- 1Department of Pharmacy, The Second Affiliated Hospital, Army Medical University, Chongqing, China
- 2Department of Microbiology, College of Basic Medical Science, Army Medical University, Chongqing, China
- 3Department of Clinical Laboratory, The 971st Hospital of Chinese People's Liberation Army Navy, Qingdao, China
Background: Membrane vesicles (MVs) are nanoscale vesicular structures produced by bacteria during their growth in vitro and in vivo. Some bacterial components can be loaded in bacterial MVs, but the roles of the loaded MV molecules are unclear.
Methods: MVs of Staphylococcus aureus RN4220 and its derivatives were prepared. Dynamic light scattering analysis was used to evaluate the size distribution, and 4D-label-free liquid chromatography–tandem mass spectrometry analysis was performed to detect protein composition in the MVs. The site-mutation S. aureus RN4220-Δhld and agrA deletion mutant RN4220-ΔagrA were generated via allelic replacement strategies. A hemolysis assay was performed with rabbit red blood cells. CCK-8 and lactate dehydrogenase release assays were used to determine the cytotoxicity of S. aureus MVs against RAW264.7 macrophages. The serum levels of inflammatory factors such as IL-6, IL-1β, and TNFα in mice treated with S. aureus MVs were detected with an enzyme-linked immunosorbent assay kit.
Results: Delta-hemolysin (Hld) was identified as a major loaded factor in S. aureus MVs. Further study showed that Hld could promote the production of staphylococcal MVs with smaller sizes. Loaded Hld affected the diversity of loaded proteins in MVs of S. aureus RN4220. Hld resulted in decreased protein diversity in MVs of S. aureus. Site-mutation (RN4220-Δhld) and agrA deletion (RN4220-ΔagrA) mutants produced MVs (ΔhldMVs and ΔagrAMVs) with a greater number of bacterial proteins than those derived from wild-type RN4220 (wtMVs). Moreover, Hld contributed to the hemolytic activity of wtMVs. Hld-loaded wtMVs were cytotoxic to macrophage RAW264.7 cells and could stimulate the production of inflammatory factor IL-6 in vivo.
Conclusion: This study presented that Hld was a major loaded factor in S. aureus MVs, and the loaded Hld played vital roles in the MV-property modification.
1. Introduction
Bacterial membrane vesicles (MVs) are nanosized bilayer lipid particles that are commonly released from bacterial cells during all their growth stages (Wang et al., 2021). Gram-negative Escherichia coli was first reported in the 1960s to secrete MVs, which are considered to bud out from the outer membrane, so they are often termed outer MVs (OMVs). Conversely, the existence of MVs released from gram-positive bacteria such as Bacillus subtilis and B. cereus was first mentioned in 1990 (Brown et al., 2015; Kaparakis-Liaskos and Ferrero, 2015; Bitto et al., 2021a). Released MVs can incorporate bacterial proteins, lipids, and genetic materials (Brown et al., 2015), and these properties confer bacterial MVs with versatile function particles for surface adhesion, cell signal transduction, virulence factor transportation, genetic material exchange, and bacterial pathogenicity, as well as for applications in vaccine candidate and drug delivery (Askarian et al., 2018; Briaud and Carroll, 2020; Escudé Martinez de Castilla et al., 2021; Kumaraswamy et al., 2021; Lee et al., 2022).
Staphylococcus aureus is a notorious pathogen causing various diseases, and the release of MVs by S. aureus was first discovered in 2009 (Lee et al., 2009). The function of S. aureus MVs may be strain-oriented. For example, MVs derived from S. aureus ATCC14458 can induce atopic dermatitis-like skin inflammation (Hong et al., 2011), clinical S. aureus isolate 06ST1048-produced MVs can induce the apoptosis of HEp-2 cells in a dose-dependent manner, and MVs prepared from ST239, ST59, and ST338 methicillin-resistant S. aureus strains could inhibit the proliferation and induce the apoptosis of epithelial cells HaCaT (Gurung et al., 2011; Chen et al., 2023); MVs of S. aureus strain NCTC6571 and clinical isolates BPH2760 and BPH 2900 can induce autophagy in A549 lung epithelial cells (Bitto et al., 2021a). MVs produced from S. aureus USA300-ΔagrA can also bind and inactivate daptomycin to protect bacteria from antibiotic killing (Pader et al., 2016). A study compared the cytopathology of host cells induced by MVs derived from different S. aureus strains and concluded that MVs with different proteomes presented distinct cytotoxic effects against host cells (Jeon et al., 2016). Importantly, delta-hemolysin (Hld) was found in the MVs from three S. aureus isolates M060, 03ST17, and 06ST1048, and the loaded Hld could induce cytotoxicity after delivery to host cells by MVs (Jeon et al., 2016). However, the biological effects of components encapsulated in S. aureus MVs such as Hld on MV properties and functions are largely unknown.
Certain components such as toxins and virulence factors can be concentrated in bacterial MVs (Rivera et al., 2010; Veith et al., 2014). Investigating the biological roles of loaded molecules in MVs has great significance for elucidating MV properties and facilitating its applications. The coding gene of Hld is located at the 85–165 bases of RNAIII, an RNA regulator involved in S. aureus agr locus carrying agrB, agrD, agrC, agrA, and RNAIII (Hodille et al., 2016). Hld is a hemolytic peptide belonging to the phenol-soluble modulin (PSM) family that contributes to S. aureus infections (Verdon et al., 2009). In the present study, Hld was identified as the most abundant component in S. aureus MVs of diverse genetic backgrounds. Hld overexpression enhanced the production of S. aureus MVs with relatively smaller sizes of MV particles, while Hld mutation or agrA deletion (RNAIII inactivation) induced differences in the composition of ΔhldMVs and ΔagrAMVs with increased protein diversity compared with that of wtMVs. Loaded Hld could modify the properties of S. aureus MVs, including hemolytic activity, cytotoxicity, and inflammatory factor stimulation.
2. Materials and methods
2.1. Bacterial strains and culture conditions
Bacterial strains and plasmids used in this study are presented in Supplementary Table 1. The reference S. aureus strains RN4220 (NCTC 8325-4) were kindly provided by Prof. Baolin Sun (University of Science and Technology of China), USA300 (ATCC BAA-1556) was a gift from Dr. Min Li (Shanghai Jiao Tong University, China), Newman (NCTC 8178) was provided by Prof. Lu Yu (Jilin University, China), MW2 was a gift from Prof. Xiaoxue Ma (China Medical University), and strains ATCC29213, ATCC4330, and ATCC25923 and two clinical isolates (TJ274 and TJ275) were kindly provided by Dr. Dongmei Wang (The 983 Hospital of PLA). S. aureus XQ was isolated from the blood sample of a 16-year-old juvenile, whose onset was a skin wound then deteriorated to a lethal S. aureus sepsis with mental disorder (Rao et al., 2015; Liu et al., 2018). S. aureus strains were grown in brain heart infusion (BHI) medium (Oxoid, UK) at 37°C with shaking (200 rpm) or cultivated on BHI agar. Escherichia coli DH5α was cultivated in Luria–Bertani (LB) medium (Oxoid, UK) at 37°C with shaking or cultured on LB agar. For plasmid pBT2 (Supplementary Table 1) maintenance, cultures were supplemented with 50 μg/ml of ampicillin (AMP) and 10 μg/ml of chloramphenicol (Cm) for E. coli and S. aureus, respectively. All strains were stored in an appropriate medium with 20% glycerol at −80°C and resuscitated by streaking on agar plates. A single colony was picked and cultured with BHI or LB medium for the subsequent experiments.
2.2. Preparation of S. aureus MVs
MV isolation was performed with some modifications as previously described (Hong et al., 2011; Yuan et al., 2018; Chen et al., 2020; Zaborowska et al., 2021). In brief, a single colony of S. aureus RN4220 from the BHI agar plate was inoculated into 1 ml BHI medium and cultured at 37°C for 6 h. Then, 100 μl of culture was transferred to 10 ml BHI broth (1:100) and cultured at 37°C overnight with shaking. On the next day, the culture was inoculated into 900 ml BHI medium (1:100) and cultured at 37°C for 22 h to achieve the highest MV production (Yuan et al., 2018). The culture supernatant was collected after centrifugation at 6,000 × g at 4°C for 20 min to remove bacterial cells and centrifugation at 10,000 × g at 4°C for 30 min to remove cell debris and then filtered through a 0.45 μm vacuum filter (Millipore, USA), to remove possible cell debris or aggregates. The filtrate was concentrated to 240 ml with a 100 kDa cutoff hollow fiber cartridge column (GE Healthcare, USA) and then filtered through a 0.22 μm filter (Millipore, USA). The supernatant was ultracentrifuged at 200,000 × g at 4°C for 3 h.
The MV pellets were washed twice with phosphate-buffered saline (PBS, pH7.2) and resuspended in sterile PBS, and the amount of protein in the MVs was measured with Bradford Protein Assay Kit (Beyotime, China) as previously described (MacDonald and Beveridge, 2002; Tartaglia et al., 2018; Bitto et al., 2021b). Prepared MVs were stored at −80°C for use.
