- 1The Second Affiliated Hospital and Yuying Children’s Hospital, Wenzhou Medical University, Wenzhou, China
- 2Key Laboratory of Medical Genetics of Zhejiang Province, Key Laboratory of Laboratory Medicine, Ministry of Education, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China
- 3Medical Molecular Biology Laboratory, School of Medicine, Jinhua Polytechnic, Jinhua, China
Background: Achromobacter is a genus of gram-negative bacteria that can act as opportunistic pathogens. Recent studies have revealed that some species of Achromobacter show inherent resistance to β-lactams, but the resistance mechanisms of Achromobacter mucicolens have rarely been reported.
Method: The bacterium was isolated using standard laboratory procedures. The agar dilution method was used to determine the minimum inhibitory concentrations (MICs). Genome sequencing was performed using the PacBio RS II and Illumina HiSeq 2500 platforms, and the Comprehensive Antibiotic Resistance Database (CARD) was used to annotate the drug resistance genes. The localization of the novel β-lactamase AMZ-1 was determined, and its characteristics were determined via molecular cloning and enzyme kinetic analysis. The phylogenetic relationship and comparative genomic analysis of the resistance gene-related sequences were also analyzed.
Result: Achromobacter mucicolens Y3, isolated from a goose on a farm in Wenzhou, showed resistance to multiple antibiotics, including penicillins and cephalosporins. BlaAMZ–1 showed resistance to amoxicillin, penicillin G, ampicillin, cephalothin and cefoxitin, and the resistance activity could be inhibited by β-lactamase inhibitors. Enzyme kinetic analysis results showed that AMZ-1 has hydrolytic activity against a wide range of substrates, including cephalothin, amoxicillin, penicillin G, and cefoxitin but not ampicillin. The hydrolytic activity of AMZ-1 was greatly inhibited by avibactam but much more weakly inhibited by tazobactam. Mobile genetic elements could not be found around the blaAMZ–1-like genes, which are conserved on the chromosomes of bacteria of the genus Achromobacter.
Conclusion: In this study, a novel AmpC gene, blaAMZ–1, from the animal-origin bacterium A. mucicolens Y3 was identified and characterized. It conferred resistance to some penicillins and first- and second-generation cephalosporins. The identification of this novel resistance gene will be beneficial for the selection of effective antimicrobials to treat associated infections.
Introduction
Achromobacter, a genus of the family Alcaligenaceae, are gram-negative and nonfermentative. This genus was originally discovered and established in 1923 by the Committee of the Society of American Bacteriologists (Isler et al., 2020). These organisms are widely distributed in the environment and have the potential to become opportunistic pathogens under certain conditions, such as cystic fibrosis, blood or solid organ malignancies, renal failure, or specific immune deficiencies (Swenson and Sadikot, 2015). Currently, there are 19 officially recognized species within the Achromobacter genus, and most members of this genus exhibit nonpathogenic characteristics, except Achromobacter xylosoxidans. A. xylosoxidans has been demonstrated to be an opportunistic pathogen causing a range of infections in immunocompromised individuals (Ahmed et al., 2009), while the pathogenic potential of Achromobacter mucicolens has been infrequently documented.
In recent years, clinical Achromobacter isolates have presented multidrug resistance, posing a challenge to effective infection management in affected patients (Isler et al., 2020). Efflux pumps and chromosome-encoded β-lactamases are expressed at a basal level, leading to reduced drug susceptibility. Despite the identification of several β-lactamases, such as TMB-1, OXA-114, OXA-285a, and OXA-285b, in A. xylosoxidans isolates (Doi et al., 2008; El Salabi et al., 2012; Papalia et al., 2018), the drug resistance mechanism of A. mucicolens has rarely been documented (Al-Asadi et al., 2022).
β-Lactams remain dominant among antibacterial drugs (Bush and Bradford, 2016), but the overuse of these antibiotics has led to increased microbial resistance. There are three main mechanisms of bacterial resistance to β-lactams: hydrolysis of drugs by β-lactamases (Li et al., 2007), inability of drug target binding (Li et al., 2007), and drug target alteration. The main resistance mechanism of gram-negative bacteria is the production of β-lactamases, which inactivate β-lactams by hydrolyzing the structural β-lactam ring (Li et al., 2007; Bush and Bradford, 2016). AmpC β-lactamases have a catalytic serine residue and, based on their structure, are assigned to class C in the Ambler classification of β-lactamases (Ambler, 1980). AmpC β-lactamases can hydrolyze penicillins and cephalosporins, including cephamycins (such as cefoxitin and cefotetan), oxyiminocephalosporins (such as ceftazidime, cefotaxime, and ceftriaxone), and monobactams (such as aztreonam) (Jacoby, 2009). AmpC β-lactamases have a molecular weight between 34 and 40 kDa, generally have isoelectric points > 8.0, and can be induced and expressed at high levels by many bacteria due to mutations (Jacoby, 2009). The increased antibiotic resistance resulting from overexpression of AmpC enzymes has become an increasingly serious problem, especially with regard to infections caused by opportunistic bacterial pathogens (Tamma et al., 2019).
In this study, we isolated an A. mucicolens strain from a goose, sequenced its complete genome sequence and designated it A. mucicolens Y3. The novel AmpC gene blaAMZ–1 identified in the chromosome of A. mucicolens Y3 conferred resistance to amoxicillin, penicillin G, ampicillin, cephalothin and cefoxitin. Finding novel resistance genes will help to further elucidate the complexity of drug resistance mechanisms.
Materials and methods
Bacterial strains and plasmids
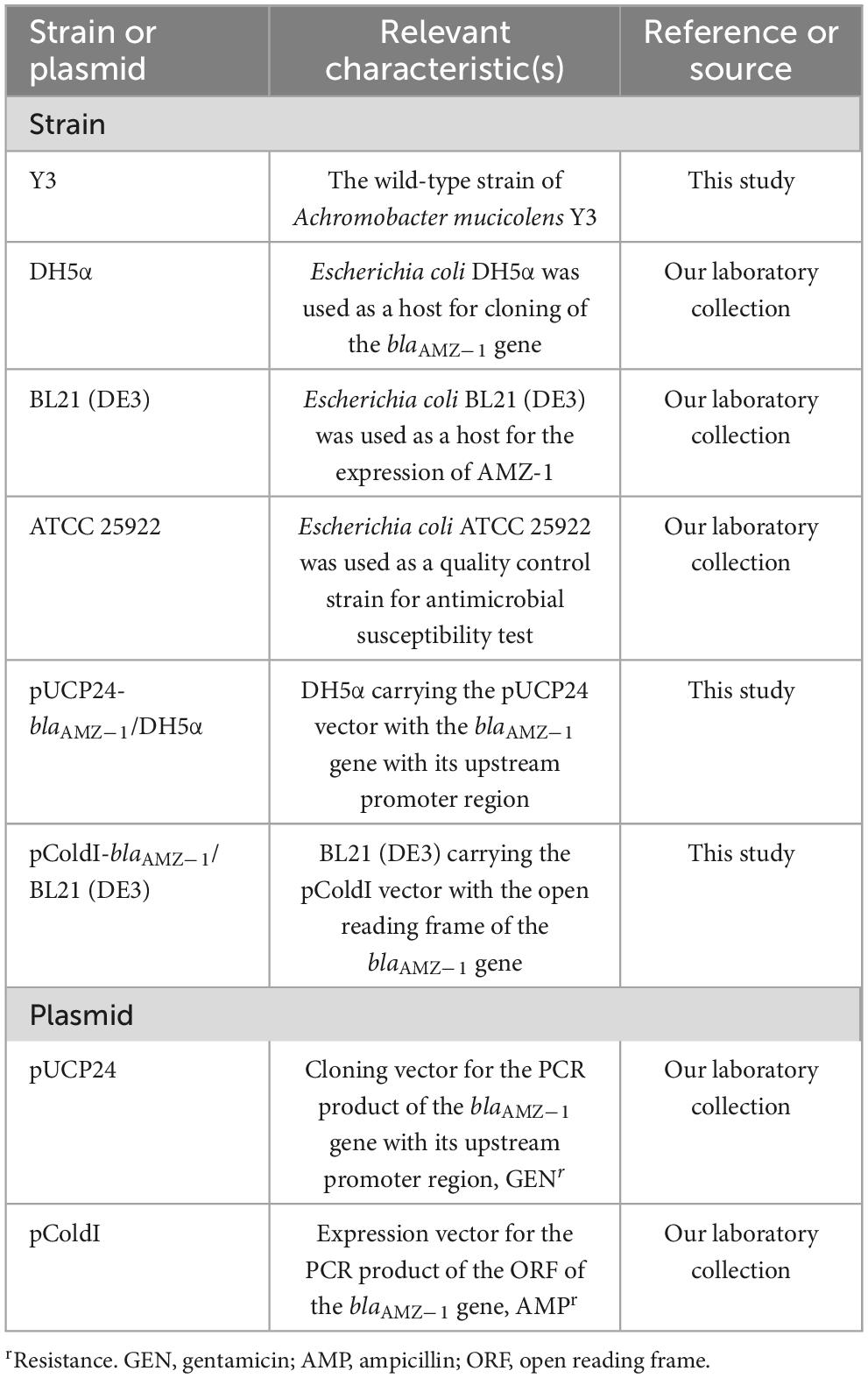
Achromobacter mucicolens Y3 was isolated from the fecal swab of a goose at a livestock farm during a survey on the antimicrobial resistance of bacteria in Wenzhou, southern China. The strain species was verified using 16S ribosomal RNA (rRNA) gene homology (Konstantinidis and Tiedje, 2005) and average nucleotide identity (ANI) analyses (Richter and Rosselló-Móra, 2009). The bacterial strains and plasmids used in this work are listed in Table 1.
