- 1College of Animal Sciences, Yangtze University, Jingzhou, Hubei, China
- 2College of Arts and Sciences, Yangtze University, Jingzhou, Hubei, China
- 3State Key Laboratory of Agricultural Microbiology, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, Hubei, China
Introduction: Actinobacillus pleuropneumoniae is an important respiratory pathogen, which can cause porcine contagious pleuropneumonia and lead to great economic losses to worldwide swine industry. High potassium is an adverse environment for bacteria, which is not conducive to providing turgor pressure for cell growth and division. Two-component system CpxAR is an important regulatory system of bacteria in response to environmental changes, which is involved in a variety of biological activities, such as antibiotic resistance, periplasmic protein folding, peptidoglycan metabolism and so on.
Methods: However, little is known about the role of CpxAR in high potassium stress in A. pleuropneumoniae. Here, we showed that CpxAR is critical for cell division of A. pleuropneumoniae under high potassium (K+) stress.
Results: qRT-PCR analysis found that CpxAR positively regulated the cell division genes ftsEX. In addition, we also demonstrated that CpxR-P could directly bind the promoter region of the cell division gene ftsE by EMSA.
Discussion: In conclusion, our results described a mechanism where CpxAR adjusts A. pleuropneumoniae survival under high-K+ stress by upregulating the expression of the cell division proteins FtsE and FtsX. These findings are the first to directly demonstrate CpxAR-mediated high-K+ tolerance, and to investigate the detailed molecular mechanism.
Introduction
Actinobacillus pleuropneumoniae is the aetiological agent of porcine pleuropneumonia, a serious respiratory disease with high morbidity, high lethality and substantial economic losses in the worldwide swine industry (Pattison et al., 1957; Frey, 1995; Sassu et al., 2018). Currently, 19 serovars of A. pleuropneumoniae have been identified based on the antigenic diversity of the capsular polysaccharides and lipopolysaccharides (Stringer et al., 2021; Scherrer et al., 2022). To successfully invade the host, A. pleuropneumoniae regulates the expression of its metabolism and virulence-related genes to adapt to adverse environments, such as cold, heat and osmotic.
In bacteria, two-component system (TCS) is a fine signal network to sense any change, and regulates the expression of bacterial effectors for adaptation and survival (Zschiedrich et al., 2016). To increase their adaptability, A. pleuropneumoniae has encoded five two-component systems (TCS) to detect and respond to diverse environments, such as CpxAR, QseBC, PhoBR, ArcAB and NarPQ (Xu et al., 2008). The CpxAR TCS is found in many bacteria, consisting of CpxA (histidine kinase) and CpxR (transcriptional regulatory protein), and has been implicated in envelope stress responses. In A. pleuropneumoniae, the CpxAR plays an important role in heat stress, cold stress, biofilm formation, capsule synthesis, and pathogenesis (Li et al., 2018; Yan et al., 2020; Yao et al., 2022; Liu et al., 2022a,b).
In A. pleuropneumoniae, the CpxAR modulates biofilm formation and virulence by regulating the expression of the pgaABCD operon through rpoE (Li et al., 2018). CpxAR is also found to mediate A. pleuropneumoniae stress resistance and virulence by activating the expression of WecA, which is essential for O-antigen biosynthesis (Yan et al., 2020). Moreover, CpxAR responds to heat stress by suppressing the expression of the type IV pilin ApfA, which is prone to misfolding and aggregation and therefore reduces bacterial survival under heat stress (Liu et al., 2022a). In addition, we demonstrated previously that CpxAR contributes to virulence by upregulating the expression of the polysaccharide capsule export system CpxDCBA (Liu et al., 2022b).
Potassium (K+) is the major cation in bacterial cytoplasm, and essential for maintaining ion homeostasis which is important for regulating pH and membrane potential (Ballal et al., 2007). But, excessive K+ is cytotoxic to bacteria, affecting turgor pressure which can lead to disturbances in cell growth and division (Sweet et al., 2021). However, most bacteria have evolved several K+-translocating systems to maintain K+ homeostasis, such as TrkAH, KtrAB, KtrCD, KdpFABC, KimA, Kup, Kef, YjbQ and KhtSTU (Stautz et al., 2021).
Schmidt and colleagues found that FtsE and FtsX are essential for cell division (Schmidt et al., 2004). In addition, previous studies have shown that FtsE contributes to the translocation of K-pump to the cell membrane including KdpA, TrkH, and Kup (Ukai et al., 1998; Mir et al., 2015). In E. coli, disruption of FtsE function exhibited growth defect under high-K+ stress (Ukai et al., 1998).
In this study, we showed that CpxAR contributes to A. pleuropneumoniae growth and cell division under high-K+ stress. In addition, we confirmed that CpxAR directly activates the expression of cell division genes ftsEX. Taken together, our study describes a new mechanism by which CpxAR enables A. pleuropneumoniae to adjust its survival by up-regulating the expression of ftsEX under high-K+ stress.
