- 1Department of Applied Microbiology, Lixiahe District Institute of Agricultural Sciences in Jiangsu/National Agricultural Experimental Station for Agricultural Microbiology in Yangzhou, Yangzhou, China
- 2Department of Biochemistry, School of Biological Sciences, University of Sussex, Brighton, United Kingdom
- 3State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
Cell wall hydrolases are ubiquitous among spore-form bacteria and essential for mother cell lysis. In this study, a novel cell wall hydrolase gene cwlE involved in mother cell lysis was characterized from Bacillus thuringiensis subsp. israelensis (Bti) strain Bt-59. cwlE was specifically expressed in Bti and located in the large plasmid carrying the insecticidal genes. The encoded CwlE protein consists of a MurNAc-LAA domain and two highly conserved catalytic residues (E26 and E151). The recombinant CwlE-His protein was able to digest the cell wall of Bti, indicating that CwlE is an N-acetylmuramoyl-L-alanine amidase. Transcriptional analysis indicated that cwlE began to express at the early stage of stationary phase and was controlled by SigE. Single mutation of cwlE gene delayed Bti mother cell lysis, while double mutation of cwlE and sigK completely blocked Bti mother cell lysis. After exposure to UV light to deactivate the crystal proteins, the level of decrease of insecticidal activity against mosquito larvae of Bt-59 (ΔcwlE-sigK) was less than that observed for Bt-59. This study elucidates the mechanism of Bti mother cell lysis and provides an effective strategy for mosquito control using Bt products with increased persistence.
Introduction
Mosquitoes are tropical disease vectors for transmitting malaria, dengue, filariasis, and yellow fever, posing an enormous public health menace (Reisen, 2010; Zhang et al., 2017). Although chemical pesticides are the most widespread way to control mosquitoes, they are losing the advantage due to pest resistance and environmental pollution (Maciel-de-Freitas et al., 2014; Omotayo et al., 2022). Bacillus thuringiensis subsp. israelensis (Bti) is a gram-positive bacterium that forms a parasporal crystal composed of protein protoxins with specific insecticidal activity against dipteran larvae (Federici et al., 2013; Ben-Dov, 2014). Long-term exposure of mosquito larvae to Bti did not evolve into resistance to this microbial larvicide, which supports its sustainable use for integrated control practices (Carvalho et al., 2018). The low probability of development of resistance to Bti is attributed to the synergistic interaction among its four main Cry toxins, namely Cry4Aa, Cry4Ba, Cry11Aa, and Cyt1A (Vasquez et al., 2009). However, the main disadvantage of Bti is its short efficacy duration against mosquito larvae, limiting its widespread use as a larvicide.
Like other B. thuringiensis (Bt) strains, the durable spore and the parasporal crystal are set free with Bti mother cell lysis at the late sporulation stage (Ben-Dov, 2014). Then, the exposed insecticidal crystal proteins (ICPs) are easily inactivated by environmental factors, especially solar irradiation (Ramirez-Lepe et al., 2003; Zogo et al., 2019). An asporogenic Bt strain that expressed a chimeric cry1C/Ab gene showed high potency against various lepidopteran pests, and parasporal crystals produced by the strain remained encapsulated within the mother cells, which protected them from UV light degradation (Sanchis et al., 1999). Therefore, it is feasible to achieve the protection of ICPs from solar inactivation by knocking down the crucial cell wall hydrolase gene to block Bti mother cell lysis.
Three cell wall hydrolase genes (cwlB, cwlC, and cwlH) were identified in Bacillus subtilis, a well-established model for mother cell lysis (Vollmer et al., 2008). Except for the expression of cwlB in vegetative growth stage, cwlC and cwlH were expressed during the sporulation phase (Foster, 1992; Kuroda et al., 1993; Nugroho et al., 1999). Deleting of a single hydrolase gene did not affect mother cell lysis, but both cwlB-cwlc and cwlB-cwlH mutations blocked mother cell lysis of B. subtilis (Nugroho et al., 1999). While in B. thuringiensis subsp. kurstaki (Btk) strain HD73, the deletion of cwlC completely blocked Btk mother cell lysis, which lead to ICPs encapsulated in the mother cell (Chen et al., 2018). Based on the previous study, although CwlC of Bti showed 97% amino acid sequence identity with that of Btk, the deletion of cwlC just delayed the lysis of mother cells in Bti (Huang et al., 2022). The role of CwlC from Bti performed during the mother cell lysis was quite different from that of Btk, indicating that the hydrolysis of Bti mother cell wall involves a complex lysase system.
In the previous study, three cell wall hydrolase genes (cwlB, cwlC, and cwlE) were identified in the genome of Bti strain Bt-59 (Huang et al., 2022). cwlB and cwlC were regulated by SigK, a critical regulatory factor in the sporulation phase regulating genes involved in the formation of the outer wall of the spore, mother cell lysis, and spore germination (Zheng and Losick, 1990; Nugroho et al., 1999; Huang et al., 2022). The sigK and cwlC mutation strains of Bti were constructed in the previous study. As the key Sigma factor in regulating spore maturation and germination, the absence of sigK inhibits spore formation in Bti (Peng et al., 2016; Xu et al., 2020). Due to the major insecticidal genes of Bti are controlled by the sporulation-specific sigma factor SigE, the deletion of sigK did not have a significant impact on the production of ICPs in Bt-59 (Deng et al., 2014; Xu et al., 2020). In addition, the deletion of sigK or cwlC just delayed but not completely blocked Bti mother cell lysis (Xu et al., 2020; Huang et al., 2022). Moreover, cwlE was the only hydrolase gene with high expression level in the sigK mutant of Bti, indicating that cwlE was not controlled by SigK and CwlE was a critical autolysin leading to Bti mother cells lysis (Huang et al., 2022). Based on these studies, the double mutation of cwlE and sigK is likely to block Bti mother cell lysis without affecting its gene expression of ICPs.
This work analyzed the function and transcriptional regulation of cwlE, a new cell wall hydrolase gene of Bti. The cwlE mutation strain was constructed and the deletion of cwlE delayed Bti mother cell lysis. In addition, an ICPs encapsulation strain of Bti was constructed by deleting both sigK and cwlE. The result of insecticidal activity analysis showed that the sigK-cwlE mutant strain had better tolerance to ultraviolet than the Bti wild strain. Based on the characteristics analysis of CwlE and its role in mother cell lysis, we elucidated the mechanism of Bti mother cell lysis and constructed an ICPs encapsulation Bti strain with the improvement of insecticidal persistence.
Materials and methods
Bacterial strains, plasmids, and culture conditions
The strains and plasmids used in this study were showed in Table 1. Escherichia coli strains Top10, ET12567, and BL21 (DE3) were used for molecular cloning, extracting non-methylated vectors, and protein expression, respectively (Wang et al., 2006, 2022). Bti strain Bt-59 (CGMCC strain number 16821) and Btk strain HD73 (BGSC strain number BGSC 4D4) were used to clone the target gene, for transformations and monitor promoter activity. E. coli strains were cultured at 37°C in Luria-Bertani (LB) broth medium with 100 μg/mL ampicillin (Amp) if needed. Bt strains were cultured at 30°C in Schaeffer’s sporulation Medium (SSM, 8 g L−1 nutrient broth, 0.12 g L−1 MgSO4, 1 g L−1 KCl, 0.5 mmol L−1 NaOH) with 25 μg/mL erythromycin (Ery) if needed (Schaeffer et al., 1965).
DNA manipulation
Primers used in this study were synthesized in GENERAL BIOL (Hefei, China) and showed in Supplementary Table S1. PCR was performed using PrimeSTAR HS DNA polymerase (TaKaRa, Beijing, China) with corresponding primers. DNA fragments were purified with the TIANgel Midi Purification Kit (TIANGEN, Beijing, China) and plasmids were purified with the TIANprep Mini Plasmid Kit (TIANGEN, Beijing, China). The ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China) was used to clone the DNA fragments into the linearized vectors. Plasmids were introduced into the Bt cells by electroporation as previously described (Huang et al., 2022).
Strain construction
The temperature-sensitive suicide plasmid pMAD with the cwlE mutation sequence was constructed to delete the cwlE gene in the Bt-59 genome (Supplementary Figure S1). A 908-bp fragment upstream of cwlE (fragment A) was amplified by PCR, and cwlE-1 and cwlE-2 were used as primers. A 1013-bp fragment downstream of cwlE (fragment B) was amplified by PCR, and cwlE-3 and cwlE-4 were used as primers. The cwlE mutation box was amplified using fragments A and B as templates and cwlE-1 and cwlE-4 as primers by overlapping PCR. The resulting fragment was cloned into the pMAD plasmid to generate pMADΔcwlE plasmid. Then, the recombinant plasmid pMADΔcwlE was transformed into Bt-59 and Bt-59 (ΔsigK) to generate the cwlE deletion mutant Bt-59 (ΔcwlE) and the sigK and cwlE double mutant Bt-59 (ΔcwlE-sigK).
