
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 12 October 2023
Sec. Extreme Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1265216
This article is part of the Research Topic 13th International Congress on Extremophiles: From Extremophilic Biomolecules and Microorganisms to Biotechnological and Sustainable Applications View all 11 articles
Thermophily is an ancient trait among microorganisms. The molecular principles to sustain high temperatures, however, are often described as adaptations, somewhat implying that they evolved from a non-thermophilic background and that thermophiles, i.e., organisms with growth temperature optima (TOPT) above 45°C, evolved from mesophilic organisms (TOPT 25–45°C). On the contrary, it has also been argued that LUCA, the last universal common ancestor of Bacteria and Archaea, may have been a thermophile, and mesophily is the derived trait. In this study, we took an experimental approach toward the evolution of a mesophile from a thermophile. We selected the acetogenic bacterium T. kivui (TOPT 66°C) since acetogenesis is considered ancient physiology and cultivated it at suboptimal low temperatures. We found that the lowest possible growth temperature (TMIN) under the chosen conditions was 39°C. The bacterium was subsequently subjected to adaptive laboratory evolution (ALE) by serial transfer at 45°C. Interestingly, after 67 transfers (approximately 180 generations), the adapted strain Adpt45_67 did not grow better at 45°C, but a shift in the TOPT to 60°C was observed. Growth at 45°C was accompanied by a change in the morphology as shorter, thicker cells were observed that partially occurred in chains. While the proportion of short-chain fatty acids increased at 50°C vs. 66°C in both strains, Adpt45_67 also showed a significantly increased proportion of plasmalogens. The genome analysis revealed 67 SNPs compared to the type strain, among these mutations in transcriptional regulators and in the cAMP binding protein. Ultimately, the molecular basis of the adaptation of T. kivui to a lower TOPT remains to be elucidated. The observed change in phenotype is the first experimental step toward the evolution of thermophiles growing at colder temperatures and toward a better understanding of the cold adaptation of thermophiles on early Earth.
Temperature is a key environmental factor that determines the biodiversity and species composition of habitats as it requires specific traits to allow organisms to thrive at distinct temperatures. The temperature range of habitats on Earth is huge, and living cells have been found from the polar environments and high-altitude regions (De Maayer et al., 2014) to terrestrial mud springs and submarine hot vents (Stetter, 2006). The growth temperature profile of every species is characterized by the cardinal temperatures, the minimal (TMIN), the maximal (TMAX), and the optimal temperature (TOPT) that allow growth (Wiegel, 1990). Organisms adapted to extremely cold or hot temperatures, or temperature extremophiles, are classified by their TOPT. In contrast to mesophiles growing at more or less ambient temperatures (TOPT 20–45°C), psychrophiles prefer a TOPT lower than 15°C (De Maayer et al., 2014). Thermophiles thrive optimally at temperatures higher than 45°C, while organisms with a TOPT >65–70°C are called extreme thermophiles, and hyperthermophiles possess a TOPT of >80°C (Wiegel, 1990; Zeldes et al., 2015). The temperature range of individual thermophiles may be very broad (Wiegel, 1990). For example, the hyperthermophilic Archaeon Pyrococcus furiosus grows optimally at 100°C, with a TMAX of 103°C and a TMIN of 65°C (38 K; Fiala and Stetter, 1986), and the methanogen Methanothermobacter thermautotrophicus even has a span of 55 K (Wiegel, 1990).
The specific adaptations of thermophiles have been extensively studied and include the modification and protection of all types of cellular macromolecules. For example, DNA in (hyper)thermophiles is protected by (positively charged) polyamines and by histones in Archaea (Imanaka, 2011). Common features seen in proteins of thermophiles are increased van der Waals interactions, hydrogen bonds, ionic interactions to stabilize the secondary or tertiary structure of the protein, a more hydrophobic interior, and an increased packing density of the protein (Berezovsky and Shakhnovich, 2005; Imanaka, 2011). The folding of proteins in thermophiles is supported by the action of particular chaperones, and proteins and DNA are stabilized within the cell by compatible solutes (Imanaka, 2011). Mechanisms to stabilize the cytoplasmic membrane include ether-linked membrane lipids in archaea, while the degree of saturation and the amount of branched-chain and longer fatty acids increase with temperature in bacteria (Siliakus et al., 2017). Despite these and many other specific traits attributed to life at high temperatures, a comprehensive understanding of “thermophily” is still lacking (Canganella and Wiegel, 2014), particularly toward its evolution. On the one hand, the aforementioned molecular traits to thrive at high temperatures may be derived, having evolved from a mesophile background. On the other hand, these traits may be ancient, and mesophiles may have evolved from ancient thermophiles. For example, life may have emerged at moderately warm hydrothermal and alkaline vent fields such as Lost City (40–90°C; Kelley et al., 2005). This environment is cold enough to allow first (bio)molecular reactions but rich in trace elements and in H2 and CO2. These gaseous substrates may have supported the formation of biomass precursors coupled with energy conservation, as found in recent acetogenic bacteria and methanogenic archaea (Martin, 2020). Interestingly, to the best of our knowledge, the hypothesis that mesophiles are derived from thermophiles has never been tested in laboratory evolution experiments.
In this study, we aimed to evolve the bacterium Thermoanaerobacter kivui as acetogen with a TOPT of 66°C is capable of growing in environments comparable to Lost City, and where the first life may have emerged, toward growth at lower temperatures. T. kivui has been described as an acetogen growing on a variety of substrates, including H2+CO2 but also on the sugars glucose, mannose, and fructose (Leigh et al., 1981). The physiology of T. kivui has recently been studied to a larger extent. For example, it has been shown to grow on the sugar alcohol mannitol (Moon et al., 2019), in bioelectrical systems on anodes (Deutzmann et al., 2022), and it has been subjected to ALE to grow on carbon monoxide (Weghoff and Müller, 2016). Genetic tools for genome modification (Basen et al., 2018) and plasmid-based protein production (Katsyv et al., 2021) were developed, allowing to study one of its energy-conserving hydrogenases (Katsyv and Müller, 2022), and the soluble hydrogen-dependent carbon dioxide reductase (Jain et al., 2020; Dietrich et al., 2022). This prompted us to pick T. kivui as a model thermophilic acetogen for ALE experiments. We grew five populations of T. kivui DSM2030 at 45°C (21 K below TOPT) and used the fastest growing population as inoculum for the next five populations cultivated at 45°C (Figure 1). After 67 serial transfers, corresponding to ~180 generations, we studied the phenotype and the genotype of the evolved strain, Adpt45_67.
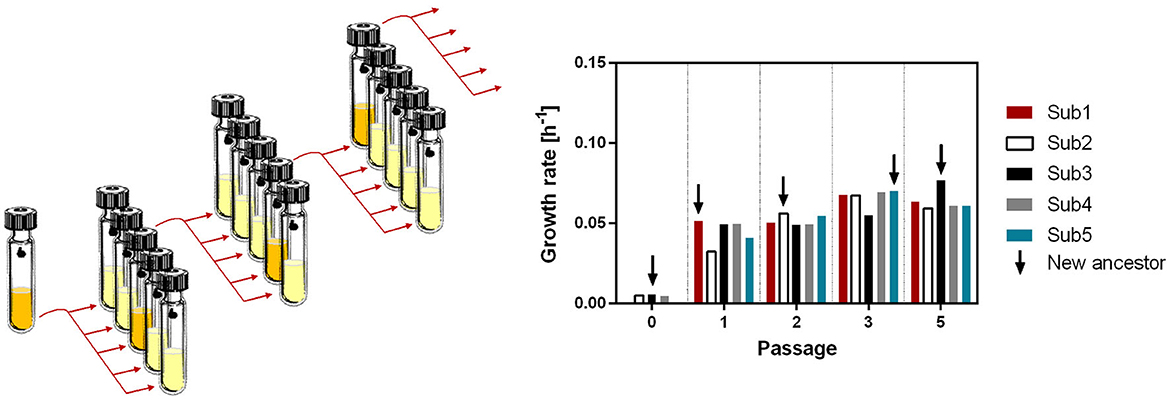
Figure 1. Serial passaging modified after Strauss et al. (2019). Five subpopulations of T. kivui were incubated for 24 h in a complex medium containing 25 mM glucose as a carbon source. The subpopulation with the highest increase in OD600 was used to generate five new subpopulations in a fresh medium.
