
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 07 September 2023
Sec. Microbe and Virus Interactions with Plants
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1257355
 Jia-Xin Lin1
Jia-Xin Lin1 Ahmad Ali1
Ahmad Ali1 Na Chu1
Na Chu1 Hua-Ying Fu1
Hua-Ying Fu1 Mei-Ting Huang1
Mei-Ting Huang1 Sylvain Ntambo Mbuya2
Sylvain Ntambo Mbuya2 San-Ji Gao1*
San-Ji Gao1* Hui-Li Zhang1*
Hui-Li Zhang1*Auxin response factor (ARF) is a critical regulator in the auxin signaling pathway, involved in a variety of plant biological processes. Here, gene members of 24 SpapARFs and 39 SpnpARFs were identified in two genomes of Saccharum spontaneum clones AP85-441 and Np-X, respectively. Phylogenetic analysis showed that all ARF genes were clustered into four clades, which is identical to those ARF genes in maize (Zea mays) and sorghum (Sorghum bicolor). The gene structure and domain composition of this ARF family are conserved to a large degree across plant species. The SpapARF and SpnpARF genes were unevenly distributed on chromosomes 1–8 and 1–10 in the two genomes of AP85-441 and Np-X, respectively. Segmental duplication events may also contribute to this gene family expansion in S. spontaneum. The post-transcriptional regulation of ARF genes likely involves sugarcane against various stressors through a miRNA-medicated pathway. Expression levels of six representative ShARF genes were analyzed by qRT-PCR assays on two sugarcane cultivars [LCP85-384 (resistant to leaf scald) and ROC20 (susceptible to leaf scald)] triggered by Acidovorax avenae subsp. avenae (Aaa) and Xanthomonas albilineans (Xa) infections and salicylic acid (SA) treatment. ShARF04 functioned as a positive regulator under Xa and Aaa stress, whereas it was a negative regulator under SA treatment. ShARF07/17 genes played positive roles against both pathogenic bacteria and SA stresses. Additionally, ShARF22 was negatively modulated by Xa and Aaa stimuli in both cultivars, particularly LCP85-384. These findings imply that sugarcane ARFs exhibit functional redundancy and divergence against stressful conditions. This work lays the foundation for further research on ARF gene functions in sugarcane against diverse environmental stressors.
Auxin is one of the crucial plant hormones participating in various biological processes such as plant growth and development as well as stress responses (Yu et al., 2022). Auxin response factors (ARFs), which are a key component of auxin signaling, function as an important transcription factor (TF) by binding directly to the auxin response elements (AuxREs: TGTCNN) in the promoters of primary/early auxin response genes (Yu et al., 2022; Rienstra et al., 2023). Most ARF proteins contain the N-terminal B3 type DNA-binding domain (DBD), the carboxyl (C)-terminal dimerization domain (CTD), and the non-conserved middle region (MR) located between the DBD and CTD (Song et al., 2023). The DBD of an ARF binds specifically to AuxREs in promoters of targeted genes to activate or repress transcriptional expression, while variations in the two last nucleotides of AuxREs (TGTCAT, TGTCAC, and TGTCGG) affect the interaction between ARFs and AuxREs and thus fine-tune the transcriptional response profile of downstream genes (Yu et al., 2022; Rienstra et al., 2023; Song et al., 2023). The CTD of an ARF includes the Phox and Bem1p (PB1) regions for dimerization, which is like the III and IV elements of Aux/IAA, facilitating protein–protein interaction through homodimerization of ARF proteins or heterodimerization of ARF and Aux/IAA proteins (Li et al., 2023). The MR of an ARF with variable sequence length and composition functions as an activation domain (AD) or repression domain (RD) (Li et al., 2023; Song et al., 2023).
The ARF gene family has been examined in several plant taxa, including flowering plants, bryophytes, and lycophytes, but ARF gene numbers are highly variable among plant species, ranging from 3 genes in Marchantia polymorpha to 51 genes in Glycine max (Chandler, 2016; Li et al., 2016, 2023). Recently, a total of 112 ARFs were identified in five genomes of orchid species, namely, Apostasia shenzhenica (17 AsARFs), Dendrobium catenatum (21 DcARFs), Phalaenopsis aphrodite (16 PaARFs), Phalaenopsis equestris (34 PeARFs), and Vanilla planifolia (24 VpARFs) (Bai et al., 2023). Sixty-seven TaARFs were identified in bread wheat (Triticum aestivum), distributed across six clades (Chaudhary et al., 2023). In addition to plant growth and development, increasing studies have shown that ARF family genes play a significant role in biotic and abiotic stress responses in many plants (Bouzroud et al., 2018; Li et al., 2023; Song et al., 2023).
Numerous ARF family genes are involved in plant defense responses to diverse environmental stresses. For example, transcript levels of most SlARF genes were deceased in tomato (Solanum lycopersicum) leaves exposed for 6 h to Pseudomonas strains including P. putida, P. fluorescens, or P. syringae DC3000 (Bouzroud et al., 2018). Rice OsARF17 played a key role in plant defense against different types of plant viruses such as fijiviruses, tenuivirus, and cytorhabdovirus through these viral proteins manipulating OsARF17 in distinct modes (Zhang et al., 2020). When leaves of two wheat cultivars with contrasting resistance to rust were infected with Puccinia triticina, distinct expression patterns of five ARF genes (TaARF4/6/12/16/27) demonstrated their opposite roles in incompatible (R-Pathogen) and compatible (S-Pathogen) interactions in response to this pathogen infection (Chandra et al., 2020). BdARF4 and BdARF8 played positive roles in response to salicylic acid (SA) and heavy metals in Brachypodium distachyon (Liu N. et al., 2018). S1ARF8A and S1ARF10A as well as their specific miRNA were involved in the regulation of salt and drought responses (Bouzroud et al., 2018), while SlARF4 was regulated by osmotic and salt stress in tomato (S. lycopersicum) (Bouzroud et al., 2020). The expression levels of TaARF8, TaARF9, and TaARF21 were significantly altered by low temperatures in wheat (T. aestivum) (Xu et al., 2020). AcARF1a and AcARF10 were negatively responsive to SA stress in kiwifruit (Actinidia chinensis) (Su et al., 2021). Three ClARF genes (ClARF4/8/12) were negatively regulated in response to abscisic acid (ABA) and SA treatments, while ClARF20 was positively regulated under Indole-3-acetic acid (IAA) and SA treatments (Zhai et al., 2023).
Modern sugarcane cultivars (Saccharum spp. hybrid, 2n = 100–130) are highly polyploids and aneuploids derived from interspecific hybridization between S. officinarum (2n = 8x = 80) and S. spontaneum (2n = 4x–16x = 40–128) (Ali et al., 2019; Zhang et al., 2022). Moreover, S. spontaneum exhibits three basic chromosome numbers (x = 8, 9, or 10) (Zhang et al., 2022). The high variation in ploidy levels of S. spontaneum genomes contributes to the study of the evolution of autopolyploidy genomes in plants (Piperidis and D’Hont, 2020; Zhang et al., 2022). The two tetraploid genomes of S. spontaneum clones AP85-441 (1n = 4x = 32, x = 8) and Np-X (2n = 4x = 40, x = 10) have been published (Zhang et al., 2018, 2022), which provides a powerful resource to identify many gene families. Our previous studies revealed that the SA-mediated signaling pathways played essential roles in sugarcane resistance to infection by Xanthomonas albilineans (Xa) causing leaf scald (Zhao et al., 2022) and Acidovorax avenae subsp. avenae (Aaa) causing red stripe (Chu et al., 2022; Zhao et al., 2023). However, the genome-wide characterization of ARFs and their expression profiles in sugarcane in response to biotic and abiotic stimuli remains unclear. Thus, the objective of this study was to systematically characterize ARF family genes from the polyploid S. spontaneum and their transcript expression profiles in sugarcane cultivars under Aaa and Xa infections as well as SA treatment, which provides an important clue for the subsequent functional identification of ARFs in sugarcane.
The genome sequences for S. spontaneum AP85-441 were obtained from The Ming Laboratory1 and those for S. spontaneum Np-X were obtained from the Zhang Laboratory.2 The BLASTp algorithm was used to search for the genes of SpapARF from AP85-441 and SpnpARF from Np-X and to verify the highest number of genes. The 35 maize ARF gene sequences (Liu et al., 2011) were acquired from the Maize Genome database3 and used as bait in a BLASTp search (score value: ≥ 100 and e-value ≤1 × 10−5). The Pfam database, which is accessible online at http://pfam.sanger.ac.uk/ was searched for the Hidden Markov Model (HMM) file corresponding to the ARF domain (PF06507). HMMER3.0 was used to retrieve ARF genes from the S. spontaneum clones AP85-441 and Np-X genome databases. False sequences were manually eliminated. The sequences of candidate genes were further confirmed by finding their functional domains through the CDD tool in NCBI,4 Pfam,5 and SMART.6 Based on more than 90% resemblance and a high bit score with identified ARF genes, these SpapARF and SpnpARF genes were further validated in two transcriptome datasets (Bio-project numbers: PRJNA579959 and PRJNA549590).
The ExPASy Server7 was used to determine the properties of the inferred ARF proteins, such as isoelectric points. The Sequence Manipulation Suite8 was used to calculate molecular weights, and the WoLF PSORT server9 was used to predict subcellular localization. The orthologous ARF proteins in S. spontaneum clones (AP85-441 and Np-X), Z. mays, and S. bicolor were discovered through orthovenn210 based on the whole genome protein sequences of these species.
The coding sequences (CDS) and protein sequences of identified ARF genes were utilized to construct the gene structure (exon-intron distribution) using the online GSDS.11 Protein sequences of ARFs were used in MEME Suite12 with default parameters to identify the conserved motifs and then visualized by TBtools v1.120 (Chen et al., 2020).
Multiple sequence alignments of all identified ARF proteins were performed by the ClustalW algorithm implemented in the MEGA6.0 program with default parameters (Kumar et al., 2018). The reference protein sequences of ARF gene family were downloaded from Z. mays (Zm)13 (Liu et al., 2011; Xing et al., 2011), Fagopyrum tataricum (Ft) (see text footnote 3) (Liu M. et al., 2018), and S. bicolor (Sb)14 (Chen et al., 2019) genome databases. The neighbor-joining method in MEGA6.0 software was used for phylogenetic analysis, and bootstrap analysis with 1,000 replicates was used to determine statistical reliability. The promoter sequences (1.5 kb upstream of the start codon) of SpapARFs and SpnpARFs were scanned for the available cis-elements at PlantCARE.15
The chromosomal location of each ARF family gene was retrieved from the general feature format 3 (gff3) file, and the chromosome number, start position, and end position of each gene were extracted and visualized using Tbtools v1.120 (Chen et al., 2020). We defined the gene duplication in accordance with the following criteria: (Ali et al., 2020) the alignment length covered >90% of the longer gene; (Ali et al., 2019) the aligned region had an identity >90%; (Ali et al., 2022) only one duplication event was counted for tightly linked genes. Comparative synteny analysis was performed to observe the evolutionary genome conservations amongst S. spontaneum clones (AP85-441 and Np-X), Z. mays, and S. bicolor. All these genomes and GFF3 files were scanned using Super-fast McScanX in TBtools, and the generated files were utilized for multiple synteny maps. The duplicated genes were investigated using an MCScanX run on each of the entire S. spontaneum genomes.