A 2693-bp fragment containing the promoter, ORF, and terminator sequences of cwlE was amplified by PCR using HFcwlE-1 and HFcwlE-2 as primers and cloned into the pHT315 shuttle vector to generate pHTHFcwlE plasmid. Then, the recombinant plasmid pHTHFcwlE was transformed into Bt-59 (ΔcwlE), Bt-59 (ΔcwlE-sigK), and HD (ΔcwlC) to generate complemented strains Bt-59 (ΔcwlE::cwlE), Bt-59 (ΔcwlE-sigK::cwlE), and HD (ΔcwlC::cwlE), respectively.
To further verify the hydrolytic activity of CwlE, an 822-bp fragment of cwlE gene was amplified by PCR using pET-cwlE-F and pET-cwlE-R as primers and cloned into the pET28a plasmid. Site-directed mutagenesis of cwlE (E24A and E151A) was performed with the Fast Mutagenesis System kit (TransGen, Beijing, China). Then, the recombinant plasmid pETcwlE, pETcwlE-E24A, and pETcwlE-E151A were transformed into BL21 to generate strains BL21(pETcwlC), BL21(pETcwlC-E24A), and BL21(pETcwlC-E151A) for protein expression. Besides, the recombinant plasmid pHTHFcwlE was transformed into HD (ΔcwlC) generating complemented strain HD (ΔcwlC::cwlE) to analyze the degradation activity of CwlE on Btk cell wall.
A 767-bp promoter region upstream of the cwlE gene (P cwlE ) was cloned into the linearized vector pHT304-18Z to construct a cwlE reporter vector. Then, the recombinant plasmids pHTP cwlE was transformed into cells of Bt-59, HD73, and HD (ΔsigE), generating strains Bt-59 (P cwlE -lacZ), HD (P cwlE -lacZ), and HD (ΔsigE) (P cwlE -lacZ), respectively. Besides, the recombinant plasmid pHTHFcwlE was transformed into HD73 and HD (ΔsigE) generating strains HD (cwlE) and HD (ΔsigE) (cwlE) to analyze the effect of sigE deletion on the expression level of cwlE.
Expression and cell wall hydrolysis of recombinant CwlE protein
After strains BL21(pETcwlE), BL21(pETcwlC-E24A), and BL21(pETcwlC-E151A) were grown in LB liquid medium (Amp, 100 μg/mL) at 37°C for 4 h with shaking at 220 rpm, 0.5 mM isopropyl-beta-D-thiogalactoside (Sangon, Shanghai, China) was added. The bacteria were cultured at 16°C for another 14 h with shaking at 160 rpm to induce protein expression. Then, the recombinant CwlE proteins with a His tag were purified according to the method described previously (Zhang et al., 2020). After grown in LB liquid medium at 30°C for 6 h with shaking at 220 rpm, Bt-59 was collected by centrifugation at 10000 rpm. Then, the bacteria were resuspended with TK buffer (0.05 M Tris–HCl, 0.05 M KCl, pH 7.0) and disrupted with a FastPrep-24 (MPBiomedicals). Unbroken cells were removed by low-speed centrifugation (3,500 rpm, 5 min). The crude cell walls were pelleted at 13500 rpm for 5 min at 4°C. Finally, the pellets were resuspended in TK buffer. The purified CwlE proteins were added to the prepared cell wall of Bt-59 for hydrolytic activity analysis (Huang et al., 2022). After incubated at 37°C, the OD540 of the mixtures were measured at the designated time point (Nugroho et al., 1999).
Analysis of the transcription start site
Total RNA was extracted from Bt-59 cultured in SSM at T15 with TRIzol reagent (Invitrogen, USA). The transcription start site of the cwlE gene was determined with 5’ RACE according to the manufacturer’s instruction of HiScript-TS 5′/3’ RACE Kit (Vazyme, Nanjing, China). The primers used for 5’ RACE are listed in Supplementary Table S1.
β-Galactosidase assays
Bt strains HD (P cwlE -lacZ) and HD (ΔsigE) (P cwlE -lacZ) were cultured in SSM (Ery, 25 μg/mL) at 30°C with shaking at 220 rpm. Two-milliliter bacterial suspensions of HD (P cwlE -lacZ) and HD (ΔsigE) (P cwlE -lacZ) were collected from T0 to T20 at 1-h intervals (T0 is the end of the exponential phase and Tn is n hours after the end of the exponential phase). The cells were harvested and the samples were prepared as previously described (Chen et al., 2018). Then, the specific β-galactosidase activity was measured and expressed as Miller units per milligram of protein (Chen et al., 2018). The values are the means for three independent experiments.
Quantitative real-time PCR (qPCR)
Total RNA was extracted from Bt strains cultured in SSM at T0, T8 and T16 with TRIzol reagent (Invitrogen, USA). T0 is the end of the exponential phase and Tn is n hours after the end of the exponential phase. The purified RNA was reverse transcribed into cDNA using HiScript IV RT SuperMix for qPCR (gDNA wiper) (Vazyme, Nanjing, China). RpsU and gatB were chosen as reference genes (Reiter et al., 2011). qPCR was performed using specific primers (Supplementary Table S1) and ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) with a 7,500 Fluorescence Quantitative PCR Instrument (ABI, California, USA). The specificity and qualified amplification efficiency of the specific primers were verified. Three biological replicates were designed. The relative expression level of each target gene was calculated according to the 2-ΔCT method (Huggett et al., 2013).
Assays of vegetative growth and Cry proteins production
The wild-type strain Bt-59, cwlE deletion mutant Bt-59 (ΔcwlE), and double mutant Bt-59 (ΔcwlE-sigK) were cultured in SSM medium at 30°C with shaking at 220 rpm. The growth curves of Bti strains were drew by measuring the OD600 values of the culture solutions every hour. After 12 h of cultivation, the number of colonies of Bt-59 and its mutants were calculated by the coating plate method. SDS-PAGE was used for the crystal protein production assay as described previously (Xu et al., 2020).
Microscopic analysis
Bt strains were cultured in SSM medium at 30°C with shaking at 220 rpm to the certain time points. Samples were collected and centrifuged, and then sediments were suspended with the appropriate volume of deionized water. Bacterial suspension was placed on a glass slide, and then cell morphology was observed by optical microscopy (ZEISS, Germany).
Bioassay of insecticidal activity
Bt-59 and Bt-59 (ΔcwlE-sigK) were cultured in a fermentation medium at 30°C with shaking at 220 rpm for 44 h as described previously (Huang et al., 2022). The fermentation media were serially diluted with water to the appropriate concentrations and used for the bioassay. Twenty larvae of Culex pipiens and Aedes albopictus in the early fourth instar were collected for bioassay in each treatment (Huang et al., 2022). Then the fermentation media were exposure to 365 nm UV light for 30 min used for the bioassay (Patel et al., 1996). The experiment was performed with three replicates at 28°C in a biochemical incubator for 24 h. The distilled water was used as a control.
Sequence and data analysis
Vector NTI 11 and GENEDOC were used for sequence analysis. The primer design was performed by Primer Premier 5.0. The phylogentic tree was constructed with MEGA Microsoft Office Excel and GraphPad Prism 6 were used for data analysis. PoloPlus statistical software was used for estimating the LC50 values and 95% confidence intervals (CIs) (Straus, 2008). Lethal dose ratios (LC50 ratio, LCR) were calculated to compare natural variation between different treatments, and the LCR value was considered to be significant (p < 0.05) when the confidence intervals did not encompass the value 1 (Straus, 2008).
Results
Bioinformatic analysis of CwlE
To identify the essential cell wall hydrolase in Bti mother cell lysis, a novel hydrolase gene named cwlE was identified from a 128 kb sized plasmid (CP039725.1) encoding multiple insecticidal protein genes of the strain Bt-59. cwlE is 816 base pairs in length and encodes a putative cell wall hydrolase with the MurNAc-LAA domain and two highly conserved catalytic residues E26 and E151 (Figure 1). The amino acid sequence identity showed CwlE shares 32% of its amino acid sequence identity with CwlM of B. licheniformis, 28% with CwlC of Bt-59, and 33% with CwlC of B. subtilis. The absence of a homologous sequence of cwlE in the genomes of bacteria other than Bti strains, as determined by the NCBI homology analysis, suggests that cwlE is a cell wall hydrolase gene uniquely expressed in Bti.

Figure 1. Bioinformatic analysis of cell wall hydrolases. MurNAc-LAA (red frame) represents the N-acetylmuramoyl-L-alanine amidase domain. The two conserved critical glutamic acid residues are marked with black asterisk. Bs and Bl are short for Bacillus subtilis and B. licheniformis, respectively. The NCBI accession number for CwlE (Bt-59), CwlC (Bt-59), CwlC (HD73), CwlC (Bs), and CwlM (Bl) are QCJ54933.1, QCJ49661.1, NC_020238.1, NP_389623.2, and P37134.1, respectively.
