
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 09 November 2023
Sec. Food Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1247211
This article is part of the Research Topic Recent advances in Campylobacter research View all 10 articles
In dynamic microbial ecosystems, bacterial communication is a relevant mechanism for interactions between different microbial species. When C. jejuni resides in the intestine of either avian or human hosts, it is exposed to diverse bacteria from the microbiome. This study aimed to reveal the influence of co-incubation with Enterococcus faecalis, Enterococcus faecium, or Staphylococcus aureus on the proteome of C. jejuni 81–176 using data-independent-acquisition mass spectrometry (DIA-MS). We compared the proteome profiles during co-incubation with the proteome profile in response to the bile acid deoxycholate (DCA) and investigated the impact of DCA on proteomic changes during co-incubation, as C. jejuni is exposed to both factors during colonization. We identified 1,375 proteins by DIA-MS, which is notably high, approaching the theoretical maximum of 1,645 proteins. S. aureus had the highest impact on the proteome of C. jejuni with 215 up-regulated and 230 down-regulated proteins. However, these numbers are still markedly lower than the 526 up-regulated and 516 down-regulated proteins during DCA exposure. We identified a subset of 54 significantly differentially expressed proteins that are shared after co-incubation with all three microbial species. These proteins were indicative of a common co-incubation response of C. jejuni. This common proteomic response partly overlapped with the DCA response; however, several proteins were specific to the co-incubation response. In the co-incubation experiment, we identified three membrane-interactive proteins among the top 20 up-regulated proteins. This finding suggests that the presence of other bacteria may contribute to increased adherence, e.g., to other bacteria but eventually also epithelial cells or abiotic surfaces. Furthermore, a conjugative transfer regulon protein was typically up-expressed during co-incubation. Exposure to both, co-incubation and DCA, demonstrated that the two stressors influenced each other, resulting in a unique synergistic proteomic response that differed from the response to each stimulus alone. Data are available via ProteomeXchange with identifier PXD046477.
Campylobacter jejuni belongs to the most frequently diagnosed bacterial gastrointestinal pathogens in humans worldwide (Acheson and Allos, 2001). In the developed world, foodborne infections most commonly occur after consumption of cross-contaminated food, prepared in parallel with poultry meat., whereas Campylobacter spp. belong to the natural commensal microbiome in poultry (Skirrow, 1991). Additional sources for infections are water, raw milk or other livestock animals (Blaser et al., 1980, 1983; Szewzyk et al., 2000). Symptoms of campylobacteriosis include severe bloody diarrhea, fever, abdominal cramps and nausea. Furthermore, Campylobacter infections are associated with severe follow-up diseases, for example the Guillain-Barré syndrome, a neural disease that can lead to paralyzes and damage of the nervous system (Rees and Hughes, 1995; Sejvar et al., 2011).
The ideal growth temperature for the Gram-negative, helical-shaped and microaerophilic bacterium lies between 37°C and 42°C. Due to its broad spectrum of virulence-associated factors that enable the survival in varying environmental conditions, C. jejuni can successfully colonize the gut of avian and mammal hosts. One of these virulence-associated factors is the ability to survive high concentrations of bile acid in the human or animal gut. Among the diverse functions of bile is the solubilization and emulsification of fat, which makes it an important biological detergent (Begley et al., 2005; Chiang, 2017). Under the exposition of bile acids, the composition of fatty acids and phospholipids of the bacterial cell membranes are altered, which leads to instabilities in the cell’s surface and consequently to the disruption of the cell (Taranto et al., 2003). Furthermore, DNA damages might be induced by the presence of bile acid in different bacteria, such as E. coli (Kandell and Bernstein, 1991; Begley et al., 2005). To overcome this stress, bacterial gut inhabitants have developed several mechanisms to cope with bile acid and are able to tolerate varying concentrations of bile.
Co-incubation can have several important positive or negative effects on the growth of different bacteria. In presence of other microbes, some pathogenic bacteria show an increase in in their virulence (Stacy et al., 2016; Fast et al., 2018). However, proteomic studies on co-incubation remain rare. A proteomic study by García-Pérez and coworkers has shown that co-incubation can reduce the number of extracellular proteins in microbial communities in wounds (García-Pérez et al., 2018). In addition, co-incubation of different bacteria with yeasts, such as C. albicans, has shown positive effects on the growth of both species, probably due to the release of nutrients into the medium or beneficial changes in pH (Ellepola et al., 2019). During co-incubation with other bacteria, C. jejuni has been shown to interact with a variety of other bacteria, for instance Bifidobacterium longum which prevents the adherence of C. jejuni to intestinal tract cells (Quinn et al., 2020a,b). A combination of different bacteria that include E. faecium can lead to a decrease of C. jejuni in the gastro-intestinal tract of poultry (Neveling and Dicks, 2021). Anis and colleagues showed that studying the co-incubation of C. jejuni with other bacteria might be an interesting topic, as the bacterial interaction might enhance C. jejuni survival when exposed to external stresses, such as the presence of oxygen (Anis et al., 2022). Further studies about co-cultivation of C. jejuni with E. coli and L. monocytogenes, showed that the adhesion potential of C. jejuni to all tested surfaces was significantly increased. In summary, this study suggests that the presence of other (foodborne) bacteria may increase the adhesion of C. jejuni, and thus, co-incubation might contribute to its pathogenicity (Klančnik et al., 2020). The only co-cultivation study that investigated transcriptomics or proteomics in C. jejuni during co-cultivation involved eukaryotic cells, specifically human INT 407 and Caco-2 epithelial cells (Negretti et al., 2019). Thus, our study aiming the proteomic adaptations of C. jejuni to bacterial co-cultivation is novel and covers a so far unexplored subject.
To our knowledge. Proteomic or transcriptomic studies of C. jejuni with other bacteria do not exist so far. The only co-cultivation studies that investigated transcriptomics or proteomics in C. jejuni during co-cultivation include eukaryotic cells, such as human INT 407 and Caco-2 epithelial cells (Negretti et al., 2019). Thus, our study aiming the proteomic adaptations of C. jejuni to bacterial co-cultivation is novel and covers a so far unexplored subject.
In this study, we aimed to observe the impact of co-incubation on the C. jejuni proteome and the possible effects of co-incubation on the bile acid response of the bacterium. Therefore, we analyzed the proteome of C. jejuni in co-incubation and under deoxycholate (DCA = deoxycholic acid) stress. DCA is a secondary bile acid, which is a product of dehydroxylation by gut microbiota and has been shown to have inhibiting effects on the growth of C. jejuni and other bacteria at a certain concentration (Lertpiriyapong et al., 2012; Vidal et al., 2021) and furthermore substantial effects on the proteome (Masanta et al., 2019). The three bacterial species chosen for co-incubation were less resistant toward DCA than C. jejuni.
One of the bacterial species chosen for the co-incubation study was E. faecalis, a Gram-positive, facultative anaerobic coccal opportunistic pathogen that belongs to the human commensal microbiome, but can also be found in environmental samples (Lebreton et al., 2014; Van Tyne and Gilmore, 2014; Fiore et al., 2019). Furthermore, we tested a close relative of E. faecalis, E. faecium, which is also an opportunistic pathogen of global importance due to its high antibiotic resistance potential (Lopes et al., 2006; Gorrie et al., 2019). The third bacterium used in this study was Staphylococcus aureus, another Gram-positive pathogen of high clinical relevance due to the high number of severe infections caused by multidrug resistant S. aureus (Klevens et al., 2007; Rasigade et al., 2014; Cheung et al., 2021).
This study aims to provide a deeper look at the co-incubation proteome of the pathogen C. jejuni with other bacteria that are usually present in the human body and the respective proteomic changes in presence of DCA. We used data-independent acquisition mass spectrometry (DIA-MS) to systematically compare the proteomic changes in co-incubation of the different bacteria with C. jejuni as well as the proteomic response to bile acid (DCA). This technique enables the quantitative analysis of every detectable compound in a sample of proteins and thus provides high reliability in the quantitative results (Huang et al., 2015). To our knowledge, this is the first proteomic co-incubation study on C. jejuni.
Campylobacter jejuni 81–176 (purchased from the American Type Culture Collection: ATCC-BAA-2151) was used for all described experiments. C. jejuni was grown overnight on CAM-agar plates from Biomérieux (Marcy-l’Étoile, France) at 42°C. Mueller-Hinton (MH) broth served as liquid medium at 37°C. To generate a microaerophilic environment, the Gas Pak™ EZ Campy Container System by BD (Franklin Lakes, NJ, USA) and an anaerobic jar for incubation were used.
Enterococcus faecalis ATCC 700802 (V583; purchased from the American Type Culture Collection), Enterococcus faecium TX0016 (also purchased from the American Type Culture Collection: ATCC BAA-472) and Staphylococcus aureus NCTC 8325 (PS 47, purchased from the BCCM/LMG: LMG 21764) were used for co-incubation experiments and grown overnight on Columbia agar plates supplemented with sheep blood purchased from Biomérieux (Marcy-l’Étoile, France).
For co-incubation experiments, the optical density at 600 nm (OD600) of C. jejuni was set to 0.5 and the OD600 of the respective other bacterium was set to 0.1. Incubation was performed in phosphate buffered saline (PBS) to avoid effects of the medium on the bile acid resistance. DCA was added to the medium at a concentration of 0.1% for E. faecalis and E. faecium and 0.075 for S. aureus. These concentrations are lethal to these Gram-positive bacteria when cultured individually. Incubation was carried out for 3 h at 37°C and shaking at 150 rpm. After three hours, a spot assay on Mueller-Hinton agar plates was done to show the survival of the bacteria after 3 h in a dilution series. Subsequently, protein extraction was done.