2.3. 4D-label-free liquid chromatography–tandem mass spectrometry analysis
The protein composition in MVs of S. aureus RN4220 (wtMVs) was identified by using 4D-label-free liquid chromatography–tandem mass spectrometry (LC-MS/MS) as previously described (Cox et al., 2014). In brief, the proteins in three biological replicate wtMV samples were digested with trypsin (Promega, USA), desalted on a C18 cartridge column (Empore™, Sigma, USA), and reconstituted in 40 μl of 0.1% (v/v) formic acid. The peptides were subjected to 4D-label-free LC-MS/MS analysis by Applied Protein Technology (Shanghai, China). The MS raw data for each sample were searched against the database of S. aureus (http://www.uniprot.org/) by using MaxQuant 1.5.3.17 software (Parianichnikov et al., 2020). Cluster 3.0 and Java TreeView software were used to perform hierarchical clustering analysis (Hoon et al., 2004; Saldanha, 2004). CELLO (http://cello.life.nctu.edu.tw/) was used to predict protein subcellular localization. Protein sequences were searched using InterProScan software to identify protein domain signatures from the InterPro member database Pfam (Quevillon et al., 2005). The gene ontology (GO) was analyzed using the protein sequences of the selected differentially expressed proteins. NCBI BLAST+ client software and InterProScan were used to find homolog sequences; the GO terms were mapped, and sequences were annotated using software Blast2GO (Conesa et al., 2005; Quevillon et al., 2005; Camacho et al., 2009). The results of GO annotation were plotted by R scripts. Kyoto Encyclopedia of Genes and Genomes (KEGG) was analyzed using the online database (http://geneontology.org/). KEGG orthology identifications were retrieved and subsequently mapped to pathways in KEGG. Enrichment analysis was applied based on Fisher's exact test, considering the whole quantified proteins as the background dataset. Benjamini–Hochberg correction for multiple testing was further applied to adjust derived P-values. Only functional categories and pathways with P-values under a threshold of 0.05 were considered significant. All raw data of 4D label-free LC-MS/MS identification was deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD039734.
2.4. SDS-PAGE and Western blot analysis
The purified MVs were mixed with 5 × SDS-PAGE loading buffer at a rate of 4:1 and incubated in boiling water for 10 min. The proteins in samples (10 μl/each) were separated by 12% (m/v) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie brilliant blue (Thermo Scientific, USA). For Western blot, the proteins after SDS-PAGE were transferred to a polyvinylidene difluoride (PVDF) membrane (GE Healthcare, USA). The PVDF membrane was blocked with PBS-T (PBS + 0.1% (v/v) Tween-20) containing 5% (m/v) skim milk (ABgent, USA) at 37°C for 60 min, followed by incubation with anti-Hld rabbit polyclonal primary antibody (1:1,000 dilution in PBS-T containing 5% (m/v) skim milk) at 4°C overnight. After five times of wash with PBS-T, the membrane was incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Abmart, China; 1:5,000 dilution) at 37°C for 1 h. After five times of wash with PBS-T, the target protein was visualized with the enhanced chemiluminescence system (Thermo Scientific) and photographed.
2.5. Gene knockout and genetic complementation
The site mutation S. aureus RN4220-Δhld and agrA deletion RN4220-ΔagrA mutants were generated via allelic replacement as previously described (Peng et al., 2017). All primers used in this study are presented in Supplementary Table 2. For the construction of RN4220-Δhld with normal RNAIII function, the 20th base “C” in the hld gene was changed to “G” to achieve a stop codon TGA and result in a truncated translation of Hld. In brief, the homologous left arm and homologous right arm were amplified with primer pairs delta-L/pBT2-delta-R (BamHI) and pBT2-delta-L (SalI)/delta-R, respectively, from the genomic DNA template of S. aureus RN4220 (GenBank accession no. NZ_CP101124) by polymerase chain reaction (PCR). The PCR fragments were purified and ligated to pBT2 plasmid via ClonExpress MultiS One Step Cloning Kit (Vazyme, China). The recombinant plasmid pBT2-Δhld was transformed into competent E. coli DH5α, extracted, and sequenced. The correct plasmid was then electrotransformed into S. aureus RN4220. The plasmid-carrying S. aureus was cultured at 30°C for 16 h, and the plasmid pBT2-Δhld was verified by double digestion with BamHI and SalI (TakaRa, Japan). The correct strain carrying pBT2-Δhld was inoculated into fresh BHI medium and cultivated at 42°C to induce the integration of the recombinant plasmid into the genome, which was verified by colony PCR with primer pairs delta-outer-L/pBT2-down and pBT2-up/delta-outer-R. The plasmid was eliminated via culturing a single colony at 25°C to achieve the mutant strain RN4220-Δhld, which was verified by PCR amplification with primer pair delta-outer-L/delta-outer-R and DNA sequencing. The construction of RN4220-ΔagrA used similar strategies mentioned above.
For genetic complementation, the tetracycline promoter tetP was introduced in front of the hld gene to obtain tetP/hld, which was inserted behind the eno gene on the genome of RN4220-Δhld. The homologous left arm, right arm, and hld gene were amplified by PCR with primer pairs up-pBT2-eno-tetP-L/up-tetP-R, down-eno-L/down-pBT2-eno-R, and tetP-hld-L/tetP-hld-R, respectively, from genomic DNA template of S. aureus RN4220. The tetP promotor fragment was amplified with primer pair tetP-L/tetP-R from the plasmid pYT3 (Supplementary Table 1). The four PCR fragments were purified and ligated to the pBT2 plasmid via ClonExpress MultiS One Step Cloning Kit. The resulting pBT2-tetP/hld was transformed into S. aureus RN4220-Δhld, and the genetic complementation strain RN4220-Δhld-tetP/hld was screened and identified using similar strategies for RN4220-Δhld construction. The hld overexpression strain RN4220-tetP/hld was generated after the transformation of pBT2-tetP/hld into the wild-type S. aureus RN4220.
2.6. Growth curve determination
The growth curves of S. aureus RN4220 and its derivatives RN4220-Δhld and RN4220-ΔagrA were determined. In brief, S. aureus strains of interest were cultured in BHI broth at 37°C with shaking overnight, and 0.2 ml of each strain culture was inoculated into 20 ml of fresh BHI medium. The optical density (OD) at 600 nm (OD600) was detected every hour for 15 h after inoculation. Growth curves were drawn using the OD600 values over culture times.
2.7. MV size evaluation
The size distribution of S. aureus MVs was evaluated by dynamic light scattering (DLS) analysis using Zetasizer Nano ZS90 (Malvern, UK). In brief, purified MVs were diluted in PBS to 0.1 mg/ml. Reads of 60-s durations were conducted in triplicate at 25°C. Data outputs were generated using Zetasizer (Nano, μV, APS) software v7.13 (Malvern, UK). The average number of particles at each binned center in the Experiment Summary output was adjusted by the dilution factor using GraphPad Prism 8.0.1 software, and the mean of three biological replicates was plotted as particles.
2.8. Hemolysis assay
A hemolysis assay was performed as previously described (Miller et al., 2021). Approximately 10 ml of whole blood was collected from the central auricular artery of a New Zealand male rabbit, which was purchased from the Laboratory Animal Center of the Third Military Medical University, and added to 1 ml heparin sodium solution, followed by three times of wash with PBS with centrifugation at 1,000 × g for 5 min at 4°C. A concentration of 2% (v/v) rabbit red blood cells (RBCs) was prepared with PBS. In total, 40 μl of PBS containing 2, 5, 10, and 20 μl, respectively, of 0.5 mg/ml MVs derived from S. aureus RN4220 (wtMVs) or its derivatives RN4220-Δhld (ΔhldMVs) and RN4220-ΔagrA (ΔagrAMVs) was added to 1 ml of 2% RBCs to the final concentration of 1, 2.5, 5, and 10 μg/ml of MVs, respectively, mixed gently, and incubated at 37°C for 30 min. After centrifuging at 8,000 × g for 10 s, the supernatant was collected, and OD540 values were determined with a spectrophotometer (Thermo Scientific). PBS-treated RBCs served as negative control, while RBC treatment with 1% (v/v) Triton X-100 represented positive control. The percentage hemolysis is calculated as follows: [(OD540 of sample−OD540 of negative control)/(D540 of positive control–OD540 of negative control)] × 100%.
2.9. Cell culture and stimulation
Macrophage RAW264.7 cells were purchased from a Chinese company Cas9X, maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, USA), and supplemented with 10% (v/v) fetal bovine serum (Zeta Life, USA) and 1% (v/v) penicillin/streptomycin (Beyotime, China) at 37°C in a humidified 5% CO2 atmosphere. RAW264.7 cells were seeded at 2.5 × 105 cells/ml in 500 μl in 24-well plates for 12 h and stimulated with 5, 10, and 20 μg/ml of wtMVs, ΔhldMVs, or ΔagrAMVs for 6 h, and the cell culture supernatants were collected for further study.
2.10. Cell viability assay and lactate dehydrogenase release detection
The cell viability was detected by a CCK-8 cell viability kit (ABclonal, China), according to the instructions provided by the manufacturer. In brief, RAW264.7 cells were seeded at 2.5 × 105 cells/ml in 100 μl RPMI1640 medium in 96-well plates for 12 h and stimulated with 10 μl of 0.05, 0.1, and 0.2 mg/ml of wtMVs, ΔhldMVs, or ΔagrAMVs (the final concentration of 5, 10, and 20 μg/mL of MVs, respectively) for 6 h at 37°C with 5% CO2. In total, 10 μl of CCK-8 solution was added to each well and incubated for 2 h, and OD450 values were determined with a spectrophotometer (Thermo Scientific). The 10 μl of PBS-treated RAW264.7 served as a negative control, and the wells with culture medium and CCK-8 solution served as blank controls. The percentage of cell-survival rate is calculated as follows: [(OD450 of sample−OD450 of blank control)/(OD450 of negative control−OD450 of blank control)] × 100%.
