Abstract
The biofilm lifestyle is critical for bacterial survival and proliferation in the fluctuating marine environment. Cyclic diguanylate (c-di-GMP) is a key second messenger during bacterial adaptation to various environmental signals, which has been identified as a master regulator of biofilm formation. However, little is known about whether and how c-di-GMP signaling regulates biofilm formation in Vibrio alginolyticus, a globally dominant marine pathogen. Here, a large set of 63 proteins were predicted to participate in c-di-GMP metabolism (biosynthesis or degradation) in a pathogenic V. alginolyticus strain HN08155. Guided by protein homology, conserved domains and gene context information, a representative subset of 22 c-di-GMP metabolic proteins were selected to determine which ones affect biofilm-associated phenotypes. By comparing phenotypic differences between the wild-type and mutants or overexpression strains, we found that 22 c-di-GMP metabolic proteins can separately regulate different phenotypic outputs in V. alginolyticus. The results indicated that overexpression of four c-di-GMP metabolic proteins, including VA0356, VA1591 (CdgM), VA4033 (DgcB) and VA0088, strongly enhanced rugose colony morphotypes and strengthened Congo Red (CR) binding capacity, both of which are indicators of biofilm matrix overproduction. Furthermore, rugose enhanced colonies were accompanied by increased transcript levels of extracellular polysaccharide (EPS) biosynthesis genes and decreased expression of flagellar synthesis genes compared to smooth colonies (WTpBAD control), as demonstrated by overexpression strains WTp4033 and ∆VA4033p4033. Overall, the high abundance of c-di-GMP metabolic proteins in V. alginolyticus suggests that c-di-GMP signaling and regulatory system could play a key role in its response and adaptation to the ever-changing marine environment. This work provides a robust foundation for the study of the molecular mechanisms of c-di-GMP in the biofilm formation of V. alginolyticus.
Introduction
Vibrio alginolyticus is one of three most abundant Vibrio species found in marine and estuarine environments worldwide, which has caused frequent outbreaks of high-mortality vibriosis in aquatic animals (e.g., fish, shrimp and shellfish) and significant economic losses to the global aquaculture industry (Xie et al., 2005; Jacobs Slifka et al., 2017; Ibangha et al., 2023; Yu et al., 2023). The transition from a planktonic to a biofilm lifestyle is key to the survival and pathogenicity of Vibrio spp. (Baker-Austin et al., 2018). Bacterial cells encased in biofilms exhibit a range of emergent properties that are quite different from free-living cells, such as enhanced social cooperation, more efficient resource capture and increased resistance to antimicrobials (Flemming et al., 2016). Therefore, biofilms provide such an important fitness advantage for Vibrio, enhancing environmental adaptations and enabling them to persist in different niches, especially within the host (Teschler et al., 2015).
Different stages of biofilm formation, including surface attachment, matrix production, biofilm maturation and dispersal, are regulated by sophisticated regulatory networks (Yildiz et al., 2001; Wang et al., 2014; Townsley and Yildiz, 2015). Evidence is mounting that the near-ubiquitous second messenger cyclic diguanylate (c-di-GMP) plays a central role in different stages of biofilm formation in a diverse group of microorganisms (Purcell et al., 2012; Romling et al., 2013; Valentini and Filloux, 2019; Ahmad et al., 2020). In the case of Vibrio spp., the best-studied example is Vibrio cholerae, the causative agent of the human disease cholera. It is now generally accepted that high levels of c-di-GMP promote extracellular polysaccharide (EPS) production and enhance biofilm formation of V. cholerae, while low levels of c-di-GMP increase cell motility and cause biofilm dispersal (Liu et al., 2010; Conner et al., 2017; Biswas et al., 2020; Wu et al., 2020). Recent evidence obtained from one of the dominant marine Vibrio spp., Vibrio parahaemolyticus, has also made it clear that high c-di-GMP favors biofilm formation (Kimbrough et al., 2020; Martinez-Mendez et al., 2021). In addition, interfering with c-di-GMP metabolism and signaling has been shown to be a promising way to control biofilm development (Liu et al., 2022; Kim et al., 2023). Therefore, modulation of the intracellular c-di-GMP pool is a key regulator of bacterial adaptation to sessile and motile lifestyles in response to complex environmental and internal cues.
The intracellular c-di-GMP pool is modulated by the competing activities of two classes of enzymes, diguanylate cyclases (DGCs) and phosphodiesterases (PDEs), which synthesize and degrade c-di-GMP, respectively. Functional DGCs typically contain a catalytically active GGDEF structural domain, while PDEs generally contain an EAL or HD-GYP active domain (Schirmer and Jenal, 2009; Romling et al., 2013). In most cases, bacterial genomes encode multiple DGCs and PDEs, however, their abundance varies among species or strains (Romling et al., 2013; Chou and Galperin, 2016). The number of genes encoding c-di-GMP metabolic proteins in Vibrio spp. is usually more than 50, e.g., the pathogens V. cholerae, V. parahaemolyticus, and V. vulnificus contain 62, 62, and 92 potential DGCs/PDEs, respectively (Shrestha et al., 2022). As recently reported, the number of c-di-GMP metabolic proteins in Vibrio spp. far exceeds that of Escherichia coli (Homma and Kojima, 2022), suggesting that the c-di-GMP signaling network in Vibrio is quite complex, which is crucial for them to sense environmental changes and initiate adaptive responses.
Notably, not all the DGCs/PDEs control the same c-di-GMP-dependent phenotypes, i.e., individual different DGCs/PDEs can generate distinct and specific output functions (Dahlstrom and O'Toole, 2017). In the case of V. cholerae, distinct c-di-GMP metabolic proteins contribute to different stages of biofilm progression (Massie et al., 2012; Zamorano-Sanchez et al., 2019). For example, among the 28 DGCs of V. cholerae, three specific DGCs, namely CdgA, CdgL and CdgO, are required for the flagellum-dependent biofilm regulatory (FDBR) response that enhances biofilm formation (Wu et al., 2020). In Pseudomonas aeruginosa PA14, the DGC SadC interacts with the type 4 pili (T4P) alignment complex protein PilO, and this interaction is an important regulatory hub for controlling early biofilm formation (Webster et al., 2021). In E. coli, the c-di-GMP metabolic proteins DgcE, DgcM, and PdeR specifically affect biofilm formation but are independent of effects on cellular c-di-GMP levels (Sarenko et al., 2017). Thus, bacteria can optimize phenotypic output to c-di-GMP levels via activation of specific c-di-GMP metabolic proteins as a response to specific environmental signals. As one of the most abundant Vibrio species in the global ocean, the ability to rapidly respond to environmental signals and transition to a biofilm lifestyle is critical for the survival of V. alginolyticus under adverse conditions (Yin et al., 2021, 2022). However, little attention has been paid to the c-di-GMP-mediated signal transduction system in V. alginolyticus, and it is unclear which specific c-di-GMP metabolic proteins play regulatory roles in the biofilm-associated phenotypes.
In this study, we first determined the number of c-di-GMP metabolic proteins encoded in the genome of V. alginolyticus HN08155, which we had previously isolated from diseased grouper and was highly virulent to Litopenaeus vannamei (Xie et al., 2020). A representative subset of c-di-GMP metabolic proteins was further selected and their individual functions in controlling biofilm-associated phenotypes were investigated. We constructed 44 derivative strains with each of the 22 DGC or PDE genes knocked out or overexpressed, and compared their colony morphology, biofilm formation, EPS production and swarming motility ability with the wild-type strain of V. alginolyticus HN08155. Overall, this study provided the first systematic bioinformatic and functional characterization of c-di-GMP metabolic proteins in V. alginolyticus and identified the specific DGCs required for biofilm formation in this dominant marine pathogen.
Results
Bioinformatic characterization of potential c-di-GMP metabolic proteins
A total of 63 proteins involved in c-di-GMP biosynthesis and degradation were identified in the genome of V. alginolyticus HN08155, of which 32 proteins contain only the GGDEF domain (hereinafter referred to as “GGDEF-only protein”), 11 proteins contain only the EAL domain (hereinafter “EAL-only protein”), 4 proteins contain only the HD-GYP domain (hereinafter “HD-GYP-only protein”), and 16 proteins contain both the GGDEF and EAL domains (hereinafter “hybrid protein”; Table 1).
Table 1
| Class | Protein | GGDEF domain | EAL /HD-GYP domain | IS | Domain architecture | Length (aa) |
|---|---|---|---|---|---|---|
| GGDEF-only proteins | VA1554 | RxGGEEF | - | √ |  | 320 |
| VA4570 | RxGGDEF | - | - |  | 320 | |
| VA0356 | RxGGEEF | - | √ |  | 626 | |
| VA0461 | RxGGEEF | - | √ |  | 582 | |
| VA4663 | RxSGDEF | - | - |  | 652 | |
| VA2975 | RxGGDEF |  | 301 | |||
| VA0931 | RxGGDEF | - | - |  | 669 | |
| VA4342 | RxGGEEF | - | - |  | 480 | |
| VA4030 | RxGGDEF | - | √ | 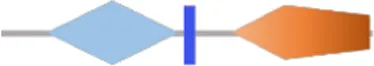 | 451 | |
| VA1591 (cdgM) | RxGGDEF | - | - |  | 513 | |
| VA4622 (pleD) | RxGGEEF | - | - |  | 319 | |
| VA3556 | RxGGEEF | - | - |  | 706 | |
| VA1419 | RxGGEEF | - | - |  | 657 | |
| VA4012 | RxGGDEF | - | - | 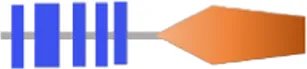 | 402 | |
| VA4624 | RxGGEEF | - | - | 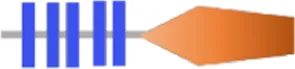 | 386 | |
| VA3192 | RxGGDEF | - | √ |  | 453 | |
| VA1626 | RxGGEEF | - | √ |  | 525 | |
| VA2058 (cdgK) | RxGGEEF | - | √ |  | 491 | |
| VA2972 | RxGGEEF | - | √ |  | 529 | |
| VA1192 | RxGGEEF | - | - |  | 445 | |
| VA4496 | RxGGEEF | - | √ |  | 637 | |
| VA3275 | RxGGEEF | - | √ |  | 682 | |
| VA3903 (cdgA) | RxGGDEF | - | - |  | 365 | |
| VA3580 | RxGGEEF | - | - |  | 527 | |
| VA0663 | RxGGEEF | - | - |  | 521 | |
| VA4033 (dgcB) | RxGGEEF | - | - |  | 337 | |
| VA4271 | RxGGEEF | - | - |  | 349 | |
| VA2336 | RxGGEEF | - | - |  | 132 | |
| VA3895 | RxAGDEF | - | - | 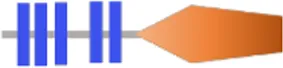 | 382 | |
| VA1875 | ExTENAS | - | - |  | 571 | |
| VA2973 | RxHSDGF | - | - |  | 271 | |
| VA3494 | QxRLGEV | - | - |  | 451 | |
| EAL-only proteins | VA1964 | - | EAL | - |  | 818 |
| VA3193 | - | EAL | - |  | 384 | |
| VA4177 | - | EAL | - |  | 681 | |
| VA1287 | - | EAL | - |  | 498 | |
| VA4408 | - | EAL | - |  | 490 | |
| VA1095 | - | ELL | - |  | 404 | |
| VA2974 | - | EVL | - |  | 268 | |
| VA3005 (cdgJ) | - | ELL | - |  | 407 | |
| VA3641 | - | HLT | - |  | 265 | |
| VA4700 | - | EAL | - |  | 262 | |
| VA4720 | - | EAL | - |  | 272 | |
| HD-GYP-only proteins | VA1608 | - | HA-GYP | - |  | 374 |
| VA1611 | - | HD-GYP | - |  | 421 | |
| VA1618 | - | HD-GYP | - |  | 405 | |
| VA2624 | - | HD-GYP | - |  | 417 | |
| Hybrid proteins | VA0068 | RxGGDEF | EIL | - |  | 1,041 |
| VA0088 | RxGGDEF | ETL | - |  | 815 | |
| VA2417 (rocS) | HxGGDEF | EAL | - |  | 679 | |
| VA3012 | SxGAGEW | ECL | - |  | 828 | |
| VA3264 | RxSEHEF | EAL | - |  | 882 | |
| VA1619 | RxHEDDF | ESL | - |  | 567 | |
| VA0108 | RxGGDEF | EAL | - |  | 848 | |
| VA1297 | RxGADEF | EAL | - |  | 719 | |
| VA3251 (mbaA) | RxSGDEF | EVL | - |  | 788 | |
| VA3814 (lapD) | RxSSDEF | EVF | - |  | 637 | |
| VA4309 | RxAGDEF | EIL | - |  | 606 | |
| VA4672 (cdgC) | RxSSNDL | EAL | - |  | 636 | |
| VA3608 | RxGGDEF | EAL | √ |  | 776 | |
| VA4418 | RxGGDEF | EAV | - |  | 512 | |
| VA2701 (csrD) | RxYDADF | ELT | - |  | 669 | |
| VA3369 | RxASDEF | EAL | - |  | 751 |
Structure prediction of 63 potential c-di-GMP metabolic proteins of V. alginolyticus HN08155 by the SMART algorithm and NCBI’s Conserved Domains Database.
