
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 20 October 2023
Sec. Infectious Agents and Disease
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1245755
Pseudomonas aeruginosa thrives in the airways of individuals with cystic fibrosis, in part by forming robust biofilms that are resistant to immune clearance or antibiotic treatment. In the cystic fibrosis lung, the thickened mucus layers create an oxygen gradient, often culminating with the formation of anoxic pockets. In this environment, P. aeruginosa can use nitrate instead of oxygen to grow. Current fluorescent reporters for studying P. aeruginosa are limited to the GFP and related analogs. However, these reporters require oxygen for the maturation of their chromophore, making them unsuitable for the study of anaerobically grown P. aeruginosa. To overcome this limitation, we evaluated seven alternative fluorescent proteins, including iLOV, phiLOV2.1, evoglow-Bs2, LucY, UnaG, Fluorescence-Activating and Absorption-Shifting Tag (FAST), and iRFP670, which have been reported to emit light under oxygen-limiting conditions. We generated a series of plasmids encoding these proteins and validated their fluorescence using plate reader assays and confocal microscopy. Six of these proteins successfully labeled P. aeruginosa in anoxia. In particular, phiLOV2.1 and FAST provided superior fluorescence stability and enabled dual-color imaging of both planktonic and biofilm cultures. This study provides a set of fluorescent reporters for monitoring P. aeruginosa under low-oxygen conditions. These reporters will facilitate studies of P. aeruginosa in biofilms or other contexts relevant to its pathogenesis, such as those found in cystic fibrosis airways. Due to the broad host range of our expression vector, the phiLOV2.1 and FAST-based reporters may be applicable to the study of other Gram-negative bacteria that inhabit similar low-oxygen niches.
Pseudomonas aeruginosa is a gram-negative opportunistic pathogen causing hospital-acquired diseases and infecting the lungs of cystic fibrosis patients. It is the most commonly isolated cystic fibrosis pathogen (Pressler et al., 2011; Winstanley et al., 2016) and is associated with worse clinical outcomes by accelerating the deterioration of pulmonary function (O’Brien and Fothergill, 2017). Therapeutic interventions against P. aeruginosa are complicated by its high level of resistance to multiple antibiotics and its ability to form biofilms (Wagner and Iglewski, 2008; Høiby et al., 2010).
Biofilms constitute a physical barrier to the host defense system (Magalhães et al., 2016). Moreover, P. aeruginosa cells within the biofilm are up to 1,000 times more resistant to antimicrobial compounds than their planktonic counterparts (Wagner and Iglewski, 2008; Hall and Mah, 2017). There is a steep O2 gradient within the cystic fibrosis airways caused by the accumulation of mucus that restrain O2 diffusion and the consumption of O2 by host cells and bacteria (Stutts et al., 1986; Worlitzsch et al., 1998, 2002). Thus, the oxygen concentration ranges from aerobic to anaerobic levels in cystic fibrosis lungs, and P. aeruginosa can thrive under all these conditions. In fact, although P. aeruginosa is an obligate aerobe, it can also grow anaerobically through denitrification using nitrate instead of O2 as an electron acceptor in the respiratory chain (Zumft, 1997; Shapleigh, 2013). Strikingly, the capacity of P. aeruginosa to form biofilms is superior in anoxia than in normoxia (Yoon et al., 2002), but tools to reliably study biofilms under low O2 are lacking.
Genetically encoded fluorescent reporters have revolutionized the imaging of dynamic processes in living cells, including in bacterial pathogens (Campbell-Valois et al., 2014; Bajunaid et al., 2020). The green fluorescent protein (GFP), with its high brightness and robustness to various environmental conditions, was instrumental in this endeavor. In addition, some variants, such as GFPmut2 (Iizuka et al., 2011), rapidly mature their chromophore, allowing for the monitoring of fast-paced cellular processes. However, GFP and related variants have drawbacks, notably their strict O2 requirement (Tsien, 1998). This property has limited their applications in living biofilms where O2 is low (Monmeyran et al., 2018).
By contrast, several alternative fluorescent proteins (FPs) are functional under oxygen-limiting conditions. However, to our knowledge, none of these FPs have been tested in P. aeruginosa. We thus selected seven FPs that spanned a wide range of excitation and emission spectra (Table 1). The first group evaluated was the light, oxygen or voltage proteins (LOV): iLOV (Chapman et al., 2008), phiLOV2.1 (Christie et al., 2012) and evoglow-Bs2, also known as EcFbFP (Drepper et al., 2007). These FPs are significantly smaller and mature faster than GFP, and they use flavin mononucleotide (FMN) as their chromophore (Chapman et al., 2008; Christie et al., 2012). One potential disadvantage of using these FPs is their low brightness (Kumagai et al., 2013). In addition, we selected LucY, which is brighter than many LOV and uses flavin adenine dinucleotide (FAD) as its chromophore (Auldridge et al., 2015). It is noteworthy that both FMN and FAD are abundant in the bacterial cytosol; thus, LOV and LucY spontaneously emit light in bacteria.
The Fluorescence-Activating and absorption-Shifting Tag (FAST) emits light when bound to a synthetic fluorogenic analog of 4-hydroxybenzylidene-rhodanine (HBR) (Plamont et al., 2016). The application of FAST in P. aeruginosa is promising because it was shown to outcompete the GFP in Escherichia coli biofilms (Monmeyran et al., 2018). UnaG and iRFP670 fluoresce when they form a complex with bilirubin (Kumagai et al., 2013) and biliverdin (Shcherbakova and Verkhusha, 2013), respectively. UnaG is characterized by its brightness and wavelengths similar to the GFP. Finally, the red-shifted emission of iRFP670 may address the auto-fluorescence of P. aeruginosa at shorter wavelengths, which is detrimental to the analyses of thick biofilms. It is important to note that bilirubin is not naturally present in P. aeruginosa and related bacteria, so it must be added to the growth medium. Similarly, free biliverdin is restricted in bacteria as its production is tightly dependent on iron availability. Therefore, it must be produced by an exogenous heme oxygenase that is co-introduced with iRFP670 into the bacterial host (Shcherbakova and Verkhusha, 2013).
Here, we report a panel of fluorescent proteins that successfully monitor P. aeruginosa in planktonic and biofilm cultures under anoxia. We demonstrate that phiLOV2.1 and FAST can be combined to track dual subpopulations of P. aeruginosa in microscopy experiments. This study provides tools that will be useful to researchers studying the behavior of P. aeruginosa when the concentration of oxygen is too low to use the GFP. The broad host range plasmid used to develop these reporters suggest that their use could be readily extended to other gram-negative bacteria.
pSMC21, a derivative of pSMC2 (Bloemberg et al., 1997), allowing the constitutive expression of the GFPmut2 under the tacp promoter, was used to derive several new plasmids. First, GFPmut2 was excised by PCR-mediated deletion mutagenesis with the High fidelity Phusion DNA polymerase (ThermoFisher) and recircularized with the T4 DNA ligase (New England Biolabs) to yield the resulting empty vector control pCFT1. Second, the coding sequences of seven alternative fluorescent proteins were amplified by PCR with the Phusion DNA polymerase and introduced independently into pCFT1 by combining PCR-mediated deletion mutagenesis and isothermal cloning with the NEB Gibson assembly master mix (NEB, #E2611), thus yielding pCFT2–8. The coding sequences of the fluorescent proteins iLOV, phiLOV2.1, evoglow-Bs2, LucY, FAST (Bio Basic Markham, ON) and UnaG (Integrated DNA Technologies, IA) were obtained by gene synthesis. iRFP670 and the heme oxygenase (HO) from Bradyrhizobium sp. ORS 278 were a kind gift from Vladislav Verkhusha (Shcherbakova and Verkhusha, 2013). The HO gene was cloned downstream of iRFP670 to generate a bicistronic gene. Both open reading frames were separated by two stop codons, a Shine-Dalgarno sequence AUCACCUCCUUA (Ma et al., 2002) and a 5-adenine spacer (Ringquist et al., 1992). The sequence of the cloned fragment in each resulting plasmid was confirmed by Sanger sequencing. The coding sequence of iLOV, phiLOV2.1, FAST and GFPmut2 were inserted by isothermal cloning into pUdO4a (Manigat et al., 2023), as described above, thus yielding pCFT3.2, pCFT4.2, pCFT7.2 and pCFT9.2 (Addgene plasmids #208806, #208807, #208808 and #208809 respectively). pUdO4a is a small plasmid (~2.8 kbp) that possesses strong transcription terminators L3S2P21, thrLABC and arcA, an ampicillin resistance gene and a BBR1 origin of replication, which can be maintained in Pseudomonas spp. (Antoine and Locht, 1992; Supplementary Figure S1 – plasmid maps). Table 2 and Supplementary Table S1 list the plasmids and primers used in this study.
