
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 12 October 2023
Sec. Virology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1258810
This article is part of the Research Topic Transmission and Infection of Arboviruses View all 14 articles
 Jennifer M. Potter-Birriel1†
Jennifer M. Potter-Birriel1† Adam R. Pollio1†
Adam R. Pollio1† Brian D. Knott2
Brian D. Knott2 Tamar Chunashvili2
Tamar Chunashvili2 Christian K. Fung1
Christian K. Fung1 Matthew A. Conte1
Matthew A. Conte1 Drew D. Reinbold-Wasson2
Drew D. Reinbold-Wasson2 Jun Hang1*
Jun Hang1*Arbovirus surveillance is fundamental for the discovery of novel viruses and prevention of febrile vector-borne illnesses. Vector-borne pathogens can rapidly expand and adapt in new geographic and environmental conditions. In this study, metagenomic surveillance was conducted to identify novel viruses in the Country of Georgia. A total of 521 mosquitoes were captured near a military training facility and pooled from species Culex pipiens (Linnaeus) (87%) and Aedes albopictus (Skuse) (13%). We decided to further analyze the Culex pipiens mosquitoes, due to the more extensive number of samples collected. Our approach was to utilize an unbiased total RNA-seq for pathogen discovery in order to explore the mosquito virome. The viral reads from this analysis were mostly aligned to Insect-specific viruses from two main families, the Iflaviridae; a positive-stranded RNA virus and the Rhabdoviridae; a negative- and single-stranded RNA virus. Our pathogen discovery analysis revealed viral reads aligning to the Merida-like virus Turkey (MERDLVT) strain among the Rhabdoviridae. To further validate this result, we conducted a BLAST sequence comparison analysis of our samples with the MERDLVT strain. Our positive samples aligned to the MERDLVT strain with 96–100% sequence identity and 99.7–100% sequence coverage. A bootstrapped maximum-likelihood phylogenetic tree was used to evaluate the evolutionary relationships among these positive pooled specimens with the (MERDLVT) strain. The Georgia samples clustered most closely with two strains from Turkey, the Merida-like virus KE-2017a isolate 139-1-21 and the Merida-like virus Turkey isolate P431. Collectively, these results show the presence of the MERDLVT strain in Georgia.
Vector-borne pathogens (VBPs) are one of the main causes of human infections worldwide (Kilpatrick and Randolph, 2012; de Almeida et al., 2021). The geographic boundaries of these vectors continue to expand and adapt around the world’s changing climate (Kilpatrick and Randolph, 2012). For this reason, pathogen discovery becomes increasingly important for arbovirus surveillance and the prevention of VBPs. The most prominent and emerging viruses belong to the Flaviviridae family. Flaviviruses are positive-stranded RNA viruses, that are recognized to have the capacity to globally infect and cause a spectrum of several diseases (Pierson and Diamond, 2020; Cuevas-Juarez et al., 2021). This family include several significant human pathogens including Yellow Fever virus (YFV), Zika virus (ZIKV), and Dengue virus (DENV) (Pierson and Diamond, 2020; Cuevas-Juarez et al., 2021). Another major vector-borne virus family, Rhabdoviridae, are single-stranded negative RNA viruses, characterized for being ubiquitous, with the ability to infect a wide range of species including plants, vertebrates, and invertebrates (Dietzgen et al., 2017). Two well-known rhabdoviruses include the rabies virus which causes disease in various animals and the infectious hematopoietic necrosis virus which causes disease in salmonid fish. Rhabdoviruses have been shown to exhibit rapid mutation rates and complex genome evolution including gains and losses of genes (Walker et al., 2015). This constant evolution of new variants has made it difficult to the scientific community to develop new vaccines. For this reason, efforts to expand mosquito-borne surveillance in developing countries must continue to be a main priority to keep up with the evolution of emerging viral diseases.
The advances of sequencing metagenomic analysis has led to the discovery of new viruses worldwide, leading to the detection and prevention of emerging viruses. A group of viruses that continues to be detected in mosquitoes are the Insect-specific viruses (ISVs). ISVs can naturally infect and replicate in arthropod hosts. An example of ISVs is the Culex Iflavi-like virus 4 which belongs to the Iflaviridae family; a positive-stranded RNA virus (Valles et al., 2017). In recent studies, researchers conducted studies to understand how these ISVs interact with pathogenic arboviruses and how it can be of benefit for vector borne disease control. Continued research will help to develop biotechnology tools that can serve as vector controls to reduce or restrict arboviral diseases (Liu et al., 2011; Bolling et al., 2012; Agboli et al., 2019). In addition, other groups are using ISVs as expression tools against arboviruses for the development of new vaccines (Erasmus et al., 2018). ISVs is gaining interest as potential vectors to restrict pathogenic hosts due to its inability to infect vertebrates (Agboli et al., 2019).