Materials and methods
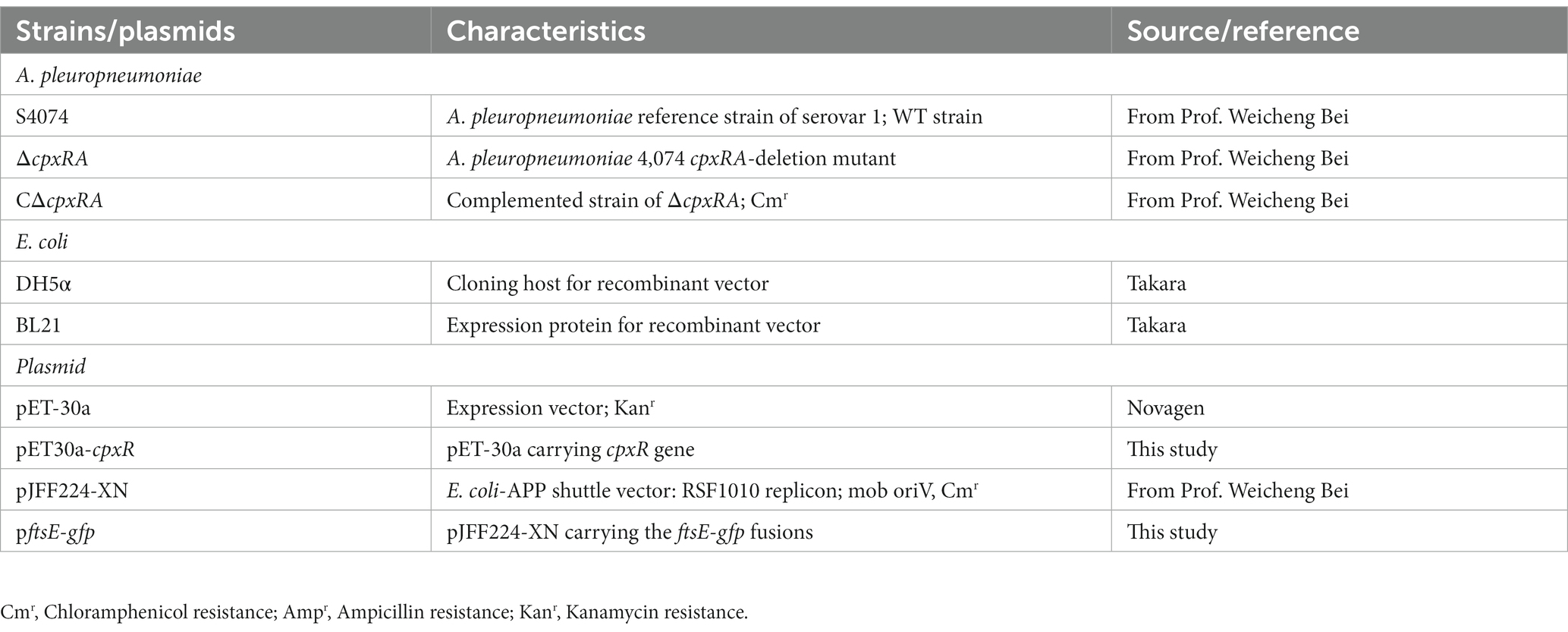
Strains, plasmid, and growth conditions
In this study, A. pleuropneumoniae strain S4074 was used as a representative strain, which was grown at 37°C in tryptic soy broth (TSB; BD, United States) added with 10% FBS and 10% (vol/vol) nicotinamide adenine dinucleotide (NAD; Biofroxx, Germany). The wild-type S4074 and in-frame mutant strain ∆cpxRA were donated by Prof. Weicheng Bei (Li et al., 2018). Escherichia coli β2155 was cultured at 37°C in Luria-Bertani (LB) with 50 μg/mL diaminopimelic acid (DAP; Sigma, USA). The bacterial strains used in this study are listed in Table 1, and primers are described in Table 2. The recombinant plasmid pJFF224-PftsEX with IPTG-inducible promoter was electrically transferred to the ∆cpxRA mutant strains to regulate the ftsEX gene expression under the different concentrations of IPTG.
SEM
For scanning electron microscopy (SEM), wild-type S4074 and its mutant derivatives were harvested after they were cultured in TSB medium supplemented with 10% FBS and 10% NAD with or without the supplementation of 0.3 M KCl. Bacterial cells were fixed with 2.5% glutaraldehyde and deposited onto copper grids (200 mesh; Zhongjingkeyi, China). The copper grids were air-dried, mounted on the sample stub and coated with gold. Subsequently, strains were observed by the SEM (VEGA3; TESCAN, Czech) and the bacterial length was measured by Image J (NIH, United States).
RNA extraction, quantitative RT-PCR, and RT-PCR
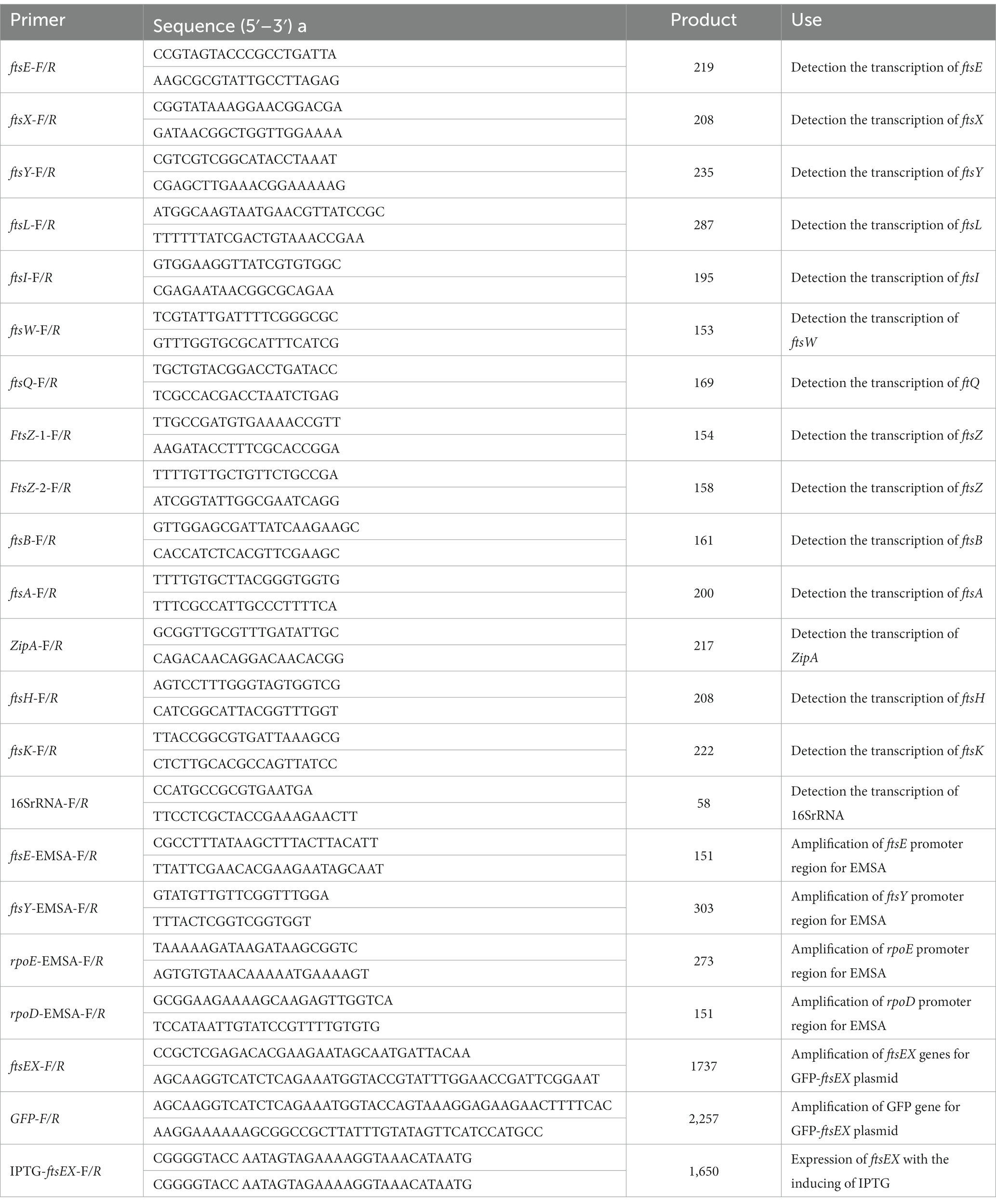
WT and ∆cpxRA mutant strains were cultivated at 37°C shaking in TSB supplemented with 10% FBS and 10% NAD to an OD600 of 0.6, centrifuged (3,000 g, 5 min) to gather cells. Total RNA of strains was extracted using Total RNA Extractor kit (Sangon Biotech, China) according to the manufacturer’s instruction. DNA impurity was eliminated using DNase I kit (Vazyme, China) according to instruction. To determine the quality, the RNA was electrophoresed on a 1% agarose gel. cDNA was synthesized using reverse transcriptase from Vazyme. Quantitative Real-Time PCR was performed using Real-Time Quantitative PCR detecting System and SYBR qPCR Mix (Vazyme, China). 16S RNA was used as the endogenous control to normalize expression of target genes. The 2−ΔΔCt method was used to calculate and analyze relative expression level of mRNA (Livak and Schmittgen, 2001). RT-PCR across the cdd-ftsX, ftsX-ftsE, ftsE-ftsY, and ftsY-rsmD junctions was performed as previously described (Liu et al., 2022a).
