- 1Department of Public Health, University of Naples “Federico II”, Naples, Italy
- 2Department of Biology, University of Oxford, Oxford, United Kingdom
- 3Department of Biology, University of Naples Federico II, Naples, Italy
- 4Department of Chemical Sciences, University of Naples Federico II, Naples, Italy
- 5Institut Pasteur, Université Paris Cité, Biodiversity and Epidemiology of Bacterial Pathogens, Paris, France
Introduction: Non-baumannii Acinetobacter species are increasingly isolated in the clinical setting and the environment. The aim of the present study was to analyze a genome database of 837 Acinetobacter spp. isolates, which included 798 non-baumannii Acinetobacter genomes, in order to define the concordance of classification and discriminatory power of 7-gene MLST, 53-gene MLST, and single-nucleotide polymorphism (SNPs) phylogenies.
Methods: Phylogenies were performed on Pasteur Multilocus Sequence Typing (MLST) or ribosomal Multilocus Sequence Typing (rMLST) concatenated alleles, or SNPs extracted from core genome alignment.
Results: The Pasteur MLST scheme was able to identify and genotype 72 species in the Acinetobacter genus, with classification results concordant with the ribosomal MLST scheme. The discriminatory power and genotyping reliability of the Pasteur MLST scheme were assessed in comparison to genome-wide SNP phylogeny on 535 non-baumannii Acinetobacter genomes assigned to Acinetobacter pittii, Acinetobacter nosocomialis, Acinetobacter seifertii, and Acinetobacter lactucae (heterotypic synonym of Acinetobacter dijkshoorniae), which were the most clinically relevant non-baumannii species of the A. baumannii group. The Pasteur MLST and SNP phylogenies were congruent at Robinson-Fould and Matching cluster tests and grouped genomes into four and three clusters in A. pittii, respectively, and one each in A. seifertii. Furthermore, A. lactucae genomes were grouped into one cluster within A. pittii genomes. The SNP phylogeny of A. nosocomialis genomes showed a heterogeneous population and did not correspond to the Pasteur MLST phylogeny, which identified two recombinant clusters. The antimicrobial resistance genes belonging to at least three different antimicrobial classes were identified in 91 isolates assigned to 17 distinct species in the Acinetobacter genus. Moreover, the presence of a class D oxacillinase, which is a naturally occurring enzyme in several Acinetobacter species, was found in 503 isolates assigned to 35 Acinetobacter species.
Conclusion: In conclusion, Pasteur MLST phylogeny of non-baumannii Acinetobacter isolates coupled with in silico detection of antimicrobial resistance makes it important to study the population structure and epidemiology of Acinetobacter spp. isolates.
Introduction
Acinetobacter spp. are aerobic, non-fermentative, Gram-negative coccobacilli that are widely distributed in the environment and are responsible for infections in animals and humans (Bouvet and Grimont, 1986; Cosgaya et al., 2017; Wong et al., 2017; Cools et al., 2019). The genus Acinetobacter includes 77 child taxa with a validly published and correct name (https://www.bacterio.net/genus/acinetobacter; accessed on June 2023). As the identification of Acinetobacter isolates at the species level has been difficult to obtain using standard phenotypic methods (Bouvet and Grimont, 1986; Cools et al., 2019), Matrix-Assisted Laser Desorption-Ionization-Time of Flight (MALDI-TOF) mass spectrometry (Marí-Almirall et al., 2017; Šedo et al., 2018) or genotypic methods, which use partial rpoB sequencing (Gundi et al., 2009) or ribosomal MLST analysis (Jolley et al., 2012), have been applied for correct Acinetobacter species assignment. The above techniques have identified Acinetobacter baumannii as the most clinically relevant species of the Acinetobacter genus, which has been demonstrated to cause community and healthcare-associated infections (Wong et al., 2017; Whiteway et al., 2022). Genomic epidemiology of A. baumannii isolates has shown the global spread of distinct clonal lineages, which have been selected because of their resistance to a broad range of antimicrobials, including carbapenems (Wong et al., 2017) and have been responsible for epidemics worldwide (Gaiarsa et al., 2019; Hamidian and Nigro, 2019). In addition to A. baumannii, A. nosocomialis, A. pittii, A. seifertii, and A. lactucae (formerly identified as A. dijkshoorniae) have been increasingly isolated from humans and reported to be responsible for infections (Cosgaya et al., 2017). A. baumannii, A. nosocomialis, A. pittii, A. seifertii, and A. lactucae showed closely related phenotypic and genotypic features and were considered members of the A. baumannii group (Cosgaya et al., 2017; Marí-Almirall et al., 2017). Epidemics caused by multidrug-resistant (MDR) and carbapenem-resistant A. nosocomialis, A. pittii, and A. seifertii have been increasingly reported (Chen et al., 2018, 2019; Chopjitt et al., 2021; Li et al., 2021).
The present study aimed to perform phylogenomic analysis of 837 isolates assigned to 72 distinct species in the Acinetobacter genus using the Pasteur MLST scheme, compare phylogenetic congruence with genome-based and ribosomal MLST (rMLST)-based phylogenies of A. baumannii group genomes, and identify antimicrobial resistance genes in Acinetobacter spp. genomes.
Materials and methods
Genome dataset
Bacterial genomes included in the analysis were manually selected from the PubMLST database1 until January 2022. In detail, we selected 39 A. baumannii complete genomes assigned to international clonal lineages ICI, ICII, and ICIII, which corresponded to Pasteur ST1, ST2, and ST3, respectively, and to additional epidemic clonal lineages assigned to Pasteur ST10, ST25, ST32, ST52, ST78, and ST79 (Gaiarsa et al., 2019). Furthermore, we collected from the National Center for Biotechnology Information (NCBI) high-quality complete genomes and, when not available, scaffolded sequences of all species into the Acinetobacter genus. The “parameters” to consider the genomes of “high quality” were: N50 ≥ 10,000 bp; number of contigs ≤1,000; identification of the correct species through rMLST (Jolley et al., 2012). The Acinetobacter selected genomes were typed by A. baumannii Pasteur MLST scheme (Diancourt et al., 2010) and rMLST (Jolley et al., 2012) using the BIGSdb software available at https://pubmlst.org/organisms/acinetobacter-baumannii/ (Jolley et al., 2018). The characteristics of the genomes were included in Supplementary Table S1 and available at https://pubmlst.org/bigsdb?db=pubmlst_abaumannii_isolates&page=project&project_id=8.
