
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 09 October 2023
Sec. Microbe and Virus Interactions with Plants
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1252039
This article is part of the Research Topic Microbiome Associated With Plant Pathogens, Pathogenesis, and Their Applications in Developing Sustainable Agriculture View all 17 articles
 Diao Zhou1,2,3
Diao Zhou1,2,3 Xingzhou Chen1,2,3
Xingzhou Chen1,2,3 Xinggang Chen1,2,3
Xinggang Chen1,2,3 Yandong Xia1,2,3
Yandong Xia1,2,3 Junang Liu1,2,3*
Junang Liu1,2,3* Guoying Zhou1,2,3*
Guoying Zhou1,2,3*Phytopathogens pose a devastating threat to the productivity and yield of crops by causing destructive plant diseases in natural and agricultural environments. Hemibiotrophic pathogens have a variable-length biotrophic phase before turning to necrosis and are among the most invasive plant pathogens. Plant resistance to hemibiotrophic pathogens relies mainly on the activation of innate immune responses. These responses are typically initiated after the plant plasma membrane and various plant immune receptors detect immunogenic signals associated with pathogen infection. Hemibiotrophic pathogens evade pathogen-triggered immunity by masking themselves in an arms race while also enhancing or manipulating other receptors to promote virulence. However, our understanding of plant immune defenses against hemibiotrophic pathogens is highly limited due to the intricate infection mechanisms. In this review, we summarize the strategies that different hemibiotrophic pathogens interact with host immune receptors to activate plant immunity. We also discuss the significant role of the plasma membrane in plant immune responses, as well as the current obstacles and potential future research directions in this field. This will enable a more comprehensive understanding of the pathogenicity of hemibiotrophic pathogens and how distinct plant immune receptors oppose them, delivering valuable data for the prevention and management of plant diseases.
Plant diseases have been a devastating threat throughout the history of agriculture. In addition to causing significant losses in global crop yields, plant diseases present major challenges to natural and agricultural systems. Phytopathogens cause devastating plant diseases by deploying infection strategies (Fisher et al., 2018). They can be classified into three main groups based on their infection strategies to extract plant nutrients: biotrophs, hemibiotrophs, and necrotrophs. Biotrophs extract nutrients from living cells and sustain host viability, whereas necrotrophs rapidly kill host cells to extract nutrients (Rajarammohan, 2021). Intermediate lifestyle hemibiotrophs begin in the biotrophic phase and subsequently transition to the necrotrophic phase (Damm et al., 2014). Hemibiotrophic pathogens (HPs) are prevalent and highly destructive phytopathogens that cause significant losses in crop quality and yield in key agricultural crops. The duration of each phase in HPs varies depending on factors such as the pathogen, host plant, temperature, secreted protein effectors, etc. (Qiu et al., 2022). For example, Phytophthora infestans has a shorter biotrophic phase than Magnaporthe oryzae, which has a shorter necrotrophic phase. Studies have also shown that hemibiotrophs utilize distinct effectors to adapt to various biotrophic/necrotic patterns. AVR3a stabilizes and targets the plant E3 ligase CMPG1 during the early stages of biotrophic infection by P. infestans to manipulate host immunity. AVR3a is subsequently downregulated, while the induction of other effectors INF1 and nep1-like proteins may facilitate the host transition to necrotrophic infection (Yaeno et al., 2011; Pirc et al., 2021). Although the term hemibiotroph was developed for pathogenic fungi, it is also sometimes used to describe the lifestyle of bacteria, oomycetes (Kraepiel and Barny, 2016; Panthapulakkal Narayanan et al., 2020). Different HPs differ in their pathogenic strategies, target hosts, etc. To induce disease in plants, HPs deploy various virulence factors to promote infection under defined environmental conditions.
Plants have evolved a sophisticated surveillance system to protect themselves from HPs. It devotes resources and energy to growth and development without threat. However, when threatened by virulence factors secreted by phytopathogens, including toxins, phytohormones, and enzymes, plants rapidly regulate gene expression to protect the host from pathogens (Jones et al., 2016). The plant surveillance system mainly relies on two classes of immune receptors to detect pathogens: membrane-anchored pattern recognition receptors (PRRs) and intracellular nucleotide-binding and leucine-rich repeat receptors (NLRs; Dangl, 2013; Van de Weyer et al., 2019). In the classical zig-zag pattern, these two classes of receptor proteins correspond to the two layers of the plant immune system (Ngou et al., 2022). In the first layer of immune surveillance, PRRs recognize pathogen- or microbe-associated molecules (PAMPs or MAMPs, respectively) present in the extracellular space, which results in PAMP-triggered immunity (PTI) or MAMP-triggered immunity (MTI; Macho and Zipfel, 2014; Wang and Chai, 2020). However, many phytopathogens can manipulate host targets to inhibit PTI signaling and successfully deliver effectors to plant cells. In the second layer of host immune surveillance, intracellular NLRs activate effector-triggered immunity (ETI) by specifically recognizing intracellular pathogen effectors (Chen et al., 2012; Kourelis and van der Hoorn, 2018). The distinction between PTI and ETI is an excellent framework for explaining the plant immune system. ETIs enhance PTI-induced defense responses by altering the expression of key genes involved in PRR signaling elements transcription and translation. Conversely, PTI also enhances ETI-triggered defense responses. PTI and ETI work together to provide robust immunity to pathogens (Ngou et al., 2021; Yuan et al., 2021b). Over the past few decades, researchers have made significant progress in studying plant immune signaling controlled by PRRs and NLRs against necrotrophic and biotrophic pathogens. In this review, we summarize the strategies by which various HPs interact with host immune receptors to activate plant immunity (Figure 1). We focus on how various immune receptors perceive characteristic molecules from different HPs. We also discuss the commonalities and differences in the pathogenicity of different HPs. This may be crucial in explaining the potential threat of pathogens attack on the host for effective defense, it can also guide the improvement and breeding of genetically diseased crops.
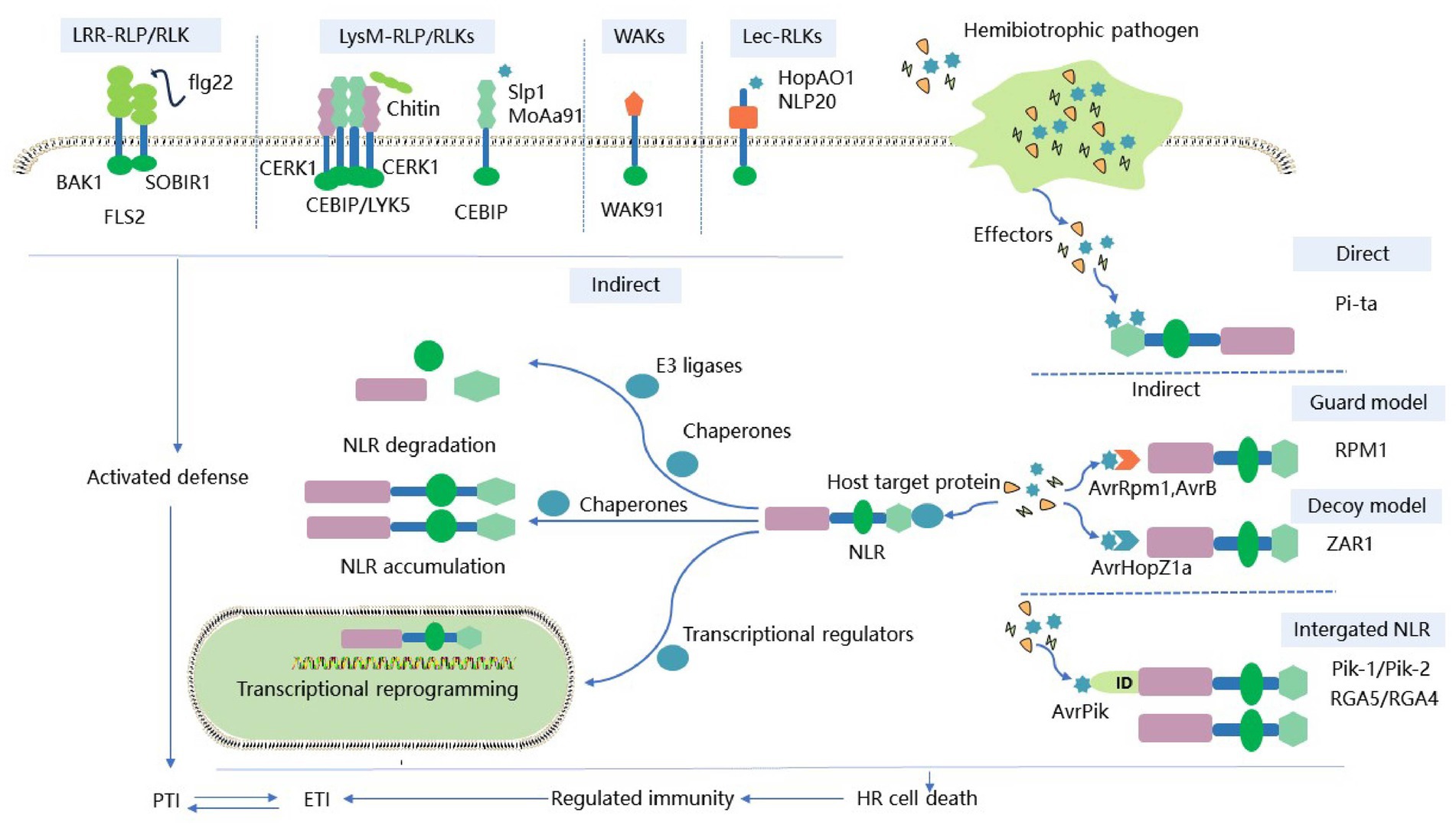
Figure 1. Schematic diagram of the two-layer immune system of plant immune receptors against HPs. Plant PRRs recognize PAMPs (e.g., bacterial flagellin, fungal chitin) or DAMPs (e.g., secretory minipeptides) to induce PTI. Adaptive HPs successfully translocate effectors into plant cells, thereby initiating a second round of plant immunity. Intracellular NLR immune receptors induce ETIs to trigger immune-related gene expression and local cell death through direct or indirect specific recognition of effector proteins. Activated PTIs enhance the defense response triggered by ETIs. While ETIs upregulate related genes that control signaling to PRRs, PTIs and ETIs together provide plants with strong immunity against HPs. ID, Integrated domain; PTI, PAMP-triggered immunity. ETI, Effector-triggered immunity; PRR, Pattern recognition receptor; HP, Hemibiotrophic pathogen; FLS2, Flagellin-sensitive 2; EFR, Elongation factor-Tu receptor; CERK1, Elicitor receptor kinase 1; CEBiP, Chitin elicitor binding protein; WAK, Wall-associated kinase; LRR, Leucine-rich repeat domain; LysM, Lysin motif domains; Lec, Lectin domain.
