
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 November 2023
Sec. Evolutionary and Genomic Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1268709
This article is part of the Research Topic Next Generation Sequencing: Applications in Foodborne Pathogens View all 8 articles
Introduction: Listeria monocytogenes is a globally distributed bacterium that exhibits genetic diversity and trait heterogeneity. The alternative sigma factor SigB serves as a crucial transcriptional regulator essential for responding to environmental stress conditions and facilitating host infection.
Method: We employed a comprehensive genetic analysis of sigB in a dataset comprising 46,921 L. monocytogenes genomes. The functional attributes of SigB were evaluated by phenotypic experiments.
Results: Our study revealed the presence of two predominant SigB factors (SigBT1 and SigBT2) in L. monocytogenes, with a robust correlation between SigBT1 and lineages I and III, as well as SigBT2 and lineage II. Furthermore, SigBT1 exhibits superior performance in promoting cellular invasion, cytotoxicity and enhancing biofilm formation and cold tolerance abilities under minimally defined media conditions compared to SigBT2.
Discussion: The functional characteristics of SigBT1 suggest a potential association with the epidemiology of lineages I and III strains in both human hosts and the natural environment. Our findings highlight the important role of distinct SigB factors in influencing the biological traits of L. monocytogenes of different lineages, thus highlighting its distinct pathogenic and adaptive attributes.
Listeria monocytogenes is a facultative pathogenic agent of listeriosis, which is a foodborne infection in the older adults, children, and immunocompromised patients. L. monocytogenes demonstrates global distribution with cases throughout Europe, North America, Oceania, Asia, South America, and Africa, highlighting its pervasive presence worldwide (Moura et al., 2021; Koopmans et al., 2023). The annual incidence of listeriosis ranges from 0.1 to 11.3 cases per million individuals, with variations caused by geographical areas and the monitoring techniques that are used (Koopmans et al., 2023). L. monocytogenes enters the host and survives a series of potentially lethal conditions in the digestive tract, including gastric acid, biliary salt, and oxidative stress, eventually causing bacteremia, meningitis, and severe maternal–fetal infections (Allerberger and Wagner, 2010). The South African listeriosis outbreak in 2017–2018, caused by ST6 L. monocytogenes, resulted in 204 fatalities and over $260 million in fatality-related costs (Olanya et al., 2019). L. monocytogenes has been found in a diverse range of sources, with a notable predominance in contaminated food products such as processed meats, soft cheeses, raw milk, and ready-to-eat foods (Ryser and Marth, 2007). This pathogen is also extensively found in natural environmental reservoirs, such as soil and polluted water systems (Ryser and Marth, 2007).
Listeria monocytogenes can be classified into 4 lineages, 13 recognized serotypes, and numerous sequence types (STs). Certain clones of L. monocytogenes have been verified and are more closely linked to clinical cases or food sources. Lineage I strains are used to form a majority of clinical isolates, while lineage II strains exhibits a more general distribution in food isolates (Maury et al., 2016). ST1, ST2, ST4, and ST6 clones are linked to clinical isolates, while ST9 and ST121 clones are common isolates in food sources (Maury et al., 2016; Muchaamba et al., 2021). Additionally, ST87 isolates have been identified as a clinical clone type in China (Wang et al., 2015; Li et al., 2018). The molecular characteristics of L. monocytogenes have been shown to be highly divergent in food surveillance, clinical cases, animals, and environment (Maury et al., 2016; Muchaamba et al., 2021). Although some genetic elements in a specific group were associated with virulence and/or stress resistance phenotypes, such as LIPI-4 in CC4, LGI-1 in CC8, SSI-2 in ST121, and pLMST6 in CC6, the underlying causes behind the observed epidemiological distribution and phenotypic variations in L. monocytogenes remain unexplained (Bergholz et al., 2018).
As a facultative bacterial pathogen, L. monocytogenes possesses remarkable regulatory networks in response to various conditions, such as abrupt environmental changes and host-induced stresses (Guariglia-Oropeza et al., 2014). The transcriptional regulation in bacteria serves as a crucial mechanism for their rapid adaptation and reproduction in an unpredictable environment. Sigma factors are initiation factors that induce the binding of RNA polymerase to promoters, thereby enabling cells to precisely regulate the timing and specificity of gene expression (Feklistov et al., 2014). As a central regulator, the sigma factor B (SigB) has been demonstrated to regulate a factor consisting of approximately 300 genes related to stress response, pathogenesis, cellular homeostasis, and metabolism in L. monocytogenes (Chaturongakul et al., 2011; Liu et al., 2017).
Analysis of sigB in 6 Listeria species comprising 4,390 isolates showed that non-synonymous mutations occurred less frequently than synonymous mutations in 164 sigB allelic types (Liao et al., 2017). The regulator sigB is a relatively stable gene in Listeria that has not significantly undergone evolution due to positive selection and homologous recombination (Liao et al., 2017). However, as the regulatory center of large functional networks, SigB has several characteristics that have not been explored yet, such as the specific types of sigB alleles and their unknown biological significance.
In this study, we conducted a large-scale genetic analysis of the sigB gene in 46,921 L. monocytogenes isolates and associated sigB with background information about its host bacteria. To evaluate the biological function of two dominant SigB factors, we further performed chromosomal mutation modification, cell culture assays, and survival phenotype analysis.
