- Clinical Microbiology Laboratory, The Second Affiliated Hospital of Zhejiang University School of Medicine, Zhejiang University, Hangzhou, China
Introduction: Ceftazidime/avibactam (CZA) is an effective alternative for the treatment of infections caused by KPC-producing carbapenem-resistant Klebsiella pneumoniae (CRKP). However, KPC variants with CZA resistance have been observed in clinical isolates, further limiting the treatment options of clinical use.
Methods: In this study, we isolated three KPC-14-producing CRKP from two patients in intensive care units without CZA therapy. The antimicrobial susceptibility was determined using the broth microdilution method. Three CRKP were subjected to whole-genome sequencing to analyze the phylogenetic relatedness and the carriage of antimicrobial resistance genes and virulence factors. Long-read sequencing was also performed to obtain the complete sequences of the plasmids. The horizontal transfer of the blaKPC-14 gene was evaluated by conjugation experiments.
Results: Three CRKP displayed resistance or reduced susceptibility to ceftazidime/avibactam, colistin, and tigecycline. Single-nucleotide polymorphism (SNP) analysis demonstrated the close phylogenetic distance between these strains. A highly similar IncFII/IncR plasmid encoding blaKPC-14 was shared by three CRKP, with blaKPC-14 located in an NTEKPC-Ib element with the core region of ISKpn27- blaKPC-14-ISKpn6. This structure containing blaKPC-14 was also observed in another tet(A)-carrying plasmid that belonged to an unknown Inc-type in two out of three isolates. The horizontal transferability of these integrated plasmids to Escherichia coli EC600 was confirmed by the cotransmission of tet(A) and blaKPC-14 genes, but the single transfer of blaKPC-14 on the IncFII/IncR plasmid failed. Three CRKP expressed yersiniabactin and carried a hypervirulence plasmid encoding rmpA2 and aerobactin-related genes, and were thus classified as carbapenem-resistant hypervirulent K. pneumoniae (hvKP).
Discussion: In this study, we reported the evolution of a mosaic plasmid encoding the blaKPC-14 gene via mobile elements in extensively drug-resistant hvKP. The blaKPC-14 gene is prone to integrate into other conjugative plasmids via the NTEKPC-Ib element, further facilitating the spread of ceftazidime/avibactam resistance.
1. Introduction
The widespread of carbapenem-resistant Klebsiella pneumoniae (CRKP) is considered an urgent threat to public health, as it complicates patient care and increases morbidity and mortality in cases of infection (Wang M. et al., 2022; Pérez-Galera et al., 2023). Available data from the China Antimicrobial Surveillance Network (CHINET1) showed that the prevalence of CRKP has rapidly increased in China, from 2.9% in 2005 to 24.2% in 2022. Colistin and tigecycline constitute some of the last resorts for the treatment of CRKP infections, however, resistance to these antibiotics in CRKP strains has also been reported recently, further reducing the repertoire of useful antibiotics (Chen et al., 2021; Tian et al., 2021).
Ceftazidime/avibactam (CZA), a novel β-lactam/β-lactamase inhibitor combination, is an effective alternative for the treatment of CRKP infections (Van Duin and Bonomo, 2016). This combination shields ceftazidime from breakdown by Ambler class A, class C, and some class D β-lactamases and thus exhibits potent inhibition of strains producing KPC and OXA-48-like carbapenemases (Criscuolo and Trecarichi, 2020). KPC-producing CRKP is widespread globally and is the predominant type of CRKP in China, which is frequently related to nosocomial outbreaks (Findlay et al., 2021; Wang L. et al., 2022; Wang M. et al., 2022). Although recent studies have shown evidence for CZA as a promising option for such infections, resistance to this antibiotic has rapidly evolved, mainly due to the production of variants of KPC-2 or KPC-3 enzymes (Humphries and Hemarajata, 2017; El-Kady et al., 2022). The single amino acid substitution that confers CZA resistance was commonly encountered in the omega loop (positions 164–179), particularly for the Asp179Tyr (D179Y) mutation in KPC-3 (KPC-31) and KPC-2 (KPC-33) (Livermore et al., 2015; Barnes et al., 2017). Additionally, KPC variants with CZA-resistance mediated by amino acid changes outside the omega loop region (e.g., KPC-41, KPC-23, KPC-14, KPC-8, KPC-123, and KPC-93) were also observed in the clinical isolates from patients following CZA therapy and those who were not treated with CZA (Bianco et al., 2021; Liu et al., 2022b; Wang L. et al., 2022). KPC-14, the variant with a deletion of two amino acids (Δ242-GT-243) of KPC-2 that exhibits CZA resistance, has been sporadically detected in clinical isolates of CRKP (Bianco et al., 2020; Niu et al., 2020; Linh et al., 2021; Jiang et al., 2022).
Here, we investigated the genetic relationship of CRKP harboring two structurally distinct blaKPC-14-encoding plasmids and analyzed the evolution of one plasmid that was able to undergo horizontal transfer between Enterobacterales.