The three Gram-positive bacterial species without presence of C. jejuni served as positive control while the approaches of the Gram-positive bacteria with the respective amount of DCA served as negative control. All samples were prepared in biological triplicates.
Cultures were centrifuged at 4,000 rpm for 10 min at 4°C. For protein-extraction from the pellet, the supernatant was discarded. For samples containing C. jejuni, pellets were resuspended in 2 mL 0.9% saline and kept on ice over the procedure. Subsequently, the Gram-negative cells were disrupted via sonification using a Branson sonifier 250 from Branson ultrasonics (Brookfield, Connecticut, USA) with the following settings: output control = 3, duty cycler = 30%. The sonification process was performed five times for 30 s followed by 30 s of cooling to avoid overheating of the proteins. Afterwards, the Gram-positive cells were disrupted using 0.75 g of 4 mm glass beads that were added to the samples and were subsequently treated in a “Fast prep 96 Homogenizer” (MP Biomedicals Germany GmbH, Eschwege, Germany) for 2 × 20 s, followed by centrifugation at 5,500 g for one minute. The supernatant was then removed and samples were centrifuged at 13,500 xg for 10 min at 4°C in a tabletop centrifuge. Finally, the supernatant was used for a Pierce assay, that was performed to determine the protein concentration of all samples. After this, the concentrations were adjusted to 1 μg/μL for DIA-MS analysis. For all samples, biological triplicates were prepared.
Protein samples were loaded onto a 4–12% NuPAGE Novex Bis-Tris Minigels (Invitrogen) and run into the gel for 1.5 cm. Following Coomassie staining, the protein areas were cut out, diced, and subjected to reduction with dithiothreitol, alkylation with iodoacetamide and finally overnight digestion with trypsin was performed. Tryptic peptides were extracted from the gel, the solution dried in a Speedvac and kept at −20°C for further analysis.
Protein digests were analyzed on a nanoflow chromatography system (nanoElute) hyphenated to a hybrid timed ion mobilityquadrupole-time of flight mass spectrometer (timsTOF Pro, all Bruker Daltonics GmbH & Co. KG, Bremen, Germany). In brief, 250 ng equivalents of peptides were dissolved in loading buffer (2% acetonitrile, 0.1% trifluoroacetic acid in water), enriched on a reversed-phase C18 trapping column (0.3 cm × 300 μm, Thermo Fisher Scientific) and separated on a reversed phase C18 column with an integrated CaptiveSpray Emitter (Aurora 25 cm × 75 μm, IonOpticks) using a 50 min linear gradient of 5–35% acetonitrile / 0.1% formic acid (v:v) at 250 nL min−1, and a column temperature of 50°C. For identification, representative samples were analyzed in PASEF acquisition mode using default manufacturer’s settings [n = 12] (Meier et al., 2018). For identification and quantification samples were analyzed in diaPASEF mode using a customized 16×2 window acquisition scheme (Skowronek et al., 2022). For each biological replicate, three technical replicates were performed in diaPASEF mode for quantitation.
The data processing was performed using the Spectronaut v16.0.220606.53000 software package (Biognosys AG, Schlieren, Switzerland). Identification of proteins as well as hybrid spectral library generation from 12×2 DDA acquisitions and 12×2 DIA acquisitions experiments were done using the Pulsar search engine against UniProtKB C. jejuni 81–176, E. faecalis ATCC 700802, E. faecium TX0016 and S. aureus NCTC 8325 proteomes using the default parameters. The False Discovery Rate (FDR) was set to 1% on the spectral, peptide and protein group levels for all samples. DIA quantification was performed with up to 6 fragments per peptide and up to 10 peptides per protein. A dynamic retention time alignment was done, as well as dynamic mass recalibration and quartile normalization, for the 1% FDR. Imputation of global data was executed for the final results table.
Perseus v1.6.2.2 was used for the statistical analysis and for generation of volcano plots to compare the different samples (Storey and Tibshirani, 2003; Tyanova et al., 2016). As significant regulation level, two-fold up- or down-expression was chosen. Proteins present in 80% of the samples were considered for further analysis. For volcano-plot generation in Perseus, a t-test was chosen with a number of randomizations = 250 and a FDR of 0.05 (Storey and Tibshirani, 2003). All proteins that are described in the following as up- or down-expressed were significantly regulated, if not otherwise stated.
Clusters of orthologous genes (COG)-categories were assigned to the proteins using the online-tool eggNOGmapper v 2.18 (Huerta-Cepas et al., 2017, 2019; Cantalapiedra et al., 2021). To identify commonly expressed proteins, Venn diagrams were generated utilizing InteractiVenn (Heberle et al., 2015). All Plots were generated using matplotlib in python3 (Van Rossum and Drake, 2009).
The interbacterial communication between C. jejuni and other bacterial species remains poorly explored to date, lacking comprehensive investigation. Our research is aimed to investigate mechanisms of this cross-talk and its potential implications in various ecological and pathogenic contexts.
We hypothesized that co-incubation of C. jejuni with other bacterial species triggers a proteomic response in C. jejuni. Three different Gram-positive species were chosen for co-incubation with C. jejuni, namely E. faecalis, E. faecium and S. aureus, which are all putative inhabitants of the intestinal host microbiome. The bacteria were incubated for three hours at 37°C in PBS, without nutrient supply since we were not interested in responses due to different degrees of nutrient competition (see scheme of the workflow in Figure 1). Instead, we aimed to target responses resulting from direct bacterial contact or from interactions with secreted molecules. Using volcano-plots generated from DIA-MS data, we compared the proteome of C. jejuni in monoculture with each of the three bacteria with C. jejuni in co-incubations.
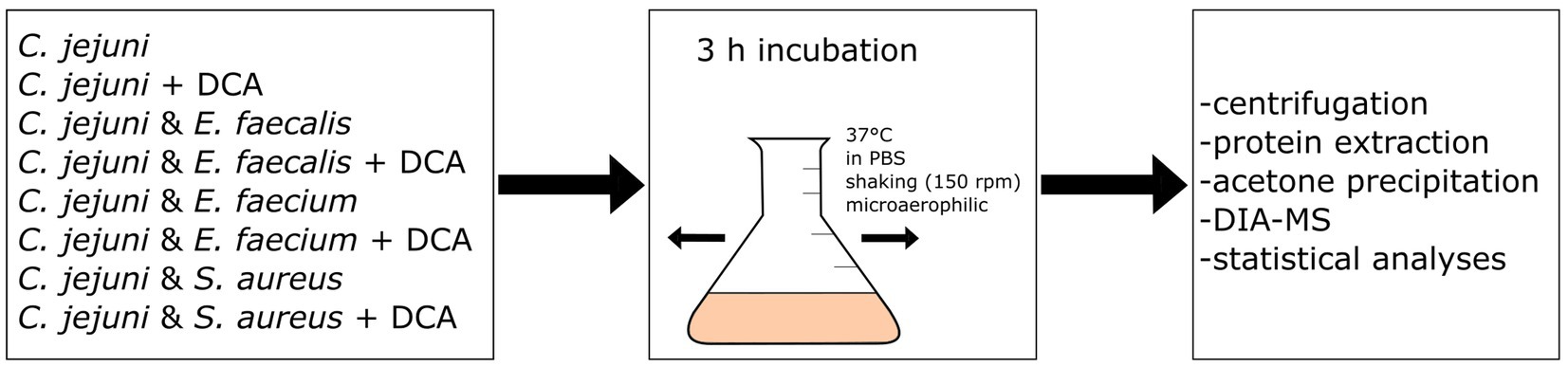
Figure 1. Workflow scheme: eight different approaches of mono- or co-incubation were prepared and incubated at 150 rpm and 37°C for three hours. After incubation, the approaches were centrifuged and the proteins were extracted and precipitated with acetone. Data-Independent Acquisition Mass Spectrometry (DIA-MS) was performed, followed by data analysis and statistical analysis.
Co-incubation resulted in all cases in an altered proteomic profile, whose dimension depends on the species used for co-incubation. With S. aureus, the changes in the proteomic profile exhibited the highest intensity with 445 differentially expressed proteins.
It is well known that S. aureus produces several toxins and hemolysins that might act against other bacteria (Shinefield, 1963; Otto, 2014). Conversely, S. aureus can also secrete beneficial substances for other microorganisms and co-exist in polymicrobial communities, which can be advantageous for infections (Nguyen and Oglesby-Sherrouse, 2016; García-Pérez et al., 2018; Karki et al., 2021). These characteristics of S. aureus might contribute to the increased number of differentially expressed proteins in the co-incubation with C. jejuni.
In the co-incubation assay with E. faecium, 405 proteins were differentially expressed and in the assay with E. faecalis, 241 proteins were differentially expressed. The ratio of up-expressed and down-expressed proteins also varied specifically.
Among the differential expressed proteins, 54 were commonly up-expressed in all three co-incubation approaches and 100 proteins were commonly down-expressed (Figure 2). The distribution of COG-categories differs between commonly up-expressed and down-expressed proteins (Figure 3). Down-expressed proteins are characterized by a higher proportion of the categories C (energy production and conversion), E (amino acid metabolism and transport), F (nucleotide metabolism and transport), I (lipid metabolism) and G (carbohydrate metabolism and transport). In contrast, up-expressed proteins are characterized by a higher proportion of the categories J (translation), L (replication and repair), M (cell wall / membrane / envelope biogenesis) and T (signal transduction).
