
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 06 October 2023
Sec. Aquatic Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1266972
This article is part of the Research Topic The Biosynthesis, Emission, and Ecophysiological Functions of Algal Volatile Organic Compounds (VOCs) View all 3 articles
Interactions between marine phytoplankton, viruses, and bacteria drive biogeochemical cycling, shape marine trophic structures, and impact global climate. Microbially produced compounds have emerged as key players in influencing eukaryotic organismal physiology, and in turn, remodel microbial community structure. This work aimed to reveal the molecular mechanism by which the bacterial quorum sensing molecule 2-heptyl-4-quinolone (HHQ), produced by the marine gammaproteobacterium Pseudoalteromonas spp., arrests cell division and confers protection from virus-induced mortality in the bloom-forming coccolithophore Emiliania huxleyi. Previous work has established alkylquinolones as inhibitors of dihydroorotate dehydrogenase (DHODH), a fundamental enzyme catalyzing the fourth step in pyrimidine biosynthesis and a potential antiviral drug target. An N-terminally truncated version of E. huxleyi DHODH was heterologously expressed in E. coli, purified, and kinetically characterized. Here, we show HHQ is a potent inhibitor (Ki of 2.3 nM) of E. huxleyi DHODH. E. huxleyi cells exposed to brequinar, the canonical human DHODH inhibitor, experienced immediate, yet reversible cellular arrest, an effect which mirrors HHQ-induced cellular stasis previously observed. However, brequinar treatment lacked other notable effects observed in HHQ-exposed E. huxleyi including significant changes in cell size, chlorophyll fluorescence, and protection from virus-induced lysis, indicating HHQ has additional as yet undiscovered physiological targets. Together, these results suggest a novel and intricate role of bacterial quorum sensing molecules in tripartite interdomain interactions in marine ecosystems, opening new avenues for exploring the role of microbial chemical signaling in algal bloom regulation and host-pathogen dynamics.
Interactions between heterotrophic bacteria and eukaryotic phytoplankton are fundamental to marine ecosystems (Buchan et al., 2014; Seymour et al., 2017). These ubiquitous interactions impact primary production on a global scale, driving biogeochemical cycles, shaping marine trophic structures, and influencing global climate (Azam and Malfatti, 2007; Buchan et al., 2014; Seymour et al., 2017). Bacterium-phytoplankton interactions are diverse, ranging from mutualist nutrient exchanges to predatory algicidal release, with specific pairs of bacteria and phytoplankton capable of both pathogenic and symbiotic relationships (Azam et al., 1983; Mayali and Azam, 2004; Barak-Gavish et al., 2023). Driving these dynamic interactions are excreted chemical compounds, which can modulate microbial metabolism and trigger shifts in community composition (Seyedsayamdost et al., 2011; Segev et al., 2016; Seymour et al., 2017; Barak-Gavish et al., 2023). By elucidating the eukaryotic molecular targets of bacterial compounds, we can establish critical mechanistic links between chemically mediated interactions and their ecological consequences, strengthening our ability to make accurate predictions about fundamental organismal interactions that form the basis of marine ecosystems.
The broad spectrum of compounds that influence microbial interactions, collectively termed infochemicals (Müller et al., 2020; Schmidt and Saha, 2021), orchestrate changes in microbial behavior (Schmidt and Saha, 2021). In bacterium-phytoplankton interactions, the exchange of infochemicals takes place primarily within the phycosphere, the area immediately surrounding the phytoplankton cell (Seymour et al., 2017). Quorum sensing (QS) signals, a class of diffusible infochemicals known to facilitate bacterial communication, have recently emerged as mediators of bacterium-phytoplankton interactions (Harvey et al., 2016) with direct influence on marine biogeochemical cycles (Urvoy et al., 2022). In bacterial communities, secreted QS signals induce community-wide shifts in gene expression, coordinating responses to changing environments (Ng and Bassler, 2009). It has also been demonstrated that eukaryotic hosts can “listen in” on QS signals, triggering a shift in host gene expression, often leading to an immune response (Lowery et al., 2008). QS-mediated interdomain interactions are well-established in human-microbial associations (Kariminik et al., 2017), but have only recently begun to be characterized in phytoplankton-bacterial associations (Dow, 2021).
Alkylquinolones are a class of QS molecules originally isolated from the human pathogen Pseudomonas aeruginosa and include both the Pseudomonas quinolone signal (PQS) and its precursor 2-heptyl-4-quinolone (HHQ) (Pesci et al., 1999). This class of QS molecules has a demonstrated impact on eukaryotic physiology (Reen et al., 2011) where they have been shown to modulate the human immune response (Kim et al., 2010a,b). More recently, HHQ was isolated from the marine gammaproteobacteria Pseudoalteromonas sp. A757 and Pseudoalteromonas galatheae (Harvey et al., 2016; Paulsen et al., 2020). Pseudoalteromonas is a globally ubiquitous genus, ranging from 0.5% to 6% of all marine bacterioplankton species worldwide (Wietz et al., 2010). Pseudoalteromonadaceae were found to have a strong association in blooms with the globally abundant coccolithophore Emiliania huxleyi, comprising up to 19% of prokaryotic community by the time the bloom collapses (Câmara dos Reis et al., 2023). Previous work has established that HHQ is capable of shifting the relative abundances of both bacteria and phytoplankton in natural populations, implicating this infochemical as a compound that mediates microbial population dynamics (Whalen et al., 2019). A subsequent investigation revealed that E. huxleyi exposure to nanomolar concentrations of HHQ could reversibly induce S-phase arrest and prevent mortality due to infection by Emiliania huxleyi virus (EhV) (Pollara et al., 2021), highlighting a new role for bacterial QS signals. However, the mechanistic basis for HHQ-induced cellular stasis remains unknown.
HHQ has been shown to modulate EhV infection dynamics; however, protection from viral induced mortality must coincide with exposure to the QS signal early [< 24 h post infection (hpi)] in infection and requires remodeling of host physiology (Pollara et al., 2021; Harvey et al., 2023). Moreover, this early window in EhV infection (within 8 hpi) corresponds to the time when EhV hijacks the host’s replication machinery to fuel viral gene synthesis involved in DNA replication, nucleotide metabolism, and lipid biosynthesis in parallel with repression of host nuclear and organelle-encoded genes (Ku et al., 2020). Moreover, within 24 h of HHQ exposure, E. huxleyi cells undergo metabolic and transcriptional changes leading to halted DNA replication, an increase in DNA lesions, and the upregulation of the DNA damage response with a notable lack of both DNA repair and apoptosis (Pollara et al., 2021). Together, these observations suggest that the cascade of physiological changes in E. huxleyi induced by HHQ exposure occurs early and may prevent the replication of EhVs.
Several of these physiological changes observed in HHQ-treated E. huxleyi may have resulted from inhibition of dihydroorotate dehydrogenase (DHODH), a key enzyme in de novo pyrimidine biosynthesis (Reis et al., 2017). Coincidently, inhibition of eukaryotic DHODH has been shown to trigger S-phase arrest via severe depletion of available nucleotide pools, leading to increased DNA lesions (Fairus et al., 2017). Nucleotide availability is a known limiting factor for viral burst size (Brown et al., 2006), as giant dsDNA viruses, including EhV, upregulate host de novo nucleotide synthesis early in the infection cycle due to their enhanced metabolic requirements (Rosenwasser et al., 2016; Vincent et al., 2021). Therefore, inhibition of E. huxleyi DHODH (EhDHODH) by HHQ may prevent the early acquisition of host nucleotides to fuel viral replication. In fact, several alkylquinolones have been established as E. coli DHODH inhibitors (Wu and Seyedsayamdost, 2017), including the structurally similar 2-heptyl-4-(1H)-quinolone N-oxide (Horwitz et al., 2022) which differs from HHQ by just a single oxygen atom.
Here, we show that HHQ is an extremely potent inhibitor of EhDHODH, exhibiting the second most potent inhibition of any DHODH recorded to date. This inhibition is attributed to competitive binding with the ubiquinone cofactor, establishing alkylquinolones as a promising reservoir for novel inhibitory agents with species specificity. Moreover, when EhDHODH was inhibited in vivo using the canonical human DHODH inhibitor brequinar, reversible cellular stasis was induced, mirroring the phenotype recorded with HHQ. However, not all of the physiological alterations induced by HHQ were observed in brequinar-treated E. huxleyi. Unexpectedly, inhibition of EhDHODH by brequinar was observed to amplify EhV201-induced mortality of E. huxleyi, while simultaneously significantly decreasing viral replication and markedly diminishing viral infectivity. These findings suggest DHODH inhibition contributes to, but does not fully explain, the protective effects of HHQ on E. huxleyi from EhV. In sum, this work provides mechanistic insight into how HHQ, a quorum sensing molecule used in bacterial communication, impacts a fundamental metabolic pathway in a global phytoplankton and presents new avenues for exploration in the molecular underpinning of HHQ-induced viral protection.
All experiments used axenic cultures of non-lith forming E. huxleyi CCMP2090 (from the National Center for Marine Algae and Microbiota, East Boothbay, ME). Phytoplankton cultures were grown in 0.2 μm-filtered, autoclaved natural seawater enriched f/2-Si medium (Guillard, 1975). Cultures were grown under a light:dark cycle of 14-h/10-h (80 ± 5 μmol photons m−2 s−1) at 18°C with a salinity of 35. MM and f/2 MB purity test broths and epifluorescence microscopy were used to confirm strain purity. Cultures were transferred every 6–7 days to maintain exponential growth.
E. huxleyi cells were enumerated via flow cytometry (Guava; Millipore). Cell abundance was determined using species-specific parameters including forward scatter, side scatter, and red fluorescence (695/50-nm) emissions. Samples of live cells were run at 0.59 μL s−1 for 3 min or until 2000 events were acquired.
Total viruses in cultures were enumerated by fixation in 1% glutaraldehyde (0.2 μm-filtered) and diluted as necessary with sterile 1X phosphate buffered saline (PBS) to ensure events were less than 500 μL−1 on the flow cytometer. Diluted samples were stained with 1X SYBR Green (Invitrogen, Waltham, MA), incubated for 10 min at 80°C in the dark, then counted on the flow cytometer as described in (Harvey et al., 2023). Media only (f/2-Si) blanks were treated in parallel to correct for background signals.