2. Materials and methods
2.1. Patients and bacterial strains
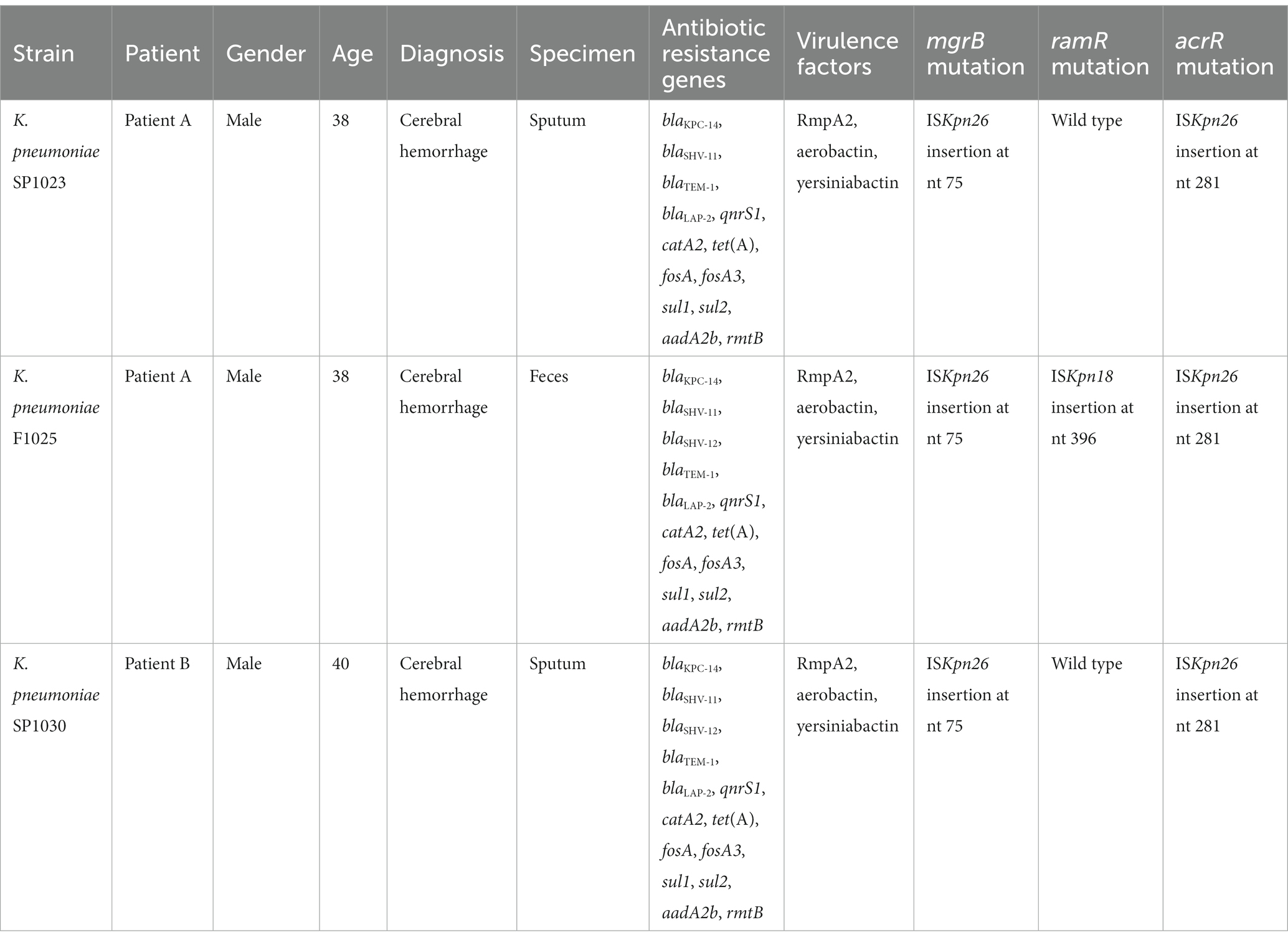
Three CRKP (strains SP1023 and F1025 from Patient A, and strain SP1030 from Patient B) were isolated from two patients admitted to the neurology intensive care unit (NICU) of a tertiary hospital in Hangzhou City in 2022. Species identification was determined by MALDI-TOF MS (Bruker Daltonik GmbH, Bremen, Germany). This study was approved by the Ethics Committee of The Second Affiliated Hospital of Zhejiang University School of Medicine.
Patient A, a 38-year-old male, had poorly controlled hypertension for several years. He underwent three consecutive intracranial hematoma evacuations at a local hospital due to cerebellar hemorrhage. Blood cultures revealed carbapenem-resistant Acinetobacter baumannii (CRAB) and a combination of cefoperazone/sulbactam (1,1, 2 g IV every 6 h) and polymyxin B (750,000 IU IV every 12 h) was administered (Figure 1A). The patient was in a coma and was transferred to the NICU of our hospital for further treatment. Upon admission, the patient received supportive treatments, including mechanical ventilation, blood transfusion, fluid replacement, and appropriate medications. The original antimicrobial therapy regimen was continued. The subsequent sputum culture revealed the growth of Klebsiella aerogenes and cefoperazone/sulbactam- and carbapenem-resistant A. baumannii. Therefore, cefoperazone/sulbactam was replaced with tigecycline (100 mg IV every 12 h) on Day 7. Nine days later (Day 16), both organisms were cleared. However, CRKP (strain SP1023) exhibiting ceftazidime/avibactam and polymyxin B resistance was isolated from the sputum sample (Figure 1A). On Day 18, fecal screening for CRE yielded pandrug-resistant K. pneumoniae (strain F1025), which was resistant to ceftazidime/avibactam, polymyxin B, and tigecycline (Figure 1A). In addition to CRKP, Pseudomonas aeruginosa and carbapenem-resistant A. baumannii (CRAB) were detected in the sputum sample on Day 32. Meropenem was used (1 g IV every 12 h) instead of the previous antimicrobials, and only CRKP was cleared after 9 days of treatment. Tigecycline was used again, and amikacin (400 mg nasogastric feeding every 12 h) was added to the treatment regimen 5 days later. However, P. aeruginosa that developed carbapenem resistance and CRAB persisted in the patient’s respiratory tract. Due to his extremely poor condition and acute exacerbation of chronic renal failure, the patient died of multiple organ failure on Day 68.
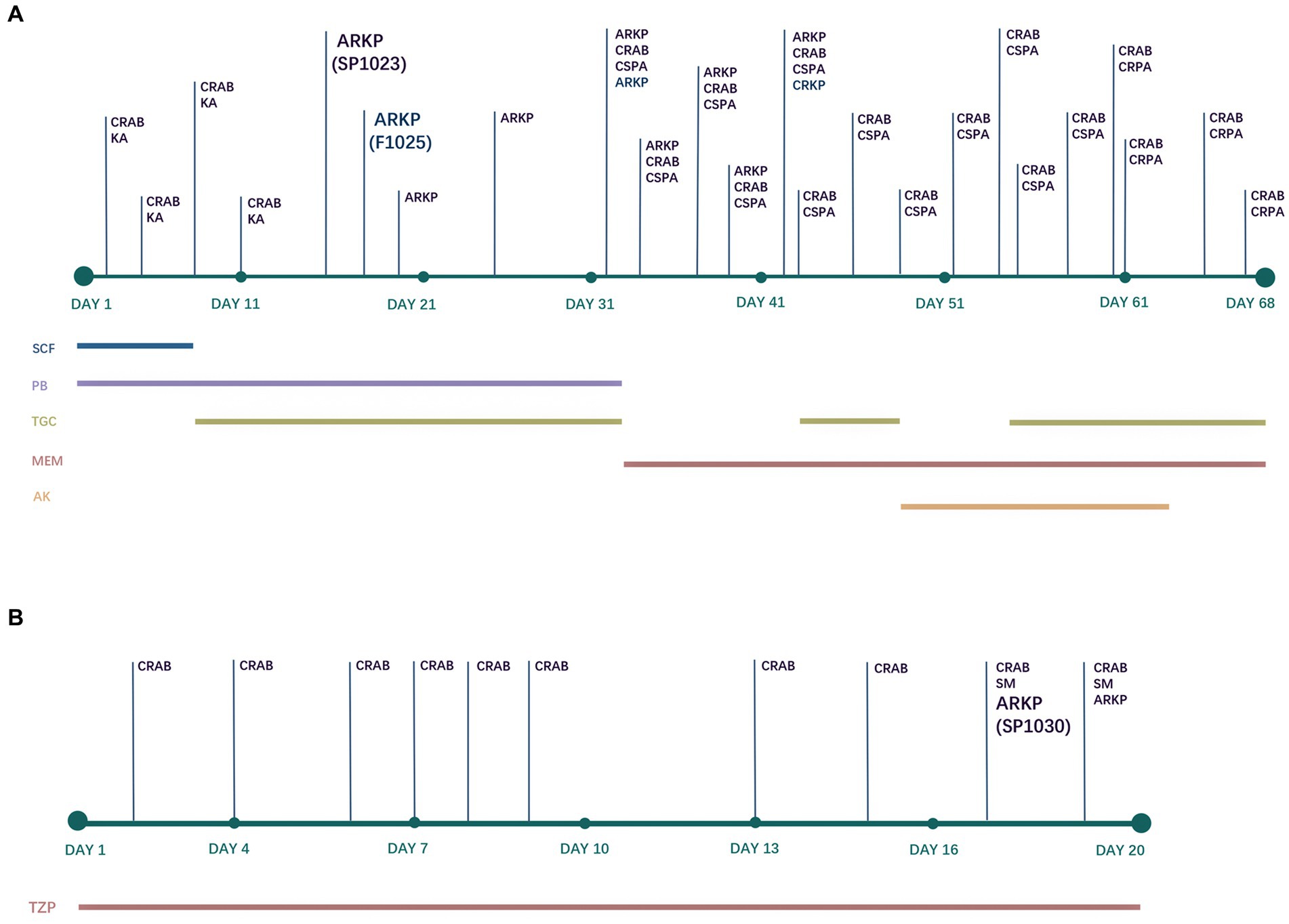
Figure 1. Antimicrobial treatments and bacterial isolation of Patient A (A) and Patient B (B). CRAB, carbapenem-resistant Acinetobacter baumannii; KA, Klebsiella aerogenes; ARKP, ceftazidime/avibactam-resistant Klebsiella pneumoniae; CSPA, carbapenem-susceptible Pseudomonas aeruginosa; CRPA, carbapenem-resistant Pseudomonas aeruginosa; CRKP, carbapenem-resistant Klebsiella pneumoniae (ceftazidime/avibactam-susceptible); SM, Stenotrophomonas maltophilia; SCF, cefoperazone/sulbactam; PB, polymyxin B; TGC, tigecycline; MEM, meropenem; AK, amikacin; TZP, piperacillin/tazobactam.