To understand the underlying regulatory mechanism of miRNAs involved in the regulation of SpapARF and SpnpARF genes, their CDS sequences were utilized to predict miRNA-targeted sites using the psRNATarget database (available at https://www.zhaolab.org) with the default parameters. Cytoscape software v.3.8.2 (available at https://cytoscape.org/) was operated to generate the schematic diagram depicting the interaction networks between miRNAs and targeted ARF genes.
The published RNA-sequencing (RNA-seq) datasets by our research group were used to examine the expression patterns of identified ARF genes. One transcriptome dataset (PRJNA579959) was derived from sugarcane cultivars, ROC22 (resistant to red stipe) vs. MT11-610 (susceptible to red stipe) (Chu et al., 2020), and another transcriptome dataset (PRJNA549590) was derived from LCP 85–384 (resistant to leaf scald) vs. ROC20 (susceptible to leaf scald) (Ntambo et al., 2019) under pathogenic bacteria Aaa and Xa infection, respectively. The FPKM values were calculated for entire ARF genes, and the log2(Fold Change) transformed values of these ARFs were used to generate and display heatmap using TBtools and the Heatmapper tool.16 The parameters of |log2(Fold Change)| > 1.0 and value of p <0.05 were set as the threshold for significantly differential expression.
Two sugarcane cultivars, LCP85-384 and ROC20, were used for stress treatments. For biotic stress, the two bacterial species, Aaa strain CNGX08 and Xa strain Xa-FJ1, were used to inoculate sugarcane young plants and sampled following the protocols developed by Zhao et al. (2023) and Ntambo et al. (2019), respectively. For SA treatment, the protocol developed by Ali et al. (2020) was used. The Trizol® reagent (Invitrogen, United States) was used to extract total RNA from leaf samples. Recombinant DNase-I (Takara, Dalian, China) was used to eliminate any remaining genomic DNA before further qRT-PCR assays. The first-strand cDNA synthesis kit from Takara (China) was used to synthesize cDNA in accordance with the manufacturer’s instructions. The qRT-PCR was carried out following the protocol developed by Ali et al. (2020). The qPCR program consisted of denaturation at 95°C for 30 s, followed by 40 cycles of 5 s at 95°C and 30 s at 60°C. Three biological and three technical replicates for each sample were performed. Primers for six candidate ARF genes (Supplementary Table S1) were designed using the GeneScript®.17 The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a reference gene and the 2-ΔΔCt quantitative method was applied to determine the relative quantitative mRNA profiling.
An analysis of variance (One-way ANOVA) was utilized for different gene expression levels at each time point. Data were gained from three biological replicates, and each biological replicate contains three technical replicates. The least significant difference (LSD) test was applied to analyze mean differences at p ≤ 0.05.
Comparative genome analysis was conducted to explore the gene space of S. spontaneum clones AP85-441 and Np-X together with S. bicolor and Z. mays B73 (Supplementary Figure S1). The maximum clusters of 35,866 (123,166 proteins) were recorded in Np-X, followed by 31,332 (131,585 proteins) clusters in Z. mays B73, and then 30,606 (83,826 proteins) clusters in AP85-441 and 25,972 (47,110 proteins) clusters in S. bicolor. Meanwhile, a total of 16,023 orthologous clusters (154,980 proteins) were common in four genomes. In addition, 5,884 clusters (16,654 proteins) were found to be unique to two S. spontaneum genomes, while 601 clusters (2,120 proteins) were identified as specific to both Z. mays and S. bicolor species. The higher number of 28,006–31,487 singletons was observed in the AP85-441 and Np-X genomes, while the lower number of 8,283 singletons was found in the Np-X genome. Furthermore, orthologous clusters among ARF gene families in four genomes were investigated (Figure 1). Overall, 17 (40 proteins), 17 (24 proteins), 21 (25 proteins), and 20 (35 proteins) orthologous ARF clusters were found in Np-X, AP85-441, S. bicolor, and Z. mays B73, respectively. Of them, a total of 16 clusters (101 proteins) of ARFs were exhibited in all four genomes, while one cluster (four proteins) was shared by two S. spontaneum genomes and the S. bicolor genome. One and seven singletons were found in S. bicolor and Z. mays B73, respectively, but none of the singletons were found in both S. spontaneum genomes.
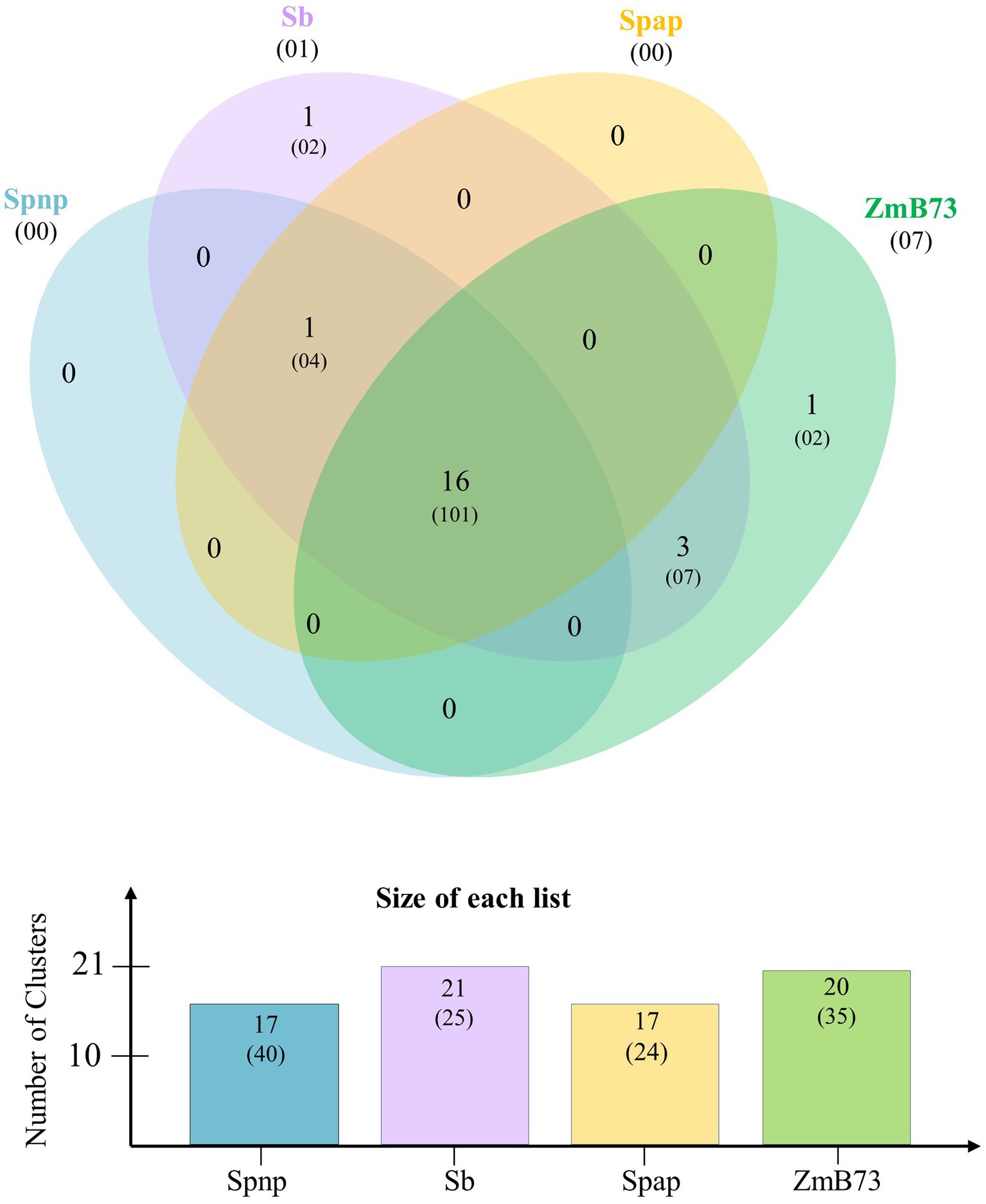
Figure 1. Orthologous ARF family gene clusters among S. spontaneum AP85-441 (Spap), S. spontaneum Np-X (Spnp), S. bicolor (Sb), and Z. mays B73 (ZmB73). The number in each sector of the diagram indicates the number of homologous clusters and the numbers in parentheses indicate the total number of genes contained within the associated clusters. The numbers in parentheses below the species names indicate the number of species-specific singletons (genes with no homologs).
A total of 24 SpapARF and 39 SpnpARF sequences were found in two genomes of S. spontaneum, AP85-441 and Np-X, respectively. Their gene characteristics were predicted, including the length of the coding sequence (CDS), the predicted molecular weight (MW) of the protein, the isoelectric point (pI), and subcellular localization (Supplementary Table S2). For SpapARF proteins, the length of proteins ranged from 583 (SpapARF07) to 1904 (SpapARF18) amino acids (aa) and the predicted MW of both proteins ranged from 65.11 kDa to 211.59 kDa. The pI ranged from 5.28 (SpapARF07) to 8.30 (SpapARF13); the number of exons varied from 3 (SpapARF04/15) to 20 (SpapARF18); all SpapARFs except SpapARF09 were predicted to be located at the nucleus. For SpnpARF proteins, the length of proteins ranged from 660 (SpnpARF14.1) to 1,647 (SpnpARF3); the predicted Mw ranged from 180.92 (SpnpARF3) to 73.24 (SpnpARF6.1/7); the pI ranged from 8.8 (SpnpARF6.6) to 5.83 (SpnpARF14.1); the exon number of SpnpARFs varied from 3 (SpnpARF3/4.1/15.2/17) to 17 (SpnpARF22.2). Of the 39 SpnpARFs, 34 genes were predicted to be located at the nucleus, while four genes were located at the chloroplast, and one gene was located at the nucleus and chloroplast.
The allelic information of ARF genes in two S. spontaneum genomes is shown in Supplementary Table S3. A total of 79 and 176 alleles were predicted in SpapARFs and SpnpARFs, respectively. The highest number of six alleles occurred in SpapARF16, while the lowest number of one allele/gene was found in SpapARF12/19/20/21. Additionally, the highest number of 15 alleles was exhibited in SpnpARF22.2 while the lowest number of one allele/gene was found in SpnpARF1.1/18/19.
A phylogenetic tree was constructed using 143 ARF protein sequences, including AP85-441 (24 SpapARFs), Np-X (39 SpnpARFs), S. bicolor (25 SbARFs), Z. mays (35 ZmARFs), and F. tataricum (20 FtARFs) (Figure 2). Based on the ARF classification in maize, all ARFs were divided into four groups (I-IV). The maximum members occurred in Group-I, comprising 56 ARF members (10 SpapARFs, 14 SpnpARFs, 11 SbARFs, 14 ZmARFs, and 7 FtARFs), followed by Group-IV with 37 ARF members (7 SpapARFs, 11 SpnpARFs, 5 SbARFs, 9 ZmARFs, and 5 FtARFs), and then Group-II and Group-III, containing 23 and 27 ARF members, respectively. Furthermore, another phylogenetic tree was constructed with ARF members from two S. spontaneum genomes (Supplementary Figure S2). All SpapARFs and SpnpARFs were distributed in four Groups (A-D), i.e., 25 genes (10 SpapARFs and 15 SpnpARFs) in Group-A, 18 genes (7 SpapARFs and 11 SpnpARFs) in Group-B, 8 genes (3 SpapARFs and 5 SpnpARFs) in Group-C, and 12 genes (4 SpapARFs and 8 SpnpARFs) in Group-D.