Phylogenetic and statistical analyses
The allelic profiles of Pasteur and ribosomal MLST schemes were extracted from all the genomes, and then the sequences were aligned using Muscle (Edgar, 2004) to generate the neighbor-joining trees using the BIGSdb software (Jolley et al., 2018). The core genome single-nucleotide polymorphisms (SNPs) were detected using the tools of PARSNP v1.1.2 (Treangen et al., 2014), and the SNP alignment was performed considering the ascertainment bias using the Lewis correction (Lewis, 2001). In detail, each genome was aligned to the reference genome NC_010611.1 of ACICU, and the alignments were then concatenated using Muscle (Edgar, 2004). The maximum-likelihood phylogenies of 574 genomes belonging to the A. baumannii group (A. baumannii, A. pittii, A. seifertii, A. lactucae, and A. nosocomialis) were performed using the concatenated alleles of the Pasteur and ribosomal MLST schemes and a reference phylogeny using genome-wide data (a core genome of 372 high-quality genes and an alignment of 17,072 SNPs). The phylogenies of Pasteur MLST, ribosomal MLST, and core SNP alignments were inferred through the GTR-GAMMA model at 100 bootstrap replicates using RAxML v.8 (Stamatakis, 2014). GTR’s GAMMA model was used for its ability to optimize the transition/transversion speed ratio and the α parameter of the gamma rate heterogeneity distribution (Stamatakis, 2014). The phylogenetic trees and annotations were visualized using the iTol v6 software.2
Statistical analyses
The statistical analyses were performed using the Robinson-Fould (R-F) and Matching clusters (M-C) topology-based tests employing TreeCmp (Bogdanowicz et al., 2012). The M-C test calculates the number of topological changes that must be made to transform a tree into a reference tree. The R-F test counts the number of splits that are unique to one of the two trees. In both cases, the two analyzed trees are identical if the value is zero. The likelihood-based Shimodaira-Hasegawa (SH) test (Shimodaira and Hasegawa, 1999) was performed with RAXML (Stamatakis, 2014). In this test, a null hypothesis assumes that two compared trees are both a correct interpretation of an alignment. The tested hypothesis is that one or more trees better represent the data. p-values lower than 0.05 indicate that the two trees are significantly different.
Analysis of antimicrobial resistance genes
The antimicrobial resistance genes were detected using abricate3 based on the ResFinder 4.0 database (Bortolaia et al., 2020). The presence of the gene and percentage of identity were indicated in the matrix in Supplementary Table S2. The threshold identity of 80%, the minimum length of 80% matches, and the coverage value of more than 90% were selected for each gene. The following acquired resistance genes were analyzed: class A, B, C, D β-lactamase, folate pathway inhibitors, rifampicin, aminoglycoside, chloramphenicol, colistin, macrolide, quaternary ammonium salts, and tetracycline resistance genes (Supplementary Table S2). The multiple alignment and phylogeny of class D β-lactamase genes were performed using Clustal W (Thompson et al., 1994) and RAXML v.8 (Stamatakis, 2014), respectively. The genomes of Acinetobacter genus were classified as MDR if they carried antimicrobial-resistant genes for at least three of the nine classes of antimicrobials considered by Magiorakos et al. (2012).
Clustering of Pasteur MLST and core SNPs phylogenies
The clusters of Pasteur sequence types (STs) were determined using the eBURST algorithm as described previously (Feil et al., 2004). Minimum spanning trees of STs were built with Phyloviz using the goeBURST algorithm (Ribeiro-Gonçalves et al., 2016). Minimum-spanning trees were generated from the seven alleles of each MLST scheme, and species were assigned based on clustering with reference STs. Additionally, SNP analysis by PARSNP was visualized using Gingr (Treangen et al., 2014), which provided an interactive display of multi-alignment variants and a phylogenetic tree estimated from the core genome alignment. Then, the values of the maximum unique matches and the data were evaluated to study the cluster phylogeny of SNPs (Treangen et al., 2014).
Results
Acinetobacter genus database
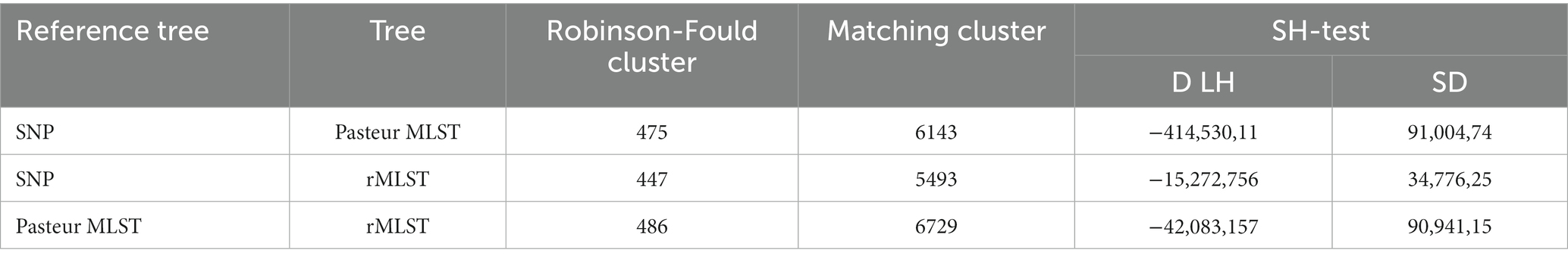
The database consisted of 837 genomes assigned to 72 distinct species in the Acinetobacter genus, which were identified at genus and species levels using the ribosomal MLST scheme (Jolley et al., 2012) and showed a genome size range of 2.85–4.85 Mpb (Supplementary Table S1). The Pasteur MLST scheme (Diancourt et al., 2010) was able to assign an allelic profile and an ST to all 837 genomes belonging to 72 species in the Acinetobacter genus; while, the ribosomal MLST scheme assigned an ST to 806 out of 837 genomes because from 1 to 20 alleles could not be identified in the genomes belonging to A. baumannii, A. nosocomialis, A. bereziniae, A. pittii, A. radioresistens, and A. seifertii. In addition, the rMLST scheme identified paralogues in 11 out of 53 loci paralogous in 57 Acinetobacter spp. (Supplementary Table S1). The genome-wide distances of the whole database analyzed using the minimum spanning tree (MST) with the Pasteur MLST scheme showed that the most abundant species were A. pittii (n = 282), A. nosocomialis (n = 175), A. seifertii (n = 61), A. baumannii (n = 39), and A. lactucae (n = 14), while 263 genomes were assigned to other 67 species of Acinetobacter genus (Figure 1). The 574 genomes assigned to the above five species have been considered the most clinically relevant species and were included in the A. baumannii group (Cosgaya et al., 2017). Among genomes within the A. baumannii group, the genetically closest species were A. pittii with A. lactucae or A. seifertii, showing 5 and 6 locus variants (LVs) genome-wide distance, respectively. The A. nosocomialis genomes assigned to ST782 and to the most frequent ST279 showed 4 and 5 LVs genome-wide distances with respect to CC2 A. baumannii genomes, respectively (Figure 1).