Patient B, a 40-year-old male with spontaneous intracerebral hemorrhage, underwent surgical evacuation of the intracranial hematoma at a local hospital. Six days later, he remained in a coma and was transferred to the NICU of our hospital. The CT scans showed postoperative hematoma in the surgical area and scattered infiltrates in both lungs. The patient received supportive treatments to reduce intracranial pressure, sedation, pain management, enteral nutrition, endotracheal intubation, and mechanical ventilation. Additionally, piperacillin/tazobactam (8:1, 4.5 g IV every 6 h) was administered for antimicrobial therapy throughout the hospitalization period (Figure 1B). On the second day of admission, CRAB was isolated from the sputum sample and persisted until discharge. On Day 17 and Day 19, CRKP (strain SP1030) was detected in the sputum culture that was resistant to ceftazidime/avibactam and colistin (Figure 1B). After 8 days of mechanical ventilation, the patient was weaned off the ventilator (Day 15). Since the patient’s vital signs stabilized and his mental function recovered, he was discharged for further treatment at a rehabilitation hospital on Day 20.
2.2. Antimicrobial susceptibility tests
The minimal inhibitory concentrations (MICs) of 18 antimicrobial agents, including imipenem, meropenem, ertapenem, ceftazidime/avibactam, ceftazidime, cefotaxime, cefepime, piperacillin/tazobactam, cefoperazone/sulbactam, cefmetazole, aztreonam, ciprofloxacin, amikacin, chloramphenicol, fosfomycin, tetracycline, tigecycline, and colistin, were determined using the broth microdilution method (Clinical and Laboratory Standards Institute, 2018) and interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (Clinical and Laboratory Standards Institute, 2021). Tigecycline susceptibility was interpreted using breakpoints recommended by the US Food and Drug Administration.2 Escherichia coli ATCC 25922, K. pneumoniae 700603, and Pseudomonas aeruginosa ATCC 27583 were used as the quality control strains in parallel.
2.3. Whole genome sequencing and genome analysis
To investigate the evolution and genetic relatedness of these CRKP, the genomic DNA of three CRKP (strains SP1023, F1025, and SP1030) was subjected to WGS by both the short-read Illumina NovaSeq 6000 platform and the hybrid long-read Oxford Nanopore PromethION 48 platform. The complete genome was assembled by Flye assembler v2.9.2 (Kolmogorov et al., 2020) and polished by Pilon v1.24 (Walker et al., 2014). The antimicrobial resistance genes and the plasmid types for the assembly scaffolds were identified by ResFinder 4.1 and PlasmidFinder 2.0, respectively, at the Center for Genomic Epidemiology.3 The sequence types and virulence factors were identified using Kleborate v0.3.0 (Lam et al., 2021). A pairwise comparison of genomes and variant callings for single-nucleotide polymorphisms (SNPs) was conducted using Snippy v4.4.5 with default settings. The plasmids encoding blaKPC-14 and virulence-associated genes were annotated by the RAST server (Overbeek et al., 2014) and BLASTN program. The comparison of plasmids was visualized and annotated by BRIG v0.95 (Alikhan et al., 2011).
2.4. Conjugation experiment
The transferability of blaKPC-14 genes was estimated by conjugation experiments with filter mating methods (Cai et al., 2008). Rifampin-resistant E. coli EC600 was used as the recipient strain. The putative transconjugants grown on selective media supplemented with 8 mg/L CZA or 30 mg/L tetracycline were identified by MALDI-TOF MS and screened for the presence of blaKPC-14 genes. The conjugation frequency equaled the number of transconjugants divided by the number of recipients.
2.5. Virulence testing in the Galleria mellonella infection model
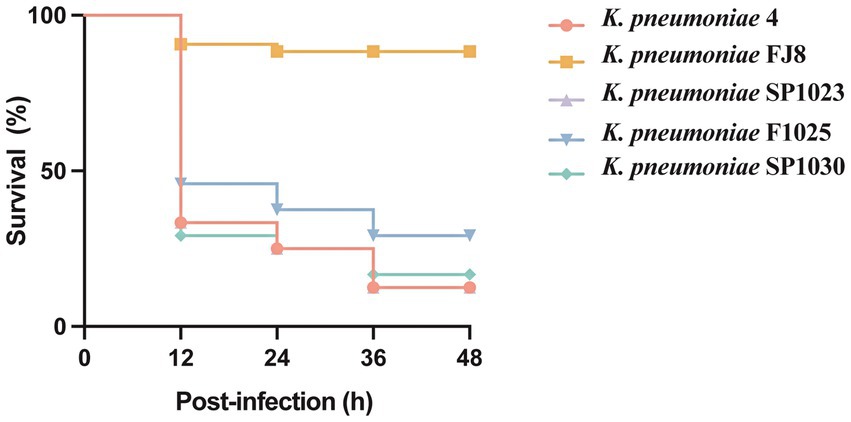
The G. mellonella (wax moth larvae) infection model was used to confirm the hypervirulent phenotype of the CRKP strains as previously described (McLaughlin et al., 2014). Overnight cultures of K. pneumoniae were diluted in sterile phosphate-buffered saline to obtain a concentration of 108 CFU/mL. Wax moth larvae weighing 250–300 mg (Tianjin Huiyude Biotech Company, Tianjin, China) were injected with 10 μL bacterial suspension and incubated for 48 h at 35°C. The survival rate of G. mellonella was recorded at 12 h, 24 h, 36 h, and 48 h. ST11 K. pneumoniae FJ8 without virulence factors and the hypervirulent K. pneumoniae 4 were used as the negative and positive controls, respectively (Gu et al., 2018). All experiments were performed in triplicate. Kaplan–Meier survival curves were plotted using Prism 9.