Construction of promoter-gfp repoter strains and promoter analyses
A ftsE-gfp fusion containing the ftsE promoter region and gfp gene, was cloned into the Xho I and Not I sites on the pJFF224-XN plasmid. Then, the ftsE-gfp reporter plasmid was electroporated into the wild-type S4074 and the ∆cpxAR mutant strain. A. pleuropneumoniae strains harboring a ftsE-gfp reporter plasmid were grown to an OD600 of 0.6 in TSB medium with or without the supplementation of 0.3 M KCl. The cells were harvested and resuspended in 1 mL of 10 mM PBS. Luminescence was measured in the Spectramax iD3 microplate reader with excitation at 485 nm and emission at 535 nm.
Expression and purification of His-CpxR
The purification of CpxR was conducted as previously described (Liu et al., 2022b). The E.coli BL21 containing the expression vector pET30a-CpxR was used to express His-CpxR protein. Expression of His-CpxR was induced by IPTG (0.5 mM) at 16°C overnight, and bacteria were harvested by centrifuging at 12000 rpm for 10 min. The cell pellet was resuspended in binding buffer (50 mM Na3PO4, pH 7.4, 500 mM NaCl, 30 mM imidazole), and lysed by sonication. The soluble fraction of the lysate was added to nickel-nitrilotriacetic acid (Ni-NTA) agarose column and mixed on Multipurpose Shaker QB-206 for 3 h. After being washed with binding buffer, the His-CpxR protein was eluted with elution buffer (20 mM Tris–HCl, pH 7.4, 500 mM NaCl, 500 mM imidazole). The purified His-CpxR was then stored at −80°C until use.
Electrophoretic mobility shift assay
The promoter regions of target genes were amplified from the A. pleuropneumoniae S4074 genome and tagged with biotin using a EMSA Probe Biotin Labeling Kit (Beyotime, China). CpxR protein was phosphorylated by acetyl phosphate (Sigma, United States) (Pogliano et al., 1997). Biotin-labeled DNA probes (1 μM) were incubated at room temperature for 20 min with 0–4 pmol CpxR protein in binding buffer (Beyotime, China). The reaction mixtures were analyzed by 4% non-denaturing polyacrylamide gel electrophoresis and transferred to nylon membrane. Next, the band was detected using a Chemiluminescent EMSA Kit (Beyotime, China).
Statistical analysis
Experimental data were analyzed by Student’s two-tailed t-tests using GraphPad Prism 7.0 (GraphPad lnc.), and shown as mean ± standard deviation (SD). Statistical significance was assumed at a p value of <0.05.