Here, we report the presence of the Merida virus in Senaki, a town within the country of Georgia. The Merida virus was discovered in Culex pipiens mosquitoes collected in various sites of a Georgia military facility. Metagenomic analysis was executed using the Chan Zuckerberg ID (CZ ID) cloud-based pipeline; results were further evaluated using phlyogenetics to determine the likely evolutionary origin of our samples. The Merida virus is classified as a rhabdovirus, a single-stranded, negative RNA virus that was originally discovered in the Yucatan Peninsula of Mexico (Charles et al., 2016).
Adult host seeking mosquitoes were collected using multiple traps: BG Sentinel 2 (Biogents AG, Regensburg, Germany), CDC light trap (Model 1,012 and 1,212, John W. Hock Company, Gainesville, FL), Stealth Trap (Model 214, John W. Hock Company, Gainesville, FL), and Fay-Prince Trap (Model 812, John W. Company, Gainesville, FL). Collection traps were placed on a tree that was either next to a pond or a pile of tires. Larval dipping collection focused on small water filled containers and larger water pools in order to collect immature mosquitoes. The larval mosquitoes were then allowed to mature in sealed containers then collected as adults. Mosquito collections were conducted between August 2018 and June 2019, in the town of Senaki, located in the Samegrelo-Zemo Svaneti region of Georgia. Mosquitoes were morphologically identified using a stereomicroscope (Leica S4E, Leica microsystems, Germany) and the ECDC MosKey Tool.1 Female mosquitoes were sorted into pools of no more than 20 individuals and stored at −80°C. Specimens were shipped frozen from Tbilisi, Georgia to Silver Spring, Maryland, United States. Where they were processed for advanced molecular characterization at the Walter Reed Army Institute of Research, Viral Detection Branch (WRAIR VDB).
Mosquitos were homogenized by bead-beating with the Bio Spec Mini-Bead Beater 16 (Bio Spec Products Inc., Bartlesville, OK, United States). The mosquitos’ homogenates were centrifuged, and the supernatant was treated with DNase I, Benzonase nuclease and RNase A as described in Sanborn et al. (2019). Lysis and RNA extraction was performed using the 5XMag MAX Pathogen RNA/DNA kit (Cat# 4462359) with the Thermo Scientific Kingfisher Flex by following the manufacturer’s user guides.
Random reverse transcription and PCR amplification (RT-PCR) was conducted on the purified nucleic acid as described by Sanborn et al. (2019). The RT-PCR amplicons were purified and quantified using the Quant-iT™ Pico Green™ dsDNA Assay (Thermo Fisher Scientific). Library prep was made using the DNA Prep library prep kit (Illumina, San Diego, CA, United States) (Product # 20060059). Libraries were quantitated using Tape Station, D5000 Screen Tape (Agilent Technologies, Inc., Santa Clara, CA, United States), pooled at equal molar concentrations, and sequenced using Illumina MiSeq system and Sequencing Reagent Kit v3 (600-cycle) (Product # MS-102-3003).
Data analysis was performed by uploading MiSeq raw sequence read data to CZID.org (Ramesh et al., 2019; Saha et al., 2019; Kalantar et al., 2020) followed by the metagenomic analysis function within the site. The data was then sorted by reference from the NCBI database. Heat maps were also generated by CZID.org.
We further verified our results by using our in-house de novo pipeline previously described (Kilianski et al., 2015).This is a preprocessing step using Cutadapt (Martin, 2011) and Prinseq (Schmieder and Edwards, 2011), followed by a de-novo assembly using RAY Meta (Boisvert et al., 2012). The resulting contigs are then further combined using Cap3 (Huang and Madan, 1999). The final scaffold is then categorized with iterative BLAST against the NCBI nucleotide database (nr/nt).
The Merida-like virus KE-2017a (accession number NC_040532.1) was used as a reference to map sequence reads on Geneious prime. The resulting sequences were then aligned to remove sequencing artifacts followed by a BLAST to show sequence similarity and identity. We then identified the individual ORFs using BLASTp for our annotation.
RASTtk (Brettin et al., 2015) with the “correct frame shift” option was used to annotate the assembled genome sequence. We referenced Merida virus (Taxon ID: 1803034) to build and identify ORFs. We then compared those results using the protein search tool BLASTp (Altschul et al., 1990). Proteins with low similarity were not reported.
