- 1Guangdong Province Key Laboratory of Microbial Signals and Disease Control, Integrative Microbiology Research Centre, South China Agricultural University, Guangzhou, China
- 2Department of Microbiology, University of Washington, Seattle, WA, United States
The Gram-negative opportunistic pathogen Pseudomonas aeruginosa possesses hierarchical quorum sensing (QS) systems. The intricate QS network of P. aeruginosa synchronizes a suite of virulence factors, contributing to the mortality and morbidity linked to the pathogenicity of this bacterium. Previous studies have revealed that variations in the lasR gene are frequently observed in chronic isolates of cystic fibrosis (CF). Specifically, LasRQ45stop was identified as a common variant among CF, lasR mutants during statistical analysis of the clinical lasR mutants in the database. In this study, we introduced LasRQ45stop into the chromosome of P. aeruginosa PAO1 through allelic replacement. The social traits of PAO1 LasRQ45stop were found to be equivalent to those of PAO1 LasR-null isolates. By co-evolving with the wild-type in caseinate broth, elastase-phenotypic-variability variants were derived from the LasRQ45stop subpopulation. Upon further examination of four LasRQ45stop sublines, we determined that the variation of T2SS-peptidase xcpA and mexT genes plays a pivotal role in the divergence of various phenotypes, including public goods elastase secretion and other pathogenicity traits. Furthermore, XcpA mutants demonstrated a fitness advantage compared to parent strains during co-evolution. Numerous phenotypic variations were associated with subline-specific genetic alterations. Collectively, these findings suggest that even within the same parental subline, there is ongoing microevolution of individual mutational trajectory diversity during adaptation.
Introduction
Pseudomonas aeruginosa is an opportunistic bacterial pathogen responsible for acute and chronic lung infections in immune-compromised individuals, including cystic fibrosis (CF) patients. This leads to high mortality and morbidity rates (Lyczak et al., 2000; Ahmed et al., 2019). The bacterium poses a significant public health challenge due to its extensive metabolic versatility, swift adaptation to diverse stresses, innate resistance, tendency to develop antibiotic resistance, biofilm formation capacity, and virulence factor secretion. Quorum sensing (QS), predominantly governs these traits, making eradication notably difficult (Huang et al., 2019; Lu et al., 2022).
P. aeruginosa features two primary acyl-homoserine lactone (AHL) QS circuits: LasIR and RhlIR (Tashiro et al., 2013). Specifically, the signal synthase LasI synthesizes 3-oxo-dodecanoyl-homoserine lactone (3OC12HSL), which the QS regulator LasR receives, activating the transcription of numerous genes. The LasR QS regulon includes over 300 genes, like lasI, rhlI, and rhlR. In contrast, the secondary QS regulator RhlR detects N-butanoyl-HSL (C4-HSL) produced by signal synthase RhlI. The RhlR regulon covers various genes, some of which overlap with the LasR regulon (Schuster et al., 2013). Genes regulated by LasR and RhlR encode essential secreted virulence factors, including protease elastase, hydrogen cyanide, and phenazines (Qin et al., 2022). The Pseudomonas quinolone signal (PQS) system, another non-AHL QS system in P. aeruginosa, involves the transcriptional regulator PqsR (also termed MvfR). This binds to 2-heptyl-3-hydroxy-4-quinolone (PQS) or its biosynthetic precursor, 2-heptyl-4-quinolone (HHQ), produced by PqsABCD and PqsH, respectively (Xiao et al., 2006).
Through genetic mutations and modifications in its quorum sensing regulatory genes, P. aeruginosa can induce phenotypic changes, preparing the bacterium for diverse environments, including the CF lung (Chugani et al., 2012; Folkesson et al., 2012). One such adaptation in chronic CF isolates is the mutational inactivation of the lasR gene. Often, LasR loss-of-function mutations trigger AHL and LasB-negative phenotypes in chronic P. aeruginosa CF isolates (Seed et al., 1995; Kiratisin et al., 2002; Wang et al., 2018). When QS is vital for growth in the P. aeruginosa PAO1 background, LasR mutants gain a fitness advantage due to reduced metabolic costs when co-cultured with wild-type P. aeruginosa PAO1. This wild-type provides shared public goods for the entire cell population (Schuster et al., 2003). Yet, if the LasR mutant population surpasses a certain threshold, it collapses (Wang et al., 2015). Introducing C4-HSL or co-evolving bacteria that produce C4-HSL with PAO1 lasR mutants swiftly leads to populations with an active RhlIR QS system (Kostylev et al., 2019). Many lasR mutant clinical isolates still respond to AHL via the RhlRI system (Chen et al., 2019; Cruz et al., 2020). Today, advancements in high-throughput DNA sequencing technologies allow millions of DNA sequences from a single microbial sample. This technology has evolved into an efficient tool for determining genetic adaptations in infected host populations (Kostylev et al., 2019).
Previous studies noted that LasR locus analysis of P. aeruginosa isolates from CF patients identified LasRQ45stop as a frequent mutation site (Cruz et al., 2020). This research aims to explore the evolutionary pathways in lab conditions focusing on the high-frequency mutation site of the LasRQ45stop variation, which has a single substitution at base 133 in lasR. Laboratory-induced mutations of LasRQ45stop in strain PAO1 indicate it as a nonfunctional LasR. When co-cultured in casein broth with cooperator PAO1, LasRQ45stop spurred the swift emergence of proteolysis-associated mutant variants. Multiple variants displayed characteristics similar to CF clinical isolates, including a functional RhlI-RhlR system due to a mexT mutation. Our study identified the xcpA mutant exhibiting numerous virulence-related attributes, including reduced extracellular protein production and twitching motility. Additionally, we present lab experiments elucidating why mexT and xcpA are prevalent in the context of LasRQ45stop evolution.
Materials and methods
Bacterial strains, media, and growth conditions
Bacterial strains and plasmids used are described in the Supplementary Table S1. P. aeruginosa was cultivated in Luria-Bertani [0.5% (w/v) yeast extract, 1% (w/v) tryptone, and 1% (w/v) sodium chloride] broth with a pH of 6.8, using 50 mM 3-(N-morpholino)propanesulfonic acid as a buffer (LB-Mops broth), PM broth with an addition of 1% sodium caseinate (casein broth) (Kostylev et al., 2019), or Pseudomonas P broth [0.14% (w/v) magnesium chloride, 2% (w/v) pancreatic digest of gelatin, and 1% (w/v) potassium sulfate]. Several types of agar were used for plating purposes, including LB agar, and skim milk agar (Sandoz et al., 2007). Typically, 100 μg/mL of gentamicin, 50 μg/mL of kanamycin, or 300 μg/mL of carboxybenzylmycin was added to the medium. Bacteria were subcultured in 14 mm Falcon tubes with fresh LB or LB-Mops broth and incubated at 37°C with shaking at 250 rpm. Escherichia coli used in the cloning process was grown in LB broth.
Strain construction
Our investigations utilized the P. aeruginosa PAO1 strain (Stover et al., 2000), which differs from the currently sequenced strain due to an 8 bp deletion at the mexT gene. Mutants with deletions of lasR, lasRQ45stop, psdR, mexT, xcpA, and lasB were derived by cloning PAO1 using the suicide vector pEXG2, as previously described (Hmelo et al., 2015). The Supplementary Table S1, lists the primers used to construct the pEXG2 knockout plasmids. Reverse-transcriptase PCR of genomic DNA confirmed all deletion mutations. To compensate for the mutations, we integrated wild-type copies of genes from pUC18-mini-Tn7-Gm, as previously reported (Choi et al., 2005).