Results
Deletion of the cpxRA genes decreases the growth of Actinobacillus pleuropneumoniae under high-K+ stress
To explore the role of CpxAR in bacterial adaptation to osmotic stress in A. pleuropneumoniae, we tested the growth traits of the wild-type S4074 and its cpxRA mutant strain grown in solid or liquid medium with or without the supplementation of 0.3 M KCl or 0.3 M NaCl. When the cells were grown in solid medium, the growth of the ∆cpxRA strain was markedly reduced with the supplementation of 0.3 M KCl, but similar to that of the WT strain with the supplementation of 0.3 M NaCl or normal medium (Figure 1A). Optical density and colony forming units showed that the growth rate of the ∆cpxRA strain was significantly lower than that of the WT strain when grown with the supplementation of 0.3 M KCl (Figures 1B,C). These findings suggested that the growth defect of the mutant strain ∆cpxRA was significantly increased compared with that of WT and C∆cpxRA strains under high-K+ stress. These results suggested that CpxAR contributes to A. pleuropneumoniae survival under high potassium stress.

Figure 1. CpxAR is required for growth under high-K+ stress. The growth traits of the WT, ∆cpxRA and C∆cpxRA strains with or without 0.3 M K+ or 0.3 M Na+ were monitored by measurement of spotting on TSA plates (A), OD600 (B), and viable cell counts (C). **p < 0.01. ***p < 0.001.
Deletion of cpxRA affects the cell division of Actinobacillus pleuropneumoniae under high-K+ stress
To further investigate how the inactivation of cpxRA genes affected the growth of A. pleuropneumoniae under high-K+ stress, we used scanning electron microscope (SEM) to observe the bacterial morphology of the WT, ∆cpxRA and C∆cpxRA strains when they were grown with or without the supplementation of 0.3 M KCl. When grown with the supplementation of 0.3 M KCl, the cell length of ∆cpxRA strain showed a 2-fold increase on average compared to the wild-type and C∆cpxRA strains (Figures 2A,B). In addition, we found that the cell length of ∆cpxRA strain was also longer than that of the wild-type and C∆cpxRA strains without K+ stress, but the difference was much smaller than that under K+ stress (Figures 2A,B). Together, these findings suggested that CpxAR regulates cell division to help A. pleuropneumoniae cope with potassium stress.
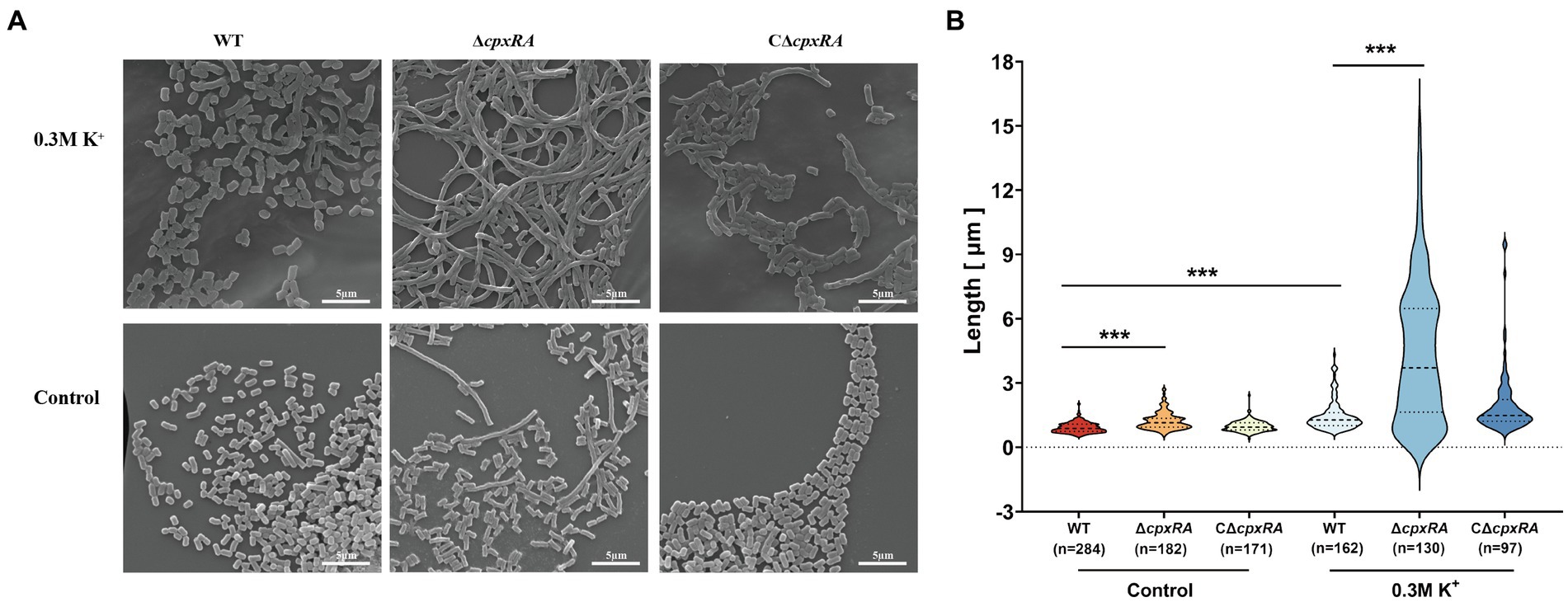
Figure 2. CpxAR impacts cell division under high-K+ stress. (A) Observation of bacterial length in the WT, ∆cpxRA, and C∆cpxRA strains grown with or without 0.3 M K+ by SEM. (B) Bacterial length measured for the WT, ∆cpxRA and C∆cpxRA strains grown with or without 0.3 M K+. The number of bacteria measured is shown in brackets. ***p < 0.001.