We compared 10 whole genome Merida-like virus (Georgia) sequences with 30 publicly available Merida-like viruses sequences, including Hirame rhabdovirus, Infectious hematopoietic necrosis virus, Zahedan rhabdovirus, Long Island tick rhabdovirus, Moussa virus, Arborteum virus, Puerto Almendras virus, Bovine ephemeral fever virus, Kimberley virus, Coastal Plain virus, Tibrogargan virus, Shayang Fly Virus 2 strain, Drosophila melanogaster sigmavirus AP30, Wuhan Louse Fly Virus 9 strain, Iriri virus nucleoprotein, European bat 1 lyssavirus, Australian bat lyssavirus, Rabies virus, Bole Tick Virus 2 strain, Culex tritaeniorhynchus rhabdovirus, Culex rhabdovirus strain CRV (Ticino, Switzerland), Merida virus isolate XY14959 nucleoprotein, Merida virus isolate CC_H, Culex rhabdovirus strain CRV (Kern, California), Merida virus isolate MERDV-2020, Merida virus isolate CMS002, Merida virus isolate MERD-Mex07, Merida virus OTU4, Merida-like virus KE-2017a isolate, and Merida-like virus isolate P431. The sequences were aligned using MAFFT version v7.475 using the high-speed setting (Katoh and Standley, 2013). This alignment quality was inspected and confirmed in Geneious, and a phylogenetic tree was constructed using IQ-TREE version 2.0.3 with Model Finder to determine the most appropriate model and the following specific settings: “-ninit 2 -bb 1,000 -nt 4” (Kalyaanamoorthy et al., 2017; Hoang et al., 2018). The resulting tree was edited using FigTree version 1.4.4.
Mosquito sampling was conducted at 10 sites in Senaki, a town in Samegrelo-Zemo Svaneti region located in western Georgia. These collections were made in 2018 and 2019 on a Georgia military training base, the coordinates of each site were recorded using the Global Positioning System (GPS) (Figure 1A; Supplementary Table S1). A total of 521 mosquitoes were captured and distributed into 45 pools from species Culex pipiens (87%) and Aedes albopictus (13%) (Figure 1B). Due to the more extensive number of samples collected, we decided to further analyze Culex pipiens mosquitoes. Figure 1C illustrates the number of Culex pipiens mosquitoes collected each year.