The LDH cytotoxicity assay kit (Beyotime, China) was used to quantify the activity of LDH released from damaged cells. RAW264.7 cells were seeded at 1 × 105 cells/ml in 200 μl of RPMI1640 medium in 96-well plates for 12 h, the culture solution was discarded, and the cells were washed once with sterile PBS. In total, 200 μl of fresh low serum culture medium (1% serum) and 20 μl of PBS containing 0.05, 0.1, and 0.2 mg/ml of wtMVs, ΔhldMVs, or ΔagrAMVs were added to each well to the final concentrations of 5, 10, and 20 μg/ml of MVs, respectively. Cell-free culture medium supplemented well served as blank control, untreated cell-seeded well served as sample control, 20 μl PBS-treated RAW264.7 served as negative control, and cells lysed with 20 μl LDH releasing reagent used as maximum enzyme activity control. After 6 h of incubation, the plate was centrifuged at 400 × g for 5 min, and 120 μl supernatant of each well was transferred to a new 96-well plate, followed by adding 60 μl of detection working solution, mixed well, and incubated at room temperature to avoid light for 30 min. OD490 and OD600 values were determined. The measured OD490 and OD600 values were each subtracted from the values of blank control. LDH activity is calculated as follows: [((OD490 − OD600 of sample) − (OD490 − OD600 of sample control))/((OD490 − OD600 of maximum enzyme activity control) − (OD490 − OD600 of sample control))] × 100%.
2.11. Detection of inflammatory factors
All animal study was conducted in accordance with the ARRIVE guidelines. In total, 6- to 8-week-old female BALB/c mice (20 ± 2 g) were purchased from Hunan Slyke Jingda Experimental Animal Co. LTD. All animal experiments were approved by the Laboratory Animal Welfare and Ethics Committee of the Army Medical University (protocol No. AMUWEC2020735).
BALB/c mice (n = 3) were intraperitoneally injected 100 μl (10 or 20 μg/ml) of wtMVs, ΔhldMVs, or ΔagrAMVs. PBS was used as the control. Mouse sera were collected 6 h post-injection. Then, the levels of inflammatory factors, including IL-6, IL-1β, and TNFα, were determined with an enzyme-linked immunosorbent assay (ELISA) kit, according to the manufacturer's instructions (ABclonal, China). The results were examined with an ELISA reader Multiskan Go (Thermo Scientific), and inflammatory factor levels were determined according to the standard curve.
2.12. Statistical analysis
All statistical analyses were performed using GraphPad Prism 8.0.1 software. One-way analysis of covariance (ANOVA) was used for comparing multiple groups. The results were presented as mean ± standard deviation (SD), and P-values of <0.05 were considered to be statistically significant.
3. Results
3.1. Abundant Hld loaded in the MVs of S. aureus with diverse genetic lineages
An accurate proteome-wide 4D-label-free LC-MS/MS (Cox et al., 2014) was performed to quantify the proteins in MVs of S. aureus RN4220 (wtMVs). A total of 704 proteins in the wtMVs were identified, and they were predicted to be membrane proteins (178, 25.3%), extracellular (147, 20.9%), and/or cytoplasmic proteins (458, 65.1% with some overlapped ones). KEGG enrichment revealed that oxidative phosphorylation was the most significant pathway involved, followed by citrate cycle, glycolysis/gluconeogenesis, pyruvate metabolism, carbon-fixation pathways in prokaryotes, and nucleotide metabolism (Supplementary Figure 1). GO analysis showed that the enriched proteins were involved in metabolic and cellular processes, catalytic activity and binding of molecular functions, and cell and cell part of cellular components (Figure 1A).
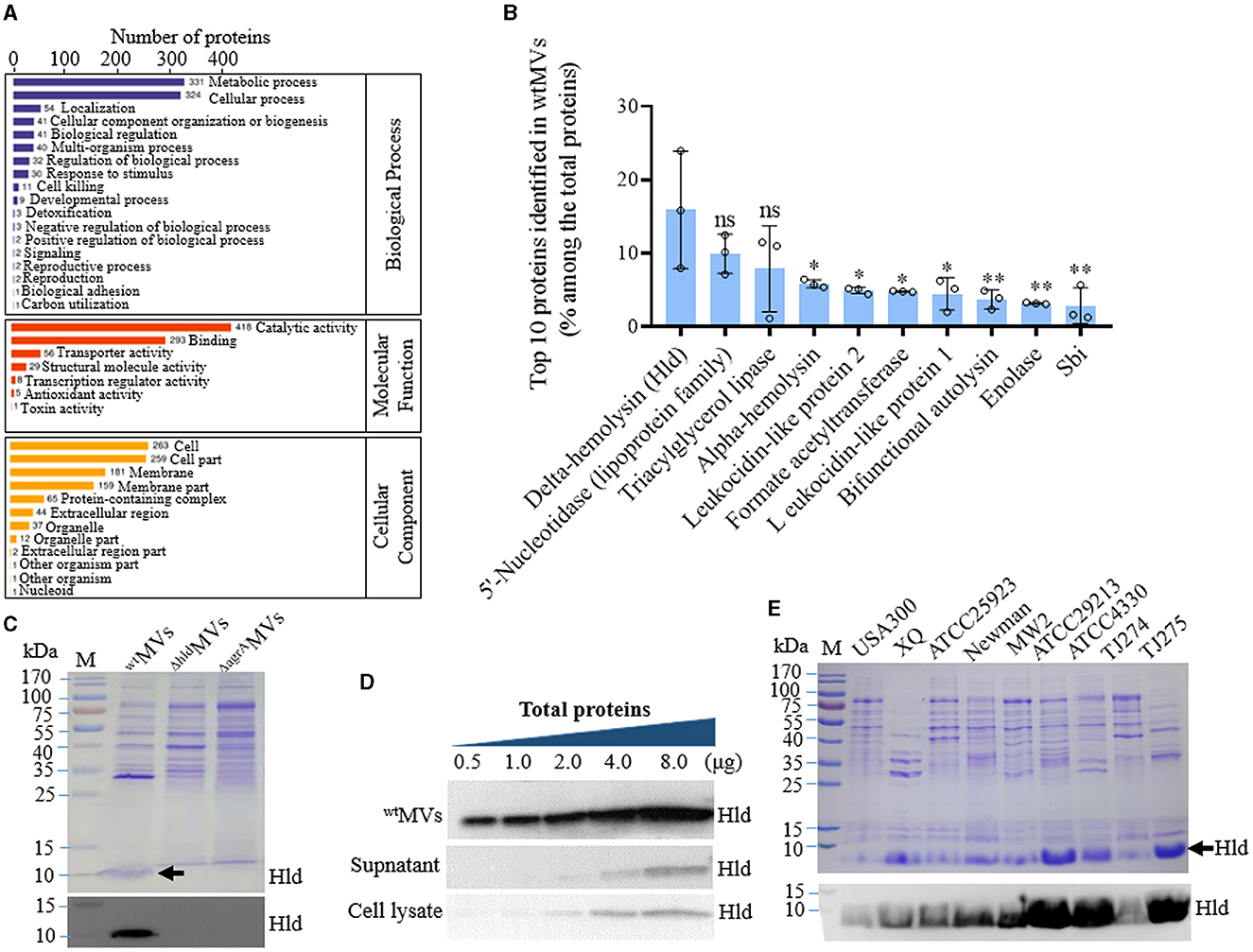
Figure 1. Hld loaded in the MVs of S. aureus with diverse genetic lineages. (A) GO function analysis of proteins identified with 4D-label-free LC-MS/MS in the MVs of S. aureus RN4220 (wtMVs). Proteins involved in representative biological processes, molecular functions, and cellular components were indicated. (B) Proportion of the top 10 proteins identified in wtMVs counted by label-free quantity (LFQ) intensity. The data were presented as mean ± SD of three biological replicates. Statistical significance was calculated by one-way ANOVA and compared with the LFQ intensity of Hld; ns indicates no statistical significance; ∗P < 0.05 and ∗∗P< 0.01. (C) Characterization of Hld in the MVs of S. aureus RN4220 and its derivatives. Comparison of protein profiles among wtMVs, ΔhldMVs derived from RN4220-Δhld, and ΔagrAMVs prepared from RN4220-ΔagrA by SDS-PAGE (top panel), the loaded Hld was indicated by a black arrow. Identification of loaded Hld in wtMVs, ΔhldMVs, and ΔagrAMVs by Western blot (bottom panel). (D) Western blot analysis of Hld in wtMVs, culture supernatant, and cell lysate of RN4220. Diverse total proteins in the samples were subjected to Hld detection and loaded Hld presented in the wtMVs. (E) Detection of loaded Hld in the MVs derived from S. aureus strains with different genetic lineages. SDS-PAGE analysis of protein profiles of MVs from indicated S. aureus strains and the loaded Hld in each bacterial MV was shown by a black arrow (top panel). Detection of loaded Hld in each bacterial MVs by Western blot (bottom panel).
Notably, Hld was the most abundant molecule in wtMVs of S. aureus RN4220, which presented a label-free quantity (LFQ) intensity of 367,430,000 and accounted for 15.6 ± 7.9% of the total protein quantity identified (Figure 1B). SDS-PAGE analysis also revealed the loaded Hld in wtMVs, which could be characterized by specific antibodies against Hld in Western blot assay (Figure 1C; Supplementary Figure 2). Compared with the protein in bacterial cell lysate and culture supernatant, Hld was remarkably loaded in the wtMVs of S. aureus RN4220 and elevated in an MV-dose-dependent manner (Figure 1D; Supplementary Figure 2). To determine whether loaded Hld occurred in MVs derived from other strains, we cultured nine S. aureus strains (USA300, XQ, ATCC25923, Newman, MW2, ATCC29213, ATCC4330, TJ274, and TJ275) and individually prepared their MVs with the same conditions to RN4220. SDS-PAGE and Western blot assays revealed the loaded Hld in all MVs tested (Figure 1E; Supplementary Figure 2), suggesting that the encapsulation of Hld in S. aureus MVs is common.