This study included whole genome data of 46,921 L. monocytogenes isolates from the NCBI database (Supplementary Table S1). The geographic information and isolation source of L. monocytogenes were obtained from the NCBI BioSample database. To determine the sequence types (STs) and lineage of L. monocytogenes, in silico MLST analysis was performed using allele scheme information from the BIGSdb database (Moura et al., 2016).
For homology analysis, BLASTN was performed to match for sigB with each L. monocytogenes genome in translated CDS format, followed by in silico extraction and translation. Corresponding to the reference strain EGD-e, the complete sigB gene was classified as having 259 amino acids. The distinct amino acid residues within the full-length SigB were identified through the alignment of protein sequences. Each dissimilar amino acid residue signifies a unique type of SigB factor. Those with fewer amino acids were categorized as premature stop codon (PMSC) variants. The protein family and conserved protein domains were performed with the UniProt and Pfam databases (Mistry et al., 2021; UniProt, 2021). The graphical representation of the amino acid probabilities at each site was generated by WebLogo (Crooks et al., 2004).
The experimental strains L. monocytogenes ICDC-LM188 (LM188) (Lineage I) and EGD-e (Lineage II) were used in this study. Strains were normally cultured in brain heart infusion (BHI, Oxoid, UK) broth or agar at 37oC. Modified Welshimer's broth (MWB, HiMedia, India) was used as a minimally defined medium (Premaratne et al., 1991). Chloramphenicol (Solarbio, China) was added to the culture medium when required at a concentration of 10 μg/ml.
Reference strain EGD-e and experimental strain LM188 are used in this study. EGD-e is a reference strain (ATCC BAA-679) of lineage II and is widely used in scientific reports, and the LM188 strain belongs to the prevalent genotype of lineage I in China (Wang et al., 2019). The sigBT1 gene of wild-type L. monocytogenes LM188 (Linage I) was replaced by sigBT2 through homologous recombination, while the sigBT2 gene of the wild-type EDG-e (Linage II) strain was replaced by sigBT1. The substituted codons, adjunct fragments from WT codons, and restriction sites (SalI and BamHI) were combined via DNA synthesis in Tsingke Biotechnology Co., Ltd. Each 1,616-bp linear product was cloned into vector pKSV7, resulting in the pKSV7::sigBT1 and pKSV7::sigBT2 vectors, which were then transformed into electrocompetent EDG-e and LM188 cells (Zhang et al., 2018). The electroporated cells were spread on BHI plates with chloramphenicol at 37°C, followed by passage cultivation at 42°C for homologous recombination. The plasmid pKSV7 was cured after growth at 37°C in BHI broth. The chromosomal sigB mutation strains LM188MT_T2 and EGD-eMT_T1 were produced through homologous recombination and verified by sequencing.
The total RNA was extracted with TRIzol (Invitrogen) and then transcribed into cDNA using Fastking GDNA Dispelling RT Supermix (Tiangen). The expression of sigB was measured by real-time PCR using SuperReal PreMix Plus (Tiangen). The expression level of sigB relative to that of the housekeeping gene (16S rDNA) was assessed by the comparative 2−ΔΔCT method.
The cytotoxic effect was evaluated on the human colon adenocarcinoma cell line HT-29, according to the previous method (Gan et al., 2020). Cytotoxicity assays were performed by measuring the release of lactate dehydrogenase (LDH) as an indicator of cell injury. HT-29 (4*104) cells were seeded in 96-well plates with an infection of bacteria at an MOI of 50 bacteria. After 4 h of incubation, the LDH concentration in the cell supernatants was determined according to the manufacturer's instructions of CytoTox 96®Non-Radioactive Cytotoxicity Assay (Promega, USA).
For invasion assay, HT-29 cells (2*105) were infected with bacteria at an MOI of 50 on 24-well plates (Gan et al., 2020). After 1 h of infection, the cells were treated with 50 μg/ml of gentamicin (Solarbio, China) for 30 min and washed with DMEM (Gibco, USA). The cells were then lysed with 0.1% Triton X100 (Sigma, USA). The intracellular bacteria were incubated on a BHI plate for colony enumeration.
For in vitro growth assays, the Listeria isolates were initially cultured in BHI broth for the activation of bacteria at a dilution ratio of 1:100. Subsequently, a bacterial suspension with a log-phase concentration of 107 CFU was transferred and incubated in MWB at 30°C. The optical density of the culture at 600 nm (OD600) was recorded at a 1-h interval using a Bioscreen C microbiology reader. The experiment was repeated three times, with five replicate wells for each trial.
Biofilm formation was quantified using the crystal violet staining method (Mao et al., 2021). Approximately 2*107 CFU of fresh bacterial suspensions were initially spread on 96-well microplates. The plates were incubated at 30°C for biofilm formation. The liquid and non-adhered cells were removed and then gently rinsed three times with PBS. The biofilm was stained with 1% crystal violet and then released with 95% ethanol. The total biofilm biomass was evaluated by measuring the OD value at 595 nm. Each experiment was replicated three times for three independent experiments.
Approximately 2*107 CFU of fresh bacterial suspension was transferred to 1 ml of MWB medium and prepared to be kept for long-term preservation at 4°C for 60 days. Following thorough vortex mixing, the bacteria were cultured on a BHI agar plate for colony enumeration. Three independent experiments were conducted with three replicates.