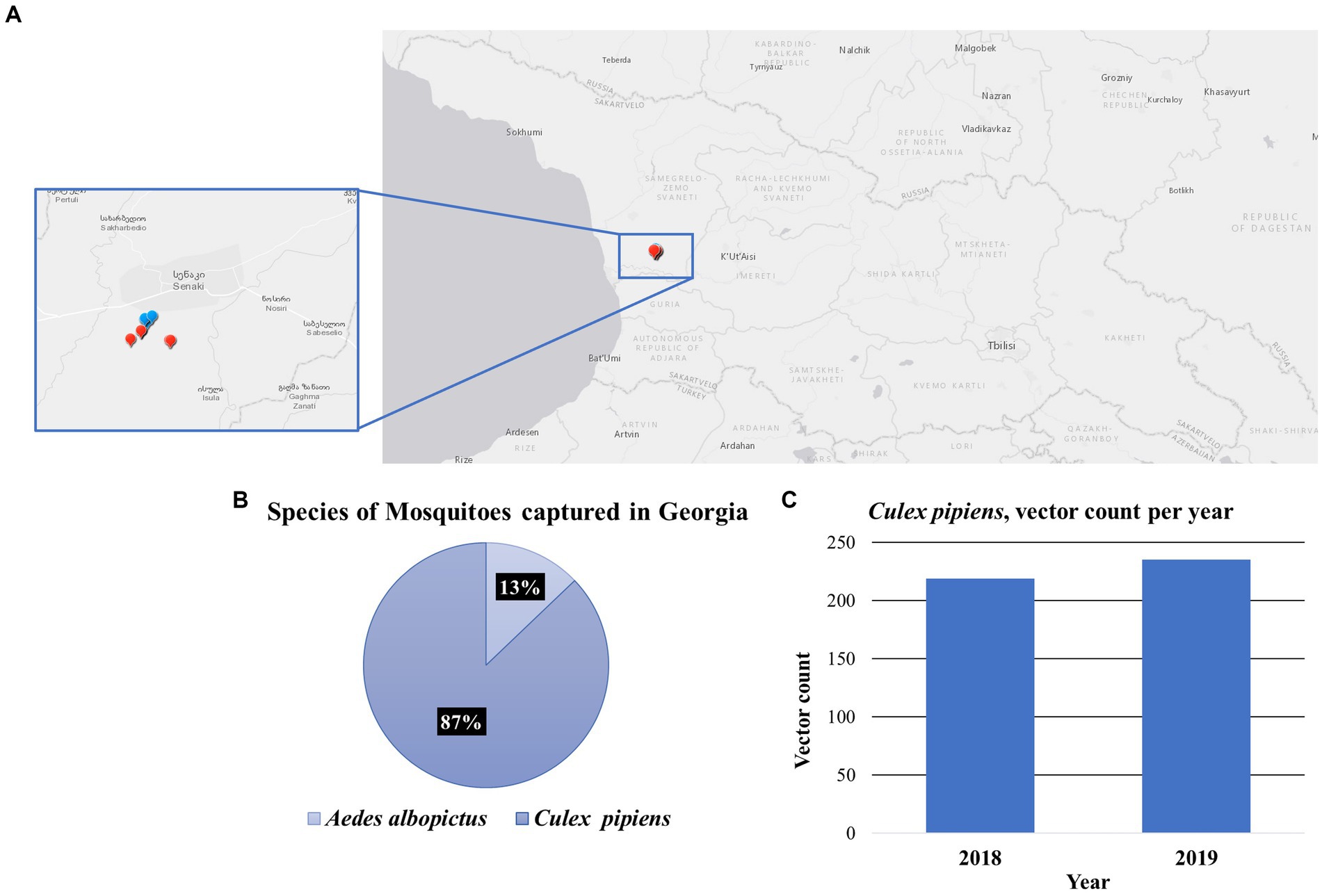
Figure 1. Mosquito collections in the Country of Georgia. (A) A map containing the location for each collection site, in the Senaki area. Each dot represents the collection site, 2018 (blue) and 2019 (red). (B) Species of mosquitoes captured in Georgia. Chart illustrates the percentage of the two species of mosquitoes captured, the Culex pipiens 87% and the Aedes albopictus 13%. (C) The vector count of Culex pipiens mosquitoes captured per year, 2018 (N = 219) and 2019 (N = 235).
To explore the viral diversity of the town of Senaki, we conducted an unbiased total RNA-seq approach to identify viruses in this specific area. This methodology was previously described and published by us (Hang et al., 2012; Sanborn et al., 2019; Pollio et al., 2022). A total of 37 pooled samples were prepared using the Illumina sequencing system. To analyze our Next-generation sequencing (NGS) raw data, we utilized the CZ ID cloud-based pipeline, a computational tool to detect microbial pathogens (Kalantar et al., 2020). With this computational tool, we obtained an average read passing filter per sample of 203,822. Sequences were aligned to the NCBI’s database of nucleotide sequences to calculate rPM (reads per million) and classified into three kingdoms. The total reads per kingdom are the following: Bacteria (1,430,993,14%), Eukaryote (445,670, 4%) and Virus (8,775,711, 82%) (Figure 2A). Most of our sequencing reads were aligned to viruses (82%); thus, we decided to further explore and compare the viral diversity between the years 2018 and 2019 (Figure 2B).
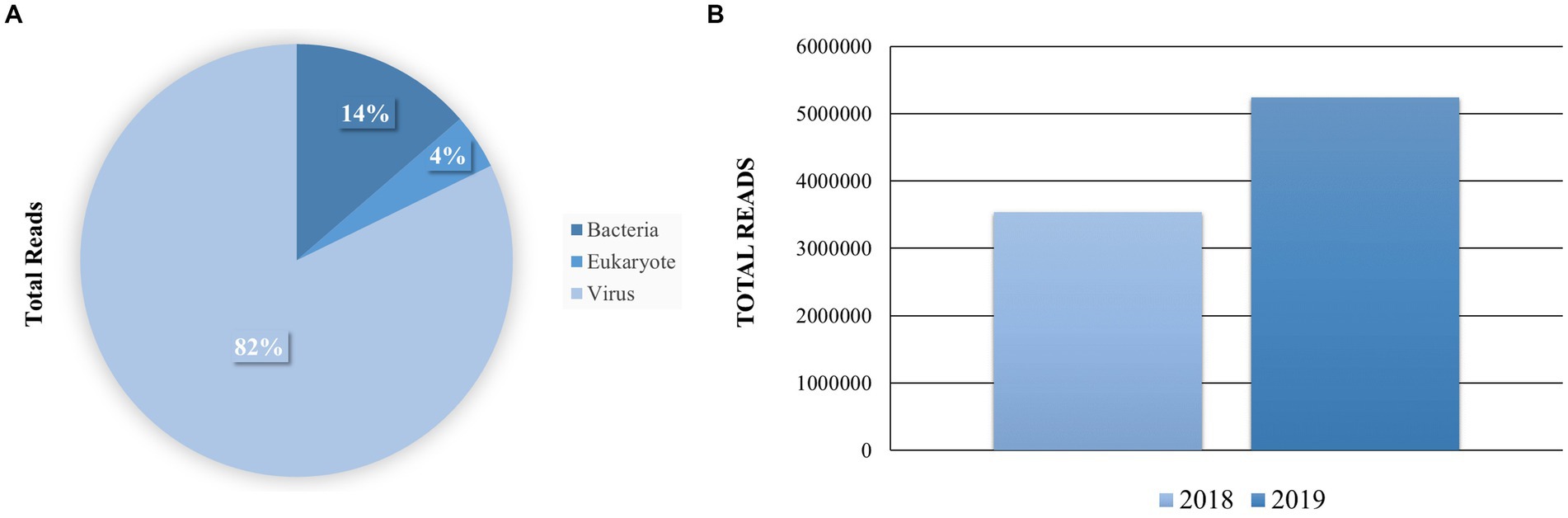
Figure 2. Distribution of the total reads per sample and taxonomy kingdom. (A) The total reads per sample were queried to the NCBI database using CZ ID and STAR. The mapped Super kingdom data is shown Bacteria 1,430,993 (14%), Eukaryote 445,670 (4%) and Virus 8,775,711 (82%). (B) Comparison of sequencing reads that were aligned to viruses per sample.
