- 1UMR545 Fromage, INRAE, VetAgro Sup, Université Clermont Auvergne, Aurillac, France
- 2Pôle Fromager AOP Massif Central, Aurillac, France
- 3Université Paris-Saclay, INRAE, AgroParisTech, UMR SayFood, Palaiseau, France
Streptococcus thermophilus is of major importance for cheese manufacturing to ensure rapid acidification; however, studies indicate that intensive use of commercial strains leads to the loss of typical characteristics of the products. To strengthen the link between the product and its geographical area and improve the sensory qualities of cheeses, cheese-producing protected designations of origin (PDO) are increasingly interested in the development of specific autochthonous starter cultures. The present study is therefore investigating the genetic and functional diversity of S. thermophilus strains isolated from a local cheese-producing PDO area. Putative S. thermophilus isolates were isolated and identified from milk collected in the Saint-Nectaire cheese-producing PDO area and from commercial starters. Whole genomes of isolates were sequenced, and a comparative analysis based on their pan-genome was carried out. Important functional properties were studied, including acidifying and proteolytic activities. Twenty-two isolates representative of the diversity of the geographical area and four commercial strains were selected for comparison. The resulting phylogenetic trees do not correspond to the geographical distribution of isolates. The clustering based on the pan-genome analysis indicates that isolates are divided into five distinct groups. A Kyoto Encyclopedia of Genes and Genomes (KEGG) functional annotation of the accessory genes indicates that the accessory gene contents of isolates are involved in different functional categories. High variability in acidifying activities and less diversity in proteolytic activities were also observed. These results indicate that high genetic and functional variabilities of the species S. thermophilus may arise from a small (1,800 km2) geographical area and may be exploited to meet demand for use as autochthonous starters.
1. Introduction
Streptococcus thermophilus is a thermophilic lactic acid bacteria (LAB) with major economic importance for the dairy industry. Due to its ability to rapidly acidify the milk, it is extensively used for the manufacture of several important fermented dairy products and is considered the second most important species among LAB after Lactococcus lactis (Harnett et al., 2022). Streptococcus thermophilus is a clonal species that has only recently emerged from a commensal ancestor of the salivarius group (Delorme, 2008). This species is part of the genus Streptococcus and is the only one that obtained the Generally Recognized as Safe (GRAS) status and the Qualified Presumption of Safety (QPS) status within this genus. Comparative genomic studies on Streptococcus thermophilus genomes highlight that this species has lost many virulence-related functions common among pathogenic streptococci. These studies suggest that the Streptococcus thermophilus genome has followed a regressive evolution process toward a specialized bacterium dedicated to growth in milk. Comparative genomics also revealed that the dairy Streptococcus genome may have been shaped mainly through loss-of-function events and horizontal gene transfer (Bolotin et al., 2004; Hols et al., 2005; Goh et al., 2011).
In the dairy industry, Streptococcus thermophilus is used as a starter culture for its capability to rapidly acidify the milk, which is a technological characteristic of major importance to guarantee a good outcome of the dairy process and to enhance food safety by preventing the development of pathogenic bacteria (Rakhmanova et al., 2018). Proteolysis is considered one of the most important enzymatic pathways in the manufacture of many fermented dairy products. In cheese production, proteolysis plays a crucial role in the flavor and texture of cheese by releasing peptides and free amino acids that could undergo secondary reactions (Fox, 1989; Smit et al., 2005).
Over the past few years, changes in the way of life of consumers have imposed requirements for uniformity in the quality of traditional dairy products with high typicality (Settanni and Moschetti, 2014). Due to this growing interest in preserving the typical sensorial properties of traditional cheeses, there is an increasing demand for autochthonous (also called local, indigenous, or wild) and specific strains to replace the well-defined industrial starter strains largely widespread and extensively used in dairy industries (Munekata et al., 2022). It is now well known that a careful selection and use of autochthonous microbial strains can contribute to obtaining high-quality, functional, and safe dairy products (Gómez et al., 2016; Campagnollo et al., 2018; Fusco et al., 2019; Ait Chait et al., 2021; Huang et al., 2021). Such strains are mostly isolated from traditionally made cheeses or raw milk (Carafa et al., 2015; Terzić-Vidojević et al., 2015; Frau et al., 2016; Fusco et al., 2019; Özkan et al., 2021).
Many studies have focused on the genetic and functional diversity of Streptococcus thermophilus strains (Giraffa et al., 2001; Morandi and Brasca, 2012; Vendramin et al., 2017; Alexandraki et al., 2019), but few have studied the genetic diversity of wild strains of this species (Andrighetto et al., 2002; Erkus et al., 2014). Few studies have been conducted on the biodiversity analysis of Streptococcus thermophilus strains using whole genome sequencing (Hu et al., 2020). All these studies reported high genetic and technological diversity within S. thermophilus strains isolated from dairy products.
Protected designation of origin (PDO) cheese producers from Massif Central (France) mostly use commercial starter cultures for the manufacture of cheeses. The use of commercial starters in cheese production results in the loss of typical characteristics of the products (Hansen, 2002). To strengthen the link with the area and improve the sensorial qualities and typicality of cheeses, PDO cheese producers are increasingly interested in the development of specific autochthonous starter cultures. To meet this demand, a first step would be to isolate new wild strains from the producing areas and investigate the genetic and functional diversity of these strains within these reservoirs. Saint-Nectaire is an uncooked pressed cheese that was awarded the PDO label in 1955. Made in one of the smallest PDO areas in Europe, between Puy-de-Dôme and Cantal (France), this is the third French PDO cheese with cow’s milk.
The present study explores the genetic and functional diversity of wild strains of Streptococcus thermophilus from natural habitats at a local scale. This diversity was explored in the Saint-Nectaire cheese-producing PDO area, one of the smallest PDO areas in Europe with only 1,800 km2. The comparative analysis of the whole genome sequences of 26 wild strains of Streptococcus thermophilus was carried out. Important functional properties of the isolates were investigated, including milk acidification and proteolytic activities.
2. Materials and methods
2.1. Isolation and identification of Streptococcus thermophilus strains
2.1.1. Isolation of strains
Presumptive Streptococcus thermophilus strains were isolated from (i) 62 milk samples (heated for 6 h at 42°C) collected on 31 different farms in the Saint-Nectaire cheese-producing PDO area (France) in summer and in winter to study the seasonal effect on biodiversity and (ii) commercial starter cultures. Isolation was carried out on M17 agar medium (Biokar Diagnostics, Beauvais, France) incubated at 42°C for 48 h. Representative colonies were picked out of this medium, purified twice, and maintained frozen at −80°C in an M17 broth medium containing 20% (v/v) glycerol. One Streptococcus thermophilus isolate was obtained from a bacterial collection of LAB isolated in 1998 from Saint-Nectaire cheeses (INRAE-ARILAIT collection). The Saint-Nectaire PDO area spreads over 69 municipalities between Puy-de-Dôme and Cantal (France), which are referenced in Supplementary Table S1. The specifications for the PDO of Saint-Nectaire cheese describe the conditions for cow breeding, milk production, cheese manufacturing, and ripening of cheeses.1
2.1.2. 16S rDNA identification
16S rDNA of Saint-Nectaire isolates was amplified with the universal primers WO2 (5′-100 GNTACCTTGTTACGACTT-3′) and W18 (5′-GAGTTTGATCMTGGCTCAG-3′), as described previously by Callon et al. (2004). Polymerase chain reaction (PCR) amplification was carried out in a final volume of 50 μL containing 1X PCR buffer with MgCl2, 1 colony of isolates, 0.25 mM each primer, 200 mM each dNTP, and 1 U Taq DNA polymerase. The thermal cycling conditions were 94°C for 3 min, followed by 25 cycles of 94°C for 30 s, 50°C for 30 s, 72°C for 1 min 30 s, and a final extension step of 72°C for 10 min. The 16S rDNA gene sequencing was performed by LGC Biosearch Technologies (Berlin, Germany).
16S rDNA of commercial strains was amplified with the universal primers A (5′-AGAGTTTGATCCTGGCTCAG-3′) and H (5′-AAGG AGGTGATCCAGCCGCA-3′; Feurer et al., 2004). PCR amplification was carried out as described above for the Saint-Nectaire isolates. The 16S rDNA gene sequencing was performed by Eurofins Genomics (Konstantz, Germany).
To identify the partial 16S rDNA sequences obtained, a search of the NCBI GenBank DNA database was conducted using the BLAST algorithm. The percentage of similarity with DNA sequences deposited in this bank was determined.
16S rDNA sequences were deposited in the NCBI GenBank database under accession numbers OR350537-OR350561.
2.1.3. Selection of representative isolates
In total, 58 isolates and 4 commercial strains were identified as Streptococcus thermophilus and were screened for their acidifying and proteolytic activity and for their growth characteristics in sterilized skimmed milk. Notably, 200 mL of sterilized skimmed milk (LACTALIS Ingredients, Bourgbarré, France) was inoculated with isolates at 106 cfu.mL−1 and incubated for 24 h in a temperature-controlled batch reactor programmed to simulate the decrease in temperature during the manufacture of Saint-Nectaire cheese type (decrease from 33°C to 9°C over 24 h). The acidification kinetics were studied by continuous pH recording (iCINAC system). Proteolysis abilities of isolates were measured using the OPA (o-phtalaldehyde) method described by Church et al. (1983). Growth characteristics of isolates were determined by enumeration of isolates on M17 agar medium every hour for 24 h.
Three groups of isolates were formed based on these results. A total of 22 isolates representative of these three groups were selected for further characterization in this study. All selected isolates collected from the farms were isolated from dairy farms with milk production and cheese manufacturing directly on the farm.
2.2. Genetic characterization of strains
2.2.1. DNA extraction and sequencing
Genomic DNA was extracted from cell cultures with Nuclospin® Tissue from Macherey Nagel according to the manufacturer’s instructions. Final concentrations were measured with a Qubit™ fluorometer using the dsDNA Broad Range Kit (Thermo Fisher Scientific). The extracted DNA was further sequenced using Illumina technology. Library preparation and sequencing were handled by Eurofins genomics (Konstantz, Germany) using a Novaseq 6000 sequencing system (Illumina, San Diego, CA, United States).
2.2.2. Genome assembly and annotation
Sequencing reads from raw fastq files were filtered at Q30 with a minimal length of 110 bp with prinSeq (Cantu et al., 2019). Reads with remaining sequencing adapters were excluded with cutadapt V4.1 (Martin, 2011). Each genome was assembled using Spades V3.13 (Bankevich et al., 2012) with the careful option and annotated with Bakta V1.5 (Schwengers et al., 2021). EBI accession numbers are referenced in Table 1.