CpxAR regulates the expression of cell division genes ftsE and ftsX under high-K+ stress
To gain insight into the mechanism of CpxAR affecting the cell division of A. pleuropneumoniae, we compared the transcript levels of cell division genes in the WT and ∆cpxRA strains by qRT-PCR. As shown in Figure 3A, the relative transcript levels of ftsE and ftsX were significantly downregulated in the ∆cpxRA strain with or without the supplementation of 0.3 M KCl, but ftsY was not (Figure 3A). However, there were no significant changes in the cell division genes ftsL, ftsI, ftsW, ftsQ, ftsZ, ftsB, ftsA, zipA, ftsH, and ftsK in the ∆cpxRA strain compared with the WT strain (Supplementary Figure S1).These results suggested that CpxAR regulates the expression of the cell division genes ftsE and ftsX in A. pleuropneumoniae.
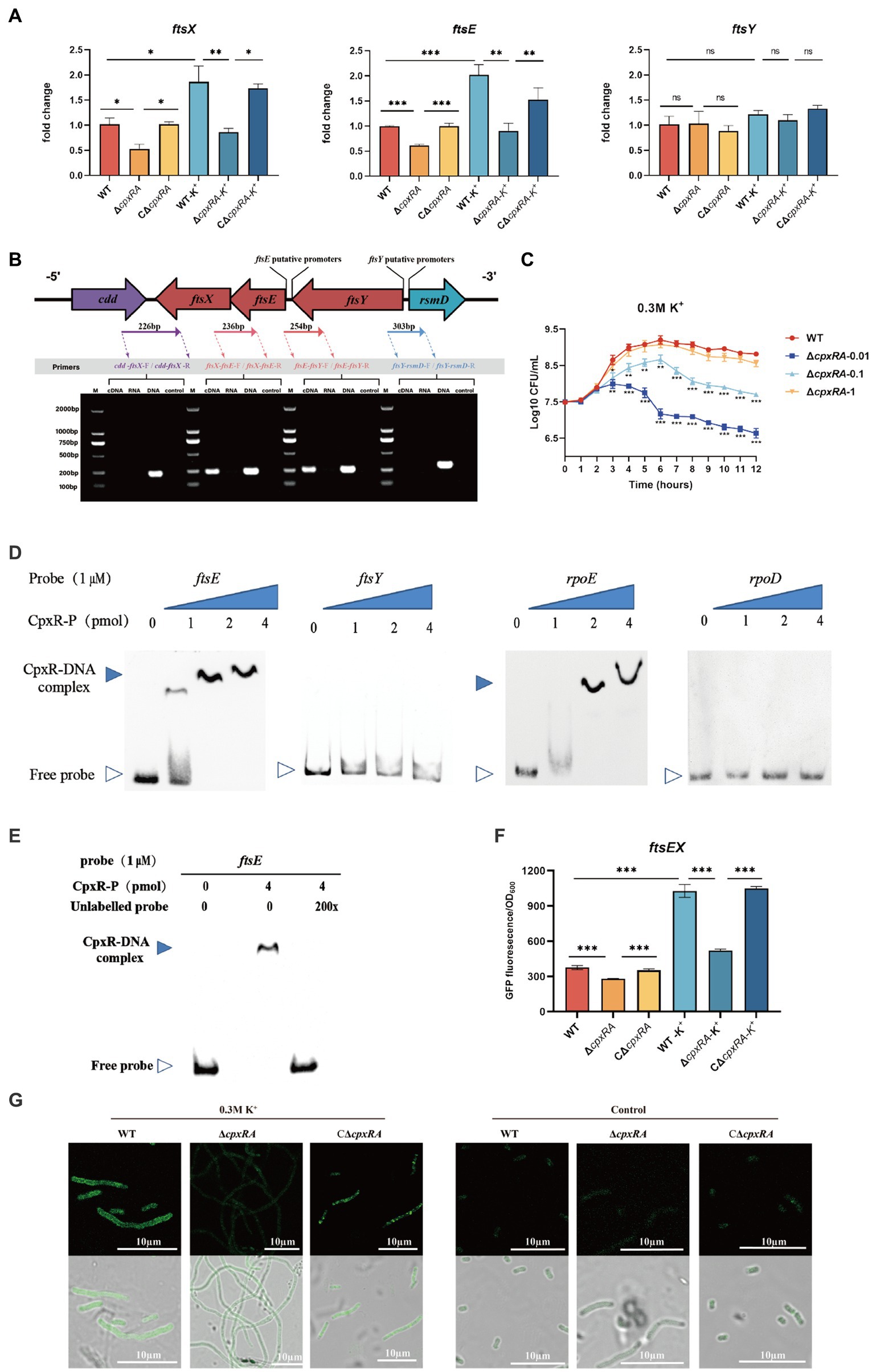
Figure 3. CpxAR regulates the expression of cell division genes ftsE and ftsX. (A) qRT-PCR analysis of ftsE, ftsX, and ftsY in WT, ∆cpxRA, and C∆cpxRA strains. (B) Schematics of organization of genes encoding the cell division proteins. Transcriptional characteristics of the ftsE, ftsX, and ftsY genes measured by RT-PCR. Lane 1, cDNA; lane 2, RNA; lane 3, DNA; and lane 4, negative control. (C) The growth analysis of strains expressing ftsEX with IPTG at indicated concentrations. (D) EMSA analysis of the binding of phosphorylated CpxR to the promoters of ftsE, ftsY, rpoE, and rpoD. rpoE and rpoD served, respectively, as positive and negative control. (E) EMSA analysis of the binding of purified CpxR to ftsE with or without unlabeled DNA probe. The promoter activity of ftsE gene in the WT and ∆cpxRA strains was analized by fluorescent microscopy (F) and measurement of relative fluorescence units (G). *p < 0.05. **p < 0.01. ***p < 0.001.