Thermoanaerobacter kivui strain LKT-1 (DSM2030), referred to as the type strain, and the adapted strain Adpt45_67 were cultivated under strict anoxic conditions at different temperatures between 40°C and 66°C in complex or defined media as described previously (Weghoff and Müller, 2016; Basen et al., 2018). Complex media contained Na2HPO4 × 2 H2O, 50 mM; NaH2PO4 × 2 H2O, 50 mM; K2HPO4, 1.2 mM; KH2PO4, 1.2 mM; NH4Cl, 4.7 mM; (NH4)2SO4, 1.7 mM; NaCl, 7.5 mM; MgSO4 × 7 H2O, 0.37 mM; CaCl2 × 2 H2O, 42 μM; Fe(II)SO4 × 7 H2O, 7.2 μM; KHCO3, 54 mM; cysteine-HCl × H2O, 3 mM; resazurin, 4.4 μM; 0.2% (w/v) yeast extract, 10 ml/l of trace element solution DSM141, and 10 ml/l of vitamin solution DSM141. Defined media were prepared similarly as complex media without the addition of yeast extract. The medium was flushed with N2:CO2 (80:20 [v:v], 1.1 x 105 Pa) before autoclaving. The pH of the medium was 7.5 after flushing. The agar medium was supplemented with 1.5% Bacto agar (BD Difco, BD Life Sciences, Heidelberg, Germany). All gases were purchased from Westfalen AG (Münster, Germany).
Growth experiments were carried out in 20 ml Hungate glass tubes with 5 ml of medium or in 100-ml or 200-ml serum bottles with 50 ml or 100 ml of medium, respectively. The glass tubes or serum bottles were sealed with butyl rubber stoppers under an atmosphere of N2:CO2 (80:20 [v:v], 1.1 x 105 Pa) unless denoted otherwise (Basen et al., 2018). Glucose was added as a carbon source from a sterile anoxic stock solution to a final concentration of 25 mM. If H2 + CO2 were used as substrates, tubes were only filled with medium to one-fourth of the volume, and the remaining headspace was replaced with H2:CO2 (80:20 [v:v], 2 x 105 Pa). For growth experiments of T. kivui at different temperatures, initial cultures were grown at 66°C to an OD600 of 1 and diluted 1/10 in fresh medium, followed by incubation at the desired temperature. Plating and cultivation of solid media were carried out according to Basen et al. (2018). The agar dilution series and the isolation of single colonies from it were performed according to Widdel and Bak (1992).
Growth in the liquid medium was monitored by measuring the optical density at 600 nm (OD600) with a spectrophotometer. Cell morphology was documented using a Nikon Eclipse Ni-U microscope equipped with a Nikon DSFi3 camera (Nikon, Tokyo, Japan). Glucose and organic acid concentrations were determined by HPLC as described previously (Zeldes et al., 2023).
The ALE approach by serial passaging was performed as described by Wein and Dagan in Strauss et al. (2019). In brief, T. kivui DSM2030 was grown on agar plates at 66°C. Three colonies were randomly sampled as the ancestral clones. The serial passage experiment was initiated by inoculating the three clones in Hungate tubes containing 2 ml of complex medium and incubating at 45°C. Subsequently, the ancestral populations were sampled into five subpopulations of each replicate. The five subpopulations were grown at 45°C, and growth was monitored as described. The subpopulations of each replicate having the highest increase in OD600 after 24–28 h were selected as the ancestors for the next subpopulations.
The genomes of adapted strains were analyzed for single-nucleotide polymorphisms as recently described (Zeldes et al., 2023).
T. kivui cultures were grown as described to an OD600 of approximately 1. Cultures were harvested for 5 min at 5,000 × g and 4°C. Cell pellets were suspended in 0.7% (w/v) aqueous MgSO4 and centrifuged again. Cell pellets were freeze-dried, and ~40 mg of cell biomass (dry weight) was sent for fatty acid (FA) analysis at the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) in Braunschweig, Germany.
Datasets were analyzed by two-way ANOVA with Tukey's test using the software Graph Pad Prism Version 6.01 (GraphPad Software, Boston, USA).
T. kivui was originally described to thrive at 50–72°C (Leigh et al., 1981). Compared to other Thermoanaerobacter species, this is a relatively narrow range (Onyenwoke and Wiegel, 2015), since, e.g., T. pseudethanolicus grows between 30°C and 80°C (Figure 2). The TOPT of T. kivui (66°C) compares favorably with that of other Thermoanaerobacter species (55°C−75°C); however, its reported TMIN of 50°C is among the highest, with many species able to grow at significantly lower temperatures between 30°C and 40°C. Wiegel also reported that they were able to cultivate T. kivui at 35°C (1990); however, growth parameters and culture conditions were not specified in the articles. These hints that the published TMIN may be too high prompted us to test the growth of T. kivui at suboptimal temperatures. Toward that, we selected a complex medium with a reduced yeast extract content (Basen et al., 2018), and glucose as a substrate over H2+CO2, since the growth rates and final optical densities in a defined medium and under autotrophic conditions are lower (Jain et al., 2020; Moon et al., 2020). Growth experiments within a temperature range of 40–66°C were performed (Figure 3A). Consistent with the literature, 66°C is the TOPT for the type strain T. kivui DSM2030 (Leigh et al., 1981), at a specific growth rate of 0.45 h−1 (Figure 3 and Supplementary Table S1). Lower temperatures resulted in decreased growth rates, as expected. The growth of T. kivui was monitored at temperatures below 50°C, and slow growth (0.033 h−1) was observed at 40°C. At temperatures below 39°C, T. kivui did not grow in our hands, in contrast to the report of Wiegel (1990); however, as described above, the author may have used different cultivation conditions. Corresponding to lower growth rates, the maximal OD600 as an indicator of the biomass yield was reached after longer incubation periods. For example, an OD600 >1 was reached after 7 h at 66°C and 60°C, after 12 h at 55°C, and after 23 h at 45°C. At a growth temperature of 40°C, an OD600 of 1 was not reached even after incubation for 108 h (Figure 3A).
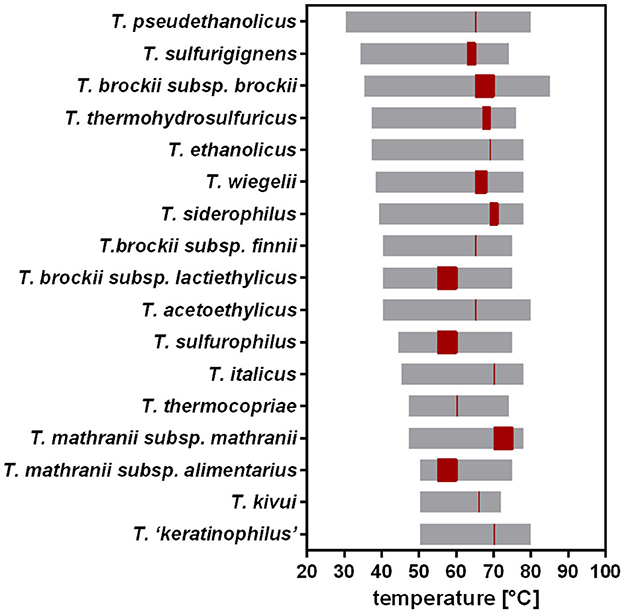
Figure 2. Previously reported temperature ranges and optimal growth temperatures for different Thermoanaerobacter species. Temperature range, gray; TOPT, red. Data from Onyenwoke and Wiegel (2015).
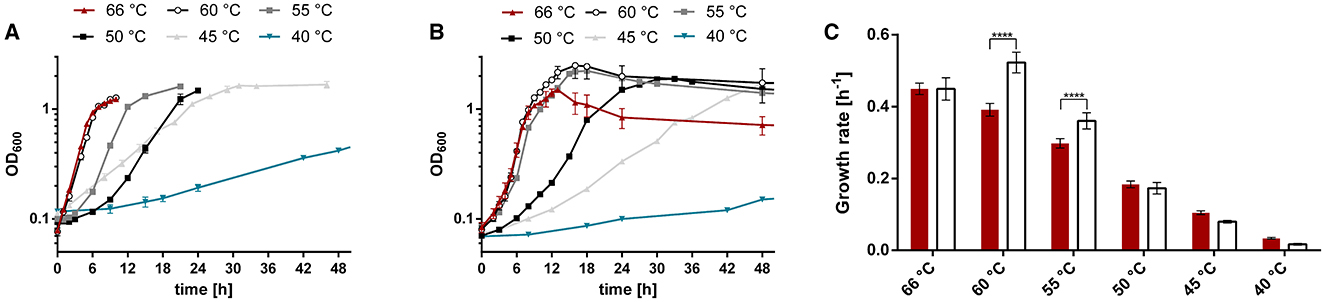
Figure 3. Growth of T. kivui strains (A) DSM2030 (type strain) and (B) Adpt45_67 at different temperatures, and (C) the corresponding growth rates. Cells were grown in 50 ml complex medium in 100 ml serum bottles with 25 mM glucose as a carbon source. Red triangles, 66°C; black open circles, 66°C; gray squares, 55°C; black squares, 50°C; gray triangles, 45°C; turquoise inverted triangles, 40°C. The error indicated shows the standard deviation from four biological replicates. Significant statistical differences (***p < 0.0005, two-way ANOVA with Tukey's test) between the type strain and the adapted strains at each temperature condition are shown.