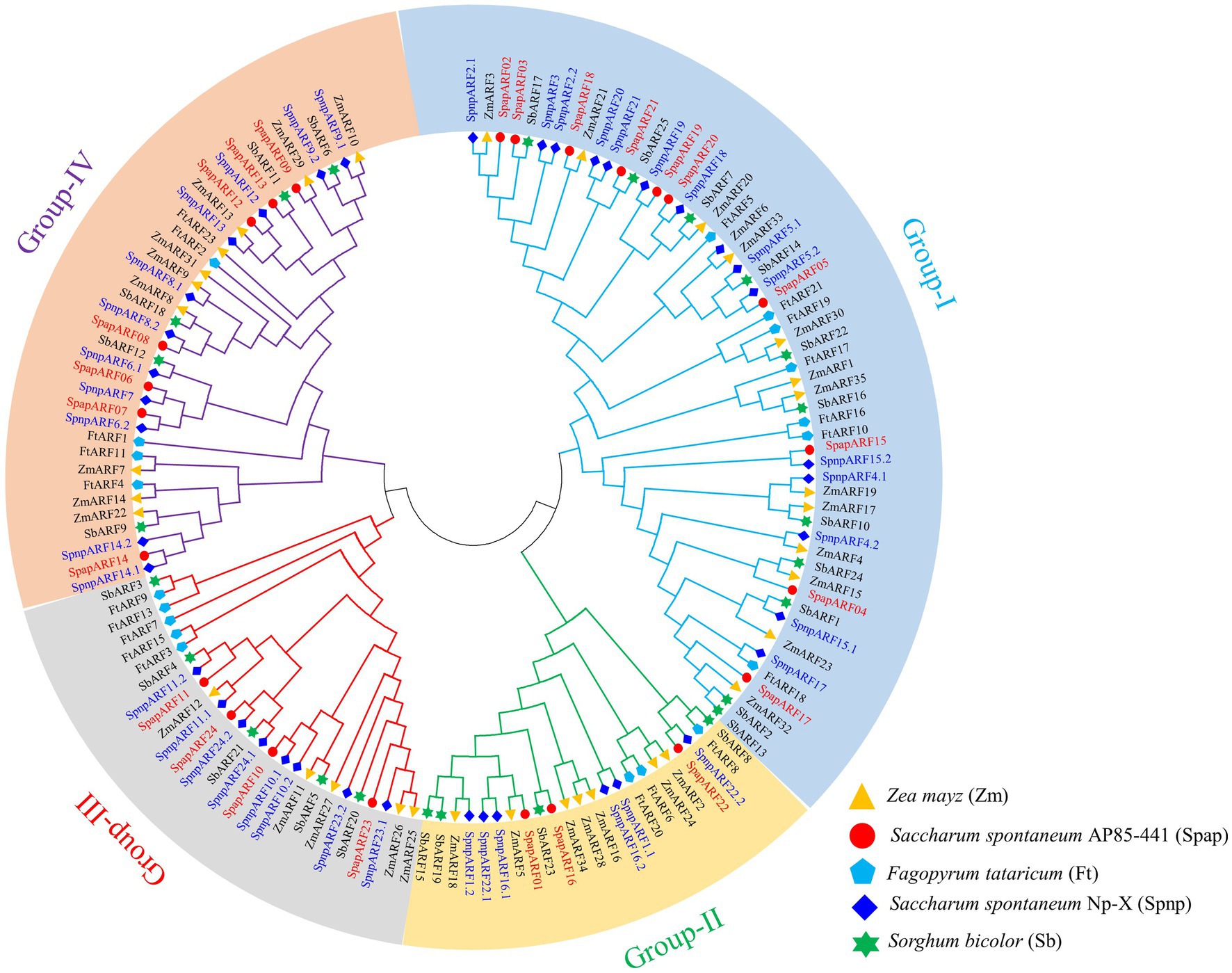
Figure 2. Phylogenetic tree of ARF family genes in S. spontaneum AP85-441 (Spap) and Np-X (Spnp), Sorghum bicolor (Sb), Fagopyrum tataricum (Ft), and Zea mays (Zm). This tree was constructed with the neighbor-joining method and 1,000 bootstraps implemented in the MEGA 6.0 software. Four phylogenetic Groups (I, II, III, and IV) are marked with different colored backgrounds and lines. SpapARF and SpnpARF genes are marked red and blue colors, respectively.
The gene structure and motif pattern of SpapARFs and SpnpARFs were examined (Figure 3). All of them had three characteristic regions: highly conserved DBD (Pfam02362) at the N-terminal, ‘Auxin-rep’ known as the ARF domain (Pfam06507) at the middle region, and the Aux/IAA domains (CTD; Pf02309) at C-terminal (Figure 3A; Supplementary Figure S3). The DBD and “Auxin_rep” domains were present in almost all identified SpapARF and SpnpARF proteins. However, the CTD domain was absent in 20 SpapARF and SpnpARF proteins, most of which belonged to Clade-III and Clade-IV; The DBD domain was lost in five SpapARF and SpnpARF proteins, distributing in Clade-I (Ali et al., 2019), Clade-II (Ali et al., 2019), and Clade-IV (Ali et al., 2020). Notably, SpapARF07 (Clade-II) and SpapARF17 (Clade-III) had two CTD domains (Figure 2A).
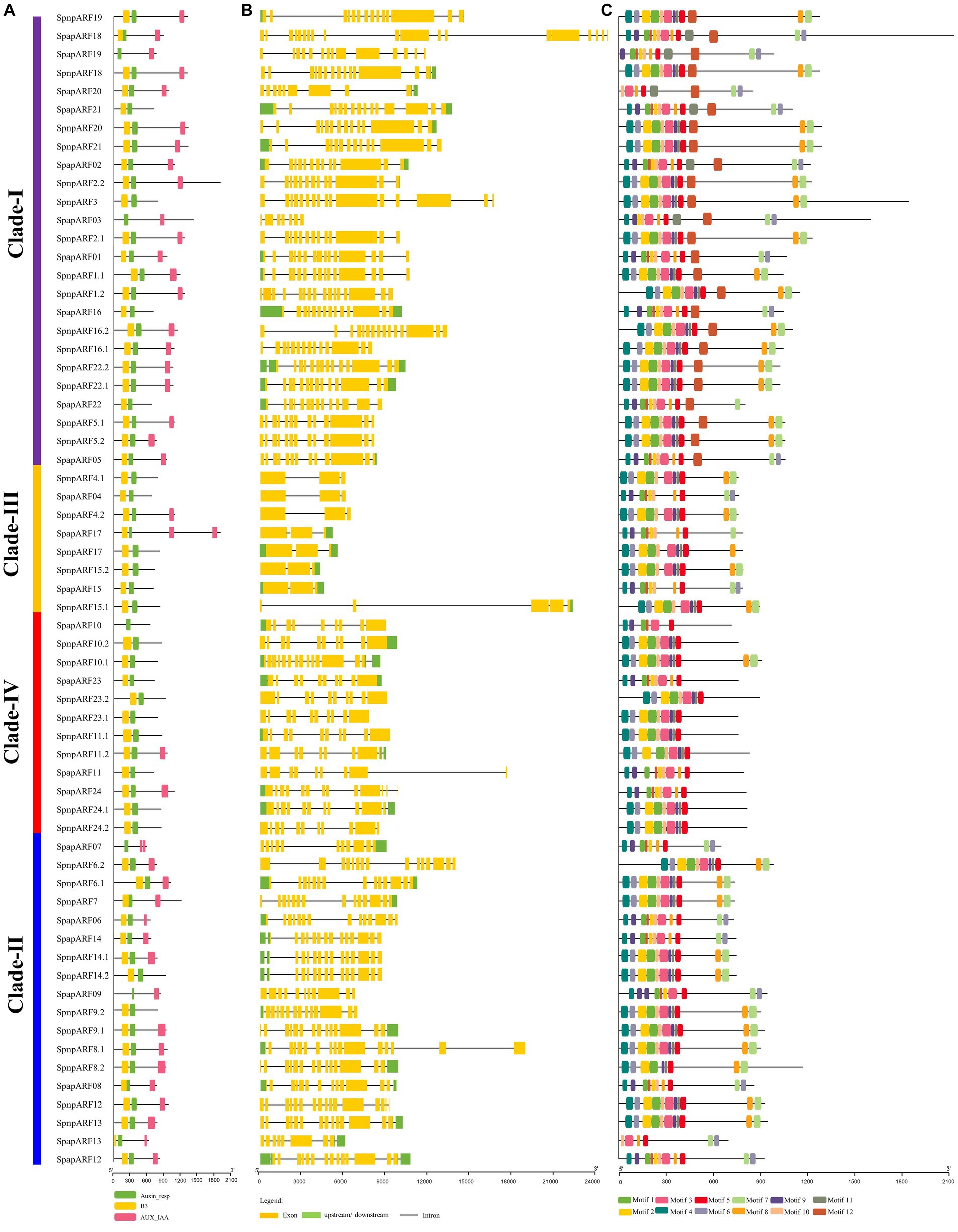
Figure 3. Conserved domains (A), exon-intron structure (B), and conserved motifs (C) of ARF genes in S. spontaneum AP85-441 (Spap) and Np-X (Spnp). (A) The Clades (I/II/III/IV) of ARF genes are identical to those phylogenetic groups (A, B, C, and D) in Supplementary Figure S2, respectively. The functional domain was obtained from model gene annotation and results of the NCBI CDD-batch search. B3: B3 DNA-binding domain; Auxin-resp: ARF domain; AUX_IAA: C-terminal dimerization domain. (B) The exons and introns in each gene are represented by lines and boxes, respectively. The sizes of exons, introns, and untranslated regions are drawn on the bottom. (C) Conserved motifs of SpapARF and SpnpARF proteins were identified using MEME (suite 4.11.4). Each motif is indicated with a colored box numbered 1 to 12 at the bottom. Protein length is estimated using the scale (amino acid) at the bottom in (A) and (C) panels, while gene length is estimated using the scale (bp) at the bottom in (B) panel.