To enumerate the concentration of viral particles actively causing cellular lysis, a most probable number assay (MPN) was used (Jarvis et al., 2010). For each treatment, equal volume aliquots of each biological replicate were pooled then serially diluted 10-fold in a stepwise fashion. A 20 μL aliquot from each dilution or undiluted pooled sample was transferred to six replicate wells of a 96-well plate containing 180 μL of axenic, exponentially growing E. huxleyi CCMP2090 at 1.0 × 106 cells mL−1. The plates were incubated under general culturing conditions for 144 h before E. huxleyi chlorophyll fluorescence, a proxy for cell abundance, was measured via SpectraMax M2e (Molecular Devices, San Jose, CA). Wells observed to have less than or equal to half of the fluorescence of the mean of control wells were designated as “cleared,” and the MPN was calculated using the EPA MPN calculator1 with a Cornish & Fisher Limit of Approximation Type.
The sequence coding for the enzyme DHODH (UniProtID: R1EHA9) from E. huxleyi CCMP1516 genome was used for heterologous expression. Class 2 DHODHs previously expressed and purified often have the N-terminal transmembrane domain and mitochondrial localization region truncated, enhancing solubility in an E. coli expression system with minimal impact on activity (Baldwin et al., 2002; Garavito et al., 2019). For EhDHODH, an appropriate start site truncating this N-terminal region was determined by MUSCLE alignment (Edgar, 2004) in MEGA11 (v11.0.13) with previously expressed and purified DHODHs from Solanum tuberosum DHODH (UniProtID: M1BCR0), Phytophthora infestans DHODH (UniProtID: I7EMP0), Arabidopsis thaliana DHODH (UniProtID: P32746), Plasmodium falciparum DHODH (UniProtID: Q08210), and Homo sapiens DHODH (UniProtID: Q02127) (Supplementary Figure S1), yielding residue 21 from EhDHODH as an appropriate start site. Classification of the excluded 20-residue sequence as a mitochondrial signal peptide was performed using DeepLoc v2.0 (Thumuluri et al., 2022). The truncated gene was incorporated into the expression vector pET24a along with an N-terminal eight-residue histidine tag (Supplementary Figure S2) and the resulting plasmid was transformed into E. coli BL21(DE3) Star (ThermoFisher Scientific, Waltham, MA).
Protein expression, purification, and refolding were performed by GenScript (Piscataway, NJ). Briefly, ΔN20EhDHODH expression was induced in 1 L of transformed E. coli at OD600 1.2 with 0.5 mM IPTG for 16 h at 15°C. The cells were harvested via centrifugation and lysed by sonication in lysis buffer (50 mM Tris–HCl, 500 mM NaCl, 1 mM TCEP, 0.5% TritonX-100, pH 8.0). The lysate was centrifuged to obtain pellets containing insoluble EhDHODH in inclusion bodies. The pellets were solubilized in 50 mM Tris–HCl, 7 M guanidine hydrochloride, 1 mM TCEP, pH 8.0, and the solubilized recombinant ΔN20EhDHODH was then purified using a Ni-NTA resin applied to a gravity flow column. Loosely bound proteins were washed from the column with 50 mM Tris–HCl, 8 M urea, pH 8.0, and ΔN20EhDHODH was eluted using 50 mM Tris–HCl, 8 M urea, with a series of increasing (20–300 mM) imidazole (pH 8.0) concentrations. The isolated ΔN20EhDHODH was refolded by dialysis into the final buffer containing 50 mM Tris–HCl, 150 mM NaCl, 0.5 M L-arginine, 10% glycerol, 1 mM DTT, pH 9.0. The yields of purified ΔN20EhDHODH protein were 7.65 mg/L of cell culture and the expected size for truncated recombinant ΔN20EhDHODH was 44.5 kDa. Yield and purity (≥90%) were confirmed via SDS-PAGE (4%–20% gradient) and Coomassie staining for visualization of isolated proteins (Supplementary Figure S3).
Because ΔN20EhDHODH was expressed and purified without flavin mononucleotide (FMN) supplementation, ΔN20EhDHODH was reconstituted with FMN by incubating equal volumes of 280 μg mL−1 EhDHODH and 37.2 μM FMN for at least 3 min on ice. The reconstituted enzyme was diluted in 50 mM Tris–HCl pH 8.0, 150 mM KCl, 0.1% Triton X-100, 10% glycerol to a 2X working stock immediately before use in downstream enzyme assays.
Protein concentration was measured using the bicinchoninic assay (Pierce, Waltham, MA) with bovine serum albumin as the standard. Absorbance was measured in 96-well plate format on a SpectraMax M2e.
In our standard assay, DHODH activity was measured by the reduction of 2,6-dichlorophenol-indophenol (DCIP) at 600 nm (ε = 19,100 M−1 cm−1) incubated at 22°C in a reaction mixture containing 50 mM Tris–HCl pH 8.0, 150 mM KCl, 0.1% Triton X-100, 10% glycerol, 1 mM L-dihydroorotate (DHO), 0.1 mM DCIP, 0.1 mM decylubiquinone (Qd) with final concentrations of 31 nM ΔN20EhDHODH or 10 nM HsDHODH (biotechne, Minneapolis, MN, Cat. No. 10062-DD-020) (Zameitat et al., 2007). To determine the kinetic constants, DHO concentration was varied from 3.1 to 1,600 μM while keeping Qd constant at 100 μM, or by varying Qd concentration from 1.2 to 600 μM with DHO fixed at 1 mM while keeping all other concentrations the same as above. Background oxidase activities due to direct reduction of DCIP by the enzyme in absence of Qd were subtracted from the activities measured in the presence of Qd (Reis et al., 2017; Garavito et al., 2019). Kinetic constants Km and vmax were calculated by fitting the data to the Michaelis–Menten equation (Eq. 1):
using GraphPad Prism v9 software. The apparent kcat was calculated by fitting the data to the equation (Eq. 2):
where is the concentration of catalytic sites (set to 31 nM, the concentration of enzyme used in the standard assay) using GraphPad Prism v9 software.
Stock solutions of 20 mM HHQ were prepared in DMSO and kept at −20°C until day of use, and 20 mM brequinar stock solutions were prepared in DMSO fresh on the day of use. For the dose–response experiments with DHODH inhibitors, the enzyme was incubated with concentrations of HHQ ranging from 0.001 to 100 μM, or brequinar ranging from 3 to 739 μM, or a volumetric equivalent solvent control. Activities were measured with the standard DCIP assay with saturating concentrations of Qd (100 μM) and DHO (1 mM), in the presence of the inhibitor. The final concentration of 0.5% DMSO used in the assay solutions did not impact the activity of either recombinant human or E. huxleyi DHODH. IC50 values were calculated, and 95% confidence intervals were estimated using GraphPad Prism v9 software by fitting the log transformation of the response variable (I; inhibitor concentration) by non-linear regression to the equation (Eq. 3):
where the Hill slope (H) is −1 and “Top” and “Bottom” are the plateaus in initial enzymatic reaction velocity. Estimates of the Ki for HHQ suggested this value would be at or below the concentration of enzyme used in our assays necessitating the use of Morrison’s equation used to determine the Ki for tightly binding inhibitors (Copeland et al., 1995; Murphy, 2004). To calculate Ki for HHQ, GraphPad Prism v9 was used to fit the data to the Morrison equation (Eq. 4) for tightly binding inhibitors (Copeland et al., 1995; Murphy, 2004).
where [E] is the enzyme concentration, [I] is the inhibitor concentration, vi is the measured initial velocity, is velocity without inhibitor, and the apparent Ki (Kiapp) is related to the Ki, substrate concentration [S], and Km for the substrate shown in Eq. 5.
For assessing the inhibition mechanism of HHQ, Dixon-plots were constructed by assaying the activity of ΔN20EhDHODH at 20, 40, and 80 nM HHQ while varying DHO concentrations from 12.5 to 1,600 μM or Qd concentrations from 1.17 to 150 μM (Copeland, 2000; Yoshino and Murakami, 2009). The reciprocal of the initial velocities was plotted against HHQ concentration, and a linear regression analysis was performed to construct trendlines (GraphPad Prism v9). Given the tight binding nature of HHQ, we recognize the limitations of this graphical method for determining inhibitory mechanisms (Copeland et al., 1995).
Google DeepMind’s AlphaFold v2.0 (Jumper et al., 2021) was queried with the non-truncated amino acid sequence of EhDHODH (UniProtID: R1EHA9), and the resulting structure was used as the basis for molecular docking. The substrate DHO, cofactors FMN and decylubiquinone, and putative inhibitors HHQ and brequinar were docked to the predicted structure using AutoDock Vina v1.1.2 (Trott and Olson, 2010; Eberhardt et al., 2021). The top output structures were visualized using UCSF ChimeraX v1.6.1 (Pettersen et al., 2021).
For the dose–response experiments, axenic E. huxleyi CCMP2090 in exponential phase at an initial concentration of 1.0 × 105 cells mL−1 were grown in triplicate in 24-well plates exposed to nine brequinar concentrations ranging from 1 to 1,000 μM for 144 h. Plates were kept under general cultivation conditions, and cell abundance was monitored daily via flow cytometry as described above. The growth rate (μ d−1) over 72 h was calculated using the exponential growth equation (Eq. 6) (Harvey et al., 2016),
where A is cell abundance at the initial and final time point (72 h), and T is days of growth over the first 3 days of the experiment. The growth rate was plotted against the concentration of brequinar to determine the concentration of compound resulting in 50% growth inhibition (IC50). The IC50 for brequinar was calculated using Eq. 3 as described above.