3.2. Hld promoted the production of S. aureus MVs with decreased size
The gene encoding Hld is within the RNAIII molecule (85–165 nt) (Figure 2A), a global RNA regulator involved in the S. aureus Agr quorum-sensing system (Hodille et al., 2016). To evaluate the function of loaded Hld in MVs, we generated an S. aureus mutant (RN4220-Δhld) by changing the 20th base of C to G in the hld gene to achieve a stop codon TGA and abolish the translation of Hld protein (Supplementary Figures 3A, B). An agrA gene knocked-out strain (RN4220-ΔagrA) was also constructed to inactivate RNAIII and Hld functions (Supplementary Figure 3C). Compared with wild-type RN4220, agrA deletion and site mutation in the hld gene did not affect the growth of RN4220-Δhld and RN4220-ΔagrA mutants (Supplementary Figure 3D). No Hld was detected in ΔhldMVs and ΔagrAMVs derived from RN4220-Δhld and RN4220-ΔagrA, respectively (Figure 1C). Western blot showed that the expressions of virulence factors such as alpha-hemolysin (Hla), gamma-hemolysin B subunit (HlgB), and Panton–Valentine leukocidin (PVL) element LukS in the RN4220-Δhld were similar to those in the wild-type strain (Supplementary Figures 3E, F). However, agrA deletion resulted in decreased expression of all virulence factors tested. These data indicated that the site-mutation strain RN4220-Δhld disabled Hld expression but had a functional RNAIII, which controlled the expression of many virulence factors, including Hla, HlgB, and PVL-LukS. By contrast, Hld expression and RNAIII function were absent in the RN4220-ΔagrA.
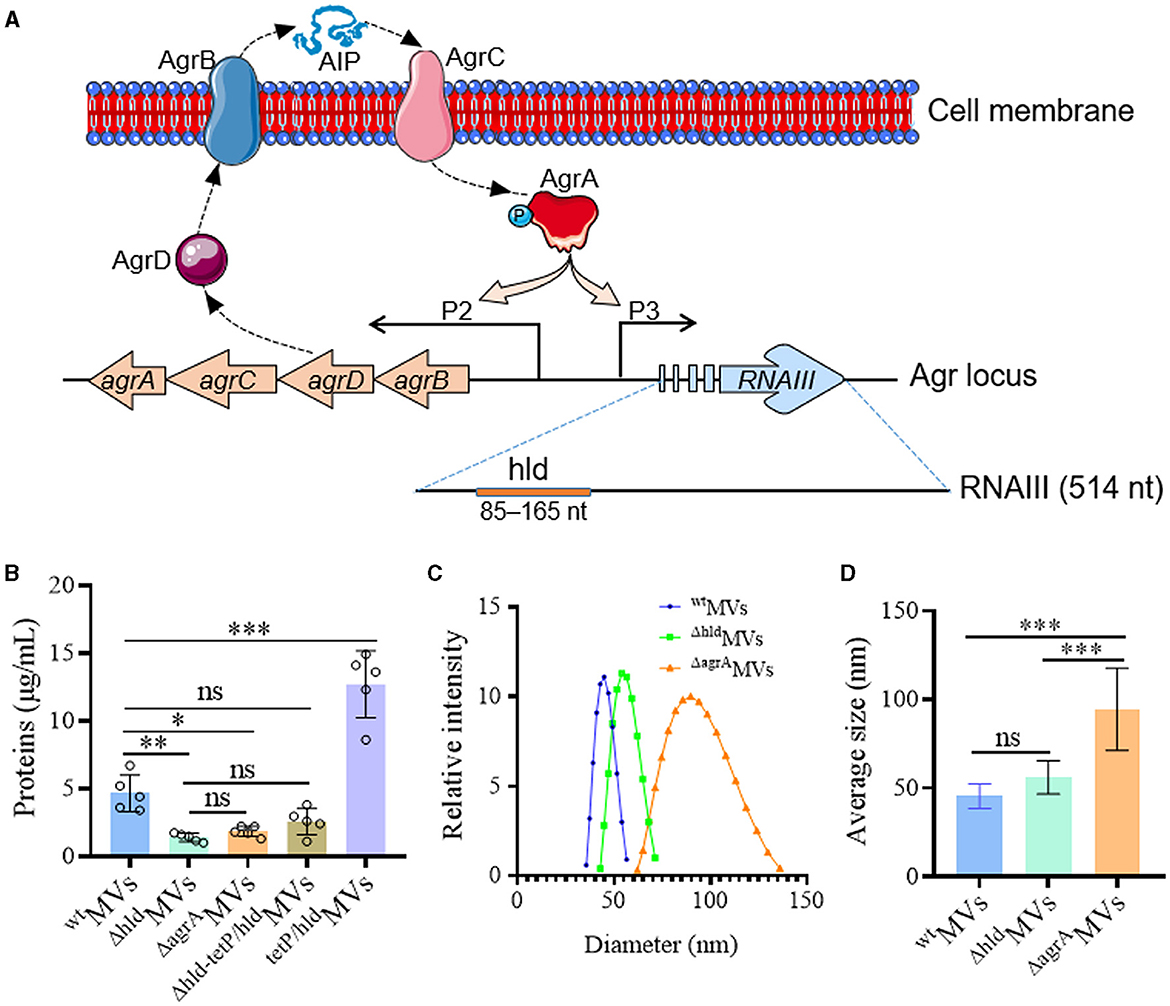
Figure 2. Hld promoted the production of S. aureus MVs with smaller sizes. (A) Schematic diagram showing the agr system and the hld gene located in RNAIII. AgrD is processed by AgrB to generate auto-inducing peptide (AIP), which can bind AgrC to activate AgrA by phosphorylation. AgrA can not only stimulate P2 promotor to promote Agr function but also activate P3 promotor to produce effective regular RNAIII, which guides Hld expression. (B) Hld enhanced MV production in S. aureus RN4220. The proteins in wtMVs (derived from RN4220), ΔhldMVs (from RN4220-Δhld), ΔagrAMVs (from RN4220-ΔagrA), Δhld−telP/hldMVs (prepared from complementation strain RN4220-Δhld-tetP/hld), and telP/hldMVs (derived from overexpression strain RN4220-tetP/hld) were quantified by Bradford assay. The data were presented as mean ± SD (n = 5). (C) The size distribution and (D) the sizes of wtMVs, ΔhldMVs, and ΔagrAMVs measured by DLS. The data were presented as mean ± SD (n = 3). Statistical significance was calculated by one-way ANOVA; ns indicates no statistical significance; ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
The MV production in RN4220-Δhld and RN4220-ΔagrA was substantially reduced compared with that in wild-type RN4220 (Figure 2B). For complementation, a tetracycline promoter TetP-controlled hld gene was constructed in RN4220-Δhld by inserting it behind the eno gene, which encoded the MV-associated enolase (Yuan et al., 2018). The restored MV production was observed in RN4220-Δhld-tetP/hld (Figure 2B). Moreover, Hld overexpression in wild-type strain RN4220 (RN4220-tetP/hld) significantly increased MV yield (P < 0.001; Figure 2B). Next, the sizes of wtMVs, ΔhldMVs, and ΔagrAMVs were measured, and the results showed that the sizes of wtMVs (45.4 ± 6.9 nm), ΔhldMVs (56.0 ± 9.3 nm), and ΔagrAMVs (94.5 ± 23.2 nm) enlarged gradually (Figure 2C). The ΔagrAMVs presented the largest particle size, which was significantly larger than those derived from RN4220-Δhld and wild-type RN4220 (P < 0.001; Figure 2D). These data suggested that Hld promoted MV production in S. aureus. Loaded Hld was associated with smaller MV particles, and other Agr-regulated factors may synergistically condense the sizes of S. aureus MVs.
3.3. Hld reduced the diversity of MV-loaded proteins
RN4220-Δhld and RN4220-ΔagrA showed lower abilities to produce MVs compared with wild-type RN4220. 4D-label-free proteomics was further performed to detect differentially loaded proteins in the ΔhldMVs and ΔagrAMVs. The results showed that 684 proteins were shared in wtMVs, ΔhldMVs, and ΔagrAMVs (Figure 3A). Compared with proteins included in wtMVs (704), Hld mutation resulted in increased proteins encapsulated in ΔhldMVs (1,381), and RNAIII inactivation even enhanced the proteins loaded by ΔagrAMVs (1,698). The differentially loaded proteins in wtMVs, ΔhldMVs, and ΔagrAMVs were primarily predicted cytoplasmic proteins (260), extracellular components (77), and membrane-associated proteins (128, Figure 3B). Compared with wtMVs, ΔhldMVs had 156 upregulated (fold change ≥ 2) and 12 downregulated (fold change ≤ 0.5) proteins, whereas ΔagrAMVs exhibited 277 upregulated and 59 downregulated proteins. However, ΔhldMVs presented 136 upregulated and 390 downregulated proteins relative to ΔagrAMVs (Figure 3C). These data indicated that loaded Hld was associated with reduced protein diversity in S. aureus MVs.