The Escherichia coli strain DH10B was used for plasmid propagation. The wild-type Pseudomonas aeruginosa str. PA14 was used in the fluorescence experiments (Rahme et al., 1995). Unless otherwise stated, the culture medium was LB supplemented with ampicillin (100 μg/mL) or carbenicillin (100 or 200 μg/mL) for DH10B and PA14 transformants, respectively. Anaerobic culturing of PA14 was performed in LB supplemented with carbenicillin, and 100 mM KNO3 (LBN) to allow anaerobic respiration. Anaerobic planktonic cultures were incubated in anaerobic Hungate culture tubes with butyl rubber stoppers and screwcaps (Hungate, #CLS-4208). Biofilm anaerobic cultures were incubated in a BactronEZ chamber (Sheldon Manufacturing. Inc., Cornelius, OR) under an anaerobic atmosphere of 5% H2, 5% CO2, 90% N2 that contained fewer than 4.7 parts per million O2 per the manufacturer specifications. All incubations were performed at 37°C.
PA14 strains harboring one of the pCFT plasmids were grown on LB agar carbenicillin plates. A single colony from each plate was used to inoculate 5 mL of LB carbenicillin. These cultures were aerobically grown for ~18 h with shaking (250 rpm), then subcultured for 5 h, and their optical density at 600 nm (OD 600) was adjusted to 1. The cultures were next transferred in the anaerobic chamber and diluted 1:100 in 5 mL LBN in anaerobic Hungate culture tubes. These sealed tubes were then incubated at 37°C for ~24 h with shaking (250 rpm) in an incubator located in a room with normal oxygen levels. Following this first incubation, the cultures were transferred back into the anaerobic chamber and diluted 1:10 in LBN. Finally, 100 μL of the resulting inoculates were dispensed in a flat black bottom 96-well plate (Greiner bio-one, #655096). The samples were overlaid with Johnson and Johnson ® oil, as described (Lam et al., 2018). Subsequently, the plate was sealed with an adhesive film (BioRad, #MSB1001), then wrapped with parafilm to prevent O2 exposure prior to exiting the anaerobic chamber. The plate was then incubated with medium shaking in a Hybrid Synergy H4 plate reader (BioTeK), where the absorbance at 600 nm was continuously tracked and recorded every hour for 24 h. The fluorescence signal of PA14 cells expressing the GFP (excitation: 480 ± 9.0 nm, emission: 511 ± 9.0 nm) was recorded at 0 and 24 h to ensure anoxia was maintained throughout the experiment. A sample of the 24 h culture was exposed to O2 for 2 h prior to the measurements to allow for the maturation of the GFP.
PA14 cultures harboring pCFT plasmids were grown anaerobically in LBN at 37°C for 24 h in anaerobic Hungate culture tubes as described above. Depending on the fluorescent proteins assayed, the samples were treated as follows. First, PA14 expressing the flavin-binding fluorescent proteins iLOV, phiLOV2.1, evoglow-Bs2 and LucY were processed immediately. Second, PA14 carrying pCFT1 (empty vector) or pCFT5 (UnaG) were grown in LBN ± 10 μM bilirubin (Merck) and processed immediately. Third, after the anaerobic incubation in the regular anaerobic medium, 1.5 mL of PA14 cultures with pCFT1 (empty vector) or pCFT7 (FAST) were transferred to a microfuge tube with their cap tightly wrapped with parafilm inside the anaerobic chamber. These cultures were then centrifuged (2,400× g, 10 min) and resuspended in PBS ± 10 μM TFAmber (HBR-3,5DM, The Twinkle Factory, Paris, France) (Li et al., 2017) as described (Streett et al., 2019). As described above, GFP-expressing cells were included in every assayed plate to confirm that anoxia was maintained during the experiment. The light emitted by the GFP-expressing cells exposed (or not) to O2 was measured along with the assayed FP at their optimal excitation and emission wavelengths (Table 1) using a Hybrid Synergy H4 Hybrid plate reader (BioTeK). The data were normalized according to the OD 600.
Experiments with PA14 planktonic cultures were carried out using the pSMC21 derivatives, whereas experiments with PA14 biofilm cultures were conducted with the pUdO4a derivatives. Aerobic PA14 cells carrying either phiLOV2.1, FAST or the empty vector were incubated in LB media supplemented with carbenicillin for 16 h to 18 h. These cultures were then diluted to an OD 600 of 1 and used as inocula for anaerobic cultures. A 1:1000 dilution of this inoculum was made for planktonic cultures that were subsequently grown anaerobically for 18 h in LBN. A 1:100 dilution of the inoculum was made for biofilm cultures grown anaerobically in a 35 mm glass-bottom dish (Cellvis, #D35-20-1.5H) underneath a 2% agarose pad in the anaerobic chamber. Biofilms were incubated in M63 minimal media supplemented with 1 mM MgSO4, 0.4% L-arginine, 100 mM KNO3 and carbenicillin (M63N) for 24 h. After the incubation, cells were prepared for imaging as follows. For planktonic cell imaging, the anaerobic cultures were centrifuged (2,400 × g, 10 min) and resuspended in M63N; for FAST imaging, this medium was supplemented with 5 μM TFCoral (HBR-3,5DOM, The Twinkle Factory) (Li et al., 2017). For biofilm imaging, the culture medium was removed, and the attached cells were washed once in PBS before adding a 200 μL solution of M63N ± 10 μM TFCoral. Both planktonic and biofilm cells were blanketed with a 2% agarose pad in a 35 mm glass-bottom dish (Cellvis, #D35-20-1.5H) for fluorescence imaging. The glass-bottom dish was then covered with a 35 mm glass-top cover (Cellvis, #D35-20-0-TOP) for differential interference contrast (DIC) imaging. To curtail O2 infiltration during imaging, the edges of the dishes were tightly wrapped in parafilm before exiting the anaerobic chamber for microscopy. It is noteworthy that although cells were incubated anaerobically and specific measures were taken to minimize O2 exposure during the imaging setup, the strict control of anoxia could not be warranted throughout the entire imaging process. Micrographs were acquired on a Zeiss LSM 880 AxioObserverZ1 confocal microscope equipped with an EC Plan-Neofluar 100×/1.3 oil M27 objective. Fluorescence was captured using the argon laser at 458 nm for phiLOV2.1 [excitation: 458 nm; emission: 466–544 nm (monocultures) and 466–522 nm (dual cultures)] and at 514 nm for FAST:TFCoral [excitation: 514 nm; emission: 555–722 nm (monocultures) and 586–722 (dual cultures)]. The photostability of iLOV and phiLOV2.1 in planktonic PA14 was evaluated by performing time-lapse acquisition every 30 s for 3 min under the conditions described above. All micrographs were collected with the ZEN software (Zeiss) and prepared with the ImageJ software (Schneider et al., 2012). The single cell quantitation of the fluorescence intensity from the planktonic micrographs was performed with ImageJ as previously described (Nigro et al., 2019). The total number of bacteria measured in these analyses was: 126 (empty vector, EV), 262 (LucY), 111 (UnaG), 218 (phiLOV2.1) and 168 (FAST:TFCoral), and were collected from four biological replicates.
For the plate assay experiments, three biological replicates are presented. The micrographs shown are representative of four biological replicates. The single-cell fluorescence intensity analyses were performed on micrographs of four biological replicates. Statistical analyses were computed with GraphPad Prism 8 (GraphPad Software, La Jolla, CA). Plate reader assay data are presented as mean ± standard deviation. The confidence level was set at 95% (p < 0.05).
To study P. aeruginosa under anaerobic conditions, we picked the FPs iLOV, phiLOV2.1, evoglow-Bs2, LucY, FAST, UnaG and iRFP670 whose fluorescence is in principle independent of O2 (Table 1). The open reading frame of each of these proteins was cloned into the vector pCFT1 to generate pCFT2–8 (Supplementary Figure S1; Table 2). pCFT6 included a bicistronic operon of iRFP670 and HO to produce the biliverdin by heme degradation, a strategy that yielded functional iRFP670 in E. coli under anaerobic conditions (Shcherbakova and Verkhusha, 2013).
