
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 08 September 2023
Sec. Infectious Agents and Disease
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1255716
This article is part of the Research Topic Reviews in Molecular Evolution of Infectious Agents and Diseases View all 3 articles
It is vital to diagnose pathogens quickly and effectively in the research and treatment of disease. Argonaute (Ago) proteins are recently discovered nucleases with nucleic acid shearing activity that exhibit specific recognition properties beyond CRISPR–Cas nucleases, which are highly researched but restricted PAM sequence recognition. Therefore, research on Ago protein-mediated nucleic acid detection technology has attracted significant attention from researchers in recent years. Using Ago proteins in developing nucleic acid detection platforms can enable efficient, convenient, and rapid nucleic acid detection and pathogen diagnosis, which is of great importance for human life and health and technological development. In this article, we introduce the structure and function of Argonaute proteins and discuss the latest advances in their use in nucleic acid detection.
Nucleic acid detection technology is an essential tool for molecular diagnosis, with crucial applications in disease diagnosis, treatment, and biosecurity. Thus, it is vital to develop nucleic acid detection technology that is simple to use, accurate, specific, and cost-effective. Currently, commonly employed diagnostic methods include polymerase chain reaction (PCR), antigen–antibody reactions, and tissue biopsies. However, PCR, the most widely used nucleic acid amplification technique, faces challenges with nonspecific amplification. To improve the precision of nucleic acid detection, innovative approaches are needed. The discovery and application of programmable nucleases, such as zinc-finger nucleases (ZFNs) (Wu et al., 2007; Shamshirgaran et al., 2022), transcription activator-like effector nucleases (TALENs) (Zhang et al., 2019), and clustered regularly interspaced short palindromic repeat systems (CRISPR-associated nuclease) (Mojica et al., 2009), have revolutionized diagnostic techniques.
Zinc finger nucleases (ZFNs) are the first generation of programmable restriction endonucleases and consist of a zinc finger DNA recognition binding part formed by multiple zinc finger units in tandem and a DNA cleavage part of the restriction endonuclease Fok I. They can generate a cut at a specific site by recognizing a particular sequence of DNA, which is then repaired using the cell’s inherent homologous recombination repair mechanism (Wu et al., 2007). Transcription activator-like effector nucleases (TALENs) are proteases inspired by ZFNs. Researchers have replaced the zinc finger recognition binding site with a transcription activator effector protein (TALE) to achieve the same recognition cleavage function (Zhang et al., 2019). Clustered regularly interspaced short palindromic repeats (CRISPR) and their associated proteins (Cas) can be recognized by PAM sequences, which are recognized and then inserted between the leader sequence and the interspaced repeats and subsequently transcribed into crRNA, which forms a complex with the Cas protein to recognize and act as a cleavage (Mojica et al., 2009; Li et al., 2019). CRISPR-Cas systems have evolved rapidly in recent years (Wang et al., 2020) and have been used in sensitive and specific pathogen nucleic acid detection and gene editing, with the development of nucleic acid detection platforms such as Cas12-DETECTOR (Broughton et al., 2020), Cas14-DETECTOR (Aquino-Jarquin, 2019), Cas9-FLASH (Kellner et al., 2019; Quan et al., 2019) and Cas13-SHERLOCK (Aman et al., 2020). SHERLOCK detection is highly specific for single nucleotide mutations and has been shown to discriminate between Zika virus SNPs (Pardee et al., 2016). Proteins such as Cas12 and Cas13 have also been used in viral detection for SARS-CoV-2 (Ding et al., 2020; Marais et al., 2020; Rabe and Cepko, 2020; Wozniak et al., 2020; Nouri et al., 2021). Apart from several programmable nucleases described above, there are several other nucleases. For example, because of their extremely specific DNA identification and nucleic acid endonucleating activity, meganucleases that are found in cellular mitochondria and chloroplasts have been developed into artificial nucleases (Maeder and Gersbach, 2016; Khalil, 2020). Besides, nucleases like S1 nuclease (Filer et al., 2019) can be utilized in nucleic acid assays to safeguard the target nucleic acid, break down unbound probes, and other heterogeneous nucleic acids. Nucleases can also be involved in the repair of clip mismatches (Tsutakawa et al., 2014). Furthermore, nucleases, such as DNAzyme (He et al., 2014), Exo III (Xu et al., 2017), can be involved in the reaction as tools for auxiliary signal amplification (Gerasimova and Kolpashchikov, 2014; Hong et al., 2017). Nucleases not only appear as a tool in the detection process, but can also be viewed as the object under detection. They have the potential to be used as a biomarker for the detection and diagnosis of clinical infections due to changes in their catalytic capacity and preferred substrate (Garcia Gonzalez and Hernandez, 2022). The recently discovered Argonaute protein (Ago) is also a class of nucleic acid endonucleases that use nucleic acids as guides and has been heavily investigated by researchers to develop more efficient diagnostic platforms for nucleic acid detection.
Argonaute proteins were first mentioned in a study of Arabidopsis mutants in 1998 and named Argonaute because of the curly leaves of the mutant plants, which resemble the tentacles of a squid (Bohmert et al., 1998). Ago protein family members are widely distributed in various organisms and can be classified according to their origin as eukaryotic Ago proteins (eAgos) and prokaryotic Ago proteins (pAgos). In general, eAgos are more structurally uniform and exhibit high conservation that only mediates RNA-directed RNA interference. In contrast, pAgos exhibit more structural and functional diversity (Sheng et al., 2014). And some bacteria and archaea Argonaute proteins can participate in host defense processes by interfering with foreign nucleic acid invasion, such as against DNA viruses and foreign plasmids (Swarts et al., 2014).
The Ago protein family is broadly similar in structure (Figure 1). pAgos can be divided into three categories according to their structural length: long pAgos, short pAgos, and PIWI-RE proteins (prokaryotic PIWI with only conserved R and E residues). The structure of long pAgos is very similar to that of eAgos and they both form a two-lobed scaffold (Kaya et al., 2016). The N-terminal lobe is connected with the PAZ domain by the L1 linker, and the C-terminal lobe is composed of the MID domain, the PIWI domain and the C-terminal. Finally, the L2 linker connects the two lobes. The structures of the short pAgos and PIWI-RE proteins are similar to that of the long pAgos at the C-terminal, except that the N-terminal end is more straightforward, without the PAZ structural domain and the L1 and L2 linker connections (Kaya et al., 2016; Sheng et al., 2017; Lisitskaya et al., 2018).

Figure 1. Schematic diagram of the structure of the Ago protein. (A) Linear representations of Ago protein structures. (B) Schematic structures of KpAgo (eAgo) and AaAgo (pAgo), including N (blue), L1 linker (purple), PAZ (green), L2 linker (purple), MID (orange), PIWI (yellow).
It has been shown that the PAZ, MID, and PIWI domains are very important parts of Argonaute protein function. PAZ and MID domains form a binding pocket that anchors and binds to the 3′ and 5′ ends of the target strand, and the target is catalyzed by the PIWI domain for shearing (Song et al., 2004; Yuan et al., 2005). The PAZ domain contains conserved aromatic residues and forms a fold that guides and binds the target DNA/RNA fragments (Cao et al., 2019), but this domain is not necessarily required for the shearing properties of Argonaute proteins. A team found that AfAgo (Ago from Archaeoglobus fulgidus), which is a short pAgos, consists of an N-terminal, MID and PIWI structural domain. AfAgo without a PAZ structural domain can also recognize and bind through the channels formed by the MID and PIWI structural domains, and perhaps the absence of the PAZ domain has an impact on binding affinity (Yan et al., 2003; Yuan et al., 2005). The PIWI domain is structurally and functionally similar to ribonuclease H (RNase H). The PIWI domain is capable of exerting nucleic acid endonuclease activity to hydrolyze RNA phosphodiester bonds and specifically catalyze the cleavage of target genes (Hur et al., 2013). This catalytic activity is achieved through the DEDX (X = N, D, H) tetrameric structure (Figure 2A), such as the DEDD tetramer present in the PIWI structural domain of CpAgo (Ago from Clostridium perfringens) found in the study, and this cleavage activity was lost with the mutation at position D614 (Parker et al., 2005). In order to gain a comprehensive understanding of the genetic relationship and characteristics of well-studied pAgos, Figures 2A,B depict the conserved active sites and a phylogenetic tree of recently studied Argonaute proteins, respectively. A summary of the biochemical properties and structural information can be found in Table 1. The primary focus of research has been on bacterial and archaeal Argonautes, which exhibit nuclease activity and primarily recognize DNA targets, unlike their eukaryotic counterparts that utilize RNA guides and targets within cells. However, some prokaryotic Argonautes, such as PliAgo and PnyAgo, can also cleave RNA targets, albeit with lower efficiency (Lisitskaya et al., 2022). While most prokaryotic Argonautes bind DNA guides, a few, like MpAgo from Marinitoga piezophila and its closest homologs, show a preference for RNA guides (Kaya et al., 2016). The functional significance of this phenomenon and the specific structural features responsible for distinguishing RNA/DNA guides and targets in prokaryotic Argonautes remain unknown (Wang et al., 2008; Kropocheva et al., 2021). It is hypothesized that these features are primarily influenced by variations in the structures of the MID and PIWI domains. Further studies investigating the activity and structures of prokaryotic Argonaute proteins are necessary to address these questions.
