
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 October 2023
Sec. Microbiological Chemistry and Geomicrobiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1253114
 Pilar C. Portela1,2
Pilar C. Portela1,2 Leonor Morgado1,2
Leonor Morgado1,2 Marta A. Silva1,2
Marta A. Silva1,2 Lukas Denkhaus3
Lukas Denkhaus3 Oliver Einsle3
Oliver Einsle3 Carlos A. Salgueiro1,2*
Carlos A. Salgueiro1,2*The recent reclassification of the strict anaerobe Geobacter sulfurreducens bacterium as aerotolerant brought attention for oxidative stress protection pathways. Although the electron transfer pathways for oxygen detoxification are not well established, evidence was obtained for the formation of a redox complex between the periplasmic triheme cytochrome PpcA and the diheme cytochrome peroxidase MacA. In the latter, the reduction of the high-potential heme triggers a conformational change that displaces the axial histidine of the low-potential heme with peroxidase activity. More recently, a possible involvement of the triheme periplasmic cytochrome family (PpcA-E) in the protection from oxidative stress in G. sulfurreducens was suggested. To evaluate this hypothesis, we investigated the electron transfer reaction and the biomolecular interaction between each PpcA-E cytochrome and MacA. Using a newly developed method that relies on the different NMR spectral signatures of the heme proteins, we directly monitored the electron transfer reaction from reduced PpcA-E cytochromes to oxidized MacA. The results obtained showed a complete electron transfer from the cytochromes to the high-potential heme of MacA. This highlights PpcA-E cytochromes’ efficient role in providing the necessary reducing power to mitigate oxidative stress situations, hence contributing to a better knowledge of oxidative stress protection pathways in G. sulfurreducens.
Bacteria from the genus Geobacter are a remarkable group of microorganisms capable of utilizing extracellular electron transfer mechanisms to harvest ATP (Lovley, 2011). For this, the microorganisms need to efficiently shuttle the electrons across the inner and outer membranes to reduce extracellular electron acceptors such as iron oxides or manganese oxides (Lovley, 2011). Contaminant compounds such as chromium, vanadium or uranium can also be used by this species to support cell growth (Ortiz-Bernad et al., 2004; Sanford et al., 2007; He et al., 2019). The seamless electron transfer (ET) is secured by a network of cytochromes that provide an effective interface between the inner and outer membranes (Reguera and Kashefi, 2019; Salgueiro et al., 2022). In fact, two distinct ET networks have been identified in Geobacter sulfurreducens: when the cell is exporting electrons to the exterior of the cell – current producing mode – or when it is receiving electrons from an external source – current consuming mode (Dumas et al., 2008; Kracke et al., 2015; Heidary et al., 2020). In the current producing mode, electrons originating from the oxidation of organic compounds are transferred to a menaquinone pool via a NADH dehydrogenase located in the inner membrane. Depending on the redox potential of the final electron acceptor, the electrons are transferred from the menaquinone pool to inner-membrane associated cytochromes ImcH, CbcL or CbcBA: ImcH is recruited when the final electron acceptor has reduction potential values above −100 mV (vs. standard hydrogen electrode, SHE), CbcL with values between −100 and − 210 mV, and CbcBA operates below −210 mV. The electrons are then transferred to periplasmic cytochromes (mainly to the cytochrome family PpcA) and from these to porin-cytochrome trans-outer membrane complexes [ExtABCD, ExtEFG, ExtHIJKL, Om(abc)B and Om(abc)C], which then convey the electrons to the extracellular electron acceptors. In addition, G. sulfurreducens is known to establish direct electrical contacts with extracellular electron acceptors via electrically conductive filaments. However, the exact mechanism of electron transport and the proteins’ nature is not yet clearly understood, with evidence on both sides: some studies identified the filaments as an assembly of repeating subunits of PilA (gene annotation number GSU1496), a type-IV pilin monomer, in which the aromatic amino acids provide a conduit for electron transfer (Reguera et al., 2005; Lovley, 2022); other studies identified that outer membrane c-type cytochromes OmcS, OmcZ and OmcE polymerize and transfer electrons through the stacked hemes in their inner core (Wang et al., 2019, 2022; Gu et al., 2023). The ET network of the current consuming mode is far less well-understood, but it is hypothesized that it shares a similar organization with the network used in the current producing mode, having redox partners poised at the outer membrane, periplasm, and inner membrane. However, comparative studies in current-consuming versus current-producing G. sulfurreducens’ biofilms showed that the abundance of the gene transcripts is considerably distinct, and hence specific proteins are recruited for each electron transfer pathway (Strycharz et al., 2011). For example, while the gene encoding for cytochrome OmcZ, essential for current-producing pathway, had much lower abundance in current-consuming biofilms, genes encoding specific electron transfer proteins, such as periplasmic monoheme cytochromes PccH and GSU2515, show the largest transcript abundance in current-consuming biofilms (Strycharz et al., 2011). In addition, G. sulfurreducens biofilms in current-consuming mode have been observed to accumulate iron oxide species in their outer membrane, possibly for facilitating the ET process in this direction (Heidary et al., 2020). This metabolic versatility has permitted Geobacter’s application in the fields of bioremediation, for the removal of toxic compounds, such as uranium, from contaminated waters (Lovley and Phillips, 1992; Anderson et al., 2003); bioenergy, for the generation of electric energy in microbial fuel cells (Santoro et al., 2017); microbial electrosynthesis, in which electrogenic bacteria provide reductive power for the production of industrially relevant compounds such as alcohols, carboxylic acids or fuels (Gong et al., 2020).
The bacteria from the genus Geobacter were initially classified as strict anaerobes (Caccavo et al., 1994). However, the complete genome sequence of G. sulfurreducens revealed the presence of proteins typically involved in aerobic respiration and protection against oxidative stress such as: superoxide dismutase (GSU1158, GSU0720), cytochrome c peroxidase (GSU2813, GSU0466), catalase (GSU2100), peroxiredoxins (GSU0066, GSU0352, GSU0893, GSU3246, and GSU3447), rubrerythrins (GSU2612, GSU2814), and hydrogenases Hya (encoded by GSU0120 to GSU0123) and Hyb (encoded by GSU0782 to GSU0785) (Methé et al., 2003; Coppi et al., 2004; Tremblay and Lovley, 2012). Subsequent studies on G. sulfurreducens supported the genomic analysis and demonstrated that it tolerates oxygen exposure up to 24 h and can utilize this molecule as electron acceptor under microaerobic conditions (10% v/v of oxygen; Lin et al., 2004; Engel et al., 2020). This characteristic brings competitive advantage to G. sulfurreducens as in the water sediments near the oxic-anoxic interface, Fe(II) produced from microbial reduction can be regenerated to Fe(III) and endow continuous extracellular electron transfer processes (Lin et al., 2004). Furthermore, this offers an opportunity for the development of oxygen-resistant G. sulfurreducens strains that are more robust for biotechnological applications (Tremblay and Lovley, 2012; Speers and Reguera, 2021). Studies carried out for the G. sulfurreducens bacterium under several oxygen conditions showed two different behaviors: with low oxygen concentration (1% oxygen provided to G. sulfurreducens culture), the bacterium overexpresses 11 type IV pilus genes (GSU2029 to GSU2039) that are hypothesized to be involved in the motility mechanism of G. sulfurreducens, endowing it to flee from the oxygen- contaminated area; in high oxygen concentration environments (5% oxygen provided to G. sulfurreducens culture), the bacterium downregulates the expression of the type IV pilus genes and instead overexpresses genes involved in cell encapsulation and biofilm production to decrease oxygen exposure (Engel et al., 2020). These pathways were observed in a genetic study that disrupted some of the important genes identified in oxidative stress response, including the macA peroxidase, and observed the mutated strains’ response to oxygen (Speers and Reguera, 2021).
To date, in G. sulfurreducens, two NiFe periplasmic hydrogenases Hya and Hyb were shown to play a role in oxidative stress protection (Tremblay and Lovley, 2012), and two diheme proteins with homology to cytochrome c peroxidases were characterized: CcpA (GSU2813; Hoffmann et al., 2009; Ellis et al., 2011) and MacA (GSU0466; Butler et al., 2004; Seidel et al., 2012). Proteomic studies have shown that MacA is relevant in the reduction pathways of Fe(III) citrate (Butler et al., 2004), Fe(III) oxide (Aklujkar et al., 2013), U(VI) (Shelobolina et al., 2007), and that it may regulate the expression of OmcB, since this protein was not expressed in a macA deletion strain (Kim and Lovley, 2008). This latter aspect might suggest an indirect role of MacA in iron reduction (Kim and Lovley, 2008). Regarding its role in oxidative stress, MacA is able to reduce hydrogen peroxide (Seidel et al., 2012), and a G. sulfurreducens strain in which this gene was disrupted displayed enhanced biofilm formation and absence of oxygen tolerance, characteristic of oxidative stress susceptibility (Speers and Reguera, 2021). In vitro, MacA was also shown to interact and exchange electrons with PpcA (Seidel et al., 2012; Dantas et al., 2017), one of the most abundant periplasmic cytochromes.