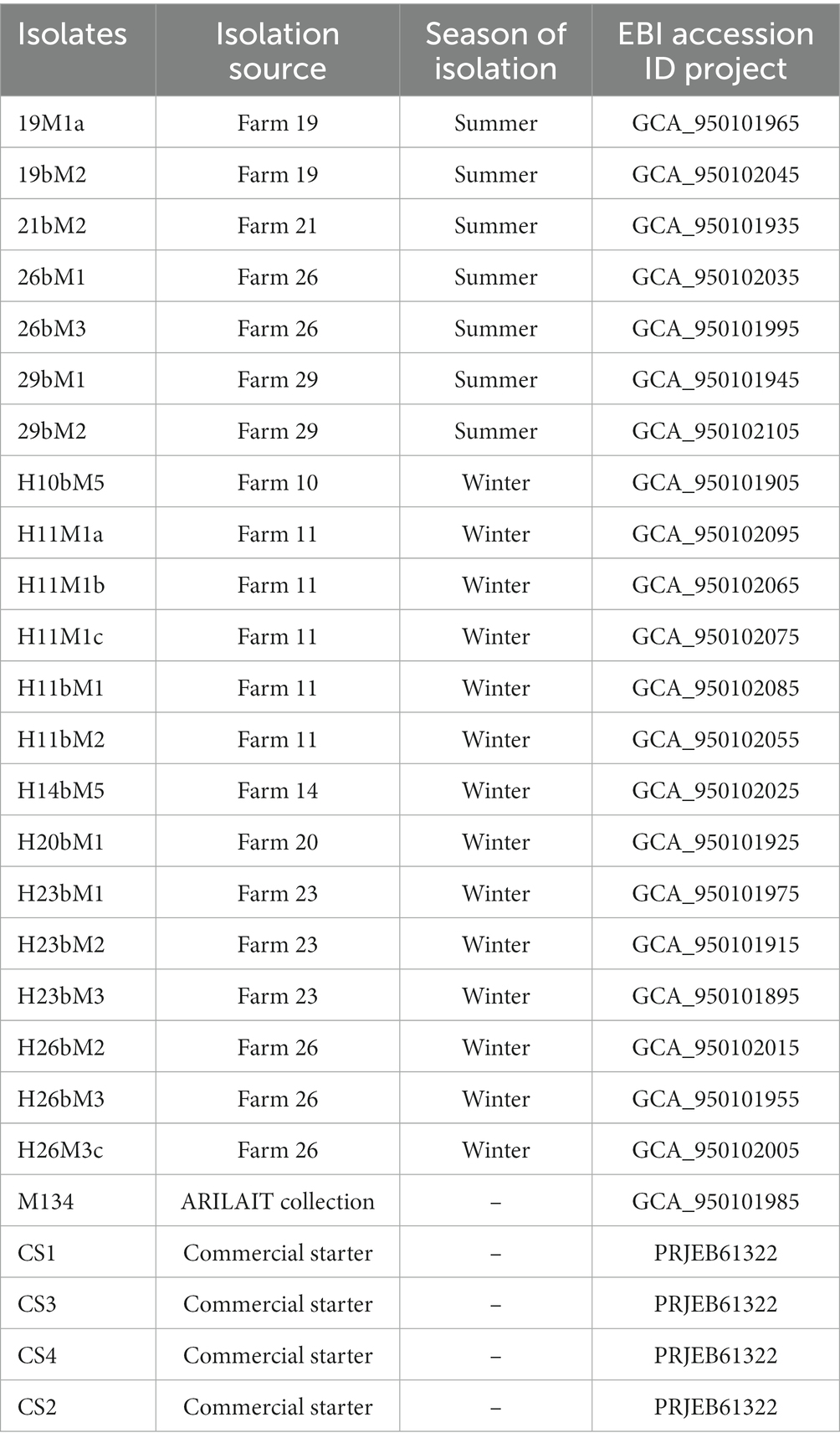
Table 1. Streptococcus thermophilus isolation sources with genomes assembly accession numbers analyzed in this study.
2.2.3. Pan-genome assembly and visualization
Annotated GFF3 files of 22 Streptococcus thermophilus isolated from the Saint-Nectaire cheese-producing PDO area and 4 strains of Streptococcus thermophilus isolated from commercial starter cultures genomes were submitted to Roary (Page et al., 2015) for pan-genome analysis using default parameters. A gene presence–absence data matrix was derived and visualized using Phandango (Hadfield et al., 2018).
2.2.4. Phylogenetic reconstruction
Nucleotide sequences of 1,350 core genes (excluding duplicated genes) were extracted from Roary results. They have been aligned with Mafft (Katoh and Standley, 2013), and distance computing and tree sampling were realized with BEAST (Suchard et al., 2018). A strict molecular clock was chosen with the HKY substitution model. BEAST (Suchard et al., 2018) is a program for Bayesian analysis of molecular sequences using MCMC (Markov chain Monte Carlo). It is entirely orientated toward rooted, time-measured phylogenies inferred using strict or relaxed molecular clock models. BEAST uses MCMC to average over tree space, so that each tree is weighted proportional to its posterior probability. We chose the strict molecular clock model with the HKY substitution model because this assumes that all branches on the tree have the same rate of evolution. This appeared reasonable considering the absence of knowledge on those rates for our new isolates.
Core-genome single nuclear polymorphism (SNP) tree was created with Parsnp (Treangen et al., 2014) on the Galaxy platform (The Galaxy Community, 2022). The resulting phylogenetic tree was visualized using iTOL.2 Parsnp is a genome multi-alignment tool designed to align genome sequences. It aligns and provides the output as the multiple sequence alignment of given sequences, SNP variations, and the core genome phylogeny. For this reconstruction, LMD-9 was used as the reference genome. The genome sequencing of Streptococcus thermophilus LMD-9 was described previously (Makarova et al., 2006), and the complete genome sequence can be accessed at GenBank under accession number CP000419.
Kyoto Encyclopedia of Genes and Genomes (KEGG) numbers (Kanehisa and Goto, 2000; Kanehisa, 2019; Kanehisa et al., 2023) for accessory genes were obtained using the eggnog-mapper v2 web tool (Iyer et al., 2010). Partial least squares discriminant analysis (PLS-DA) was performed using the mixOmics package of R software V 4.2.3.3
2.3. Technological characterization of isolates
2.3.1. Preparation of model cheese curd
The model cheese curds (MCC) were prepared according to Callon et al. (2016) with some modifications. In total, 40 mL of pasteurized milk (Ferme des Peupliers, Normandie, France) was incubated at 33°C and inoculated with isolates at 106 cfu.mL−1. The milk was coagulated with 12 μL of calf rennet for 45 min at 33°C and then centrifuged for 20 min at 20,000 g at 33°C. The supernatant (lactoserum) was discarded, and the curd was incubated for 24 h in a temperature-controlled batch reactor programmed to simulate the decrease in temperature during the manufacture of Saint-Nectaire cheese type (decrease from 33°C to 9°C over 24 h). Each strain was tested three times.
2.3.2. Growth characteristics
To determine the growth ability of isolates, MCC samples were collected at 0, 6, and 24 h. MCC samples were blended two times for 2 min in a phosphate buffer of pH 7.5 using a Stomacher blender (Interscience, St Nom la Bretèche, France) for dissociating all the cell aggregates. After appropriate dilution in Ringer’s solution, S. thermophilus isolates were enumerated on M17 agar and incubated at 30°C for 48 h.
2.3.3. Acidifying activity
To analyze the acidifying activities of isolates, MCC samples were collected at 0, 6, and 24 h, and pH was measured in the core of the MCC samples using a WTW pH 526 pH meter with a LOT406-M6-DXK electrode (Mettler-Toledo S.A, Viroflay, France). A classification of isolates based on their acidifying activities was constructed according to that previously reported by Morandi and Brasca (2012), with some modifications. The pH unit target values were modified because we worked on a model cheese curd with a temperature decrease mimicking Saint-Nectaire cheesemaking (33°C to 9°C), while precedent studies worked on fermented milk at a constant temperature of 37°C.
2.3.4. Proteolytic activity
The extraction of water-soluble nitrogen from the MCC samples was carried out according to the method described by Myagkonosov et al. (2021) with some modifications. Five grams of MCC samples were mixed with 5 cm3 of deionized water and homogenized with a stomacher for 4 min. The resulting mixture was transferred to a volumetric flask, and the volume was made up to 100 mL with deionized water. The mixture was kept at 40°C for 1 h with continuous shaking. The samples were centrifuged at 3,000 g for 30 min. After centrifugation, the samples were cooled to 4°C, and the upper fat layer was removed. The supernatant was separated and filtered with a pore size of 0.45 μm. The resulting filtrate was mixed with deionized water at a ratio of 1:5. Next, 3 mL of OPA (o-phtalaldehyde) reagent prepared according to Church et al. (1983) was added to 300 μL of the solution, and after 2 min, absorbance was measured at 340 nm with a 7,200 spectrophotometer Jenway (Dutscher, France). The results have been expressed in mmol.L−1 of glycine based on a calibration curve. The proteolytic activity of isolates was determined at 24 h of fermentation by subtracting the results at 0 h of fermentation.
2.3.5. Statistical analysis
Statistical analysis of biochemical data was performed using the XLSTAT software (Addinsoft, Paris, France). The results are reported as means ± standard deviation. The normality of the data was checked using a Shapiro–Wilk test (p < 0.05). The test rejected the hypothesis Ha of normality when the value of p was ≤0.05. A large number of variables did not have a normal distribution, and therefore a non-parametric test (Kruskal-Wallis) and post-hoc comparison (Conover-Iman procedure) were used to compare the concentrations obtained. Differences between the mean values were considered significant at p < 0.05. Pearson’s correlations at the 5% significance level (p < 0.05) were used to explain the relationship between functional properties.
3. Results and discussion
3.1. Isolation and identification
In this study, strains belonging to the Streptococcus thermophilus species were isolated from the Saint-Nectaire cheese-producing PDO area (France).
All isolates were identified on the basis of 16S rDNA sequence alignment using the NCBI blast algorithm. Isolates showing a percentage of similarity equal to or higher than 99% with Streptococcus thermophilus DNA sequences available in this database were considered to be Streptococcus thermophilus. In total, 58 isolates were identified as Streptococcus thermophilus.
Twenty-two isolates collected throughout the geographical producing area and representative of diversity, together with four commercial strains, were selected and characterized for their genetic and technological properties. Conditions of isolation and the EBI accession number are referenced in Table 1. The genome sequencing and assembly-related information are shown in Supplementary Table S2.
3.2. Genetic diversity
3.2.1. Phylogenetic reconstruction
Two phylogenetic analyses were performed (Figure 1). The first one was based on core-gene alignment using the Bayesian method and allowed to identify seven clades (Figure 1A), namely, A–G. The other was based on single nucleotide polymorphisms (SNPs) detected (Figure 1B) and allowed the distinction of seven clades, namely, 1–7.
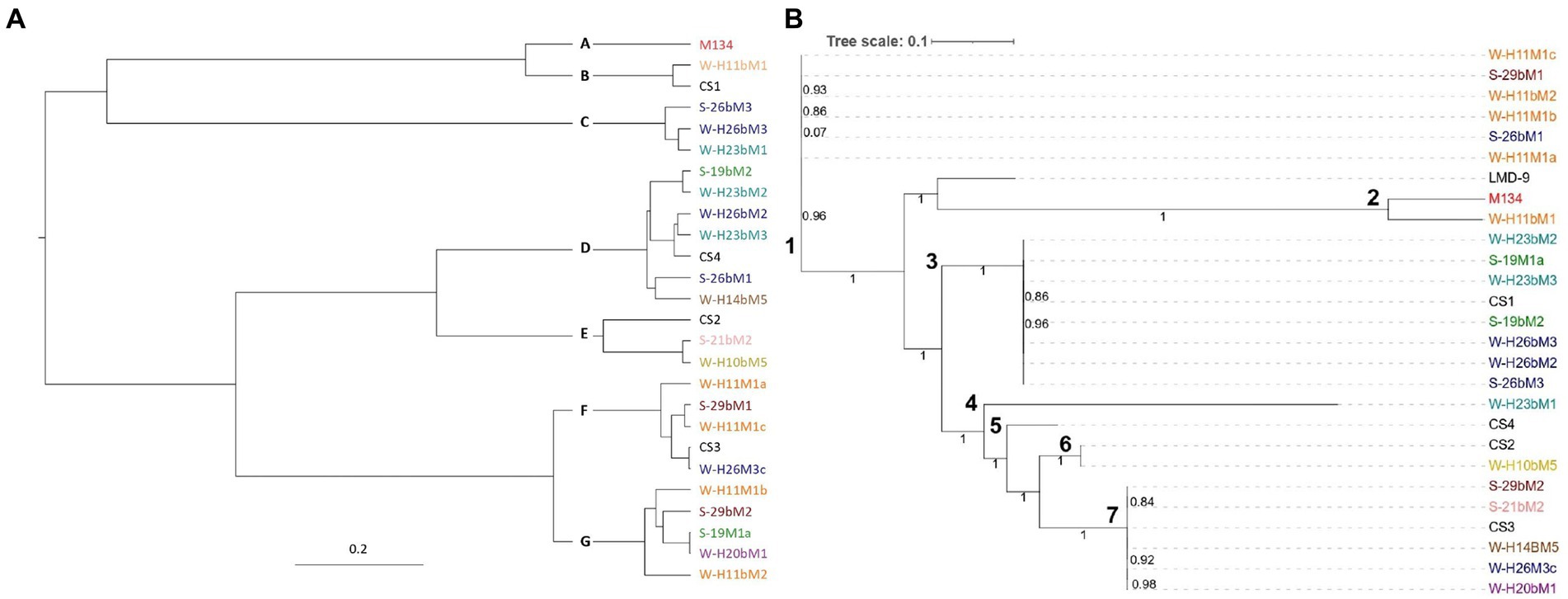
Figure 1. Phylogenetic trees of the 26 Streptococcus thermophilus isolates based on (A) core genome and (B) single nucleotide polymorphisms. Isolates from summer milks are annotated with an “S” and isolates from winter milks are annotated with a “W.” Isolate names with the same colors were isolated from the same farm.