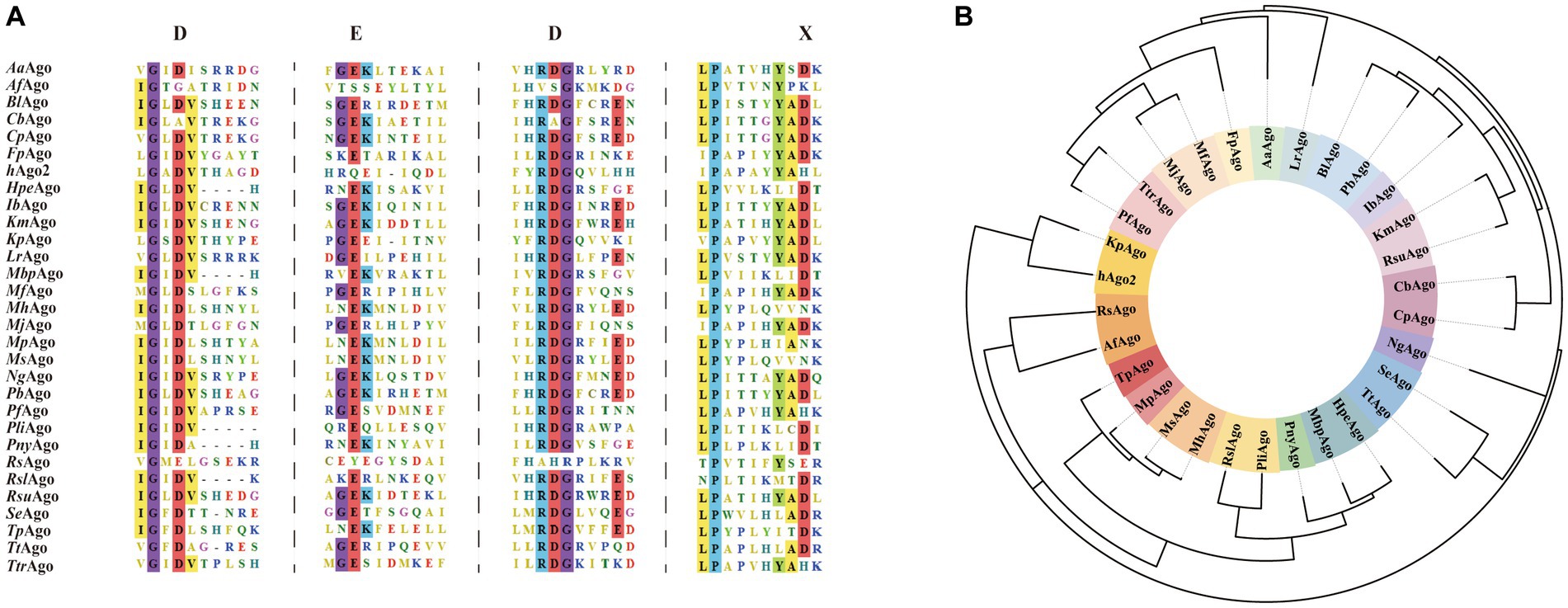
Figure 2. (A) Multiple sequence alignments of Argonaute proteins. The sequences include AaAgo (Aquifex aeolicus Argonaute, WP_010880937.1), AfAgo (Archaeoglobus fulgidus Argonaute, WP_010878815.1), BlAgo (Brevibacillus laterosporus Argonaute, WP_277546270.1), CbAgo (Clostridium butyricum Argonaute, WP_058142162.1), CpAgo (Clostridium perfringens Argonaute, WP_003477422.1), FpAgo (Ferroglobus placidus Argonaute, WP_012966655.1), hAgo2 (Homo sapiens Argonaute, NP_036286.2), HpeAgo (H. penzbergensis Argonaute, WP_139173808.1), IbAgo (Intestinibacter bartlettii Argonaute, WP_007287731.1), KmAgo (Kurthia massiliensis Argonaute, WP_010289662.1), KpAgo (Vanderwaltozyma polyspora Argonaute, XP_001644461.1), LrAgo (Limnothrix rosea Argonaute, WP_075892274.1), MbpAgo (Mucilaginibacter paludism Argonaute, WP 008504757.1), MfAgo (Methanocaldococcus fervens Argonaute, WP_015791216.1), MhAgo (Marinitoga hydrogenitolerans Argonaute, UniProtKBSwiss-Prot A0A1M5A5Z8.1), MjAgo (Methanocaldococcus jannaschii Argonaute, WP_010870838.1), MpAgo (Marinitoga piezophila Argonaute, WP_014295921.1), MsAgo (Marinitoga sp. 1155 Argonaute, WP_047265940.1), NgAgo (Natronobacterium gregoryi Argonaute, WP_005580376.1), PbAgo (Paenibacillus borealis Argonaute, WP_042211195.1), PfAgo (Pyrococcus furiosus Argonaute, WP_011011654.1), PliAgo (Pseudooceanicola lipolyticus pAgo, WP_100161590.1), PnyAgo (Pedobacter nyackensis Argonaute, WP_084286803.1), RsAgo (Rhodobacter sphaeroides Argonaute, UniProtKB A4WYU7), RslAgo (R. slithyformis Argonaute, WP_013921749.1), RsuAgo (Rummeliibacillus suwonensis Argonaute, WP_146547607.1), SeAgo (Synechococcus elongatus Argonaute, WP_011244830.1), TpAgo (Thermotoga profunda Argonaute, WP_041081268.1), TtAgo (Thermus thermophilus Argonaute, WP_011174533.1), TtrAgo (Thermococcus thioreducens Argonaute, WP_055429304.1). The catalytic tetrad DEDX of Argonautes is highlighted red in the sequence alignment. (B) Schematic phylogenetic tree of above described Argonaute proteins.
The Ago protein is also found to play an essential role in the formation of the RNA silencing complex (RISC), as a critical protein in the silencing pathway, and is bound to another class of glycine- and tryptophan-rich (GW/WG) proteins known as the “Ago hook,” which are involved in maintaining the maintenance of genomic stability (Zander et al., 2014), such as RsAgo (Ago from Rhodobacter sphaeroides), which was discovered to bind to foreign nucleic acids and thus perform host defense (Hegge et al., 2019).
While polymerase chain reaction (PCR) has long reigned in molecular diagnostics in recent years, people have urgent demands, such as the SARS-CoV-2 pandemic, for newer technologies that can provide more direct and convenient detection methods. This inspires researchers to explore new molecular diagnostic methods. Researchers have combined CRISPR-Cas or Argonaute proteins with biosensing technologies such as fluorescence and microfluidic control devices to develop new nucleic acid detection technologies (Santiago-Frangos et al., 2022). Nucleic acid biosensing systems such as DETECTR and HOLMES have been established based on CRISPR-Cas12a and CRISPR-Cas9 to meet various clinical testing requirements (Jiang and Doudna, 2017; He et al., 2019). At the same time, these new biosensing platforms also show good sensitivity and specificity. Similar to Cas proteins, Argonaute proteins can recognize and cleave target sequences under the guidance of gDNA. However, this kind of protein requires no additional specific sequences (such as PAM sequences) to recognize the target sequences, and the proteins are widely found in a variety of organisms (Kropocheva et al., 2022). Based on this, researchers have developed a range of nucleic acid detection platforms.