Assays for LasR activity, Cyanide and pyocyanin production
We measured promoter activity in cells cultured in LB-MOPS broth, as previously reported (Feltner et al., 2016), using lasI-gfp reporter strains in a 96-well plate. These were incubated at 37°C with continuous shaking in a Synergy H1 microplate reader (Biotec, America). GFP fluorescence (excitation at 485 nm, emission at 535 nm) and cell density (OD600) were recorded at 2 h intervals for 28 h (refer to Supplementary Table S1 in the Supplementary material). For the detection of cyanide, we employed a cyanogenic test paper approach detailed previously (Chen et al., 2019). Pyocyanin activity was estimated using the modified protocol of O’Loughlin et al. (2013). The P. aeruginosa PAO1 strain was cultured overnight and subsequently diluted to an OD600 of 0.01, followed by incubation in 3 mL of P broth medium for 24 h.
Motility assays
This study assessed swarming motility using a medium composed of nutritional broth [0.8% (w/v)] supplemented with D-glucose [0.5% (w/v)] and solidified with 0.5% (w/v) Bacto Agar. Overnight liquid broth cultures were adjusted to an optical density of 1.0 before inoculation onto plates. Growth was evaluated after 18–24 h of incubation at 37°C. After 24 h of growth in LB medium containing 1% solidified Bacto Agar at 37°C, the agar was gently detached, revealing a twitching motility zone adhered to the petri dish. A 10 min stain with 1% (wt/vol) crystal violet made the twitching zones visible, and their diameters were subsequently measured (Liu et al., 2022).
QS signal measurements
Quorum sensing signal extraction was carried out as described (Dong et al., 2008). The bacterium was cultivated in LB-Mops broth for 18 h, using 3 mL. Subsequently, an equal volume of acidified ethyl acetate (0.1% acetic acid) was combined with 3 mL of culture supernatant for signal extraction. Dried samples were reconstituted in 200 μL methanol and immediately filtered before LC-MS/MS analysis. We isolated and selected the prevalent signaling molecules for quantitative LC-MS/MS investigation. Modifications to the LC-MS procedure were made (Liu et al., 2022). HPLC was conducted using a Waters X Select HSS T3 1.8 μm phase column (2.1 × 100 mm) on the Dionex Ultimate 3000 system (Thermo Fisher Scientific). Metabolites identified included 3-oxo-C12-HSL (m/z 298.20128) and C4-HSL (m/z 172.09682). Data evaluation was executed with TraceFinder and Thermo Xcalibur, both from Thermo Fisher Scientific.
Assessment of extracellular protease production
Bacteria grown on LB broth milk agar plates were incubated for 30 h at 37°C. The amount of extracellular protease produced was quantified by measuring the area of the proteolytic zone surrounding the bacterial colony.
Competition experiments
Under these conditions, the gentamicin-resistance marker is neutral. We carried out competitive studies using P. aeruginosa strains both with and without this marker. The proportion of the competitor bearing the gentamicin-resistance marker was set at 1%, 10%, 50%, or 90% in casein broth for 48 h. To ascertain cell yields, plate counting, both with and without 10 μg/mL gentamicin, was employed to gauge proportional abundances after 48 h of incubation. The relative fitness was calculated by dividing the ratio of mutant to wild type after 48 h by the ratio at the time of inoculation.
Social evolution experiments
For evolution experiments (Supplementary Figure S2C), log-phase LB-Mops cultures of both cooperator and competitor were prepared, adding the competitor to the casein broth at a ratio of 0.1 (total 100 μL). Two days post primary inoculation, once visible proliferation was observed, a 60 μL volume of the transfer inoculum was introduced daily into a 3 mL casein broth. Serial dilutions of isolates were grown on LB agar plates both with and without 10 μg/mL gentamicin to determine the percentage of protease-deficient cells. Cultures were sampled from LB agar plates containing 10 μg/mL gentamicin, and colonies were then streaked daily onto skim milk agar plates to detect protease-altering variations.
Genome variant analysis
Bacteria were isolated from individual colonies and cultured in LB broth with a buffer for a duration of 24 h. Cells were collected for analysis via centrifugation. Bacteria were cultivated in LB-Mops broth, and genomic DNA was extracted using the Easy Pure Bacteria Genomic DNA Kit (TRAN). The genomic DNA of the four sublines (LasRQ45stop-1, LasRQ45stop-2, LasRQ45stop-3, and LasRQ45stop-4) was sequenced with the Illumina NovaSeq S4 PE-150 (Novogene, China). Whole Genomics Solution Corporation conducted the whole-genome sequencing and assembly. PCR testing was employed to validate the genomic sequencing results.
Statistical analysis
Results are depicted as mean ± SD. Experimental groups were evaluated using an unpaired student’s t-test (one-tailed, unequal variance). Differences with a p-value <0.05 were considered significant. Experiments were repeated to confirm their consistency.
Results
The high-frequency mutant LasRQ45stop presents fitness advantages over WT
The lasRC133T substitution has been identified as a recurring mutation site in P. aeruginosa CF isolates (Feltner et al., 2016). This single-base substitution introduces a premature stop codon into the lasR sequence, terminating protein translation prematurely. This mutation was henceforth referred to as LasRQ45stop. To validate the functionality of LasRQ45stop, we generated a knock-out deletion mutant LasR, an allelic replacement mutant LasRQ45stop, and a LasR complementation strain C-LasRQ45stop in the laboratory model strain P. aeruginosa PAO1. Comparing the QS-associated traits of the strains mentioned, we found that the homologous LasRQ45stop mutation in P. aeruginosa acts as a nonfunctional LasR in QS-associated phenotypes, such as elastase secretion, pyocyanin production, and 3OC12-HSL and C4-HSL levels (Figures 1A–F).