The relationship between the L. monocytogenes lineage and the type of SigB factor was analyzed by Fisher's exact test. The OD600 was assessed by repetitive measurement deviation analysis. Invasion efficiency, LDH concentration, and biofilm biomass used t-tests for statistical analysis. Significant differences were judged at a P < 0.05, P < 0.01, or P < 0.001.
The sigB gene is composed of 780 base pairs (Bp) that code for 259 amino acids of full-length protein. Through the sequence analysis of 46,921 L. monocytogenes isolates, 46,773 isolates (99.79%) contain a single full-length sigB gene, while 86 isolates contain premature stop codons (PMSCs) in the sigB gene. Additionally, the sigB gene could not be aligned with the genomic sequences of 12 isolates, and 4 isolates harbored two copies of the sigB gene. Furthermore, the sigB gene in 46 isolates was fragmented and not included in the analysis (Supplementary Table S1).
Among full-length sigB genes, 112 distinct protein types of SigB factor were identified and characterized by 88 variant sites and 171 completely conserved sites, and the predominant types were SigBT1 and SigBT2 (Figure 1A, Supplementary Tables S1, S2). In addition, the sigB gene of 86 L. monocytogenes isolates was truncated with deletion mutations in 43 isolates, insertion mutations in 20 isolates, and nonsense point mutations in 23 isolates (Supplementary Table S1). Finally, one strain of an unknown lineage carried both SigBT1 and SigBT2 factors, and three isolates belonging to lineage II carried two identical SigBT2 factors (Supplementary Table S1).
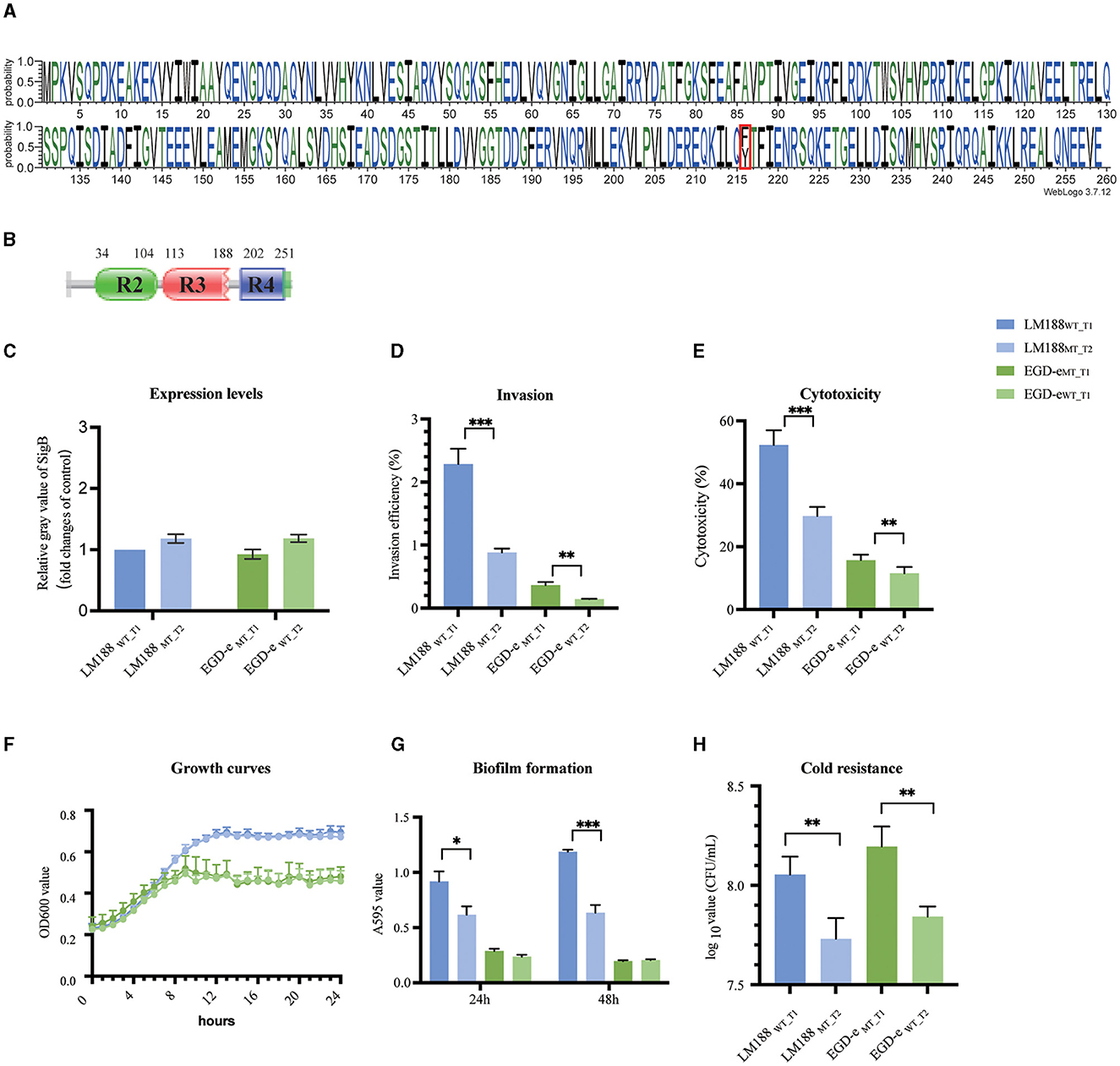
Figure 1. (A) Weblogo built using 46,773 sequences of full-length SigB. (B) Conserved protein domains. (C) Expression levels. (D) Invasion. (E) Cytotoxicity. (F) Growth curves. (G) Biofilm formation. (H) Cold resistance (*P < 0.05, **P < 0.01, ***P < 0.001).
