
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Microbiol. , 03 October 2023
Sec. Systems Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1259287
This article is part of the Research Topic Plastisphere in Aquatic Environments: Microbial Diversity, Biofilm Structure, and Implications for Ecosystems and Human Health View all 7 articles
Microplastics quickly become colonised by diverse microbial communities, known as the Plastisphere. There is growing concern that microplastics may support the enrichment and spread of pathogenic or antimicrobial resistant microorganisms, although research to support the unique role of microplastics in comparison to control particles remains inconclusive. Limitations to this research include the microbiological methods available for isolating adhered microbes. Culture-based methods provide some of the most established, accessible and cost-effective microbiological protocols, which could be extremely useful in helping to address some of the remaining key questions in Plastisphere research. Previous works have successfully cultured bacteria from plastics, but these have not yet been reviewed, nor compared in efficiency. In this study, we compared four common biofilm extraction methods (swabbing, sonication, vortexing, sonication followed by vortexing) to extract and culture a mixed community of bacteria from both microplastic (polyethylene, polypropylene and polystyrene) and control (wood and glass) particles. Biofilm extraction efficiency and viability of bacterial suspension was determined by comparing CFU/mL of four different groups of bacteria. This was verified against optical density and 16S rRNA qPCR. Overall, we found that all tested methods were able to remove biofilms, but to varying efficiencies. Sonicating particles with glass beads for 15 min, followed by vortexing for a further minute, generated the highest yield and therefore greatest removal efficiency of culturable, biofilm-forming bacteria.
Microplastics (0.1 μm–5 mm; Thompson et al., 2004) are environmentally prevalent pollutants that can support a diverse array of microbes, known as the Plastisphere (Zettler et al., 2013). These microplastic-associated biofilms have also been shown to harbour pathogenic or antimicrobial resistant bacteria, potentially increasing human or animal exposure to disease-causing, drug-resistant microbes (Arias-Andres et al., 2018; Bowley et al., 2021). Therefore, it is crucial for research efforts to determine the risk posed by microplastics to ecological systems and human, animal or environmental health. However, it is not yet clear whether the Plastisphere includes significantly more bacteria of concern than naturally existing particles.
There are a number of different techniques that have been used to investigate the microbial community profiles associated with microplastics (De Tender et al., 2017; Bartkova et al., 2021). These methods can be divided into molecular (e.g., DNA extraction followed by downstream applications such as sequencing), and culture-based (e.g., cultivation on agar plates) techniques. Molecular methods can be used to provide a holistic and sensitive picture of community composition, virulence and/or antimicrobial resistance (AMR). The first step of almost all molecular protocols is the extraction of DNA from the community, and the efficiency of DNA extraction methods from microplastics were previously evaluated by Debeljak et al. (2017). Culture-based methods have also been adopted in previous Plastisphere research (Table 1), though, these methods have not yet been compared in efficiency.
Culture-based methods, though less sensitive than molecular methods and limited to the study of culturable organisms, are cost-effective, replicable and widely accessible (e.g., to less specialised, smaller or lower-income laboratories). Perhaps more importantly, working with cultured isolates allows measurement of key phenotypes, such as AMR and pathogenicity, that can otherwise only be inferred from sequence-based methods.
When investigating the Plastisphere, the first step in most culture-based methods is to remove the biofilm from the particle, creating a bacterial suspension. Whilst the methods that have previously been adopted to achieve this have shown success in biofilm removal (Table 1), there has been no measure of removal efficiency. In some cases, biofilms are not removed but are instead used to produce an enrichment culture, whereby particles harbouring biofilms are added to a sterile, nutrient rich broth. The resulting liquid culture is then used for downstream approaches. However, this method may arguably only select for strains capable of free-living growth, biasing the community against strictly biofilm-forming phenotypes. In addition, inducing a free-living phenotype may alter the expression of virulence factors or AMR that is associated with biofilm formation. Therefore, we have not included this style of isolation in our study, and have instead focused on entire biofilm extraction, with the aim of retaining viable cells.
Without assessment of removal efficiency, culture-based research efforts are not currently optimised, comparable or consistent. This study therefore aimed to compare the efficiency of some of the most used protocols to extract viable bacterial cells from microplastics. The methods selected for testing were: swabbing, sonication with glass beads, vortexing with glass beads, and sonication with glass beads followed by vortexing. In additional to testing removal efficiency from microplastics, natural (wood) and inert (glass) particles were also included, to ensure the extraction method we propose is the most efficient for both microplastic and control particles. This is crucial, given that future research should not only focus on the microbial communities present on microplastics, but also in comparison to communities present on natural control substrates.
Microplastic and wood particles sampled from the environment, 4 mm glass beads (Novagen ColiRollers, LOT: D00136263), Iso-Sensitest broth (Oxoid; LOT: 3177183), glycerol (ThermoFisher, LOT: P01H051), NaCl (Sigma-Aldrich, PCode: 1003326144), Luria-Bertani (LB) agar (Fisher Bioreagents, LOT: 188453), CM1205 Chromogenic Coliform agar (ISO; Oxoid, LOT: 3004362), DNeasy Ultra-Clean Microbial kit (Qiagen, LOT: 169011985), DNeasy PowerBiofilm kit (Qiagen, LOT: 169048148), custom synthetic gBlocks and primers (Table 2) provided by IDTDNA, PrimerDesign Precision Plus SYBR Green Master Mix (Z-PPLUS-R-SY-10ML) and bovine serum albumin (20 mg/mL, Fisher Scientific, LOT, 170419-0461).
PerkinElmer 208 Spotlight 400 (Perkin Elmer, United Kingdom), CAT-II safety cabinet, sterile cotton swab (DeltaLab, LOT: 191114), vortex (Scientific Industries, serial number A6. 1130), sonication bath (VWR Ultrasonic Cleaner, Model: USC100T), Varioskan Flash plate reader (serial number 3001-1778), BioTek Synergy plate reader (serial number 254.462), colony counter (Stuart, serial number RCC0221P160) and Applied Biosystems QuantStudio 7 Flex (serial number 278871498).