PfAgo (Pyrococcus furiosus Argonaute), a DNA-guided nucleic acid endonuclease derived from thermophilic archaea with an optimal temperature between 80 and 100°C, is the first Ago protein with a complete three-dimensional structure. It is reckoned that PfAgo uses the 15–31 nt 5′ phosphorylation of siDNAs to shear ssDNA. The optimal temperature for this shearing activity is 87–99°C. The reaction requires the involvement of Mn2+, Mg2+, and preferably the NaCl concentration of 50–250 mM (Swarts et al., 2015; He et al., 2021).
A research team established a novel nucleic acid detection platform named PAND (PfAgo-mediated Nucleic acid Detection) (He et al., 2019). The platform combines PCR with PfAgo, which has the DNA-targeted nucleic acid shearing property, to enable multiplexed nucleic acid detection of DNA. The recreation uses the three given gDNA sequences to instruct PfAgo to perform nucleic acid shearing, which has shown high specificity for clinical samples. In 2021, Wang’s team used the PAND platform in the detection of SARS-CoV-2 (Wang et al., 2021), and recently, Chen’s team used it in the detection of Parvovirus B19 (B19V) (Chen et al., 2023). Based on the PAND detection platform, researchers have chosen to build a PLCR detection platform by combining ligase chain reaction (LCR) with PfAgo, increasing the specificity for distinguishing single base mutations in gene sequences and requiring only two temperature shifts to complete the amplification cycle (Wang et al., 2021). Compared to PAND, PLCR uses a modified LCR product for gDNA, rather than adding additional gDNA after the target amplification, which also greatly simplifies the equipment requirements. It is noteworthy that amplification of non-target sequences should be avoided by controlling the number of cycles and Tm value conditions. LCR reactions are usually performed under conditions such as 30 cycles and oligonucleotides with Tm values close to 50°C, which avoid amplification independent of the template. This platform can reach detection limits of 10 aM in 70 min and 1 aM in 100 min. To some extent, this method achieves the accuracy comparable to that of qPCR in a much shorter time, enabling susceptible, multichannel, precise single-base mutant detection (Liu et al., 2021; Wang et al., 2021).
Based on the high activity exhibited by PfAgo at 95°C, this nucleic acid biosensing platform was combined with usPCR to establish a USPCRP platform. This method amplifies the target nucleic acid in the sample by PCR to obtain the appropriate guide DNA, thus reducing the time and human resources from designing the gDNA. This also allows the USPCRP system to show higher sensitivity, up to 10 aM, and high specific single-base resolution (He et al., 2019, 2021). In order to reduce the annealing temperature and the cycle time, the length of the double-stranded DNA product should be designed to be 22–28 bp, and two primers need to be designed: prime-g and prime-h. Primer-g is phosphorylated at the 5′ end and is used to generate the guide DNA from the target DNA. Primer-h is the conventional primer for higher yields. Surprisingly, the study mapped the temperature and time of denaturation and annealing in PCR and found that at a temperature combination of 70°C/40°C, substantial yields of guide DNA could be obtained in 30 min at a time as short as 1 s, which yielded a substantial time benefit (He et al., 2021).
At the same time, another one-pot multiplex detection platform, MULAN (Multiplex Argonaute-based Nucleic acid detection system), also based on PfAgo, was developed. It is a platform that integrates the amplification-cleavage-assay steps in a single platform for portable, sealed-tube operation, avoiding the risk of splitting and contamination. The specific cleavage properties of the PfAgo protein are exploited to increase the amplification product and fluorescence signal by designing two corresponding gDNAs for the amplification target. Primary cleavage of one strand of the gDNA generates a new gDNA that directs secondary specific division with different fluorescence modifications. The presence of the target nucleic acid sequence is determined by detecting the fluorescence signal from various channels. In addition, this single-tube system contains a separate space that allows amplification products to be mixed with PfAgo, avoiding contamination from exposure to the external environment. This mini isothermal fluorescence detector (MIFD) design makes the whole operation more convenient, providing a new avenue for POCT. More notably, this device enables the simultaneous detection of multiple viruses by loading three sets of primers, gDNA, and fluorescent agents designed for SARS-CoV-2, Influenza A virus, and Influenza B virus multiplexes into a single jar of the detection system. In the absence of observed cross-reactivity, the MULAN system can specifically produce fluorescent signals for the respective targets, and no dissimilar signs are observed for viruses that are not present (Li et al., 2022; Ye et al., 2022). This result indicates that the MULAN system can achieve accurate viral multiplex detection in the presence of a single Ago protease. The schematic of MULAN、PLCR and USPCRP system was shown in Figure 3. CRISPR detection systems have also been successful in detecting different targets in a single reaction, but each requires an extra Cas protein. This reflects the great promise of Ago proteins in the development of nucleic acid detection technologies (Gootenberg et al., 2018; Myhrvold et al., 2018; Ooi et al., 2021). Recently, a team studying Enterocytozoon hepatopenaei (EHP) combined PfAgo with recombinase polymerase amplification (RPA) to establish a method for the detection of pathogenic bacteria. This method has a satisfactory performance in that it does not require strict laboratory conditions, which provides a new method for the multiplex identification of pathogenic bacteria (Yang et al., 2023).
In addition to developing PfAgo for use in nucleic acid detection platforms, other Ago proteins are being increasingly exploited. Mengjun Fang’s team mined a new Argonaute from the archaeon Thermococcus thioreducens (TtrAgo) and used it for the detection of HBV DNA. Another TtAgo-based platform named NAVIGATER (Nucleic Acids of Clinical Interest Via DNA-Guided Argonaute from Thermus thermophilus) has been developed and used in the detection of single nucleotide mutations by designing a guide complementary to the predominant wild-type nucleic acid sequence in the sample (Song et al., 2020). The nuclease activity of TtAgo declines when a rare mutation occurs at position 10 or 11 of the target nucleic acid. The researchers designed a guide sequence complementary to the predominant wild-type nucleic acid sequence in the sample. The number of mutants increases when detected and the wild-type target nucleic acid sequence is cleaved by Argonaute, allowing the detection of the presence of mutants. Then SNVs were enriched by PCR amplification, and this method can specifically enrich multiple mutant alleles in a single sample. Compared to previously reported rare allele enrichment methods, NAVIGATER exhibits great strengths. For example, TtAgo behaves flexibly because it does not require a PAM motif or a specific recognition site and a single TtAgo-guide complex can cleave several targets (Sternberg et al., 2014; Swarts et al., 2014).
During RNA detection, it is also possible to link PfAgo to the ligase chain reaction (RT-LCR), which requires an additional step in reverse transcription during the reaction of PLCR. However, this method does not require RNA for guidance, avoids the instability and higher costs associated with RNA, and allows for multichannel detection. In a trial of clinical samples, HPV genomic DNA from a cervical swab and SARS-CoV-2 genomic RNA from a throat swab were tested in separeate channels. The N gene and the ORF 1ab gene of SARS-CoV-2 and HPV subtypes were tested separately, and it has been found that this method could achieve high sensitivity for DNA or RNA with a sensitivity of up to 10 aM in 70 min. Although absolute quantification is not yet possible, the advantages of sensitivity, specificity, and high channelization are already considerable (Wang et al., 2021).
