- 1Department of Bioscience, College of Bioresource Sciences, Nihon University, Fujisawa, Kanagawa, Japan
- 2Department of Applied Biological Sciences, College of Bioresource Sciences, Nihon University, Fujisawa, Kanagawa, Japan
- 3Department of Chemistry for Life Sciences and Agriculture, Faculty of Life Sciences, Tokyo University of Agriculture, Tokyo, Japan
A novel aluminum-tolerant bacterial strain CA42 was isolated from the aquatic plant Eleocharis dulcis, which grows in a highly acidic swamp in Vietnam. Inoculation with CA42 allowed Oryza sativa to grow in the presence of 300 μM AlCl3 at pH 3.5, and biofilms were observed around the roots. Using 16S rRNA gene sequencing analysis, the strain was identified as Pullulanibacillus sp. CA42. This strain secreted large amounts of an extracellular polysaccharide (CA42 EPS). Results from structural analyses on CA42 EPS, namely methylation analysis and nuclear magnetic resonance (NMR), indicated that the chemical structure of CA42 EPS was a glycogen-like α-glucan. Purified CA42 EPS and the commercially available oyster glycogen adsorbed aluminum ions up to 15–30 μmol/g dry weight. Digestion treatments with α-amylase and pullulanase completely attenuated the aluminum ion-adsorbing activity of purified CA42 EPS and oyster glycogen, suggesting that the glycogen-like structure adsorbed aluminum ions and that its branching structure played an important role in its aluminum adsorbing activity. Furthermore, the aluminum tolerance of CA42 cells was attenuated by pullulanase treatment directly on the live CA42 cells. These results suggest that CA42 EPS adsorbs aluminum ions and is involved in the aluminum tolerance mechanism of Pullulanibacillus sp. CA42. Thus, this strain may be a potential plant growth-promoting bacterium in acidic soils. In addition, this study is the first to report a glycogen-like polysaccharide that adsorbs aluminum ions.
1. Introduction
Acid sulfate soil is the common name given to soils containing iron sulfides (pyrites). It is estimated that this kind of soil covers approximately 500,000 km2 worldwide (Michael et al., 2017) and is especially abundant in Australia and Asia. When acid sulfate soil is dug up, pyrite becomes exposed to the air and is oxidized to sulfuric acid; consequently, both soil and water show a lower pH. Such soil is known as actual acid sulfate soil (AASS) (Dent and Pons, 1995). Below pH 4.5, aluminum becomes more soluble and toxic to plants, and a few micronutrients, such as manganese (Lasat, 2002) and iron (McGrath et al., 2001), become more soluble and toxic. Most plant nutrients, especially phosphorus, become more limited in acidic soils (Kochian et al., 2004), leading to serious environmental destruction and significant economic problems.
However, few plants can survive in such extreme environments. Since it has become clear that acid- and aluminum–resistant microorganisms coexist in the rhizosphere of such plants, such microorganisms may be used for AASS bioremediation and crop cultivation. The use of these microorganisms in protecting the microenvironment of the plant surface and rhizosphere may yield a new and effective bioremediation method that can be used to recover AASS. Furthermore, protecting only the plant surfaces and rhizosphere is economical and has a minimal environmental impact.
In our previous study on developing bioremediation measures for AASS, we isolated several bacteria associated with plants grown in the highly acidic aquatic environments of AASS in Southeast Asia and reported new bacterial species (Aizawa et al., 2007, 2008, 2010a,b). For instance, we isolated a new strain of Acidocella aluminiidurans AL46 from Panicum repens grown in a highly acidic swamp in the AASS of Vietnam (Kimoto et al., 2010). AL46 grows at pH 3.0 and is tolerant to 500 mM aluminum sulfate or 200 mM aluminum chloride, which are higher than those observed in other bacteria. Furthermore, AL46 produces an aluminum-binding capsular polysaccharide that may be involved in resistance to high concentrations of aluminum (Aizawa and Urai, 2020). An aluminum-tolerant bacterium, CA42, was recently isolated from the Chinese water chestnut Eleocharis dulcis, growing in a highly acidic swamp in the AASS of Vietnam. CA42 was selected for plant growth promotion in acidic soils based on its ability to restore rice growth in inoculation experiments in the presence of aluminum ions. CA42 also produces a large amount of extracellular polysaccharide (EPS).
In the present study, we identified and characterized CA42 and determined the structure and aluminum-adsorbing ability of the EPS it produces. Furthermore, we validated the involvement of EPS in the aluminum tolerance of CA42.
2. Materials and methods
2.1. Isolation of the aluminum-tolerant strain CA42
CA42 was isolated from E. dulcis by using diluted tryptic soy broth 1:10 (1/10 TS) agar plates, which were prepared by diluting 2.75 g tryptic soy without glucose broth (BD, Franklin Lakes, NJ, United States) per liter of water and solidified with 15.0 g of agar (BD) per liter (pH 4.0). Acidic plates were prepared by mixing double-strength TS medium component without agar and 3% agar solution that had been autoclaved separately to prevent agar hydrolysis. The growth of CA42 in the presence of AlCl3 was tested using modified TS (MTS) medium (2.75 g·l−1 tryptic soy without glucose, 0.2 g·l−1 MgCl2·7H2O, 0.1 g·l−1 CaCl2·2H2O, 0.1 g·l−1 NaCl, 0.02 g·l−1 FeCl2·6H2O, 0.5 g·l−1 (NH4)2SO4; pH 4.0).
Furthermore, the AlCl3 tolerance of CA42 was examined through a growth promotion test (inoculation experiments) on rice plants (IR36) in Yoshida’s solution (pH 3.5) containing 300 μM AlCl3. Briefly, IR36 seeds were surface-sterilized as described by Kochian and Shaff (1991). Inoculation experiments were performed as described by Elbeltagy et al. (2001). Inoculated plants were cultivated in Yoshida’s solution (pH 3.2) (Yoshida, 1976) for 60 days in growth chambers (LPH-350S; Nippon Medical & Chemical Instruments Co. Ltd., Osaka, Japan) with 100 μmol m−2 s−1 illumination from cool-white fluorescent lamps at a 12-h photoperiod at 30°C.