To investigate the impact of brequinar on EhV infection dynamics, triplicate 20 mL cultures containing exponentially growing axenic E. huxleyi CCMP2090 at an initial concentration of 9.0 × 104 cells mL−1 were inoculated with EhV201 (1:1 virus:host ratio). Concurrently, cells were exposed to brequinar at a concentration shown to induce cellular stasis for 72 h (135 μM brequinar) and just below that concentration (75 μM brequinar), allowing for continued E. huxleyi growth. Triplicate control cultures were prepared with both viral lysate and a volumetric equivalent of DMSO. Cultures were harvested daily for E. huxleyi cell counts and EhV enumeration as described above. A pooled sample of all three replicates for each treatment were used to perform an MPN assay to enumerate the concentration of virus actively causing cellular lysis beginning at 24 hpi and performed every subsequent 24 h until 144 hpi.
All statistical analyses were done using GraphPad Prism v9. Experiments conducted to examine differences in cell or viral abundances over time were tested for significance using repeated measures ANOVA with Sidak’s multiple comparison post hoc tests. Any p-values less than or equal to 0.05 were considered statistically significant in all tests.
EhDHODH can be categorized as a class 2 DHODH, as the enzyme contains both an N-terminal mitochondrial signal peptide and an N-terminal helical domain facilitating binding of a ubiquinone cofactor. Following published literature on expressing class 2 DHODHs (Baldwin et al., 2002; Zameitat et al., 2007; Garavito et al., 2019), the N-terminal mitochondrial signal peptide (first 20 residues) was truncated based on amino acid alignments with previously expressed and purified DHODHs including S. tuberosum DHODH, P. infestans DHODH, A. thaliana DHODH, P. falciparum DHODH, and H. sapiens DHODH (Supplementary Figure S1). To evaluate the potential inhibitory effects of HHQ on EhDHODH, ΔN20EhDHODH was heterologously expressed and purified from E. coli BL21. This truncation, a common method to remove the hydrophobic transmembrane anchor (Garavito et al., 2019), was insufficient to solubilize ΔN20EhDHODH in our E. coli expression system, necessitating purification and refolding from inclusion bodies formed during expression. SDS-PAGE confirmed the successful purification and expected molecular weight of ΔN20EhDHODH (Supplementary Figure S3).
Saturating substrate and cofactor concentrations, as well as kinetic parameters of ΔN20EhDHODH were established and compared to other DHODHs (Figure 1, Table 1). ΔN20EhDHODH demonstrated a specific activity of 31 ± 1 μmol min−1 mg−1, a kcatapp of 23 ± 1 s−1, a Kmapp of 170 ± 30 μM for DHO, and a Kmapp of 8 ± 1 μM for Qd.
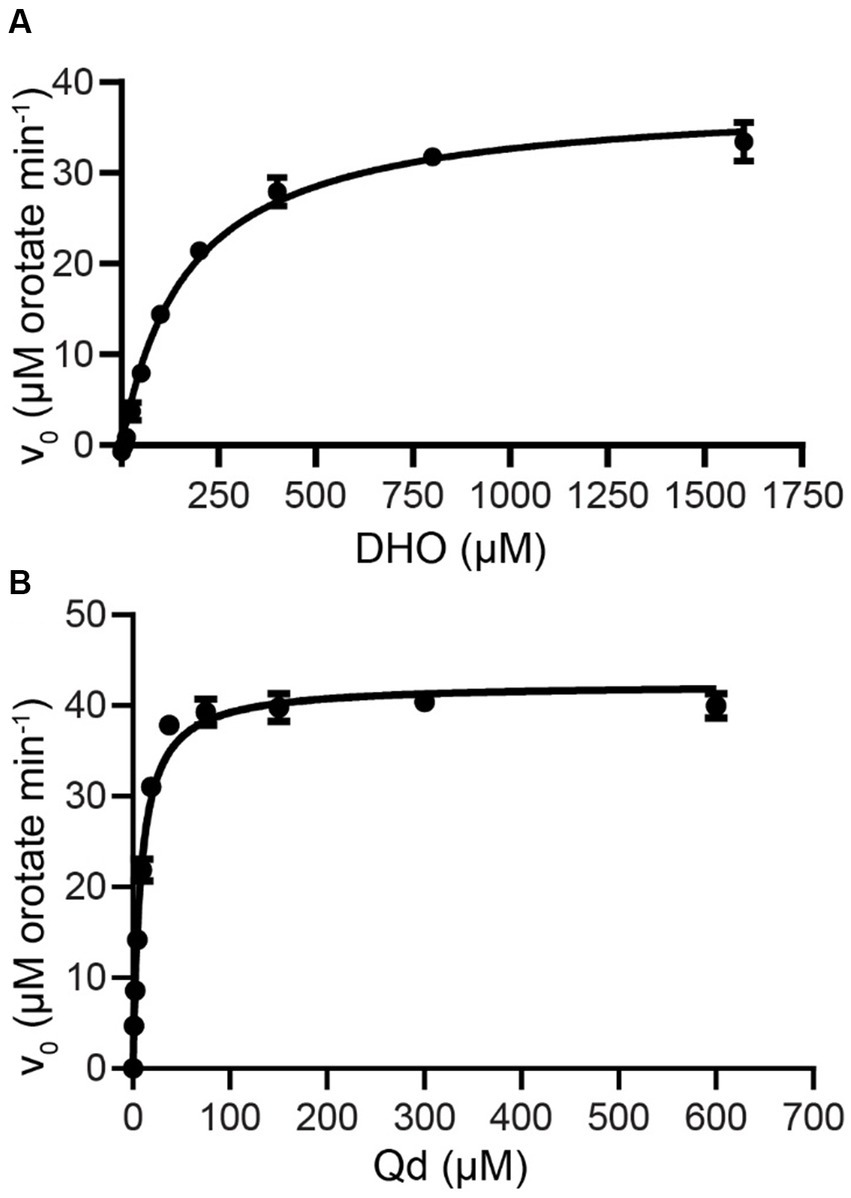
Figure 1. Steady-state kinetics of purified truncated recombinant E. huxleyi DHODH. ΔN20EhDHODH saturation curves for substrate (A) L-dihydroorotate (DHO) and cofactor (B) decylubiquinone (Qd) are shown. Non-linear regression analysis was used to fit the curves to the Michaelis–Menten equation (Eq. 1). Mean and standard deviation of triplicate experiments shown.
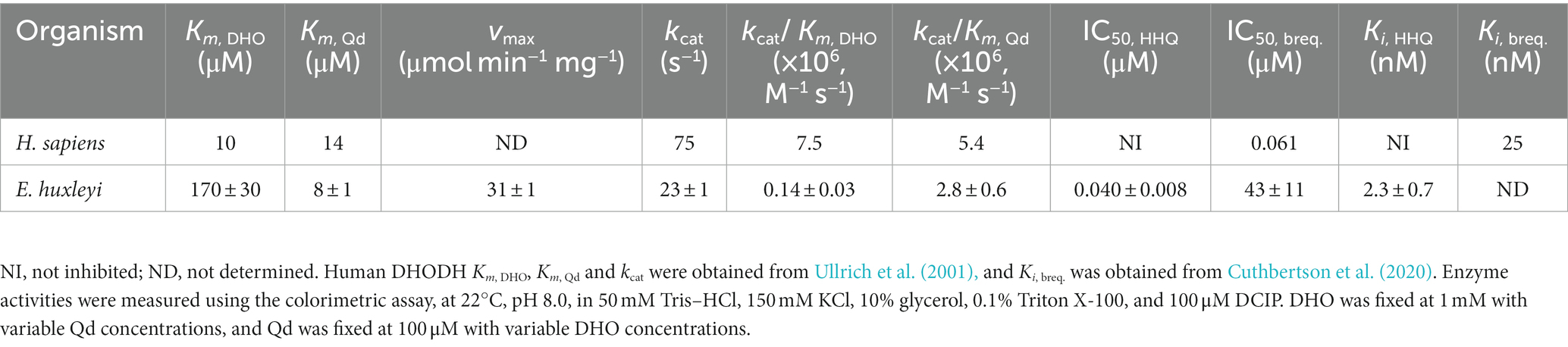
Table 1. Apparent kinetic and inhibitory parameters of E. huxleyi DHODH, human DHODH and their inhibitors.
Using saturating concentrations of DHO and Qd (1 mM and 100 μM, respectively), we constructed a dose–response curve to determine the IC50 of HHQ (Figure 2). HHQ exhibits potent inhibition of ΔN20EhDHODH, with an IC50 of 40 nM against 31 nM of ΔN20EhDHODH. Estimates of the Ki of HHQ suggested this value would be at or below the concentration of enzyme used in our assays necessitating the use of Morrison’s equation for tightly binding inhibitors to determine the Ki (Copeland et al., 1995; Murphy, 2004). Fitting the dose–response data to the Morrison’s equation yielded a Ki of 2.3 ± 0.7 nM (Table 1). For comparison, the inhibitory effect of brequinar, the canonical inhibitor of human DHODH, was substantially less potent against ΔN20EhDHODH, with an IC50 of 43 μM (Figure 2), a thousand-fold higher than the IC50 for HHQ. The inhibitory effect of HHQ was also tested against human DHODH, and no measurable dose-dependent inhibitory effect was observed up to 100 μM HHQ (data not shown). A summary of the inhibitor constants and their counterparts against human DHODH are summarized in Table 1.
