- 1School of Food Science and Engineering, Ningxia University, Yinchuan, Ningxia, China
- 2College of Life Sciences, Northwest A and F University, Yangling, Shaanxi, China
- 3School of Life Sciences, Ningxia University, Yinchuan, Ningxia, China
- 4College of Enology and Horticulture, Ningxia University, Yinchuan, Ningxia, China
- 5Wine Institution of Ningxia Region, Yinchuan, Ningxia, China
The organoleptic profile and quality of wine are affected by the presence of different non-Saccharomyces species and strains. Therefore, the identification and characterization of non-Saccharomyces yeasts are the first step to understand their function, and to develop a better strain selection program for winemaking. This study investigated the biodiversity of non-Saccharomyces yeasts associated with spontaneous fermentation of Cabernet Sauvignon wines from five sub-regions (Shi Zuishan, Yinchuan, Yu Quanying, Qing Tongxia and Hong Sibu) in Ningxia, China. Yeast species were identified by sequencing the 26S rRNA D1/D2 region, and strains at the subspecies level were discriminated using tandem repeat-tRNA (TRtRNA) PCR analysis. A total of 524 yeast colonies were isolated, and 19 non-Saccharomyces yeast species belonging to 10 genera were identified, including Aureobasidium pullulans, Cryptococcus albidus, Cryptococcus sp., C. flavescens, C. terrestris, C. magnus, Cystofilobasidium ferigula, Candida zemplinina, Filobasidium magnum, Filobasidium sp., F. elegans, Hanseniaspora uvarum, Metschnikowia pimensis, M. pulcherrima, Naganishia albida, Pichia kluyveri, P. kudriavzevii, Rhodotorula glutinis and R. graminis. Hanseniaspora uvarum, C. zemplinina, and M. pulcherrima were the three most dominated species, while other non-Saccharomyces species were only present in the early stage of spontaneous fermentations at different levels. Further, for the yeast discrimination at strain level, 34 profiles were obtained by amplification with primer pairs TtRNASC/5CAG, while 40 profiles were obtained with primer pairs TtRNASC/ISSR-MB. This study explored the diversity of non-Saccharomyces species in Ningxia, China, and made an important contribution of genetic resources for further strain development.
1. Introduction
During wine fermentation, yeasts could convert sugar to ethanol and carbon dioxide, glycerol, higher alcohols, aldehydes and esters, etc. According to the strength of fermentation capacity, yeasts are classified as Saccharomyces cerevisiae and non-Saccharomyces. In the past, non-Saccharomyces yeasts were considered as undesired or spoilage yeasts during wine fermentation. However, recent studies have showed the positive contributions of non-Saccharomyces yeasts to wine quality, such as generating aromatic compounds through enzymatic reactions, increasing glycerol production, decreasing ethanol content and stabilizing wine color (Ciani et al., 2016; Padilla et al., 2016; Medina et al., 2017; Benito et al., 2019; Morata et al., 2020; Maicas and Mateo, 2023). Moreover, most of these enological characteristics are species and strain dependent (Padilla et al., 2016; Binati et al., 2019). Therefore, the discrimination of different non-Saccharomyces yeast strains is a step of primary importance to their biotechnological application. In recent years, the regional flavor peculiarity of non-Saccharomyces strains has drawn widespread attention, and there are increasing demands for wine style diversification and local characteristics. Therefore, growing studies worldwide begin to focus on the development and utilization of wine-related non-Saccharomyces yeasts (Binati et al., 2019; Zhao et al., 2021; Lai et al., 2022; Morata et al., 2022; Tamang and Lama, 2023).
The research on the isolation, identification and enological characteristics of non-Saccharomyces yeasts is one of the research hot spots at present, which provides a reliable reference for the exploring of local non-Saccharomyces strains with great industrial potential (Casas-Godoy et al., 2021). Considering the great diversity and potential applications of different non-Saccharomyces yeast strains within the same species, it is important to discriminate them at both species and subspecies level for subsequent study. Although there are several well documented molecular biological techniques for identifying yeast species such as sequencing of 26S rRNA D1/D2 (Kurtzman and Robnett, 1998) and restriction analysis of the 5.8S rDNA ITS region (Guillamón et al., 1998; Lorca et al., 2018), there are limited studies available for discriminating non-Saccharomyces yeasts at subspecies level. In recent years, various technologies of molecular fingerprinting have been used for distinguishing non-Saccharomyces strains, such as random amplified polymorphic DNA (RAPD) (Binati et al., 2019), fourier transform infrared spectroscopy (FTIR) (Nieuwoudt et al., 2006; Grangeteau et al., 2016), tandem repeat-tRNA (TRtRNA) (Barquet et al., 2012), amplified fragment length polymorphisms (AFLP) (Esteve-Zarzoso et al., 2010; Albertin et al., 2016; Baselga et al., 2017), and microsatellites (Albertin et al., 2016). However, most of these methods have the drawbacks of laborious procedures and are not suitable for handling large quantity of samples. In addition, RAPD technology is susceptible to various factors, such as Mg2+, dNTP, template, and reaction temperature. FTIR cannot complete qualitative and quantitative identification of the cell components in a practical situation due to the effects of environmental changes. AFLP data are difficult to interpret, and requires complex bioinformatic programs. Compared with those methods, TRtRNA-PCR offers simplicity, reproducibility, and high discrimination power for typing non-Saccharomyces yeast strains (Barquet et al., 2012). According to the trend of simplification and automation, TRtRNA is a promising method to discriminate non-Saccharomyces yeast strains.