The plasma membrane (PM) serves as the frontline of defense against pathogens in plants and is essential for pathogen detection, signal transduction, and cellular homeostasis maintenance. Several PRRs are present in plant PMs that detect PAMPs, DAMPs, or effector proteins that induces PTI response. In the zig-zag immune model, the PTI response prevents most HPs from invasion and reproduction. Adapted pathogens secrete large amounts of effectors to evade or inhibit PTI. Correspondingly, plants have also evolved intracellular NLR receptors that directly or indirectly recognize effectors and trigger a robust ETI response. This ultimately leads to localized plant cell death (Ngou et al., 2022). The co-resistance of PTI and ETI determines many plant defense responses to pathogen infection, such as protein phosphorylation, changes in ion flux, generation of reactive oxygen species (ROS), activation of mitogen-activated protein kinases (MAPK), and pathogenesis associated with cell wall strengthening (Tsuda and Katagiri, 2010). The activation of the MAPK signaling pathway by PRR at the PM results in the phosphorylation of target proteins in plant immunity. In Arabidopsis, the HP effector protein AvrRpt2 specifically inhibits the phosphorylation of MPK4 and MPK11 induced by the PM-localized receptor FLAGELLIN-SENSITIVE 2 (FLS2; Tsuda et al., 2013; Eschen-Lippold et al., 2016). It has also been shown that MPK3 and MPK6 can regulate malate metabolism to promote PM-mediated stomatal immunity during pathogen infection (Su et al., 2017).
Although HPs primarily activate the PM-anchored protein PRR, plants can also utilize lipids on the PM to sense HPs. These lipids trigger immune signals independent of PRR interactions. For example, the effector NLP of HPs is sensed by glycosphingolipids (GIPCs) on the PM, and it is speculated that a conformational changes in the GIPC-NLP complex induces pore formation in the PM and thus cell death (Mamode Cassim et al., 2018). In the basic defense process, recognition of PAMP or DAMP by the PRR of the PM induces cell wall modification that activates endocytosis of PRR and PAMP/DAMP, followed by degradation in the vacuole. This process initiates and amplifies immune signaling (Mbengue et al., 2016). For example, Arabidopsis RLCK BIK1, BSK1 interacts with FLS2 and is rapidly phosphorylated in an FLS2-dependent manner upon recognition of the bacterial flagellin peptide flg22. FLS2’s sustained anchoring to the PM is mediated via the BFA-sensitive endocytotic pathway under steady-state conditions (Beck et al., 2012). These findings contribute to a comprehensive understanding of the vital role of plant immunity in HP interference. Collectively, plant PM regulates immune responses by detecting HPs, activating signaling cascades, controlling the cellular entry and exit of molecules, and enabling PM endocytosis.
Pattern recognition receptors (PRRs) play an important role in plant growth, development, reproduction, abiotic stress, and disease resistance, and many of them exhibit lineage-specific expansion to adapt to different pathogens within the innate immune system (Schellenberger et al., 2019; Ngou et al., 2022). Several plant PRRs have already been identified, such as FLS2, ELONGATION FACTOR-Tu RECEPTOR (EFR), ELICITOR RECEPTOR KINASE 1 (CERK1), and CHITIN ELICITOR BINDING PROTEIN (CEBiP; Tang et al., 2017). They recognize the bacterial flagellin epitope flg22, the EF-Tu epitope elf18, the plant elicitor polypeptide, and chitin released during pathogen infection, respectively (Chen et al., 2020). These PRRs form complexes with their corresponding ligands. These complexes activate downstream immune signals such as calcium influx, ROS production, MAPK signaling cascade, and defense responses (Couto and Zipfel, 2016; Zhou and Zhang, 2020). Based on the presence or absence of intracellular kinase domains, the PRR family is classified into receptor-like kinases (RLKs) and receptor-like proteins (RLPs), which act on the first layer of the plant immune system. RLKs consist of a variable N-terminal extracellular domain (ECD), a transmembrane region (TM), and a conserved cytoplasmic kinase domain (KD; Wang and Chai, 2020); In contrast, RLPs have only short domains and lack distinct kinase domains that require interaction with other kinase domain-containing proteins such as BAK1 and SOBIR1 to activate downstream signaling (Gust and Felix, 2014; Liebrand et al., 2014). RLKs and RLPs are classified into several subfamilies based on their ECDs, which include leucine-rich repeat (LRR) domains, lysin motif (LysM) domains, lectin (Lec) domains, and epidermal growth factor-like (EGF) repeat domains (Macho and Zipfel, 2014). Various subfamilies of PRRs exhibit commonalities and differences in detecting diverse HPs (Table 1; Figure 1).
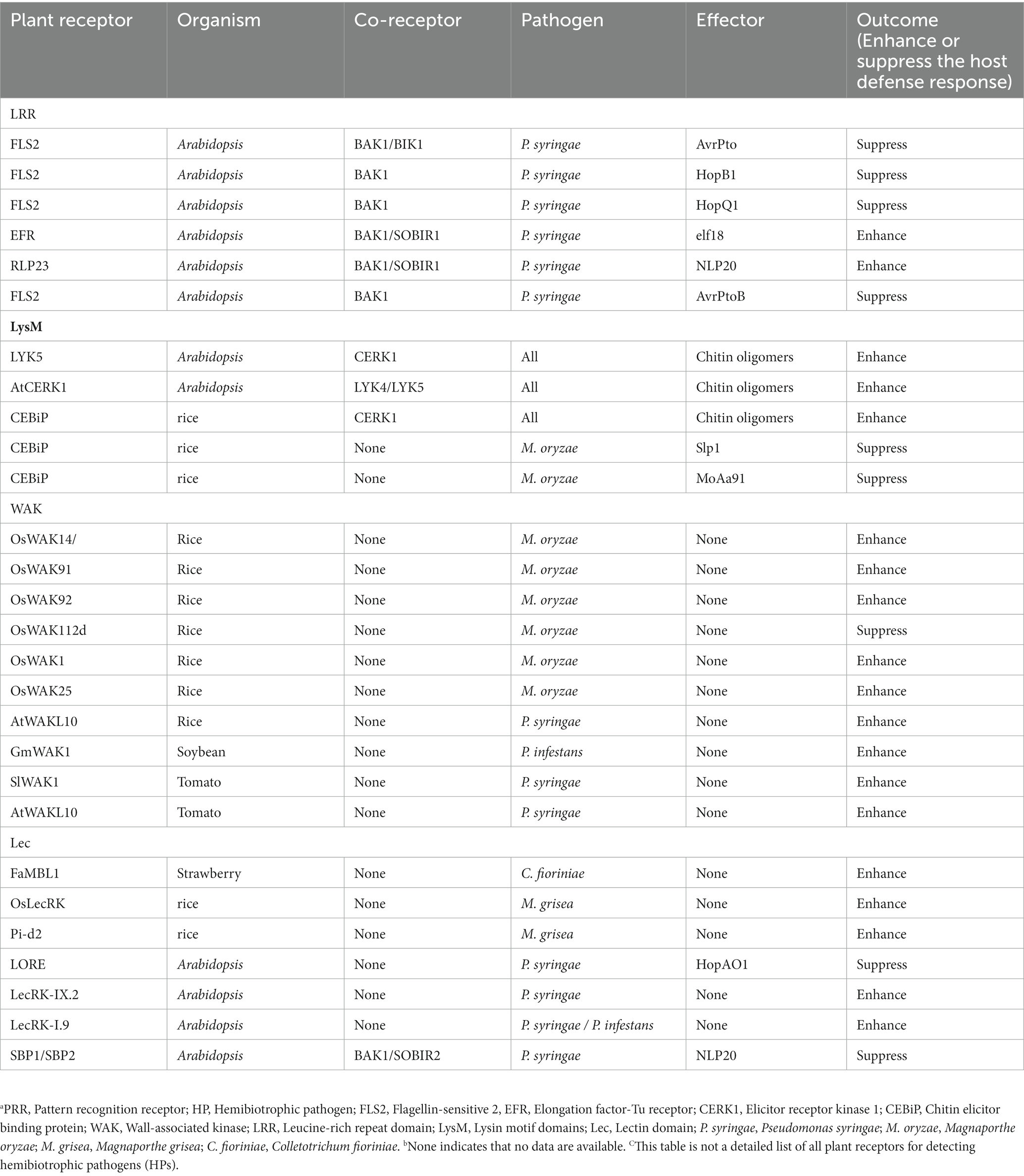
Table 1. Overview of PRRs and the outcome of interactions with typical HPs to mediate plant immunity.
Leucine-rich repeat-containing PRRs are the largest subfamily, including LRR-RLP and LRR-RLK family members. LRR-RLP/RLKs detect HPs by interacting with shorter ECD co-receptors of the same family to enhance immune signaling (Smakowska-Luzan et al., 2018). BRASSINOSTEROID INSENSITIVE 1 (BRI1)-ASSOCIATED RECEPTOR KINASE 1 (BAK1) is one of the most versatile co-receptors, also known as SOMATIC EMBRYOGENESIS RECEPTOR KINASES 3 (SERK3). It has only five LRRs in its ECD, which are centrally involved in various PTI signaling pathways (Wu et al., 2020). When hemibiotrophic bacteria interact with plants, a well-studied PRR is the LRR-RK FLS2 in most higher plants, which detects a 22-amino acid peptide derived from the N-terminus of bacterial flg22 (Lee et al., 2021). Recognition of flg22 by FLS2 and its co-receptor BAK1 is accompanied by rapid heterodimerization and phosphorylation, which activate plant immunity. To prevent the host immune responses, P. syringae secrete effectors to interfere with immune signals, such as AvrPto, AvrPtoB, HopB1, etc. AvrPto and AvrPtoB interact with FLS2 to prevent the formation of the FLS2-BAK1 complex and the phosphorylation of BIK1 (Gravino et al., 2017; Lei et al., 2020). HopB1 constitutively interacts with FLS2 prior to the activation of flg22. Upon activation, BAK1 is recruited to the FLS2-HopB1 complex. HopB1 cleaves BAK1 and its analogs via genetic transformation or bacterial delivery to inhibit FLS2 signaling and enhance pathogen virulence (Li et al., 2016; Wu et al., 2020). Like FLS2, another extensively studied LRR is the Arabidopsis EFR, which recognizes the N-terminal N-acetylated elongation factor Tu (EF-Tu) peptide of hemibiotrophic bacteria. Upon ligand binding, BAK1 is also recruited by EFR to participate in host immune signaling. Resistance to the hemibiotrophic bacteria P. syringae is activated by recognition of elf18C by EFR-Cf-9 in conjunction with SOBIR1 and BAK (Wu et al., 2019). In addition, rice LRR-RK XA21 can sense tyrosine sulfonate proteins derived from Xanthomonas oryzae to induce effective resistance to Xoo (Wei et al., 2016). These results suggest that under hemibiotrophic bacterial attack, plant LRR-RLKs participate in immune defense and self-development by phosphorylation upon binding to the corresponding co-receptors. Plant RLKs such as FLS2, EFR, and XA21 all belong to the LRR-XII subfamily, which suggests that RLKs of this subfamily may induce immunity by recognizing various protein ligands of hemibiotrophic bacteria. Similar to RLKs, BAK1 is also recruited to the two-component RLP or SOBIR1 complex by ligand recognition by RLP. BAK1 and SOBIR1 can transphosphorylate each other to activate immune signaling pathways. The Arabidopsis RLP23 forms a receptor complex by interacting with SOBIR1. Upon recognizing NLP20, it recruits the co-receptor BAK1 to the complex to enhance the LRR-mediated plant immune response to HPs (Albert et al., 2015; van der Burgh et al., 2019). Several effectors of hemibiotrophic bacteria, fungi, and oomycetes can prevent host LRR-RLP resistance to pathogens by inhibiting NLP-induced cell death, such as suppressor of necrosis 1 (SNE1) from P. infestans (Kelley et al., 2010), Colletotrichum higginsianum effector candidate (ChECs) from C.higginsianum (Kleemann et al., 2012), and MoNLP 1 (Chen et al., 2021), M. oryzae hypothetical effector gene 13 (MoHEG13; Mogga et al., 2016), and suppressors of plant cell death (SPDs) from M. oryzae (Sharpee et al., 2017). Regardless of whether hemibiotrophic bacteria, fungi, or oomycetes infect plants, although the immune response outcomes are different, FLS, EFR, and RLP23 all rely on kinase domain-containing proteins such as BAK1 or SOBIR1 receptor complexes for immune signaling. Different receptor complexes may elicit diverse immune responses by influencing intracellular structural domain phosphorylation and downstream immune signaling. Further studies of the phosphorylation properties of the different receptor complexes may reveal how these receptors are effective against localized HP infection.