A team has developed an isothermal nucleic acid detection platform for RNA using the nucleic acid detection properties of the pAgo protein in combination with a reverse transcription reaction, named MAIDEN (Mesophilic Ago-based Isothermal Detection method) (Ye et al., 2022). The target nucleic acid sequence is reverse transcribed by reverse transcriptase. The RNA in the DNA–RNA hybrid strand produced by reverse transcription is hydrolyzed with RNase H, and the resulting ssDNA is used for recognition. The secondary gDNA is produced by reacting the designed primary gDNA with the Ago-gDNA complex. The ssDNA is cleaved by recognition of the Ago-secondary gDNA complex. The experiments showed that the reverse transcription reaction could be simplified with RNase H hydrolysis digestion. The reverse transcription process could be carried out simultaneously with the Ago stepwise cleavage reaction under the same moderate temperature conditions, which further simplified the reaction process. KmAgo from the thermophilic bacterium Kurthia massiliensis, combined with a 16–20 nt long 5′-phosphorylation-guided nucleic acid sequence capable of specific cleavage of DNA/RNA target sequences under DNA/RNA guidance (Kropocheva et al., 2021). Recently, researchers have used KmAgo (Ago from Kurthia massiliensis) and PbAgo (Ago from Paenibacillus borealis) in this detection system and have achieved entirely satisfactory detection limits in a one-pot assay, with detection limits of 4.0 nM for KmAgo and 12.5 nM for PbAgo (Li et al., 2022). However, compared to thermophilic PfAgo, the activity of mesophilic KmAgo is low and a large number of enzymes need to be added in the experiment. In the future, it is necessary to apply a protein engineering strategy to improve the activity of mesophilic KmAgo at moderate temperatures to improve the sensitivity of mesophilic Ago-based detection.
Combining PfAgo with reverse transcriptase-based isothermal amplification (RT-LAMP) has resulted in a rapid, portable assay system (SPOT) (Xun et al., 2021). This system consists of an assay system capable of precisely controlled temperature and fluorescence detection and a device capable of a mobile power supply. The system involves optimizing the GC content and length of the reagents to reduce unwanted background fluorescence and separates the LAMP and PfAgo reaction components with paraffin wax. This convenient and all-in-one assay system offers a new approach to responding to and deploying large-scale epidemics and does not require a large number of specialist personnel, requiring only simple sample pre-processing to operate. Changjing Yuan’s team developed an isothermal amplification detection platform based on TtAgo, called the Thermus thermophilus Argonaute-based thermostable exponential amplification reaction (TtAgoEAR), which has shown rewarding reliability and sensitivity for the detection of viral RNA, lncRNA, and mRNA in various specimens (Yuan et al., 2023). This platform can detect RNA targets at 66°C with attomolar sensitivity and single-nucleotide resolution.
Although researchers have invented various nucleic acid detection platforms, the overall principle is similar, as shown in Figure 4. These methods are based on the classical two-step cleavage method of Argonaute. The corresponding primary guide is designed according to the target DNA, and the target is cut by the Argonaute protein to obtain the secondary guide. Then the designed reporter is cut by the Argonaute protein under the guidance of a secondary guide to obtain a fluorescence signal, and in this process, PCR or isothermal amplification can be combined to improve the detection sensitivity. In a study of foodborne pathogens, researchers provided suggestions for developing a one-step cleavage assay called a Novel and One-step cleavage method based on Argonaute by integrating Tag-specific primer extension and Exonuclease I (Exo I) (NOTE-Ago) (Li et al., 2023). The amplicons were served as the guide DNA for PfAgo. Consequently, the fluorophore-quencher reporter can be cleaved by PfAgo, leading to alterations in fluorescent intensity. Notably, Argonaute 2 (Ago2) serves as a crucial constituent within the RNA-induced silencing complex (RISC), contributing substantively to a spectrum of physiological phenomena. Conversely, aberrant modulation of Ago2 functionality exhibits a strong association with an array of human maladies, encompassing malignancies. Elucidating the actions of Argonaute 2 has prompted scholars to devise diverse techniques aimed at discerning its operations. These methodologies encompass the utilization of a dual signal amplification-assisted DNAzyme biosensor and a singular-molecule biosensor featuring gold nanoparticles. These innovative approaches have been devised with the intent of diagnosing pathologies linked to Ago2, thereby unveiling promising avenues for extensive clinical applications (Zhang et al., 2018; Wang et al., 2020; Jiang et al., 2022; Zhao et al., 2023). Table 2 provides a comparison of various CRISPR-Cas and Ago-based assays. The SHERLOCK methodology demonstrated proficiency in identifying the Zika virus (ZIKV) and dengue virus (DENV). This accomplishment resulted in a remarkable level of sensitivity at the attomolar scale (10−18 M) along with the capacity to discern single-base distinctions. The maturity of this technique is highly evident, as evidenced by its extensive application in the detection of pathogenic bacteria (Gootenberg et al., 2017). It is evident that the nucleic acid detection system utilizing Ago protein is comparable to CRISPR in terms of sensitivity. However, its notable advantage lies in not requiring an additional PAM sequence, making it highly applicable. Furthermore, it is worth noting that the majority of detection platforms rely on thermophilic Ago proteins (PfAgo and TtAgo). While these proteins offer high detection sensitivity, they necessitate the use of PCR or other heating equipment, thereby restricting their portability. Therefore, the CRISPR based detection system is more widely used because it can be detected at room temperature. Consequently, there is significant potential in developing nucleic acid detection technology based on mesophilic Ago proteins, as it would provide a highly sensitive alternative without the limitations of requiring extensive heating equipment.
By establishing various Argonaute-based nucleic acid detection platforms, we can (Wu et al., 2007) identify disease-causing pathogens for disease diagnosis and (Shamshirgaran et al., 2022) perform single-base mutation detection for pathogen identification and typing (Zhang et al., 2019). Single base mutation testing in human genomic DNA to predict the probability of disease by predicting the risk of single-base mutations (Mojica et al., 2009). Multiplex nucleic acid testing for use in clinical testing. This is based on the fact that argonaute can be used to cut arbitrary nucleic acid sequences to obtain “sticky” ends of the desired length and nucleotide composition compared to traditional restriction endonucleases. In comparison to CRISPR-Cas9, Ago protein functions as a nucleic acid enzyme capable of cleaving target sites. Unlike CRISPR-Cas9, Ago protein is not restricted by the presence of a protospacer adjacent motif (PAM) sequence. By meticulous design of the nucleic acid guiding strand, precise cleavage of arbitrary target nucleic acids can be achieved. Typically utilizing shorter lengths of genomic DNA (ranging from 15 to 24 nucleotides), the synthesis of short gDNA is more cost-effective than longer crRNA. This may potentially facilitate high-throughput guide RNA production, thereby enabling efficient high-throughput genome editing screening. Moreover, the size of pAgo is approximately 75–85 kDa, approximately half the size of Cas9 and Cas12a, which could enhance the efficient delivery of pAgo to the desired host. This provides excellent conditions for its more comprehensive application (He et al., 2019; Kropocheva et al., 2022). In addition, both MpAgo (Marinitoga piezophila Argonaute) and KmAgo can be used to probe the high-level structure of RNA. The Ago protein uses RNA as a guide and binds complementary target RNA. The target sequence recognized by nucleotide-specific sites, which connects structured RNA to guides loaded with different sequence sites of argonautes, is incubated. Cleavage sites can be detected either directly or by reverse transcription, and differences in the cleavage efficiency of single and double-stranded RNAs can be used to study the high-level structure of RNA (Schirle et al., 2014; Lapinaite et al., 2018; Kropocheva et al., 2021; Liu et al., 2021).
Recently, DNA-PAINT (DNA-Point Accumulation In Nanoscale Topology) applies Argonaute proteins to super-resolution microscopy, allowing structures to be observed beyond the microscopic fraction (Filius et al., 2020). This super-resolution method relies on the binding and unbinding of the DNA imaging strand, and the key to this technique is the rate of DNA binding. Generally, the binding rate of this technique is 106 M−1 s−1, and it takes several hours to obtain images with a high spatial resolution (5 nm). In contrast, Argonaute proteins can pre-arrange the target region into a helical conformation (Chandradoss et al., 2015; Yao et al., 2015), allowing a double helix to form between the guide nucleic acid and the target nucleic acid sequence, resulting in a binding rate close to approximately 107 M−1 s−1. CbAgo (Clostridium butyricum Argonaute) is employed in this technique. Its well-targeted properties allow Ago-PAINT to generate super-resolution images of diffraction-limited structures as much as 10 times faster than conventional DNA-PAINT, which provides a more favorable tool for visualizing cellular networks during the study of disease development (Cui et al., 2019; Hegge et al., 2019; Filius et al., 2020). In addition, a research team has applied Ago protein to detect microRNAs (miRNAs) and developed a highly reliable miRNA profiling technique, named Ago-FISH (Argonaute-based Fluorescence In Situ Hybridization) (Shin et al., 2020). TtAgo, which is more stable under experimental conditions, is preloaded with DNA probes. This method accelerates the targeted binding of DNA probes, maintaining a high specificity while significantly improving the speed of miRNA detection. Similarly, some researchers have used Ago proteins for dynamic observation of transcriptional processes in vivo. By fluorescently labeling the Ago protein NRDE-3, the dsRNA was programmed to observe the transcriptional process in the organism. This method avoids the in vitro synthesis of antisense RNA and transfer, which makes the whole observation process faster and easier, and provides a new idea for transcriptome engineering (Toudji-Zouaz et al., 2021).