MacA is a 35 kDa membrane-associated diheme cytochrome peroxidase located in the periplasm (Butler et al., 2004). The two hemes in this protein are functionally distinct: the His-Met axially coordinated high-potential (HP) heme receives electrons from putative electron donors and, upon reduction, triggers conformational changes that detach the distal histidine of the low-potential (LP) heme, endowing this heme with peroxidase activity (Seidel et al., 2012). The five members of the periplasmic family of triheme cytochromes (PpcA-E) are responsible for mediating ET in the periplasm (Dantas et al., 2015). Each member of the family has approximately 9.5 kDa of molecular weight and three low-spin hemes coordinated by two histidines (Dantas et al., 2015). Despite the high sequence identity, the individual heme redox properties of the five cytochromes are different (Morgado et al., 2010).
Proteomic and gene knockout studies in G. sulfurreducens revealed different expression levels for PpcA-E cytochromes depending on the final electron acceptor [Fe(III) citrate (Ding et al., 2008), Fe(III) oxides (Ding et al., 2008), Mn(IV) oxides (Aklujkar et al., 2013)]. Even though different expression levels were detected, a very recent study suggested that the absence of a particular triheme periplasmic cytochrome could be compensated by the overexpression of another member of the family, highlighting the versatility of these proteins (Choi et al., 2022). Furthermore, G. sulfurreducens strains for which each one or all triheme periplasmic cytochromes were knocked out showed more susceptibility to hydrogen peroxide, suggesting also a possible role of these proteins in radical detoxification mechanisms (Choi et al., 2022).
In this work, we directly probed ET reactions between the peroxidase MacA and the PpcA-E cytochrome family with the aim of determining the most relevant interacting pairs and elucidating the detoxification pathways involving MacA.
The five triheme cytochromes (PpcA-E) and MacA were overexpressed in E. coli BL21(DE3)::pEC86, in which the pEC86 plasmid contains the cytochrome maturation cluster ccmABCDEFGH for aerobic overexpression of c-type cytochromes (Arslan et al., 1998), and purified according to the protocols previously described (Londer et al., 2002; Morgado et al., 2007; Pokkuluri et al., 2010; Seidel et al., 2012) in an ÄKTA Prime Plus Chromatography System (GE Healthcare).
Briefly, for the triheme cytochromes, the E. coli BL21(DE3)::pEC86 cells were transformed with the plasmid pCK32 (encoding for each PpcA-E mature cytochrome) and grown in 2xYT media supplemented with chloramphenicol (34 μg/mL) and ampicillin (100 μg/mL) at 30°C, 200 rpm. Upon reaching an OD600nm value of ∼1.5, 10 μM isopropyl β-D-thiogalactoside (IPTG) was added and the culture grew overnight at 30°C, 180 rpm. Cells were harvested by centrifugation (6,400 xg, 20 min, 4°C) and the cell pellet was gently resuspended in 30 mL lysis buffer [20% sucrose (Fisher scientific), 100 mM Tris–HCl (NZYTtech) pH 8.0 and 0.5 mM EDTA (Sigma), 0.5 mg/mL lysozyme (Fluka)] per liter of initial cell culture. The lysate was centrifuged at 15,000 xg, 20 min, 4°C and then ultracentrifuged at 150,000 xg, 90 min, 4°C. The clear supernatant was dialyzed twice against 10 mM Tris–HCl, pH 8.5. Purification encompassed a cation exchange chromatography step using 2 × 5 mL Bio-Scale™ Mini UNOsphere™ S Cartridges (BioRad) equilibrated with the same dialysis buffer. The protein was eluted with a 150 mL gradient of 0–300 mM NaCl. The red fractions containing PpcA-E were selected and concentrated to 1.5 mL, and then loaded onto a Superdex 75 XK16/60 molecular exclusion column (GE Healthcare) equilibrated with 100 mM sodium phosphate buffer, pH 8.0.
In the case of MacA, transformed E. coli cells, containing the plasmid pETSN22 that includes macA gene (gsu0466) and a N-terminal Strep-Tag(II),were grown overnight at 30°C, 200 rpm in a media supplemented with ampicillin (100 μg/mL) and chloramphenicol (20 μg/mL). No induction with IPTG was needed as the plasmid’s T7 promoter was leaky enough to obtain good protein yields. The cells were collected by centrifugation and resuspended in 2 mL per gram of cells of lysis buffer containing 20 mM Tris–HCl, pH 8.0, 250 mM NaCl. The cells were disrupted in a microfluidizer (Microfluidics) at 1,100 bar with three cycles and centrifuged a first time at 20,000 xg, 20 min, 4°C, and a second time at 100,000 xg, 1 h, 4°C. The supernatant was then purified using two steps: firstly using a streptactin superflow column (IBA), with elution of bound protein in a single step with 2.5 mM D-desthiobiotin in running buffer; secondly, by loading the eluate onto a size exclusion column (Superdex 200, 16/60, GE Healthcare) equilibrated with buffer containing 20 mM Tris–HCl, pH 8.0, 100 mM NaCl.
For the NMR studies, all proteins’ buffer was exchanged to 8 mM sodium phosphate buffer (pH 7) with sodium chloride (final ionic strength of 20 mM) followed by three cycles of lyophilization with 2H2O.
All NMR spectra were acquired in a Bruker Avance 600 MHz spectrometer at 298 K with 32 k data points, sweep width of 96.2 kHz and 1 k scans. 1H chemical shifts were calibrated using 2,2-dimethyl-2-silapentane-5-sulphonate as reference at 0 ppm (Wishart et al., 1995) and TOPSPIN software (Bruker Biospin) was used for spectra processing and analysis.
A lyophilized sample was resuspended in 500 μL of degassed 2H2O (final concentration 50 μM) inside an anaerobic glovebox (MBraun) with oxygen levels kept below 0.1 ppm and the NMR tube was sealed with a rubber cap. Spectra were acquired in the oxidized state (as purified), ascorbate-reduced state (1 mM sodium ascorbate in the presence of 5 μM mediator tetramethyl-1,4-phenylenediamine), and fully reduced state (50 μM sodium dithionite).
The experimental procedure previously described (Morgado and Salgueiro, 2022) was followed to assess the redox reactions between PpcA-E and MacA.
NMR tubes containing each triheme cytochrome were prepared separately by resuspending lyophilized proteins in 350 μL 2H2O (for a final concentration of 50 μM in the experiment). Each PpcA-E sample was reduced by first flushing out all oxygen from the NMR tube with hydrogen, and then by adding catalytic amounts of hydrogenase from Desulfovibrio vulgaris (Hildenborough; Morgado et al., 2008). After complete reduction, hydrogen was removed with argon and samples were taken into the anaerobic glovebox. These procedures ensure that the PpcA-E samples do not contain oxygen. For MacA’s stock solution, the lyophilized protein was resuspended in 150 μL of degassed 2H2O (for a 200 μM final concentration in the experiment). MacA’s stock solution was added to each of PpcA-E cytochromes in molar ratios (PpcA-E:MacA) 1:1, 1:2, 1:3, and 1:4. After each MacA addition, the NMR tube was sealed again and a 1D 1H-NMR spectrum was immediately acquired. For the control experiment, degassed 2H2O was added to a reduced PpcA-E sample instead of MacA and a 1D 1H-NMR spectrum was acquired to ensure no oxygen was present in the degassed 2H2O that would be used for resuspending MacA. This step is essential to ensure that there is no oxygen contamination in the MacA sample and the observed oxidation phenomena are due to the presence of MacA and not by the presence of oxygen. At the end of the experiment (PpcA-E:MacA molar ratio 1:4), the NMR tube rubber cap was removed and the sample was exposed to atmospheric oxygen, and a final 1D 1H-NMR spectrum was acquired.
To study the interacting interface regions between PpcB-E and MacA and to complement the existing data for the redox pair PpcA-MacA (Dantas et al., 2017), NMR chemical shift perturbation of the heme methyl signals was measured by comparing 1D 1H-NMR spectra of isolated PpcB-E or MacA in the oxidized state with those obtained for a 1:4 PpcB-E:MacA ratio in the oxidized state.