Hydrolytic activity against cell wall of CwlE
To investigate the function of CwlE in Bti, the recombinant CwlE-His protein was purified using nickel column affinity chromatography. To determine whether the conserved glutamic acids are essential for enzymatic catalytic activity, two distinct single amino acid substitutions (E26A and E151A) were separately introduced into the CwlE-expressing strains using site-directed mutagenesis. SDS-PAGE showed that the molecular weight of the recombinant CwlE and its mutant proteins is approximately 35 kDa (Figure 2A). Then, mixtures of the recombinant proteins and the Bti cell wall were incubated at 37°C to determine the cell wall lytic ability of the CwlE and its mutant proteins. After incubation of CwlE with the cell wall for 60 min, the optical density (OD) of the mixture decreased by about 24% (Figure 2B). In contrast to the wild-type CwlE protein, the two mutant CwlE proteins (E26A and E151A) exhibited no catalytic activity against the Bti cell wall (Figure 2B).
Figure 2. Expression of CwlE and its hydrolytic activity of cell wall. (A) SDS-PAGE analysis of recombinant CwlE and its mutant proteins purified by nickel column affinity chromatography. M, protein molecular size marker. CwlE, wild-type CwlE protein; E26A, replacement of glutamic acid with alanine of CwlE protein at position 24; E151A, replacement of glutamic acid with alanine of CwlE protein at position 151. (B) Digestion of Bacillus thuringiensis subsp. israelensis cell walls by purified CwlE and its mutant proteins. Cell walls incubated with addition of BSA protein were used as control. Each value represents the mean of at least three independent replicates. Error bars show standard deviations. (C) Mother cell lysis of HD (ΔcwlC::cwlE) was observed by optical microscopy after treatment with biological staining agent basic red at T18, T22, T26, and T30. T0 is the end of the exponential phase and Tn is n hours after the end of the exponential phase. Green arrows, red arrows, and blue arrows represent vegetative cells, spores, and crystal proteins, respectively. Scale bars, 5 μm.
After 15 days of growth in SSM medium the mother cells of Btk mutant strain HD (ΔcwlC) did not autolyze in a previous study (Chen et al., 2018). The cwlE gene containing its promoter region sequence was transferred into HD (ΔcwlC) strain to further determine the cell wall lytic ability of CwlE. Cells of HD (ΔcwlC::cwlE) began to autolyze at T22 and completely autolyzed after 8 h of continuous culture (Figure 2C). The result indicated that the promoter of cwlE gene showed transcriptional activity in Btk, and CwlE could hydrolyze cell wall of HD73 and restore the phenotype of HD (ΔcwlC).
Transcriptional regulation of the cwlE gene
The result of 5’ RACE-PCR experiment showed that the transcription start site of cwlE gene is a T located 97 nucleotides upstream of its translational start codon ATG (Figure 3A). The putative cwlE promoter contains the sequences TAATATCT and AATAAAAT, which are located −35 and − 10 nucleotides upstream of the cwlE transcription start site, respectively (Figure 3A). These sequences are similar to those in the transcriptional regulatory factor SigE dependent promoters (KHATANHT and MATANNHT; H is A, C, or T; K is G or T; M is A or C; N is A, C, G, or T.) (Du et al., 2012), suggested that the cwlE promoter might be recognized and the expression of the cwlE gene was regulated by SigE.
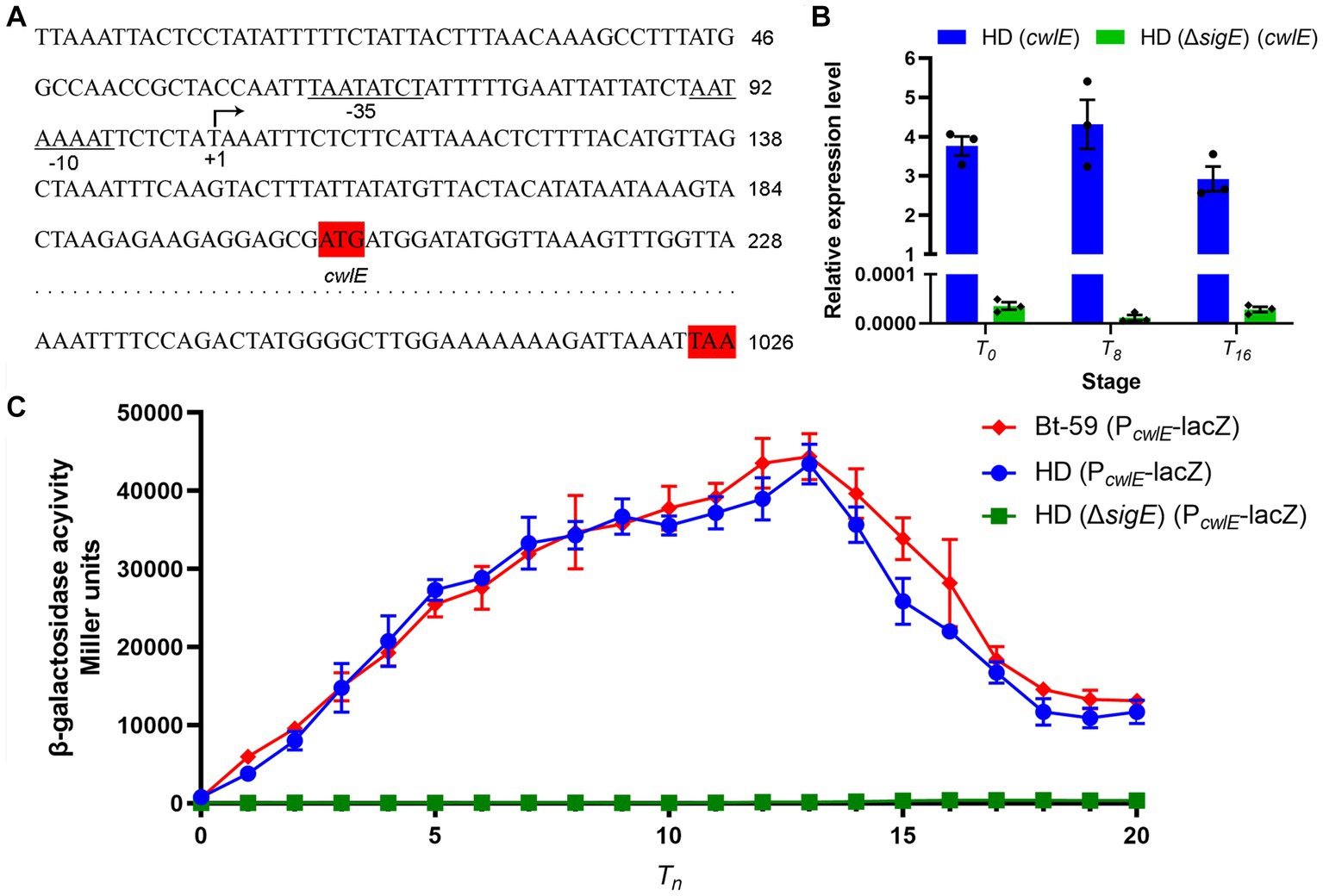
Figure 3. Analysis of promoter sequence and transcriptional regulation of the cwlE gene. (A) Sequence analysis of the promoter regions from Bt-59 cwlE. The predicted transcription start site T is denoted with arrow, while the putative −35 and − 10 motifs are underlined and denoted. The translation start codon and ending codon of cwlE are marked shaded in red. (B) Expression level of cwlE gene in HD (cwlE) and HD (ΔsigE) (cwlE) at T0, T8, and T16. Each value represents the mean of at least three independent replicates. Error bars represent the standard error of the mean. T0 is the end of the exponential phase and Tn is n hours after the end of the exponential phase. (C) Assays of β-galactosidase activity of the cwlE promoter in Bt-59, HD73, and HD (ΔsigE). T0 is the end of the exponential phase and Tn is n hours after the end of the exponential phase. Each value represents the mean of at least three independent replicates. The error bars represent standard deviations.
The results of qPCR showed that the cwlE gene containing its promoter could be expressed in HD73, while it was almost undetectable in HD (ΔsigE) (Figure 3B). To further determine whether cwlE is regulated in a SigE-dependent manner, the Bt-59 (P cwlE -lacZ), HD (P cwlE -lacZ), and HD (ΔsigE) (P cwlE -lacZ) strains were constructed. β-Galactosidase activity assays showed that reporter vector was expressed both in Bt-59 and HD73, but not in sigE mutant strain of HD73 (Figure 3C). The result showed that the cwlE promoter could be recognized by SigE in Bti and Btk, and the expression of cwlE was controlled by SigE. In Bt-59, the expression of the reporter vector started at T0, reached a maximum at T13, and then decreased in the following time (Figure 3C). Furthermore, the transcriptional activity of the cwlE promoter did not differ significantly between Bt-59 and HD73 (Figure 3C).