2.6. Nucleotide sequence accession numbers
The complete genome of the chromosome and plasmids for K. pneumoniae SP1023, F1025, and SP1030 were downloaded with BioSample accession numbers SAMN36464959, SAMN36465167, and SAMN36465169, respectively.
3. Results
3.1. Antimicrobial susceptibility results
As Table 1 illustrated, three CRKP shared a high-level resistance to CZA with MIC values of >64/4 mg/L and showed resistance or decreased susceptibility to meropenem and ertapenem. These strains also shared an extensive drug resistance (XDR) profile (Magiorakos et al., 2012) to ceftazidime, cefotaxime, cefepime, aztreonam, ciprofloxacin, amikacin, chloramphenicol, fosfomycin, and tetracycline but retained susceptibility to imipenem. More worrisome, resistance to colistin was observed in these CRKP, and K. pneumoniae F1024 exhibited additional resistance to another clinically important antibiotic, tigecycline, which was interpreted as resistance to almost all the antimicrobial agents frequently used in clinical settings.
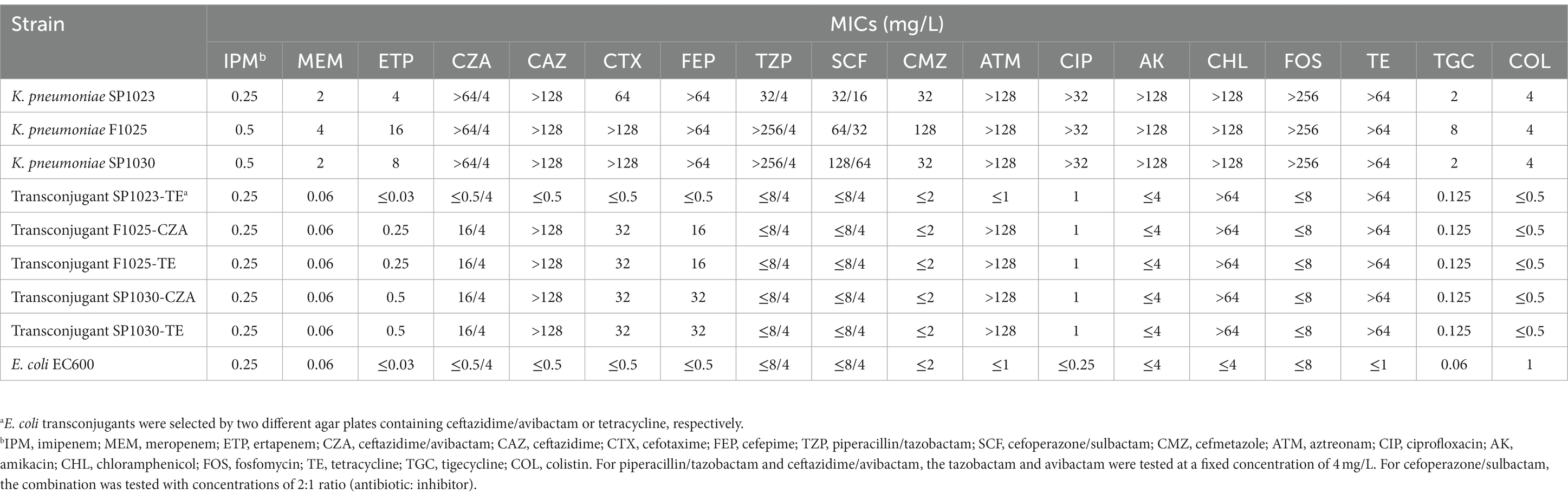
Table 1. Antimicrobial susceptibility results of K. pneumoniae isolates and their E. coli transconjugants.
3.2. Whole genome analysis of KPC-14-producing isolates
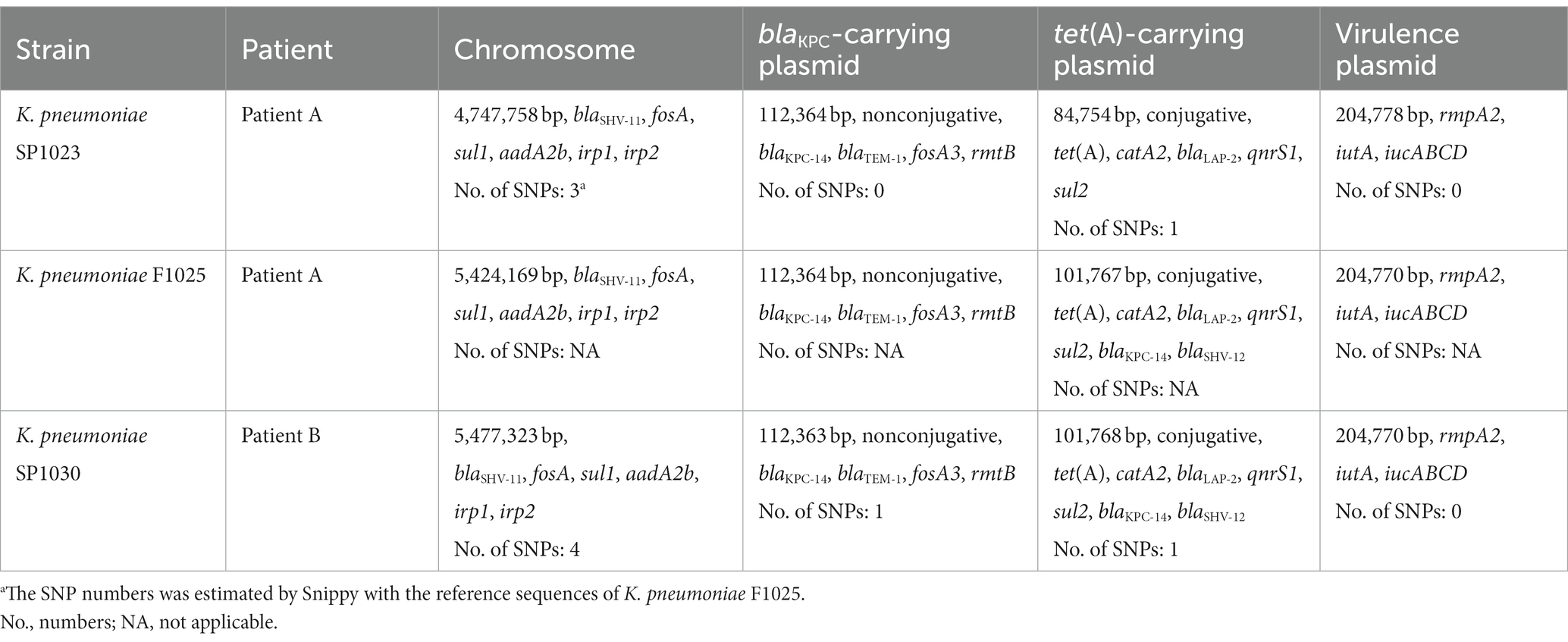
All three CRKP were identified as the ST11 type, which was the predominant ST type of CRKP in China (Liu et al., 2022a). Moreover, these three strains all belonged to the K64 serotype. Genome-based phylogenetic analysis suggested that these CRKP were closely related within 27 SNPs (with strain F1025 as the reference genome), indicating that these XDR strains originated from the same clone.