We decided to use 45°C as the temperature for serial passaging of cultures for ALE, since it seems to provide a good balance of strong selective pressure (only a few degrees above the lowest temperature at which growth is observed) and fast enough growth to achieve a high number of generations through daily passaging. In ALE, organisms are observed under controlled laboratory conditions and specific growth conditions for extended periods of time, allowing the isolation of adapted phenotypes. Many different strategies for ALE have been developed and further improved (Dragosits and Mattanovich, 2013; Strauss et al., 2019), from simple long-term passaging experiments with Escherichia coli over several thousand generations (Cooper and Lenski, 2000), to ALE using increased mutation rate and targeted genetic engineering (Luan et al., 2013; Badran and Liu, 2015; Suzuki et al., 2015; Kang et al., 2019). In this study, a variant of the serial passaging strategy (Strauss et al., 2019) was selected, in which five populations were evolved in parallel to increase the number of genotypes tested (Figure 1). Targeted selection was performed continuously on the fastest growing population, defined by the highest increase in OD600 within a growth period of 24–28h. Within that time frame, T. kivui was still in the exponential growth phase, excluding a more complex adaptation to the stationary growth phase (Vasi and Lenski, 1999). After ~180 generations (passage 67—Supplementary Figure S3), the T. kivui population with the highest biomass increase (adapted strain Adpt45_67) was again selected, and a detailed study of the phenotype was performed.
We compared the growth of the type strain (DSM2030; acclimated, three transfers at 45°C) and the adapted strain Adpt45_67 (~180 generations at 45°C) during growth on the “adaptation” medium, complex medium plus 25 mM glucose, at temperatures between 40°C and 66°C (Figure 3B). We found that under these conditions, Adpt45_67 grows as fast as the type strain at 66°C (0.45 h−1 vs. 0.45 h−1, Figure 3C, Supplementary Table S1). At 45°C, the temperature chosen for adaptation, Adpt45_67, unexpectedly does not grow faster than the type strain (0.08 h−1 vs. 0.104 h−1). At 40°C, the lowest temperature tested, growth rates were reduced to below 10% of the rates at 66°C (0.017 h−1 and 0.033 h−1), demonstrating the challenge of ALE at lower temperatures. Interestingly, the only significant changes were observed at 60°C and 55°C. At both temperatures, Adpt45_67 grew significantly faster than the type strain (0.523 h−1 vs. 0.391 h−1 at 60°C and 0.360 h−1 vs. 0.297 h−1 at 55°C), leading to a shift in the TOPT from 66°C to 60°C (see also Supplementary Figure S1).
We then tested whether the effect is medium-dependent, since adaptations to medium types and compounds have been reported from the Lenski lab (Cooper and Lenski, 2000). We grew the type strain and Adpt45_67 at 66°C and 60°C in a defined medium, which is identical to the complex medium except for the omission of yeast extract (2 g l−1). Since T. kivui is an acetogen, we initially confirmed that Adpt45_67 still grows on H2+CO2, forming acetate. Since all ALE experiments and growth rate comparisons (Figures 1, 3) were performed under heterotrophic growth conditions, we again used glucose as a substrate for the comparison of the growth phenotypes. We observed that Adpt45_67 has a significantly lower growth rate than the type strain (Figure 4) at either temperature without yeast extract. This phenotype in the non-adaptive environment was somewhat surprising, since in E. coli this has not been observed in short-term evolution experiments (~800 generations) (Kang et al., 2019), but fitness tradeoffs were rather observed in long-term ALE (Leiby and Marx, 2014). However, the observed decreased fitness in the non-adaptive condition (defined medium) described here may be the result of a single or few mutations rather than accumulated mutations, or of a single mutation that favors growth at low temperatures but not in the defined medium. Adpt45_67 still grew without yeast extract and maintained the ability to synthesize amino acids and putative other growth factors. Second, Adpt45_67 grew better at 60°C (0.176 h−1) than at 66°C (0.134 h−1), confirming the observed shift in TOPT from 66°C to 60°C.
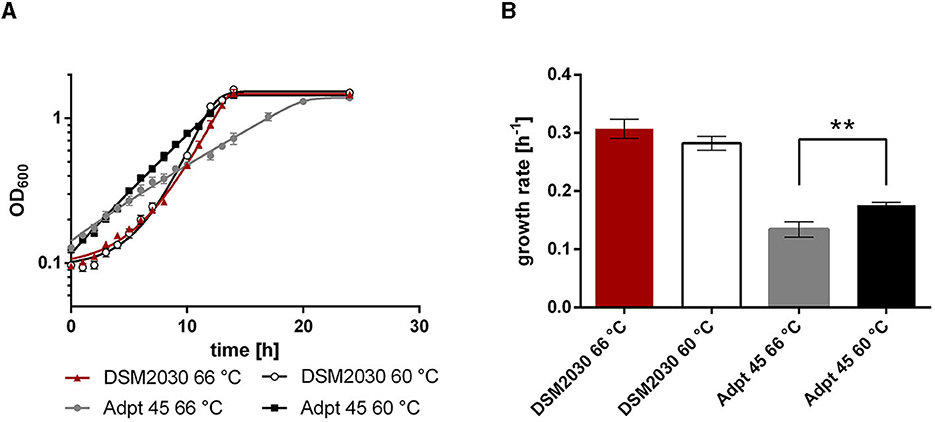
Figure 4. Growth of T. kivui strains (A) DSM2030 (type strain) and Adpt45_67 in the defined medium at 66°C and 60°C, and (B) the corresponding growth rates. Red, type strain at 66°C; white, Type strain at 60°C; gray, Adpt45_67 at 66°C; black, Adpt45_67 at 60°C. Cells were grown in 50 ml of medium in 100 ml serum bottles with 25 mM glucose as a carbon source. The error indicated shows the standard deviation from four biological replicates. Significant statistical differences (**p < 0.005, two-way ANOVA with Tukey's test) between the type strain and the adapted strains at each temperature condition are shown.
The type strain of T. kivui DSM2030 is characterized by long slender rod-shaped cells during growth on glucose of a size of ~0.7 μm in width and up to 7.5 μm in length (Leigh et al., 1981), which we similarly observed after growth at 66°C for 20 h in the type strain (Figure 5A). Growth of the type strain and the Adpt45_67 at 45°C resulted in an increase in cell diameter and a shortening of cell length to 2–3 μm, as well as a striking number of long cell chains (Figures 5B, C). Interestingly, this morphology appeared to be reversible, since the incubation of Adpt45_67 at 66°C for 20 h resulted in a type strain such as morphology (Figure 5D).
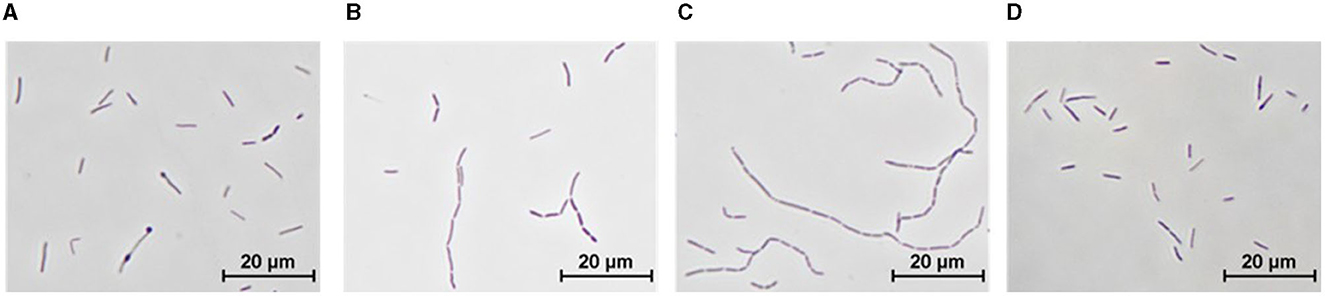
Figure 5. Morphology of T. kivui strains as observed by phase-contrast microscopy. T. kivui strains were grown in 50 ml of complex medium in 100 ml serum bottles with 25 mM glucose as a carbon source. (A) DSM2030 (type strain), 20 h at 66°C (TOPT), (B) DSM2030 after five transfers at 45°C, 24 h at 45°C, (C) Adpt45_67, 24 h at 45°C, and (D) Adpt45_67, 24 h at 60°C (TOPT).