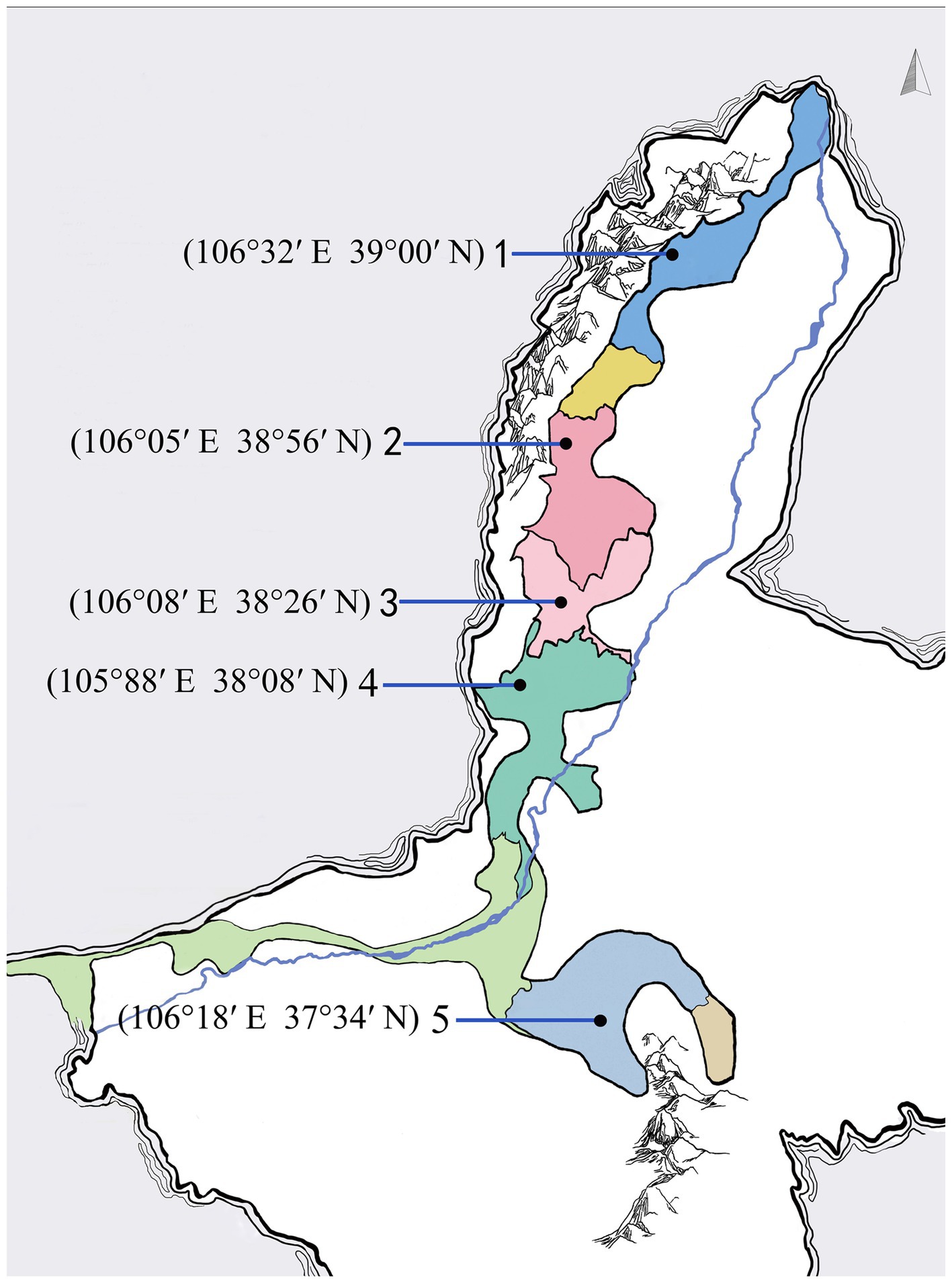
Ningxia is one of the oldest and promising wine producing regions in China (Sun et al., 2017). It can be divided into five sub-regions named Shi Zuishan, Yinchuan, Yu Quanying, Qing Tongxia and Hong Sibu (Figure 1), according to their different geographical location and climatic condition (Li et al., 2022). The typical geographical and climatic characteristics have facilitated to form rich and diverse yeast communities. In recent years, several case studies have explored the density and diversity of indigenous yeast community in Ningxia. Yeast species identified in Ningxia are generally associated with genera Candida, Hanseniaspora, Issatchenkia, Metschnikowia, Pichia, Saccharomyces, Torulaspora and Zygosaccharomyces (Li et al., 2011; Sun et al., 2017; Zhang et al., 2020). Although the enological characteristics of a few non-Saccharomyces yeast species were investigated, the wealth of non-Saccharomyces yeast biodiversity with still hidden potential in wine fermentation needs further investigation.
The aims of the present work were to evaluate the genetic diversity and relatedness among non-Saccharomyces strains of five sub-regions of Ningxia, to establish a strain collection to preserve the non-Saccharomyces genetic resources of China, and to provide reference and scientific basis for their further development for regional wine production. Firstly, this study analyzed the phylogenetic diversity of indigenous non-Saccharomyces yeasts isolated from Cabernet Sauvignon spontaneous fermentations. Then, the dynamics of different non-Saccharomyces species associated with fermentation stage and sub-regions were evaluated. Finally, TRtRNA-PCR was used to discriminate non-Saccharomyces yeasts at subspecies level. Dendrograms were constructed based on similarity among different patterns of bands, and the genetic relationships of all strains were also evaluated.
2. Materials and methods
2.1. Isolation of indigenous non-Saccharomyces yeast strains
The Cabernet Sauvignon grapes for this study were harvested in 5 sub-areas of Eastern Foot of Helan Mountain named Shi Zuishan, Yinchuan, Yu Quanying, Qing Tongxia and Hong Sibu, Ningxia, China (Figure 1). In 2020, Ripe and physically undamaged grapes were manually destemmed and crushed, and naturally fermented with the skin under aseptic conditions. The initial sugar content, total acidity, pH of grapes and residual sugars, alcohol content of final wines were shown in Supplementary Table S1. All the fermentations were conducted in 10.0 L fermenters, and the fermentations were performed in duplicate. 60 mg/L of potassium metabisulfite and 20 mg/L of pectinase were added to the must after crushing. The fermentation temperature was controlled at 25–28°C. The progress of the fermentations was monitored by measuring CO2 loss every 12 h (Supplementary Figure S1). Samples were collected during spontaneous fermentations at 1 d (grape must after crushing), 2 d, 3 d and 5 d.
At each stage, aliquots of several dilutions (from 10−1 to 10−6) were spread in triplicate on WLN agar (Hopebiol, China) and supplemented with 100 mg/L chloramphenicol to inhibit bacterial growth (Pallmann et al., 2001). For each sample, based on the colony morphology and frequency, 15 to 25 non-Saccharomyces colonies were proportionately isolated from the selected dilution plate, thus yielding 524 isolates for the subsequent identification. After purification on YPD agar (containing 2% glucose, 2% peptone, and 1% w/v yeast extract). All the strains were stored in 20% glycerol at −80°C for subsequent identification.
2.2. DNA extraction
The DNA extraction protocol for Non-Saccharomyces yeasts was adapted from previous studies (Renouf et al., 2007; Sun and Liu, 2014). 19 species in 10 genera of yeasts, Aureobasidium pullulans, Cryptococcus albidus, Cryptococcus sp., C. flavescens, C. terrestris, C. magnus, Cystofilobasidium ferigula, Candida zemplinina, Filobasidium magnum, Filobasidium sp., F. elegans, Hanseniaspora uvarum, Metschnikowia pimensis, M. pulcherrima, Naganishia albida, Pichia kluyveri, P. kudriavzevii, Rhodotorula glutinis and R. graminis were included. Briefly, the biomass grown on Petri dishes were collected in TE buffer and washed once. Then, 500 μL of nuclei lysis (Promega) and 300 μL of protein precipitation solution (Promega) were used for the DNA extraction. Residual polyphenols were precipitated after addition of 100 μL of a 10% polyvinyl-pyrrolidone (PVP; Sigma-Aldrich). Visible mass of DNA was obtained by addition 500 μL of isopropanol, and then 70% ethanol solution was added to re-suspend the pellet. Finally, DNA was qualified using DN-1000 Spectro-photomter (NanoDrop, United States). All DNA samples were stored at −20°C before use.
2.3. Identification of non-Saccharomyces species
All 524 yeast isolates were identified by sequence analysis of the 26S rRNA D1/D2 domain with primers NL1 and NL4 according to Kurtzman and Robnett (1998). PCR was performed in a final volume of 25 μL containing 0.2 mM of each dNTP, 1 μL DNA template, 1.5 mM MgCl2, 0.2 μM of each primer, 0.1 U Easy Taq Buffer Polymerase (TransGen Biotechnology, Beijing, China) and 1 × Easy Taq Buffer. Samples were subjected to the following thermal profile for amplification in FlexCycler2 PCR Thermal Cyclers (Model 844–00069, Analytik Jena AG, Jena, Germany): initial denaturation at 95°C for 5 min, 36 cycles of 94°C for 1 min, 52°C for 1 min and 72°C for 80 s, and a final extension at 72°C for 8 min. PCR products were separated by 2% (w/v) agarose gel electrophoresis in 0.5 × TBE buffer for 40 min at 110 V. The products gave positive results were sent to Sangon Biotech (Shanghai) Co. Ltd. for purification and sequencing. The sequences were analyzed using the Blast method of NCBI.1 DNA sequences were deposited in GenBank under the accession numbers: OP445815 Aureobasidium pullulans (NXU 21–01), OP445816 Cryptococcus albidus (NXU 21–02), OQ857299 Cryptococcus sp. (NXU 21–18), OP445817 Cryptococcus flavescens (NXU 21–03), OP445818 Cryptococcus terrestris (NXU 21–04), OP445819 Cryptococcus magnuse (NXU 21–05), OP445820 Cystofilobasidium ferigula (NXU 21–06), OP445821 Candida zemplinina (NXU 21–07), OP445822 Filobasidium magnum (NXU 21–08), OQ857300 Filobasidium sp. (NXU 21–19), OP445823 Filobasidium elegans (NXU 21–09), OP445824 Hanseniaspora uvarum (NXU 21–10), OP445825 Metschnikowia pimensis (NXU 21–11), OP445826 Metschnikowia pulcherrima (NXU 21–12), OP445827 Naganishia albida (NXU 21–13), OP445828 Pichia kluyveris (NXU 21–14), OP445829 Pichia kudriavzevii (NXU 21–15), OP445830 Rhodotorula glutinis (NXU 21–16) and OP445831 Rhodotorula graminis (NXU 21–17).