A heat map was built, to analyze the presence and abundance of viral reads in our samples. The heat map was set to NT rPM > =10 which shows viruses with at least 10 reads per million aligned reads to simplify our overall analysis (Figure 3). The most abundant viruses aligned to ISVs including: the Culex Iflavi-like virus 4 (2,167,511.86), and the Merida-like virus KE-2017a (1,304,966.73) (Supplementary Figure S1). These results indicate the abundance of two main families in our pooled samples: the Iflaviridae; a positive-stranded RNA virus and the Rhabdoviridae; a single-stranded, negative RNA virus. We found the Merida-like virus KE-2017a in most of the collected pooled samples from 2018 (Figure 3). The Merida virus was first detected in the Culex quinquefasciatus species in the Yucatan Peninsula of Mexico and classified as a Rhabdovirus (Charles et al., 2016). This virus has been detected in other places around the world (Charles et al., 2016; Ergunay et al., 2017; Sadeghi et al., 2018; da Silva Neves et al., 2021). Airborne surveillance conducted in Thrace and Anatolia, two regions from Turkey recently identified the Merida-like virus Turkey (MERDLVT) (Ergunay et al., 2017). This virus is closely related to the Merida virus detected in Mexico. Due to the proximity of Georgia to Turkey, these results could imply a migration of the Merida virus.
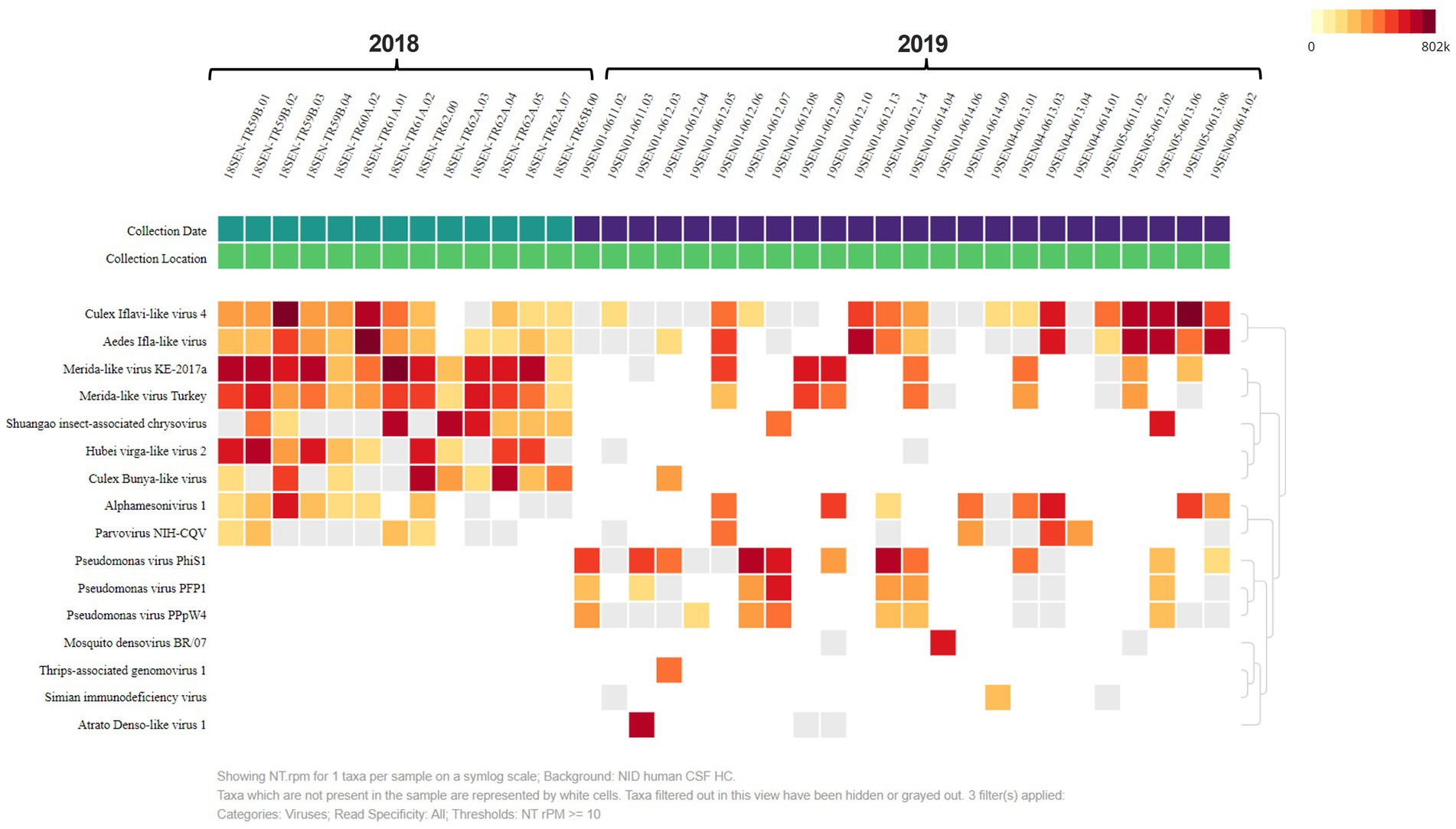
Figure 3. Heat Map analysis shows virus diversity in collected samples. The heat map was set to NT rPM > =10 which only shows viruses with at least 10 reads per million. Samples are divided by collection date. The most abundant viruses aligned to insect-specific viruses including: the Culex Iflavi-like virus 4, Merida-like virus KE-2017a and the Merida-like virus Turkey.
We used phylogenetic tools to further evaluate the relationship of these positive pooled specimens with the Merida virus. The phylogenetic analysis indicates that our positive samples are closely related to two viruses from mosquitoes collected in Turkey, the Merida-like virus KE-2017a and the MERDLVT (Figure 4). 81% of ultrafast bootstrap replicates supported the common ancestor node that places the two samples from Turkey with two samples from Georgia (Merida_virus_Georgia_OQ725976 and Merida_virus_Georgia_OQ725975). To further confirm the previous analysis, we used the BLAST sequencing alignment tool to evaluate and compare the sequence identity between the Georgia samples and the MERLDVT strain. Our positive samples aligned to the MERDLVT strain with 96%–100% sequence identity with 99.7%–100% aligned genome sequence coverage.