From the two resulting trees, it could be noticed that SN-isolates from the same farms were distributed in various clades. For example, in Figure 1A, isolates H11M1a, H11bM1, and H11M1b were all isolated on farm 26, but were not located in the nearby clade. A similar situation is observed in Figure 1B for isolates 26bM1, 26bM3, and H26M3c. Moreover, strains from different farms may be clustered together.
These results indicate that the phylogenetic reconstruction of isolates did not correspond to their geographical distribution, despite a small isolation area (1,800 km2) and closed milk production. Vendramin et al. (2017) and Hu et al. (2020) reported similar results with strains isolated from China and Italy, respectively, compared to strains isolated from other continents. This distinction indicated that the isolation source did not exert an important impact on the evolution of Streptococcus thermophilus, although the farms are quite close to one another. It is also notable that isolates from summer and winter milk were distributed into different clades and not clustered together, suggesting that the genetic variability of the isolates did not depend on the season of isolation.
3.2.2. Pan-genome analysis
The pan-genome of the 22 Saint-Nectaire isolates and the 4 commercial strains was performed by clustering the genes encoding complete protein sequences into core and accessory genomes using Roary (Page et al., 2015).
The core genome is the set of genes shared by all the genomes studied and can be divided into hard-core genes, which are defined to be present in >99% of the genomes, and soft-core genes, which are present in 95%–99% of the genomes. The accessory genome is shared by a subset of the genomes tested and is subdivided into shell genes, which are present in 15%–95% of genomes, and cloud genes, which are found in less than 15% of genomes. From the 3,269 genes constituting the pan-genome of the 26 Streptococcus thermophilus isolates studied here, we found 1,450 (44%) core genes (present in 24/26 genomes), including 1,355 (41%) hard core genes (present in 25/26 genomes) and 95 (3%) soft core genes (present in 24/25 genomes), and 1,819 (56%) accessory genes (present in less than 24 genomes), which might be responsible for the fundamental differences in phenotypic characteristics between the isolates (Daughtry et al., 2018), including 776 (24%) shell genes (present in 3–24 genomes) and 1,053 (32%) cloud genes (present in less than 3 genomes; Figure 2). These results are similar to those reported by Alexandraki et al. (2019), who analyzed the pan-genome of 23 strains of S. thermophilus and identified 1,089 core genes (52%) and 997 accessory genes (48%).
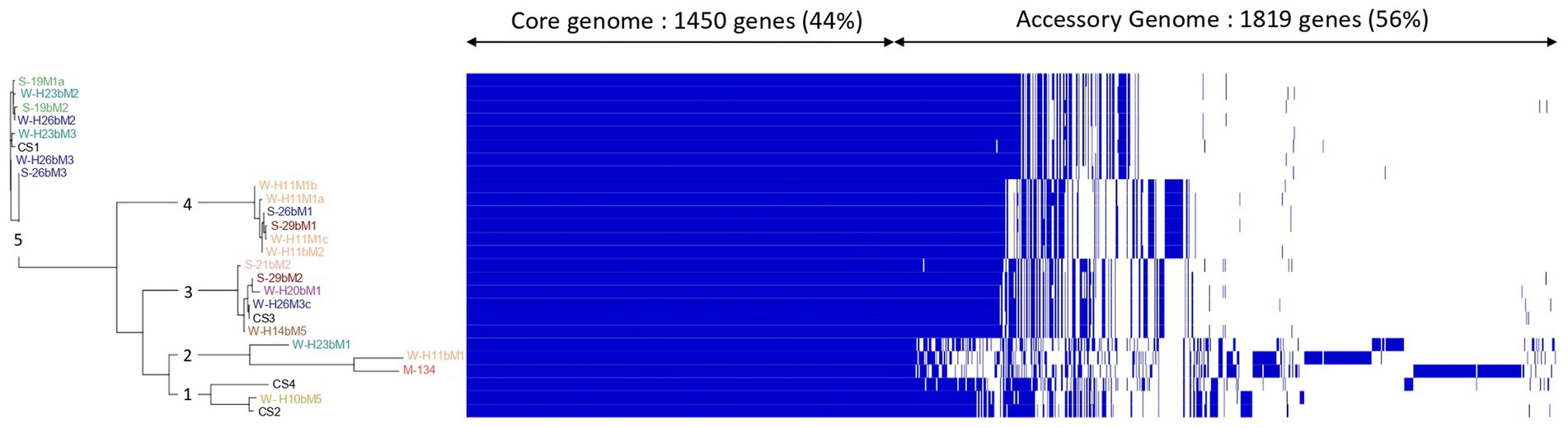
Figure 2. Clustering of isolates associated with the visualization of the Streptococcus thermophilus isolate pan-genome. The pan-genome was visualized based on the Phandandgo software (Hadfield et al., 2018). In the Roary matrix, genomes are shown as rows, and homologous gene clusters are depicted as columns. The presence of a gene cluster in a genome is indicated by blue. Core gene clusters that are found in all genomes are shown on the left side of the matrix. Isolates from summer milks are annotated with an “S” and isolates from winter milks are annotated with a “W.” Isolate names with the same colors were isolated from the same farm.
The generated clustering based on the pan-genome was concordant with the phylogenetic reconstruction based on SNPs (Figure 1B) in defining the relationships among isolates. It showed five main clusters with obvious differences in accessory genes for each of them (Figure 2). The first cluster consisted of SN-isolate H10bM5 and commercial strains CS4 and CS2. Cluster 2 included SN-isolates H11bM1, M134, and H23bM1. SN-isolates 21bM2, 29bM2, H20bM1, H26M3c, H14bM5, and commercial strain CS3 were clustered together. The fourth cluster comprised SN-isolates H11M1b, H11M1a, 26bM1, 29bM1, H11M1c, and H11bM2. Finally, the fifth cluster consisted of SN-isolates 19M1a, H23bM2, 19bM2, H26bM2, H23bM3, H26bM3, 26bM3, and commercial strain CS1. These results reveal a high genetic variation level probably related to the accessory gene content across isolates, which are involved in different functional categories. These findings are in accordance with Giraffa et al. (2001), Andrighetto et al. (2002), and Morandi and Brasca (2012), who also reported a high genetic variability of Streptococcus thermophilus with the RAPD-PCR method.
3.2.3. KEGG functional analysis
A hierarchical clustering of the isolates was performed on the accessory genes annotated with the KEGG pathway (Figure 3). Almost 77% of the accessory genes were positively annotated with a KEGG ko number. These genes were divided into 23 functional KEGG pathways, the most important being “Protein families: signaling and cellular processes” (18% of the total annotated genes), “Unclassified: genetic information processing” (16%), “Membrane transport” (10%), “Carbohydrate metabolism” (7%), “Protein families: genetic information processing” (7%), “Protein families: metabolism” (6%), and amino acid metabolism (6%). These results are in accordance with Vendramin et al. (2017), who performed a genome comparison of eight Streptococcus thermophilus strains of dairy origin isolated in Italy. They investigated the strain-specific features by assigning functional categories of the SEED subsystem to the gene content of the strains. Among the functional categories identified, four accounted for a large part of the strain diversity, including “amino acids and derivatives,” “carbohydrates,” “DNA metabolism,” and “membrane transport,” covering almost 50% of the specific genes. Although the annotations are different due to the databases used, their results are similar to the present study.
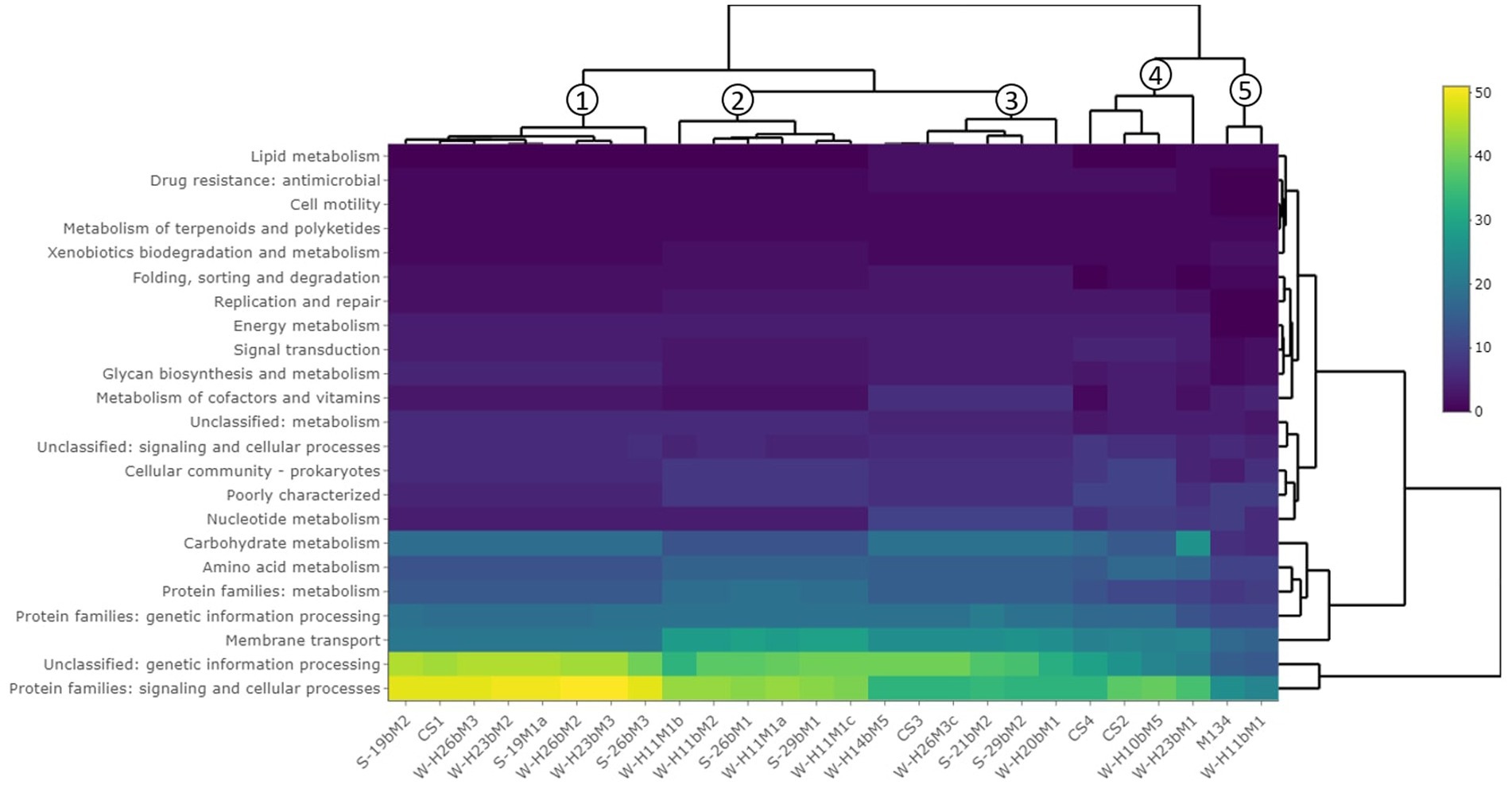
Figure 3. Hierarchical clustering of isolates based on the gene content summarized by KEGG pathways. The clustering on top of the figure shows similarities between isolates.