Antimicrobial susceptibility testing
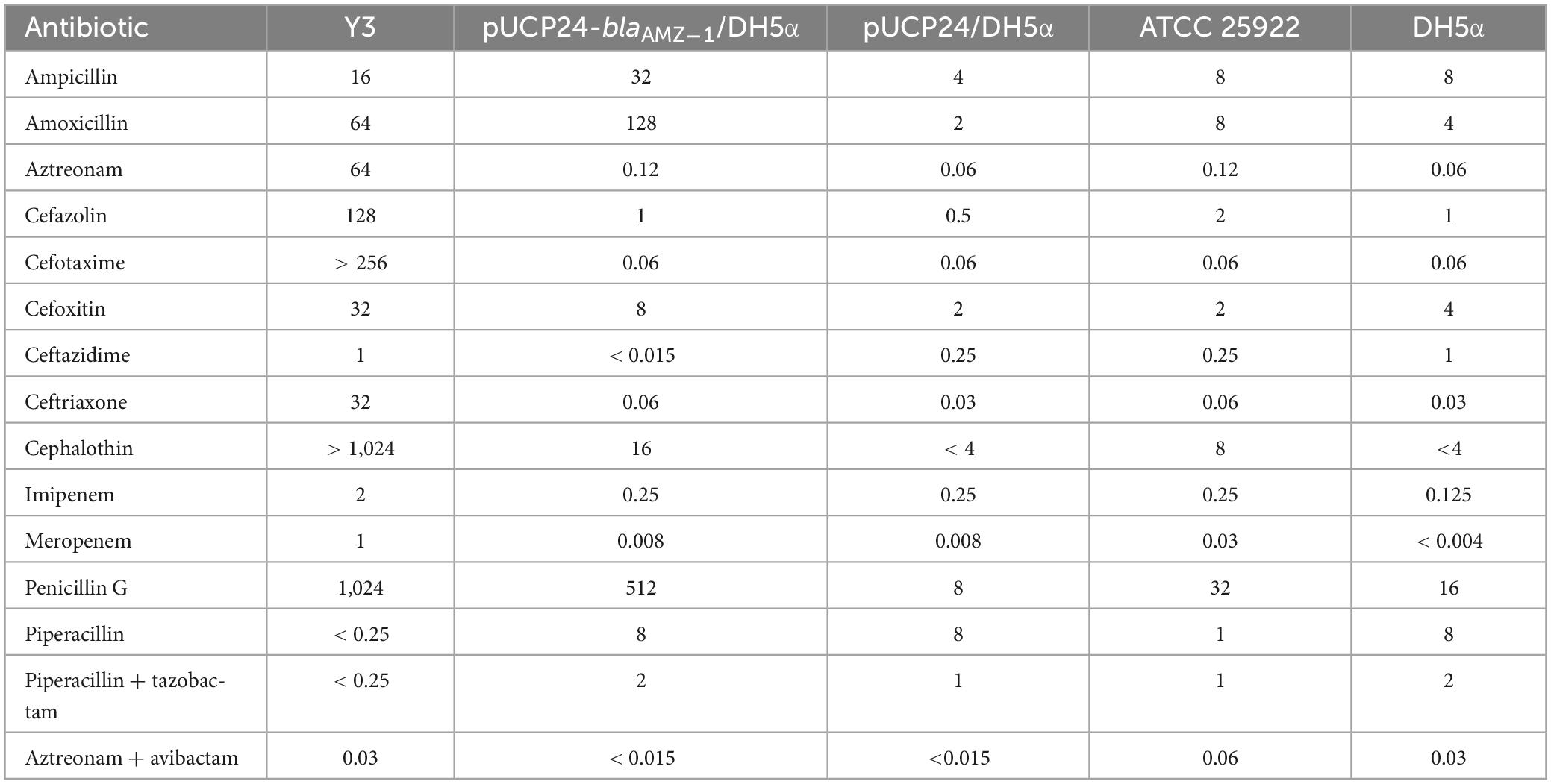
The standard broth dilution method was used to determine minimum inhibitory concentrations (MICs) with Mueller Hinton (MH) agar plates according to the Clinical and Laboratory Standards Institute (CLSI, 2021) guidelines. Thirteen antimicrobials, including ampicillin, amoxicillin, aztreonam, and so on were tested (Table 2). The bacterial solution cultured overnight was diluted ten-fold with normal saline after adjusting the bacterial concentration to the 0.5 McFarland’s standard and then used to determine the MICs of the antimicrobials. The CLSI (2021) guidelines were followed to determine the antibiotic sensitivity pattern based on the measured antibiotic MIC levels. Escherichia coli ATCC25922 was used as a standard quality control.