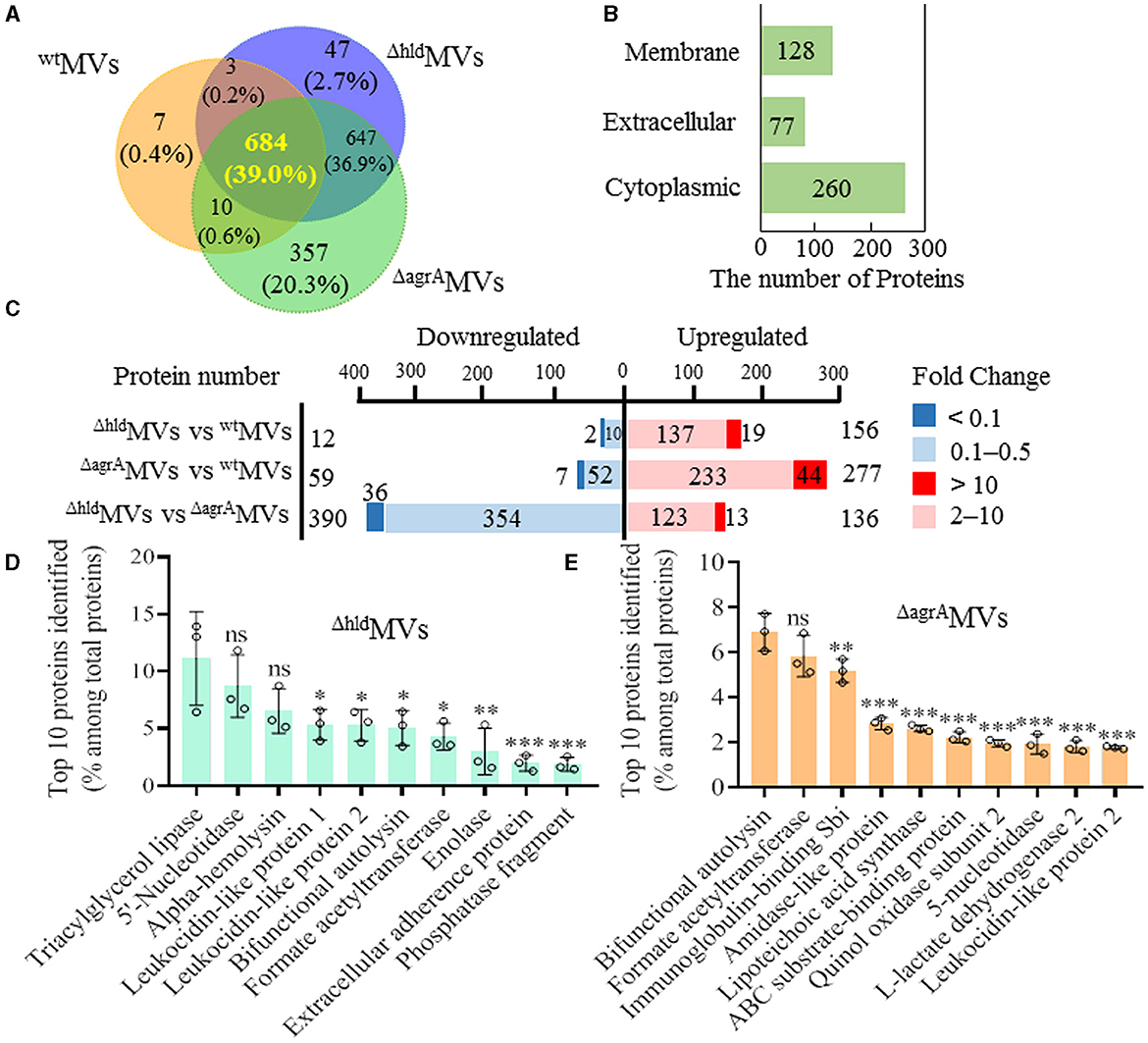
Figure 3. Comparative proteomic analysis of MVs from S. aureus RN4220, RN4220-Δhld, and RN4220-ΔagrA. (A) Venn diagram showing protein overlap among wtMVs, ΔhldMVs, and ΔagrAMVs. (B) Subcellular localization of differentially expressed proteins in ΔhldMVs and ΔagrAMVs. (C) Differentially expressed proteins between paired MVs indicated. The fold change ≥2 and ≤0.5 (P < 0.05) were considered as significantly upregulated and downregulated proteins, respectively. The protein number with fold change >10 or <0.1 was separately indicated. The top 10 proteins identified in (D) ΔhldMVs and (E) ΔagrAMVs. The proportion of the predominant proteins in MVs was characterized by LFQ intensity. The data were presented as mean ± SD (n = 3). Statistical significance was calculated by one-way ANOVA and compared with the LFQ intensity of the top one protein; ns indicates no statistical significance; ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
The top 10 MV proteins dominant in ΔhldMVs and ΔagrAMVs were analyzed. Out of the top 10 proteins, 8 were shared by ΔhldMVs and wtMVs (Figures 1B, 3D). Among the top 10 MV proteins, Hld and immunoglobulin-binding protein Sbi were unique in wtMVs, whereas extracellular adherence protein and 30 kDa neutral phosphatase fragment were specific for ΔhldMVs. By contrast, ΔagrAMVs had diverse abundant proteins (Figures 1B, 3E). KEGG analysis revealed that the upregulated proteins in ΔhldMVs vs. wtMVs were involved in ABC transporters, bacterial secretion, photosynthesis, and protein export, whereas the downregulated proteins were enriched in purine metabolism, quorum sensing, biosynthesis of cofactors, and nucleotide metabolism (Supplementary Figure 4A). GO analysis indicated that the most abundant differentially loaded proteins in ΔhldMVs vs. wtMVs were involved in cellular metabolic process and localization for biological process; catalytic activity, binding, and transporter activity for molecular function; and membrane, membrane part, cell, and cell part for cellular component (Supplementary Figure 4B). These results demonstrated that the loading of Hld in wtMVs may inhibit the encapsulation of other proteins and affect the diversity of MV-loaded proteins.
3.4. Loaded Hld contributed to the hemolytic activity of S. aureus MVs
Hld plays crucial roles in staphylococcal pathogenicity, such as causing hemolysis and allergy (Nakamura et al., 2013). The hemolytic activities of wtMVs, ΔhldMVs, and ΔagrAMVs were detected. Rabbit RBCs (2%) were mixed with 1, 2.5, 5, and 10 μg/ml of MVs, respectively, and incubated at 37°C for 30 min. The values of OD540 were detected, and the results showed that wtMVs presented higher hemolytic activity than ΔhldMVs and ΔagrAMVs (Figure 4A). The hemolytic activity of ΔhldMVs prepared from Hld mutant RN4220-Δhld significantly increased in an MV-dose-dependent manner (P < 0.001, Figure 4B). However, different concentrations of ΔagrAMVs derived from the agrA deletion strain RN4220-ΔagrA presented comparable hemolytic activity to PBS control. The content of hemolytic proteins in MVs was analyzed by proteomics, and the results showed that wtMVs loaded a large amount of hemolytic proteins with the most Hld, which accounted for 15.9% of the total proteins in wtMVs. The proportion of hemolytic proteins in ΔhldMVs was reduced, and Hla occupied the most in ΔhldMVs (6.8%). By contrast, the content of hemolytic proteins in ΔagrAMVs was the lowest (Supplementary Figure 5). These results indicated that Hld was an important component that contributed to the hemolytic activity of wtMVs, and other factors such as Hla and leukocidin-like proteins may also play important roles in the hemolytic activity of S. aureus MVs.
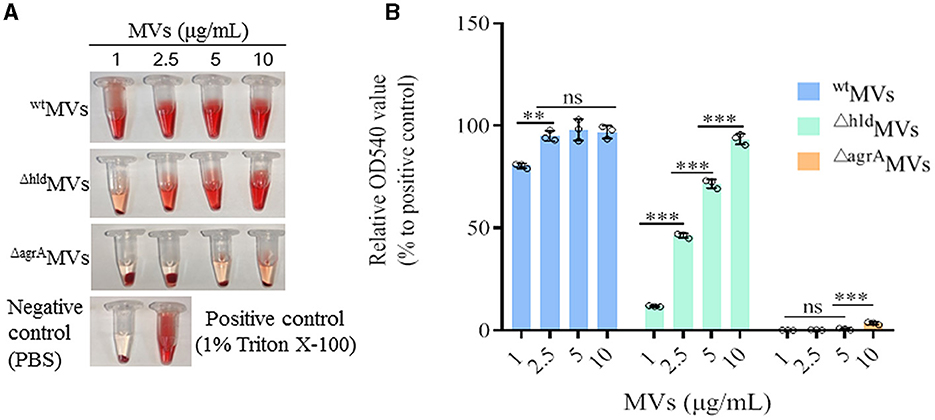
Figure 4. Hemolytic activity of wtMVs, ΔhldMVs, and ΔagrAMVs. (A) Hemolysis tested in tubes. Representative tubes carried 2% rabbit red blood cells after treatment with diverse concentrations of wtMVs, ΔhldMVs, and ΔagrAMVs. PBS-treated cells served as negative control, and 1% Triton X-100-treated cells served as positive control. (B) Hemolysis rate determination. The OD540 value of each hemolysis tube was determined. The OD540 value relative to the positive control was calculated. Data are presented as the mean ± SD of three independent experiments. Statistical significance was calculated by one-way ANOVA; ns represents no significance, ∗∗P < 0.01 and ∗∗∗P < 0.001.