During the game between HPs and plants, plant LysM ectodomains induce immune responses by recognizing N-acetylglucosamine molecules, such as fungal chitin oligomers and bacterial peptidoglycan (PGN; McCombe et al., 2022). The LysM family consists of LysM-RLK and LysM-RLP. LysM-RLKs have a LysM outer domain, a single channel transmembrane domain and a cytoplasmic kinase domain. LysM-RLP has only one outer domain connected to the outer membrane by GPI anchors (Buendia et al., 2018). CERK1 is a typical member of the LysM-RLK family consisting of three LysM structural domains. In rice and Arabidopsis, autophosphorylation of specific amino acids in CERK1 regulates chitin-induced immune signals secreted by HPs (Fliegmann et al., 2011; Suzuki et al., 2016; Yu et al., 2021). CERK1 not only recognizes hemibiotrophic fungal chitin, but is also a crucial co-receptor for bacterial PGN. CERK1, together with lysin motif receptor kinase 1 (LYM1) and LYM3, is required for the perception of bacterial PGN and for the development of basal resistance to P. syringae. Studies have also shown that deletion of CERK1 increases susceptibility to hemibiotrophic fungi and bacteria (Giovannoni et al., 2021). In Arabidopsis, LYK5 exhibits a greater affinity for chitin than CERK1. Once LYK5 externally detects chitin, CERK1 forms a dimer and transmits chitin signals to LYK5. Afterward, CERK1 phosphorylates LYK5 and itself in vesicles, which are then internalized (Erwig et al., 2017). LYK4 and AtCERK1 can form a complete complex with LYK5. LYK4 serves as a scaffolding protein or a co-receptor for LYK5, whereas AtCERK1 senses chitin and mediates homodimerization and phosphorylation, all of which promotes chitin triggering signaling (Wan et al., 2012; Cao et al., 2014; Xue et al., 2019). Unlike the chitin-sensing mechanism in Arabidopsis, the rice LysM-RLP CEBiP recognizes chitin and forms a dimer with OsCERK1 to activate plant disease resistance to HP and initiates downstream immune signaling pathways (Hayafune et al., 2014; Desaki et al., 2018; Hu et al., 2021). The results suggest that dimerization of the LysM receptor plays a vital role in ligand detecting, receptor activation, and immune signal transduction during HP infection. Aggregated PGN from hemibiotrophic bacteria is recognized as PAMP by the LysM receptor in Arabidopsis and activates immunity against hemibiotrophic bacteria in plants. Chitin is a major component of fungal cell walls. It plays a crucial molecular role in LysM-mediated host defense responses. Recognition and immune stimulation of chitin secreted by hemibiotrophic fungi in rice or Arabidopsis depend on the LysM-type PRRs OsCEBiP/OsCERK1, LYK4/LYK5, or AtCERK1, respectively. While plants use different sensing systems for bacterial-secreted PGN and fungal-secreted chitin, the chitin sensing system employed by fungi is structurally similar to the carbohydrate portion of bacterial-secreted PGN.
Plant LysM proteins recognize HPs to induce immune responses. HPs have evolved multiple mechanisms to evade plant immune recognition (Wang et al., 2022; Zhao L. et al., 2023). HPs secrete effectors with LysM domains that compete with high affinity for plant LysM receptors. Alternatively, HPs secrete effectors that modify plant LysM receptors to sequester, mask, alter, or prevent the host from degrading pathogen cell walls. These behaviors would regulate plant cytoplasmic signaling, suppress plant immunity, and regulate the transition of HPs from the biotrophic to the necrotrophic stage (Hu et al., 2021; Tian et al., 2022b). The secreted effector protein Slp1 of M. oryzae contains a LysM domain that accumulates at the early stage of infection between fungi and rice cells. This protein disrupts host chitin-triggered immunity by utilizing the LysM structural domain to competitively bind chitin oligosaccharides with CEBiP (Sanchez-Vallet et al., 2013). Similarly, M. oryzae depends not only on LysM Slp1 but also on MoAa91 of M. oryzae. This protein is vital for surface recognition and inhibition of chitin-induced plant immune responses. Further studies shown that MoAa91 competitively binds chitin to the rice immune receptor CEBiP to inhibit chitin-induced plant immune responses (Li et al., 2020). Many HPs-secreted LysM effectors remove chitin oligomers from the host infection site by intermolecular LysM dimerization, or prevent host recognition of chitin by the formation of polymeric precipitates, such as C.higginsianum ChElp1 and ChElp2 (Takahara et al., 2016; Sanchez-Vallet et al., 2020; Tian et al., 2022a). Intermolecular interactions such as homodimerization and phosphorylation demonstrate the significant implications of precisely management of host-pathogen processes, which are also essential for enhancing disease resistance in crops. However, it is unclear whether there are secreted proteins in HPs that more broadly regulate plant defense responses and cell death, potentially mediating the transition from biotrophy to necrotrophy.
Besides PRRs with LRR structures and LysM structures, there is increasing evidence that WAK receptors and lectin receptors play a significant role in plant-microbe interactions. A number of many immune-related WAKs have also been cloned recently. WAKs, a receptor-like kinase required for recognizing oligogalacturonides (OGs), often possess an extracellular EGF-like domain, and are found in both dicots and monocots (Stephens et al., 2022). They can identify pathogens with different lifestyles and regulate HP resistance by modifying host cell walls and regulating hormone fluctuations within host cells. WAKs play a crucial role in plant signaling pathways for immune and abiotic stress responses (Yue et al., 2022). OsWAKs in rice have been discovered to regulate the basic defense against M. oryzae either positively or negatively. Quantitative resistance has been positively affected by OsWAK14, OsWAK91, and OsWAK92, whereas resistance to rice blast has been negatively affected by OsWAK112d. OsWAK91 participates in intercellular signal transduction by generating ROS with possess antibacterial properties (Delteil et al., 2016). AtWAKL10 is thought to enhance plant resistance against P. syringae. Transgenic Arabidopsis lacking WAK exhibits increased susceptibility to P. syringae compared to the wild type (Bot et al., 2019). WAK receptors may regulate host immune resistance by modulating hormone fluctuations during HP infection of different host plants. Overexpression of OsWAK1 and OsWAK25 can enhance host resistance to M. oryzae, and salicylic acid (SA) treatment can up-regulate the expression of OsWAK1 and OsWAK25 genes (Li et al., 2009; Harkenrider et al., 2016). Recently, it was discovered that GmWAK1 relies on the SA pathway to alleviate oxidative stress-induced damage in soybeans resistant to P. infestans (Zhao M. et al., 2023). Immune-related WAK also prevents pathogen penetration during HP attack by altering cell wall composition to enhance cell wall strength. Upon infection of tomato by P. syringae, SlWAK1 strengthens the cell wall through callose deposition to restrict pathogen penetration and spread (Zhang et al., 2020). This ability of WAKs to regulate the cell wall indicates their potential role in plant growth, development, and response to abiotic stresses. OsWAK91/OsDees1 knockout rice has increased susceptibility to M. oryzae and inhibited growth (Delteil et al., 2016). In tomato, AtWAKL10 exhibits resistance to P. syringae while also upregulated when treated with the abiotic stress signaling factor S-nitroso-L-cysteine. Additionally, its knockout gene mutant displays increased tolerance to drought stress, but lower tolerance to salt stress (Bot et al., 2019). The WAK receptors not only enhance or inhibit host resistance when different HPs infect various hosts, but also affect plant growth and development. The mechanisms by which WAK receptors recognize or transduce pathogen signals and their impact on the transition of HPs from the biotrophic to necrotrophic stage remain unclear.
Lectin receptor-like kinases (LecRKs) are unique PRRs that specifically recognize carbohydrates such as mannose induced by elicitors or pathogens (De Coninck and Van Damme, 2022). LecRKs appear to constitute a vital recognition system on the surface of plant cells during plant-microbe interactions, and may play a vital role in plant immunity or stress responses (Wang and Gou, 2022). Furthermore, LecRKs are classified into L-type, C-type, and G-type. G-type and L-type LecRKs are activated by PAMP signaling perception, which triggers PTI to HPs. G-type LecRKs Pi-d2 and OsLecRK have been found to trigger plant defense against rice blast and leaf blight, as well as activate various signal responses in plant innate immunity (Li et al., 2015). Among them, the OsLecRK mutant is more susceptible to Magnaporthe grisea infection than the wild type, with a reduction in mRNA levels of PR1, LOX2, and CHS defense-related genes (Cheng et al., 2013). Similar to LRR receptors, G-type SBP1 and SBP2 can specifically activate immunity by positively regulating the interaction between RLP23 receptors and BAK1 co-receptors (Bao et al., 2023). G-type LecRK LORE was identified as a target site for the effector HopAO1 of P. syringae. During the initial stages of infection, LORE detects bacterial lipopolysaccharides and triggers autophosphorylation to activate the immune response. In the advanced stage of infection, HopAO1 interacts with LORE within host cells, leading to the dephosphorylation of LORE. This effectively suppresses the immune response and makes the host more susceptible to the infection (Chen et al., 2017; Luo et al., 2020). HPs that secrete various molecules recognized by the same host PRR receptors lead to diverse plant immune reactions. Phytohormones play a vital role in plant-pathogen interactions, such as JA, which activate plant defense responses against pathogens. The G-type Lec-RLK FaMBL1 from strawberry can bind to mannose from the cell wall of Colletotrichum fioriniae. Overexpression of FaMBL1 leads to a reduction in JA content (Ma et al., 2023). The L-types of LecRK-IX.1, LecRK-IX.2, and LecRK-I.9 in Arabidopsis are considered to have defense responses against Phytophthora. Overexpression of LecRK-IX.2 phosphorylates RBOHD to enhance ROS production and SA accumulation in PTI response (Wang Y. et al., 2015; Wang Y. et al., 2016; Luo et al., 2017). Similar to LecRK-IX.2, LecRK-I.9 is another L-type LecRK in Arabidopsis, known as DORN1, which also confers plant resistance to P. syringae DC3000 and Phytophthora resistance (Balague et al., 2017; Sun Y. L. et al., 2020). DORN1 recognizes extracellular ATP signals and directly phosphorylates RBOHD. This induces Ca2+ influx, MAPK activation, ROS accumulation, and defense gene expression, and host stomatal closure to restrict HPs invasion (Luo et al., 2017; Wang et al., 2018). These studies indicate that LecRLKs are involved in PTI, ETI, SA, and JA signaling pathways and could enhance plant defense against HPs. In molecular breeding, manipulation of one pathway may impact other signaling responses, due to the interconnected nature of these pathways.