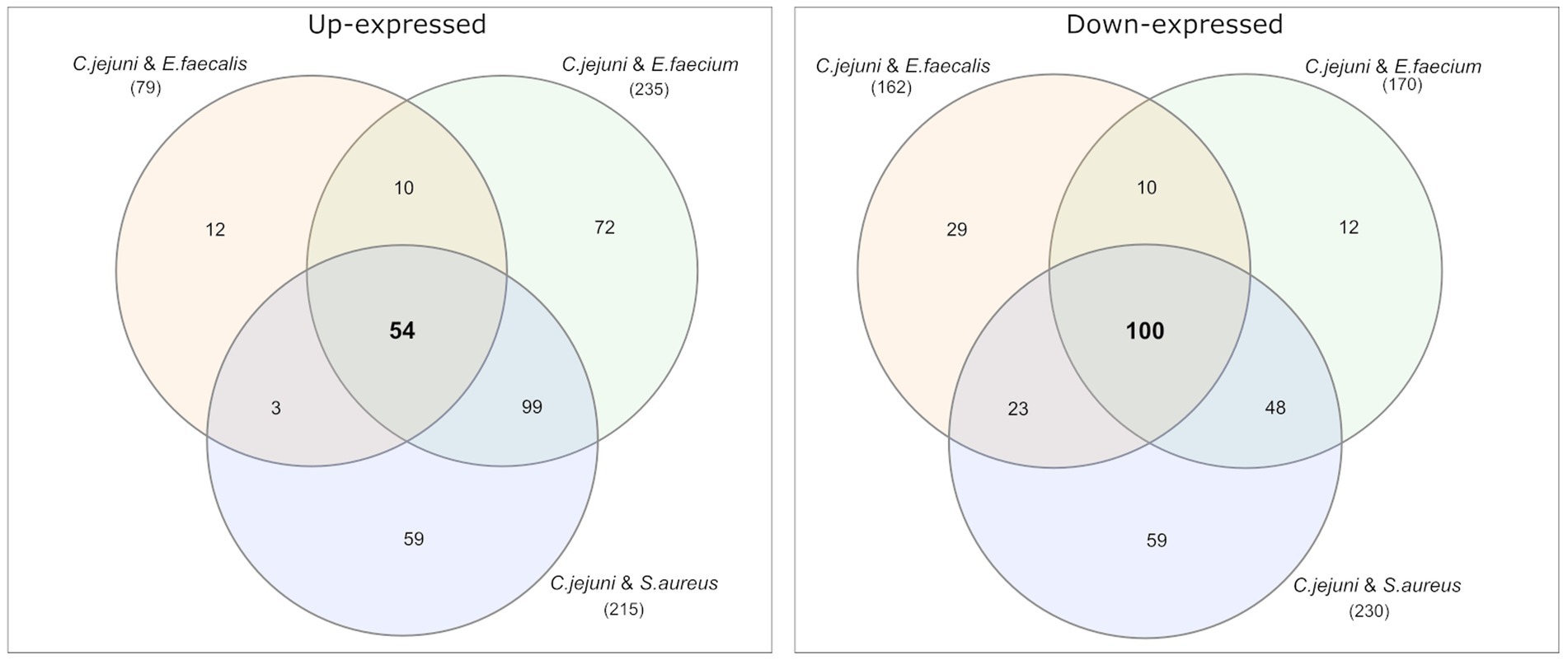
Figure 2. Venn diagrams were used to show the commonly up- and down-expressed proteins in C. jejuni during co-incubation with E. faecalis, E. faecium, and S. aureus in the pellet. The analysis revealed that 54 proteins were commonly up-expressed in all three co-incubation approaches, while 100 proteins were commonly down-expressed.
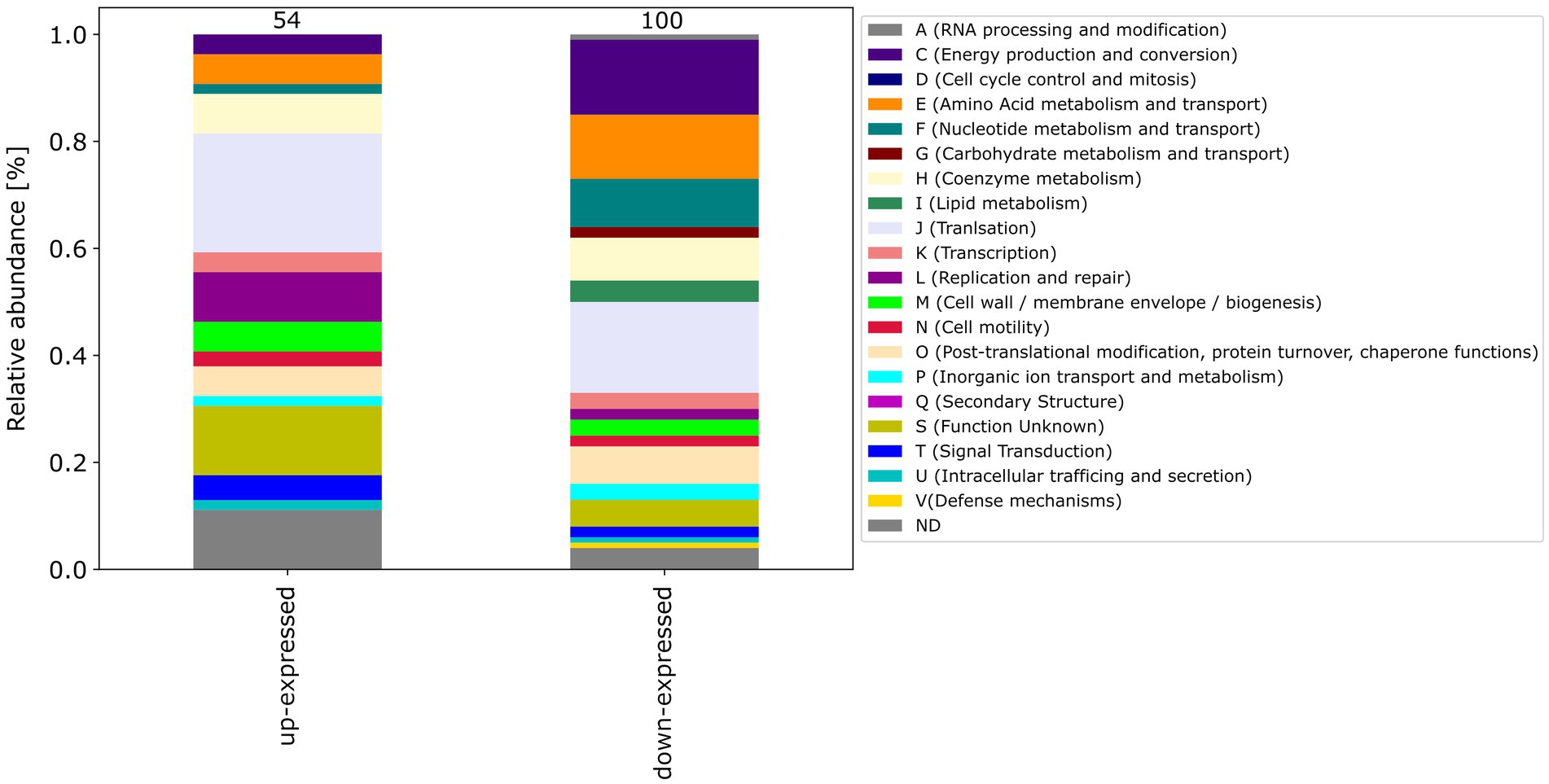
Figure 3. This figure shows the COG-categories of the 54 proteins commonly up-expressed and 100 proteins commonly down-expressed in all three co-incubation approaches. The samples were normalized before analysis. The stacked bar plot displays the percentage distribution of the COG-categories, with different colors representing different categories.
The differentially expressed proteins in all approaches were sorted according to their difference expression level. We compared the top 20 up- and down-expressed proteins of each co-incubation proteome (see Supplementary Excel file), in order to identify commonly regulated proteins with a high degree of regulation. Four commonly up-expressed proteins were found in the top 20 up-expressed proteins: hemolysin A (A0A0H3PEK7_CAMJJ), a DNA/RNA non-specific endonuclease (A0A0H3PJE6_CAMJJ), a putative lipoprotein (A0A0H3PA71_CAMJJ), and a putative membrane protein (A0A0H3PDB2_CAMJJ). Hemolysin A is associated with the lysis of cells via disruption of the cell membrane. This phenomenon is particularly observed for red blood cells (erythrocytes), as certain pathogens have the ability to cause lysis of these cells to access an iron source (Skals et al., 2010; Wiles and Mulvey, 2013). Therefore hemolysis is considered as an important virulence factor (Elliott et al., 1998; Kielian et al., 2001; Vandenesch et al., 2012). The putative lipoprotein can be associated with cell–cell contact with host epithelial cells, but probably also with other bacteria (Speare et al., 2022). Putative lipoproteins can be considered as virulence factors, as well. Many outer membrane proteins in pathogenic bacteria are virulence factors that enable or facilitate bacterial attachment to host cell surfaces (Gao et al., 2021). The exact function of the putative membrane protein remains unknown. However, this membrane associated protein might also play a role in intercellular communication. Lipoproteins are a diverse group of membrane proteins that play crucial roles in various biological processes, including cellular physiology, cell division, and virulence. They have significant importance and impact on these phenomena (Kovacs-Simon et al., 2011).