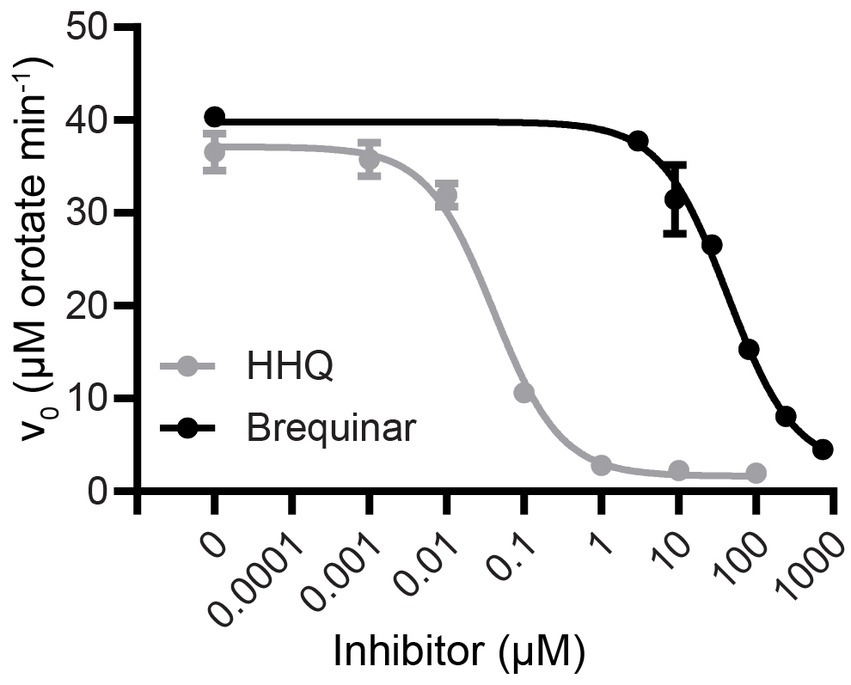
Figure 2. Dose–response inhibition curves of HHQ and brequinar for truncated recombinant E. huxleyi DHODH. Activities were measured with the standard colorimetric DHODH activity assay with saturating concentrations of Qd (100 μM) and DHO (1 mM) in the presence of 0–100 μM HHQ or 0–729 μM brequinar for ΔN20EhDHODH. Non-linear regression analysis was used to generate the trendline. Mean and standard deviation of triplicate experiments shown.
To further understand the interaction between EhDHODH and HHQ, Dixon plots were constructed (Supplementary Figure S4), which indicated HHQ demonstrates competitive inhibition with respect to Qd and uncompetitive inhibition against DHO. Molecular docking of HHQ and brequinar to an AlphaFold 2.0-predicted 3D structure of EhDHODH supports the model whereby HHQ and brequinar bind to the decylubiquinone binding site (Supplementary Figure S5). In the docked structure, HHQ slots into the ubiquinone access tunnel with its alkyl chain pointing inward toward the catalytic center of the enzyme, and HHQ forms a hydrogen bond between the carbonyl of HHQ and S63. The benzene ring of HHQ is positioned at the entrance of the ubiquinone access tunnel which is comprised primarily of hydrophobic residues. Mid-way through the tunnel, the pyridine ring lines up with the charged / polar residues including S63. At the end of the tunnel, the hydrophobic alkyl chain of HHQ associates with more hydrophobic residues toward the catalytic center.
To mechanistically connect enzymatic DHODH inhibition established by our in vitro studies and the cellular stasis characteristic of in vivo HHQ exposure (Pollara et al., 2021) we conducted a dose–response experiment with E. huxleyi exposed to brequinar concentrations ranging from 0 to 1,000 μM. Cellular stasis was induced at 135 μM brequinar with little to no growth or cell death over 72 h (Figure 3). For axenic E. huxleyi (CCMP2090), the IC50 of brequinar was determined to be 345 μM, while HHQ was nearly a thousand-fold lower at 363 nM (Pollara et al., 2021). To examine whether the growth inhibitory effect of brequinar was reversible, E. huxleyi cultures exposed to brequinar for 120-h were diluted 20-fold in fresh f/2-Si media to yield a final concentration of 6.75 μM brequinar, a concentration determined not to exhibit any inhibitory effect on E. huxleyi growth (Figure 3). Under these conditions, exponential growth was restored by 72 h post-dilution (Supplementary Figure S6). Unlike HHQ (Pollara et al., 2021), brequinar did not induce an increase in E. huxleyi cell size or chlorophyll fluorescence (Supplementary Figure S7).
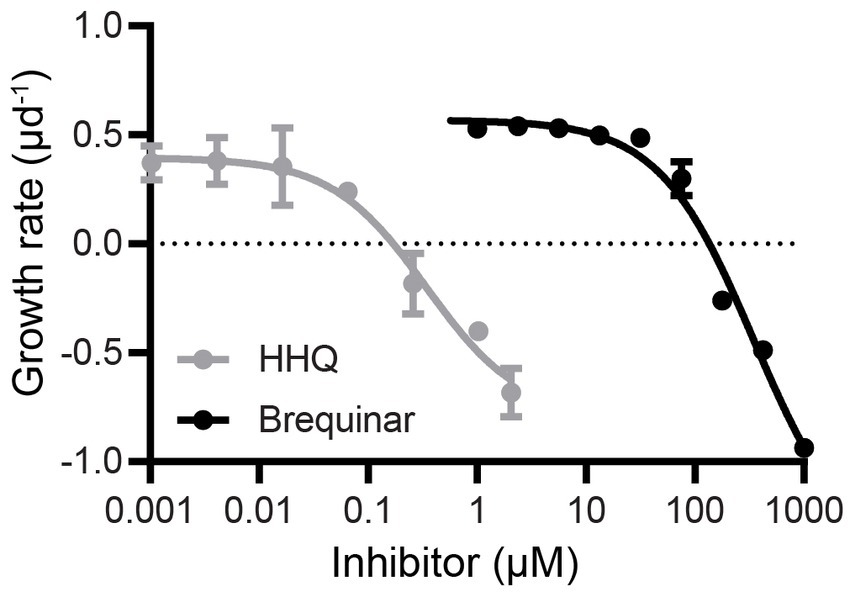
Figure 3. Dose–response comparison of E. huxleyi growth rate during exposure to HHQ and brequinar. Exponentially growing axenic E. huxleyi (CCMP2090) were dosed with brequinar (0–1,000 μM) or HHQ (0–0.54 μM) previously [HHQ data from Pollara et al. (2021)], then monitored for 72 h. The growth rate was calculated over 72 h and the mean and standard deviation of biological triplicates are shown.
To test whether DHODH inhibition alone was contributing to cellular protection from virus-induced mortality, E. huxleyi cultures were exposed to 75 or 135 μM brequinar, then infected with EhV201 at a 1:1 host-to-virus ratio. Brequinar did not protect E. huxleyi from virus-induced mortality; rather, cell lysis was enhanced by brequinar in infected cultures (Figure 4A). Beginning at 96 hpi, total viral abundance in brequinar-exposed infected cultures was significantly (p-value ≤ 0.0001) less relative to infected cultures with no brequinar addition (Figure 4B), indicating viral replication was impacted. Moreover, at 135 μM brequinar, the concentration of virus actively causing cellular lysis was zero at all time points examined compared to control, with the exception of 120 hpi (Figure 4B). At a concentration of 75 μM brequinar, a concentration which still allowed for E. huxleyi growth, a significant (p-value ≤ 0.05) decrease in infectious viral particles was observed for 24 hpi and 48 hpi compared to control (Figure 4B).
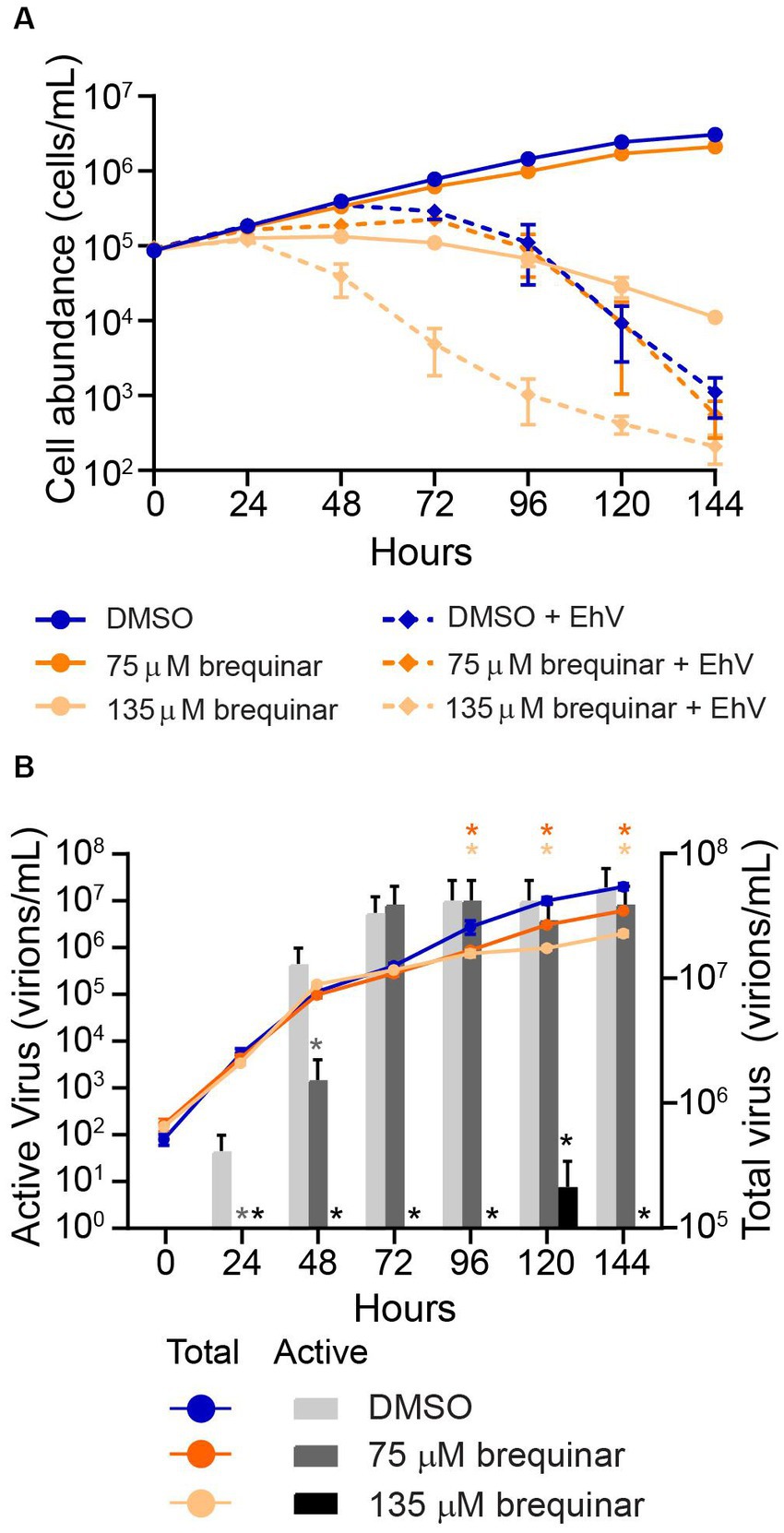
Figure 4. Effect of brequinar on E. huxleyi growth rate and viral dynamics. (A) E. huxleyi (CCMP2090) cultures were dosed with brequinar with and without E. huxleyi virus (EhV) 201 (1:1 host:virus ratio). E. huxleyi cell abundance was measured every 24 h over 144 hpi. (B) Infectious viral titer (left Y-axis) and total viral abundance (right Y-axis) were measured over 144 hpi. In both (A,B), cell abundance and total viral abundance are shown as the mean and standard deviation of triplicates. Total virion abundance between brequinar treatments and DMSO controls were significantly different (p-value ≤ 0.0001, denoted with an asterisk with colors corresponding to respective brequinar treatment) at 96, 120, and 144 hpi using repeated measures ANOVA and Sidak’s post hoc test for significance. Infectious or active viral particles were quantified using the MPN assay from 24 to 144 hpi, and the mean and 95% confidence interval are shown. Cultures dosed with 75 μM brequinar had significantly reduced (p-value ≤ 0.05, denoted with an asterisk in dark grey) infectious virion abundance at 24 and 48 hpi relative to DMSO controls. Cultures dosed with 135 μM brequinar had significantly reduced (p-value ≤ 0.05, denoted with an asterisk in black) infectious virion abundance relative to DMSO controls at all timepoints measured.