2.2. Characterization of CA42
The 16S rRNA gene of strain CA42 was amplified via PCR using universal primers (Tamura et al., 2001), and the nearly complete 16S rRNA gene nucleotide sequence (1,546 bp) was obtained. Sequence similarity was determined using the featured identification service of EzBioCloud (16S-based ID)1 (Yoon et al., 2017). Multiple alignments of the sequence data were performed using ClustalX (Thompson et al., 1997). Subsequently, phylogenetic relationships with closely related species were determined using MEGA version 11 (Tamura et al., 2021). Evolutionary distances were computed as previously described (Jukes and Cantor, 1969). Phylogenetic trees were constructed using the maximum parsimony (Kluge and Farris, 1969), maximum likelihood (Felsenstein, 1981), and neighbor-joining (Saitou and Nei, 1987) methods. The reliabilities of these tree topologies were evaluated using bootstrap analysis with 1,000 replicates (Felsenstein, 1985).
The production of indole acetic acid was tested by using 1/10 TS containing 1% (w/v) glucose and 5 mM tryptophan (pH 3.5 and 6.0).
2.3. Extraction and purification of CA42 EPS
CA42 EPS was extracted from cells cultivated on 1/10 TS agar plates and purified using enzymatic treatments, phenol-chloroform treatment, and ethanol precipitation, as previously described (Urai et al., 2006).
2.4. Monosaccharide analysis
CA42 EPS (50 μg) was completely hydrolyzed using 2 M trifluoroacetic acid (TFA) at 100°C for 3 h. The obtained monosaccharides were labeled with 4-aminobenzoic acid ethyl ester (ABEE) and analyzed using high-performance liquid chromatography (HPLC; Nexera System; Shimadzu, Kyoto, Japan), as described previously (Urai et al., 2006). To determine the absolute configuration of CA42 EPS, the TFA hydrolysate of EPS was converted into acetylated (−)-2-butyl glycoside and analyzed using gas–liquid chromatography (GLC) (Leontein et al., 1978; Gerwig et al., 1979).
2.5. Nuclear magnetic resonance (NMR) analysis
NMR spectra were recorded at 500 MHz (1H) and 125 MHz (13C) using an ECA 500 instrument (JEOL Ltd., Tokyo, Japan) or at 600 MHz (1H) and 150 MHz (13C) with an ECZ 600 instrument (JEOL Ltd.). Chemical shifts were administered in parts per million (ppm), with acetone (δ 1H 2.23 ppm, δ 13C 31.1 ppm) used as an internal reference for samples measured in D2O solutions. Signals were assigned based on the results of the heteronuclear single-quantum coherence (HMQC) and heteronuclear multiple-bond coherence (HMBC) experiments. 1H NMR chemical shifts of the overlapping signals were obtained from the center of the cross peaks in the 2D spectra.
2.6. Methylation of CA42 EPS
Methylation of CA42 EPS was performed as described previously (Ciucanu and Kerek, 1984). Subsequently, the methylated polysaccharide was hydrolyzed, reduced with sodium borodeuteride, acetylated, and analyzed using GLC–mass spectrometry (MS) (GCMS-QP2020NX; Shimadzu, Kyoto, Japan).
2.7. Enzymatic digestion of CA42 EPS
CA42 EPS was digested with pullulanase (Sigma-Aldrich Co. LLC, St. Louis, MO, United States) or α-amylase (Tokyo Chemical Industry Co., Ltd., Tokyo, Japan) in 0.1 M acetate buffer (pH 5.0) at 37°C overnight. After denaturation of the enzyme via heat treatment at 100°C for 5 min, the products were recovered via freeze-drying. Oyster glycogen (FUJIFILM Wako Pure Chemical Corporation, Tokyo, Japan) and amylose (Sigma-Aldrich Co. LLC) were added using the same procedure.
Gel filtration column chromatography was performed using the Bio-Gel P-2 gel filtration column (900 mm × 15 mm ϕ; Bio-Rad Laboratories, Inc., CA, United States), with 0.2 M acetic acid as the eluent. Fractions containing saccharides were monitored using the phenol-H2SO4 method (Dubois et al., 1956).
2.8. Evaluation of aluminum ion-adsorbing activity of CA42 EPS
The aluminum ion-adsorbing ability of CA42 was measured according to methods reported by Kerven et al. (1989) and Maejima et al. (2017). A colorimetric method using the pyrocatechol violet (PCV) reagent was used to measure the free aluminum ions in the glucan samples containing the aluminum solution. An aqueous solution of CA42 EPS or standard glucan (final concentration of 1 mg/mL) was mixed with AlCl3 to a final concentration of 25 μM, and the concentration of free aluminum ions was measured by determining the adsorbance of PCV at 585 nm. The reduction in free Al3+ ions observed in the presence of glucans was considered as the amount of Al3+ adsorbed by the glucan.
2.9. Effect of pullulanase treatment on the aluminum tolerance of CA42
CA42 was cultured in 1/10 TS broth at 28°C for 2 d, and cells were recovered using centrifugation at 10,000 × g for 10 min. Cells were resuspended in 1 mL of 0.1 M acetate buffer (pH 5.0). Then, 30 μL of pullulanase was added, and the cells were incubated at 37°C for 4 h. Pullulanase-treated cells were recovered using centrifugation at 10,000 × g for 10 min, and the cells were rinsed with acetate buffer. Then, 5 μL of 1 M AlCl3 solution was added, and the cells were incubated at 28°C for 4 h. The cells exposed to aluminum ions were rinsed with 0.85% (w/v) saline, while the number of living cells was measured by calculating the number of colony-forming units on the 1/10 TS agar plates. Escherichia coli BW25113 was used as a control to evaluate aluminum toxicity. The survival rate of each strain was compared to that of cells that were not exposed to aluminum ions.
2.10. Statistical analysis
SPSS software (version 20.0; IBM Japan, Tokyo, Japan) was used. Normality of distribution and equality of variance were assessed using the Shapiro–Wilk’ and Levene’s tests. p values <0.01 were considered significant.