Among the top 20 up-expressed proteins in co-incubation were three membrane-interactive proteins, which might indicate an increased adherence to other bacteria through contact with other bacterial species. Enhanced adherence is only one possible reason for the high expression of membrane associated proteins in co-incubation. Whether these effects are due to co-incubation remains unclear. Our study employed in vitro co-incubation experiments to investigate proteomic changes. While these experiments allow for controlled conditions and precise analysis, they may not fully replicate the complex interactions that occur within host environments. A genomic assessment could validate this hypothesis. Nevertheless, it is known that in presence of other commensal microbes, some (pathogenic) bacteria can enhance their virulence (Stacy et al., 2016; Fast et al., 2018).
Moreover, four commonly down-expressed proteins were found in the top 20 down-expressed proteins, namely the translation initiation factor IF-3 (IF3_CAMJJ), a DNA-directed RNA polymerase subunit omega (RPOZ_CAMJJ), an ATP synthase subunit beta (ATPB_CAMJJ) and a 6,7-dimethyl-8-ribityllumazine synthase (RISB_CAMJJ).
Some Campylobacter strains possess the capability to employ a type 6 secretion system (T6SS), which can be used for communication with their surrounding environment including other bacteria (Chen et al., 2015; Gallique et al., 2017). However, C. jejuni 81–176 does not harbor a type 6 secretion system (Liaw et al., 2019), which implies the utilization of alternative mechanisms for bacterial communication. However, other C. jejuni strains, for example the strains C. jejuni 488, 43,431 or RC039 utilize a type 6 secretion system (Liaw et al., 2019), indicating that cross-talk via a type 6 secretion system-dependent protein secretion would be possible in some C. jejuni strains.
In order to identify proteins that are specifically regulated during co-incubation, we compared the changes in the proteomic profile after co-incubation with the stress response during incubation with bile acids, which was previously shown to trigger a strong proteomic stress response in C. jejuni (Masanta et al., 2019). After 3 h incubation with 0.1% DCA, a substantial proportion of C. jejuni proteins were differentially expressed. A total of 526 proteins were identified among the up-expressed proteins, which is ~10-fold more than the 54 up-expressed proteins during co-incubation with Gram-positve bacteria. Likewise, 516 proteins were down-expressed after DCA incubation, which is ~5-fold more than the number during co-incubation with Gram-positve bacteria. This leads to the assumption that the exposure to DCA provokes a significantly more pronounced proteomic response compared to the co-incubation scenarios.
Venn diagrams show the overlapping proteins between both approaches (Figure 4 and Supplementary Figures S2, S3). Out of the 54 commonly up-expressed proteins during co-incubation, 36 proteins were also found in C. jejuni monoculture with DCA. This indicates that only the 18 remaining proteins are specific for co-incubation (see Supplementary Table S1). Moreover, from the 516 down-expressed proteins in C. jejuni in presence of DCA, 78 were shared with the 100 down-expressed proteins in the co-incubation approach (Figure 4), indicating that the 22 remaining proteins are specifically down-expressed in co-incubation (see Supplementary Table S6). In total, 343 proteins were commonly up-expressed and 277 proteins were commonly down-expressed in presence of DCA and co-incubation (Figure 5).
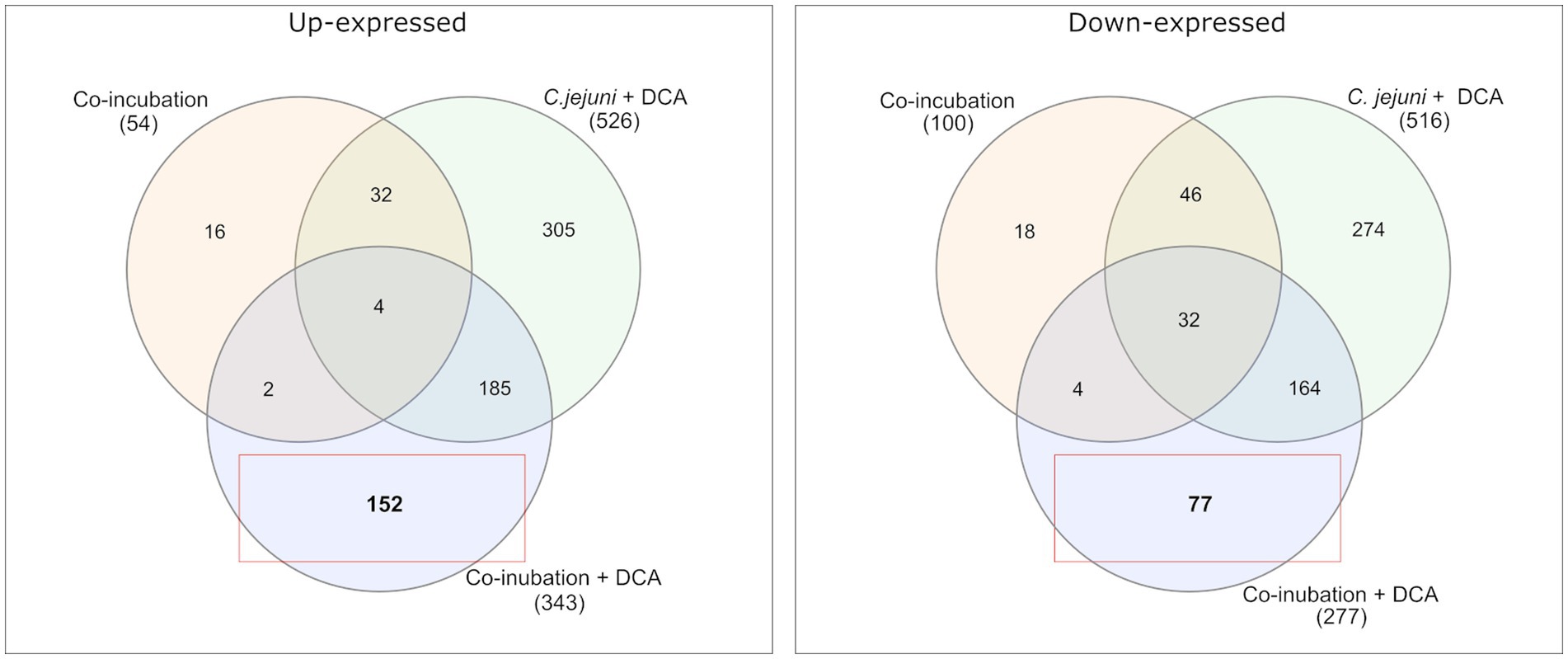
Figure 4. A Venn diagram is presented to compare the commonly up-expressed proteins (left) of C. jejuni in co-incubation with E. faecalis, E. faecium, and S. aureus with and without DCA, and the up-expressed proteins of C. jejuni with DCA in monoculture. A total of 152 proteins were specifically detected in co-incubation with DCA and not in the other approaches. The down-expressed proteins are shown on the right. The red boxes highlight proteins that are uniquely and specifically expressed in the co-incubation with DCA approach.
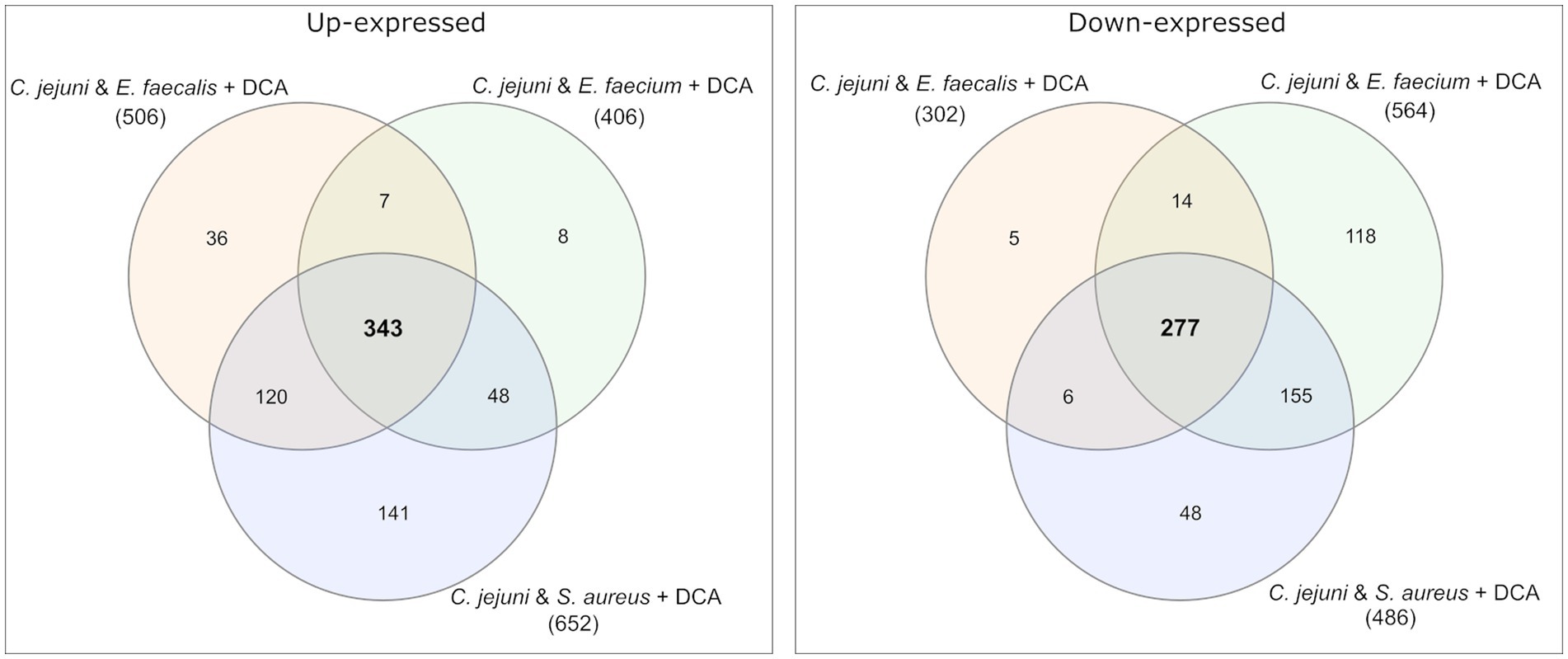
Figure 5. Venn diagrams were used to illustrate the commonly up- and down-expressed proteins of C. jejuni in co-incubation with E. faecalis, E. faecium, and S. aureus in the pellet after the addition of DCA. The analysis revealed that 343 proteins were commonly up-expressed in all three co-incubation approaches, while 277 proteins were commonly down-expressed.