Microplastic and wood particles were collected by-hand in October 2021 from a riverbank in Truro, Cornwall (United Kingdom; 50.260048, −5.045549). Microplastics were separated into sterile 50 mL falcon tubes using forceps, with a separate tube for ‘bio-beads’ (bio-media from sewage treatment), ‘nurdles’ (pre-production pellets), expanded polystyrene beads and wood. All samples were transported in cool boxes and processed within 4 h. In the laboratory, all particles were rinsed in deionised water to remove any remaining sediment or natural debris from the surface, then left to air dry. Microplastic and wood particles that were 4 mm in size were selected for further processing. Additionally, 4 mm glass beads (Novagen ColiRollers, LOT: D00136263) were purchased.
Bio-beads, nurdles, glass beads and wood particles were autoclaved at 121°C for 15 min. To validate sterility, 6 of each particle were placed into sterile 15 mL falcon tubes containing 10 mL Iso-Sensitest broth (Oxoid; LOT: 3177183) and shaken (180 rpm) at 37°C overnight. Sterility was confirmed for all particles where there were no visual changes in optical density (OD) of the media. As the polystyrene particles were expanded polystyrene, autoclaving was not suitable for sterilisation. Therefore, gamma irradiation was outsourced to Becton Dickinson (Plymouth, United Kingdom): 500 polystyrene particles were separated into 5 mL Eppendorf tubes and gamma irradiated (10.2–10.6 kGy delivered for 3,600 s x2, followed by a further 900 s). As before, to validate sterility, 6 of the particles were placed into 10 mL Iso-Sensitest broth and shaken (180 rpm) at 37°C overnight. Sterility was confirmed for all particles where there were no visual changes in optical density (OD) of the media.
To ensure all nurdles, bio-beads and polystyrene particles were the same polymer type within their morphotypes, we performed attenuated total reflectance Fourier-transform infrared spectroscopy (ATR-FTIR) using a PerkinElmer 208 Spotlight 400 (Perkin Elmer, United Kingdom). Spectra were recorded as a mean of 4 scans at 4,000 nm wavelengths. Samples which produced spectra with a match less than 60% were automatically excluded. From this, bio-beads were identified as polyethylene and nurdles were identified as polypropylene. Any bio-beads or nurdles that did not match these polymers were excluded. As the machine applied force when scanning the particle, the shape of the polystyrene particles changed following identification. We therefore analysed a subset of these particles to ascertain that the remaining particles are highly likely to be expanded polystyrene.
Particles (bio-beads/polyethylene, nurdles/polypropylene, expanded polystyrene, wood and glass) were sampled and processed as described (see section 3.1).