Genome sequencing
The Generay Genomic DNA Miniprep Kit (Shanghai Generay Biotech Co., Ltd., Shanghai, China) was used to extract the total genomic DNA of A. mucicolens Y3, which was sequenced with both the Illumina HiSeq 2500 and PacBio RS II platforms by Shanghai Sunny Biotechnology Co., Ltd. (Shanghai, China), and the read lengths were pE150 and 10 kb with coverage of 150× each, respectively. The PacBio long reads were assembled by Unicycler v0.4.8 (Wick et al., 2017), and the quality of the draft genome assembly was enhanced by Pilon by mapping Illumina short reads to the assembly to correct any misassembled bases (Walker et al., 2014). Genes were predicted and annotated by using Prokka v1.14.6 (Seemann, 2014). DIAMOND v2.0.11 (Buchfink et al., 2021) with an e-value cutoff of 1e-5 was utilized to search the predicted proteins in the NCBI nonredundant protein database. The Comprehensive Antibiotic Resistance Database (CARD)1 and Resistance Gene Identifier v5.2.02 were used to annotate the resistance genes (McArthur et al., 2013). Using the NCBI 16S ribosomal RNA database2, 16S rRNA gene homology analysis was carried out. The NCBI database3 and the gcType database4 (Shi et al., 2021) were used to determine the ANI using FastANI (Jain et al., 2018). Multiple sequence alignment of AMZ-1 and other β-lactamases was carried out using MAFFT v7.490 (Katoh and Standley, 2013), and the results were visualized using the msa package in R (Bodenhofer et al., 2015). A neighbor-joining phylogenetic tree including AMZ-1 and other functionally characterized β-lactamases from the Beta-Lactamase DataBase5 and UniProtKB/Swiss-Prot Database6 was reconstructed using MEGA11 (Kumar et al., 2018). The phylogenetic tree was visualized using the online tool iTol (Letunic and Bork, 2007). The figures of the genetic environments surrounding the blaAMZ–1 and blaAMZ–1(-like) genes were generated by clinker v0.0.24 (Gilchrist and Chooi, 2021). The molecular weight and pI value were predicted using ProtParam. GNU Parallel (Tange, 2021).
Cloning of the blaAMZ–1 gene and the expression and purification of AMZ-1
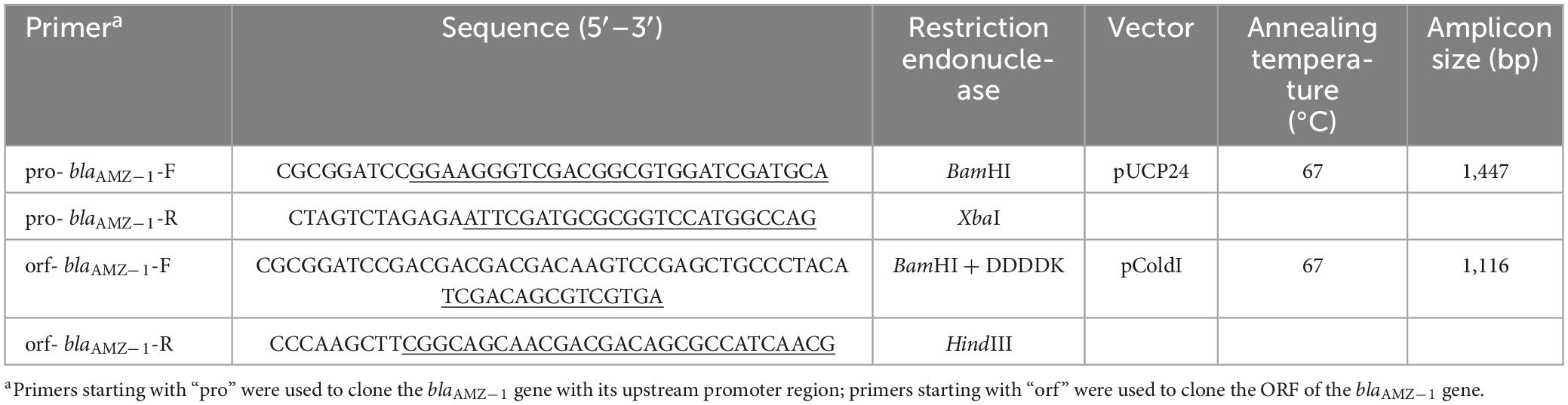
Using A. mucicolens Y3 genomic DNA as a template for polymerase chain reaction (PCR), the blaAMZ–1 gene was cloned. The nucleotide sequence of blaAMZ–1 with its upstream promoter region was amplified by 2× Phanta Max Master Mix (Vazyme, Nanjing, China) (Table 3). The cloning vector pUCP24 was digested with both BamHI and XbaI (Takara Bio Inc., Dalian, China) and purified by the FastPure Gel DNA Extraction Mini Kit (Vazyme, Nanjing, China), and the PCR product was also treated with the same procedures as the vector pUCP24. The ligation of the target gene into the linear vector was performed using a T4 DNA ligase cloning kit (Takara Bio Inc., Dalian, China). The recombinant plasmid (pUCP24-pro-blaAMZ–1) was transformed into E. coli DH5α by the calcium chloride method. Bacterial colonies were cultured on LB agar plates supplemented with gentamicin (40 μg/mL). The recombinant plasmid was verified by both PCR and PCR product Sanger sequencing (Shanghai Sunny Biotechnology Co., Ltd., Shanghai, China).
Similar to the cloning procedures, the ORF of blaAMZ–1 without a signal peptide sequence was cloned (Table 3). Using pColdI as an expression vector, the recombinant plasmid (pColdI-orf-blaAMZ–1) was transformed into E. coli BL21 (DE3). The transformants were cultured on LB agar plates supplemented with ampicillin (100 μg/mL). For the expression of AMZ-1, the recombinant [pColdI-blaAMZ–1/BL21 (DE3)] was cultured overnight in LB medium, diluted 100-fold in fresh medium with ampicillin (100 μg/mL) and incubated at 37°C and 220 rpm for approximately 2–3 h, until the OD600 reached 0.6. After incubation in an ice bath for > 30 min, isopropyl-β-d-thiogalactopyranoside (IPTG, Sigma Chemicals Co., St. Louis, MO, USA) was added at a final concentration of 1 mM, and then another incubation was performed at 16°C for 16 h. The bacteria were collected by centrifugation (8,000 rpm, 4°C, 10 min) using an ultrasonic bacterial crusher, followed by centrifugation (12,000 rpm, 4°C, 30 min) for supernatant collection. The AMZ-1 protein with a his-tag was purified by nickel affinity chromatography with BeyoGold his-tag purification resin (Beyotime, Shanghai, China) and then digested with Enterokinase (GenScript, Nanjing, China) for 36 h at 37°C to remove the his-tag. The protein was identified by SDS-PAGE using a 12% acrylamide separation gel and Coomassie blue G-250 staining. The protein concentration was determined by a BCA kit (Beyotime, Shanghai, China) at 562 nm.
Enzyme kinetic analysis
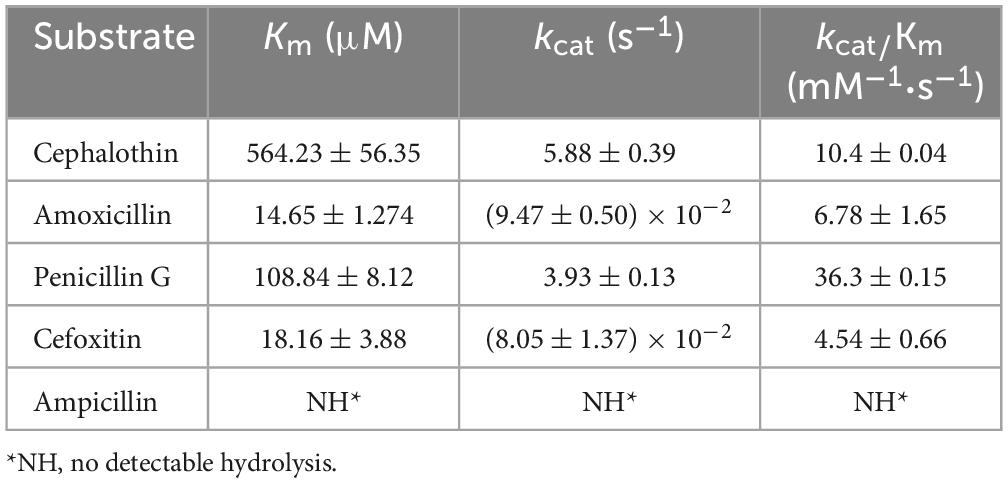
Kinetic analysis was performed as previously reported (Bush et al., 1993). Purified AMZ-1 β-lactamase and β-lactam antibiotics were mixed and examined using a UV–VIS spectrophotometer (SpectraMax M5, Molecular Devices, USA) at 37°C in 10 mM phosphate buffer (pH 7.0) in a final reaction volume of 200 μL. The detection wavelength was derived from the maximum absorption wavelength reported in the literature. The steady-state kinetic parameters (kcat and Km) were determined by nonlinear regression of the initial reaction rates with the Michaelis-Menten equation in Prism (version 8) software (GraphPad Software, San Jose, CA, USA).