Argonaute proteins also play an essential role in maintaining homeostatic mechanisms with RNA interference. It has been shown that when viral infection occurs in eukaryotes, siRNA can form RNA-induced silencing complexes (RISC) with Ago proteins and direct the degradation of the associated viral nucleic acids to maintain homeostasis in the organism (Lisitskaya et al., 2018; Zhang et al., 2022). This provides new ideas for studying the progress and treatment of diseases.
Argonaute proteins still have a long way to go from nucleic acid detection technology to a full-fledged molecular diagnostic method. Although some countries have granted emergency use authorizations and Ago proteins have been rapidly developed for use as a product in molecular diagnostics, their future development will continue to require new explorations (Qin et al., 2022). In addition, the activity and stability of the same Ago protein vary in different experiments and must be further investigated and standardized. Detection limits, intra-experimental reproducibility, clinical sensitivity, and diagnostic specificity in the report for nucleic acid testing platforms involving Ago proteins must be further standardized. Currently, fewer Ago proteins can be used to develop nucleic acid detection platforms, and researchers need to mine more Ago proteins with nucleic acid endonuclease activity that can be used in actual detection systems. In recent years, mesophilic pAgos with DNA cleavage activity have been mined, including Cb (Clostridium butyricum) Ago (Hegge et al., 2019; Kuzmenko et al., 2019), Lr (Limnothrix rosea) Ago (Kuzmenko et al., 2019), Se (Syne-chococcuselongatus) Ago (Olina et al., 2020), Km (Kurthia massiliensis) Ago (Kropocheva et al., 2021; Liu et al., 2021), and pAgos with RNA shearing activity include Tt (Thermus thermophilus) Ago, Mp (Marinitoga piezophile) Ago, Cp (Clostridium perfringens) Ago (Cao et al., 2019), and Km (Kurthia massiliensis) Ago (Wang et al., 2008). It has recently been shown that Mbp (Mucilaginibacter paludism) Ago exhibits RNA cleavage properties under a wide range of temperature conditions and is active against both 14–21 nt 5’P-gDNA and 15–18 nt 5’OH-gDNA, which is similar to the previously discovered KmAgo protein that cleaves not only RNA targets but also DNA targets precisely (Liu et al., 2021). This broadly targeted Ago protein provides a new idea to develop a platform for nucleic acid detection with a broader range of applications (Li et al., 2022).
Hence, it is postulated that an amalgamation of diverse fields encompassing the Argonaute system, engineering, microelectronics, and miniaturization holds the potential to amplify both detection throughput and automation. An illustrative instance of this is the isothermal amplification, which stands as a transformative advancement compared to the conventional polymerase chain reaction (PCR), offering augmented adaptability and an innovative array of methodologies for on-site detection applications. Furthermore, the combination of isothermal amplification with the latest CRISPR Cas13 collateral cleavage technology profoundly enhances the precision and rapidity of detection. However, the cleavage activity of mesophilic Ago is much lower than that of thermophilic Ago. Therefore, in order to improve the sensitivity of mesophilic, protein engineering strategy to generate an evolved Ago with higher activity at moderate temperature is necessary. At the same time, the cleavage efficiency and accuracy of different Ago proteases depend on temperature, divalent ions, gDNA terminal phosphorylation, and length. Therefore, we should pay attention to these influencing factors when developing new reliable nucleic acid detection platforms to achieve higher sensitivity and accuracy of nucleic acid detection. The Argonaute protein has shown great potential as a new programmable nucleic acid binding protein to participate in the next generation of molecular diagnostic technologies. Continued innovative research will bring its function to a greater extent.
ZW: Conceptualization, Writing – original draft, Writing – review & editing, Funding acquisition. LY: Formal analysis, Writing – original draft. WS: Funding acquisition, Supervision, Writing – review & editing. JM: Supervision, Writing – review & editing.
This work was supported by grants from Jiangsu Innovative and Entrepreneurial Talent Programme (JSSCBS20211594) and the Changzhou Science and Technology Project (Applied Based Research, No. CJ20220102).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aman, R., Mahas, A., and Mahfouz, M. (2020). Nucleic acid detection using CRISPR/Cas biosensing technologies. ACS Synth. Biol. 9, 1226–1233. doi: 10.1021/acssynbio.9b00507
Aquino-Jarquin, G. (2019). CRISPR-Cas 14 is now part of the artillery for gene editing and molecular diagnostic. Nanomedicine 18, 428–431. doi: 10.1016/j.nano.2019.03.006
Bohmert, K., Camus, I., Bellini, C., Bouchez, D., Caboche, M., and Benning, C. (1998). AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17, 170–180. doi: 10.1093/emboj/17.1.170
Broughton, J. P., Deng, X., Yu, G., Fasching, C. L., Servellita, V., Singh, J., et al. (2020). CRISPR-Cas 12-based detection of SARS-CoV-2. Nat. Biotechnol. 38, 870–874. doi: 10.1038/s41587-020-0513-4
Cao, P., Cao, D., Si, L., Su, X., Tian, L., Chang, W., et al. (2020). Structural basis for energy and electron transfer of the photosystem I-Isi A-flavodoxin supercomplex. Nat. Plants 6, 167–176. doi: 10.1038/s41477-020-0593-7
Cao, Y., Sun, W., Wang, J., Sheng, G., Xiang, G., Zhang, T., et al. (2019). Argonaute proteins from human gastrointestinal bacteria catalyze DNA-guided cleavage of single- and double-stranded DNA at 37 °C. Cell Discov. 5:38. doi: 10.1038/s41421-019-0105-y
Chandradoss, S. D., Schirle, N. T., Szczepaniak, M., Mac Rae, I. J., and Joo, C. (2015). A dynamic search process underlies micro RNA targeting. Cells 162, 96–107. doi: 10.1016/j.cell.2015.06.032
Chen, W., Qiu, L., Luo, T., Lu, Z., Wang, X., Hong, Q., et al. (2023). Novel nucleic acid detection for human parvovirus B19 based on Pyrococcus furiosus Argonaute protein. Viruses 15:595. doi: 10.3390/v15030595
Cui, T. J., Klein, M., Hegge, J. W., Chandradoss, S. D., van der Oost, J., Depken, M., et al. (2019). Argonaute bypasses cellular obstacles without hindrance during target search. Nat. Commun. 10:4390. doi: 10.1038/s41467-019-12415-y
Ding, X., Yin, K., Li, Z., Lalla, R. V., Ballesteros, E., Sfeir, M. M., et al. (2020). Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas 12a assay. Nat. Commun. 11:4711. doi: 10.1038/s41467-020-18575-6