To test the effects of the expression of the FPs on the growth of PA14, strains carrying the different plasmid were incubated in LB supplemented with carbenicillin and nitrate (LBN) under anaerobic conditions. Most strains carrying the plasmids containing the FPs grew similarly to the empty vector (Figure 1A). A notable exception was the strain carrying pCFT6, which contained iRFP670 and the HO, and appeared to grow at a slower rate. However, the statistical analyses of the growth rate during the exponential phase indicated this change was not significant (Supplementary Table S2). There was no increase in the fluorescence of the GFP between the beginning and the end of the measurements (Figure 1B), indicating that anoxia was maintained throughout the experiment. By contrast, exposure of the 24 h sample to O2 during 2 h increased fluorescence by approximately 20-fold, confirming that immature GFP had indeed accumulated during anaerobic growth (Figure 1B). Taken together, these results suggest that when cultured anaerobically, the viability and the growth of PA14 are unaffected by the expression of our collection of FPs, except perhaps for the iRFP670-HO bicistronic gene.
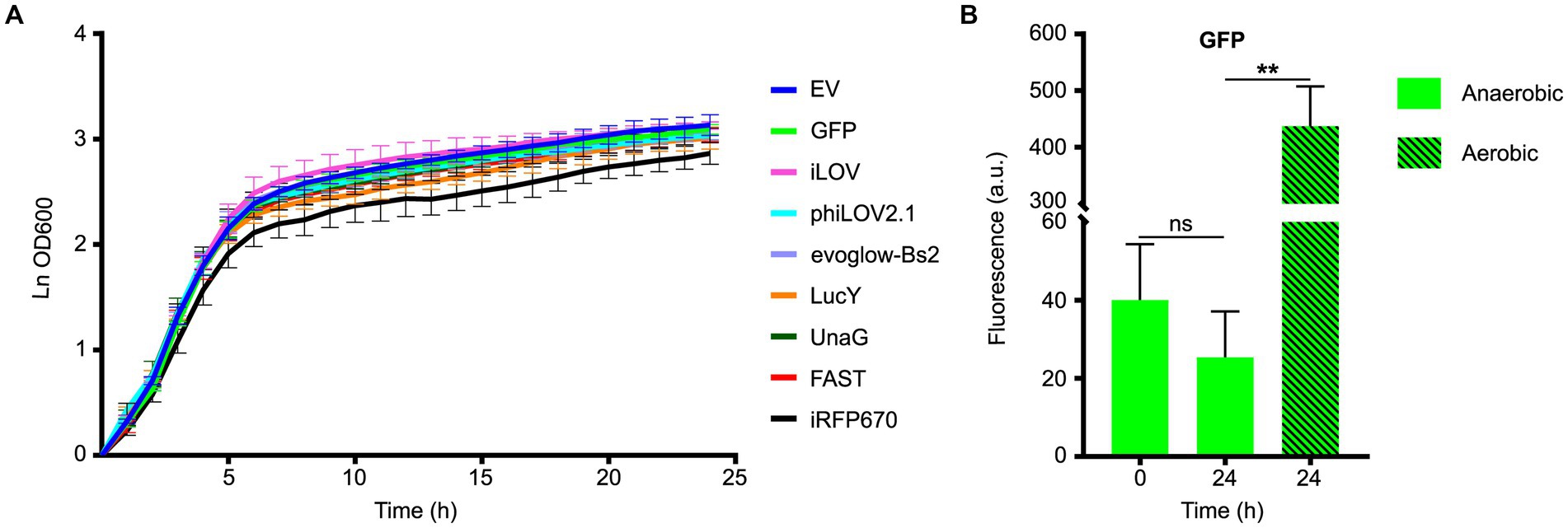
Figure 1. Fitness evaluation of Pseudomonas aeruginosa PA14 expressing different fluorescent proteins under anoxia. (A) Growth curves of PA14 carrying the plasmids pCFT1-8 and pSCM21 (Table 2), and incubated in LB supplemented with KNO3 and carbenicillin at 37°C under anaerobic conditions. The growth rate constant for each strain during the exponential phase was calculated and compared to the control empty vector (EV). The statistical significance of the change in exponential growth rate between the control EV and the strains expressing the fluorescent proteins was assessed by a two-tailed unpaired Student’s t-test (p < 0.05); none of the variations observed were significant. (B) The fluorescence of PA14 cells expressing GFP was measured at the beginning (0 h, anaerobic) and the end of the experiment (24 h, anaerobic), and after a subsequent exposure to oxygen (24 h, aerobic). Statistical analysis was performed using a paired Student’s t-test. Fluorescence was normalized with the OD 600, and the means with standard deviations as error bars are shown (ns = nonsignificant; n = 3).
Since the fluorescent proteins had little to no impact on growth, we proceeded to compare their signal intensity. We assumed that their fluorescent signal would be representative of their overall behavior in PA14 and used it as a proxy that integrated their unique critical properties such as their expression level, maturation and brightness. This is a practical solution that circumvented the challenges arising from comparing a diverse set of proteins. Therefore, to evaluate the functionality of the FPs under anoxia, we first measured their fluorescence in PA14 using a plate reader assay.
In this context, we grew planktonic PA14 cultures harboring the plasmids pCFT1–8 under anoxia and distributed them in 96-well plates under the anoxia-preserving conditions described above. PA14 expressing the LOV FPs showed a robust fluorescence signal compared to cells carrying the empty vector (Figure 2A), with iLOV showing the highest signal, followed by evoglow-Bs2 and phiLOV2.1. PA14 cells expressing LucY also displayed strong fluorescence relative to those carrying the empty vector (Figure 2B). Similarly, PA14 cells expressing UnaG (Figure 3A) and FAST (Figure 3B) exhibited a substantial fluorescence increase in the presence of 10 μM bilirubin and 10 μM TFAmber, respectively. In contrast, the fluorescence of cells harboring the empty vector was negligible.
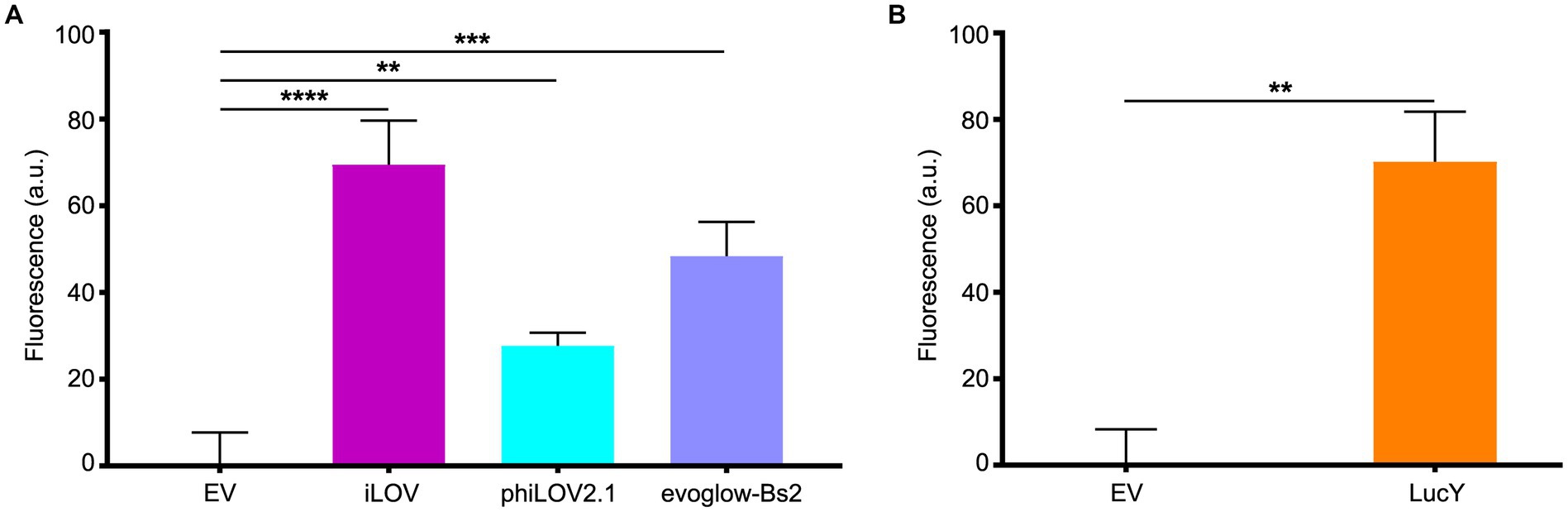
Figure 2. Fluorescence of PA14 expressing the flavin-based fluorescent proteins in anoxia. (A) Fluorescence of PA14 cells carrying plasmids allowing the expression of iLOV (pCFT3), phiLOV2.1 (pCFT4) and evoglow-Bs2 (pCFT2) compared with those carrying the empty vector (pCFT1). Fluorescence was recorded at excitation 450 nm and emission 495 nm. Statistical analysis was performed using one-way analysis of variance (ANOVA) followed by Bonferroni’s multiple comparisons test. (B) Fluorescence of PA14 cells carrying plasmids allowing the expression of LucY (pCFT8) expressed in PA14 cells compared with those carrying the empty vector (pCFT1). Fluorescence was recorded at excitation 460 nm and emission 530 nm. Statistical analysis was done using an unpaired Student’s t-test. Fluorescence was normalized with the OD 600, and the means with standard deviations as error bars are shown (**p < 0.01; ***p < 0.001; ****p < 0.001; n = 3).