2.4. Phylogenetic analysis
The sequences of different non-Saccharomyces species were aligned using MAFFT version 7 (Katoh and Standley, 2013), and the sequence alignment was subjected to Maximum Likelihood analysis using IQ-TREE version 2.2.0 (Minh et al., 2020). Bootstrap values were calculated from 1,000 iterations.
2.5. Tandem repeat-tRNA PCR analysis
Tandem repeat-tRNA fingerprinting (TRtRNA) analysis of all non-Saccharomyces strains was carried out with TtRNASc (5′- GCTTCTATGGCCAAGTTG-3′), ISSR-MB (5′-CTCACAACAACAACAACA-3′), and 5CAG (5′- CAGCAGCAGCAGCAG-3′) according to Barquet et al. (2012). Amplification was performed as follows: volume of 20 μL, 2.5 mM MgCl2, 0.2 mM dNTPs, 1 μM each primer, 2 uL 10 × Easy Buffer and 1 U Taq DNA polymerase and 1 μL genomic DNA. Amplification was procedure as follows: Predenaturation 95°C for 5 min; Denaturation at 95°C for 60s and annealing at 50°C for 60s, 35 cycles of 90s at 72°C, and a final extension of 10 min at 72°C. The PCR products were separated by electrophoresis on 1.2% agarose gels which was applied with 100 V for 45 min in 0.5 × TBE buffer and photographed under UV light. TRtRNA sequence types of Angel yeast NS-D (T. delbrueckii), Excellence B-Nature (M. pulcherrima) and Excellence X-Fresh (L. thermotolerans) were used as references to compare the genetic profiles of the tested non-Saccharomyces strains.
2.6. Cluster analysis of the strains
The TRtRNA sequence patterns obtained after gel electrophoresis were used for the construction of a presence/absence matrix, taking into account the total number of different bands observed (Sun et al., 2017). Similarities based on the Dice coefficient were calculated and UPGMA clustering was obtained using NTSYS software (Mercado et al., 2010).
3. Results
3.1. Phylogenetic analysis of yeast isolates from different sub-regions of Ningxia
A total of 524 non-Saccharomyces isolates were collected during spontaneous fermentation of Cabernet Sauvignon wines from 5 sub-regins of Ningxia, China. As showed in Table 1, 19 species in 10 genera of non-Saccharomyces yeasts were identified by sequencing the 26S rRNA D1/D2 domain. The identified yeast species include Aureobasidium pullulans, Cryptococcus albidus, Cryptococcus sp., C. flavescens, C. terrestris, C. magnus, Cystofilobasidium ferigula, Candida zemplinina, Filobasidium magnum, Filobasidium sp., F. elegans, Hanseniaspora uvarum, Metschnikowia pimensis, M. pulcherrima, Naganishia albida, Pichia kluyveri, P. kudriavzevii, Rhodotorula glutinis and R. graminis. Table 1 shows the distribution, and origin of these yeast species. Regarding the different yeast species, H. uvarum and C. zemplinina were the two most abundant species. The proportion of H. uvarum was 73.66% (386/524) followed by C. zemplinina, 11.26% (59/524), while the proportions of other 17 species were less than 4.20% (22/524).
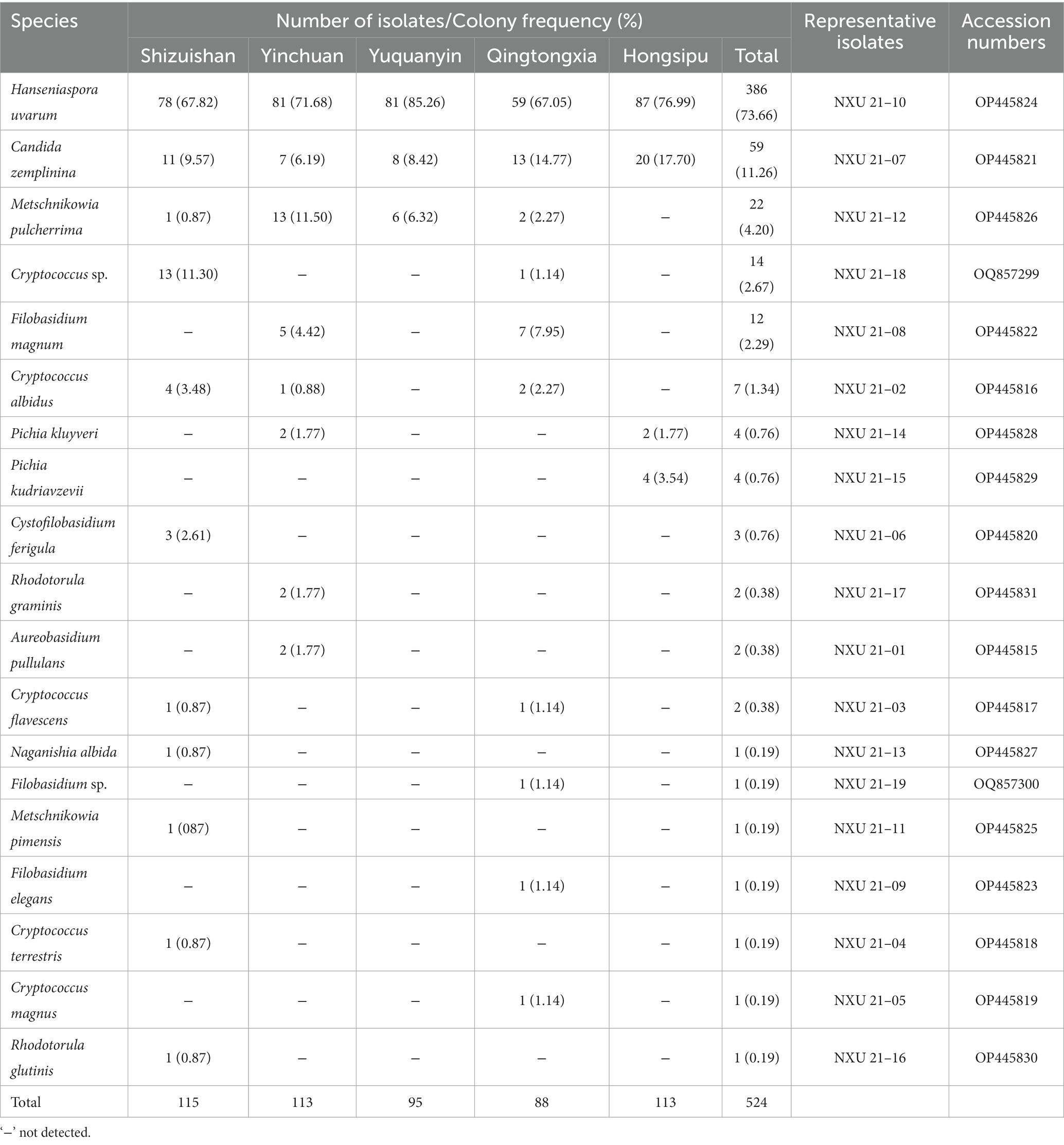
Table 1. The yeast species and their frequency isolated from spontaneous fermentations of Cabernet Sauvignon wines in different sub-regions.