Previous work established that the alkylquinolone quorum sensing signal HHQ has significant effects on modulating growth and viral infection dynamics in the globally abundant coccolithophore E. huxleyi (Harvey et al., 2016; Pollara et al., 2021; Harvey et al., 2023), but the mechanistic basis of these effects was unknown. Alkylquinolones have been shown to inhibit E. coli DHODH (Wu and Seyedsayamdost, 2017; Horwitz et al., 2022), a fundamental enzyme in de novo pyrimidine synthesis. Moreover, the lack of available nucleotide pools is known to induce S-phase arrest in eukaryotic cells (Fairus et al., 2017), which could explain the cellular stasis previously observed in HHQ-treated E. huxleyi (Harvey et al., 2016; Pollara et al., 2021). Having a sufficient quantity of host-supplied nucleotides is also of critical importance during viral infection and propagation, and the resulting loss of pyrimidine production by DHODH inhibition could explain the decrease in viral abundance observed in EhV-infected E. huxleyi cultures treated with HHQ (Harvey et al., 2023).
Here, we establish HHQ to be a potent inhibitor of recombinant EhDHODH with a Ki of 2.3 nM, placing this inhibitor second only to 2-biphenyl-4-yl-3-methyl-quinoline-4-carboxylic acid (Batt et al., 1995) as the most potent inhibitor of any DHODH reported on BindingDB (Liu et al., 2007). There has been significant interest in developing synthetic inhibitors for human and human pathogen DHODHs. These inhibitors include brequinar derivatives (Batt et al., 1995; Madak et al., 2018), leflunomide derivatives (Davies et al., 2009), and pyrazoles (Haque et al., 2002), the most potent of which having inhibitory constants ranging from 4.0 to 1.7 nM. HHQ represents the first naturally produced compound to have an inhibitory constant for a eukaryotic DHODH with potency comparable to those inhibitors developed for therapeutic use in humans.
This study provides evidence of the structural basis for HHQ’s potent and species-specific inhibition. Similar to other alkylquinolones (Wu and Seyedsayamdost, 2017; Horwitz et al., 2022), Dixon plots and in silico modeling both provide evidence that HHQ likely binds the ubiquinone binding site (Supplementary Figures S4, S5), thus preventing the ubiquinone-mediated regeneration of oxidized FMN (Fagan et al., 2006). DHODH inhibitors that competitively bind to the ubiquinone binding site are often species specific due to high variability in the N-terminal region of DHODH, which contains residues that line the ubiquinone channel (Hurt et al., 2006; Garavito et al., 2019). Consequently, the degree to which inhibitor structural motifs carry over between distantly related DHODHs is often limited. Indeed, HHQ’s inhibitory qualities do not extend to human DHODH. Likewise, brequinar is 1,000-fold less potent of an inhibitor for EhDHODH compared to HHQ. Previous studies found that brequinar derivatives with enhanced affinity for human DHODH formed new electrostatic interactions, including strategically positioned hydrogen-bonds within the ubiquinone channel (Madak et al., 2018). Docking results indicate a unique hydrogen bond between the carbonyl on HHQ with the hydroxyl on S63, which may stabilize HHQ in the ubiquinone channel and account for HHQ’s high affinity specific for EhDHODH.
Furthermore, when comparing the sequence of EhDHODH with DHODHs from human and the two algal species Phaeodactylum tricornutum and Dunaliella tertiolecta, shown previously to be insensitive to HHQ (Harvey et al., 2016), there is substantial variation in the residues forming the EhDHODH ubiquinone channel (Supplementary Figure S8). In docking studies with EhDHODH, only one (L36) of the 15 residues shown to be within 5 Å of decylubiquinone, HHQ, or brequinar is conserved with HsDHODH, while two residues (V14 and L47) are conserved with P. tricornutum, and two residues (L47 and L59) are conserved with D. tertiolecta (Supplementary Figure S8). Likewise, residues lining the channel within human DHODH (R136, Y356, V134, V143, and H55), deemed crucial for the binding of brequinar (Davis and Copeland, 1997; Hurt et al., 2006; Madak et al., 2018), are not conserved in the protein alignment with E. huxleyi, likely explaining the reduced potency of brequinar observed against EhDHODH. Together, these data indicate that HHQ inhibition of DHODH is highly species specific, likely resulting from the unique structure of EhDHODH’s ubiquinone channel.
Exposure of E. huxleyi cells to brequinar was used to estimate the influence of DHODH inhibition on host cell division and viral propagation dynamics. While brequinar did induce cellular stasis in E. huxleyi, albeit at micromolar concentrations, this inhibitor failed to mimic all of HHQ’s cellular effects, notably the increased cell size and chlorophyll fluorescence seen with HHQ treatment (Pollara et al., 2021) (Supplementary Figure S7). Following brequinar exposure at 135 μM, a decline in cell abundance was observed after 72 h in contrast to complete cellular stasis recorded for 504 h under HHQ (410 nM) treatment (Pollara et al., 2021). The loss of cells after 72 h of cellular stasis following brequinar exposure parallels studies showing DHODH inhibition eventually leading to programmed cell death (PCD) (Arnould et al., 2017; Fairus et al., 2017), which is in stark contrast to HHQ-exposed E. huxleyi cells, which exhibit no physiological hallmarks of PCD (Pollara et al., 2021). This raises the possibility that HHQ may interfere with additional, yet unidentified, cellular stress signaling pathway(s), which might otherwise lead to PCD.
The application of brequinar to infected cells was insufficient to protect cells from virus-induced mortality. At 75 μM brequinar, a concentration that allows for E. huxleyi growth, infected cultures tracked with paired controls, with no inhibition or enhancement of viral lysis observed. At 135 μM brequinar, a cellular stasis-inducing concentration analogous to 410 nM HHQ treatment used previously (Pollara et al., 2021), E. huxleyi cell death in response to EhV infection was enhanced. This result was unexpected due to the wealth of literature reporting DHODH inhibitors as promising antivirals, including brequinar (Qing et al., 2010; Hoffmann et al., 2011; Wang et al., 2011; Lucas-Hourani et al., 2013; Luthra et al., 2018; Demarest et al., 2022; Zheng et al., 2022). Our observations prompt questions of whether the increased cell death is a result of DHODH inhibition in E. huxleyi or non-DHODH-related cytotoxic effects of brequinar that synergize with the stress of viral infection. In human cell lines, the dose of brequinar capable of decreasing dengue viral infectivity in human cell lines ranged from 0.1 to 3 μM (Qing et al., 2010), corresponding to 33- to 1,000-fold below the cytotoxicity threshold concentration (approximately 100 μM) following 24 h of drug exposure (Hoffmann et al., 2011). Because brequinar is a poor inhibitor of EhDHODH, the concentration that would likely inhibit viral lysis in E. huxleyi cells overlaps with the cytotoxicity threshold of brequinar. It is therefore difficult to resolve whether the enhanced lysis during infection was due to DHODH inhibition or cytotoxic effects of brequinar.
Though lysis was not prevented by brequinar in infected cultures, there were significant changes in total virion and infectious virion production over the course of infection. Both concentrations of brequinar tested resulted in a significant decrease in total virion abundance relative to infected controls starting at 96 hpi (Figure 4B). However, the brequinar treatments reached a maximum total virion abundance comparable to the infected controls, while HHQ treatment resulted in a 10-fold decrease in total virion abundance relative to the control (Harvey et al., 2023), suggesting HHQ is more effective than brequinar at preventing viral DNA replication. In HHQ-exposed cells, infectious virion production saw a 1,000-fold reduction compared to controls (Harvey et al., 2023). Similarly, 135 μM brequinar treatment showed complete elimination of infectious virion production with the exception of 120 hpi. These findings suggest that DHODH inhibition contributes to a decrease in both total and infectious virion production and may largely act on post-viral DNA replication processes.
DHODH inhibitors have previously been shown to limit the titer of infectious negative-sense RNA viruses, positive-sense RNA viruses, DNA viruses, and retroviruses (Qing et al., 2010; Hoffmann et al., 2011; Wang et al., 2011; Lucas-Hourani et al., 2013; Luthra et al., 2018; Demarest et al., 2022; Zheng et al., 2022). This broad-spectrum antiviral activity is thought to have resulted from insufficient nucleotide pools available for viral replication (Hoffmann et al., 2011; Wang et al., 2011; Luthra et al., 2018; Demarest et al., 2022). However, DHODH inhibition also appears to activate innate cellular immunity, including processes regulated by the interferon regulatory factor-1 (IRF1), a transcription factor known to maintain an “antiviral state” in mammalian cells (Lucas-Hourani et al., 2013; Luthra et al., 2018; Panda et al., 2019). It is therefore possible that DHODH inhibition in algal cells triggers an analogous antiviral pathway(s) such as induction of a hypersensitive PCD response (Heath, 2000; Bidle, 2016), downregulation of viral lipid biosynthesis (Rosenwasser et al., 2014), or RNAi-mediated gene silencing (De Riso et al., 2009).
