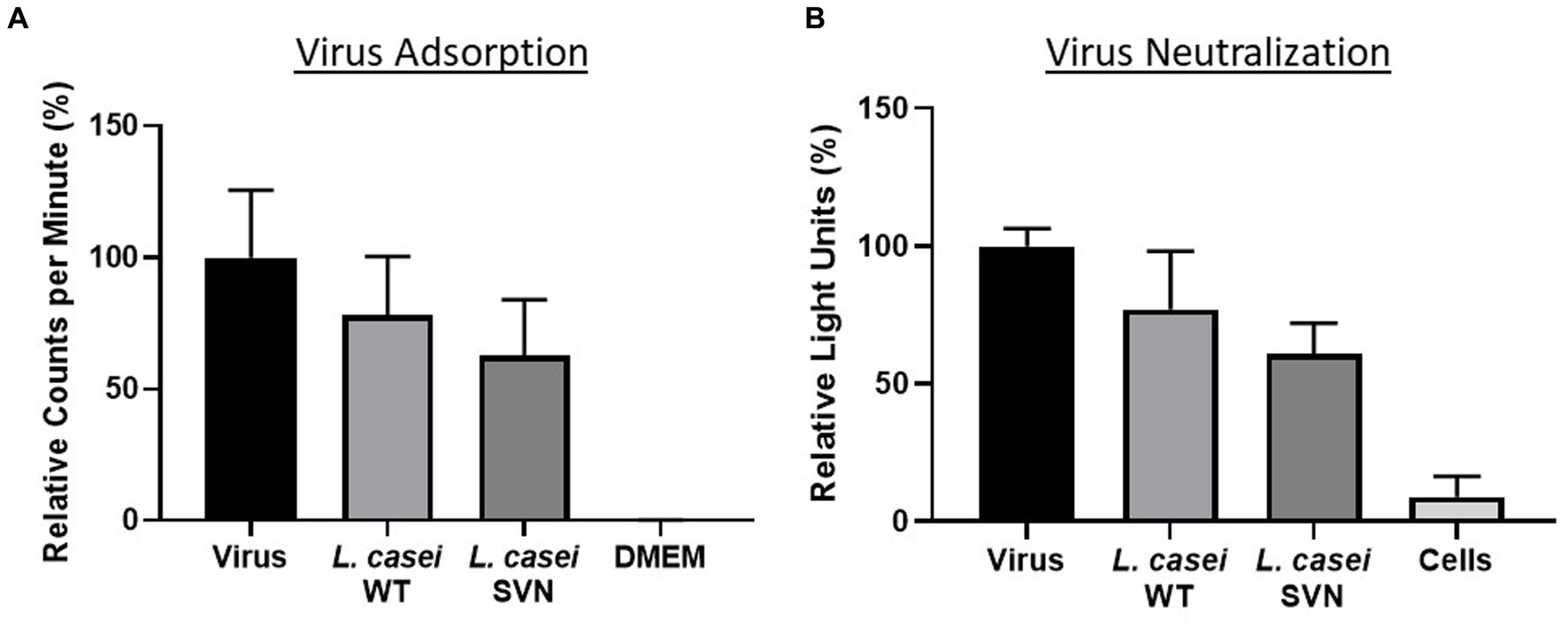
Figure 3. Functional studies of SVN-engineered L. casei bacteria. (A) Virus adsorption assay. (B) Virus neutralization assay. Wild-type (WT) L. casei as the negative bacterial control; virus only used as the positive control; DMEM medium or cells as the negative control.