This hierarchical clustering (Figure 3) allowed an evaluation of the similarity between isolates based on the number of accessory genes associated with a functional KEGG pathway. Five groups were identified and are similar to those observed in the phylogenetic tree based on the pan-genome with a difference (Figure 2).
The first group is composed of isolates H23bM2, 19bM2, 19M1a, H26bM3, 26bM3, and CS1. The second group includes isolates H11M1c, 29bM1, H11M1a, 26bM1, H11bM2, and H11M1b. Isolates 21bM2, H14bM5, H26M3c, 29bM2, H20bM1, and CS3 formed the third group. Isolates H10bM5, H23bM1, CS4, and CS2 were clustered in the fourth group, and the fifth group consisted of isolates M134 and H11bM1.
The different groups stand out for their functional KEGG class. A PLS-DA analysis was performed (Figure 4) to identify KEGG classes that differentiate isolates into these groups. PLS-DA results showed that the groups were easily recognized based on the accessory gene content of each KEGG class. These KEGG classes may be the indicators to distinguish these three clades. Figure 5 shows the variable’s contribution from components 1 (A) and 2 (B) of the PLS-DA analysis, and Figure 6 represents the clustered image map (CIM) associated with it. These figures showed the functional KEGG pathways overrepresented in each of the clades that allowed their differentiation. Heatmaps, representing the lowest level of the KEGG pathway database (gene ko level), were constructed for each KEGG pathway identified as characteristic of a group (Supplementary Figures S1–S4).
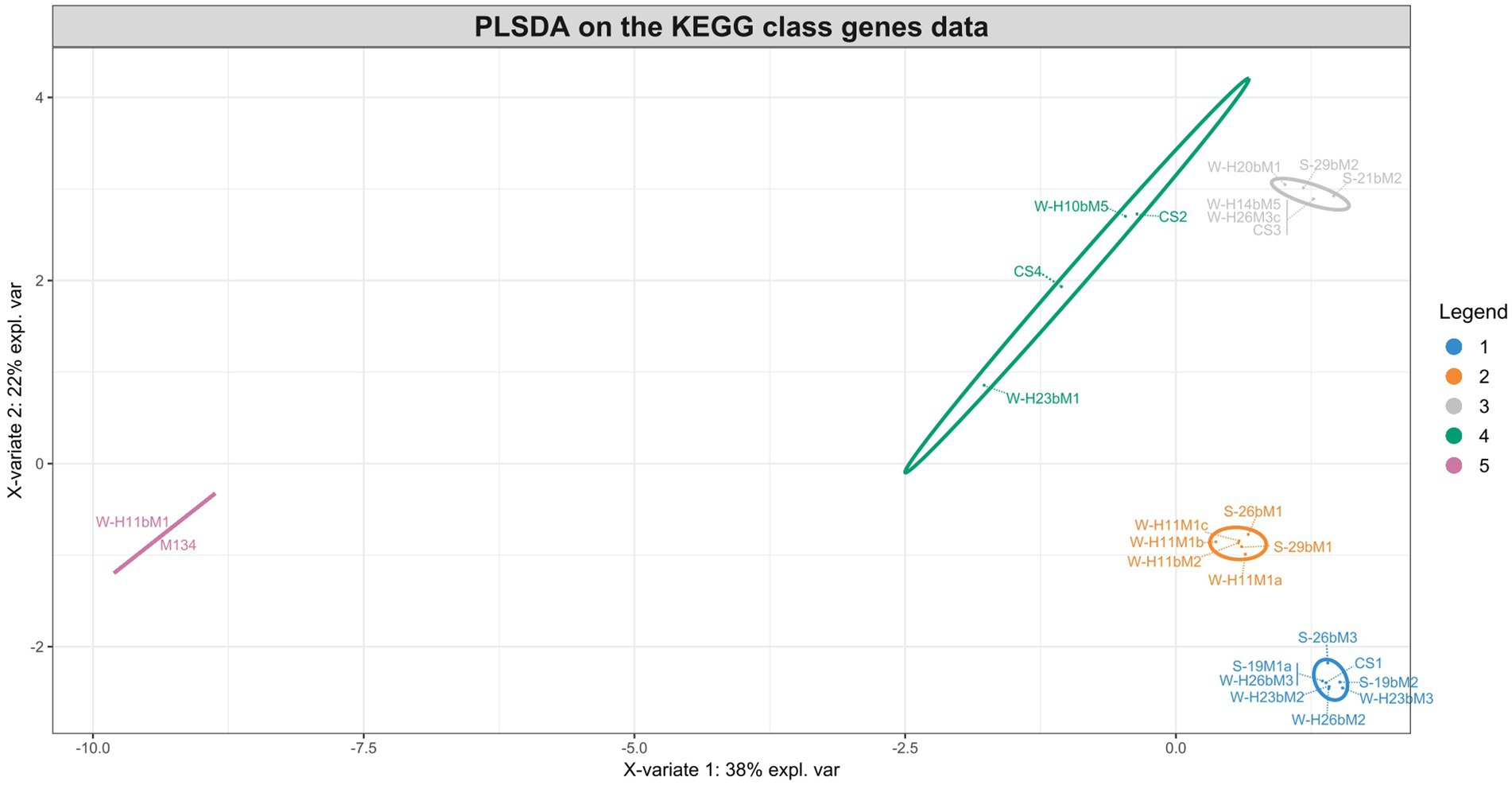
Figure 4. Sample plots from the PLS-DA analysis performed on accessory gene numbers of the KEGG class for groups 1 (blue), 2 (orange), 3 (gray), 4 (green), and 5 (pink).
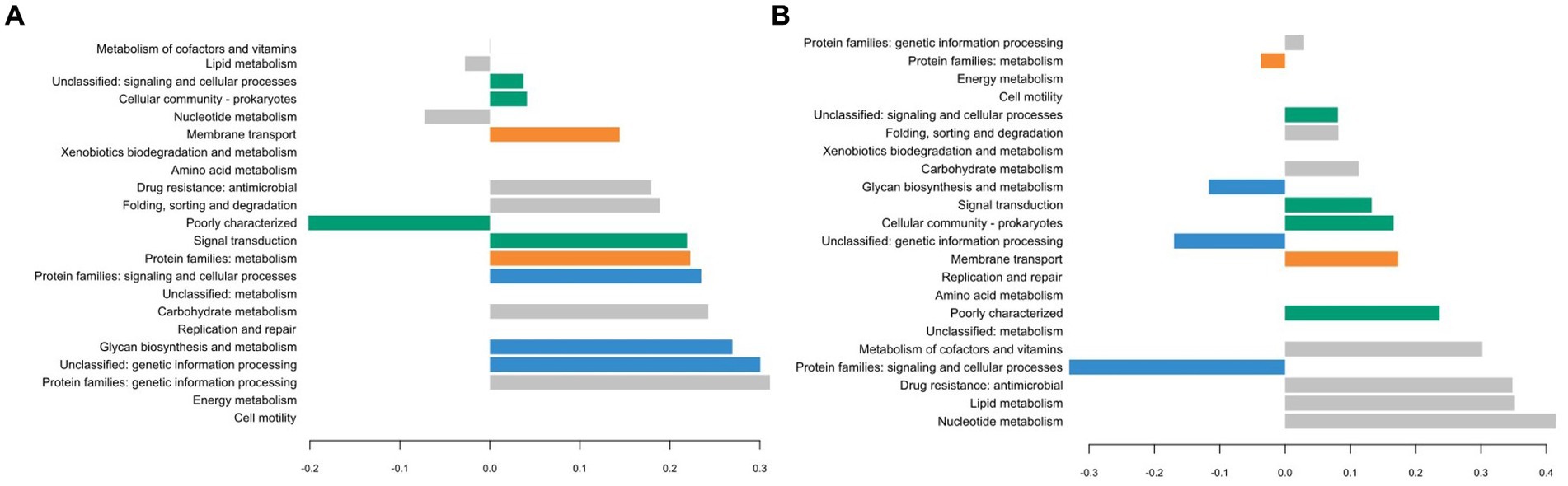
Figure 5. Plot loadings representing the variable’s contribution on component 1 (A) and on component 2 (B) of the PLS-DA analysis. Group 1 is colored in blue, 2 in orange, 3 in gray, 4 in green, and 5 in pink.
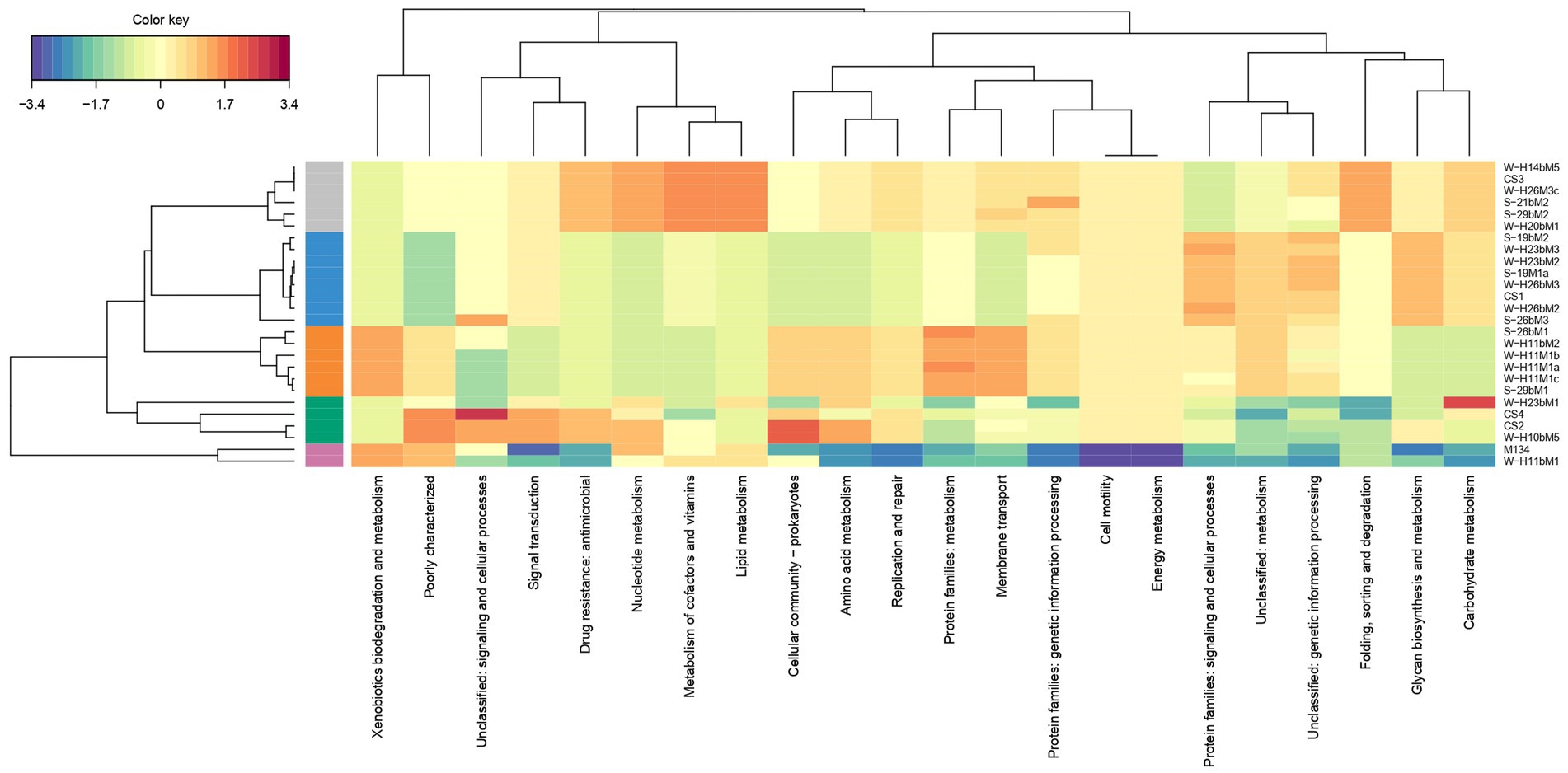
Figure 6. Clustered image map (CIM) associated with PLS-DA analysis. Groups are distinguished by colors on the left side of the map. Group 1 is colored in blue, 2 in orange, 3 in gray, 4 in green, and 5 in pink.