These observations indicated, not unexpectedly, that the cells restructured their outer cell envelope in response to a temperature much below TOPT. We next determined the fatty acid composition of the cytoplasmic membrane of T. kivui DSM2030 and Adpt45_67 at 66°C and 50°C (Figure 6 and Supplementary Figure S2). In the type strain, the major fatty acids with the largest fractions were i-C15:0, i-C17:0, and C16:0, with branched fatty acids representing ~93% of total fatty acids. At the first glance, the membrane fatty acid composition of Adpt45_67 appears to be extremely similar; it also contains high amounts of the same fatty acids. A closer look revealed a much higher fraction of dimethyl acetals, indicative of plasmalogens (~37% vs. 17% in the type strain at 66°C, Figure 6B), including the plasmalogens i-C16:0 P (at 66°C) and a-C15:0 P (at 50°C) that were only detected in Adpt45_67. A surprising observation was the absence of unsaturated fatty acids in the fatty acid content of both strains, except for the type strain at 60°C, where a small amount of C(18:1) w7c was detected. In general, the fatty acid composition varies between bacterial species and can even be strain-specific, with nutrient availability and growth phase also showing an influence (Suutari and Laakso, 1992; Siliakus et al., 2017). Accordingly, bacteria have a number of options to adapt their fatty acid composition and thus membrane fluidity to environmental conditions. For the genus Bacillus and Clostridia, it is known that the major fatty acids are branched chained and that the regulation of fatty acid composition is mainly driven by the type of branching and chain length (Chan et al., 1971; Oshima and Miyagawa, 1974; Sikorski et al., 2008). When comparing thermophilic with mesophilic representatives, the utilization of iso-branched FAs in favor of anteiso-branched FAs was shown to decrease with decreasing temperature (Suutari and Laakso, 1992; Sikorski et al., 2008; Siliakus et al., 2017). This change in ratio was also observed between the type strain and Adpt45_67 as well as the reduction in chain length. Growth at a suboptimal temperature of 50°C resulted in a general shortening of fatty acids in the profiles of the strains studied, as expected, with an increase in C11 to C15 fatty acids and a decrease in C16 and C17 fatty acids. Notable among these were a sharp decrease in the plasmalogen i-C17:0 P (from 11.4 to 3.5%) and a significant increase in i-C15:0 (from 42.9 to 58.8%) in the type strain. Since their overall concentration decreased at 50°C in the type strain, plasmalogens likely do not represent a short-term response to growth at suboptimal temperatures in T. kivui. Plasmalogens are typical components of the cell membrane of strictly anaerobic bacteria (Goldfine, 2010; Jackson et al., 2021). They are characterized by the presence of a vinyl ether linkage at the sn-1 position and an ester linkage at the sn-2 position. Plasmalogens have a lower transition temperature than their diacyl counterparts, which is why they are considered to play a role in shaping the biophysical properties of cellular membranes (Goldfine, 2010; Koivuniemi, 2017; Vítová et al., 2021). Thus, the increased use of plasmalogens could be a way for T. kivui to fine-tune membrane fluidity at lower temperatures. In this study, Adpt45_67 shows a significantly higher concentration of plasmalogens than the type strain. It should be noted, however, that the plasmalogens are also subject to the aforementioned reduction in chain length with decreasing temperature.
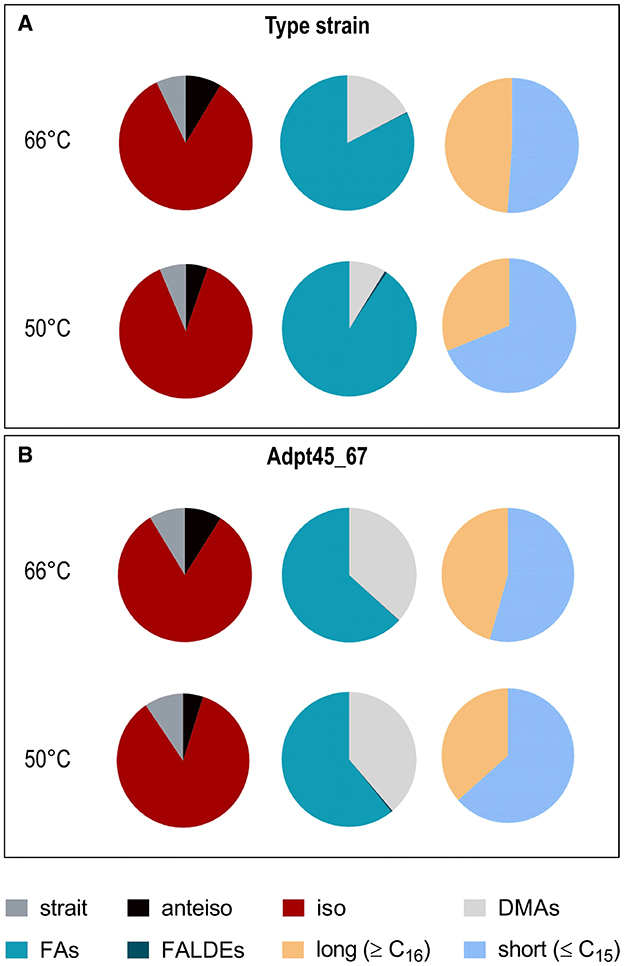
Figure 6. Effect of growth temperature on the total fatty acid composition of T. kivui. T. kivui strains DSM2030 (A) and Adpt45_67 (B) were grown in a complex medium with 25 mM glucose at optimal (66°C and 60°C) and suboptimal (50°C) temperatures to an OD600 of ~1. Left panels: red, iso-branched; gray, anteiso-unbranched; black, anteiso-branched. Middle panels: turquoise, fatty acids; light gray, plasmalogens; dark blue, fatty aldehydes. Right panels: yellow, long-chain >C16; light blue, short-chain <C16.
In order to elucidate the genomic changes responsible for the phenotype observed, the genome of T. kivui Adpt45_67 was sequenced and compared to that of the type strain (Hess et al., 2014). Altogether, 67 single-nucleotide polymorphisms were observed across the 2.397 Mbp genome (Supplementary Table S2). Thirty-three of the SNPs were also in genomes of strains adapted to other environmental stresses in our laboratory (unpublished data). Among these was a mutation (P216L; CCA → CTA) in fabG, a gene essential for fatty acid metabolism that encodes the 3-oxoacyl-[acyl-carrier-protein] reductase, which may have contributed to changes in fatty acid composition. Among the 34 remaining unique mutations, nine were found to be silent. Another 14 were attributed to genes with hypothetical or putative functions, leaving 11 unique, non-silent SNPs in genes or intergenic regions affecting gene expression with annotated functions (Table 1). Four of the SNPs were only found in part of the analyzed DNA, meaning that the majority of the population still contained the original base. This is likely as Adpt45_67 may be considered an adapted population, since it was not re-isolated from solid media, and therefore, the partial SNPs may be interpreted as transient toward a cold-adapted phenotype, and it will be studied whether they carry through in later generations of the ongoing ALE of T. kivui Adp45 at 45°C. Among those SNPs are two that affect the same transposase, TKV_c7000, indicating that this transposase may be involved in temperature response. Moreover, 5.7% of Adpt45_67 led to the mutation G28V in the alternative sigma factor H. This is of interest, since SigH has been described to be involved in several bacteria, including the Gram-positive Corynebacterium glutamicum, where it is involved in heat shock and oxygen stress responses (Kim et al., 2005), though the deletion of sigH in Synechococcus PCC 7002 does not affect growth at a suboptimal temperature (Inoue-Sakamoto et al., 2007). In Staphylococcus aureus, the gene is involved in competence for DNA uptake (Morikawa et al., 2012). Seven SNPs were found in 100% of the reads. Two of them are located in the intragenic regions and may affect the expression level of the genes encoding an ATPase and an alanine racemase, the latter of which is involved in cell wall biogenesis. Two more are located in transcriptional regulators that have relatively low expression levels at 66°C (unpublished data), but may be of importance in cold adaptation. Since T. kivui easily takes up and integrates environmental DNA at 66°C (Basen et al., 2018; Zeldes et al., 2023), the SNP in Cas6-1, a CRISPR RNA maturation endonuclease (Jesser et al., 2019), may modulate the ability to acquire foreign DNA as a strategy for the adaptation of the environment. Another SNP was detected in the gene encoding the potassium ATPase kdpC N146K (AAT → AAG). It is tempting to speculate that potassium has an influence on cold adaptation in T. kivui since, recently, the respective potassium uptake system has been shown to be involved in the cold adaptation of the mesophilic oligotrophic bacterium Caulobacter crescentus (de Araujo et al., 2021). Finally, an SNP R136T (AGG → ACG) was found in the gene of the cAMP-binding protein, Crp (TKV_c24530). Crp is a global regulator (Ma et al., 2004) that, on the one hand, may be involved in temperature response, providing Adpt45_67 a fitness advantage at lower temperatures, and that is linked to sugar uptake via PTS systems, which, on the other hand, may explain the observed weaker growth of Adpt45_67 in a defined medium.