Sewage influent samples were collected from a wastewater treatment plant in Falmouth, United Kingdom (serving a population of approximately 43,000) in June 2021. Samples were transported in cool boxes, then mixed 1:1 with 40% glycerol (ThermoFisher, LOT: P01H051) and stored at −70°C until use. Aliquots were then thawed, spun down at 14,800 rpm for 1 min, supernatant discarded and pellet resuspended in 1 mL sterile 0.85% NaCl (Sigma-Aldrich, PCode: 1003326144) twice to remove chemical and nutrient carryover (Murray et al., 2020). The resuspended pellet was used to inoculate at 10% (vol/vol) in 10 mL Iso-Sensitest broth in 50 mL sterile falcon tubes containing sterile particles (Figure 1). Tubes were shaken at 50 rpm for 20 h at 37°C. Within each tube (i.e., biological replicate, of which there were 6), there were 6 individual particle replicates of each particle type, to account for individual variability of particle shapes and sizes (Figure 1). This inoculation procedure was repeated 5 times using the same starting inoculum to provide a set of biofilms for each of the extraction methods and a set for 16S rRNA qPCR.
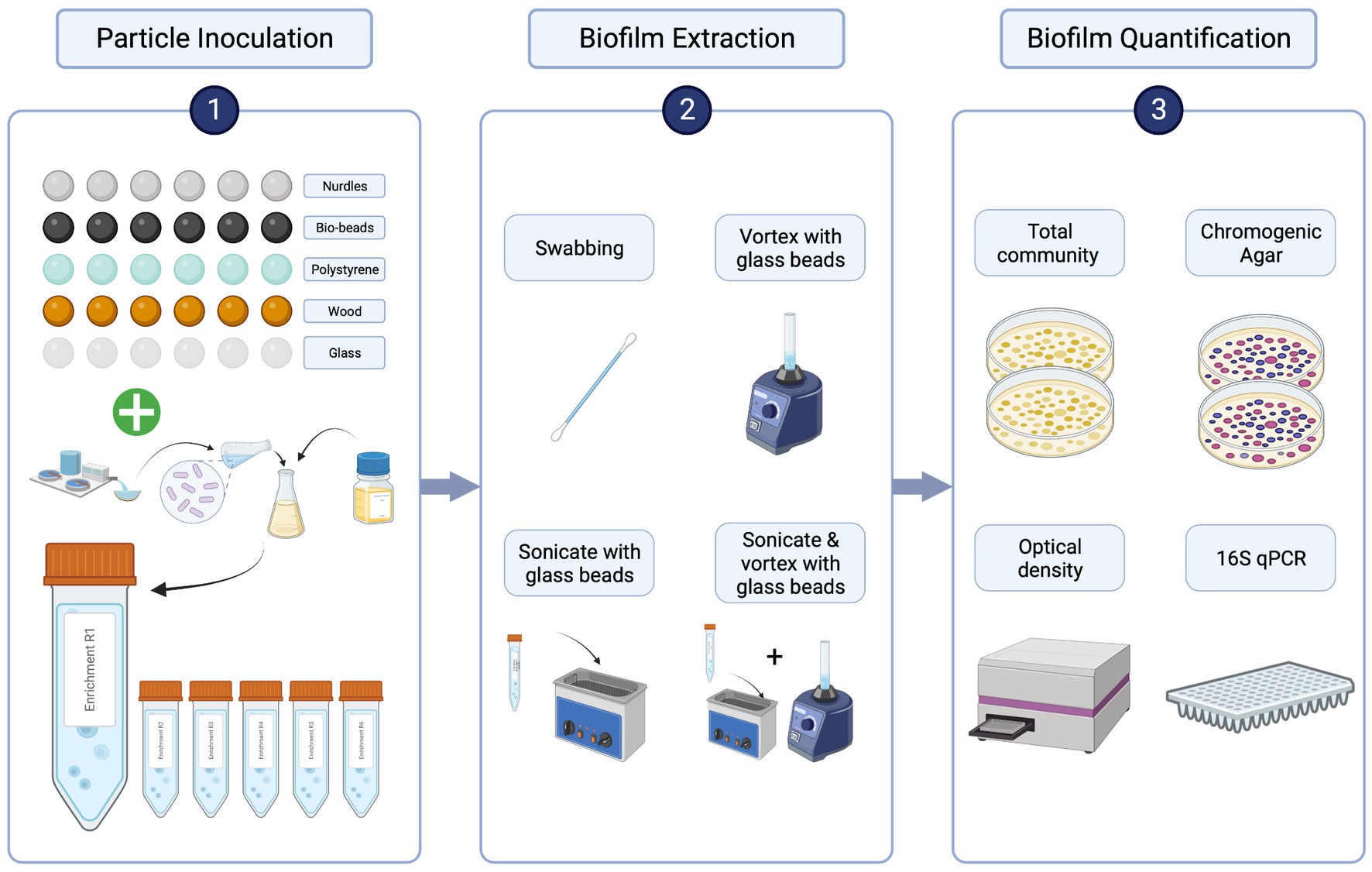
Figure 1. Comparing biofilm extraction protocols workflow. 1. Particles were inoculated with a washed sewage community in a nutrient rich broth overnight [x5 for each extraction method (4) and molecular validation (1)]. 2. Particles were rinsed with sterile NaCl then each of the 4 extraction methods were used to suspend biofilms from each particle type in sterile NaCl. 3. Biofilm extractions were quantified by plating biofilm suspensions on LB and chromogenic selective agar. Colony counts were validated using OD and genotypic quantification: DNA was extracted directly from the 5th set of inoculated particles using the DNeasy PowerBiofilm Kit (Qiagen), and the DNA was extracted from the biofilm suspensions for each extraction method using DNeasy UltraClean Microbial Kit (Qiagen). From these DNA samples, 16S rRNA quantitative-polymerase chain reaction (qPCR) was performed to assess the extraction efficiency of the biofilm extraction techniques compared with total biofilm community as obtained by the direct DNA extraction from the particles. Created with BioRender.com.
Following particle inoculation (see section 3.2.2), 500 μL of liquid culture was taken and cryogenically stored with glycerol. The remaining, surrounding liquid culture was decanted and particles were rinsed twice with sterile 0.85% NaCl to remove any loosely attached bacteria and left to air dry in sterile conditions inside a CAT-II safety cabinet. Each set of particles were then processed according to the different extraction methods, and biofilm suspension were produced as follows (Figure 2):
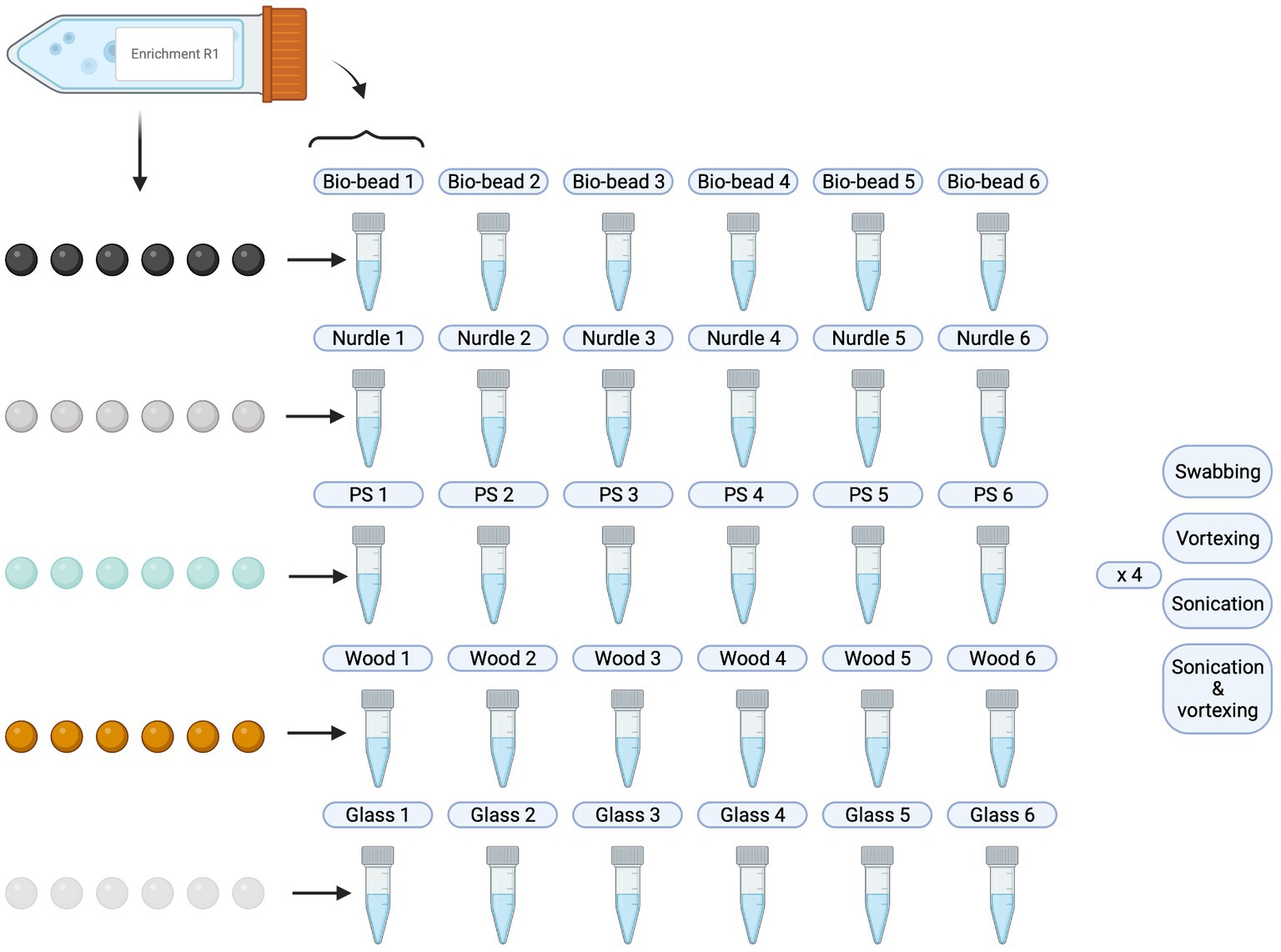
Figure 2. Schematic overview of biofilm suspensions produced. PS, polystyrene. Created with BioRender.com.