Whole-genome analysis demonstrated that three CRKP exhibited a similar carriage profile of β-lactamase genes, including blaKPC-14, blaSHV-11, blaTEM-1, and blaLAP-2, while K. pneumoniae F1025 and SP1030 additionally expressed SHV-12 (Table 2). However, the amplification of the blaKPC gene failed both in A.baumannii and P. aeruginosa isolated from Patient A. Multiple antimicrobial resistance genes were also identified in three CRKP, conferring resistance to quinolones (qnrS1), phenicols (catA2), sulfonamides (sul1 and sul2), tetracyclines [tet(A)], fosfomycin (fosA and fosA3), and aminoglycosides (aadA2b and rmtB). Detailed analysis showed three CRKP shared that mutations for type 1 Tet(A) variants (I5R, V55M, I75V, T84A, S201A, F202S, and V203F) and the insertion of ISKpn26 elements at position 75 in the acrR gene, both of which contributed to the elevated MICs of tigecycline (Chiu et al., 2017). Insertional inactivation by ISKpn18 was also detected in ramR, another tigecycline resistance determinant gene, further driving the generation of resistance to this antibiotic in K. pneumoniae SP1025. The mgrB genes was interrupted by ISKpn26 at the same position (nucleotide 75) in three CRKP, thus accounting for the resistance to colistin.
3.3. Transferability of blaKPC-14-carrying plasmids
The blaKPC-14 gene could be conjugated into E. coli EC600 from K. pneumoniae F1025 and SP1030 with similar conjugation efficiencies of 2.1 × 10−5 and 3.5 × 10−5, respectively, but conjugation failed in strain SP1023, suggesting a different location of the blaKPC-14 gene among these strains. Elevated CZA and several β-lactam MIC values were observed in E. coli transconjugants for K. pneumoniae F1025 and SP1030, confirming the functionality of blaKPC-14 (Table 1). Notably, these E. coli transconjugants also showed resistance to tetracycline and slightly higher tigecycline MIC values, indicating the possible cotransfer of tet(A) and blaKPC-14 genes. Therefore, selective media containing tetracycline were used to screen for putative transconjugants. Each of the three CRKP was able to transfer the tet(A) gene to the recipient E. coli EC600 with a similar efficiency at approximately 10−5; however, E. coli transconjugants with different donors displayed heterogeneity in antimicrobial susceptibility profiles. Unlike the E. coli transconjugants F1025-TE and SP1030-TE (with K. pneumoniae F1025 and SP1030 as donors, respectively), which displayed similar profiles to those of their counterparts for blaKPC-14, the E. coli transconjugant SP1023-TE (with K. pneumoniae SP1023 as the donor) showed decreased susceptibility to tetracycline and tigecycline but retained the same MIC values for CZA and other β-lactams as the recipient strain. The amplification of blaKPC-14 was also carried out in E. coli transconjugants F1025-TE and SP1030-TE, but failed in SP1023-TE, further supporting the dissimilar blaKPC-14 gene locations and plasmid carriage of the three CRKP.
3.4. Molecular analysis of blaKPC-14-carrying plasmids
To clarify the location and genetic platforms of blaKPC-14 genes, hybrid long-read sequencing of all three strains was performed. This yielded the complete genome for three CRKP with similar sizes of approximately 6 Mbp, consisting of one chromosome and varied numbers of plasmids (Table 3). A similar IncFII/IncR hybrid plasmid encoding blaKPC-14 was harbored by all three CRKP, designated as plasmids pSP1023-KPC, pF1025-KPC, and pSP1030-KPC for K. pneumoniae SP1023, F1025, and SP1030, respectively. These plasmids carried a variety of additional antimicrobial resistance determinants, including the blaTEM-1, fosA3, and rmtB genes, with sizes ranging from 112,363–112,364 bp. Blast analysis demonstrated that the backbone of this plasmid showed the highest similarity to plasmid pCRKP66R-3 with 100% identity (100% coverage, GenBank accession CP063835), which harbored the blaKPC-2 gene. These plasmids also displayed 100% identity to two other blaKPC-2-encoding plasmids named plasmid pC76-KPC (86% coverage, GenBank accession CP080299) and pCT77-KPC (87% coverage, GenBank accession CP080305), both of which were previously discovered in CRKP in our hospital (Figure 2) (Chen et al., 2021). The blaKPC-14 gene was in the non-Tn4401 element as an NTEKPC-Ib-like transposon with a genetic array of “IS26-ΔTn3-ISKpn8-blaKPC-14-ISKpn6-korC-klcA-rep-orf-IS26,” which was commonly and uniquely identified in ST11 KPC-producing CRKP (Yang et al., 2021). Consistent with many IncFIIK/IncR blaKPC-harboring plasmids widely disseminated in China, this plasmid was nonconjugative, which was probably due to the absence of relaxase, thus explaining the failure of the single transfer of blaKPC-14 in the conjugation experiment.