Lactococcus lactis is another type of LAB which has a round shape and is widely used in the production of buttermilk and cheese. Thus, it is a very safe host strain candidate for anti-Ebola infection. We created the construct to remove the protein marker GFP but keep the E-tag marker from the construct pLSVN3, and this new construct was designated as pLSVN7 (Figure 2A). The construct pLSVN7 was transformed into the L. lactis using electroporation method. Positive bacterial colonies in the erythromycin selection plates were picked for further evaluation. The PCR method was first used to confirm the pLSVN7 plasmid was in the bacterium of L. lactis using primers 048 and 199. The expected ~700 bp band was identified (Figure 4A). Western blotting further demonstrated that SVN fusion protein (E-tag-SVN-ANC) was shown and in the correct molecular weight of 55kD both in anti-E-tag and anti-SVN specific antibodies. The 16kD SVN-E-tag fusion protein from the E. coli BL-21 (DE3) lysate as the positive control was also shown in the Western blots (Figures 4B,C). These Western blotting data demonstrated that the E-tag-SVN-ANC fusion protein was expressed in L. lactis. Next, the Confocal microscopy method was used to verify the surface display. Two colors of fluorescence were used for labeling E-tag (green) and SVN (red), respectively. The results demonstrated that the SVN fusion protein is clearly expressed on the surface of the bacterium. The single color, green or red and the merged color of green and red indicated the overlapping presence of fusion protein surface expression (Figure 5). Furthermore, Flow cytometry analysis was also conducted, and the data presented in Figure 6, shows that the positive rate of bacteria has reached 92.4%. The results demonstrated that the SVN-fusion protein is unambiguously expressed on the surface of these bacteria.
Figure 4. Characterizations of SVN expression in L. lactis. (A) PCR verification of SVN plasmid in L. lactis. PCR verified a positive band of ~700 bp from primers of 048 and SVN reverse primer 199. (B) Western blots showing anti-E-tag and Anti-SVN positive bands. 1. L. lactis WT, 2, L. lactis SVN. 3. E. coil SVN (positive control). SVN specific polyclonal antibody A64238-050 (Epigentek).
