
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 11 October 2023
Sec. Food Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1247348
This article is part of the Research TopicImpact of Dairy Farming on the Safety of Raw Milk and Milk ProductsView all 6 articles
 Boxue Si1,2†
Boxue Si1,2† Kaizhen Liu1,2,3†
Kaizhen Liu1,2,3† Guoxin Huang1,2
Guoxin Huang1,2 Meiqing Chen1,2
Meiqing Chen1,2 Jiyong Yang1,2
Jiyong Yang1,2 Xufang Wu1,2
Xufang Wu1,2 Ning Li1,2
Ning Li1,2 Wenhao Tang1,2
Wenhao Tang1,2 Shengguo Zhao1,2
Shengguo Zhao1,2 Nan Zheng1,2
Nan Zheng1,2 Yangdong Zhang1,2*
Yangdong Zhang1,2* Jiaqi Wang1,2*
Jiaqi Wang1,2*Introduction: Milk fat is the most variable nutrient in milk, and recent studies have shown that rumen bacteria are closely related to milk fat. However, there is limited research on the relationship between rumen bacteria and milk fatty. Fatty acids (FAs) are an important component of milk fat and are associated with various potential benefits and risks to human health.
Methods: In this experiment, forty-five healthy Holstein dairy cows with alike physiological and productive conditions were selected from medium-sized dairy farms and raised under the same feeding and management conditions. The experimental period was two weeks. During the experiment, raw milk and rumen fluid were collected, and milk components were determined. In this study, 8 high milk fat percentage (HF) dairy cows and 8 low milk fat percentage (LF) dairy cows were selected for analysis.
Results: Results showed that the milk fat percentage in HF group was significantly greater than that of the dairy cows in the LF group. 16S rRNA gene sequencing showed that the rumen bacterial abundance of HF dairy cows was significantly higher than that in LF dairy cows; at the genus level, the bacterial abundances of Prevotellaceae_UCG-001, Candidatus_Saccharimonas, Prevotellaceae_UCG-003, Ruminococcus_1, Lachnospiraceae_XPB1014_group, Lachnospiraceae_AC2044_group, probable_genus_10 and U29-B03 in HF group were significantly higher than those in the LF group. Spearman rank correlation analysis indicated that milk fat percentage was positively related to Prevotellaceae_UCG-001, Candidatus_Saccharimonas, Prevotellaceae_UCG-003, Ruminococcus_1, Lachnospiraceae_XPB1014_group, Lachnospiraceae_AC2044_group, probable_genus_10 and U29-B03. Furthermore, Prevotellaceae_UCG-001 was positively related to C14:0 iso, C15:0 iso, C18:0, Ruminococcus_1 with C18:1 t9, Lachnospiraceae_AC2044_group with C18:1 t9 and C18:1 t11, U29-B03 with C15:0 iso.
Discussion: To sum up, rumen bacteria in dairy cows are related to the variation of milk fat, and some rumen bacteria have potential effects on the deposition of certain fatty acids in raw milk.
Milk fat is one of the main nutrient component in milk, and it is also a key indicator for assessing milk quality. The content of milk fat is generally 3% ~ 5% in milk, and the main component is triglyceride, which account for about 98% of milk fat. The remaining components of milk fat include diacylglycerol, monoacylglycerol, phospholipid, glycolipid, cholesterol, free fatty acids and fat-soluble vitamins (Liu et al., 2018; Amores and Virto, 2019). Fatty acids (FAs) are an important component of milk fat and are associated with various potential benefits and risks to human health. Previous studies have shown that unsaturated fatty acids reduce hypercholesterolemia and risk of cardiovascular diseases, whereas saturated fatty acids and trans fatty acids have opposite effects (Huth and Park, 2012;Valenzuela et al., 2019; Guo et al., 2023). Bovine milk contains many different fatty acids, from essential FA such as linoleic (C18:2) and α-linolenic (18,3) FA to human health promoting FA such as con-jugated linoleic acid (CLA) (C18:2) (German and Dillard, 2006; Calder, 2015). These unsaturated fatty acids have potential benefits in preventing cardiovascular disease, anticancer, anti-inflammatory, and antioxidant properties (Gómez-Cortés et al., 2018; Lawrence, 2021). Recent scientific research has shown that Branched chain fay acids and Odd-chain fatty acids are trace but important bioactive components in food. Which are gradually garnering attention from scientists for their protective effect against inflammatory and cancer (Gomez-Cortes and de la Fuente, 2020). In addition, several studies have shown that Medium chain fatty acids (MCFAs) are useful in the treatment of a variety of neurological and metabolic disorders [e.g., Alzheimer’s disease (Courchesne-loyer et al., 2017; Croteau et al., 2017), cancer (Jansen and Walach, 2016), diabetes (Geng et al., 2016), and obesity (St-Onge and Jones, 2002)] due to their immunoactivities and intestinal probiotic effects。MCFAs can be directly absorbed by intestinal epithelial cells and rapidly transferred to the liver through the portal vein, and MCFAs can cross the blood–brain barrier and preferentially undergo β-oxidation. Therefore, MCFAs are highly efficient in absorption and transportation.
Fatty acid is the main component of milk fat, and its type, composition and content have been widely concerned in the current years. The milk fatty acid was mainly synthesised by rumen microbia and exogenous uptake (Parodi, 2004). Butyric acid manufactured by microbial fermentation is absorbed and converted into β-hydroxybutyric acid by rumen epithelial cells, and acetic acid and β-hydroxybutyric acid are transported to the mammary gland to synthesize short and medium chain fatty acids. However long chain fatty acids are mainly derived from dietary lipids and body fat (Buitenhuis et al., 2019). In the context of the rumen microbial community digesting feed, it is interesting to study how much changes in milk FA can be explained by changes in the rumen microbiome.
The milk fat in milk is affected by numerous factors, such as rumen bacteria, diet, feeding management, health status, season, parity and lactation stage of dairy cows (Latham et al., 1974; Yang et al., 2013; Buitenhuis et al., 2019). In the rumen of dairy cows, bacteria can degrade plant fibers to volatile fatty acids (VFAs) by fermentation (Patil et al., 2018). Rumen bacteria have been reported to have significant influence on rumen fermentation, as well as the variation of milk yield and milk composition (Bainbridge et al., 2016; Hooman et al., 2016). The ratio of Firmicutes to Bacteroidetes was found to be positively related to milk fat yield (Jami et al., 2014; Scharen et al., 2018). Hassan et al. found that dietary regulation can increase the relative richness of Firmicutes and decrease the relative richness of Bacteroidetes, thereby increasing milk fat yield (Hassan et al., 2020). Milk fat percentage was reported the association with the abundance of Dialister, Megasphaera, Lachnospira and Sharpea in rumen (Pitta et al., 2020).
Previous studies have shown a relationship between milk fat and rumen microbiota. We speculated that the rumen bacterial composition of HF and LF groups was different, and there were differences in rumen fermentation, rumen metabolites, milk fat composition and fatty acid composition. The purpose of this study was to analyze the differences in rumen bacteria between HF and LF groups by 16S rRNA gene sequencing technology, and the relationship between differential bacteria and milk fat.
All dairy cows in this experiment were conformed to the research method, guaranteed animal welfare, and were approved by the Animal Care Committee of Institute of Animal Sciences of Chinese Academy of Agricultural Sciences (No: IAS 2019–28).
This experiment was carried out in Tianjin Mengde Dairy Farm, with more than 1,500 Holstein dairy cows in spring. The dairy farm has advanced facilities and records the milk yield of individual dairy cows every day. From more than 1,500 lactating dairy cows, 45 healthy dairy cows with similar body weight, parity (1.3 ± 0.74), day in milk (153 ± 16d) and daily milk yield (34 ± 6 kg) were selected. After collecting milk samples, MilkoScan FT120 (Foss, Hillerod) was used to determine the content of milk protein, milk fat, lactose, and total solids. The data were subjected to quality control through the mean and standard deviation (SD), and split into HF group (n = 8) and LF group (n = 8) according to the milk fat percentage. The feed intake and milk yield between the two groups were similar, which avoided the large difference in milk fat percentage caused by the huge difference in milk yield, and met the requirements of experiment.
All 45 dairy cows were raised in the same enclosure, drinking water freely and fed with TMR three times a day for two weeks. The daily feeding time was 7:00, 13:00, and 18:00, respectively, and the feeding amount and remaining feed amount of each dairy cow were recorded, and the individual feed intake was calculated. Milking took place half an hour before feeding and individual milk production was recorded by the milking system (Liu et al., 2022).