Gene structure analysis demonstrated that the exon number in ARF genes of S. spontaneum ranged from three (all genes except for SpnpARF15.1 in Clade-III) to 20 (SpapARF18 in Clade-I; Figure 3B). In Clade-1, most SpnpARFs and SpapARFs contained 10–20 exons, except for seven exons in SpapARF03. SpnpARFs and SpapARFs in Clade-II contained 8–16 exons, while those ARF genes in Clade-IV held 8–15 exons. The majority of ARF genes in Clade-III had only three exons, except for SpnpARF15.1 with five exons. The longest exon structures of SpnpARFs and SpapARFs were found in Clade-1, while the longest intron structure was observed in SpnpARF15.1 (Clade-III). Overall, similar exon/intron numbers and their positions in SpnpARFs and SpapARFs occurred in the same clade. However, variable exon/intron numbers were present in some genes clustered in the same clade. For example, SpnpARF15.1 in Clade-III had four introns (two longest introns) and five exons, whereas the rest of the members only had two introns and three exons.
Conserved motif analysis revealed that 12 conserved motifs (Motifs 1–12) were identified (Figure 3C), namely, Motifs 1, 2, and 10 that were annotated as DBD; Motifs 6, 8, and 12 were CTD; Motifs 3, 5, 9, and 11 were Auxin-rep, and Motifs 4 and 7 belonged to unknown domains. The length of conserved motifs varied from 15 (Motif 11) to 50 (Motifs 1–4 and 12) amino acids. Among the 12 motifs, six motifs (Motifs 1–5 and 10) were commonly found in SpnpARFs and SpapARFs. Most genes in Clade-I contained all 12 motifs, while Motifs 7 and 8 were absent in all genes except SpnpARF10.8 in Clade-IV. However, Motif 7 was specifically present in almost all SpnpARFs and SpapARFs of Clade-I/II/III. Motif 6 was unavailable in most genes in Clade-II/III and IV.
Twenty-two cis-elements responsive to biotic and abiotic stresses were predicted to be present in the 1.5 kb promoter regions of SpapARFs and SpnpARFs (Supplementary Figure S4). These elements were classified into three classes based on their functions (hormone-related, stress-related, and growth and development-related elements). Among them, 18.2% of the elements belonged to the hormone-related class, including ABRE (involved in abscisic acid responsiveness), AE-Box (light-responsive element), TGA-element (auxin-responsive element), and TGACG-motif (involved in MeJA responsiveness); 63.6% elements were related to biotic and abiotic stresses, including ARE (essential for anaerobic induction), W-box (responsive to wounding or pathogens), MYB (stress-responsive), MBS (involved in drought-inducibility), WUN-motif (wound-responsive element), as-1 (oxidative stress-responsive), DRE core (dehydration-responsive), STRE (stress-responsive), and TATA-box (stress-responsive element); 18.2% elements were associated with the development-related class, including CAAT-box (growth-related), GT1-motif (light-responsive), CAT-box (related to meristem expression), and G-box (involved in light responsiveness). Notably, SpnpARF3 and SpapARF15 had the highest number of 33 and 30 copies of the TATA-box element in their promotor regions, while seven genes (SpapARF06/17/18/19 and SpnpARF10.1/11.1/22.2) possessed all the stress-related elements in their promoter regions. Three elements (MYC, STRE, and TATA-box) were present in all SpapARF and SpnpARF genes, while CAAT-box and MYB-binding sites specifically existed in the promoter regions of SpapARFs and SpnpARFs, respectively. All hormone-related cis-elements were found in the promoters of nine genes (SpapARF02/08/17/18 and SpnpARF1.1/3/6.1/8.2/11.1).
The distribution of SpapARF and SpnpARF genes in two S. spontaneum genomes (Np-X and AP85-441) is shown in Supplementary Figure S5. The majority of SpapARF and SpnpARF genes are located on the proximate or distal ends of the chromosomes. All 24 SpapARF genes were unevenly distributed on 16 chromosomes of AP85-441 (Supplementary Figure S5A), namely, three SpapARFs in chromosome Chr04B, two SpapARFs in six chromosomes (Chr02B, Chr03D, Chr04C, Chr05B, Chr06C, and Chr08C) each, and one SpapARF in nine individual chromosomes (Chr01A, Chr03B, Chr04D, Chr05A, Chr06A/D, Chr07A/C, and Chr08D). However, none of the SpapARF genes were distributed in the remaining 16 chromosomes of AP85-441. Meanwhile, all 39 SpnpARF genes were unequally distributed on 26 chromosomes of Np-X genome (Supplementary Figure S5B), i.e., four SpnpARFs in chromosome Chr04C, two SpnpARFs in 10 chromosomes (Chr03A/B, Chr04B, Chr06C, Chr07A, Chr08A/C, Chr09D, and Chr10A/D) each, and one SpnpARF in 15 chromosomes. However, none of the SpnpARF genes were found in the other 14 chromosomes of Np-X.
Collinearity and gene duplication analyses in all SpapARFs uncovered that 11/24 (45.8%) pairs of SpapARF genes were possibly involved in allelic and non-allelic segmental duplications (Figure 4A). Gene duplication was found in two pairs of non-allelic duplicated genes, including SpapARF15-SpapARF16 on Chr04B-Chr04C and SpapARF22-SpapARF03 on Chr06A-Chr06C. Nine allelic segmental duplications were found on different chromosomes, while seven SpapARF genes (SpapARF04/07/10/14/15/16/17) were duplicated on the unassembled scaffolds. Notably, chromosomes Chr4A/B/C/D were hotspots for gene duplication, as evidenced by four segmentally duplicated pairs. A singular duplication gene pair was identified on chromosomes Chr03A/B and Chr01A/D, while no duplicated gene pair was found on chromosomes Chr2A/B/C/D. Regarding SpnpARF genes in the Np-X genome, 36/39 (92.3%) gene pairs were involved in segmental duplication (Figure 4B). Eleven gene pairs including SpnpARF-SpnpARF24.1/24.2, SpnpARF2.1-SpnpARF2.2-SpnpARF3, SpnpARF12-SpnpARF13, SpnpARF20-SpnpARF21, SpnpARF22.1-SpnpARF16.1, SpnpARF1.1-SpnpARF1.2, SpnpARF15.1-SpnpARF15.2, SpnpARF10.1-SpnpARF23.1-SpnpARF2.2, and SpnpARF4.1-SpnpARF4.2 were duplicated segmentally on various chromosomes. The remaining 25 gene pairs were segmentally duplicated on the unknown scaffold regions of different chromosomes. The most abundance of segmentally duplicated genes was present on chromosome Chr04A/B/C/D, followed by Chr03A/B/C/D and Chr10A/B/D. However, only a single duplication was observed on Chr05A/C. No tandem duplication event was present on both S. spontaneum genomes.
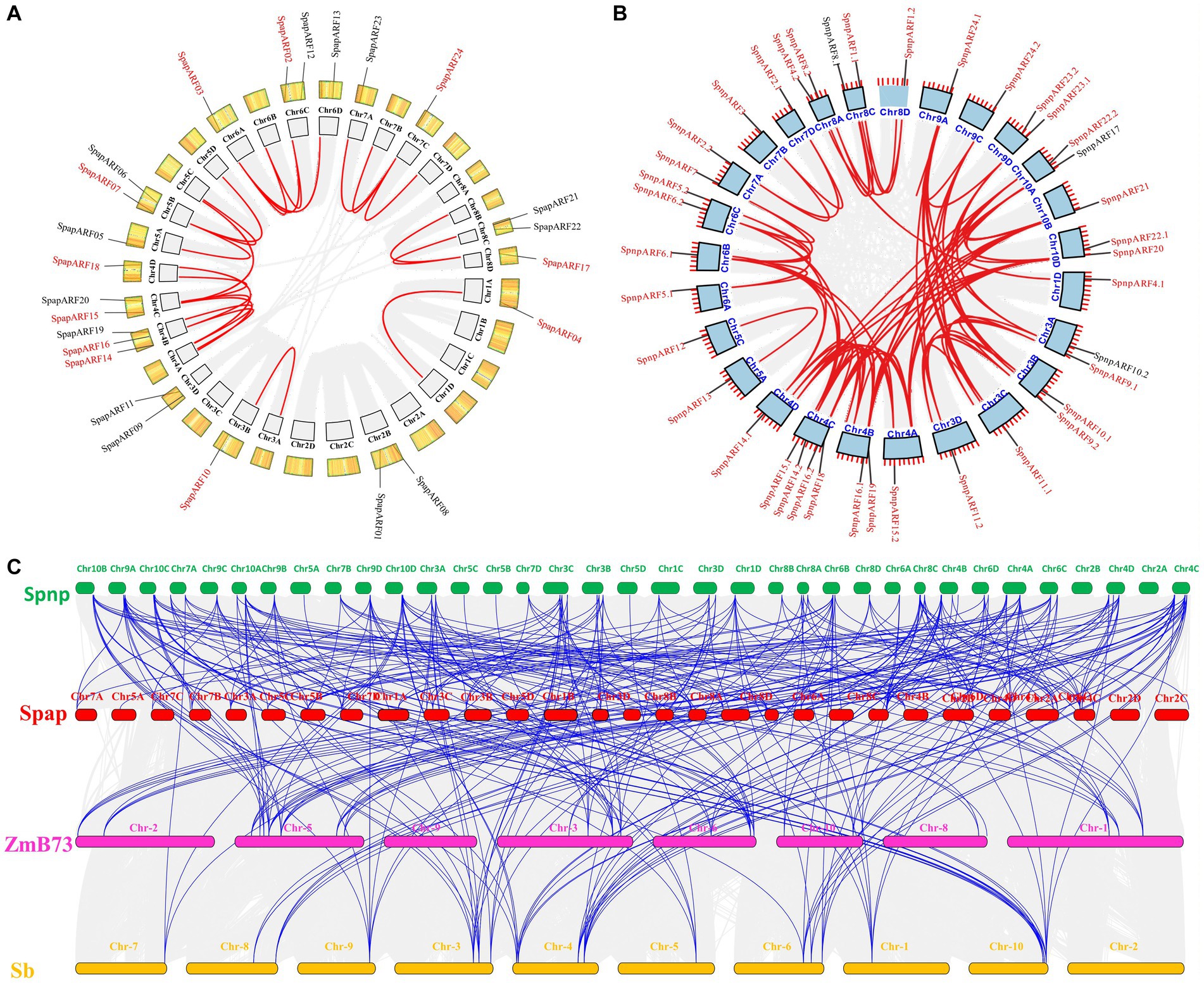
Figure 4. Circos illustration of duplicated ARF genes on chromosomes of S. spontaneum AP85-441 (Spap) (A) and Np-X (Spnp) (B), and multicollinearity analysis of ARF genes in four genomes including two S. spontaneum clones, Z. mays B73 (ZmB73), and S. bicolor (Sb) (C). The grey ribbon indicates a collinear relationship among blocks in the whole genome and the red ribbon shows ARF paralogs in (A) and (B) panels. The chromosomes are arranged in a colored arc. Genes with red color represent segmentally duplicated events. Blue lines represent syntenic ARFs in the (C) panel and gray lines in the background represent all orthologous genes among four genomes.
Comparative synteny analysis among S. spontaneum clones (Np-X and AP85-441), Z. mays B73, and S. bicolor showed that Np-X (SpnpARFs) and AP85-411 (SpapARFs) had remarkable syntenic associations with each other and Z. mays B73. A total of 76 SpnpARFs shared syntenic relationships in the AP85-441 genome, and 72 SpnpARFs had syntenic relationships in the Z. mays B73 genome. In addition, 51 SpnpARFs had syntenic relationships in the S. bicolor genome. Notably, no synthetic relationship was observed between SpnpARF4.2 and SpnpARF10.2 (Figure 4C; Supplementary Table S4).