Dong, H., Huang, F., Guo, X., Xu, X., Liu, Q., Li, X., et al. (2021). Characterization of Argonaute nucleases from mesophilic bacteria Paenibacillus borealis and Brevibacillus laterosporus. Bioresour. Bioprocess. 8, 1–12. doi: 10.1186/s40643-021-00478-z
Doxzen, K. W., and Doudna, J. A. (2017). DNA recognition by an RNA-guided bacterial Argonaute. PLoS One 12:e0177097. doi: 10.1371/journal.pone.0177097
Fang, M., Xu, Z., Huang, D., Naeem, M., Zhu, X., and Xu, Z. (2022). Characterization and application of a thermophilic Argonaute from archaeon Thermococcus thioreducens. Biotechnol. Bioeng. 119, 2388–2398. doi: 10.1002/bit.28153
Filer, J. E., Channon, R. B., Henry, C. S., and Geiss, B. J. (2019). A nuclease protection ELISA assay for colorimetric and electrochemical detection of nucleic acids. Anal. Methods 11, 1027–1034. doi: 10.1039/C8AY02729C
Filius, M., Cui, T. J., Ananth, A. N., Docter, M. W., Hegge, J. W., van der Oost, J., et al. (2020). High-speed super-resolution imaging using protein-assisted DNA-PAINT. Nano Lett. 20, 2264–2270. doi: 10.1021/acs.nanolett.9b04277
Garcia Gonzalez, J., and Hernandez, F. J. (2022). Nuclease activity: an exploitable biomarker in bacterial infections. Expert. Rev. Mol. Diagn. 22, 265–294. doi: 10.1080/14737159.2022.2049249
García-Quintans, N., Bowden, L., Berenguer, J., and Mencía, M. (2019). DNA interference by a mesophilic Argonaute protein, Cbc Ago. F1000Res 8:321. doi: 10.12688/f1000research.18445.1
Gerasimova, Y. V., and Kolpashchikov, D. M. (2014). Enzyme-assisted target recycling (EATR) for nucleic acid detection. Chem. Soc. Rev. 43, 6405–6438. doi: 10.1039/C4CS00083H
Gootenberg, J. S., Abudayyeh, O. O., Kellner, M. J., Joung, J., Collins, J. J., and Zhang, F. (2018). Multiplexed and portable nucleic acid detection platform with Cas 13, Cas 12a, and Csm 6. Science 360, 439–444. doi: 10.1126/science.aaq0179
Gootenberg, J. S., Abudayyeh, O. O., Lee, J. W., Essletzbichler, P., Dy, A. J., Joung, J., et al. (2017). Nucleic acid detection with CRISPR-Cas 13a/C2c2. Science 356, 438–442. doi: 10.1126/science.aam9321
Guo, X., Sun, Y., Chen, L., Huang, F., Liu, Q., and Feng, Y. (2021). A hyperthermophilic Argonaute from Ferroglobus placidus with specificity on guide binding pattern. Front. Microbiol. 12:654345. doi: 10.3389/fmicb.2021.654345
He, R., Wang, L., Wang, F., Li, W., Liu, Y., Li, A., et al. (2019). Pyrococcus furiosus Argonaute-mediated nucleic acid detection. Chem. Commun. 55, 13219–13222. doi: 10.1039/C9CC07339F
He, R., Wang, L., Wang, F., Yang, J., Yu, X., Wang, Y., et al. (2021). Combination of ultrashort PCR and Pyrococcus furiosus Argonaute for DNA detection. Analyst 147, 35–39. doi: 10.1039/D1AN01521D
He, J. L., Zhu, S. L., Wu, P., Li, P. P., Li, T., and Cao, Z. (2014). Enzymatic cascade based fluorescent DNAzyme machines for the ultrasensitive detection of Cu (II) ions. Biosens. Bioelectron. 60, 112–117. doi: 10.1016/j.bios.2014.03.065
Hegge, J. W., Swarts, D. C., Chandradoss, S. D., Cui, T. J., Kneppers, J., Jinek, M., et al. (2019). DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute. Nucleic Acids Res. 47, 5809–5821. doi: 10.1093/nar/gkz306
Hong, M., Wang, M., Wang, J., Xu, X., and Lin, Z. (2017). Ultrasensitive and selective electrochemical biosensor for detection of mercury (II) ions by nicking endonuclease-assisted target recycling and hybridization chain reaction signal amplification. Biosens. Bioelectron. 94, 19–23. doi: 10.1016/j.bios.2017.02.031
Hur, J. K., Zinchenko, M. K., Djuranovic, S., and Green, R. (2013). Regulation of Argonaute slicer activity by guide RNA 3′ end interactions with the N-terminal lobe. J. Biol. Chem. 288, 7829–7840. doi: 10.1074/jbc.M112.441030
Jiang, F., and Doudna, J. A. (2017). CRISPR-Cas 9 structures and mechanisms. Annu. Rev. Biophys. 46, 505–529. doi: 10.1146/annurev-biophys-062215-010822
Jiang, X., Liu, Y., Liu, Q., and Ma, L. (2022). Characterization of a programmable Argonaute nuclease from the mesophilic bacterium Rummeliibacillus suwonensis. Biomol. Ther. 12:355. doi: 10.3390/biom12030355
Jiang, S., Zhang, X., Li, D.-l., Wang, T.-t., Ma, F., and Zhang, C.-Y. (2022). Construction of a gold nanoparticle-based single-molecule biosensor for simple and sensitive detection of Argonaute 2 activity. J. Mater. Chem. B 10, 5594–5601. doi: 10.1039/D2TB00802E
Jin, S., Zhan, J., and Zhou, Y. (2021). Argonaute proteins: structures and their endonuclease activity. Mol. Biol. Rep. 48, 4837–4849. doi: 10.1007/s11033-021-06476-w
Kaya, E., Doxzen, K. W., Knoll, K. R., Wilson, R. C., Strutt, S. C., Kranzusch, P. J., et al. (2016). A bacterial Argonaute with noncanonical guide RNA specificity. Proc. Natl. Acad. Sci. U. S. A. 113, 4057–4062. doi: 10.1073/pnas.1524385113
Kellner, M. J., Koob, J. G., Gootenberg, J. S., Abudayyeh, O. O., and Zhang, F. (2019). SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 14, 2986–3012. doi: 10.1038/s41596-019-0210-2
Khalil, A. M. (2020). The genome editing revolution: review. J. Genet. Eng. Biotechnol. 18:68. doi: 10.1186/s43141-020-00078-y
Kropocheva, E., Kuzmenko, A., Aravin, A. A., Esyunina, D., and Kulbachinskiy, A. (2021). A programmable pAgo nuclease with universal guide and target specificity from the mesophilic bacterium Kurthia massiliensis. Nucleic Acids Res. 49, 4054–4065. doi: 10.1093/nar/gkab182
Kropocheva, E. V., Lisitskaya, L. A., Agapov, A. A., Musabirov, A. A., Kulbachinskiy, A. V., and Esyunina, D. M. (2022). Prokaryotic Argonaute proteins as a tool for biotechnology. Mol. Biol. 56, 854–873. doi: 10.1134/S0026893322060103
Kuzmenko, A., Oguienko, A., Esyunina, D., Yudin, D., Petrova, M., Kudinova, A., et al. (2020). DNA targeting and interference by a bacterial Argonaute nuclease. Nature 587, 632–637. doi: 10.1038/s41586-020-2605-1
Kuzmenko, A., Yudin, D., Ryazansky, S., Kulbachinskiy, A., and Aravin, A. A. (2019). Programmable DNA cleavage by ago nucleases from mesophilic bacteria Clostridium butyricum and Limnothrix rosea. Nucleic Acids Res. 47, 5822–5836. doi: 10.1093/nar/gkz379
Lapinaite, A., Doudna, J. A., and Cate, J. H. D. (2018). Programmable RNA recognition using a CRISPR-associated Argonaute. Proc. Natl. Acad. Sci. U. S. A. 115, 3368–3373. doi: 10.1073/pnas.1717725115
Li, X., Dong, H., Guo, X., Huang, F., Xu, X., Li, N., et al. (2022). Mesophilic Argonaute-based isothermal detection of SARS-CoV-2. Front. Microbiol. 13:957977. doi: 10.3389/fmicb.2022.957977
Li, Y., Kou, J., Han, X., Qiao, J., Zhang, W., Man, S., et al. (2023). Argonaute-triggered visual and rebuilding-free foodborne pathogenic bacteria detection. J. Hazard. Mater. 454:131485. doi: 10.1016/j.jhazmat.2023.131485
Li, W., Liu, Y., He, R., Wang, L., Wang, Y., Zeng, W., et al. (2022). A programmable pAgo nuclease with RNA target preference from the psychrotolerant bacterium Mucilaginibacter paludis. Nucleic Acids Res. 50, 5226–5238. doi: 10.1093/nar/gkac315