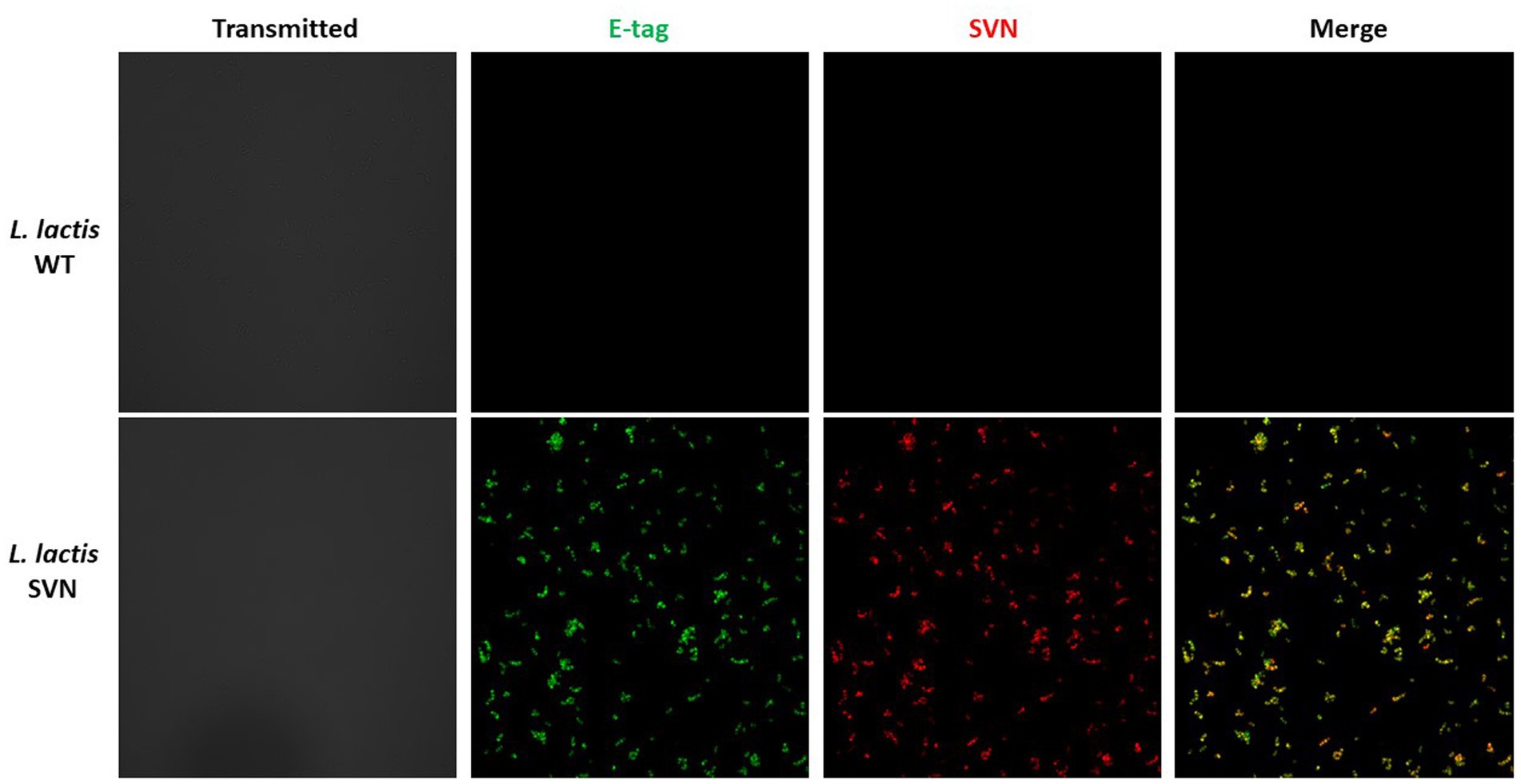
Figure 5. Confocal analysis of SVN-engineered Lactococcus lactis. Antibodies used for staining: AF-488 (green) for E-tag; AF-594 (red) for SVN. For more details, please see the Method section.
Figure 6. Flow cytometry analysis of SVN-engineered Lactococcus lactis. The 1st antibody (AF-488 conjugated, green) used is for staining E-tag; The 2nd antibody used (AF-594, red) is for staining SVN.
Functional studies were performed against the pseudotyped Ebola viruses. First, we used the viral particles adsorption method to evaluate the bacterial ability for capturing the viruses. We then tested the engineered strain activity to inhibit Ebola virus infection using TZM-bl cells. The virus adsorption and neutralization data are shown in Figure 7. Compared to the wild-type control bacteria, L. lactis, SVN engineered bacteria demonstrated improved functional abilities, absorbing 22.1% more pseudoviral particles and reducing infection by 28.5%. Overall, the SVN expressing L. lactis was able to adsorb 37.6% and neutralize 53.5% of the total pseudotyped Ebola virus, reducing infection in half compared to the virus only positive control (Figures 7A,B). In comparison, the wild-type L. lactis also demonstrated some ability to bind pseudotyped Ebola virus and reduce infection (15.5% adsorption and 24.7% neutralization) through non-specific binding, but the specific binding of the engineered bacteria to the viruses and are more than twice as effective, suggesting the SVN-expressing L. lactis bacteria can offer better protection against Ebola virus infection.
Figure 7. Functional studies of SVN-engineered L. lactis. (A) Virus adsorption assay. (B) Virus neutralization assay. Wild-type (WT) L. lactis as the negative bacterial control; virus only used as the positive control; DMEM medium or cells as the negative control. Significances were determined by using unpaired two-tailed Student’s t-test at p-value ≤0.05.