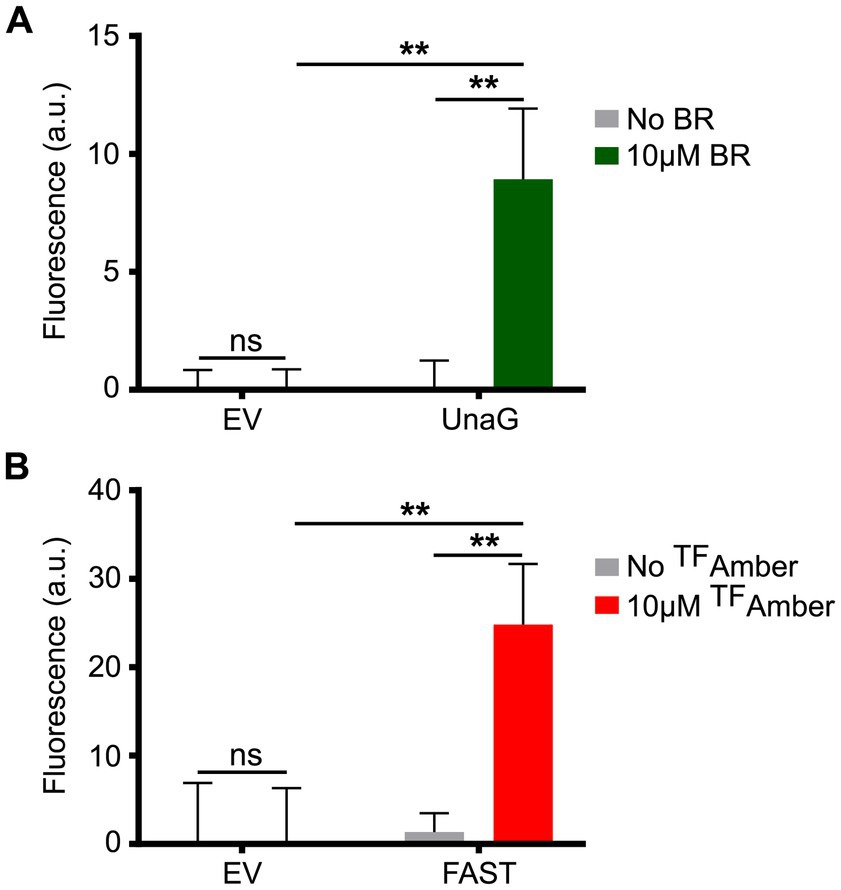
Figure 3. Fluorescence of PA14 expressing UnaG and FAST in anoxia. (A) Fluorescence of PA14 cells carrying plasmids allowing the expression of UnaG (pCFT5) compared with those carrying the empty vector (pCFT1) in absence and presence of 10 μM bilirubin (BR). Fluorescence was recorded at excitation 498 nm and emission 527 nm. (B) Fluorescence of PA14 cells carrying plasmids allowing the expression of FAST (pCFT7) compared with those carrying the empty vector (pCFT1) in the presence and the absence of 10 μM TFAmber. Fluorescence was recorded at excitation 482 nm and emission 536 nm. In both panels, statistical analysis was executed using two-way analysis of variance (ANOVA) followed by Bonferroni’s multiple comparisons test. Fluorescence was normalized with the OD 600, and the means with standard deviations as error bars are shown (ns, nonsignificant; **p < 0.01; n = 3).
iRFP670 did not fluoresce when co-expressed with the HO in PA14 under anoxia (Supplementary Figure S2). In order to emit light, iRFP670 must form a complex with its chromophore biliverdin, which can be derived from heme in the presence of HO. Although this enzymatic reaction requires O2, previous data suggested that fluorescent protein related to iRFP670 can emit light in anaerobically grown E. coli, simultaneously expressing the HO (Rumyantsev et al., 2015). Our data indicated this is not possible in PA14. We reasoned that providing biliverdin, excess hemin or both in the growth medium might rescue the fluorescence. Unfortunately, these additives did not increase iRFP670 emission under anoxia. By contrast, iRFP670 fluoresced when co-expressed with the HO in aerobically incubated PA14, independent of exogenous additives (Supplementary Figure S2). Altogether, these data suggested that HO and O2 are required to yield enough biliverdin to produce functional iRFP670 in PA14. Hence, we did not investigate iRFP670 further. On the other hand, we pursued our investigation of the six FPs that emitted light under anoxia according to this plate reader assay.
As previously reported (Christie et al., 2012), we found phiLOV2.1 to be more photostable than iLOV (Supplementary Figure S3) and thus selected it for microscopic analyses of PA14. Next, we evaluated the other FPs to identify a suitable co-imaging partner for phiLOV2.1. Bacteria expressing LucY and UnaG were both visible in fluorescent microscopy (Supplementary Figure S4). The fluorescence of bacteria expressing LucY was bright and homogenous (Supplementary Figure S4A). By contrast, the fluorescence of bacteria expressing UnaG was heterogenous, with only a small subpopulation of strongly fluorescent cells (Supplementary Figure S4B). Subsequent single-cell analyses of the fluorescent intensity confirmed these observations (Supplementary Figure S4C). Thus, the use of UnaG in PA14 was complicated by the heterogeneity of its signal. Our attempts to resolve this issue failed and led us to discard UnaG.
While the use of LucY seemed promising, it was not ideal for use in combination with phiLOV2.1 because their emission spectra overlap. The emission spectrum of FAST:TFAmber used in the plate reader assay also overlaps with phiLOV2.1. Hence, we turned our attention to FAST:TFCoral to cover the red region left unoccupied by the exclusion of iRFP670 as we reasoned it would combine well with phiLOV2.1. Indeed, planktonic PA14 cells carrying the empty vector were invisible (Figure 4A), whereas those expressing phiLOV2.1 or FAST:TFCoral were fluorescent (Figure 4B). The single-cell analyses of the fluorescent intensity showed that phiLOV2.1 yielded a homogenous and strong signal. By contrast, FAST:TFCoral resulted in a heterogenous and overall lower signal (Supplementary Figure S5). The heterogeneity of the signal observed with FAST:TFCoral is reminiscent of that of UnaG and may be attributable, at least in part, to their use of an exogenous chromophore. This hypothesis is supported by the homogenous signal of LucY and phiLOV2.1 who use an endogenous chromophore. Next, we tested the combined use of phiLOV2.1 and FAST:TFCoral to image dual cultures of P. aeruginosa. In brief, the strains used in monocultures were mixed and imaged. To avoid spectral bleed through, the span of wavelengths collected was narrowed for both fluorescent proteins. Once analyzed with these new acquisition parameters, the phiLOV2.1 and FAST:TFCoral subpopulations were distinguishable (Figure 4C). However, due to adjusting the acquisition parameters, the signal of both fluorescent proteins was dimmer than under the acquisition parameters used in the monocultures.