The analysis revealed two predominant types of SigB factors, namely, SigBT1 (50.42%) and SigBT2 (47.09%). Both types shared a variant site at codon 216. Furthermore, SigBT1 encodes phenylalanine at this codon, while SigBT2 encodes tyrosine (Figure 1A). The SigB protein belongs to the Sigma 70 family and contains three domains, including Sigma 70 r2, r3, and r4, which are predicted by the UniProt and Pfam databases (Figure 2B). The variant site of codon 216 is located on the r4 domain.
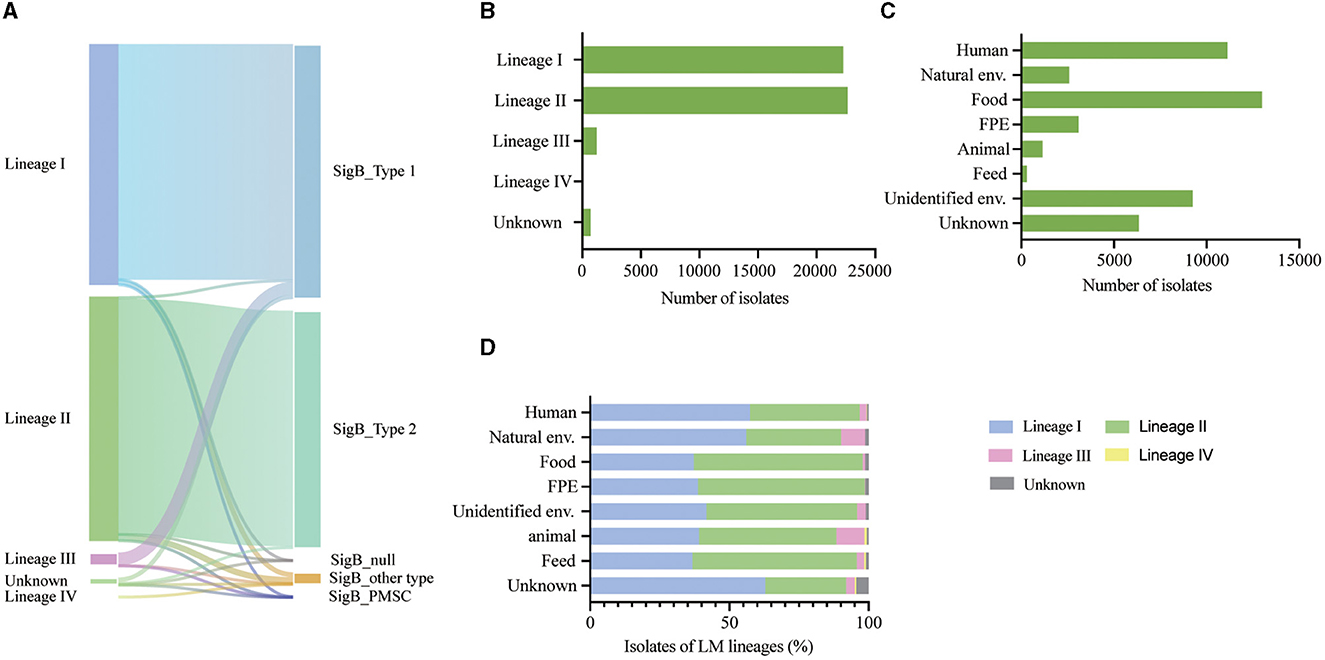
Figure 2. (A) Sankey diagram illustrating the correlation between variation type of SigB and lineages. (B) Prevalence of L. monocytogenes categorized by lineages. (C) Prevalence of L. monocytogenes based on different sources. (D) Proportional distribution of lineages among L. monocytogenes (LM) from diverse sources.
To assess the difference between the biological functions of SigBT1 and SigBT2, we constructed sigB mutants in chromosomal loci by homologous recombination. Four strains of L. monocytogenes, namely, wild-type LM188WT_T1, mutant-type LM188MT_T2, wild-type EGD-eWT_T2, and mutant-type EGD-eMT_T1, were used for conducting phenotypic experiments. Additionally, there was no significant difference in sigB gene expression levels between LM188WT_T1 and LM188MT_T2, as well as between EGD-eWT_T2 and EGD-eMT_T1 under BHI conditions (Figure 1C).
Cellular invasion and cytotoxicity assays in the HT-29 cell line were assessed to evaluate the effect of two SigB types on virulence. LM188WT_T1 strain exhibited significantly higher invasion ability (P < 0.001) and cytotoxicity (P < 0.001) compared to the LM188MT_T2 strain. Additionally, the EGD-eMT_T1 strain also showed significantly higher invasion capacity (P < 0.01) and cytotoxicity (P < 0.01) than the EGD-eWT_T2 strain (Figures 1D, E). These results suggest that SigB plays a significant role in the virulence of L. monocytogenes, and SigBT1 outperforms SigBT2 in enhancing cellular invasion and cytotoxicity.