Bacterial therapy is a promising approach for human health and the common use of probiotics demonstrates its great potential (Yadav et al., 2020; Tegegne and Kebede, 2022). Using commensal bacteria for treating viral diseases has received broad attention since the bacterial microbicide has its advantages such as easy-to-use, cost-effective, and long-term efficacy (Ramachandran and Shanmughavel, 2009; Obiero et al., 2012). Especially, natural lectins as inhibitors can reduce unnecessary immune responses and enhance specific inhibiting activities for therapeutic applications (Akkouh et al., 2015; Mitchell et al., 2017; El-Maradny et al., 2021; Fernandez Romero et al., 2021; Mazur-Marzec et al., 2021; Naik and Kumar, 2022). For developing this method to use bacteria for antiviral diseases, bacterial engineering is a critical step. In this report, we engineered two types of Lactic acid bacteria (LAB) for surface display of the Ebola inhibitor scytovirin (SVN). Both bacterial strains successfully displayed scytovirin on the surface, suggesting the applicability of our constructs for surface expression of SVN fusion protein in lactic acid bacteria. The fluorescence of the SVN lectin fusion molecules when stained with GFP or SVN antibodies perfectly overlapped on the surface of the bacterium indicating a high display of inhibitors. The positive rate of engineered bacteria is higher in L. lactis (92.4%) than in L. casei (33.5%), suggesting that lower positive rate in L. casei is due to the higher genetic instability because certain bacteria may lose the SVN plasmid or SVN gene during the replication, especially when the antibiotic pressure is reduced. Another possibility is the genetic recombination between the SVN plasmid and host genome by which the plasmid composition could be changed or damaged. We found that LAB were capable of quickly developing resistance to erythromycin (5 μg/mL) selective pressure, especially when grown in stationary, liquid culturing conditions, suggesting that the bacteria lose the plasmid (data not shown). Thus, increased erythromycin concentration is usually needed to get more genetically stable strains. In general, using a genomic integration method for engineering would be better than plasmid transformation for making genetically stable strains, which is essential for clinical or therapeutic use. In this research, L. lactis was found to be highly stable and would be assessed for in vivo (mice) colonization, and for protective efficacy against challenge with infectious Ebola viruses conducted in the BSL-4 containment.
In the construction of fusion protein plasmids, the GFP protein marker seems unnecessary. Small protein tags such as E-tag appear to be adequate for detection and evaluation during the studies. Removing GFP reduced the size of the fusion protein by ~27kD, nearly 3x the size of SVN, maximizing the exposure of SVN inhibitor and preventing possible steric hindrance from GFP. Eliminating GFP from the fusion protein also presents another advantage for in vivo applications by avoiding the potential for immunogenicity and cytotoxicity caused by the GFP protein (Ansari et al., 2016).
In this report, our data from the in vitro study with pseudotyped Ebola virus serves as a proof of concept of using engineered commensal bacteria for blocking Ebola infection. Commensal bacteria have previously been demonstrated to inhibit HIV-1 infection (Nahui Palomino et al., 2017), and it is likely they provide a similar baseline level of protection at mucosal surfaces against other viral infections. This study indicates that this baseline level of protection is around 20–25% for WT L. lactis. Our data shows that lectin displaying bacteria can bind and neutralize significantly more viruses compared to the wild-type commensal bacteria, suggesting that engineered bacteria could be used prophylactically to improve upon the health benefit already provided by commensal bacteria by decreasing the likelihood of contracting a viral disease. This is contingent upon the engineered bacteria successfully colonizing various mucosal surfaces in the body and stably producing recombinant protein. To investigate this, we will test different routes of administration in mice (oral, nasal, rectal, etc.) to determine the stability of the L. lactis strain under different physiological conditions, as well as determining the efficacy of the bacteria to prevent in vivo infection prior to advancing to clinical trials. In conclusion, the commensal bacterial based anti-Ebola approach is promising and will be beneficial for combating this deadly viral disease.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
JW: Data curation, Methodology, Writing – review & editing, Formal analysis, Investigation, Validation, Visualization. NN: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – review & editing, Visualization. WW: Data curation, Formal Analysis, Investigation, Methodology, Writing – review & editing. LW: Formal Analysis, Investigation, Methodology, Writing – review & editing. HH: Investigation, Writing – review & editing. S-HX: Data curation, Investigation, Writing – review & editing, Conceptualization, Formal Analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. JW was supported by the NIH T-32 grant (5T32NS105594). This project was supported by the NIH R21 grant (1R21AI126299-01A1).