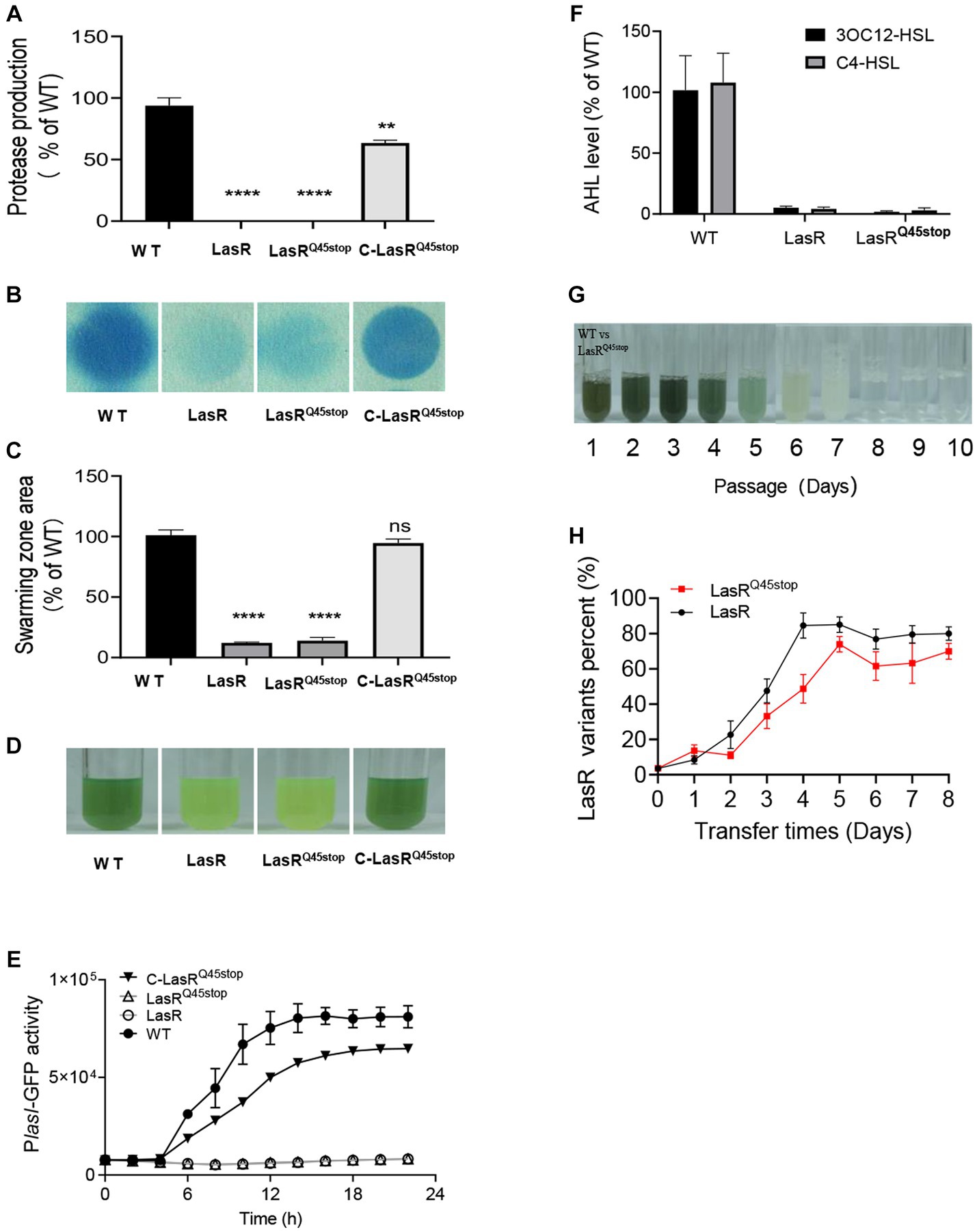
Figure 1. Characteristics in QS regulon of LasRQ45stop in solitary presence and co-culture states. (A) Protease production on casein agar. The PAO1 LasR mutant and the LasRQ45stop mutant produce relatively little protease. All data shown are the average values of three independent experiments ± SD. (B) Cyanide production by P. aeruginosa strains and mutants. Cyanide was monitored as described in Materials and methods. The blue coloration of the filter paper reflects cyanide production. (C) Swarming motility phenotype of PAO1 wild-type and LasR mutants. All data shown are the average values of three independent experiments ± SD. (D) Color changes of P. aeruginosa cultures in the presence of LasR mutant after 24 h, a reflection of the effect of LasR mutant on phenazine production. (E) LasR activity in the LasR mutant and PAO1 measured as GFP fluorescence (RFU). All strains contained the PlasI-gfp fusion plasmid pProbe-GT-PlasIgfp. (F) Relative AHL levels of the indicated strain. The AHL levels produced by strain PAO1 are defined as 100%. AHL was measured after 18 h of growth in LB-Mops broth. All data shown are the average values of three independent experiments ± SD. (G) An example of one serial transfer experiment. Each photographic image was captured after 1 day of growth. The relative abundance of LasR mutant (as a percentage of the total population) in culture samples just before transfer (60 μL) is distinguished by resistance markers, and the daily passage number is shown below the tubes. Cultures that only partially degrade casein have a milky white appearance due to protein precipitation. (H) Competition between WT PAO1 and LasR mutant (black), or LasRQ45stop mutant (red). The initial frequency of LasR mutant was 0.01. Three biological replicates were performed. All data shown are the average values of three independent experiments ± SD. Two-tailed p-values were determined using unpaired t-tests. Statistical significance: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001; ns, no significant difference (p ≥ 0.05).
To ascertain earlier studies indicated that LasR mutants possess fitness advantages over WT cells when co-cultured in minimal medium with casein (casein broth) as the sole carbon source. However, this advantage was negated with the addition of adenosine to the casein medium (Feng et al., 2019). To determine if LasRQ45stop exhibited superior fitness to WT, we co-cultured the gentamicin-resistant PAO1 LasRQ45stop mutant alongside WT in casein broth. LasRQ45stop mutants were then distinguished and counted from WT using plates with or without gentamicin resistance. The schematic diagram (Supplementary Figure S2C) illustrates that 1% competitors (LasRQ45stop or LasR) were individually combined with 99% of WT in the initial casein broth, then passaged daily, and the frequency of mutant strains was subsequently monitored. Pyocyanin production during the initial days of the evolution process hinted at the robust growth of the PAO1 co-culture. Pyocyanin levels tapered off from day 4 to day 6, culminating in a milky culture by day 7. No noticeable alteration in the casein medium post-day 8 signified a population collapse (Figure 1G). Consistent with expectations, the proportion of LasRQ45stop mutants surged, peaking at nearly 70% in cultures (Figure 1H). This shift in frequency suggests that the LasRQ45stop mutant can act as a social cheater when co-cultured with WT in a casein-based medium.
Protease-change mutants rapidly emerge during growth of LasRQ45stop in caseinate
LasR mutations are prevalent in P. aeruginosa isolated from chronic infections (Feltner et al., 2016). Interestingly, many LasR-null clinical isolates partially restored the QS regulon over extended infections (Feltner et al., 2016). This suggests that post-LasR-deficient evolution might confer diverse benefits. We were intrigued by the evolvability and evolutionary trajectory of the high-frequency mutant LasRQ45stop in minimal medium with casein as the exclusive carbon and energy source. A competitive assay between the LasRQ45stop variant and WT was conducted as described above (Supplementary Figure S2C). LasRQ45stop variants were isolated using LB Gm-resistant plates, and their protease activity was assessed on skim milk agar plates (Supplementary Figure S2D). Variants exhibiting notable protease alterations were selected for further analysis (Supplementary Figure S2D). We identified four protease-altered LasRQ45stop variants from these evolutionary experiments. Variants LasRQ45stop-1, LasRQ45stop-2, and LasRQ45stop-4 demonstrated reduced protease activity relative to the parental strain LasRQ45stop. In contrast, LasRQ45stop-3 exhibited increased protease activity, approaching that of the wild-type strain (Figure 2A). We also evaluated the influence of these four isolates on virulence-associated traits, such as cell motility and pyocyanin production. Consequently, the twitching motility of LasRQ45stop-1, LasRQ45stop-2, and LasRQ45stop-4 was diminished compared to the parental strain LasRQ45stop (Figure 2B). Remarkably, LasRQ45stop-3 amplified swarming motility, while LasRQ45stop-1, LasRQ45stop-2, and LasRQ45stop-4 did not exhibit significant deviations from their parental strain LasRQ45stop (Figure 2C). Additionally, pyocyanin production was elevated in LasRQ45stop-3 (Figure 2D). Of note, LasRQ45stop-4 maintained functional pyocyanin production, independent of its protease activity phenotype (Figures 2A,D).