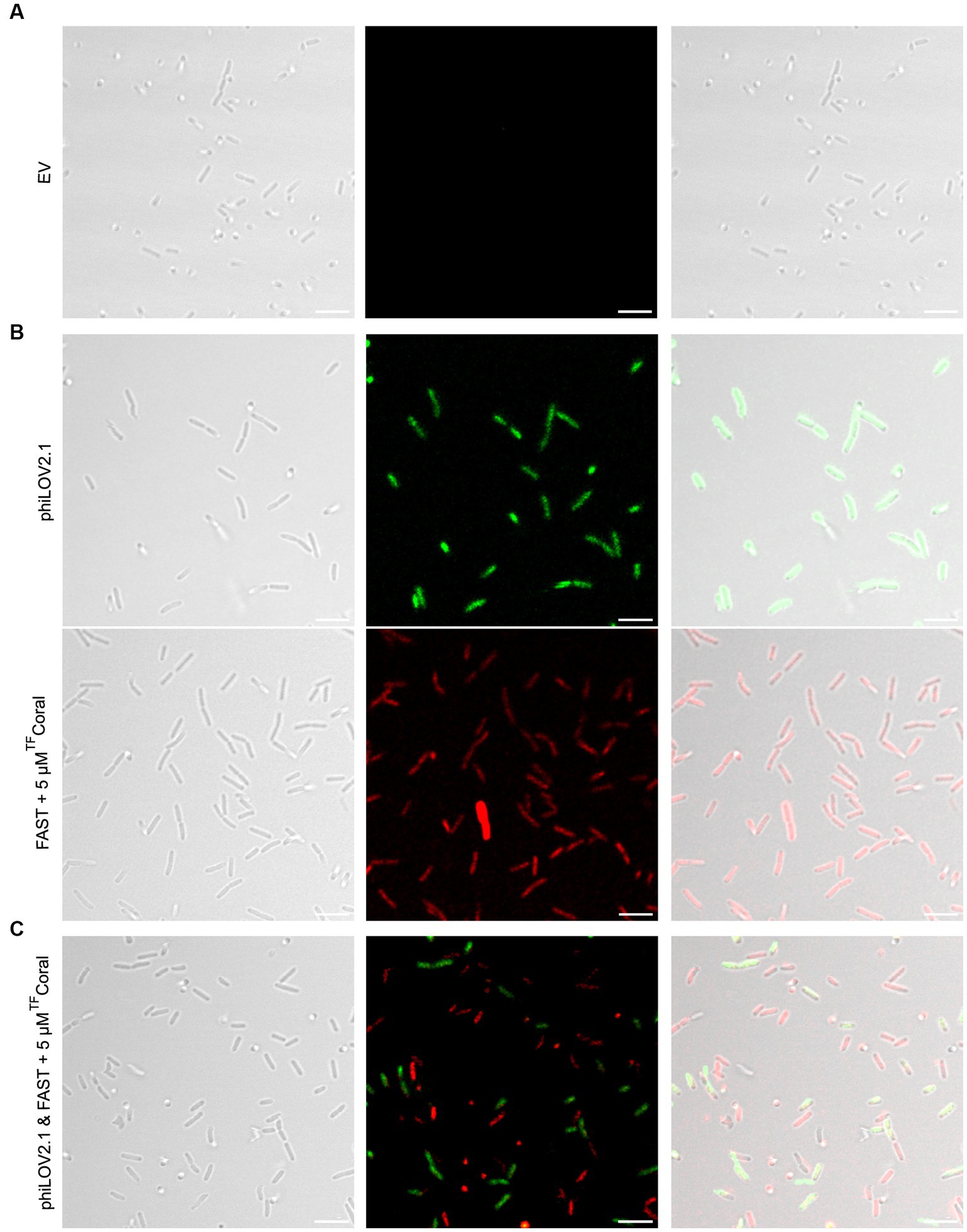
Figure 4. Fluorescence imaging of planktonic PA14 with phiLOV2.1 and FAST. (A) Micrograph of PA14 harboring the empty vector (pCFT1). (B) Micrograph of PA14 expressing phiLOV2.1 (pCFT4) and FAST:TFCoral (pCFT7) in the top and bottom panel, respectively. (C) Micrograph of a mix of PA14 cells expressing phiLOV2.1 or FAST:TFCoral (1:1 inoculation ratio). From left to right: differential interference contrast (DIC), single channel fluorescence and overlay (scale bar = 5 μm).
Preliminary microscopy observations indicated that light emitted by phiLOV2.1 and FAST:TFCoral decreased beyond detection limits in PA14 biofilms (data not shown). This decrease may be due to a reduction of the activity of the tac promoter or a reduction in the number of plasmid copies in biofilms. To address this pitfall, we subcloned the original tacp::phiLOV2.1 and tacp::FAST reporters into pUdO4a (Manigat et al., 2023). This synthetic plasmid harbors a broad host range BBR1 origin, which is functional in Pseudomonas spp. and differs from that used in pSMC21 (Supplementary Figure S1B; Antoine and Locht, 1992). In addition, the strong transcriptional terminators flanking the multiple cloning sites of pUdO4a may contribute to increasing gene expression (Manigat et al., 2023). Indeed, we found that the pUdO4a derivative pCFT4.2 expressing phiLOV2.1 produced higher fluorescence levels in planktonic PA14 than its pSMC21-derived counterpart pCFT4 (Supplementary Figure S5).
Fortunately, the increased fluorescent signal of the FPs from the pUdO4a derivatives allowed the imaging of PA14 biofilms (Figure 5). Biofilm cultures carrying the empty vector were included as negative control (Figure 5A). In comparison with this control, biofilm monocultures of PA14 cells expressing phiLOV2.1 and FAST:TFCoral emitted light upon excitation at the expected wavelength (Figure 5B). Akin to the mixed planktonic cultures described above (Figure 4C), PA14 subpopulations expressing either of the two FPs were distinguishable in biofilm micrographs (Figure 5C). However, the signal distribution in the dual cultures appeared more scattered, with the appearance of a higher proportion of weakly non-fluorescent cells than in the monoculture. This could result from the adjustment of the acquisition parameters required for dual imaging, as described above. It is noteworthy that the optimal concentration of the TFCoral chromophore was higher in biofilm than in planktonic cultures. This is likely due to the higher cell density in biofilms.
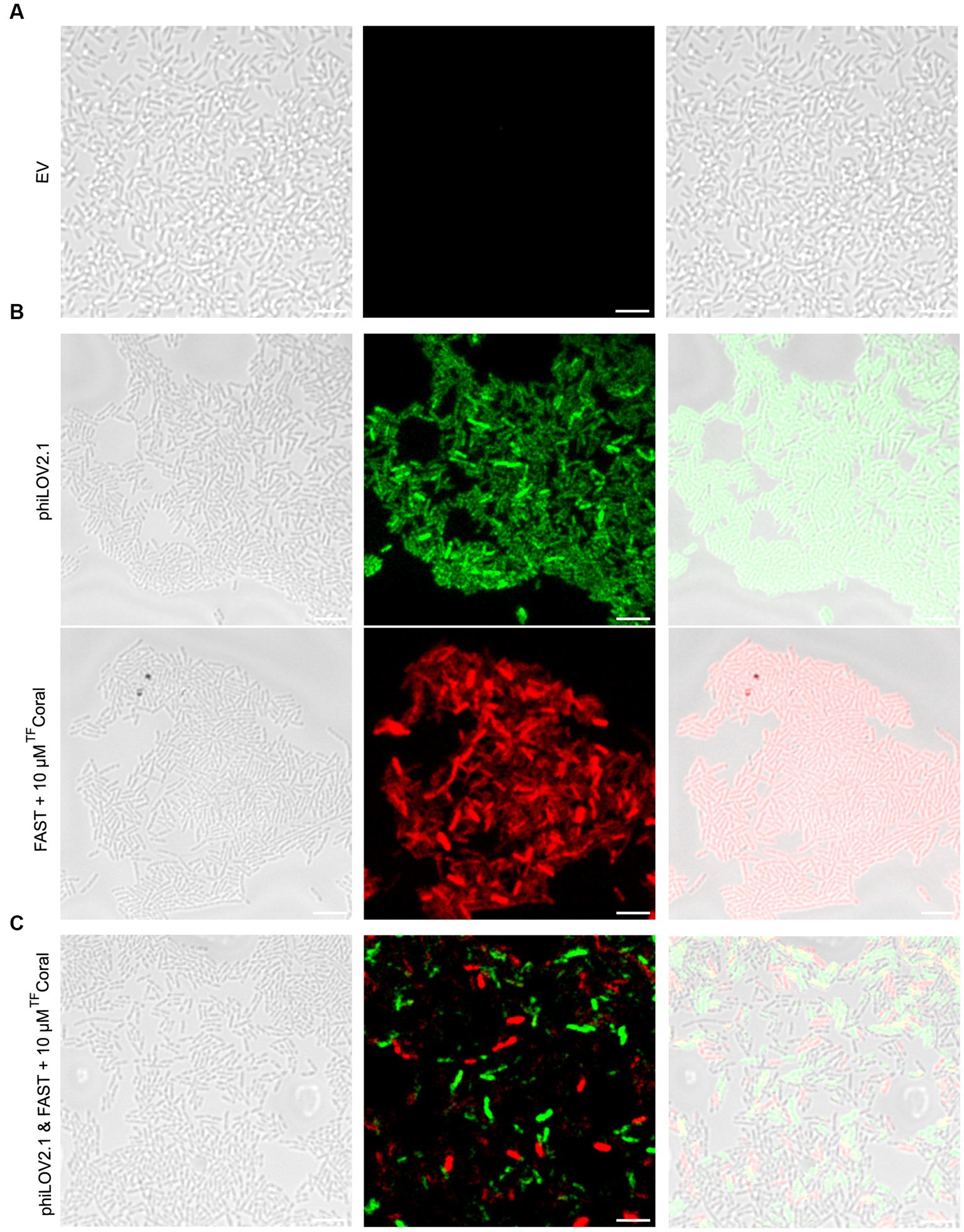
Figure 5. A new expression system allows fluorescence imaging of PA14 biofilms with phiLOV2.1 and FAST. (A) Micrograph of PA14 harboring the empty vector (pUdO4a). (B) Micrograph of PA14 expressing phiLOV2.1 (pCFT4.2) and PA14 expressing FAST:TFCoral (pCFT7.2) in the top and bottom panel, respectively. (C) Micrograph of co-cultured PA14 cells expressing phiLOV2.1 or FAST:TFCoral (1:1 mix ratio). From left to right: differential interference contrast (DIC), single channel fluorescence and overlay (scale bar = 5 μm).