We thank the University of Nebraska - Lincoln Microscopy Core facility (You Zhou, Terri Fangman) and Flow Cytometry Core facility (Dirk Anderson) for helping with sample analyses.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdool Karim, S. S., and Baxter, C. (2012). Overview of microbicides for the prevention of human immunodeficiency virus. Best Pract. Res. Clin. Obstet. Gynaecol. 26, 427–439. doi: 10.1016/j.bpobgyn.2012.01.010
Akkouh, O., Ng, T. B., Singh, S. S., Yin, C., Dan, X., Chan, Y. S., et al. (2015). Lectins with anti-HIV activity: a review. Molecules 20, 648–668. doi: 10.3390/molecules20010648
Alexandre, K. B., Gray, E. S., Lambson, B. E., Moore, P. L., Choge, I. A., Mlisana, K., et al. (2010). Mannose-rich glycosylation patterns on HIV-1 subtype C gp120 and sensitivity to the lectins, Griffithsin, Cyanovirin-N and Scytovirin. Virology 402, 187–196. doi: 10.1016/j.virol.2010.03.021
Alexandre, K., Malatji, K., and Mulaudzi, T. (2020). Comparison of the antiviral activity of the microbicide candidate griffithsin and its tandemers derivatives against different modes of HIV-1 transmission. Virology 544, 12–20. doi: 10.1016/j.virol.2020.01.017
Ansari, A. M., Ahmed, A. K., Matsangos, A. E., Lay, F., Born, L. J., Marti, G., et al. (2016). Cellular GFP toxicity and immunogenicity: potential confounders in in vivo cell tracking experiments. Stem Cell Rev. Rep. 12, 553–559. doi: 10.1007/s12015-016-9670-8
Barrientos, L. G., and Gronenborn, A. M. (2005). The highly specific carbohydrate-binding protein cyanovirin-N: structure, anti-HIV/Ebola activity and possibilities for therapy. Mini Rev. Med. Chem. 5, 21–31. doi: 10.2174/1389557053402783
Bokesch, H. R., O'Keefe, B. R., McKee, T. C., Pannell, L. K., Patterson, G. M., Gardella, R. S., et al. (2003). A potent novel anti-HIV protein from the cultured cyanobacterium Scytonema varium. Biochemistry 42, 2578–2584. doi: 10.1021/bi0205698
Bowen, E. T., Platt, G. S., Lloyd, G., Raymond, R. T., and Simpson, D. I. (1980). A comparative study of strains of Ebola virus isolated from southern Sudan and northern Zaire in 1976. J. Med. Virol. 6, 129–138. doi: 10.1002/jmv.1890060205
Brichacek, B., Lagenaur, L. A., Lee, P. P., Venzon, D., and Hamer, D. H. (2013). In vivo evaluation of safety and toxicity of a Lactobacillus jensenii producing modified cyanovirin-N in a rhesus macaque vaginal challenge model. PLoS One 8:e78817. doi: 10.1371/journal.pone.0078817
Cenciarelli, O., Pietropaoli, S., Malizia, A., Carestia, M., D'Amico, F., Sassolini, A., et al. (2015). Ebola virus disease 2013-2014 outbreak in West Africa: an analysis of the epidemic spread and response. Int J Microbiol 2015:769121
Colleluori, D. M., Tien, D., Kang, F., Pagliei, T., Kuss, R., McCormick, T., et al. (2005). Expression, purification, and characterization of recombinant cyanovirin-N for vaginal anti-HIV microbicide development. Protein Expr. Purif. 39, 229–236. doi: 10.1016/j.pep.2004.10.009
El-Maradny, Y. A., El-Fakharany, E. M., Abu-Serie, M. M., Hashish, M. H., and Selim, H. S. (2021). Lectins purified from medicinal and edible mushrooms: insights into their antiviral activity against pathogenic viruses. Int. J. Biol. Macromol. 179, 239–258. doi: 10.1016/j.ijbiomac.2021.03.015
Emau, P., Tian, B., O’keefe, B. R., Mori, T., McMahon, J. B., Palmer, K. E., et al. (2007). Griffithsin, a potent HIV entry inhibitor, is an excellent candidate for anti-HIV microbicide. J. Med. Primatol. 36, 244–253. doi: 10.1111/j.1600-0684.2007.00242.x