Swabbing: for each biological replicate and each particle type, individual particle replicates were secured using sterile forceps and, using a sterile cotton swab (DeltaLab, LOT: 191114), the entire surface of the particle was swabbed for 1 min. The cotton swab was then placed into an Eppendorf tube containing 600 μL sterile 0.85% NaCl and mixed in a circular motion for 10 s to encourage detachment of bacteria from the swab into the liquid.
Vortexing with glass beads: all 6 individual particle replicates for each of the 6 biological replicates for each particle type were placed into 600 μL sterile 0.85% NaCl in Eppendorf tubes. 5 sterile 4 mm glass beads were added to each tube and vortexed (2,500 rpm, Scientific Industries, serial number A6. 1130) for 1 min [as Lear et al. (2022)].
Sonicating with glass beads: all 6 individual particle replicates for each of the 6 biological replicates for each particle type were placed into 600 μL sterile 0.85% NaCl in Eppendorf tubes. 5 sterile 4 mm glass beads were added to each tube and placed into a sonication bath (VWR Ultrasonic Cleaner, Model: USC100T) for 15 min at 45 kHz.
Sonication with glass beads, followed by vortexing: all 6 individual particle replicates for each of the 6 biological replicates for each particle type were placed into 600 μL sterile 0.85% NaCl in Eppendorf tubes. 5 sterile 4 mm glass beads were added to each tube and placed into a sonication bath for 15 min at 45 kHz. Each tube was then vortexed (2,500 rpm, Scientific Industries, serial number A6. 1130) for 1 min.
From the suspensions from all the above methods, 100 μL was taken and used to generate OD (600 nm) readings, then diluted and plated onto agar plates. The remaining 500 μL of biofilm extraction was taken from the tube and placed into a fresh Eppendorf containing 500 μL 40% glycerol and stored at-70°C until DNA extraction.
DNA extraction directly from the biofilm: all 6 individual particle replicates for each of the 6 biological replicates for each particle type were placed into 1 mL 20% glycerol and stored at −70°C until DNA extraction.
OD readings were taken at 600 nm using either a Varioskan Flash (serial number 3001-1778) or BioTek Synergy (serial number 254.462). Absorbance values were not significantly affected by the model used (p > 0.05; Supplementary material 1.2), and only single timepoint measurements were obtained, thus reducing variability encountered by additional parameters, e.g., shaking speed and/or motion, and temperature variations. Sterile 0.85% NaCl was used for blanks/controls (6 replicates).
To enumerate total culturable bacterial community, LB agar (Fisher Bioreagents, LOT: 188453) was used. To distinguish and enumerate Escherichia coli (E. coli), coliforms and non-coliforms, CM1205 Chromogenic Coliform agar (ISO; Oxoid, LOT: 3004362) was used. Biofilm suspensions were diluted prior to plating according to a 10-fold dilution series performed a priori to detect the optimum dilution which achieved 20–80 CFUs of E. coli. 100 μL of each diluted biofilm suspension was plated in duplicate onto both types of agar using glass beads (Worthington et al., 2001). Plates were inverted and incubated at 37°C for 18–24 h. Colonies were then counted using a colony counter (Stuart, serial number RCC0221P160) and CFU/mL was generated for each colony phenotype: total (LB), E. coli (blue/purple colonies), non-E. coli coliforms (pink colonies) and non-coliforms (white colonies).
Once thawed, DNA extraction was performed on the biofilm suspensions using the DNeasy Ultra-Clean Microbial kit (Qiagen, LOT: 169011985) according to the manufacturer’s instructions. To extract DNA directly from the particles, cryogenic aliquots of inoculated particles were thawed, and DNA extraction was performed using the DNeasy PowerBiofilm kit (Qiagen, LOT: 169048148) according to the manufacturer’s instructions. These DNA samples were then used as template DNA for 16S rRNA quantitative polymerase chain reaction (qPCR).
Standard curves were generated with custom synthetic gBlocks (Table 2) provided by IDTDNA, prepared according to the manufacturer’s instructions and stored in single-use aliquots at-20°C. gBlocks used to generate standard curves were diluted 10-fold from 106 to 102. Efficiency of qPCR reactions ranged from 91% to 95%, with R2 values ranging from 0.997–0.999. qPCR was performed using the PrimerDesign Precision Plus SYBR Green Master Mix (Z-PPLUS-R-SY-10ML), on the Applied Biosystems QuantStudio 7 Flex (serial number 278871498). Reactions comprised of 10 μL Master Mix, 5 μL template, 2 μL primer (1 μL of each forward and reverse primers, Table 2), 0.2 μL bovine serum albumin (20 mg/mL, Fisher Scientific, LOT, 170419–0461), and nuclease free water up to a final volume of 20 μL. Cycling parameters used were a 2-min initial Hot Start activation at 95°C, followed by 40 cycles of data collection with 10 s at 95°C and 60 s at 60°C. Primers are listed in Table 2 and were provided by IDTDNA.
All statistics were performed in RStudio. All data (CFU/mL, OD readings and 16 s rRNA gene copy number, inclusive) were tested for normality using a Shapiro Wilks test. Where data was normally distributed, a t-test was used to compare biofilm extraction methods. Where data was non-normally distributed, a non-parametric Kruskal Wallis test was used. Where p < 0.05, a Dunn’s test was further used to identify significant differences. p-values were adjusted for multiple testing.
For the different particle types, CFU/mL were compared for each biofilm extraction method and verified against OD readings and 16S rRNA qPCR. Specifically, we investigated which extraction method significantly yielded the greatest E. coli, coliform, non-coliform and LB (‘total bacteria’) CFU/mL and highest OD readings. For 16S rRNA data, we determined which extraction method achieved an average 16S rRNA gene copy number that was not significantly different to (or greater than) the direct biofilm DNA extraction control. Where an extraction method, or multiple extraction methods, were found to be the most efficient, this was noted in Table 3. For example, data generated for polystyrene (Figure 3), revealed that sonication followed by vortexing, and vortexing alone yielded coliform counts significantly higher than sonication alone, or swabbing (p < 0.05). In this case, sonication followed by vortexing and vortexing alone were allocated as the most efficient extraction methods for coliforms (Table 3).