To evaluate the evolutionary relationship of these species, a phylogenetic tree reflecting the genome homology and genetic distance of the yeasts was constructed (Figure 2). It is evident from the phylogenetic tree that A. pullulans formed a single clade while the other 18 species showing two main clades. In the phylogenetic tree, C. flavescens and C. terrestris, R. glutinis and R. graminis, P. kluyveri and P. kudriavzevii, M. pimensis and M. pulcherrima were clustered together, respectively.
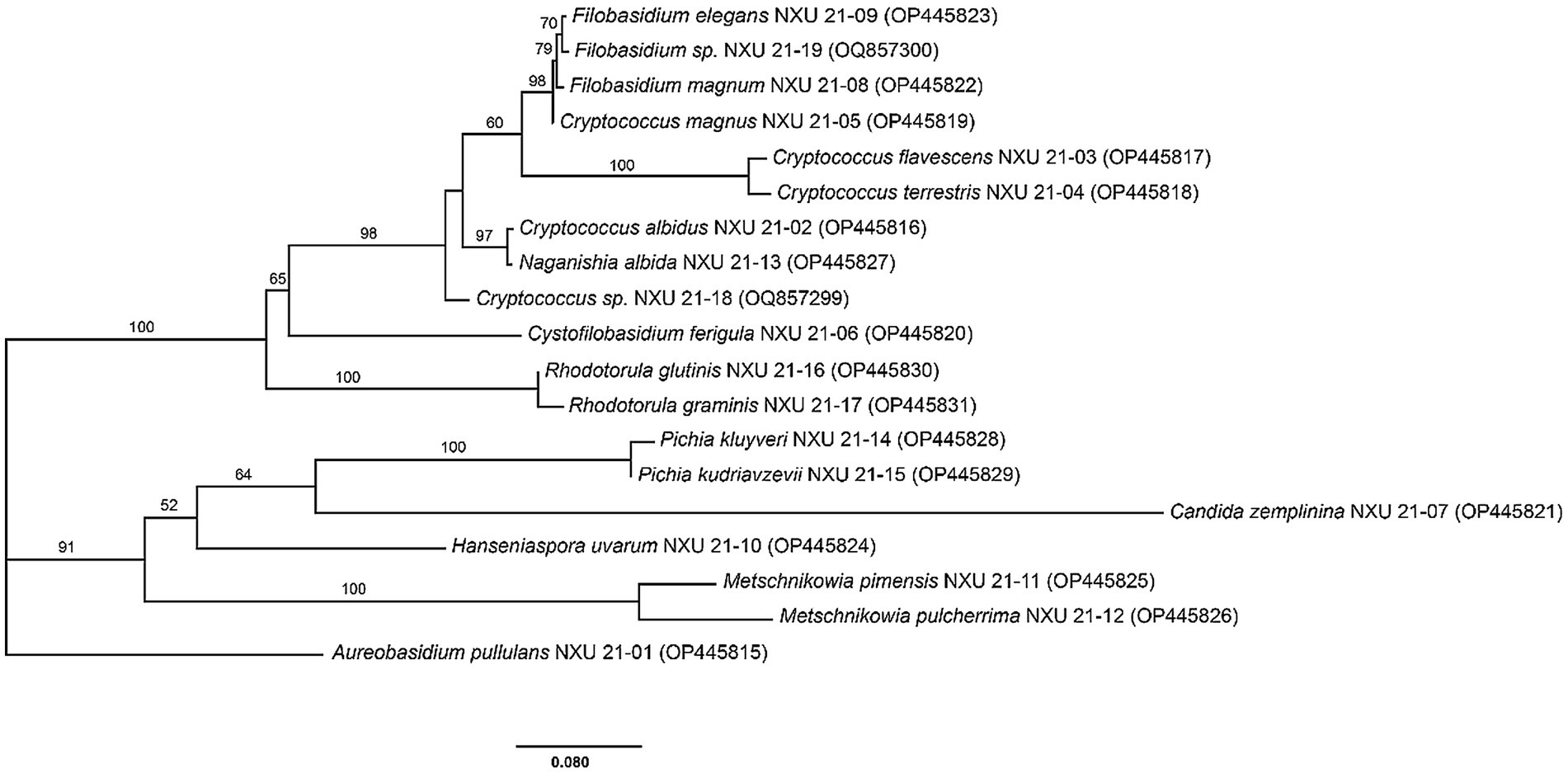
Figure 2. Phylogenetic tree of representative non-Saccharomyces yeast isolates obtained from five sub-regions of Ningxia based on the sequence analysis of the 26S rRNA D1/D2 region using the Maximum-Likelihood method. The scale bar shows 0.080, Bootstrap support values were estimated based on 1,000 replicates and are showed above the branches (> 50%).
3.2. Dynamics of non-Saccharomyces species during fermentations in different sub-regions
To explore the dynamic changes of non-Saccharomyces species during the spontaneous fermentation of Cabernet Sauvignon in different sub-regions, the numbers of yeasts at 1 d, 2 d, 3 d and 5 d were analyzed in this study. The yeast diversity and community changes at the species level during spontaneous fermentations are showed in Figure 3. An obvious change of yeast community composition was observed during the whole process in all five sub-regions. In general, day 1 showed the highest yeast diversity, and the numbers of yeast species decreased during fermentation process. More specifically, the decrement of yeast species in Shi Zuishan and Qing Tongxia was particularly evident, which decreased from 10 species at day 1 to 2 and 3 species, respectively, at day 2. However, the lowest yeast diversity was found in Yu Quanying with 3 species of H. uvarum, C. zemplinina and M. pulcherrima at day 1. On day 5, only Hong Sibu could be isolated 2 different yeast species compared with 1 species isolated in other four sub-regions. It was noted that H. uvarum showed an increasing trend throughout the fermentation process, especially in Yu Quanying, and the proportion of H. uvarum achieved up to 100% on day 2 until day 5. C. zemplinina was the secondly abundant species on day 2 to day 5. N. albida, R. glutinis, C. terrestris, C. ferigula and M. pimensis were only found in Shi Zuishan and were not detected in the other four sub-regions.