The cell division genes ftsY, ftsE, and ftsX are adjacent on the A. pleuropneumoniae chromosome (Figure 3B). To characterize the ftsYEX locus, we performed RT-PCR across the cdd-ftsX, ftsX-ftsE, ftsE-ftsY, and ftsY-rsmD junctions. The RT-PCR analysis indicated that the ftsY, ftsE, and ftsX genes are co-transcribed as a single mRNA and comprise an operon. To verify whether the ftsEX operon contributes to A. pleuropneumoniae survival under high-K+ stress, the growth of ∆cpxRA/PftsEX was assayed. When the expression of FtsE and FtsX increased with the increase of IPTG concentration, the growth defects of the ∆cpxRA strain were significantly rescued (Figure 3C). These observations indicated that the ftsEX operon plays an important role in A. pleuropneumoniae response to high-K+ stress.
Here, the prediction results (BDGP)1 showed that the ftsYEX operon has two putative promoter regions, respectively, located upstream of ftsY and ftsE. To explore the mechanism by which CpxR regulates the ftsYEX operon expression, we tested whether CpxR-P binds to the promoter regions of ftsY and ftsE using gel shift analysis. EMSA analysis showed that CpxR was capable to bind with the promoter regions of ftsE and rpoE (positive control), but not to the promoter regions of ftsY and rpoD (negative control) (Figure 3D). In addition, EMSAs showed that phosphorylated CpxR did not bind to the promoter sequence of ftsE when 200X of unlabeled probes were added (Figure 3E). To examine the link between CpxR and the expression of the ftsEX operon, the GFP reporter strains for ftsE gene promoter were constructed. As shown in Figures 3F,G, fluorescence intensities and confocal microscopy analysis showed that the transcrition activity of the promoter was significantly decreased in ∆cpxRA strain with or without high-K+ stress. Together, these data demonstrated that CpxAR directly regulates the expression of ftsEX operon.
Discussion
The two-component system is an important group of signal transduction systems in bacteria that interacts with sudden stimulus and responds accordingly to survive in adverse environment (Zhao et al., 2022). Prototypical TCS is composed of a membrane-bound sensor kinase and a cytoplasmic response regulator (Eguchi and Utsumi, 2008). In these systems, the histidine kinase will autophosphorylate when sensing a stimulus, and transfers the phosphoryl group to the response regulator, which then regulates the expression of specific genes by binding these promoters. Previous studies showed that CpxAR plays multiple regulatory roles in A. pleuropneumoniae virulence, biofilm formation and stress resistance (Yan et al., 2020). However, the function of the CpxAR system adaptation to other surrounding stresses remains unknown.
Potassium is the major cation in the cytoplasm for bacteria, which is essential for providing turgor pressure for cell growth and division (Sweet et al., 2021). When external K+ concentrations are high, bacteria adjust their survival by maintaining cellular K+ homeostasis. However, the mechanism of bacterial survival under high-K+ stress requires further exploration. In this study, the growth rate of WT strain under K+ stress was significantly lower than that of strain without K+ stress, indicating that high-K+ stress inhibits cell growth and division of A. pleuropneumoniae. Furthermore, we showed that the CpxAR system plays an important role in growth and division of A. pleuropneumoniae during high-K+ stress.
FtsEX is a putative ABC transporter type complex that facilitates the assembly of division proteins and other proteins to the cytoplasmic membrane and is thought to play an important role in cell division (Schmidt et al., 2004; Reddy, 2007). In E. coli, disruption of the ftsE gene prevents the localization of the K+-pump proteins to the inner membrane (Ukai et al., 1998). In the present study, qRT-PCR analysis showed that cell division genes ftsE and ftsX are controlled by CpxAR in A. pleuropneumoniae. Indeed, it seemed that FtsE and FtsX are related to the mechanism of CpxAR-mediated potassium stress. In E. coli, cell division genes ftsY, ftsE, and ftsX comprise an operon (Gill et al., 1986), which is consistent with our RT-PCR analysis in A. pleuropneumoniae. But, we found that the ftsYEX operon in A. pleuropneumoniae contains two promoter regions by sequence analysis, oneupstream of ftsY and the other upstream of ftsE.
Previous studies have shown that CpxR can regulate the transcription of a wide range of genes by directly binding to their promoter regions (De Wulf et al., 2002; Vogt and Raivio, 2012). Here, EMSA analysis showed that CpxR can directly interact with ftsE promoter. Combined with qRT-PCR analysis, these results confirmed that the ftsEX operon is directly and positively regulated by the CpxAR system.
In summary, our findings gain new and important insights into the function of A. pleuropneumoniae CpxAR in response to extreme environments, and show that the CpxAR system directly regulates cell division genes to maintain potassium homeostasis (Figure 4). Because the CpxAR system is ubiquitous in many bacteria, our study contribute to the understanding of bacterial environmental adaptability. Future studies will aim to identify more CpxR-regulated genes, which could expand the knowledge of CpxAR function.