Figure 4. Maximum likelihood tree of rhabdovirus family including Merida virus and Merida-like virus samples. Merida virus genomes from Georgia samples are highlighted in red and closely related MERDLVT samples are highlighted in blue. Node confidence values represent ultrafast bootstrap replicate support.
Our aim was to explore unique viral communities to expand our knowledge in mosquito virome populations. To explore the viral component of the town of Senaki we used an unbiased metagenomic approach. Pooled specimens of Culex pipiens mosquitoes were studied to reveal mosquito virome. In both years, most viral reads belonged to the Culex Iflavi-like virus 4. In addition, we found most samples containing the Merida-like virus KE-2017a in pooled specimens from 2018. A phylogenetic analysis revealed that these positive pooled samples shared a common ancestor with the Merida-like virus KE-2017a isolate 139-1-21 and the Merida-like virus Turkey isolate P431. The known Merida viruses shown in this phylogenetic analysis cluster geographically.
The Merida virus was initially detected in Culex quinquefasciatus mosquitoes and assigned to the genus Merhavirus within the Rhabdoviridae family (Charles et al., 2016). Since then, the virus has been detected in metagenomic analyses conducted in other mosquito species and around the world (Charles et al., 2016; Ergunay et al., 2017). However, the pathology of the Merida virus has not been further explored.
Controlling vector populations has become widely implemented approach to prevent the endemic spread of VBPs. With the continuous expansion of vector habitats, it is critical to monitor vector populations for potential emerging viral diseases to better prepare for new threats. Our work has added contributed NGS data and curated genome assemblies of novel Merida-like viruses that can be further used to better understand viral communities within the mosquito population. These data have the potential in combination with additional surveys to identify new and emerging diseases.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, OQ725971; https://www.ncbi.nlm.nih.gov/genbank/, OQ725972; https://www.ncbi.nlm.nih.gov/genbank/, OQ725973; https://www.ncbi.nlm.nih.gov/genbank/, OQ725974; https://www.ncbi.nlm.nih.gov/genbank/, OQ725975; https://www.ncbi.nlm.nih.gov/genbank/, OQ725976; https://www.ncbi.nlm.nih.gov/genbank/, OQ725977; https://www.ncbi.nlm.nih.gov/genbank/, OQ725978; https://www.ncbi.nlm.nih.gov/genbank/, OQ725979; https://www.ncbi.nlm.nih.gov/genbank/, OQ725980.