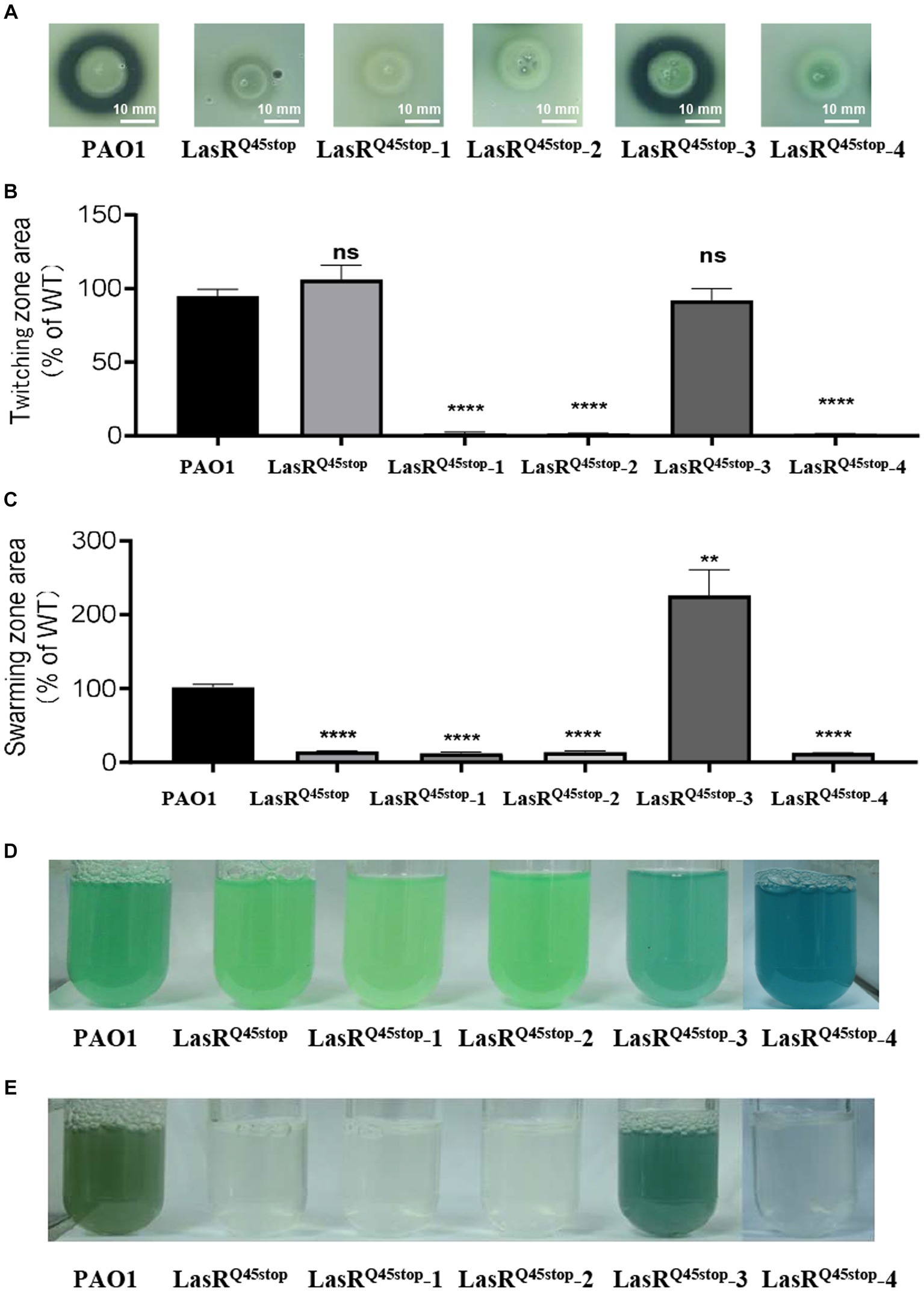
Figure 2. Characteristics of three protease-negative LasRQ45stop mutant variants (LasRQ45stop-1, LasRQ45stop-2, and LasRQ45stop-4) and protease-producing LasRQ45stop (LasRQ45stop-3) mutant variants derived from the LasRQ45stop mutant. (A) Protease production of isolates from co-cultured casein broth was measured on plates containing 1% (m/v) skimmed milk for 24 h. (B,C) Motility phenotypes of P. aeruginosa PAO1 and its isogenic QS LasRQ45stop strains. Image showing the comparative zone of twitching (B) and swarming motility (C) of PAO1 and its isogenic QS LasRQ45stop strains. All data shown are the average values of three independent experiments ± SD. (D,E) Cultures of P. aeruginosa PAO1, a LasRQ45stop mutant, and the evolved LasRQ45stop variants, grown in LP medium for 24 h (D) and casein broth for 48 h (E). The culture is visibly blue as a result of pyocyanin overproduction. Three biological replicates were performed. All data shown are the average values of three independent experiments ± SD. Two-tailed p-values were determined using unpaired t-tests. Statistical significance: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001; ns, no significant difference (p ≥ 0.05). Scale bar = 10 mm.
Mutations in mexT and xcpA are associated with the post-lasR-deficient evolution phenotype
To uncover the genetic variations responsible for the observed differences in virulence-associated phenotypes, we conducted a genome-wide association study among the parent LasRQ45stop and four protease-altered mutants (LasRQ45stop-1, LasRQ45stop-2, LasRQ45stop-3, and LasRQ45stop-4). This analysis identified three genes: mexT (PA2492), xcpA (PA4528), and psdR (PA4499) (Table 1). The single base indel or in-frame deletion in the xcpA gene (Table 1) was found in LasRQ45stop-1, LasRQ45stop-2, and LasRQ45stop-4. Conversely, a missense variant in the mexT gene was identified in LasRQ45stop-3 (Table 1). The LasRQ45stop-4 subline displayed additional structural variations (SVs), culminating in the deletion of the entire mexT gene and adjacent genes spanning from PA2259 to PA2544. This range is part of the 38.3 kb deletion observed in LasRQ45stop-4. Furthermore, we noted that all four sequenced isolates carried a nonsynonymous mutation in psdR, aligning with prior research that identified frequent psdR mutations in casein evolution experiments (Asfahl et al., 2015; Kostylev et al., 2019). Nevertheless, psdR has been demonstrated to enhance growth on casein independently of QS (Asfahl et al., 2015). We thus infer that MexT and XcpA are instrumental in the observed virulence-associated phenotypic variations in the evolved isolates.
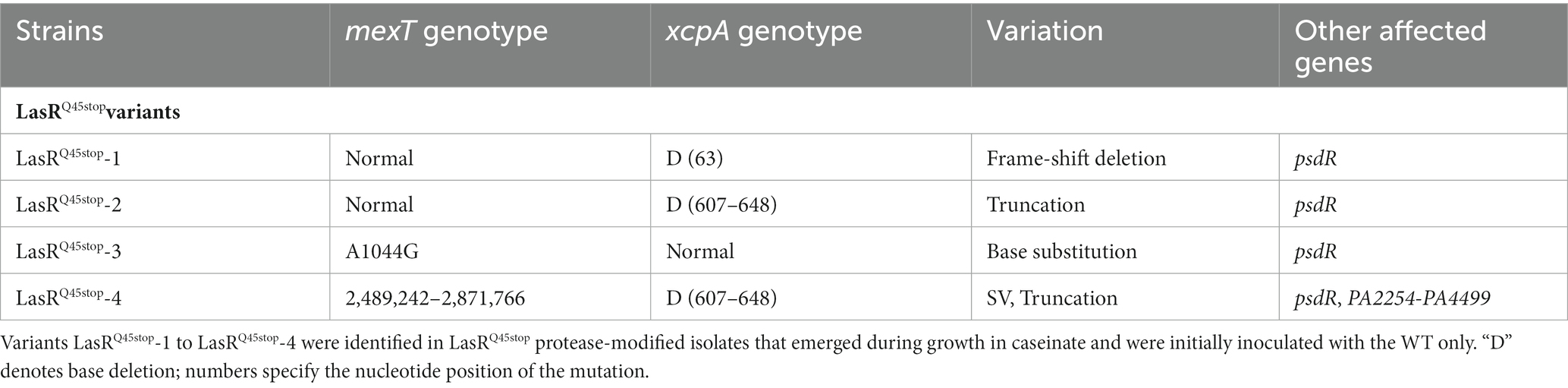
Table 1. Summary of xcpA and mexT variants found in in protease-modified variants of PAO1 LasRQ45stop isolates that emerged during growth in caseinate.