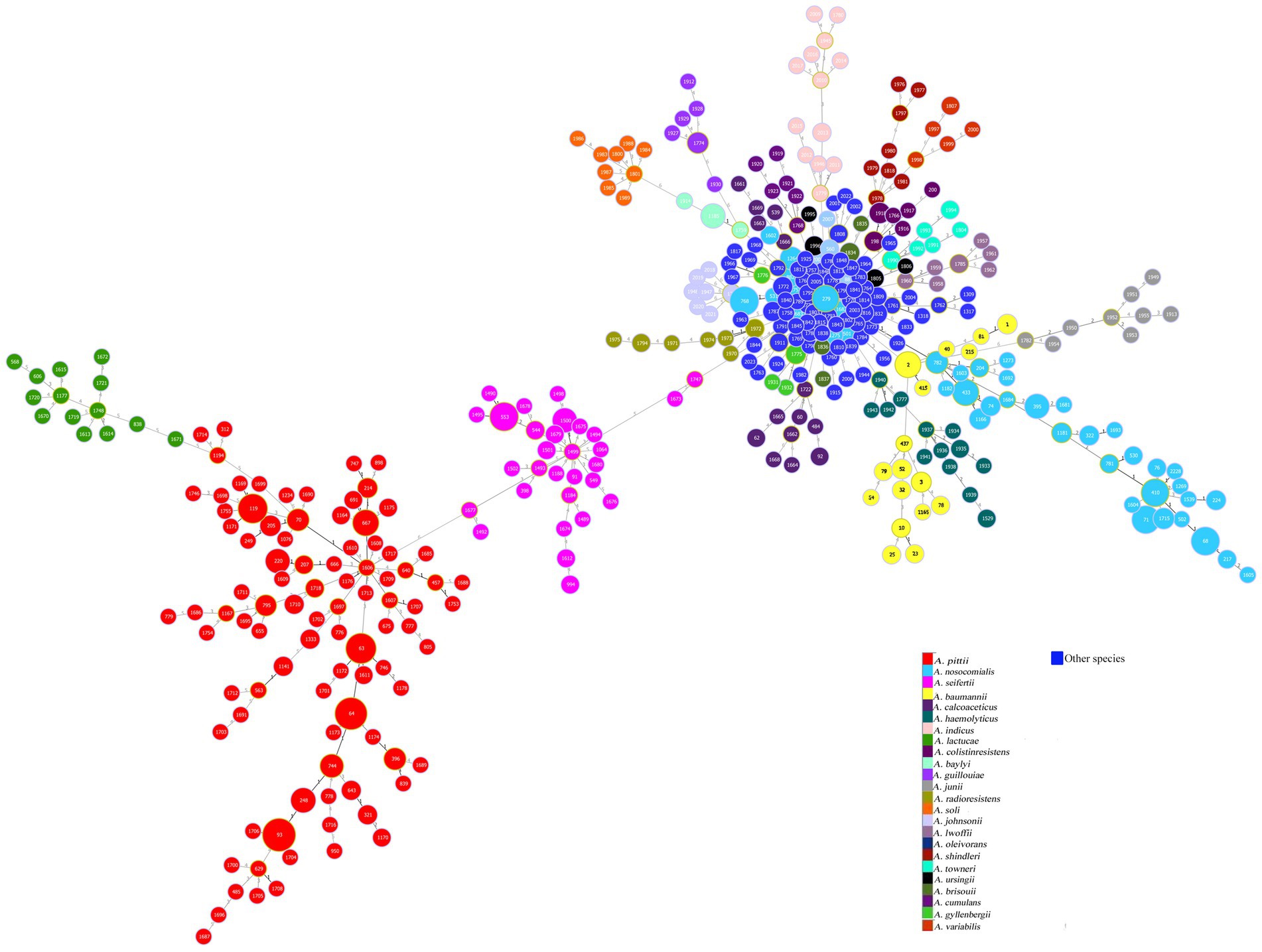
Figure 1. Minimum spanning tree of 837 genomes inferred by the Pasteur MLST scheme. The different colors indicate the 24 Acinetobacter species that are represented by more than 6 isolates, while the blue color indicates the 48 Acinetobacter species that are represented by less than 6 genomes. The numbers within each circle indicate the ST. The size of the circle is proportional to the number of genomes belonging to the same ST. The figure was obtained using the eBURST algorithm with the Phyloviz software (Ribeiro-Gonçalves et al., 2016).
Maximum-likelihood phylogeny of Acinetobacter baumannii group
The core genome-SNP, Pasteur MLST, and rMLST maximum-likelihood phylogenies of 574 genomes belonging to the A. baumannii group (A. baumannii, A. pittii, A. seifertii, A. lactucae, and A. nosocomialis) showed similar inter- and intra-species distributions of branch lengths and nodes (Figure 2). In particular, the branch lengths of SNPs and rMLST phylogenies had values between ~10−6 and ~ 10−2 (Figures 2A,C), while the branch lengths of Pasteur MLST phylogeny were between ~10−6 and ~ 10−3 (Figure 2B). The core genome SNPs and Pasteur MLST phylogenies showed bootstrap values greater than 50 for ancestral nodes of all species belonging to the A. baumannii group (Figures 2A,B). In addition, rMLST phylogeny assigned bootstrap values greater than 50 to the ancestral nodes of A. baumannii, A. seifertii, and A. lactucae species, while bootstrap values of 36 for A. pittii ancestral node and more uneven values ranging from 21 to 94 were assigned across A. nosocomialis genomes (Figure 2C). The above data demonstrated that all three phylogenies identified A. baumannii, A. pittii, A. seifertii, A. lactucae, and A. nosocomialis as distinct species in the A. baumannii group and a strong genomic similarity between A. lactucae and A. pittii species. Moreover, all three maximum-likelihood phylogenies showed high genomic heterogeneity among A. nosocomialis genomes (Figure 2). The statistical comparison between core genome SNP phylogeny and Pasteur MLST or rMLST phylogenies using the R-F and M-C tests showed similar statistical values and concordance among phylogenies (Table 1).