To better understand the roles of miRNAs in managing the post-transcriptional regulation of ARFs, the CDS regions of 24 SpapARFs and 39 SpnpARFs were examined in the psRNATarget database against the published miRNAs of S. officinarum. A total of 17 genes (7 SpapARFs and 10 SpnpARFs) were targeted by six miRNAs that belong to four different families (Figure 5A; Supplementary Table S5). Two miRNAs, ‘sof-miR167a and sof-miR167b’ targeted nine genes (six SpnpARFs and three SpapARFs). The miRNA ‘sof-miR168a’ targeted nine other genes (four SpnpARFs and five SpapARF). The miRNA ‘sof-miR396’ targeted four SpnpARFs and two SpapARFs. The miRNA ‘sof-miR408e’ targeted two SpnpARFs and one SpapARFs. The miRNA ‘sof-miR168b’ targeted two SpnpARFs. On the other hand, SpnpARF22.1/22.2 were targeted by four miRNAs each, while SpnpARF16.1/16.2 and SpapARF22 were targeted by three miRNAs each. Four genes, SpnpARF18, SpnpARF19, SpapARF19, and SpapARF20, were targeted by only one miRNA (sof-miR168a) (Figure 5A; Supplementary Table S5). The representative miRNA-targeted sites in SpnpARF1.1/16.1/18/22.1 and SpapARF01/16/18/22 are illustrated in Figure 5B.
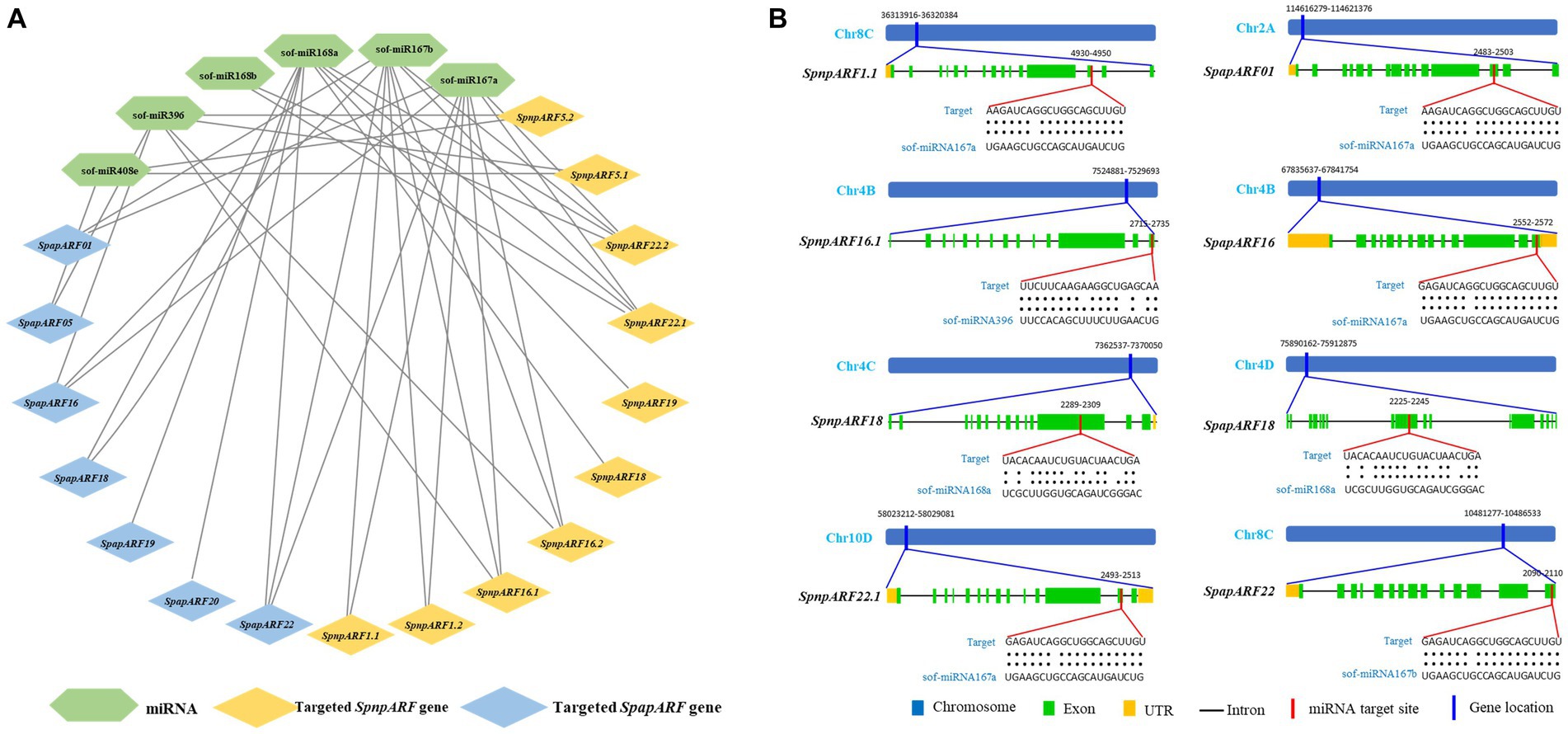
Figure 5. Network map of predicted miRNAs targeting SpapARFs and SpnpARFs (A) and schematic diagram of miRNAs targeting sites in some ARFs (B). (A) Green, blue, and orange boxes stand for miRNAs and their targeted genes of SpapARFs and SpnpARFs, respectively. (B) The thick blue bar indicates the location of the ARF gene harbored at the specific chromosome. The thick red bar indicates the location of the miRNA targeted at the gene sequence. The RNA sequence of each complementary site from 5′ to 3′ and the predicted miRNA sequence from 3′ to 5′ are shown with red lines below the gene sequences. All miRNAs predicted to target SpapARF and SpnpARF genes are shown in Supplementary Table S5. Sof: S. officinarum.
The published transcriptome dataset of MT11-610 vs. ROC22 inoculated by Aaa was retrieved and used to access the transcript profiles of SpapARF and SpnpARF genes (Figure 6A; Supplementary Table S6). Upon Aaa infection, RNA-seq data showed that eight SpapARF genes (SpapARF04/10/11/23 and SpnpARF4.1/10.1/11.1/23.1) were upregulated in ROC22 (resistant to red stripe), while they were depressed in MT11-610 (susceptible to red stripe) across all time points. Transcript expression levels [log2(Fold Change)] of SpapARF21 and SpnpARF21/24.1 were increased in both cultivars across all time points, while their expression levels were higher in ROC22 than in MT11-610 at each time point. Meanwhile, expression levels of SpapARF01/02/07/16/17/19 and SpnpARF1.1/3/16.1/17/18 decreased in both cultivars across all time points. On the other hand, six representative ShARF genes were selected from different phylogenetic groups (ShARF02/04/17 in Group I, and ShARF07/10/22 in Group IV, III, and II, respectively) to analyze transcript levels in two sugarcane cultivars (LCP85-384 and ROC20) infected by Aaa (Figure 6B). Overall, the six genes except for ShARF22 were upregulated to some degree in LCP85-384 at 24–72 h post inoculation (hpi). For example, the expression levels of ShARF02/07/17 were significantly enhanced with increases of 1.2–6.1 folds in LCP85-384 but were decreased or non-significantly changed across all time points, compared with the control group (mock-inoculation at 0 hpi). ShARF04 was positively induced in both cultivars under Aaa stimuli at 24–72 hpi. However, ShARF10 was upregulated in the two cultivars at some time points, while ShARF22 was upregulated only in ROC20 at 72 hpi, compared with the control group.
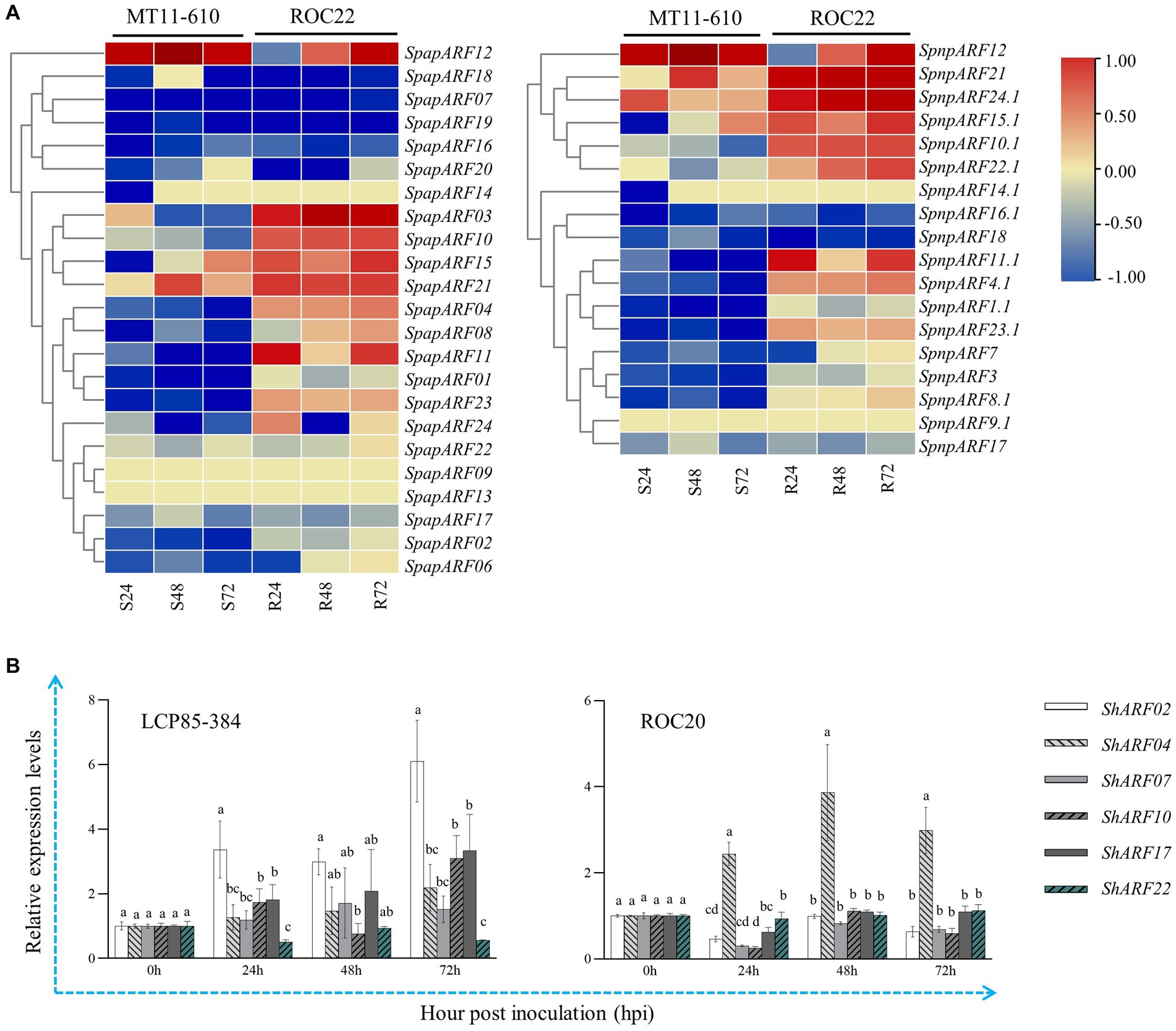
Figure 6. Transcript expression profiles of SpapARF and SpnpARF genes in sugarcane cultivars infected by Acidovorax avenae subsp. avenae (Aaa). (A) Transcriptome dataset of two sugarcane cultivars MT11-610 (S: susceptible to red stripe) and ROC22 (R: resistant to red stripe) triggered by Aaa infection. RNA-seq data were recorded for each cultivar at 0, 24, 48, and 72 h post-inoculation (hpi). Colored boxes in each column represent relative expression levels of individual genes with log2(Fold Change). (B) qRT-PCR-based expression profiling of six candidate ARF genes in sugarcane cultivars (ROC20 and LCP85-384) under Aaa infection. Leaf samples were collected at 0, 24, 48, and 72 hpi. All data are shown as the mean ± standard error (SE).