 GGDEF;
GGDEF;  EAL;
EAL;  HAMP;
HAMP;  REC;
REC;  GAF;
GAF;  CHASE;
CHASE;  PAS;
PAS;  PAC;
PAC;  PBP2;
PBP2;  TM;
TM;  TPR;
TPR;  CBS;
CBS;  FIST;
FIST;  HD-GYP. The individual structural domains represented by different colors and shapes are shown at the bottom. The shapes of all representative domains are not drawn to scale. For proteins containing GGDEF or EAL domain, the corresponding amino acid residues are written in black if they are not mutated, otherwise they are written in bold black. For predicted allosteric inhibition site (Is) of DGCs, √ means Is included, otherwise not included.
HD-GYP. The individual structural domains represented by different colors and shapes are shown at the bottom. The shapes of all representative domains are not drawn to scale. For proteins containing GGDEF or EAL domain, the corresponding amino acid residues are written in black if they are not mutated, otherwise they are written in bold black. For predicted allosteric inhibition site (Is) of DGCs, √ means Is included, otherwise not included.
We analyzed the neighboring genes of all 63 c-di-GMP metabolic proteins, since the possible transcriptional co-regulation at adjacent or proximate locations may affect their regulatory roles (Seshasayee et al., 2010; Schäper et al., 2017). On the basis of genome mapping and functional annotation, we identified 8 c-di-GMP metabolic proteins (VA3192, VA3193, VA4663, VA1419, VA3614, VA1297, VA3814 and VA2701) with adjacent or proximity operons encoding proteins that may be associated with biofilm formation or motility (Supplementary Figure S1). Notably, the neighboring genes of the GGDEF-only proteins VA4663 and VA1419 were predicted to be involved in biofilm formation, whereas the neighboring genes of the EAL-only protein VA3614 and the hybrid protein VA1297 (with an incomplete GGDEF) were predicted to be associated with motility. Furthermore, we found that some of the c-di-GMP metabolic proteins, including VA2972, VA2973, VA2974 and VA2975, VA1618 and VA1619, VA4622 and VA4624, as well as VA4030 and VA4033, are genomically encoded in close proximity to each other (Supplementary Figure S1). Previous studies have demonstrated that certain protein interactions between GGDEF/EAL domain proteins can specifically affect distinct output functions, and these proteins have to be co-expressed (Lindenberg et al., 2013; Sarenko et al., 2017). Based on these data, we speculate that these c-di-GMP metabolic proteins, which are closely encoded in the genome of V. alginolyticus, may benefit in co-regulating specific output functions through protein–protein interaction patterns.
The N-terminal sensory input domains located in the GGDEF or EAL has been shown to have a significant effect on the activity of DGCs or PDEs in response to environmental cues (Townsley and Yildiz, 2015). Thus, we analyzed the N-terminal sensory domains of all 63 c-di-GMP metabolic proteins. The PAS domain, which acts as a signal transducer coupling ligand binding to cellular signaling responses (Hutchin et al., 2021), was the most abundant N-terminal sensory domains and was observed in 12 c-di-GMP metabolic proteins (Table 1; Supplementary Table S3). Some even contain three PAS domains, such as the protein VA0461. Conserved domains with important roles in signal transduction, including HAMP, CHASE, REC and GAF, were also observed at the N-terminal of several c-di-GMP signaling proteins. Other N-terminal sensory domains detected in only one or two c-di-GMP metabolic proteins include PBPb, TPR, CBS, and FIST (Table 1; Supplementary Table S3).
The presence of transmembrane (TM) helices may be one of the reasons why distinct GGDEF (or EAL) proteins have different functions owing to their different intracellular localization (Guvener and Harwood, 2007; Collins et al., 2020; Hengge, 2021). The results showed that a total of 32 proteins contain TM domain(s) (Supplementary Figure S2; Supplementary Table S3), suggesting that these 32 proteins may be directly involved in the sensing of extracellular signals.
the highly conserved amino acid residues based on RxGGDE/EF motif are essential for DGC activity, while ExLxR or HD-GYP are required for PDE activity (Romling et al., 2013). Based on this proven theory, 29 of the 32 GGDEF-only proteins were predicted to have DGC activity (Supplementary Figure S3), while 10 of the 11 EAL-only proteins (Supplementary Figure S4) and 3 of the 4 HD-GYP-only proteins (Supplementary Figure S5) were predicted to possess PDE activity. In addition, 8 and 13 of the 16 hybrid proteins were predicted to have DGC and PDE activities, respectively, with 7 of them (VA0068, VA0088, VA0108, VA3608, VA3251, VA4309 and VA2417) having dual DGC and PDE activities. The remaining 2 hybrid proteins were predicted to be non-catalytic at the GGDEF and EAL sites, with VA3814 (LapD) and VA2701 (CsrD) predicted to be c-di-GMP receptors that may positively contribute to biofilm formation (Kitts et al., 2019; Kharadi and Sundin, 2022). For GGDEF domain-containing proteins with the RxxD motif, the presence of an allosteric product inhibition site (I-site) and its binding to c-di-GMP will result allosteric inhibition of DGC activity (Christen et al., 2006). Our results showed that nearly one-third of the GGDEF-only proteins contain the I-site, while only 1 of the 16 hybrid proteins contains the I-site (Table 1). Sequence alignment of all 63 c-di-GMP metabolic proteins with other GGDEF/EAL/HD-GYP domain-containing proteins revealed that the majority of residues in the active sites are conserved (Supplementary Figures S3–S6).
Selection of 22 representative c-di-GMP metabolic proteins for functional characterization
To investigate the regulatory role of c-di-GMP signaling in the control of biofilm-associated phenotypes in V. alginolyticus, 22 representative c-di-GMP metabolic genes (Figure 1) were selected for further study by the following three criteria. Firstly, given that some homologous genes have been reported to have regulatory roles in biofilm-associated phenotypes in other bacteria, 10 genes were selected based on this principle, including genes VA1591 (cdgM), VA2058 (cdgK), VA3903 (cdgA), VA4033 (dgcB), VA3005 (cdgJ), VA2417 (rocS), VA2701 (csrD), VA4672 (cdgC), VA3251 (mbaA), and VA4622 (pleD). Secondly, we selected 4 genes (VA3192, VA3193, VA4663 and VA3814) whose neighboring genes encode proteins that may be associated with EPS production, biofilm formation, or motility (Supplementary Figure S1). In particular, the gene VA3814 was also selected because it encodes a potential c-di-GMP receptor protein that shares 66.51% identity with the LapD protein of V. cholerae (Figure 1A), which has been demonstrated to regulate biofilm formation by controlling the adhesion protein LapA (Kitts et al., 2019). Finally, 3 genes encoding GGDEF-only proteins (VA4342, VA0356 and VA0663), 2 genes encoding EAL-only proteins (VA4700 VA4408), and 3 genes encoding hybrid proteins (VA0068, VA0108 and VA0088) were also selected based on the active or sensor domains they contain. The 22 c-di-GMP metabolic proteins of V. alginolyticus are conserved among closely related Vibrio species, including V. parahaemolyticus, V. harveyi, V. owensii, V. vulnificus, V. cholerae, and V. anguillarum, however, the conservativeness was significantly decreased in some phylogenetically distantly related bacterial species, such as P. aeruginosa, P. fluorescens, Xanthomonas campestris pv. oryza, Burkholderia cenocepacia and E. coli (Figure 1). Therefore, these 22 c-di-GMP metabolic proteins were selected as representatives and their individual functions in regulating biofilm-associated phenotypes were explored.