Li, Y., Liu, L., and Liu, G. (2019). CRISPR/Cas multiplexed biosensing: a challenge or an insurmountable obstacle? Trends Biotechnol. 37, 792–795. doi: 10.1016/j.tibtech.2019.04.012
Lisitskaya, L., Aravin, A. A., and Kulbachinskiy, A. (2018). DNA interference and beyond: structure and functions of prokaryotic Argonaute proteins. Nat. Commun. 9:5165. doi: 10.1038/s41467-018-07449-7
Lisitskaya, L., Shin, Y., Agapov, A., Olina, A., Kropocheva, E., Ryazansky, S., et al. (2022). Programmable RNA targeting by bacterial Argonaute nucleases with unconventional guide binding and cleavage specificity. Nat. Commun. 13:4624. doi: 10.1038/s41467-022-32079-5
Liu, Q., Guo, X., Xun, G., Li, Z., Chong, Y., Yang, L., et al. (2021). Argonaute integrated single-tube PCR system enables supersensitive detection of rare mutations. Nucleic Acids Res. 49:e75. doi: 10.1093/nar/gkab274
Liu, Y., Li, W., Jiang, X., Wang, Y., Zhang, Z., Liu, Q., et al. (2021). A programmable omnipotent Argonaute nuclease from mesophilic bacteria Kurthia massiliensis. Nucleic Acids Res. 49, 1597–1608. doi: 10.1093/nar/gkaa1278
Maeder, M. L., and Gersbach, C. A. (2016). Genome-editing technologies for gene and cell therapy. Mol. Ther. 24, 430–446. doi: 10.1038/mt.2016.10
Marais, G., Naidoo, M., Hsiao, N. Y., Valley-Omar, Z., Smuts, H., and Hardie, D. (2020). The implementation of a rapid sample preparation method for the detection of SARS-CoV-2 in a diagnostic laboratory in South Africa. PLoS One 15:e0241029. doi: 10.1371/journal.pone.0241029
Mojica, F. J. M., Diez-Villasenor, C., Garcia-Martinez, J., and Almendros, C. (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740. doi: 10.1099/mic.0.023960-0
Myhrvold, C., Freije, C. A., Gootenberg, J. S., Abudayyeh, O. O., Metsky, H. C., Durbin, A. F., et al. (2018). Field-deployable viral diagnostics using CRISPR-Cas 13. Science 360, 444–448. doi: 10.1126/science.aas8836
Nouri, R., Tang, Z., Dong, M., Liu, T., Kshirsagar, A., and Guan, W. (2021). CRISPR-based detection of SARS-CoV-2: a review from sample to result. Biosens. Bioelectron. 178:113012. doi: 10.1016/j.bios.2021.113012
O'Geen, H., Ren, C., Coggins, N. B., Bates, S. L., and Segal, D. J. (2018). Unexpected binding behaviors of bacterial Argonautes in human cells cast doubts on their use as targetable gene regulators. PLoS One 13:e0193818. doi: 10.1371/journal.pone.0193818
Olina, A., Kuzmenko, A., Ninova, M., Aravin, A. A., Kulbachinskiy, A., and Esyunina, D. (2020). Genome-wide DNA sampling by ago nuclease from the cyanobacterium Synechococcus elongatus. RNA Biol. 17, 677–688. doi: 10.1080/15476286.2020.1724716
Ooi, K. H., Liu, M. M., Tay, J. W. D., Teo, S. Y., Kaewsapsak, P., Jin, S., et al. (2021). An engineered CRISPR-Cas 12a variant and DNA-RNA hybrid guides enable robust and rapid COVID-19 testing. Nat. Commun. 12:1739. doi: 10.1038/s41467-021-21996-6
Pardee, K., Green, A. A., Takahashi, M. K., Braff, D., Lambert, G., Lee, J. W., et al. (2016). Rapid, low-cost detection of zika virus using programmable biomolecular components. Cells 165, 1255–1266. doi: 10.1016/j.cell.2016.04.059
Parker, J. S., Roe, S. M., and Barford, D. (2005). Structural insights into mRNA recognition from a PIWI domain-si RNA guide complex. Nature 434, 663–666. doi: 10.1038/nature03462
Qi, J., Dong, Z., Shi, Y., Wang, X., Qin, Y., Wang, Y., et al. (2016). NgAgo-based fabp 11a gene knockdown causes eye developmental defects in zebrafish. Cell Res. 26, 1349–1352. doi: 10.1038/cr.2016.134
Qin, Y., Li, Y., and Hu, Y. (2022). Emerging Argonaute-based nucleic acid biosensors. Trends Biotechnol. 40, 910–914. doi: 10.1016/j.tibtech.2022.03.006
Quan, J., Langelier, C., Kuchta, A., Batson, J., Teyssier, N., Lyden, A., et al. (2019). FLASH: a next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Nucleic Acids Res. 47:e83. doi: 10.1093/nar/gkz418
Rabe, B. A., and Cepko, C. (2020). SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc. Natl. Acad. Sci. U. S. A. 117, 24450–24458. doi: 10.1073/pnas.2011221117
Rashid, U. J., Paterok, D., Koglin, A., Gohlke, H., Piehler, J., and Chen, J. C. (2007). Structure of Aquifex aeolicus argonaute highlights conformational flexibility of the PAZ domain as a potential regulator of RNA-induced silencing complex function. J. Biol. Chem. 282, 13824–13832. doi: 10.1074/jbc.M608619200
Santiago-Frangos, A., Nemudryi, A., Nemudraia, A., Wiegand, T., Nichols, J. E., Krishna, P., et al. (2022). CRISPR-Cas, Argonaute proteins and the emerging landscape of amplification-free diagnostics. Methods 205, 1–10. doi: 10.1016/j.ymeth.2022.06.002
Schirle, N. T., Sheu-Gruttadauria, J., and Mac Rae, I. J. (2014). Structural basis for micro RNA targeting. Science 346, 608–613. doi: 10.1126/science.1258040
Shamshirgaran, Y., Liu, J., Sumer, H., Verma, P. J., and Taheri-Ghahfarokhi, A. (2022). Tools for efficient genome editing; ZFN, TALEN, and CRISPR. Methods Mol. Biol. 2495, 29–46. doi: 10.1007/978-1-0716-2301-5_2
Sheng, G., Gogakos, T., Wang, J., Zhao, H., Serganov, A., Juranek, S., et al. (2017). Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes. Nucleic Acids Res. 45, 9149–9163. doi: 10.1093/nar/gkx547
Sheng, G., Zhao, H., Wang, J., Rao, Y., Tian, W., Swarts, D. C., et al. (2014). Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. U. S. A. 111, 652–657. doi: 10.1073/pnas.1321032111
Shin, S., Jung, Y., Uhm, H., Song, M., Son, S., Goo, J., et al. (2020). Quantification of purified endogenous mi RNAs with high sensitivity and specificity. Nat. Commun. 11:6033. doi: 10.1038/s41467-020-19865-9
Song, J., Hegge, J. W., Mauk, M. G., Chen, J., Till, J. E., Bhagwat, N., et al. (2020). Highly specific enrichment of rare nucleic acid fractions using Thermus thermophilus argonaute with applications in cancer diagnostics. Nucleic Acids Res. 48:e19. doi: 10.1093/nar/gkz1165
Song, J. J., Smith, S. K., Hannon, G. J., and Joshua-Tor, L. (2004). Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305, 1434–1437. doi: 10.1126/science.1102514
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C., and Doudna, J. A. (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas 9. Biophys. J. 106:695A-A. doi: 10.1016/j.bpj.2013.11.3848
Swarts, D. C., Hegge, J. W., Hinojo, I., Shiimori, M., Ellis, M. A., Dumrongkulraksa, J., et al. (2015). Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res. 43, 5120–5129. doi: 10.1093/nar/gkv415
Swarts, D. C., Jore, M. M., Westra, E. R., Zhu, Y., Janssen, J. H., Snijders, A. P., et al. (2014). DNA-guided DNA interference by a prokaryotic Argonaute. Nature 507, 258–261. doi: 10.1038/nature12971
Swarts, D. C., Szczepaniak, M., Sheng, G., Chandradoss, S. D., Zhu, Y., Timmers, E. M., et al. (2017). Autonomous generation and loading of DNA guides by bacterial Argonaute. Mol. Cell 65, 985–998.e6. doi: 10.1016/j.molcel.2017.01.033