3. Results
3.1. Isolation and characterization of the aluminum-tolerant strain CA42
Among the bacteria isolated from E. dulcis using 1/10 TS agar plates containing 5 mM AlCl3 (pH 3.5), strain CA42 restored the growth of rice plants in Yoshida’s solution (pH 3.5) containing 300 μM AlCl3 (Figure 1A). In addition, biofilm was observed around the roots of the rice plants inoculated with CA42 (Figure 1B). The organism formed yellowish, round, smooth, and flat colonies with entire margins. The strain showed good growth on 1/10 TS agar plates at 20–37°C, with optimal growth at 30°C. Strain CA42 showed good growth at pH 4–6, with optimal growth at pH 5 when cultured at 30°C for 5 d.
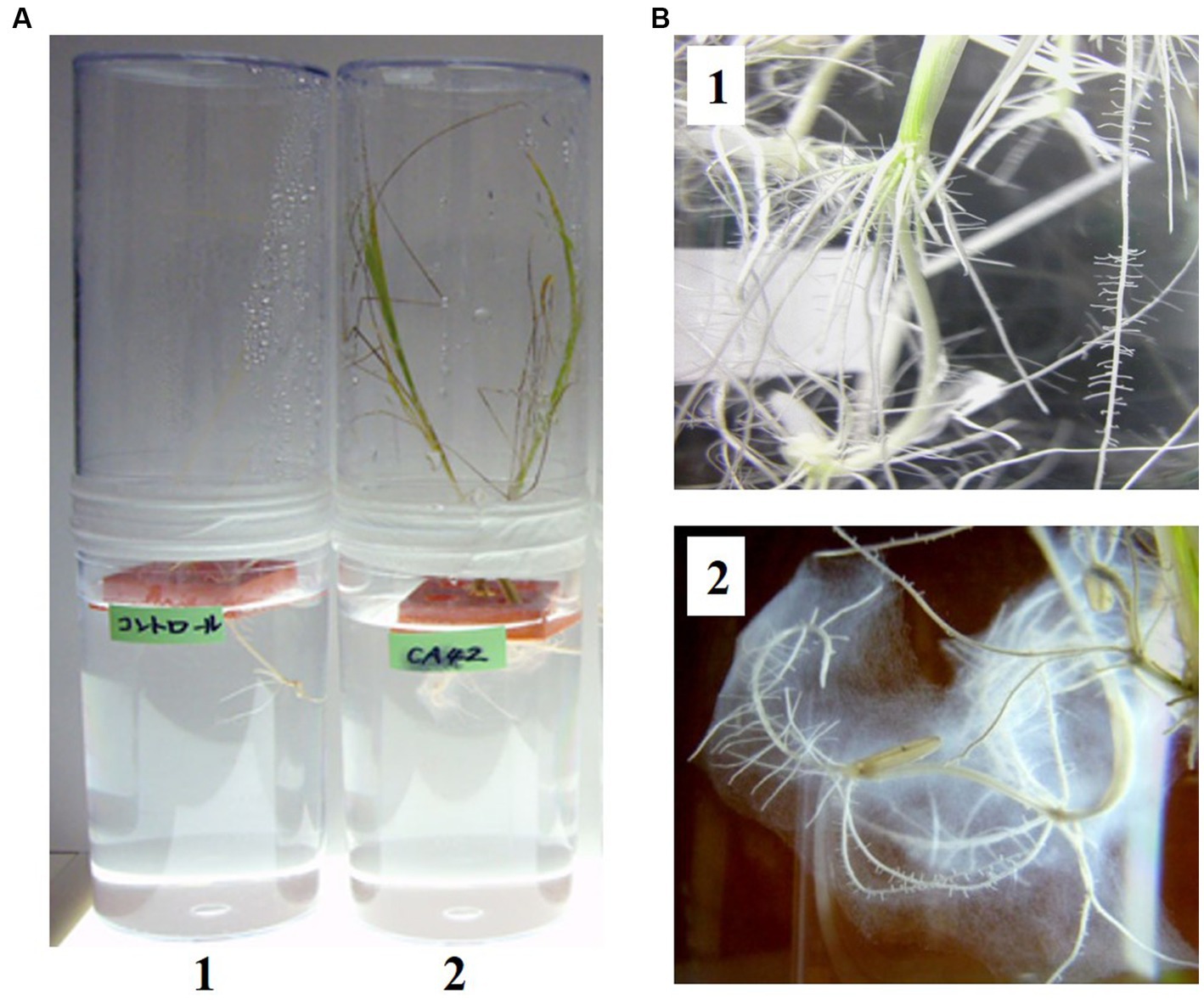
Figure 1. Inoculation of Oryza sativa cultivar indica IR36 with Pullulanibacillus sp. CA42 in Yoshida’s solution (pH 3.5) containing 300 μM aluminum chloride (A). 1, Non-inoculated rice. 2, Rice inoculated with Pullulanibacillus sp. CA42. Biofilm around the roots of the rice inoculated with Pullulanibacillus sp. CA42 was observed. Enlarged photo of each root (B).
The 16S rRNA gene sequence of CA42 showed 98.38 and 97.95% similarity to those of P. naganoensis (AB021193) and P. uraniitolerans (AM931441), respectively. The partial 16S rRNA gene sequence of CA42 was submitted to the GenBank/EMBL/DDBJ databases (Accession number: AB520692). Furthermore, phylogenetic analysis results revealed that strain CA42 belonged to the genus Pullulanibacillus (Figure 2). These and other physiological and biochemical data (unpublished) suggest that the isolate represents a novel species of the genus Pullulanibacillus.