The NLRs perceive effector proteins in host cells to activate an ETI immune response against these pathogens (Barragan and Weigel, 2021). It is a significant member of the plant resistance R protein family with a conserved modular structure consisting of a central NB-ARC domain (nucleotide-binding adapter, APAF-1, R protein, and CED-4), a C-terminal leucine-rich repeat (LRR) domain, and a unique N-terminal domain (Duxbury et al., 2021). Based on their variable N-terminal structure, it is predominantly categorized as either the coil-coil (CC) type or Toll/interleukin-1 receptor type (TIR), known as CNLs and TNLs (Monteiro and Nishimura, 2018; Yuan et al., 2021a; Maruta et al., 2022). Typically, the CC or TIR in the N-terminal domain is considered the signaling domain. The central NB-ARC domain acts as a molecular switch that regulates the binding or hydrolysis of ADP or ATP to determine the signaling state of NLR. LRR domains may play a significant role in ligand recognition and regulatory activity (Jubic et al., 2019; Ma et al., 2020). NLRs often exist in an inactive state when not infected by pathogens due to various intramolecular and intermolecular interactions. Upon recognition of hemibiotrophic effectors, these interactions are disrupted, which activates the NLR to trigger programmed cell death (Sun Y. et al., 2020).
Nucleotide-binding and leucine-rich repeat receptors recognize pathogen effectors through various strategies, including direct recognition, indirect recognition, and paired NLR recognition (Kroj et al., 2016). Some NLRs recognize effector protein patterns directly, which are extensively characterized. HPs are mainly recognized by NLR receptors indirectly. The indirect interaction between ligands and receptors is well-described in the “guard” and “decoy” recognition models (van der Hoorn and Kamoun, 2008). Other effector targets, including transcriptional regulators, molecular chaperones, and ubiquitin ligases, could serve as potential “guards” or “decoys.” These effector targets regulate transcriptional reprogramming and host protein stability, respectively (Sun Y. et al., 2020; Duxbury et al., 2021). There is also a small portion of plant NLR that includes an additional “integrated” domain (ID). This ID recognizes HPs through integrated decoy patterns and activates downstream immune responses (Sarris et al., 2016). The strategies used to activate immunity by various interactions between NLRs and different HPs are discussed in detail below (Figure 1; Table 2).
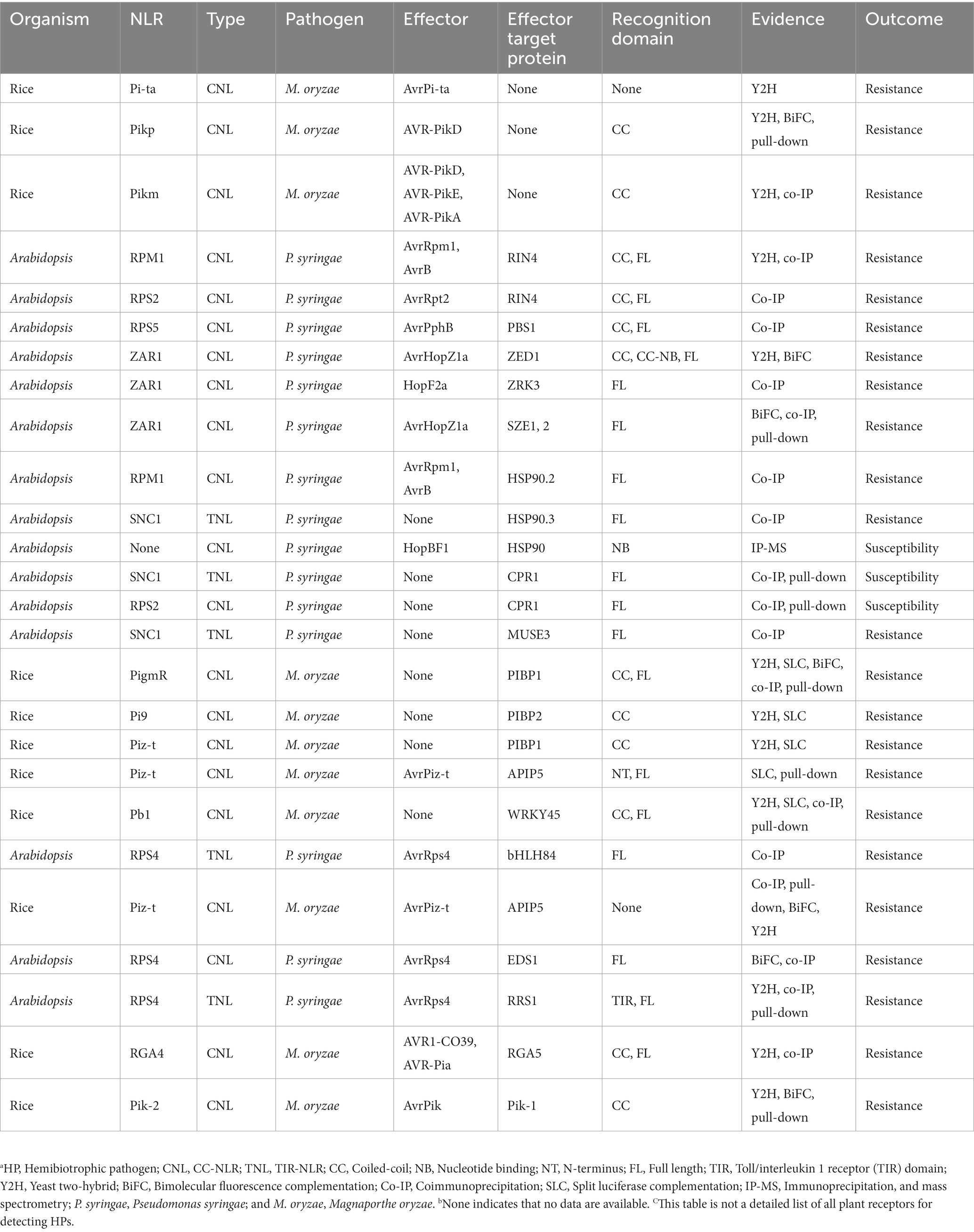
Table 2. Overview of intracellular recognition receptors and the outcome of interactions with typical HPs to mediate immune results.
Most of the proteins encoded by R genes in each plant genome are NLRs. NLRs directly or indirectly recognize effectors secreted by HPs and activate ETI response (Barragan and Weigel, 2021). Direct interaction between hemibiotrophic effectors and plant NLRs is the most intuitive and simple method. Most CNLs act as effector receptors, called sensor CNL. For example, plant CNLs detect their cognate effectors via direct interactions. Rice NLR Pi-ta binds to the M. oryzae effector AVR-Pita (Jia et al., 2000). Intriguingly, the CNL protein encoded by the Pik allele in rice can also perceive multiple AVR-Pik effectors of M. oryzae via physical interaction. Pikm can recognize three AvrPik effectors of M. oryzae, while Pikp can recognize only one (De la Concepcion et al., 2018).
Hemibiotrophic pathogens are mainly recognized by NLR receptors indirectly. Two recognition models can well describe the indirect interaction between ligands and receptors, namely the “guard” model and the “decoy” model (van der Hoorn and Kamoun, 2008). Modification of guards or decoys by effectors may cause changes in the conformation of NLRs, which leads to the activation of ETI (Schreiber et al., 2016). Since many effectors targeted by NLRs are unknown, and it is often unclear whether these effector targets are guardees or decoys (Kapos et al., 2019). The “guard” model suggests that NLR proteins monitor the integrity of target proteins in plant cells and activate immune responses upon perturbation or modification by pathogen effectors. For example, P. syringae effectors AvrRpm1, AvrB, and AvrRpt2 specifically target the guardee protein RPM1-interacting 4 (RIN4), while CNLs RPM1 and RPS2 monitor RIN4 steric hindrance or post-translational modification to exert disease resistance (Day et al., 2005; El Kasmi et al., 2017). In soybean, CNL RPG1-B monitors the homolog RIN4 in a comparable way (Selote and Kachroo, 2010). Another well-studied example is that of the guardee CNL RPS5 monitors the host target kinase PBS1, where the P. syringae effector protease AvrPphB cleaves PBS1 to activate RPS5-mediated immunity (Ade et al., 2007). It is generally established that all kinases and pseudokinases serve as “guards” or “decoys,” and interact with CNLs, but not with TNLs. Decoy proteins have no defined biological, cellular, or physiological role in host defense. Instead, they imitate toxic targets to activate the host surveillance system and detect effector molecules. Decoys probably evolved by duplicating ancestral guardians (van Wersch et al., 2020). As a decoy protein, the pseudokinase RLCK XII family ZED1 interacts with the acetyltransferase HopZ1a effector secreted by P. syringae. ZED1 is acetylated to activate CNL ZAR1 (Wang G. X. et al., 2015). Recent research revealed that ZED1 forms a complex with ZAR1 after acetylation by HopZ1a, triggering the assembly of higher-order complexes in plants that form a resistosome similar to the ZAR1-RKS1-PBL2UMP complex (Hu et al., 2020). Other RLCKs are targeted by HopZ1a and are recognized by the ZAR1-ZED1 complex. RKS1 performs an adapter function similar to ZED1 (Bastedo et al., 2019). The pseudokinase ZRK3 and the RLCK family SZE1 and SZE2 also bind to ZAR1 (Liu et al., 2019). It appears that indirect interactions may expand the capacity of certain plant immune receptors to detect additional pathogen effectors. Furthermore, indirect interactions may offer more avenues to enhance pathogen control by regulating receptors.
Plant NLRs’ assembly, activity, and stability are tightly regulated to ensure appropriate host defense responses against HPs. Studies have shown that molecular chaperones and ubiquitin ligases are essential for NLRs’ assembly, activity, and stability (Duxbury et al., 2021; Huang et al., 2021). There are several chaperones in the HSP90 family that play a role in NLR-mediated defenses in Arabidopsis. HSP90.2 interacts with CNL RPM1 to strengthen RPM1 protein stability. HSP90.3 interacts with TNL SNC1 to negatively regulate SNC1 accumulation (Hubert et al., 2003; Huang S. et al., 2014). Recent studies have shown that the P. syringae effector HopBF1 phosphorylates HSP90 to trigger hypersensitivity in plants. This finding uncovers a previously unidentified mechanism by which hemibiotrophic bacteria regulate host immunity (Lopez et al., 2019). And HSP90 may assist the suppressor of G-two allele of SKP1 (SGT1) in forming the Skp1-Cul1-F-box (SCF) E3 ubiquitin ligase complex, which targets immune receptors for degradation. This process is critical for maintaining appropriate levels of immune receptor proteins to avoid autoimmunity. Similarly, ubiquitin ligases interact with NLRs to help regulate their levels without pathogen infection. Upon P. syringae infection, the proteasome effector destroys the E3 ligase F-box protein CPR1, interacts with Arabidopsis TNL SNC1 and CNL RPS2, and reduces their protein accumulation, thus inducing a defense response (Gou et al., 2012). Knocking down the E4 ligase MUSE3 in Arabidopsis causes increased levels of TNL SNC1 and CNL RPS2. Overexpression of MUSE3 and CPR1 enhanced polyubiquitination and protein degradation of these immune receptors (Huang Y. et al., 2014). Host molecular chaperones and ubiquitin indirectly control NLR activity and stability to modulate immune responses when various HP infect the host. This suggests that NLR activity and homeostasis are critical for plant disease resistance.