The first group was characterized by three KEGG pathways, namely, “Protein families: signaling and cellular process,” “Unclassified: genetic information processing,” and “Glycan biosynthesis and metabolism.” More accessory genes encoded for transposases and putative transposases were identified in the “Unclassified: genetic information processing” KEGG pathway for the first group in comparison with others (Supplementary Figure S1). Transposases are typically present within bacterial genomes as small, mobile elements solely comprising their own transposition genes; they are important factors for horizontal gene transfer between strain genomes and are therefore important for the safety evaluation of strains for their use as starter cultures. However, many of the accessory genes of the first group, grouping in the “Protein families: signaling and cellular processes,” were involved in the clustered regularly interspaced short palindromic repeat (CRISPR) and CRISPR-associated (Cas) proteins immune defense system in comparison to other groups (Supplementary Figure S2). It is now well known that the CRISPR/Cas system confers resistance against foreign genetic elements, such as phages (Bolotin et al., 2004; Mojica et al., 2005; Lillestøl et al., 2006). As phage infection of starters is the most common cause of incomplete fermentation in the dairy industry, such genes could represent a great advantage for the use of these isolates as starter cultures.
Then, the KEGG pathways “Protein families: metabolism” and “Membrane transport” allowed the differentiation of the second group, which might confer benefits to isolates during the manufacture of cheeses since accessory genes associated with these functional KEGG pathways encoded for dipeptidases (pepDA, pepDB; Supplementary Figure S3) and peptide transport systems (oligopeptide transport system Opp; Supplementary Figure S4), among other things, which are involved in proteolysis and thus are important for aromatic and textural properties of cheeses (Savijoki et al., 2006; Rodríguez-Serrano et al., 2018).
The third group stands out for the KEGG pathways “Protein families: genetic information processing,” “Nucleotide metabolism,” “Metabolism of cofactors and vitamins,” “Drug resistance: antimicrobial,” “Folding, sorting, and degradation,” and important functional categories for cheese manufacturing, including “carbohydrate metabolism” and “Lipid metabolism.” Carbohydrate metabolism is the most important metabolism pathway for starter cultures because of their primary role in producing lactic acid from lactose (Hutkins and Goh, 2014). Lipid metabolism is also of high interest because lipolysis plays an essential role in the development of flavors in cheeses (McSweeney and Sousa, 2000).
The KEGG pathways “Cellular community: prokaryotes,” “Signal transduction,” and “Unclassified: signaling and cellular process” characterized the fourth group and might facilitate the survival of cells under stressful environmental conditions. These functional traits may be of great advantage as the manufacture of cheeses exposes starters to various environmental stresses such as low pH, osmotic stress, and high pressure (Zotta et al., 2008).
The PLS-DA analysis did not identify the characteristic KEGG functional class of the fifth group, probably due to their lower content of accessory genes in comparison with other isolates, with only 210 accessory genes for isolate M134 and 207 for isolate H11bM1. However, these isolates were distantly separated from commercial strains based on the phylogenetic reconstruction based on core genes (Figure 1) and on the clustering associated with the pan-genome (Figure 2), which could be an added value for potential use as autochthonous starter cultures to strengthen the typical characteristics of cheeses.
Interestingly, commercial strains CS1, CS2, and CS3 were not significantly distantly separated from the centroid of their respective clusters, unlike the CS4 strain (Figure 2), indicating that commercial strains are representative of Streptococcus thermophilus genetic diversity. According to the PLS-DA analysis (Figures 4–6), the main differences between CS4 and SN-isolates H10bM5 and CS2 were the number of genes implicated in several KEGG categories, including “Protein families: metabolism” and “Carbohydrate metabolism.” In these categories, isolate CS4 had a higher number of accessory genes encoding dipeptidases (Supplementary Figure S3) and a higher content of accessory genes implicated in carbohydrate transport and metabolism (Supplementary Figure S5) than CS2 and H10bM5 isolates. However, these results focused only on accessory genes. To avoid this bias and study the carbohydrate metabolism of each isolate, a search for specific genes in their whole genomes was performed.
3.2.4. Carbohydrate metabolism and sugar transport system
The sugar utilization ability of S. thermophilus strains is of major importance in dairy fermentation as it directly affects the rate of milk acidification. Various studies suggest that the utilization of glucose, lactose, and fructose by S. thermophilus is consistently observed, whereas the utilization of sucrose, galactose, mannose, and maltose exhibits variable profiles (O’Leary and Woychik, 1976; Thomas et al., 2011; Erkus et al., 2014).
The lactose, glucose, galactose, sucrose, and fructose transport and utilization systems were analyzed in the genomes of the 22 SN-isolates and the 4 commercial strains (Figure 7), and the genes of each isolate implicated in these systems are listed in Supplementary Table S3.
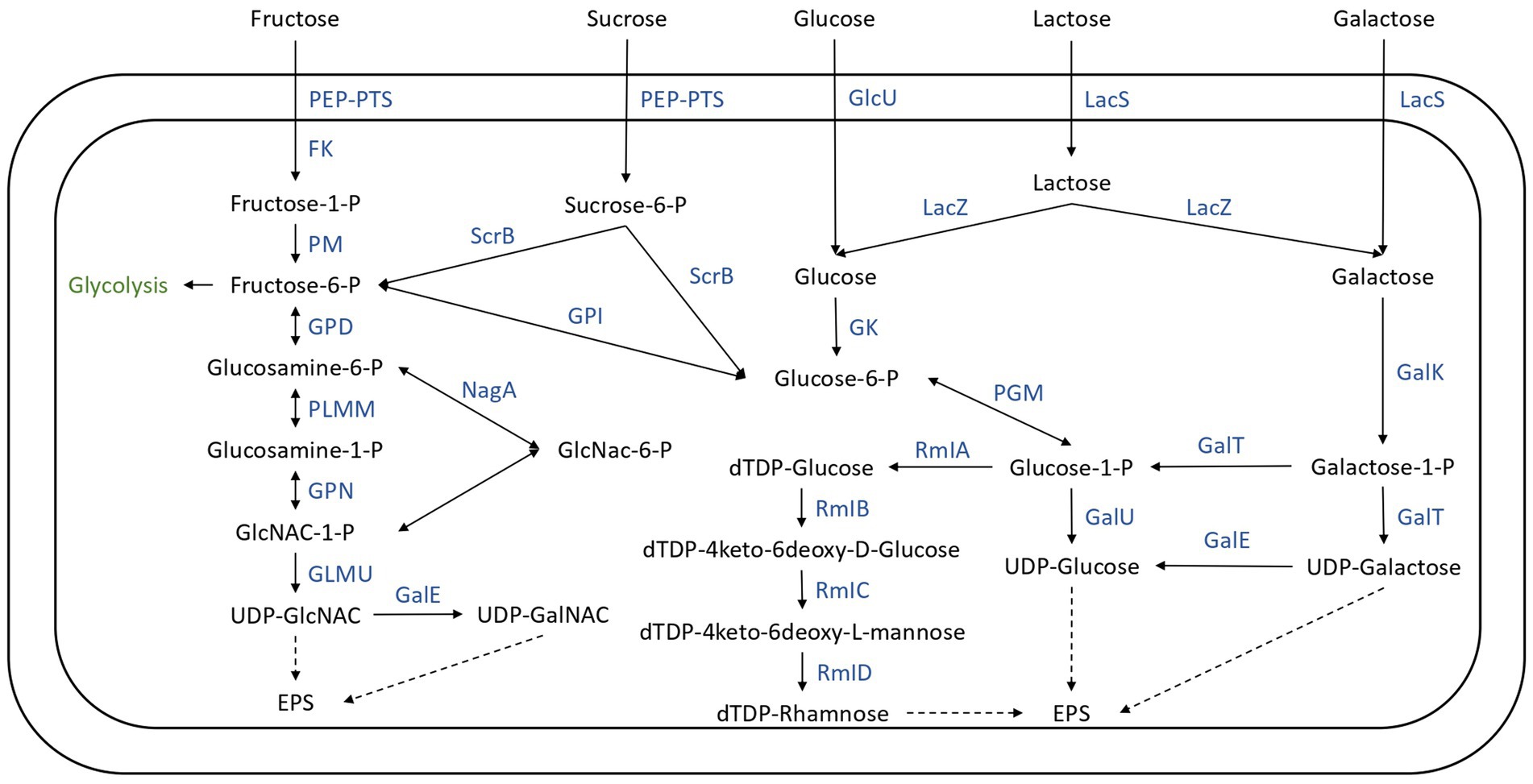
Figure 7. Carbohydrate transport and metabolism in the 26 Streptococcus thermophilus isolates. FK, fructokinase; GalE, UDP-galactose-4-epimerase; GalK, galactokinase; GalT, UTP-galactose-1-phosphate uridyltransferase; GalU, UDP-glucose pyrophosphorylase; GK, glucokinase; GlcU, glucose permease; GLMU, N-acetylglucosamine-1-phosphate uridyltransferase; GPD, glucosamine-6-phosphate deaminase; GPI, glucose-6-phosphate isomerase; GPN, glucosamine-1-phosphate N-acetyltransferase; LacS, lactose permease; LacZ, beta-glucosidase; Nag, N-acetyl-alpha-D-galactosaminidase; PEP-PTS, phosphoenolpyruvate-sugar phosphotransferase system; PGM, α-phosphoglucomutase; PLMM, phosphoglucosamine mutase; PM, phosphomutase; RMIa, glucose-1-phosphate thimidyltransferase; RmIb, dTDP-glucose 4, 6 dehydratase; RmIC, dTDP-4-deshydrorhamnose 3,5-epimerase; RmID, DTDP-4-keto-L-rhamnose reductase; ScrB, sucrose-6-phosphate hydrolase.
Lactose represents the principal carbohydrate in milk and is the preferred carbon and energy source of Streptococcus thermophilus (Xiong et al., 2019), probably due to the adaptation of the species to the milk (Goh et al., 2011; Alexandraki et al., 2019). In this species, lactose transport and hydrolysis are controlled by the lac operon. It encodes a lactose permease (LacS) and a cytoplasmic β-galactosidase (LacZ) which cleave the lactose into glucose and galactose (Xiong et al., 2019). The 26 genomes all harbored the LacZ and LacS genes (Figure 7; Supplementary Table S3). The resulting glucose moiety is phosphorylated to glucose-6-P by glucokinase and is further utilized through the glycolysis pathway (Figure 7). The Leloir pathway is the most common route for galactose utilization in S. thermophilus. Galactose is converted to glucose-1-phosphate by the galRKTEM gene cluster, which consists of the regulator GalR, galactokinase (GalK), galactose-1-phosphate uridyltransferase (GalT), UDP-glucose-4-epimerase (GalE), and galactosemutarotase (GalM; Xiong et al., 2019; Figure 7). However, most strains of S. thermophilus are unable to ferment galactose despite having intact galRKTEM gene clusters for the Leloir pathway (Vaughan et al., 2001; de Vin et al., 2005). The most probable explanation for this observed phenotype is a deficient promoter leading to insufficient transcription (Vaughan et al., 2001).
Gal-positive strains are of technological importance principally for their ability to completely ferment lactose, which results in a reduced amount of galactose being present, which cannot serve as a carbon source for spoilage or pathogenic bacteria (Hutkins and Morris, 1987).
The DNA fragments of the galR-galK intergenic region of the 26 isolates were picked out from their genome sequences (Supplementary Figure S6). Four different types of fragments were detected in the 26 genomes analyzed. Isolates 19M1a, H23bM2, 19bM2, 26bM3, H23bM3, H26bM2, H26bM3, and CS1 are type A; 21bM2, 29bM2, H20bM1, H26M3c, CS3, H14bM5, H10bM5, CS2, CS4, and H23bM1 are type B; H11M1a, H11M1b, H11M1c, 26bM1, 29bM1, and H11bM2 are type C; and isolates M134 and H11bM1 belong to type D.