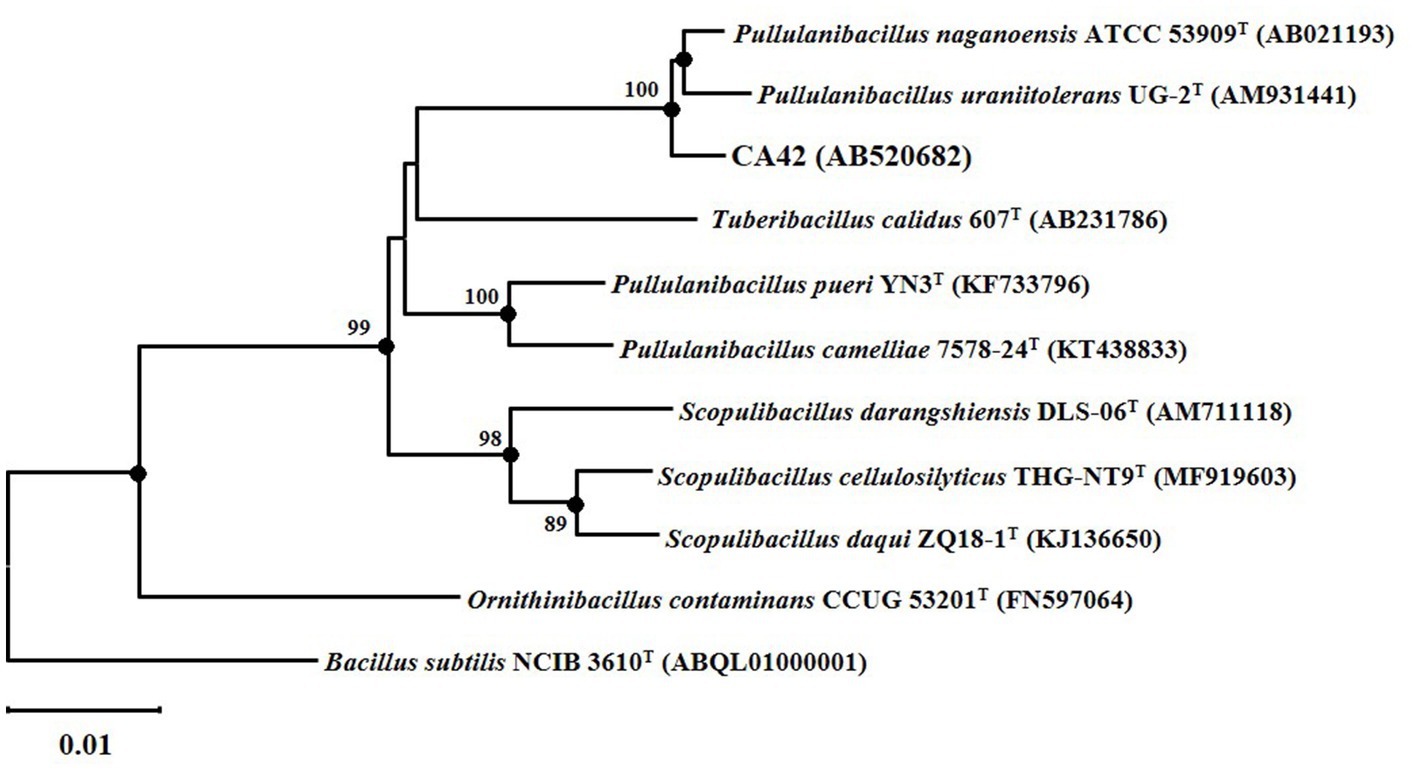
Figure 2. Neighbor-joining tree based on a nearly complete 16S rRNA gene sequence, showing the positions of strain CA42 and its phylogenetic neighbors. Filled circles indicate the corresponding nodes (groupings) that were also recovered in the maximum-likelihood and maximum-parsimony trees. Numbers at the nodes are percentages of bootstrap values based on 1,000 resampled datasets; only those above 70% are indicated. The sequence of Bacillus subtilis NCIB 3610T was used as an outgroup. Bar, 0.01 nucleotide substitutions per nucleotide position.
CA42 produced indole acetic acid in tryptophan-containing medium, indicating that CA42 belongs to plant-growth promoting rhizobacteria. Furthermore, CA42 showed good growth in the presence of 5 mM AlCl3 in MTS medium and produced a large amount of EPS on 1/10 TS agar plates.
3.2. CA42 produced a glycogen-like EPS
Purified CA42 EPS was observed to be a white fibrous compound soluble in hot water and dimethyl sulfoxide, but not in cold water, methanol, or ethanol. At 280 or 255 nm, little or no adsorption was detected, suggesting that the CA42 EPS did not contain proteins or nucleic acids. Furthermore, the monosaccharide content of CA42 EPS was determined via TFA hydrolysis, followed by HPLC analysis. The results showed the presence of glucose as the sole polysaccharide component. The absolute configuration of glucose was determined using GLC with the acetylated (−)-2-butyl derivatives. The results showed that the glucose had the D-configuration.
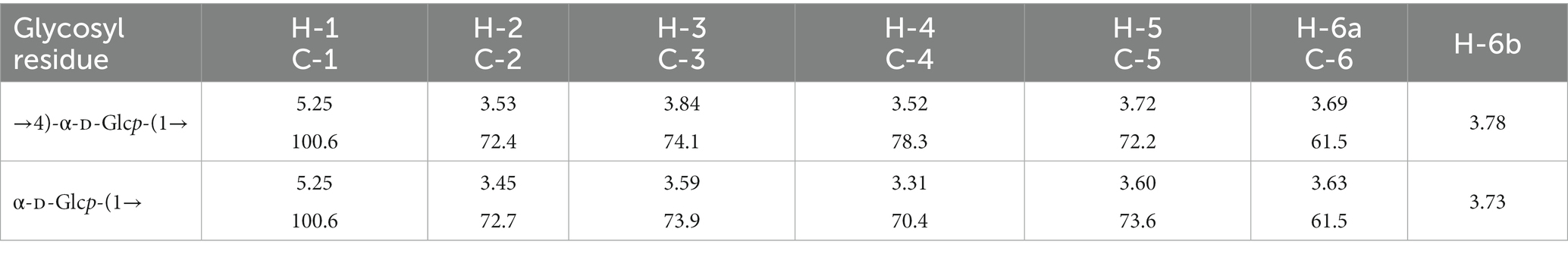
One resonance signal was observed at δ 5.25 and δ 100.6 in the anomeric regions of the 1H and 13C NMR spectra, respectively (Figure 3). These anomeric signals were assigned to the α configuration based on the observed chemical shift values (Bubb, 2003). In addition, the 13C NMR spectrum showed one signal at δ 61.5 (C-6) and eight at δ 70–79 (C-2–C-5). The 1H NMR spectrum displayed several signals at δ 3.31–3.84 (H-2–H-6). The 1H chemical shifts of the CA42 EPS were assigned using two-dimensional total correlation spectroscopy (TOCSY) and two-dimensional double quantum filtered-correlation spectroscopy (DQF-COSY) experiments and showed two spin systems (Table 1). The 13C chemical shifts of CA42 EPS were assigned using HSQC and HMBC (Table 1). The results of the NMR analyses clearly showed that the CA42 EPS mainly consisted of α-1,4-linked residues of D-glucose.