To substantiate this, we engineered MexT-and XcpA-related mutants using LasRQ45stop-PsdR as the progenitor strain. Our results show that LasRQ45stop-PsdR-MexT and LasRQ45stop-PsdR-MexTA1044G mutants exhibited markedly elevated pyocyanin and elastase production (Figure 3), as well as augmented swarming motility (Figure 3). Deletion of xcpA from both LasRQ45stop-PsdR and LasRQ45stop-PsdR-MexT mutants led to reduced QS-dependent elastase phenotypes and twitching motility (Figure 3). Alterations in mexT have been highlighted as pivotal in reshaping the QS circuits and the fitness of P. aeruginosa PAO1 (Kostylev et al., 2019). XcpA is a prepilin peptidase that is part of the Xcp type II secretion system (T2SS) in P. aeruginosa. Furthermore, the Xcp T2SS is governed by the QS pathway, which P. aeruginosa utilizes for secreting major virulence factors, including LasA and LasB elastases (Li and Lee, 2019). Therefore, we conclude that the xcpA and mexT mutations underpin the evolved phenotype, at least under laboratory conditions.
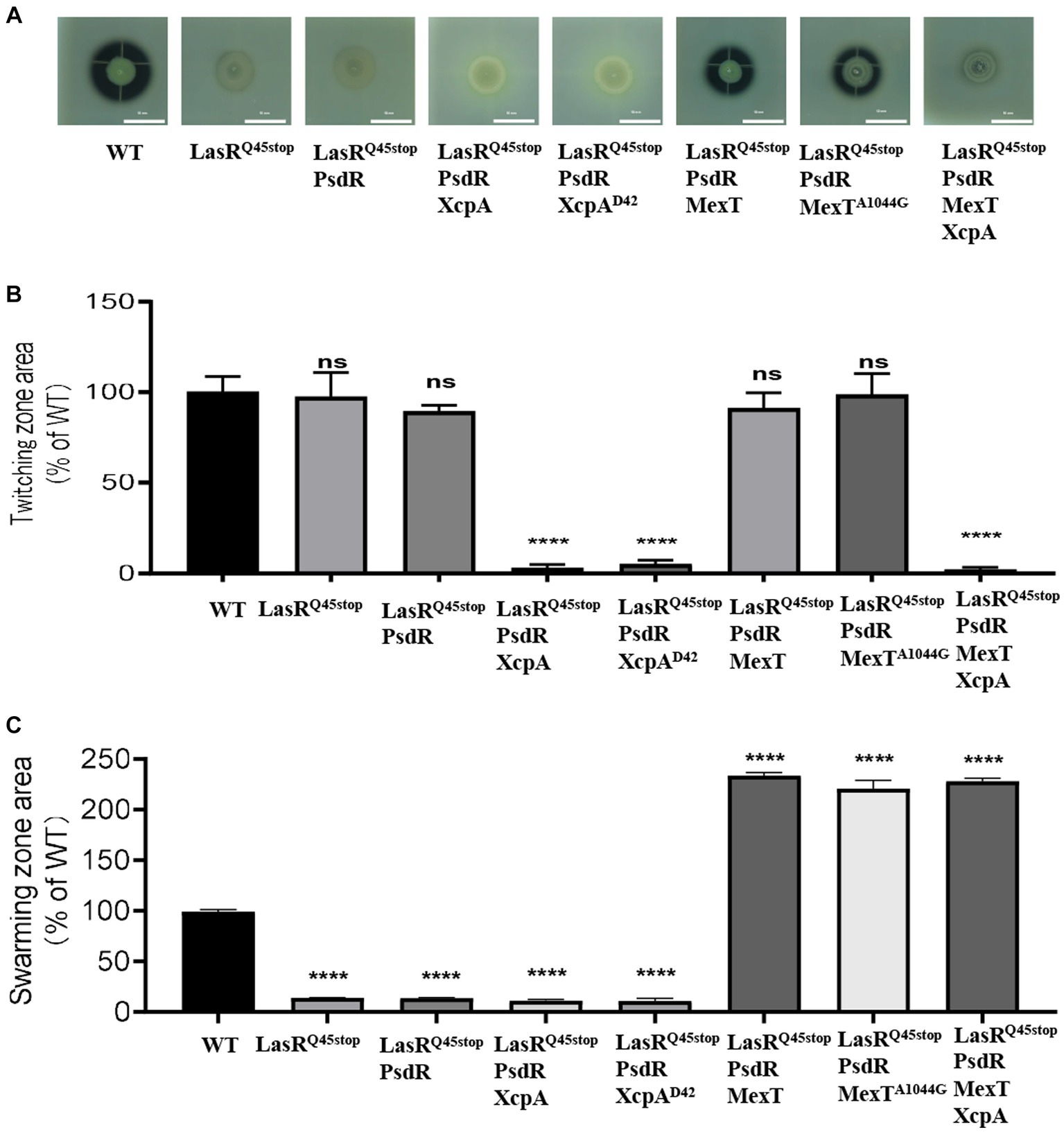
Figure 3. mexT and xcpA was related to the phenotype changes. (A) The influence of PsdR, MexT, and XcpA mutations on protease production in the P. aeruginosa LasRQ45stop mutant. The images are of colonies after about 24 h of incubation in milk agar plate. The influence of PsdR, MexT, and XcpA mutations on twitching (B) and swarming (C) motility phenotypes in the P. aeruginosa LasRQ45stop mutant. Three biological replicates were performed. All data shown are the average values of three independent experiments ± SD. Two-tailed p-values were determined using unpaired t-tests. Statistical significance: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001; ns, no significant difference (p ≥ 0.05). Scale bar = 10 mm.
The xcpA mutant gains a fitness advantage in caseinate
Mutants devoid of the metabolic cost of QS could possess a negative frequency-dependent fitness advantage over the parental strain when co-cultured in an environment where QS function is essential for growth. In previous research, LasR-MexT mutants exhibited a competitive advantage over the parental LasR mutant (Kostylev et al., 2019). Similarly, we postulated that XcpA and LasB mutants might also harbor a competitive edge. We co-cultured each identified mutant with the parental strain, starting at a frequency of 10%, to evaluate their relative fitness in PM-caseinate. Distinguished mutants were marked as gentamicin-resistant (GmR). Relative fitness was ascertained by taking the ratio of mutant-to-parent bacteria at the endpoint and dividing it by the initial mutant-to-parent bacteria ratio (Chen et al., 2019). A control was set up with unlabeled and labeled parental strains, yielding a relative fitness of 1 (Figure 4A). Conversely, the relative fitness of the XcpA mutant, when pitted against LasRQ45stop-PsdR-MexT or WT, exceeded 1, indicating a survival advantage in the XcpA mutant. However, the LasB mutant, which only lacked elastase production, exhibited a fitness comparable to the LasRQ45stop-PsdR-MexT parent strain (Figure 4A). Additionally, varying proportions of XcpA mutants were co-cultured with their parent strains in PM-caseinate for 48 h to determine their frequency-dependent fitness. When XcpA mutants were more abundant relative to LasRQ45stop-PsdR-MexT in the culture, their relative fitness was less than 1. However, when LasRQ45stop-PsdR-MexT bacteria predominated over the mutant in the starting inoculum, the relative fitness for XcpA exceeded 1 (Figure 4B). The relative fitness of the mutants declines as the mutant subpopulation expands (Diggle et al., 2007). These observations underscore that XcpA mutants can effectively invade their ancestral populations under conditions that require QS.