Transcriptional reprogramming is a frequent occurrence in plant immunity that involves numerous transcriptional regulators. The coordination and nuclear localization of NLRs and immune transcription factors in the transcriptional machinery is crucial to selectively activate plant defense genes during HPs infection. The transcription factor RRM shows CNL-dependent nuclear localization and is unaffected by HPs. RRM binds to the CNL encoded by the resistance genes PigmR, Pi9, and Piz-t, and establishes a direct link between transcriptional activation of the immune response and NLR-mediated pathogen perception by directly binding to the A/T-rich cis-element DNA in the target gene (Zhai et al., 2019). This is in contrast to the nuclear localization of the constitutive transcription factor WRKY45. This transcription factor induces resistance through the SA signaling pathway regulated by the ubiquitin-proteasome system. The CNL protein encoded by the rice blast resistance gene Pb1 interacts with WRKY45 in the nucleus to regulate broad-spectrum resistance to M. oryzae (Inoue et al., 2013). EDS1 regulates the defense signaling pathway. The interactions between Arabidopsis NLR RSP4 and EDS1 result in similar but distinct nuclear NLR-dependent translocations. The RPS4 and EDS1 complex is predominantly located in the cytoplasm, but can also be observed in the nucleus during homeostasis or upon AvrRPS4 infection. Additionally, RPS4 binds to EDS1 in the cytoplasm without relying on another NLR RRS1. Instead, when RRS1 is present, an RPS4-EDS1 complex is observed in the nucleus. This suggests the existence of pre- and post-activation states for the nuclear localization of RPS4, RRS1 and EDS1 complexes. The RPS4-EDS1 binding in the nucleus may be unaffected by HP effectors such as AvrRps4 in the presence of RRS1 (Wang R. Y. et al., 2016). It has also been shown that the effector Avrpiz-t from M. oryzae interacts with the bzip-type transcription factor APIP5 in the cytoplasm, inhibiting its transcriptional activity and protein accumulation during the necrotic stage. Additionally, the rice NLR Piz-t inhibits plant ETI necrosis by interacting with APIP5 (Wang R. Y. et al., 2016). These showing how the host utilizes transcription factors as imitation substances for effectors to prevent host immune responses induced by various HPs.
Many NLRs paired with additional domains or motifs are also critical for plant protection against HPs compared to regular NLRs. The paired NLR contains one NLR with an ID at its C-terminus that mimics the virulence target of an effector protein, and thus acts as a sensor for detecting effector proteins. Another NLR acts as a classical executive NLR that performs signal transduction functions (Grund et al., 2019). Some NLR gene pairs are frequently close in the same locus on the chromosome. These pairs share a promoter. The two genes that encode RPS4 and RRS1 are located adjacent to each other and are arranged in opposite directions. This suggests they may co-regulate transcription, with an interval of approximately 300 bp between them (Narusaka et al., 2009). RRS1 has an extra structural domain called WRKY at the C-terminus. RPS4 and RRSI together form a heterodimeric complex that recognizes the effector AvrRps4 and confers resistance to P. syringae (Guo et al., 2020). In rice, the C-terminus of the NLR-paired RGA5 contains an HMA structural domain that acts as an ID that interacts with the effectors AVR1-CO39 and AVR-Pia in M. oryzae. The NLR-paired RGA4 acts as an NLR executor that induces robust HR in tobacco leaves (Cesari et al., 2013; De la Concepcion et al., 2021). The co-expression of Pikp-1, Pikp-2, and M. oryzae effector AVR-PikD, where Pikp-1 possesses an HMA, and Pikp-2 conducts signal transduction, induces a significant HR in tobacco (Maqbool et al., 2015; Cesari et al., 2022). Guardees or decoys allows plants to detect a wide range of pathogen effectors with a relatively small repertoire of NLRs (Frailie and Innes, 2021; Liu et al., 2021). In the RPS4/RRS1 and RGA4/RGA5 heterodimers, one NLR is involved in effector identification while the other is involved in defense signaling. Understanding the extent of heterodimerization in plant NLRs is crucial in gaining insights into plant NLR evolution and diversity. Moreover, pairing NLR sensors and actuators in plant design could improve effector recognition specificity and resistance profiles.
Plant diseases caused by phytopathogens are a major threat to global food security and can lead to significant economic losses, such as HPs. Notably, M. oryzae, Colletotrichum spp., and P. syringae are considered important HPs due to their ease of cultivation, genetic modification, and typical hemibiotrophic characteristics (Doehlemann et al., 2017). Therefore, they are commonly utilized as models for studying plant immunity activation by HPs. Several other species from broad genera are also classified as HPs, including significant plant pathogens like Fusarium (Ma et al., 2013), Verticillium (Fradin and Thomma, 2006), Mycosphaerella (Churchill, 2011), and others. All of these species have an asymptomatic stage of varying lengths. In most cases, they do not develop into typical biotrophic specialized organs and do not make close contact with the host cells. Therefore, the pathogenic lifestyle of HPs comprises asymptomatic, quiescent, latent or endogenous stages and requires different forms of control. To effectively manage plant diseases caused by HPs, it is essential to understand the interaction between HPs and host plants, as well as their strategies for activating plant immunity. Currently, genetic control of plant diseases aims to improve plant resistance. This is achieved through techniques like genome editing, which targets specific genes, and resistance gene enrichment sequencing. Plant disease resistance is mostly determined by genes that have receptors detect when pathogens enter the plant and trigger immune responses. In this review, we discuss plant cell surface PRR and intracellular NLR immunoreceptors that detect various HP and initiate appropriate immune responses. Furthermore, we discuss effector target proteins, including transcription factors, ubiquitin ligases, and molecular chaperones. These proteins could potentially function as models of “guards” and “decoys” for the indirect molecular immunity against HP in the ongoing arms race between plants and pathogens. Over time, a complex recognition system has developed between the immune receptors and HP. By summarizing the interaction between HPs and plant immune receptors that trigger host immune responses, it was found that PAMPs may be conserved and prevalent in various microorganisms, and the cell-surface PRR complexes have similar co-receptors, including BAK1, SOBIR1, or CERK1. By combining genomics, transcriptomics, effectomics, and high-throughput phenotypic analysis, it is anticipated that that crops will achieve better and sustainable protection against various diseases through the utilization of stacked plant immune receptors. For example, multiple plant immune receptors may be designed to recognize the same type of pathogen, or they may be designed for different types of pathogens, such as oomycetes and fungi. We may discover novel defense mechanisms and corresponding pathogen factors. This results in improved disease management and control.
However, it is unclear how these simple conceptual models allow PRRs and NLRs to adapt to the virtually unlimited of immune signaling space. Moreover, the Agrobacterium transient assay is useful for enhancing analysis of plant receptor function in solanaceous hosts. However, it might not work well for studying receptor function in soybean or other monocotyledonous plants. This difficulty has also been a major constraint in finding different pathogen effector proteins. Plant PTI and ETI signaling systems may appear simple, but they are rather complex. One immune receptor like EDSI could regulate a single target protein of the host and may be targeted by multiple effectors like RIN4 (Sun Y. L. et al., 2020). Each signal directly or indirectly coordinates with each other to jointly regulate plant immunity, growth and development. Elimination of host susceptibility to HPs through gene editing techniques poses potential risks. New susceptibility genes may be introduced while the desired traits are being engineered for transfer to other species. A recent study of cell trajectory analysis of based on single-cell omics technology found that, at the early stages of immune cell trajectory, the progression of disease from the immune state to the susceptible state is a continuous process (Zhu et al., 2023). In the future, advancements in spatial omics with high resolution, next-generation sequencing technology, and new bioinformatics algorithms and pipelines will provide new genome-wide data for HPs. This will enable researchers to gain greater insight into intricate plant immune responses and their dynamic interactions with pathogens in specific spatial contexts. However, with so many candidate effector genes or genomes, it is unclear how HP effectors shift from biotrophy to necrotrophy, manipulate host targets, and interfere with plant immune activation.
DZ, GZ, and JL contributed to the conception of the manuscript. DZ wrote the original draft. XZC, XGC, and YX provided editing assistance. All authors contributed to the article and approved the submitted version.
This research was supported by grants from the National Natural Science Foundation of China (32271900), the National Natural Science Foundation of China (31971661), the Hunan Provincial Innovation Foundation for Postgraduate (CX20230748), and the Scientific Innovation Fund for Postgraduates of Central South University of Forestry and Technology (2023CX01002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ade, J., DeYoung, B. J., Golstein, C., and Innes, R. W. (2007). Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc. Natl. Acad. Sci. U. S. A. 104, 2531–2536. doi: 10.1073/pnas.0608779104