Effect of the cwlE and cwlE-sigK deletion on the vegetative growth and Cry gene expression
Both the cwlE deletion mutant Bt-59 (ΔcwlE) and the cwlE-sigK double deletion mutant Bt-59 (ΔcwlE-sigK) have been constructed and verified (Figure 4A; Supplementary Figure S2). The growth curve of Bti strains during the nutritional period in SSM medium revealed that cwlE deletion or cwlE and sigK double deletion did not affect the nutritional growth of Bt-59 (Figure 4B). After being cultured in SSM medium for 12 h, the total number of cells of Bt-59, Bt-59 (ΔcwlE), Bt-59 (ΔsigK), and Bt-59 (ΔcwlE-sigK) were 2.3 × 108 CFU/mL, 2.4 × 108 CFU/mL, 1.5 × 108 CFU/mL, and 1.3 × 108 CFU/mL, respectively (Figure 4C). Unlike cwlE single mutation, cwlE and sigK double mutation affected the cells number of Bt-59, similar to sigK single mutation constructed previously (Xu et al., 2020).
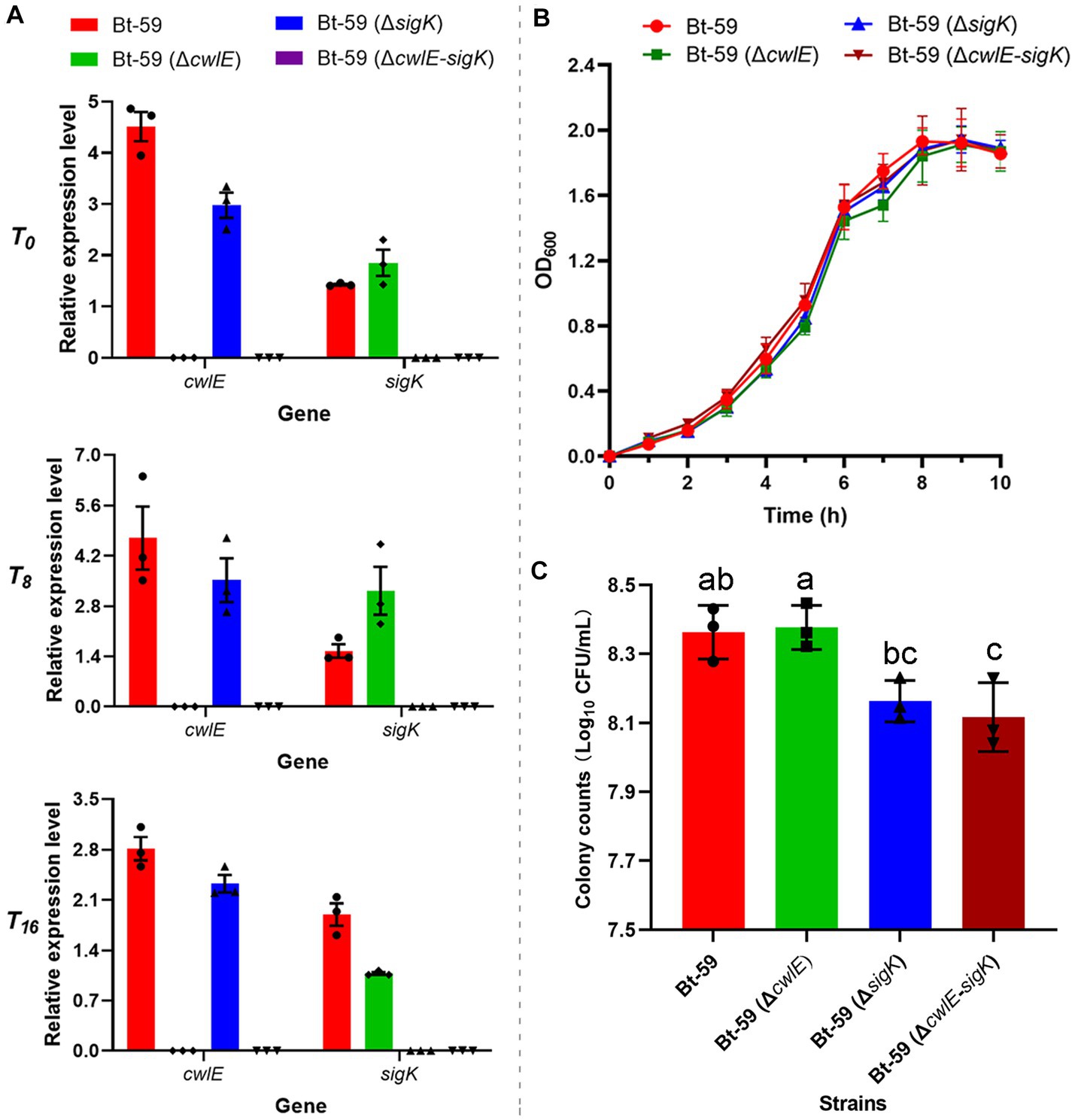
Figure 4. Effect of the cwlE and cwlE-sigK deletion on the vegetative growth and Cry proteins production of Bt-59. (A) The cwlE deletion mutants were verified by qPCR. Each value represents the mean of at least three independent replicates. Error bars represent the standard error of the mean. T0 is the end of the exponential phase and Tn is n hours after the end of the exponential phase. (B) Growth curves of Bt-59, Bt-59 (ΔcwlE), and Bt-59 (ΔcwlE-sigK). Each value represents the mean of at least three independent replicates. The error bars represent standard deviations. (C) After culturing in SSM medium for 10 h, the number of Bt-59, Bt-59 (ΔcwlE), and Bt-59 (ΔcwlE-sigK) cells was detected by viable count. Each value represents the mean of at least three independent replicates. The error bars represent standard deviations. Statistically significant difference analysis was performed by ordinary one-way ANOVA (p < 0.05).
The expression levels of four main insecticidal protein genes (cry4Aa, cry4Ba, cry11Aa, and cyt1Aa) in Bt-59, Bt-59 (ΔcwlE), Bt-59 (ΔsigK), and Bt-59 (ΔcwlE-sigK) were revealed by qPCR (Figure 5). Compared with the wild strain, the deletion of cwlE or sigK did not affect the expression of cry4Aa and cry4Ba at T0, T8, and T16. cry11Aa showed similar expression levels in the four strains at T0 and T8, while its expression was reduced in the mutants Bt-59 (ΔsigK) and Bt-59 (ΔcwlE-sigK) at T16. The deletion of cwlE did not affect the expression of cyt1Aa, while the expression of cyt1Aa was reduced in cwlE and sigK double mutation strain Bt-59 (ΔcwlE-sigK). Besides, Cry proteins production of Bt-59 and its mutants were examined by SDS-PAGE after being cultured in SSM medium for 24 h. The results showed that deletion of cwlE had no effect on Cry proteins production in Bti, while the cwlE-sigK deletion decreased the expression of Cry11Aa (Supplementary Figure S3).
Figure 5. The expression levels of cry4Aa, cry4Ba, cry11Aa, and cyt1Aa in Bt-59, Bt-59 (ΔcwlE), Bt-59 (ΔsigK), and Bt-59 (ΔcwlE-sigK) at T0, T8, and T16 were revealed by qPCR. Each value represents the mean of at least three independent replicates. Error bars represent the standard error of the mean. T0 is the end of the exponential phase and Tn is n hours after the end of the exponential phase. Statistically significant difference analysis was performed by ordinary one-way ANOVA (p < 0.05).
Deletion of the cwlE gene delayed mother cell lysis
Optical microscopy was used to investigate the morphology of the Bt-59 and Bt-59 (ΔcwlE) grown in SSM medium to various growth phases (Figure 6). The result showed that cells of Bt-59 began to lyse at T16, while cells of Bt-59 (ΔcwlE) began to lyse at T20. Up to T24, the cells of Bt-59 were completely lysed, and the most of the cells of Bt-59 (ΔcwlE) were lysed. After another 4 hours of culture, the cells of Bt-59 (ΔcwlE) were completely lysed. In addition, the cells of cwlE gene restoration strain Bt-59 (ΔcwlE::cwlE) began to autolyze at T16, and virtually all of the cells were lysed at T24, which was similar to Bt-59.