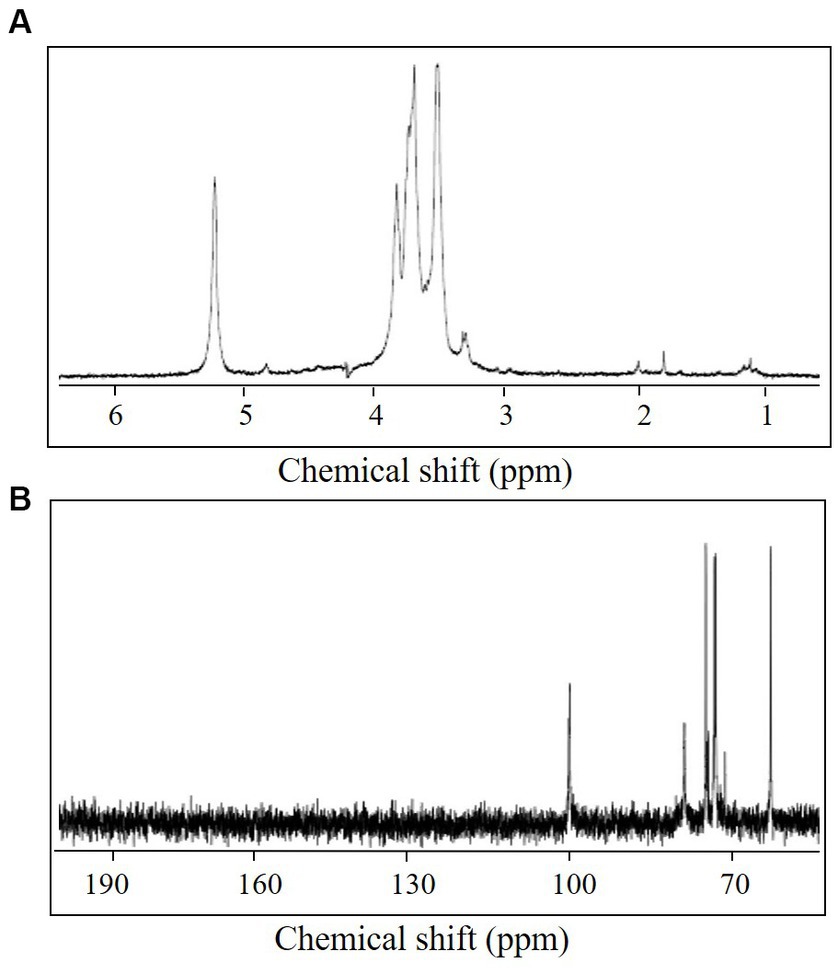
Figure 3. 1H nuclear magnetic resonance (NMR) spectrum (A) and 13C NMR spectrum (B) of CA42 extracellular polysaccharide, recorded in D2O at 70°C. ppm, parts per million.
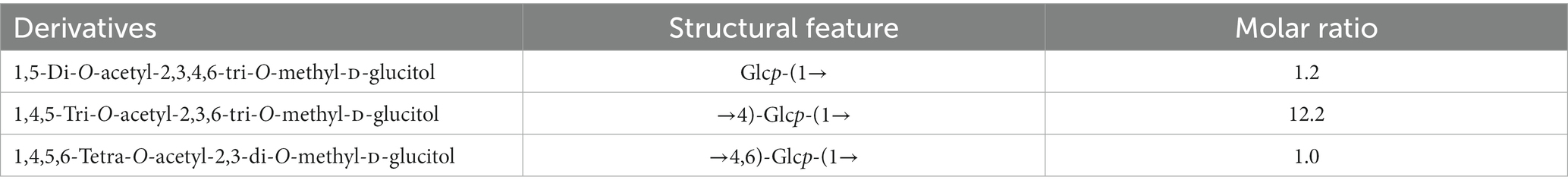
Next, the CA42 EPS was methylated, and the derived alditol acetates were analyzed using GLC-MS (Table 2). Three peaks were observed, which were identified as 1,5-di-O-acetyl-2,3,4,6-tri-O-methyl-D-glucitol; 1,4,5-tri-O-acetyl-2,3,6-tri-O-methyl-D-glucitol; and 1,4,5,6-tetra-O-acetyl-2,3-di-O-methyl-D-glucitol. The presence of 1,4,5,6-tetra-O-acetyl-2,3-di-O-methyl-D-glucitol indicated that CA42 EPS is branched at C-6 of the D-glucose residues.
CA42 EPS was digested by pullulanase, specifically degrading α-1,6-glucosidic bonds at the branching point of glycogen, and was fractionated using gel filtration column chromatography. Glucose, maltobiose, and maltotriose were detected using the phenol–H2SO4 method (Figure 4). When oyster glycogen was treated using the same procedure, almost the same elution profile was obtained (Figure 4). These results indicate that CA42 EPS has a side-chain structure similar to that of oyster glycogen. In summary, CA42 EPS is glycogen-like polysaccharide consisting of a repeating unit (Figure 5).