Albert, I., Bohm, H., Albert, M., Feiler, C. E., Imkampe, J., Wallmeroth, N., et al. (2015). An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat Plants 1:15140. doi: 10.1038/nplants.2015.140
Balague, C., Gouget, A., Bouchez, O., Souriac, C., Haget, N., Boutet-Mercey, S., et al. (2017). The Arabidopsis thaliana lectin receptor kinase LecRK-I.9 is required for full resistance to Pseudomonas syringae and affects jasmonate signalling. Mol. Plant Pathol. 18, 937–948. doi: 10.1111/mpp.12457
Bao, Y., Li, Y., Chang, Q., Chen, R., Wang, W., Zhang, Q., et al. (2023). A pair of G-type lectin receptor-like kinases modulates nlp20-mediated immune responses by coupling to the RLP23 receptor complex. J. Integr. Plant Biol. 65, 1312–1327. doi: 10.1111/jipb.13449
Barragan, A. C., and Weigel, D. (2021). Plant NLR diversity: the known unknowns of pan-NLRomes. Plant Cell 33, 814–831. doi: 10.1093/plcell/koaa002
Bastedo, D. P., Khan, M., Martel, A., Seto, D., Kireeva, I., Zhang, J. F., et al. (2019). Perturbations of the ZED1 pseudokinase activate plant immunity. PLoS Pathog. 15:e1007900. doi: 10.1371/journal.ppat.1007900
Beck, M., Zhou, J., Faulkner, C., MacLean, D., Robatzek, S., and Zhou, J. (2012). Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 24, 4205–4219. doi: 10.1105/tpc.112.100263
Bot, P., Mun, B. G., Imran, Q. M., Hussain, A., Lee, S. U., Loake, G., et al. (2019). Differential expression of AtWAKL10 in response to nitric oxide suggests a putative role in biotic and abiotic stress responses. PeerJ 7:e7383. doi: 10.7717/peerj.7383
Buendia, L., Girardin, A., Wang, T. M., Cottret, L., and Lefebvre, B. (2018). LysM receptor-like kinase and LysM receptor-like protein families: an update on phylogeny and functional characterization. Front. Plant Sci. 9:1531. doi: 10.3389/fpls.2018.01531
Cao, Y. R., Liang, Y., Tanaka, K., Nguyen, C. T., Jedrzejczak, R. P., Joachimiak, A., et al. (2014). The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. Elife 3:e03766. doi: 10.7554/eLife.03766
Cesari, S., Thilliez, G., Ribot, C., Chalvon, V., Michel, C., Jauneau, A., et al. (2013). The Rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481. doi: 10.1105/tpc.112.107201
Cesari, S., Xi, Y. X., Declerck, N., Chalvon, V., Mammri, L., Pugniere, M., et al. (2022). New recognition specificity in a plant immune receptor by molecular engineering of its integrated domain. Nat. Commun. 13:1524. doi: 10.1038/s41467-022-29196-6
Chen, J. B., Bao, S. W., Fang, Y. L., Wei, L. Y., Zhu, W. S., Peng, Y. L., et al. (2021). An LRR-only protein promotes NLP-triggered cell death and disease susceptibility by facilitating oligomerization of NLP in Arabidopsis. New Phytol. 232, 1808–1822. doi: 10.1111/nph.17680
Chen, D., Cao, Y., Li, H., Kim, D., Ahsan, N., Thelen, J., et al. (2017). Extracellular ATP elicits DORN1-mediated RBOHD phosphorylation to regulate stomatal aperture. Nat. Commun. 8:2265. doi: 10.1038/s41467-017-02340-3
Chen, Q., Dong, C., Sun, X., Zhang, Y., Dai, H., and Bai, S. (2020). Overexpression of an apple LysM-containing protein gene, MdCERK1-2, confers improved resistance to the pathogenic fungus, Alternaria alternata, in Nicotiana benthamiana. BMC Plant Biol. 20:146. doi: 10.1186/s12870-020-02361-z
Chen, Y., Liu, Z. Y., and Halterman, D. A. (2012). Molecular determinants of resistance activation and suppression by Phytophthora infestans effector IPI-O. PLoS Pathog. 8:e1002595. doi: 10.1371/journal.ppat.1002595
Cheng, X. Y., Wu, Y., Guo, J. P., Du, B., Chen, R. Z., Zhu, L. L., et al. (2013). A rice lectin receptor-like kinase that is involved in innate immune responses also contributes to seed germination. Plant J. 76, 687–698. doi: 10.1111/tpj.12328
Churchill, A. C. (2011). Mycosphaerella fijiensis, the black leaf streak pathogen of banana: progress towards understanding pathogen biology and detection, disease development, and the challenges of control. Mol. Plant Pathol. 12, 307–328. doi: 10.1111/j.1364-3703.2010.00672.x
Couto, D., and Zipfel, C. (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552. doi: 10.1038/nri.2016.77
Damm, U., O'Connell, R. J., Groenewald, J. Z., and Crous, P. W. (2014). The Colletotrichum destructivum species complex—hemibiotrophic pathogens of forage and field crops. Stud. Mycol. 79, 49–84. doi: 10.1016/j.simyco.2014.09.003
Dangl, J. L. (2013). Pivoting the plant immune system from dissection to deployment (vol 341, pg 746, 2013). Science 341:1175. doi: 10.1126/science.123601
Day, B., Dahlbeck, D., Huang, J., Chisholm, S. T., Li, D., and Staskawicz, B. J. (2005). Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell 17, 1292–1305. doi: 10.1105/tpc.104.030163
De Coninck, T., and Van Damme, E. J. M. (2022). Plant lectins: Handymen at the cell surface. Cell Surf, 8:100091. doi: 10.1016/j.tcsw.2022.100091
De la Concepcion, J. C., Franceschetti, M., Maqbool, A., Saitoh, H., Terauchi, R., Kamoun, S., et al. (2018). Polymorphic residues in rice NLRs expand binding and response to effectors of the blast pathogen (vol 4, pg 576, 2018). Nat. Plants 4:734. doi: 10.1038/s41477-018-0248-0
De la Concepcion, J. C., Maidment, J. H. R., Longya, A., Xiao, G., Franceschetti, M., and Banfield, M. J. (2021). The allelic rice immune receptor Pikh confers extended resistance to strains of the blast fungus through a single polymorphism in the effector binding interface. PLoS Pathog. 17:e1009368. doi: 10.1371/journal.ppat.1009368
Delteil, A., Gobbato, E., Cayrol, B., Estevan, J., Michel-Romiti, C., Dievart, A., et al. (2016). Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol. 16:17. doi: 10.1186/s12870-016-0711-x
Desaki, Y., Miyata, K., Suzuki, M., Shibuya, N., and Kaku, H. (2018). Plant immunity and symbiosis signaling mediated by LysM receptors. Innate Immun. 24, 92–100. doi: 10.1177/1753425917738885
Doehlemann, G., Okmen, B., Zhu, W., and Sharon, A. (2017). Plant pathogenic fungi. Microbiol. Spectr. 5. doi: 10.1128/microbiolspec.FUNK-0023-2016
Duxbury, Z., Wu, C. H., and Ding, P. (2021). A comparative overview of the intracellular guardians of plants and animals: NLRs in innate immunity and beyond. Annu. Rev. Plant Biol. 72, 155–184. doi: 10.1146/annurev-arplant-080620-104948
El Kasmi, F., Chung, E. H., Anderson, R. G., Li, J., Wan, L., Eitas, T. K., et al. (2017). Signaling from the plasma-membrane localized plant immune receptor RPM1 requires self-association of the full-length protein. Proc. Natl. Acad. Sci. U. S. A. 114, E7385–E7394. doi: 10.1073/pnas.1708288114
Erwig, J., Ghareeb, H., Kopischke, M., Hacke, R., Matei, A., Petutschnig, E., et al. (2017). Chitin-induced and CHITIN ELICITOR RECEPTOR KINASE1 (CERK1) phosphorylation-dependent endocytosis of Arabidopsis thaliana LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE5 (LYK5). New Phytol. 215, 382–396. doi: 10.1111/nph.14592
Eschen-Lippold, L., Jiang, X., Elmore, J. M., Mackey, D., Shan, L., Coaker, G., et al. (2016). Bacterial AvrRpt2-like cysteine proteases block activation of the Arabidopsis mitogen-activated protein kinases, MPK4 and MPK11. Plant Physiol. 171, 2223–2238. doi: 10.1104/pp.16.00336
Fisher, M. C., Hawkins, N. J., Sanglard, D., and Gurr, S. J. (2018). Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 360, 739–742. doi: 10.1126/science.aap7999
Fliegmann, J., Uhlenbroich, S., Shinya, T., Martinez, Y., Lefebvre, B., Shibuya, N., et al. (2011). Biochemical and phylogenetic analysis of CEBiP-like LysM domain-containing extracellular proteins in higher plants. Plant Physiol. Biochem. 49, 709–720. doi: 10.1016/j.plaphy.2011.04.004
Fradin, E. F., and Thomma, B. P. (2006). Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 7, 71–86. doi: 10.1111/j.1364-3703.2006.00323.x
Frailie, T. B., and Innes, R. W. (2021). Engineering healthy crops: molecular strategies for enhancing the plant immune system. Curr. Opin. Biotechnol. 70, 151–157. doi: 10.1016/j.copbio.2021.04.006
Giovannoni, M. A.-O., Lironi, D., Marti, L., Paparella, C., Vecchi, V., Gust, A. A.-O., et al. (2021). The Arabidopsis thaliana LysM-containing receptor-like kinase 2 is required for elicitor-induced resistance to pathogens. Plant Cell Environ. 44, 3545–3562. doi: 10.1111/pce.14192
Gou, M. Y., Shi, Z. Y., Zhu, Y., Bao, Z. L., Wang, G. Y., and Hua, J. (2012). The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J. 69, 411–420. doi: 10.1111/j.1365-313X.2011.04799.x
Gravino, M., Locci, F., Tundo, S., Cervone, F., Savatin, D. V., and De Lorenzo, G. (2017). Immune responses induced by oligogalacturonides are differentially affected by AvrPto and loss of BAK1/BKK1 and PEPR1/PEPR2. Mol. Plant Pathol. 18, 582–595. doi: 10.1111/mpp.12419
Grund, E., Tremousaygue, D., and Deslandes, L. (2019). Plant NLRs with integrated domains: Unity makes strength. Plant Physiol. 179, 1227–1235. doi: 10.1104/pp.18.01134
Guo, H., Ahn, H. K., Sklenar, J., Huang, J., Ma, Y., Ding, P., et al. (2020). Phosphorylation-regulated activation of the Arabidopsis RRS1-R/RPS4 immune receptor complex reveals two distinct effector recognition mechanisms. Cell Host Microbe 27, 769–781 e766. doi: 10.1016/j.chom.2020.03.008
Gust, A. A., and Felix, G. (2014). Receptor like proteins associate with SOBIR1-type of adaptors to form bimolecular receptor kinases. Curr. Opin. Plant Biol. 21, 104–111. doi: 10.1016/j.pbi.2014.07.007
Harkenrider, M., Sharma, R., De Vleesschauwer, D., Tsao, L., Zhang, X. T., Chern, M., et al. (2016). Overexpression of rice wall-associated kinase 25 (OsWAK25) alters resistance to bacterial and fungal pathogens. PLoS One 11:e0147310. doi: 10.1371/journal.pone.0147310
Hayafune, M., Berisio, R., Marchetti, R., Silipo, A., Kayama, M., Desaki, Y., et al. (2014). Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc. Natl. Acad. Sci. U. S. A. 111, E404–E413. doi: 10.1073/pnas.1312099111
Hu, S. P., Li, J. J., Dhar, N., Li, J. P., Chen, J. Y., Jian, W., et al. (2021). Lysin motif (LysM) proteins: interlinking manipulation of plant immunity and fungi. Int. J. Mol. Sci. 22:3114. doi: 10.3390/ijms22063114
Hu, M. J., Qi, J. F., Bi, G. Z., and Zhou, J. M. (2020). Bacterial effectors induce oligomerization of immune receptor ZAR1 in vivo. Mol. Plant 13, 793–801. doi: 10.1016/j.molp.2020.03.004