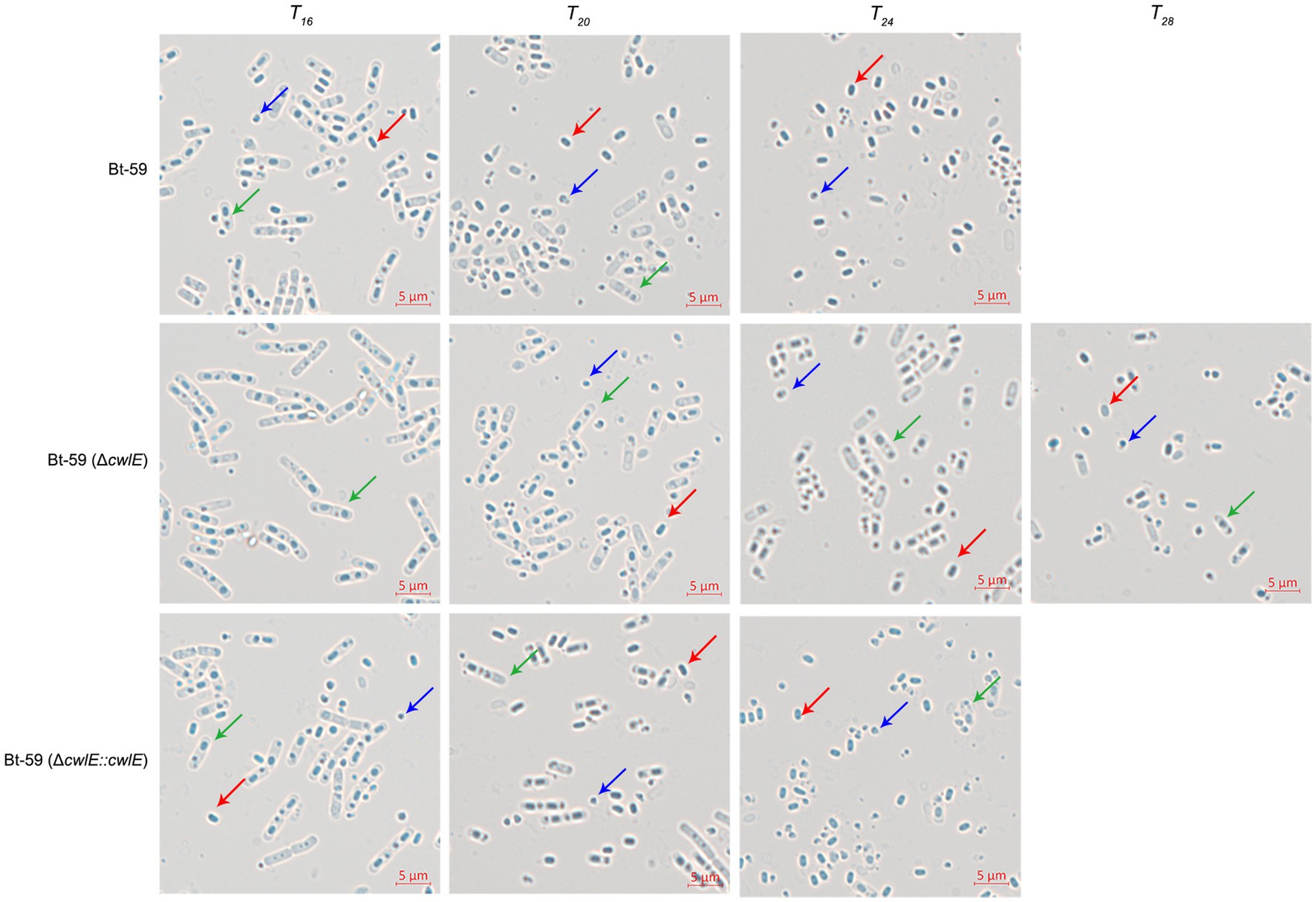
Figure 6. Effect of cwlE deletion on mother cell lysis observed by optical microscopy. Mother cell lysis of Bt-59 and Bt-59 (ΔcwlE::cwlE) was observed at T16, T20, and T24. Mother cell lysis of mutant strain Bt-59 (ΔcwlE) was observed at T16, T20, T24 and T28. T0 is the end of the exponential phase and Tn is n hours after the end of the exponential phase. Green arrows, red arrows, and blue arrows represent vegetative cells, spores, and crystal proteins, respectively. Scale bars, 5 μm.
cwlE and sigK double mutant completely blocked mother cell lysis
To further verify the role of cwlE in mother cell lysis of Bti, the cell wall hydrolase gene cwlE and the transcription regulatory factor gene sigK double deletion mutant Bt-59 (ΔcwlE-sigK) was constructed based on the sigK mutant strain. Optical microscopy was used to examine the morphology of Bt-59 (ΔsigK) and Bt-59 (ΔcwlE-sigK) grown in SSM medium to various growth phases (Figure 7A). The result showed that parts of cells of Bt-59 (ΔsigK) were lysed at T48. Up to T72, the cells of Bt-59 (ΔsigK) were completely lysed, whereas mother cell lysis of Bt-59 (ΔcwlE-sigK) could not be observed. The cell morphology of the complemented strain Bt-59 (ΔcwlE-sigK::cwlE) was comparable to that of the sigK mutant strain. Furthermore, Bt-59 (ΔcwlE-sigK) was cultured in SSM for 7 days to further examine the mother cell lysis phenotype. The results showed that the cells of Bt-59 (ΔcwlE-sigK) still did not autolyze after 7 days of growth (Figure 7B).
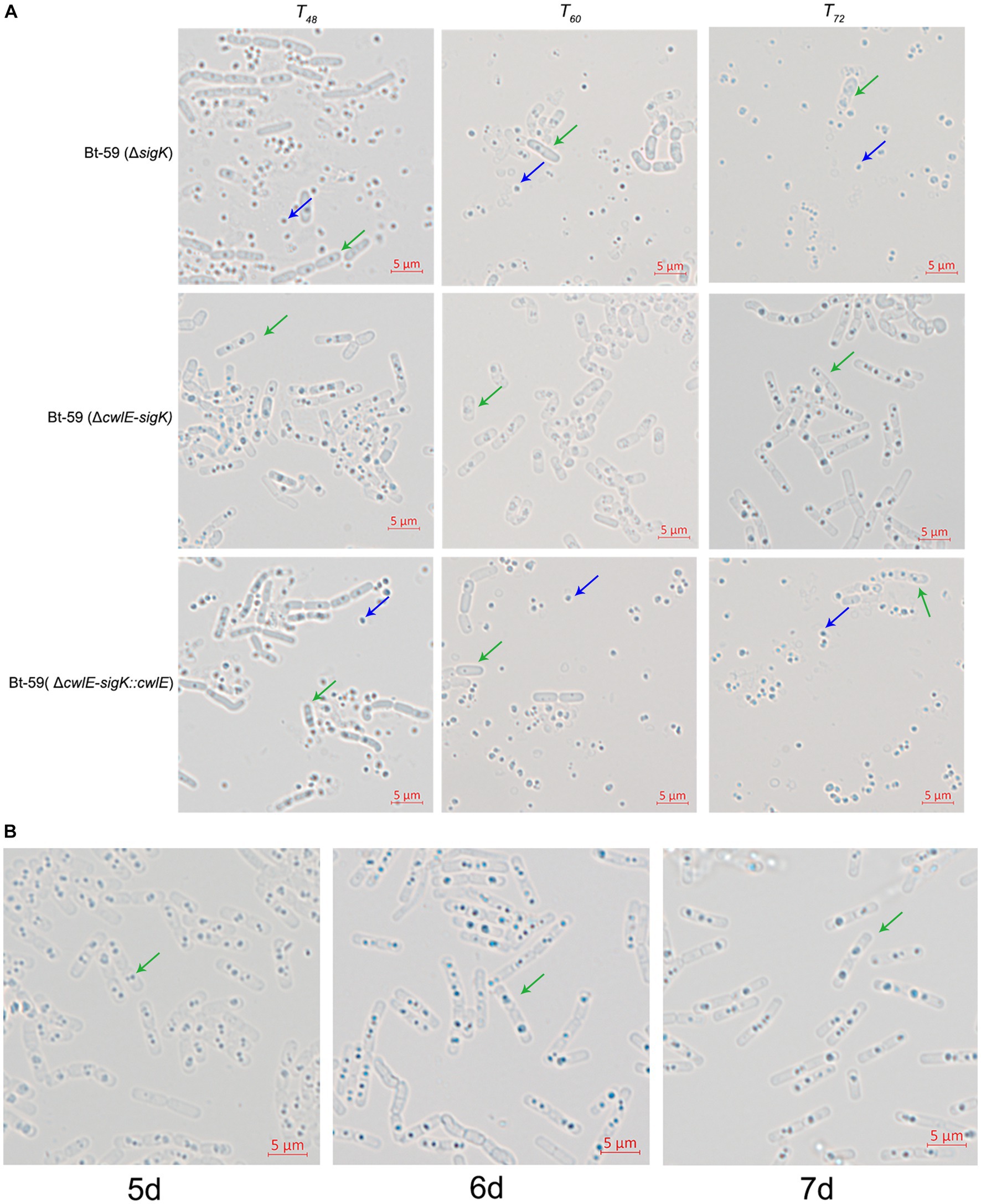
Figure 7. Effect of cwlE and sigK double deletion on mother cell lysis observed by optical microscopy. (A) Mother cell lysis of Bt-59 (ΔsigK), Bt-59 (ΔcwlE-sigK), and Bt-59 (ΔcwlE-sigK::cwlE) were observed at T48, T60, and T72. T0 is the end of the exponential phase and Tn is n hours after the end of the exponential phase. Green arrows and blue arrows represent vegetative cells and crystal proteins, respectively. Scale bars, 5 μm. (B) Mother cell lysis of mutant strain Bt-59 (ΔcwlE-sigK) was observed at 5 d, 6 d, and 7d. Green arrows represent vegetative cells. Scale bars, 5 μm.