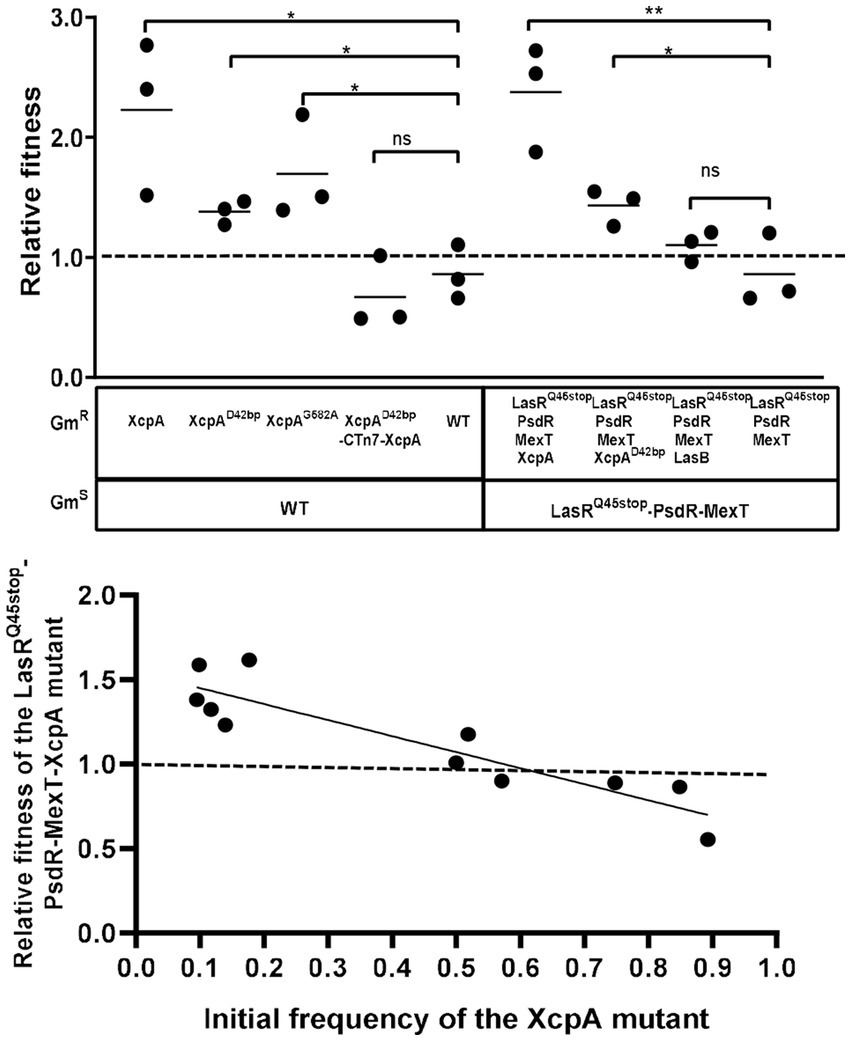
Figure 4. Growth and competition of the MexT mutant, the XcpA mutant, the LasB mutant and the parent strain. (A) Competition experiments showing the competitive indexes of competitor strains against the parent strain (WT or LasRQ45stop-PsdR-MexT) after 24 h in PM casein broth. Each symbol represents the outcome of an individual experiment; the solid lines represent means for each group. The starting percentage of the competitors was 10%. The LasB mutant was not fit than the parent, and the XcpA mutant was more fit than the parent. (B) The relative fitness of a XcpA mutant exhibits a negative frequency dependence. Data are from 24 h casein broth cultures. The outcomes above the dashed line indicate the competitor had a fitness advantage and below indicated the parent LasRQ45stop-PsdR-MexT had a fitness advantage. All data shown are the average values of three independent experiments ± SD. Two-tailed p-values were determined using unpaired t-tests. Statistical significance: *p < 0.05 and **p < 0.01; ns, no significant difference (p ≥ 0.05).
Discussion
P. aeruginosa, a well-known human pathogen, is particularly adept at infecting immunocompromised individuals, leading to severe infections (Moradali et al., 2017). Early work highlighted the complexity of interconnected quorum sensing (QS) regulatory circuits in controlling virulence phenotypes and adapting to complex environments (Willcox et al., 2008). The QS transcription factor LasR sits atop this hierarchy, regulating the expression of the RhlIR and PQS pathways (Cruz et al., 2020). LasR mutations have been shown to be prevalent in P. aeruginosa during both in vitro and in vivo evolution. Analysis of the associated lasR sequences from CF isolates (Feltner et al., 2016) indicated that LasRQ45stop is a high-frequency mutant (Supplementary Figure S1 and Supplementary Table S2). QS-regulated biological phenotypic assessments were diminished in LasRQ45stop mutants. However, these assessments were restored to wild-type levels upon the addition of functional LasR (Figure 1). This suggests that LasRQ45stop might function as a QS antagonist, inhibiting P. aeruginosa infection. Paradoxically, inactivating LasR mutations are frequently detected in isolates from chronic CF infections, suggesting an evolutionary selection favoring LasR mutants (Cruz et al., 2020). Recent research indicates that defects in the LasIR system can result in the evolution of MexT and PqsR mutations under laboratory conditions, leading to reactivation of P. aeruginosa QS regulatory circuits through rewiring (Chen et al., 2019; Kostylev et al., 2019). Phenotypic and genotypic microevolution result from niche adaptation to the CF lung environment and competition among different microbiomes. Micro-evolutionary diversity changes dynamically over time, complicating the accurate diagnosis and treatment of P. aeruginosa infections (Hwang et al., 2021).
The inherent genetic diversity of P. aeruginosa has been elucidated due to its suitability for adaptive laboratory evolution experiments. We identified LasRQ45stop evolutionary trajectories caused by acquiring stable mutant phenotypes, leading to rapid adaptation in stressful and fluctuating environments. Our findings substantiate the implications of LasRQ45stop microevolution. We employed a combination of molecular biology and bioinformatics analyses to study the evolution of LasRQ45stop cessation. We demonstrated that LasRQ45stop promotes an increased frequency of psdR, mexT, and xcpA mutations when co-cultured in our caseinate broth (Table 1). The psdR mutation enhances individual fitness by alleviating repression of intracellular metabolism, limiting growth. This mutation has been frequently observed in evolution experiments involving casein (Asfahl et al., 2015; Kostylev et al., 2019). Even with psdR deletion in both cooperators and cheaters, the pattern of invasion by the LasR mutant remains largely unaffected (Feng et al., 2019). The mexT mutation isolates have been identified both in the lab and clinical settings (Liu et al., 2022), while xcpA mutations frequently appear in environmental isolates (Supplementary Table S3). Moreover, our data highlight the role of MexT and XcpA in the adaptation of P. aeruginosa. Microbial cells exhibit substantial adaptation to environmental stress (Hogardt and Heesemann, 2010). Certain pivotal mutations in the genome can significantly bolster bacterial populations in securing a competitive edge in hostile environments (Feng et al., 2019).