Biofilm formation and proliferation ability were evaluated for culture in a minimally defined MWB medium. No significant disparity was observed in the growth profiles between LM188WT_T1 and LM188MT_T2, as well as between EGD-eMT_T1 and EGD-eWT_T2 (Figure 1E), suggesting that the variation in SigB factor has no significant effect on bacterial growth. We found that the LM188WT_T1 strain exhibited higher biofilm formation ability at 24 h (P < 0.05) and 48 h (P < 0.001) than the LM188MT_T2 strain, and both EGD-eMT_T1 and EGD-eWT_T2 strains had weak growth and biofilm formation ability in MWB medium (Figures 1F, G). Furthermore, the results also showed that LM188WT_T1 and EGD-eMT_T1 strains with SigBT1 exhibited stronger cold tolerance compared to LM188MT_T2 and EGD-eWT_T2 carrying SigBT2 at 4°C (P < 0.01) (Figure 1H). These results suggest that the SigB type has a significant effect on biofilm formation and cold tolerance of L. monocytogenes, and SigBT1 exhibits superior performance compared to SigBT2 in terms of biofilm formation and cold tolerance.
We analyzed 46,921 L. monocytogenes genomes from 6 continents, 66 countries, and various sources, including 22,288 lineage I isolates, 22,644 lineage II isolates, 1,227 lineage III isolates, 65 lineage IV isolates, and 697 unknown lineage isolates (Figures 2B, C).
After analyzing 46,921 L. monocytogenes isolates from the NCBI database, it was observed that SigBT1 was present in 21,791 isolates (97.77%) of lineage I, 1,188 isolates (96.82%) of lineage III, 207 (0.91%) isolates of lineage II, and 400 (57.38%) isolates of an unknown lineage. SigBT2 existed in 21,786 (96.21%) isolates of lineage II and 235 (33.72%) isolates of unknown lineage. Therefore, SigBT1 is highly correlated with lineages I and III (P < 0.001), and SigBT2 is highly correlated with lineage II (P < 0.001) (Figure 2A).
The L. monocytogenes isolates were distributed globally and there was an association between L. monocytogenes lineages and different sources. Isolates from human and natural environmental sources were distributed across the lineages, with lineage I being the most prevalent (Figure 2D). Similar proportions of each lineage were distributed in food and food production environment (FPE)-sourced isolates, with lineage II being the most prevalent (Figure 2D).
L. monocytogenes is an important foodborne pathogen that can colonize and penetrate the gastrointestinal tract, blood–brain barrier, and placental barrier. L. monocytogenes develops numerous stress response mechanisms in order to survive and persist in a variety of harsh environments, both outside and within the host. SigB, an alternative general stress sigma factor, modulates the transcriptional landscapes of stress response, pathogenesis, and homeostasis in L. monocytogenes (Garner et al., 2006; Toledo-Arana et al., 2009; van Der Veen and Abee, 2010; Gahan and Hill, 2014; Dorey et al., 2019). Transcription factors bind to the specific DNA sequences of the target gene and then control the initiation or inhibition of their transcription, regulating the gene expression in bacteria (Balleza et al., 2009). Mutations of bacterial transcription factors, such as K259 lysine acetylation of HrdB, F318A mutation of σ54, I48S in region 1.1 of E. coli σ70, and dynamic variation of MarR transcription factors, affect transcription regulation by altering their interaction with the RNAP core enzyme and its binding activity toward target promoters (Wigneshweraraj et al., 2002; Deochand and Grove, 2017; Kim et al., 2020; Pletnev et al., 2020). The novel SigB (Q225P) mutation in Staphylococcus aureus maintains virulence but enhances biofilm formation by affecting the promoter activity (Liu et al., 2018). The study of variation in transcription factors in bacteria can offer important insights into gene expression regulation and bacterial responses.
In this study, we demonstrated that the correlation between SigBT1 and lineage I/III as well as SigBT2 and lineage II, and codon 216 of SigB play an important role in the invasion and environmental stress adaptation of L. monocytogenes. The 216 variant site of SigB is located in domain sigma70 r4, which forms the second largest interface with RNA polymerase and serves as a key contact point for transcriptional activators that recognize the upstream sequences of the -35 promoter region (Paget, 2015).
In this study, 92.39% of isolates were classified as lineage I in L. monocytogenes isolates carrying SigBT1, and 98.93% of the isolates were identified as lineage II among the isolates harboring SigBT2. We found that L. monocytogenes with SigBT1 had a higher invasive ability to human colon cancer cells than isolates with SigBT2, suggesting that SigBT1 is associated with the high virulence of L. monocytogenes. When L. monocytogenes reaches the host intestinal lumen, transcriptional reshaping is activated by SigB-mediated virulence genes (Toledo-Arana et al., 2009). SigB-mediated control of inlA and inlB expression is critical for L. monocytogenes invasion of host epithelial cells, as demonstrated by a significantly reduced invasion in a sigB deletion mutant strain (Kazmierczak et al., 2003; Kim et al., 2005; Garner et al., 2006; Bierne et al., 2007). InlA is essential for the pathogenesis of L. monocytogenes as it mediates the identification and invasion of epithelial cells through its specific interaction with E-cadherin, which is a fundamental step to crossing the intestinal barrier (Bierne et al., 2007; Disson et al., 2008). Among 15 transcriptional regulators, SigB is a key factor in the invasion of human epithelial colorectal adenocarcinoma cells by L. monocytogenes (Rukit et al., 2022). Therefore, SigBT1 is associated with the prevalence of lineage I and lineage III of L. monocytogenes in patients and animals.