It is now well documented that a single point of mutation with a G-to-A substitution in the −9 position of the −10 box in the galK promoter results in improved galK-promoter activity and could be responsible for the ability to ferment galactose (Vaughan et al., 2001; van den Bogaard et al., 2004; de Vin et al., 2005; Giaretta et al., 2018; Xiong et al., 2019). The presence of this mutation was examined in the 26 genomes studied here. Consequently, among the isolates, only M134 and H11bM1 appeared capable of galactose utilization as they owned the relevant mutation related to the Gal-positive phenotype. However, de Vin et al. (2005) and Hu et al. (2020) demonstrated that the gal promoter does not exclusively determine the gal-positive phenotype of Streptococcus thermophilus strains. Experimental verification is therefore required to validate this prediction.
The utilization of sucrose and fructose by S. thermophilus required a phosphoenolyruvate-phosphotransferase system (PEP-PTS), consisting of a PEP-dependent phosphotransferase (enzyme I, EI), a histidine-containing phosphocarrier protein (HPr), and a sugar-specific permease (enzyme II, EII). The 26 genomes investigated all harbored a PEP-dependent phosphotransferase and the HPr, as well as genes responsible for sucrose and fructose PTS transporter sugar-specific permease enzymes (Figure 7; Supplementary Table S3). Furthermore, genes responsible for sucrose and fructose utilization detailed in Figure 7 were detected in these genomes (Supplementary Table S3). However, experimental procedures are required to identify the ability of these isolates to ferment these carbohydrates.
3.3. Technological diversity
The acidifying abilities of 22 Streptococcus thermophilus isolated from the Saint-Nectaire cheese-producing PDO area (SN-isolates) and 4 strains of Streptococcus thermophilus isolated from commercial starter cultures were evaluated in model cheese curd. From the pH values obtained, ΔpH6h and ΔpH24h were calculated (Figure 8).
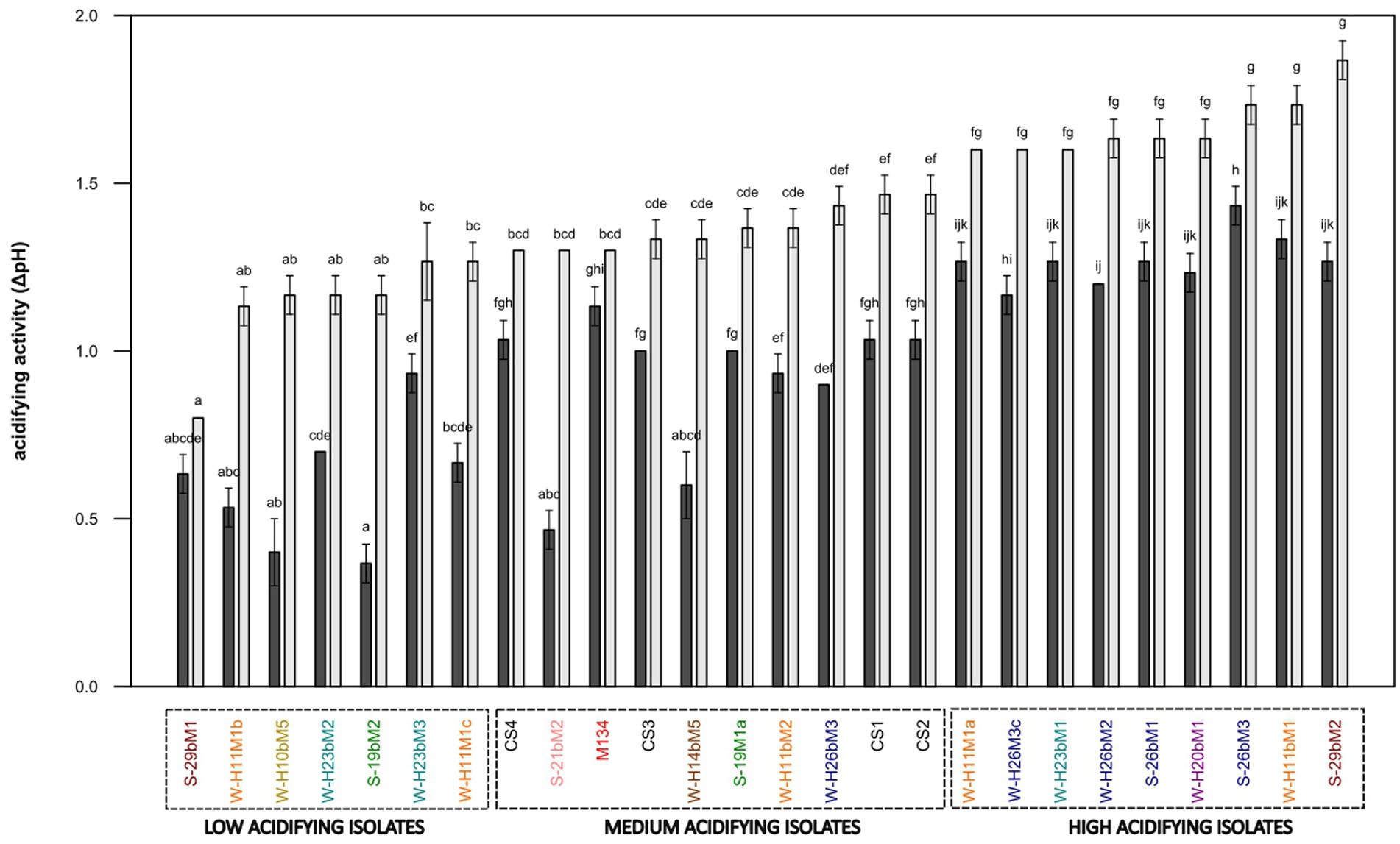
Figure 8. Acidifying activity of Streptococcus thermophilus isolates (a–k): mean values of the same time of fermentation without a common superscript are significantly different (p < 0.05) according to the Conover-Iman test. Isolates from summer milks are annotated with an “S” and isolates from winter milks are annotated with a “W.” Isolate names with the same colors were isolated from the same farm.
Half of the 22 SN-isolates lowered the pH by more than one pH unit after 6 h of fermentation, suggesting fast acidifying ability. As expected, all commercial strains presented fast acidifying abilities, which is one of the most important criteria for cheese manufacturing (Tidona et al., 2020).
Concerning the ability to reduce the pH of the model cheese curds at 24 h, the following three main groups of isolates were distinguished: (i) low acidifying isolates with a pH decrease below 1.30 pH units, including 32% of the SN-isolates (19bM2, H10bM5, H11M1b, H11M1c, H23bM2, H23bM3, and 29bM1); (ii) medium acidifying isolates, showing a pH decrease between 1.30 and 1.50 pH units, comprising 27% of total SN-isolates and 100% of the commercial strains (CS1, 19M1a, 21bM2, H11bM2, H14bM5, H26bM3, M134, CS3, CS4, and CS2); (iii) high acidifying isolates, causing a pH decrease higher than 1.50 pH units, regrouping 41% of the SN-isolates (H11M1a, H26bM2, H26M3c, 26bM1, 26bM3, 29bM2, H11bM1, H20bM1, and H23bM1). It could be noticed that SN-isolates from the same farms and SN-isolates from summer and winter milk are distributed into the three acidifying activity categories, and that the four commercial strains were clustered in the group of medium acidifying activity, while the autochthonous SN-isolates showed various acidifying activities.
This clustering reveals a high variability in acidifying activity observed within Streptococcus thermophilus SN-isolates, suggesting that this feature is strain-dependent, in accordance with Giraffa et al. (2001). This diversity of acidifying activities of the SN-isolates does not depend on the season or the farms. In contrast, commercial strains showed homogeneous acidifying activity. This homogeneity is probably due to the specific selection of the strains based on their functional characteristics to be used as starter cultures (Gibbons and Rinker, 2015).
The proteolytic activities of the 22 SN-isolates and the 4 commercial strains of Streptococcus thermophilus were evaluated in a model cheese curd at 24 h of fermentation, and the results are presented in Figure 9. The proteolytic activities of Streptococcus thermophilus SN-isolates and commercial strains varied slightly, from 0.046 to 0.077 mmoleqGlycine.L−1 for 29bM1 SN-isolates and CS2 commercial strains, respectively.
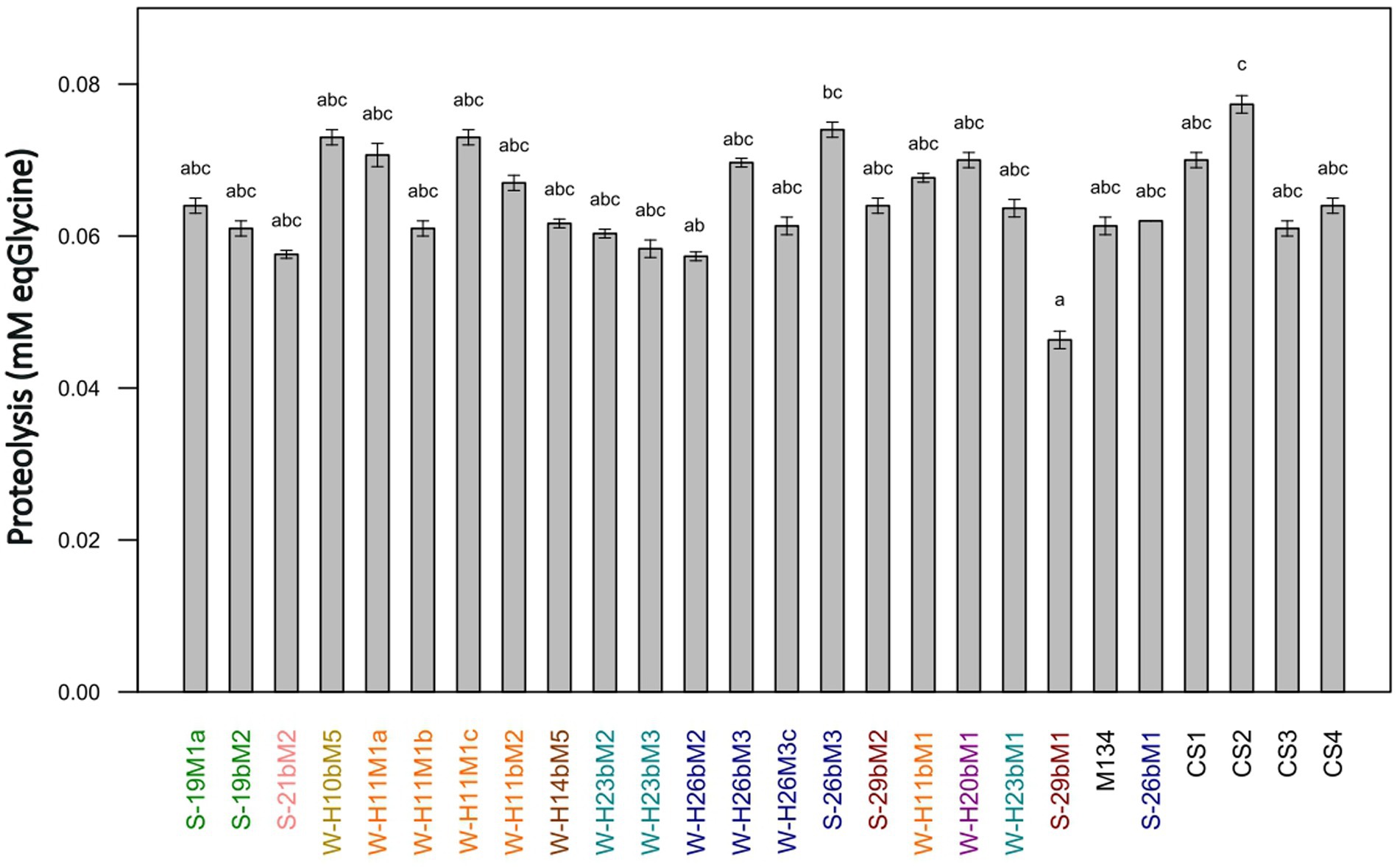
Figure 9. Proteolytic activities of Streptococcus thermophilus isolates (a–c): mean values without a common superscript are significantly different (p < 0.05) according to the Conover-Iman test. Isolates from summer milks are annotated with an “S” and isolates from winter milks are annotated with a “W.” Isolate names with the same colors were isolated from the same farm.