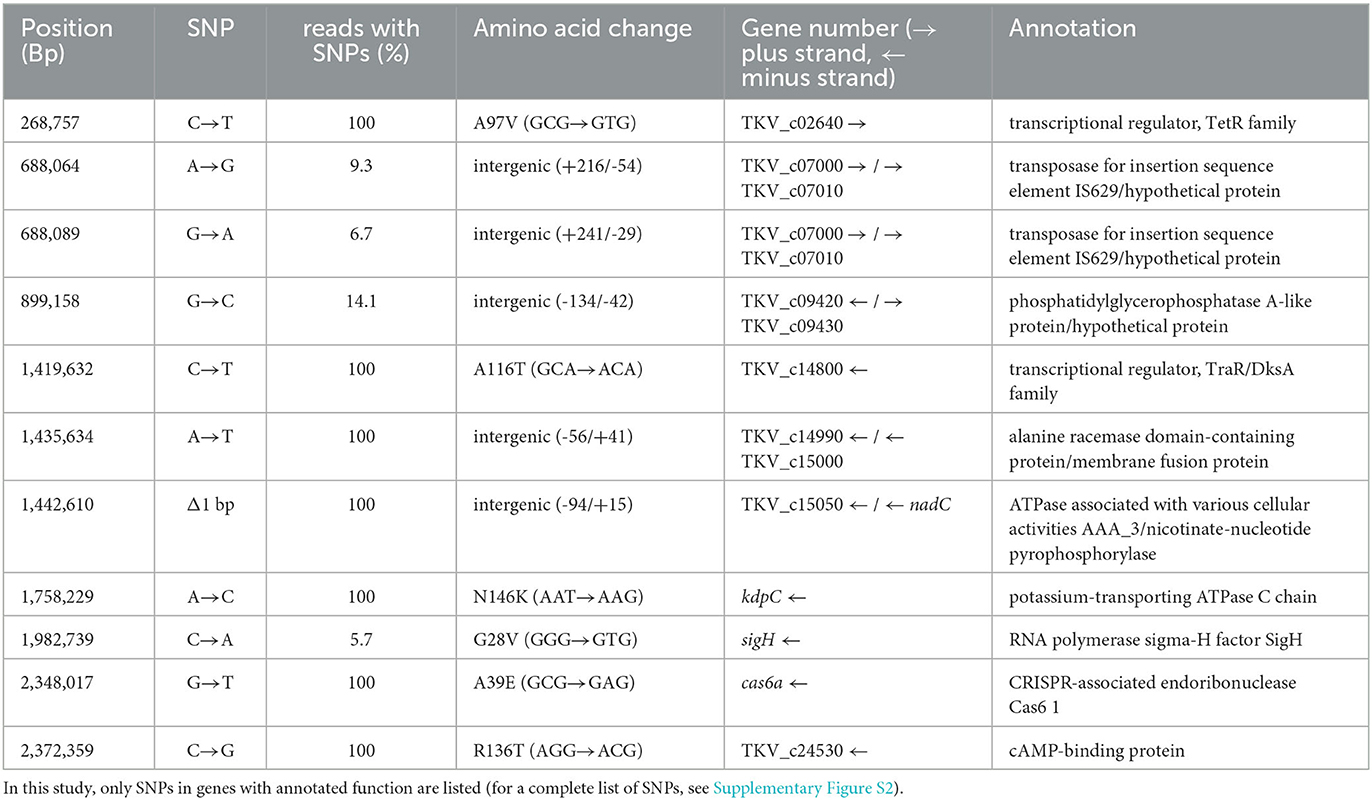
Table 1. Non-silent or intergenic single-nucleotide polymorphisms (SNPs) unique to the genome of T. kivui Adpt45_67 (i.e., not yet observed in other T. kivui adapted strains).
In conclusion, the genotype of T. kivui changed in response to prolonged incubation at 45°C in complex media at several loci that have been attributed to regulation or temperature adaptation in bacteria.
There is an ongoing controversy about whether the first cells were (hyper)thermophilic or mesophilic. On the one hand, and from a human perspective, hot temperatures are extreme, and it seems straightforward to think that thermophiles must have evolved from mesophiles; on the other hand, ~4 billion years ago, when life arose, the Earth or at least parts of it such as the Hadean Ocean were still relatively warm (Lunine, 2006). One of the pro-arguments for a hot-start, however, is the existence of a reverse gyrase in both domains of life, Bacteria, and Archaea, which has been argued based on its phylogeny (Catchpole and Forterre, 2019), and beyond that, it might be acquired through horizontal gene transfer by either domain. It has also been suggested that the progenitor may have been a moderate thermophile that evolved at a submarine alkaline, moderately hot vent such as the Lost City hydrothermal vent field (Russell et al., 2010; Sousa et al., 2013). Moreover, the progenitor, or LUCA, the last universal ancestor of Archaea and Bacteria, was proposed to share features of recent acetogenic bacteria and methanogenic archaea (Martin and Sousa, 2016). Both acetogenesis and methanogenesis are dependent on energy conservation from the conversion of H2+CO2. Genes toward H2+CO2 utilization, the respective essential cofactor biosynthesis pathways and the terminal electron accepting pathway of acetogens and CO2 fixation pathway in methanogens, the Wood–Ljungdahl pathway (WLP; Ljungdahl, 1986; Wood et al., 1986), were suggested to be present in the genome of LUCA (Weiss et al., 2016). Thus, acetogens are likely ancient (Basen and Müller, 2017; Martin, 2020). It may ultimately be unresolvable whether the progenitor was a thermophile or a mesophile. Undisputable, however, thermophiles are ancient, with many archaeal and bacterial phyla at the root of phylogenetic trees (Pace, 1997; Stetter, 2006), and it is likely that at least in some cases mesophilic microorganisms evolved from thermophilic ancestors, as reported for the ancient bacterial phylum Thermotogales (Zhaxybayeva et al., 2009) and for mesophilic Methanococcus species (Lecocq et al., 2021) based on phylogenetic analyses. It still remains intriguing how this evolution happened since it required a stepwise molecular adaptation based on modifications to the genome.