During the experiment, the management conditions were the same. Milk samples and rumen fluid samples were taken on the last day of the experiment. Raw milk samples were collected every day according to the milking time, three times a day, 50 mL each time, stored at 4°C, and finally blended in a 4:3:3 ratio as a milk sample for subsequent milk composition determination.
The collection time of rumen fluid was before the first feeding in the morning. About 200 mL rumen fluid samples were collected before the morning feeding from each cow using oral stomach tube. The rumen fluid samples were immediately filtered using 4 layers of gauze, and then separated it into two parts. One was immediately measured for pH value, the other aliquot was poured into a 50 mL sterilized tube and stored frozen for subsequent determination of rumen bacterial composition and rumen fermentation parameters.
Determination of NH3-N and VFAs were performed after rumen fluid thawing. Analyze 16 samples for each indicator. The concentration of NH3-N was determined by phenol-sodium hypochlorite colorimetric method referenced the previously paper (Wang et al., 2019). The concentration of VFAs was determined by gas chromatography FID detector.
The collected rumen fluid was thawed, and then DNA extraction was performed using the cetyltrimethylammonium bromide (CTAB) reagent. The purity of extracted DNA was checked using 1% agarose gel, and the DNA concentration was determined using a NanoDrop 2000 UV–vis spectrophotometer (Thermo Scientific, Wilmington, United States). The purified DNA as template, 341F (5’-CCTACGGGNGGCWGCAG-3′) and 806R (5’-GGACTACHVGGGTWTCTAAT-3′) as primers, the V3 to V4 of bacterial 16S rRNA gene was amplified using an ABI GeneAmp 9,700 PCR thermocycler (ABI, CA, United States). The PCR amplification conditions and the specific steps of sequencing all adopt the method of Liu et al. (2022). Briefly, The PCR protocol was 94°C for 2 min, followed by 30 cycles at 98°C for 10 s and 62°C for 30 s. The PCR reaction solution consisted of 10 × KOD Buffer 5 mL, 2 mmol/L dNTP 5 mL, 25 mmol/L MgSO4 3 mL, 10 mmol/L primer 1.5 mL respectively, KOD Polymerase (TOYOBO, Japan) 1 mL, and the template DNA 100 ng. The purified products were quantified using a Quantus Fluorometer (Promega, United States), and pooled in equimolar, then pairedend sequenced (2 × 300 bp) at Illumina Hiseq 2,500 PE250 platform (Illumina, San Diego, United States) under the standard protocols by GENE DENOVO (Guangzhou, China). The raw reads of 16S rRNA gene sequencing were deposited into the NCBI Sequence Read Archive database (accession number: PRJNA722820).
All raw reads analysis and the quality control were referenced to the methods as reported by Liu et al. (2022). UPARSE (7.1 version, http://drive5.com/uparse/) was used to cluster the operational taxonomic unit (OTU) with 97% similarity cutoff, then identified, and removed the chimeric sequences. The longest read was deemed as a representative sequence of the taxonomy for each OTU. Further, those representative sequences were identified against to the Silva SSU128 database using RDP Classifier.
The test was carried out by the method independently developed by our laboratory. Draw 2 mL of liquid milk sample, add 25 μL of internal standard C19:0 fatty acid methyl ester (10,000 μg/mL), add 4 mL of n-hexane/isopropanol (V/V = 3/2) mixed solution, after vortexing for 30s, centrifugation (4°C, 12,000 rpm) for 3 min. Pipette the n-hexane phase into a high temperature test tube with a lid, add n-hexane, centrifuge, take the upper n-hexane phase, repeat twice. To the n-hexane phase containing the lipid extract, 2 mL of NaOH-CH3OH solution (20 g/L) was added, and saponification and alkali-catalyzed methyl esterification at 50°C for 20 min in a water bath. Subsequently, Acid-catalyzed methyl esterification was carried out by adding 2 mL of acetyl chloride-methanol solution (100 mL/L) at 90°C for 120 min in a water bath. After cooling, add ultrapure water, transfer the n-hexane phase, and dilute to 10 mL. Add about 1 g of anhydrous sodium sulfate, vortex for 30 s, and let stand for 10 min. Pipette 200 μL of the n-hexane phase into the bottle and dilute the volume to 1 mL, so that the dilution factor of liquid milk to sample bottle is 25. Analysis with GC–MS.
Testing equipment: Gas chromatography tandem mass spectrometer 7890A/7000B (G-150). The conditions of use of the instrument are as follows, chromatographic column: CP-Sil 88; programmed temperature; gas: helium; constant pressure: 38 psi; inlet temperature: 250°C; quadrupole temperature: 150°C; injection volume: 1 μL.
Import the experimental data into Excel 2016 for preliminary statistical. Significance analysis was performed using SPSS (version 26.0). Non-parametric tests (Kruskal-Wallis) were used to compare the differences in milk composition, rumen fermentation indexes, and fatty acid composition between the two groups of cows (Nikoloudaki et al., 2019). The omicsmart1 microbial analysis platform was used to analyze and compare rumen bacteria, including alpha diversity analysis, PCA analysis, species composition analysis, and indicator species analysis. Correlation analysis was performed using Spearman’s rank analysis method. The relationship between rumen bacteria and milk components, NH3-N and VFAs, and the relationship between rumen bacteria and raw milk fatty acid composition were mainly analyzed.
Table 1 indicates that the day in milk and milk yield of the dairy cows in two groups were similar, and there was no statistical significance (p = 0.958). The CP, NDF, ADF, and DMI intakes of cows in HF group were significantly higher than that in the LF group (P<0.05). The milk fat percentage is the most concerned indicator. The milk fat percentage of the dairy cows in the HF group was 4.42, which was significantly greater than that in the LF group (p < 0.01).
The analysis results of the rumen fermentation indexes of dairy cows in the two groups were shown in Table 2. The pH and NH3-N concentration of rumen fluid between the two groups of dairy cows were similar, and the difference was not significant (p > 0.05). At the same time, the difference of total VFAs and individual VFAs between the two groups of dairy cows was also not significant (p > 0.05). However, it was worth noting that the concentration of VFAs in the rumen fluid of dairy cows in the HF group was higher than those in the LF group. In addition, the ratio of acetic acid to propionic acid and the mole percentage of each VFA between the two groups of dairy cows were not significant, and there was no statistical significance.
16S rRNA gene sequencing analysis of rumen fluid samples from 16 cows obtained a total of 1,624,665 sequences, with an average of 10,1,541 ± 4,613 sequences per sample. After the sequencing results were processed for quality control, the bacterial richness index was obtained by using the alpha diversity analysis of rumen bacteria. The results in Figure 1 showed that the bacterial abundance indices Sobs, Simpson and Shannon in the HF group were significantly higher than LF group (p < 0.05, p < 0.05, p < 0.01). It indicated that there is a higher abundance of bacteria in the HF rumen.
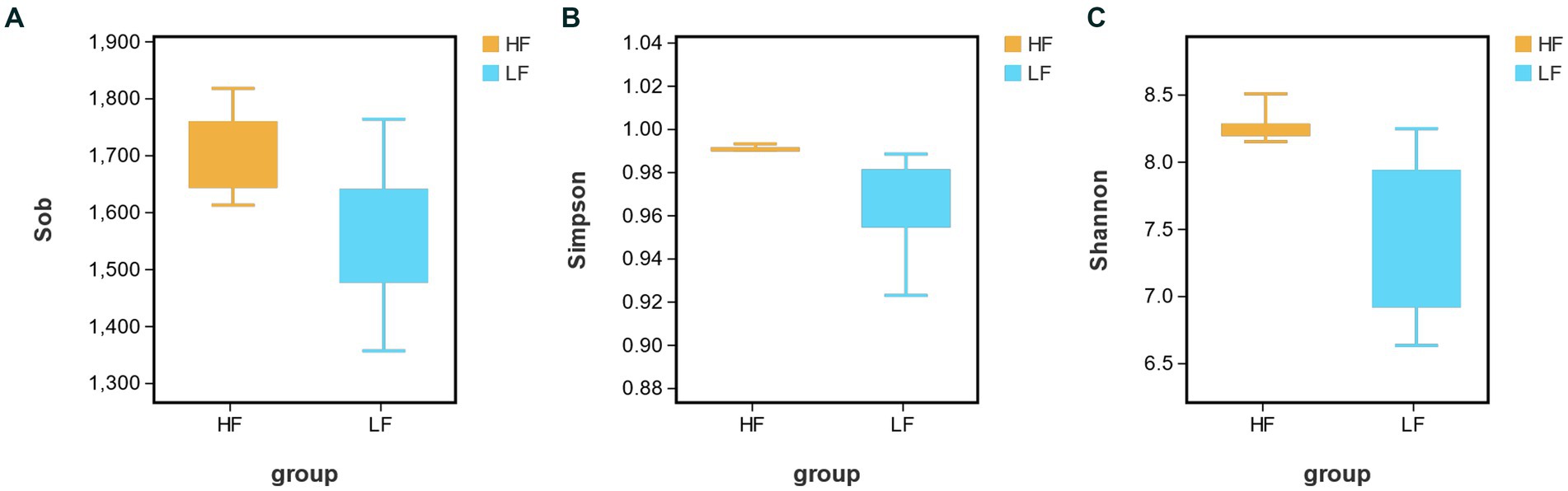
Figure 1. Analysis of rumen bacterial richness index [(A) Sobs; (B) Simpson; and (C) Shannon] in HF group and LF group. HF means high milk fat percentage; LF means low milk fat percentage.