Galia et al. (2009) and Dandoy et al. (2011) have highlighted a link between high acidifying activities and the presence of an efficient proteolytic system, whereas our results showed low proteolytic activities and a limited diversity on this criteria in comparison with other studies on the characterization of Streptococcus thermophilus strains (Urshev et al., 2014; Hu et al., 2020; Tidona et al., 2020) or other species (Zaaraoui et al., 2021; Abarquero et al., 2022; Sugajski et al., 2022). These findings are in accordance with Harnett et al. (2022), who reported Streptococcus thermophilus as a LAB species generally considered poorly proteolytic. To ensure sufficient proteolytic activity in dairy products, Streptococcus thermophilus is often associated with other species for the development of starter cultures, such as Lactobacillus delbrueckii subsp. bulgaricus. The symbiotic growth of both species leads to protocooperation, inducing higher proteolytic and acidifying rates (Hervé-Jiménez, 2008).
In addition, a PLS-DA analysis was performed on the KEGG annotation of accessory genes with acidification groups (Supplementary Figure S7) and indicated that the KEGG annotations of accessory genes did not allow for distinguishing the acidification groups. The lack of diversity in proteolytic activities also indicates that no link could be found between these functional properties and groups obtained by hierarchical clustering based on the KEGG annotation of the accessory genes.
The literature does not often discuss the gene expression of LAB in relation to its acidifying capabilities. The variations in acidifying potential among different strains, besides the presence/absence of genes, could be due to differential expression of multiple genes related to metabolism. For instance, in a study conducted by Galia et al. (2016), two strains harboring the same allele of the prtS gene, encoding a cell-wall-anchored proteinase linked to fast acidifying capabilities, presented different acidification rates. The authors demonstrated that this difference could be due to the differential expression of several metabolic-related genes (prtS, codY, ilvE, livJ, and relA) during growth in milk, with an over-expression of these genes in the most acidifying strain compared to the other. They also reported that the transcriptional regulator CodY could also play an important role in the acidifying capacity of Streptococcus thermophilus through the regulation of nitrogen and carbon metabolism. These conclusions support the results of Lu et al. (2015), who reported that CodY plays an important role in the regulation of cellular processes in Streptococcus thermophilus and showed that the global regulator CodY controlled amino acid metabolism and lactose utilization processes. To better understand the phenotype differences of our genetically closed isolates, a thorough study of the expression and regulation of genes involved in carbon and nitrogen metabolism should be conducted.
The lack of a link between the technological capacities and the presence/absence of the genes of the 26 isolates could also simply be due to the low number of studied isolates. Indeed, when it comes to analyzing correlations between phenotypic and genotypic traits of bacteria, the number of samples (or strains) sequenced plays a crucial role in the power to detect correlations. By increasing the number of samples, it would be possible to significantly improve the ability to identify statistically significant correlations.
4. Conclusion
Wild strains of Streptococcus thermophilus could represent a source of genetic and functional variability from which novel strains or properties might be selected for cheese production. This diversity exists within Streptococcus thermophilus isolated at the same location and is not dependent on the season of isolation. A specific technological characterization and safety evaluation of isolates should be carried out to validate the use of this diversity as an autochthonous starter culture.
Data availability statement
The data presented in this study are deposited in online repositories. Genomes can be found in the EBI repository under the project accession number PRJEB61322. All input and output files of the bioinformatic analysis are available at https://doi.org/10.57745/MSGWXC.
Author contributions
AG conceived the study, carried out experiments, analyzed the data, performed genome analysis, and wrote the manuscript. ST conceived the study, performed genome analysis, analyzed the data, and participated in the writing of the manuscript. CCa designed the experiments and conceived the project. SH and ED-B supervised the experiments and provided guidance. PG conceived the project and supervised the study. PB and CCh conceived the project, participated in the writing of the manuscript, and supervised the study. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by “Interprofession du Saint-Nectaire” and “Association Nationale Recherche Technologie” (convention CIFRE N° 2020/0839).
Acknowledgments
We thank Mathilde LACALMONTIE, Béatrice DESSERRE, and François ALLEGRETTI for their advice and technical assistance in microbiology.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1245510/full#supplementary-material
Footnotes
References
Abarquero, D., Renes, E., Combarros-Fuertes, P., Fresno, J. M., and Tornadijo, M. E. (2022). Evaluation of technological properties and selection of wild lactic acid bacteria for starter culture development. LWT 171:114121. doi: 10.1016/j.lwt.2022.114121
Ait Chait, Y., Gunenc, A., Hosseinian, F., and Bendali, F. (2021). Antipathogenic and probiotic potential of Lactobacillus brevis strains newly isolated from Algerian artisanal cheeses. Folia Microbiol. 66, 429–440. doi: 10.1007/s12223-021-00857-1
Alexandraki, V., Kazou, M., Blom, J., Pot, B., Papadimitriou, K., and Tsakalidou, E. (2019). Comparative genomics of Streptococcus thermophilus support important traits concerning the evolution, biology and technological properties of the species. Front. Microbiol. 10:2916. doi: 10.3389/fmicb.2019.02916
Andrighetto, C., Borney, F., Barmaz, A., Stefanon, B., and Lombardi, A. (2002). Genetic diversity of Streptococcus thermophilus strains isolated from Italian traditional cheeses. Int. Dairy J. 12, 141–144. doi: 10.1016/S0958-6946(01)00134-0
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bolotin, A., Quinquis, B., Renault, P., Sorokin, A., Ehrlich, S. D., Kulakauskas, S., et al. (2004). Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22, 1554–1558. doi: 10.1038/nbt1034
Callon, C., Arliguie, C., and Montel, M.-C. (2016). Control of Shigatoxin-producing Escherichia coli in cheese by dairy bacterial strains. Food Microbiol. 53, 63–70. doi: 10.1016/j.fm.2015.08.009
Callon, C., Millet, L., and Montel, M.-C. (2004). Diversity of lactic acid bacteria isolated from AOC Salers cheese. J. Dairy Res. 71, 231–244. doi: 10.1017/S0022029904000159
Campagnollo, F. B., Margalho, L. P., Kamimura, B. A., Feliciano, M. D., Freire, L., Lopes, L. S., et al. (2018). Selection of indigenous lactic acid bacteria presenting anti-listerial activity, and their role in reducing the maturation period and assuring the safety of traditional Brazilian cheeses. Food Microbiol. 73, 288–297. doi: 10.1016/j.fm.2018.02.006
Cantu, V. A., Sadural, J., and Edwards, R. (2019). PRINSEQ++, a multi-threaded tool for fast and efficient quality control and preprocessing of sequencing datasets. PeerJ 7:e27553v1. doi: 10.7287/peerj.preprints.27553v1
Carafa, I., Nardin, T., Larcher, R., Viola, R., Tuohy, K., and Franciosi, E. (2015). Identification and characterization of wild lactobacilli and pediococci from spontaneously fermented mountain cheese. Food Microbiol. 48, 123–132. doi: 10.1016/j.fm.2014.12.003
Church, F. C., Swaisgood, H. E., Porter, D. H., and Catignani, G. L. (1983). Spectrophotometric assay using o-Phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins. J. Dairy Sci. 66, 1219–1227. doi: 10.3168/jds.S0022-0302(83)81926-2
Dandoy, D., Fremaux, C., Henry de Frahan, M., Horvath, P., Boyaval, P., Hols, P., et al. (2011). The fast milk acidifying phenotype of Streptococcus thermophilus can be acquired by natural transformation of the genomic island encoding the cell-envelope proteinase PrtS. Microb. Cell Fact. 10:S21. doi: 10.1186/1475-2859-10-S1-S21
Daughtry, K. V., Johanningsmeier, S. D., Sanozky-Dawes, R., Klaenhammer, T. R., and Barrangou, R. (2018). Phenotypic and genotypic diversity of Lactobacillus buchneri strains isolated from spoiled, fermented cucumber. Int. J. Food Microbiol. 280, 46–56. doi: 10.1016/j.ijfoodmicro.2018.04.044
de Vin, F., Rådström, P., Herman, L., and De Vuyst, L. (2005). Molecular and biochemical analysis of the galactose phenotype of dairy Streptococcus thermophilus strains reveals four different fermentation profiles. Appl. Environ. Microbiol. 71, 3659–3667. doi: 10.1128/AEM.71.7.3659-3667.2005
Delorme, C. (2008). Safety assessment of dairy microorganisms: Streptococcus thermophilus. Int. J. Food Microbiol. 126, 274–277. doi: 10.1016/j.ijfoodmicro.2007.08.014
Erkus, O., Okuklu, B., Yenidunya, A. F., and Harsa, S. (2014). High genetic and phenotypic variability of Streptococcus thermophilus strains isolated from artisanal Yuruk yoghurts. LWT 58, 348–354. doi: 10.1016/j.lwt.2013.03.007
Feurer, C., Irlinger, F., Spinnler, H. E., Glaser, P., and Vallaeys, T. (2004). Assessment of the rind microbial diversity in a farmhouse-produced vs a pasteurized industrially produced soft red-smear cheese using both cultivation and rDNA-based methods. J. Appl. Microbiol. 97, 546–556. doi: 10.1111/j.1365-2672.2004.02333
Fox, P. F. (1989). Proteolysis during cheese manufacture and Ripening1. J. Dairy Sci. 72, 1379–1400. doi: 10.3168/jds.S0022-0302(89)79246-8
Frau, F., Nuñez, M., Gerez, L., Pece, N., and Font, G. (2016). Development of an autochthonous starter culture for spreadable goat cheese. Food Sci. Technol 36, 622–630. doi: 10.1590/1678-457X.08616
Fusco, V., Quero, G. M., Poltronieri, P., Morea, M., and Baruzzi, F. (2019). Autochthonous and probiotic lactic acid Bacteria employed for production of "advanced traditional cheeses". Foods 8:412. doi: 10.3390/foods8090412
Galia, W., Jameh, N., Perrin, C., Genay, M., and Dary-Mourot, A. (2016). Acquisition of PrtS in Streptococcus thermophilus is not enough in certain strains to achieve rapid milk acidification. Dairy Sci. Technol. 96, 623–636. doi: 10.1007/s13594-016-0292-3
Galia, W., Perrin, C., Genay, M., and Dary, A. (2009). Variability and molecular typing of Streptococcus thermophilus strains displaying different proteolytic and acidifying properties. Int. Dairy J. 19, 89–95. doi: 10.1016/j.idairyj.2008.08.004
Giaretta, S., Treu, L., Vendramin, V., da Silva Duarte, V., Tarrah, A., Campanaro, S., et al. (2018). Comparative transcriptomic analysis of Streptococcus thermophilus TH1436 and TH1477 showing different capability in the use of galactose. Front. Microbiol. 9:1765. doi: 10.3389/fmicb.2018.01765
Gibbons, J. G., and Rinker, D. C. (2015). The genomics of microbial domestication in the fermented food environment. Curr. Opin. Genet. Dev. 35, 1–8. doi: 10.1016/j.gde.2015.07.003
Giraffa, G., Paris, A., Valcavi, L., Gatti, M., and Neviani, E. (2001). Genotypic and phenotypic heterogeneity of Streptococcus thermophilus strains isolated from dairy products. J. Appl. Microbiol. 91, 937–943. doi: 10.1046/j.1365-2672.2001.01464.x
Goh, Y. J., Goin, C., O’Flaherty, S., Altermann, E., and Hutkins, R. (2011). Specialized adaptation of a lactic acid bacterium to the milk environment: the comparative genomics of Streptococcus thermophilus LMD-9. Microb. Cell Fact. 10:S22. doi: 10.1186/1475-2859-10-S1-S22
Gómez, N. C., Ramiro, J. M. P., Quecan, B. X. V., and de Melo Franco, B. D. G. (2016). Use of potential probiotic lactic acid Bacteria (LAB) biofilms for the control of Listeria monocytogenes, salmonella typhimurium, and Escherichia coli O157:H7 biofilms formation. Front. Microbiol. 7:863. doi: 10.3389/fmicb.2016.00863
Hadfield, J., Croucher, N. J., Goater, R. J., Abudahab, K., Aanensen, D. M., and Harris, S. R. (2018). Phandango: an interactive viewer for bacterial population genomics. Bioinformatics 34, 292–293. doi: 10.1093/bioinformatics/btx610
Hansen, E. B. (2002). Commercial bacterial starter cultures for fermented foods of the future. Int. J. Food Microbiol. 78, 119–131. doi: 10.1016/S0168-1605(02)00238-6
Harnett, J., Patrick, A., Caddick, C., Pearce, L., and Davey, G. (2022). “Lactic acid Bacteria: Streptococcus thermophilus☆” in Encyclopedia of dairy sciences. eds. P. L. H. McSweeney and J. P. McNamara. 3rd ed (Oxford: Academic Press), 256–262.