Another published transcriptome dataset of LCP85-384 vs. ROC20 inoculated with Xa was retrieved and used to explore the transcript profiles of SpapARF and SpnpARF genes (Figure 7A; Supplementary Table S7). Eight genes (SpapARF01/03/20/22 and SpnpARF1.1/2.1/8.2/22.1) were upregulated in both cultivars across all time points, but nine genes (SpapARF02/06/10/14/15 and SpnpARF7/10.1/14.1/15.1) were downregulated (Figure 7A). The transcript levels of SpapARF11/23 and SpnpARF23.1 were increased in LCP85-384, whereas they were decreased in ROC20 in response to Xa infection during 24–72 hpi. Furthermore, the transcript profiles of ShARF02/04/07/10/17/22 were identified by qRT-PCR assay in both cultivars inoculated with Xa (Figure 7B). Compared with mock-inoculation control, transcript levels of ShARF07/17 were obviously increased by 13.5–64.5% in LCP585-384 and 20.7–99.2% in ROC20 at 24–72 hpi. However, ShARF2/10/22 were downregulated in both cultivars across all time points except for a specific time point (24 hpi). ShARF04 was significantly upregulated in LCP85-384 at 72 hpi, whereas it was dramatically depressed in ROC20 across all time points.
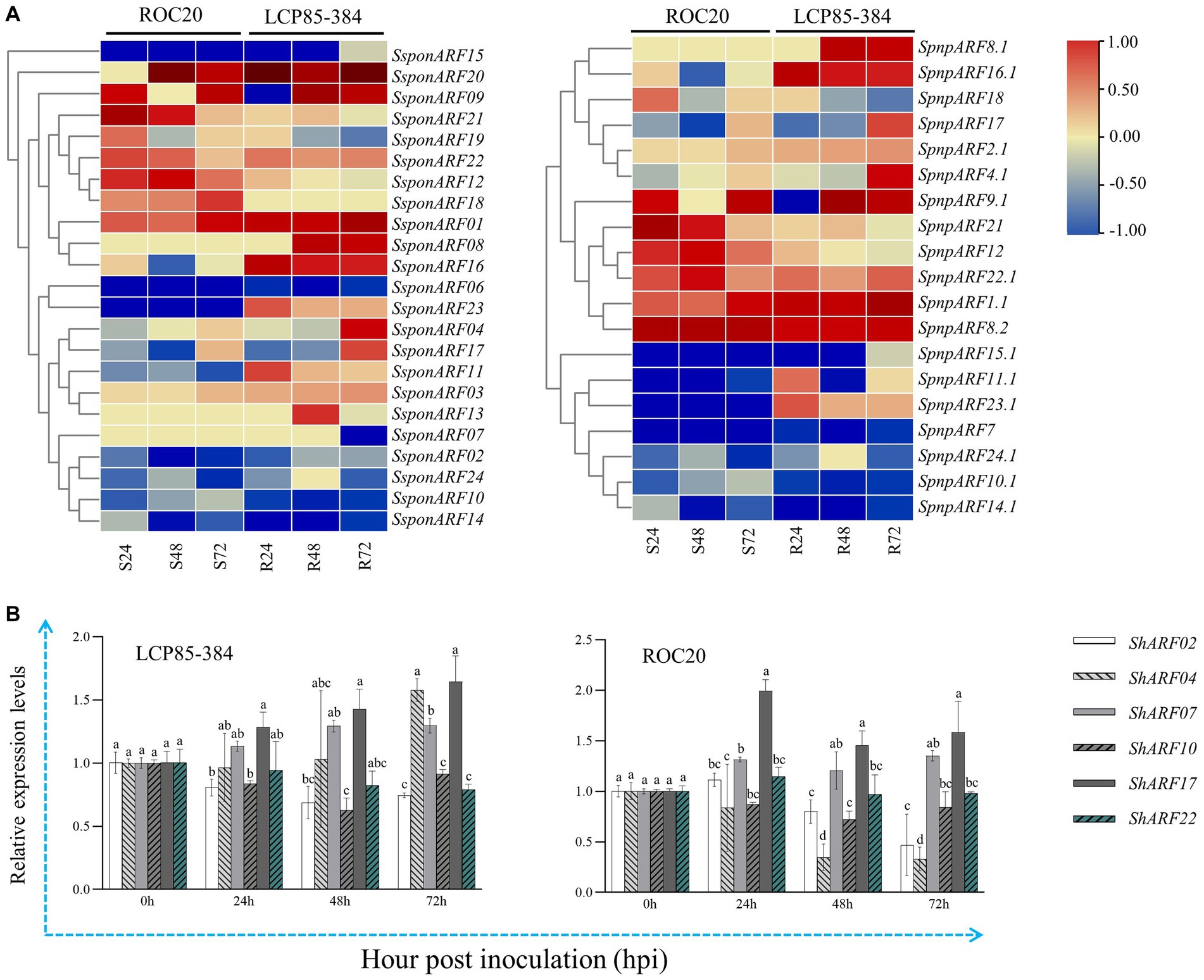
Figure 7. Transcript expression profiles of SpapARF and SpnpARF genes in sugarcane cultivars infected by Xanthomonas albilineans (Xa). (A) Transcriptome dataset of two sugarcane cultivars ROC20 (S: susceptible to leaf scald) and LCP85-384 (R: resistant to leaf scald) triggered by Xa infection. RNA-seq data were recorded for each cultivar at 0, 24, 48, and 72 h post-inoculation (hpi). Colored boxes in each column represent relative expression levels of individual genes with log2(Fold Change). (B) qRT-PCR-based expression profiling of six candidate ARF genes in sugarcane cultivars (ROC20 and LCP85-384) under Xa infection. Leaf samples were collected at 0, 24, 48, and 72 h post treatment (hpt). All data are shown as the mean ± standard error (SE).
Transcript profiles of six genes (ShARF02/04/07/10/17/22) were explored in ROC20 and LCP85-384 under SA treatment based on qRT-PCR assays (Figure 8). Overall, expression levels of six genes except for ShARF04 were increased in both cultivars, particularly at 12 and 24 h post-treatment (hpt). Compared with the mock-treatment (0 hpt), ShARF02/07/17 were upregulated with increases of 1.5–2.5-fold in LCP85-384 and 1.1–2.2-fold in ROC20 at 12–24 hpt. ShARF10 was obviously upregulated in LCP85-384 but non-significantly changed in ROC20 under SA treatment at 12–24 hpt. The expression level of ShARF22 was significantly enhanced in LCP85-384 at 12–24 hpt and in ROC20 at 6–12 hpt. Notably, ShARF04 was significantly decreased in both cultivars across all time points.
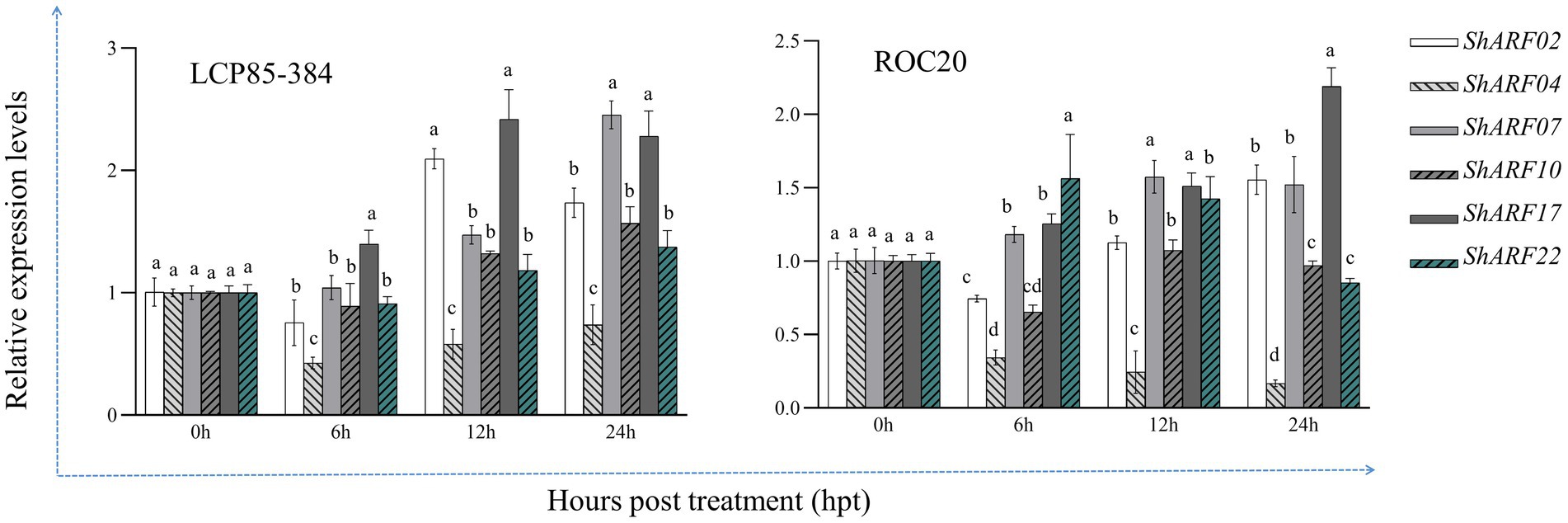
Figure 8. The qRT-PCR-based expression profiling of six candidate ARF genes in two sugarcane cultivars ROC20 (susceptible to leaf scald) and LCP85-384 (resistant to leaf scald) under exogenous salicylic acid (SA) stress. Leaf samples were collected at 0, 24, 48, and 72 h post treatment (hpt). All data are shown as the mean ± standard error (SE).