Figure 1
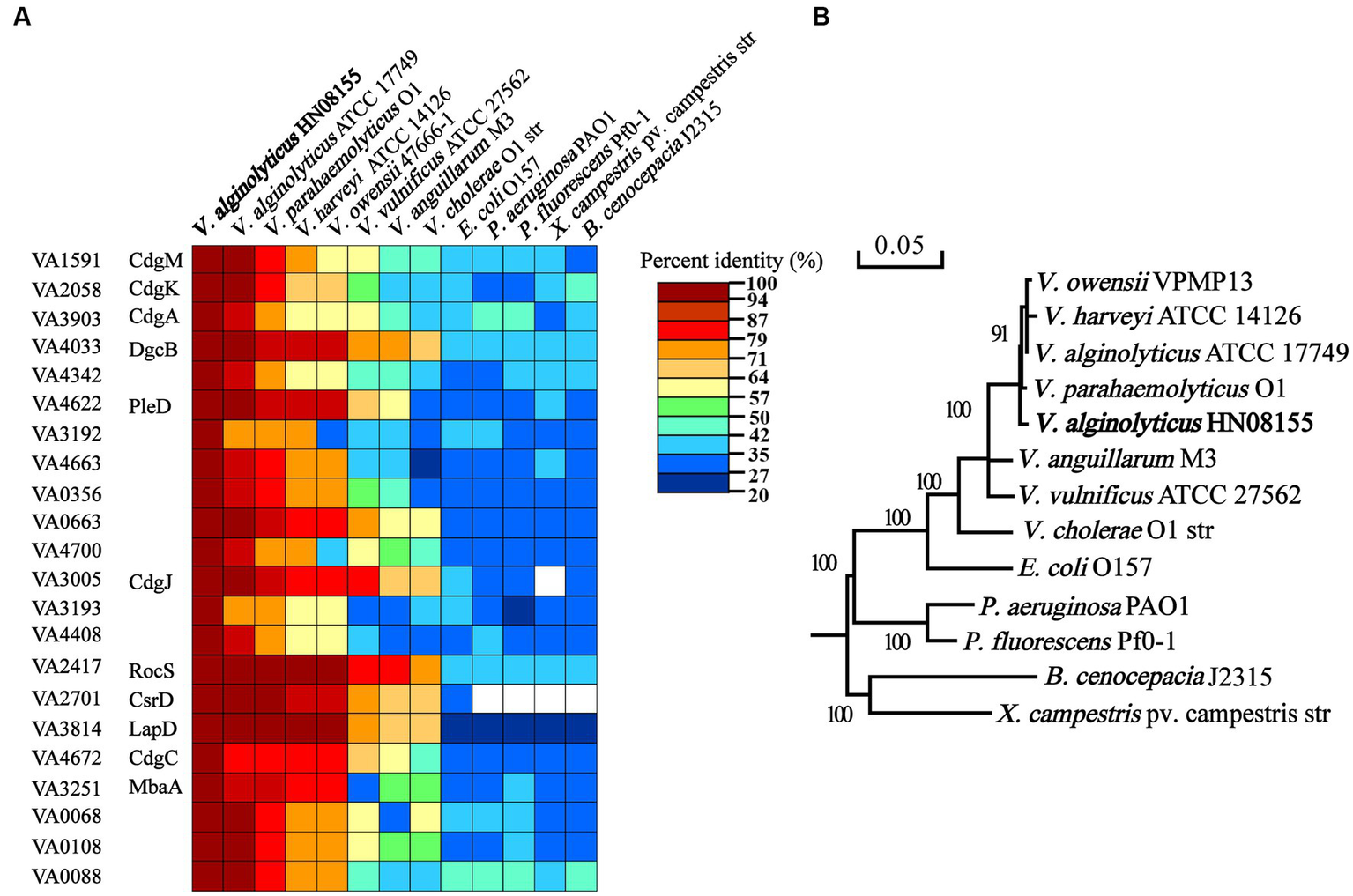
Comparisons of the 22 c-di-GMP metabolic proteins of Vibrio alginolyticus containing a GGDEF or EAL domain with those of closely related Vibrio species and other bacterial species (A). Phylogenetic tree analysis of V. alginolyticus and other species based on 16S rRNA gene sequences (B).
A total of 44 derivative strains with each of the 22 DGC or PDE genes knocked out or overexpressed were generated. As shown in Supplementary Figure S7, neither individual mutations nor overexpression of these 22 genes affected the growth of strain HN08155, so the phenotypic differences between the derivative and wild-type strains described below should be attributed to the regulatory role of the corresponding c-di-GMP metabolic genes.
Effect of different c-di-GMP metabolic genes on colony morphology
Changes in colony morphology have been widely used to reflect changes in biofilm matrix production levels, as rugose (also termed wrinkled or corrugated) colony morphologies often correlate with increased EPS production (Yildiz and Visick, 2009). Thus, the regulatory effect of these 22 c-di-GMP metabolic genes on rugose colony formation was examined on LBS (containing 3% NaCl) and LB (containing 1% NaCl) plates (Supplementary Figure S8). We found that only one overexpression strain ∆VA4033p4033 exhibited rugose colony morphotypes on both LBS and LB plates (Figure 2A). Unlike strain ∆VA4033p4033, the overexpression strains ∆VA1591p1591, ∆VA0356p0356, and ∆VA0088p0088 only formed enhanced rugose colonies on LB plates (Figure 2B). The above results indicated that the regulatory effects of different c-di-GMP metabolic genes on the rugose colony morphology of V. alginolyticus are influenced by the salinity concentration in the medium. It is notable that all knockout mutant strains of the c-di-GMP metabolic gene had a smooth colony morphology similar to that of the wild-type (Supplementary Figure S8), indicating that the mutation of a c-di-GMP metabolic gene does not lead to changes in the colony morphology of V. alginolyticus under the assay conditions of this work.
Figure 2

Representative images of colony morphology of the wild-type strain (or wild-type carrying pBAD vector) and 4 c-di-GMP metabolic gene deletion mutants and corresponding 4 overexpression strains on LBS (A) and LB (B) plates. Experiments were performed in three independent biological replicates and representative images are shown (scaled to equal diameter; bars = 5 mm).
Correlation analysis of colony rugosity with EPS production and flagellar biosynthesis
The DGC VA4033 (DgcB) was the only one of the 22 representative c-di-GMP metabolic proteins that enhanced colony rugosity on both LBS and LB plates. For this reason, correlations between colony rugosity with EPS production and flagellar analysis were determined by quantitative PCR (qPCR) under conditions of overexpression of VA4033. As shown in Figure 3, overexpression of VA4033 significantly increased the transcript levels of all tested genes that are involved in EPS production, including 6 genes of the EPS biosynthetic gene cluster (vpsQ-wcaJ), as well as the vpsT gene encoding a helix-turn-helix transcriptional regulator (p < 0.05). On the contrary, most of the genes in the flagellar synthesis gene cluster, including class I (flgBFGH1I2K-flaB), class II (fliA-flhA-fliPON), class III (fliFES-flaABF) and lafAT, were significantly down-regulated in the overexpression strains WTp4033 and ∆4033p4033 compared to strain WTpBAD (p < 0.05; Figure 3). The above results indicate that rugosity morphology is accompanied by increased transcript levels of EPS biosynthesis genes and decreased expression levels of flagellar synthesis genes, suggesting that c-di-GMP inversely regulates EPS production and flagellar biogenesis in V. alginolyticus.
Figure 3
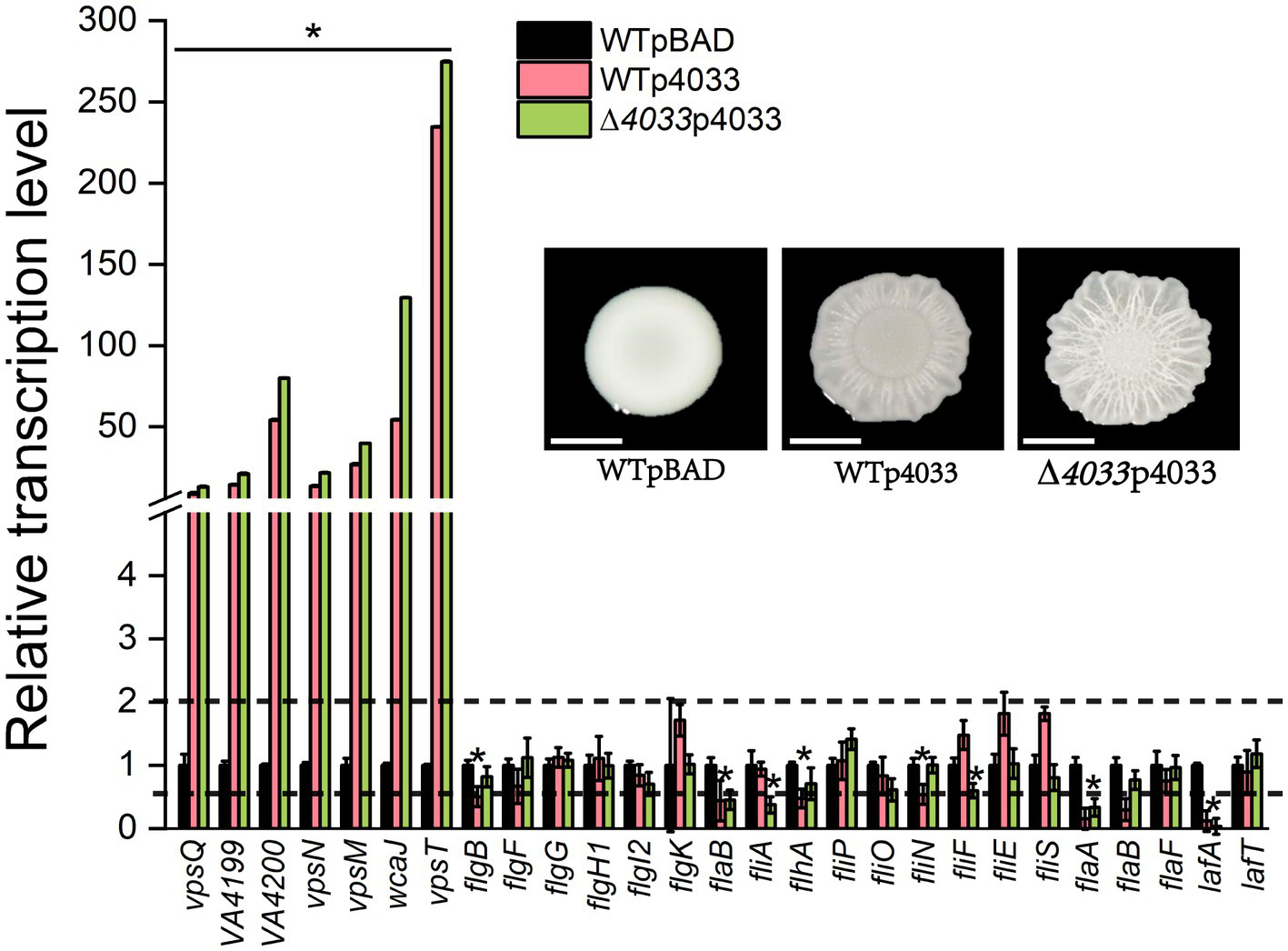
Relative expression levels of genes involved in EPS production and flagellar synthesis between strain WTpBAD and overexpression strains WTp4033 (or ∆VA4033p4033) on LBS plates. The transcript levels of genes in WTpBAD were set to a value of 1 as a reference, and the upper and lower dashed lines indicate expression levels of 2-fold and 50% compared to the WTpBAD, respectively. The results presented are the mean of triplicate experiments and error bars represent SDs. *, p < 0.05.