The β-lactamase inhibitors avibactam and tazobactam were used at various concentrations, and nitrocefin (final concentration at 100 μM) was added as a substrate. The samples were preincubated with the purified AMZ-1 β-lactamase for 5 min at 37°C before the addition of substrate, and immediately after the addition of nitrocefin, they were scanned at the optimum wavelength for nitrocefin for 2 min. The inhibitor concentration required to reduce the hydrolysis of 100 μM nitrocefin by 50% (IC50) was determined by nonlinear regression with the log (inhibitor) vs. response (three parameters) in Prism (version 8) software (GraphPad Software, San Jose, CA, USA).
Accession numbers
The complete chromosome and blaAMZ–1 gene sequences of A. mucicolens Y3 have been submitted to GenBank, and the accession numbers are CP123364 and OQ948417, respectively.
Results and discussion
Genome characteristics and resistance profile of A. mucicolens Y3
The isolate A. mucicolens Y3 was obtained from an anal swab of a goose on a farm in Wenzhou, Zhejiang Province, China. The 16S RNA gene homology analysis showed that the 16S RNA gene of A. mucicolens Y3 shared the closest identity (100% coverage and 99.87% identity) with that of A. mucicolens LMG 26685 (GCA_902859725.1). Moreover, the ANI analysis revealed that isolate Y3 shared the highest ANI (96.96%) with A. mucicolens LMG 26685. ANI is the gold standard for determining species classification, and the cutoff score (≥ 95%) is the threshold to classify the species of a certain bacterium using a comparison between the unclassified bacterium genome and all the bacterial genomes available in public databases (Richter and Rosselló-Móra, 2009). Ultimately, isolate Y3 was assigned to the species A. mucicolens and designated A. mucicolens Y3.
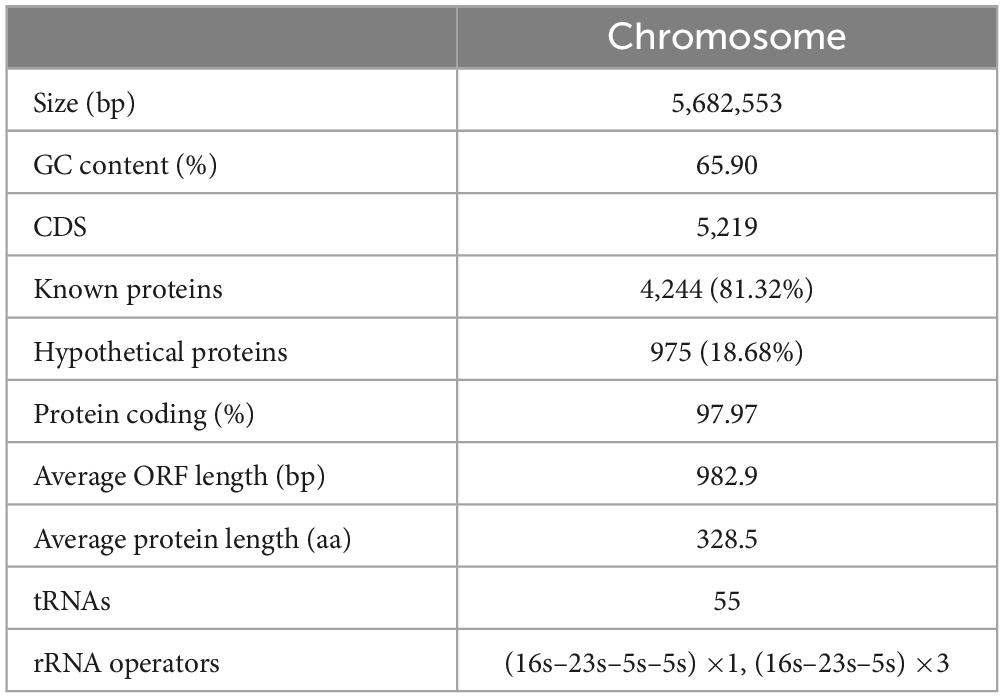
The A. mucicolens Y3 genome consists of a circular chromosome (plasmid-free). The chromosome is approximately 5.68 Mb in length, encoding 5,219 ORFs with an average GC content of 65.90% (Table 4). A search of the NCBI nucleotide database showed that there are 400 genome sequences of the genus Achromobacter, of which 14 belong to species A. mucicolens (in addition to A. mucicolens Y3 in this study). Among the 14 A. mucicolens genome sequences, five are from humans, eight are from the environment (including four from sinks, three from soils and one from sediment), and only one is from an animal (cattle). The genome sizes range from 5.6 to 6.4 Mb, with most between 5.9 and 6.3 Mb, and most of the genomes are larger than that of A. mucicolens Y3 (Supplementary Table 1).
In this study, antibiotic susceptibility testing showed that A. mucicolens Y3 exhibited high MIC levels for several β-lactam antibiotics, such as cephalothin (> 1,024 μg/mL), penicillin G (1,024 μg/mL), cefotaxime (> 256 μg/mL), cefazolin (128 μg/mL), amoxicillin (64 μg/mL), and aztreonam (64 μg/mL) (Table 2). A. mucicolens Y3 was susceptible to piperacillin (< 0.25 μg/mL), which is consistent with the susceptibility of other Achromobacter strains (Liu et al., 2016). Achromobacter strains have exhibited resistance to narrow-spectrum penicillins, first- and second-generation cephalosporins, ceftriaxone, cefotaxime, and aztreonam, whereas they may still be susceptible to ceftazidime, cefepime, piperacillin, and carbapenem (Isler et al., 2020). Multidrug efflux pumps and chromosomal OXA-114-like β-lactamases are recognized as the primary resistance mechanisms of Achromobacter species (Isler et al., 2020). Recently, with whole-genome sequencing, the molecular resistance mechanism of an A. mucicolens isolate was determined, wherein 208 drug resistance-related genes, including 25 putative β-lactamase genes, were predicted in the genome (Al-Asadi et al., 2022).
In the annotation results of the whole genome of A. mucicolens Y3, none of the predicted ORFs with ≥ 80% amino acid identity with the functionally characterized β-lactamase genes were identified. Instead, three predicted β-lactamase genes with identities between 50.0 and 30.0% with functionally characterized β-lactamase genes were present. They included a blaSRT–C2-like gene, a blaOXA–656-like gene, and a blaNmcR-like gene that shared amino acid identities of 50.0, 39.8, and 32.4% with SRT-C2 (AAQ90024), OXA-656 (WP_122630843), and NmcR (CAA79966), respectively. To determine whether any of them was a functional β-lactam antibiotic resistance gene, the gene with the highest identity (blaSRT–C2-like) was cloned first, and its resistance function was determined. It was interesting to find that this blaSRT–C2-like gene (designated blaAMZ–1 in this work) was indeed functional.