3.5. Hld-loaded MVs were cytotoxic to macrophages
Macrophage RAW264.7 cells were treated with 5, 10, and 20 μg/ml of wtMVs, ΔhldMVs, or ΔagrAMVs for 6 h, and the cell death was evaluated. Microscopy revealed that RAW264.7 treated with 20 μg/ml of wtMVs presented greater cell disruption than those treated with ΔhldMVs (Figure 5A), whereas the morphology of RAW264.7 treated with 20 μg/ml ΔagrAMVs was similar to that of the PBS control group. CCK-8 assay and LDH release detection demonstrated that wtMV was highly toxic, even at the low concentration of 5 μg/ml (Figures 5B, C). However, the toxicity of ΔhldMVs was significantly reduced, and only a high concentration of ΔhldMVs (20 μg/ml) presented toxicity to RAW264.7 cells compared with the PBS control. No significant difference in cell survival and LDH release was observed between ΔagrAMVs and PBS treatment. The proteomic data showed that wtMVs loaded a large amount of virulence proteins (33.7%), whereas 19.36% of the total proteins in ΔhldMVs were counted to be virulence factors, and only 5.1% of virulence proteins were loaded in ΔagrAMVs (Supplementary Figure 5). These data suggested that Hld-loaded wtMVs were more toxic to RAW264.7 macrophages.
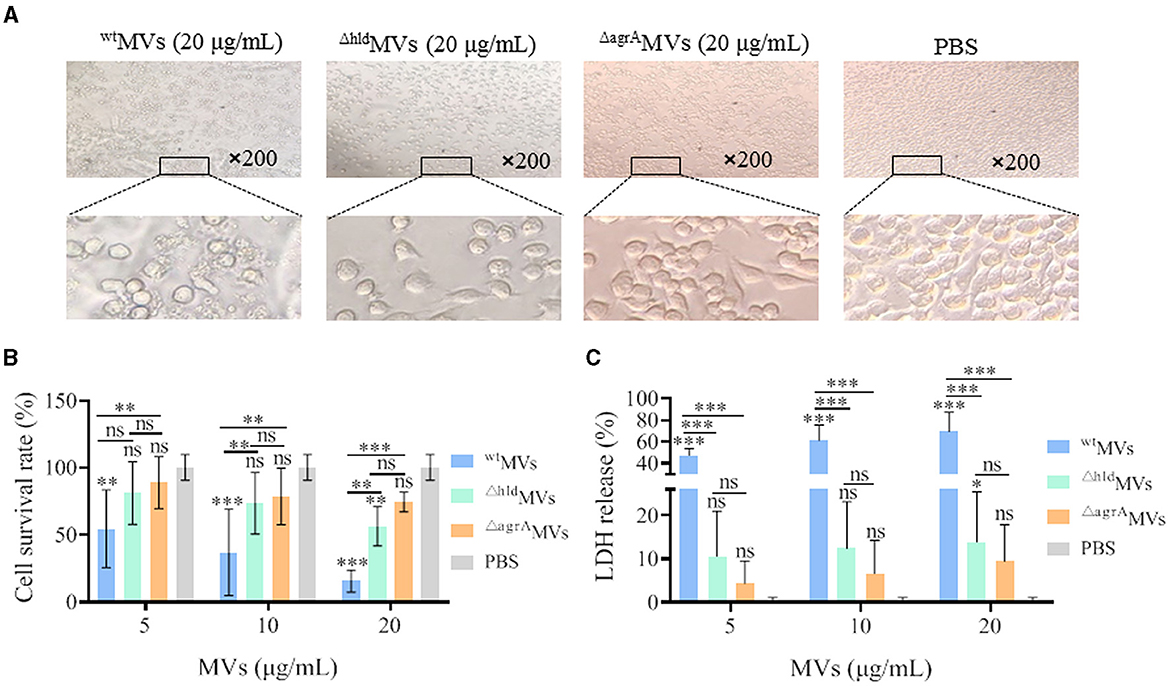
Figure 5. Effect of MVs on macrophage RAW264.7 cells. (A) Morphological changes of RAW264.7 cells after treatment with wtMVs, ΔhldMVs, and ΔagrA MVs at 20 μg/ml concentration. (B) Cell-survival rate of RAW264.7 cells. Macrophage cells were treated with different concentrations of wtMVs, ΔhldMVs, or ΔagrAMVs for 6 h, and the cell-survival rate was measured by CCK-8 assay. (C) LDH release of RAW264.7 cells. Cells were treated with diverse concentrations of wtMVs, ΔhldMVs, or ΔagrAMVs for 6 h, and LDH release was measured by a Cytotoxicity Assay Kit. PBS served as a negative control. Data are presented as mean ± SD of three independent experiments. Statistical significance was calculated by one-way ANOVA relative to that of PBS control in each group, and the other comparisons were also indicated; ns represents no significance; ∗P < 0.1, ∗∗P < 0.01, and ∗∗∗P < 0.001.
3.6. Loaded Hld MVs promoted the production of inflammatory factors in vivo
The levels of inflammatory factor production represent the activation of the immune system caused by bacterial virulence factors in vivo. BALB/c mice were intraperitoneally injected with different concentrations of wtMVs, ΔhldMVs, and ΔagrAMVs (n = 3 per group). The sera were collected at 6 h post-injection and subjected to quantification of IL-6, IL-1β, and TNFα levels through ELISA. As shown in Figure 6A, the IL-6 levels in mice treated with 20 μg/ml of wtMVs significantly increased compared with those in ΔhldMV- and ΔagrAMV-challenged mice (P < 0.001). However, comparable serum IL-1β and TNFα levels were presented between MV- and PBS-treated mice (Figures 6B, C). These results indicated that loaded Hld was a potent virulence factor for S. aureus MV pathogenicity.
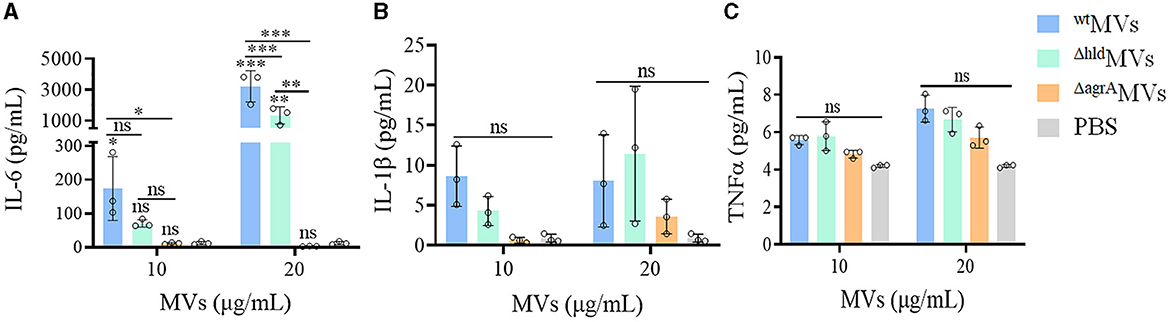
Figure 6. Effects of MVs on inflammatory factor production in vivo. Mice were injected with wtMVs, ΔhldMVs, or ΔagrAMVs for 6 h, and the levels of inflammatory factors (A) IL-6, (B) IL-1β, and (C) TNFα in the mouse sera were determined with an ELISA kit. PBS served as a negative control. Data are presented as the mean ± SD (n = 3 for each group). Statistical significance was calculated by one-way ANOVA relative to that of PBS control in each group, and the other comparisons were also indicated; ns represents no significance, ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
4. Discussion
MV release is a ubiquitous process of bacteria during their growth in vitro and in vivo (Beveridge, 1999). The proteins identified in MVs vary due to bacterial species, strains, culture conditions, MV preparation processes, and characterization methods used (Bitto et al., 2021b). A total of 431 proteins have been identified from MVs of the gram-positive pathogen Clostridium perfringens strain CP4 by using 1D gel separation and LC-MS/MS analysis (Jiang et al., 2014). With label-free quantitative LC-MS/MS determination, 543 proteins are characterized from MVs of a clinical isolate Cutibacterium acnes (Jiang et al., 2022). Lee et al. (2009) first discovered MV release in S. aureus ATCC14458 and identified approximately 90 proteins in the MVs through nano-LC-ESI-MS/MS. A total of 222 proteins are consistently characterized in MVs derived from S. aureus N305, a bovine mastitis isolate (Tartaglia et al., 2018), and ~253 proteins are associated with MVs derived from S. aureus strains O11, O46, RF122, and MW2 after nano-LC-ESI-MS/MS analysis (Tartaglia et al., 2020). In the current study, 4D-label-free LC-MS/MS was used to identify proteins encapsulated in MVs of S. aureus RN4220 (Cox et al., 2011; Meier et al., 2018). A total of 704 proteins were consistently identified in all three parallel biological samples of wtMVs derived from wild-type RN4220. The high sensitivity and deep coverage of 4D-label-free LC-MS/MS analysis resulted in 612 more proteins determined in wtMVs derived from the same S. aureus strain (Yuan et al., 2018).
The function of certain MV-loaded proteins is seldom investigated, but the loading of some components such as cytoplasmic proteins and virulence factors has been characterized in bacterial MVs (Yuan et al., 2018). Herein, Hld presented the highest LFQ intensity in the 4D-label-free LC-MS/MS analysis of proteins loaded in wtMVs (Figure 1B). SDS-PAGE and Western blot assays revealed that Hld was the most abundant protein in MVs prepared from S. aureus strains with different genetic lineages (Figures 1C, E), which was confirmed by another study (Jeon et al., 2016). Our data and other findings suggested that loaded Hld in staphylococcal MVs is a common feature. As one of the major virulence factors, Hld is involved in staphylococcal infections by targeting an extensive variety of mammalian cells, such as human monocytes, polymorphonuclear leukocytes, macrophages, mast cells, and erythrocytes (Hodille et al., 2016; Rao et al., 2022). MV-assisted release and transportation may be very important for Hld function. Our results showed that loaded Hld contributed to the hemolytic activity of S. aureus MVs, and Hld mutation remarkably reduced the hemolytic activity of ΔhldMVs (Figure 4). Moreover, Hld-loaded wtMVs were more cytotoxic to macrophage RAW264.7 cells than ΔhldMVs and ΔagrAMVs (Figures 5A, C). Importantly, Hld-loaded wtMVs increased the stimulatory effect of the inflammatory factor production in vivo (Figure 6), indicating that Hld-loaded wtMVs had strong virulence.