Identification of c-di-GMP metabolic genes involved in static biofilm formation
To determine which c-di-GMP metabolic genes play a role in biofilm formation in V. alginolyticus, we quantified the static biofilm formation ability of c-di-GMP metabolic gene deficient and overexpression strains under LB culture condition by crystal violet staining. Our previous results showed that the biofilm formation ability of the mutant strain ∆cdgH was significantly lower than that of the wild-type, and therefore the biofilm-deficient strain ∆cdgH was used as a negative control in all subsequent biofilm assays.
Among the 10 mutants of GGDEF-only gene, 6 of them (∆VA1591, ∆VA3903, ∆VA4033, ∆VA4663, ∆VA3192 and ∆VA0663) formed biofilms at approximately 60% ~ 90% of the level of the wild-type, 1 mutant (∆VA0356) formed biofilms at about 40% of the level of the wild-type, and 3 mutants (∆VA2058, ∆VA4342 and ∆VA4622) formed biofilms at levels exceeding that of the wild-type (Figure 4). The biofilm formation levels of the corresponding overexpression strains of the 10 c-di-GMP metabolic genes recovered to varying degrees to that of the wild-type, with strains ∆VA1591p1591 and ∆VA0356p0356 being almost comparable to the wild-type, strains ∆VA0663p0663 and ∆VA4342p4342 producing slightly lower biofilms than the wild-type, and strains ∆VA2058p2058, ∆VA3903p3903, ∆VA4033p4033, ∆VA4622p4622 and ∆VA4663p4663 formed biofilms slightly or even significantly higher than the wild-type (Figure 4). Unexpectedly, despite the fact that VA3192 was predicted to be a DGC and its GGDEF domain was intact, the biofilm formation level of strain ∆VA3192p3192 was as low as that of the negative control ∆cdgH. We speculate that a possible explanation is that the protein VA3192 may not be involved in the regulation of biofilm formation by c-di-GMP metabolism, as previous study has indicated that the regulation of biofilm formation by the protein GdpS in Staphylococcus was independently of c-di-GMP (Holland et al., 2008). In conclusion, these results indicate that most of the predicted DGCs can promote static biofilm formation, and their mode of action is likely to be through the synthesis of c-di-GMP.
Figure 4
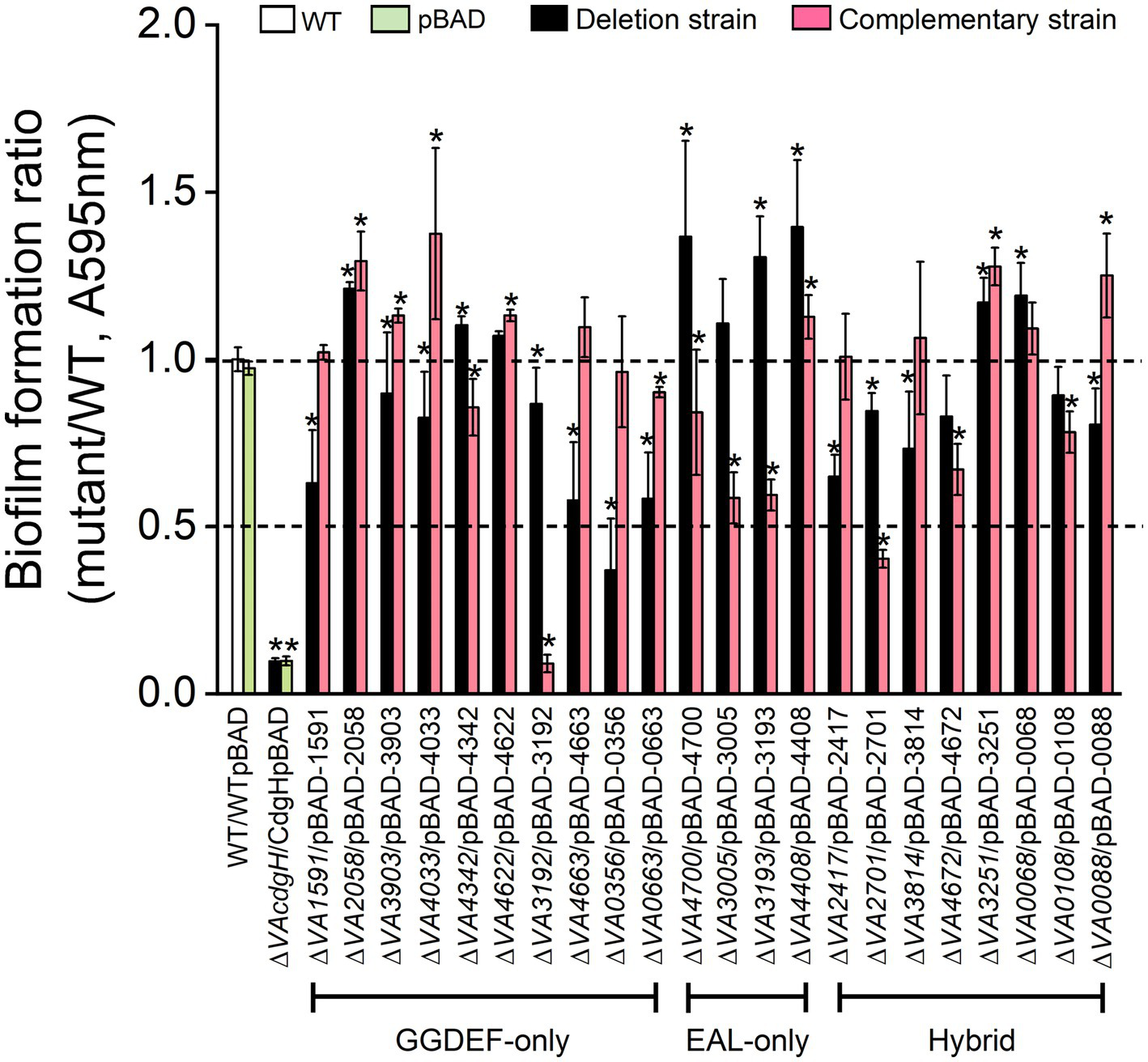
Static biofilm formation assay. The wild-type, 22 c-di-GMP metabolic gene deletion mutants, and the corresponding 22 overexpression strains were cultured in LB liquid medium in 96-well plate for 6 h. All measurements of biofilm biomass were normalized to the value of the wild-type. The results presented are the mean of triplicate experiments and error bars represent SDs. *, p < 0.05.
All of the 4 mutants of EAL-only gene, including ∆VA4700, ∆VA3005, ∆VA3193 and ∆VA4408, showed significantly increased biofilm formation compared to the wild-type, with mutant ∆VA3005 producing a lower biofilm than the other three mutants. The corresponding overexpression strains ∆VA4700p4700, ∆VA3005p3005 and ∆VA3193p3193 formed biofilms at approximately 50% of the level of the wild-type, however, strain ∆VAVA4408p4408 showed no deficiency in biofilm formation (Figure 4).
Six of the 8 mutants of GGDEF-EAL gene (∆VA2417, ∆VA2701, ∆VA3814, ∆VA4672, ∆VA0108 and ∆VA0088) formed biofilms at 50% ~ 90% of the level of the wild-type. The level of biofilms formed by the overexpression strains ∆VA2417p2417 and ∆VA3814p3814 was close to the wild-type, and the biofilm formation ability of strain ∆VA0088p0088 was significantly higher than that of the wild-type (p < 0.05), probably because the DGC activity of protein VA0088 of V. alginolyticus is prevalent under the assay conditions of this work. The level of biofilms formed by the overexpression strains ∆VA2701p2701, ∆VA4672p4672 and ∆VA0108p0108 was significantly lower than that of the wild-type (p < 0.05). The remaining two mutants ∆VA3251 and ∆VA0068 exhibited significantly higher biofilm formation ability than the wild-type (p < 0.05), and their corresponding overexpression strains ∆VA3251p3251 and ∆VA0068p0068 also showed enhanced biofilm formation ability compared to the wild-type (Figure 4).
Identification of c-di-GMP metabolic genes involved in EPS production
The intracellular c-di-GMP level is positively correlated with EPS production (Rashid et al., 2003; Casper-Lindley and Yildiz, 2004; Srivastava et al., 2013), and the ability of EPS to bind Congo Red (CR) has been shown to be an effective and intuitive way to detect EPS (Schäper et al., 2017). Therefore, we identified the c-di-GMP metabolic genes involved in EPS production in V. alginolyticus by CR binding method.
Among the 10 mutants of GGDEF-only gene, none of them showed a significant difference in CR binding intensity from that of the wild-type (Figure 5), suggesting that deletion of a single c-di-GMP metabolic gene cannot significantly cause changes in CR binding activity. However, overexpression strains of these 10 DGCs resulted in significant changes in CR binding ability, with strains ∆VA1591p1591, ∆VA4033p4033, and ∆VA0356p0356 showing strong CR binding ability and rugose colony morphologies, and strain ∆VA3192p3192 showing lower CR binding than the above three strains, followed by ∆VA2058p2058, ∆VA4622p4622, and ∆VA4663p4663, and the remaining 3 overexpression strains ∆VA3903p3903, ∆VA4342p4342, ∆VA0663p0663 had similar CR binding ability to the wild-type (Figure 5).
Figure 5

EPS production determined with Congo red plates. Representative images of the wild-type, 22 c-di-GMP metabolic gene deletion mutants and corresponding 22 overexpression strains on TSA plates staining with Congo red were shown. Experiments were performed in three independent biological replicates (scaled to equal diameter; bars = 5 mm).
Similarly, none of the 4 mutants of EAL-only gene (∆VA4700, ∆VA3005, ∆VA3193, and ∆VA4408) displayed a significant difference in CR binding ability from that of the wild-type (Figure 5). However, unlike most of the DGC overexpression strains showed stronger CR binding than the wild-type, none of the 4 PDE overexpression strains (∆VA4700p4700, ∆VA3005p3005, ∆VA3193p3193, and ∆VA4408p4408) had weaker CR binding compared to the wild-type (Figure 5).
All of the 8 mutants of GGDEF-EAL gene (∆VA2417, ∆VA2701, ∆VA3814, ∆VA4672, ∆VA3251, ∆VA0068, ∆VA0108, and ∆VA0088) had indistinguishable CR binding compared to the wild-type (Figure 5). Compared to the wild-type, the overexpression strain ∆VA0088p0088 exhibited stronger CR binding and enhanced colony rugosity (Figure 5), which was consistent with its increased static biofilm formation ability (Figure 4). The overexpression strains ∆VA2701p2701 and ∆VA3251p3251 also showed higher CR binding ability than the wild-type, but displayed smooth colony morphology (Figure 5). The remaining overexpression strains ∆VA2417p2417, ∆VA4672p4672, ∆VA0068p0068, and ∆VA0108p0108 showed similar CR binding ability to the wild-type (Figure 5).