Docking of the redox pairs was performed with the HADDOCK2.4 webserver (van Zundert et al., 2016; Honorato et al., 2021) using as input the default parameters and the information on the most affected hemes from the chemical shift perturbation experiments: HP heme for MacA, heme IV for PpcB and PpcE, heme I and heme IV for PpcC, heme III and IV for PpcD. Atomic coordinates of PpcB (PDB 3BXU), PpcC (PDB 3H33), PpcD (PDB 3H4N, chain B), PpcE (PDB 3H34), MacA (PDB 4AAL, chain A) were used for the docking. The procedure was performed with MacA’s chain A to match the conditions on the previously reported docking PpcA-MacA (Dantas et al., 2017).
The previous electrochemical and structural characterization of MacA (Seidel et al., 2012) showed that it can adopt three main redox states – fully oxidized, partially reduced (from hereafter referred as ascorbate-reduced), and fully reduced (by the addition of sodium dithionite) – and, as such, NMR spectra were acquired for each condition (Figure 1). In the fully oxidized state, on one hand, the HP heme (His-Met coordinated) undergoes a rapid high/low-spin equilibrium due to the weekly bound methionine (Seidel et al., 2012) and, thus, in the 1D 1H-NMR spectrum the heme methyl signals are broad and found in the characteristic region ranging from 60 to 80 ppm. On the other hand, those from the low-spin, LP heme (His-His coordinated) are observable in the 20–35 ppm region. Addition of ascorbate reduces the HP heme, which changes to low-spin state, causing the disappearance of 60–80 ppm signals and the appearance of a signal at −2.77 ppm, a hallmark of low-spin His-Met coordination that corresponds to the methionine’s CH3ε group (Seidel et al., 2012). Conformational changes in MacA’s backbone associated to HP heme reduction detach the LP heme distal histidine, and this heme is now in high-spin state, which causes broadening of its signals and a shift to the 40–60 ppm region. Upon addition of sodium dithionite, both hemes are fully reduced: the HP heme maintains its low-spin state, corroborated by the observation of the axial methionine signal at −2.88 ppm, and the LP heme maintains its high-spin state with broad signals observed in the characteristic region of 10–25 ppm (Keller et al., 1972). This behavior is similar to the one observed for a cytochrome c peroxidase from Pseudomonas stutzeri (ATCC 11607) that was previously characterized (Villalaín et al., 1984). The slight variation of the chemical shift of the HP’s axial methionine CH3ε group may be explained by local redox-linked conformation changes triggered by the LP heme (oxidized in the ascorbate-reduced state, and reduced in the fully reduced state; Villalaín et al., 1984).
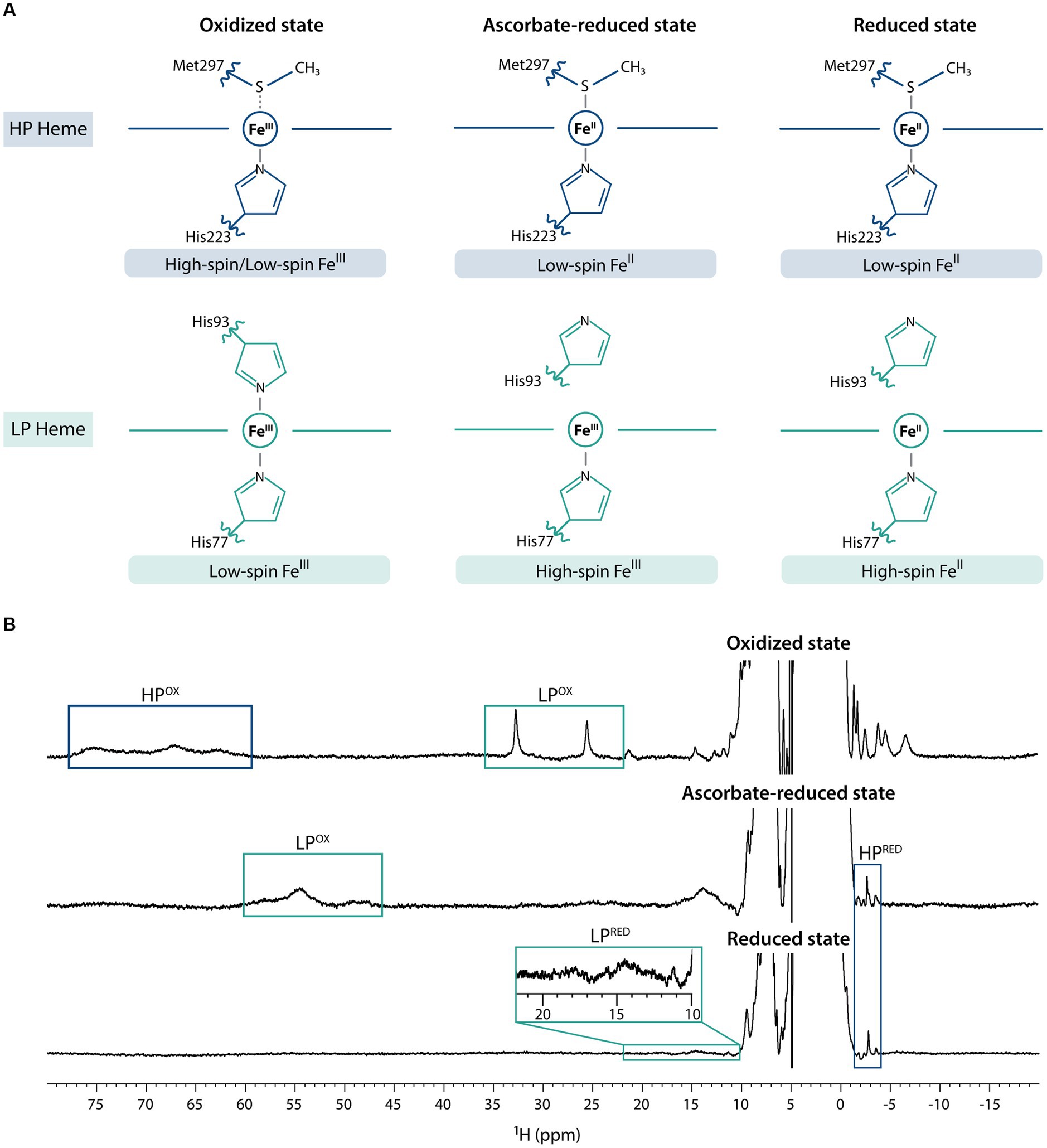
Figure 1. Fingerprints of MacA in the three redox states. (A) MacA’s heme axial coordination in the oxidized, ascorbate-reduced state, and fully reduced state with the indication of respective iron spin states, (B) 1D 1H-NMR spectra of MacA in the three different redox states (pH 7 and 298 K) with labeling of the hallmark signals. Expansion of the 10–22 ppm range in the reduced spectra highlights the high-spin signals of the LP heme.
In classical bacterial peroxidases the LP heme has a negative reduction potential value, whereas the HP heme has a positive one (Nóbrega and Pauleta, 2019). In the case of MacA, the redox potential value of the LP heme has been experimentally determined (−241 mV), but the value for the HP heme has not. However, since this protein is reduced by sodium ascorbate (reduction potential value of +330 mV; Njus et al., 2020) and analysis of the HP heme’s reduction potential values determined for other classical bacterial peroxidases, places MacA’s HP heme reduction potential value within the range + 320 mV (Pseudomonas aeruginosa; Ellfolk et al., 1983) to +450 mV (Nitrosomonas europaea; Arciero and Hooper, 1994). Together with the data obtained for the redox properties of the PpcA-E periplasmic triheme cytochromes (Table 1), this suggests that the latter have the necessary thermodynamic properties to reduce MacA’s HP heme (Dantas et al., 2017). The thermodynamic data is in accordance with the mechanistic description of MacA, as the HP heme is responsible for providing reducing power to the LP heme for its peroxidase activity. In fact, the lower reduction potential value of the LP heme (− 241 mV) compared to those of the PpcA-E proteins makes the reduction of the MacA’s LP heme by any triheme cytochrome very unlikely.
To test this hypothesis, ET was probed by NMR after the addition of oxidized MacA to reduced cytochromes (PpcA-E), in anaerobic conditions, at molar ratios PpcA-E:MacA 1:0, 1:1, 1:2, 1:3, 1:4 (Figure 2). The ET studies monitored by NMR rely on the different spectral features displayed by cytochromes in their different oxidation states (Morgado and Salgueiro, 2022). The signal dispersion in the oxidized state is generally wider than in the reduced state due to the paramagnetic contribution of the unpaired electron(s) to the observable chemical shifts of the heme substituents and nearby residues (Salgueiro et al., 2019). Additionally, the signal dispersion in the oxidized state is more variable, since the paramagnetic effect is determined by the specific geometry of the heme axial ligands (Turner et al., 1995; Bertini et al., 1999). In the case of MacA, the presence of a high-spin group in the oxidized state [Fe(III), S = 5/2] makes the spectral window wider (from −10 to 80 ppm) than for PpcA-E’s, whose heme groups are low-spin state [Fe(III), S = 1/2] with a typical signal dispersion from −10 to 30 ppm. In the reduced state, the spectral width is narrower and partially superimposable due to the presence of low-spin groups: PpcA-E proteins are low-spin with a spectral width from −5 to 12 ppm, and MacA possesses a low-spin (HP heme) and a high-spin (LP heme) group, thus presenting a spectral width from −4 to 25 ppm.
