- Department of Biology, Davidson College, Davidson, NC, United States
Bacteriophages constitute a ubiquitous threat to bacteria, and bacteria have evolved numerous anti-phage defense systems to protect themselves. These systems include well-studied phenomena such as restriction endonucleases and CRISPR, while emerging studies have identified many new anti-phage defense systems whose mechanisms are unknown or poorly understood. Some of these systems involve overcoming lysogenization defect (OLD) nucleases, a family of proteins comprising an ABC ATPase domain linked to a Toprim nuclease domain. Despite being discovered over 50 years ago, OLD nuclease function remained mysterious until recent biochemical, structural, and bioinformatic studies revealed that OLD nucleases protect bacteria by functioning in diverse anti-phage defense systems including the Gabija system and retrons. In this review we will highlight recent discoveries in OLD protein function and their involvement in multiple discrete anti-phage defense systems.
1. Introduction
Bacteria are under constant threat from viruses termed bacteriophages, or phages. It is estimated that there are 1031 phage particles in nature (Hendrix et al., 1999; Mushegian, 2020), making them the most abundant biological agent on the planet. To protect themselves from this ubiquitous threat, bacteria have evolved numerous systems to ward off phage infections (Hampton et al., 2020; Georjon and Bernheim, 2023). A recurring theme in many of these defense systems is the targeted cleavage of phage nucleic acid. The restriction-modification (R-M) system constitutes a classic example (Loenen et al., 2014), while more recently CRISPR sequences were discovered to generate immunological memory of previous infections and ultimately generate acquired defense (Mojica et al., 2005; Barrangou et al., 2007). Similar to how the R-M system once ushered in the modern era of recombinant DNA technology, the CRISPR/Cas9 system has likewise revolutionized biological and industrial research.
The impact of R-M and CRISPR/Cas9 systems demonstrates the fruitfulness of basic biological research into anti-phage defense mechanisms, and recent years have witnessed the discovery of myriad anti-phage defense systems, many of which remain relatively uncharacterized. Some anti-phage defense systems feature the activity of prokaryotic Argonaute proteins that employ DNA endonuclease activity as the driving mechanism of anti-phage defense (Swarts et al., 2014; Kuzmenko et al., 2020). Others, like BREX (bacteriophage exclusion; Goldfarb et al., 2015), involve methylation to distinguish self from non-self DNA but do not rely on nucleolytic degradation to achieve cell defense (Gordeeva et al., 2019). Other systems do not achieve defense through the preservation of the cell but rather through abortive infection, in which infected cells effect cell death before the phage can complete its replicative cycle (Lopatina et al., 2020). Systems resulting in abortive infection include CBASS (cyclic oligonucleotide-based antiphage signaling system) and PYCSAR (pyrimidine cyclase system for antiphage resistance) systems, which use cyclic dinucleotides as signaling molecules (Cohen et al., 2019; Tal et al., 2021). Systematic surveys of genomes, in particular focusing on genomic defense islands, continue to uncover new defense systems (Doron et al., 2018; Gao et al., 2020; Millman et al., 2022), most of which remain poorly understood. A close look at several systems, including the Gabija system (Doron et al., 2018; Cheng et al., 2021) and retrons (Millman et al., 2020), illuminate a recurring appearance of overcoming lysogenization defect (OLD) family nucleases. A classification scheme has been proposed in which OLD proteins can be assigned to different classes depending on their surrounding genetic context (Dot et al., 2023). In this review we will focus on the composition, structure, and function of Class 1, Class 2, and Class 3 OLD nucleases across diverse anti-phage defense systems.
2. Class 1 OLD proteins: phage-phage interference and structure overview
Although defense islands have been increasingly observed to harbor anti-phage defense systems (Makarova et al., 2011), Class 1 OLD systems belie this trend as they are instead composed of single genes not found proximal to other candidate defense genes (Figure 1; Schiltz et al., 2019). The archetype for understanding Class 1 OLD proteins, and indeed the original discovery and namesake for the entire OLD family of proteins, arises from early phage genetics experiments. Gianpiero Sironi showed that P2 phage was unable to lysogenize E. coli mutants he named lyd (lysogenization defective), and subsequently identified P2 mutants that could lysogenize lyd mutants (Sironi, 1969). Sironi named this P2 mutant phenotype old for overcoming lysogenization defect. Subsequent work determined that lambda phage is unable to lysogenize a wild-type P2 prophage but that this phage interference is eliminated by mutations in old (Lindahl et al., 1970). Lyd mutants turned out to reside in recB and recC (Lindahl et al., 1970) components of the RecBCD helicase-nuclease complex that plays critical roles both in homologous recombination and in defense against phages via double-stranded DNA degradation (Dillingham and Kowalczykowski, 2008). Expression of the P2 Old protein is sufficient to kill recBC− cells (Sironi et al., 1971), a phenotype that continues to be used to assess P2 Old function (Schiltz et al., 2020). P2 Old’s interference with lambda phage was an early example of prophage-encoded anti-phage defense, a phenomenon that is now recognized as widespread (Patel and Maxwell, 2023). Indeed, the P2 old locus has been observed to encode other anti-phage systems at this position (Rousset et al., 2022; Vassallo et al., 2022).