By listing species abundance information based on OTUs, a PCA plot was carried out to show the similarity between and within the two groups of dairy cows. The PCA diagram indicated that the variation degrees of the PC1 and PC2 were 56.67 and 21.93%, respectively. Moreover, the rumen bacterial composition of HF group and LF group dairy cows was clearly distinguished, indicating the rumen bacterial structure was significantly different between HF group and LF group (Figure 2). Samples with high community structure similarity in PCA plot tended to cluster together, whereas the greater the community difference, the farther the distance. It could be seen that the structural similarity of the rumen flora of dairy cows in the HF group was significantly greater than that in the LF group.
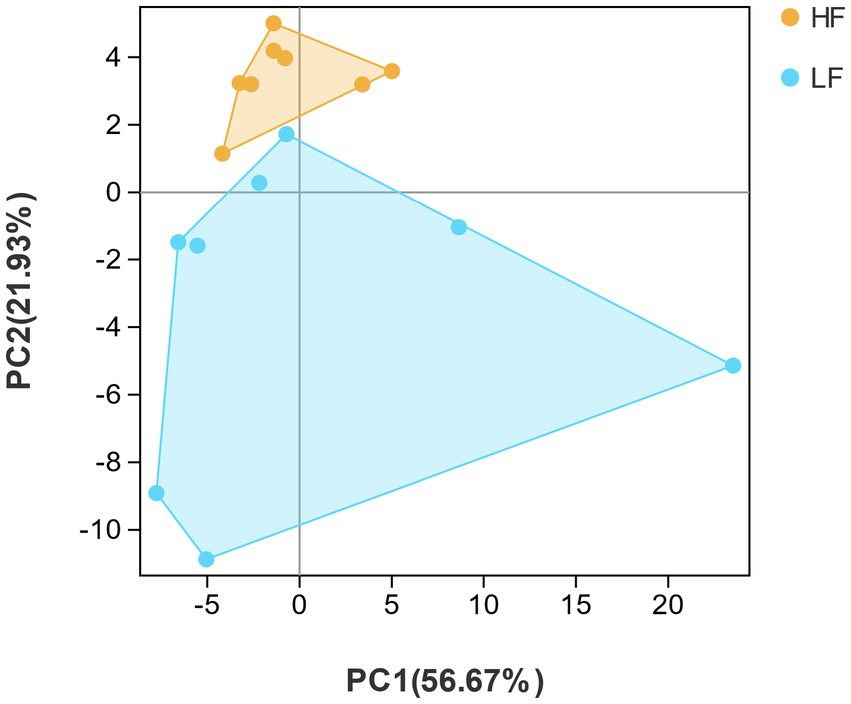
Figure 2. Principal component analysis (PCA). PCA analysis of rumen bacteria in HF group and LF group. HF means high milk fat percentage; LF means low milk fat percentage.
A total of 25 bacterial phyla were discovered at the phylum level after classification analysis. Among them, the relative abundance of Bacteroidetes and Firmicutes accounted for more than 93.0% (Figure 3A). The ratio of Firmicutes to Bacteroidetes was 0.873 in the HF group and 0.724 in the LF group. As shown in Figure 3B, a total of 225 bacteria were detected at the genus level, and the relative abundance of Prevotella_1 was the highest, and both groups account for more than 20% of the total bacteria, followed by Succiniclasticum, and Prevotella-7. LF had 1.69 times more Succiniclasticum than HF (HF 11.39%, LF 19.22), Prevotella_7 was 3.14 times higher than HF (HF 2.83%, LF 8.89%).
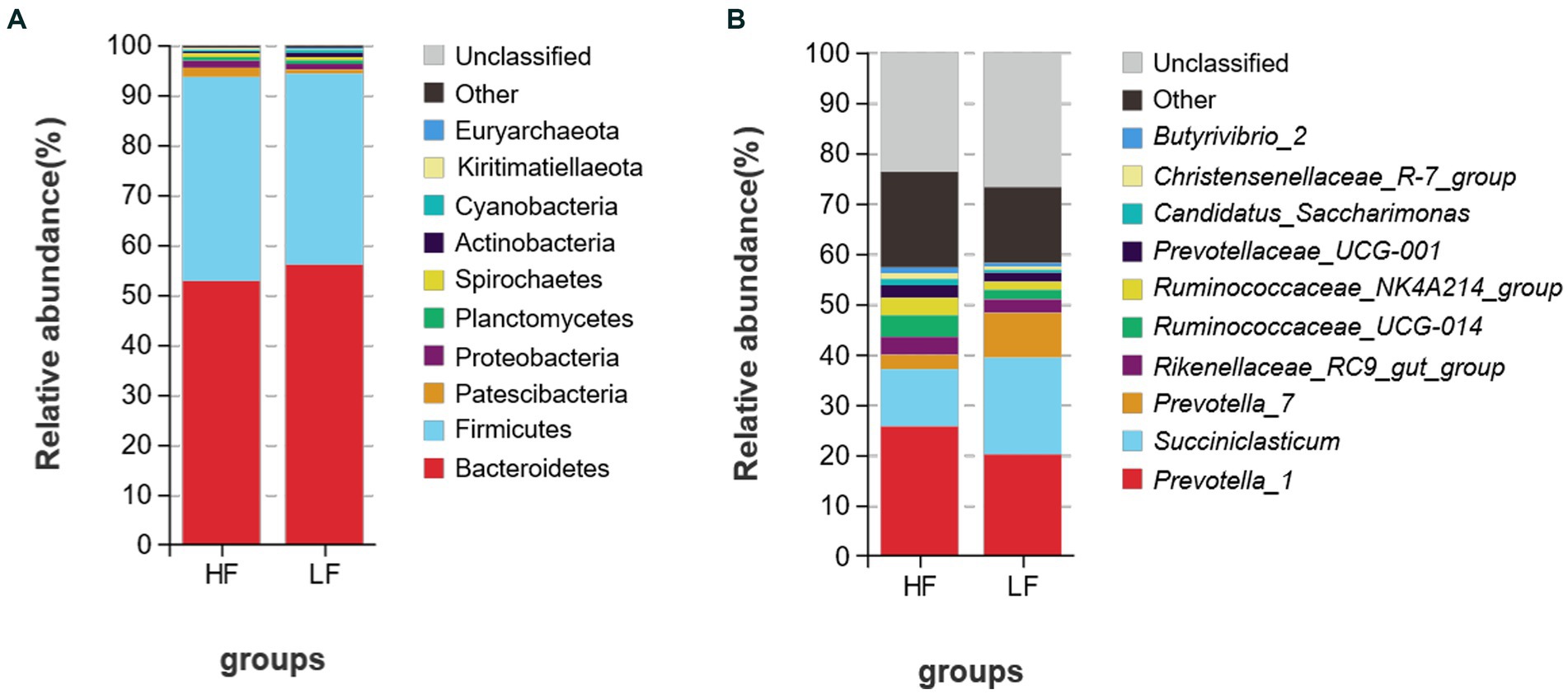
Figure 3. Relative abundance of rumen bacteria at the phylum (A) and genus (B) levels of dairy cows in HF group and LF group. HF means high milk fat percentage; LF means low milk fat percentage.