While the aforementioned studies are based on comparative and phylogenetic analyses of genomes, the ALE of a thermophile toward a lower TOPT has never been tested experimentally, to the best of our knowledge. This triggered us to grow and transfer the acetogen Thermoanaerobacter kivui at 45°C, a temperature 5–10°C above its TMIN and 21°C below its TOPT. The choice fell on an acetogen since acetogenesis—as thermophily—is an ancient trait (Basen and Müller, 2017), as described above. After ~180 generations/67 transfers at 45°C, the T. kivui population (strain Adpt45_67) was surprisingly not better adapted to 45°C, but to temperatures 5–10 K below the TOPT (55–60°C). This is somewhat in line with an observation and hypothesis published over 30 years ago by Jürgen Wiegel. He reported on a temperature plateau below the TOPT of certain anaerobic thermophiles, including Thermoanaerobacter species, allowing them to relatively quickly adapt to slightly lower temperatures. Moreover, their growth temperature (tolerance) range is large (35–78°C) compared to that of many mesophiles (Onyenwoke and Wiegel, 2015). Considering a scenario in which single Thermoanaerobacter sp. cells were suddenly exposed to a suboptimal temperature—in the ancient world or recently—their capability to cope at much lower temperatures may have prevented their rapid extinction in the environment, and that may equally hold true for other thermophiles with a broad temperature span (Wiegel, 1990). In that regard, it is worth noticing that there is increasing evidence for thermophiles in cold habitats far below their TMIN, such as Arctic sediments, soils, seawater, and even ice (Milojevic et al., 2022). The perseverance of thermophiles in arctic habitats correlates well with their broad temperature range. These populations are likely transported from populations in warmer habitats such as deep sediments, subsurface petroleum reservoirs, or ocean ridges (Hubert et al., 2009), and they may resemble ancestral thermophiles in their development toward mesophile.
Considering the evolution of a mesophile to a thermophile, mesophiles possess a narrower temperature range, ~30–35°C (Wiegel, 1990). Particularly, they face the challenge that temperatures slightly exceeding the upper-temperature limit (TMAX) may quickly lead to protein denaturation. Nonetheless, ALE experiments in mesophiles toward higher TOPT have been carried out, with mixed results. A successful evolution of E. coli in a continuous system with gradually increasing temperatures (from 44°C to 49.7°C) was reported (Blaby et al., 2012). The resulting strain had a significantly increased TOPT of 46°C, demonstrating that (moderate) thermophiles may have evolved from mesophilic ancestors. The adapted strain carried 31 single-nucleotide substitutions, among these putative critical mutations in the glycerol facilitator gene glpF and in the fatty acid desaturase/isomerase fabA. Interestingly, a mutation in the kdpD gene was recognized, a sensor kinase associated with the potassium ATPase gene kdpC, which we observed to carry a mutation. In another experiment, sequential transfer of a mutator strain Zymomonas mobilis and of E. coli with stepwise temperature increase for ~400 and 550 generations, respectively, resulted in an increased TMAX of 2–3 K (to 41°C and to 47°C), with cells dying if the temperature was further increased (Kosaka et al., 2019). The number of mutations observed in the thermally adapted strains was surprisingly small (24 and 9, respectively), in the same range as observed in this study. While most studies of thermal adaptations aimed to increase TOPT or TMAX, often in the context of adaptation to climate change or toward a biotechnological application, recently, a community approach was carried out to evolve Escherichia coli (TOPT 37°C) and Saccharomyces cerevisiae (TOPT 30°C) to 20°C and 15°C, respectively, ~15 K below their TOPT (Strauss et al., 2019). More than 20 groups from all over the world tried different approaches for a thermal adaptation of the two organisms to a suboptimal temperature. Most ALE approaches were successful; however, the two organisms behaved differently. In E. coli, the most common phenotype was a decrease in the lag phase, while in S. cerevisiae, the yield at the lower temperature increased. Improvement of growth rate was not observed or only to a limited extent, similar to what we observed with T. kivui Adpt45_67; however, we did not observe a lag phase in T. kivui of >1 h when transferred to 45°C. Unfortunately, the authors do not report on whether TOPT or TMIN were affected.
Microorganisms persevering in an unfavorable environment, even if they grow at low rates, allow time for a population to “gather” mutations, some of which are selected for growth and adaptation to the unfavorable environment. Adpt45_67 obviously is not yet adapted for better growth at 45°C, but sustains at 39°C, and, compared to the wild type, showed a TOPT shifted to 60°C. It is not clear yet, whether this adaptation halts on the plateau of 60–55°C (Wiegel, 1990), or whether more generations at 45°C will further shift the TOPT and finally increase the growth rate at 45°C. In that case, the observed genotype here may be interpreted as transient. Only a few unique and non-silent mutations were identified (Table 1), with some of them potentially related to temperature adaptation or gene regulation. Targeted point mutations may identify which of these SNPs are essential to the adapted phenotype. Continuation of the ALE may identify further mutations and allow insights into their order of occurrence. We conclude that Thermoanaerobacter and many thermophiles are well prepared to survive or even thrive at ambient temperatures (Wiegel, 1990), which may have facilitated not only a slight transition of the TOPT but a complete thermophile-to-mesophile transition over large evolutionary time scales.
The evolution of thermal adaptation has been linked to a generally different amino acid composition. From a pan-genome analysis, it is evident that the evolution of mesophilic Methanococcus sp. from thermophilic ancestors was directly linked to the replacement of lysine and an increase in the percentage of threonine, glutamine, serine, aspartate, and asparagine (Lecocq et al., 2021). The authors conclude that amino acid replacement via single mutations rather than horizontal gene transfer (HGT) events caused the adaptation to the lower temperature. This reflects, however, large evolutionary time scales, and cannot explain adaptations to slightly lower temperatures in a shorter time period as observed here. On a shorter time scale, it may be conceivable that thermophiles profit from HGT events in adaptation to lower temperatures. T. kivui, the organism used in this study, has recently been shown to efficiently take up and integrate foreign DNA from a laboratory environment into its genome, which allowed the strain to adapt to a certain medium type (Zeldes et al., 2023).
Further attempts to adapt thermophiles to lower temperatures in the laboratory are warranted to elucidate whether the observed shift toward a lower TOPT can be reproduced or increased beyond the temperature plateau and whether growth also improves at the lower temperature end. A difficulty that we encountered was certainly the low growth rates at much lower temperatures, which decrease the number of generations (and thus the number of possible mutations) in ALE experiments. Nonetheless, experimental approaches toward a cold adaptation of thermophiles will be essential to ultimately prove considerations from genome comparisons. It will be of interest to see whether the ALE of thermophiles toward a lower TOPT will lead to an increase in the GC content (Hu et al., 2022) or a change in the amino acid pattern (Sauer and Wang, 2019; Lecocq et al., 2021) in the long term. In the short term, these experiments may resolve the order of single evolutive events of particular importance, such as single gene (e.g., transcriptional regulators) inactivation by point mutations or HGT events (in case DNA of mesophiles is supplied), and enable the study of the respective phenotypic effects in single organisms, such as in the acetogen T. kivui.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/sra/, SRR25301688.
ML: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing—original draft. CP: Validation, Writing—original draft, Data curation, Formal analysis, Investigation, Methodology, Visualization. BZ: Investigation, Validation, Data curation, Formal analysis, Methodology, Visualization, Writing—review and editing. AP: Resources, Software, Writing—review and editing, Data curation, Formal analysis, Investigation. RD: Writing—review and editing, Resources, Software. MB: Writing—original draft, Validation, Conceptualization, Funding acquisition, Project administration, Resources, Supervision.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by a grant from VolkswagenStiftung (Volkswagen Foundation, A123851) in the Experiment! program (exploratory phase). The authors acknowledge financial support from the Deutsche Forschungsgemeinschaft and Universität Rostock within the funding program Open Access Publishing.