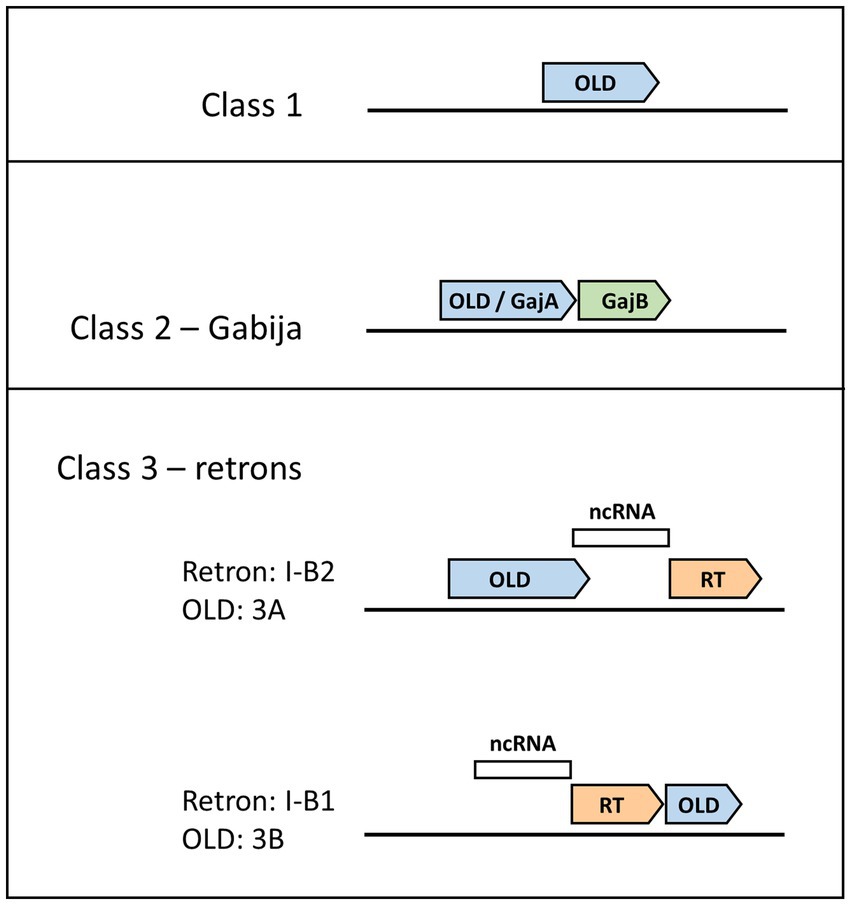
Figure 1. Genomic layouts of OLD protein classes. Gene neighborhood organizations for Class 1, 2, and 3 OLD proteins are shown, with OLD proteins in blue. Class 1 OLD proteins appear as single genes. Class 2 OLD proteins are synonymous with GajA, and are found together with GajB (green), which shows homology to UvrD/PcrA/Rep-like helicases. Class 3 OLD proteins are found in retron cassettes and have two possible genomic layouts depending on OLD positioning relative to the reverse transcriptase (RT, orange) and non-coding RNA (ncRNA, white). Class 3A OLD proteins are found in type I-B2 retrons, while Class 3B OLD proteins are found in type I-B1 retrons.
After early phage genetics defined the protein name and function in P2, studies of OLD nucleases mostly disappeared for many years. One notable exception was the purification and characterization of a P2 Old construct fused to maltose-binding protein (MBP; Myung and Calendar, 1995). This study established that P2 Old-MBP displayed 5’to 3′ exonuclease activity on dsDNA. Furthermore, ATP was found to enhance but not be required for DNA cleavage, and the ATPase activity was not stimulated by the addition of DNA. Whether these activities were unique to P2 Old or were common to OLD proteins would remain unknown until recently (Table 1). A few years later, bioinformatic analysis showed that OLD proteins were composed of an N-terminal ATP-binding cassette (ABC)-family ATPase and a C-terminal Toprim domain (Figure 2; Aravind et al., 1998). ABC ATPases are found in diverse proteins ranging from membrane transporters to nuclear structural maintenance of chromosomes (SMC) proteins including condensins and cohesins (Krishnan et al., 2020). The Toprim domain is a divalent metal-binding domain found in topoisomerases, DnaG-type primases, RecR proteins, and OLD family nucleases (Aravind et al., 1998). Toprim domains possess a conserved acidic motif that binds to divalent cations which promote phosphoryl transfer reactions (Keck et al., 2000; Kato et al., 2003; Schmidt et al., 2010), suggesting that DNA cleavage by OLD proteins may follow canonical two-metal DNA cleavage mechanisms (Steitz and Steitz, 1993).