C-di-GMP metabolic proteins were essential for flagellar-mediated swarming motility
The swarming motility and biofilm formation or EPS production are inversely regulated by c-di-GMP-mediated signaling (Kuchma et al., 2007, 2015). Semi-solid motility plates containing 0.6% agar were used to investigate whether the 10 proteins involved in the synthesis of c-di-GMP play a role in flagellar-mediated swarming motility. The ∆flhF strain cannot synthesis flagella as a negative control. Of the 10 mutants of GGDEF-only gene, 4 of them (∆VA1591, ∆VA4033, ∆VA4342 and ∆VA3192) showed a significant increase in swarming diameter compared to the wild type. In contrast, mutants ∆VA2058, ∆VA3903, ∆VA4622, ∆VA0356, ∆VA0663, and ∆VA4663 showed a slight decrease in swarming diameter compared to the wild type or were comparable to the wild type (Figure 6A). The respective overexpression strains of these mutants produced varying degrees swarming inhibition, with strains ∆VA1591p1591 and ∆VA0356p0356 producing less than 50% of the swarming phenotype compared to the wild type, strains ∆VA2058p2058 and ∆VA4033p4033 showing 50% ~ 60% of the swarming ability level of the wild type, and strain ∆VA4033p4033 producing a rugose phenotype (Figure 6B). Strains ∆VA3903p3903, ∆VA3192p3192, ∆VA4342p4342, ∆VA4663p4663, and ∆VA0663p0663 exhibited swarming ability at 70% ~ 90% level of the wild type, while strain ∆VA4622p4622 had comparable swarming ability to the wild type (Figure 6A). These results suggest that these 10 predicted DGCs have different degrees of inhibition on the swarming ability of V. alginolyticus.
Figure 6
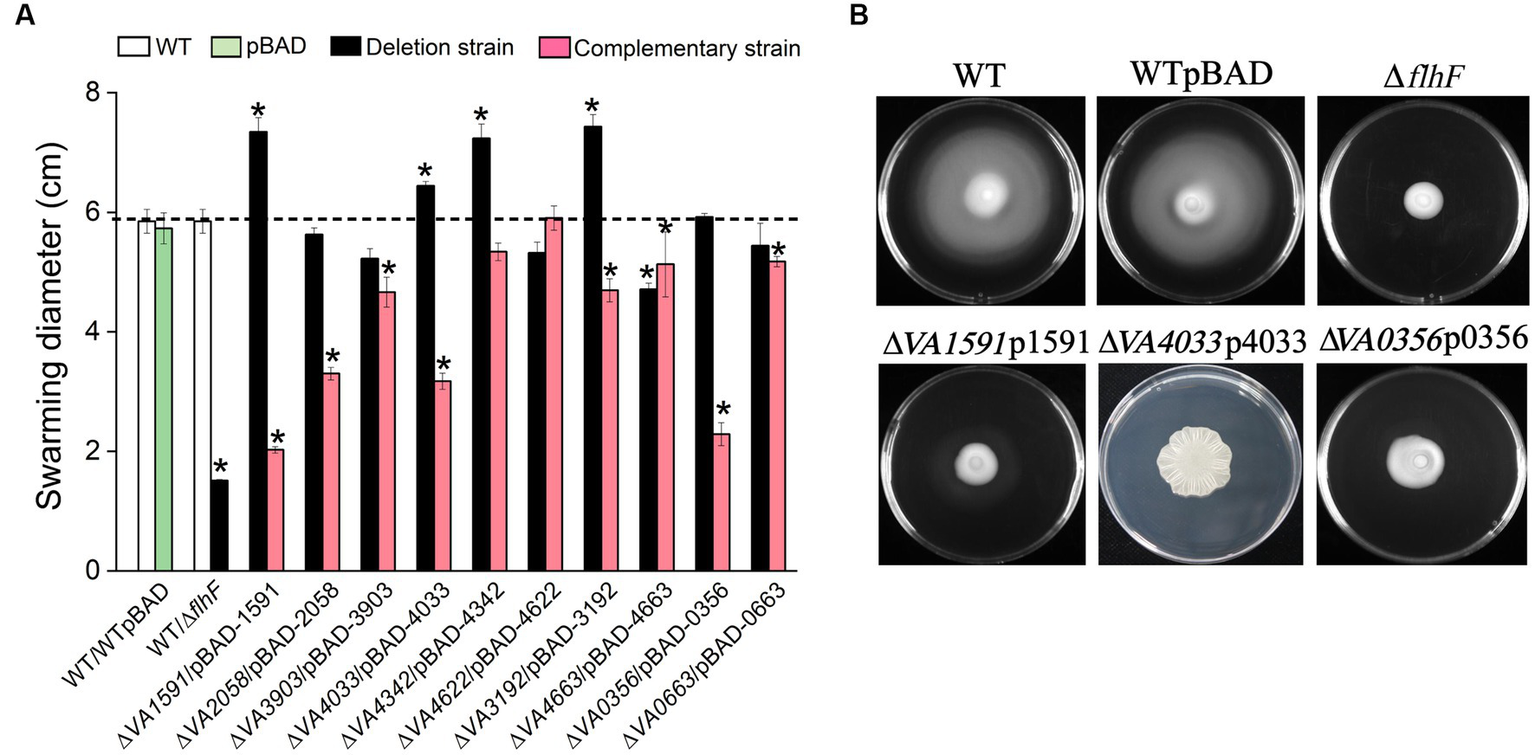
Swarming motility assay. The swarming diameter of the wild type, 10 c-di-GMP metabolic gene deletion mutants, and corresponding 10 overexpression strains on LBS plates with 0.6% agar, the results presented are the mean of triplicate experiments and error bars represent SDs. *, p < 0.05. (A) Representative images of swarming motility reduction in strains ∆1591p1591, ∆4033p4033 and ∆0356p0356. The nonmotile (no clearly visible movement outside the colony) ∆flhF mutant served as a negative control (B).
Discussion
Currently, it is still unclear about the role of c-di-GMP signaling in V. alginolyticus and how the different c-di-GMP metabolic proteins regulate biofilm formation. In this work, we identified a total of 63 c-di-GMP metabolic proteins with putative GGDEF/EAL/HD-GYP domains in the genome of V. alginolyticus HN08155. By systematic bioinformatic analyses, we selected 22 representative c-di-GMP metabolic proteins and explored their regulatory roles in biofilm-associated phenotypes, including colony rugosity, static biofilm formation, EPS production and motility. To this end, we constructed knockout mutants and overexpression strains of these 22 c-di-GMP metabolic genes, respectively, and found that several key c-di-GMP metabolic genes (VA1591, VA0356, VA0088, VA4033, VA3192 and VA4408) had significant influences on the biofilm-associated phenotypes of V. alginolyticus (Figure 7).
Figure 7
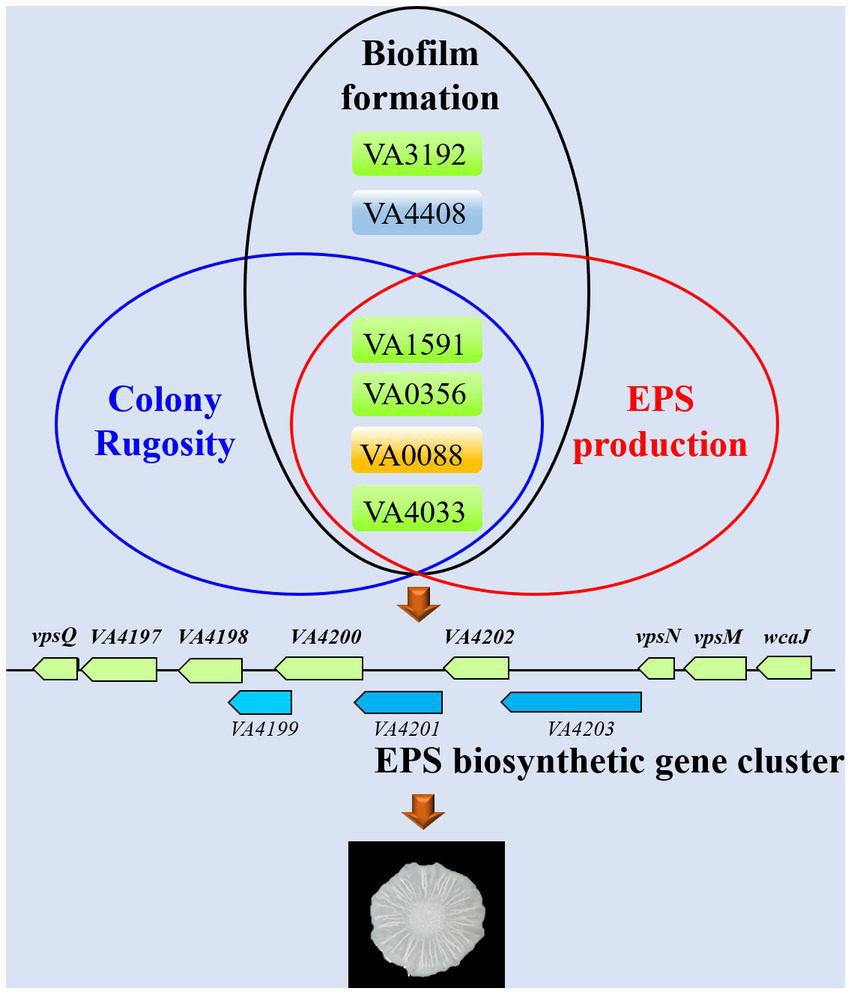
Schematic for proposed conceptual model for key c-di-GMP metabolic genes that required for biofilm-associated phenotypes in V. alginolyticus.
We identified 4 c-di-GMP metabolic genes, including VA4033, VA1591, VA0356, and VA0088, all with intact active sites for the GGDEF domain (Table 1), involved in the enhanced rugose colony phenotypes on LB agar plates (Figure 2). It is well known that there is a strong correlation between the phenotypes of colony rugosity, biofilm formation and EPS production, and moreover, these phenotypes are often accompanied by elevated intracellular c-di-GMP levels (Nakhamchik et al., 2008; Romling et al., 2013; Wu et al., 2020; Xiao et al., 2022). In general, enhanced EPS production often results in rugose colony phenotypes (Casper-Lindley and Yildiz, 2004; Beyhan et al., 2008), a phenomenon also demonstrated in the present work, for example, overexpression strains ∆VA1591p1591, ∆VA4033p4033, ∆VA0356p0356, and ∆VA0088p0088 produced rugose colonies that also had strong CR binding capacities (Figures 2, 5). However, in some strains, these phenotypes were only weakly correlated. For example, overexpression strains ∆VA1591p1591 and ∆VA0356p0356 had a corrugated phenotype and strong CR binding but did not produce a hyper-biofilm phenotype, in contrast, some strains (e.g., ∆VA2058p2058, ∆VA4700, ∆VA3005, ∆VA3193, and ∆VA4408) had significantly increased biofilm formation but produced smooth colony morphology with weak binding to CR (Supplementary Figure S8; Figures 4, 5). Furthermore, overexpression strains ∆VA3192p3192 and ∆VA2701p2701 showed relatively stronger binding ability to CR than the wild-type, but produced a significant deficiency in biofilm formation (Figures 4, 5). The deficiency of biofilm formation in strain ∆VA3192p3192 was inconsistent with the predicted DGC activity of the protein VA3192, although this phenomenon has been previously identified (Sperling et al., 2019), however, whether this inconsistency is mediated by c-di-GMP signaling will be further investigated. In summary, colony rugosity and CR binding can be largely indicative of each other, especially strong CR binding that concomitantly characterizes the phenotype of super rugose. However, biofilm formation does not always correspond to the above two phenotypes. As previously reported, the functional diversity of c-di-GMP metabolic proteins in other Gram-negative bacteria has also been demonstrated (Ha et al., 2014; Chen et al., 2020; Shrestha et al., 2022).