There were 8 different genera of bacteria between the two groups, which basically belong to Bacteroidetes and Firmicutes (Figure 4). The relative abundance of Prevotellaceae_UCG-001, Candidatus_Saccharimonas, Prevotellaceae_UCG-003, Ruminococcus_1, Lachnospiraceae_XPB1014_group, Lachnospiraceae_AC2044_group, probable_genus_10 and U29-B03 of rumen fluid in HF dairy cows were significantly greater than those in LF dairy cows (p < 0.05).
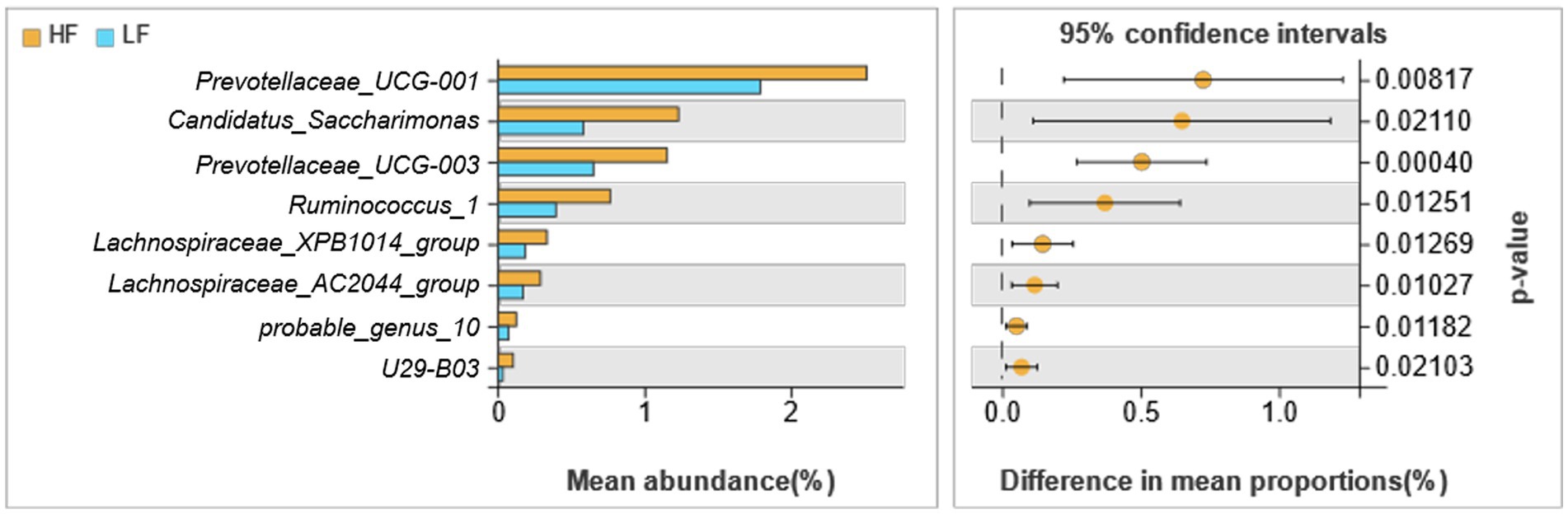
Figure 4. Comparison of rumen bacteria in dairy cows in HF group and LF group. HF means high milk fat percentage; LF means low milk fat percentage.
Correlations between milk composition, rumen fermentation indexes, and relative abundance of rumen bacteria were analyzed in HF and LF dairy cows using Spearman rank correlation analysis. The result is shown in Figure 5, Prevotellaceae_UCG-001, Candidatus_Saccharimonas, Prevotellaceae_UCG-003, Ruminococcus_1, Lachnospiraceae_XPB1014_group, Lachnospiraceae_AC2044_group, probable_genus_10 and U29-B03 bacterial relative abundance were significantly positive correlated with milk fat percentage, and all of them were negatively correlated with non-dairy solids and lactose content. Among the rumen fermentation indexes, the relative abundance of 8 rumen differential bacteria were all positively related to pH, but inversely correlated with NH3-N. The relative abundance of Prevotellaceae_UCG-003 was positively related to VFAs.
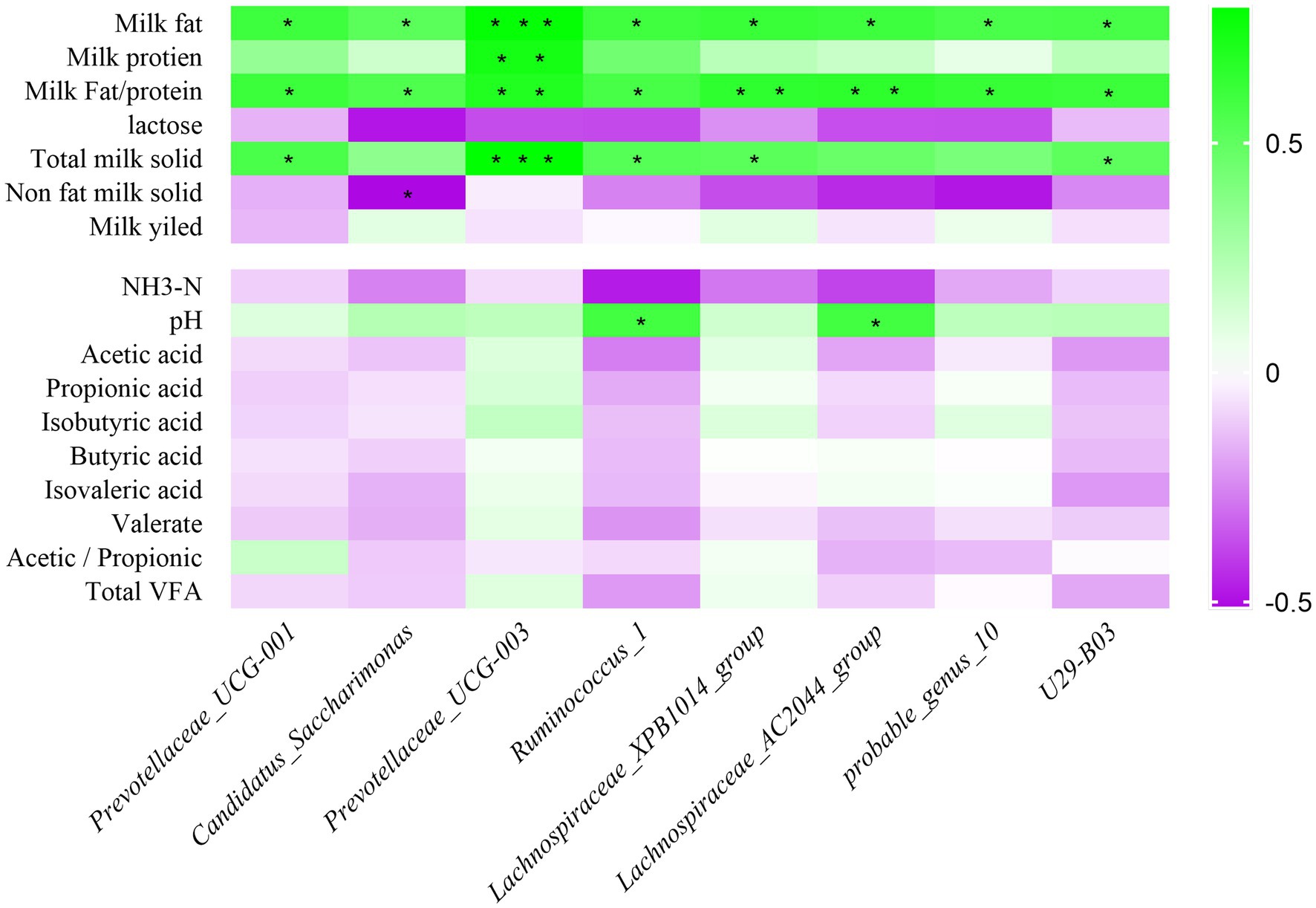
Figure 5. Correlation analysis of rumen bacteria with milk components and fermentation parameters. Green represents positive correlation and purple represents negative correlation. The color depth represents the correlation coefficient value, and the darker the color, the larger the value. NH3-N means ammonia nitrogen; VFA means volatile fatty acid; *represents a significant difference (0.01 < p < 0.05). ** represents (0.001 < p < 0.01), *** represents (p < 0.001).