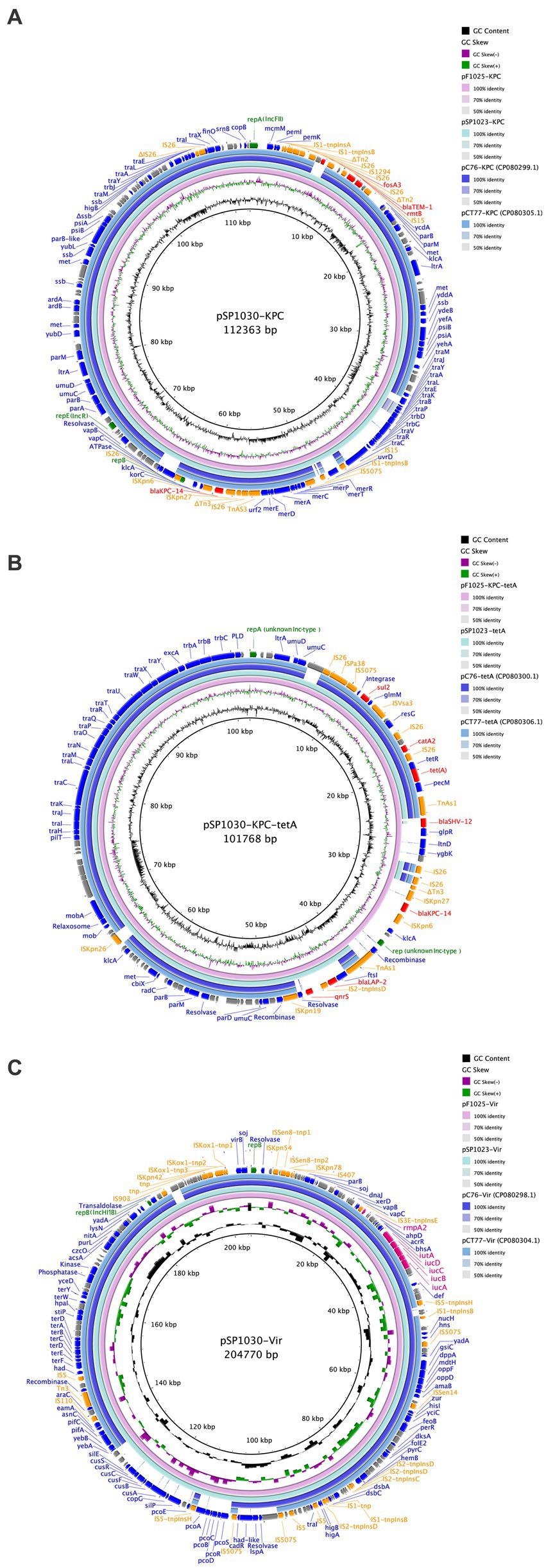
Figure 2. Comparisons of blaKPC-carrying plasmid (A), tet(A)-carrying plasmid (B), and the virulence plasmid (C) with the reference plasmids. The circles from inside to outside display the scale in kilobase pairs, the GC skew, the GC content, the similarity to the reference plasmids, and the annotation of the plasmid in our study, respectively. All insertion sequences are labeled orange, antimicrobial resistance genes are labeled red, genes encoding proteins with known functions are labeled blue, rep genes are labeled green, and the hypothetical proteins are labeled gray.
The type 1 Tet(A) variant colocalized with the catA2, blaLAP-2, qnrS1, and sul2 genes on unknown Inc-type plasmids with sizes of 84,754 bp, 101,767 bp, and 101,768 bp for K. pneumoniae SP1023, F1025, and SP1030, respectively (Figure 2A). This backbone was also observed in tet(A)-harboring plasmids in K. pneumoniae C76 and K. pneumoniae CT77 with 100% identity (91% coverage) but integrated with a 4,956 bp DNA fragment (based on K. pneumoniae SP1030) containing the blaLAP-2 and qnrS1 genes. Unlike plasmid pSP1023-tetA in K. pneumoniae SP1023, the blaKPC-14 locus of 8,248 bp was inserted into the tet(A)-carrying plasmids in K. pneumoniae F1025 and SP1030, which were further designated as plasmid pF1025-KPC-tetA and pSP1030-KPC-tetA, respectively. Another resistance locus harboring the blaSHV-12 gene was also observed in these two plasmids but was absent in plasmid pSP1023-tetA. The structural discrepancy in these tet(A)-carrying plasmids echoed the differences in the genetic and phenotypic profiles of the transconjugants we noticed. These observations indicated that the plasmids carrying both tet(A) and blaKPC-14 genes possibly evolved from those with a single occurrence of the tet(A) gene, such as pSP1023-tetA, which could also be further traced back to previously identified plasmids. The blaKPC-14 genes were flanked by an NTEKPC-Ib-like transposon similar to that on the IncFII/IncR plasmids in our study, indicating that these nonconjugative plasmids might be the source of the blaKPC-14-carrying fragments for plasmids pF1025-KPC-tetA and pSP1030-KPC-tetA. As previously reported, the diversity of mobile elements and transposons in NTEKPC-I elements actively promoted the transposition of the blaKPC genes to various genetic locations (Yang et al., 2021); thus, we speculated that the mobilization event mediated the integration of the blaKPC-14 locus into the backbones of tet(A)-carrying plasmids, thus creating the binary carriage profile of blaKPC-14-harboring plasmids in K. pneumoniae F1025 and SP1030. The integrated plasmids retained the fully functional conjugative genes homologous to the previous tet(A)-carrying plasmids (Figure 2), further facilitating the cotransfer and dissemination of KPC-14 and the type 1 Tet(A) variant among Enterobacterales.
3.5. Virulence analysis
The hypervirulent phenotype of three CRKP was observed in the G. mellonella infection model (Figure 3). At 48 h post-infection, the survival rates of larvae infected by strains SP1023, F1025, and SP1030 were 12.5, 29.2, and 16.7%, respectively, which were lower than that of larvae infected by the negative control strain K. pneumoniae FJ8 at 79.2%. WGS analysis showed that these strains expressed the same virulence factor profile specific to hypervirulent K. pneumoniae (hvKP), including RmpA2, aerobactin, and yersiniabactin (Table 2); thus, the CRKP in our study were classified as CR-hvKP. Long-read sequencing revealed that the rmpA2 and the iutAiucABCD gene cluster (aerobactin siderophore) were located on the IncHI1B/IncFIB-type pLVPK-like plasmid with sizes ranging from 204,770 to 204,778 bp (Table 3, Figure 2C). These plasmids showed high identity (>99%) to a variety of other hypervirulence plasmids of K. pneumoniae in the NCBI database, indicating the wide dissemination of these plasmids conferring hypervirulent phenotypes.
4. Discussion
The rapid and wide dissemination of KPC-producing CRKP represents a serious threat to public health and a serious challenge for healthcare workers. Most of the KPC-producing CRKP also harbor determinants that confer resistance to a variety of antimicrobial agents thus further limiting the clinical options for treatment. CZA exhibited great activity against these multidrug-resistant pathogens; however, many concerns have been raised over the emergence of resistant KPC variants with various genetic landscapes, demonstrating the substantial evolutionary potential of this enzyme (Findlay et al., 2021; Liu et al., 2022b; Wu et al., 2022).
In this study, we described the emergence of the CZA-resistant CRKP harbored the blaKPC-14 gene on two structurally different plasmids. Three CRKP collectively harbored a blaKPC-14-carrying IncFII/IncR plasmid, which was widespread and commonly identified in KPC-producing CRKP in China (Chen et al., 2014; Dong et al., 2018). There were a few studies that described the emergence and in vivo selection of KPC-14 rendering resistance following CZA treatment (Bianco et al., 2020; Niu et al., 2020). However, due to the absence of CZA therapeutic regimens during the hospitalization of the two patients, the source of the blaKPC-14 gene in our hospital was still unclear. As recently shown, the KPC-14 enzyme demonstrated the loss of carbapenemase activity (Compain and Arthur, 2017), which was true for the low-level resistance or susceptibility to the carbapenems of KPC-14 producers in our study. This finding reminded us that these resistance determinants may silently spread and be easily ignored during routine surveillance in clinical settings. Although these blaKPC-14-encoding plasmids were nonconjugative, they could provide translocatable fragments as reservoirs for the mobilization of the blaKPC-14 gene into other plasmid backbones. In this study, we also identified a conjugative plasmid integrated with the blaKPC-14-containing NTEKPC-I element, which was relatively prevalent among clinical strains in China (Cerdeira et al., 2019). Structurally similar plasmids were previously reported in our hospital with the colocation of the type 1 Tet(A) variant (Chen et al., 2021), which was associated with resistance to another clinically important antibiotic, tigecycline. Moreover, the evolution of these plasmids was observed, as they could additionally capture various other resistance loci, further contributing to the coselection and persistence of these determinants of resistance to last-line antibiotics. The considerable genetic plasticity of these plasmids enabled the acquisition of further resistance-encoding and hypervirulence-encoding genetic elements; thus, these isolates can better adapt to various environments to stimulate the spread of blaKPC-14 among Enterobacterales.