This study was dedicated to Jürgen Wiegel, whose study inspired the authors to start working on thermophilic anaerobic bacteria and their evolution. The authors gratefully acknowledge Monika Timm, Ilona Boldt (both University of Rostock), and Melanie Heinemann (Georg-August University Göttingen) for technical support in the laboratory. Moreover, MB is grateful to Volker Müller for fruitful discussions. For the special feature on the 11th International Congress on Extremophiles in 2016, we wrote a review together, arguing about likely ancient acetogens thriving at high temperatures (Basen and Müller, 2017). This current report, for the Frontiers Research Topic on the 13th International Congress on Extremophiles, may be seen as a practical continuation of some of our thoughts in the abovementioned publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1265216/full#supplementary-material
Badran, A. H., and Liu, D. R. (2015). Development of potent in vivo mutagenesis plasmids with broad mutational spectra. Nat. Commun. 6, 8425. doi: 10.1038/ncomms9425
Basen, M., Geiger, I., Henke, L., and Müller, V. (2018). A genetic system for the thermophilic acetogenic bacterium Thermoanaerobacter kivui. Appl. Environ. Microbiol. 84, e02210–02217. doi: 10.1128/AEM.02210-17
Basen, M., and Müller, V. (2017). “Hot” acetogenesis. Extremophiles 21, 15–26. doi: 10.1007/s00792-016-0873-3
Berezovsky, I. N., and Shakhnovich, E. I. (2005). Physics and evolution of thermophilic adaptation. Proc. Natl. Acad. Sci. USA 102, 12742–12747. doi: 10.1073/pnas.0503890102
Blaby, I. K., Lyons, B. J., Wroclawska-Hughes, E., Phillips, G. C., Pyle, T. P., Chamberlin, S. G., et al. (2012). Experimental evolution of a facultative thermophile from a mesophilic ancestor. Appl. Environ. Microbiol. 78, 144–155. doi: 10.1128/AEM.05773-11
Catchpole, R. J., and Forterre, P. (2019). The evolution of reverse gyrase suggests a non-hyperthermophilic last universal common ancestor. Mol. Biol. E36, 2737–2747. doi: 10.1093/molbev/msz180
Chan, M., Himes, R. H., and Akagi, J. M. (1971). Fatty acid composition of thermophilic, mesophilic, and psychrophilic clostridia. J. Bacteriol. 106, 876–881. doi: 10.1128/jb.106.3.876-881.1971
Cooper, V. S., and Lenski, R. E. (2000). The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407, 736–739. doi: 10.1038/35037572
de Araujo Martins, H. L., Vicente, B. P., Lorenzetti, A. M., Koide, A. P. R. T., and Marques, M.V. (2021). Cold regulation of genes encoding ion transport systems in the oligotrophic bacterium Caulobacter crescentus. Microbiol. Spectr. 9, 21. doi: 10.1128/Spectrum.00710-21
De Maayer, Anderson, P., Cary, D. C., and Cowan, D .A. (2014). Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep. 15, 508–517. doi: 10.1002/embr.201338170
Deutzmann, J. S., Kracke, F., Gu, W. Y., and Spormann, A. M. (2022). Microbial electrosynthesis of acetate powered by Intermittent electricity. Environ. Sci. Technol. 56, 16073–16081. doi: 10.1021/acs.est.2c05085
Dietrich, H. M., Righetto, R. D., Kumar, A., Wietrzynski, W., Trischler, R., Schuller, S. K., et al. (2022). Membrane-anchored HDCR nanowires drive hydrogen-powered CO2 fixation. Nature 607, 823–830. doi: 10.1038/s41586-022-04971-z
Dragosits, M., and Mattanovich, D. (2013). Adaptive laboratory evolution—Principles and applications for biotechnology. Microb. Cell. Fact. 12, 64. doi: 10.1186/1475-2859-12-64
Fiala, G., and Stetter, K. O. (1986). Pyrococcus furiosus sp.nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch. Microbiol. 145, 56–61. doi: 10.1007/BF00413027
Goldfine, H. (2010). The appearance, disappearance and reappearance of plasmalogens in evolution. Prog. Lipid Res. 49, 493–498. doi: 10.1016/j.plipres.2010.07.003
Hess, V., Poehlein, A., Weghoff, M. C., Daniel, R., and Müller, V. (2014). A genome-guided analysis of energy conservation in the thermophilic, cytochrome-free acetogenic bacterium Thermoanaerobacter kivui. BMC Genom. 15, 1139. doi: 10.1186/1471-2164-15-1139
Hu, E-. Z., Lan, X-. R., Liu, Z-. L., Gao, J., and Niu, D-. K. (2022). A positive correlation between GC content and growth temperature in prokaryotes. BMC Genom. 23, 110. doi: 10.1186/s12864-022-08353-7
Hubert, C., Loy, A., Nickel, M., Arnosti, C., Baranyi, C., Brüchert, V., et al. (2009). A constant flux of diverse thermophilic bacteria into the cold arctic seabed. Science 325, 1541–1544. doi: 10.1126/science.1174012
Imanaka, T. (2011). Molecular bases of thermophily in hyperthermophiles. Proc. Jpn. Acad., Ser. B. 87, 587–602. doi: 10.2183/pjab.87.587
Inoue-Sakamoto, K., Gruber, T. M., Christensen, S. K., Arima, H., Sakamoto, T., Bryant, D. A., et al. (2007). Group 3 sigma factors in the marine cyanobacterium Synechococcus sp. strain PCC 7002 are required for growth at low temperature. J. Gen. Appl. Microbiol. 53, 89–104. doi: 10.2323/jgam.53.89
Jackson, D. R., Cassilly, C. D., Plichta, D. R., Vlamakis, H., Liu, H., Melville, S. B., et al. (2021). Plasmalogen biosynthesis by anaerobic bacteria: identification of a two-gene operon responsible for plasmalogen production in Clostridium perfringens. Acs. Chem. Biol. 16, 6–13. doi: 10.1021/acschembio.0c00673
Jain, S., Dietrich, H. M., Müller, V., and Basen, M. (2020). Formate Is required for growth of the thermophilic acetogenic bacterium Thermoanaerobacter kivui lacking hydrogen-dependent carbon dioxide reductase (HDCR). Front. Microbiol. 11, 59. doi: 10.3389/fmicb.2020.00059
Jesser, R., Behler, J., Benda, C., Reimann, V., and Hess, W. R. (2019). Biochemical analysis of the Cas6-1 RNA endonuclease associated with the subtype I-D CRISPR-Cas system in Synechocystis sp. PCC 6803. RNA Bio. 16, 481–491. doi: 10.1080/15476286.2018.1447742
Kang, M., Kim, K., Choe, D., Cho, S., Kim, S. C., Palsson, B., et al. (2019). Inactivation of a mismatch-repair system diversifies genotypic landscape of Escherichia coli during adaptive laboratory evolution. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01845
Katsyv, A., and Müller, V. (2022). A purified energy-converting hydrogenase from Thermoanaerobacter kivui demonstrates coupled H+ -translocation and reduction in vitro. J. Biol. Chem. 298. doi: 10.1016/j.jbc.2022.102216
Katsyv, A., Schoelmerich, M. C., Basen, M., and Müller, V. (2021). The pyruvate:ferredoxin oxidoreductase of the thermophilic acetogen, Thermoanaerobacter kivui. FEBS Open Bio. 11, 1332–1342. doi: 10.1002/2211-5463.13136
Kelley, D. S., Karson, J. A., Fruh-Green, G. L., Yoerger, D. R., Shank, T. M., Butterfield, D. A., et al. (2005). A serpentinite-hosted ecosystem: the lost city hydrothermal field. Science 307, 1428–1434. doi: 10.1126/science.1102556
Kim, T. H., Kim, H. J., Park, J. S., Kim, Y., Kim, P., Lee, H. S., et al. (2005). Functional analysis of sigH expression in Corynebacterium glutamicum. Biochem. Biophys. Res. Commun. 331, 1542–1547. doi: 10.1016/j.bbrc.2005.04.073
Koivuniemi, A. (2017). The biophysical properties of plasmalogens originating from their unique molecular architecture. FEBS Lett. 591, 2700–2713. doi: 10.1002/1873-3468.12754
Kosaka, T., Nakajima, Y., Ishii, A., Yamashita, M., Yoshida, S., Murata, M., et al. (2019). Capacity for survival in global warming: adaptation of mesophiles to the temperature upper limit. PLoS ONE 14, e0215614. doi: 10.1371/journal.pone.0215614
Lecocq, M., Groussin, M., Gouy, M., and Brochier-Armanet, C. (2021). The molecular determinants of thermoadaptation: Methanococcales as a case study. Mol. Biol. Evol. 38, 1761–1776. doi: 10.1093/molbev/msaa312