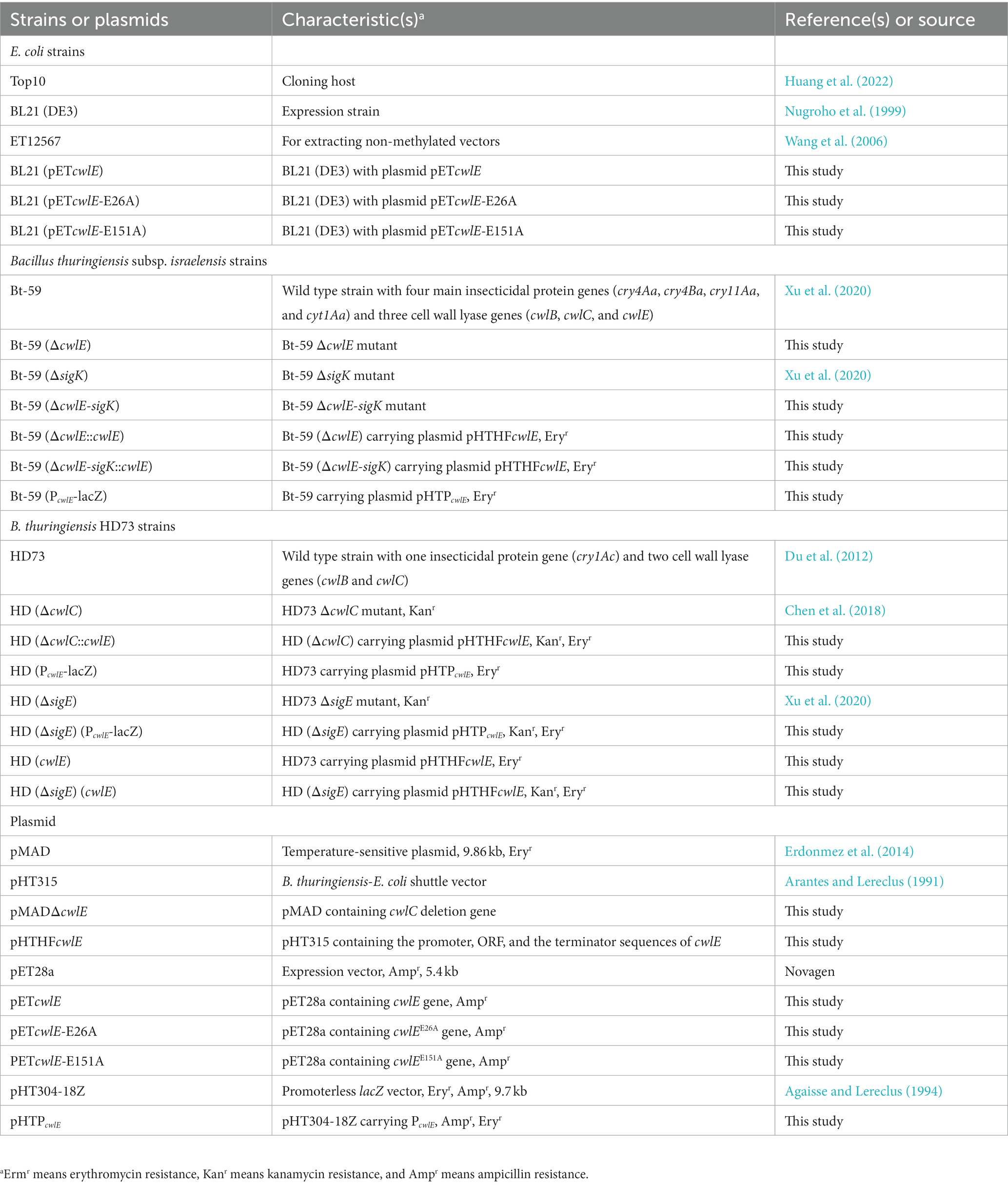
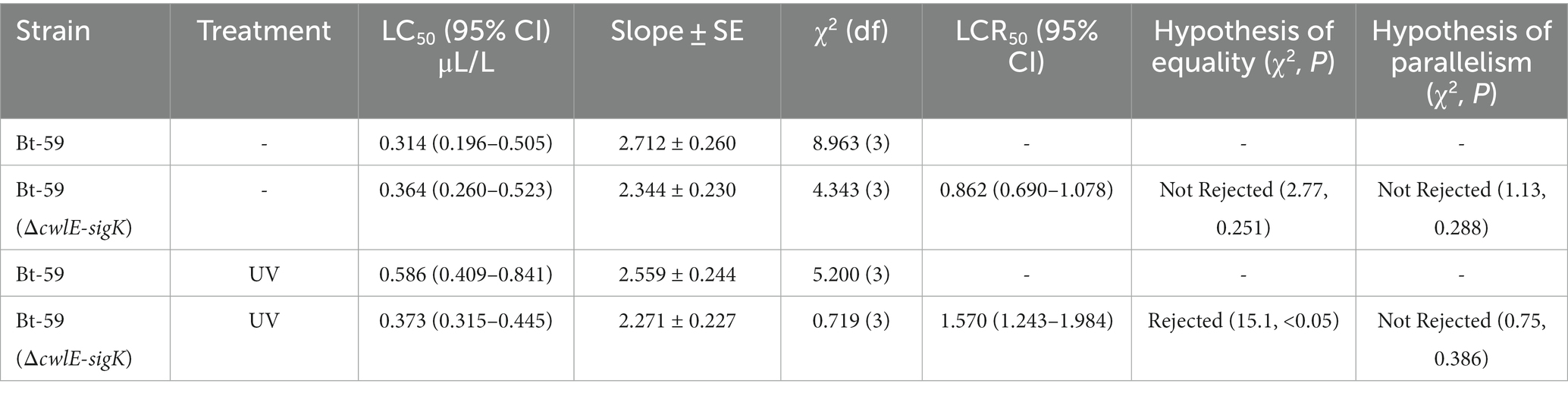
Insecticidal activity against mosquito larvae
The early fourth-instar larvae of Culex pipiens and Aedes albopictus were fed with fermentation suspension serially diluted with water to the appropriate concentrations to determine the insecticidal activity of the cwlE and sigK double deletion mutant Bt-59 (ΔcwlE-sigK). The LC50 values of Bt-59 and Bt-59 (ΔcwlE-sigK) against C. pipiens were 0.314 μL/L and 0.364 μL/L, respectively (Table 2). After exposure to UV light, the insecticidal activity of Bt-59 and Bt-59 (ΔcwlE-sigK) against C. pipiens reduced by 86.62 and 2.47%, respectively (Table 2). The LC50 values of Bt-59 and Bt-59 (ΔcwlE-sigK) against A. albopictus were 2.066 μL/L and 2.075 μL/L, respectively (Table 3). After exposure to UV light, the insecticidal activity of Bt-59 and Bt-59 (ΔcwlE-sigK) against A. albopictus reduced by 52.76 and 12.43%, respectively (Table 3). The hypothesis of equality and parallelism showed that the cwlE and sigK double mutant did not affect the insecticidal activity of Bti against mosquito larvae (Figures 8A,C). In contrast, the level of decrease of insecticidal activity against mosquito larvae of Bt-59 (ΔcwlE-sigK) was less than that observed for Bt-59 after UV treatment (Figures 8B,D).
Figure 8. Insecticidal activity of Bt-59 and Bt-59 (ΔcwlE-sigK) against mosquito larvae. (A) Insecticidal activity of Bt-59 and Bt-59 (ΔcwlE-sigK) against Culex pipiens. (B) After exposure to UV light, the insecticidal activity of Bt-59 and Bt-59 (ΔcwlE-sigK) against C. pipiens. (C) Insecticidal activity of Bt-59 and Bt-59 (ΔcwlE-sigK) against Aedes albopictus. (D) After exposure to UV light, the insecticidal activity of Bt-59 and Bt-59 (ΔcwlE-sigK) against A. albopictus.