The pattern of the COG categories of differentially proteins in the monoculture approach with DCA differs from commonly expressed proteins in co-incubation (Figures 67). The percentage of up-expressed proteins assigned to the categories J (translation), L (replication and repair) and T (signal transduction) is higher in the co-incubation proteome, while categories C (energy production and conversion), G (carbohydrate metabolism and transport), M (cell wall / membrane / envelope / biogenesis) and V (defense mechanisms) are more present in the monoculture of C. jejuni and DCA. Categories C, E, F and J are more down-expressed in the co-incubation approach.
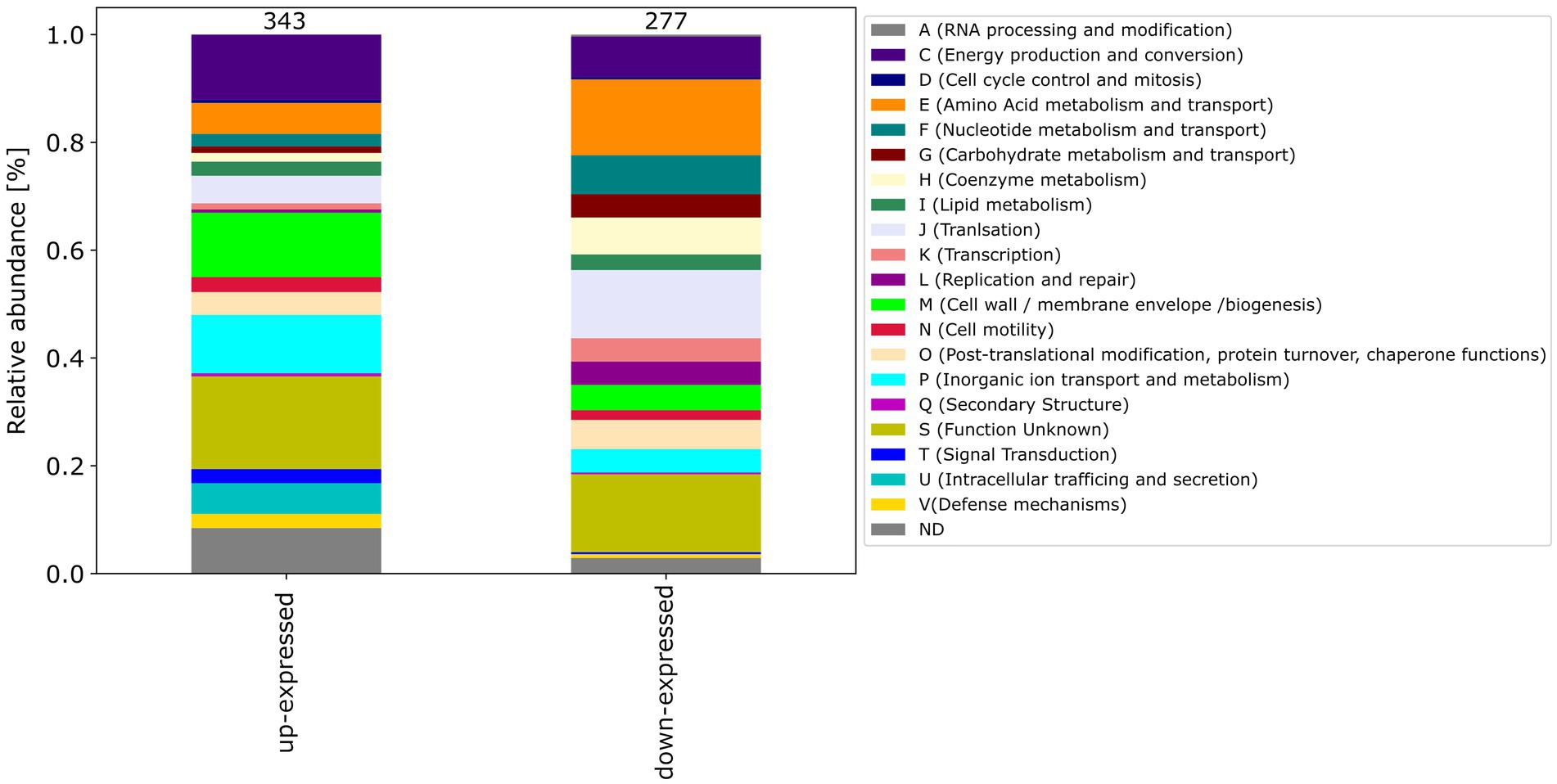
Figure 6. Stacked bar plots were generated to visualize the up- and down-expressed proteins of C. jejuni during co-incubation with DCA. A total of 343 up-expressed and 277 down-expressed proteins were assigned to their respective COG categories. The stacked bar plot displays the percentage distribution of COG categories in different colors.
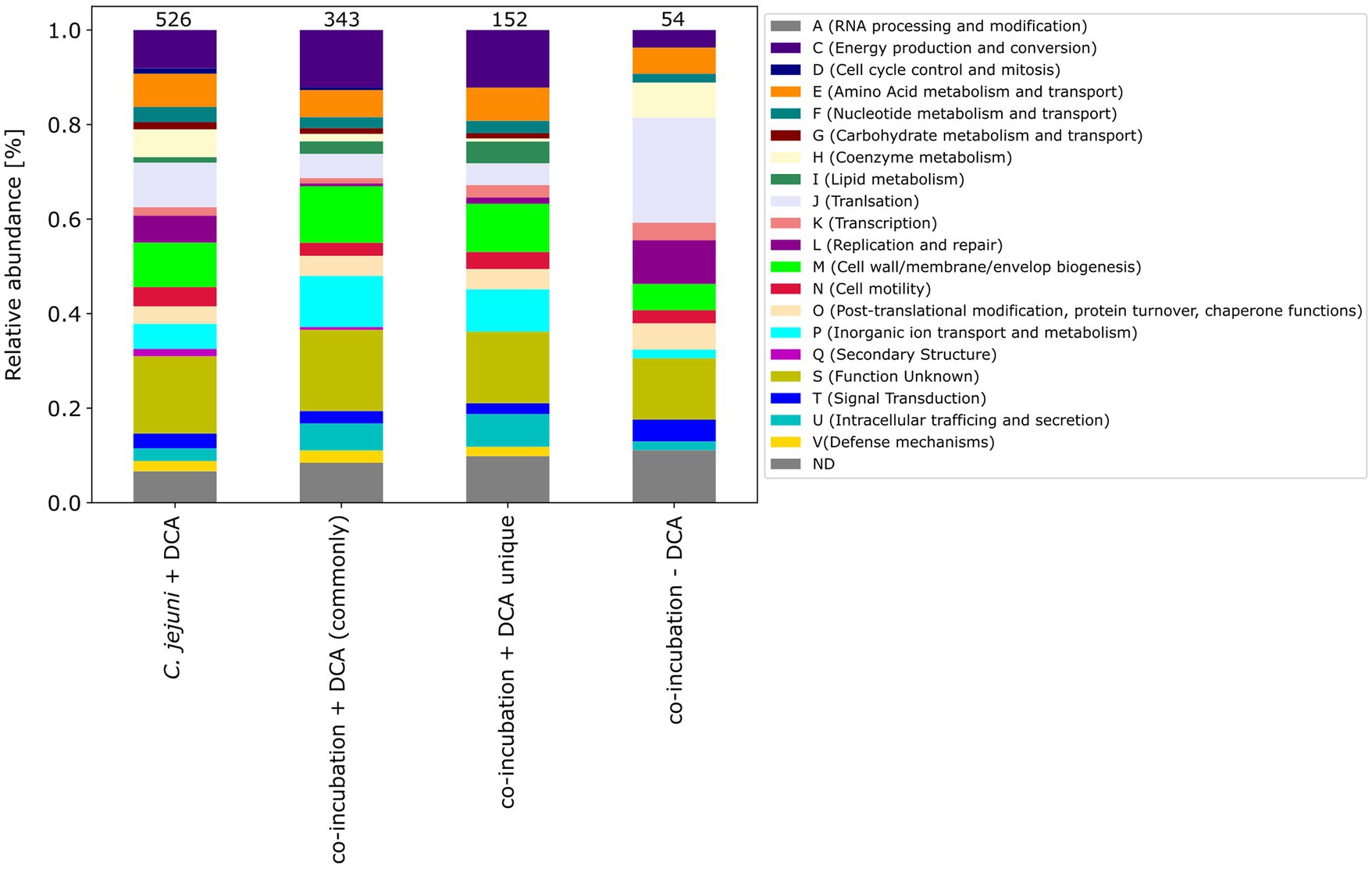
Figure 7. The COG categories of the proteins that were up-expressed in the C. jejuni mono-culture approach with DCA, the proteins that were commonly expressed in co-incubation with DCA, and the unique up-expressed proteins of the co-cultivation approach are shown. For comparison, the approach of the 54 up-expressed proteins in co-incubation without DCA is also depicted on the right.