In this study, we demonstrated that the fluorescent proteins iLOV, phiLOV2.1, evoglow-Bs2, LucY, FAST and UnaG allowed for the monitoring of live P. aeruginosa in plate reader assays under anaerobic conditions. In addition, we showed that phiLOV2.1, LucY and FAST:TFCoral are practical solutions for confocal microscopy. We characterized further the use of phiLOV2.1 and FAST:TFCoral and showed that they allowed for co-monitoring of dual subpopulations of P. aeruginosa by fluorescence microscopy. In summary, this study identified alternative fluorescent proteins for plate reader and microscopy assays when O2 levels are insufficient for the maturation of the GFP chromophore.
Our initial analyses included iRFP670. However, co-expressing this fluorescent protein and the heme oxygenase required to produce its biliverdin chromophore yielded no fluorescence under anaerobic growth. In mammalian tissue culture cells, biliverdin-binding fluorescent proteins such as iRFP670 could work under low oxygen conditions by scavenging biliverdin, which is ubiquitous in their cytosol (Ponka, 1999; Shcherbakova and Verkhusha, 2013). In several pathogenic bacteria, including P. aeruginosa, heme oxygenases are expressed under iron starvation conditions (Schmitt, 1997; Wilks and Schmitt, 1998; Zhu et al., 2000; Ratliff et al., 2001; Skaar et al., 2004; Wegele et al., 2004), limiting the natural production of biliverdin in the growth conditions used here. Therefore, a Bradyrhizobium heme oxygenase was introduced to increase the endogenous production of biliverdin (Shcherbakova and Verkhusha, 2013). However, in anaerobically grown P. aeruginosa, these heme oxygenases did not yield sufficient biliverdin to produce a detectable iRFP670 signal. This issue was not resolved by exogenous biliverdin, probably because of its lack of membrane permeability or accumulation in the cytosol. Conversely, the same strain grown in normoxia fluoresced. This suggests that cytosolic iRFP670 can function in P. aeruginosa, but the paucity of intracellular biliverdin precludes its use under anoxia.
UnaG has been used under aerobic and anaerobic conditions in Bacteroides thetaiotaomicron, Bacteroides ovatus and E. coli by adding bilirubin in the growth media (Chia et al., 2020, 2021). Here, we observed that P. aeruginosa expressing UnaG was also fluorescent when grown in the presence of bilirubin. Nevertheless, the signal was heterogeneous, with only a small fraction of the population emitting a high fluorescent signal. When averaged over the whole population in the plate reader assay, the signal of UnaG seemed inferior to the LOV proteins. Poor membrane permeability (Hancock and Woodruff, 1988) and efflux pumps, such as MexAB-OprM (Michea Hamzehpour et al., 1995; Plesiat et al., 1997), may contribute to limiting the amount of bilirubin available to UnaG in the cytosol of P. aeruginosa.
Regarding the LOV fluorescent proteins, our data were congruent with previous studies. The signal of iLOV was the strongest, whereas the signal of phiLOV2.1 was the weakest, and the signal of evoglow-Bs2 was intermediate (Buckley et al., 2015; Mordaka and Heap, 2018; Streett et al., 2021). However, the photostability of phiLOV2.1 (Christie et al., 2012) likely explains its superior performance in fluorescence microscopy of P. aeruginosa, which has also been observed in Clostridium species (Buckley et al., 2016). Although the fluorescence intensity of LucY was similar to iLOV as reported (Drepper et al., 2007; Auldridge et al., 2015), we did not investigate LucY in depth because phiLOV2.1 was a more practical option to use in combination with FAST. Indeed, the efficiency of FAST:HBR as fluorescent reporters has been previously described in several microorganisms, including E. coli (Monmeyran et al., 2018; Streett et al., 2019), Clostridium acetobutylicum (Streett et al., 2019), Listeria monocytogenes (Peron-Cane Id et al., 2020) and in the archaea Methanococcus maripaludis (Hernandez and Costa, 2022). Likewise, we showed that FAST:HBR is suitable for monitoring P. aeruginosa in plate reader assays (TFAmber) and by confocal light microscopy (TFCoral). Taken together, these data suggest that phiLOV2.1 and FAST:HBR are currently the best candidates for imaging dual cultures of P. aeruginosa under anaerobic conditions.
The dual imaging of P. aeruginosa subpopulations with phiLOV2.1 and FAST:TFCoral was straightforward in planktonic cells. Upon switching to biofilms, we observed a substantial reduction in the fluorescence signal that was detrimental to imaging. We hypothesized that this phenomenon could stem from the downregulation of the promoter or a reduction of the number of copies of the plasmid when P. aeruginosa formed biofilms. This first generation of reporters was derived from pSMC21, a historical P. aeruginosa expression plasmid (Bloemberg et al., 1997; Cowan et al., 2000). It previously worked to monitor biofilm in aerobic conditions with the bright GFPmut2 (Davey et al., 2003). The low brightness of phiLOV2.1 and FAST:TFCoral revealed previously hidden suboptimal performance of pSMC21 as an expression vector. We, thus, switched these reporter genes to a novel plasmid called pUdO4a. This plasmid is maintained by a BBR1 origin, its sequence is fully known, much smaller than that of pSMC21, and its multiple cloning site is flanked by strong transcription terminators that may contribute to increasing the expression levels of a gene of interest. Indeed, the brightness of P. aeruginosa harboring the pUdO4a derivatives pCFT4.2 (phiLOV2.1) and pCFT7.2 (FAST:TFCoral) allowed the imaging of individual cells in the biofilms. Moreover, it also distinguished the two subpopulations in dual-culture biofilms. It is noteworthy that during co-imaging, the span of wavelengths collected was narrowed to avert signal overlap. This put some cells under the threshold of detection, thereby unveiling the limitation in the compatibility of phiLOV2.1 and FAST:TFCoral. Fluorescent proteins with brighter emission and more widely separated emission peaks would likely improve the imaging performance. Nevertheless, this study paves the way to monitoring dual P. aeruginosa populations in a variety of genetic background and environmental conditions. The broad host range of the BBR1 origin suggests that the second generation of pCFT reporters derived from pUdO4a could be seamlessly transferred to several other gram-negative bacteria.
This study identified six fluorescent proteins that are functional under anoxia in P. aeruginosa. However, their brightness is only a small fraction of that of the GFP under aerobic conditions. Therefore, discovering brighter and complementary O2-independent fluorescent proteins will drive future development by improving our capacity to monitor multiple P. aeruginosa subpopulations. For example, three brighter variants of phiLOV2.1 were recently described – phiLOV3, mini-GFP1 and mini-GFP2 (Babakhanova et al., 2022; Liang et al., 2022). Secondly, FAST derivatives such as FAST2 and tdFAST2 have improved brightness in their HBR-bound state (Tebo et al., 2018). Moreover, frFAST binds a different chromophore, which shifts the fluorescence emission palette toward the farred (Li et al., 2020). Alternative self-labeling proteins not tested here, such as SNAPf, CLIPf and Halo-tag, yielded excellent anaerobic reporters (Los et al., 2008; Sun et al., 2011). Finally, pUdO4a and its derivatives described above provide an excellent platform to expand the fluorescent tools for studying microbes under oxygen-limited conditions in a variety of environmental and physiological settings.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
CFT contributed to the design of the study, performed the experiments, analyzed the data, and wrote the manuscript. FXCV and TFM obtained the funding, designed the study, and critically revised the manuscript. All authors approved the submitted version.
This work was funded by the Faculty of Science of the University of Ottawa, Discovery grants 05587 and 05650, and a research tool and instrument equipment grant 00835 from the Natural Sciences and Engineering Research Council of Canada (NSERC), the CIHR grant 159517, and the CFI John R. Evans Leaders fund grant 34789 that allowed the purchase of the confocal microscope used in this study. CFT received a stipend from the NSERC CREATE in the Technologies for Microbiome Science and Engineering (TechnoMiSE) program and the Ontario Graduate Scholarship program.