Researchers in the field of the molecular epidemiology of L. monocytogenes have focused on investigating the source of human infection as well as its presence in food and food processing environments, with lineage I being frequently associated with human infection and lineage II being prevalent in food and environmental sources (Maury et al., 2016; Koopmans et al., 2023). However, little attention has been paid to its prevalence in natural environments. This study revealed that L. monocytogenes isolates in natural vs. food environments have distinct molecular characteristics, with lineage I being more prevalent in natural environments and lineage II being more prevalent in food environments. Additionally, in this study, a substantial proportion of lineage III strains originated from natural environments and animal sources. These are likely due to two distinct environmental conditions: natural environments have limited nutrition, while food environments have abundant nutrition.
The sigB gene plays a crucial role in biofilm formation and has increased expression levels in both static and continuous-flow biofilms (van Der Veen and Abee, 2010; Hsu et al., 2020). Our findings suggest that SigBT1 enhances biofilm formation in low-nutrient settings in the MWB culture medium. The effect of SigB factor types on clinical strain LM188 is significantly apparent compared to that on strain EGD-e. This might be attributed to weaker phenotypes of EGD-e, which is a strain of unclear source, specifically biofilm formation and reduced growth in defined minimal media. Therefore, SigBT1 contributes to the higher biofilm-forming ability observed in lineage I strains under such conditions, which may partially explain its prevalence and persistence in natural environments.
The sigB gene is involved in survival from chill stress in L. monocytogenes, with varying degrees of impact influenced by different serotypes (Moorhead and Dykes, 2004). Our finding also indicates that SigBT1 isolates exhibit a greater capacity for cold tolerance as compared to SigBT2 isolates when cultured in a minimal medium. This observation suggests that SigBT1 would help lineage I and lineage III isolates endure low temperatures under nutrient-poor conditions and increase their likelihood of survival and persistence in frigid natural environments. L. monocytogenes ubiquitously exist in various sources and can survive in the presence of high osmolarity, low-temperature conditions, and extreme environments (Chan and Wiedmann, 2009). Different microbial lineages have adapted to different environments and sources, and their proportions vary depending on the environment they inhabit.
In conclusion, our study demonstrated that lineage-specific SigB types influenced the survival, virulence, and adaptation of L. monocytogenes under specific conditions. Contrary to SigBT2, SigBT1 exhibited a more pronounced role in promoting invasiveness and virulence as well as improving biofilm formation and long-term cold survival under limited nutritional conditions. SigBT1 could contribute to the prevalence and persistence of L. monocytogenes linage I and linage III strains in clinical settings and the natural environment, highlighting its pivotal role in the epidemiology of this pathogen. These findings offer a new insight into the molecular mechanisms underlying the phenotypic variations and epidemiological distribution of L. monocytogenes, paving the way for future research on the development of targeted interventions to prevent and control infections caused by this pathogen.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
CY: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing—review & editing. PM: Conceptualization, Formal analysis, Methodology, Software, Visualization, Writing—original draft. YW: Supervision, Project administration, Writing—review & editing. LG: Data curation, Writing—review & editing. LLiu: Methodology, Writing—original draft. JC: Methodology, Writing—original draft. LLi: Methodology, Writing—original draft. HS: Project administration, Writing—review & editing. XL: Project administration, Writing—review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from the National Institute for Communicable Disease Control and Prevention, China CDC (2021ZZKT003) (102393220020020000029).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1268709/full#supplementary-material
Allerberger, F., and Wagner, M. (2010). Listeriosis: a resurgent foodborne infection. Clin. Microbiol. Infect 16, 16–23. doi: 10.1111/j.1469-0691.2009.03109.x
Balleza, E., Lopez-Bojorquez, L. N., Martinez-Antonio, A., Resendis-Antonio, O., Lozada-Chavez, I., Balderas-Martinez, Y. I., et al. (2009). Regulation by transcription factors in bacteria: beyond description. FEMS Microbiol. Rev. 33, 133–151. doi: 10.1111/j.1574-6976.2008.00145.x