Discussion
N-acetylmuramyl-L-alanine amidase contains the MurNAc-LAA domain and hydrolyzes the β-1,4 glycosidic bond between N-acetylglucosamine and N-acetylmuramic acid in polysaccharide chain, which is ubiquitous among spore-form bacteria and essential for the mother cell lysis (Vollmer et al., 2008; Bhagwat et al., 2020). CwlC is an essential cell wall hydrolase in Btk mother cell lysis and was distributed among the members of the B. cereus group (Chen et al., 2018). The CwlC exhibited the identical critical catalytic residues and conservative domains as Btk, whereas the deletion of cwlC delayed the lysis of mother cells in Bti (Huang et al., 2022). Although the mutation of sigK could inhibit the expression of the cell wall hydrolase genes cwlC and cwlB, it could not block cell wall lysis in Bti (Xu et al., 2020; Huang et al., 2022). This evidence demonstrates conclusively that Bti contains additional crucial cell wall hydrolases associated with mother cell lysis.
Bti’s novel cell wall hydrolase gene, cwlE, which is located on the large plasmid carrying the insecticidal genes, was characterized in this study. The absence of a cwlE homolog in other bacterial genomes, as determined by the NCBI homology analysis, indicates that cwlE is exclusively expressed in Bti. The CwlE is a new N-acetylmuramyl-L-alanine amidase, consisting of a MurNAc-LAA domain and two highly conserved catalytic residues (E26 and E151). Similar to the cwlC deletion mutant, the cwlE deletion mutant was unable to entirely prevent Bti mother cell lysis. In B. subtillis, cell wall lytic enzymes were functionally redundant because none of the single autolysin gene deletions affected mother cell lysis (Smith and Foster, 1995; Nugroho et al., 1999). However, the single mutant of autolysin genes delayed mother cell lysis in Bti, which differed from that in B. subtillis.
Sigma factors influence physiological process by regulating the transcription of various peptidoglycan hydrolase genes at various growth phases in Bacillus (Smith et al., 2000). Until now, the cell wall hydrolases involved in mother cell lysis were regulated by SigK in Bt strains (Yang et al., 2013; Chen et al., 2018; Huang et al., 2022). Analysis of promoter transcription activity showed that cwlE was regulated by SigE, which is the key Sigma factor regulating the expression of major insecticidal genes during the early sporulation stage in Bti (Deng et al., 2014). Our finding indicated that CwlE might be the key cell wall hydrolase causing the mother cell lysis in the sigK deletion mutant. To further verify the role of CwlE in Bti mother cell lysis, a double mutant strain was constructed by deleting of cwlE and sigK in this study. The results showed that deletion of cwlE and sigK could completely block Bti mother cell lysis, and cell wall lysis of double mutant could not be observed even after 7 days of culture in SSM medium. Therefore, we hypothesized that the cell wall lysis of Bti was accomplished through the synergy of the three hydrolases, CwlC, CwlB, and CwlE, and the absence of any cell wall hydrolases could delay mother cell lysis.
When Bt formulation is applied in the field, ultraviolet radiation is a key factor leading to crystal protein inactivation and reducing its insecticidal activity (Bravo et al., 2011). In the previous study, the mature crystal was produced and encapsulated in mother cells of a sigK deletion mutant of Bt strain, resulting in increased insecticidal activity against lepidoptera pests and greater resistance against UV radiation (Sanchis et al., 1999). In our study, the double mutant strain Bt-59 (ΔcwlE-sigK) showed increased insecticidal activity against the mosquito larva than the wild-type strain after exposure to UV light. By blocking mother cell lysis, it is possible to engineer a Bt strain with enhanced insecticidal persistence according to the current research.
We elucidated the function of CwlE in Bti mother cell lysis by deleting the hydrolase gene cwlE in this study. However, due to the lack of experimental data, we are not yet clear about the physiological function of cwlE expression at the early stage of stationary phase. Furthermore, compared to Btk, the reason why Bti requires more cell wall hydrolases to participate in mother cell lysis is also unclear. We speculate that due to the different midgut environments of different target insects, the synergistic effect of multiple cell wall hydrolases may allow Bti to release spores and crystal proteins faster in the mosquito midgut to complete population reproduction. Further investigations are needed to confirm this hypothesis.
Conclusion
cwlE is a novel N-acetylmuramyl-L-alanine amidase gene in Bti strain Bt-59 and its expression is dependent on transcriptional regulation factor SigE. Deletion of cwlE delayed Bti mother cell lysis mother without impacting the vegetative growth and Cry proteins production. As a cell wall hydrolase uniquely expressed in Bti, CwlE is the key hydrolase causing the mother cell lysis in the sigK deletion mutant. The double mutation of cwlE and sigK completely blocked Bti mother cell lysis even after 7 days of growth. Due to the impact of the double mutation on the expression level of the major crystal protein genes were not significant, Bt-59 (ΔcwlE-sigK) still maintains high insecticidal activity against mosquito larvae. In addition, the level of decrease of insecticidal activity against mosquito larvae of Bt-59 (ΔcwlE-sigK) was less than that observed for Bt-59 after UV treatment. In summary, this study identified a novel and unique cell wall hydrolase gene cwlE, elucidated the mechanism of Bti mother cell lysis, and provided an effective strategy for mosquito control using crystal encapsulation of Bt strain with the increased resistance against UV light inactivation.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
JX conceived and designed research. LH, CL, and YX conducted experiments. NC and FS contributed new experimental idea. LH and GH analyzed data. LH wrote the manuscript. All authors read and approved the manuscript.
Funding
This work was funded by grants from the National Natural Science Foundation (31530095), the National Key Research and Development Program of China (2017YFD0200400), Jiangsu Province Independent Innovation of Agricultural Science [CX(21)3087, CX(22)1009] and Yangzhou Science and Technology Project (YZ2021049).
Acknowledgments
We thank Qin Liu, Qi Peng, Linghuan Xu, Nan Zhang, Yurong Lu, and Yangsheng Cai for participating in some of the work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1250542/full#supplementary-material
References
Agaisse, H., and Lereclus, D. (1994). Structural and functional analysis of the promoter region involved in full expression of the cryIIIA toxin gene of Bacillus thuringiensis. Mol. Microbiol. 13, 97–107. doi: 10.1111/j.1365-2958.1994.tb00405.x
Arantes, O., and Lereclus, D. (1991). Construction of cloning vectors for Bacillus thuringiensis. Gene 108, 115–119. doi: 10.1016/0378-1119(91)90495-W
Ben-Dov, E. (2014). Bacillus thuringiensis subsp. israelensis and its dipteran-specific toxins. Toxins (Basel) 6, 1222–1243. doi: 10.3390/toxins6041222
Bhagwat, A., Mixon, M., Collins, C. H., and Dordick, J. S. (2020). Opportunities for broadening the application of cell wall lytic enzymes. Appl. Microbiol. Biotechnol. 104, 9019–9040. doi: 10.1007/s00253-020-10862-y
Bravo, A., Likitvivatanavong, S., Gill, S. S., and Soberón, M. (2011). Bacillus thuringiensis: a story of a successful bioinsecticide. Insect Biochem. Mol. Biol. 41, 423–431. doi: 10.1016/j.ibmb.2011.02.006
Carvalho, K. D. S., Crespo, M. M., Araújo, A. P., da Silva, R. S., de Melo-Santos, M. A. V., de Oliveira, C. M. F., et al. (2018). Long-term exposure of Aedes aegypti to Bacillus thuringiensis var. israelensis did not involve altered susceptibility to this microbial larvicide or to other control agents. Parasit. Vectors 11:673. doi: 10.1186/s13071-018-3246-1
Chen, X., Gao, T., Peng, Q., Zhang, J., Chai, Y., and Song, F. (2018). Novel cell wall hydrolase CwlC from Bacillus thuringiensis is essential for mother cell lysis. Appl. Environ. Microbiol. 84, e02640–e02617. doi: 10.1128/AEM.02640-17
Deng, C., Peng, Q., Song, F., and Lereclus, D. (2014). Regulation of cry gene expression in Bacillus thuringiensis. Toxins (Basel) 6, 2194–2209. doi: 10.3390/toxins6072194