Toudji-Zouaz, A., Bertrand, V., and Barrière, A. (2021). Imaging of native transcription and transcriptional dynamics in vivo using a tagged Argonaute protein. Nucleic Acids Res. 49:e86-e. doi: 10.1093/nar/gkab469
Tsutakawa, S. E., Lafrance-Vanasse, J., and Tainer, J. A. (2014). The cutting edges in DNA repair, licensing, and fidelity: DNA and RNA repair nucleases sculpt DNA to measure twice, cut once. DNA Repair 19, 95–107. doi: 10.1016/j.dnarep.2014.03.022
Wang, L., He, R., Lv, B., Yu, X., Liu, Y., Yang, J., et al. (2021). Pyrococcus furiosus Argonaute coupled with modified ligase chain reaction for detection of SARS-CoV-2 and HPV. Talanta 227:122154. doi: 10.1016/j.talanta.2021.122154
Wang, Y., Juranek, S., Li, H., Sheng, G., Tuschl, T., and Patel, D. J. (2008). Structure of an Argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456, 921–926. doi: 10.1038/nature07666
Wang, Y., Juranek, S., Li, H., Sheng, G., Wardle, G. S., Tuschl, T., et al. (2009). Nucleation, propagation and cleavage of target RNAs in ago silencing complexes. Nature 461, 754–761. doi: 10.1038/nature08434
Wang, Z.-y., Li, P., Cui, L., Qiu, J.-G., Jiang, B., and Zhang, C.-Y. (2020). Integration of nanomaterials with nucleic acid amplification approaches for biosensing. TrAC Trends Anal. Chem. 129:115959. doi: 10.1016/j.trac.2020.115959
Wang, Y., Sheng, G., Juranek, S., Tuschl, T., and Patel, D. J. (2008). Structure of the guide-strand-containing argonaute silencing complex. Nature 456, 209–213. doi: 10.1038/nature07315
Wang, L., Xie, X., Lv, B., Liu, Y., Li, W., Zhang, Z., et al. (2022). A bacterial Argonaute with efficient DNA and RNA cleavage activity guided by small DNA and RNA. Cell Rep. 41:111533. doi: 10.1016/j.celrep.2022.111533
Wang, F., Yang, J., He, R., Yu, X., Chen, S., Liu, Y., et al. (2021). PfAgo-based detection of SARS-CoV-2. Biosens. Bioelectron. 177:112932. doi: 10.1016/j.bios.2020.112932
Wang, M., Zhang, R., and Li, J. (2020). CRISPR/cas systems redefine nucleic acid detection: principles and methods. Biosens. Bioelectron. 165:112430. doi: 10.1016/j.bios.2020.112430
Willkomm, S., Oellig, C. A., Zander, A., Restle, T., Keegan, R., Grohmann, D., et al. (2017). Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein. Nat. Microbiol. 2:17035. doi: 10.1038/nmicrobiol.2017.35
Wozniak, A., Cerda, A., Ibarra-Henríquez, C., Sebastian, V., Armijo, G., Lamig, L., et al. (2020). A simple RNA preparation method for SARS-CoV-2 detection by RT-qPCR. Sci. Rep. 10:16608. doi: 10.1038/s41598-020-73616-w
Wu, J., Kandavelou, K., and Chandrasegaran, S. (2007). Custom-designed zinc finger nucleases: what is next? Cell. Mol. Life Sci. 64, 2933–2944. doi: 10.1007/s00018-007-7206-8
Wu, Z., Tan, S., Xu, L., Gao, L., Zhu, H., Ma, C., et al. (2017). NgAgo-gDNA system efficiently suppresses hepatitis B virus replication through accelerating decay of pregenomic RNA. Antivir. Res. 145, 20–23. doi: 10.1016/j.antiviral.2017.07.005
Xu, L., Shen, X., Li, B., Zhu, C., and Zhou, X. (2017). G-quadruplex based Exo III-assisted signal amplification aptasensor for the colorimetric detection of adenosine. Anal. Chim. Acta 980, 58–64. doi: 10.1016/j.aca.2017.05.015
Xun, G., Lane, S. T., Petrov, V. A., Pepa, B. E., and Zhao, H. (2021). A rapid, accurate, scalable, and portable testing system for COVID-19 diagnosis. Nat. Commun. 12:2905. doi: 10.1038/s41467-021-23185-x
Yan, K. S., Yan, S., Farooq, A., Han, A., Zeng, L., and Zhou, M. M. (2003). Structure and conserved RNA binding of the PAZ domain. Nature 426, 468–474. doi: 10.1038/nature02129
Yang, L., Guo, B., Wang, Y., Zhao, C., Zhang, X., Wang, Y., et al. (2023). Pyrococcus furiosus Argonaute combined with recombinase polymerase amplification for rapid and sensitive detection of Enterocytozoon hepatopenaei. J. Agric. Food Chem. 71, 944–951. doi: 10.1021/acs.jafc.2c06582
Yao, C., Sasaki, H. M., Ueda, T., Tomari, Y., and Tadakuma, H. (2015). Single-molecule analysis of the target cleavage reaction by the drosophila RNAi enzyme complex. Mol. Cell 59, 125–132. doi: 10.1016/j.molcel.2015.05.015
Ye, X., Zhou, H., Guo, X., Liu, D., Li, Z., Sun, J., et al. (2022). Argonaute-integrated isothermal amplification for rapid, portable, multiplex detection of SARS-CoV-2 and influenza viruses. Biosens. Bioelectron. 207:114169. doi: 10.1016/j.bios.2022.114169
Yuan, C., Fang, J., and Fu, W. (2023). Thermus thermophilus Argonaute-based isothermal amplification assay for ultrasensitive and specific RNA detection. Anal. Chem. 95, 8291–8298. doi: 10.1021/acs.analchem.3c00599
Yuan, Y. R., Pei, Y., Chen, H. Y., Tuschl, T., and Patel, D. J. (2006). A potential protein-RNA recognition event along the RISC-loading pathway from the structure of A. aeolicus Argonaute with externally bound si RNA. Structure 14, 1557–1565. doi: 10.1016/j.str.2006.08.009
Yuan, Y. R., Pei, Y., Ma, J. B., Kuryavyi, V., Zhadina, M., Meister, G., et al. (2005). Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell 19, 405–419. doi: 10.1016/j.molcel.2005.07.011
Zander, A., Holzmeister, P., Klose, D., Tinnefeld, P., and Grohmann, D. (2014). Single-molecule FRET supports the two-state model of Argonaute action. RNA Biol. 11, 45–56. doi: 10.4161/rna.27446
Zander, A., Willkomm, S., Ofer, S., van Wolferen, M., Egert, L., Buchmeier, S., et al. (2017). Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii. Nat. Microbiol. 2:17034. doi: 10.1038/nmicrobiol.2017.34
Zhang, D., Ma, F., Leng, J., and Zhang, C.-y. (2018). A dual signal amplification-assisted DNAzyme biosensor for ultrasensitive detection of Argonaute 2 activity. Chem. Commun. 54, 13678–13681. doi: 10.1039/C8CC08553F
Zhang, S., Zhang, X., Bie, Y., Kong, J., Wang, A., Qiu, Y., et al. (2022). STUB1 regulates antiviral RNAi through inducing ubiquitination and degradation of dicer and AGO2 in mammals. Virol. Sin. 37, 569–580. doi: 10.1016/j.virs.2022.05.001
Zhang, H. X., Zhang, Y., and Yin, H. (2019). Genome editing with mRNA encoding ZFN, TALEN, and Cas 9. Mol. Ther. 27, 735–746. doi: 10.1016/j.ymthe.2019.01.014
Keywords: nucleic acid detection technology, pathogen diagnosis, Argonaute, nucleases, biosensor
Citation: Wu Z, Yu L, Shi W and Ma J (2023) Argonaute protein-based nucleic acid detection technology. Front. Microbiol. 14:1255716. doi: 10.3389/fmicb.2023.1255716
Received: 09 July 2023; Accepted: 28 August 2023;
Published: 08 September 2023.
Edited by:
Jens Andre Hammerl, Bundesinstitut für Risikobewertung, GermanyReviewed by:
Guohui Li, Jiangsu University, ChinaCopyright © 2023 Wu, Yu, Shi and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiyun Wu, emhpeXVud3VAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.