Bergholz, T. M., Shah, M. K., Burall, L. S., Rakic-Martinez, M., and Datta, A. R. (2018). Genomic and phenotypic diversity of Listeria monocytogenes clonal complexes associated with human listeriosis. Appl. Microbiol. Biotechnol. 102, 3475–3485. doi: 10.1007/s00253-018-8852-5
Bierne, H., Sabet, C., Personnic, N., and Cossart, P. (2007). Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect 9, 1156–1166. doi: 10.1016/j.micinf.2007.05.003
Chan, Y. C., and Wiedmann, M. (2009). Physiology and genetics of Listeria monocytogenes survival and growth at cold temperatures. Crit. Rev. Food Sci. Nutr. 49, 237–253. doi: 10.1080/10408390701856272
Chaturongakul, S., Raengpradub, S., Palmer, M. E., Bergholz, T. M., Orsi, R. H., Hu, Y., et al. (2011). Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative sigma factors sigmaB, sigmaC, sigmaH, and sigmaL in Listeria monocytogenes. Appl. Environ. Microbiol. 77, 187–200. doi: 10.1128/AEM.00952-10
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Deochand, D. K., and Grove, A. (2017). MarR family transcription factors: dynamic variations on a common scaffold. Crit. Rev. Biochem. Mol. Biol. 52, 595–613. doi: 10.1080/10409238.2017.1344612
Disson, O., Grayo, S., Huillet, E., Nikitas, G., Langa-Vives, F., Dussurget, O., et al. (2008). Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature 455, 1114–1118. doi: 10.1038/nature07303
Dorey, A., Marinho, C., Piveteau, P., and O'byrne, C. (2019). Role and regulation of the stress activated sigma factor sigma B (sigma(B)) in the saprophytic and host-associated life stages of Listeria monocytogenes. Adv. Appl. Microbiol. 106, 1–48. doi: 10.1016/bs.aambs.2018.11.001
Feklistov, A., Sharon, B. D., Darst, S. A., and Gross, C. A. (2014). Bacterial sigma factors: a historical, structural, and genomic perspective. Annu. Rev. Microbiol. 68, 357–376. doi: 10.1146/annurev-micro-092412-155737
Gahan, C. G., and Hill, C. (2014). Listeria monocytogenes: survival and adaptation in the gastrointestinal tract. Front. Cell Infect Microbiol. 4, 9. doi: 10.3389/fcimb.2014.00009
Gan, L., Mao, P., Jiang, H., Zhang, L., Liu, D., Cao, X., et al. (2020). Two prevalent Listeria ivanovii subsp. ivanovii clonal strains with different virulence exist in wild rodents and pikas of China. Front. Vet. Sci. 7, 88. doi: 10.3389/fvets.2020.00088
Garner, M. R., Njaa, B. L., Wiedmann, M., and Boor, K. J. (2006). Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect Immun. 74, 876–886. doi: 10.1128/IAI.74.2.876-886.2006
Guariglia-Oropeza, V., Orsi, R. H., Yu, H., Boor, K. J., Wiedmann, M., Guldimann, C., et al. (2014). Regulatory network features in Listeria monocytogenes-changing the way we talk. Front. Cell Infect Microbiol. 4, 14. doi: 10.3389/fcimb.2014.00014
Hsu, C. Y., Cairns, L., Hobley, L., Abbott, J., O'byrne, C., and Stanley-Wall, N. R. (2020). Genomic differences between Listeria monocytogenes EGDe isolates reveal crucial roles for sigb and wall rhamnosylation in biofilm formation. J. Bacteriol. 202, 19. doi: 10.1128/JB.00692-19
Kazmierczak, M. J., Mithoe, S. C., Boor, K. J., and Wiedmann, M. (2003). Listeria monocytogenes sigma B regulates stress response and virulence functions. J. Bacteriol. 185, 5722–5734. doi: 10.1128/JB.185.19.5722-5734.2003
Kim, H., Marquis, H., and Boor, K. J. (2005). SigmaB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology 151, 3215–3222. doi: 10.1099/mic.0.28070-0
Kim, J. E., Choi, J. S., Kim, J. S., Cho, Y. H., and Roe, J. H. (2020). Lysine acetylation of the housekeeping sigma factor enhances the activity of the RNA polymerase holoenzyme. Nucleic Acids Res. 48, 2401–2411. doi: 10.1093/nar/gkaa011
Koopmans, M. M., Brouwer, M. C., Vazquez-Boland, J. A., and Van De Beek, D. (2023). Human listeriosis. Clin. Microbiol. Rev. 36, e0006019. doi: 10.1128/cmr.00060-19
Li, W., Bai, L., Fu, P., Han, H., Liu, J., Guo, Y., et al. (2018). The epidemiology of Listeria monocytogenes in China. Foodborne Pathog. Dis. 15, 459–466. doi: 10.1089/fpd.2017.2409
Liao, J., Wiedmann, M., and Kovac, J. (2017). Genetic stability and evolution of the sigB allele, used for listeria sensu stricto subtyping and phylogenetic inference. Appl. Environ. Microbiol. 83. doi: 10.1128/AEM.00306-17
Liu, H., Shang, W., Hu, Z., Zheng, Y., Yuan, J., Hu, Q., et al. (2018). A novel SigB(Q225P) mutation in Staphylococcus aureus retains virulence but promotes biofilm formation. Emerg. Microbes Inf. 7, 72. doi: 10.1038/s41426-018-0078-1
Liu, Y., Orsi, R. H., Boor, K. J., Wiedmann, M., and Guariglia-Oropeza, V. (2017). Home alone: elimination of all but one alternative sigma factor in Listeria monocytogenes allows prediction of new roles for sigma(B). Front. Microbiol. 8, 1910. doi: 10.3389/fmicb.2017.01910