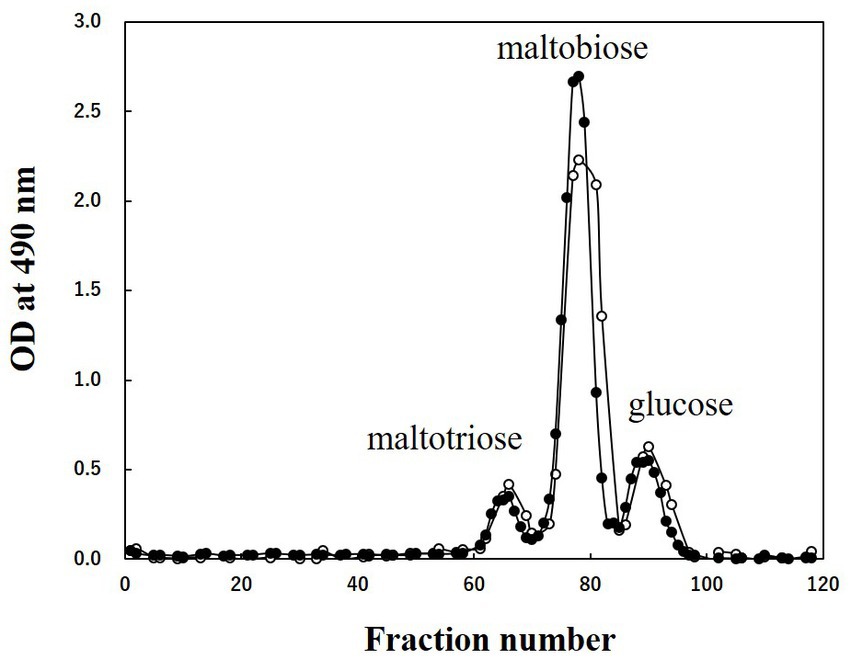
Figure 4. Results of gel filtration column chromatography of CA42 extracellular polysaccharide (EPS) and oyster glycogen treated with pullulanase. Oligosaccharides were applied to the Bio-Gel P-2 gel filtration column (900 mm × 15 mm ϕ), and 0.2 M acetic acid was used as the eluate. Glucose, maltobiose, and maltotriose were used as size markers. Closed circles, CA42 EPS; opened circles, oyster glycogen; OD, optical density.
3.3. Adsorption of aluminum ion by CA42 EPS
To analyze the role of CA42 EPS in aluminum tolerance, we examined whether the polysaccharide adsorbed aluminum ions (Figure 6). CA42 EPS adsorbed 14.2 ± 0.2 μmol/g (dry weight) aluminum ion at a low pH. Commercially available oyster glycogen also adsorbed 22.8 ± 0.1 μmol/g (dry weight) aluminum ion, while amylose showed little to no adsorption (0.5 ± 0.4 μmol/g dry weight). In addition, enzymatic treatment with either α-amylase or pullulanase completely attenuated the aluminum ion adsorption activity of CA42 EPS, as well as that of oyster glycogen. These results suggest that the glycogen-adsorbed aluminum ion and its branching structure play important roles in the aluminum tolerance of CA42.
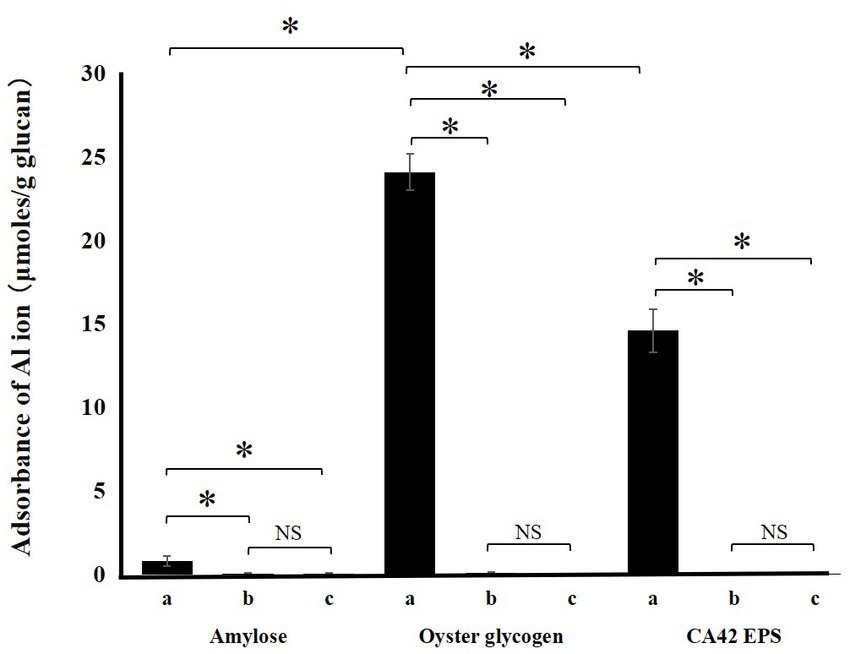
Figure 6. Adsorption of aluminum ion by CA42 extracellular polysaccharides (EPS) and standard glucans. Adsorbed ammonium ion in μmol was divided by the samples in grams. (a), native sample; (b), α-amylase-treated sample; (c), pullulanase-treated sample. *p < 0.01; NS, not significant.
3.4. Pullulanase treatment attenuated the aluminum tolerance of CA42 cells
Since pullulanase treatment attenuated the aluminum ion-adsorption activity of CA42 EPS, we validated the effect of pullulanase treatment on the aluminum tolerance of CA42 cells. E. coli was used to evaluate the aluminum toxicity. The survival rate of each strain was compared with that in the absence of aluminum. The survival rate of E. coli was approximately 2%, regardless of pullulanase treatment. CA42 showed 53% survival in the presence of aluminum and 16% survival after pullulanase treatment (Figure 7). These results suggest that CA42 EPS is mainly involved in the aluminum tolerance of Pullulanibacillus sp. CA42.