A total of 22 fatty acids were significantly different (p < 0.05) between the two groups of cows (Table 3; Figure 6). C7:0, C8:0, C9:0, C10:0, C11:0, C12:0, C13:0, and MCFA were significantly higher (p < 0.05) in cows in the high milk fat group than in cows in the low milk fat group. In contrast, cows in the high milk fat group had C16:1 c7, C17:1 t10, C18:1 c9, C18:3 c6, c9, c12, C20:1 c11, C20:2 c11, c14, C22:0, C22:1 c13, C20:3 c11, c14, c17, C20:4 c5, c8, c11, c14, C20:5 c5, c8, c11, c14, c17, C24:0, C22:4 c7, c10, c13, c16, and C22:5 c7, c10, c13, c16, c19 were significantly lower (p < 0.05) than cows in the low milk fat group. SCFA, LCFA, SFA, MUFA, PUFA, and Trans contents were not significantly different (p > 0.05) between the two groups.
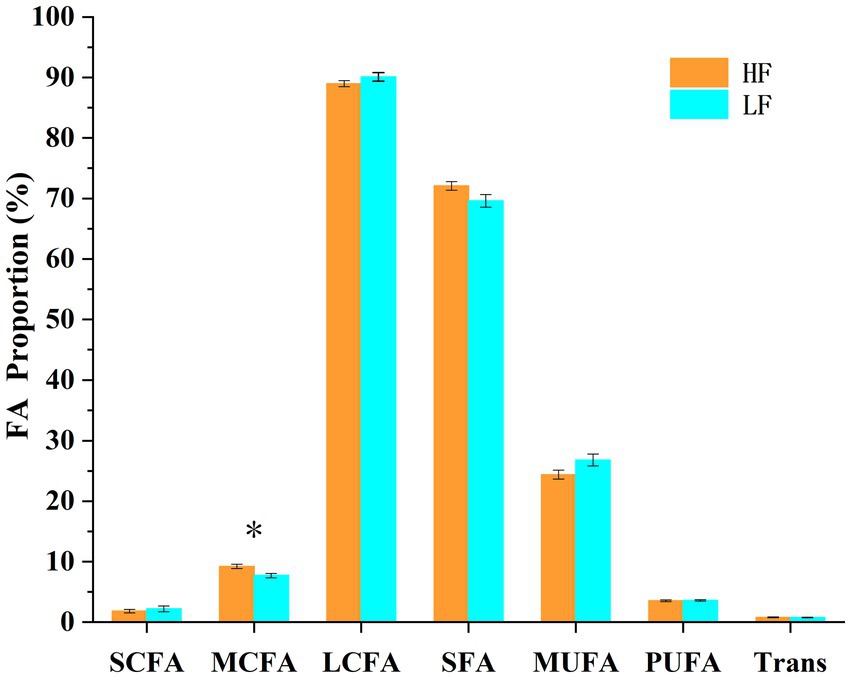
Figure 6. Different fatty acid groups in the high and low milk fat rate groups. SCFA: short chain fatty acids (<6 carbons), MCFA: medium chain fatty acids (6–12 carbons), LCFA: long chain fatty acids (>12 carbons), SFA: saturated fatty acids, MUFA: monounsaturated fatty acids, PUFA: polyunsaturated fatty acids, Trans: total trans fatty acids. The error line is standard error of the mean. * represents a significant difference (p < 0.05).
Detecting fatty acids in raw milk by GC–MS, we determined a total of 59 fatty acids, and carried out correlation research with fatty acids. The result was shown in Figure 7, the relative abundance of Prevotellaceae_UCG-001 was positively related to C14:0 iso, C15:0 iso, and C18:0 (p < 0.05); the relative abundance of Ruminococcus_1 was positively related to C18:1 t9 (p < 0.05); the relative abundance of Lachnospiraceae_XPB1014_group was positively related to C7:0 (p < 0.05); The relative abundance of Lachnospiraceae_AC2044_group was positively related to C18:1 t9 and C18:1 t11 (p < 0.05); The bacterial abundance of U29-B03 was positively correlated with the C15:0 iso (p < 0.05). Summarily, these 8 differential bacteria were positively correlated with short and medium chain fatty acids. However, it was inversely correlated with long chain fatty acids.
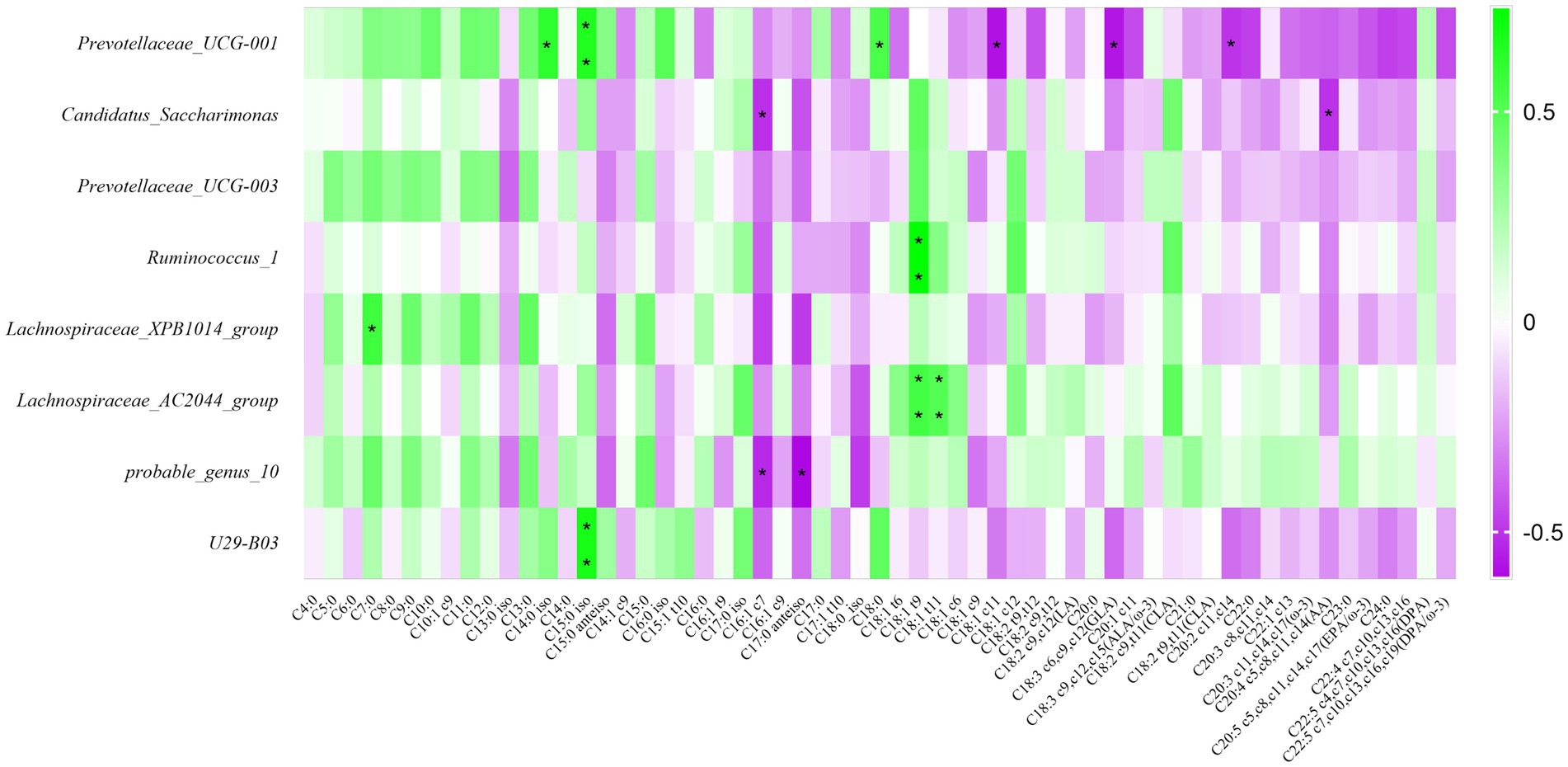
Figure 7. Correlation analysis of rumen bacteria with milk components and fermentation parameters. Green represents positive correlation and purple represents negative correlation. The color depth represents the correlation coefficient value, and the darker the color, the larger the value. *represents a significant difference (0.01 < p < 0.05). **represents very significant difference (p < 0.01).