Several previous studies have demonstrated that changes in the expression patterns of genes associated with EPS biosynthetic cluster are of significance for the biofilm structure of Vibrio species (Fong and Yildiz, 2007; Fong et al., 2017). We identified a cluster of 11 genes that encode proteins predicted to be associated with the production of EPS in V. alginolyticus HN08155, and several proteins in this cluster are homologous to the vps cluster of V. cholerae (Figures 3, 7). VpsT is a master transcriptional regulator that activates the expression of genes in the vps cluster by binding to the promoter sequence of vpsL in a c-di-GMP-dependent manner (Casper-Lindley and Yildiz, 2004; Tischler and Camilli, 2004; Krasteva et al., 2010). In this study, we demonstrated that the transcript levels of several genes in the EPS biosynthetic cluster were significantly increased in rugose colonies (WTp4033 and ∆VA4033p4033) compared to smooth colonies (WTpBAD), which is consistent with previous findings that EPS production leads to rugose morphology (Yildiz et al., 2001; Rashid et al., 2003; Casper-Lindley and Yildiz, 2004; Beyhan et al., 2008; Teschler et al., 2015).
This study showed that most of the mutants of the GGDEF-only proteins had increased or equivalent swarming motility phenotypes to the wild type, in contrast, 9 out of 10 DGC overexpression strains were inhibited in swarming motility (Figure 6). This was in accordance with the established research findings that high c-di-GMP levels suppress motility (Liu et al., 2010; Romling et al., 2013; Conner et al., 2017). Interestingly, the overexpression strain ∆VA4033p4033 exhibited a unique phenotype of weak and flat corrugated colonies on plate containing 0.6% agar, which was totally different from the smooth extended colonies of the other 9 overexpression strains on the same soft agar plate (Figure 6B; Supplementary Figure S9). In Caulobacter crescentus, the DGC activity of DgcB is stimulated upon surface sensing, a process mediated by flagellar motor, which ultimately leads to the rapid synthesis of polysaccharide adhesins called the holdfast (Hug et al., 2017). In this study, the qPCR results showed that the transcript levels of EPS biosynthesis genes were significantly increased when the function of protein VA4033 (DgcB) was fully complemented (data not shown). In addition, Hershey et al. found that DgcB could produce c-di-GMP at an instantaneous burst rate and immediately stimulate the production of polysaccharide adhesins in C. crescentus (Hershey et al., 2021). Therefore, although the plate containing 0.6% agar is favorable for flagellar-mediated motility (Supplementary Figure S9), overexpression strain ∆VA4033p4033 can also produce flat corrugated colonies, which we speculate is due to its “fast nature” in producing c-di-GMP and polysaccharide adhesins. However, the specific regulatory mechanism of protein VA4033 on the flat colony rugosity of V. alginolyticus under semi-solid condition needs further investigation.
In summary, this work provides the first insight into the role of c-di-GMP metabolic genes in controlling colony morphology, biofilm formation, EPS production and motility in V. alginolyticus. We found that each c-di-GMP metabolic gene can regulate multiple phenotypes, with 4 key c-di-GMP metabolic genes having important roles in the control of colony rugosity and EPS production. The large number of c-di-GMP metabolic proteins encoded by V. alginolyticus suggests that this marine pathogen may utilize a complex c-di-GMP signaling regulatory network to modulate its lifestyle changes in response to various environmental cues.
Materials and methods
Bioinformatic analyses
The functional annotation results of V. alginolyticus HN08155 were obtained from 11 databases, including VFDB, ARDB, CAZY, IPR, SWISSPROT, COG, CARD, GO, KEGG, NR, and T3SS, from the genome sequencing dataset. The c-di-GMP metabolic proteins were retrieved from the result annotation file based on “search for conserved domains.” For all searches: the entry “GGDEF” refers to GGDEF domain, “EAL” refers to EAL domain, and “HD” refers to HD-GYP domain. The transmembrane signaling and sensory partner domains of all c-di-GMP metabolic proteins were analyzed using the SMART algorithm1 and NCBI’s Conserved Domain Database (CDD).2 Conservation analysis of the GGDEF and EAL domain proteins of V. alginolyticus with other bacterial species using the NCBI Blastp Database. The phylogenetic tree was constructed with DNAMAN software.
Sequence alignments of the GGDEF, EAL and HD-GYP domains of V. alginolyticus and other related species were generated with ClustalW3 and formatted with ESPript 3.04 to determine the positions of the corresponding conserved site residues. Sequence logos were created based on the conserved domains of GGDEF, EAL and HD-GYP using the DNAMAN comparison tool and WebLogo 3: Create.5 The intactness of the catalytic sites of GGDEF, EAL and HD-GYP was determined according to previous research (Galperin et al., 1999; Christen et al., 2006; Seshasayee et al., 2010; Romling et al., 2013; Shrestha et al., 2022). Briefly, the active site (AGGDEF site) in the GGDEF domain was considered functional if the RxGGD/EEF signature motif was unmutated, and the inhibitory site (IGGDEF site) that can bind c-di-GMP was considered functional if an RxxD motif was present in 5 residues upstream of the AGGDEF site. The AEAL site was considered functional if the ExLxR signature motif was unmutated. The AHD-GYP site was considered functional if the HHExxDGxxGYP motif was conserved.
Strains and growth conditions
Bacterial strains and plasmids used in this study are listed Supplementary Table S1. V. alginolyticus strains were grown in Luria-Bertani (LB) medium (1% tryptone, 0.5% yeast extract, and 1% NaCl) or LBS medium (1% tryptone, 0.5% yeast extract, and 1% NaCl) at 30°C. E. coli used for DNA manipulation (strains DH5α and Top10) and for conjugational transfer (strain β2163) were routinely cultured in LB medium at 37°C. Solid medium was prepared using 1.5% agar. When needed, antibiotics were added at the following concentrations: for E. coli, ampicillin at 100 μg/mL and chloramphenicol at 50 μg/mL; and for V. alginolyticus, ampicillin at 100 μg/mL and chloramphenicol at 25/50 μg/mL for liquid/solid medium, respectively. L-arabinose at 0.02% (vt/vol) in growth medium was used for overexpression strains, and the final concentration of 2,6-Diaminopimelic acid (DAP) used for strain β2163 was 0.3 mM. Unless stated otherwise, all V. alginolyticus strains were pre-cultured in LBS medium containing appropriate antibiotics.
Construction of mutants and overexpression strains
All DNA manipulations were performed by standard molecular protocols. Deletion mutants of interest genes in V. alginolyticus were constructed via allelic exchange system. Briefly, the up-and down-stream flanking regions of the desired deletion gene, each approximately 600 bp in length, were amplified separately from the genomic DNA of V. alginolyticus using the primers shown in Supplementary Table S2. The two PCR products were ligated by overlapping extension PCR technique and cloned into the suicide plasmid pDM4 via designed restriction enzyme sites using T4 DNA ligase. The recombinant plasmid was transformed into a conjugative strain β2163 containing the pir gene, which was then conjugated with V. alginolyticus. Transconjugants were selected on LBS agar plates containing both ampicillin and chloramphenicol. Finally, V. alginolyticus mutant strains that had undergone homologous recombination were selected with 10% sucrose and validated by PCR.
For the construction of overexpression strains, the coding region of interest genes (excluding the stop codon) was amplified and ligated into the plasmid pBAD/Myc-HisA (the original ampicillin resistance site was replaced by chloramphenicol) by One Step Cloning Kit (Vazyme, Nanjing, China). The recombinant plasmid was transformed into E. coli Top10, and after sequencing, the plasmid was transformed into the conjugative strain β2163. All of these constructs were expressed to produce myc-6xHis tagged recombinant proteins. All PCR products were validated by DNA sequencing.
Analysis of bacterial growth
Freshly pre-cultured bacterial cultures were adjusted to the same cell density and inoculated 1:100 into fresh LBS or LB medium without antibiotics (supplemented with L-arabinose when needed), and then transferred 800 μL into each well of a 48-well microtiter plate. Fresh LBS or LB medium without bacterial inoculation served as a negative control. Bacterial growth was monitored by measuring OD600 every 30 min for 24 h at 30°C, using a multilabel plate reader (Biotek Winooski, Vermont, United States).
Analysis of colony morphology
Colony morphology assays were performed on LBS and LB plates. A single colony of freshly cultured V. alginolyticus strains was inoculated and grown in liquid LBS medium with shaking for 12 h (supplemented with antibiotics when appropriate). Bacterial cultures in LBS medium were diluted 1:100 into 20 mL of fresh LBS medium (supplemented with antibiotics and 0.02% L-arabinose when appropriate), and shaken for 3–4 h, then cell density was measured and normalized to an OD600 of 0.7. Afterwards, 4 μL of the resulting culture was spotted on LBS or LB agar plates (containing 1.5% agar) without antibiotics, allowed to dry and incubated at 30°C for 24 h. The diameter of the colonies was measured using a vernier caliper and imaged using a Canon PowerShot G7 X Mark III camera.
RNA extraction and qPCR
Colony biofilms of strains WTpBAD, WTp4033, and ∆VA4033p4033 were collected separately at 24 h and total RNA was extracted using Trizol reagent according to standard procedures. First-strand cDNA was synthesized using the HiScript® II Q RT SuperMix for qPCR (+gDNA wiper; Vazyme). Quantitative PCR (qPCR) was performed using the ChamQ Universal SYBR qPCR Master Mix (Vazyme) on a real-time PCR instrument (Roche, Basel, Switzerland). The relative expression levels of the target genes were calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001). The gyrB gene was used as the internal reference gene. The data were analyzed based on three independent biological replicates.
Biofilm formation assay
Biofilm formation assays were performed in LB medium according to previous research with minor modifications (Wilksch et al., 2011). Freshly pre-cultured bacterial cultures (adjusted to an OD600 of 0.75) were inoculated 1:100 into fresh LB medium without antibiotics but supplemented with L-arabinose, and then transferred 200 μL into each well of a 96-well microtiter plate. Fresh LB medium without bacterial inoculation served as a negative control. The plates were incubated at 30°C for 6 h under static conditions, at which point the cultures were discarded from the wells and washed twice with PBS buffer. Wells were dried and then stained with 0.1% crystal violet for 20 min, followed by slow washing with distilled water and subsequently allowed to dry. Wells were destained with 200 μL of 33% (vol/vol) acetic acid for 20 min, and then the solution was transferred to a new 96-well plate, followed by quantification by measuring the optical density at 595 nm on a multilabel plate reader (BioTek).