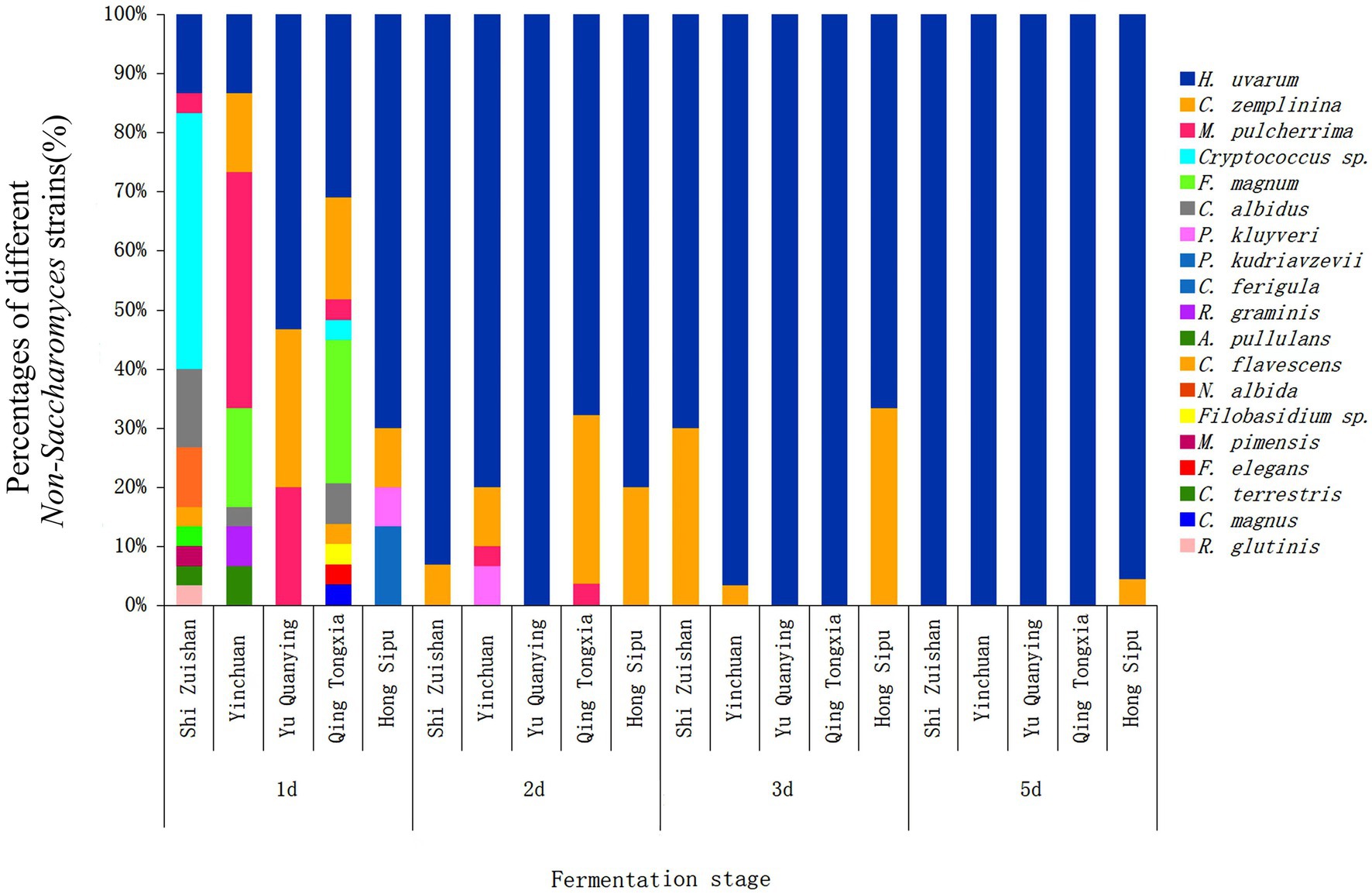
Figure 3. The relative abundance of non-Saccharomyces species isolated from different stages during Cabernet Sauvignon spontaneous fermentation in Shi Zuishan, Yin Chuan, Yu Quanying, Qing Tongxia, Hong Sibu.
3.3. Intraspecific genetic diversity in non-Saccharomyces
TRtNASC/5CAG and TRtNASC/ISSR-MB discriminated the 524 non-Saccharomyces isolates into 34 (Figure 4) and 40 profiles (Figure 5), respectively. Among them, neither primer pair could discriminate F. elegans, Filobasidium sp., M. pimensis and N. albida. Compared with TRtNASC/5CAG, pairs of primers TRtNASC/ISSR-MB resulted in a higher discrimination power with the exception of R. graminis. More specifically, TRtNASC/ISSR-MB could discriminate H. uvarum (386 isolates), C. zemplinina (59 isolates), M. pulcherrima (22 isolates), Cryptococcus sp., (14 isolates), and R. glutinis (1 isolate) into 6, 7, 4, 3 and 1 profiles, respectively (Figure 5). However, TRtNASC/5CAG could discriminate these yeast species into 5, 4, 2, 2 and 0 profiles, respectively (Figure 4). For the other 9 yeast species, both the two pairs showed the same discrimination power for the same species.
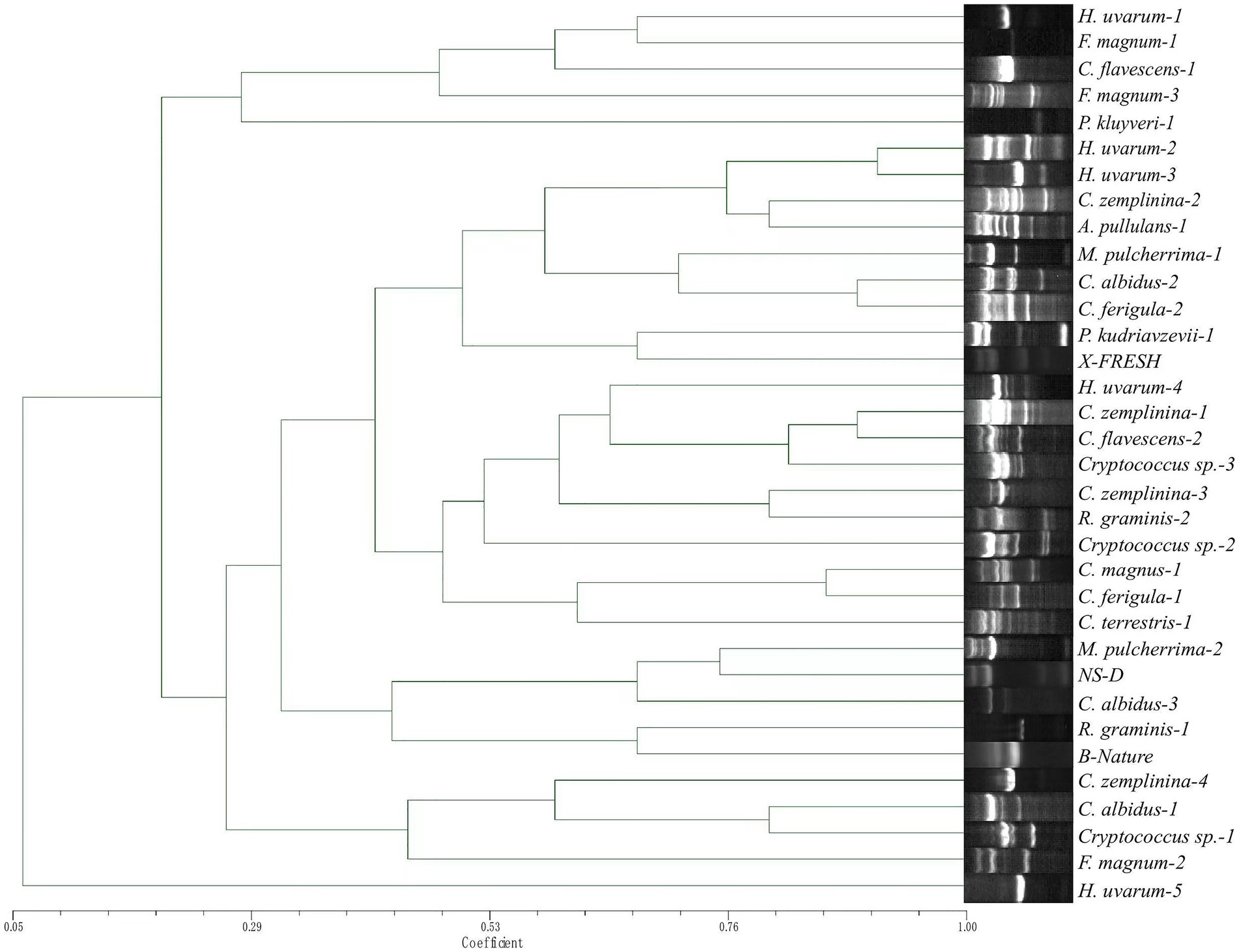
Figure 4. Unweighted pair group method with arithmetic mean dendrogram showing genetic relatedness of the non-Saccharomyces isolates obtained from Eastern foot of Helan Mountain distinguished by primer pairs TRtNASC and 5CAG.