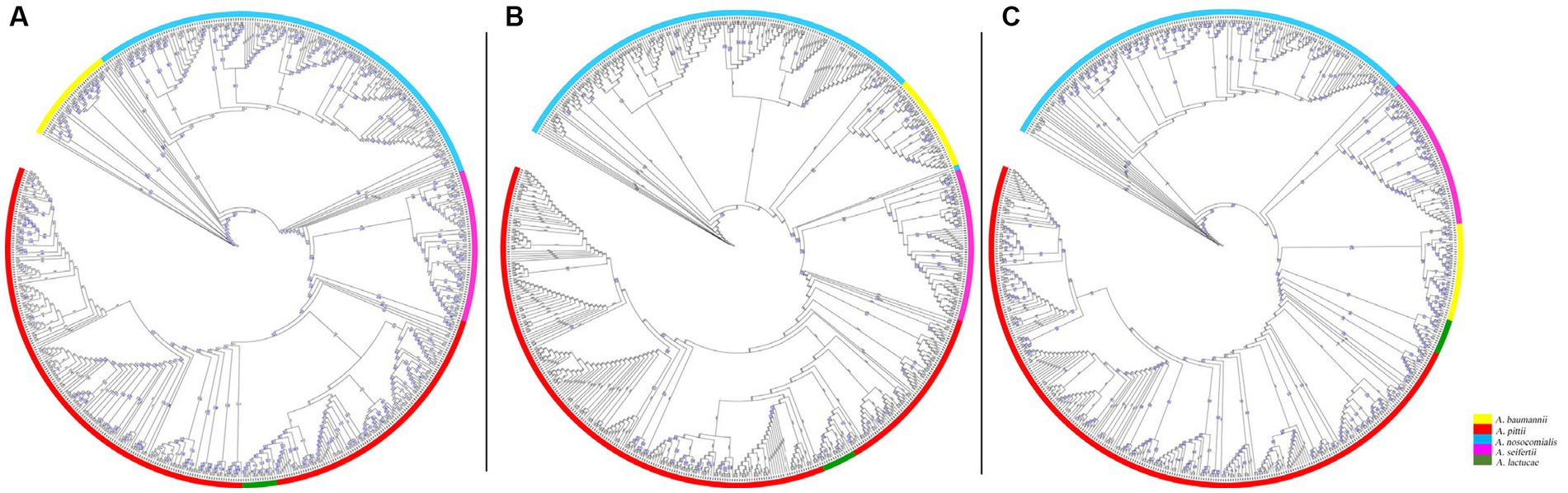
Figure 2. Phylogeny of A. baumannii group genomes. Maximum-likelihood phylogeny of 574 A. baumannii group genomes, including 39 A. baumannii (yellow labels), 285 A. pittii (red labels), 175 A. nosocomialis (blue labels), 61 A. seifertii (pink labels), and 14 A. lactucae genomes (green labels), was inferred on coreSNPs (A), Pasteur MLST (B), and rMLST (C) using RAxML. Bootstrap values are indicated in blue on tree branches, while the lengths of the branches are indicated in black. The figures were obtained using iTol v6 (https://itol.embl.de/).
Clustering of core SNP and Pasteur MLST phylogenies of Acinetobacter baumannii group
To evaluate the discriminatory power of the core genome SNP phylogeny and the Pasteur MLST phylogeny, we analyzed and compared the clusters identified by the two phylogenies. Clustering of core SNP phylogeny identified three clusters (1–3) in A. pittii genomes, one cluster (4) in A. seifertii genomes, and no clusters in A. nosocomialis genomes (Figure 3A). Cluster analysis of Pasteur MLST phylogeny showed four clusters in A. pittii having ST63, ST119, ST207, and ST396 as ancestral STs; two clusters in A. nosocomialis with ST410 and ST279 as ancestral STs; and one cluster in A. seifertii with ancestral ST553. Acinetobacter baumannii genomes showed three clades assigned to ST1, ST2, and ST10; and A. lactucae genomes counted 14 singletons and no clades (Figure 3B). Interestingly, A. pittii clusters 1, 2, and 3 of the core SNP phylogeny corresponded to A. pittii ST396, ST119, and ST207 clusters in the Pasteur MLST phylogeny, respectively; A. seifertii cluster 4 of the core SNP phylogeny corresponded to A. seifertii ST553 cluster of the Pasteur MLST phylogeny (Figure 3).
Figure 3. Cluster analysis of core genome SNP and Pasteur MLST phylogenies. Cluster analysis of core genome SNP (A) and Pasteur MLST (B) phylogenies of 574 genomes belonging to Acinetobacter baumannii group. (A) The four main clusters identified in the core genome SNP phylogeny are highlighted in violet (cluster 1), fuxia (cluster 2), brown (cluster 3), and green (cluster 4). (B) The seven main clusters identified in the Pasteur MLST phylogeny are highlighted in blue (cluster ST63), brown (cluster ST207), pink (cluster ST119), violet (cluster ST296), green (cluster ST553), orange (cluster ST410), and red (cluster ST279). The phylogenies were analyzed with RAxML, and the figures were obtained using iTol v6 (https://itol.embl.de/).