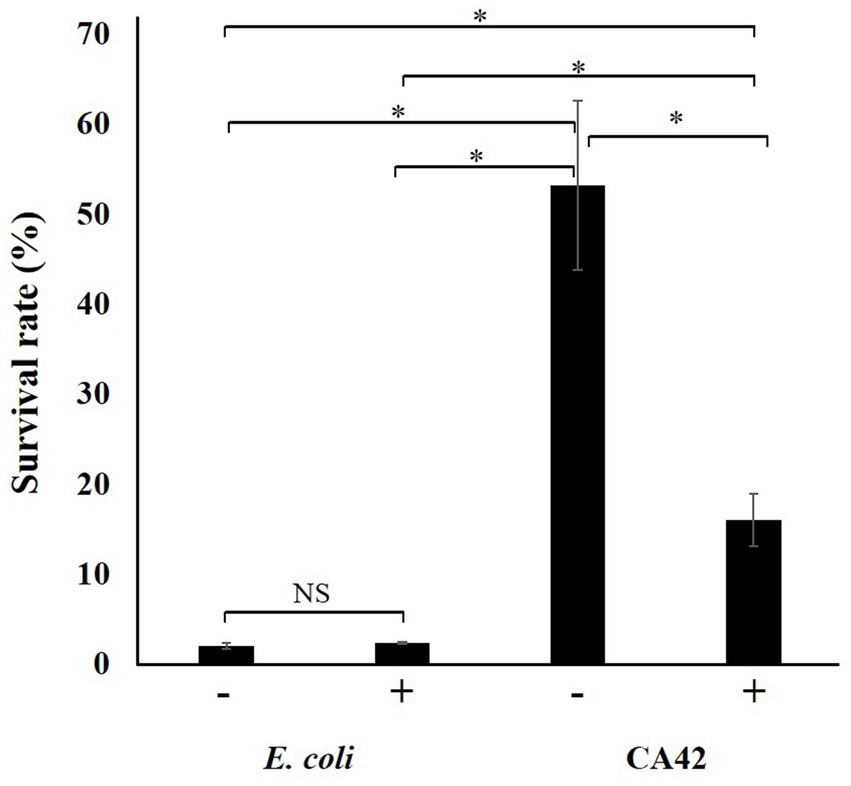
Figure 7. Effect of pullulanase treatment on the aluminum tolerance of CA42. The survival rate of each strain was compared to that of cells not exposed to aluminum ions. E. coli was used as the control to evaluate the aluminum toxicity. −, no treatment; +, pullulanase treatment. *p < 0.01; NS, not significant.
4. Discussion
In the present study, we aimed to characterize the novel aluminum-tolerant strain CA42, isolated from AASS, which allows Oryza sativa to grow in the presence of 300 μM AlCl3 at pH 3.5. The strain was identified as Pullulanibacillus sp. CA42, and this is the first strain belonging to the genus Pullulanibacillus that showed aluminum tolerance. CA42 produced a large amount of EPS, a glycogen-like α-glucan, that adsorbs aluminum ions. To the best of our knowledge, this study is the first to characterize EPS produced by a strain belonging to the genus Pullulanibacillus. The branching structure of CA42 EPS plays an important role in its aluminum adsorption activity. This study is the first report of glycogen-like EPS that adsorb aluminum ions. Furthermore, aluminum tolerance of CA42 was attenuated via pullulanase treatment. Our findings suggest that CA42 can act as a plant growth-promoting bacterium in acidic soils, and that CA42 EPS may protect plants growing in AASS. Currently, we are conducting cloning experiments using CA42 EPS synthetic genes to construct EPS-deficient mutants. With these knockout mutants, we intend to clarify the roles of CA42 EPS in aluminum tolerance and plant growth promotion. The mechanism underlying the aluminum adsorption capacity of CA42 EPS and its application as a new aluminum-adsorbing substance that functions in acidic environments should be elucidated in future studies. This study proposes the new mechanism of aluminum tolerance using a bacterial extracellular polysaccharide. The strain isolated in this study may be a potential plant growth-promoting bacterium in actual acid sulfate soils.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, AB520692.
Author contributions
TA and MU contributed equally to the concept, design, and analysis of the data, as well as the writing of the manuscript. JS and SS contributed to the majority of experiments. TY and YM contributed to the structural polysaccharide analysis. All authors contributed to the article and approved the submitted version.
Funding
This work was partially supported by the MEXT KAKENHI (grant numbers 20 K05735 and 21 K12298) and Nihon University College of Bioresource Sciences Research Grants for 2018, 2019, and 2020.
Acknowledgments
We would like to thank S. Sasaki and M. Nakajima for providing us the opportunity to conduct this study. We thank V. B. Nguyen, N. M. Do, H. Sumida, H. Uchiyama, M. Sunairi, and other members of the “Center of Excellence in 21st Century Projects” of the Ministry of Education, Culture, Sports, Science, and Technology of Japan. We thank K. Ueda at the Life Science Center for permitting the use of the instrument facilities. We thank the members of the Integrated Research Institute for providing technical assistance. We thank T. Asai, K. Kimoto, and other members of our laboratory for their technical assistance and encouragement. We would also like to thank Editage (www.editage.com) for English language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Aizawa, T., Nguyen, B. V., Kimoto, K. I., Iwabuchi, N., Sumida, H., Hasegawa, I., et al. (2007). Curtobacterium ammoniigenes sp. nov., an ammonia-producing bacterium isolated from plants inhabiting acidic swamps in actual acid sulfate soil areas of Vietnam. Int. J. Syst. Evol. Microbiol. 57, 1447–1452. doi: 10.1099/ijs.0.64731-0
Aizawa, T., Nguyen, B. V., Nakajima, M., and Sunairi, M. (2010a). Burkholderia heleia sp. nov., a nitrogen-fixing bacterium isolated from an aquatic plant, Eleocharis dulcis, that grows in highly acidic swamps in actual acid sulfate soil areas of Vietnam. Int. J. Syst. Evol. Microbiol. 60, 1152–1157. doi: 10.1099/ijs.0.015198-0
Aizawa, T., Nguyen, B. V., Vijarnsorn, P., Kimoto, K., Sasaki, S., Nakajima, M., et al. (2008) in Application of symbiotic bacteria isolated from plants adapted to actual acid sulfate soil, in development of new bioremediation Systems of Acid Sulfate Soil for agriculture and forestry. eds. S. Sasaki, et al. (Kyoto, Japan: Nakanishi Printing Co, Ltd), 57–62.