For survival in the environment and adaptation to hosts for nourishment and pathogenicity, P. aeruginosa deploys a diverse array of virulence factors (Vidaillac and Chotirmall, 2021). Among these, the T2SS-peptidase XcpA is involved in the secretion of guanylate cyclase ExoA, proteases LasA/B, and several other factors, many of which have been earmarked as potential therapeutic targets (Swietnicki et al., 2019). Importantly, the lasB gene encodes elastase, a pivotal virulence component of P. aeruginosa. Its synthesis is orchestrated by the QS system, while its secretion modus operandi is overseen by the T2SS, also referred to as the general secretory pathway (GSP) (Filloux, 2004). The 12 identified Gsp components of the P. aeruginosa machinery were initially labeled as XcpA and XcpP to-Z and later renamed as XcpAO and XcpPC to-ZM, aligning with other GSP system nomenclature (Bleves et al., 2010). XcpA, recognized as a prepilin peptidase, was initially discovered and described in P. aeruginosa. It is believed to excise leader peptides endopeptidically, methylate the N-terminal amino acid residue, and cleave Xcp pre-pseudopilin substrates (Filloux, 2011). Few studies have explored the essential biological activities of prepilin peptidase in bacteria (LaPointe and Taylor, 2000). Tn insertions in the xcpA genes of P. aeruginosa strain PA14 manifested positive selection during fitness for gastrointestinal (GI) colonization but predominantly displayed attenuated fitness for systemic dissemination amidst neutropenia. This observation stemmed from a reliable mouse model mirroring the trajectory of human infections in cancer and bone marrow transplant patients (Koh et al., 2005). In Methanococcus voltae, FlaK operates as a preflagellin peptidase; membranes of the flaK mutant exhibited inability to process preflagellin in vitro (Bardy and Jarrell, 2003). In Bacillus subtilis, ComC functions as a prepilin peptidase; its inactivation results in the accumulation of pre-ComGC, likely due to the unprocessed protein’s inability to translocate outside the membrane (Chung and Dubnau, 1995). The significance of XcpA, a class of prepilin peptidase, in processing secretion systems has been documented. Pathogenic bacteria and environmental entities could capitalize on the extracellular dispatch of an array of toxins, lipases, and enzymes adept at breaking down complex materials, mediated through the secretory system (Korotkov and Sandkvist, 2019).
This study sheds light on the role of XcpA in P. aeruginosa evolutionary paths. Our findings indicate that the XcpA mutation confers a fitness advantage. When mutant XcpA is present in PsdR-LasRQ45stop-MexT, it reduces exoprotein secretion and enhances individual fitness. Although the physiological value of xcpA-associated microevolution is not fully detailed in this study, we delve into its role in fitness and intraspecific competition in casein broth. Our evaluation scope is limited to studying individual LasR variants and their evolution with casein proteins under laboratory conditions. Notably, pathogenic adaptation mutations are not consistent across studies, suggesting diverse evolutionary paths of pathogenic adaptation in P. aeruginosa. Still, our findings pave the way for understanding LasR’s evolutionary paths under lab conditions and potential further studies on XcpA in population evolution.
Data availability statement
Genomic sequences of LasRQ45stop-1, LasRQ45stop-2, LasRQ45stop-3, and LasRQ45stop-4 have been deposited at NCBI (BioProject PRJNA967237).
Author contributions
MC: Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. RC: Methodology, Resources, Supervision, Writing – review & editing. LL: Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from Guangdong Basic and Applied Basic Research Foundation of Natural Science Foundation (Grant Nos. 2022A1515010564 and 2020A1515110111), the Key Realm R&D Program of Guangdong Province (2020B0202090001), National Natural Science Foundation of China (32200025).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1256785/full#supplementary-material
References
Ahmed, S. A. K. S., Rudden, M., Smyth, T. J., Dooley, J. S. G., Marchant, R., and Banat, I. M. (2019). Natural quorum sensing inhibitors effectively downregulate gene expression of Pseudomonas aeruginosa virulence factors. Appl. Microbiol. Biotechnol. 103, 3521–3535. doi: 10.1007/s00253-019-09618-0
Asfahl, K. L., Walsh, J., Gilbert, K., and Schuster, M. (2015). Non-social adaptation defers a tragedy of the commons in Pseudomonas aeruginosa quorum sensing. ISME J. 9, 1734–1746. doi: 10.1038/ismej.2014.259
Bardy, S. L., and Jarrell, K. F. (2003). Cleavage of preflagellins by an aspartic acid signal peptidase is essential for flagellation in the archaeon Methanococcus voltae. Mol. Microbiol. 50, 1339–1347. doi: 10.1046/j.1365-2958.2003.03758.x
Bleves, S., Viarre, V., Salacha, R., Michel, G. P. F., Filloux, A., and Voulhoux, R. (2010). Protein secretion systems in Pseudomonas aeruginosa: a wealth of pathogenic weapons. Int. J. Med. Microbiol. 300, 534–543. doi: 10.1016/j.ijmm.2010.08.005
Chen, R., Déziel, E., Groleau, M., Schaefer, A. L., and Greenberg, E. P. (2019). Social cheating in a Pseudomonas aeruginosa quorum-sensing variant. Proc. Natl. Acad. Sci. U.S.A. 116, 7021–7026. doi: 10.1073/pnas.1819801116
Choi, K., Gaynor, J. B., White, K. G., Lopez, C., Bosio, C. M., Karkhoff-Schweizer, R. R., et al. (2005). A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2, 443–448. doi: 10.1038/nmeth765
Chugani, S., Kim, B. S., Phattarasukol, S., Brittnacher, M. J., Choi, S. H., Harwood, C. S., et al. (2012). Strain-dependent diversity in the Pseudomonas aeruginosa quorum-sensing regulon. Proc. Natl. Acad. Sci. U.S.A. 109, E2823–E2831. doi: 10.1073/pnas.1214128109
Chung, Y. S., and Dubnau, D. (1995). ComC is required for the processing and translocation of ComGC, a pilin-like competence protein of Bacillus subtilis. Mol. Microbiol. 15, 543–551. doi: 10.1111/j.1365-2958.1995.tb02267.x
Cruz, R. L., Asfahl, K. L., Van den Bossche, S., Coenye, T., Crabbé, A., Dandekar, A. A., et al. (2020). RhlR-regulated acyl-Homoserine lactone quorum sensing in a cystic fibrosis isolate of Pseudomonas aeruginosa. mBio 11:e00532. doi: 10.1128/mBio.00532-20
Diggle, S. P., Griffin, A. S., Campbell, G. S., and West, S. A. (2007). Cooperation and conflict in quorum-sensing bacterial populations. Nature 450, 411–414. doi: 10.1038/nature06279
Dong, Y. H., Zhang, X. F., An, S. W., Xu, J. L., and Zhang, L. H. (2008). A novel two-component system BqsS-BqsR modulates quorum sensing-dependent biofilm decay in Pseudomonas aeruginosa. Commun. Integr. Biol. 1, 88–96. doi: 10.4161/cib.1.1.6717
Feltner, J. B., Wolter, D. J., Pope, C. E., Groleau, M. C., Smalley, N. E., Greenberg, E. P., et al. (2016). LasR variant cystic fibrosis isolates reveal an adaptable quorum-sensing hierarchy in Pseudomonas aeruginosa. mBio 7:e01513. doi: 10.1128/mBio.01513-16