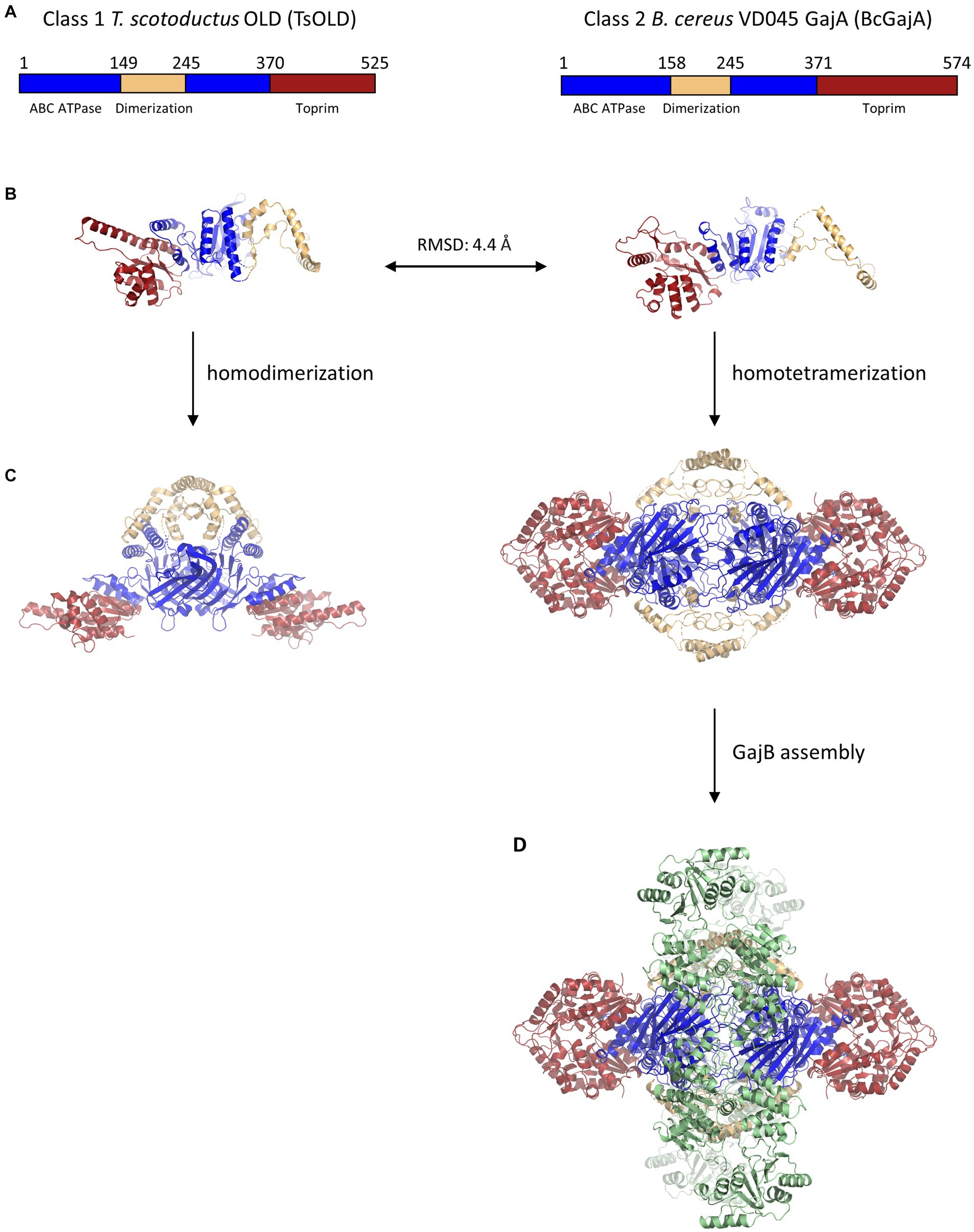
Figure 2. Structural similarity and multimeric assembly of Class 1 and Class 2 OLD proteins. (A) Schematics showing domain organization within Class 1 T. scotoductus OLD and Class 2 B. cereus GajA OLD proteins. A dimerization domain (tan) is inserted into each ABC ATPase domain (blue). The larger size of BcGajA arises from a helical insert into the Toprim domain (red) that is conserved among Class 2 OLD proteins and makes them larger than Class 1 OLD proteins by about 50 amino acids on average. (B) Structural superposition of TsOLD (PDB: 6P74) and BcGajA (PDB: 8SM3) full-length monomers shows structural similarity (RMSD = 4.4 Å) despite only 22.2% sequence identity. (C) TsOLD dimerizes and BcGajA tetramerizes in crystal structures, but in neither structure are the ATPases poised for ATP hydrolysis. (D) Two separate GajB dimers (green) bind to a GajA homotetramer to assemble an octameric GajAB complex.
Decades would pass until the field of OLD nuclease research was reignited recently with detailed studies of OLD nuclease structure and function, as well as bioinformatic and genetic analyses of their function in anti-phage defense systems. A critical breakthrough was the first structural determination of a full-length OLD protein, the Class 1 OLD in Thermus scotoductus (TsOLD; Figure 2; Schiltz et al., 2020). The structure revealed the protein to adopt a homodimeric structure in which dimerization was mediated through a dimerization domain inserted into the ABC ATPases. Although the ATPases are docked in a conformation not competent to achieve ATP hydrolysis, the structure confirmed that the ATPase domain has structural homology with the ATP-binding cassette (ABC) family of ATPases. Further structural studies will be required to determine whether Class 1 OLD proteins may exist in higher multimeric complexes or associate with other proteins. Biochemical characterization of TsOLD showed that, like P2 Old, TsOLD displays robust 5’to 3′ exonuclease activity on linear dsDNA, as well as the ability to linearize circular plasmid DNA substrates. However, TsOLD is unlike P2 OLD in that the addition of ATP has essentially no effect on DNA cleavage by TsOLD (Table 1; Schiltz et al., 2020).
Mutagenesis studies in P2 Old showed that the conserved acidic metal-binding residues of the Toprim domain, as well as the conserved Walker A lysine critical for ATP binding, were all required for killing recBC− cells, implicating both the ATPase and Toprim domains as essential for at least some in vivo activities (Schiltz et al., 2020). However, although the structural determination of TsOLD was a watershed moment propelling OLD protein research forward, the only Class 1 OLD protein established to provide defense against a phage remains P2 Old’s defense of prophages against lambda. The anti-phage defense functions of OLD proteins are better understood from studies of multicomponent systems featuring Class 2 or Class 3 OLD proteins.