Figure 5. Unweighted pair group method with arithmetic mean dendrogram showing genetic relatedness of the non-Saccharomyces isolates obtained from Eastern foot of Helan Mountain distinguished by primer pairs TRtNASC and ISSR-MB.
Genetic relatedness was also evaluated by constructing dendrograms compiled from all Tandem repeat-tRNA (TRtRNA) fingerprinting. For primer pairs TRtNASC and ISSR-MB (Figure 5), an obvious difference was observed in the relationships of different non-Saccharomyces species. All 8 strains of H. uvarum were clustered together when the Dice coefficient is 0.482. Cystofilobasidium ferigula formed a single clade, while most of the Cryptococcus species except for Cryptococcus sp.-2, C. magnus-1 and C. zemplinina were cluster together with a Dice coefficient of 0.35. Six groups are indicated in Figure 4, when the Dice coefficient is 0.434. Pichia kudriavzevii-1 and H. uvarum-5 formed a single clade respectively, while other groups contain at least four species.
4. Discussion
Non-Saccharomyces yeasts are essential in the development of important metabolites that determine the wine flavor and aroma, greatly influencing the characteristics of wine (Jolly et al., 2013). Understanding the genetic diversity of non-Saccharomyces strains can make an important contribution toward delineating the genetic distance of these strains, as well as providing genetic material for further strain development.
4.1. Non-Saccharomyces yeast species and their function
In this study, the genetic diversity of indigenous non-Saccharomyces strains was investigated during the spontaneous fermentations of Cabernet Sauvignon grown in 5 sub-areas of Ningxia wine region, China. 19 yeast species in 10 genera were isolated and identified in this study. Compared to previous studies (Li et al., 2011; Sun et al., 2017; Wang et al., 2017; Zhang et al., 2020, 2021), the yeast species of C. zemplinina, H. uvarum, M. pulcherrima and P. kluyveri were the frequently found species but with different distribution. However, some of these species like A. pullulans, C. albidus, C. flavescens, C. terrestris, C. ferigula, F. magnum, F. elegans, F. magnum, M.pimensis, N. albida, R. glutinis and R. graminis have not been characterized previously in this region. The findings from these different studies suggest the time- and location-dependent variations in yeast communities. In addition, this study revealed that in different sub-regions the yeast distributions were different. Specifically, 10 species in 7 genera were found in Shizuishan compared to 3 species in 3 genera found in Yuquanying. The same observations were made in the Shangri-La wine region, China (Zhao et al., 2021), which showed that different non-Saccharomyces associated with spontaneous fermentation of Cabernet Sauvignon were present at different geographical location with different microclimate characteristics. Likewise, in our study, differences in location and microclimate were responsible for the large effect on the variation in yeast diversity during spontaneous fermentation of Cabernet Sauvignon wines in different sub-regions. Since yeast community variations during spontaneous fermentation are attributed to the location, vintage year, cultivar., enological practices (such as SO2 addition and yeast inoculation) (Andorrà et al., 2008; Sun and Liu, 2014; Wang et al., 2021), our findings represent only a ‘snapshot in time’. Garofalo et al. (2016) found significant differences in non-Saccharomyces biodiversity among locations and vintages. Consequently, in order to obtain a more reliable result of non-Saccharomyces yeast community, more research on yeast isolation and preservation should be performed consistently for long periods of time.
Recently, the enological behaviors and potential application of indigenous non-Saccharomyces have drawn widespread attention (Zhao et al., 2022; Castrillo and Blanco, 2023). Although H. uvarum, M. pulcherrima and C. zemplinina are the abundant yeast species during wine fermentation in this study. The function of minor species such as N. albida and F. magnum were also of special interest in our study, and their current use in winemaking applications is rather limited. N. albida and F. magnum were the dominant species isolated from Qaidam Basin Desert, China, with Mars-like extreme environments such as hyper-arid, salt deposits, high UV, similar mineral compositions and so on (Wei et al., 2022). In our study, F. magnum maintained a larger population at day 1 in YinChuan (16.67%) and Qing Tongxia (24.14%). Generally, Ningxia belongs to arid and semiarid climate with saline soil. It is understandable that this environment has contributed to their occurrence in the grape must (day 1). F. magnum was found as an endophytic fungus in the vineyard environment and was associated with the formation of grape flavor (Sayed et al., 2021), making it a candidate for enhancing wine flavor in our future study of yeast selection program. Other yeasts like A. pullulans, C. albidus and C. magnus possess biological control activity against fungal pathogen of grape and other fruits (De Capdeville et al., 2007; Pantelides et al., 2015; Türkoğlu et al., 2022). These yeasts have the potential to suppress phytopathogens by competition for space and the production of substances that affect hyphae integrity (niche competition). In addition, species having potential roles in wine aroma, flavor and texture during the early stages of fermentation and finished wines were of special interest in our study. Perhaps of most importance to Ningxia wine makers, R. graminis and R. glutinis has the potential as a flavor producer due to their lipid production (Martinez-Silveira et al., 2022; Zhao and Li, 2022). Future studies will investigate their potential in winemaking.
4.2. Intraspecific genetic diversity in non-Saccharomyces
Non-Saccharomyces excreted enzymes which have an impact on the technological traits are species and strain dependent. Therefore, it is necessary to discriminate each isolate before characterizing its potential as a producer or starter for industrial application. Although the discrimination within S. cerevisiae species was well reported by interdelta method and microsatellites, there are limited studies available for different non-Saccharomyces strains. In this study, we used TRtRNA to discriminate the non-Saccharomyces yeast strains. 34 and 40 distinct TRtRNA profiles were found using 5CAG and ISSR-MB, respectively. The primer ISSR-MB showed higher discriminatory capacity than primer 5CAG, as was also observed previously (Barquet et al., 2012; Nisiotou et al., 2022). This study achieved a lower discrimination power within H. uvarum and M. pulcherrima isolates. Four M. pulcherrima strains out of 22 strains were discriminated in this study using TRtRNA PCR method. The degree of variability (18.18%) was low compared to previous study (70.59%,12/17) (Barquet et al., 2012), which possibly due to the low diversity of this species in Ningxia.
4.3. Evaluation the colonizing of commercial non-Saccharomyces yeasts
The application of active dry yeast in wine fermentation is very common in the winemaking industry in China. Currently, Angel yeast NS-D (T. delbrueckii), Excellence B-Nature (M. pulcherrima) and Excellence X-Fresh (L.thermotolerans) were commonly used in Ningxia for increasing the complexity of the wines. Fortunately, the commercial non-Saccharomyces yeast strains were not detected during all the spontaneous fermentations in this study. However, it has been reported that commercial S. cerevisiae RC212 colonizing of the winery in Ningxia (Sun et al., 2017). It is understandable that the results could have contributed to the utiliation of Lalvin RC212 by local winery for more than 20 years, while the commercial non-Saccharomyces has been used for wine production in recent years. The application of active dry yeast in wine fermentation could reduce the diversity of indigenous yeasts and affect the regional characteristics of wines. Ningxia is one of the best known viticulture regions in China. Therefore, maintaining the biodiversity and preserving the genetic resources of indigenous non-Saccharomyces yeasts have become an urgent step in this region.