Du, L., Qiu, L., Peng, Q., Lereclus, D., Zhang, J., Song, F., et al. (2012). Identification of the promoter in the intergenic region between orf1 and cry8Ea1 controlled by sigma H factor. Appl. Environ. Microbiol. 78, 4164–4168. doi: 10.1128/AEM.00622-12
Erdonmez, D., Mosayyebi, S., Erkan, K., Salimi, K., Nagizade, N., Saglam, N., et al. (2014). Nanofabrication and characterization of PVA-organofiller/ag nanocoatings on pMAD plasmids. Appl. Surf. Sci. 318, 127–131. doi: 10.1016/j.apsusc.2014.02.007
Federici, B. A., Park, H. W., and Bideshi, D. K. (2013). Overview of the basic biology of Bacillus thuringiensis with emphasis on genetic engineering of bacterial larvicides for mosquito control. Open Toxinology J. 3, 83–100. doi: 10.2174/1875414701003010083
Foster, S. J. (1992). Analysis of the autolysins of Bacillus subtilis 168 during vegetative growth and differentiation by using renaturing polyacrylamide gel electrophoresis. J. Bacteriol. 174, 464–470. doi: 10.1128/jb.174.2.464-470.1992
Huang, L., Xu, L., Han, G., Crickmore, N., Song, F., and Xu, J. (2022). Characterization of CwlC, an autolysin, and its role in mother cell lysis of Bacillus thuringiensis subsp. israelensis. Lett. Appl. Microbiol. 74, 92–102. doi: 10.1111/lam.13590
Huggett, J. F., Foy, C. A., Benes, V., Emslie, K., Garson, J. A., Haynes, R., et al. (2013). The digital MIQE guidelines: minimum information for publication of quantitative digital PCR experiments. Clin. Chem. 59, 892–902. doi: 10.1373/clinchem.2013.206375
Kuroda, A., Asami, Y., and Sekiguchi, J. (1993). Molecular cloning of a sporulation-specific cell wall hydrolase gene of Bacillus subtilis. J. Bacteriol. 175, 6260–6268. doi: 10.1128/jb.175.19.6260-6268.1993
Maciel-de-Freitas, R., Avendanho, F. C., Santos, R., Sylvestre, G., Araújo, S. C., Lima, J. B., et al. (2014). Undesirable consequences of insecticide resistance following Aedes aegypti control activities due to a dengue outbreak. PLoS One 9:e92424. doi: 10.1371/journal.pone.0092424
Nugroho, F. A., Yamamoto, H., Kobayashi, Y., and Sekiguchi, J. (1999). Characterization of a new sigma-K-dependent peptidoglycan hydrolase gene that plays a role in Bacillus subtilis mother cell lysis. J. Bacteriol. 181, 6230–6237. doi: 10.1128/JB.181.20.6230-6237.1999
Omotayo, A. I., Dogara, M. M., Sufi, D., Shuaibu, T., Balogun, J., Dawaki, S., et al. (2022). High pyrethroid-resistance intensity in Culex quinquefasciatus (say) (Diptera: Culicidae) populations from Jigawa, north-west, Nigeria. PLoS Negl. Trop. Dis. 16:e0010525. doi: 10.1371/journal.pntd.0010525
Patel, K. R., Wyman, J. A., Patel, K. A., and Burden, B. J. (1996). A mutant of Bacillus thuringiensis producing a dark-brown pigment with increased uv resistance and insecticidal activity. J. Invertebr. Pathol. 67, 120–124. doi: 10.1006/jipa.1996.0018
Peng, Q., Kao, G., Qu, N., Zhang, J., Li, J., and Song, F. (2016). The regulation of exosporium-related genes in Bacillus thuringiensis. Sci. Rep. 6:19005. doi: 10.1038/srep19005
Ramirez-Lepe, M., Aguilar, O., Ramírez-Suero, M., and Escudero, B. (2003). Protection of the spore-toxin complex of Bacillus thuringiensis serovar israelensis from ultraviolet irradiation with aluminum-cmc encapsulation and photoprotectors. Southwest. Entomol. 28, 137–143.
Reisen, W. K. (2010). Landscape epidemiology of vector-borne diseases. Annu. Rev. Entomol. 55, 461–483. doi: 10.1146/annurev-ento-112408-085419
Reiter, L., Kolsto, A.-B., and Piehler, A. P. (2011). Reference genes for quantitative, reverse-transcription PCR in Bacillus cereus group strains throughout the bacterial life cycle. J. Microbiol. Methods 86, 210–217. doi: 10.1016/j.mimet.2011.05.006
Sanchis, V., Gohar, M., Chaufaux, J., Arantes, O., Meier, A., Agaisse, H., et al. (1999). Development and field performance of a broad-spectrum nonviable asporogenic recombinant strain of Bacillus thuringiensis with greater potency and UV resistance. Appl. Environ. Microbiol. 65, 4032–4039. doi: 10.1128/AEM.65.9.4032-4039.1999
Schaeffer, P., Millet, J., and Aubert, J. P. (1965). Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. U. S. A. 54, 704–711. doi: 10.1073/pnas.54.3.704
Smith, T. J., Blackman, S. A., and Foster, S. J. (2000). Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146, 249–262. doi: 10.1099/00221287-146-2-249
Smith, T. J., and Foster, S. J. (1995). Characterization of the involvement of two compensatory autolysins in mother cell lysis during sporulation of Bacillus subtilis 168. J. Bacteriol. 177, 3855–3862. doi: 10.1128/jb.177.13.3855-3862.1995
Straus, D. L. (2008). Copper sulfate toxicity to channel catfish fry: yolk sac versus swim-up fry. N. Am. J. Aqualcult. 70, 323–327. doi: 10.1577/A06-092.1
Vasquez, M. I., Violaris, M., Hadjivassilis, A., and Wirth, M. C. (2009). Susceptibility of Culex pipiens (Diptera: Culicidae) field populations in Cyprus to conventional organic insecticides, Bacillus thuringiensis subsp. israelensis, and methoprene. J. Med. Entomol. 46, 881–887. doi: 10.1603/033.046.0421
Vollmer, W., Blanot, D., and de Pedro, M. A. (2008). Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167. doi: 10.1111/j.1574-6976.2007.00094.x
Vollmer, W., Joris, B., Charlier, P., and Foster, S. (2008). Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32, 259–286. doi: 10.1111/j.1574-6976.2007.00099.x
Wang, J., Yu, Q., Peng, Q., Slamti, L., Zhang, R., Hou, S., et al. (2022). Deletion of the novel gene mother cell lysis X results in Cry1Ac encapsulation in the Bacillus thuringiensis HD73. Front. Microbiol. 13:951830. doi: 10.3389/fmicb.2022.951830
Wang, G., Zhang, J., Song, F., Wu, J., Feng, S., and Huang, D. (2006). Engineered Bacillus thuringiensis GO33A with broad insecticidal activity against lepidopteran and coleopteran pests. Appl. Microbiol. Biotechnol. 72, 924–930. doi: 10.1007/s00253-006-0390-x
Xu, L., Han, G., Fan, X., Lv, J., Zhang, X., Peng, Q., et al. (2020). Characteristics of the sigK deletion mutant from Bacillus thuringiensis var. israelensis strain Bt-59. Curr. Microbiol. 77, 3422–3429. doi: 10.1007/s00284-020-02150-9
Yang, J., Peng, Q., Chen, Z., Deng, C., Shu, C., Zhang, J., et al. (2013). Transcriptional regulation and characteristics of a novel N-acetylmuramoyl-L-alanine amidase gene involved in Bacillus thuringiensis mother cell lysis. J. Bacteriol. 195, 2887–2897. doi: 10.1128/JB.00112-13
Zhang, Q., Hua, G., and Adang, M. J. (2017). Effects and mechanisms of Bacillus thuringiensis crystal toxins for mosquito larvae. Insect Sci. 24, 714–729. doi: 10.1111/1744-7917.12401
Zhang, R., Slamti, L., Tong, L., Verplaetse, E., Ma, L., Lemy, C., et al. (2020). The stationary phase regulator CpcR activates cry gene expression in non-sporulating cells of Bacillus thuringiensis. Mol. Microbiol. 113, 740–754. doi: 10.1111/mmi.14439
Zheng, L. B., and Losick, R. (1990). Cascade regulation of spore coat gene expression in Bacillus subtilis. J. Mol. Biol. 212, 645–660. doi: 10.1016/0022-2836(90)90227-D
Zogo, B., Tchiekoi, B. N., Koffi, A. A., Dahounto, A., Ahoua Alou, L. P., Dabiré, R. K., et al. (2019). Impact of sunlight exposure on the residual efficacy of biolarvicides Bacillus thuringiensis israelensis and Bacillus sphaericus against the main malaria vector, Anopheles gambiae. Malar J. 18:55. doi: 10.1186/s12936-019-2687-0
Keywords: Bacillus thuringiensis subsp. israelensis, mother cell lysis, cell wall hydrolase, transcriptional regulation, encapsulation
Citation: Huang L, Han G, Crickmore N, Li C, Xia Y, Song F and Xu J (2023) Characterization of a novel cell wall hydrolase CwlE involved in Bacillus thuringiensis subsp. israelensis mother cell lysis. Front. Microbiol. 14:1250542. doi: 10.3389/fmicb.2023.1250542
Edited by:
Mitsuo Ogura, Tokai University, JapanReviewed by:
Vipin Rana, University of Maryland, College Park, United StatesSandra M. Ruzal, University of Buenos Aires, Argentina
Christopher Cote, United States Army Medical Research Institute of Infectious Diseases (USAMRIID), United States
Copyright © 2023 Huang, Han, Crickmore, Li, Xia, Song and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Xu, YmlvLXhqQDE2My5jb20=
 Lixin Huang
Lixin Huang Guangjie Han
Guangjie Han Neil Crickmore
Neil Crickmore Chuanming Li1
Chuanming Li1 Fuping Song
Fuping Song