Antimicrobial resistance genes in Acinetobacter spp. genomes
We analyzed the presence of antimicrobial resistance genes in the 837 genomes assigned to 72 distinct Acinetobacter species. The ResFinder software identified 188 genes encoding for resistance to 12 antimicrobial categories (Supplementary Table S2). The presence of class A, B, C, and D β-lactamases in the neighbor-joining tree of the 837 genomes assigned to 72 distinct Acinetobacter species is shown in Figure 4. The class A β-lactamase genes blaCARB-8,-2,-14,-16 were found in 1 A. nosocomialis, 4 A. pittii, 1 A. towneri, and 1 A. bereziniae genomes; the class A β-lactamase gene blaSCO-1 was found in 1 A. radioresistens genome. In addition, class A β-lactamase genes blaTEM-1A,-1B,-1D were found in 9 of 39 A. baumannii genomes; blaPER-1,-2 ESBL genes were found in 1 A. baumannii, 2 A. nosocomialis, 1 A. pittii, and 1 A. radioresistens genomes; blaVEB-1,-7 ESBL genes were found in 1 A. baumannii and 2 A. pittii genomes. The class B metallo-β-lactamase (MBL) genes blaGIM-1, blaIMP-1,-4,-14,-19,-34 blaNDM-1,-16, and blaVIM-2,-4 were identified in 12 of 72 species of Acinetobacter genus. Among species, 34 of 282 A. pittii genomes showed at least one MBL gene. The blaNDM-1 was the most frequent MBL gene and was found in A. nosocomialis, A. pittii, A. lactucae, A. junii, A. bereziniae, A. cumulans, A. wuhouensis, A. sichuanensis, A. rongchengensis, A. indicus, and A. variabilis genomes. The blaADC-25 class C β-lactamase was found in the five species belonging to A. baumannii group and A. calcoaceticus genomes, but not in other Acinetobacter spp. (Figure 4; Supplementary Table S2). The class D oxacillinase, which is a naturally occurring enzyme in several Acinetobacter species, was found in 503 isolates assigned to 35 Acinetobacter species (Figure 4; Supplementary Table S2). In all, 94 class D β-lactamase genes belonging to 11 distinct blaOXA family genes (blaOXA-211, blaOXA-134, blaOXA-214, blaOXA 294, blaOXA-51, blaOXA-213, blaOXA-274, blaOXA-286, blaOXA-58, blaOXA-40, and blaOXA-23) were identified (Supplementary Figure S1). Among the five species of A. baumannii group, A. baumannii, A. pittii, and A. lactucae genomes showed intrinsic class D β-lactamase belonging to blaOXA-51 and blaOXA-213 family genes, respectively, while A. nosocomialis and A. seifertii genomes did not show any intrinsic class D β-lactamase. Similarly, intrinsic class D β-lactamases were identified in other Acinetobacter species, such as blaOXA-134-like in A. lwoffii and A. schindleri, blaOXA-211 family in A. johnsonii, blaOXA-213 family in A. calcoaceticus, blaOXA-214 family in A. haemolyticus, blaOXA-228 family in A. bereziniae, blaOXA-286 family in A. viviani, A. disperses, and A. courvalini genomes, and blaOXA-294 family gene in A. proteoliticus, A. gyllenbergii, and A. colistinresistens (Figure 4). Also, the presence of class D carbapenemase genes belonging to blaOXA-23, blaOXA-40, or blaOXA-58 family genes were found in 16 of 39 A. baumannii genomes, 27 of 282 A. pittii genomes, 8 of 145 A. nosocomialis genomes, 2 of 2 A. seifertii genomes, 9 of 9 A. radioresistens genomes, 5 of 6 A. cumulans genomes, and 3 of 11 A. colistinresistens genomes (Figure 4). Moreover, 46 of 72 Acinetobacter species showed at least one antimicrobial resistance gene (Supplementary Table S2), and 91 isolates assigned to 17 species of the Acinetobacter genus showed at least three genes encoding for resistance to three different antimicrobial classes and were classified as MDR isolates (Magiorakos et al., 2012) (Supplementary Table S2; Figure 4).
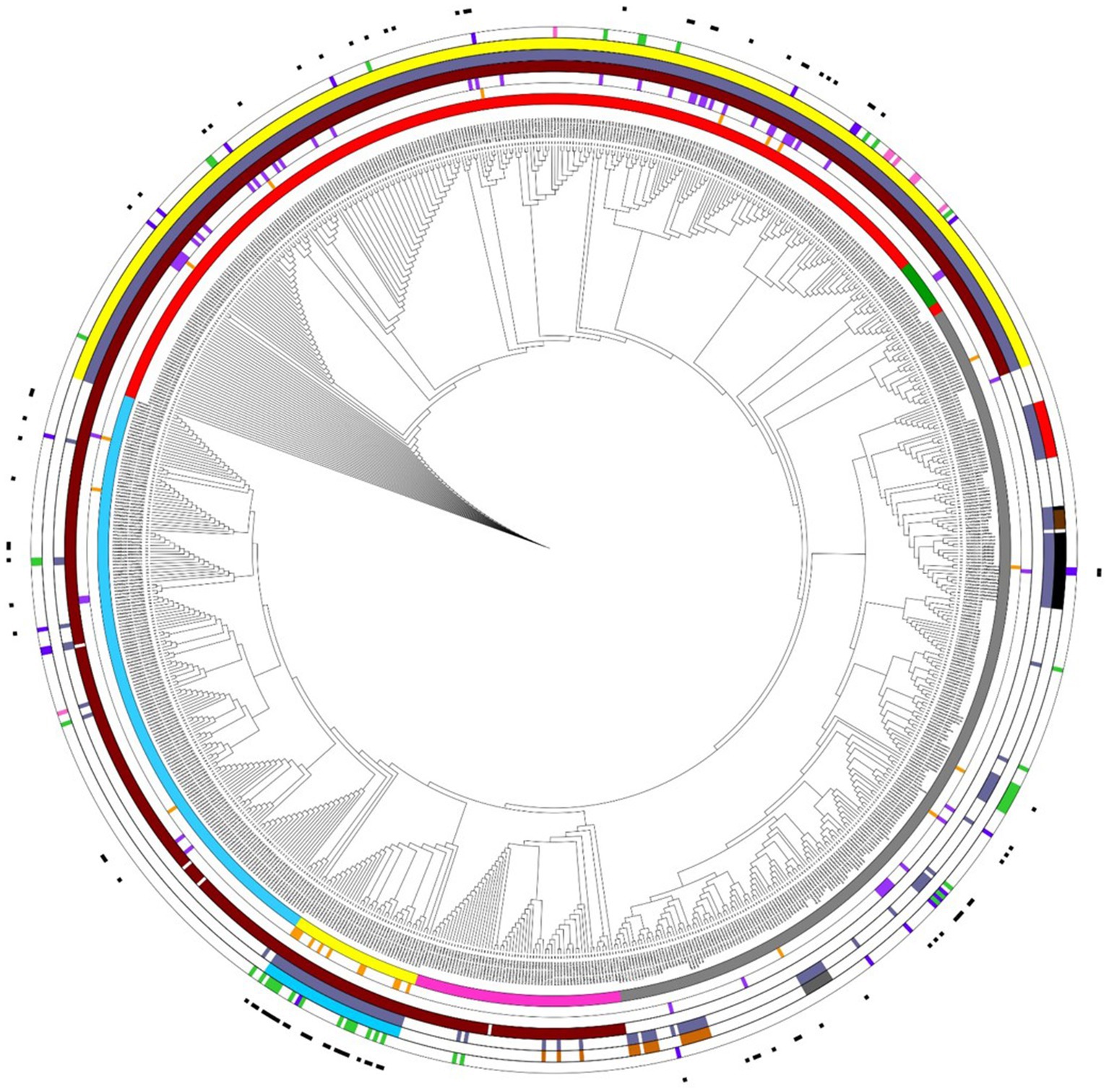
Figure 4. Antimicrobial resistance genes in a neighbor-joining tree of 837 Acinetobacter spp. genomes inferred on the Pasteur MLST scheme. The inner ring indicates the following species: A. baumannii (yellow labels), A. pittii (red labels), A. nosocomialis (blue labels), A. seifertii (pink labels), A. lactucae (green labels), and other 263 Acinetobacter spp. genomes (gray labels). The second inner ring (orange) indicates class A β-lactamase. The third inner ring (purple) indicates class B β-lactamase. The fourth inner ring (claret) indicates class C β-lactamase. The fifth inner ring (violet) indicates class D β-lactamase. The two external rings indicate the presence of at least one or two class D β-lactamases using the colored groups shown in Supplementary Figure S1. The external black rectangles indicate the isolates classified as MDR. The figure was obtained using iTol v6 (https://itol.embl.de/).