In C. jejuni the most relevant mechanism to survive bile acid stress is the CmeABC multidrug efflux pump, that belongs to the resistance nodulation-division (RND) type multidrug efflux systems (Lin et al., 2003). CmeABC consists of a three-gene operon encoding for a membrane fusion protein - CmeA, the efflux pump membrane transporter - CmeB and CmeC, which is the outer membrane lipoprotein (Lin et al., 2002). Knockout mutants of these genes led to significant loss of bile acid resistance (Lin et al., 2003). In a proteomic study, Masanta et al. showed that the proteins belonging to the CmeABC multidrug efflux pump were up-expressed under bile acid stress exposure (Masanta et al., 2019). Furthermore, Malik-Kale et al. found CmeABC and other virulence genes up-regulated under DCA stress in a microarray study (Malik-Kale et al., 2008).
Both publications explored the response of C. jejuni to bile acids, but they differ in their methods and focus. Malik-Kale et al. investigated the effect of DCA on the virulence gene expression of C. jejuni such as ciaB, cmeABC, dccR, and tlyA using microarray analyses, while Masanta et al. used label-free mass spectrometry to analyze the proteome of C. jejuni in response to sublethal concentrations of DCA. However, both studies focused on the response of C. jejuni to bile acids and suggest that bile acids can alter the behavior of C. jejuni. In contrast to Malik-Kale and colleagues, who focused on the kinetics of cell invasion, Masanta et al. focused on the downregulation of basic biosynthetic pathways and the transcription machinery.
As in Masanta et al.’s study, we used data independent mass spectrometry to obtain the data. However, the focus of our study was the proteome during co-incobation under bile acid stress. Due to the findings in previous work on the reaction of C. jejuni to DCA, the presence of CmeA, B or C in all our samples with DCA served as indicator that the proteome under bile acid stress is detected and depicted. In the co-incubation approach without DCA, none of the the CmeABC proteins was detected (Supplementary Table S2).
Among the 22 specifically down-expressed proteins during co-incubation were mostly general metabolic proteins. In the 18 commonly up-expressed proteins during co-incubation, we found proteins that might play a role in the interaction between C. jejuni with other bacteria. For example, a conjugative transfer regulon protein (Q9KIR9_CAMJJ) was detected among the up-expressed proteins in all three samples. The presence of this protein indicates that horizontal gene transfer may be occurring between these bacteria, whereby genetic material can be exchanged between different species (Llosa et al., 2002). This mechanism of genetic exchange could allow for the acquisition of novel genetic traits, such as antibiotic resistance or other beneficial genes and indicates a potential for cross-communication between bacteria.
Additionally, the chaperone protein DnaJ was found among these proteins (DNAJ_CAMJJ), indicating an active response toward stress. DnaJ and related Hsp proteins are highly conserved among species and play a role in diverse processes such as folding and unfolding of proteins, translation and ATPase activity of specific chaperones (Qiu et al., 2006). This indicates that the bacteria might be stressed by either the presence of other bacteria or the absence of nutrients.
We also studied the proteomic response in the presence of both triggers, DCA plus co-incubation with Gram-positive bacteria. This should reveal the relative influence of the individual triggers on the common response. Among the 18 up-expressed proteins that were specific to co-incubation, only two were up-expressed in the approach of co-incubation with DCA (Figure 4). These proteins were a Histidine kinase (A0A0H3PE96_CAMJJ) and a tRNA modification GTPase MnmE (MNME_CAMJJ) (Supplementary Table S1). Furthermore, of 22 down-expressed proteins that were specific for co-incubation, only four proteins remained down-expressed when DCA was added. The limited number of commonly regulated proteins in co-incubation with and without DCA indicates that DCA seems to suppress the specific co-incubation response to a large extent.
Comparing the co-incubation plus DCA approach to the monoculture of C. jejuni with DCA, 185 proteins occurred commonly among the up-expressed candidates, which corresponded to ~37.8% of the 490 proteins that were up-expressed in the monoculture with DCA, excluding the 36 proteins, that also occurred in co-incubation without DCA (Figures 45). This led to the assumption that the additional trigger of co-incubation might also inhibit the expression of a certain amount of the DCA response specific proteins in C. jejuni. Moreover, 77 proteins were uniquely down-expressed in the approach of co-incubation plus DCA (Figure 4), while 196 of the 277 down-expressed proteins in this approach were shared with the C. jejuni monoculture with DCA.
The proteomes in co-incubation with and without DCA exhibit significant dissimilarities. In total, 152 proteins were found to be specifically up-expressed when both triggers, co-incubation plus DCA, are present. Due to the fact that these 152 proteins occurred only in the approach co-incubation plus DCA, and were not a combination of both triggers, it can be assumed, that the proteomic response in presence of both, DCA and another bacterium possesses a unique character (Figure 4).
Moreover, the respective COG-categories were assigned to these 152 proteins (Figure 7). Compared to the monoculture proteome with DCA, the categories M (cell wall / membrane envelope / biogenesis), P (inorganic ion transport and metabolism) and U (intracellular trafficking) were increased in co-incubation with DCA. A detailed analysis of these 152 proteins revealed a high number of ABC-transporter associated proteins, proteins related to antibiotic resistance, efflux and transport proteins and general membrane proteins (Supplementary Table S5).
Furthermore, the COG categories of the 77 proteins commonly exclusively down-expressed in the approach of co-incubation with DCA were determined. When compared to the 516 down-expressed proteins in C. jejuni with DCA and the 277 commonly down-expressed proteins in co-incubation with DCA, the pattern of the 77 proteins shows similarities but also differences (Figure 8). An increase of proteins belonging to the category E (amino acid metabolism and transport) was observed and a decrease of proteins belonging to the category J (translation) was observed when compared to the other samples.
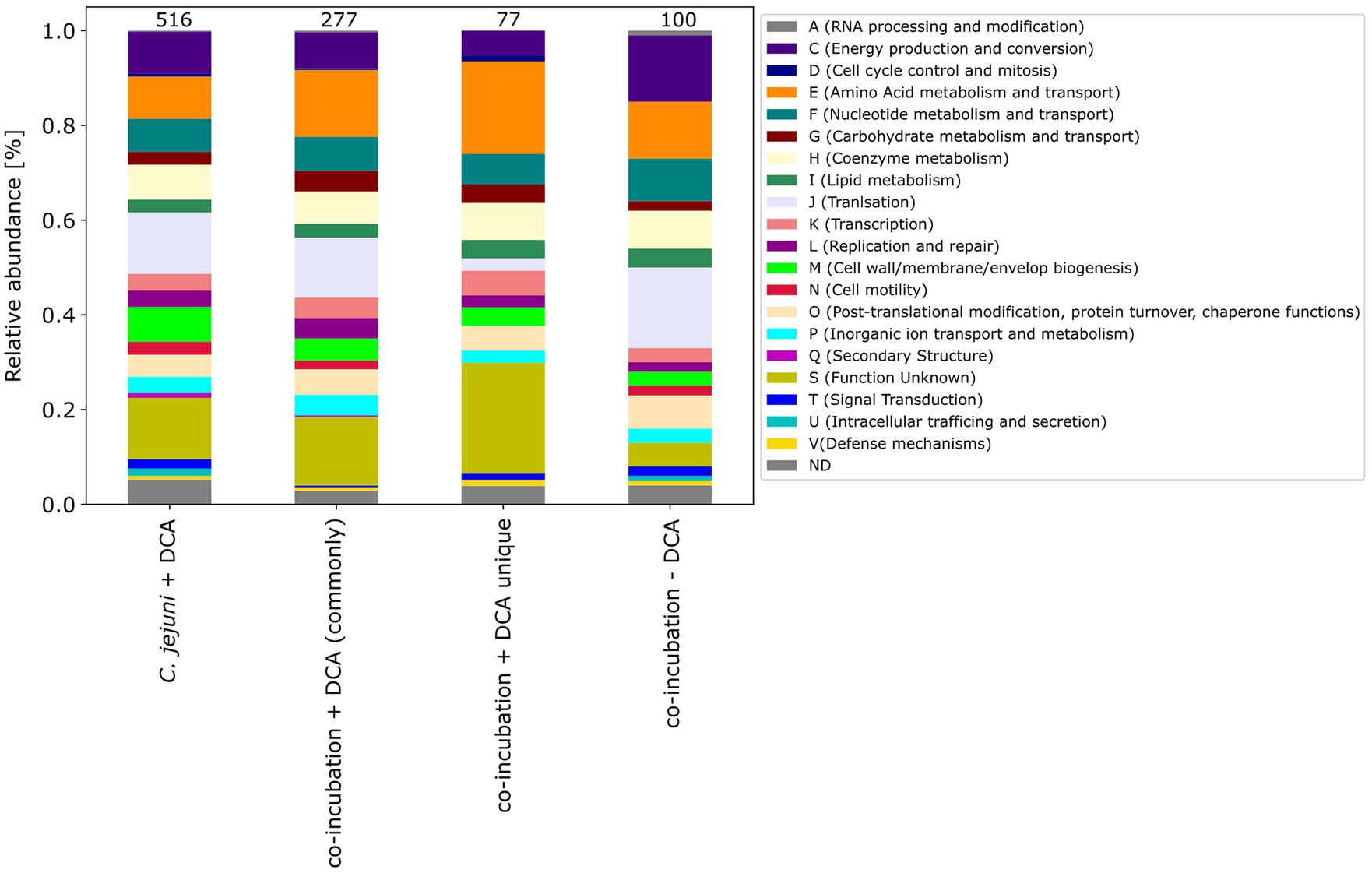
Figure 8. The COG categories of proteins that were down-expressed in the C. jejuni mono-culture approach with DCA, as well as the commonly expressed proteins in co-incubation with DCA and the unique up-expressed proteins of the co-cultivation approach are depicted. For comparison, the approach of the 100 down-expressed proteins in co-incubation without DCA is shown on the right.