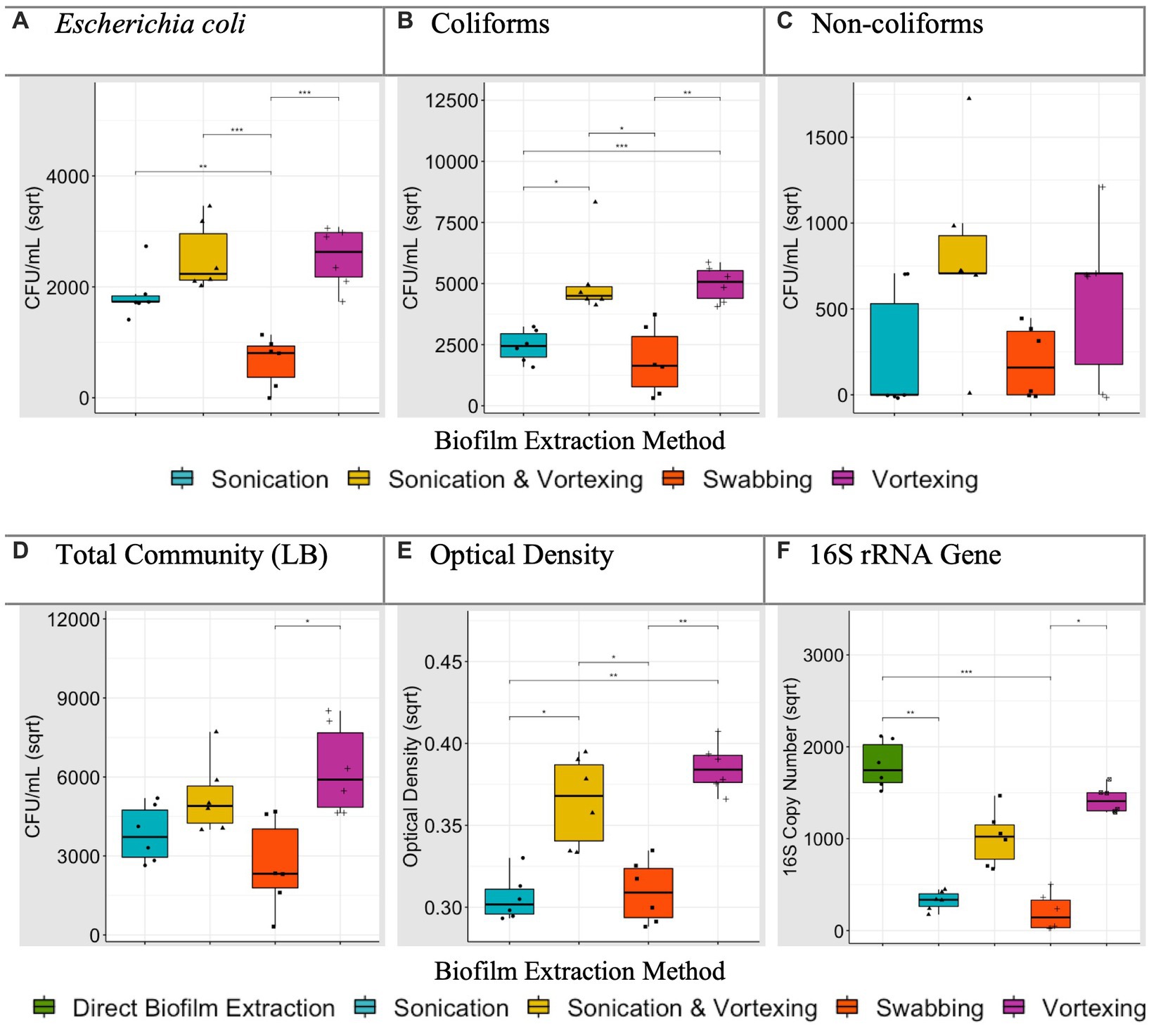
Figure 3. Average (biological replicate = 6) polystyrene biofilm extraction quantification for each extraction technique. *p < 0.05, t-test or Dunn’s test according to normality (adjusted for multiple comparisons). Plots (A–D) present square root transformed CFU/mL data. Plot (E) presents square root transformed OD (600 nm) data. Plot (F) presents square root transformed 16S rRNA gene copy number data, including the direct biofilm DNA extraction treatment.
After analysing biofilm extraction data for all particles (all Figures for remaining particles can be found in Supplementary material, summarised in Table 3), swabbing was the most efficient extraction method 1 time (3.3%), vortexing with glass beads was the most efficient 15 times (50%), sonicating with glass beads was the most efficient 14 times (47%) and sonicating followed by vortexing with glass beads was the most efficient 24 times (80%; Table 3). With this, and taking the processing time (Table 4) into account, we concluded that sonication followed by vortexing was the most efficient method for extracting the biofilms from our study particles.
This study aimed to identify a simple and repeatable method which efficiently extracted particle-associated biofilm communities into a culturable, bacterial suspension. To do this, we used four different extraction methods, which had been previously adopted (Table 1) to extract complex bacterial biofilms from inoculated microplastics and control particles. By comparing CFU/mL, OD readings and 16S rRNA qPCR and factoring in time constraints, we concluded that sonication with glass beads followed by vortexing was the most efficient way to extract the biofilms from our test particles (bio-beads, nurdles, polystyrene, wood and glass).
Supporting our finding, Kobayashi et al. (2009) performed a similar study which aimed to improve the detection of biofilm-forming Staphylococcus aureus strains on polymethylmethacrylate coupons. The combination of sonication and vortexing enhanced the yield of culturable bacteria. In addition, whilst the present study is designed with environmental microbiology in mind where bacteria grow in complex communities, Kobayashi et al. (2009) proposed that their finding may be useful in the clinical setting for dislodging biofilm forming bacteria from orthopaedic devices or other implants.