An increasing number of complete sequences of plant genomes, particularly polyploid crops, is available, providing the opportunity to identify the members of the ARF gene family at the genome-wide level. Herein, we identified 24 SpapARF (79 alleles) and 39 SpnpARFs (176 alleles) family members in two genomes of S. spontaneum clones AP85-441 (genome size; 3.13Gb) (Zhang et al., 2018) and Np-X (genome size; 2.83 Gb) (Zhang et al., 2022). A high variation in the gene numbers of SpapARF and SpnpARFs was observed between two S. spontaneum genomes. This can be explained by the fact that different chromosome numbers and variations in ploidy levels are present in both S. spontaneum clones. AP85-441 is a haploid (1n = 4x = 32, x = 8) generated from a culture of the octoploid SES208 (2n = 8x = 64) (Zhang et al., 2018), whereas Np-X (2n = 4x = 40, x = 10) is a tetraploid (Zhang et al., 2022). The numbers of ARF family genes were obviously varied among genomes of higher plants, such as 23 in A. thaliana (125 Mb) (Okushima et al., 2005), and 25 ARF in O. sativa (450 Mb, 2n = 24) (Wang et al., 2007), 36 in Z. mays (2.3 Gb, 2n = 20) (Wang et al., 2012), 51 in G. max (978.9 Mb, 2n = 40) (Ali et al., 2022), and 67 in T. aestivum (17.0 Gb, 2n = 6x = 42) (Chaudhary et al., 2023). The base number of chromosomes (x), chromosome size, and ploidy level may be primary contributors to genome expansion in plants such as ferns and lycophytes (Wang et al., 2022).
Gene duplication includes whole genome duplication (polyploidization), segmental duplication, and tandem duplication, which is a very important mechanism that increases gene copy number and contributes to the generation of new genes (Goldtzvik et al., 2023). Our findings showed that fragmental duplication events of SpapARFs and SpnpARFs were detected in both genomes, especially in Np-X, but no tandem duplication event occurred, suggesting that segmental duplication events are mainly promoting the expansion of the ARF gene family. Zhai et al. (2023) also proposed that segmental duplication, rather than tandem duplications, contributed to the expansion of the ARF gene family in Coix lacryma-jobi. Additionally, the whole genome duplication may also contribute to this gene expansion in S. spontaneum due to the ploidy level of this plant species, which ranges from tetraploid (4x) to hexaploid (16x) (Zhang et al., 2022). The intron-exon patterns showed that the exon number in each SpapARF and SpnpARF gene greatly varied. Similar phenomena were reported in numerous plants, such as the dicotyledon A. thaliana (Okushima et al., 2005) and the monocotyledon T. aestivum (Chaudhary et al., 2023). These results suggested that the speciation of the ARF gene family may facilitate various biological functions across dicot and monocot plant species.
The miRNAs which are a class of small non-coding RNAs regulate gene expression through post-transcriptional modifications such as mRNA cleavage or translational inhibition, and they play critical roles in plant basic development and defense responses to environmental signals (Raza et al., 2023). In this study, six different miRNAs were predicted to target 17 ARFs (7 SpapARFs and 10 SpnpARFs). For instance, nine ARFs from S. spontaneum were targeted by sof-miR167a, sof-miR167b, and sof-miR168a. It seems that these ARFs could be regulated by sof-miR167 and sof-miR168, which may act as a crucial role in biological processes via miRNA-ARF modules involved in auxin signaling in sugarcane. A previous study showed that tobacco NtMIR167a-NtARF6/8 was critical for tobacco regulating plant growth and inorganic phosphorus (Pi) starvation tolerance (Chen et al., 2018). Additionally, maize Zma-miR167-ZmARF3/30 regulated polyamine oxidase 1 (ZmPAO1) expression and then modulated H2O2 production, conferring resistance to maize chlorotic mottle virus (Liu et al., 2022).
In this study, the temporal expression dynamics of SpapARFs and SpnpARFs were investigated in two transcriptome datasets in response to bacterial infection. Our data showed that some genes shared similar expression profiles, but some genes had different expression profiles under either Aaa or Xa infection, suggesting that sugarcane ARF family genes underwent functional divergence and redundancy in evolution. This phenomenon is frequent in plants, particularly in complex polyploidy crops such as sugarcane (Li et al., 2022). Notably, some ARF genes (such as SpapARF02) showed similar functions in sugarcane defense responses to Aaa and Xa infections, while others (such as SpapARF01) played the opposite role in responses to both bacterial infections. Similar appearance of functional divergence and redundancy was present in tomato SlARFs, as evidenced by transcript expressions of SlARFs in plant leaves exposed for 6 h to six biotic stressors, including flagellin, P. putida, P. syringae, P. fluorescens, Agrobacterium tumefaciens, and yellow curl virus (YCV) Bouzroud et al. (2018). For instance, SlARF8B (homolog of SpapARF01) was upregulated against P. syringae but downregulated under other stressors. SlARF7B (homolog of SpapARF02) was upregulated against YCV, but downregulated or unchanged under other stressors. SlARF16A (homolog of SpnpARF4.1) was downregulated in response to all biotic stress. The transcript level of SlARF3 (homolog of SpapARF10 and SpnpARF10.1) was increased under infections by P. syringae, A. tumefaciens, and YCV, while it was decreased against biotic stresses by P. putida, P. fluorescens, and Flagellin (Bouzroud et al., 2018).
It was noted that there was a discrepancy in the transcript expression profiles of six representative ShARFs between qRT-PCR and RNA-seq datasets, particularly in two experiments with Aaa infection. One reason would be that different sugarcane cultivars and Aaa strains were used in both datasets. For the qRT-PCR assay, leaf samples collected from two cultivars (LCP85-384 and ROC20) infected by the Aaa strain CNGX08 (Zhao et al., 2023) were used in this study. However, leaf samples collected from two cultivars (MT11-610 and ROC22) under the Aaa strain SC-026 infection (Chu et al., 2020) were used for RNA-seq. Another reason would be that these tested genes were less expressed. Everaert et al. (2017) proposed that approximately 85% of the genes showed consistent results between RNA-seq and qPCR data, and a small but specific gene set showed inconsistent results between both methods. The feature of these genes with inconsistent expression measurements was that they were typically smaller, had fewer exons, and had lower expressed levels (Everaert et al., 2017).
SA is a key plant hormone, which acts as a crucial signaling molecule for establishing defense responses in plants subjected to stressful cues (Kaya et al., 2023). Exogenous application of SA in plants can induce immune-like responses (Ding and Ding, 2020). Our results showed that ShARF04 played a negative regulator role, whereas ShARF07/17 acted as positive regulators under SA treatment. Similarly, BdARF18 (homolog of ShARF04) was negatively induced, but BdARF06 (homolog of ShARF07) and BdARF17 (homolog of ShARF17) were positively regulated in B. distachyon in response to SA treatment (Liu N. et al., 2018). In contrast, AcARF10 (homolog of ShARF04) was downregulated, but AcARF19a (homolog of ShARF07) and AcARF26a (homolog of ShARF17) were upregulated across all time points in A. chinensis against SA stress (Su et al., 2021). In addition, transcript levels of ClARF5 (homolog of ShARF04), ClARF (homolog of ShARF07), and ClARF7 (homolog of ShARF17) were upregulated by SA application during all time points in C. lacryma-jobi (Zhai et al., 2023). Taken together, these homologous ARF genes may act as positive or negative regulators against exogenous SA stress, depending on plant species. However, the regulatory mechanism of these ARFs involved in the SA-mediated plant immune response remains unclear.
In this study, genome-wide identification showed the presence of 24 SpapARFs and 39 SpnpARFs genes in S. spontaneum clones AP85-441 and Np-X, respectively. The characteristics of these ARF family genes uncovered that they had a highly divergent gene structure and a considerable number of cis-regulatory elements associated with biotic and abiotic stress, suggesting that these ARF genes possess diverse functions in sugarcane encountering environmental stress. Additionally, 17 ARFs from S. spontaneum were predicted to be targeted by numerous miRNAs, such as “sof-miR167a/b and sof-miR168a/b,” indicating that post-transcriptional regulation may participate in sugarcane in response to various stressors through the auxin signaling pathway. Furthermore, gene expression analysis revealed that redundant and divergent functions occurred among sugarcane ARFs, including fragmental duplication gene pairs (ShARF17 vs. ShARF22) in response to bacterial infection and SA stress. Overall, this study demonstrated for the first time the genome-wide identification of the ARF gene family and its defense responses to bacterial infection and SA treatment in sugarcane, which provides an important fundament for further identification of which candidate ARFs are curial regulators involved in biotic and abiotic stress responses.
The data presented in the study are deposited in the NCBI repository, BioProject Accession No: PRJNA549590 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA549590) and PRJNA579959 (https://www.ncbi.nlm.nih.gov/bioproject/ PRJNA579959).