JP-B: Formal analysis, Methodology, Writing – original draft, Writing – review & editing, Conceptualization, Investigation, Visualization. AP: Formal analysis, Methodology, Writing – original draft, Writing – review & editing, Conceptualization, Investigation. BK: Methodology, Project administration, Writing – review & editing, Investigation, Formal analysis. TC: Methodology, Writing – review & editing, Investigation. CF: Formal analysis, Methodology, Software, Visualization, Writing – review & editing, Validation. MC: Formal analysis, Methodology, Writing – review & editing, Software, Visualization, Validation. DR-W: Formal analysis, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing, Investigation, Resources. JH: Formal analysis, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing, Investigation, Resources, Validation, Visualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Funding was provided by the Global Emerging Infections Surveillance and Response System (GEIS), Division by the Armed Forces Health Surveillance Branch, projects P0092_18_GA, P0031_19_GA, P0171_21_WR, P0175_22_WR.
We thank James S. Hilaire, Nicole R. Nicholas, Tuan K. Nguyen, and April N. Griggs for their assistance in project management and sample tracking, storage, and retrieval.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Material has been reviewed by the authors’ respective institutions. There is no objection to its presentation and/or publication. The views expressed here are those of the authors and do not reflect the official policy of the Department of the Army, Department of Defense or US Government. This is the work of US government employees and may not be copyrighted (17 USC 105).
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1258810/full#supplementary-material
Agboli, E., Leggewie, M., Altinli, M., and Schnettler, E. (2019). Mosquito-specific viruses-transmission and interaction. Viruses 11:873. doi: 10.3390/v11090873
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Boisvert, S., Raymond, F., Godzaridis, E., Laviolette, F., and Corbeil, J. (2012). Ray Meta: scalable de novo metagenome assembly and profiling. Genome Biol. 13:R122. doi: 10.1186/gb-2012-13-12-r122
Bolling, B. G., Olea-Popelka, F. J., Eisen, L., Moore, C. G., and Blair, C. D. (2012). Transmission dynamics of an insect-specific flavivirus in a naturally infected Culex pipiens laboratory colony and effects of co-infection on vector competence for West Nile virus. Virology 427, 90–97. doi: 10.1016/j.virol.2012.02.016
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Charles, J., Firth, A. E., Lorono-Pino, M. A., Garcia-Rejon, J. E., Farfan-Ale, J. A., Lipkin, W. I., et al. (2016). Merida virus, a putative novel rhabdovirus discovered in Culex and Ochlerotatus spp. mosquitoes in the Yucatan peninsula of Mexico. J. Gen. Virol. 97, 977–987. doi: 10.1099/jgv.0.000424
Cuevas-Juarez, E., Pando-Robles, V., and Palomares, L. A. (2021). Flavivirus vaccines: virus-like particles and single-round infectious particles as promising alternatives. Vaccine 39, 6990–7000. doi: 10.1016/j.vaccine.2021.10.049
da Silva Neves, N. A., Pinto, A. Z. L., Melo, F. L., Maia, L. M. S., da Silva Ferreira, R., de Carvalho, M. S., et al. (2021). Sialovirome of Brazilian tropical anophelines. Virus Res. 302:198494. doi: 10.1016/j.virusres.2021.198494
de Almeida, J. P., Aguiar, E. R., Armache, J. N., Olmo, R. P., and Marques, J. T. (2021). The virome of vector mosquitoes. Curr. Opin. Virol. 49, 7–12. doi: 10.1016/j.coviro.2021.04.002
Dietzgen, R. G., Kondo, H., Goodin, M. M., Kurath, G., and Vasilakis, N. (2017). The family Rhabdoviridae: mono- and bipartite negative-sense RNA viruses with diverse genome organization and common evolutionary origins. Virus Res. 227, 158–170. doi: 10.1016/j.virusres.2016.10.010
Erasmus, J. H., Seymour, R. L., Kaelber, J. T., Kim, D. Y., Leal, G., Sherman, M. B., et al. (2018). Novel insect-specific Eilat virus-based chimeric vaccine candidates provide durable, mono- and multivalent, single-dose protection against lethal alphavirus challenge. J. Virol. 92, e01274–e01217. doi: 10.1128/JVI.01274-17