3. Class 2 OLD proteins: the Gabija system
Recent discoveries of diverse anti-phage defense systems show that OLD proteins not only function on their own as Class 1 proteins but also are found in multicomponent anti-phage defense systems such as Class 2 OLDs found in the Gabija system (Doron et al., 2018). The genetic organization of the Gabija system was discovered independently by two groups. In the first instance, as part of a systematic discovery of anti-phage defense systems, the Sorek group identified a system composed of an OLD protein (GajA), and a UvrD/PcrA/Rep-like helicase (GajB), a system they would name Gabija after the Lithuanian mythology goddess of fire (Figure 1; Doron et al., 2018). They estimated the Gabija system is present in 8.5% of a set of 38,167 microbial genomes, while more recent bioinformatic tools designed to identify anti-phage defense systems place Gabija frequency closer to 15% (Payne et al., 2022; Tesson et al., 2022). The second Gabija system discovery came from the Chappie group which was studying OLD nucleases and termed them as Class 1 or Class 2 depending on whether they were found alone (Class 1) or in tandem with a UvrD/PcrA/Rep-like helicase (Class 2; Schiltz et al., 2019).
The first structural work on Class 2 OLD nucleases was performed on isolated Toprim domains from Burkholderia pseudomallei (BpOLD) and Xanthomonas campestris p.v. campestris (XccOLD; Schiltz et al., 2019). Their Toprim domains contain an extra helical domain insert that differentiates them from Class 1 OLD proteins and makes Class 2 OLD proteins about 50 amino acids longer on average (Figure 2). Structural analysis of the BpOLD Toprim domain shows its conserved acidic motif binds two Mg2+ ions and suggests a canonical two-metal mechanism for DNA cleavage. Biochemical assays with full-length BpOLD and XccOLD show nonspecific endonuclease and exonuclease activity on lambda phage DNA which is unaffected by ATP concentration (Table 1). The authors proposed that the BpOLD ATPase domain, which is competent to hydrolyze ATP (Cheng et al., 2021), plays a regulatory role in the cleavage of substrates by the Toprim domain (Schiltz et al., 2019).
The most studied Gabija system is from Bacillus cereus VD045 and is found to offer protection against phages of the Siphoviridae family including the SPβ, Φ105, and rho14, as well as phages from the Podoviridae family such as Φ29 (Table 1; Doron et al., 2018). Although initial biochemical characterization described the B. cereus GajA (BcGajA) as functionally distinct from OLD nucleases (Cheng et al., 2021), structural superposition of full-length monomers from T. scotoductus OLD and BcGajA reveals an RMSD of 4.4 Å despite a sequence identity of only 22.2% (Figure 2; Schiltz et al., 2020; Antine et al., 2023). On the basis of their structural similarity and their shared genomic proximity to UvrD/PcrA/Rep-like helicases, we suggest that all GajA proteins are Class 2 OLD proteins, and vice versa.
Like all Class 2 OLD proteins, BcGajA comprises an N-terminal ABC ATPase domain and a C-terminal Toprim domain. BcGajA is currently the only OLD protein known to cleave DNA with sequence specificity (Table 1), as it nicks DNA at a site found both in lambda and T7 phage dsDNA (Cheng et al., 2021). Interestingly, in lambda DNA the two cut sites overlap, resulting in apparent dsDNA cleavage activity. It is worth noting that BcGajA resides in a gram-positive bacteria that is not infected by lambda or T7 phages, and thus the significance of this cut site being found in lambda and T7 phage DNA is not clear. BcGajA endonuclease activity is robust under low ATP concentrations, while high nucleotide concentrations inhibit BcGajA DNA binding and cleavage (Cheng et al., 2021, 2023). Interestingly, BcGajA is the first OLD protein shown to lack ATP hydrolysis activity (Table 1). These results led the authors to propose that the ATPase domains regulate the Toprim domain by inhibiting its DNA cleavage activity in the presence of high nucleotide concentrations and that GajA becomes activated upon nucleotide depletion resulting from phage invasion, replication, and transcription. Recent studies have reported nucleotide depletion mechanisms in other anti-phage defense systems (Hsueh et al., 2022; Tal et al., 2022), supporting the idea that nucleotide depletion may be common in abortive infection mechanisms.
The second component of the Gabija system, GajB, is predicted to be a UvrD/PcrA/Rep-like helicase (Doron et al., 2018). UvrD helicases translocate along ssDNA in the 3′ to 5′ direction and couple the binding and hydrolysis of one ATP with the unwinding of one base-pair of duplex DNA (Matson, 1986; Lee and Yang, 2006). Recent structural and biochemical data have shed light on the function of GajB and its interactions with GajA. Structural studies show that BcGajB binds to a pre-formed BcGajA tetramer to assemble a 4:4 octameric complex in which with two sets of GajB dimers flank a centralized GajA tetramer (Figure 2; Antine et al., 2023). Although BcGajA purified in the absence of BcGajB displays robust endonuclease nicking activity (Cheng et al., 2021), BcGajB is required for anti-phage defense via an abortive infection mechanism (Doron et al., 2018; Cheng et al., 2023). A recent study shows that BcGajB, surprisingly, does not exhibit any helicase activity but instead functions as a (d)ATP/(d)GTPase (Cheng et al., 2023). Furthermore, the addition of either ssDNA or dsDNA stimulates nucleotide hydrolysis by GajB. The authors propose that BcGajB senses 3′ termini, possibly originating from BcGajA DNA cleavage, which activates its (d)ATP/(d)GTPase hydrolytic activity, thereby driving nucleotide depletion and contributing to cell death (Cheng et al., 2023). Further studies will be required to see whether these results, including a lack of GajB helicase activity, are generalizable to other Gabija systems.