5. Conclusion
This study investigated the genetic diversity of non-Saccharomyces yeast at both species and strain level in five sub-regions of Ningxia, China, which has not previously been examined. The results of this study showed that different non-Saccharomyces strains were associated with different sub-regions. TRtRNA method is inadequate to discriminate species of F. elegans, Filobasidium sp., M. pimensis and N. albida. In addition, pairs of primers TRtNASC/ISSR-MB resulted in a higher discrimination power of H. uvarum, C. zemplinina, M. pulcherrima, Cryptococcus sp. and R. glutinis compared to TRtNASC/5CAG. This study is an important step before embarking on more time-consuming and labor-intensive methods to determine their performance in technological applications of interest. Their enological characteristics and interaction with S. cerevisiae worth further investigation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
RL: Conceptualization, Formal analysis, Writing-original draft, Writing - review & editing. DF: Formal analysis, Investigation, Methodology, Writing – review & editing. HW: Formal analysis, Writing - review & editing. ZZ: Experiment, Data Curation, Writing - review & editing. NL: Data curation, Formal analysis, Writing-original draft. YS: Conceptualization, Funding acquisition, Methodology, Supervision, Writing - review & editing.
Funding
This study was financially supported by the Key Research and Development Project of Ningxia Hui Autonomous Region (2022BBF02015, 2021BEF02014), National Natural Science Foundation of China (31960473), and Innovation project for Graduate Students of Ningxia University (GIP2021042).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1253969/full#supplementary-material
Abbreviations
TRtRNA, tandem repeat-tRNA.
Footnotes
References
Albertin, W., Setati, M. E., Miot-sertier, C., Bely, M., Mostert, T. T., Colonna-Ceccaldi, B., et al. (2016). Hanseniaspora uvarum from winemaking environments show spatial and temporal genetic clustering. Front. Microbiol. 6:1569. doi: 10.3389/fmicb.2015.01569
Andorrà, I., Landi, S., Mas, A., Guillamón, J. M., and Esteve-Zarzoso, B. (2008). Effect of oenological practices on microbial populations using culture independent techniques. Food Microbiol. 25, 849–856. doi: 10.1016/j.fm.2008.05.005
Barquet, M., Valentina, M., Medina, K., Pérez, G., Carrau, F., and Gaggero, C. (2012). Tandem repeat-tRNA (TRtRNA) PCR method for the molecular typing of non-Saccharomyces subspecies. Appl. Microbiol. Biotechnol. 93, 807–814. doi: 10.1007/s00253-011-3714-4
Baselga, I., Zafra, O., Pérez Lago, E., Francisco-Álvarez, R., Rodriguez-Tarduchy, G., and Santos, C. (2017). An AFLP based method for the detection and identification of indigenous yeast in complex must samples without a microbiological culture. Int. J. Food Microbiol. 241, 89–97. doi: 10.1016/j.ijfoodmicro.2016.09.014
Benito, Á., Calderón, F., and Benito, S. (2019). The influence of non-Saccharomyces species on wine fermentation quality parameters. Fermentation 5:54. doi: 10.3390/fermentation5030054
Binati, R. L., Innocente, G., Gatto, V., Celebrin, A., Polo, M., Felis, E. G., et al. (2019). Exploring the diversity of a collection of native non-Saccharomyces yeasts to develop co-starter cultures for winemaking. J. Food Res. Int. 122, 432–442. doi: 10.1016/j.foodres.2019.04.043
Casas-Godoy, L., Arellano-Plaza, M., Kirchmayr, M., Barrera-Martínez, I., and Gschaedler-Mathis, A. (2021). Preservation of non-Saccharomyces yeasts: current technologies and challenges. Compr. Rev. Food Sci. Food Saf. 20, 3464–3503. doi: 10.1111/1541-4337.12760
Castrillo, D., and Blanco, P. (2023). Characterization of indigenous non-Saccharomyces yeast strains with potential use in winemaking. Front. Biosci. (Elite Ed.) 15:1. doi: 10.31083/j.fbe1501001
Ciani, M., Morales, P., Comitini, F., Tronchoni, J., Canonico, L., Curiel, J. A., et al. (2016). Non-conventional yeast species for lowering ethanol content of wines. Front. Microbiol. 7:642. doi: 10.3389/fmicb.2016.00642
Capdeville, G.De., Souza Júnior, M. T., Santos, J. R. P., Miranda, S. P., Caetano, A. R., Falcão, R., et al. (2007). Scanning electron microscopy of the interaction between Cryptococcus magnus and Colletotrichum gloeosporioides on papaya fruit. Pesqui. Agropecu. Bras. 42:1537–1544. doi: 10.1590/s0100-204x2007001100004
Esteve-Zarzoso, B., Hierro, N., Mas, A., and Guillamón, J. M. (2010). A new simplifed AFLP method for wine yeast strain typing. LWT - food. Sci. Technol. 43, 1480–1484. doi: 10.1016/j.lwt.2010.05.016
Garofalo, C., Russo, P., Beneduce, L., Massa, S., Spano, G., and Capozzi, V. (2016). Non-Saccharomyces biodiversity in wine and the ‘microbial terroir’: a survey on Nero di Troia wine from the Apulian region, Italy. Ann. Microbiol. 66, 143–150. doi: 10.1007/s13213-015-1090-5
Grangeteau, C., Gerhards, D., Terrat, S., Dequiedt, S., Alexandre, H., Guilloux-Benatier, M., et al. (2016). FT-IR spectroscopy: a powerful tool for studying the inter- and intraspecific biodiversity of cultivable non-Saccharomyces yeasts isolated from grape must. J. Microbiol. Methods 121, 50–58. doi: 10.1016/j.mimet.2015.12.009
Guillamón, J. M., Sabaté, J., Barrio, E., Cano, J., and Querol, A. (1998). Rapid identification of wine yeast species based on RFLP analysis of the ribosomal internal transcribed spacer (ITS) region. Arch. Microbiol. 169, 387–392. doi: 10.1007/s002030050587
Jolly, N. P., Varela, C., and Pretorius, I. S. (2013). Not your ordinary yeast: non- Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 14, 215–237. doi: 10.1111/1567-1364.12111
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kurtzman, C. P., and Robnett, C. J. (1998). Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73, 331–371. doi: 10.1023/A:1001761008817
Lai, Y.-T., Hsieh, C.-W., Lo, Y.-C., Liou, B.-K., Lin, H.-W., Hou, C.-Y., et al. (2022). Isolation and identification of aroma-producing non-Saccharomyces yeast strains and the enological characteristic comparison in wine making. LWT - Food Sci. Technol. 154:112653. doi: 10.1016/j.lwt.2021.112653
Li, E. H., Liu, A. G., Xue, B., and Liu, Y. L. (2011). Yeast specie sassociated with spontaneous wine fermentation of cabernet sauvignon from Ningxia, China. World J. Microbiol. Biotechnol. 27, 2475–2482. doi: 10.1007/s11274-011-0711-9
Li, F. H., Zhang, X. Y., Feng, R., Chen, R. W., Zhang, Y. H., and Wei, J. G. (2022). Microclimate characteristics of vineyards in east foothills region of Helan Mountain in Ningxia. J. Arid Environ. 40, 284–295. doi: 10.11755/j.issn.1006-7639(2022)-02-0284
Lorca, G., Uribe, S., Martinez, C., Godoy, L., and Ganga, M. A. (2018). Screening of native S. cerevisiae strains in the production of pajarete wine: a tradition of atacama region, Chile. J. Wine Res. 29, 130–142. doi: 10.1080/09571264.2018.1465900