Feldmann, H., and Geisbert, T. W. (2011). Ebola haemorrhagic fever. Lancet 377, 849–862. doi: 10.1016/S0140-6736(10)60667-8
Fernandez Romero, J. A., Paglini, M. G., Priano, C., Koroch, A., Rodriguez, Y., Sailer, J., et al. (2021). Algal and cyanobacterial lectins and their antimicrobial properties. Mar. Drugs 19. doi: 10.3390/md19120687
Garrison, A. R., Giomarelli, B. G., Lear-Rooney, C. M., Saucedo, C. J., Yellayi, S., Krumpe, L. R., et al. (2014). The cyanobacterial lectin scytovirin displays potent in vitro and in vivo activity against Zaire Ebola virus. Antivir. Res. 112, 1–7. doi: 10.1016/j.antiviral.2014.09.012
Girard, L., Birse, K., Holm, J. B., Gajer, P., Humphrys, M. S., Garber, D., et al. (2018). Impact of the griffithsin anti-HIV microbicide and placebo gels on the rectal mucosal proteome and microbiome in non-human primates. Sci. Rep. 8:8059. doi: 10.1038/s41598-018-26313-8
Huskens, D., and Schols, D. (2012). Algal lectins as potential HIV microbicide candidates. Mar. Drugs 10, 1476–1497. doi: 10.3390/md10071476
Jacob, S. T., Crozier, I., Fischer, W. A. 2nd, Hewlett, A., Kraft, C. S., Vega, M. A., et al. (2020). Ebola virus disease. Nat. Rev. Dis. Primers. 6:13. doi: 10.1038/s41572-020-0147-3
Koharudin, L. M., and Gronenborn, A. M. (2014). Antiviral lectins as potential HIV microbicides. Curr. Opin. Virol. 7, 95–100. doi: 10.1016/j.coviro.2014.05.006
Kuhn, J. H., Adachi, T., Adhikari, N. K. J., Arribas, J. R., Bah, I. E., Bausch, D. G., et al. (2019). New filovirus disease classification and nomenclature. Nat. Rev. Microbiol. 17, 261–263. doi: 10.1038/s41579-019-0187-4
Lagenaur, L. A., Sanders-Beer, B. E., Brichacek, B., Pal, R., Liu, X., Liu, Y., et al. (2011). Prevention of vaginal SHIV transmission in macaques by a live recombinant Lactobacillus. Mucosal Immunol. 4, 648–657. doi: 10.1038/mi.2011.30
Li, M., Patton, D. L., Cosgrove-Sweeney, Y., Ratner, D., Rohan, L. C., Cole, A. M., et al. (2011). Incorporation of the HIV-1 microbicide cyanovirin-N in a food product. J. Acquir. Immune Defic. Syndr. 58, 379–384. doi: 10.1097/QAI.0b013e31823643fe
Li, Y., Zhang, X., Chen, G., Wei, D., and Chen, F. (2008). Algal lectins for potential prevention of HIV transmission. Curr. Med. Chem. 15, 1096–1104. doi: 10.2174/092986708784221421
Mazur-Marzec, H., Ceglowska, M., Konkel, R., and Pyrc, K. (2021). Antiviral Cyanometabolites-A review. Biomol. Ther. 11. doi: 10.3390/biom11030474
Mitchell, C. A., Ramessar, K., and O'Keefe, B. R. (2017). Antiviral lectins: selective inhibitors of viral entry. Antivir. Res. 142, 37–54. doi: 10.1016/j.antiviral.2017.03.007
Mori, T., O'Keefe, B. R., Sowder, R. C. 2nd, Bringans, S., Gardella, R., Berg, S., et al. (2005). Isolation and characterization of griffithsin, a novel HIV-inactivating protein, from the red alga Griffithsia sp. J. Biol. Chem. 280, 9345–9353. doi: 10.1074/jbc.M411122200
Moulaei, T., Botos, I., Ziolkowska, N. E., Bokesch, H. R., Krumpe, L. R., McKee, T. C., et al. (2007). Atomic-resolution crystal structure of the antiviral lectin scytovirin. Protein Sci. 16, 2756–2760. doi: 10.1110/ps.073157507
Nahui Palomino, R. A., Zicari, S., Vanpouille, C., Vitali, B., and Margolis, L. (2017). Vaginal lactobacillus inhibits HIV-1 replication in human tissues ex vivo. Front. Microbiol. 8:906. doi: 10.3389/fmicb.2017.00906