In this study, the rumen bacterial composition, milk composition and rumen fermentation indexes of dairy cows in HF and LF groups were investigated. Few previous studies in this area have been reported. A recent study showed that dairy cows in the high milk solids group had a significantly higher percentage of milk fat than cows in the low milk solids group (Liu et al., 2022). We also acquired the same research results and similar findings were also found by Nichols et al. (2018). Milk fat is affected by many factors, among which the interaction of diet structure and rumen fermentation was one of the main reasons (Buitenhuis et al., 2019; Wang et al., 2019; Xue et al., 2020). In our results, the pH value and NH3-N concentration indicated that the rumen maintained a stable acid–base environment, and the molar percentage of VFAs composition indicated that the rumen fermentation model of the dairy cows did not change. And, no differences were found in rumen fermentation indicators between HF and LF group dairy cows. This was difficult to explain the difference in milk fat between the two groups of cows. But research has found that the NDF and ADF feed intake of HF was significantly higher than that in LF dairy cows (13.4% and 14.4%, respectively). Plant fibers are decomposed by rumen bacteria into VFAs and other precursors for synthetic milk fat (Patil et al., 2018). This may explain why the milk fat percentage of dairy cows in the HF was greater than that in the LF.
Recently, it has been reported that rumen bacteria affect milk composition in dairy cows (Zeng et al., 2019; Zou et al., 2019). In this study, 16S rRNA gene sequencing technology was used to investigate rumen bacteria in dairy cows. It was found that the rumen bacterial richness of dairy cows in the HF was obviously higher than that of the dairy cows in the LF. Xue et al. (2018) found that the lower the richness of the rumen microflora, the lower the milk fat percentage, which is in line with the current study. It was reported that rumen microbial abundance was negatively correlated with feed efficiency and milk fat percentage (Shabat et al., 2016; Li and Le, 2017; Xue et al., 2020). This indicated that the concentration of total VFAs and individual VFAs in LF rumen should be higher than HF, however the results were opposite. We speculated that this may be related to the high number and structural similarity of rumen bacteria in HF dairy cows, resulting in higher rumen fermentation efficiency and fermentation products. In addition, the concentration of rumen VFAs was the common result of microbial fermentation and rumen absorption, and individual absorption differences may also affected. Future studies on volatile fatty acids absorption and its relationship to rumen bacteria should be investigated, which has implications for improving milk fat.
It was found that there were differences of the rumen bacteria between the two groups through the analysis of rumen bacterial composition (Jami et al., 2014). Jami et al., found that the ratio of Firmicutes and Bacteroidetes had extremely significant positive correlation with milk fat, which exhibited the same result as this study. The mean value of Firmicutes/Bacteroidetes in HF group was 0.873, which was greater than that in LF group, 0.724, and there are significant differences. Prevotella was reported to be the most abundant genus in the rumen of dairy cows at the genus level (Lima et al., 2014; Xue et al., 2019; Liu et al., 2022), and the same result was found in this study. The same was found in buffalo, where Prevotella was also the predominant genus (Lin et al., 2015; Iqbal et al., 2018). In addition, Succiniclasticum was the second most abundant. Succiniclasticum ferments carbohydrates to produce succinate, which was the precursor of propionic acid (Wallace et al., 2015). Based on our results, the propionate concentration of HF dairy cows was higher than that of LF, thus we speculated that the abundance of Succiniclasticum in HF dairy cows was greater than that of LF. However, the rumen bacterial results showed that Vibrio succinates in low-LF was 1.69 times higher than HF, which was inconsistent with our speculation. Xue et al., also found the same result in their study (Xue et al., 2019). This difference may be due to imprecise taxonomic assessments only at the genus level.
In-depth research on rumen microorganisms not only improve rumen digestion and feed utilization efficiency, but also provide a theoretical basis for improving milk fat and milk yield (Firkins and Yu, 2015). We focused on the analysis on differences at the genus level, specific bacteria that may be associated with milk fat. In previous reports, Lachnospiraceae and Ruminococcaceae were important bacteria for fermentative production of volatile fatty acids (Biddle et al., 2013). We found that Lachnospiraceae_XPB1014_group, Lachnospiraceae_AC2044_group, probable_genus_10 were positively related to milk fat percentage. In addition, Lachnospiraceae_XPB1014_group, probable_genus_10, Prevotellaceae_UCG-003 were positively related to isobutyric acid and propionic acid, Liu et al., also had the same findings (Liu et al., 2022). Zou et al.’s study on buffalo found that Ruminococcus was negatively related to the content of butyric acid, and we also had the same finding (Zou et al., 2019). Jami et al., and Jiang et al., found that the relative abundance of Prevotella was inversely related to milk fat (Jami et al., 2014; Jiang et al., 2017). However, Xue et al., found that Prevotella could produce higher VFAs (Xue et al., 2019). Our results found that Prevotellaceae_UCG-003 was positively correlated with VFAs, while Prevotellaceae_UCG-001 was negatively correlated, which may be due to the more detailed classification. Jewell et al. had found that there were functional differences between bacteria of the same genus (Kelsea et al., 2015). In this study, it was also found that Prevotellaceae_UCG-001, Candidatus_Saccharimonas, Prevotellaceae_UCG-003, Ruminococcus_1, U29-B03 were all positively correlated with milk fat. Acetate is reported to be the main substrate for milk fat synthesis in the rumen of dairy cows (Xue et al., 2020). These bacteria may be positively correlation with acetate production (Xue et al., 2020; Liu et al., 2022).
Milk quality is closely related to the composition and content of fatty acids. High quality milk should be rich in unsaturated fatty acids, short chain fatty acids and medium chain fatty acids, with a corresponding reduction in saturated fatty acids and trans fatty acids (Chen et al., 2023). Saturated fatty acids can reduce people’s risk of cardiovascular disease, but unsaturated and trans fatty acids can increase the risk. Short chain and medium chain fatty acids both have anti-inflammatory, tumor growth inhibition, and obesity alleviation functions (Gómez-Cortés et al., 2018; Prado et al., 2019). In this study, it was found that the medium chain fatty acid content of cows in the HF group was significantly higher than that of the LF group, which is favorable for the production of high quality milk. Rumen bacteria break down plant fibers into volatile fatty acids (especially acetic acid and butyric acid) to synthesize medium chain fatty acids (Prado et al., 2019). The high abundance of rumen bacteria in the HF group provided the basis for the production of large amounts of medium chain fatty acids. In addition, although there was no significant difference between saturated and unsaturated fatty acids, cows in the HF group had higher levels of saturated fatty acids than those in the LF group and lower levels of unsaturated fatty acids than those in the LF group. This may be due to the conversion of unsaturated fatty acids to saturated fatty acids by rumen bacterial hydrogenation (Koch and Elascano, 2018).
To reveal the effect of rumen bacteria on milk fat, we further analyzed the association of rumen bacteria with the fatty acid composition of raw milk. Our results showed that Prevotellaceae_UCG-001, Candidatus_Saccharimonas, Prevotellaceae_UCG-003, Ruminococcus_1, Lachnospiraceae_XPB1014_group, Lachnospiraceae_AC2044_group, probable_genus_10 and U29-B03 were favorable for the deposition of short and medium chain fatty acids in raw milk, but not for long chain fatty acids. The reason may be due to the potential biohydrogenation of these rumen bacteria. It had been reported that Lachnospiraceae and Prevotellaceae were the major biohydrogenation bacteria in the rumen of dairy cows. It was noteworthy that, Prevotellaceae_UCG-001 promoted deposition of C14:0 iso, C15:0 iso, C18:0; Ruminococcus_1 could promote deposition of C18:1 t9; Lachnospiraceae_AC2044_group could promote the deposition of C18:1 t9 and C18:1 t11; U29-B03 could promote the deposition of C15:0 iso. Bart et al. studied the relationship between rumen microbes and fatty acid composition of raw milk. The results showed that bacteria such as Prevotella, Lachnospiraceae, Ruminococcus could affect the synthesis of C18 unsaturated fatty acids and odd-chain fatty acids (Buitenhuis et al., 2019), and we also had the same result. Branched-chain fatty acids have the same active function of inhibiting inflammation and suppressing tumor growth (Mika et al., 2016). It was of great significance to human health. Prevotellaceae_UCG-001 and U29-B03 can be considered as rumen bacteria that improve milk quality. It was of practical significance to study the relationship between rumen bacterial composition and fatty acid synthesis, especially beneficial fatty acid synthesis, for improving milk quality and protecting human health.