Hervé-Jiménez, L. (2008). Post-genomic analysis of Streptococcus thermophilus and Lactobacillus delbruekii ssp. bulgaricus cooperative growth in milk. Appl. Environ. Microbiol. 75:2062. doi: 10.1128/AEM.01984-08
Hols, P., Hancy, F., Fontaine, L., Grossiord, B., Prozzi, D., Leblond-Bourget, N., et al. (2005). New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol. Rev. 29, 435–463. doi: 10.1016/j.fmrre.2005.04.008
Hu, T., Cui, Y., Zhang, Y., Qu, X., and Zhao, C. (2020). Genome analysis and physiological characterization of four Streptococcus thermophilus strains isolated from Chinese traditional fermented milk. Front. Microbiol. 11:184. doi: 10.3389/fmicb.2020.00184
Huang, L., Goda, H. A., Abdel-Hamid, M., Renye, J. A. Jr., Yang, P., Huang, Z., et al. (2021). Partial characterization of probiotic lactic acid bacteria isolated from Chinese dairy products. Int. J. Food Prop. 24, 446–456. doi: 10.1080/10942912.2021.1900233
Hutkins, R., and Goh, Y. J. (2014). Streptococcus: Streptococcus thermophilus In Encyclopedia of Food Microbiology: Second Edition (Elsevier Inc.), 554–559.
Hutkins, R. W., and Morris, H. A. (1987). Carbohydrate metabolism by Streptococcus thermophilus: a review. J. Food Prot. 50, 876–884. doi: 10.4315/0362-028X-50.10.876
Iyer, R., Tomar, S. K., Uma Maheswari, T., and Singh, R. (2010). Streptococcus thermophilus strains: multifunctional lactic acid bacteria. Int. Dairy J. 20, 133–141. doi: 10.1016/j.idairyj.2009.10.005
Kanehisa, M. (2019). Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. doi: 10.1002/pro.3715
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M., and Ishiguro-Watanabe, M. (2023). KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592. doi: 10.1093/nar/gkac963
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Lillestøl, R., Redder, P., Garrett, R. A., and Brügger, K. (2006). A putative viral defense mechanism in archaeal cells. Archaea 2, 59–72. doi: 10.1155/2006/542818
Lu, W. W., Wang, Y., Wang, T., and Kong, J. (2015). The global regulator CodY in Streptococcus thermophilus controls the metabolic network for escalating growth in the milk environment. Appl. Environ. Microbiol. 81, 2349–2358. doi: 10.1128/AEM.03361-14
Makarova, K., Slesarev, A., Wolf, Y., Sorokin, A., Mirkin, B., Koonin, E., et al. (2006). Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U. S. A. 103, 15611–15616. doi: 10.1073/pnas.0607117103
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17, 10–12. doi: 10.14806/ej.17.1.200
McSweeney, P. L. H., and Sousa, M. J. (2000). Biochemical pathways for the production of flavour compounds in cheeses during ripening: a review. Lait 80, 293–324. doi: 10.1051/lait:2000127
Mojica, F. J. M., Díez-Villaseñor, C., García-Martínez, J., and Soria, E. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60, 174–182. doi: 10.1007/s00239-004-0046-3
Morandi, S., and Brasca, M. (2012). Safety aspects, genetic diversity and technological characterisation of wild-type Streptococcus thermophilus strains isolated from north Italian traditional cheeses. Food Control 23, 203–209. doi: 10.1016/j.foodcont.2011.07.011
Munekata, P. E. S., Chaves-Lopez, C., Fernandez-Lopez, J., Viuda-Martos, M., Sayas-Barbera, M. E., Perez-Alvarez, J. A., et al. (2022). Autochthonous starter cultures in cheese production – a review. Food Rev. Intl. 39, 5886–5904. doi: 10.1080/87559129.2022.2097691
Myagkonosov, D. S., Abramov, D. V., Ovchinnikova, E. G., and Krayushkina, V. N. (2021). Express method for assessing proteolysis in cheese and aromatic additives with cheese flavor. Food Syst 3, 4–10. doi: 10.21323/2618-9771-2020-3-4-4-10
O’Leary, V. S., and Woychik, J. H. (1976). Utilization of lactose, glucose, and galactose by a mixed culture of Streptococcus thermophilus and Lactobacillus bulgaricus in Milk treated with lactase enzyme. Appl. Environ. Microbiol. 32, 89–94. doi: 10.1128/aem.32.1.89-94.1976
Özkan, E. R., Demirci, T., Öztürk, H. İ., and Akın, N. (2021). Screening Lactobacillus strains from artisanal Turkish goatskin casing Tulum cheeses produced by nomads via molecular and in vitro probiotic characteristics. J. Sci. Food Agric. 101, 2799–2808. doi: 10.1002/jsfa.10909
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Rakhmanova, A., Khan, Z. A., and Shah, K. (2018). A mini review fermentation and preservation: role of lactic acid Bacteria. MOJFPT 6, 414–417. doi: 10.15406/mojfpt.2018.06.00197
Rodríguez-Serrano, G. M., Garcia-Garibay, J. M., Cruz-Guerrero, A. E., Gomez-Ruiz, L., Ayala-Nino, A., Castaneda-Ovando, A., et al. (2018). Proteolytic System of Streptococcus thermophilus. J. Microbiol. Biotechnol. 28, 1581–1588. doi: 10.4014/jmb.1807.07017
Savijoki, K., Ingmer, H., and Varmanen, P. (2006). Proteolytic systems of lactic acid bacteria. Appl. Microbiol. Biotechnol. 71, 394–406. doi: 10.1007/s00253-006-0427-1
Schwengers, O., Jelonek, L., Dieckmann, M. A., Beyvers, S., Blom, J., and Goesmann, A. (2021). Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microbial Genomics 7:000685. doi: 10.1099/mgen.0.000685
Settanni, L., and Moschetti, G. (2014). New trends in technology and identity of traditional dairy and fermented meat production processes: preservation of typicality and hygiene. Trends Food Sci. Technol. 37, 51–58. doi: 10.1016/j.tifs.2014.02.006
Smit, G., Smit, B. A., and Engels, W. J. M. (2005). Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 29, 591–610. doi: 10.1016/j.femsre.2005.04.002
Suchard, M. A., Lemey, P., Baele, G., Ayres, D. L., Drummond, A. J., and Rambaut, A. (2018). Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4:vey016. doi: 10.1093/ve/vey016
Sugajski, M., Maślak, E., Złoch, M., Rafińska, K., Pomastowski, P., Białczak, D., et al. (2022). New sources of lactic acid bacteria with potential antibacterial properties. Arch. Microbiol. 204:349. doi: 10.1007/s00203-022-02956-0
Terzić-Vidojević, A., Tonković, K., Leboš Pavunc, A., Beganović, J., Strahinić, I., Kojić, M., et al. (2015). Evaluation of autochthonous lactic acid bacteria as starter cultures for production of white pickled and fresh soft cheeses. LWT 63, 298–306. doi: 10.1016/j.lwt.2015.03.050
The Galaxy Community (2022). The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 50, W345–W351. doi: 10.1093/nar/gkac247
Thomas, M., Wrzosek, L., Ben-Yahia, L., Noordine, M.-L., Gitton, C., Chevret, D., et al. (2011). Carbohydrate metabolism is essential for the colonization of Streptococcus thermophilus in the digestive tract of Gnotobiotic rats. PloS One 6:e28789. doi: 10.1371/journal.pone.0028789
Tidona, F., Francolino, S., Ghiglietti, R., Locci, F., Carminati, D., Laforce, P., et al. (2020). Characterization and pre-industrial validation of Streptococcus thermophilus strains to be used as starter cultures for Crescenza, an Italian soft cheese. Food Microbiol. 92:103599. doi: 10.1016/j.fm.2020.103599
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524. doi: 10.1186/s13059-014-0524-x
Urshev, Z., Ninova-Nikolova, N., Ishlimova, D., Pashova-Baltova, K., Michaylova, M., and Savova, T. (2014). Selection and characterization of naturally occurring high acidification rate Streptococcus thermophilus strains. Biotechnol. Biotechnol. Equip. 28, 899–903. doi: 10.1080/13102818.2014.966233
van den Bogaard, P. T. C., Hols, P., Kuipers, O. P., Kleerebezem, M., and de Vos, W. M. (2004). Sugar utilisation and conservation of the gal-lac gene cluster in Streptococcus thermophilus. Syst. Appl. Microbiol. 27, 10–17. doi: 10.1078/0723-2020-00258
Vaughan, E. E., van den Bogaard, P. T. C., Catzeddu, P., Kuipers, O. P., and de Vos, W. M. (2001). Activation of silent gal genes in thelac-gal regulon of Streptococcus thermophilus. J. Bacteriol. 183, 1184–1194. doi: 10.1128/jb.183.4.1184-1194.2001
Vendramin, V., Treu, L., Campanaro, S., Lombardi, A., Corich, V., and Giacomini, A. (2017). Genome comparison and physiological characterization of eight Streptococcus thermophilus strains isolated from Italian dairy products. Food Microbiol. 63, 47–57. doi: 10.1016/j.fm.2016.11.002
Xiong, Z.-Q., Kong, L.-H., Meng, H.-L., Cui, J.-M., Xia, Y.-J., Wang, S.-J., et al. (2019). Comparison of gal–lac operons in wild-type galactose-positive and -negative Streptococcus thermophilus by genomics and transcription analysis. J. Ind. Microbiol. Biotechnol. 46, 751–758. doi: 10.1007/s10295-019-02145-x
Zaaraoui, L., Bouksaim, A., Elhamdani, M., Benali, A., Oukassou, M., Ounine, K., et al. (2021). Typical Moroccan goat lactic acid Bacteria and their assay as starters. Nova Biotec Chim. 20:e958. doi: 10.36547/nbc.958
Keywords: Streptococcus thermophilus, diversity, genome comparison, functional properties, autochthonous starter, cheese
Citation: Grizon A, Theil S, Callon C, Gerber P, Helinck S, Dugat-Bony E, Bonnarme P and Chassard C (2023) Genetic and technological diversity of Streptococcus thermophilus isolated from the Saint-Nectaire PDO cheese-producing area. Front. Microbiol. 14:1245510. doi: 10.3389/fmicb.2023.1245510
Edited by:
Photis Papademas, Cyprus University of Technology, CyprusReviewed by:
Vinícius Da Silva Duarte, Norwegian University of Life Sciences, NorwayLi Weicheng, Inner Mongolia Agricultural University, China
Copyright © 2023 Grizon, Theil, Callon, Gerber, Helinck, Dugat-Bony, Bonnarme and Chassard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna Grizon, YW5uYS5ncml6b25AaW5yYWUuZnI=; Sebastien Theil, c2ViYXN0aWVuLnRoZWlsQGlucmFlLmZy
 Anna Grizon
Anna Grizon Sebastien Theil
Sebastien Theil Cecile Callon
Cecile Callon Pauline Gerber2
Pauline Gerber2 Eric Dugat-Bony
Eric Dugat-Bony Christophe Chassard
Christophe Chassard