Aizawa, T., Nguyen, B. V., Vijarnsorn, P., Nakajima, M., and Sunairi, M. (2010b). Burkholderia acidipaludis sp. nov., aluminum-tolerant bacteria isolated from Chinese water chestnut (Eleocharis dulcis) growing in highly acidic swamps in South-East Asia. Int. J. Syst. Evol. Microbiol. 60, 2036–2041. doi: 10.1099/ijs.0.018283-0
Aizawa, T., and Urai, M. (2020). Structural analysis of an aluminum-binding capsular polysaccharide produced by Acidocella aluminiidurans strain AL46, an aluminum-tolerant bacterium isolated from plant roots in a highly acidic swamp in actual acid sulfate soil. Carbohydr. Res. 498:108163. doi: 10.1016/j.carres.2020.108163
Bubb, W. A. (2003). NMR spectroscopy in the study of carbohydrates: characterizing the structural complexity. Concepts. Magn. Reson. 19A, 1–19. doi: 10.1002/cmr.a.10080
Ciucanu, I., and Kerek, F. (1984). A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131, 209–217. doi: 10.1016/0008-6215(84)85242-8
Dent, D. L., and Pons, L. J. (1995). A world perspective on acid sulphate soils. Geoderma 67, 263–276. doi: 10.1016/0016-7061(95)00013-E
Dubois, M., Gilles, K. A., Hamilton, J. K., Rebers, P. A., and Smith, F. (1956). Colorimetric method for determination of sugars and related substances. Anal. Chem. 28, 350–356. doi: 10.1021/ac60111a017
Elbeltagy, A., Nishioka, K., Sato, T., Suzuki, H., Ye, B., Hamada, T., et al. (2001). Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Appl. Environ. Microbiol. 67, 5285–5293. doi: 10.1128/AEM.67.11.5285-5293.2001
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376. doi: 10.1007/BF01734359
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Gerwig, G. J., Kamerling, J. P., and Vliegenthart, J. F. (1979). Determination of the absolute configuration of mono-saccharides in complex carbohydrates by capillary G.L.C. Carbohydr. Res. 77, 10–17. doi: 10.1016/s0008-6215(00)83788-x
Jukes, T. H., and Cantor, C. R. (1969). “Evolution of protein molecules” in Mammalian protein metabolism. ed. H. N. Munro (New York: Academic Press), 21–132. doi: 10.1016/B978-1-4832-3211-9.50009-7
Kerven, G. L., Edwards, D. G., Asher, C. J., Hallman, P. S., and Kokot, S. (1989). Aluminum determination in soil solution. II. Short-term colorimetric procedures for the measurement of inorganic monomeric aluminum in the presence of organic acid ligands. Aust. J. Soil Res. 27, 91–102. doi: 10.1071/SR9890091
Kimoto, K. I., Aizawa, T., Urai, M., Bao Ve, N., Suzuki, K. I., Nakajima, M., et al. (2010). Acidocella aluminiidurans sp. nov., an aluminium-tolerant bacterium isolated from Panicum repens grown in a highly acidic swamp in actual acid sulfate soil area of Vietnam. Int. J. Syst. Evol. Microbiol. 60, 764–768. doi: 10.1099/ijs.0.011569-0
Kluge, A. G., and Farris, F. S. (1969). Quantitative phyletics and the evolution of anurans. Syst. Zool. 18, 1–32. doi: 10.2307/2412407
Kochian, L. V., Hoekenga, O. A., and Pineros, M. A. (2004). How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 55, 459–493. doi: 10.1146/annurev.arplant.55.031903.141655
Kochian, L. V., and Shaff, J. E. (1991). “Investigating the relationship between aluminium toxicity, root growth and root-generated ion currents, in plant-soil interactions at low pH” in Developments in plant and soil sciences. eds. R. J. Wright, V. C. Baligar, and R. P. Murrmann (Dordrecht: Springer), 769–778.
Lasat, M. M. (2002). Phytoextraction of toxic metals: a review of biological mechanisms. J. Environ. Qual. 31, 109–120. doi: 10.2134/jeq2002.0109
Leontein, K., Lindberg, B., and Lönngren, J. (1978). Assignment of absolute configuration of sugars by g.l.c. of their acetylated glycosides formed from chiral alcohols. Carbohydr. Res. 62, 359–362. doi: 10.1016/S0008-6215(00)80882-4
Maejima, E., Osaki, M., Wagatsuma, T., and Watanabe, T. (2017). Contribution of constitutive characteristics of lipids and phenolics in roots of tree species in myrtales to aluminum tolerance. Physiol. Plantarum. 160, 11–20. doi: 10.1111/ppl.12527
McGrath, S. P., Zhao, F. J., and Lombi, E. (2001). Plant and rhizosphere processes involved in phytoremediation of metal-contaminated soils. Plant Soil 232, 207–214. doi: 10.1023/A:1010358708525
Michael, P. S., Fitzpatrick, R. W., and Reid, R. J. (2017). Effects of live wetland plant macrophytes on acidification, redox potential and sulphate content in acid sulphate soils. Soil Use Manag. 33, 471–481. doi: 10.1111/sum.12362
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Tamura, T., Hatano, K., and Hatano, K. (2001); Phylogenetic analysis of the genus Actinoplanes and transfer of Actinoplanes minutisporangius Ruan). 1986 and ‘Actinoplanes aurantiacus’ to Cryptosporangium minutisporangium comb.nov. and Cryptosporangium aurantiacum sp. nov. Int. J. Syst. Evol. Microbiol. 51, 2119–2125. doi: 10.1099/00207713-51-6-2119
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The clustal_x windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Urai, M., Anzai, H., Ogihara, J., Iwabuchi, N., Harayama, S., Sunairi, M., et al. (2006). Structural analysis of an extracellular polysaccharide produced by Rhodococcus rhodochrous strain S-2. Carbohydr. Res. 341, 766–775. doi: 10.1016/j.carres.2005.12.013
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Keywords: Pullulanibacillus, extracellular polysaccharide, aluminum tolerance, plant-growth promoting rhizobacteria, structure, NMR spectroscopy
Citation: Aizawa T, Sato J, Saito S, Yasuda T, Maruyama Y and Urai M (2023) An extracellular polysaccharide is involved in the aluminum tolerance of Pullulanibacillus sp. CA42, a newly isolated strain from the Chinese water chestnut growing in an actual acid sulfate soil area in Vietnam. Front. Microbiol. 14:1241244. doi: 10.3389/fmicb.2023.1241244
Edited by:
Mohammad Ali Amoozegar, University of Tehran, IranCopyright © 2023 Aizawa, Sato, Saito, Yasuda, Maruyama and Urai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomoko Aizawa, YWl6YXdhLnRvbW9rb0BuaWhvbi11LmFjLmpw; Makoto Urai, bXUyMDYxNzdAbm9kYWkuYWMuanA=
†These authors have contributed equally to this work
 Tomoko Aizawa
Tomoko Aizawa Junki Sato2
Junki Sato2 Makoto Urai
Makoto Urai