There were significant differences of the rumen microbial flora diversity and the abundance in dairy cows between HF and LF. The richness of rumen bacteria in the HF was significantly greater than that in the LF, and the HF had a similar microbial community structure and little difference in colonies. Prevotellaceae_UCG-001, Candidatus_Saccharimonas, Prevotellaceae_UCG-003, Ruminococcus_1, Lachnospiraceae_XPB1014_group, Lachnospiraceae_AC2044_group, probable_genus_10 and U29-B03 bacterial were significantly positive correlated with milk fat percentage. And, Prevotellaceae_UCG-003 could increase the production of VFAs. In addition, we also found that Prevotellaceae_UCG-001 had significant deposition effects on C14:0 iso, C15:0 iso, C18:0, and Lachnospiraceae_AC2044_group on long-chain fatty acids C18:1 t9, C18:1 t11.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA722820.
The animal studies were approved by Animal Care Committee of Institute of Animal Sciences of CAAS (Beijing, China). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
BS: investigation, data curation, and writing – original draft. KL and GH: methodology and validation. MC and XW: formal analysis. JY, NL, and WT: investigation. YZ: conceptualization and writing – review. SZ: conceptualization and project administration. NZ: investigation and writing – review and editing. JW: supervision, conceptualization, and writing – review and editing. All authors contributed to the article and approved the submitted version.
This study was supported by the National Key R&D Program of China (2022YFD1301004), the State Key Laboratory of Animal Nutrition and Feeding (2004DA125184G2108), the Agricultural Science and Technology Innovation Program (ASTIP-IAS12) and Modern Agro-Industry Technology Research System of the PR China (CARS-36).
We would like to thank all authors for their suggestions and assistance in experimental design and article writing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Amores, G., and Virto, M. D. (2019). M. Total and free fatty acids analysis in milk and dairy fat. Separations 6, 1–14. doi: 10.3390/separations6010014
Bainbridge, M. L., Cersosimo, L. M., Wright, A., and Kraft, J. (2016). Rumen bacterial communities shift across a lactation in Holstein, Jersey and Holstein × Jersey dairy cows and correlate to rumen function, bacterial fatty acid composition and production parameters. FEMS Microbiol. Ecol. 92:fiw059. doi: 10.1093/femsec/fiw059
Biddle, A., Stewart, L., Blanchard, J., and Leschine, S. (2013). Untangling the genetic basis of fibrolytic specialization by lachnospiraceae and ruminococcaceae in diverse gut communities. Diversity 5, 627–640. doi: 10.3390/d5030627
Buitenhuis, B., Lassen, J., Noel, S. J., Plichta, D. R., Sørensen, P., Difford, G. F., et al. (2019). Impact of the rumen microbiome on milk fatty acid composition of Holstein cattle. Genet. Sel. Evol. 51, 23–29. doi: 10.1186/s12711-019-0464-8
Calder, P. C. (2015). Functional roles of fatty acids and their effects on human health. J. Parenter. Enter. Nutr. 39, 18S–32S. doi: 10.1177/0148607115595980
Chen, M., Wang, F., Wu, X., Si, B., Pan, J., Zheng, N., et al. (2023). Updating the fatty acid profiles of retail bovine milk in China based on an improved GC-MS method: implications for nutrition. Front. Nutr. 10:1204005. doi: 10.3389/fnut.2023.1204005
Courchesne-loyer, A., Croteau, E., Castellano, C. A., St-Pierre, V., Hennebelle, M., and Cunnane, S. C. (2017). Inverse relationship between brain glucose and ketone metabolism in adults during short-term moderate dietary ketosis: a dual tracer quantitative positron emission tomography study. J. Cereb. Blood Flow Metab. 37, 2485–2493. doi: 10.1177/0271678X16669366
Croteau, E., Castellano, C. A., Fortier, M., Bocti, C., Fulop, T., Paquet, N., et al. (2017). A cross-sectional comparison of brain glucose and ketone metabolism in cognitively healthy older adults, mild cognitive impairment and early Alzheimer's disease. Exp. Gerontol. 107, 18–26. doi: 10.1016/j.exger.2017.07.004
Firkins, J. L., and Yu, Z. (2015). Ruminant nutrition symposium: how to use data on the rumen microbiome to improve our understanding of ruminant nutrition. J. Anim. Sci. 93, 1450–1470. doi: 10.2527/jas.2014-8754
Geng, S., Zhu, W., Xie, C., Li, X., Wu, J., Liang, Z., et al. (2016). Medium-chain triglyceride ameliorates insulin resistance and inflammation in high fat diet-induced obese mice. Eur. J. Nutr. 55, 931–940. doi: 10.1007/s00394-015-0907-0
German, J. B., and Dillard, C. J. (2006). Composition, structure and absorption of milk lipids: a source of energy, fat-soluble nutrients and bioactive molecules. Crit. Rev. Food Sci. Nutr. 46, 57–92. doi: 10.1080/10408690590957098
Gomez-Cortes, P., and de la Fuente, M. A. (2020). Metabolic origin and bioactive properties of odd and branched-chain fatty acids in ruminants' milk. Revista Mexicana De Ciencias Pecuarias. 11, 1174–1191. doi: 10.22319/rmcp.v11i4.5171
Gómez-Cortés, P., Juárez, M., and de la Fuente, M. A. (2018). Milk fatty acids and potential health benefits: an updated vision. Trends Food Sci. Technol. 81, 1–9. doi: 10.1016/j.tifs.2018.08.014
Guo, Q., Li, T., Qu, Y., Liang, M., Ha, Y., Zhang, Y., et al. (2023). New research development on trans fatty acids in food: biological effects, analytical methods, formation mechanism, and mitigating measures. Prog. Lipid Res. 89:101199. doi: 10.1016/j.plipres.2022.101199
Hassan, F. U., Ebeid, H. M., Tang, Z. H., Li, M. W., Peng, L. J., and Peng, K. P. (2020). A mixed phytogenic modulates the rumen bacteria composition and milk fatty acid profile of water buffaloes. Front. Vet. Sci. 7:569. doi: 10.3389/fvets.2020.00569
Hooman, D., Tun, H. M., Cardoso, F. C., Plaizier, J. C., Ehsan, K., and Loor, J. J. (2016). Linking peripartal dynamics of ruminal microbiota to dietary changes and production parameters. Front. Microbiol. 7:2143. doi: 10.3389/fmicb.2016.02143
Huth, P. J., and Park, K. M. (2012). Influence of dairy product and Milk fat consumption on cardiovascular disease risk: a review of the evidence. Adv. Nutr. 3, 266–285. doi: 10.3945/an.112.002030
Iqbal, M. W., Zhang, Q., Yang, Y. B., Li, L. L., Zou, C. X., Huang, C. H., et al. (2018). Comparative study of rumen fermentation and microbial community differences between water buffalo and Jersey cows under similar feeding conditions. J. Appl. Anim. Res. 46, 740–748. doi: 10.1080/09712119.2017.1394859
Jami, E., White, B. A., Itzhak, M., and Heimesaat, M. M. (2014). Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS One 9:e85423. doi: 10.1371/journal.pone.0085423
Jansen, N., and Walach, H. (2016). The development of tumours under a ketogenic diet in association with the novel tumour marker TKTL1: a case series in general practice. Oncol. Lett. 11, 584–592. doi: 10.3892/01.2015.3923
Jiang, Y., Ogunade, I. M., Arriola, K. G., Qi, M., Vyas, D., and Staples, C. R. (2017). Effects of the dose and viability of saccharomyces cerevisiae. 2. Ruminal fermentation, performance of lactating dairy cows, and correlations between ruminal bacteria abundance and performance measures. J. Dairy Sci. 100, 8102–8118. doi: 10.3168/jds.2016-12371
Kelsea, A., Caroline, A., Christine, L., Paul, J., and Garret, S. (2015). Ruminal bacterial community composition in dairy cows is dynamic over the course of two lactations and correlates with feed efficiency. Appl. Environ. Microbiol. 81, 4697–4710. doi: 10.1128/aem.00720-15