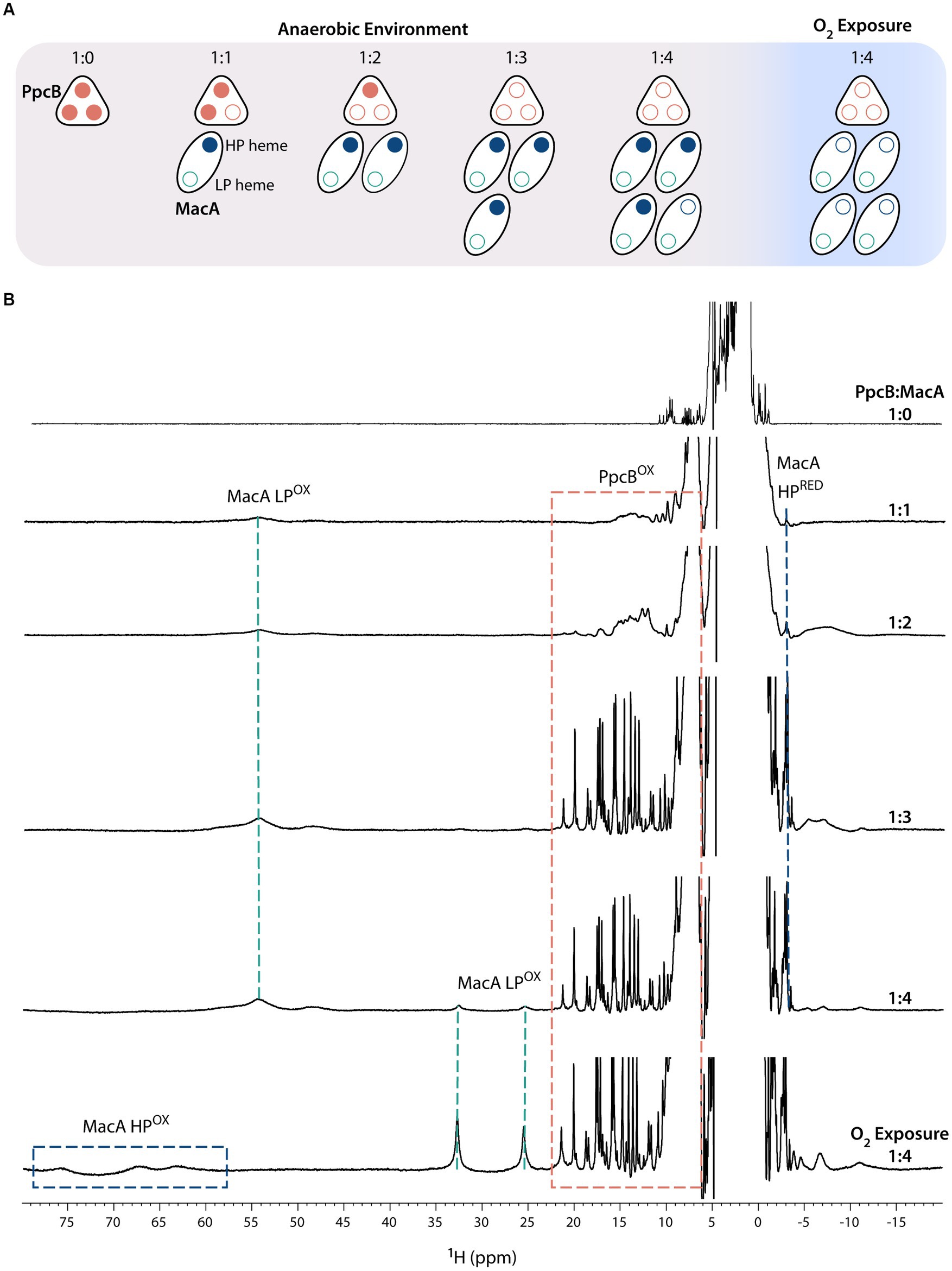
Figure 2. Electron transfer reaction between PpcB and MacA (pH 7 and 298 K). (A) Schematic representation of the experimental setup in which all additions of MacA to PpcB were done in anaerobic environment. At the end of the experiment, the proteins were exposed to atmospheric oxygen. Reduced and oxidized hemes are represented by closed and open circles, respectively. (B) 1D-1H NMR spectra acquired at the different PpcB:MacA molar ratios. The PpcB:MacA 1:0 molar ratio spectrum corresponds to the fully reduced PpcB state. The NMR spectral features of both proteins are indicated by dashed rectangles.
Figure 2 illustrates the ET reaction between PpcB and MacA. The data obtained for the other redox pairs are indicated in Supplementary Figures S1–S4. For all PpcA-E cytochromes, at 1:1 and 1:2 molar ratios, we observe the replacement of their sharp and typical reduced signals by broad signals dispersed over larger spectral widths, indicating partial oxidation of each of the triheme cytochrome. If ET occurred to both of MacA’s hemes, by the 1:2 ratio the triheme cytochromes would be fully oxidized. Instead, the observed partial oxidation suggests that only one MacA heme is participating in ET, corroborated by MacA’s spectral signature like the ascorbate-reduced state, highlighted by the presence of the HP’s reduced axial methionine CH3ε signal and LP oxidized signals in the region 40–60 ppm. At 1:3 molar ratio, the triheme cytochromes are fully oxidized, indicating that an equimolar concentration of oxidized (triheme cytochromes) and reduced hemes (MacA’s HP heme) co-exist. Upon addition of excess MacA (1:4 ratio), while the triheme cytochromes maintain their oxidized NMR fingerprints, MacA’s LP oxidized signals start to appear, indicating that the triheme cytochromes have been depleted of reducing power and the MacA added in excess remains fully oxidized (MacA’s HP oxidized signals are not evident due to their broadness and much lower intensity than LP ones). Sample exposure to atmospheric oxygen did not affect the triheme cytochrome signals, which remained oxidized, whereas MacA’s spectral features changed to the ones indicating full oxidation, further confirming its semi-reduced state during all the anaerobic additions.
In conclusion, the ET reaction between the redox pairs is complete and only involves the HP heme. In fact, at an equimolar concentration of HP heme (PpcA-E:MacA 1:3 molar ratio) all the triheme cytochromes are fully oxidized and no intermediate oxidation state is observed. This contrasts with the redox pair CbcL:PpcA, in which the proteins maintain an intermediate oxidation state even at high molar ratios (Antunes et al., 2022). CbcL is an inner membrane associated cytochrome, with a periplasmic domain containing nine c-type heme groups, that plays a role in the ET to extracellular electron acceptors with low redox potential values. It was proposed that CbcL mediates the ET from the menaquinone pool to the periplasmic cytochrome PpcA. It was shown that the periplasmic domain of CbcL donates electrons to PpcA, as expected from its lower midpoint reduction potential. However, the difference of only 56 mV between the reduction potential values of the two proteins ensures an overlap of their redox windows in a way that they remain in equilibrium, acting as a reservoir of electrons to permit a constant flow whenever CbcL is loaded with electrons by the menaquinone pool and PpcA oxidized by its redox partner.
On the other hand, the results obtained for the PpcA-E:MacA complexes are in line with the physiological description of bacterial peroxidases in the literature (Nóbrega and Pauleta, 2019), as the HP heme is responsible for relaying the electrons to the LP heme with peroxidase activity. Although distinct, the midpoint and individual heme reduction potential values of PpcA-E are comparable (midpoint of, approximately, −130 mV cf. Table 1). Thus, considering the considerable difference between this value and that of MacA’s HP heme (in the range + 320 to +450 mV), no significant thermodynamic priorities are expected. This is indeed corroborated by our experiments, as no periplasmic cytochrome displays preferential electron transfer to MacA. Furthermore, the in vivo study by Choi et al. (2022) suggests that the transcription level might play a more preponderant role in oxidative stress protection: the presence of a single periplasmic cytochrome in G. sulfurreducens mutated strains is enough to provide oxygen stress protection, although the strains which presented less cytochrome concentration – such as the ones expressing only PpcC, PpcE, or in which all PpcA-E cytochromes were deleted – were the most susceptible ones to hydrogen peroxide.
The ability of all triheme cytochromes to reduce MacA corroborates the pivotal role of this family in providing reducing power to a great array of redox partners as observed in G. sulfurreducens (Choi et al., 2022). This emphasizes that any periplasmic cytochrome from the PpcA-E family may be immediately recruited for oxidative stress response, ensuring that the peroxidase MacA has all the necessary reductive power available for a prompt elimination of radical species.