Discussion
Acinetobacter baumannii frequently causes healthcare-associated infections and is considered the most relevant clinical species of the Acinetobacter genus (Wong et al., 2017; Cools et al., 2019). Moreover, non-baumannii Acinetobacter species, such as A. nosocomialis, A. pittii, A. seifertii, and A. lactucae, showing phenotypic and genotypic characteristics similar to those of A. baumannii and included in the A. baumannii group, are increasingly reported as responsible for infections in humans (Cosgaya et al., 2017). Furthermore, novel taxa are currently isolated into the Acinetobacter genus (https://www.bacterio.net/genus/acinetobacter; accessed on June 2023), which are difficult to identify using standard phenotypic (Cools et al., 2019) and molecular techniques (Marí-Almirall et al., 2017). The present study analyzed the genomic features of 837 isolates assigned to 72 distinct species in the Acinetobacter genus. Our data demonstrated that the rMLST and Pasteur MLST schemes were able to genotype and identify at species levels isolates assigned to 72 distinct species in the Acinetobacter genus, thus providing useful and validated tools for the identification and characterization of Acinetobacter spp. genomes.
In addition, we analyzed the phylogeny of non-baumannii genomes of the A. baumannii group using core genome SNPs, or concatenated alleles of the Pasteur MLST and rMLST schemes, and compared clusters identified by core genome SNP and Pasteur MLST phylogeny. In keeping with previous data (Gaiarsa et al., 2019), core genome SNP phylogeny and Pasteur MLST phylogeny concordantly identified distinct clades and clonal lineages in A. baumannii genomes. The phylogenies of A. pittii genomes showed clusters 1, 2, and 3 identified by core genome SNPs, which corresponded to ST396, ST119, and ST207 clusters identified by the Pasteur MLST scheme, respectively, and ST63 cluster identified only by the Pasteur MLST scheme. This is in agreement with previous data showing the prevalence of ST119, ST63, and ST207 genotypic profiles among A. pittii isolates (Yang et al., 2012; Kamolvit et al., 2015; Sung et al., 2015; Zhang and Zhou, 2018; Chopjitt et al., 2021) and the identification of a monophyletic bacterial population and distinct clusters among A. pittii genomes (Chopjitt et al., 2021). Our data also showed that core genome SNP, Pasteur MLST, and rMLST-based phylogenies all included A. lactucae genomes into A. pittii genomes and identified A. lactucae genomes as singletons. Although A. lactucae (formerly A. dijkshoorniae) was identified as a distinct species of the Acinetobacter genus (Cosgaya et al., 2017; Marí-Almirall et al., 2017), maximum-likelihood phylogenies indicate that A. lactucae genomes cannot be distinguished from A. pittii genomes. Furthermore, core genome SNP phylogeny of A. seifertii genomes identified one single cluster (cluster 4) corresponding to Pasteur ST553, which emerged as a dominant clonal lineage in Asia (Li et al., 2021). As for A. nosocomialis genomes, no clades were identified by core genome SNP phylogeny, while one prevalent clade, ST410, was identified by Pasteur MLST phylogeny. This finding is in agreement with previous studies showing the selection of the ST410 genotype among A. nosocomialis epidemics (Chen et al., 2018, 2019; Jing et al., 2022). The data shown in this study are also in agreement with previous data showing that the population structure of A. nosocomialis genomes is highly heterogeneous (Jing et al., 2022).
The spread of epidemic A. baumannii clonal lineages has been favored by their carbapenem resistance and multidrug resistance (Zarrilli et al., 2013; Gaiarsa et al., 2019; Hamidian and Nigro, 2019). Likewise, A. nosocomialis, A. pittii, A. lactucae, and A. seifertii responsible for epidemics are carbapenem-resistant and MDR (Cosgaya et al., 2017; Wong et al., 2017). The analysis of antimicrobial resistance genes in the genomes of the 72 species of the Acinetobacter genus confirmed the presence of carbapenem resistance genes in A. baumannii, A. nosocomialis, A. pittii, and A. seifertii genomes belonging to A. baumannii group, while only 1 of 14 A. lactucae genomes carried blaNDM-1 carbapenemase and several antimicrobial resistance genes. In keeping with previous publications (Evans and Amyes, 2014; Cosgaya et al., 2017), the data reported herein showed that A. baumannii, A. lactucae, and A. pittii, but not A. nosocomialis and A. seifertii, were the species included in the A. baumannii group possessing a naturally occurring oxacillinase. The presence of class D beta-lactamases was found in 32 additional species of Acinetobacter genomes, thus reinforcing the evidence that this is a characteristic of the Acinetobacter genus (Evans and Amyes, 2014). In agreement with previous studies (Cosgaya et al., 2017; Hamidian and Nigro, 2019), we found the presence of blaOXA-23, blaOXA-40, and blaOXA-58 carbapenemase genes in A. baumannii, A. nosocomialis, A. pittii, A. seifertii, and other Acinetobacter species. Notably, all nine A. radioresistens genomes in our database showed the blaOXA-23 gene, which reinforced the evidence that A. radioresistens is the progenitor of the blaOXA-23 gene and the source of carbapenem resistance for A. baumannii (Poirel et al., 2008). Moreover, in agreement with previous data showing that the blaNDM type is the most common type of metallo-beta-lactamase contributing to carbapenem resistance in clinical isolates of A. baumannii (Zarrilli et al., 2013; Gaiarsa et al., 2019) and other Acinetobacter spp. (Yang et al., 2012; Yamamoto et al., 2013; Sung et al., 2015; Pfeifer et al., 2020; Alattraqchi et al., 2021), we found blaNDM type genes in the genomes of 12 species of the Acinetobacter genus.
Conclusion
The data presented herein analyze a genome collection of isolates assigned to 72 distinct species of Acinetobacter genus. The non-baumannii Acinetobacter genomes database, which has been validated by rMLST and Pasteur MLST, represents a useful tool for genome sequencing-based identification at the species level and typing of Acinetobacter spp. isolates.
The phylogenies of A. nosocomialis, A. lactucae, A. pittii, and A. seifertii genomes belonging to the A. baumannii group demonstrate the presence of distinct clades in the A. pittii and A. seifertii genomes, while the A. nosocomialis genomes are highly heterogeneous. “In silico” analysis of antimicrobial resistance in isolates assigned to 72 distinct species of the Acinetobacter genus shows the presence of carbapenemases and resistance genes to several antimicrobial classes in A. baumannii, A. nosocomialis, A. pittii, A. seifertii, and other Acinetobacter spp.