4. Class 3 OLD proteins: retron-driven anti-phage defense
Retrons are genetic elements found in bacteria comprising a reverse transcriptase and an adjacent non-coding RNA. Their discovery stems from a 1984 study identifying a small multi-copy, single-stranded DNA (msDNA; Yee et al., 1984), which subsequent studies determined was composed of a ssDNA covalently linked to a non-coding RNA used as a template by the reverse transcriptase (Dhundale et al., 1987; Lampson et al., 1989). Retron function remained mysterious until a recent breakthrough study that demonstrated retrons belong to anti-phage defense systems triggering cell death through abortive infection (Millman et al., 2020). These defense systems contain three components: the aforementioned reverse transcriptase and non-coding RNA, as well as an effector protein (Figure 1). Effector proteins vary tremendously and include cold shock proteins, zinc finger nucleases, and proteases. Out of 4,802 genomes analyzed, the retron effector protein was an OLD nuclease 4% of the time (Millman et al., 2020). OLD nucleases found as effector proteins in retron defense systems have been classified as Class 3 OLD enzymes, with further subdivision depending on whether the old gene lies upstream (Class 3A) or downstream (Class 3B) of the reverse transcriptase (Dot et al., 2023). Within the retron naming system, these genomic organizations have been described as type I-B2 or type I-B1, respectively (Mestre et al., 2020).
Class 3 OLD enzymes have not yet been characterized through biochemical and structural means like either Class 1 or Class 2 OLD enzymes, but AlphaFold models show that, like their Class I and Class 2 counterparts, they comprise an N-terminal ABC ATPase and a C-terminal Toprim domain. Most of what is known about Class 3 OLD proteins arises from genetic studies of anti-phage defense. The best characterized Class 3 OLD protein belongs to Retron-Eco8, which provides defense against T4, T7, SECΦ4, SECΦ6, and SECΦ18 through an abortive infection mechanism (Table 1; Millman et al., 2020). Mutational analysis shows that mutations in the conserved aspartates in the YADD motif of the reverse transcriptase, or in the conserved guanosine branching point in the non-coding RNA, or in the Walker A lysine of the OLD nuclease each eliminate anti-phage defense activity (Millman et al., 2020). These data demonstrate that all three retron components are necessary for anti-phage defense by Retron-Eco8.
The mechanisms by which retrons in general and Class 3 OLD enzymes in particular lead to either phage recognition or abortive infection remain elusive. A recent study shows that T7, SECΦ4, SECΦ6, and SECΦ18 phages were all able to escape anti-phage defense by Retron-Eco8 through mutations in phage single-stranded binding (SSB) protein (Stokar-Avihail et al., 2023). This study showed that expressing the phage SSB in cells that express Retron-Eco8 was sufficient to drive cell toxicity and that mutations in the SSB were sufficient to alleviate that toxicity. Furthermore, pull-down experiments showed that wild-type but not mutant phage SSB was pulled down with Retron-Eco8 msDNA. While these data demonstrate a direct association between retron msDNA and phage SSB, they do not explain the role of the Class 3 OLD enzyme, which was already established to be essential for anti-phage defense (Millman et al., 2020). Further studies of Class 3 OLD enzymes will be required to understand their role in mediating retron-dependent anti-phage defense.
5. Future considerations
The last several years have seen a marked increase in studies examining the structure, biochemistry, and in vivo function of OLD proteins. The results of these studies reveal a complicated landscape in which the biochemical properties and functions of OLD proteins can vary from one another and depend heavily on the surrounding genetic, cellular, and chemical context. Many outstanding questions in the field remain, and below we highlight just a few of them:
Do Class 1 OLD proteins provide broad-spectrum anti-phage defense on their own? The only known example of a Class 1 OLD protein offering protection against a phage is the idiosyncratic example of P2 prophages providing Old-dependent defense only against lambda phage. Although the broad-spectrum anti-phage defense by Class 2 (Gabija) and Class 3 (retron) OLD proteins has been definitively established, there remains no known example of a bacterial or archaeal Class 1 OLD protein sufficient to drive broad-spectrum anti-phage defense. Multicomponent systems like Gabija and retrons have shown that each genetic component is necessary for anti-phage defense, and so it would be valuable to learn of cellular systems where an OLD protein alone is sufficient to drive anti-phage defense.
What are the regulatory relationships between OLD ATPases, Toprim domains, and DNA engagement? It may be that the answer depends on the specific OLD protein in question. For example, ATP has been reported to increase DNA cleavage for P2 Old, have minimal effect on DNA cleavage in TsOLD, and strongly inhibit DNA cleavage activity in BcGajA (Table 1). More work will be required to understand the regulatory relationships between the ATPase and Toprim domains. Moreover, although structural studies of OLD proteins have been critical breakthroughs, there remains a paucity of structural data of OLD proteins bound to DNA. Such studies will be required to understand how the protein engages with DNA, the physical basis for any sequence- or structural specificity OLD proteins may have, and what regulatory mechanisms might prevent DNA binding or cleavage depending on the surrounding biochemical context.