Although redox interactions are characterized by their transiency, we aimed to identify whether a preferential interface existed when triheme cytochromes transfer electrons to MacA by probing the chemical shift variation of the heme methyl groups. Analysis of the most perturbed signals (Figure 3) shows that the interacting interface in the triheme cytochromes involves, in larger extent, heme IV – except for PpcC in which heme I’s methyls are the most affected ones – and MacA’s HP heme. PpcC also has an affected substituent in heme IV and PpcD has one in heme III.
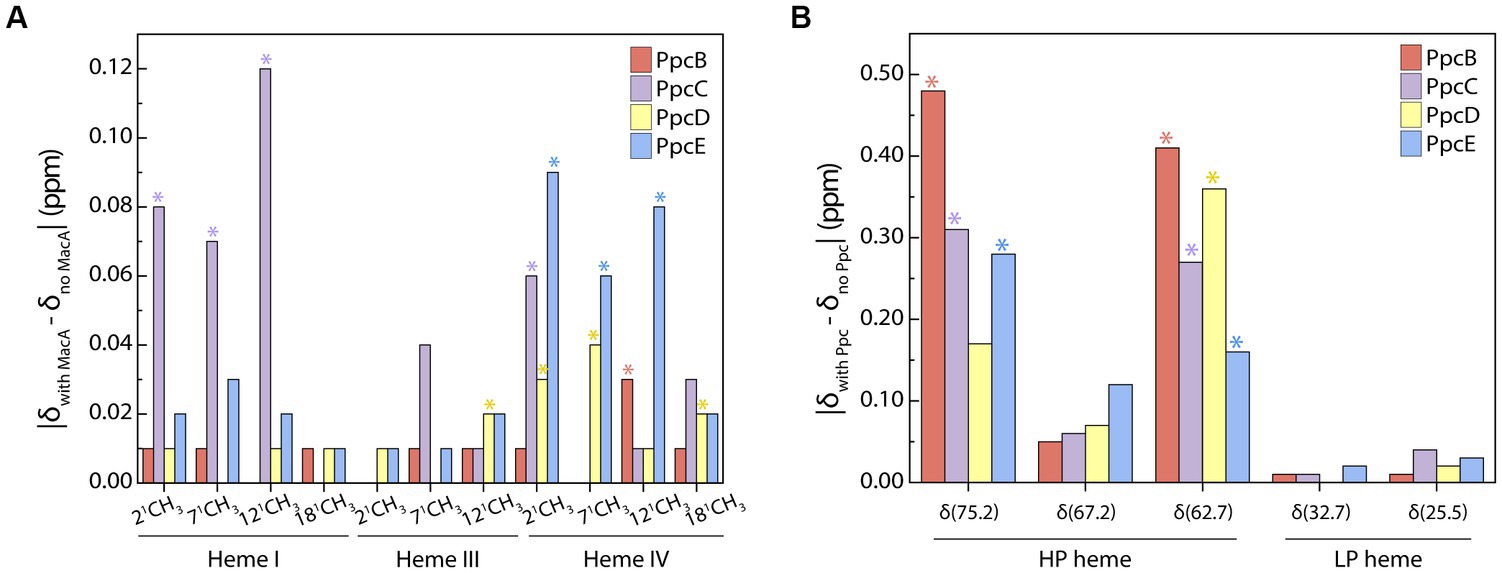
Figure 3. Interacting interface of triheme cytochromes and MacA. Comparison of PpcB-E’s (A) and MacA’s (B) heme substituents’ chemical shift variations in the oxidized state calculated from the data obtained in the PpcB-E:MacA 1:4 ratio. The most affected groups are indicated with an asterisk and were selected by calculating the cut-off value for each set of experiments following the procedure reported by Schumann and co-workers (Schumann et al., 2007). The cut-off values were the following: (A) Δδcut-off (PpcB) 0.01 ppm, Δδcut-off (PpcC) 0.05 ppm, Δδcut-off (PpcD) 0.02 ppm, Δδcut-off (PpcE) 0.04 ppm; (B) Δδcut-off (PpcB) 0.28 ppm, Δδcut-off (PpcC) 0.19 ppm, Δδcut-off (PpcD) 0.18 ppm, Δδcut-off (PpcE) 0.15 ppm.
Docking of the triheme cytochromes to MacA (Figure 4) showed that the interaction between the redox pairs is, most likely, driven by electrostatic charge, as the triheme cytochromes’ hemes relevant for the interaction are located within a positive electrostatic surface and MacA’s HP heme is in a negative electrostatic patch. The interaction results obtained are in accordance with a hypothesis proposed by Pokkuluri et al. (2010) that pointed out that all triheme cytochrome homologs have the highest structural similarity and positive electrostatic surface potential near heme IV, thus suggesting that ET to the redox partners occurred in this region. In the case of PpcC there is a partial positive charge surrounding heme I, as well, that may favor electrostatic attraction. However, redox interactions may be also governed by hydrophobic interactions (Nóbrega and Pauleta, 2019) and, as such, the combination of these two elements may explain the different interaction interface in this cytochrome.
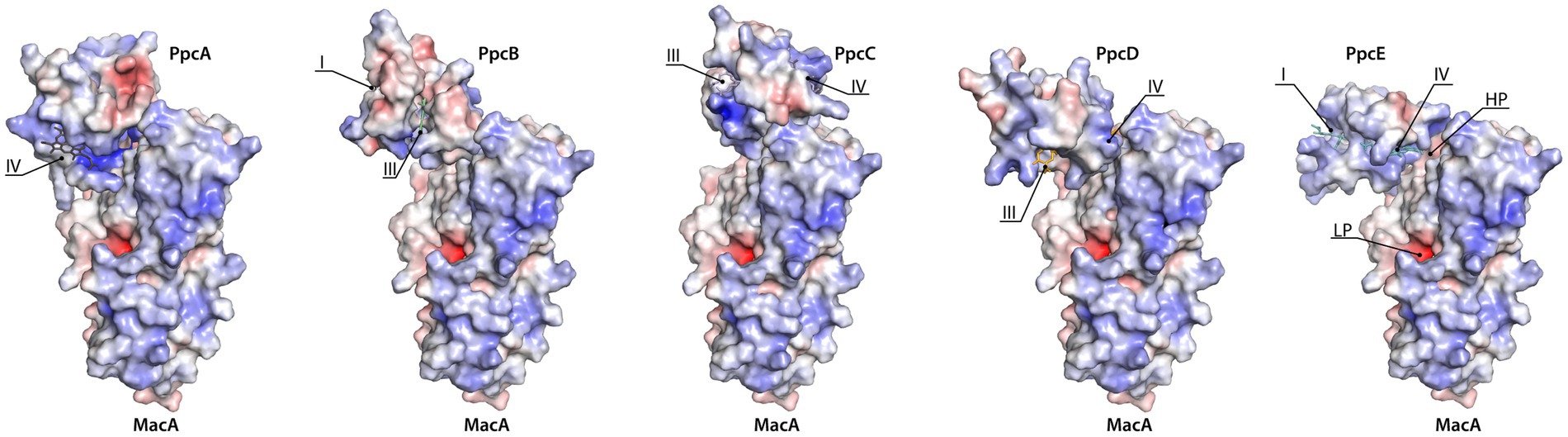
Figure 4. Docking of triheme cytochromes with MacA peroxidase monomer. The best docking solutions found by HADDOCK2.4 (van Zundert et al., 2016; Honorato et al., 2021) for the pairs PpcB-E with MacA are represented. Docking of PpcA with MacA is shown for the sake of completeness since it had been determined previously (Dantas et al., 2017). MacA is represented in the same orientation. The molecules’ surfaces depict the electrostatic potential, −7kBT (red) to +7kBT (blue), calculated by APBS (Dolinsky et al., 2007; Konecny et al., 2013). HP and LP stand for MacA’s HP heme and LP heme and the triheme cytochromes’ hemes are indicated by roman numbers. Structures rendered with PyMOL (Schrodinger, 2015).
The chemical shift perturbation values observed in these interactions are within the range of reported labile complexes (Worrall et al., 2003). Interestingly, the values obtained for triheme cytochromes are lower than MacA’s and this may be explained in terms of the possible orientation of the molecule in the transient complex: for MacA, the productive encounter only happens near the HP heme, thus the favorable interaction interface is much smaller than for the triheme cytochromes, as they may adopt a higher number of orientations that still allow for a productive encounter since all hemes possess a suitable reduction potential for donating an electron. To investigate this hypothesis, we aligned the structures from the best docking solutions either through MacA or the triheme cytochromes (Figure 5). Then, using Chimera (Pettersen et al., 2004) we determined the RMSD of the structure that was not aligned, i.e., the triheme cytochrome’s RMSD in the first case and MacA’s RMSD in the latter, to quantify the variability of docking positions. The results show that the RMSD values for the triheme cytochromes’ positioning on MacA are lower than the reverse situation, illustrating that the spatial solutions for a productive encounter on MacA are lower than for the triheme cytochromes.