Mao, P., Wang, Y., Gan, L., Sun, H., Wang, Y., Li, L., et al. (2021). Function and distribution of the conjugative plasmid pLM1686 in foodborne Listeria monocytogenes in China. Int. J. Food Microbiol. 352, 109261. doi: 10.1016/j.ijfoodmicro.2021.109261
Maury, M. M., Tsai, Y. H., Charlier, C., Touchon, M., Chenal-Francisque, V., Leclercq, A., et al. (2016). Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 48, 308–313. doi: 10.1038/ng.3501
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Moorhead, S. M., and Dykes, G. A. (2004). Influence of the sigB gene on the cold stress survival and subsequent recovery of two Listeria monocytogenes serotypes. Int. J. Food Microbiol. 91, 63–72. doi: 10.1016/S0168-1605(03)00332-5
Moura, A., Criscuolo, A., Pouseele, H., Maury, M. M., Leclercq, A., Tarr, C., et al. (2016). Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2, 16185. doi: 10.1038/nmicrobiol.2016.185
Moura, A., Lefrancq, N., Wirth, T., Leclercq, A., Borges, V., Gilpin, B., et al. (2021). Emergence and global spread of Listeria monocytogenes main clinical clonal complex. Sci. Adv. 7, eabj9805. doi: 10.1126/sciadv.abj9805
Muchaamba, F., Eshwar, A. K., Stevens, M. J. A., Stephan, R., and Tasara, T. (2021). Different Shades of Listeria monocytogenes: strain, serotype, and lineage-based variability in virulence and stress tolerance profiles. Front. Microbiol. 12, 792162. doi: 10.3389/fmicb.2021.792162
Olanya, O. M., Hoshide, A. K., Ijabadeniyi, O. A., Ukuku, D. O., Mukhopadhyay, S., Niemira, B. A., et al. (2019). Cost estimation of listeriosis (Listeria monocytogenes) occurrence in South Africa in 2017 and its food safety implications. Food Control 102, 231–239. doi: 10.1016/j.foodcont.2019.02.007
Paget, M. S. (2015). Bacterial sigma factors and anti-sigma factors: structure, function and distribution. Biomolecules 5, 1245–1265. doi: 10.3390/biom5031245
Pletnev, P., Pupov, D., Pshanichnaya, L., Esyunina, D., Petushkov, I., Nesterchuk, M., et al. (2020). Rewiring of growth-dependent transcription regulation by a point mutation in region 1.1 of the housekeeping sigma factor. Nucleic Acids Res. 48, 10802–10819. doi: 10.1093/nar/gkaa798
Premaratne, R. J., Lin, W. J., and Johnson, E. A. (1991). Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl. Environ. Microbiol. 57, 3046–3048. doi: 10.1128/aem.57.10.3046-3048.1991
Rukit, J., Boonmee, A., Kijpornyongpan, T., Tulsook, K., Baranyi, J., Chaturongakul, S., et al. (2022). Roles of alternative sigma factors in invasion and growth characteristics of Listeria monocytogenes 10403s into human epithelial colorectal adenocarcinoma Caco-2 cell. Front. Microbiol. 13, 901484. doi: 10.3389/fmicb.2022.901484
Toledo-Arana, A., Dussurget, O., Nikitas, G., Sesto, N., Guet-Revillet, H., Balestrino, D., et al. (2009). The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956. doi: 10.1038/nature08080
UniProt, C. (2021). UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489. doi: 10.1093/nar/gkaa1100
van Der Veen, S., and Abee, T. (2010). Importance of SigB for Listeria monocytogenes static and continuous-flow biofilm formation and disinfectant resistance. Appl. Environ. Microbiol. 76, 7854–7860. doi: 10.1128/AEM.01519-10
Wang, Y., Jiao, Y., Lan, R., Xu, X., Liu, G., Wang, X., et al. (2015). Characterization of Listeria monocytogenes isolated from human Listeriosis cases in China. Emerg. Microbes. Infect. 4, e50. doi: 10.1038/emi.2015.50
Wang, Y., Luo, L., Li, Q., Wang, H., Wang, Y., Sun, H., et al. (2019). Genomic dissection of the most prevalent Listeria monocytogenes clone, sequence type ST87, in China. BMC Genom. 20, 1014. doi: 10.1186/s12864-019-6399-1
Wigneshweraraj, S. R., Casaz, P., and Buck, M. (2002). Correlating protein footprinting with mutational analysis in the bacterial transcription factor sigma54 (sigmaN). Nucleic Acids Res. 30, 1016–1028. doi: 10.1093/nar/30.4.1016
Keywords: Listeria monocytogenes, SigB factor, genetic analysis, lineage, biological trait
Citation: Mao P, Wang Y, Gan L, Liu L, Chen J, Li L, Sun H, Luo X and Ye C (2023) Large–scale genetic analysis and biological traits of two SigB factors in Listeria monocytogenes: lineage correlations and differential functions. Front. Microbiol. 14:1268709. doi: 10.3389/fmicb.2023.1268709
Received: 03 August 2023; Accepted: 13 October 2023;
Published: 13 November 2023.
Edited by:
Alejandro Garrido-Maestu, International Iberian Nanotechnology Laboratory (INL), PortugalReviewed by:
Sabino Pacheco, National Autonomous University of Mexico, MexicoCopyright © 2023 Mao, Wang, Gan, Liu, Chen, Li, Sun, Luo and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changyun Ye, eWVjaGFuZ3l1bkBpY2RjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.