We thank Jeffrey W. Keillor for sharing his plate reader, Christopher Boddy for sharing his gel documenting system, and Alain Stintzi for allowing us to perform initial tests in his anaerobic chamber.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1245755/full#supplementary-material
Antoine, R., and Locht, C. (1992). Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from gram-positive organisms. Mol. Microbiol. 6, 1785–1799. doi: 10.1111/j.1365-2958.1992.tb01351.x
Auldridge, M. E., Cao, H., Sen, S., Franz, L. P., Bingman, C. A., Yennamalli, R. M., et al. (2015). LucY: a versatile new fluorescent reporter protein. PLoS One 10:e0124272. doi: 10.1371/journal.pone.0124272
Babakhanova, S., Jung, E. E., Namikawa, K., Zhang, H., Wang, Y., Subach, O. M., et al. (2022). Rapid directed molecular evolution of fluorescent proteins in mammalian cells. Protein Sci. 31, 728–751. doi: 10.1002/pro.4261
Bajunaid, W., Haidar-Ahmad, N., Kottarampatel, A. H., Ourida Manigat, F., Silué, N., Tchagang, C. F., et al. (2020). The T3SS of Shigella: expression, structure, function, and role in vacuole escape. Microorganisms 8:1933. doi: 10.3390/microorganisms8121933
Bloemberg, G. V., O’Toole, G. A., Lugtenberg, B. J., and Kolter, R. (1997). Green fluorescent protein as a marker for Pseudomonas spp. Appl. Environ. Microbiol. 63, 4543–4551. doi: 10.1128/aem.63.11.4543-4551.1997
Buckley, A. M., Jukes, C., Candlish, D., Irvine, J. J., Spencer, J., Fagan, R. P., et al. (2016). Lighting up Clostridium difficile: reporting gene expression using fluorescent LOV domains. Sci. Rep. 6, 6, 1–11. doi: 10.1038/srep23463
Buckley, A. M., Petersen, J., Roe, A. J., Douce, G. R., and Christie, J. M. (2015). LOV-based reporters for fluorescence imaging. Curr. Opin. Chem. Biol. 27, 39–45. doi: 10.1016/j.cbpa.2015.05.011
Campbell-Valois, F. X., Schnupf, P., Nigro, G., Sachse, M., Sansonetti, P. J., and Parsot, C. (2014). A fluorescent reporter reveals on/off regulation of the Shigella type III secretion apparatus during entry and cell-to-cell spread. Cell Host Microbe 15, 177–189. doi: 10.1016/j.chom.2014.01.005
Chapman, S., Faulkner, C., Kaiserli, E., Garcia-Mata, C., Savenkov, E. I., Roberts, A. G., et al. (2008). The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proc. Natl. Acad. Sci. 105, 20038–20043. doi: 10.1073/pnas.0807551105
Chia, H. E., Koebke, K. J., Rangarajan, A. A., Koropatkin, N. M., Marsh, E. N. G., and Biteen, J. S. (2021). New Orange ligand-dependent fluorescent reporter for anaerobic imaging. ACS Chem. Biol. 16, 2109–2115. doi: 10.1021/acschembio.1c00391
Chia, H. E., Zuo, T., Koropatkin, N. M., Marsh, E. N. G., and Biteen, J. S. (2020). Imaging living obligate anaerobic bacteria with Bilin-binding fluorescent proteins. Curr. Res. Microb. Sci. 1, 1–6. doi: 10.1016/j.crmicr.2020.04.001
Christie, J. M., Hitomi, K., Arvai, A. S., Hartfield, K. A., Mettlen, M., Pratt, A. J., et al. (2012). Structural tuning of the fluorescent protein iLOV for improved photostability. J. Biol. Chem. 287, 22295–22304. doi: 10.1074/jbc.M111.318881
Cowan, S. E., Gilbert, E., Khlebnikov, A., and Keasling, J. D. (2000). Dual labeling with green fluorescent proteins for confocal microscopy. Appl. Environ. Microbiol. 66, 413–418. doi: 10.1128/AEM.66.1.413-418.2000
Davey, M. E., Caiazza, N. C., and O’Toole, G. A. (2003). Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185, 1027–1036. doi: 10.1128/jb.185.3.1027-1036.2003
Drepper, T., Eggert, T., Circolone, F., Heck, A., Krauß, U., Guterl, J.-K. K., et al. (2007). Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 25, 443–445. doi: 10.1038/nbt1293
Hall, C. W., and Mah, T.-F. (2017). Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 41, 276–301. doi: 10.1093/femsre/fux010
Hancock, R. E. W., and Woodruff, W. A. (1988). Roles of porin and beta-lactamase in beta-lactam resistance of Pseudomonas aeruginosa. Rev. Infect. Dis. 10, 770–775. doi: 10.1093/clinids/10.4.770
Hernandez, E., and Costa, K. C. (2022). The fluorescence-activating and absorption-shifting tag (FAST) enables live-cell fluorescence imaging of Methanococcus maripaludis. J. Bacteriol. 204:e0012022. doi: 10.1128/jb.00120-22
Høiby, N., Ciofu, O., and Bjarnsholt, T. (2010). Pseudomonas aeruginosa biofilms in cystic fibrosis. Fut. Microbiol. 5, 1663–1674. doi: 10.2217/fmb.10.125
Iizuka, R., Yamagishi-Shirasaki, M., and Funatsu, T. (2011). Kinetic study of de novo chromophore maturation of fluorescent proteins. Anal. Biochem. 414, 173–178. doi: 10.1016/j.ab.2011.03.036
Kumagai, A., Ando, R., Miyatake, H., Greimel, P., Kobayashi, T., Hirabayashi, Y., et al. (2013). A bilirubin-inducible fluorescent protein from eel muscle. Cells 153, 1602–1611. doi: 10.1016/j.cell.2013.05.038
Lam, K. L., Lin, S., Liu, C., Wu, X., Tang, S., Kwan, H. S., et al. (2018). Low-cost method generating in situ anaerobic conditions on a 96-well plate for microbial fermentation in food research. J. Agric. Food Chem. 66, 11839–11845. doi: 10.1021/acs.jafc.8b04888
Li, C., Plamont, M.-A., Sladitschek, H. L., Rodrigues, V., Aujard, I., Neveu, P., et al. (2017). Dynamic multicolor protein labeling in living cells. Chem. Sci. 8, 5598–5605. doi: 10.1039/c7sc01364g
Li, C., Tebo, A. G., Thauvin, M., Plamont, M.-A., Volovitch, M., Morin, X., et al. (2020). A far-red emitting fluorescent chemogenetic reporter for in vivo molecular imaging. Angew. Chem. 132, 18073–18079. doi: 10.1002/ange.202006576
Liang, G. T., Lai, C., Yue, Z., Zhang, H., Li, D., Chen, Z., et al. (2022). Enhanced small green fluorescent proteins as a multisensing platform for biosensor development. Front. Bioeng. Biotechnol. 10, 1–16. doi: 10.3389/fbioe.2022.1039317
Los, G. V., Encell, L. P., McDougall, M. G., Hartzell, D. D., Karassina, N., Zimprich, C., et al. (2008). HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 3, 373–382. doi: 10.1021/cb800025k
Ma, J., Campbell, A., and Karlin, S. (2002). Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J. Bacteriol. 184, 5733–5745. doi: 10.1128/jb.184.20.5733-5745.2002
Magalhães, A. P., Azevedo, N. F., Pereira, M. O., and Lopes, S. P. (2016). The cystic fibrosis microbiome in an ecological perspective and its impact in antibiotic therapy. Appl. Microbiol. Biotechnol. 100, 1163–1181. doi: 10.1007/s00253-015-7177-x
Manigat, F. O., Connell, L. B., Stewart, B. N., LePabic, A.-R., Tessier, C. J. G., Emlaw, J. R., et al. (2023). pUdOs: concise plasmids for bacterial and mammalian cells. bioRxiv 2023.07.05.547852. doi: 10.1101/2023.07.05.547852
Michea Hamzehpour, M., Pechere, J., and Plesiat, P. (1995). OprK and OprM define two genetically distinct multidrug efflux systems in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39, 2392–2396. doi: 10.1128/AAC.39.11.2392
Monmeyran, A., Thomen, P., Jonquière, H., Sureau, F., Li, C., Plamont, M. A., et al. (2018). The inducible chemical-genetic fluorescent marker FAST outperforms classical fluorescent proteins in the quantitative reporting of bacterial biofilm dynamics. Sci. Rep. 8, 10336–10311. doi: 10.1038/s41598-018-28643-z