Adding additional complexity are reports of HHQ suppressing human immune responses by inhibition of macrophage activation and nitric oxide (NO) production (Kim et al., 2010b), as well as suppression of innate immunity by the inhibition of the transcription factor NF-κB and its downstream target genes (Kim et al., 2010a). Parallels can be drawn between mammalian and E. huxleyi cells in their response to HHQ. Recent findings have shown that E. huxleyi exposure to HHQ limits production of NO during viral infection (Harvey et al., 2023), which is well established to play a role in regulating innate cellular immunity in plants (Yoshioka et al., 2011). Indeed, co-induction of NO and reactive oxygen species (ROS) leads to a hypersensitive PCD immune response in plants (Delledonne et al., 2001), and in phytoplankton both NO and ROS are considered hallmarks of PCD in general (Bidle, 2016), though their specific roles in regulating innate immunity have yet to be explored.
Though suppression of innate immunity by HHQ at first seems contradictory to viral protection, when we consider the role of PCD in EhV infection, the utility of this suppression becomes evident. During EhV infection, the virus tightly controls host PCD activation pathways to both prevent the host from activating PCD prematurely as a strategy to rid itself of the virus and facilitate viral release later in the infection cycle (Bidle and Falkowski, 2004). EhV infection coincides with NO and ROS production (Evans et al., 2006; Schieler et al., 2019), as well as induction of an autophagy-like process (Schatz et al., 2014), each of which are typically referred to in the context of immune responses in mammalian and plant systems, but here are hijacked by the virus and are critical for completion of the EhV infection cycle (Schatz et al., 2014; Sheyn et al., 2016; Schieler et al., 2019). Therefore, HHQ-mediated suppression of these “immune” responses could cripple EhV’s ability to complete its infection and propagation cycle. In combination with potent DHODH inhibition limiting the availability of nucleotide pools, HHQ likely has additional complementary pathways that lead to reduction of both viral DNA replication and viral-mediated host lysis.
A possible secondary target of HHQ is poly (ADP-ribose) polymerase (PARP), a master regulator of the DNA damage response (DDR) (Jagtap and Szabó, 2005). The accumulation of DNA lesions characteristic of HHQ exposure (Pollara et al., 2021), and DHODH more broadly, would ordinarily overstimulate the PARP-regulated DDR, depleting ATP and NAD+ pools, thereby initiating apoptotic pathways (Yu et al., 2002). Moreover, PARP has also been extensively implicated in innate cellular immunity of both mammalian and plant cells (Chang and Alvarez-Gonzalez, 2001; Hassa et al., 2003; Adams-Phillips et al., 2010). Expression of genes regulated by NF-κB are known to be dependent on PARP1 activity (Chang and Alvarez-Gonzalez, 2001; Hassa et al., 2003), and inhibition of PARP in Arabidopsis has been shown to inhibit plant immune responses downstream of pathogen-induced ROS production (Adams-Phillips et al., 2010). Furthermore, benzimidazole derivatives, which share an internal structural motif with HHQ, are known dual inhibitors of DHODH and PARP (Abdullah et al., 2015), and HHQ has been reported to inhibit human PARP at a concentration of 50 μM (Pollara et al., 2021). It would therefore be of interest to determine if HHQ could inhibit E. huxleyi PARP, thereby averting the PCD response that would typically be realized by DHODH inhibition and / or viral infection.
Fundamental questions remain about the ecological impact of HHQ-mediated DHODH inhibition. What incentive is there for Pseudoalteromonas spp. to inhibit de novo pyrimidine synthesis in E. huxleyi, thereby inducing cellular stasis? Accumulation of DNA lesions, such as those characteristic of HHQ-exposure and DHODH inhibition in general, causes a remodeling of cellular metabolism that results in an increase in ATP production to fuel DNA repair (Milanese et al., 2019). When DNA repair is faulty, this ATP accumulates and in turn inhibits glycolysis, which is thought to prevent superfluous ATP production (Milanese et al., 2019). In line with this metabolic remodeling, previous work established that E. huxleyi exposure to HHQ results in an increased transcript abundance of enzymes associated with the tricarboxylic acid cycle, which would correlate with production of reducing equivalents for ATP production, as well as reduced transcript abundance of hexokinase, the first step in glycolysis (Pollara et al., 2021). Together these findings provide important supporting evidence for our hypothesis that an HHQ-exposed E. huxleyi cell converts to a fuel cell, incapable of cell division yet producing a surplus of energy storage molecules, all the while preserving glucose reserves. It is conceivable that such a metabolic profile shift could benefit bacterial growth and may be preferable to bacterially-mediated predation of the phytoplankton cell. However, metabolomic characterization of HHQ-induced changes in metabolic exchange between the algal host and bacterium are needed to validate this hypothesis.
The discovery that a bacterial communication molecule, HHQ, is a potent inhibitor of a fundamental enzyme in eukaryotic nucleotide synthesis establishes a novel avenue through which microbial chemical communication can shape algal physiology and, in turn, bloom dynamics and microbial communities more broadly. The high affinity and specificity of HHQ for EhDHODH suggests an intimate co-evolutionary relationship between the host and bacterium. Future work elucidating how HHQ might alter the chemical currency between Pseudoalteromonas spp. and its algal host E. huxleyi may undercover a species-specific metabolic incentive for the bacterium to induce cellular stasis in the algal host, even during viral assault. The evolution of this HHQ-mediated tripartite interdomain interaction remains elusive but unraveling the mechanistic underpinnings will undoubtably reveal a novel three-way interaction between a phytoplankton host, a bacterium, and a virus.
The data presented in this study are deposited in the STRENDA database, https://www.beilstein-strenda-db.org/, accession number V4LL20.
OG: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Writing – original draft, Writing – review & editing. KW: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by NSF OCE 1657818 and NSF IOS 2041748 and by the Charles E. Kaufman Foundation Integrated Research Education Grant (KA2021-121932) to KW. In addition, OG was supported by the Arnold and Mable Beckman Fellowship.
We thank Kay Bidle for sharing E. huxleyi virus. We also thank Joanne Brown, Nicole Cunningham, and Zak Kerrigan for research support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1266972/full#supplementary-material
Abdullah, I., Chee, C. F., Lee, Y.-K., Thunuguntla, S. S. R., Satish Reddy, K., Nellore, K., et al. (2015). Benzimidazole derivatives as potential dual inhibitors for PARP-1 and DHODH. Bioorg. Med. Chem. 23, 4669–4680. doi: 10.1016/j.bmc.2015.05.051
Adams-Phillips, L., Briggs, A. G., and Bent, A. F. (2010). Disruption of poly(ADP-ribosyl)ation mechanisms alters responses of Arabidopsis to biotic stress. Plant Physiol. 152, 267–280. doi: 10.1104/pp.109.148049
Arnould, S., Rodier, G., Matar, G., Vincent, C., Pirot, N., Delorme, Y., et al. (2017). Checkpoint kinase 1 inhibition sensitises transformed cells to dihydroorotate dehydrogenase inhibition. Oncotarget 8, 95206–95222. doi: 10.18632/oncotarget.19199
Azam, F., Fenchel, T., Field, J. G., Gray, J. S., Meyer-Reil, L. A., and Thingstad, F. (1983). The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263. doi: 10.3354/meps010257
Azam, F., and Malfatti, F. (2007). Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5:Article 10. doi: 10.1038/nrmicro1747
Baldwin, J., Farajallah, A. M., Malmquist, N. A., Rathod, P. K., and Phillips, M. A. (2002). Malarial dihydroorotate dehydrogenase. Substrate and inhibitor specificity. J. Biol. Chem. 277, 41827–41834. doi: 10.1074/jbc.M206854200
Barak-Gavish, N., Dassa, B., Kuhlisch, C., Nussbaum, I., Brandis, A., Rosenberg, G., et al. (2023). Bacterial lifestyle switch in response to algal metabolites. elife 12:e84400. doi: 10.7554/eLife.84400
Batt, D. G., Copeland, R. A., Dowling, R. L., Gardner, T. L., Jones, E. A., Orwat, M. J., et al. (1995). Immunosuppressive structure-activity relationships of Brequinar and related cinchoninic acid derivatives. Bioorg. Med. Chem. Lett. 5, 1549–1554. doi: 10.1016/0960-894X(95)00252-O
Bidle, K. D. (2016). Programmed cell death in unicellular phytoplankton. Curr. Biol. 26, R594–R607. doi: 10.1016/j.cub.2016.05.056
Bidle, K. D., and Falkowski, P. G. (2004). Cell death in planktonic, photosynthetic microorganisms. Nat. Rev. Microbiol. 2, 643–655. doi: 10.1038/nrmicro956
Brown, C. M., Lawrence, J. E., and Campbell, D. A. (2006). Are phytoplankton population density maxima predictable through analysis of host and viral genomic DNA content? J. Mar. Biol. Assoc. U. K. 86, 491–498. doi: 10.1017/S0025315406013397
Buchan, A., LeCleir, G. R., Gulvik, C. A., and González, J. M. (2014). Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698. doi: 10.1038/nrmicro3326
Câmara dos Reis, M., Romac, S., Le Gall, F., Marie, D., Frada, M. J., Koplovitz, G., et al. (2023). Exploring the phycosphere of Emiliania huxleyi: from bloom dynamics to microbiome assembly experiments. Mol. Ecol. 1–16. doi: 10.1111/mec.16829
Chang, W.-J., and Alvarez-Gonzalez, R. (2001). The sequence-specific DNA binding of NF-κB is reversibly regulated by the automodification reaction of poly (ADP-ribose) polymerase 1 *. J. Biol. Chem. 276, 47664–47670. doi: 10.1074/jbc.M104666200
Copeland, R. A. (2000). Enzymes: a practical introduction to structure, mechanism, and data analysis. New York, NY: John Wiley & Sons.