BlaAMZ–1 confers resistance to penicillins and cephalosporins
For analysis of the potential function of blaAMZ–1, the blaAMZ–1 gene with its promoter region was cloned, and a recombinant plasmid (pUCP24-blaAMZ–1) was transformed into E. coli DH5α. The transformant (pUCP24-blaAMZ–1/DH5α) exhibited resistance to some penicillins and first- and second-generation cephalosporins. Compared with the control strain (pUCP24/DH5α), the transformant exhibited an increase in the MICs of a variety of β-lactam antimicrobials, including amoxicillin, penicillin G, ampicillin, cephalothin and cefoxitin, the MICs of which increased 64-, 64-, 8-, > 4- and 4-fold, respectively. In addition, the MICs of some antimicrobials, such as aztreonam, cefazolin, and ceftriaxone, increased two-fold each for the transformant compared the control strain. When used in combination with the β-lactamase inhibitors tazobactam and avibactam, both piperacillin (piperacillin + tazobactam) and aztreonam (aztreonam + avibactam) exhibited a 4- and > 8-fold decrease in MIC. The results suggested that the novel β-lactamase resistance gene blaAMZ–1 may play a role in the β-lactam antibiotic resistance of A. mucicolens Y3.
As mentioned above, among all the functionally characterized β-lactamases, blaAMZ–1 showed 49.0% similarity (50.0% identity and 98.0% coverage) with blaSRT–C2. Similar to blaAMZ–1 and compared to the control, blaSRT–C2 increased MICs by more than four-fold for amoxicillin, cefoxitin and cephalothin (Mammeri et al., 2004). However, blaSRT–C2 had a wider antibiotic resistance spectrum for second- and third-generation cephalosporins, and it also conferred resistance to cefotaxime, ceftazidime and ceftriaxone and showed much higher MICs for some β-lactam antimicrobials than blaAMZ–1, such as cefotaxime (16 vs. 0.06 μg/mL) or ceftazidime (512 vs. < 0.015 μg/mL). BlaSRT–C1 showed much higher MICs for cefazolin (512 vs. 2 μg/mL) and piperacillin (32 vs. 1 μg/mL) than blaAMZ–1 (Raimondi et al., 2001). BlaLR13–1 showed the same MICs increase for ampicillin (eight-fold) and cefoxitin (four-fold) as blaAMZ–1 and a lower MIC for amoxicillin than blaAMZ–1 (8 vs. 64 μg/mL) (Matsumura et al., 1998). The reason for the difference in MICs may the structural variations between the gene sequences.
To further validate the enzyme’s function, the kinetic parameters of AMZ-1 β-lactamase were analyzed (Table 5). The purified AMZ-1 from the recombinant [pColdI-blaAMZ–1/BL21(DE3)] exhibited hydrolytic activities against some penicillins (penicillin G and amoxicillin) and some cephalosporins (cephalothin and cefoxitin). Cephalothin showed the highest Km value among all the tested substrates, while amoxicillin showed the lowest. However, the results of the enzyme kinetic hydrolytic activity test were not completely consistent with the MIC changes of the recombinant strain (pUCP24-blaAMZ–1/DH5α) in the antimicrobial susceptibility test (Table 2). Although the recombinant strain (pUCP24-blaAMZ–1/DH5α) exhibited an eight-fold increase in the MIC of ampicillin, the hydrolytic activity of AMZ-1 for this antimicrobial was not detectable. This result may be attributed to differences in activity between in vitro and in vivo conditions. β-Lactamase may be overproduced in vivo under antibiotic pressure and in in vitro experiments; despite several attempts, sufficient amounts of β-lactamase could not be enriched to observe the corresponding antibiotic hydrolysis (Pérez-Llarena et al., 2014). A similar situation has been reported in a previous study for CTX-M-37. Although the recombinant strain exhibited a four-fold increase in the MIC of ceftazidime, the hydrolytic activity of the β-lactamase could not be reliably determined with enzyme concentrations up to 253 nM in the reaction mixture (El Salabi et al., 2012). The turnover (kcat) values of penicillin G (3.93 s–1) and cephalothin (5.88 s–1) were approximately 50-fold higher than those of amoxicillin (9.47 × 10–2 s–1) and cefotaxime (8.05 × 10–2 s–1). Compared with SRT-C2, AMZ-1 showed stronger hydrolytic activities (kcat/Km) for cefoxitin (4.54 vs. 2 mM–1⋅s–1), while SRT-C2 showed higher hydrolysis activity than AMZ-1 for other antibiotics, such as cephalothin (48,000 vs. 10.4 mM–1⋅s–1), amoxicillin (10 vs. 6.78 mM–1⋅s–1) and penicillin G (5,000 vs. 36.3 mM–1⋅s–1) (Mammeri et al., 2004). In addition, SRT-C2 and AMZ-1 had the same kcat values for amoxicillin (0.01 s–1) and cefoxitin (0.01 s–1). The IC50 (50% inhibitory concentration) of β-lactamase inhibitors showed that the AmpC β-lactamase inhibitor avibactam had a conspicuous inhibitory effect on AMZ-1 (IC50: 819.67 × 10–6 μM); however, the class A β-lactamase inhibitor tazobactam showed a much weaker inhibitory effect on AMZ-1 (IC50: 1,094.97 μM) (Bush and Bradford, 2016).
Molecular characteristics and phylogenetic relationship of AMZ-1 to its relatives
The novel β-lactamase gene blaAMZ–1 is 1,191 bp in size and encodes a premature protein of 396 amino acids. The protein has a predicted molecular weight of 42.82 kDa and a predicted pI of 9.42. There is a signal peptide cleavage site between glycine and arginine at amino acid residues 26 and 27. The molecular weight of the mature protein without the signal peptide is 40.06 kDa.
To analyze the structural characteristics of the novel β-lactamase AMZ-1, 16 functionally characterized homologs (identity > 47.0%) were retrieved from the β-lactamase database. Alignment of the deduced amino acid sequence of AMZ-1 against the 16 β-lactamases revealed that four structural elements that are essential for the activity of the class C β-lactamases are conserved in AMZ-1 and the other β-lactamases. These include the characteristic catalytic residues of the serine active site of β-lactamases, S-I-S-K (serine-isoleucine-serine-lysine) at positions 89–92 and the typical class C β-lactamase motifs Y-A-N (tyrosine-alanine-asparagine) at positions 175–177, A-A-E-A (alanine-alanine-glutamic-alanine) at positions 243–246, and K-T-G (lysine-threonine-glycine) at positions 340–342 (Figure 1).