The mechanisms underlying S. aureus MV production and regulation are largely unknown (Qiao et al., 2021). The present study revealed that Hld played an important role in the production of S. aureus MVs (Figure 2B). In view of MV application, the large-scale preparation of bacterial MVs is valuable (Wang et al., 2018). However, Hld utilization in promoting staphylococcal MVs may raise safety concerns, such as Hld loading and its cytotoxicity. Interestingly, our data demonstrated that Hld promoted the production of S. aureus MVs with relatively smaller sizes, Hld mutation considerably enlarged the size of ΔhldMVs, and Agr inactivation resulted in even larger ΔagrAMVs (Figures 2C, D). These data indicated that Agr-associated virulence factors may reduce the size of S. aureus MV particles. Other factors involved in S. aureus MV size compression are worthy of further investigation.
Apart from the yield increase and size reduction of Hld-loaded wtMVs, the proteins included in wtMVs (704) were fewer than those encapsulated in ΔhldMVs (1,381) and ΔagrAMVs (1,698) after 4D-label-free LC-MS/MS analysis (Figure 3A). The encapsulation of Hld in wtMVs decreased the diversity of proteins loaded in S. aureus MVs, but the reason for this phenomenon is unclear. Smaller sizes and a competitive mechanism may contribute to the encapsulation of certain proteins into S. aureus MVs. The top 10 MV proteins dominant in wtMVs, ΔhldMVs, and ΔagrAMVs were analyzed (Figures 1B, 3D, E). Among the top 10 loaded proteins, Hld and immunoglobulin-binding protein Sbi were unique for wtMVs (Al Kindi et al., 2021), whereas extracellular adherence protein and 30 kDa neutral phosphatase fragment dominated in ΔhldMVs. KEGG analysis showed that upregulated proteins in ΔhldMVs vs. wtMVs primarily functioned in ABC transporters, bacterial secretion, and protein export, whereas the downregulated ones were enriched in purine metabolism, quorum sensing, and nucleotide metabolism (Supplementary Figure 4A). These results suggested that the inhibition of protein loading into S. aureus MVs by Hld may be selective, with a decrease in transport-related proteins. The significance and mechanism underlying the Hld modification of S. aureus MV protein encapsulation require further investigation.
In conclusion, our data shed light on the role of Hld in the property modification of S. aureus MVs. Hld was identified as the most abundant toxin in MVs of S. aureus strains with diverse genetic lineages. It could promote the release of staphylococcal MVs with smaller sizes than those derived from Hld mutant. Moreover, loaded Hld resulted in reduced protein diversity in S. aureus MVs. The loaded Hld contributed to the hemolytic activity of wtMVs, which exhibited greater toxicity to macrophage RAW264.7 cells than ΔhldMVs and ΔagrAMVs. Furthermore, Hld-loaded wtMVs promoted the production of inflammatory factor IL-6 in vivo. The loading and multifaceted roles of Hld in the vesicle property modification of S. aureus MVs suggested that a rational construction was needed to prepare MVs for application.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was approved by Laboratory Animal Welfare and Ethics Committee of the Army Medical University (Protocol No. AMUWEC2020735). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
XR: Conceptualization, Project administration, Supervision, Writing—review and editing. JC: Funding acquisition, Data curation, Investigation, Methodology, Software, Writing—original draft. YL: Methodology, Data curation, Investigation, Writing—review and editing. WS: Data curation, Investigation, Methodology, Software, Writing—original draft. YY: Formal analysis, Methodology, Writing—review and editing. YW: Data curation, Writing—original draft. ZH: Formal analysis, Writing—original draft. XH: Formal analysis, Methodology, Writing—original draft. RZ: Project administration, Supervision, Writing—review and editing. JY: Conceptualization, Funding acquisition, Visualization, Writing—review and editing. JH: Investigation, Resources, Writing—review and editing.
Funding
This study was supported by the National Natural Science Foundation of China (grant numbers 82302063 to JC and 32000652 to JY).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1254367/full#supplementary-material
References
Al Kindi, A., Williams, H., Matsuda, K., Alkahtani, A. M., Saville, C., Bennett, H., et al. (2021). Staphylococcus aureus second immunoglobulin-binding protein drives atopic dermatitis via IL-33. J. Allergy Clin. Immun. 147, 1354–1368.e3. doi: 10.1016/j.jaci.2020.09.023
Askarian, F., Lapek, J. D., Dongre, M., Tsai, C., Kumaraswamy, M., Kousha, A., et al. (2018). Staphylococcus aureus membrane-derived vesicles promote bacterial virulence and confer protective immunity in murine infection models. Front. Microbiol. 9, 262. doi: 10.3389/fmicb.2018.00262
Beveridge, T. J. (1999). Structures of Gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181, 4725–4733. doi: 10.1128/JB.181.16.4725-4733.1999
Bitto, N. J., Cheng, L., Johnston, E. L., Pathirana, R., Phan, T. K., Poon, I. L. H., et al. (2021a). Staphylococcus aureus membrane vesicles contain immunostimulatory DNA, RNA and peptidoglycan that activate innate immune receptors and induce autophagy. J. Extracell. Vesic. 10, e12080. doi: 10.1002/jev2.12080
Bitto, N. J., Zavan, L., Johnston, E. L., Stinear, T. P., Hill, A. F., and Kaparakis-Liaskos, M. (2021b). Considerations for the analysis of bacterial membrane vesicles: methods of vesicle production and quantification can influence biological and experimental outcomes. Microbiol. Spectr. 9, e01273–e01221. doi: 10.1128/Spectrum.01273-21
Briaud, P., and Carroll, R. K. (2020). Extracellular vesicle biogenesis and functions in Gram-positive bacteria. Infect. Immun. 88, e00433–e00420. doi: 10.1128/IAI.00433-20
Brown, L., Wolf, J. M., Prados-Rosales, R., and Casadevall, A. (2015). Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 13, 620–630. doi: 10.1038/nrmicro3480
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformat. 10, 421. doi: 10.1186/1471-2105-10-421
Chen, G., Bai, Y., Li, Z., Wang, F., Fan, X., and Zhou, X. (2020). Bacterial extracellular vesicle-coated multi-antigenic nanovaccines protect against drug-resistant Staphylococcus aureus infection by modulating antigen processing and presentation pathways. Theranostics 10, 7131–7149. doi: 10.7150/thno.44564
Chen, X, Zhang, J., Yang, M., Du, G., and Chen, F. (2023). Methicillin-resistant Staphylococcus aureus membrane vesicles inhibit the proliferation and induce the apoptosis of epithelial cells. Pathogens 11, 1429. doi: 10.3390/pathogens11121429
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., Robles, M., et al. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Cox, J., Hein, M. Y., Luber, C. A., Paron, I., Nagaraj, N., and Mann, M. (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 13, 2513–2526. doi: 10.1074/mcp.M113.031591
Cox, J., Neuhauser, N., Michalski, A., Scheltema, R. A., Olsen, J. V., and Mann, M. (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805. doi: 10.1021/pr101065j
Escudé Martinez de Castilla, P., Tong, L., Huang, C., Sofias, A. M., Pastorin, G., Chen, X., et al. (2021). Extracellular vesicles as a drug delivery system: a systematic review of preclinical studies. Adv. Drug Deliver. Rev. 175, 113801. doi: 10.1016/j.addr.2021.05.011
Gurung, M., Moon, D. C., Choi, C. W., Lee, J. H., Bae, Y. C., Kim, J., et al. (2011). Staphylococcus aureus produces membrane-derived vesicles that induce host cell death. PLoS ONE 6, e27958. doi: 10.1371/journal.pone.0027958
Hodille, E., Cuerq, C., Badiou, C., Bienvenu, F., Steghens, J. P., Cartier, R., et al. (2016). Delta hemolysin and phenol-soluble modulins, but not alpha hemolysin or panton-valentine leukocidin, induce mast cell activation. Front. Cell. Infect. Microbiol. 6, 180. doi: 10.3389/fcimb.2016.00180
Hong, S. W., Kim, M. R., Lee, E. Y., Kim, J. H., Kim, Y. S., Jeon, S. G., et al. (2011). Extracellular vesicles derived from Staphylococcus aureus induce atopic dermatitis-like skin inflammation. Allergy 66, 351–359. doi: 10.1111/j.1398-9995.2010.02483.x