Congo red assay
Congo red staining was performed on TSA agar plates containing 120 μg/mL Congo red (supplemented with 0.02% L-arabinose when appropriate). Five microliters of freshly pre-cultured bacterial cultures (adjusted to an OD600 of 0.7) was dropped onto TSA agar plates without antibiotics. The plates were incubated at 30°C for 24 h and left to stand at room temperature for about 12 h, and then imaged using a Canon PowerShot G7 X Mark III camera.
Swarming motility assay
All V. alginolyticus strains were tested for swarming motility on LBS plates supplemented with 0.02% L-arabinose when appropriate. Two microliters of freshly pre-cultured bacterial cultures (adjusted to an OD600 of 0.7) was spotted on swarming plates containing 0.6% agar and incubated at 30°C for 24 h. The mutant strain ∆flhF without swarming ability was used as a negative control. The swarming motility assay was performed in at least six replicates of three independent experiments.
Statistical analyses
Statistical analyses were carried out using the SPSS 18.0 statistical software. All assays were performed in triplicate, and the results were presented as the mean ± standard deviation. Analyses of statistical differences were conducted with the paired two-tailed Student’s t test.
Data availability
The GenBank accession numbers of the 22 c-di-GMP metabolic genes, including VA1591 (cdgM), VA2058 (cdgK), VA3903 (cdgA), VA4033 (dgcB), VA4342, VA4622 (pleD), VA3192, VA4663, VA0356, VA0663, VA4700, VA3005 (cdgJ), VA3193, VA4408, VA2417 (rocS), VA2701 (csrD), VA3814 (lapD), VA4672 (cdgC), VA3251 (mbaA), VA0068, VA0108 and VA0088 were OP893990, OP893991, OP893992, OP893993, OP893994, OP893995, OP893996, OP893997, OP893998, OP893999, OP894000, OP894001, OP894002, OP894003, OP894004, OP894005, OP894006, OP894007, OP894008, OP894009, OP894010, and OP894011, respectively.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the National Natural Science Foundation of China (no. 32260927 and no. 32060835), Hainan Province Science and Technology Special Fund (ZDYF2022XDNY349), and Natural Science Foundation of Hainan Province (2019RC106).
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The GenBank accession numbers of the 22 c-di-GMP metabolic genes, including VA1591 (cdgM), VA2058 (cdgK), VA3903 (cdgA), VA4033 (dgcB), VA4342, VA4622 (pleD), VA3192, VA4663, VA0356, VA0663, VA4700, VA3005 (cdgJ), VA3193, VA4408, VA2417 (rocS), VA2701 (csrD), VA3814 (lapD), VA4672 (cdgC), VA3251 (mbaA), VA0068, VA0108 and VA0088 were OP893990, OP893991, OP893992, OP893993, OP893994, OP893995, OP893996, OP893997, OP893998, OP893999, OP894000, OP894001, OP894002, OP894003, OP894004, OP894005, OP894006, OP894007, OP894008, OP894009, OP894010, and OP894011, respectively.
Author contributions
X-XG: Conceptualization, Investigation, Supervision, Writing – original draft. Y-HZ: Writing – original draft, Writing – review & editing. H-MC: Writing – original draft, Data curation. NZ: Data curation, Writing – original draft. YH: Data curation, Writing – original draft. HL: Methodology, Writing – original draft. Z-YX: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1258415/full#supplementary-material
Footnotes
1.^http://smart.embl-heidelberg.de/
2.^https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml
3.^https://www.genome.jp/tools-bin/clustalw
References
1
AhmadI.NygrenE.KhalidF.MyintS. L.UhlinB. E. (2020). A cyclic-di-GMP signalling network regulates biofilm formation and surface associated motility of Acinetobacter baumannii 17978. Sci. Rep.10:1991. doi: 10.1038/s41598-020-58522-5
2
Baker-AustinC.OliverJ. D.AlamM.AliA.WaldorM. K.QadriF.et al. (2018). Vibrio spp. infections. Nat. Rev. Dis. Primers.4:8. doi: 10.1038/s41572-018-0005-8
3
BeyhanS.OdellL. S.YildizF. H. (2008). Identification and characterization of cyclic diguanylate signaling systems controlling rugosity in Vibrio cholerae. J. Bacteriol.190, 7392–7405. doi: 10.1128/JB.00564-08
4
BiswasS.ChouhanO. P.BandekarD. (2020). Diguanylate Cyclases in Vibrio cholerae: essential regulators of lifestyle switching. Front. Cell. Infect. Microbiol.10:582947. doi: 10.3389/fcimb.2020.582947
5
Casper-LindleyC.YildizF. H. (2004). Vps T is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol.186, 1574–1578. doi: 10.1128/JB.186.5.1574-1578.2004
6
ChenY.ZhouJ.LvM.LiangZ.ParsekM. R.ZhangL. H. (2020). Systematic analysis of c-di-GMP signaling mechanisms and biological functions in Dickeya zeae EC1. mBio11:e02993-20. doi: 10.1128/mBio.02993-20
7
ChouS. H.GalperinM. Y. (2016). Diversity of cyclic Di-GMP-binding proteins and mechanisms. J. Bacteriol.198, 32–46. doi: 10.1128/JB.00333-15
8
ChristenB.ChristenM.PaulR.SchmidF.FolcherM.JenoeP.et al. (2006). Allosteric control of cyclic di-GMP signaling. J. Biol. Chem.281, 32015–32024. doi: 10.1016/S0021-9258(19)84115-7
9
CollinsA. J.SmithT. J.SondermannH.O'TooleG. A. (2020). From input to output: the lap/c-di-GMP biofilm regulatory circuit. Annu. Rev. Microbiol.74, 607–631. doi: 10.1146/annurev-micro-011520-094214
10
ConnerJ. G.Zamorano-SanchezD.ParkJ. H.SondermannH.YildizF. H. (2017). The ins and outs of cyclic di-GMP signaling in Vibrio cholerae. Curr. Opin. Microbiol.36, 20–29. doi: 10.1016/j.mib.2017.01.002
11
DahlstromK. M.O'TooleG. A. (2017). A symphony of Cyclases: specificity in Diguanylate cyclase signaling. Annu. Rev. Microbiol.71, 179–195. doi: 10.1146/annurev-micro-090816-093325
12
FlemmingH. C.WingenderJ.SzewzykU.SteinbergP.RiceS. A.KjellebergS. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol.14, 563–575. doi: 10.1038/nrmicro.2016.94
13
FongJ. C.RogersA.MichaelA. K.ParsleyN. C.CornellW. C.LinY. C.et al. (2017). Structural dynamics of RbmA governs plasticity of Vibrio cholerae biofilms. elife6:e26163. doi: 10.7554/eLife.26163
14
FongJ. C.YildizF. H. (2007). The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J. Bacteriol.189, 2319–2330. doi: 10.1128/JB.01569-06
15
GalperinM. Y.NataleD. A.AravindL.KooninE. V. (1999). A specialized version of the HD hydrolase domain implicated in signal transduction. J. Mol. Microbiol. Biotechnol.1, 303–305. PMID:
16
GuvenerZ. T.HarwoodC. S. (2007). Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol. Microbiol.66, 1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x
17
HaD. G.RichmanM. E.O'TooleG. A. (2014). Deletion mutant library for investigation of functional outputs of cyclic diguanylate metabolism in Pseudomonas aeruginosa PA14. Appl. Environ. Microbiol.80, 3384–3393. doi: 10.1128/AEM.00299-14
18
HenggeR. (2021). High-specificity local and global c-di-GMP signaling. Trends Microbiol.29, 993–1003. doi: 10.1016/j.tim.2021.02.003
19
HersheyD. M.FiebigA.CrossonS. (2021). Flagellar perturbations activate adhesion through two distinct pathways in Caulobacter crescentus. MBio12:e03266-20. doi: 10.1128/mBio.03266-20
20
HollandL. M.O'DonnellS. T.RyjenkovD. A.GomelskyL.SlaterS. R.FeyP. D.et al. (2008). A staphylococcal GGDEF domain protein regulates biofilm formation independently of cyclic dimeric GMP. J. Bacteriol.190, 5178–5189. doi: 10.1128/JB.00375-08
21
HommaM.KojimaS. (2022). Roles of the second messenger c-di-GMP in bacteria: focusing on the topics of flagellar regulation and Vibrio spp. Genes Cells27, 157–172. doi: 10.1111/gtc.12921
22
HugI.DeshpandeS.SprecherK. S.PfohlT.JenalU. (2017). Second messenger-mediated tactile response by a bacterial rotary motor. Science358, 531–534. doi: 10.1126/science.aan5353
23
HutchinA.CorderyC.WalshM. A.WebbJ. S.TewsI. (2021). Phylogenetic analysis with prediction of cofactor or ligand binding for Pseudomonas aeruginosa PAS and cache domains. Microbiol. Spectr.9:e0102621. doi: 10.1128/spectrum.01026-21
24
IbanghaI. I.DigwoD. C.OzochiC. A.EnebeM. C.AtebaC. N.ChigorV. N. (2023). A meta-analysis on the distribution of pathogenic Vibrio species in water sources and wastewater in Africa. Sci. Total Environ.881:163332. doi: 10.1016/j.scitotenv.2023.163332
25
Jacobs SlifkaK. M.NewtonA. E.MahonB. E. (2017). Vibrio alginolyticus infections in the USA, 1988-2012. Epidemiol. Infect.145, 1491–1499. doi: 10.1017/S0950268817000140
26
KharadiR. R.SundinG. W. (2022). CsrD regulates amylovoran biosynthesis and virulence in Erwinia amylovora in a novel cyclic-di-GMP dependent manner. Mol. Plant Pathol.23, 1154–1169. doi: 10.1111/mpp.13217
27
KimH. S.HamS. Y.RyooH. S.KimD. H.YunE. T.ParkH. D.et al. (2023). Inhibiting bacterial biofilm formation by stimulating c-di-GMP regulation using citrus peel extract from Jeju Island. Sci. Total Environ.872:162180. doi: 10.1016/j.scitotenv.2023.162180
28
KimbroughJ. H.CribbsJ. T.McCarterL. L. (2020). Homologous c-di-GMP-binding Scr transcription factors orchestrate biofilm development in Vibrio parahaemolyticus. J. Bacteriol.202:e00723-19. doi: 10.1128/JB.00723-19
29
KittsG.GiglioK. M.Zamorano-SánchezD.ParkJ. H.TownsleyL.CooleyR. B.et al. (2019). A conserved regulatory circuit controls large Adhesins in Vibrio cholerae. mBio10:e02822-19. doi: 10.1128/mBio.02822-19