How do phages escape these defense systems? While bacteria have evolved complex mechanisms to impede viral infection, phages continually develop mechanisms to overcome them (Gao and Feng, 2023). One such mechanism involves the release of the Gabija anti-defense 1 (Gad1) protein by phage Φ3T to thwart the Gabija system of B. cereus VD045 (Yirmiya et al., 2023). To resist anti-phage defense, Gad1 binds to the GajA dimerization domain of the GajAB complex and prevents DNA binding and cleavage (Antine et al., 2023). Similar phage proteins called Thoeris anti-defense 1 and 2 (Tad1 and Tad2) have been recently discovered in the Thoeris system, another widely distributed bacterial anti-phage defense system (Leavitt et al., 2022). Future studies will no doubt illuminate other escape mechanisms, the understanding of which may be critical for the development of successful phage therapy (Kortright et al., 2019).
How many classes of OLD proteins exist, and in how many different systems? Class 4 OLD proteins have been proposed to contribute to the function of the PARIS system (Rousset et al., 2022; Dot et al., 2023). Unlike the other three classes, in the PARIS system the ATPase and Toprim domains are encoded on separate genes (designated ariA and ariB, respectively; Rousset et al., 2022). However, the authors noted that sometimes AriAB is encoded as a single-gene fusion, in which case it is not clear whether the system comprises a separate OLD class, or whether the system operates differently from Class 1 OLD proteins. Further studies will be required to determine whether the PARIS system constitutes a separate OLD class. Another recent study noted that a component of the anti-plasmid Wadjet system (Doron et al., 2018), jetD, has homology with the Toprim domain of OLD nucleases, while the jetABC components include an ABC ATPase with homology to the bacterial condensin complex MukBEF (Deep et al., 2022). These studies suggest we have much to learn about the myriad systems in which an ABC ATPase and a Toprim domain work in concert in defense systems. Future studies will surely uncover in more detail how OLD proteins have been leveraged by organisms to defend themselves.
Author contributions
KA: Writing – original draft, Writing – review and editing. BT-S: Conceptualization, Visualization, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Davidson College.
Acknowledgments
We thank Joshua Chappie and Philip Kranzusch for helpful discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Antine, S. P., Johnson, A. G., Mooney, S. E., Leavitt, A., Mayer, M. L., Yirmiya, E., et al. (2023). Structural basis of Gabija anti-phage defense and viral immune evasion. bioRxiv :2023.05.01.538945. doi: 10.1101/2023.05.01.538945
Aravind, L., Leipe, D. D., and Koonin, E. V. (1998). Toprim--a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 26, 4205–4213. doi: 10.1093/nar/26.18.4205
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Cheng, R., Huang, F., Lu, X., Yan, Y., Yu, B., Wang, X., et al. (2023). Prokaryotic Gabija complex senses and executes nucleotide depletion and DNA cleavage for antiviral defense. Cell Host Microbe 31, 1331–1344.e5. doi: 10.1016/j.chom.2023.06.014
Cheng, R., Huang, F., Wu, H., Lu, X., Yan, Y., Yu, B., et al. (2021). A nucleotide-sensing endonuclease from the Gabija bacterial defense system. Nucleic Acids Res. 49, 5216–5229. doi: 10.1093/nar/gkab277
Cohen, D., Melamed, S., Millman, A., Shulman, G., Oppenheimer-Shaanan, Y., Kacen, A., et al. (2019). Cyclic GMP-AMP signalling protects bacteria against viral infection. Nature 574, 691–695. doi: 10.1038/s41586-019-1605-5
Deep, A., Gu, Y., Gao, Y.-Q., Ego, K. M., Herzik, M. A. J., Zhou, H., et al. (2022). The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA. Mol. Cell 82, 4145–4159.e7. doi: 10.1016/j.molcel.2022.09.008
Dhundale, A., Lampson, B., Furuichi, T., Inouye, M., and Inouye, S. (1987). Structure of msDNA from Myxococcus xanthus: evidence for a long, self-annealing RNA precursor for the covalently linked, branched RNA. Cells 51, 1105–1112. doi: 10.1016/0092-8674(87)90596-4
Dillingham, M. S., and Kowalczykowski, S. C. (2008). RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72, 642–671. doi: 10.1128/MMBR.00020-08
Doron, S., Melamed, S., Ofir, G., Leavitt, A., Lopatina, A., Keren, M., et al. (2018). Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359:eaar4120. doi: 10.1126/science.aar4120
Dot, E. W., Thomason, L. C., and Chappie, J. S. (2023). Everything OLD is new again: how structural, functional, and bioinformatic advances have redefined a neglected nuclease family. Mol. Microbiol. 120, 122–140. doi: 10.1111/mmi.15074