Maicas, S., and Mateo, J. J. (2023). The life of Saccharomyces and non-Saccharomyces yeasts in drinking wine. Microorganisms 11:1178. doi: 10.3390/microorganisms11051178
Martinez-Silveira, A., Garmendia, G., Rufo, C., and Vero, S. (2022). Production of microbial oils by the oleaginous yeast Rhodotorula graminis S1/2R in a medium based on agro-industrial by-products. World J. Microbiol. Biotechnol. 38:46. doi: 10.1007/s11274-022-03236-1
Medina, K., Boido, E., Dellacassa, E., and Carrau, F. (2017). Effects of non-Saccharomyces yeasts on color, anthocyanin, and anthocyanin- derived pigments of Tannat grapes during fermentation. Am. J. Enol. Vitic. 69, 148–156. doi: 10.5344/ajev.2017.17055
Mercado, L., Jubany, S., Gaggero, C., Masuelli, R. W., and Combina, M. (2010). Molecular relationships between Saccharomyces cerevisiae strains involved in winemaking from Mendoza Argentina. Curr. Microbiol. 61, 506–514. doi: 10.5344/ajev.2017.17055
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Morata, A., Arroyo, T., Bañuelos, M. A., Blanco, P., Briones, A., Cantoral, J. M., et al. (2022). Wine yeast selection in the Iberian Peninsula: Saccharomyces and non-Saccharomyces as drivers of innovation in Spanish and Portuguese wine industries. Crit. Rev. Food Sci. Nutr. 10, 1–29. doi: 10.1080/10408398.2022.2083574
Morata, A., Escott, C., Banuelos, M. A., Loira, I., Fresno, J. M. D., Gonzalez, C., et al. (2020). Contribution of non-Saccharomyces yeasts to wine freshness. A review. Biomolecules 10:34. doi: 10.3390/biom10010034
Nieuwoudt, H. H., Pretorius, I. S., Bauer, F. F., Nel, D. G., and Prior, B. A. (2006). Rapid screening of the fermentation profiles of wine yeasts by fourier transform infrared spectroscopy. J. Microbiol. Methods 67, 248–256. doi: 10.1016/j.mimet.2006.03.019
Nisiotou, A., Gyftogianni, E., and Banilas, G. (2022). Evaluation of different molecular markers for genotyping non-Saccharomyces wine yeast species. Microbiol. Res. 13, 643–654. doi: 10.3390/microbiolres13030046
Padilla, B., Gil, J. V., and Manzanares, P. (2016). Past and Future of Non-Saccharomyces Yeasts: From Spoilage Microorganisms to Biotechnological Tools for Improving Wine Aroma Complexity. Front. Microbiol. 7:411. doi: 10.3389/fmicb.2016.00411
Pallmann, C. L., Brown, J. A., Olineka, T. L., Cocolin, L., Mills, D. A., and Bisson, L. F. (2001). Use of WL medium to profile native flora fermentations. Am. J. Enol. Vitic. 52, 198–203. doi: 10.1016/S0065-2164(01)49013-7
Pantelides, I. S., Christou, O., Tsolakidou, M. D., Tsaltas, D., and Ioannou, N. (2015). Isolation, identification and in vitro screening of grapevine yeasts for the control of black aspergilli on grapes. Biol. Control 88, 46–53. doi: 10.1016/j.biocontrol.2015.04.021
Renouf, V., Claisse, O., and Lonvaud-Funel, A. (2007). Inventory and monitoring of wine microbial consortia. Appl. Microbiol. Biotechnol. 75, 149–164. doi: 10.1007/s00253-006-0798-3
Sayed, S. M., El-Shehawi, A. M., Elarnaouty, S. A., Al-Otaibi, S. A., El-Shazly, S. S., Alotaibi, S. S., et al. (2021). Molecular characterization of endophytic fungal communities associated with Vitis vinifera L. at Taif region of Saudi Arabia. J. Environ. Biol. 42, 177–185. doi: 10.22438/jeb/42/2/MRN-1577
Sun, Y., and Liu, Y. L. (2014). Investigating of yeast species in wine fermentation using terminal restriction fragment length polymorphism method. Int. J. Food Microbiol. 38, 201–207. doi: 10.1016/j.fm.2013.09.001
Sun, Y., Qin, Y., Pei, Y., Wang, G., Joseph, C. M. L., Bisson, L. F., et al. (2017). Evaluation of Chinese Saccharomyces cerevisiae wine strains from different geographical origins. Am. J. Enol. Vitic. 68, 73–80. doi: 10.5344/ajev.2016.16059
Tamang, J. P., and Lama, S. (2023). Diversity of yeasts in Indian fermented foods and alcoholic beverages. FEMS Yeast Res. 23:foad011. doi: 10.1093/femsyr/foad011
Türkoğlu, S., Zengin, A., Ozturk, M., and Çağrı-Mehmetoğlu, A. (2022). Mathematical modelling of biocontrol effect of Cryptococcus albidus on the growth of Penicillium expansum on Fuji apple fruits. Biol. Control 170:104908. doi: 10.1016/j.biocontrol.2022.104908
Wang, Z. H., Liu, Y. Q., Feng, C. E., Li, R. J., and Chen, Q. P. (2017). Isolation and identification of Saccharomyces cerevisiae strain of east Helan Mountain area in Ningxia. Food Res Dev 38, 176–180. doi: 10.3969/j.issn.1005-6521.2017.11.039
Wang, X. F., Schlatter, D. C., Glawe, D. A., Edwards, C. G., Weller, D. M., Paulitz, T. C., et al. (2021). Native yeast and non-yeast fungal communities of cabernet sauvignon berries from two Washington state vineyards, and persistence in spontaneous fermentation. Int. J. Food Microbiol. 350:109225. doi: 10.1016/j.ijfoodmicro.2021.109225
Wei, X. Y., Zhu, H. Y., Song, L., Zhang, R. P., Li, A. H., Niu, Q. H., et al. (2022). Yeast diversity in the Qaidam Basin desert in China with the description of five new yeast species. J. Fungi 8:858. doi: 10.3390/jof8080858
Zhang, J. J., Shang, Y. M., Chen, J. Y., Brunel, B., Peng, S. S., Li, S., et al. (2021). Diversity of non-Saccharomyces yeasts of grape berry surfaces from representative cabernet sauvignon vineyards in Henan Province, China. FEMS Microbiol. Lett. 368:fnab142. doi: 10.1093/femsle/fnab142
Zhang, W. X., Tan, Y. N., Sun, Y., and Zhang, X. Y. (2020). Diversity of yeasts in spontaneous fermentation must of eastern Helan Mountain areas. China Brewing 39, 30–35. doi: 10.11882/j.issn.0254-5071.2020.12.007
Zhao, D., and Li, C. J. (2022). Multi-omics profiling reveals potential mechanisms of culture temperature modulating biosynthesis of carotenoids, lipids, and exopolysaccharides in oleaginous red yeast Rhodotorula glutinis ZHK. LWT 171:114103. doi: 10.1016/j.lwt.2022.114103
Zhao, Y., Sun, Q. Y., Tian, B., Zhu, S. S., Du, F., Mao, R. Z., et al. (2022). Evaluation of four indigenous non-Saccharomyces yeasts isolated from the Shangri-La wine region (China) for their fermentation performances and aroma compositions in synthetic grape juice fermentation. J Fungi (Basel) 8:146. doi: 10.3390/jof8020146
Keywords: non-Saccharomyces, diversity, wine fermentation, molecular fingerprinting, TRtRNA
Citation: Li R, Feng D, Wang H, Zhang Z, Li N and Sun Y (2023) Genetic diversity of non-Saccharomyces yeasts associated with spontaneous fermentation of Cabernet Sauvignon wines from Ningxia, China. Front. Microbiol. 14:1253969. doi: 10.3389/fmicb.2023.1253969
Edited by:
Uelinton Manoel Pinto, University of São Paulo, BrazilReviewed by:
Alessandra Ribeiro, University of São Paulo, BrazilLiliana Godoy Olivares, Pontificia Universidad Católica de Chile, Chile
Copyright © 2023 Li, Feng, Wang, Zhang, Li and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yue Sun, eXVlc3VuQG54dS5lZHUuY24=
†These authors have contributed equally to this work
 Ruirui Li1†
Ruirui Li1† Yue Sun
Yue Sun