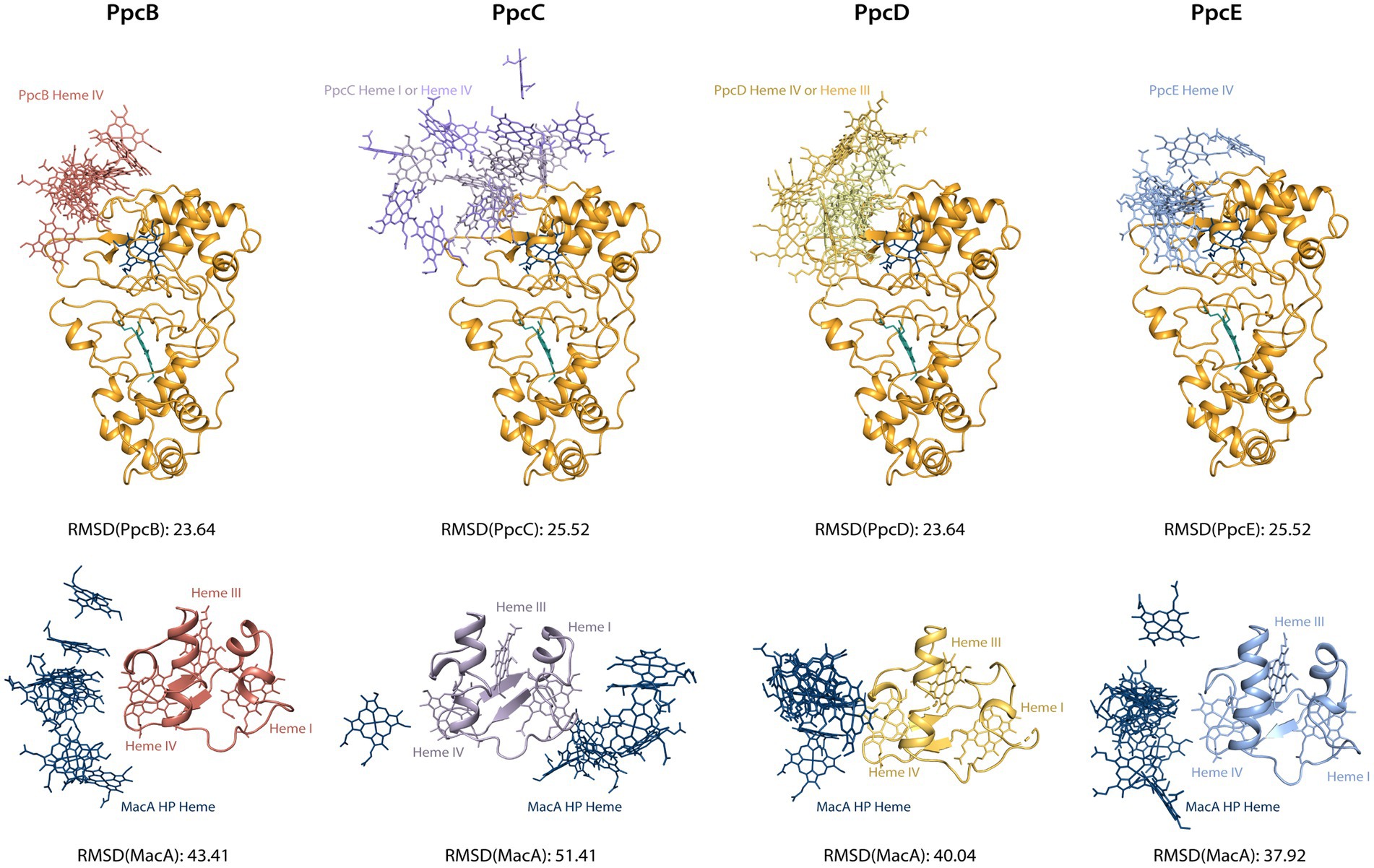
Figure 5. Docking solutions of triheme cytochromes and MacA peroxidase. The alignment of the complexes was made by forcing alignment of all structures to the best structure determined by HADDOCK2.4 either through MacA (top structures) or triheme cytochrome (bottom structures). The best structure of each calculated cluster is represented, as well as the RMSD value. Only the relevant interacting hemes are represented: HP heme for MacA, heme IV for PpcB and PpcE, hemes I and IV for PpcC, hemes III and IV for PpcD. Structures generated with PyMOL (Schrodinger, 2015).
Recent studies suggested that MacA and the PpcA family of periplasmic triheme cytochromes participated in the oxidative stress protection pathways in G. sulfurreducens (Speers and Reguera, 2021; Choi et al., 2022). In this work, direct ET from the periplasmic triheme cytochromes (PpcA-E) to MacA was probed by NMR and the results obtained showed that all the triheme cytochromes undergo complete oxidation and provide all their reducing power specifically to MacA’s HP heme. Monitorization of each redox complex interaction interface shows a higher chemical shift perturbation of MacA’s heme substituents than the triheme cytochrome ones. This indicated that, while for MacA it is necessary that the interaction occurs near the HP heme, in the case of triheme cytochromes there is a higher degree of conformations that they can adopt, thus facilitating an efficient ET. Nevertheless, this interaction is most likely driven by electrostatic complementarity of the triheme cytochrome’s positively charged patch around heme IV or heme I (in the case of PpcC), and MacA’s negatively charged patch flanking the HP heme.
Redox complexes with PpcA were also observed for cytochromes OmcF (Morgado and Salgueiro, 2022) and CbcL (Antunes et al., 2022). While the ET reaction observed in the redox pair PpcA:OmcF resembles the one observed in the present work between the PpcA-E family and MacA, in the redox pair PpcA:CbcL the two proteins remain in a semi-reduced state. This behavior illustrates the metabolic advantage of the periplasmic triheme cytochrome family, as its redox properties are versatile enough to regulate redox networks with either upstream or downstream redox partners. Finally, the results from this study provide an important contribution in the characterization of G. sulfurreducens’ electron flow in oxidative stress conditions, highlighting the ubiquitous role of triheme periplasmic cytochromes.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
CS supervised the project. CS, LM, and PP designed the experiments. PP, LM, MS, and LD performed the experiments. PP, LM, and CS performed the data analysis. PP, LM, CS, and OE wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by Fundação para a Ciência e Tecnologia (FCT – Portugal) through the following grants: PTDC/BIA-BQM/4967/2020 (to CS), 2020.04717.BD (to PP), EXPL/BIA-BQM/0770/2021 (to LM), UIDP/04378/2020 and UIDB/04378/2020 (UCIBIO), LA/P/0140/2020 (Associate Laboratory i4HB), and Deutsche Forschungsgemeinschaft (CRC 1381, project ID 403222702; to OE). The NMR spectrometers are part of the National NMR Network (PTNMR) and are supported by FCT-MCTES (ROTEIRO/0031/2013 – PINFRA/22161/2016) co-funded by FEDER through COMPETE 2020, POCI, and PORL and FCT through PIDDAC.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1253114/full#supplementary-material
Aklujkar, M., Coppi, M. V., Leang, C., Kim, B. C., Chavan, M. A., Perpetua, L. A., et al. (2013). Proteins involved in electron transfer to Fe(III) and Mn(IV) oxides by Geobacter sulfurreducens and Geobacter uraniireducens. Microbiology 159, 515–535. doi: 10.1099/mic.0.064089-0
Anderson, R. T., Vrionis, H. A., Ortiz-Bernad, I., Resch, C. T., Long, P. E., Dayvault, R., et al. (2003). Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69, 5884–5891. doi: 10.1128/AEM.69.10.5884-5891.2003
Antunes, J. M. A., Silva, M. A., Salgueiro, C. A., and Morgado, L. (2022). Electron flow from the inner membrane towards the cell exterior in Geobacter sulfurreducens: biochemical characterization of cytochrome CbcL. Front. Microbiol. 13:8015. doi: 10.3389/fmicb.2022.898015
Arciero, D. M., and Hooper, A. B. (1994). A di-heme cytochrome c peroxidase from Nitrosomonas europaea catalytically active in both the oxidized and half-reduced states. J. Biol. Chem. 269, 11878–11886. doi: 10.1016/S0021-9258(17)32655-8
Arslan, E., Schulz, H., Zufferey, R., Künzler, P., and Thöny-Meyer, L. (1998). Overproduction of the Bradyrhizobium japonicum c-type cytochrome subunits of the cbb3 oxidase in Escherichia coli. Biochem. Biophys. Res. Commun. 251, 744–747. doi: 10.1006/bbrc.1998.9549
Bertini, I., Luchinat, C., Parigi, G., and Walker, F. A. (1999). Heme methyl 1H chemical shifts as structural parameters in some low-spin ferriheme proteins. J. Biol. Inorg. Chem. 4, 515–519. doi: 10.1007/s007750050337
Butler, J. E., Kaufmann, F., Coppi, M. V., Núñez, C., and Lovley, D. R. (2004). MacA, a diheme c-type cytochrome involved in Fe(III) reduction by Geobacter sulfurreducens. J. Bacteriol. 186, 4042–4045. doi: 10.1128/JB.186.12.4042-4045.2004
Caccavo, F. Jr., Lonergan, D. J., Lovley, D. R., Davis, M., Stolz, J. F., and McInerney, M. J. (1994). Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl. Environ. Microbiol. 60, 3752–3759. doi: 10.1128/aem.60.10.3752-3759.1994