Copeland, R. A., Lombardo, D., Giannaras, J., and Decicco, C. P. (1995). Estimating KI values for tight binding inhibitors from dose-response plots. Bioorg. Med. Chem. Lett. 5, 1947–1952. doi: 10.1016/0960-894X(95)00330-V
Cuthbertson, C. R., Guo, H., Kyani, A., Madak, J. T., Arabzada, Z., and Neamati, N. (2020). The dihydroorotate dehydrogenase inhibitor Brequinar is synergistic with ENT1/2 inhibitors. ACS Pharmacol. Transl. Sci. 3, 1242–1252. doi: 10.1021/acsptsci.0c00124
Davies, M., Heikkilä, T., McConkey, G. A., Fishwick, C. W. G., Parsons, M. R., and Johnson, A. P. (2009). Structure-based design, synthesis, and characterization of inhibitors of human and plasmodium falciparum dihydroorotate dehydrogenases. J. Med. Chem. 52, 2683–2693. doi: 10.1021/jm800963t
Davis, J. P., and Copeland, R. A. (1997). Histidine to alanine mutants of human dihydroorotate dehydrogenase: identification of a brequinar-resistant mutant enzyme. Biochem. Pharmacol. 54, 459–465. doi: 10.1016/S0006-2952(97)00197-4
De Riso, V., Raniello, R., Maumus, F., Rogato, A., Bowler, C., and Falciatore, A. (2009). Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res. 37:e96. doi: 10.1093/nar/gkp448
Delledonne, M., Zeier, J., Marocco, A., and Lamb, C. (2001). Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. 98, 13454–13459. doi: 10.1073/pnas.231178298
Demarest, J. F., Kienle, M., Boytz, R., Ayres, M., Kim, E. J., Patten, J. J., et al. (2022). Brequinar and dipyridamole in combination exhibits synergistic antiviral activity against SARS-CoV-2 in vitro: rationale for a host-acting antiviral treatment strategy for COVID-19. Antivir. Res. 206:105403. doi: 10.1016/j.antiviral.2022.105403
Dow, L. (2021). How do quorum-sensing signals mediate algae–Bacteria interactions? Microorganisms. Microorganisms 9:Article 7. doi: 10.3390/microorganisms9071391
Eberhardt, J., Santos-Martins, D., Tillack, A. F., and Forli, S. (2021). AutoDock Vina 1.2.0: new docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 61, 3891–3898. doi: 10.1021/acs.jcim.1c00203
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Evans, C., Malin, G., Mills, G. P., and Wilson, W. H. (2006). Viral infection of Emiliania Huxleyi (prymnesiophyceae) leads to elevated production of reactive oxygen Species1. J. Phycol. 42, 1040–1047. doi: 10.1111/j.1529-8817.2006.00256.x
Fagan, R. L., Nelson, M. N., Pagano, P. M., and Palfey, B. A. (2006). Mechanism of flavin reduction in class 2 dihydroorotate dehydrogenases. Biochemistry 45, 14926–14932. doi: 10.1021/bi060919g
Fairus, A. K. M., Choudhary, B., Hosahalli, S., Kavitha, N., and Shatrah, O. (2017). Dihydroorotate dehydrogenase (DHODH) inhibitors affect ATP depletion, endogenous ROS and mediate S-phase arrest in breast cancer cells. Biochimie 135, 154–163. doi: 10.1016/j.biochi.2017.02.003
Garavito, M. F., Narvaez-Ortiz, H. Y., Pulido, D. C., Löffler, M., Judelson, H. S., Restrepo, S., et al. (2019). Phytophthora infestans dihydroorotate dehydrogenase is a potential target for chemical control – a comparison with the enzyme from Solanum tuberosum. Front. Microbiol. 10:1479. doi: 10.3389/fmicb.2019.01479
Guillard, R. R. L. (1975). “Culture of phytoplankton for feeding marine invertebrates” in Culture of marine invertebrate animals: proceedings—1st conference on culture of marine invertebrate animals Greenport. eds. W. L. Smith and M. H. Chanley (New York, NY: Springer US), 29–60.
Haque, T. S., Tadesse, S., Marcinkeviciene, J., Rogers, M. J., Sizemore, C., Kopcho, L. M., et al. (2002). Parallel synthesis of potent, Pyrazole-based inhibitors of Helicobacter pylori dihydroorotate dehydrogenase. J. Med. Chem. 45, 4669–4678. doi: 10.1021/jm020112w
Harvey, E. L., Deering, R. W., Rowley, D. C., El Gamal, A., Schorn, M., Moore, B. S., et al. (2016). A bacterial quorum-sensing precursor induces mortality in the marine Coccolithophore, Emiliania huxleyi. Front. Microbiol. 7:59. doi: 10.3389/fmicb.2016.00059
Harvey, E. L., Yang, H., Castiblanco, E., Coolahan, M., Dallmeyer-Drennen, G., Fukuda, N., et al. (2023). Quorum sensing signal disrupts viral infection dynamics in the coccolithophore Emiliania huxleyi. Aquat. Microb. Ecol. 89, 75–86. doi: 10.3354/ame01998
Hassa, P. O., Buerki, C., Lombardi, C., Imhof, R., and Hottiger, M. O. (2003). Transcriptional coactivation of nuclear factor-κB-dependent gene expression by p300 is regulated by poly(ADP)-ribose polymerase-1. J. Biol. Chem. 278, 45145–45153. doi: 10.1074/jbc.M307957200
Heath, M. C. (2000). Hypersensitive response-related death. Plant Mol. Biol. 44, 321–334. doi: 10.1023/a:1026592509060
Hoffmann, H.-H., Kunz, A., Simon, V. A., Palese, P., and Shaw, M. L. (2011). Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc. Natl. Acad. Sci. 108, 5777–5782. doi: 10.1073/pnas.1101143108
Horwitz, S. M., Blue, T. C., Ambarian, J. A., Hoshino, S., Seyedsayamdost, M. R., and Davis, K. M. (2022). Structural insights into inhibition of the drug target dihydroorotate dehydrogenase by bacterial hydroxyalkylquinolines. RSC Chem. Biol. 3, 420–425. doi: 10.1039/D1CB00255D
Hurt, D. E., Sutton, A. E., and Clardy, J. (2006). Brequinar derivatives and species-specific drug design for dihydroorotate dehydrogenase. Bioorg. Med. Chem. Lett. 16, 1610–1615. doi: 10.1016/j.bmcl.2005.12.029
Jagtap, P., and Szabó, C. (2005). Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat. Rev. Drug Discov. 4, 421–440. doi: 10.1038/nrd1718
Jarvis, B., Wilrich, C., and Wilrich, P. -T. (2010). Reconsideration of the derivation of most probable numbers, their standard deviations, confidence bounds and rarity values. J. Appl. Microbiol. 109, 1660–1667. doi: 10.1111/j.1365-2672.2010.04792.x
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kariminik, A., Baseri-Salehi, M., and Kheirkhah, B. (2017). Pseudomonas aeruginosa quorum sensing modulates immune responses: an updated review article. Immunol. Lett. 190, 1–6. doi: 10.1016/j.imlet.2017.07.002
Kim, K., Kim, Y. U., Koh, B. H., Hwang, S. S., Kim, S.-H., Lépine, F., et al. (2010a). HHQ and PQS, two Pseudomonas aeruginosa quorum-sensing molecules, down-regulate the innate immune responses through the nuclear factor-κB pathway. Immunology 129, 578–588. doi: 10.1111/j.1365-2567.2009.03160.x
Kim, K., Kim, S.-H., Lépine, F., Cho, Y.-H., and Lee, G. R. (2010b). Global gene expression analysis on the target genes of PQS and HHQ in J774A.1 monocyte/macrophage cells. Microb. Pathog. 49, 174–180. doi: 10.1016/j.micpath.2010.05.009
Ku, C., Sheyn, U., Sebé-Pedrós, A., Ben-Dor, S., Schatz, D., Tanay, A., et al. (2020). A single-cell view on alga-virus interactions reveals sequential transcriptional programs and infection states. Sci. Adv. 6:eaba4137. doi: 10.1126/sciadv.aba4137
Liu, T., Lin, Y., Wen, X., Jorissen, R. N., and Gilson, M. K. (2007). BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res. 35, D198–D201. doi: 10.1093/nar/gkl999
Lowery, C., Dickerson, T. J., and Janda, K. D. (2008). Interspecies and interkingdom communication mediated by bacterial quorum sensing. Chem. Soc. Rev. 37, 1337–1346. doi: 10.1039/B702781H
Lucas-Hourani, M., Dauzonne, D., Jorda, P., Cousin, G., Lupan, A., Helynck, O., et al. (2013). Inhibition of pyrimidine biosynthesis pathway suppresses viral growth through innate immunity. PLoS Pathog. 9:e1003678. doi: 10.1371/journal.ppat.1003678
Luthra, P., Naidoo, J., Pietzsch, C. A., De, S., Khadka, S., Anantpadma, M., et al. (2018). Inhibiting pyrimidine biosynthesis impairs Ebola virus replication through depletion of nucleoside pools and activation of innate immune responses. Antivir. Res. 158, 288–302. doi: 10.1016/j.antiviral.2018.08.012
Madak, J. T., Cuthbertson, C. R., Miyata, Y., Tamura, S., Petrunak, E. M., Stuckey, J. A., et al. (2018). Design, synthesis, and biological evaluation of 4-Quinoline carboxylic acids as inhibitors of dihydroorotate dehydrogenase. J. Med. Chem. 61, 5162–5186. doi: 10.1021/acs.jmedchem.7b01862
Mayali, X., and Azam, F. (2004). Algicidal bacteria in the sea and their impact on algal Blooms1. J. Eukaryot. Microbiol. 51, 139–144. doi: 10.1111/j.1550-7408.2004.tb00538.x
Milanese, C., Bombardieri, C. R., Sepe, S., Barnhoorn, S., Payán-Goméz, C., Caruso, D., et al. (2019). DNA damage and transcription stress cause ATP-mediated redesign of metabolism and potentiation of anti-oxidant buffering. Nat. Commun. 10:4887. doi: 10.1038/s41467-019-12640-5