Gao, L., Altae-Tran, H., Böhning, F., Makarova, K. S., Segel, M., Schmid-Burgk, J. L., et al. (2020). Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 369, 1077–1084. doi: 10.1126/science.aba0372
Gao, Z., and Feng, Y. (2023). Bacteriophage strategies for overcoming host antiviral immunity. Front. Microbiol. 14:1211793. doi: 10.3389/fmicb.2023.1211793
Georjon, H., and Bernheim, A. (2023). The highly diverse antiphage defence systems of bacteria. Nat. Rev. Microbiol. 21, 686–700. doi: 10.1038/s41579-023-00934-x
Goldfarb, T., Sberro, H., Weinstock, E., Cohen, O., Doron, S., Charpak-Amikam, Y., et al. (2015). BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 34, 169–183. doi: 10.15252/embj.201489455
Gordeeva, J., Morozova, N., Sierro, N., Isaev, A., Sinkunas, T., Tsvetkova, K., et al. (2019). BREX system of Escherichia coli distinguishes self from non-self by methylation of a specific DNA site. Nucleic Acids Res. 47, 253–265. doi: 10.1093/nar/gky1125
Hampton, H. G., Watson, B. N. J., and Fineran, P. C. (2020). The arms race between bacteria and their phage foes. Nature 577, 327–336. doi: 10.1038/s41586-019-1894-8
Hendrix, R. W., Smith, M. C. M., Burns, R. N., Ford, M. E., and Hatfull, G. F. (1999). Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc. Natl. Acad. Sci. 96, 2192–2197. doi: 10.1073/pnas.96.5.2192
Hsueh, B. Y., Severin, G. B., Elg, C. A., Waldron, E. J., Kant, A., Wessel, A. J., et al. (2022). Phage defence by deaminase-mediated depletion of deoxynucleotides in bacteria. Nat. Microbiol. 7, 1210–1220. doi: 10.1038/s41564-022-01162-4
Kato, M., Ito, T., Wagner, G., Richardson, C. C., and Ellenberger, T. (2003). Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol. Cell 11, 1349–1360. doi: 10.1016/s1097-2765(03)00195-3
Keck, J. L., Roche, D. D., Lynch, A. S., and Berger, J. M. (2000). Structure of the RNA polymerase domain of E. coli primase. Science 287, 2482–2486. doi: 10.1126/science.287.5462.2482
Kortright, K. E., Chan, B. K., Koff, J. L., and Turner, P. E. (2019). Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25, 219–232. doi: 10.1016/j.chom.2019.01.014
Krishnan, A., Burroughs, A. M., Iyer, L. M., and Aravind, L. (2020). Comprehensive classification of ABC ATPases and their functional radiation in nucleoprotein dynamics and biological conflict systems. Nucleic Acids Res. 48, 10045–10075. doi: 10.1093/nar/gkaa726
Kuzmenko, A., Oguienko, A., Esyunina, D., Yudin, D., Petrova, M., Kudinova, A., et al. (2020). DNA targeting and interference by a bacterial Argonaute nuclease. Nature 587, 632–637. doi: 10.1038/s41586-020-2605-1
Lampson, B. C., Inouye, M., and Inouye, S. (1989). Reverse transcriptase with concomitant ribonuclease H activity in the cell-free synthesis of branched RNA-linked msDNA of Myxococcus xanthus. Cells 56, 701–707. doi: 10.1016/0092-8674(89)90592-8
Leavitt, A., Yirmiya, E., Amitai, G., Lu, A., Garb, J., Herbst, E., et al. (2022). Viruses inhibit TIR gc ADPR signalling to overcome bacterial defence. Nature 611, 326–331. doi: 10.1038/s41586-022-05375-9
Lee, J. Y., and Yang, W. (2006). UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cells 127, 1349–1360. doi: 10.1016/j.cell.2006.10.049
Lindahl, G., Sironi, G., Bialy, H., and Calendar, R. (1970). Bacteriophage lambda; abortive infection of bacteria lysogenic for phage P2. Proc. Natl. Acad. Sci. U. S. A. 66, 587–594. doi: 10.1073/pnas.66.3.587
Loenen, W. A. M., Dryden, D. T. F., Raleigh, E. A., Wilson, G. G., and Murray, N. E. (2014). Highlights of the DNA cutters: a short history of the restriction enzymes. Nucleic Acids Res. 42, 3–19. doi: 10.1093/nar/gkt990
Lopatina, A., Tal, N., and Sorek, R. (2020). Abortive infection: bacterial suicide as an antiviral immune strategy. Annu. Rev. Virol. 7, 371–384. doi: 10.1146/annurev-virology-011620-040628
Makarova, K. S., Wolf, Y. I., Snir, S., and Koonin, E. V. (2011). Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J. Bacteriol. 193, 6039–6056. doi: 10.1128/JB.05535-11
Matson, S. W. (1986). Escherichia coli helicase II (urvD gene product) translocates unidirectionally in a 3′ to 5′ direction. J. Biol. Chem. 261, 10169–10175. doi: 10.1016/S0021-9258(18)67506-4
Mestre, M. R., González-Delgado, A., Gutiérrez-Rus, L. I., Martínez-Abarca, F., and Toro, N. (2020). Systematic prediction of genes functionally associated with bacterial retrons and classification of the encoded tripartite systems. Nucleic Acids Res. 48, 12632–12647. doi: 10.1093/nar/gkaa1149
Millman, A., Bernheim, A., Stokar-Avihail, A., Fedorenko, T., Voichek, M., Leavitt, A., et al. (2020). Bacterial retrons function in anti-phage defense. Cells 183, 1551–1561.e12. doi: 10.1016/j.cell.2020.09.065
Millman, A., Melamed, S., Leavitt, A., Doron, S., Bernheim, A., Hör, J., et al. (2022). An expanded arsenal of immune systems that protect bacteria from phages. Cell Host Microbe 30, 1556–1569.e5. doi: 10.1016/j.chom.2022.09.017