Koch, L., and Elascano, G. J. (2018). Milk fat depression: etiology, theories, and soluble carbohydrate interactions. J. Anim. Res. Nut. 3:2. doi: 10.21767/2572-5459.100046
Latham, M. J., Sutton, J. D., and Sharpe, M. E. (1974). Fermentation and microorganisms in the rumen and the content of fat in the milk of cows given low roughage rations. J. Dairy Sci. 57, 803–810. doi: 10.3168/jds.s0022-0302(74)84968-4
Lawrence, G. D. (2021). Perspective: the saturated fat-unsaturated oil dilemma: relations of dietary fatty acids and serum cholesterol, atherosclerosis, inflammation, Cancer, and all-cause mortality. Adv. Nutr. 12, 647–656. doi: 10.1093/advances/nmab013
Li, F., and Le, L. G. (2017). Metatranscriptomic profiling reveals linkages between the active rumen microbiome and feed efficiency in beef cattle. Appl. Environ. Microbiol. 83, e00061–e00017. doi: 10.1128/aem.00061-17
Lima, F. S., Oikonomou, G., Lima, S. F., Bicalho, M., and Bicalho, R. C. (2014). Prepartum and postpartum rumen fluid microbiomes: characterization and correlation with production traits in dairy cows. Appl. Environ. Microbiol. 81, 1327–1337. doi: 10.1128/aem.03138-14
Lin, B., Henderson, G., Zou, C. X., Cox, F., Liang, X. W., Janssen, P. H., et al. (2015). Characterization of the rumen microbial community composition of buffalo breeds consuming diets typical of dairy production systems in southern China. Appl. Environ. Microbiol. 81, 1327–1337. doi: 10.1016/j.anifeedsci.2015.06.013
Liu, Z., Rochfort, S., and Cocks, B. (2018). Milk lipidomics: what we know and what we don't. Prog. Lipid Res. 71, 70–85. doi: 10.1016/j.plipres.2018.06.002
Liu, K., Zhang, Y., Huang, G., Zheng, N., Zhao, S., and Wang, J. (2022). Ruminal bacterial community is associated with the variations of total milk solid content in Holstein lactating cows. Anim. Nutr. 9, 175–183. doi: 10.1016/j.aninu.2021.12.005
Mika, A., Stepnowski, K., Proczko, M., Wisniewski, P., Sledzinski, M., and Sledzinski, T. (2016). A comprehensive study of serum odd- and branched-chain fatty acids in patents with excess weight. Obesity (Ssiver Sring) 24, 1669–1676. doi: 10.1002/oby.21560
Nichols, K., Bannink, A., Pacheco, S., Van, V., Dijkstra, J., and Van, L. H. (2018). Feed and nitrogen efficiency are affected differently but milk lactose production is stimulated equally when isoenergetic protein and fat is supplemented in lactating dairy cow diets. J. Dairy Sci. 101, 7857–7870. doi: 10.3168/jds.2017-14276
Nikoloudaki, O., Lemos, W. J. F., and Junior, L. B. (2019). How multiple farming conditions correlate with the composition of the raw cow's milk lactic microbiome. Environ. Microbiol. 23, 1702–1716. doi: 10.1111/1462-2920.15407
Patil, R. D., Ellison, M. J., Wolff, S. M., Shearer, C., Wright, A. M., and Cockrum, R. R. (2018). Poor feed efficiency in sheep is associated with several structural abnormalities in the community metabolic network of their ruminal microbes. J. Anim. Sci. 96:2113. doi: 10.1093/jas/sky096
Pitta, D. W., Indugu, N., Vecchiarelli, B., Hennessy, M., Baldin, M., and Harvatine, K. J. (2020). Effect of 2-hydroxy-4-(methylthio) butanoate (hmtba) supplementation on rumen bacterial populations in dairy cows when exposed to diets with risk for milk fat depression. J. Dairy Sci. 103, 2718–2730. doi: 10.3168/jds.2019-17389
Prado, L. A., Schmidely, P., Nozière, P., and Ferlay, A. (2019). Milk saturated fatty acids, odd- and branched-chain fatty acids, and isomers of C18:1, C18:2, and C18:3n-3 according to their duodenal flows in dairy cows: a meta-analysis approach. J. Dairy Sci. 102, 3053–3070. doi: 10.3168/jds.2018-15194
Scharen, M., Frahm, J., Kersten, S., Meyer, U., Hummel, J., Breves, G., et al. (2018). Interrelations between the rumen microbiota and production, behavioral, rumen fermentation, metabolic, and immunological attributes of dairy cows. J. Dairy Sci. 101, 4615–4637. doi: 10.3168/jds.2017-13736
Shabat, S., Sasson, G., Doron-Faigenboim, A., Durman, T., Yaacoby, S., Miller, M. B., et al. (2016). Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 10, 2958–2972. doi: 10.1038/ismej.2016.62
St-Onge, M. -P., and ones, P. J. H.. (2002). Physiological effects of medium-chain triglycerides: potential agents in the prevention of obesity. J. Nutr. 132: 329–332.DOl: 329, doi: 10.1093/jn/132.3.329
Valenzuela, C. A., Baker, E. J., Miles, E. A., and Calder, P. C. (2019). Eighteen-carbon trans fatty acids and inflammation in the context of atherosclerosis. Prog. Lipid Res. 76:101009. doi: 10.1016/j.plipres.2019.101009
Wallace, R. J., Mckain, N., Duthie, C. A., Hyslop, J. J., Ross, D. W., Waterhouse, A., et al. (2015). The rumen microbial metagenome associated with high methane production in cattle. BMC Genomics 16:839. doi: 10.1186/s12864-015-2032-0
Wang, L. Z., Liu, K. Z., Wang, Z. S., Bai, X., Peng, Q. H., and Jin, L. (2019). Bacterial community diversity associated with different utilization efficiencies of nitrogen in the gastrointestinal tract of goats. Front. Microbiol. 10:239. doi: 10.3389/fmicb.2019.00239
Xue, M. Y., Sun, H. Z., Wu, X. H., Guan, L. L., and Liu, J. X. (2018). Assessment of rumen microbiota from a large dairy cattle cohort reveals the pan and core bacteriomes contributing to varied phenotypes. Appl. Environ. Microbiol. 84, e00970–e00918. doi: 10.1128/aem.00970-18
Xue, M. Y., Sun, H. Z., Wu, X. H., Guan, L. L., and Liu, J. X. (2019). Assessment of rumen bacteria in dairy cows with varied milk protein yield. J. Dairy Sci. 102, 5031–5041. doi: 10.3168/jds.2018-15974
Xue, M. Y., Sun, H. Z., Wu, X. H., Liu, J. X., and Guan, L. L. (2020). Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome 8:64. doi: 10.1186/s40168-020-00819-8
Yang, L., Yang, Q., Yi, M., Pang, Z. H., and Xiong, B. H. (2013). Effects of seasonal change and parity on raw milk composition and related indices in chinese Holstein cows in northern China. J. Dairy Sci. 96, 6863–6869. doi: 10.3168/jds.2013-6846
Zeng, H. B., Guo, C. Z., Sun, D. M., Seddik, H. E., and Mao, S. Y. (2019). The ruminal microbiome and metabolome alterations associated with diet-induced milk fat depression in dairy cows. Meta 9:154. doi: 10.3390/metabo9070154
Zou, C. X., Gu, Q. C., Zhou, X. K., Xia, Z. S., Muhammad, W. I., and Tang, Q. F. (2019). Ruminal microbiota composition associated with ruminal fermentation parameters and milk yield in lactating buffalo in Guangxi, China—a preliminary study. J. Anim. Physiol. Anim. Nutr. (Berl) 103, 1374–1379. doi: 10.1111/jpn.13154
Keywords: milk fat, 16S rRNA gene sequencing, rumen bacteria, fatty acid composition, key bacteria
Citation: Si B, Liu K, Huang G, Chen M, Yang J, Wu X, Li N, Tang W, Zhao S, Zheng N, Zhang Y and Wang J (2023) Relationship between rumen bacterial community and milk fat in dairy cows. Front. Microbiol. 14:1247348. doi: 10.3389/fmicb.2023.1247348
Received: 28 June 2023; Accepted: 22 September 2023;
Published: 11 October 2023.
Edited by:
Armin Tarrah, University of Guelph, CanadaReviewed by:
Wilson José Fernandes Lemos Junior, Austrian Institute of Technology (AIT), AustriaCopyright © 2023 Si, Liu, Huang, Chen, Yang, Wu, Li, Tang, Zhao, Zheng, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yangdong Zhang, emhhbmd5YW5nZG9uZ0BjYWFzLmNu; Jiaqi Wang, d2FuZ2ppYXFpQGNhYXMuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.