Pasteur MLST phylogeny of non-baumannii Acinetobacter isolates coupled with in silico detection of antimicrobial resistance is important to study the population structure and epidemiology of Acinetobacter spp. isolates.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
AM: Formal analysis, Writing – original draft, Data curation, Investigation, Methodology. JB: Data curation, Formal analysis, Writing – original draft, Software. MI: Software, Methodology, Writing – original draft. MS: Formal analysis, Investigation, Writing – original draft. GS: Methodology, Software, Writing – original draft. KJ: Methodology, Conceptualization, Data curation, Formal analysis, Writing – review & editing. SB: Conceptualization, Formal analysis, Methodology, Writing – review & editing. RZ: Conceptualization, Formal analysis, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the Italian Ministry of University and Research (MUR): PRIN2017 (grant no. 2017SFBER to RZ) and PRIN2022 (grant no. 202288EJ8B to RZ).
Acknowledgments
We thank the team of curators of the Pasteur Acinetobacter MLST and ribosomal MLST schemes for curating the data and making them publicly available at http://pubmlst.org/abaumannii/.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1264030/full#supplementary-material
Footnotes
References
Alattraqchi, A. G., Mohd Rani, F., Rahman, A., Ismail, S., Cleary, D. W., Clarke, S. C., et al. (2021). Complete genome sequencing of Acinetobacter baumannii AC1633 and Acinetobacter nosocomialis AC1530 unveils a large multidrug-resistant plasmid encoding the NDM-1 and OXA-58 Carbapenemases. mSphere. 6, e01076–e01020. doi: 10.1128/mSphere.01076-20
Bogdanowicz, D., Giaro, K., and Wróbel, B. (2012). TreeCmp: comparison of trees in polynomial time. Evol. Bioinformatics Online 8, EBO.S9657–EBO.S9487. doi: 10.4137/EBO.S9657
Bonnin, R. A., Ocampo-Sosa, A. A., Poirel, L., Guet-Revillet, H., and Nordmann, P. (2012). Biochemical and genetic characterization of carbapenem-hydrolyzing β-lactamase OXA-229 from Acinetobacter bereziniae. Antimicrob. Agents Chemother. 56, 3923–3927. doi: 10.1128/AAC.00257-12
Bortolaia, V., Kaas, R. F., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Bouvet, P. J. M., and Grimont, P. A. D. (1986). Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Bacteriol. 36, 228–240. doi: 10.1099/00207713-36-2-228
Chen, F. J., Huang, W. C., Liao, Y. C., Wang, H. Y., Lai, J. F., Kuo, S. C., et al. (2019). Molecular epidemiology of emerging Carbapenem resistance in Acinetobacter nosocomialis and Acinetobacter pittii in Taiwan, 2010 to 2014. Antimicrob. Agents Chemother. 63, e02007–e02018. doi: 10.1128/AAC.02007-18
Chen, H. Y., Yang, Y. S., Hsu, W. J., Chou, Y. C., Huang, L. S., Wang, Y. C., et al. (2018). Emergence of carbapenem-resistant Acinetobacter nosocomialis strain ST410 harbouring plasmid-borne blaOXA-72 gene in Taiwan. Clin. Microbiol. Infect. 24, 1023–1024. doi: 10.1016/j.cmi.2018.04.009
Chopjitt, P., Putthanachote, N., Ungcharoen, R., Hatrongjit, R., Boueroy, P., Akeda, Y., et al. (2021). Genomic characterization of clinical extensively drug-resistant Acinetobacter pittii isolates. Microorganisms. 9:242. doi: 10.3390/microorganisms9020242
Cools, P., Nemec, A., Kämpfer, P., and Vaneechoutte, M. (2019). Acinetobacter, Chryseobacterium, Moraxella, and other nonfermentative gram-negative rods. Manual of clinical microbiology., 12th ed. 829–857. http://hdl.handle.net/1854/LU-8605267
Cosgaya, C., Ratia, C., Marí-Almirall, M., Rubio, L., Higgins, P. G., Seifert, H., et al. (2017). In vitro and in vivo virulence potential of the emergent species of the Acinetobacter baumannii (ab) group. Front. Microbiol. 10:2429. doi: 10.3389/fmicb.2019.02429
Diancourt, L., Passet, V., Nemec, A., Dijkshoorn, L., and Brisseet, S. (2010). The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi: 10.1371/journal.pone.0010034
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Evans, B. A., and Amyes, S. G. B. (2014). OXA β-Lactamases. Clin. Microbiol. Rev. 27, 241–263. doi: 10.1128/CMR.00117-13
Feil, E. J., Li, B. C., Aanensen, D. M., Hanage, W. P., and Spratt, B. G. (2004). eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186, 1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004
Figueiredo, S., Bonnin, R. A., Poirel, L., Duranteau, J., and Nordmann, P. (2012). Identification of the naturally occurring genes encoding carbapenem-hydrolysing oxacillinases from Acinetobacter haemolyticus, Acinetobacter johnsonii, and Acinetobacter calcoaceticus. Clin. Microbiol. Infect. 18, 907–913. doi: 10.1111/j.1469-0691.2011.03708.x
Figueiredo, S., Poirel, L., Seifert, H., Mugnier, P., Benhamou, D., and Nordmann, P. (2010). OXA-134, a naturally occurring carbapenem-hydrolyzing class D beta-lactamase from Acinetobacter lwoffii. Antimicrob. Agents Chemother. 54, 5372–5375. doi: 10.1128/AAC.00629-10
Gaiarsa, S., Biffignandi, G. B., Esposito, E. P., Castelli, M., Jolley, K. A., Brisse, S., et al. (2019). Comparative analysis of the two Acinetobacter baumannii multilocus sequence typing (MLST) schemes. Front Microbiol. 10:930. doi: 10.3389/fmicb.2019.00930
Graña-Miraglia, L., Evans, B. A., López-Jácome, L. E., Hernández-Durán, M., Colín-Castro, C. A., Volkow-Fernández, P., et al. (2020). Origin of OXA-23 variant OXA-239 from a recently emerged lineage of Acinetobacter baumannii international clone V. mSphere 5:e00801–19. doi: 10.1128/mSphere.00801-19
Gundi, V. A., Dijkshoorn, L., Burignat, S., Raoult, D., and La Scola, B. (2009). Validation of partial rpoB gene sequence analysis for the identification of clinically important and emerging Acinetobacter species. Microbiology 155, 2333–2341. doi: 10.1099/mic.0.026054-0