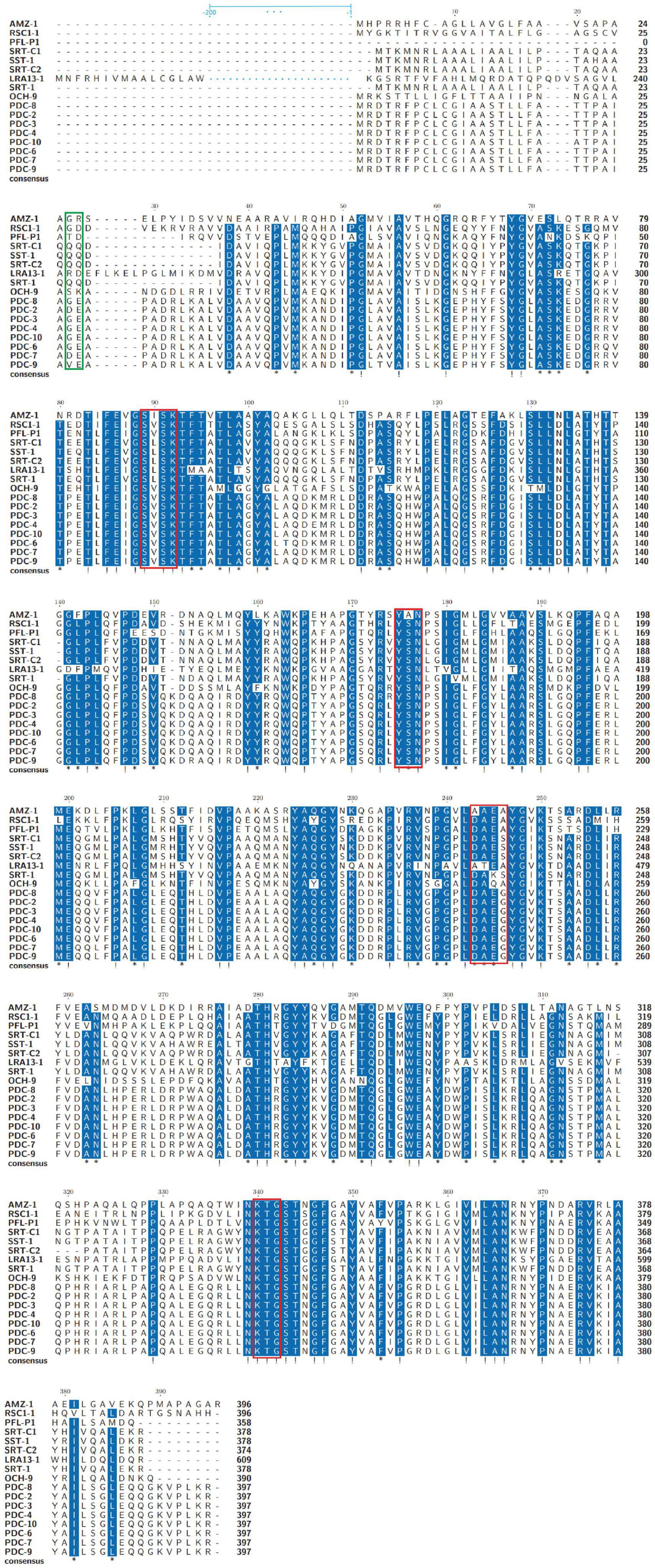
Figure 1. Multiple amino acid sequence alignment of AMZ-1 and other class C β-lactamases. Exclamations indicate fully conserved residues; asterisks indicate strongly similar residues; gaps are represented using hyphens. The green frame indicates the peptide cleavage site, and the red frames indicate four conserved motifs. Two hundred amino acids are omitted and are indicated by blue ellipses for LRA13-1.
Further phylogenetic analysis of AMZ-1 against the 16 functionally characterized β-lactamases revealed that AMZ-1 appeared as a new branch and was located relatively close to the SST-1 and SRT proteins (including SRT-1, SRT-C1 and SRT-C2) (Figure 2A). This result suggested that AMZ-1 constitutes a novel lineage of the β-lactamase family. Analysis of the distribution of the blaAMZ–1 homologous genes revealed that 31 functionally uncharacterized proteins with > 80% global amino acid sequence similarity with AMZ-1 were present in the NCBI database, and they were all from the genus Achromobacter. Apart from 13 sequences without specific species classification, most sequences were from A. mucicolens (7 sequences). The closest relative to AMZ-1 was a putative class C β-lactamase (WP_175155855) from A. mucicolens sharing an amino acid sequence similarity of 99.49% (99.49% identity and 100% coverage). The phylogenetic analysis also confirmed that this protein shared the closest evolutionary relationship with AMZ-1 (Figure 2B).
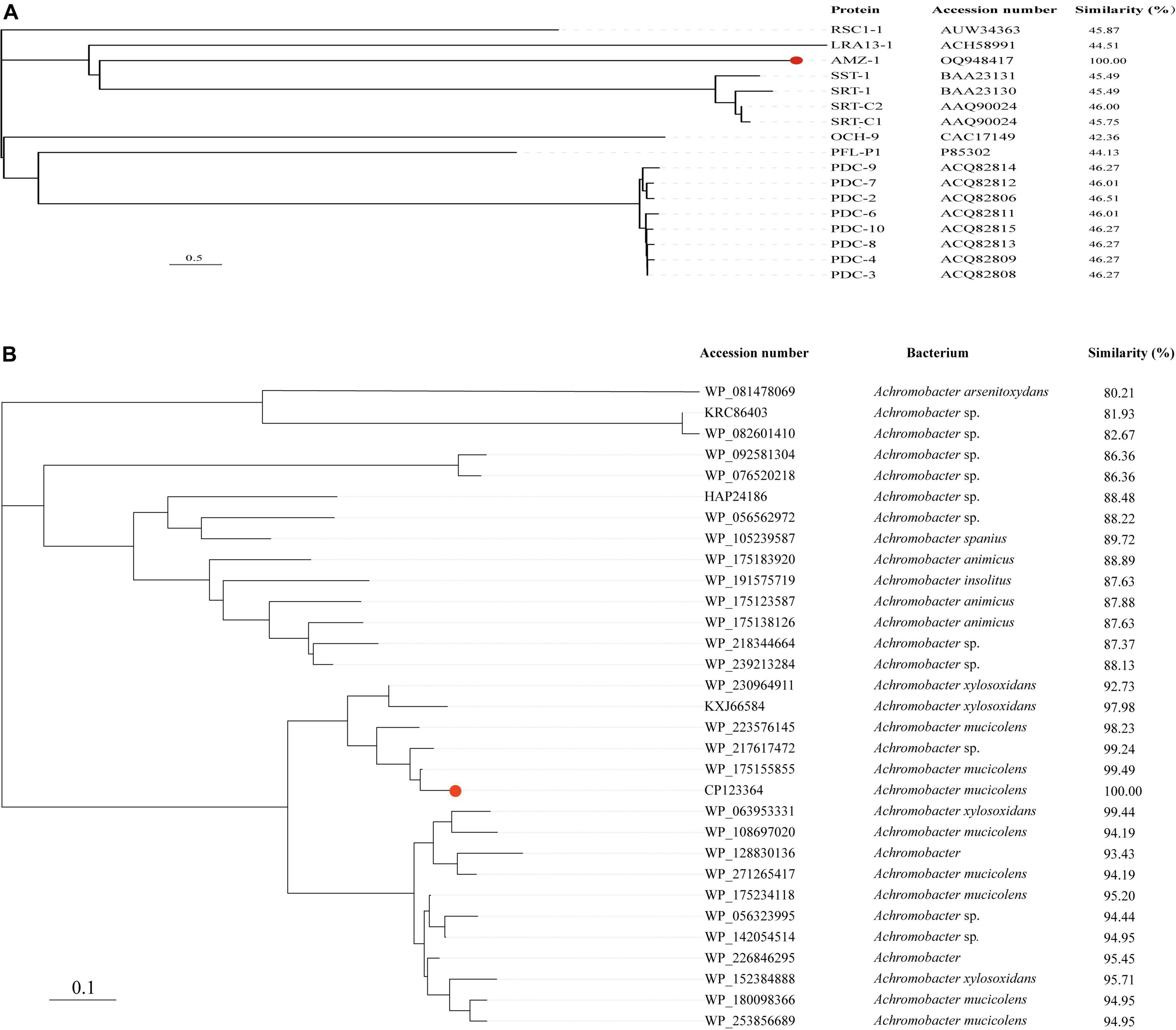
Figure 2. A phylogenetic tree showing the relationship of AMZ-1 to other functionally characterized AmpC β-lactamases (A) and to other putative β-lactamases (> 80% similarity) (B). AMZ-1 is highlighted in red.