Mordaka, P. M., and Heap, J. T. (2018). Stringency of synthetic promoter sequences in Clostridium revealed and circumvented by tuning promoter library mutation rates. ACS Synth. Biol. 7, 672–681. doi: 10.1021/acssynbio.7b00398
Nigro, G., Arena, E. T., Sachse, M., Moya-Nilges, M., Marteyn, B. S., Sansonetti, P. J., et al. (2019). Mapping of Shigella flexneri’s tissue distribution and type III secretion apparatus activity during infection of the large intestine of guinea pigs. Pathog. Dis. 77, 1–9. doi: 10.1093/femspd/ftz054
O’Brien, S., and Fothergill, J. L. (2017). The role of multispecies social interactions in shaping Pseudomonas aeruginosa pathogenicity in the cystic fibrosis lung. FEMS Microbiol. Lett. 364, 1–10. doi: 10.1093/femsle/fnx128
Peron-Cane Id, C., Fernandez Id, J.-C., Leblanc, J., Wingertsmann, L., Gautierid, A., Despratid, N., et al. (2020). Fluorescent secreted bacterial effectors reveal active intravacuolar proliferation of Listeria monocytogenes in epithelial cells. PLoS Pathog. 16:e1009001. doi: 10.1371/journal.ppat.1009001
Plamont, M.-A., Billon-Denis, E., Maurin, S., Gauron, C., Pimenta, F. M., Specht, C. G., et al. (2016). Small fluorescence-activating and absorption-shifting tag for tunable protein imaging in vivo. Proc. Natl. Acad. Sci. 113, 497–502. doi: 10.1073/pnas.1513094113
Plesiat, P., Aires, J. R., Godard, C., and Köhler, T. (1997). Use of steroids to monitor alterations in the outer membrane of Pseudomonas aeruginosa. J. Bacteriol. 179, 7004–7010. doi: 10.1128/jb.179.22.7004-7010.1997
Ponka, P. (1999). Cell biology of heme. Am. J. Med. Sci. 318, 241–256. doi: 10.1016/S0002-9629(15)40628-7
Pressler, T., Bohmova, C., Conway, S., Dumcius, S., Hjelte, L., Høiby, N., et al. (2011). Chronic Pseudomonas aeruginosa infection definition: EuroCareCF working group report. J. Cyst. Fibros. 10, S75–S78. doi: 10.1016/S1569-1993(11)60011-8
Rahme, L. G., Stevens, E. J., Wolfort, S. F., Shao, J., Tompkins, R. G., and Ausubel, F. M. (1995). Common virulence factors for bacterial pathogenicity in plants and animals. Science 268, 1899–1902. doi: 10.1126/science.7604262
Ratliff, M., Zhu, W., Deshmukh, R., Wilks, A., and Stojiljkovic, I. (2001). Homologues of neisserial heme oxygenase in gram-negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J. Bacteriol. 183, 6394–6403. doi: 10.1128/jb.183.21.6394-6403.2001
Ringquist, S., Shinedling, S., Barrick, D., Green, L., Binkley, J., Stormo, G. D., et al. (1992). Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol. Microbiol. 6, 1219–1229. doi: 10.1111/J.1365-2958.1992.tb01561.X
Rumyantsev, K. A., Shcherbakova, D. M., Zakharova, N. I., Emelyanov, A. V., Turoverov, K. K., and Verkhusha, V. V. (2015). Minimal domain of bacterial phytochrome required for chromophore binding and fluorescence. Sci. Rep. 5:18348. doi: 10.1038/srep18348
Schmitt, M. P. (1997). Utilization of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J. Bacteriol. 179, 838–845. doi: 10.1128/jb.179.3.838-845.1997
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Shapleigh, J. P. (2013). “Denitrifying prokaryotes” in The prokaryotes: prokaryotic physiology and biochemistry. eds. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer), 405–425.
Shcherbakova, D. M., and Verkhusha, V. V. (2013). Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods 10, 751–754. doi: 10.1038/nmeth.2521
Skaar, E. P., Gaspar, A. H., and Schneewind, O. (2004). IsdG and IsdI, Heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 279, 436–443. doi: 10.1074/jbc.M307952200
Streett, H., Charubin, K., and Papoutsakis, E. T. (2021). Anaerobic fluorescent reporters for cell identification, microbial cell biology and high-throughput screening of microbiota and genomic libraries. Curr. Opin. Biotechnol. 71, 151–163. doi: 10.1016/j.copbio.2021.07.005
Streett, H. E., Kalis, K. M., and Papoutsakis, E. T. (2019). A strongly fluorescing anaerobic reporter and protein-tagging system for Clostridium organisms based on the fluorescence-activating and absorption-shifting tag protein (FAST). Appl. Environ. Microbiol. 85, 1–15. doi: 10.1128/AEM.00622-19
Stutts, M. J., Knowles, M. R., Gatzy, J. T., and Boucher, R. C. (1986). Oxygen consumption and ouabain binding sites in cystic fibrosis nasal epithelium. Pediatr. Res. 20, 1316–1320. doi: 10.1203/00006450-198612000-00026
Sun, X., Zhang, A., Baker, B., Sun, L., Howard, A., Buswell, J., et al. (2011). Development of SNAP‐tag fluorogenic probes for wash‐free fluorescence imaging. Chembiochem 12, 2217–2226. doi: 10.1002/cbic.201100173
Tebo, A. G., Pimenta, F. M., Zhang, Y., and Gautier, A. (2018). Improved chemical-genetic fluorescent markers for live cell microscopy. Biochemistry 57, 5648–5653. doi: 10.1021/acs.biochem.8b00649
Tsien, R. Y. (1998). The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544. doi: 10.1146/annurev.biochem.67.1.509
Wagner, V. E., and Iglewski, B. H. (2008). Pseudomonas aeruginosa biofilms in CF infection. Clin. Rev. Allergy Immunol. 35, 124–134. doi: 10.1007/s12016-008-8079-9
Wegele, R., Tasler, R., Zeng, Y., Rivera, M., and Frankeinberg-Dinkel, N. (2004). The heme oxygenase(s)-phytochrome system of Pseudomonas aeruginosa. J. Biol. Chem. 279, 45791–45802. doi: 10.1074/jbc.M408303200
Wilks, A., and Schmitt, M. P. (1998). Expression and characterization of a heme oxygenase (Hmu O) from Corynebacterium diphtheriae. J. Biol. Chem. 273, 837–841. doi: 10.1074/jbc.273.2.837
Winstanley, C., O’Brien, S., and Brockhurst, M. A. (2016). Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol. 24, 327–337. doi: 10.1016/j.tim.2016.01.008
Worlitzsch, D., Herberth, G., Ulrich, M., and Doring, G. (1998). Catalase, myeloperoxidase and hydrogen peroxide in cystic fibrosis. Eur. Respir. J. 11, 377–383. doi: 10.1183/09031936.98.11020377
Worlitzsch, D., Tarran, R., Ulrich, M., Schwab, U., Cekici, A., Meyer, K. C., et al. (2002). Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109, 317–325. doi: 10.1172/JCI13870
Yoon, S. S., Hennigan, R. F., Hilliard, G. M., Ochsner, U. A., Parvatiyar, K., Kamani, M. C., et al. (2002). Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3, 593–603. doi: 10.1016/S1534-5807(02)00295-2
Zhu, W., Hunt, D. J., Richardson, A. R., and Stojiljkovic, I. (2000). Use of heme compounds as Iron sources by pathogenic Neisseriae requires the product of the hemO gene. J. Bacteriol. 182, 439–447. doi: 10.1128/jb.182.2.439-447.2000
Keywords: Pseudomonas aeruginosa, anaerobic, biofilm, fluorescence microscopy, LOV, Fluorescence-Activating and Absorption-Shifting Tag (FAST), GFP, LucY
Citation: Tchagang CF, Mah T-F and Campbell-Valois F-X (2023) Anaerobic fluorescent reporters for live imaging of Pseudomonas aeruginosa. Front. Microbiol. 14:1245755. doi: 10.3389/fmicb.2023.1245755
Received: 23 June 2023; Accepted: 25 September 2023;
Published: 20 October 2023.
Edited by:
Christophe Bordi, Aix Marseille Université, FranceReviewed by:
Monica Serrano, Universidade Nova de Lisboa, PortugalCopyright © 2023 Tchagang, Mah and Campbell-Valois. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: François-Xavier Campbell-Valois, ZmNhbXBiZWxAdW90dGF3YS5jYQ==; Thien-Fah Mah, dG1haEB1b3R0YXdhLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.