In summary, our investigation highlights the proteomic response of C. jejuni to co-incubation as well as bile acid stress. We cover a high percentage of the total proteome of C. jejuni in our DIA-MS analysis, which demonstrates a small but distinct interaction potential between C. jejuni and the other bacteria via membrane-interactive proteins, indicating that the other bacteria contribute to an increased virulence in the environment.
We also report a remarkable overlap between the proteomic response of C. jejuni in co-incubation in presence of DCA and the approach of C. jejuni monoculture with bile acid.
However, we were able to identify a unique response when both triggers (co-incubation and DCA) are present. This distinct response highlights the complexity of cellular interactions and shows the potential role of C. jejuni in proteomic response pathways under these specific conditions and enables future research in the field of proteomic analyses under different influences.
One limitation of this study is the labor-intensive nature of DIA MS analysis, which makes it difficult to undertake additional research in this experimental setup. Furthermore, the findings may not be generalizable to other Campylobacter strains since only a single strain (C. jejuni 81–176) was included. Although the study primarily focused on the C. jejuni proteome, there is a lack of comprehensive analysis regarding the proteomic responses of the other bacteria involved in the co-incubation. To provide a more holistic understanding of the interactions and proteomic dynamics within the complex co-incubation system, future research should aim to explore this aspect. We acknowledge that our study primarily focused on proteomic changes in C. jejuni during co-incubation with other bacteria. While our results provide valuable insights into proteomic alterations, we did not perform a genomic assessment of C. jejuni. Therefore, we are unable to definitively conclude that the observed effects in communication and potential virulence enhancement are solely due to co-incubation.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD046477.
CL, WB, UG, and AZ: conceptualization. CL, WB, and AZ: methodology. CL and AD: software. AD, WB, and CL: validation. AD and CL: formal analysis and investigation. AD performed growth curve analysis and prepared bacterial samples. CL performed mass-spectrometric analysis and resources. UG and AZ: data curation. AD and CL: writing—original draft preparation. AD: writing—review and editing. AD, CL, UG, WB, and AZ: visualization. AD prepared all figures. WB and AZ: supervison. AZ: project administration. AZ and UG: funding acqusition. All authors have read and agreed to the published version of the manuscript.
This work was funded by the Deutsche Forschungsgemeinschaft (DFG) (grant number ZA 697/6–1). The APC of the paper was funded by the Open Access support program of the Deutsche Forschungsgemeinschaft and the publication fund of the Georg August Universität Göttingen.
We thank Lisa Neuenroth and Fabio Trebini (UMG) for the implementation of DIA-MS. This publication is part of AD’s doctoral study. This publication is part of Annika Dreyer’s doctoral study “Proteomic Profiles of Campylobacter jejuni and Enterococci during Co-incubation and under Bile Acid Stress” (2023).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1247211/full#supplementary-material
Acheson, D., and Allos, B. M. (2001). Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32, 1201–1206. doi: 10.1086/319760
Anis, N., Bonifait, L., Quesne, S., Baugé, L., Yassine, W., Guyard-Nicodème, M., et al. (2022). Survival of Campylobacter jejuni co-cultured with Salmonella spp. in aerobic conditions. Pathogens 11:812. doi: 10.3390/pathogens11070812
Begley, M., Gahan, C. G. M., and Hill, C. (2005). The interaction between bacteria and bile. FEMS Microbiol. Rev. 29, 625–651. doi: 10.1016/j.femsre.2004.09.003
Blaser, M. J., LaForce, F. M., Wilson, N. A., and Wang, W. L. L. (1980). Reservoirs for human Campylobacteriosis. J. Infect. Dis. 141, 665–669. doi: 10.1093/infdis/141.5.665
Blaser, M. J., Taylor, D. N., and Feldman, R. A. (1983). Epidemiology of Campylobacter jejuni infections. Epidemiol. Rev. 5, 157–176. doi: 10.1093/oxfordjournals.epirev.a036256
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P., and Huerta-Cepas, J. (2021). eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829. doi: 10.1093/molbev/msab293
Chen, L., Zou, Y., She, P., and Wu, Y. (2015). Composition, function, and regulation of T6SS in Pseudomonas aeruginosa. Microbiol. Res. 172, 19–25. doi: 10.1016/j.micres.2015.01.004
Cheung, G. Y. C., Bae, J. S., and Otto, M. (2021). Pathogenicity and virulence of Staphylococcus aureus. Virulence 12, 547–569. doi: 10.1080/21505594.2021.1878688
Chiang, J. Y. (2017). Recent advances in understanding bile acid homeostasis. F1000Res 6:2029. doi: 10.12688/f1000research.12449.1
Ellepola, K., Truong, T., Liu, Y., Lin, Q., Lim, T. K., Lee, Y. M., et al. (2019). Multi-omics analyses reveal synergistic carbohydrate metabolism in Streptococcus mutans-Candida albicans mixed-species biofilms. Infect. Immun. 87, e00339–e00319. doi: 10.1128/IAI.00339-19
Elliott, S. J., Srinivas, S., Albert, M. J., Alam, K., Robins-Browne, R. M., Gunzburg, S. T., et al. (1998). Characterization of the roles of Hemolysin and other toxins in enteropathy caused by alpha-hemolytic Escherichia coli linked to human diarrhea. Infect. Immun. 66, 2040–2051. doi: 10.1128/IAI.66.5.2040-2051.1998
Fast, D., Kostiuk, B., Foley, E., and Pukatzki, S. (2018). Commensal pathogen competition impacts host viability. Proc. Natl. Acad. Sci. U. S. A. 115, 7099–7104. doi: 10.1073/pnas.1802165115
Fiore, E., Van Tyne, D., and Gilmore, M. S. (2019). Pathogenicity of Enterococci. Microbiol. Spectr. 7. doi: 10.1128/microbiolspec.GPP3-0053-2018
Gallique, M., Bouteiller, M., and Merieau, A. (2017). The type VI secretion system: a dynamic system for bacterial communication? Front. Microbiol. 8:1454. doi: 10.3389/fmicb.2017.01454
Gao, Q., Lu, S., Wang, M., Jia, R., Chen, S., Zhu, D., et al. (2021). Putative Riemerella anatipestifer outer membrane protein H affects virulence. Front. Microbiol. 12:708225. doi: 10.3389/fmicb.2021.708225
García-Pérez, A. N., de Jong, A., Junker, S., Becher, D., Chlebowicz, M. A., Duipmans, J. C., et al. (2018). From the wound to the bench: exoproteome interplay between wound-colonizing Staphylococcus aureus strains and co-existing bacteria. Virulence 9, 363–378. doi: 10.1080/21505594.2017.1395129
Gorrie, C., Higgs, C., Carter, G., Stinear, T. P., and Howden, B. (2019). Genomics of vancomycin-resistant Enterococcus faecium. Microb. Genomics 5:e000283. doi: 10.1099/mgen.0.000283
Heberle, H., Meirelles, G. V., da Silva, F. R., Telles, G. P., and Minghim, R. (2015). InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics 16:169. doi: 10.1186/s12859-015-0611-3
Huang, Q., Yang, L., Luo, J., Guo, L., Wang, Z., Yang, X., et al. (2015). SWATH enables precise label-free quantification on proteome scale. Proteomics 15, 1215–1223. doi: 10.1002/pmic.201400270
Huerta-Cepas, J., Forslund, K., Coelho, L. P., Szklarczyk, D., Jensen, L. J., von Mering, C., et al. (2017). Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122. doi: 10.1093/molbev/msx148
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi: 10.1093/nar/gky1085
Kandell, R. L., and Bernstein, C. (1991). Bile salt/acid induction of DNA damage in bacterial and mammalian cells: implications for colon cancer. Nutr. Cancer 16, 227–238. doi: 10.1080/01635589109514161
Karki, A. B., Ballard, K., Harper, C., Sheaff, R. J., and Fakhr, M. K. (2021). Staphylococcus aureus enhances biofilm formation, aerotolerance, and survival of Campylobacter strains isolated from retail meats. Sci. Rep. 11:13837. doi: 10.1038/s41598-021-91743-w
Kielian, T., Cheung, A., and Hickey, W. F. (2001). Diminished virulence of an alpha-toxin mutant of Staphylococcus aureus in experimental brain abscesses. Infect. Immun. 69, 6902–6911. doi: 10.1128/IAI.69.11.6902-6911.2001
Klančnik, A., Gobin, I., Jeršek, B., Smole Možina, S., Vučković, D., Tušek Žnidarič, M., et al. (2020). Adhesion of Campylobacter jejuni is increased in association with foodborne Bacteria. Microorganisms 8:201. doi: 10.3390/microorganisms8020201
Klevens, R. M., Morrison, M. A., Nadle, J., Petit, S., Gershman, K., Ray, S., et al. (2007). Invasive methicillin-resistant Staphylococcus aureus infections in the United States. Jama 298, 1763–1771. doi: 10.1001/jama.298.15.1763
Kovacs-Simon, A., Titball, R. W., and Michell, S. L. (2011). Lipoproteins of bacterial pathogens. Infect. Immun. 79, 548–561. doi: 10.1128/IAI.00682-10
Lebreton, F., Willems, R. J. L., and Gilmore, M. S. (2014). “Enterococcus Diversity, Origins in Nature, and Gut Coloni-zation”, in Enterococci: From Commensals to Leading Causes of Drug Resistant Infection, eds. M. S. Gilmore, D. B. Clewell, Y. Ike, and N. Shankar (Boston: Massachusetts Eye and Ear Infirmary). Available at: http://www.ncbi.nlm.nih.gov/books/NBK190427/ (Accessed October 28, 2023).