Genetic environments of the blaAMZ–1 and blaAMZ–1-like genes
To analyze the genetic environments of blaAMZ–1 and its relatives, the sequences containing a blaAMZ–1-like gene at the center that shared ≥ 85.0% amino acid identity and ≥ 95% coverage with blaAMZ–1 were retrieved from the NCBI nucleotide database. Finally, a total of five sequences approximately 20 kb in length with blaAMZ–1-like genes at the centers were retained for further analysis. All five sequences were from the genus Achromobacter, including one (A. mucicolens IA) from the same species as the one characterized in this work, A. mucicolens. The other four included A. xylosoxidans (A. xylosoxidans DN002), A. spanius (A. spanius MYB73), and 2 strains with unclassified species (Achromobacter sp. 77 and Achromobacter sp. MFA1 R4) (Figure 3). No mobile genetic elements were predicted in the regions surrounding these sequences. Comparative genomic analysis of the 6 sequences (including the sequence characterized in this work) revealed that the structure of one sequence from the same species (A. mucicolens IA) was most similar to the structure of the sequence in this work, with 100% coverage and 97.39% identity. Among the other four sequences, three shared similar downstream regions with the sequence characterized in this work; however, the downstream region of the last sequence was completely different from the others. In the upstream regions, the four sequences showed similar structures but were different from the other two (the sequence characterized in this work and A. mucicolens IA), and they had 4 to 5 extra ORFs inserted adjacent to the blaAMZ–1-like genes.
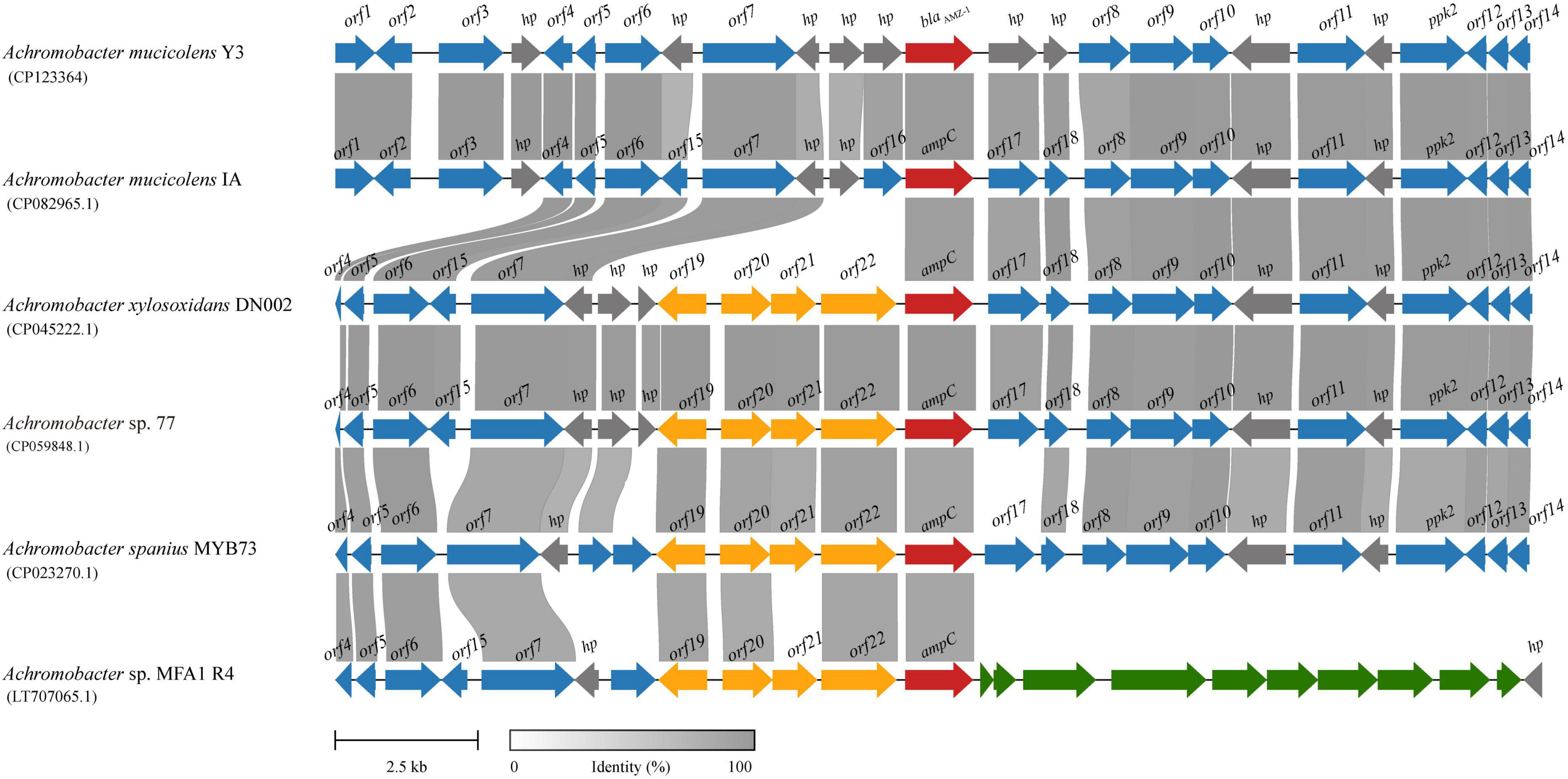
Figure 3. Genetic environment of the blaAMZ–1 and blaAMZ–1-like genes. The regions with ≥ 80% amino acid identity are colored gray. AmpC β-lactamases are colored red and the other genes are colored based on gene function. hp, hypothetical protein; ppk2, polyphosphate kinase 2; other orfs without direct gene names are listed in Supplementary Table 2.
Conclusion
In this work, a novel β-lactamase gene designated blaAMZ–1 encoded in the chromosome of A. mucicolens Y3 isolated from a goose was characterized. AMZ-1 shared the highest amino acid identity of 50.0% with the functionally characterized AmpC enzyme SRT-C2. The cloned blaAMZ–1 gene showed resistance against some β-lactam antibiotics, including amoxicillin and cephalothin, and the β-lactamase AMZ-1 exhibited a similar substrate spectrum response. Similar to other AmpC β-lactamases, AMZ-1 is strongly inhibited by avibactam but much less strongly inhibited by tazobactam. Closer homologs of blaAMZ–1 tend to appear in the different species of the genus Achromobacter. The identification and characterization of the novel resistance gene will help in the selection of effective antimicrobials to treat infections caused by opportunistic pathogens that carry the novel resistance gene.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in this article/Supplementary material.
Ethics statement
This study used strains obtained from an anal swab of a goose on an animal farm in Wenzhou. The owner of the farm was informed of the study and expressed approval for the sampling of animals. All experimental procedures involving animals were approved by the Animal Welfare and Ethics Committee of Wenzhou Medical University, Zhejiang Province, China.
Author contributions
HZ, KL, QB, and DL: conceived and designed the experiments. YZ, JZ, GZ, NL, YS, JL, and TZ: performed the experiments. YZ, XZ, QL, and XL: data analysis and interpretation. YZ, QB, and DL: drafting of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Science and Technology Project of Wenzhou City, China (N20210001), the Science and Technology Project of Jinhua City, China (2022-2-013 and 2022-4-017), and the Zhejiang Provincial Natural Science Foundation of China (LY19C060002 and LQ17H190001).
Acknowledgments
The authors would like to acknowledge all study participants and individuals who contributed to this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1252427/full#supplementary-material
Footnotes
- ^ http://arpcard.mcmaster.ca
- ^ https://blast.ncbi.nlm.nih.gov
- ^ https://www.ncbi.nlm.nih.gov/
- ^ https://gctype.wdcm.org/
- ^ http://www.bldb.eu/
- ^ https://www.uniprot.org/
References
Ahmed, M. S., Nistal, C., Jayan, R., Kuduvalli, M., and Anijeet, H. K. I. (2009). Achromobacter xylosoxidans, an emerging pathogen in catheter-related infection in dialysis population causing prosthetic valve endocarditis: A case report and review of literature. Clin. Nephrol. 71, 350–354. doi: 10.5414/cnp71350
Al-Asadi, S. A., Al-Kahachi, R. E. S., Alwattar, W. M. A., Bootwala, J., and Sabbah, M. A. (2022). Genomic insights into Achromobacter mucicolens IA antibiotic resistance. Microbiol. Spectr. 10, e0191621. doi: 10.1128/spectrum.01916-21
Ambler, R. P. (1980). The structure of beta-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 289, 321–331. doi: 10.1098/rstb.1980.0049
Bodenhofer, U., Bonatesta, E., Horejš-Kainrath, C., and Hochreiter, S. (2015). MSA: An R package for multiple sequence alignment. Bioinformatics 31, 3997–3999. doi: 10.1093/bioinformatics/btv494
Buchfink, B., Reuter, K., and Drost, H.-G. (2021). Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368. doi: 10.1038/s41592-021-01101-x
Bush, K., and Bradford, P. A. (2016). β-Lactams and β-Lactamase inhibitors: An overview. Cold Spring Harb. Perspect. Med. 6, a025247. doi: 10.1101/cshperspect.a025247
Bush, K., Macalintal, C., Rasmussen, B. A., Lee, V. J., and Yang, Y. (1993). Kinetic interactions of tazobactam with beta-lactamases from all major structural classes. Antimicrob. Agents Chemother. 37, 851–858.