Ergunay, K., Brinkmann, A., Litzba, N., Gunay, F., Kar, S., Oter, K., et al. (2017). A novel rhabdovirus, related to Merida virus, in field-collected mosquitoes from Anatolia and Thrace. Arch. Virol. 162, 1903–1911. doi: 10.1007/s00705-017-3314-4
Hang, J., Forshey, B. M., Kochel, T. J., Li, T., Solorzano, V. F., Halsey, E. S., et al. (2012). Random amplification and pyrosequencing for identification of novel viral genome sequences [research support, U.S. Gov't, non-P.H.S.]. J. Biomol. Tech. 23, 4–10. doi: 10.7171/jbt.12-2301-001
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Huang, X., and Madan, A. (1999). CAP3: a DNA sequence assembly program. Genome Res. 9, 868–877. doi: 10.1101/gr.9.9.868
Kalantar, K. L., Carvalho, T., de Bourcy, C. F. A., Dimitrov, B., Dingle, G., Egger, R., et al. (2020). IDseq-an open source cloud-based pipeline and analysis service for metagenomic pathogen detection and monitoring. Gigascience 9:giaa111. doi: 10.1093/gigascience/giaa111
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kilianski, A., Carcel, P., Yao, S., Roth, P., Schulte, J., Donarum, G. B., et al. (2015). Pathosphere.org: pathogen detection and characterization through a web-based, open source informatics platform. BMC Bioinformatics 16:416. doi: 10.1186/s12859-015-0840-5
Kilpatrick, A. M., and Randolph, S. E. (2012). Drivers, dynamics, and control of emerging vector-borne zoonotic diseases. Lancet 380, 1946–1955. doi: 10.1016/S0140-6736(12)61151-9
Liu, S., Vijayendran, D., and Bonning, B. C. (2011). Next generation sequencing technologies for insect virus discovery. Viruses 3, 1849–1869. doi: 10.3390/v3101849
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal 17, 10–12. doi: 10.14806/ej.17.1.200
Pierson, T. C., and Diamond, M. S. (2020). The continued threat of emerging flaviviruses. Nat. Microbiol. 5, 796–812. doi: 10.1038/s41564-020-0714-0
Pollio, A. R., Jiang, J., Lee, S. S., Gandhi, J. S., Knott, B. D., Chunashvili, T., et al. (2022). Discovery of Rickettsia spp. in mosquitoes collected in Georgia by metagenomics analysis and molecular characterization. Front. Microbiol. 13:961090. doi: 10.3389/fmicb.2022.961090
Ramesh, A., Nakielny, S., Hsu, J., Kyohere, M., Byaruhanga, O., de Bourcy, C., et al. (2019). Metagenomic next-generation sequencing of samples from pediatric febrile illness in Tororo. Uganda. PLoS One 14:e0218318. doi: 10.1371/journal.pone.0218318
Sadeghi, M., Altan, E., Deng, X., Barker, C. M., Fang, Y., Coffey, L. L., et al. (2018). Virome of > 12 thousand Culex mosquitoes from throughout California. Virology 523, 74–88. doi: 10.1016/j.virol.2018.07.029
Saha, S., Ramesh, A., Kalantar, K., Malaker, R., Hasanuzzaman, M., Khan, L. M., et al. (2019). Unbiased metagenomic sequencing for pediatric meningitis in Bangladesh reveals Neuroinvasive chikungunya virus outbreak and other unrealized pathogens. MBio 10, e02877–e02819. doi: 10.1128/mBio.02877-19
Sanborn, M. A., Klein, T. A., Kim, H. C., Fung, C. K., Figueroa, K. L., Yang, Y., et al. (2019). Metagenomic analysis reveals three novel and prevalent mosquito viruses from a single Pool of Aedes vexans nipponii collected in the Republic of Korea. Viruses 11:222. doi: 10.3390/v11030222
Schmieder, R., and Edwards, R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864. doi: 10.1093/bioinformatics/btr026
Valles, S. M., Chen, Y., Firth, A. E., Guerin, D. M. A., Hashimoto, Y., Herrero, S., et al. (2017). ICTV virus taxonomy profile: Iflaviridae. J. Gen. Virol. 98, 527–528. doi: 10.1099/jgv.0.000757
Keywords: mosquito, Culex pipiens, pathogen discovery, vector control, Merida like-virus, NGS
Citation: Potter-Birriel JM, Pollio AR, Knott BD, Chunashvili T, Fung CK, Conte MA, Reinbold-Wasson DD and Hang J (2023) Metagenomics analysis reveals presence of the Merida-like virus in Georgia. Front. Microbiol. 14:1258810. doi: 10.3389/fmicb.2023.1258810
Received: 04 August 2023; Accepted: 27 September 2023;
Published: 12 October 2023.
Edited by:
Ke Liu, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Mel C. Melendrez, Anoka-Ramsey Community College, United StatesCopyright © 2023 Potter-Birriel, Pollio, Knott, Chunashvili, Fung, Conte, Reinbold-Wasson and Hang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Hang, anVuLmhhbmcuY2l2QGhlYWx0aC5taWw=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.