Leiby, N., and Marx, C. J. (2014). Metabolic erosion primarily through mutation accumulation, and not tradeoffs, drives limited evolution of substrate specificity in Escherichia coli. PLoS Biol. 12, e1001789. doi: 10.1371/journal.pbio.1001789
Leigh, J. A., Mayer, F., and Wolfe, R. S. (1981). Acetogenium kivui, a new thermophilic hydrogen-oxidizing, acetogenic bacterium. Arch. Microbiol. 129, 275–280. doi: 10.1007/BF00414697
Ljungdahl, L. G. (1986). The autotrophic pathway of acetate synthesis in acetogenic bacteria. Annu. Rev. Microbiol. 40, 415–450. doi: 10.1146/annurev.mi.40.100186.002215
Luan, G., Cai, Z., Li, Y., and Ma, Y. (2013). Genome replication engineering assisted continuous evolution (GREACE) to improve microbial tolerance for biofuels production. Biotechnol. Biofuels 6, 137. doi: 10.1186/1754-6834-6-137
Lunine, J. I. (2006). Physical conditions on the early Earth. Philos. T R Soc. B 361, 1721–1731. doi: 10.1098/rstb.2006.1900
Ma, H. W., Kumar, B., Ditges, U., Gunzer, F., Buer, J., Zeng, A. P., et al. (2004). An extended transcriptional regulatory network of Escherichia coli and analysis of its hierarchical structure and network motifs. Nuc. Acids Res. 32, 6643–6649. doi: 10.1093/nar/gkh1009
Martin, W. F. (2020). Older than genes: the acetyl CoA pathway and origins. Front. Microbiol. 11, 817. doi: 10.3389/fmicb.2020.00817
Martin, W. F., and Sousa, F. L. (2016). Early microbial evolution: the age of anaerobes. Cold Spring Harbor Perspect. Biol. 8, 18. doi: 10.1101/cshperspect.a018127
Milojevic, T., Cramm, M. A., Hubert, C. R. J., and Westall, F. (2022). Freezingandrdquo, thermophiles: from one temperature extreme to another. Microorganisms 10, 2417. doi: 10.3390/microorganisms10122417
Moon, J., Henke, L., Merz, N., and Basen, M. (2019). A thermostable mannitol-1-phosphate dehydrogenase is required in mannitol metabolism of the thermophilic acetogenic bacterium Thermoanaerobacter kivui. Environ. Microbiol. 21, 3728–3736. doi: 10.1111/1462-2920.14720
Moon, J., Jain, S., Müller, V., and Basen, M. (2020). Homoacetogenic conversion of mannitol by the thermophilic acetogenic bacterium Thermoanaerobacter kivui requires external CO2. Front. Microbiol. 11, 571736. doi: 10.3389/fmicb.2020.571736
Morikawa, K., Takemura, A. J., Inose, Y., Tsai, M., Nguyen Thi, L. T., Ohta, T., et al. (2012). Expression of a cryptic secondary sigma factor gene unveils natural competence for DNA transformation in Staphylococcus aureus. PLoS Pathog. 8, e1003003. doi: 10.1371/journal.ppat.1003003
Onyenwoke, R. U., and Wiegel, J. (2015). “Thermoanaerobacter,” in Bergey's Manual of Systematics of Archaea and Bacteria, eds M. E. Trujillo, S. Dedysh, P. DeVos, B. Hedlund, P. Kämpfer, F. A. Rainey et al. (Hoboken, NJ: Bergey's Manual Trust and John Wiley and Sons, Ltd), doi: 10.1002/9781118960608.gbm00751
Oshima, M., and Miyagawa, A. (1974). Comparative studies on the fatty acid composition of moderately and extremely thermophilic bacteria. Lipids 9, 476–480. doi: 10.1007/BF02534274
Pace, N. R. (1997). A molecular view of microbial diversity and the biosphere. Science 276, 734–740. doi: 10.1126/science.276.5313.734
Russell, M. J., Hall, A. J., and Martin, W. (2010). Serpentinization as a source of energy at the origin of life. Geobiol. 8, 355–371. doi: 10.1111/j.1472-4669.2010.00249.x
Sauer, D. B., and Wang, D-. N. (2019). Predicting the optimal growth temperatures of prokaryotes using only genome derived features. Bioinformatics 35, 3224–3231. doi: 10.1093/bioinformatics/btz059
Sikorski, J., Brambilla, E., Kroppenstedt, R. M., and Tindall, B. J. (2008). The temperature-adaptive fatty acid content in Bacillus simplex strains from ‘Evolution Canyon', Israel. Microbiol.-SGM 154, 2416–2426. doi: 10.1099/mic.0.2007/016105-0
Siliakus, M. F., Van Der Oost, J., and Kengen, S. W. M. (2017). Adaptations of archaeal and bacterial membranes to variations in temperature, pH and pressure. Extremophiles 21, 651–670. doi: 10.1007/s00792-017-0939-x
Sousa, F. L., Thiergart, T., Landan, G., Nelson-Sathi, S., Pereira, I.a.C., Allen, J.F., Lane, N., et al. (2013). Early bioenergetic evolution. Philos T R Soc. B 36, 8. doi: 10.1098/rstb.2013.0088
Stetter, K. O. (2006). History of discovery of the first hyperthermophiles. Extremophiles 10, 357–362. doi: 10.1007/s00792-006-0012-7
Strauss, K. S., Schirman, D., Jona, G., Brooks, A.N., Kunjapur, A.M., Nguyen Ba, A.N., et al. (2019). Evolthon: a community endeavor to evolve lab evolution. PLoS Biol. 17, e3000182. doi: 10.1371/journal.pbio.3000182
Suutari, M., and Laakso, S. (1992). Changes in fatty acid branching and unsaturation of Streptomyces griseus and Brevibacterium fermentans as a response to growth temperature. Appl. Environ. Microbiol. 58, 2338–2340. doi: 10.1128/aem.58.7.2338-2340.1992
Suzuki, H., Kobayashi, J., Wada, K., Furukawa, M., and Doi, K. (2015). Thermoadaptation-directed enzyme evolution in an error-prone thermophile derived from Geobacillus kaustophilus HTA426. Appl. Environ. Microbiol. 81, 149–158. doi: 10.1128/AEM.02577-14
Vasi, F. K., and Lenski, R. E. (1999). Ecological strategies and fitness tradeoffs in Escherichia coli mutants adapted to prolonged starvation. J. Gen. 78, 43–49. doi: 10.1007/BF02994702
Vítová, M., Palyzová, A., and Rezanka, T. (2021). Plasmalogens—Ubiquitous molecules occurring widely, from anaerobic bacteria to humans. Prog. Lipid Res. 83, 101111. doi: 10.1016/j.plipres.2021.101111
Weghoff, M. C., and Müller, V. (2016). CO metabolism in the thermophilic acetogen Thermoanaerobacter kivui. Appl. Environ. Microbiol. 82, 2312–2319. doi: 10.1128/AEM.00122-16
Weiss, M. C., Sousa, F. L., Mrnjavac, N., Neukirchen, S., Roettger, M., Nelson-Sathi, S., et al. (2016). The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 1, 16116. doi: 10.1038/nmicrobiol.2016.116
Widdel, F., and Bak, F. (1992). “Gram-negative mesophilic sulfate-reducing bacteria,” in The Prokaryotes, eds. A.T. Balows, H.G. Dworkin, M. Harder, K. H. Schleifer (New York: Springer), 3352-3378. doi: 10.1007/978-1-4757-2191-1_21
Wiegel, J. (1990). Temperature spans for growth: hypothesis and discussion. FEMS Microbiol. Lett. 75, 155–169. doi: 10.1111/j.1574-6968.1990.tb04092.x
Wood, H. G., Ragsdale, S. W., and Pezacka, E. (1986). The acetyl-CoA pathway of autotrophic growth. FEMS Microbiol. Lett. 39, 345–362. doi: 10.1111/j.1574-6968.1986.tb01865.x
Zeldes, B., Poehlein, A., Jain, S., Baum, C., Daniel, R., Müller, V., et al. (2023). DNA uptake from a laboratory environment drives unexpected adaptation of a thermophile to a minor medium component. ISME Commun. 3, 2. doi: 10.1038/s43705-022-00211-7
Zeldes, B. M., Keller, M. W., Loder, A. J., Straub, C. T., Adams, M. W. W., Kelly, R. M., et al. (2015). Extremely thermophilic microorganisms as metabolic engineering platforms for production of fuels and industrial chemicals. Front. Microbiol. 6, 1209. doi: 10.3389/fmicb.2015.01209
Keywords: adaptive laboratory evolution, origin of life, cold adaptation, acetogens, thermophiles, Thermoanaerobacter kivui
Citation: Lehmann M, Prohaska C, Zeldes B, Poehlein A, Daniel R and Basen M (2023) Adaptive laboratory evolution of a thermophile toward a reduced growth temperature optimum. Front. Microbiol. 14:1265216. doi: 10.3389/fmicb.2023.1265216
Received: 22 July 2023; Accepted: 14 September 2023;
Published: 12 October 2023.
Edited by:
Melina Kerou, University of Vienna, AustriaReviewed by:
Byung-Kwan Cho, Korea Advanced Institute for Science and Technology, Republic of KoreaCopyright © 2023 Lehmann, Prohaska, Zeldes, Poehlein, Daniel and Basen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mirko Basen, bWlya28uYmFzZW5AdW5pLXJvc3RvY2suZGU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.