Naik, S., and Kumar, S. (2022). Lectins from plants and algae act as anti-viral against HIV, influenza and coronaviruses. Mol. Biol. Rep. 49, 12239–12246. doi: 10.1007/s11033-022-07854-8
Obiero, J., Mwethera, P. G., Hussey, G. D., and Wiysonge, C. S. (2012). Vaginal microbicides for reducing the risk of sexual acquisition of HIV infection in women: systematic review and meta-analysis. BMC Infect. Dis. 12:289. doi: 10.1186/1471-2334-12-289
O'Keefe, B. R., Murad, A. M., Vianna, G. R., Ramessar, K., Saucedo, C. J., Wilson, J., et al. (2015). Engineering soya bean seeds as a scalable platform to produce cyanovirin-N, a non-ARV microbicide against HIV. Plant Biotechnol. J. 13, 884–892. doi: 10.1111/pbi.12309
Platt, E. J., Bilska, M., Kozak, S. L., Kabat, D., and Montefiori, D. C. (2009). Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J. Virol. 83, 8289–8292. doi: 10.1128/JVI.00709-09
Ramachandran, R., and Shanmughavel, P. (2009). Role of microbicides in the prevention of HIV and sexually transmitted diseases – a review. Curr. HIV Res. 7, 279–286. doi: 10.2174/157016209788347921
Tegegne, B. A., and Kebede, B. (2022). Probiotics, their prophylactic and therapeutic applications in human health development: a review of the literature. Heliyon 8:e09725. doi: 10.1016/j.heliyon.2022.e09725
Vamvaka, E., Evans, A., Ramessar, K., Krumpe, L. R., Shattock, R. J., O'Keefe, B. R., et al. (2016). Cyanovirin-N produced in rice endosperm offers effective pre-exposure prophylaxis against HIV-1BaL infection in vitro. Plant Cell Rep. 35, 1309–1319. doi: 10.1007/s00299-016-1963-5
Wang, L. L., Estrada, L., Wiggins, J., Anantpadma, M., Patten, J. J., Davey, R. A., et al. (2022). Ligand-based design of peptide entry inhibitors targeting the endosomal receptor binding site of filoviruses. Antivir. Res. 206:105399. doi: 10.1016/j.antiviral.2022.105399
Wei, W., Wiggins, J., Hu, D., Vrbanac, V., Bowder, D., Mellon, M., et al. (2019). Blocking HIV-1 infection by chromosomal integrative expression of human CD4 on the surface of Lactobacillus acidophilus ATCC 4356. J. Virol. 93. doi: 10.1128/JVI.01830-18
Welker, D. L., Coburn, B. M., McClatchy, J. H., and Broadbent, J. R. (2019). Multiple pulse electroporation of lactic acid bacteria Lactococcus lactis and Lactobacillus casei. J. Microbiol. Methods 166:105741. doi: 10.1016/j.mimet.2019.105741
WHO (n.d.). Available at: https://www.who.int/health-topics/ebola/
Xiong, C., O'Keefe, B. R., Botos, I., Wlodawer, A., and McMahon, J. B. (2006). Overexpression and purification of scytovirin, a potent, novel anti-HIV protein from the cultured cyanobacterium Scytonema varium. Protein Expr. Purif. 46, 233–239. doi: 10.1016/j.pep.2005.09.019
Keywords: scytovirin, lectins, lactic acid bacteria, Ebola virus, bacterial engineering, surface display
Citation: Wiggins J, Nguyen N, Wei W, Wang LL, Hollingsead Olson H and Xiang S-H (2023) Lactic acid bacterial surface display of scytovirin inhibitors for anti-ebolavirus infection. Front. Microbiol. 14:1269869. doi: 10.3389/fmicb.2023.1269869
Received: 31 July 2023; Accepted: 20 October 2023;
Published: 23 November 2023.
Edited by:
Gaëtan Ligat, Université Toulouse III Paul Sabatier, FranceReviewed by:
Joseph Atia Ayariga, Alabama State University, United StatesCopyright © 2023 Wiggins, Nguyen, Wei, Wang, Hollingsead Olson and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shi-Hua Xiang, c3hpYW5nMkB1bmwuZWR1
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.