30
KrastevaP. V.FongJ. C.ShikumaN. J.BeyhanS.NavarroM. V.YildizF. H.et al. (2010). Vibrio cholerae Vps T regulates matrix production and motility by directly sensing cyclic di-GMP. Science327, 866–868. doi: 10.1126/science.1181185
31
KuchmaS. L.BrothersK. M.MerrittJ. H.LiberatiN. T.AusubelF. M.O'TooleG. A. (2007). BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol.189, 8165–8178. doi: 10.1128/JB.00586-07
32
KuchmaS. L.DelalezN. J.FilkinsL. M.SnavelyE. A.ArmitageJ. P.O'TooleG. A. (2015). Cyclic di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa PA14 requires the MotAB stator. J. Bacteriol.197, 420–430. doi: 10.1128/JB.02130-14
33
LindenbergS.KlauckG.PesaventoC.KlauckE.HenggeR. (2013). The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control. EMBO J.32, 2001–2014. doi: 10.1038/emboj.2013.120
34
LiuX.BeyhanS.LimB.LiningtonR. G.YildizF. H. (2010). Identification and characterization of a phosphodiesterase that inversely regulates motility and biofilm formation in Vibrio cholerae. J. Bacteriol.192, 4541–4552. doi: 10.1128/JB.00209-10
35
LiuX.CaoB.YangL.GuJ. D. (2022). Biofilm control by interfering with c-di-GMP metabolism and signaling. Biotechnol. Adv.56:107915. doi: 10.1016/j.biotechadv.2022.107915
36
LivakK. J.SchmittgenT. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods25, 402–408. doi: 10.1006/meth.2001.1262
37
Martinez-MendezR.Camacho-HernandezD. A.Sulvaran-GuelE.Zamorano-SanchezD. (2021). A trigger phosphodiesterase modulates the global c-di-GMP Pool, motility, and biofilm formation in Vibrio parahaemolyticus. J. Bacteriol.203:e0004621. doi: 10.1128/JB.00046-21
38
MassieJ. P.ReynoldsE. L.KoestlerB. J.CongJ. P.AgostoniM.WatersC. M. (2012). Quantification of high-specificity cyclic diguanylate signaling. Proc. Natl. Acad. Sci.109, 12746–12751. doi: 10.1073/pnas.1115663109
39
NakhamchikA.WildeC.Rowe-MagnusD. A. (2008). Cyclic-di-GMP regulates extracellular polysaccharide production, biofilm formation, and rugose colony development by Vibrio vulnificus. Appl. Environ. Microbiol.74, 4199–4209. doi: 10.1128/AEM.00176-08
40
PurcellE. B.McKeeR. W.McBrideS. M.WatersC. M.TamayoR. (2012). Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. J. Bacteriol.194, 3307–3316. doi: 10.1128/JB.00100-12
41
RashidM. H.RajannaC.AliA.KaraolisD. K. (2003). Identification of genes involved in the switch between the smooth and rugose phenotypes of Vibrio cholerae. FEMS Microbiol. Lett.227, 113–119. doi: 10.1016/S0378-1097(03)00657-8
42
RomlingU.GalperinM. Y.GomelskyM. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev.77, 1–52. doi: 10.1128/MMBR.00043-12
43
SarenkoO.KlauckG.WilkeF. M.PfifferV.RichterA. M.HerbstS.et al. (2017). More than enzymes that make or break cyclic Di-GMP-local signaling in the Interactome of GGDEF/EAL domain proteins of Escherichia coli. mBio8:e01639-17. doi: 10.1128/mBio.01639-17
44
SchäperS.SteinchenW.KrolE.AltegoerF.SkotnickaD.Søgaard-AndersenL.et al. (2017). AraC-like transcriptional activator CuxR binds c-di-GMP by a PilZ-like mechanism to regulate extracellular polysaccharide production. Proc. Natl. Acad. Sci.114, E4822–E4831. doi: 10.1073/pnas.1702435114
45
SchirmerT.JenalU. (2009). Structural and mechanistic determinants of c-di-GMP signalling. Nat. Rev. Microbiol.7, 724–735. doi: 10.1038/nrmicro2203
46
SeshasayeeA. S.FraserG. M.LuscombeN. M. (2010). Comparative genomics of cyclic-di-GMP signalling in bacteria: post-translational regulation and catalytic activity. Nucleic Acids Res.38, 5970–5981. doi: 10.1093/nar/gkq382
47
ShresthaP.RazviA.FungB. L.EichingerS. J.VisickK. L. (2022). Mutational analysis of Vibrio fischeri c-di-GMP-modulating genes reveals complex regulation of motility. J. Bacteriol.204:e0010922. doi: 10.1128/jb.00109-22
48
SperlingL.Mulero AlegriaM. D.KaeverV.CurtisP. D. (2019). Analysis of Brevundimonas subvibrioides developmental signaling systems reveals inconsistencies between phenotypes and c-di-GMP levels. J. Bacteriol.201:e00447-19. doi: 10.1128/jb.00447-19
49
SrivastavaD.HsiehM. L.KhataokarA.NeiditchM. B.WatersC. M. (2013). Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol. Microbiol.90, 1262–1276. doi: 10.1111/mmi.12432
50
TeschlerJ. K.Zamorano-SanchezD.UtadaA. S.WarnerC. J.WongG. C.LiningtonR. G.et al. (2015). Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat. Rev. Microbiol.13, 255–268. doi: 10.1038/nrmicro3433
51
TischlerA. D.CamilliA. (2004). Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol.53, 857–869. doi: 10.1111/j.1365-2958.2004.04155.x
52
TownsleyL.YildizF. H. (2015). Temperature affects c-di-GMP signalling and biofilm formation in Vibrio cholerae. Environ. Microbiol.17, 4290–4305. doi: 10.1111/1462-2920.12799
53
ValentiniM.FillouxA. (2019). Multiple roles of c-di-GMP signaling in bacterial pathogenesis. Annu. Rev. Microbiol.73, 387–406. doi: 10.1146/annurev-micro-020518-115555
54
WangH.AyalaJ. C.BenitezJ. A.SilvaA. J. (2014). The LuxR-type regulator VpsT negatively controls the transcription of rpoS, encoding the general stress response regulator, in Vibrio cholerae biofilms. J. Bacteriol.196, 1020–1030. doi: 10.1128/JB.00993-13
55
WebsterS. S.LeeC. K.SchmidtW. C.WongG. C. L.O'TooleG. A. (2021). Interaction between the type 4 pili machinery and a diguanylate cyclase fine-tune c-di-GMP levels during early biofilm formation. Proc. Natl. Acad. Sci.118:e2105566118. doi: 10.1073/pnas.2105566118
56
WilkschJ. J.YangJ.ClementsA.GabbeJ. L.ShortK. R.CaoH.et al. (2011). MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog.7:e1002204. doi: 10.1371/journal.ppat.1002204
57
WuD. C.Zamorano-SanchezD.PagliaiF. A.ParkJ. H.FloydK. A.LeeC. K.et al. (2020). Reciprocal c-di-GMP signaling: incomplete flagellum biogenesis triggers c-di-GMP signaling pathways that promote biofilm formation. PLoS Genet.16:e1008703. doi: 10.1371/journal.pgen.1008703
58
XiaoY.LiangQ.HeM.WuN.NieL.ChenW.et al. (2022). Second messenger c-di-GMP modulates exopolysaccharide pea-dependent phenotypes via regulation of eppA expression in Pseudomonas putida. Appl. Environ. Microbiol.88:e0227021. doi: 10.1128/aem.02270-21
59
XieZ. Y.GongX. X.XuX. D.MeiB.XuanX. Z.LongH.et al. (2020). Identification of Vibrio alginolyticus virulent strain-specific DNA regions by suppression subtractive hybridization and PCR. J. Appl. Microbiol.129, 1472–1485. doi: 10.1111/jam.14739
60
XieZ. Y.HuC. Q.ChenC.ZhangL. P.RenC. H. (2005). Investigation of seven Vibrio virulence genes among Vibrio alginolyticus and Vibrio parahaemolyticus strains from the coastal mariculture systems in Guangdong. China. Lett. Appl. Microbiol.41, 202–207. doi: 10.1111/j.1472-765X.2005.01688.x
61
YildizF. H.DolganovN. A.SchoolnikG. K. (2001). VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol.183, 1716–1726. doi: 10.1128/JB.183.5.1716-1726.2001
62
YildizF. H.VisickK. L. (2009). Vibrio biofilms: so much the same yet so different. Trends Microbiol.17, 109–118. doi: 10.1016/j.tim.2008.12.004
63
YinW. L.XieZ. Y.ZengY. H.ZhangJ.LongH.RenW.et al. (2022). Two (p)ppGpp Synthetase genes, relA and spoT, are involved in regulating cell motility, exopolysaccharides production, and biofilm formation of Vibrio alginolyticus. Front. Microbiol.13:858559. doi: 10.3389/fmicb.2022.858559
64
YinW. L.ZhangN.XuH.GongX. X.LongH.RenW.et al. (2021). Stress adaptation and virulence in Vibrio alginolyticus is mediated by two (p)ppGpp synthetase genes, relA and spoT. Microbiol. Res.253:126883. doi: 10.1016/j.micres.2021.126883
65
YuY.TangM.WangY.LiaoM.WangC.RongX.et al. (2023). Virulence and antimicrobial resistance characteristics assessment of Vibrio isolated from shrimp (Penaeus vannamei) breeding system in South China. Ecotoxicol. Environ. Saf.252:114615. doi: 10.1016/j.ecoenv.2023.114615
66
Zamorano-SanchezD.XianW.LeeC. K.SalinasM.ThongsomboonW.CegelskiL.et al. (2019). Functional specialization in Vibrio cholerae diguanylate cyclases: distinct modes of motility suppression and c-di-GMP production. mBio10:e00670-19. doi: 10.1128/mBio.01960-20
Summary
Keywords
Vibrio alginolyticus, c-di-GMP, rugose, biofilm, extracellular polysaccharide
Citation
Gong X-X, Zeng Y-H, Chen H-M, Zhang N, Han Y, Long H and Xie Z-Y (2023) Bioinformatic and functional characterization of cyclic-di-GMP metabolic proteins in Vibrio alginolyticus unveils key diguanylate cyclases controlling multiple biofilm-associated phenotypes. Front. Microbiol. 14:1258415. doi: 10.3389/fmicb.2023.1258415
Received
14 July 2023
Accepted
05 September 2023
Published
21 September 2023
Volume
14 - 2023
Edited by
Haike Antelmann, Freie Universität Berlin, Germany
Reviewed by
Yiquan Zhang, Affiliated Nantong Hospital 3 of Nantong University, China; Sumit Biswas, Birla Institute of Technology and Science, India; Ute Römling, Karolinska Institutet (KI), Sweden
Updates
Copyright
© 2023 Gong, Zeng, Chen, Zhang, Han, Long and Xie.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen-Yu Xie, xiezyscuta@163.com
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.