Though the primary focus of this study was to compare the efficiency of different extraction methods to collect viable cells from microplastic-associated biofilms, as a result, we also compared amount of the 16S rRNA gene. Our results suggest that a biofilm suspension step, prior to DNA extraction, is unnecessary. This is because the 16S rRNA copy numbers within the direct DNA extractions was consistently greater than most of the extraction methods we tested, irrespective of particle type. Additionally, if the biofilm suspensions are plated on agar and the cultured bacteria are used for downstream molecular work, it is important to recognise that the majority of naturally occurring bacteria are non-culturable, and therefore this would not give a full community representation.
As for the limitations in the present study, it should be noted that the methods described here may only be suitable for larger microplastic particles, greater than 2 mm, and of an appropriate size for sonication and vortexing. Additional modifications may be required for smaller particles, and for larger particles, methods like swabbing and scraping may be more suitable. Finally, culturing bacteria from nanoplastics remains largely uninvestigated and requires further research attention. When investigating the role of nanoplastics in supporting Plastisphere communities, unique challenges arise, largely owing to the smaller dimensions of these particles, which may be similar to or even smaller than microorganisms (Shi et al., 2023). Therefore, though nanoplastics are proposed to pose greater levels of toxicity than microplastics (Sharma et al., 2023), their ability to support microbial communities or influence interactions between microbes is unclear, and methods to investigate this remain under development.
In summary, we demonstrate that using sonication in combination with vortexing produces an efficient yield of culturable cells, comparable to the quantity that can be obtained by extracting the DNA directly from the biofilm using a specifically designed, biofilm extraction kit. We therefore propose that sonication in combination with vortexing should be adopted when isolating culturable bacteria from the Plastisphere. By standardising our methods across laboratories and study designs, it will increase our understanding of the role of microplastics in supporting distinct, pathogenic or AMR communities, and the subsequent ecological threats they pose.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
ES: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. AB: Conceptualization, Funding acquisition, Methodology, Supervision, Validation, Writing – review & editing. MC: Conceptualization, Funding acquisition, Methodology, Supervision, Validation, Writing – review & editing. PL: Conceptualization, Funding acquisition, Methodology, Supervision, Validation, Writing – review & editing. AM: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. We acknowledge support and give thanks to the generous philanthropic donations made to support this work from Melissa Murdoch, the Barnsbury Trust and Beach Guardian. Thank you also to the University of Exeter and Plymouth Marine Laboratory for providing funding for this research. AB acknowledges support from NERC and BBSRC. AM acknowledges support from NERC (NE/W006251/1). PL and MC acknowledge support from NERC (NE/V007351/1).
Thank you to William Hugo Gaze (University of Exeter) for scientific insight during the planning of this work. Thank you to Becton Dickinson (Plymouth, United Kingdom) for their assistance in particle sterilisation. Thank you to the Laboratory Technicians at the University of Exeter’s Environment and Sustainability Institute and Plymouth Marine Laboratory for experimental assistance and laboratory training. For the purpose of open access, the authors have applied a “Creative Commons Attribution (CC BY)” to any Author Accepted Manuscript version arising.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1259287/full#supplementary-material
Arias-Andres, M., Klümper, U., Rojas-Jimenez, K., and Grossart, H.-P. (2018). Microplastic pollution increases gene exchange in aquatic ecosystems. Environ. Pollut. 237, 253–261. doi: 10.1016/j.envpol.2018.02.058
Bartkova, S., Kahru, A., Heinlaan, M., and Scheler, O. (2021). Techniques used for analyzing microplastics, antimicrobial resistance and microbial community composition: a mini-review. Front. Microbiol. 12:603967. doi: 10.3389/fmicb.2021.603967
Bowley, J., Baker-Austin, C., Porter, A., Hartnell, R., and Lewis, C. (2021). Oceanic hitchhikers–assessing pathogen risks from marine microplastic. Trends Microbiol. 29, 107–116. doi: 10.1016/j.tim.2020.06.011
Carpenter, E. J., Anderson, S. J., Harvey, G. R., Miklas, H. P., and Peck, B. B. (1972). Polystyrene spherules in coastal waters. Science 178, 749–750. doi: 10.1126/science.178.4062.749
De Tender, C., Schlundt, C., Devriese, L., Mincer, T., Zettler, E., and Amaral-Zettler, L. (2017). A review of microscopy and comparative molecular-based methods to characterize “plastisphere” communities. Anal. Methods 9, 2132–2143. doi: 10.1039/C7AY00260B
Debeljak, P., Pinto, M., Proietti, M., Reisser, J., Ferrari, F. F., Abbas, B., et al. (2017). Extracting DNA from ocean microplastics: a method comparison study. Anal. Methods 9, 1521–1526. doi: 10.1039/C6AY03119F