Choi, S., Chan, C. H., and Bond, D. R. (2022). Lack of specificity in Geobacter periplasmic electron transfer. J. Bacteriol. 204:e00322. doi: 10.1128/jb.00322-22
Coppi, M. V., O'Neil, R. A., and Lovley, D. R. (2004). Identification of an uptake hydrogenase required for hydrogen-dependent reduction of Fe(III) and other electron acceptors by Geobacter sulfurreducens. J. Bacteriol. 186, 3022–3028. doi: 10.1128/JB.186.10.3022-3028.2004
Dantas, J. M., Brausemann, A., Einsle, O., and Salgueiro, C. A. (2017). NMR studies of the interaction between inner membrane-associated and periplasmic cytochromes from Geobacter sulfurreducens. FEBS Lett. 591, 1657–1666. doi: 10.1002/1873-3468.12695
Dantas, J. M., Morgado, L., Aklujkar, M., Bruix, M., Londer, Y. Y., Schiffer, M., et al. (2015). Rational engineering of Geobacter sulfurreducens electron transfer components: a foundation for building improved Geobacter-based bioelectrochemical technologies. Front. Microbiol. 6:752. doi: 10.3389/fmicb.2015.00752
Ding, Y.-H. R., Hixson, K. K., Aklujkar, M. A., Lipton, M. S., Smith, R. D., Lovley, D. R., et al. (2008). Proteome of Geobacter sulfurreducens grown with Fe(III) oxide or Fe(III) citrate as the electron acceptor. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1784, 1935–1941. doi: 10.1016/j.bbapap.2008.06.011
Dolinsky, T. J., Czodrowski, P., Li, H., Nielsen, J. E., Jensen, J. H., Klebe, G., et al. (2007). PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525. doi: 10.1093/nar/gkm276
Dumas, C., Basseguy, R., and Bergel, A. (2008). Microbial electrocatalysis with Geobacter sulfurreducens biofilm on stainless steel cathodes. Electrochim. Acta 53, 2494–2500. doi: 10.1016/j.electacta.2007.10.018
Ellfolk, N., Rönnberg, M., Aasa, R., Andréasson, L.-E., and Vänngård, T. (1983). Properties and function of the two hemes in Pseudomonas cytochrome c peroxidase. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 743, 23–30. doi: 10.1016/0167-4838(83)90413-2
Ellis, K. E., Seidel, J., Einsle, O., and Elliott, S. J. (2011). Geobacter sulfurreducens cytochrome c peroxidases: electrochemical classification of catalytic mechanisms. Biochemistry 50, 4513–4520. doi: 10.1021/bi200399h
Engel, C. E. A., Vorländer, D., Biedendieck, R., Krull, R., and Dohnt, K. (2020). Quantification of microaerobic growth of Geobacter sulfurreducens. PLoS One 15:e0215341. doi: 10.1371/journal.pone.0215341
Gong, Z., Yu, H., Zhang, J., Li, F., and Song, H. (2020). Microbial electro-fermentation for synthesis of chemicals and biofuels driven by bi-directional extracellular electron transfer. Synth. Systems Biotechnol. 5, 304–313. doi: 10.1016/j.synbio.2020.08.004
Gu, Y., Guberman-Pfeffer, M. J., Srikanth, V., Shen, C., Giska, F., Gupta, K., et al. (2023). Structure of Geobacter cytochrome OmcZ identifies mechanism of nanowire assembly and conductivity. Nat. Microbiol. 8, 284–298. doi: 10.1038/s41564-022-01315-5
He, Y., Gong, Y., Su, Y., Zhang, Y., and Zhou, X. (2019). Bioremediation of Cr (VI) contaminated groundwater by Geobacter sulfurreducens: environmental factors and electron transfer flow studies. Chemosphere 221, 793–801. doi: 10.1016/j.chemosphere.2019.01.039
Heidary, N., Kornienko, N., Kalathil, S., Fang, X., Ly, K. H., Greer, H. F., et al. (2020). Disparity of cytochrome utilization in anodic and cathodic extracellular electron transfer pathways of Geobacter sulfurreducens biofilms. J. Am. Chem. Soc. 142, 5194–5203. doi: 10.1021/jacs.9b13077
Hoffmann, M., Seidel, J., and Einsle, O. (2009). CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase. J. Mol. Biol. 393, 951–965. doi: 10.1016/j.jmb.2009.09.001
Honorato, R. V., Koukos, P. I., Jiménez-García, B., Tsaregorodtsev, A., Verlato, M., Giachetti, A., et al. (2021). Structural biology in the clouds: the WeNMR-EOSC ecosystem. Front. Mol. Biosci. 8:9513. doi: 10.3389/fmolb.2021.729513
Keller, R. M., Wüthrich, K., and Debrunner, P. G. (1972). Proton magnetic resonance reveals high-spin iron (II) in ferrous cytochrome P450cam from Pseudomonas putida. Proc. Natl. Acad. Sci. 69, 2073–2075. doi: 10.1073/pnas.69.8.2073
Kim, B.-C., and Lovley, D. R. (2008). Investigation of direct vs. indirect involvement of the c-type cytochrome MacA in Fe(III) reduction by Geobacter sulfurreducens. FEMS Microbiol. Lett. 286, 39–44. doi: 10.1111/j.1574-6968.2008.01252.x
Konecny, R., Baker, N. A., and McCammon, J. A. (2013). iAPBS: a programming interface to the adaptive Poisson–Boltzmann solver. Computational Sci. Discovery 5:015005. doi: 10.1088/1749-4699/5/1/015005
Kracke, F., Vassilev, I., and Krömer, J. O. (2015). Microbial electron transport and energy conservation – the foundation for optimizing bioelectrochemical systems. Front. Microbiol. 6:0575. doi: 10.3389/fmicb.2015.00575
Lin, W. C., Coppi, M. V., and Lovley, D. R. (2004). Geobacter sulfurreducens can grow with oxygen as a terminal electron acceptor. Appl. Environ. Microbiol. 70, 2525–2528. doi: 10.1128/AEM.70.4.2525-2528.2004
Londer, Y. Y., Pokkuluri, P. R., Tiede, D. M., and Schiffer, M. (2002). Production and preliminary characterization of a recombinant triheme cytochrome c7 from Geobacter sulfurreducens in Escherichia coli. Biochimica et Biophysica Acta (BBA) - Bioenergetics 1554, 202–211. doi: 10.1016/S0005-2728(02)00244-X
Lovley, D. R. (2011) in Advances in microbial physiology. ed. R. K. Poole, vol. 59 (Cambridge: Academic Press), 1–100.