Lertpiriyapong, K., Gamazon, E. R., Feng, Y., Park, D. S., Pang, J., Botka, G., et al. (2012). Campylobacter jejuni type VI secretion system: roles in adaptation to Deoxycholic acid, host cell adherence, invasion, and in vivo colonization. PLoS One 7:e42842. doi: 10.1371/journal.pone.0042842
Liaw, J., Hong, G., Davies, C., Elmi, A., Sima, F., Stratakos, A., et al. (2019). The Campylobacter jejuni type VI secretion system enhances the oxidative stress response and host colonization. Front. Microbiol. 10:2864. doi: 10.3389/fmicb.2019.02864
Lin, J., Michel, L. O., and Zhang, Q. (2002). CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46, 2124–2131. doi: 10.1128/AAC.46.7.2124-2131.2002
Lin, J., Sahin, O., Michel, L. O., and Zhang, Q. (2003). Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect. Immun. 71, 4250–4259. doi: 10.1128/IAI.71.8.4250-4259.2003
Llosa, M., Gomis-Ruth, F. X., Coll, M., and Cruz, F. D. L. (2002). Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45, 1–8. doi: 10.1046/j.1365-2958.2002.03014.x
Lopes, M. de F. S., Simões, A. P., Tenreiro, R., Marques, J. J. F., and Crespo, M. T. B. (2006). Activity and expression of a virulence factor, gelatinase, in dairy enterococci. Int. J. Food Microbiol. 112, 208–214. doi: 10.1016/j.ij-foodmicro.2006.09.004
Malik-Kale, P., Parker, C. T., and Konkel, M. E. (2008). Culture of Campylobacter jejuni with sodium Deoxycholate induces virulence gene expression. J. Bacteriol. 190, 2286–2297. doi: 10.1128/JB.01736-07
Masanta, W. O., Zautner, A. E., Lugert, R., Bohne, W., Gross, U., Leha, A., et al. (2019). Proteome profiling by label-free mass spectrometry reveals differentiated response of Campylobacter jejuni 81–176 to sublethal concentrations of bile acids. Prot. Clin. Appl. 13:e1800083. doi: 10.1002/prca.201800083
Meier, F., Brunner, A.-D., Koch, S., Koch, H., Lubeck, M., Krause, M., et al. (2018). Online parallel accumulation–serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteomics 17, 2534–2545. doi: 10.1074/mcp.TIR118.000900
Negretti, N. M., Clair, G., Talukdar, P. K., Gourley, C. R., Huynh, S., Adkins, J. N., et al. (2019). Campylobacter jejuni demonstrates conserved proteomic and transcriptomic responses when co-cultured with human INT 407 and Caco-2 epithelial cells. Front. Microbiol. 10:755. doi: 10.3389/fmicb.2019.00755
Neveling, D. P., and Dicks, L. M. T. (2021). Probiotics: an antibiotic replacement strategy for healthy broilers and productive rearing. Probiotics & Antimicro. Prot. 13, 1–11. doi: 10.1007/s12602-020-09640-z
Nguyen, A. T., and Oglesby-Sherrouse, A. G. (2016). Interactions between Pseudomonas aeruginosa and Staphylococcus aureus during co-cultivations and polymicrobial infections. Appl. Microbiol. Biotechnol. 100, 6141–6148. doi: 10.1007/s00253-016-7596-3
Otto, M. (2014). Staphylococcus aureus toxins. Curr. Opin. Microbiol. 17, 32–37. doi: 10.1016/j.mib.2013.11.004
Qiu, X.-B., Shao, Y.-M., Miao, S., and Wang, L. (2006). The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 63, 2560–2570. doi: 10.1007/s00018-006-6192-6
Quinn, E. M., Kilcoyne, M., Walsh, D., Joshi, L., and Hickey, R. M. (2020a). A whey fraction rich in immunoglobulin G combined with Bifidobacterium longum subsp. infantis ATCC 15697 exhibits synergistic effects against Campylobacter jejuni. IJMS 21:4632. doi: 10.3390/ijms21134632
Quinn, E. M., Slattery, H., Walsh, D., Joshi, L., and Hickey, R. M. (2020b). Bifidobacterium longum subsp. infantis ATCC 15697 and goat Milk oligosaccharides show synergism in vitro as anti-Infectives against Campylobacter jejuni. Foods 9:348. doi: 10.3390/foods9030348
Rasigade, J.-P., Dumitrescu, O., and Lina, G. (2014). New epidemiology of Staphylococcus aureus infections. Clin. Microbiol. Infect. 20, 587–588. doi: 10.1111/1469-0691.12718
Rees, J. H., and Hughes, R. A. C. (1995). Campylobacter jejuni infection and Guillain–Barré syndrome. N. Engl. J. Med. 333, 1374–1379. doi: 10.1056/NEJM199511233332102
Sejvar, J. J., Baughman, A. L., Wise, M., and Morgan, O. W. (2011). Population incidence of Guillain-Barré syndrome: a systematic review and Meta-analysis. Neuroepidemiology 36, 123–133. doi: 10.1159/000324710
Shinefield, H. R. (1963). V. An analysis and interpretation. Arch. Pediatr. Adolesc. Med. 105:683. doi: 10.1001/archpedi.1963.02080040685019
Skals, M., Jensen, U. B., Ousingsawat, J., Kunzelmann, K., Leipziger, J., and Praetorius, H. A. (2010). Escherichia coli α-Hemolysin triggers shrinkage of erythrocytes via KCa3.1 and TMEM16A channels with subsequent phosphatidylserine exposure. J. Biol. Chem. 285, 15557–15565. doi: 10.1074/jbc.M109.082578
Skirrow, M. B. (1991). Epidemiology of Campylobacter enteritis. Int. J. Food Microbiol. 12, 9–16. doi: 10.1016/0168-1605(91)90044-P
Skowronek, P., Thielert, M., Voytik, E., Tanzer, M. C., Hansen, F. M., Willems, S., et al. (2022). Rapid and in-depth coverage of the (Phospho-)proteome with deep libraries and optimal window design for dia-PASEF. Mol. Cell. Proteomics 21:100279. doi: 10.1016/j.mcpro.2022.100279
Speare, L., Woo, M., Dunn, A. K., and Septer, A. N. (2022). A putative lipoprotein mediates cell-cell contact for type VI secretion system-dependent killing of specific competitors. MBio 13:e0308521. doi: 10.1128/mbio.03085-21
Stacy, A., Fleming, D., Lamont, R. J., Rumbaugh, K. P., and Whiteley, M. (2016). A commensal bacterium promotes virulence of an opportunistic pathogen via cross-respiration. MBio 7, e00782–e00716. doi: 10.1128/mBio.00782-16
Storey, J. D., and Tibshirani, R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 100, 9440–9445. doi: 10.1073/pnas.1530509100
Szewzyk, U., Szewzyk, R., Manz, W., and Schleifer, K.-H. (2000). Microbiological safety of drinking water. Annu. Rev. Microbiol. 54, 81–127. doi: 10.1146/annurev.micro.54.1.81
Taranto, M. P., Fernandez Murga, M. L., Lorca, G., and Valdez, G. F. (2003). Bile salts and cholesterol induce changes in the lipid cell membrane of Lactobacillus reuteri. J. Appl. Microbiol. 95, 86–91. doi: 10.1046/j.1365-2672.2003.01962.x
Tyanova, S., Temu, T., Sinitcyn, P., Carlson, A., Hein, M. Y., Geiger, T., et al. (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740. doi: 10.1038/nmeth.3901
Vandenesch, F., Lina, G., and Henry, T. (2012). Staphylococcus aureus Hemolysins, bi-component Leukocidins, and Cytolytic peptides: a redundant arsenal of membrane-damaging virulence factors? Front. Cell. Inf. Microbio. 2:12. doi: 10.3389/fcimb.2012.00012
Van Tyne, D., and Gilmore, M. S. (2014). Friend turned foe: evolution of enterococcal virulence and antibiotic resistance. Annu. Rev. Microbiol. 68, 337–356. doi: 10.1146/annurev-micro-091213-113003
Vidal, J. E., Wier, M. N., Angulo-Zamudio, U. A., McDevitt, E., Vidal, A. G. J., Alibayov, B., et al. (2021). Prophylactic inhibition of colonization by Streptococcus pneumoniae with the secondary bile acid metabolite Deoxycholic acid. Infect. Immun. 89:e0046321. doi: 10.1128/IAI.00463-21
Keywords: Campylobacter jejuni, co-incubation, Enterococcus faecalis, Enterococcus faecium, Staphylococcus aureus, bile acids, proteomics
Citation: Dreyer A, Lenz C, Groß U, Bohne W and Zautner AE (2023) Characterization of Campylobacter jejuni proteome profiles in co-incubation scenarios. Front. Microbiol. 14:1247211. doi: 10.3389/fmicb.2023.1247211
Received: 28 June 2023; Accepted: 24 October 2023;
Published: 09 November 2023.
Edited by:
Odile Tresse, INRA Centre Angers-Nantes Pays de la Loire, FranceReviewed by:
Vathsala Mohan, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaCopyright © 2023 Dreyer, Lenz, Groß, Bohne and Zautner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andreas Erich Zautner, YXphdXRuZUBnd2RnLmRl
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.