Forero-López, A., Brugnoni, L., Abasto, B., Rimondino, G., Lassalle, V., Ardusso, M., et al. (2022). Plastisphere on microplastics: in situ assays in an estuarine environment. J. Hazard. Mater. 440:129737. doi: 10.1016/j.jhazmat.2022.129737
Grogan, A. E., Mallin, M. A., and Cahoon, L. B. (2021). Investigation of polyethylene terephthalate (PET) drinking bottles as marine reservoirs for fecal bacteria and phytoplankton. Mar. Pollut. Bull. 173:113052. doi: 10.1016/j.marpolbul.2021.113052
Hernández-Sánchez, C., Pestana-Ríos, Á. A., Villanova-Solano, C., Domínguez-Hernández, C., Díaz-Peña, F. J., Rodríguez-Álvarez, C., et al. (2023). Bacterial colonization of microplastics at the beaches of an Oceanic Island, Tenerife, Canary Islands. Int. J. Environ. Res. Public Health 20:3951. doi: 10.3390/ijerph20053951
Joseph, B., Otta, S. K., Karunasagar, I., and Karunasagar, I. (2001). Biofilm formation by salmonella spp. on food contact surfaces and their sensitivity to sanitizers. Int. J. Food Microbiol. 64, 367–372. doi: 10.1016/S0168-1605(00)00466-9
Junker, L. M., and Hay, A. G. (2004). Effects of triclosan incorporation into ABS plastic on biofilm communities. J. Antimicrob. Chemother. 53, 989–996. doi: 10.1093/jac/dkh196
Kapetanović, D., Vardić Smrzlić, I., Kazazić, S., Omanović, D., Cukrov, N., Cindrić, A.-M., et al. (2023). A preliminary study of the cultivable microbiota on the plastic litter collected by commercial fishing trawlers in the South-Eastern Adriatic Sea, with emphasis on vibrio isolates and their antibiotic resistance. Mar. Pollut. Bull. 187:114592. doi: 10.1016/j.marpolbul.2023.114592
Kim, M., and Han, M. (2014). Characteristics of biofilm development in an operating rainwater storage tank. Environ. Earth Sci. 72, 1633–1642. doi: 10.1007/s12665-014-3067-2
Kirstein, I. V., Kirmizi, S., Wichels, A., Garin-Fernandez, A., Erler, R., Löder, M., et al. (2016). Dangerous hitchhikers? Evidence for potentially pathogenic vibrio spp. on microplastic particles. Mar. Environ. Res. 120, 1–8. doi: 10.1016/j.marenvres.2016.07.004
Kobayashi, H., Oethinger, M., Tuohy, M. J., Procop, G. W., and Bauer, T. W. (2009). Improved detection of biofilm-formative bacteria by vortexing and sonication: a pilot study. Clin. Orthop. Relat. Res. 467, 1360–1364. doi: 10.1007/s11999-008-0609-5
Laganà, P., Caruso, G., Corsi, I., Bergami, E., Venuti, V., Majolino, D., et al. (2019). Do plastics serve as a possible vector for the spread of antibiotic resistance? First insights from bacteria associated to a polystyrene piece from King George Island (Antarctica). Int. J. Hyg. Environ. Health 222, 89–100. doi: 10.1016/j.ijheh.2018.08.009
Lear, L., Padfield, D., Dowsett, T., Jones, M., Kay, S., Hayward, A., et al. (2022). Bacterial colonisation dynamics of household plastics in a coastal environment. Sci. Total Environ. 838:156199. doi: 10.1016/j.scitotenv.2022.156199
Lehtola, M. J., Miettinen, I. T., Keinänen, M. M., Kekki, T. K., Laine, O., Hirvonen, A., et al. (2004). Microbiology, chemistry and biofilm development in a pilot drinking water distribution system with copper and plastic pipes. Water Res. 38, 3769–3779. doi: 10.1016/j.watres.2004.06.024
Liang, H., de Haan, W. P., Cerdà-Domènech, M., Méndez, J., Lucena, F., García-Aljaro, C., et al. (2023). Detection of faecal bacteria and antibiotic resistance genes in biofilms attached to plastics from human-impacted coastal areas. Environ. Pollut. 319:120983. doi: 10.1016/j.envpol.2022.120983
Liu, S., Deng, W.-K., Niu, S.-H., Mo, C.-H., Liao, X.-D., and Xing, S.-C. (2023). Doxycycline combined manure microbes to enhances biofilm formation of the soil plastisphere and increases the surface bio-risk of microplastics vehicle. Chem. Eng. J. 454:140530. doi: 10.1016/j.cej.2022.140530
Lobelle, D., and Cunliffe, M. (2011). Early microbial biofilm formation on marine plastic debris. Mar. Pollut. Bull. 62, 197–200. doi: 10.1016/j.marpolbul.2010.10.013
Metcalf, R., White, H. L., Moresco, V., Ormsby, M. J., Oliver, D. M., and Quilliam, R. S. (2022). Sewage-associated plastic waste washed up on beaches can act as a reservoir for faecal bacteria, potential human pathogens, and genes for antimicrobial resistance. Mar. Pollut. Bull. 180:113766. doi: 10.1016/j.marpolbul.2022.113766
Metcalf, R., White, H. L., Ormsby, M. J., Oliver, D. M., and Quilliam, R. S. (2023). From wastewater discharge to the beach: survival of human pathogens bound to microplastics during transfer through the freshwater-marine continuum. Environ. Pollut. 319:120955. doi: 10.1016/j.envpol.2022.120955
Moore, R. E., Millar, B. C., and Moore, J. E. (2020). Antimicrobial resistance (AMR) and marine plastics: can food packaging litter act as a dispersal mechanism for AMR in oceanic environments? Mar. Pollut. Bull. 150:110702. doi: 10.1016/j.marpolbul.2019.110702
Morfin, A. P. S. (2021). Microbial biofilms on microplastics: A look into the estuarine plastisphere of the Chesapeake Bay. University of Maryland, College Park.
Munir, E., Suryanto, D., Pasaribu, Y., Mubtasima, S., Hartanto, A., Lutfia, A., et al. (2022). "Occurrence of microbial community on plastic wastes in Terjun landfill, North Sumatra", In: IOP conference series: Earth and environmental science IOP Publishing), 012080.