Huang, Y., Minaker, S., Roth, C., Huang, S., Hieter, P., Lipka, V., et al. (2014). An E4 ligase facilitates Polyubiquitination of plant immune receptor resistance proteins in Arabidopsis. Plant Cell 26, 485–496. doi: 10.1105/tpc.113.119057
Huang, S., Monaghan, J., Zhong, X., Lin, L., Sun, T., Dong, O. X., et al. (2014). HSP90s are required for NLR immune receptor accumulation in Arabidopsis. Plant J. 79, 427–439. doi: 10.1111/tpj.12573
Huang, J., Wu, X., and Gao, Z. (2021). A nucleocytoplasmic-localized E3 ligase affects the NLR receptor stability. Biochem. Biophys. Res. Commun. 583, 1–6. doi: 10.1016/j.bbrc.2021.10.052
Hubert, D. A., Tornero, P., Belkhadir, Y., Krishna, P., Takahashi, A., Shirasu, K., et al. (2003). Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 22, 5679–5689. doi: 10.1093/emboj/cdg547
Inoue, H., Hayashi, N., Matsushita, A., Liu, X. Q., Nakayama, A., Sugano, S., et al. (2013). Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein-protein interaction. Proc. Natl. Acad. Sci. U. S. A. 110, 9577–9582. doi: 10.1073/pnas.1222155110
Jia, Y., McAdams, S. A., Bryan, G. T., Hershey, H. P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. doi: 10.1093/emboj/19.15.4004
Jones, J. D. G., Vance, R. E., and Dangl, J. L. (2016). Intracellular innate immune surveillance devices in plants and animals. Science 354:aaf6395. doi: 10.1126/science.aaf6395
Jubic, L. M., Saile, S., Furzer, O. J., El Kasmi, F., and Dangl, J. L. (2019). Help wanted: helper NLRs and plant immune responses. Curr. Opin. Plant Biol. 50, 82–94. doi: 10.1016/j.pbi.2019.03.013
Kapos, P., Devendrakumar, K. T., and Li, X. (2019). Plant NLRs: from discovery to application. Plant Sci. 279, 3–18. doi: 10.1016/j.plantsci.2018.03.010
Kelley, B. S., Lee, S. J., Damasceno, C. M. B., Chakravarthy, S., Kim, B. D., Martin, G. B., et al. (2010). A secreted effector protein (SNE1) from Phytophthora infestans is a broadly acting suppressor of programmed cell death. Plant J. 62, 357–366. doi: 10.1111/j.1365-313X.2010.04160.x
Kleemann, J., Rincon-Rivera, L. J., Takahara, H., Neumann, U., van Themaat, E. V. L., van der Does, H. C., et al. (2012). Sequential delivery of host-induced virulence effectors by Appressoria and intracellular hyphae of the Phytopathogen Colletotrichum higginsianum. PLoS Pathog. 8:e1002643. doi: 10.1371/journal.ppat.1002643
Kourelis, J., and van der Hoorn, R. A. L. (2018). Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30, 285–299. doi: 10.1105/tpc.17.00579
Kraepiel, Y., and Barny, M. A. (2016). Gram-negative phytopathogenic bacteria, all hemibiotrophs after all? Mol. Plant Pathol. 17, 313–316. doi: 10.1111/mpp.12345
Kroj, T., Chanclud, E., Michel-Romiti, C., Grand, X., and Morel, J. B. (2016). Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 210, 618–626. doi: 10.1111/nph.13869
Lee, D. H., Lee, H. S., and Belkhadir, Y. (2021). Coding of plant immune signals by surface receptors. Curr. Opin. Plant Biol. 62:102044. doi: 10.1016/j.pbi.2021.102044
Lei, L., Stevens, D. M., and Coaker, G. (2020). Phosphorylation of the Pseudomonas effector AvrPtoB by Arabidopsis SnRK2.8 is required for bacterial virulence. Mol. Plant 13, 1513–1522. doi: 10.1016/j.molp.2020.08.018
Li, L., Kim, P., Yu, L., Cai, G., Chen, S., Alfano, J. R., et al. (2016). Activation-dependent destruction of a co-receptor by a Pseudomonas syringae effector dampens plant immunity. Cell Host Microbe 20, 504–514. doi: 10.1016/j.chom.2016.09.007
Li, Y., Liu, X., Liu, M., Wang, Y., Zou, Y., You, Y., et al. (2020). Magnaporthe oryzae auxiliary activity protein MoAa91 functions as chitin-binding protein to induce Appressorium formation on artificial inductive surfaces and suppress plant immunity. mBio 11, e03304–19. doi: 10.1128/mbio.03304-03319
Li, J. B., Sun, Y. D., Liu, H., Wang, Y. Y., Jia, Y. L., and Xu, M. H. (2015). Natural variation of rice blast resistance gene pi-d2. Genet. Mol. Res. 14, 1235–1249. doi: 10.4238/2015.February.13.2
Li, H., Zhou, S. Y., Zhao, W. S., Su, S. C., and Peng, Y. L. (2009). A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol. Biol. 69, 337–346. doi: 10.1007/s11103-008-9430-5
Liebrand, T. W. H., van den Burg, H. A., and Joosten, M. H. A. J. (2014). Two for all: receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci. 19, 123–132. doi: 10.1016/j.tplants.2013.10.003
Liu, X. R., Ao, K., Yao, J., Zhang, Y. L., and Li, X. (2021). Engineering plant disease resistance against biotrophic pathogens. Curr. Opin. Plant Biol. 60:101987. doi: 10.1016/j.pbi.2020.101987
Liu, C., Cui, D. Y., Zhao, J. B., Liu, N., Wang, B., Liu, J., et al. (2019). Two Arabidopsis receptor-like cytoplasmic kinases SZE1 and SZE2 associate with the ZAR1-ZED1 complex and are required for effector-triggered immunity. Mol. Plant 12, 967–983. doi: 10.1016/j.molp.2019.03.012
Lopez, V. A., Park, B. C., Nowak, D., Sreelatha, A., Zembek, P., Fernandez, J., et al. (2019). A bacterial effector mimics a host HSP90 client to undermine immunity. Cells 179:205. doi: 10.1016/j.cell.2019.08.020
Luo, X., Wu, W., Liang, Y., Xu, N., Wang, Z., Zou, H., et al. (2020). Tyrosine phosphorylation of the lectin receptor-like kinase LORE regulates plant immunity. EMBO J. 39:e102856. doi: 10.15252/embj.2019102856
Luo, X. M., Xu, N., Huang, J. K., Gao, F., Zou, H. S., Boudsocq, M., et al. (2017). A lectin receptor-like kinase mediates pattern-triggered salicylic acid signaling. Plant Physiol. 174, 2501–2514. doi: 10.1104/pp.17.00404
Ma, L. J., Geiser, D. M., Proctor, R. H., Rooney, A. P., O'Donnell, K., Trail, F., et al. (2013). Fusarium pathogenomics. Annu. Rev. Microbiol. 67, 399–416. doi: 10.1146/annurev-micro-092412-155650
Ma, L., Haile, Z. M., Sabbadini, S., Mezzetti, B., Negrini, F., and Baraldi, E. (2023). Functional characterization of MANNOSE-BINDING LECTIN 1, a G-type lectin gene family member, in response to fungal pathogens of strawberry. J. Exp. Bot. 74, 149–161. doi: 10.1093/jxb/erac396
Ma, S., Lapin, D., Liu, L., Sun, Y., Song, W., Zhang, X., et al. (2020). Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 370:eabe3069. doi: 10.1126/science.abe3069
Macho, A. P., and Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272. doi: 10.1016/j.molcel.2014.03.028
Mamode Cassim, A., Gouguet, P., Gronnier, J., Laurent, N., Germain, V., Grison, M., et al. (2018). Plant lipids: key players of plasma membrane organization and function. Prog. Lipid Res. 73, 1–27. doi: 10.1016/j.plipres.2018.11.002
Maqbool, A., Saitoh, H., Franceschetti, M., Stevenson, C. E. M., Uemura, A., Kanzaki, H., et al. (2015). Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. Elife 4:e08709. doi: 10.7554/eLife.08709
Maruta, N., Burdett, H., Lim, B. Y. J., Hu, X., Desa, S., Manik, M. K., et al. (2022). Structural basis of NLR activation and innate immune signalling in plants. Immunogenetics 74, 5–26. doi: 10.1007/s00251-021-01242-5
Mbengue, M., Bourdais, G., Gervasi, F., Beck, M., Zhou, J., Spallek, T., et al. (2016). Clathrin-dependent endocytosis is required for immunity mediated by pattern recognition receptor kinases. Proc. Natl. Acad. Sci. U. S. A. 113, 11034–11039. doi: 10.1073/pnas.1606004113
McCombe, C. L., Greenwood, J. R., Solomon, P. S., and Williams, S. J. (2022). Molecular plant immunity against biotrophic, hemibiotrophic, and necrotrophic fungi. Essays Biochem. 66, 581–593. doi: 10.1042/Ebc20210073
Mogga, V., Delventhal, R., Weidenbach, D., Langer, S., Bertram, P. M., Andresen, K., et al. (2016). Magnaporthe oryzae effectors MoHEG13 and MoHEG16 interfere with host infection and MoHEG13 counteracts cell death caused by Magnaporthe-NLPs in tobacco (vol 35, pg 1169, 2016). Plant Cell Rep. 35:1187. doi: 10.1007/s00299-016-1968-0
Monteiro, F., and Nishimura, M. T. (2018). Structural, functional, and genomic diversity of plant NLR proteins: An evolved resource for rational engineering of plant immunity. Annu. Rev. Phytopathol. 56, 243–267. doi: 10.1146/annurev-phyto-080417-045817
Narusaka, M., Shirasu, K., Noutoshi, Y., Kubo, Y., Shiraishi, T., Iwabuchi, M., et al. (2009). RRS1 and RPS4 provide a dual resistance-gene system against fungal and bacterial pathogens. Plant J. 60, 218–226. doi: 10.1111/j.1365-313X.2009.03949.x
Ngou, B. P. M., Ahn, H. K., Ding, P. T., and Jones, J. D. G. (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592:110. doi: 10.1038/s41586-021-03315-7
Ngou, B. P. M., Ding, P. T., and Jones, J. D. G. (2022). Thirty years of resistance: zig-Zag through the plant immune system. Plant Cell 34, 1447–1478. doi: 10.1093/plcell/koac041
Panthapulakkal Narayanan, S., Lung, S. C., Liao, P., Lo, C., and Chye, M. L. (2020). The overexpression of OsACBP5 protects transgenic rice against necrotrophic, hemibiotrophic and biotrophic pathogens. Sci. Rep. 10:14918. doi: 10.1038/s41598-020-71851-9
Pirc, K., Hodnik, V., Snoj, T., Caserman, S., Podobnik, M., Bohm, H., et al. (2021). Nep1-like proteins as a target for plant pathogen control. PLoS Pathog. 17:e1009477. doi: 10.1371/journal.ppat.1009477
Qiu, J. H., Xie, J. H., Chen, Y., Shen, Z. N., Shi, H. B., Naqvi, N. I., et al. (2022). Warm temperature compromises JA-regulated basal resistance to enhance Magnaporthe oryzae infection in rice. Mol. Plant 15, 723–739. doi: 10.1016/j.molp.2022.02.014
Rajarammohan, S. (2021). Redefining plant-Necrotroph interactions: the thin line between Hemibiotrophs and Necrotrophs. Front. Microbiol. 12:673518. doi: 10.3389/fmicb.2021.673518
Sanchez-Vallet, A., Saleem-Batcha, R., Kombrink, A., Hansen, G., Valkenburg, D. J., Thomma, B. P. H. J., et al. (2013). Fungal effector Ecp6 outcompetes host immune receptor for chitin binding through intrachain LysM dimerization. Elife 2:e00790. doi: 10.7554/eLife.00790
Sanchez-Vallet, A., Tian, H., Rodriguez-Moreno, L., Valkenburg, D. J., Saleem-Batcha, R., Wawra, S., et al. (2020). A secreted LysM effector protects fungal hyphae through chitin-dependent homodimer polymerization. PLoS Pathog. 16:e1008652. doi: 10.1371/journal.ppat.1008652
Sarris, P. F., Cevik, V., Dagdas, G., Jones, J. D. G., and Krasileva, K. V. (2016). Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 14:8. doi: 10.1186/s12915-016-0228-7