CLSI (2021). Performance standards for antimicrobial susceptibility testing, M100, 31st Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Doi, Y., Poirel, L., Paterson, D. L., and Nordmann, P. (2008). Characterization of a naturally occurring class D β-Lactamase from Achromobacter xylosoxidans. Antimicrob. Agents Chemother. 52, 1952–1956. doi: 10.1128/AAC.01463-07
El Salabi, A., Borra, P. S., Toleman, M. A., Samuelsen, Ø, and Walsh, T. R. (2012). Genetic and biochemical characterization of a novel metallo-β-lactamase, TMB-1, from an Achromobacter xylosoxidans strain isolated in Tripoli, Libya. Antimicrob. Agents Chemother. 56, 2241–2245. doi: 10.1128/AAC.05640-11
Gilchrist, C. L. M., and Chooi, Y.-H. (2021). clinker and clustermap.js: Automatic generation of gene cluster comparison figures. Bioinformatics 37, 2473–2475. doi: 10.1093/bioinformatics/btab007
Isler, B., Kidd, T. J., Stewart, A. G., Harris, P., and Paterson, D. L. (2020). Achromobacter infections and treatment options. Antimicrob. Agents Chemother. 64, e1025–e1020. doi: 10.1128/AAC.01025-20
Jacoby, G. A. (2009). AmpC β-Lactamases. Clin. Microbiol. Rev. 22, 161–182. doi: 10.1128/CMR.00036-08
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114. doi: 10.1038/s41467-018-07641-9
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Konstantinidis, K. T., and Tiedje, J. M. (2005). Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 102, 2567–2572. doi: 10.1073/pnas.0409727102
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Letunic, I., and Bork, P. (2007). Interactive tree of life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. doi: 10.1093/bioinformatics/btl529
Li, X.-Z., Mehrotra, M., Ghimire, S., and Adewoye, L. (2007). β-Lactam resistance and β-lactamases in bacteria of animal origin. Vet. Microbiol. 121, 197–214. doi: 10.1016/j.vetmic.2007.01.015
Liu, C., Pan, F., Guo, J., Yan, W., Jin, Y., Liu, C., et al. (2016). Hospital acquired pneumonia due to Achromobacter spp. in a geriatric ward in China: Clinical characteristic, genome variability, biofilm production, antibiotic resistance and integron in isolated strains. Front. Microbiol. 7:621. doi: 10.3389/fmicb.2016.00621
Mammeri, H., Poirel, L., Bemer, P., Drugeon, H., and Nordmann, P. (2004). Resistance to cefepime and cefpirome due to a 4-amino-acid deletion in the chromosome-encoded AmpC beta-lactamase of a Serratia marcescens clinical isolate. Antimicrob. Agents Chemother. 48, 716–720. doi: 10.1128/AAC.48.3.716-720.2004
Matsumura, N., Minami, S., and Mitsuhashi, S. (1998). Sequences of homologous beta-lactamases from clinical isolates of Serratia marcescens with different substrate specificities. Antimicrob. Agents Chemother. 42, 176–179. doi: 10.1128/AAC.42.1.176
McArthur, A. G., Waglechner, N., Nizam, F., Yan, A., Azad, M. A., Baylay, A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357. doi: 10.1128/AAC.00419-13
Papalia, M., Traglia, G., Ruggiero, M., Almuzara, M., Vay, C., Gutkind, G., et al. (2018). Characterisation of OXA-258 enzymes and AxyABM efflux pump in Achromobacter ruhlandii. J. Glob. Antimicrob. Resist. 14, 233–237. doi: 10.1016/j.jgar.2018.03.015
Pérez-Llarena, F. J., Zamorano, L., Kerff, F., Beceiro, A., García, P., Miró, E., et al. (2014). Genetic and kinetic characterization of the novel AmpC β-Lactamases DHA-6 and DHA-7. Antimicrobial Agents and Chemotherapy 58, 6544. doi: 10.1128/AAC.03144-14
Raimondi, A., Sisto, F., and Nikaido, H. (2001). Mutation in Serratia marcescens AmpC beta-lactamase producing high-level resistance to ceftazidime and cefpirome. Antimicrob. Agents Chemother. 45, 2331–2339. doi: 10.1128/AAC.45.8.2331-2339.2001
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Seemann, T. (2014). Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shi, W., Sun, Q., Fan, G., Hideaki, S., Moriya, O., Itoh, T., et al. (2021). gcType: A high-quality type strain genome database for microbial phylogenetic and functional research. Nucleic Acids Res. 49, D694–D705. doi: 10.1093/nar/gkaa957
Swenson, C. E., and Sadikot, R. T. (2015). Achromobacter respiratory infections. Ann. Am. Thorac. Soc. 12, 252–258. doi: 10.1513/AnnalsATS.201406-288FR
Tamma, P. D., Doi, Y., Bonomo, R. A., Johnson, J. K., and Simner, P. J. (2019). A primer on AmpC β-Lactamases: Necessary knowledge for an increasingly multidrug-resistant world. Clin. Infect. Dis. 69, 1446–1455. doi: 10.1093/cid/ciz173
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Keywords: blaAMZ–1, AmpC, Achromobacter mucicolens, resistance gene, kinetic analysis
Citation: Zhang Y, Zhao J, Zhang G, Lin N, Sha Y, Lu J, Zhu T, Zhang X, Li Q, Zhang H, Lin X, Li K, Bao Q and Li D (2023) Identification and characterization of a novel β-lactamase gene, blaAMZ–1, from Achromobacter mucicolens. Front. Microbiol. 14:1252427. doi: 10.3389/fmicb.2023.1252427
Received: 03 July 2023; Accepted: 04 September 2023;
Published: 21 September 2023.
Edited by:
Longzhu Cui, Jichi Medical University, JapanReviewed by:
Asad Mustafa Karim, Tsinghua University, ChinaJozsef Soki, University of Szeged, Hungary
Copyright © 2023 Zhang, Zhao, Zhang, Lin, Sha, Lu, Zhu, Zhang, Li, Zhang, Lin, Li, Bao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiyu Bao, YmFvcXlAZ2Vub21pY3MuY24=; Dong Li, bGlkb25nQHdtdS5lZHUuY24=
 Yuan Zhang
Yuan Zhang Jingxuan Zhao
Jingxuan Zhao Guozhi Zhang
Guozhi Zhang Naru Lin
Naru Lin Yuning Sha
Yuning Sha Junwan Lu
Junwan Lu Tingting Zhu
Tingting Zhu Xueya Zhang
Xueya Zhang Qiaoling Li
Qiaoling Li Hailin Zhang
Hailin Zhang Xi Lin
Xi Lin Kewei Li
Kewei Li Qiyu Bao
Qiyu Bao Dong Li1,2*
Dong Li1,2*