Hamidian, M., and Nigro, S. J. (2019). Emergence, molecular mechanisms and global spread of carbapenem resistant Acinetobacter baumannii. Microb Genom. 5:e000306. doi: 10.1099/mgen.0.000302
Héritier, C., Poirel, L., Fournier, P. E., Claverie, J. M., Raoult, D., and Nordmann, P. (2005). Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49, 4174–4179. doi: 10.1128/AAC.49.10.4174-4179.2005
Jing, L., Xu, Z., Zhang, Y., Li, D., Song, Y., Hu, H., et al. (2022). Metagenomic insights into pathogenic characterization of ST410 Acinetobacter nosocomialis prevalent in China. Pathogens. 11:838. doi: 10.3390/pathogens11080838
Jolley, K. A., Bliss, C. M., Bennett, J. S., Bratcher, H. B., Brehony, C., Colles, F. M., et al. (2012). Ribosomal multilocus sequence typing: universal characterization of bacteria from domain to strain. Microbiology 158, 1005–1015. doi: 10.1099/mic.0.055459-0
Jolley, K. A., Bray, J. E., and Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
Kamolvit, W., Derrington, P., Paterson, D. L., and Sidjabat, H. E. (2015). A case of IMP-4-, OXA-421, OXA-96-, and CARB-2-producing Acinetobacter pittii sequence type 119 in Australia. J. Clin. Microbiol. 53, 727–730. doi: 10.1128/JCM.02726-14
Lewis, P. O. (2001). A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 50, 913–925. doi: 10.1080/106351501753462876
Li, L. H., Yang, Y. S., Sun, J. R., Huang, T. W., Huang, W. C., Chen, F. J., et al. (2021). Clinical and molecular characterization of Acinetobacter seifertii in Taiwan. J. Antimicrob. Chemother. 76, 312–321. doi: 10.1093/jac/dkaa432
Magiorakos, A. P., Srinivasan, A. R., Carey, B., Carmeli, Y., Falagas, E. M., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Marí-Almirall, M., Cosgaya, C., Higgins, P. G., Van Assche, A., Telli, M., Huys, G., et al. (2017). MALDI-TOF/MS identification of species from the Acinetobacter baumannii (ab) group revisited: inclusion of the novel A. Seifertii and A. Dijkshoorniae species. Clin. Microbiol. Infect. 23, 210.e1–210.e9. doi: 10.1016/j.cmi.2016.11.020
Pfeifer, Y., Trifonova, A., Pietsch, M., Brunner, M., Todorova, I., Gergova, I., et al. (2020). Clonal transmission of gram-negative bacteria with Carbapenemases NDM-1, VIM-1, and OXA-23/72 in a Bulgarian hospital. Microb. Drug Resist. 23, 301–307. doi: 10.1089/mdr.2016.0059
Poirel, L., Figueiredo, S., Cattoir, V., Carattoli, A., and Nordmann, P. (2008). Acinetobacter radioresistens as a silent source of carbapenem resistance for Acinetobacter spp. Antimicrob. Agents Chemother. 52, 1252–1256. doi: 10.1128/AAC.01304-07
Ribeiro-Gonçalves, B., Francisco, A. P., Vaz, C., Ramirez, M., and Carriço, J. A. (2016). PHYLOViZ online: web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic Acids Res. 44, W246–W245. doi: 10.1093/nar/gkw359
Šedo, O., Radolfová-Křížová, L., Nemec, A., and Zdráhal, Z. (2018). Limitations of routine MALDI-TOF mass spectrometric identification of Acinetobacter species and remedial actions. J. Microbiol. Methods 154, 79–85. doi: 10.1016/j.mimet.2018.10.009
Shimodaira, H., and Hasegawa, M. (1999). Multiple comparison of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16, 1114–1116. doi: 10.1093/oxfordjournals.molbev.a026201
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Sung, J. Y., Koo, S. H., Kim, S., and Kwon, G. C. (2015). Emergence of Acinetobacter pittii harboring New Delhi metallo-beta-lactamase genes in Daejeon. Korea. Ann Lab Med. 35, 531–534. doi: 10.3343/alm.2015.35.5.531
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524. doi: 10.1186/s13059-014-0524-x
Whiteway, C., Breine, A., Philippe, C., and Van der Henst, C. (2022). Acinetobacter baumannii. Trends Microbiol. 30, 199–200. doi: 10.1016/j.tim.2021.11.008
Wong, D., Nielsen, T. B., Bonomo, R. A., Pantapalangkoor, P., Luna, B., and Spellberg, B. (2017). Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin. Microbiol. Rev. 30, 409–447. doi: 10.1128/CMR.00058-16
Yamamoto, M., Nagao, M., Matsumura, Y., Hotta, G., Matsushima, A., Ito, Y., et al. (2013). Regional dissemination of Acinetobacter species harbouring metallo-β-lactamase genes in Japan. Clin. Microbiol. Infect. 19, 729–736. doi: 10.1111/1469-0691.12013
Yang, J., Chen, Y., Jia, X., Luo, Y., Song, Q., Zhao, W., et al. (2012). Dissemination and characterization of NDM-1-producing Acinetobacter pittii in an intensive care unit in China. Clin. Microbiol. Infect. 18, E506–E513. doi: 10.1111/1469-0691.12035
Zarrilli, R., Pournaras, S., Giannouli, M., and Tsakris, A. (2013). Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int. J. Antimicrob. Agents 41, 11–19. doi: 10.1016/j.ijantimicag.2012.09.008
Keywords: Acinetobacter baumannii group, Acinetobacter spp., Pasteur Multilocus Sequence Typing, ribosomal Multilocus Sequence Typing, maximum-likelihood phylogeny, antimicrobial resistance genes
Citation: Migliaccio A, Bray J, Intoccia M, Stabile M, Scala G, Jolley KA, Brisse S and Zarrilli R (2023) Phylogenomics of Acinetobacter species and analysis of antimicrobial resistance genes. Front. Microbiol. 14:1264030. doi: 10.3389/fmicb.2023.1264030
Edited by:
Maria Soledad Ramirez, California State University, United StatesReviewed by:
German Matias Traglia, Universidad de La Republica, UruguaySonia Alejandra Gomez, National Scientific and Technical Research Council (CONICET), Argentina
Copyright © 2023 Migliaccio, Bray, Intoccia, Stabile, Scala, Jolley, Brisse and Zarrilli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raffaele Zarrilli, cmFmemFycmlAdW5pbmEuaXQ=
 Antonella Migliaccio
Antonella Migliaccio James Bray
James Bray Michele Intoccia
Michele Intoccia Maria Stabile
Maria Stabile Giovanni Scala
Giovanni Scala Keith A. Jolley
Keith A. Jolley Sylvain Brisse
Sylvain Brisse Raffaele Zarrilli
Raffaele Zarrilli