The multidimensional transmission and potential silent spread of blaKPC-14 in CRKP is concerning, especially in those that also carry hypervirulent phenotypes. The KPC-14-producing CRKP in our study originated from the same ST11 clone, which was the dominant clone of CRKP in China and served as a salient example of the evolutionary acquisition of resistance genes and virulence factors for a newly emerged superbug (Liao et al., 2020). The pLVPK-like virulence plasmids commonly converted normal ST11 strains to ST11 hvKP and were first reported in our hospital in 2018 (Gu et al., 2018). These plasmids enhanced the environmental survival and the rapid dissemination of the hypervirulent phenotype with a limited fitness cost in ST11 CRKP (Zhou et al., 2020), consistent with the persistence of these plasmids during spread and evolution observed in our study. More worrisome, these superbugs still have the exceptional ability to attain extra resistance to clinically important drugs such as colistin and tigecycline via in vivo selection following antibiotic treatment. This could have affected K. pneumoniae F1025 in our study, which displayed resistance to almost all antibiotics, including CZA, colistin, and tigecycline. The emergence of such XDR strains may cause severe infections that are difficult to treat with current antibiotics, especially for ICU patients with complicated diseases.
5. Conclusion
In this study, we described the evolution of a conjugative mosaic plasmid encoding the blaKPC-14 gene via mobile elements in CR-hvKP. The nonconjugative IncFII/IncR plasmid could serve as the reservoir of the mobilizable fragment of the blaKPC-14 gene, similar to NTEKPC-I elements, allowing it to integrate into other conjugative plasmid backbones, further facilitating the spread of blaKPC-14. Therefore, constant surveillance to control the development and further spread of CZA resistance is of great significance.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The studies involving humans were approved by the Ethics Committee of The Second Affiliated Hospital of Zhejiang University School of Medicine. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from primarily isolated as part of your previous study for which ethical approval was obtained. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
LW: Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Writing – original draft. WS: Data curation, Formal analysis, Investigation, Validation, Writing – original draft, Software, Visualization. JC: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The work was supported by the Zhejiang Provincial Natural Science Foundation of China (LY22H200001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Alikhan, N.-F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Barnes, M. D., Winkler, M. L., Taracila, M. A., Page, M. G., Desarbre, E., Kreiswirth, B. N., et al. (2017). Klebsiella pneumoniae Carbapenemase-2 (KPC-2), substitutions at ambler position Asp179, and resistance to ceftazidime-avibactam: unique antibiotic-resistant phenotypes emerge from β-lactamase protein engineering. mBio 8, e00528–e00517. doi: 10.1128/mBio.00528-17
Bianco, G., Boattini, M., Iannaccone, M., Bondi, A., Ghibaudo, D., Zanotto, E., et al. (2021). Carbapenemase detection testing in the era of ceftazidime/avibactam-resistant KPC-producing Enterobacterales: a 2-year experience. J. Glob. Antimicrob. Resist. 24, 411–414. doi: 10.1016/j.jgar.2021.02.008
Bianco, G., Boattini, M., Iannaccone, M., Cavallo, R., and Costa, C. (2020). Bloodstream infection by two subpopulations of Klebsiella pneumoniae ST1685 carrying KPC-33 or KPC-14 following ceftazidime/avibactam treatment: considerations regarding acquired heteroresistance and choice of carbapenemase detection assay. J. Antimicrob. Chemother. 75, 3075–3076. doi: 10.1093/jac/dkaa283
Cai, J. C., Zhou, H. W., Zhang, R., and Chen, G.-X. (2008). Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli isolates possessing the plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob. Agents Chemother. 52, 2014–2018. doi: 10.1128/AAC.01539-07
Cerdeira, L. T., Lam, M. M. C., Wyres, K. L., Wick, R. R., Judd, L. M., Lopes, R., et al. (2019). Small IncQ1 and col-like plasmids harboring blaKPC-2 and non-Tn 4401 elements (NTEKPC-IId) in high-risk lineages of Klebsiella pneumoniae CG258. Antimicrob. Agents Chemother. 63, e02140–e02118. doi: 10.1128/AAC.02140-18
Chen, L., Mathema, B., Chavda, K. D., DeLeo, F. R., Bonomo, R. A., and Kreiswirth, B. N. (2014). Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 22, 686–696. doi: 10.1016/j.tim.2014.09.003
Chen, J., Zeng, Y., Zhang, R., and Cai, J. (2021). In vivo emergence of colistin and tigecycline resistance in carbapenem-resistant hypervirulent Klebsiella pneumoniae during antibiotics treatment. Front. Microbiol. 12:702956. doi: 10.3389/fmicb.2021.702956
Chiu, S.-K., Huang, L.-Y., Chen, H., Tsai, Y.-K., Liou, C.-H., Lin, J.-C., et al. (2017). Roles of ramR and tet(a) mutations in conferring tigecycline resistance in carbapenem-resistant Klebsiella pneumoniae clinical isolates. Antimicrob. Agents Chemother. 61, e00391–e00317. doi: 10.1128/AAC.00391-17
Clinical and Laboratory Standards Institute (2018). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 11. Wayne, PA: CLSI standard M07.
Clinical and Laboratory Standards Institute (2021). Performance standards for antimicrobial susceptibility Ttesting, 31. Wayne, PA: CLSI supplement M100.