J-XL: Writing – original draft. AA: Writing – review & editing. NC: Writing – review & editing and data curation. H-YF: Writing – review & editing and resources. M-TH: Resources, writing – review & editing. SM: Writing – review & editing. S-JG: Supervision, writing – review & editing, funding acquisition, and project administration. H-LZ: Writing – review & editing and supervision.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the China Agriculture Research System of MOF and MARA (grant no. CARS-170302) and the Special Projects for the Central-guided Local Science and Technology Development (grant no. 2022L3086).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1257355/full#supplementary-material
1. ^http://www.life.illinois.edu
2. ^http://sugarcane.zhangjisenlab.cn
3. ^http://www.maizegenome.org
4. ^https://www.ncbi.nlm.nih.gov
6. ^http://smart.emblheidelberg.de/
8. ^https://www.bioinformatics.org
10. ^https://orthovenn2.bioinfotoolkits.net
14. ^https://plants.ensembl.org
Ali, A., Chu, N., Ma, P., Javed, T., Zaheer, U., Huang, M. T., et al. (2020). Genome-wide analysis of mitogen-activated protein (MAP) kinase gene family expression in response to biotic and abiotic stresses in sugarcane. Physiol. Plant. 171, 86–107. doi: 10.1111/ppl.13208
Ali, A., Khan, M., Sharif, R., Mujtaba, M., and Gao, S. J. (2019). Sugarcane omics: an update on the current status of research and crop improvement. Plants. 8:344. doi: 10.3390/plants8090344
Ali, S., Wang, W., Zhang, Z., Xie, L., Boer, D. R., and Khan, N. (2022). Genome-wide identification, expression and interaction analysis of ARF and AUX/IAA gene family in soybean. Front Biosci-Landmrk. 27:251. doi: 10.31083/j.fbl2708251
Bai, Y., Ma, Y., Chang, Y., Zhang, W., Deng, Y., Zhang, N., et al. (2023). Identification and transcriptome data analysis of ARF family genes in five Orchidaceae species. Plant Mol. Biol. 112, 85–98. doi: 10.1007/s11103-023-01354-4
Bouzroud, S., Gasparini, K., Hu, G., Barbosa, M. A. M., Rosa, B. L., Fahr, M., et al. (2020). Down regulation and loss of auxin response factor 4 function using CRISPR/Cas9 alters plant growth, stomatal function and improves tomato tolerance to salinity and osmotic stress. Gen. Dent. 11:272. doi: 10.3390/genes11030272
Bouzroud, S., Gouiaa, S., Hu, N., Bernadac, A., Mila, I., Bendaou, N., et al. (2018). Auxin response factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum). PLoS One 13:0193517. doi: 10.1371/journal.pone.0193517
Chandler, J. W. (2016). Auxin response factors. Plant Cell Environ. 39, 1014–1028. doi: 10.1111/pce.12662
Chandra, S., Satapathy, L., Basu, S., Jha, S. K., Kumar, M., and Mukhopadhyay, K. (2020). Characterization of the leaf rust responsive ARF genes in wheat (Triticum aestivum L.). Plant Cell Rep. 39, 1639–1654. doi: 10.1007/s00299-020-02591-3
Chaudhary, C., Sharma, N., and Khurana, P. (2023). Genome-wide identification of aux/IAA and ARF gene families in bread wheat (Triticum aestivum L.). Protoplasma 260, 257–270. doi: 10.1007/s00709-022-01773-y
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, X., Liu, Z. P., Shi, G. Q., Bai, Q. Q., Guo, C. J., and Xiao, K. (2018). MIR167a transcriptionally regulates ARF6 and ARF8 and mediates drastically plant pi-starvation response via modulation of various biological processes. Plant Cell Tiss. Org. 133, 177–191. doi: 10.1007/s11240-017-1371-8
Chen, D., Wang, W., Wu, Y., Xie, H., Zhao, L., Zeng, Q., et al. (2019). Expression and distribution of the auxin response factors in Sorghum bicolor during development and temperature stress. Int. J. Mol. Sci. 20:4816. doi: 10.3390/ijms20194816
Chu, N., Zhou, J. R., Fu, H. Y., Huang, M. T., Zhang, H. L., and Gao, S. J. (2020). Global gene responses of resistant and susceptible sugarcane cultivars to Acidovoraxavenae subsp. avenae identified using comparative transcriptome analysis. Microorganisms 8:10. doi: 10.3390/microorganisms8010010
Chu, N., Zhou, J. R., Rott, P. C., Li, J., Fu, H. Y., Huang, M. T., et al. (2022). ScPR1 plays a positive role in the regulation of resistance to diverse stresses in sugarcane (Saccharum spp.) and Arabidopsis thaliana. Ind. Crop. Prod. 180:114736. doi: 10.1016/j.indcrop.2022.114736
Ding, P., and Ding, Y. (2020). Stories of salicylic acid: a plant defense hormone. Trends Plant Sci. 25, 549–565. doi: 10.1016/j.tplants.2020.01.004
Everaert, C., Luypaert, M., Maag, J. L., Cheng, Q. X., Dinger, M. E., Hellemans, J., et al. (2017). Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci. Rep. 7:1559. doi: 10.1038/s41598-017-01617-3
Goldtzvik, Y., Sen, N., Lam, S. D., and Orengo, C. (2023). Protein diversification through post-translational modifications, alternative splicing, and gene duplication. Curr. Opin. Struct. Biol. 81:102640. doi: 10.1016/j.sbi.2023.102640
Kaya, C., Ugurlar, F., Ashraf, M., and Ahmad, P. (2023). Salicylic acid interacts with other plant growth regulators and signal molecules in response to stressful environments in plants. Plant Physiol. Biochem. 196, 431–443. doi: 10.1016/j.plaphy.2023.02.006
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, Y., Han, S., and Qi, Y. (2023). Advances in structure and function of auxin response factor in plants. J. Integr. Plant Biol. 65, 617–632. doi: 10.1111/jipb.13392
Li, S. B., Xie, Z. Z., Hu, C. G., and Zhang, J. Z. (2016). A review of auxin response factors (ARFs) in plants. Front. Plant Sci. 7:47. doi: 10.3389/fpls.2016.00047
Li, J., Zhao, J. Y., Shi, Y., Fu, H. Y., Huang, M. T., Meng, J. Y., et al. (2022). Systematic and functional analysis of non-specific lipid transfer protein family genes in sugarcane under Xanthomonas albilineans infection and salicylic acid treatment. Front. Plant Sci. 13:1014266. doi: 10.3389/fpls.2022.1014266
Liu, N., Dong, L., Deng, X., Liu, D., Liu, Y., Li, M., et al. (2018). Genome-wide identification, molecular evolution, and expression analysis of auxin response factor (ARF) gene family in Brachypodium distachyon L. BMC Plant Biol. 18, 336–315. doi: 10.1186/s12870-018-1559-z
Liu, Y., Jiang, H., Chen, W., Qian, Y., Ma, Q., Cheng, B., et al. (2011). Genome-wide analysis of the auxin response factor (ARF) gene family in maize (Zea mays). Plant Growth Regul. 63, 225–234. doi: 10.1186/1471-2164-12-178
Liu, X., Liu, S., Chen, X., Prasanna, B. M., Ni, Z., Li, X., et al. (2022). Maize miR167-ARF3/30-polyamine oxidase 1 module-regulated H2O2 production confers resistance to maize chlorotic mottle virus. Plant Physiol. 189, 1065–1082. doi: 10.1093/plphys/kiac099
Liu, M., Ma, Z., Wang, A., Zheng, T., Huang, L., Sun, W., et al. (2018). Genome-wide investigation of the auxin response factor gene family in tartary buckwheat (Fagopyrum tataricum). Int. J. Mol. Sci. 19:3526. doi: 10.1155/2021/3102399
Ntambo, M. S., Meng, J. Y., Rott, P. C., Henry, R. J., Zhang, H. L., and Gao, S. J. (2019). Comparative transcriptome profiling of resistant and susceptible sugarcane cultivars in response to infection by Xanthomonas albilineans. Int. J. Mol. Sci. 20:6138. doi: 10.3390/ijms20246138
Okushima, Y., Overvoorde, P. J., Arima, K., Alonso, J. M., Chan, A., Chang, C., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444–463. doi: 10.1105/tpc.104.028316
Piperidis, N., and D’Hont, A. (2020). Sugarcane genome architecture decrypted with chromosome-specific oligo probes. The Plant J 103, 2039–2051. doi: 10.1111/tpj.14881
Raza, A., Charagh, S., Karikari, B., Sharif, R., Yadav, V., Mubarik, M. S., et al. (2023). miRNAs for crop improvement. Plant Physiol. Biochem. 201:107857. doi: 10.1016/j.plaphy.2023.107857
Rienstra, J., Hernández-García, J., and Weijers, D. (2023). To bind or not to bind: how auxin response factors select their target genes. J. Exp. Bot. 11:erad259. doi: 10.1093/jxb/erad259
Song, X., Xiong, Y., Kong, X., and Huang, G. (2023). Roles of auxin response factors in rice development and stress responses. Plant Cell Environ. 46, 1075–1086. doi: 10.1111/pce.14494
Su, L., Xu, M., Zhang, J., Wang, Y., Lei, Y., and Li, Q. (2021). Genome-wide identification of auxin response factor (ARF) family in kiwifruit (Actinidia chinensis) and analysis of their inducible involvements in abiotic stresses. Physiol. Mol. Biol. Plants 27, 1261–1276. doi: 10.1007/s12298-021-01011-4
Wang, D., Pei, K., Fu, Y., Sun, Z., Li, S., Liu, H., et al. (2007). Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene. 394, 13–24. doi: 10.1016/j.gene.2007.01.006
Wang, Y. J., Deng, D. X., Shi, Y. T., Miao, N., Bian, Y. L., and Yin, Z. T. (2012). Diversification, phylogeny and evolution of auxin response factor (ARF) family: insights gained from analyzing maize ARF genes. Mol. Biol. Rep. 39, 2401–2415. doi: 10.1007/s11033-011-0991-z
Wang, F. G., Wang, A. H., Bai, C. K., Jin, D. M., Nie, L. Y., Harris, A. J., et al. (2022). Genome size evolution of the extant lycophytes and ferns. Plant Divers. 44, 141–152. doi: 10.1016/j.pld.2021.11.007
Xing, H. Y., Pudake, R. N., Guo, G. G., Xing, G. F., Hu, Z. R., Zhang, Y. R., et al. (2011). Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genomics. 12:178. doi: 10.1186/1471-2164-12-178
Xu, L., Wang, D., Liu, S., Fang, Z., Su, S., Guo, C., et al. (2020). Comprehensive atlas of wheat (Triticum aestivum L.) AUXIN RESPONSE FACTOR expression during male reproductive development and abiotic stress. Front. Plant Sci. 11:586144. doi: 10.3389/fpls.2020.586144
Yu, Z., Zhang, F., Friml, J., and Ding, Z. (2022). Auxin signaling: research advances over the past 30 years. J. Integr. Plant Biol. 64, 371–392. doi: 10.1111/jipb.13225
Zhai, Y., Shen, X., Sun, Y., Liu, Q., Ma, N., Zhang, X., et al. (2023). Genome-wide investigation of ARF transcription factor gene family and its responses to abiotic stress in Coix (Coix lacryma-jobi L.). Protoplasma 260, 1389–1405. doi: 10.1007/s00709-023-01855-5
Zhang, H. H., Li, L. L., He, Y. Q., Qin, Q. Q., Chen, C. H., Wei, Z. Y., et al. (2020). Distinct modes of manipulation of rice auxin response factor OsARF17 by different plant RNA viruses for infection. P. Natl. Acad. Sci. USA 117, 9112–9121. doi: 10.1073/pnas.1918254117
Zhang, Q., Qi, Y., Pan, H., Tang, H., Wang, G., Hua, X., et al. (2022). Genomic insights into the recent chromosome reduction of autopolyploid sugarcane Saccharum spontaneum. Nat. Genet. 54, 885–896. doi: 10.1038/s41588-022-01084-1
Zhang, J., Zhang, X., Tang, H., Zhang, Q., Hua, X., Ma, X., et al. (2018). Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet. 50, 1565–1573. doi: 10.1038/s41588-018-0237-2
Zhao, J. Y., Chen, J., Hu, Z. T., Li, J., Fu, H. Y., Rott, P. C., et al. (2023). Genetic and morphological variants of Acidovorax avenae subsp. avenae cause red stripe of sugarcane in China. Front. Plant Sci. 14:1127928. doi: 10.3389/fpls.2023.1127928
Zhao, J. Y., Chen, J., Shi, Y., Fu, H. Y., Huang, M. T., Rott, P. C., et al. (2022). Sugarcane responses to two strains of Xanthomonas albilineans differing in pathogenicity through a differential modulation of salicylic acid and reactive oxygen species. Front. Plant Sci. 13:1087525. doi: 10.3389/fpls.2022.1087525
Keywords: auxin response factor, biotic and abiotic stress, expression pattern, sugarcane, stress response
Citation: Lin J-X, Ali A, Chu N, Fu H-Y, Huang M-T, Mbuya SN, Gao S-J and Zhang H-L (2023) Identification of ARF transcription factor gene family and its defense responses to bacterial infection and salicylic acid treatment in sugarcane. Front. Microbiol. 14:1257355. doi: 10.3389/fmicb.2023.1257355
Received: 14 July 2023; Accepted: 18 August 2023;
Published: 07 September 2023.
Edited by:
Dean Gabriel, University of Florida, United StatesReviewed by:
Michael Butterfield, Sugarcane Research Center (CTC), BrazilCopyright © 2023 Lin, Ali, Chu, Fu, Huang, Mbuya, Gao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: San-Ji Gao, Z2Fvc2FuamlAZmFmdS5lZHUuY24=; Hui-Li Zhang, aGx6aGFuZ0BmYWZ1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.