Müller, C., Caspers, B. A., Gadau, J., and Kaiser, S. (2020). The power of infochemicals in mediating individualized niches. Trends Ecol. Evol. 35, 981–989. doi: 10.1016/j.tree.2020.07.001
Murphy, D. J. (2004). Determination of accurate KI values for tight-binding enzyme inhibitors: an in silico study of experimental error and assay design. Anal. Biochem. 327, 61–67. doi: 10.1016/j.ab.2003.12.018
Ng, W.-L., and Bassler, B. L. (2009). Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43, 197–222. doi: 10.1146/annurev-genet-102108-134304
Panda, D., Gjinaj, E., Bachu, M., Squire, E., Novatt, H., Ozato, K., et al. (2019). IRF1 maintains optimal constitutive expression of antiviral genes and regulates the early antiviral response. Front. Immunol. 10:1019. doi: 10.3389/fimmu.2019.01019
Paulsen, S. S., Isbrandt, T., Kirkegaard, M., Buijs, Y., Strube, M. L., Sonnenschein, E. C., et al. (2020). Production of the antimicrobial compound tetrabromopyrrole and the Pseudomonas quinolone system precursor, 2-heptyl-4-quinolone, by a novel marine species Pseudoalteromonas galatheae sp. Nov. Sci. Rep. 10:21630. doi: 10.1038/s41598-020-78439-3
Pesci, E. C., Milbank, J. B. J., Pearson, J. P., McKnight, S., Kende, A. S., Greenberg, E. P., et al. (1999). Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. 96, 11229–11234. doi: 10.1073/pnas.96.20.11229
Pettersen, E. F., Goddard, T. D., Huang, C. C., Meng, E. C., Couch, G. S., Croll, T. I., et al. (2021). UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82. doi: 10.1002/pro.3943
Pollara, S. B., Becker, J. W., Nunn, B. L., Boiteau, R., Repeta, D., Mudge, M. C., et al. (2021). Bacterial quorum-sensing signal arrests phytoplankton cell division and impacts virus-induced mortality. MSphere 6, e00009–e00021. doi: 10.1128/mSphere.00009-21
Qing, M., Zou, G., Wang, Q.-Y., Xu, H. Y., Dong, H., Yuan, Z., et al. (2010). Characterization of dengue virus resistance to brequinar in cell culture. Antimicrob. Agents Chemother. 54, 3686–3695. doi: 10.1128/aac.00561-10
Reen, F. J., Mooij, M. J., Holcombe, L. J., McSweeney, C. M., McGlacken, G. P., Morrissey, J. P., et al. (2011). The Pseudomonas quinolone signal (PQS), and its precursor HHQ, modulate interspecies and interkingdom behaviour. FEMS Microbiol. Ecol. 77, 413–428. doi: 10.1111/j.1574-6941.2011.01121.x
Reis, R. A. G., Calil, F. A., Feliciano, P. R., Pinheiro, M. P., and Nonato, M. C. (2017). The dihydroorotate dehydrogenases: past and present. Arch. Biochem. Biophys. 632, 175–191. doi: 10.1016/j.abb.2017.06.019
Rosenwasser, S., Mausz, M. A., Schatz, D., Sheyn, U., Malitsky, S., Aharoni, A., et al. (2014). Rewiring host lipid metabolism by large viruses determines the fate of Emiliania huxleyi, a bloom-forming alga in the ocean. Plant Cell 26, 2689–2707. doi: 10.1105/tpc.114.125641
Rosenwasser, S., Ziv, C., Creveld, S. G.Van, and Vardi, A. (2016). Virocell metabolism: metabolic innovations during host–virus interactions in the ocean. Trends Microbiol., 24, 821–832. doi: 10.1016/j.tim.2016.06.006
Schatz, D., Shemi, A., Rosenwasser, S., Sabanay, H., Wolf, S. G., Ben-Dor, S., et al. (2014). Hijacking of an autophagy-like process is critical for the life cycle of a DNA virus infecting oceanic algal blooms. New Phytol. 204, 854–863. doi: 10.1111/nph.13008
Schieler, B. M., Soni, M. V., Brown, C. M., Coolen, M. J. L., Fredricks, H., Van Mooy, B. A. S., et al. (2019). Nitric oxide production and antioxidant function during viral infection of the coccolithophore Emiliania huxleyi. ISME J. 13, 1019–1031. doi: 10.1038/s41396-018-0325-4
Schmidt, R., and Saha, M. (2021). Infochemicals in terrestrial plants and seaweed holobionts: current and future trends. New Phytol. 229, 1852–1860. doi: 10.1111/nph.16957
Segev, E., Wyche, T. P., Kim, K. H., Petersen, J., Ellebrandt, C., Vlamakis, H., et al. (2016). Dynamic metabolic exchange governs a marine algal-bacterial interaction. elife 5:e17473. doi: 10.7554/eLife.17473
Seyedsayamdost, M. R., Case, R. J., Kolter, R., and Clardy, J. (2011). The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat. Chem. 3, 331–335. doi: 10.1038/nchem.1002
Seymour, J. R., Amin, S. A., Raina, J.-B., and Stocker, R. (2017). Zooming in on the phycosphere: the ecological interface for phytoplankton–bacteria relationships. Nat. Microbiol. 2, 1–12. doi: 10.1038/nmicrobiol.2017.65
Sheyn, U., Rosenwasser, S., Ben-Dor, S., Porat, Z., and Vardi, A. (2016). Modulation of host ROS metabolism is essential for viral infection of a bloom-forming coccolithophore in the ocean. ISME J. 10, 1742–1754. doi: 10.1038/ismej.2015.228
Thumuluri, V., Almagro Armenteros, J. J., Johansen, A. R., Nielsen, H., and Winther, O. (2022). DeepLoc 2.0: multi-label subcellular localization prediction using protein language models. Nucleic Acids Res. 50, W228–W234. doi: 10.1093/nar/gkac278
Trott, O., and Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 31, 455–461. doi: 10.1002/jcc.21334
Ullrich, A., Knecht, W., Fries, M., and Löffler, M. (2001). Recombinant expression of N-terminal truncated mutants of the membrane bound mouse, rat and human flavoenzyme dihydroorotate dehydrogenase. A versatile tool to rate inhibitor effects? Eur. J. Biochem. 268, 1861–1868. doi: 10.1046/j.1432-1327.2001.02061.x
Urvoy, M., Labry, C., L’Helguen, S., and Lami, R. (2022). Quorum sensing regulates bacterial processes that play a major role in marine biogeochemical cycles. Front. Mar. Sci. 9:834337. doi: 10.3389/fmars.2022.834337
Vincent, F., Sheyn, U., Porat, Z., Schatz, D., and Vardi, A. (2021). Visualizing active viral infection reveals diverse cell fates in synchronized algal bloom demise. Proc. Natl. Acad. Sci. 118:e2021586118. doi: 10.1073/pnas.2021586118
Wang, Q.-Y., Bushell, S., Qing, M., Xu, H. Y., Bonavia, A., Nunes, S., et al. (2011). Inhibition of dengue virus through suppression of host pyrimidine biosynthesis. J. Virol. 85, 6548–6556. doi: 10.1128/jvi.02510-10
Whalen, K. E., Becker, J. W., Schrecengost, A. M., Gao, Y., Giannetti, N., and Harvey, E. L. (2019). Bacterial alkylquinolone signaling contributes to structuring microbial communities in the ocean. Microbiome 7:93. doi: 10.1186/s40168-019-0711-9
Wietz, M., Gram, L., Jørgensen, B., and Schramm, A. (2010). Latitudinal patterns in the abundance of major marine bacterioplankton groups. Aquat. Microb. Ecol. 61, 179–189. doi: 10.3354/ame01443
Wu, Y., and Seyedsayamdost, M. R. (2017). Synergy and target promiscuity drive structural divergence in bacterial Alkylquinolone biosynthesis. Cell Chem. Biol. 24, 1437–1444.e3. doi: 10.1016/j.chembiol.2017.08.024
Yoshino, M., and Murakami, K. (2009). A graphical method for determining inhibition constants. J. Enzyme Inhib. Med. Chem. 24, 1288–1290. doi: 10.3109/14756360902829766
Yoshioka, H., Mase, K., Yoshioka, M., Kobayashi, M., and Asai, S. (2011). Regulatory mechanisms of nitric oxide and reactive oxygen species generation and their role in plant immunity. Nitric Oxide 25, 216–221. doi: 10.1016/j.niox.2010.12.008
Yu, S.-W., Wang, H., Poitras, M. F., Coombs, C., Bowers, W. J., Federoff, H. J., et al. (2002). Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science 297, 259–263. doi: 10.1126/science.1072221
Zameitat, E., Freymark, G., Dietz, C. D., Löffler, M., and Bölker, M. (2007). Functional expression of human dihydroorotate dehydrogenase (DHODH) in pyr4 mutants of ustilago maydis allows target validation of DHODH inhibitors in vivo. Appl. Environ. Microbiol. 73, 3371–3379. doi: 10.1128/AEM.02569-06
Keywords: HHQ, Pseudoalteromonas, dihydroorotate dehydrogenase, phytoplankton, quorum sensing, virus-host interactions
Citation: Garrett O and Whalen KE (2023) A bacterial quorum sensing signal is a potent inhibitor of de novo pyrimidine biosynthesis in the globally abundant Emiliania huxleyi. Front. Microbiol. 14:1266972. doi: 10.3389/fmicb.2023.1266972
Received: 25 July 2023; Accepted: 08 September 2023;
Published: 06 October 2023.
Edited by:
Zhaojiang Zuo, Zhejiang Agriculture and Forestry University, ChinaReviewed by:
Zhang Huiying, Fujian Agriculture and Forestry University, ChinaCopyright © 2023 Garrett and Whalen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kristen E. Whalen, a3doYWxlbjFAaGF2ZXJmb3JkLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.