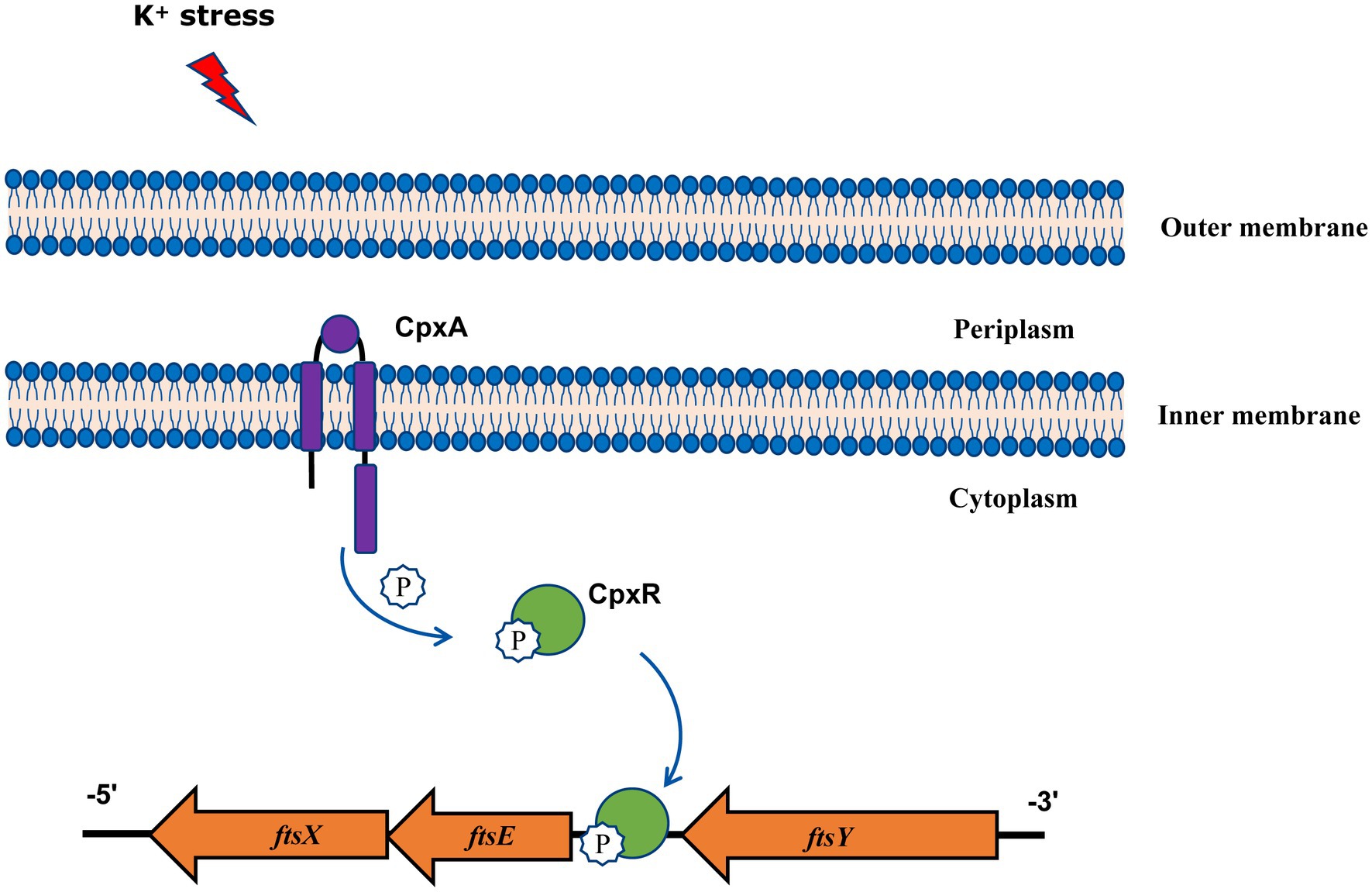
Figure 4. Model of CpxAR-ftsEX regulatory circuit in A. pleuropneumoniae. The high-K+ stress activates the TCS CpxAR which directly induces expression of the ftsEX operon.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
JW: Data curation, Investigation, Project administration, Software, Writing – original draft. RZ: Investigation, Writing – original draft. YJ: Investigation, Writing – original draft. TX: Investigation, Writing – original draft. LD: Investigation, Writing – original draft. QY: Investigation, Writing – original draft. WZ: Investigation, Writing – original draft. HX: Investigation, Writing – original draft. XG: Software, Supervision, Writing – original draft. JH: Writing – review & editing. WB: Funding acquisition, Resources, Supervision, Writing – original draft. FL: Conceptualization, Formal analysis, Funding acquisition, Methodology, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Natural Science Foundation of China (32002252) and National Key Research and Development Program of China (2022YFD1800905).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Correction note
A correction has been made to this article. Details can be found at: 10.3389/fmicb.2025.1645185.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1259935/full#supplementary-material
Footnotes
References
Ballal, A., Basu, B., and Apte, S. K. (2007). The Kdp-ATPase system and its regulation. J. Biosci. 32, 559–568. doi: 10.1007/s12038-007-0055-7
De Wulf, P., McGuire, A. M., Liu, X., and Lin, E. C. (2002). Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 277, 26652–26661. doi: 10.1074/jbc.M203487200
Eguchi, Y., and Utsumi, R. (2008). Introduction to bacterial signal transduction networks. Adv. Exp. Med. Biol. 631, 1–6. doi: 10.1007/978-0-387-78885-2_1
Frey, J. (1995). Virulence in Actinobacillus pleuropneumoniae and RTX toxins. Trends Microbiol. 3, 257–261. doi: 10.1016/s0966-842x(00)88939-8
Gill, D. R., Hatfull, G. F., and Salmond, G. P. (1986). A new cell division operon in Escherichia coli. Mol. Gen. Genet. 205, 134–145. doi: 10.1007/BF02428043
Li, H., Liu, F., Peng, W., Yan, K., Zhao, H., Liu, T., et al. (2018). The CpxA/CpxR two-component system affects biofilm formation and virulence in Actinobacillus pleuropneumoniae. Front. Cell. Infect. Microbiol. 8:72. doi: 10.3389/fcimb.2018.00072
Liu, F., Peng, W., Yan, K., Huang, J., Yuan, F., He, Q., et al. (2022a). CpxAR of Actinobacillus pleuropneumoniae contributes to heat stress response by repressing expression of type IV pilus gene apfA. Microbiol Spectr 10:e0252322. doi: 10.1128/spectrum.02523-22
Liu, F., Yao, Q., Huang, J., Wan, J., Xie, T., Gao, X., et al. (2022b). The two-component system CpxA/CpxR is critical for full virulence in Actinobacillus pleuropneumoniae. Front. Microbiol. 13:1029426. doi: 10.3389/fmicb.2022.1029426
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mir, M. A., Arumugam, M., Mondal, S., Rajeswari, H. S., Ramakumar, S., and Ajitkumar, P. (2015). Mycobacterium tuberculosis cell division protein, FtsE, is an ATPase in dimeric form. Protein J. 34, 35–47. doi: 10.1007/s10930-014-9593-7
Pattison, I. H., Howell, D. G., and Elliot, J. (1957). A haemophilus-like organism isolated from pig lung and the associated pneumonic lesions. J. Comp. Pathol. 67, 320–IN37. doi: 10.1016/s0368-1742(57)80031-9
Pogliano, J., Lynch, A. S., Belin, D., Lin, E. C., and Beckwith, J. (1997). Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 11, 1169–1182. doi: 10.1101/gad.11.9.1169
Reddy, M. (2007). Role of FtsEX in cell division of Escherichia coli: viability of ftsEX mutants is dependent on functional SufI or high osmotic strength. J. Bacteriol. 189, 98–108. doi: 10.1128/JB.01347-06
Sassu, E. L., Bosse, J. T., Tobias, T. J., Gottschalk, M., Langford, P. R., and Hennig-Pauka, I. (2018). Update on Actinobacillus pleuropneumoniae-knowledge, gaps and challenges. Transbound. Emerg. Dis. 65, 72–90. doi: 10.1111/tbed.12739