Mojica, F. J. M., Díez-Villaseñor, C., García-Martínez, J., and Soria, E. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60, 174–182. doi: 10.1007/s00239-004-0046-3
Mushegian, A. R. (2020). Are there 1031 virus particles on earth, or more, or fewer? J. Bacteriol. 202:e00052-20. doi: 10.1128/JB.00052-20
Myung, H., and Calendar, R. (1995). The old exonuclease of bacteriophage P2. J. Bacteriol. 177, 497–501. doi: 10.1128/jb.177.3.497-501.1995
Patel, P. H., and Maxwell, K. L. (2023). Prophages provide a rich source of antiphage defense systems. Curr. Opin. Microbiol. 73:102321. doi: 10.1016/j.mib.2023.102321
Payne, L. J., Meaden, S., Mestre, M. R., Palmer, C., Toro, N., Fineran, P. C., et al. (2022). PADLOC: a web server for the identification of antiviral defence systems in microbial genomes. Nucleic Acids Res. 50, W541–W550. doi: 10.1093/nar/gkac400
Rousset, F., Depardieu, F., Miele, S., Dowding, J., Laval, A.-L., Lieberman, E., et al. (2022). Phages and their satellites encode hotspots of antiviral systems. Cell Host Microbe 30, 740–753.e5. doi: 10.1016/j.chom.2022.02.018
Schiltz, C. J., Adams, M. C., and Chappie, J. S. (2020). The full-length structure of Thermus scotoductus OLD defines the ATP hydrolysis properties and catalytic mechanism of class 1 OLD family nucleases. Nucleic Acids Res. 48, 2762–2776. doi: 10.1093/nar/gkaa059
Schiltz, C. J., Lee, A., Partlow, E. A., Hosford, C. J., and Chappie, J. S. (2019). Structural characterization of class 2 OLD family nucleases supports a two-metal catalysis mechanism for cleavage. Nucleic Acids Res. 47, 9448–9463. doi: 10.1093/nar/gkz703
Schmidt, B. H., Burgin, A. B., Deweese, J. E., Osheroff, N., and Berger, J. M. (2010). A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature 465, 641–644. doi: 10.1038/nature08974
Sironi, G. (1969). Mutants of Escherichia coli unable to be lysogenized by the temperate bacteriophage P2. Virology 37, 163–176. doi: 10.1016/0042-6822(69)90196-2
Sironi, G., Bialy, H., Lozeron, H. A., and Calendar, R. (1971). Bacteriophage P2: interaction with phage lambda and with recombination-deficient bacteria. Virology 46, 387–396. doi: 10.1016/0042-6822(71)90040-7
Steitz, T. A., and Steitz, J. A. (1993). A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. U. S. A. 90, 6498–6502. doi: 10.1073/pnas.90.14.6498
Stokar-Avihail, A., Fedorenko, T., Hör, J., Garb, J., Leavitt, A., Millman, A., et al. (2023). Discovery of phage determinants that confer sensitivity to bacterial immune systems. Cells 186, 1863–1876.e16. doi: 10.1016/j.cell.2023.02.029
Swarts, D. C., Jore, M. M., Westra, E. R., Zhu, Y., Janssen, J. H., Snijders, A. P., et al. (2014). DNA-guided DNA interference by a prokaryotic Argonaute. Nature 507, 258–261. doi: 10.1038/nature12971
Tal, N., Millman, A., Stokar-Avihail, A., Fedorenko, T., Leavitt, A., Melamed, S., et al. (2022). Bacteria deplete deoxynucleotides to defend against bacteriophage infection. Nat. Microbiol. 7, 1200–1209. doi: 10.1038/s41564-022-01158-0
Tal, N., Morehouse, B. R., Millman, A., Stokar-Avihail, A., Avraham, C., Fedorenko, T., et al. (2021). Cyclic CMP and cyclic UMP mediate bacterial immunity against phages. Cells 184, 5728–5739.e16. doi: 10.1016/j.cell.2021.09.031
Tesson, F., Hervé, A., Mordret, E., Touchon, M., d’Humières, C., Cury, J., et al. (2022). Systematic and quantitative view of the antiviral arsenal of prokaryotes. Nat. Commun. 13:2561. doi: 10.1038/s41467-022-30269-9
Vassallo, C. N., Doering, C. R., Littlehale, M. L., Teodoro, G. I. C., and Laub, M. T. (2022). A functional selection reveals previously undetected anti-phage defence systems in the E. coli pangenome. Nat. Microbiol. 7, 1568–1579. doi: 10.1038/s41564-022-01219-4
Yee, T., Furuichi, T., Inouye, S., and Inouye, M. (1984). Multicopy single-stranded DNA isolated from a gram-negative bacterium, Myxococcus xanthus. Cells 38, 203–209. doi: 10.1016/0092-8674(84)90541-5
Keywords: OLD nuclease, overcoming lysogenization defect, Gabija, retrons, Toprim, ABC ATPase, anti-phage defense, abortive infection
Citation: Akritidou K and Thurtle-Schmidt BH (2023) OLD family nuclease function across diverse anti-phage defense systems. Front. Microbiol. 14:1268820. doi: 10.3389/fmicb.2023.1268820
Edited by:
Alicja Wegrzyn, Polish Academy of Sciences, PolandReviewed by:
Peter Weigele, New England Biolabs, United StatesKamil Steczkiewicz, Polish Academy of Sciences, Poland
Copyright © 2023 Akritidou and Thurtle-Schmidt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bryan H. Thurtle-Schmidt, YnJ0aHVydGxlc2NobWlkdEBkYXZpZHNvbi5lZHU=
 Konstantina Akritidou
Konstantina Akritidou Bryan H. Thurtle-Schmidt
Bryan H. Thurtle-Schmidt