Schellenberger, R., Touchard, M., Clement, C., Baillieul, F., Cordelier, S., Crouzet, J., et al. (2019). Apoplastic invasion patterns triggering plant immunity: plasma membrane sensing at the frontline. Mol. Plant Pathol. 20, 1602–1616. doi: 10.1111/mpp.12857
Schreiber, K. J., Baudin, M., Hassan, J. A., and Lewis, J. D. (2016). Die another day: molecular mechanisms of effector-triggered immunity elicited by type III secreted effector proteins. Semin. Cell Dev. Biol. 56, 124–133. doi: 10.1016/j.semcdb.2016.05.001
Selote, D., and Kachroo, A. (2010). RPG1-B-derived resistance to AvrB-expressing Pseudomonas syringae requires RIN4-like proteins in soybean. Plant Physiol. 153, 1199–1211. doi: 10.1104/pp.110.158147
Sharpee, W., Oh, Y., Yi, M., Franck, W., Eyre, A., Okagaki, L. H., et al. (2017). Identification and characterization of suppressors of plant cell death (SPD) effectors from Magnaporthe oryzae. Mol. Plant Pathol. 18, 850–863. doi: 10.1111/mpp.12449
Smakowska-Luzan, E., Mott, G. A., Parys, K., Stegmann, M., Howton, T. C., Layeghifard, M., et al. (2018). An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature 553, 342–346. doi: 10.1038/nature25184
Stephens, C., Hammond-Kosack, K. E., and Kanyuka, K. (2022). WAKsing plant immunity, waning diseases. J. Exp. Bot. 73, 22–37. doi: 10.1093/jxb/erab422
Su, J. A.-O., Zhang, M. A.-O. X., Zhang, L., Sun, T., Liu, Y. A.-O., Lukowitz, W. A.-O., et al. (2017). Regulation of stomatal immunity by interdependent functions of a pathogen-responsive MPK3/MPK6 Cascade and abscisic acid. Plant Cell 29, 526–542. doi: 10.1105/tpc.16.00577
Sun, Y. L., Qiao, Z. Z., Muchero, W., and Chen, J. G. (2020). Lectin receptor-like kinases: the sensor and mediator at the plant cell surface. Front. Plant Sci. 11:596301. doi: 10.3389/fpls.2020.596301
Sun, Y., Zhu, Y. X., Balint-Kurti, P. J., and Wang, G. F. (2020). Fine-tuning immunity: players and regulators for plant NLRs. Trends Plant Sci. 25, 695–713. doi: 10.1016/j.tplants.2020.02.008
Suzuki, M., Shibuya, M., Shimada, H., Motoyama, N., Nakashima, M., Takahashi, S., et al. (2016). Autophosphorylation of specific threonine and tyrosine residues in Arabidopsis CERK1 is essential for the activation of chitin-induced immune signaling. Plant Cell Physiol. 57, 2312–2322. doi: 10.1093/pcp/pcw150
Takahara, H., Hacquard, S., Kombrink, A., Hughes, H. B., Halder, V., Robin, G. P., et al. (2016). Colletotrichum higginsianum extracellular LysM proteins play dual roles in appressorial function and suppression of chitin-triggered plant immunity. New Phytol. 211, 1323–1337. doi: 10.1111/nph.13994
Tang, D. Z., Wang, G. X., and Zhou, J. M. (2017). Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29, 618–637. doi: 10.1105/tpc.16.00891
Tian, H., Fiorin, G. L., Kombrink, A., Mesters, J. R., and Thomma, B. (2022a). Fungal dual-domain LysM effectors undergo chitin-induced intermolecular, and not intramolecular, dimerization. Plant Physiol. 190, 2033–2044. doi: 10.1093/plphys/kiac391
Tian, H., MacKenzie, C. I., and Rodriguez-Moreno, L. (2022b). Three LysM effectors of Zymoseptoria tritici collectively disarm chitin-triggered plant immunity (vol 22, pg 683, 2021). Mol. Plant Pathol. 23:1830. doi: 10.1111/mpp.13271
Tsuda, K., and Katagiri, F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. doi: 10.1016/j.pbi.2010.04.006
Tsuda, K., Mine, A., Bethke, G., Igarashi, D., Botanga, C. J., Tsuda, Y., et al. (2013). Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana. PLoS Genet. 9:e1004015. doi: 10.1371/journal.pgen.1004015
Van de Weyer, A. L., Monteiro, F., Furzer, O. J., Nishimura, M. T., Cevik, V., Witek, K., et al. (2019). A species-wide inventory of NLR genes and alleles in Arabidopsis thaliana. Cells 178:1260-+. doi: 10.1016/j.cell.2019.07.038
van der Burgh, A. M., Postma, J., Robatzek, S., and Joosten, M. H. A. J. (2019). Kinase activity of SOBIR1 and BAK1 is required for immune signalling. Mol. Plant Pathol. 20, 410–422. doi: 10.1111/mpp.12767
van der Hoorn, R. A., and Kamoun, S. (2008). From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20, 2009–2017. doi: 10.1105/tpc.108.060194
van Wersch, S., Tian, L., Hoy, R., and Li, X. (2020). Plant NLRs: the whistleblowers of plant immunity. Plant Commun. 1:100016. doi: 10.1016/j.xplc.2019.100016
Wan, J. R., Tanaka, K., Zhang, X. C., Son, G. H., Brechenmacher, L., Tran, H. N. N., et al. (2012). LYK4, a Lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol. 160, 396–406. doi: 10.1104/pp.112.201699
Wang, J. Z., and Chai, J. J. (2020). Structural insights into the plant immune receptors PRRs and NLRs(1)([OPEN]). Plant Physiol. 182, 1566–1581. doi: 10.1104/pp.19.01252
Wang, Y., Cordewener, J. H. G., America, A. H. P., Shan, W. X., Bouwmeester, K., and Govers, F. (2015). Arabidopsis lectin receptor kinases LecRK-IX.1 and LecRK-IX.2 are functional analogs in regulating Phytophthora resistance and plant cell death. Mol. Plant-Microbe Interact. 28, 1032–1048. doi: 10.1094/Mpmi-02-15-0025-R
Wang, Z., and Gou, X. P. (2022). The first line of defense: receptor-like protein kinase-mediated stomatal immunity. Int. J. Mol. Sci. 23:343. doi: 10.3390/ijms23010343
Wang, R. Y., Ning, Y. S., Shi, X. T., He, F., Zhang, C. Y., Fan, J. B., et al. (2016). Immunity to Rice blast disease by suppression of effector-triggered necrosis. Curr. Biol. 26, 2399–2411. doi: 10.1016/j.cub.2016.06.072
Wang, Y., Nsibo, D. L., Juhar, H. M., Govers, F., and Bouwmeester, K. (2016). Ectopic expression of Arabidopsis L-type lectin receptor kinase genes LecRK-I.9 and LecRK-IX.1 in Nicotiana benthamiana confers Phytophthora resistance. Plant Cell Rep. 35, 845–855. doi: 10.1007/s00299-015-1926-2
Wang, Y., Pruitt, R. N., Nurnberger, T., and Wang, Y. C. (2022). Evasion of plant immunity by microbial pathogens. Nat. Rev. Microbiol. 20, 449–464. doi: 10.1038/s41579-022-00710-3
Wang, G. X., Roux, B., Feng, F., Guy, E., Li, L., Li, N. N., et al. (2015). The decoy substrate of a pathogen effector and a Pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe 18, 285–295. doi: 10.1016/j.chom.2015.08.004
Wang, L. M., Wilkins, K. A., and Davies, J. M. (2018). Arabidopsis DORN1 extracellular ATP receptor; activation of plasma membrane K+-and Ca2+−permeable conductances. New Phytol. 218, 1301–1304. doi: 10.1111/nph.15111
Wei, T., Chern, M., Liu, F., and Ronald, P. C. (2016). Suppression of bacterial infection in rice by treatment with a sulfated peptide. Mol. Plant Pathol. 17, 1493–1498. doi: 10.1111/mpp.12368
Wu, Y., Gao, Y., Zhan, Y., Kui, H., Liu, H., Yan, L., et al. (2020). Loss of the common immune coreceptor BAK1 leads to NLR-dependent cell death. Proc. Natl. Acad. Sci. U. S. A. 117, 27044–27053. doi: 10.1073/pnas.1915339117
Wu, J., Reca, I. B., Spinelli, F., Lironi, D., De Lorenzo, G., Poltronieri, P., et al. (2019). An EFR-Cf-9 chimera confers enhanced resistance to bacterial pathogens by SOBIR1- and BAK1-dependent recognition of elf18. Mol. Plant Pathol. 20, 751–764. doi: 10.1111/mpp.12789
Xue, D. X., Li, C. L., Xie, Z. P., and Staehelin, C. (2019). LYK4 is a component of a tripartite chitin receptor complex in Arabidopsis thaliana. J. Exp. Bot. 70, 5507–5516. doi: 10.1093/jxb/erz313
Yaeno, T., Li, H., Chaparro-Garcia, A., Schornack, S., Koshiba, S., Watanabe, S., et al. (2011). Phosphatidylinositol monophosphate-binding interface in the oomycete RXLR effector AVR3a is required for its stability in host cells to modulate plant immunity. Proc. Natl. Acad. Sci. U. S. A. 108, 14682–14687. doi: 10.1073/pnas.1106002108
Yu, T. Y., Sun, M. K., and Liang, L. K. (2021). Receptors in the induction of the plant innate immunity. Mol. Plant-Microbe Interact. 34, 587–601. doi: 10.1094/Mpmi-07-20-0173-Cr
Yuan, M., Jiang, Z., Bi, G., Nomura, K., Liu, M., Wang, Y., et al. (2021a). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109. doi: 10.1038/s41586-021-03316-6
Yuan, M., Ngou, B. P. M., Ding, P., and Xin, X. F. (2021b). PTI-ETI crosstalk: an integrative view of plant immunity. Curr. Opin. Plant Biol. 62:102030. doi: 10.1016/j.pbi.2021.102030
Yue, Z. L., Liu, N., Deng, Z. P., Zhang, Y., Wu, Z. M., Zhao, J. L., et al. (2022). The receptor kinase OsWAK11 monitors cell wall pectin changes to fine-tune brassinosteroid signaling and regulate cell elongation in rice. Curr. Biol. 32, 2454–2466.e2457. doi: 10.1016/j.cub.2022.04.028
Zhai, K. R., Deng, Y. W., Liang, D., Tang, J., Liu, J., Yan, B. X., et al. (2019). RRM transcription factors interact with NLRs and regulate broad-Spectrum blast resistance in Rice. Mol. Cell 74:996. doi: 10.1016/j.molcel.2019.03.013
Zhang, N., Pombo, M. A., Rosli, H. G., and Martin, G. B. (2020). Tomato Wall-associated kinase SlWak1 depends on Fls2/Fls3 to promote Apoplastic immune responses to Pseudomonas syringae. Plant Physiol. 183, 1869–1882. doi: 10.1104/pp.20.00144
Zhao, M., Li, N. H., Chen, S. M., Wu, J. J., He, S. F., Zhao, Y. X., et al. (2023). GmWAK1, Novel Wall-associated protein kinase, positively regulates response of soybean to Phytophthora sojae infection. Int. J. Mol. Sci. 24:798. doi: 10.3390/ijms24010798
Zhao, L., Liao, Z. W., Feng, L. P., An, B., He, C. Z., Wang, Q. N., et al. (2023). Colletotrichum gloeosporioides Cg2LysM contributed to virulence toward rubber tree through affecting invasive structure and inhibiting chitin-triggered plant immunity. Front. Microbiol. 14:1129101. doi: 10.3389/fmicb.2023.1129101
Zhou, J. M., and Zhang, Y. (2020). Plant immunity: danger perception and signaling. Cells 181, 978–989. doi: 10.1016/j.cell.2020.04.028
Keywords: hemibiotrophic pathogen, cell surface pattern recognition receptor, intracellular immune receptor, effector, plant immunity
Citation: Zhou D, Chen X, Chen X, Xia Y, Liu J and Zhou G (2023) Plant immune receptors interact with hemibiotrophic pathogens to activate plant immunity. Front. Microbiol. 14:1252039. doi: 10.3389/fmicb.2023.1252039
Received: 03 July 2023; Accepted: 20 September 2023;
Published: 09 October 2023.
Edited by:
Jian-Wei Guo, Kunming University, ChinaReviewed by:
Satyabrata Nanda, Centurion University of Technology and Management, IndiaCopyright © 2023 Zhou, Chen, Chen, Xia, Liu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoying Zhou, emd5aW5ncXFAMTYzLmNvbQ==; Junang Liu, a2pjOTYyMEAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.