Compain, F., and Arthur, M. (2017). Impaired inhibition by avibactam and resistance to the ceftazidime-avibactam combination due to the D179Y substitution in the KPC-2 β-lactamase. Antimicrob. Agents Chemother. 61, e00451–e00417. doi: 10.1128/AAC.00451-17
Criscuolo, M., and Trecarichi, E. M. (2020). Ceftazidime/avibactam and ceftolozane/tazobactam for multidrug-resistant gram hegatives in patients with hematological malignancies: current experiences. Antibiotics 9:58. doi: 10.3390/antibiotics9020058
Dong, N., Yang, X., Zhang, R., Chan, E. W.-C., and Chen, S. (2018). Tracking microevolution events among ST11 carbapenemase-producing hypervirulent Klebsiella pneumoniae outbreak strains. Emerg. Microbes Infect. 7, 1–8. doi: 10.1038/s41426-018-0146-6
El-Kady, R. A. E.-H., Elbaiomy, M. A., and Elnagar, R. M. (2022). Molecular mechanisms mediating ceftazidime/avibactam resistance amongst carbapenem-resistant Klebsiella pneumoniae isolates from cancer patients. Infect. Drug Resist. 15, 5929–5940. doi: 10.2147/IDR.S384972
Findlay, J., Poirel, L., Juhas, M., and Nordmann, P. (2021). KPC-mediated resistance to ceftazidime-avibactam and collateral effects in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 65:e0089021. doi: 10.1128/AAC.00890-21
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2018). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46. doi: 10.1016/S1473-3099(17)30489-9
Humphries, R. M., and Hemarajata, P. (2017). Resistance to ceftazidime-avibactam in Klebsiella pneumoniae due to porin mutations and the increased expression of KPC-3. Antimicrob. Agents Chemother. 61, e00537–e00517. doi: 10.1128/AAC.00537-17
Jiang, M., Sun, B., Huang, Y., Liu, C., Wang, Y., Ren, Y., et al. (2022). Diversity of ceftazidime-avibactam resistance mechanism in KPC-2-producing Klebsiella pneumoniae under antibiotic selection pressure. Infect. Drug Resist. 15, 4627–4636. doi: 10.2147/IDR.S371285
Kolmogorov, M., Bickhart, D. M., Behsaz, B., Gurevich, A., Rayko, M., Shin, S. B., et al. (2020). metaFlye: scalable long-read metagenome assembly using repeat graphs. Nat. Methods 17, 1103–1110. doi: 10.1038/s41592-020-00971-x
Lam, M. M. C., Wick, R. R., Watts, S. C., Cerdeira, L. T., Wyres, K. L., and Holt, K. E. (2021). A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 12:4188. doi: 10.1038/s41467-021-24448-3
Liao, W., Liu, Y., and Zhang, W. (2020). Virulence evolution, molecular mechanisms of resistance and prevalence of ST11 carbapenem-resistant Klebsiella pneumoniae in China: a review over the last 10 years. J. Glob. Antimicrob. Resist. 23, 174–180. doi: 10.1016/j.jgar.2020.09.004
Linh, T. D., Thu, N. H., Shibayama, K., Suzuki, M., Yoshida, L., Thai, P. D., et al. (2021). Expansion of KPC-producing Enterobacterales in four large hospitals in Hanoi, Vietnam. J. Glob. Antimicrob. Resist. 27, 200–211. doi: 10.1016/j.jgar.2021.09.007
Liu, C., Dong, N., Chan, E. W. C., Chen, S., and Zhang, R. (2022a). Molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in China, 2016–20. Lancet Infect. Dis. 22, 167–168. doi: 10.1016/S1473-3099(22)00009-3
Liu, C., Wu, Y., Huang, L., Zhang, Y., Sun, Q., Lu, J., et al. (2022b). The rapid emergence of ceftazidime-avibactam resistance mediated by KPC variants in carbapenem-resistant Klebsiella pneumoniae in Zhejiang province, China. Antibiotics 11:731. doi: 10.3390/antibiotics11060731
Livermore, D. M., Warner, M., Jamrozy, D., Mushtaq, S., Nichols, W. W., Mustafa, N., et al. (2015). In vitro selection of ceftazidime-avibactam resistance in Enterobacteriaceae with KPC-3 carbapenemase. Antimicrob. Agents Chemother. 59, 5324–5330. doi: 10.1128/AAC.00678-15
Magiorakos, A.-P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
McLaughlin, M. M., Advincula, M. R., Malczynski, M., Barajas, G., Qi, C., and Scheetz, M. H. (2014). Quantifying the clinical virulence of Klebsiella pneumoniae producing carbapenemase Klebsiella pneumoniae with a galleria mellonellamodel and a pilot study to translate to patient outcomes. BMC Infect. Dis. 14:31. doi: 10.1186/1471-2334-14-31
Niu, S., Chavda, K. D., Wei, J., Zou, C., Marshall, S. H., Dhawan, P., et al. (2020). A ceftazidime-avibactam-resistant and carbapenem-susceptible Klebsiella pneumoniae strain harboring blaKPC-14 isolated in New York City. mSphere 5, e00775–e00720. doi: 10.1128/mSphere.00775-20
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Pérez-Galera, S., Bravo-Ferrer, J. M., Paniagua, M., Kostyanev, T., De Kraker, M. E. A., Feifel, J., et al. (2023). Risk factors for infections caused by carbapenem-resistant Enterobacterales: an international matched case-control-control study (EURECA). eClinicalMedicine 57:101871. doi: 10.1016/j.eclinm.2023.101871
Tian, Y., Zhang, Q., Wen, L., and Chen, J. (2021). Combined effect of polymyxin B and tigecycline to overcome heteroresistance in carbapenem-resistant Klebsiella pneumoniae. Microbiol. Spectr. 9, e00152–e00121. doi: 10.1128/Spectrum.00152-21
Van Duin, D., and Bonomo, R. A. (2016). Ceftazidime/avibactam and ceftolozane/tazobactam: second-generation β-lactam/β-lactamase inhibitor combinations. Clin. Infect. Dis. 63, 234–241. doi: 10.1093/cid/ciw243
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Wang, M., Earley, M., Chen, L., Hanson, B. M., Yu, Y., Liu, Z., et al. (2022). Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): a prospective, multicentre, cohort study. Lancet Infect. Dis. 22, 401–412. doi: 10.1016/S1473-3099(21)00399-6
Wang, L., Shen, W., Zhang, R., and Cai, J. (2022). Identification of a novel ceftazidime-avibactam-resistant KPC-2 variant, KPC-123, in Citrobacter koseri following ceftazidime-avibactam treatment. Front. Microbiol. 13:930777. doi: 10.3389/fmicb.2022.930777
Wu, Y., Yang, X., Liu, C., Zhang, Y., Cheung, Y. C., Chan, W. C., et al. (2022). Identification of a KPC variant conferring resistance to ceftazidime-avibactam from ST11 carbapenem-resistant Klebsiella pneumoniae strains. Microbiol. Spectr. 10, e02655–e02621. doi: 10.1128/spectrum.02655-21
Yang, X., Dong, N., Chan, E. W.-C., Zhang, R., and Chen, S. (2021). Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. 29, 65–83. doi: 10.1016/j.tim.2020.04.012
Keywords: blaKPC-14 gene, ceftazidime/avibactam, CR-hvKp, Klebsiella pneumoniae, gene transfer
Citation: Wang L, Shen W and Cai J (2023) Mobilization of the blaKPC-14 gene among heterogenous plasmids in extensively drug-resistant hypervirulent Klebsiella pneumoniae. Front. Microbiol. 14:1261261. doi: 10.3389/fmicb.2023.1261261
Edited by:
Benjamin Andrew Evans, University of East Anglia, United KingdomCopyright © 2023 Wang, Shen and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiachang Cai, Y2FpamlhY2hhbmdAemp1LmVkdS5jbg==
†These authors have contributed equally to this work
 Lin Wang†
Lin Wang† Jiachang Cai
Jiachang Cai