Murray, A. K., Stanton, I. C., Wright, J., Zhang, L., Snape, J., and Gaze, W. H. (2020). The selection end points in communities of bacteria SELECT method: a novel experimental assay to facilitate risk assessment of selection for antimicrobial resistance in the environment. Environ. Health Perspect. 128:107007. doi: 10.1289/EHP6635
Murray, A. K., Zhang, L., Yin, X., Zhang, T., Buckling, A., Snape, J., et al. (2018). Novel insights into selection for antibiotic resistance in complex microbial communities. MBio 9:00969-00918. doi: 10.1128/mbio
Ormsby, M. J., White, H. L., Metcalf, R., Oliver, D. M., and Quilliam, R. S. (2023). Clinically important E. coli strains can persist, and retain their pathogenicity, on environmental plastic and fabric waste. Environ. Pollut. 326:121466. doi: 10.1016/j.envpol.2023.121466
Pang, G., Li, X., Ding, M., Jiang, S., Chen, P., Zhao, Z., et al. (2023). The distinct plastisphere microbiome in the terrestrial-marine ecotone is a reservoir for putative degraders of petroleum-based polymers. J. Hazard. Mater. 453:131399. doi: 10.1016/j.jhazmat.2023.131399
Perveen, S., Pablos, C., Reynolds, K., Stanley, S., and Marugán, J. (2023). Growth and prevalence of antibiotic-resistant bacteria in microplastic biofilm from wastewater treatment plant effluents. Sci. Total Environ. 856:159024. doi: 10.1016/j.scitotenv.2022.159024
Quilliam, R. S., Jamieson, J., and Oliver, D. M. (2014). Seaweeds and plastic debris can influence the survival of faecal indicator organisms in beach environments. Mar. Pollut. Bull. 84, 201–207. doi: 10.1016/j.marpolbul.2014.05.011
Radisic, V., Nimje, P. S., Bienfait, A. M., and Marathe, N. P. (2020). Marine plastics from Norwegian west coast carry potentially virulent fish pathogens and opportunistic human pathogens harboring new variants of antibiotic resistance genes. Microorganisms 8:1200. doi: 10.3390/microorganisms8081200
Rasool, F. N., Saavedra, M. A., Pamba, S., Perold, V., Mmochi, A. J., Maalim, M., et al. (2021). Isolation and characterization of human pathogenic multidrug resistant bacteria associated with plastic litter collected in Zanzibar. J. Hazard. Mater. 405:124591. doi: 10.1016/j.jhazmat.2020.124591
Rodrigues, A., Oliver, D. M., McCarron, A., and Quilliam, R. S. (2019). Colonisation of plastic pellets (nurdles) by E. coli at public bathing beaches. Mar. Pollut. Bull. 139, 376–380. doi: 10.1016/j.marpolbul.2019.01.011
Sharma, V. K., Ma, X., Lichtfouse, E., and Robert, D. (2023). Nanoplastics are potentially more dangerous than microplastics. Environ. Chem. Lett. 21, 1933–1936. doi: 10.1007/s10311-022-01539-1
Shi, X., Chen, Z., Wei, W., Chen, J., and Ni, B.-J. (2023). Toxicity of micro/nanoplastics in the environment: roles of plastisphere and eco-corona. Soil Environ Health 1:100002. doi: 10.1016/j.seh.2023.100002
Silva, M. M., Maldonado, G. C., Castro, R. O., de Sá Felizardo, J., Cardoso, R. P., Anjos, R. M., et al. (2019). Dispersal of potentially pathogenic bacteria by plastic debris in Guanabara Bay, RJ, Brazil. Mar. Pollut. Bull. 141, 561–568. doi: 10.1016/j.marpolbul.2019.02.064
Song, J., Jongmans-Hochschulz, E., Mauder, N., Imirzalioglu, C., Wichels, A., and Gerdts, G. (2020). The travelling particles: investigating microplastics as possible transport vectors for multidrug resistant E. coli in the Weser estuary (Germany). Sci. Total Environ. 720:137603. doi: 10.1016/j.scitotenv.2020.137603
Sudhakar, M., Trishul, A., Doble, M., Kumar, K. S., Jahan, S. S., Inbakandan, D., et al. (2007). Biofouling and biodegradation of polyolefins in ocean waters. Polym. Degrad. Stab. 92, 1743–1752. doi: 10.1016/j.polymdegradstab.2007.03.029
Suzuki, M. T., Taylor, L. T., and DeLong, E. F. (2000). Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66, 4605–4614. doi: 10.1128/AEM.66.11.4605-4614.2000
Syranidou, E., Karkanorachaki, K., Amorotti, F., Repouskou, E., Kroll, K., Kolvenbach, B., et al. (2017). Development of tailored indigenous marine consortia for the degradation of naturally weathered polyethylene films. PloS One 12:e0183984. doi: 10.1371/journal.pone.0183984
Szabó, I., al-Omari, J., Szerdahelyi, G. S., Farkas, M., al-Omari, Y., Szabó, P. M., et al. (2021). In situ investigation of plastic-associated bacterial communities in a freshwater lake of Hungary. Water Air Soil Pollut. 232:493. doi: 10.1007/s11270-021-05445-0
Tan, B., Li, Y., Xie, H., Dai, Z., Zhou, C., Qian, Z.-J., et al. (2022). Microplastics accumulation in mangroves increasing the resistance of its colonization vibrio and Shewanella. Chemosphere 295:133861. doi: 10.1016/j.chemosphere.2022.133861
Thompson, R. C., Olsen, Y., Mitchell, R. P., Davis, A., Rowland, S. J., John, A. W., et al. (2004). Lost at sea: where is all the plastic? Science 304:838. doi: 10.1126/science.1094559
Vidal-Verdú, À., Latorre-Pérez, A., Molina-Menor, E., Baixeras, J., Peretó, J., and Porcar, M. (2022). Living in a bottle: bacteria from sediment-associated Mediterranean waste and potential growth on polyethylene terephthalate. Microbiol Open 11:e1259. doi: 10.1002/mbo3.1259
Worthington, M. T., Luo, R. Q., and Pelo, J. (2001). Copacabana method for spreading E. coli and yeast colonies. Biotechniques 30, 738–742. doi: 10.2144/01304bm05
Zettler, E. R., Mincer, T. J., and Amaral-Zettler, L. A. (2013). Life in the “plastisphere”: microbial communities on plastic marine debris. Environ. Sci. Technol. 47, 7137–7146. doi: 10.1021/es401288x
Zhang, Y., Lu, J., Wu, J., Wang, J., and Luo, Y. (2020). Potential risks of microplastics combined with superbugs: enrichment of antibiotic resistant bacteria on the surface of microplastics in mariculture system. Ecotoxicol. Environ. Saf. 187:109852. doi: 10.1016/j.ecoenv.2019.109852
Zhurina, M. V., Bogdanov, K. I., Mendeleev, D. I., Tikhomirov, V. A., Pleshko, E. M., Gannesen, A. V., et al. (2023). Phylogenetic constitution and survival of microbial biofilms formed on the surface of polyethylene composites protected with Polyguanidine biocides. Coatings 13:987. doi: 10.3390/coatings13060987
Keywords: biofilm, microplastics, Plastisphere, bacterial culture, biofilm extraction, microbiology
Citation: Stevenson EM, Buckling A, Cole M, Lindeque PK and Murray AK (2023) Culturing the Plastisphere: comparing methods to isolate culturable bacteria colonising microplastics. Front. Microbiol. 14:1259287. doi: 10.3389/fmicb.2023.1259287
Received: 15 July 2023; Accepted: 04 September 2023;
Published: 03 October 2023.
Edited by:
George Tsiamis, University of Patras, GreeceReviewed by:
João Pedro Rueda Furlan, University of São Paulo, BrazilCopyright © 2023 Stevenson, Buckling, Cole, Lindeque and Murray. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aimee K. Murray, YS5rLm11cnJheUBleGV0ZXIuYWMudWs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.