Feng, X., Kostylev, M., Dandekar, A. A., and Greenberg, E. P. (2019). Dynamics of cheater invasion in a cooperating population of Pseudomonas aeruginosa. Sci. Rep. 9:10190. doi: 10.1038/s41598-019-46651-5
Filloux, A. (2004). The underlying mechanisms of type II protein secretion. Biochim. Biophys. Acta 1694, 163–179. doi: 10.1016/j.bbamcr.2004.05.003
Filloux, A. (2011). Protein secretion systems in Pseudomonas aeruginosa: an essay on diversity, evolution, and function. Front. Microbiol. 2:155. doi: 10.3389/fmicb.2011.00155
Folkesson, A., Jelsbak, L., Yang, L., Johansen, H. K., Ciofu, O., Høiby, N., et al. (2012). Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat. Rev. Microbiol. 10, 841–851. doi: 10.1038/nrmicro2907
Hmelo, L. R., Borlee, B. R., Almblad, H., Love, M. E., Randall, T. E., Tseng, B. S., et al. (2015). Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat. Protoc. 10, 1820–1841. doi: 10.1038/nprot.2015.115
Hogardt, M., and Heesemann, J. (2010). Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int. J. Med. Microbiol. 300, 557–562. doi: 10.1016/j.ijmm.2010.08.008
Huang, H., Shao, X., Xie, Y., Wang, T., Zhang, Y., Wang, X., et al. (2019). An integrated genomic regulatory network of virulence-related transcriptional factors in Pseudomonas aeruginosa. Nat. Commun. 10:2931. doi: 10.1038/s41467-019-10778-w
Hwang, W., Yong, J. H., Min, K. B., Lee, K., Pascoe, B., Sheppard, S. K., et al. (2021). Genome-wide association study of signature genetic alterations among Pseudomonas aeruginosa cystic fibrosis isolates. PLoS Pathog. 17:e1009681. doi: 10.1371/journal.ppat.1009681
Kiratisin, P., Tucker, K. D., and Passador, L. (2002). LasR, a transcriptional activator of Pseudomonas aeruginosa virulence genes, functions as a multimer. J. Bacteriol. 184, 4912–4919. doi: 10.1128/JB.184.17.4912-4919.2002
Koh, A. Y., Priebe, G. P., and Pier, G. B. (2005). Virulence of Pseudomonas aeruginosa in a murine model of gastrointestinal colonization and dissemination in neutropenia. Infect. Immun. 73, 2262–2272. doi: 10.1128/IAI.73.4.2262-2272.2005
Korotkov, K. V., and Sandkvist, M. (2019). Architecture, function, and substrates of the type II secretion system. EcoSal Plus 8. doi: 10.1128/ecosalplus.ESP-0034-2018
Kostylev, M., Kim, D. Y., Smalley, N. E., Salukhe, I., Greenberg, E. P., and Dandekar, A. A. (2019). Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy. Proc. Natl. Acad. Sci. U.S.A. 116, 7027–7032. doi: 10.1073/pnas.1819796116
LaPointe, C. F., and Taylor, R. K. (2000). The type 4 prepilin peptidases comprise a novel family of aspartic acid proteases. J. Biol. Chem. 275, 1502–1510. doi: 10.1074/jbc.275.2.1502
Li, X., and Lee, J. (2019). Quorum sensing-dependent post-secretional activation of extracellular proteases in Pseudomonas aeruginosa. J. Biol. Chem. 294, 19635–19644. doi: 10.1074/jbc.RA119.011047
Liu, Y., Ahator, S. D., Wang, H., Feng, Q., Xu, Y., Li, C., et al. (2022). Microevolution of the mexT and lasR reinforces the bias of quorum sensing system in laboratory strains of Pseudomonas aeruginosa PAO1. Front. Microbiol. 13:821895. doi: 10.3389/fmicb.2022.821895
Lu, Y., Liu, Y., Zhou, C., Liu, Y., Long, Y., Lin, D., et al. (2022). Quorum sensing regulates heteroresistance in Pseudomonas aeruginosa. Front. Microbiol. 13:1017707. doi: 10.3389/fmicb.2022.1017707
Lyczak, J. B., Cannon, C. L., and Pier, G. B. (2000). Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2, 1051–1060. doi: 10.1016/s1286-4579(00)01259-4
Moradali, M. F., Ghods, S., and Rehm, B. H. A. (2017). Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front. Cell Infect. Microbiol. 7:39. doi: 10.3389/fcimb.2017.00039
O’Loughlin, C. T., Miller, L. C., Siryaporn, A., Drescher, K., Semmelhack, M. F., and Bassler, B. L. (2013). A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 110, 17981–17986. doi: 10.1073/pnas.1316981110
Qin, S., Xiao, W., Zhou, C., Pu, Q., Deng, X., Lan, L., et al. (2022). Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target Ther. 7:199. doi: 10.1038/s41392-022-01056-1
Sandoz, K. M., Mitzimberg, S. M., and Schuster, M. (2007). Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl. Acad. Sci. U.S.A. 104, 15876–15881. doi: 10.1073/pnas.0705653104
Schuster, M., Joseph Sexton, D., Diggle, S. P., and Peter Greenberg, E. (2013). Acyl-Homoserine lactone quorum sensing: from evolution to application. Annu. Rev. Microbiol. 67, 43–63. doi: 10.1146/annurev-micro-092412-155635
Schuster, M., Lostroh, C. P., Ogi, T., and Greenberg, E. P. (2003). Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185, 2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003
Seed, P. C., Passador, L., and Iglewski, B. H. (1995). Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J. Bacteriol. 177, 654–659. doi: 10.1128/jb.177.3.654-659.1995
Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964. doi: 10.1038/35023079
Swietnicki, W., Czarny, A., Antkowiak, L., Zaczynska, E., Kolodziejczak, M., Sycz, J., et al. (2019). Identification of a potent inhibitor of type II secretion system from Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 513, 688–693. doi: 10.1016/j.bbrc.2019.04.055
Tashiro, Y., Yawata, Y., Toyofuku, M., Uchiyama, H., and Nomura, N. (2013). Interspecies interaction between Pseudomonas aeruginosa and other microorganisms. Microbes Environ. 28, 13–24. doi: 10.1264/jsme2.me12167
Vidaillac, C., and Chotirmall, S. H. (2021). Pseudomonas aeruginosa in bronchiectasis: infection, inflammation, and therapies. Expert Rev. Resp. Med. 15, 649–662. doi: 10.1080/17476348.2021.1906225
Wang, Y., Gao, L., Rao, X., Wang, J., Yu, H., Jiang, J., et al. (2018). Characterization of lasR-deficient clinical isolates of Pseudomonas aeruginosa. Sci. Rep. 8:13344. doi: 10.1038/s41598-018-30813-y
Wang, M., Schaefer, A. L., Dandekar, A. A., and Greenberg, E. P. (2015). Quorum sensing and policing of Pseudomonas aeruginosa social cheaters. Proc. Natl. Acad. Sci. U.S.A. 112, 2187–2191. doi: 10.1073/pnas.1500704112
Willcox, M. D. P., Zhu, H., Conibear, T. C. R., Hume, E. B. H., Givskov, M., Kjelleberg, S., et al. (2008). Role of quorum sensing by Pseudomonas aeruginosa in microbial keratitis and cystic fibrosis. Microbiology 154, 2184–2194. doi: 10.1099/mic.0.2008/019281-0
Keywords: microevolution, bacterial evolution, virulence factors, Pseudomonas aeruginosa , quorum sensing, pathogenicity traits
Citation: Cheng M, Chen R and Liao L (2023) T2SS-peptidase XcpA associated with LasR evolutional phenotypic variations provides a fitness advantage to Pseudomonas aeruginosa PAO1. Front. Microbiol. 14:1256785. doi: 10.3389/fmicb.2023.1256785
Edited by:
Nityananda Chowdhury, Medical University of South Carolina, United StatesReviewed by:
Karishma Bisht, Princeton University, United StatesAnton Hartmann, Ludwig Maximilian University of Munich, Germany
Copyright © 2023 Cheng, Chen and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruiyi Chen, Y2hlbnJlZUB1dy5lZHU=; Lisheng Liao, bGlzaGVuZ2xpYW9Ac2NhdS5lZHUuY24=
 Mengmeng Cheng
Mengmeng Cheng Ruiyi Chen
Ruiyi Chen Lisheng Liao
Lisheng Liao