Scherrer, S., Peterhans, S., Neupert, C., Rademacher, F., Bartolomei, G., Sidler, X., et al. (2022). Development of a novel high resolution melting assay for identification and differentiation of all known 19 serovars of Actinobacillus pleuropneumoniae. Microbiology 11:e1272. doi: 10.1002/mbo3.1272
Schmidt, K. L., Peterson, N. D., Kustusch, R. J., Wissel, M. C., Graham, B., Phillips, G. J., et al. (2004). A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J. Bacteriol. 186, 785–793. doi: 10.1128/JB.186.3.785-793.2004
Stautz, J., Hellmich, Y., Fuss, M. F., Silberberg, J. M., Devlin, J. R., Stockbridge, R. B., et al. (2021). Molecular mechanisms for bacterial potassium homeostasis. J. Mol. Biol. 433:166968. doi: 10.1016/j.jmb.2021.166968
Stringer, O. W., Bosse, J. T., Lacouture, S., Gottschalk, M., Fodor, L., Angen, O., et al. (2021). Proposal of Actinobacillus pleuropneumoniae serovar 19, and reformulation of previous multiplex PCRs for capsule-specific typing of all known serovars. Vet. Microbiol. 255:109021. doi: 10.1016/j.vetmic.2021.109021
Sweet, M. E., Larsen, C., Zhang, X., Schlame, M., Pedersen, B. P., and Stokes, D. L. (2021). Structural basis for potassium transport in prokaryotes by KdpFABC. Proc. Natl. Acad. Sci. U. S. A. 118:e2105195118. doi: 10.1073/pnas.2105195118
Ukai, H., Matsuzawa, H., Ito, K., Yamada, M., and Nishimura, A. (1998). ftsE(Ts) affects translocation of K+-pump proteins into the cytoplasmic membrane of Escherichia coli. J. Bacteriol. 180, 3663–3670. doi: 10.1128/JB.180.14.3663-3670.1998
Vogt, S. L., and Raivio, T. L. (2012). Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol. Lett. 326, 2–11. doi: 10.1111/j.1574-6968.2011.02406.x
Xu, Z., Zhou, Y., Li, L., Zhou, R., Xiao, S., Wan, Y., et al. (2008). Genome biology of Actinobacillus pleuropneumoniae JL03, an isolate of serotype 3 prevalent in China. PLoS One 3:e1450. doi: 10.1371/journal.pone.0001450
Yan, K., Liu, T., Duan, B., Liu, F., Cao, M., Peng, W., et al. (2020). The CpxAR two-component system contributes to growth, stress resistance, and virulence of Actinobacillus pleuropneumoniae by upregulating wecA transcription. Front. Microbiol. 11:1026. doi: 10.3389/fmicb.2020.01026
Yao, Q., Xie, T., Fu, Y., Wan, J., Zhang, W., Gao, X., et al. (2022). The CpxA/CpxR two-component system mediates regulation of Actinobacillus pleuropneumoniae cold growth. Front. Microbiol. 13:1079390. doi: 10.3389/fmicb.2022.1079390
Zhao, Z., Xu, Y., Jiang, B., Qi, Q., Tang, Y. J., Xian, M., et al. (2022). Systematic identification of CpxRA-regulated genes and their roles in Escherichia coli stress response. mSystems 7:e0041922. doi: 10.1128/msystems.00419-22
Keywords: Actinobacillus pleuropneumoniae, two-component system, CpxAR, high-K+ stress, cell division, ftsEX
Citation: Wan J, Zhang R, Jia Y, Xie T, Dai L, Yao Q, Zhang W, Xiao H, Gao X, Huang J, Bei W and Liu F (2023) The two-component system CpxAR is required for the high potassium stress survival of Actinobacillus pleuropneumoniae. Front. Microbiol. 14:1259935. doi: 10.3389/fmicb.2023.1259935
Edited by:
Lei Deng, Harvard Medical School, United StatesReviewed by:
Sébastien Bontemps-Gallo, Institut Pasteur de Lille, FranceVictor H. Bustamante, National Autonomous University of Mexico, Mexico
Abraham Loera Muro, Centro de Investigación Biológica del Noroeste (CIBNOR), Mexico
Ricardo Oropeza, National Autonomous University of Mexico, Mexico
Jorge Antonio Yañez, Meritorious Autonomous University of Puebla, Mexico
Copyright © 2023 Wan, Zhang, Jia, Xie, Dai, Yao, Zhang, Xiao, Gao, Huang, Bei and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Liu, bGl1ZmVuZzY4NDMxQDE2My5jb20=
 Jiajia Wan1
Jiajia Wan1 Lu Dai
Lu Dai Qing Yao
Qing Yao Xuejun Gao
Xuejun Gao Weicheng Bei
Weicheng Bei Feng Liu
Feng Liu