Lovley, D. R. (2022). On the existence of pilin-based microbial nanowires. Front. Microbiol. 13:2610. doi: 10.3389/fmicb.2022.872610
Lovley, D. R., and Phillips, E. J. P. (1992). Bioremediation of uranium contamination with enzymatic uranium reduction. Environ. Sci. Technol. 26, 2228–2234. doi: 10.1021/es00035a023
Methé, B. A., Nelson, K. E., Eisen, J. A., Paulsen, I. T., Nelson, W., Heidelberg, J. F., et al. (2003). Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science 302, 1967–1969. doi: 10.1126/science.1088727
Morgado, L., Bruix, M., Londer, Y. Y., Pokkuluri, P. R., Schiffer, M., and Salgueiro, C. A. (2007). Redox-linked conformational changes of a multiheme cytochrome from Geobacter sulfurreducens. Biochem. Biophys. Res. Commun. 360, 194–198. doi: 10.1016/j.bbrc.2007.06.026
Morgado, L., Bruix, M., Orshonsky, V., Londer, Y. Y., Duke, N. E. C., Yang, X., et al. (2008). Structural insights into the modulation of the redox properties of two Geobacter sulfurreducens homologous triheme cytochromes. Biochimica et Biophysica Acta (BBA) - Bioenergetics 1777, 1157–1165. doi: 10.1016/j.bbabio.2008.04.043
Morgado, L., Bruix, M., Pessanha, M., Londer, Y. Y., and Salgueiro, C. A. (2010). Thermodynamic characterization of a triheme cytochrome family from Geobacter sulfurreducens reveals mechanistic and functional diversity. Biophys. J. 99, 293–301. doi: 10.1016/j.bpj.2010.04.017
Morgado, L., and Salgueiro, C. A. (2022). Elucidation of complex respiratory chains: a straightforward strategy to monitor electron transfer between cytochromes. Metallomics 14:mfac012. doi: 10.1093/mtomcs/mfac012
Njus, D., Kelley, P. M., Tu, Y.-J., and Schlegel, H. B. (2020). Ascorbic acid: the chemistry underlying its antioxidant properties. Free Radic. Biol. Med. 159, 37–43. doi: 10.1016/j.freeradbiomed.2020.07.013
Nóbrega, C. S., and Pauleta, S. R. (2019) in Advances in microbial physiology. ed. R. K. Poole, vol. 74 (Cambridge: Academic Press), 415–464.
Ortiz-Bernad, I., Anderson, R. T., Vrionis, H. A., and Lovley, D. R. (2004). Vanadium respiration by Geobacter metallireducens: novel strategy for in situ removal of vanadium from groundwater. Appl. Environ. Microbiol. 70, 3091–3095. doi: 10.1128/AEM.70.5.3091-3095.2004
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Pokkuluri, P. R., Londer, Y. Y., Yang, X., Duke, N. E. C., Erickson, J., Orshonsky, V., et al. (2010). Structural characterization of a family of cytochromes c7 involved in Fe(III) respiration by Geobacter sulfurreducens. Biochimica et Biophysica Acta (BBA) - Bioenergetics 1797, 222–232. doi: 10.1016/j.bbabio.2009.10.007
Reguera, G., and Kashefi, K. (2019) in Advances in microbial physiology. ed. R. K. Poole, vol. 74 (Cambridge: Academic Press), 1–96.
Reguera, G., McCarthy, K. D., Mehta, T., Nicoll, J. S., Tuominen, M. T., and Lovley, D. R. (2005). Extracellular electron transfer via microbial nanowires. Nature 435, 1098–1101. doi: 10.1038/nature03661
Salgueiro, C. A., Dantas, J. M., and Morgado, L. (2019). Principles of nuclear magnetic resonance and selected biological applications. Rad. Bioanal.: Spectroscop. Techn. Theor.l Methods 8:247. doi: 10.1007/978-3-030-28247-9_9
Salgueiro, C. A., Morgado, L., Silva, M. A., Ferreira, M. R., Fernandes, T. M., and Portela, P. C. (2022). From iron to bacterial electroconductive filaments: exploring cytochrome diversity using Geobacter bacteria. Coord. Chem. Rev. 452:214284. doi: 10.1016/j.ccr.2021.214284
Sanford, R. A., Wu, Q., Sung, Y., Thomas, S. H., Amos, B. K., Prince, E. K., et al. (2007). Hexavalent uranium supports growth of Anaeromyxobacter dehalogenans and Geobacter spp. with lower than predicted biomass yields. Environ. Microbiol. 9, 2885–2893. doi: 10.1111/j.1462-2920.2007.01405.x
Santoro, C., Arbizzani, C., Erable, B., and Ieropoulos, I. (2017). Microbial fuel cells: from fundamentals to applications. Rev. J. Power Sources 356, 225–244. doi: 10.1016/j.jpowsour.2017.03.109
Santos, T. C., Silva, M. A., Morgado, L., Dantas, J. M., and Salgueiro, C. A. (2015). Diving into the redox properties of Geobacter sulfurreducens cytochromes: a model for extracellular electron transfer. Dalton Trans. 44, 9335–9344. doi: 10.1039/C5DT00556F
Schumann, F. H., Riepl, H., Maurer, T., Gronwald, W., Neidig, K. P., and Kalbitzer, H. R. (2007). Combined chemical shift changes and amino acid specific chemical shift mapping of protein-protein interactions. J. Biomol. NMR 39, 275–289. doi: 10.1007/s10858-007-9197-z
Seidel, J., Hoffmann, M., Ellis, K. E., Seidel, A., Spatzal, T., Gerhardt, S., et al. (2012). MacA is a second cytochrome c peroxidase of Geobacter sulfurreducens. Biochemistry 51, 2747–2756. doi: 10.1021/bi300249u
Shelobolina, E. S., Coppi, M. V., Korenevsky, A. A., DiDonato, L. N., Sullivan, S. A., Konishi, H., et al. (2007). Importance of c-type cytochromes for U(VI) reduction by Geobacter sulfurreducens. BMC Microbiol. 7:16. doi: 10.1186/1471-2180-7-16
Speers, A. M., and Reguera, G. (2021). Competitive advantage of oxygen-tolerant bioanodes of Geobacter sulfurreducens in bioelectrochemical systems. Biofilms 3:100052. doi: 10.1016/j.bioflm.2021.100052
Strycharz, S. M., Glaven, R. H., Coppi, M. V., Gannon, S. M., Perpetua, L. A., Liu, A., et al. (2011). Gene expression and deletion analysis of mechanisms for electron transfer from electrodes to Geobacter sulfurreducens. Bioelectrochemistry 80, 142–150. doi: 10.1016/j.bioelechem.2010.07.005
Tremblay, P.-L., and Lovley, D. R. (2012). Role of the NiFe hydrogenase Hya in oxidative stress defense in Geobacter sulfurreducens. J. Bacteriol. 194, 2248–2253. doi: 10.1128/JB.00044-12
Turner, D. L., Costa, H. S., Coutinho, I. B., Legall, J., and Xavier, A. V. (1997). Assignment of the ligand geometry and redox potentials of the trihaem ferricytochrome c3 from Desulfuromonas acetoxidans. Eur. J. Biochem. 243, 474–481. doi: 10.1111/j.1432-1033.1997.0474a.x
Turner, D. L., Salgueiro, C. A., Schenkels, P., LeGall, J., and Xavier, A. V. (1995). Carbon-13 NMR studies of the influence of axial ligand orientation on haem electronic structure. Biochimica et Biophysica Acta (BBA)-Protein Struc. Molec.r Enzymol. 1246, 24–28. doi: 10.1016/0167-4838(94)00175-G
van Zundert, G. C. P., Rodrigues, J. P. G. L. M., Trellet, M., Schmitz, C., Kastritis, P. L., Karaca, E., et al. (2016). The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725. doi: 10.1016/j.jmb.2015.09.014
Villalaín, J., et al. (1984). NMR and electron-paramagnetic-resonance studies of a dihaem cytochrome from Pseudomonas stutzeri (ATCC 11607) (cytochrome c peroxidase). Eur. J. Biochem. 141, 305–312. doi: 10.1111/j.1432-1033.1984.tb08192.x
Wang, F., Gu, Y., O’Brien, J. P., Yi, S. M., Yalcin, S. E., Srikanth, V., et al. (2019). Structure of Microbial Nanowires Reveals Stacked Hemes that Transport Electrons over Micrometers. Cell 177, 361–369.e10. doi: 10.1016/j.cell.2019.03.029
Wang, F., Mustafa, K., Suciu, V., Joshi, K., Chan, C. H., Choi, S., et al. (2022). Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing. Nat. Microbiol. 7, 1291–1300. doi: 10.1038/s41564-022-01159-z
Wishart, D. S., Bigam, C. G., Yao, J., Abildgaard, F., Dyson, H. J., Oldfield, E., et al. (1995). 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 6, 135–140. doi: 10.1007/BF00211777
Keywords: Geobacter, oxidative stress, cytochrome, peroxidase, electron transfer, protein–protein interactions, NMR
Citation: Portela PC, Morgado L, Silva MA, Denkhaus L, Einsle O and Salgueiro CA (2023) Exploring oxidative stress pathways in Geobacter sulfurreducens: the redox network between MacA peroxidase and triheme periplasmic cytochromes. Front. Microbiol. 14:1253114. doi: 10.3389/fmicb.2023.1253114
Received: 04 July 2023; Accepted: 11 September 2023;
Published: 04 October 2023.
Edited by:
Yiran Dong, China University of Geosciences Wuhan, ChinaReviewed by:
Xing Liu, Fujian Agriculture and Forestry University, ChinaCopyright © 2023 Portela, Morgado, Silva, Denkhaus, Einsle and Salgueiro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carlos A. Salgueiro, Y3NhbGd1ZWlyb0BmY3QudW5sLnB0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.