Hoon, M. J., Imoto, S., Nolan, J., and Miyano, S. (2004). Open source clustering software. Bioinformatics 20, 1453–1454. doi: 10.1093/bioinformatics/bth078
Jeon, H., Oh, M. H., Jun, S. H., Kim, S. I., Choi, C. W., Kwon, H. I., et al. (2016). Variation among Staphylococcus aureus membrane vesicle proteomes affects cytotoxicity of host cells. Microb. Pathog. 93, 185–193. doi: 10.1016/j.micpath.2016.02.014
Jiang, M., Fan, X., Jiang, Z., Chen, H., Liu, Y., Yu, T., et al. (2022). Comparative proteomic analysis of membrane vesicles from clinical C. acnes isolates with differential antibiotic resistance. Clin. Cosmet. Investig. Dermatol. 15, 703–712. doi: 10.2147/CCID.S363537
Jiang, Y., Kong, Q., Roland, K. L., and Curtiss, R. III. (2014). Membrane vesicles of Clostridium perfringens type A strains induce innate and adaptive immunity. Int. J. Med. Microbiol. 304, 431–443. doi: 10.1016/j.ijmm.2014.02.006
Kaparakis-Liaskos, M., and Ferrero, R. L. (2015). Immune modulation by bacterial outer membrane vesicles. Nat. Rev. Immunol. 15, 375–387. doi: 10.1038/nri3837
Kumaraswamy, M., Wiull, K., Joshi, B., Sakoulas, G., Kousha, A., Vaaje-Kolstad, G., et al. (2021). Bacterial membrane-derived vesicles attenuate vancomycin activity against methicillin-resistant Staphylococcus aureus. Microorganisms 9, 2055. doi: 10.3390/microorganisms9102055
Lee, A. R., Park, S. B., Kim, S. W., Jung, J. W., Chun, J. H., Kim, J., et al. (2022). Membrane vesicles from antibiotic-resistant Staphylococcus aureus transfer antibiotic-resistance to antibiotic-susceptible Escherichia coli. J. Appl. Microbiolol. 132, 2746–2759. doi: 10.1111/jam.15449
Lee, E. Y., Choi, D. Y., Kim, D. K., Kim, J. W., Park, J. O., Kim, S., et al. (2009). Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 9, 5425–5436. doi: 10.1002/pmic.200900338
Liu, H., Shang, W., Hu, Z., Zheng, Y., Yuan, J., Hu, Q., et al. (2018). A novel SigB(Q225P) mutation in Staphylococcus aureus retains virulence but promotes biofilm formation. Emerg. Microbes Infect. 7, 72. doi: 10.1038/s41426-018-0078-1
MacDonald, K. L., and Beveridge, T. J. (2002). Bactericidal effect of gentamicin-induced membrane vesicles derived from Pseudomonas aeruginosa PAO1 on gram-positive bacteria. Can. J. Microbiol. 48, 810–820. doi: 10.1139/w02-077
Meier, F., Brunner, A. D., Koch, S., Koch, H., Lubeck, M., Krause, M., et al. (2018). Online parallel accumulation-serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteom. 17, 2534–2545. doi: 10.1074/mcp.TIR118.000900
Miller, R. P., Berlois, M. E., Hall, A. G., Simpson, A. H. R. W., Smith, I. D. M., and Hall, A. C. (2021). Effects of antibiotics on alpha-toxin levels during Staphylococcus aureus culture: Implications for the protection of chondrocytes in a model of septic arthritis. Cartilage 12, 362–376. doi: 10.1177/1947603519828433
Nakamura, Y., Oscherwitz, J., Cease, K. B., Chan, S. M., Muñoz-Planillo, R., Hasegawa, M., et al. (2013). Staphylococcus δ-toxin induces allergic skin disease by activating mast cells. Nature 503, 397–340. doi: 10.1038/nature12655
Pader, V., Hakim, S., Painter, K. L., Wigneshweraraj, S., Clarke, T. B., and Edwards, A. M. (2016). Staphylococcus aureus inactivates daptomycin by releasing membrane phospholipids. Nat. Microbiol. 2, 16194. doi: 10.1038/nmicrobiol.2016.194
Parianichnikov, N., Koch, H., Koch, S., Lubeck, M., Heilig, R., Brehmer, S., et al. (2020). MaxQuant software for ion mobility enhanced shotgun proteomics. Mol. Cell. Proteom. 19, 1058–1069. doi: 10.1074/mcp.TIR119.001720
Peng, H., Hu, Q., Shang, W., Yuan, J., Zhang, X., Liu, H., et al. (2017). WalK(S221P), a naturally occurring mutation, confers vancomycin resistance in VISA strain XN108. J. Antimicrob. Chemother. 72, 1006–1013. doi: 10.1093/jac/dkw518
Qiao, L., Rao, Y., Zhu, K., Rao, X., and Zhou, R. (2021). Engineered remolding and application of bacterial membrane vesicles. Front. Microbiol. 12, 729369. doi: 10.3389/fmicb.2021.729369
Quevillon, E., Silventoinen, V., Pillai, S., Harte, N., Mulder, N., Apweiler, R., et al. (2005). InterProScan: protein domains identifier. Nucl. Acids Res. 33, W116–120. doi: 10.1093/nar/gki442
Rao, Q., Shang, W., Zhou, K., Zhou, R., and Rao, X. (2015). Fatal multiple organ failure in an adolescent due to community-acquired methicillin-susceptible Staphylococcus aureus ST121/agrIV lineage: case report and review. Rev. Med. Microbiol. 26, 163–167. doi: 10.1097/MRM.0000000000000050
Rao, Y., Peng, H., Shang, W., Hu, Z., Yang, Y., Tan, L., et al. (2022). A vancomycin resistance-associated WalK(S221P) mutation attenuates the virulence of vancomycin-intermediate Staphylococcus aureus. J. Adv. Res. 40, 167–178. doi: 10.1016/j.jare.2021.11.015
Rivera, J., Cordero, R. J. B., Nakouzi, A. S., Frases, S., Nicola, A., and Casadevall, A. (2010). Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc. Natl. Acad. Sci. U. S. A. 107, 19002–19007. doi: 10.1073/pnas.1008843107
Saldanha, A. J. (2004). Java treeview–extensible visualization of microarray data. Bioinformatics 20, 3246–3248. doi: 10.1093/bioinformatics/bth349
Tartaglia, N. R., Breyne, K., Meyer, E., Cauty, C., Jardin, J., Chretien, D., et al. (2018). Staphylococcus aureus extracellular vesicles elicit an immunostimulatory response in vivo on the murine mammary gland. Front. Cell. Infect. Microbiol. 8, 277. doi: 10.3389/fcimb.2018.00277
Tartaglia, N. R., Nicolas, A., Rodovalho, V. R., Luz, B. S. R., Briard-Bion, V., Krupva, Z., et al. (2020). Extracellular vesicles produced by human and animal Staphylococcus aureus strains share a highly conserved core proteome. Sci. Rep. 10, 8467. doi: 10.1038/s41598-020-64952-y
Veith, P. D., Chen, Y. Y., Gorasia, D. G., Chen, D., Glew, M. D., O'Brien-Simpson, N. M., et al. (2014). Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J. Proteome Res. 13, 2420–2432. doi: 10.1021/pr401227e
Verdon, J., Girardin, N., Lacombe, C., Berjeaud, J. M., and Hechard, Y. (2009). Delta-hemolysin, an update on a membrane-interacting peptide. Peptides 30, 817–823. doi: 10.1016/j.peptides.2008.12.017
Wang, X., Koffi, P. F., English, O. F., and Lee, J. C. (2021). Staphylococcus aureus extracellular vesicles: a story of toxicity and the stress of 2020. Toxins 13, 75. doi: 10.3390/toxins13020075
Wang, X., Thompson, C. D., Weidenmaier, C., and Lee, J. C. (2018). Release of Staphylococcus aureus extracellular vesicles and their application as a vaccine platform. Nat. Commun. 9, 1379. doi: 10.1038/s41467-018-03847-z
Yuan, J., Yang, J., Hu, Z., Yang, Y., Shang, W., Hu, Q., et al. (2018). Safe staphylococcal platform for the development of multivalent nanoscale vesicles against viral infections. Nano Lett. 18, 725–733. doi: 10.1021/acs.nanolett.7b03893
Zaborowska, M., Vazirisani, F., Shah, F. A., Firdaus, R., Omar, O., Ekström, K., et al. (2021). Immunomodulatory effects exerted by extracellular vesicles from Staphylococcus epidermidis and Staphylococcus aureus isolated from bone-anchored prostheses. Biomaterials 278, 121158. doi: 10.1016/j.biomaterials.2021.121158
Keywords: Staphylococcus aureus, membrane vesicle, Hld, agrA, inflammatory factor
Citation: Chen J, Lv Y, Shang W, Yang Y, Wang Y, Hu Z, Huang X, Zhang R, Yuan J, Huang J and Rao X (2023) Loaded delta-hemolysin shapes the properties of Staphylococcus aureus membrane vesicles. Front. Microbiol. 14:1254367. doi: 10.3389/fmicb.2023.1254367
Received: 07 July 2023; Accepted: 05 September 2023;
Published: 06 October 2023.
Edited by:
Maria Filippa Addis, University of Milan, ItalyReviewed by:
Francis E. Jenney Jr., Philadelphia College of Osteopathic Medicine Georgia, United StatesQian Liu, Shanghai Jiao Tong University, China
Copyright © 2023 Chen, Lv, Shang, Yang, Wang, Hu, Huang, Zhang, Yuan, Huang and Rao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jizhen Yuan, eXVhbmppemhlbkAxNjMuY29t; Jingbin Huang, aGpiMjAwOTEzNjRAMTI2LmNvbQ==; Xiancai Rao, cmFveGlhbmNhaUAxMjYuY29t
†These authors have contributed equally to this work
 Juan Chen
Juan Chen Yuhuan Lv2,3†
Yuhuan Lv2,3† Weilong Shang
Weilong Shang Yi Yang
Yi Yang Yuting Wang
Yuting Wang Zhen Hu
Zhen Hu Rong Zhang
Rong Zhang Jizhen Yuan
Jizhen Yuan Jingbin Huang
Jingbin Huang Xiancai Rao
Xiancai Rao